Significance
The evolution of a body plan in multicellular organisms is a central problem in biology. Here, we provide genetic evidence of the involvement of the Hippo cascade, a key growth regulating pathway in axis formation in Hydra, an early emerging metazoan. We demonstrate that the Hippo kinases regulate YAP upstream of Wnt signaling in axis formation and show that YAP and Hippo kinase pathway regulation of cell proliferation and control of the actin cytoskeleton were established early in metazoan evolution. Based on our results we speculate that Hippo signaling and nuclear Yap served to link continuous cell division, cell density, and axis formation early in metazoan evolution.
Keywords: Hydra, Hippo, Yap, Wnt, axis formation
Abstract
How did cells of early metazoan organisms first organize themselves to form a body axis? The canonical Wnt pathway has been shown to be sufficient for induction of axis in Cnidaria, a sister group to Bilateria, and is important in bilaterian axis formation. Here, we provide experimental evidence that in cnidarian Hydra the Hippo pathway regulates the formation of a new axis during budding upstream of the Wnt pathway. The transcriptional target of the Hippo pathway, the transcriptional coactivator YAP, inhibits the initiation of budding in Hydra and is regulated by Hydra LATS. In addition, we show functions of the Hippo pathway in regulation of actin organization and cell proliferation in Hydra. We hypothesize that the Hippo pathway served as a link between continuous cell division, cell density, and axis formation early in metazoan evolution.
How animals establish and pattern their primary body axis is a fundamental problem in biology. Data from a wide range of bilaterian animals suggest that Wnt signaling controls posterior identity during body plan formation (1–3). Wnt also drives axis formation in Cnidaria, the sister phylum to Bilateria (4–7). Thus, an axial patterning role for Wnt signaling was present in the last common ancestor of Cnidaria and Bilateria, which diverged ∼650 million years ago.
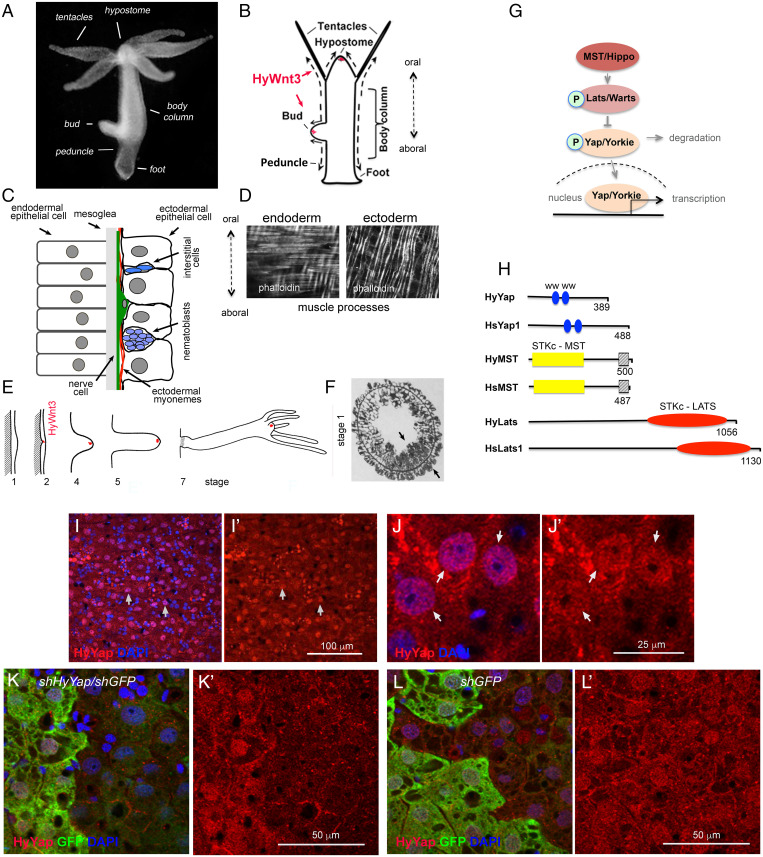
Hydra, a small freshwater cnidarian with a simple body plan, exhibits amazing regenerative and budding capabilities, as described in 1744 by Trembley (8). The Hydra body is essentially a tube of epithelial cells, aligned along the oral–aboral axis (Fig. 1A and B). Hydra has only two epithelial layers, an ectoderm and endoderm, separated by an extracellular matrix called the mesoglea (Fig. 1C). Epithelial cells of both layers are attached to the mesoglea and exhibit actin–myosin contractile elements called muscle processes on their basal sides (Fig. 1C and D). Cells of the interstitial cell lineage, stem cells, which give rise to nerves, nematocytes, gland cells, and germ cells, are intermingled among the epithelial cells (Fig. 1C). Epithelial cells continuously divide and are displaced along the oral–aboral axis toward the head and foot. The balance between cell production and loss of cells via sloughing from the ends and bud formation determines the size of the animal (9, 10) (Fig. 1B).
Fig. 1.
Hydra homolog of YAP is expressed in ectodermal epithelial cells. (A) Photo of a live Hydra. (B) Schematic of Hydra body plan; arrows indicate directions of cell displacement along the oral/aboral axis. (C) Schematic of a section through the Hydra body column. (D) Hydra endodermal and ectodermal muscle processes visualized with phalloidin. (E) Schematic of Hydra budding (adapted from ref. 11). The area of expression of HyWnt3 is confined to about 50 ectodermal epithelial cells marked in red (12). (F) Stage 1: Transverse section through the budding zone, arrows indicate increased cell density in the ectoderm and endoderm (adapted from ref. 13). (G) Schematic of the Hippo pathway. (H) Schematic of Hydra and mammalian homologs of Yap, MST, and LATS proteins; WW - proline-rich sequences binding domain; STKc–MST1/2 - catalytic domain of MST family of serine/threonine kinases;  - MST1-SARAH–apoptosis-mediating domain; STKc–LATS - catalytic domain of LATS family of serine/threonine kinases. (I–J′) Apical view of Hydra ectoderm immunostained with anti-HyYap serum. Arrows point to the nuclei of ectodermal epithelial cells. (K–L′) Apical view of ectoderm of GFP polyp electroporated with shGFP/HyYap (K and K′), shGFP alone (L and L′) hairpins and immunostained with anti-GFP and anti-HyYap antibodies; animals were fixed 6 d after electroporation.
- MST1-SARAH–apoptosis-mediating domain; STKc–LATS - catalytic domain of LATS family of serine/threonine kinases. (I–J′) Apical view of Hydra ectoderm immunostained with anti-HyYap serum. Arrows point to the nuclei of ectodermal epithelial cells. (K–L′) Apical view of ectoderm of GFP polyp electroporated with shGFP/HyYap (K and K′), shGFP alone (L and L′) hairpins and immunostained with anti-GFP and anti-HyYap antibodies; animals were fixed 6 d after electroporation.
Tentacles and buds are formed by tissue evagination. Budding, Hydra’s asexual form of reproduction, generates a new body axis (14) (Fig. 1B and E). Bud induction depends on the size of the mother polyp and its epithelial growth rate (15). Budding becomes visible by a thickening of the ectoderm at a site in the lower half of the body column (stage 1 in Fig. 1E and F) and continues as evagination of both layers. The formation of a new axis in Hydra is also controlled by the Wnt/β-catenin pathway (4, 6). Experimental activation of Wnt signaling in the body column results in formation of ectopic axes (4, 6, 12, 16, 17). Expression of HyWnt3 is detected early in budding, but only after thickening of the ectoderm is visible (12, 18). It is unknown what pathway(s) leads to the thickening of the ectoderm and induction of HyWnt3.
The Hippo pathway is a key regulator of cell proliferation, differentiation, and apoptosis (19). It consists of a cascade of kinases that controls nuclear localization of the transcription factor Yap (Yorkie in Drosophila) (Fig. 1G). MST (Drosophila Hippo) kinase phosphorylates and activates LATS (Drosophila Warts) that, in turn, phosphorylates Yap (Yorkie) leading to its binding to 14-3-3 and cytoplasmic retention or ubiquitination and degradation (20). Nuclear Yap/Yorkie, which promotes the expression of proproliferative and antiapoptotic genes, is involved in regulation of organ size (20, 21) and morphogenesis (22).
The Hippo pathway is remarkably conserved in metazoans and their unicellular ancestors (23). Functional studies using Hippo, Warts, and Yorkie homologs from the unicellular holozoan Capsaspora owczarzaki proteins ectopically expressed in Drosophila indicate the growth-regulating capabilities of the Hippo pathway were established before the emergence of metazoans (23). The full repertoire of proteins making up the Hippo cascade has been identified in Ctenophora and Cnidaria (24, 25). Immunostaining analysis of Yap in the cnidarian Clytia hemisphaerica suggested that regulation of nuclear/cytoplasmic localization of Yap could be a mechanism halting cell division and triggering differentiation programs (24). However, the function of the Hippo kinases has not been elucidated in cnidarians.
Here, we investigate the role of the Hippo pathway in morphogenesis and axis formation in Hydra. Using shRNA-mediated knockdown as well as a gain-of-function transgenic approach, we show that the Hippo pathway components LATS and YAP regulate axis formation and morphogenesis in Hydra. Our studies indicate that the Hippo pathway affects epithelial growth and acts upstream of HyWnt3 during axis formation in Hydra, suggesting that linkage between these two signaling pathways occurred early in metazoan evolution.
Results
HyYap Regulates Proliferation in Hydra.
Transcriptomic and genomic data identified single homologs of Yap, Lats, and MST in Hydra vulgaris, which we refer to as HyYap, HyLATS, and HyMST (Fig. 1H and SI Appendix, Fig. S1 A–C), consistent with recent studies (26). To explore HyYap function in vivo, we generated antiserum against residues 1 to 159 of the protein. Immunostaining of Hydra polyps revealed HyYap in the nuclei and cytoplasm of ectodermal epithelial cells (Fig. 1I–J′). Both nuclear and cytoplasmic staining were specifically removed by preabsorption of anti-HyYap serum with HyYap-GST antigen (SI Appendix, Fig. S1E). The distribution of HyYap was similar to the subcellular distribution of mammalian and Drosophila Yap homologs (27, 28). Immunoblotting of Hydra lysates with the anti-HyYap serum revealed a single strong band of ∼52 kDa, which was lost by preabsorption with the HyYap-GST antigen (SI Appendix, Fig. S1D) and by shRNA knockdown (see below).
To explore the function of HyYap, we combined shRNA knockdown protocols developed for Hydra (29, 30) and for the sea anemone Nematostella vectensis (31). To optimize the protocol, we used a transgenic Hydra line expressing GFP in the ectodermal epithelial cells (32). Transgenic polyps were electroporated with shGFP, leading to mosaic down-regulation of GFP. Down-regulation of GFP was seen on one side of 60 to 70% of the electroporated polyps 4 to 5 d after electroporation and was visible 4 wk after electroporation (SI Appendix, Fig. S2A). These data indicated that electroporation is an effective way to generate mosaic loss of function in Hydra. Mosaic loss of function has been a powerful tool in Drosophila, and we show here that this is also the case in Hydra.
To knock down HyYap, GFP-expressing polyps were electroporated with a mixture of shGFP hairpins and two different shHyYap hairpins, shHyYap1 and sHyhYap2 (SI Appendix, Table S1, shHyYap further in text). Electroporation with a combination of shGFP and shHyYap dramatically reduced both nuclear and cytoplasmic HyYap staining (Fig. 1K and K′). Importantly, HyYap staining was not affected by electroporation with a combination of shGFP and shHyYapscr (1:1 mixture of shHyYap1 scrambled and shHyYap2 scrambled, SI Appendix, Table S2) or shGFP alone (Fig. 1L and L′ and SI Appendix, Fig. S2B). Knocking down of HyYap was confirmed by qPCR and immunoblot analyses (SI Appendix, Fig. S2 C–E). Importantly, all GFP-negative cells were also HyYap-negative, i.e., all affected cells received both shGFP and shHyYap hairpins, leading to a reduction in the level of both proteins in the cell (Fig. 1K and K′).
Mammalian and Drosophila Yap homologs promote proliferation (20, 33). To determine whether Yap regulates proliferation in Hydra, we performed 5-ethynyl-2′-deoxyuridine (EdU) incorporation assays on GFP-expressing polyps electroporated with shGFP alone, with shGFP and shHyYap, or with shGFP and shHyYapscr. The graph (SI Appendix, Fig. S2H) shows the ratio of EdU-positive GFP+ cells to EdU-positive GFP− cells determined for each individual polyp 5 to 6 d after electroporation. These results demonstrate that reduction of HyYap significantly slows the cell cycle, consistent with a role for HyYap in promoting cell proliferation.
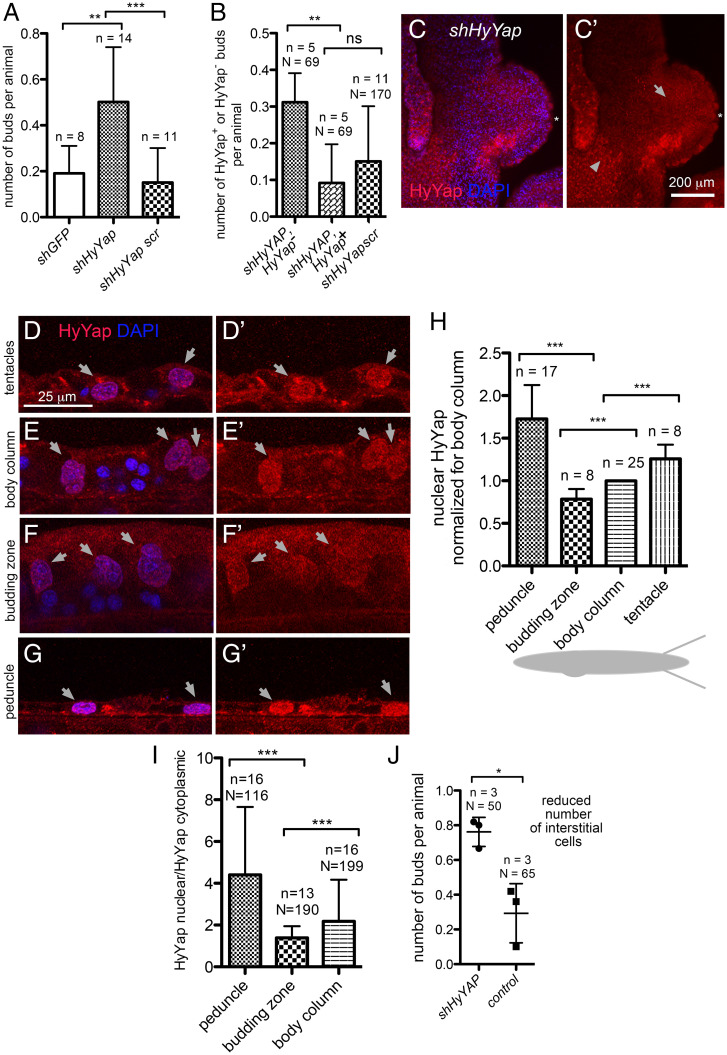
HyYap Represses Bud Formation.
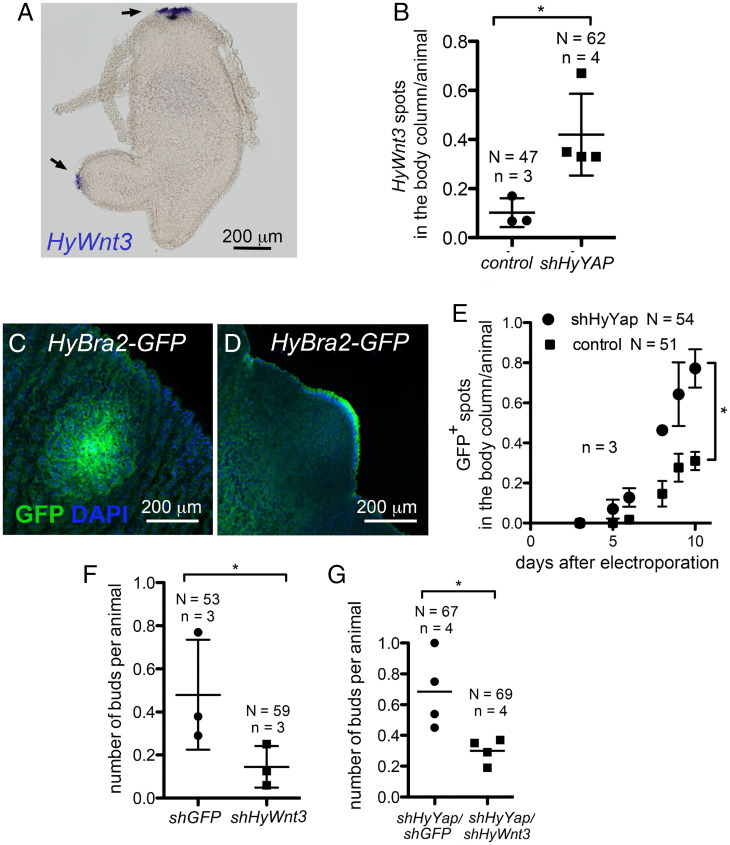
Unexpectedly, knockdown of HyYap caused a significant increase in the number of polyps with buds (Fig. 2A). All buds formed at the normal location, the budding zone. Budding normally starts with an evagination of both ectoderm and endoderm, with the epithelial cells where budding is initiated forming the tip of the bud (11). Staining for GFP and HyYap revealed that the majority of bud tips formed from cells lacking HyYap, indicating that budding was initiated in cells that had lost HyYap (Fig. 2B–C′).
Fig. 2.
HyYap is a negative regulator of Hydra budding. (A) Graph showing an increased rate of budding in polyps electroporated with shHyYap compared to controls; n, number of experiments, each experiment included 10 to 20 polyps; two-tailed unpaired t test. (B) Graph shows that a significant majority of buds developed in polyps electroporated with shHyYap originated from HyYap− tissue; n, number of experiments; N, total number of polyps used in analysis; two-tailed unpaired t test. (C and C′) Lateral view of a bud developing from HyYap− tissue; asterisk points to the tip of the bud; arrow, to HyYap− tissue; arrowhead, to HyYap+ tissue. (D–G′) Lateral view of the ectoderm of tentacles (D and D′), body column (E and E′), budding zone (F and F′), and a peduncle (G and G′) immunostained with anti-HyYap serum; arrows point to nuclei of ectodermal epithelial cells. (H) Graph shows the intensities of HyYap immunostaining in nuclei of tentacles, body column, budding zone, and a peduncle normalized for intensity in the nuclei of a body column and superimposed on a schematic drawing of Hydra; for each animal, the average intensity of immunostaining was measured in the nuclei of ectodermal epithelial (10 to 20 nuclei for each area); n, number of polyps; two-tailed unpaired t test. (I) Graph shows nuclear/cytoplasmic ratio of HyYap in the body column, budding zone, and peduncle determined by immunostaining for each cell; N, number of cells; n, number of polyps; two-tailed unpaired t test. (J) Graph shows the budding rate in animals treated with 10 mM hydroxyurea for 48 h prior to electroporation; n, number of experiments; N, number of polyps; two-tailed unpaired t test. P values are: ns (P > 0.05), * (P ≤ 0.05), **(P ≤ 0.01), *** (P ≤ 0.001).
Since down-regulation of HyYap led to bud formation, we hypothesized that budding normally occurs in areas of low HyYap expression. We quantified HyYap staining in ectodermal epithelial cells along the body column of the polyp (Fig. 2D–G′). Staining intensities were measured in the nucleus and cytoplasm for each cell. Interestingly, both the lowest level of nuclear HyYap (Fig. 2H) and the lowest nuclear/cytoplasmic ratio of HyYap were observed in the budding zone (Fig. 2I). In contrast, the highest level of nuclear HyYap, and highest nuclear/cytoplasmic HyYap ratio were observed in tentacles and the peduncle (Fig. 2H and I). The observed differences in the intensities of HyYap nuclear immunostaining could not be accounted for by the differences in the area of nuclei (SI Appendix, Fig. S2I).
Single-cell transcriptome data indicate that HyYap is expressed in the interstitial cell lineage (26, 34). To determine whether ectopic budding is caused by loss of HyYap in ectodermal epithelial cells or interstitial cells, polyps were treated with 10 mM hydroxyurea (HU) for 48 h prior to electroporation. This treatment eliminates at least 50% of interstitial cells from the body column, without affecting the number of epithelial cells (35). Increased budding was still observed upon HyYap knockdown (Fig. 2J), implying that HyYap functions in epithelial cells to restrict budding. The results are consistent with the observation that pharmacological inhibition of HyYap leads to increased budding (26). Together these data indicate that HyYap acts as a negative regulator of budding and suggested that control of the nuclear/cytoplasmic distribution of HyYap may be an important mechanism for regulating bud initiation.
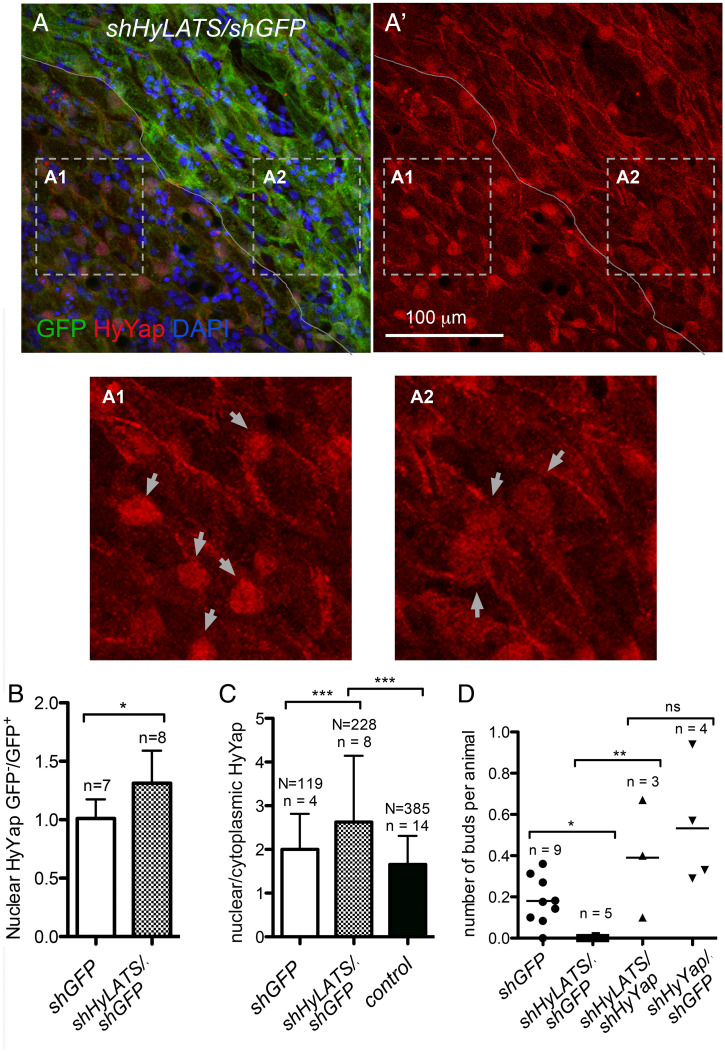
HyLATS Regulates HyYAP Localization and Bud Formation.
LATS kinases phosphorylate YAP, promoting its retention in the cytoplasm and degradation (Fig. 1G) (20, 21, 36). Thus, reduced LATS promotes active, nuclear YAP. We generated polyps mosaic for HyLATS knockdown by electroporating GFP-expressing polyps with shHyLATS/shGFP (SI Appendix, Table S1). Electroporation resulted in a significant decrease of HyLATS mRNA (SI Appendix, Fig. S2J). Importantly, immunоstaining revealed increased nuclear accumulation of HyYap in epithelial cells electroporated with shHyLATS and shGFP (Fig. 3A, A′, A1, A2, and B). The nuclear/cytoplasmic ratio of HyYap was also significantly higher in cells electroporated with shHyLATS (Fig. 3C). Thus, LATS regulates Yap nuclear localization in Hydra, as it does in bilaterians.
Fig. 3.
HyLATS regulates cellular localization of HyYap and the budding rate. (A, A′, A1, and A2) Apical view of Hydra ectoderm electroporated with shHyLATS/shGFP and immunostained for GFP and HyYap; the border between GFP+ and GFP− areas is marked; arrows point to nuclei; A1 and A2 are high magnification of GFP− and GFP+ areas. Nuclear abundance of HyYap increases in GFP− ectodermal epithelial cells. (B) Graph shows the ratio of nuclear HyYap intensities between GFP− and GFP+ areas of GFP polyps electroporated with either shGFP or shGFP/shHyYap. Note, that electroporation with shGFP alone does not affect the nuclear abundance of HyYap (GFP−/GFP+ ∼1). The average intensities of immunostaining were measured and the ratios were calculated individually for each animal; n, number of polyps; two-tailed unpaired t test. (C) Graph shows the ratio between nuclear and cytoplasmic HyYap in nonelectroporated GFP+ ectodermal epithelial cells, cells electroporated with shGFP alone, and cells electroporated with shGFP/shHyLats. Nuclear/cytoplasmic ratio was measured and calculated for each individual cell. N, number of cells; n, number of polyps; two-tailed unpaired t test. (D) Graph shows the budding rate of hydras electroporated with either shGFP (control), shHyLats/shGFP, shHyLats/shHyYap, or shHyYap/shGFP. n, number of experiments; 10 to 20 polyps were used in each experiment for each condition; two-tailed unpaired t test. P values are: ns (P > 0.05), *(P ≤ 0.05), **(P ≤ 0.01), ***(P ≤ 0.001).
Since knocking down HyLATS resulted in increased nuclear accumulation of HyYap, we expected a concomitant increase in epithelial cell proliferation. However, incorporation of EdU was not significantly higher in HyLATS knockdown cells than in GFP knockdown controls (SI Appendix, Fig. S2H). This may be explained by incomplete knockdown of HyLATS. Significantly, knocking down HyLATS halted production of buds (Fig. 3D). Critically, bud formation was rescued when animals were electroporated with a combination of shHyYap and shHyLATS (Fig. 3D). These data show that the phenotype caused by the loss of LATS (which results in higher nuclear Yap levels) can be rescued by loss of Yap. Taken together, these data indicate that the amount of nuclear HyYap in ectodermal epithelial cells of the budding zone is a controlling factor for bud formation.
To test the effects of overexpression of nuclear HyYap, we generated a transgenic Hydra line that constitutively expressed a gain-of-function mutant HyYap in ectodermal epithelial cells (SI Appendix, Fig. S3 A–C). HyYapS72A bore a S72A substitution that should make it resistant to LATS phosphorylation and subsequent cytoplasmic retention and degradation (28). The use of an operon expression construct marked HyYapS72A-expressing cells with DsRed2. We were able to establish a culture of mosaic (30 to 70% transgenic cells) HyYapS72A animals. These animals had markedly reduced budding (SI Appendix, Fig. S3D) and eventually lost the ability to propagate. Thus, loss of HyYap increases budding and loss of HyLATS or constitutive HyYap gain of function suppresses budding. These data indicate that the Hippo pathway acts as a regulator of bud formation.
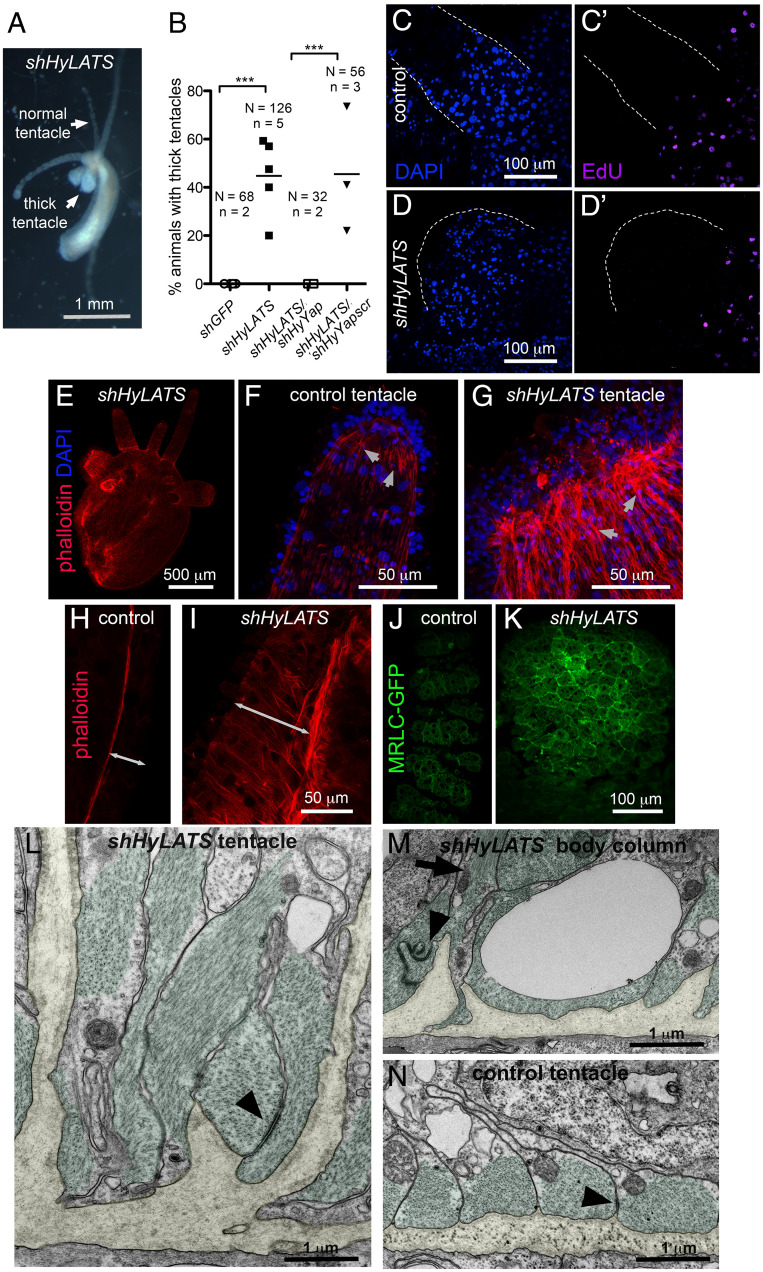
Inhibition of the Hippo Pathway Alters Hydra Morphology.
In addition to suppression of budding, knockdown of HyLATS had a strong effect on morphology. Tentacles containing cells in which HyLATS was knocked down became shorter and thicker (Fig. 4A and B). We wondered if these dramatic alterations in morphology were due to overactivation of HyYap. Significantly, double electroporation of shHyLATs and shHyYap rescued the morphological defects caused by loss of HyLATS (Fig. 4B), indicating that the thick tentacles were the result of excess HyYap activity.
Fig. 4.
Inhibition of the Hippo pathway changes Hydra morphology. (A) Live polyp electroporated with shHyLats, 14 d after electroporation. (B) Graph shows the percentage of thick tentacle formation in polyps electroporated with shGFP (control), shHyLATS alone, shHyLATS/shHyYap, and shHyLATS/shHyYapscr. N, number of polyps; n, number of experiments; two-tailed unpaired t test. (C–D′) EdU is not detected in either control (C and C′) or thick (D and D′) tentacles. Edges of tentacles are outlined by dotted lines. (E) Hydra polyp electroporated with shHyLATS and stained with phalloidin, 12 d after electroporation. Visible tear along the body column is an artifact of fixation and is common when shortened shHyLats electroporated polyps are fixed. (F and G) Ends of normal (F) and thick (G) tentacles stained with phalloidin. Arrows point to the ectodermal muscle processes that are filled with actin fibers and oriented along the length of the tentacle. (H and I) Lateral view of the ectoderm of the body columns underneath normal tentacle (H) and thick (I) tentacle stained with phalloidin. Double-headed arrows indicated the thickness of the ectoderm. (J and K) Apical view of the normal (J) and thick (K) tentacles of MRLC-GFP polyps immunostained for GFP. (L–N) Transmission electron microscopy of cross-sections visualizing the basal compartment of ectodermal epithelial cells in shHyLats tentacles (L), shHyLats body column (M), and wild-type tentacles (N). (L and M) Muscle processes (green) exhibit abnormal elongation along the apical–basal axis of the cells and sometimes ectopic positioning distant from the mesoglea. Muscle processes remain connected by normal numbers of spot desmosome-like junctions (arrowheads), but they show a dramatic loss of their parallel alignment along the polyp’s oral–aboral body axis as shown in wild-type controls (N). (L and M) At positions where the disrupted planar array of muscle fibers had gaps, the mesoglea (yellow) folded into the cytoplasm of the ectodermal epithelial cells without losing the hemidesmosome-like junctions usually located at the basal membrane surface of epithelial cells. (N) A representative image with an ectopic muscle process running along the apical–basal axis (arrow) pointing toward the mesoglea folding. ***(P ≤ 0.001)
The epithelial cells of the body column of Hydra are continually displaced into the tentacles, where they arrest in the G2 phase of the cell cycle and terminally differentiate (Fig. 1B) (9). Since the Hydra epithelial cell cycle lacks a G1 phase (37), the absence of incorporated EdU indicated that both control and LATS knockdown cells in the tentacles underwent G2 arrest (Fig. 4C–D′). HyWi, the Hydra homolog of piwi (10, 38), marks undifferentiated interstitial cells (i.e., cells that have not been displaced into the tentacles or the basal disk). Immunostaining with HyWi antibodies showed no change compared to controls (SI Appendix, Fig. S3 E–F′).
Examination of the thick tentacles and the body column below them revealed thickening of the ectoderm and an accumulation of ectopic actin fibers (Fig. 4E–I) and accumulation of myosin along the apical surface of ectodermal epithelial cells (Fig. 4J and K).
Consistent with the results of HyLATS knockdown, mosaic HyYapS72A transgenic polyps also often developed short thick tentacles (SI Appendix, Fig. S3B). HyYapS72A transgenic cells were elongated in the apicobasal direction and had ectopic actin fibers (SI Appendix, Fig. S3 G–K) as in HyLATS knockdown polyps. These data support the proposal that HyLATS and HyYAP act together in a pathway to regulate body morphology. Eventually, cells expressing HyYapS72A took over the animal and due to morphological defects the animals were unable to feed themselves and died.
To understand the cellular basis for the altered morphology, we analyzed ultrathin sections of shHyLATS thick tentacles and adjacent body column tissue using transmission electron microscopy. While the apical compartment of ectodermal epithelial cells and the endodermal layer did not exhibit obvious defects in cytoplasmic organization, there was a dramatic loss of normal structure in the basal part of ectodermal epithelial cells (Fig. 4L–N and SI Appendix, Fig. S4). HyLATS knockdown affected shape, positioning, and orientation of muscle processes. We observed gaps in the planar array of these processes, individual processes elongated along the cells apical–basal axis, and some detached from the mesoglea. The parallel alignment of neighboring processes was strongly disrupted and randomized, sometimes even the parallel alignment of actin filaments within a muscle process was lost (Fig. 4L and M). There was a clear loss of planar polarity in this tissue layer. Interestingly, the mesoglea adjacent to the shHyLATS knockdown cells showed folding toward the apical surface of the ectoderm (Fig. 4L and M and SI Appendix, Fig. S4 C and D). In the body column, we detected muscle processes apical to the tip of mesoglea folding running perpendicular to their normal planar orientation (Fig. 4M). Thus, disruption of the normal, nonfolded mesoglea sheet may be a result of contraction of these ectopic muscle fibers along the epithelial cell’s apical–basal axis. shHyLATS tentacle and body column tissue both showed these defects, but knockdown phenotypes were clearly stronger in the tentacles than in the body column.
The Hippo Pathway Acts Upstream of the Canonical Wnt Pathway during Budding.
The canonical Wnt pathway is activated early in budding and induces axis formation in Hydra (4, 6, 12). Experimental activation of the Wnt pathway in the Hydra body column leads to induction of ectopic axes, and inhibition of Wnt signaling blocks axis formation (4, 17). Since knocking down of HyYap led to increased bud formation, we hypothesized that the Hippo pathway lies upstream of Wnt/β-catenin signaling. In early buds, HyWnt3 expression is seen in a patch of 15 to 20 cells, soon after thickening of the ectoderm and expression, and remains at the tip of the growing bud (12) (Figs. 1E and 5A). To test the effects of the Hippo pathway on canonical Wnt signaling, we first examined expression of HyWnt3 upon loss of HyYap. Significantly, more polyps electroporated with shHyYap had HyWnt3 patches in the budding zone than controls (Fig. 5B).
Fig. 5.
HyYap acts upstream of canonical Wnt signaling during budding. (A). Whole mount in situ hybridization of Hydra with anti-HyWnt3 probe. Arrows point to areas of HyWn3t expression. (B) Graph shows the increased number of HyWnt3 spots in the body column of polyps electroporated with shHyYap; two-tailed unpaired t test. (C and D) Anti-GFP immunostaining of HyBra2-GFP polyps electroporated with shHyYap, 6 d after electroporation. (C) Apical view. (D) Lateral view of GFP+ bud. (E) Graph shows the increased number of GFP+ spot in the body column of HyBra2-GFP polyps electroporated with shHyYap; two-tailed unpaired t test. (F) Graph shows decreased budding rate in polyps electroporated with shHyWnt compared to Hydras electroporated with shGFP. N, number of animals; n, number of experiments; one-tailed paired t test. (G) Graph shows the decreased budding rate in polyps electroporated with shHyYap/shHyWnt compared with Hydras electroporated with shHyYap alone; two-tailed unpaired t test. *(P ≤ 0.05)
We also examined expression of HyBra2, a hypostome-specific gene, which is induced by Wnt expression (39). HyBra2 is expressed early in budding and expression persists in the hypostome of the growing bud (39). We used a transgenic Hydra line that expresses GFP under the control of the HyBra2 promoter (40) to assay induction of budding upon loss of HyYap (Fig. 5 C and D). GFP patches were observed in the budding zone in a significantly higher number of animals upon electroporation with shHyYap compared to controls (Fig. 5E), indicating that HyBra2 expression is activated upon loss of HyYap.
Knockdown of shHyWnt3 alone results in reduced production of buds, as expected from the key role of HyWnt3 in axis initiation (41) (Fig. 5F). To test whether budding by loss of HyYap is mediated by increased HyWnt3, we simultaneously knocked down HyYap and HyWnt3. Importantly, the increased budding that resulted from down-regulation of HyYap was suppressed when shHyWnt3 was electroporated along with shHyYap (Fig. 5G). These data indicate that HyYAP acts upstream of HyWnt3 expression to suppress bud formation in Hydra. The Hippo pathway thus integrates growth and axis formation upstream of Wnt signaling.
Discussion
The canonical Wnt (Wnt/β-catenin) pathway is both necessary and sufficient for axis formation in cnidarians: Transplantation of the hypostome, the organizer, into a body column of a host, or experimental induction of canonical Wnt signaling, leads to formation of a new axis (4, 5, 7). However, in Hydra, the onset of HyWnt3 expression occurs after the first signs of bud formation (12) pointing to events regulating budding upstream of HyWnt3. Early studies connected budding to the cell cycle and cell displacement in Hydra (9, 10, 42); however, molecular mechanisms connecting these processes were never illuminated.
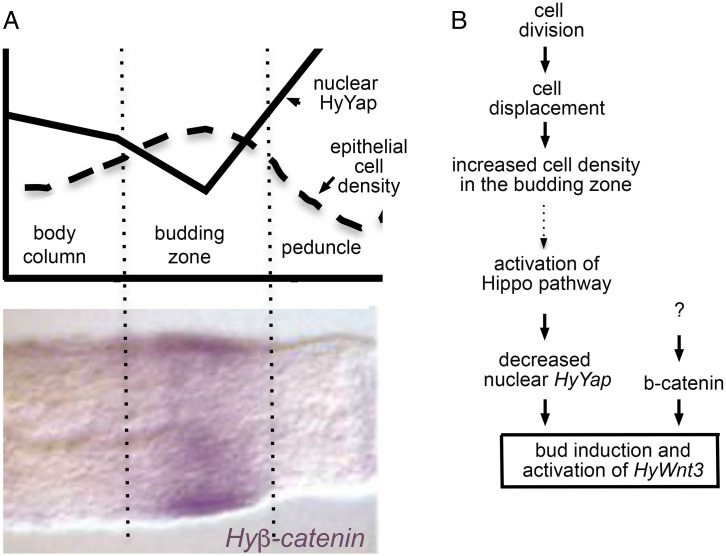
In a normally fed Hydra polyp, the rate of cell division is similar along the body column (10). Due to continuous cell division, cells below the subtentacle zone are being pushed down the body column (10). In the budding zone, excess cells are forced out of the maternal axis to form a new axis, a bud (9). Buds do not form in starved Hydra, since the rate of cell division cannot compensate for the rate of cell loss at the ends (15). Ectodermal epithelial cells that are about to form a bud are packed tighter than cells of the body column: they elongate in an apicobasal direction and reduce the area of attachment to mesoglea, the basement membrane (Fig. 1F) (13). Nuclear localization of mammalian and Drosophila Yap is negatively regulated by cell density (21, 28). Similarly, we find that in Hydra, highly packed cells of the budding zone have less nuclear HyYap than body column cells and especially less than the flat cells of the peduncle and tentacles (Figs. 2E–G′ and 6A). Since down-regulation of HyYap leads to induction of budding, we suggest that HyYap might act as a molecular link between continuous cell division and budding (Fig. 6B). A seeming paradox is that loss of HyYap reduces proliferation. However, knocking down of HyYap does not completely block epithelial cell division as we have shown here. In addition, when HyYap is knocked down locally, the rest of the body column cells continue to divide, pushing electroporated cells into the budding zone and aboral end.
Fig. 6.
The proposed role of the Hippo pathway in axis formation in Hydra. (A) Schematic drawings of the axial distributions of epithelial cell density (adapted from ref. 10) and nuclear HyYap superimposed over Hyβ-catenin in situ (adapted from ref. 12). (B) Hypothetical model identifying HyYap as a molecular link between cell division and axis formation in Hydra. Dotted arrow indicates causal connection based on analogy with Drosophila and mammals that yet has to be demonstrated in Hydra.
We show that the Hippo pathway regulates budding upstream of canonical Wnt signaling. Yap and its homologs Yorkie and TAZ also bind and inhibit Wnt signaling by binding Disheveled (43, 44). Yap and TAZ also bind and inhibit β-catenin (45). Interestingly, buds that were induced by knocking down of HyYap always formed at the normal location, the budding zone, despite the larger knockdown area. Intriguingly, expression of Hyβ-catenin is also higher in the budding zone than in surrounding body column tissue (Fig. 6A) (12). Hyβ-catenin can directly activate transcription of HyWnt3 (16). Thus, induction of HyWnt3 through release of Hyβ-catenin upon knocking down of HyYap is a possible scenario. Also required for budding is noncanonical Wnt signaling. Noncanonical Wnt activation occurs in the densely packed ectodermal cells early in budding, before the onset of HyWnt3 expression, and depends on Hyβ-catenin (17).
Our data clearly show that HyYap acts upstream of HyWnt3 expression; however, it is not clear how HyYAP affects HyWnt3 transcription. One potential hypothesis is that HyYAP could bind to HyWnt3 regulatory regions. We examined the upstream region of the HyWnt3 gene for canonical YAP/TEAD binding sites (SI Appendix, Fig. S5). We found several such binding sites, raising the possibility that YAP/TEAD complexes could bind directly to the regulatory regions of the HyWnt3 gene. YAP has been shown to have both coactivator and corepressor functions. However, it is also quite possible that increasing bud formation by reducing YAP indirectly affects Wnt3 expression. To determine whether these sites are responsible for the induction of HyWnt3 expression during budding will require mutation of these sites and testing them in vivo during budding (using approaches similar to those in ref. 16) and determining the effects of gain and loss of HyYap and HyLATS on reporter expression.
Budding is not the only mode for axis formation in Hydra. Axis formation, which is controlled by the canonical Wnt signaling, also occurs during head regeneration (14) and during development of the aggregate, a self-organization of a clump of dissociated Hydra cells into multiple axes (13). Development of the aggregate is a great experimental system to study the mechanisms of Hydra axis formation, but does not occur in nature. Here, we focus our studies on the role of the Hippo pathway during animal budding, a major form of Hydra reproduction.
Inhibition of the Hippo pathway by knocking down HyLATS results in dramatic shortening and thickening of tentacles. Epithelial cell cycle arrest and the absence of undifferentiated interstitial cells suggest proper differentiation of the HyLATS mutant tentacle tissue. However, dramatic change of epithelial cell morphology in HyLATS knockdown tentacles leaves a possibility of the axial patterning being altered and calls for a more detailed investigation. Immunofluorescence (IF) and electron microscopy (EM) analyses show that HyLATS knockdown leads to major changes in the actin cytoskeleton and epithelial cell shape. Interestingly, inhibition of Wart (LATS) in Drosophila stimulates polymerization of F-actin in a Yorkie-independent manner, by acting on the capping protein (46, 47). In contrast, in mammalian cells, activation of F-actin requires the transcription activity of Yap (48). In Hydra, knocking down of HyYap rescues the shLATS phenotype, suggesting a role for HyYap transcriptional control of actin. Abnormal polymerization of actin could explain the randomized orientation of the muscle processes, misfolding of mesoglea, and thickening of the ectoderm seen in the HyLATS knockdown.
To summarize, we show that the conserved LATS-Yap-Hippo signaling pathway plays a major role in morphogenesis in the cnidarian Hydra and acts upstream of expression of Wnt3 and canonical Wnt signaling during axis initiation. Our findings demonstrate that, similar to Drosophila and mammals, in Hydra the amount of nuclear Yap is reduced in the area of high cell density, and that reduction of nuclear Yap leads to activation of Wnt/β-catenin signaling and budding (Fig. 6B). We speculate that Hippo signaling, and nuclear Yap serve to link continuous cell division, cell density, and axis formation early in metazoan evolution (Fig. 6B).
Materials and Methods
Animal and Culture Condition.
The AEP strain of H. vulgaris was cultured at 18 °C in Hydra medium (1.0 mM CaCl2, 1 mM NaHCO3, 0.1 mM MgCl2, 0.03 mM KNO3, 1 mM Tris HCl pH 7.8, and fed with Artemia nauplii every 2 d.
In Situ Hybridization.
All procedures at room temperature were carried out with rotation on a nutator. Animals starved for 48 h were relaxed in 2% urethane for 2 min and fixed in 4% paraformaldehyde O/N at 4 °C. Samples were washed for 5 min each in 100% ethanol, three times; ethanol:PBT 3:1, once; ethanol:PBT 1:1, once; ethanol:PBT 1:3, once; and PBT, three times, following by treatment with proteinase K (10 μg/mL) and 4 mg/mL glycine for 10 min each. Next, samples were treated with 0.1 M triethanolamine (pH 7.8) twice and 0.25% (vol/vol) acetic anhydride in 0.1 M triethanolamine (pH 7.8) twice for 5 min each, washed twice with PBT for 5 min, and postfixed with 4% PFA for 20 min. Fixator was removed by fives washes with PBT for 5 min each wash. Next, the endogenous alkaline phosphatase was removed by heating samples at 80 °C for 30 min and washed sequentially once in PBT, PBT:hybridization buffer (HB) once, and HB once for 10 min each. Samples were prehybridized in HB at 55 °C for 2 h. Then digoxygenin-labeled RNA probe was added and hybridization was carried out for 48 to 60 h at 55 °C. Hybridization solution (HS) was composed of 50% formamide, 5×SSC (750 mM NaCl, 75 mM sodium citrate), 0.02% (wt/vol) each Ficoll, bovine serum albumin (BSA, fraction V), and polyvinylpyrolidone, 200 mg/mL yeast tRNA, 100 mg/mL heparin, 0.1% Tween-20, and 0.1% CHAPS. To remove unhybridized probe, samples were washed at 55 °C for 10 min each in HS, HS:2×SSC 3:1, HS:2×SSC 1:1, HS:2×SSC 1:3, following by two 30-min washes with 0.1% CHAPS in 2×SSC. In a preparation for binding with the anti-dioxygenin antibody, samples were moved at room temperature and washed twice for 10 min in MAB (100 mM maleic acid, 150 mM NaCl, pH 7.5), then for 30 min in 1% BSA in MAB, and then blocked for 2 h in blocking solution (BS) (80% MAB-BSA, 20% heat-inactivated sheep serumn). Alkaline phosphatase-conjugated anti-digoxy genin Fab fragments were diluted 1:400 in BS and preabsorbed for at least 2 h against fixed Hydra. The preabsorbed Fab fragments were diluted to a final dilution of 1:2,000 in BS and incubated with samples overnight at 4 °C. The next day, the unbound antibodies were removed by eight washes with MAB for 30 min each, samples were equilibrated with the alkaline phosphatase staining buffer NTMT (100 mM NaCl, 100 mM Tris, pH 9.5, 50 mM MgCl2, 0.1% Tween-20) in three 5-min washes, with the final wash also containing 1 mM levamisole. Alkaline phosphatase reaction was carried out at 37 °C in NTMT in the presence of 5 μL/mL p-nitroblue tetrazolium chloride (NBT) and 3.75 l/mL 5-bromo-4-chloro-3-indolyl phosphate (BCIP) in the dark. Reaction was stopped with EtOH, refixed in 3.7% formaldehyde, dehydrated in ethanol series, 2 min each (70% EtOH once, 95% EtOH once, and 100% EtOH twice) and mounted in Euparal. A 128- to 663-bp segment of Hydra Wnt3 coding sequence (accession No. AF272673) was used to make an in situ probe. Digoxygenin-labeled RNA probes were made according to the protocol supplied by Roche.
Database Search and Phylogenetic Analysis.
To identify cnidarian homologs of Fat-like, Ds and CELSR proteins, we searched The National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov) and National Human Genome Research Institute (NHGRI) (https://research.nhgri.nih.gov) databases. We have identified H. vulgaris homologs of Yap (NM_001309649, NP_001296578), Hippo (MST) (XM_004212124, MW650879). and LATS (XM_012698864, MW650881). Sequences used in the analysis and their accession Nos. are as follows: DmYorkie (DQ099897), MmYap1 (BC094313), HsYap2 (AAP92710), HsTAZ (AJ299431), NvYap (XM_001627445.2), DrLATS1 (XM_005160312), DrLATS2 (NM_001128256), DmLATS (Warts) (U29608), HsLATS1 (AF104413), NvLATS (XM_001628046), HsMST (U18297), DmMST (Hippo) (NM_001274163), NvMST (XM_032384310), and DrMST (BC164215). For generation of the phylogenetic tree, the sequences were aligned using MAFFT (Multiple Alignment using Fast Fourier Transform) (https://www.ebi.ac.uk/Tools/msa/mafft/) or Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) and analyzed using Akaike Information Criterion (www.atgc-montpellier.fr).
Production of Antibodies.
A peptide corresponding to residues 1 to 159 of HyYap was expressed as GST-fusions using the GEX4t-1 vector (Millipore) in Escherichia coli strain BL21 and purified on glutathione-agarose (Thermo Scientific). Purified protein was used to immunize guinea pigs (Cocalico Biologicals).
Immunoblot and Immunofluorescence Analysis.
For immunoblotting, polyps were dissolved in lysis buffer (1% Sodium Dodecyl Sulfate (SDS); 10% glycerol; 30 mM Tris⋅HCl, pH 6.8) containing 2% beta-mercaptoethanol, boiled for 5 min, chilled on ice for 5 min, and electrophoresed in a 10% Sodium dodecyl-sulfate polyacrylamide gel. Transfer of the proteins onto a polyvinylidene difluoride (PVDF) membrane was done in transfer buffer (20% methanol, 25 mM Tris base, 192 mM glycine) overnight at 4 °C. The membranes were incubated with antibodies (total anti-HyYap serum was used at 1:1,000 dilution, anti-actin [clone C4, Millipore] at 1:2,000 dilution, anti-GAPDH [Sigma] at 1:1,000 dilution) in blocking buffer (TBS-Tween 0.1% containing 5% powdered milk) for 90 min at room temperature (RT). After 3 × 10 min washes with Tris-buffered saline with 0.1% Tween (TBS-T), membranes were incubated with Horseradish Peroxidase HRP-conjugated secondary antibody (GE Healthcare) diluted 1:10,000 in blocking buffer for 1 h at RT. Visualization was done by enhanced chemiluminescence (ECL) detection (Thermo Scientific).
To perform immunocytochemistry on whole mounts, animals were relaxed in 2% urethane for 2 min and then fixed in either Lavdovski’s fixative (ethanol∶formaldehyde∶acetic acid∶H2O 50∶10∶4∶36) for HyYap and GFP antibodies, or in 4% paraformaldehyde for phalloidin staining overnight at 4 °C. Then, animals were washed 3 × 10 min in PBT Phosphate-buffered saline (PBS) with 0.1% Triton); animals fixed with 4% paraformaldehyde were permeabilized in PBS with 1.0% Triton for 30 min and incubated in PBT with 2% Bovine serum albumin (BSA) for 1 h. Samples were incubated with primary antibodies overnight at 4 °C. Then, animals were washed 3 × 10 min PBT, incubated with secondary antibodies for 30 min, washed with PBT 3 × 10 min and mounted using Vectashield mounting medium containing DAPI (Vector Laboratories, Inc). Primary antibody dilutions were as follows: anti-HyYap total serum, 1:1,000; anti-GFP (Abcam, ab13970). Fluor-conjugated secondary antibodies (The Jackson Laboratory) were used at 1:400 dilution. Alexa 555-phalloidin (Abcam) was used at 1:2,000 dilution.
Measurement of Immunofluorescent Intensity.
To measure the intensity of immunofluorescence 1.5 μm z-stack confocal images were analyzed by NIS-Elements AR Analysis. To compare intensities of immunostaining in the nuclei and the cytoplasm of the one cell, the intensities of the area covering about half of the nucleus and the area of a similar size just outside of the nucleus were measured. Normalization of the intensity of immunofluorescence in nuclei for the nuclear size was calculated individually for each animal using the equation (<ν> × <Α>)/(<ν>bc × <Α>bc), where <ν> is an average intensity of immunofluorescence, <Α> is the average projection area of nuclei, and <ν>bc and <Α>bc, values obtained in the body column.
shRNA Production and Electroporation.
shRNAs were designed and made according to ref. 31. For each gene, two hairpins were synthesized (SI Appendix, Table S1) and both were used for electroporation at 1:1 ratio. Electroporation procedure was performed according to ref. 29.
Transmission Electron Microscopy.
Transmission electron microscopy was done according to standard protocols (49). shHyLats polyps and wild-type controls were relaxed with 2% urethane in Hydra medium for 3 min and then fixed with a mixture of glutaraldehyde and osmium tetroxide in phosphate buffer according to ref. 50 on ice for 2 h. Samples were dehydrated in an increasing series of acetone and embedded into EMBed812 epoxy resin. The 80-nm ultrathin sections were cut with an ultracut UCT mictrotome (Leica) using a Diatome Diamond knife (Diatome). Sections were mounted on grids and stained with lead citrate and examined with Libra 120 energy filter transmission electron microscope (Zeiss). Images were acquired with a 2 × 2k high speed camera and an ImageSP software (Tröndle).
Supplementary Material
Acknowledgments
We thank Joe Culotti (Lunenfeld Tanenbaum Research Institute, Toronto) for invaluable support of the project and help in writing the manuscript, Leonid Brown (University of Guelph) for help with culturing Hydra, Marina Gertsenstein (Toronto Center for Phenogenomics) for help with the electroporation procedure, Thomas Bosch and Alexander Klimovich (University of Kiel) for providing GFP transgenic Hydra, Taylor Skokan (University of California, San Francisco) for providing the MRLC-GFP transgenic Hydra, and Celina Juliano (University of California, Davis) for providing anti-HyWi antibody. H.M. is supported by funding from the Barnes-Jewish/Christian investigator program and is a Larry J. Shapiro and Carol-Ann Uetake-Shapiro Professor. B.H. is supported by the European Commission H2020 Marie Sklodowska-Curie COFUND research grant No. 847681 Ageing, Regeneration and Drug Research. R.S. is supported by Grant 1R24GM080537-01A1 from the National Institute of General Medical Sciences.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2203257119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information. All sequences used in the study are available through NCBI and NHGRI databases (51–53).
References
- 1.Meinhardt H., Models of biological pattern formation: From elementary steps to the organization of embryonic axes. Curr. Top. Dev. Biol. 81, 1–63 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Niehrs C., On growth and form: A Cartesian coordinate system of Wnt and BMP signaling specifies bilaterian body axes. Development 137, 845–857 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Petersen C. P., Reddien P. W., Wnt signaling and the polarity of the primary body axis. Cell 139, 1056–1068 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Broun M., Gee L., Reinhardt B., Bode H. R., Formation of the head organizer in hydra involves the canonical Wnt pathway. Development 132, 2907–2916 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Browne E. N., The production of new hydrants in hydra by the insertion of small grafts. J. Exp. Zool. 7, 1–37 (1909). [Google Scholar]
- 6.Gee L., et al. , beta-catenin plays a central role in setting up the head organizer in hydra. Dev. Biol. 340, 116–124 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Trevino M., Stefanik D. J., Rodriguez R., Harmon S., Burton P. M., Induction of canonical Wnt signaling by alsterpaullone is sufficient for oral tissue fate during regeneration and embryogenesis in Nematostella vectensis. Dev. Dyn. 240, 2673–2679 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trembley A., Mémoires pour servir à l'histoire d'un genre de polypes d'eau douce, à bras en forme de cornes (J. & H. Verbeek, Leiden, 1744), p. xv, 324 p. [Google Scholar]
- 9.Campbell R. D., Tissue dynamics of steady state growth in Hydra littoralis. II. Patterns of tissue movement. J. Morphol. 121, 19–28 (1967). [DOI] [PubMed] [Google Scholar]
- 10.Campbell R. D., Tissue dynamics of steady state growth in Hydra littoralis. I. Patterns of cell division. Dev. Biol. 15, 487–502 (1967). [DOI] [PubMed] [Google Scholar]
- 11.Otto J. J., Campbell R. D., Budding in Hydra attenuata: Bud stages and fate map. J. Exp. Zool. 200, 417–428 (1977). [DOI] [PubMed] [Google Scholar]
- 12.Hobmayer B., et al. , WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature 407, 186–189 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Graf L., Gierer A., Size, shape and orientation of cells in budding hydra and regulation of regeneration in cell aggregates. Wilehm Roux Arch Dev Biol 188, 141–151 (1980). [DOI] [PubMed] [Google Scholar]
- 14.Bode H. R., Axial patterning in hydra. Cold Spring Harb. Perspect. Biol. 1, a000463 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otto J. J., Campbell R. D., Tissue economics of hydra: Regulation of cell cycle, animal size and development by controlled feeding rates. J. Cell Sci. 28, 117–132 (1977). [DOI] [PubMed] [Google Scholar]
- 16.Nakamura Y., Tsiairis C. D., Özbek S., Holstein T. W., Autoregulatory and repressive inputs localize Hydra Wnt3 to the head organizer. Proc. Natl. Acad. Sci. U.S.A. 108, 9137–9142 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Philipp I., et al. , Wnt/beta-catenin and noncanonical Wnt signaling interact in tissue evagination in the simple eumetazoan Hydra. Proc. Natl. Acad. Sci. U.S.A. 106, 4290–4295 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aufschnaiter R., Wedlich-Söldner R., Zhang X., Hobmayer B., Apical and basal epitheliomuscular F-actin dynamics during Hydra bud evagination. Biol. Open 6, 1137–1148 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng Z., Moroishi T., Guan K. L., Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang J., Wu S., Barrera J., Matthews K., Pan D., The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122, 421–434 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Zhao B., et al. , Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reginensi A., et al. , A critical role for NF2 and the Hippo pathway in branching morphogenesis. Nat. Commun. 7, 12309 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sebé-Pedrós A., Zheng Y., Ruiz-Trillo I., Pan D., Premetazoan origin of the hippo signaling pathway. Cell Rep. 1, 13–20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coste A., Jager M., Chambon J. P., Manuel M., Comparative study of Hippo pathway genes in cellular conveyor belts of a ctenophore and a cnidarian. Evodevo 7, 4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hilman D., Gat U., The evolutionary history of YAP and the hippo/YAP pathway. Mol. Biol. Evol. 28, 2403–2417 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Unni M., Reddy P. C., Pal M., Sagi I., Galande S., Identification of components of the Hippo pathway in Hydra and potential role of YAP in cell division and differentiation. Front. Genet. 12, 676182 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basu S., Totty N. F., Irwin M. S., Sudol M., Downward J., Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol. Cell 11, 11–23 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Oh H., Irvine K. D., In vivo regulation of Yorkie phosphorylation and localization. Development 135, 1081–1088 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogg M. C., et al. , An evolutionarily-conserved Wnt3/β-catenin/Sp5 feedback loop restricts head organizer activity in Hydra. Nat. Commun. 10, 312 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe H., et al. , Nodal signalling determines biradial asymmetry in Hydra. Nature 515, 112–115 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Karabulut A., He S., Chen C. Y., McKinney S. A., Gibson M. C., Electroporation of short hairpin RNAs for rapid and efficient gene knockdown in the starlet sea anemone, Nematostella vectensis. Dev. Biol. 448, 7–15 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Wittlieb J., Khalturin K., Lohmann J. U., Anton-Erxleben F., Bosch T. C., Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 6208–6211 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Overholtzer M., et al. , Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc. Natl. Acad. Sci. U.S.A. 103, 12405–12410 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siebert S., et al. , Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science 365, eaav9314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bode H. R., Flick K. M., Smith G. S., Regulation of interstitial cell differentiation in Hydra attenuata. I. Homeostatic control of interstitial cell population size. J. Cell Sci. 20, 29–46 (1976). [DOI] [PubMed] [Google Scholar]
- 36.Hao Y., Chun A., Cheung K., Rashidi B., Yang X., Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 283, 5496–5509 (2008). [DOI] [PubMed] [Google Scholar]
- 37.David C. N., Campbell R. D., Cell cycle kinetics and development of Hydra attenuata. I. Epithelial cells. J. Cell Sci. 11, 557–568 (1972). [DOI] [PubMed] [Google Scholar]
- 38.Teefy B. B., Siebert S., Cazet J. F., Lin H., Juliano C. E., PIWI-piRNA pathway-mediated transposable element repression in Hydra somatic stem cells. RNA 26, 550–563 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bielen H., et al. , Divergent functions of two ancient Hydra Brachyury paralogues suggest specific roles for their C-terminal domains in tissue fate induction. Development 134, 4187–4197 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Glauber K. M., et al. , A small molecule screen identifies a novel compound that induces a homeotic transformation in Hydra. Development 140, 4788–4796 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lengfeld T., et al. , Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev. Biol. 330, 186–199 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Webster G., Hamilton S., Budding in hydra: The role of cell multiplication and cell movement in bud initiation. J. Embryol. Exp. Morphol. 27, 301–316 (1972). [PubMed] [Google Scholar]
- 43.Barry E. R., et al. , Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature 493, 106–110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varelas X., et al. , The Hippo pathway regulates Wnt/beta-catenin signaling. Dev. Cell 18, 579–591 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Imajo M., Miyatake K., Iimura A., Miyamoto A., Nishida E., A molecular mechanism that links Hippo signalling to the inhibition of Wnt/β-catenin signalling. EMBO J. 31, 1109–1122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernández B. G., et al. , Actin-Capping Protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development 138, 2337–2346 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Lucas E. P., et al. , The Hippo pathway polarizes the actin cytoskeleton during collective migration of Drosophila border cells. J. Cell Biol. 201, 875–885 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calvo F., et al. , Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat. Cell Biol. 15, 637–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holstein T. W., Hess M. W., Salvenmoser W., Preparation techniques for transmission electron microscopy of Hydra. Methods Cell Biol. 96, 285–306 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Shigenaka Y., Roth L. E., Pihlaja D. J., Microtubules in the heliozoan axopodium. 3. Degradation and reformation after dilute urea treatment. J. Cell Sci. 8, 127–151 (1971). [DOI] [PubMed] [Google Scholar]
- 51.Brooun M., et al. , The Hippo pathway regulates axis formation and morphogenesis in Hydra. HyYap. https://research.nhgri.nih.gov/HydraAEP/genewiki/gene_page.cgi?gene=HVAEP1.G001931&sp=hvaep. [DOI] [PMC free article] [PubMed]
- 52.Brooun M., et al. , The Hippo pathway regulates axis formation and morphogenesis in Hydra. HyMST. https://research.nhgri.nih.gov/HydraAEP/genewiki/gene_page.cgi?gene=HVAEP4.G007579&sp=hvaep. [DOI] [PMC free article] [PubMed]
- 53.Brooun M., et al. , The Hippo pathway regulates axis formation and morphogenesis in Hydra. HyLATS. https://research.nhgri.nih.gov/HydraAEP/genewiki/gene_page.cgi?gene=HVAEP3.G006729&sp=hvaep. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information. All sequences used in the study are available through NCBI and NHGRI databases (51–53).