Abstract
Objective
This study aimed to identify genetic variants enriched in Southwest American Indian (SWAI) individuals that associate with BMI.
Methods
Whole genome sequencing data (n = 296) were used to identify potentially functional variants that are common in SWAI individuals (minor allele frequency ≥10%) but rare in other ethnic groups (minor allele frequency < 0.1%). Enriched variants were tested for association with BMI in 5,870 SWAI individuals. One variant was studied using a luciferase reporter, and haplotypes that included this variant were analyzed for association with various measures of obesity (n = 917‐5,870), 24‐hour energy expenditure (24‐h EE; n = 419), and skeletal muscle biopsy expression data (n = 207).
Results
A 5′ untranslated region variant in cytochrome b5 type A (CYB5A), rs548402150, met the enrichment criteria and associated with increased BMI (β = 2%, p = 0.004). Functionally, rs548402150 decreased luciferase expression by 30% (p = 0.003) and correlated with decreased skeletal muscle CYB5A expression (β = −0.5 SD, p = 0.0008). Combining rs548402150 with two splicing quantitative trait loci in CYB5A identified a haplotype carried almost exclusively in SWAI individuals that associated with increased BMI (β = 3%, p = 0.0003) and decreased CYB5A expression, whereas the most common haplotype in all ethnic groups associated with lower BMI and percentage of body fatness, increased 24‐h EE, and increased CYB5A expression.
Conclusions
Further studies on the effects of CYB5A on 24‐h EE and BMI may provide insights into obesity‐related physiology.
Study Importance.
What is already known?
-
►
Southwest American Indian (SWAI) individuals are disproportionally affected by obesity compared with other ethnic groups.
-
►
Obesity has a heritable component.
What does this study add?
-
►
This study provides a set of potentially functional variants that are enriched in SWAI individuals compared with other ethnic groups.
-
►
Haplotype analysis of an enriched expression quantitative trait locus rs548402150 in cytochrome b5 type A (CYB5A), and two splicing quantitative trait loci, rs7238987 and rs7238784, in the same locus, identified an obesity‐risk haplotype (ATA) that was enriched in SWAI individuals and an obesity‐protective haplotype (CCG) that was prevalent in all other ethnic groups.
How might these results change the direction of research?
-
►
The influence of genetic variants on obesity can vary based on ethnic backgrounds, as we have identified the CYB5A variant rs548402150 alone and in haplotype with rs7238987 and rs7238784 to be enriched in SWAI individuals compared with other ethnic groups.
-
►
Our study demonstrates associations between CYB5A haplotypes and expression levels, as well as associations between CYB5A haplotypes and energy expenditure and BMI; however, future studies are needed to demonstrate causality.
INTRODUCTION
Southwest American Indian (SWAI) individuals are at a higher risk for obesity than most other ethnic groups; however, most of the large‐scale studies to identify genetic variants that contribute to polygenic obesity have been conducted predominantly in European, Asian, and African populations (1, 2, 3). We previously assessed established BMI genes and/or variants (identified in other ethnic groups) for an association with BMI in our sample of SWAI individuals, and several variants replicated (4, 5, 6, 7, 8, 9). Melanocortin 4 receptor (MC4R) appeared to have the largest impact, as, on a population level, we found that 2.4% of SWAI individuals carried a missense variation shown to increase BMI (8). However, MC4R alone does not explain the high prevalence of severe obesity in SWAI individuals.
Similar to SWAI individuals, isolated populations in the Pacific Islands have been found to have a high prevalence of obesity (10, 11, 12). A genome‐wide study in Samoan individuals identified a variant in the CREB3 regulatory factor (CREBRF) gene that had a very large effect on risk for obesity (11). Interestingly, this variant was common in Samoan individuals but extremely rare in non‐Pacific Island populations. The association with obesity was later replicated in other Polynesian populations such as Guam and Saipan (12).
To determine whether SWAI individuals similarly have a founder effect resulting in enrichment of a variant that contributes to their high prevalence of obesity, the current study used the following strategy: 1) identify all variation in whole‐genome sequencing (WGS) data from 296 full‐heritage Pima Indian individuals and select all variants likely to be functional based on their Combined Annotation Dependent Depletion (CADD) score; 2) among the potentially functional variants, identify those that are unique or highly enriched in these 296 Pima Indian individuals compared with other ethnic groups (1000 Genome Project and Genome Aggregation Database [gnomAD]); and 3) determine whether any of these highly enriched variants may have a role in obesity by genotyping them in a larger sample of SWAI individuals (n = 5870) that are informative for BMI.
METHODS
Identification of enriched functional variants
WGS of DNA was performed by Illumina (Illumina, Inc., San Diego, California) with 30× average coverage for 296 SWAI participants who were full‐heritage Pima Indian (defined as all of their parents and grandparents being full Pima Indian). Among this group of 296 full‐heritage Pima Indian individuals, none were first‐degree relatives. Sequencing reads were analyzed by using human genome reference sequence build 37 for alignment, and the resulting variant calls were annotated for predicted pathogenicity by CADD scores using CADD version 1.0 to assess all nonsynonymous substitutions (13). Functional variants with a Phred‐scale CADD score >10 (top 10% of the most deleterious single‐nucleotide polymorphisms [SNPs]) were analyzed for enrichment, in which an enriched allele was defined by being common (frequency ≥10%) among the 296 full‐heritage Pima Indian individuals but exceedingly rare to absent (minor allele frequency [MAF] < 0.1%) in global populations represented in the 1000 Genomes Project Phase 3 (14) and gnomAD version 2 (15) databases.
Genotyping and association analysis
Twelve potentially functional variants that were enriched in WGS data from 296 full‐heritage Pima Indian individuals were genotyped in a population‐based sample of 5,870 SWAI individuals, informative for longitudinal measures of BMI. Genotyping was done using TaqMan SNP Genotyping Assays (Thermo Fisher Scientific, Waltham, Massachusetts). However, for several SNPs, the genotypes on these 5,870 individuals already existed because the SNP had, by chance, been included on a previously genotyped custom Affymetrix Axiom array (Affymetrix, Santa Clara, California).
Clinical measurements
The SWAI volunteers are from a community in Arizona where most residents are of Pima Indian heritage. Volunteers participated in a longitudinal study from 1965 to 2007 with biennial measurements of height and weight to calculate BMI and a 75‐g oral glucose tolerance test for determining type 2 diabetes (T2D) status by 1997 American Diabetes Association (ADA) criteria (16). Because T2D is very common in this population, and T2D progression and its treatment can affect BMI, the measure of BMI analyzed in this study was the highest BMI recorded at any examination at age ≥15 years when the volunteer was determined to be nondiabetic (n = 5,870; mean BMI = 35.2 [8.4] kg/m2, 43.8% male, mean age = 29.6 [11.4] years) (17). A subset of nondiabetic participants also had measures of percentage of body fatness (%fat, n = 917) and 24‐h energy expenditure (EE) (n = 419). Percentages of body fat mass and fat‐free mass were obtained by the underwater weighing method until August 1993 and thereafter by total body dual‐energy x‐ray absorptiometry (DPX‐1; Lunar Radiation Corp., Madison, Wisconsin), with values between the two methods made comparable by using a unit‐specific conversion equation (18). EE was continuously measured inside a whole‐room indirect calorimeter (respiratory chamber) during energy balance and weight stability, as previously described in detail (17, 19, 20). Briefly, the volunteers entered the chamber around 0800 and remained in the chamber for 23 hours and 15 minutes. Three meals were provided at 1100, 1600, and 1900. The rate of EE was measured continuously, calculated for each 15‐minute interval, and then extrapolated to the 24‐hour interval (24‐h EE). Written informed consent was provided by all study participants. Protocols for the study were approved by the review board of the NIDDK.
Statistical analyses
Statistical analysis for the variants was performed using SAS version 9.4 (SAS Institute Inc., Cary, North Carolina) and SOLAR (Sequential Oligogenic Linkage Analysis Routines, University of Maryland, Baltimore, Maryland). Associations with clinical measures were analyzed by a linear mixed model fitted with a variance components covariance structure to account for genetic relatedness among individuals. To avoid bias, the genetic relatedness matrix has been previously estimated as the proportion of the genome shared identically by descent (IBD) between each pair of 5,870 individuals included in this study (21). Genomic segments shared IBD were identified with the fastIBD function of Beagle package (22) using 482,616 autosomal markers with MAF > 0.05. For the association analysis, BMI data were log‐transformed, and the p values were adjusted for age, sex, birth year, and the first five genetic principal components. The SWAI‐enriched variants analyzed for association with BMI were considered significant after Bonferroni correction (p < 0.05/N functional enriched variants). P values for 24‐h EE were adjusted for age, sex, fat mass, fat‐free mass, and spontaneous physical activity level.
Haplotype analysis
Haplotype reconstruction was performed from all genotypes in the 1000 Genomes Project Phase 3 and 5,870 SWAI participants using PHASE version 2.1.1 software (23). PHASE implements a Bayesian method that allows use of these a priori expectations to inform haplotype reconstruction. This method reduces error rate in haplotype estimation for population genotypic data. PHASE also estimates the uncertainty associated with each phase call and avoids inappropriate overconfidence in statistically reconstructed haplotypes. The probability that each individual carried one or two copies of each haplotype was calculated and then analyzed in regression models in a fashion analogous to individual variants.
Expression quantitative trait loci
Genome‐wide mRNA levels were previously measured using the Affymetrix 1.0 Human Exon microarray in skeletal muscle (n = 207) biopsies from the SWAI population (24). Linear regression analysis was used to evaluate the association between genotypes (considering an additive model) and batch/sex normalized gene expression levels after adjustment for age and genetic admixture. β coefficients in batch‐ and sex‐standardized values (expressed as SD) were used to present the differences in gene expression relative to the risk allele. Linear regression analyses were also conducted for body fat and standardized gene expression levels as an independent variable and adjusting for covariates (age, sex, and genetic admixture).
Functional analysis of the CYB5A rs548402150 5′ untranslated region variant
Immortalized human skeletal muscle cells (Applied Biological Materials, Richmond, British Columbia, Canada) were maintained in Prigrow III medium (Applied Biological Materials) containing 4.5 g/L of glucose, 1.5 g/L of sodium bicarbonate, 10% fetal bovine serum (FBS), and 1% penicillin‐streptomycin. To construct the CYB5A 5′ untranslated region (UTR)‐promoter vectors, minus‐strand DNA fragments containing the rs548402150 non‐risk (C) or obesity‐risk (T) allele were polymerase chain reaction (PCR) amplified using the following primers: forward (5′ TGTAGGTACCGGAGCTCACATTCATCAGACACC‐3′) and reverse (5′ ACGTAAGCTTCAGCTCCACCCGGGACATTC‐3′). Amplicons were inserted at Acc65I and HindIII sites (underlined in primers) in the pGL4.10 basic vector, which contains the firefly luciferase reporter gene (Promega, Madison, Wisconsin). All constructs were sequenced to confirm alleles. One microgram of either the pGL4.10 basic vector or CYB5A 5′ UTR‐promoter vectors carrying the non‐risk and obesity‐risk alleles were cotransfected with 50 ng of pGL4.74(hRluc/TK) containing the Renilla luciferase reporter gene (Promega) into human skeletal muscle cells with Lipofectamine 3000 per manufacturer’s instructions (Invitrogen, Life Technologies Corp., Carlsbad, California). The cells were harvested 72 hours after transfection, and luciferase activities were measured on a TD‐20/20 luminometer (Turner BioSystems, Inc., Sunnyvale, California) using the dual‐luciferase assay kit (Promega) per manufacturer’s instructions. Relative luciferase activities were calculated as a ratio of the firefly to Renilla luciferase activity. The ratios were then normalized to the non‐risk promoter luciferase activity. Transfections were done in triplicate, and the experiments were performed three times on separate days. Difference in activity was determined using a one‐sample t test.
RESULTS
Assessing functional variants enriched in SWAI individuals for an association with BMI
Twelve potentially functional (CADD > 10) variants were found to be enriched in WGS data from 296 full‐heritage Pima Indian individuals, where the variant allele was common (MAF ≥ 10%) in full‐heritage Pima Indian individuals but was exceedingly rare (MAF < 0.1%) in other ethnic groups (Table 1). Genotyping of these 12 enriched variants in a clinically phenotyped, population‐based sample of 5,870 SWAI individuals, representing 2,857 full‐heritage Pima Indian individuals and 3,013 individuals who were not full‐heritage (on average, individuals were half Pima Indian), showed that only rs548402150 (G/A) in the 5′ UTR of CYB5A significantly associated with BMI (significance requires a Bonferroni corrected p ≤ 0.004), in which the enriched allele (A) associated with an increase in BMI (β = 2% increase per A‐allele copy, p = 0.004). Among all 27 ethnic groups represented in the 1000 Genomes Project database, the rs548402150 A allele was observed only in the FIN (Finnish of Finland; frequency A = 0.005) and MXL (people of Mexican Ancestry in Los Angeles, California; frequency A = 0.0156) cohorts (Supporting Information Table S1).
TABLE 1.
Potentially functional (CADD > 10) variants enriched in full‐heritage Pima Indian individuals (MAF ≥ 10%) compared with other ethnic groups (MAF < 0.1%)
| Gene | Variant | Alleles | CADD | Frequency of alt alleles (%) | ||||
|---|---|---|---|---|---|---|---|---|
| Ref | Alt | Full‐heritage Pima Indian (n = 256) | SWAI sample a (n = 5870) | 1KGP (n = 2504) | gnomAD (n = 15,708) | |||
| SELENOO | rs200026401 | C | T | 11.08 | 18 | 11 | 0.02 | 0.03 |
| CYB5A | rs548402150 | G | A | 10.37 | 17 | 11 | 0.06 | 0.04 |
| CORO1B | rs561441501 | C | T | 13.20 | 16 | 10 | 0.02 | 0.03 |
| PARP16 | rs183702278 | T | C | 10.19 | 14 | 10 | 0.02 | 0.03 |
| RACGAP1 | rs191158076 | A | G | 25.30 | 14 | 10 | 0.04 | 0.05 |
| MPP4 | rs199986951 | T | C | 14.07 | 13 | 7 | 0.02 | 0.03 |
| CLK4 | rs146962272 | C | G | 10.62 | 12 | 8 | 0.04 | 0.04 |
| ZNF821 | rs77481038 | A | G | 24.10 | 12 | 7 | 0.08 | 0.04 |
| GFI1B | rs78837507 | C | T | 19.53 | 11 | 7 | 0.06 | 0.01 |
| INHBB | rs200554894 | G | A | 10.27 | 11 | 8 | 0.04 | 0.02 |
| ARHGEF11 | rs190557237 | G | A | 14.26 | 10 | 6 | 0.04 | 0.02 |
| SCUBE3 | rs199585730 | T | C | 10.83 | 10 | 6 | 0.02 | 0.003 |
Abbreviations: alt, alternative; ARHGEF11, Rho guanine nucleotide exchange factor 11; CADD, Combined Annotation Dependent Depletion score; CLK4, CDC like kinase 4; CORO1B, coronin 1B; CYB5A, cytochrome b5 type A; GFI1B, growth factor independent 1B transcriptional repressor; gnomAD, Genome Aggregation Database; INHBB, inhibin subunit β B; MAF, minor allele frequency; MPP4, MAGUK p55 scaffold protein 4; PARP16, poly(ADP‐ribose) polymerase family member 16; RACGAP1, Rac GTPase activating protein 1; Ref, reference; SCUBE3, signal peptide, CUB domain and EGF like domain containing 3; SELENOO, selenoprotein O; SWAI, Southwest American Indian; ZNF821, zinc finger protein 821; 1KGP, 1000 Genomes Project.
Variants were genotyped in the entire SWAI sample (2,857 full‐heritage Pima Indian individuals and 3,013 mixed‐heritage Pima Indian individuals) to assess association with BMI.
rs548402150 is an expression quantiative trait locus in skeletal muscle
Expression quantitative trait locus (eQTL) analysis using prior global gene expression from skeletal muscle biopsies isolated from SWAI individuals (n = 207) showed that the rs548402150 genotype was correlated with CYB5A expression, in which the higher‐BMI‐risk A allele associated with a decrease in CYB5A expression (β = 0.5‐SD decrease of gene expression levels per allele copy, p = 0.0008). In vitro luciferase reporter assays in human skeletal muscle cells supported this functional effect with a significant 30% decrease (p = 0.003) in luciferase activity for the CYB5A 5′ UTR‐promoter vector containing the A allele (Figure 1).
FIGURE 1.
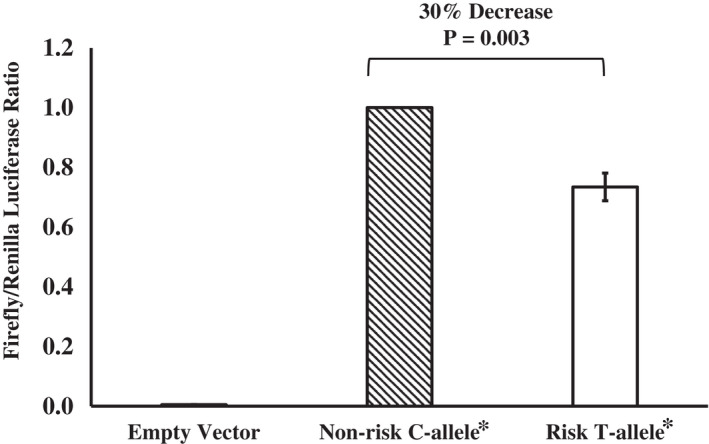
The effect of the CYB5A 5′ untranslated region variant rs548402150 on luciferase activity in human skeletal muscle cells. *Although databases report rs548402150 as G/A, CYB5A is transcribed from the minus‐strand DNA; therefore, a functional effect of rs548402150 was assessed using alleles C/T. Each bar represents the mean ± SD of luciferase activity, normalized to the non‐risk C allele from three replicates in one experiment. A total of three independent experiments were performed. Significant changes (percentage) in activity were determined by a one‐sample t test. CYB5A, cytochrome b5 type A
CYB5A haplotype analysis for obesity‐related traits
In a prior analysis of exome variation in SWAI individuals, we reported that a Proline96Proline, rs7238987, in exon 3 of CYB5A, nominally associated with BMI (25). This rs7238987 variant has subsequently been identified as a splicing QTL (sQTL) in several Genotype‐Tissue Expression Project (GTEx) tissues. When analyzed in the current sample set, rs7238987 was similarly associated with BMI (β = 2% increase per T‐allele copy, p = 0.0002) and was found to be common in both SWAI individuals (MAF = 29%) and global populations (MAF = 15%). More recently, we have detected another GTEx sQTL within intron 3 of CYB5A, rs7238784, that also has a trend for increased BMI (β = 1% increase per C‐allele copy, p = 0.057) and that is common in SWAI individuals (MAF = 46%), as well as globally (MAF = 34%). Therefore, haplotypes were constructed with these three functional variants: rs548402150 (G/A), rs7238987 (C/T), and rs7238784 (A/C). Frequencies and error rates for the haplotype construction with SWAI individuals and 1000 Genomes Project genotypic data are available in Supporting Information Table S2. These haplotypes were analyzed for associations with BMI, %fat, and 24‐h EE in SWAI individuals (Table 2). As shown in Table 2, the ATA haplotype associated with increased BMI (β = 3%; p = 0.0003), and, in a much smaller sample size, nominally associated with an increase in %fat (β = 0.8% increase; p = 0.04). This haplotype has a frequency of 13% in SWAI individuals. However, in the 1000 Genomes Project database, the ATA haplotype was observed only in one individual from Finland and two individuals of Mexican ancestry (MXL study), giving an overall ATA frequency in 1000 Genomes Project populations of 0.06%. In contrast, the most common haplotype in the 1000 Genomes Project populations was CCG, with a 68% frequency. This CCG haplotype was less common in SWAI individuals (frequency = 45%), in whom it associated with lower BMI (β = −2%; p = 0.0016), lower %fat (β = −1.2%; p = 0.0001), and higher 24‐h EE (β = 30 kcal/d; p = 0.005). The haplotypes were further analyzed for association with CYB5A expression in skeletal muscle biopsies from 207 SWAI individuals (Table 3). The ATA haplotype associated with lower expression of CYB5A when compared with all other haplotypes. Conversely, the CCG haplotype associated with greater expression of CYB5A compared with all other haplotypes.
TABLE 2.
Haplotypes for rs7238784 (A/C), rs7238987 (C/T), and rs548402150 (G/A) and their association with BMI, %fat, and 24‐h EE in SWAI individuals
| Mean BMI based on haplotype copy | BMI (n = 5870) | %fat (n = 917) | 24‐h EE (n = 419) | Number of individuals by haplotype copy for BMI, %fat, and 24‐h EE | Haplotype frequencies (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype | 0 copy | 1 copy | 2 copies | β | p value | β | p value | β | p value | 0 copy | 1 copy | 2 copies | SWAI (n = 5870) | 1KGP (n = 2504) |
| CCG | 36.1 | 35.1 | 34.1 | −0.016 | 0.0016 | −1.2 | 0.0001 | 30.3 | 0.0053 | 1664; 266; 121 | 3061; 450; 225 | 1145; 171; 73 | 46 | 68 |
| ACG | 35.2 | 35.1 | 35.0 | −0.007 | 0.23 | 0.3 | 0.37 | −23.2 | 0.06 | 3304; 507; 236 | 2228; 355; 164 | 338; 55; 19 | 25 | 18 |
| ATG | 35.0 | 35.6 | 35.1 | 0.014 | 0.048 | 0.8 | 0.04 | 2.4 | 0.87 | 4075; 646; 292 | 1680; 256; 122 | 115; 15; 5 | 16 | 12 |
| ATA | 34.7 | 36.3 | 38.6 | 0.027 | 0.0003 | 0.8 | 0.04 | −25.0 | 0.09 | 4428; 649; 291 | 1338; 248; 123 | 104; 20. 5 | 13 | 0.06 |
| CTG | 35.2 | 35.9 | – | 0.071 | 0.31 | – | – | – | – | 5855; 916; 419 | 15; 1; 0 | 0; 0; 0 | 0.12 | 0.64 |
| ACA | 35.2 | 39.4 | – | 0.120 | 0.14 | – | – | – | – | 5859; 916; 419 | 11; 1; 0 | 0; 0; 0 | 0.09 | 0.0 |
Statistical associations were analyzed by linear regression using an additive model for haplotypes and the generalized estimating equation procedure to account for sibship. P values for BMI were adjusted for age, sex, birth year, and the first five genetic principal components. P values for %fat were adjusted for age, sex, and the first five genetic principal components. P values for 24‐h EE were adjusted for age, sex, fat mass, fat‐free mass, spontaneous physical activity level, and the first five genetic principal components. Data for haplotypes that associated with either low BMI or high BMI with p < 0.05 are shaded.
Abbreviations: SWAI, Southwest American Indian; 1KGP, 1000 Genomes Project; 24‐h EE, 24‐hour energy expenditure; %fat, percentage of body fatness.
TABLE 3.
Association of CYB5A haplotypes with CYB5A expression levels in skeletal muscle tissue biopsies
| Haplotype | 0 copy | 1 copy | 2 copies | β | p value |
|---|---|---|---|---|---|
| CCG | 57 | 107 | 43 | 0.21 | 0.03 |
| ACG | 116 | 82 | 9 | 0.10 | 0.39 |
| ATG | 151 | 54 | 2 | −0.09 | 0.55 |
| ATA | 147 | 57 | 3 | −0.49 | 0.0005 |
Data for haplotypes that associate with either low BMI or high BMI are shaded. Batch‐ and sex‐standardized expression levels are expressed in SD units. P values were adjusted for age and first principal component.
Abbreviations: CYB5A, cytochrome b5 type A.
CYB5A expression in skeletal muscle has an inverse relationship with %fat
At the same inpatient visit from which the skeletal muscle biopsy was obtained to assess gene expression, individuals also underwent a dual‐energy x‐ray absorptiometry measurement of %fat. There was an inverse relationship between %fat and CYB5A gene expression level in muscle, where higher %fat correlated with lower CYB5A expression. However, in this small sample (n = 207), the relationship did not achieve statistical significance (β = −0.80% of body fat per 1‐SD difference in gene expression level, p = 0.06).
DISCUSSION
The present study identified rs548402150, a 5′ UTR variant in CYB5A highly enriched in SWAI individuals, to be associated with measures of obesity. A functional effect of rs548402150 was demonstrated in vitro, in which a luciferase reporter gene in human skeletal muscle cells showed that the BMI‐risk T allele (minus strand) decreased gene expression. Parallel with this observation, eQTL analysis in skeletal muscle biopsies from SWAI individuals showed that the risk‐allele carriers had decreased CYB5A expression. The effect of rs548402150 on gene expression may be explained by the variant being positioned in a CCCTC‐binding factor (CTCF) consensus site. CTCF is a zinc finger protein that functions as a transcription factor, with the ability to activate or repress gene expression (26).
Haplotype analysis of rs548402150 and two sQTL variants, rs7238987 and rs7238784, identified that the most enriched haplotype in SWAI individuals (ATA), compared with other ethnic groups, associated with higher BMI and %fat, whereas the haplotype most frequent globally (CCG) associated with lower BMI, lower %fat, and increased 24‐h EE in SWAI individuals. The ATA haplotype associated with lower CYB5A expression in muscle biopsies from SWAI volunteers; whereas the CCG haplotype associated with increased CYB5A expression levels. It should be noted that although the obesity‐risk haplotype ATA did not reach statistical significance in its association with 24‐h EE, it was directionally consistent (carriers of the high BMI haplotype had lower 24‐h EE; β = −25.0; p = 0.09) and the lack of significance could be due to low power, as the sample size for this phenotype was small.
CYB5A encodes a hemoprotein that functions as an electron carrier to enhance the function of membrane‐bound oxygenases involved in lipid biosynthesis (27) and fatty acid oxidation (28). Dysregulation of these processes has been linked to weight gain. Specifically, epidemiological evidence in developing nations has suggested that a reduction in fat oxidation is associated with obesity (29). We hypothesize that the reduction in CYB5A expression may contribute to obesity in SWAI individuals by reducing the ability to use fat stores for energy.
CYB5A is expressed in several tissues, such as adipose, liver, brain, and skeletal muscle, that may have an important role in influencing energy balance and thereby may affect body weight and composition. However, we have previously collected only skeletal muscle and subcutaneous adipose biopsies from SWAI volunteers. Because our eQTL analysis supported a relationship between genotype and CYB5A expression in skeletal muscle but not adipose tissue, and our haplotype analysis identified an association with EE for which skeletal muscle is a key tissue, we hypothesize that CYB5A may exert a role in this tissue. However, we cannot exclude that this gene may influence body composition via an effect in other tissues.
To our knowledge, we are the only group who has proposed that CYB5A affects body weight. Both a previous genome‐wide association study for BMI and a whole‐exome sequencing study in SWAI individuals (25, 30) have reported variation at this locus that associated with BMI. Although it is clear that associations cannot prove causality, other studies included in the T2D Knowledge Portal have also reported associations between CYB5A variants and BMI. The T2D Knowledge Portal contains at least 18 different data sets of varying ethnicities for genetic associations with BMI, with the largest cohort coming from the Genetic Investigation of ANthropometric Traits (GIANT)‐UK Biobank (>700,000 samples). From these data sets, there are 374 variants in or near CYB5A that associate with BMI (p ≤ 0.05). The direction of the BMI associations varies by SNP, but, overall, this gene associates only with BMI and no other traits. Therefore, considering the association of CYB5A genotypic data with both BMI and CYB5A expression levels in the current study, and the association of variation at the CYB5A locus with BMI in these other studies, we propose that more in‐depth analyses of CYB5A, such as animal knockout studies, are warranted.
CONCLUSION
In conclusion, our study identified rs548402150 to be enriched in SWAI individuals and associated with measures of obesity, potentially through the reduction of CYB5A. However, further investigation into this mechanism is warranted. Specifically, future studies should do the following: 1) confirm CTCF binding to the variant region; and 2) assess changes in fatty acid oxidation by knocking down CYB5A in skeletal muscle.O
CONFLICT OF INTEREST
The authors declared no conflict of interest.
Supporting information
Table S1
Table S2
ACKNOWLEDGMENTS
We thank volunteers for their participation in the studies. This research was supported by the Intramural Research Program of the NIH and used the computational resources of the Biowulf system at the NIH, Bethesda, Maryland.
Day SE, Traurig M, Kumar P, et al. Functional variants in cytochrome b5 type A (CYB5A) are enriched in Southwest American Indian individuals and associate with obesity. Obesity (Silver Spring). 2022;30:546–552. doi: 10.1002/oby.23359
Funding information
This work was supported by the intramural research program of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), an institute within the National Institutes of Health (NIH).
REFERENCES
- 1. Knowler WC, Pettitt DJ, Saad MF, et al. Obesity in the Pima Indians: its magnitude and relationship with diabetes. Am J Clin Nutr. 1991;53:1543S‐S1551. [DOI] [PubMed] [Google Scholar]
- 2. Ingelsson E, McCarthy MI. Human genetics of obesity and type 2 diabetes mellitus: past, present, and future. Circ Genom Precis Med. 2018;11:e002090. doi: 10.1161/CIRCGEN.118.002090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nair AK, Baier LJ. Complex genetics of type 2 diabetes and effect size: what have we learned from isolated populations? Rev Diabet Stud. 2015;12:299‐319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Muller YL, Hanson RL, Piaggi P, et al. Assessing the role of 98 established loci for BMI in American Indians. Obesity (Silver Spring). 2019;27:845‐854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Traurig M, Mack J, Hanson RL, et al. Common variation in SIM1 is reproducibly associated with BMI in Pima Indians. Diabetes. 2009;58:1682‐1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Traurig MT, Perez JM, Ma L, et al. Variants in the LEPR gene are nominally associated with higher BMI and lower 24‐h energy expenditure in Pima Indians. Obesity (Silver Spring). 2012;20:2426‐2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Muller YL, Thearle MS, Piaggi P, et al. Common genetic variation in and near the melanocortin 4 receptor gene (MC4R) is associated with body mass index in American Indian adults and children. Hum Genet. 2014;133:1431‐1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thearle MS, Muller YL, Hanson RL, et al. Greater impact of melanocortin‐4 receptor deficiency on rates of growth and risk of type 2 diabetes during childhood compared with adulthood in Pima Indians. Diabetes. 2012;61:250‐257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim HI, Gosalia N, Ye B, et al. Characterization of exome variants and their metabolic impact in 6,716 American Indians from Southwest US. Am J Hum Genet. 2020;107:251‐264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zimmet P, Taft P, Guinea A, Guthrie W, Thoma K. The high prevalence of diabetes mellitus on a central pacific island. Diabetologia. 1977;13:111‐115. [DOI] [PubMed] [Google Scholar]
- 11. Minster RL, Hawley NL, Su C‐T, et al. A thrifty variant in CREBRF strongly influences body mass index in Samoans. Nat Genet. 2016;48:1049‐1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hanson RL, Safabakhsh S, Curtis JM, et al. Association of CREBRF variants with obesity and diabetes in Pacific Islanders from Guam and Saipan. Diabetologia. 2019;62:1647‐1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886‐D894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. The 1000 Genomes Project Consortium ; Auton A, Brooks LD, Durbin RM, et al. A global reference for human genetic variation. Nature. 2015;526:68‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434‐443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. American Diabetes Association Expert Committee on the Diagnosis and Classification of Diabetes Mellitus . Report of the Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Diabetes Care. 1997;20:1183‐1197. [DOI] [PubMed] [Google Scholar]
- 17. Piaggi P, Masindova I, Muller YL, et al. A Genome‐wide association study using a custom genotyping array identifies variants in GPR158 associated with reduced energy expenditure in American Indians. Diabetes. 2017;66:2284‐2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tataranni PA, Ravussin E. Use of dual‐energy x‐ray absorptiometry in obese individuals. Am J Clin Nutr. 1995;62:730‐734. [DOI] [PubMed] [Google Scholar]
- 19. Ravussin E, Lillioja S, Anderson TE, Christin L, Bogardus C. Determinants of 24‐hour energy expenditure in man. Methods and results using a respiratory chamber. J Clin Invest. 1986;78:1568‐1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Piaggi P, Thearle MS, Bogardus C, Krakoff J. Lower energy expenditure predicts long‐term increases in weight and fat mass. J Clin Endocrinol Metab. 2013;98:E703‐E707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsueh WC, Nair AK, Kobes S, et al. Identity‐by‐descent mapping identifies major locus for serum triglycerides in Amerindians largely explained by an APOC3 founder mutation. Circ Cardiovasc Genet. 2017;10:e001809. doi: 10.1161/CIRCGENETICS.117.001809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Browning BL, Browning SR. A fast, powerful method for detecting identity by descent. Am J Hum Genet. 2011;88:173‐182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978‐989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mason CC, Hanson RL, Ossowski V, et al. Bimodal distribution of RNA expression levels in human skeletal muscle tissue. BMC Genomics. 2011;12:98. doi: 10.1186/1471-2164-12-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang K, Nair AK, Muller YL, et al. Whole exome sequencing identifies variation in CYB5A and RNF10 associated with adiposity and type 2 diabetes. Obesity (Silver Spring). 2014;22:984‐988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim S, Yu NK, Kaang BK. CTCF as a multifunctional protein in genome regulation and gene expression. Exp Mol Med. 2015;47:e166. doi: 10.1038/emm.2015.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paton CM, Ntambi JM. Biochemical and physiological function of stearoyl‐CoA desaturase. Am J Physiol Endocrinol Metab. 2009;297:E28‐E37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bhatt MR, Khatri Y, Rodgers RJ, Martin LL. Role of cytochrome b5 in the modulation of the enzymatic activities of cytochrome P450 17alpha‐hydroxylase/17,20‐lyase (P450 17A1). J Steroid Biochem Mol Biol. 2017;170:2‐18. [DOI] [PubMed] [Google Scholar]
- 29. Frisancho AR. Reduced rate of fat oxidation: a metabolic pathway to obesity in the developing nations. Am J Hum Biol. 2003;15:522‐532. [DOI] [PubMed] [Google Scholar]
- 30. Bian L, Traurig M, Hanson RL, et al. MAP2K3 is associated with body mass index in American Indians and Caucasians and may mediate hypothalamic inflammation. Hum Mol Genet. 2013;22:4438‐4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2


