Summary
Background
Subjective or objective subtle cognitive decline (SCD) is considered the preclinical manifestation of Alzheimer's disease (AD), which is a potentially crucial window for preventing or delaying the progression of the disease.
Methods
To explore the potential mechanism of disease progression and identify relevant biomarkers, we comprehensively assessed the peripheral blood transcriptomic alterations in SCD, covering lncRNA, mRNA, and miRNA.
Findings
Dysregulated protein-coding mRNA at both gene and isoform levels implicated impairment in the type I interferon signaling pathway in SCD. Specifically, this pathway was regulated by the transcription factor STAT1 and ncRNAs NRIR and has-miR-146a-5p. The miRNA-mRNA-lncRNA co-expression network revealed hub genes for the interferon module. Individuals with lower interferon signaling activity and lower expression of a hub gene STAT1 exhibited a higher conversion rate to mild cognitive impairment (MCI).
Interpretation
Our findings illustrated the down-regulation of interferon signaling activity would potentially increase the risk of disease progression and thus serve as a pre-disease biomarker.
Funding
This work was partly supported by National Key R&D Program of China (2020YFA0712403), National Natural Science Foundation of China (61932008), Shanghai Municipal Science and Technology Major Project (2018SHZDZX01), the 111 Project (No. B18015) of China, Greater Bay Area Institute of Precision Medicine (Guangzhou) (Grand No. IPM21C008), Natural Science Foundation of Shanghai (21ZR1403200), and Shanghai Center for Brain Science and Brain-Inspired Technology.
Keywords: Subjective or objective subtle cognitive decline (SCD), Alzheimer's disease (AD), Peripheral blood transcriptome, Type I interferon signaling, Progression biomarkers
Research in context.
Evidence before this study
We searched PubMed, Google Scholar, and Web of Science for studies published between database inception and Oct 15, 2021, using the search terms ((Alzheimer's * OR preclinical Alzheimer's OR dementia OR cognitive impairment OR cognitive decline OR cognitive complaints) AND (blood OR peripheral) AND (transcript* OR RNA OR expression OR microarray) AND (biomarker OR alteration OR changes)), without language restrictions. Most of these studies focused on the identification of expression changes in Alzheimer's disease (AD) or mild cognitive impairment. The preclinical stage of AD (e.g., subjective or objective subtle cognitive decline) is a crucial window to prevent the disease progression. There is a lack of systemic research about transcriptomic profiling and biomarkers for preclinical AD.
Added value of this study
In this comprehensive peripheral blood transcriptomic analysis for preclinical AD, we found that the interferon stimulated genes and isoforms were substantially downregulated in subjective or objective subtle decline patients (SCDs). The activity of type I interferon signaling was significantly inhibited in SCDs whereas it was activated in individuals with mild cognitive decline and AD. We further identified dysregulated regulators for the interferon signaling including transcription factors, microRNAs, lncRNAs, and hub genes. Moreover, we noticed that SCDs with progression to MCI had a lower interferon signaling activity than those stable NC and SCD individuals in an independent blood transcriptomic AD cohort.
Implications of all the available evidence
Individuals with lower type I interferon activity had a significantly higher progression risk in both short and long-term follow-up visits, suggesting the impairment of interferon signaling that is involved in the antiviral process contributes to disease progression. Our findings provide support to the virus infection hypothesis and provide candidate progression biomarkers for preclinical AD.
Alt-text: Unlabelled box
Introduction
Alzheimer's disease (AD), the most common cause of dementia, is a progressive neurodegenerative disorder that causes deficits in memory, thinking, decision making, and social skills.1 The pathophysiological events of AD begin years before the onset of clinical dementia.2 The National Institute on Aging-Alzheimer's Association (NIA-AA) proposed three subsequent stages of AD, including the preclinical stage of AD, mild cognitive impairment (MCI), and AD dementia.3, 4, 5 The primary hypothesis on the progression of AD pathologies stated that the initial deposition of amyloid-β peptide (Aβ) leads to subsequent tau hyperphosphorylation, neuron and synaptic loss, and cognitive decline.6 However, the continued failure of clinical trials aimed at decreasing Aβ plaques implies that there may be alternative causes of disease progression or the neuronal injury of the late MCI and AD dementia may be irreversible.7 The long preclinical phase with mild neuronal damage provides a potentially crucial opportunity for exploring the potential pathogenic mechanisms of AD and preventing or delaying the progression of this disease.5,8
Subjective cognitive decline characterized by a self-reported persistent decline in cognitive capacity compared to a previously normal status prior to the appearance of objective cognitive impairment, occurs at the preclinical stage of AD and may serve as a symptomatic indicator of preclinical AD.9 Compared with cognitively normal, objective subtle cognitive decline was associated with faster amyloid accumulation and neurodegeneration.10 The occurrence of biomarker abnormalities associated with AD in subjective or objective subtle cognitive decline (SCD), including decreased concentrations of Aβ42 and increased concentrations of tau in cerebrospinal fluid (CSF),11 provides supportive evidence for SCD as the earliest manifestation of AD. Moreover, SCD is associated with an increased risk for future cognitive decline12, 13, 14–longitudinal data analyses revealed that individuals with subjective cognitive concerns carried a four-fold and a six-fold increased risk of incident MCI and dementia, respectively, compared to those without memory complaints.14,15 The cumulative conversion rate of SCD progression to MCI and dementia is about 20.76% and 7.23% with high heterogeneity.16 Therefore, exploring the early changes of subjective cognitive decline in preclinical Alzheimer's disease and predicting the conversion risk are urgently required.
The transcriptome represents a quantitative phenotype that reveals the biological processes disrupted in disorders. Despite the blood-brain barrier, blood expression data captured the majority of predictive molecular pathways for neurodegenerative progression as identified in brain expression data for AD.17 Compared with brain and CSF biomarkers, blood-based biomarkers are more obtainable and require less invasive collection methods. However, to our knowledge, no previous study has comprehensively evaluated the blood transcriptomic profiling for SCD in preclinical Alzheimer's disease.
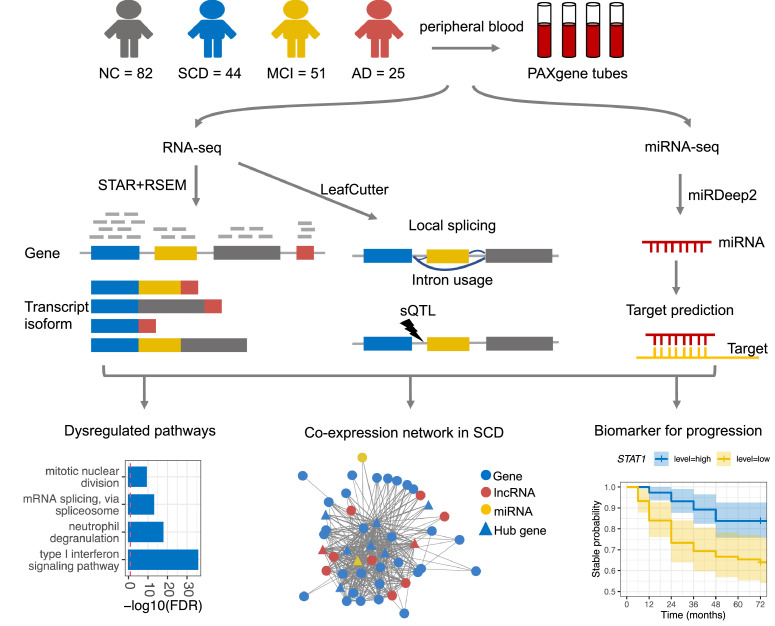
Here, to explore the peripheral transcriptomic dysregulation in SCD, we performed RNA sequencing and microRNA sequencing on peripheral blood samples of SCD subjects and age-matched elderly control subjects with normal cognition (NC). The same procedures were also performed on MCI and AD blood samples as a comparison. We characterized the transcriptomic changes across multiple levels, including gene expression, transcript isoform expression, splicing, lncRNA and miRNA regulation networks, and the co-expression network (Figure 1). We found that the type I interferon signaling pathway was remarkably downregulated in SCD while upregulated in later stages, possibly due to a comprehensive orchestration in multiple levels of transcriptional regulation. Outside data showed that normal samples with lower expression of hub genes in the co-expression network module for the type I interferon signaling pathway exhibited a higher disease conversion rate to MCI. The dysregulated molecular pathways and hub genes in SCD would provide insights into the etiology of disease conversion and serve as candidate biomarkers.
Figure 1.
Study design and overview of the study.
Methods
Participants
The participants in disease groups were recruited from outpatients visiting the Department of Geriatric Medicine of the Shanghai Sixth People's Hospital between March 2019 and March 2020. For normal elderly control group, we recruited the elderly people (Age: 50–80 years) living nearby with normal cognition function. A total of 222 participants were primarily enrolled in this study, including 49 SCD samples, 57 MCI samples, 28 AD samples, and 88 age-matched subjects with normal cognition (NC). All procedures were approved by the Ethics Committee of the Shanghai Sixth People's Hospital (Approval No: 2019-032), and written informed consents were collected from all participants prior to actions and analyses. The additional recruitment criteria include: (1) without a disease history or family history of other neurologic or psychiatric diseases, such as Parkinson's disease, depression, epilepsy, and neuron developmental delay; (2) without serious somatic diseases; (3) with adequate vision and hearing. To avoid the effect of comorbidity on blood transcriptome profiles, samples with disease histories of hematological disease and tumor were excluded. After excluding individuals according to the above criteria, 202 participants, comprising 82 NC, 44 SCD, 51 MCI, and 25 AD samples, were finally enrolled in this study.
The details of neuropsychological assessment and diagnostic criteria can be found in Supplementary materials.
RNA library preparation, sequencing, mapping, and quantification
The details of RNA and miRNA library preparation, sequencing, mapping, and quantification can be found in Supplementary materials.
Differential expression analysis and enrichment analysis
Expected counts were compiled from gene and isoform-level RSEM quantifications and imported into R for downstream analyses. Genes/isoforms were filtered to include those with TPM (transcripts per million) > 1 in at least 25% of samples. MiRNA samples were filtered to include those with counts > 3 in at least 50% of samples. Removing low expressed genes/miRNAs resulted in 12,664 genes and 36,005 isoforms, and 811 miRNAs of 202 participants.
Differentially expressed genes (DEG), differentially expressed transcript isoforms (DET), and differentially expressed miRNA (Wald test, P-value < 0.05, |log2(Fold Change, FC)| > log2(1.3)) were identified using R package DESeq218 with adjusting for covariates (age, gender, and RIN (RNA integrity number)) in the generalized linear model, where no significant sequencing batch was found (Supplementary Figure 1a). Normalized expression matrices after correcting for library size were obtained for downstream analysis.
To gain further insight into the biological functions of differentially expressed genes, we performed enrichment analysis of gene ontology (GO) biological processes, KEGG, and Reactome pathways using R package gProfileR with the hypergeometric test model (v0.7.0; https://biit.cs.ut.ee/gprofiler/).19 IPA20 (Ingenuity Pathway Analysis; QIAGEN) and gene set enrichment analysis (GSEA) (V4.1.0)21,22 were also applied to further predict whether the pathway is activated or inhibited. Moreover, miRNA targets were extracted from miRTarBase v7.0 (http://mirtarbase.mbc.nctu.edu.tw/)23 and miRDB v6.0 (http://mirdb.org).24,25 More details can be found in Supplementary materials.
Differential local splicing (DS) analysis and splicing quantitative trait loci (sQTL) discovery
Differential local splicing events were identified using LeafCutter26 and schematically visualized using the LeafCutter shiny app. DS intron clusters and domains were mapped onto transcripts using gViz (v1.32) and ensembldb (v2.12.1) R packages. FastQTL27 was used to identify cis-splicing QTLs (sQTLs) within regions of +-100kb of the intron clusters. See the supplementary materials and methods for full details.
Weighted gene coexpression network analysis (WGCNA)
To explore the pattern of correlations between all possible pairs of genes (including protein-coding genes, lncRNA, and miRNA) and identify their modules, signed networks were constructed with the R package WGCNA28 using a library-size normalized expression matrix of SCD and NC samples at gene and isoform levels (see parameters in Supplementary materials). Modules were summarized by module eigengene (ME) that was defined as the first principal component (PC1) of the module. Module (eigengene)-disease associations were evaluated using Pearson's correlation. For each gene in the modules, the module membership (MM, correlation of its gene expression profile with the module eigengene of a given module) and trait-based gene significance/relationship (GS, correlation between gene expression and the disease status (i.e., SCD = 1, NC = 0)) were evaluated. Hub genes were identified based on the cut-off criteria (|MM| > 0.8 and |GS| > 0.2).
Differential expression analysis and biomarkers identification in ADNI
An independent AD blood gene expression data was obtained from the ADNI (Alzheimer's Disease Neuroimaging Initiative, http://adni.loni.usc.edu).29 Among participants who were not diagnosed as MCI or AD, we further identified NC and SCD were identified using the participant self-report everyday cognition and neuropsychological tests. See the supplementary materials and methods for full details. ADNI transcriptome data was sequenced using the Affymetrix Human Genome U219 Array. Limma30 was applied to identify differentially expressed genes while adjusting for covariates of age, gender and RIN.
To determine whether a gene could be a candidate biomarker for disease progression from SCD to MCI or not, SCD samples were divided into two groups based on its expression level, one group higher than the median level and the other lower than the median. To compare the conversion rate of the two groups, Kaplan-Meier survival curves were fitted and plotted using survival and survminer R packages.
Statistics
Statistical analysis was performed using R (version 4.0.2). Demographics were compared using ANOVA (analysis of variance) test for continuous variables and a Chi-Square test for categorical variables. The missing values (one NC and one AD samples) were imputed as the mean value of corresponding groups. Age was categorized as an ordered variable based on the world health organization. Comparisons of gene expression and PSI (percent-spliced-in) between disorders and NC were performed using two-tailed Student's t-test and Wilcoxon test, respectively. Correlations between module eigengenes and phenotype traits were assessed with the Pearson's correlation. Overlaps of DE genes between two sets were assessed using the hypergeometric test. To correct for multiple comparisons, the Benjamini-Hochberg method was applied. We performed logistic regression to select features the type I interferon signature and module genes and to evaluate the diagnosis power of the expression of these selected genes for detecting SCD from NC in our dataset and the ADNI dataset.
Role of funding source
None of the funders had any roles in the study design, data collection, data analyses, data interpretation, writing of the report, or decision to publish the study.
Results
Participant characteristics
To explore the potential mechanism of disease progression and identify candidate blood transcriptome biomarkers, we performed RNA sequencing and microRNA sequencing on peripheral blood samples from 44 participants with SCD and 82 individuals with normal cognition (NC), and assessed the peripheral blood transcriptomic dysregulation for SCD (Figure 1). To observe the dynamics of these changes as the disease progresses, 51 MCI, and 25 AD samples were also sequenced following the same procedures as the SCD samples. To reduce the impact of comorbidities on the transcriptome, individuals with a history of hematological diseases, tumor and brain diseases (e.g., Parkinson's disease) were removed. Table 1 showed the demographic and cognitive characteristics of the enrolled participants from the four groups. The characteristics of the RNA sequencing data for the participants can be found in Supplementary Figure 2. Compared with the NC group, the SCD and MCI groups did not show any significant difference with respect to age, gender, or education, whereas the AD patients were much older and had a higher proportion of APOE4 carriers than other groups. Both MCI and AD groups were significantly different from the NC group according to their cognition assessments, i.e. MMSE (Mini-Mental State Examination), MoCA_B (Montreal cognitive assessment-basic), and ACEIII (Addenbrooke's Cognitive Examination - III) scores. The matched confounding factors between SCD and NC ensured that the transcriptomic changes identified in SCD mainly resulted from the disease state.
Table 1.
The characteristics of participants of each group.
| NC (n = 82) | SCD (n = 44) |
MCI (n = 51) |
AD (n = 25) |
P-valued | ||||
|---|---|---|---|---|---|---|---|---|
| Mean (SD) or % | Mean (SD) or % | P-valuea | Mean (SD) or % | P-valueb | Mean (SD) or % | P-valuec | ||
| Age | 64.37 (8) | 64.73 (7.18) | 0.995 | 66.06 (7.72) | 0.62 | 71.8 (8.6) | 2.83e−4 | 0.00055 |
| Gender (Male %) | 32.93% | 31.82% | 1 | 23.53% | 0.34 | 36% | 0.97 | 0.62 |
| Education | 11.67 (4.31) | 12.07 (3.49) | 0.94 | 10.16 (2.68) | 0.13 | 10.08 (4.76) | 0.28 | 0.029 |
| APOE4 + (%) | 23.17% | 11.36% | 0.17 | 25.49% | 0.92 | 40% | 0.16 | 0.056 |
| MMSE | 27.76 (2.22) | 27.59 (1.63) | 0.99 | 26.51 (2.05) | 0.032 | 17.24 (4.79) | 4.11e−14 | 5.07e−44 |
| MoCA_B | 25.45 (2.95) | 24.43 (2.94) | 0.38 | 22 (3.21) | 3.06e−07 | 12.32 (5.46) | 4.12e−14 | 3.22e−39 |
| ACEIII | 80.51 (11.14) | 78.98 (7.54) | 0.88 | 70.86 (9.36) | 9.04e−06 | 51.08 (16.71) | 4.65e−14 | 1.89e−24 |
NC = normal cognition; SCD = subjective or subtle objective cognitive decline; MCI = mild cognitive impairment; AD = Alzheimer's disease; MMSE = Mini-Mental State Examination; MoCA_B = Montreal cognitive assessment-basic; ACEIII = Addenbrooke's Cognitive Examination - III; SD = standard deviation.
Results presented as mean ± SD or frequencies with proportions. Quantitative and categorical characteristics differences were assessed with ANOVA and Chi-Square test, respectively.
P-value for comparison between SCD and NC.
P-value for comparison between MCI and NC.
P-value for comparison between AD and NC.
P-value for comparison among NC, SCD, MCI and AD.
The type I interferon signaling pathway is down-regulated in SCD
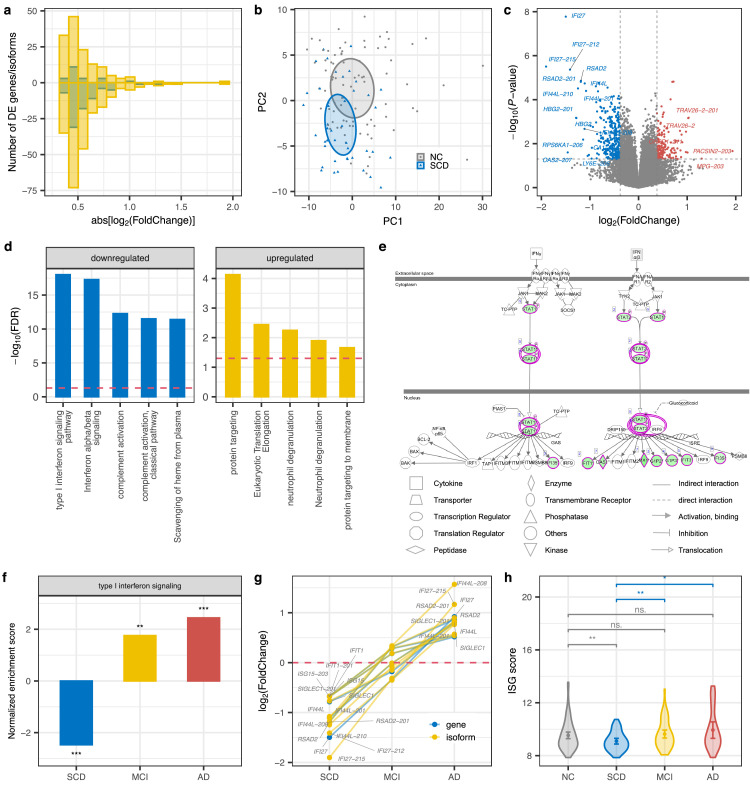
Transcript isoform diversity and dysregulation show higher disease specificity and are being increasingly implicated in psychiatric and neurodegenerative diseases,31,32 we therefore quantified gene expression at both gene and isoform-level. With the transcriptome data, we identified 101 protein-coding genes and 360 transcript isoforms that were differentially expressed (P < 0.05 & |log2(Foldchange, FC)| > log2(1.3)) in the SCD group as compared with the NC group (Figure 1a and Supplementary Table 1). Notably, although there was a substantial overlap (P = 4.4e−155, hypergeometric tests) between differentially expressed (DE) genes and isoforms, isoform-level alterations exhibited larger fold changes (Figure 2a) and disease specificity (Supplementary Figure 1c-d) than the gene-level, highlighting the importance of splicing dysregulation in preclinical AD pathogenesis. In accordance with SCD as the earliest manifestation of AD, some previously reported AD-associated genes were identified here, such as NR3C1 and GSK3B.33,34 Consistent with previous results,35 principal component analysis (PCA) based on the DE genes and isoforms (Figure 2b, Supplementary Figure 1b and 1e-f) revealed moderately separated clusters between disorders and normal controls. We also noticed a scattered distribution of samples in each group. Both of which indicate a high heterogeneity of expression profile of peripheral blood.
Figure 2.
Downregulation of the interferon signaling pathway in SCD. (a) Fold change (FC) histograms for up (positive number) or down-regulated (negative number) protein-coding genes (blue) and isoforms (yellow) in SCD. Abs: absolute. (b) PCA plot on differentially expressed genes and isoforms of SCD. (c) Volcano plot displaying the statistical significance (P-value) versus magnitude of change (fold change) of protein-coding genes and transcripts in SCD compared with NC. DE genes/isoforms with |log2(FC)| > 1 are labeled. (d) Pathway enrichment analysis for genes that were differentially expressed at gene or isoform level in SCD. The top 5 enriched pathways are shown. Blue and yellow represent down and up-regulated pathways, respectively. The red dashed line indicates the significance threshold (FDR = 0.05). (e) IPA canonical pathway for interferon signaling. Green shading represents down-regulated genes. (f) GSEA in type I interferon signaling pathways across disorders. Barplot shows the normalized enrichment score (NES) and significance. A positive/negative NES value indicates up/down-regulation of this gene set in disease compared with NC. ***, P < 0.001; **, P < 0.01. (g) Fold changes of ISG genes (IFI44L, IFI27, RSAD2, SIGLEC1, IFIT1, and IS15) defining the IFN signature at gene and isoform level across disorders. (h) ISG score of each group (NC = 82, SCD = 44, MCI = 51, AD = 25) based on the mean expression of these six signature genes. Error bar indicates 95% confidence interval of ISG score. P values were determined with the two-tailed t-test. ***, P < 0.001; **, P < 0.01; *, P < 0.05; ·, P < 0.1; ns., P > 0.1. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
As shown in Figure 2c, some interferon (IFN) stimulated genes and their isoforms were remarkably downregulated (log2(FC) < −1) in SCD, e.g. IFI27, OAS2, IFI44L, and RSAD2. Consistent with this finding, the interferon signaling pathway activity was significantly inhibited in SCD (Supplementary Figure 3a), as confirmed by ingenuity pathway analysis (IPA),20 especially for the type I interferon signaling pathway (Figure 2d, Supplementary Table 2). Notably, STAT1, a key mediator of IFN signaling,36 was downregulated in SCD (Figure 2e, Supplementary Figure 3b). STAT1 mediates cellular response to interferons, cytokines, and other growth factors and activates the transcription of IFN-stimulated genes that target almost any step in a virus life cycle.37 Therefore, the downregulation of STAT1 and IFN stimulated genes and type I IFN signaling in SCD would increase the replication of viral, like Herpesviridae. The bacterial and viral infections were reported to contribute to the pathophysiology of AD or to cognitive decline, most frequently implicating Herpesviridae.38
Notably, consistent with the up-regulation of type I interferon pathway in AD brains,39 this signaling pathway was activated in AD peripheral blood (Supplementary Figure 3c-d, Supplementary Table 2), which may arise from the feedback regulation of antiviral response. Furthermore, gene set enrichment analysis (GSEA), takes all of the genes into consideration rather than only DE genes, which also gives validation to type I IFN signaling being down-regulated in SCD while up-regulated in MCI and AD compared with NC (Figure 2f). The ISG (IFN-stimulated gene) score defining type I IFN signaling signature based on the mean of expression of six ISGs (IFI44L, IFI27, RSAD2, SIGLEC1, IFIT1, and IS15) in this pathway,40,41 was significantly reduced in SCD compared with NC and gradually increased in MCI and AD (Figure 2g and 2h). Consistent with previous findings on sex difference in the interferon pathway,42 the interferon pathway tends to have higher activity in females than males in each group (Supplementary Figure 4). To avoid the effects of gender on our result, we have compared the type I interferon signaling activity between SCD and NC stratified by gender and found that the signaling activity was lower in SCD than NC either in men or women. Together, these results demonstrated that in contrast to the MCI and AD stages, the SCD stage exhibited a down-regulation of type I interferon signaling pathways.
STAT1, a key transcription regulator of type I IFN signaling, is differentially spliced in SCD
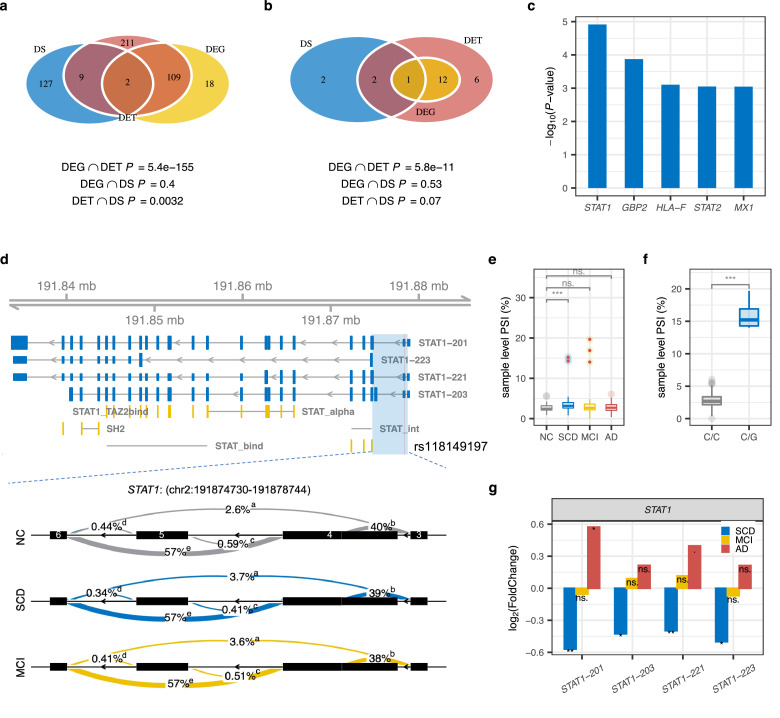
Alternative splicing is an important post-transcriptional regulation mechanism, contributing to isoform diversity and protein complexity.43,44 Here, we identified 138 genes with differentially local splicing (DS, P < 0.001) in SCD compared with NC (Figure 3a, Supplementary Table 3). DS genes in SCD overlapped significantly (P = 0.0032, hypergeometric tests) with DE transcript isoforms, indicating that local splicing could partially explain isoform dysregulation. There were significant pairwise overlaps of DS genes between three stages of AD (Supplementary Figure 5a). For example, differentially excised introns of NRF1, a transcription factor contributing to the pathogenesis of neurodegenerative diseases via perturbation of diverse mitochondrial and extra-mitochondrial functions,45 were found in SCD and AD (Supplementary Figure 5d). However, DS genes in AD exhibited few overlaps with those previously identified in brain transcriptome data (Supplementary Figure 5b-c),46,47 highlighting the tissue-specificity of splicing events.
Figure 3.
Differential local splicing in SCD compared to NC. (a) Overlaps between significant DE genes (DEG), DE transcript isoforms (DET), and genes with significant differentially spliced (DS) intron clusters in SCD. P values for hypergeometric tests of pairwise overlaps between data types are shown at the bottom. Blue, yellow, and red indicate DS, DEG, and DET, respectively. (b) Overlaps between DEG, DET, and DS of IFN signaling genes in SCD. (c) The significance of DS IFN signaling genes in SCD. (d) Whole-gene view of STAT1 highlighting the intron cluster (blue block, chr2:191874730-191878744) with significant DS in SCD (P = 1.2e−5, Chi-Square test) and MCI (P = 7.4e−4, Chi-Square test), as well as transcripts STAT1-201, 203, 221 and STAT1-223 that were differentially expressed in SCD. Protein domain mappings from the Pfam database are shown in yellow. STAT_int, STAT protein, protein interaction domain; STAT_alpha, STAT protein, all-alpha domain; STAT_bind, STAT protein, DNA binding domain; SH2, SH2 domain; STAT1_TAZ2bind, STAT1 TAZ2 binding domain. The red line denotes the sQTL rs118149197. The numbers inside the exon of the bottom intron cluster represent the exon numbers of the gene model. (e) The distribution of individual PSI (percent spliced in) for the intron cluster in (d) for each group (NC = 82, SCD = 44, MCI = 51, AD = 25). Samples with rs118149197 were denoted with red points. P-values were determined with the two-tailed Wilcoxon test. Boxplot spans the first to the third quartile with the line inside the box representing the median value. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ·, P < 0.1; ns., P > 0.1. (f) PSI distribution for samples with different rs118149197 genotypes. P-values and quartiles were determined and presented in the same way as (e). (g) Fold Changes of differentially expressed transcripts of STAT1 in SCD. P-values were determined using Wald test in DEseq2. ***, P < 0.001; **, P < 0.01; *, P < 0.05; ·, P < 0.1; ns., P > 0.1. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Differentially spliced type I IFN signaling genes in SCD significantly overlapped with those that were differentially expressed at the isoform level, including STAT1, STAT2, and MX1 (Figures 3b-c and Supplementary Figure 6), which may partially resolve the dysregulation of the type I IFN signaling pathway. Notably, a DS intron cluster (chr2:191874730-191878744) in STAT1 exhibited significantly increased exon (i.e., exon 4 in the gene model) skipping in SCD (P = 1.22e−05, Chi-Square test) and MCI (P = 7.4e−4, Chi-Square test) compared with NC (Figure 3d). We identified splicing quantitative trait locus (sQTLs) (Supplementary Figure 7 and Supplementary Table 4) driving or contributing to this DS using FastQTL27 after adjusting for known and inferred covariates (see Supplementary Methods). The most significant SNP rs118149197 (P = 4.3e−20, linear regression in FastQTL, Figure 3d-e), located in the 5’UTR region of STAT1, with a higher mutation burden in SCD (n = 2) and MCI (n = 3) than NC (n = 0), was predicted to affect RNA splicing by SPIDEX.48 Individuals with the mutation had a significantly higher PSI (percent spliced in) than those without the mutation (Figure 3f). The above findings indicate that the splicing differences between SCD and CN may arise from different genotypes and other factors (e.g., sQTLs in Supplementary Figure 7). In line with a previous observation that DS events might predict the aberrant expression of isoforms,31 some transcripts of STAT1 showed significantly decreased expression and transcript usages in SCD, such as STAT1-201 and STAT1-223 (Figure 3g, Supplementary Figure 8). As described above, STAT1 is one of the key transcription regulators of type I interferon signaling and activates the transcription of IFN-stimulated genes. Therefore, the dysregulated splicing and expression of STAT1 in SCD patients may reduce their response to interferons and the defense to viruses (e.g, herpes simplex).
NRIR and has-miR-146a-5p, upstream regulators of type I IFN signaling genes, are dysregulated in SCD
Long noncoding RNAs (lncRNAs) and miRNAs regulate gene expression at epigenetic, transcriptional, and post-transcriptional levels.49,50 They play key roles in neurogenesis, neuronal maturation, neuronal function, and neuronal survival, and thus are involved in many neurological diseases like epilepsy and AD.51, 52, 53 Here, we sought to explore the regulation of ncRNAs for the IFN signaling genes in SCD.
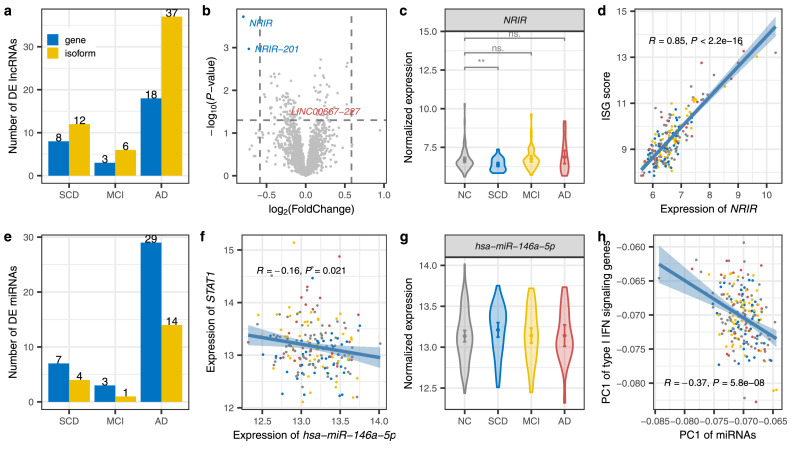
Based on GENCODE annotation, we identified 8 lncRNAs exhibiting differential expression in SCD compared with NC (Figure 4a, Supplementary Table 5). Notably, as shown in the volcano plot (Figure 4b), NRIR was remarkably down-regulated (log2FC = −0.80; P = 1.9e−4, Wald test in DESeq218) in SCD. The expression of this lncRNA was slightly increased in MCI (log2FC = 0.2), and significantly elevated in AD (log2FC = 0.6; P = 0.06, Wald test in DESeq218) (Figure 4c). Consistent with NRIR driving interferon response in human monocytes,54 NRIR exhibited significantly positive correlation with the ISG score (Pearson's correlation, R = 0.85, P < 2.2e−16, Figure 4d) for the type I IFN signaling and STAT1 (Pearson's correlation, R = 0.68, P < 2.2e−16, Supplementary Figure 9), indicating that decreased expression of NRIR might play a regulatory role in the impaired interferon activity of SCD.
Figure 4.
Dysregulation of ncRNAs regulated type I interferon signaling. (a) The number of differentially expressed long non-coding genes at gene (blue) and isoform (yellow) levels in SCD, MCI, and AD. (b) Volcano plot displaying the statistical significance (P-value, Wald test in DEseq2) versus magnitude of change (FC) of long noncoding genes and transcripts in SCD compared with NC. DE lncRNAs with |log2(FC)| > log2(1.5) are labeled. (c) Violin plots with means and 95% confidence intervals of the normalized expression level of NRIR in each group. P values were determined with the two-tailed Wilcoxon test. (d) Pearson's correlation between ISG scores and normalized expression of NRIR. Shadow represents the 95% confidence interval. (e) Up and down-regulated miRNAs among disorders. (f) Pearson's correlation between normalized expression of has-miR-146a-5p and STAT1.(g) Distribution of normalized expression level of has-miR-146a-5p in each group. (h) Pearson's correlation between the PC1 of type I IFN signaling genes and the PC1 of miRNAs that target these genes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
To assess the potential role of miRNAs dysregulation in transcriptomic alteration, we performed genome-wide miRNA expression profiling in samples with mRNA sequencing. We identified differently expressed miRNAs (Figure 4e, Supplementary Table 6) and predicted their target genes. We noticed that has-miR-125b, a miRNA has been consistently reported as altered in blood, CSF, and the brain in AD compared with controls in at least two studies,55, 56, 57, 58 was dysregulated in both SCD and AD. Several miRNAs significantly targeted genes in the type I IFN signaling pathway (FDR < 0.05, Supplementary Table 7). Notably, the targets of has-miR-146a-5p showed the most significant enrichment in type I IFN signaling pathway (FDR = 6.3e−15, hypergeometric test), significantly negatively regulated down-expressed type I IFN genes, such as STAT1 (Figure 4f). Compared with NC, it was slightly up-regulated in SCD (P = 0.089, one-tailed t-test; P = 0.17, linear regression with adjusting covariates) and back to normal in MCI and AD (Figure 4g). The negative correlation (Pearson's correlation, R = −0.37, P = 5.8e−8) between the PC1 of type I IFN signaling genes and the PC1 of miRNAs that targeted these genes (Figure 4h) suggests that the accumulation of slightly upregulated miRNA negatively regulates the type I IFN signaling pathway.
Co-expression network module negatively associated with SCD is enriched for IFN signaling pathway
To further gain a systematic understanding of the relationship between expression changes and disease status and regulatory interactions among molecules, we performed integrated weighted gene co-expression network analysis (WGCNA) for protein-coding mRNAs and lncRNAs at gene and isoform levels, and miRNA to assign individual RNAs into network modules.59
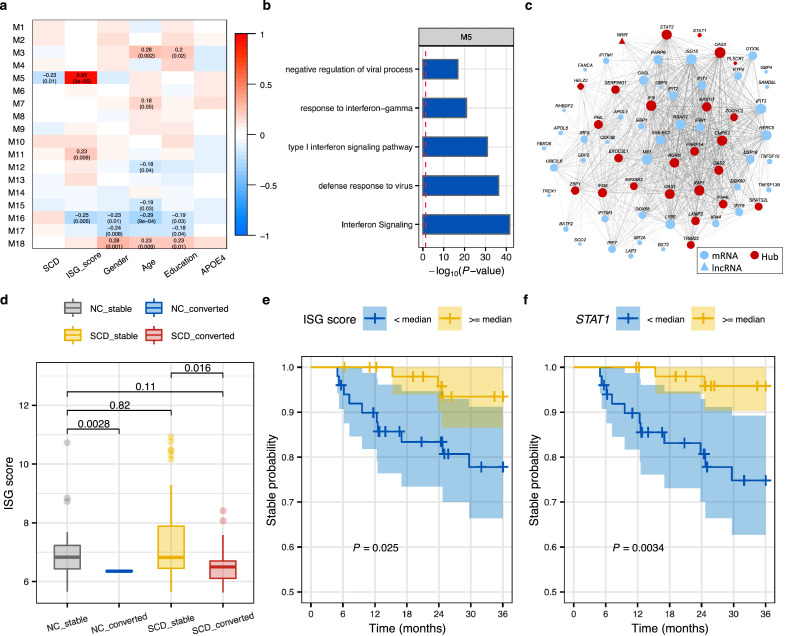
We identified 18 gene modules (Figure 5a) summarized by eigengenes (i.e., PC1) in SCD and NC individuals, and assessed the association between them and disease status and covariates. The module M5 was significantly negatively correlated with disease status (Pearson's correlation, R = −0.23, P = 0.01) but not with any confounding factors such as age, gender, RIN, or sequencing batch, suggesting that this module was primarily driven by the SCD status (Figure 5a and Supplementary Figure 10a). It showed a remarkably positive correlation with the ISG score (Pearson's correlation, R = 0.93, P = 3.0e−55) and was enriched for the interferon signaling pathway (Figure 5b). WGCNA permits screening for the hub genes that may be promising biomarkers for diagnosis and prediction of outcomes of disease.59 We evaluated the module membership (MM) and trait-based gene significance/relationship (GS) for each gene in the identified modules.59 Several hub genes with both higher correlation with diagnosis (|GS| > 0.2) and higher module connectivity (|MM| > 0.80) were identified in module M5 (Figure 5c, Supplementary Figure 10c), including the transcription factor STAT1 and lncRNA regulator NRIR of IFN signaling identified above. Besides, STAT2, another key mediator of the JAK-STAT pathway, had the highest within-module connectivity (MM = 0.94) and a strong correlation with SCD (GS = −0.22), suggesting an association with SCD and a key role in the regulation of the interferon signaling. We further performed preservation analysis59 for these modules using the ADNI expression data (Supplementary Figure 10b). We found that 13 out of 18 modules had high preservation scores (Zsummary >10), including the M5 interferon module, suggesting that the modules were strongly preserved between ours and ADNI's datasets. The isoform-level network of SCD (Supplementary Figure 11) captured the generally equivalent interferon module (i.e., module M5), as well as a module M12 related to neutrophil degranulation, mRNA metabolism, and mRNA splicing, demonstrating the importance of splicing dysregulation. In contrast to what we found in SCD, modules of virus infection (module M11) and interferon signaling (module M19) in the AD co-expression network were significantly upregulated (Supplementary Figure 12).
Figure 5.
Hub genes of the co-expression interferon signaling module serve as candidate biomarkers for conversion to MCI. (a) Pearson's correlation between module eigengenes and phenotypes. Left module-blocks represent the miRNA-lncRNA-mRNA co-expression modules at the gene level defined in SCD and NC samples. Correlation coefficients (top) and P-value (bottom) are shown in the grid, where red and blue colors indicate positive and negative correlations, respectively. Only significant associations (P < 0·05) are displayed. (b) Top 5 enriched (hypergeometric test) pathways of genes in the module M5. (c) The network of genes in the module M5, where only interaction edges with correlation coefficient > 0.15 are plotted. Edge thickness and node size correspond to pair-wise correlation and node connectivity, respectively. Node shapes represent different biotypes. Hub genes are highlighted with the red nodes. (d) The distribution of the ISG score in NC and SCD by conversion status (Stable NC = 24, converted NC = 2, Stable SCD = 88, converted SCD = 13). P values were determined with one-tailed t-test. (e-f) The Kaplan-Meier curves showing the 3-year disease conversion of SCD participants in ADNI (Alzheimer's Disease Neuroimaging Initiative) grouped by the median of the ISG score (e) and the expression of STAT1(f). Shadow indicates the 95% confidence interval. P-values of conversion difference were determined by Log-rank test. Yellow and blue denote higher or lower than the median level, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
STAT1 and IFN signature may serve as candidate biomarkers for conversion of SCD to MCI
We sought to validate whether interferon signaling is repeatedly dysregulated in SCD or associated with the progression of SCD to MCI in a longitudinal dataset from ADNI.29 Among participants who did not meet Jak/Bondi criteria for MCI, 26 NC and 101 SCD individuals were further diagnosed using neuropsychological testing (see Methods). Compared with NC (73.8 ± 5.34 years; 11 males), there was a similar gender composition but an older age distribution in the SCD group (76.0 ± 6.88 years; 41 males). Two NC and 13 SCD samples progressed to MCI within short-term (36-month) follow-up visits. Our dataset and the ADNI dataset had a limited overlap of top-ranked 500 DE genes between SCD vs. NC (Supplementary Figure 13a). There was a moderate performance (Supplementary Figure 14, AUC (the area under the curve) = 0.70 ± 0.09) in SCD diagnosis using the interferon signature and module genes in the ADNI dataset. Interestingly, we noticed that interferon-stimulated genes were consistently down-regulated in SCD with progression to MCI or AD compared with the stable ones (without progression), including STAT1, IFI6, IFIT2, OAS3, and IRF9 (Supplementary Figures 13b and 15). Moreover, SCD with conversion exhibited a lower ISG score than those without progression in both NC and SCD groups (Figure 5d), indicating the potential of interferon signaling in predicting disease progression.
We next sought to determine whether type I IFN signaling activity or the hub genes in the IFN module could be used as biomarkers for disease conversion. Firstly, the SCD participants were divided into two groups with one group having ISG scores higher than the median and the other group having ISG scores lower than the median, where the median is calculated based on all SCDs. As expected, the samples with low interferon activity were found to have a higher risk of conversion to MCI (Figure 5e). The SCD individuals were then stratified by the median expression level of each hub gene. For 20 out of 23 hub genes (Supplementary Table 8), individuals of the lower-expression group carried a higher risk of conversion to MCI compared with the higher-expression group, particularly for STAT1 (P = 0.0034, log-rank test in Kaplan-Meier survival curves, Figure 5f). In the group with lower expression of STAT1, 11 out of 50 SCD samples progressed to MCI, whereas only 2 individuals progressed to MCI in the higher-expression group. STAT1 mediates the actions of IFNs and cytokines and upregulates genes causing pathogen response.60 It was one of the most significantly downregulated genes in SCD with conversion compared with the stable ones (P = 8.73e−5, log2(FC) = −0.43, empirical Bayes statistics in limma30; Supplementary Figure 15a). Long-term follow ups of 6-year and 10-year showed similar trends as the 3-year (Supplementary Figure 16). These results were consistent with Yang et al.’s findings that higher baseline plasma interferon-γ was associated with slower cognitive decline.61 We suppose the downregulation of STAT1 and interferon signaling may weaken the cellular antiviral ability and increase virus replication (like replication of Herpseviridae),62 and therefore increase the risk of disease conversion. Taken together, the above findings indicate that STAT1, a hub gene of the interferon signaling module, as well as the IFN signature, could be used as the candidate biomarkers for disease conversion.
Discussion
We present a comprehensive transcriptome analysis for preclinical AD, including protein-coding and long noncoding genes at gene and isoform levels, local splicing, and miRNAs, as well as an integrative miRNA-mRNA-lncRNA co-expression network. We noticed that transcript isoforms exhibited larger effect sizes and disease specificity, highlighting the importance of splicing dysregulation in disease pathogenesis. We found that the activity of interferon signaling pathways was significantly down-regulated in SCD but up-regulated in MCI and AD. The impaired IFN activity in SCD may arise from differential splicing, the regulation of lncRNA (e.g., NRIR) and miRNA (e.g., has-miR-146a-5), and regulation of hub transcription factors (e.g., STAT1 and STAT2). We also identified ISG score for type I IFN signaling and a positive regulatory hub gene (STAT1) of the IFN signaling module to be candidate biomarkers for conversion to MCI in SCD. SCD individuals with lower interferon signaling activity and expression of STAT1 in ADNI exhibited a significantly higher conversion rate to MCI in both short and long-term follow-up visits.
The accumulating evidence supports the long-standing infectious hypothesis for AD etiology, such as the presence of the herpes virus in brains and amyloid plaques of AD patients, and the increased risk of dementia in samples with herpes infection.63, 64, 65 The inflammation for AD hallmarks has long been recognized that inflammation leads to the aggregation of Aβ plaques and tangles, which in turn can lead to more inflammation.66 This cascade could be initiated by microbial pathogens. Alternatively, the presence of Aβ and tau in brain is the protective response against infection.67 IFNs are a group of cytokines released by host cells to protect the cells from viral infections. Individuals with declined interferon signaling activity might have a higher risk of viral infections like Herpseviridae, and thus increased risk of progression to AD. Increased viral infection or replication would cause a feedback up-regulation of the antiviral response,68 which explains the up-regulation of defence response pathways in MCI and AD.
Antiviral treatment was associated with decreased risk of dementia.69 A pilot trial of IFNβ1a for early mild-to-moderate Alzheimer's patients showed the treatment group revealed significant improvements in the instrumental activities of daily living and physical self-maintenance scale compared with the placebo group, indicating a potential protective role for antiviral treatment against dementia development.70 Therefore, interferon supplementation for preclinical AD patients with impaired interferon activity might prevent or delay disease progression. Nevertheless, the epidemiological evidence indicated treatment of patients with non-steroidal anti-inflammatory drugs prior to the development of AD reduced the possibility of developing the disease.71 Therefore, it's also crucial to assess disease heterogeneity, the time point of initiation, and the duration of treatment.
Although we show the value of our integrative analysis presented here, there are some limitations in our study. First, although blood-based biomarkers are attractive for their easy availability, gene expression data from peripheral blood could be easily affected by underlying factors. There was a limited repeatability of the dysregulated interferon genes for SCD vs. NC in ADNI, and a moderate diagnostic power of type I interferon signature and module genes for ADNI SCD vs. NC samples (AUC = 0.7 and 0.66 in the internal and external validation, respectively). This might be due to the high heterogeneity of the SCD status, as well as the differences in diagnostic criteria (different cognitive tests for different populations), sequencing methods (RNA-seq in our dataset vs. Microarray in the ADNI), age distributions (64.73 ± 7.18 years old in our dataset vs. 76.0 ± 6.88 in ADNI), and the ethnicities (Asians in our dataset vs. non-Hispanic White Americans in ADNI). Second, there was no assessment of amyloid and tau pathology in our study, which would further help confirm the risk of conversion of SCD. Third, Alzheimer's disease is a heterogeneous disease with diverse pathophysiologic mechanisms,68 the current sample size is not enough to capture molecular subtypes, which is critical for precision medicine. In addition, the study was designed as a cross-sectional analysis, although several dysregulated pathways and candidate biomarker features were proposed, further longitudinal large-scale studies are needed to support the dynamics of interferon signaling and the validity of high-risk features.
Collectively, our integrative analysis of different transcriptome biotypes at multiple levels revealed the blood transcriptional changes across progression stages of AD, particularly in SCD. These data consistently demonstrate that interferon signaling pathways are significantly down-regulated in SCD. We propose candidate biomarkers, STAT1 and the interferon signature, for conversion from SCD to MCI. Our findings provide an alternative hypothesis of disease progression mechanisms and guidance for disease prevention and intervention.
Contributors
Zhao XM. conceived and designed the experiments. Song L. carried out data processing and analyses. Song L. and Chen J. have accessed and verified the data. Song L. wrote the manuscript. Zhao XM., Chen J. and Feng J. revised the manuscript. Guo Q. and Lo CY. collected samples. All authors read and approved the final version of the manuscript. Zhao XM. was responsible for the decision to submit the manuscript.
Data sharing statement
The mRNA- and miRNA-sequencing data generated in this study have been deposited in the Genome Sequence Archive for Human (GSA-Human) in the National Genomics Data Center (NGDC) under accession no. HRA000942. The ADNI data used in this study were obtained from the ADNI database (https://adni.loni.usc.edu). All custom code used in this work is available at the following GitHub repository: https://github.com/ZhaoXM-Lab/SCD_preAD.
Declaration of interests
The authors report no competing interests.
Acknowledgments
This work was partly supported by National Key R&D Program of China (2020YFA0712403), National Natural Science Foundation of China (61932008), Shanghai Municipal Science and Technology Major Project (2018SHZDZX01), the 111 Project (No. B18015) of China, Greater Bay Area Institute of Precision Medicine (Guangzhou) (Grand No. IPM21C008), Natural Science Foundation of Shanghai (21ZR1403200), and Shanghai Center for Brain Science and Brain-Inspired Technology. Part of the data used here is from the ZIB (Zhangjiang Internaitional Brain Bank) Consortium.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104175.
Contributor Information
Liting Song, Email: ltsong18@fudan.edu.cn.
Jingqi Chen, Email: jingqichen@fudan.edu.cn.
Chun-Yi Zac Lo, Email: zaclocy@gmail.com.
Qihao Guo, Email: qhguo@sjtu.edu.cn.
Jianfeng Feng, Email: jffeng@fudan.edu.cn.
Xing-Ming Zhao, Email: xmzhao@fudan.edu.cn.
Appendix. Supplementary materials
References
- 1.Ballard C, Gauthier S, Corbett A, Brayne C, Aarsland D, Jones E. Alzheimer's disease. Lancet. 2011;377(9770):1019–1031. doi: 10.1016/S0140-6736(10)61349-9. [DOI] [PubMed] [Google Scholar]
- 2.Morris JC. Early-stage and preclinical Alzheimer disease. Alzheimer Dis Assoc Disord. 2005;19(3):163–165. doi: 10.1097/01.wad.0000184005.22611.cc. [DOI] [PubMed] [Google Scholar]
- 3.McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer's disease: recommendations from the national institute on aging-Alzheimer's association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7(3):263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albert MS, DeKosky ST, Dickson D, et al. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the national institute on aging-Alzheimer's association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7(3):270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sperling RA, Aisen PS, Beckett LA, et al. Toward defining the preclinical stages of Alzheimer's disease: recommendations from the national institute on aging-Alzheimer's association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7(3):280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hardy JA, Higgins GA. Alzheimer's disease: the amyloid cascade hypothesis. Science. 1992;256(5054):184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 7.Sperling RA, Jack CR, Aisen PS. Testing the right target and right drug at the right stage. Sci Transl Med. 2011;3(111) doi: 10.1126/scitranslmed.3002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mantovani E, Zucchella C, Schena F, Romanelli MG, Venturelli M, Tamburin S. Towards a redefinition of cognitive frailty. J Alzheimers Dis. 2020;76(3):831–843. doi: 10.3233/JAD-200137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jessen F, Amariglio RE, van Boxtel M, et al. A conceptual framework for research on subjective cognitive decline in preclinical Alzheimer's disease. Alzheimers Dement. 2014;10(6):844–852. doi: 10.1016/j.jalz.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas KR, Bangen KJ, Weigand AJ, et al. Objective subtle cognitive difficulties predict future amyloid accumulation and neurodegeneration. Neurology. 2020;94(4):e397–e406. doi: 10.1212/WNL.0000000000008838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visser PJ, Verhey F, Knol DL, et al. Prevalence and prognostic value of CSF markers of Alzheimer's disease pathology in patients with subjective cognitive impairment or mild cognitive impairment in the DESCRIPA study: a prospective cohort study. Lancet Neurol. 2009;8(7):619–627. doi: 10.1016/S1474-4422(09)70139-5. [DOI] [PubMed] [Google Scholar]
- 12.Dufouil C, Fuhrer R, Alperovitch A. Subjective cognitive complaints and cognitive decline: consequence or predictor? The epidemiology of vascular aging study. J Am Geriatr Soc. 2005;53(4):616–621. doi: 10.1111/j.1532-5415.2005.53209.x. [DOI] [PubMed] [Google Scholar]
- 13.Glodzik-Sobanska L, Reisberg B, De Santi S, et al. Subjective memory complaints: presence, severity and future outcome in normal older subjects. Dement Geriatr Cogn. 2007;24(3):177–184. doi: 10.1159/000105604. [DOI] [PubMed] [Google Scholar]
- 14.Donovan NJ, Amariglio RE, Zoller AS, et al. Subjective cognitive concerns and neuropsychiatric predictors of progression to the early clinical stages of Alzheimer disease. Am J Geriatr Psychiatry. 2014;22(12):1642–1651. doi: 10.1016/j.jagp.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abner EL, Kryscio RJ, Caban-Holt AM, Schmitt FA. Baseline subjective memory complaints associate with increased risk of incident dementia: the PREADVISE trial. J Prev Alzheimers Dis. 2015;2(1):11–16. doi: 10.14283/jpad.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parfenov VA, Zakharov VV, Kabaeva AR, Vakhnina NV. Subjective cognitive decline as a predictor of future cognitive decline: a systematic review. Dement Neuropsychol. 2020;14(3):248–257. doi: 10.1590/1980-57642020dn14-030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iturria-Medina Y, Khan AF, Adewale Q, Shirazi AH. Alzheimer's disease neuroimaging I. Blood and brain gene expression trajectories mirror neuropathology and clinical deterioration in neurodegeneration. Brain. 2020;143(2):661–673. doi: 10.1093/brain/awz400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reimand J, Arak T, Adler P, et al. g:Profiler-a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Res. 2016;44(W1):W83–W89. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kramer A, Green J, Pollard J, Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30(4):523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1 alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 23.Huang HY, Lin YCD, Li J, et al. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020;48(D1):D148–D154. doi: 10.1093/nar/gkz896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu WJ, Wang XW. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20:1–10. doi: 10.1186/s13059-019-1629-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen YH, Wang XW. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li YI, Knowles DA, Humphrey J, et al. Annotation-free quantification of RNA splicing using LeafCutter. Nat Genet. 2018;50(1):151–158. doi: 10.1038/s41588-017-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ongen H, Buil A, Brown AA, Dermitzakis ET, Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics. 2016;32(10):1479–1485. doi: 10.1093/bioinformatics/btv722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol. 2005;4:1–45. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 29.Mueller SG, Weiner MW, Thal LJ, et al. Alzheimer's disease neuroimaging initiative. Adv Behav Biol. 2008;57:183–189. [Google Scholar]
- 30.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gandal MJ, Zhang P, Hadjimichael E, et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362(6420) doi: 10.1126/science.aat8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vuong CK, Black DL, Zheng S. The neurogenetics of alternative splicing. Nat Rev Neurosci. 2016;17(5):265–281. doi: 10.1038/nrn.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Quervain DJF, Poirier R, Wollmer MA, et al. Glucocorticoid-related genetic susceptibility for Alzheimer's disease. Hum Mol Genet. 2004;13(1):47–52. doi: 10.1093/hmg/ddg361. [DOI] [PubMed] [Google Scholar]
- 34.Llorens-Martin M, Jurado J, Hernadez F, Avila J. GSK-3 beta, a pivotal kinase in Alzheimer disease. Front Mol Neurosci. 2014;7 doi: 10.3389/fnmol.2014.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shigemizu D, Mori T, Akiyama S, et al. Identification of potential blood biomarkers for early diagnosis of Alzheimer's disease through RNA sequencing analysis. Alzheimers Res Ther. 2020;12(1):87. doi: 10.1186/s13195-020-00654-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Au-Yeung N, Mandhana R, Horvath CM. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT. 2013;2(3):e23931. doi: 10.4161/jkst.23931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Shea JJ, Schwartz DM, Villarino AV, Gadina M, McInnes IB, Laurence A. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu Rev Med. 2015;66:311–328. doi: 10.1146/annurev-med-051113-024537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lovheim H, Gilthorpe J, Johansson A, Eriksson S, Hallmans G, Elgh F. Herpes simplex infection and the risk of Alzheimer's disease: a nested case-control study. Alzheimers Dement. 2015;11(6):587–592. doi: 10.1016/j.jalz.2014.07.157. [DOI] [PubMed] [Google Scholar]
- 39.Roy ER, Wang B, Wan YW, et al. Type I interferon response drives neuroinflammation and synapse loss in Alzheimer disease. J Clin Investig. 2020;130(4):1912–1930. doi: 10.1172/JCI133737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hadjadj J, Yatim N, Barnabei L, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369(6504):718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeremiah N, Neven B, Gentili M, et al. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J Clin Investig. 2014;124(12):5516–5520. doi: 10.1172/JCI79100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol. 2016;16(10):626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 43.Kornblihtt AR, Schor IE, Allo M, Dujardin G, Petrillo E, Munoz MJ. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol. 2013;14(3):153–165. doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- 44.Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Satoh J, Kawana N, Yamamoto Y. Pathway analysis of ChIP-Seq-based NRF1 target genes suggests a logical hypothesis of their involvement in the pathogenesis of neurodegenerative diseases. Gene Regul Syst Biol. 2013;7:139–152. doi: 10.4137/GRSB.S13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li HD, Funk CC, McFarland K, et al. Integrative functional genomic analysis of intron retention in human and mouse brain with Alzheimer’s disease. Alzheimers Dement. 2021;17(6):984–1004. doi: 10.1002/alz.12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raj T, Li YI, Wong G, et al. Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility. Nat Genet. 2018;50(11):1584–1592. doi: 10.1038/s41588-018-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mendes de Almeida R, Tavares J, Martins S, et al. Whole gene sequencing identifies deep-intronic variants with potential functional impact in patients with hypertrophic cardiomyopathy. PLoS One. 2017;12(8) doi: 10.1371/journal.pone.0182946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genom Proteom Bioinform. 2009;7(4):147–154. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gil N, Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet. 2020;21(2):102–117. doi: 10.1038/s41576-019-0184-5. [DOI] [PubMed] [Google Scholar]
- 51.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kopp F. Molecular functions and biological roles of long non-coding RNAs in human physiology and disease. J Gene Med. 2019;21(8):e3104. doi: 10.1002/jgm.3104. [DOI] [PubMed] [Google Scholar]
- 53.Delay C, Mandemakers W, Hebert SS. MicroRNAs in Alzheimer's disease. Neurobiol Dis. 2012;46(2):285–290. doi: 10.1016/j.nbd.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 54.Mariotti B, Servaas NH, Rossato M, et al. The long non-coding RNA NRIR drives IFN-response in monocytes: implication for systemic sclerosis. Front Immunol. 2019;10 doi: 10.3389/fimmu.2019.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagaraj S, Zoltowska KM, Laskowska-Kaszub K, Wojda U. MicroRNA diagnostic panel for Alzheimer's disease and epigenetic trade-off between neurodegeneration and cancer. Ageing Res Rev. 2019;49:125–143. doi: 10.1016/j.arr.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 56.Banzhaf-Strathmann J, Benito E, May S, et al. MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer's disease. EMBO J. 2014;33(15):1667–1680. doi: 10.15252/embj.201387576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lusardi TA, Phillips JI, Wiedrick JT, et al. MicroRNAs in human cerebrospinal fluid as biomarkers for Alzheimer's disease. J Alzheimers Dis. 2017;55(3):1223–1233. doi: 10.3233/JAD-160835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burgos K, Malenica I, Metpally R, et al. Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer's and Parkinson's diseases correlate with disease status and features of pathology. PLoS One. 2014;9(5):e94839. doi: 10.1371/journal.pone.0094839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Casanova JL, Holland SM, Notarangelo LD. Inborn errors of human JAKs and STATs. Immunity. 2012;36(4):515–528. doi: 10.1016/j.immuni.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang HS, Zhang C, Carlyle BC, et al. Plasma IL-12/IFN-gamma axis predicts cognitive trajectories in cognitively unimpaired older adults. Alzheimers Dement. 2022;18(4):645–653. doi: 10.1002/alz.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crameri M, Bauer M, Caduff N, et al. MxB is an interferon-induced restriction factor of human herpesviruses. Nat Commun. 2018;9:1–16. doi: 10.1038/s41467-018-04379-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carbone I, Lazzarotto T, Ianni M, et al. Herpes virus in Alzheimer's disease: relation to progression of the disease. Neurobiol Aging. 2014;35(1):122–129. doi: 10.1016/j.neurobiolaging.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 64.Sochocka M, Zwolinska K, Leszek J. The infectious etiology of Alzheimer's disease. Curr Neuropharmacol. 2017;15(7):996–1009. doi: 10.2174/1570159X15666170313122937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Middleton PJ, Petric M, Kozak M, Rewcastle NB, Crappermclachlan DR. Herpes-simplex viral genome and senile and presenile dementias of Alzheimer and pick. Lancet. 1980;1(8176):1038. doi: 10.1016/s0140-6736(80)91490-7. [DOI] [PubMed] [Google Scholar]
- 66.Akiyama H, Barger S, Barnum S, et al. Inflammation and Alzheimer's disease. Neurobiol Aging. 2000;21(3):383–421. doi: 10.1016/s0197-4580(00)00124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bourgade K, Le Page A, Bocti C, et al. Protective effect of amyloid-beta peptides against herpes simplex virus-1 infection in a neuronal cell culture model. J Alzheimers Dis. 2016;50(4):1227–1241. doi: 10.3233/JAD-150652. [DOI] [PubMed] [Google Scholar]
- 68.Neff RA, Wang MH, Vatansever S, et al. Molecular subtyping of Alzheimer’s disease using RNA sequencing data reveals novel mechanisms and targets. Sci Adv. 2021;7(2) doi: 10.1126/sciadv.abb5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lopatko Lindman K, Hemmingsson ES, Weidung B, et al. Herpesvirus infections, antiviral treatment, and the risk of dementia-a registry-based cohort study in Sweden. Alzheimers Dement (NY) 2021;7(1):e12119. doi: 10.1002/trc2.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grimaldi LME, Zappala G, Iemolo F, et al. A pilot study on the use of interferon beta-1a in early Alzheimer’s disease subjects. J Neuroinflamm. 2014;11:1–6. doi: 10.1186/1742-2094-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.in t' Veld BA, Ruitenberg A, Hofman A, et al. Nonsteroidal antiinflammatory drugs and the risk of Alzheimer's disease. N Engl J Med. 2001;345(21):1515–1521. doi: 10.1056/NEJMoa010178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.