Summary
Thermogenic brown and beige adipocytes counteract obesity by enhancing energy dissipation via uncoupling protein-1 (Ucp1). However, the effect of genetic variation on these cells, a major source of disease susceptibility, has been less well studied. Here we examined beige adipocytes from obesity-prone C57BL/6J (B6) and obesity-resistant 129X1/SvJ (129) mouse strains and identified a cis-regulatory variant rs47238345 that is responsible for differential Ucp1 expression. The alternative T allele of rs47238345 at the Ucp1 -12kb enhancer in 129 facilitates the allele-specific binding of nuclear factor I-A (NFIA) to mediate allele-specific enhancer-promoter interaction and Ucp1 transcription. Furthermore, CRISPR-Cas9/Cpf1-mediated single nucleotide polymorphism (SNP) editing of rs47238345 resulted in increased Ucp1 expression. We also identified Lim homeobox protein 8 (Lhx8), whose expression is higher in 129 than in B6, as a trans-acting regulator of Ucp1 in mice and humans. These results demonstrate the cis- and trans-acting effects of genetic variation on Ucp1 expression that underlie phenotypic diversity.
Subject areas: Physiology, Molecular physiology, Molecular biology, Epigenetics
Graphical abstract

Highlights
-
•
NFIA in adipocytes determines Ucp1 expression between obesity-prone and -resistant mouse strains
-
•
Allele-specific binding of NFIA at the Ucp1 -12kb enhancer mediates differential Ucp1 expression
-
•
Editing of a SNP at the Ucp1 -12kb enhancer is sufficient to increase Ucp1 in obesity-prone strain
Physiology; Molecular physiology; Molecular biology; Epigenetics
Introduction
While white adipose tissue (WAT) stores excess energy in the form of lipid, brown adipose tissue (BAT) dissipates energy in the form of heat via mitochondrial uncoupling protein-1 (Ucp1) as well as through other pathways. A subset of white adipocytes in WAT depots undergo “browning” in response to cold and β-adrenergic stimulation. These types of thermogenic, Ucp1-positive adipocytes are called beige adipocytes. We previously identified a transcription factor nuclear factor I-A (NFIA) as a regulator of brown and beige adipocyte differentiation. NFIA activates the brown-fat-specific enhancers and facilitates the binding of peroxisome proliferator-activated receptor γ (PPARγ), the master transcriptional regulator of adipogenesis, to control the brown fat gene program (Hiraike et al., 2017). Since the re-discovery of human thermogenic adipose tissue (Cypess et al., 2009; van Marken Lichtenbelt et al., 2009; Nedergaard et al., 2007; Saito et al., 2009; Virtanen et al., 2009), these tissues have attracted interest as a promising target for the treatment of obesity and its complications including type 2 diabetes. Thermogenic brown and beige adipocyte activity is inversely correlated with body mass index (van Marken Lichtenbelt et al., 2009) and age (Yoneshiro et al., 2011). However, its activity is highly variable even among lean and young individuals (Yoneshiro et al., 2013). Interestingly, inbred mouse strains also show differential vulnerability to diet-induced obesity and exhibit concordant adipocyte browning capability (Almind and Kahn, 2004; Guerra et al., 1998; Soccio et al., 2017), suggesting the effect of genetic variation on thermogenic adipocyte activity in mice and humans.
Genetic variation is a major source of diversity in phenotypes and disease susceptibility. Although coding variants are obviously important in disease pathogenesis, more than 80% of disease-associated variants identified by genome-wide association studies (GWAS) lie within non-coding regions (Maurano et al., 2012), suggesting that these variants work by regulating target gene expression. Non-coding variants affect target gene expression by altering transcription factor binding motif and also by long-range chromatin interaction in a tissue-specific manner (Claussnitzer et al., 2015; Musunuru et al., 2010; Smemo et al., 2014). Recently, inbred mouse strains have been used to dissect the effects of genetic variation (Heinz et al., 2013; Soccio et al., 2015). Here, we comprehensively analyzed the effect of genetic variation on adipocyte browning using obesity-prone C57BL/6J (B6) mice, obesity-resistant 129X1/SvJ (129) mice, and F1 offspring of these two strains as a model system. We focused on beige adipocytes because Ucp1 expression in beige adipocytes is strongly affected by genetic variation among inbred mice strains, while that in classical brown adipocytes is relatively constant across strains (Guerra et al., 1998). We observed an extensive cis effect of genetic variation on expression levels of Ucp1, the most representative effector gene of thermogenic adipocytes. Moreover, we identified a cis-regulatory variant, rs47238345, at the Ucp1 -12kb enhancer that is responsible for differential Ucp1 expression through allele-specific binding analysis of nuclear factor I-A (NFIA). We also identified Lim homeobox protein 8 (Lhx8), whose expression is higher in 129 than in B6, as a trans-acting positive regulator of Ucp1 expression.
Results
The difference in browning potential between C57BL/6J and 129X1/SvJ is cell-autonomous
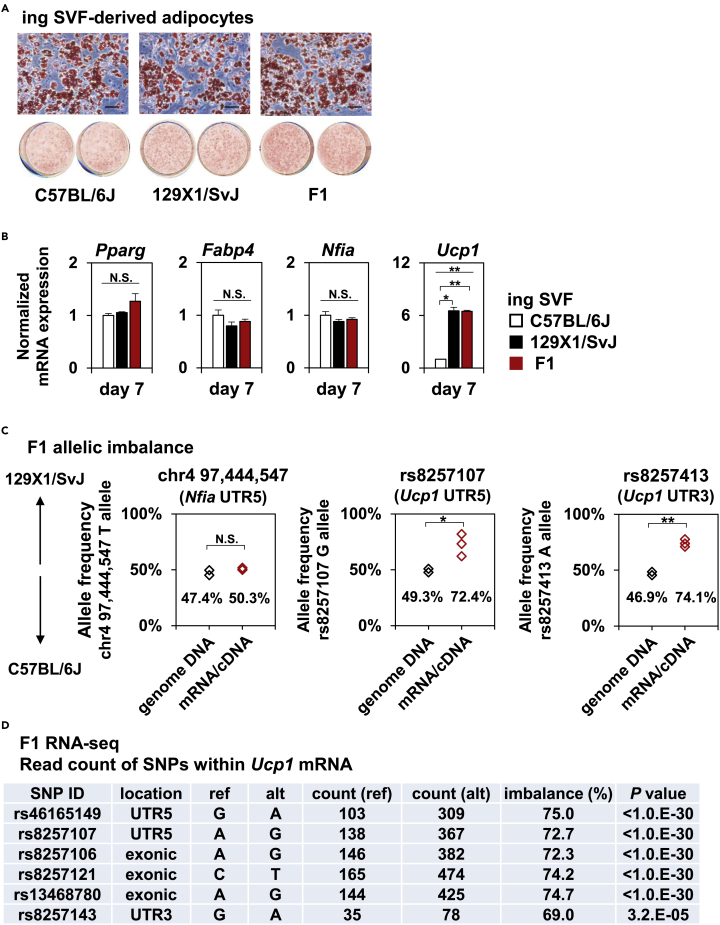
While B6 is the most commonly used inbred mouse strain in metabolism research, this strain is more prone to diet-induced obesity than other strains such as 129. Consistently, the 129 strain is known to exhibit higher browning capability compared to B6. To examine whether this difference is cell-autonomous, we isolated stromal vascular fraction (SVF) from inguinal white adipose tissue (iWAT) of B6 and 129 mice, immortalized the cells and induced adipocyte differentiation using cocktails for thermogenic adipocytes. B6- and 129-derived cells exhibited a comparable degree of adipocyte differentiation when evaluated by Oil red O staining and mRNA expression of Pparg and its target Fabp4 (Figures 1A and 1B). Nevertheless, 129-derived cells showed significantly higher Ucp1 expression at both the mRNA and the protein levels (Figures 1C and 1D). Among well-known canonical thermogenic genes, we found that RNA expression of Ppargc1a was also moderately up-regulated, while that of Dio2 was unchanged, in 129-derived cells (Figure 1C). We found no difference in mRNA expression of Tle3, a negative regulator of thermogenic gene program (Pearson et al., 2019; Villanueva et al., 2013). We observed that mRNA expression of genes implicated in Ucp1-independent thermogenesis (Chouchani et al., 2019) was comparable between B6 and 129 or even lower in 129 cells (in the case of Ckmt1, Figure 1E), suggesting that the difference in browning capability between B6 and 129 cells is predominantly, if not entirely, attributable to Ucp1-dependent thermogenesis. Of note, we did not find a difference in mRNA expression of known transcriptional regulators of thermogenic adipocytes including Nfia, Ebf2, and Prdm16 (Figure 1F), or for protein expression of NFIA (Figure 1D), suggesting that differences in browning capability are not explained by the abundance of these known regulators. These results indicate that 129-derived adipocytes show higher Ucp1 expression than B6-derived cells in a cell-autonomous manner; this difference is not explained by differential expression of known transcriptional regulators including NFIA.
Figure 1.
The difference in browning potential between C57BL/6J and 129X1/SvJ is cell-autonomous
(A) Stromal vascular fraction (SVF) from inguinal white adipose tissue (iWAT) of C57BL/6J and 129X1/SvJ mice were immortalized and induced to differentiate into thermogenic adipocytes. The cells were stained with Oil Red O at day 7 of differentiation. Scale bar, 100μm.
(B and C) Common adipocyte genes Pparg and Fabp4 (B) and genes involved in canonical thermogenesis such as Ucp1, Ppargc1a, Dio2, and Tle3 (C) were quantified by RT-qPCR at the indicated time points (mean ± SEM; n = 3 independent samples; ∗∗p < 0.01; N.S., not significant; significance was determined by two-tailed Student’s t test).
(D) Western blot analysis of UCP1 and NFIA in C57BL/6J and 129X1/SvJ cells. β actin was used as a loading control.
(E and F) Genes indicated in Ucp1-independent thermogenesis (E) and transcriptional regulator of brown and beige adipocytes (F) were quantified by RT-qPCR at the indicated time points (mean ± SEM; n = 3 independent samples; ∗∗p < 0.01; N.S., not significant; significance was determined by two-tailed Student’s t test).
Cis effect of genetic variation is a major driver of difference in Ucp1 expression between B6 and 129
To address the mechanism(s) by which genetic variation determines differences in browning potential and Ucp1 expression between B6- and 129-derived cells, we performed a whole-genome sequencing analysis of 129 cells and identified 5,434,592 single nucleotide variants (SNVs) compared to the ncbi37/mm9 C57BL/6J genome. As most GWAS signals map to non-coding regions and work as cis-regulatory variants, to address the involvement of a cis effect of genetic variation on Ucp1 expression between B6 and 129, we made F1 offspring of B6 and 129 mice and again isolated SVFs from iWAT of B6, 129 and F1 mice. The degree of adipocyte differentiation was comparable among the three groups as evaluated by Oil Red O staining (Figure 2A). mRNA expression of Pparg, Fabp4, and Nfia were also comparable among the three groups (Figure 2B). In this condition, 129 cells and F1 cells exhibited significantly higher Ucp1 expression than B6 cells (Figure 2B).
Figure 2.
Cis effect on Ucp1 mRNA expression in F1 offspring of B6 and 129
(A) SVF from iWAT of B6, 129, and F1 mice were immortalized and induced to differentiate into thermogenic adipocyte differentiation. The cells were stained with Oil Red O at day 7 of differentiation. Scale bar, 100μm.
(B) Ppag, Fabp4, Nfia and Ucp1 were quantified by RT-qPCR at the indicated time points. One-way ANOVA followed by Bonferroni’ post-hoc test. (mean ± SEM; n = 3 independent samples; ∗p < 0.05, ∗∗p < 0.01; N.S., not significant).
(C) Allelic imbalance of the mRNA as well as genomic DNA at the indicated locus were evaluated by qPCR analysis (mean ± SEM; n = 3 independent samples; ∗p < 0.05, ∗∗p < 0.01; N.S., not significant; significance was determined by two-tailed Student’s t test).
(D) A table showing the read count as well as allelic imbalance of all the six SNPs that lies within Ucp1 mRNA. Raw mapped counts of RNA-seq to reference allele (B6) and alternative allele (129) are shown. Imbalance means the rate of the alternative count. p values were calculated using the binomial test. See also Figure S1.
To directly examine the possible cis effect on gene expression, we evaluated imbalanced gene expression in F1 cells. We did not find an imbalanced expression of Nfia mRNA, whose expression was comparable between B6 and 129, in F1 cells (Figure 2C, left). On the other hand, we did find a 129-favoring, significant, and strong (>70%) imbalance in Ucp1 mRNA when examined using two independent single nucleotide polymorphisms (SNPs, rs8257107 and rs8257143) using qPCR assays (Figure 2C, middle and right). Consistently, we found a significant and strong imbalance in all the six SNPs that lie within Ucp1 mRNA in RNA-seq analysis of F1 cells (Figure 2D). Multiple genes located near Ucp1 also exhibited 129-favoring imbalance, suggesting that genes within the same topologically associated domain (TAD) are coordinately regulated in cis (Figure S1A). Publicly available Hi-C analysis of differentiating 3T3-L1 adipocytes suggests that these genes are, indeed, located within the same TAD (Figure S1B).
Allelic imbalance of NFI binding at the Ucp1 -12kb enhancer
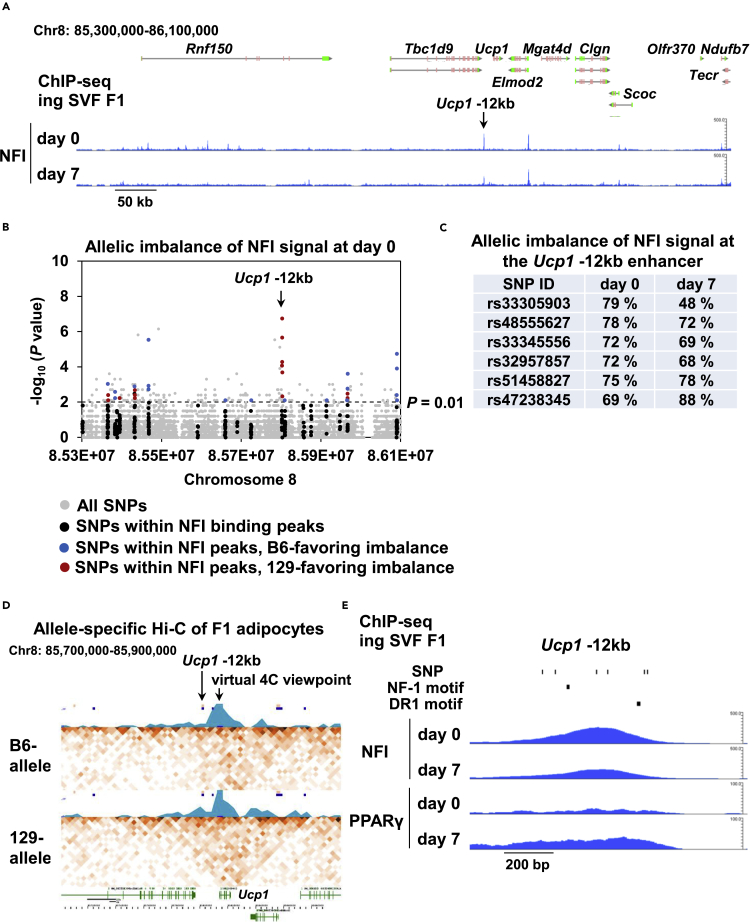
Imbalanced Ucp1 expression is predicted to result from imbalanced transcription factor binding at the Ucp1 enhancer. To address this, we performed chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq) analysis in F1 cells using the NFI antibody, which reacts primarily with NFIA (Figure 3A). We have shown previously that NFIA activates the brown-fat-specific enhancers even before differentiation and facilitates the binding of PPARγ to control the brown fat gene program (Hiraike et al., 2017). Therefore, we sought to evaluate imbalanced NFI binding between the B6-derived allele and 129-derived allele in F1 cells using SNPs at the Ucp1 locus at day 0 of differentiation. Quite strikingly, we observed six consecutive SNPs that exhibited significant and 129-favoring strong allelic imbalance at the Ucp1 -12kb enhancer (Figure 3B). This allelic imbalance was observed throughout the differentiation (Figure 3C). Furthermore, Hi-C analysis of differentiated F1 adipocytes showed 129-favoring allele-specific long-range chromatin interaction between the Ucp1 -12kb enhancer and its promoter (Figure 3D), strongly indicating that 129-favoring allelic imbalance of NFI binding at the Ucp1 -12kb enhancer results in allele-specific enhancer-promoter interaction, in turn leading to allele-specific Ucp1 expression. Of note, because B6-derived and 129-derived alleles are equally exposed to trans-acting effects in F1 cells, these results demonstrate that cis effects determine differential binding of NFI at the Ucp1 enhancer in F1 cells. Having said that, we did not find an overlap between six SNPs and the NF-1 motif nor the DR-1 motif, a binding motif for PPARγ that is recruited to the Ucp1 enhancer by NFIA during adipocyte differentiation (Figure 3E); this result suggests that differential NFI binding at the Ucp1 -12kb enhancer cannot be explained by the alteration of motif for NFI itself nor that for PPARγ.
Figure 3.
Allelic imbalance of NFI binding at the Ucp1 -12kb enhancer
(A) A ChIP-seq track for NFI in F1 cells at the Ucp1 loci before and after adipocyte differentiation.
(B) A plot showing the -log10 (p value) of the allelic imbalance of NFI binding signal at day 0 of adipocyte differentiation near the Ucp1 loci. p values for the allelic imbalance were calculated using the binomial test.
(C) A table showing the 129-favoring allelic imbalance of NFI binding signal (the rate of ChIP-seq tag count for 129 alleles) of six SNPs located at the Ucp1 -12kb enhancer.
(D) Hi-C matrix and virtual 4C view near Ucp1 locus. Hi-C interaction matrix for B6-allele and 129-allele is shown, respectively, with 3kb bin. A virtual 4C view (light blue graph) summarizes the interaction counts viewed from the transcription start site (TSS) of Ucp1.
(E) A ChIP-seq track for NFI and PPARγ before and after differentiation at the Ucp1 -12kb enhancer. Location of six SNPs at the Ucp1 -12kb enhancer as well as the location of NF-1 as well as DR-1 motifs are also shown.
rs47238345 at the Ucp1 enhancer disrupts the motif for vitamin D receptor (VDR) in the 129 allele, inhibits VDR binding, and facilitates reciprocal NFIA binding to increase Ucp1 expression
To explore a transcription factor whose binding motif is affected by the aforementioned six SNPs in an unbiased manner, we searched for binding motifs that overlap with these six SNPs at the Ucp1 -12kb enhancer (Figure 4A). An overlap between rs47238345 and the binding motif for RXRα:VDR heterodimer (DR-3 motif) particularly raised our attention because the alternative T allele of rs47238345 in 129 cells disrupts second direct repeats of the motif and was predicted to decrease the binding affinity of RXRα:VDR to the genome (Figure 4B, note that reverse complementary sequence of the genome is shown to indicate canonical direct repeat three motif, or two directly repeated AGGTCA-like motifs spaced by three base pairs). Moreover, VDR has been reported to be a negative regulator of UCP1 expression in human adipocytes in a ligand-independent manner (Malloy and Feldman, 2013). In addition, VDR knockout in B6 mice resulted in increased Ucp1 expression and protection from diet-induced obesity (Narvaez et al., 2009), and the previously published ChIP-seq analysis for VDR in 3T3-L1 preadipocytes 4 hours after adding DMI (dexamethasone, methylisobutylxanthine, and insulin) cocktail revealed VDR binding at the Ucp1 -12kb enhancer (Siersbæk et al., 2014) (Figure 4C). Protein expression of VDR was comparable between B6- and 129-derived preadipocytes (Figure 4D).
Figure 4.
rs47238345 at the Ucp1 enhancer disrupts the VDR motif in the 129 alleles, inhibits VDR binding, and facilitates reciprocal NFIA binding to increase Ucp1 expression
(A) A table showing the overlap between six SNPs at the Ucp1 enhancer and known transcription factor binding motifs. Overlap between rs48555627 or rs51458827 and known transcription factor binding motifs were not observed in our computational analysis. The FIMO online software converted log-odds scores into p values, assuming a zero-order background model.
(B) A consensus RXR:VDR motif logo is shown above the motif sequence in B6 and 129 strains at the Ucp1 -12kb enhancer. The location of rs47238345 is indicated. Note that reverse complementary sequence of the genome is shown to indicate canonical direct repeat three motif (DR3 motif), or two directly repeated AGGTCA-like motifs spaced by three base pairs.
(C) A ChIP-seq track showing the overlap of NFI binding sites in F1 cells and VDR binding sites in 3T3-L1 cells at the Ucp1 -12kb enhancer.
(D) Western blot analysis of VDR in B6 and 129 cells. β actin was used as a loading control.
(E) Control siRNA or siRNA for VDR was transfected into B6 cells and the cells were stained with Oil Red O six days after inducing adipocyte differentiation. Scale bar, 100μm.
(F) Nr1i1 was quantified by RT-qPCR at the indicated time points (mean ± SEM; n = 3 independent samples; ∗∗p < 0.01; significance was determined by two-tailed Student’s t test).
(G) Western blot analysis of VDR in control and VDR knockdown cells. Β actin was used as a loading control.
(H and I) Common adipocyte genes Pparg and Fabp4 (H) and Nfia and Ucp1 (I) were quantified by RT-qPCR at the indicated time points (mean ± SEM; n = 3 independent samples; ∗∗p < 0.01; N.S., not significant; significance was determined by two-tailed Student’s t test).
(J) ChIP-qPCR analysis of NFI. Ucp1 9.5kb and Pparg2 53kb are background sites (mean ± SEM; n = 2 independent samples). The representative result of multiple independent experiments is shown. See also Figure S2.
To address the functional role of VDR on adipocyte differentiation as well as Ucp1 expression in our model system, we performed a small interfering RNA (siRNA)-mediated knockdown experiment of VDR in B6 cells (Figure 4E). Compared with 129-derived cells, B6 cells were predicted to possess a higher binding affinity to VDR at the Ucp1 -12kb enhancer owing to their reference C allele of rs47238345. We observed a significant knockdown of VDR at both the RNA and protein levels (Figures 4F and 4G). Although the degree of adipocyte differentiation was comparable between control and knockdown cells when evaluated by Oil red O staining (Figure 4E) and mRNA expression of Pparg and Fabp4 (Figure 4H), expression levels of Ucp1 were significantly up-regulated by knockdown of VDR (Figure 4I). On the other hand, expression levels of Nfia as well as other well-known canonical thermogenic genes Ppargc1a and Dio2 were unaltered (Figures 4I and S2A). Furthermore, ChIP-qPCR analysis showed that the binding of NFI to the Ucp1 -12kb enhancer was facilitated in knockdown cells, suggesting a competitive relationship between NFIA and VDR at the Ucp1 -12kb enhancer (Figure 4J, left). The binding of NFIA to the Pparg2 77kb enhancer was unchanged upon VDR knockdown (Figure 4J, right).
We also focused on an overlap between rs32957857 and the binding motif for BCL6b (B-cell CLL/lymphoma six member B). Although the role of BCL6b in adipocyte differentiation has not yet been described, BCL6b is known as transcriptional repressor and the alternative C allele of rs3295787 in 129 cells was predicted to decrease the binding affinity of BCL6b to the genome (Figure S2B). siRNA-mediated knockdown of BCL6b (Figure S2C) resulted in the up-regulation of Fabp4 mRNA and down-regulation of Pparg mRNA, while Nfia mRNA was comparable between control and knockdown cells (Figure S2D). In this condition, we found significantly up-regulated expression of Ucp1 mRNA (Figure S2E), suggesting that the de-repression of Ucp1 by BCL6b in 129-alelle also contributes to 129-favoring allelic imbalance of Ucp1 mRNA expression in F1 cells.
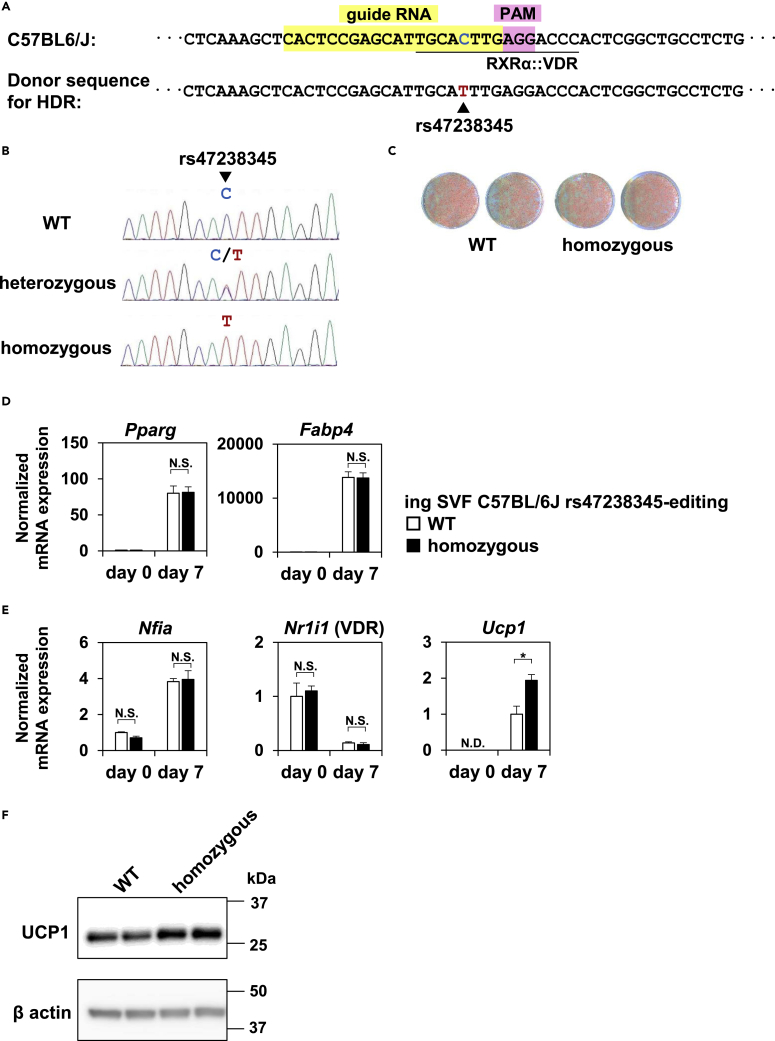
SNP editing of rs47238345 at the Ucp1 -12kb enhancer in mice resulted in increased Ucp1 expression
To directly examine the functional consequences of the alternative T allele of rs47238345 on the transcriptional regulation of Ucp1, we performed SNP editing of rs47238345 in mice using clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9/Cpf1. We designed guide RNA to edit rs47238345 from reference C (B6) to alternative T (129) in the B6 background (Figure 5A) and achieved successful editing (Figure 5B). We isolated SVF from iWAT of wild-type and homozygously rs47238345-edited mice, immortalized the cells, and induced thermogenic adipocyte differentiation. The degree of adipocyte differentiation was comparable between wild-type and rs47238345-edited cells when evaluated by Oil red O staining (Figure 5C) and mRNA expression of Pparg and Fabp4 (Figure 5D). mRNA expression of Nfia and Nr1i1 (which encodes VDR) were also comparable (Figure 5E). Nevertheless, we found significantly up-regulated expression of Ucp1 in rs47238345-edited cells (Figure 5E), demonstrating the physiological importance of rs47238345 in the transcriptional regulation of Ucp1 expression. Expression levels of other canonical thermogenic genes such as Ppargc1a and Dio2 were unchanged (Figure S3A), suggesting the specific effect of rs47238345 editing on Ucp1 expression. We also observed that protein expression of Ucp1 was up-regulated in rs47238345-edited cells (Figure 5F). However, mRNA expression of Ucp1 in rs47238345-edited cells was lower than that in 129 adipocytes (Figure S3B), suggesting that variant(s) other than rs47238345, including rs32957857 (Figures S2A-S2D), also contributes to the effect of genetic variation on Ucp1 expression between B6 and 129. Indeed, no difference was observed in body weight between WT mice and rs47238345-edited mice on a high-fat diet at thermoneutrality (Figure S3C), indicating that editing of rs47238345 alone is not sufficient to protect mice from obesity.
Figure 5.
SNP editing of rs47238345 at the Ucp1 -12kb enhancer in mice resulted in increased Ucp1 expression
(A) A guide RNA sequence, PAM site, and donor sequence for homology-directed repair (HDR) to edit rs47238345 from C to T in the B6 background is shown.
(B) A sanger sequence showing successful editing of rs47238345 in wild-type, heterozygously and homozygously edited mice.
(C) SVF from iWAT of wild-type and homozygously edited mice were immortalized and induced to differentiate into thermogenic adipocytes. The cells were stained with Oil Red O at day 7 of differentiation.
(D and E) Common adipocyte genes Pparg and Fabp4 (D) and Nfia and Ucp1 (E) were quantified by RT-qPCR at the indicated time points (mean ± SEM; n = 3 independent samples; ∗p < 0.05; N.S., not significant; significance was determined by two-tailed Student’s t test).
(F) Western blot analysis of UCP1 and NFIA in WT and rs47238345 homozygously edited cells. β actin was used as a loading control. See also Figure S3.
Protein-coding single nucleotide polymorphisms have a limited effect on differences in browning capability between B6 and 129
To gain further insights into the mechanism(s) by which genetic variation affects differential Ucp1 expression between B6 and 129, we also explored possible trans-acting regulator. In this regard, we identified the protein-coding, nonsynonymous variant rs32942538, which causes arginine (R) to glutamine (Q) substitution at codon 761 of PRDM16 (Figure S4A, NM_001177995). We did not find nonsynonymous variants in other representative regulators of thermogenic adipocytes such as PPARγ, EBF2, and NFIA.
To examine the effect of the R761Q mutant on the function of PRDM16, we introduced wild-type and R761Q mutant PRDM16 into C2C12 myoblasts using retroviral vectors (Figures S4B and S4C). We also included ΔZF-1 mutant of PRDM16 which lacks a binding domain for PPARγ and therefore lacks the ability to induce brown adipogenesis (Kajimura et al., 2009; Seale et al., 2008), as a negative control (Figure S4C). Ectopically introduced wild-type and R761Q PRDM16 exhibited similar protein expression, while ΔZF-1 PRDM16 appeared to have a slightly lower expression (Figure S4D). In this condition, wild-type and R761Q, but not ΔZF-1 mutant, were able to induce adipocyte differentiation as evaluated by Oil red O staining and mRNA expression of Pparg and Fabp4 (Figures S3C and S3E). R761Q mutant was also able to induce Ucp1 expression as wild-type PRDM16 (Figure S2E); this suggests that R761Q mutation does not affect the brown adipogenic function of PRDM16, at least in this experimental condition.
Lim homeobox protein-8 (Lhx8) is a trans-acting browning regulator
We also examined the possibility that changes in the abundance of regulatory proteins might contribute to differences in browning capability between B6 and 129. To this end, we performed RNA-seq analysis of B6- and 129-derived adipocytes and found fourteen transcription factors whose expression levels were significantly higher in 129 cells than in B6 cells (>2-fold, p value <0.05, Figure 6A). To prioritize candidate transcription factors that positively regulate adipocyte browning, we used a publicly available RNA-seq dataset of inguinal WAT (iWAT) and epididymal WAT (eWAT)(Soccio et al., 2015) and hypothesized that positive regulator of adipocyte browning exhibits higher expression in iWAT compared to eWAT because iWAT is known to possess higher browning capacity. This led us to focus on a transcription factor Lim homeobox protein-8 (Lhx8), and independently performed qPCR analysis validated the significantly higher expression of Lhx8 in 129 compared to B6 cells (Figure 6B). We also confirmed that expression levels of Lhx8 were significantly higher in iWAT compared to eWAT, of 16 weeks old, male B6 mice on a normal chow diet (Figure 6C). Of note, Lhx8 has been proposed as a marker of classical brown adipocytes rather than beige adipocytes (Petrovic et al., 2010), although this was later modulated by the same group (de Jong et al., 2015).
Figure 6.
Lim homeobox protein-8 (Lhx8) is a trans-acting browning regulator
(A) A table showing the list of up-regulated transcription factors (TDs) in 129X1/SvJ cells compared to C57BL/6J cells in RNA-seq (>2-fold, p < 0.05).
(B) Lhx8 were quantified by RT-qPCR in B6 and 129 cells (mean ± SEM; n = 3 independent samples; ∗p < 0.05; significance was determined by two-tailed Student’s t test).
(C) Lhx8 were quantified by RT-qPCR in iWAT and eWAT, of 16 weeks old, male B6 mice on a normal chow diet (n = 4 independent samples; ∗p < 0.05; significance was determined by two-tailed Student’s t test).
(D) Control siRNA or siRNA for Lhx8 was transfected into 129 cells and the cells were stained with Oil Red O seven days after inducing adipocyte differentiation. Scale bar, 100μm.
(E-G) Lhx8 (E), common adipocyte genes Pparg and Fabp4 (F), and Nfia and Ucp1 (G) were quantified by RT-qPCR at the indicated time points (mean ± SEM; n = 3 independent samples; ∗p < 0.05, ∗∗p < 0.01; significance was determined by two-tailed Student’s t test).
(H) Control and 3xFLAG-tagged Lhx8 expressing B6 cells were stained with Oil Red O seven days after inducing adipocyte differentiation. Scale bar, 100μm.
(I) Western blot analysis of 3xFLAG-Lhx8 in control and 3xFLAG-Lhx8-expressing cells. β actin was used as a loading control.
(J and K) Common adipocyte genes Pparg and Fabp4 (J), and genes involved in the thermogenic gene program Nfia, Ucp1, Ppargc1a, and Dio2 (K) were quantified by RT-qPCR at the indicated time points (mean ± SEM; n = 3 independent samples; ∗p < 0.05; N.S., not significant; significance was determined by two-tailed Student’s t test). See also Figure S4.
To examine the endogenous role of Lhx8 on adipocyte differentiation and adipocyte browning, we performed siRNA-mediated loss-of-function experiment using 129 cells (Figure 6D). siRNA for Lhx8 achieved approximately 80% knockdown compared with control siRNA throughout the differentiation (Figure 6E). Knockdown of Lhx8 resulted in severely impaired adipocyte differentiation both in terms of lipid accumulation as evaluated by Oil red O staining (Figure 6D) and mRNA expression of Pparg as well as Fabp4 (Figure 6F). Accordingly, mRNA expression of Nfia and Ucp1 was severely impaired (Figure 6G). We also performed a gain-of-function experiment using a retroviral vector that expresses 3xFLAG-tagged Lhx8 in B6 cells (Figure 6H). The introduction of 3xFLAG-tagged Lhx8 (Figure 6I) did not affect the degree of lipid accumulation (Figure 6H) or mRNA expression of Pparg and Fabp4 (Figure 6J). However, we found significantly up-regulated expression of Ucp1 in Lhx8-expressing cells, while expression levels of Nfia were unaltered (Figure 6K). Expression levels of Ppargc1a were somewhat decreased and that of Dio2 were unaltered, suggesting that the effect of Lhx8 on the thermogenic gene program would be specific to Ucp1 (Figure 6K). Together, these results indicate that Lhx8, whose expression is higher in 129 than in B6, is required for adipocyte differentiation and also a positive regulator of Ucp1 expression.
Expression levels of LHX8 and UCP1 in human perirenal brown adipose tissue were concurrently higher in patients with pheochromocytoma
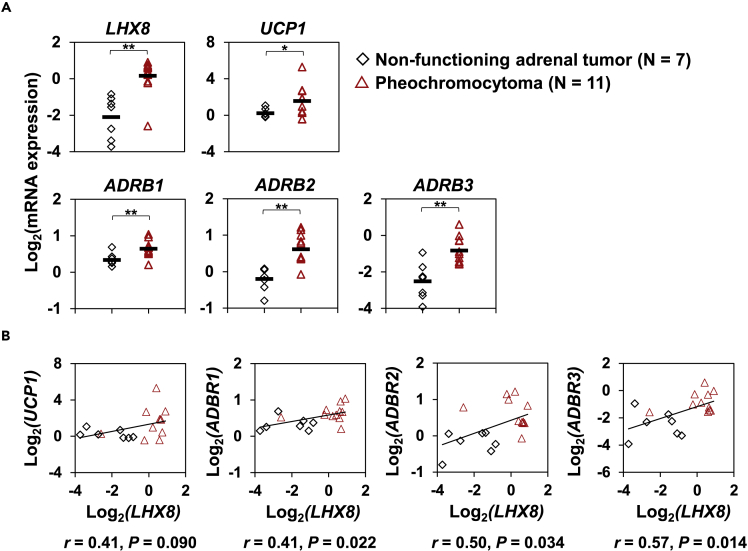
Finally, to explore the translational relevance of the role of Lhx8 on thermogenic adipocytes in humans, we analyzed the perirenal BAT of human patients with pheochromocytoma and non-functioning adrenal tumors. Perirenal BAT is activated in patients with pheochromocytoma (Nagano et al., 2015). We observed that expression levels of LHX8 and UCP1 in perirenal BAT were concurrently higher in patients with pheochromocytoma than in those who had non-functioning adrenal tumors, although the association between expression levels of LHX8 and UCP1 did not reach significance (Figures 7A and 7B). We also found that expression levels of ADRB1, ADRB2, and ADRB3 were up-regulated in patients with pheochromocytoma (Figure 7A). Moreover, expression levels of these genes were significantly and positively correlated with LHX8 expression (Figure 7B). These results suggest LHX8 may act as a trans-acting positive regulator of thermogenic adipocytes also in humans.
Figure 7.
Expression levels of LHX8 and UCP1 in human perirenal BAT were concurrently higher in patients with pheochromocytoma
(A) Expression levels of LHX8, UCP1, ADRB1, ADRB2, and ADRB3 in perirenal brown fat of human patients with pheochromocytoma or non-functioning adrenal tumors were quantified by RT-qPCR (mean ± SEM; N = 7 independent samples for non-functioning adrenal tumors and N = 11 independent samples for pheochromocytoma; ∗p < 0.05, ∗∗p < 0.01; significance was determined by two-tailed Student’s t test). Expression levels of UCP1 and LHX8 mRNA were re-analyzed by a qPCR run for this work using the same tissues used in Nagano et al. (2015).
(B) Correlation of mRNA expression between LHX8 and indicated genes. Pearson’s correlation coefficients and p values are shown.
Discussion
Understanding how non-coding regulatory variants modulate target gene expression is key to translating GWAS results into mechanistic insights and understanding the interindividual difference in disease susceptibility. Using beige adipocytes from obesity-prone B6 and obesity-resistant 129 mouse strains as a model system, we identified the causal SNP rs47238345 that is responsible for differential Ucp1 expression between these two strains. We showed that the alternative T allele of rs47238345 in 129-allele precludes the binding of VDR, a negative regulator of Ucp1, by disrupting its binding motif and reciprocally facilitating the binding of NFIA, leading to the up-regulation of Ucp1 transcription. We directly demonstrated the functional importance of rs47238345 through CRISPR-Cas9/Cpf1-mediated SNP editing in mice. Thermogenic beige adipocytes derived from rs47238345-edited mice exhibited higher Ucp1 expression than cells derived from wild-type mice. However, no difference was observed in body weight between WT mice and rs47238345-edited mice on a high-fat diet at thermoneutrality, suggesting that other variant(s) including rs32957857 also contribute to the effect of genetic variation on Ucp1 expression between B6 and 129. We also found that Lhx8, whose expression is higher in 129 than in B6, is a trans-acting positive regulator of Ucp1 expression. Finally, we observed that LHX8 and UCP1 in human perirenal BAT were concurrently up-regulated in patients with pheochromocytoma compared with non-functioning adrenal tumors.
In this work, we thoroughly investigated the allele-specific regulation of Ucp1, the most representative effector gene of thermogenic adipocytes. An alternative approach would include looking at allele-specific transcription factor binding and allele-specific chromatin interaction in a genome-wide manner, focusing on loci that exhibit significant 129-favored imbalance, and exploring previously unappreciated effector genes that determine thermogenic adipocyte function. Accumulating evidence suggests that thermogenic brown and beige adipocytes possess Ucp1-independent thermogenic pathways and that these cells also exhibit physiological roles beyond heat generation (Chouchani et al., 2019; Kajimura et al., 2015). A genome-wide allele-specific analysis will further identify previously unappreciated pathways that contribute to the beneficial effect of thermogenic adipocytes to counteract obesity and diabetes.
The differential browning capability among various inbred mouse strains has been known for decades. Although 129 and A/J strains exhibit higher browning capacity, B6 strain exhibits lower browning capacity: accordingly, B6 strain is prone to obesity and insulin resistance (Almind and Kahn, 2004; Guerra et al., 1998; Surwit et al., 1998). Efforts have been made to understand the mechanism(s) underlying the differential browning capability and differential Ucp1 expression among different strains (Li et al., 2019; Soccio et al., 2017; Xue et al., 2005). However, causal SNP(s) that explain these differences have remained elusive. In this study, using comprehensive genomics, epigenomics, transcriptomics, and CRISPR-Cas9/Cpf1-mediated SNP editing in mice, we demonstrated that rs47238345 is responsible for, at least partly, the differential Ucp1 expression between B6 and 129 strains. We assert that our strategy for identifying the causal SNP(s) between B6 and 129 is directly applicable also to human studies. In this context, although UCP1 has not been implicated in obesity GWAS nor type 2 diabetes GWAS so far, a UCP1 -3826 G to A variant rs1800592 is known to be associated with UCP1 mRNA abundance (Esterbauer et al., 1998) as well as BMI (Heilbronn et al., 2000). Additionally, a UCP1 5′ UTR A to C variant that alters promoter activity is reported to be associated with type 2 diabetes (Mori et al., 2001). Unbiased and comprehensive analyses to identify and dissect regulatory variant(s) associated with obesity and diabetes via modulating UCP1 expression in human thermogenic adipocytes represent an attractive direction for future study.
In conclusion, here we identified rs47238345 as a causal variant that determines differential Ucp1 expression between obesity-prone B6 and obesity-resistant 129 mouse strains. rs47238345 works as a cis-regulatory variant and modulates the binding motif for VDR, resulting in a 129-favoring allele-specific binding of NFIA. We also identified trans-acting regulator Lhx8 and its relevance to human thermogenic adipocytes. These results would provide a platform for understanding the mechanism(s) by which genetic variation affects human thermogenic adipocyte activity and opens a door toward implementing precision medicine in anti-obesity therapy.
Limitations of the study
Here we identified the effect of genetic variation between C57BL/6J and 129X1/SvJ mouse strains on Ucp1 expression in a comprehensive manner. However, the effect of genetic variation on other functional genes in thermogenic adipocytes remains unexplored. Also, allele-specific analysis using human samples would be required to identify and dissect regulatory variant(s) that controls UCP1 expression in human thermogenic adipocytes. These experiments would establish thermogenic adipocytes as a target for the treatment of obesity and diabetes via a precision medicine approach.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-NFIA | Sigma | HPA006111; RRID: AB_1854422 |
| anti-UCP1 | Abcam | ab10983; RRID: AB_2241462 |
| anti-VDR | Santa Cruz Biotechnology | sc-13133; RRID: AB_628040 |
| anti-FLAG M2 | Sigma | F3165; RRID: AB_259529 |
| anti-β actin | Sigma | A3854; RRID: AB_262011 |
| anti-NFI | Santa Cruz Biotechnology | sc-30198; RRID: AB_2287441 |
| anti-PPARγ | Santa Cruz Biotechnology | sc-7273; RRID: AB_628115 |
| anti-PPARγ | Perseus Proteomics | A3409A; RRID: AB_843370 |
| Biological samples | ||
| Human tissue samples | Nagano et al., 2015 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Dexamethasone | Wako | 041-18861 |
| IBMX | Sigma | 17018-1G |
| Rosiglitazone | GlaxoSmithKline | BRL49653C |
| Indomethacin | Sigma | I7378 |
| Humulin R U-100 | Eli Lily | N/A |
| T3 | Fluka | 91990 |
| Forskolin | Sigma | F3917 |
| Critical commercial assays | ||
| KOD mutagenesis kit | TOYOBO | SMK-101 |
| Lipofectamine RNAiMAX | Invitrogen | 13778-150 |
| Lipofectamine 2000 | Invitrogen | 11668-019 |
| TruSeq Stranded mRNA Library Prep Kit | Illumina | RS-122-2101 |
| KAPA hyper prep kit | KAPA Biosystems | KK8502 |
| Protein A Sepharose 4 Fast Flow | GE | 17-1279-02 |
| Protein G Sepharose 4 Fast Flow | GE | 17-0618-02 |
| MboI | NEB | R0147 |
| biotin-14-dATP | Life Technologies | 19524-016 |
| DNA Polymerase I, Large (Klenow) Fragment | NEB | M0210 |
| T4 DNA Ligase | NEB | M0202 |
| AMPure XP beads | Beckman Coulter | A63882 |
| Dynabeads MyOne Streptavidin T1 beads | Life Technologies | 65602 |
| Nextera DNA Sample Prep kit | Illumina | FC-121-1030 |
| Deposited data | ||
| RNA-seq analysis of beige adipocytes from obesity prone C57BL/6J (B6), obesity-resistant 129X1/SvJ (129) and F1 offspring. | This paper | GSE188263 |
| Whole genome-seq analysis of beige adipocytes from obesity-resistant 129X1/SvJ (129). | This paper | GSE188263 |
| ChIP-seq analysis of beige adipocytes from F1 offspring of obesity prone C57BL/6J and obesity-resistant 129X1/SvJ (129). | This paper | GSE188263 |
| HiC analysis of beige adipocytes from F1 offspring of obesity prone C57BL/6J and obesity-resistant 129X1/SvJ (129). | This paper | GSE188263 |
| Experimental models: Cell lines | ||
| ing WAT-derived SVF, C57BL/6J | This paper | N/A |
| ing WAT-derived SVF, 129X1/SvJ | This paper | N/A |
| ing WAT-derived SVF, F1 | This paper | N/A |
| ing WAT-derived SVF, C57BL/6J, rs47238345 homozygously edited | This paper | N/A |
| C2C12 | ATCC | CRL-1772 |
| Plat E | Cell Biolabs | RV-101 |
| Experimental models: Organisms/strains | ||
| C57BL/6J rs47238345-edited mice | This paper | N/A |
| Oligonucleotides | ||
| Guide RNA sequence, donor sequence for HDR and qPCR primers used in this study | This paper | Sequences are shown in Table S1 |
| Control siRNA | Santa Cruz Biotechnology | sc-37007 |
| siVDR | Santa Cruz Biotechnology | sc-36811 |
| siBCL6b | Santa Cruz Biotechnology | sc-141670 |
| Recombinant DNA | ||
| pMXs-3xFLAG-LHX8 | This paper | N/A |
| pMSCV-PRDM16 WT | Hiraike et al., 2017 | N/A |
| pMSCV-PRDM16 R761Q | This paper | N/A |
| pMSCV-PRDM16 ΔZF-1 | This paper | N/A |
| Software and algorithms | ||
| BWA-MEM 0.7.10 | Li and Durbin, 2009 | N/A |
| Picard software ver 2.9.4 | Broad institute | N/A |
| Novosort ver.1.03.01 | Novocraft Technologies | N/A |
| GATK HaplotypeCaller ver 3.7-0-gcfedb67 | Depristo et al., 2011 | N/A |
| batman aligner | Tennakoon et al., 2012 | N/A |
| MACS ver. 1.4.2 | Zhang et al., 2008 | N/A |
| STAR aligner ver. 2.7.3a | Dobin et al., 2013 | N/A |
| Hi-C pro pipeline ver. 2.9.0 | Servant et al., 2015 | N/A |
| FIMO ver. 4.12.0 | Grant et al., 2011 | N/A |
| Other | ||
| High Fat Diet 32 | Clea Japan | HFD32 |
Resource availability
Lead contact
Further information and request for resources and reagents should be directed to and would be fulfilled by the lead contact, Yuta Hiraike.
Materials availability
Unique materials generated in this study is available upon complete materials transfer agreement.
Experimental model and subject details
Cell culture
Stromal vascular fraction (SVF) of inguinal white adipose tissue from 8 weeks-old male mice was isolated as reported previously(Hiraike et al., 2020) using gentleMACS™ Octo Dissociator with Heaters (Miltenyi Biotec). The cells were then immortalized using retroviral vector expressing SV-40 large T antigen. For adipocyte differentiation of immortalized SVF cells, at the confluence, cells were treated for 48 hours in medium containing 10% FBS, 0.5 mM isobutylmethylxanthine, 125 nM indomethacin, 1 μM dexamethasone, 850 nM insulin, 1 nM T3 and 1 μM rosiglitazone. After 48 hours, cells were switched to medium containing 10% FBS, 850 nM insulin, 1 nM T3 and 1 μM rosiglitazone. Recombinant DNA experiments was approved by the committee on genetically modified organisms of the Graduate School of Medicine, the University of Tokyo and conducted according to the institutional guidelines at the University of Tokyo.
Mice studies
SNP editing of rs47238345 in mice using clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9/Cpf1 was performed in Laboratory of Animal Resources, Center for Disease Biology and Integrative Medicine, Graduate School of Medicine, The University of Tokyo. We designed a guide RNA to edit rs47238345 from C (reference, B6) to T (alternative, 129) in C57BL/6J background. Cas9 protein (Guide-it recombinant Cas9 protein), sgRNA (Integrated DNA technologies) and ssDNA (FASMAC) were delivered to C57BL/6J embryos at the pronuclear stage by microinjection. Briefly, embryos were transferred to M2 medium (M7167, SIGMA-ALDRICH), and microinjected with the mixture containing Cas9 protein (50 ng/μL), sgRNA (25 ng/μL) and ssDNA (5 ng/μL). After microinjection, survived embryos were cultured in mWM for a night and were transferred into oviducts of 0.5-day-post-coitum recipients. Mutant mosaic mice were crossed with C57BL/6J mice to obtain mice with germline transmission. Male mice were used for all the experiment in this study, including primary cell isolation. A guide RNA sequence and a donor sequence used for homology directed repair (HDR) in shown in Table S1A. All animal work was approved by Institutional Animal Care and Use Committees (IACUC) of the University of Tokyo (P14-056, H17-014 and P18-030) and conducted according to the institutional guidelines at the University of Tokyo.
Human studies
Perirenal BAT samples were obtained from eleven patients with pheochromocytoma (52.2 ± 4.8 years old, 4 subjects were male and 7 were female) and seven with non-functioning adrenal tumors (50.7 ± 5.0 years old, 2 subjects were male and 5 were female), as previously described (Nagano et al., 2015). All procedures were approved by the Hiroshima University Ethics Committee and also by the research ethics committee of the Graduate School of Medicine, the University of Tokyo. All the procedures were conducted according to the Declaration of Helsinki, and all the patients gave written informed consent before taking part in the study.
Method details
Retroviral expression system
For gain-of-function experiments, we used the pMXs retroviral expression system as previously described (Hiraike et al., 2017). Retroviral vectors expressing PRDM mutants were constructed using the KOD mutagenesis kit (TOYOBO) according to the manufacturer’s instructions.
siRNA-mediated gene knockdown
For VDR or BCL6b knockdown experiments by lipofection, a control siRNA and a siRNA for VDR or BCL6b was purchased from Santa Cruz Biotechnology (sc-37007, sc-36811 and sc-141670). The siRNA was transfected using lipofectamine RNAiMAX (Invitrogen) 2 days before confluence, according to the manufacture’s instruction.
RNA expression analysis
Total RNA from cultured cells or tissues was isolated using TRIzol reagent (Invitrogen) and RNeasy Mini columns (QIAGEN). Isolated RNA was reverse-transcribed using ReverTra Ace qPCR RT Master Mix kit (Takara). Real-time quantitative PCR (SYBR green) analysis was performed on QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems). Rplp0 was used as an internal normalization control. For RNA-seq, libraries were prepared using TruSeq Stranded mRNA Library Prep Kit (Illumina) according to the manufacturer’s instructions. A list of primers used for RT-qPCR analysis is shown in Tables S1B and S1C.
Western blotting
Tissues were lysed in radioimmunoprecipitation assay (RIPA) buffer containing 0.1% SDS, 1% NP-40, 0.5% Na deoxycholate, 150 mM NaCl, 50 mM Tris-Cl (pH 8.0), 1 mM EDTA supplemented with protease inhibitor (Roche). Proteins were separated by SDS-PAGE, transferred to nitrocellulose membrane, and detected with the antibodies anti-NFIA (1:1000 dilution, Sigma HPA006111), anti-UCP1 (1:2000 dilution, Abcam, ab10983), anti-VDR (1:100 dilution, Santa Cruz Biotechnology, sc-13133), anti-FLAG M2 (Sigma F3165), and anti-β actin (Sigma A3854).
ChIP
ChIP was performed as described previously(Hiraike et al., 2017, 2020) with some modifications. Briefly, samples were treated by nuclear extraction buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2 and 0.1% IGEPAL CA-630) for 10 minutes and immediately cross-linked with 1% formaldehyde for 7.5 minutes at room temperature. Cross-linking was quenched using 125 mM glycine for 5 minutes. The chromatin was sheared by a probe sonicator (Branson) and was spun at 15,000 rpm for 5 minutes. Antibodies were added for overnight incubation at 4°C. Mixes of Protein A and Protein G Sepharose (GE) added to samples for 4 hours at 4°C. Subsequent procedures were performed as described previously. The antibodies used were NFI (Santa Cruz Biotechnology, sc-30198) and PPARγ (mix of Santa Cruz Biotechnology, sc-7273, and Perseus Proteomics, A3409A). ChIP-seq libraries were prepared using KAPA hyper prep kit (KAPA Biosystems) according to the manufacturer’s instructions. A list of primers used for ChIP-qPCR analysis is shown in Table S1D.
Hi-C
Hi-C experiments were performed following the previously published in-situ Hi-C protocol(Rao et al., 2014). Briefly, five million cells were cross-linked with 1% formaldehyde following quenching with 0.2 M of glycine. Cells were lysed with Hi-C lysis buffer (10 mM Tris-HCl pH8.0, 10 mM NaCl, 0.2% Igepal CA-630) and the chromatin was digested by MboI (NEB, R0147) restriction enzyme. The both ends of the digested chromatin were filled in and marked with biotin-14-dATP (Life Technologies, 19524-016) by DNA Polymerase I, Large (Klenow) Fragment (NEB, M0210), and ligated by T4 DNA Ligase (NEB, M0202). The DNA was purified by ethanol precipitation and sheared to 300–500 bp using Covaris E220 following size selection by AMPure XP beads (Beckman Coulter, A63882). Then the DNA fragments were enriched by Dynabeads MyOne Streptavidin T1 beads (Life Technologies, 65602). After repair of sheared DNA ends, the Illumina indexed adapters were ligated to them. Hi-C libraries were amplified by 8–12 cycles of PCR before high-throughput sequencing.
High-throughput sequencing
High-throughput sequencing was performed by using the HiSeq 2500, HiSeqX or MiSeq sequencer (Illumina).
Whole genome sequencing (WGS) and variant calls
Whole genome sequencing libraries were prepared from genomic DNA using the Nextera DNA Sample Prep kit (Illumina). The sequencing libraries were sequenced on a HiSeqX using TruSeq SBS kit v3 at Takara Bio Inc, according to the manufacturer’s instructions. The one hundred and fifty cycles of single-end sequencing was performed and the primary base call files were de-multiplexed and converted into FASTQ format using bcl2fastq (ver1.8.4) pipeline. FASTQ reads were alignfed to reference mouse genome build mm9 (canonical) by using BWA-MEM 0.7.10 with default parameters as previously described (Li and Durbin, 2009; Mandai et al., 2017). The mapped reads were assigned to read groups and sorted using Picard software (ver 2.9.4). Presumed PCR duplicate reads were eliminated using Novosort ver.1.03.01 (Novocraft Technologies). The variant calling of single nucleotide variants (SNVs) was performed using Genome Analysis Toolkit (GATK) Best Practice Workflow pipeline using HaplotypeCaller (ver 3.7-0-gcfedb67) according to the GATK Best Practices for somatic short variant discovery(Depristo et al., 2011). This analysis identified 5,434,592 SNVs in 129X1/SvJ compared to reference mouse genome build mm9 (C57BL/6J).
ChIP-seq data processing
High-throughput sequence data were checked with fastqc (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The trimmed data were mapped to mm9 genome using batman aligner – “BatMis” version 3.0.0 with option “-n2 -m1” (using only uniquely mapped and allowing mismatch up to 2)(Tennakoon et al., 2012). Peak calling was performed using MACS(Zhang et al., 2008) ver. 1.4.2 with p values 10−5. Regarding the allelic imbalance analysis of ChIP-seq in F1 cells, we summarized the count for A (C57BL/6J, B6) and B (129X1/SvJ,129) allele at the positions of 5,434,592 SNPs from the mapped BAM file. We applied the null-hypothesis that both alleles were equally bound by the NFI transcription factor. Then the count of A and B should follow binomial distribution. The probabilities of allelic imbalance for each SNP position were calculated. Known transcription factor motifs that overlap with SNPs in the Ucp1 -12kb enhancer was searched using the FIMO online software(Grant et al., 2011) version 4.12.0, using “JASPAR2018_CORE_vertebrates_non-redundant” as input motifs.
RNA-seq data processing
The sequenced data were mapped to mm9 genome and Refseq information derived at UCSC web site (https://genome.ucsc.edu/). The STAR aligner(Dobin et al., 2013) version 2.7.3a was applied to make RNA-seq count data with “FilterMultimapNmax 1”. The RPKM data for RNA-seq count were used for further analyses. For the allelic imbalance analysis of RNA-seq in F1 cells, we followed the same strategy as the imbalance analysis of ChIP-seq in F1 cells.
Hi-C data processing
Hi-C data were processed by Hi-C pro pipeline(Servant et al., 2015) vers. 2.9.0 considered with restriction fragment information of MboI and mm9 mouse genome with default option. Briefly, this pipeline was able to exclude low-quality mapped pairs. The valid pairs were used for further analyses. The allele-specific analysis was also processed by Hi-C pro with SNPs information between B6 and 129. Paired fragments were mapped to 129 and 129, or B6 and B6 with approximately rate 3–4%. Those mapped to B6 and 129 respectively were 0.1–0.2%. The Hi-C data were visualized using in-house program. Virtual 4C views were summarized at any viewpoint of genomic position with any resolution.
Quantification and statistical analysis
Statistics and reproducibility
Data were shown as mean ± s.e.m unless otherwise specified. The exact numbers of replicates were shown in each legend. Two-tailed student's t-test was performed to determine the statistical significance between two groups unless otherwise specified, with a p value of less than 0.05 considered significant. One-way ANOVA analysis followed by Bonferroni’ post-hoc test was performed for multi-group comparison. Binomial test was performed for the allelic imbalance analysis. The FIMO online software(Grant et al., 2011) was used for the analysis of the overlap between known transcription factor motifs and SNPs, and the software converted log-odds scores into p values, assuming a zero-order background model. We checked that the data met the assumption of the statistic tests, and variances were similar between the groups being tested.
Acknowledgments
We thank Kaori Shiina and Shiro Fukuda (The University of Tokyo, Tokyo, Japan) for their help in high-throughput sequencing. This work is funded by a research grant from the University of Tokyo Excellent Young Researcher Program to Y.H.; by a Japan Society for the Promotion of Science (JSPS) KAKENHI Grant-in-Aid for Early-Career Scientists, grant number 19K17976 to Y.H.; by a grant for Front Runner of Future Diabetes Research (FFDR) from the Japan Foundation for Applied Enzymology, grant number 17F005 to Y.H.; by a grant from Pharmacological Research Foundation to Y.H.; by a grant from Mochida Memorial Foundation for Medical and Pharmaceutical Research to Y.H.; by a grant from MSD Life Science Foundation to Y.H.; by a grant from Daiwa Securities Health Foundation to Y.H.; by a grant from Tokyo Biochemical Research Foundation to Y.H.; by a JSPS KAKENHI Grant-in-Aid for Scientific Research (B), grant number 20H04101 to H.W.; by a JSPS KAKENHI Grant-in-Aid for Scientific Research (C), grant number 17K09818 to H.W.; by a grant from the Cell Science Research Foundation to H.W.; by a grant from Takeda Science Foundation to H.W.; by a grant from The Naito Foundation to H.W.; by a JSPS KAKENHI Grant-in-Aid for Scientific Research on Innovative Areas (Research in a proposed research area), grant number 19H05744 to H.A.; by an AMED-FORCE research grant from the Japan Agency for Medical Research and Development (AMED), grant number 21gm4010014h0001 to T.Y.; by an AMED-CREST research grant from AMED, grant number JP18gm0510018 to T.Y.; by a Takeda Hosho Grant for Research in Medicine from Takeda Science Foundation to T.Y.
Author contributions
Y.H. and H.W. designed the study. Y.H., T.W., M.O., K.S., and S.O. performed experiments. S.T., M.N., and Y.H. performed computational analysis under the supervision of H.A. M.K., H.N., and A.A. generated rs47238345-edited mice. G.N., H.O., K.O., and M.Y. performed the analysis of human perirenal BAT. Y.H. wrote the article with inputs from T.K., H.A., H.W., and T.Y. All authors reviewed and approved the final article.
Declaration of interests
We declare that none of the authors have a financial interest related to this work.
Published: August 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104729.
Contributor Information
Yuta Hiraike, Email: hiraike-tky@umin.net.
Hiroyuki Aburatani, Email: haburata-tky@umin.ac.jp.
Hironori Waki, Email: wakih@gipc.akita-u.ac.jp.
Toshimasa Yamauchi, Email: tyamau@m.u-tokyo.ac.jp.
Supplemental information
Data and code availability
-
•
High-throughput sequencing data have been deposited at the Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. The accession number is listed in the key resources table.
-
•
This work does not report original code.
-
•
Any additional data or information required to reanalyze the findings shown here are available from the lead contact upon request.
References
- Almind K., Kahn C.R. Genetic determinants of energy expenditure and insulin resistance in diet-induced obesity in mice. Diabetes. 2004;53:3274–3285. doi: 10.2337/diabetes.53.12.3274. [DOI] [PubMed] [Google Scholar]
- Chouchani E.T., Kazak L., Spiegelman B.M. New advances in adaptive thermogenesis: UCP1 and beyond. Cell Metab. 2019;29:27–37. doi: 10.1016/j.cmet.2018.11.002. [DOI] [PubMed] [Google Scholar]
- Claussnitzer M., Dankel S.N., Kim K.-H., Quon G., Meuleman W., Haugen C., Glunk V., Sousa I.S., Beaudry J.L., Puviindran V., et al. FTO obesity variant circuitry and adipocyte browning in humans. N. Engl. J. Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess A.M., Lehman S., Williams G., Tal I., Rodman D., Goldfine A.B., Kuo F.C., Palmer E.L., Tseng Y.-H., Doria A., et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., Del Angel G., Rivas M.A., Hanna M., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterbauer H., Oberkofler H., Liu Y.M., Breban D., Hell E., Krempler F., Patsch W. Uncoupling protein-1 mRNA expression in obese human subjects: the role of sequence variations at the uncoupling protein-1 gene locus. J. Lipid Res. 1998;39:834–844. [PubMed] [Google Scholar]
- Grant C.E., Bailey T.L., Noble W.S. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C., Koza R.A., Yamashita H., Walsh K., Kozak L.P. Emergence of brown adipocytes in white fat in mice is under genetic control effects on body weight and adiposity. J. Clin. Invest. 1998;102:412–420. doi: 10.1172/JCI3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilbronn L.K., Kind K.L., Pancewicz E., Morris A.M., Noakes M., Clifton P.M. Association of -3826 G variant in uncoupling protein-1 with increased BMI in overweight Australian women. Diabetologia. 2000;43:242–244. doi: 10.1007/s001250050036. [DOI] [PubMed] [Google Scholar]
- Heinz S., Romanoski C.E., Benner C., Allison K.A., Kaikkonen M.U., Orozco L.D., Glass C.K. Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503:487–492. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraike Y., Waki H., Yu J., Nakamura M., Miyake K., Nagano G., Nakaki R., Suzuki K., Kobayashi H., Yamamoto S., et al. NFIA co-localizes with PPARγ and transcriptionally controls the brown fat gene program. Nat. Cell Biol. 2017;19:1081–1092. doi: 10.1038/ncb3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraike Y., Waki H., Miyake K., Wada T., Oguchi M., Saito K., Tsutsumi S., Aburatani H., Yamauchi T., Kadowaki T. NFIA differentially controls adipogenic and myogenic gene program through distinct pathways to ensure brown and beige adipocyte differentiation. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1009044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong J.M.A., Larsson O., Cannon B., Nedergaard J. A stringent validation of mouse adipose tissue identity markers. Am. J. Physiol. Endocrinol. Metab. 2015;308:E1085–E1105. doi: 10.1152/ajpendo.00023.2015. [DOI] [PubMed] [Google Scholar]
- Kajimura S., Seale P., Kubota K., Lunsford E., Frangioni J.V., Gygi S.P., Spiegelman B.M. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S., Spiegelman B.M., Seale P. Brown and beige fat: physiological roles beyond heat generation. Cell Metab. 2015;22:546–559. doi: 10.1016/j.cmet.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Schwalie P.C., Bast-Habersbrunner A., Mocek S., Russeil J., Fromme T., Deplancke B., Klingenspor M. Systems-genetics-Based inference of a core regulatory network underlying white fat browning. Cell Rep. 2019;29:4099–4113.e5. doi: 10.1016/j.celrep.2019.11.053. [DOI] [PubMed] [Google Scholar]
- Malloy P.J., Feldman B.J. Cell-autonomous regulation of brown fat identity gene UCP1 by unliganded vitamin D receptor. Mol. Endocrinol. 2013;27:1632–1642. doi: 10.1210/me.2013-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandai M., Watanabe A., Kurimoto Y., Hirami Y., Morinaga C., Daimon T., Fujihara M., Akimaru H., Sakai N., Shibata Y., et al. Autologous induced stem-cell-derived retinal cells for macular degeneration. N. Engl. J. Med. 2017;376:1038–1046. doi: 10.1056/NEJMoa1608368. [DOI] [PubMed] [Google Scholar]
- van Marken Lichtenbelt W.D., Vanhommerig J.W., Smulders N.M., Drossaerts J.M.A.F.L., Kemerink G.J., Bouvy N.D., Schrauwen P., Teule G.J.J. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J., et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H., Okazawa H., Iwamoto K., Maeda E., Hashiramoto M., Kasuga M. A polymorphism in the 5′ untranslated region and a Met229→ Leu variant in exon 5 of the human UCP1 gene are associated with susceptibility to Type II diabetes mellitus. Diabetologia. 2001;44:373–376. doi: 10.1007/s001250051629. [DOI] [PubMed] [Google Scholar]
- Musunuru K., Strong A., Frank-Kamenetsky M., Lee N.E., Ahfeldt T., Sachs K.V., Li X., Li H., Kuperwasser N., Ruda V.M., et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano G., Ohno H., Oki K., Kobuke K., Shiwa T., Yoneda M., Kohno N. Activation of classical brown adipocytes in the adult human perirenal depot is highly correlated with PRDM16-EHMT1 complex expression. PLoS One. 2015;10:e0122584. doi: 10.1371/journal.pone.0122584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narvaez C.J., Matthews D., Broun E., Chan M., Welsh J. Lean phenotype and resistance to diet-induced obesity in vitamin D receptor knockout mice correlates with induction of uncoupling protein-1 in white adipose tissue. Endocrinology. 2009;150:651–661. doi: 10.1210/en.2008-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedergaard J., Bengtsson T., Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am. J. Physiol. Endocrinol. Metab. 2007;293:E444–E452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- Pearson S., Loft A., Rajbhandari P., Simcox J., Lee S., Tontonoz P., Mandrup S., Villanueva C.J. Loss of TLE3 promotes the mitochondrial program in beige adipocytes and improves glucose metabolism. Genes Dev. 2019;33:747–762. doi: 10.1101/gad.321059.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic N., Walden T.B., Shabalina I.G., Timmons J.A., Cannon B., Nedergaard J. Chronic peroxisome proliferator-activated receptor γ (PPARγ) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J. Biol. Chem. 2010;285:7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M., Okamatsu-ogura Y., Matsushita M., Watanabe K., Yoneshiro T., Nio-kobayashi J., Iwanaga T., Miyagawa M., Kameya T., Nakada K., et al. High incidence of metabolically active Brown adipose effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P., Bjork B., Yang W., Kajimura S., Chin S., Kuang S., Scimè A., Devarakonda S., Conroe H.M., Erdjument-Bromage H., et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant N., Varoquaux N., Lajoie B.R., Viara E., Chen C.J., Vert J.P., Heard E., Dekker J., Barillot E. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbæk R., Rabiee A., Nielsen R., Sidoli S., Traynor S., Loft A., Poulsen L.L.C., Rogowska-Wrzesinska A., Jensen O.N., Mandrup S. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep. 2014;7:1443–1455. doi: 10.1016/j.celrep.2014.04.042. [DOI] [PubMed] [Google Scholar]
- Smemo S., Tena J.J., Kim K.H., Gamazon E.R., Sakabe N.J., Gómez-Marín C., Aneas I., Credidio F.L., Sobreira D.R., Wasserman N.F., et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soccio R.E., Chen E.R., Rajapurkar S.R., Safabakhsh P., Marinis J.M., Dispirito J.R., Emmett M.J., Briggs E.R., Fang B., Everett L.J., et al. Genetic variation determines PPARγ function and anti-diabetic drug response in vivo. Cell. 2015;162:33–44. doi: 10.1016/j.cell.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soccio R.E., Li Z., Chen E.R., Foong Y.H., Benson K.K., Dispirito J.R., Mullican S.E., Emmett M.J., Briggs E.R., Peed L.C., et al. Targeting PPARγ in the epigenome rescues genetic metabolic defects in mice. J. Clin. Invest. 2017;127:1451–1462. doi: 10.1172/JCI91211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surwit R.S., Wang S., Petro A.E., Sanchis D., Raimbault S., Ricquier D., Collins S. Diet-induced changes in uncoupling proteins in obesity-prone and obesity-resistant strains of mice. Proc. Natl. Acad. Sci. USA. 1998;95:4061–4065. doi: 10.1073/pnas.95.7.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennakoon C., Purbojati R.W., Sung W.K. BatMis: a fast algorithm for k-mismatch mapping. Bioinformatics. 2012;28:2122–2128. doi: 10.1093/bioinformatics/bts339. [DOI] [PubMed] [Google Scholar]
- Villanueva C.J., Vergnes L., Wang J., Drew B.G., Hong C., Tu Y., Hu Y., Peng X., Xu F., Saez E., et al. Adipose subtype-selective recruitment of TLE3 or Prdm16 by PPARγ specifies lipid storage versus thermogenic gene programs. Cell Metab. 2013;17:423–435. doi: 10.1016/j.cmet.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen K.A., Lidell M.E., Orava J., Heglind M., Westergren R., Niemi T., Taittonen M., Laine J., Savisto N.J., Enerbäck S., et al. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- Xue B., Coulter A., Rim J.S., Koza R.A., Kozak L.P. Transcriptional synergy and the regulation of Ucp1 during Brown adipocyte induction in white fat depots. Mol. Cell Biol. 2005;25:8311–8322. doi: 10.1128/MCB.25.18.8311-8322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneshiro T., Aita S., Matsushita M., Okamatsu-Ogura Y., Kameya T., Kawai Y., Miyagawa M., Tsujisaki M., Saito M. Age-related decrease in cold-activated brown adipose tissue and accumulation of body fat in healthy humans. Obesity. 2011;19:1755–1760. doi: 10.1038/oby.2011.125. [DOI] [PubMed] [Google Scholar]
- Yoneshiro T., Aita S., Matsushita M., Kayahara T., Kameya T., Kawai Y., Iwanaga T., Saito M. Recruited brown adipose tissue as an antiobesity agent in humans. J. Clin. Invest. 2013;123:3404–3408. doi: 10.1172/JCI67803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
High-throughput sequencing data have been deposited at the Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. The accession number is listed in the key resources table.
-
•
This work does not report original code.
-
•
Any additional data or information required to reanalyze the findings shown here are available from the lead contact upon request.