Abstract
Mitochondrial DNA (mtDNA) encodes protein subunits and translational machinery required for oxidative phosphorylation (OXPHOS). Using repurposed whole-exome sequencing data, in the present study we demonstrate that pathogenic mtDNA mutations arise in tumours at a rate comparable to those in the most common cancer driver genes. We identify OXPHOS complexes as critical determinants shaping somatic mtDNA mutation patterns across tumour lineages. Loss-of-function mutations accumulate at an elevated rate specifically in complex I and often arise at specific homopolymeric hotspots. In contrast, complex V is depleted of all non-synonymous mutations, suggesting that impairment of ATP synthesis and mitochondrial membrane potential dissipation are under negative selection. Common truncating mutations and rarer missense alleles are both associated with a pan-lineage transcriptional programme, even in cancer types where mtDNA mutations are comparatively rare. Pathogenic mutations of mtDNA are associated with substantial increases in overall survival of colorectal cancer patients, demonstrating a clear functional relationship between genotype and phenotype. The mitochondrial genome is therefore frequently and functionally disrupted across many cancers, with major implications for patient stratification, prognosis and therapeutic development.
Somatic mutations are the underlying drivers of cancer, with the discovery and characterization of recurrent, functional somatic events constituting the capstone goal of cancer genomics. Genomic searches for recurrent driver mutations have focused on the nuclear exome, motivated by the concentration of recurrent mutations in the coding regions of a subset of nuclear DNA-encoded genes. This targeted approach has powered the discovery of common and rare driver mutations in exonic regions, but by corollary has left the overwhelming majority of the genome underexplored, and the driver events it may harbour unidentified. Numerous examples now exist of the prevalence and function of oncogenic mutations beyond the nuclear exome, including mutations to the TERT promoter, non-coding RNAs, including ribosomal (r)RNAs, small nuclear RNAs and enhancers1. A fundamental challenge therefore is to discover new functional somatic alterations beyond the nuclear exome with a fixed and limited sequencing capacity.
Somatic mutations in tumours commonly affect mtDNA2-6, arising in both protein-coding genes and non-coding RNA genes required for translation of essential, membrane-bound subunits of four complexes required for OXPHOS (Fig. 1a). Despite abundant pharmacological, genetic and clinical data demonstrating that perturbation of different OXPHOS complexes (referred to from here on as complexes) produce distinct cellular adaptations7,8, the importance of each complex in shaping mtDNA mutation patterns in cancer is unknown. As mtDNA is not commonly targeted by whole-exome sequencing (WES) panels, previous analyses of mtDNA mutations have relied on cohorts profiled by whole-genome sequencing (WGS), with consequently diminished statistical power to detect recurrent patterns of mutation relative to exome sequencing studies8. However, due to the extremely high copy number and off-target hybridization rate of mtDNA, mtDNA reads are abundant in widely available exome sequencing of tumours9. Therefore, mtDNA represents an opportunity for discovery through repurposing of existing exome sequencing data.
Fig. 1 ∣. MtDNA mutations are among the most frequent genomic alterations in cancer.
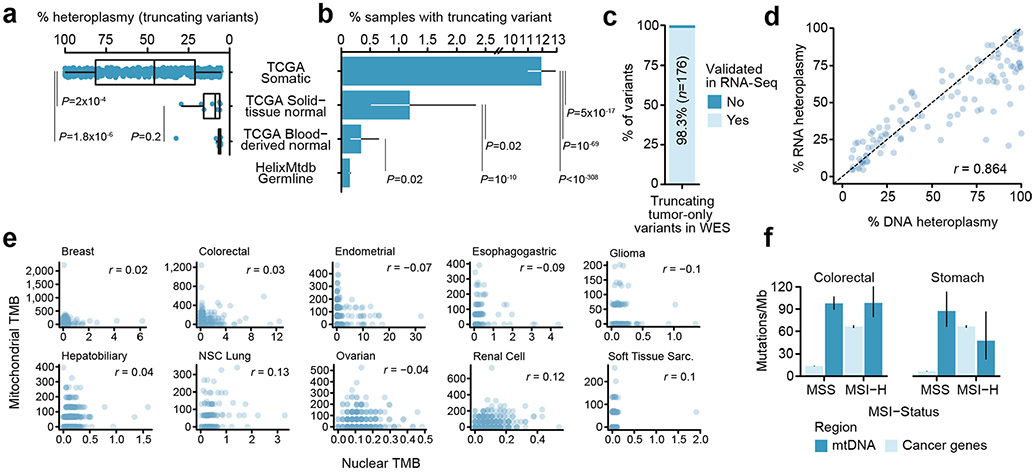
a, Schematic of OXPHOS system and project workflow. Top row: CI–CV and their reactions; centre row: mtDNA genomic regions encoding protein subunits of the associated OXPHOS complex; bottom row: overview of project workflow, in which somatic mutations in mtDNA genes are used to explore intercomplex differences, mutational recurrence and transcriptional phenotype associated with mitochondrial dysfunction. b, Average number of tumours with sufficient coverage to call variants at a mtDNA position. Truncating mutations were assumed to be somatic and therefore allowed for tumour-only variant calling (dark blue), whereas non-truncating (protein-coding, non-truncating tRNA and rRNA mutations) required sufficient coverage in both tumour and matched-normal samples (light blue). Grey shows the number of WGS samples from PCAWG for comparison. c, The percentages of variants called from off-target reads, which were validated in either RNA-seq or WGS data from the same tumours. d, The correlation between variant heteroplasmy as observed in RNA- and DNA-seq (n = 2,575 mutations with coverage ≥30 reads in both DNA and RNA). e, The correlation between TMB (mutations per Mb) among mtDNA (y axis) and nuclear-encoded, cancer-associated genes (referred to simply as cancer genes; x axis), (n = 3,624 well-covered pan-cancer tumours). f, Mutation rates (mutations per Mb) of individual mtDNA-encoded genes (blue) and nuclear-encoded, cancer-associated genes (grey). Inset plot: mutation rates among 504 genes with mtDNA genes highlighted. Outer plot: close-up of the inset plot in the region containing all 37 mtDNA genes; commonly mutated nuclear cancer genes in this region are labelled for reference. g, Comparison of truncating mutation rates (truncating variants per Mb) between 13 mtDNA-encoded protein-coding genes and 185 nuclear-encoded TSGs. The P value was from a two-sided, Wilcoxon’s rank-sum test. h, Same as in g but comparing non-truncating mutation rate (non-synonymous, non-truncating variants per Mb) between 13 mtDNA protein-coding genes and 168 nuclear oncogenes. i, Percentage of patients with truncating mtDNA variants either somatically (in TCGA tumour samples) or germline (among ~200,000 normal samples). Error bars are 95% binomial CIs; the P value is from a two-sided, two-sample z-test.
In the present study, we assessed the determinants, functional consequences and clinical outcomes associated with mtDNA mutations in cancer. We report three discoveries: first, we observe that respiratory complex is a fundamental determinant of the burden and functional consequence of tumour-associated mtDNA mutations. Complex I (CI, NADH:ubiquinone oxidoreductase) subunits are strongly enriched for highly pathogenic mutations in specific tissue lineages, whereas complex V (CV, ATP synthase) subunits are broadly depleted of all non-synonymous mutations. Complex III (CIII, ubiquinol:cytochrome c oxidoreductase) demonstrates increased rates of missense, but not truncating mutations. Second, we find that specific mutant alleles in mtDNA arise recurrently as hotspots, with six highly recurrent mtDNA mutation hotspots evident at specific homopolymeric loci encoding CI subunits, whereas rarer but recurrent mutations affect both protein-coding genes and RNA elements. Third, we report that specific mutant mtDNA alleles produce phenotypes of functional and clinical significance. Truncating mtDNA mutations are associated with a lineage-agnostic transcriptional programme implicating both metabolism and genes related to the immune response. Furthermore, in colorectal cancer, where both truncating and non-truncating mtDNA variants are common, we find that their presence is associated with superior clinical outcomes. These results argue that mitochondrial respiration is commonly and functionally perturbed in well-defined contexts across cancer, and that reanalysis of existing genomic data can yield new discoveries in underexplored genomic terrain.
Results
MtDNA mutations in tumours from off-target reads.
To study patterns of mtDNA mutations in tumours, we reasoned that the sheer amount of off-target reads aligning to mtDNA in WES data would be sufficient to call somatic mtDNA mutations in a large proportion of samples. To study mtDNA mutations in tumours with WES, we assembled a dataset of pan-cancer paired tumour and matched-normal exome sequencing samples from The Cancer Genome Atlas (TCGA; n = 10,132; Extended Data Fig. 1a). Individual cancer types varied widely in their mtDNA coverage from off-target reads, which we found to be driven by differences in the exome sequencing capture protocol implemented by different sequencing centres (Extended Data Fig. 1b). Such inconsistent sequencing coverage is an inherent limitation to mtDNA variant calling from exome sequencing, because variants located in regions without adequate sequencing coverage are not identifiable. We therefore developed a methodology to be cognizant of the sequencing coverage at each position in each sample (Methods), focusing our analysis on regions of mtDNA in protein-coding genes and genes coding for mitochondrial rRNAs and transfer (t)RNAs (Methods and Supplementary Table 1).
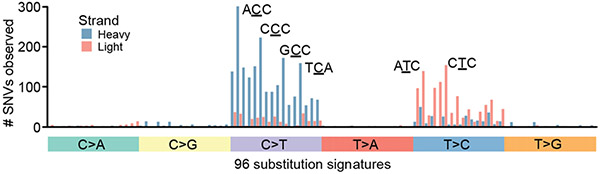
We implemented a variant-calling approach modelled after state-of-the-art methodologies for exome sequencing, in which we took the intersection of two variant callers (MuTect2 (ref. 10)) and an in-house variant caller based on the SAMtools mpileup utility11 (Methods). Variants in mtDNA exhibited a strand-specific enrichment for C>T mutations on the heavy strand and T>C mutations on the light strand (Extended Data Fig. 2). Based on 789 tumour samples from TCGA with whole-genome sequences in the Pan-Cancer Analysis of Whole Genomes (PCAWG) cohort3, 95.6% of mutation calls from WES were validated against published mutation calls from the PCAWG data (Fig. 1c). We also evaluated the possibility that nuclear-encoded mitochondrial pseudogenes (NUMTs) could corrupt variant calling. As NUMTs do not show evidence of appreciable transcription, unlike mtDNA genes12, we reasoned that recapitulating mtDNA variants in RNA-sequencing (RNA-seq) from the same sample would be evidence that they arose in mtDNA and not in NUMTs. Indeed, we found that 96.9% of variants in samples with both DNA-seq and RNA-seq were validated in RNA. In addition, we observed a strong correlation between DNA and RNA heteroplasmy overall (Pearson’s r = 0.918; Fig. 1d), confirming that the vast majority of observed mutations are expressed and providing further evidence that the mutations called by our approach are not attributable to NUMTs.
In total, we identified 4,381 mtDNA mutations from 10,132 tumour samples. Among a subset of 3,264 paired tumour/normal samples, with sufficient coverage to call mtDNA mutations in at least 90% of the mitochondrial genome (32% of tumour/normal pairs in our dataset overall, referred to throughout as ‘well-covered’ samples), 57% (95% confidence interval (CI) = 56–59%) had at least one mtDNA variant, in agreement with previous estimates for mtDNA mutation incidence in pan-cancer-sequencing data2. Consistent with independent mutagenic processes operating in the nuclear and mitochondrial genomes, we observed no correlation between nuclear and mitochondrial mutation burdens pan-cancer or within individual cancer types (Fig. 1e and Extended Data Fig. 3e). Furthermore, in colorectal and stomach cancers where microsatellite instability (MSI) is common, the presence of MSI affected mutation burden in the nuclear but not the mitochondrial genome (Extended Data Fig. 3f). Mitochondrial tumour mutation burden (TMB) was positively correlated with patient age at the time of diagnosis in multiple cancer types, including leukaemia, endometrial and renal cell cancers, and soft-tissue sarcomas; however, no correlation was observed with tumour pathological stage (Extended Data Fig. 4a,b).
The mutation rate in the coding region of mtDNA is roughly 67.8 mutations per Mb, roughly sixfold higher than the rate in 468 cancer-associated genes in the Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK–IMPACT) panel13 of 11.3 mutations per Mb (P < 10−308 (computational limit of detection), two-sided Poisson’s test). Only two genes (TP53, KRAS) exhibited rates higher than that of the most mutated mtDNA-encoded genes (Fig. 1f). Furthermore, the 13 protein-coding mtDNA genes exhibited a 4.2-fold higher rate of truncating variants that disrupt the reading frame (that is, nonsense mutations and frameshift insertions and deletions (indels)) compared with truncating mutations among 185 known tumour-suppressor genes (TSGs) in the MSK–IMPACT panel (P = 9 × 10−5, two-sided Wilcoxon rank-sum test; Fig. 1g), and a 6.7-fold higher rate of non-truncating, non-synonymous than 168 MSK–IMPACT oncogenes (P = 6 × 10−9; Fig. 1h). Notably, considering only variants with exceptionally high heteroplasmies of >80%, MT-ND4 and MT-ND5 exhibited truncating mutation rates of ~5 mutations per Mb, comparable to or exceeding that of most TSGs.
In total, 11.9% of tumours across all cancers (95% CI = 11.0–12.9%) harboured a truncating mtDNA variant absent in the patient’s matched-normal sample. In contrast, only 0.15% of normal blood samples exhibited a truncating variant (95% CI = 0.13–0.17%) based on a recent analysis of ~200,000 mtDNA genomes14 (Fig. 1i). The rate of truncating mutations in mtDNA genes in tumours therefore represents an 80-fold increase compared with truncating mutations observed in normal human genomes (Supplementary Table 2). Of the 619 truncating mutations we observed, 196 (32%, 95% binomial CI = 28–35%) had >80% heteroplasmy despite underlying tumour impurity, indicating that systemic mitochondrial dysfunction is a common feature of tumours. Furthermore, high-heteroplasmy truncating variants were significantly more common than high-heteroplasmy silent mutations, expected to be generally subject to neutral selection (139/555, 25%, 95% CI = 21–29%; P = 0.01, two-sided Fisher’s exact test).
Truncating mutations preferentially target CI at homopolymeric hotspots.
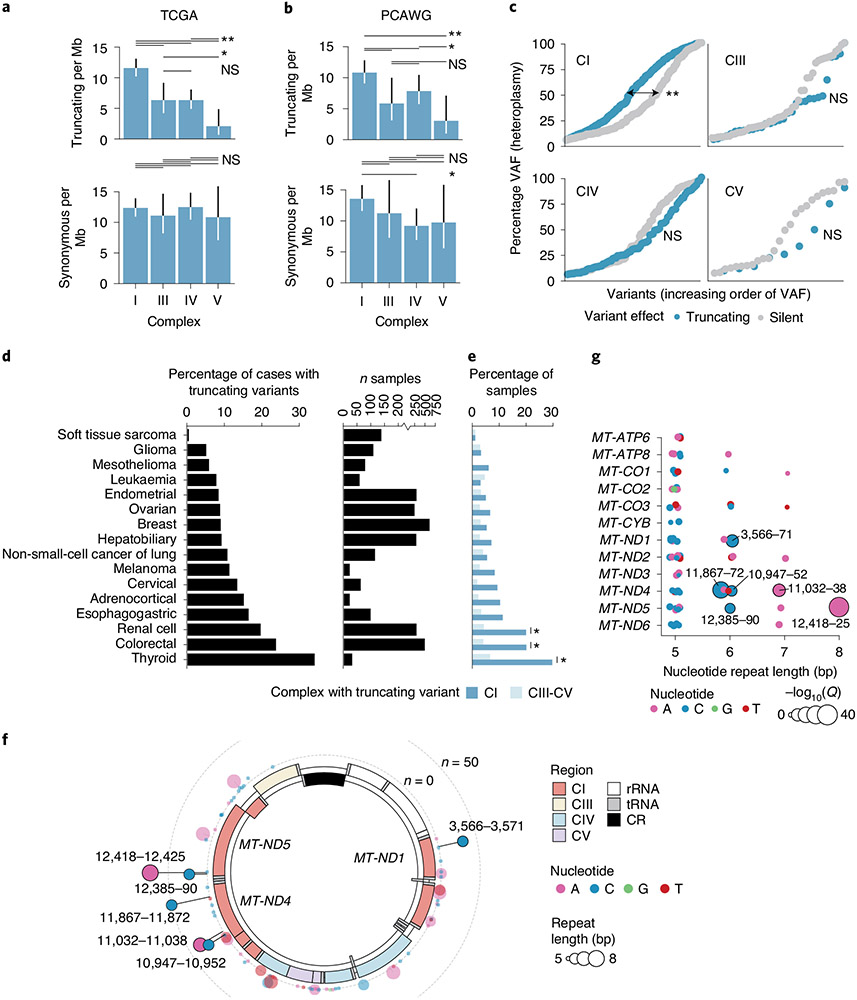
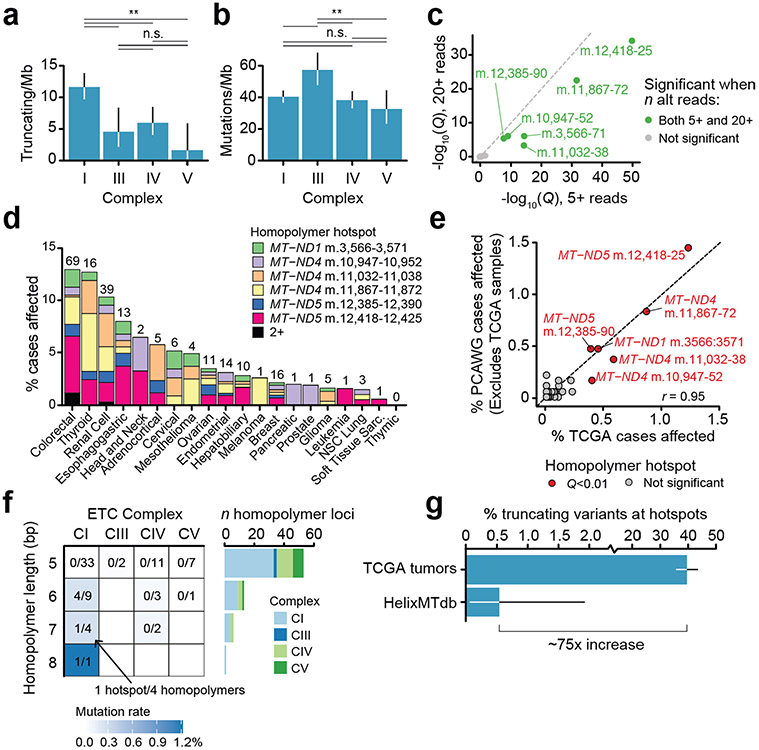
The physiological response to genetic or pharmacological inhibition of mitochondrial respiration is determined by which mtDNA-encoded complex is disrupted, implicating OXPHOS complex as a potential determinant of selective pressure for mutation. We therefore investigated the somatic mutation rate by complex, controlling for the relative length of mtDNA coding for genes in each complex and uneven coverage within each sequenced sample. Truncating variants arose at a twofold or greater rate in CI relative to the other complexes (P = 0.001 for least significant comparison, two-sided Poisson’s test; Fig. 2a). No difference in mutation rate between complexes was observed for silent mutations (P = 0.5 for the most significant comparison). Unlike variants in other complexes, truncating variants in CI demonstrated higher heteroplasmy than silent variants (P = 1 × 10−6, CI; most significant for other complexes, P = 0.4, two-sided Wilcoxon’s rank-sum test), suggestive of specific positive selective pressure for truncating variants in CI subunits (Fig. 2c). Finally, CV genes (MT-ATP6 and MT-ATP8) demonstrated significantly lower rates of truncating but not synonymous mutations. The findings above were recapitulated in n = 1,951 tumours from the PCAWG WGS dataset, after excluding samples overlapping with our own cohort (Fig. 2b), and were recapitulated with more stringent mutation-calling thresholds (Extended Data Fig. 5a).
Fig. 2 ∣. Truncating variants preferentially target CI.
a, Comparison of truncating mutation rate (truncating variants per Mb) across OXPHOS complexes CI, CIII, CIV and CV. Synonymous mutation rates are shown below for comparison (truncating mutations, n = 352; synonymous mutations, n = 475). The P values are from two-sided Poisson’s exact test. *P < 0.1; **P < 0.01; NS, not significant. b, Validation of analysis in a using data from n = 1,951 WGS tumours from ICGC/PCAWG after removing samples that are also in TCGA (truncating mutations, n = 198; synonymous mutations, n = 263; P values as in a). c, Distributions of truncating and silent mutation heteroplasmy (estimated by VAF) among variants in OXPHOS CI, CIII, CIV or CV. The difference in heteroplasmy between truncating and silent mutations is calculated by two-sided Wilcoxon’s rank-sum test (CI, P = 1 × 10−6, not significant for other complexes). d, Percentage of tumours with truncating mtDNA variants per cancer type, among well-covered samples. Right: number of well-covered samples per cancer type. e, Percentage of samples per cancer type with truncating variants affecting OXPHOS CI or CIII–CV. The asterisk indicates cancer types with enriched truncating variants targeting CI compared with CIII–CV (Q < 0.01, two-sided McNemar’s test). f, Circular mtDNA genome annotated with 73 homopolymer repeat loci ≥5 bp in length. Dot height from the circular mtDNA genome indicates the number of affected samples, dot colour indicates the identity of the repeated nucleotide (A, C, G, T) and dot width indicates the length of the repeat region (5–8 bp). It includes putatively somatic truncating variants with tumour-only sequencing coverage. The six solid-colour homopolymer loci highlighted were found to be statistically enriched hotspots for frameshift indels in tumours. g, The 73 homopolymer repeat loci arranged by gene and repeat size. Dot width indicates −log10(Q value) for enriched frameshift indels in tumours. The six hotspot loci are labelled.
Tumours of different lineages exhibited wide variability in the incidence of truncating mutations, with ≤5% of some cancer types affected by truncating mutations (sarcomas, gliomas), to ≥20% of other cancer types (renal cell, colorectal, thyroid) in a manner that is consistent with previous work3 (Fig. 2d). In renal, thyroid and colorectal cancers, the truncating variant burden was defined by specific enrichment for mutations to CI but not other complexes (Q value < 0.01, two-sided McNemar’s test; Fig. 2e). Truncating variants in these three cancers affected ~20–30% of all samples, corresponding to a prevalence akin to common tumour suppressors in these diseases. Taken together with Fig. 2a-c, these data point to lineage-specific positive selective pressure for CI loss-of-function variants, and suggest that selection against disruption of CV, which could irreparably impair mitochondrial ATP production, cristae morphology and dissipation of membrane potential, is not tolerated. These findings indicate that the functional consequence of mtDNA variants is a key determinant of somatic mtDNA mutational patterns.
Unexpectedly, we observed that truncating mutations frequently arose at the same genomic locus, analogous to hotspot mutations that accumulate in cancer driver genes and often reflect selective pressure15,16. These recurrent alleles were exclusively indels characterized by a homopolymeric sequence context. We therefore developed an approach to detect recurrent mutations at homopolymeric loci by modelling incidence of frameshift indels at each locus as a function of their base-pair length (Methods). Six single-nucleotide repeat loci (out of 73 loci of ≥5 bp in length) in MT-ND1 (m.3566–3571, n = 32), MT-ND4 (m.10947–10952, n = 25; m.11032–11038, n = 34; and m.11867–11872, n = 50) and MT-ND5 (m.12385–12390, n = 23 and m.12418–12425, n = 73) accumulated mutations at a rate above null expectation (Q value < 0.01, Fig. 2f). Homopolymer hotspots arose only at single-nucleotide loci of at least 6 nt in length (P = 0.0002, two-sided Fisher’s exact test), which were composed of A or C homopolymer repeats, and exclusively encoded subunits of CI. Importantly, other homopolymers of equivalent length (≥6) and nucleotide content exist both in CI and in CIII/CIV/CV but did not exhibit recurrent mutations, indicating a high degree of specificity to hotspot positions (Fig. 2g and Extended Data Fig. 5f). These six homopolymeric repeat loci collectively accounted for 40% of all truncating variants observed in our data (95% binomial CI = 36–44%) and 57% (95% CI = 52–62%) of frameshift indels overall, and were a pervasive phenomenon across tumour lineages (Extended Data Fig. 5d). These hotspots overlapped with 100-bp-long windows previously reported to be enriched for frameshift mutations17 and with indels in rare, often benign, renal oncocytomas18. Homopolymeric hotspot mutations arose in the PCAWG WGS cohort (after excluding any samples overlapping with our cohort) at a rate highly consistent with TCGA cohort (Pearson’s r=0.95), and were ~75-fold more common in TCGA tumour samples than in the HelixMTdb database of 200,000 saliva-derived normal samples (Extended Data Fig. 5g), indicating that the indels detected in TCGA at hotspot loci were not artefacts due to calling variants in microsatellite regions with poor coverage. To further evaluate the recurrence of homopolymeric hotspots, we studied an independent cohort of 34,052 tumour samples from 30,575 patients with advanced and heavily treated pan-cancer tumours profiled by the MSK–IMPACT targeted sequencing platform13,19. We observed that five of six hotspots from TCGA were also significantly enriched for indels in the MSK–IMPACT dataset. This analysis revealed an additional two homopolymers with significantly enriched indels unique to MSK–IMPACT samples (Extended Data Fig. 6a), including one in CIV (MT-CO3, m.9532–9537), which arose in characteristically different cancer types (for example, prostate and non-small-cell lung cancer) than the CI hotspots that mainly arose in kidney, colorectal and thyroid cancers (Extended Data Fig. 6b). Although mutations at homopolymeric tracts have not been widely described in the germline literature, the most recurrent hotspot (MT-ND5 m.12418–12425) has been previously reported as the site of a germline frameshift deletion (A12425del) in a mitochondrial disease patient20.
Non-synonymous and RNA variants arise as rare recurrent pathogenic alleles.
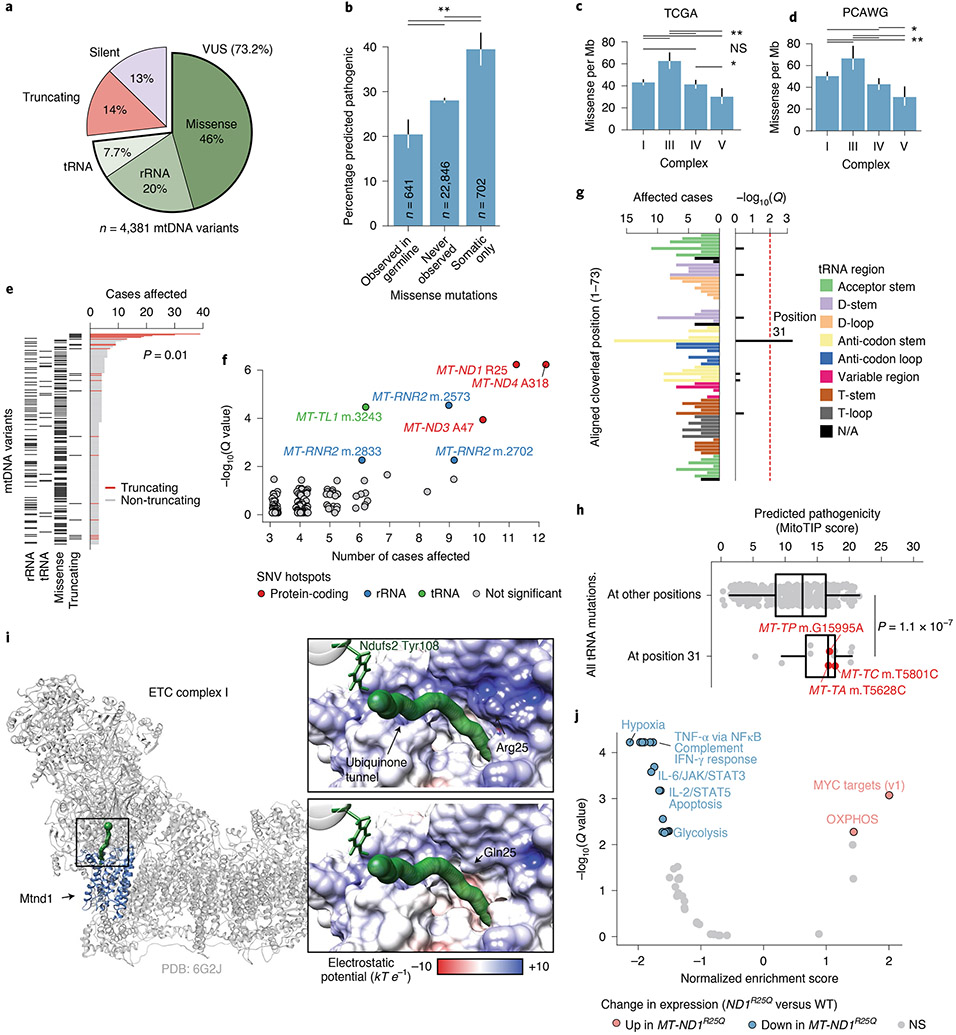
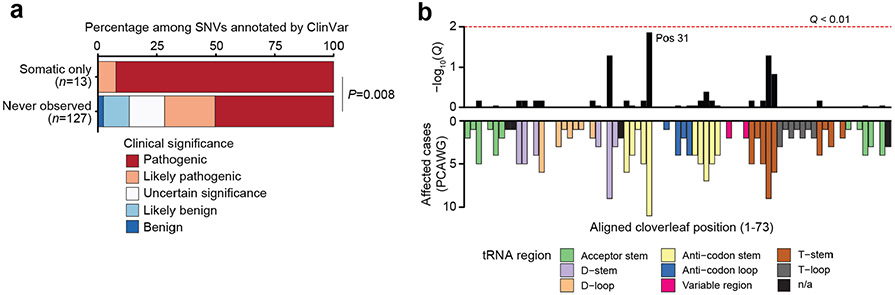
The bulk of somatic variants in mtDNA were non-truncating, non-synonymous mutations, including tRNA/rRNA mutations, missense mutations, in-frame indels, translation start site mutations and non-stop mutations (collectively referred to as variants of unknown significance (VUSs), 73.2% of n = 4,381 variants, Fig. 3a). Somatic VUSs were twice as likely to be predicted pathogenic compared with germline polymorphisms observed among ~200,000 normal samples from the HelixMTdb dataset (APOGEE21 score, 39.5% of somatic-only variants compared with 20.4% of germline-arising; P=6 × 10−14, two-sided Wilcoxon’s rank-sum test; Fig. 3b). In addition VUSs that arose only as somatic mutations in tumours were predicted to be more pathogenic than those that never arose in tumours (P = 5 × 10−11). Finally, considering only the subset of possible somatic-only single-nucleotide variants (SNVs) with annotated clinical significance in ClinVar22, somatic-only VUSs were annotated as having significantly elevated pathogenicity compared to that of those never observed in tumours (P = 0.008, two-sided Cochran–Armitage trend test; Extended Data Fig. 7a). Together, these data suggest that somatic VUSs exhibit elevated pathogenicity relative to null expectation.
Fig. 3 ∣. Non-truncating mtDNA mutations arise as rare recurrent alleles in protein-coding and RNA elements.
a, The proportion of truncating, synonymous and VUS somatic mtDNA mutations in the present study (VUSs further classified by gene type). b, The percentage of unique VUSs predicted to be pathogenic by APOGEE21, among variants that: (1) were ever germline variants among ~200,000 normal samples from HelixMTdb; (2) were never observed somatically mutated in tumours; or (3) were observed only as somatic mutations (P values from two-sided, two-sample z-tests (top to bottom): 5 × 10−11, 6 × 10−14, 3 × 10−5). c, Comparison of missense mutation rate across OXPHOS CI, CIII, CIV and CV (n = 1,718 missense mutations; P values from two-sided Poisson’s exact tests; *P < 0.1; **P < 0.01; NS, not significant). d, Validation of a among n = 1,951 WGS tumours from ICGC/PCAWG after removing samples also in TCGA (n = 1,073 missense mutations; P values and asterisks as in a). e, Top: number of samples with each unique mutation arising in three or more tumours. Bottom: variant consequence. f, Individual positions in mtDNA with SNVs in three or more tumours, and their enrichment (statistical test described in Methods). Positions with Q < 0.01 are coloured by gene type. g, The number of samples with SNVs at the equivalent position of the tRNA’s folded-cloverleaf structure across all tRNAs (left), and the position’s statistical enrichment (right; statistical test described in Methods). N/A, not available. h, Predicted pathogenicity (based on MitoTIP50) of position 31 variants compared with all possible mutations at other positions (only 5% are shown to reduce image size). Variants affecting three or more tumours are highlighted (P value from a two-sided Wilcoxon’s rank-sum test calculated using all mutations). i, The structure of mammalian CI (grey) highlighting Mtnd1 (blue), and the ubiquinone-binding tunnel (green); the black box indicates a close-up region. Close-ups: the predicted surface electrostatic potential of Mtnd1 wild-type (top) and Arg25Gln mutant (bottom) samples, leading to its binding site at Ndufs2 Tyr108. j, Differentially expressed mSigDB Hallmark genesets between colorectal tumours with MT-ND1R25Q and those without non-silent somatic mtDNA variants (that is, wild-type (WT)). Normalized enrichment score and adjusted P values based on gene set enrichment analysis using the fgsea R package55. IFN, interferon.
We next evaluated the tendency for VUSs to target specific complexes of the OXPHOS system. In contrast to truncating variants, protein-coding VUSs were most frequent in CIII (P = 1 × 10−7 for the least significant comparison, two-sided Poisson’s test; Fig. 3c), the functional integrity of which as a site for ubiquinol oxidation has recently been described as essential for tumour cell proliferation23. Consistent with the pattern in truncating variants, VUSs to CV subunits were still depleted compared with the other complexes (P = 0.01 for least significant comparison). These observations were validated using data from the PCAWG (Fig. 3d) and were robust to a more conservative read-support threshold for variant calling (Extended Data Fig. 5b). Together, these findings suggest that tumours preferentially accumulate somatic missense mtDNA mutations in a manner dictated by the OXPHOS complex, possibly driven by their capacity to disrupt mitochondrial function. Furthermore, they support the hypothesis for purifying selection against variants that compromise the physiological functions of CV.
Specific alleles produced by SNVs were far less recurrent than homopolymer indels (P = 0.01, two-sided Wilcoxon’s rank-sum test among distinct variants mutated in three or more tumours; Fig. 3e). However, we still observed a number of loci with weakly recurrent non-truncating variants. We developed a statistical test for recurrence of these loci that controls for coverage and mutation sequence context, identifying seven SNV hotspots in the mitochondrial genome (Q < 0.01; Fig. 3f), including three in protein-coding genes (all in CI), three in rRNA (all in MT-RNR2) and one in a tRNA (MT-TL1; see Methods). In contrast to the high fraction of truncating mutations that are explained by a relatively small number of hotspot alleles, hotspot SNV mutations accounted for only 1.6% of all VUSs; the vast majority of VUSs were non-recurrent, usually arising in a single sample. Furthermore, 0 of 33 mutations arising at the three protein-coding hotspot positions identified was a nonsense mutation, introducing an early stop codon, suggesting that either the mutagenic mechanism generating homopolymeric indel hotspots has a high degree of specificity (for example, replicative slippage) or truncating hotspots themselves may engender unique phenotypes beyond conventional loss of function.
Mitochondrial tRNAs (mt-tRNAs) are commonly mutated in the context of germline mitochondrial disease. It is of interest that the somatic hotspot m.3243 A>G in MT-TL1 (somatically mutated in six patients) is also the causative variant of approximately 30% of all mtDNA disease24,25. We additionally observed mutations clustered in adjacent positions m.3242 (n = 5) and m.3244 (n = 4, recently described as a recurrent mutation in Hürthle cell carcinoma of the thyroid26), suggesting that recurrent mutations in MT-TL1 could affect a common secondary structure element required for either tRNA processing or regulation of transcription via mTERF1 binding27. The mt-tRNAs, with the exception of mt-tRNASer(AGY), adopt a relatively conserved cloverleaf structure on folding, and mutations to mt-tRNAs are known to disrupt specific secondary structure elements with downstream impacts on, for example, stability or amino acid charging. We therefore statistically tested each position of the aligned canonical mt-tRNA structure for enriched somatic mutations (Methods). This analysis identified position 31 in the anti-codon stem of the folded tRNA molecule as a site of recurrent mutation across mt-tRNAs (Q = 4.7 × 10−4; Fig. 3g), which we further validated using the non-TCGA subset of PCAWG samples (Q = 0.014, Extended Data Fig. 7b). It is interesting that position 31 was observed to be mutated at an eightfold higher rate in tRNAs encoded on the light strand (for example, MT-TC, n = 5; MT-TP, n = 4; MT-TA, n = 3) compared with heavy-strand-encoded tRNAs (P = 2 × 10−4; two-sided Fisher’s exact test). As a group, mutations at structural position 31 were predicted to be more pathogenic by MitoTIP relative to mutations at other tRNA positions (Fig. 3h), and in the case of m.5628 T>C in MT-TA (n = 3) are associated with the mitochondrial disease chronic progressive external ophthalmoplegia28. These data suggest that specific structural features of mt-tRNAs may undergo recurrent mutation and impair mitochondrial function.
To understand the potential function of rare protein-coding SNV hotspots in mtDNA, we focused on a recurrent mutation at MT-ND1R25, which was identified somatically in 11/10,132 TCGA patients (0.11%) and 5/2,836 PCAWG patients (0.18%). All 16 instances resulted in a substitution of arginine with glutamine, encoded by a G>A substitution at position m.3380. MT-ND1R25Q was previously described in a case report as the causative variant in the development of severe mitochondrial disease29. Consistent with its pathogenicity, the Arg25Gln variant was never observed as a germline polymorphism among ~200,000 normal samples in the HelixMTdb database, where the mutant alleles at residue Arg25 always produced synonymous mutations (m.3381 A>G, n = 57). Residue Arg25 is conserved across vertebrates29, and is part of a cluster of charged residues in CI that form a structural bottleneck in the ubiquinone-binding tunnel leading to the binding site30. We therefore modelled the effect of MT-ND1R25Q using a recent, high-resolution structure of mammalian CI, which revealed gross changes to the local charge environment due to loss of the relatively bulky, positively charged arginine side chain. Due to the location of this substitution within the Q-binding tunnel, this is predicted to substantially impact function (Fig. 3i). Focusing on colorectal cancer, which demonstrated the largest numbers of tumours harbouring MT-ND1R25Q (n = 8), we examined whether the presence of MT-ND1R25Q was associated with a particular transcriptional signature. Relative to mtDNA wild-type tumours, we observed that MT-ND1R25Q tumours were characterized by upregulation of MYC targets and OXPHOS genes, and downregulation of gene signatures associated with hypoxia, interleukin (IL)-2/STAT5 signalling, tumour necrosis factor α (TNF-α) signalling via nuclear factor κ-light-chain-enhancer of activated B cells (NFκB; Fig. 3j). These data suggest that MT-ND1R25Q promotes a transcriptional phenotype characterized by increased mitochondrial metabolism and suppressed expression of innate immune genes.
Mitochondrial genotype underlies a lineage-agnostic transcriptional programme.
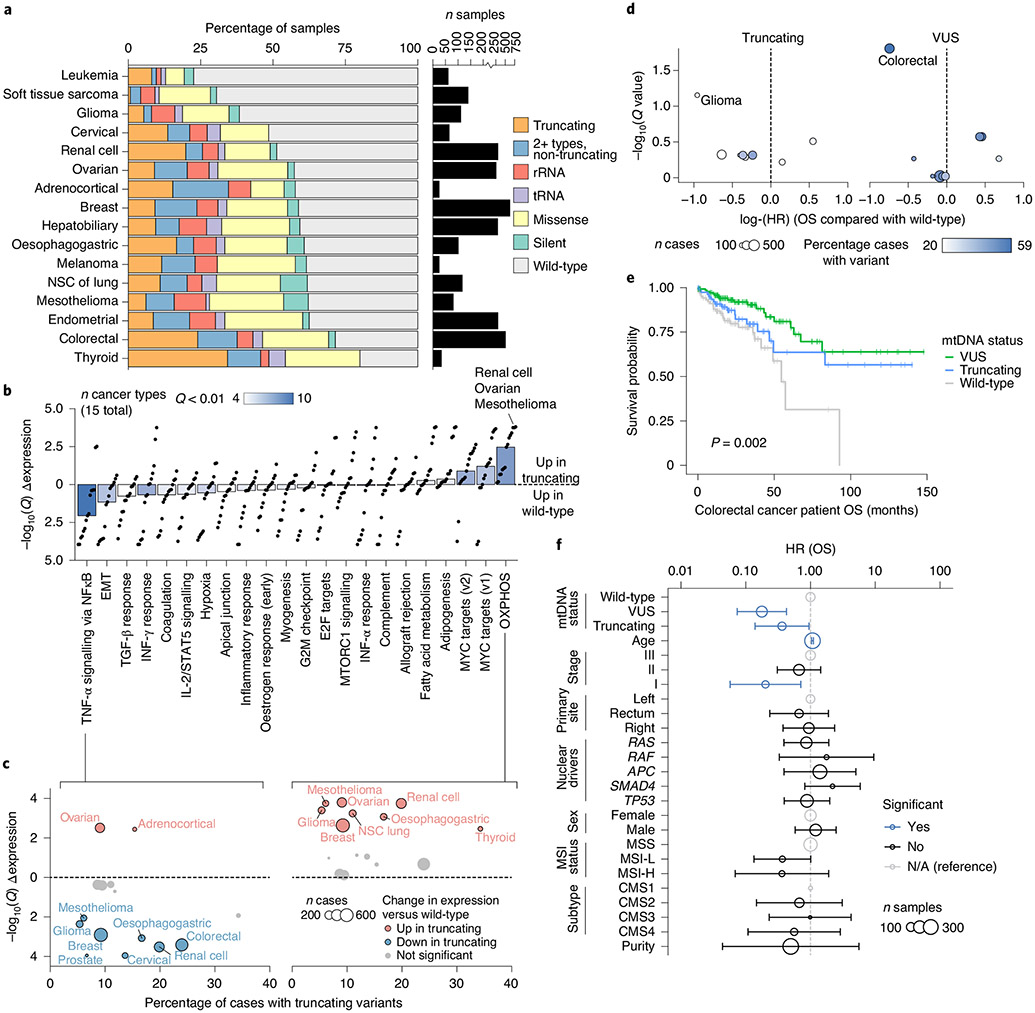
Given the lineage specificity underlying both truncating variants and truncating/SNV hotspots, we studied the overall burden of distinct classes of mtDNA variants (that is, producing a truncating, missense, synonymous tRNA or rRNA variant) across cancer types. Restricting our analysis to well-covered samples (and in addition requiring coverage of all homopolymeric hotspots; see Methods), we found that the fraction of mutant samples across cancer types ranged from approximately 23% of leukaemias (95% binomial CI = 13–35%) to as high as 80% of thyroid cancers (95% CI = 63–92%; Fig. 4a). There was no correlation between the fraction of well-covered samples in a cancer type and the proportion of samples with a somatic mtDNA mutation (Extended Data Fig. 1d), indicating that the highly variable incidence of different somatic variants across cancer types was not biased by their differing coverage.
Fig. 4 ∣. Mitochondrial genotypes associated with transcriptional and clinical phenotypes.
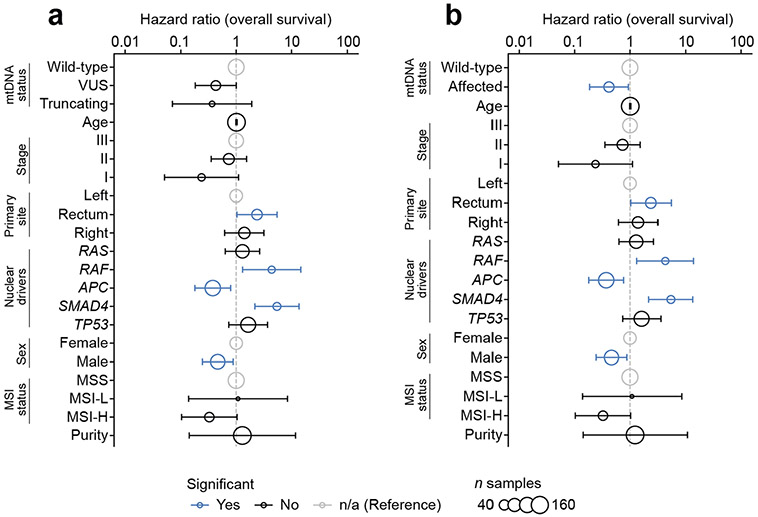
a, Percentage of well-covered tumours with different types of somatic mtDNA variants per cancer type. Right: number of well-covered samples per cancer type. NSC, non-small-cell cancer. b, Differential expression of mSigDB Hallmarks genesets, between samples with truncating mtDNA variants and those with no non-synonymous somatic mutations (that is, wild-type samples). Differential expression is quantified by directional −log10(Q value): >0 denotes upregulation in samples with truncating variants; <0 denotes downregulation. Each dot is a single cancer type’s level of dysregulation. Bars show the median level of dysregulation across 15 cancer types; bar shading shows the number of cancer types with significant dysregulation (Q < 0.01) in either direction. IFN, interferon; TGF, transforming growth factor. c, Differential expression of TNF-α via NFκB signalling (left) and OXPHOS (right) genesets in individual cancer types. The x axis: percentage of samples with truncating variants; y axis matches the y axis in b. Dot width denotes number of well-covered samples. d, Effect size and statistical significance of mtDNA truncating variants (left) and VUSs (right) on OS among individual cancer types. Effect sizes (quantified as log(hazard ratios)) are from univariate Cox proportional hazards models run for each cancer type independently. The Q values are adjusted P values from the model coefficients for each cancer type. e, Kaplan–Meier plot showing difference in OS among n = 344 TCGA colorectal cancer patients with somatic VUSs (n = 152), truncating variants (n = 84) or no non-synonymous mutations (that is, wild-type, n = 108). f, Multivariate analysis of the effect of mtDNA variants on OS among n = 344 TCGA colorectal cancer patients (stages 1–3). Truncating variants and VUSs are each compared with wild-type samples, while controlling for known prognostic clinical and genomic covariates using Cox proportional hazards model. Hazard ratios are shown on a log scale and error bars are 95% CIs from Cox proportional hazards regression. Point size indicates the number of samples with the associated covariate value (except for age, which was coded as a continuous variable, and therefore the size corresponds to the total number of samples). Blue points are statistically significant (P < 0.05); black points are not significant; grey points are reference categories. EMT, epithelial-to-mesenchymal transition.
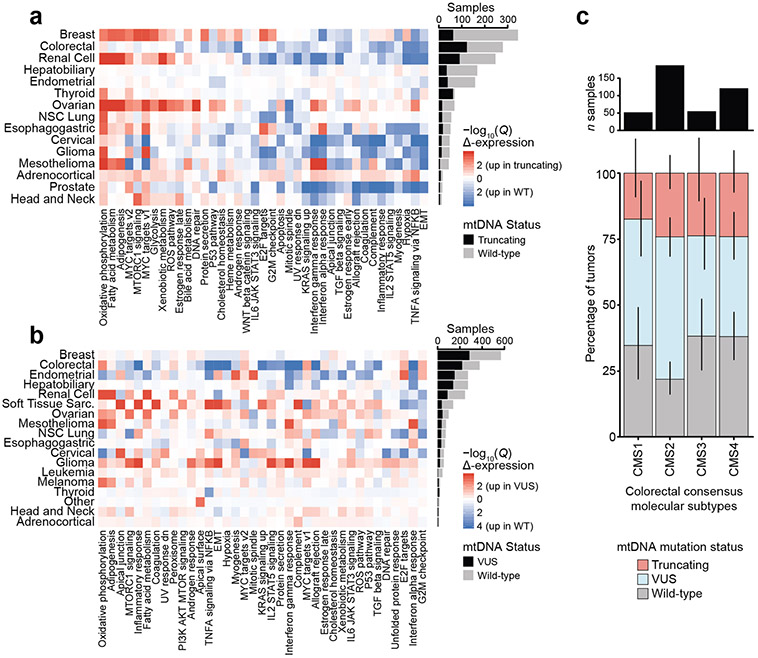
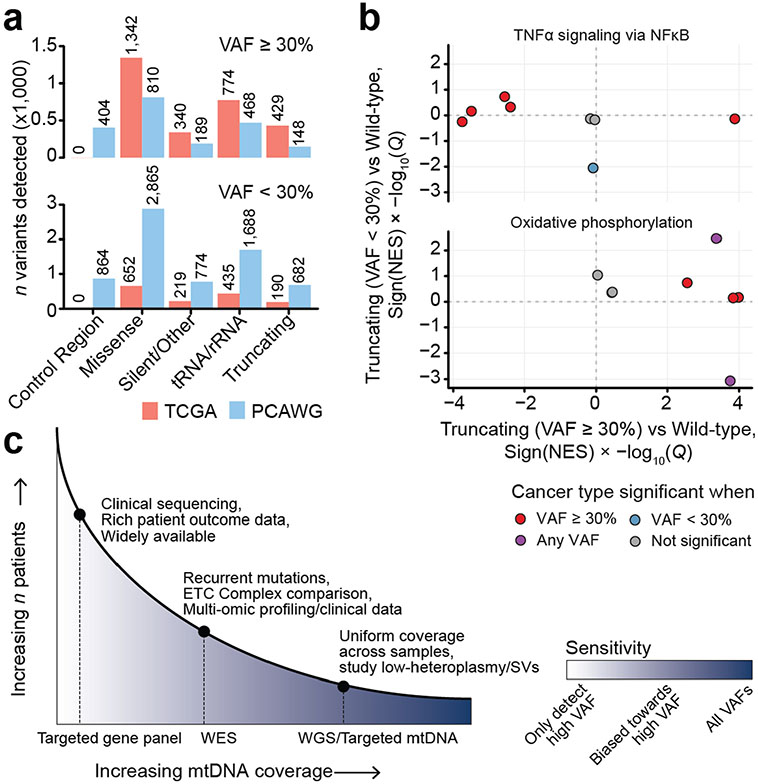
Truncating mtDNA mutations approaching homoplasmy (>80% heteroplasmy) were identified in nearly all cancer types, suggesting that even cancers in which mtDNA mutations are uncommon may still contain rare instances of individual tumours with highly mutant mitochondria. In renal and thyroid tumours, truncating mutations are known to induce oncocytic neoplasia, whereby tumour cells accumulate dysfunctional mitochondria31,32. We therefore sought to evaluate whether truncating mutations induced functionally similar consequences across different tumour lineages, by comparing the gene expression profiles of tumour samples with truncating mtDNA variants with tumour samples with wild-type mtDNA (harbouring no non-synonymous somatic mutations in protein-coding or RNA genes; see Methods). In half of all cancer types, tumours harbouring truncating mutations exhibited a conserved expression programme characterized by upregulation of genes associated with OXPHOS and downregulation of genes associated with TNF-α via NFκB signalling (Fig. 4b and Extended Data Fig. 8a) in a manner that was robust to variation in tumour purity. Critically, these expression programmes were evident in cancer types such as glioma and mesothelioma, where the proportion of samples with a truncating variant was comparatively low (Fig. 4c). We then evaluated the degree to which this observation was dependent on heteroplasmy, by repeating the analysis using only truncating variants with variant allele frequency (VAF) ≥30% compared with wild-type tumours, or VAF < 30% compared with wild-type. This revealed that higher-VAF truncating variants were more commonly associated with a change in expression of genes related to OXPHOS and TNF-α via NFκB compared with low-VAF variants (Extended Data Fig. 10b), suggesting the presence of a dosage effect by which an increase in the proportion of mitochondria in cells with pathogenic variants increases the transcriptional dysregulation observed in bulk tissue.
Given that the hotspot MT-ND1R25Q exhibited an expression programme resembling truncating variants, we investigated the generic transcriptional consequences of mtDNA VUSs (Methods). Compared with truncating variants, fewer genesets demonstrated lineage-agnostic changes in samples with VUSs. As with truncating variants, the most upregulated gene set in VUS-harbouring tumours was OXPHOS (increased in 5/18 cancer types; Extended Data Fig. 8b), but the magnitude of this enrichment was attenuated relative to truncating variants. Notably, several cancer types, such as colorectal cancer, demonstrated a lineage-specific pattern of gene expression changes, suggesting that mtDNA VUSs are capable of eliciting a phenotype in specific cancer types.
To examine the translational value of mtDNA genotype, we determined the association between mtDNA mutation status and clinical outcome (overall survival (OS)) across cancer types. Using univariate Cox proportional hazards regression, for each cancer type we determined the effect size and significance of both mtDNA truncating variants and VUSs compared with samples with no somatic mtDNA variants (wild-type). Colorectal cancer demonstrated the largest (by effect size) significant association between OS time and mtDNA genotype (colorectal patients with VUSs had a hazard ratio (HR) of 0.47 (95% CI = 0.03–0.75)) compared with those with wild-type mtDNA (Q value = 0.02, Cox proportional hazards regression; Fig. 4d). Notably, VUSs in colorectal cancer are also associated with a unique transcriptional downregulation of multiple genesets including TNF-α via NFκB, hypoxia and complement (Fig. 3j and Extended Data Fig. 8b), further suggesting a cryptic phenotype of these variants in affected tumours. We additionally observed a weak association between mitochondrial genotype and underlying molecular subtype33, with some enrichment of mtDNA mutations in the canonical consensus molecular subtype (CMS) 2 of colorectal tumours (Extended Data Fig. 8c).
We therefore further evaluated whether mtDNA mutations may be prognostically meaningful in colorectal cancer. Among 344 stage I-III colorectal cancer patients in the TCGA, we found mtDNA VUSs to correlate with improved OS compared with wild-type (truncating variants had an intermediate effect), based on both a univariate analysis (P = 0.002, Kaplan–Meier test; Fig. 4e) and a multivariate test controlling for clinically relevant prognostic covariates33,34 (VUS: HR = 0.18, 95% CI = 0.07–0.43, P = 1 × 10−4; truncating: HR = 0.36, 95% CI = 0.14–0.95; P = 0.04, Cox proportional hazards regression; Fig. 4f). This finding was validated in an independent set of 172 well-covered stage I–III colorectal cancer patients from the MSK–IMPACT cohort34, controlling for the same set of covariates (excluding CMS, which was not available): compared with patients with no somatic mtDNA variants, those affected with either VUS or truncating variants had improved OS (HR = 0.42; 95% CI = 19–93%; P = 0.03; Extended Data Fig. 9b). As individual categories, VUSs and truncating variants trended towards improved OS compared with wild-type, but did not reach statistical significance (VUS: HR = 0.43; 95% CI = 0.18–1.00; P = 0.05; truncating: HR = 0.37; 95% CI = 0.07-1.9; P = 0.2; Extended Data Fig. 9a). Taken together with emerging data on the proliferative consequences of age-associated mtDNA mutations in colonic crypts35, these findings suggest that somatic mtDNA mutations are associated with a clinically and biologically distinct class of colorectal tumours.
Discussion
Although recent evolutionary data suggest that mtDNA mutations may be under positive selection in cancers of the kidney and thyroid5, the broader relevance of somatic mtDNA mutations in cancer remains a point of confusion and debate. Drawing inspiration from analyses describing hotspots of somatic mutations in the nuclear DNA of tumours, we studied the recurrence of mutant mtDNA alleles. The discovery that the OXPHOS complex shapes mtDNA mutation patterns in a manner that produces mutation hotspots, in connection with orthogonal data on the structural consequences, transcriptomic effects and clinical importance of these alleles in patients with germline mtDNA disease, supports the hypothesis that mitochondrial respiration is the target of mutations across many tumours.
Our results indicate that the OXPHOS complex, tissue lineage and mutation consequence collectively shape the incidence and putative function of mtDNA mutations. We find that truncating mutations preferentially impact CI and non-synonymous mutations of all classes are depleted in CV. Furthermore, the apparent selection for CI mutations is specific to thyroid, kidney and colorectal tumour lineages. This suggests that cancer cells in these lineages can better tolerate, or perhaps even utilize, loss of CI and associated metabolic consequences (for example, NAD+:NADH changes), whereas loss of capacity for ATP synthesis, membrane potential dissipation and/or cristae morphology through CV mutations appears to be universally selected against. That CIII demonstrates elevated rates (relative to other complexes) of missense mutations, but not truncating mutations, is consistent with its essential role in ubiquinol oxidation, suggesting that mild disruption of CIII is preferential for clonal expansion in tumour cells23. Broad metabolic plasticity, permitting anabolism in the presence of loss-of-function mutations to various nuclear DNA-encoded tricarboxylic acid cycle enzymes and other metabolic challenges has been described previously in several cancer lineages36-38. Whether the mtDNA mutations described in this work result in similarly plastic metabolic outcomes is a major outstanding question, with the potential for development of rational, targeted therapies39. For example, it is probable that an altered cellular redox balance caused by CI loss-mediated decreases in NAD+:NADH ratio will affect cellular metabolism and phenotype differently to CIII loss-mediated changes in Q/cytochrome c redox status and reactive oxygen species signalling23,40.
There is substantial evidence that, in particular subtypes of thyroid and kidney cancer, mtDNA mutations are the root cause of metabolic adaptations and morphological (oncocytic) changes associated with suppression of mitochondrial respiration41. What remains unclear is how to extrapolate the function of truncating mutations in otherwise essential mtDNA genes to cancer types where oncocytic tumours are rarely, if ever, observed, but in which the fraction of samples harbouring these mutations is nevertheless substantial (for example, colorectal cancers). Critically, our transcriptional data suggest that, even in cancer types where truncating mtDNA mutations are rare, they nevertheless promote a transcriptional programme characterized by increased expression of OXPHOS genes and downregulation of genes related to innate immune pathways. Extensive previous work has demonstrated that the integrity of mtDNA and mitochondrial respiration is essential for tumour growth and metastasis. In light of this, we interpret the lineage-agnostic upregulation of OXPHOS genes in response to truncating mutations as a compensatory, but likely inadequate, mechanism to maintain adequate respiratory capacity. The net quantitative effect on respiratory capacity is still uncertain.
The analytical approach we have employed utilizes off-target reads from large cohorts of exome and targeted sequencing data. Doing so vastly expands the number of tumour samples available for analysis, but comes at the cost of low coverage across the cohort (Extended Data Fig. 10). Indeed, although our study covers substantially more tumour samples than a recent analysis of mtDNA mutations in PCAWG (WGS based), the overall rate of mutations detected per sample is larger in the PCAWG analysis (1.28 mutations per sample (95% CI = 1.24–1.32) in PCAWG versus 0.43 (0.42–0.45) in TCGA). The high sensitivity of PCAWG is well suited to analysis of subclonal mtDNA mutations, tumour evolution and copy-number/structural variants. In contrast, mtDNA analysis from WES is more likely to identify rare tumour subtypes with elevated rates of mtDNA variants and/or recurrent variants, but with a sensitivity biased towards variants with elevated heteroplasmy. As pathogenic mtDNA variants elicit phenotypes in a dosage-dependent manner, with heteroplasmic loads of ~50% or higher potentially necessary for mitochondrial dysfunction to manifest, a bias towards high-heteroplasmy variants is more likely to enrich for variants of clinical and translational importance. Given these differences, the relative strengths and weaknesses of the WGS and WES approaches should be carefully considered when designing future studies of somatic mitochondrial genetics.
Despite recent advances that have been made towards defining the pattern and prevalence of mtDNA mutations in cancer2-4, major challenges remain in determining the functional relevance of mtDNA mutations in promoting tumour development. Progress has been hindered by a lack of precise methods for engineering of mammalian mtDNA42. As a result, emphasis has been placed on limited mutagenesis approaches, such as the use of a mutant, error-prone form of the replicative mtDNA polymerase γ35,43. Beyond this, more refined tools to genetically engineer mtDNA are now finally emerging, which might permit the creation of defined mtDNA mutations to further interrogate these observations in appropriate model systems of cancer44.
The nuclear genome has, historically, been the focus of research into the genetic basis of cancer. Somatic alterations to mtDNA are, however, among the most common mutational events across tumour lineages, and specific patterns of mtDNA mutations define and prognostically stratify patient cohorts. These observations motivate a holistic investigation of the relationship between the spatially, heritably and evolutionarily distinct nuclear and mitochondrial genomes, and a redefinition of our understanding of cancer—not a disease of the genome, but a disease of the genomes.
Methods
Tumour and normal-sample sequencing cohorts.
Tumour and matched-normal sequencing data for TCGA samples were obtained from the Genomic Data Commons (GDC) Data Portal (https://portal.gdc.cancer.gov). Briefly, all tumour and matched-normal barcodes included in the MC3 MAF45 (https://gdc.cancer.gov/about-data/publications/pancanatlas) file were converted to universally unique identifiers (UUIDs) using the TCGAutils R package (v.1.9.3), and these UUIDs were queried for WES BAM files sliced for chrM using the GDC Application Programming Interface. We then queried the GDC Data Portal for RNA-seq BAM files for TCGA tumours already with WES data. This process yielded paired tumour and matched-normal WES BAMs for 10,132 TCGA patients, of whom 9,455 had additional RNA-seq data. In addition to the raw sequencing data for TCGA samples, from which we called mtDNA mutations (Calling mitochondrial variants), we obtained somatic mitochondrial mutation calls for 2,836 WES tumours from International Cancer Genome Consortium (ICGC)/PCAWG3, of which 885 also had TCGA sequencing data. Nuclear somatic mutations for TCGA samples were obtained from the MC3 MAF subset for the samples for which mtDNA WES BAMs were available. Finally, mtDNA mutation calls for 195,983 normal samples were obtained from the HelixMTdb cohort of sequenced saliva samples from healthy individuals14.
MSK–IMPACT patient cohort and mutational data.
An additional 34,052 pan-cancer paired tumour and matched-normal samples were obtained from 30,575 patients with advanced and pre-treated solid cancers in the MSK–IMPACT clinical sequencing cohort. All MSK–IMPACT patients provided written informed consent and were prospectively sequenced as part of their active care at Memorial Sloan Kettering Cancer Center (MSKCC) between January 2014 and July 2019 as part of an Institutional Review Board-approved research protocol (NCT01775072)13,46. Briefly, each patient’s solid tumour and blood specimens were sequenced using a customized hybridization, capture-based, next-generation sequencing assay called MSK–IMPACT, which targeted 341, 410 or 468 known clinically relevant cancer-associated genes, depending on the version of the assay. Then, 498 distinct cancer histological subtypes were grouped into primary cancer diagnoses according to the OncoTree structured disease classifications (http://oncotree.mskcc.org). Primary diagnoses into which fewer than 50 tumours were classified, as well as histological subtypes without primary diagnosis classifications on OncoTree, were combined in an ‘Other’ classification, resulting in 42 main cancer types.
Annotating mtDNA regions included in our analysis.
Each mitochondrially encoded gene’s name, start/end positions and DNA strand were obtained from Biomart for human reference genome GRCh38 (release 95). Subsequently, each mtDNA position (1–16569) was annotated with its associated genetic information. Any mtDNA positions located at the overlap of two genes were annotated only as associated with whichever gene started first in numerical genomic position. Variants in non-genic mtDNA regions were excluded in our analyses. To this end, we excluded any variants in the mtDNA control region (positions 1–576, 16024–16569), as well as 89 other non-genic positions. We similarly excluded variants in hypermutated regions of mtDNA, including 302–316, 514–524 and 3106–3109. After these measures, the genomic length of mtDNA retained in our analyses was 15,354 bp. (The complete list of 16,569 mtDNA positions and their annotated reasons for exclusion is provided in Supplementary Table 1.)
On average 6,100 tumours were sequenced at sufficient depth to call mutations at each mtDNA position (mean ± s.d. for 5,399–6,800 samples covered at a given position; Fig. 1b), compared with 2,836 whole-genome tumour sequences from the PCAWG WGS dataset. When further restricted to regions sequenced at sufficient depth in both tumour and matched-normal samples, each position was covered in 4,769 tumour/normal pairs on average (mean ± s.d. for 4,148–5,390 samples). Each genomic position was sequenced to sufficient depth in comparable numbers of tumour and normal tissue, indicating that differential sequencing coverage between tumour and normal samples did not result in a biased mutation-calling sensitivity in specific regions of the genome (Extended Data Fig. 1c).
Calling mitochondrial variants.
Mutations to the mitochondrial genome were obtained from variants called by both of two independent variant-calling pipelines. In the first pipeline, Mutect2 (GATK v.4.1.2.0)47 was used to call variants in chrM in tumour and normal samples individually, the results of which were subsequently intersected to obtain variants called that were supported in a given patient’s tumour and matched-normal samples. Briefly, Mutect2 was run in mitochondrial mode for each patient’s tumour and normal sample independently against human reference genome GRCh38 (with minimum base quality score = 20, minimum mapping quality = 10, aggressive pcr-indel model, and other standard-quality control arguments for paired-end reads). Artefacts were subsequently removed using GATK FilterMutectCalls (GATK v.4.1.2.0)47 and multi-allelic sites were split into individual variants using the norm function from bcftools (v.1.9)48. The resulting tumour and normal Variant Call Format (VCF) files were then merged using gatk HaplotypeCaller (GATK v.4.1.2.0)47, to annotate variants in the tumour VCF with their coverage in the normal sample. The resulting VCF was converted to a MAF file using vcf2maf (v.1.6.17, https://github.com/mskcc/vcf2maf). Finally, variants from the generated MAF file were then filtered out unless the variant allele was supported by at least one read in both forward and reverse directions. In the second pipeline, samtools mpileup (v.1.9)11 was used to generate a pileup file using variant-supporting reads with a minimum mapping quality of 20 and base alignment quality of 10. Reads failing quality checks or marked as PCR duplicates were removed. Variants were required to contain at least two variant-supporting reads in the forward and reverse directions. In each pipeline, variants were additionally filtered to ensure ≥5% VAFs in the tumour and ≥5 reads supporting the alternative allele. Variants identified by both pipelines were retained for further analysis. In rare cases, multiple indels were called in a sample within a homopolymeric region (single-nucleotide repeats of ≥5 bp), with distinct alt-read counts and VAF values and identical read-depth values. These multiple indels were collapsed to a single representative indel call. Briefly, using the Mutect2 variant calls, whichever indel had the highest VAF in the tumour sample was taken as the representative indel. The count of alt-reads in both tumour and normal samples was replaced with their corresponding summed counts across the original multiple indels, and the VAFs in both tumour and normal samples were re-calculated from the new summed alt-read counts divided by the original read-depth.
We investigated whether protein-truncating variants were detectable in patient germlines, hypothesizing that these should be significantly depleted due to the essentiality of functional OXPHOS in normal human cells. Indeed, whereas 11.9% (95% binomial CI = 11.0–12.9%) of pan-cancer tumour samples harboured truncating variants, only 0.34% (0.16–0.65%) of patients’ blood-derived, matched-normal samples harboured truncating variants (P = 1 × 10−69, two-sided, two-sample z-test; Extended Data Fig. 3b), which was also similar to the rate seen in the HelixMTdb dataset (0.15% (0.13–0.17%); P = 0.02) and consistent with the hypothesis that polyclonal haematopoiesis and negative selection against pathogenic mtDNA mutations in certain haematopoietic lineages hinders clonal expansion of truncating variants in the blood. It is interesting that solid tissue-derived normal samples had a slightly elevated rate of truncating variants to blood-derived normals (1.2% (0.5–2.3%); P = 0.02), possibly due to somatic expansions of normal cells harbouring these variants heteroplasmically. Similarly, we observed normal tissue-derived truncating variants to have substantially depleted heteroplasmies compared with tumour-derived truncating variants (Extended Data Fig. 3a). These findings suggested that truncating mutations identified in tumours without concomitant coverage of the matched-normal tissue (that is, ‘rescued’ truncating mutations) could be assumed to be of somatic origin. We subsequently tested whether rescued truncating variants could represent technical artefacts by examining their incidence in a complementary sequencing modality, matched RNA-seq data. We observed that 98.3% of rescued truncating mutations validated in RNA (among n = 176 truncating mutations in samples with matched RNA-seq; Extended Data Fig. 3c), and that the heteroplasmies in DNA and RNA demonstrated strong correlation (Pearson’s r = 0.864; Extended Data Fig. 3d). Based on these analyses, all protein-truncating mtDNA mutations of unknown somatic status due to insufficient normal sample coverage were retained as probable somatic mutations.
Mutations were therefore classified as of somatic origin according to the following criteria: non-truncating variants (that is, all variant classifications other than nonsense mutations and frameshift indels) were classified as somatic if the matched-normal sample had a minimum coverage of 5 reads and 0 normal reads called the alternative allele. Truncating variants in tumour samples were assumed to be of somatic origin. All other variants were not classified as somatic and were excluded from the present study, unless otherwise noted. As a result of inherently low mtDNA coverage in the MSK–IMPACT dataset (owing to its use of targeted sequencing), in the MSK–IMPACT clinical analysis we additionally included mutations on the basis of their likelihood to be somatic, if there was insufficient coverage in the matched-normal sample to determine somatic status. Mutations were classified as probably somatic if they were never observed as germline mutations in any TCGA or HelixMTdb samples, and arose with heteroplasmy <100%.
We then investigated the degree to which variant calling with off-target sequencing reads biased our sensitivity to detect mutations at lower heteroplasmies. We compared the absolute number of variants called in either the TCGA or the PCAWG datasets between mutations with heteroplasmy <30% and ≥30%. This revealed that, when considering mutations in genic regions with VAFs ≥ 30%, our approach (applied to a larger number of samples than PCAWG) increased the overall number of mutations called in all categories by at least 65% and, in the case of truncating variants, by nearly 300% (Extended Data Fig. 10a). In contrast, PCAWG detected far more variants at heteroplasmies <30% despite a markedly smaller number of samples sequenced. This analysis indicates that the sensitivity of our approach is inherently biased to detect mutations at higher heteroplasmy, which are more likely to detect dysfunctional phenotypes due to increased mutant load. This comes at the expense of decreased sensitivity for low-heteroplasmy variants, which are more appropriately studied using WGS-based approaches3 (Extended Data Fig. 10c).
Nuclear mutational data and annotation.
Somatic mutations in nuclear-encoded, cancer-associated genes for TCGA samples were obtained from the PanCanAtlas MC3 MAF file. Mutations in this file were subset for those among the 468 genes on the MSK–IMPACT clinical sequencing panel13. The MAF file was annotated for known, likely and predicted oncogenic driver mutations using the MAF-Annotator tool provided by OncoKB49 (https://github.com/oncokb/oncokb-annotator). Mutations annotated by OncoKB as ‘Oncogenic’, ‘Likely Oncogenic’ or ‘Predicted Oncogenic’ previously determined cancer hotspot mutations15,16, or truncating variants to TSGs (that is, frameshift indels, splice site and nonsense mutations) were classified as potential driver alterations.
Calculating TMB in mtDNA or nuclear DNA.
TMB was calculated for cohorts of tumour subsets for various genomic regions, including: (1) individual mitochondrially or nuclear-encoded genes; (2) mtDNA genes grouped by OXPHOS CI, CIII, CIV or CV; (3) the entire mitochondrial genome (excluding non-genic and polymorphic regions); (4) a set of known nuclear-encoded TSGs; and (5) a set of known nuclear-encoded oncogenes. In each case, the TMB was calculated as the total number of somatic mutations among the relevant collection of tumours divided by the total genomic length sequenced in these tumours (in Mb s−1). For TMBs calculated from mutations called in off-target sequencing data (that is, mtDNA variants in TCGA samples), the total genomic length sequenced was the number of genomic positions with sufficient coverage to call somatic variants (5+ read coverage in both tumour and normal sample), summed across all samples. For TMBs calculated from targeted regions (nuclear DNA, mtDNA in PCAWG samples), the total genomic length sequenced was the length of the targeted region (entire gene for mtDNA, exonic regions for nuclear DNA) multiplied by the number of samples. Error bars for TMBs were calculated as 95% Poisson’s exact CIs for rates, using the total number of mutations as the count of events, and the genomic length sequenced in megabases as the time at risk.
Identifying hotspot positions for mitochondrial variants.
We identified mtDNA positions with statistically recurrent SNVs by comparing the observed proportion of mutations at an individual position (out of the total number of mutations acquired in its gene) with a rate of mutations at the position expected by chance with a one-sided binomial test. The probability for SNVs at each position of a gene, Ppos,gene, was modelled as a Bernoulli trial, where the likelihood of a mutation arising at a given position by its mutability (μ) relative to the mutability of all other bases in the gene is: . Consistent with previous work15, we estimated the mutability for each position as a function of its trinucleotide context, that is, for each position, its mutability, μpos, was calculated as the count of SNVs matching the trinucleotide context of the position of interest, spos, out of the total count of SNVs anywhere in the mitochondrial genome, stotal (after excluding the control region and other blacklisted regions). Due to the highly strand-specific mutation signatures we observed for SNVs in mtDNA (Extended Data Fig. 1c), we used the complete set of 64 unique trinucleotides to retain this information when calculating the mutability for each position, rather than collapsing the central nucleotide to C or T, resulting in the conventional 32 unique trinucleotides. As the proportion of patients for whom a given position had sequencing coverage in paired tumour and normal samples linearly affects the likelihood of observing a somatic mutation at the position, the mutability of a position was adjusted to control for this by multiplying it by the ratio of the number of samples with paired tumour–normal sequencing coverage at the position, Cpos, out of the total number of samples, Nsamples so that . The mutability associated with the gene was calculated as the sum of each position’s trinucleotide mutability. Therefore, for a gene L bp in length: . The final parameter for the binomial test (that is, the likelihood of a mutation in a gene arising at the given position by chance) was therefore . Each position mutated in five or more samples in each gene was subsequently tested for statistically enriched mutations by comparing its observed number of mutations out of the total number of mutations in the gene with this binomial parameter, using a right-tailed binomial test. The full list of generated P values across all genes was then corrected for multiple hypothesis testing.
Homo polymer hotspots for indels.
To identify homopolymer regions with statistically enriched rates of indels, we modelled the proportion of samples with indels across all homopolymers as a function of the homopolymer region’s length (that is, the number of repeated nucleotides, from five to eight). To this end, all single-nucleotide repeats of ≥5 bp were identified in the mitochondrial reference genome, resulting in n = 73 unique homopolymer loci in whitelisted coding mtDNA. We then modelled the fraction of frameshift indels across 73 homopolymers observed to arise at a specific homopolymer locus, h, as a binomial process, dictated by the length of the homopolymer, lh, divided by the summed length of all homopolymers, such that the expected likelihood of a frameshift indel arising at a homopolymer by chance is given by: . We then tested each homopolymer locus for enriched mutations with a one-sided binomial test, that is, for each homopolymer locus, the number of Bernoulli trials was the number of samples with complete sequencing coverage for the homopolymer region and two flanking base pairs; the number of successes was the number of samples with frameshift indels at (or immediately adjacent to) the given homopolymer, and the fraction of successful trials was compared with the expected probability, ph. Identical results for hotspot analysis were observed when considering indels with a minimum of 20 reads of support, ensuring that these results were not artefactually driven by low sequencing coverage (Extended Data Fig. 5c).
Hotspot positions in tRNA cloverleaf structure.
Positions of the tRNA cloverleaf secondary structure were individually tested for an enriched rate of SNVs at the equivalent aligned positions of the 22 mitochondrially encoded tRNAs. A map of genomic positions in mt-tRNAs to cloverleaf structure positions was provided by MitoTIP50 (https://github.com/sonneysa/MitoTIP/blob/master/Output/tRNA%20data%20and%20scoring_scored.xlsx) and used to assign SNVs at tRNAs to structural positions. Under the null hypothesis that mutations accumulate at structurally aligned positions randomly, the proportion of SNVs aligning to a specific position in the tRNA cloverleaf should be approximately equal to the number of times the aligned position was sequenced at a sufficient depth in both tumour and matched-normal samples to call somatic mutations, out of the total number of tRNA base pairs sequenced at sufficient depth across all samples and at all structural positions. Therefore, for a given position of the tRNA cloverleaf structure, p, the number of SNVs observed across all tRNAs at this aligned position, tp, out of T SNVs across all positions of all tRNAs was tested for enrichment using a one-sided binomial test, compared with an expected rate equal to the number of tRNA bases aligned to this position sequenced at sufficient depth, bp, out of B tRNA bases sequenced at sufficient depth across all positions of all tRNAs.
Classifying sample mtDNA variant status.
Each tumour sample was classified according to the presence and type of its somatic mitochondrial variants. As gaps in sequencing coverage may make existing variants undetectable and result in the incorrect classification of such samples as ‘wild-type’ for somatic variants, we attempted to classify only samples with sequencing coverage in both tumour and matched-normal samples of at least 90% of the included region of mtDNA (referred to as ‘well covered’ throughout). Furthermore, given the high incidence of truncating indels that we observed at six hotspot loci, we additionally required that these six loci be sequenced at sufficient coverage in the tumour sample, to ensure that samples potentially harbouring recurrent indels would be excluded and not misclassified. Samples not meeting either of these conditions were classified as having ‘Unknown’ mtDNA mutation status. The remaining samples were then classified according to a decision tree as follows: samples with any protein-truncating variants were classified as ‘Truncating’; remaining samples still unclassified with multiple mtDNA variants of different types (among missense, rRNA and tRNA variants) were classified as ‘2+ non-truncating types’; remaining samples with tRNA mutations were classified as ‘tRNA’; remaining samples with rRNA mutations were classified as ‘rRNA’; remaining samples with non-truncating, non-synonymous, protein-coding mutations were classified as ‘missense’; remaining samples with silent mutations were classified as ‘silent’; and finally samples still unclassified were classified as ‘wild-type’. This logic prioritizes minimizing annotation bias over conserving sample size, to meaningfully compare the incidence of different variant types across samples. However, in our analysis of the effect of mtDNA variants on differential gene expression or survival, we modified the logic to prioritize conservation of sample size. To this end, in RNA-seq and survival analyses, samples with any observed truncating variants were classified as truncating, regardless of their sequencing coverage. For samples from the MSK–IMPACT cohort, a modified procedure was used to annotate sample mtDNA status: in the present study, we first excluded samples with tumour coverage of <60% of mtDNA (due to our inclusion of’likely somatic’ mutations (Calling mitochondrial variants), we required only tumour coverage rather than both tumour and matched normal). Among the remaining samples, we categorized samples with any truncating variants as ‘Truncating’, samples with any somatic or likely somatic VUSs but no truncating variants and no indel hotspots with missing coverage as ‘VUS’ and samples with no truncating or somatic/likely somatic VUS mutations as ‘wild-type’.
Testing genesets for transcriptional dysregulation due to mtDNA variants.
A matrix of estimated gene expression counts (RNA-seq by expectation-maximization values normalized to correct for batch effects) for TCGA samples was downloaded from TCGA PanCanAtlas45 supplementary data (http://api.gdc.cancer.gov/data/3586c0da-64d0-4b74-a449-5ff4d9136611). Gene expression estimates were rounded to integer values, and subsequently genes with zero estimated counts in all samples were removed, as were genes with unknown gene symbols. To evaluate differentially expressed genes between two groups of samples with different mtDNA variant type (that is, truncating versus wild-type colorectal samples), the rounded gene expression matrix was subset for the relevant samples and input into the DESeq2 (ref.51) package in R using the DESeqDataSetFromMatrix utility, along with a table of tumour sample barcodes with their associated mtDNA classification. Differentially expressed genes were tested and their log(fold-change) (log(FC)) values were shrunk using the apeglm52 package. P values for all genes tested were corrected for multiple hypothesis testing using the Benjamini–Hochberg method54. The resulting data from this analysis were used to calculate a statistic for each gene equal to log10(Q value) × sign(log(FC)). All genesets from the mSigDB Hallmark gene set collection54 (v.7.1) were then tested for significant up- or downregulation based on this statistic for each gene using the fgsea package55 in R, with a minimum gene set size of 10 genes, a maximum size of 500 genes and 100,000 permutations.
Genomic and clinical annotations for colorectal cancer survival analysis.
Clinical data for TCGA colorectal cancer patients, including OS time/status, American Joint Committee on Cancer (AJCC) pathological tumour stage, age at diagnosis, sex and tumour tissue site were obtained from TCGA Firehose legacy data on cbioportal (https://www.cbioportal.org/study/summary?id=coadread_tcga). Clinical data were subset for patients with sequencing data in the MC3 MAF. These data were then annotated with MSI status (MSS, MSI-low, MSI-high) based on published data for patients where this was available56. AJCC pathological tumour staging data were collapsed into stages I, II, III and IV, and then stage IV patients were excluded. The tumour site was encoded as ‘right-colon’ if the primary site was: ascending colon, caecum, hepatic flexure or transverse colon; or encoded as ‘left-colon’ for: descending colon, sigmoid colon or splenic flexure. Patients with tumour tissue from the rectum were encoded as ‘rectum’ for their tumour site. Tumour purity was obtained from the GDC for the PanCancer Atlas publication (http://api.gdc.cancer.gov/data/4f277128-f793-4354-a13d-30cc7fe9f6b5) and merged to the clinical data by tumour barcode. The clinical data for each sample were then annotated for the presence of known or probable nuclear-encoded driver alterations in KRAS/HRAS/NRAS, BRAF, APC, SMAD4 and TP53, as based on mutation calls from the TCGA MC3 MAF57 (Nuclear mutational data and annotation). Each patient in the clinical data was then annotated as having a known/probable driver alteration in each of KRAS/HRAS/NRAS (grouped into RAS), BRAF, APC, SMAD4 or TP53. The complete multivariate model used in the Cox proportional hazards regression was therefore: OS ~ mtDNA status + Age + Stage + Site + RAS + RAF + APC + SMAD4 + TP53 + Sex + MSI status + Tumour purity + CMS type; it was used to analyse the OS of n = 341 patients with non-missing values for all covariates. Multivariate survival analysis for the MSK-IMPACT cohort was performed using a similar workflow: curated clinical data for patient age at diagnosis, tumour stage, tumour location, sex and MSI status were obtained from a previously published subset of colorectal cancer patients from the MSK–IMPACT cohort34 (https://www.cbioportal.org/study/summary?id=crc_msk_2017). OS time and status data were obtained from the MSK–IMPACT clinical sequencing cohort (queried March 2019). Samples were annotated as having driver mutations in KRAS/HRAS/NRAS, BRAF, APC, SMAD4 or TP53, as described above for TCGA. Tumour purity estimates were calculated using FACETS (v.0.5.6) with a C value of 100, and tumours for which the algorithm did not converge on a purity estimate were excluded from the survival analysis. For consistency with TCGA (which included only one tumour sample per patient), MSK–IMPACT patients with multiple colorectal cancer tumour samples were randomly assigned a single representative sample (among those with non-missing values for the aforementioned covariates), resulting in n = 172 tumours for the same number of patients. CMS data were unavailable for MSK–IMPACT, so the complete Cox proportional hazards regression model was OS ~ mtDNA status + Age + Stage + Site + RAS + RAF+ APC + SMAD4 + TP53 + Sex + MSI status + Tumour purity.
Structural impact of MT-ND1R25Q variant on CI.
The structural impact of the MT-ND1R25Q variant was investigated using an electron-microscopy-derived structure of mitochondrial CI in Mus musculus (PDB accession no. 6G2J)30. The UCSF Chimera software (v.1.13.1)58 was used to insert the Arg25Gln mutation using the swapaa command. The ubiquinone-binding tunnel was predicted using the CAVER Analyst (v.2.0b)59 software run on the wild-type Protein Data Bank (PDB) structure, starting from the side-chain oxygen atom in Ndufs2Y108, and using a minimum probe radius of 1.4 Å (0.14 nm) as described by the authors30. The surface electrostatic charge for wild-type and mutant structures was determined using the APBS software60 (http://server.poissonboltzmann.org/pdb2pqr) with default parameters, after subsetting the PDB structure for Mtnd1 (chain H), and converting the resulting PDB file to PQR using PDB accession no. 2PQR (ref. 61). All structure visualizations were generated using UCSF Chimera.
Statistical analyses and figures.
All statistical analyses were performed using the R statistical programming environment (v.3.6.1). Protein structure figures were generated using UCSF Chimera. Cox proportional hazards regression was performed using the Survival library, and the Kaplan–Meier and multivariate survival forest plots were generated using the survminer library in R. The Cochran–Armitage test for trend was calculated using the DescTools R library. ETC schematic (Fig. 1a) was generated in Adobe Illustrator. All other figures were generated using the ggplot2 library in R. Unless otherwise noted, error bars for proportions are 95% binomial CIs calculated using the Pearson–Klopper method; error bars for rates (for example, mutations per Mb) are Poisson’s exact 95% CIs calculated with the pois.exact function from the epitools library in R. In all boxplots, boxes show 25th and 75th percentile values centred at the median; upper and lower whiskers are the most extreme values within 1.5× the interquartile range above the 75th percentile and below the 25th percentile values, respectively. Unless otherwise noted, P values for difference in proportions were calculated using Fisher’s exact tests or two-sample z-tests, and for difference in rates using Poisson’s exact tests. P values were corrected for multiple comparisons using the Benjamini–Hochberg method53 and reported as Q values when applicable.
Extended Data
Extended Data Fig. 1 ∣. Baseline demographics of cohort and aspects of sequencing coverage.
a, The demographic distributions of patient age, race, gender, mtDNA somatic mutation status, history of neoadjuvant treatment, mtDNA coverage, and tumor sample type for each of the cancer types included in our analysis. Somatic mutation status is annotated among the subset of samples with ≥90% paired tumor-normal mtDNA sequencing coverage (see Methods: classifying sample mtDNA variant status); mtDNA status distributions are shown for cancer types >10 such samples. Cancer types are ordered by increasing proportions of samples with VUS or truncating mtDNA mutations. b, Cancer type mtDNA coverage variation based on sequencing center. Center, the average percentage of mtDNA (among regions considered in our study) with sufficient coverage for calling mutations, compared between different cancer types in our cohort. Dot color indicates the sequencing center from which the exome sequencing data originate. Top, density histograms of the average % mtDNA coverage for each sequencing center. Samples sequenced at the Broad Institute are uniquely depleted for mtDNA off-target coverage. c, mtDNA coverage from off-target reads at each position. The number of samples for which the given mtDNA position was sequenced to at least 5 reads (top, the depth threshold used in our analyses) and 20 reads (bottom, for comparison). Red, the number of samples using unpaired tumor-only data, applicable only for protein-truncating variants which were always assumed to be of somatic origin; blue, the number using only matched-normal samples; green, the number of samples with coverage in both tumor and matched-normal samples at the given position (applicable for all non-truncating variants which required evidence that the variant was absent in the matched normal to be classified as somatic). Purple, the number of whole-genome sequenced samples available from ICGC/PCAWG for comparison. d, Proportion of samples with detectable mutations is not biased by cancer type sequencing coverage. There is no correlation between the fraction of well-covered samples in a cancer type and the proportion of well-covered samples with a detectable somatic mtDNA mutation. Cancer types with ≥30 well-covered samples shown, P-value and 95% confidence intervals from linear regression.
Extended Data Fig. 2 ∣. Strand-specific mutational signatures in our dataset.
The frequency of somatic SNVs on the light or heavy mtDNA strand with each of the 96 possible mutational signatures with trinucleotide contexts (among n = 3,872 SNVs). Blue bars indicate the prevalence of mutational signatures for heavy-strand encoded SNVs (substitutions at C or T central nucleotides); red bars indicate those for light-strand encoded SNVs (substitutions at G or T nucleotides, which were standardized to their C or T complementary nucleotide). The most prevalent mutational signatures are labeled. The underlined central position is mutated with the single nucleotide substitution labeled in the tile below.
Extended Data Fig. 3 ∣. Analysis of mutation burden in normal tissues and of tumor mtDNA mutation burden with nuclear mutagenic processes.
a, Comparison of heteroplasmies between truncating variants detected in tumor tissue, adjacent normal tissue, and blood. P-values from two-sided Wilcoxon-rank sum test. Boxes are centered at the median and extended to from 25th-percentile to 75th-percentile; whiskers extend from 25/75th-percentiles to the largest value within 1.5 × IQR (interquartile range, 75th-percentile - 25th-percentile. b, Rate of truncating variants in TCGA tumors compared to matched non-malignant tissue, matched blood, and unmatched saliva samples from HelixMTdb. Truncating variants arise at 10-80-fold higher rate in tumors relative to normal tissues. Error bars are exact binomial 95% confidence intervals. P-values are from two-sided two-sample z-tests. c, The percentage of rescued truncating variants in TCGA that are recapitulated in orthogonal RNA sequencing from the same tumor sample. d, Correlation between heteroplasmies of rescued truncating variants in DNA and orthogonal RNA sequencing. Pearson correlation coefficient as shown. e, Mitochondrial and nuclear tumor mutation burdens (TMB, mutations/Mb) are shown for each well-covered tumor, among cancer types with n ≥ 100 samples. Nuclear TMBs are calculated based on mutations to 468 cancer-associated genes and their total exonic-sequence length. Pearson correlation coefficients r indicate no linear correlation between mitochondrial and nuclear TMBs were observed for any cancer type tested. f, TMBs for somatic mtDNA mutations and mutations to cancer-associated genes are compared between microsatellite stable (MSS) and microsatellite unstable (MSI-High) tumors, for both (n colorectal cancer: MSI=65, MSS=318; n stomach adenocarcinomas: MSI=75, MSS=256). Although MSI-High tumors have elevated TMB for nuclear cancer genes, there is no effect on mtDNA TMB. Moreover, mtDNA TMB is similar to (or exceeds) that of nuclear cancer associated genes in both cancer types. Error bars are 95% exact Poisson confidence intervals.
Extended Data Fig. 4 ∣. Age- and tumor stage-associations of somatic mtDNA mutations across cancer types.
Heatmap shows tumor mutation burden (total mutations/total covered Mbps) for samples of each tumor type (a) combined across varying patient age at time of diagnosis and (b) tumor pathologic stage. Gray tiles indicate cancer type/age combinations with fewer than 3 patients; cancer types shown had at least 2 non-gray tiles. Right column: Spearman correlation coefficient r indicating correlation between age or pathologic stage and tumor mutation burden. Asterisks denote statistically significant correlations based on FDR-corrected P-values from a Student’s t-distribution.
Extended Data Fig. 5 ∣. Molecular features of truncating variants at homopolymeric loci.
a,b, Enrichment for truncating variants in CI and non-truncating in CIII when restricted to mutations with 20+ reads supporting the alternate allele. Error bars are 95% Poisson exact confidence intervals; P-values from two-sided Poisson tests. c, Comparison of frameshift indel homopolymer hotspots detected among indels supported by a minimum of 20 alt-reads (Y-axis) to those with a minimum support of 5 alt-reads (X-axis). d, Percentage of cases per cancer type with truncating frameshift indels at any of 6 indel hotspot loci. Plotted cancer types had ≥ 20 well-covered samples (n=4,432 paired tumor and matched-normal samples total). Bar height indicates the fraction of samples with any indels at homopolymer hotspot out of the total number of well-covered samples for the given cancer type; numbers above bars indicate the total number of cases. e, Validation of homopolymeric indel hotspot loci. The proportion of samples in TCGA (X-axis) or PCAWG (excluding samples also in TCGA, Y-axis) with frameshift indels at 73 homopolymeric regions. The 6 indel hotspot loci are colored red and labeled. y=x is shown as a dashed line. Pearson correlation coefficient r as indicated. f, Breakdown of homopolymer loci and their hotspot incidence rates by mitochondrial complex. Heatmap tile shading indicates overall mutation rate (total number of mutants across homopolymer loci divided by the total number of samples with sufficient sequencing coverage). Fractions in tile labels are the number of homopolymer hotspots divided by the total number of homopolymer loci. Right, histogram of the total number of loci with each homopolymer length. g, The percentage of all truncating variants which arose at 6 homopolymer hotspot loci in TCGA tumor samples and in saliva-derived normal samples from HelixMTdb. Error bars are 95% binomial confidence intervals.
Extended Data Fig. 6 ∣. Homopolymer hotspots for frameshift indels in TCGA and MSK-IMPACT cohorts.
a, Analysis of 73 homopolymer loci for enrichment of protein-truncating indels in TCGA samples in shown on X-axis, and in MSK-IMPACT samples on Y-axis. 5 out of 6 originally reported hotspot loci are enriched in both TCGA and MSK-IMPACT (green), while 1 was only enriched in TCGA samples (orange). Two additional candidate hotspot loci are unique to the MSK-IMPACT dataset (recurrently observed in TCGA, but not reaching statistical significance). b, Heatmap indicates the proportion of truncating indels at each of the 8 homopolymer hotspots to affect samples of different cancer types (that is rows are proportion of samples with indels at a given homopolymer summing to 1; clustered by hierarchical clustering). Right, histogram of the total number of affected samples in TCGA and MSK-IMPACT data. Note that the CIV hotspot discovered in MSK-IMPACT data preferentially arises in lung and prostate cancers.
Extended Data Fig. 7 ∣. Validation of VUS pathogenicity and tRNA mutation recurrence.
a, VUSs only observed in tumors are more likely to be pathogenic. Bars compare the clinical significance (annotated by ClinVar) of SNVs observed somatically in tumors but never in patients’ matched-normal samples against SNVs never observed in either tumor or matched-normal samples. P-value from a two-sided Cochran-Armitage trend test. b, Validation of tRNA structural hotspots in PCAWG. The number of samples with SNVs in tRNAs at the indicated cloverleaf structural position, bottom; top, the statistical enriched of the given position for mutations. Position 31 Q-value=0.014, n=196 tRNA mutations among 1,951 PCAWG samples.
Extended Data Fig. 8 ∣. mtDNA mutations produce transcriptional phenotypes.
a,b, Transcriptional dysregulation attributed to truncating (a) and VUS (b) mtDNA variants. Heatmaps shows directional significance of dysregulation of a given geneset in tumors with truncating or VUS mtDNA variants among the given cancer type; −log10(Q-value) > 2 indicates significant up-regulation in mutated compared to wild-type samples, < −2 indicates significant down-regulation. Histograms on the right show the number of wild-type samples and mutated samples used in calculating differentially expressed genes and dysregulated genesets. c, Difference in mtDNA mutation status between colorectal cancer consensus molecular subtypes. Left, the proportion of samples with wild-type mtDNA (that is no somatic mutations), VUS (any non-truncating) or truncating variants among colorectal tumors with each consensus molecular subtype (CMS) is shown. Right, histogram of the number of well-covered colorectal tumors. There was a statistically significant difference in mtDNA mutation status between different CMS classifications (P=0.03, Chi-squared test, n=415 samples total, error bars are 95% exact binomial confidence intervals).
Extended Data Fig. 9 ∣. mtDNA mutations are protective in colorectal cancer patients in the MSK-IMPACT cohort.
a, Multivariate survival analysis based on Cox proportional hazards regression demonstrating the effect of VUS or truncating mtDNA mutations (relative to wild-type) on colorectal cancer patient overall survival in the MSK-IMPACT cohort. b, Same as in (a) but treating VUS and truncating mtDNA mutations as a single class compared to wild-type. Error bars are 95% confidence intervals from Cox proportional-hazards regression, n=172 MSK-IMPACT patients.
Extended Data Fig. 10 ∣. Repurposing whole-exome and clinical sequencing data optimizes sample size at the expense of sensitivity for low-heteroplasmy variants.
a, The number of different classes of mtDNA variants detected from either repurposed TCGA samples using our approach, or using only whole-genome sequenced tumors from PCAWG, stratified by heteroplasmy < 30% or ≥ 30%. Labels above bars indicate the exact number. b, Comparison of the difference in gene expression between (1) high-heteroplasmy truncating mutations and wild-type tumors (X-axis) and (2) low-heteroplasmy truncating mutations and wild-type tumors (Y-axis). c, Strengths and use-cases of three common tumor DNA sequencing modalities for mtDNA mutation analysis. WGS-based approaches are optimal for studying low-heteroplasmy variants and identifying structural variants and mtDNA copy number, while whole-exome and targeted gene-panel-based approaches optimize sample size, detection of recurrent variants, and clinical associations.
Supplementary Material
Acknowledgements
We thank the members of the Reznik, Gammage and Taylor laboratories for discussion and support. We also thank L. Finley, K. Birsoy, J. Blaza, C. Winchester and N. Rusk for their feedback. A.N.G., M.K., W.K.C., A.A.H., M.F.B, B.S.T. and E.R. were supported by the National Cancer Institute (NCI) Cancer Center Support (grant no. P30 CA008748). W.K.C. was supported by a National Institutes of Health (NIH) award (no. T32 GM132083). K.C.L. was supported by an F31 Predoctoral Fellowship from the NCI (award no. 7F31CA247528-02). B.S.T. was supported by the NIH (award nos. U54 OD020355, R01 CA207244, R01 CA204749 and R01 CA245069), as well as the American Cancer Society, Anna Fuller Fund and the Josie Robertson Foundation. E.R. was supported by the Geoffrey Beene Cancer Research Center Grant Award, Department of Defense Kidney Cancer Research Program (no. W81XWH-18-1-0318), and a Kidney Cancer Association Young Investigator Award. P.A.G. was supported by core funding from CRUK BI (nos. A17196 and A31287).
Footnotes
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Code availability
R code to regenerate all figures is available on github (https://github.com/reznik-lab/mtdna-mutations) with the relevant data and instructions to execute the code.
Competing interests
B.S.T. reports receiving honoria and research funding from Genentech and Illumina, and advisory board activities for Boehringer Ingelheim and Loxo Oncology, a wholly owned subsidiary of Eli Lilly, Inc. All stated activities were outside the work described in the present study. He is currently an employee of Loxo Oncology. P.A.G. is a shareholder of Pretzel Therapeutics Inc. The remaining authors declare no competing interests.
Extended data is available for this paper at https://doi.org/10.1038/s42255-021-00378-8.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s42255-021-00378-8.
Data availability
All data not available for download on public repositories as described in Methods are available on GitHub (https://github.com/reznik-lab/mtdna-mutations).
References
- 1.Hornshøj H et al. Pan-cancer screen for mutations in non-coding elements with conservation and cancer specificity reveals correlations with expression and survival. NPJ Genom. Med 3, 1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ju YS et al. Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. eLife 3, e02935 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan Y et al. Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat. Genet 52, 342–352 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stewart JB et al. Simultaneous DNA and RNA mapping of somatic mitochondrial mutations across diverse human cancers. PLoS Genet. 11, e1005333 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grandhi S et al. Heteroplasmic shifts in tumor mitochondrial genomes reveal tissue-specific signals of relaxed and positive selection. Hum. Mol. Genet 26, 2912–2922 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hopkins JF et al. Mitochondrial mutations drive prostate cancer aggression. Nat. Commun 8, 656 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.To T-L et al. A compendium of genetic modifiers of mitochondrial dysfunction reveals Intra-organelle buffering. Cell 179, 1222–1238.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birsoy K et al. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162, 540–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samuels DC et al. Finding the lost treasures in exome sequencing data. Trends Genet. 29, 593–599 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cibulskis K et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H et al. The sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collura RV, Auerbach MR & Stewart CB A quick, direct method that can differentiate expressed mitochondrial genes from their nuclear pseudogenes. Curr. Biol 6, 1337–1339 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Cheng DT et al. Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J. Mol. Diagn 17, 251–264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolze A et al. Selective constraints and pathogenicity of mitochondrial DNA variants inferred from a novel database of 196,554 unrelated individuals. Preprint at biorxiv 10.1101/798264 (2019). [DOI] [Google Scholar]
- 15.Chang MT et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol 34, 155–163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang MT et al. Accelerating discovery of functional mutant alleles in cancer. Cancer Discov. 8, 174–183 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Triska P et al. Landscape of germline and somatic mitochondrial DNA mutations in pediatric malignancies. Cancer Res. 79, 1318–1330 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gopal RK et al. Early loss of mitochondrial complex I and rewiring of glutathione metabolism in renal oncocytoma. Proc. Natl Acad. Sci. USA 115, E6283–E6290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zehir A et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med 23, 703–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alston CL et al. A novel mitochondrial MTND5 frameshift mutation causing isolated complex I deficiency, renal failure and myopathy. Neuromuscul. Disord 20, 131–135 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Castellana S et al. High-confidence assessment of functional impact of human mitochondrial non-synonymous genome variations by APOGEE. PLoS Comput. Biol 13, e1005628 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landrum MJ et al. Clinvar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D1067 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marténez-Reyes I et al. Mitochondrial ubiquinol oxidation is necessary for tumour growth. Nature 585, 288–292 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Hattab AW, Adesina AM, Jones J & Scaglia F MELAS syndrome: clinical manifestations, pathogenesis, and treatment options. Mol. Genet. Metab 116, 4–12 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Gorman GS et al. Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann. Neurol 77, 753–759 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gopal RK et al. Widespread chromosomal losses and mitochondrial DNA alterations as genetic drivers in Hürthle cell carcinoma. Cancer Cell 34, 242–255.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terzioglu M et al. MTERF1 binds mtDNA to prevent transcriptional interference at the light-strand promoter but is dispensable for rRNA gene transcription regulation. Cell Metab. 17, 618–626 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Spagnolo M et al. A new mutation in the mitochondrial tRNA(Ala) gene in a patient with ophthalmoplegia and dysphagia. Neuromuscul. Disord 11, 481–484 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Horváth R, Reilmann R, Holinski-Feder E, Ringelstein EB & Klopstock T The role of complex I genes in MELAS: a novel heteroplasmic mutation 3380G>A in ND1 of mtDNA. Neuromuscul. Disord 18, 553–556 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Agip A-NA et al. Cryo-EM structures of complex I from mouse heart mitochondria in two biochemically defined states. Nat. Struct. Mol. Biol 25, 548–556 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joshi S et al. The genomic landscape of renal oncocytoma identifies a metabolic barrier to tumorigenesis. Cell Rep. 13, 1895–1908 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganly I et al. Integrated genomic analysis of Hürthle cell cancer reveals oncogenic drivers, recurrent mitochondrial mutations, and unique chromosomal landscapes. Cancer Cell 34, 256–270.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guinney J et al. The consensus molecular subtypes of colorectal cancer. Nat. Med 21, 1350–1356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yaeger R et al. Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell 33, 125–136.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith AL et al. Age-associated mitochondrial DNA mutations cause metabolic remodelling that contributes to accelerated intestinal tumorigenesis. Nat. Cancer 1, 976–989 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan H et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med 360, 765–773 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baysal BE et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science 287, 848–851 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Tomlinson IPM et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat. Genet 30, 406–410 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Fendt S-M, Frezza C & Erez A Targeting metabolic plasticity and flexibility dynamics for cancer therapy. Cancer Discov. 10, 1797–1807 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gammage PA & Frezza C Mitochondrial DNA: the overlooked oncogenome? BMC Biol. 17, 53 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Priolo C et al. Impairment of gamma-glutamyl transferase 1 activity in the metabolic pathogenesis of chromophobe renal cell carcinoma. Proc. Natl Acad. Sci. USA 115, E6274–E6282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gammage PA, Moraes CT & Minczuk M Mitochondrial genome engineering: the revolution may not be CRISPR-ized. Trends Genet. 34, 101–110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trifunovic A et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429, 417–423 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Mok BY et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature 583, 631–637 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cancer Genome Atlas Research Network et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet 45, 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hyman DM et al. Precision medicine at Memorial Sloan Kettering Cancer Center: clinical next-generation sequencing enabling next-generation targeted therapy trials. Drug Discov. Today 20, 1422–1428 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKenna A et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakravarty D et al. Oncokb: a precision oncology knowledge base. JCO Precis. Oncol 10.1200/pO.17.00011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sonney S et al. Predicting the pathogenicity of novel variants in mitochondrial tRNA with MitoTIP. PLoS Comput. Biol 13, e1005867 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu A, Ibrahim JG & Love MI Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35, 2084–2092 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benjamini Y et al. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. stat. Soc. B 57, 289–300 (1995). [Google Scholar]
- 54.Liberzon A et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sergushichev A An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. Preprint at bioRxiv 10.1101/060012 (2016). [DOI] [Google Scholar]
- 56.Liu Y et al. Comparative molecular analysis of gastrointestinal adenocarcinomas. Cancer Cell 33, 721–735.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ellrott K et al. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst. 6, 271–281.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pettersen EF et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Jurcik A et al. CAVER Analyst 2.0: analysis and visualization of channels and tunnels in protein structures and molecular dynamics trajectories. Bioinformatics 34, 3586–3588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baker NA, Sept D, Joseph S, Holst MJ & McCammon JA Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dolinsky TJ, Nielsen JE, McCammon JA & Baker NA PDB2PQR: an automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data not available for download on public repositories as described in Methods are available on GitHub (https://github.com/reznik-lab/mtdna-mutations).