Significance
Protein translation is essential and highly conserved in all organisms. Consequently, the two essential components of the protein translation machinery, transfer RNAs (tRNAs) and ribosomes, are the main targets of ribotoxins for cell killing. While it is reasonable to assume that damaged tRNAs can be repaired by RNA repair systems, it is unclear whether the ribosomal damage can be reversed. Here, we demonstrate that the bacterial RtcB2–PrfH pair is a ribosome rescue and repair system, specifically reversing the lethal ribosomal damage in the decoding center inflicted by at least two families of ribotoxins. Our study provides insight into the arms race among bacteria by acquiring or evolving new offensive or defensive weaponry to survive in an environment of limited nutrition.
Keywords: ribosome rescue, RNA repair, ribotoxin, cryo-EM
Abstract
RtcB is involved in transfer RNA (tRNA) splicing in archaeal and eukaryotic organisms. However, most RtcBs are found in bacteria, whose tRNAs have no introns. Because tRNAs are the substrates of archaeal and eukaryotic RtcB, it is assumed that bacterial RtcBs are for repair of damaged tRNAs. Here, we show that a subset of bacterial RtcB, denoted RtcB2 herein, specifically repair ribosomal damage in the decoding center. To access the damage site for repair, however, the damaged 70S ribosome needs to be dismantled first, and this is accomplished by bacterial PrfH. Peptide-release assays revealed that PrfH is only active with the damaged 70S ribosome but not with the intact one. A 2.55-Å cryo-electron microscopy structure of PrfH in complex with the damaged 70S ribosome provides molecular insight into PrfH discriminating between the damaged and the intact ribosomes via specific recognition of the cleaved 3′-terminal nucleotide. RNA repair assays demonstrated that RtcB2 efficiently repairs the damaged 30S ribosomal subunit but not the damaged tRNAs. Cell-based assays showed that the RtcB2–PrfH pair reverse the damage inflicted by ribosome-specific ribotoxins in vivo. Thus, our combined biochemical, structural, and cell-based studies have uncovered a bacterial defense system specifically evolved to reverse the lethal ribosomal damage in the decoding center for cell survival.
Conflicts between organisms are fundamental biological phenomena. In an environment of limited nutrition, some bacteria produce toxins to kill their neighbors to eliminate competition. Since the protein translation machinery is essential and universally conserved in all organisms, it is the main target of both small-molecule and protein toxins. Most protein toxins targeting protein translation are ribotoxins, inflicting damage on essential RNAs required for protein translation. The major sites targeted by ribotoxins include the sarcin–ricin loop in the large ribosomal subunit (1, 2), the decoding center in the small ribosomal subunit (3–6), and the anticodon loops of transfer RNAs (tRNAs) (7–11). Of particular relevance to this work is the damage done in the decoding center. To date, two different families of ribotoxins, the C-terminal toxin domain of colicin E3 (ColE3-CT) and the C-terminal toxin domain of CdiA effector from Enterobacter cloacae (CdiA-CTECL), are known to cleave 16S rRNA at the same site (between nucleotides A1493 and G1494), demonstrating convergent evolution of different lineages of ribotoxins targeting one of the most conserved sites in the ribosome. The highly conserved nucleotides near the cleavage site are critical for decoding. Consequently, ribosomal damage by ColE3-CT and CdiA-CTECL result in stalled protein translation, ultimately leading to cell death (6, 12).
To survive, the bacteria targeted by toxins employ a variety of defensive mechanisms to neutralize the toxins imposed on them. The majority appear to employ a head-on approach to directly confront the invading toxins. Here, we propose that if the damage inflicted by an invading toxin is reversible, an alternative approach of indirect confrontation could also be employed. The clash between a ribotoxin and an RNA repair system appears to be such an example, as the conflict is carried out via an RNA substrate. We have previously reported two bacterial RNA repair systems that are able to repair ribotoxin-cleaved tRNAs in vitro (13, 14). Here, we explore possible RNA repair by bacterial RtcBs (15–18). Compared with other known RNA repair systems, bacterial RtcBs are approximately 10-fold more abundant. Despite extensive in vitro characterization of bacterial RtcBs, their in vivo biological targets remain unknown. In this work, we show that while EcRtcB1 efficiently repairs damaged tRNAs, EcRtcB2 specifically repairs damaged ribosome resulting from specific cleavage of 16S rRNA in the decoding center by at least two families of ribosome-specific ribotoxins.
Results
Bioinformatic Analysis of RtcB Family and a Hypothesis of RtcB2-PrfH Functions.
To provide insight into biological functions of RtcBs, we first performed bioinformatic analysis of Pfam PF01139, the RtcB protein family. Our analysis revealed that the vast majority of RtcBs are, indeed, found in bacteria (SI Appendix, Fig. S1A). We also constructed a sequence similarity network (SSN) of PF01139 (SI Appendix, Fig. S1B). SSN revealed that a subset of RtcBs, denoted RtcB2 here, forms a separate cluster (SI Appendix, Fig. S1B, cluster 2), suggesting that RtcB2 might be functionally distinct from the majority of RtcBs from cluster 1. Furthermore, the overwhelming majority of bacterial RtcB2s are encoded in a two-component operon that also encodes PrfH (19) (SI Appendix, Fig. S2A). In Escherichia coli K12 and its derivative strains, both RtcB2 and PrfH are presumably inactive due to a significant deletion within the rtcB2-prfH operon that encompasses both coding genes (SI Appendix, Fig. S2B). To our knowledge, both RtcB2 and PrfH have not been experimentally characterized before.
PrfH was first reported by Atkins et al. as a homolog of bacterial RF1 and RF2 (19). Because of high conservation of the GGQ domain compared with RF1 and RF2, PrfH was suggested to catalyze peptide release during protein translation. On the other hand, the N-terminal domain of PrfH is significantly divergent from the stop-codon recognition domains of RF1 and RF2. Therefore, it was suggested that PrfH recognizes a certain unknown mRNA feature rather than the stop codons. At the time, the function of the neighboring gene was unknown; it was later shown to be a member of RNA ligase family, RtcB (15, 20, 21). These analyses and revelations led us to hypothesize that a damaged ribosome might be the biological substrate of the two proteins encoded in the operon.
The 70S Ribosome Damaged in the Decoding Center Is the Substrate of PrfH.
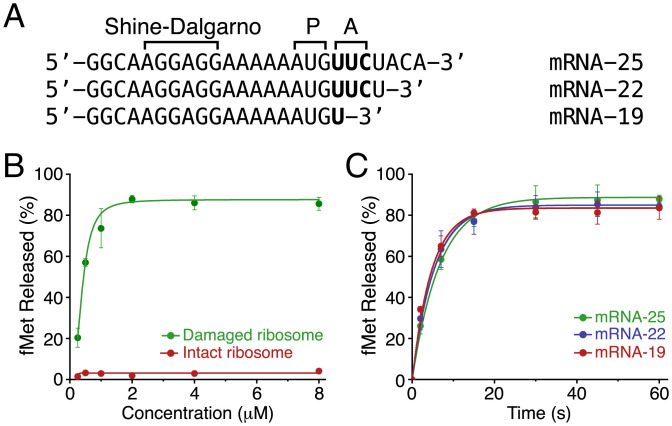
To elucidate biological function of bacterial PrfH, we cloned, overexpressed, and purified recombinant E. coli PrfH (EcPrfH). We first used the intact E. coli 70S ribosome and a 25-nucleotide mRNA (Fig. 1A, mRNA-25) as the substrates, and no peptide release was detected (Fig. 1B, red curve). We next prepared and purified E. coli 70S ribosome with the 16S rRNA cleaved by CdiA-CTECL. The assays using the damaged 70S ribosome showed robust peptide release activity of EcPrfH (Fig. 1B, green curve), indicating that 70S ribosome with a specific damage in the decoding center is likely the biological substrate of PrfH.
Fig. 1.
Substrate specificity of peptide release by EcPrfH. (A) Sequences of three mRNAs with different length employed for the assays. P, P site of ribosome; A, A site of ribosome. (B) Concentration-dependent activities of PrfH using the damaged 70S ribosome (green) and the intact 70S ribosome (red) as substrates. mRNA-25 was employed for the assays, and the reactions were terminated after 60 s. (C) Time course of the activities of PrfH with three different mRNAs. The concentration of PrfH employed for the assays was 2 μM.
Bacterial RF1 and RF2 require the presence of mRNA with stop codons in the A site. To assess whether the activity of PrfH also requires mRNA in the A site, we prepared two additional mRNAs with systematic truncations at their 3′ ends that encompass the A site (Fig. 1A). PrfH is approximately equally active with all three mRNAs (Fig. 1C), indicating that, unlike RF1 and RF2, the activity of PrfH does not depend on the presence of mRNA in the A site. We carried out additional peptide release assays, allowing us to obtain kinetic parameters of the PrfH-catalyzed reaction (SI Appendix, Fig. S3).
Cryo-EM Structure of the Damaged E. coli 70S•PrfH•tRNA•mRNA Complex.
To provide molecular insight into the recognition of the damaged 70S ribosome by PrfH, we determined the cryo-electron microscopy (cryo-EM) structure of EcPrfH in complex with the damaged E. coli 70S ribosome, uncharged tRNAfMet, and mRNA-25 at 2.55-Å resolution (Fig. 2A and SI Appendix, Figs. S4 and S6 and Table S1). The structure revealed the presence of mRNA (Fig. 2A, dark blue) as well as tRNAfMet occupying both the P and E sites of the ribosome (Fig. 2A, green and light blue). Importantly, EcPrfH is found to occupy the A site approximately where RF1 or RF2 should locate during normal translation termination (22, 23) (Fig. 2A, red). We also carried out a similar structural study with the intact E. coli 70S ribosome, and no EcPrfH could be found in the A site (SI Appendix, Fig. S5). This is consistent with the result of peptide-release assays showing that the intact 70S ribosome is not a substrate of EcPrfH (Fig. 1B).
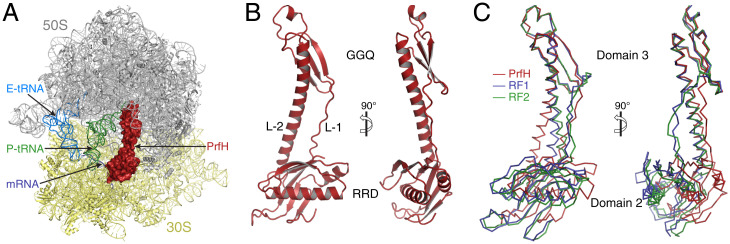
Fig. 2.
Structure of EcPrfH in complex with the damaged E. coli 70S ribosome, P-site tRNA (P-tRNA) and E-site tRNAs (E-tRNA), and mRNA-25. (A) Overview of the structure of the complex. Two subunits of the ribosome are colored gray and yellow, respectively. P-site tRNA is colored green, E-site tRNA is in light blue, and mRNA is dark blue. PrfH occupies the A site of the damaged ribosome and is depicted at the surface and colored red. Some 23S rRNA residues at the front of the image were not shown to have a better view of the decoding center. (B) Cartoon representation of the PrfH structure. GGQ refers to the peptide-hydrolase domain that contains the strictly conserved GGQ motif. L-1, linker-1; L-2, linker-2. (C) Ribbon representation of the structures of PrfH (red), RF1 (blue), and RF2 (green), aligned based on the superimpositions of the structures of 70S ribosomes associated with these factors.
The folding of EcPrfH is similar to the structures of domain 2, domain 3, and part of domain 4 of RF1 and RF2 (Fig. 2 B and C and SI Appendix, Fig. S7). This is further confirmed by the Dali structural search (24) with EcPrfH, as the top scores are all structures of RF1 and RF2. To facilitate structural analysis of EcPrfH, we divided the structure of EcPrfH into three portions: the top and bottom domains and the linkers connecting these two domains (Fig. 2B and SI Appendix, Fig. S8). The top domain is composed of residues 108 through 161, and this domain is highly homologous to domain 3 of RF1 and RF2 (SI Appendix, Fig. S7A). This domain is, therefore, named GGQ domain (Fig. 2B, Top). The bottom is composed of residues 2 through 98 and residues180 through 204. As described in the next section, this domain is responsible for the recognition of ribosomal RNAs, and it is therefore named rRNA-recognition domain (RRD) (Fig. 2B, Bottom). These two domains are connected by two linkers, a loop composed of residues 99 through 107 (linker 1) and part of a long helix composed of residues 162 through 179 (linker 2) (SI Appendix, Fig. S8).
The structure of PrfH is highly homologous to the ones of RF1 and RF2 when individual domains are compared (SI Appendix, Fig. S7 A and B), but less so when the entire structures are compared (SI Appendix, Fig. S7C). The cause of this disparity is the changes of relative position and orientation of the two domains in PrfH compared with their counterparts in RF1 and RF2. This is clearly demonstrated when PrfH, RF1, and RF2 are aligned based on the superpositions of the 70S ribosome structures that are associated with these factors (Fig. 2C). While the GGQ domain of PrfH and domain 3 of RF1 and RF2 are in the same position, the RRD of PrfH shifts ∼8 Å vertically and ∼16 Å horizontally, and rotates ∼25° vertically, relative to domain 2 of RF1 and RF2 (Fig. 2C). As a result, the RRD of PrfH does not contact the mRNA (SI Appendix, Fig. S9). Instead, it interacts extensively with rRNAs as well as S12 protein.
Molecular Recognition of the Damaged 70S Ribosome by PrfH.
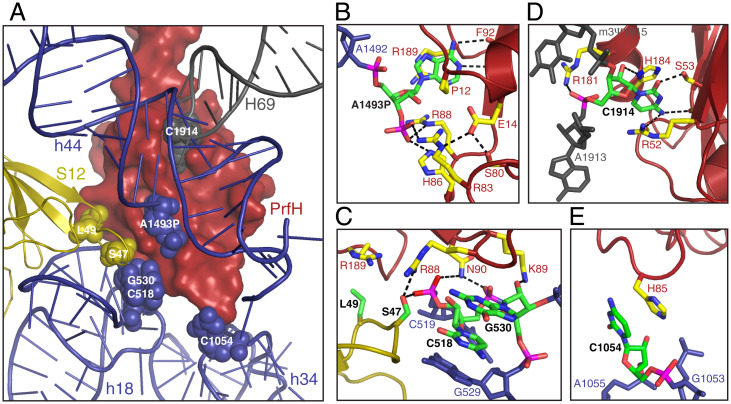
In the decoding center of bacterial ribosome, several universally conserved residues from the 30S subunit are responsible for monitoring codon–anticodon interactions (25). They are A1492 and A1493 from helix 44, C518 and G530 from helix 18 (h18), C1054 from helix 34 (h34), and S47 from ribosomal protein S12. Except for A1492, all these residues make direct contact with PrfH (Fig. 3A). In addition, residue C1914 from helix 69 of 23S rRNA also interacts with PrfH (Fig. 3 A, gray).
Fig. 3.
Molecular recognition of the damaged 70S ribosome by PrfH. (A) Overview of the nucleotides and amino acids in the damaged 70S ribosome making direct contact with PrfH. PrfH is at the surface, and the ribosome is in cartoon. The residues that make direct contact with PrfH are highlighted in spheres. (B) Recognition of the 3′-terminal A1493P of the cleaved 16S rRNA by PrfH. Atoms in A1493P and in the residues from PrfH are colored individually, with carbon in green (A1493P) and yellow (PrfH), oxygen in red, nitrogen in blue, and phosphate in magenta. (C) Specific interactions between PrfH and residues from h18 of 16S rRNA and S12 protein. (D) Recognition of C1914 of 23S rRNA by PrfH. (E) Observed single interaction between H85 of PrfH and C1054 from h34 of 16S rRNA.
The center of the RRD is a pocket specifically recognizing the 3′-terminal nucleotide of the cleaved 16S rRNA. The reaction carried out by CdiA-CTECL should formally produce 2′,3′-cyclic phosphate. However, the cryo-EM density fits 3′-phosphate significantly better than 2′,3′-phosphate (SI Appendix, Fig. S6D). Therefore, at least the majority of cryo-EM particles have 3′-phosphorylated A1493 (denoted A1493P herein) as the cleaved 3′-terminal nucleotide. The A1493P-recognition pocket is mainly surrounded by two walls (SI Appendix, Fig. S10D). The base of A1493P is sandwiched between the side chain of R189 and residues G10 through P12, and it is recognized via two hydrogen bonds with the main chain of F9 (Fig. 3B). The ribose of A1493P stacks on top of the side chain of R88. The most extensive interactions occur at the 3′-phosphate group that originally belongs to G1494 before cleavage, forming hydrogen bonds with the side chains of R83, H86, and R88.
On the outside of the first wall is a highly conserved surface patch that is responsible for interactions with residues from h18 and S12 protein (Fig. 3C and SI Appendix, Fig. S10D). Specifically, the side chain of K89 hydrogen bonds with the 2′-OH group of G530, the side chain of N90 hydrogen bonds with the phosphate backbones of both C518 and C519, and the side chain of S47 of S12 protein hydrogen bonds with the phosphate of C519.
On the outside of the second wall is a second pocket for specific recognition of C1914 from helix 69 (Fig. 3D). Specifically, the base of C1914 is sandwiched between the side chains of H184 and R52, and it is recognized by hydrogen bonding with the side chain of S53 and the carbonyl groups of A9 and R52. The side chains of H184 and R181 hydrogen bond with the 2′-OH and phosphate groups of C1914, respectively. In addition, PrfH also makes a single contact with C1054 from h34 via stacking of the side chain of H85 on the base of C1054 (Fig. 3E).
RNA Substrates of EcRtcB2 and EcRtcB1 Are Mutually Exclusive.
All E. coli strains encode EcRtcB1 and EcRtcB2. These two RtcBs only share 28% sequence identity, implying their distinct biological functions. While EcRtcB1 is encoded in a two-component operon that also encodes RtcA (26), EcRtcB2 is encoded in a two-component operon together with EcPrfH (SI Appendix, Fig. S2). Approximately 60 E. coli strains also encode a third RtcB that is distinct from both EcRtcB1 and EcRtcB2.
To provide insight into substrate specificity of EcRtcB2, we prepared three ribotoxin-cleaved E. coli RNA substrates for qualitative RNA repair assays (SI Appendix, Fig. S11). They are 30S subunits having 16S rRNA cleaved by CdiA-CTECL, tRNAAsp cleaved by the C-terminal toxin domain of colicin E5 (ColE5-CT) between nucleotides 34 and 35, and tRNAArg cleaved by the C-terminal toxin domain of colicin D (ColD-CT) between nucleotides 38 and 39. In addition to recombinant EcRtcB2 potentially repairing these three RNA substrates, we also carried out identical sets of experiments using recombinant EcRtcB1 for comparison.
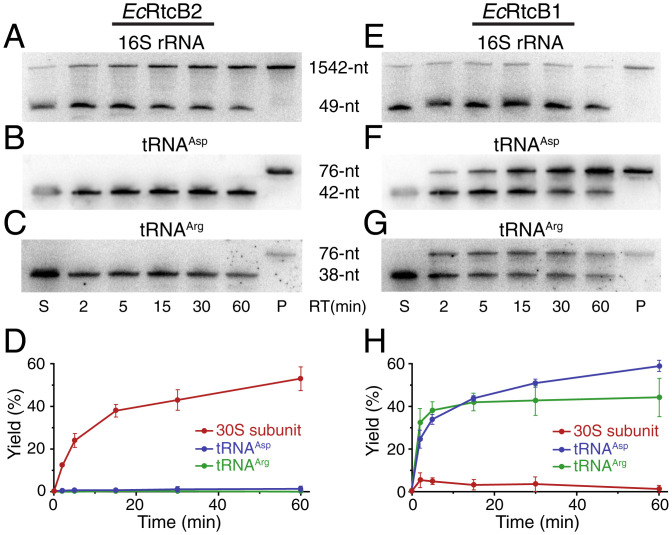
EcRtcB2 efficiently repairs the damaged 30S subunit (Fig. 4 A and D, the red curve in the latter). On the other hand, less than 2% of damaged tRNAs were repaired by EcRtcB2 based on quantitation of the gels shown in Fig. 4 B and C. The calculated yield might be caused with the uncertainty of quantitation. We argue that if it were from the repaired product of damaged tRNAs, one would have expected the increase of repair yield over reaction time. This was not what was observed (Fig. 4D, blue and green curves). Based on these analyses, we conclude that EcRtcB2 does not repair damaged tRNAs.
Fig. 4.
Substrate specificity of RNA repair by EcRtcB2 and EcRtcB1. (A–C) Northern blotting of denaturing polyacrylamide gels analyzing RNA repair of three different RNA substrates by EcRtcB2. S, substrates; P, total RNAs isolated from E. coli cells without the exposure to ribotoxins; RT, reaction time. (D) Quantitation of the RNA-repair reactions carried out by EcRtcB2 (error bars represent the mean ± SD; n = 2 biologically independent experiments). (E–G) Northern blotting of denaturing polyacrylamide gels analyzing RNA repair of three different RNA substrates by EcRtcB1. (H) Quantitation of the RNA-repair reactions carried out by EcRtcB1 (error bars represent the mean ± SD; n = 2 biologically independent experiments).
The results from EcRtcB1 are just the opposite. EcRtcB1 appears to not repair the damaged ribosome (Fig. 4 E and H, the red curve in the latter) but repairs damaged tRNAs efficiently (Fig. 4 F and G). Based on the quantitation of full-length 16S rRNA and 3′-cleaved fragment (Fig. 4E), less than 6% of the damaged 30S subunit has been repaired by EcRtcB1 at its maximum (Fig. 4H, red curve with the reaction time point of 5 min). The number might represent EcRtcB1 repairing some minor, nonspecific ribosomal damages outside the decoding center where EcRtcB1 might have access. EcRtcB1 has previously been shown to reduce nonspecific rRNA fragmentation caused by cell stress (27). Again, we argue that since the yield of repair did not increase over reaction time (Fig. 4H, red curve), EcRtcB1 does not repair the damaged 30S subunit with the specific 16S rRNA cleavage by CdiA-CTECL.
EcRtcB2 and EcPrfH Reverse the Damage Inflicted by ColE3-Like Ribotoxins In Vivo.
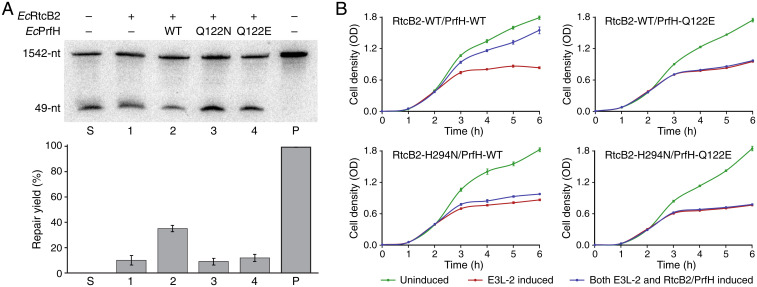
To dissect biological roles of RtcB2 and PrfH, we carried out additional in vitro and in vivo experiments. Ribosomal repair described in the previous section were carried out in the absence of EcPrfH by using the low concentration of Mg2+ ion to dissociate the damaged 70S ribosome into 50S and 30S subunits. Here, we performed similar experiments but with high concentration of Mg2+ ion, which keeps the damaged 70S ribosome intact. In the absence of EcPrfH, EcRtcB2 carried out modest repair of the damaged ribosome (Fig. 5A, lane 1). This was presumably due to dissociation of some damaged 70S ribosomes during cell growth or sample preparation, making a small amount of the damaged 30S subunit available for repair. Addition of EcPrfH significantly increases the repair of the damaged ribosome by EcRtcB2 (Fig. 5A, lane 2), indicating that efficient ribosomal repair requires EcPrfH to rescue and dismantle the damaged 70S ribosome. This is further supported by the assays with EcPrfH mutants, as addition of EcPrfH Q122N or Q122E mutant (mutation of the catalytic Q in the GGQ motif) reduced the repair levels comparable to the one that lacks EcPrfH (Fig. 5A, lanes 3 and 4, and compare with lane 1).
Fig. 5.
Elucidating biological functions of EcRtcB2 and EcPrfH via in vitro and in vivo assays. (A) (Top) Northern blotting of denaturing polyacrylamide gel analyzing RNA repair of the damaged 70S ribosome by EcRtcB2 without or with the addition of EcPrfH. S, substrate; P, total RNAs isolated from E. coli cells without the exposure to ribotoxin. (Bottom) Quantitation of the RNA-repair reactions carried out by EcRtcB2 (error bars represent the mean ± SD; n = 3 biologically independent experiments). (B) Growth curves of E. coli cells with various combinations of expressions of E3L-2 and RtcB2–PrfH (error bars represent the mean ± SD; n = 6 biologically independent experiments). OD, optical density.
To evaluate biological functions of RtcB2–PrfH in vivo, we developed an in vivo system employing pBAD and pQE60 (28, 29) to express a ribotoxin and RtcB2–PrfH, respectively, in E. coli cells. However, cloning the gene encoding ColE3-CT into pBAD plasmid was not successful. We reasoned that, in the wild, only a few copies of ColE3 might be sufficient to kill an E. coli cell. Therefore, ColE3 is too toxic for an in vivo assay in a laboratory setting because the number of the expressed ribotoxin is significantly higher. To compensate for the number of a ribotoxin in a laboratory setting but with a similar biological effect, we searched for colicin E3-like (E3L) ribotoxins that are less toxic. Therefore, we first constructed an SSN of the cytotoxic protein family (Pfam PF09000) to which ColE3 belongs, followed by selecting E3Ls that are distant from ColE3 (SI Appendix, Fig. S12 A and B). Four selected E3Ls are all toxic to E. coli cells, but the degree of toxicity differs from one another judged by their ability to inhibit cell growth (SI Appendix, Fig. S12C). Northern blotting of total RNAs isolated from E3L-expressed cells confirmed that these four E3Ls carry out the same ribosomal damage as ColE3-CT and CdiA-CTECL (SI Appendix, Fig. S12D).
Expression of the RtcB2–PrfH pair reversed the cell growth inhibited by the expression of all four E3Ls (SI Appendix, Fig. S12E), indicating that the RtcB2–PrfH pair is able to neutralize the toxicity of E3Ls. However, the ability of RtcB2–PrfH to counteract E3Ls differs among the four E3Ls, with E3L-1 being counteracted the most and E3L-3 the least. The outcomes based on E3L-2 provide the best compromise between the toxicity of E3L and the neutralizing effect of RtcB2–PrfH, allowing us to investigate both the toxicity of a ribotoxin and the effect of repair by RtcB2–PrfH accurately. Therefore, the E3L-2 system was selected for further study, which includes more accurate data as well as additional assays with the RtcB2–PrfH mutants (Fig. 5B). We employed the same EcPrfH-Q122E mutant for the in vivo assays. For EcRtcB2, we used H294N mutation. The equivalent histidine in other RtcBs has been shown to be the essential catalytic residue (16, 18, 30).
Expression of the wild-type enzymes of both RtcB2 and PrfH recovers most of the cell growth inhibited due to the expression of E3L-2 (Fig. 5B, Top Left). Disabling the enzymatic activity of both RtcB2 and PrfH completely abolish their biological function in vivo (Fig. 5B, Bottom Right). Mutation of PrfH alone also produced the same effect (Fig. 5B, Top Right). This observation also indicates that other endogenous ribosome rescue factors in E. coli, such as tmRNA-SmpB (31–34), ArfA•RF2 (35–39), and ArfB (40–42), are not able to replace the biological role played by PrfH. Disabling RtcB2 also significantly reduced the ability of RtcB2–PrfH to neutralize the toxicity of E3L-2 (Fig. 5B, Bottom Left). However, RtcB2-H294N/PrfH-WT clearly retains a small but reproducible biological activity. Further investigation is required to uncover the cause of the small difference between RtcB2 and PrfH mutants. Collectively, our in vivo assays indicate that the reversal of damage inflicted by ribosome-specific ribotoxins in bacterial cells requires the biological functions of both RtcB2 and PrfH, which cannot be replaced by RtcB1 and other ribosome rescue factors.
Discussion
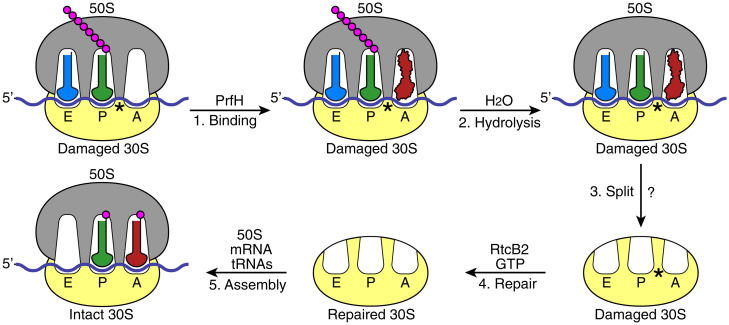
Based on in vitro and in vivo data presented in this study, we propose a model depicting the following biological events occurring in bacteria that have been invaded by a ribotoxin such as ColE3-CT or CdiA-CTECL. Ribosomal damage in the decoding center results in the stalled 70S ribosome (Fig. 6, Top Left). Unlike the stalled ribosomes due to 3′-truncated mRNA, the stalled ribosome caused by ribosomal damage should still have mRNA occupying the A site and beyond. Furthermore, the population of the stalled ribosome is likely to be significantly higher than the one due to 3′-truncated mRNAs, which was estimated to be 2 to 4% (43). RNA repair cannot occur with the damaged 70S ribosome, as RtcB2 is not able to access the damage site. PrfH enters the empty A site (Fig. 6, step 1). Specific interactions of PrfH with rRNAs and S12 protein of the damaged ribosome orient its GGQ domain to catalyze hydrolysis of the nascent peptide attached to P-site tRNA (Fig. 6, step 2). The removal of the nascent peptide allows 70S ribosome to split into 50S and 30S subunits (Fig. 6, step 3). It is unclear whether PrfH alone is sufficient for the split of the 70S ribosome or if additional cellular factors are required. RtcB2 accesses the damage site of the dissociated 30S subunit, performing RNA repair using GTP as the cofactor (Fig. 6, step 4). The repaired 30S subunit re-enters the ribosome pool, assembling into a new 70S ribosome to start a new round of protein translation (Fig. 6, step 5).
Fig. 6.
A proposed model of biological events occurring in a bacterial cell invaded by a ribotoxin such as ColE3-CT or CdiA-CTECL. The protein-translation machinery is schematically depicted, including 50S ribosomal subunit (gray), 30S ribosomal subunit (yellow), messenger RNA (mRNA; blue), A-, P-, and E-site tRNAs (red, green, and light blue, respectively), amino acids (magenta), and PrfH (red). The ribosomal damage caused by a ribotoxin is marked with an asterisk.
Because of extensive and precise interactions between the RRD of PrfH and the damaged 70S ribosome revealed by the cryo-EM structure (Fig. 3), it is difficult to envision that PrfH is capable of rescuing other stalled 70S ribosomes. Therefore, PrfH appears to have evolved to deal with only one issue: the biological consequence resulting from specific cleavage of 16S rRNA between A1493 and G1494.
Colicin E3 was the first ribotoxin to be characterized, 50 years ago (3, 4), that disables 70S ribosome for protein translation via specific cleavage of 16S rRNA between A1493 and G1494. Over the last half century, it was unclear whether such lethal ribosomal damage could be reversed to allow the cell to survive. Here, we demonstrate that it might be accomplished through the combined effort of PrfH and RtcB2. In addition, our RNA repair assays revealed that RtcB2 is strictly specific for repairing 30S subunit with a specific cleavage of 16S rRNA between A1493 and G1494. Therefore, although we cannot rule out the possibility that RtcB2 might have enzymatic activity on other RNA substrates in vivo, its biological role appears to repair the ribosomal damage only occurring in the decoding center. This argument is certainly consistent with the biological role of PrfH discussed above. Conversely, the fact that many bacteria require the presence of an operon encoding RtcB2 and PrfH underscores the severity of the threat posed by ribosome-specific ribotoxins such as ColE3-CT and CdiA-CTECL to bacteria.
In addition to bacterial RtcB2s, 98 RtcB2s are also present in eukaryotic organisms, based on our bioinformatic analysis (SI Appendix, Fig. S1), and most of those organisms are fungi and plants. Since fungi and plants employ Trl1 for tRNA splicing (44, 45), we predict that eukaryotic RtcB2s carry out repair of the 40S subunit with the damage similar to the one observed here in bacteria. This raises two interesting questions regarding how the eukaryotic ribosome is damaged and how the damaged 80S ribosome is rescued, as ribotoxins equivalent to ColE3-CT and CdiA-CTECL have not been reported in eukaryotes and PrfH is only found in bacteria. Our analysis also indicates that bacterial RtcBs are more diverse than we anticipated. For example, SSN of the entire RtcB family classifies RtcBs into eight clusters (SI Appendix, Fig. S1B). The present study, together with previous characterization of archaeal and human RtcBs, only allows us to tentatively assign RNA substrates of RtcBs from clusters 1a, 1b, and 2. Therefore, it would be interesting to investigate possible biological substrates of RtcBs from other five clusters, which might yield surprises.
Materials and Methods
Bioinformatic Analysis of Pfam PF01139, the RtcB Protein Family.
Bioinformatic analyses were performed on the database of UniProt 2020_04 and InterPro 81. Calculations were carried out at the Enzyme Function Initiative website (https://efi.igb.illinois.edu/) (46). PF01139 (Pfam of RtcB) was submitted for the initial calculation for the SSN of RtcB. After the initial calculation was complete, SSN was finalized with the setting of the alignment score threshold at 200 and the minimum sequence length at 350. The SSN file with 100% identification (e.g., 100% representative node) was displayed with Cytoscape (47) to produce the initial SSN. The nodes corresponding to Bacteria, Archaea, Eukaryota, and viruses were selected separately, and SSNs corresponding to these four classifications were generated. These four additional SSNs allowed us to obtain the numbers of unique sequences of all RtcB for Bacteria, Archaea, Eukaryota, and viruses as shown in SI Appendix, Fig. S1A. To obtain the corresponding numbers for RtcB2s, similar analysis was carried out, but only with RtcB sequences that belong to cluster 2 of the SSN.
To generate the SSN shown in SI Appendix, Fig. S1B, it was necessary to perform the calculation using only 25% of total RtcB sequences, randomly selected. This is because if all RtcB sequences were used, the SSN file would have been too big to be calculated and displayed by Cytoscape using the parameters shown in SI Appendix, Fig. S1B. Again, the SSN file was processed and displayed with Cytoscape, and yFiles Organic Layout was used for the layout of nodes and edges. Minor adjustments of the positions of a couple of clusters were made to make nodes fit better within the space of the figure. The entire SSN was arbitrarily divided into eight clusters, with bacterial RtcB from cluster 2 being the main focus of the study.
Cloning, Overexpression, and Purification of Recombinant Proteins.
The gene encoding EcPrfH (from E. coli ATCC 25922) was cloned into pRSF-1 vector, which carries a N-terminal 6xHis tag followed by a SUMO tag. To obtain EcPrfH with the side chain of glutamine in the GGQ motif methylated, EcPrfH was coexpressed with methyltransferase PrmC in an E. coli BL21 (DE3) strain at 18 °C overnight induced with 0.5% lactose (48). Cells were harvested with centrifugation and the pellets were resuspended in lysis buffer (20 mM Tris⋅HCl, pH 8.0, 500 mM NaCl, 5% glycerol). Cells were lysed using a French press, and cell debris was removed by centrifugation at 20,000g for 40 min at 4 °C. The supernatant was filtered with a 0.45-μm filter, and the filtered solution was loaded into a HisTrap column (GE Healthcare). The proteins were eluted with the imidazole gradient. The fractions containing the SUMO-tagged PrfH were combined and incubated with Ulp1 protease to cleave the SUMO tag. The untagged EcPrfH was obtained by passing through the second HisTrap column.
The genes encoding EcRtcB1 and EcRtcB2 were cloned into a pETDuet-1 vector without the N-terminal His tag. The two proteins were overexpressed in E. coli BL21 (DE3) at 18 °C overnight induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). The harvested cells were resuspended in diethylaminoethyl-A buffer (20 mM Tris⋅HCl, pH 8.0, 10 mM NaCl, 5% glycerol) and lysed using a French press. Cell debris was removed by centrifugation at 20,000g for 40 min at 4 °C, followed by filtration through a 0.45-μm filter to clarify the lysate. The proteins were purified from the clarified lysate using a fast protein liquid chromatography system with diethylaminoethyl, heparin, and Superdex 200 size-exclusion chromatography. Overexpression and purification of ColD-CT were carried out by following the published procedure (49). Mutants for EcPrfH (Q122N and Q122E) were generated using QuikChange site directed mutagenesis (Agilent), followed by overexpression and protein purification as described above.
Purification of the Intact and the Damaged E. coli 70S Ribosome.
The intact 70S ribosome was purified using the method described previously (50). For the damaged 70S ribosome, E. coli MG1655 harboring plasmid pCH450::cdiA-CTECL and pTrc99a::cdiIECL (both plasmids were generous gifts from C. S. Hayes, University of California, Santa Barbara) was grown in Luria-Bertini (LB) medium to an optical density at 600-nm wavelength (OD600) of 0.4. To induce the expression of CdiA-CTECL, 0.2% arabinose was added. After 5 min of expression, cells were harvested, and the damaged 70S ribosome was purified analogous to the purification of the intact 70S ribosome.
Peptide-Release Assays.
The kinetic experiments on peptide release were carried out as previously described (48) with minor modifications. First, the intact 70S ribosomal complexes or the damaged 70S ribosomal complexes, containing intact or cleaved E. coli 70S ribosome, f-[35S]-Met-tRNAfMet, and one of the three mRNAs shown in Fig. 1A, was assembled as previously described. The complex (50 nM) was then reacted with EcPrfH of various concentrations at 37 °C. The reactions were quenched by addition of 5% ice-cold trichloroacetic acid at different time points, and the precipitants were removed with centrifugation at 18,000g for 10 min at 4 °C to separate f-[35S]-Met from f-[35S]-Met-tRNAfMet. The supernatant was recovered, and the released f-[35S]-Met was counted in 2 mL of Bio-Safe II Complete Counting Mixture (Research Products International). The maximum releasable fMet (fMetMax) was determined by incubating the 70S ribosomal complexes (50 nM) with 200 μM puromycin (Sigma-Aldrich) at 37 °C for 30 s (51). The fraction of f-[35S]-Met was determined by the ratio between the released f-[35S]-Met from the reaction and fMetMax.
Electron Microscopy, Data Collection, and Image Processing.
The experimental conditions of cryo-EM sample preparation are similar to the ones of peptide release assays. For the damaged 70S•PrfH, ribosomal complexes were formed by incubating 0.5 μM damaged 70S ribosome with mRNA (1 μM), tRNAfMet, and PrfH (5 μM) in buffer A (20 mM HEPES, pH 7.4, 15 mM magnesium acetate, 150 mM potassium acetate, 4 mM β-mercaptoethanol, 2 mM spermidine, 0.05 mM spermine) for 30 min at 37 °C. Sample preparation for cryo-EM was carried out as described (38). Complexes were first diluted to 80 nM and aliquots of 2.5 μL were incubated for 30 s on glow-discharged holey carbon grids with thin-layer carbon film (Quantifoil R1.2/1.3, 300 mesh, copper). Grids were blotted for 3 s in 100% humidity at 4 °C and plunge frozen with a Vitrobot (FEI). Data were collected in vitreous ice using a Titan Krios G3i electron microscope operating at 300 keV and equipped with a Falcon III direct electron detector (FEI). We used 4,106 micrographs showing Thon rings beyond 3.5 Å. The drifts of movie frames were corrected using MotionCor2 (52), and the contrast transfer functions were determined using CTFFIND4 (53).
Subsequently, a total of 997,751 particles were picked automatically by Relion3 software using Laplacian-of-Gaussian, extracted, and subjected to reference-free two-dimensional classifications to remove nonribosomal particles (54). Three-dimensional classification was employed to remove the ribosomal subunits. We further analyzed 243,376 particles with PrfH in the A site and tRNA in the P and E sites, using focused classification with signal subtraction. A total of 207,600 particles were subjected to the final refinement and yielded a global three-dimensional reconstruction with an overall resolution of 2.55 Å.
To determine whether PrfH is able to bind intact ribosome, 0.5 μM intact 70S ribosome, mRNA (1 μM), tRNAfMet, and PrfH (5 μM) were mixed in buffer A and incubated at 37 °C for 30 min. We acquired 3,750 micrographs using the same parameters as the damaged 70S•PrfH complex. Data processing was also carried out in Relion3. No particle with PrfH was found. Instead, most ribosomes were complexed with E-site tRNA, P-site tRNA, and mRNA. A total of 87,508 particles were selected for final refinement and yielded a 3.30-Å reconstruction.
Resolutions were reported based on the gold-standard Fourier shell correlation of 0.143 criterion (55, 56). The final map was sharpened by applying a negative B-factor estimated using Relion3 (54, 55). Local resolution was estimated using ResMap (57).
Model Building and Refinement.
The recently reported cryo-EM structure of the intact E. coli 70S•ArfA•RF2•tRNA complex, excluding ArfA, RF2, and tRNAs, was used as a starting model for structure refinement (35). The structure of the RRD of PrfH was built de novo according to the density. The GGQ domain of PrfH was built using domain 3 of RF2 in the 70S•ArfA•RF2•tRNA complex as the starting model (35). Initial fitting of protein, tRNA, and ribosome subunits were performed in Chimera (58). The structures were refined in Phenix (59). The final model was validated using MolProbity (60). SI Appendix, Table S1 summarizes refinement statistics for the structure. Maps were visualized using Chimera, and figures were generated using Chimera and PyMOL.
Preparation of tRNA and Ribosome Substrates for RNA Repair.
Cleaved tRNAAsp was prepared in vivo with the expression of ColE5-CT. Thus, the gene encoding ColE5-CT was cloned into arabinose-inducible pBAD33 vector and transformed into E. coli MG1655 cells. When the OD600 of the E. coli cells reached 0.4, expression of ColE5-CT was induced with the addition of 0.2% l-arabinose. After 10 min of induction, cells were harvested with centrifugation, and total tRNAs were isolated using acid guanidinium thiocyanate-phenol-chloroform extraction. The cleaved tRNAs, which include cleaved tRNAAsp, were further purified by denaturing polyacrylamide gel electrophoresis (DPAGE).
Our previous experience indicated that, for reasons we still do not understand, tRNAArg substrate prepared in vivo was not a good substrate. Therefore, cleaved tRNAArg was prepared in vitro using the recombinant ColD-CT. Total tRNAs (70 μM) isolated from E. coli MG1655 were incubated with recombinant ColD-CT (0.21 μM) at 37 °C for 10 min in the presence of cleavage buffer (25 mM HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid], pH 7.0, 25 mM NaCl, 5 mM MgCl2, 0.05 mg/mL bovine serum albumin), and the cleaved products were purified by DPAGE.
Preparation of cleaved 70S ribosome for repair assay was similar to the procedure of obtaining the damaged E. coli 70S ribosome described previously, with minor modification. First, CdiA-CTECL was expressed for 30 min instead of 5 min. Second, following the expression of CdiA-CTECL, the immunity protein inhibiting CdiA-CTECL, CdiIECL, was also expressed for 3 min with the induction of 1 mM IPTG. The cells were then harvested, and the pellets were resuspended in ribosome dissociation buffer (20 mM HEPES, pH 7.5, 1 mM MgCl2, 200 mM NH4Cl, 4 mM β-mercaptoethanol) supplemented with 0.5 mM phenylmethylsulfonyl fluoride and 5.5 μg/mL DNase I. The cells were lysed using a French press, and the lysate was clarified with centrifugation at 15,000g for 30 min at 4 °C. This clarified lysate was used directly for the ribosome repair assays.
Cleaved 70S substrate for the sequential reactions by EcPrfH and EcRtcB2 was prepared in a similar manner. However, the cells were lysed in ribosome lysis buffer (20 mM HEPES, pH 7.5, 6 mM MgCl2, 30 mM NH4Cl, 4 mM β-mercaptoethanol) supplemented with 0.5 mM phenylmethylsulfonyl fluoride and 5.5 μg/mL DNase I. Subsequent sample processing was performed as described above.
Repair of Cleaved RNA Substrates and Northern Blotting Analysis.
Repair of cleaved tRNAAsp was carried out with either recombinant EcRtcB1 or EcRtcB2 in a reaction mix containing 50 mM Tris⋅HCl, pH 7.4, 2 mM GTP, 5 mM MnCl2, 20 mM MgCl2, 6 μM enzyme, and 2 μM of the cleaved tRNA substrate at 37 °C. Repair of the cleaved tRNAArg was carried out under identical conditions except the concentrations of the enzyme and the tRNA substrate were five times greater. The enzyme was preincubated with all components in the reaction mix except the tRNA substrate at 37 °C for 5 min before the addition of tRNA substrate. We removed 8-μL aliquots at specific time intervals: 2 min, 5 min, 15 min, 30 min, and 60 min. The “no-enzyme” control was prepared by taking an aliquot of the reaction mix before the addition of the enzyme, followed by addition of the tRNA substrates. The reactions were quenched by addition of 8 μL DPAGE loading buffer. The samples were heated at 95 °C for 3 min, followed by 12% DPAGE analysis for 25 min. After the gel analysis, RNAs were transferred to a nylon membrane, followed by hybridization with 5′-32P-radiolabeled DNA probes targeting the 3′ half of E. coli tRNAAsp and tRNAArg, respectively.
For the repair of the damaged 30S subunit, an estimation of ribosomal RNA concentration was made based on the absorbance of the clarified lysate at 260 nm. The reaction mix consisted of 50 mM Tris⋅HCl, pH 7.4, 2 mM guanosine-5′-triphosphate (GTP), 5 mM MnCl2, 10 μM enzyme, and 1 μM of the damaged or the intact 30S subunit. We removed 75-μL aliquots at the specific time intervals and quenched the reactions by addition of TRIzol reagent (ThermoFisher Scientific). This was followed by extraction of total RNA. We analyzed 3 μg of RNA from each reaction by 4% DPAGE gel for 25 min, followed by Northern blotting using a 5′-32P-radiolabeled DNA probe complementary to the 3′ end of E. coli 16S rRNA (6).
The concentration of the cleaved 70S substrate for the sequential reactions by EcPrfH and EcRtcB2 was estimated on the basis of the absorbance of the clarified lysate at 260 nm. The substrate was preincubated with EcPrfH at 37 °C for 10 min in a reaction mix consisting of 50 mM Tris⋅HCl, pH 7.4, 2 mM GTP, 5 mM MnCl2, 6 mM MgCl2, 30 mM NH4Cl, 50 µM EcPrfH, and 1 µM cleaved 70S substrate to allow dissociation of the ribosomal subunits. Subsequently, EcRtcB2 was added at a final concentration of 10 µM, followed by incubation at 37 °C for 20 min. Identical reactions were set up for the EcPrfH mutants (Q122N and Q122E). All reactions were quenched by adding TRIzol reagent, followed by total RNA extraction. Northern Blotting analysis was performed as described above.
All Northern blots were exposed to phosphor screen, imaged using STORM Imager (GE), and the bands quantified using ImageQuant software. The intensities of all bands were adjusted with the bands of background. The percentage of repair for tRNAAsp and tRNAArg was determined by calculating the intensity of the full-length tRNA band as a percentage of the total band intensity (full-length and cleaved) for each sample. Unlike tRNA substrates, intact 16S rRNA is already present in the repair substrate. Therefore, calculation of the percentage of repair of 30S subunit is a bit more complicated. Thus, a total intensity of each lane was obtained with the addition of the intensities of the intact or repaired 16S rRNA and the 49-nt cleavage product. The ratio of this value was calculated for each sample relative to the no-enzyme control (see the sample in the S lane shown in Figs. 4 C and G and 5A), and the band intensities in each lane were adjusted with this ratio. Next, the band intensity of the intact 16S rRNA band of the no-enzyme control was subtracted from each of the adjusted repaired 16S rRNA bands. Finally, the extent of repair was determined by calculating the percentage of this repaired 16S rRNA–band intensity with respect to the total adjusted band intensities (repaired band and cleavage product).
In Vivo Assays.
The gene encoding E3L-1, E3L-2, E3L-3, and E3L-4 (UniProt numbers A0A2W6YY66, A0A2S3UWT3, A0A132N0E3, and A0A132MI77, respectively) was purchased from IDT. The synthetic genes were cloned into pBAD33 plasmid with SacI and HindIII restriction sites. Similarly, the RtcB2–PrfH operon was amplified with EcoRI and BamHI restriction sites and cloned into a pQE60 plasmid. The single- and double-point mutations were introduced into RtcB2 (H294N) and PrfH (Q122E) plasmids by quick-change mutagenesis according to the protocol provided by Agilent. All mutations were verified by DNA sequencing of the entire gene prior to use. The resulting plasmids, either toxin (namely, pBAD33-E3L-1, pBAD33-E3L-2, pBAD33-E3L-3, and pBAD33-E3L-4) alone or together with the repair operon (namely, pQE60-RtcB2-WT/PrfH-WT, pQE60-RtcB2-H294N/PrfH-WT, pQE60-RtcB2-WT/PrfH-Q122E, and pQE60-RtcB2-H294N/PrfH-Q122E) were transformed into E. coli MG1655 strain. Chloramphenicol (25 μg/mL) and ampicillin (100 μg/mL) for pBAD33 and pQE60, respectively, were added to the media to retain the plasmids in the E. coli cells. To induce expression of the toxin in pBAD33 plasmid, l-(+)-arabinose (Sigma-Aldrich) was added at a concentration of 3 mM, and 1 mM IPTG was added to induce the expression of RtcB2-PrfH in the pQE60 plasmid. LB medium (Fisher) was used to culture bacteria at 37 °C.
For growth experiments, overnight culture was grown from a single colony, subcultured, and normalized to an OD600 of ∼0.02 in fresh LB medium. Subsequently, the culture was split in two: E3L uninduced and induced. Induction occurred when cells reached an OD600 of ∼0.3 to 0.4. Growth was monitored for 6 h by measuring OD600 of cultures every 1 h. The experiment was done in triplicate and error bars indicate SEs (n = 3). To test RNA repair, the culture was split into three after subculturing: uninduced, E3L induced, and both E3L and RtcB2–PrfH induced. The induction and subsequent measurements were the same as the ones for toxin only. For further experiments with E3L-2 depicted in Fig. 5B, each experiment was performed six times and error bars indicate SEs (n = 6).
Supplementary Material
Acknowledgments
We thank the cryo-EM center of Southern University of Science and Technology for cryo-EM data collection, C. S. Hayes (University of California Santa Barbara) for providing us the plasmids that encodes CdiA-CTECL and its immunity protein, and P. Wang for carrying out the initial stage of research. This work was supported by the NIH (Grants GM145060 and GM120764 to R.H.H.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2202464119/-/DCSupplemental.
Data Availability
Electron microscopy maps have been deposited in the Electron Microscopy Data Bank under accession codes EMD-24944 (61) for the damaged 70S•PrfH complex and EMD-24945 (62) for the intact 70S complex. Coordinates have been deposited in the Protein Data Bank under accession codes 7SA4 (63) for the damaged 70S•PrfH complex.
References
- 1.Endo Y., Wool I. G., The site of action of alpha-sarcin on eukaryotic ribosomes. The sequence at the alpha-sarcin cleavage site in 28 S ribosomal ribonucleic acid. J. Biol. Chem. 257, 9054–9060 (1982). [PubMed] [Google Scholar]
- 2.Endo Y., Mitsui K., Motizuki M., Tsurugi K., The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. The site and the characteristics of the modification in 28 S ribosomal RNA caused by the toxins. J. Biol. Chem. 262, 5908–5912 (1987). [PubMed] [Google Scholar]
- 3.Bowman C. M., Dahlberg J. E., Ikemura T., Konisky J., Nomura M., Specific inactivation of 16S ribosomal RNA induced by colicin E3 in vivo. Proc. Natl. Acad. Sci. U.S.A. 68, 964–968 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senior B. W., Holland I. B., Effect of colicin E3 upon the 30S ribosomal subunit of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 68, 959–963 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng C. L., et al. , Structural basis for 16S ribosomal RNA cleavage by the cytotoxic domain of colicin E3. Nat. Struct. Mol. Biol. 17, 1241–1246 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck C. M., et al. , CdiA from Enterobacter cloacae delivers a toxic ribosomal RNase into target bacteria. Structure 22, 707–718 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amitsur M., Morad I., Kaufmann G., In vitro reconstitution of anticodon nuclease from components encoded by phage T4 and Escherichia coli CTr5X. EMBO J. 8, 2411–2415 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogawa T., et al. , A cytotoxic ribonuclease targeting specific transfer RNA anticodons. Science 283, 2097–2100 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Tomita K., Ogawa T., Uozumi T., Watanabe K., Masaki H., A cytotoxic ribonuclease which specifically cleaves four isoaccepting arginine tRNAs at their anticodon loops. Proc. Natl. Acad. Sci. U.S.A. 97, 8278–8283 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu J., Huang B., Esberg A., Johansson M. J. O., Byström A. S., The Kluyveromyces lactis γ-toxin targets tRNA anticodons. RNA 11, 1648–1654 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michalska K., et al. , Functional plasticity of antibacterial EndoU toxins. Mol. Microbiol. 109, 509–527 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lancaster L. E., Savelsbergh A., Kleanthous C., Wintermeyer W., Rodnina M. V., Colicin E3 cleavage of 16S rRNA impairs decoding and accelerates tRNA translocation on Escherichia coli ribosomes. Mol. Microbiol. 69, 390–401 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan C. M., Zhou C., Huang R. H., Reconstituting bacterial RNA repair and modification in vitro. Science 326, 247 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Wang P., Selvadurai K., Huang R. H., Reconstitution and structure of a bacterial Pnkp1-Rnl-Hen1 RNA repair complex. Nat. Commun. 6, 6876 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka N., Shuman S., RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J. Biol. Chem. 286, 7727–7731 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakravarty A. K., Subbotin R., Chait B. T., Shuman S., RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc. Natl. Acad. Sci. U.S.A. 109, 6072–6077 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maughan W. P., Shuman S., Characterization of 3′-phosphate RNA ligase paralogs RtcB1, RtcB2, and RtcB3 from Myxococcus xanthus highlights DNA and RNA 5′-phosphate capping activity of RtcB3. J. Bacteriol. 197, 3616–3624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maughan W. P., Shuman S., Distinct contributions of enzymic functional groups to the 2′,3′-cyclic phosphodiesterase, 3′-phosphate guanylylation, and 3′-ppG/5′-OH ligation steps of the Escherichia coli RtcB nucleic acid splicing pathway. J. Bacteriol. 198, 1294–1304 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baranov P. V., et al. , Diverse bacterial genomes encode an operon of two genes, one of which is an unusual class-I release factor that potentially recognizes atypical mRNA signals other than normal stop codons. Biol. Direct 1, 28 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popow J., et al. , HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science 331, 760–764 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Englert M., Sheppard K., Aslanian A., Yates J. R. III, Söll D., Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc. Natl. Acad. Sci. U.S.A. 108, 1290–1295 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laurberg M., et al. , Structural basis for translation termination on the 70S ribosome. Nature 454, 852–857 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Weixlbaumer A., et al. , Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322, 953–956 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holm L., DALI and the persistence of protein shape. Protein Sci. 29, 128–140 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogle J. M., et al. , Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Genschik P., Drabikowski K., Filipowicz W., Characterization of the Escherichia coli RNA 3′-terminal phosphate cyclase and its ς54-regulated operon. J. Biol. Chem. 273, 25516–25526 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Engl C., Schaefer J., Kotta-Loizou I., Buck M., Cellular and molecular phenotypes depending upon the RNA repair system RtcAB of Escherichia coli. Nucleic Acids Res. 44, 9933–9941 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee N. L., Gielow W. O., Wallace R. G., Mechanism of araC autoregulation and the domains of two overlapping promoters, Pc and PBAD, in the l-arabinose regulatory region of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 78, 752–756 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guzman L. M., Belin D., Carson M. J., Beckwith J., Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Englert M., et al. , Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3′-terminal phosphate and 5′-OH. Proc. Natl. Acad. Sci. U.S.A. 109, 15235–15240 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janssen B. D., Hayes C. S., The tmRNA ribosome-rescue system. Adv. Protein Chem. Struct. Biol. 86, 151–191 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neubauer C., Gillet R., Kelley A. C., Ramakrishnan V., Decoding in the absence of a codon by tmRNA and SmpB in the ribosome. Science 335, 1366–1369 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keiler K. C., Mechanisms of ribosome rescue in bacteria. Nat. Rev. Microbiol. 13, 285–297 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Rae C. D., Gordiyenko Y., Ramakrishnan V., How a circularized tmRNA moves through the ribosome. Science 363, 740–744 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.James N. R., Brown A., Gordiyenko Y., Ramakrishnan V., Translational termination without a stop codon. Science 354, 1437–1440 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huter P., et al. , Structural basis for ArfA-RF2-mediated translation termination on mRNAs lacking stop codons. Nature 541, 546–549 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Ma C., et al. , Mechanistic insights into the alternative translation termination by ArfA and RF2. Nature 541, 550–553 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Zeng F., et al. , Structural basis of co-translational quality control by ArfA and RF2 bound to ribosome. Nature 541, 554–557 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demo G., et al. , Mechanism of ribosome rescue by ArfA and RF2. eLife 6, e14874 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gagnon M. G., Seetharaman S. V., Bulkley D., Steitz T. A., Structural basis for the rescue of stalled ribosomes: Structure of YaeJ bound to the ribosome. Science 335, 1370–1372 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan K.-H., et al. , Mechanism of ribosome rescue by alternative ribosome-rescue factor B. Nat. Commun. 11, 4106–4111 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carbone C. E., Demo G., Madireddy R., Svidritskiy E., Korostelev A. A., ArfB can displace mRNA to rescue stalled ribosomes. Nat. Commun. 11, 5552 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito K., et al. , Nascentome analysis uncovers futile protein synthesis in Escherichia coli. PLoS One 6, e28413 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phizicky E. M., Schwartz R. C., Abelson J., Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J. Biol. Chem. 261, 2978–2986 (1986). [PubMed] [Google Scholar]
- 45.Englert M., Beier H., Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res. 33, 388–399 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zallot R., Oberg N., Gerlt J. A., The EFI web resource for genomic enzymology tools: Leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. Biochemistry 58, 4169–4182 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shannon P., et al. , Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng F., Jin H., Peptide release promoted by methylated RF2 and ArfA in nonstop translation is achieved by an induced-fit mechanism. RNA 22, 49–60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garza-Sánchez F., Gin J. G., Hayes C. S., Amino acid starvation and colicin D treatment induce A-site mRNA cleavage in Escherichia coli. J. Mol. Biol. 378, 505–519 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaw J. J., Green R., Two distinct components of release factor function uncovered by nucleophile partitioning analysis. Mol. Cell 28, 458–467 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Freistroffer D. V., Kwiatkowski M., Buckingham R. H., Ehrenberg M., The accuracy of codon recognition by polypeptide release factors. Proc. Natl. Acad. Sci. U.S.A. 97, 2046–2051 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zivanov J., et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, 163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosenthal P. B., Henderson R., Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Scheres S. H. W., Chen S., Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tian Y., et al., Data from “Sequential rescue and repair of stalled and damaged ribosome by bacterial PrfH and RtcB.” Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-24944. Deposited 24 September 2021.
- 62.Tian Y., et al., Data from “Sequential rescue and repair of stalled and damaged ribosome by bacterial PrfH and RtcB.” Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-24945. Deposited 24 September 2021.
- 63.Tian Y., et al., Data from “Sequential rescue and repair of stalled and damaged ribosome by bacterial PrfH and RtcB.” Protein Data Bank. https://www.rcsb.org/structure/7SA4. Deposited 24 September 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Electron microscopy maps have been deposited in the Electron Microscopy Data Bank under accession codes EMD-24944 (61) for the damaged 70S•PrfH complex and EMD-24945 (62) for the intact 70S complex. Coordinates have been deposited in the Protein Data Bank under accession codes 7SA4 (63) for the damaged 70S•PrfH complex.