Abstract
Background
Pinellia Tenore (Araceae) is a genus of perennial herbaceous plants, all of which have medicinal value. The chloroplast (cp) genome data of Pinellia are scarce, and the phylogenetic relationship and gene evolution remain unclear.
Methods and results
We sequenced and annotated the Pinellia pedatisecta cp genome and combined it with previously published genomes for other Pinellia species. We used bioinformatics methods to analyse the genomic structure, repetitive sequences, interspecific variation, divergence hotspots, phylogenetic relationships, divergence time estimation and selective pressure of four Pinellia plastomes. Results showed that the cp genomes of Pinellia varied in length between 168,178 (P. pedatisecta MN046890) and 164,013 bp (P. ternata KR270823). A total of 68–111 SSR loci were identified as candidate molecular markers for further genetic diversity study. Eight mutational hotspot regions were determined, including psbI-trnG-UCC, psbM-rpoB, ndhJ-trnT-UGU, trnP-UGG-trnW-CCA, ndhF-trnN-GUU, ndhG-ndhE, ycf1-rps15 and trnR-ycf1. Gene selection pressure suggested that four genes were subjected to positive selection. Phylogenetic inferences based on the complete cp genomes revealed a sister relationship between Pinellia and Arisaema plants whose divergence was estimated to occur around 22.48 million years ago. All Pinellia species formed a monophyletic evolutionary clade in which P. peltata, rather than P. pedatisecta, earlier diverged, indicating that P. pedatisecta is not the basal taxon of Pinellia but P. peltata may be.
Conclusions
The cp genomes of Pinellia will provide valuable information for species classification, identification, molecular breeding and evolutionary exploration of the genus Pinellia.
Supplementary Information
The online version of this article (10.1007/s11033-022-07617-5) contains supplementary material, which is available to authorized users.
Keywords: Pinellia, Phylogeny, Evolution, Divergence time estimation, Chloroplast genome
Introduction
Pinellia Tenore is a small eastern Asian genus in the Araceae family. Although there are only seven perennial herbaceous species in Pinellia genus [1], every member it contains is important traditional Chinese medicinal plant and was recorded in Chinese herb classics more than 2000 years ago; the most famous among them is P. ternata, with an annual demand of 5500–6000 tons [2]. They have been used for the treatment of viper bites, lumbago, allergic reaction and externally to treat traumatic injury, abscesses, neck lymphosarcoma, breast mastitis and uterine cancer [3, 4].
In recent years, the phylogeny and evolution of monocots have come under intense scrutiny with the rapid development of molecular phylogenetic systematics, and multiple studies have highlighted the phylogeny of Pinellia as problematic [5, 6]. The phylogenetic position of the Pinellia genus in Araceae has been controversial, and sister groups of Pinellia show discrepancy under different classification systems and studies [7, 8]. The six major taxonomic systems of Araceae, namely, Schott system [9], Engler system [10], Hutchinson system [11], Grayum system [12], Bogner and Nicolson system [13] and Mayo et al. system [14], have different views on the sister genus of Pinellia, with four genera (Crytocoryne, Langenandra, Ambrosina and Arisaema) as candidates. With the development of molecular systematics, analyses of the plastome gene and restriction-site sequences suggested that the Arisaema genus is strongly related to Pinellia and is the sister genus of Pinellia [7, 15, 16]. Contrary to the above classification, Keating [17] and Bogner and Petersen [18] based on morpho-anatomical data argued that Arisaema and Pinellia cannot gather into a unique clade for their morphological discrepancy of stamens.
Members of Pinellia exhibit wide variations in flower, leaf, bulblet, spathe and ovule characteristics [19, 20]. The low-level taxonomy and interspecific phylogeny of Pinellia are difficult to address based on morphological traits. The most comprehensive below-genus phylogenetic analysis to date has been provided by Yin, a study using a matrix of all seven Pinellia species and ITS and trnL-F DNA barcode sequences [21]. Combined with the morphological traits, Yin concluded that P. pedatisecta is the basal taxon of Pinellia and suggested that its taxonomic rank should be elevated to a section. However, our group previously conducted interspecific phylogenetic analyses of Pinellia by using four DNA barcode sequences (ITS, matK, rbcL and trnL-F) with the same method as Yin. The topologies of four barcodes were not consistent, with P. pedatisecta in the outer layer of ITS and matK trees and in the inner layer of rcbL and trnL-F trees, receiving considerably lower bootstrap values (Supplementary Fig. S1) probably due to insufficient sequence length and interspecific variations.
The chloroplast (cp) is an important self-replicating organelle that plays a crucial role in photosynthesis and in the synthesis of pigment, protein and starch [22]. The cp contains its own circular double-stranded genome, which is inherited maternally in most angiosperms or paternally in some gymnosperms [23]. Unlike the nuclear genome, the cp lacks meiotic recombination. These properties, along with adequate levels of polymorphism, make it a suitable molecule for studies on phylogeny and evolution [24]. Scientists, especially Henriquez CL and Abdullah research group, have long been devoted to the plastome phylogeny of the Araceae family [25–28], and sequenced the cp genome of P. pedatisecta (MN046890) in 2020 [22]. Nevertheless, their studies focused more on the backbone phylogeny at the taxonomic level of the entire or subfamilies of Araceae, and local phylogenetic relationships of Pinellia genus were rarely paid attention to. Moreover, we previously performed a comparative genomics analysis within Pinellia genus, and found that the published P. pedatisecta cp genome was relatively different from those of two other species, P. peltata (NC052862) [29] and P. ternata (KR270823) [30] in sequence length, gene content, GC content, etc. We determined to identify, sample and sequence the cp genome of P. pedatisecta independently for a reliable phylogeny of Pinellia species. The plastome evolution of Pinellia was also discussed, including the estimations of gene selection pressure and divergence time which have not been studied before.
To reveal the interspecific diversification pattern of Pinellia, we determined the complete cp genome of P. pedatisecta and compared its sequence features with three other Pinellia plastomes. The main goals of this study were to (1) characterize and compare the cp genomes of Pinellia and detect the sequence differences between Pinellia species and between published and newly assembled cp genomes of P. pedatisecta; (2) identify simple sequence repeats (SSRs), long repeats and genetically variable regions and select divergence hotspots as candidate DNA barcodes; (3) reconstruct phylogenetic relationships of Pinellia species based on the cp genome alignments and verify their phylogenetic position within Araceae; and (4) estimate genes selection pressure and divergence time for determining the relative order and spacing of speciation events of Pinellia. Comparative cp genomic analysis could provide theoretical basis to further understand the evolution of the Araceae family and additional insights into the long-standing controversial intergenus and intragenus phylogeny of Pinellia.
Materials and methods
Plant material, DNA extraction, sequencing, assembly and annotation
In our study, the cp genome of P. pedatisecta was sequenced to explore the phylogeny and evolution of Pinellia. Fresh leaves of P. pedatisecta from Linyi City, Shandong Province were sampled. Voucher specimens were deposited in Shandong Academy of Chinese Medicine. Total genomic DNA was extracted from 100 mg of silica-dried leaf by using a DNeasy Plant MiniKit (Qiagen, CA, USA) according to the manufacturer’s instructions [31]. The quantity and quality of genomic DNA were examined by using ND-2000 spectrometer (ThermoFisher Scientific, Wilmington, DE, USA) and 0.8% agarose gel electrophoresis. The chloroplast genome of Salvia plebeia (NC050929) was assembled in our previous study [31]. Taking this work as a guidance, the DNA sample pre-treatment (Covaris M220 [Covaris, US; 250 bp] and VAHTS™ Universal DNA Library Prep Kit [Vazyme, China]), whole genome sequencing (Illumina Hiseq 1500 platform [Illumina Inc., USA]), cp genome assembly (Skewer, Basic Local Alignment Search Tool [BLAST], SOAPdenovo v.2.04, GapCloser and MUMmer), junction validation, and annotation (Plann v. 1.1.2, BLAST and Apollo) were performed in turn with the cp genome of P. peltata (NC052862) as a reference sequence. The primers for junction validation are listed in Supplementary Table S1. The cp genome obtained in this study has been submitted to the NCBI database (www.ncbi.nlm.nih.gov). The physical map of P. pedatisecta cp genome was produced with Chloroplot (https://irscope.shinyapps.io/Chloroplot/).
Repeat structure identification
SSRs were identified by MISA [32], and the minimum thresholds for mono-, di-, tri-, tetra-, penta- and hexa-nucleotides were set to 10, 6, 5, 5, 5 and 5, respectively. REPuter [33] was used to detect two kinds of long repeats: forward and palindromic repeats which have been reported relatively more prevalent in the cp genomes of Araceae family [22, 26]. Detection parameter settings were used as follows: repfind -d -p -h 3 -l 30 -best 50. Tandem Repeats Finder (http://tandem.bu.edu/trf/trf.html) was used to find tandem repeats with the default settings.
Genome comparative analysis
In addition to the newly sequenced cp genome, 26 available cp genome sequences of Pinellia and related species were downloaded from the NCBI database: P. pedatisecta (MN046890), P. ternata (KR270823), P. peltata (NC052862), Arisaema franchetianum (MN046885), Arisaema ringens (MK111107), Arisaema erubescens (MT676834), Arisaema nepenthoides (MW338731), Typhonium blumei (NC051872), Sauromatum giganteum (NC050648), Alocasia navicularis (MN046882), Colocasia esculenta (JN105689), Pistia stratiotes (MN885890), Amorphophallus konjac (MK611803), Calla palustris (MN046887), Epipremnum aureum (KR872391), Epipremnum amplissimum (MN477424), Aglaonema costatum (MN046881), Monstera adansonii (MN046888), Zantedeschia aethiopica (KY792991), Pothos scandens (MN046891), Symplocarpus renifolius (KY039276), Symplocarpus nipponicus (MK341566), Lemna minor (DQ400350), Acorus calamus (AJ879453), Acorus americanus (EU273602) and Acorus gramineus (KP099646). The multiple sequence alignment of the 27 cp genome sequences was performed using MAFFT v.7 with the default settings and adjusted manually where necessary with BioEdit v.7.2.5 software. On the basis of the aligned sequence matrix of the cp genomes of Pinellia, interspecific nucleotide diversity (K) was evaluated by sliding window analysis with a step size of 1000 bp and window length of 2000 bp in DnaSP v.5.10 [34]. The evolutionary divergences of the four Pinellia cp genomes were evaluated using nucleotide differences and p-distance by MEGA v.10.0.4. The protein-coding sequences (CDSs) were extracted using Geneious v.2019.1.3 [35].
Phylogenetic analyses
Two datasets (cp genome and coding sequence) were used to construct the phylogenetic topology of 27 Pinellia and related species with maximum likelihood (ML) methods, respectively. ML analyses were performed using RAxML 8.2.9 under the GTRGAMMA model with 1000 rapid bootstrap replicates. Three species of Acorus (A. calamus, A. americanus and A. gramineus) were used as outgroups.
Divergence time estimation
Accurate estimation of the divergence time in a taxon is important to understand its evolutionary history. Divergence times were estimated using PAML mcmctree (PAML v.4.9j) [36] with the approximate likelihood calculation method. The analysis was performed on 27 complete cp genome sequences used in the phylogenetic analysis with three known calibration times: (1) divergence between P. pedatisecta and T. blumei was 36–32 million years ago (Mya); (2) divergence between A. navicularis and P. stratiotes was 85–46 Mya; and (3) all species except the genus Acorus from the Araceae family arose 122–117 Mya inferred from the published knowledge-based TimeTree (timetree.org). Posterior distributions of parameters were approximated using two independent mcmctree analyses of 10,000,000 generations with 20% burn-in. Tracer v.1.4.1 was used for checking the convergence of the chains through adequate effective sample sizes (ESSs).
Gene selection site analysis
A total of 74 single-copy protein-coding genes shared by four Pinellia plastid genomes (Table 1) were extracted and aligned by Geneious v.9.0.5 [35] and MAFFT v.7. The ML tree was constructed using RAxML V.8.2.9 with the GTRGAMMA model based on complete cp genomes. Protein-coding exon and each value of dN, dS and ω were calculated using the site-specific model in the Codeml program (seqtype = 1, model = 0, Nsites = 0, 1, 2, 3, 7, 8) of PAML v.4.9j [36]. To determine the selected sites, we compared the model M0 (one ratio) versus M3 (discrete), M1 (neutral) versus M2 (positive selection) and M7 (beta) versus M8 (beta and ω) and carried out the three likelihood ratio tests (LRTs). Only consistent sites of positive selection with significant support from posterior probability (p of (ω > 1) ≥ 0.99; Bayes Empirical Bayes approach [BEB]) were identified. BEB recognized by Models M2 and M8 were further considered.
Table 1.
Summary of features of four Pinellia chloroplast genomes
| Taxon | Accession number | Length (bp) | Number of genes | GC content (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genome | LSC | SSC | IR | Total | Protein coding | tRNA | rRNA | Genome | LSC | SSC | IR | ||
| P. pedatisecta | MZ702636 | 164,682 | 90,947 | 22,523 | 25,606 | 129 | 85 | 36 | 8 | 35.71 | 33.92 | 29.43 | 42.09 |
| P. pedatisecta | MN046890 | 168,178 | 92,963 | 23,981 | 25,617 | 130 | 85 | 37 | 8 | 35.08 | 33.35 | 26.85 | 42.09 |
| P. peltata | NC052862 | 164,923 | 90,089 | 24,871 | 24,981 | 130 | 86 | 36 | 8 | 36.51 | 34.53 | 31.77 | 42.44 |
| P. ternata | KR270823 | 164,013 | 89,783 | 22,980 | 25,625 | 131 | 86 | 37 | 8 | 36.66 | 34.6 | 31.66 | 42.53 |
Results
Chloroplast genome features in Pinellia
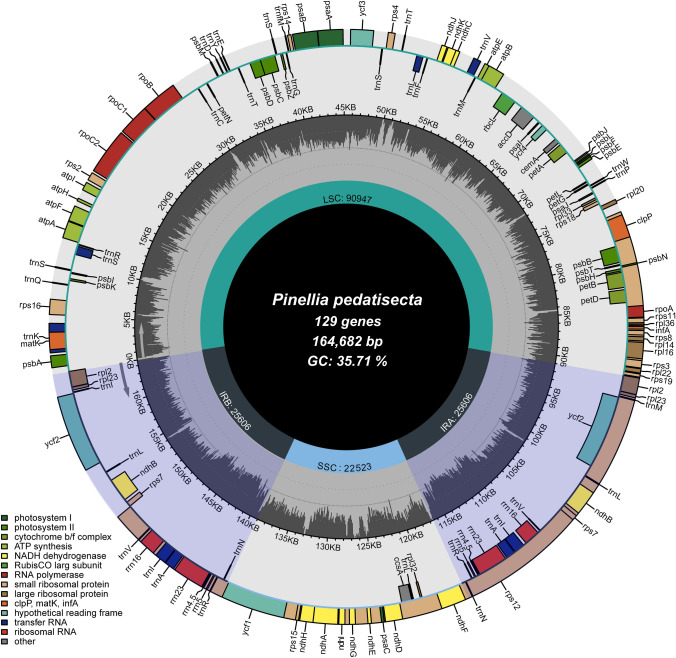
In our study, the cp genome of P. pedatisecta was sequenced, assembled and validated (Fig. S2). A total of 49,904,104 PE raw reads were generated using the Illumina Sequencing System. The novel cp genome sequence has been preserved in GenBank (MZ702636, Table 1). The cp genome of P. pedatisecta was circular double-stranded DNA and displayed a quadripartite structure (Fig. 1). The cp genome assembled in this study was 164,682 bp in length, which was 3,496 bp shorter than the published one (MN046890, 168,178 bp) mainly due to the contraction of the LSC region. Moreover, the GC content in the SSC region of the cp genome MZ702636 was 2.58% higher than that of MN046890. The cp genome MN046890 had the same rRNA gene content as that of MZ702636 but contained one more tRNA gene (tRNA-His) (Supplementary Table S2).
Fig. 1.
Chloroplast genome map of P. pedatisecta assembled in this study. The centre of the figure provides length information of the cp genome. In the first inner circle, the proportion of the shaded parts represents the GC content of each part. The gene names are labelled on the outermost layer. The transcription directions for the inner and outer genes are listed clockwise and anticlockwise, respectively
After downloading from the NCBI database, protein-coding gene number variations of 24 cp genomes of the Araceae family, including four Pinellia cp genomes, were analysed (Fig. S3). Compared with the cp genome of the model plant Arabidopsis thaliana, two genes, infA and ycf68, were inserted among most Araceae plants, while one-copy ycf1 gene was missing. In the Araceae family, there were significant differences in the number of ycf68 genes among species. The ycf68 genes in 60% of Araceae cp genomes were completely missing, while the remaining 40% of the cp genomes had two copies. Even within the Pinellia genus, the number of ycf68 genes was also inconsistent. Nevertheless, there was no discrepancy in the type and number of protein-coding genes between our newly assembled cp genome (MZ702636) and the published one (MN046890).
SSR and repeat sequence analyses
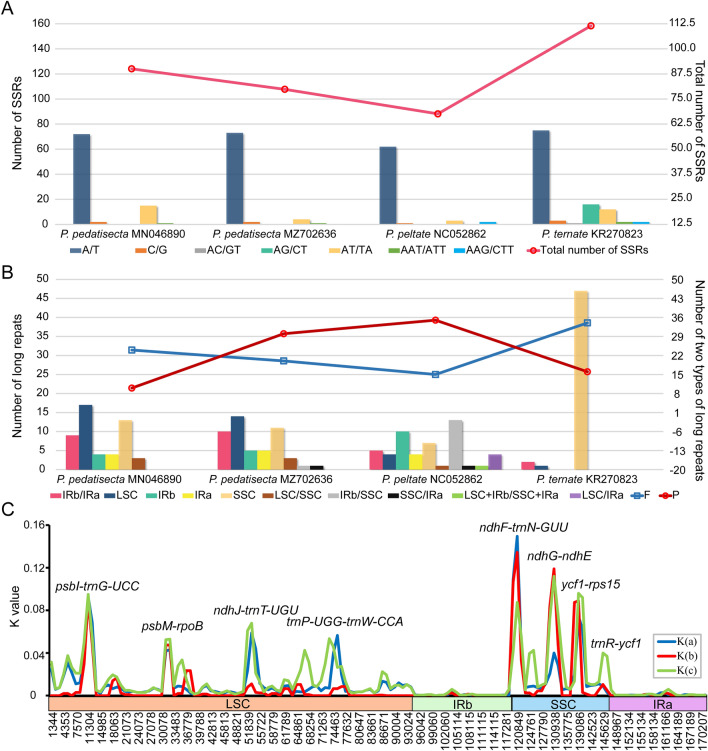
The number of SSRs in the four Pinellia cp genomes ranged from 68 (P. peltata) to 111 (P. ternata, Fig. 2A and Supplementary Table S3). Three kinds of SSRs were discovered, namely, mononucleotide, dinucleotide and trinucleotide. Among each Pinellia species, mononucleotide repeats were the most common, whereas trinucleotide repeats accounted for the lowest proportion of SSRs. The number of A/T mononucleotide repeats exceeded that of the other three types combined (Fig. 2A). Interspersed repeated sequences were identified by using REPuter for four plastomes (Fig. 2B and Supplementary Table S4). Except for IR regions, the repeats’ length ranged from 31 to 328 bp with forward (F) repeats as the relative prevalent type. Most interspersed repeats of two P. pedatisecta cp genomes were located in the LSC regions, while the long repeats of P. peltata and P. ternata were predominantly in the IRb/SSC and SSC regions, respectively. The total number of tandem repeats for four plastid genomes was in the range of 34–274. P. peltata had the least repeats, and P. pedatisecta (MN046890) had the most (Supplementary Table S5).
Fig. 2.
The SSRs (A), interspersed repeated sequences (B), and interspecific nucleotide diversity (C) among the chloroplast genomes of Pinellia. X-axis in Fig. 3C: position of a window. Y-axis in Fig. 3C: sequence divergence (K values) between species of each window. K(a): K values between P. pedatisecta and P. peltata; K(b): K values between P. pedatisecta and P. ternata; K(c): K values between P. ternata and P. peltata
Comparative genomic analyses
The differences and evolutionary divergences among four Pinellia cp genomes were compared using nucleotide substitutions and sequence distance (Supplementary Table S6). Across all four Pinellia cp genomes, the p-distance was 0.000402–0.013668, and the value of nucleotide differences was 66–2459. The comparison results of nucleotide substitution and genetic distance were consistent. The sequence divergence level of two cp genomes of P. pedatisecta was the lowest. The differentiation between P. pedatisecta and P. ternata was less than that between P. pedatisecta and P. peltata. Among three Pinellia species, P. ternata and P. peltata were the most genetically distant. Moreover, the K value (sequence divergence between species) was calculated, and the sliding windows of the K values were constructed by DnaSP (Fig. 2C and Supplementary Table S7). Figure 2C showed that the sequence divergence between P. pedatisecta and P. ternata was much lower than the two other K values. The sequence divergence between P. pedatisecta and P. peltata was not so different from that between P. ternata and P. peltata. Eight highly variable regions with great K values were detected, namely, psbI-trnG-UCC, psbM-rpoB, ndhJ-trnT-UGU, trnP-UGG-trnW-CCA, ndhF-trnN-GUU, ndhG-ndhE, ycf1-rps15 and trnR-ycf1. Four of these regions were located in the LSC region, and the remaining four were located in the SSC region, all of which were present in the non-coding regions.
Phylogenetic analyses
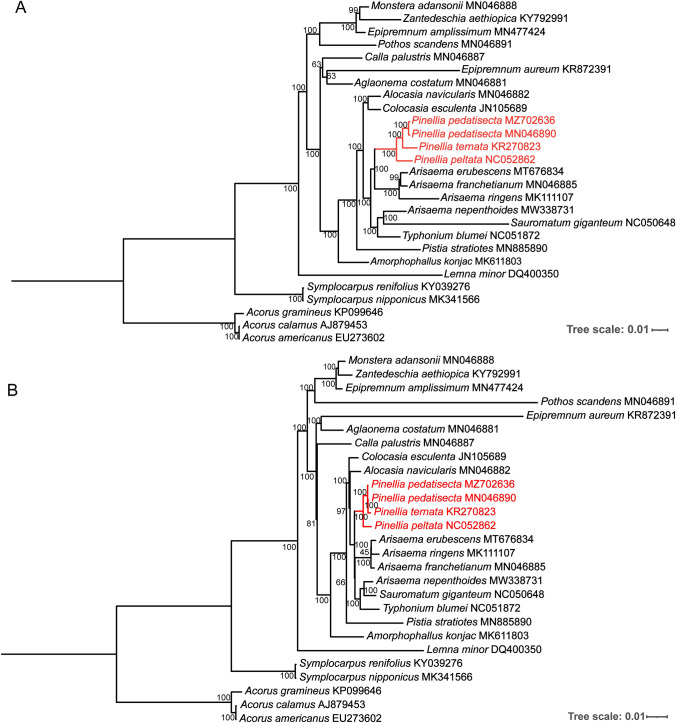
To analyze the phylogenetic relationship of Pinellia species, we constructed phylogenetic trees using whole cp genome sequences and their CDSs by ML methods. For Pinellia species, the topological structure obtained by either complete cp genome or CDSs was roughly identical (Fig. 3). The topology based on entire cp genomes showed that Pinellia species were monophyletic and clustered into three clades: one for two P. pedatisecta plastomes, one for P. ternata and one for P. peltata with strong support (bootstrap value 100%). P. pedatisecta and P. ternata clustered together, exhibiting the highest genetic similarity among the studied representatives of Pinellia genus. In contrast to the CDSs phylogeny (Fig. 3B), in which Pinellia species were placed most closely to Arisaema, Sauromatum and Typhonium clade, the whole-length plastome phylogeny placed some Arisaema species as sister to the genus Pinellia with 100% bootstrap support (Fig. 3A).
Fig. 3.
Phylogenetic trees constructed with the whole cp genomes (A) and their protein-coding sequences (B) by using ML method. Numbers near each branch are bootstrap values
Divergence time estimation
In this study, we used full-length sequences of cp genomes of 23 Araceae family plants (including four Pinellia cp genomes) and three outgroups to estimate the divergence times of major clades in the Araceae family. Our dating analysis resulted in estimates for the crown node of the Araceae family of 91.99 Mya (95% highest posterior density [HPD] = 46.69–125.17 Mya; node 1 in Fig. S4; Supplementary Table S8) in the early Cretaceous. The divergence time of the four genera Pinellia, Arisaema, Sauromatum and Typhonium was estimated at 25.65 Mya (95% HPD = 12.93–36.35 Mya; node 2) in the Oligocene. The age estimation for the crown node of Pinellia and most species of Arisaema was dated to 22.48 Mya (95% HPD = 11.39–33.32 Mya; node 3) in the late Oligocene and early Miocene. Pinellia diverged from its sister clade at 13.09 Mya (95% HPD = 4.91–21.96 Mya; node 4 in Fig. S4; Supplementary Table S8) in the Miocene.
Gene selection pressure analysis of protein sequences
The site-specific selective pressure on four Pinellia plastomes was assumed using the site model in PAML program. Three pairs of site model comparisons (M0 vs M3, M1 vs M2a and M7 vs M8) showed four protein-coding genes subjected to positive selection (LRT of the three comparisons all p < 0.05; Supplementary Tables S9 and S10), namely, petB, rpoC1, rps12 and ycf1. Of these genes, ycf1 had the highest number of sites (21 sites; Table S9), followed by rpoC1 (4 sites), petB (1 site) and rps12 (1 site).
Discussion
Phylogenetic position of the Pinellia genus in Araceae
In the previous classification systems, the Pinellia genus belonged to the Araceae family, but its sister genus remains controversial. In the Schott system [9], which is the first classification system of the Araceae family, the genus Pinellia was placed in the tribus Alleluehieae, related to two genera of Crytocoryne and Langenandra. In the Engler system [10], Pinellia belonged to the subtrib. Pinelliinae, trib. Areae, subfam. Aroideae, in the same tribus as the Arisaema genus. In the Hutchinson system [11], this genus was placed in trib. Areae, related to two genera of Crytocoryne and Ambrosina. In the classification system of Grayum [12], Pinellia belonged to trib. Pinellia, subfam. Aroideae. After modifying the Engler system by Bogner and Nicolson [13], the Pinellia genus changed to the subtrib. Atherurinae, trib. Pinellia, subfam. Aroideae. Mayo et al. [14] conducted taxonomic analysis of 106 taxa in the Araceae family using 63 morphological and anatomical traits and proposed the latest classification system of the Araceae family. In this system, Pinellia was placed in trib. Arisaemateae, subfam. Aroideae with the Arisaema genus as sister group. Since the 21st century, with the development of molecular systematics, many studies have investigated the phylogenetic position of the Pinellia genus in Araceae [7, 15–18], yet controversy regarding molecular systematics and morphological classification persists. French et al. [16], Cabrera et al. [7] and Cusimano et al. [15] performed molecular phylogenetic analyses by applying different types and numbers of DNA marker sequence data, and they discovered that the Arisaema genus is strongly related to Pinellia among Araceae plants, and is the sister genus of Pinellia. Contrary to the above classification, Keating [17] and Bogner and Petersen [18] based on morpho-anatomical data argued that Arisaema and Pinellia cannot be grouped into a unique clade. One significant morphological discrepancy between them was that almost all Arisaema species have at least partially fused stamens, whereas Pinellia and other related genera (e.g. Sauromatum and Typhonium) have free stamens.
The cp genome is one of three subcellular compartments in the plant genome, and it is mainly inherited from the maternal parent [37]. Given its high conservation and abundant interspecific variation, the cp genome has the potential to provide distinguishing differences that can help molecularly classify closely related species [38]. With advances in high-throughput sequencing, achieving the cp genome is easily acquirable at a large scale with low costs. Researchers have proposed the entire cp genome as a super barcode to discriminate and classify closely related species [39]. To date, phylogenies of several genus-level taxa have been further clarified by using cp genome sequences, such as Epimedium [40], Paris [41] and Sanguisorba [42]. Hence, we compared 24 cp genomes of the Araceae family, including four plastomes of Pinellia, to explore the cp genome molecular phylogeny of Pinellia. The resulting phylogeny based on entire cp genomes in this study showed that Pinellia and Arisaema plants were gathered into one branch and sister groups to each other with a well-supported bootstrap value (100%; Fig. 3A). Although the Araceae family is a major group of monocotyledons, there is still a limited number of cp genomes available from Araceae species, which may result in some congeneric species, such as Arisaema plants, not clustering together in one phylogenetic branch (Fig. 3). The absence of plastid genomes from three other candidates of the Pinellia sister groups, Crytocoryne, Langenandra and Ambrosina [21], could also hinder our discovery of the intergenus phylogeny of Pinellia.
Phylogeny within the Pinellia genus
Despite recent advances in molecular phylogenetic studies, deep evolutionary relationships and below-genus taxonomic classification of Pinellia remain unresolved. Yin [21] made the first comprehensive phylogenetic analyses of all seven Pinellia species by using the sequences of ITS and trnL-F DNA barcodes. The resulting phylogeny of both ITS and trnL-F sequences supported that P. pedatisecta and the six remaining Pinellia species were sister species, and P. pedatisecta served as a basal taxon in Pinellia. Moon et al. [43] reconstructed the phylogenetic trees of three species, P. ternata, P. tripartita and P. pedatisecta, with matK and rbcL sequences; they found that the topology of two trees is inconsistent. The basal taxon in the matK tree was P. pedatisecta, congruent with that by Yin [21], while the basal taxon in the rbcL tree was P. ternata. Furthermore, after morphological investigation, Yin found that P. pedatisecta has many characteristics of the Pinellia genus. These traits include perennial herb with a small tuber at the top of the main tuber, persistent spathe, female inflorescence adnate to the spathe, unisexual flowers, no perianth, one straight ovule and green fruit. Hence, combined with the molecular phylogeny, Yin concluded that the basal taxon of the Pinellia genus is P. pedatisecta, the species native to shady woodland areas, forested slopes and valleys in northern and western China; he suggested that the taxonomic rank of P. pedatisecta should be elevated to a section of Pinellia [21], which was the first and only below-genus taxonomic recommendation of Pinellia.
In our opinion, P. pedatisecta indeed has some of the characteristic traits of Pinellia, but it also possesses so many characteristics significantly different from other congeneric species. For example, lanceolate spathe, no constriction in the tube and eaves of spathe, no diaphragm between the female inflorescence and male inflorescence and no bulblet on the petiole [15, 19, 20]. Our group previously conducted phylogenetic analyses of Pinellia by using four different barcode sequences with maximum parsimony (MP) method. The topologies of the four barcodes were not consistent, with poor bootstrap values (Fig. S1) probably due to insufficient sequence length and interspecific variation. Hence, plastid genomes, an ideal model for evolutionary and comparative genomic studies of related species, were necessary here for a molecular phylogeny of Pinellia with a significantly higher resolution.
After comparing the structural organization of three previously published cp genomes of Pinellia, we found that the sequence similarity between the published P. pedatisecta plastome (MN046890) and the cp genomes of P. peltata (NC052862) and P. ternata (KR270823) was clearly lower than expected in the following five aspects.
The length of the P. pedatisecta plastome was significantly longer than that of the two other species (Table 1), specifically 3.2 kb longer than that of P. peltata and even 4.1 kb longer than that of P. ternata. The length of the LSC region in P. pedatisecta was 3.18 kb longer than that of P. ternata.
With regard to protein-coding gene content, all two ycf68 genes were missing in the published cp genome of P. pedatisecta (Fig. S3), compared with P. peltata and P. ternata, which could have the potential to affect the physiological functions of plants, although some authors suggested that the ycf68 gene likely does not encode a protein [44].
The GC content of published P. pedatisecta was lower than that in P. peltata and P. ternata (Table 1), especially in the SSC region, which was 4.92% lower than that in P. peltata and 4.81% lower than that in P. ternata.
After the alignment of three published cp genomes and further estimation of locally collinear blocks (LCBs) with MAUVE 2.4.0, the gene order comparison revealed one rearrangement (~ 128 bp) between the published plastomes of P. pedatisecta and P. ternata (Fig. S5A and S5B).
A significant insertion/deletion variation was noted across three cp genomes in this group, located between tRNA-Ser (GCU) and tRNA-Ser (CGA) genes in the LSC region with the length of ~ 1.2 kb (10,714–11,950 bp in the published P. pedatisecta cp genome and 10,192–10,234 bp in the self-assembled one; Fig. S5C and S5D).
These five sequence discrepancies do not follow the rule of “chloroplast genomes of related species, especially those within the same genus, are generally highly conserved” [45, 46]. Therefore, we determined to identify, sample and sequence the plastome of P. pedatisecta independently for further phylogenetic analyses of Pinellia.
In this study, the phylogenetic tree based on whole cp genomes showed that the species of Pinellia were clustered into three clades: one for two P. pedatisecta plastomes, one for P. ternata and one for P. peltata with high support values (Fig. 3). P. pedatisecta and P. ternata, which possess pedate and three full-lobed leaves, respectively, grouped together in topology, exhibiting the highest genetic similarity among the studied representatives of Pinellia genus. In contrast, P. peltata, characterized by undivided and peltata leaf and development of stem tuber, was placed in the outer layer of Pinellia topology. P. pedatisecta is buried inside the cp genome phylogeny, incongruent with the phylogeny from ITS and trnL-F sequences by Yin [21]. Furthermore, comparative genomic analyses showed that the nucleotide substitutions and sequence distance between P. pedatisecta and P. ternata were the smallest (1275 and 0.007206; Table S6), while that between P. ternata and P. peltata was the largest (2459 and 0.013668). The interspecific nucleotide diversity (K value) between P. pedatisecta and P. ternata was considerably lower than the two other K values (Fig. 2C), both of which confirmed that P. peltata shared less sequence similarity with P. pedatisecta and P. ternata. Limited by the number of Pinellia cp genomes published so far, we cannot determine whether P. peltata is the ancestor of Pinellia species, but it is possible. P. peltata has the same floral morphological characteristics as other Pinellia species, except for P. pedatisecta [29]. For example, solitary inflorescence, persistent spathe, with constriction in the tube and eaves of spathe, female inflorescence adnate to the spathe, with diaphragm between the female inflorescence and male inflorescence, unisexual flowers, no perianth, one locular ovary and one straight ovule [21]. However, the peltata leaf characteristics of P. peltata are considerably different. Thus, the below-genus taxonomic classification of Pinellia is suggested to be based on differences in leaves or other tissue morphological characteristics between species, rather than flowers.
Variations and evolution of Pinellia cp genomes
In this study, the cp genome of P. pedatisecta was sequenced (MZ702636) with the length of 164,682 bp, which fell within the cp genome size range for angiosperms but much smaller than its published one (MN046890, 168,178 bp; Table 1). Among four Pinellia plastid genomes, LSC regions showed the most difference in size, with the shortest 89,783 bp of P. ternata KR270823 and the longest 92,963 bp of P. pedatisecta MN046890. Additionally, the inferred structures and protein-coding gene contents were in accordance except for the infA and ycf68 genes (Fig. 1 and S3). Analysis with DnaSP inferred that some of the most divergent regions of psbI-trnG-UCC, psbM-rpoB, ndhJ-trnT-UGU, trnP-UGG-trnW-CCA, ndhF-trnN-GUU, ndhG-ndhE, ycf1-rps15 and trnR-ycf1, as shown in Fig. 2C, were found for further related species identification of Pinellia.
SSRs in the cp genome are an efficient marker tool for population genetic structure and phylogeography [47]. In this study, 68–111 SSR loci were identified between Pinellia species (Supplementary Table S3). These SSR loci could provide candidate molecular markers for the genetic diversity study of Pinellia. The composition of SSR loci in the cp genomes of three Pinellia species was similar to that of most angiosperms, with A/T mononucleotide repeats dominating all the repeat units. This phenomenon may be one of the reasons for the abundance of A/T bases in cp genomes. Repeat units in the cp genome can cause sequence polymorphism, providing information for further genetic diversity study of Pinellia.
At present, specific studies on the divergence time estimation of Pinellia by using molecular data, particularly plastid genomes, are lacking. Li et al. [48] first supposed that species with simple leaves (e.g. P. peltata) are less evolved than those with pedate or lobed leaves (e.g. P. pedatisecta and P. ternata) on the basis of the palynology and isozyme characteristics of Pinellia species. Yin [21] speculated that the Pinellia genus first appeared in the Paleogene according to geographic distribution and the origin time of its related genus (Arisaema). Here, based on whole cp genome sequences, our divergence time estimation indicated that Pinellia species originated at ~ 22.48 Mya (95% HPD = 11.39–33.32 Mya; node 3 in Fig. S4; Supplementary Table S8) in the late Oligocene and early Miocene, congruent with that estimated by Yin and further refined, and diverged diversely at ~ 13.09 Mya (95% HPD = 4.91–21.96 Mya; node 4 in Fig. S4) in the Miocene. Furthermore, within the Pinellia genus, P. peltata diverged first at ~ 5.34 Mya earlier than two other species, P. pedatisecta and P. ternata; these findings were consistent with the results of Li et al. [48]. The present estimation of divergence times based on plastid genomes and fossil data provides new insights and a hypothetical foundation for future studies on the origin and earlier evolution of Pinellia.
Genes subjected to positive selection have a significant impact on the creative effects of populations, changes under selection stress, and genetic drift led to the rapid transformation of genes into new common adaptive combinations [49]. In this study, we calculated the non-synonymous/synonymous substitution rate ratio (ω = dN/dS) for each of the 74 single copy protein-coding genes shared by the analyzed plastid genomes of Pinellia. Four genes with high posterior probability of codon sites in the BEB test were acquired and considered as genes under positive selection (Fig. 4 and Tables S9 and S10). These genes included one gene for cytochrome b/f complex subunit protein in the photosystem II reaction (petB) [50], one DNA-dependent RNA polymerase gene (rpoC1), one gene for ribosome small subunit protein (rps12) and ycf1 gene. All these genes were also detected in other plants [51–54], and these genes may have played a significant role in the adaptive evolution of Pinellia. The specific role needs to be further studied.
Fig. 4.
Positive selection sites logo of four Pinellia plastomes
The genus Pinellia includes seven species in total. Only three Pinellia species were studied here, and as a consequence, it might not fully show the real effect of the phylogeny of Pinellia. However, our study was the first comprehensive phylogenetic analysis of Pinellia at the cp genome level, and the resulting topologies based on either whole cp genome or CDS datasets both definitively show that P. pedatisecta cannot be the ancestor of Pinellia species. The main habitat of all three Pinellia species in our study is in the southeastern portion of Asia, where hybridization and introgression have been reported [55, 56], which may cause interspecific gene flow in response to ecological selection and lead to difficulties in molecular phylogenetic analysis. Given that the cp and nuclear genomes evolve independently, the phylogeny of cp genomes alone is insufficient in making taxonomic decisions about Pinellia. Therefore, more methods and more species are needed in further study to improve our ability to better understand the phylogeny and evolution of Pinellia.
Conclusions
The genus Pinellia Tenore is comprised of seven species, all of which are important medicinal plant resources. Species phylogenetic classification is vital for protecting species diversity and selecting high quality germplasm resources. But due to members of Pinellia exhibit rather wide variations in morphological structures, the low-level taxonomy and interspecific phylogeny of Pinellia are difficult to address based on morphology. With the development of next-generation sequencing technology, complete chloroplast genomes have been widely employed to explore phylogenetic relationships of intra-or inter-genus. However, the variation and evolution of the whole chloroplast genomes in the genus Pinellia have been ignored. Here, we sequenced the chloroplast genome of P. pedatisecta, compared it with previously published plastid genomes, and reconstructed phylogenetic relationships of Pinellia based on chloroplast genomes and their CDSs. The divergence time estimation and selective pressure of Pinellia plastomes were also investigated. The results showed some variations and adaptive evolution between Pinellia complete chloroplast genomes including size, structure and nucleotide diversity, which provided valuable information for species classification and evolution. In addition, our results revealed a sister relationship between Pinellia and Arisaema plants whose divergence was estimated to occur around 22.48 million years ago. All Pinellia species formed a monophyletic evolutionary clade in which P. peltata, rather than P. pedatisecta, earlier diverged, indicating that P. pedatisecta is not the basal taxon of Pinellia but P. peltata may be. In conclusion, our results could provide insight into the chloroplast genome evolution and phylogeny of Pinellia genus and even of Araceae family, which would be useful for selecting high quality Pinellia germplasm resources in the future.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Baosheng Liao for his support in data analysis.
Abbreviations
- cp
Chloroplast
- SSRs
Simple sequence repeats
- WGS
Whole-genome sequencing
- PE
Paired-end
- BLAST
Basic Local Alignment Search Tool
- LSC
Large single copy
- SSC
Small single copy
- IR
Inverted repeat
- indel
Insertion-deletion
- Pi
Nucleotide diversity
- CDS
Protein-coding sequence
- ML
Maximum likelihood
- Mya
Million years ago
- ESS
Effective sample size
- LRT
Likelihood ratio test
- BEB
Bayes Empirical Bayes approach
- HPD
Highest posterior density
- MP
Maximum parsimony
- LCBs
Locally collinear blocks
Author contributions
NC and XL conceived and designed the study; NC and WC collected and analyzed the data; NC drafted the initial version of the manuscript; NC, XL and PW contributed to later versions of the manuscript.
Funding
This study was supported by the Major Special Project of Scientific and Technological Cooperation of Bijie City (Grant No. 2021-02), Research Incubation Fund Project of Shandong Academy of Chinese Medicine (Grant No. 2021SACM-3), Medical and Health Science and Technology Development Project of Shandong Province (Grant No. 202102041135) and Key Research and Development Program of Shandong Province (Grant No. 2020CXGC010505-04).
Data availability
The chloroplast genome sequence of Pinellia pedatisecta assembled here is accessible via GenBank with the accession number of MZ702636 and Global Pharmacopoeia Genome Database (GPGD,
http://www.gpgenome.com/species/40314). Raw sequencing data is available at NCBI SRA database with the accession number of SRR15328795.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiwen Li, Email: XWLi@icmm.ac.cn.
Ping Wang, Email: wangpingjinan@163.com.
References
- 1.Liu Y, Hui RK, Deng RN, Wang JJ, Wang M, Li ZY. Abnormal male meiosis explains pollen sterility in the polyploid medicinal plant Pinellia ternata (Araceae) Genet Mol Res. 2012;11(1):112–120. doi: 10.4238/2012.January.17.1. [DOI] [PubMed] [Google Scholar]
- 2.Zhu Y, Zhu G, Guo Q, Zhu Z, Wang C, Liu Z. A comparative proteomic analysis of Pinellia ternata leaves exposed to heat stress. Int J Mol Sci. 2013;14(10):20614–20634. doi: 10.3390/ijms141020614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozlov VA, Zhuravkin IN, Tsyrlova IG. Comparative analysis of the migratory capacity of hematopoietic stem cells and of immunocompetent precursors after acute hypoxia and the administration of testosterone. Dokl Akad Nauk SSSR. 1978;239(3):747–749. [PubMed] [Google Scholar]
- 4.Nauheimer L, Metzler D, Renner SS. Global history of the ancient monocot family Araceae inferred with models accounting for past continental positions and previous ranges based on fossils. New Phytol. 2012;195(4):938–950. doi: 10.1111/j.1469-8137.2012.04220.x. [DOI] [PubMed] [Google Scholar]
- 5.Renner SS, Zhang LB. Biogeography of the Pistia clade (Araceae): based on chloroplast and mitochondrial DNA sequences and Bayesian divergence time inference. Syst Biol. 2004;53(3):422–432. doi: 10.1080/10635150490445904. [DOI] [PubMed] [Google Scholar]
- 6.Renner SS, Zhang LB, Murata J. A chloroplast phylogeny of Arisaema (Araceae) illustrates tertiary floristic links between Asia, North America, and East Africa. Am J Bot. 2004;91(6):881–888. doi: 10.3732/ajb.91.6.881. [DOI] [PubMed] [Google Scholar]
- 7.Cabrera LI, Salazar GA, Chase MW, Mayo SJ, Bogner J, Davila P. Phylogenetic relationships of aroids and duckweeds (Araceae) inferred from coding and noncoding plastid DNA. Am J Bot. 2008;95(9):1153–1165. doi: 10.3732/ajb.0800073. [DOI] [PubMed] [Google Scholar]
- 8.Boyce PC. The Genus Hapaline (Araceae: Aroideae: Caladieae) Kew Bull. 1996;51(1):63–82. doi: 10.2307/4118745. [DOI] [Google Scholar]
- 9.Boyce P, Schott H (1860) Schott, H.W. Prodromus Systematis Aroidearum (1860).
- 10.Engler A (1920) Araceae. Pars generalis et Index familiae generalis. In. Edited by Engler A, vol. 74 (IV. 23A). Leipzig, Wilhelm Engelmann.
- 11.Hutchinson J. The families of flowering plants, arranged according to a new system based on their probable phylogeny. 2. London: Internet Archive; 1959. [Google Scholar]
- 12.Grayum MH. Evolution and phylogeny of the Araceae. Ann Missouri Bot Gard. 1990;77:628–697. doi: 10.2307/2399668. [DOI] [Google Scholar]
- 13.Bogner J, Nicolson DH. A revised classification of Araceae with dichotomous keys. Willdenowia. 1991;21:35–50. [Google Scholar]
- 14.Mayo SJ, Bogner J, Boyce PC. Araceae. In: Kubitzki K, editor. Flowering plants monocotyledons. Berlin: Springer; 1998. pp. 26–74. [Google Scholar]
- 15.Cusimano N, Bogner J, Mayo SJ, Boyce PC, Wong SY, Hesse M, Hetterscheid WL, Keating RC, French JC. Relationships within the Araceae: comparison of morphological patterns with molecular phylogenies. Am J Bot. 2011;98(4):654–668. doi: 10.3732/ajb.1000158. [DOI] [PubMed] [Google Scholar]
- 16.French JC, Chung MG, Hur YK. Chloroplast DNA phylogeny of the Ariflorae. In: Rudall PJ, Cribb P, Cutler DF, Humphries CJ, editors. Monocotyledons: systematics and evolution. Kew: Royal Botanic Gardens; 1995. pp. 255–275. [Google Scholar]
- 17.Keating RC. Vegetative anatomical data and its relationship to a revised classification of the genera of Araceae. Ann Missouri Bot Gard. 2004;91:485–494. [Google Scholar]
- 18.Fontana ME, Cavalcante CAV. Índices baseados no número de clientes para localização de itens em armazéns. Production. 2013;23(3):561–569. doi: 10.1590/s0103-65132013005000002. [DOI] [Google Scholar]
- 19.Li YW, Zhou FQ, Zhang SP, Li ZY, Kong QY. Studies on morphology and histology of Pinellia pedatisecta. Zhong Yao Cai. 2008;31(2):206–209. [PubMed] [Google Scholar]
- 20.Guo QS, Wang QY, Shi HZ. Study on morphological characteristics of pollen grains of Pinellia ternata in different populations. Zhongguo Zhong Yao Za Zhi. 2006;31(1):27–30. [PubMed] [Google Scholar]
- 21.Yin T. Phylogeny and biogeography of Pinellia (Araceae) with special reference to phylogenetic problems of Remusatia. Beijing: Kunming Institute of Botany Chinese Academy of Sciences; 2002. [Google Scholar]
- 22.Henriquez CL, Abdullah AI, Carlsen MM, Zuluaga A, Croat TB, McKain MR. Evolutionary dynamics of chloroplast genomes in subfamily Aroideae (Araceae) Genomics. 2020;112(3):2349–2360. doi: 10.1016/j.ygeno.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 23.Henriquez CL, Abdullah AI, Carlsen MM, Zuluaga A, Croat TB, McKain MR. Molecular evolution of chloroplast genomes in Monsteroideae (Araceae) Planta. 2020;251(3):72. doi: 10.1007/s00425-020-03365-7. [DOI] [PubMed] [Google Scholar]
- 24.Zhai W, Duan X, Zhang R, Guo C, Li L, Xu G, Shan H, Kong H, Ren Y. Chloroplast genomic data provide new and robust insights into the phylogeny and evolution of the Ranunculaceae. Mol Phylogenet Evol. 2019;135:12–21. doi: 10.1016/j.ympev.2019.02.024. [DOI] [PubMed] [Google Scholar]
- 25.Abdullah HCL, Mehmood F, Shahzadi I, Ali Z, Waheed MT, Croat TB, Poczai P, Ahmed I. Comparison of chloroplast genomes among species of unisexual and bisexual clades of the monocot family Araceae. Plants (Basel) 2020;9(6):737. doi: 10.3390/plants9060737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abdullah HCL, Mehmood F, Carlsen MM, Islam M, Waheed MT, Poczai P, Croat TB, Ahmed I. Complete chloroplast genomes of Anthurium huixtlense and Pothos scandens (Pothoideae, Araceae): unique inverted repeat expansion and contraction affect rate of evolution. J Mol Evol. 2020;88(7):562–574. doi: 10.1007/s00239-020-09958-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdullah HCL, Croat TB, Poczai P, Ahmed I. Mutational dynamics of aroid chloroplast genomes II. Front Genet. 2020;11:610838. doi: 10.3389/fgene.2020.610838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdullah HCL, Mehmood F, Hayat A, Sammad A, Waseem S, Waheed MT, Matthews PJ, Croat TB, Poczai P, et al. Chloroplast genome evolution in the Dracunculus clade (Aroideae, Araceae) Genomics. 2021;113(1 Pt 1):183–192. doi: 10.1016/j.ygeno.2020.12.016. [DOI] [PubMed] [Google Scholar]
- 29.Cui N, Liao BS, Liang CL, Li SF, Zhang H, Xu J, Li XW, Chen SL. Complete chloroplast genome of Salvia plebeia: organization, specific barcode and phylogenetic analysis. Chin J Nat Med. 2020;18(8):563–572. doi: 10.1016/S1875-5364(20)30068-6. [DOI] [PubMed] [Google Scholar]
- 30.Cai Z, Wang H, Wang G (2020) Complete chloroplast genome sequence of Pinellia ternata (Thunb.) Breit, a medicinal plants to China. Mitochondrial DNA B Resour 5(3):2107–2108. 10.1080/23802359.2020.1765207 [DOI] [PMC free article] [PubMed]
- 31.Cui N, Liao BS, Liang CL, Li SF, Zhang H, Xu J, Li XW, Chen SL (2020) Complete chloroplast genome of Salvia plebeia: organization, specific barcode and phylogenetic analysis. Chin J Nat Medicines 18(8):563–572. 10.1016/S1875-5364(20)30068-6 [DOI] [PubMed]
- 32.Beier S, Thiel T, Munch T, Scholz U, Mascher M (2017) MISA-web: a web server for microsatellite prediction. Bioinformatics 33(16):2583–2585. 10.1093/bioinformatics/btx198 [DOI] [PMC free article] [PubMed]
- 33.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R (2001) REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res 29(22):4633–4642. 10.1093/nar/29.22.4633 [DOI] [PMC free article] [PubMed]
- 34.Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11):1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed]
- 35.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C et al (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed]
- 36.Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24(8):1586–1591. [DOI] [PubMed]
- 37.McCauley DE, Sundby AK, Bailey MF, Welch ME (2007) Inheritance of chloroplast DNA is not strictly maternal in Silene vulgaris (Caryophyllaceae): evidence from experimental crosses and natural populations. Am J Bot 94(8):1333–1337. 10.3732/ajb.94.8.1333 [DOI] [PubMed]
- 38.Wu L, Wu M, Cui N, Xiang L, Li Y, Li X, Chen S (2021) Plant super-barcode: a case study on genome-based identification for closely related species of Fritillaria. Chin Med 16(1):52. 10.1186/s13020-021-00460-z [DOI] [PMC free article] [PubMed]
- 39.Li X, Yang Y, Henry RJ, Rossetto M, Wang Y, Chen S (2015) Plant DNA barcoding: from gene to genome. Biol Rev Camb Philos Soc 90(1):157–166. 10.1111/brv.12104 [DOI] [PubMed]
- 40.Zhang Y, Du L, Liu A, Chen J, Wu L, Hu W, Zhang W, Kim K, Lee SC, Yang TJ et al (2016) The Complete Chloroplast Genome Sequences of Five Epimedium Species: Lights into Phylogenetic and Taxonomic Analyses. Front Plant Sci 7:306. 10.3389/fpls.2016.00306 [DOI] [PMC free article] [PubMed]
- 41.Huang Y, Li X, Yang Z, Yang C, Yang J, Ji Y (2016) Analysis of Complete Chloroplast Genome Sequences Improves Phylogenetic Resolution in Paris (Melanthiaceae). Front Plant Sci 7:1797. 10.3389/fpls.2016.01797 [DOI] [PMC free article] [PubMed]
- 42.Meng XX, Xian YF, Xiang L, Zhang D, Shi YH, Wu ML, Dong GQ, Ip SP, Lin ZX, Wu L et al (2018) Complete Chloroplast Genomes from Sanguisorba: Identity and Variation Among Four Species. Molecules 23(9):2137. 10.3390/molecules23092137 [DOI] [PMC free article] [PubMed]
- 43.Moon BC, Kim WJ, Ji Y, Lee YM, Kang YM, Choi G (2016) Molecular identification of the traditional herbal medicines, Arisaematis Rhizoma and Pinelliae Tuber, and common adulterants via universal DNA barcode sequences. Genet Mol Res 15(1). 10.4238/gmr.15017064 [DOI] [PubMed]
- 44.Raubeson LA, Peery R, Chumley TW, Dziubek C, Fourcade HM, Boore JL, Jansen RK (2007) Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics 8:174. 10.1186/1471-2164-8-174 [DOI] [PMC free article] [PubMed]
- 45.Daniell H, Lin CS, Yu M, Chang WJ (2016) Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol 17(1):134. 10.1186/s13059-016-1004-2 [DOI] [PMC free article] [PubMed]
- 46.Wang Y, Wang S, Liu Y, Yuan Q, Sun J, Guo L (2021) Chloroplast genome variation and phylogenetic relationships of Atractylodes species. BMC Genomics 22(1):103. 10.1186/s12864-021-07394-8 [DOI] [PMC free article] [PubMed]
- 47.Angioi SA, Desiderio F, Rau D, Bitocchi E, Attene G, Papa R (2009) Development and use of chloroplast microsatellites in Phaseolus spp. and other legumes. Plant Biol (Stuttg) 11(4):598–612. 10.1111/j.1438-8677.2008.00143.x [DOI] [PubMed]
- 48.Li L (1999) Micromorphological Characteristics and Enzyme Analysis of Some Chinese Pinellias and Their Taxonomic Significance. Acta Botanica Yunnanica 21(4):442–448
- 49.Xie DF, Yu Y, Deng YQ, Li J, Liu HY, Zhou SD, He XJ (2018) Comparative Analysis of the Chloroplast Genomes of the Chinese Endemic Genus Urophysa and Their Contribution to Chloroplast Phylogeny and Adaptive Evolution. Int J Mol Sci 19(7):1847. 10.3390/ijms19071847 [DOI] [PMC free article] [PubMed]
- 50.Gui L, Jiang S, Xie D, Yu L, Huang Y, Zhang Z, Liu Y (2020) Analysis of complete chloroplast genomes of Curcuma and the contribution to phylogeny and adaptive evolution. Gene 732:144355. 10.1016/j.gene.2020.144355 [DOI] [PubMed]
- 51.Liu ML, Fan WB, Wang N, Dong PB, Zhang TT, Yue M, Li ZH (2018) Evolutionary Analysis of Plastid Genomes of Seven Lonicera L. Species: Implications for Sequence Divergence and Phylogenetic Relationships. Int J Mol Sci 19(12):4039. 10.3390/ijms19124039 [DOI] [PMC free article] [PubMed]
- 52.Chen Y, Zhong H, Zhu Y, Huang Y, Wu S, Liu Z, Lan S, Zhai J (2020) Plastome structure and adaptive evolution of Calanthe s.l. species. PeerJ 8:e10051. 10.7717/peerj.10051 [DOI] [PMC free article] [PubMed]
- 53.Gao C, Deng Y, Wang J (2018) The Complete Chloroplast Genomes of Echinacanthus Species (Acanthaceae): Phylogenetic Relationships, Adaptive Evolution, and Screening of Molecular Markers. Front Plant Sci 9:1989. 10.3389/fpls.2018.01989 [DOI] [PMC free article] [PubMed]
- 54.Petersen G, Darby H, Lam VKY, Pedersen HAE, Merckx V, Zervas A, Seberg O, Graham SW (2019) Mycoheterotrophic Epirixanthes (Polygalaceae) has a typical angiosperm mitogenome but unorthodox plastid genomes. Ann Bot 124(5):791–807. 10.1093/aob/mcz114 [DOI] [PMC free article] [PubMed]
- 55.Carlhoff S, Duli A, Nagele K, Nur M, Skov L, Sumantri I, Oktaviana AA, Hakim B, Burhan B, Syahdar FA et al (2021) Genome of a middle Holocene hunter-gatherer from Wallacea. Nature 596(7873):543–547. 10.1038/s41586-021-03823-6 [DOI] [PMC free article] [PubMed]
- 56.Kinoshita G, Nunome M, Kryukov AP, Kartavtseva IV, Han SH, Yamada F, Suzuki H (2019) Contrasting phylogeographic histories between the continent and islands of East Asia: Massive mitochondrial introgression and long-term isolation of hares (Lagomorpha: Lepus). Mol Phylogenet Evol 136:65–75. 10.1016/j.ympev.2019.04.003 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The chloroplast genome sequence of Pinellia pedatisecta assembled here is accessible via GenBank with the accession number of MZ702636 and Global Pharmacopoeia Genome Database (GPGD,
http://www.gpgenome.com/species/40314). Raw sequencing data is available at NCBI SRA database with the accession number of SRR15328795.