Significance
Alterations in the epigenome play a critical role in the malignant progression of brain tumors. Different subtypes of brain tumors exhibit distinct patterns of epigenomic remodeling; however, the underlying mechanisms that regulate these context-specific epigenetic states are poorly defined. Using native mouse models of high-grade glioma and ependymoma, we discovered that the developmental transcription factor Sox9 plays context-specific roles in these subtypes of brain tumors. These tumor-specific roles for Sox9 are driven by distinct interactions with the histone deacetylase complex in high-grade glioma and ZFTA-RELA protein in ependymoma. Our findings indicate that the roles of developmental transcription factors in brain tumor can vary based on distinct epigenetic functions and have critical implications for the clinical use of epigenetic therapies.
Keywords: high-grade glioma, ependymoma, epigenetics, histone, transcription
Abstract
Epigenetic dysregulation is a universal feature of cancer that results in altered patterns of gene expression that drive malignancy. Brain tumors exhibit subtype-specific epigenetic alterations; however, the molecular mechanisms responsible for these diverse epigenetic states remain unclear. Here, we show that the developmental transcription factor Sox9 differentially regulates epigenomic states in high-grade glioma (HGG) and ependymoma (EPN). Using our autochthonous mouse models, we found that Sox9 suppresses HGG growth and expands associated H3K27ac states, while promoting ZFTA-RELA (ZRFUS) EPN growth and diminishing H3K27ac states. These contrasting roles for Sox9 correspond with protein interactions with histone deacetylating complexes in HGG and an association with the ZRFUS oncofusion in EPN. Mechanistic studies revealed extensive Sox9 and ZRFUS promoter co-occupancy, indicating functional synergy in promoting EPN tumorigenesis. Together, our studies demonstrate how epigenomic states are differentially regulated in distinct subtypes of brain tumors, while revealing divergent roles for Sox9 in HGG and EPN tumorigenesis.
Epigenetic dysregulation is a hallmark feature of cancer that contributes to the pathophysiology of a host of malignant brain tumors and intersects with distinct developmental programs (1, 2). High-grade glioma (HGG) is a universally lethal form of brain cancer, where nearly half of adult HGGs contain mutations in chromatin modifying genes (3, 4), in addition to somatic mutations, copy number alterations, and gene fusions. Pediatric HGGs are defined by mutations in histone variant H3.3 that co-occur with mutations in p53 and chromatin remodeler ATRX (5). Other forms of pediatric brain tumors like ependymoma (EPN) have very low mutation rates and yet are characterized by patterns of epigenetic dysfunction, including global DNA hypomethylation, CpG island hypermethylation, and loss of repressive histone mark (6). Together, these observations highlight the epigenetic changes prevalent in brain tumors and the varying degrees that epigenetic programs contribute to tumorigenesis in different tumor subtypes. Despite these findings, the molecular mechanisms that lead to distinct forms of epigenetic dysregulation in specific subtypes of brain tumors remain largely unknown.
Given the significant role of epigenomic dysregulation in brain tumor pathogenesis, several studies have mapped the active chromatin mark histone H3 lysine K27 acetylation (H3K27ac) in clinical samples of HGG (7–10) and EPN (11, 12). These studies have revealed that unique H3K27ac profiles accompany these different subtypes of brain tumors (12) and identified core transcription factors predicted to govern tumor gene expression programs (9, 11). Among these, the transcription factor Sox9 emerged as one of the most active at H3K27ac occupancy across pediatric HGG and adult HGG and was the highest in EPN (9, 11). Critically, we have shown that Sox9 plays an essential role in early glial development and adult astrocyte function (13, 14). Sox9 expression is elevated in both human HGG and EPN tumors (7, 15), and our previous reports of Sox9 association with H3K27ac occupancy across HGG and EPN (9, 11) raise two key questions. The first question is whether Sox9 is an essential component of the molecular machinery that shapes tumor-specific epigenomic states in HGG and EPN. The second question stems from these distinct H3K27ac profiles and deciphering how Sox9 transcriptional programs diversify between HGG and EPN. Indeed, a loss of Sox9 has been shown to delay tumorigenesis in both HGG xenografts and EPN in vitro (7, 16, 17). However, mechanisms of how Sox9 influences epigenetic states and tumorigenesis in different brain tumor subtypes remain undefined.
In this study, we manipulated Sox9 expression in two autochthonous, immunocompetent mouse models (18–21), finding that it suppresses tumor growth in HGG, while promoting tumorigenesis in EPN. Molecular analyses revealed differential regulation of H3K27ac states by Sox9, which are coupled with unique protein interactions between Sox9 and epigenomic regulators in HGG and EPN. Together, this study demonstrates that Sox9 differentially contributes to tumorigenesis across distinct subtypes of brain tumors, while further revealing that it functions as a key regulator of diverse chromatin states and gene expression programs in brain tumors.
Results
HGG and EPN Models Exhibit Distinct H3K27ac Epigenomic States.
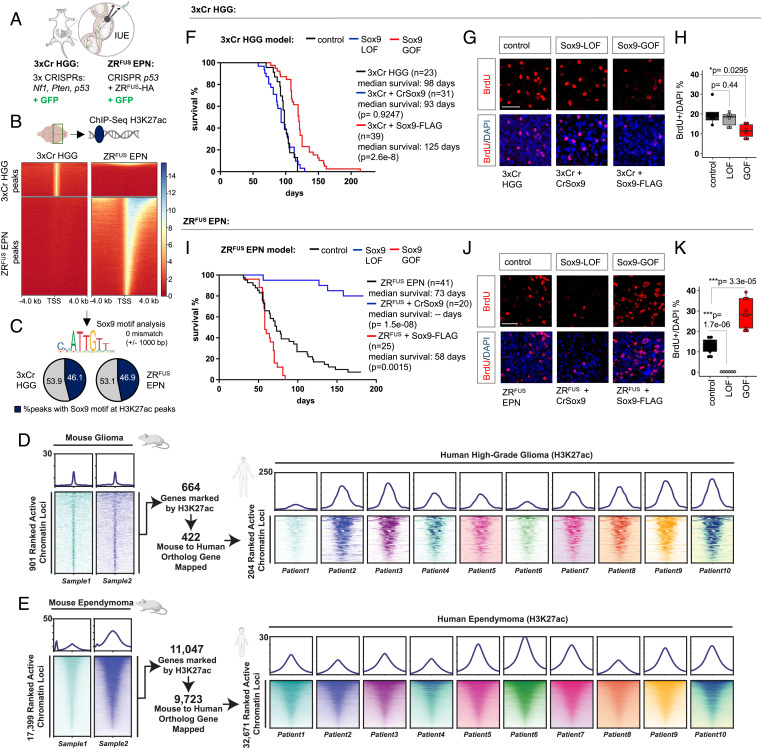
To dissect subtype-specific epigenomic states in brain tumors, we performed studies on two different native mouse models of HGG and EPN. Our mouse model of HGG, hereafter termed as 3xCr HGG (Fig. 1A), uses in utero electroporation (IUE) for CRISPR-Cas9-mediated knockout of prevalent tumor suppressors Nf1, Pten, and Trp53 to induce brain tumors that can be traced by a GFP reporter (18–20). Our mouse model of EPN, hereafter termed as ZRFUS EPN, uses IUE-based overexpression of the oncogenic fusion protein ZFTA-RELA (ZRFUS), which is the primary genetic driver in supratentorial EPNs, along with CRISPR-Cas9-mediated knockout of Trp53 (21) (Fig. 1A).To interrogate epigenomic states in these two distinct mouse models, we dissected GFP-positive tumor tissue and performed chromatin immunoprecipitation coupled with next generation sequencing (ChIP-Seq) of the H3K27ac-activating histone mark. This revealed that ZRFUS EPN contained threefold more H3K27ac peaks compared with 3xCr HGG (10,791 peaks in 3xCr HGG and 33,292 peaks in ZRFUS EPN) and these peaks are largely unique (Fig. 1B), as there was minimal overlap between these H3K27ac peaks and the associated gene ontology (GO) between these models (SI Appendix, Fig. S1A). These observations demonstrate that our 3xCr HGG and ZRFUS EPN models reflect the distinct epigenomic states present in human HGG and EPN (12). In addition, it aligns with our previous findings that ZRFUS directly binds DNA and facilitates tumor-specific transcriptional programs (21).
Fig. 1.
Epigenetic states of H3K27ac and the effect of Sox9 manipulation in 3xCr HGG and ZRFUS EPN tumor subtypes. (A) Schematic of IUE to generate HGG and EPN in a native mouse model. (B) Comparison showing heatmaps of ChIP-H3K27ac signal at 4 kb from peak center at transcription start site (TSS) in 3xCr HGG and ZRFUS EPN tumors. (C) Pie charts showing the percentage of total H3K27ac sites that carries the Sox9 motif allowing 0 mismatch at 1,000 bp from peak center. (D) Comparison of active H3K27ac peaks between mouse 3xCr HGG and 10 human HGG patient tumors. (E) Comparison of active H3K27ac peaks between mouse ZRFUS EPN and 10 human Rela-fusion-positive EPN patient tumors. (F) Kaplan–Meier survival curves of 3xCr HGG control (n = 23), Sox9-LOF (n = 31), and Sox9-GOF (n = 39). (G and H) Representative images of BrdU staining on P90 3xCr HGG tumors and box plots showing quantification of BrdU over DAPI-labeled cells (n = 3 mice per group, 2 sections each; scale bar: 50 μm). (I) Kaplan–Meier survival curves of ZRFUS EPN control (n = 41), Sox9-LOF (n = 20), and Sox9-GOF (n = 25). (J and K) Representative images of BrdU staining on P70 ZRFUS EPN tumors and box plots of quantification of BrdU over DAPI-labeled cells (n = 3 mice per group, 3 sections each; scale bar: 50 μm). P values in Kaplan–Meier curves were calculated using the log-rank test. P values in box plots were calculated using one-way ANOVA with Tukey’s test (*P < 0.05, ***P < 0.001).
To identify transcription factors associated with H3K27ac epigenetic states in 3xCr HGG and ZRFUS EPN, we performed unbiased motif analysis on identified ChIP-Seq peaks, revealing 16 transcription factors associated with H3K27ac in both HGG and EPN (SI Appendix, Fig. S1B). Of these, Sox9 motifs have previously been reported to be enriched at sites of H3K27ac occupancy in both HGG (9) and EPN (11), indicating that our models are suitable for examining the role of Sox9 as a core transcription factor in these tumors. Subsequent motif analysis of Sox9-specific motifs further revealed that nearly half of all H3K27ac peaks carry Sox9 motifs in both 3xCr HGG and ZRFUS EPN (Fig. 1C). Furthermore, a comparison of active chromatin (H3K27ac) states across both our 3xCr HGG and ZRFUS EPN mouse models with human primary tumor tissue revealed significant conservation between our mouse models and human HGG (11) and ZFTA-RELA fusion–positive EPN patient tumors (12) (Fig. 1D and E and Dataset S1). Coupled with the above observations, these results suggest that HGG and EPN are associated with distinct H3K27ac profiles across mouse and human tumors and Sox9 is associated with these distinct epigenetic states in both of these tumors.
Sox9 Plays Distinct Roles in HGG and EPN Tumorigenesis.
Sox9 expression has been shown to be elevated in both adult HGG (7) and in pediatric EPN (15). Therefore, we evaluated whether Sox9 influences tumorigenesis in our tumor models by performing gain-of-function (GOF) and loss-of-function (LOF) studies. For Sox9-LOF studies, we used a CRISPR-Cas9 approach to delete Sox9, where we coelectroporated the corresponding IUE mixture with a CRISPR-Cas9 construct containing guide RNA (gRNA) against Sox9 (SI Appendix, Fig. S1C–F). Similarly, we coelectroporated a C-terminal FLAG-tagged Sox9 construct in our mouse models to achieve Sox9 overexpression in the GOF studies (SI Appendix, Fig. S1G–I). LOF and GOF were confirmed by RT-qPCR to quantify Sox9 transcript levels in addition to immunostaining or Western blots for an assessment of Sox9 protein levels. Using overall animal survival as a proxy for tumor burden, we found that mice bearing HGG Sox9-LOF tumors showed no significant difference in survival compared with 3xCr HGG controls, while HGG Sox9-GOF significantly prolonged survival (Fig. 1F). Bromodeoxyuridine (BrdU) labeling complemented the survival studies, revealing no significant change in cellular proliferation in HGG Sox9-LOF, whereas HGG Sox9-GOF tumors demonstrated a significant reduction in BrdU-labeled cells compared with 3xCr HGG controls (Fig. 1G and H).
Next, we performed a parallel analysis on our ZRFUS EPN model, finding that mice bearing EPN Sox9-LOF (SI Appendix, Fig. S1J and K) tumors demonstrated significantly prolonged animal survival where more than 50% survived beyond 6 mo (Fig. 1I). In contrast, mice bearing EPN Sox9-GOF tumors (SI Appendix, Fig. S1L and M) showed significantly reduced survival compared with the ZRFUS EPN control group (Fig. 1I). BrdU-labeling studies were consistent with survival data, where Sox9-LOF and Sox9-GOF showed significantly decreased and increased BrdU incorporation, respectively, when compared with the control ZRFUS EPN (Fig. 1J and K). Taken together, our findings indicate that Sox9 plays distinct roles in HGG and EPN tumors, where it promotes tumorigenesis in our ZRFUS EPN model and suppresses tumorigenesis in our 3xCr HGG model.
Sox9 Differentially Regulates H3K27ac Epigenetic States in HGG and EPN.
The diverse roles of Sox9 in HGG and EPN, coupled with its association with distinct epigenomic states in these tumors, led us to examine how it regulates epigenomic states in these tumors. Toward this, we performed ChIP-Seq experiments to assess H3K27ac deposition across the genome in 3xCr HGG and ZRFUS EPN tumors after Sox9 manipulation.
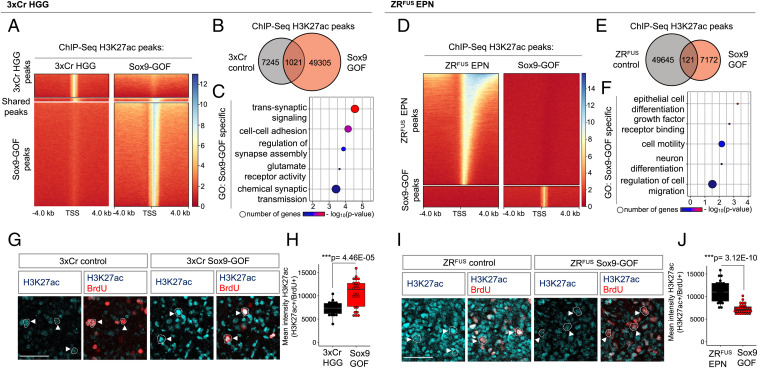
In 3xCr HGG, Sox9-GOF tumors exhibited 6.8-fold more H3K27ac peaks compared with 3xCr HGG control, whereas Sox9-LOF tumors demonstrated a marginal reduction in global H3K27ac peaks compared with the 3xCr HGG control (Fig. 2A and B and SI Appendix, Fig. S2A and B). An analysis of the Sox9-GOF enriched H3K27ac peak GO terms revealed a regulation of synaptic signaling (Fig. 2C), whereas both HGG control and Sox9-LOF-specific terms demonstrate an enrichment of cell–cell adhesion–associated genes (SI Appendix, Fig. S2C and D). Strikingly, parallel analysis in ZRFUS EPN revealed that Sox9-GOF led to a 6.9-fold reduction in H3K27ac peaks compared with ZRFUS EPN control (Fig. 2D and E). A subsequent GO analysis of genes at the Sox9-GOF-specific peaks revealed regulation of RNA binding and cellular differentiation genes (Fig. 2F and SI Appendix, Fig. S2E). Together, these data demonstrate that Sox9 has a profound effect on H3K27ac epigenetic states in both 3xCr HGG and ZRFUS EPN and suggests that Sox9 regulates epigenetic states in these tumor subtypes via distinct mechanisms.
Fig. 2.
Sox9 differentially regulates H3K27ac states in 3xCr HGG and ZRFUS EPN tumor subtypes. (A) Comparison showing heatmaps of ChIP-H3K27ac signal at 4 kb from peak center in 3xCr control versus Sox9-GOF HGG. (B) Venn diagram of the number peaks unique to HGG control and Sox9-GOF and overlapping across two independent biological replicates. (C) GO analysis of genes carrying H3K27ac peaks unique to Sox9-GOF compared with HGG control. (D) Comparison showing heatmaps of ChIP-H3K27ac signal at 4 kb from peak center in control versus Sox9-GOF EPN. (E) Venn diagram of the number peaks unique to EPN control and Sox9-GOF and overlapping across two independent biological replicates. (F) GO analysis of genes carrying H3K27ac peaks unique to Sox9-GOF compared with EPN control. (G) Representative images of H3K27ac colabeled with BrdU staining in P90 control and Sox9-GOF HGG tumors. (H) Box plots showing quantification of H3K27ac fluorescence intensity in BrdU-positive cells (n = 3 mice per group, 20 to 30 cells each; scale bar: 50 μm). (I) Representative images of H3K27ac colabeled with BrdU staining in P70 control and Sox9-GOF EPN tumors. (J) Box plots showing quantification of H3K27ac fluorescence intensity in BrdU-positive cells (n = 3 mice per group, 25 to 30 cells each; scale bar: 50 μm). P values in box plots were calculated using one-way ANOVA with Tukey’s test (*P < 0.05, ***P < 0.001).
The above observations also suggest that Sox9 differentially influences total H3K27ac levels in HGG and EPN. To confirm enhanced or diminished H3K27ac levels in 3xCr HGG and ZRFUS EPN, respectively, we determined H3K27ac levels specifically in BrdU-labeled tumor cells. Consistent with the ChIP-Seq results, we observed significantly higher H3K27ac levels in Sox9-GOF HGG compared with 3xCr HGG control (Fig. 2G and H), coupled with no changes in the Sox9-LOF HGG (SI Appendix, Fig. S2F–G). In contrast to HGG, we observed significantly reduced H3K27ac levels in Sox9-GOF tumor cells compared with the ZRFUS EPN control (Fig. 2I and J), further establishing that Sox9 differentially induces overall shifts in total H3K27ac levels in these two tumor subtypes.
Sox9 Suppresses Gene Expression in ZRFUS EPN.
To better understand Sox9-mediated disease mechanisms in HGG, we dissected GFP-positive 3xCr HGG tumor tissue and performed RNA sequencing (RNA-Seq) to identify differentially expressed genes (DEGs). First, we observed that in Sox9-LOF, 39.8% of DEGs (362 genes) were up-regulated while a greater proportion of 60.2% DEGs were down-regulated (548 genes) (SI Appendix, Fig. S3A). GO analysis further revealed an association with inflammatory response transcriptional programs representing mostly the down-regulated DEGs in HGG Sox9-LOF (SI Appendix, Fig. S3B and C). Given that H3K27ac epigenomic signatures correspond to regions of active transcription, we hypothesized that increased levels of H3K27ac in HGG Sox9-GOF will lead to the up-regulation of genes, while in EPN Sox9-GOF, the decreased H3K27ac levels will correspond with down-regulation of genes. To test this, we performed RNA-Seq on HGG Sox9-GOF and EPN Sox9-GOF along with respective controls. Analysis in the HGG Sox9-GOF identified 1,170 DEGs (Fig. 3A), a slightly larger proportion of which were up-regulated (53.3%, 624 genes) in comparison with down-regulated genes (46.7%, 546 genes). A parallel analysis with EPN Sox9-GOF revealed 824 DEGs, and strikingly only 6.9% of these were up-regulated (57 genes), while 93.1% were down-regulated (767 genes) (Fig. 3B and Dataset S2), supporting the hypothesis that Sox9-GOF in ZRFUS EPN will lead to transcriptional repression (Fig. 3C and D). In addition, correlation of these DEGs with ChIP-Seq data from the Epigenomics Roadmap Project revealed that the DEGs up-regulated in HGG Sox9-GOF are associated with histone acetylation, specifically H3K27ac (SI Appendix, Fig. S3D). In contrast, the DEGs in the EPN Sox9-GOF are associated with histone methylation, specifically H3K27me3 (SI Appendix, Fig. S3E), a repressive histone mark. Together, these data support the notion that Sox9 functions to suppress gene expression in ZRFUS EPN.
Fig. 3.
Sox9 differentially regulates gene expression in 3xCr HGG and ZRFUS EPN tumor subtypes. (A and B) Volcano plots depicting RNA-Seq data comparing (A) 3xCr control versus Sox9-GOF HGG and (B) ZRFUS control versus Sox9-GOF EPN. RNA-Seq experiments were performed in independent biological triplicates (P < 0.05 and fold change of >2). (C) Expression heatmap analysis of up-regulated genes in Sox9-GOF HGG and down-regulated genes in Sox9-GOF EPN. (D) Bar graph showing the number of Sox9 GOF DEGs in 3xCr HGG compared with ZRFUS EPN; note the suppressed gene expression in Sox9-GOF EPN. (E) Venn diagram and (F and G) GO terms of DEGs from RNA-Seq data that overlap with genes acquiring or losing H3K27ac peaks in Sox9-GOF (F) 3xCr HGG and (G) ZRFUS EPN, respectively. (H and I) RT-qPCR shows fold enrichment of transcripts in Sox9-GOF relative to control in (H) 3xCr HGG and (I) ZRFUS EPN after normalization using Gapdh (n = 3 mice per group). Data shown as mean ± SEM; P values were calculated using Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.00001). RT-qPCR primer sequences are listed in Dataset S8.
To understand the nature of Sox9 gene regulation and how it differentially influences tumorigenesis in HGG and EPN, we next evaluated the GO of the DEGs from the Sox9-GOF RNA-Seq from these tumor models. GO analyses revealed that up-regulated DEGs in 3xCr HGG are associated with protein phosphorylation (SI Appendix, Fig. S3F and G), while down-regulated DEGs in ZRFUS EPN are associated with synaptic signaling gene sets (SI Appendix, Fig. S3H and I). We also evaluated a series of stem cell, neuronal, glial, and microglial markers to assess if different cell lineages and cellular components are differentially affected by Sox9-GOF and observed no significant differences in the majority of these markers (SI Appendix, Fig. S3J). Next, we further investigated the H3K27ac status of the actively transcribed or repressed DEGs in Sox9-GOF HGG and EPN by overlaying our RNA-Seq and ChIP-Seq datasets. This analysis revealed that 116 genes up-regulated in the RNA-Seq dataset also acquire H3K27ac peaks in Sox9-GOF HGG (Fig. 3E and SI Appendix, Fig. S3K), representing GO terms associated with cell adhesion and synaptic signaling (Fig. 3F). We further selected a subset of these genes based on expression levels from The Cancer Genome Atlas database and validated their up-regulation through RT-qPCR (Fig. 3H). In parallel, for the EPN Sox9-GOF dataset, we found that 240 of the down-regulated genes lose H3K27ac peaks (Fig. 3E and SI Appendix, Fig. S3L and Dataset S3) and represent GO terms associated with synaptic signaling and cellular differentiation (Fig. 3G); reduced expression on a subset of genes was further validated by RT-qPCR (Fig. 3I). While GO categories associated with synapses were observed for both of these gene sets, there was an overlap of only 5 genes between these sets of 116 and 240 genes, indicating distinct Sox9-H3K27ac regulatory mechanisms in these 2 types of brain tumors. Together with the ChIP-Seq analysis, these data indicate that Sox9 induces distinct gene expression programs via H3K27ac regulation in HGG and EPN.
Sox9 Interacts with the Histone Deacetylation Complex in HGG.
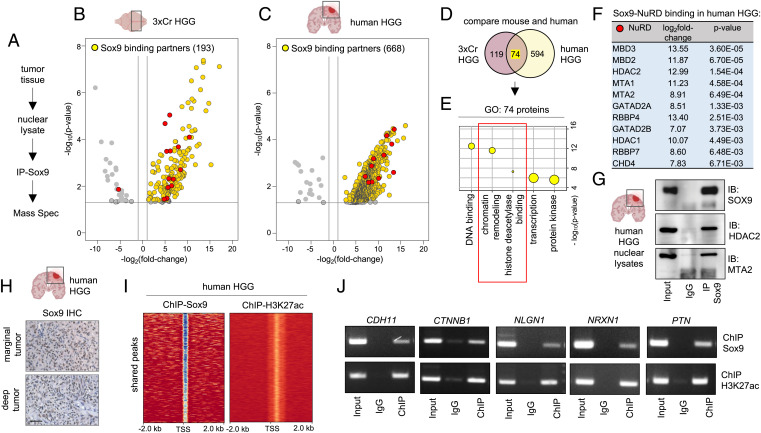
The foregoing data suggest that manipulation of Sox9 selectively regulates H3K27ac states in HGG and EPN that led us to investigate whether Sox9 directly interacts with the histone acetylation machinery. Toward this, we isolated GFP tumor tissue from 3xCr HGG brains, purified nuclear lysates, and performed immunoprecipitation of Sox9 followed by mass spectrometry (IP-MS) (Fig. 4A), identifying 193 interacting partners of Sox9 in 3xCr HGG (Fig. 4B). In parallel, we performed IP-MS of Sox9 from human HGG patient samples, identifying 668 Sox9 interacting partners (Fig. 4C). A comparison of the mouse and human HGG Sox9 interactome showed 74 common binding partners (Fig. 4D), with GO analysis demonstrating an enrichment of categories for DNA binding and transcriptional regulation. In addition, we also observed GO terms associated with histone acetylation (Fig. 4E and SI Appendix, Fig. S4A), with the majority of these proteins belonging to a well-characterized nucleosome remodeling and deacetylase complex (NuRD) in both human HGG (Fig. 4F) and 3xCr HGG (SI Appendix, Fig. S4B–F). In both mouse and human HGG, the three most strongly associated NuRD members were among the top 25 binding partners, and in human HGG, this represents the top 3.7% of Sox9 interactome (Dataset S4). In addition, among the Sox9 binding partners in both mouse and human HGG, ontology related to NuRD complex function was the top statistically significant category (Dataset S5). We validated Sox9 binding to a subset of these NuRD complex members by IP of Sox9 in human and 3xCr HGG samples, followed by Western blot analysis (Fig. 4G and SI Appendix, Fig. S4G).
Fig. 4.
Sox9 interacts with histone deacetylation complex in HGG. (A) Schematic of IP-Sox9 and MS proteomic experiment to identify Sox9 binding partners. (B and C) Volcano plots depicting IP-MS data of Sox9 interactome in (B) 3xCr HGG and (C) human HGG. Fold change was calculated over control samples of nuclear lysates incubated with beads only without antibody. IP-MS experiments were performed in independent biological triplicates for 3xCr HGG and two independent human patient HGG samples (P < 0.05 and fold change of >2). (D) Venn diagram showing the number of unique and overlapping Sox9 binding partners in mouse and human HGG. (E) GO terms associated with the 74 shared Sox9 interactors in mouse and human HGG showing enrichment of histone deacetylation. (F) Table showing Sox9 binding fold change and P values with NuRD complex members. Note all NuRD members are depicted by red circles in volcano plots shown in B and C. (G) Sox9 coimmunoprecipitation with NuRD members Hdac2 and Mta2 from human HGG nuclear lysates. (H) IHC of Sox9 in human HGG marginal and deep tumors (brown: Sox9; blue: hematoxylin counterstain; scale bar: 50 μm). (I) ChIP heatmaps at 2 kb from peak center of ChIP experiments with Sox9 and H3K27ac from human HGG tissue, showing that 9,539 of identified H3K27ac peaks are also co-occupied by Sox9 peaks. (J) ChIP-PCR validation of a subset of Sox9 and H3K27ac cotargets in human HGG. ChIP-PCR primer sequences are listed in Dataset S8.
The IP-MS data suggest that Sox9 regulates H3K27ac states in HGG by interacting with members of the NuRD complex; therefore, we sought to determine the extent to which Sox9 is associated with H3K27ac epigenomic states in clinical samples of human HGG. Consistent with previous reports, we observed elevated Sox9 in human HGG (7) (Fig. 4H). Next, we performed ChIP-Seq experiments with Sox9 and H3K27ac on human HGG samples, identifying 22,627 Sox9 peaks and 16,644 H3K27ac peaks (SI Appendix, Fig. S4H and I). A comparison of these two datasets revealed that among the H3K27ac peaks, 57%, representing 9,539 peaks, were also associated with Sox9 peak sites (Fig. 4I and Dataset S6) and that GO terms linked to these genes showed categories of cell–cell adhesion and synapse assembly (SI Appendix, Fig. S4J). To independently validate Sox9 binding to the regulatory regions of these genes, we performed ChIP-PCR validation on a subset of these genes (Fig. 4J). Collectively, these observations indicate that Sox9 interacts with proteins that modulate histone acetylation in HGG and that its gene regulatory network is tightly associated with these epigenomic states.
Sox9 Interacts with ZFTA-RELA and Coregulates Oncogenic Programs in EPN.
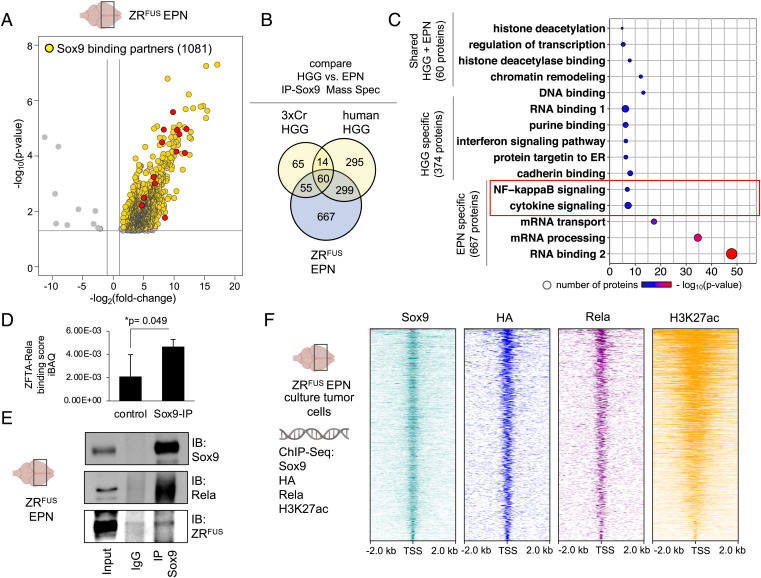
Interactions between Sox9 and the NuRD complex in HGG prompted us to examine whether these same interactions occur in EPN. Therefore, we performed IP-MS for Sox9 in our mouse ZRFUS EPN model, identifying 1,081 interacting proteins (Fig. 5A). To identify tumor-specific protein interactions across HGG and EPN, we compared the IP-MS data for Sox9 between these datasets, which revealed that 667 proteins specifically associate with Sox9 in ZRFUS EPN tissue and 374 proteins are specific to HGG in both 3xCr mouse and human tissues (Fig. 5B). Shared binding partners consisted of 60 proteins, which were predominantly NuRD complex members, further establishing these interactions with Sox9 across both subtypes of brain tumors (Fig. 5C and SI Appendix, Fig. S5A–D). We further searched for EPN-specific Sox9 interactors and identified NF b signaling (Fig. 5C). A subsequent analysis of these proteins revealed the ZRFUS fusion itself, which is a genetic driver in EPN and has recently been reported to be associated with H3K27ac transcriptional activation in our ZRFUS EPN model (21), associates with Sox9 in this context. Furthermore, among the Sox9 interactions that are exclusive to ZRFUS EPN, we specifically searched for known histone acetylation proteins. This identified 19 binding partners that have previously been reported to be associated with histone acetylation (Dataset S7). Of these 19 members, the ZFTA-RELA protein was the only member that is involved in H3K27ac epigenetic regulation in ZRFUS EPN (21).
Fig. 5.
Sox9 interacts with ZFTA-RELA and coregulates oncogenic programs in ZRFUS EPN. (A) Volcano plot depicting IP-MS data of Sox9 interactome in ZRFUS EPN. Fold change was calculated over control samples of nuclear lysates incubated with beads only without antibody. IP-MS experiments were performed in independent biological triplicates (P < 0.05 and fold change of >2). (B) Venn diagram showing the number of unique and overlapping Sox9 binding partners in 3xCr HGG, human HGG and ZRFUS EPN, and (C) GO terms associated with these interaction partners. NF-κB signaling unique to the ZRFUS EPN-specific Sox9 interactome is highlighted in red. (D) Quantification of Sox9 binding to ZFTA-RELA in ZRFUS EPN from IP-MS data. (E) Sox9 co-IP experiments with Rela proteins from ZRFUS EPN lysates. (F) Schematic of ChIP-Seq experiments from ZRFUS EPN cells and heatmap profiles demonstrating co-occupancy between Sox9, HA, Rela, and H3K27ac.
Using Sox9-IP and immunoblot analysis, we validated Sox9 interactions with ZRFUS in EPN tumor tissue (Fig. 5D and E). To determine the extent of these interactions in the context of transcriptional regulation, we performed ChIP-Seq studies in mouse ZRFUS EPN cells for Sox9, ZRFUS, Rela, and H3K27ac, finding an abundance of genomic regions that contain Sox9 and ZRFUS co-occupancy, that are coupled with H3K27ac states (Fig. 5F and Dataset S6). An analysis of these genes that are coregulated by Sox9 and ZRFUS revealed an enrichment of cell adhesion genes (SI Appendix, Fig. S5E). Importantly, Sox9 and ZRFUS cotargets in EPN were found to have minimal overlap of 10 to 12% genes with Sox9 and H3K27ac targets in HGG (SI Appendix, Fig. S5F), further supporting distinct Sox9 control in HGG and ZRFUS EPN. Furthermore, we also observed an absence of a Sox9 and Rela interaction in HGG tumor tissue (SI Appendix, Fig. S5G). This analysis of the Sox9 interactome in EPN highlights key interactions with the ZFTA-RELA fusion while also demonstrating conserved interactions with the NuRD complex and other components of the epigenomic machinery.
Discussion
Aberrant epigenetic states are mediated by the reorganization of histone marks, DNA methylation patterns, or incorporation of histone variants that critically influence tumor biology and is a hallmark feature of numerous pediatric and adult brain cancers (2, 3, 5, 22, 23). While these findings have directed interest in developing therapies targeting epigenetic modifiers, a fundamental challenge lies in the fact that epigenetic modifiers control hundreds of genes, which are often context dependent (2). Therefore, it is essential to understand context-specific mechanisms of epigenetic dysregulation in cancer. Our unbiased active chromatin mapping in mouse and human tumors pointed to Sox9 as a lead core transcription factor that governs a substantial proportion of the transcriptome (9, 11). Indeed, transcriptional programs mediated by developmental transcription factors like Sox9 are often recapitulated in tumorigenesis (24, 25). Therefore, in this study, we used Sox9 as an entry point to interrogate H3K27ac in the context of HGG and EPN, which represent pediatric and adult brain tumors, respectively. We show that Sox9 differentially controls tumorigenesis and transcriptional programs in HGG and EPN, and the mechanism by which Sox9 oversees these H3K27ac states is through differential interactions with epigenetic modifiers.
Sox9 DNA binding motifs are enriched in H3K27ac occupancy sites in human HGG (9), in EPN (11), and in our native mouse models 3xCr HGG and ZRFUS EPN. Sox9 has also been reported to be a pioneering transcription factor regulating active chromatin in skin stem cells (26). Our study shows increased Sox9-induced opposing effects on animal survival in 3xCr HGG versus ZRFUS EPN, where it prolongs survival in 3xCr HGG and acts as an oncogene in ZRFUS EPN. While prior Sox9-LOF studies have reported that it functions to promote proliferation in HGG and EPN cell lines (11, 16, 17), its role in native brain tumor models using systemic functional analyses in two disease subtypes in parallel has not been previously examined. This also raises an important issue that Sox9-LOF studies in human tumors using in vitro methods may involve artifacts of using cell lines. Although, it is also possible that LOF studies in mouse tumors do not reflect human diseases, and parallels from human tumor samples need to be drawn whenever possible. In addition, a previous study interrogated the effects of Sox9 deletion on tumor progression using mouse xenografts, showing that it promotes tumorigenesis in HGG lines carrying EGFR mutations by collaborating with another transcription factor to drive oncogenic gene regulatory programs (7). In contrast, our study used a native mouse HGG model without EGFR mutations and demonstrated that the overexpression of Sox9 prolongs survival and, in this genotype, Sox9 collaborates with histone deacetylases to regulate epigenomic programs. Put together with prior studies, our results reinforce context-dependent roles of Sox9. Interestingly, in melanoma, Sox9 has also been implicated as a suppressor of tumorigenesis (27). Context-dependent roles also extend to other transcription factors, where Zbtb7a and Arid1a have been shown to function as an oncogene or tumor suppressor in different contexts (28, 29).
Our comprehensive molecular analysis of Sox9’s role in 3xCr HGG and ZRFUS EPN revealed that it not only modulates distinct H3K27ac states in these two tumors but also substantially shifts overall H3K27ac levels in contrasting ways. While increasing Sox9 led to enhanced H3K27ac in 3xCr HGG, in ZRFUS EPN, it led to diminished H3K27ac. This signifies that Sox9 acts a transcriptional activator in HGG and a transcriptional repressor in EPN, as demonstrated by our observation of drastically reduced gene expression in EPN Sox9-GOF tumors. In HGG, Sox9 has been shown to activate c-Myc in the context of EGFR mutations (7) and stem cell markers in patient-derived glioma stem cells (17). In contrast, our results of transcriptional repression by Sox9 in EPN is a surprising observation that has not been previously characterized. In addition, contrasting roles of transcription factors in different contexts have also been demonstrated for the extensively studied transcriptional activator Brd4 (30), which has recently been reported to also operate as a transcriptional repressor of p53 in the context of acute myeloid leukemia (31).
To determine the biochemical mechanisms of how Sox9 regulates H3K27ac, our proteomic studies established that Sox9 associates with the histone deacetylating NuRD complex in 3xCr and human HGG. This observation, when coupled with our epigenomic studies, illustrates that protein–protein interactions between transcription factors and epigenetic modifiers are critical regulators of context-dependent epigenetic states. The last decade has witnessed extensive efforts dedicated to large scale-sequencing studies exploring the complex interplay between the genome and epigenome in brain cancer (22). Critically, while these studies have led to significant advancements, our study highlights the importance of including proteomic studies to characterize protein–protein interactions that are responsible for epigenetic dysregulation in cancer. Our finding that overexpression of Sox9 in 3xCr HGG led to enhanced H3K27ac provides a model wherein the Sox9-NuRD interaction inhibits NuRD binding to DNA, thereby increasing H3K27ac, since NuRD removes histone acetyl groups (SI Appendix, Fig. S6). In addition, this model also explains the prolonged survival of Sox9-GOF HGG since NuRD inhibition is well characterized to afford survival advantage. Indeed, Hdac inhibitors were the first class of approved epigenetic therapy in cancers (2). In HGG, knockdown of several NuRD members like Rbbp4, Mbd3, and Mbd2 suppress tumorigenesis (32–34) and Hdac2 inhibitors are currently undergoing clinical trials.
Our proteomic studies also revealed striking differences in Sox9 protein interactions between ZRFUS EPN and HGG. Although Sox9-NuRD interactions exist in both HGG and ZRFUS EPN, the expanded Sox9 interactome in EPN may titrate out the effect of Sox9 on NuRD DNA binding in ZRFUS EPN. Excitingly, we also observed that Sox9 associates with ZFTA-RELA, the fusion oncoprotein driving EPN that has been recently reported to recruit transcriptional coactivators at H3K27ac active sites (21). Therefore, our findings that increased Sox9 in ZRFUS EPN led to diminished H3K27ac further provides a model wherein the Sox9-ZRFUS interaction inhibits ZRFUS binding to DNA, thereby reducing H3K27ac in ZRFUS EPN (SI Appendix, Fig. S6). Furthermore, the Sox9-ZRFUS association may also direct activation at a subset of ZRFUS targets promoting functional synergy for EPN tumorigenesis. More broadly, these results also establish that oncofusion proteins may provide venues for protein–protein interactions, which can lead to epigenomic remodeling in tumors. Indeed, the Ewing’s sarcoma EWS/FLI1 and the acute myeloid leukemia PML/RARα oncofusions can also remodel the epigenome (35–38). Furthermore, we observed significant interactions between Sox9 and RNA binding proteins associated with the maintenance of RNA stability, RNA transport, and RNA splicing in ZRFUS EPN shown in Fig. 5C. Indeed, RNA binding proteins are well known to be associated with transcription factors at active chromatin (39). Nevertheless, how Sox9 is involved in EPN tumorigenesis through interactions with RNA binding proteins will need further study. Taken together, we have identified tumor subtype–specific Sox9-dependent epigenetic states that translate into contrasting animal survival outcomes and are driven by differential interactions with epigenetic modifiers.
Materials and Methods
Patient HGG Samples.
HGGs were obtained from patients undergoing surgical resection at Baylor College of Medicine. Informed consent was obtained from all patients. This study was approved by Baylor College of Medicine Institutional Review Board protocol H-35355. Diagnoses were confirmed by a neuropathologist. For histology, samples were paraffin embedded. For proteomic experiments, samples were snap frozen. For ChIP experiments, fresh tumor samples were used for subsequent processing as described later.
IUE and Mouse Models of 3xCr HGG and ZRFUS EPN.
All mouse tumors were generated in the CD-1 IGS mouse background at embryonic day 16. To generate mouse 3xCr HGG and ZRFUS EPN tumors, we performed IUE as previously described (19–21). Briefly, the uterine horns were exposed, and the plasmid mixture was injected into the embryonic lateral ventricles along with Fast Green dye as the indicator. Electroporation was accomplished with BTW Tweezertrodes connected to the pulse generator (BTX 8300), set at 33 V and 55 ms per pulse six times at 100-ms intervals. In the 3xCr HGG model, the plasmid mixture was composed of helper plasmid pGLAST-PBase (2.0 μg/μL) and all other DNA (1.5 μg/μL), including PBCAG-GFP and the CRISPR-Cas9 construct with gRNAs against Nf1, Pten, and Trp53. In the ZRFUS EPN model, the plasmid mixture was composed of helper plasmid pGLAST-PBase (2.0 μg/μL) and others (1.0 μg/μL) including PBCAG-GFP, PBCAG-ZRFUS-HA, and CRISPR-Cas9 construct with gRNA against Trp53. Since the helper plasmid encoding piggyBac transposase is under a GLAST promoter and the IUE procedure was performed during early gliogenesis (embryonic day 14.5 to 16.5), brain tumor cells generated in our mouse models arose from the radial glia lineage (40). Coelectroporation of PBCAG-GFP allowed a fluorescent visualization of tumors. The IUE schematic shown in Fig. 1A is adapted from ref. 41. GOF and LOF studies of Sox9 were performed by coelectroporating PBCAG-mSox9-Flag (1.0 μg/μL) and a CRISPR-Cas9 construct with gRNA against Sox9 exon 1 (1.5 μg/μL, gRNA sequence: 5′- GGTGTTCTCCGTGTCCGAGC-3′), respectively. The Sox9 gRNA sequence was generated through the Broad Institute Genetic Perturbation Platform portal and validated by the mismatch-cleavage SURVEYOR assay (IDT, 706020) on genomic DNA obtained from tumors. Primer sequences for SURVEYOR are 5′-GGCTCGCGTATGAATCTCCT-3′ (forward), 5′-CAGTCATATTCACGCCCCCA-3′ (reverse), 5′-AACTTCTGTGGGAGCGACAA-3′ (forward), and 5′-GCTTGACGTGTGGCTTGTTC-3′ (reverse). Primer sequences for evaluation of Sox9-GOF were directed at Sox9-FLAG, namely, 5′-GGCTCCTACTACAGTCACGC-3′ (forward) and 5′-CGTCGTCATCCTTGTAATCGG-3′ (reverse). Primer sequences for evaluation of Sox9-LOF were directed at exon 1, namely, 5′-CCTGGACCCCTTCATGAAG-3′ (forward) and 5′-CTTCCTCGCTCTCCTTCTTC-3′ (reverse). All animals were euthanized when moribund and survival dates were recorded for Kaplan–Meier survival curve analyses. All procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at Baylor College of Medicine and conform to the US Public Health Service Policy on Human Care and Use of Laboratory Animals.
Mouse Brain Tumor Collection.
Mice with tumors were observed for symptoms suggestive of tumors including lethargy, hunched posture, decreased grooming, trembling, partial limb paralysis, and abnormal gait, representing the IACUC permitted end point. Unless a time point was specified, mice were humanely euthanized after demonstration of symptoms. Both male and female mice were used in this study. To examine tumor cell proliferation, mouse brains at postnatal day P90 for 3xCr HGG set and P70 for ZRFUS EPN set were collected 4 h after BrdU pulsing (100 μg/g body weight in phosphate-buffered saline [PBS]) by intraperitoneal injection.
ChIP.
Mice brains were harvested between P90 and 120 for the 3xCr HGG sample set and P65 to 95 for the ZRFUS EPN set. Tumor tissues labeled with GFP were dissected and dissociated in cold PBS using a pellet homogenizer on ice. Chromatin was crosslinked using a freshly prepared 1.1% formaldehyde solution with rocking at room temperature for 10 min, followed by addition of 0.1 M glycine. Cell pellets were collected by centrifugation at 3,500 rpm for 5 min at 4 °C, washed with PBS, and frozen at −80 °C until further processing. Pellets were resuspended with PBS/PMSF containing 0.5% Igepal to release nuclei followed by washing with cold ChIP-Buffer (0.25% TritonX, 10 mM EDTA, 0.5 mM EGTA, 10 mM Hepes [pH 6.5]), and nuclei were lysed with ChIP lysis buffer (0.5% SDS, 5 mM EDTA, 25 mM Tris⋅HCl [pH 8]) for 15 to 20 min at room temperature. Lysates were sonicated to 250 to 350 bp using the Diagenode Bioruptor. Chromatin was quantified using the Quant-iT double-stranded DNA (dsDNA) assay kit (Thermo Fisher, Q33120), diluted fivefold (2 mM EDTA, 150 mM NaCl, 1% Triton X-100, 20 mM Tris⋅HCl; pH 8.0; with protease inhibitors), and incubated with antibody overnight at 4 °C with rotation. For ChIP-H3K27ac, 15 μg of chromatin was incubated with rabbit anti-H3K27ac (2 μg; Abcam, ab4729) and for ChIP-Sox9, 100 μg of chromatin was incubated with rabbit anti-Sox9 (10 μg; EMDMillipore, ab5535). The next day, lysates were incubated with Protein A/G magnetic beads (Thermo, 88802) for 5 to 6 h at 4 °C, followed by washing with Tris-SDS-EDTA-I buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris⋅HCl; pH 8.0), Tris-SDS-EDTA-II buffer (TSEI buffer with 500 mM NaCl), LiCl buffer (250 mM LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris⋅HCl; pH 8.0), and Tris-EDTA buffer (10 mM Tris-HCl; pH 8.0, 1 mM EDTA). To release DNA fragments, samples were incubated in freshly prepared elution buffer (1% SDS, 0.1 M NaHCO3) for 20 min at 65 °C twice. Elutions were treated with proteinase K (0.4 mg/mL; ThermoFisher, AM2546) and NaCl (0.125M) overnight at 65 °C for reverse crosslinking. Subsequently, ChIP-DNA was purified using a PCR purification kit (Qiagen, 28104) and quantified using the Quant-iT dsDNA assay kit. For ChIP-Seq experiments, 10 to 12 ng of ChIP-DNA was used for library preparation as described below. For ChIP-PCR validation, ChIP-DNA was obtained in the same way as above except for additional control samples, wherein sonicated lysates were incubated with rabbit anti-IgG (R&D Systems, AB-105-C). Subsequently, purified ChIP-DNA were analyzed in PCR reactions using AccuPrime Pfx DNA polymerase (ThermoFisher, 12344-032) that were carried out at 95 °C for 5 min; 40 cycles of 95 °C for 30 s, 55 °C for 1 min and 68 °C for 30 s; followed by 68 °C for 2 min. Primers used for ChIP-PCR were designed at Sox9 binding sites 1,000 bp from the transcription start site of candidate genes and are listed in Dataset S8.
ChIP-Seq Library Preparation, Sequencing, and Bioinformatic Analysis.
ChIP libraries were prepared using the TruSeq ChIP Library Preparation Kit (Illumina, IP-202-1012), according to the manufacturer’s instructions. Libraries ranging from 250 to 350 bp were extracted from gel incisions using the QIAquick Gel Extraction Kit (QIAGEN, 28706), PCR amplified, and purified using AMPure XP beads (Beckman Coulter Life Science, A63882). The quality of the resulting libraries was analyzed on the Standard Sensitivity NGS Fragment Analysis Kit (Agilent formerly AATI, DNF-473-0500) on a 12-Capillary Fragment Analyzer. Libraries were quantified using the Quant-iT dsDNA assay kit (Q33120), and equal concentrations (2 nM) of libraries were pooled and subjected to single-end (1 × 150) sequencing of ∼60 to 80 million reads per sample using the High Output v2 kit (Illumina, FC-404-2002) on a NextSeq550 following the manufacturer’s instructions. All ChIP-Seq experiments in 3xCr HGG and ZRFUS EPN were performed on two independent biological replicates.
Sequencing files from each flow cell lane were downloaded, and the resulting fastq files were merged. Quality control was performed using fastQC (v0.11.17) and MultiQC (v0.9). Reads were mapped to the mouse genome mm10 or human genome hg38 assembly using bowtie2 (v 2.2.6) (42). Using the HOMER (v4.10) software suite (43), bedgraph files and tag directories were made. The findPeaks command in histone (for H3K27ac) or factor (for Sox9) mode was used to filter ChIP peaks enriched over input control. Annotation of enriched peaks was performed using annotatePeaks with mm10 or hg38 assembly. Integrated Genome Browser–compatible files were made using samtools (v1.9), sort and index, deepTools (v3.2.0), and bamCompare (44, 45). Motif analysis for transcription factors was performed using findMotifsGenome.pl, and Sox9-specific motif analysis was done using seq2profile.pl at 100 bp from the peak center. Overlapping and differentially bound peaks were obtained using mergePeaks or getDifferentialPeaks, and peaks were visualized using computeMatrix and plotHeatmap. GOs were determined by submitting genes associated with ChIP peaks at Enrichr, and significant GO terms with P values of <0.01 were selected for visualization using ggplot. For ChIP-Seq analysis of Sox9, HA, Rela, and H3K27ac in ZRFUS EPN cells, raw paired-end reads were trimmed using trimGalore (v0.6.7) and subsequently aligned to the mm10 reference genome using Bowtie2. The resulting alignments were sorted and marked for duplicates using picard tools (MarkDuplicates, v2.21.1) and reads per kilobase of transcript, per million mapped reads–normalized BigWig tracks files were generated using deeptools (v3.4.3). Peaks were called using MACS2 (v2.2.7.1). To perform comparative analyses, ZRFUS Cut&Run sequencing data as described in Arabzade et al. were used (21). The peaks shared by ZRFUS and Sox9 and the Sox9-specific peaks were inferred using intersectBed, and the resulting regions were plotted using deeptools computeMatrix. To find pathways enriched in the ZRFUS shared and Sox9-specific sites, Kyoto Encyclopedia of Genes and Genomes enrichment analysis was performed using clusterProfiler, with a P value cutoff of 0.05 (46).
For comparison with human ChIP-Seq H3K27ac, we collected two mouse H3K27ac samples, mapped to mm10, each for HGG and EPN. For human samples, we collected 10 samples each, mapped to hg19. Human HGG and EPN patient ChIP-H3K27ac data have been previously published (11, 12). For each sample, we called enhancer using ROSE (47) and filtered to a set of enhancers present in all samples in the respective group. The genes associated with the filtered set of mouse H3K27ac active regions were then mapped to human (hg19) orthologous genes using the R package gprofiler. The enhancers associated with the orthologous genes in human were plotted in a heatmap.
Immunohistochemistry (IHC) and Immunofluorescent (IF) Staining.
For immunostaining, mouse brains were fixed through intracardial perfusion of 4% paraformaldehyde and further fixed overnight in fresh 4% paraformaldehyde followed by dehydration with 70% EtOH and paraffin embedding at the Pathology Core of Breast Center in Baylor College of Medicine. For IHC, 10-μm paraffin-embedded sections were deparaffinized as follows: 3 × 3 min in xylene, 3 × 3 min in 100% EtOH, 3 × 3 min in 95% EtOH, 3 min in 80% EtOH, 3 min in 70% EtOH, 3 min in 50% EtOH, 5 min in ddH2O, and 5 min in PBS. Antigen retrieval was performed by incubating the sections in 10 mM sodium citrate (pH 6.0; 0.05% Tween20) at 95 °C for 20 min. Endogenous peroxidases were blocked with 3% H2O2 for 15 min at room temperature. After 1 h serum blocking, slides were incubated with primary antibody overnight at 4 °C. The next day, slides were rinsed with PBS and incubated with secondary antibody at room temperature for 1 h. DAB chromogen and hematoxylin were applied for the color matrix and counterstaining. Finally, the slides were dehydrated by three rounds of 5 min each in 95% EtOH, 100% EtOH, and xylene. Staining was preserved with Permount Mounting Media (Electron Microscope Sciences, 17986-01) under a coverslip. For IF staining, 10-μm sections were deparaffinized followed by antigen retrieval as above. After 1 h of serum blocking and overnight primary antibody incubation at 4 °C, slides were incubated with species-specific secondary antibodies tagged with Alexa Fluor 488, 568, or 647 for 1 h at room temperature. The following primary antibodies were used: rabbit anti-Sox9 (IHC: 1:1,000; IF: 1:500; EMDMillipore, AB5535), rabbit anti-FLAG (1:500; Cell Signaling Technologies, 14793S), rat anti-BrdU (1:200; Abcam, ab6326), rat anti-HA (1:100; Roche, 11867431001), and rabbit anti-H3K27ac (1:500; Abcam, ab4729). The following secondary antibodies were used at 1:1,000 dilution: goat anti-rabbit (ThermoFisher, A11036/A2124S) and goat anti-rat (ThermoFisher, A11077/A11006). After Hoechst nuclear counterstaining (1:10,000; ThermoFisher, H3570), coverslips were mounted with VECTASHIELD antifade mounting medium. Images were acquired at a 20× objective using a Zeiss Axio Imager M2 Fluorescent microscope for IHC and a Zeiss LSM 980 confocal microscope for IF images. For quantification of H3K27ac, z-stacked images were taken across replicates and mean intensity was evaluated using ImageJ.
RNA Extraction, Library Preparation, Sequencing, and Bioinformatic Analysis.
Mouse brains were harvested between P90 and 120 for the 3xCr HGG sample set and between P65 and 95 for the ZRFUS EPN set. Tissues were rinsed with PBS and dissociated in TRIZOL (ThermoFisher, 15596018). Total RNA was extracted using the RNeasy Mini Kit (Qiagen, 74106) according to the manufacturer’s instructions. Illumina sequencing libraries with 6-bp single indices were constructed from 300 ng total RNA using the TruSeq Stranded mRNA kit (Illumina, RS-122-2101). Equal concentrations (2 nM) of libraries were pooled and subjected to paired-end sequencing of ∼20 to 30 million reads per sample using the Mid Output v2 kit (Illumina, 20024904) on an Illumina NextSeq550 following the manufacturer’s instructions.
Sequencing files from each flow cell lane were downloaded in fastq files and merged. Quality control was performed using fastQC (v0.11.7) and MultiQC (v1.11). Reads were mapped to the mouse genome mm10 assembly using STAR (v2.5.0a) (48). In R (v4.1.2), mapped reads were used to build count matrices using Bioconductor packages GenomicAlignments (v1.26.0) and GenomicFeatures (v1.42.2) (49). University of California Santa Cruz transcripts were downloaded from Illumina iGenomes in the GTF file format. DESeq2 (v1.30.1) (50) was used for normalization and differential gene expression analysis. GOs were determined using Enrichr, and significant GO terms with P values of <0.01 were selected for visualization using ggplot2 (v3.3.5) and GOplot (v1.0.2). Gene expression heatmaps were generated using ComplexHeatmap (v2.6.2).
RT-qPCR.
RNA was extracted as described above for sample preparation of RNA-seq. For RT-qPCR, 500 ng of RNA was converted to complementary DNA (cDNA) using iScript Reverse Transcriptase Supermix (BioRad, 1708841). RT-qPCR was performed using PerfeCTa SYBR Green Fast Mix (Quantabio, 95072-012) on a Roche Light Cycler 480 instrument. Reactions were set up using 2 ng cDNA, 250 nM primers, and 1× SYBR mix. qPCR was carried out at 95 °C for 30 s, 40 cycles of 95 °C for 5 s and 60 °C for 30 s, with subsequent melting curve analysis. The expression of transcripts of target genes was normalized to Gapdh. Primers used for qPCR are listed in Dataset S8.
IP-MS.
Tumor tissues labeled with GFP were dissected and washed with cold PBS three times, followed by dissociation using a pellet homogenizer and extraction of nuclear lysates using NE-PER Nuclear and Cytoplasmic Extraction Reagents (ThermoFisher, 78833) according to the manufacturer’s instructions. The nuclear fraction was ultracentrifuged at 100,000 g for 20 min at 4 °C followed by incubation with a protein A Sepharose slurry (GE Healthcare Life Sciences, 17-0780-01) for 30 min. The precleared lysate was then incubated with Sox9 antibody (EMDMillipore, ab5535) for 1 h at 4 °C followed by ultracentrifugation and secondary incubation with a protein A Sepharose slurry for 1 h. The precleared and IP’ed beads were briefly washed with NETN buffer (50 mM Tris pH 7.3, 170 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40), boiled in 2× NUPAGE LDS sample buffer (Life Technologies), and resolved on a NuPAGE 10% Bis-Tris Gel (Life Technologies). The eluted proteins were visualized with Coomassie Brilliant blue stain, excised into gel pieces according to the molecular weight, and in-gel digested with trypsin. The liquid chromatography-tandem mass spectrometry (MS) analysis was carried out using the nanoLC1000 system coupled to an Orbitrap Fusion mass spectrometer (Thermo Scientific). The peptides were loaded on a two-column setup with a precolumn (2 cm × 100 µm internal diameter [I.D.]) and analytical column (5 cm × 150 µm I.D. for 3xCr HGG samples; 20 cm × 75 µm I.D. for human HGG, ZRFUS EPN samples) filled with Reprosil-Pur Basic C18 (1.9 µm; Dr. Maisch GmbH). The peptide elution was done using a discontinuous gradient of 90% acetonitrile buffer (B) in 0.1% formic acid (5 to 28% B, 800 nL/min: 45-min gradient for 3xCr HGG samples; 2 to 30% B, 200 nL/min: 110-min gradient for human HGG, ZRFUS EPN samples). The MS instrument was operated in data-dependent mode with MS1 acquisition in Orbitrap (120,000 resolution, AGC 5e5, 50-ms injection time) followed by MS2 in Ion Trap (Rapid Scan, HCD 30%, AGC 5e4). The MS raw data were searched using Proteome Discoverer 2.0 software (Thermo Scientific) with Mascot algorithm against the mouse or human National Center for Biotechnology Information (NCBI) refseq database updated 24 March 2020. The precursor ion tolerance and product ion tolerance were set to 20 ppm and 0.5 Da, respectively. A maximum cleavage of two with trypsin enzyme, dynamic modification of oxidation on methionine, protein N-term acetylation, and destreak on cysteine were allowed. The peptides identified from the mascot result file were validated with 5% false discovery rate (FDR). The gene product inference and quantification were done with the label-free iBAQ approach using the gpGrouper algorithm (PubMed identifier 30093420). For statistical assessment, missing value imputation was employed through sampling a normal distribution N (μ-1.8 σ, 0.8σ), where μ, σ are the mean and SD of the quantified values. For differential analysis, we used the moderated t test as implemented in the R package limma, and multiple-hypothesis testing correction was performed with the Benjamini–Hochberg procedure. For Venn diagrams of Sox9 interactors, all proteins with a fold change of >2 and P value of <0.05 were considered for analysis.
Co-IP and Western Blot.
Nuclear lysates were prepared as described above for sample preparation for MS. IP was performed by adding anti-Sox9 antibody at 4 °C, and the supernatant was used for IP with rabbit anti-Sox9 (EMDMillipore, ab5535) or rabbit anti-IgG (R&D Systems, AB-105-C) overnight at 4 °C. Subsequent pull-down was performed by adding protein A agarose beads (ThermoFisher, 15918-014) for an additional 5 h at 4 °C. The beads were collected, washed, and boiled in 2× SDS gel loading dye to elute immunoprecipitated proteins, which were analyzed by Western blot. Inputs (10% of lysate) and immunoprecipitated proteins from the anti-IgG and anti-Sox9 IPs were run on a 10% SDS polyacrylamide gel, followed by wet transfer to nitrocellulose membrane at 400 mA for 45 min. The membrane was blocked by 5% milk in Tris-buffered saline with Tween20 (TBST), followed by incubation overnight at 4 °C in the primary antibodies anti-Sox9 (EMDMillipore, ab5535), anti-Hdac2 (Abcam, ab32117), and anti-Mta2 (Abcam, ab8106) at 1:1,000 dilution. The next day, membranes were washed three times with TBST, incubated at room temperature for 1 h in horseradish peroxidase–conjugated anti-rabbit IgG at 1:2,000 dilution in 5% milk, washed again three times with TBST, and developed using luminol reagent (Santa Cruz Biotechnology, sc2048).
Mouse ZRFUS EPN Cell Line Generation and Cell Culture.
The mouse ZRFUS cell line was generated as described in our previous study (21). Briefly, mouse ZRFUS EPN tumors were collected based on the GFP signal, rinsed with PBS, and dissociated into single cells using the Brain Tumor Dissociation Kit (MACS, 130-095-942) following the manufacturer’s instructions. The cells were then grown in neural basal medium supplemented with sodium pyruvate, glutamine, B-27 (ThermoFisher, 12587010), N-2 (ThermoFisher, 17502048), rhEGF (R&D system, 236-EG), and rhFGF (R&D system, 4114-TC). Cells were cultured on Matrigel (Corning, 354277)-coated cultured dishes.
Quantification and Statistical Analyses.
Sample sizes and statistical tests are provided in the figure legends. For Kaplan–Meier survival analysis, the log-rank test was used to compare survival differences across groups. One way ANOVA was used for BrdU proliferation and H3K27ac quantification, followed by Tukey’s test to compare individual means. For RT-qPCR, two-tailed Student’s t test was used for statistical calculations. Significant differences are denoted by asterisks in associated graphs. The distribution of data was assumed to be normal, but this was not formally tested. Data are presented as ± SEM. Levels of statistical significance are indicated as follows: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Mice were excluded from analyses if they did not demonstrate GFP reporter activity indicating unsuccessful electroporation. Box plots with significance were generated using ggsignif (v0.6.0) with ggplot2 (v3.3.2).
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (R01-NS071153 to B.D., R01-NS124093 to B.D. and G.R., R01NS094615 to G.R., and K08-NS110976 to A. Jalali), the National Cancer Institute–Cancer Therapeutic Discovery (U01-CA217842 to B.D.), and the Diana Helis Henry and Adrienne Helis Malvin Medical Research Foundation (to B.D. and S.C.M.). S.C.M. is supported by grants from the NIH (1R01NS116361), an Alex’s Lemonade Stand Foundation A Award, a Cancer Prevention and Research Institute of Texas (CPRIT) Scholar in Cancer Research Award, the Pediatric Brain Tumor Foundation, the V Scholar Foundation, and the American Lebanese Syrian Associated Charities St Jude Children’s Research Hospital. D.S. is supported by a K99 grant from the NIH (1K99-DC019668). We thank Anthony Grichuk for embedding and sectioning human HGG samples. The Baylor College of Medicine Mass Spectrometry Proteomics Core is supported by a Dan L. Duncan Comprehensive Cancer Center NIH award (P30 CA125123), a CPRIT Core Facility Award (RP210227), and an NIH High End Instrument award (S10OD026804). Images in schematics were created using Biorender.com.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2202015119/-/DCSupplemental.
Data Availability
The RNA-Seq and ChIP-Seq datasets generated during this study are available at the NCBI Gene Expression Omnibus (GEO) website (series GSE202961) (51) under RNA-Seq GEO: GSE202958 (52) and ChIP-Seq GEO: GSE202960 (53). The MS datasets generated during this study have been deposited to the ProteomeXchange Consortium via the Proteomics Identification Database (PRIDE) partner repository with the dataset identifier PXD033863 (54). All other study data are included in the article and/or supporting information.
References
- 1.Plass C., et al. , Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat. Rev. Genet. 14, 765–780 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Phillips R. E., Soshnev A. A., Allis C. D., Epigenomic reprogramming as a driver of malignant glioma. Cancer Cell 38, 647–660 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennan C. W., et al. ; TCGA Research Network, The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallo M., et al. , MLL5 orchestrates a cancer self-renewal state by repressing the histone variant H3.3 and globally reorganizing chromatin. Cancer Cell 28, 715–729 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Schwartzentruber J., et al. , Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Mack S. C., et al. , Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature 506, 445–450 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu F., et al. , EGFR mutation promotes glioblastoma through epigenome and transcription factor network remodeling. Mol. Cell 60, 307–318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kundaje A., et al. ; Roadmap Epigenomics Consortium, Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gimple R. C., et al. , Glioma stem cell–specific superenhancer promotes polyunsaturated fatty-acid synthesis to support EGFR signaling. Cancer Discov. 9, 1248–1267 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X., et al. , MYC-regulated mevalonate metabolism maintains brain tumor–initiating cells. Cancer Res. 77, 4947–4960 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mack S. C., et al. , Therapeutic targeting of ependymoma as informed by oncogenic enhancer profiling. Nature 553, 101–105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mack S. C., et al. , Chromatin landscapes reveal developmentally encoded transcriptional states that define human glioblastoma. J. Exp. Med. 216, 1071–1090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang P., et al. , Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron 74, 79–94 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ung K., et al. , Olfactory bulb astrocytes mediate sensory circuit processing through Sox9 in the mouse brain. Nat. Commun. 12, 5230 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Bont J. M., et al. , Differential expression and prognostic significance of SOX genes in pediatric medulloblastoma and ependymoma identified by microarray analysis. Neuro-oncol. 10, 648–660 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z., et al. , SOX9-PDK1 axis is essential for glioma stem cell self-renewal and temozolomide resistance. Oncotarget 9, 192–204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacLeod G., et al. , Genome-wide CRISPR-Cas9 screens expose genetic vulnerabilities and mechanisms of temozolomide sensitivity in glioblastoma stem cells. Cell Rep. 27, 971–986.e9 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Glasgow S. M., et al. , Glia-specific enhancers and chromatin structure regulate NFIA expression and glioma tumorigenesis. Nat. Neurosci. 20, 1520–1528 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.John Lin C. C., et al. , Identification of diverse astrocyte populations and their malignant analogs. Nat. Neurosci. 20, 396–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu K., et al. , PIK3CA variants selectively initiate brain hyperactivity during gliomagenesis. Nature 578, 166–171 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arabzade A., et al. , ZFTA-RELA dictates oncogenic transcriptional programs to drive aggressive supratentorial ependymoma. Cancer Discov. 11, 2200–2215 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sturm D., et al. , Paediatric and adult glioblastoma: Multiform (epi)genomic culprits emerge. Nat. Rev. Cancer 14, 92–107 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mack S. C., Hubert C. G., Miller T. E., Taylor M. D., Rich J. N., An epigenetic gateway to brain tumor cell identity. Nat. Neurosci. 19, 10–19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laug D., Glasgow S. M., Deneen B., A glial blueprint for gliomagenesis. Nat. Rev. Neurosci. 19, 393–403 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song H. R., et al. , Nuclear factor IA is expressed in astrocytomas and is associated with improved survival. Neuro-oncol. 12, 122–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adam R. C., et al. , Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 521, 366–370 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Passeron T., et al. , Upregulation of SOX9 inhibits the growth of human and mouse melanomas and restores their sensitivity to retinoic acid. J. Clin. Invest. 119, 954–963 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang G., et al. , Zbtb7a suppresses prostate cancer through repression of a Sox9-dependent pathway for cellular senescence bypass and tumor invasion. Nat. Genet. 45, 739–746 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun X., et al. , Arid1a has context-dependent oncogenic and tumor suppressor functions in liver cancer. Cancer Cell 32, 574–589.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi J., Vakoc C. R., The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol. Cell 54, 728–736 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Latif A. L., et al. , BRD4-mediated repression of p53 is a target for combination therapy in AML. Nat. Commun. 12, 241 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu D., Hunter S. B., Vertino P. M., Van Meir E. G., Overexpression of MBD2 in glioblastoma maintains epigenetic silencing and inhibits the antiangiogenic function of the tumor suppressor gene BAI1. Cancer Res. 71, 5859–5870 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitange G. J., et al. , Retinoblastoma binding protein 4 modulates temozolomide sensitivity in glioblastoma by regulating DNA repair proteins. Cell Rep. 14, 2587–2598 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moon B. S., et al. , Epigenetic modulator inhibition overcomes temozolomide chemoresistance and antagonizes tumor recurrence of glioblastoma. J. Clin. Invest. 130, 5782–5799 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grignani F., et al. , Fusion proteins of the retinoic acid receptor-α recruit histone deacetylase in promyelocytic leukaemia. Nature 391, 815–818 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Martens J. H. A., et al. , PML-RARα/RXR alters the epigenetic landscape in acute promyelocytic leukemia. Cancer Cell 17, 173–185 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Sankar S., et al. , Mechanism and relevance of EWS/FLI-mediated transcriptional repression in Ewing sarcoma. Oncogene 32, 5089–5100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riggi N., et al. , EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell 26, 668–681 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao R., et al. , Pervasive chromatin-RNA binding protein interactions enable RNA-based regulation of transcription. Cell 178, 107–121.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen F., LoTurco J., A method for stable transgenesis of radial glia lineage in rat neocortex by piggyBac mediated transposition. J. Neurosci. Methods 207, 172–180 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding X. F., Fan M., Nonviral gene therapy of the nervous system: Electroporation. Methods Mol. Biol. 1382, 297–305 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heinz S., et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H., et al. ; 1000 Genome Project Data Processing Subgroup, The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramírez F., et al. , deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu T., et al. , clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whyte W. A., et al. , Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawrence M., et al. , Software for computing and annotating genomic ranges. PLOS Comput. Biol. 9, e1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D. Sardar, et al., Sox9 directs divergent epigenomic states in brain tumor subtypes. GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE202961. Deposited 13 May 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D. Sardar, et al., Sox9 directs divergent epigenomic states in brain tumor subtypes (RNA-Seq). GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE202958. Deposited 13 May 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.D. Sardar, et al., Sox9 directs divergent epigenomic states in brain tumor subtypes (ChIP-Seq). GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE202960. Deposited 13 May 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D. Sardar, et al., Sox9 directs divergent epigenomic states in brain tumor subtypes. PRIDE. https://www.ebi.ac.uk/pride/archive/projects/PXD033863. Deposited 12 May 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq and ChIP-Seq datasets generated during this study are available at the NCBI Gene Expression Omnibus (GEO) website (series GSE202961) (51) under RNA-Seq GEO: GSE202958 (52) and ChIP-Seq GEO: GSE202960 (53). The MS datasets generated during this study have been deposited to the ProteomeXchange Consortium via the Proteomics Identification Database (PRIDE) partner repository with the dataset identifier PXD033863 (54). All other study data are included in the article and/or supporting information.