Significance
Pathogens can drive rapid evolution in the species they infect, providing a model for how adaptations arise. We found that some genotypes of the fruit fly Drosophila melanogaster are resistant to a common viral pathogen called Drosophila A virus. Resistance is caused by a transposable element that has inserted into the gene Veneno, resulting in the gene encoding a shortened protein. The original form of the protein has no effect on virus resistance, but when truncated it gains a new and potent antiviral function, providing a large fitness advantage to infected flies. This demonstrates a novel mechanism by which transposable elements can generate adaptive phenotypes.
Keywords: Drosophila, transposable element, adaptation, virus, Tudor domain
Abstract
Hosts are continually selected to evolve new defenses against an ever-changing array of pathogens. To understand this process, we examined the genetic basis of resistance to the Drosophila A virus in Drosophila melanogaster. In a natural population, we identified a polymorphic transposable element (TE) insertion that was associated with an ∼19,000-fold reduction in viral titers, allowing flies to largely escape the harmful effects of infection by this virulent pathogen. The insertion occurs in the protein-coding sequence of the gene Veneno, which encodes a Tudor domain protein. By mutating Veneno with CRISPR-Cas9 in flies and expressing it in cultured cells, we show that the ancestral allele of the gene has no effect on viral replication. Instead, the TE insertion is a gain-of-function mutation that creates a gene encoding a novel resistance factor. Viral titers remained reduced when we deleted the TE sequence from the transcript, indicating that resistance results from the TE truncating the Veneno protein. This is a novel mechanism of virus resistance and a new way by which TEs can contribute to adaptation.
Pathogen infection is an important determinant of fitness in natural populations, and the nature of this selection pressure is continually changing as new pathogens appear or existing pathogens evolve to escape host defenses. This can result in rapid evolutionary change and continual innovation in the immune defenses of animals. Identifying these new defenses can therefore provide insights into the genetic basis of adaptation and the mechanisms by which hosts counter infection in nature.
Transposable elements (TEs) can be an important source of genetic variation that natural selection can act on to generate new adaptations. In many species, including Drosophila melanogaster, TEs are highly dynamic within populations (1). Studies of both mutation accumulation lines and the frequency of TEs in populations have provided compelling evidence that TE insertions tend to be deleterious (2–4). However, TEs are also an important source of beneficial mutations that give rise to adaptations (5). A common mechanism of this involves the TE inserting upstream of genes and altering their expression. This is especially prevalent in the evolution of insecticide resistance, where TEs can lead to the up-regulation of detoxification enzymes (6–8). In other cases, TEs or retroviruses have been domesticated and play important roles in host biology (9). For example, Syncytin genes, which play a role in nutrient transfer across the placenta, have arisen through multiple independent domestication events across mammals and a species of placental lizard (10, 11). In mammals, TEs and endogenous retroviruses have played an important role in the evolution of the immune system, giving rise to enhancers regulating the expression of genes in response to interferon, and the RAG proteins that cleave DNA during V(D)J recombination (12, 13). In D. melanogaster TEs affect the expression of immunity genes (14), and we have reported a Doc element insertion that is associated with resistance to the virus DMelSV (15, 16).
In insects and other invertebrates, the absence of an adaptive immune system means infection must be controlled by innate immune defenses (17), with the RNA interference (RNAi) pathway being a key defense against viruses (18). While these core immune pathways evolve rapidly (19, 20), it is unclear what role they play in the evolution of resistance. In humans, a key component of antiviral defenses is provided by a diverse collection of proteins that can inhibit viral replication by targeting almost any step of the viral replication cycle (21). Many of these are dominantly acting cell-autonomous molecules known as restriction factors. Viruses have frequently evolved mechanisms to escape restriction factors and the restriction factors are often under positive selection, suggesting they are involved in an evolutionary arms race (21). Restriction factors have been little-studied in insects, but we have described several polymorphic genes in Drosophila that have large effects on viral replication (15, 16, 22–24). It is likely these play a critical role in host-virus evolution in insects, analogous to restriction factors in mammals.
Despite approximately a third of flies in wild populations of D. melanogaster being infected with one or more viruses (25), many studies of antiviral immunity have used viruses that are rare or absent in nature. For this reason, we investigated resistance to DAV (Drosophila A virus), which typically infects about 5% of flies in natural populations of D. melanogaster (25, 26). DAV is a positive-sense single-stranded RNA virus that is related to the Permutotetraviridae (25, 27). Little is known about its interaction with Drosophila or its effects on the health of infected insects. To understand the evolution of virus resistance, we used a panel of inbred fly lines with publicly available whole genome sequences (28) to investigate the genetic factors that cause variation in susceptibility to DAV in nature.
Results
Some Genotypes in a Natural Population Are Resistant to DAV.
We investigated genetic variation in susceptibility to DAV, which is a common virus in natural populations of D. melanogaster (25). We used 182 inbred lines of flies from the Drosophila Genetic Reference Panel (DGRP) collection, that were derived from flies collected from a natural population in North Carolina (28). Across these fly lines, we infected a total of 11,985 female flies, extracted RNA from groups of 15 flies 3 d postinfection (dpi), and estimated viral titers by qPCR.
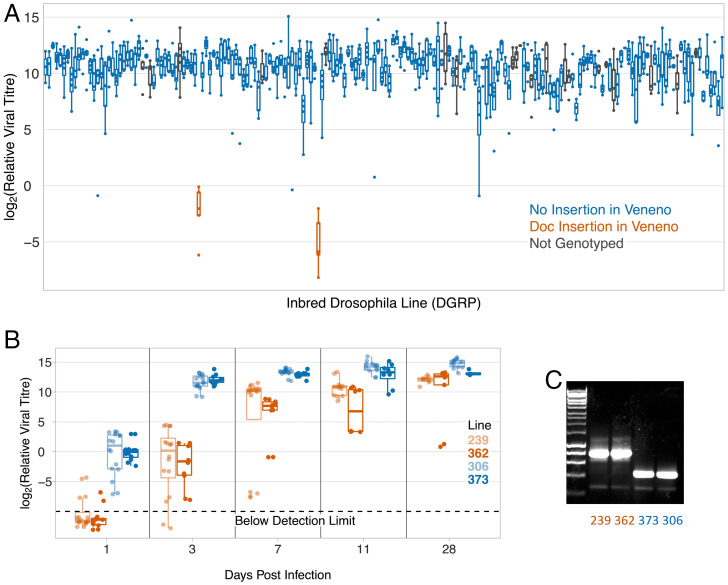
We found that there was substantial genetic variation in susceptibility to DAV, with 50% of the variance in viral titer being explained by genetic differences between the lines (Fig. 1A; 95% CI, 43 to 57%). Two lines were extremely resistant, having a mean viral titer ∼19,000 times lower than the rest of the lines (Fig. 1A). These lines explain the majority of the genetic variance in DAV resistance, and if they are excluded from the dataset only 20% of the variance in viral titer is explained by genetic effects. Over a time-course the reduction in viral titer was greatest shortly after infection, and by 28 dpi the viral titer in the resistant flies was only slightly lower than in the susceptible flies (Fig. 1B; ANOVA, days postinfection × resistance status: F = 14.7, df = 1,226, P = 0.0002).
Fig. 1.
Genetic variation in susceptibility to DAV in a natural population. (A) Viral titer in 182 inbred lines of D. melanogaster 3 dpi. (B) Viral titer over a time-course. In (A) and (B), each point is from ∼15 female flies infected by intrathoracic inoculation. Titer was estimated using qPCR relative to mRNA from the Drosophila gene EF1α100E. Points below the detection limit are jittered vertically. The box shows the median and interquartile range. (C) PCR genotyping for the presence of the Doc insertion.
We have previously measured the susceptibility of these lines to three other viruses: the positive-strand RNA viruses DCV (Dicistroviridae) and FHV (Nodaviridae) and the negative strand virus DMelSV (Rhabdoviridae) (16). However, the two lines that are highly resistant to DAV are not resistant to these viruses (SI Appendix, Fig. S1A; DMelSV, linear model including CHKov1 and ref(2)P genotype as cofactor: F = 1.13, df = 1,185, P = 0.29; DCV: F = 0.70, df = 1,153, P = 0.41; FHV: F = 0.002, df = 1,180, P = 0.96). Therefore, this mechanism of resistance is likely virus-specific.
Increased Fecundity of Resistant Genotypes Upon Infection.
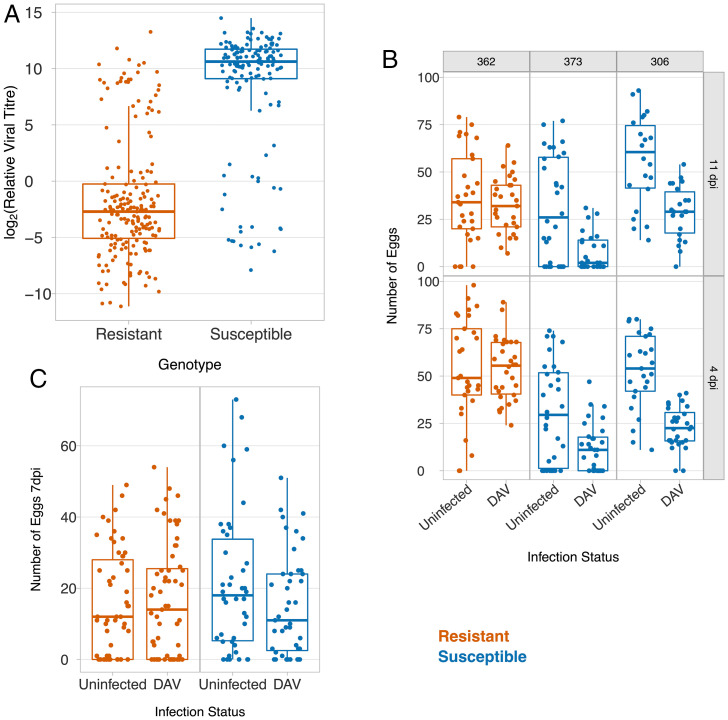
As DAV is common in nature, we investigated how resistance affects the fitness of infected flies. It is known that different mechanisms protect flies against viral infection through different routes (29). Therefore, we first mimicked natural infection by feeding adults with live yeast paste that was contaminated with the virus. In an attempt to obtain an ecologically realistic dose, the virus was extracted from infected flies, mixed with yeast and water, and added to the fly vials such that the virus extracted from a single fly would be split between five vials. Seven days postinfection, the median viral titer in the susceptible flies was ∼10,260 times greater than in the resistant flies (Fig. 2A; t = 17.04, df = 307, P < 10−16). Therefore, resistance protects flies against oral infection with the virus.
Fig. 2.
The effect of resistance on host fitness after infection. (A) Viral titer relative to EF1α100E mRNA 7 d after oral infection. (B and C) The effect of DAV infection on the number of eggs laid by single females over 20 h following infection by intrathoracic inoculation (B) or oral infection (C). In (A and C), the resistant and susceptible genotypes are recombinants between resistant and susceptible DGRP lines (see Fig. 4 for details). In (B), the three Drosophila lines are the F1 progeny of a cross between a line from the DGRP panel and an isogenic w1118 line.
Little is known about the effects of DAV on the fitness of Drosophila, so we measured the effects of DAV infection on fecundity. To avoid the effects of inbreeding depression, we crossed the inbred lines to a standard susceptible laboratory line and inoculated the F1 progeny with DAV or saline solution. Four days postinfection, females from two susceptible lines laid 58 to 63% fewer eggs than uninfected controls. In contrast, there was no reduction in the number of eggs laid by a resistant line (Fig. 2B; Quasipoisson GLM, resistance status × infection status interaction: χ2 = 25.3, df = 1, P = 5 × 10−7). The results were similar 11 dpi (Fig. 2B; Quasipoisson GLM, resistance status × infection status interaction: χ2 = 12.2, df = 1, P = 0.0005).
To investigate the effects of infection through a natural route, we examined the fecundity of flies after they had been fed food contaminated with DAV. DAV infection caused a 28% reduction in the fecundity of the susceptible flies but no reduction in the fecundity of the resistant genotypes (Fig. 2C; Quasipoisson GLM, resistance status × infection status interaction: χ2 = 3.8, df =1, P = 0.05).
To investigate the survival of flies we inoculated flies with two different doses of the virus and followed their survival for 65 d. DAV substantially reduced lifespan, but regardless of the dose mortality only increased from ∼30 dpi (SI Appendix, Fig. S1). Despite this large effect on survival, there was no consistent difference in the survival of resistant and susceptible flies after infection at either dose (SI Appendix, Fig. S1). A possible explanation is that at the time when infected flies start to die there is little difference in the viral load of the resistant and susceptible flies (Fig. 1B and SI Appendix, Fig. S1).
Increased resistance to infection is sometimes genetically correlated with reduced fitness in uninfected animals (30). As a measure of reproductive success, we measured the number of adult offspring produced by homozygous resistant and susceptible flies (data obtained by allowing the eggs laid over 20 h by 72 uninfected females in Fig. 2C to develop into adults). We found there was no significant difference between the reproductive success of resistant and susceptible flies (Poisson GLM: χ2 = 1.63, df = 1, P = 0.20). In a separate experiment we measured survival from first instar larvae to adulthood in a subset of these lines, and again there was no difference between resistant and susceptible genotypes (N = 185 larvae, Quasibinomial GLM: χ2 = 0.001, df = 1, P = 0.97). Furthermore, we found that there were no significant correlations between DAV titer (Fig. 1A) and published measurements of lifespan and fecundity made on these inbred lines (DGRP panel; SI Appendix, Table S1).
Resistance Is Caused by a Single Dominant Major-Effect Locus.
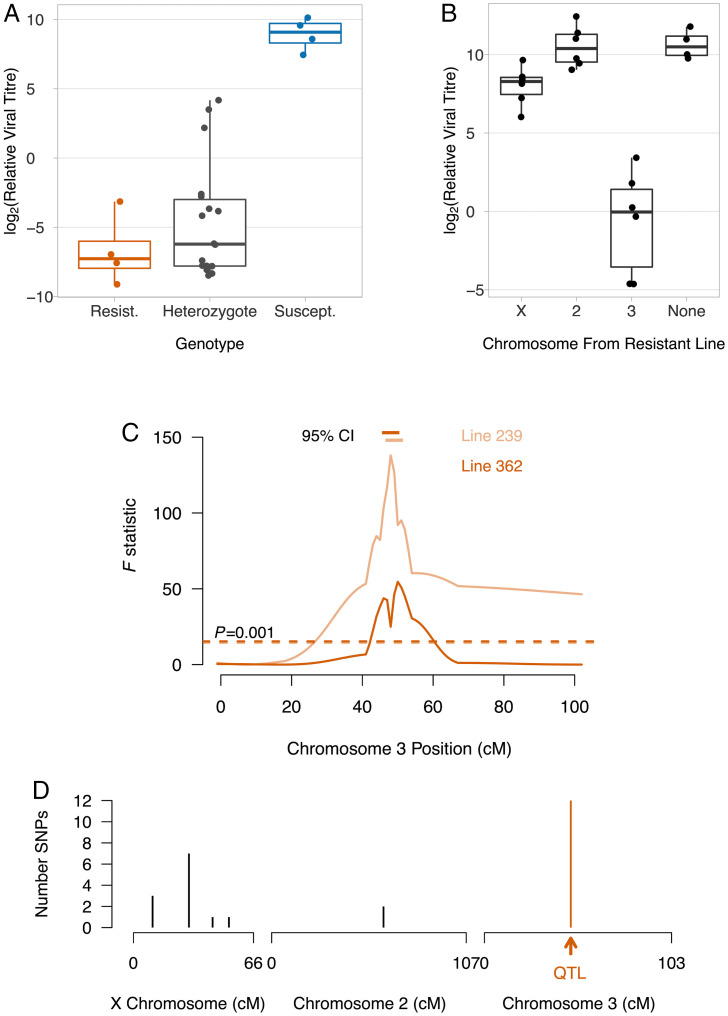
Our next goal was to characterize the genetic basis of resistance to DAV. We found that resistance is genetically dominant, with the F1 progeny of a cross having the phenotype of the resistant parent (Fig. 3A; Tukey honest significant difference (HSD) test, susceptible vs. heterozygote: P < 0.00001, resistant vs. heterozygote: P = 0.59). To identify which chromosomes affect susceptibility, we generated flies which carry a single chromosome from a resistant line and two chromosomes from a susceptible line (Fig. 3B). When these flies were infected, viral titers were strongly reduced in lines where chromosome 3 came from the resistant parent (Fig. 3B; Tukey HSD test, chromosome 2 vs. 3: P < 0.00001, chromosome X vs. 3: P = 0.00001). There was also a small effect of the X chromosome (Fig. 3B; Tukey HSD test, chromosome 2 vs. X: P = 0.04).
Fig. 3.
Genetic mapping of DAV resistance. (A) Dominance in resistant flies (DGRP 239 and 362), susceptible flies (DGRP 48 and 91), and F1 progeny. Pairs of lines were combined as they did not differ significantly. Each point is 20 females. (B) Resistance was mapped to chromosome by substituting chromosomes between lines. (C) The resistance QTL was mapped by crossing DGRP 239 × 373 and 362 × 306, to generate 84 and 72 recombinant lines, respectively. DAV titer was measured in a single female from each line. Ten molecular markers were genotyped across chromosome 3, and genotypes between markers inferred (31). The dashed line is a significance threshold obtained by permutation. The horizontal bars are 95% bootstrap intervals on the QTL location. In (A–C) flies were infected by inoculation. Viral titer was estimated 3 dpi relative to EF1α100E mRNA. (D) The genomic location of SNPs for which the two resistant lines one allele and the 180 susceptible lines had a different allele.
To map the region of chromosome 3 that controls resistance, we crossed two pairs of resistant and susceptible lines and created two panels of lines carrying recombinant third chromosomes. We genotyped molecular markers across the chromosome, and inferred genotype probabilities between these markers (31). In both crosses we identified a single quantitative trait locus (QTL) associated with resistance, and in both cases this mapped to the same region (Fig. 3C; line 239 cross: 47 to 49 cM; line 362 cross: 46 to 50 cM).
The genetic mapping results suggest that the same allele might be responsible for DAV resistance in the two different lines. We therefore searched the genome sequences of the 182 inbred lines that we used in our infection experiments (Fig. 1A) for consistent single nucleotide polymorphism (SNP) differences between resistant and susceptible lines. We found 26 such polymorphisms, 12 of which were on chromosome 3 (Fig. 3D). All of these fell within the QTL we identified in our genetic mapping experiments (Fig. 3 C and D). Together these results demonstrate that a single major-effect polymorphism on chromosome 3 controls susceptibility to DAV.
High-Resolution Genetic Mapping Identifies the Region Controlling Resistance.
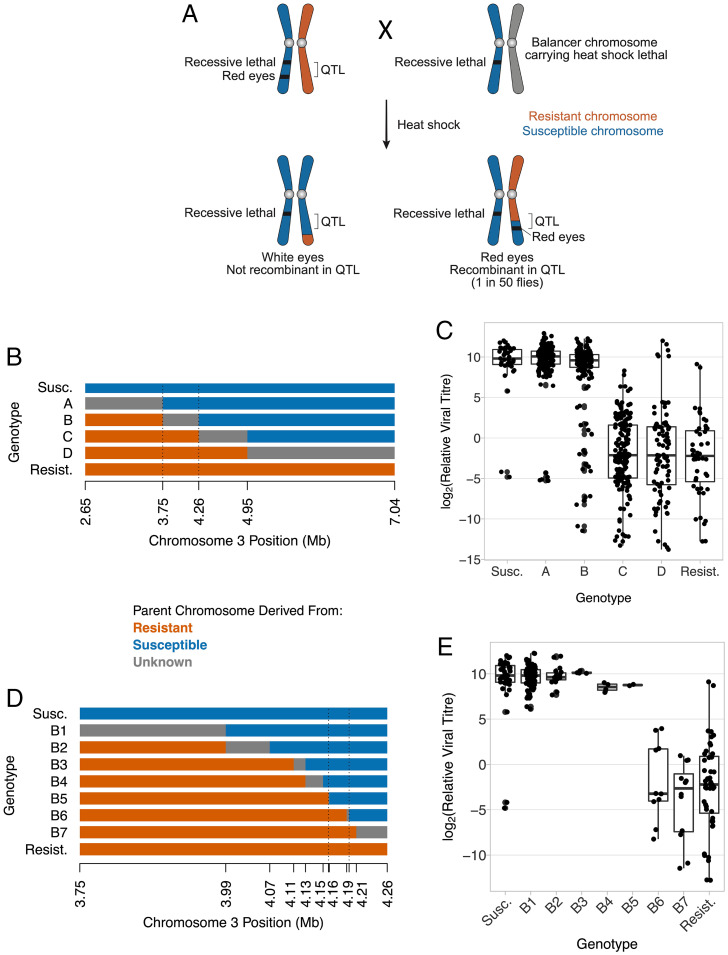
The QTL controlling resistance contained many genes, and the recombination rate in this region of the genome made it impractical to continue using simple genetic crosses to identify the causative gene. To overcome this, we devised a genetic cross to identify recombinants within the QTL based on eye color (Fig. 4A). In this cross, nonrecombinant flies either had white eyes, or died as a result of a homozygous recessive lethal allele or the expression of a lethal gene controlled by a heat shock promoter. Using this approach, we generated 643 lines that were recombinant within the QTL.
Fig. 4.
High resolution genetic mapping of DAV resistance. (A) The genetic cross used to select recombinants within the QTL controlling susceptibility to DAV. (B) The recombinant genotypes generated in the first phase of mapping with the location of five molecular markers used to genotype the recombinants. The dashed lines mark the region controlling susceptibility inferred from (C). (C) The viral titer of the genotypes in (B). Each point is an independent recombinant line (mean, 16.6 flies/line). (D) Recombinants between the dotted lines in (B) with the location of 12 markers. Dashed lines mark the region controlling susceptibility inferred in (E). (E) The viral titer of the genotypes shown in (D). The points are replicates of the infection assay (5 to 20 flies), and some lines are represented by multiple replicates. Flies were infected by inoculation; titer was estimated 3 dpi relative to EF1α100E. Titer measurements in (E) are a subset of (C).
We scored five molecular markers across the QTL, which allowed us to identify six genotypes with different recombination breakpoints (Fig. 4B). We retained 219 lines for phenotyping and found that they varied considerably in susceptibility to infection (Fig. 4C; ANOVA: F = 129, df = 5,258, P < 0.00001). Within genotype B there was a mixture of resistant and susceptible flies (Fig. 4C), indicating that the gene was within the recombinant region of these lines (between the dotted lines in Fig. 4B).
To identify the region affecting DAV resistance more precisely we carried out additional phenotyping and genotyping of lines that had recombined in this region. First, we genotyped a high density of molecular markers to precisely define recombination breakpoints in the lines (Fig. 4D; all recombinants in this region were retained from our panel of 643 lines). Second, we performed additional infection experiments to ensure we had accurate estimates of DAV susceptibility in recombinant lines whose breakpoints defined the location of the gene (Fig. 4E). Combining these two datasets, we found that the polymorphism controlling DAV susceptibility was in a region of 33,847 bp encompassing 11 genes (Fig. 4D; 3R:4163320.4197167 in genome v5).
A Transposable Element Insertion in Veneno Is Associated with Resistance.
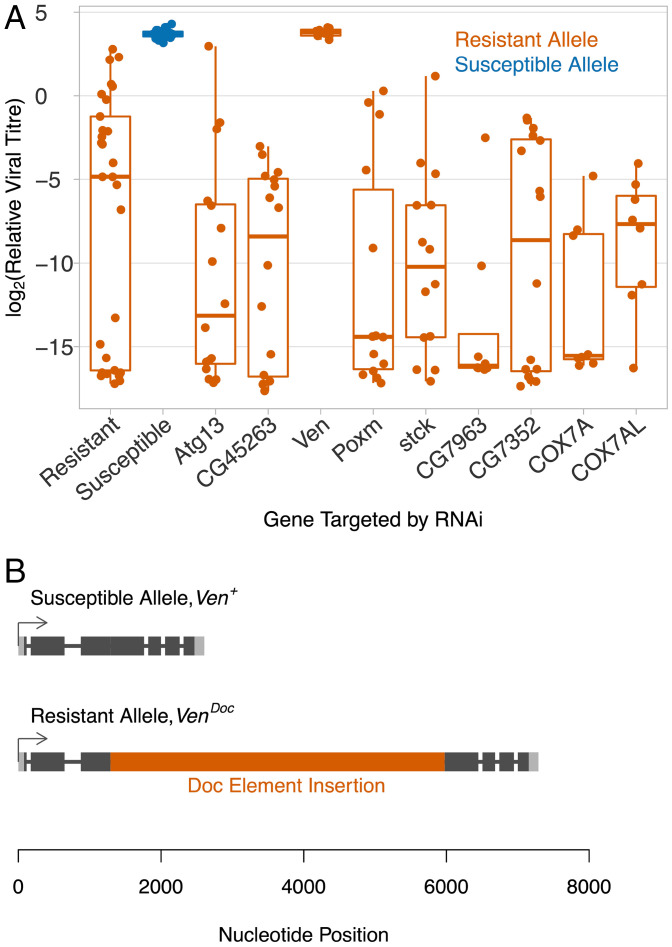
To test which of these genes underlies virus resistance, we knocked down the expression of the allele of these genes found in resistant flies by RNAi. Of the nine genes where this was successful, eight remained resistant to DAV (Fig. 5A). However, when we knocked down the expression of the gene CG9684, which we named Veneno (Ven) after its ortholog in Aedes aegypti (32), there was an ∼2,000-fold increase in viral titer compared to the resistant control, resulting in similar viral titers to susceptible flies (Fig. 5A; Welch’s t test, susceptible vs. Ven knock-down: t = 0.77, df = 11.0, P = 0.46, resistant vs. Ven knock-down: t = 7.88, df = 28.2, P = 10−8). Alongside this experiment we measured the transcription of the candidate genes in adult females. Again, Veneno was the only gene where there was a substantial difference in expression between the resistant and susceptible lines (SI Appendix, Fig. S2A; Welch’s t test: t = 12.51, df = 5.81, P = 0.00002). In line with this, using published microarray expression data from the DGRP panel (33), we found that Ven expression had a higher correlation with DAV resistance than any other gene in females, but showed no relationship in males (Pearson’s r = 0.5, false discovery rate (FDR) = 7.5 × 10−12 in females and r = 0.04, FDR = 0.99 in males, SI Appendix, Fig. S2B and Datasets S1 and S2).
Fig. 5.
Veneno is associated with susceptibility to DAV. (A) The effect on DAV titer of knocking down the expression of nine genes in the region associated with DAV resistance. Viral titer was estimated 3 dpi using qPCR relative to EF1α100E. (B) The structure of Veneno in resistant and susceptible lines.
The published genome sequences of the resistant and susceptible lines were generated with short read sequencing (28) and therefore may not include structural variants such as insertions. We therefore amplified Veneno from genomic DNA by PCR. While the susceptible line yielded PCR products of the expected size, the resistant allele contained a large insertion. Through a series of PCR reactions, we amplified and Sanger-sequenced a 4,685 bp insertion in the protein-coding sequence of exon 3 of Veneno (Fig. 5B; GenBank Accession: MZ047782). Using Basic Local Alignment Search Tool (BLAST), we identified the insertion as a Doc element, which is a non-LTR (long tandem repeat) retrotransposon. Compared to the published full-length Doc element sequence (34), our sequence has a 37 bp deletion at the 5′ end, three single nucleotide mismatches, and a single nucleotide deletion (DGRP-362 sequence; Dataset S3). We named the allele of Veneno carrying the Doc element VenDoc and the allele without the insertion Ven+.
To test whether the Doc insertion is associated with DAV resistance, we checked for the presence of the insertion in 162 DGRP lines. We genotyped these lines using two primers on either side of the Doc element insertion and one within, which results in different sized PCR products when amplifying VenDoc and Ven+ (Fig. 1C). We found that the Doc element insertion is perfectly associated with the DAV resistance (Fig. 1A; Fisher’s exact test: P = 0.00008). By sequencing both breakpoints between the Doc element and Veneno, we confirmed that the insertion was in the identical location in the two resistant lines. Therefore, in this experiment VenDoc is associated with an ∼19,000-fold reduction in DAV titer.
Truncation of Veneno Created a Novel Resistance Factor.
Many TE insertions associated with adaptive traits result from the insertion altering gene expression. In the experiments above we found that Ven expression is correlated with DAV titer in females. This correlation is entirely caused by the VenDoc allele having reduced expression (SI Appendix, Fig. S2A), as when these lines are removed there is no correlation between Ven+ expression and DAV titer (Pearson’s correlation in females: r = 0.06, P = 0.43). Furthermore, using qPCR we found that VenDoc expression was only reduced in in female abdomens; in males and the female thorax the two alleles were expressed at similar levels, or the resistant allele had slightly higher expression (SI Appendix, Fig. S2C). The epigenetic silencing of TE insertions in the Drosophila genome can reduce the expression of nearby genes in the female germline (35), which may explain the reduced expression of VenDoc in female abdomens. Regardless of its causes, because VenDoc confers DAV resistance in both males and females (SI Appendix, Fig. S2D), it is unlikely that resistance results from the Doc element insertion altering gene expression.
This is the second Doc element insertion that is associated with virus resistance in Drosophila. We previously reported a Doc insertion that is associated with resistance to the rhabdovirus DMelSV (15, 16). This led us to hypothesize that Doc elements may have intrinsic antiviral activity. To test this hypothesis, we created transgenic flies expressing RNAi constructs targeting two different regions of Doc. However, despite successfully knocking down Doc expression, this did not affect DAV titers (SI Appendix, Fig. S3).
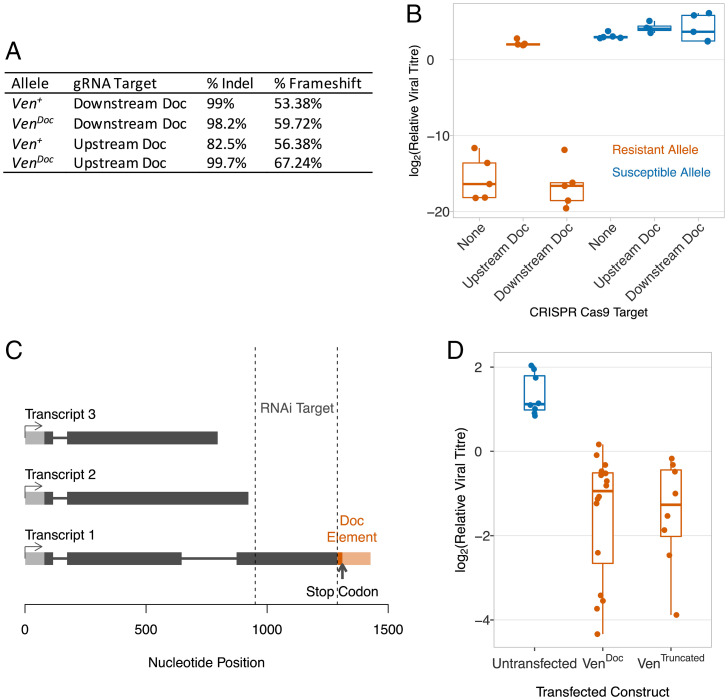
To investigate the effect of Ven+ and VenDoc on susceptibility to DAV, we mutated the sequence upstream and downstream of the Doc insertion using CRISPR/Cas9. We created flies that express Cas9 and either Ven+ or VenDoc. Alongside this, we generated two transgenic fly lines which expressed a CRISPR/Cas9 guide RNA molecule targeting either a Veneno exon before the Doc insertion or after the insertion. Again, each of these transgenes was crossed into a line carrying either Ven+ or VenDoc. We crossed these lines, producing F1 progeny that express Cas9 and guide RNAs throughout somatic cells, with the aim of producing somatic mutations. By amplifying the Cas9 cutting sites by PCR and Illumina sequencing the products, we estimated that this strategy mutated 82.3 to 99.7% of the chromosomes (Fig. 6A and Dataset S4). These flies were then infected by DAV. Mutation of VenDoc upstream of the insertion resulted in an ∼200,000-fold higher viral titer than control VenDoc flies (Fig. 6B, Tukey’s HSD test: P < 10−7). Mutation of the same region of Ven+ had no effect on viral titer, indicating that the Doc element insertion is a gain-of-function mutation (Fig. 6B, Tukey’s HSD test: P = 0.95). In contrast, when either allele of Veneno was mutated downstream of the insertion there was no effect on viral titers (Fig. 6B; Tukey’s HSD tests: P > 0.95).
Fig. 6.
The Doc element insertion in Veneno creates a novel transcription factor. (A and B) Exons of Veneno were mutated by expressing gRNAs and Cas9 in somatic cells, targeting sequences upstream and downstream of the Doc insertion. (A) The gRNA targets were Illumina sequenced to estimate the proportion mutated chromosomes. (B) DAV titer estimated 3 dpi relative to RpL32. (C) Oxford Nanopore Technologies transcriptome sequencing identified three polyadenylated transcripts of VenDoc. (D) Clonal stably transfected cell lines expressing V5-tagged Transcript 1 of VenDoc, or the equivalent construct where sequence from the Doc element has been deleted (VenTruncated). Viral titers relative to RpL32 were estimated 3 dpi.
These results suggest that VenDoc encodes a recently evolved antiviral molecule. As the Doc element is found in locations throughout the genome, we were unable to reconstruct transcripts encoded by VenDoc from short sequence reads. We therefore used the long-read Oxford Nanopore Technologies platform to sequence the transcriptome of a line carrying VenDoc. This allows us to sequence the full-length transcripts in a single read, and we mapped 8.8 million reads (NCBI SRA: SRR15541957) to a genome sequence which we had modified to include the Doc insertion in Ven. We identified three polyadenylated transcripts: transcript 1 (4 reads), transcript 2 (39 reads), and transcript 3 (4 reads) (Fig. 6C). We orientated the reads using the poly-A tail sequence, and all the reads were transcribed from the sense strand of the gene. Only transcript 1 contained regions which were targeted by the Veneno RNAi knock-down that eliminated the resistant phenotype (Figs. 5A and 6C). Furthermore, this was the only transcript containing an open reading frame ending with a stop codon, and with the same intron structure as Ven+ (Fig. 6C). VenDoc transcript 1 contains the Veneno sequence upstream of the Doc element, and the first 137 bp of the Doc element, including a stop codon 18 bp downstream from the site of insertion (Fig. 6C).
To test whether this transcript encodes an antiviral molecule, we expressed VenDoc transcript 1 in cultured Drosophila cells. DAV readily infects and replicates in DL2 cells (SI Appendix, Fig. S4A). We therefore stably transfected this cell line with a construct expressing V5-tagged VenDoc transcript 1 under the control of an inducible promoter. To ensure all the cells expressed the construct, we then established a clonal cell line from the transfected cells. When these cells were infected, expressing VenDoc transcript 1 led to an approximately sevenfold reduction in DAV titer 3 dpi (Fig. 6D, VenDoc vs. untransfected; Tukey’s HSD test: P = 0.00003). Similar results were obtained using nonclonal cells expressing VenDoc transcript 1 without the tag (SI Appendix, Fig. S4B). Both a Western blot and flow cytometry confirmed that these cells were expressing the truncated protein (SI Appendix, Fig. S4 C–E).
The antiviral effects of VenDoc could either result from the Doc insertion truncating the Veneno protein, or from a new function that requires a chimeric Veneno-Doc protein. To distinguish these possibilities, we established a clonal cell line that was stably transfected with a construct expressing VenDoc transcript 1 from which we had deleted the sequence derived from the Doc element. When we induced expression of this transgene, these cells were resistant to DAV (Fig. 6D, Ventruncated vs. untransfected; Tukey’s HSD test: P = 0.0003). Therefore, the Doc insertion has created a new antiviral molecule by truncating Veneno, and resistance does not require a chimeric Veneno-Doc protein.
Resistance Does Not Require the RNAi Pathway or the Tudor or MYND Domains in Veneno.
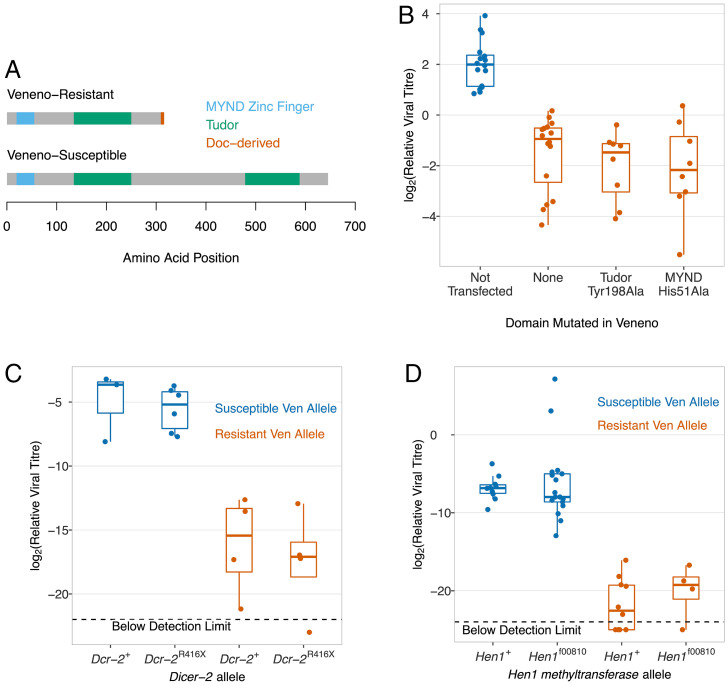
The Doc element insertion alters the domain structure of Veneno. The susceptible allele of Veneno (Ven+) encodes a protein with two Tudor domains, which are predicted to bind methylated arginine or lysine residues (Fig. 7A) (36). There is also an MYND (myeloid, Nervy, and DEAF-1) zinc finger domain, which is another domain normally involved in protein-protein interactions (37). These domains are also present in the ortholog of this gene in the mosquito Aedes aegypti, and here the protein acts as an adaptor protein that interacts with proteins in the antiviral piRNA pathway found in this species (32). The resistant allele (VenDoc) encodes a molecule that has lost the Tudor domain at the carboxyl-terminal end of the protein (Fig. 7A).
Fig. 7.
DAV resistance is independent of the RNAi pathway, and Tudor and MYND domains in Veneno. (A) Protein domains encoded by Ven+ and Vendoc Transcript 1. (B) Clonal stably transfected cell lines expressing V5-tagged Transcript 1 of VenDoc with the mutations His51Ala and Tyr198Ala that disrupt the MYND and Tudor domains respectively. Two susceptible controls (blue) are combined (untransfected cells and cells transfected with GFP; same data as in Fig. 6D). (C and D) In flies, Dcr-2 and Hen1 mutants were combined with Ven+ and Vendoc. Viral titer was estimated 3 dpi using qPCR relative RpL32.
One hypothesis is that Veneno has an adaptor function which facilitates the formation of a protein complex, and resistance results from changes to this complex. We tested whether the Tudor or MYND zinc finger domains were necessary for DAV resistance. We used site directed mutagenesis to target residues essential for the function of these domains in the constructs used to transfect cells. In the MYND zinc finger domain we mutated the histidine at residue 51 in the protein to an alanine (His51Ala). This histidine is required to chelate zinc and is therefore required for the correct functioning and folding of this domain (38). In the Tudor domain we mutated the tyrosine at position 198 to alanine (Tyr198Ala). The tyrosine is involved in forming the binding pocket for the symmetric demethylation of arginine, which allows protein-protein interactions (32, 39). We then stably transfected cells with these plasmids and generated clonal cell lines to ensure all cells were transfected. Neither the Tudor nor MYND zinc finger domains are necessary for resistance, as cell lines expressing both VenDoc, His51Ala and VenDoc, Tyr198Ala had substantially reduced DAV titers (Fig. 7B; Tukey’s HSD tests, P < 10−7).
The main antiviral defense of insects is RNAi, and Tudor domain proteins frequently function as adaptor proteins in small RNA pathways. Furthermore, Veneno physically interacts with Hen1, which methylates siRNAs (40), and R2D2 (41), which loads small interfering RNAs (siRNAs) into the RNA-induced silencing complex to guide the enzymatic shearing of viral RNA. We therefore tested the hypothesis that resistance requires the RNAi pathway by combining mutations in RNAi pathway genes Dicer-2 (Dcr-2R416X) and Hen1 (Hen1f00810) with the two alleles of Veneno. We found that VenDoc still made flies resistant to DAV when combined with these mutations, indicating that resistance caused by Veneno does not require the siRNA pathway (Fig. 7 C and D). We also found that knocking out the RNAi pathway did not increase the viral titer, which was unexpected as this pathway is thought to protect flies against a broad spectrum of viruses (Fig. 7 C and D). We confirmed this by infecting three further RNAi pathway mutant lines (Dcr-2L811fsx, Ago251B and Ago2414), again finding no increase in viral titer (SI Appendix, Fig. S5A). Together these results demonstrate that RNAi is not an important defense against DAV, and the antiviral effects of VenDoc do not rely on this pathway.
In Aedes aegypti, Veneno acts as an adaptor protein that assembles a protein complex involved in the production of piRNAs from viral RNA (32). To test whether the mechanism of VenDoc resistance depends on the piRNA pathway, we knocked down Ago3, vret and vas expression by RNAi in flies carrying both the resistant and susceptible allele of Veneno. In all three cases there was no effect on DAV titers (SI Appendix, Fig. S5B). This is consistent with previous results showing that in Drosophila the piRNA pathway is restricted to the germline and plays no role in antiviral immunity (42, 43). Together our results demonstrate that neither small RNA pathways nor the domains involved in protein-protein interactions are necessary for resistance to DAV.
Discussion
As new pathogens appear in populations and existing pathogens evolve to escape immunity, there is continual natural selection favoring novel host defenses. We have found that a TE insertion into the protein-coding sequence of the Tudor domain protein Veneno has resulted in the gene gaining an antiviral function. The resistant allele of the gene encodes a truncated protein that acts as a potent resistance factor that massively reduces titers of DAV, while the ancestral susceptible form of the protein has no effect on the virus. As DAV is common in nature (25) and causes large reductions in the fecundity of susceptible flies, this allele protects flies against a virulent pathogen. This adds to a growing body of evidence from multiple species of animals that much of the genetic variation in susceptibility to naturally occurring pathogens is explained by a small number of major-effect polymorphisms (16, 44–46). This contrasts with most quantitative traits which tend to be controlled by many variants with small phenotypic effects (45, 46). The simple genetic architecture likely results from the evolutionary arms race between hosts and their pathogens driving major-effect alleles up in frequency (45).
The study of viral immunity in invertebrates has been dominated by investigations of broad-spectrum and conserved antiviral defenses such as RNAi and autophagy. However, our discovery of VenDoc adds to the list of major-effect polymorphisms which cause virus resistance in Drosophila, such as CHKov1, ref(2)P, pastrel, and Ge-1 (15, 16, 22–24). The mechanism by which these genes protect flies against viral infection is unclear, but they may encode restriction factors, analogous to those that play a critical role in defending mammals against viruses. Regardless of mechanism, these genes are central to the defenses of Drosophila against viruses as they can have large effects on susceptibility. They differ from conserved antiviral pathways such as RNAi in two ways. First, they mostly protect their hosts against a narrow range of viral taxa. Therefore, despite their central importance to antiviral defense, they may be missed by studies using a single virus that may have been isolated from a different species. Second, they are mostly recent evolutionary innovations and are polymorphic in populations. These antiviral factors therefore arise because natural selection is continually generating new defenses against the viruses encountered in nature.
TEs frequently underlie adaptations novel selection pressures (8, 47, 48). In many cases these adaptive TE insertions alter gene expression, often by inserting upstream of the gene. For example, insecticide resistance has repeatedly evolved when TE insertions up-regulate the expression of detoxification enzymes (8, 48, 49). TE insertions can also generate new adaptations when the element itself is recruited to a host function, a process known as domestication. This can involve the element fusing to another gene in the genome. For example, the gene SETMAR in primates is a chimera of the gene SET and the transposon Hsmar1, which retains functions of both a TE domain and the original gene (50–52). In a striking parallel with VenDoc, we have previously reported how a Doc element insertion into the Drosophila gene CHKov1 is associated with resistance to a rhabdovirus (15, 16). This raised the possibility that Doc element sequences may be recruited to a new antiviral function in these gene-TE chimeras. However, we did not find support for the gene-TE chimera hypothesis, nor for resistance resulting from a change in gene expression.
Resistance is instead caused by the TE-dependent truncation of Veneno, resulting from the TE insertion shortening the transcript and prematurely terminating translation. To our knowledge, this is the first demonstration of such a mechanism giving rise to a protein with a novel function. Other cases of TEs introducing stop codons into the coding sequence of genes are thought to be simple loss-of-function mutations. For example, a Hel-1 LTR retrotransposon insertion into the HevCalP gene in the moth Heliothis virescens introduces a stop codon (53). This allele is resistant to Bacillus thuringiensis (Bt) toxins used in pest control, but resistance is recessive as the truncation results in a loss of function of the host protein (53). In contrast, DAV resistance is a gain of function mutation.
The mechanism of resistance may rely on Veneno playing a role in RNA biology. Ortholog of Veneno of Veneno (32) plays a role in piRNA biogenesis, while Veneno physically interacts with components of the siRNA pathway (41). However, VenDoc resistance did not require a functional siRNA or piRNA pathway. Veneno also has three domains involved in protein-protein interactions, but when we mutated residues that are essential for the function of these domains, VenDoc still conferred virus resistance. The molecular mechanism by which Veneno protects Drosophila against viral infection therefore remains uncertain. However, the observation that Veneno interacts with proteins involved in RNA biology suggests that some currently unknown function of the protein has been recruited to a novel antiviral function.
Despite effectively protecting flies against a virulent pathogen, the resistant allele of Veneno is found at a frequency of just over 1%, so only about 1 in 50 flies in this population will be resistant. This could result from resistance having arisen recently, so there is insufficient time for the allele to reach a high frequency. Alternatively, the benefits of resistance may be balanced by costs. In Drosophila, the Tudor domain proteins Qin, Krimper, and Tudor-SN are all expressed in the germline where they play a role in the piRNA pathway (36, 54), and the high expression of the susceptible allele of Veneno in ovaries suggests it may have a related function. As the resistant allele has greatly reduced expression in ovaries and has lost a Tudor domain, it is likely its original function will have been changed. While our data suggests that in infected flies the benefits of resistance likely outweigh the costs, we lack the statistical power to detect more modest costs of VenDoc. If these exist, they could result in the resistant allele being under balancing selection, either due to heterozygote advantage or negative frequency dependent selection.
The strong and rapidly changing selection pressures that pathogens impose on host populations provide a model to study the genetics of adaptation. Our work demonstrates a mechanism by which TEs can generate these adaptations. The role of TEs in adaptation to DAV infection mirrors studies of how populations have adapted to the selection pressures that humans impose populations when they use pesticides or alter the environment (8, 47–49). The importance of TEs in evolution likely stems not simply from them being a major cause of mutation, but also because of features of these mutations such as the magnitude of the change in DNA sequence and biases in where the elements insert. It is clear that alongside the harm they cause their hosts, TEs can allow populations to evolve to overcome a diverse range of challenges.
Materials and Methods
Resistant and susceptible lines were identified by infecting the DGRP lines with DAV and measuring viral loads by qPCR. We then mapped resistance by using balancer chromosomes to create chromosome substitution lines, followed by the generation of recombinant inbred lines, and finally using the cross in Fig. 4A. Genes within the QTL were knocked down using transgenic RNAi constructs, and Ven was mutated by expressing cas9 and gRNAs in transgenic flies. To identify protein domains involved in resistance we transfected Drosophila cells with plasmids that express modified alleles of Ven. Detailed methods are in SI Appendix.
Supplementary Material
Acknowledgments
We thank Karyn Johnson for providing DAV. This work was funded by grants from the Natural Environment Research Council (NE/P00184X/1) and the Leverhulme Trust (RPG-2020-236) to F.J. R.C. is funded by the São Paulo Research Foundation (FAPESP) (2013/25991-0 and 2015/08307-3), the National Council for Scientific and Technological Development (CNPq) (307447/2018-9) and a Newton Advanced Fellowship from the Royal Society (NAF\R1\180244). O.B. is funded by the Dr. Herchel Smith Fellowship.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122026119/-/DCSupplemental.
Data Availability
The raw data from experiments and scripts to reproduce the figures and statistical analyses have been deposited in the Cambridge Data Repository (https://doi.org/10.17863/CAM.84829) (55). The Oxford Nanopore RNA sequence reads are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRR15541957) (56). DNA Sequence data have been deposited in the NCBI GenBank (MZ047782) (57).
References
- 1.Kofler R., Nolte V., Schlötterer C., Tempo and mode of transposable element activity in Drosophila. PLoS Genet. 11, e1005406 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houle D., Nuzhdin S. V., Mutation accumulation and the effect of copia insertions in Drosophila melanogaster. Genet. Res. 83, 7–18 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Mackay T. F., Lyman R. F., Jackson M. S., Effects of P element insertions on quantitative traits in Drosophila melanogaster. Genetics 130, 315–332 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charlesworth B., Sniegowski P., Stephan W., The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371, 215–220 (1994). [DOI] [PubMed] [Google Scholar]
- 5.González J., Petrov D. A., The adaptive role of transposable elements in the Drosophila genome. Gene 448, 124–133 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daborn P. J., et al. , A single p450 allele associated with insecticide resistance in Drosophila. Science 297, 2253–2256 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Schmidt J. M., et al. , Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 6, e1000998 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlenke T. A., Begun D. J., Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc. Natl. Acad. Sci. U.S.A. 101, 1626–1631 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malik H. S., Henikoff S., Positive selection of Iris, a retroviral envelope-derived host gene in Drosophila melanogaster. PLoS Genet. 1, e44 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malik H. S., Retroviruses push the envelope for mammalian placentation. Proc. Natl. Acad. Sci. U.S.A. 109, 2184–2185 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornelis G., et al. , An endogenous retroviral envelope syncytin and its cognate receptor identified in the viviparous placental Mabuya lizard. Proc. Natl. Acad. Sci. U.S.A. 114, E10991–E11000 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chuong E. B., Elde N. C., Feschotte C., Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 351, 1083–1087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carmona L. M., Schatz D. G., New insights into the evolutionary origins of the recombination-activating gene proteins and V(D)J recombination. FEBS J. 284, 1590–1605 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ullastres A., Merenciano M., González J., Regulatory regions in natural transposable element insertions drive interindividual differences in response to immune challenges in Drosophila. Genome Biol. 22, 265 (2021). [DOI] [PMC free article] [PubMed]
- 15.Magwire M. M. M., Bayer F., Webster C. L. C. L., Cao C., Jiggins F. M. F. M., Successive increases in the resistance of Drosophila to viral infection through a transposon insertion followed by a duplication. PLoS Genet. 7, e1002337 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magwire M. M. M., et al. , Genome-wide association studies reveal a simple genetic basis of resistance to naturally coevolving viruses in Drosophila melanogaster. PLoS Genet. 8, e1003057 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabian D. K., Fuentealba M., Dönertaş H. M., Partridge L., Thornton J. M., Functional conservation in genes and pathways linking ageing and immunity. Immun. Ageing 18, 23 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obbard D. J., Gordon K. H. J., Buck A. H., Jiggins F. M., The evolution of RNAi as a defence against viruses and transposable elements. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 99–115 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Obbard D. J., Jiggins F. M., Halligan D. L., Little T. J., Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr. Biol. 16, 580–585 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Obbard D. J., Welch J. J., Kim K.-W., Jiggins F. M., Quantifying adaptive evolution in the Drosophila immune system. PLoS Genet. 5, e1000698 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kluge S. F., Sauter D., Kirchhoff F., SnapShot: Antiviral restriction factors. Cell 163, 774 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Cao C., Magwire M. M. M., Bayer F., Jiggins F. M., A polymorphism in the processing body component Ge-1 controls resistance to a naturally occurring rhabdovirus in Drosophila. PLoS Pathog. 12, e1005387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bangham J., Obbard D. J., Kim K.-W., Haddrill P. R., Jiggins F. M., The age and evolution of an antiviral resistance mutation in Drosophila melanogaster. Proc. Biol. Sci. 274, 2027–2034 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao C., Cogni R., Barbier V., Jiggins F. M., Complex coding and regulatory polymorphisms in a restriction factor determine the susceptibility of Drosophila to viral infection. Genetics 206, 2159–2173 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webster C. L., et al. , The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster. PLoS Biol. 13, e1002210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cogni R., Ding S. D., Pimentel A. C., Day J. P., Jiggins F. M., Wolbachia reduces virus infection in a natural population of Drosophila. Commun. Biol. 4, 1327 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ambrose R. L., et al. , Drosophila A virus is an unusual RNA virus with a T=3 icosahedral core and permuted RNA-dependent RNA polymerase. J. Gen. Virol. 90, 2191–2200 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackay T. F. C., et al. , The Drosophila melanogaster genetic reference panel. Nature 482, 173–178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferreira Á. G., et al. , The Toll-dorsal pathway is required for resistance to viral oral infection in Drosophila. PLoS Pathog. 10, e1004507 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKean K. A., Yourth C. P., Lazzaro B. P., Clark A. G., The evolutionary costs of immunological maintenance and deployment. BMC Evol. Biol. 8, 76 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haley C. S., Knott S. A., A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69, 315–324 (1992). [DOI] [PubMed] [Google Scholar]
- 32.Joosten J., et al. , The Tudor protein Veneno assembles the ping-pong amplification complex that produces viral piRNAs in Aedes mosquitoes. Nucleic Acids Res. 47, 2546–2559 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang W., et al. , Genetic basis of transcriptome diversity in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 112, E6010–E6019 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Hare K., Alley M. R. K., Cullingford T. E., Driver A., Sanderson M. J., DNA sequence of the Doc retroposon in the white-one mutant of Drosophila melanogaster and of secondary insertions in the phenotypically altered derivatives white honey and white-eosin. MGG Mol. Gen. Genet. 225, 17–24 (1991). [DOI] [PubMed] [Google Scholar]
- 35.Lee Y. C. G., Karpen G. H., Pervasive epigenetic effects of Drosophila euchromatic transposable elements impact their evolution. eLife 6, e25762 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siomi M. C., Mannen T., Siomi H., How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev. 24, 636–646 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gamsjaeger R., Liew C. K., Loughlin F. E., Crossley M., Mackay J. P., Sticky fingers: Zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 32, 63–70 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Liu Y., et al. , Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO’s activity. Cancer Cell 11, 483–497 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H., et al. , Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev. 24, 1876–1881 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guruharsha K. G., et al. , A protein complex network of Drosophila melanogaster. Cell 147, 690–703 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Majzoub K., The Antiviral siRNA Interactome in Drosophila melanogaster (Universite de Strasbourg, 2013). [Google Scholar]
- 42.Petit M., et al. , piRNA pathway is not required for antiviral defense in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 113, E4218–E4227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewis S. H., et al. , Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat. Ecol. Evol. 2, 174–181 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zamparini A. L., et al. , Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila. Development 138, 4039–4050 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duxbury E. M., et al. , Host-pathogen coevolution increases genetic variation in susceptibility to infection. eLife 8, e46440 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hill A. V. S., Evolution, revolution and heresy in the genetics of infectious disease susceptibility. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 840–849 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van’t Hof A. E., et al. , The industrial melanism mutation in British peppered moths is a transposable element. Nature 534, 102–105 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Rostant W. G., Wedell N., Hosken D. J., Transposable elements and insecticide resistance. Adv. Genet. 78, 169–201 (2012). [DOI] [PubMed] [Google Scholar]
- 49.French-Constant R., Daborn P., Feyereisen R., Resistance and the jumping gene. BioEssays 28, 6–8 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Cordaux R., Udit S., Batzer M. A., Feschotte C., Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc. Natl. Acad. Sci. U.S.A. 103, 8101–8106 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tellier M., Chalmers R., Human SETMAR is a DNA sequence-specific histone-methylase with a broad effect on the transcriptome. Nucleic Acids Res. 47, 122–133 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu D., et al. , The human SETMAR protein preserves most of the activities of the ancestral Hsmar1 transposase. Mol. Cell. Biol. 27, 1125–1132 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gahan L. J., Gould F., Heckel D. G., Identification of a gene associated with Bt resistance in Heliothis virescens. Science (80-.). 293 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Sato K., Iwasaki Y. W., Siomi H., Siomi M. C., Tudor-domain containing proteins act to make the piRNA pathways more robust in Drosophila. Fly (Austin) 9, 86–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.F. Jiggins, O. Brosh, D. Fabian, D. Cogni, Data for "A novel transposable element-mediated mechanism causes antiviral resistance in Drosophila through truncating the Veneno protein." Cambridge Data Repository. https://www.repository.cam.ac.uk/handle/1810/337512. Deposited 26 May 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.University of Cambridge, Oxford Nanopore RNA sequence reads from "Transposable Elements and the Evolution of Virus Resistance in Drosophila melanogaster" accession SRR15541957. NCBI Sequence Read Archive. https://www.ncbi.nlm.nih.gov/sra/?term=SRR15541957. Accessed 24 June 2022. [Google Scholar]
- 57.O. Brosh, D. Fabian, F. Jiggins, Drosophila melanogaster Veneno (Ven) gene, complete cds; and non-LTR retrotransposon Doc putative reverse transcriptase and putative RNA binding protein genes, complete cds. Accession MZ047782. NCBI GenBank. https://www.ncbi.nlm.nih.gov/nuccore/MZ047782. Deposited 24 April 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data from experiments and scripts to reproduce the figures and statistical analyses have been deposited in the Cambridge Data Repository (https://doi.org/10.17863/CAM.84829) (55). The Oxford Nanopore RNA sequence reads are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRR15541957) (56). DNA Sequence data have been deposited in the NCBI GenBank (MZ047782) (57).