Abstract
Background & Aims
Upon hepatitis B virus (HBV) infection, partially double-stranded viral DNA converts into a covalently closed circular chromatinized episomal structure (cccDNA). This form represents the long-lived genomic reservoir responsible for viral persistence in the infected liver. Although the involvement of host cell DNA damage response in cccDNA formation has been established, this work investigated the yet-to-be-identified histone dynamics on cccDNA during early phases of infection in human hepatocytes.
Methods
Detailed studies of host chromatin-associated factors were performed in cell culture models of natural infection (ie, Na+-taurocholate cotransporting polypeptide (NTCP)-overexpressing HepG2 cells, HepG2hNTCP) and primary human hepatocytes infected with HBV, by cccDNA-specific chromatin immunoprecipitation and loss-of-function experiments during early kinetics of viral minichromosome establishment and onset of viral transcription.
Results
Our results show that cccDNA formation requires the deposition of the histone variant H3.3 via the histone regulator A (HIRA)-dependent pathway. This occurs simultaneously with repair of the cccDNA precursor and independently from de novo viral protein expression. Moreover, H3.3 in its S31 phosphorylated form appears to be the preferential H3 variant found on transcriptionally active cccDNA in infected cultured cells and human livers. HIRA depletion after cccDNA pool establishment showed that HIRA recruitment is required for viral transcription and RNA production.
Conclusions
Altogether, we show a crucial role for HIRA in the interplay between HBV genome and host cellular machinery to ensure the formation and active transcription of the viral minichromosome in infected hepatocytes.
Keywords: HBV, cccDNA, chromatin, HIRA, H3.3
Abbreviations used in this paper: BrdU, bromodeoxyuridine; BSA, bovine serum albumin; CAF-1, chromatin assembly complex; cccDNA, covalently closed circular DNA; ChIP, chromatin immunoprecipitation; CMV, cytomegalovirus; Ct, threshold cycle; DIG, digoxigenin; DMEM, Dulbecco’s modified Eagle medium; DMSO, dimethylsulfoxide; E2F, E2 Factor; FCS, fetal calf serum; HBc, HBV core protein; HBV, hepatitis B virus; HBx, HBV x protein; HepG2hNTCP, Na+-taurocholate cotransporting polypeptide (NTCP)-overexpressing HepG2 cells; HIRA, histone regulator A; hpi, hours postinfection; mcHBV, minicircle hepatitis B virus; PBS, phosphate-buffered saline; PF, protein-free; pgRNA, pregenomic RNA; PHH, primary human hepatocyte; PLA, proximity ligation assay; pMC, plasmid minicircle; PMSF, phenylmethylsulfonyl fluoride; qPCR, quantitative polymerase chain reaction; rcDNA, relaxed circular DNA; RNAP II, RNA polymerase II; SDS, sodium dodecyl sulfate; siRNA, small interfering RNA; SMC, Structural maintenance of chromosomes; WT, wild-type
Graphical abstract
Summary.
We describe the involvement of the histone chaperone histone regulator A in the formation of the chromatinized form of hepatitis B virus episome in the nucleus of infected hepatocytes. Histone regulator A is also required for full transcriptional activity of the established hepatitis B virus minichromosome.
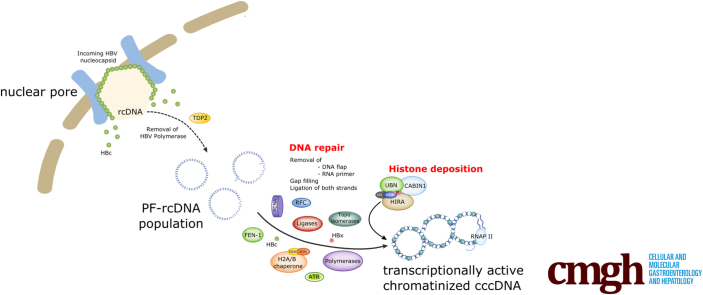
Hepatitis B virus (HBV) is a hepadnavirus that necessitates a mandatory nuclear phase to convert the partially double-stranded relaxed circular DNA (rcDNA) covalently attached to the viral polymerase protein contained in the virion into a covalently closed circular DNA (cccDNA) episome.1 cccDNA represents not only the unique transcriptional template for viral protein expression and genome replication, but also a long-lived repository for viral genetic information. Similar to other DNA viruses that persist as episomes in host nuclei, the HBV genome must adopt a chromatin structure to maintain the stability and accessibility of its genome and thus relies on host cellular molecular machinery for its replication.2,3 HBV can be assimilated to a pararetrovirus because viral genome replication requires the reverse transcription of the pregenomic RNA (pgRNA) into new rcDNA by the viral polymerase within newly assembled nucleocapsids in the cytoplasm.1 Unaffected by current antiviral treatments inhibiting viral DNA synthesis, cccDNA is responsible for viral persistence in infected hepatocytes and its elimination remains the utmost goal for curing chronic HBV infections.4
Unlike SV40 and papilloma viruses, which already package their circular DNA genomes in a nucleosomal state in the virion,5 HBV rcDNA enters the host cell nucleus devoid of nucleosomes and must divert the host cell machinery to build a de novo chromatin structure in the newly infected cells. A number of recent studies have indicated that the conversion of rcDNA into cccDNA necessitates events linked to DNA repair.3,6, 7, 8, 9, 10, 11, 12, 13 However, the factors involved in histone deposition on the nucleosome-free HBV genome in the hepatocyte nuclei have been largely overlooked to date.
The histone regulator A (HIRA) promotes H3.3 deposition and nucleosome assembly independently of DNA synthesis by exploiting its capacity to bind nonspecifically to H3.3–H4 histone dimers and naked DNA.14, 15, 16 HIRA recently was involved in the deposition of H3.3 onto foreign herpes simplex virus and cytomegalovirus (CMV) genomes.17 H3.3 histone variant is expressed constitutively throughout the cell cycle in proliferating and quiescent cells. It is distinguished from the replicative variants H3.1/2 by a single amino acid substitution in the amino-terminal tail, a serine at position 31 in place of an alanine in H3.1/2 and in the regions recognized by distinct chaperones.18 The importance of phosphorylation at S31 in H3.3 has been shown recently for major transitions in transcription programs.18, 19, 20
In this study, we analyzed the cccDNA nucleoprotein structure in human hepatocytes very early after de novo infection. Both histone deposition on incoming HBV genome and viral transcript expression began within hours after viral entry and the histone chaperone complex HIRA-dependent H3.3 deposition was essential for the formation of supercoiled cccDNA. Although HBV protein neosynthesis was dispensable for HIRA and H3.3 recruitment to cccDNA, the viral core protein delivered by incoming virions was temporarily and physically associated to cccDNA-bound HIRA. The observation that HIRA depletion after the establishment of cccDNA severely decreased the expression of viral RNA uncovered a dual role for HIRA complex in both HBV chromatin assembly and transcriptional activation by promoting deposition and recycling of serine 31 phosphorylated H3.3.
Results
Supercoiled and Transcriptionally Active cccDNA Appears Early After Viral Infection
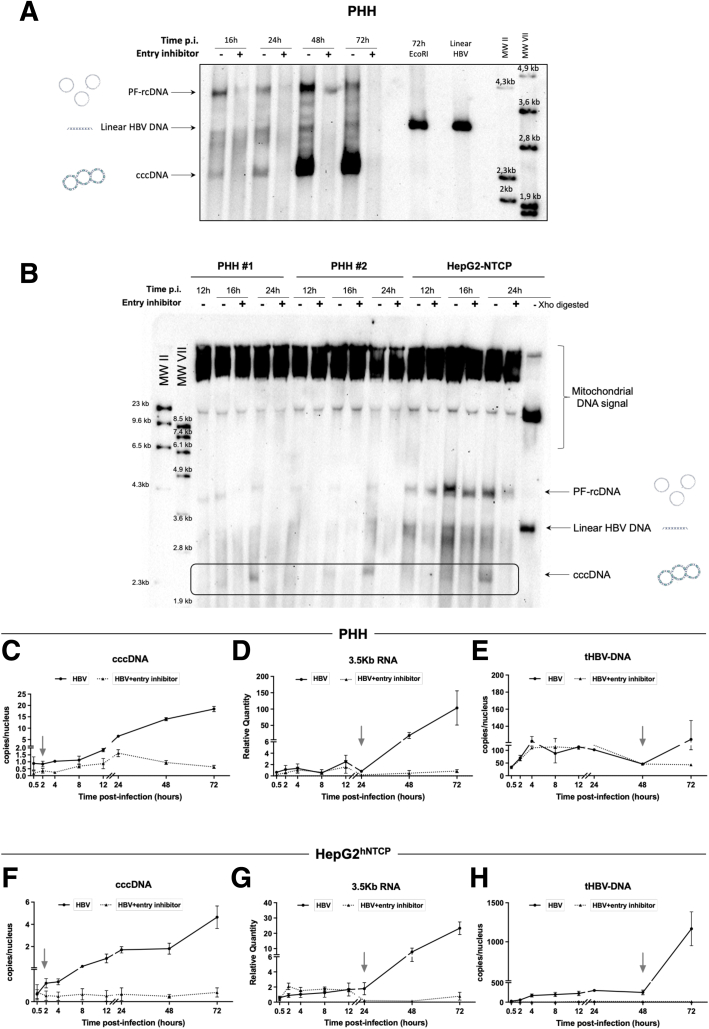
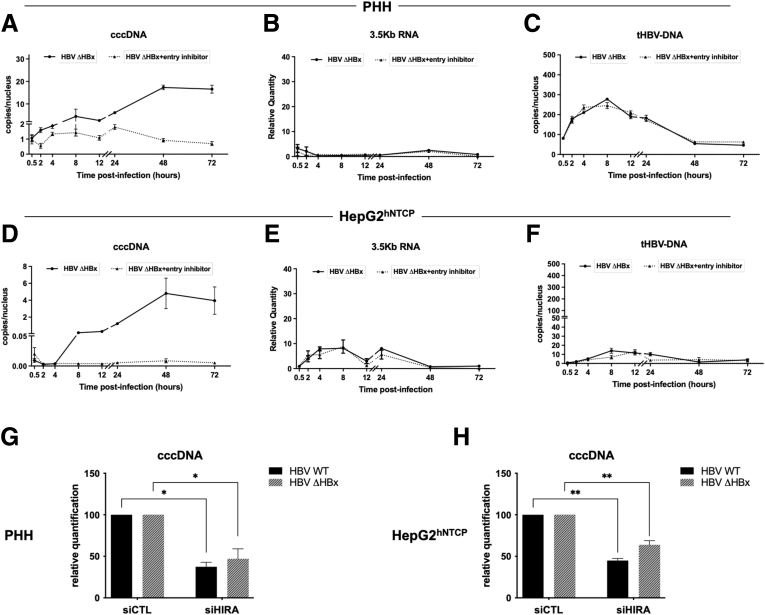
Kinetics of cccDNA formation in Na+-taurocholate cotransporting polypeptide (NTCP)-overexpressing HepG2 cells (HepG2hNTCP) cells and in primary human hepatocytes (PHHs) were analyzed by Southern blot and quantitative polymerase chain reaction (qPCR) after infection with HBV inoculum in the presence or not of a specific entry inhibitor that served to set the detection background. To specifically enrich for viral episomal genome in the nucleus of infected hepatocytes, an adapted Hirt extraction procedure was performed to select for genomic forms already deprived of the covalently attached viral polymerase.21 This viral genome population presumably consists of a mixture of at least 3 distinct DNA species, one with both strands remaining open; another with a covalently closed minus strand rcDNA, collectively called protein-free rcDNA (PF-rcDNA); and the double-strand covalently closed cccDNA.22,23 cccDNA can be specifically distinguished from the other species by virtue of the difference in electrophoresis mobility associated to its supercoiled conformation (Figure 1A and B). The incoming viral genome was composed mostly of PF-rcDNA (Figure 1B) (12 hours postinfection [hpi] and entry inhibitor treated samples), while cccDNA appeared afterward (16 hpi). Interestingly, PF-rcDNA levels decreased along with a cccDNA increase from 48 to 72 hpi, in agreement with its role as a direct precursor of cccDNA formation24 (Figure 1A). Consistently, using the more sensitive real-time qPCR method, cccDNA was detected as soon as 2 hpi and began to increase at 4 hpi (Figure 1C and F). The levels of intracellular 3.5-kb HBV RNA, detecting the replicative intermediate pgRNA, increased exponentially from 24 hpi, indicating the initiation of viral transcription from cccDNA (Figure 1D and G). Quantification of total HBV DNA, including rcDNA delivered by incoming virions, cccDNA and replicative intermediates generated by pgRNA reverse transcription, showed a basal level as a result of the input virions entering the cells that remained stable until 48 hpi, followed by an increase indicating the initiation of pgRNA reverse transcription and active viral replication (Figure 1E and H). Taken together, these data show that PF-rcDNA to cccDNA conversion occurs early after infection and precedes a phase in which it occurs concomitantly with viral transcription (from 24 hpi onward) of established cccDNA pool and total DNA accumulation (from 48 hpi onward) with similar kinetics in HepG2hNTCP cells and primary human hepatocytes.
Figure 1.
HBV minichromosome establishment occurs very rapidly after infection. (A and B) Southern blot analysis of cccDNA appearance kinetic. PHHs or HepG2hNTCP cells were infected with HBV in the presence or not of 100 nmol/L preS1-mimicking peptide for up to 16 hours and then harvested at the indicated times points. Mitochondrial DNA was used as an internal loading control. The specificity of the cccDNA band is shown by linearization after digestion with EcoRI or XhoI restriction enzymes. (C–H) qPCR quantification of viral cccDNA, 3.5-kb RNA, and total HBV DNA (tHBV-DNA) in (C–E) PHHs and (F–H) HepG2hNTCP-infected cells. Cells were inoculated with HBV for up to 16 hours and then harvested at the indicated time points. cccDNA and tHBV-DNA quantification were normalized over β-globin quantity, while the relative 3.5-kb RNA amount was normalized over the housekeeping gene GUSb expression. Graphs represent the means ± SEM of at least 3 independent experiments. MW, molecular weight; p.i., post infection.
HIRA Is Required for Full cccDNA Supercoiling in De Novo Infected Cells
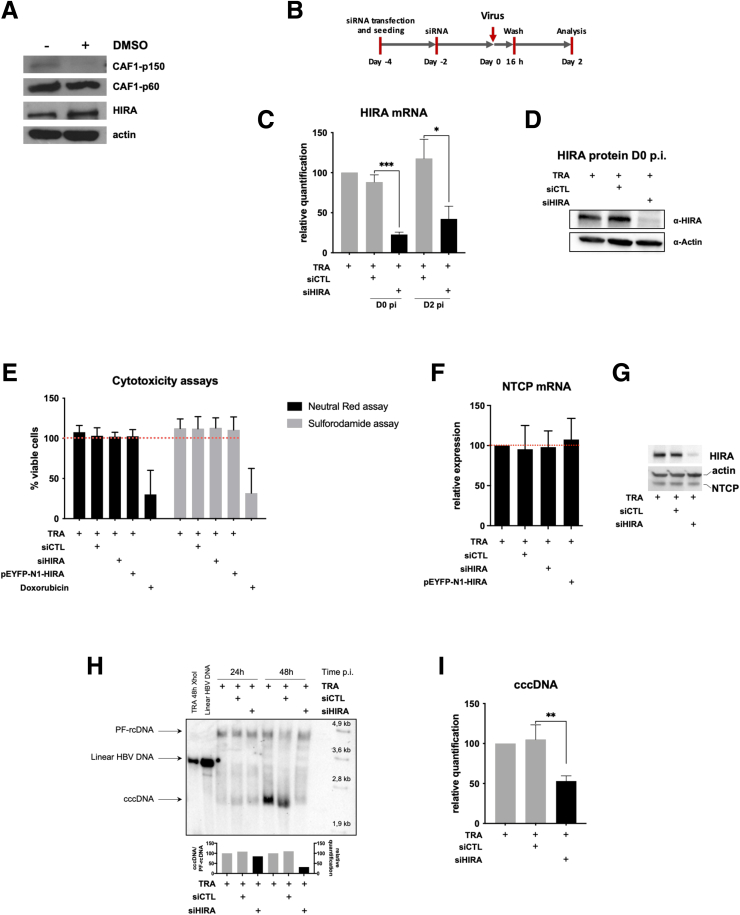
De novo histone deposition is required to form nucleosomes on the naked cccDNA precursor.25 A number of biological reactions are required to convert partially double-stranded, viral polymerase–associated rcDNA into cccDNA and most of them could be ascribed to DNA repair pathways.6 Interestingly, histone H3 variants H3.1/2 and H3.3 and their respective histone chaperone complexes, chromatin assembly complex (CAF-1) and HIRA, have been involved in histone deposition linked to DNA damage,26,27 placing them as attractive candidates to promote nucleosome assembly on cccDNA. However, after dimethylsulfoxide (DMSO) treatment, which favors cell differentiation and viral infection, while HIRA levels remained stable, expression levels of the p150 subunit of CAF-1 decreased in nondividing HepG2hNTCP (Figure 2A), as already observed for other differentiated cells.28 These data, along with the observation that only HIRA could deposit H3.3 on foreign viral DNA,17 prompted us to investigate the contribution of HIRA in cccDNA formation within the first 72 hpi in HepG2hNTCP cells. Two sequential small interfering RNA (siRNA) transfections (Figure 2B) were performed to achieve a significant reduction of HIRA expression, both at the messenger RNA and protein levels (Figure 2C and D), without significant effects on cell viability (Figure 2E) and HBV-receptor hNTCP RNA and protein expression (Figure 2F and G). HBV infection was performed 48 hours after the second transfection, when HIRA protein was depleted more than 70% (Figure 2C and D). Southern blot and qPCR analysis at 48 hpi showed a sharp decrease of cccDNA amounts in HIRA-depleted cells compared with controls (Figure 2H and I). Importantly, HIRA knockdown did not affect the levels of PF-rcDNA, suggesting that HIRA is not required for processes preceding PF-rcDNA to cccDNA conversion, but is necessary for the subsequent steps leading to cccDNA formation and maintenance (Figure 2H).
Figure 2.
Full cccDNA supercoiling in de novo infected cells requires HIRA protein expression. (A) Detection by Western blot analysis of CAF-1 (subunits p150 and p60) and HIRA before and after 72 hours of 2.5% DMSO addition to HepG2hNTCP cells. (B) HepG2hNTCP cells were transfected twice with siRNA anti-HIRA or a nontargeting siRNA (siCTL) and then inoculated for 16 hours with HBV. Cells were harvested for analysis 2 dpi. (C and D) HIRA messenger RNA (mRNA) and protein expression after siRNA transfection was determined by (B) real-time qPCR and (C) Western blot, respectively. β-actin served as Western blot loading control. HIRA mRNA was normalized over housekeeping GUSb gene levels and expressed as relative to the control condition treated only with the transfection reagent (TRA). (E–G) HepG2hNTCP cells were transfected with siRNA against HIRA according to the timelines shown in Figure 6A and inoculated for 16 hours with HBV at 250 viral genome equivalents/cell. The cells were harvested for analysis 2 dpi. (E) Neutral red and sulforhodamide cytotoxicity assay analysis. Doxorubicin treatment served as positive control for cell mortality. (F) mRNA and (G) protein levels of hNTCP receptor. β-actin served as Western blot loading control. (H and I) cccDNA amount at 2 dpi was measured by (H) Southern blot and (I) qPCR. cccDNA quantification was normalized over β-globin quantity and expressed as relative to TRA control. The 2-tailed P value was calculated for a risk threshold of .05 using the 2/K sample permutation test with Monte Carlo resampling approximation. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. p.i., post infection.
A Naked, Not Fully Double-Stranded Precursor Is the Substrate of HIRA During cccDNA Formation
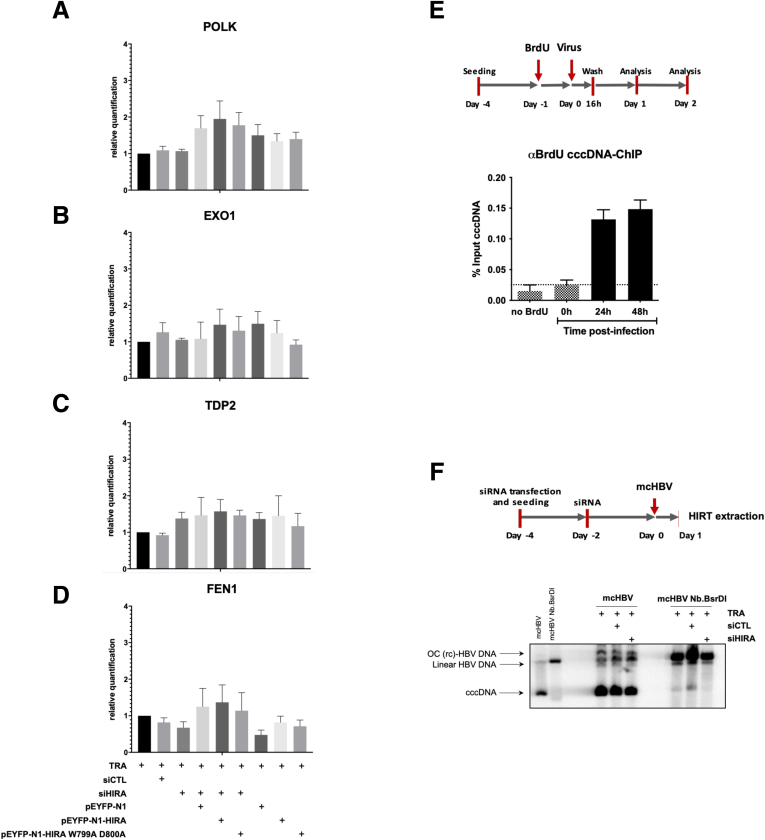
Next, we asked whether requirement for HIRA is linked to repair events associated with conversion of PF-rcDNA toward cccDNA (ie, a complete circular double-stranded DNA). First, we verified if the levels of transcripts of genes involved in rcDNA to cccDNA conversion3,9 were affected upon loss of HIRA. We could not show significant differences in DNA Polymerase Kappa (POLK), Exonuclease 1 (EXO1), tyrosyl-DNA phosphodiesterase-2, and flap structure-specific endonuclease 1 transcript expression (Figure 3A–D) after HIRA silencing and transcomplementation, suggesting that repair activities involved in the first processing events of HBV genome are not affected by HIRA down-regulation. Next, we monitored kinetics of bromodeoxyuridine (BrdU) incorporation in the nuclear HBV genome to assess the DNA repair activity during the early phases of HBV infection by performing cccDNA–chromatin immunoprecipitation (ChIP) analysis using an anti–BrdU-specific antibody (Figure 3E). BrdU incorporation was detected in cccDNA at 48 and 72 hpi indicating that repair and gap filling processes are involved in cccDNA formation in living cells (Figure 3E). Taking advantage of the minicircle technology,29 we introduced nicks in 1 strand of the circular double-stranded minicircle HBV (mcHBV) by digestion with the nicking endonuclease Nb.BsrDI, triggering mcHBV relaxation and mimicking a covalently closed minus strand rcDNA structure, recently described as the main precursor of cccDNA22,23 (Figure 3F). After HIRA knock-down, either a double-stranded or a nicked mcHBV was transfected and the formation of supercoiled cccDNA was monitored by Southern blot (Figure 3F). The levels of cccDNA arising from the double-stranded circular mcHBV were not affected by the absence of HIRA. In contrast, when the damaged mcHBV was transfected, less supercoiled cccDNA formation was detected in the absence HIRA (Figure 3F). This suggests that HIRA is critical for cccDNA formation when DNA repair is required, such as conversion of PF-rcDNA after its entry in the nucleus of infected cells.
Figure 3.
Full cccDNA supercoiling in de novo infected cells occurs concomitantly to HBV genome repair. (A–D) Messenger RNA levels of (A) POLK, (B) EXO1, (C) TDP2, and (D) FEN1 were quantified by real-time qPCR assay and expressed as a percentage of TRA-treated cells after normalization over GUSb housekeeping gene expression. Data represent the means ± SEM of at least 3 independent experiments. (E) ChIP analysis of BrdU-containing cccDNA molecules. HepG2hNTCP cells were treated with 20 μmol/L for 24 hours before HBV infection and cultured for the indicated time points. cccDNA-ChIP qPCR using no antibody or anti-E2F antibody served as ChIP technical negative controls (Figure 8A and B), and the signal at 0.5 hpi was considered a specific qPCR background for cccDNA quantification (Figure 1F). Data are expressed as a percentage of enrichment with respect to initial input chromatin. (F) mcHBV constructs digested or not with Nb.BsrDI were transfected into HepG2hNTCP cells 48 hours after transfection with siHIRA or siCTL. Cells were harvested 24 hours after mcHBV transfection and the cccDNA amount was measured by Southern blot. Graphs represent the means ± SEM of at least 3 independent experiments. EXO1, Exonuclease 1; FEN-1, flap structure-specific endonuclease 1; OC (rc)-HBV DNA, open circular (relaxed circular) HBV DNA; POLK, DNA polymerase kappa; TDP2, tyrosyl-DNA phosphodiesterase-2.
HIRA-Dependent cccDNA Formation Does Not Require HBV Protein Neosynthesis
To investigate a putative role of early translated HBV proteins in HIRA-dependent cccDNA formation, we took advantage of an HBV x protein (HBx)-deficient HBV virus (ΔHBx-HBV).30 The HBx protein plays a pivotal role for the full transcriptional activity of established cccDNA, thus ΔHBx-HBV infection allows the formation of cccDNA, which, however, remains transcriptionally inactive.30,31 After infection with a ΔHBx-HBV genome containing virus, cccDNA appeared with similar kinetics as WT-HBV virus, while no 3.5-kb RNA and DNA replicative intermediates could be detected in PHHs (compare Figure 1C–E with Figure 4A–C) and HepG2hNTCP (compare Figure 1F–H with Figure 4D–F). HIRA knock-down before ΔHBx-HBV infection decreased cccDNA levels to a similar extent as in WT HBV infection conditions in PHHs (Figure 4G) and HepG2hNTCP (Figure 4H).
Figure 4.
HBV protein neosynthesis is not required for HIRA-dependent cccDNA formation. (A–F) qPCR quantification of viral (A and D) cccDNA, (B and E) 3.5-kb RNA, and (C and F) total HBV DNA (tHBV-DNA) in (A–C) PHH and (B–F) HepG2hNTCP cells infected with ΔHBx-HBV in the presence or not of 100 nmol/L pre-S1 mimicking peptide for up to 16 hours and then harvested at the indicated times points. cccDNA and tHBV-DNA quantification was normalized over β-globin quantity, while the relative 3.5-kb RNA amount was normalized over the housekeeping gene GUSb expression. (G) PHH and (H) HepG2hNTCP cells were transfected with siRNA against HIRA according to the experimental timeline shown in Figure 2B and inoculated for 16 hours with either WT or ΔHBx HBV. Cells were harvested for analysis 2 dpi. cccDNA amount was measured by qPCR, normalized over β-globin quantity, and expressed as a percentage of siCTL-treated cells. Graphs represent the means ± SEM of at least 3 independent experiments. The 2-tailed P value was calculated for a risk threshold of .05 using the 2/K sample permutation test with Monte Carlo resampling approximation. ∗P < .05 and ∗∗P < .01.
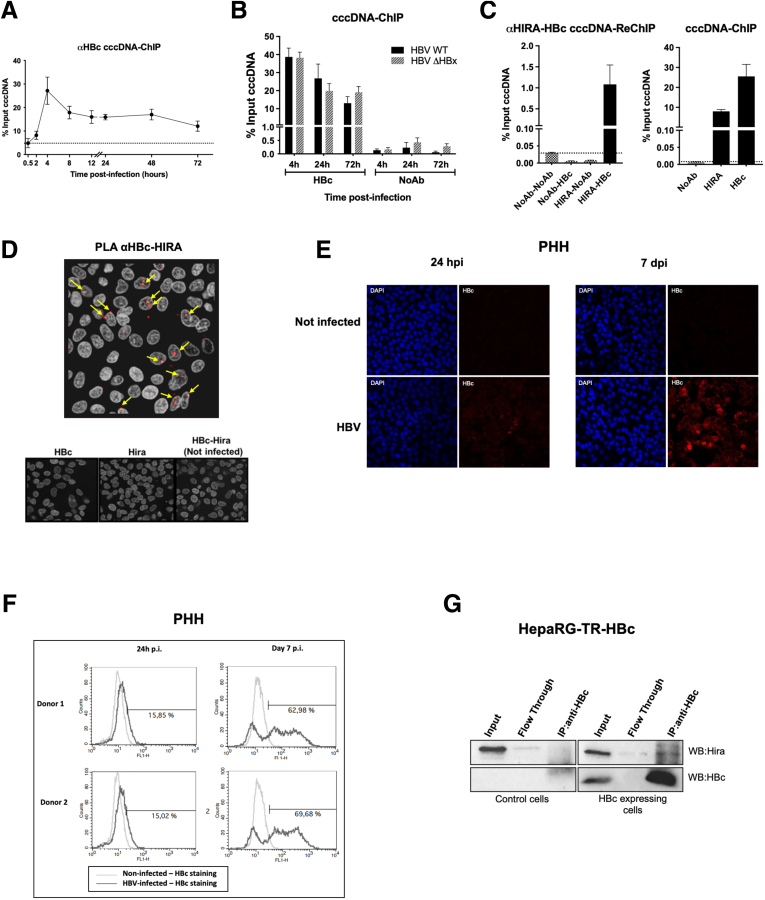
In experimental models of nuclear import,32,33 the naked nucleocapsids were shown to enter the nuclear pore to release the rcDNA, together with the viral capsid HBV core protein (HBc), into the nucleoplasm. Nonetheless, the role of HBc in cccDNA formation, if any, remains to be elucidated. First, we investigated the kinetics of HBc binding to cccDNA by ChIP-qPCR experiments, finding that HBc was bound to cccDNA at all time points investigated (Figure 5A). The binding was not affected when infection was performed with a ΔHBx mutant genome deficient for de novo viral protein expression, thus implying that HBc from the viral inoculum migrated to the nucleus and bound to cccDNA at early time points after infection (Figure 5B). Sequential ChIP experiments performed 24 hpi in HepG2hNTCP cells indicated that HBc and HIRA could bind to the same cccDNA molecule (Figure 5C). This co-occupancy is in line with proximity ligation assay (PLA) experiments evidencing a high proximity between the 2 proteins in HBV-infected PHHs at 24 hpi (Figure 5D), when approximately 15% of cells are positive for anti-HBc staining (Figure 5E and F). Moreover, using a HepaRG cell line expressing HBc (HepaRG-TR-HBc), we showed that HBc and HIRA not only can be located in close proximity but also can co-immunoprecipitate (Figure 5G). Taken together, these data exclude the need of HBV protein neosynthesis for HIRA-dependent cccDNA formation, but suggest a yet-to-be investigated involvement of the viral capsid protein in the early events leading to cccDNA formation.
Figure 5.
Incoming HBV core protein associates to cccDNA and HIRA early after infection. (A and B) HepG2hNTCP cells were infected with either WT or ΔHBx HBV for up to 16 hours and then extensively washed and cultured for the indicated time points before ChIP analysis using an antibody against HBc. The levels of HBc on cccDNA were analyzed through the infection kinetics by ChIP-qPCR and expressed as the percentage of input chromatin. cccDNA-ChIP qPCR using no antibody (NoAb) or anti-E2F antibody served as technical negative controls (Figure 8A–D), and the signal at 0.5 hpi was considered a specific qPCR background for cccDNA quantification (Figure 1F). (C) the simultaneous presence of HIRA and HBc on the same cccDNA molecule was assessed by sequential ChIP-qPCR 24 hpi using an antibody against HIRA first and then an antibody directed against anti-HBc for immunoprecipitation. NoAb-NoAb, NoAb-HBc, and HIRA-NoAb combinations of sequential immunoprecipitation served as negative controls, and IP with single HBc and HIRA served as positive controls. Graphs represent the means ± SEM of at least 3 independent experiments. (D) Proximity between HBc and HIRA was assessed by PLA in HBV-infected PHHs at 24 hpi. The PLA signal is indicated by arrows. Uninfected and infected PHHs stained with only HBc or HIRA antibodies were used as negative controls (lower panels). (E) Immunofluorescence and (F) flow cytometry analysis of HBc-positive PHHs at 24 hpi and 7 dpi. Immunofluorescence was performed with antibody against HBc (red) and nuclei are stained by 4′,6-diamidino-2-phenylindole (DAPI) signal (blue). (G) Western blot analysis of HIRA-HBc immunoprecipitation in HepaRG cells inducible for HBc expression. Immunoprecipitation was performed with an anti-HBc antibody, using HepaRG-TR-HBc noninduced cells as a control (left panel). Western blot with anti-HIRA antibody showed a specific band in the immunoprecipitated fraction in the presence of HBc (right panel).
HIRA Trimerization Is Required for cccDNA Formation
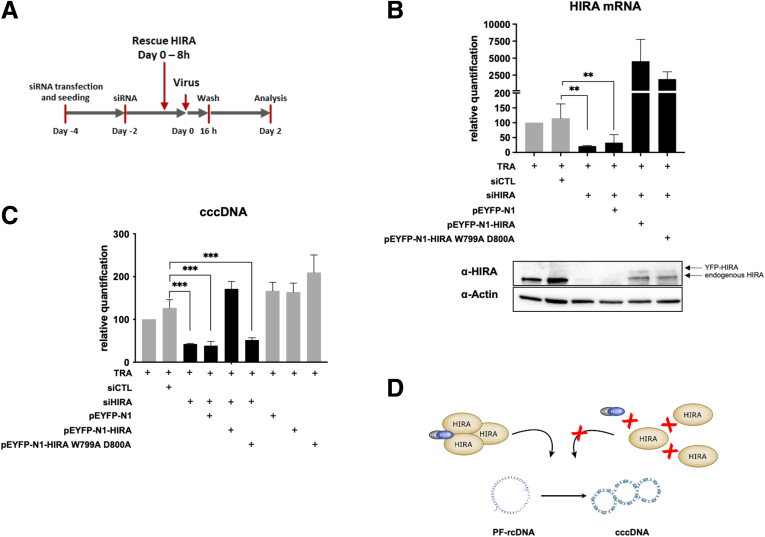
To further confirm the involvement of HIRA in histone loading during cccDNA formation, we rescued HIRA expression after silencing in HepG2hNTCP cells by transfecting a construct containing either a wild-type (WT) (pEYFP-N1-HIRA) or a HIRA mutated sequence (pEYFP-N1-HIRA W799A D800A) 8 hours before HBV infection (Figure 6A). The latter encodes a protein unable to homotrimerize, a property that is essential for new H3.3 deposition.16,34 As shown in Figure 6, reverse-transcription qPCR and Western blot analysis confirmed the restoration of HIRA messenger RNA levels and the appearance of the exogenous protein isoforms upon transfection (Figure 6B). In parallel, the cccDNA amount evaluated by qPCR went back to the level of nonsilenced controls when HIRA depletion was transcomplemented with wild-type HIRA, but not with the W799A D800A HIRA mutant (Figure 6C). These data argue for the requirement of H3.3 deposition function of the HIRA trimer during the cccDNA formation process in the nucleus of living hepatocytes (Figure 6D).
Figure 6.
HIRA trimerization is required for HBV minichromosome establishment. (A) HepG2hNTCP cells were transfected twice with siHIRA or siCTL and then transfected with plasmids encoding for either WT HIRA (pEYFP-N1-HIRA) or for a trimerization-incompetent HIRA mutant (pEYFP-N1-HIRA W799A D800A) before inoculation with HBV. The cells were harvested for analysis 2 dpi. (B) HIRA messenger RNA (mRNA) and protein expression after siRNA transfection and transcomplementation was determined by real-time qPCR and Western blot. β-actin served as Western blot loading control. HIRA mRNA was normalized over housekeeping GUSb gene expression and expressed as relative to the control treated only with the TRA. (C) cccDNA levels at 2 dpi were measured by qPCR. The cccDNA amount was normalized over β-globin quantity and then expressed as relative to the control treated only with TRA. (D) Schematic representation indicating that HIRA trimerization, required for H3.3 deposition, is necessary for full PF-rcDNA to cccDNA conversion in living infected hepatocytes. Graphs represent the means ± SEM of at least 3 independent experiments. The 2-tailed P value was calculated for a risk threshold of .05 using the 2/K sample permutation test with Monte Carlo resampling approximation. ∗∗P < .01, and ∗∗∗P < .001.
Histone Variant H3.3 Is Deposited on cccDNA During its Chromatinization
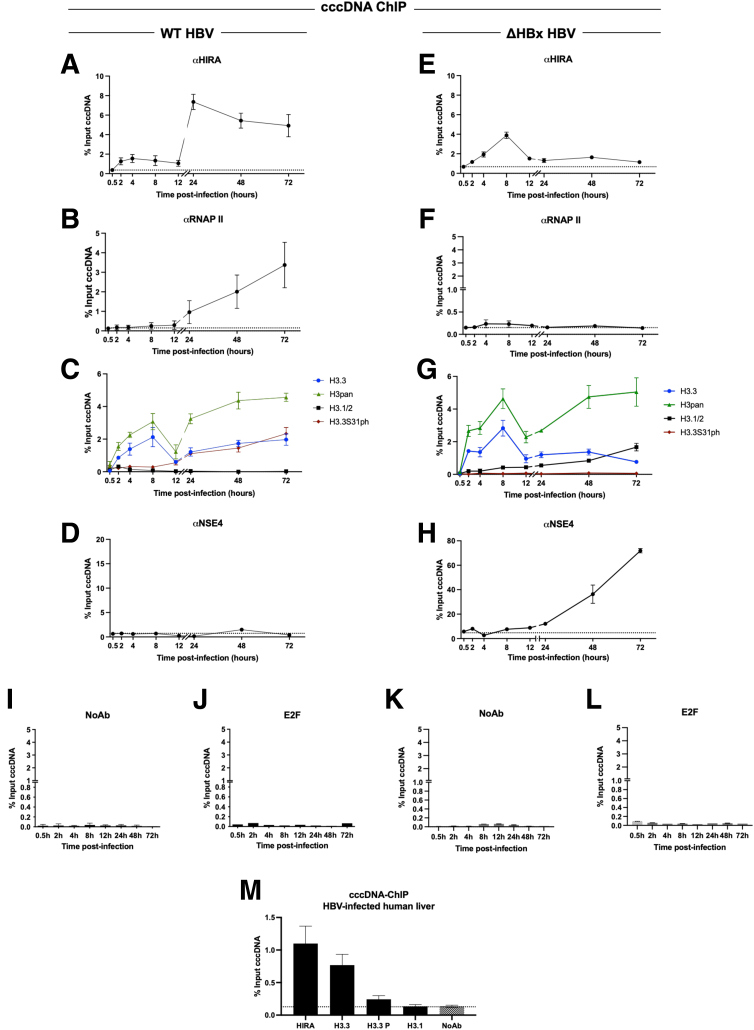
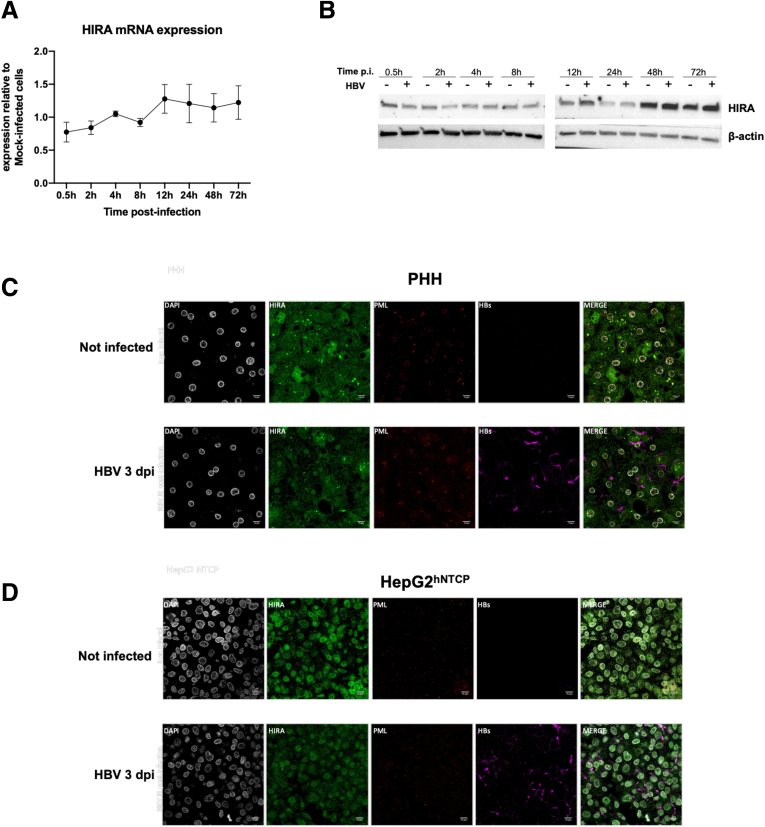
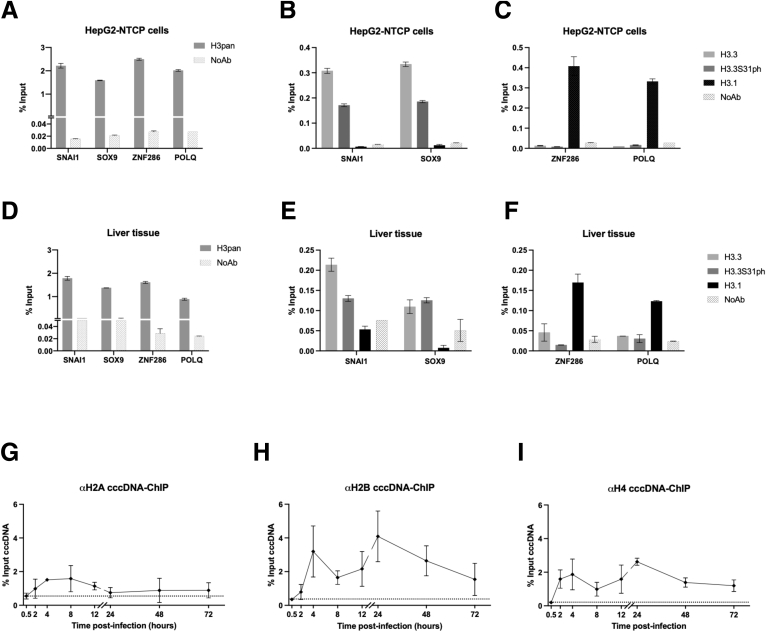
As shown in Figure 1, the appearance of cccDNA spans at least the first 72 hpi, precedes viral transcription by 24 hours, and overlaps with it afterward (Figure 1C, D, F, and G). HIRA cccDNA-ChIP experiments performed between 0.5 and 72 hpi in HepG2hNTCP cells showed that HIRA association to cccDNA was detected as soon as the HBV minichromosome was detected (ie, 2 hpi) (Figure 1F), remained stable until 12 hpi, and then increased (Figure 7A), in parallel with the recruitment of RNA polymerase II (RNAP II) on cccDNA (Figure 7B) and the beginning of viral transcription (Figure 1G). Importantly, cccDNA-ChIP enrichment appears specific, given that negative controls, including unrelated protein E2 Factor (E2F) or lack of primary antibody, did not generate any cccDNA signal (Figure 7I and J). HIRA increased binding to cccDNA was not correlated to any significant modulation of HIRA expression or localization in infected cells compared with uninfected ones (Figure 8). HIRA association with cccDNA thus appears to follow a 2-step behavior with a recruitment at 2 hpi that remains constant up to 12 hpi, followed by a sharp 6-fold increase at 24 hpi that lasts until 72 hpi. In line with HIRA ChIP signal, H3.3 was detected associated to cccDNA, also showing a 2-phase recruitment kinetic, with a peak at 8 hpi and then a progressive increase beginning at 12 hpi (Figure 7C). Across the same time points, no specific enrichment for H3.1/3.2 was detected (Figure 7C). ChIP signal from an antibody recognizing all H3 variants (H3pan) showed a cccDNA enrichment profile similar to that of H3.3, suggesting that the histone variant might represent most of H3 protein associated to cccDNA within 72 hpi. The specificity of H3.3 and H3.1/3.2 antibodies in ChIP was confirmed by analyzing preferential recruitment of H3.3 and H3.1/3.2 to their respective targets on host cell genome (Figure 9A–F). In parallel, nucleosomal core histones (H2A, H2B, and H4) also were found associated to cccDNA from 2 hpi onward (Figure 9G–I), further indicating nucleosome assembly on cccDNA.
Figure 7.
Dynamics of HIRA and H3.3 recruitment and phosphorylation of H3.3S31 during de novo cccDNA formation and transcription of established cccDNA pool. HepG2hNTCP cells were infected with (A–D) WT or (E–H) ΔHBx-HBV for up to 16 hours and then extensively washed and cultured for the indicated time points before ChIP-qPCR analysis using antibodies against (A and E) histone chaperone HIRA, (B and F) RNAP II, (C and G) total H3 (H3pan) and histone variants H3.3 and H3.1/2 and H3.3S31ph, and (D and H) SMC5/6 complex subunit NSE4. (I–L) cccDNA-ChIP qPCR using no antibody (NoAb) or anti-E2F antibody served as technical negative controls. The signal at 0.5 hpi was considered a specific qPCR background for cccDNA quantification (Figure 1F). (M) Snap-frozen liver samples from 3 chronically infected male patients were subjected to cccDNA-ChIP with antibodies against HIRA, H3.3, H3.3S31ph, and H3.1/2. NoAb immunoprecipitation served as ChIP negative control. Data are expressed as a percentage of enrichment with respect to initial input chromatin and represent the means ± SEM of at least 3 independent experiments.
Figure 8.
HIRA expression levels and cellular localization are not affected by HBV infection. HepG2hNTCP cells or PHHs were cultured in 2.5% DMSO containing medium for 72 hours, HBV infected at 250 viral genome equivalents/cell for up to 16 hours and then extensively washed and cultured for the indicated time points. HIRA messenger RNA (mRNA) and protein expression were analyzed throughout the infection kinetic by (A) real-time qPCR, (B) Western blot, and (C and D) immunofluorescence. HIRA mRNA levels were normalized over the housekeeping gene GUSb. β-actin signal served as loading control for Western blot analysis. Graphs represent the means ± SEM of at least 3 independent experiments. Immunofluorescence was performed with antibodies against HIRA (green), promyelocytic leukemia protein (PML, red), and HBV S protein (violet). Nuclei were stained by 4′,6-diamidino-2-phenylindole (DAPI) signal in grey.
Figure 9.
Recruitment of the nucleosome core histones onto cccDNA. ChIP-qPCR analysis of genomic target sites shown to be preferentially bound by either H3.3 (SNAI1 and SOX9 promoters) or H3.1/2 (ZNF286 promoter and POLQ gene body) by using antibodies recognizing all H3 variants (H3pan), H3.3, H3.3S31ph, and H3.1/2 in (A–C) HBV-infected HepG2hNTCP cells at 72 hpi and (D–F) human liver tissue derived from 3 chronically HBV-infected patients. No antibody (NoAb) immuniprecipitation served as ChIP technical negative control. (G–I) HepG2hNTCP cells were infected at 250 viral genome equivalents/cell for up to 16 hours and then extensively washed and cultured for the indicated time points before ChIP-qPCR analysis using antibodies against histones H2A (G), H2B (H) and H4 (I). Data are expressed as a percentage of enrichment with respect to initial input chromatin and represent the means ± SEM of at least 3 independent experiments.
Transcription of the Established cccDNA Pool Is Associated With the Recruitment of HIRA and Phosphorylation of H3.3S31
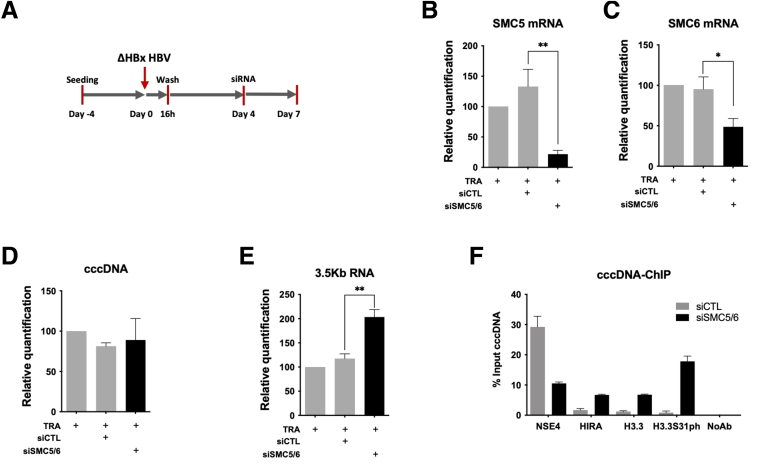
Recent data in an artificial transfection-based model of HBV replication indicated that exogenously expressed H3.3 affected HBV RNA levels.35 H3.3 tail differs from canonical histones H3.1/H3.2 for a serine in place of an alanine in position 31.19 Paramount evidence supports the possibility that H3.3S31 could regulate through phosphorylation key activities driving specific transcription programs.18, 19, 20 In our model of natural HBV infection, H3.3S31ph was detected by cccDNA-ChIP from 12 hpi onward and showed an enrichment kinetic superimposable to that of H3.3 (Figure 7C). To determine if H3.3 recruitment to cccDNA and serine 31 phosphorylation (S31ph) after 12 hpi could be linked to transcriptional activity, HepG2hNTCP cells were infected with the ΔHBx viral strain (Figure 4). Interestingly, kinetics of recruitment of HIRA and H3.3 did not differ significantly between WT and ΔHBx infections within the first 12 hpi (Figure 7A, C, E, and G) and, as expected, lack of viral transcription was associated with no specific enrichment of RNAP II on cccDNA (Figure 7F) and the appearance of the Structural maintenance of chromosomes (SMC)5/6 repressive complex31,36 on cccDNA derived from ΔHBx HBV, as shown by the recruitment of the subunit NSE431,36 (Figure 7H), which was barely associated to transcriptionally active cccDNA (WT HBV condition) (Figure 7D). However, in contrast to WT, we did not detect increased recruitment of HIRA and H3.3 in the ΔHBx condition, or specific H3.3S31ph after 12 hpi. Nevertheless, we could detect an increasing enrichment for H3.1/3.2 recruitment to cccDNA that was not detected in the WT (Figure 7G), as if a lack of H3.3 was compensated by H3.1/2 deposition. Negative controls, including unrelated protein E2F or lack of primary antibody, did not generate any cccDNA signal, confirming the specificity of cccDNA-ChIP enrichment (Figure 7K and L). These data suggest that HIRA is involved in both cccDNA formation in the very early phases after PF-rcDNA entry into the host cell and transcription from the established viral minichromosome pool via the deposition of H3.3, which was found to be phosphorylated on S31 on transcriptionally active cccDNA. This conclusion is supported by the observation that, after SMC5/6 complex knockdown and cccDNA transcription recovery, HIRA, H3.3, and H3.3S31ph recruitment to ΔHBx-HBV cccDNA was increased (Figure 10). Notably, the association between cccDNA, HIRA, and H3.3 in its S31 phosphorylated form was confirmed in vivo in liver tissues derived from chronic HBV-infected patients (Figure 7M).
Figure 10.
ΔHBx-HBV transcription recovery is associated to enhanced HIRA and H3.3 recruitment to cccDNA. (A) Experimental timeline for SMC5/6 knock-down after HBV infection. HepG2hNTCP cells were infected for 16 hours and then extensively washed and transfected with siSMC5 and siSMC6 or siCTL at 4 dpi. Cells were harvested for analysis at 7 dpi. Messenger RNA (mRNA) expression of (B) SMC5 and (C) SMC6 after siRNA transfection was determined by real-time qPCR. (D) cccDNA and (E) 3.5-kb RNA amount was measured by qPCR at 7 dpi. cccDNA quantification was normalized over β-globin quantity, while relative 3.5-kb RNA amount was normalized over the housekeeping gene GUSb expression. (F) cccDNA-ChIP analysis using anti-NSE4, HIRA, H3.3, and H3.3S31ph antibodies at 9 dpi in CTL vs SMC5/6-depleted conditions. NoAb served as ChIP technical negative control. Graphs represent the means ± SEM of at least 3 independent experiments. The 2-tailed P value was calculated for a risk threshold of .05 using the 2/K sample permutation test with Monte Carlo resampling approximation. ∗P < .05 and ∗∗P < .01.
HIRA Is Required for the Maintenance of cccDNA Transcriptional Activity
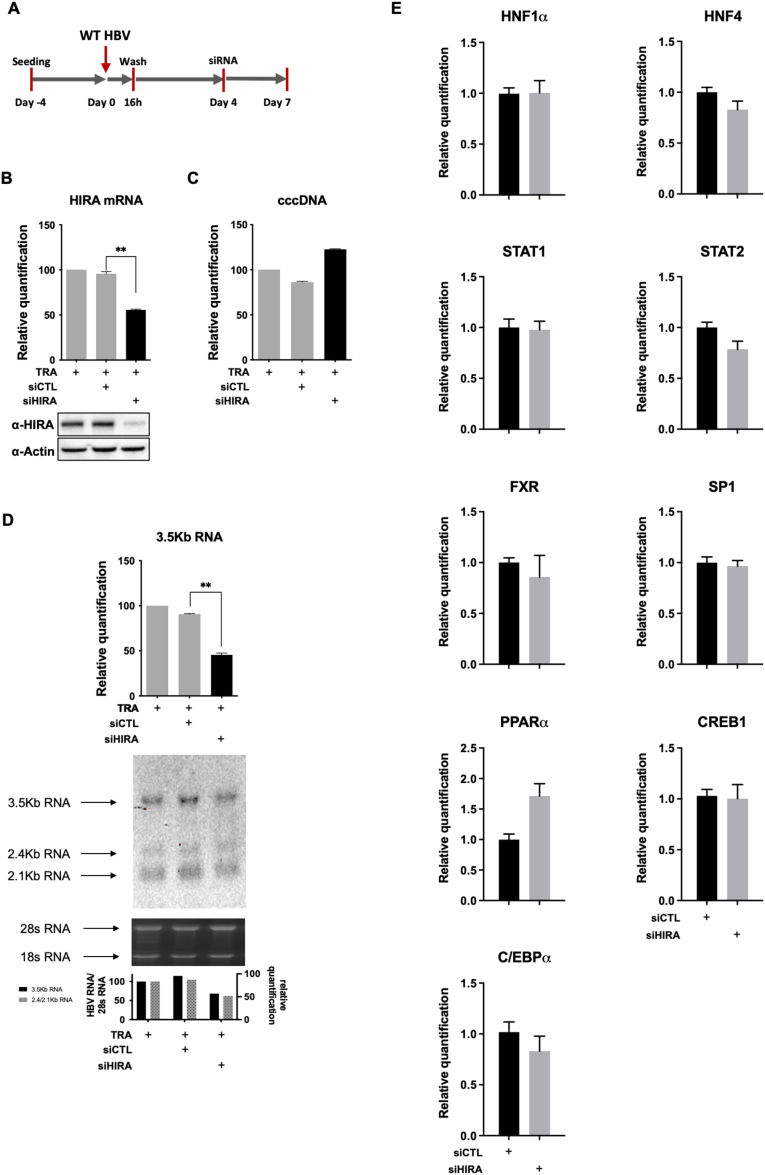
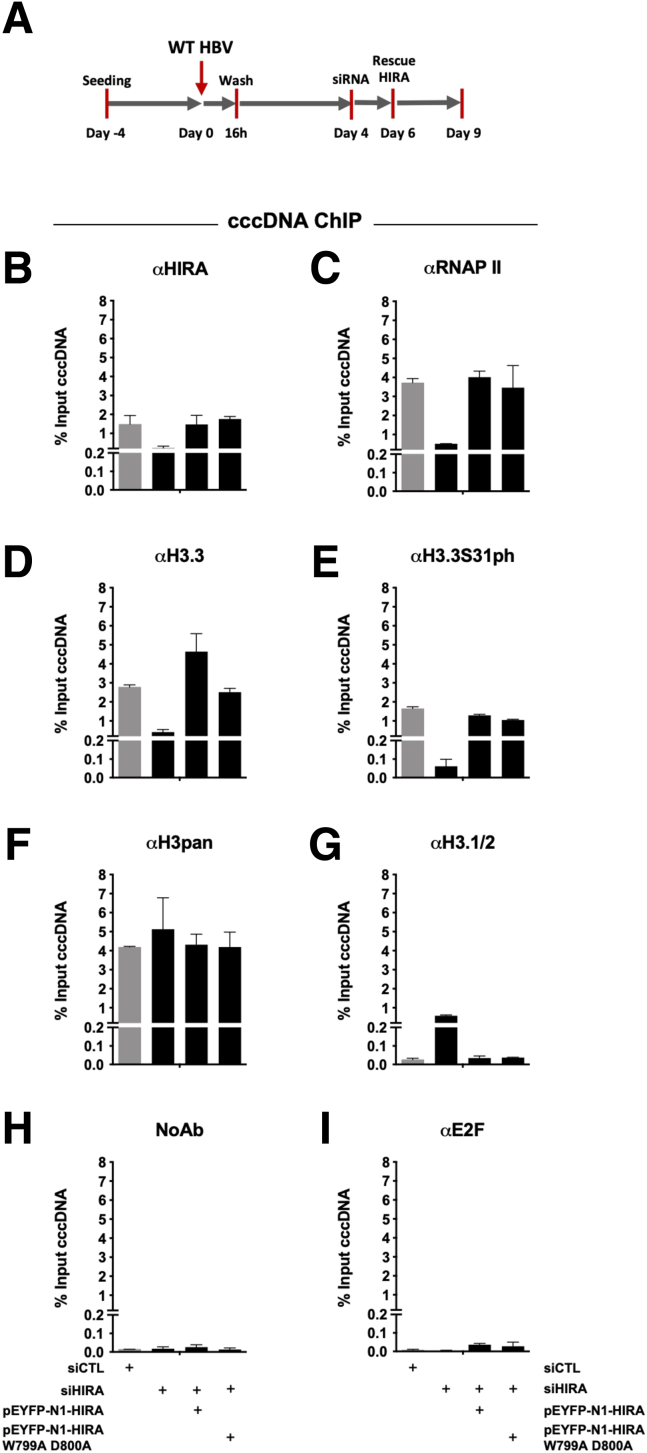
To further confirm HIRA involvement in cccDNA transcriptional activity, we depleted HIRA once the cccDNA pool was established (ie, 4 dpi)23,37 (Figure 11A). Down-regulation of HIRA RNA and protein expression (Figure 11B) did not affect the global levels of cccDNA (Figure 11C), but was associated with reduced HBV RNA levels, including 3.5-, 2.4-, and 2.1-kb species (Figure 11D), in the absence of any significant difference in the expression of known HBV-regulating transcription factors3 (Figure 11E). cccDNA-ChIP analysis indicated a decrease in cccDNA-associated H3.3, its S31 phosphorylated form, and cellular RNAP II levels in HIRA-depleted conditions (Figure 12A–E). Levels of cccDNA-associated H3pan were not affected, while H3.1/2 were slightly increased (Figure 12F and G). Interestingly, transcomplementation of HIRA with both WT and W799A D800A mutant constructs was able to restore cccDNA recruitment levels of HIRA, H3.3, RNAP II, and H3.3S31 phosphorylated forms similar to the Control siRNA (siCTL) condition (Figure 12B–G). Negative controls, including unrelated protein E2F or lack of primary antibody, did not generate any cccDNA signal, confirming the specificity of cccDNA-ChIP enrichment (Figure 12H and I). Overall, these results suggest that HIRA, but not its trimerization, is necessary to promote optimal viral transcription of the pool of already established cccDNA.
Figure 11.
HIRA is required for the maintenance of cccDNA transcriptional activity. (A) Experimental timeline for HIRA knock-down after HBV infection. HepG2hNTCP cells were infected for 16 hours and then extensively washed and transfected with siHIRA or siCTL at 4 dpi. Cells were harvested for analysis at 7 dpi. (B) HIRA messenger RNA (mRNA) and protein expression after siRNA transfection was determined by real-time qPCR and Western blot. β-actin served as Western blot loading control. (C) cccDNA and (D) 3.5-kb RNA amount was measured by qPCR and Northern blot at 7 dpi. cccDNA quantification was normalized over β-globin quantity, while relative 3.5-kb RNA amount was normalized over the housekeeping gene GUSb expression. (E) HepG2hNTCP cells were transfected with siRNA against HIRA according to the timeline shown in Figure 2B and inoculated for 16 hours with HBV at 250 viral genome equivalents/cell. The cells were harvested for analysis at 7 dpi. mRNA levels of hepatocyte nuclear factor-1α (HNF1α), hepatocyte nuclear factor-4α (HNF4α), signal transducer and activator of transcription 1 and 2 (STAT1, STAT2), nuclear receptor farnesoid X (FXR), specificity protein 1 (SP1), peroxisome proliferator activated receptor α (PPARα), CAMP responsive element binding protein 1 (CREB1), and CCAAT/enhancer-binding protein α (C/EBPα) were quantified by real-time qPCR assay and expressed as a percentage of TRA-treated cells after normalization over GUSb housekeeping gene expression. Data represent the means ± SEM of at least 3 independent experiments. Graphs represent the means ± SEM of at least 3 independent experiments. The 2-tailed P value was calculated for a risk threshold of .05 using the 2/K sample permutation test with Monte Carlo resampling approximation. ∗∗P < .01.
Figure 12.
HIRA is required for recruitment of H3.3, RNAP II, and H3.3S31ph to established cccDNA. (A–I) cccDNA-ChIP analysis at 9 dpi in CTL vs HIRA-depleted and transcomplemented conditions (WT or W799A D800A mutant constructs). cccDNA ChIP was performed with (B) antibodies against HIRA, (C) cellular RNAP II, (D) H3.3, and its (E) phosphorylated form H3.3S31ph, (F) H3pan, and (G) H3.1/2. (H and I) No antibody (NoAb) or anti-E2F antibody served as ChIP technical negative controls. Graphs represent the means ± SEM of at least 3 independent experiments.
Discussion
The HBV cccDNA must adopt a chromatin structure similar to that of the host cell chromosomes for its transcription6,38,39 in the infected hepatocyte nuclei. Coupling Southern blot and ChIP-qPCR analysis, we found that histone loading occurred simultaneously with repair of the partially double-stranded cccDNA precursor (PF-rcDNA). Although our data were generated from a population of cccDNA containing infected hepatocytes, we can assume that the breaks into PF-rcDNA strands, evoking a target for the DNA repair machinery, represent a trigger for HIRA recruitment enabling subsequent histone deposition coupled to DNA repair, in agreement with the role of HIRA in priming damaged chromatin early after damage recognition.40
Previous work conducted in cell-free systems have reproduced steps leading from PF-rcDNA to supercoiled cccDNA without the need for histone chaperones.8,9 Interestingly, we showed that in infected hepatocytes, a defective H3.3 deposition owing to HIRA depletion or the lack of HIRA homotrimerization severely impacted the appearance of supercoiled cccDNA. Therefore, it can be argued that nucleosome formation is not required for the conversion of PF-rcDNA to a complete double-stranded circular DNA molecule per se, but that HIRA (and H3.3 deposition) is required for the stabilization of the HBV genome as an episome in the nucleus of living hepatocytes. This also suggests that HIRA is involved in preventing degradation of naked DNA and/or shaping the viral genome into a chromatin structure that can be handled by the host transcriptional machinery for the benefit of the viral genome. It will be interesting to determine the kinetics of events on single cccDNA molecules, when the yet unresolved technical limitations in analyzing single HBV genomes in infected nuclei will be overcome.
In our experimental conditions, from 24 hpi onward, formation of new cccDNA molecules and transcription occurred concomitantly. The use of a modified HBV lacking the regulatory HBx protein (ΔHBx) resulting in transcriptionally inactive cccDNA in infected hepatocytes30,31 provided evidence for transcription-coupled HIRA recruitment to established cccDNA and for the association between H3.3 S31 phosphorylation and active viral transcription. H3.3S31ph recently was related to increased in-cis acetylation along with a decrease of trimethylation of its lysine in position 27, thus providing chromatin access to regulatory factors at selected transcribed regions.19 Notably, cccDNA-associated H3 histone tails have been shown to be highly acetylated on K27, while no specific trimethylation was detected in infected hepatocytes and human liver samples.39 Our results highlighted a preferential deposition of H3.3 over H3.1/2 on cccDNA in liver samples from HBV-infected patients. Whether this would be instrumental for H3.3S31ph to happen and to regulate access to the viral genome of either viral or host cell chromatin regulators warrants further investigation. The demonstration that HIRA depletion decreased H3.3, its S31 phosphorylated form, and RNA Polymerase II enrichment on established cccDNA and affected viral RNA production further suggests a pivotal role for HIRA in maintaining cccDNA active transcription. Interestingly, homotrimerization of HIRA does not seem to be required for its binding to established cccDNA, suggesting a possible difference in HIRA-associated protein partners during de novo H3.3 deposition vs recycling on cccDNA, as already observed on the human genome. However, we cannot rule out a possible involvement of other histone chaperones in cccDNA activity at later time points, as indicated for CAF-1 by Yang et al41 at 12 days after infection.
Differently from the observation made with herpes viruses and cytomegalovirus,17,42 we did not see any specific HIRA relocalization to promyelocytic leukemia bodies after HBV infection either in HepG2hNTCP cells or in primary human hepatocytes (Figure 8C and D). This is consistent with the observation that HBV infection might be established without significant perturbation of host cell innate immune response.43 Indeed, the HBV genome remains shielded from cytoplasmic innate immune sensors thanks to its capsid shell, which shuttles the viral DNA until the inner face of the nuclear basket.33 Given the association of the HBV capsid protein (HBc) to cccDNA at all investigated time points and its close proximity with HIRA in infected cells, it is tempting to speculate that HBc may be required by HBV to evade host cell immune defense mechanisms before the initial chromatinization of the viral genome and might participate in the building of the cccDNA nucleosomal structure together with H3.3 deposited by the HIRA complex. Whether HBc is required for HIRA recruitment onto incoming HBV genome remains to be investigated. Thanks to its C-terminal arginine-rich domain, HBc could be assimilated to protamine-like protein VII of adenoviruses,44 but instead of being replaced by host cell histones on the viral genome, HBc would integrate into the HBV chromatin structure to regulate nucleosomal spacing.38
Altogether, our results indicate that the HIRA complex is a crucial proviral factor for HBV infection, determining the formation of its stable viral minichromosome and sustaining its transcriptional activity by promoting H3.3 deposition and recycling (Figure 13). Detection of H3.3S31ph bound to active cccDNA molecules opens exciting perspectives for the investigation of the role of the host chromatin regulatory machinery in determining the fate of HBV infection.
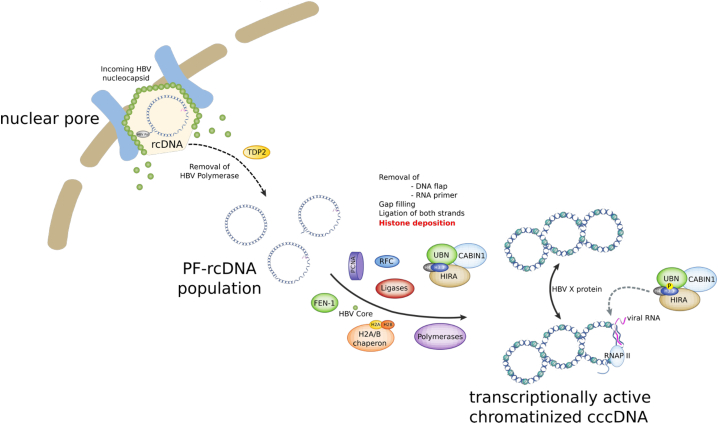
Figure 13.
Schematic representation of HIRA involvement in cccDNA formation and transcriptional activity in infected hepatocytes. Once entered in the cell, the HBV nucleocapsid is shuttled to the nuclear pore, where the viral genome, the naked, partially double-stranded rcDNA covalently attached to the HBV polymerase, is released in the nucleoplasm together with the HBV core protein (HBc). Host DNA repair cellular machinery components, comprising tyrosyl-DNA phosphodiesterase-2 (TDP2), flap structure-specific endonuclease 1 (FEN-1), replication factor C (RFC), proliferating cell nuclear antigen (PCNA), translesion DNA polymerases, topoisomerases, and components of the ATR-CHK1 pathway are heavily involved in the biological reactions leading to viral polymerase and RNA primer eviction from rcDNA, as well as in viral DNA strand completion and ligation. The HIRA complex, through de novo H3.3 deposition, first ensures the building of HBV genome chromatin structure, allowing the establishment of the cccDNA pool, and then contributes to cccDNA active transcription, which is associated to H3.3 phosphorylation on S31. CABIN1, Calcineurin Binding Protein 1; UBN, Ubinuclein 1.
Methods
Production of WT and Mutated HBV Viral Inoculum
HBV (genotype D, subtype ayw) inoculum was prepared from filtered HepAD3845 (WT HBV) or HepG2-H1.3-K6 cells (ΔHBx-HBV)30 supernatants by polyethylene-glycol-MW-8000 (PEG8000; Sigma, St. Louis, MO) precipitation (8% final) as previously described.46 Viral stock with a titer reaching at least 1 × 1010 viral genome equivalents/mL was tested endotoxin free and used for infection. HepG2-H1.3-K6 cells were established by stable integration of a 1.3-fold HBV genome (genotype D, subtype ayw) carrying premature stop codon mutations at amino acid position 7 after the ATG in both the 5’ and 3’ HBx open reading frames, without affecting the other viral open reading frames (S, precore, core, and polymerase).
PHH Isolation
PHHs were isolated from surgical liver resections, after informed consent of patients (institutional review board agreements DC-2008-99 and DC-2008-101) as previously described47 and plated in complete William's medium supplemented with 1% penicillin/streptomycin (Life Technologies, Carlsbad, CA), 1% glutamine (Life Technologies), 5 μg/mL human insulin (Sigma-Aldrich, St. Louis, MO), 25 μg/mL hydrocortisone hemisuccinate UPJOHN (SERB, Paris, France), and 5% fetal calf serum (FCS) (Fetalclone II; PERBIO, Helsingborg, Sweden). PHHs were maintained in William’s medium supplemented with 1.8% DMSO (Sigma-Aldrich) and infected with HBV within 72 hours after plating.
Cell Culture and HBV Infection
HepG2hNTCP cells were seeded at 105 cells/cm2 in Dulbecco’s modified Eagle medium (DMEM) supplemented with penicillin (Life Technologies), streptomycin (Life Technologies), sodium pyruvate (Life Technologies), and 5% FCS (Fetalclone II). The day after, medium was renewed and complemented with 2.5% DMSO (Sigma). After 72 hours, cells were infected at a multiplicity of infection of 250 in the presence of 4% PEG8000 for up to 16 hours and then extensively washed and cultured for the indicated time points in complete DMEM containing 2.5% DMSO until harvesting. HepaRG-TR-HBc cells were maintained in complete William's medium supplemented with 1% penicillin/streptomycin (Life Technologies), 1% glutamine (Life Technologies), 5 μg/mL human insulin (Sigma-Aldrich), 25 μg/mL hydrocortisone hemisuccinate (UPJOHN, SERB, Paris, France), and 5% FCS (Fetalclone II). Tetracycline was added to the medium for 72 hours to induce HBc protein expression. BrdU was purchased from Sigma (B9285) and used at a concentration of 20 μmol/L to treat HepG2hNTCP cells 24 hours before HBV infection. Myristoylated preS1-myr peptide (sequence: GTNLSVPNPLGFFPDHQLDPAFRANSNNPDWDFNPNKDHWPEANKVG, synthesized by GeneScript) was used at 100 nmol/L to prevent virus entry into the cells.48,49
Analysis of Viral Parameters During Replication
Total DNA was purified from infected cells using the MasterPure Complete DNA Purification Kit (Lucigen, MIddleton, WI) and quantified using the TaqMan Gene Expression assay (ID: Pa03453406_s1). To increase the specificity of HBV cccDNA detection, qPCR was preceded by a nuclease digestion using 10 U of T5 exonuclease (Lucigen) for 500 ng total DNA. Selective cccDNA qPCR was performed using primers and probes spanning the gap region in the HBV rcDNA (Table 1).50,51 Serial dilutions of an HBV monomer plasmid (pHBV-EcoRI) were used as standard for quantification. For normalization, the number of human hepatocytes was estimated by measuring human β-globin copies (TaqMan assay ID: Hs00758889_s1) while human genomic DNA (Roche Applied Science, Mannheim, Germany) was used as a standard curve for quantification.
Table 1.
Primers and Probes Sequences Used for TaqMan and SYBRgreen qPCR
| Target | Sequence |
|---|---|
| cccDNA_for | CCGTGTGCACTTCGCTTCA |
| cccDNA_rev | GCACAGCTTGGAGGCTTGA |
| cccDNA_probe | [6FAM]CATGGAGACCACCGTGAACGCCC[BBQ] |
| 3.5-kb RNA_for | GGAGTGTGGATTCGCACTCCT |
| 3.5-kb RNA_rev | AGATTGAGATCTTCTGCGAC |
| 3.5-kb RNA_probe | [6FAM]AGGCAGGTCCCCTAGAAGAAGAACTCC[BBQ] |
| HIRA_for | GGCCTCGGAAGGACTCTC |
| HIRA_rev | AGACAGACACATGGCCTCCT |
| NTCP_for | GAAGGACAAGGTGCCCTATAAA |
| NTCP_rev | GGATTTGAGGACGATCCCTATG |
| EXO1_for | TGAGGAAGTATAAAGGGCAGGT |
| EXO1_rev | AGTTTTTCAGCACAAGCAATAGC |
| TDP2_for | TGGAGTTTGCCTCGGTCGCA |
| TDP2_rev | TGGTTTCAGGTCGGCGTTCCA |
| FEN1_for | ATGACATCAAGAGCTACTTTGGC |
| FEN1_rev | GGCGAACAGCAATCAGGAACT |
| POLk_for | TGAGGGACAATCCAGAATTGAAG |
| POLk_rev | CTGCACGAACACCAAATCTCC |
| SNAI1 promoter_for | CGCTCCGTAAACACTGGATAA |
| SNAI1 promoter_rev | GCACATCACTGGGGAGGAAG |
| SOX9 promoter_for | CAGGAGGCAAAGACCAAAAC |
| SOX9 promoter_rev | CACATCGACCTTGAGCTCTG |
| ZNF268 promoter_for | CCTGTTGATCCTGCTCTTCTG |
| ZNF268 promoter_rev | GAATAAAGACCCCTTGGATTCAG |
| POLQ gene body_for | GCAGTCCCTTACTGGCAATG |
| POLQ gene body_rev | TGGAGAGTGAGAACCCCTTC |
| STAT1_for | GTGGAAAGACAGCCCTGCAT |
| STAT1_rev | ACTGGACCCCTGTCTTCAAGAC |
| STAT2_for | CCCCATCGACCCCTCATC |
| STAT_rev | GAGTCTCACCAGCAGCCTTGT |
| Sp1_for | GGTGGGCAGTATGTTGTGG |
| Sp1_rev | CCAGGTAGTCCTGTCAGAACTT |
| PPAR_for | ACGCGAGAATGAGGCAGATCCCA |
| PPARa_rev | GGCCTGTGTGGCAGTTCCTACT |
| FXR (NR1H4)_for | GTGAGGGGTGTAAAGGTTTCT |
| FXR (NR1H4)_rev | GCCTGTATACATACATTCAGCCA |
| HNF4a_for | TCTTACGATTTAGCCGGCAGT |
| HNF4a_rev | TTCCTTCTTCATGCCAGCCC |
| HNF1a_Ex2_for | CTCAACCAGTCCCACCTGTC |
| HNF1a_Ex3_rev | GCTCTTCAATCAGCCCTCCC |
| CREB_for | GTGTGTTACGTGGGGGAGAG |
| CREB_rev | GCATCTCCACTCTGCTGGTT |
| CEBP γ_for | TTTCGTAACCGTCGCTCCTC |
| CEBP γ_rev | ATGTTGTTCCTCTCTCGGCG |
| SMC6_Ex14_for | AGCAACGCAACTTGAACAGAT |
| SMC6_Ex15_rev | TCTCTACACACTGGCGCTT |
| SMC5_Ex3_for | GAGGATGTTCTAGAGGCATGGT |
| SMC5_Ex4_rev | AGGCTGCAACTTTCTCTTC |
CEBP, CCAAT/enhancer-binding protein; CREB, CAMP responsive element binding protein; FEN1, flap structure-specific endonuclease 1; for, forward; FXR, nuclear receptor farnesoid X; HNF, hepatocyte nuclear factor; POLk, DNA polymerase kappa; PPAR, peroxisome proliferator activated receptor; rev, reverse; SMC, Structural maintenance of chromosomes; SNAI1, Snail Family Transcriptional Repressor 1; SOX9, SRY-Box Transcription Factor 9; Sp1, specificity protein 1; STAT, signal transducer and activator of transcription; TDP2, tyrosyl-DNA phosphodiesterase-2; ZNF, zinc finger protein.
Total RNA was extracted from infected cells using the NucleoSpin RNA kit (Macherey-Nagel, Düren, Germany) and retrotranscribed using SuperScript III reverse transcriptase according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). A total of 3.5-kb viral RNA (comprising pgRNA and precore RNA) was amplified using previously described primers and probes (Table 1).51 The expression of the human housekeeping gene GUSb (TaqMan assay ID: Hs00939627_m1) was used for normalization. Real-time PCRs were performed using an Applied QuantStudio 7 machine and TaqMan Advanced Fast Master Mix or SYBR Green Master Mix (ThermoFisher, Waltham, MA).
Cytotoxicity Assays
Neutral red uptake assay and sulforhodamine staining were performed to assess cell viability after siRNA transfection and HIRA overexpression. For the neutral red uptake assay, the protocol was adapted from Repetto et al.52 After neutral red uptake assay, cells were washed 3 times with deionized water, dried, and processed for sulforhodamine staining.53
Plasmid and siRNA Cell Transfection
pEYFP-N1-HIRA and pEYFP-N1-HIRA W799A D800A constructs were kindly provided by Ray-Gallet et al16 and were transfected in HepG2NTCP cells and PHH using TransIT-2020 (Mirus Bio LLC, Madison, WI) following the manufacturer’s protocol. siRNA targeting HIRA (ON-TARGETplus HIRA siRNA GGAUAACACUGUCGUCAUC)40 and the corresponding siRNA mutated in position 9–11 (siRNA CTL: GGAUAACAGACUCGUCAUC) to serve as negative control were transfected at 10 nmol/L concentration in HepG2hNTCP cells and PHH using Lipofectamine RNAiMAX reagent (ThermoFisher), following the manufacturer’s instructions. siRNA against human SMC5 (ON-TARGETplus SMARTPool L-014117-01; Horizon Discovery, Waterbeach, UK) and SMC6 (ON-TARGETplus SMARTPool L-018408-01; Horizon Discovery) were transfected at a 10 nmol/L concentration in HepG2hNTCP cells using Lipofectamine RNAiMAX reagent (ThermoFisher), in parallel with ON-TARGETplus nontargeting pool (D-001810-10; Horizon Discovery) as negative control.
Western Blot
Cells were lysed in RIPA buffer supplemented with protease inhibitor cocktail (PIC) 1× and phenylmethylsulfonyl fluoride (PMSF) 1×. Proteins were migrated in 4%–20% mini-PROTEAN TGX stain-Free Precast Gel (Bio-Rad Laboratories, Hercules, CA) and transferred onto a nitrocellulose membrane (Bio-Rad Laboratories). Blots were blocked for 1 hour with 5% milk in 1× Tris buffered saline (Sigma) and stained with primary antibody in blocking buffer overnight at 4°C. After primary antibody incubation, blots were washed 3 times with 1× Tris buffered saline with 0.1% Tween 20, stained with horseradish peroxidase–conjugated secondary antibodies for 1 hour at room temperature, and washed again 3 times with 1× Tris buffered saline with 0.1% Tween 20. Detection occurred using the Bio-Rad Clarity Western ECL and the ChemiDoc XRS system (Bio-Rad). The antibodies used are listed in Table 2.
Table 2.
List of Antibodies Used in this Study
| Target | Company | Dilution |
|---|---|---|
| HBc for PLA and ChIP | Invitrogen SC2362651, Waltham, MA | 4 μg |
| HBc for IF and FACS | Invitrogen MA1-7607, Waltham, MA | 1:500 |
| HIRA for PLA, ChIP, and WB | Abcam Ab20655, Cambridge UK | 2.5 μg/1:500 |
| H3.3 for ChIP | Abcam Ab62642, Cambridge UK | 5 μg |
| H3.3S31ph for ChIP | Abcam ab92628, Cambridge UK | 5 μg |
| RNA polymerase II for ChIP | Diagenode C15200004, Liège, Belgium | 1 μg |
| BrdU for ChIP | BD Pharmingen 555627, San Diego, CA | 5 μg |
| E2F for ChIP | Santa Cruz Biotechnology, Dallas, TX | 5 μg |
| β-actin for WB | Abcam Ab6276, Cambridge UK | 1:10,000 |
| H3pan for ChIP | Diagenode C15410324, Liège, Belgium | 2 μg |
| H2A for ChIP | Diagenode C15410166, Liège, Belgium | 2 μg |
| H2B for ChIP | Diagenode C15410157, Liège, Belgium | 1 μg |
| H4 for ChIP | Diagenode C15410156, Liège, Belgium | 2 μg |
| H3.1/3.2 for ChIP | Active motif AB_2793710, Carlsbad, CA | 4 μg |
| HBc for WB | Dako B0586, Santa Clara, CA | 1:500 |
| HBs for IF | Abcam ab8636, Cambridge UK | 1:100 |
| Goat anti-mouse IgG (H+L) secondary antibody, HRP | Invitrogen 62-6520 Waltham, MA | 1:20,000 |
| Goat anti-rabbit IgG (H+L) secondary antibody, HRP | Invitrogen 31460, Waltham, MA | 1:10,000 |
| PML for IF | Abcam ab179466, Cambridge UK | 1:500 |
| NSE4 for ChIP | Abcepta AP9909a, San Diego, CA | 4 μg |
FACS, Fluorescence-activated cell sorting; H+L, heavy and light chains of the IgG molecule; HBs, hepatitis B surface; HRP, horseradish peroxidase; IF, immunofluorescence; PML, promyelocytic leukemia; WB, western blotting.
Northern Blot
Purified RNA was denatured at 50°C for 1 hour with glyoxal reagent (Life Technologies) and then subjected to electrophoresis through 1× phosphate buffer in a 1.2% agarose gel and transferred to a positively charged nylon membrane (Amersham N+; GE, Dornstadt, Germany). Membrane-bound RNA was hybridized overnight at 42°C to digoxigenin (DIG)-labeled HBV-specific probes (Table 3, forward probes 1–8). The membrane was washed twice in low-stringency wash buffer (1× saline-sodium citrate [SSC], 0.1% sodium dodecyl sulfate [SDS]) for 30 minutes at room temperature and twice in high-stringency wash buffer (0.1× SSC, 0.1% SDS) for 30 minutes at 65°C. Detection was performed using anti-DIG alkaline phosphatase (1:20,000 dilution) and CDP-Star reagent (Roche) according to the manufacturer’s recommendation and imaged using the ChemiDoc MP imaging system (Bio-Rad). 18S and 28S ribosomal RNA signal was used as loading and quality control.
Table 3.
DIG-Labeled HBV DNA Probe Sequences
| Target | Name | Sequence |
|---|---|---|
| HBV | HBV-F1 | TAGCGCCTCATTTTGTGGGT |
| HBV-R1 | CTTCCTGTCTGGCGATTGGT | |
| HBV-F2 | TAGGACCCCTTCTCGTGTTA | |
| HBV-R2 | CCGTCCGAAGGTTTGGTACA | |
| HBV-F3 | ATGTGGTATTGGGGGCCAAG | |
| HBV-R3 | GGTTGCGTCAGCAAACACTT | |
| HBV-F4 | TGGACCTTTTCGGCTCCTC | |
| HBV-R4 | GGGAGTCCGCGTAAAGAGAG | |
| HBV-F5 | GTCTGTGCCTTCTCATCTG | |
| HBV-R5 | AGGAGACTCTAAGGCTTCC | |
| HBV-F6 | TACTGCACTCAGGCAAGCAA | |
| HBV-R6 | TGCGAATCCACACTCCGAAA | |
| HBV-F8 | AGACGAAGGTCTCAATCGCC | |
| HBV-R8 | ACCCACAAAATGAGGCGCTA |
Hirt Extraction and Southern Blot Analysis
DNA was extracted following an adapted Hirt procedure.21 A total of 90 ug DNA was subjected to Southern blot analyses using a mix of DIG-labeled probes (synthesized using primers listed in Tables 3 and 4 and the PCR DIG probe synthesis kit [Roche]), an Alkaline phosphatase-conjugated anti-DIG antibody (Roche) and CDP-Star (Roche) according to the manufacturer’s instructions. Mitochondrial DNA detection served as internal loading control.
Table 4.
DIG-Labeled Mitochondrial DNA Probe Sequences
| Target | Name | Sequence |
|---|---|---|
| Mitochondrial DNA | Forward-human NADH dehydrogenase 1 | CCCTACTTCTAACCTCCCTGTTCTTAT |
| Reverse-human NADH dehydrogenase 1 | CATAGGAGGTGTATGAGTTGGTCGTA | |
| Forward-human NADH dehydrogenase 5 | ATTTTATTTCTCCAACATACTCGGATT | |
| Reverse-human NADH dehydrogenase 5 | GGGCAGGTTTTGGCTCGTA | |
| Forward-human ATP synthase membrane subunit 6 | CATTTACACCAACCACCCAACTATC | |
| Reverse-human ATP synthase membrane subunit 6 | CGAAAGCCTATAATCACTGTGCC |
Immunofluorescence
Cells were fixed with 4% paraformaldehyde and washed with phosphate-buffered saline (PBS) 1×. After permeabilization in 0.5% Triton X-100 (Sigma Aldrich, St. Louis, MO), the cells were blocked with 3% bovine serum albumin (BSA) and then incubated with HIRA, promyelocytic leukemia, hepatitis B surface, or hepatitis B core protein antibody overnight (Table 2). The next day, the cells were incubated with secondary antibody anti-rabbit IgG Alexa Fluor (488/555/594/647) (dilution 1:1000; Invitrogen). Coverslips were mounted onto slips with Duolin In Situ Mounting Medium with 4′,6-diamidino-2-phenylindole (DUO82040-5ML; Sigma). Images were acquired with the Zeiss (Jena, Germany) LSM 780 NLO confocal microscope.
PLA
PLA was performed on infected HepG2hNTCP cells according to the manufacturer’s instructions (DUO92014, DUO92004, DUO92002, and DUO82040; Duolink; Sigma-Aldrich, St Louis, MO). Briefly, after fixation by 4% paraformaldehyde for 30 minutes at room temperature, and quenching by 1 mol/L glycine, cells were permeabilized for 30 minutes at room temperature with PBS-Triton 0.3%, and then blocked for 30 minutes at 37°C with blocking buffer. Cells were incubated with the 2 primary antibodies overnight at 4°C (Table 2). PLA probes were diluted and added to the coverslips for 1 hour at 37°C. After a ligation step of 30 minutes at 37°C, the amplification was performed during 100 minutes at 37°C. Finally, coverslips were mounted onto slips with Duolink In Situ Mounting Medium with 4′,6-diamidino-2-phenylindole (DUO82040-5ML; Sigma). Images were acquired with the Zeiss LSM 780 NLO confocal microscope.
Flow Cytometry Analysis
After trypsinization, cells were pelleted and washed in PBS 1× and then fixed in 4% formaldehyde (pH 7.4) in PBS 1× for 10 minutes at room temperature. After centrifugation, permeabilization was performed in PBS supplemented with 0.1% Triton X-100 and 3% BSA. Cell pellets were collected and resuspended in PBS supplemented with 3% BSA to proceed with the immunostaining. The primary antibody (Table 2) was added directly into the PBS/BSA 3% solution for 60 minutes on ice. PBS supplemented with 0.1% Triton X-100 and 3% BSA was used to wash the cells 2 times before centrifugation and incubation with the fluorochrome-conjugated secondary antibody (Table 2) for 30 minutes on ice in the dark. After 2 washes in PBS supplemented with 0.1% Triton X-100 and 3% BSA, the cells were resuspended in ice-cold PBS, 3% BSA, and 1% sodium azide, and analyzed directly on the flow cytometer (FacsCalibur; BD Biosciences, San Jose, CA).
ChIP
ChIP experiments were performed at the indicated time points (from 30 minutes to 72 hours) postinfection as previously described.51 Briefly, after a 15-minute cross-linking step with 1% formaldehyde at 37°C, nuclei were extracted in lysis buffer (5 mmol/L PIPES, 85 mmol/L KCl, 0.5% NP-40, 1 mmol/L PMSF, and 1× PIC) and resuspended in sonication buffer (1% SDS, 10 mmol/L EDTA, 50 mmol/L Tris-HCl pH 8, 1 mmol/L PMSF, and 1× PIC) after centrifugation. After sonication and a preclearing step, chromatin was subjected to overnight immunoprecipitation at 4°C using 2–5 μg of antibodies indicated in Table 2 or no antibody. Immune complexes then were incubated for 2 hours with protein G agarose beads at 4°C, washed, and eluted in 10 mmol/L Tris-HCl pH 8, 5 mmol/L EDTA, 50 mmol/L NaCl, 1% SDS, 50 μg proteinase K, and 1× PIC. Immunoprecipitated DNA was extracted and quantified by qPCR using cccDNA-specific primers (Table 1). Samples were normalized to input DNA using the Δ threshold cycle (Ct) method where ΔCt = Ct (input) − Ct (immunoprecipitation) and expressed as a percentage of the input after normalization over no antibody signal (Figure 7I and J). As an additional negative control, ChIP experiments were performed with the cccDNA unrelated protein E2F (Figure 7K and L).
Specificity of antibodies used to immunoprecipitate H3.3 vs H3.1/2 variants was confirmed by analyzing the differential enrichment on genomic target sites shown to be preferentially bound by either H3.3 (SNAI1 and SOX9 promoters) or H3.1/2 (ZNF286 promoter and POLQ gene body)54 both in HepG2hNTCP cells and human liver tissue (Figure 9A–F).
Sequential Chromatin Immunoprecipitation
Cells were processed as for ChIP experiments until overnight immunoprecipitation. Immune complexes then were incubated for 2 hours with protein G agarose beads at 4°C, washed, and eluted in 10 mmol/L dithiothreitol. Eluted samples then were re-exposed to overnight immunoprecipitation at 4°C using 2–5 μg of antibodies in Re-ChIP buffer (1% Triton X-100, 2 mmol/L EDTA, 150 mmol/L NaCl, and 20 mmol/L Tris-HCl pH 8) and further processed as per the classic ChIP protocol described earlier.
Minicircle HBV Production and Transfection in HepG2-NTCP Cells
The plasmid plasmid minicircle (pMC)-HBV containing the full-length 3182-bp genotype D strain ayw HBV sequence was inserted into a parental plasmid vector generated from pMC.CMV-MCS-SV40polyA with the deletion of CMV promoter and polyA sequences as described by Yan et al.29 The 39-nucleotide attR site insertion in pMC-HBV was located immediately before the start codon of the preS1 gene, corresponding to the middle of the terminal protein domain and spacer region of the polymerase gene to minimize the disruption of the HBV genome.29 ZYCY10P3S2T competent bacteria (System Biosciences, Palo Alro, CA) then were transformed with the pMC-HBV and a single colony amplified in Terrific Broth (Sigma Aldrich, St. Louis, MO over 12 hours at 42°C under 200 rpm agitation. Amplification was performed by adding 2 volumes of Lysogeny broth medium overnight at 42°C under 200 rpm agitation. Arabinose induction was performed at 32°C for 4 hours by adding Lysogeny broth medium supplemented with 0.1% L-arabinose and NaOH to ensure a pH of 7. After centrifugation at 4500 rpm for 10 minutes at 4°C, plasmid DNA was extracted using the Nucleobond Xtra Maxi endotoxin free kit according to the manufacturer’s instructions (Macherey-Nagel) and digested by the NdeI (New England Biolabs) restriction enzyme for 2 hours at 37°C and by a plasmid-safe DNase (System Biosciences) overnight at 37°C to linearize the parental construct and get rid of it. After purification, the resulting plasmid was observed on agarose gel to check for the elimination of the parental plasmid. Eighty percent to 90% confluent HepG2hNTCP cells were transfected with the pMC-HBV plasmids and the TransIT-2020 (Mirus) transfecting agent according to the manufacturer’s instructions in DMEM supplemented with 5% FCS, 1% GlutaMAX (ThermoFischer, Waltham, MA), and 1% sodium pyruvate. The following day, cells were washed once with 1× PBS and cultured for 3 more days in DMEM supplemented with 5% FCS, 1% GlutaMAX, 1% sodium pyruvate, and 1% penicillin-streptomycin.
Human Liver Samples
Human liver samples derived from 3 untreated chronic hepatitis B male patients belonging to a historical cohort collected at Hospices Civils de Lyon (Lyon University, Lyon, France). These patients underwent liver biopsy as part of their clinical follow-up evaluation, a fragment was preserved for research purposes and stored at -80°C. The protocol was approved by the competent Institutional Ethics Committee (Comité de Protection de Personnes Sud est IV 11/040, authorization number DC-2008-235). Written informed consent was obtained from all patients and/or their legal guardians to undergo a liver biopsy. No patients were co-infected with human immunodeficiency virus, hepatitis C virus, or hepatitis delta virus.
Statistical Analysis
Statistics were performed using R (R Foundation for Statistical Computing, Vienna, Austria; http://www.R-project.org). Reverse-transcription-qPCR and ChIP enrichment values obtained from the different drug treatments were compared using the 2/K sample permutation test with Monte Carlo resampling approximation and a 2-tailed P value was calculated for a risk threshold of .05.
All authors had access to the study data and reviewed and approved the final manuscript.
CRediT Authorship Contributions
Maëlle Locatelli, PhD (Conceptualization: Equal; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Visualization: Lead; Writing – original draft: Equal)
Jean-Pierre Quivy, PhD (Conceptualization: Equal; Formal analysis: Equal; Writing – review & editing: Supporting)
Fleur Chapus, PhD (Investigation: Supporting; Methodology: Supporting)
Maud Michelet (Investigation: Equal; Methodology: Equal)
Judith Fresquet, Master (Investigation: Equal; Methodology: Equal)
Sarah Maadadi, master (Investigation: Supporting; Methodology: Supporting)
Amel Neila Aberkane, Master (Investigation: Supporting; Methodology: Supporting)
Audrey Diederichs, Master (Investigation: Supporting; Methodology: Supporting)
Julie Lucifora, PhD (Methodology: Supporting)
Michel Rivoire, MD (Resources: Supporting)
Genevieve Almouzni, PhD (Writing – review & editing: Supporting)
Barbara Testoni, PhD (Conceptualization: Lead; Formal analysis: Equal; Funding acquisition: Lead; Investigation: Supporting; Methodology: Supporting; Visualization: Equal; Writing – original draft: Lead; Writing – review & editing: Lead)
Fabien Zoulim, MD, PhD (Conceptualization: Lead; Funding acquisition: Lead; Supervision: Lead; Writing – review & editing: Lead)
Acknowledgments
The authors thank Dr D. Ray-Gallet for expert advice and for the pEYFP-N1-HIRA and pEYFP-N1-HIRA W799A D800A constructs, and Maud Michelet, Jennifer Molle, Anaëlle Dubois, and Océane Floriot for their help in the isolation of primary human hepatocytes, as well as Professor Michel Rivoire's surgical staff for providing liver resections.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by the Agence Nationale de Recherche (ANR) program “Investissement d'avenir” Laboratoires d'Excellence (LabEx) DEVweCAN (Cancer Development and Targeted Therapies) grant ANR-10-LABX-61, and by the Agence Nationale de Recherches sur le Sida et les hépatites virales |Maladies Infectieuses Emergentes (ANRS|MIE, grant ECTZ93319) (F.Z. and B.T.) and for the Almouzni's team by la Ligue Nationale Contre le Cancer (Equipe Labellisée Ligue), LabEx DEEP (Development, Epigenetics, Epigenetics and lifetime Potential) grants ANR-11-LABX-0044_DEEP, ANR-10-IDEX- 0001-02, Université Paris Science & Lettres (PSL), and European Research Council (ERC) grant ERC-2015-ADG-694694 ChromADICT (Chromatin Adaptations through Interactions of Chaperones in Time).
Data Transparency Statement The authors declare that all the data supporting the findings of this study are available within the article or from the corresponding authors upon reasonable request.
References
- 1.Seeger C., Mason W.S. Molecular biology of hepatitis B virus infection. Virology. 2015;479–480C:672–686. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lieberman P.M. Chromatin regulation of virus infection. Trends Microbiol. 2006;14:132–140. doi: 10.1016/j.tim.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Xia Y., Guo H. Hepatitis B virus cccDNA: formation, regulation and therapeutic potential. Antiviral Res. 2020;180 doi: 10.1016/j.antiviral.2020.104824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fanning G.C., Zoulim F., Hou J., Bertoletti A. Therapeutic strategies for hepatitis B virus infection: towards a cure. Nat Rev Drug Discov. 2019;18:827–844. doi: 10.1038/s41573-019-0037-0. [DOI] [PubMed] [Google Scholar]
- 5.Saper G., Kler S., Asor R., Oppenheim A., Raviv U., Harries D. Effect of capsid confinement on the chromatin organization of the SV40 minichromosome. Nucleic Acids Res. 2013;41:1569–1580. doi: 10.1093/nar/gks1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schreiner S., Nassal M. A role for the host DNA damage response in hepatitis B virus cccDNA formation-and beyond? Viruses. 2017;9:125. doi: 10.3390/v9050125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitamura K., Que L., Shimadu M., Koura M., Ishihara Y., Wakae K., Nakamura T., Watashi K., Wakita T., Muramatsu M. Flap endonuclease 1 is involved in cccDNA formation in the hepatitis B virus. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long Q., Yan R., Hu J., Cai D., Mitra B., Kim E.S., Marchetti A., Zhang H., Wang S., Liu Y., Huang A., Guo H. The role of host DNA ligases in hepadnavirus covalently closed circular DNA formation. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei L., Ploss A. Core components of DNA lagging strand synthesis machinery are essential for hepatitis B virus cccDNA formation. Nat Microbiol. 2020;5:715–726. doi: 10.1038/s41564-020-0678-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Königer C., Wingert I., Marsmann M., Rösler C., Beck J., Nassal M. Involvement of the host DNA-repair enzyme TDP2 in formation of the covalently closed circular DNA persistence reservoir of hepatitis B viruses. Proc Natl Acad Sci U S A. 2014;111:E4244–E4253. doi: 10.1073/pnas.1409986111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi Y., Gao Z., Xu G., Peng B., Liu C., Yan H., Yao Q., Sun G., Liu Y., Tang D., Song Z., He W., Sun Y., Guo J.-T., Li W. DNA polymerase κ is a key cellular factor for the formation of covalently closed circular DNA of hepatitis B virus. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheraz M., Cheng J., Tang L., Chang J., Guo J.-T. Cellular DNA topoisomerases are required for the synthesis of hepatitis B virus covalently closed circular DNA. J Virol. 2019;93 doi: 10.1128/JVI.02230-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo J., Luckenbaugh L., Hu H., Yan Z., Gao L., Hu J. Involvement of host ATR-CHK1 pathway in hepatitis B virus covalently closed circular DNA formation. mBio. 2020;11 doi: 10.1128/mBio.03423-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 15.Ray-Gallet D., Woolfe A., Vassias I., Pellentz C., Lacoste N., Puri A., Schultz D.C., Pchelintsev N.A., Adams P.D., Jansen L.E.T., Almouzni G. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell. 2011;44:928–941. doi: 10.1016/j.molcel.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Ray-Gallet D., Ricketts M.D., Sato Y., Gupta K., Boyarchuk E., Senda T., Marmorstein R., Almouzni G. Functional activity of the H3.3 histone chaperone complex HIRA requires trimerization of the HIRA subunit. Nat Commun. 2018;9:3103. doi: 10.1038/s41467-018-05581-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rai T.S., Glass M., Cole J.J., Rather M.I., Marsden M., Neilson M., Brock C., Humphreys I.R., Everett R.D., Adams P.D. Histone chaperone HIRA deposits histone H3.3 onto foreign viral DNA and contributes to anti-viral intrinsic immunity. Nucleic Acids Res. 2017;45:11673–11683. doi: 10.1093/nar/gkx771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sitbon D., Boyarchuk E., Dingli F., Loew D., Almouzni G. Histone variant H3.3 residue S31 is essential for Xenopus gastrulation regardless of the deposition pathway. Nat Commun. 2020;11:1256. doi: 10.1038/s41467-020-15084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armache A., Yang S., Martínez de Paz A., Robbins L.E., Durmaz C., Cheong J.Q., Ravishankar A., Daman A.W., Ahimovic D.J., Klevorn T., Yue Y., Arslan T., Lin S., Panchenko T., Hrit J., Wang M., Thudium S., Garcia B.A., Korb E., Armache K.-J., Rothbart S.B., Hake S.B., Allis C.D., Li H., Josefowicz S.Z. Histone H3.3 phosphorylation amplifies stimulation-induced transcription. Nature. 2020;583:852–857. doi: 10.1038/s41586-020-2533-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martire S., Gogate A.A., Whitmill A., Tafessu A., Nguyen J., Teng Y.-C., Tastemel M., Banaszynski L.A. Phosphorylation of histone H3.3 at serine 31 promotes p300 activity and enhancer acetylation. Nat Genet. 2019;51:941–946. doi: 10.1038/s41588-019-0428-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lucifora J., Salvetti A., Marniquet X., Mailly L., Testoni B., Fusil F., Inchauspé A., Michelet M., Michel M.-L., Levrero M., Cortez P., Baumert T.F., Cosset F.-L., Challier C., Zoulim F., Durantel D. Detection of the hepatitis B virus (HBV) covalently-closed-circular DNA (cccDNA) in mice transduced with a recombinant AAV-HBV vector. Antiviral Res. 2017;145:14–19. doi: 10.1016/j.antiviral.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 22.Gao W., Hu J. Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. J Virol. 2007;81:6164–6174. doi: 10.1128/JVI.02721-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo J., Cui X., Gao L., Hu J. Identification of an intermediate in hepatitis B virus covalently closed circular (CCC) DNA formation and sensitive and selective CCC DNA detection. J Virol. 2017;91 doi: 10.1128/JVI.00539-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo H., Mao R., Block T.M., Guo J.-T. Production and function of the cytoplasmic deproteinized relaxed circular DNA of hepadnaviruses. J Virol. 2010;84:387–396. doi: 10.1128/JVI.01921-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bock C.T., Schranz P., Schröder C.H., Zentgraf H. Hepatitis B virus genome is organized into nucleosomes in the nucleus of the infected cell. Virus Genes. 1994;8:215–229. doi: 10.1007/BF01703079. [DOI] [PubMed] [Google Scholar]
- 26.Avvakumov N., Nourani A., Côté J. Histone chaperones: modulators of chromatin marks. Mol Cell. 2011;41:502–514. doi: 10.1016/j.molcel.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 27.Soria G., Polo S.E., Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell. 2012;46:722–734. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Polo S.E., Theocharis S.E., Klijanienko J., Savignoni A., Asselain B., Vielh P., Almouzni G. Chromatin assembly factor-1, a marker of clinical value to distinguish quiescent from proliferating cells. Cancer Res. 2004;64:2371–2381. doi: 10.1158/0008-5472.can-03-2893. [DOI] [PubMed] [Google Scholar]
- 29.Yan Z., Zeng J., Yu Y., Xiang K., Hu H., Zhou X., Gu L., Wang L., Zhao J., Young J.A.T., Gao L. HBVcircle: a novel tool to investigate hepatitis B virus covalently closed circular DNA. J Hepatol. 2017;66:1149–1157. doi: 10.1016/j.jhep.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Lucifora J., Arzberger S., Durantel D., Belloni L., Strubin M., Levrero M., Zoulim F., Hantz O., Protzer U. Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. J Hepatol. 2011;55:996–1003. doi: 10.1016/j.jhep.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 31.Decorsière A., Mueller H., van Breugel P.C., Abdul F., Gerossier L., Beran R.K., Livingston C.M., Niu C., Fletcher S.P., Hantz O., Strubin M. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature. 2016;531:386–389. doi: 10.1038/nature17170. [DOI] [PubMed] [Google Scholar]
- 32.Panté N., Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rabe B., Delaleau M., Bischof A., Foss M., Sominskaya I., Pumpens P., Cazenave C., Castroviejo M., Kann M. Nuclear entry of hepatitis B virus capsids involves disintegration to protein dimers followed by nuclear reassociation to capsids. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torné J., Ray-Gallet D., Boyarchuk E., Garnier M., Le Baccon P., Coulon A., Orsi G.A., Almouzni G. Two HIRA-dependent pathways mediate H3.3 de novo deposition and recycling during transcription. Nat Struct Mol Biol. 2020;27:1057–1068. doi: 10.1038/s41594-020-0492-7. [DOI] [PubMed] [Google Scholar]
- 35.Alvarez-Astudillo F., Garrido D., Varas-Godoy M., Gutiérrez J.L., Villanueva R.A., Loyola A. The histone variant H3.3 regulates the transcription of the hepatitis B virus. Ann Hepatol. 2021;21 doi: 10.1016/j.aohep.2020.09.005. [DOI] [PubMed] [Google Scholar]
- 36.Allweiss L., Giersch K., Pirosu A., Volz T., Muench R.C., Beran R.K., Urban S., Javanbakht H., Fletcher S.P., Lütgehetmann M., Dandri M. Therapeutic shutdown of HBV transcripts promotes reappearance of the SMC5/6 complex and silencing of the viral genome in vivo. Gut. 2022;71:372–381. doi: 10.1136/gutjnl-2020-322571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ko C., Chakraborty A., Chou W.-M., Hasreiter J., Wettengel J.M., Stadler D., Bester R., Asen T., Zhang K., Wisskirchen K., McKeating J.A., Ryu W.-S., Protzer U. Hepatitis B virus genome recycling and de novo secondary infection events maintain stable cccDNA levels. J Hepatol. 2018;69:1231–1241. doi: 10.1016/j.jhep.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bock C.T., Schwinn S., Locarnini S., Fyfe J., Manns M.P., Trautwein C., Zentgraf H. Structural organization of the hepatitis B virus minichromosome. J Mol Biol. 2001;307:183–196. doi: 10.1006/jmbi.2000.4481. [DOI] [PubMed] [Google Scholar]
- 39.Tropberger P., Mercier A., Robinson M., Zhong W., Ganem D.E., Holdorf M. Mapping of histone modifications in episomal HBV cccDNA uncovers an unusual chromatin organization amenable to epigenetic manipulation. Proc Natl Acad Sci U S A. 2015;112:E5715–E5724. doi: 10.1073/pnas.1518090112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adam S., Polo S.E., Almouzni G. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell. 2013;155:94–106. doi: 10.1016/j.cell.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 41.Yang G., Feng J., Liu Y., Zhao M., Yuan Y., Yuan H., Yun H., Sun M., Bu Y., Liu L., Liu Z., Niu J.-Q., Yin M., Song X., Miao Z., Lin Z., Zhang X. HAT1 signaling confers to assembly and epigenetic regulation of HBV cccDNA minichromosome. Theranostics. 2019;9:7345–7358. doi: 10.7150/thno.37173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen C., Corpet A., Roubille S., Maroui M.A., Poccardi N., Rousseau A., Kleijwegt C., Binda O., Texier P., Sawtell N., Labetoulle M., Lomonte P. Promyelocytic leukemia (PML) nuclear bodies (NBs) induce latent/quiescent HSV-1 genomes chromatinization through a PML NB/histone H3.3/H3.3 chaperone axis. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niu C., Livingston C.M., Li L., Beran R.K., Daffis S., Ramakrishnan D., Burdette D., Peiser L., Salas E., Ramos H., Yu M., Cheng G., Strubin M., Delaney W.E.I.V., Fletcher S.P. The Smc5/6 complex restricts HBV when localized to ND10 without inducing an innate immune response and is counteracted by the HBV X protein shortly after infection. PLoS One. 2017;12 doi: 10.1371/journal.pone.0169648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross P.J., Kennedy M.A., Christou C., Risco Quiroz M., Poulin K.L., Parks R.J. Assembly of helper-dependent adenovirus DNA into chromatin promotes efficient gene expression. J Virol. 2011;85:3950–3958. doi: 10.1128/JVI.01787-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ladner S.K., Otto M.J., Barker C.S., Zaifert K., Wang G.H., Guo J.T., Seeger C., King R.W. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother. 1997;41:1715–1720. doi: 10.1128/aac.41.8.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luangsay S., Gruffaz M., Isorce N., Testoni B., Michelet M., Faure-Dupuy S., Maadadi S., Ait-Goughoulte M., Parent R., Rivoire M., Javanbakht H., Lucifora J., Durantel D., Zoulim F. Early inhibition of hepatocyte innate responses by hepatitis B virus. J Hepatol. 2015;63:1314–1322. doi: 10.1016/j.jhep.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 47.Lecluyse E.L., Alexandre E. Isolation and culture of primary hepatocytes from resected human liver tissue. Methods Mol Biol. 2010;640:57–82. doi: 10.1007/978-1-60761-688-7_3. [DOI] [PubMed] [Google Scholar]
- 48.Stadelmayer B., Diederichs A., Chapus F., Rivoire M., Neveu G., Alam A., Fraisse L., Carter K., Testoni B., Zoulim F. Full-length 5’RACE identifies all major HBV transcripts in HBV-infected hepatocytes and patient serum. J Hepatol. 2020;73:40–51. doi: 10.1016/j.jhep.2020.01.028. [DOI] [PubMed] [Google Scholar]
- 49.Ni Y., Lempp F.A., Mehrle S., Nkongolo S., Kaufman C., Fälth M., Stindt J., Königer C., Nassal M., Kubitz R., Sültmann H., Urban S. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology. 2014;146:1070–1083. doi: 10.1053/j.gastro.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 50.Allweiss L., Volz T., Giersch K., Kah J., Raffa G., Petersen J., Lohse A.W., Beninati C., Pollicino T., Urban S., Lütgehetmann M., Dandri M. Proliferation of primary human hepatocytes and prevention of hepatitis B virus reinfection efficiently deplete nuclear cccDNA in vivo. Gut. 2018;67:542–552. doi: 10.1136/gutjnl-2016-312162. [DOI] [PubMed] [Google Scholar]
- 51.Lebossé F., Inchauspé A., Locatelli M., Miaglia C., Diederichs A., Fresquet J., Chapus F., Hamed K., Testoni B., Zoulim F. Quantification and epigenetic evaluation of the residual pool of hepatitis B covalently closed circular DNA in long-term nucleoside analogue-treated patients. Sci Rep. 2020;10 doi: 10.1038/s41598-020-78001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Repetto G., del Peso A., Zurita J.L. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat Protoc. 2008;3:1125–1131. doi: 10.1038/nprot.2008.75. [DOI] [PubMed] [Google Scholar]
- 53.Vichai V., Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1:1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 54.Gomes A.P., Ilter D., Low V., Rosenzweig A., Shen Z.-J., Schild T., Rivas M.A., Er E.E., McNally D.R., Mutvei A.P., Han J., Ou Y.-H., Cavaliere P., Mullarky E., Nagiec M., Shin S., Yoon S.-O., Dephoure N., Massagué J., Melnick A.M., Cantley L.C., Tyler J.K., Blenis J. Dynamic incorporation of histone H3 variants into chromatin is essential for acquisition of aggressive traits and metastatic colonization. Cancer Cell. 2019;36:402–417.e13. doi: 10.1016/j.ccell.2019.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]