Abstract
Mutations in the SLC35C1 gene encoding the Golgi GDP-fucose transporter are known to cause leukocyte adhesion deficiency II. However, improvement of fucosylation in leukocyte adhesion deficiency II patients treated with exogenous fucose suggests the existence of an SLC35C1-independent route of GDP-fucose transport, which remains a mystery. To investigate this phenomenon, we developed and characterized a human cell–based model deficient in SLC35C1 activity. The resulting cells were cultured in the presence/absence of exogenous fucose and mannose, followed by examination of fucosylation potential and nucleotide sugar levels. We found that cells displayed low but detectable levels of fucosylation in the absence of SLC35C1. Strikingly, we show that defects in fucosylation were almost completely reversed upon treatment with millimolar concentrations of fucose. Furthermore, we show that even if fucose was supplemented at nanomolar concentrations, it was still incorporated into glycans by these knockout cells. We also found that the SLC35C1-independent transport preferentially utilized GDP-fucose from the salvage pathway over the de novo biogenesis pathway as a source of this substrate. Taken together, our results imply that the Golgi systems of GDP-fucose transport discriminate between substrate pools obtained from different metabolic pathways, which suggests a functional connection between nucleotide sugar transporters and nucleotide sugar synthases.
Keywords: cell metabolism, glycoprotein biosynthesis, Golgi, membrane protein, N-linked glycosylation, SLC35C1, SLC35C2, GDP-fucose synthesis, fucose supplementation, LADII
Abbreviations: 2-AB, 2-aminobenzamide; AAL, Aleuria aurantia lectin; Cas9, CRISPR-associated protein 9; cDNA, complementary DNA; DMEM, Dulbecco’s modified Eagle's medium; FPGT, fucose-1-phosphate guanylyltransferase; FUK, fucokinase; HA, hemagglutinin; HEK293T, human embryonic kidney 293T cell line; LADII, leukocyte adhesion deficiency II; NST, nucleotide sugar transporter; qPCR, quantitative PCR; SEAP, secreted alkaline phosphatase; TBS, Tris-buffered saline
Fucose is an abundant component of many N- and O-glycans as well as some glycolipids. Except for O-fucosylation, where it is the first sugar in the sequence, fucose is always the terminal sugar in the structure of oligosaccharides. Fucose can be attached to other sugars or proteins via one of four types of glycosidic bonds: α-1,2, α-1,3, α-1,4, and α-1,6. In N-glycans, fucose is predominantly α-1,6-linked to the first (Asn-bound) N-acetylglucosamine residue. This type of fucose is referred to as a core fucose, and it is incorporated into the oligosaccharide structure only after the attachment of at least one N-acetylglucosamine residue to mannose. For a broader overview of the consecutive steps of N-glycan processing, see the study by Varki et al. (1).
Fucosylated oligosaccharides have many biologically relevant functions. AB0 blood group antigens are among the best known fucosylated glycans. O-linked fucose may affect certain ligand–receptor interactions involved in signal transduction. Fucose decorates certain oligosaccharides that are exposed on the surface of leukocytes and serve as ligands for selectins. Binding of the latter to the former initiates the leukocyte adhesion cascade, a multistep process of leukocyte migration to the site of injury or infection.
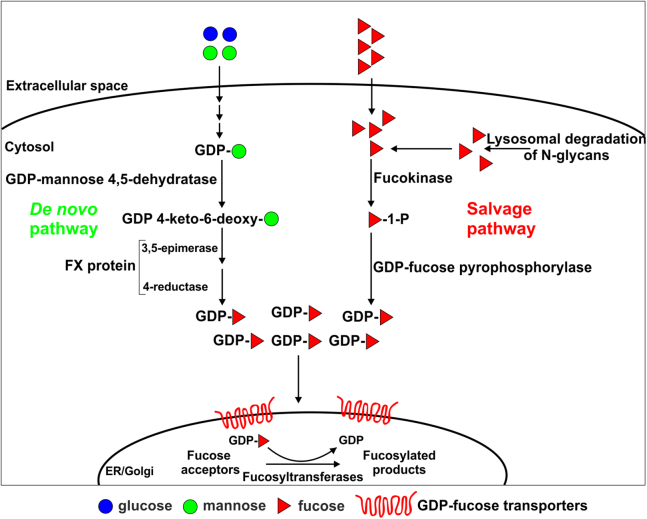
Incorporation of fucose into glycoconjugates is mediated by fucosyltransferases, which are predominantly Golgi-resident type II membrane proteins. Fucosyltransferases use the active form of fucose, that is, GDP-fucose, as a substrate. GDP-fucose is synthesized in the cytoplasm by two different pathways (shown in Fig. 1). The main source of this nucleotide sugar is the so-called de novo pathway, which is estimated to provide 90 to 95% of the total GDP-fucose pool in the cell (2, 3). The primary substrate for this three-step pathway is mannose. First, this monosugar is converted into GDP-mannose and then GDP-D-mannose 4,6-dehydratase converts GDP-mannose to GDP-4-keto-6-deoxymannose. This keto intermediate is then converted into GDP-fucose by an epimerase/reductase enzyme complex termed the FX protein or GDP-l-fucose synthase (also known as GFUS, FCL, SDR4E1, or TSTA3) (4).
Figure 1.
Simplified representation of GDP-fucose metabolism in mammalian cells. GDP-fucose is synthesized in two independent pathways, that is, de novo (from GDP-mannose) and salvage (from fucose).
The second alternative way of GDP-fucose synthesis is the so-called salvage pathway. This route uses the cytoplasmic pool of free fucose, which on the one hand is recovered from glycoconjugates by lysosomal α-fucosidase activity and on the other hand is supplied from the environment. Little is known about the fucose transport system from the extracellular space to the interior of the cell, but it appears to function through facilitated diffusion through specific channel(s) (5). The first step of the salvage pathway involves ATP-dependent synthesis of fucose-1-phosphate by the enzyme fucokinase (FUK; also known as FCSK). Fucose-1-phosphate guanylyltransferase (FPGT; GDP-fucose pyrophosphorylase), the second enzyme acting in the pathway, catalyzes the conversion of fucose-1-phosphate and GTP to GDP-fucose (6). As already mentioned, it is estimated that only 5 to 10% of the total GDP-fucose pool is synthesized by this pathway (2, 3).
To reach lumenally oriented catalytic centers of fucosyltransferases, GDP-fucose has to be transported across the Golgi membrane. This function is thought to be played by GDP-fucose–specific nucleotide sugar transporters (NSTs), which are hydrophobic type III membrane proteins with 6 to 10 transmembrane domains. NSTs are believed to act as antiporters; an imported nucleotide sugar is exchanged with the corresponding nucleoside monophosphate (7). After reaching the Golgi lumen, the activated monosugar constitutes a substrate for a respective glycosyltransferase, which attaches the monosugar to an acceptor, whereas the released nucleoside diphosphate is broken down into a nucleoside monophosphate and an inorganic phosphate (8).
In mammals, the role of the main GDP-fucose transporter is attributed to the product of the SLC35C1 gene, which was identified by two independent groups in 2001 (9, 10). Although SLC35C1 is thought to be the major transporter in mammals, GDP-fucose has also been shown to translocate through its homolog, SLC35C2. SLC35C2 was shown to localize predominantly in the Golgi, but a small subset of it was found in the endoplasmic reticulum and endoplasmic reticulum–Golgi intermediate compartment. SLC35C2 is specifically required for O-fucosylation of certain proteins, including the Notch receptor, which in turn does not require SLC35C1 activity (11, 12).
Leukocyte adhesion deficiency II (LADII) is a rare autosomal recessive genetic disease characterized by an overall reduction in fucosylation of glycoconjugates. This syndrome is caused by mutations in the SLC35C1 gene (13). The characteristic symptoms of this disease include psychomotor retardation, facial dysmorphism, Bombay phenotype, short stature, immunodeficiency, leukocytosis, as well as recurrent and frequent bacterial infections (14). LADII was first diagnosed in 1992 (15). To date, 19 individuals bearing inactivating mutations in the SLC35C1 gene have been reported (14, 15, 16, 17, 18, 19, 20, 21, 22, 23), with a predominance of point mutations.
The concept of using exogenous fucose to improve fucosylation in LADII patients was first proposed in 1998. Pioneering experiments were performed with lymphoblastic cells obtained from one of the patients bearing the Arg147Cys mutation in the SLC35C1 amino acid sequence. Supplementation of the cell culture with 10 mM fucose for 5 days triggered an increase in the number of fucosylated structures (24). These results paved the way to the concept of using fucose supplementation in patients themselves. Since 1999, several LADII patients were treated with oral fucose. For some of them, such treatment was successful, resulting in an improvement in psychomotor development and a reduction in neutrophil count (22, 25). However, in the case of two LADII patients, fucose supplementation failed to improve their condition (20, 26). The administered doses of fucose varied from 25 to 2000 mg/kg body weight (20, 25, 26). An improvement in fucosylation was also achieved when the LADII patient–derived fibroblasts were cultured in the presence of 0.1 to 10 mM fucose (18).
To explain the effectiveness of the fucose treatment in responsive LADII patients, it was postulated that the mutant SLC35C1 variants display some residual transport activity (9). Therefore, a therapy causing an increase in the cytosolic concentration of this nucleotide sugar could potentially allow the insufficient transporting activity of the defective SLC35C1 variants to be overcome. However, direct proof of an increase in the cytosolic GDP-fucose concentration in the fucose-fed cells bearing mutations in the SLC35C1 gene has not been presented to date. An alternative hypothesis to explain the efficiency of the oral fucose therapy could be the existence of an accessory GDP-fucose transport system. However, no such route has been identified so far.
In 2007, a knockout-based study conducted in mice revealed that the fucose treatment works even in the absence of SLC35C1 (27). These findings for the first time suggested the existence of an SLC35C1-independent GDP-fucose transport into the Golgi, as the observed effect could no longer be explained by a residual activity of the SLC35C1 mutants. It was hypothesized that the SLC35C1 deficiency could be overcome by the SLC35C2 activity, but this has never been proved. In general, no studies that could possibly explain this phenomenon in greater detail have been undertaken so far.
Since the responsiveness of the cells completely devoid of the SLC35C1 activity to fucose treatment has not been elucidated to date, in this study, we carried out a more in-depth investigation of this intriguing effect. Within our general interest in the metabolism of fucose and protein fucosylation, specific goals included (i) generation of single SLC35C1 and double SLC35C1/SLC35C2 knockouts, (ii) optimization of fucose-feeding conditions, (iii) establishment of a method allowing quantification of the extent of N-glycan fucosylation, (iv) structural characterization of N- and O-glycans produced by the wildtype and knockout cells, (v) investigation of N-glycan fucosylation in response to various ranges of exogenous fucose concentrations, (vi) single-point and time-course analyses of intracellular GDP-fucose concentration in control and fucose-fed cells, (vii) determination of the contribution of de novo and salvage GDP-fucose biosynthetic pathways to fucose incorporation into N-glycans in the wildtype and knockout cells, and (viii) assessment of a potential cellular genetic response to fucose supplementation.
In this study, we developed an approach coupling exoglycosidase digestion with HPLC to quantify the percentage of the core-fucosylated N-glycans with great precision. We found that in the knockout cells, the core fucosylation of N-glycans is greatly reduced but can be nearly completely restored by supplementing the cells with millimolar concentrations of fucose. Supplementation with 5 mM fucose caused a significant increase in the intracellular GDP-fucose concentration, regardless of the presence/absence of GDP-fucose transporters. Next, by feeding low (nanomolar) concentrations of radioactive fucose and mannose into the culture medium, we were able to follow the metabolic fate of the supplemented compounds and attribute radioactivity to fucosylated structures using our HPLC-based method. Surprisingly, the SLC35C1 knockouts showed a dramatic impairment of the ability to incorporate mannose-derived radioactivity into the fucosylated N-glycans. In sharp contrast, they were able to incorporate even higher quantities of radiolabeled fucose than the wildtype cells. To explain these observations, we hypothesize that in the SLC35C1 knockouts, the salvage pathway is preferred over the de novo pathway as a source of GDP-fucose for fucosylation of N-glycans. We also investigated the potential genetic response of the SLC35C1 knockouts to fucose treatment. No major gene upregulation/downregulation was observed. Finally, we excluded SLC35C2 as an alternative supplier of GDP-fucose for N-glycan fucosylation.
Results
Generation of the single SLC35C1, SLC35C2, and double SLC35C1/SLC35C2 knockouts
To develop research models for our study, SLC35C1 knockouts (C1KO) were generated in two human cell lines, that is, human embryonic kidney 293T (HEK293T) and HepG2, and SLC35C2 knockout (C2KO) was generated in HEK293T cell line using the CRISPR–CRISPR-associated protein 9 (Cas9) approach. In addition, a double SLC35C1/SLC35C2 knockout (C1/C2KO) was developed in HEK293T cells to evaluate the influence of SLC35C2 protein on the N- and O-glycosylation in the SLC35C1 knockout cells and to examine the effect of fucose treatment in cells lacking both GDP-fucose transporters.
The knockouts were confirmed by both RT–PCR performed on total RNA and PCR performed on genomic DNA (Fig. S1, A–C). The forward/reverse primers were designed to align upstream/downstream of the CRISPR/Cas9-mutated regions of the SLC35C1/SLC35C2 genes and to yield 133 to 236 bp fragments (Table 1). As the relative electrophoretic mobility changes are significant, such short fragments allow the observation of even small (in terms of base pair) effects of CRISPR–Cas9 activity on a gel.
Table 1.
Primers used in this study
| No. | Primer name | Primer sequence 5ʹ-3ʹ | Product length (bp) |
|---|---|---|---|
| Primers used in RT–PCR analysis with total RNA as a template | |||
| 1 | F_C1KO | CTTCCCCAGCTTGCGCCTG | 133 |
| 2 | R_C1KO | GCGGCCCACATTGTAGAAGGC | 133 |
| 3 | F_C1/C2KO | GCTTCTCTACTACTGCTTCTC | 233 |
| 4 | R_C1/C2KO | TCAAGCGCCGTCGCCAGAGCT | 233 |
| Primers used in PCR analysis with genomic DNA as a template | |||
| 5 | F_C1KO | CTTCCCCAGCTTGCGCCTG | 236 |
| 6 | R_C1KO | TTGTTCTTCACTGCTTCAGTC | 236 |
| 7 | F_C1/C2KO | ATTTCCCCCTCTTCATGACG | 165 |
| 8 | R_C1/C2KO | TTCTCATCCTATGGTTCCCCA | 165 |
| Primer used in gene amplification and site-directed mutagenesis | |||
| 9 | F_HAC1 | AAAAAGTCGACATGGCATACCCATACGACGTACCAGACT ACGCAATGAATAGGGCCCCTCTGAAGCGGT | NA |
| 10 | R_HAC1 | AAAAAGTCGACTCACACCCCCATGGCGCTCTT | NA |
| 11 | F_C1_420_PAM | ACGTCGGTGTAGCCTTCTACA | NA |
| 12 | R_C1_420_PAM | ACTTGAGGCAGAGGTTATTG | NA |
Electrophoretic separation of the RT–PCR/PCR products on 2% agarose gel (Fig. S1, A–C) showed multiple bands corresponding to the CRISPR–Cas9-mutated SLC35C1/SLC35C2 gene fragments. The fact that no bands corresponding to the wildtypes were observed even after 35 (genomic DNA) or 40 (mRNA) RT–PCR/PCR cycles excludes the presence of the wildtype allele.
The SLC35C1 knockout was in addition verified at the protein level using an anti-SLC35C1 antibody by Western blotting, where no band was detected in the knockouts in contrast to the wildtype cells (Fig. S1D), as well as by double staining with the latter antibody and fucose-specific Aleuria aurantia lectin (AAL; Vector Laboratories; Fig. S1E). Unfortunately, because of the lack of a suitable antibody, a similar confirmation was not possible in the case of the SLC35C2 gene. Hence, the SLC35C2 knockout was in addition verified by genomic DNA sequencing (Figs. S2 and S3 for single SLC35C2 and double SLC35C1/SLC35C2 knockouts, respectively). All the resulting sequences contained deletions that either cause a frameshift or introduce a premature stop codon, whereas the wildtype SLC35C2 sequence was not detected in any of the analyzed clones.
To show that the knockout phenotype was specifically associated only with the disruption of the SLC35C1 gene, we also developed SLC35C1-deficient clones stably overexpressing a recombinant hemagglutinin (HA)-tagged SLC35C1 (in both HEK293T and HepG2 cell lines). The overexpression was confirmed by Western blotting performed on cell lysates using an anti-HA antibody (Fig. S1F). The obtained stable transfectants were also subjected to immunostaining to confirm the Golgi localization of the overexpressed protein (Fig. S1G).
Optimization of fucose supplementation conditions
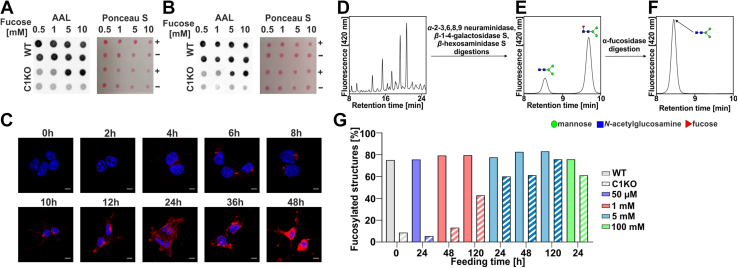
In order to establish a system to study the effect of fucose supplementation on the restoration of fucosylation in the SLC35C1 knockouts, feeding parameters, that is, fucose concentration in the culture medium as well as duration of the treatment, had to be optimized. In the first stage, we employed a dot blot analysis using AAL (Fig. 2, A and B). Several concentrations of fucose were tested, that is, 0.5, 1, 5, and 10 mM (concentration range inspired by Helmus et al. (18)). We found that after 5 days, 5 mM concentration was sufficient to substantially restore fucosylation of glycoconjugates in the SLC35C1 knockouts. AAL staining performed on the SLC35C1 knockouts cultured in the presence of 5 mM fucose for 0 to 48 h demonstrated that the level of fucosylation gradually increased over time and an optimum (i.e., fucosylation compared with the wildtype) was reached after 24 h. After another 12 h, however, the cells became overloaded with aggregated fucose-containing structures (Fig. 2C).
Figure 2.
Optimization of the conditions of fucose supplementation. AAL dot blot analysis of the wildtype and SLC35C1 knockout HEK293T (A) and HepG2 (B) cells after 5 days of supplementation with (+) or without (−) different fucose concentrations. C, AAL staining (red) of HEK293T SLC35C1 knockout supplemented with 5 mM fucose during 48 h. 0 h—control cells (no fucose supplementation, image reused in 6E panel). Cell nuclei were counterstained with DAPI. The scale bar represents 10 μm. D–F, schematic representation of our HPLC-based method for quantification of-α1,6-fucosylation of N-glycans. G, quantitative analysis of α-1,6-fucosylation of N-glycans in HEK293T wildtype and SLC35C1 knockout cells supplemented with different fucose concentrations for indicated periods. AAL, Aleuria aurantia lectin; DAPI, 4',6-diamidino-2-phenylindole; HEK293T, human embryonic kidney 293T cell line.
Since the lectin-based analysis does not allow for quantification of the observed fucosylation defects/rescue effects, we established a new robust method, based on exoglycosidase digestions followed by HPLC separation, that allows quantification of the degree of α-1,6-fucosylation (core fucose). In our approach, we used specific glycosidases to reduce all multibranched complex-type N-glycan species into a simple biantennary (GlcNAc)2(Man)3 structure, either fucosylated or nonfucosylated (Fig. 2, D and E). Then by taking a ratio of the area of the fucosylated peak over the sum of areas of both peaks (fucosylated and nonfucosylated), we determined the percentage of fucosylated structures for each sample. To confirm that the fucosylated peak contains only the (GlcNAc)2(Man)3Fuc structure, α-fucosidase digestion was performed (Fig. 2F).
By employing our method, we tested supplementation of selected fucose concentrations (50 μM–100 mM) for different periods (1–5 days) in wildtype and SLC35C1 knockout HEK293T cells. Feeding the cells with 50 μM or 1 mM fucose even for as long as 5 days was not sufficient to restore the wildtype α-1,6-fucosylation level in the SLC35C1 knockout cells. Only supplementation of the SLC35C1 knockout cells with 5 mM fucose restored the fucosylation phenotype to a level comparable to the wildtype cells, but at the same time, it did not affect core fucosylation in the wildtype cells. At the same time, supplementation with a very high fucose concentration (100 mM) did not increase the level of core fucosylation in the wildtype cells (Fig. 2G). Therefore, for the final experiments, the conditions of 5 mM fucose and 24 h were selected.
Supplementation of fucose in the knockout cells significantly increases α-1,6-fucosylation of N-glycans
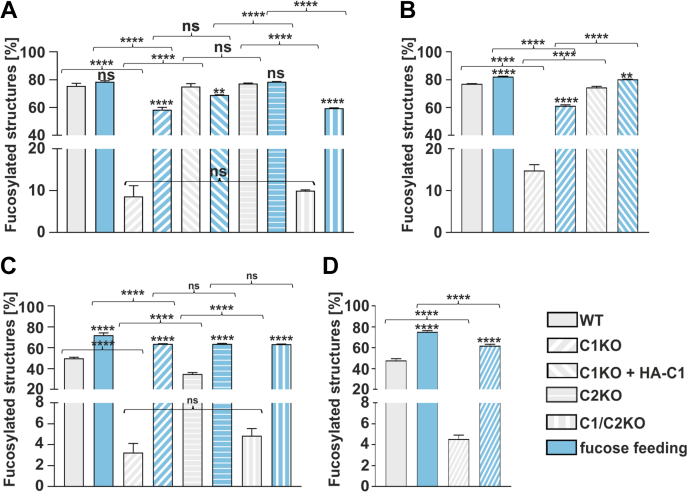
We quantified α-1,6-fucosylation of N-glycans in all modified cell lines (Fig. 3). From the analysis of the cellular N-glycans from HEK293T cells (Fig. 3A), we concluded that (i) the native level of core fucosylation in the nonfed wildtype cells varies around ∼75% and is compromised to ∼8 to 10% in the single SLC35C1 and double SLC35C1/SLC35C2 knockouts, (ii) ectopic expression of the HA-tagged SLC35C1 in the single SLC35C1 knockouts restores the wildtype phenotype, (iii) nearly complete α-1,6 fucosylation (∼60%) can be achieved in both single SLC35C1 and double SLC35C1/SLC35C2 knockouts upon administration of 5 mM fucose for 24 h, and (iv) inactivation of the SLC35C2 gene has no influence on the α-1,6-fucosylation of N-glycans. As anticipated, similar observations were made for the cellular N-glycans from HepG2 cells (Fig. 3B). MALDI-TOF analysis of N-glycan structures confirmed those results, showing the presence of a fucosylated structure in the SLC35C1 knockouts and restoration of fucosylated structures after fucose feeding (Figs. S4, S5 and Table S1).
Figure 3.
Quantification of the percentage of the core-fucosylated N-glycan structures. HPLC quantification of N-glycans derived from endogenous HEK293T (A) and HepG2 (B) glycoproteins. N-glycans decorating SEAP glycoprotein overexpressed by HEK293T (C) and HepG2 (D) cell lines. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; and ∗∗∗∗p < 0.0001 as determined using one-way ANOVA with the Tukey post hoc test. Data are presented as mean ± SD. Each sample was run in three technical replicates. HEK293T, human embryonic kidney 293T cell line; ns, not significant, SEAP, secreted alkaline phosphatase.
During the separation and purification of cellular N-glycans, there is a possibility of contamination with the oligosaccharides derived from serum glycoproteins contained in the culture medium. To address this problem, the N-glycans derived from an engineered ectopically expressed and secreted reporter glycoprotein, secreted alkaline phosphatase (SEAP), were also analyzed. The method for SEAP isolation efficiently prevents contamination with serum glycoproteins (28). From the analysis of α-1,6-fucosylated N-glycans derived from the SEAP overexpressed in the wildtype and knockout HEK293T cells (Fig 3, C and D), we concluded that (i) in the SLC35C1 and SLC35C1/C2 knockouts, α-1,6-fucosylation of SEAP-derived N-glycans is still present (∼3–5%) and supplementation of 5 mM fucose for 24 h restores α-1,6-fucosylation in these cells, (ii) SLC35C2 knockout has no influence on the α-1,6-fucosylation of SEAP-derived N-glycans. Interestingly, the level of α-1,6-fucosylation observed in the fucose-fed wildtype and knockout cells was higher than it was in the wildtype cells before supplementation, which could be a specific response of the overexpressed SEAP to the supplementation (Fig. 3C). As anticipated, similar observations were made for the HepG2 cells (Fig. 3D).
Supplementation of fucose restores fucosylation of O-glycans in the knockout cells
Certain mucin-type O-glycans are also fucosylated. The effect of GDP-fucose transport deficiency on this type of oligosaccharides was only investigated using lectins (27). Therefore, we in addition analyzed cellular mucin-type O-glycans using the CORA (Cellular O-Glycome Reporter/Amplification) strategy (29).
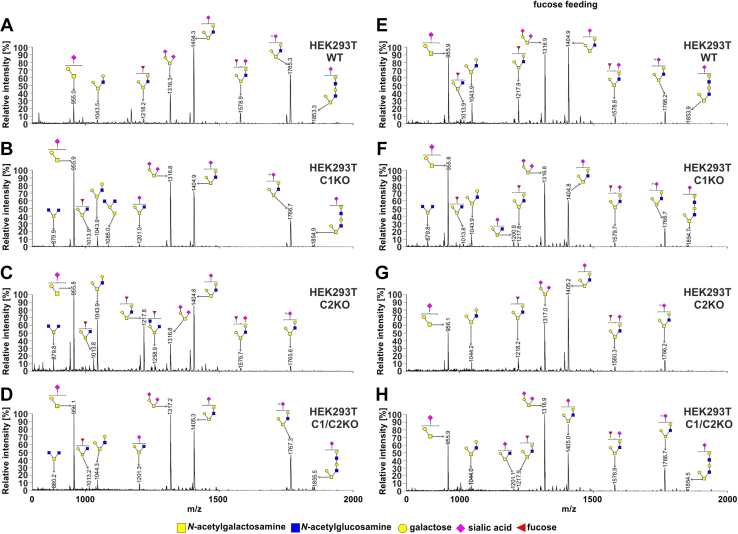
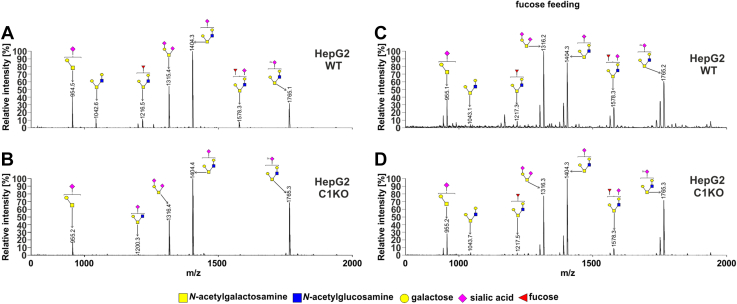
From the analysis of the HEK293T cells (Fig. 4 and Table S2), we concluded that (i) in the wildtype cells, two among eight of the identified glycans were fucosylated (m/z values of ∼1218 and ∼1578; Fig. 4A), (ii) both fucosylated species were absent in the SLC35C1 and SLC35C1/C2 knockouts, in which only their nonfucosylated counter partners were detected (m/z values of ∼1044 and 1404), (iii) a new fucosylated species was observed in the knockouts (m/z of ∼1013, Fig. 4, B–D), (iv) both fucosylated species were present in the SLC35C2 knockout, and (v) supplementation with 5 mM fucose for 24 h led to reappearance of the fucosylated species in the knockouts. Surprisingly, after fucose supplementation, additional fucosylated species were observed in the wildtype and SLC35C1 knockout cells but not in the SLC35C2 and SLC35C1/SLC35C2 knockouts (Fig. 4, E–H). In the case of HepG2 cells, the fucosylated O-glycan species were not produced by the SLC35C1 knockout. However, in contrast to HEK293T cells, fucose supplementation of the wildtype HepG2 cells did not have an influence on fucosylated O-glycans (Fig. 5). The observed differences could be cell line specific.
Figure 4.
O-glycosylation fingerprinting of HEK293T cells. MALDI-TOF mass spectra of permethylated mucin-type Bn-O-glycans secreted to the culture medium were permethylated and analyzed in a positive-ion mode. Structural assignments based on biosynthetic knowledge were prepared using the GlycoWorkBench tool (2.1; EuroCarbDB). HEK293T, human embryonic kidney 293T cell line.
Figure 5.
O-glycosylation fingerprinting of HepG2 cells. MALDI-TOF mass spectra of permethylated mucin-type Bn-O-glycans secreted to the culture medium were permethylated and analyzed in a positive-ion mode. Structural assignments based on biosynthetic knowledge were prepared using the GlycoWorkBench tool (2.1; EuroCarbDB).
These results prove the role of SLC35C1 in the process of mucin-type O-glycan fucosylation and are in line with the observations made for the N-glycans. Importantly, supplementation with 5 mM fucose for 24 h restored fucosylation of O-glycans in the single SLC35C1 and double SLC35C1/SLC35C2 knockout cells.
Supplementation of fucose increases intracellular GDP-fucose concentration
Although it is anticipated that in the SLC35C1-deficient cells, the exogenous fucose efficiently enters the cells and becomes converted into GDP-fucose, to our best knowledge, this phenomenon has never been quantified in the cells lacking SLC35C1 activity.
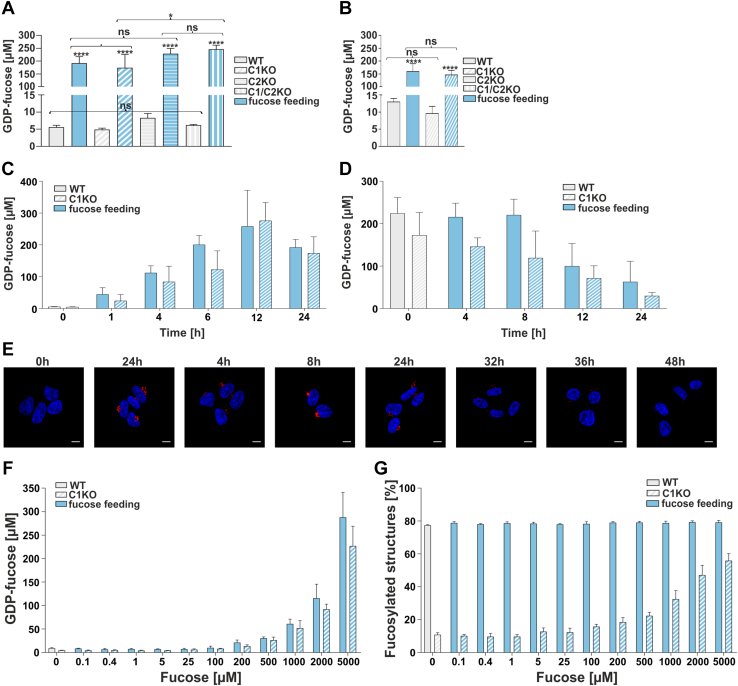
For separation and quantitative analysis of nucleotide sugars, we employed a modified ion-pair solid-phase extraction and HPLC strategy developed by Räbinä et al. (30). Our results demonstrated that in the nonfed HEK293T cells, the intracellular concentration of GDP-fucose was around ∼5 μM, regardless of the genotype (wildtype or single/double knockout; Fig 6A). This shows that GDP-fucose transporter deficiency of the SLC35C1 knockout did not cause GDP-fucose accumulation. This is in contrast to the research conducted on the SLC35A1 (CMP–sialic acid transporter) knockout, for which such an effect was observed (31). Importantly, upon fucose supplementation, GDP-fucose concentration increased ∼40- to 50-fold in all HEK293T cell lines, up to ∼200 to 250 μM (Fig. 6A). A similar experiment was performed for HepG2 cells, where the basal concentrations of GDP-fucose were slightly higher (∼10 μM), and the effect of supplementation was somewhat less pronounced (∼15-fold, up to ∼150 μM) (Fig. 6B). Based on these findings, we concluded that the externally supplemented fucose efficiently enters the cytoplasm and is converted to GDP-fucose in all tested cell lines.
Figure 6.
Quantification of intracellular GDP-fucose concentration. Intracellular GDP-fucose concentration in HEK293T (A) and HepG2 (B) cell lines. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; and ∗∗∗∗p < 0.0001 as determined using one-way ANOVA with the Tukey post hoc test. Data are represented as mean ± SD. Each sample was run in three biological replicates. C, time-course analysis (0–24 h) of GDP-fucose synthesis in wildtype and SLC35C1 knockout HEK293T cells fed with 5 mM fucose. Each sample was run in three biological replicates. D, analysis of GDP-fucose degradation in wildtype and SLC35C1 knockout HEK293T cells. The cells were first cultured for 24 h in the presence of 5 mM fucose and then for another 24 h in a fucose-free medium. Each sample was run in three biological replicates. E, disappearance of the fucosylation phenotype visualized by AAL staining (red). 0 h—control cells (no fucose supplementation, image reused in 2C panel). Cell nuclei were counterstained with DAPI. The scale bar represents 10 μm. F, dependence of the intracellular GDP-fucose concentration on the concentration of the fucose supplemented to the culture medium (HEK293T). G, dependence of the percentage of the core fucosylated N-glycans on the concentration of the fucose supplemented to the culture medium (HEK293T). Each sample was run in three biological replicates. AAL, Aleuria aurantia lectin; DAPI, 4',6-diamidino-2-phenylindole; HEK293T, human embryonic kidney 293T cell line; ns, not significant.
Supplementation of fucose causes dynamic changes in fucose metabolism
As a follow-up on these remarkable increases in the GDP-fucose concentration in response to fucose supplementation, measurement of the intracellular concentration of fucose was undertaken. Using an enzymatic assay, we determined the intracellular concentration of fucose at a level of ∼1 mM in the SLC35C1 knockouts supplemented with 5 mM fucose. From these numbers, one can postulate an approximate equilibrium of 5 mM/∼1 mM/∼0.2 mM for the extracellular fucose/intracellular fucose/intracellular GDP-fucose.
Subsequently, in order to study the GDP-fucose turnover, time-course evaluation of the intracellular GDP-fucose concentration was performed in the HEK293T wildtype and SLC35C1 knockout cells over the course of 24 h. As shown in Figure 6C, GDP-fucose concentrations close to the maximum levels of ∼250 μM were reached as soon as 6 to 12 h after starting supplementation. However, when the supplementation was terminated, GDP-fucose concentration remained stable for ∼8 h and then started to gradually decline toward basal levels (Fig. 6D). These observations were then correlated to the glycophenotype investigated using AAL fluorescent staining (Fig. 6E). After 24 h of supplementation, fucosylation was strongly pronounced, which was manifested by efficient lectin binding (Fig. 6E, 24 h). Withdrawal of the monosaccharide from the culture medium caused a gradual reduction of fucosylation, which was no longer detectable after 48 h (Fig. 6E, 48 h).
Having discovered that supplementation of 5 mM fucose for 24 h led to a drastic increase in the intracellular GDP-fucose concentration, we next examined how different fucose concentrations would affect the intracellular GDP-fucose concentration and core fucosylation of N-glycans. Therefore, we cultured the wildtype and SLC35C1 knockout HEK293T cells in the presence of 0 to 5 mM fucose for 24 h and determined the GDP-fucose concentrations for each of the tested fucose concentrations. A visible increase in the GDP-fucose concentration required the supplementation of at least 0.2 to 1 mM fucose (Fig. 6F). Then, we observed that in the SLC35C1 knockouts, core fucosylation of N-glycans increased proportionally to the concentration of intracellular GDP-fucose (Fig. 6G). The finding that increasing GDP-fucose concentration in the wildtype cells does not have any effect on the core fucosylation of N-glycan is in line with our previous results. Our data demonstrate that a significant increase in the intracellular GDP-fucose concentration is necessary to restore core fucosylation of N-glycans in the SLC35C1 knockouts.
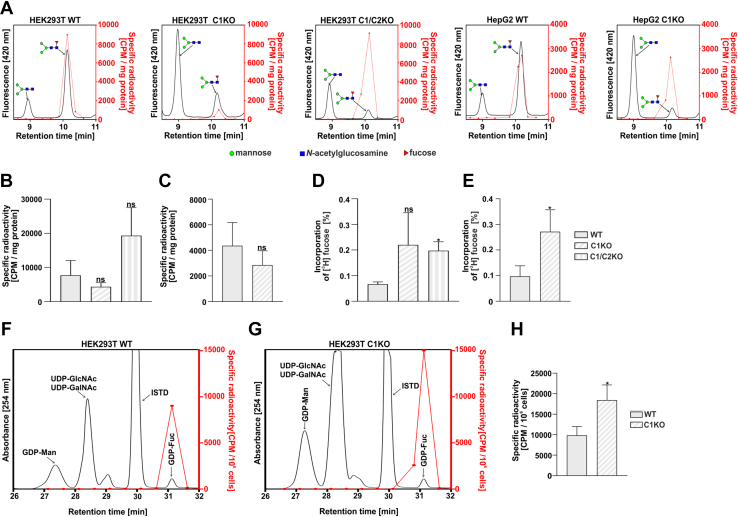
Our data showed that GDP-fucose derived from the salvage pathway is efficiently utilized in the SLC35C1 and SLC35C1/C2 knockout cells. By supplementation of cells with a low concentration of radiolabeled [6-3H]fucose, we were able to track the biosynthesis of GDP-fucose via the salvage pathway and incorporation of fucose into N-glycans. We found that the radioisotope was incorporated into core fucosylated N-glycans produced by all tested cell lines (Fig. 7, A–C). It means that even small amounts of exogenous fucose can be incorporated into core fucosylated N-glycans in the SLC35C1 and SLC35C1/C2 knockouts. Based on the known total radioactivity of the fucose substrate fed to the cells and on the measured radioactivity in the isolated fraction of the core fucosylated N-glycans, one can estimate the efficiency of the radioisotope incorporation (Fig. 7, D and E). The fact that a higher percentage of core fucose was incorporated into N-glycans by the knockout cells may suggest a greater contribution of the GDP-fucose derived from the salvage pathway in the knockout cells as compared with the wildtype cells. We also observed that the biosynthesis of GDP-fucose in the salvage pathway occurs in both the wildtype and knockout cells (Fig. 7, F–H). Altogether, these results may suggest the preferential usage of the salvage pathway–produced GDP-fucose for the N-glycan fucosylation in the SLC35C1 knockout cells.
Figure 7.
Metabolic labeling of cellular N-glycans with radioactive fucose.A, exemplary representative HPLC chromatograms of digested N-glycans isolated from indicated cell lines. The fluorescence signal from the 2-AB-labeled N-glycans (black solid lines) was overlaid with the radioactivity data (red dots and solid lines). Total radioactivity of the fucosylated N-glycans normalized for the amount of the starting material (total protein) from HEK293T (B) and HepG2 (C) cell lines. Percentage of the radioactive fucose incorporated into the fucosylated N-glycans isolated from HEK293T (D) and HepG2 (E) cell lines. To calculate the values, the sum of radioactivity measured in the HPLC fractions corresponding to the fucosylated species was divided by the total radioactivity of the fucose added to the culture medium. Comparison of the relative incorporation of the radioactive fucose derived from the salvage pathway in the wildtype (F) and SLC35C1 knockout (G) HEK293T cells. H, total radioactivity of the GDP-fucose normalized for total number of HEK293T cells. ∗p < 0.05 as determined using two-tailed unpaired t test with Welch’s correction. Data are presented as mean ± SD. Each sample was run in three biological replicates. 2-AB, 2-aminobenzamide; ISTD, internal standard (GDP-glucose); ns, not significant.
Supplementation of millimolar mannose increases the intracellular GDP-mannose but not the intracellular GDP-fucose concentration
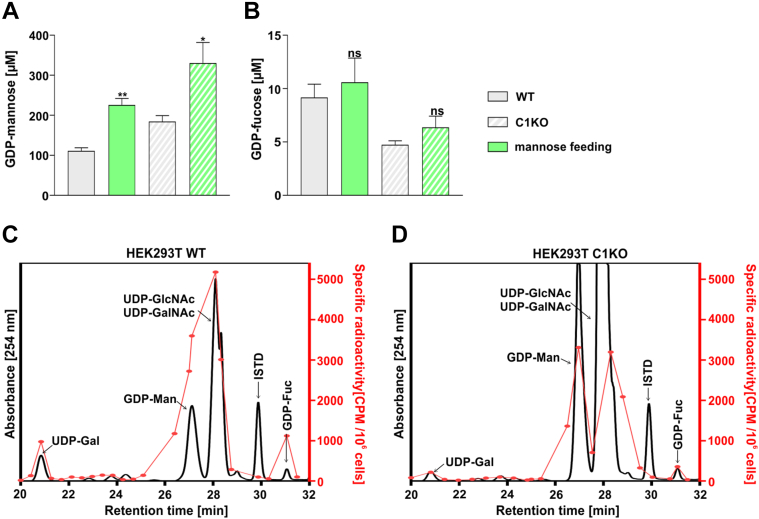
The data presented previously clearly suggest that the restoration of the fucosylation of N-glycans in the SLC35C1 knockout cells requires elevated intracellular GDP-fucose concentration (Fig. 6, F and G), which can be efficiently achieved by fucose supplementation to the culture medium. However, fucose to GDP-fucose conversion constitutes only up to ∼10% of the total GDP-fucose production, whereas the rest is thought to come from mannose to GDP-fucose conversion. Hence, we have investigated whether supplementation of 5 mM mannose to the culture medium for 48 h would cause the intracellular GDP-fucose concentration to increase (Fig. 8).
Figure 8.
Quantification of intracellular GDP-mannose and GDP-fucose concentration in cells fed with mannose. Intracellular concentrations of GDP-mannose (A) and GDP-fucose (B) in the wildtype and SLC35C1 knockout HEK293T cells cultured for 48 h in the absence and presence of 5 mM mannose. ∗p < 0.05; ∗∗p < 0.01 as determined using two-tailed unpaired t test with Welch’s correction. Data are presented as mean ± SD. Each sample was run in three biological replicates. Comparison of relative incorporation of the radioactive fucose derived from the de novo pathway in wildtype (C) and SLC35C1 knockout (D) HEK293T cells. HEK293T, human embryonic kidney 293T cell line; ISTD, internal standard (GDP-glucose); ns, not significant.
First, we have tested whether the exogenously supplied mannose is intracellularly converted into GDP-mannose (Fig. 8A). Here, in both the wildtype and knockout cells, 5 mM mannose supplementation caused an increase in the intracellular GDP-mannose concentration from ∼100 to ∼220 μM in the wildtype and from ∼180 to ∼330 μM in the knockout. However, no significant increase in GDP-fucose concentration was observed, regardless of the cell line (Fig. 8B).
As a follow-up and also to verify whether the de novo pathway works properly in the analyzed cells, we have examined whether radiolabeled mannose supplemented in low (nanomolar) concentrations would be converted into GDP-fucose. As evident from the radioactive signal, both the wildtype and SLC35C1 knockout cells efficiently converted the supplemented radiolabeled mannose into GDP-fucose (Fig. 8, C and D). It needs to be noted that except the GDP-mannose and GDP-fucose, radioactivity signal could be also assigned to other metabolites.
The de novo and salvage pathways differentially contribute into N-glycan fucosylation in the SLC35C1 knockout cells
To compare the relative contribution of different GDP-fucose biosynthesis pathways into the N-glycan fucosylation, the wildtype and SLC35C1-deficient HEK293T cells were cultured in the presence of nanomolar concentrations of either radioactive fucose (salvage pathway) or radioactive mannose (de novo pathway).
Apart from conversion into GDP-fucose, mannose can also be processed into other nucleotide sugars, for example, GDP-mannose, and as such incorporated into the complex-type N-glycans. To address this problem, we established a new approach in which the N-glycans are first digested with α-fucosidase, then the postreaction mixture is passed through a graphite column, and finally the radioactivity of the flow-through is measured. This strategy allows us to specifically assign the detected radioactivity to fucose present in N-glycans.
First, the contribution of the de novo pathway was assessed by comparing the specific radioactivity of the flow-through obtained from the wildtype and SLC35C1-deficient cells fed with radioactive mannose. The incorporation of radioactive precursor into the N-glycans derived from the knockout was reduced to only ∼6% as compared with the wildtype’s sample. This indicates that incorporation of fucose derived from the GDP-fucose synthesized via the de novo pathway (i.e., from mannose) turned out to be nearly completely abolished (Fig. 9B).
Figure 9.
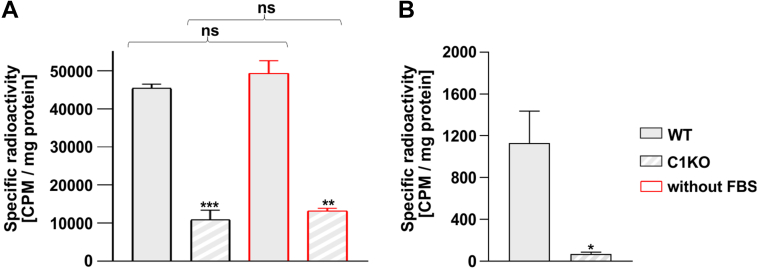
Relative contribution of the de novo and salvage pathways in glycan fucosylation.A, incorporation of the radioactive fucose derived from the GDP-fucose produced in the salvage pathway into N-glycans measured for the wildtype (full bars) and SLC35C1 knockout (dashed bars) HEK293T cells cultured in the presence (black contours) and absence (red contours) of fetal bovine serum (FBS). B, incorporation of the radioactive fucose derived from the GDP-fucose produced in the de novo pathway into N-glycans measured for the wildtype (full bar) and SLC35C1 knockout (dashed bar) HEK293T cells. Radioactivity was normalized for the amount of the starting material (total protein). ∗p < 0.05; ∗∗∗p < 0.001 as determined using two-tailed unpaired t test with Welch’s correction. Data are presented as mean ± SD. Each sample was run in three biological replicates. HEK293T, human embryonic kidney 293T cell line; ns, not significant.
Mammalian cell lines are routinely cultured in media supplemented with 10% fetal bovine serum, which may contain free fucose. Since the radiolabelling experiments described previously utilize minute concentrations of the radioactive precursor, the potential influence of the serum-derived fucose cannot be neglected. For this reason, using a colorimetric assay, we have determined the concentration of fucose in the serum to be ∼20 μM, which corresponds to ∼2 μM concentration of fucose in the complete culture medium. To exclude the potential influence of the serum-derived fucose on the obtained results, the experiment with radioactive fucose was conducted using the cells cultured with and without serum. In both cases, the SLC35C1 knockout cells incorporated about a quarter of the radioactivity as compared with the wildtype (Fig. 9A).
Although it was shown previously that the free fucose present in the serum does not affect the level of incorporation of low amount of radioactive fucose, it cannot be excluded that an additional pool of free fucose is being produced as a result of the activity of α-l-fucosidase secreted by the cells themselves during culturing (32). Hence, two more experiments were conducted to assess the potential effect of the presence of free fucose in the cell culture medium (Fig. S6).
In the first experiment, the wildtype and SLC35C1 knockout cells cultured in the presence of 4 μCi/ml of radioactive fucose were supplemented with a series of concentrations of nonradioactive fucose. Neither the percentage of fucosylation (Fig. S6A) nor the radioactivity of the isolated glycans (Fig. S6B) were influenced by the increasing nonradioactive fucose in the tested range of 0 to 0.2 μM. In the second experiment, the cells were grown in a complete culture medium supplemented with low increasing concentrations of radiolabeled fucose (1, 4, and 8 μCi/ml; Fig. S6C). Here, the incorporation of radiolabeled fucose into N-glycans was proportional to the amount of supplemented radioactive fucose.
Quantitative PCR analysis of expression levels of selected glycosylation-related genes
There is a possibility that some fucosylation-related genes become upregulated or downregulated by the exogenously added fucose. Therefore, to complement our metabolic studies, expression levels of selected glycosylation-related genes were analyzed in the control and fucose-fed SLC35C1-deficient cell lines. For the analysis of a larger subset of genes, a Universal Probe Library–based assay was designed (Fig. S7). Here, 91 different genes, including NSTs, glycosyltransferases, and biosynthetic enzymes, involved in metabolism of different monosaccharides (galactose, N-acetylglucosamine, mannose, xylose, and fucose) were investigated. No statistically significant differences could be assigned for any of the tested genes.
To analyze differences in gene expression between the wildtype and SLC35C1 knockouts in HEK293T and HepG2 cells, a SYBR Green–based quantitative PCR (qPCR) assay was performed for six selected genes, that is, FCL, GMDS, FUK, FPGT1, FPGT2, and SLC35C2 (Tables S4 and S5). Similarly to the previous findings, no statistically significant differences between the wildtype and SLC35C1 knockout HEK293T and HepG2 cells (both control and supplemented with exogenous fucose) were observed.
Discussion
This study confirmed the existence of SLC35C1-independent GDP-fucose transport routes in the Golgi complex and determined the intracellular fate of the exogenously supplied fucose. Moreover, the results showed that the SLC35C1-dependent route mainly transports GDP-fucose provided by the de novo biosynthetic pathway, whereas the SLC35C1-independent route nearly exclusively translocates GDP-fucose derived from the salvage pathway.
The SLC35C1-deficient mammalian model was for the first time developed and characterized by Hellbusch et al. in 2007 (27). In that study, treatment of cells from different mouse organs with exogenously supplied fucose partially restored glycoprotein fucosylation, which encouraged the authors to propose the existence of an alternative SLC35C1-independent GDP-fucose transport mechanism. However, the improvement in fucosylation was demonstrated using only a single method, that is, lectin-based flow cytometry, and no intermediate metabolic steps, for example, conversion of fucose into GDP-fucose, were investigated.
The study presented here in multiple aspects extends the previously conducted research. First of all, the improvement in fucosylation caused by supplementation of the culture medium with millimolar concentrations of fucose was precisely determined to be ∼77% by our self-developed quantitative HPLC-based approach. Moreover, we also found that nanomolar concentrations of exogenous fucose are sufficient to observe its incorporation into N-glycans in the knockout cells. Furthermore, we determined the extent to which exogenous fucose is transported into the cytoplasm and how it contributes to the increase of the concentration of the intracellular GDP-fucose (the extracellular fucose, intracellular fucose, and intracellular GDP-fucose were shown to exist in a 5 mM/1 mM/0.2 mM equilibrium). Finally, we found that the SLC35C1-independent Golgi GDP-fucose transport systems preferentially use the nucleotide sugar pool derived from the salvage pathway, which we consider the main outcome of this study.
In this study, we developed a quantitative HPLC-based approach allowing for very precise determination of the level of core fucosylation of N-glycans. This form of fucosylation, mediated by a single Fut8 enzyme, is present in many cell types including HEK293T and HepG2, which were selected as our model. Moreover, it is easy to study because of the well-established procedures of N-glycan isolation and HPLC analysis. As shown by our data, the level of core-fucosylated N-glycans in the wildtype HEK293T and HepG2 cell lines is high (∼80%) but does not reach 100%, giving a possibility to potentially observe a further increase in fucosylation, that is, overfucosylation, upon fucose treatment. On the other hand, in these cell lines, the level of core-fucosylated structures is high enough to make a clear distinction between the wildtype and the SLC35C1 knockout phenotypes. To conclude, the choice of cell lines was dictated by the high percentage of core-fucosylated N-glycans they produce, which in turn was demanded by the quantitative HPLC-based approach we employed.
Our quantitative HPLC-based approach revealed that the core N-glycan fucosylation is significantly compromised in the SLC35C1-deficient cells. However, some residual fucosylation could still be detected in these cell lines. The SLC35C2 protein is also specific for GDP-fucose. Thus, its activity could potentially compensate for the lack of SLC35C1 in the single knockout cells. This study demonstrated that this is not the case, as knocking out the SLC35C2 gene in the SLC35C1-deficient cells did not make the fucosylation defect any more severe. These results suggest that there must be yet another system responsible for the SLC35C1-independent GDP-fucose import into the Golgi lumen.
Strikingly, supplementation of the culture medium with millimolar concentrations of fucose caused a substantial improvement of core N-glycan fucosylation in both single SLC35C1 and double SLC35C1/SLC35C2 knockout cells. In contrast, in the wildtype cells, fucose treatment did not cause any further improvement in fucosylation, which is in line with the results obtained by Moriwaki et al. (33), who did not observe any increase in AAL reactivity with glycoproteins produced by the wildtype Hep3B cells supplemented with up to 5 mM fucose. This finding in addition supports our assumption that SLC35C1 mainly utilizes GDP-fucose produced by the de novo pathway, whereas GDP-fucose synthesized via the salvage pathway does not appear to be an optimal source of this nucleotide sugar in the wildtype cells, even if the latter is present in a large excess. We also hypothesize that the residual fucosylation detectable in the knockout cells is derived from GDP-fucose produced by the salvage pathway, which is not the main source of this nucleotide sugar in the wildtype cells. We believe that if the SLC35C1-independent GDP-fucose transport system used the nucleotide sugar substrate produced by the main biosynthetic pathway (i.e., de novo), the level of core fucosylated N-glycans in the knockout cells would be much higher.
It should be emphasized that fucose treatment of the LADII patients and SLC35C1-deficient cells can only be successful if the following requirements are fulfilled: (i) fucose must efficiently enter the cells, (ii) after entering the cell, fucose must be readily converted into GDP-fucose via the salvage pathway, and (iii) GDP-fucose must reach the Golgi lumen to become available for the catalytic centers of fucosyltransferases. Importantly, quantification of intracellular GDP-fucose concentration in fucose-fed SLC35C1 knockout cells was not attempted in the previous studies. To our best knowledge, the changes in the GDP-fucose content in response to exogenously supplied fucose were not widely examined. The few reports addressing this issue include the studies by: (i) Moriwaki et al. (33) performed on the wildtype Hep3B cells, (ii) Kanda et al. (34) on the several CHO sublines, and (iii) Sosicka et al. (35) on the wildtype HepG2, CHO, and Huh7 cells. Importantly, neither of these studies attempted to determine the intracellular GDP-fucose concentration in fucose-fed SLC35C1 knockout cells, which is crucial for our understanding of the responsiveness of these cells to exogenous fucose.
Here, we determined GDP-fucose concentrations in the control and fucose-fed wildtype and knockout cells. We found that the baseline concentrations of GDP-fucose were similar in the wildtype and knockout cells, whereas a remarkable increase could be observed upon fucose treatment in all the cell lines analyzed. In native conditions, the salvage pathway was shown to play only a negligible role in the overall GDP-fucose production. We found, however, that the capacity of this pathway is very high, that is, the corresponding enzymes are able to process significantly greater amounts of the primary substrate than normally present in the cell. qPCR data exclude the possibility that the exogenously added fucose triggers strong upregulation of expression of the genes encoding enzymes acting in this pathway as we did not observe any significant changes in the relative levels of the corresponding transcripts between the control and fucose-fed SLC35C1 knockout cells.
Our results clearly show that the therapeutic approaches for congenital disorders of glycosylations associated with impaired NST activity should aim for a drastic increase in the intracellular concentration of the corresponding nucleotide sugar substrates. It should be noted that treatment of these congenital disorders of glycosylations by oral monosaccharide supplementation can only be successful if the Km of an enzyme initiating the corresponding salvage pathway is sufficiently high. Otherwise, the treatment would simply be ineffective. FUK is the first enzyme acting in the salvage pathway of GDP-fucose biosynthesis. Importantly, in the case of FUK purified from pig kidney, Km for fucose was determined to be as high as 27 μM (36). This explains why submillimolar and millimolar concentrations of exogenous fucose are required to efficiently stimulate GDP-fucose synthesis and rescue N-glycan fucosylation in SLC35C1-deficient cells.
Given the fact that fucose treatment nearly completely rescued the fucosylation defect in the SLC35C1-deficient cells, it appears surprising that fucose-based therapy turned out to be ineffective for some of the LADII patients. There are two possible explanations for this phenomenon. First, the nonresponsive LADII patients might bear additional mutations outside the SLC35C1 gene that affect the performance of the fucosylation machinery. The other possible explanation is that the presence of certain mutant SLC35C1 variants is somehow more deleterious to the cells than the absence of the protein.
Regardless of the improvement of fucosylation in the SLC35C1-deficient cells treated with millimolar concentrations of fucose, we also found that these cells are able to incorporate fucose supplemented in nanomolar concentrations. This proves that in the knockout cells, the salvage pathway is able to fuel the SLC35C1-independent GDP-fucose transport system even at extremely low fucose concentrations.
Since increasing the intracellular concentration of GDP-fucose via fucose treatment improved core fucosylation of N-glycans, we wondered if it would be possible to boost the production of this nucleotide sugar by feeding the cells with millimolar concentrations of mannose given the fact that the de novo pathway of GDP-fucose synthesis utilizes GDP-mannose as a substrate. However, regardless of the cell line analyzed, mannose treatment did not cause any significant changes in the GDP-fucose concentration. This can be partially explained by the fact that mannose is converted to a variety of different metabolites (Fig. 8). Although in mannose-fed cells, a statistically significant increase in the GDP-mannose concentration was demonstrated, this did not translate into a similar increase in the GDP-fucose level. We therefore hypothesize that both in the wildtype and knockout cells, the de novo pathway operates with nearly maximum efficiency, and it is impossible to improve its performance by increasing the amount of its primary substrate, that is, GDP-mannose. From these findings, it can be concluded that treatment of LADII patients with mannose would not be an attractive alternative to fucose feeding.
Based on our results, we propose the existence of three different GDP-fucose transport systems in the Golgi membrane. The first one is SLC35C1-dependent and mainly utilizes the nucleotide sugar pool derived from the de novo pathway. The other two are not dependent on SLC35C1 and mainly use the nucleotide sugar pool synthesized in the salvage pathway. However, the first of them works even under very low (nanomolar) concentrations of exogenous fucose that do not cause any increase in the GDP-fucose concentrations, whereas the other requires much higher concentrations of this nucleotide sugar that can only be obtained by feeding the cells with submillimolar and millimolar concentrations of fucose. The existence of the latter SLC35C1-independent transport system is strongly supported by the nolinear dependence of the GDP-fucose content on the concentration of exogenous fucose. We hypothesize that this route may not be physiological and is not exclusively specific for GDP-fucose.
The hypothesis raised previously assumes the existence of distinct, independent, and separate cytosolic pools of GDP-fucose. Such a phenomenon was recently proposed by Sosicka et al. (35), who elegantly demonstrated that different fucosyltransferases utilize distinct GDP-fucose pools derived from distinct fucose sources. We believe that this effect could be achieved by selective cooperation of the different GDP-fucose transport systems with distinct fucosyltransferases (similar to, e.g., SLC35C2, which was shown to specifically support O-fucosylation (11, 12)). But the question arises how different Golgi transport systems could discriminate between the GDP-fucose pools derived from distinct metabolic pathways. This might require physical proximity (or even an association) between the enzymes acting in the individual pathways and the corresponding transport systems. Although the predominant Golgi localization of NSTs is well established, little is known about the precise subcellular distribution of nucleotide sugar synthases. Coates et al. (37) postulated cytoplasmic localization of the GDP-mannose and UDP-glucose pyrophosphorylases. This appears to hold true for all the other nucleotide sugar synthases except for the CMP–sialic acid synthetase, whose nuclear localization was demonstrated in the same study. However, it is widely believed that cytoplasmic enzymes catalyzing consecutive reactions in the individual pathways are not randomly distributed. Instead, they are sequestered to specific cytoplasmic subcompartments where they form functional assemblies that support substrate channeling (38). It cannot be excluded that the enzymes acting in the GDP-fucose biosynthetic pathways also display such a tendency.
Importantly, GDP-fucose was shown to inhibit the de novo pathway in humans (39) and bacteria (40). At the same time, this pathway is thought to be the main source of GDP-fucose in mammalian cells. Therefore, an immediate delivery of the GDP-fucose synthesized by the de novo pathway to the site of its ultimate utilization (i.e., the Golgi lumen) would be highly beneficial as it would ensure the efficient course of the GDP-fucose biosynthesis by shifting the equilibrium of the reaction. Such a scenario could be possible if the enzymes acting in the de novo pathway and the main GDP-fucose transporter, that is, SLC35C1, were located nearby. Although the mammalian FUK was shown to be nearly completely inhibited by 60 μM GDP-fucose (36), we did not observe such a phenomenon, as we were able to increase the intracellular concentration of GDP-fucose up to ∼200 μM by fucose treatment. This may suggest that the GDP-fucose formed in the salvage pathway does not accumulate in the site of its synthesis but instead is immediately channeled to the subcompartments in which it is subsequently utilized. Interestingly, the interactions between a plant UDP-glucose 4-epimerase and two UDP-galactose transporters were recently reported (41). Therefore, it is highly likely that also the enzymes acting in the de novo pathway of GDP-fucose biosynthesis are near to the SLC35C1 protein in mammalian cells.
This study confirmed the existence of alternative SLC35C1-independent GDP-fucose transport systems in the Golgi complex. It also demonstrated that SLC35C2 activity is dispensable for this transport to occur. We observed that the exogenously supplemented fucose efficiently enters the cells and is readily converted to GDP-fucose via the salvage pathway. Strikingly, the SLC35C1-deficient cells were virtually unable to incorporate fucose derived from GDP-fucose produced by the de novo pathway. There are several aspects that still need to be revealed, for example, the molecular identity of the SLC35C1-independent GDP-fucose transport systems and how different Golgi transport systems discriminate between the nucleotide sugar pools derived from different biosynthetic pathways. Nevertheless, we strongly believe that the findings obtained in this study initiate a brand new chapter in our perception of fucosylation.
Experimental procedures
Cell culturing and gene inactivation
HEK293T cells purchased from American Type Culture Collection (catalog no.: CRL-3216), and HepG2 cells purchased from the collection of the Department of Cancer Immunology (Hirszfeld Institute of Immunology and Experimental Therapy, Polish Academy of Sciences) were cultured in Dulbecco’s modified Eagle's medium (DMEM high glucose; Sigma–Aldrich) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were kept at 37 °C under 5% CO2.
A ready-to-use Santa Cruz Biotechnology CRISPR–Cas9 kit was used to inactivate the SLC35C1 gene in HEK293T and HepG2 cell lines. Cells were transfected with a mixture of human SLC35C1 double nickase plasmids (catalog no.: sc-410008-NIC-2) using the FuGENE HD transfection reagent (Promega) according to the manufacturer's protocol. Next, cells were cultured in DMEM complete medium supplemented with 1 μg/ml of puromycin for 3 weeks to select transfected cells. After that time, clones were isolated and checked for the presence of the SLC35C1 transcript using one-step RT–PCR with the HEK293T and HepG2 wildtype cells as the controls. Analysis of genomic DNA derived from the SLC35C1-deficient cells was also performed. Primers used in the reactions are listed in Table 1. The next step in confirmation of SLC35C1 gene knockout was Western blotting.
Inactivation of the SLC35C2 gene in the SLC35C1 knockout HEK293T cells was performed in the same manner. A Santa Cruz Biotechnology CRISPR–Cas9 kit with a mixture of human SLC35C2 double nickase plasmids (catalog no.: sc-409264-NIC-2) was applied. Confirmation of SLC35C2 gene knockout was performed at both RNA and genomic DNA levels using the HEK293T wildtype cells as a control. Used primers are listed in Table 1.
Generation of cell lines expressing HA-tagged recombinant SLC35C1 protein
A complementary DNA (cDNA) encoding human SLC35C1 protein (National Center for Biotechnology Information accession number: NM_018389.5) was generated from total RNA isolated from the HEK293T wildtype cells. The forward primer contained the nucleotide sequence of the HA tag. The amplified sequence was cloned into a pSelect-zeo-mcs plasmid vector (InvivoGen) using the SalI (New England Biolabs) restriction enzyme. Subsequently, HA-SLC35C1 mRNA was modified within the protospacer adjacent motif region recognized by Cas9 protein by introducing a silent mutation using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs) according to the manufacturer's protocol. This step was necessary to protect the HA-SLC35C1 mRNA from the activity of the Cas9 protein in the knockout cells. The primers used for mutagenesis are listed in Table 1.
The SLC35C1 knockout HEK293T and HepG2 cells were transfected with the obtained vector using the FuGENE HD transfection reagent according to the manufacturer’s instructions. Cells were then cultured in DMEM complete medium with the addition of zeocin (400 μg/ml for HEK293T and 200 μg/ml for HepG2) until stable clones were isolated. Cells stably expressing the HA-tagged SLC35C1 were identified by immunofluorescent staining with an anti-HA primary antibody followed by a secondary antibody conjugated with an Alexa Fluor dye. In addition, confirmation of expression of the desired protein was done by Western blotting.
Western blotting
Cells were harvested by scrapping and washed twice with PBS. Collected cells were lysed using CelLytic M (Sigma–Aldrich) supplemented with protease inhibitor cocktail (Bimake) and 0.15% EDTA. Then, protein concentration was determined using Roti-Nanoquant assay (Carl Roth) and adjusted to the same values for all samples. Cell lysates were separated in 10% SDS-PAGE gels and transferred onto nitrocellulose membranes (Amersham). After that, the membranes were blocked. Then, specific fragments of membranes were incubated with appropriate primary and secondary antibodies (listed in Table 2). Steps of blocking, antibody incubation, washing, and signal detection were performed as described previously (42).
Table 2.
Antibodies used in Western blotting analyses
| Antibody | Catalog no. | Origin | Dilution | Manufacturer |
|---|---|---|---|---|
| Anti-SLC35C1 | PA564146 | Rabbit | 1:1000 | Thermo Fisher Scientific |
| Anti-HA | ab9110 | Rabbit | 1:1000 | Abcam |
| Anti-HSP60 | sc-376261 | Mouse | 1:50,000 | Santa Cruz Biotechnology |
| Anti-GAPDH | ab8245 | Mouse | 1:10,000 | Abcam |
| Anti-mouse/rat immunoglobulin G—horseradish peroxidase | W402B | Goat | 1:10,000 | Promega |
| Anti-rabbit immunoglobulin G—horseradish peroxidase | A0545 | Goat | 1:10,000 | Sigma–Aldrich |
Fluorescence staining
Subcellular localization of the endogenous GDP-fucose transporter and HA-tagged recombinant SLC35C1 protein was determined using primary and secondary antibodies listed in Table 3. Cells were immunostained as described previously (43). In the case of determination of fucosylation level with biotinylated lectin in the wildtype and SLC35C1-deficient cells, the procedure was modified. After fixation, cells were permeabilized for 5 min at room temperature using 0.1% Triton X-100 in Tris-buffered saline (TBS). Nonspecific binding sites were blocked with 3% bovine serum albumin in TBS for 1 h at room temperature. Then, biotinylated AAL (catalog no.: B-1395-1; 1:300 dilution) diluted in blocking solution containing 1 mM CaCl2 and 1 mM MnCl2 was added for 1 h at 37 °C. Cells were washed with TBS. Slides were then incubated with streptavidin-Cy3 (Sigma–Aldrich; 1:500 dilution) solution for 1 h at 37 °C. The rest of the protocol remained unchanged. The resulting samples were analyzed using a Leica SP8 confocal microscope, and the obtained images were processed using ImageJ software (National Institutes of Health (NIH)).
Table 3.
Antibodies used in immunofluorescence staining experiments
| Antibody | Catalog no. | Origin | Dilution | Manufacturer |
|---|---|---|---|---|
| Anti-GM130 | 610823 | Mouse | 1:100 | BD Biosciences |
| Anti-calnexin | ab75801 | Rabbit | 1:100 | Abcam |
| Anti-SLC35C1 | PA564146 | Rabbit | 1:100 | Thermo Fisher Scientific |
| Anti-HA | ab9110 | Rabbit | 1:100 | Abcam |
| Anti-HA | 26183 | Mouse | 1:100 | Thermo Fisher Scientific |
| Anti-rabbit immunoglobulin G [IgG]—Alexa Fluor 488 | A21206 | Donkey | 1:200 | Life Technologies |
| Anti-mouse/rat IgG—Alexa Fluor 568 | A10037 | Donkey | 1:200 | Life Technologies |
| Anti-mouse/rat IgG—Alexa Fluor 488 | A21202 | Donkey | 1:200 | Life Technologies |
| Anti-rabbit IgG—Alexa Fluor 568 | A10042 | Donkey | 1:200 | Life Technologies |
N-glycan analysis
Control and fucose-fed (5 mM, 24 h) cells were harvested and lysed as described previously. Protein concentration was brought up to 2 mg/ml for each sample. Obtained lysates were treated with acetone to concentrate proteins. Proteins were then dissolved and enzymatically deglycosylated. Released glycans were purified, labeled with 2-aminobenzamide (2-AB), and analyzed as described previously (42). Obtained glycans were subjected to MALDI-TOF analysis.
O-glycan analysis
Control and fucose-fed (5 mM, 24 h) cells were cultured in medium with 5% fetal bovine serum with the addition of peracetylated O-glycan precursor (Ac3GalNAcBn) for 3 days. Subsequently, O-glycans contained in the culture medium were purified according to an adopted method described by Kudelka et al. (29). The obtained glycans were subjected to MALDI-TOF analysis.
MALDI-TOF mass spectroscopy analysis of N- and O-glycans
Prior to MALDI-TOF analysis, 2-AB-labeled N-glycans were desialylated using α2-3,6,8,9-neuraminidase A (New England Biolabs) and purified in the manner reported previously (42). Both N- and O-glycans were analyzed in a positive-ion mode as described previously (44).
Analysis of SEAP-derived N-glycans
Cells were transiently transfected with the 6xHis-SEAP construct using the FuGENE HD transfection reagent according to the manufacturer's recommendations. Medium containing secreted protein was collected. The SEAP reporter glycoprotein was subjected to further purification following the protocol described by Olczak and Szulc (28). Then, isolated N-glycans were labeled with 2-AB and analyzed as described previously (42).
Preparation of RNA
Total RNA was extracted from ∼3.2 × 106 cells (HEK293T, HepG2) using the Total RNA Mini Kit (A&A Biotechnology). Isolated RNA was treated with DNase I (A&A Biotechnology) and purified using the Clean-Up RNA Concentrator Kit (A&A Biotechnology). RNA integrity was verified spectrophotometrically.
SYBR Green–based qRT–PCR
Reverse transcription was carried out on 1 μg of total RNA using a SensiFAST cDNA Synthesis Kit (Bioline). PCR was carried out using the Luna Universal qPCR Master Mix (New England Biolabs) and a LightCycler 96 instrument (Roche). The amplification reaction comprised initial denaturation at 95 °C for 1 min and followed by 45 amplification steps (denaturation at 95 °C for 15 s; primer annealing and extension at 60 °C for 30 s). The melting curves were analyzed to monitor the quality of PCR products. Relative quantification of expression of respective genes was determined by the ΔΔCq method using human GADPH (NM_002046.7) as a reference. Three independent experiments (biological replicates) were performed to test each experimental condition, and each sample was run in three technical replicates. Hence, in total 3 × 3 = 9 runs were executed per one experimental condition. No template controls were included on each reaction plate to check for contamination. Negative controls consisting of untranscribed RNA (no-reverse transcription controls) were performed to check for genomic DNA contamination. All primers used in this study are listed in Table 4.
Table 4.
List of primers used in SYBR Green–based quantitative RT–PCR
| Gene | National Center for Biotechnology Information accession | Forward primer | Reverse primer |
|---|---|---|---|
| FX | NM_003313.3 | GACAAGACGACCTACCCGAT | GTTCTGCACGTCGATCATCC |
| FUK | NM_145059.2 | GACTGTGGCAGGGCTTTCA | CAGCCGATAGGTCATGATGG |
| FPGT1 | NM_003838.4 | GGGTGACATTGCCGATCTTA | CCAAAGCCTGCAGAAAGTCA |
| FPGT2 | NM_001199328.2 | GGAGTCTGTTTCCTGTCATGC | CAAACTGGGAAAATGCGTG |
| GAPDH | NM_002046.7 | AGGTCGGAGTCAACGGATTT | TGACAAGCTTCCCGTTCTCA |
| GMDS | NM_001253846.1 | GCCATGCCAAGGACTATGTG | TCTCGACAAATTCCCGGACA |
Universal Probe Library-based qRT–PCR
Preparation of cDNA
cDNA was produced using a Transcriptor First Strand cDNA Synthesis Kit (Hoffmann-La Roche) according to the manufacturer's instructions. Total RNA (4 μg) extracted from HEK293T cells was mixed with 2 μl of 50 pmol/μl anchored-oligo(dT)18 primer and supplemented with PCR-grade water up to a final volume of 26 μl. The template–primer mixture was incubated in a thermal block cycler with a heated lid for 10 min at 65 °C to denature the RNA secondary structures and cooled on ice. Subsequently, 5× concentrated Transcriptor Reverse Transcriptase Reaction Buffer (8 μl), 40 U/μl Protector RNase Inhibitor (1 μl), and Deoxynucleotide Mix (10 mM each; 4 μl) and 20 U/μl Transcriptor Reverse Transcriptase were added. The final reaction mixture (40 μl) was incubated in a thermal block cycler with a heated lid for 30 min at 55 °C and inactivated for 5 min at 85 °C.
qPCR procedure
qPCR mixture was assembled at room temperature by mixing cDNA (40 μl) obtained in the previous step with PCR-grade water (960 μl) and 2× concentrated LightCycler 480 Probes Master (Roche; 1 ml) and pipetted into a 96-well RealTime ready Custom Panel plate (Roche; 20 μl per well). The plate was sealed with LightCycler 480 Sealing Foil (Roche) and rotated in a horizontal position for 5 min at 1000 rpm to dissolve primers and probes lyophilized on the bottom of the Custom Panel plate wells. The plate was spun down in a centrifuge equipped with a 96-well plate swing-bucket rotor to remove any potential air bubbles and subjected to the qPCR experiment using the LightCycler 96 instrument. The PCR was initiated by a 10-min preincubation at 96 °C followed by 45 amplification steps (95 °C for 10 s; 60 °C for 30 s; and 72 °C for 1 s).
Custom Panel qPCR plates
qPCR assays (primer pairs and probes) were purchased from Roche and supplied in a lyophilized form in 96-well plates. The exact configuration of the plate including assay positions on the plate, assay IDs and gene names, sequences of forward and reverse primers, and Universal Probe Library probe numbers is provided in Table S3. Assays were verified by the supplier to fulfill a number of quality criteria including (i) PCR efficiency 2.0 ± 0.2 (equals 100 ± 10%), (ii) Cq of highest cDNA concentration ≤34, (iii) linear dynamic range of at least three logs, (iv) high amplification specificity, no side products in gel analysis, (v) sigmoidal amplification curve, and (vi) fluorescence intensity of amplification curves between 5 and 50 fluorescence units.
Selection of a reference gene
The entire dataset, that is, the data from all eight Custom Panel plates that were run (four experiments with cDNA from SLC35C1-deficient HEK293T cells and four experiments from SLC35C1-deficient HEK293T cells grown in fucose-supplemented medium), was used to reveal the optimal gene to reference any potential changes in expression level of all other tested genes. The list of candidates included 85 unique assays as out of 96 wells, five were technical, whereas for six genes, the criterion of giving a product in all eight experiments was not met. The analysis was performed using four different algorithms, that is, GeNorm, NormFinder, BestKeeper, and delta-Cq methods. The most stable consensus gene was beta-1,4-glucuronyltransferase 1 (B4GAT1).
qPCR data analysis
To test the null hypothesis that feeding cells with 5 mM fucose does not affect the expression of the tested glycosylation-related genes, the SLC35C1-deficient HEK293T cells grown in fucose-supplemented medium (N = 4, i.e., four biological repetitions or four Custom Panel qPCR plates) were compared with the SLC35C1-deficient HEK293T cells supplemented with PBS (also N = 4). Relative expression folds were calculated using the ΔΔCq method following the study by Taylor et al. (45). The results were subjected to the t test and Benjamini–Hochberg procedure to assess the significance of the gene expression differences in the nonfed and fed cells.
Optimization of fucose supplementation conditions
Three supplementation parameters were optimized, that is, time of supplementation, frequency of medium exchange, and concentration of l-fucose. The first optimizations were carried out in two variants. In the first variant, the wildtype and SLC35C1 knockout cells were cultured in complete medium (DMEM high glucose, 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin) with addition of 0.5, 1, 5, and 10 mM l-fucose dissolved in PBS for 5 and 12 days, and the medium was exchanged every day. In the second variant, the wildtype and SLC35C1 knockout cells were cultured in complete medium with addition of 0.5, 1, 5, and 10 mM l-fucose dissolved in PBS for 5 and 12 days, but the medium was replaced every other day. As the control, the wildtype and SLC35C1-deficient cells were kept in complete medium supplemented with PBS. Control cells were also cultured in two variants as the fucose-fed cells. After that, cells were collected, lysed, and subjected to dot blotting with AAL to visualize the fucosylation level. The procedure of dot blotting is described in the later section.
In the next step, SLC35C1 knockout cells were cultured on glass 8-well microscope slides (Merck) in complete medium with 5 mM l-fucose for 48 h. Cells were fixed after 2, 4, 6, 8, 10, 24, 36, and 48 h after starting supplementation. Fixed cells were subjected to fluorescent staining with AAL according to the procedure mentioned previously.
As the last experiment, the wildtype and SLC35C1 knockout cells were treated with different l-fucose concentrations for different times. Cells were collected and lysed. Cell lysates were subjected to N-glycan extraction performed as described previously. Purified and 2-AB-labeled N-glycans were analyzed for α-1,6-fucosylation as described later.
Dot blotting
Whole cell lysates obtained from untreated and fucose-fed cells were applied directly on nitrocellulose membrane (Amersham). Glycans containing fucose were detected using biotinylated AAL (catalog no.: B-1395-1; 1:300 dilution) as described previously (42). Lectin attached to glycans was visualized by streptavidin conjugated to horseradish peroxidase (Vector Laboratories; catalog no.: SA-5014-1; 1:50,000 dilution) by chemiluminescence reaction using a Western Lightning Plus-ECL kit (PerkinElmer). Equal protein loading was demonstrated by staining proteins on nitrocellulose membrane with Ponceau S solution.
Determination of l-fucose concentration in fucose-fed SLC35C1-deficient cells
Concentration of l-fucose in fucose-fed SLC35C1-deficient HEK293T cells was determined using a commercial kit (l-fucose, Megazyme). Briefly, ∼120 × 10⁶ cells were resuspended in MilliQ and then sonicated. Next, in order to get rid of proteins from the lysate, ice-cold perchloric acid was added. The mixture was spun down, and the supernatant was collected. Subsequently, an assay sample was prepared by mixing the supernatant with the detection buffer and NADP⁺ solution (the latter two were supplied with the kit) in volumes recommended by the manufacturer. The mixture was incubated for 3 min. After that, an l-fucose dehydrogenase suspension (supplied with the kit) was added to the mixture in the volume recommended by the manufacturer. Finally, the absorbance was measured at 340 nm using a Beckman DU-640 spectrophotometer. Concentration of fucose was determined using a calibration curve assuming that the volume of a single HEK293T cell is 3.5 pL.
Radioactive labeling of N-glycans and nucleotide sugars
Cells were cultured in complete medium supplemented with 4 μCi/ml L-[6-3H]-fucose (American Radiolabeled Chemicals; specific activity = 60 Ci/mmol) for 24 h or 20 μCi/ml d-[1-3H]-mannose (American Radiolabeled Chemicals; specific activity = 20 Ci/mmol) for 48 h to analyze N-glycans. Then cells were harvested and subjected to N-glycan preparation followed by analysis of N-glycan α-1,6 fucosylation. For analysis of nucleotide sugars, cells were incubated in complete medium with addition of 4 μCi/ml l-[6-3H]-fucose (American Radiolabeled Chemicals; specific activity = 60 Ci/mmol) for 24 h or 167 μCi/ml D-[1-3H]-mannose (American Radiolabeled Chemicals; specific activity = 20 Ci/mmol) for 48 h. Cells were collected, and nucleotide sugars were extracted as described later.
Analysis of N-glycan α-1,6-fucosylation
Purified, 2-AB-labeled, and dried N-glycan pools were dissolved in 25 μl of certified, exoglycosidase-free GlycoBuffer 1 (50 mM sodium citrate, pH 6.0; New England Biolabs). Neuraminidase A (40 units per reaction), β-1-4 galactosidase S (16 units per reaction), and β-N-acetyl-glucosaminidase S (8 units per reaction) were added to start digestion. All enzymes were purchased from New England Biolabs. Reactions were performed overnight at 37 °C in an air-heated incubator to avoid evaporation. In these conditions, virtually, all complex-type N-glycans were converted to two conserved trimannosyl core structures (fucosylated at the first N-acetylglucosamine and nonfucosylated). The postreaction mixtures were separated on a GlycoSep N Plus column (Prozyme) at 40 °C. High-resolution gradient 1 (Prozyme manual for the column) was applied, and detection was performed with a fluorescence detector (330 nm excitation and 420 nm emission) connected to a PerkinElmer Series 200 HPLC gradient system. The percentage of N-glycan core fucosylation was calculated from peak area of the fucosylated trimannosyl core compared with area of the peak eluted as a nonfucosylated trimannosyl structure, using TotalChrom software (PerkinElmer).
The same procedure of separation was applied for N-glycans released from cells labeled with 3H-fucose. In these experiments, fractions of eight drops (approximately 230 μl) were collected manually and directly transferred to scintillation vials.
Calculations of the extent of radioactive fucose incorporation were based on radioactivity (expressed in counts per minute) of a radioisotope of known concentration and known specific activity compared with peak areas of standard, 2-AB-labeled, and trimannosyl core structures (fucosylated and nonfucosylated) of known concentrations (all standard glycans were purchased from Oxford GlycoSystems).
Analysis of N-glycan fucosylation after 3H-mannose and 3H-fucose labeling
Purified, 2-AB-labeled, and dried N-glycan pools released from cells labeled with 3H-fucose or 3H-mannose were resuspended in 25 μl of certified exoglycosidase-free GlycoBuffer 1. Then α1-2,4,6 fucosidase O (New England Biolabs; four units per reaction) was added to start defucosylation. The reaction was performed overnight at 37 °C. After digestion, the mixtures were complemented with 250 μl of 1% TFA, mixed, and applied to the Graphite Spin Columns (Pierce), previously washed twice with 100 μl of 1 M ammonia, primed with 100 μl of acetonitrile, and finally equilibrated twice with 100 μl of 1% TFA; each step was completed by centrifugation at 2000g for 1 min. Applied samples were bound to the graphite resin in closed spin column for 20 min, with periodic vortex mixing. After 20 min of incubation, the columns were centrifuged at 2000g for 3 min and washed three times with 100 μl of 5% acetonitrile/water/1% TFA solution. The flow-through and washes (approximately 550 μl of total volume) containing released free fucose were combined and transferred to scintillation vials. Glycans bound to the column were released from spin columns with three elutions, each with 150 μl of 60% acetonitrile/water/0.5% TFA. Fractions were combined and transferred to scintillation vials. The last step confirmed the 100% rate of fucosidase digestion with no detectable radioactivity in released N-glycans.
Control reactions for the same procedure were also performed, without fucosidase added to GlycoBuffer 1. This confirmed lack of nonenzyme defucosylation during overnight incubations.
HPLC separation of nucleotide sugars
Nucleotide sugars were purified from collected and frozen cells using a protocol previously published (30). Enriched nucleotide sugar pools were separated in ion-pairing reverse-phase HPLC, as described previously (46) with some changes. The column was replaced with Interstil ODS-4, 250 × 3 mm, 3 μm particle size; flow rate was set to 0.3 ml/min. Buffer A (100 mM potassium phosphate, pH 6.4 supplemented with 8 mM tetrabutylammonium hydrogen sulphate) was used for equilibration, and buffer B (70% of A plus 30% acetonitrile) was used as an eluent. Separations were performed using the following gradient: 0% for 20 min, 0% to 72% for 25 min, 72–77% for 10 min, and, finally, equilibration at 0% of buffer A for 25 min. Nucleotide sugar samples were resuspended in Milli-Q water.
Sample volumes of 5 μl or less were injected to start separations. The column was connected to the Nexera Shimadzu HPLC system. Detection was performed at 254 nm with an SPD_M30A Diode Array Detector equipped with a high-sensitivity quartz cell. Calculations of nucleotide sugar concentrations were performed with LabSolutions Software (Shimadzu). Standard high-purity nucleotide sugars were purchased from Sigma–Aldrich. The absolute quantification of nucleotide sugars was performed by comparison of the detected signals to the externally added reference compounds. Next, by referring to the starting number of cells and to the anticipated volume of a single cell (3 and 3.5 pl per cell for HepG2 and HEK293T, respectively), it was possible to estimate the intracellular nucleotide sugar concentrations.
The same procedure of ion-pairing reverse-phase separation was applied for nucleotide sugar pools, previously purified from cells labeled with 3H-fucose and 3H-mannose (as described before). In these experiments, fractions of five drops (approximately 150 μl) between the 26th and 33rd minute of the HPLC gradient were collected manually and directly transferred to scintillation vials. Retention times under separation conditions described previously were approximately 27.2 and 31.1 min, for GDP-mannose and GDP-fucose, respectively.
Statistical analysis
Statistical parameters including data plotted (mean ± SD), p values, and statistical tests used are detailed in figure legends. Statistical analyses were performed using GraphPad Prism 8 (GraphPad Software, Inc). Data were analyzed by Student’s t test and the Benjamini–Hochberg procedure or Welch’s correction and by one-way ANOVA with the Tukey post hoc test.
Data availability
The MS data have been deposited in the GlycoPost under ID GPST000261. Some unpublished data are available upon request from the corresponding author, e-mail: mariusz.olczak@uwr.edu.pl, on reasonable request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Auhen Shauchuk for the help in generation of SLC35C1-deficient HEK293T and HepG2 cells.
Author contributions
M. O. conceptualization; E. S., B. S., M. W., W. W., A. M., and M. O. methodology; E. S., B. S., M. W., W. W., A. M., and M. O. investigation; E. S., B. S., M. W., and W. W. formal analysis; D. M.-S. and M. W. writing–original draft; E. S., B. S., D. M.-S., W. W., and M. O. writing–review and editing; B. S., M. W., and W. W. visualization; M. O. supervision; M. O. funding acquisition.
Funding and additional information
This work is supported by the National Science Centre (Cracow, Poland), (grant no.: 2016/21/B/NZ5/00144; to M. O.).
Edited by Chris Whitfield
Footnotes
Present address for Aleksandra Makowiecka: Department of Chemical Biology and Bioimaging, Wroclaw University of Science and Technology, Wrocław, Poland
Supporting information
References
- 1.Varki A., Cummings R.D., Esko J.D., Stanley P., Hart G.W., Aebi M., et al., editors. Essentials of Glycobiology, Cold Spring Harbor Laboratory Press. Cold Spring Harbor; New York: 2015-2017. [PubMed] [Google Scholar]
- 2.Yurchenco P.D., Atkinson P.H. Equilibration of fucosyl glycoprotein pools in HeLa cells. Biochemistry. 1977;16:944–953. doi: 10.1021/bi00624a021. [DOI] [PubMed] [Google Scholar]
- 3.Yurchenco P.D., Atkinson P.H. Fucosyl-glycoprotein and precursor polls in HeLa cells. Biochemistry. 1975;14:3107–3114. doi: 10.1021/bi00685a011. [DOI] [PubMed] [Google Scholar]
- 4.Moriwaki K., E. Miyoshi E. Fucosylation and gastrointestinal cancer. World J. Hepatol. 2010;2:151–161. doi: 10.4254/wjh.v2.i4.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiese T.J., Dunlap J.A., Yorek M.A. L-fucose is accumulated via a specific transport system in eukaryotic cells. J. Biol. Chem. 1994;269:22705–22711. [PubMed] [Google Scholar]
- 6.Pastuszak I., Ketchum C., Hermanson G., Sjoberg E.J., Drake R., Elbein A.D. GDP-L-fucose pyrophosphorylase. Purification, cDNA cloning, and properties of the enzyme. J. Biol. Chem. 1998;273:30165–30174. doi: 10.1074/jbc.273.46.30165. [DOI] [PubMed] [Google Scholar]
- 7.Capasso J.M., Hirschberg C.B. Mechanisms of glycosylation and sulfation in the Golgi apparatus: Evidence for nucleotide sugar/nucleoside monophosphate and nucleotide sulfate/nucleoside monophosphate antiports in the Golgi apparatus membrane. Proc. Natl. Acad. Sci. U. S. A. 1984;81:7051–7055. doi: 10.1073/pnas.81.22.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn N.J., White A. The role of nucleoside diphosphatase in a uridine nucleotide cycle associated with lactose synthesis in rat mammary-gland Golgi apparatus. Biochem. J. 1977;168:423–433. doi: 10.1042/bj1680423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lübke T., Marquardt T., Etzioni A., Hartmann E., von Figura K., Körner C. Complementation cloning identifies CDG-IIc, a new type of congenital disorders of glycosylation, as a GDP-fucose transporter deficiency. Nat. Genet. 2001;28:73–76. doi: 10.1038/ng0501-73. [DOI] [PubMed] [Google Scholar]
- 10.Lühn K., Wild M.K., Eckhardt M., Gerardy-Schahn R., Vestweber D. The gene defective in leukocyte adhesion deficiency II encodes a putative GDP-fucose transporter. Nat. Genet. 2001;28:69–72. doi: 10.1038/ng0501-69. [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa H.O., Ayukawa T., Nakayama M., Higashi S., Kamiyama S., Nishihara S., et al. Two pathways for importing GDP-fucose into the endoplasmic reticulum lumen function redundantly in the O-fucosylation of Notch in Drosophila. J. Biol. Chem. 2010;285:4122–4129. doi: 10.1074/jbc.M109.016964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu L., Hou X., Shi S., Körner C., Stanley P. Slc35c2 promotes Notch1 fucosylation and is required for optimal Notch signaling in mammalian cells. J. Biol. Chem. 2010;285:36245–36254. doi: 10.1074/jbc.M110.126003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma B., Simala-Grant J.L., Taylor D.E. Fucosylation in prokaryotes and eukaryotes. Glycobiology. 2006;16:158–184. doi: 10.1093/glycob/cwl040. [DOI] [PubMed] [Google Scholar]
- 14.Gazit Y., Mory A., Etzioni A., Frydman M., Scheuerman O., Gershoni-Baruch R., et al. Leukocyte adhesion deficiency type II: Long-term follow-up and review of the literature. J. Clin. Immunol. 2010;30:308–313. doi: 10.1007/s10875-009-9354-0. [DOI] [PubMed] [Google Scholar]
- 15.Etzioni A., Frydman M., Pollack S., Avidor I., Phillips M.L., Paulson J.C., et al. Brief report: Recurrent severe infections caused by a novel leukocyte adhesion deficiency. N. Engl. J. Med. 1992;327:1789–1792. doi: 10.1056/NEJM199212173272505. [DOI] [PubMed] [Google Scholar]
- 16.Dauber A., Ercan A., Lee J., James P., Jacobs P.P., Ashline D.J., et al. Congenital disorder of fucosylation type 2c (LADII) presenting with short stature and developmental delay with minimal adhesion defect. Hum. Mol. Genet. 2014;23:2880–2887. doi: 10.1093/hmg/ddu001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hüllen A., Falkenstein K., Weigel C., Huidekoper H., Naumann-Bartsch N., Spenger J., et al. Congenital disorders of glycosylation with defective fucosylation. J. Inherit. Metab. Dis. 2021;44:1441–1452. doi: 10.1002/jimd.12426. [DOI] [PubMed] [Google Scholar]
- 18.Helmus Y., Denecke J., Yakubenia S., Robinson P., Lühn K., Watson D.L., et al. Leukocyte adhesion deficiency II patients with a dual defect of the GDP-fucose transporter. Blood. 2006;107:3959–3966. doi: 10.1182/blood-2005-08-3334. [DOI] [PubMed] [Google Scholar]
- 19.van de Vijver E., Maddalena A., Sanal Ö., Holland S.M., Uzel G., Madkaikar M., et al. Hematologically important mutations: Leukocyte adhesion deficiency (first update) Blood Cells Mol. Dis. 2012;48:53–61. doi: 10.1016/j.bcmd.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hidalgo A., Ma S., Peired A.J., Weiss L.A., Cunningham-Rundles C., Frenette P.S. Insights into leukocyte adhesion deficiency type 2 from a novel mutation in the GDP-fucose transporter gene. Blood. 2003;101:1705–1712. doi: 10.1182/blood-2002-09-2840. [DOI] [PubMed] [Google Scholar]
- 21.Cagdas D., Yilmaz M., Kandemir N., Tezcan I., Etzioni A., Sanal Ö. A novel mutation in leukocyte adhesion deficiency type II/CDGIIc. J. Clin. Immunol. 2014;34:1009–1014. doi: 10.1007/s10875-014-0091-7. [DOI] [PubMed] [Google Scholar]
- 22.Cooper N., Li Y.T., Möller A., Schulz-Weidner N., Sachs U.J., Wagner H., et al. Incidental diagnosis of leukocyte adhesion deficiency type II following ABO typing. Clin. Immunol. 2020;221:108599. doi: 10.1016/j.clim.2020.108599. [DOI] [PubMed] [Google Scholar]
- 23.Knapp K.M., Luu R., Baerenfaenger M., Zijlstra F., Wessels H., Jenkins D., et al. Biallelic variants in SLC35C1 as a cause of isolated short stature with intellectual disability. J. Hum. Genet. 2020;65:743–750. doi: 10.1038/s10038-020-0764-4. [DOI] [PubMed] [Google Scholar]
- 24.Karsan A., Cornejo C.J., Winn R.K., Schwartz B.R., Way W., Lannir N., et al. Leukocyte Adhesion Deficiency Type II is a generalized defect of de novo GDP-fucose biosynthesis. Endothelial cell fucosylation is not required for neutrophil rolling on human nonlymphoid endothelium. J. Clin. Invest. 1998;101:2438–2445. doi: 10.1172/JCI905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marquardt T., Lühn K., Srikrishna G., Freeze H.H., Harms E., Vestweber D. Correction of leukocyte adhesion deficiency type II with oral fucose. Blood. 1999;94:3976–3985. [PubMed] [Google Scholar]
- 26.Sturla L., Rampal R., Haltiwanger R.S., Fruscione F., Etzioni A., Tonetti M. Differential terminal fucosylation of N-linked glycans versus protein O-fucosylation in leukocyte adhesion deficiency type II (CDG IIc) J. Biol. Chem. 2003;278:26727–26733. doi: 10.1074/jbc.M304068200. [DOI] [PubMed] [Google Scholar]
- 27.Hellbusch C.C., Sperandio M., Frommhold D., Yakubenia S., Wild M.K., Popovici D., et al. Golgi GDP-fucose transporter-deficient mice mimic congenital disorder of glycosylation IIc/leukocyte adhesion deficiency II. J. Biol. Chem. 2007;282:10762–10772. doi: 10.1074/jbc.M700314200. [DOI] [PubMed] [Google Scholar]
- 28.Olczak M., Szulc B. Modified secreted alkaline phosphatase as an improved reporter protein for N-glycosylation analysis. PLoS One. 2021;16 doi: 10.1371/journal.pone.0251805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kudelka M.R., Antonopoulos A., Wang Y., Duong D.M., Song X., Seyfried N.T., et al. Cellular O-Glycome Reporter/Amplification to explore O-glycans of living cells. Nat. Methods. 2016;13:81–86. doi: 10.1038/nmeth.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Räbinä J., Mäki M., Savilahti E.M., Järvinen N., Penttilä L., Renkonen R. Analysis of nucleotide sugars from cell lysates by ion-pair solid-phase extraction and reversed-phase high-performance liquid chromatography. Glycoconjugate J. 2001;18:799–805. doi: 10.1023/a:1021107602535. [DOI] [PubMed] [Google Scholar]
- 31.van Tol W. Radboud University Nijmegen; the Netherlands: 2020. Discovery of the Sugar Supply Pathways for the O-Mannosylation of Dystroglycan. On the Road for Treating Muscular Dystrophy-Dystroglycanopathy (Doctoral Dissertation) [Google Scholar]
- 32.Liu T.W., Ho C.W., Huang H.H., Chang S.M., Popat S.D., Wang Y.T., et al. Role for α-l-fucosidase in the control of Helicobacter pylori-infected gastric cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14581–14586. doi: 10.1073/pnas.0903286106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moriwaki K., Noda K., Nakagawa T., Asahi M., Yoshihara H., Taniguchi N., et al. A high expression of GDP-fucose transporter in hepatocellular carcinoma is a key factor for increases in fucosylation. Glycobiology. 2007;17:1311–1320. doi: 10.1093/glycob/cwm094. [DOI] [PubMed] [Google Scholar]
- 34.Kanda Y., Imai-Nishiya H., Kuni-Kamochi R., Mori K., Inoue M., Kitajima-Miyama K., et al. Establishment of a GDP-mannose 4,6-dehydratase (GMD) knockout host cell line: A new strategy for generating completely non-fucosylated recombinant therapeutics. J. Biotechnol. 2007;130:300–310. doi: 10.1016/j.jbiotec.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 35.Sosicka P., Ng B.G., Pepi L.E., Shajahan A., Wong M., Scott D.A., et al. Metabolic heritage mapping: Heterogenous pools of cytoplasmic nucleotide sugars are selectively utilized by various glycosyltransferases. bioRxiv. 2021 doi: 10.1101/2021.11.03.467160. preprint. [DOI] [Google Scholar]
- 36.Park S.H., Pastuszak I., Drake R., Elbein A.D. Purification to apparent homogeneity and properties of pig kidney L-fucose kinase. J. Biol. Chem. 1998;273:5685–5691. doi: 10.1074/jbc.273.10.5685. [DOI] [PubMed] [Google Scholar]
- 37.Coates S.W., Gurney T., Jr., Sommers L.W., Yeh M., Hirschberg C.B. Subcellular localization of sugar nucleotide synthetases. J. Biol. Chem. 1980;255:9225–9229. [PubMed] [Google Scholar]
- 38.Pareek V., Sha Z., He J., Wingreen N.S., Benkovic S.J. Metabolic channeling: predictions, deductions, and evidence. Mol. Cell. 2021;81:3775–3785. doi: 10.1016/j.molcel.2021.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullivan F.X., Kumar R., Kriz R., Stahl M., Xu G.Y., Rouse J., et al. Molecular cloning of human GDP-mannose 4,6-dehydratase and reconstitution of GDP-fucose biosynthesis in vitro. J. Biol. Chem. 1998;273:8193–8202. doi: 10.1074/jbc.273.14.8193. [DOI] [PubMed] [Google Scholar]
- 40.Kornfeld R.H., Ginsburg V. Control of synthesis of guanosine 5'-diphosphate D-mannose and guanosine 5'-diphosphate L-fucose in bacteria. Biochim. Biophys. Acta. 1966;117:79–87. doi: 10.1016/0304-4165(66)90154-1. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y., Wang Y., Wang C., Rautengarten C., Duan E., Zhu J., et al. BRITTLE PLANT1 is required for normal cell wall composition and mechanical strength in rice. J. Integr. Plant Biol. 2021;63:865–877. doi: 10.1111/jipb.13050. [DOI] [PubMed] [Google Scholar]
- 42.Maszczak-Seneczko D., Olczak T., Wunderlich L., Olczak M. Comparative analysis of involvement of UGT1 and UGT2 splice variants of UDP-galactose transporter in glycosylation of macromolecules in MDCK and CHO cell lines. Glycoconjugate J. 2011;28:481–492. doi: 10.1007/s10719-011-9348-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bazan B., Wiktor M., Maszczak-Seneczko D., Olczak T., Kaczmarek B., Olczak M. Lysine at position 329 within a C-terminal dilysine motif is crucial for the ER localization of human SLC35B4. PLoS One. 2018;13 doi: 10.1371/journal.pone.0207521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szulc B., Sosicka P., Maszczak-Seneczko D., Skurska E., Shauchuk A., Olczak T., et al. Biosynthesis of GlcNAc-rich N- and O-glycans in the Golgi apparatus does not require the nucleotide sugar transporter SLC35A3. J. Biol. Chem. 2020;295:16445–16463. doi: 10.1074/jbc.RA119.012362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor S.C., Nadeau K., Abbasi M., Lachance C., Nguyen M., Fenrich J. The ultimate qPCR experiment: producing publication quality, reproducible data the first time. Trends Biotechnol. 2019;37:761–774. doi: 10.1016/j.tibtech.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Nakajima K., Kitazume S., Angata T., Fujinawa R., Ohtsubo K., Miyoshi E., et al. Simultaneous determination of nucleotide sugars with ion-pair reversed-phase HPLC. Glycobiology. 2010;20:865–871. doi: 10.1093/glycob/cwq044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS data have been deposited in the GlycoPost under ID GPST000261. Some unpublished data are available upon request from the corresponding author, e-mail: mariusz.olczak@uwr.edu.pl, on reasonable request.