Abstract
Mitochondria are the critical organelles involved in various cellular functions. Mitochondrial biogenesis is activated by multiple cellular mechanisms which require a synchronous regulation between mitochondrial DNA (mtDNA) and nuclear DNA (nDNA). The mitochondrial DNA copy number (mtDNA-CN) is a proxy indicator for mitochondrial activity, and its alteration reflects mitochondrial biogenesis and function. Despite the precise mechanisms that modulate the amount and composition of mtDNA, which have not been fully elucidated, mtDNA-CN is known to influence numerous cellular pathways that are associated with cancer and as well as multiple other diseases. In addition, the utility of current technology in measuring mtDNA-CN contributes to its extensive assessment of diverse traits and tumorigenesis. The present review provides an overview of mtDNA-CN variations across human cancers and an extensive summary of the existing knowledge on the regulation and machinery of mtDNA-CN. The current information on the advanced methods used for mtDNA-CN assessment is also presented.
Keywords: mitochondrial DNA, mtDNA copy number alterations, mtDNA replication, biomarker, cancer
1. Introduction
Cancer is known as one of the underlying causes of most public health threats and is considered the leading cause of death after stroke and coronary heart disease on a global scale (1). Based on reports from the World Health Organisation (WHO), cancer is the first or second leading cause of cancer-related deaths before the age of 70 years in 112 countries. As reported by GLOBOCAN, there were approximately 19.3 million new cancer cases and 10.0 million cancer-related deaths in 2020, and new cases are estimated to increase to 28.4 million in 2040 (1).
Cancer occurs mainly due to dynamic genetic alterations, and the resultant genome deformities may promote the uncontrolled growth of abnormal cells and spreading throughout the body. It is well accepted that the generation of abnormal cells accumulates mutations in essential genes, leading to cancer cell formation. Accumulated mutations can further increase genomic instability, including rearrangements and chromosomal changes (2,3). Thus, genomic instability is the main element and one of the underlying mechanisms in the initiation and progression of human cancer (4).
The growing interest in the mechanistic understanding of the specific factors in cancer development allows researchers to investigate the role of the second human genome, which is mitochondrial DNA (mtDNA). It is widely known that the role of mitochondria is implicated in tumor promotion and development. In the 1920s, Dr Otto Warburg observed that most cancer cells have an altered metabolism characterized by high glycolytic activity. This condition is referred to as ‘aerobic glycolysis’ and is considered to be linked with a constant diminished mitochondrial oxidative phosphorylation (OXPHOS) (5). Since then, a considerable amount of research has been conducted, which is devoted to the dysfunction of mitochondrial respiration in cancer development (6,7). Previous studies have revealed that cancer cells exhibit a highly glycolytic phenotype and have the capacity to utilize glucose at higher rates than their origin cells (8,9). Moreover, it was observed that cancer cell lines are incapable of adequately upregulating OXPHOS, thus, increasing their dependency on glycolysis (10).
mtDNA is recognized to be highly unstable and liable to damage compared to the nuclear genome, due to the persistent exposure to oxidative attack coupled with deficient DNA repair machinery (11). As a result, mtDNA suffers from the formation of sequence alterations or depletion of mtDNA content, consequently leading to various disorders and cancer tumorigenesis. The levels of mtDNA copy number (mtDNA-CN) may reflect the biogenesis and pathology of mitochondria, which are responsible for the phenotypic changes in malignant tumors. It implies that mtDNA-CN may serve as a potential marker in a broad spectrum of cancers, including breast, lung, colorectal, head and neck, and gastric cancers (10–14).
In the present review, the current knowledge concerning the mtDNA-CN alterations found in human cancer were investigated. Relevant literature was comprehensively searched (between 2004 and 2022) using the online electronic databases: PubMed, Web of Science, Scopus as well as other appropriate resources. The key words searched for included ‘mtDNA-CN alterations’ or ‘mtDNA content’ and ‘cancer’ or ‘neoplasm’. Of the 84 published articles, 76 were included and considered to meet the inclusion criteria of the case-control studies that investigated the association between mtDNA-CN alterations and various types of human cancer. In this review, an update on the roles and mechanisms of mtDNA-CN in tumorigenesis were outlined. In addition, available evidence on the incidence of mtDNA content variations in a wide range of human cancers were provided.
2. mtDNA
Mitochondria are eukaryotic organelles involved in a broad spectrum of cellular processes, including the production of ATP, cellular metabolism, reactive oxygen species (ROS) production, activation of apoptosis, and calcium homeostasis. The generation of mitochondrial energy is entirely dependent on five multi-subunit protein complexes (complex I–V) of the OXPHOS system together with the tricarboxylic acid (TCA) cycle (15). Protein complexes I, III, IV, and V have contributions from both mtDNA and nuclear DNA (nDNA) genes, but complex II is completely nuclear-encoded. Therefore, a proper interaction between both the nucleus and mitochondria is needed to achieve sufficient levels of a functional OXPHOS system.
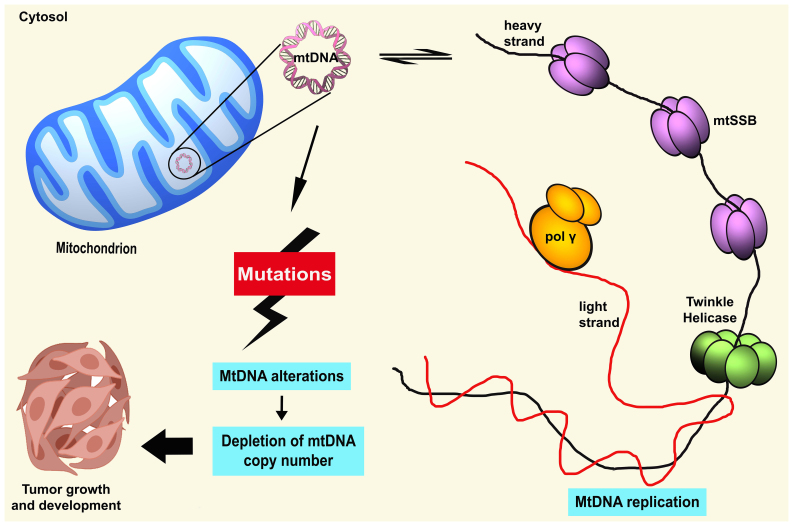
Human mtDNA is maternally inherited due to the higher number of mtDNA copies found in the egg of a female compared to sperm which resides in the mitochondrial matrix (16). mtDNA is organized into nucleoids that allocate across the mitochondrial networks (17). These nucleoids are composed of an accumulation of mtDNA molecules and several elements of protein factors, for instance, mitochondrial transcription factor A (TFAM), mitochondrial single-stranded binding protein (mtSSB), polymerase γ (pol γ), and helicase Twinkle (18). mtDNA is a close, double membrane and circular form of 16,569 nucleotide pairs. mtDNA contains 37 genes, coding for two ribosomal RNAs, 22 transfer RNAs, and 13 elements consisting of seven subunits of complex I (ND 1 to 6 and ND4L), one subunit of complex III (cytochrome b), three subunits of complex IV (COX I, COX II and COX III), and two subunits of complex V (ATPase 6 and ATPase 8) (18). mtDNA contains two distinct strands based on the density differences in caesium chloride gradient (19). The heavy strand (H-strand) has a greater level of GC content and encodes for 12S and 16S rRNAs, 14 tRNAs, and 12 subunits of mitochondrial respiratory complexes. In addition, the light strand (L-strand) encodes eight tRNAs and ND6 (18).
mtDNA carries only two non-coding regions and is almost entirely composed by coding sequences (>90%), and their genes do not present introns. The main non-coding region is termed a displacement loop (D-loop) region covering 1,122 base pairs of the genome (20). A D-loop has all the essential materials for the transcription and replication process: The origin of H-strand replication (OH), the promoters for H- and L-strand transcription (HSP1, HSP2, and LSP), and three conserved sequence blocks (CSB I, CSB II, and CSB III) (18). Therefore, D-loop variations may affect the promoter sequences and consequent changes of protein binding affinities of the inducers or modulators of mtDNA transcription, which could induce replication error. On the other hand, the origin of L-strand replication (OL) appears in the second non-coding region with only 30 nucleotide pairs (18).
3. mtDNA-CN
Regulation of mtDNA-CN
Notably, the mtDNA-CN may represent the amount of mitochondria in a cell. Almost hundreds or thousands of mtDNA copies are available in each cell and vary considerably depending on cell types and tissue origins (21). Of note, mtDNA-CN remains consistent within each cell based on its energy requirement but may markedly change during the aging process, cell differentiation, hormone therapy, and exercise (13). Tissues with high energy demands, such as the brain, and skeletal and cardiac muscles have more mtDNA-CNs than other tissues, as for example, kidney and liver cells (22,23). It has been determined that mtDNA-CN in oocytes reaches up to 100 thousand copies, and recent analyses have shown that there are ~4–6 thousand copies in the heart and 0.5–2×103 copies in the lungs, liver, and kidneys (24,25).
In general, each mitochondrial molecule per cell shares similar mtDNA sequences referred to as homoplasmy. However, due to the highly polymorphic nature of mtDNA, a mixture of wild-type and mutant mtDNA coexist within the same cell, termed heteroplasmy. This discrepancy could be attributed to numerous somatic mtDNA mutations typically caused by oxidative stress, high ROS exposure, replication errors, and aging (26). Heteroplasmy threshold occurs when the frequency of heteroplasmy attains 60–90% of mutant mtDNA and reflects the human mitochondrial pathology (6,27). Conceivably, the heterogeneity of the mtDNA genome may act as an essential participant in mitochondrial dysfunction, which is responsible for the development of various complex diseases.
mtDNA replication
In 1972, Robberson et al studied mammalian mtDNA replication by applying cesium chloride-purified mitochondrial DNA and observed it under an electron microscope (28). With the combination of numerous research studies, three models of mtDNA replication have been proposed, including the asymmetric strand-displacement model, strand-coupled bidirectional replication, and RNA incorporated throughout the lagging strand (RITOLS) (20,29,30). Over the last 40 years, the strand-displacement model has been the central reference in the knowledge of mtDNA replication. Based on this model, the production of a new leading strand (H strand) commences at the OH site within the D-loop region. The synthesis proceeds continuously until 70% (~11 kb) of the mtDNA genome circle, and when the OL origin of a lagging strand (L strand) is exposed, the initiation of the lagging strand occurs in the opposite direction (20).
Although several models of mtDNA replication have been previously proposed, it is critical to highlight that the understanding of the exact mechanism is still under debate and not fully elucidated at present. However, it should be noted that the mtDNA replication process occurs by a molecular machinery that is different from the nuclear replisome. Nonetheless, nDNA is still involved in the regulation of mtDNA-CN since all trans-acting factors engaged in mtDNA replication are encoded by nDNA (31). Mitochondrial replication is implemented by the onset of proteins which is initiated by mitochondrial transcription factor A (TFAM) and aided by mtDNA pol γ, hexameric Twinkle helicase, mtSSB, mitochondrial RNA polymerase (POLRMT), transcription elongation factor (TEFM), transcription factor B2 (TFB2M), exonuclease MGME1, DNA ligase III, and RNAse H1 (27).
Mitochondrial TFAM is a member of the high mobility group (HMG) box domain family and is present at a ratio of 1 molecule per 16–17 base pairs of mtDNA (32). It is considered that TFAM is sufficient to coat the mitochondrial genome, in addition to playing a major role in the initiation of transcription and replication of mtDNA. Furthermore, TFAM regulates protein binding at the cis-regulatory region of the mitochondrial D-loop (31). It has been demonstrated that TFAM plays a critical part in mtDNA-CN regulation and maintenance in vivo (33,34). A previous study revealed that loss of mtDNA was caused by mouse-TFAM deficiency, whereas the elevated mtDNA-CN was driven by the overexpression of TFAM (35). Moreover, the levels of mtDNA molecules needed for the transcription and replication process are dependent on TFAM concentrations available (36). Therefore, the expression level of TFAM was hypothesized to be directly proportional to the level of mtDNA molecules (35).
mtDNA pol γ is the sole DNA polymerase of mitochondria responsible for replicating and repairing the mtDNA genome (32). It has a high proofreading ability to eradicate the misincorporation of DNA bases. mtDNA pol γ also appears to function with Twinkle helicase and mtSSB protein to perform, to some extent, complete mtDNA replication. Korhonen et al discovered that the synthesis of ssDNA molecules during replication is achieved with the cooperation of mtDNA pol γ, Twinkle, and mtSSB (37). Twinkle is a 72-kDa monomer and is greatly homologous to bacteriophage T7 gene 4 primase/helicase (T7gp4) (38). A preliminary study revealed that Twinkle has a 5′ to 3′ DNA helicase activity and unwinds the dsDNA at the mtDNA replication fork (39). Milenkovic et al reported the significance of Twinkle in nascent D-loop strand synthesis during mtDNA replication (40). However, the activity of both mtDNA pol γ and Twinkle is stimulated by mtSSB (39). mtSSB was found to be more abundant in human HeLa cells than mtDNA. mtSSB can support the unwinding activity of Twinkle helicase and improve mtDNA pol γ activity (41,42). The processivity of Twinkle helicase increased when bound to mtSSB and could unwind the duplexes longer than needed for strand separation (43). Furthermore, mtSSB binds to ssDNA to preserve and maintain ssDNA in an active single-stranded state (44). The regulation of mtSSB has a possible mechanistic link with TFAM for D-loop stabilization and mtDNA maintenance (41).
Mutations of regulatory factors involved in mtDNA replication
It has been proven that the accumulation of somatic mutations in critical regions, for instance, in conservative sequences areas, replication sites, promoter, or transcription factor binding sites, can lead to repression of mtDNA gene expression and subsequently damage mitochondrial biosynthesis (45) (Fig. 1). These alterations substantially alter mitochondrial respiration capacity, other than stimulating a higher rate of oxidative stress and apoptosis (45). mtDNA mutations frequently occur within the hypervariable D-loop region, particularly in a homopolycytidine stretch or near the replication origins of the mtDNA H-strand (11). This crucial region possesses a triple-stranded DNA structure, which is severely liable to the persistent effect of oxidative insults, thus, possibly increasing the rate of mutations. It was theorized that aberration in the D-loop could alter the binding affinities of several regulators of nuclear-encoded DNA on the regulatory site of mtDNA (20). Thus, mutations in this particular region may modify mtDNA transcription and subsequently cause disturbances in mtDNA respiration and ROS production (46). The damaged mtDNA is removed by mitophagy in the cell, which causes the reduction of mtDNA-CN (47). Therefore, as suggested, the alterations in the D-loop region could critically affect the regulation of mtDNA at the heteroplasmy level and mtDNA-CN (48).
Figure 1.
Overview of the occurrence of aberrant mtDNA copy number. MtDNA mutations caused by endogenous attack can impair the regulation of mitochondrial replication followed by the aberration of mtDNA copy number level. Due to this, overall mtDNA biogenesis could be affected which may further facilitate tumor progression and development of human cancers. mtDNA, mitochondrial DNA; mtSSB, mitochondrial single-stranded binding protein; pol γ, polymerase γ.
Available literature has revealed that somatic mutations in the D-loop region are markedly associated with a decreased level of mtDNA in hepatocellular carcinoma, invasive breast cancer, and Ewing's sarcoma (49,50). Moreover, decreased mtDNA-CN was correlated with tumors that harbored somatic mutations, which were located close to replication origins of the H-strand or at the homopolymeric C-stretch in the D-loop (50–52). Sequence mutations in the D-loop region, specifically, in the polycytidine stretch caused by continuous oxidative stress, could lead to slippage error and/or mispairing during mtDNA replication or repair by mtDNA pol γ (53). mtDNA pol γ has low frameshift fidelity and is susceptible to mistakes, resulting in decreased proofreading efficiency (54). Defects in mtDNA pol γ can arise from increased oxidative damage, and in turn, this may induce further mtDNA errors. Previously, published studies delineated that transgenic mice with high-level proofreading-deficient pol γ expression induce mitochondrial mutations in cardiac tissues (55). However, the homozygous knock-in mice carrying-expressed proofreading-deficient pol γ developed a mutator phenotype with a higher rate of mutated and deleted mtDNA (56). Thus, the faulty mtDNA pol γ may reflect the biosynthesis of mtDNA, which affects the regulation of mtDNA-CN.
A plethora of evidence has revealed the close association of mtDNA pol γ and the amount of mtDNA-CN in human cancers. Spelbrink et al identified that transient expression of pol γ-myc mutants suppressed mtDNA polymerase activity and was associated with depleted mtDNA in vivo (57). Furthermore, Singh et al revealed that 63% of pol γ mutations in breast cancer tumors led to mtDNA depletion (58). Data from a previous study, indicated that mtDNA-CN was decreased in patients with colorectal cancer, and exhibited somatic mutations in pol γ or germline nucleotide mutations were found in the region encoding pol γ polymerase domain (59). Another previous study revealed that pol γ SNP genotype was markedly associated with the increased level of mtDNA-CN and improved survival among patients with hepatocellular carcinoma (60). Indeed, aberrant mtDNA pol γ is considered to contribute to repair system deficiency, which leads to mtDNA-CN alterations.
The presence of Twinkle helicase is also critical in mtDNA-CN, as the interactions of helicase-primase during mtDNA replication play a part in controlling mtDNA levels in cells. Theoretically, Twinkle has been suggested to be responsible for the switching between abortive and full-length mtDNA replication during the pre-termination events of triple-stranded D-loop structure formation (38). This switching thus determines the regulation of mtDNA levels in the cell. In a preliminary study, Twinkle was identified as the causative gene of autosomal dominant progressive external ophthalmoplegia (adPEO) correlated with several mtDNA deletions (61). To date, >40 point mutations have been detected in the TWNK gene encoding Twinkle, which is associated with multiple diseases (38).
Previous research has touted that the role of Twinkle helicase can be inferred from the regulation of the amount of mtDNA. One study in 2004 reported that overexpression of the Twinkle helicase in transgenic mouse lines led to an increase in mtDNA-CN (62). Twinkle helicase overexpression may also act on anti-helicase blocking, thus permitting increased mtDNA-CN and successful mtDNA replication (40). However, studies of mutant Twinkle variants in vivo demonstrated that mutations of this helicase lead to extreme mtDNA depletion and increased apoptosis in flies (63,64). Therefore, this suggests that Twinkle is the sole replicative helicase for mitochondrial biogenesis.
In general, it appears that the knockout of these regulatory elements in mtDNA replication could affect the mtDNA genome, particularly within the non-coding region. Although any defects or absence of these elements may perturb the regulation of mtDNA copies, overexpression of a protein involved in this machinery does not constantly increase mtDNA-CN. Moreover, mtDNA depletion and secondary multiple deletions/duplications may be the outcomes of the flaws in mtDNA machinery proteins with the consequent mitochondrial dysregulation (38). Thus, common regulation of mtDNA-CN may be limited, thus increasing the inability of mitochondria to maintain normal function, subsequently establishing a background liability of tumor development.
4. Effects of aberrant mtDNA-CN
It is well-recognized that regulation of mtDNA-CN is significant to govern the cellular energy needs. mtDNA-CN was shown to be markedly associated with the level of oxidative insults in the mtDNA genome and involved in ROS production (up to 85%) to modulate apoptosis or cell differentiation (65). Aberrant mtDNA-CN can lead to elevation of mtDNA oxidative stress and disruption of mtDNA gene expressions. Subsequently, this may affect overall mtDNA functions, including the OXPHOS system, ROS production, signal transduction, cell apoptosis, cell growth, and mitochondria-to-nucleus retrograde signaling (66,67). Certainly, disturbances in the OXPHOS system may result in a reduction of intracellular ATP generation (6). Depleted mtDNA causes a decrease in OXPHOS capacity, which triggers a high compensatory increase in glycolysis to cover the total ATP (7). Thus, the aberrant mtDNA-CN can reduce the rate of mitochondrial biogenesis and normal cellular function that eventually confer the emergence of tumors.
Previous findings have described the functional significance of oxidative stress correlated with the mtDNA-CN. Intrinsic oxidative stress induced by noxious ROS was revealed to be closely associated to cell toxicity, DNA injury, copy number changes, and malignant transformation in human cancer cells (68). A previous study on mtDNA-CN alterations conducted in human leukocytes revealed that oxidative stress in blood circulation influences the level of mtDNA-CN via the alterations of plasma antioxidants/oxidants and an oxidative insult to DNA (69). In addition, Al-Kafaji and Golbahar revealed the effects of the glucose-induced oxidative stress and mtDNA-CN in human mesangial cells, in an in vitro study. The results revealed a higher mtDNA-CN in the mesangial cells, as a response following higher oxidative stress stimulated by high glucose activity (70). In addition, mtDNA-CN and oxidative stress are associated with environmental exposure to tobacco, pollutants, smoke, drugs, xenobiotics, and radiation (66).
It is also important to emphasize the functional impact of mtDNA-CN alteration on nDNA instability. Notably, aberrant mtDNA-CN could potentially interrupt mitochondrial membrane potential, contributing to mitochondrial dysfunction (71). As a result, the initiation of retrograde signaling occurs and mitochondria communicate their changing functional and metabolic state to the nucleus. This response causes the impairment of the expression profile of nuclear genes and modifies cell physiology and morphology (71). In 1989, Corral et al found that chemically-induced rat hepatomas and the HT-1080 fibrosarcoma cell line exhibited increased copy numbers of COI, COII, and COIII pseudogenes in nDNA compared with normal cells (72). In addition, it was demonstrated that mtDNA depletion in HeLa cells showed higher lipid peroxidation and oxidative damage to nDNA (73).
Furthermore, it has been shown that aberrant mtDNA-CN is associated with resistance to apoptosis and the promotion of cancer (7). mtDNA-depleted cells were demonstrated to activate the Akt pathway, which could inhibit the apoptosis of cells (74). Furthermore, nuclear factor-κB/Rel signaling, which is involved in apoptosis resistance and cancer development, can be activated by depleted mtDNA content (75,76). By contrast, recent data revealed that mtDNA-CN was increased in apoptotic tumor cell lines, suggesting that increased mtDNA-CN acts as a defense mechanism of tumor cells to prevent apoptosis (77). Decreased mtDNA-CN in tumor cells increased ROS levels, the rate of apoptosis, and sensitivity against chemotherapeutic drugs (77). This finding is in line with a previous study that demonstrated that increased mtDNA-CN induces survival and apoptosis resistance in colorectal cancer cells in vivo and in vitro (78).
Previously, published studies have revealed increased or decreased mtDNA-CN in some tumor entities. Several studies have assessed the quantitative changes of mtDNA-CN in peripheral blood specimens and the resected tumorous tissues compared with non-tumorous tissues (65). Increased mtDNA-CN was observed in breast, bladder, esophageal, head and neck squamous cell, kidney, and liver cancers (except in lung carcinoma) compared with non-tumorous tissues (79). Moreover, decreased mtDNA-CN was revealed in kidney clear cell carcinoma, hepatocellular carcinoma, and myeloproliferative tumors (80). The association of mtDNA-CN variation with the risk of cancer progression has also been reported. A meta-analysis by Hu et al (66) discovered that increased mtDNA-CN was markedly associated with the risk of lymphoma, melanoma and breast cancer, but inversely associated with hepatic carcinoma. This discrepant result of mtDNA-CN in different cancer types implies that mtDNA-CN occurs in a cancer-specific manner, as a consequence of different needs of energy metabolism in human cancer tissues. Notably, knowledge of mtDNA-CN in tumorigenesis remains elusive and requires elucidation.
5. Restoration of mtDNA-CN
The current understanding of mtDNA-CN regulation is still unclear, although evidence of mtDNA replication and maintenance has accumulated and improved over the years. Several theories and models have been proposed to explain mtDNA-CN regulation. It is hypothesized that all the binding proteins involved in mtDNA replication, may be key factors in the maintenance and regulation of mtDNA levels (79). Furthermore, a study performed by Tang et al suggested that the regulation of mtDNA-CN could depend on the availability of nucleotide pools (80). Subsequently, these authors introduced another theory, which proposed that several replication origins may affect the regulation of mtDNA-CN (81). Additionally, different levels of mtDNA-CN (wild-type, duplicated, or deleted mtDNA) were shown to be independent of mtDNA size, indicating that the copy numbers were not controlled by the number of molecules or mtDNA genomes present (81). In 2009, a threshold model for mtDNA-CN control was proposed (31). The model suggested that upregulation of mtDNA replication machinery could be the cause of a lower mtDNA-CN threshold, whereas mtDNA degradation could be the result of a higher mtDNA-CN threshold (31).
mtDNA-CN is important in early development and differentiation, as mature cells need enough copies to meet their specific demands (82). Premature cells exhibit a high level of copy numbers during oogenesis and achieve maximal levels in mature oocytes. Copy number is subsequently decreased during the development of preimplantation before gastrulation (83). A low copy number in these undifferentiated cells indicates the mtDNA set point, which allows these cells to acquire sufficient copies of mtDNA to sustain their energy demands (18). Cells that utilize high ATP consumption, including the heart, muscle, and brain cells contain high mtDNA-CN, while cells that retain low copies such as endothelial cells depend on glycolysis (84).
The mtDNA set point of a cell requires synchrony between nDNA and mtDNA genomes to establish a cooperative process of mtDNA differentiation and replication (84). However, cancer cells that contain depleted mtDNA-CN have to re-establish their mtDNA set point (85). It has been demonstrated that levels of mtDNA-CN influence the frequency and progression of GBM cells. The number and duration of tumor formation were proportional to the mtDNA depletion level. Thus, mtDNA-depleted GBM cells restored their mtDNA-CN for their tumor growth process (84). In addition, mtDNA-CN can be recovered by the mtDNA replication machinery of a cell and mitochondrial horizontal gene transfer from adjacent cells (85,86). The migration of mtDNA horizontal gene transfer from normal cells to tumor cells with deficient mtDNA results in the restoration of respiration, initiation of tumors, and metastasis (87).
It is also plausible that mtDNA variations are implicated in mtDNA-CN restoration. mtDNA with impaired respiration function or carrying mutations deals with this damage by increasing mtDNA-CN in accordance with the proportion of mtDNA-CN in the cell (85). A preliminary study reported that a high mtDNA-CN found in aging tissue cells acts as a feedback mechanism to counterbalance the metabolic injury in mitochondria carrying mutated mtDNA and a defective respiratory chain (88). Additionally, a previous study by Yeung et al revealed that GBM cells harbored the accumulation of mtDNA variants in ND6 and ND4 genes, that encoded the subunits of complex I of the respiratory chain, resulting in impaired mtDNA function and consequently affecting mtDNA levels (89). A more recent study demonstrated a marked increase of mtDNA-CN in samples with high-allele-frequency truncating mutations encoded by nDNA. The authors suggested that the increased mtDNA-CN was necessary to compensate for the damage induced by truncating mutations (90).
6. Assessment of mtDNA-CN
To date, numerous studies have determined the levels of mtDNA-CN in tissues and peripheral blood samples. Notably, peripheral blood has been proposed as a reliable source, due to the easily obtainable and non-invasive approach in estimating mtDNA-CN in human cancer. Moreover, mtDNA-CN alterations in peripheral blood are proposed as a potential indicator, to study mitochondrial function and aerobic metabolism and have been revealed to be significantly correlated with the risk of cancer (91,92). However, since mitochondrial numbers and mtDNA copies in a cell differ within tissue types, it is crucial to use accurate methods in order to prevent any compromised variables in assessing the copy numbers.
Several methods currently exist for the quantification of mtDNA-CN (93–95). One of the most well-known methods for assessing mtDNA-CN in human samples and model organisms is quantitative real-time PCR (qPCR) which assesses the ratio of an mtDNA gene to a reference gene (nuclear genome). Previous studies have used several genes for mtDNA level assessment, including MTATP8, MTND1, MTCOX1, 16S rRNA, and MTCYB for mtDNA, while ACTB, B2M, HGB, and GAPDH have been used as nuclear genomes (10,14,66,96–98). Recently, droplet digital PCR (ddPCR) was developed as a high-efficiency method to measure mtDNA-CN without applying external standards (99,100). The ddPCR utilizes a similar workflow as in qPCR technology, and a sample can be segmented into thousands of oil emulsion droplets (99). The amplification occurs in individual droplets to avoid bias in PCR efficacy and inhibitors (100). However, the restricted efficacy of ddPCR may affect quantification accuracy when measuring a large copy number (100,101).
Other available methods to quantify mtDNA-CN include genotyping microarray probe intensities, whole genome sequencing (WGS), whole exome sequencing (WES), and DNA sequencing read counts (102). It has been shown that WGS data could provide a more sensitive and accurate assessment of mtDNA levels compared with the qPCR method (101,102). The WGS data provide complete genome sequencing reads including mtDNA and nDNA (101), and WGS serves as the only current method which concurrently measures mtDNA-CN and mutations (103).
It is conceivable that methods used in purifying DNA are also critical in assessing mtDNA content. Methods utilized for DNA extraction or isolation may affect the yield of mtDNA integrity due to the circular and compact size of mtDNA (102,104). Any deviation from a specific protocol may have a critical impact on the accuracy of mtDNA-CN estimation. A number of studies with different extraction techniques have been performed to establish a more accurate method for measuring mtDNA-CN (93,95,105–108). The column kit isolation method is often used to extract DNA fragments ≥50 kb and highly depends on chaotropic salts and low pH to induce the DNA binding capacity. This method potentially decreases total DNA, which may increase the bias in the measurement of the DNA ratio (106). Previously, organic solvents containing precipitated ethanol were demonstrated to be more accurate compared to column-based methods (106). The organic solvent method is time-consuming and mostly relies on technical skill, however, it offers a high DNA yield (106). Furthermore, a method with less usage of spin columns or without spin columns may be able to diminish GC-dependent bias (109). Recently, it was revealed that the lysis-based method has low variability and is more accurate than other traditional methods. Cells are directly lysed without any isolation or spin columns, ensuring minimal hands-on time in addition to reducing biased mtDNA-CN estimation (102).
7. Involvement of mtDNA-CN in human cancers
With the numerous types of cancers that have been assessed for mtDNA content alterations, it is necessary to examine the molecular mechanism of mtDNA-CN occurrence in carcinogenesis. With this aim, an extensive search in Google Scholar, PubMed, online databases, and published journal articles was performed, to summarize the involvement of mtDNA-CN alterations in various types of cancer (Table I).
Table I.
Distribution of mtDNA copy number levels in selected cancer types of different countries.
| Type of cancer | No. of samples | Laboratory methods | MthNA gene | Nuclear gene | MthNA levels | Country | (Refs.) |
|---|---|---|---|---|---|---|---|
| Breast | 302 | qPCR | tRNALeu | B2M | Decreased | USA | (52) |
| 59 | qPCR | D˗loop | ACTB | Decreased | China | (51) | |
| 102 | qPCR | MTATP8 | GADPH | Decreased | Switzerland | (108) | |
| 183 | qPCR | MT˗ND1 | 18S RNA | Increased | Singapore | (113) | |
| 103 | qPCR | MT˗ND1 | HGB | Increased | USA | (92) | |
| 1000 | qPCR | MT˗ND1 | HGB | Increased | USA | (96) | |
| 1,108 | qPCR | MT˗ND1 | ALB | Increased | UK | (112) | |
| 506 | qPCR | mtDNA | B2M | Increased | China | (91) | |
| 570 | qPCR | MT˗ND1 | ACTB | N/A | EPIC | (12) | |
| 82 | qPCR | tRNALeu | 18S rRNA | Decreased | Mexico | (111) | |
| 60 | qPCR | MT˗ND1 | ACTB | Decreased | Taiwan | (107) | |
| 60 | qPCR | MT˗ND1 | ACTB | Decreased | Taiwan | (110) | |
| Colorectal | 60 | qPCR | MT˗ND1 | ACTB | Decreased | China | (121) |
| 444 | qPCR | MT˗ND1 | BRCA1 | Decreased | China | (122) | |
| 736 | qPCR | MT˗ND2 | FASLG | Decreased | Canada | (120) | |
| 74 | qPCR | D˗loop | B2M | Decreased | Netherlands | (123) | |
| 422 | qPCR | MT˗ND1 | 18S rRNA | Increased | Singapore | (117) | |
| 320 | qPCR | MT˗ND1 | HGB | Increased | China | (116) | |
| 126 | qPCR | 16S rRNA | B2M | Increased | North India | (14) | |
| 24 | qPCR | mtDNA | ACTB | Increased | China | (114) | |
| 104 | qPCR | COXI | ACTB | Increased | China | (115) | |
| 324 | qPCR | MT˗ND2 | AluYb8 | Decreased | USA | (119) | |
| Gliomas | 28 | qPCR | MT˗ND2 | FALSG | Increased | Italy | (124) |
| 124 | qPCR | MT˗ND1 | ACTB | Increased | China | (122) | |
| 336 | qPCR | MT˗ND1 | HGB | Increased | China | (127) | |
| 390 | qPCR | MT˗ND1 | HGB | Increased | USA | (126) | |
| 35 | qPCR | D˗loop & COXII | ACTB | Decreased | Australia | (131) | |
| 162 | qPCR | MT˗ND1 | RNase P | Decreased | India | (132) | |
| 67 | qPCR | N/A | N/A | Increased | France | (128) | |
| 414 | qPCR | MT˗ND1 | ACTB | Increased | China | (129) | |
| Gastric | 76 | qPCR | mtDNA | ACTB | Decreased | China | (137) |
| 20 | PAGE | D˗loop | ACTB | Decreased | China | (133) | |
| 31 | qPCR | MT˗ND1 | ACTB | Decreased | Taiwan | (134) | |
| 109 | qPCR | COXI | ACTB | Increased | Korea | (67) | |
| 103 | qPCR | MT˗ND1 | ACTB | Decreased | China | (135) | |
| 984 | qPCR | MT˗ND1 | HGB | Increased | China | (16) | |
| 162 | qPCR | mtDNA | HBB | Decreased | China | (136) | |
| 109 | qPCR | COXI | HBB | Decreased | Korea | (98) | |
| Prostate | 9 | qPCR | MT˗ND1 | HBB | Increased | USA | (138) |
| 196 | qPCR | MT˗ND1 | HGB | Decreased | USA | (139) | |
| 102 | qPCR | MT˗ND1 | HBB | Increased | India | (140) | |
| 793 | qPCR | MT˗ND1 | HBB | Increased | USA | (141) | |
| 1,751 | qPCR | MT˗ND1 | HGB | Decreased | USA | (142) | |
| 46 | qPCR | MT˗ND1 | B2M | Decreased | Australia | (143) | |
| 317 | qPCR | MT˗ND1 | HGB | Decreased | USA | (144) | |
| Esophageal | 20 | qPCR | mtDNA | B2M | Increased/Decreased | USA | (145) |
| 42 | qPCR | N/A | N/A | Increased | China | (146) | |
| 72 | qPCR | MT˗ND1 | 18S rRNA | Increased | Taiwan | (147) | |
| 80 | qPCR | COXI | COXIV | Increased | Japan | (149) | |
| 141 | qPCR | MT˗ND1 | ACTB | Increased | China | (148) | |
| Lung | 29 | qPCR | MT˗ND1 | 18S rRNA | Decreased | Taiwan | (65) |
| 122 | qPCR | MT˗ND1 | HBB | Increased | China | (156) | |
| 874 | qPCR | MT˗ND1 | HBB | N/A | China | (158) | |
| 128 | qPCR | MT˗ND1 | 36B4 | Decreased | China | (155) | |
| 227 | qPCR | MT˗ND1 | HBB | Increased | Finland | (157) | |
| 37 | qPCR | MT˗HVI | HBB | Decreased | China | (13) | |
| Renal cell | 37 | Southern blot | mtDNA | 18S rRNA | Decreased | Austria | (159) |
| 375 | qPCR | MT˗ND1 | HGB | Decreased | USA | (160) | |
| 1,217 | qPCR | MT˗ND1 | HBB | Decreased | USA | (161) | |
| 252 | qPCR | MT˗ND1 | HBB | Increased | USA | (163) | |
| 5 | qPCR | tRNALeu | 18S rRNA | Decreased | Taiwan | (162) | |
| 57 | qPCR | MT˗ND1 | HBB | Increased | Egypt | (164) | |
| Head and neck | 76 | qPCR | COXI & II | ACTB | Decreased | USA | (152) |
| 91 | qPCR | COXI | ACTB | Increased | USA | (151) | |
| 75 | qPCR | tRNALeu | 18S rRNA | Increased | Taiwan | (153) | |
| 570 | qPCR | MT˗ND1 | HGB | Increased/Decreased | China | (95) | |
| 50 | qPCR | D˗loop | GADPH | Increased | India | (154) | |
| Pancreatic | 43 | qPCR | MT˗ND1 | SLCO1B1 | Decreased | Poland | (172) |
| 406 | qPCR | MT˗ND1 | HBB | Increased | Taiwan | (171) | |
| 476 | qPCR | MT˗ND1 | ALB | Decreased | EPIC | (173) | |
| Endometrial | 20 | qPCR | MT˗ND1 | ACTB | Increased | Italy | (166) |
| 65 | qPCR | MT˗ND1 | HBB | Increased | China | (165) | |
| 139 | qPCR | MT˗ND1 | HGB | Decreased | USA | (167) | |
| Oral | 35 | qPCR | COXI & II | ACTB | Decreased | Japan | (170) |
| 124 | qPCR | D˗loop | GADPH | Decreased | India | (168) | |
| 143 | qPCR | MT˗ND1 | HGB | Increased | USA | (169) | |
| Ovarian | 42 | qPCR | MT˗ND1 | HBB | Increased | China | (174) |
| SLCO2B1/SERPINA | |||||||
| 24 | qPCR | MT˗ND1/ND5 | 1 | Increased | Hungary | (175) | |
| Melanoma | 136 | qPCR | MT˗ND1 | HBB | Increased | USA | (176) |
| 500 | qPCR | MT˗ND1 | HGB | Increased | USA | (177) | |
| Laryngeal | 40 | qPCR | COXII | ACTB | Increased | China | (179) |
| 204 | qPCR | MT˗ND1 | ACTB | Increased | China | (178) |
ACTB, β-actin; ALB, albumin; B2M, β-2-microglobulin; COXI, mitochondrially encoded cytochrome c oxidase I; COXII, mitochondrially encoded cytochrome c oxidase II; D-loop, displacement loop; EPIC, European Prospective Investigation into Cancer and Nutrition; FASLG, Fas ligand; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HBB, hemoglobin subunit β; HGB, human globulin; MTATP8, mitochondrially encoded ATP synthase membrane subunit 8; MT-HVI, mitochondrial hypervariable loop-1; MT-ND1, mitochondrially encoded NADH dehydrogenase 1; MT-ND2, mitochondrially encoded NADH dehydrogenase 2; nCOXIV, nuclear DNA-coded cytochrome c oxidase IV; RNase P, Ribonuclease P; SERPINA1, serpin family A member 1; SLCO1B1, solute carrier organic anion transporter family member 1B1; SLCO2B1, solute carrier organic anion transporter family member 2B1; tRNALeu, tRNA leucine; 16S rRNA, 16S ribosomal RNA; 18S rRNA, 18S ribosomal RNA; N/A, not applicable.
Breast cancer
Several experimental approaches investigating mtDNA-CN in patients with breast cancer have been aggressively performed that demonstrated a varying spectrum of mtDNA alterations. To date, most of the analyzed patients who suffered from breast cancer exhibited a significant change in mtDNA-CN, which was positively correlated with the risk of breast cancer.
The alterations of mtDNA-CN in breast cancer were first reported in 2006, and the results revealed decreased mtDNA-CN in 38 breast cancer samples compared with the paired non-tumorous tissues (107). According to an analysis of mtDNA-CN in 59 paired breast tumor tissues and non-tumorous tissues in 2007, the mtDNA-CN level appeared to be lower in tumor tissues, advanced age, and advanced tumor stage (51). The authors also observed that tumors harboring mutations that occurred in the D-loop region had less mtDNA-CN. Consistent with this finding, the studies by Bai et al and Jiang et al, also observed that somatic mutations that occurred in that area may facilitate the reduction of the level of mtDNA in breast tumorigenesis (52,91). Moreover, decreased mtDNA-CN was found in 82% of invasive breast tissue samples compared with the normal counterparts from a total of 102 tumor tissue samples (108).
In 2010, Hsu et al investigated the depleted mtDNA content in breast cancer in response to anthracycline treatment in vivo and vitro (110). The results revealed that decreased disease-free survival of patients was associated with higher mtDNA content compared with patients with decreased mtDNA content. These authors also demonstrated that mtDNA-depleted breast cancer cells had greater sensitivity to doxorubicin treatment and higher ROS production. These findings indicate that the level of copies in mtDNA may serve as a valuable biomarker in predicting response to anthracycline treatment in breast cancer. A more recent analysis reported that mtDNA-CN was markedly decreased in breast tumor tissue samples among 82 tumor cases (111). The results also revealed that mtDNA-CN was decreased in the sequences with three deletions at A249del, A290del, and A291del or C16327T, while the copy number was increased in sequences containing C16111T, G16319A, or T16362C (111).
Notably, Shen et al examined the mtDNA-CN in connection to certain endogenous antioxidants and oxidants, using peripheral blood specimens (92). The study concluded that an increase in the level of mtDNA content was associated with the development of breast cancer. However, mtDNA-CN was inversely associated with changes in antioxidant and oxidant status. In a subsequent study, these authors also revealed that higher mtDNA content was associated with a higher risk of breast cancer, in addition to the presence of mitochondrial length heteroplasmy in hypervariable I (HV1) and hypervariable II (HV2) regions (96). Thus, it was hypothesized that the appearance of HV1 and HV2 length heteroplasmy may be involved in the initiation and promotion of breast cancer.
Furthermore, a comprehensive study of mtDNA-CN in peripheral blood cells by Lemnrau et al provided evidence that an increased level in mtDNA-CN was shown to be associated with the increased risk of breast cancer (112). This study also claimed that the results revealed a stronger association with the risk of breast cancer as mtDNA-CN was more accurately measured using prospectively collected blood specimens and a large sample size. These data are in line with a study by Thyagarajan et al which revealed that the increase of mtDNA-CN in peripheral blood samples was associated with a higher risk of breast cancer (113). In another study, the assessment of leukocyte telomere length (LTL) and mtDNA-CN were investigated in 570 breast tumor cases and 538 controls from the EPIC cohort, using real-time qPCR analysis (12). The study, with collected samples obtained 15 years apart, revealed a link between telomere length and mtDNA-CN. Additionally, it was observed that longer LTL was strongly associated with an increased risk of breast cancer, while mtDNA content was not found to be associated with the risk of breast cancer.
Colorectal cancer
mtDNA content was assessed in 24 patients with colon cancer and 20 patients with rectal cancer by Feng et al (114). This study revealed a significant increase in mtDNA-CN changes associated with tumor stage, mainly in stages I and II, suggesting that mtDNA-CN is involved in the early progression of colorectal cancer (114). Moreover, an analysis of mtDNA 4,977 bp deletion and mtDNA-CN was conducted in the same year, and the results revealed that increased mtDNA-CN was associated with the levels of the 4,977 bp deletion and with tumor stage (115). In a different study, Qu et al demonstrated that higher mtDNA content was found in patients with colorectal cancer than in the paired controls, and was shown to be markedly associated with higher risk of colorectal cancer, similar to a study by Kumar et al (14,116). However, Thyagarajan et al observed a U-shaped association of mtDNA-CN and colorectal cancer risk among Singaporean Chinese patients, indicating that patients with increased and decreased mtDNA-CNs were at increased risk for colorectal cancer (117).
A 2017 study by Tong et al investigated the effect of D-loop demethylation on mtDNA content using five colorectal cancer cell lines (118). In this study, it was determined that a DNA hypomethylating agent caused an increase of mtDNA-CN and alterations in the cell biology of colorectal cancer (118). Furthermore, Sun et al reported that increased mtDNA-CN was correlated with the regulation of the survival and metastasis of microsatellite-stable colorectal cancer cells (78). Recently, a study of mtDNA-CN using blood specimens from 324 female patients and 658 paired controls was performed by Yang et al. In this study, it was demonstrated that mtDNA-CN was inversely associated with the risk of colorectal cancer in a dose-dependent manner (119). In addition, previous research identified the mtDNA-CN increase and reduction (39.6 and 60.4%) in colorectal tissues compared with the non-cancerous rectum or colon tissues, respectively (120). This study also revealed that there was no statistically significant association between the mtDNA-CN variable with the overall survival and disease-free survival of the patients.
Several studies on colorectal cancer with reduced mtDNA-CNs have also been published. As demonstrated by Cui et al, mtDNA-CN was shown to be lower in colorectal cancer tissues than its corresponding counterparts (121). Moreover, Huang et al also observed lower mtDNA-CNs in blood specimens of female patients who suffered from colorectal cancer than in those of controls (122). In addition, a subsequent study revealed a significantly decreased mtDNA-CN in tumorous tissue than in adjacent tissue (123). The authors also revealed that the mtDNA-CN was markedly lower in mutated BRAF and microsatellite instability (MSI) tumors but increased in KRAS mutated tumors. These findings suggest that the mtDNA-CN plays a significant role in tumorigenesis.
Gliomas
In 2013, Marucci et al observed the correlation between mtDNA content with morphology and survival using an immunohistochemical method in a group of patients with glioblastoma (GBM) (124). The findings revealed that 10 cases exhibited oncocytic changes, and nine of these cases had markedly increased mtDNA content compared with control tissues. In the same year, Dickinson et al determined whether GBM cells can regulate mtDNA-CN and chromosomal gene expression during differentiation compared with human neural stem cells (hNSCs) (84). The results revealed that GBM cells did not upregulate mtDNA-CN, respiratory capacity, and the expression of nuclear-encoded mtDNA replication factors during differentiation compared with hNSCs. It was also determined that tumors originating from mtDNA-depleted GBM cells retrieved mtDNA-CNs for tumor development, indicating that mtDNA may play a crucial role in tumor progression (84).
An increased mtDNA content was also reported in a case-control study of patients with glioma using peripheral blood lymphocytes (PBLs) (125). This study also revealed that increased mtDNA content in PBLs was associated with the risk of glioma. This data is also in agreement with a previous study by Shen et al that observed a higher mtDNA-CN in glioma cases compared with control subjects and was significantly associated with glioma risk (126). Chen et al reported that increased mtDNA-CN was markedly correlated with a worse prognosis in patients with glioma (127). However, two separate studies demonstrated that increased mtDNA was associated with an improved prognosis in patients with GBM (128,129).
Conversely, a previous study revealed decreased mtDNA-CN in temozolomide-resistant glioma cells (130). More recently, Shen et al analyzed mtDNA alterations as a therapeutic method in pediatric patients with high-grade gliomas. In this study, the reduction of mtDNA-CN in glioma cases compared with normal brains, was reported (131). Furthermore, Sravya et al observed that decreased mtDNA-CN was associated with worse prognosis and treatment resistance in GBM (132).
Gastric cancer
The incidence of mtDNA-CN in gastric cancer was originally observed by Li et al in 2004 (133). In this study, a decreased mtDNA-CN was observed in 20 cases of gastric cancer compared with matched paracancerous tissues using the polyacrylamide gel electrophoresis method (133). Subsequently, Wu et al reported a significantly decreased mtDNA-CN in 17 out of 31 cases of gastric cancer (134). In another study, Zhang et al determined that most patients with gastric cancer had a low mtDNA-CN compared with the non-tumorous gastric group (135). Moreover, it was found that mtDNA content variation was involved in cancer-related deaths in patients with advanced gastric cancer, as well as the risk of lymph node metastasis.
A previous study revealed that leukocyte mtDNA-CN was not associated with the risk of gastric cancer (136). Nevertheless, the authors noted a possible early disease potential with a low level of mtDNA-CN in gastric cancer. Furthermore, an analysis of D-loop demethylation in association with mtDNA-CN was conducted in 76 gastric cancer and the respective non-cancerous stomach tissues (137). The results revealed markedly decreased mtDNA-CN in cancer tissues, specifically in advanced stages, suggesting this mtDNA-CN decrease as a late molecular event during gastric cancer development. However, mtDNA-CN was shown to be increased in gastric cells following demethylation treatment. Thus, it was inferred that demethylation in the D-loop region may act as one of the factors that affect the relative mtDNA content level in gastric cancer (137).
Conversely, Lee et al observed increased mtDNA content in 64.2% of gastric cancer tissues compared with non-tumorous tissues (67). Another previous study also demonstrated increased mtDNA content in gastric cancer compared with the control group and suggested that mtDNA content and relative telomere length were the independent factors for predicting the risk of gastric cancer (16). Additionally, Jung et al analyzed the correlation between telomere length and mtDNA-CN. The authors determined that telomere length was positively associated with mtDNA-CN in gastric cancer tissues and corresponding non-tumorous tissues (98).
Prostate cancer
A higher proportion of mtDNA-CNs in prostate cancer has been reported in some previous studies. Based on research conducted by Mizumachi et al, an increased mtDNA-CN (78%) was found in prostate cancer tissue compared with adjacent normal tissues (138). Similarly, a study by Zhou et al revealed that patients who suffered from prostate cancer exhibited a significantly increased mtDNA-CN compared with healthy subjects, which was associated with a higher risk of prostate cancer and advanced tumor stage (139). In 2017, Abhishek et al analyzed the relationship between cadmium, zinc, and mtDNA-CN with the clinopathological characteristics of patients with prostate cancer (140). In this study, it was revealed that higher mtDNA-CN, as well as zinc and cadmium compounds were correlated with Gleason scores in prostate cancer cases compared with normal controls (140). In another study, the authors observed that a high mtDNA-CN was associated with and increased risk of non-aggressive prostate cancer (141).
A decrease in mtDNA-CN was identified in the peripheral blood of patients with prostate cancer (142). The study demonstrated that low mtDNA-CN was correlated with aggressive prostate cancer, which indicated poor progression-free survival among the patients (142). In addition, a previous study performed by Kalsbeek et al demonstrated a significantly decreased mtDNA-CN in prostate tumor tissue compared with non-tumorous tissue (143). Recently, Xu et al revealed that patients with prostate cancer who exhibited reduced mtDNA-CN were associated with a lower risk of biochemical recurrence compared with those who harbored increased mtDNA-CN (144).
Esophageal cancer
The first report of mtDNA-CN in esophageal cancer was described by Tan et al (145). In this study, high and low mtDNA-CNs were found in esophageal tumor tissue, and no correlation was established between mtDNA-CN and mutations (145). The following year, a study conducted by Liu et al examined mtDNA content in 42 tissue samples and discovered an increased mtDNA-CN in esophageal cancer tissues (146). In 2010, Lin et al observed a gradual increase of mtDNA-CN among cigarette smokers and wine drinkers in patients with esophageal squamous cell carcinoma (147). In a previous study, Li et al observed an elevated mtDNA-CN in patients with esophageal squamous cell carcinoma compared with control subjects, and this increase in mtDNA-CN was significantly associated with cancer-associated mortality risk (148).
Relatively few studies, by contrast, have revealed a low mtDNA-CN in esophageal tumor cases compared with controls. In a previous study, Masuike et al identified a reduced mtDNA-CN (56.0%) in resected tumorous samples compared with non-tumorous specimens (149). Decreased mtDNA-CN was shown to be correlated with the depth of tumor invasion and tumor stage (149). In this study, it was also revealed that patients with reduced mtDNA-CNs had a significantly poor 5-year overall survival compared with patients with increased mtDNA-CNs. Moreover, a study on mtDNA-CNs in esophageal adenocarcinoma was carried out using peripheral blood leukocytes of 218 esophageal adenocarcinoma cases and the corresponding control samples (150). The results demonstrated a significantly decreased mtDNA-CN in tumor cases compared with the controls, this decrease in mtDNA-CN was markedly associated with a high risk of esophageal adenocarcinoma (150).
Head and neck cancer
mtDNA-CN alterations in head and neck cancer was revealed in a study by Kim et al (151). In this study an evident increase in mtDNA-CN was demonstrated from normal tissue to malignant head and neck tumors. The study also identified that a markedly elevated mtDNA-CN was associated with tumor grade. By contrast, another study revealed a decreased mtDNA-CN in postoperative salivary rinse samples of head and neck cancer (152). Reduced mtDNA-CN was associated with never-smoker status and in response to post-treatment radiation therapy after the first surgery (152).
A case-control study of head and neck cancer of Taiwanese patients showed a two-fold increase of mtDNA-CN, compared with the control cohort (153). It was also identified that patients with increased mtDNA-CN and advanced cancer stage were associated with a higher mortality rate (153). Similar outcomes were also reported by Kumar et al, who analyzed the association of mtDNA-CNs with smoke and smokeless tobacco, betel quid chewing, and alcohol-using cell-free mtDNA samples (154). The findings revealed the increased mtDNA-CN in patients with head and neck squamous cell carcinoma with these observed habits compared with the healthy control cohort. A different study that involved 570 head and neck squamous cell carcinoma cases and 597 controls among the Chinese population revealed an increase of mtDNA-CN in patients with head and neck squamous cell carcinoma compared with the control group (95). Moreover, the authors also found a U-shaped association of mtDNA-CN and the risk of cancer, which suggested the importance of mtDNA-CN in the progression of head and neck cancer.
Lung cancer
A number of studies have investigated the association between mtDNA-CN and lung cancer progression. In a previous study, Lin et al found decreased mtDNA-CN and low levels of oxidative mtDNA damage in lung tumor tissues after chemotherapy (65). Moreover, another study revealed that lower mtDNA-CN was associated with the late stage of lung cancer, which harbored a poorer prognosis (155). In a separate study, mitochondrial MSI (mtMSI) and mtDNA-CN were examined in 37 lung carcinoma tissues and paired with non-cancerous tissue specimens (13). This study demonstrated that the mtDNA-CN in tumor tissue was lower than that in non-cancerous tissue, and the mtDNA-CN in tumor tissue harboring mtMSI was significantly reduced compared with the other lung carcinoma specimens (13).
In 2009, Bonner et al determined that mtDNA-CN was associated with a higher risk of lung cancer among older patients (156). Furthermore, Hosgood et al revealed that mtDNA-CN was markedly associated with age and observed a dose-dependent association between mtDNA-CN and increased lung cancer risk in heavy smokers (157). In a pooled analysis of three study populations, there was no association between mtDNA-CN and the risk of lung cancer (158).
Renal cell cancer
In 2004, Meierhofer et al analyzed mtDNA-CN changes in 37 patients with renal cell cancer using Southern blot analysis. The results of this study revealed a decreased mtDNA-CN in 92% of renal carcinoma tissues compared with the non-cancerous tissues (159). Previous studies also showed a reduced mtDNA-CN in resected tissues of renal cell cancer compared with normal tissues. These studies identified a significant association between mtDNA-CN and the high risk of renal cell cancer (160,161). Moreover, Xing et al determined a high heritability of mtDNA content (65%) in this cancer, suggesting a vital role of mtDNA-CN in renal cell cancer development (160). A study by Lin et al revealed that lower mtDNA-CN with the addition of reduced mitochondrial enzyme activity may be involved in a drug-resistance phenotype and the progression of renal cell cancer (162). However, mtDNA-CN was revealed to be higher in peripheral blood samples of renal cell carcinoma patients (163,164). These studies also revealed a significant association between the increased levels of mtDNA-CN and the increased risk of renal cell cancer (163,164).
Endometrial cancer
In a study by Wang et al mtDNA-CNs were examined in 65 endometrial cancer and 41 non-cancer cases. In this study, a significant increase in mtDNA-CN in patients with endometrial cancer compared with the non-cancer subjects, was reported. In addition, it was discovered that patients who exhibited mtMSI at nucleotide position 303 carried a markedly higher level of mtDNA-CN (165). In addition, a previous study revealed increased mtDNA-CN and citrate synthase activity in endometrial cancer cases compared with those in the healthy group (166).
By contrast, a cancer-based study which included 139 peripheral blood samples of patients with endometrial cancer and 139 paired controls conducted in 2016 discovered decreasing numbers of copies of mtDNA in the endometrial cancer cases. The authors claimed that reduced mtDNA-CN had significant combined effects with smoking status, obesity, hypertension, and diabetes history in the increased risk of endometrial cancer (167).
Oral cancer
Studies of mtDNA-CNs in patients with oral cancer have also been performed. In a study by Mondal et al, mtDNA-CN was examined in 124 patients with oral cancer and 140 subjects as controls (168). In this study, mtDNA-CN was significant in tobacco-betel quid chewers than tobacco-betel quid non chewers, while reduced mtDNA-CN was markedly associated with higher tumor stage (168).
Conversely, a study in 2014 revealed that individuals with elevated mtDNA-CNs were markedly associated with an increased risk of oral cancer compared with individuals with a decreased mtDNA-CN (169). Furthermore, another previous study that involved real-time PCR and immunohistochemistry revealed lower PGC-1α/TFAM expression and mtDNA-CN in oral tumorous tissues compared with the corresponding non-tumorous tissues. The authors suggest that the reduced mitochondrial PGC-1α/TFAM pathway is involved in oral cancer (170).
Pancreatic cancer
According to a study by Lynch et al, elevated mtDNA-CN was found in patients with pancreatic cancer compared with the controls (171). The findings also revealed a significant association between increased mtDNA-CN and the increased risk of pancreatic cancer (171). By contrast, another study performed by Tuchalska-Czuroń et al demonstrated a markedly lower mtDNA-CN in tumor tissues compared with corresponding normal pancreatic tissues (172). However, the study failed to identify a significant difference in the overall survival of the patients, which indicated that mtDNA-CN was not a prognostic indicator of pancreatic cancer (172). This result is in line with a more recent study which revealed lower mtDNA-CN associated with older age and high body mass index among EPIC patients (173).
Ovarian cancer
There are relatively few studies available on the assessment of mtDNA-CN in human ovarian cancer. The earliest report of mtDNA-CN in ovarian cancer was described by Wang et al in 2006 (174). In this study, an increased mtDNA-CN was observed in tumorous tissues of ovarian cancer compared with non-tumorous tissues. It was also observed that the early stages of cancer harbored markedly higher mtDNA copies compared with the late stages (174). In a relatively recent study, a group of researchers performed mtDNA-CN assessment in blood and plasma samples from 24 ovarian cancer patients and matched samples (175). The results revealed higher mtDNA copies in patients with advanced cancer stages than in healthy subjects. In addition, the authors detected elevated levels of mtDNA-CN in exosomes as well as in plasma, and peripheral blood of patients with advanced-stage ovarian cancer, indicating that mtDNA-CN varies depending on the needs of cell types and physiological conditions (175).
Other cancers
Studies of the mtDNA-CN alterations have also been documented previously in several other types of cancers. In a study by Hyland et al the relationship between mtDNA-CN and the risk of melanoma cancer in blood samples was investigated using quantitative PCR. In this study, it was observed that increased mtDNA-CN in melanoma cases were associated with CDKN2A mutations (176). This data is consistent with another study that demonstrated a higher proportion of mtDNA copies in melanoma cases compared with controls, and that a high number of mtDNA copies was significantly correlated with a higher risk melanoma (177).
Furthermore, elevated mtDNA-CN has been reported in laryngeal cancer cases (178,179). Guo et al determined mtDNA-CN in 40 resected tissue specimens of laryngeal cancer and matched blood controls. The results showed increased mtDNA copies in tumor tissues compared with the peripheral blood controls. Additionally, patients that exhibited D-loop mutations carried a higher mtDNA-CN, indicating significant effects of unstable mtDNA D-loop region with mutational loads and polymorphisms in larynx tumorigenesis (179). A separate study also revealed a greater mtDNA-CN in paraffin-embedded tissues of laryngeal cancer than in non-tumorous tissues (178). In addition, it was also revealed that reduced mtDNA copies were markedly correlated with smoking status, tumor invasion, and tumor stage (178).
8. Future approaches in determining mtDNA-CN changes
mtDNA-CN has been proposed as a reliable biomarker in predicting cancer prognosis. It is worth mentioning that control groups, solid tumors, and other diseases can be distinguished by the level of mtDNA-CN (100). To date, there are still no standardized methods used for the relative assessment of mtDNA-CNs in clinical settings.
Certainly, various quantification methods or pre-analytical and analytical factors can influence final estimation and produce varying data of mtDNA content among laboratories (180). In this review, the variations of mtDNA-CN were presented in tumorous tissues compared with non-tumorous tissue specimens, or in body fluids, including circulating blood cells, saliva, and cell-free serum. Since tissues and organs are not easily accessible, body fluids have been frequently used in numerous studies as indicators in determining mtDNA content.
Notably, various DNA extraction methods produce conflicting results, which lead to incorrect copy number assessment. Extraction methods can influence the yield of total mtDNA, and the failure to dilute genomic DNA appropriately due to the varying genome sizes of the nuclear and mitochondrial genome may affect the mtDNA-CN values (181). Therefore, new methods are essential for measuring mtDNA integrity, since with the available methods there is difficulty in differentiating between intact mtDNA and damaged mtDNA fragments. Currently, mtDNA-CN can be readily measured from cell lysates, which have less downstream utility and are advantageous for small samples, except that they are restricted only to cultured cells (102). However, the first quantification of mtDNA-CN based on dried blot spot samples has been developed, and has shown promising results compared to conventional samples (182). The study demonstrated that the average mtDNA-CN was markedly higher in dried blot spots than in the whole blood specimens collected 5 to 10 years apart. A dried blood spot sample may be a viable alternative as it is a more stable, less costly, and less invasive method. It is easy to transport and available for long-term storage at room temperature (182). With all the features mentioned, a dried blood spot sample is optimal for large-scale studies of multiple research fields. However, future additional research is still warranted in assessing mtDNA-CNs with the usage of dried blood spot specimens.
Furthermore, the qPCR-based method serves as the current gold standard in mtDNA-CN quantification and has been extensively used in published findings. Generally, the use of both targeted mtDNA and nDNA genes and external standards may vary among laboratories (100). This may lead to diverse data outcomes, and a challenge in standardizing methods for clinical setup. To date, there is no gold standard for reference genes as they are usually selected based on sample use and diseases, with numerous analyses mostly using the repetitive and altered regions (100). Notably, the nuclear genome can duplicate >97% of the mtDNA genome, giving rise to pseudogenes which are nuclear insertions of mitochondrial origin (183). The mtDNA genes used in the mtDNA-CN measurement may be capable of co-amplifying the nuclear pseudogenes (181). In addition, mtDNA genome duplication is inconsistent among individuals and diseases, hence, it is difficult to recognize the regions in the mtDNA genome which were not duplicated in the nuclear genome (100).
Presently, next-gene sequencing methods, including WES and WGS quantification, provide great effectiveness and comprehensive assessment of mtDNA changes. These methods may provide better results of mtDNA-CN than the gold standard qPCR method as they could detect changes in various samples, including paraffin-embedded tissues, formalin-fixed specimens, and body fluids (103). A recent study optimized the detection of circulating cell-free mtDNA in patients with cancer using a low-depth WGS approach, which revealed a markedly decreased mtDNA-CN in plasma DNA samples compared with fresh tumor tissues (184). Plasma samples contain a very low abundance of mtDNA, and therefore, the usage of qPCR is not applicable for mtDNA-CN estimation in plasma samples due to its low quality (184). Thus, the advent of a capture-based NGS method such as WGS is advantageous for mtDNA content detection using challenging samples. As suggested by a previous study, the NGS approach is anticipated to become a gold standard in measuring mtDNA-CNs in the future (101).
Nonetheless, standardization of all parameters involved in assessing mtDNA-CN such as sample preservation, isolation methods, PCR inhibitors, and quantification methods is significant to avoid bias and false assessment. While these variables are normalized, reference values are crucial for each measurement process, and research findings need to be compared carefully (180). Thus, a thorough appraisal of various approaches and factors involved in mtDNA-CN assessment has yet to be completed (102).
9. Conclusion
In conclusion, emerging evidence suggests that variations in mtDNA-CNs can contribute to the initiation and progression of tumorigenesis. Despite the comprehensive findings of mtDNA-CN alterations in various human cancers, there is still limited understanding of the mechanisms of how cells modulate mtDNA-CN. Although the relevant mechanism has yet to be clarified, it is undeniable that mtDNA-CN is a very promising potential target in cancer research. Therefore, more efforts are required to study the undetermined mechanism of mtDNA-CN alterations in cancer development.
Acknowledgements
Not applicable.
Funding Statement
This present review was supported by the Ministry of Higher Education Malaysia under the Fundamental Research Grant Scheme (FRGS) with Project Code: FRGS/1/2021/SKK0/USM/02/2 (203.PPSP.6171310).
Availability of data and materials
Not applicable.
Authors' contributions
SMAR performed the literature search and wrote the manuscript. AAMY contributed to conceptualization, designing, drafting, and editing of the manuscript. SZNMK and SMAR designed the figure and prepared the table. AAMY, AP, FA, ZI critically reviewed and revised the manuscript. All authors have read and approved the final manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Moon JJ, Lu A, Moon C. Role of genomic instability in human carcinogenesis. Exp Biol Med (Maywood) 2019;244:227–240. doi: 10.1177/1535370219826031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pikor L, Thu K, Vucic E, Lam W. The detection and implication of genome instability in cancer. Cancer Metastasis Rev. 2013;32:341–352. doi: 10.1007/s10555-013-9429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yusoff AAM, Abdullah WSW, Khair SZNM, Radzak SMA. A comprehensive overview of mitochondrial DNA 4977-bp deletion in cancer studies. Oncol Rev. 2019;13:409. doi: 10.4081/oncol.2019.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Otto AM. Warburg effect(s)-a biographical sketch of Otto Warburg and his impacts on tumor metabolism. Cancer Metab. 2016;4:5. doi: 10.1186/s40170-016-0145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Modica-Napolitano JS, Kulawiec M, Singh K. Mitochondria and human cancer. Curr Mol Med. 2007;7:121–131. doi: 10.2174/156652407779940495. [DOI] [PubMed] [Google Scholar]
- 7.Chen EI. Mitochondrial dysfunction and cancer metastasis. J Bioenerg Biomembr. 2012;44:619–622. doi: 10.1007/s10863-012-9465-9. [DOI] [PubMed] [Google Scholar]
- 8.Pedersen PL. Bioenergetics of cancer cells-a brief orientation to this minireview series. J Bioenerg Biomembr. 1997;29:301–302. doi: 10.1023/A:1022417911796. [DOI] [Google Scholar]
- 9.Rempel A, Mathupala SP, Griffin CA, Hawkins AL, Pedersen PL. Glucose catabolism in cancer cells: Amplification of the gene encoding type II hexokinase. Cancer Res. 1996;56:2468–2471. [PubMed] [Google Scholar]
- 10.Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, Armistead S, Lemire K, Orrell J, Teich J, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292:125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 11.Radzak S, Khair Z, Ahmad F, Idris Z, Yusoff A. Accumulation of mitochondrial DNA microsatellite instability in Malaysian patients with primary central nervous system tumors. Turk Neurosurg. 2021;31:99–106. doi: 10.5137/1019-5149.JTN.27893-20.4. [DOI] [PubMed] [Google Scholar]
- 12.Campa D, Barrdahl M, Santoro A, Severi G, Baglietto L, Omichessan H, Tumino R, Bueno-de-Mesquita HBA, Peeters PH, Weiderpass E, et al. Mitochondrial DNA copy number variation, leukocyte telomere length, and breast cancer risk in the European prospective investigation into cancer and nutrition (EPIC) study. Breast Cancer Res. 2018;20:29. doi: 10.1186/s13058-018-0955-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai JG, Zhang ZY, Liu QX, Min JX. Mitochondrial genome microsatellite instability and copy number alteration in lung carcinomas. Asian Pacific J Cancer Prev. 2013;14:2393–2399. doi: 10.7314/APJCP.2013.14.4.2393. [DOI] [PubMed] [Google Scholar]
- 14.Kumar B, Bhat ZI, Bansal S, Saini S, Naseem A, Wahabi K, Burman A, Kumar GT, Saluja SS, Rizvi MMA. Association of mitochondrial copy number variation and T16189C polymorphism with colorectal cancer in North Indian population. Tumour Biol. 2017;39:1010428317740296. doi: 10.1177/1010428317740296. [DOI] [PubMed] [Google Scholar]
- 15.Zhao RZ, Jiang S, Zhang L, Yu ZB. Mitochondrial electron transport chain, ROS generation and uncoupling (Review) Int J Mol Med. 2019;44:3–15. doi: 10.3892/ijmm.2019.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu X, Mao Y, Huang T, Yan C, Yu F, Du J, Dai J, Ma H, Jin G. High mitochondrial DNA copy number was associated with an increased gastric cancer risk in a Chinese population. Mol Carcinog. 2017;56:2593–2600. doi: 10.1002/mc.22703. [DOI] [PubMed] [Google Scholar]
- 17.Gilkerson R, Bravo L, Garcia I, Gaytan N, Herrera A, Maldonado A, Quintanilla B. The mitochondrial nucleoid: Integrating mitochondrial DNA into cellular homeostasis. Cold Spring Harb Perspect Biol. 2013;5:a011080. doi: 10.1101/cshperspect.a011080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Facucho-Oliveira JM, St John JC. The relationship between pluripotency and mitochondrial DNA proliferation during early embryo development and embryonic stem cell differentiation. Stem Cell Rev Rep. 2009;5:140–158. doi: 10.1007/s12015-009-9058-0. [DOI] [PubMed] [Google Scholar]
- 19.Montoya J, Christianson T, Levens D, Rabinowitz M, Attardi G. Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proc Natl Acad Sci USA. 1982;79:7195–7199. doi: 10.1073/pnas.79.23.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clayton DA. Transcription and replication of mitochondrial DNA. Hum Reprod. 2000;15((Suppl 2)):S11–S17. doi: 10.1093/humrep/15.suppl_2.11. [DOI] [PubMed] [Google Scholar]
- 21.Turnbull HE, Lax NZ, Diodato D, Ansorge O, Turnbull DM. The mitochondrial brain: From mitochondrial genome to neurodegeneration. Biochim Biophys Acta. 2010;1802:111–121. doi: 10.1016/j.bbadis.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chabi B, Mousson De Camaret B, Duborjal H, Issartel JP, Stepien G. Quantification of mitochondrial DNA deletion, depletion, and overreplication: Application to diagnosis. Clin Chem. 2003;49:1309–1317. doi: 10.1373/49.8.1309. [DOI] [PubMed] [Google Scholar]
- 23.Castellani CA, Longchamps RJ, Sun J, Guallar E, Arking DE. Thinking outside the nucleus: Mitochondrial DNA copy number in health and disease. Mitochondrion. 2020;53:214–223. doi: 10.1016/j.mito.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D'Erchia AM, Atlante A, Gadaleta G, Pavesi G, Chiara M, De Virgilio C, Manzari C, Mastropasqua F, Prazzoli GM, Picardi E, et al. Tissue-specific mtDNA abundance from exome data and its correlation with mitochondrial transcription, mass and respiratory activity. Mitochondrion. 2015;20:13–21. doi: 10.1016/j.mito.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Prosser R, Simonetti S, Sadlock J, Jagiello G, Schon EA. Rearranged mitochondrial genomes are present in human oocytes. Am J Hum Genet. 1995;57:239–247. [PMC free article] [PubMed] [Google Scholar]
- 26.Yusoff AA, Ahmad F, Idris Z, Jaafar H, Abdullah JM. Understanding mitochondrial DNA in brain tumorigenesis. Mol Considerations Evol Surg Manag Issues Treat Patients with a Brain Tumor. 2015:3–28. [Google Scholar]
- 27.Fontana GA, Gahlon HL. Mechanisms of replication and repair in mitochondrial DNA deletion formation. Nucleic Acids Res. 2020;48:11244–11258. doi: 10.1093/nar/gkaa804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robberson DL, Kasamatsu H, Vinograd J. Replication of mitochondrial DNA. Circular replicative intermediates in mouse L cells. Proc Natl Acad Sci USA. 1972;69:737–741. doi: 10.1073/pnas.69.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holt IJ, Lorimer HE, Jacobs HT. Coupled leading-and lagging-strand synthesis of mammalian mitochondrial DNA. Cell. 2000;100:515–524. doi: 10.1016/S0092-8674(00)80688-1. [DOI] [PubMed] [Google Scholar]
- 30.Yasukawa T, Reyes A, Cluett TJ, Yang MY, Bowmaker M, Jacobs HT, Holt IJ. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006;25:5358–5371. doi: 10.1038/sj.emboj.7601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clay Montier LL, Deng JJ, Bai Y. Number matters: Control of mammalian mitochondrial DNA copy number. J Genet Genomics. 2009;36:125–131. doi: 10.1016/S1673-8527(08)60099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falkenberg M. Mitochondrial DNA replication in mammalian cells: Overview of the pathway. Essays Biochem. 2018;62:287–296. doi: 10.1042/EBC20170100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 34.Kanki T, Ohgaki K, Gaspari M, Gustafsson CM, Fukuoh A, Sasaki N, Hamasaki N, Kang D. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol Cell Biol. 2004;24:9823–9834. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson NG. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 36.Farge G, Mehmedovic M, Baclayon M, van den Wildenberg SM, Roos WH, Gustafsson CM, Wuite GJ, Falkenberg M. In vitro-reconstituted nucleoids can block mitochondrial DNA replication and transcription. Cell Rep. 2014;8:66–74. doi: 10.1016/j.celrep.2014.05.046. [DOI] [PubMed] [Google Scholar]
- 37.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peter B, Falkenberg M. Twinkle and other human mitochondrial DNA helicases: Structure, function and disease. Genes (Basel) 2020;11:408. doi: 10.3390/genes11040408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korhonen JA, Gaspari M, Falkenberg M. Twinkle has 5′ ->3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J Biol Chem. 2003;278:48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 40.Milenkovic D, Matic S, kühl I, Ruzzenente B, Freyer C, Jemt E, Park CB, Falkenberg M, Larsson NG. Twinkle is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication. Hum Mol Genet. 2013;22:1983–1993. doi: 10.1093/hmg/ddt051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takamatsu C, Umeda S, Ohsato T, Ohno T, Abe Y, Fukuoh A, Shinagawa H, Hamasaki N, Kang D. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 2002;3:451–456. doi: 10.1093/embo-reports/kvf099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miralles Fusté J, Shi Y, Wanrooij S, Zhu X, Jemt E, Persson Ö, Sabouri N, Gustafsson CM, Falkenberg M. In vivo occupancy of mitochondrial single-stranded DNA binding protein supports the strand displacement mode of DNA replication. PLoS Genet. 2014;10:e1004832. doi: 10.1371/journal.pgen.1004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Datta A, Brosh RM., Jr New insights into DNA helicases as druggable targets for cancer therapy. Front Mol Biosci. 2018;5:59. doi: 10.3389/fmolb.2018.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruhanen H, Borrie S, Szabadkai G, Tyynismaa H, Jones AW, Kang D, Taanman JW, Yasukawa T. Mitochondrial single-stranded DNA binding protein is required for maintenance of mitochondrial DNA and 7S DNA but is not required for mitochondrial nucleoid organisation. Biochim Biophys Acta. 2010;1803:931–939. doi: 10.1016/j.bbamcr.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 45.Grzybowska-Szatkowska L, Slaska B. Mitochondrial DNA and carcinogenesis (review) Mol Med Rep. 2012;6:923–930. doi: 10.3892/mmr.2012.1027. [DOI] [PubMed] [Google Scholar]
- 46.Reddy TV, Govatati S, Deenadayal M, Sisinthy S, Bhanoori M. Impact of mitochondrial DNA copy number and displacement loop alterations on polycystic ovary syndrome risk in south Indian women. Mitochondrion. 2019;44:35–40. doi: 10.1016/j.mito.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 47.Li HL, Slone J, Fei L, Huang T. Mitochondrial DNA variants and common diseases: A mathematical model for the diversity of age-related mtDNA mutations. Cells. 2019;8:608. doi: 10.3390/cells8060608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicholls TJ, Minczuk M. In D-loop: 40 years of mitochondrial 7S DNA. Exp Gerontol. 2014;56:175–181. doi: 10.1016/j.exger.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 49.Lee HC, Li SH, Lin JC, Wu CC, Yeh DC, Wei YH. Somatic mutations in the D-loop and decrease in the copy number of mitochondrial DNA in human hepatocellular carcinoma. Mutat Res. 2004;547:71–78. doi: 10.1016/j.mrfmmm.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 50.Yu M, Wan Y, Zou Q. Decreased copy number of mitochondrial DNA in Ewing's sarcoma. Clin Chim Acta. 2010;411:679–683. doi: 10.1016/j.cca.2010.01.035. [DOI] [PubMed] [Google Scholar]
- 51.Yu M, Zhou Y, Shi Y, Ning L, Yang Y, Wei X, Zhang N. Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB Life. 2007;59:450–457. doi: 10.1080/15216540701509955. [DOI] [PubMed] [Google Scholar]
- 52.Bai R, Chang J, Yeh K, Lou MA, Lu JF, Tan DJ, Liu H, Wong LJ. Mitochondrial DNA content varies with pathological characteristics of breast cancer. J Oncol. 2011;2011:496189. doi: 10.1155/2011/496189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yusoff AAM, Radzak SMA, Khair SZNM. Research highlights on contributions of mitochondrial DNA microsatellite instability in solid cancers - an overview. Contemp Oncol (Pozn) 2022;26:8–26. doi: 10.5114/wo.2022.115674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kazak L, Reyes A, Holt IJ. Minimizing the damage : Repair pathways keep mitochondrial DNA intact. Nat Rev Mol Cell Biol. 2012;13:659–671. doi: 10.1038/nrm3458. [DOI] [PubMed] [Google Scholar]
- 55.Zhang D, Mott JL, Chang SW, Denniger G, Feng Z, Zassenhaus HP. Construction of transgenic mice with tissue-specific acceleration of mitochondrial DNA mutagenesis. Genomics. 2000;69:151–161. doi: 10.1006/geno.2000.6333. [DOI] [PubMed] [Google Scholar]
- 56.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly-Y M, Gidlöf S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 57.Spelbrink JN, Toivonen JM, Hakkaart GAJ, Kurkela JM, Cooper HM, Lehtinen SK, Lecrenier N, Back JW, Speijer D, Foury F, Jacobs HT. In vivo functional analysis of the human mitochondrial DNA polymerase POLG expressed in cultured human cells. J Biol Chem. 2000;275:24818–24828. doi: 10.1074/jbc.M000559200. [DOI] [PubMed] [Google Scholar]
- 58.Singh KK, Ayyasamy V, Owens KM, Koul MS, Vujcic M. Mutations in mitochondrial DNA polymerase-gamma promote breast tumorigenesis. J Hum Genet. 2009;54:516–524. doi: 10.1038/jhg.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Linkowska K, Jawien A, Marszałek A, Skonieczna K, Grzybowski T. Searching for association of the CAG repeat polymorphism in the mitochondrial DNA polymerase gamma gene (POLG) with colorectal cancer. Acta Biochim Pol. 2015;62:625–627. doi: 10.18388/abp.2014_935. [DOI] [PubMed] [Google Scholar]
- 60.Long X, Wang X, Chen Y, Guo X, Zhou F, Fan Y, Ge N, Guo M, Zhang Z, Dong G. Polymorphisms in POLG were associated with the prognosis and mtDNA content in hepatocellular carcinoma patients. Bull Cancer. 2017;104:500–507. doi: 10.1016/j.bulcan.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 61.Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 62.Tyynismaa H, Sembongi H, Bokori-Brown M, Granycome C, Ashley N, Poulton J, Jalanko A, Spelbrink JN, Holt IJ, Suomalainen A. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum Mol Genet. 2004;13:3219–3227. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- 63.Sanchez-Martinez A, Calleja M, Peralta S, Matsushima Y, Hernandez-Sierra R, Whitworth AJ, Kaguni LS, Garesse R. Modeling pathogenic mutations of human twinkle in Drosophila suggests an apoptosis role in response to mitochondrial defects. PLoS One. 2012;7:e43954. doi: 10.1371/journal.pone.0043954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsushima Y, Kaguni LS. Differential phenotypes of active site and human autosomal dominant progressive external ophthalmoplegia mutations in Drosophila mitochondrial DNA helicase expressed in Schneider cells. J Biol Chem. 2007;282:9436–9444. doi: 10.1074/jbc.M610550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin CS, Wang LS, Tsai CM, Wei YH. Low copy number and low oxidative damage of mitochondrial DNA are associated with tumor progression in lung cancer tissues after neoadjuvant chemotherapy. Interact Cardiovasc Thorac Surg. 2008;7:954–958. doi: 10.1510/icvts.2008.177006. [DOI] [PubMed] [Google Scholar]
- 66.Hu L, Yao X, Shen Y. Altered mitochondrial DNA copy number contributes to human cancer risk: Evidence from an updated meta-analysis. Sci Rep. 2016;6:35859. doi: 10.1038/srep35859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee H, Lee JH, Kim DC, Hwang I, Kang YN, Gwon GJ, Choi IJ, Kim S. Is mitochondrial DNA copy number associated with clinical characteristics and prognosis in gastric cancer? Asian Pacific J Cancer Prev. 2015;16:87–90. doi: 10.7314/APJCP.2015.16.1.87. [DOI] [PubMed] [Google Scholar]
- 68.Han Y, Chen JZ. Oxidative stress induces mitochondrial DNA damage and cytotoxicity through independent mechanisms in human cancer cells. Biomed Res Int. 2013;2013:825065. doi: 10.1155/2013/825065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu CS, Tsai CS, Kuo CL, Chen HW, Lii CK, Ma YS, Wei YH. Oxidative stress-related alteration of the copy number of mitochondrial DNA in human leukocytes. Free Radic Res. 2003;37:1307–1317. doi: 10.1080/10715760310001621342. [DOI] [PubMed] [Google Scholar]
- 70.Al-Kafaji G, Golbahar J. High glucose-induced oxidative stress increases the copy number of mitochondrial DNA in human mesangial cells. Biomed Res Int. 2013;2013:754946. doi: 10.1155/2013/754946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guha M, Avadhani NG. Mitochondrial retrograde signaling at the crossroads of tumor bioenergetics, genetics and epigenetics. Mitochondrion. 2013;13:577–591. doi: 10.1016/j.mito.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Corral M, Paris B, Baffet G, Tichonicky L, Guguen-Guillouzo C, Kruh J, Defer N. Increased level of the mitochondrial ND5 transcript in chemically induced rat hepatomas. Exp Cell Res. 1989;184:158–166. doi: 10.1016/0014-4827(89)90374-1. [DOI] [PubMed] [Google Scholar]
- 73.Delsite RL, Rasmussen LJ, Rasmussen AK, Kalen A, Goswami PC, Singh KK. Mitochondrial impairment is accompanied by impaired oxidative DNA repair in the nucleus. Mutagenesis. 2003;18:497–503. doi: 10.1093/mutage/geg027. [DOI] [PubMed] [Google Scholar]
- 74.Suzuki S, Naito A, Asano T, Evans TT, Reddy SAG, Higuchi M. Constitutive activation of AKT pathway inhibits TNF-induced apoptosis in mitochondrial DNA-deficient human myelogenous leukemia ML-1a. Cancer Lett. 2008;268:31–37. doi: 10.1016/j.canlet.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Biswas G, Anandatheerthavarada HK, Zaidi M, Avadhani NG. Mitochondria to nucleus stress signaling: A distinctive mechanism of NFkappaB/Rel activation through calcineurin-mediated inactivation of IkappaBbeta. J Cell Biol. 2003;161:507–519. doi: 10.1083/jcb.200211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu T, Stark GR. Cytokine overexpression and constitutive NFkappaB in cancer. Cell Cycle. 2004;3:1114–1117. doi: 10.4161/cc.3.9.1130. [DOI] [PubMed] [Google Scholar]
- 77.Mei H, Sun S, Bai Y, Chen Y, Chai R, Li H. Reduced mtDNA copy number increases the sensitivity of tumor cells to chemotherapeutic drugs. Cell Death Dis. 2015;6:e1710. doi: 10.1038/cddis.2015.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sun X, Zhan L, Chen Y, Wang G, He L, Wang Q, Zhou F, Yang F, Wu J, Wu Y, et al. Increased mtDNA copy number promotes cancer progression by enhancing mitochondrial oxidative phosphorylation in microsatellite-stable colorectal cancer. Signal Transduct Target Ther. 2018;3:8. doi: 10.1038/s41392-018-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moraes CT. What regulates mitochondrial DNA copy number in animal cells? Trends Genet. 2001;17:199–205. doi: 10.1016/S0168-9525(01)02238-7. [DOI] [PubMed] [Google Scholar]
- 80.Tang Y, Schon EA, Wilichowski E, Vazquez-Memije ME, Davidson E, King MP. Rearrangements of human mitochondrial DNA (mtDNA): New insights into the regulation of mtDNA copy number and gene expression. Mol Biol Cell. 2000;11:1471–1485. doi: 10.1091/mbc.11.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tang Y, Manfredi G, Hirano M, Schon EA. Maintenance of human rearranged mitochondrial DNAs in long-term cultured transmitochondrial cell lines. Mol Biol Cell. 2000;11:2349–2358. doi: 10.1091/mbc.11.7.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kelly RD, Mahmud A, McKenzie M, Trounce IA, St John JC. Mitochondrial DNA copy number is regulated in a tissue specific manner by DNA methylation of the nuclear-encoded DNA polymerase gamma A. Nucleic Acids Res. 2012;40:10124–10138. doi: 10.1093/nar/gks770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee W, Johnson J, Gough DJ, Donoghue J, Cagnone GL, Vaghjiani V, Brown KA, Johns TG, St John JC. Mitochondrial DNA copy number is regulated by DNA methylation and demethylation of POLGA in stem and cancer cells and their differentiated progeny. Cell Death Dis. 2015;6:e1664. doi: 10.1038/cddis.2015.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dickinson A, Yeung KY, Donoghue J, Baker MJ, Kelly RD, McKenzie M, Johns TG, St John JC. The regulation of mitochondrial DNA copy number in glioblastoma cells. Cell Death Differ. 2013;20:1644–1653. doi: 10.1038/cdd.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun X, St John JC. Modulation of mitochondrial DNA copy number in a model of glioblastoma induces changes to DNA methylation and gene expression of the nuclear genome in tumours. Epigenetics Chromatin. 2018;11:53. doi: 10.1186/s13072-018-0223-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tan AS, Baty JW, Dong LF, Bezawork-Geleta A, Endaya B, Goodwin J, Bajzikova M, Kovarova J, Peterka M, Yan B, et al. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab. 2015;21:81–94. doi: 10.1016/j.cmet.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 87.Berridge MV, Crasso C, Neuzil J. Mitochondrial genome transfer to tumor cells breaks the rules and establishes a new precedent in cancer biology. Mol Cell Oncol. 2018;5:e1023929. doi: 10.1080/23723556.2015.1023929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee HC, Wei YH. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int J Biochem Cell Biol. 2005;37:822–834. doi: 10.1016/j.biocel.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 89.Yeung KY, Dickinson A, Donoghue JF, Polekhina G, White SJ, Grammatopoulos DK, McKenzie M, Johns TG, St John JC. The identification of mitochondrial DNA variants in glioblastoma multiforme. Acta Neuropathol Commun. 2014;2:1. doi: 10.1186/2051-5960-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yuan Y, Ju YS, Kim Y, Li J, Wang Y, Yoon CJ, Yang Y, Martincorena I, Creighton CJ, Weinstein JN, et al. Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat Genet. 2020;52:342–352. doi: 10.1038/s41588-019-0557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jiang H, Zhao H, Xu H, Hu L, Wang W, Wei Y, Wang Y, Peng X, Zhou F. Peripheral blood mitochondrial DNA content, A10398G polymorphism, and risk of breast cancer in a Han Chinese population. Cancer Sci. 2014;105:639–645. doi: 10.1111/cas.12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shen J, Platek M, Mahasneh A, Ambrosone CB, Zhao H. Mitochondrial copy number and risk of breast cancer: A pilot study. Mitochondrion. 2010;10:62–68. doi: 10.1016/j.mito.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fazzini F, Schöpf B, Blatzer M, Coassin S, Hicks AA, Kronenberg F, Fendt L. Plasmid-normalized quantification of relative mitochondrial DNA copy number. Sci Rep. 2018;8:15347. doi: 10.1038/s41598-018-33684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Castellani CA, Longchamps RJ, Sumpter JA, Newcomb CE, Lane JA, Grove ML, Bressler J, Brody JA, Floyd JS, Bartz TM, et al. Mitochondrial DNA copy number can influence mortality and cardiovascular disease via methylation of nuclear DNA CpGs. Genome Med. 2020;12:84. doi: 10.1186/s13073-020-00778-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang L, Lv H, Ji P, Zhu X, Yuan H, Jin G, Dai J, Hu Z, Su Y, Ma H. Mitochondrial DNA copy number is associated with risk of head and neck squamous cell carcinoma in Chinese population. Cancer Med. 2018;7:2776–2782. doi: 10.1002/cam4.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shen J, Wan J, Song R, Zhao H. Peripheral blood mitochondrial DNA copy number, length heteroplasmy and breast cancer risk: A replication study. Carcinogenesis. 2015;36:1307–1313. doi: 10.1093/carcin/bgv130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xia P, An HX, Dang CX, Radpour R, Kohler C, Fokas E, Engenhart-Cabillic R, Holzgreve W, Zhong XY. Decreased mitochondrial DNA content in blood samples of patients with stage I breast cancer. BMC Cancer. 2009;9:454. doi: 10.1186/1471-2407-9-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jung SJ, Cho JH, Park WJ, Heo YR, Lee JH. Telomere length is correlated with mitochondrial DNA copy number in intestinal, but not diffuse, gastric cancer. Oncol Lett. 2017;14:925–929. doi: 10.3892/ol.2017.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.O'Hara R, Tedone E, Ludlow A, Huang E, Arosio B, Mari D, Shay JW. Quantitative mitochondrial DNA copy number determination using droplet digital PCR with single-cell resolution. Genome Res. 2019;29:1878–1888. doi: 10.1101/gr.250480.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Memon AA, Zöller B, Hedelius A, Wang X, Stenman E, Sundquist J, Sundquist K. Quantification of mitochondrial DNA copy number in suspected cancer patients by a well optimized ddPCR method. Biomol Detect Quantif. 2017;13:32–39. doi: 10.1016/j.bdq.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Filograna R, Mennuni M, Alsina D, Larsson NG. Mitochondrial DNA copy number in human disease: The more the better? FEBS Lett. 2021;595:976–1002. doi: 10.1002/1873-3468.14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Longchamps RJ, Castellani CA, Yang SY, Newcomb CE, Sumpter JA, Lane J, Grove ML, Guallar E, Pankratz N, Taylor KD, et al. Evaluation of mitochondrial DNA copy number estimation techniques. PLoS One. 2020;15:e0228166. doi: 10.1371/journal.pone.0228166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhou K, Mo Q, Guo S, Liu Y, Yin C, Ji X, Guo X, Xing J. A novel next-generation sequencing-based approach for concurrent detection of mitochondrial DNA copy number and mutation. J Mol Diagn. 2020;22:1408–1418. doi: 10.1016/j.jmoldx.2020.09.005. [DOI] [PubMed] [Google Scholar]
- 104.Picard M. Blood mitochondrial DNA copy number: What are we counting? Mitochondrion. 2021;60:1–11. doi: 10.1016/j.mito.2021.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ware SA, Desai N, Lopez M, Leach D, Zhang Y, Giordano L, Nouraie M, Picard M, Kaufman BA. An automated, high-throughput methodology optimized for quantitative cell-free mitochondrial and nuclear DNA isolation from plasma. J Biol Chem. 2020;295:15677–15691. doi: 10.1074/jbc.RA120.015237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Guo W, Jiang L, Bhasin S, Khan SM, Swerdlow RH. DNA extraction procedures meaningfully influence qPCR-based mtDNA copy number determination. Mitochondrion. 2009;9:261–265. doi: 10.1016/j.mito.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW, Lee LM, Wei YH, Lee HC. Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosom Cancer. 2006;45:629–638. doi: 10.1002/gcc.20326. [DOI] [PubMed] [Google Scholar]
- 108.Fan AX, Radpour R, Haghighi MM, Kohler C, Xia P, Hahn S, Holzgreve W, Zhong XY. Mitochondrial DNA content in paired normal and cancerous breast tissue samples from patients with breast cancer. J Cancer Res Clin Oncol. 2009;135:983–989. doi: 10.1007/s00432-008-0533-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nacheva E, Mokretar K, Soenmez A, Pittman AM, Grace C, Valli R, Ejaz A, Vattathil S, Maserati E, Houlden H, et al. DNA isolation protocol effects on nuclear DNA analysis by microarrays, droplet digital PCR, and whole genome sequencing, and on mitochondrial DNA copy number estimation. PLoS One. 2017;12:e0180467. doi: 10.1371/journal.pone.0180467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hsu CW, Yin PH, Lee HC, Chi CW, Tseng LM. Mitochondrial DNA content as a potential marker to predict response to anthracycline in breast cancer patients. Breast J. 2010;16:264–270. doi: 10.1111/j.1524-4741.2010.00908.x. [DOI] [PubMed] [Google Scholar]
- 111.Domínguez-de-la-Cruz E, Muñoz ML, Pérez-Muñoz A, García-Hernández N, Moctezuma-Meza C, Hinojosa-Cruz JC. Reduced mitochondrial DNA copy number is associated with the haplogroup, and some clinical features of breast cancer in Mexican patients. Gene. 2020;761:145047. doi: 10.1016/j.gene.2020.145047. [DOI] [PubMed] [Google Scholar]
- 112.Lemnrau A, Brook MN, Fletcher O, Coulson P, Tomczyk K, Jones M, Ashworth A, Swerdlow A, Orr N, Garcia-Closas M. Mitochondrial DNA copy number in peripheral blood cells and risk of developing breast cancer. Cancer Res. 2015;75:2844–2850. doi: 10.1158/0008-5472.CAN-14-1692. [DOI] [PubMed] [Google Scholar]
- 113.Thyagarajan B, Wang R, Nelson H, Barcelo H, Koh W, Yuan M. Mitochondrial DNA copy number is associated with breast cancer risk. PLoS One. 2013;8:e65968. doi: 10.1371/journal.pone.0065968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Feng S, Xiong L, Ji Z, Cheng W, Yang H. Correlation between increased copy number of mitochondrial DNA and clinicopathological stage in colorectal cancer. Oncol Lett. 2011;2:899–903. doi: 10.3892/ol.2011.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen T, He J, Shen L, Fang H, Nie H, Jin T, Wei X, Xin Y, Jiang Y, Li H, et al. The mitochondrial DNA 4,977-bp deletion and its implication in copy number alteration in colorectal cancer. BMC Med Genet. 2011;12:8. doi: 10.1186/1471-2350-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Qu F, Liu X, Zhou F, Yang H, Bao G, He X, Xing J. Association between mitochondrial DNA content in leukocytes and colorectal cancer risk: A case-control analysis. Cancer. 2011;117:3148–3155. doi: 10.1002/cncr.25906. [DOI] [PubMed] [Google Scholar]
- 117.Thyagarajan B, Wang R, Barcelo H, Koh WP, Yuan JM. Mitochondrial copy number is associated with colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2012;21:1574–1581. doi: 10.1158/1055-9965.EPI-12-0138-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tong H, Zhang L, Gao J, Wen S, Zhou H, Feng S. Methylation of mitochondrial DNA displacement loop region regulates mitochondrial copy number in colorectal cancer. Mol Med Rep. 2017;16:5347–5353. doi: 10.3892/mmr.2017.7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang K, Li X, Forman MR, Monahan PO, Graham BH, Joshi A, Song M, Hang D, Ogino S, Giovannucci EL, et al. Pre-diagnostic leukocyte mitochondrial DNA copy number and colorectal cancer risk. Carcinogenesis. 2019;40:1462–1468. doi: 10.1093/carcin/bgz159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mohideen AM, Dicks E, Parfrey P, Green R, Savas S. Mitochondrial DNA polymorphisms, its copy number change and outcome in colorectal cancer. BMC Res Notes. 2015;8:272. doi: 10.1186/s13104-015-1250-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cui HH, Huang P, Wang ZJ, Zhang Y, Zhang Z, Xu W, Wang X, Han Y, Guo X. Association of decreased mitochondrial DNA content with the progression of colorectal cancer. BMC Cancer. 2013;13:110. doi: 10.1186/1471-2407-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Huang B, Gao YT, Shu XO, Wen W, Yang G, Li G, Courtney R, Ji BT, Li HL, Purdue MP, et al. Association of leukocyte mitochondrial DNA copy number with colorectal cancer risk: Results from the Shanghai women's health study. Cancer Epidemiol Biomarkers Prev. 2014;23:2357–2365. doi: 10.1158/1055-9965.EPI-14-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.van Osch FH, Voets AM, Schouten LJ, Gottschalk RW, Simons CC, van Engeland M, Lentjes MH, van den Brandt PA, Smeets HJ, Weijenberg MP. Mitochondrial DNA copy number in colorectal cancer: Between tissue comparisons, clinicopathological characteristics and survival. Carcinogenesis. 2015;36:1502–1510. doi: 10.1093/carcin/bgv151. [DOI] [PubMed] [Google Scholar]
- 124.Marucci G, Maresca A, Caporali L, Farnedi A, Betts CM, Morandi L, de Biase D, Cerasoli S, Foschini MP, Bonora E, et al. Oncocytic glioblastoma: A glioblastoma showing oncocytic changes and increased mitochondrial DNA copy number. Hum Pathol. 2013;44:1867–1876. doi: 10.1016/j.humpath.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 125.Zhang J, Li D, Qu F, Chen Y, Li G, Jiang H, Huang X, Yang H, Xing J. Association of leukocyte mitochondrial DNA content with glioma risk: Evidence from a Chinese case-control study. BMC Cancer. 2014;14:680. doi: 10.1186/1471-2407-14-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shen J, Song R, Lu Z, Zhao H. Mitochondrial DNA copy number in whole blood and glioma risk: A case control study. Mol Carcinog. 2016;55:2089–2094. doi: 10.1002/mc.22453. [DOI] [PubMed] [Google Scholar]
- 127.Chen Y, Zhang J, Huang X, Zhang J, Zhou X, Hu J, Li G, He S, Xing J. High leukocyte mitochondrial DNA content contributes to poor prognosis in glioma patients through its immunosuppressive effect. Br J Cancer. 2015;113:99–106. doi: 10.1038/bjc.2015.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dardaud LM, Bris C, Desquiret-Dumas V, Boisselier B, Tabouret E, Mokhtari K, Figarella-Branger D, Rousseau A, Procaccio V. High mitochondrial DNA copy number is associated with longer survival in young patients with glioblastoma. Neuro Oncol. 2019;21:1084–1085. doi: 10.1093/neuonc/noz072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang Y, Qu Y, Gao K, Yang Q, Shi B, Hou P, Ji M. High copy number of mitochondrial DNA (mtDNA) predicts good prognosis in glioma patients. Am J Cancer Res. 2015;5:1207–16. [PMC free article] [PubMed] [Google Scholar]
- 130.Oliva CR, Nozell SE, Diers A, McClugage SG, III, Sarkaria JN, Markert JM, Darley-Usmar VM, Bailey SM, Gillespie GY, Landar A, Griguer CE. Acquisition of temozolomide chemoresistance in gliomas leads to remodeling of mitochondrial electron transport chain. J Biol Chem. 2010;285:39759–39767. doi: 10.1074/jbc.M110.147504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shen H, Yu M, Tsoli M, Chang C, Joshi S, Liu J, Ryall S, Chornenkyy Y, Siddaway R, Hawkins C, Ziegler DS. Targeting reduced mitochondrial DNA quantity as a therapeutic approach in pediatric high-grade gliomas. Neuro Oncol. 2020;22:139–151. doi: 10.1093/neuonc/noz140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sravya P, Nimbalkar VP, Kanuri NN, Sugur H, Verma BK, Kundu P, Rao S, Uday Krishna AS, Somanna S, Kondaiah P, et al. Low mitochondrial DNA copy number is associated with poor prognosis and treatment resistance in glioblastoma. Mitochondrion. 2020;55:154–163. doi: 10.1016/j.mito.2020.10.001. [DOI] [PubMed] [Google Scholar]
- 133.Li F, Wang X, Han C, Lin J. Decreased mtDNA copy number of gastric cancer: A new tumor marker? Chin J Clin Oncol. 2004;1:250–255. doi: 10.1007/BF02739809. [DOI] [Google Scholar]
- 134.Wu CW, Yin PH, Hung WY, Li AF, Li SH, Chi CW, Wei YH, Lee HC. Mitochondria DNA mutations and mitochondrial DNA depletion in gastric cancer. Genes Chromosom Cancer. 2005;44:19–28. doi: 10.1002/gcc.20213. [DOI] [PubMed] [Google Scholar]
- 135.Zhang G, Qu Y, Dang S, Yang Q, Shi B, Hou P. Variable copy number of mitochondrial DNA (mtDNA) predicts worse prognosis in advanced gastric cancer patients. Diagn Pathol. 2013;8:173. doi: 10.1186/1746-1596-8-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liao LM, Baccarelli A, Shu XO, Gao YT, Ji BT, Yang G, Li HL, Hoxha M, Dioni L, Rothman N, et al. Mitochondrial DNA copy number and risk of gastric cancer: A report from the Shanghai women's health study. Cancer Epidemiol Biomarkers Prev. 2011;20:1944–1949. doi: 10.1158/1055-9965.EPI-11-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wen SL, Zhang F, Feng S. Decreased copy number of mitochondrial DNA: A potential diagnostic criterion for gastric cancer. Oncol Lett. 2013;6:1098–1102. doi: 10.3892/ol.2013.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mizumachi T, Muskhelishvili L, Naito A, Furusawa J, Fan CY, Siegel ER, Kadlubar FF, Kumar U, Higuchi M. Increased distributional variance of mitochondrial DNA content associated with prostate cancer cells as compared with normal prostate cells. Prostate. 2008;68:408–417. doi: 10.1002/pros.20697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhou W, Zhu M, Gui M, Huang L, Long Z, Wang L, Chen H, Yin Y, Jiang X, Dai Y, et al. Peripheral blood mitochondrial DNA copy number is associated with prostate cancer risk and tumor burden. PLoS One. 2014;9:e109470. doi: 10.1371/journal.pone.0109470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Abhishek A, Singh V, Sinha RJ, Ansari NG, Siddiqe MK, Verma M, Kumar M. To study the relationship between cadmium, zinc and mtDNA copy number in North Indian patients suffering from prostate cancer: A case control study. Afr J Urol. 2017;23:126–132. doi: 10.1016/j.afju.2016.05.011. [DOI] [Google Scholar]
- 141.Moore A, Lan Q, Hofmann JN, Liu CS, Cheng WL, Lin TT, Berndt SI. A prospective study of mitochondrial DNA copy number and the risk of prostate cancer. Cancer Causes Control. 2017;28:529–538. doi: 10.1007/s10552-017-0879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tu H, Gu J, Meng QH, Kim J, Davis JW, He Y, Wagar EA, Thompson TC, Logothetis CJ, Wu X. Mitochondrial DNA copy number in peripheral blood leukocytes and the aggressiveness of localized prostate cancer. Oncotarget. 2015;6:41988–41996. doi: 10.18632/oncotarget.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kalsbeek AM, Chan EK, Grogan J, Petersen DC, Jaratlerdsiri W, Gupta R, Lyons RJ, Haynes AM, Horvath LG, Kench JG, et al. Altered mitochondrial genome content signals worse pathology and prognosis in prostate cancer. Prostate. 2018;78:25–31. doi: 10.1002/pros.23440. [DOI] [PubMed] [Google Scholar]
- 144.Xu J, Chang WS, Tsai CW, Bau DT, Davis JW, Thompson TC, Logothetis CJ, Gu J. Mitochondrial DNA copy number in peripheral blood leukocytes is associated with biochemical recurrence in prostate cancer patients in African Americans. Carcinogenesis. 2020;41:267–273. doi: 10.1093/carcin/bgz139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tan DJ, Chang J, Liu LL, Bai RK, Wang YF, Yeh KT, Wong LJC. Significance of somatic mutations and content alteration of mitochondrial DNA in esophageal cancer. BMC Cancer. 2006;6:93. doi: 10.1186/1471-2407-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liu Z, Zhao Z, Zhao Q. Change in the mitochondrial DNA copy number and its significance in esophageal squamous cell carcinoma. Chin J Clin Oncol. 2007;34:7–9. [Google Scholar]
- 147.Lin CS, Chang SC, Wang LS, Chou TY, Hsu WH, Wu YC, Wei YH. The role of mitochondrial DNA alterations in esophageal squamous cell carcinomas. J Thorac Cardiovasc Surg. 2010;139:189–197.e4. doi: 10.1016/j.jtcvs.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 148.Li H, Tian Z, Zhang Y, Yang Q, Shi B, Hou P, Ji M. Increased copy number of mitochondrial DNA predicts poor prognosis of esophageal squamous cell carcinoma. Oncol Lett. 2018;15:1014–1020. doi: 10.3892/ol.2017.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Masuike Y, Tanaka K, Makino T, Yamasaki M, Miyazaki Y, Takahashi T, Kurokawa Y, Nakajima K, Mori M, Doki Y. Esophageal squamous cell carcinoma with low mitochondrial copy number has mesenchymal and stem-like characteristics, and contributes to poor prognosis. PLoS One. 2018;13:e0193159. doi: 10.1371/journal.pone.0193159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Xu E, Sun W, Gu J, Chow WH, Ajani JA, Wu X. Association of mitochondrial DNA copy number in peripheral blood leukocytes with risk of esophageal adenocarcinoma. Carcinogenesis. 2013;34:2521–2524. doi: 10.1093/carcin/bgt230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kim MM, Clinger JD, Masayesva BG, Ha PK, Zahurak ML, Westra WH, Califano JA. Mitochondrial DNA quantity increases with histopathologic grade in premalignant and malignant head and neck lesions. Clin Cancer Res. 2004;10:8512–8515. doi: 10.1158/1078-0432.CCR-04-0734. [DOI] [PubMed] [Google Scholar]
- 152.Jiang WW, Rosenbaum E, Mambo E, Zahurak M, Masayesva B, Carvalho AL, Zhou S, Westra WH, Alberg AJ, Sidransky D, et al. Decreased mitochondrial DNA content in posttreatment salivary rinses from head and neck cancer patients. Cancer Ther Clin. 2006;12:1564–1569. doi: 10.1158/1078-0432.CCR-05-1471. [DOI] [PubMed] [Google Scholar]
- 153.Cheau-Feng Lin F, Jeng YC, Huang TY, Chi CS, Chou MC, Chin-Shaw Tsai S. Mitochondrial DNA copy number is associated with diagnosis and prognosis of head and neck cancer. Biomarkers. 2014;19:269–274. doi: 10.3109/1354750X.2014.902101. [DOI] [PubMed] [Google Scholar]
- 154.Kumar M, Srivastava S, Singh SA, Das AK, Das GC, Dhar B, Ghosh SK, Mondal R. Cell-free mitochondrial DNA copy number variation in head and neck squamous cell carcinoma: A study of non-invasive biomarker from Northeast India. Tumor Biol. 2017 Oct 26; doi: 10.1177/1010428317736643. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 155.Chen J, Zhang L, Yu X, Zhou H, Luo Y, Wang W, Wang L. Clinical application of plasma mitochondrial DNA content in patients with lung cancer. Oncol Lett. 2018;16:7074–7081. doi: 10.3892/ol.2018.9515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bonner MR, Shen M, Liu CS, DiVita M, He X, Lan Q. Mitochondrial DNA content and lung cancer risk in Xuan Wei, China. Lung Cancer. 2009;63:331–334. doi: 10.1016/j.lungcan.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hosgood HD, III, Liu CS, Rothman N, Weinstein SJ, Bonner MR, Shen M, Lim U, Virtamo J, Cheng WL, Albanes D, Lan Q. Mitochondrial DNA copy number and lung cancer risk in a prospective cohort study. Carcinogenesis. 2010;31:847–849. doi: 10.1093/carcin/bgq045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kim C, Bassig BA, Seow WJ, Hu W, Purdue MP, Shu XO, Huang WY, Liu CS, Cheng WL, Lin TT, et al. Pooled analysis of mitochondrial DNA copy number and lung cancer risk in three prospective studies. Cancer Epidemiol Biomarkers Prev. 2014;23:2977–2980. doi: 10.1158/1055-9965.EPI-14-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Meierhofer D, Mayr JA, Foetschl U, Berger A, Fink K, Schmeller N, Hacker GW, Hauser-Kronberger C, Kofler B, Sperl W. Decrease of mitochondrial DNA content and energy metabolism in renal cell carcinoma. Carcinogenesis. 2004;25:1005–1010. doi: 10.1093/carcin/bgh104. [DOI] [PubMed] [Google Scholar]
- 160.Xing J, Chen M, Wood CG, Lin J, Spitz MR, Ma J, Amos CI, Shields PG, Benowitz NL, Gu J, et al. Mitochondrial DNA content: Its genetic heritability and association with renal cell carcinoma. J Natl Cancer Inst. 2008;100:1104–1112. doi: 10.1093/jnci/djn213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Purdue MP, Hofmann JN, Colt JS, Hoxha M, Ruterbusch JJ, Davis FG, Rothman N, Wacholder S, Schwartz KL, Baccarelli A, Chow WH. A case-control study of peripheral blood mitochondrial DNA copy number and risk of renal cell carcinoma. PLoS One. 2012;7:e43149. doi: 10.1371/journal.pone.0043149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lin CS, Lee HT, Lee MH, Pan SC, Ke CY, Chiu AW, Wei YH. Role of mitochondrial DNA copy number alteration in human renal cell carcinoma. Int J Mol Sci. 2016;17:814. doi: 10.3390/ijms17060814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hofmann JN, Hosgood HD, III, Liu CS, Chow WH, Shuch B, Cheng WL, Lin TT, Moore LE, Lan Q, Rothman N, Purdue MP. A nested case-control study of leukocyte mitochondrial DNA copy number and renal cell carcinoma in the prostate, lung, colorectal and ovarian cancer screening trial. Carcinogenesis. 2014;35:1028–1031. doi: 10.1093/carcin/bgt495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Elsayed ET, Hashad MM, Elgohary IE. Mitochondrial DNA copy number variation as a potential predictor of renal cell carcinoma. Int J Biol Markers. 2017;32:e313–e318. doi: 10.5301/ijbm.5000268. [DOI] [PubMed] [Google Scholar]
- 165.Wang Y, Liu VW, Xue WC, Tsang PC, Cheung AN, Ngan HY. The increase of mitochondrial DNA content in endometrial adenocarcinoma cells: A quantitative study using laser-captured microdissected tissues. Gynecol Oncol. 2005;98:104–110. doi: 10.1016/j.ygyno.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 166.Cormio A, Guerra F, Cormio G, Pesce V, Fracasso F, Loizzi V, Resta L, Putignano G, Cantatore P, Selvaggi LE, Gadaleta MN. Mitochondrial DNA content and mass increase in progression from normal to hyperplastic to cancer endometrium. BMC Res Notes. 2012;5:279. doi: 10.1186/1756-0500-5-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sun Y, Zhang L, Ho SS, Wu X, Gu J. Lower mitochondrial DNA copy number in peripheral blood leukocytes increases the risk of endometrial cancer. Mol Carcinog. 2016;55:1111–1117. doi: 10.1002/mc.22373. [DOI] [PubMed] [Google Scholar]
- 168.Mondal R, Ghosh SK, Choudhury JH, Seram A, Sinha K, Hussain M, Laskar RS, Rabha B, Dey P, Ganguli S, et al. Mitochondrial DNA copy number and risk of oral cancer: A report from Northeast India. PLoS One. 2013;8:e57771. doi: 10.1371/journal.pone.0057771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.He Y, Gong Y, Gu J, Lee JJ, Lippman SM, Wu X. Increased leukocyte mitochondrial DNA copy number is associated with oral premalignant lesions: an epidemiology study. Carcinogenesis. 2014;35:1760–1764. doi: 10.1093/carcin/bgu093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Takeda D, Hasegawa T, Ueha T, Sakakibara A, Kawamoto T, Minamikawa T, Sakai Y, Komori T. Decreased mitochondrial copy numbers in oral squamous cell carcinoma. Head Neck. 2016;36:1170–1175. doi: 10.1002/hed.24194. [DOI] [PubMed] [Google Scholar]
- 171.Lynch SM, Weinstein SJ, Virtamo J, Lan Q, Liu CS, Cheng WL, Rothman N, Albanes D, Stolzenberg-Solomon RZ. Mitochondrial DNA copy number and pancreatic cancer in the alpha-tocopherol beta-carotene cancer prevention study. Cancer Prev Res (Phila) 2011;4:1912–1919. doi: 10.1158/1940-6207.CAPR-11-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tuchalska-Czuroń J, Lenart J, Augustyniak J, Durlik M. Is mitochondrial DNA copy number a good prognostic marker in resectable pancreatic cancer? Pancreatology. 2019;19:73–79. doi: 10.1016/j.pan.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 173.Gentiluomo M, Katzke VA, Kaaks R, Tjønneland A, Severi G, Perduca V, Boutron-Ruault MC, Weiderpass E, Ferrari P, Johnson T, et al. Mitochondrial DNA copy-number variation and pancreatic cancer risk in the prospective EPIC cohort. Cancer Epidemiol Biomarkers Prev. 2020;29:681–686. doi: 10.1158/1055-9965.EPI-19-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wang Y, Liu VW, Xue WC, Cheung AN, Ngan HY. Association of decreased mitochondrial DNA content with ovarian cancer progression. Br J Cancer. 2006;95:1087–1091. doi: 10.1038/sj.bjc.6603377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Keserű JS, Soltész B, Lukács J, Márton É, Szilágyi-Bónizs M, Penyige A, Póka R, Nagy B. Detection of cell-free, exosomal and whole blood mitochondrial DNA copy number in plasma or whole blood of patients with serous epithelial ovarian can. J Biotechnol. 2019;298:76–81. doi: 10.1016/j.jbiotec.2019.04.015. [DOI] [PubMed] [Google Scholar]
- 176.Hyland PL, Pfeiffer RM, Rotunno M, Hofmann JN, Liu CS, Cheng WL, Yuenger J, Lan Q, Tucker MA, Goldstein AM, Yang XR. Constitutive mitochondrial DNA copy number in peripheral blood of melanoma families with and without CDKN2A mutations. J Carcinog Mutagen. 2014;2014((Suppl 4)):S006. doi: 10.4172/2157-2518.S4-006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Shen J, Gopalakrishnan V, Lee JE, Fang S, Zhao H. Mitochondrial DNA copy number in peripheral blood and melanoma risk. PLoS One. 2015;10:e0131649. doi: 10.1371/journal.pone.0131649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dang S, Qu Y, Wei J, Shao Y, Yang Q, Ji M, Shi B, Hou P. Low copy number of mitochondrial DNA (mtDNA) predicts worse prognosis in early-stage laryngeal cancer patients. Diagn Pathol. 2014;9:28. doi: 10.1186/1746-1596-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Guo W, Yang D, Xu H, Zhang Y, Huang J, Yang Z, Chen X, Huang Z. Mutations in the D-loop region and increased copy number of mitochondrial DNA in human laryngeal squamous cell carcinoma. Mol Biol Rep. 2013;40:13–20. doi: 10.1007/s11033-012-1939-7. [DOI] [PubMed] [Google Scholar]
- 180.Hurtado-Roca Y, Ledesma M, Gonzalez-Lazaro M, Moreno-Loshuertos R, Fernandez-Silva P, Enriquez JA, Laclaustra M. Adjusting MtDNA quantification in whole blood for peripheral blood platelet and leukocyte counts. PLoS One. 2016;11:e0163770. doi: 10.1371/journal.pone.0163770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Malik AN, Czajka A. Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction? Mitochondrion. 2013;13:481–492. doi: 10.1016/j.mito.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 182.Anderson C, Fry RC, Hartwell H, Kleeberger C, Sandler DP, Nichols HB. Measurement of mitochondrial DNA copy number in dried blood spots: A pilot study. Mitochondrion. 2021;56:35–39. doi: 10.1016/j.mito.2020.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Parr RL, Dakubo GD, Thayer RE, McKenney K, Birch-Machin MA. Mitochondrial DNA as a potential tool for early cancer detection. Hum Genomics. 2006;2:252–257. doi: 10.1186/1479-7364-2-4-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Liu Y, Zhou K, Guo S, Wang Y, Ji X, Yuan Q, Su L, Guo X, Gu X, Xing J. NGS-based accurate and efficient detection of circulating cell-free mitochondrial DNA in cancer patients. Mol Ther Nucleic Acids. 2021;23:657–666. doi: 10.1016/j.omtn.2020.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.