Significance
The G protein–coupled bile acid receptor (GPBAR) is the native bile acid receptor and therapeutic target for treating metabolic diseases. However, specific bile acids are suspected to promote cancer progression. We showed that different Gs/arrestin-biased signaling of GPBAR played key roles in non–small cell lung cancer (NSCLC) cell viability. Specifically, β-arrestin–biased GPBAR agonist R399 stimulated cell growth through promoting YAP (Yes-associate Protein) activation. Conversely, GPBAR activation triggered by deoxycholic acid and INT-777, which showed a preference to Gs signaling, inhibited cancer cell progression. Moreover, we delineated key structural determinants of β-arrestin bias and protumor activity of GPBAR and identified that phosphorylation of GPBAR by GRK2/GRK5 contributed to β-arrestin–biased signaling. These results may facilitate rational design of GPBAR-targeting drugs with anticancer benefits.
Keywords: GPBAR, NSCLC, YAP, INT-777, R399
Abstract
The G protein–coupled bile acid receptor (GPBAR) is the membrane receptor for bile acids and a driving force of the liver–bile acid–microbiota–organ axis to regulate metabolism and other pathophysiological processes. Although GPBAR is an important therapeutic target for a spectrum of metabolic and neurodegenerative diseases, its activation has also been found to be linked to carcinogenesis, leading to potential side effects. Here, via functional screening, we found that two specific GPBAR agonists, R399 and INT-777, demonstrated strikingly different regulatory effects on the growth and apoptosis of non–small cell lung cancer (NSCLC) cells both in vitro and in vivo. Further mechanistic investigation showed that R399-induced GPBAR activation displayed an obvious bias for β-arrestin 1 signaling, thus promoting YAP signaling activation to stimulate cell proliferation. Conversely, INT-777 preferentially activated GPBAR-Gs signaling, thus inactivating YAP to inhibit cell proliferation and induce apoptosis. Phosphorylation of GPBAR by GRK2 at S310/S321/S323/S324 sites contributed to R399-induced GPBAR–β-arrestin 1 association. The cryoelectron microscopy (cryo-EM) structure of the R399-bound GPBAR-Gs complex enabled us to identify key interaction residues and pivotal conformational changes in GPBAR responsible for the arrestin signaling bias and cancer cell proliferation. In summary, we demonstrate that different agonists can regulate distinct functions of cell growth and apoptosis through biased GPBAR signaling and control of YAP activity in a NSCLC cell model. The delineated mechanism and structural basis may facilitate the rational design of GPBAR-targeting drugs with both metabolic and anticancer benefits.
Bile acids are ampholytic metabolic products generated by hepatocytes and secreted into the intestine, where they are diversified through further modification by the microbiota (1). The main bile acids in humans include cholic acid (CA), chenodeoxycholic acid (CDCA), deoxycholic acid (DCA), lithocholic acid (LCA), and their corresponding conjugated forms. Owing to their amphipathic properties, bile acids play key roles in facilitating the digestion and absorption of dietary lipids and fat-soluble vitamins. In addition to their functions as powerful emulsifiers, accumulating evidence has revealed that bile acids, as important signaling molecules in the liver–bile acid–microbiota axis, exert profound and diverse regulatory effects on multiple pathophysiological processes, from the maintenance of glucose, lipid, and energy homeostasis to involvement in inflammation and cancer (2). Consistent with their diverse signaling roles, numerous natural bile acids and bile acid derivatives have been found to have therapeutic effects on primary sclerosing cholangitis, nonalcoholic steatohepatitis, type 2 diabetes, and neurodegenerative diseases such as amyotrophic lateral sclerosis (2–5). However, certain bile acids or bile acid derivatives have also been reported to promote cancer progression. Therefore, understanding the exact mechanisms of bile acid–induced cellular signaling will provide important guidance for therapeutic utilization and optimization of this group of signaling molecules.
In cells, bile acids perform their functions mostly through two receptors, the nuclear farnesoid X receptor (FXR) and the seven-transmembrane receptor G protein–coupled bile acid receptor (GPBAR), also called TGR5 or GPR131 (2). Whereas FXR mediates the transcriptional function of many bile acids, the membrane receptor GPBAR is responsible for both quick reactions and long-term genomic changes downstream of G proteins or arrestins (2). Bile acids that are antagonists of FXR and agonists of GPBAR have shown great potential to treat a spectrum of metabolic diseases. Many of these bile acids can induce the coupling of GPBAR to Gs, which contributes to their beneficial effects, including prevention of type 2 diabetes and obesity and amelioration of inflammation, thereby providing possibilities for drug discovery for metabolic diseases and related disorders (2). Recent studies have shown that in addition to Gs coupling, bile acids can also initiate β-arrestin activity through GPBAR activation to regulate the antiviral immune response (6), highlighting the complexity of bile acid–GPBAR signaling. However, little is currently known about the implications of GPBAR–β-arrestin coupling in many other pathophysiological processes.
Emerging evidence has suggested that GPBAR is implicated in cancer development (7, 8). For example, 23(S)-mCDCA was shown to suppress the proliferation and migration of human kidney cancer cells through activation of GPBAR (9). In contrast, CDCA-induced GPBAR activation was reported to stimulate the proliferation of endometrial cancer cells (10). These data indicate that GPBAR activation triggered by different agonists may occur through distinct signaling mechanisms, for example, Gs versus Arrestin signaling bias. Determining how different signaling pathways downstream of GPBAR contribute to carcinogenesis is highly valuable for eliminating the unwanted side effects of drugs targeting GPBAR in the treatment of metabolic syndromes.
Recently, we solved the cryoelectron microscopy (cryo-EM) structures of GPBAR-Gs in complex with the bile acid derivative INT-777 and the synthetic compound P395, not only providing a fingerprint for the recognition of different bile acids by GPBAR but also offering a preliminary understanding of the bias of GPBAR in response to the binding of different ligands (11). However, the mechanism by which these different GPBAR conformational states induced by the binding of diverse GPBAR agonists associated with distinct cellular outcomes remains elusive. In the present study, we explored the biological consequences, detailed signaling cascades, and structural basis of GPBAR activation in response to specific agonists of GPBAR in a non–small cell lung cancer (NSCLC) model. Our present findings may facilitate the rational design of GPBAR-targeting drugs with both metabolic and anticancer benefits.
Results
Differential Contributions of GPBAR Activation by a Panel of Bile Acids or Synthetic Agonists to NSCLC Cell Growth and Apoptosis.
To investigate the biological roles of different agonists to GPBAR activation, we examined the expression level of GPBAR in different NSCLC cell lines with different genetic background and histological types by Western blot (SI Appendix, Table S1). Cell lines H1299 and H1975, which have a relatively high GPBAR expression level, and cell lines A549 and PC9, which have a relatively low GPBAR expression level, were selected for subsequent experiments (SI Appendix, Fig. S1 A and B). We then examined the effects of eight native bile acids, one semisynthetic bile acid INT-777, and two synthetic chemicals R399 and P395 on cell growth using an MTT (3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide) assay in NSCLC cell line H1299, which has high endogenous GPBAR expression (12). Interestingly, different agonists triggered different growth responses in H1299 cells at least partially through GPBAR activation (Fig. 1A). More specifically, several tested agonists (e.g., CA, LCA, DCA, ursodeoxycholic acid, and INT-777) inhibited H1299 cell proliferation, whereas others (e.g., CDCA, taurocholic acid, taurodeoxycholic acid, glycocholic acid, P395, and R399) increased H1299 cell proliferation in a dose-dependent manner (Fig. 1A, Upper). To elucidate the roles of GPBAR in these ligand-induced responses, two specific small interfering RNAs (siRNAs) targeting GPBAR were exploited to knock down GPBAR expression in H1299 cells. The RNA interference (RNAi)–induced reduction in GPBAR expression level was verified by Western blot analysis and biotinylation of cell surface proteins (SI Appendix, Fig. S1 C–E). Then siGPBAR-2, which showed a higher knockdown efficiency than siGPBAR-1, was introduced into H1299 cells in subsequent experiments. Notably, GPBAR silencing almost completely abolished the promotive effects of P395 and R399 and the suppressive effects of INT-777 and partially blocked the regulatory effects of the other agonists on cell growth (Fig. 1A, Lower). We therefore selected INT-777 and R399 (two selective and potent GPBAR agonists that showed strong differential regulatory roles in H1299 cell growth via binding to GPBAR) and DCA (a native GPBAR agonist that showed partial effects on cell growth by interacting with GPBAR) for further exploration. We next examined the effects of DCA, INT-777, and R399 on cell growth using an MTT assay and flow cytometric analysis. Consistent with the cell growth screening results, the results of the MTT assay showed that whereas both DCA and INT-777 inhibited H1299 cell proliferation, R399 increased H1299 cell proliferation in a time-dependent manner (SI Appendix, Fig. S1F). Moreover, the effects of DCA, INT-777, and R399 on cell proliferation were explored in two additional NSCLC cell lines, A549 and PC9, which showed relatively low endogenous GPBAR expression levels. Consistently, DCA and INT-777 exerted antiproliferative effects while R399 had proproliferative effects on A549 and PC9 cells, but at a higher concentration than they did in H1299 and H1975 cells (SI Appendix, Fig. S2 A–D). We next examined the effects of R399, INT-777, and DCA on apoptosis. Flow cytometric analysis with fluorescein isothiocyanate–conjugated Annexin V and propidium iodide staining showed that H1299 cells treated with INT-777 and DCA, but not H1299 cells treated with R399, exhibited a higher apoptosis rate than those treated with vehicle (Fig. 1B and SI Appendix, Fig. S1G). The effects of DCA, INT-777, and R399 on apoptosis were further confirmed by a TUNEL (TdT-mediated dUTP nick end labeling) assay (Fig. 1C and SI Appendix, Fig. S1H). In parallel, Western blot analysis showed that DCA and INT-777 treatment led to significant increases in the levels of the apoptosis markers, Cleaved PARP1 and Cleaved Caspase-3, while R399 treatment had little effect on the levels of these proteins compared with those in the vehicle group (Fig. 1D and SI Appendix, Fig. S1I). These data suggested that R399 and INT-777 played distinct roles in regulating apoptosis of H1299 cells. Such differential roles of R399 and INT-777 in regulating cell proliferation and apoptosis were further verified in another NSCLC cell line (H1975) with high GPBAR expression (SI Appendix, Fig. S3 A–F).
Fig. 1.
R399 enhances cell growth while INT-777 inhibits cell growth and promotes apoptosis GPBAR dependently in NSCLC in vitro. (A) H1299 cells transfected with scramble siRNA (siCTRL) and siGPBAR-2 were treated with different concentrations of the indicated GPBAR agonists or dimethyl sulfoxide (DMSO) for 72 h (n = 6). Cell viability was assessed using the MTT assay. (B–D) H1299 cells were treated with DMSO, DCA (80 μM), INT-777 (2 μM), and R399 (700 nM) for 24 h, respectively (n = 3). Apoptotic cells were detected by flow cytometry analysis (B) and TUNEL staining (C). Cleaved-PARP1 (poly(ADP-ribose) polymerase 1) and Cleaved Caspase-3 were detected by Western blot (D). All data are presented as the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 based on the Student’s t-test. CTRL, control; siCTRL, small interfering control; FITC, fluorescein isothiocyanate–conjugated; GCA, glycocholic acid; ns, not significant; PI, propidium iodide; TCA, taurocholic acid; TDCA, taurodeoxycholic acid.
In addition to activate membrane receptor GPBAR, the bile acids were regulators of nuclear receptor FXR. We therefore knocked down GPBAR or pharmaceutically inhibited FXR using an FXR-specific inhibitor (T-βMCA) in H1299 and H1975 cells (13–15), both of which have relatively high endogenous GPBAR and FXR expression, and detected the alterations of cell proliferation and apoptosis in response to stimulations of DCA, INT-777, and R399. The results showed that both GPBAR silencing and FXR inhibition attenuated DCA-induced suppression of cell proliferation and apoptosis. In contrast, only GPBAR silencing, but not FXR inhibition, abrogated the growth-inhibitory and apoptosis-inducing effect of INT-777 and the growth-promotive effect of R399 (SI Appendix, Fig. S3 G–J). These data indicated the importance of GPBAR in mediating the effects of INT-777 and R399 as well as the possible synergistic effects of GPBAR and FXR in mediating DCA function. Our results are consistent with previous studies indicating that INT-777 is a GPBAR-specific agonist without FXR activity (16) and little structural similarity lies between the synthetic GPBAR agonist R399 and the bile acids.
Taken together, these results indicated that R399 and INT-777 differentially regulated the proliferation and apoptosis of NSCLC cells via their actions on GPBAR.
R399 Enhances But INT-777 Inhibits NSCLC Cell Tumorigenesis In Vivo through Activation of GPBAR.
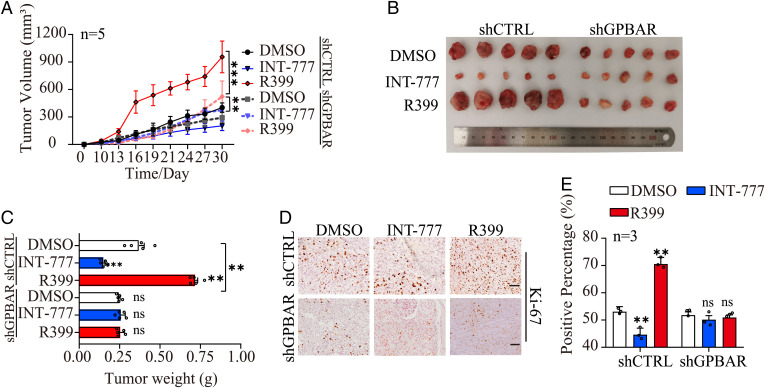
We next investigated whether R399 and INT-777 can differentially regulate the tumorigenic capacity of NSCLC cells in vivo. Two specific short hairpin RNAs (shRNAs) targeting GPBAR and one control shRNA were introduced into H1299 cells by lentivirus infection (SI Appendix, Fig. S4A). H1299-shGPBAR cells and the corresponding control cells were subcutaneously inoculated unilaterally into the axilla of athymic mice, which were then treated with R399, INT-777, or vehicle. Consistent with the in vitro data, R399 administration accelerated but INT-777 administration inhibited the formation and growth of tumors originating from the control cells at the implantation site compared with the tumors in vehicle-treated mice. However, these effects of R399 and INT-777 on tumorigenesis were abrogated in mice inoculated with H1299-shGPBAR cells (Fig. 2A and SI Appendix, Fig. S4B). This finding was further supported by the measured volumes and weights of tumors originating from the above-mentioned cells with the indicated treatments at the end of the experiment (Fig. 2 A–C). However, the weights of the mice in each group did not differ significantly (SI Appendix, Fig. S4C). Moreover, we performed immunohistochemical (IHC) staining to determine the expression levels of the proliferation marker Ki67 and the apoptosis marker Cleaved Caspase-3 in the xenografts, and we performed a TUNEL assay on the xenografts. Strikingly, elevated Ki67-positive staining was observed in H1299 cells treated with R399 and decreased Ki67-positive staining was seen in H1299 cells treated with INT-777, but knockdown of GPBAR in H1299-shGPBAR cells abolished these effects (Fig. 2 D and E). Similarly, the numbers of TUNEL-positive and Cleaved Caspase-3–positive cells in the INT-777 treatment group were significantly increased compared with those in the vehicle treatment group (SI Appendix, Fig. S4 D–G). In contrast, the numbers of TUNEL-positive and Cleaved Caspase-3–positive cells in mice inoculated with H1299-shGPBAR cells and treated with INT-777 were similar to those in mice inoculated with H1299-shGPBAR cells and treated with vehicle (SI Appendix, Fig. S4 D–G). These results indicated that INT-777–induced apoptosis is dependent on GPBAR. Collectively, these results confirmed that R399 and INT-777 triggered completely opposite (promotive and suppressive, respectively) effects on tumorigenesis in vivo through GPBAR.
Fig. 2.
R399 promotes but INT-777 inhibits tumorigenesis in vivo in a GPBAR-dependent manner. (A) H1299-shGPBAR and its control cells were subcutaneously injected into BALB/c-nu/nu mice (n = 5) with treatments described in Materials and Methods. The volume of the subcutaneous tumors was measured every 3 d from the 10th day following implantation. (B) Representative images of the dissected tumors are shown. A ruler is used to demonstrate the size of the tumor (the length of the ruler is 20 cm and the minimum scale is 1 mm). (C) Quantifications of tumor weights with different treatments were calculated. (D and E) Cell proliferation was detected by Ki-67 staining of the tumor sections. Quantitative analyses of the positive cells are shown in the graphs. Data are presented as the mean ± SEM. **P < 0.01 and ***P < 0.001 based on the Student’s t-test. CTRL, control; DMSO, dimethyl sulfoxide; ns, not significant.
The roles of GPBAR in different cancer types are not clearly defined (8–10, 17–19). We then investigated the contributions of the endogenous activity of GPBAR to cell viability of NSCLC cells both in vitro and in the mouse model. We silenced GPBAR expression in H1299 and H1975 cells using GPBAR-specific siRNAs and overexpressed GPBAR in A549 and PC9 cells (SI Appendix, Fig. S5 A–C). MTT assay analysis showed that GPBAR silencing inhibited cell proliferation of H1299 and H1975 (SI Appendix, Fig. S5A), whereas overexpression of GPBAR markedly promoted cell growth of A549 and PC9 cells compared to their parental control cells (SI Appendix, Fig. S5 B and C). Consistent with these in vitro data, in the xenograft tumor model in nude mice, the growth rates and weights of excised tumors were significantly reduced in the GPBAR-knockdown group compared with the control group (Fig. 2 A and C).
Moreover, we explored the clinical relevance of GPBAR expression in NSCLC. We detected the expression of GPBAR in 28 NSCLC tissues paired with adjacent noncancerous tissues (ANTs) by IHC. IHC analysis showed that compared to ANT tissues (average IHC score, 3.6), GPBAR were highly expressed in NSCLC tissues (25 of 28 [89.28%]; average IHC score, 8.08) and the expression of GPBAR was increased with the development of NSCLC (SI Appendix, Fig. S6 A and B). In addition, the expression of Ki67 was remarkably higher in NSCLC than in ANT tissues and was significantly associated with tumor grade (SI Appendix, Fig. S6C). Further correlation analysis revealed that the expression of GPBAR was positively correlated with Ki67, indicating the possible promotive roles of GPBAR in NSCLC cell proliferation (SI Appendix, Fig. S6D). Given that many other components such as endothelial and stromal cell populations presented in ANT tissues, we evaluated the expression of cytokeratin 7 (CK7), thyroid transcription factor-1 (TTF-1), and Napsin A, three representative lung epithelium markers, to inspect the presence of epithelial cells in ANT tissues by IHC staining. The results showed that the adjacent tissues exhibited relatively extensive and high expression of CK7, TTF-1, and Napsin A, indicating the presence of epithelial cells in ANT tissues (SI Appendix, Fig. S6 E and F). Although our present data revealed that the expression of GPBAR in ANT tissues was very low and almost undetectable in epithelial cells, the global expression profiling of GPBAR deserves an in-depth investigation using single-cell–based analysis in the future.
In addition, our observations were consistent with previous studies showing that the GPBAR expression level was positively correlated with an advanced clinical stage of NSCLC (12), suggesting that GPBAR may have a tumor-promotive function in NSCLC. Analyses of the size and composition of the bile acid pool orchestrating with GPBAR expression by increasing the clinical sample size and using GPBAR gene-modified NSCLC organoids stimulated with differential bile acids are required to define the exact roles of the contribution of GPBAR and its endogenous ligands in NSCLC.
Transcriptome Profiling Identified YAP Signaling to Be Activated by R399 But Inhibited by INT-777.
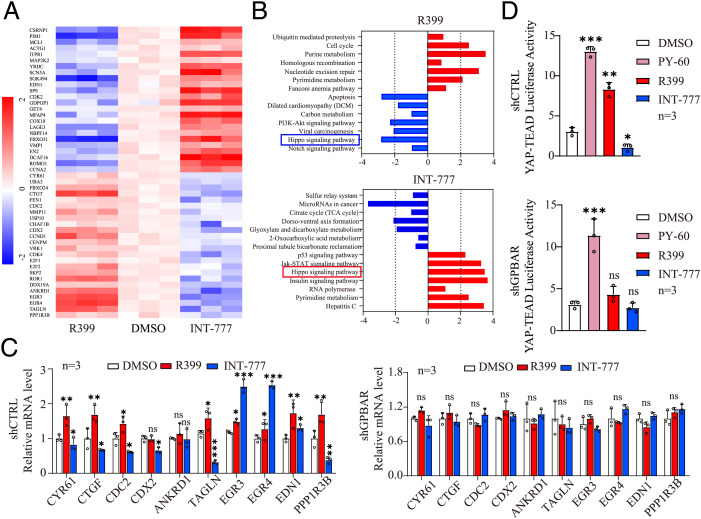
To further investigate the downstream intracellular signaling modulated by R399 and INT-777, we performed transcriptome microarray analysis of H1299 cells exposed to R399, INT-777, and vehicle. Hierarchical clustering of transcriptome data was performed, identifying 274 up-regulated genes and 295 down-regulated genes in INT-777–treated cells compared to vehicle-treated cells as well as 707 up-regulated genes and 875 down-regulated genes in R399-treated cells compared to vehicle-treated cells (P < 0.05, |log2 fold change| > 1) (Fig. 3A). Pathway enrichment analysis showed that the differentially expressed genes were most strongly enriched in six signaling pathways (cell cycle, purine metabolism, nucleotide excision repair, Hippo signaling, apoptosis, and PI3K (Phosphatidylinositol-3-kinase-Protein Kinase B)-AKT signaling) after R399 treatment and in seven pathways (microRNAs in cancer, P53 signaling, Jak-STAT signaling, Hippo signaling, insulin signaling, pyrimidine metabolism, and hepatitis C) in response to INT-777 treatment (Fig. 3B). Among the pathways differentially regulated by R399 and INT-777, Hippo signaling attracted our attention, considering the importance of this signaling pathway in controlling cell growth and apoptosis. Notably, Hippo signaling was down-regulated in H1299 cells after R399 treatment but up-regulated after INT-777 treatment (Fig. 3B). YAP is a key downstream effector of the Hippo signaling cascade and functions as a transcriptional coactivator to bind mainly with TEA domain (TEAD) transcription factors to enhance transcription (20–24). In the context of the “inactive” Hippo pathway, decreased phosphorylation of YAP at serine 127 promotes its nuclear translocation, thereby enhancing the transcription of its target genes (25). We then verified the effect of R399 and INT-777 on the expression of a series of YAP-TEAD target genes, including connective tissue growth factor (CTGF) and cysteine rich angiogenic inducer 61 (CYR61), two representative YAP-TEAD target genes that usually serve as a readout of YAP activity (26). Consistent with the transcriptome data, 7 of the 10 selected YAP-TEAD target genes, including CYR61 and CTGF, showed increased expression after treatment with R399 and decreased expression after treatment with INT-777 (Fig. 3C, Left). Strikingly, GPBAR knockdown abolished the alterations in the expression of the above-mentioned genes induced by R399 or INT-777 (Fig. 3C, Right). We then performed a TEAD luciferase reporter assay to examine the effects of R399 and INT-777 on YAP signaling, using the known specific YAP activator, PY-60, as a reference. Notably, PY-60 and R399 induced ∼ 13- and 8 fold of the YAP transcriptional activity increase, but INT-777 inhibited YAP transcriptional activity by approximately twofold compared to control vehicle in H1299-shCTRL cells (Fig. 3 D, Upper). GPBAR knockdown abolished the alterations in YAP transcriptional activity induced by R399 or INT-777 but had no significant effects on YAP activation in response to PY-60 stimulation, suggesting essential roles of GPBAR in the modulation of YAP-TEAD transcriptional activity by R399 or INT-777 (Fig. 3 D, Lower). In addition, we investigated the influence of R399 and INT-777 on the phosphorylation of YAP on serine 127 (pYAP-S127) and the subcellular distribution of YAP using Western blot analysis and an immunofluorescence (IF) assay. R399 inhibited but INT-777 promoted phosphorylation of YAP at serine 127 in a time- and GPBAR-dependent manner (SI Appendix, Fig. S7 A–D). In parallel, R399 treatment led to significant nuclear accumulation of YAP, while INT-777 treatment resulted in cytosolic accumulation of YAP, implying the different roles of R399 and INT-777 in regulating YAP phosphorylation and nuclear shuttling (SI Appendix, Fig. S7 E–I).
Fig. 3.
Transcriptome analysis reveals that R399 activates but INT-777 inhibits Hippo-YAP signaling in H1299 cells. (A) Heat map of differentially expressed genes (DEGs) in R399-treated and INT-777–treated H1299 cells compared to vehicle-treated cells, respectively. Red indicates up-regulated genes (log2 fold change > 1 and P < 0.05). Blue indicates down-regulated genes (log2 fold change < −1 and P < 0.05). (B) Pathway analysis of DEGs in R399-treated and INT-777–treated H1299 cells. (C) Expression analysis (qRT-PCR) of the indicated YAP target genes in H1299-shGPBAR cells and its control cells stimulated with R399 (700 nM) or INT-777 (2 μM) for 24 h (n = 3). (D) Effects of PY-60 (10 μM), R399 (700 nM), or INT-777 (2 μM) on YAP-TEAD transcriptional activity were determined by the TEAD luciferase reporter assay in H1299-shGPBAR cells and control cells for 24 h (n = 3). Data are presented as the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 based on the Student’s t-test. CTRL, control; DMSO, dimethyl sulfoxide; mRNA, messenger RNA; ns, not significant; TCA, taurocholic acid.
YAP Signaling Is a Key Switch Controlling Different Cellular Functions Regulated by R399 and INT-777.
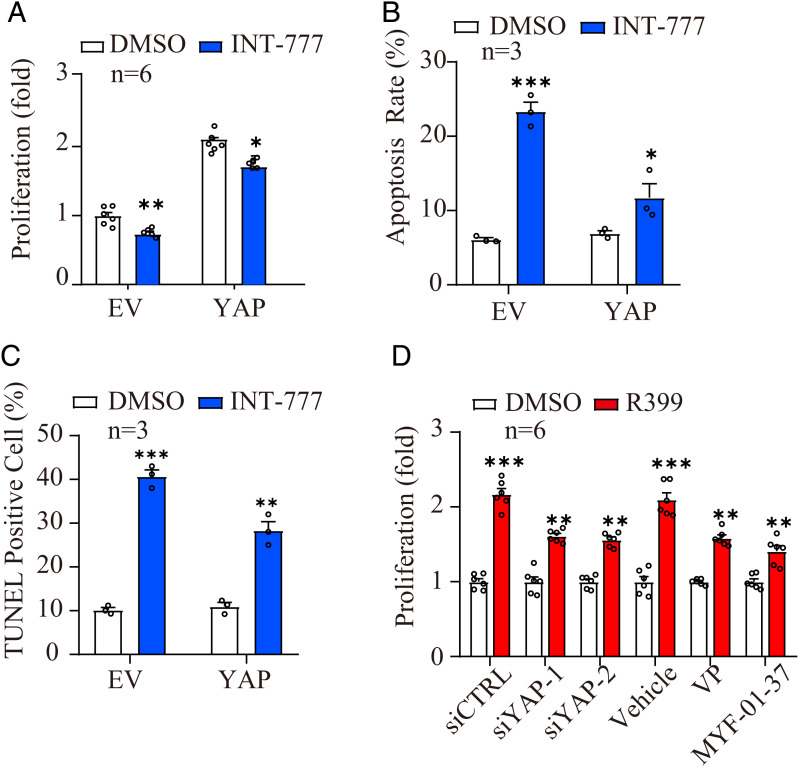
To further investigate contributions of YAP signaling to the cellular responses elicited by R399 and INT-777, we overexpressed the YAP in H1299 cells and knocked down YAP expression or blocked YAP activity by siRNA or two widely used YAP-TEAD inhibitors, Verteporfin and MYF-01-37, respectively. Importantly, the results showed that YAP overexpression blocked the inhibitory effects of INT-777 on cell proliferation as well as significantly suppressed the apoptosis of H1299 cells induced by INT-777 (Fig. 4 A–C and SI Appendix, Fig. S8 A–C). Similarly, both YAP silencing and YAP inhibition attenuated the proliferation-promoting effect of R399 on H1299 cells by MTT analysis (Fig. 4D and SI Appendix, Fig. S8 D and E). Collectively, these data suggested that YAP signaling played critical roles in the different effects of R399 and INT-777 on the growth and apoptosis of H1299 cells.
Fig. 4.
YAP signaling mediates the biological roles of R399 and INT-777 in H1299 cells. (A) MTT analysis was performed to detect the effects of YAP overexpression on INT-777 (2 μM)–induced cell growth inhibition of H1299 cells (n = 6). (B) Flow cytometry was performed to detect the effects of YAP overexpression on INT-777 (2 μM)–induced cell apoptosis of H1299 cells. Quantitative analyses of the cell apoptosis ratio of each group (n = 3). (C) TUNEL analysis were performed to detect the effects of YAP overexpression on INT-777 (2 μM)–induced cell apoptosis of H1299 cells. (Scale bars, 50 µm.) (E) Quantitative analyses of the TUNEL-positive cell ratio of each group (n = 3). (D) MTT analysis was performed to detect the effects of YAP-specific siRNAs, YAP inhibitor, Verteporfin (VP, 4 µM), and MYF-01-37 (10 µM) on R399 (700 nM)–induced cell growth promotion of H1299 cells (n = 6). All data are presented as the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 based on the Student’s test. DMSO, dimethyl sulfoxide; FITC, fluorescein isothiocyanate–conjugated; ns, not significant; PI, propidium iodide.
β-Arrestin 1 Is Required for R399-Induced YAP Activation, and Gs Signaling Is Responsible for INT-777–Triggered YAP Inactivation.
GPBAR can activate both G protein and arrestin signaling to exert diverse biological effects. For example, GPBAR activation can stimulate glucagon-like peptide-1 secretion as well as cystic cholangiocyte proliferation through Gs/cyclic adenosine monophosphate (cAMP) signaling (1, 2, 27). Moreover, GPBAR stimulation has been reported to contribute to the innate antiviral response via β-arrestin–mediated SRC (SRC Proto-Oncogene, Non-Receptor Tyrosine Kinase) activation (6, 11). Notably, in previous studies, both G protein and arrestin signaling were shown to separately connect with YAP signaling (28–31). Therefore, R399 and INT-777 may elicit different activities of G proteins and arrestins, a phenomenon called the Gs versus Arrestin signaling bias, to differentially control YAP activity in the growth and apoptosis of H1299 cells.
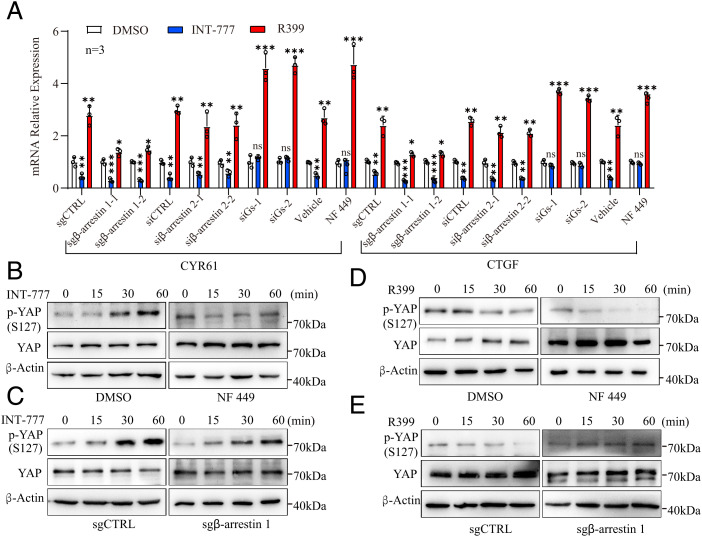
We therefore used two specific single guide RNAs targeting β-arrestin 1, two specific siRNAs targeting β-arrestin 2, two specific siRNAs targeting Gs, and a Gs inhibitor (NF 449) to investigate the contributions of β-arrestin 1/2 and Gs to the regulation of YAP activity by INT-777 and R399 (Fig. 5A and SI Appendix, Fig. S9 A–C). Consequently, the INT-777–induced reduction in YAP transcriptional activity was reversed by NF 449 treatment or siRNA-mediated Gs knockdown but was exacerbated after β-arrestin 1 knockdown (Fig. 5A and SI Appendix, Fig. S9 D). By contrast, β-arrestin 1 knockdown, but not Gs silencing or inhibition, slightly widened the window of increased YAP transcriptional activity induced by R399 (Fig. 5A and SI Appendix, Fig. S9 D). The contributions of β-arrestin 1 and Gs to alterations of YAP activity induced by R399 and INT-777 were further confirmed by analyses of YAP phosphorylation and subcellular distribution (Fig. 5 B–E and SI Appendix, Fig. S9 E–N). However, in contrast to the phenotype of β-arrestin 1 knockdown, β-arrestin 2 silencing had no significant effects on R399-induced YAP activation by TEAD luciferase reporter assay or by examination of the known target gene expression of YAP (Fig. 5A and SI Appendix, Fig. S9 D). Collectively, these findings indicated that β-arrestin 1, but not β-arrestin 2, signaling promoted YAP activation in response to R399, whereas Gs signaling mediated the inhibition of YAP activity in response to INT-777 stimulation.
Fig. 5.
R399-induced YAP activation is β-arrestin dependent, while INT-777–triggered YAP inhibition is Gs dependent. (A) Effects of β-arrestin 1/2 silencing, Gs silencing, or Gs inhibitor NF 449 (10 μM) on changes in YAP target gene expression induced by R399 (700 nM) or INT-777 (2 μM). (B–E) Effects of NF 449 (10 μM) and β-arrestin 1 silencing on the alterations of YAP S127 phosphorylation triggered by INT-777 (2 μM) (B and C) or R399 (700 nM) (D and E) were analyzed using Western blot. All data are presented as the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 based on the Student’s t-test. CTRL, control; DMSO, dimethyl sulfoxide; IB, immunoblotting; mRNA, messenger RNA; ns, not significant.
Importantly, previous studies have indicated that β-arrestin 1 facilitates YAP nuclear localization through physical interaction with YAP (28). We therefore evaluated whether R399-driven YAP nuclear shuttling is also dependent on the interaction between β-arrestin 1 and YAP. Coimmunoprecipitation (Co-IP) analysis revealed that R399 promoted the interaction between β-arrestin 1 and YAP in a time-dependent manner (SI Appendix, Fig. S9 O and P). We then performed Co-IP analysis in cytoplasmic and nuclear fractions derived from H1299 cells and double IF staining to further examine the comigration of YAP and β-arrestin 1 to the nucleus after R399 exposure. Strikingly, Co-IP analysis revealed that the interaction between β-arrestin 1 and YAP in the cytoplasm was increased after 15 min of R399 treatment. At 30 min after R399 stimulation, the co-increased β-arrestin 1/YAP expression in the nucleus was observed, although the presence of YAP and β-arrestin 1 protein was decreased in the cytoplasm (SI Appendix, Fig. S10 A and B). Similar results were obtained from the IF analysis (SI Appendix, Fig. S10C). These data indicated that R399 promoted the interaction between β-arrestin 1 and YAP, thereby facilitating their nuclear translocation. In contrast, R399 showed no significant effects on the association of β-arrestin 2 and YAP (SI Appendix, Fig. S10 D and E).
Collectively, these data suggested that β-arrestin 1 mediated the effects of R399 on YAP activation (SI Appendix, Fig. S9Q), while Gs signaling contributed to YAP inhibition in response to INT-777 stimulation. However, the detailed mechanism needs further exploration.
The Functional Differences of R399 and INT-777 in H1299 Cells Are Attributed to Their Different Biased Properties.
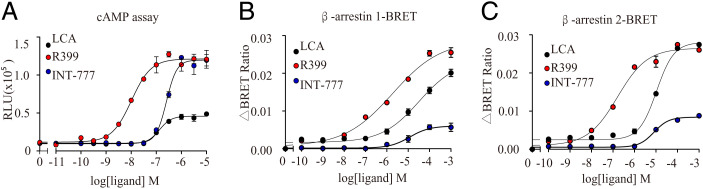
Because the β-arrestin pathway and Gs signaling differentially regulate YAP activity, which plays important roles in regulating NSCLC cell growth and apoptosis, we then characterized the biased properties of GPBAR activation in H1299 cells in response to R399 or INT-777 treatment. We performed the Gα-Gγ dissociation assay to examine the Gs, Gi, Gq, and G12/13 signaling of GPBAR in response to INT-777 and R399, using the endogenous bile acid LCA as a reference. Notably, GPBAR showed only Gs coupling activity but not detectable Gi, Gq, or G12/13 activities in response to INT-777 or R399 stimulation under our experimental situation (SI Appendix, Fig. S11 A–E). Further, the GloSensor cAMP assay and the β-arrestin recruitment assay revealed that INT-777 showed much weaker activity than R399 in the β-arrestin 1 and β-arrestin 2 recruitment assay but showed comparable Gs coupling ability (Fig. 6 A–C). Importantly, whereas R399 was shown to be a β-arrestin 1/2–biased ligand using the endogenous bile acid LCA as a reference, INT-777 was a Gs-biased ligand. The calculated differential bias values (Δβ) between INT-777 and R399 were 1.04 ± 0.23 (β-arrestin 1 vs. Gs) and 0.73 ± 0.09 (β-arrestin 2 vs. Gs) (SI Appendix, Fig. S11E). Consistent with this finding, functional analysis showed that knockdown of either β-arrestin 1 or β-arrestin 2 markedly suppressed but blockade of Gs activity slightly enhanced R399-induced cell growth, suggesting the selective involvement of β-arrestin 1 and β-arrestin 2 rather than Gs in R399-triggered cell behavior (SI Appendix, Fig. S12A). In contrast, Gs silencing or inhibition of Gs activity markedly reduced the antiproliferative and proapoptotic effects of INT-777, whereas knockdown of β-arrestin 1 or β-arrestin 2 had minimal effects on these phenotypic changes triggered by INT-777, indicating that the effects of INT-777 were dependent on Gs but not β-arrestin 1 or β-arrestin 2 (SI Appendix, Fig. S12 B and C). These results suggested that Gs activity was associated with the effects of INT-777 on cell growth and apoptosis, whereas the arrestin pathway contributed to the increased growth of H1299 cells in response to R399 stimulation.
Fig. 6.
Gs bias for INT-777 and β-arrestin bias for R399 contributed to the distinct biological consequences elicited in NSCLC. (A–C) Comparison of the biased properties of INT-777 and R399. Both INT-777 and R399 were assessed for cAMP signaling (A) and β-arrestin 1/2 recruitment (B and C). BRET, bioluminescence resonance energy transfer; CTRL, control; ns, not significant; RLU, Renilla Luciferase.
Studies have suggested that β-arrestin 1 and β-arrestin 2 have distinct or even antagonistic roles in specific pathophysiological contexts (32–35). For example, in a mouse model of myocardial infarction, β-arrestin 1 was demonstrated to be detrimental, whereas β-arrestin 2 was beneficial for the recovery of cardiac structure and function (36). β-arrestin 2 effectively augments but β-arrestin 1 represses nuclear factor-κB activity in immune cells (37). In addition, multiple lines of evidence have underscored the diverse and profound role of β-arrestin 1 in regulating cell proliferation and apoptosis in different cellular contexts or different pathophysiological conditions. For example, β-arrestin 1 has been reported to promote malignant proliferation and transmit antiapoptotic signals in different cancer cell models, including prostate cancer, hepatocellular carcinoma, and breast cancer (38–40). However, β-arrestin 1 has also been proven to exert proapoptotic roles in several cell types, such as in cardiac myocytes and intestinal stem cells, which may be dependent on contexts of specific cell or receptor types (33, 36, 41). In our present study, Western blot analysis showed that the expression level of β-arrestin 2 was comparable to that of β-arrestin 1 in H1299 cells (SI Appendix, Fig. S13 A and B). MTT assay and flow cytometry analysis showed that β-arrestin 1 knockdown inhibited cell proliferation and induced cell apoptosis of H1299 cells (SI Appendix, Fig. S13 C and D), whereas β-arrestin 1 overexpression stimulated cell growth and had no significant effects on cell apoptosis (SI Appendix, Fig. S13 E–H). These data indicate that β-arrestin 1 may exert proproliferation and antiapoptotic roles in NSCLC cells, which is consistent with the results of a previous study (38). Notably, either β-arrestin 1 knockdown or β-arrestin 2 silencing significantly alleviated R399-induced cell growth, suggesting that β-arrestin 1 and β-arrestin 2 played similar roles in mediating the cellular responses downstream of R399-induced GPBAR activation in H1299 cells.
To explore the possible biased properties of GPBAR activation in H1299 cells in response to other endogenous agonists, we examined the downstream signaling of GPBAR activated by DCA using the GloSensor cAMP assay and the β-arrestin recruitment assay, using the endogenous agonist LCA as a reference. Notably, DCA and LCA showed similar efficacies in cAMP accumulation assays (SI Appendix, Fig. S14A). However, no detectable recruitment of β-arrestin 1/2 to GPBAR was observed in response to DCA stimulation, indicating that DCA is a Gs-biased ligand (SI Appendix, Fig. S14 B and C). Previous studies have suggested that G protein activation is commonly earlier than arrestin-mediated ERK (extracellular regulated protein kinases) activation (42, 43). However, no detectable arrestin recruitment to GPBAR could be observed in the time course from 2 to 30 min in response to DCA stimulation using R399 as a control. These data suggested that DCA is a more Gs bias ligand of GPBAR compared with R399 (SI Appendix, Fig. S15A). In addition, similar cAMP activity but decreased arrestin recruitment in GPBAR overexpressed cells induced by INT-777 than that induced by R399 was observed at all time points across 1 to 45 min (SI Appendix, Fig. S15 A and B). Consistently, MTT assay and flow cytometry analysis revealed that silencing of Gs by RNAi or application of Gs inhibitor NF 449, but not silencing of β-arrestin 1/2, partially impaired DCA-induced cell growth inhibition and apoptosis. These results collectively supported that DCA modulated H1299 cell growth and apoptosis through a Gs-dependent signaling (SI Appendix, Fig. S14 D and E).
G Protein–Coupled Receptor Kinases GRK2 and GRK5 Are Essential for R399-Induced GPBAR–β-arrestin 1 Recruitment and Downstream Signaling.
G protein–coupled receptor kinases (GRKs) play important roles in β-arrestin–dependent signaling through mediating receptor phosphorylation (44–46). However, little is known about the specific roles of individual GRKs for modulation of GPBAR function and phosphorylation. Expression profiling indicated that GRK2, GRK5, and GRK6 were enriched in the H1299 cell line (SI Appendix, Fig. S16A). Moreover, R399 induced the association between GPBAR and GRK2/GRK5, but not GRK6, in a dose-dependent manner in a bioluminescence resonance energy transfer assay (Fig. 7A). More importantly, the β-arrestin recruitment assay revealed that siRNA silencing of GRK2 and GRK5 dramatically decreased the recruitment of β-arrestin 1 to GPBAR in response to R399 stimulation, suggesting the important contributions of GRK2 and GRK5 to R399-induced β-arrestin 1 recruitment to GPBAR (Fig. 7B). Moreover, GRK2 or GRK5 silencing significantly attenuated R399-induced YAP activation and cell proliferation, consistent with their important roles in mediating the GPBAR/β-arrestin 1 association (Fig. 7 C and D and SI Appendix, Fig. S16 B and C).
Fig. 7.
GRK2 and GRK5 were essential for R399-induced GPBAR–β-arrestin 1 association. (A) Effects of GRK2, GRK5, or GRK6 on the R399-induced bioluminescence resonance energy transfer (BRET) assay. (B) Effects of knockdown of GRK2, GRK5, or GRK2 and GRK5 on R399-induced β-arrestin 1 recruitment. (C and D) Effects of GRK2 and GRK5 silencing on changes in TEAD-relative luciferase activity induced by R399 (700 nM) and on R399-induced cell growth were evaluated by MTT analysis (n = 6). All data are presented as the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 based on the Student’s t-test. CTRL, control; DMSO, dimethyl sulfoxide; ns, not significant.
We next exploited mass spectrometry to analyze the exact phosphorylation sites of GPBAR by GRK2. Flag-tagged GPBARs were transiently transfected in the H1299 cell line and were immunoprecipitated by Flag antibody after stimulation with R399 (700 nM). Importantly, phosphorylation at the S310, S321, S323, and S324 sites of GPBAR was observed in R399-stimulated cells, but only significantly less enriched phosphorylation of S321 could be detected after GRK2 knockdown. These results suggested that GRK2 mediated phosphorylation of GPBAR at the S310, S321, S323 and S324 sites, which contribute to the R399-induced GPBAR–β-arrestin 1 association (SI Appendix, Fig. S17 A and B).
Together, these findings revealed that GRK2- and GRK5-mediated GPBAR phosphorylation might promote β-arrestin 1 recruitment to the receptor, thus facilitating downstream YAP activation to stimulate cell growth.
Cryo-EM Structure of the R399-GPBAR-Gs Complex.
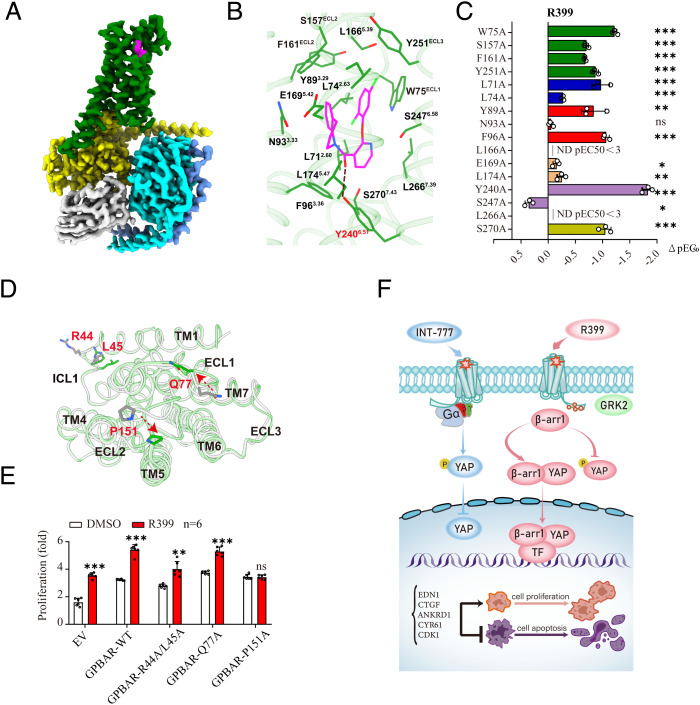
Recently, we solved the cryo-EM structure of INT-777–bound GPBAR-Gs, which provides important insights into the mechanisms by which GPBAR recognizes agonists with structures similar to those of bile acids (11). To provide the GPBAR structural basis underlying the differential regulation of tumorigenesis by different GPBAR agonists, we sought to determine the cryo-EM structure of the R399-activated GPBAR-Gs complex. We coexpressed BRIL (a thermostable apocytochrome b562RIL)-GPBAR, Gβ1/Gγ2, and Gαs in Spodoptera frugiperda (Sf9) cells. R399 at 50 μM was added to form the signaling complex and the conformation-selective nanobody Nb35 was used to facilitate complex formation. The complex was then solubilized in 0.5% lauryl maltose neopentyl glycol and 0.1% cholesteryl hemisuccinate and subjected to tandem purification via an anti-Flag affinity column and size-exclusion chromatography (SI Appendix, Fig. S18A). The homogenous samples generated were used for further cryo-EM analysis. We then collected 4,884 cryo-EM movies and selected 2,941,406 particle projections to construct the final EM density map of the R399-GPBAR-Gs complex at an overall resolution of 3.2 Å (SI Appendix, Fig. S18 B–E). The EM density map enabled confident assignment of most GPBAR residues, including all seven-transmembrane helices with both the intracellular and extracellular loops and most side chains from the receptor and G protein (Fig. 8A and SI Appendix, Fig. S19 A and B). In particular, a well-defined density was observed for R399, which was used to analyze the detailed interactions between R399 and GPBAR (Fig. 8A).
Fig. 8.
Binding of R399 to GPBAR and the structural basis of biased agonism by R399. (A) Cryo-EM density of the GPBAR-Gs-R399 complex. R399 is shown in magenta, GPBAR in green, Gαs in yellow, Gβ in cyan, Gγ in light blue, and Nb35 in gray. (B) Structural view of the insertion of R399 into the ligand pocket. (C) Effects of mutations of the GPBAR ligand binding pocket on the ligand binding of R399. Values are the mean ± SEM of three independent experiments for the wild type (WT) and mutants (P values are as follows: <0.0001, <0.0001, <0.0001, <0.0001, <0.0001, <0.0001, 0.0028, 0.1612, <0.0001, no detectable singnal [ND], 0.0162, 0.0067, <0.0001, 0.0016, ND, and <0.0001 from Top to Bottom for the R399 group). (D) Extracellular view of the GPBAR transmembrane bundle showing the location of the residues with different root mean square deviation between R399 and INT-777–bound GPBAR, colored in green and gray, respectively. Residues with significant conformational changes are marked in red. (E) A549 cells were transfected with empty vector (EV), GPBAR-WT, and GPBAR mutants (R44A/L45A, Q77A, and P151A), respectively. Then the above-mentioned cells were treated with R399 (700 nM) or dimethyl sulfoxide (DMSO) for 72 h (n = 6). (F) Schematic model of molecular mechanism underlying biased GPBAR signaling elicited by INT-777 and R399 through YAP activity in NSCLC cells. GPBAR activation by INT-777 and R399 bidirectionally regulated the YAP pathway through selective activation of Gs and β-arrestin 1 signaling effectors. All data are presented as the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 based on the Student’s t-test. All results are representative of three independent experiments. ns, not significant.
Binding of R399 to GPBAR.
R399 assumes a “V-shaped” configuration inside the ligand binding pocket of GPBAR. Whereas the central pyridinyl ring of R399 sits at the bottom, the 2,5-dichlorocyclohexyl and 3,4-dihydroquinolin-1(2H)-yl groups constitute two hydrophobic wings of the V shape and face each other (Fig. 8B and SI Appendix, Fig. S19C). Unlike the contacts of INT-777 but similar to those of another synthetic compound (P395), the interaction between R399 and GPBAR is dominated by hydrophobic interactions, with only one polar contact (SI Appendix, Fig. S19D). These hydrophobic interactions can be divided into three parts (Fig. 8B and SI Appendix, Fig. S19 C and E). Whereas Y893.29 and F161ECL2 create a hydrophobic patch to hold the 3,4-dihydroquinolin-1(2H)-yl group, L712.60, L742.63, W75ECL1, and Y251ECL3 constitute another hydrophobic strip to accommodate the 2,5-dichlorocyclohexyl group of R399. In particular, the two chlorides in the dichlorocyclohexyl group of R399 directly contact L712.60, L742.63, and Y251ECL3, and this interaction may contribute to the high binding affinity of R399. L1665.39 contacts both the 3,4-dihydroquinolin-1(2H)-yl and dichlorocyclohexyl groups of R399 (SI Appendix, Fig. S19F). Consistent with this finding, mutations in residues surrounding the two hydrophobic wings, including the L71A, W75A, F161A, and L166A mutations, significantly decreased R399-induced cAMP accumulation (Fig. 8C and SI Appendix, Fig. S19G).
At the center of R399, the pyridinyl ring is seated in the hydrophobic bottom formed by the residues L712.60, F963.36, Y2406.51, and L2667.39. In particular, the special ketone linker in R399, which bridges the 3,4-dihydroquinolin-1(2H)-yl group and pyridinyl ring, forms a hydrogen bond with the hydroxyl group of Y2406.51 (Fig. 8B and SI Appendix, Fig. S19 C and D). In our recent studies, GPBAR agonists, including a panel of bile acids and the synthetic compound P395, were shown to form a similar H-bond with Y2406.51, an interaction that was a key step in GPBAR activation (11). Consistent with previous studies, the Y240A mutation significantly decreased the potency of R399 in inducing cAMP accumulation.
Structural Basis of the Functional Biased Property of R399.
Using the endogenous bile acid LCA as a reference, we found that whereas INT-777 was a Gs-biased ligand, R399 showed arrestin-biased properties (Fig. 6 A–C). In contrast to INT-777, R399 lacked interactions with the hydrophobic patch formed by L2446.55, L2627.35, and L2637.36 but participated in new interactions with L1745.47 and S2707.43 of GPBAR ( SI Appendix, Fig. S19D and SI Appendix, Table S2).
To further investigate the structural basis of the GPBAR activation bias in response to R399 and INT-777, we inspected the conformational differences between the R399- and INT-777–bound GPBAR structures via root mean square deviation over Cα atoms (SI Appendix, Fig. S20A). Notably, two extracellular loops (ECL1 and ECL2) as well as the intracellular loop (ICL1) showed the most significant differences between the R399-bound GPBAR structure and the INT-777–bound GPBAR structure (Fig. 8D). In particular, P151 was shifted by 5.1 Å toward TM1 and TM4 in the R399-GPBAR-Gs structure compared to the INT-777–GPBAR–Gs complex structure. We therefore examined alanine mutations of the residues R44ICL1 and L45ICL1, Q77 ECL1, and P151ECL2, which exhibited significant conformational differences between the two structures, to examine the effects of these residues on Gs- and arrestin-coupled downstream signaling. The R44ICL1A, L45ICL1A, Q77 ECL1A, and P151ECL2A mutations resulted in a significant decrease in arrestin recruitment that exceeded the decrease in cAMP accumulation (SI Appendix, Fig. S20 B and C). Moreover, subsequent functional analysis revealed that the P151ECL2A mutation abolished the R399-induced proliferation of A549 cells overexpressing wild-type GPBAR or the corresponding mutants (Fig. 8E and SI Appendix, Fig. S20D and E). Collectively, these findings suggested that the different interaction patterns and conformational changes in GPBAR in response to R399 and INT-777 binding—for instance, the conformational changes at the P151 position in ECL1—contributed to the biased property of R399 and its ability to promote cancer cell growth.
Discussion
GPBAR, as one of the membrane bile acid–sensing receptors, activates various intracellular signaling pathways upon interaction with bile acids and has been reported to be closely associated with the cancer development (7, 8, 10, 17). However, the roles of GPBAR in carcinogenesis remain controversial and little is known about the contribution of GPBAR to lung cancer. For example, whereas GPBAR activity might suppress kidney and gastric cancer cell proliferation and migration (9, 18), activation of GPBAR was also reported to promote endometrial cancer cell proliferation and cholangiocarcinoma progression (10, 19), indicating that GPBAR may exert distinct roles in different physiological or cellular contexts. Importantly, a recent study suggested that GPBAR was highly expressed in NSCLC, which positively correlated with an advanced clinical stage of NSCLC (12). GPBAR knockdown inhibited cell proliferation, migration, and invasion of NSCLC cells (12). Consistent with the previous study, functional analysis in vitro showed that GPBAR overexpression promoted NSCLC cell growth in several cell lines, whereas GPBAR knockdown inhibited cell growth and induced cell apoptosis, suggesting a tumor-promotive function of GPBAR. Further, an in vivo xenograft tumor model in nude mice, along with the clinical relevance analysis, supported the tumor-promotive activity of GPBAR in NSCLC.
Importantly, several groups have reported that even in the same cancer types, GPBAR activated by different agonists may exert different pro- or antitumor effects. For instance, 23(S)-mCDCA–triggered GPBAR activation suppresses (17) but taurolithocholic acid enhances the migration of gastric cancer cells (18, 47), implying the complexity of the net effects after GPBAR activation and potential involvement of the biased signaling of GPBAR. In our present study, we therefore explored the cellular outcomes of GPBAR activation induced by different agonists in NSCLC cells. Interestingly, at least two different GPBAR-specific agonists, INT-777 and R399, showed opposite regulatory roles in NSCLC cell growth via GPBAR activation. Similar to the effects of the endogenous agonist DCA, INT-777 treatment suppressed cell proliferation in a dose-dependent manner and induced cell apoptosis. In contrast, R399 treatment promoted cell proliferation but did not show significant effects on cell apoptosis. These distinct effects of GPBAR activation induced by INT-777 and R399 were further supported by in vivo studies, confirming the hypothesis that different GPBAR agonists have distinct regulatory effects on cancer cell growth. Such findings that different endogenous or exogenous agonists of GPBAR elicit distinct or even opposing biological outcomes in regulating NSCLC cell viability indicated the importance of the size and composition of the bile acid pool orchestrating with GPBAR expression in NSCLC carcinogenesis. The net effect of GPBAR especially related to the bile acid homeostasis disruption in NSCLC is worth in-depth exploration in the future.
Moreover, in our present study, we deciphered that distinct signaling cascades and the structural basis underlying the different cellular responses elicited by different agonists triggered GPBAR activation through inducing biased downstream signaling (Gs vs. Arrestin pathways). Global transcriptome sequencing and subsequent pathway enrichment analysis revealed that Hippo signaling was inhibited by R399 treatment but enhanced by INT-777 treatment. Further mechanistic investigation showed that GPBAR activation by R399 displayed an obvious bias for β-arrestin 1, which subsequently activated downstream YAP signaling. Conversely, binding of INT-777 to GPBAR preferentially activated Gs signaling, which inhibited YAP signaling. The opposite regulatory effects of different GPBAR agonists on YAP signaling were the key switch for the different effects of these agonists on cell viability. In particular, we have delineated that GRK2 and GRK5 are essential for the recruitment of β-arrestin 1 to GPBAR in response to R399 stimulation, and we identified potential GPBAR phosphorylation sites by GRK2 using mass spectrometry. Our results indicated the important roles of GRKs in the modulation of GPBAR biology.
Accumulating evidence has demonstrated that dysregulation of the Hippo-YAP signaling pathway is tightly related to the development and progression of different cancers (29, 48–53). Understanding the mechanisms by which G protein–coupled receptors (GPCRs) regulate YAP signaling is highly valuable for the design of therapeutics targeting this important group of drug targets. Importantly, previous studies have shown that GPCR signaling acts as an upstream signal that can regulate Hippo-YAP signaling in a distinctive G protein–dependent or β-arrestin–dependent manner. For instance, epinephrine- or glucagon-mediated activation of Gs-coupled receptors was shown to inhibit YAP activation by increasing Lats1/2 kinase activity (29). Conversely, lysophosphatidic acid or sphingosine 1-phosphate–induced activation of G12/13- or Gq/11-coupled receptors was shown to result in YAP activation by inhibiting Lats1/2 kinases (29). Moreover, endothelin-1 (ET-1)–triggered ET-1 receptor activation was found to promote the nuclear accumulation of YAP by enhancing the physical interaction between β-arrestin 1 and YAP (28). Consistent with these observations but more interestingly, our data herein indicated that GPBAR activation by different agonists bidirectionally regulated the Hippo-YAP pathway through selective activation of distinct downstream signaling effectors (i.e., Gs and β-arrestin 1) (Fig. 8F). Although the precise mechanisms underlying the regulation of YAP signaling by GPBAR need in-depth exploration and the effects of GPBAR-biased signaling on cancer progression need verification in other tumor systems (especially considering the high heterogeneity of carcinomas and dispensability of YAP for the progression of certain cancers), our study provides not only a rationale for developing potentially synergistic therapies targeting the GPBAR-YAP axis in NSCLC but also useful guidance for designing GPBAR agonists to treat metabolic diseases and prevent the potential side effect of tumor progression mediated by these ligands.
To provide the structural basis for the differential regulation of GPBAR function by INT-777 and R399, we solved the cryo-EM structure of the R399-GPBAR-Gs complex at 3.2-Å resolution. Compared with INT-777, R399 lost hydrophobic interactions with TM6 and TM7 residues L2446.55, L2627.35, and L2637.36 but formed new interactions with TM5 residues, which caused significant conformational changes in ICL1, ECL1, and ECL2. Most importantly, mutation of one of these residues, P151ECL2A, significantly reduced the β-arrestin 1 biased property of R399 and abolished the R399-induced growth of H1299 cells. Therefore, the structural study of GPBAR in complex with R399 and other ligands afforded a structural understanding of distinct GPBAR functions and provided useful guidance for further design of GPBAR agonists with both metabolic and anticancer benefits.
In summary, our study revealed the distinct functions of GPBAR activation in NSCLC in response to INT-777 and R399 binding and identified the molecular and structural determinants of the signaling bias of INT-777 and R399. Our results indicate that fine-tuning of GPBAR agonists may provide valuable tools for modulating one specific intracellular signaling pathway associated with disease, thus achieving individualized precision medicine and reducing adverse side effects.
Materials and Methods
Image Processing and Three-Dimensional Reconstruction.
Image stacks were subjected to motion correction using MotionCor2.1. Contrast transfer function parameters were determined by Gctf (54, 55). The particles were subjected to two rounds of three-dimensional classification using the map of the P395-bound GPBAR-Gs complex (Electron Microscopy Data Bank accession no. EMD-30344) as the initial reference model (11). Local resolution was determined using the Bsoft package with half maps as input maps (56).
Model Building and Refinement.
The structure of the P395-bound GPBAR-Gs complex (Protein Data Bank accession no. 7CFM) was used as the initial template for model building (11). Agonist coordinates and geometry restraint were generated using phenix.elbow (57). This initial model was then subjected to iterative rounds of manual adjustment and automated refinement in Coot (58) and Phenix (57), respectively. The final refinement statistics are provided in SI Appendix, Table S7.
For other methods, see SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Tao Zhang for providing the plasmid p-3×Myc-CMV-YAP in this study. We also thank Dr. Wenwen Liu for the YAP signaling inhibitor, Verteporfin. We are grateful to Dr. Weiping Tang (Cnkingbio Company Ltd.) for bioinformatics assistance. The cryo-EM data were collected at the Cryo-Electron Microscopy Center of Zhejiang University. This work was supported by grants from the National Key R&D Program of China (2018YFC1003600 to X.Y. and J.-P.S., 2019YFA0904200 to J.-P.S. and P.X., and 2019YFA0508800 to Y.Z. and P.X.), the National Science Fund for Excellent Young Scholars (82122070 to F.Y.), the National Science Fund for Distinguished Young Scholars (81773704 to J.-P.S.), the National Natural Science Foundation of China (31870781 to P.-Z., 31900936 to F.Y., 81922071 to Y.Z., 32100959 to C.M., and 92057121 to X.Y.), the Key Research Project of the Natural Science Foundation of Beijing, China (Z200019 to J.-P.S.), the Major Fundamental Research Program of the Natural Science Foundation of Shandong Province, China (ZR2020ZD39 to J.-P.S.), the Key Research and Development Program of Shandong Province (2021CXGC011105 to J.-P.S.), the Zhejiang Province Science Fund for Distinguished Young Scholars (LR19H310001 to Y.Z. and LR22C050002 to C.M.), and Key R&D Projects of Zhejiang Province (2021C03039 to Y.Z.). We thank the Translational Medicine Core Facility of Shandong University for consultation and instrument availability that supported this work.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117054119/-/DCSupplemental.
Data Availability
The cryo-EM density map and corresponding atomic coordinates of the R399-bound GPBAR-G complex data have been deposited in the Electron Microscopy Data Bank (accession no. EMD-33452) (59) and the Protein Data Bank (accession no. 7XTQ) (60). All other study data are included in the article and/or SI Appendix.
References
- 1.Qi X., et al. , Gut microbiota-bile acid-interleukin-22 axis orchestrates polycystic ovary syndrome. Nat. Med. 25, 1225–1233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng X., et al. , Hyocholic acid species improve glucose homeostasis through a distinct TGR5 and FXR signaling mechanism. Cell Metab. 33, 791–803.e7 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Poropat G., Giljaca V., Stimac D., Gluud C., Bile acids for primary sclerosing cholangitis. Cochrane Database Syst. Rev. 1, CD003626 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arab J. P., Karpen S. J., Dawson P. A., Arrese M., Trauner M., Bile acids and nonalcoholic fatty liver disease: Molecular insights and therapeutic perspectives. Hepatology 65, 350–362 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiriyama Y., Nochi H., The biosynthesis, signaling, and neurological functions of bile acids. Biomolecules 9, E232 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu M. M., et al. , Virus-induced accumulation of intracellular bile acids activates the TGR5-β-arrestin-SRC axis to enable innate antiviral immunity. Cell Res. 29, 193–205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen W. D., Yu D., Forman B. M., Huang W., Wang Y. D., Deficiency of G-protein-coupled bile acid receptor Gpbar1 (TGR5) enhances chemically induced liver carcinogenesis. Hepatology 57, 656–666 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Jia W., Xie G., Jia W., Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 15, 111–128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su J., et al. , The G-protein-coupled bile acid receptor Gpbar1 (TGR5) protects against renal inflammation and renal cancer cell proliferation and migration through antagonizing NF-κB and STAT3 signaling pathways. Oncotarget 8, 54378–54387 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casaburi I., et al. , Chenodeoxycholic acid through a TGR5-dependent CREB signaling activation enhances cyclin D1 expression and promotes human endometrial cancer cell proliferation. Cell Cycle 11, 2699–2710 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Yang F., et al. , Structural basis of GPBAR activation and bile acid recognition. Nature 587, 499–504 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Liu X., et al. , The membrane bile acid receptor TGR5 drives cell growth and migration via activation of the JAK2/STAT3 signaling pathway in non-small cell lung cancer. Cancer Lett. 412, 194–207 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Sayin S. I., et al. , Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17, 225–235 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Wahlström A., et al. , Induction of farnesoid X receptor signaling in germ-free mice colonized with a human microbiota. J. Lipid Res. 58, 412–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu T., et al. , FXR regulates intestinal cancer stem cell proliferation. Cell 176, 1098–1112.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas C., et al. , TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 10, 167–177 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao R. Y., et al. , High expression of TGR5 predicts a poor prognosis in patients with pancreatic cancer. Int. J. Clin. Exp. Pathol. 11, 3567–3574 (2018). [PMC free article] [PubMed] [Google Scholar]
- 18.Guo C., et al. , The G-protein-coupled bile acid receptor Gpbar1 (TGR5) suppresses gastric cancer cell proliferation and migration through antagonizing STAT3 signaling pathway. Oncotarget 6, 34402–34413 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reich M., et al. , TGR5 is essential for bile acid-dependent cholangiocyte proliferation in vivo and in vitro. Gut 65, 487–501 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Moya I. M., Halder G., Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 20, 211–226 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Ma S., Meng Z., Chen R., Guan K. L., The hippo pathway: Biology and pathophysiology. Annu. Rev. Biochem. 88, 577–604 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Misra J. R., Irvine K. D., The Hippo signaling network and its biological functions. Annu. Rev. Genet. 52, 65–87 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin K. C., Park H. W., Guan K. L., Regulation of the Hippo pathway transcription factor TEAD. Trends Biochem. Sci. 42, 862–872 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Z., Guan K. L., Hippo signaling in embryogenesis and development. Trends Biochem. Sci. 46, 51–63 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang P., et al. , The alteration of Hippo/YAP signaling in the development of hypertrophic cardiomyopathy. Basic Res. Cardiol. 109, 435 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Nguyen C. D. K., Yi C., YAP/TAZ signaling and resistance to cancer therapy. Trends Cancer 5, 283–296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maczewsky J., et al. , TGR5 activation promotes stimulus-secretion coupling of pancreatic β-cells via a PKA-dependent pathway. Diabetes 68, 324–336 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Tocci P., et al. , β-arrestin1/YAP/mutant p53 complexes orchestrate the endothelin A receptor signaling in high-grade serous ovarian cancer. Nat. Commun. 10, 3196 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu F. X., et al. , Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780–791 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu F. X., Zhao B., Guan K. L., Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163, 811–828 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meng Z., Moroishi T., Guan K. L., Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shukla A. K., Xiao K., Lefkowitz R. J., Emerging paradigms of β-arrestin-dependent seven transmembrane receptor signaling. Trends Biochem. Sci. 36, 457–469 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Srivastava A., Gupta B., Gupta C., Shukla A. K., Emerging functional divergence of β-arrestin isoforms in GPCR function. Trends Endocrinol. Metab. 26, 628–642 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Kohout T. A., Lin F. S., Perry S. J., Conner D. A., Lefkowitz R. J., Beta-arrestin 1 and 2 differentially regulate heptahelical receptor signaling and trafficking. Proc. Natl. Acad. Sci. U.S.A. 98, 1601–1606 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang Y., et al. , Opposing functions of β-arrestin 1 and 2 in Parkinson’s disease via microglia inflammation and Nprl3. Cell Death Differ. 28, 1822–1836 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bathgate-Siryk A., et al. , Negative impact of β-arrestin-1 on post-myocardial infarction heart failure via cardiac and adrenal-dependent neurohormonal mechanisms. Hypertension 63, 404–412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao H., et al. , Identification of beta-arrestin2 as a G protein-coupled receptor-stimulated regulator of NF-kappaB pathways. Mol. Cell 14, 303–317 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Kong Z., et al. , β-arrestin1-medieated inhibition of FOXO3a contributes to prostate cancer cell growth in vitro and in vivo. Cancer Sci. 109, 1834–1842 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y., et al. , β-arrestin1 enhances hepatocellular carcinogenesis through inflammation-mediated Akt signalling. Nat. Commun. 6, 7369 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Shenoy S. K., et al. , β-arrestin1 mediates metastatic growth of breast cancer cells by facilitating HIF-1-dependent VEGF expression. Oncogene 31, 282–292 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhan Y., et al. , β-Arrestin1 inhibits chemotherapy-induced intestinal stem cell apoptosis and mucositis. Cell Death Dis. 7, e2229 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nobles K. N., et al. , Distinct phosphorylation sites on the β(2)-adrenergic receptor establish a barcode that encodes differential functions of β-arrestin. Sci. Signal. 4, ra51 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jain R., Watson U., Vasudevan L., Saini D. K., ERK activation pathways downstream of GPCRs. Int. Rev. Cell Mol. Biol. 338, 79–109 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Liggett S. B., Phosphorylation barcoding as a mechanism of directing GPCR signaling. Sci. Signal. 4, pe36 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Ren X. R., et al. , Different G protein-coupled receptor kinases govern G protein and beta-arrestin-mediated signaling of V2 vasopressin receptor. Proc. Natl. Acad. Sci. U.S.A. 102, 1448–1453 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Q., et al. , Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1. Nature 595, 600–605 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carino A., et al. , The bile acid receptor GPBAR1 (TGR5) is expressed in human gastric cancers and promotes epithelial-mesenchymal transition in gastric cancer cell lines. Oncotarget 7, 61021–61035 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng X., et al. , A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol. Cancer 18, 47 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao B., Li L., Lei Q., Guan K. L., The Hippo-YAP pathway in organ size control and tumorigenesis: An updated version. Genes Dev. 24, 862–874 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He C., et al. , The Hippo/YAP pathway interacts with EGFR signaling and HPV oncoproteins to regulate cervical cancer progression. EMBO Mol. Med. 7, 1426–1449 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou X., et al. , Regulation of Hippo/YAP signaling and esophageal squamous carcinoma progression by an E3 ubiquitin ligase PARK2. Theranostics 10, 9443–9457 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang S., et al. , Hippo signaling suppresses cell ploidy and tumorigenesis through Skp2. Cancer Cell 31, 669–684.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Z., et al. , EIF3H orchestrates Hippo pathway-mediated oncogenesis via catalytic control of YAP stability. Cancer Res. 80, 2550–2563 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heymann J. B., Guidelines for using Bsoft for high resolution reconstruction and validation of biomolecular structures from electron micrographs. Protein Sci. 27, 159–171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 59.L. Ma et al., EMD-33452, Cryo-EM structure of the R399-bound GPBAR-Gs complex. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-33452. Deposited 17 May 2022. [Google Scholar]
- 60.L. Ma et al., 7XTQ, Cryo-EM structure of the R399-bound GPBAR-Gs complex. Protein Data Bank. https://www.rcsb.org/structure/7XTQ. Deposited 17 May 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM density map and corresponding atomic coordinates of the R399-bound GPBAR-G complex data have been deposited in the Electron Microscopy Data Bank (accession no. EMD-33452) (59) and the Protein Data Bank (accession no. 7XTQ) (60). All other study data are included in the article and/or SI Appendix.