Abstract
Background and Aims
Current management of inflammatory bowel disease leaves a clear unmet need to treat the severe epithelial damage. Modulation of Wnt signaling might present an opportunity to achieve histological remission and mucosal healing when treating IBD. Exogenous R-spondin, which amplifies Wnt signals by maintaining cell surface expression of Frizzled (Fzd) and low-density lipoprotein receptor-related protein receptors, not only helps repair intestine epithelial damage, but also induces hyperplasia of normal epithelium. Wnt signaling may also be modulated with the recently developed Wnt mimetics, recombinant antibody-based molecules mimicking endogenous Wnts.
Methods
We first compared the epithelial healing effects of RSPO2 and a Wnt mimetic with broad Fzd specificity in an acute dextran sulfate sodium mouse colitis model. Guided by Fzd expression patterns in the colon epithelium, we also examined the effects of Wnt mimetics with subfamily Fzd specificities.
Results
In the DSS model, Wnt mimetics repaired damaged colon epithelium and reduced disease activity and inflammation and had no apparent effect on uninjured tissue. We further identified that the FZD5/8 and LRP6 receptor–specific Wnt mimetic, SZN-1326-p, was associated with the robust repair effect. Through a range of approaches including single-cell transcriptome analyses, we demonstrated that SZN-1326-p directly impacted epithelial cells, driving transient expansion of stem and progenitor cells, promoting differentiation of epithelial cells, histologically restoring the damaged epithelium, and secondarily to epithelial repair, reducing inflammation.
Conclusions
It is feasible to design Wnt mimetics such as SZN-1326-p that impact damaged intestine epithelium specifically and restore its physiological functions, an approach that holds promise for treating epithelial damage in inflammatory bowel disease.
Keywords: Inflammatory Bowel Diseases, IBD, Ulcerative Colitis, UC, Epithelial Repair, Wnt, Frizzled
Abbreviations used in this paper: AltEntero, alternative enterocyte; AltEnteroPC, alternative enterocyte progenitor cell; βGal, β-galactosidase; DAI, Disease Activity Index; DSS, dextran sulfate sodium; FACS, fluorescence-activated cell sorter; FBS, fetal bovine serum; FZD, Frizzled; GFP, green fluorescent protein; GSEA, gene set enrichment analysis; H&E, hematoxylin and eosin; IBD, inflammatory bowel disease; IgG, immunoglobulin; IL, interleukin; IP, intraperitoneally; LGR, leucine-rich repeat-containing G protein–coupled receptor; LRP, lipoprotein receptor-related protein; mRNA, messenger RNA; PBS, phosphate-buffered saline; PBST, Triton X-100 in phosphate-buffered saline; RSPO, R-spondin; RT-qPCR, quantitative reverse-transcription polymerase chain reaction; SA, streptavidin; scRNA-seq, single-cell RNA-sequencing; SNN, shared nearest neighbor; STF, Super TopFlash; TA, transit amplifying; TNF-α, tumor necrosis factor α; UC, ulcerative colitis
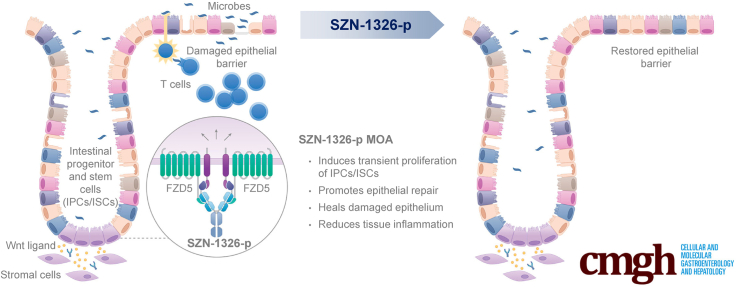
Graphical abstract
Summary.
Our antibody-based receptor-specific Wnt mimetic, SZN-1326-p, repaired damaged colon epithelium in a severe mouse colitis model. SZN-1326-p induced transient expansion of stem and progenitor cells, promoted cell differentiation to restore tissue histology and improved epithelial barrier, leading to inflammation reduction.
Inflammatory bowel disease (IBD) affects 8 million patients globally.1 Disruption of the colon epithelial barrier is a defining characteristic of IBD such as in ulcerative colitis (UC), allowing the exposure of the luminal microbes to the intestinal stromal and immune cells, resulting in inflammation and further damage to the epithelium, creating a vicious cycle.2,3 Although histological remission is arising as a new target for UC treatment,4 all available treatments are anti-inflammatory agents that only indirectly help heal the epithelium by reducing tissue inflammation, and the rates of mucosal healing and histological remission remain low.5, 6, 7, 8
Wnts are secreted, lipid-modified glycoproteins that function as key regulators of stem cells in many tissues.9,10 In the intestine, Wnt signaling plays an important role in maintaining integrity of the epithelium under tissue homeostasis and during injury repair. Canonical Wnt signaling involves 2 receptors, Frizzled (FZD) and low-density lipoprotein receptor-related protein (LRP). Binding of Wnt to FZD and LRP triggers stabilization and translocation of β-catenin into the nucleus, where β-catenin associates with T cell factor or lymphoid enhancer factors to activate Wnt target genes. The 10 FZDs (FZD1–10) are coexpressed in different but overlapping sets across tissues, and there are limited data on the specificity between Wnts and FZDs.11 Synthetic Wnt mimetics with different FZD subfamily specificities have been explored and were shown to bear β-catenin signaling inducing activities in vitro and in vivo.12, 13, 14, 15, 16, 17, 18
Wnt signaling can be regulated by a family of secreted factors, the R-spondins (RSPOs) (RSPO1–4), which enhance Wnt signaling by binding to the cell surface leucine-rich repeat-containing G protein–coupled receptor 4–6 (LGR4–6) and the 2 E3 ubiquitin ligases, RNF43 and ZNRF3. This binding prevents E3 ligase–mediated ubiquitination of the FZD and LRP receptors at the plasma membrane, thereby enhancing the effect of endogenous Wnt ligands on the cell.19, 20, 21 RSPOs also play an important role in intestine epithelial biology where they were shown to enhance proliferation of normal intestinal epithelium18,22,23 and facilitate injury repair in a dextran sulfate sodium (DSS) model.24
In this work, we first showed that Wnt mimetics repaired the injured colon epithelium in a DSS colitis model, reducing the Disease Activity Index (DAI) and histology damage score with better efficacy than RSPO2 or the combination of RSPO2 and Wnt mimetics. Consistent with previous reports, Wnt mimetics alone did not affect normal epithelium, while RSPO2 caused hyperplasia. Next, guided by profiling the expression of Fzds in normal and disease conditions, we narrowed in on Fzd5 and showed that a subfamily Fzd5,8-specific Wnt mimetic, SZN-1326-p, was effective both in vitro in organoid culture and in vivo in intestine injury repair. Using single-cell RNA-sequencing (scRNA-seq), we demonstrated that in the mouse DSS model, SZN-1326-p activated Wnt signaling on colon epithelial cells, including multiple stem and progenitor cells, leading to transient proliferation followed by accelerated epithelial differentiation and healing with a concomitant reduction in inflammation. This effect was limited to the injured region, with no impact on normal intestine tissue. Our work suggests that SZN-1326-p could be a new therapeutic option for the treatment of UC.
Results
Treatment With Wnt Mimetics Rapidly Repaired DSS-Damaged Colon Epithelium
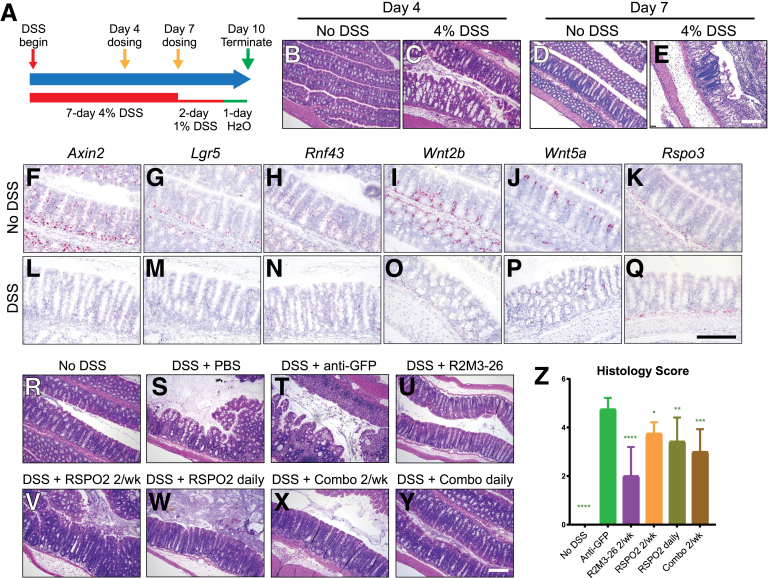
DSS-induced colon colitis mouse models share important disease features with human UC, including the disruption of the intestinal epithelial barrier. We established an acute severe DSS mouse model to study the impact of Wnt signal activation on epithelial repair (Figure 1A). In this model, mice were given 4% (w/v) DSS in the first 7 days to trigger damage to the colon epithelium. Animals were then given 1% (w/v) DSS until takedown at day 10 to maintain the damage and to minimize spontaneous repair of the epithelium. Consistent with previously reported DSS studies,25 damage by DSS to the colon epithelium was visible by hematoxylin and eosin (H&E) stain at day 4 and continued to progress to day 7 (compare Figure 1B–E). RNAscope in situ analyses showed a reduction of messenger RNA (mRNA) expression of Wnt target genes Axin2, Lgr5, and Rnf43 (compare Figure 1F–H to 1L–N). The mRNA expression of Wnt ligands Wnt2b and Wnt5a was also reduced in the colon epithelium and the surrounding mesenchymal cell layers, respectively (compare Figure 1I and J to 1O and P). The mRNA expression of the predominant mouse intestinal RSPO, Rspo3, in the mesenchymal cells underneath the colon crypts was not affected by DSS (compare Figure 1K–Q).
Figure 1.
Wnt mimetics alone repaired the damaged colon epithelium in an acute DSS colitis model. (A) A diagram illustrating the acute DSS model used. DSS was included in drinking water at the indicated percentage. Wnt mimetics and RSPO were dosed twice on day 4 and day 7 (yellow arrows) or daily starting on day 4. Animals were terminated on day 10. (Animals were switched to plain drinking water on the last day for compliance with Institutional Animal Care and Use Committee protocol.) (B–D) H&E stain of transverse colon sections of normal colon and the damaged colon on day 4 and day 7. (F–Q) RNAscope in situ showing expression of Axin2, Lgr5, Rnf43, Wnt2b, Wnt5a, and Rspo3 in normal and DSS colon tissues. (R–Y) Representative H&E stain of transverse colon of the indicated treatment groups: (R) no DSS, (S) DSS with PBS, (T) with 2 treatments of 3 mg/kg anti-GFP, (U) with 2 treatments of 10 mg/kg R2M3-26, (V, W) with 2 or daily treatments of 3 mg/kg RSPO2, and (X, Y) with 2 or daily treatments of 1 mg/kg RSPO2 + 0.3 mg/kg R2M3-26 combo. (Z) Colon histology score of acute DSS conditions after the indicated treatments. Statistical analysis was performed as described in the Methods with all comparisons made to the anti-GFP group. Scale bars = 200 μm.
In the established DSS model, we injected 2 doses of R2M3-26, a Wnt mimetic targeting FZD1,2,5,7,8 and LRP6, referred to as FA-L6 in Fowler et al,14 starting at day 4 when DSS damage to the epithelium was already visible followed by another dose at day 7, and evaluated its effect on epithelial repair at day 10, a 6-day treatment. As shown in Figure 1U, the R2M3-26–treated colon restored crypt architecture with less tissue inflammation as compared with phosphate-buffered saline (PBS) or anti-green fluorescent protein (anti-GFP) immunoglobulin (IgG) control treatments (Figure 1R–U). The histology score consistently showed that R2M3-26 effectively repaired the DSS-damaged colon tissue, reducing the colitis score from 4.75 to 2.0 (Figure 1Z). Because RSPO was previously reported to ameliorate DSS-induced colitis in mice,24 we also examined the effect of RSPO2 in our DSS mouse model. RSPO2 was injected intraperitoneally (IP) either twice weekly or daily starting on day 4 of DSS treatment. While repair to the damaged colon epithelium was observed with RSPO2 treatments, the effect was less compared with the that of R2M3-26 (Figure 1V, W, and Z). Similarly, the combined treatment of R2M3-26 and RSPO2, whether twice weekly or daily, restored colon crypt architecture and improved the colon histology but to a lesser extent than R2M3-26 alone (Figure 1X–Z).
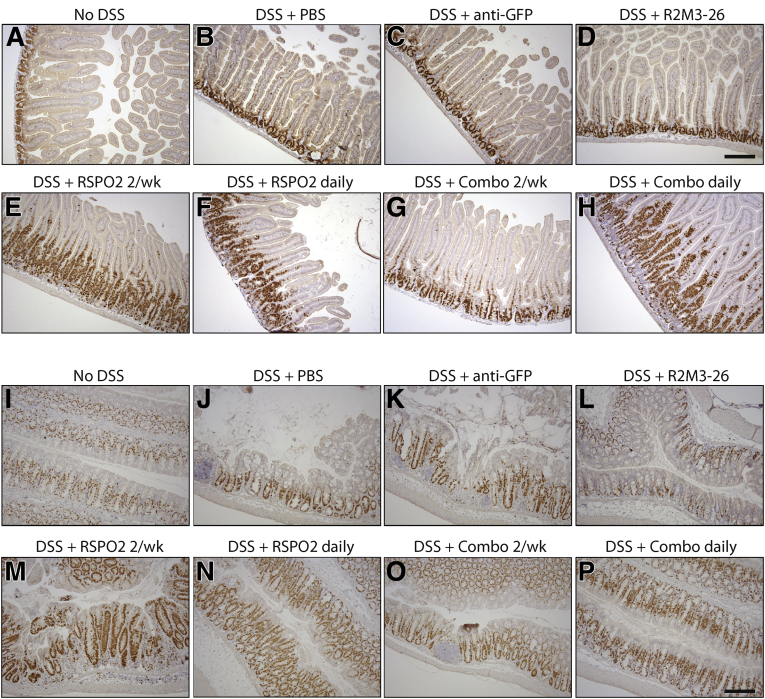
RSPO alone or combined treatment of RSPO and the Wnt mimetic, 18R5-DKK1c, was shown to stimulate hyperproliferation of the small intestine stem cells and transit-amplifying (TA) cells, leading to overgrowth of small intestine crypt and villi length in normal mice.18 The effects of the treatments on epithelial cell proliferation were assessed by Ki67 immunohistochemistry in our DSS model. At day 10, we also observed an expansion of Ki67 expression by RSPO2 treatments or by the combination treatment of R2M3-26 and RSPO2 in the duodenum (Figure 2E–H) and the colon (Figure 2M–P). However, R2M3-26 alone did not lead to an expansion of Ki67, either in the duodenum or in the colon epithelium at day 10 of the DSS model (Figure 2D and L), consistent with a previous study expressing Wnt agonists in uninjured animals.18 Therefore, Wnt agonist treatment alone was able to repair the DSS-damaged colon epithelium without causing hyperproliferation of the intestinal epithelium.
Figure 2.
Wnt mimetics alone did not cause hyperproliferation in the damaged colon or in the small intestine. (A–H) Ki67 staining of duodenum sections of animals treated with (A) no DSS, (B) DSS with PBS injections, (C) DSS with 2 IP injections of 3 mg/kg anti-GFP, (D) DSS with 2 IP injections of 10 mg/kg R2M3-26, (E) DSS with 2 IP injections of 3 mg/kg RSPO2, (F) DSS with daily IP injections of 3 mg/kg RSPO2, (G) DSS with 2 IP injections of 1 mg/kg RSPO2 + 0.3 mg/kg R2M3-26 combo, and (H) DSS with daily IP injections of 1 mg/kg RSPO2 + 0.3 mg/kg R2M3-26 combo. (I–P) Ki67 staining of transverse colon sections of animals treated with Ki67 staining of duodenum sections of animals treated with (I) no DSS, (J) DSS with PBS injections, (K) DSS with 2 IP injections of 3 mg/kg anti-GFP, (L) DSS with 2 IP injections of 10 mg/kg R2M3-26, (M) DSS with 2 IP injections of 3 mg/kg RSPO2, (N) DSS with daily IP injections of 3 mg/kg RSPO2, (O) DSS with 2 IP injections of 1 mg/kg RSPO2 + 0.3 mg/kg R2M3-26 combo, and (P) DSS with daily IP injections of 1 mg/kg RSPO2 + 0.3 mg/kg R2M3-26 combo. Scale bars = 100 μm.
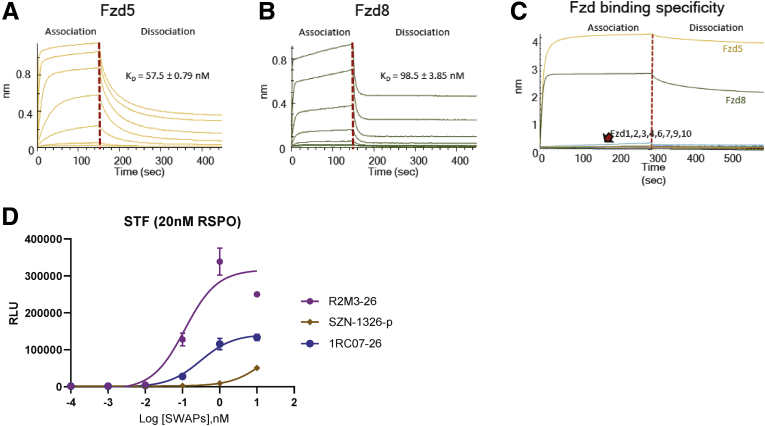
SZN-1326-p, a Fzd5,8-Targeted Wnt Mimetic, Stimulated Growth of Mouse Intestinal Organoids
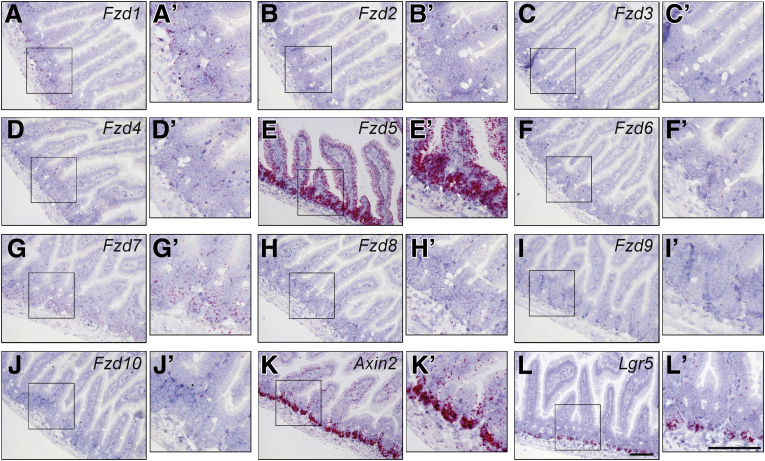
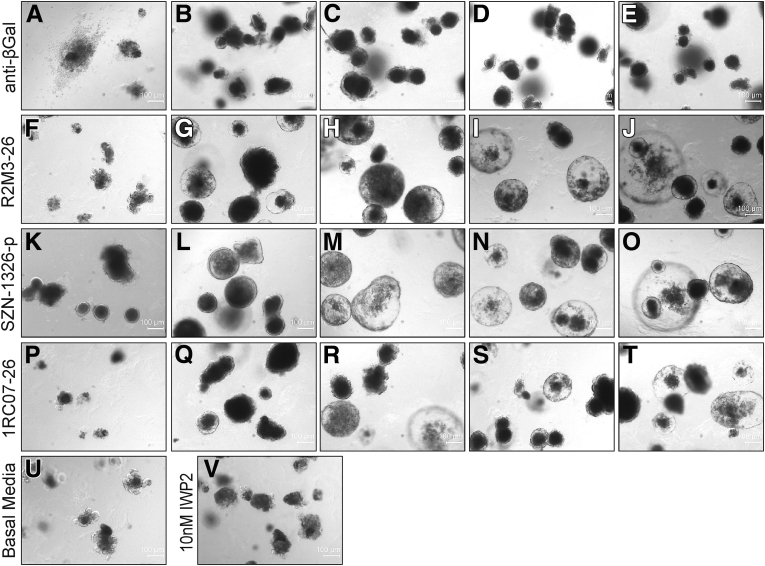
Wnt agonists can be tailored to bind individual Fzd receptors or a subfamily of Fzd receptors to achieve better tissue or cell-type specificity.12,16,17 RNAscope in situ analyses showed that, in the mouse small intestinal epithelium, Fzd5 was expressed at the highest level, followed by Fzd1 and Fzd7 (Figure 3A–J). Fzd1 and Fzd7 are expressed primarily near the crypt bottom where Lgr5 positive stem cells reside (Figure 3L). Expression of Fzd5 was concentrated near the crypt-villi border and in the crypt bottom columnar stem cells in the duodenum overlapping with the strong Axin2 positive domain, which was also positive for the stem cell marker Lgr5 (Figure 3K). We then tested whether stimulating Wnt signaling with a Wnt agonist that is specific to the Fzd5 and Fzd8 subfamily (Figure 4A–C) or to the Fzd1, Fzd2, and Fzd7 subfamily was sufficient to stimulate epithelial cell proliferation in a mouse small intestinal organoid culture. The subfamily specific Wnt mimetics are active in vitro in the Super TopFlash (STF) assay (Figure 4D). Mouse small intestinal organoids were treated with the Porcupine inhibitor IWP2 to inhibit endogenous Wnt ligand secretion in the cultured organoid. When these organoids were subjected to no protein treatment (Figure 5V) or were treated with a control anti-β-galactosidase (βGal) IgG (Figure 5A–E), the organoids were not maintained and quickly degenerated. In contrast, treatment with R2M3-26, the Fzd1,2,5,7,8 pan-specific Wnt mimetic, at a wide dose range was able to stimulate cell proliferation, producing growing transparent sphere-shaped organoids (Figure 5F–J). Both a Fzd5,8-specific Wnt mimetic, SZN-1326-p, and a Fzd1,2,7-specific Wnt mimetic, 1RC07-26 (referred to as FB-L6 in Fowler et al),14 were able to stimulate organoid proliferation and growth (Figure 5K–T). The effects of the subfamily specific Wnt mimetics were comparable to the effect of the pan-specific agonist.
Figure 3.
The FZD family of receptors showed differential expression pattern in the small intestinal epithelium. (A–L) expression of each of the 10 Fzd receptors Fzd1–10, Axin2 and Lgr5 in the normal duodenum was examined by RNAscope in situ hybridization. (A’–L’) insets with zoom in view showing Fzd expression in the small intestinal crypts. Arrows in E’ indicate intestinal stem cells. Scale bars = 100 μm.
Figure 4.
In vitro activity of Fzd5,8 subfamily–specific Wnt mimetic SZN-1326-p. (A) The binding affinity of the Fzd5,8 binder IgG of SZN-1326-p to its target Fzd5 CRD measured on Octet. equilibrium dissociation constant (KD) indicated on the graph. (B) The binding affinity of the Fzd5,8 binder IgG of SZN-1326-p to its target Fzd8 CRD measured on Octet. KD indicated on the graph. (C) The binding specificity of the Fzd5,8 binder IgG of SZN-1326-p to each of the 10 Fzd CRDs examined on Octet. (D) The dose-dependent STF activities of SZN-1326-p, of the Fzd1,2,7-specific mimetic 1RC07-26 and of the Fzd1,2,5,7,8 pan specific mimetic R2M3-26 in the presence of 20 nM RSPO2 measured in Huh-7 cells. RLU, relative luminescence units.
Figure 5.
The Fzd5,8-specific Wnt mimetic, SZN-1326-p, effectively promoted intestinal organoid growth. (A–T) Representative mouse small intestinal organoids treated with a dose titration of the indicated Wnt mimetic with different Fzd specificity. For each protein, organoids were treated with, from left to right, (A, F, K, P) 10 pM, (B, G, L, Q) 100 pM, (C, H, M, R) 1 nM, (D, I, N, S) 10 nM, or (E, J, O, T) 100 nM of the negative control anti-βGal or the indicated Wnt mimetic in the presence of 10 nM IWP2 in basal media. (U) Organoids grew in basal media. (V) Organoids treated with only 10 nM IWP2 in basal media. Scale bars = 100 μm.
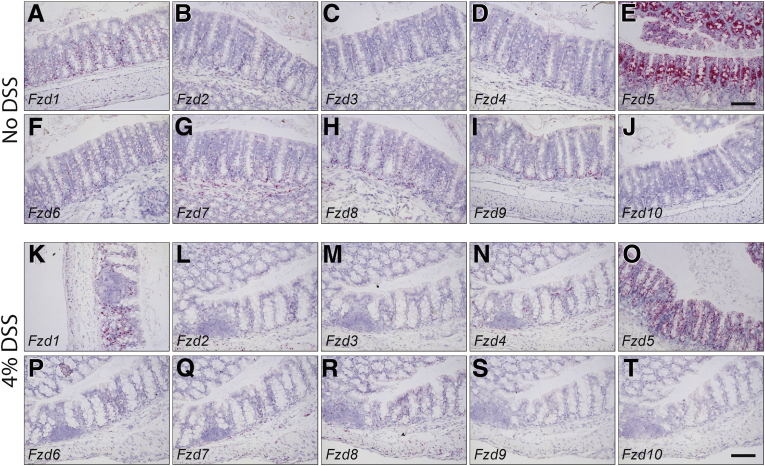
The Fzd5,8-Specific Wnt Mimetic, SZN-1326-p, Was Efficacious in Repairing the DSS-Damaged Colon Epithelium
In situ analysis demonstrated that the colon epithelium had a Fzd expression pattern similar to the small intestine (Figure 6A–J) and that Fzd5 was also expressed at the highest level among all Fzds in the colon epithelium. This differential expression of Fzds was maintained in the DSS condition, albeit the expression of all Fzds was reduced by DSS (Figure 6K–T). We next examined if the Fzd subfamily–specific mimetics were able to repair the DSS-damaged colon epithelium. In the DSS model, 2 doses of control anti-GFP IgG treatment or Wnt mimetic treatment were injected IP on days 4 and 7 and the animals were sacrificed on day 10 for histology and serum analyses (Figure 7A). In contrast to the severe tissue damage and inflammation observed in the no protein treatment or the anti-GFP-treated colon (Figure 7C and D), both SZN-1326-p (Fzd5,8) and 1RC07-26 (Fzd1,2,7) treatments resulted in repair of the colon epithelium (Figure 7F and G). The effect on colon histology from the 2 Fzd subfamily–specific Wnt mimetics was comparable to that of the Fzd1,2,5,7,8 pan-specific mimetic, R2M3-26 (Figure 7E). Similar to R2M3-26 treatment, fecal score and DAI also improved with SZN-1326-p and 1RC07-26 treatment (Figure 7H and I). Improvement in fecal score and DAI was greater with SZN-1326-p as compared with R2M3-26 or 1RC07-26. Consistent with the DAI, the overall histology score of SZN-1326-p–treated DSS colon was significantly improved and was better than that of the 1RC07-26-treated colon (Figure 7J), suggesting more efficacious colitis reduction and epithelial repair from the Fzd5,8-specific Wnt mimetic, SZN-1326-p, than the Fzd1,2,7-specific Wnt mimetic, 1RC07-26.
Figure 6.
The FZD family of receptors were expressed at different levels in the colon. (A-J) colon expression of the 10 Fzd receptors in naïve mice was examined by RNAscope in situ hybridization. (K–T) colon expression of the 10 Fzd receptors in mice treated with 7 days of 4% DSS. Scale bars = 100 μm.
Figure 7.
Fzd5,8-specific SZN-1326-p repaired DSS damaged colon and improved DAI and inflammation. (A) Diagram illustrating the acute DSS study design. (B–G) H&E stain of transverse colon sections of animals treated (B) with no DSS or (C) with DSS with PBS, (D) with anti-GFP, (E) with 2 doses of 10 mg/kg R2M3-26, (F) with 2 doses of 10 mg/kg SZN-1326-p, and (G) with 2 doses of 3 mg/kg 1RC07-26. Scale bar = 200 μm. (H) Daily animal fecal score of each treatment group. (I) Disease activity (DAI) progression for each treatment group. (J) Composite histology score reading by an independent pathologist, on colon samples harvested at termination. (K–M) Effect of Wnt mimetics on serum level of proinflammatory cytokines, (K) TNF-α, (L) IL-6, (M) IL-8. (N-P) Effect of a wide dose range of SZN-1326-p, both as a single dose (N) or as twice weekly doses (O), on DAI and colon histology score (P). Statistical analysis was performed as described in the Materials and Methods with all comparisons made with the anti-GFP group.
DSS-induced colitis damage causes luminal pathogen infiltrate and tissue immune response, resulting in elevation of serum cytokines. We then asked whether the colitis reduction observed with the Wnt mimetics would be accompanied by reduced serum cytokine levels. Indeed, treatment with each of the 3 Wnt mimetics reduced the DSS-induced serum levels of the proinflammatory cytokines, tumor necrosis factor α (TNF-α), interleukin (IL)-6, and IL-8 (Figure 7K–M).
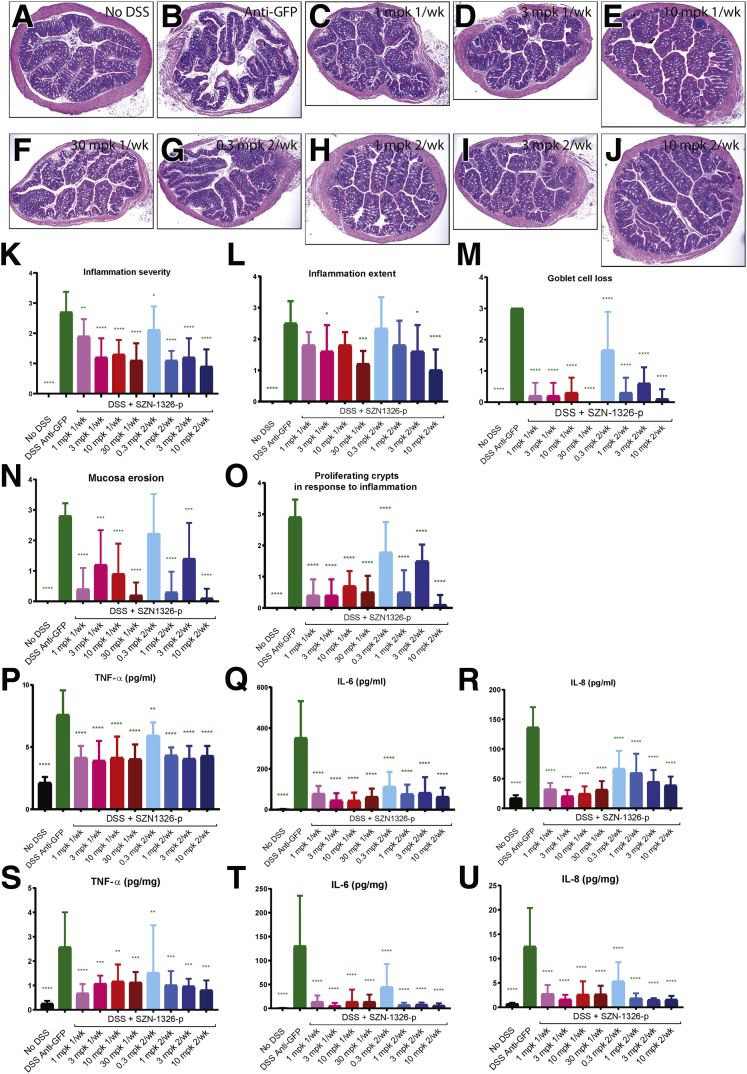
Efficacy of SZN-1326-p in the DSS model was further tested with a dose-ranging study, where SZN-1326-p was injected IP either once on day 4 at 1, 3, 10, and 30 mg/kg or twice on day 4 and day 7 at 0.3, 1, 3, and 10 mg/kg. Significant improvement of tissue histology, DAI and histology scores were observed for all dose groups (Figures 8A–O and 7N–P). All dose groups also showed significant reduction in serum and tissue levels of proinflammatory cytokines TNF-α, IL-6, and IL-8 (Figure 8P–U).
Figure 8.
The Fzd5,8-specific mimetic, SZN-1326-p, showed dose response efficacy in the acute DSS model. (A–J) H&E stain of cross sections of transverse colon of animals treated with (A) no DSS, (B) DSS + anti-GFP, (C) DSS + 1 dose of 1mpk SZN-1326-p, (D) DSS + 1 doses of 3 mpk SZN-1326-p, (E) DSS + 1 dose of 10 mpk SZN-1326-p, (F) DSS + 1 dose of 30 mpk SZN-1326-p, (G) DSS + 2 doses of 0.3 mpk SZN-1326-p, (H) DSS + 2 doses of 1mpk SZN-1326-p, (I) DSS + 2 doses of 3 mpk SZN-1326-p, and (J) DSS + 2 doses of 10 mpk SZN-1326-p. (K–N) Histology score reading of the 5 indicated parameters by an independent pathologist, on colon samples harvested at termination on day 10. (P–R) Serum levels of proinflammatory cytokines TNF-α, IL-6, and IL-8 measured in the SZN-1326-p dose response study. (S-U) Colon tissue cytokine levels of TNF-α, IL-6, and IL-8 measured in the SZN-1326-p dose response study.
DSS Injury Induced Expression of Inflammatory Pathway Genes in the Epithelium
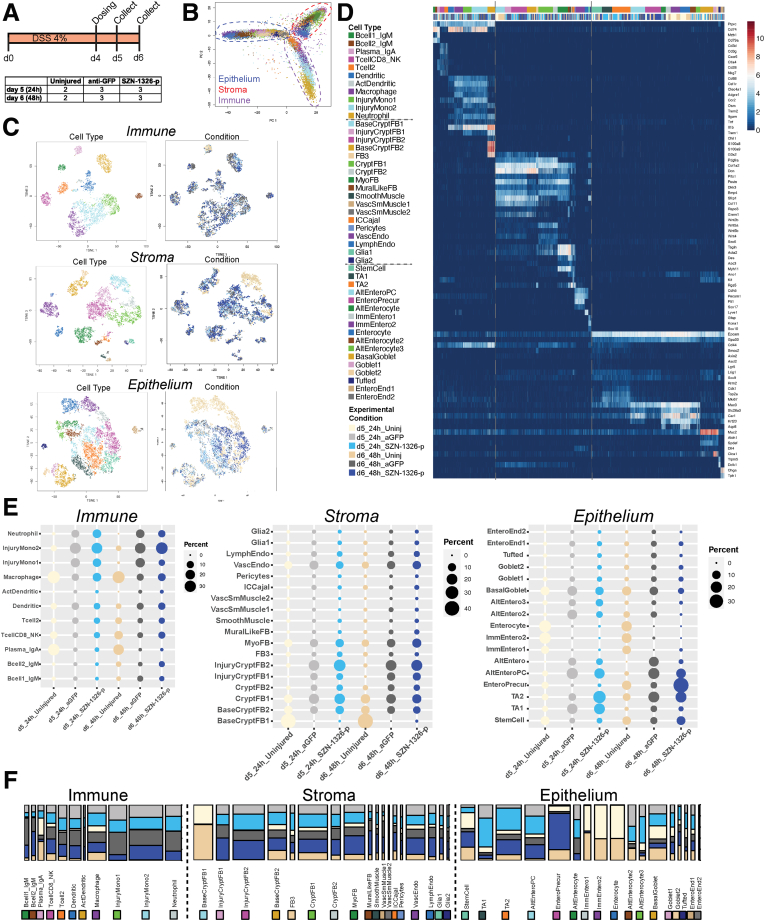
Our tissue-level analysis in the acute DSS model led to several key questions. First, what cells responded first to treatment by SZN-1326-p? Second, how did SZN-1326-p impact differentiation of epithelial cells? Third, was the effect on reducing inflammatory cytokines directly on immune cells or indirectly through restoration of the epithelium? To address these questions, we applied scRNA-seq to investigate the early transcriptome response of the SZN-1326-p–treated colon in the acute DSS mouse model. As in the previous studies, 4% (w/v) DSS was administered in the water, and mice were injected IP on day 4 with 10 mg/kg of either the anti-GFP control protein or SZN-1326-p. Colon was collected for scRNA-seq at days 5 and 6, 24 and 48 hours postinjection, respectively (Figure 9A). After filtering, our dataset contained 22,717 total cells. We normalized and applied cluster analysis to the complete dataset to identify each lineage and group, subsequently subdivided each lineage and group, and applied dimensionality reduction and cluster analysis on the subset of cells in each (Figure 9B–D). There were 3 major cell groups, immune (4835 cells), mesenchyme and stroma (7509), and the epithelium (10373) (Figure 9B–D).
Figure 9.
scRNA-seq revealed a range of cell types across all tissue layers, including several that were injury specific. (A) Experimental design of the scRNA-seq experiment. (B) Plot of the first 2 principal components showing the 3 tissue layers radiating from the center. (C) t-SNE projections showing the annotated clusters (cell type), and experimental condition of the cells. The legend in D applies to C. (D) Log2 normalized UMI counts for all cell types across all 3 tissue layers or lineages. The top color bar refers to cell type; the bottom bar denotes experimental condition. Color annotation for experimental conditions also applies to E and F. (E) For the immune, stromal, and epithelial cells, the percentage of cells from each experimental condition (indicated on the bottom) comprises each cluster (indicated on the left). The columns sum to 100% for that condition. (F) Barplots representing the composition of each cell type by experimental condition: each bar represents 100% of the cells in that cell type; the wider the bar, the more cells comprise that cell type. See D for color annotation of experimental conditions.
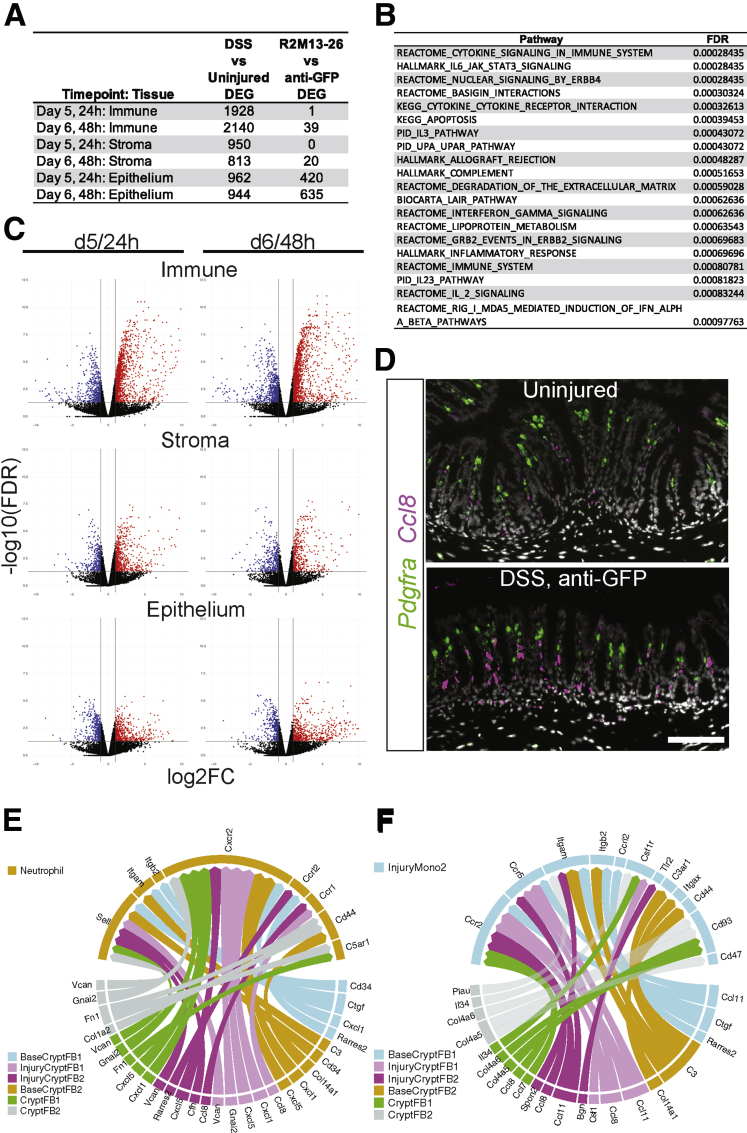
We first assessed the effect of DSS injury by comparing the DSS, anti-GFP condition with the uninjured condition. DSS induced distinct cell types in each tissue layer or lineage, and this was responsible for a large portion of the lineage level differential gene expression. In the immune lineage, no cell types disappeared upon injury. Rather, by day 5 of DSS treatment, several cell types appeared in the damaged colon samples. In the stromal cells, DSS resulted in the appearance of new populations of fibroblasts expressing inflammatory cytokines and chemokines, consistent with recent reports of proinflammatory fibroblasts in UC patients and the DSS mouse model.26,27 For a detailed discussion of the inflammation-associated impact of DSS on the stromal and immune cells, please refer to the respective lineage and layer cell annotations in the Materials and Methods, Figure 10 and Supplementary Tables 1 and 2.
Figure 10.
DSS damage induced large-scale inflammatory responses, including potential stromal to immune cell signaling. (A) The number of genes that are significantly differentially expressed (false discovery rate [FDR] < 0.05, log2FC ≥ |1|) in each tissue layer or lineage by time point in 1 of the 2 comparisons: DSS damage (anti-GFP) vs uninjured and SZN-1326-p/DSS damage vs anti-GFP/DSS damage. (B) The top 20 gene sets or pathways identified by GSEA that were enriched in the stromal cells upon DSS treatment compared with the uninjured state at day 5. (C) Volcano plots of the differentially expressed genes between the DSS-damage and uninjured conditions for the indicated tissue lineage or layer and time point. Genes that display at least a 2-fold change and have an FDR of <0.05 are indicated in red (increased) and blue (decreased). Any gene with an FDR lower than 10E-12 was floored at 10E-12. (D) RNA in situ hybridization of the chemokine Ccl8 and the fibroblast marker Pdgfra in the colon of uninjured, and DSS/anti-GFP samples at day 5. Scale bar represents 100 mm. (E, F) Cell-cell interaction plots displaying the top 5% of potential signaling interactions between the cell types represented in the bottom half of the circle with the target cell type represented by the top half of the circle. The arrows represent the ligand toward the receptor.
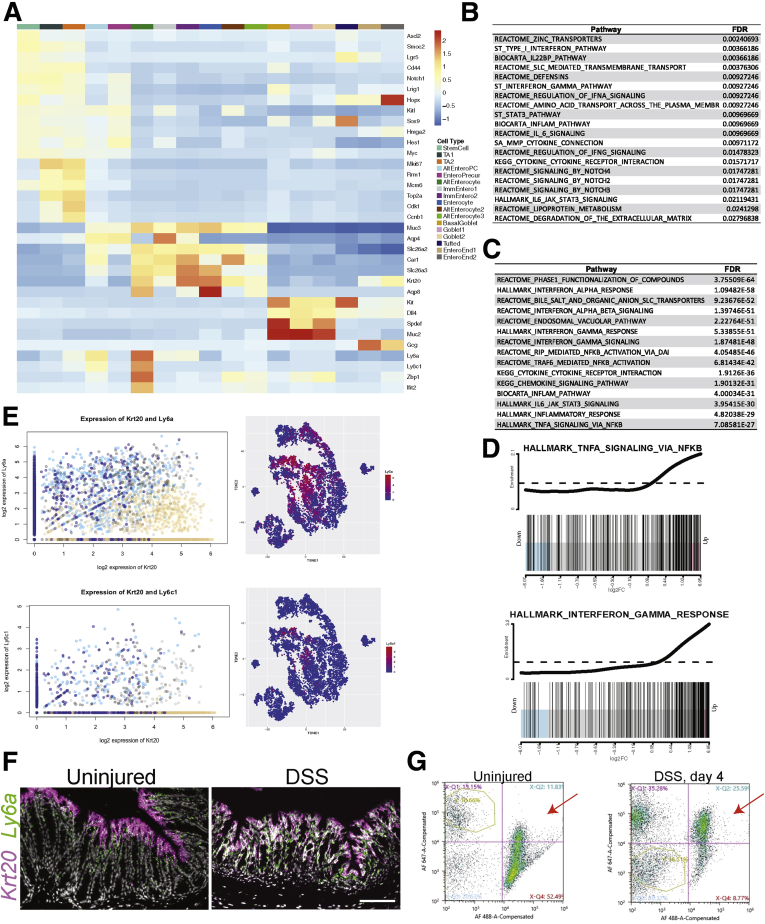
When we compared the epithelial lineage in the DSS-damaged condition to the uninjured epithelium, inflammatory pathways were enriched (Figure 11B), and several cytokines and immune signaling molecules were enriched in the epithelium upon DSS treatment (Supplementary Table 3). A census of the cell types present on day 5 showed that DSS damage to the epithelium led to the alteration of normal, differentiated absorptive cells. The main enterocyte lineage in the uninjured state was composed of 3 cell populations in our data from both time points (immature enterocyte 1 and 2 and enterocyte) (Figure 9C–F). However, in the DSS-injured epithelium, there were cells with enterocyte gene expression signatures that coexpressed genes associated with immune signaling. This was most obvious in 2 groups of cells, annotated as alternative enterocytes (AltEnteros) and AltEntero progenitor cells (AltEnteroPCs). These 2 populations are almost exclusively derived from DSS-treated animals (Figure 9E and 9F). Owing to the expression of genes such as Ly6a, Ly6c1, Zbp1, and Ifit2, these cells appeared to have a gene expression profile similar to the inflammatory signaling observed in fetal-like stem cells (Figure 11A).28,29 By gene set enrichment analysis (GSEA), these cells showed strong enrichment for inflammatory signaling, such as Interferon Gamma and TNFα-mediated nuclear factor kappa B signaling (Figure 11C and D). The AltEnteros and AltEnteroPCs appeared to be de-differentiating enterocytes, coexpressing mature enterocyte genes such as Krt20 with high levels of inflammatory and fetal-like stem cell genes such as Ly6a and Ly6c1, a phenomenon specific to the injury condition (Figure 11E). We confirmed the expression of Ly6a in the Krt20 expressing cells by RNA in situ hybridization, where they were coexpressed in the apical region of the crypt (Figure 11F), and we detected a higher percentage of LY6A expressing epithelial cells by fluorescence-activated cell sorting (FACS) upon DSS treatment (Figure 11G). The AltEnteroPCs had begun to initiate the cell cycle (Figure 11A), suggesting that at least some of these cells were injury-induced progenitors derived from enterocytes. Additionally, injury condition cells that clustered in the stem cell and TA1 and TA2 groups also showed elevated expression of immune genes such as Ly6a (Figure 11E), suggesting that even though they were similar to the uninjured stem and progenitor cells, they were altered to some degree by the injury. In sum, DSS damage leads to injury-specific, inflammatory epithelial cell states that include progenitors.
Figure 11.
DSS injury induces specific inflammatory cell types/states in the epithelium. (A) Row-scaled Z scores for each gene across the aggregate of each cell type in the epithelium. The color bar corresponds to the cell type. (B) Selected top gene sets or pathways enriched in the epithelium at day 5 upon DSS damage determined by GSEA. (C) Selected top gene sets enriched in the AltEnteros injuryiinduced cell type at day 5 (24 hours) relative to the other cell types in the epithelium. (D) The distribution of gene expression for the 2 indicated gene sets in the AltEnteros. (E) (top left) The log2-normalized expression of Krt20 and Ly6a in the epithelial cells showing coexpression of high levels of Ly6a with Krt20 in the DSS injury conditions. (Top right) t-SNE plot colored by Ly6a expression in the epithelial cells. (Bottom left) The log2-normalized values showing coexpression of Krt20 and Ly6c1 in the epithelial cells. (Bottom right) t-SNE plot of the epithelial cells colored by the expression of Ly6c1. (F) RNA in situ of the enterocyte marker, Krt20, and Ly6a. Scale bar = 100 mm. (G) FACS plots showing the degree of enrichment of EPCAM-positive, LY6A-positive cells (upper right quadrant of each plot) in the uninjured condition (left) or upon 4 days of DSS damage (right).
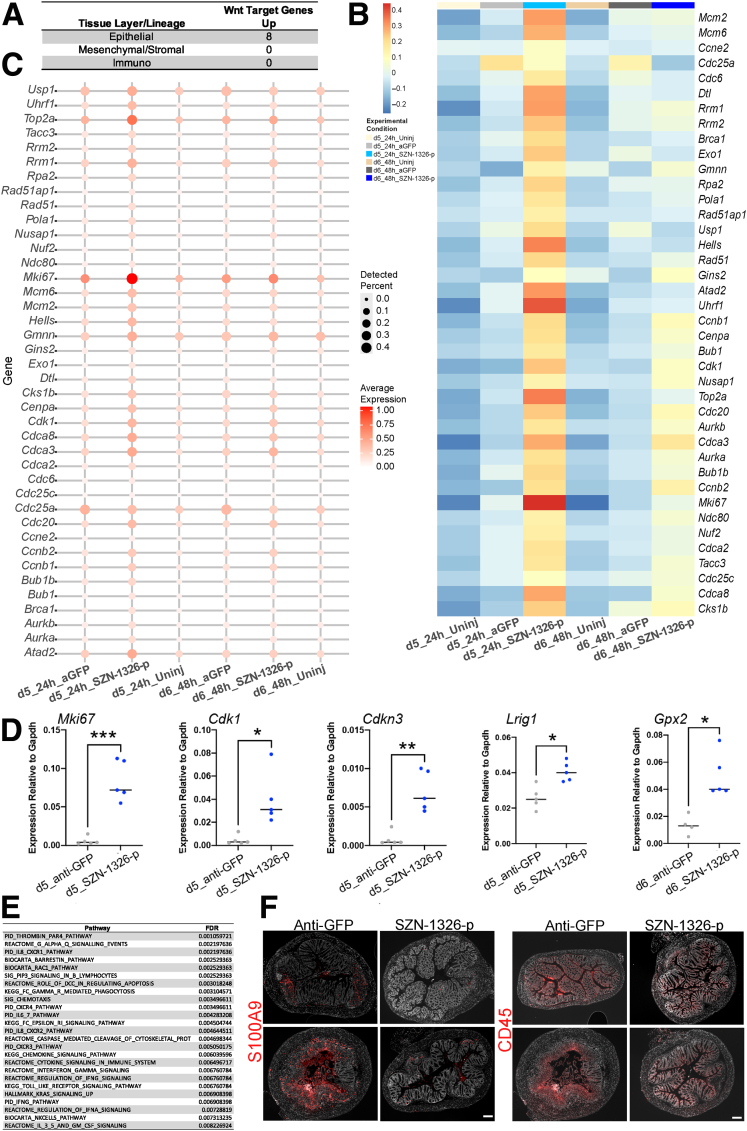
SZN-1326-p Promoted Wnt Target and Cell Cycle Gene Expression and Expanded the PC Populations in the Epithelium Immediately Following Dosing
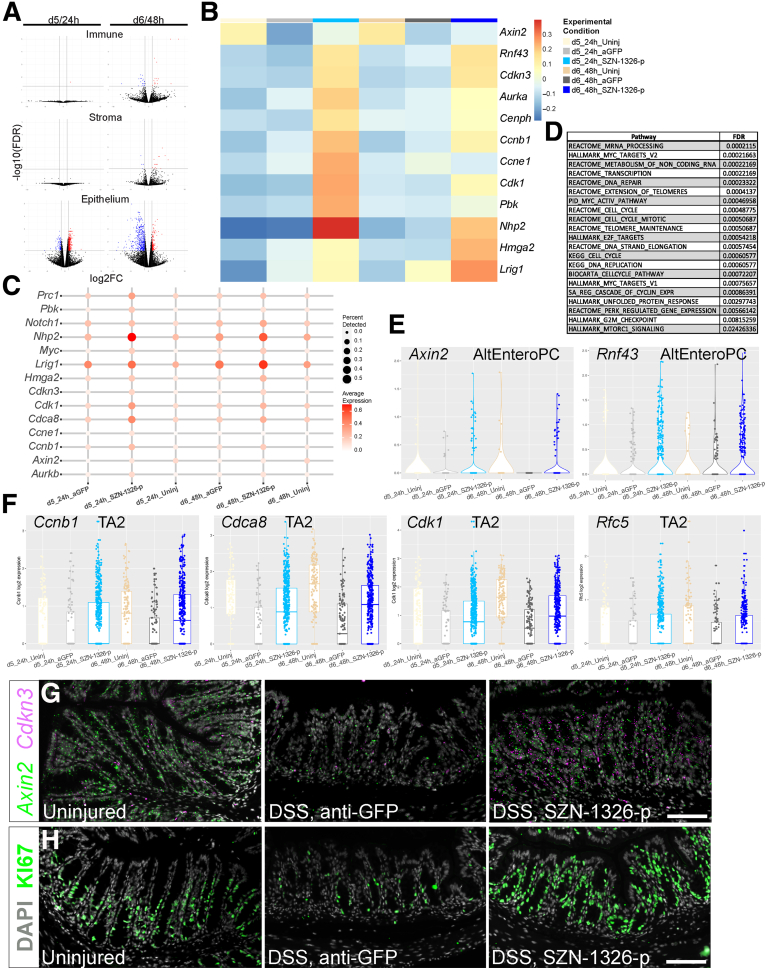
The direct effect of SZN-1326-p was predominately on the epithelium. At a global level, at 24 hours after dosing, SZN-1326-p led to the differential expression of over 400 genes in the epithelium but almost no significant effect on the immune and stromal cells (Figures 10A and 12A). SZN-1326-p increased expression of a wide range of Wnt target and cell cycle genes in the epithelium, both by increasing expression levels and by expanding the percentage of cells expressing the genes (Figures 12B and C and 13B and C; Supplementary Tables 4 and 5). GSEA on the epithelium comparing SZN-1326-p to anti-GFP treatment showed that the cell cycle, telomere maintenance, the Wnt target Myc pathway, mammalian target of rapamycin complex signaling, and the unfolded protein response stress response were strongly upregulated in the epithelium by SZN-1326-p (Figure 12D; Supplementary Table 6).
Figure 12.
SZN-1326-p increased Wnt target and cell cycle gene expression and expanded the progenitors in the epithelium. (A) Volcano plots of gene expression differences between the SZN-1326-p treatment and the anti-GFP treatment in the DSS damage model: the x-axis represents log2 expression, and the y-axis represents the -log10 of the adjusted P value (FDR) from differential expression analysis. Genes that show greater than absolute log2-fold change of 1 and FDR <0.05 are indicated in blue (decreased) and red (increased) in the indicated lineage or tissue layer and time point. Any gene with an FDR lower than 10E-5 was floored at 10E-5. (B) Average Z-scores of Wnt target, cell cycle, and progenitor gene expression aggregated by experimental condition within the epithelial lineage. (C) Relevant Wnt target, cell cycle, and stem/progenitor gene expression; size and intensity represent the percentage of cells with detectable expression and expression level, respectively. (D) Selected top gene sets or pathways (from GSEA) enriched in the SZN-1326-p treatment relative to the anti-GFP treated DSS-injured epithelium. (E) Expression of the Wnt target genes, Axin2 and Rnf43, in the AltEnteroPCs. (F) Boxplots showing expression of the cell cycle genes, Ccnb1, Cdca8, Cdk1 and Rfc5, that were enriched in the TA2 cells upon SZN-1326-p treatment. (G) RNA in situ hybridization of 2 Wnt target genes, Axin2 and Cdkn3 at day 5; nuclei labeled with DAPI. (H) Immunohistochemistry for the proliferative cell marker Ki67 at day 6; nuclei labeled with DAPI. Scale bars = 100 mm.
Figure 13.
SZN-1326-p predominately impacted the epithelium by promoting expression of Wnt target and cell cycle genes with a concomitant reduction in inflammatory pathway expression. (A) The number of significantly enriched intestinal epithelial organoid Wnt target genes in the indicated colon tissue layers or lineages. (B) Heatmap of canonical cell cycle gene expression in the epithelium displaying average Z scores for the genes. (C) Gene expression dot plot of canonical cell cycle genes in the epithelium across the 6 different experimental conditions. (D) RT-qPCR of bulk colon samples in the DSS model at the day 5 time point except Gpx2, which is from day 6. Unpaired, 2-tailed t tests were applied with the Holm multiple comparisons correction: ∗P < .05, ∗∗P < .01, ∗∗∗P = .001. (F) Selected top pathways reduced (derived from GSEA) in the epithelium after SZN-1326-p treatment relative to the anti-GFP treatment in the DSS model at 48 hours (day 6). (G) Expression of S100A9 and CD45 at day 10 (6 days after treatment); 2 example images each from the indicated treatment condition. Scale bar = 200 μm.
Although effects of SZN-1326-p were observed on the stromal and immune cells, both the total number of genes (Figure 10A) and the number of reported intestine-specific Wnt target genes30 that were affected relative to the anti-GFP treatment was notably lower (Figure 13A). Importantly, we did not detect Axin2 enrichment at either the lineage or cell type level in any stromal or immune cells (Supplementary Tables 2). Furthermore, there were very few, if any, pathways significantly enriched in the stromal or immune cells by GSEA when we compared SZN-1326-p with anti-GFP treatment (Supplementary Table 6), again confirming that the predominant, direct impact of SZN-1326-p was on the epithelium at 24 hours after dosing. Although we did not see a major impact of SZN-1326-p on the stromal or immune cells at day 5 or even at day 6, there was a reduction in immune cells and cytokine levels over time that was detectable by day 10 (Figures 7K–M and 13F), likely secondary to the direct impact of SZN-1326-p on the epithelium.
The predominant cell types impacted by SZN-1326-p were the progenitor and precursor populations, including the injury-induced, AltEntero cell types. Differential expression analysis revealed a significant increase in Wnt target genes in several distinct cell types (eg, TA2, AltEnteroPC, AltEnteros, enterocyte precursors) (Figure 12E; Supplementary Table 4). SZN-1326-p also significantly increased expression of many cell cycle genes in multiple cell types in the epithelium (Supplementary Table 5), especially the TA2 and injury-specific progenitors (AltEnteroPC) (Figure 12F). We validated the increase in Wnt target gene expression, detecting an expansion of Axin2 and Cdkn3 expression in the colon crypts of the SZN-1326-p–treated samples (Figure 12G). Furthermore, the TA1 and TA2 progenitor cells had the highest expression of genes associated with cell cycle (Figure 11A), and there was an expanded contribution of the SZN-1326-p–treated samples in these groups at 24 hours after treatment (Figure 9E), consistent with expansion of the progenitors early after dosing. We validated the increase in cell cycle gene expression by quantitative reverse transcription-polymerase chain reaction (RT-qPCR) on colon samples (Figure 13D). Furthermore, immunohistochemistry for Ki67 detected a robust increase in the number of proliferative cells in the colonic epithelium upon SZN-1326-p treatment when compared with the anti-GFP treatment group by 48 hours after dosing (Figure 12H).
In addition to increasing expression of genes directly involved in the cell cycle, SZN-1326-p also increased expression of several stem or progenitor cell genes such as Lrig1,31 Hmga2,32 Hes1,33 and Nhp2 (Figures 12B and C and 13D; Supplementary Table 3).34 In summary, immediately after dosing, SZN-1326-p increased Wnt target and cell cycle gene expression in multiple epithelial cell types, predominantly in the different subtypes of stem and progenitor cells including the injury-induced, AltEntero cell types, leading to an expansion of the progenitor pool.
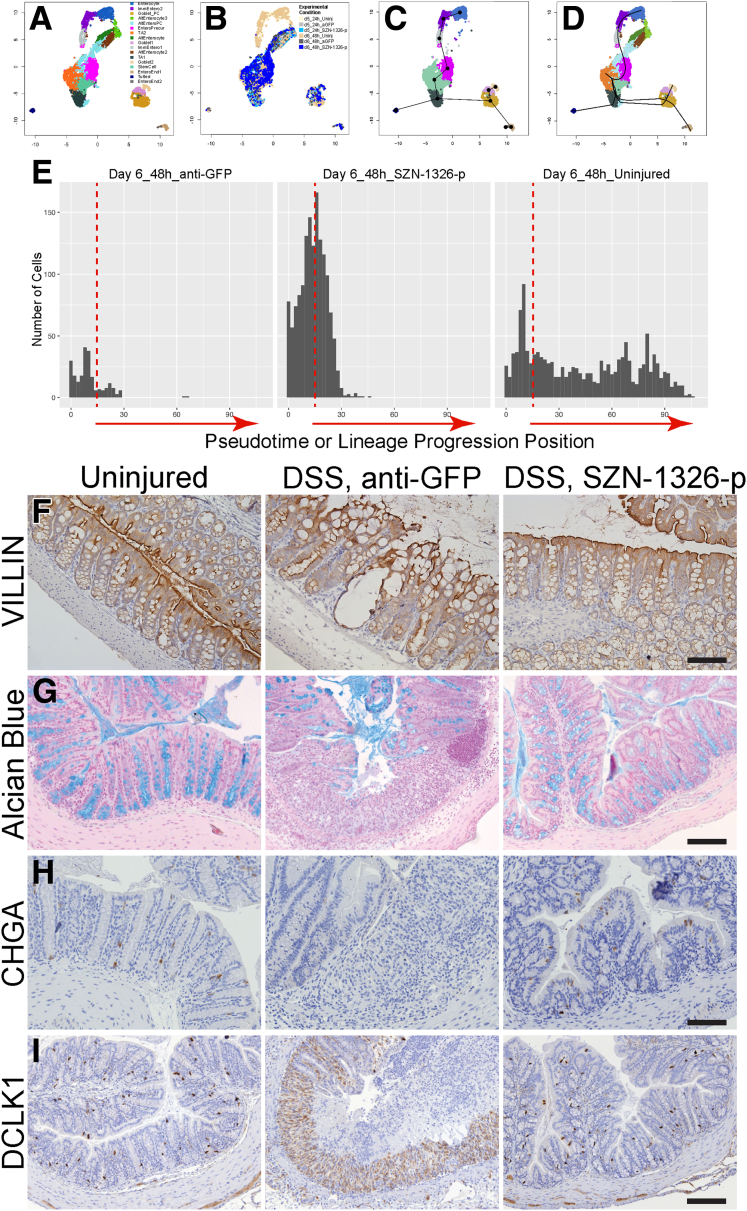
SZN-1326-p–Treated Epithelial Cells Differentiated More Quickly After Proliferation
Time-stamping allowed us to detect where the day 6 (48-hour) cells were enriched relative to the day 5 (24-hour) cells for all 3 treatment conditions: uninjured, injured or anti-GFP, and injured or SZN-1326-p. The day 5 and day 6 uninjured cells were approximately equally represented in all clusters as expected (Figures 9C–F and 14A and B). However, we noticed apparent differences in the cell types that were preferentially enriched for the SZN-1326-p– or anti-GFP–treated day 5 and day 6 injured samples. For the anti-GFP samples, there were more cells in the AltEntero groups (AltEntero2, AltEntero3) and the TA1 groups at day 5 relative to the day 6 time point, and there were about equal percentages of cells in the AltEnteroPCs at both time points. In the SZN-1326-p samples, there were more TA1 and TA2 cells at day 5 relative to day 6, and a higher percentage of stem cells at day 6 relative to day 5. Importantly, there was a substantial enrichment of SZN-1326-p–treated cells in the enterocyte precursors at day 6 relative to the anti-GFP–treated samples. These together suggested that the SZN-1326-p–treated cells appeared accelerated in differentiating toward enterocytes by day 6.
Figure 14.
SZN-1326-p treatment caused accelerated differentiation and improved barrier marker expression in the DSS model. (A–D) Uniform manifold approximation and projection (UMAP) of the epithelial cells. (A) UMAP of epithelial cells colored by cluster or cell type. (B) UMAP colored by experimental condition. (C) The lineage trajectory prediction algorithm, slingshot, was applied to the epithelial lineage. The stem cell and TA2 cell types were merged and set as the starting cluster. (D) The lineage trajectory indicating a transition from the stem cell or TA cells to the enterocyte precursor (EnteroPrecur) cells on the way to the immature and mature enterocytes (going up) and from the stem cell or TA cells down and bifurcating either toward tufted cells or goblet and enteroendocrine cells. (E) The number of cells at the 48-hour or day 6 time point along the trajectory of the enterocyte lineage in D. The vertical dashed lines represent the same position along the axis in all 3 plots. (F–I) Staining of differentiated cell type markers: (F) VILLIN for enterocytes, (G) Alcian blue for goblet cells, (H) CHGA for enteroendocrine cells, and (I) DCLK for tuft cells at day 10 in the acute DSS injury model. Experimental condition indicated at the top. Scale bars = 100 μm.
To complement our timestamp-based observations, we employed the lineage trajectory inference tool, slingshot. Because we had evidence that some enterocytes were de-differentiating upon DSS injury, we removed the apparent de-differentiating or altered state enterocyte clusters and applied slingshot to the cell clusters that include at least 5% of cells from the uninjured condition. We set the combined stem cell and TA2 cells as the starting point (Figure 14A and C), and slingshot predicted that from the initial starting group, cells would progress toward TA1, goblet, tufted, and enteroendocrine in one direction and toward the enterocytes in the other (Figure 14D). Based on the predicted lineage trajectory pseudotime values, there was a higher percentage of SZN-1326-p–treated samples that were further along in the enterocyte lineage trajectory by day 6 (48 hours) relative to the control-treated cells (Figure 14E), consistent with the actual previous timestamping data. The ultimate validation of improved differentiation was that expression of markers of the differentiated cell types, enterocytes, goblet cells, enteroendocrine and tuft cells, resembled that of uninjured colon in the SZN-1326-p treatment group relative to the anti-GFP control group on day 10 (6-days after SZN-1326-p treatment) (Figure 14F–I).
SZN-1326-p Treatment Led to Epithelial Barrier Marker Restoration
In our studies looking at day 10 after injury, we observed that SZN-1326-p treatment led to repair of the epithelium at a histological level (Figure 7). There was an increase in mucin and barrier-associated gene expression in the SZN-1326-p–treated samples relative to anti-GFP in a few stem and progenitor cell types (Figure 15A). When we assessed expression of the tight junction marker, TJP1 (ZO1), its expression was restored in the SZN-1326-p–treated colon at day 10 (Figure 15B), consistent with re-establishment of tight junctions and reduced tissue inflammation (Figure 8S–U).
Figure 15.
SZN-1326-p increased barrier gene expression and led to barrier marker restoration in the DSS injury model. (A) Differential gene expression values for significantly enriched mucin and barrier associated genes in the indicated epithelial cell type, comparing DSS/SZN-1326-p with DSS/anti-GFP. (B) Immunofluorescence staining of the tight junction marker TJP1 (ZO-1) (green) in transverse colon samples of the indicated treatment at day 10. Nuclei were counterstained with DAPI (blue). Scale bar = 100 μm.
Discussion
Mucosal healing is a key unmet need in current IBD treatment. Physiological Wnt signaling is fundamental to intestine epithelial homeostasis and renewal. Therefore, modulation of Wnt signaling might present an opportunity to help IBD patients achieve histological remission and the ultimate goal of mucosal healing. We reported recently the creation of synthetic Wnt mimetics with druglike properties, particularly in the form of recombinant, bispecific antibodies that bring together Fzd and Lrp to stimulate signaling, mimicking endogenous Wnt ligands.14,35 In this work, we showed that treatment with Wnt mimetics alone is capable of repairing damaged intestine epithelium in a severe UC disease model. Importantly, unlike RSPO, the Wnt mimetics did not induce hyperplasia of normal intestine epithelium. We narrowed down this robust efficacy in the intestine to Fzd5,8- and Lrp6-specific Wnt mimetics, SZN-1326-p. SZN-1326, a clinical candidate molecule derived from SZN-1326-p, is equally efficacious in the DSS model (Surrozen, 2020, unpublished data).
In the intestine, Wnt signaling is most active in the basal half of the epithelium, affecting stem cell renewal, promoting progenitor proliferation, and guiding epithelial cells through the TA stages where lineage commitment and differentiation initiates.36 However, FZD receptors are differentially expressed at various levels in unique patterns across tissues, and the functional relevance of receptor-specific signaling remains unclear, even in the intestine epithelium where Wnt biology has been extensively investigated. For example, when protein antagonists against different Fzd subclasses were added to intestinal organoids, it was suggested that the Fzd7 subclass (Fzd1,2,7) was essential in maintaining intestinal stem/progenitor cells in organoids,37 consistent with an earlier study that showed by genetic ablation the predominant role of Fzd7 in intestinal stem cell maintenance and epithelial homeostasis.38 Alternatively, a conditional loss of Fzd5 suggested its requirement for Paneth cell maturation in the small intestine.39 Recently, however, Fzd5,8-specific Wnt agonism with Wnt mimetics constructed by different designs was argued to be sufficient to drive proliferation of intestinal organoids,12,17 and when combined with RSPO, to expand crypts in the uninjured small intestine.16 Throughout the intestine, including the colon, we found Fzd5 to be highly enriched in and fairly specific to the epithelium, including after injury. Furthermore, in an intestinal organoid proliferation assay, the Fzd5,8-specific Wnt mimetic, SZN-1326-p, showed equivalent or stronger activity compared with the pan Fzd1,2,7,5,8-specific or Fzd1,2,7-specific Wnt mimetics. The differences in these seemingly contradictory findings on the roles of Fzd5/8 vs Fzd1,2,7 not only may lie in the different approaches used in the studies, but also suggest there may be redundancy in the function or capability of Fzd5 and Fzd7 during uninjured tissue homeostasis. Importantly, the much higher level of Fzd5 expression in the epithelium coupled with the potent activity of a Fzd5,8-specific Wnt mimetic in organoids renders it a more appealing target for Wnt receptor agonism as a therapeutic option for intestinal damage.
The Fzd5,8-specific SZN-1326-p treatment resulted in rapid healing of the mucosa, improving tissue histology and disease activity in a few days with a concomitant reduction in inflammation and colitis symptoms. We investigated the mechanism of action of SZN-1326-p in this injury model and found that SZN-1326-p predominately impacted the epithelium shortly after dosing. Wnt target genes such as Axin2 were only increased in the epithelium at 24 hours posttreatment, suggesting that utilizing FZD receptor specificity is a viable option for directing tissue layer–specific pathway activation. Coinciding with the induction of Wnt target genes, SZN-1326-p caused a robust increase in cell cycle gene expression in a broad spectrum of progenitor cells, whether normal stem or progenitors responding to injury or in altered cell states consistent with de-differentiation. These transcriptome changes manifested in the transient expansion of the progenitor pool and accelerated differentiation into the proper secretory and absorptive lineages of the colonic epithelium and re-establishment of the epithelial barrier. Critically, this direct impact on epithelial regeneration and barrier restoration secondarily led to a reduction in inflammatory signals and infiltrating immune cells.
The injury and damage context appears to set the stage for epithelial progenitor expansion. In addition to impacting developmental signaling pathways such as epidermal growth factor and Notch (see Materials and Methods), injury caused an inflammatory response in all tissue layers. In the epithelium, interferon gamma and nuclear factor kappa B pathways were active after injury, and recent work in other stem cell niches has shown that inflammatory signaling can facilitate the initial proliferative response to injury.40,41 Activation of the nuclear factor kappa B and Wnt pathways together may even promote the process of de-differentiation toward progenitors in the intestine.42 In the DSS model, Wnt signaling was drastically reduced in the colonic epithelium, possibly resulting from a reduction in expression of specific Wnts and an increase in several Wnt antagonists. SZN-1326-p was able to overcome this Wnt signaling deficiency and activate Wnt signaling in the epithelium to synergize with these inflammatory signals to enhance progenitor proliferation, albeit transiently.
Different from the effects of RSPO, which impacts both the uninjured and damaged epithelium,18,23,24 targeted, receptor-level Wnt signaling agonism with a Wnt mimetic only promotes crypt proliferation in the damaged tissue context. In the uninjured epithelium, while RSPO by itself caused proliferation of intestinal stem cells, Wnt mimetics only induced proliferation and expansion of intestinal crypts when coupled with exogenous RSPO.16,18 We also observed extensive proliferation in the small intestine and colon when the Wnt mimetic SZN-1326-p was introduced together with RSPO2 in our DSS model. Although we observed amelioration of DSS-induced colitis in mice by RSPO as previously reported,24 we also observed hyperproliferation with RSPO treatment in the DSS model. Our finding that a Wnt mimetic by itself was able to induce expression of β-catenin target genes, and proliferation of epithelial cells only in the injured colon in the DSS colitis model is paramount.
Critically, Wnt pathway activation by SZN-1326-p did not lead to crypt hyperproliferation or expansion. This stands in stark contrast to not only RSPO treatment, but also the effects of heritable, genetic mutants. When the negative regulator Apc is genetically ablated or constitutively active mutants of β-catenin are expressed, crypts proliferate in an uncontrolled manner, failing to differentiate.36,43 However, SZN-1326-p avoids these outcomes by mimicking endogenous Wnt signaling and initiating pathway activation at the receptor level, in contrast to the permanent genetic alterations that circumvent negative feedback. By impacting the pathway at the level of the receptor, SZN-1326-p allows negative feedback mechanisms to take effect. For example, Axin2 itself is induced, contributing to the destruction complex. Expression of the E3 ubiquitin ligase Rnf43, also a Wnt target gene, is increased, and it promotes the removal of FZD receptors from the cell surface. Furthermore, SZN-1326-p increased expression of some inhibitors of cyclin dependent kinases, potentially limiting proliferation.
Multiple lines of evidence suggested that Wnt mimetic molecules have the desired properties for restoring diseased intestine tissue back to normal physiology. First, short treatment of Wnt mimetics (eg, SZN-1326-p) induced rapid restoration of epithelial tissue: in a severe DSS model, a single injection of SZN-1326-p at various doses restored normal histology of the damaged colon epithelium within 6 days of treatment. Secondary to the epithelial healing, SZN-1326-p reduced inflammatory cytokines and DAI, indicating elimination of the vicious cycle of barrier breach, microbial pathogen invasion, tissue inflammation, and damage. SZN-1326-p restored Wnt signals and the stem cell niche in damaged colon tissue, without additional effects on the crypts after repair. Last, Wnt mimetics alone do not have effects on normal intestine epithelium while RSPO induced hyperplasia. Therefore, we developed a Fzd5,8- and Lrp6-specific Wnt activator with optimal tissue repair and physiological activities, and molecules like SZN-1326-p hold great promise for directly improving histological remission for better long-term outcome in UC.
Materials and Methods
Critical regents used in the study are listed in Table 1.
Table 1.
Reagents and Resources
| Reagent or Resource | Source | Identifier |
|---|---|---|
| ACD probes | ||
| RNAscope Probe—Mm-Axin2 | ACD #400331 | |
| RNAscope Probe—Mm-Lgr5 | ACD #312171 | |
| RNAscope Probe—Mm-Rnf43 | ACD #400371 | |
| RNAscope Probe—Mm-Wnt2b | ACD #405031 | |
| RNAscope Probe—Mm-Wnt5a | ACD #316791 | |
| RNAscope Probe—Mm-RSPO3 | ACD #402011 | |
| RNAscope Probe—Mm-Fzd1 | ACD #404871 | |
| RNAscope Probe—Mm-Fzd2 | ACD #404881 | |
| RNAscope Probe—Mm-Fzd3 | ACD #404891 | |
| RNAscope Probe—Mm-Fzd4 | ACD #404901 | |
| RNAscope Probe—Mm-Fzd5 | ACD #404911 | |
| RNAscope Probe—Mm-Fzd6 | ACD #404921 | |
| RNAscope Probe—Mm-Fzd7 | ACD #404931 | |
| RNAscope Probe—Mm-Fzd8 | ACD #404941 | |
| RNAscope Probe—Mm-Fzd9 | ACD #404951 | |
| RNAscope Probe—Mm-Fzd10 | ACD #315781 | |
| RNAscope Probe—Mm-Krt20 | ACD #402301 | |
| RNAscope Probe—Mm-Ly6a | ACD #427571 | |
| RNAscope Probe—Mm-Pdgfra | ACD #480661 | |
| RNAscope Probe—Mm-Ccl8 | ACD #546211 | |
| RNAscope Probe—Mm-Cdkn3 | ACD #401701 | |
| Antibodies, enzymatic kits | ||
| Rabbit anti-Villin (SP145) | Abcam | ab130751 |
| Rabbit anti-DCLK/DCAMKL1 (D2U3L) | Cell Signaling Technology | CST 62257 |
| Rabbit anti-chromogranin A | Abcam | ab15160 |
| Rabbit anti-ZO-1 (clone 1A12) | Thermo Fisher Scientific | 33-9100 |
| Rabbit anti-Ki67 | Abcam | 15580 |
| Rat anti-KI67 (clone SolA15) | Thermo Fisher Scientific | 14-5698-82 |
| Rat anti-EPCAM-Alexa-488 (clone G8.8) | BioLegend | 118210 |
| Rat anti-LY6A-Alexa-647 (clone E13-161.7) | BioLegend | 122518 |
| Rat IgG2 Isotype control-Alexa-488 | BioLegend | 400525 |
| FcR blocking Reagent | Miltenyi Biotec | 130-092-575 |
| Donkey anti-rat IgG (H&L), highly cross-adsorbed secondary antibody, Alexa Fluor 488 | Thermo Fisher Scientific | A-21208 |
| Anti-Green Fluorescence Protein (Anti-GFP) human IgG | Surrozen | |
| hFc-RSPO2 | Surrozen | |
| R2M3-26, bi-specific appended human IgG effector-less format | Surrozen | |
| SZN-1326-p, bi-specific appended human IgG effector-less format (parental molecule of a clinical candidate SZN-1326) | Surrozen | |
| 1RC07-26, bi-specific appended human IgG effector-less format | Surrozen | |
| RNAscope 2.5 HD Assay-Red | ACD Bio | |
| RNAscope Mulitplex Fluorescent Reagent Kit, v2 Assay | ACD Bio | |
| Zymo Direct-zol RNA Microprep | Zymo | R2062 |
| MagMAX mirVana Total RNA Isolation Kit | Thermo Fisher Scientific | A27828 |
| Applied Biosystems High-Capacity cDNA Reverse Transcription Kit | Thermo Fisher Scientific | 4368814 |
| Applied Biosystems TaqMan Fast Advanced Master Mix | Thermo Fisher Scientific | 4444557 |
| Chemicals, peptides, proteins | ||
| DMEM/F12 | Thermo Fisher Scientific | 12634-010 |
| DAPI | Thermo Fisher Scientific | D1306 |
| Fetal bovine serum | Thermo Fisher Scientific | 10438-026 |
| Liberase | Sigma | 05401127001 |
| DNAse1 | Sigma | 04716728001 |
| EDTA | Sigma | 03609 |
| Phosphate-buffered saline | Thermo Fisher Scientific | 10010-023 |
| HEPES | Thermo Fisher Scientific | J16924-AE |
| Sodium pyruvate | Thermo Fisher Scientific | 11360-070 |
| Pen-Strep | Thermo Fisher Scientific | 15140-122 |
| Antibiotic/antimycotic 100× | Thermo Fisher Scientific | 15240-062 |
| Hank’s Balanced Salt Solution | Thermo Fisher Scientific | 14175-079 |
| TrypLE | Thermo Fisher Scientific | 12604-013 |
| Triton X-100 | ACROS Organics | 21568-2500 |
| TSA Plus Cyanine 3 System | Akoya Bioscience | NEL744001KT |
| TSA Plus cyanine 5 System | Akoya Bioscience | NEL745001KT |
| Vectashield Vibrance antifade mounting medium with DAPI | Vector Laboratories | H-1800 |
Animal Husbandry
Mice were obtained from the Jackson Laboratory (Bar Harbor, ME). All animal experimentation was in accordance with the Guide for the Care and Use of Laboratory Animals prepared by the National Academy of Sciences. Protocols for animal experimentation were approved by the Surrozen Institutional Animal Care and Use Committee. Seven-week-old C57Bl/6J female mice were acclimatized 4–5 per cage a minimum of 2 days prior to the experiment. Mice were kept 12-hour light–dark cycle in a 30%–70% humidity environment and room temperature ranging from 20°C to 26°C.
DSS-Induced Acute Colitis
Seven- to 8-week-old female C57BL6/J mice were fed with 4% (w/v) DSS (MP Biomedicals, Irvine, CA; #160110) in drinking water from day 1 to day 7 to induce colitis and were switched to 1% DSS from day 8. Proteins were injected IP once on day 4, twice on day 4 and 7, or daily on days 4–9. Animals were terminated on day 10, allowing a 6-day course of protein treatment, and the colon was harvested for histology and RT-qPCR. In one of the studies, DSS induced mouse body weight loss for animals treated with anti-GFP was nearly 25% on day 9, so animals were switched to drinking water with no DSS for compliance with Institutional Animal Care and Use Committee rules (Figure 1A). DAI was calculated based on the average score of weight loss, stool consistency, and degree of intestinal bleeding.44 Scoring system by grading was on a scale of 0–4 using the following parameters: loss of body weight (0 = 0%–1%; 1 = 1%–6%; 2 = 6%–12%; 3 = 12%–18%; 4 = >18%), stool consistency (0 = normal; 1 = soft but still formed; 2 = soft; 3 = very soft, wet; 4 = watery diarrhea) and intestinal bleeding (0–1 = negative hemoccult; 2 = positive hemoccult; 3 = blood traces in stool visible; 4 = gross rectal bleeding). Statistical significance was determined by 1-way analysis of variance, Holm-Sidak test (GraphPad Prism Version 9.3.1; GraphPad Software, San Diego, CA) in all the plots. All comparisons were made with the anti-GFP group. Error bars indicate mean ± SD. In the figures, ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001.
Tissue Histology
Small intestine and colon were extracted and, after removing fecal content, weighted and their length measured. The desired small intestine segments (duodenum, jejunum, ileum) and colon segments (ascending, transverse, and descending colon) were cut out and fixed directly in 10% neutral buffered formalin overnight. Tissues were then transferred to 70% ethanol before paraffin embedding. Paraffin tissue blocks were then sectioned to 5-μM thickness and stained with H&E for histology analysis. Pathology reading was performed by an independent pathologist, who was blinded to the treatment groups. Briefly, 2–3 sections were obtained from each segment of the colon and evaluated after H&E stain. Grading was done on a scale of 0 (normal) to 4 (most severe lesion) with the following 5 parameters: inflammation severity (presence of mixed leukocytes [ie, macrophages, neutrophils, mononuclear cells, lymphocytes]), inflammation extent (location of inflammatory cell infiltrate extending from the mucosa to the serosa), mucosa erosion (alteration of mucosal architecture), crypt proliferation (presence of crypt epithelial cell proliferation in mucosa adjacent to areas of injury [eg, erosion/ulceration]), and goblet cell loss (decrease in relative number of goblet cells as compared with normal). The overall histopathological scores were determined by averaging scores of the 5 parameters listed previously.
Immunohistochemistry and Indirect Immunofluorescence
Slides with 5-μM-thick formalin fixed paraffin embedded tissue sections were deparaffinized followed by citrate buffer (pH 6) antigen retrieval in a steamer. Slides were then washed thoroughly in tap water followed by 1× wash in 0.1% Triton X-100 in PBS (PBST). Subsequently, tissue sections were blocked with serum free protein block (Agilent, Santa Clara, CA; X090930-2) for 1 hour at room temperature before incubation in primary antibodies. Tissue sections were then washed in PBST at least 3 times followed by incubation in secondary antibody. Afterwards, tissue sections were washed with PBST and mounted with Vectashield Vibrance antifade mounting medium with DAPI (H-1800; Vector Laboratories, Burlingame, CA).
RNA In Situ Hybridization
Expression of mRNA was detected by RNAscope in situ hybridization (ACD Bio, Newark, CA). RNAscope probes used are listed in Table 1. For colorimetric visualization, standard RNAscope 2.5 HD Assay-Red protocol (ACD Bio; Document #322360) was followed, and images were acquired on a Leica DMi8 microscope equipped with a DFC7000T camera (Leica, Wetzlar, Germany). For fluorescent RNAscope in situ hybridization, standard RNAscope Multiplex Fluorescent Reagent Kit v2 Assay protocol was followed (ACD Bio; Document #323100) and coupled with the TSA Plus Fluorescein, Cyanine 3, or Cyanine 5 systems. Fluorescent images were acquired on a Leica Thunder imaging system.
RNA Isolation and RT-qPCR
The MagMAX mirVana (Thermo Fisher Scientific, Waltham, MA; A27828) Total RNA Isolation Kit was used for RNA isolation on a KingFisher (Thermo Fisher Scientific) sample purification system. Reverse transcription was done with the Applied Biosystems High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific; 4368814), followed by qPCR using the TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific; 4444557).
Affinity Measurements
Binding kinetics of R2M13, the Fzd binding portion of SZN-1326-p, Fab to each cysteine-rich domain (CRD) of Fzd5,8 was determined by biolayer interferometry using Octet Red 96 (Pall FortéBio, Fremont, CA) instrument at 30°C, 1000 rpm with streptavidin (SA) biosensors. Biotinylated CRDs of Fzds diluted to 25 nM in the running buffer (PBS, 0.05% Tween 20, 0.5% bovine serum albumin, pH 7.2) were captured to the SA biosensor followed by dipping into wells containing the R2M13 Fab protein at different concentrations in running buffer or into running buffer only as a reference channel. Equilibrium dissociation constant (KD) for each binder was calculated based on fitting to a 1:1 binding model. Binding specificities of R2M13 IgG to 10 Fzds were examined by the biolayer interferometry assay. Biotinylated Fzd CRDs35 diluted to 50 nM in running buffer were captured to the SA biosensor followed by dipping into wells containing R2M13 IgG at 200 nM in running buffer.
STF Assay
Signaling activity of the Wnt mimetics was measured using the Huh7 human liver cells containing a luciferase gene controlled by a Wnt-responsive promoter (STF assay) following an established protocol.35
Organoid Culture and Proliferation Assay
Mouse small intestinal organoids (STEMCELL Technologies, Vancouver, British Columbia, Canada) were maintained in mouse IntestiCult Organoid Growth Medium (STEMCELL Technologies) and passaged once a week until the date of assay. Assay for Wnt mimetic activity was performed following the reported protocol.35 Each condition included 5–6 repeats. Media and treatments were changed once on day 4 after plating. Images of the 3-dimensional cultured organoids were acquired on day 7.
Fluorescence-Activated Cell Sorting
Mouse colon was dissociated as described below and resuspended in FACS buffer (Hank’s Balanced Salt Solution, 2% fetal bovine serum [FBS], 10 mM HEPES, 1 mM sodium pyruvate, and 1% Pen-Strep or antibiotic/antimycotic solution). Prior to FACS, cells were passed through a 40-μm filter, and DAPI was added to distinguish live/dead cells. Prior to target antibody incubation, FcR blocking reagent (Miltenyi Biotec, Bergisch Gladbach, Germany; 130-092-575) was added to the samples and incubated for 10 minutes. To detect epithelial LY6A-positive cells, samples were incubated with fluorophore conjugated monoclonal antibodies to EPCAM (Rat anti-EPCAM-Alexa-488) and LY6A (Rat anti-LY6A-Alexa-647) for 20 minutes on ice followed by washing with chilled FACS buffer. We used minus antibody controls and an isotype control antibody to establish gates.
scRNA-seq: Tissue Dissociation, Cell Isolation, Library Preparation, Sequencing
For the acute DSS model, mice were treated with 4% DSS in their drinking water throughout the duration of the experiment. DSS-treated animals were dosed with 10-mg/kg SZN-1326-p or an anti-GFP antibody on day 4 of the DSS treatment. On days 5 and 6, we collected cells from 2 uninjured mice and from 3 replicates each for anti-GFP– and SZN-1326-p–treated DSS animal. Each animal was considered a replicate.
The transverse colon was isolated from each animal and feces were removed. After a brief wash in cold PBS, the colon was cut longitudinally, and the tissue was cut into 3- to 4-mm-long fragments. Tissue fragments were incubated in prewarmed (37°C) PBS with 5 mM EDTA in a shaker at 37°C at 150 rpm for 15 minutes. After 15 minutes, the tubes containing the samples were vigorously shaken for 10 seconds to release more epithelial cells. The epithelial cells floating in suspension were removed to a new tube and centrifuged at 200 rcf for 2 minutes.
The residual tissue containing the remaining epithelia and stroma/lamina propria was then incubated in 8–12.5 mL of lamina propria dissociation buffer (AdvDMEM/F12 with 10 mM HEPES, 0.2% FBS, DNAse1 (80 U/mL), Liberase (0.2 mg/mL), and 1% antibiotic/antimycotic) at 37°C for 30 minutes with horizontal shaking at 150 rpm. After pelleting, the epithelial cells were resuspended in 1 mL of TrypLE with DNase1, incubated at 37°C for 5 minutes, and then triturated with a P1000 pipette for 30 seconds. After trituration, 10 mL of PBS plus 50 U/mL DNAse1 were added to the epithelial cells, and they were centrifuged at 500 rcf, 4°C, and the supernatant was removed. Epithelial cells were then washed 1 time in FACS buffer (Hank’s Balanced Salt Solution, 2% FBS, 10 mM HEPES, 1 mM sodium pyruvate, and 1% Pen-Strep or antibiotic/antimycotic solution) before another round of centrifugation and final resuspension in 0.5 mL of FACS buffer. Following 30 minutes of dissociation in LP dissociation buffer, the remaining tissue fragments and suspension were centrifuged at 500 rcf for 5 minutes. Supernatant was removed down to 1 mL, and the sample was triturated with a P1000 until the solution was homogeneous and all tissue fragments had dissociated. After trituration, the sample was centrifuged at 500 rcf for 5 minutes at 4°C and washed in FACS buffer prior to resuspension in 1 mL of FACS buffer in preparation for FACS.
All cells were passed through a 40-μm filter prior to FACS. DAPI was used to assess viability, and only DAPI-negative cells were collected. Cells were collected from the epithelial fraction and then from the epithelial/lamina propria fraction and combined (1:5 ratio) and counted on a hemocytometer prior to cell capture. Standard 10x Genomics Chromium 3′ v3 scRNA-seq reagents (PN1000075; 10x Genomics, Pleasanton, CA) were used. Approximately 4000–4500 cells were loaded per channel. Cells from 1 animal replicate were captured per channel. Standard 10x Genomics Chromium 3′ v3 scRNA-seq RT, cDNA amplification, and sequencing library preparation protocols were followed. Multiplexed sequencing libraries were sequenced on Illumina Nova Seq 6000 S1 lanes (Illumina, San Diego, CA), averaging about 50,000 reads per cell.
scRNA-seq Analysis
Illumina read data was processed using the 10x Genomics Cell Ranger (version 3.0.2) pipeline, which runs the STAR aligner, on the mm10-3.0.0 version of the mouse transcriptome. Demultiplexed UMI count data was then assessed, and following exploratory data analysis, low-quality cells and low-expression genes were removed in part by using the R package scone (version 1.14.0) (R Foundation for Statistical Computing, Vienna, Austria) and dataset-specific filtering cutoffs: only cells with >1000 UMIs and with ≥500 and ≤6500 genes and ≤60,000 UMIs were retained to remove presumably empty droplets and limit doublets. We filtered cells with a mitochondrial gene percentage more than 1 SD above the mean. We retained only those genes expressed in the upper quartile of at least 3 cells, yielding 16,039 genes. UMI count data was normalized using deconvolution scaling from the R package scran (version 1.18.5).45 After normalization, we observed neither batch-specific cell groups when assessed in reduced dimension space nor strong correlations between quality control metrics and gene expression principal components within each lineage or tissue layer.
To the complete, filtered dataset, we applied a shared nearest neighbor (SNN) graph–based clustering method46 by using the wrapper function (buildSNNGraph) from the R package scran (version 1.18.5) with k equal to 40 coupled with the cluster_louvain function from the R package igraph (version 1.2.6) to the first 10 principal components derived from the top 2000 most variable genes across the dataset. This allowed us to broadly group the cells and identify cell types within the 3 tissue layers or lineages (immune, stromal, epithelial). Based on the initial clustering, the data were subsetted into 3 smaller datasets, and the cells within each layer or lineage were clustered using the SNN graph–based method and the walktrap algorithm implemented with the cluster_walktrap function from the igraph package, applied to the first 15 principal components derived from the top 2000 most variable genes within that subsetted layer or lineage (immune, stromal, epithelial). Cell type or subtype identities were determined using established marker genes and published literature.
Within each layer or lineage dataset, differential gene expression analysis was performed at the single cell level for each cluster in one vs all and pairwise comparisons within each layer or lineage by using wrapper functions within the R package clusterExperiment (version 2.10.1)47 to run edgeR (version 3.32.1).48,49 Differential gene expression analysis between experimental conditions was performed with the R package edgeR (version 3.32.1) on pseudobulk samples following aggregation of single cells within biological replicate samples. This type of differential gene expression analysis was implemented at the lineage level and at the cell type or cluster level. Differential expression comparisons were performed between experimental conditions (DSS injury vs uninjured and within the DSS injury samples for SZN-1326-p–treated vs anti-GFP–treated) within each of the 3 layers or lineages (epithelial, stromal, immune) and within individual cluster or cell types within each lineage for each time point (24 or 48 hours). We applied GSEA, also called pathway analysis, by implementing the fry function from the R package limma (version 3.46.0).50 Gene sets were obtained from the Broad Institute’s Molecular Signature Database (MSigDB) and included Hallmark and curated (C2) gene sets of the KEGG, Biocarta, PID, Reactome, ST, SIG, and SA types. We also implemented the kegga function of the limma package, which only uses KEGG pathways, and observed similar results. We applied GSEA pseudobulk samples aggregated by replicate.
We applied cell-to-cell or intercellular network (connectivity) analysis to the entire dataset. We went back to the full dataset and provided the more granular cluster/cell type assignments. We applied the R packages, SingleCellSignalR (version 1.2.0)51 and Connectome (version 1.0.1).52 With Connectome, for the paracrine signaling reported in the paper between the stromal cells to immune cells, we set the major fibroblasts cell types as the source cells (CryptBaseFB1,2; InjuryFB1,2; CryptFB1,2) and the receiving cells as either Neutrophil or InjuryMono2, and we selected the top 5 signaling vectors for each cell-cell combination. Lineage trajectory inference was performed using the R package slingshot (version 1.8.0).53
To ascertain the ability of SZN-1326-p to impact Wnt target gene expression, we started with a Wnt signaling target gene list,54 added additional genes with support from the literature, and intersected this list with the differentially expressed genes by tissue layer as presented in Supplementary Table 4.
Epithelial Cell Annotation
We describe the cell type annotation for the epithelial lineage subsequently. The epithelial stem cell cluster was defined as such because they had the highest expression of Ascl2 and Smoc2, and they expressed Lgr5, Cd44, Notch1, Hopx, and Lrig1.31,55, 56, 57, 58 There were 2 clusters of progenitor cells defined by their high expression of the cell cycle, indicating that they were actively proliferating cells, and they also expressed Lgr5, Cd44, Smoc2, and Hopx and lower levels of Ascl2.These 2 groups of cells were labeled as TA progenitor cells TA1 and TA2. There is much overlap between the stem cell, TA1, and TA2 groups, but we called TA1 and TA2 TA because of their higher level of the cell cycle and lower percentage of Ascl2-expressing cells. These 3 stem or progenitor cell groups contained cells from all experimental conditions, but there was clear heterogeneity, with some cells derived from injured samples expressing higher levels of some of the fetal-like immune signature genes, Ly6a and Ly6c1.
There were 2 groups of cells that were almost exclusively derived from injured cells, and they expressed relatively high levels of genes previously reported to be expressed in injured or infected intestinal tissue and associated with inflammatory signaling and a fetal-like stem cell signature such as Ly6a and Ly6c1 (Figure 11A and E).28,29 One of these groups of cells appeared to be enterocytes that were expressing injury associated immune and inflammatory signaling genes. These were labeled AltEnteros. The other group was very similar, but these cells were beginning to express cell cycle genes, suggesting that they were de-differentiating into progenitors from enterocytes, and these were labeled AltEnteroPCs. It is important to note that a few of these cells do express high levels of Clusterin, a gene shown to be expressed in a rare subset of epithelial cells with functional stem cell properties,59 but the Ly6 genes show much higher enrichment in these cells. At a minimum, this suggests that these injury-induced cell types or states include these Clusterin-positive cells. There were 2 other groups of enterocytes (AltEntero2, AltEntero3) that were mainly populated by cells from the injury condition, although there was a low percentage of cells from the uninjured condition in these 2 groups. They expressed elevated levels of immune or inflammatory genes and appeared to be showing a damage response. The AltEntero2 and AltEntero3 cells have lower expression of immune signaling related genes relative to the AltEnteros and AltEnteroPCs, suggesting that they may be earlier in the process of initiating the immune signaling gene expression. Our observation of coexpression neither proves that these cells are de-differentiating enterocytes nor rules out plasticity of other cell types, but it demonstrates that there are cells with altered transcription states upon injury.
There was a group of cells that expressed genes such as Slc26a2, Slc26a3, Ces2a, Car1, and Muc3 that are enriched or specific to the enterocytes, and these cells had only low-level cell cycle gene expression. They were also closely positioned (and thus similar) to immature enterocytes in reduced dimension plots (Figures 9E and 10E), and we labeled them as enterocyte precursors. Notably, these cells express Hopx. They contained cells from all conditions but were enriched for the 48-hour SZN-1326-p treatment. There were 3 clusters of immature and mature enterocytes (ImmEnterocyte1,2, enterocyte) that were populated with cells almost exclusively from the uninjured cells (Figure 11E), and they represent the normal enterocyte lineage, expressing Slc26a2 and Slc26a3, with the mature enterocytes having enriched expression of Krt20, Car4, and Aqp8.
There were several goblet cell clusters, all expressing goblet cell markers Atoh1, Spdef, and Muc2. One group expressed Kit and Reg4 and relatively higher levels of the transcription factor Spdef and the mucin gene Muc2. These cells also had low-level cell cycle gene expression, and they were labeled BasalGoblet. The fact that several of these cells express low levels of the cell cycle may indicate that some of them are progenitors or recently derived from progenitors. These cells appear to be basal secretory cells reported to express high levels of Kit and Reg4 and to reside at the crypt base.60,61 These cells also show some enrichment for Hopx, as the enterocyte precursors and unlike the 2 more differentiated goblet clusters described next. There were 2 other clusters of goblet cells that had relatively higher expression of Dll4, Fcgbp, Clca1, Tff3, and Gal3st2c, and the relative expression level of these genes distinguishes them: the Goblet1 cluster had relatively lower levels of Spdef and higher Fcgbp and Clca1 relative to the Goblet2 cells. All goblet cell clusters had high expression of Muc2.
There were also tuft cells, which expressed high levels of Trpm5, Nrgn, Dclk1, and Hck. We also identified enteroendocrine cells (enteroendocrine1,2), characterized by expression of Tph1 and the chromogranin genes Chga and Chgb. They also expressed Olfr78 and Piezo2, 2 genes enriched in different colonic L cells.62 These clusters displayed a fair amount of heterogeneity, with cells expressing varying levels of a range of small, secreted peptides, including Gcg, Pyy, and Insl5. Based on a recent analysis of enteroendocrine cells in proximal and distal colon by Neurod1 lineage tracing,62 these 2 clusters likely represent a heterogeneous mix of enterochromaffin and L cell subtypes. Discriminating heterogeneity within these cells extracted from the transverse colon is not the focus of this work but could be worth future efforts.
Notch signaling was upregulated in the epithelium by the DSS damage. When we applied GSEA pathway analysis to the epithelium, 4 of the top 10 enriched pathways centered on Notch signaling (Figure 11B). Stem and progenitor cells in the epithelium expressed Notch1 and Hes1 (Figure 11A). We detected an enrichment of Notch1 in the epithelium in the DSS-injured condition relative to the uninjured controls (Supplementary Table 3) and an enrichment for Hes1 expression in the SZN-1326-p vs anti-GFP treatment groups. Differential gene expression analysis between the stromal clusters revealed enrichment of Dll1 expression in the 2 injury-induced fibroblasts, InjuryCryptFB1,2, and it was enriched in the DSS condition cells within the CryptFB1 subtype (Supplementary Tables 1 and 2). The DSS treatment impacted the expression of modulators of Wnt signaling in the underlying fibroblasts. In normal, uninjured colon, the crypt base fibroblasts (BaseCryptFB1, BaseCryptFB2) expressed Rspo3 and lower levels of Rspo1.22 The injury-induced InjuryCryptFB1,2 fibroblasts continue to express Rspo3 and have elevated expression of Rspo1, consistent with our RNA in situ hybridization for Rspo3 (Figure 1K and Q) and a report of Rspo1-positive cells being involved in the damage associated response in the intestine.63 In line with these results, a recent study reported that Rspo3 expression in the underlying stromal cells is necessary for the delayed endogenous repair in the acute DSS model after cessation of injury.64 At the lineage level, Wnt10b decreased and Wnt5a increased upon DSS damage (Supplementary Table 3), and Wnt2 and Wnt10b showed a slight decrease in the cells from the DSS-treatment condition in the BaseCryptFB2 cells (Supplementary Table 2). In contrast, there was a robust increase in expression of secreted Wnt antagonists in the stromal cells. Sfrp1,2,5, Wif1, Dkk2, and Grem1 were all significantly increased in the DSS condition relative to the uninjured stroma (Supplementary Table 3). The injury-induced fibroblasts (InjuryCryptFB1, FB2) and the DSS-treated cells displayed relatively strong enrichment of Sfrp1,2 and the DSS-treatment cells within the CryptFB1 fibroblasts showed enrichment for Wif1 and within the BaseCryptFB2 for Sfrp1 (Supplementary Table 1 and 2). In sum, our analysis predicts that DSS-induced changes in the stroma could limit Wnt signaling in the epithelium, consistent with the robust reduction in Axin2 expression in the crypts observed by day 7 of DSS treatment by RNA in situ analysis (Figure 1F and L). Likewise, in our scRNA-seq data, we observed a reduction of Axin2 expression by day 5 of DSS treatment (Supplementary Table 3; Figure 12B and C), while the Fzd receptors (mainly Fzd5) were still expressed in the epithelium.
Stromal Cell Annotation
Cluster analysis of the stromal or mesenchymal cells identified 18 clusters, spanning a range of fibroblasts and fibroblast-like cells, smooth muscle cells, interstitial cells of Cajal, vascular and lymphatic endothelial cells, pericytes, and glia. There were 2 major classes of fibroblasts that are normally present in the colon based on the contributions from the uninjured animals, all of which were Col1a1, Dpt, Fbln1, Lum, and Pdgfra positive, consistent with the broad fibroblast genes identified in Muhl et al.65 One subset (BaseCryptFB1,2) matched the profile for fibroblasts positioned at or near the base of the crypt, having relatively lower Pdgfra expression and relatively higher expression of Wnt2, Wnt2b, Rspo3, Grem1, Dcn, and Col14a1, while expressing relatively lower levels of Wnt5a and Bmp3, Bmp5, Bmp7 ligands.22,66 The BaseCryptFB1 cells expressed higher levels of Adamdec1 and Bmp4, Bmp5, Wnt4, and Wnt5b than the BaseCryptFB2 cells, and the BaseCryptFB2 cells express higher levels of Col8a1, Lum, and Il33 than the BaseCryptFB1 group. While expressed at a low-level, the BaseCryptFB2 fibroblasts also express Wnt9a, Wnt10b, and Wnt11. Both BaseCryptFB1,2 express relatively higher levels of Cd34, Thy1, and Sfrp1 than the CryptFB1,2 cells described subsequently, also consistent with the BaseCryptFB1,2 cells being positioned near the base of the epithelial crypts.63,66
The second major fibroblast subtype are the CryptFB1,2 cells. These appear to be fibroblasts positioned more apically along the crypt, and they express higher levels of Wnt5a, Bmp2, Bmp3, Bmp5, Bmp7, Fgf9, and Col4a5 relative to the BaseCryptFB1,2 cells. More apical fibroblasts express higher levels of Wnt5a and these Bmps.66 The CryptFB1,2 fibroblasts also express higher levels of F3 and Sox6, recently reported to be expressed in fibroblasts in close proximity to the epithelium,26 and they express relatively high levels of Foxl1, a gene reported to mark subepithelial telocytes or mesenchymal cells that provide at least some of the critical Wnts that support the epithelial crypts.67,68 However, it should be noted that few genes are absolutely specific to one fibroblast subtype. For example, the CryptFB1,2 cells express relatively high levels of Foxl1, but the BaseCryptFB1 also express some Foxl1. Furthermore, the CryptFB1,2 fibroblasts, express higher levels of the secreted Wnt antagonist, Wif1, and the secreted LRP receptor antagonist Dkk3 is expressed in all fibroblasts.
Interestingly, Gli1 expression has been associated with fibroblasts that are essential for the Wnt-secreting niche,69 and we find Gli1 to be expressed at a low level in both the BaseCryptFB1, 2 and CryptFB1,2 cells. Based on the necessity of Gli1-expressing cells for secreting Wnts to maintain the colonic stem cells,69 our data would suggest that these conditional genetic studies have actually shown that a range of crypt fibroblasts are important because the tools likely label cells from multiple fibroblast types. We identified another fibroblast-like cell type that we termed FB3. These cells appear to be fibroblast-like cells that have been reported to be embedded in smooth muscle cells near motor neuron terminals and express the P2ry1 receptor and the Kcnn3 channel.70,71 They also express Nog and high levels of Lum, Dcn, and Fibin.
We identified a myofibroblast cell type, termed MyoFB, that shows enrichment for Postn, Hhip, Mfap5, Tpm2, Acta2, Tagln, Myl9, Myhll, Lmod1, Cnn1, Des, and Actg2. We also identified a myofibroblast-like cell type that we termed MuralLikeFB, based on recent findings from a cross-tissue fibroblast comparison.65 These cells express many of the genes enriched in the MyoFB, but they express higher levels of smooth muscle associated genes such as Acta2, Tpm2, Actg2, Cnn1, Sorbs2, and Des, and show more specific enrichment for Igfbp2, Chrdl1, Npnt, and Ano1. They express high levels of Rspo3, Grem1, and Grem2. Both the MyoFB and MuralLikeFB cells express very low levels of Pdgfra and Dpt. We identified a group of smooth muscle cells (SmoothMuscle) that express high levels of Tpm2, Acta2, Tagln, Myl9, and Myh11 and relatively specific expression of Kcnab1, Lrrc10b, and Pln. They also express Rgs4, Notch3, Jag1, and Olfr558, similar to the 2 groups of vascular smooth muscle cells (VasSmMuscle1,2). The VascSmMuscle1,2 cells express high levels of Pdgfrb, Dcn, and Des, but they express relatively lower levels of Tpm2 and Acta2 relative to the SmoothMuscle, MyoFB, and MuralLikeFB cells. The interstitial cells of Cajal (ICCajal) express Kit, Ano1, Grem2, and Rspo3, and they also display fairly specific expression of Hand1 and Nog. The pericytes display relatively high expression of Pdgfrb and Dcn, and weak expression of Des and several myosin and actin genes. Like the SmoothMuscle and VascSmMuscle1,2 cells, they express Olfr558, Rgs4, and Notch3, and like the vascular endothelial cells (VascEndo), they express Cd34, Fabp4, Pecam1, Sox17, Notch4, Cd93, Egfl7, Jag2, Col13a1, and Cdh5. The lymphatic endothelial cells (LymphEndo) express Pecam1, Egfl7, and Cdh5 as well as Reln, Flt4, and Lyve1. Last, we identified 2 groups of glia cells, labeled Gfap, S100b, Sox10, Sfrp5, and Kcna1.
The InjuryCryptFB1,2 cells are almost exclusively populated by cells derived from DSS-injured animals. They share characteristics of both the BaseCryptFB1,2 and the CryptFB1,2 fibroblasts. For example, like the BaseCryptFB1,2, they express relatively high levels of Dcn, Mgp, Fbln1, and Sfrp1. Like the BaseCryptFB1 and the CryptFB1,2, they express high levels of Adamdec1 and relatively higher levels of Bmp4 and Bmp5. They also express Wnt2 and Wnt2b at similar levels to BaseCryptFB1 cells, and they express low levels of Wnt5a. As we will discuss subsequently, these show an enrichment for expression of many inflammation-associated genes, for example, Cxcl13, Cxcl5, and Ccl8.
The injury-associated fibroblast populations express several genes that would be expected to impact epithelial repair. They have a very similar gene expression profile as the FB1 population, but they also express several of the immune cell mediators and Sfrp1 and Grem1, suggesting that the FB1 cells transition to the InjuryFB1 and InjuryFB2. The injury-specific fibroblasts (InjuryFB1, InjuryFB2) express the Wnt signaling potentiator Rspo1, the antagonists Sfrp1 and Grem1, and several Wnt molecules, consistent with cells reported to be involved in damage associated response.63 Importantly, Rspo3 expression in the InjuryFB1 and InjuryFB2 appears very similar to the level in the CryptBaseFB1 population, consistent with our RNA in situ hybridization results in Figure 1. DSS damage caused a reduction in Wnt signaling in the epithelium. It has been reported that prostaglandin E2 is required for canonical Wnt signaling activity.72 However, there is an increase in Ptgs2 (Cox2) expression in both the stroma and epithelium upon DSS treatment, suggesting that prostaglandin E2 levels do not underlie the reduction in Wnt signaling. The injury-induced fibroblast subtypes (InjuryCryptFB1, InjuryCryptFB2) and the DSS-damaged condition fibroblasts remaining in the CryptFB1 and CryptFB2 subtypes display increased expression of the epidermal growth factor receptor signaling ligands Ereg and Nrg1 (Supplementary Tables 2 and 3).
One or both of the InjuryCryptFB1,2 were enriched for expression of Ccl2, Ccl4, Ccl7, Ccl8, Ccl11, Cxcl2, Cxcl5, Cxcl12, Cxcl13, Cxcl14, Cxcl16, Ptgs2, and Saa3 (Supplementary Table 1). These observations are consistent with recent human scRNA-seq studies of human IBD samples that report fibroblasts expressing chemokines and other inflammation associated genes.26,27 While these 2 populations appeared upon injury, the BaseCryptFB1 population was almost undetectable in the DSS samples. Even in some of the fibroblasts subtypes that included cells from all treatment groups, DSS-induced damage caused a significant increase in expression of a range of inflammatory and immune response molecules relative to the uninjured cells of the same subtype: in BaseCryptFB2 (Ccl2, Ccl7, Ccl8, Cxcl1, Cxcl2, Cxcl5, Cxcl12, Cxcl13, Il6, Il11, Csf3, Lcn2, Ptgs2, Saa3) and in CryptFB1 (Ccl2, Ccl7, Ccl8, Ccl11, Ccl12, Cxcl1, Cxcl2, Cxcl5, Cxcl9, Cxcl10, Cxcl13, Il6, Il11, Csf3, Lcn2, Saa3). (Supplementary Table 3). While the macrophages, monocytes, and neutrophils expressed chemokines that would recruit incoming neutrophils and monocytes,73 our data provided evidence that the fibroblasts also contributed to the recruitment of infiltrating immune cells to the damaged colon. GSEA revealed that stromal cells enrichment for pathways associated with modifying the extracellular matrix and immune signaling (Figure 10B), and cell-cell interaction network analysis detected many ligand-receptor pairs potentially mediating paracrine signaling (both secreted and extracellular matrix (ECM) based) between multiple fibroblast subpopulations and the infiltrating neutrophils and monocytes (Figure 10E and F). We confirmed the increase in expression of the chemokine Ccl8 in the stromal fibroblasts by RNA in situ hybridization (Figure 10D), validating this response in the fibroblasts adjacent to the crypts.
Immune Cell Annotation
Cluster analysis identified 11 immune cell types. We identified 2 groups of B cells, termed Bcell1_IgM and Bcell2_IgM. These B cells were enriched for cells from the DSS-injured animals and nearly absent from the colon in the uninjured mice, and they had already migrated to the tissue by day 5 and remained at day 6 of DSS. Both show gene enrichment for Cd79a, Ebf1, Ms4a1, Ighd, Cd19, Ccr7, and Ighm. These 2 groups of B cells are very similar, and Bcell2_IgM has higher expression of Iglc2, Iglc3, Vpreb3, Pax5, Pou2af1, and Mzb1 relative to Bcell1_IgM. Our analysis revealed a very distinct plasma cell population expressing Igha, and we termed these cells Plasma_IgA. They express Cd28, Igkc, Iglc2, Iglv1, Igha, and Mzb1, and they express lower levels of Cd79a relative to the Bcell1,2_IgM cells.
There are 2 heterogeneous groups of T cells in our data. Both express Cd3d, Cd3g, Cxcr6, Ctla4, Trbc1, Trbc2, and Il2rb. The first group, TcellCD8_NK, contains Cd8a-positive T cells and natural killer cells, and shows gene enrichment for S100a4, Nkg7, Prf1, Ncr1, Ifng, Ccr2, Ccr5, and Ccl5. The Tcell2 cells show an enrichment in some cells for Il22. We identified 2 groups of dendritic cells, dendritic cells, labeled Dendritic, and what appear to be activated dendritic cells, labeled ActDendritic. Both show enrichment for Flt3, a dendritic-specific marker,74 Ifitm1, S100a4, and Tnfrsf18. The Dendritic cluster displays higher expression of Itgae, Cd209a, and Lyz2, while the ActDendritic cells are enriched for Cacnb3, Ccl5, Ccr7 and Fscn1. Cacnb3 is expressed in stimulated dendritic cells,75 and Ccr7 is involved in their homing and migration.76 There was 1 major macrophage cluster in our data (Macrophage), characterized by expression of Csf1r, Cd68, and Adgre1. The macrophages expressed relatively high levels of C1qa, C1qb, C1qc, Ms4a7, and Apoe. The infiltration of monocytes in the DSS colitis model is expected,73 and there were 2 populations of monocytes, and both were strongly enriched with cells from the DSS-injured animals. There were almost no uninjured cells in the InjuryMono1 cells and only a few in the InjuryMono2 group. Both populations of monocytes expressed Cd68, Adgre1, Csf1r, Trem1, Il1b, Ccr2, and Ccr5. The InjuryMono1 showed an enrichment for Trem1, Osm, S100a4, and Tgm2, and Vcan relative to the InjuryMono2 cells, and they even showed weak expression of genes that were robustly expressed by the neutrophils (eg, S100a8, S100a9, and G0s2). The InjuryMono2 more closely resembled the macrophages, and they expressed relatively higher levels of C1qa, C1qb, C1qc, and Cd74 than the InjuryMono1 cells. Likewise, the arrival of neutrophils has been well documented in the DSS model and in patients, in which the degree of neutrophil infiltration correlates with the severity of UC.77, 78, 79 Accordingly, the last group of cells was a population of neutrophils (Neutrophil) that is injury specific and expressed Csf3r and Cxcr2, 2 genes involved in neutrophil differentiation and recruitment into tissues, respectively.80,81 These cells expressed high levels of proinflammatory genes such as S100a8, S100a9, Trem1, Nlrp3, and Il1b. It was recently reported that inflammatory monocytes are present in UC and express high levels of Osm, but they did not observe neutrophils in their data.27 In our acute DSS model data, we see that the neutrophils express equal or higher levels of proinflammatory genes than the infiltrating monocytes, including Osm, Il1b, and Tnf (Supplementary Table 1).
scRNA-seq Data Access
The scRNA-seq data reported in this paper are available at GEO accession GSE201723. Analysis scripts will be made available for noncommercial use upon request.
Acknowledgments
The authors thank Vince Meador for histopathology expertise and Davide Risso for advice on the scRNA-seq analysis. They thank the CEO of Surrozen, Craig Parker, for scientific discussions and reviewing the manuscript. They also thank the scientific founders of Surrozen, Chris Garcia, Claudia Janda, Calvin Kuo, Roel Nusse, and Hans Clevers, for helpful discussions over the last few years.
Data Availability Statement
Data and analytic methods included in the manuscript will be made available for noncommercial use upon request. Surrozen proprietary materials used in the study will be made available to academic institutions only, under a Material Transfer Agreement.
CRediT authorship contributions
Liqin Xie, Ph.D. (Conceptualization: Equal; Investigation: Equal; Methodology: Equal; Writing – review & editing: Supporting)
Russell Fletcher, Ph.D. (Conceptualization: Equal; Investigation: Equal; Methodology: Equal; Writing – original draft: Equal; Writing – review & editing: Equal)
Diksha Bhatia, M.Sc. (Investigation: Supporting; Methodology: Supporting)
Darshini Shah, M.Sc. (Investigation: Supporting; Methodology: Supporting)
Jacqueline Phipps, B.Sc. (Investigation: Supporting; Methodology: Supporting)
Shalaka Deshmukh, M.Sc. (Investigation: Supporting; Methodology: Supporting)
Haili Zhang, Ph.D. (Investigation: Supporting; Methodology: Supporting)
Jingjing Ye, Ph.D. (Investigation: Supporting; Methodology: Supporting)
Sungjin Lee, Ph.D. (Investigation: Supporting; Methodology: Supporting)
Lucas Le, B.Sc. (Investigation: Supporting; Methodology: Supporting)
Maureen Newman, Ph.D. (Investigation: Supporting; Methodology: Supporting)
Hui Chen, Ph.D. (Investigation: Supporting; Methodology: Supporting)
Asmiti Sura, M.Sc. (Investigation: Supporting; Methodology: Supporting)
Suhani Gupta, M.Sc. (Investigation: Supporting; Methodology: Supporting)
Laura Sanman, Ph.D. (Investigation: Supporting; Methodology: Supporting)
Fan Yang, Ph.D. (Investigation: Supporting; Methodology: Supporting)
Weixu Meng, Ph.D. (Investigation: Supporting; Methodology: Supporting)
Helene Baribault, Ph.D. (Investigation: Supporting; Methodology: Supporting)
Geertrui Vanhove, M.D. Ph.D. (Supervision: Equal; Writing – review & editing: Supporting)
Wen-Chen Yeh, M.D. Ph.D. (Conceptualization: Supporting; Supervision: Equal; Writing – review & editing: Supporting)
Yang Li, Ph.D. (Conceptualization: Supporting; Supervision: Equal; Writing – review & editing: Supporting)
Chenggang Lu, Ph.D. (Conceptualization: Lead; Investigation: Lead; Methodology: Lead; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of Interest All authors are current or former employees of Surrozen and shareholders of Surrozen stock.
Supplementary Material
References
- 1.Jairath V., Feagan B.G. Global burden of inflammatory bowel disease. Lancet Gastroenterol Hepatol. 2020;5:2–3. doi: 10.1016/S2468-1253(19)30358-9. [DOI] [PubMed] [Google Scholar]
- 2.Chang J.T. Pathophysiology of inflammatory bowel diseases. N Engl J Med. 2020;383:2652–2664. doi: 10.1056/NEJMra2002697. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi T., Siegmund B., Le Berre C., Wei S.C., Ferrante M., Shen B., Bernstein C.N., Danese S., Peyrin-Biroulet L., Hibi T. Ulcerative colitis. Nat Rev Dis Primers. 2020;6:74. doi: 10.1038/s41572-020-0205-x. [DOI] [PubMed] [Google Scholar]
- 4.Chateau T., Feakins R., Marchal-Bressenot A., Magro F., Danese S., Peyrin-Biroulet L. Histological remission in ulcerative colitis: under the microscope is the cure. Am J Gastroenterol. 2020;115:179–189. doi: 10.14309/ajg.0000000000000437. [DOI] [PubMed] [Google Scholar]
- 5.Feagan B.G., Sandborn W.J., Lazar A., Thakkar R.B., Huang B., Reilly N., Chen N., Yang M., Skup M., Mulani P., Chao J. Adalimumab therapy is associated with reduced risk of hospitalization in patients with ulcerative colitis. Gastroenterology. 2014;146:110–118.e3. doi: 10.1053/j.gastro.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 6.Reinisch W., Sandborn W.J., Hommes D.W., D'Haens G., Hanauer S., Schreiber S., Panaccione R., Fedorak R.N., Tighe M.B., Huang B., Kampman W., Lazar A., Thakkar R. Adalimumab for induction of clinical remission in moderately to severely active ulcerative colitis: results of a randomised controlled trial. Gut. 2011;60:780–787. doi: 10.1136/gut.2010.221127. [DOI] [PubMed] [Google Scholar]
- 7.Rutgeerts P., Sandborn W.J., Feagan B.G., Reinisch W., Olson A., Johanns J., Travers S., Rachmilewitz D., Hanauer S.B., Lichtenstein G.R., de Villiers W.J., Present D., Sands B.E., Colombel J.F. Infliximab for induction and maintenance therapy for ulcerative colitis. N Engl J Med. 2005;353:2462–2476. doi: 10.1056/NEJMoa050516. [DOI] [PubMed] [Google Scholar]
- 8.Sands B.E., Peyrin-Biroulet L., Loftus E.V., Jr., Danese S., Colombel J.F., Törüner M., Jonaitis L., Abhyankar B., Chen J., Rogers R., Lirio R.A., Bornstein J.D., Schreiber S. Vedolizumab vs adalimumab for moderate-to-severe ulcerative colitis. N Engl J Med. 2019;381:1215–1226. doi: 10.1056/NEJMoa1905725. [DOI] [PubMed] [Google Scholar]
- 9.Nusse R., Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 10.Whyte J.L., Smith A.A., Helms J.A. Wnt signaling and injury repair. Cold Spring Harb Perspect Biol. 2012;4:a008078. doi: 10.1101/cshperspect.a008078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janda C.Y., Waghray D., Levin A.M., Thomas C., Garcia K.C. Structural basis of Wnt recognition by Frizzled. Science. 2012;337:59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H., Lu C., Ouyang B., Zhang H., Huang Z., Bhatia D., Lee S.J., Shah D., Sura A., Yeh W.C., Li Y. Development of potent, selective surrogate WNT molecules and their application in defining Frizzled requirements. Cell Chem Biol. 2020;27:598–609.e4. doi: 10.1016/j.chembiol.2020.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Dang L.T., Miao Y., Ha A., Yuki K., Park K., Janda C.Y., Jude K.M., Mohan K., Ha N., Vallon M., Yuan J., Vilches-Moure J.G., Kuo C.J., Garcia K.C., Baker D. Receptor subtype discrimination using extensive shape complementary designed interfaces. Nat Struct Mol Biol. 2019;26:407–414. doi: 10.1038/s41594-019-0224-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fowler T.W., Mitchell T.L., Janda C.Y., Xie L., Tu S., Chen H., Zhang H., Ye J., Ouyang B., Yuan T.Z., Lee S.J., Newman M., Tripuraneni N., Rego E.S., Mutha D., Dilip A., Vuppalapaty M., Baribault H., Yeh W.C., Li Y. Development of selective bispecific Wnt mimetics for bone loss and repair. Nat Commun. 2021;12:3247. doi: 10.1038/s41467-021-23374-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janda C.Y., Dang L.T., You C., Chang J., de Lau W., Zhong Z.A., Yan K.S., Marecic O., Siepe D., Li X., Moody J.D., Williams B.O., Clevers H., Piehler J., Baker D., Kuo C.J., Garcia K.C. Surrogate Wnt agonists that phenocopy canonical Wnt and β-catenin signalling. Nature. 2017;545:234–237. doi: 10.1038/nature22306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miao Y., Ha A., de Lau W., Yuki K., Santos A.J.M., You C., Geurts M.H., Puschhof J., Pleguezuelos-Manzano C., Peng W.C., Senlice R., Piani C., Buikema J.W., Gbenedio O.M., Vallon M., Yuan J., de Haan S., Hemrika W., Rösch K., Dang L.T., Baker D., Ott M., Depeille P., Wu S.M., Drost J., Nusse R., Roose J.P., Piehler J., Boj S.F., Janda C.Y., Clevers H., Kuo C.J., Garcia K.C. Next-generation surrogate Wnts support organoid growth and deconvolute Frizzled pleiotropy in vivo. Cell Stem Cell. 2020;27:840–851.e6. doi: 10.1016/j.stem.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tao Y., Mis M., Blazer L., Ustav M.J., Steinhart Z., Chidiac R., Kubarakos E., O'Brien S., Wang X., Jarvik N., Patel N., Adams J., Moffat J., Angers S., Sidhu S.S. Tailored tetravalent antibodies potently and specifically activate Wnt/Frizzled pathways in cells, organoids and mice. Elife. 2019;8:e46134. doi: 10.7554/eLife.46134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan K.S., Janda C.Y., Chang J., Zheng G.X.Y., Larkin K.A., Luca V.C., Chia L.A., Mah A.T., Han A., Terry J.M., Ootani A., Roelf K., Lee M., Yuan J., Li X., Bolen C.R., Wilhelmy J., Davies P.S., Ueno H., von Furstenberg R.J., Belgrader P., Ziraldo S.B., Ordonez H., Henning S.J., Wong M.H., Snyder M.P., Weissman I.L., Hsueh A.J., Mikkelsen T.S., Garcia K.C., Kuo C.J. Non-equivalence of Wnt and R-spondin ligands during Lgr5(+) intestinal stem-cell self-renewal. Nature. 2017;545:238–242. doi: 10.1038/nature22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carmon K.S., Gong X., Lin Q., Thomas A., Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A. 2011;108:11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Lau W., Barker N., Low T.Y., Koo B.K., Li V.S., Teunissen H., Kujala P., Haegebarth A., Peters P.J., van de Wetering M., Stange D.E., van Es J.E., Guardavaccaro D., Schasfoort R.B., Mohri Y., Nishimori K., Mohammed S., Heck A.J., Clevers H. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 21.Glinka A., Dolde C., Kirsch N., Huang Y.L., Kazanskaya O., Ingelfinger D., Boutros M., Cruciat C.M., Niehrs C. LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signalling. EMBO Rep. 2011;12:1055–1061. doi: 10.1038/embor.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greicius G., Kabiri Z., Sigmundsson K., Liang C., Bunte R., Singh M.K., Virshup D.M. PDGFRα(+) pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc Natl Acad Sci U S A. 2018;115:E3173–E3181. doi: 10.1073/pnas.1713510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim K.A., Kakitani M., Zhao J., Oshima T., Tang T., Binnerts M., Liu Y., Boyle B., Park E., Emtage P., Funk W.D., Tomizuka K. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science. 2005;309:1256–1259. doi: 10.1126/science.1112521. [DOI] [PubMed] [Google Scholar]
- 24.Zhao J., de Vera J., Narushima S., Beck E.X., Palencia S., Shinkawa P., Kim K.A., Liu Y., Levy M.D., Berg D.J., Abo A., Funk W.D. R-spondin1, a novel intestinotrophic mitogen, ameliorates experimental colitis in mice. Gastroenterology. 2007;132:1331–1343. doi: 10.1053/j.gastro.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Cooper H.S., Murthy S.N., Shah R.S., Sedergran D.J. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab Invest. 1993;69:238–249. [PubMed] [Google Scholar]
- 26.Kinchen J., Chen H.H., Parikh K., Antanaviciute A., Jagielowicz M., Fawkner-Corbett D., Ashley N., Cubitt L., Mellado-Gomez E., Attar M., Sharma E., Wills Q., Bowden R., Richter F.C., Ahern D., Puri K.D., Henault J., Gervais F., Koohy H., Simmons A. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell. 2018;175:372–386.e17. doi: 10.1016/j.cell.2018.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smillie C.S., Biton M., Ordovas-Montanes J., Sullivan K.M., Burgin G., Graham D.B., Herbst R.H., Rogel N., Slyper M., Waldman J., Sud M., Andrews E., Velonias G., Haber A.L., Jagadeesh K., Vickovic S., Yao J., Stevens C., Dionne D., Nguyen L.T., Villani A.C., Hofree M., Creasey E.A., Huang H., Rozenblatt-Rosen O., Garber J.J., Khalili H., Desch A.N., Daly M.J., Ananthakrishnan A.N., Shalek A.K., Xavier R.J., Regev A. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell. 2019;178:714–730.e22. doi: 10.1016/j.cell.2019.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nusse Y.M., Savage A.K., Marangoni P., Rosendahl-Huber A.K.M., Landman T.A., de Sauvage F.J., Locksley R.M., Klein O.D. Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature. 2018;559:109–113. doi: 10.1038/s41586-018-0257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yui S., Azzolin L., Maimets M., Pedersen M.T., Fordham R.P., Hansen S.L., Larsen H.L., Guiu J., Alves M.R.P., Rundsten C.F., Johansen J.V., Li Y., Madsen C.D., Nakamura T., Watanabe M., Nielsen O.H., Schweiger P.J., Piccolo S., Jensen K.B. YAP/TAZ-dependent reprogramming of colonic epithelium links ECM remodeling to tissue regeneration. Cell Stem Cell. 2018;22:35–49.e7. doi: 10.1016/j.stem.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boonekamp K.E., Heo I., Artegiani B., Asra P., van Son G., de Ligt J., Clevers H. Identification of novel human Wnt target genes using adult endodermal tissue-derived organoids. Dev Biol. 2021;474:37–47. doi: 10.1016/j.ydbio.2021.01.009. [DOI] [PubMed] [Google Scholar]
- 31.Powell A.E., Wang Y., Li Y., Poulin E.J., Means A.L., Washington M.K., Higginbotham J.N., Juchheim A., Prasad N., Levy S.E., Guo Y., Shyr Y., Aronow B.J., Haigis K.M., Franklin J.L., Coffey R.J. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parisi S., Piscitelli S., Passaro F., Russo T. HMGA proteins in stemness and differentiation of embryonic and adult stem cells. Int J Mol Sci. 2020;21:362. doi: 10.3390/ijms21010362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goto N., Ueo T., Fukuda A., Kawada K., Sakai Y., Miyoshi H., Taketo M.M., Chiba T., Seno H. Distinct roles of HES1 in normal stem cells and tumor stem-like cells of the intestine. Cancer Res. 2017;77:3442–3454. doi: 10.1158/0008-5472.CAN-16-3192. [DOI] [PubMed] [Google Scholar]
- 34.Fong Y.W., Ho J.J., Inouye C., Tjian R. The dyskerin ribonucleoprotein complex as an OCT4/SOX2 coactivator in embryonic stem cells. Elife. 2014;3:e03573. doi: 10.7554/eLife.03573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H., Lu C., Lee S.J., Li Y. Protocol to generate and characterize potent and selective WNT mimetic molecules. STAR Protoc. 2020;1:100043. doi: 10.1016/j.xpro.2020.100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mah A.T., Yan K.S., Kuo C.J. Wnt pathway regulation of intestinal stem cells. J Physiol. 2016;594:4837–4847. doi: 10.1113/JP271754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nile A.H., de Sousa E.M.F., Mukund S., Piskol R., Hansen S., Zhou L., Zhang Y., Fu Y., Gogol E.B., Kömüves L.G., Modrusan Z., Angers S., Franke Y., Koth C., Fairbrother W.J., Wang W., de Sauvage F.J., Hannoush R.N. A selective peptide inhibitor of Frizzled 7 receptors disrupts intestinal stem cells. Nat Chem Biol. 2018;14:582–590. doi: 10.1038/s41589-018-0035-2. [DOI] [PubMed] [Google Scholar]
- 38.Flanagan D.J., Phesse T.J., Barker N., Schwab R.H., Amin N., Malaterre J., Stange D.E., Nowell C.J., Currie S.A., Saw J.T., Beuchert E., Ramsay R.G., Sansom O.J., Ernst M., Clevers H., Vincan E. Frizzled7 functions as a Wnt receptor in intestinal epithelial Lgr5(+) stem cells. Stem Cell Rep. 2015;4:759–767. doi: 10.1016/j.stemcr.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Es J.H., Jay P., Gregorieff A., van Gijn M.E., Jonkheer S., Hatzis P., Thiele A., van den Born M., Begthel H., Brabletz T., Taketo M.M., Clevers H. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat Cell Biol. 2005;7:381–386. doi: 10.1038/ncb1240. [DOI] [PubMed] [Google Scholar]
- 40.Chen M., Reed R.R., Lane A.P. Acute inflammation regulates neuroregeneration through the NF-κB pathway in olfactory epithelium. Proc Natl Acad Sci U S A. 2017;114:8089–8094. doi: 10.1073/pnas.1620664114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kyritsis N., Kizil C., Zocher S., Kroehne V., Kaslin J., Freudenreich D., Iltzsche A., Brand M. Acute inflammation initiates the regenerative response in the adult zebrafish brain. Science. 2012;338:1353–1356. doi: 10.1126/science.1228773. [DOI] [PubMed] [Google Scholar]
- 42.Schwitalla S., Fingerle A.A., Cammareri P., Nebelsiek T., Göktuna S.I., Ziegler P.K., Canli O., Heijmans J., Huels D.J., Moreaux G., Rupec R.A., Gerhard M., Schmid R., Barker N., Clevers H., Lang R., Neumann J., Kirchner T., Taketo M.M., van den Brink G.R., Sansom O.J., Arkan M.C., Greten F.R. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152:25–38. doi: 10.1016/j.cell.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 43.Krausova M., Korinek V. Wnt signaling in adult intestinal stem cells and cancer. Cell Signal. 2014;26:570–579. doi: 10.1016/j.cellsig.2013.11.032. [DOI] [PubMed] [Google Scholar]
- 44.Wirtz S., Popp V., Kindermann M., Gerlach K., Weigmann B., Fichtner-Feigl S., Neurath M.F. Chemically induced mouse models of acute and chronic intestinal inflammation. Na. Protoc. 2017;12:1295–1309. doi: 10.1038/nprot.2017.044. [DOI] [PubMed] [Google Scholar]
- 45.Lun A.T., Bach K., Marioni J.C. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu C., Su Z. Identification of cell types from single-cell transcriptomes using a novel clustering method. Bioinformatics. 2015;31:1974–1980. doi: 10.1093/bioinformatics/btv088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Risso D., Purvis L., Fletcher R.B., Das D., Ngai J., Dudoit S., Purdom E. clusterExperiment and RSEC: A Bioconductor package and framework for clustering of single-cell and other large gene expression datasets. PLoS Comput Biol. 2018;14 doi: 10.1371/journal.pcbi.1006378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y., Lun A.T., Smyth G.K. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res. 2016;5:1438. doi: 10.12688/f1000research.8987.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu D., Lim E., Vaillant F., Asselin-Labat M.L., Visvader J.E., Smyth G.K. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics. 2010;26:2176–2182. doi: 10.1093/bioinformatics/btq401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cabello-Aguilar S., Alame M., Kon-Sun-Tack F., Fau C., Lacroix M., Colinge J. SingleCellSignalR: inference of intercellular networks from single-cell transcriptomics. Nucleic Acids Res. 2020;48:e55. doi: 10.1093/nar/gkaa183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raredon MSB, Yang J, Garritano J, Wang M, Kushnir D, Schupp JC, Adams TS, Greaney AM, Leiby KL, Kaminski N, Kluger Y, Levchenko A, Niklason LE. Connectome: computation and visualization of cell-cell signaling topologies in single-cell systems data. bioRxiv, https://doi.org/10.1101/2021.01.21.427529. [DOI] [PMC free article] [PubMed]
- 53.Street K., Risso D., Fletcher R.B., Das D., Ngai J., Yosef N., Purdom E., Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gougelet A., Torre C., Veber P., Sartor C., Bachelot L., Denechaud P.D., Godard C., Moldes M., Burnol A.F., Dubuquoy C., Terris B., Guillonneau F., Ye T., Schwarz M., Braeuning A., Perret C., Colnot S. T-cell factor 4 and β-catenin chromatin occupancies pattern zonal liver metabolism in mice. Hepatology. 2014;59:2344–2357. doi: 10.1002/hep.26924. [DOI] [PubMed] [Google Scholar]
- 55.Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 56.Gracz A.D., Fuller M.K., Wang F., Li L., Stelzner M., Dunn J.C., Martin M.G., Magness S.T. Brief report: CD24 and CD44 mark human intestinal epithelial cell populations with characteristics of active and facultative stem cells. Stem Cells. 2013;31:2024–2030. doi: 10.1002/stem.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schuijers J., Junker J.P., Mokry M., Hatzis P., Koo B.K., Sasselli V., van der Flier L.G., Cuppen E., van Oudenaarden A., Clevers H. Ascl2 acts as an R-spondin/Wnt-responsive switch to control stemness in intestinal crypts. Cell Stem Cell. 2015;16:158–170. doi: 10.1016/j.stem.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 58.Shvab A., Haase G., Ben-Shmuel A., Gavert N., Brabletz T., Dedhar S., Ben-Ze'ev A. Induction of the intestinal stem cell signature gene SMOC-2 is required for L1-mediated colon cancer progression. Oncogene. 2016;35:549–557. doi: 10.1038/onc.2015.127. [DOI] [PubMed] [Google Scholar]
- 59.Ayyaz A., Kumar S., Sangiorgi B., Ghoshal B., Gosio J., Ouladan S., Fink M., Barutcu S., Trcka D., Shen J., Chan K., Wrana J.L., Gregorieff A. Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature. 2019;569:121–125. doi: 10.1038/s41586-019-1154-y. [DOI] [PubMed] [Google Scholar]
- 60.Rothenberg M.E., Nusse Y., Kalisky T., Lee J.J., Dalerba P., Scheeren F., Lobo N., Kulkarni S., Sim S., Qian D., Beachy P.A., Pasricha P.J., Quake S.R., Clarke M.F. Identification of a cKit(+) colonic crypt base secretory cell that supports Lgr5(+) stem cells in mice. Gastroenterology. 2012;142:1195–1205.e6. doi: 10.1053/j.gastro.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sasaki N., Sachs N., Wiebrands K., Ellenbroek S.I., Fumagalli A., Lyubimova A., Begthel H., van den Born M., van Es J.H., Karthaus W.R., Li V.S., López-Iglesias C., Peters P.J., van Rheenen J., van Oudenaarden A., Clevers H. Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon. Proc Natl Acad Sci U S A. 2016;113:E5399–E5407. doi: 10.1073/pnas.1607327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Billing L.J., Larraufie P., Lewis J., Leiter A., Li J., Lam B., Yeo G.S., Goldspink D.A., Kay R.G., Gribble F.M., Reimann F. Single cell transcriptomic profiling of large intestinal enteroendocrine cells in mice—Identification of selective stimuli for insulin-like peptide-5 and glucagon-like peptide-1 coexpressing cells. Mol Metab. 2019;29:158–169. doi: 10.1016/j.molmet.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stzepourginski I., Nigro G., Jacob J.M., Dulauroy S., Sansonetti P.J., Eberl G., Peduto L. CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc Natl Acad Sci U S A. 2017;114:E506–E513. doi: 10.1073/pnas.1620059114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cox C.B., Storm E.E., Kapoor V.N., Chavarria-Smith J., Lin D.L., Wang L., Li Y., Kljavin N., Ota N., Bainbridge T.W., Anderson K., Roose-Girma M., Warming S., Arron J.R., Turley S.J., de Sauvage F.J., van Lookeren Campagne M. IL-1R1-dependent signaling coordinates epithelial regeneration in response to intestinal damage. Sci Immunol. 2021;6 doi: 10.1126/sciimmunol.abe8856. [DOI] [PubMed] [Google Scholar]
- 65.Muhl L., Genové G., Leptidis S., Liu J., He L., Mocci G., Sun Y., Gustafsson S., Buyandelger B., Chivukula I.V., Segerstolpe Å., Raschperger E., Hansson E.M., Björkegren J.L.M., Peng X.R., Vanlandewijck M., Lendahl U., Betsholtz C. Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat Commun. 2020;11:3953. doi: 10.1038/s41467-020-17740-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brügger M.D., Valenta T., Fazilaty H., Hausmann G., Basler K. Distinct populations of crypt-associated fibroblasts act as signaling hubs to control colon homeostasis. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aoki R., Shoshkes-Carmel M., Gao N., Shin S., May C.L., Golson M.L., Zahm A.M., Ray M., Wiser C.L., Wright C.V., Kaestner K.H. Foxl1-expressing mesenchymal cells constitute the intestinal stem cell niche. Cell Mol Gastroenterol Hepatol. 2016;2:175–188. doi: 10.1016/j.jcmgh.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shoshkes-Carmel M., Wang Y.J., Wangensteen K.J., Tóth B., Kondo A., Massasa E.E., Itzkovitz S., Kaestner K.H. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature. 2018;557:242–246. doi: 10.1038/s41586-018-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Degirmenci B., Valenta T., Dimitrieva S., Hausmann G., Basler K. GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature. 2018;558:449–453. doi: 10.1038/s41586-018-0190-3. [DOI] [PubMed] [Google Scholar]
- 70.Kurahashi M., Zheng H., Dwyer L., Ward S.M., Koh S.D., Sanders K.M. A functional role for the 'fibroblast-like cells' in gastrointestinal smooth muscles. J Physiol. 2011;589:697–710. doi: 10.1113/jphysiol.2010.201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vanderwinden J.M., Rumessen J.J., de Kerchove d'Exaerde A., Jr., Gillard K., Panthier J.J., de Laet M.H., Schiffmann S.N. Kit-negative fibroblast-like cells expressing SK3, a Ca2+-activated K+ channel, in the gut musculature in health and disease. Cell Tissue Res. 2002;310:349–358. doi: 10.1007/s00441-002-0638-4. [DOI] [PubMed] [Google Scholar]
- 72.Goessling W., North T.E., Loewer S., Lord A.M., Lee S., Stoick-Cooper C.L., Weidinger G., Puder M., Daley G.Q., Moon R.T., Zon L.I. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell. 2009;136:1136–1147. doi: 10.1016/j.cell.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jones G.R., Bain C.C., Fenton T.M., Kelly A., Brown S.L., Ivens A.C., Travis M.A., Cook P.C., MacDonald A.S. Dynamics of colon monocyte and macrophage activation during colitis. Front Immunol. 2018;9:2764. doi: 10.3389/fimmu.2018.02764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu K., Nussenzweig M.C. Origin and development of dendritic cells. Immunol Rev. 2010;234:45–54. doi: 10.1111/j.0105-2896.2009.00879.x. [DOI] [PubMed] [Google Scholar]
- 75.Bros M., Dexheimer N., Ross R., Trojandt S., Höhn Y., Tampe J., Sutter A., Jährling F., Grabbe S., Reske-Kunz A.B. Differential gene expression analysis identifies murine Cacnb3 as strongly upregulated in distinct dendritic cell populations upon stimulation. Gene. 2011;472:18–27. doi: 10.1016/j.gene.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 76.Riol-Blanco L., Sánchez-Sánchez N., Torres A., Tejedor A., Narumiya S., Corbí A.L., Sánchez-Mateos P., Rodríguez-Fernández J.L. The chemokine receptor CCR7 activates in dendritic cells 2 signaling modules that independently regulate chemotaxis and migratory speed. J Immunol. 2005;174:4070–4080. doi: 10.4049/jimmunol.174.7.4070. [DOI] [PubMed] [Google Scholar]
- 77.Marchal Bressenot A., Riddell R.H., Boulagnon-Rombi C., Reinisch W., Danese S., Schreiber S., Peyrin-Biroulet L. Review article: the histological assessment of disease activity in ulcerative colitis. Aliment Pharmacol Ther. 2015;42:957–967. doi: 10.1111/apt.13375. [DOI] [PubMed] [Google Scholar]
- 78.Vidal-Lletjós S., Andriamihaja M., Blais A., Grauso M., Lepage P., Davila A.M., Gaudichon C., Leclerc M., Blachier F., Lan A. Mucosal healing progression after acute colitis in mice. World J Gastroenterol. 2019;25:3572–3589. doi: 10.3748/wjg.v25.i27.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wéra O., Lancellotti P., Oury C. The dual role of neutrophils in inflammatory bowel diseases. J Clin Med. 2016;5:118. doi: 10.3390/jcm5120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eash K.J., Greenbaum A.M., Gopalan P.K., Link D.C. CXCR2 and CXCR4 antagonistically regulate neutrophil trafficking from murine bone marrow. J Clin Invest. 2010;120:2423–2431. doi: 10.1172/JCI41649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Touw I.P., Beekman R. Severe congenital neutropenia and chronic neutrophilic leukemia: an intriguing molecular connection unveiled by oncogenic mutations in CSF3R. Haematologica. 2013;98:1490–1492. doi: 10.3324/haematol.2013.090571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and analytic methods included in the manuscript will be made available for noncommercial use upon request. Surrozen proprietary materials used in the study will be made available to academic institutions only, under a Material Transfer Agreement.