Abstract
Numerous key biological processes rely on the concept of multivalency, where ligands achieve stable binding only upon engaging multiple receptors. These processes, like viral entry or immune synapse formation, occur on the diffusive cellular membrane. One crucial, yet underexplored aspect of multivalent binding is the mobility of coupled receptors. Here, we discuss the consequences of mobility in multivalent processes from four perspectives: (I) The facilitation of receptor recruitment by the multivalent ligand due to their diffusivity prior to binding. (II) The effects of receptor preassembly, which allows their local accumulation. (III) The consequences of changes in mobility upon the formation of receptor/ligand complex. (IV) The changes in the diffusivity of lipid environment surrounding engaged receptors. We demonstrate how understanding mobility is essential for fully unravelling the principles of multivalent membrane processes, leading to further development in studies on receptor interactions, and guide the design of new generations of multivalent ligands.
Keywords: drug design, membranes, multivalency, receptors, targeting
Multivalent membrane processes result in numerous receptors being coupled on a diffusive surface. As such, these binding mechanisms depend heavily on the intrinsic mobility on the cell membrane. The Minireview discusses the importance of mobility in multivalent binding, demonstrating that it is crucial for the deeper understanding of receptor interactions, and to guide the design of new generations of multivalent ligands.

1. Introduction
The kinetics of membrane components is a central parameter in receptor/ligand binding. The diffusive lipid bilayer provides a dynamic platform for embedded proteins (receptors), whose mobility was shown to play a pivotal role in various biological processes, including T cell activation [1] and the formation of stable focal adhesions. [2] In particular, membrane mobility is often relevant for multivalent interactions, where a ligand with multiple binding sites engages a number of receptors present on the surface,[ 3 , 4 ] while monovalent interactions are too weak to ensure stable binding (Figure 1 a). In such systems, a matching geometry between the ligand and receptors is required (Figure 1 b). This binding dependency on the spatial distribution forms the basis of a unique, sharp discrimination between surfaces of different densities of receptors.[ 3 , 4 , 5 , 6 ] Such superselectivity is utilised to target particular cell populations by highly‐specific synthetic ligands, proposed as sensing nanodevices [7] or therapeutic platforms.[ 8 , 9 ] Thus, the spatial tolerance of multivalent binding—and by connection the mobility of complex‐forming molecules—are highly significant.
Figure 1.
Multivalent interactions on cell membranes require spatial matching between binding molecules. Since monovalent interactions are too weak to ensure stable binding (a), multivalent ligands cannot bind to immobile surfaces with mismatched spacing (b). To overcome this restriction, the synthetic ligands are often modified with an excess of binders to overcome the spatial requirements (c). Often in nature the valency of interactions is significantly more controlled (d), and so is ligand's geometry (e). As the ligand becomes more rigid and matching, the penalties related to conformational ΔS conf and combinatorial ΔS comb entropies decrease. In biologically relevant scenario, the receptors are embedded in a diffusive lipid bilayer. The mobility of receptors allows them to adjust to the ligand's geometry (f) and provides another control mechanism for the cell (g). However, the entropy loss related to demixing and restriction of translational movement of the receptors (ΔS r) is higher than on less mobile surfaces (h).
In some cases the diffusivity effects are less crucial for ensuring binding; synthetic ligands, as well as natural multivalent constructs (e.g., enveloped viruses[ 10 , 11 ]) frequently carry an excess of binding sites (Figure 1 c) to statistically ensure matching with the spatial distribution of receptors. [12] However, for many natural systems the ligand design features a well‐defined valency (Figure 1 d), as well as a constrained geometry (Figure 1 e): [13] one of the most prominent examples is the architecture of specific bivalent antibodies, featuring a limited flexibility of their signature Y‐shape.[ 14 , 15 ] In such cases multivalent binding occurs when the membrane components can be organised in a particular spatial arrangement, matching the geometry of the ligand.[ 16 , 17 ] The mobility of the membrane thus plays a significant role in fulfilling the spacing requirements of the embedded receptors, in processes as crucial as, e.g., the activation of adaptive immune cells, where receptor clustering was identified as a key regulatory mechanism. [18] Understanding its role will deepen our knowledge of the natural binding processes and guide the design of novel multivalent ligands, relying not on statistical “brute force”, but rather a more sophisticated, tailored approach. [19]
Considering the surface dynamics of a multivalent system, three new aspects of binding emerge: (I) changes to spatial tolerance, as receptor positions are not fixed but could be adjusted (Figure 1 f), (II) an additional mechanism through which the cell can control signalling processes (Figure 1 g), and (III) since coupling of mobile receptors will considerably change their degrees of freedom, additional entropic costs may determine the favourability of the binding process (Figure 1 h). These costs are critical for the design of multivalent therapeutics with respect to their flexibility.
All reactions, including binding, can be described with respect to the energetic changes they induce. In general, the reaction is favourable if it results in a decrease of the Gibbs free energy ΔG<0. This change is defined as ΔG=ΔH−TΔS, determined by the temperature of the reaction (T), as well as changes in the enthalpy (ΔH) and entropy (ΔS). Constraint of molecules involved in multivalent binding (e.g., coupling of mobile receptors) results in a decrease of entropy (ΔS<0). However, we note that other phenomena occurring during binding might result in an increase of entropy, e.g., release of counterions. [20] The larger the entropic decrease is, the less favourable the binding reaction. Various components of the analysed system will contribute to these entropic costs, as they get restricted during the reaction.
This notion is widely discussed with respect to ligand design,[ 21 , 22 , 23 ] particularly in terms of conformational and combinatorial (ΔS comb) entropic costs of binding (Figure 1 c–e).[ 24 , 25 , 26 , 27 ] Similar considerations can be applied to study the mobility of receptors embedded in a lipid membrane. Their initial diffusion will dictate the magnitude of an unfavourable loss of the translational freedom upon multivalent binding (Figure 1 h). Additionally, their clustering resulting from recruitment contributes to the loss of the entropy of demixing. As here we consider solely the impact of membrane mobility, this entropy loss contributed by the receptors (ΔS r) is the only one we will discuss. More broad analyses of the thermodynamic aspect of multivalent tethering have previously been reviewed by Martinez‐Veracoechea and Leunissen. [24]
Here, we discuss the various aspects of cellular membrane mobility and its effects on multivalent binding, considering a geometrically constrained ligand. With this work, we want to emphasise how a deeper understanding of the dynamics involved in multivalent binding will broaden our insight into biological processes of highest importance: from viral entry to immune synapse formation. [28] In a focused rather than exhaustive manner, we will present four perspectives on the dynamic multivalency, as schematically summarised in Figure 2.
Figure 2.
Four aspects of membrane mobility in multivalent receptor/ligand binding discussed in this work: receptor mobility during (a) and before (b) binding, as well as binding‐dependent changes in the mobility of the receptor/ligand complex (c) and surrounding lipids (d). The thickness of the “movement” arrows indicates the magnitude of the diffusivity. Here, we do not discuss the flexibility of the ligand and assume the distance between its binding sites to remain unchanged throughout the binding process.
Starting with the first aspect of membrane mobility, diffusion of receptors allows their recruitment by the constrained ligand. Receptor diffusion makes coupling possible, even though their spacing is not initially adjusted to the geometry of a ligand (Figure 2 a). On the other hand, receptor preassembly may also occur, introducing an additional control mechanism for signalling processes inside‐out. This may result in a more favourable ligand binding due to smaller entropic penalty and increased local concentration (Figure 2 b). Thirdly, upon binding the diffusivity and localisation of the receptors in membrane compartments may change (Figure 2 c). Binding processes will also affect lipids within the bilayer, particularly the ones directly associated with embedded proteins, so‐called annular lipids. [29] This effect may give rise to the emergence of lipid–protein compartments of distinct physical properties (Figure 2 d).
The description of dynamic components of receptor binding is often highly complex. For example, proteins do not only undergo discrete changes between on/off mobility states, but develop various diffusion profiles. [30] Here we will not dissect these intricate mobility changes, but focus on the universal importance of kinetics in multivalent binding. In this Minireview we argue a simple, generalised idea behind dynamic multivalency: finding a balance between well‐defined, energetically favourable interactions, while allowing for a certain range of adjustability to reduce its dependency on the non‐significant environmental variations, [31] as well as provide the means for the cell to control vital signalling processes.[ 1 , 2 , 32 ]
2. Receptor Mobility During Binding
In the first scenario discussed here (Figure 2 a), the ligand approaches a surface on which receptor arrangement is neither highly concentrated nor preassembled to match the spacing of its binding sites. If the surface is immobile, no multivalent bonds could be formed, considering a geometrically restricted ligand and weak monovalent interactions. But what happens if the receptors on the surface can freely diffuse?
Binding often results in reorganisation of membrane components [33] and the presence of a ligand, even a monovalent one, can drive a change in receptor spacing. Any phenomenon modulating receptor/receptor interactions will change their clustering profile. For example, a clustering of lipid‐anchored zinc‐metalloporphyrins was studied in the presence of zinc‐binding ligands varying in chemical structure.[ 34 , 35 ] Even though clustering in the presence of a multivalent ligand was much stronger than for monovalent ones, [35] the studies showed that even monovalent binding influences clustering behaviour of the receptors by modifying their chemical and physical properties, [34] particularly hydrophobicity and charge distribution.
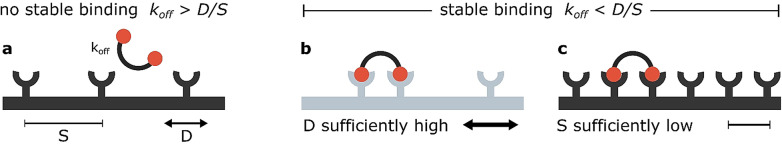
With clustering profiles changing upon binding, the receptors and partially bound ligands can exhibit increased affinity towards each other. However, such change is not necessary for receptor recruitment on a mobile surface. If the mobile receptor will find itself in close proximity to the pre‐formed complex before the monovalent binding dissociates, it can be recruited by a ligand,[ 36 , 37 , 38 , 39 , 40 ] forming a bivalently‐linked complex, as illustrated in Figure 3.
Figure 3.
Binding‐driven receptor clustering takes place when the pre‐binding event is slower than the timescale at which two receptors will diffuse in relevant proximity to each other. When the diffusion is too slow and the initial receptor spacing does not match the ligand's, no stable multivalent binding can be observed (a). However, if the receptors are quickly diffusing (b) or their concentration facilitates binding even on the immobile surfaces (c), the ligand can couple multiple receptors. Mobility of the membrane and the embedded receptors has been colour‐coded as previously: mobile (pale blue), immobile (black). The parameters mentioned are as follows: k off—the rate of dissociation of the pre‐bound receptor/ligand complex, D—diffusion coefficient of the receptors, S—surface area which the receptor needs to cover before arriving in proximity of the pre‐bound complex to enable a second stabilising binding event.
Three parameters are of primary importance in this case: k off [s−1]—the rate of dissociation of the pre‐bound receptor/ligand complex, D [μm2 s−1]—diffusion coefficient of the receptors, S [μm2]—surface area which the receptor needs to cover before arriving in proximity of the pre‐bound complex to enable a second stabilising binding event, strongly correlated with the receptor density. Assuming a diffusion‐limited reaction, the formation of multivalent connection depends on the interplay of these values, as presented by Equation 1.
| (1) |
Note, that Equation (1) presents a simplified case, introduced to illustrate the coupled importance of diffusivity and surface coverage of the receptors, and is not a complete mathematical model of binding. The equation neglects the influence of linker length and flexibility,[ 41 , 42 , 43 , 44 ] as well as the mobility of the complex which would change the statistical analysis of the problem. Virus entry involves one example of such mobility, where viruses were shown to diffuse upon the formation of an initial bond with the cell membrane.[ 45 , 46 ]
The approach described by Equation (1) and schematically illustrated in Figure 3 allows to conclude that with an increasing surface coverage of receptors their mobility becomes less important—even immobile receptors will efficiently bind the rigid ligand at appropriately high concentrations. The interplay between diffusion rates and surface density of receptors was therefore a topic of numerous research reports, as described below.
The interplay of surface density and mobility in determining efficiency of multivalent binding was widely studied, employing various model systems.[ 36 , 37 , 38 ] For example, binding efficiency between mobile and immobile surfaces was compared for biotin/streptavidin, [36] as well as with adamantane binding with multiple β‐cyclodextrins conjugated to a hyaluronan polymer acting as a multivalent scaffold. [37] The reports show that on immobile surfaces the efficiency and stability of binding was impaired compared to on mobile ones, however only below a particular (system‐specific) density of receptors. This suggests that the diffusion of receptors might allow for a spacing adjustment to the matching distance, while on an immobile, uniformly covered surface the distances between receptors are too large for a given multivalent ligand to allow for binding.
Similarly, liposomes functionalised with an inhibitory peptide bound by heptameric anthrax toxin [38] are reported to be more potent antitoxic agents when formed of a fluid‐phased (highly diffusive) lipid membrane. However, the selectivity of the binding was reported to be higher for immobile gel‐phased bilayers. This also suggests that the adjustment in receptor distances takes place on fluid bilayers, enabling more efficient binding in a wider range of surface coverages. Yet, the mobility effects on selectivity should not be disregarded, particularly in light of the importance of superselectivity offered by multivalency.
Analysis of physical aspects of surface binding is most often performed on model lipid bilayers,[ 36 , 37 , 38 , 39 ] yet reported biological observations have led to similar conclusions. For example, integrins at focal adhesion points were demonstrated to cycle between pre‐binding diffusive state and immobilisation upon their activation, [2] with the free diffusion prior to ligand binding being crucial for their activity.[ 47 , 48 , 49 ]
While experiments on biological systems indicated that pre‐binding diffusivity of receptors is often critical for their functionality, model membranes and simulations allowed better control over diffusion coefficient (D) and surface coverage (coupled with S). Together, this range of studies suggests that mobility of receptors embedded in the bilayer is of significance in many signalling cellular pathways involving multivalent processes, e.g., formation of immune synapses. [18] Clustering and cross‐linking of receptors often requires them to be mobile in the bilayer and can be strongly correlated with their activity.[ 30 , 50 , 51 , 52 , 53 , 54 ] This mobility, however, plays a bigger role at low receptor concentrations.[ 43 , 55 , 56 ] In sufficiently high surface coverage the receptors can find themselves in the binding range of a multivalent ligand with minimal translational movement. Similarly, the effects of receptor density are also manifested after ligand‐induced clustering, where a higher local density within the complex favours rebinding, but results in lower number of receptors available outside the contact area. [57]
Importantly, in physiological conditions high receptor concentrations are indeed possible to obtain. [43] In model systems this concentration was ensured globally,[ 58 , 59 , 60 ] yet in a compartmentalised cell membrane changes in receptor concentrations can be induced locally. Receptors confined in less diffusive lipid domains could be tightly spaced to better match the ligand geometry. Additionally, their dynamic behaviour upon binding will not change drastically, therefore the entropic cost of complex formation is effectively lower. Local increase in the concentration of receptors may also constitute a triggering mechanism for the cell signalling pathways, particularly in a view of concentration‐dependent superselectivity. With this number of advantages, such a solution was indeed adopted by the cellular membranes in the form of distinct lipid domains in a compartmentalised bilayer.
3. Pre‐Binding Receptor Assembly
In the previous section we analysed systems where spontaneous diffusion of membrane‐anchored receptors is crucial for the formation of multivalent receptor/ligand complexes. However, receptor clustering can also occur pre‐binding and act as a trigger for the complexation process (Figure 2 b), as reported, for example, in T cell receptor (TCR) nanoclustering, [61] illustrated in Figure 4. This mechanism is particularly significant when the architecture of the ligand features a distinct and spatially confined geometry, as highlighted in this Minireview. Such preassembly results in an increased local concentration of receptors and higher proximity of subsequent binding sites, therefore increasing association constant k on′′ of secondary binding following the formation of the first (monovalent) connection. Additionally, an even more significant effect will be observed in the decrease of the dissociation constant k off, [62] as rebinding will be facilitated with quickly accessible receptors. Furthermore, the decrease of receptors’ mobility pre‐binding reduces the entropic cost of coupling. Thus, their local assembly makes multivalent binding more energetically favourable, and as such has indeed been observed in various biological processes.[ 61 , 63 , 64 , 65 , 66 , 67 , 68 , 69 ] One of the most significant mechanisms employed by the cell to prearrange and spatially couple surface components is the compartmentalisation of the membrane.
Figure 4.
T cell receptors (TCR) form nanoclusters before binding to an antigenic peptide presented by major histocompatibility complex molecules (pMHC) on the surface of antigen‐presenting cells (APCs). Subsequent microcluster formation is critical to sustain the lifetime of signalling. The illustration was created after Schamel et al., Immunol. Rev. (2013). [61]
Compartmentalisation can be achieved by different mechanisms, such as particular protein–protein interactions (e.g. tetraspanin‐enriched microdomains)[ 70 , 71 ] or the phase separation of saturated lipids and cholesterol from the unsaturated lipid bilayers. [72] The latter is a most well‐known example of membrane domains, termed “rafts”. Here we will cover all the compartmentalisation mechanisms with an umbrella term “lipid domains”, which refers to parts of a membrane with distinct (diffusive) properties. For a recent analysis of the “raft” concept we direct the reader to the review by Levental et al., [73] which particularly emphasises that the formation of compartments in a membrane is a result of an interplay between both proteins and lipids.
Domains in the lipid bilayer are characterised by distinct properties, especially their diffusive behaviour. [73] An important role of these compartments is to concentrate membrane components, which is significant for many multivalent mechanisms [74] and is particularly worth studying with the concept of concentration‐dependent superselectivity in mind. It has been observed that in various signalling processes membrane receptors aggregate prior to ligand attachment,[ 66 , 67 , 68 ] supporting the idea of multivalent binding depending on preassembly of surface receptors: at locally increased concentrations recruitment of multiple binders is facilitated and the rebinding of a dissociated ligand is significantly more probable.[ 69 , 75 ] We suggest that the pre‐binding clustering additionally increases selectivity towards densely covered membrane compartments, as the probability of receptor/ligand encounter is shifted towards more populated regions at non‐uniformly covered surfaces. [69]
Proteins, including receptors, can be sorted into localised domains via lipidation: posttranslational modifications with various lipid moieties, guiding their localisation in particular membrane parts. [76] For example, lipidation of TCRs targets them towards specific lipid domains and it was shown that when this affinity is lost their signalling activity is compromised. [77] Although protein segregation receives a lot of attention in biological systems,[ 63 , 64 , 65 ] proteins themselves can also induce the segregation of bilayer components. The mobility and distribution of membrane units, both lipids and proteins, are strongly coupled and change in a co‐dependent manner. [73] For instance, during T cell activation, nanoscale lipid domains coalesce into larger patches, where associated proteins have strongly reduced mobility. [78]
Hence, various membrane components cooperate to find the most energetically stable system for multivalent binding. In case of prearrangement guided by lipid compartments, constrain within a low‐mobility domain decreases the translational entropic penalty of the receptors, as the restriction of their diffusivity is less drastic. Furthermore, their preassembly leads to an increase in their local concentration, suggesting the possible action of superselectivity mechanisms. As a result of such density increase, the association constant k on′′ of the secondary coupling is higher, but more significantly k off is lower due to facilitated rebinding. This shifts the overall binding equilibrium towards a lower K d=k off/k on. Additionally, the cell benefits from the control over preassembly‐dependent binding, being now determined by the membrane, not the environment or the ligand. Thus, the metabolism of the cell can guide the inside‐out signalling by changing the local arrangement of the membrane receptors.
Compartmentalisation of the membrane plays a role not only in triggering binding, but also after the establishment of the multivalent interaction. In a broad spectrum of membrane‐binding viruses, some receptors are found in low‐mobility lipid domains before ligand encounter, while in some cases they are only translocated there upon the viral attachment. [79] This leads to another question considered in this work: how does the mobility of membrane‐anchored structures change upon multivalent coupling?
4. Receptor/Ligand Complex Mobility
Mobility is a crucial parameter determining multivalent binding, but in turn, binding will also affect the dynamic properties of the engaged molecules. Shortly, upon bond formation the kinetics of the complex will invariably change (Figure 2 c).
In a first approximation, the formation of a receptor/ligand bond changes the molecular volume of the diffusing entity, both upon monovalent ligand binding to a single receptor, as well as consecutive increase in valency of interactions. As the radius of the inclusion increases, its diffusivity in a membrane will decrease. These changes in the translational diffusion D t of a membrane‐embedded complex—approximated as cylinder of radius R—is described by two models.
The Saffman–Delbrück (SD) model, valid only for membrane inclusions that are small compared with the characteristic length scale (l m), indicates scaling of translational diffusion with R as D t∝ln(2l m/R). In the intermediate and large inclusions, the Hughes‐Pailthorpe‐White (HPW) model predicts D t∝1/R scaling.[ 80 , 81 ] Both membrane proteins[ 82 , 83 ] and lipid domains[ 84 , 85 ] were shown to follow the SD model, where an increase in size causes a weak logarithmic decrease of their diffusion. However, SD model fails at describing mobility of large constructs, and is therefore dissuaded from being used in membrane domain analysis. [80]
Note that both models are only valid for free‐standing, rather than supported lipid bilayers. While these models consider the friction components of the flow surrounding the embedded structure, on the bilayer close to a solid substrate another factor will affect the diffusive behaviour: interactions between the lipids and the surface. This model, named after Evans and Sackmann (ES), predicts much sharper dependency of the diffusivity on the inclusion's radius: D t∝1/R 2. Even though supported lipid membranes are not found in natural systems, fundamental studies often employ supported model bilayers, and so the ES model should not be disregarded. Further details of these hydrodynamic models can be found in the review by Block. [81]
Following the hydrodynamic analysis, we consider a general statement that the mobility of a receptor/ligand system is reduced upon multivalent coupling. This is indeed observed, for example for a virus binding with increasing copies of receptors on the host membrane. For the description of changes in virus dynamics upon surface attachment we direct the reader to the review by Boulant et al. [86]
The mobility decrease was also reported in the previously mentioned studies on multivalent binding on model bilayers using adamantane/β‐cyclodextrins complex. [37] Using fluorescence recovery after photobleaching (FRAP), a strong reduction in adamantane diffusion coefficient, from 0.86±0.01 μm2 s−1 to 0.15±0.01 μm2 s−1, was observed after multivalent probe was introduced to the system. Similar observations were made by studying a biological system of GABAA receptors binding with muscimol ligand on hippocampal neurons with a fluorescence correlation spectroscopy (FCS). [87] Diffusion coefficient of a freely diffusing muscimol ligand (approx. 230 μm2 s−1) was found to be two orders of magnitude higher than for the bound receptor/ligand complex (approx. 3 μm2 s−1). Importantly, in both cases binding processes gave rise to the emergence of a population of receptors with much more strongly reduced diffusivity. This is suggested to be the result of association with immobile lipid domains, but could also hint on post‐binding clustering or the presence of immobile fraction of receptor proteins. These findings imply a more complex biophysical behaviour, perhaps also guided by biological phenomena, although we highlight that the cited reports were investigating both cellular, as well as model membranes.
A significant aspect of complex formation to be unravelled is how the mobilities of individual receptors are coupled after multivalent ligand binding. Model DNA‐based multivalent systems were employed to investigate this question.[ 88 , 89 ] In one of the studies, vesicles were used as multivalent ligands carrying cholesterol‐anchored oligonucleotides, binding to complementary DNA strands tethered to the surface. The distribution of their diffusion coefficients studied with total internal reflection fluorescence (TIRF) presented clearly distinct peaks after eliminating transient binders. These were attributed to vesicles bound with a defined number of DNA anchors, as schematically presented in Figure 5 a. This analysis showed that the diffusion coefficient (D) of multivalently bound ligands on a mobile surface depends on a valency of anchorage (n) as D∝1/n. Linear scaling of diffusivity with n shows that despite being cross‐linked by a multivalent ligand, the receptors continue to move independently, following the free draining model, rather than as an aggregated ensemble. Independent anchor mobilities have also been demonstrated by other reports.[ 90 , 91 ]
Figure 5.
The diffusivity of receptors changes upon multivalent binding. a) The DNA‐tethered vesicles (red) used as a model multivalent system, [88] showing that diffusion coefficient D is inversely proportional to the valency of anchorage n. The D∝1/n dependency suggests independent mobility of each bound “receptor”. b) In resting B cells, the B‐cell receptor (BCR) is excluded from low mobility lipid domains (rafts). Upon binding, multivalent antigen oligomerises the BCR, increasing its affinity for the domains, where it undergoes phosphorylation which initiates the signalling cascade. The illustration was sketched after Pierce, Nat. Rev. Immunol. (2002). [93]
Complex formation may change receptor affinity towards particular lipid domains in cellular membranes. Certain membrane proteins are not associated with less mobile compartments when unbound, but they move towards them upon their multivalent coupling. [92] For example, cross‐linking of B cell receptors results in their increased affinity towards lipid rafts, where the complex becomes stably incorporated, [93] as illustrated in Figure 5 b. Similarly, integrins pre‐organise into nanoclusters which are spatially linked with lipid domains, yet remaining mobile before binding. Upon ligand association, larger microclusters of ligand/integrin receptor complexes are formed. [47] Binding of a multivalent Fc receptor (Fc RI) to immunoglobulin E (IgE) is another example where the complex loses its lateral mobility upon multivalent binding, compared with a monomeric interaction. [94]
To investigate this in more detail, experiments were performed on integrins embedded in model bilayers formed with coexisting domains: cholesterol‐deficient liquid disordered (Ld) phases of higher mobility and cholesterol‐rich liquid ordered (Lo) phases of lower mobility. [95] Studied integrins showed affinity for liquid disordered phases in the absence of a ligand, yet after the addition of binding vitronectin their preference towards less mobile liquid ordered domains is more pronounced. This transition was shown to occur with no changes in integrin conformation, suggesting that it is guided by a biophysical mechanism.
In summary, not only the mobility impacts binding—the binding can in turn change the mobility of the membrane components. The expected decrease in diffusivity following an increase of size can be accompanied by changes in complex affinity towards a particular lipid environment. In some cases, receptors’ transfer towards low‐mobility domains upon binding might be more thermodynamically favourable. Importantly, it is suggested that the receptors coupled by a multivalent ligand exhibit independent movements of each anchor in the bilayer, although more nuanced studies of this phenomenon are yet to be performed.
5. Binding‐Dependent Lipid Mobility
So far, we have discussed the effects of lipid bilayer mobility on multivalent surface binding. Since the cell membrane relies on reciprocal interactions between its components, membrane proteins also have a modulating effect on the lipid environment (Figure 2 d), e.g., by inducing changes of bilayer thickness [96] or curvature. [97] Thus, membrane‐spanning proteins interact and influence lipids surrounding them.
Particularly, the lateral diffusions of membrane‐embedded proteins and their annular lipids are correlated. It has been shown that diffusion of lipids in the proximity of proteins is noticeably decreased compared to freely diffusing ones.[ 98 , 99 , 100 ] A protein determines the formation of a nanometre‐scale lipid patch of reduced mobility, revoking the compartmentalisation of membranes described above. On the other hand, the mobility of the formed receptor/ligand complex was shown to depend on the strength of protein–lipid association, [101] further illustrating the reciprocal action of proteins and surrounding lipids.
The strong correlation between receptors and their lipid environment manifests also in the changes in lipid mobility upon ligand binding. Since a receptor protein is coupled with the annular lipids, its binding and clustering will influence membrane organisation. Studies on pentameric cholera toxin binding of membrane‐anchored monosialoganglioside GM1 hint at the importance of the lipid phase in how binding affects the lipid mobility; only the background lipids (not participating in the binding process) with the phase‐transition temperature close to the room temperature showed noticeable reduction of the lateral diffusion rate upon receptor/ligand association. [102] These findings suggest that the changes in lipid mobility determined by the embedded proteins may be of similar nature to the phase‐dependent membrane compartmentalisation.
For ligands with mobile binding sites, adhesion to (also mobile) membrane receptors can promote the coalescence of less diffusive lipid domains.[ 103 , 104 ] Simulation‐based studies showed that binding occurring within the multiple nanodomains “clips in” the membrane in scattered positions. This restrains its spontaneous fluctuations considerably. Upon domains coalescing into a bigger patch of less mobile lipids, a large portion of the membrane remains unrestrained, making such arrangement energetically favourable, as illustrated in Figure 6.
Figure 6.
Schematic illustration of coalescence of ligand‐associated mobile lipid nanodomains. Here, lipids surrounding the receptor are spatially coupled with it, but the whole domain remains diffusive. Initial binding in random positions (a) results in largely restrained fluctuation of the membrane. Upon establishing the connections, demixing of the annular domains (b) is energetically favoured, as it allows for less restrictions of the system.
Coalescence upon multivalent binding was indeed observed experimentally, also in the case of geometrically constrained ligand, e.g., the pentameric cholera toxin, whose presence in the system resulted in lipid sections clustering into larger domains. [105] We propose that the described phenomenon may play a role in the emergence of larger patches of less mobile lipids upon binding, observed also in other biological systems.[ 61 , 73 , 106 ] Additionally, studies on cholera toxin showed that the lipids surrounding coupled receptors rearrange to adapt to their newly forced positions. [107] This gives rise to an emergence of a textured lipid pattern, with the main bilayer reorganisation formed in the vicinity of the centre of the bound protein. This in turn was correlated with the biological function of the protein: cholera toxin induces membrane perturbation in its centre to allow DNA translocation inside the cell. This example illustrates how the changes observed in the molecule conformation, distribution and mobility, even if they are forced by laws of physics, influence and benefit biological processes.
6. Conclusion
As the actions of lipids and proteins are strongly reciprocal, multivalent binding on a membrane links physical properties of the lipid bilayer with the biological coupling of surface components. Nature often makes use of geometrically constrained ligands as a highly controllable binder. Consequently, the spatial distribution of targeted receptors becomes an important parameter determining the binding. We took a critical look at significant phenomena and variables that guide the dynamic aspects of multivalent binding. Particularly, we discussed how the spontaneous mobility of membrane structures allows recruitment of multiple receptors by multivalent ligand, which upon initial monovalent binding engages further receptors depending on their diffusion and surface coverage. Alternatively, preassembly of receptors facilitates binding by increasing their local concentration. There, intracellular processes can guide bilayer compartmentalisation, which provides local variations in concentration and diffusivity of the interacting molecules, particularly important for cell‐guided superselectivity manifesting below a specific receptor spacing. Furthermore, upon coupling, the mobility of the formed receptor/ligand complex is dependent on the valency of binding and can regulate the complex's localisation in a particular lipid environment. Finally, while the properties of the bilayer strongly determine the binding processes, membrane‐embedded proteins also influence the dynamics of surrounding (annular) lipids, and their multivalent coupling may result in lipid domains coalescing into larger patches. In short, all aspects of dynamic multivalency are strongly interdependent, as schematically summarised in Figure 7.
Figure 7.
Graphic summary of mobility/multivalency interplay as discussed in this work. The effects of lipids and proteins on their diffusivity are reciprocal, and similarly binding will influence not only receptor spacing and mobility, but the dynamics of the surrounding lipids as well.
Multivalent binding is one of the processes that gain a lot from both biological and physical insights. The mechanical properties of the bilayer and thermodynamics‐driven behaviour are strongly coupled with the signalling processes and chemistry of embedded proteins. An important remaining challenge is to deconvolute these aspects: provide a general description of the dynamic systems and assess which phenomena are specific to a particular receptor/ligand pair and a cellular binding process. In that respect, multivalent coupling calls for a close interdisciplinary collaboration in order to fully unravel the foundations of receptor/ligand binding in the context of dynamic signalling. In the light of increasing application of superselectivity in synthetic biodevices, a closer look at its dependency on membrane mobility will guide the design of novel biosensors, drug delivery systems or vaccines. Importantly, it can also give rise to an innovative generation of therapeutics, targeting not just the specific biochemical features of the molecular interactions, but make use of their dynamics and biophysical properties to control the selectivity of binding.
Conflict of interest
The authors declare no conflict of interest.
Biographical Information
Diana Morzy obtained her PhD at the Cavendish Laboratory, University of Cambridge, holding scholarships from EPSRC and the Winton Programme for the Physics of Sustainability. Since then, she has been working at EPFL in Lausanne as a postdoctoral researcher, modelling the effects of dynamic components in multivalent binding on lipid membranes.

Biographical Information
Maartje Bastings is a Dutch biomaterials engineer leading the Programmable Biomaterials Laboratory at EPFL. Her lab specialises in DNA‐based supramolecular materials using multivalency as driving force for molecular design. She has emerged as specialist in bridging supramolecular materials with biology, taking an engineering approach with a focus on biophysical quantification of interactions. Her goal is to use multivalent patterns to control the properties of surfaces and soft matter, and explore the role of spatial tolerance in molecular biology.

Acknowledgements
The authors acknowledge funding from Volkswagen Stiftung “Life?” (Grant No. 98200) and thank Dr. Jorieke Weiden and Vincenzo Caroprese for their help with the manuscript. Open access funding provided by Ecole Polytechnique Federale de Lausanne.
D. Morzy, M. Bastings, Angew. Chem. Int. Ed. 2022, 61, e202114167; Angew. Chem. 2022, 134, e202114167.
References
- 1. Urbančič I., Schiffelers L., Jenkins E., Gong W., Santos A. M., Schneider F., O'Brien-Ball C., Vuong M. T., Ashman N., Sezgin E., Eggeling C., FEBS Lett. 2021, 595, 2127–2146. [DOI] [PubMed] [Google Scholar]
- 2. Rossier O., Octeau V., Sibarita J. B., Leduc C., Tessier B., Nair D., Gatterdam V., Destaing O., Albigès-Rizo C., Tampé R., Cognet L., Choquet D., Lounis B., Giannone G., Nat. Cell Biol. 2012, 14, 1057–1067. [DOI] [PubMed] [Google Scholar]
- 3. Curk T., Dobnikar J., Frenkel D., in Multivalency, Wiley, Chichester, 2017, pp. 75–101. [Google Scholar]
- 4. Martinez-Veracoechea F. J., Frenkel D., Proc. Natl. Acad. Sci. USA 2011, 108, 10963–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Scheepers M. R. W., van IJzendoorn L. J., Prins M. W. J., Proc. Natl. Acad. Sci. USA 2020, 117, 22690–22697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Christy A. T. R., Kusumaatmaja H., Miller M. A., Phys. Rev. Lett. 2021, 126, 028002. [DOI] [PubMed] [Google Scholar]
- 7. Curk T., Brackley C. A., Farrell J. D., Xing Z., Joshi D., Direito S., Bren U., Angioletti-Uberti S., Dobnikar J., Eiser E., Frenkel D., Allen R. J., Proc. Natl. Acad. Sci. USA 2020, 117, 8719–8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang Z., Ali M. M., Eckert M. A., Kang D. K., Chen Y. Y., Sender L. S., Fruman D. A., Zhao W., Biomaterials 2013, 34, 9728–9735. [DOI] [PubMed] [Google Scholar]
- 9. Carlson C. B., Mowery P., Owen R. M., Dykhuizen E. C., Kiessling L. L., ACS Chem. Biol. 2007, 2, 119–127. [DOI] [PubMed] [Google Scholar]
- 10. Delguste M., Zeippen C., Machiels B., Mast J., Gillet L., Alsteens D., Sci. Adv. 2018, 4, 1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cuellar-Camacho J. L., Bhatia S., Reiter-Scherer V., Lauster D., Liese S., Rabe J. P., Herrmann A., Haag R., J. Am. Chem. Soc. 2020, 142, 12181–12192. [DOI] [PubMed] [Google Scholar]
- 12. Chittasupho C., Ther. Delivery 2012, 3, 1171–1187. [DOI] [PubMed] [Google Scholar]
- 13. Liese S., Netz R. R., ACS Nano 2018, 12, 4140–4147. [DOI] [PubMed] [Google Scholar]
- 14. Li T., Tracka M. B., Uddin S., Casas-Finet J., Jacobs D. J., Livesay D. R., PLoS Comput. Biol. 2015, 11, e1004327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zimmermann J., Romesberg F. E., Brooks C. L., Thorpe I. F., J. Phys. Chem. B 2010, 114, 7359–7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shaw A., Hoffecker I. T., Smyrlaki I., Rosa J., Grevys A., Bratlie D., Sandlie I., Michaelsen T. E., Andersen J. T., Högberg B., Nat. Nanotechnol. 2019, 14, 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schvartzman M., Palma M., Sable J., Abramson J., Hu X., Sheetz M. P., Wind S. J., Nano Lett. 2011, 11, 1306–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li M., Yu Y., J. Cell Sci. 2021, 134, 2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nie C., Parshad B., Bhatia S., Cheng C., Stadtmüller M., Oehrl A., Kerkhoff Y., Wolff T., Haag R., Angew. Chem. Int. Ed. 2020, 59, 15532–15536; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 15662–15666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Achazi K., Haag R., Ballauff M., Dernedde J., Kizhakkedathu J. N., Maysinger D., Multhaup G., Angew. Chem. Int. Ed. 2021, 60, 3882–3904; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 3926–3950. [Google Scholar]
- 21. Krishnamurthy V. M., Estroff L. A., Whitesides G. M., Fragment-Based Approaches in Drug Discovery, Wiley-VCH, Weinheim, 2006, pp. 11–53. [Google Scholar]
- 22. Kane R. S., Langmuir 2010, 26, 8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bandlow V., Liese S., Lauster D., Ludwig K., Netz R. R., Herrmann A., Seitz O., J. Am. Chem. Soc. 2017, 139, 16389–16397. [DOI] [PubMed] [Google Scholar]
- 24. Martinez-Veracoechea F. J., Leunissen M. E., Soft Matter 2013, 9, 3213–3219. [Google Scholar]
- 25. Nangreave J., Yan H., Liu Y., J. Am. Chem. Soc. 2011, 133, 4490–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. von Krbek L. K. S., Achazi A. J., Schoder S., Gaedke M., Biberger T., Paulus B., Schalley C. A., Chemistry 2017, 23, 2877–2883. [DOI] [PubMed] [Google Scholar]
- 27. Dubel N., Liese S., Scherz F., Seitz O., Angew. Chem. Int. Ed. 2019, 58, 907–911; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2019, 131, 918–923. [Google Scholar]
- 28. Kiessling L. L., Lamanna A. C., in Chem. Probes Biol., Springer, Dordrecht, 2003, pp. 345–357. [Google Scholar]
- 29. Warren G. B., Houslay M. D., Metcalfe J. C., Birdsall N. J. M., Nature 1975, 255, 684–687. [DOI] [PubMed] [Google Scholar]
- 30. Dustin M. L., Bivona T. G., Philips M. R., Nat. Immunol. 2004, 5, 363–372. [DOI] [PubMed] [Google Scholar]
- 31. Bastings M. M. C., Hermans T. M., Spiering A. J. H., Kemps E. W. L., Albertazzi L., Kurisinkal E. E., Dankers P. Y. W., Macromol. Biosci. 2019, 19, 1800296. [DOI] [PubMed] [Google Scholar]
- 32. Abram C. L., Lowell C. A., Annu. Rev. Immunol. 2009, 27, 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jung H., Robison A. D., Cremer P. S., J. Struct. Biol. 2009, 168, 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tomas S., Milanesi L., Nat. Chem. 2010, 2, 1077–1083. [DOI] [PubMed] [Google Scholar]
- 35. Grochmal A., Ferrero E., Milanesi L., Tomas S., J. Am. Chem. Soc. 2013, 135, 10172–10177. [DOI] [PubMed] [Google Scholar]
- 36. Dubacheva G. V., Araya-Callis C., Geert Volbeda A., Fairhead M., Codeé J., Howarth M., Richter R. P., J. Am. Chem. Soc. 2017, 139, 4157–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dubacheva G. V., Curk T., Frenkel D., Richter R. P., J. Am. Chem. Soc. 2019, 141, 2577–2588. [DOI] [PubMed] [Google Scholar]
- 38. Rai P., Padala C., Poon V., Saraph A., Basha S., Kate S., Tao K., Mogridge J., Kane R. S., Nat. Biotechnol. 2006, 24, 582–586. [DOI] [PubMed] [Google Scholar]
- 39. Di Iorio D., Lu Y., Meulman J., Huskens J., Chem. Sci. 2020, 11, 3307–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Albertazzi L., Martinez-Veracoechea F. J., Leenders C. M. A., Voets I. K., Frenkel D., Meijer E. W., Proc. Natl. Acad. Sci. USA 2013, 110, 12203–12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huskens J., Curr. Opin. Chem. Biol. 2006, 10, 537–543. [DOI] [PubMed] [Google Scholar]
- 42. Fasting C., Schalley C. A., Weber M., Seitz O., Hecht S., Koksch B., Dernedde J., Graf C., Knapp E. W., Haag R., Angew. Chem. Int. Ed. 2012, 51, 10472–10498; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2012, 124, 10622–10650. [Google Scholar]
- 43. Szklarczyk O. M., González-Segredo N., Kukura P., Oppenheim A., Choquet D., Sandoghdar V., Helenius A., Sbalzarini I. F., Ewers H., PLoS Comput. Biol. 2013, 9, e1003310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Overeem N. J., Hamming P. H. E., Grant O. C., Di Iorio D., Tieke M., Bertolino M. C., Li Z., Vos G., De Vries R. P., Woods R. J., Tito N. B., Boons G. J. P. H., Van Der Vries E., Huskens J., ACS Cent. Sci. 2020, 6, 2311–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Barrow E., Nicola A. V., Liu J., Virol. J. 2013, 10, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mercer J., Schelhaas M., Helenius A., Annu. Rev. Biochem. 2010, 79, 803–833. [DOI] [PubMed] [Google Scholar]
- 47. Bakker G. J., Eich C., Torreno-Pina J. A., Diez-Ahedo R., Perez-Samper G., Van Zanten T. S., Figdor C. G., Cambi A., Garcia-Parajo M. F., Proc. Natl. Acad. Sci. USA 2012, 109, 4869–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Eich C., Manzo C., De Keijzer S., Bakker G. J., Reinieren-Beeren I., Garcia-Parajo M. F., Cambi A., Sci. Rep. 2016, 6, 20693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mehrbod M., Mofrad M. R. K., PLoS Comput. Biol. 2013, 9, 1002948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chan P. Y., Lawrence M. B., Dustin M. L., Ferguson L. M., Golan D. E., Springer T. A., J. Cell Biol. 1991, 115, 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Irannejad R., Von Zastrow M., Curr. Opin. Cell Biol. 2014, 27, 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Metzger H., Kinet J., FASEB J. 1988, 2, 3–11. [DOI] [PubMed] [Google Scholar]
- 53. Courtney A. H., Puffer E. B., Pontrello J. K., Yang Z. Q., Kiessling L. L., Proc. Natl. Acad. Sci. USA 2009, 106, 2500–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tzlil S., Ben-Shaul A., Biophys. J. 2005, 89, 2972–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. English T. J., Hammer D. A., Biophys. J. 2005, 88, 1666–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dwyer J. D., Bloomfield V. A., Biopolymers 1981, 20, 2323–2336. [DOI] [PubMed] [Google Scholar]
- 57. Overeem N. J., van der Vries E., Huskens J., Small 2021, 17, 2007214. [DOI] [PubMed] [Google Scholar]
- 58. Thomas G. B., Rader L. H., Park J., Abezgauz L., Danino D., DeShong P., English D. S., J. Am. Chem. Soc. 2009, 131, 5471–5477. [DOI] [PubMed] [Google Scholar]
- 59. Jiang H., Smith B. D., Chem. Commun. 2006, 16, 1407–1409. [DOI] [PubMed] [Google Scholar]
- 60. Doyle E. L., Hunter C. A., Phillips H. C., Webb S. J., Williams N. H., J. Am. Chem. Soc. 2003, 125, 4593–4599. [DOI] [PubMed] [Google Scholar]
- 61. Schamel W. W. A., Alarcón B., Immunol. Rev. 2013, 251, 13–20. [DOI] [PubMed] [Google Scholar]
- 62. Huskens J., Prins L. J., Haag R., Ravoo B. J., Multivalency: Concepts, Research & Applications, Wiley, Hoboken, 2018. [Google Scholar]
- 63. Brown D. A., Rose J. K., Cell 1992, 68, 533–544. [DOI] [PubMed] [Google Scholar]
- 64. Stone M. B., Shelby S. A., Nńñez M. F., Wisser K., Veatch S. L., Elife 2017, 6, e19891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Varshney P., Yadav V., Saini N., Immunology 2016, 149, 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Qin K., Dong C., Wu G., Lambert N. A., Nat. Chem. Biol. 2011, 7, 740–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Volkó J., Kenesei Á., Zhang M., Várnai P., Mocsár G., Petrus M. N., Jambrovics K., Balajthy Z., Müller G., Bodnár A., Tóth K., Waldmann T. A., Vámosi G., Proc. Natl. Acad. Sci. USA 2019, 116, 21120–21130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Su Z., Dhusia K., Wu Y., J. Chem. Phys. 2021, 154, 0055101. [DOI] [PubMed] [Google Scholar]
- 69. Caré B. R., Soula H. A., BMC Syst. Biol. 2011, 5, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Halova I., Draber P., Front. Cell Dev. Biol. 2016, 4, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yáñez-Mó M., Barreiro O., Gordon-Alonso M., Sala-Valdés M., Sánchez-Madrid F., Trends Cell Biol. 2009, 19, 434–446. [DOI] [PubMed] [Google Scholar]
- 72. Sengupta P., Baird B., Holowka D., Semin. Cell Dev. Biol. 2007, 18, 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Levental I., Levental K. R., Heberle F. A., Trends Cell Biol. 2020, 30, 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Simons K., Toomre D., Nat. Rev. Mol. Cell Biol. 2000, 1, 31–39. [DOI] [PubMed] [Google Scholar]
- 75. Gopalakrishnan M., Forsten-Williams K., Nugent M. A., Täuber U. C., Biophys. J. 2005, 89, 3686–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Levental I., Lingwood D., Grzybek M., Coskun Ü., Simons K., Proc. Natl. Acad. Sci. USA 2010, 107, 22050–22054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kabouridis P. S., Mol. Membr. Biol. 2006, 23, 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tanimura N., Nagafuku M., Minaki Y., Umeda Y., Hayashi F., Sakakura J., Kato A., Liddicoat D. R., Ogata M., Hamaoka T., Kosugi A., J. Cell Biol. 2003, 160, 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ripa I., Andreu S., López-Guerrero J. A., Bello-Morales R., Front. Microbiol. 2021, 12, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Petrov E., Schwille P., Biophys. J. 2008, 94, L41–L43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Block S., Biomolecules 2018, 8, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Weiß K., Neef A., Van Q., Kramer S., Gregor I., Enderlein J., Biophys. J. 2013, 105, 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ramadurai S., Holt A., Krasnikov V., Van Den Bogaart G., Killian J. A., Poolman B., J. Am. Chem. Soc. 2009, 131, 12650–12656. [DOI] [PubMed] [Google Scholar]
- 84. Stanich C. A., Honerkamp-Smith A. R., Putzel G. G., Warth C. S., Lamprecht A. K., Mandal P., Mann E., Hua T. A. D., Keller S. L., Biophys. J. 2013, 105, 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cicuta P., Keller S. L., Veatch S. L., J. Phys. Chem. B 2007, 111, 3328–3331. [DOI] [PubMed] [Google Scholar]
- 86. Boulant S., Stanifer M., Lozach P. Y., Viruses 2015, 7, 2794–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Meissner O., Häberlein H., Biochemistry 2003, 42, 1667–1672. [DOI] [PubMed] [Google Scholar]
- 88. Block S., Zhdanov V. P., Höök F., Nano Lett. 2016, 16, 4382–4390. [DOI] [PubMed] [Google Scholar]
- 89. Merminod S., Edison J. R., Fang H., Hagan M. F., Rogers W. B., Nanoscale 2021, 13, 12602–12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Camley B. A., Brown F. L. H., Soft Matter 2013, 9, 4767–4779. [Google Scholar]
- 91. Knight J. D., Lerner M. G., Marcano-Velá Zquez J. G., Pastor R. W., Falke J. J., Biophys. J. 2010, 99, 2879–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Füllekrug J., Simons K., Ann. N. Y. Acad. Sci. 2004, 1014, 164–169. [DOI] [PubMed] [Google Scholar]
- 93. Pierce S. K., Nat. Rev. Immunol. 2002, 2, 96–105. [DOI] [PubMed] [Google Scholar]
- 94. Holowka D., Baird B., Annu. Rev. Biophys. Biomol. Struct. 1996, 25, 79–112. [DOI] [PubMed] [Google Scholar]
- 95. Ge Y., Gao J., Jordan R., Naumann C. A., Biophys. J. 2018, 114, 158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kaiser H. J., Orłowski A., Róg T., Nyholm T. K. M., Chai W., Feizi T., Lingwood D., Vattulainen I., Simons K., Proc. Natl. Acad. Sci. USA 2011, 108, 16628–16633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Corradi V., Mendez-Villuendas E., Ingólfsson H. I., Gu R. X., Siuda I., Melo M. N., Moussatova A., Degagné L. J., Sejdiu B. I., Singh G., Wassenaar T. A., Delgado Magnero K., Marrink S. J., Tieleman D. P., ACS Cent. Sci. 2018, 4, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Pluhackova K., Gahbauer S., Kranz F., Wassenaar T. A., Böckmann R. A., PLoS Comput. Biol. 2016, 12, e1005169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Niemelä P. S., Miettinen M. S., Monticelli L., Hammaren H., Bjelkmar P., Murtola T., Lindahl E., Vattulainen I., J. Am. Chem. Soc. 2010, 132, 7574–7575. [DOI] [PubMed] [Google Scholar]
- 100. Ebersberger L., Schindler T., Kirsch S. A., Pluhackova K., Schambony A., Seydel T., Böckmann R. A., Unruh T., Front. Cell Dev. Biol. 2020, 8, 579388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kalli A. C., Rog T., Vattulainen I., Campbell I. D., Sansom M. S. P., J. Membr. Biol. 2017, 250, 337–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yamazaki V., Sirenko O., Schafer R. J., Groves J. T., J. Am. Chem. Soc. 2005, 127, 2826–2827. [DOI] [PubMed] [Google Scholar]
- 103. Li L., Wang X., Wu H., Shao Y., Wu H., Song F., Front. Mol. Biosci. 2021, 8, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Li L., Hu J., Shi X., Różycki B., Song F., Soft Matter 2021, 17, 1912–1920. [DOI] [PubMed] [Google Scholar]
- 105. Van Zanten T. S., Gómez J., Manzo C., Cambi A., Buceta J., Reigada R., Garcia-Parajo M. F., Proc. Natl. Acad. Sci. USA 2010, 107, 15437–15442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Viola A., Trends Immunol. 2001, 22, 322–327. [DOI] [PubMed] [Google Scholar]
- 107. Watkins E. B., Miller C. E., Majewski J., Kuhl T. L., Proc. Natl. Acad. Sci. USA 2011, 108, 6975–6980. [DOI] [PMC free article] [PubMed] [Google Scholar]