Abstract
In this study, we succeeded in differentiating Lactobacillus plantarum, Lactobacillus pentosus, and Lactobacillus paraplantarum by means of recA gene sequence comparison. Short homologous regions of about 360 bp were amplified by PCR with degenerate consensus primers, sequenced, and analyzed, and 322 bp were considered for the inference of phylogenetic trees. Phylograms, obtained by parsimony, maximum likelihood, and analysis of data matrices with the neighbor-joining model, were coherent and clearly separated the three species. The validity of the recA gene and RecA protein as phylogenetic markers is discussed. Based on the same sequences, species-specific primers were designed, and a multiplex PCR protocol for the simultaneous distinction of these bacteria was optimized. The sizes of the amplicons were 318 bp for L. plantarum, 218 bp for L. pentosus, and 107 bp for L. paraplantarum. This strategy permitted the unambiguous identification of strains belonging to L. plantarum, L. pentosus, and L. paraplantarum in a single reaction, indicating its applicability to the speciation of isolates of the L. plantarum group.
The species Lactobacillus plantarum, Lactobacillus pentosus, and Lactobacillus paraplantarum are genotypically closely related and show highly similar phenotypes. The genetic heterogeneity of the L. plantarum group has been demonstrated by Dellaglio et al. (8) on the basis of DNA-DNA hybridization data: three groups were identified which were later classified as L. plantarum sensu stricto (2), L. pentosus (28), and L. paraplantarum (7).
Despite the importance of these species for fermented foods of plant, animal, and fish origin, their correct identification is complicated by the ambiguous response of traditional physiological tests and molecular methods: Fourier transform infrared spectroscopy of lactobacilli from breweries was not able to differentiate spectra from L. plantarum and L. pentosus (6). Randomly amplified polymorphic DNA-PCR and related numerical analysis gave satisfying results, but the methods were applied only to L. plantarum and L. pentosus (27). Finally, two papers reported the selection of species-specific PCR primers for the L. plantarum group; however, in one case specificity towards L. paraplantarum was not checked (21), and in the other the primers did not guarantee sufficient specificity (3). Satisfying results have been obtained by Southern-type hybridization with a pyrDFE probe (4), randomly amplified polymorphic DNA-PCR, and AFLP (Keygene NV, Wageningen, The Netherlands) (S. Torriani, F. Clementi, F. Dellaglio, B. Hoste, M. Vancanneyt, and J. J. Swings, Proc. Sixth Symp. Lactic Acid Bacteria, poster B2, 1999), but such methods are not suitable for routine identification requirements. The difficulty of correct identification of these species and the increasing interest in some of their properties, e.g., probiotic activities (13) and tannin degradation (19), indicates the need for a simple and reliable molecular method for the definite differentiation of L. plantarum, L. paraplantarum, and L. pentosus.
PCR using species-specific oligonucleotides designed based on phylogenetic molecular markers could be a useful approach, since these molecules are ubiquitous and relatively highly conserved. For this purpose, 16S ribosomal DNA sequences are not suitable because of the high identity value (>99%) shared by L. plantarum and L. pentosus (5, 21). Consequently, the definition of phylogenetic distances is also not feasible by such a classical approach for L. plantarum group species. It has been proposed that the recA gene could be used as a phylogenetic marker (11, 17), and it has already given satisfying results for many bacterial genera, including bifidobacteria (15).
RecA is a small protein (352 amino acids in Escherichia coli) implicated in homologous DNA recombination, SOS induction, and DNA damage-induced mutagenesis (11). This panoply of functions implies multiple biochemical activities, including DNA binding (double and single stranded), pairing and exchange of homologous DNA, ATP hydrolysis, and coproteolitic cleavage of the LexA, λcI, and UmuD proteins (11). Due to its fundamental role, the recA gene is ubiquitous, and its gene product has been proposed as a phylogenetic marker for distantly related species (11, 17).
In this study, short recA gene sequences were obtained and analyzed in order to infer a phylogenetic classification scheme, still lacking for the three species of the L. plantarum group. Moreover, species-specific primers were designed based on the obtained sequences and were used in a multiplex PCR assay.
MATERIALS AND METHODS
Bacterial strains and cultivation.
The bacterial strains tested for inferring phylogeny were L. paraplantarum LMG 16673T, L. paraplantarum LMG 18402, L. paraplantarum NIRD P2, L. pentosus LMG 10755T, L. pentosus LMG 18401, L. plantarum ATCC 14917T, L. plantarum B41, and L. plantarum NCFB 340.
The strains used as positive or negative controls in the multiplex PCR assay were L. plantarum strain A, L. pentosus strain 3 and strain 6 from our collection, Acetobacter aceti LMG 1261T, Bifidobacterium breve LMG 11042T, Enterococcus faecium LMG 8147T, Lactobacillus casei ATCC 393T, L. casei ATCC 334, Lactobacillus paracasei subsp. paracasei NCFB 151T, Lactobacillus rhamnosus LMG 6400T, Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842T, Lactobacillus helveticus ATCC 15009T, Lactococcus lactis subsp. lactis LMG 6890T, and Streptococcus thermophilus strain S203 (our collection).
E. coli strain XL1-Blue, used for cloning procedures, was grown in Luria-Bertani broth (0.1% tryptone, 0.1% sodium chloride, 0.05% yeast extract, pH 7.0) for 16 h at 37°C. Transformed E. coli XL1-Blue was grown on Luria-Bertani agar plates plus 100 μg of ampicillin/ml, 0.5 mM isopropyl-β-d-thiogalactopyranoside, and 80 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside/ml.
Lactobacilli and E. faecium were grown in MRS broth (Oxoid) for 16 h at 37°C, B. breve was grown in MRS broth with 0.05% cysteine added for 48 h at 37°C in anaerobiosis obtained with Anaerocult A (Merck, Darmstadt, Germany), A. aceti was grown in YPM broth (0.5% yeast extract, 0.3% peptone, 2.5% mannitol; pH not adjusted) for 3 days at 30°C, and L. lactis and S. thermophilus were grown in M17 broth (Oxoid) for 24 h at 37°C.
DNA preparation.
Genomic DNA of lactic acid bacteria was extracted as described by Marmur (18). In genomic DNA extraction from A. aceti the method was modified by the addition of 2× hexadecyltrimethylammonium bromide solution (2% hexadecyltrimethylammonium bromide, 100 mM Tris [pH 8], 20 mM EDTA [pH 8], 1.4 mM NaCl, and 1% polyvinylpyrrolidone), with incubation at 65°C for 10 min.
recA gene fragment amplifications.
For the amplification of recA gene regions, 300 ng of chromosomal DNA from each Lactobacillus strain was added to a 50-μl PCR mixture (2.5 U of Sigma-Aldrich REDTaq DNA polymerase in 1× REDTaq reaction buffer, 50 μM each primer, 100 μM deoxynucleotide triphosphates) and submitted to initial denaturation of 1 min at 94°C followed by 30 cycles of 1 min at 94°C, 1 min and 30 s at 55°C, and 1 min at 72°C in a GeneAmp PCR System 2400 (Perkin-Elmer). The primers used for the amplification were 5′-TT(C,T) AT(A,T,C) GA(C,T) GC(A,T,C,G) GA(A,G) CA(C,T) GC-3′ (forward primer, corresponding to amino acid region 92 to 98 of the E. coli RecA protein [10]) and 5′-CC(A,T) CC(A,T) G(T,G)(A,T) GT(A,T,C) GT(C,T) TC(A,T,C,G) GG-3′(reverse primer, corresponding to amino acid region 206 to 211 of the E. coli RecA protein [9]). The expected size of the amplicons was about 360 bp.
Amplification products were purified from 2% agarose gel by the QIAEX II Gel Extraction System (Qiagen GmbH, Hilden, Germany).
Cloning of PCR products.
Three microliters of a suspension containing 30 ng of purified amplicons of recA gene fragments (360 bp) was added to a ligation mixture containing 50 ng of pGEM-T vector (Promega Corp., Madison, Wis.) and 3 U of T4 DNA ligase (Promega) in 1× ligation buffer and incubated overnight at 4°C. Two microliters of the ligation mixture was used to transform E. coli XL1-Blue by electroporation in an E. coli Pulser apparatus (Bio-Rad Laboratories, Richmond, Calif.). Recombinant clones carrying the insert were selected by blue-white screening. Plasmids containing DNA fragments were extracted with the Quiaprep Spin Miniprep kit (Qiagen). The presence of the insert was verified by restriction analysis with enzyme BglII (Takara Shuzo Co., Ltd., Otsu, Shiga, Japan). Restriction was carried out for 1 h at 37°C in 1× reaction buffer with 10 U of enzyme in a final volume of 20 μl.
Sequencing.
Sequencing reactions were performed at Centro Genoma Vegetale-ENEA CR Casaccia, Rome, Italy, on purified plasmids with primers T7 and SP6 (both strands for each sequence).
Analysis of sequence data and construction of phylogenetic trees.
The identities of sequences obtained were verified by a BLASTX (1) search against major molecular databases. Putative amino acid sequences were deduced by the GeneDoc program version 2.5.000 (K. B. Nicholas and H. B. Nicholas, unpublished data).
The nucleotide and putative amino acid sequences of the recA gene fragments were aligned with the CLUSTAL W program (26). Phylogenetic trees were constructed using programs in the PHYLIP software package version 3.5c. Calculation of distance matrices was carried out by DNADIST and PROTDIST programs on nucleotide and putative amino acid sequences, respectively, using the default models. Trees were inferred using the NEIGHBOR program, which implements the neighbor-joining method of Saitou and Nei (22). Phylogenetic trees from gene sequences were also constructed with the DNAML (maximum likelihood) and DNAPARS (parsimony) programs. The “jumble” option in program menus was always selected to test different sequence orders in inferring phylogenies to minimize the influence of sequence order on the tree topology (16). A bootstrap confidence analysis was performed by generating 1,000 replicated data sets with the SEQBOOT program, constructing trees with DNADIST and NEIGHBOR, and generating the consensus tree with the CONSENSE program (all in the PHYLIP software package version 3.5c).
Reference sequences used in phylogenetic analysis.
The following bacterial recA gene sequences were tested as outgroups in phylogenetic analysis: accession number M81465 (Acholeplasma laidlawii), AJ006705 (Acidothermus cellulolyticus), M29680 (Anabaena variabilis), AF214780 (Arthrobacter globiformis), X52132 (Bacillus subtilis), AF094756 (B.breve), U61497 (Clostridium perfringens), AJ006707 (Frankia alni), AJ006706 (Geodermatophilus obscurus), AJ286124 (L. paracasei subsp. paracasei NCFB 151T), AJ286116 (Lactobacillus zeae LMG 17315T), M88106 (L. lactis), L22073 (Mycoplasma mycoides), U30293 (Ruminococcus albus), U33924 (Spirulina platensis), L25893 (Staphylococcus aureus), U21934 (Streptococcus pyogenes), and M29495 (Synechococcus sp.).
Multiplex PCR assay.
A multiplex PCR assay (20 μl) was performed with the recA gene-based primers paraF (5′-GTC ACA GGC ATT ACG AAA AC-3′), pentF (5′-CAG TGG CGC GGT TGA TAT C-3′), planF (5′-CCG TTT ATG CGG AAC ACC TA-3′), and pREV (5′-TCG GGA TTA CCA AAC ATC AC-3′). The PCR mixture was composed of 1.5 mM MgCl2, the primers paraF, pentF, and pREV (0.25 μM each), 0.12 μM primer planF, 12 μM deoxynucleotide triphosphates (3 μM each), 0.025 U of Taq (Polymed, Florence, Italy)/μl, 1× PCR buffer (Polymed), and 5 ng of DNA/μl. PCRs were performed on a Perkin-Elmer GeneAmp PCR System 2400 with initial denaturation at 94°C for 3 min, 30 cycles of denaturation at 94°C (30 s), annealing at 56°C (10 s), and elongation at 72°C (30 s), and final extension at 72°C for 5 min. The PCR products were visualized on a 2% agarose gel in 1× TAE (40 mM Tris-acetate, 1 mM EDTA, pH 8.2) buffer. Purified amplicons were sequenced using the designed multiplex species-specific primers to confirm their identities.
Nucleotide sequence accession numbers.
The EMBL-GenBank-DDBJ accession numbers assigned to the recA internal region for the lactic acid bacterial strains investigated in this study are AJ286120 (L. paraplantarum LMG 16673T), AJ271963 (L. paraplantarum LMG 18402), AJ271964 (L. paraplantarum NIRD P2), AJ286118 (L. pentosus LMG 10755T), AJ292254 (L. pentosus LMG 18401), AJ286119 (L. plantarum ATCC 14917T), AJ271962 (L. plantarum B41), and AJ292255 (L. plantarum NCFB 340).
RESULTS
Identification and alignment of recA sequences.
Sequence identities were verified by a BLASTX (1) search against available molecular databases. Moreover, the presence of Asp-100, Tyr-103, Asp-144, and Ser-145 (E. coli numbering) in the putative translation fragments further confirms their identity, since those residues are considered functionally important (25).
In the phylogenetic analysis, sequence fragments of 322 bp were considered, excluding regions of primer annealing. In these regions, point mutations, likely due to primer degeneration, were indeed observed.
Sequence alignment did not introduce any gaps, and there were no ambiguous positions that could bias the inference of phylogeny. This was not surprising, since the sequences were derived from closely related species.
Phylogenetic analysis.
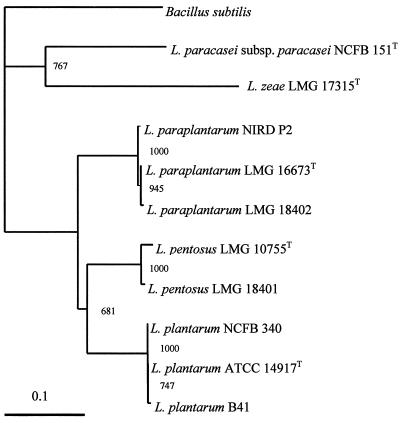
Different factors known to influence the phylogenetic analysis, i.e., the choice of outgroup sequences (24) and bioinformatic methods (14), were considered. recA gene sequences of several reference organisms, which are available in major databases, were selected as outgroups to evaluate the resulting classification scheme. Congruent results were obtained with all reference sequences tested (data not shown). An example of a phylogenetic tree obtained with B. subtilis as the outgroup and L. paracasei subsp. paracasei NCFB 151T and L. zeae LMG 17315T as reference sequences is shown in Fig. 1.
FIG. 1.
Phylogenetic tree showing the relative positions of L. plantarum, L. paraplantarum, and L. pentosus as inferred by the neighbor-joining method with the recA gene from B. subtilis as the outgroup sequence. Bootstrap values for a total of 1,000 replicates are given. Trees generated by the maximum likelihood and parsimony methods have the same topology. The bar indicates 10% sequence divergence. In this tree topology, the phylogenetic distance between organisms is the sum of the horizontal segments.
Three different methods (parsimony, maximum likelihood, and distance matrix data with neighbor-joining) were applied for the inference of the tree. The phylogenetic trees obtained had the same topology (Fig. 1), proving the accuracy of the classification scheme. The three species under study were unambiguously differentiated by the comparative sequence analysis of the short fragment of the recA gene, as indicated by the bootstrap values in Fig. 1.
Since analysis of RecA protein sequences has been proposed for reconstructing bacterial phylogenetic trees (11), a comparison between the resolution powers of gene sequences and those of the corresponding putative amino acid sequences was carried out. The putative amino acid sequences share very high identity, so the lengths of branches in the amino acid tree are practically null (data not shown). Percentage identity values for the nucleotide (upper right) and amino acid (lower left) sequences are given in Table 1.
TABLE 1.
Identity values calculated for 322-bp nucleotide sequences of recA gene fragments and putative 107-residue amino acid sequences
| Strain | Identity (%)a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| L. paraplantarum LMG 16673T | L. paraplantarum LMG 18402 | L. paraplantarum NIRD P2 | L. pentosus LMG 10755T | L. pentosus LMG 18401 | L. plantarum ATCC 14917T | L. plantarum B41 | L. plantarum NCFB 340 | |
| L. paraplantarum LMG 16673T | 99.7 | 99.4 | 83.9 | 84.8 | 85.4 | 85.1 | 85.1 | |
| L. paraplantarum LMG 18402 | 99.1 | 99.1 | 83.5 | 84.5 | 85.1 | 84.8 | 84.8 | |
| L. paraplantarum NIRD P2 | 100 | 99.1 | 84.2 | 85.1 | 85.4 | 85.1 | 85.1 | |
| L. pentosus LMG 10755T | 97.2 | 98.1 | 99.1 | 100 | 86 | 85.7 | 85.7 | |
| L. pentosus LMG 18401 | 97.2 | 98.1 | 99.1 | 100 | 86.7 | 86.3 | 86.3 | |
| L. plantarum ATCC 14917T | 98.1 | 98.1 | 99.1 | 100 | 100 | 99.7 | 99.7 | |
| L. plantarum B41 | 97.2 | 97.2 | 98.1 | 99.1 | 99.1 | 99.1 | 99.7 | |
| L. plantarum NCFB 340 | 98.1 | 98.1 | 99.1 | 100 | 100 | 100 | 99.1 | |
Identity values are shown for recA gene fragments (bold face) and amino acid sequences.
Primer design and multiplex PCR assay.
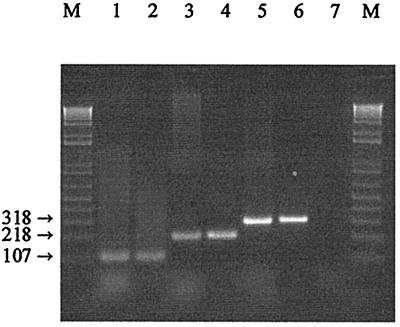
The 322-bp recA fragment sequences were aligned, and four regions were selected for the design of species-specific primers. A single reverse primer (pREV) and three forward primers (paraF, pentF, and planF) were used in the PCR tests. The designed pREV primer partially overlapped (5 bases; replacing an N with a G) the degenerate primer used by Duwat et al. (9). Numbering the fragment sequences from 1 to 322, the forward specific primers were located as follows: planF, the specific primer for L. plantarum, nucleotide positions 9 to 28; pentF, the specific primer for L. pentosus, nucleotide positions 109 to 127; and paraF, the specific primer for L. paraplantarum, nucleotide positions 220 to 239. Consequently, the expected sizes of the amplicons were 318 bp for L. plantarum, 218 bp for L. pentosus, and 107 bp for L. paraplantarum. To avoid the presence of aspecific bands, the multiplex PCR protocol was optimized in relation to the temperature and time of annealing and the relative concentrations of the primers. The amplification of DNA from all of the L. plantarum, L. pentosus, and L. paraplantarum strains produced only the expected product, while none of the negative controls produced any amplicon. The identities of the three fragments were confirmed by sequencing. An example of the results is shown in Fig. 2.
FIG. 2.
Amplification products obtained from the recA multiplex assay. Lane M contained a 1-kb PLUS DNA ladder (Life Technology Inc., Gaithersburg, Md.). Lanes 1 and 2, PCR amplification products from L. paraplantarum LMG 16673T and NIRD P2, respectively; lanes 3 and 4, PCR amplification products from L. pentosus LMG 10755T and LMG 18401, respectively; lanes 5 and 6, PCR amplification products from L. plantarum ATCC 14917T and NCFB 340, respectively; lane 7, L. casei ATCC 334 (negative control).
DISCUSSION
Phylogenetic analysis of partial recA gene sequences permitted a clear distinction among L. plantarum, L. pentosus, and L. paraplantarum. This assignment relies only on sequence properties, since results produced by several outgroups and different methods prove the robustness of recA phylogenetic analysis. The high bootstrap values obtained are much more significant considering that the recA sequences of two facultatively heterofermentative lactobacilli belonging to the same 16S rRNA phylogenetic group were included in the analysis.
From the comparative analysis of gene sequences and those of the corresponding amino acids, some particularities emerged concerning nucleotide substitutions and identity values. Nearly all nucleotide mutations are synonymous substitutions; consequently, identity values for gene sequences are variable and intraspecific groups are clearly distinguishable, while for amino acid sequences there is little variability. A maximum of three nonsynonymous nucleotide substitutions occurred, which produced the following amino acid replacements: isoleucine (L. paraplantarum strains) instead of valine at amino acid position 4, isoleucine (L. paraplantarum LMG 18402) instead of threonine at position 23, and alanine (L. plantarum B41) instead of valine at position 66. The sole nonconservative substitution was the introduction of isoleucine with a nonpolar side chain instead of threonine with a polar one. The remaining substitutions involved exchanges between nonpolar amino acids.
From a taxonomic point of view, our results confirm that short recA gene sequences have high discriminating power for species difficult to differentiate, as already demonstrated for Bifidobacterium infantis and Bifidobacterium longum (15). At present, the most reliable method for the definition of a species is DNA-DNA hybridization, and it has also been stated that there is a correspondence between these data and the degree of identity of 16S rRNA sequences (23). It is generally accepted that species having 70% or greater DNA similarity usually have more than 97% 16S rRNA sequence identity (23). On the other hand, the identity of 16S rRNA sequences may not be sufficient to guarantee that two taxa belong to the same species (12). This suggests that 16S ribosomal DNA sequence analysis is not the appropriate method to replace DNA reassociation to delineate species and measure intraspecies relationships (23). In the present study, the species rank was indicated by a range of recA identity values of 83.5 to 86.7%, which corresponds to DNA hybridization homologies of 15 to 56% (7). In light of our results and those of previous studies (e.g., reference 15), the recA gene can be proposed as a new method for inferring relationships among very closely related species. recA gene analysis can have a double potential as a phylogenetic marker: its amino acid sequence can be used to infer phylogenies between distantly related taxa, thus avoiding the disadvantages of rRNAs, e.g., overestimation of the relatedness of species with similar nucleotide frequencies, nonindependence of substitution patterns at different sites, and bias derived from different GC contents of the organisms (11). Furthermore, the nucleotide sequence can be used to evaluate phylogenetic relationships and relative taxonomic positions of recently diverged species with similar GC contents and nucleotide frequencies. In fact, at the nucleotide level, recA can tolerate mutations which do not or only slightly alter its product. These mutations give us information about recent evolutionary history, which is too recent to be fixed in slowly diverging sequences like 16S rRNAs (20).
By exploiting the variable nucleotide regions of recA gene fragments, we have also succeeded in designing a set of species-specific PCR primers. These can be used separately as primer pairs in different species-specific PCRs but also in a multiplex assay. The optimization of this multiplex PCR makes it possible to assign one isolate to a definite species of the L. plantarum group in a single step rather than with three distinct reactions. This assay is rapid (less than 2 h), reliable for its high-specificity reaction conditions, and complete, since all three species of the group are investigated.
The clear distinction obtained with short gene sequences validates the possibility of using the recA gene as a phylogenetic-taxonomic marker for closely related species and opens new possibilities for a rapid and reliable identification of lactic acid bacteria of importance for food.
ACKNOWLEDGMENTS
We are grateful to Luca Mizzi, Dipartimento di Genetica e di Biologia dei Microrganismi, Università di Milano, Milan, Italy, for his skillful assistance in phylogenetic analysis and to Franca Rossi for valuable revision of the manuscript.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergey D H, Harrison F C, Breed R S, Hammer B W, Huntoon F M. Lactobacillus plantarum, In: Buchanan R E, Gibbons N E, editors. Bergey's manual of determinative bacteriology. 1st ed. Baltimore, Md: The Williams & Wilkins Co.; 1923. p. 250. [Google Scholar]
- 3.Berthier F, Ehrlich S D. Rapid species identification within two groups of closely related lactobacilli using PCR primers that target the 16S/23S rRNA spacer region. FEMS Microbiol Lett. 1998;161:97–106. doi: 10.1111/j.1574-6968.1998.tb12934.x. [DOI] [PubMed] [Google Scholar]
- 4.Bringel F, Curk M-C, Hubert J-C. Characterization of lactobacilli by Southern-type hybridization with a Lactobacillus plantarum pyrDFE probe. Int J Syst Bacteriol. 1996;46:588–594. doi: 10.1099/00207713-46-2-588. [DOI] [PubMed] [Google Scholar]
- 5.Collins M D, Rodrigues U M, Ash C, Aguirre M, Farrow J A E, Martinez-Murcia A, Phillips B A, Williams A M, Wallbanks S. Phylogenetic analysis of the genus Lactobacillus and related lactic acid bacteria as determined by reverse transcriptase sequencing of 16S rRNA. FEMS Microbiol Lett. 1991;77:5–12. [Google Scholar]
- 6.Curk M-C, Peladan F, Hubert J-C. Fourier transform infrared (FTIR) spectroscopy for identifying Lactobacillus species. FEMS Microbiol Lett. 1994;123:241–248. [Google Scholar]
- 7.Curk M-C, Hubert J-C, Bringel F. Lactobacillus paraplantarum sp. nov., a new species related to Lactobacillus plantarum. Int J Syst Bacteriol. 1996;46:595–598. doi: 10.1099/00207713-46-2-595. [DOI] [PubMed] [Google Scholar]
- 8.Dellaglio F, Bottazzi V, Vescovo M. Deoxyribonucleic acid homology among Lactobacillus species of the subgenus Streptobacterium Orla-Jensen. Int J Syst Bacteriol. 1975;25:160–172. [Google Scholar]
- 9.Duwat P, Ehrlich S D, Gruss A. Use of degenerate primers for polymerase chain reaction cloning and sequencing of the Lactococcus lactis subsp. lactis recA gene. Appl Environ Microbiol. 1992;58:2674–2678. doi: 10.1128/aem.58.8.2674-2678.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dybvig K, Hollingshead S K, Heath D G, Clewell D B, Sun F, Woodard A. Degenerate oligonucleotide primers for enzymatic amplification of recA sequences from gram-positive bacteria and mycoplasmas. J Bacteriol. 1992;174:2729–2732. doi: 10.1128/jb.174.8.2729-2732.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisen J A. The RecA protein as a model molecule for the molecular systematic studies of bacteria: comparison of trees of RecAs and 16S RNA from the same species. J Mol Evol. 1995;41:1105–1123. doi: 10.1007/BF00173192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox G E, Wisotzkey J D, Jurtshuk P. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 13.Herias M V, Hessle C, Telemo E, Midtvedt T, Hanson L A, Wold A E. Immunomodulatory effects of Lactobacillus plantarum colonizing the intestine of gnotobiotic rats. Clin Exp Immunol. 1999;116:283–290. doi: 10.1046/j.1365-2249.1999.00891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J. Improving the accuracy of phylogenetic estimation by combining different methods. Syst Biol. 1993;42:331–340. [Google Scholar]
- 15.Kullen M J, Brady L J, O'Sullivan D J. Evaluation of using a short region of the recA gene for the rapid and sensitive speciation of dominant bifidobacteria in the human large intestine. FEMS Microbiol Lett. 1997;154:377–383. doi: 10.1111/j.1574-6968.1997.tb12670.x. [DOI] [PubMed] [Google Scholar]
- 16.Lake J A. The order of sequence alignment can bias the selection of tree topology. Mol Biol Evol. 1991;8:378–385. doi: 10.1093/oxfordjournals.molbev.a040654. [DOI] [PubMed] [Google Scholar]
- 17.Lloyd A T, Sharp P M. Evolution of the recA gene and the molecular phylogeny of bacteria. J Mol Evol. 1993;37:399–407. doi: 10.1007/BF00178869. [DOI] [PubMed] [Google Scholar]
- 18.Marmur J. A procedure for the isolation of DNA from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 19.Osawa R, Kuroiso K, Goto S, Shimizu A. Isolation of tannin-degrading lactobacilli from humans and fermented foods. Appl Environ Microbiol. 2000;66:3093–3097. doi: 10.1128/aem.66.7.3093-3097.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palys T, Nakamura L K, Cohan F M. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int J Syst Bacteriol. 1997;47:1145–1156. doi: 10.1099/00207713-47-4-1145. [DOI] [PubMed] [Google Scholar]
- 21.Quere F, Deschamps A, Urdaci M C. DNA probe and PCR-specific reaction for Lactobacillus plantarum. J Appl Microbiol. 1997;82:783–790. doi: 10.1046/j.1365-2672.1997.00157.x. [DOI] [PubMed] [Google Scholar]
- 22.Saitou N, Nei M. Reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 24.Stackebrandt E, Ludwig W. The importance of using outgroup reference organisms in phylogenetic studies: the Atopobium case. Syst Appl Microbiol. 1994;17:39–43. [Google Scholar]
- 25.Story R M, Steitz T A. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 26.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Reenen C A, Dicks L M T. Evaluation of numerical analysis of Random Amplified Polymorphic DNA (RAPD)-PCR as a method to differentiate Lactobacillus plantarum and Lactobacillus pentosus. Curr Microbiol. 1996;32:183–187. doi: 10.1007/s002849900033. [DOI] [PubMed] [Google Scholar]
- 28.Zanoni P, Farrow J A E, Phillips B A, Collins M D. Lactobacillus pentosus (Fred, Peterson and Anderson) sp. nov., nom. rev. Int J Syst Bacteriol. 1987;37:339–341. [Google Scholar]