Significance
The main finding reported here is that acetylation converts superoxide dismutase-2 (SOD2) from a mitochondrial antioxidant to a nuclear histone demethylase. The change in function involves the binding of iron instead of the canonical cofactor manganese. Unlike well-characterized histone demethylases, FeSOD2 uses H2O2, which is typically increased in hypoxic tumor niches, as substrate. Along these lines, we found that nuclear FeSOD2 promotes lineage plasticity largely by facilitating the reactivation of gene expression associated with epithelial-to-mesenchymal transition and stemness reprogramming. Together, our results provide evidence in support of an acetylation-dependent nucleus-specific function of SOD2 associated with the emergence of more tumorigenic and metastatic cancer cell phenotypes.
Keywords: SOD2, iron, manganese, epigenetic, breast cancer
Abstract
The dichotomous behavior of superoxide dismutase-2 (SOD2) in cancer biology has long been acknowledged and more recently linked to different posttranslational forms of the enzyme. However, a distinctive activity underlying its tumor-promoting function is yet to be described. Here, we report that acetylation, one of such posttranslational modifications (PTMs), increases SOD2 affinity for iron, effectively changing the biochemical function of this enzyme from that of an antioxidant to a demethylase. Acetylated, iron-bound SOD2 localizes to the nucleus, promoting stem cell gene expression via removal of suppressive epigenetic marks such as H3K9me3 and H3K927me3. Particularly, H3K9me3 was specifically removed from regulatory regions upstream of Nanog and Oct-4, two pluripotency factors involved in cancer stem cell reprogramming. Phenotypically, cells expressing nucleus-targeted SOD2 (NLS-SOD2) have increased clonogenicity and metastatic potential. FeSOD2 operating as H3 demethylase requires H2O2 as substrate, which unlike cofactors of canonical demethylases (i.e., oxygen and 2-oxoglutarate), is more abundant in tumor cells than in normal tissue. Therefore, our results indicate that FeSOD2 is a demethylase with unique activities and functions in the promotion of cancer evolution toward metastatic phenotypes.
Cancer cell evolution toward developing metastatic potential depends in large part on the loss of lineage commitment simultaneous to the acquisition of phenotypic plasticity (1, 2). Overwhelming evidence indicates that the reactivation of pluripotency genes normally silenced in adult somatic cells promotes lineage plasticity acquisition in tumor cells (3, 4), enabling metastatic dissemination. In particular, the reactivation of Oct-4, SOX2, and Nanog, a triad of stemness transcription factors involved in tumor progression toward metastasis (5), requires demethylation of repressive epigenetic marks associated with heterochromatin maintenance, including H3K9me3 and H3K27me3 (6–8). Removal of these marks is canonically mediated by iron/2-oxoglutarate–dependent members of the JmJC domain–containing histone demethylase family (KDM) that require oxygen as an indispensable cofactor (9, 10). As tumors grow, poor vascularization and increasing metabolic activity drive tumors into hypoxia, a condition that paradoxically promotes pluripotency as well as the emergence of stem-like cells in the evolving tumor hypoxic niches (11). Hence, how cancer cells reactivate pluripotency genes under low oxygen availability is a matter of significant biochemical and clinical interest. Superoxide dismutase-2 (SOD2) is a primarily mitochondrial enzyme with established roles in detoxification of reactive oxygen species (ROS) produced by the electron transport chain. It has been widely acknowledged that in cancer, SOD2 assumes functional roles that are irreconcilable with its known antioxidant/tumor suppressor activities (12, 13). These roles include promoting the acquisition of capacities consistent with those of cancer stem cells, including anoikis and therapy resistance, lineage plasticity, clonogenicity, and increased metastatic potential (13). Several recent studies have linked these noncanonical activities of SOD2 with an acetylated form that drastically changes SOD2 structure from that of the antioxidant tetramer to a monomer that displays hydrogen peroxide–driven, iron-dependent oxidase activity (13–15). We report here that FeSOD2 is a major form of SOD2 populating the nucleus of matrix-detached cancer cells. We also report that acetylated FeSOD2 is the predominant form of SOD2 in the nucleus of stem-like cancer cells where it antagonizes epithelial cell lineage commitment via reshaping the chromatin epigenetic landscape. To that effect, results presented below indicate that FeSOD2 interacts with members of chromatin remodeler complexes, such as SWI/SNF, while displaying direct H3K9me3/H3K27me3 demethylase activity. Directing SOD2 to the nucleus promotes chromatin opening at regions encoding genes associated with lineage plasticity, dedifferentiation, and stemness reprogramming, resulting in increased metastatic potential in breast cancer cells. Hence, results are consistent with a model where FeSOD2 confers H3 demethylase activity to chromatin remodeling complexes thereby promoting lineage plasticity acquisition in cancer.
Results
Cancer Cells Grown in Suspension Have Acetylated SOD2 in the Nucleus.
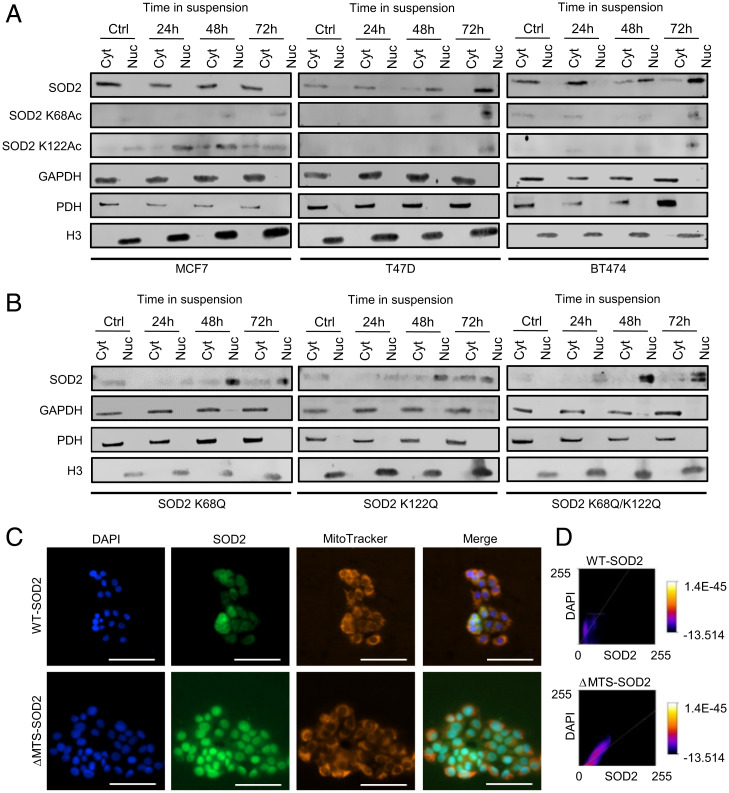
Breast cancer cells were grown in ultra-low adherence plates for up to 72 h, a condition that favors the expansion of cancer cells with stem cell–like characteristics. Results in Fig. 1A show the gradual transition of SOD2 from exclusively mitochondrial to nuclear localization parallel to an increase in acetylation of both K68 and K122 in at least three different breast cancer cell lines. Acetylated SOD2 was detected in the nucleus of all cell lines used in this study but was least evident in MCF7 cells that also retained SOD2 localization predominantly in the mitochondria/cytosol. Differently, in T47D and BT474 breast cancer cells, SOD2 nearly completely transferred to the nucleus within 72 h in suspension. Hence, to minimize the possible confounding effects of loss of mitochondrial SOD2, MCF7 cells were chosen for additional studies. To determine whether acetylation played a role in facilitating nuclear localization K68, K122, or both, simultaneously, were replaced by glutamine (Q), a mimetic of acetyllysine. Data shown in Fig. 1B indicate that acetylation of either or both K68 and K122, as modeled by mutation to glutamine significantly increased localization of SOD2 in the nucleus of MCF7 cells, suggesting that acetylation, which disrupts the tetrameric structure of SOD2, promotes nuclear localization. Surprisingly, we observed that deletion of the mitochondrial targeting sequence (MTS) promotes strong nuclear localization of ΔMTS-SOD2 (Fig. 1 C and D), suggesting that extramitochondrial SOD2 naturally relocates to the nucleus. Taken together, results in Fig. 1 indicate that structural modifications such as those caused by acetylation switch SOD2 localization from mitochondrial to nuclear in detached cancer cells.
Fig. 1.
Acetylated SOD2 localizes to the nucleus. (A) Acetylated SOD2 accumulates in the nuclear fraction of breast cancer cell lines cultured in suspension. (B) SOD2 accumulates in the nuclear fraction of MCF7 cells expressing SOD2 K68Q, K122Q, and K68Q/K122Q mutants. (C) SOD2 localizes to the nucleus in the absence of MTS. (D) Quantification of WT- and ΔMTS-SOD2 colocalization with nuclear staining. Figures are representative of at least three independent experiments. Cyt, extranuclear fraction; Nuc, nuclear fraction; GAPDH, glyceraldehyde 3-phosphate dehydrogenase–cytosolic subcellular localization marker; PDH, pyruvate dehydrogenase–mitochondrial subcellular localization marker; H3, histone 3–nuclear subcellular localization marker. (White scale bars, 100 µm.)
SOD2 Interactome in the Nucleus Indicates Involvement in Chromatin Remodeling.
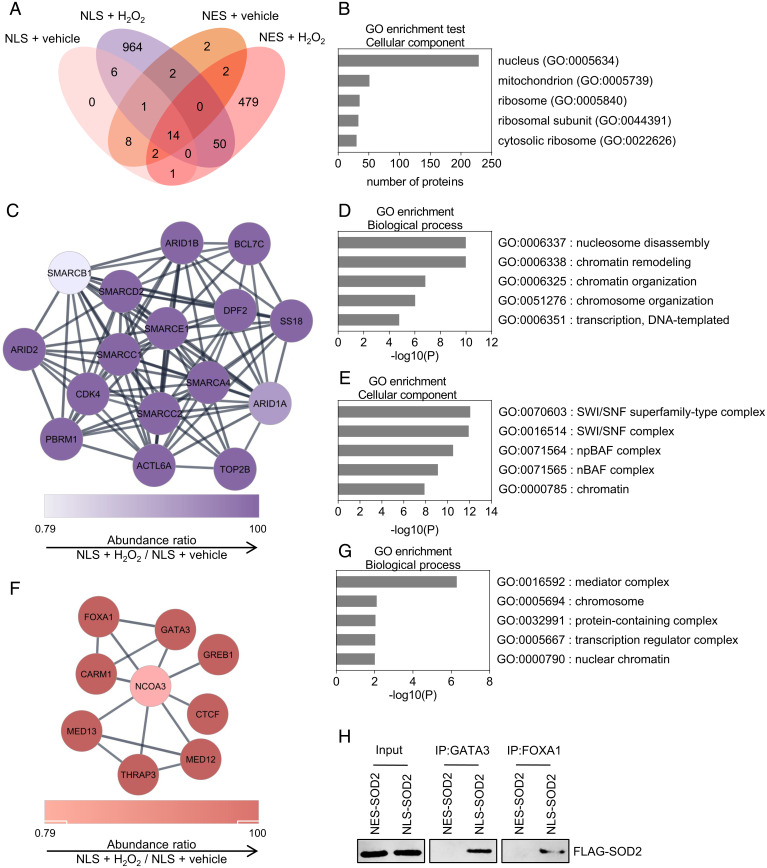
The finding of SOD2 in the nucleus of breast cancer cells prompted us to design studies aimed at determining the biological function of nuclear SOD2. To do that, we focused on identifying binding partners that could help identify potential biologic functions. Proximity biotin labeling was performed using an ascorbate peroxidase (APEX) construct fused to SOD2 bearing a nuclear localization sequence (NLS-SOD2, bait) followed by proteomics identification of biotinylated nuclear proteins. Results shown in Fig. 2A indicated that NLS-SOD2-APEX produced labeling of a total of 1,040 targets with the vast majority (964) being exclusively detected in the experimental group and not in controls such as SOD2 fused to leucine-rich nuclear export sequence (NES-SOD2) (Fig. 2A). As expected, the majority of proteins labeled by NLS-SOD2-APEX were predominantly nuclear as well (Fig. 2B). Clustering analysis of proteins labeled by NLS-SOD2-APEX indicated a disproportional enrichment in components of the SWI/SNF complex (Fig. 2 C–E) involved in chromatin remodeling via nucleosome repositioning (16). In fact, 10 members of the SWI/SNF complex including SMARCA4 (BRG1), SMARCB1, SMARCC1, SMARCC2, SMARCD2, SMACE1, ARID1A, ARID1B, ARID2, and BCL7C were identified as proximal ligands to nuclear SOD2 indicating a particularly close interaction between SOD2 and SWI/SNF ATP-dependent chromatin remodeling enzymes. In addition, our study identified a secondary cluster of proteins involved in the regulation of oncogenic transcription via chromatin remodeling (Fig. 2 F and G). These included nuclear receptor coactivator-3 (NCOA3), a transcriptional coactivator involved in breast cancer progression (17); CTCF, a chromatin remodeler known to interact with members of the SWI/SNF complex (18, 19); FOXA1, a pioneer transcription factor frequently mutated in progressing estrogen receptor positive (ER+) breast cancer (20); and GATA3, a transcription factor recently shown to regulate mammary epithelial cell differentiation in association with FOXA1 and ERα (21). To confirm interaction of nuclear SOD2 with chromatin remodeler proteins, we performed immunoprecipitation (IP) assays targeting GATA3 and FOXA1. Results in Fig. 2H demonstrated that only NLS-SOD2, but not mitochondrial wild-type (WT)-SOD2, was coimmunoprecipitated with the IP targets, confirming that this interaction occurred in the nucleus. Overall, results summarized in Fig. 2 indicate a role for SOD2 in the coregulation of chromatin epigenetic remodeling and epithelial cell lineage differentiation at the nucleosomal level.
Fig. 2.
Nuclear SOD2 interacts with proteins involved in chromatin remodeling and organization. (A) Venn diagram displaying the number of proteins interacting with NLS-SOD2 and NES-SOD2 identified by APEX and proteomic analysis. (B) Gene ontology annotation (cellular component) for identified NLS-SOD2 interacting proteins. (C and F) Chromatin remodeling–enriched networks of NLS-SOD2 interacting proteins. (D) Biological processes associated with the network identified in C. (E) Gene ontology annotation (cellular component) for the network identified in C. (G) Biological processes associated with the network identified in F. (H) Immunoprecipitation of GATA Binding Protein-3 (GATA3) and Forkhead box A-1 (FOXA1). Figures are representative of at least three independent experiments.
Nuclear SOD2 Promotes Nucleosomal Histone Lysine Demethylation.
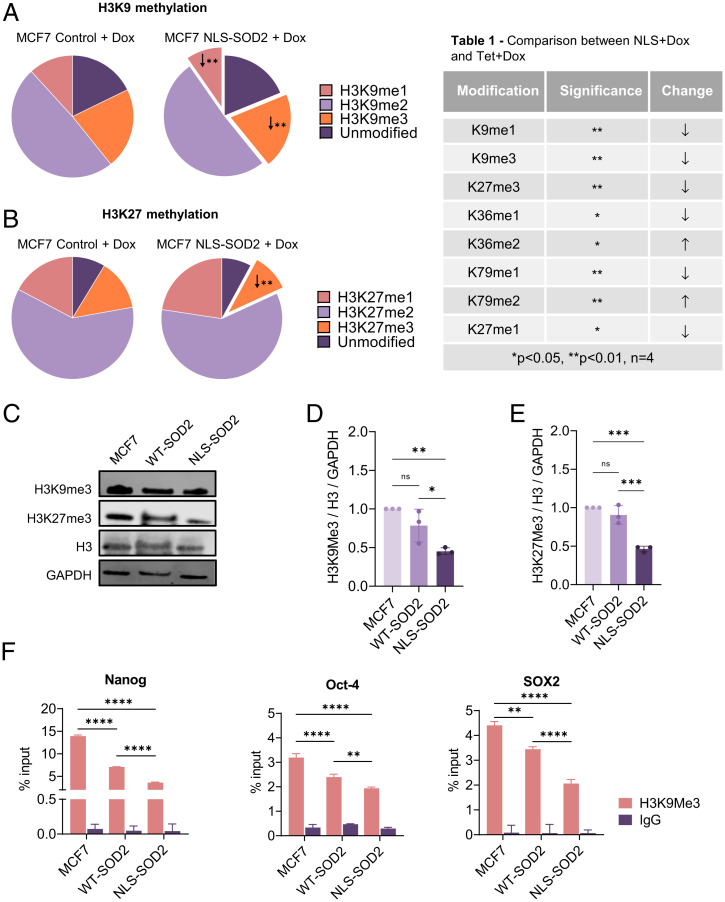
ATP-dependent chromatin remodeling complexes like the SWI/SNF superfamily are specialized protein machineries dedicated to facilitating transcription via repositioning nucleosomes to increase DNA accessibility. It is becoming increasingly clear that higher order chromatin reorganization is linked to subnucleosomal changes required to optimize transcription including histone modifications that directly impact transcriptional efficiency. Because none of the SWI/SNF complex components found to interact with NLS-SOD2 display histone modifying activity, we hypothesized that nuclear SOD2 could be involved in the direct posttranslational modification (PTM) of nucleosomal histones. To investigate this possibility, we generated MCF7 cells expressing NLS-SOD2 under a tetracycline-inducible promoter (tet-ON) to produce isogenic doxycycline (Dox)-dependent NLS-SOD2–expressing models. Dox-induced NLS-SOD2–expressing MCF7 cells, as well as Dox-treated controls, were compared using proteomic and immunochemical approaches. Results shown in Fig. 3 A and B, Table 1) summarize the main finding of a generalized reduction in H3 methylation induced by NLS-SOD2 expression. Importantly, two repressive epigenetic marks H3K9me3 and H3K27me3 with well-established roles in silencing the transcription of genes involved in lineage plasticity acquisition (i.e., Nanog, Oct-4, and SOX2) were reduced according to mass spectrometry quantification, suggesting a potential role for nuclear SOD2 in the epigenetic regulation of genes involved in dedifferentiation and stemness reprogramming. Using MCF7 constitutively expressing similar levels of SOD2 mutants (SI Appendix, Fig. S1), we confirmed by Western blot that only NLS-SOD2–expressing cells significantly reduced the H3K9me3 and H3K27me3 marks compared to parent cells (Fig. 3 C–E). To further test the idea that nuclear SOD2 activates genes involved in lineage plasticity acquisition, chromatin immunoprecipitation (ChiP) experiments were performed to determine whether NLS-SOD2 decreases deposition of repressive H3K9me3 marks onto the promoter regions of three core stemness genes. Results shown in Fig. 3F indicate that NLS-SOD2 significantly reduced H3K9me3 associated with the promoters of Nanog, Oct-4, and SOX2, suggesting that nuclear SOD2 promotes dedifferentiation in cancer cells by activating the expression of stemness genes epigenetically silenced via histone H3 lysine methylation.
Fig. 3.
Nuclear SOD2 induces H3 histones demethylation. (A) Mass spectrometry analysis of H3K9 methylation status (unmodified, mono, di-, or trimethyl) identified in MCF7 cells conditionally expressing NLS-SOD. (B) Mass spectrometry analysis of H3K27 methylation status (unmodified, mono, di-, or trimethyl) identified in MCF7 cells conditionally expressing NLS-SOD. (C) Western blot analysis of H3K9me3 and H3K27me3 in MCF7 cells constitutively expressing WT SOD2 (Tet+Dox) and NLS-SOD2 (NLS+Dox). (D) Quantification of H3K9me3 protein levels relative to total H3. (E) Quantification of H3K27me3 protein levels relative to total H3. (F) H3K9me3 enrichment on promoter regions of Nanog, Oct-4, and SOX2 (stemness genes) measured by ChIP-qPCR. Figures are representative of at least three independent experiments. ns, not significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; error bars indicate mean ± SD.
Transcriptome Analysis Indicates Nuclear SOD2 Promotes the Activation of Oncogenic Programs Associated with Lineage Plasticity Acquisition.
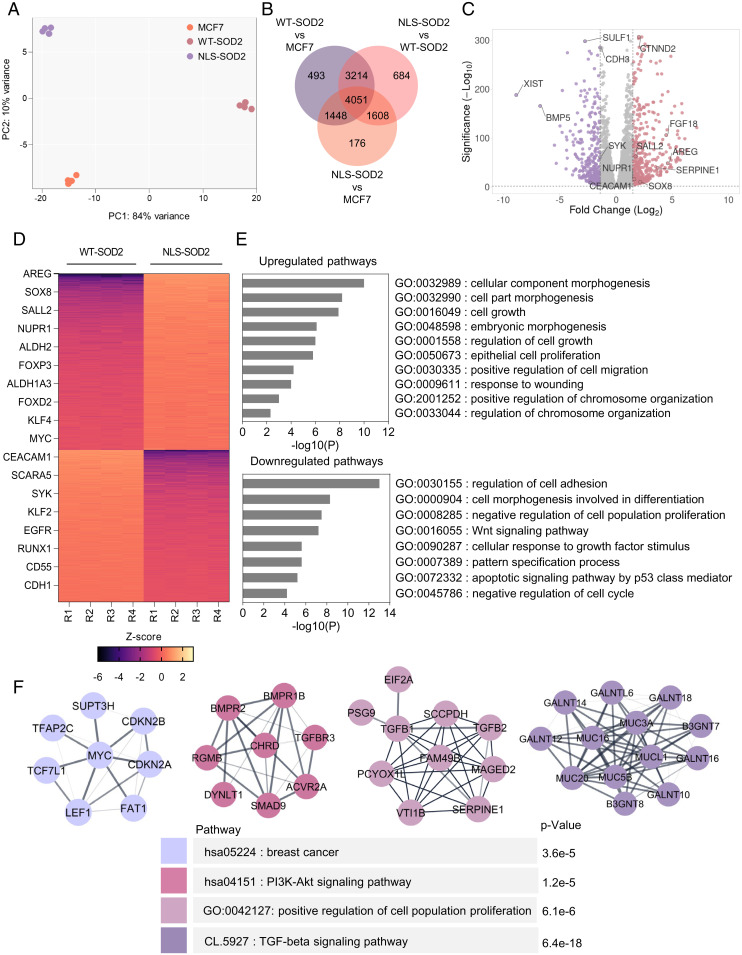
The finding of a potential role for nuclear SOD2 in promoting the transcription of a few genes involved in lineage plasticity acquisition, such as Nanog, Oct4, SOX2, and SOX9 (SI Appendix, Fig. S2A), led us to perform more detailed studies comparing the transcriptome of cells expressing either WT-SOD2 or NLS-SOD2. Parental MCF7 cells were used as baseline reference. Results in Fig. 4A indicate that the localization of SOD2 to the nucleus was a major component of cluster separation. The Venn diagram in Fig. 4B show 9,557 differentially expressed genes in NLS-SOD2 compared to WT-SOD2, represented in the form of a volcano plot shown in Fig. 4C. Interestingly, the analysis of differentially expressed genes shown in Fig. 4D indicate an up-regulation of lineage plasticity genes, including KLF4 and Myc, two core stemness transcription factors (22); Sall4, an Oct-4 target gene involved in lineage plasticity acquisition; SOX8, a member of the SOXE network involved in epithelial-to-mesenchymal transition (EMT); as well as numerous prometastasis oncogenes, including FOXP3, NUPR1, and AREG. In fact, gene set enrichment analysis (GSEA) of Hallmark genes showed up-regulation of genes associated with EMT (SI Appendix, Fig. S2 B and C) and KRAS signaling (SI Appendix, Fig. S2 D and E) in cells expressing NLS-SOD2. We also found that NLS-SOD2 antagonizes gene expression programs that enforce epithelial lineage commitment as indicated by a reduction in the expression of epithelial cell markers such as CEACAM1, EGFR, E-cadherin (CDH1), as well as suppressors of breast cancer progression like KLF2 and RUNX1. Gene ontology analysis of transcriptomic data further highlighted the role of NLS-SOD2 in up-regulating programs associated with proliferation, migration, and lineage plasticity while downregulating the expression of tumor suppressor genes (Fig. 4E). Finally, complementary gene network analysis shown in Fig. 4F identified at least three functional clusters involved in EMT, Myc signaling, and SMAD9/TGFβ signaling, as well as an up-regulation of mucin markers associated with breast cancer progression. Taken together, results in Fig. 4 indicate nuclear SOD2 facilitates chromatin remodeling associated with the progression of epithelioid breast cancer toward metastatic mesenchymal phenotypes.
Fig. 4.
Nuclear SOD2 increases expression of genes associated with EMT and stemness. (A) Principal component analysis (PCA) plot of MCF7 cells expressing WT- and NLS-SOD2. (B) Overlap of all differentially expressed genes in NLS-SOD2 vs. MCF7, WT-SOD2 vs. MCF7, and NLS-SOD2 vs. WT-SOD2, considering significance P < 0.01 cutoff. (C) Volcano plot of up-regulated (in red) and down-regulated (in purple) mRNAs identified in NLS-SOD2 vs. WT-SOD2. Dotted lines in the x axis represent Log2fold change (FC) = 1.5 cutoff. Dotted line in the y axis represents significance (–Log10 = 2) cutoff. (D) Heatmap of core genes differentially enriched in NLS-SOD2 vs. WT-SOD2 considering Log2FC = 1.5 cutoff. (E) GO term enrichment analysis of genes significantly up-regulated (Top) and down-regulated (Bottom) in NLS-SOD2–expressing cells compared to WT-SOD2. (F) Protein–protein interaction network analysis of NLS-SOD2 up-regulated genes based on STRING database. n = 4.
Phenotypic Analysis of NLS-SOD2 Expressing Cells Confirm Mesenchymal/Invasive Behavior.
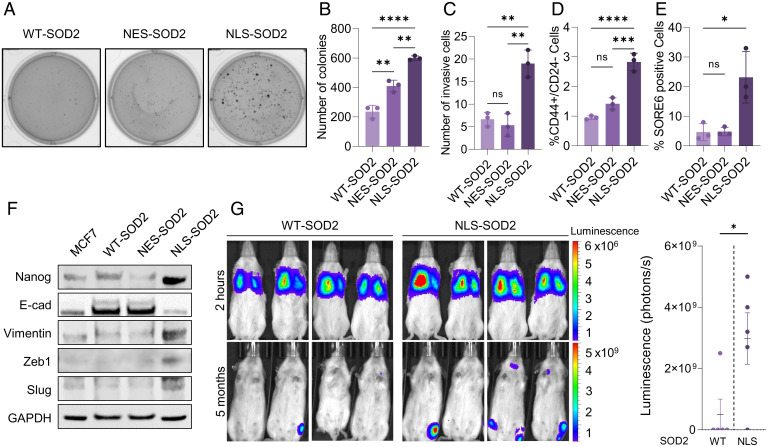
Findings shown in Fig. 4 motivated additional studies focused on phenotyping MCF7 cells expressing NLS-SOD2. Results from these studies using single clones with matched levels of SOD2 overexpression indicated that cells expressing NLS-SOD2 were more capable of clonogenic growth in soft agar (Fig. 5 A and B) and displayed significantly more invasive behavior when compared with cells overexpressing either WT-SOD2 or NES-SOD2 (Fig. 5C). Consistent with more aggressive behavior, NLS-SOD2–expressing cells were also enriched in CD44+/CD24− cells that identify breast cancer stem cells (Fig. 5D) (23). This observation was further confirmed using SORE6, a functional genetic stem cell biosensor that reports Oct-4 and SOX2 transcriptional activation (24). Fig. 5E shows a marked increase in the number of SORE6+ cells within NLS-SOD2 cells relative to WT-SOD2– and NES-SOD2–expressing controls. Additionally, Western blot data shown in Fig. 5F indicate increased expression of both stemness transcription factors (i.e., Nanog) as well as changes to EMT markers (i.e., E-cadherin, vimentin, ZEB1, and Snai2/Slug) only in NLS-SOD2–expressing cells, compared to WT and NES controls. Similar results were obtained at the level of Nanog, Oct-4, SOX2, and SOX9 mRNA expression using qRT-PCR (SI Appendix, Fig. S2A). To test whether NLS-SOD2–induced up-regulation of EMT genes promotes transformation of noninvasive MCF7 cells into a metastatic phenotype, luciferin-expressing WT-SOD2 or NLS-SOD2 MCF7 cell lines were injected into the tail vein of immune-deficient NOD-Scid IL2Rgammanull (NSG) mice. The formation of metastatic lesions in these mice was then assessed by bioluminescence in vivo using an IVIS/LAGO imaging system after intraperitoneal (i.p.) injection of luciferin. Results in Fig. 5G show that incidence of metastasis was higher in animals injected with MCF7 cells expressing NLS-SOD2, indicating that accumulation of SOD2 in the nucleus of noninvasive luminal breast cancer cells increases their metastatic potential. Collectively, these results confirm nuclear SOD2 functions as an oncoprotein promoting the activation of programs involved in breast cancer progression toward mesenchymal/invasive phenotypes.
Fig. 5.
Nuclear SOD2 activates epithelial-to-mesenchymal transformation and stemness reprogramming. (A) Anchorage-independent soft-agar colony formation in WT-, NES-, NLS-SOD2–expressing cells. (B) Quantification of colony formation assay. (C) Transwell invasion assay of WT-, NES-, and NLS-SOD2–expressing cells. (D) FACS analysis of CD44+/CD24− membrane markers. (E) Quantification of cancer stem cells using SORE6 reporter. (F) Western blot analysis of stemness (Nanog) and epithelial-to-mesenchymal transition (E-cadherin, vimentin, Zeb1, and Slug) markers. (G) Metastatic dissemination in NSG mice injected with WT-SOD2 or NLS-SOD2 MCF7 cells. Figures are representative of at least three independent experiments. ns, not significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; error bars indicate mean ± SD.
Nuclear SOD2 Displays Fe/H2O2-Dependent H3 Demethylase Activity.
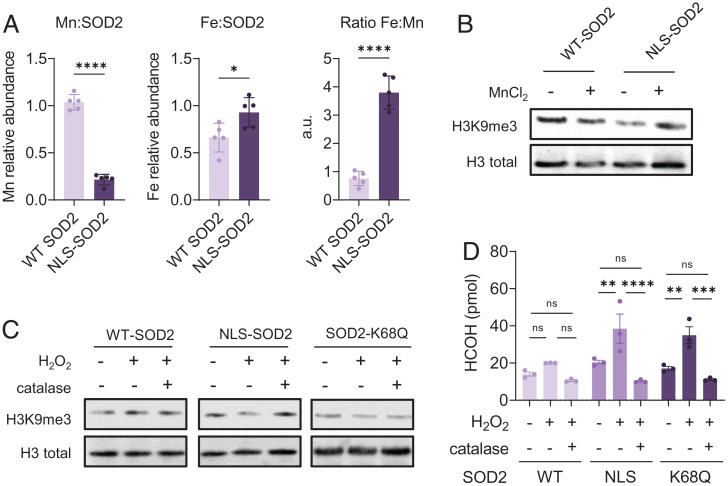
Though results shown above indicated nuclear SOD2 reduces H3 lysine methylation, it remained unclear whether SOD2 had direct demethylase activity. It has been previously described that iron incorporation, as well as lysine acetylation, promote the switch of SOD2 function from Mn-dependent superoxide dismutase to a Fe-dependent peroxidase (13–15). The observation that translocation of SOD2 to the nucleus is associated with K68 acetylation led us to the hypothesis that H3 lysine demethylation is catalyzed by the peroxidase activity of FeSOD2 in the nucleus. Because the peroxidase activity of SOD2 is Fe-dependent, we first tested whether nuclear SOD2 is bound to Fe instead of Mn. For this, we assessed the metal composition of purified Flag-tagged WT- or NLS-SOD2 from MCF7 cells by inductively coupled plasma mass spectrometry (ICP-MS). Results shown in Fig. 6A indicate that NLS-SOD2 is depleted of Mn and enriched with Fe, compared to WT-SOD2, consistent with the idea that nuclear SOD2 functions as an Fe-dependent peroxidase. Additionally, MnCl2 supplementation of MCF7 cells expressing NLS-SOD2 inhibited H3K9me3 demethylation but had little impact on cells expressing WT-SOD2 (Fig. 6B), suggesting that histone demethylase activity is mediated by FeSOD2, but not MnSOD2. To address the hypothesis that H3 lysine demethylation is catalyzed by Fe-dependent peroxidase activity of SOD2, in vitro demethylase assays were performed by incubating histones isolated from calf thymus with purified recombinant SOD2 in the presence of hydrogen peroxide (H2O2). Recombinant SOD2 was obtained from BL21 (DE3) cells expressing Strep-tagged human SOD2 cultured in minimum media supplemented with ferric citrate or manganese chloride to enhance the formation of FeSOD2 or MnSOD2, respectively. Results in SI Appendix, Fig. S3 show that FeSOD2, but not MnSOD2, reduces H3K9me3 levels in a way that depends on H2O2 as indicated by the effect of catalase in suppressing H3 demethylation. Likewise, Flag-tagged NLS-SOD2 purified from MCF7 cells, as well as recombinant FeSOD2-K68Q that mimics the acetylated FeSOD2 form in the nucleus, also exhibited H2O2-dependent demethylase activity in vitro as measured by Western blotting while WT-SOD2 did not (Fig. 6C). Additionally, we quantified formaldehyde (HCHO) generation in these reactions. Formaldehyde is a product of lysine demethylation and was observed when either NLS-SOD2 or FeSOD2-K68Q were incubated with H2O2 and histones but not when WT-SOD2 (Fig. 6D) was used. This indicates that nuclear FeSOD2 can directly demethylate H3. Combined, results shown in Fig. 6 indicate that acetylated FeSOD2 displays H2O2-dependent demethylase activity directed toward repressive trimethyl epigenetic marks on nucleosomal H3 histones.
Fig. 6.
Iron-bound SOD2 acquires peroxidase-dependent histone demethylase activity. (A) Mn and Fe abundance in WT-SOD2 or NLS-SOD2 purified from MCF7 cells quantified by ICP-MS. (B) NLS-SOD2 expression in MCF7 cells decreases H3K9me3 compared to WT-SOD2 expression and WT phenotype is rescued by supplementation with MnCl2. (C) NLS-SOD2, but not WT-SOD2, purified from MCF7 cells, as well as recombinant SOD2-K68Q purified from bacterial culture promote H3 K9 demethylation in vitro when incubated with total histones isolated from calf thymus in the presence of 10 µM H2O2. (D) Quantification of HCOH generation from purified NLS-SOD2 and SOD2-K68Q incubated with isolated histones in the presence of 10 µM H2O2. A total of 20 µM catalase was used as control. Figures are representative of at least three independent experiments. ns, not significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; error bars indicate mean ± SD.
Nuclear SOD2 Positivity Is Increased in Metastatic Breast Cancer Showing Hormone Receptor Status Conversion.
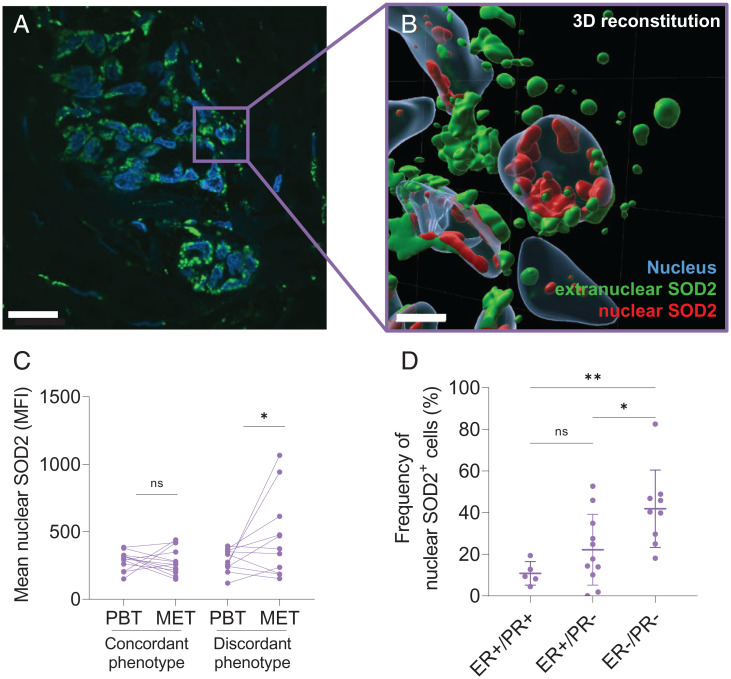
Changes in hormone receptor status, such as estrogen receptor (ER) and progesterone receptor (PR), between primary and metastatic tumors result from dynamic clonal remodeling and are frequently observed in breast cancer. Receptor conversion introduces significant complications for the clinical management of the disease and impacts therapy outcomes (25). Based on the finding that nuclear SOD2 promotes lineage plasticity acquisition, we hypothesized that there is an association between an increase in the frequency of primary tumor cells with positive nuclear SOD2 staining and the incidence of metastasis with discordant hormone receptor status. To test this idea, we analyzed biopsies of tumor metastasis and matching primary tumor samples from 25 patients representing different breast cancer molecular subtypes and their phenotypically discordant metastasis. Nuclear localization of SOD2 was examined in primary tissue and metastasis by staining SOD2 (pseudocolored green) and nucleus (DAPI, pseudocolored blue). Results in Fig. 7A show a representative fluorescence micrograph of core A7 displaying a cluster of cells with clear positive nuclear SOD2 staining. To ascertain nuclear localization clusters with overlaid nuclear (DAPI) and SOD2 (anti-SOD2, green) stains were subjected to three-dimensional (3D) digital reconstruction based on z-stack images acquired by confocal microscopy (Fig. 7B). A representative result from these reconstructions indicated the unequivocal presence of SOD2 both in the cytoplasm (pseudocolored green) and in the nucleus (pseudocolored red) of cells showing double positive nuclear staining. As hypothesized, the comparison between primary breast tumors (PBTs) with their phenotypically concordant metastasis (MET) showed no increase in the mean intensity of nuclear SOD2 stain. Meanwhile, PBTs with phenotypically discordant metastasis showed a statistically significant increase in nuclear SOD2 staining in the metastatic lesion compared to the primary tumor tissue sample (Fig. 7C). In addition, when primary tumors were stratified by molecular subtype, a significant increase in the frequency of cells with nuclear SOD2 positivity was detected in the more aggressive ER− subtypes compared to ER+. Collectively, results in Fig. 7 are consistent with the idea that nuclear SOD2 promotes clonal heterogeneity in breast cancer, contributing to the metastatic dissemination of more aggressive and less differentiated subtypes of breast cancer.
Fig. 7.
SOD2 localizes to the nucleus of tumor cells in patient-derived tumor biopsies. (A) Representative immunofluorescence staining of breast tumor biopsy (core A7) showing colocalization of SOD2 (green) and nuclear marker DAPI (blue) in 100× confocal magnification. (White scale bars, 20 µm.) (B) A 3D reconstruction of breast cancer cell nuclei (blue) showing cytoplasmic SOD2 (green) and nuclear SOD2 (red). (White scale bars, 5 µm.) (C) Nuclear SOD2 MFI in breast cancer tissue microarray indicates nuclear SOD2 signal is higher in samples with discordant hormone receptor status in PBT and corresponding MET. Lines connect PBT and MET from the same patients. (D) Increased number of nuclear SOD2 positive cells (nucSOD2+) in PBT correlates with aggressive breast cancer phenotype. ns, not significant. *P < 0.05, **P < 0.01; n = 25, error bars indicate mean ± SD.
Discussion
Recent research from our group as well as studies of others have established that the biochemical function of SOD2 transitions with tumor progression from that of a mitochondrial antioxidant at earlier stages to that of an oncoprotein facilitating the emergence of phenotypes that are considerably more aggressive (13, 14, 26, 27). The opposing context-dependent activities of SOD2 in cancer have suggested the existence of different forms of the enzyme associated with tumor suppression as well as oncogenic transformation. More recently, the acetylated form of SOD2 that is inactive as a dismutase of superoxide, but exhibits a H2O2-dependent oxidase activity (28), has emerged as the likely form of the enzyme involved in the evolution of breast cancer cells toward stem-like (13), chemoresistant (14) phenotypes. Though the peroxidase activity has been linked to an iron-bound monomeric form of the enzyme (14, 15, 28), molecular mechanisms underlying the effect of SOD2 in promoting cancer cell progression toward these more aggressive phenotypes were still lacking. After observing SOD2 nuclear localization both in cell cultures grown in suspension as well as in biopsies from breast cancer patients, we designed studies to determine the biochemical functions of nuclear SOD2. Studies shown here indicate that acetylation promotes both the accumulation of the enzyme in a monomeric form (14) as well as facilitates its localization to the nucleus. They also indicate that the nuclear form of SOD2 is heavily acetylated and predominantly bound to iron (Fe) instead of manganese (Mn). In addition, data shown in Fig. 1C indicate that SOD2 deprived of its MTS strongly localizes in the nucleus. Taken together with previously published studies, SI Appendix, Fig. S4 summarizes an emerging model whereby acetylation destabilizes the tetrameric mitochondrial form of SOD2, which possesses antioxidant activity, while promoting the accumulation of a monomer that displays Fe-dependent peroxidase activity (14) and localizes in the nucleus (35). Based on interactome as well as histone PTM proteomic studies, a biochemical function of nuclear SOD2 identified in the present study was that of a H3 lysine demethylase particularly involved in the remodeling of the epigenetic landscape. We found that this activity allows cancer cells to explore genetic programs associated with dedifferentiation, stemness, and stress resilience. Surprisingly, but consistent with the overall findings, we discovered that SOD2 associates with chromatin remodeling complexes involved in the regulation of gene transcription. However, neither SWI/SNF nor NCOA complexes have the ability to directly alter histone PTM associated with transcriptional activation or repression of critical genes involved in lineage plasticity acquisition. This suggested that acetylated FeSOD2 monomers could be recruited by the complexes to fulfill such a role. Consistently, we found that nuclear FeSOD2 removes repressive H3 methyl marks (i.e., H3K9me3 and H3K27me3) associated with promoter regions of pluripotency genes (i.e., Oct-4, SOX2, and Nanog), hence indicating a role for nuclear SOD2 in tumor progression toward metastatic disease. Nuclear-localized SOD2 was, in fact, increased in metastatic lesions showing hormone receptor variation, which is indicative of increased phenotypic plasticity. In addition, primary tumors lacking ER and PR expression that are normally less differentiated showed the highest frequency of cells with nuclear-localized SOD2. To that effect, we had already reported that SOD2 acetylation is higher in metastatic lesions compared to primary tumors, though a biochemical function for this modification beyond the activation of a peroxidase function was not evident at the time. Directing SOD2 to the nucleus caused significant changes to the cell transcriptome promoting the expression of oncogenes and EMT-associated genes, while antagonizing the expression of genes enforcing epithelial lineage commitment. This finding indicates that nuclear SOD2 may serve as a biomarker of tumors that are undergoing transformation toward more invasive phenotypes. As FeSOD2 demethylase activity depends on H2O2 instead of the canonical KDM cofactors O2 or 2-oxo-glutarate, our results also suggest that nuclear FeSOD2 may function as an alternative demethylase particularly important for tumor cells adapting to hypoxic niches where O2 is low and paradoxically steady-state H2O2 is higher than in normal tissue. Interestingly, these hypoxic niches provide ideal conditions for dedifferentiation and acquisition of stem cell–like properties by cancer cells.
In summary, results presented here indicate that nuclear SOD2 is a Fe/H2O2-dependent histone demethylase associated with the transition of epithelium-like breast cancer cells to mesenchymal, more metastatic phenotypes. The very different structural and functional features of FeSOD2 compared to mitochondrial SOD2, as well as the apparent uniqueness of its localization in the nucleus of transformed cancer cells suggest that novel diagnostic tools and therapeutic opportunities may exist.
Materials and Methods
Experimental Model and Subject Details.
Cell lines and culture.
All cell lines were maintained in minimum essential medium (MEM) (Gibco, 11095080) supplemented with 10% fetal bovine serum (FBS) (HyClone, SH30070.03) and 1% penicillin and streptomycin (Thermo Fischer, 15140122). MCF7, T47D, and BT474 parental cell lines were obtained from ATCC. All cell lines were authenticated and negative for mycoplasma contamination. When indicated, cells were cultivated in an ultralow adherence culture dish (Corning, 3262). MCF7 cell lines constitutively expressing SOD2 mutants were obtained by lentiviral transduction and selected using appropriate antibiotics. SOD2 mutants were cloned into lentiviral vector pLenti-CMV-MCS-GFP-SV-puro (29) (Addgene, 73582), by using an existing SOD2 sequence as previously described (13) with the addition of nuclear localization signal (NLS), nuclear exclusion signal (NES), or Flag-tag. HEK-293T cells were transfected using Lipofectamine 3000 (Invitrogen, L3000001) together with lentiviral packaging constructs 8.91 and VSVG. Stably transduced MCF7 cells were selected using 1 µM/mL puromycin (Gibco, A1113803).
Method Details.
Protein extraction and Western blot.
Cells lines were washed three times with PBS and lysed at 0 °C for 15 min using radio-immunoprecipitation assay (RIPA) buffer (Thermo Scientific, 89900) supplemented with protease inhibitor mixture (Roche, 04693116001). Cell lysate was centrifuged at 15,000 × g for 15 min and supernatant was removed. Protein content in the supernatant was measured using the BCA Protein Assay Kit (Thermo Scientific, 23225). Sodium dodecyl-sulfate polacrylamide gel electrophoresis (SDS-PAGE) analyses were performed in NuPage 4 to 12% Bis-Tris Protein Gels (Invitrogen, NP0335) in Novex 2-(N-morpholino)ethanesulfonic acid (MES) buffer (Invitrogen, B0002). After electrophoresis, proteins were transferred to nitrocellulose membranes in NuPage Transfer buffer (Invitrogen, NP0006) containing 20% methanol (Fisher Scientific, A4524). Membranes were blocked with Intercept Blocking Buffer (LI-COR, 92760001) and incubated with primary antibodies as described in each experimental design and in the figure legends. Then, membranes were washed three times with TBS-T (20 mM Tris HCl pH 7.4, 137 mM NaCl, 2.7 mM KCl, 0.1% Tween-20) and incubated with secondary antibodies IRDye goat anti-rabbit (LI-COR, 92632211) or IRDye goat anti-mouse (LI-COR, 92568070). Western blots were imaged in the Odyssey Fc Imaging System (LI-COR).
Nuclear fractionation.
Nuclear fractionation protocols were adapted from Suzuki et al. (30). Western blots confirming the subcellular localization of SOD2 were performed using the following primary antibodies: anti-SOD2 (Abcam, ab13533), anti-SOD2 K68Ac (Abcam, ab137037), anti-SOD2 K122Ac (Abcam, ab214675), anti-GAPDH (Cell Signaling, 5174), anti-PDH (Cell Signaling, 3205), anti-H3 (Cell Signaling, 14269). All the primary antibodies were incubated overnight at 4 °C at 1:1,000 dilution. For full details, see SI Appendix.
Immunofluorescence microscopy.
Cells were plated in black/clear-bottom 96-well plates and incubated for 16 h at 37 °C with 5% CO2 for adhesion. Subsequently, cells were fixed with 4% formaldehyde solution (Sigma-Aldrich, F8775) for 15 min at room temperature (RT) and permeabilized with ethanol for 1 min. Then, cells were washed with PBS pH 7.4 and blocked with 5% (wt/vol) bovine serum albumin (BSA) (Sigma-Aldrich, A9648) for 2 h at 4 °C and stained for 16 h at 4 °C. Cellular sublocalization of SOD2 was confirmed using primary antibodies anti-SOD2 (Abcam, ab13533; 1:100 dilution), or anti-Flag (anti-DYKDDDK Tag) (Cell Signaling, 2368; 1:100 dilution), followed by secondary antibody Alexa488 anti-IgG (Invitrogen, A21441). Nuclear and mitochondrial staining was performed with DAPI (Thermo Fischer, D1306) and MitoTracker Red CMXRos (Invitrogen, M7512) for 30 min at 4 °C. Images were collected on a BioTek LionheartFX automated microscope using the following parameters: zoom 1, laser autofocus, light-emitting diode (LED) 390 (1225009), LED 465 (1225001), LED 623 (1225005), filters DAPI (390/447, 1225115), GFP (469/525, 1225101), and CY5 (628/685, 1225105). The 16-bit raw TIFF imaging data were analyzed and quantified by ImageJ. Nuclear localization of SOD2 was calculated using the Colocalization Threshold plugin that generated two-dimensional (2D) scatterplot of pixel intensity correlation and linear regression fit.
Interactome assay.
Protein–protein interactions were identified using the biotin-based proximity labeling technique described by Rhee et al. (31). Biotin-tagged proteins were isolated from MCF7 cells expressing NES- and NLS-SOD2 fused to APEX in the presence or absence of H2O2. Protein digestion and mass spectrometry analysis were performed by the Center for Biomedical Mass Spectrometry Research at Medical College of Wisconsin (Milwaukee, WI). For full details, see SI Appendix and (34).
Coimmunoprecipitation.
Coimmunoprecipitation (co-IP) was performed using the Thermo Scientific Pierce co-IP kit (Thermo Fisher, 26149). Briefly, FOXA1 (Abcam, ab170933) and GATA3 (Proteintech, 66400-1 Ig) antibodies were immobilized for 2 h using AminoLink Plus coupling resin. The resins were washed and incubated with NLS-SOD2 or NES-SOD2 cell lysate overnight. After incubation, proteins were eluted, quantified, and analyzed by Western blotting using anti-Flag (anti-DYKDDDK Tag) (Cell Signaling, 2368).
Histone posttranslational modification analysis by mass spectrometry.
MCF7 cells were transduced with NLS-SOD2 coding sequence under the Dox-induced Tet-ON system. Tet-ON control and Tet-ON NLS-SOD2 MCF7 cells were treated with 100 ng/mL Dox for 48 h to induce the expression of NLS-SOD2 before histone isolation according to protocol described by Lin and Garcia (32). Soluble histones were quantified by BCA Protein Assay (Pierce, 23228) and 50 µg was submitted to mass spectrometry analysis in the Mass Spectrometry Core at the University of Illinois Chicago. Full details in SI Appendix.
Histone posttranslational modification analysis by Western blot.
Western blots were performed as described above. The following primary antibodies were used: anti-H3K9me3 (Cell Signaling, 13969), anti-H3K27me3 (Cell Signaling, 9733), anti-GAPDH (Cell Signaling, 5174), and anti-H3 (Cell Signaling, 14269). All the primary antibodies were incubated overnight at 4 °C and at 1:1,000 dilution.
RNA isolation and qRT-PCR.
Total RNA was isolated from ∼1 × 106 cells by using the RNeasy Mini Kit (QIAGEN, 74106) with additional DNase treatment using the RNase-Free DNase Set (QIAGEN, 1023460). cDNA was synthetized using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, 4368813) according to the manufacturer’s specifications. qPCR was performed using Fast SYBR Green (Applied Biosystems, 4385612) on a Quant Studio 6 Flex PCR system (Applied Biosystems). Gene expression in each sample was normalized to GAPDH. For primer sequences, see SI Appendix.
RNA sequencing.
RNA was isolated as described above from each sample. Sequencing libraries were generated using Illumina Novaseq platform. Sample quality, library complexity, and alignment statistics were checked using an established pipeline at the NUSeq Core from the Center for Genetic Medicine at Northwestern University.
Sequencing data analysis.
All RNA-sequencing analysis was performed with four biological replicates. The sequencing reads were aligned against the reference human genome hg19. Differentially expressed genes (DEGs) were identified using EdgeR in NLS-SOD2 compared to WT-SOD2 with a cutoff of ≥1.5-fold increase and false discovery rate (FDR) threshold of 0.01, by generalized linear model (GLM) approach. Volcano plots and heatmap were generated using normalized expression levels of differentially expressed genes in pairwise comparison using VolcaNoseR (https://huygens.science.uva.nl/VolcaNoseR/) and GraphPad Prism 9. STRING network was generated using the list of DEGs uploaded to Cytoscape 3 against reference species (Homo sapiens) with confidence cutoff of 0.8 and zero additional interactions. STRING networks were clustered using the Markov cluster (MCL) algorithm with granularity of 4. STRING functional enrichment was retrieved for network using the whole genome as background. GSEA (Hallmark genes analysis) was performed using the NLS-SOD2 × WT-SOD2 dataset.
Chromatin immunoprecipitation and ChIP-qPCR.
Approximately 5 × 106 cells were fixed with 1% methanol-free formaldehyde solution (Thermo Scientific, 28908) for 10 min at RT. Then, reaction was quenched with the addition of 125 mM glycine (Sigma-Aldrich, G8898) for 5 min. Chromatin isolation and immunoprecipitation were carried out using the Magna ChIP A/G kit (Millipore, 1710085). For full details and primer sequences, see SI Appendix.
Colony formation assay.
Monodispersed MCF7 cells (n = 5,000) were mixed with 0.6% noble agar (Sigma-Aldrich, A5431) in Minimum Essential Medium (MEM) medium containing 10% (vol/vol) FBS and placed on top of 1% noble agar and medium layer in six-well plates. Cells were cultured for 21 d at 37 °C and then stained with 1 mg/mL nitrotetrazolium blue chloride solution (Sigma-Aldrich, N6639).
Transwell invasion assay.
Matrigel matrix (BD, 354234) was mixed with Dulbeccos's Modified Eagle Medium (DMEM) (1:8) and 100 µL of the mixture was laid over the cell culture insert (Falcon, 353182) placed on top of a 12-well companion plate. The mix was left undisturbed overnight in a CO2 incubator. A total of 5,000 cells in 700 µL medium were added on top of the insert and an additional 500 µL of medium containing 200% FBS was added into the companion plate well below the insert. The cells were incubated for 16 h in a CO2 incubator. Then, the supernatant and the Matrigel matrix in the insert were removed and the invading cells on the membrane of the insert were stained with DAPI and counted using an automatic microscope (Lionheart Fx Biotek).
CD44+/CD24− analysis.
Approximately 1 × 106 cells were incubated for 15 min with allophycocyanin (APC)-conjugated mouse anti-CD44 (BD Biosciences, 560890) and phycoerythrin ( PE)-conjugated mouse anti-CD24 (BD Biosciences, 560991) according to instructions provided by the manufacturer. Cells were analyzed using a flow cytometer instrument (BD FACSymphony A5-Laser Analyzer) equipped with FACSDiva software.
SORE6+ assay.
MCF7 cells were transduced for 72 h with SORE6+ lentiviral reporter made from plasmids obtained from Lalage M. Wakefield, Laboratory of Cancer Biology and Genetics, National Cancer Institute, Bethesda, MD. The expression of associated GFP (under SORE6 promoter) was analyzed using an automatic microscope (Lionheart Fx Biotek).
Stemness and EMT marker levels.
Western blots were performed as described above. The following primary antibodies were used: anti-Nanog (Cell Signaling, 8022), anti–E-cad (Cell Signaling, 14472), anti-vimentin (Cell Signaling, 5741), anti-Zeb1 (Cell Signaling, 70512), anti-Slug (Cell Signaling, 9585), and anti-GAPDH (Cell Signaling, 5174). All the primary antibodies were incubated overnight at 4 °C and with 1:1,000 dilution.
In vivo metastasis studies.
All mouse experimentation was conducted in accordance with standard operating procedures approved by the Institutional Animal Care and Use Committee at Northwestern University. NSG mice were acquired from The Jackson Laboratory (005557). For tail-vein injection, 2 million MCF7 cells expressing WT- or NLS-SOD2 transduced with lentiviral vector expressing luciferase were resuspended in 200 μL PBS and injected into female NSG mice at 6 to 8 wk of age. Imaging of grafted tumors was performed using an IVIS in vivo imager (PerkinElmer) after i.p. injection of RediJect D-Luciferin bioluminescent substrate (PerkinElmer, 770504) (150 mg/kg body weight) within 2 h and 5 mo after cell injection. The signal was measured as radiance (photons/second/square centimeter/steradian) and analyzed with Living Image software.
Recombinant human SOD2 purification from BL21 (DE3).
Escherichia coli BL21 (DE3) cells were transfected with pET21 vector cloned with human SOD2-encoding sequence fused to Strep-tag as described in Ganini et al. (33). For full details, see SI Appendix.
Histone demethylase assay.
Bulk histones isolated from calf thymus (Sigma-Aldrich, 10223565001) were used to test demethylase activity of purified SOD2 variants. For this, 20 ng/µL bulk histones were incubated with 4 ng/µL SOD2 in TBS (50 mM Tris HCl pH 7.5, 150 mM NaCl) with or without addition of 10 µM H2O2 (Fisher Chemical, H325500) or 20 µM catalase (Sigma-Aldrich, C1345). The reaction was incubated at 37 °C for 2 h and histone lysine methylation status was assessed by Western blot.
Formaldehyde quantification.
A PicoProbe Formaldehyde Assay Kit (BioVision, K805) was used to quantify formaldehyde (HCHO) produced by demethylase activity of SOD2. For this, 20 ng/µL of purified WT-SOD2, NLS-SOD2, or SOD2-K68Q were incubated with 500 ng/µL bulk histones isolated from calf thymus (Sigma-Aldrich, 10223565001) with or without addition of 10 µM H2O2 or 20 µM catalase in HCHO assay buffer containing 1× enzyme mix provided by the kit manufacturer. Reaction was incubated for 4 h at RT and developed with the addition of 1× HCHO developer and 1× PicoProbe for 20 min at RT. Fluorescence (Ex: 535 nm, Em: 587 nm) was measured by Spectra Max M5 (Molecular Devices) and subtracted from sample background control. HCHO concentration was calculated relative to a standard curve generated with HCHO standard provided by manufacturer.
Metal quantification by ICP-MS.
For metal quantification, WT-SOD2 and NLS-SOD2 mutants were purified using buffers pretreated with Chellex 100 (Sigma-Aldrich, C7901). Total iron and manganese were quantified by ICP-MS by the Quantitative Bioelement Imaging Center (Northwestern University). For full details, see SI Appendix.
Nuclear localization of SOD2 in tumor microarray.
Breast carcinoma with matched metastatic carcinoma tissue microarray (TMA) was purchased from US Biomax, Inc (BRM961a). Samples were stained for immunofluorescence microscopy according to standard procedures using anti-SOD2 (Abcam, ab13533) and nucleus (DAPI) by the Mouse Histology and Phenotyping Laboratory (Northwestern University). TMA cores corresponding to patient-matched PBT and MET were imaged in a Nikon W1 Dual Cam Spinning Disk Confocal automated microscope using 10× and 100× objectives, 50-µm pinhole, and GFP and DAPI channels at the Center for Advanced Microscopy (Northwestern University). The 16-bit images generated at 10× magnification were analyzed using FIJI ImageJ (1.53c). Nuclei were detected in DAPI channel from 10× images using StarDist2D plugin in a versatile model (fluorescent nuclei), normalized image option, 0.5% score threshold, 0.15 overlap threshold, and automatic region of interest (ROI) position. ROI generated with area between 20 and 80 square pixels were overlapped to GFP channel and the mean fluorescent intensity (MFI) correspondent to nuclear SOD2 was measured in each cell identified. Cells were considered nuclear SOD2-positive (SOD2+) if MFI was higher than the value of the 75th percentile of all nuclei detected in the entire TMA. Frequency of nuclear SOD2+ cells was calculated in each PBT core as the number of SOD2+ divided by total number or nuclei identified in the same core. The 16-bit z-stack images obtained with 100× magnification were processed by IMARIS Software 9.7 (Oxford Instruments) for 3D image reconstructions.
Quantification and Statistical Analysis.
Statistical analyses were carried out as described in the relevant method sections above. Where applicable, statistical significance is reported in the figures and corresponding legends.
Supplementary Material
Acknowledgments
We acknowledge the technical assistance of David Kirchebeuchler from Center of Advanced Microscopy. Recombinant SOD2-K68Q was expressed and purified by Dr. Hyun Lee and Dr. Robel D. Demissie (Biophysics Core, University of Illinois Chicago). Elemental analysis was performed at the Northwestern University Quantitative Bio-Element Imaging Center generously supported by NASA Ames Research Center Grant NNA04CC36G. We are grateful for funding from the NIH, National Institute of Allergy and Infectious Diseases (NIAID) R01AI131267 (to M.G.B.); National Institute of Environmental Health Sciences (NIEHS) R01028149 (to M.G.B.); National Cancer Institute (NCI) R01CA216882 (to M.G.B.); the Liz and Eric Lefkofsky Foundation Innovation Award (to M.G.B.); and the Department of Defense/Army Research Office (DOD/ARO) Grant 72983 (to M.G.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110348119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Marjanovic N. D., et al. , Emergence of a high-plasticity cell state during lung cancer evolution. Cancer Cell 38, 229–246.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lourenco A. R., et al. , Differential contributions of pre- and post-EMT tumor cells in breast cancer metastasis. Cancer Res. 80, 163–169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedmann-Morvinski D., Verma I. M., Dedifferentiation and reprogramming: Origins of cancer stem cells. EMBO Rep. 15, 244–253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dravis C., et al. , Epigenetic and transcriptomic profiling of mammary gland development and tumor models disclose regulators of cell state plasticity. Cancer Cell 34, 466–482.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gkountela S., et al. , Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell 176, 98–112.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Julienne H., Audit B., Arneodo A., Embryonic stem cell specific “master” replication origins at the heart of the loss of pluripotency. PLOS Comput. Biol. 11, e1003969 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loh Y. H., Zhang W., Chen X., George J., Ng H. H., Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 21, 2545–2557 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu H., et al. , Chemotherapy-induced S100A10 recruits KDM6A to facilitate OCT4-mediated breast cancer stemness. J. Clin. Invest. 130, 4607–4623 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glaser S. F., et al. , The histone demethylase JMJD2B regulates endothelial-to-mesenchymal transition. Proc. Natl. Acad. Sci. U.S.A. 117, 4180–4187 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klose R. J., Kallin E. M., Zhang Y., JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7, 715–727 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Keith B., Simon M. C., Hypoxia-inducible factors, stem cells, and cancer. Cell 129, 465–472 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhar S. K., Tangpong J., Chaiswing L., Oberley T. D., St Clair D. K., Manganese superoxide dismutase is a p53-regulated gene that switches cancers between early and advanced stages. Cancer Res. 71, 6684–6695 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He C., et al. , SOD2 acetylation on lysine 68 promotes stem cell reprogramming in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 116, 23534–23541 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y., et al. , Lysine 68 acetylation directs MnSOD as a tetrameric detoxification complex versus a monomeric tumor promoter. Nat. Commun. 10, 2399 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganini D., Santos J. H., Bonini M. G., Mason R. P., Switch of mitochondrial superoxide dismutase into a prooxidant peroxidase in manganese-deficient cells and mice. Cell Chem. Biol. 25, 413–425.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pazin M. J., Kadonaga J. T., SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell 88, 737–740 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Truong T. H., et al. , Cancer stem cell phenotypes in ER+ breast cancer models are promoted by PELP1/AIB1 complexes. Mol. Cancer Res. 16, 707–719 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michel B. C., et al. , A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat. Cell Biol. 20, 1410–1420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marino M. M., et al. , Interactome mapping defines BRG1, a component of the SWI/SNF chromatin remodeling complex, as a new partner of the transcriptional regulator CTCF. J. Biol. Chem. 294, 861–873 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arruabarrena-Aristorena A., et al. , FOXA1 mutations reveal distinct chromatin profiles and influence therapeutic response in breast cancer. Cancer Cell 38, 534–550.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takaku M., Grimm S. A., De Kumar B., Bennett B. D., Wade P. A., Cancer-specific mutation of GATA3 disrupts the transcriptional regulatory network governed by Estrogen Receptor alpha, FOXA1 and GATA3. Nucleic Acids Res. 48, 4756–4768 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi K., Yamanaka S., Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Al-Hajj M., Wicha M. S., Benito-Hernandez A., Morrison S. J., Clarke M. F., Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 100, 3983–3988 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang B., et al. , A flexible reporter system for direct observation and isolation of cancer stem cells. Stem Cell Reports 4, 155–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao W., et al. , Receptor conversion impacts outcomes of different molecular subtypes of primary breast cancer. Ther. Adv. Med. Oncol. 13, 17588359211012982 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hemachandra L. P., et al. , Mitochondrial superoxide dismutase has a protumorigenic role in ovarian clear cell carcinoma. Cancer Res. 75, 4973–4984 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hart P. C., et al. , MnSOD upregulation sustains the Warburg effect via mitochondrial ROS and AMPK-dependent signalling in cancer. Nat. Commun. 6, 6053 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ansenberger-Fricano K., et al. , The peroxidase activity of mitochondrial superoxide dismutase. Free Radic. Biol. Med. 54, 116–124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Witwicka H., et al. , Studies of OC-STAMP in osteoclast fusion: A new knockout mouse model, rescue of cell fusion, and transmembrane topology. PLoS One 10, e0128275 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki K., Bose P., Leong-Quong R. Y., Fujita D. J., Riabowol K., REAP: A two minute cell fractionation method. BMC Res. Notes 3, 294 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhee H. W., et al. , Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin S., Garcia B. A., Examining histone posttranslational modification patterns by high-resolution mass spectrometry. Methods Enzymol. 512, 3–28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganini D., Petrovich R. M., Edwards L. L., Mason R. P., Iron incorporation into MnSOD A (bacterial Mn-dependent superoxide dismutase) leads to the formation of a peroxidase/catalase implicated in oxidative damage to bacteria. Biochim. Biophys. Acta 1850, 1795–1805 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marley J., Lu M., Bracken C., A method for efficient isotopic labeling of recombinant proteins. J. Biomol. NMR 20, 71–75 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Zemanovic S., et al. , Dynamic phosphorylation of the C terminus of Hsp70 regulates the mitochondrial import of SOD2 and redox balance. Cell Rep. 25, 2605–2616.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.