Significance
Essential physiological processes such as cell migration and endocytosis require the assembly and catalyzed disassembly of the cytoskeletal branched networks composed of actin filaments and the Arp2/3 complex. We used single-molecule fluorescence microscopy to directly observe the molecular mechanisms of branch disassembly. The two proteins glial maturation factor (GMF) and cofilin both carry out branch pruning, releasing the Arp2/3 complex and converting the branched network into linear actin filaments. Although GMF and cofilin are evolutionarily related, they work by different pathways that can function independently but also additively in debranching, suggesting that they play collaborative roles in taking apart branched actin networks.
Keywords: Arp2/3 complex, branched actin networks, tropomyosin, actin depolymerizing factor homology, glial maturation factor
Abstract
Eukaryotic cells contain branched actin networks that are essential for endocytosis, motility, and other key cellular processes. These networks, which are formed by filamentous actin and the Arp2/3 complex, must subsequently be debranched to allow network remodeling and to recycle the Arp2/3 complex. Debranching appears to be catalyzed by two different members of the actin depolymerizing factor homology protein family: cofilin and glial maturation factor (GMF). However, their mechanisms of debranching are only partially understood. Here, we used single-molecule fluorescence imaging of Arp2/3 complex and actin filaments under physiological ionic conditions to observe debranching by GMF and cofilin. We demonstrate that cofilin, like GMF, is an authentic debrancher independent of its filament-severing activity and that the debranching activities of the two proteins are additive. While GMF binds directly to the Arp2/3 complex, cofilin selectively accumulates on branch–junction daughter filaments in tropomyosin-decorated networks just prior to debranching events. Quantitative comparison of debranching rates with the known kinetics of cofilin–actin binding suggests that cofilin occupancy of a particular single actin site at the branch junction is sufficient to trigger debranching. In rare cases in which the order of departure could be resolved during GMF- or cofilin-induced debranching, the Arp2/3 complex left the branch junction bound to the pointed end of the daughter filament, suggesting that both GMF and cofilin can work by destabilizing the mother filament–Arp2/3 complex interface. Taken together, these observations suggest that GMF and cofilin promote debranching by distinct yet complementary mechanisms.
Branched actin networks nucleated by the Arp2/3 complex generate forces used to drive membrane protrusion, cell motility, endocytosis, phagocytosis, autophagy, and cell–cell adhesion (1–8). Extensive effort over the last twenty years has provided us with a detailed understanding of how actin filament branches are assembled, but comparatively less is known about how the branches are turned over (i.e., disassembled). In cells, branches turn over in seconds at the leading edge and at sites of endocytosis (9, 10). Rapid debranching in cells enables dynamic remodeling of the architecture of actin networks, recycling of the Arp2/3 complex for new rounds of actin assembly, and destabilization and disassembly of actin networks. In contrast to branches in vivo, branches formed in vitro are highly stable, lasting from tens of minutes to hours (11–13), which indicates that additional cellular factors are required to promote rapid debranching in vivo.
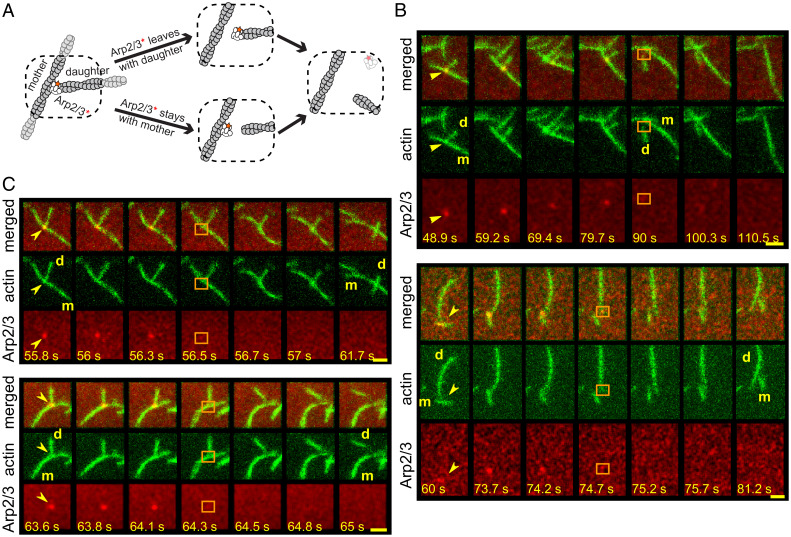
To form a branch, the Arp2/3 complex binds to the side of an existing actin filament (the “mother” filament), and together with a nucleation-promoting factor (NPF) such as the verprolin homolgy/cofilin homology/acidic (VCA) fragment of Wiskott-Aldrich syndrome protein (WASP) nucleates the polymerization of a “daughter” filament at a characteristic ∼70° angle (Fig. 1A). Before a daughter filament forms, the Arp2/3 complex alone binds and dissociates rapidly from the sides of mother filaments [in < 1 s (14)]. However, once a daughter filament begins to form, the conformation of the Arp2/3 complex changes, producing the stable branch structure (15–18). The Arp2/3 complex remains in this structure as an integral component, using distinct surfaces to bind the side of the mother filament and the pointed end of the daughter filament. Cellular mechanisms for disassembling these intrinsically stable branch junctions are still not well understood.
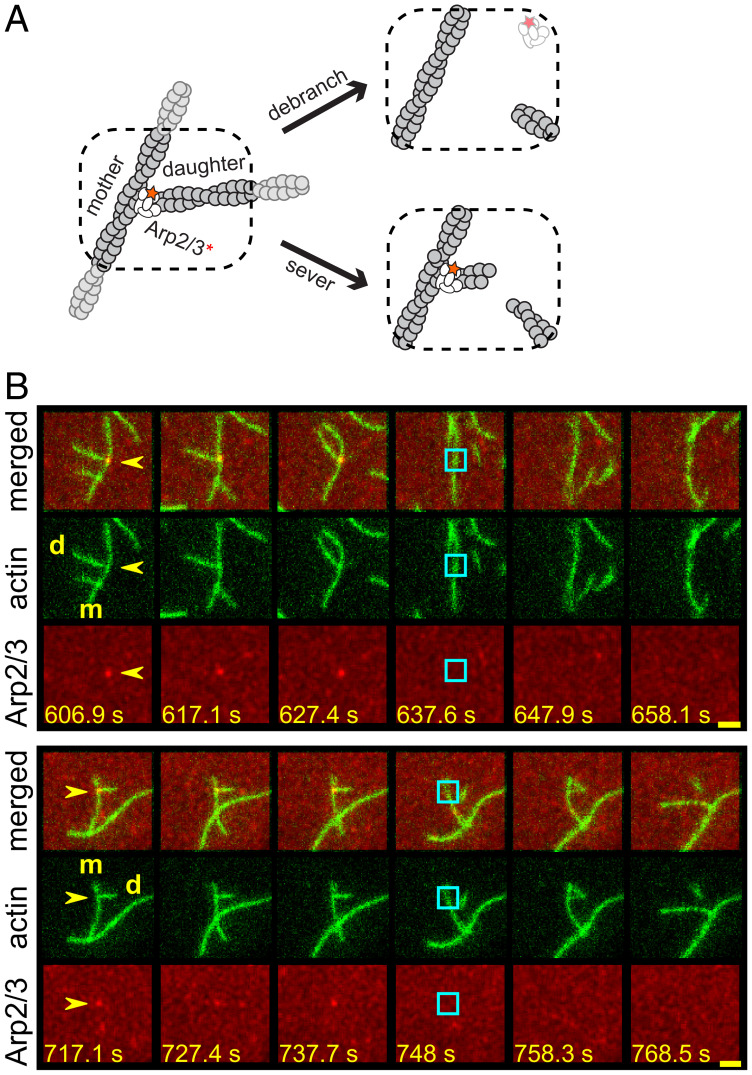
Fig. 1.
Debranching of Arp2/3-mediated actin branches by GMF. (A) Cartoon illustrating the two-color debranching assay with dye (red star/asterisk)-labeled Arp2/3 complex (white) and actin (gray). (B) Examples of debranching induced by 50 nM GMF in physiological buffer in the two-color assay. Each triplet of rows of images depicts a sequence of consecutive video frames showing a branch and its subsequent debranching in merged view (Top), actin channel (Middle), and Arp2/3 channel (Bottom). The time stamp (seconds) for each image is indicated (Scale bars:, 2.4 µm). Yellow arrows indicate the branches of interest; cyan squares mark the former branch site on the mother (m) filament immediately after the dissociation of the daughter (d) filament and the disappearance of the Arp2/3 complex.
Three different conserved proteins—glial maturation factor (GMF), cofilin, and coronin—each have been implicated (individually) in promoting debranching in cells. There is genetic and biochemical evidence for mammalian coronin-1B promoting debranching, and its mechanism of action appears to involve direct binding to the Arp2/3 complex (19, 20). There is also clear biochemical and genetic evidence support for the role for GMF in debranching. GMF is a member of the larger actin depolymerizing factor/cofilin homology (ADF-H) protein family (21, 22). While most ADF-H family members bind directly to filamentous and/or monomeric actin, GMF does not. Instead, GMF binds with high affinity to the Arp2/3 complex (13, 23, 24). In vitro, GMF catalyzes daughter filament dissociation without affecting other aspects of actin filament organization or dynamics (13). Thus, GMF is considered to be a factor specialized for debranching (22). Consistent with this view, the debranching activity of GMF is required in vivo for proper actin network turnover and lamellipodial retraction dynamics in migrating cells (25, 26).
In contrast to GMF, cofilin binds directly to actin filaments and has multiple activities in promoting actin network turnover. These include filament severing and pointed-end depolymerization (21, 27), and a proposed role in debranching. The role of cofilin in debranching is supported by studies in vitro using total internal reflection fluorescence (TIRF) microscopy to monitor the dissociation of daughter filament branches from mother filaments (12, 28). However, because cofilin also severs actin filaments, it has remained unclear whether it prunes daughter filaments by destabilization of the branch junction (i.e., authentic debranching, as with GMF) or whether the apparent debranching is due to severing events too close to the branch junction to be resolved by light microscopy. Chan et al. (28) favored the former view, but the interpretation of some of their data is controversial (see Discussion). A more direct test, such as direct single-molecule observations using fluorescently labeled Arp2/3 complex and cofilin, has the potential to clarify this proposed function of cofilin.
In this study, we have directly compared the debranching activities of GMF and cofilin using a 2-color single-molecule fluorescence technique that simultaneously monitors fluorescently labeled Arp2/3 complex and actin. This approach allows clear differentiation between authentic debranching events and severing events near branch junctions. Using this assay, we found that GMF and cofilin each induce debranching under physiological ionic conditions; previous studies only tested debranching at lower ionic strength. In addition, cofilin further increases debranching rates at a near-saturating GMF concentration, suggesting that they have distinct and complementary mechanisms of action. We observed cofilin accumulating on the daughter filament near the branch site preceding debranching and, upon dissociation of the daughter filament by either protein, we could sometimes detect the Arp2/3 complex remaining transiently bound to the pointed end of the daughter filament. Taken together, the results suggest that cofilin and GMF are independently capable of destabilizing branch junctions and likely do so by binding to different locations in the branch.
Results
Single-Molecule Experiments Distinguish Authentic Debranching Events from Severing near Branch Junctions.
GMF induces debranching (i.e., daughter filament dissociation) without severing actin filaments at other locations (13). In contrast, cofilin is thought to stimulate debranching in addition to promoting the severing of actin filaments (28). Chan and coworkers (28) proposed that cofilin’s debranching and severing activities are independent, as suggested by these two activities having distinct cofilin concentration dependences. However, this prior work lacked the resolution to distinguish between authentic debranching events (branch junction dissociation) versus the severing of daughter filaments near branch junctions, leaving the Arp2/3 complex and a very short daughter branch at this site (Fig. 1 A, Bottom Right). In contrast, an authentic debranching event completely removes the Arp2/3 complex and the daughter filament from the mother filament (Fig. 1 A, Top Right).
To resolve the cofilin debranching mechanism, we developed a single-molecule fluorescence assay using differentially labeled Saccharomyces cerevisiae Arp2/3 complex and actin. In this assay, non-tethered branched filaments were first assembled in the TIRF chamber by mixing monomeric actin (partially labeled with Alexa Fluor 488 (AF488), Janelia Fluor 464 (JF646)-labeled Arp2/3 complex, and an unlabeled VVCA fragment of human N-WASP. Then the assembly ingredients were washed out, and S. cerevisiae GMF, cofilin, or control buffer was introduced. Debranching was monitored over time by scoring the separation of the daughter filament from the mother and the concomitant loss of the single labeled Arp2/3 complex from the branch junction (examples in Fig. 1B, showing GMF-induced debranching). In this manner, our assay distinguishes between authentic debranching events and severing near branch junctions.
We also considered how ionic conditions affect debranching. Previous in vitro studies on GMF- and cofilin-mediated debranching were performed using low ionic strength buffers (typically containing only 50 mM KCl) optimized for actin polymerization (12, 13, 23, 28, 29). However, physiological ionic strength is considerably higher. Therefore, we developed a more physiological buffer (SI Appendix, Table S1) based on elemental analysis of yeast cytoplasm (30, 31) and directly compared debranching by GMF in the low-salt (50 mM KCl) and physiological buffers (SI Appendix, Fig. S1A). In the absence of GMF, branches were highly stable (debranching rates < 5 × 10−5 s−1) in both the low-salt and physiological buffers (SI Appendix, Fig. S1 B and C). Under both conditions, the addition of 50 nM GMF stimulated daughter filament dissociation by more than 50-fold, producing debranching rates of ∼2.5 × 10−3 s−1. These rates of GMF-induced debranching agree well with previous reports (13, 23). Since yeast GMF promotes debranching equally well in low-salt and physiological buffers, we performed all subsequent experiments in physiological buffer except where noted.
Cofilin Induces Authentic Debranching.
Using the assay described above, we investigated whether cofilin, like GMF, is capable of inducing bona fide debranching events. At cofilin concentrations of 100 nM or higher, authentic debranching (validated by the concomitant loss of the daughter filament and Arp2/3 complex) was readily observed (examples in Fig. 2A).
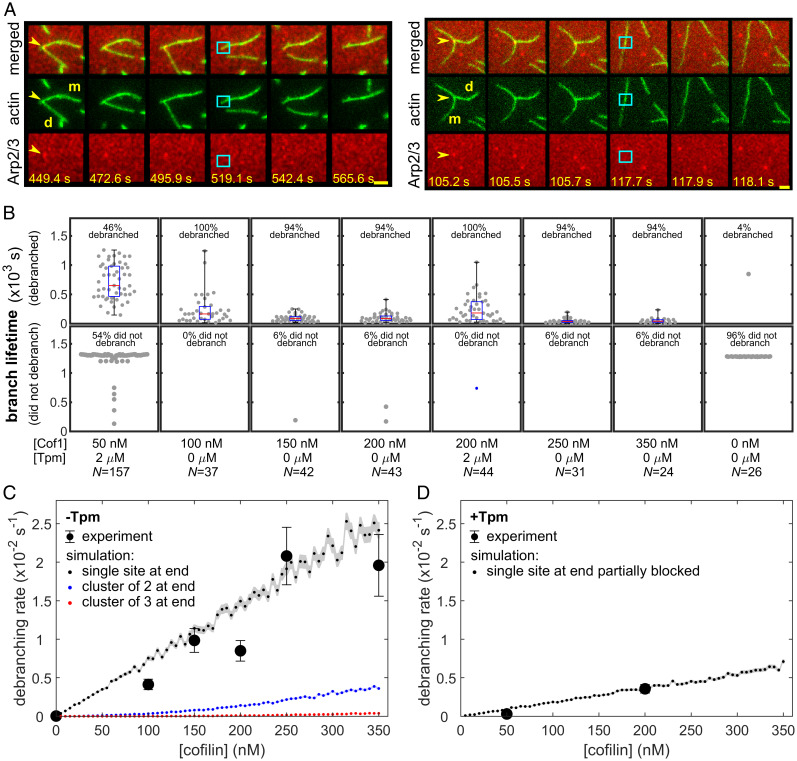
Fig. 2.
Cofilin-induced debranching. (A) Examples of debranching mediated by 200 nM cofilin in physiological buffer. Each triplet of image rows depicts a sequence of consecutive frames showing a branch and its subsequent debranching in merged view (Top), actin channel (Middle), and Arp2/3 channel (Bottom). The time stamp (seconds) for each image is indicated (Scale bars, 2.4 µm). Yellow arrows indicate the branches of interest; cyan squares mark the former branch site on the mother (m) filament immediately after the dissociation of the daughter (d) filament and the simultaneous disappearance of the Arp2/3 complex. (B) Debranching at varying concentrations of cofilin in physiological buffer in the presence or absence of 2 µM Tpm1.7. Points indicate lifetimes of individual branches seen to debranch (Top) or total time of observation for branches that lasted until the end of the experiment or diffused away without being seen to debranch (Bottom). Whisker plots indicate median (red), 25th and 75th percentiles (blue), and maximum and minimum (black). Lifetime distributions and fits are presented in SI Appendix, Fig. S2, and censoring-corrected debranching rates are given in SI Appendix, Table S2. (C) Debranching rates (± SE) calculated from the data in B for experiments without (black circles) Tpm1.7. Also shown are debranching rates predicted by a model in which cofilin binding to only a single site at the end of an actin filament (for example, to the last actin subunit at the daughter filament pointed end) is enough to trigger debranching (black points). Also shown are results for analogous models in which debranching requires simultaneous binding of cofilin molecules to at least two (blue points) or three (red points) consecutive binding sites at the filament end. Analogous results are obtained (SI Appendix, Fig. S3) for models in which the cofilin binding site(s) that triggers debranching is in the middle of a filament (e.g., on the mother filament adjacent to Arp2/3) rather than at the end. Note that the simulations are not fits to the data; they are predictions with no free parameters calculated from known cofilin binding/dissociation rate constants (see Materials and Methods). Shading shows the SE of the model predictions. (D) The same as for C, but for debranching rates measured in the presence of 2 µM Tpm1.7. In this case, the simulated model includes the additional assumption that the single actin site is obstructed by Tpm 75% of the time, a value that was chosen for good agreement with the data.
Cofilin binding to actin filaments is cooperative. At low nanomolar concentrations of cofilin, isolated individual cofilin molecules sparsely decorate the sides of filaments (32–34). In contrast, at micromolar concentrations of cofilin, intermittent segments of dense cofilin decoration form (35, 36). In the nanomolar range, we found that debranching scaled roughly linearly with cofilin concentration (Fig. 2C, black). Monte Carlo kinetic simulations based on rate constants for cofilin binding and release from actin filaments (32) showed that the observed debranching rates were consistent with a simple model (Fig. 2C, black points; SI Appendix, Fig. S3, black points) in which the binding of cofilin to a particular single site on the actin filament (for example, the last actin subunit at the daughter filament pointed end, or a specific subunit on the mother filament adjacent to the Arp2/3 complex) was sufficient to induce debranching. In contrast, models in which debranching was induced only by the simultaneous binding of cofilin to two (or more) consecutive sites on a filament did not match the data (Fig. 2C, blue or red; SI Appendix, Fig. S3, blue or red).
At 50 nM cofilin, the severing of actin filaments was prevalent, interfering with the accurate measurement of debranching rates. Therefore, to further differentiate between severing and authentic debranching, we measured debranching rates on filaments in the presence of tropomyosin (Tpm), which decorates the sides of filaments and protects them against severing (37–39). We incubated preformed branches with 2 µM human Tpm1.7, a concentration previously reported to almost completely block cofilin-induced severing (38), and then exposed filaments to 50 or 200 nM cofilin in the continued presence of 2 µM Tpm1.7. As expected, Tpm1.7 strongly suppressed severing. On the other hand, it only moderately reduced the rate of debranching (Fig. 2B; compare Fig. 2 C and D). The observation that Tpm1.7 blocks severing along the lengths of filaments without blocking cofilin-mediated debranching could indicate that the sites that must be bound by cofilin to trigger debranching are only weakly obstructed by Tpm. This observation agrees well with our simulation data above.
Independent Stimulation of Debranching by Cofilin and GMF.
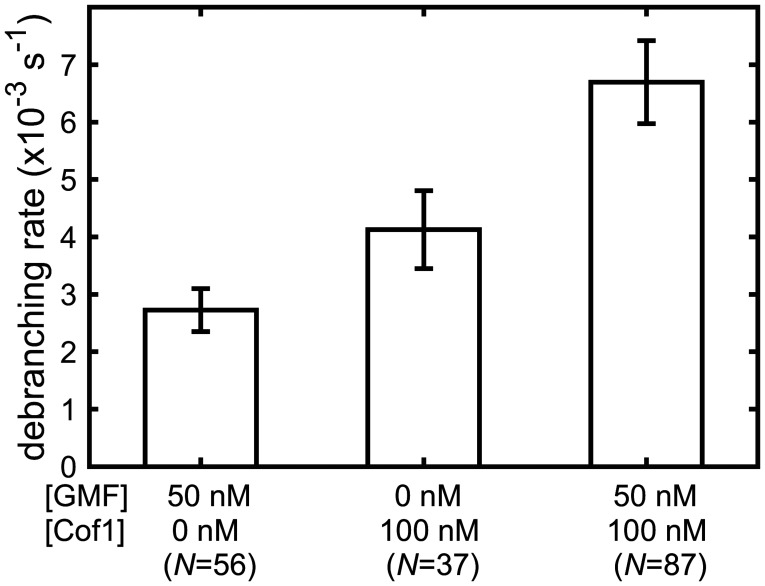
While previous work has shown that GMF directly interacts with the Arp2/3 complex to stimulate debranching (13, 23, 24), our data suggest a mechanism in which cofilin stimulates debranching by binding to actin. Therefore, we tested whether cofilin and GMF might work together to stimulate debranching by comparing their individual and combined activities (Fig. 3). For these measurements, we used a concentration of GMF (50 nM) that nearly saturated its debranching activity (13). Specifically, we investigated whether the simultaneous presence of 100 nM cofilin further enhances the rate of debranching over that seen at near-saturating GMF. The measured debranching rate for 50 nM GMF plus 100 nM cofilin was (6.7 ± 0.7) × 10−3 s−1 and was nearly identical to the sum of the individual debranching rates for 50 nM GMF alone and 100 nM cofilin alone, (6.8 ± 0.8) × 10−3 s−1. The results show that cofilin and GMF have additive effects in stimulating debranching under these conditions. These data are consistent with a mechanism in which cofilin and GMF bind independently to distinct (non-overlapping) sites at branch junctions to promote daughter filament dissociation.
Fig. 3.
GMF and cofilin independently promote debranching. Debranching rates (± SE) mediated by 50 nM GMF and 100 nM cofilin individually and together were measured and analyzed as in SI Appendix, Figs. S1 and S2 in physiological buffer. Experiments were performed in the absence of Tpm. Lifetime distributions and fits are presented in SI Appendix, Fig. S2C, and censoring-corrected debranching rates are given in SI Appendix, Table S2.
Debranching Is Stimulated by Cofilin Binding to the Daughter Filament at Branch Sites.
The foregoing data suggest that cofilin stimulates debranching by binding to actin filaments but that cofilin-mediated debranching is only weakly inhibited by Tpm1.7, which can decorate actin filaments and block most cofilin binding. Together, these data suggest that debranching results from cofilin binding to regions of the filament that are poorly protected by Tpm. Interestingly, previous studies have shown that Tpm decoration is weaker at filament ends (38, 40), raising the possibility that Tpm may poorly protect daughter filament pointed ends at branch junctions. To investigate the essential locations for cofilin binding during debranching events, we again performed debranching assays in the presence of Tpm1.7 but this time used fluorescently labeled cofilin (Cy3-Cof1) at 200 nM (Fig. 4 and SI Appendix, Fig. S4 and Movies S1–S4). In these experiments, branched AF488-labeled actin filaments were assembled and then decorated with Tpm1.7 (2 µM). Next, Cy3-Cof1 was introduced (along with 2 µM Tpm1.7), and debranching was monitored over time. To allow fast simultaneous imaging of Cy3-Cof1 and JF646-Arp2/3 (0.2 s per frame), we imaged actin filaments only intermittently in these experiments. Cy3-Cof1 decoration was apparent at the pointed ends of many filaments, as previously described (38). In addition, a clearly visible Cy3-Cof1 spot appeared at branch junctions prior to most (10 out of 11) of the observed debranching events. This spot often displayed a gradual increase in fluorescence intensity approaching the time of debranching (e.g., black traces in Fig. 4B and SI Appendix, Fig. S4 B, D, and F), suggesting that it represented the formation of a short cofilin-decorated segment proximal to the branch junction. Upon debranching, Cy3-Cof1 fluorescence was seen to depart with the pointed end of the daughter filament. In all (11 out of 11) of these debranching events, Cy3-Cof1 fluorescence at the daughter pointed end continued to increase even after debranching. In contrast, in cases where the former branch site on the mother filament could be unambiguously tracked just after debranching, we observed little or no detectable Cy3-Cof1 fluorescence remaining at that location. The rapid divergence of black and blue traces in Fig. 4B and SI Appendix, Fig. S4 B, D, and F shows that the departure of the daughter filament–cofilin complex from the former branch site on the mother filament occurred within a fraction of a second of debranching.
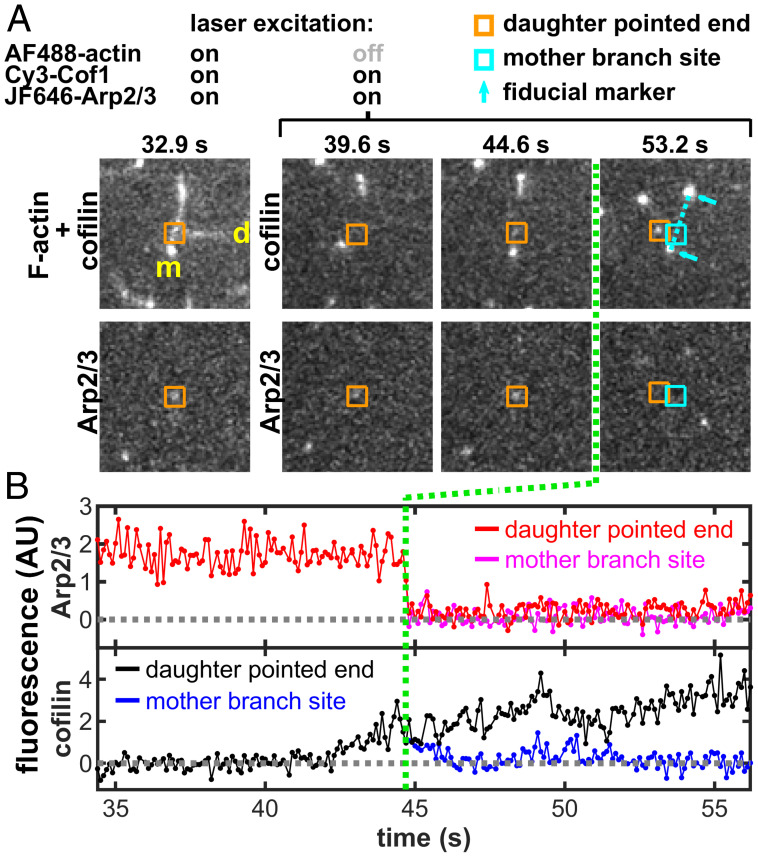
Fig. 4.
Example of a debranching event with fluorescently labeled cofilin and Arp2/3 complex in the presence of 2 µM Tpm1.7, which occurs at the designated time (green line). The same event is shown in Movie S1; other examples are shown in SI Appendix, Fig. S4 and corresponding Movies S2–S4. The 3-color experiment used fluorescently labeled actin (blue-excited AF488-actin), cofilin (green-excited Cy3-Cof1), and Arp2/3 complex (red-excited JF646-Arp2/3) in physiological buffer. In this experiment, green and red lasers were on continuously, whereas the blue laser was only on intermittently (only at 32.9 s, in the images shown here). (A) Image pairs at selected time points showing cofilin (Top) and the Arp2/3 complex (Bottom) before debranching (at 39.6 s and 44.6 s) and after debranching (at 53.2 s). The top panel of the first image pair (32.9 s) shows superimposed actin and cofilin fluorescence. The position of the pointed end of the daughter filament is inferred (orange square) from the position of the JF646-Arp2/3 fluorescence spot (before debranching), and from tracking the daughter pointed end Cy3-Cof1 decoration (after debranching). After debranching, the position on the mother filament at which the branch had been present (cyan square) is inferred by interpolating between the Cy3-Cof1 decorated mother filament ends. A 9.7 µm × 9.7 µm area centered at the mother branch site of each image is displayed. The images displayed here were spatially filtered by a 2 × 2 matrix of ones for clarity. (B) Fluorescence recorded over a time period spanning the debranching event in A. Plots show Cy3-Cof1 and JF646-Arp2/3 intensities integrated over 3 × 3 pixel areas tracking with the daughter pointed end and mother branch site. For each protein, only a single record is shown to the left of the green line since the locations of the daughter pointed end and mother branch site are identical before debranching.
In the above experiments, actin was imaged less frequently than the other color channels. Nevertheless, immediately after debranching the relative movement of the daughter filament pointed end away from the former branch site on the mother filament could be inferred from the movement of fiducial marks on the filaments visible in the Cy3-Cof1 and/or JF646-Arp2/3 channels. In addition, we also performed two kinds of confirmatory experiments: 1) we continuously acquired images of actin alternating with images of cofilin + Arp2/3 (e.g., Movie S5, corresponding to SI Appendix, Fig. S5A), and 2) we continuously acquired images of cofilin alternating with images of actin + Arp2/3 (e.g., Movies S6 and S7, corresponding to SI Appendix, Fig. S5 B and C). These experimental designs allowed higher time–resolution imaging of the daughter and mother filaments to better confirm their positions immediately after debranching. The results from these experiments again almost always (16 of 17 debranching events) showed Cy3-Cof1 accumulating at the daughter filament pointed end before debranching and then departing with the daughter filament upon debranching. In the examples shown, the former branch site on the mother filament appeared to have little or no detectable Cy3-Cof1 fluorescence remaining just after debranching. While we cannot unambiguously exclude the possibility that debranching was caused by mother-associated cofilin (e.g., binding that is too dim or transient to be detected), debranching in all the 3-color experiments was almost invariably associated with a visible Cy3-cofilin signal that departed with the daughter filament. The most straightforward interpretation of the data is that cofilin stimulates debranching in the presence of Tpm by binding to a Tpm-unprotected (or weakly protected) segment of the daughter filament near the branch junction.
Single-Molecule Imaging of Intermediate States in Debranching.
GMF and cofilin each can accelerate debranching by ∼50-fold or more (Fig. 2C and SI Appendix, Fig. S1B). However, which of the two Arp2/3–filament interfaces is first disrupted during debranching events has remained an open question. In principle, the Arp2/3 complex could initially leave with the pointed end of the daughter filament, or alternatively it could remain on the side of the mother filament (Fig. 5A). These debranching intermediates are inherently difficult to detect because the diffusive separation of mother and daughter filaments after debranching is slow compared to the diffusion of the released Arp2/3 complex away from filaments. Likely for these reasons, the intermediates were not seen in prior studies (29).
Fig. 5.
GMF and cofilin can both debranch by breaking the Arp2/3–mother filament interaction. (A) Cartoon illustrating alternative possible debranching pathways. (B) Two examples of frame sequences showing that Arp2/3 is transiently retained on the daughter filament after GMF-mediated debranching recorded in physiological buffer (Top) or imidazole low-salt buffer (Bottom). The yellow arrows mark branches of interest with indicated mother (m) and daughter (d) filaments; squares mark the position of the daughter pointed end in the first frame after debranching showing the retention of Arp2/3. (C) Two examples of Arp2/3 retention on the daughter filament after cofilin-mediated debranching in imidazole low-salt buffer. The yellow arrows mark branches of interest with indicated mother (m) and daughter (d) filaments; squares mark the position of the daughter pointed end in the first frame after debranching showing the retention of Arp2/3 (Scale bars, 2.4 µm).
Using single-molecule analysis (with labeled Arp2/3 complex) we attempted to “catch” the short-lived intermediate states during debranching stimulated by cofilin and GMF by following ∼100 debranching events (for cofilin and GMF each) using high time resolution (0.1 to 0.5 s). Even under these optimized conditions, in most cases the Arp2/3 complex departure and relative movement of mother and daughter filaments occurred in the same frame of the recording. However, we captured a few examples (five for GMF and four for cofilin) at various time resolutions that resolved the transient intermediate. In each of these cases, the Arp2/3 complex left the branch junction attached to the pointed end of the daughter filament (Fig. 5 B and C). The interaction was short-lived, with the Arp2/3 complex disappearing from the pointed end of the daughter filament in the video frame immediately following the frame in which debranching was observed. While this evidence is not conclusive, it hints at a common mechanistic feature shared by GMF and cofilin, in which both proteins induce destabilization of the mother filament–Arp2/3 complex interaction.
Discussion
Actin filament branch junctions assembled by the Arp2/3 complex are inherently stable structures, spontaneously dissociating in vitro on a time scale of tens of minutes to hours (11–13) (Fig. 2 and SI Appendix, Fig. S1 B and C). The formation of branches is coupled to conformational changes in the Arp2/3 complex that alter both the mother filament side-binding and daughter filament pointed-end–binding interfaces (15–18). The Arp2/3 complex in solution exists in a spectrum of conformational states, which can be influenced by binding of NPF activators, mother filament side-binding, and daughter filament polymerization (18, 41–43). Arp2/3 complex in solution (with or without NPF bound) makes only transient interactions with the side of a mother filament (14). However, the mature branch junction conformational state of the Arp2/3 complex is achieved only after binding to the mother filament (or the mother analog protein Dip1), association of an NPF and actin subunits, dissociation of the NPF, and polymerization of the daughter filament (17, 44). This conformation has highly stable binding interactions with both the mother side and daughter pointed end.
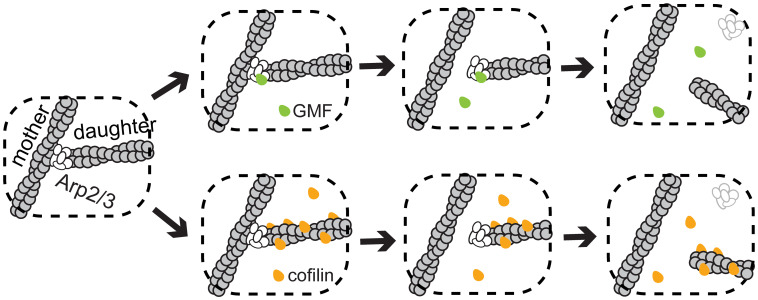
Debranching-promoting factors drive the reverse pathway. In Arp2/3 complex in solution (in the absence of actin), GMF binds to a single high-affinity site on the ARPC1 and Arp2 subunits (23, 24) and possibly an additional lower-affinity site on Arp3. The site(s) at which GMF binds to a branch junction to promote its disassembly is not unambiguously established and is proposed to involve contacts with Arp2/3 complex subunits and possibly the daughter filament but not the mother filament (24). In the ∼5% of cases in which we could resolve the order of departure, the Arp2/3 complex was transiently retained on the daughter pointed end after GMF-stimulated mother-daughter detachment. Therefore, we propose the simple 2-step model for GMF-induced debranching shown in Fig. 6 (Top). In this mechanism, GMF binding to its site(s) at the branch junction drives a conformational change in the Arp2/3 complex that allosterically destabilizes the mother filament–Arp2/3 complex interface and promotes Arp2/3 complex release from the side of the mother filament. Since we cannot resolve the order of departure in most cases, we cannot exclude the possibility that in some debranching events the Arp2/3 complex dissociates from the daughter filament pointed end before it leaves the mother filament.
Fig. 6.
Proposed independent pathways of GMF- and cofilin-stimulated debranching. GMF (green) triggers branch dissociation by binding to Arp2/3 complex (white), while cofilin (orange) acts by binding to a site at the pointed end of the daughter filament.
We show here that under physiological ionic conditions, cofilin, like GMF, is an authentic actin network debrancher that disrupts the interaction between the Arp2/3 complex and the mother filament rather than merely severing the daughter filament near the branch junction. The existence of a cofilin-induced debranching pathway, distinct from severing, is consistent with previous observations showing that in the absence of Tpm, severing rates are highest on filaments that are only partially occupied by cofilin, whereas debranching is maximal on fully cofilin-occupied networks (28).
Our demonstration of the additive effects of cofilin and near-saturating concentrations of GMF on the rate of debranching suggest that the two proteins could act by binding to distinct, nonoverlapping sites. While it is possible that the cofilin site is partially or completely on the Arp2/3 complex, the cofilin concentration dependence of the debranching rate is quantitatively consistent with cofilin binding to a single, unperturbed binding site on either the mother or daughter filament. An early study (28) implicated mother filament binding by cofilin in debranching, but this conclusion was based on a pyrene–Arp2/3 complex fluorescence assay that may underestimate the relevant Arp2/3–filament interaction kinetics by 2 to 3 orders of magnitude (14). With fluorescently labeled cofilin, we see that the protein is present at the branch site prior to debranching and then usually departs with the daughter filament. The data support a simple mechanism (lower branch in Fig. 6) in which cofilin needs only bind to a single site on the daughter filament near or at its pointed end to induce debranching. The quantitative model predicts that this site becomes occupied either by the binding of a single isolated cofilin molecule or by the growth of a cofilin-decorated segment of actin (events that are more likely to occur at low and high concentrations of cofilin, respectively). We depict cofilin-induced debranching as proceeding via a 2-step process in which the Arp2/3 complex leaves with the daughter filament. As above with GMF, this is a provisional conclusion based on extrapolation from the small fraction of debranching events in which the two steps can be observed in sequence.
Cofilin-stimulated debranching has been proposed to occur by destabilizing the Arp2/3 complex interfaces with either the daughter (24) or the mother (28) filament. Our data most closely support a model in which cofilin decoration on the daughter filament triggers debranching by destabilizing the Arp2/3–mother filament interface. The binding of cofilin induces structural rearrangements in the actin filament that propagate more toward the pointed end than the barbed end (45). It is possible that the binding of a cofilin molecule at (or near) the daughter filament pointed end can cause an analogous structural rearrangement of the Arp2 and Arp3 subunits, along with other conformational changes in the Arp2/3 complex, promoting the dissociation of daughter and Arp2/3 from the side of the mother filament.
Finally, our data suggest how GMF and cofilin, by their analogous but complementary debranching mechanisms, might work in concert in cells to promote the turnover of branched actin networks and recycling of the Arp2/3 complex. This is likely true even for branched networks decorated by Tpm, on which severing is substantially inhibited but debranching is not.
Materials and Methods
Actin Purification and Labeling.
Rabbit skeletal muscle actin (RMA) and labeled RMA were purified as follows. Actin monomers were purified from acetone powder as described in ref. 46: “Rabbit skeletal muscle actin was purified from acetone powder [(47)] generated from frozen ground hind leg muscle tissue of young rabbits (Pel-Freez). Lyophilized acetone powder stored at −80°C was mechanically sheared in a coffee grinder, resuspended in G-buffer (5 mM Tris-HCl pH 7.5, 0.5 mM DTT, 0.2 mM ATP, and 0.1 mM CaCl2), and then cleared by centrifugation for 20 min at 50,000 × g. Supernatant was collected and further filtered with Whatman paper. Actin was polymerized by the addition of 2 mM MgCl2 and 50 mM NaCl to the filtrate and overnight incubation at 4°C with slow stirring. Next morning, NaCl powder was added to a final concentration of 0.6M, and stirring was continued for another 30 min at 4°C. F-actin was pelleted by centrifugation for 150 min at 120,000 × g, and the pellet was solubilized by Dounce homogenization and dialyzed against G-buffer for 48 h at 4°C. Monomeric actin was then precleared at 435,000 × g and loaded onto an S200 (16/60) gel-filtration column (GE Healthcare) equilibrated in G-buffer. Fractions containing actin were stored at 4°C.”
Fluorescently labeled actin monomers were obtained by polymerization cycling of F-actin labeled with Alexa Fluor 488 tetrafluorophenyl ester (ThermoFisher Scientific) as described in ref. 14: “To fluorescently label actin, it was first polymerized overnight at 4°C in Pipes buffer [50 mM KCl, 0.1 mM MgCl2, 0.2 mM ATP, 25 mM K+-Pipes (pH 8.3)], reacted with Alexa Fluor 488 tetrafluorophenyl ester (AF488-TFPE; Molecular Probes) for 3 h at 22°C, dialyzed against G-buffer [3 mM Tris·Cl−, 0.1 mM CaCl2, 0.2 mM ATP, 0.5 mM DTT (pH 8)] for 2 d at 4°C to depolymerize, and gel filtered [(47)]”.
Purification and Labeling of SNAP-Tagged Arp2/3 Complex.
SNAP-tagged S. cerevisiae Arp2/3 complex was purified and labeled using a protocol modified from a previously published method (48). Yeast strain BGY1430, expressing ARC18-SNAP-TEV-3HA (14), was grown in 4 × YPD in Fernbach flasks with shaking to OD600 ∼ 20, washed in water, resuspended at 1:1 wt/wt in a modified HEK buffer (MHEK: 50 mM Hepes pH 7.4, 1 mM EGTA, 50 mM KCl, 2 mM MgCl2), flash-frozen in droplets into liquid nitrogen, and stored at −80 °C. Frozen cells were mechanically sheared in a coffee grinder at liquid nitrogen temperature and thawed by the addition of MHEK buffer plus an EDTA-free CØmplete protease inhibitor tablet (Roche, EDTA-free). The lysate was clarified by centrifugation at 300,000 × g for 30 min. An anti-HA antibody Sepharose column was constructed by conjugating 100 µL ascites fluid of an HA monoclonal antibody (Clone HA-7, H9658, Sigma) to 1 g cyanogen bromide–activated Sepharose 4B (C9142, Sigma) according to the manufacturer’s protocol. The clarified yeast lysate was passed through a 4 mL anti-HA antibody Sepharose column 3 times at 4 °C. The column was washed with five column volumes (20 mL) each of MHEK buffer plus 0.25 mM ATP (MHEKATP) and 500 mM KCl in MHEKATP (MHEK500ATP) buffer, and again with MHEKATP. After the last wash, 5 mL 1 μM BG-JF646 dye substrate (49) in MHEKATP buffer supplemented with 0.5 mM DTT was washed into the column to label the bound HA-TEV-SNAP-Arp2/3 complex at room temperature in the dark for 1.5 h. Excess dye substrate was washed away at 4 °C with five column volumes each of MHEKATP and MHEK500ATP buffer. The column was then equilibrated with five column volumes of storage buffer (MHEK + 20% sucrose + 0.5 mM ATP + 0.5 mM DTT). SNAP-Arp2/3 complex was then released from the column by overnight digestion at 4 °C with 500 units of ProTEV plus (Sigma V6101) in 5 mL storage buffer and eluted with ∼6 mL storage buffer. This sample was concentrated (100K Amicon Ultra) to ∼1 mL final volume, aliquoted, flash-frozen, and stored at −80 °C. Labeling efficiency was estimated by counting the fraction of branch junctions that had labeled Arp2/3 complexes by TIRF microscopy and was typically between 70% -80%. Arp2/3 concentration was estimated by Coomassie-stained SDS gel using bovine serum albumin as a standard.
VVCA Purification.
A monomeric VVCA protein corresponding to amino acids 393 to 503 of human N-WASP protein was expressed and purified using a GST-VVCA construct (pBG1579, pCOOL-VVCA human N-WASP) (50). GST-VVCA was expressed in Escherichia coli strain BL21(DE3)pLysS for 4 h at 37 °C after induction with 1 mM IPTG. Cells were harvested and resuspended in buffer V (50 mM Hepes, pH 7.5, 50 mM KCl), flash-frozen, and stored at −80 °C. Frozen cells were thawed, and at the same time a CØmplete protease inhibitor tablet (Roche, EDTA-free) and 1 mm DTT were added. Cells were then lysed by sonication and clarified by centrifugation at 300,000 × g for 30 min. Glutathione agarose (Pierce 16100) was washed 4 times by pelleting and resuspension with an equal volume of buffer V plus 1 mM DTT. After a final pelleting, 0.75 mL washed glutathione agarose pellet was mixed with 3 mL clarified lysate on a rotator at 4 °C for 1.5 h. The VVCA-bound resin was again washed 4 times and pelleted. VVCA was then released by mixing the pellet with 0.75 mL buffer V plus 1 mM DTT containing 1 unit of biotinylated-thrombin (MilliporeSigma 696723) and incubating at 4 °C for 2 h. Released VVCA was recovered by pelleting the agarose. VVCA concentration was measured by its absorbance at 280 nm (ε280 = 5,500 M−1 cm−1). Glycerol (20% final) was added, and then the protein was aliquoted and flash-frozen for storage at −80 °C.
Cofilin Purification.
A plasmid for expressing GST-tagged S. cerevisiae cofilin (GST-Cof1) with a preScission protease cut site was engineered by ligating the Cof1 coding region from pGEX2T-Cof1 (51) into pGEX6P1 using BamHI/EcoRI sites. GST-Cof1 was expressed in E. coli BL21(DE3)pLysS induced with 1 mM IPTG at 37 °C for 4 h. Cells were harvested, resuspended in buffer V, and stored frozen at −80 °C. Cells were thawed and resuspended in buffer V supplemented with a CØmplete protease inhibitor tablet (Roche, EDTA-free) and 1 mM DTT, lysed by sonication, and clarified by centrifugation at 30,000 g for 20 min. Commercial glutathione agarose was washed as described above before use. The clarified lysate was mixed with the washed glutathione agarose, at a ratio of 5 mL lysate to 3 mL glutathione agarose, on a rotator at 4 °C for 2 to 3 h. The cofilin-bound resin was washed 5 times with an equal volume of buffer V and 1 mM DTT. Cofilin was released by cleavage with 4.5 unit/mL of preScision protease (Pierce HRV 3C protease) at 4 °C overnight, and then the agarose beads were removed by centrifugation. Released cofilin was further purified through a prepacked 1 mL HiTrapQ column (GE Healthcare) using a 50 mM to 1 M NaCl linear gradient (20 min run time at 1 mL/min flow rate) in 50 mM Hepes buffer, pH 7.5. The concentration of purified cofilin was determined by its absorbance at 280 nm (ε280 = 15,930 M−1 cm−1). Then glycerol (20% final) was added before aliquoting and flash-freezing for storage at −80 °C.
Cy3-Cof1 Purification and Labeling.
A single-cysteine Cof1 mutant (T46C/C62A) (52) was purified and labeled as described in ref. 36: “The protein was expressed fused to a glutathione-S-transferase (GST) tag with a thrombin cleavage site and expressed in E. coli BL21 DE3. Cells were grown to log phase at 37°C in TB medium, then induced with 1 mM IPTG at 18°C overnight. Cells were harvested by centrifugation and pellets were stored at −80°C. Frozen pellets were resuspended in lysis buffer (20 mM NaPO4 pH 7.8, 300 mM NaCl, 1 mM DTT, 1 mM PMSF + protease inhibitors [0.5 μM each of pepstatin A, antipain, leupeptin, aprotinin, and chymostatin]). Cells were lysed by sonication with a tip sonicator while keeping the tubes on ice. The lysate was cleared by centrifugation at 150,000 × g for 30 min at 4°C. The supernatant was then incubated with glutathione-agarose beads for 1 h on a rotator at 4°C. The beads were first washed thoroughly with washing buffer (50 mM Tris pH 7.5, 150 mM NaCl and 1 mM DTT) to remove unbound protein and then incubated with thrombin (0.05 mg/mL) to cleave Cofilin from bead-bound GST. The cleaved protein was recovered by centrifugation. The supernatant containing the protein was concentrated and loaded on to a Superose 12 gel-filtration column (GE Healthcare, Pittsburgh, PA) pre-equilibrated with 10 mM Tris pH 7.5, 50 mM NaCl and 1 mM DTT. The fractions containing Cofilin were pooled, concentrated, snap frozen in liquid N2 and stored at −80°C. To prepare fluorescently labeled Cofilin, Cof1(T46C/C62A) was purified as described above and dialyzed overnight against 10 mM Tris-HCl pH 7.5, 50 mM NaCl, and 0.2 mM Tris(2-carboxyethyl)phosphine (TCEP) at 4°C. The dialyzed protein was then mixed with a 10-fold molar excess of Cy3-maleimide (GE Healthcare, Pittsburgh, PA) and incubated overnight in the dark at 4°C. Free dye was removed using a PD-10 desalting column. The labelled protein was then aliquoted, snap frozen in liquid N2 and stored at −80°C.”
GMF Purification.
S. cerevisiae GMF (Gmf1) was a gift from Dr. Siyang Guo and was prepared as described in ref. 48: “S. cerevisiae Gmf1 was expressed as cleavable glutathione S-transferase (GST) fusions in E. coli strain BL21(DE3) T1R. Cells were grown to mid log phase at 37°C and expression was induced for 16 h at 20°C by addition of 1mM IPTG. Cells were harvested by centrifugation, and each 1 L of culture was resuspended in 30 mL of Buffer GSH-WB1 (200 mM NaCl, 20 mM Tris/HCl pH 8.0, 2 mM EDTA, 2mM DTT). During resuspension, 1 mM PMSF was added. Cells were lysed by extrusion and clarified by centrifugation at 46,000 × g in a JA25.50 rotor (Beckman Coulter Inc.) for 30 min at 4°C. Supernatants were mixed with glutathione Sepharose 4B beads (GE Healthcare) and rocked at 4°C for 40 min, then collected in a 1.2 cm wide disposable column. Beads were washed three times with 10 column volumes of Buffer GSH-WB1. GST tags were removed by overnight, in-column digestion at 4°C with HRV3C protease (Novagen Inc., Madison, WI). Released Gmf1... [was] collected and loaded on a homemade 4 mL SOURCE15Q anion exchange column. The column was run with QA7I buffer (10 mM Imidazole pH 7.0, 1 mM DTT) and QB7I buffer (10 mM Imidazole pH 7.0, 1 mM DTT, 1 M NaCl). The column was equilibrated at 3% QB7I. The collected protein sample was diluted fourfold with QA7I and applied to the column, and then proteins were eluted with a 25 column-volume linear gradient of CB71 (10–60%). Peak fractions containing Gmf1 were collected and flash frozen in 0.6 mL aliquots. Prior to use, an aliquot was thawed, centrifuged for 10 min at 15,000 × g at 4°C, and the upper 0.5 mL was applied to a Superdex 200 (10/300) column (GE Healthcare) equilibrated in 10 mM Na+-HEPES pH 7.5, 50 mM KCl, 1 mM EGTA pH 8.0, 2 mM MgCl2, 0.5mM DTT, and 0.1 mM ATP. The concentration of Gmf1 was assessed by absorbance at 280 nm.”
Tpm1.7 Purification.
Human Tpm1.7 was a gift from Dr. Sylvia Jansen and was prepared as described in ref. 38: “Human TPM isoforms were expressed in BL21 (DE3) E. coli by growing cells at 37°C in TB medium to log phase and then inducing expression with 1 mM IPTG at 18°C for 16 h. Cells were harvested by centrifugation, and lysed by sonication in 20 mM Tris (pH 7.5), 0.5 M NaCl, and 5 mM MgCl2. The lysate was then incubated at 80°C for 10 min in a water bath, cooled for 10 min at −20°C, and cleared by centrifugation at 30,000 × g for 20 min. Next, the supernatant was isoelectrically precipitated by dropwise addition of 0.3 M HCl to pH ∼4.7 and centrifuged at 5000 rpm for 15 min at 4°C, after which the pellet was resuspended in 100 mM Tris (pH 7.5), 0.5 M NaCl, 5 mM MgCl2, and 1 mM DTT. This precipitation step was repeated one more time, and the resuspended pellet was dialyzed to 20 mM HEPES (pH 6.8), 50 mM NaCl, and 0.5 mM DTT. Next, dialyzed protein was applied to a 5 mL HiTrap Q HP column (GE Healthcare Biosciences) and eluted with a linear gradient of NaCl (50–600 mM). The fractions containing Tpm1.7 were concentrated and further purified on a Superose 6 gel-filtration column (GE Healthcare Biosciences) equilibrated in 20 mM Tris (pH 7.5), 50 mM KCl, 2 mM MgCl2, and 1 mM DTT). Aliquots were flash-frozen and stored at −80°C.“
Visualization of Single-Arp2/3 Complex Debranching.
Multiwavelength debranching assays with visualization of individual Arp2/3 complexes were performed on a custom-built TIRF microscope (53). Sample chambers were assembled from cleaned glass coverslips that were passivated by a silane derivative of methoxy polyethylene glycol (Laysan Bio, mPEG-silane MW 2,000). Coverslips (No. 1.5, 64 mm × 24 mm and 25 mm × 25 mm) were first cleaned by sonication at room temperature for 1 h each in 2% Micro-90 (Cole Parmer), ethanol, and 0.1 mM KOH. The coverslips were rinsed thoroughly with purified water after each of these cleaning steps. Next, they were dried thoroughly in a stream of filtered nitrogen, placed in contact on one side with 20 mg/mL mPEG-silane dissolved in 1:4 (vol/vol) water adjusted to pH 1 with HCl:ethanol (13, 54), and dried overnight at 70 °C. Prior to use, residual silane was removed by rinsing with water or with 10 mM Tris buffer (pH 8.0), and the coverslips were dried again with nitrogen. The sample chambers were constructed by sandwiching thin lines of silicone vacuum grease (Dow Corning), forming the boundaries of the sample lanes, between the passivated surfaces of a 25 mm × 25 mm and a 64 mm × 24 mm coverslip.
TIRF experiments were performed in either low-salt or physiological ionic conditions (SI Appendix, Table S1). The imaging solutions were prepared just before use from 3× or 10× buffer stocks (SI Appendix, Table S1), to yield final concentrations of 1× ions, 0.5 mM ATP, 1 mM DTT, 0.2% methylcellulose (Sigma M0512, 4000 cP), O2-scavenging components (4.5 mg/mL glucose [Fisher D16], 0.22 mg/mL glucose oxidase [Sigma G2133], and 0.036 mg/mL catalase [Sigma C3155]), and triplet state quenchers (55) (1 mM propyl gallate [Fluka 02370], 2 mM 4-nitrobenzyl alcohol [Aldrich N12821], and 2 mM Trolox [Aldrich 23881]).
To perform debranching experiments, branches were first assembled on the slide by combining monomeric actin (0.5 to 1 µM, 15% labeled), VVCA (100 nM), and Arp2/3 (60 nM) in low-salt buffer. (Branches formed in physiological buffer contained fluorescent actin aggregates, so this condition was not used for branch assembly.) After a suitable number of branches formed (typically 5 to 10 min), branch formation was stopped by flushing the free Arp2/3, VVCA, and monomeric actin from the chamber with low-salt or physiological imaging solution. The preformed branches were then exposed to flowed-in GMF and/or cofilin in imaging solution, or imaging solution alone for the buffer control.
Low-time-resolution (typically 10 to 20 s) image sequences (e.g., Fig. 1B) were collected by cycling through square arrays of 2 × 2 (4 fields of view) or 3 × 3 (9 fields of view) in order to increase the number of branches observed and acquiring images with 0.2 s integration time. High-time-resolution image sequences (0.1 to 0.5 s integration time) were collected continuously from a single field of view. Samples were simultaneously excited by 488 and 633 nm laser lines at 0.03 to 0.1 mW and 1.0 to 1.5 mW, respectively [powers measured incident to the final mirror upstream of the micromirrors (53)]. Experiments with Cy3-Cof1 used 0.2 s integration with simultaneous 532 nm (0.1 mW) and 633 nm (1.0 mW) excitation, with occasional added 488 nm excitation (0.03 mW). Emission was recorded with dual-view optics with a wavelength cutover of 635 nm. Subpixel mapping between points in the long- and short-wavelength dual-view channels was determined by single-molecule imaging of calibration samples containing oligonucleotides labeled with both Cy3 and Cy5 (53) using locally weighted–mean mapping (56).
Debranching Rate Measurement.
Filaments and branches in our experiments were not tethered to the surface and were free to move about. The recorded images were inspected frame by frame to identify and track branches in the recording. In all experiments, we selected for analysis branches with a daughter and mother filament joined at a branch point that colocalized with a fluorescently labeled Arp2/3 complex. The fluorescent spot from the Arp2/3 complex at the branch junction and the coordinated movements of the mother and daughter filaments were used to distinguish true branches from two filaments coming together at a single point briefly by chance.
A debranching event was scored when the disappearance of the Arp2/3 complex was accompanied by relative movement of the daughter filament pointed end and the mother filament that was detected within one video frame (10 to 20 s time resolution experiments) or within five frames (0.1 to 0.5 s time resolution experiments).
Under experimental conditions in which debranching was fast enough that most debranching events occurred before the end of the experimental recording, the distribution of branch lifetimes appeared exponential (e.g., SI Appendix, Figs. S1C and S2) as expected for a single rate-limiting step in debranching. For some branches, the branch lifetime ti was measured directly from the video recording. For other branches, debranching was not observed either because the branch drifted out of the field of view or the branch was still intact at the end of recording. In these cases, ti is the lifetime over which the branch could be observed, which represents a lower limit on the true branch lifetime. To account for the bias caused by this statistical censoring of some of the measurements, we calculated the Bayesian estimate of the mean debranching rate and its SE as [equation 21 in Ensign and Pande (57)]
and
where n is the number of observed debranching events and N is the total number of branches followed.
Tracking of Cofilin and Arp2/3 Complex During Debranching.
In debranching experiments using Cy3-Cof1 (Fig. 4 and SI Appendix, Fig. S4), we tracked the intensities at branch junctions and at the pointed ends of daughter filaments in both Cy3-Cof1 and JF646-Arp2/3 channels. Before debranching, the pointed ends of daughter filaments and the branch junctions were coincident. These locations were determined by fitting a 2-dimensional Gaussian to the Arp2/3 fluorescence spot image in each frame. After debranching events, the positions of the pointed ends of daughter filaments and the former branch junctions would separate. Cy3-Cof1 that had accumulated at the pointed end of the departed daughter filament was fitted to a 2-dimensional Gaussian to determine the location of the filament end. The former branch sites on mother filaments (after debranching events) were estimated by 2-dimensional Gaussian fitting of fiducial marks on the mother filaments: These could be point(s) of Cy3-Cof1 accumulation (mostly at the ends of mother filaments or at a position near a branch) or Arp2/3 complex fluorescence from another branch junction on the same mother filament. The position of the branch site relative to these fiducial marks before debranching was used to approximate the location of the former branch junction after debranching. All Gaussian fitting was performed on unprocessed images. Cofilin and Arp2/3 fluorescence intensity time records were measured as the integrated intensity of a 3 × 3 pixel area centered at the branch junction and/or daughter filament pointed end in each channel. The images containing Arp2/3 fluorescence displayed in the debranching examples shown in Figs. 1B, 2A, and 5 B and C were spatially filtered with a 4 × 4 matrix of ones .
Monte Carlo Simulation of Cofilin Binding.
Monte Carlo kinetic simulation of a debranching model in which cofilin binding to the final actin subunit at the daughter pointed end triggers debranching was performed by modeling a linear filament of 500 actin subunits in the presence of free cofilin at concentration L. Binding to each unoccupied actin subunit was simulated for each Δt = 0.1 s time interval using the binding probabilities Δt kf L, Δt ωf kf L, and Δt ωf2 kf L, respectively, for subunits with 0, 1, or 2 neighboring subunits bound by cofilin. ωf is the unitless cooperativity factor for binding, and kf is the second-order binding rate constant. In an analogous manner, dissociation from each occupied actin subunit during Δt was simulated using the dissociation probabilities Δt kr, Δt ωr kr, and Δt ωr2 kr, where ωr is the unitless cooperativity factor for dissociation and kr is the first-order dissociation rate constant. For comparison with the experimental data, each simulation was carried out to 1,200 s, the typical length of our debranching experiments. Debranching was considered to have occurred when a chosen cofilin site was occupied (Fig. 2C and SI Appendix, Fig. S3, black points), or when the chosen site and 1 additional (Fig. 2C and SI Appendix, Fig. S3, blue points), or 2 additional (Fig. 2C and SI Appendix, Fig. S3, red points) adjacent subunits were simultaneously occupied. Debranching rates were calculated from 500 simulations at each cofilin concentration using the same Bayesian method used for the experimental data. The simulations used the values of wr, kr, wf, and kf measured in single-molecule experiments (32) performed at lower ionic strength but otherwise under conditions (no pyrene actin; surface-proximal filaments) that closely correspond to those of our experiments.
Supplementary Material
Acknowledgments
We thank Professor Douglas Theobald for advice on analyzing censored lifetime data, Dr. Greg Hoeprich for RMA preparation, and Dr. Faisal Chaudhry for labeled cofilin. We thank members of the B.L.G. laboratory and J.G. laboratory for helpful discussions, and Drs. Greg Hoeprich and Shashank Shekhar for comments on the manuscript. This work was supported by grants from the NIH to B.L.G. (R35 GM134895) and J.G. (R01 GM081648), and by the Brandeis Materias Science Research and Engineering Center, Bioinspired Soft Materials (NSF DMR-2011846).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. B.N. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2115129119/-/DCSupplemental.
Data Availability
Source data for all figures, consisting of images, experimental and simulated branch lifetimes, and MATLAB code used to generate the figures and perform the Monte Carlo simulations, is available at Zenodo repository (https://zenodo.org DOI: 10.5281/zenodo.5207693) (58).
References
- 1.Senju Y., Lappalainen P., Regulation of actin dynamics by PI(4,5)P2 in cell migration and endocytosis. Curr. Opin. Cell Biol. 56, 7–13 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Bieling P., et al. , Force feedback controls motor activity and mechanical properties of self-assembling branched actin networks. Cell 164, 115–127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kage F., et al. , FMNL formins boost lamellipodial force generation. Nat. Commun. 8, 14832 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papalazarou V., Machesky L. M., The cell pushes back: The Arp2/3 complex is a key orchestrator of cellular responses to environmental forces. Curr. Opin. Cell Biol. 68, 37–44 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaumouillé V., Waterman C. M., Physical constraints and forces involved in phagocytosis. Front. Immunol. 11, 1097 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kast D. J., Dominguez R., The cytoskeleton-autophagy connection. Curr. Biol. 27, R318–R326 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Efimova N., Svitkina T. M., Branched actin networks push against each other at adherens junctions to maintain cell-cell adhesion. J. Cell Biol. 217, 1827–1845 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu C., et al. , Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell 148, 973–987 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lacy M. M., Baddeley D., Berro J., Single-molecule turnover dynamics of actin and membrane coat proteins in clathrin-mediated endocytosis. eLife 8, e52355 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyoshi T., et al. , Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: Evidence of frequent filament severing. J. Cell Biol. 175, 947–955 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amann K. J., Pollard T. D., Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc. Natl. Acad. Sci. U.S.A. 98, 15009–15013 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blanchoin L., Pollard T. D., Mullins R. D., Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr. Biol. 10, 1273–1282 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Gandhi M., et al. , GMF is a cofilin homolog that binds Arp2/3 complex to stimulate filament debranching and inhibit actin nucleation. Curr. Biol. 20, 861–867 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith B. A., Daugherty-Clarke K., Goode B. L., Gelles J., Pathway of actin filament branch formation by Arp2/3 complex revealed by single-molecule imaging. Proc. Natl. Acad. Sci. U.S.A. 110, 1285–1290 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson R. C., et al. , Crystal structure of Arp2/3 complex. Science 294, 1679–1684 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Shaaban M., Chowdhury S., Nolen B. J., Cryo-EM reveals the transition of Arp2/3 complex from inactive to nucleation-competent state. Nat. Struct. Mol. Biol. 27, 1009–1016 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fäßler F., Dimchev G., Hodirnau V.-V., Wan W., Schur F. K. M., Cryo-electron tomography structure of Arp2/3 complex in cells reveals new insights into the branch junction. Nat. Commun. 11, 6437 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmet A., et al. , Cryo-EM structure of NPF-bound human Arp2/3 complex and activation mechanism. Sci. Adv. 6, eaaz7651 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai L., Marshall T. W., Uetrecht A. C., Schafer D. A., Bear J. E., Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell 128, 915–929 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humphries C. L., et al. , Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J. Cell Biol. 159, 993–1004 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poukkula M., Kremneva E., Serlachius M., Lappalainen P., Actin-depolymerizing factor homology domain: A conserved fold performing diverse roles in cytoskeletal dynamics. Cytoskeleton (Hoboken) 68, 471–490 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Goode B. L., Sweeney M. O., Eskin J. A., GMF as an actin network remodeling factor. Trends Cell Biol. 28, 749–760 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ydenberg C. A., et al. , GMF severs actin-Arp2/3 complex branch junctions by a cofilin-like mechanism. Curr. Biol. 23, 1037–1045 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luan Q., Nolen B. J., Structural basis for regulation of Arp2/3 complex by GMF. Nat. Struct. Mol. Biol. 20, 1062–1068 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haynes E. M., et al. , GMFβ controls branched actin content and lamellipodial retraction in fibroblasts. J. Cell Biol. 209, 803–812 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poukkula M., et al. , GMF promotes leading-edge dynamics and collective cell migration in vivo. Curr. Biol. 24, 2533–2540 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ono S., Functions of actin-interacting protein 1 (AIP1)/WD repeat protein 1 (WDR1) in actin filament dynamics and cytoskeletal regulation. Biochem. Biophys. Res. Commun. 506, 315–322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan C., Beltzner C. C., Pollard T. D., Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr. Biol. 19, 537–545 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pandit N. G., et al. , Force and phosphate release from Arp2/3 complex promote dissociation of actin filament branches. Proc. Natl. Acad. Sci. U.S.A. 117, 13519–13528 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Eunen K., Bakker B. M., The importance and challenges of in vivo-like enzyme kinetics. Perspectives in Science 1, 126–130 (2014). [Google Scholar]
- 31.van Eunen K., et al. , Measuring enzyme activities under standardized in vivo-like conditions for systems biology. FEBS J. 277, 749–760 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Hayakawa K., Sakakibara S., Sokabe M., Tatsumi H., Single-molecule imaging and kinetic analysis of cooperative cofilin-actin filament interactions. Proc. Natl. Acad. Sci. U.S.A. 111, 9810–9815 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wioland H., et al. , ADF/cofilin accelerates actin dynamics by severing filaments and promoting their depolymerization at both ends. Curr. Biol. 27, 1956–1967.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bibeau J. P., Gray S., De La Cruz E. M., Clusters of a few bound cofilins sever actin filaments. J. Mol. Biol. 433, 166833 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suarez C., et al. , Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr. Biol. 21, 862–868 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shekhar S., Chung J., Kondev J., Gelles J., Goode B. L., Synergy between cyclase-associated protein and cofilin accelerates actin filament depolymerization by two orders of magnitude. Nat. Commun. 10, 5319 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christensen J. R., et al. , Competition between tropomyosin, fimbrin, and ADF/cofilin drives their sorting to distinct actin filament networks. eLife 6, e23152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jansen S., Goode B. L., Tropomyosin isoforms differentially tune actin filament length and disassembly. Mol. Biol. Cell 30, 671–679 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gateva G., et al. , Tropomyosin isoforms specify functionally distinct actin filament populations in vitro. Curr. Biol. 27, 705–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsiao J. Y., Goins L. M., Petek N. A., Mullins R. D., Arp2/3 complex and cofilin modulate binding of tropomyosin to branched actin networks. Curr. Biol. 25, 1573–1582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodal A. A., et al. , Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat. Struct. Mol. Biol. 12, 26–31 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Espinoza-Sanchez S., Metskas L. A., Chou S. Z., Rhoades E., Pollard T. D., Conformational changes in Arp2/3 complex induced by ATP, WASp-VCA, and actin filaments. Proc. Natl. Acad. Sci. U.S.A. 115, E8642–E8651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narvaez-Ortiz H. Y., Nolen B. J., Unconcerted conformational changes in Arp2/3 complex integrate multiple activating signals to assemble functional actin networks. Curr. Biol. 32, 975–987.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rouiller I., et al. , The structural basis of actin filament branching by the Arp2/3 complex. J. Cell Biol. 180, 887–895 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hocky G. M., Sindelar C. V., Cao W., Voth G. A., De La Cruz E. M., Structural basis of fast- and slow-severing actin-cofilactin boundaries. J. Biol. Chem. 296, 100337 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shekhar S., Hoeprich G. J., Gelles J., Goode B. L., Twinfilin bypasses assembly conditions and actin filament aging to drive barbed end depolymerization. J. Cell Biol. 220, e202006022 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spudich J. A., Watt S., The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871 (1971). [PubMed] [Google Scholar]
- 48.Guo S., et al. , Abp1 promotes Arp2/3 complex-dependent actin nucleation and stabilizes branch junctions by antagonizing GMF. Nat. Commun. 9, 2895 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grimm J. B., et al. , A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith B. A., et al. , Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. eLife 2, e01008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lappalainen P., Fedorov E. V., Fedorov A. A., Almo S. C., Drubin D. G., Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 16, 5520–5530 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chaudhry F., et al. , Srv2/cyclase-associated protein forms hexameric shurikens that directly catalyze actin filament severing by cofilin. Mol. Biol. Cell 24, 31–41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Friedman L. J., Chung J., Gelles J., Viewing dynamic assembly of molecular complexes by multi-wavelength single-molecule fluorescence. Biophys. J. 91, 1023–1031 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ticau S., Friedman L. J., Ivica N. A., Gelles J., Bell S. P., Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading. Cell 161, 513–525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dave R., Terry D. S., Munro J. B., Blanchard S. C., Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys. J. 96, 2371–2381 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friedman L. J., Gelles J., Multi-wavelength single-molecule fluorescence analysis of transcription mechanisms. Methods 86, 27–36 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ensign D. L., Pande V. S., Bayesian single-exponential kinetics in single-molecule experiments and simulations. J. Phys. Chem. B 113, 12410–12423 (2009). [DOI] [PubMed] [Google Scholar]
- 58.J. Chung, B.L. Goode, J. Gelles, Single-molecule analysis of actin filament debranching by cofilin and GMF. Zenodo. https://zenodo.org/record/5207693#.YsO3C3ZByM8. Deposited 24 June 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for all figures, consisting of images, experimental and simulated branch lifetimes, and MATLAB code used to generate the figures and perform the Monte Carlo simulations, is available at Zenodo repository (https://zenodo.org DOI: 10.5281/zenodo.5207693) (58).