Abstract
At least five genes of the gibberellin (GA) biosynthesis pathway are clustered on chromosome 4 of Gibberella fujikuroi; these genes encode the bifunctional ent-copalyl diphosphate synthase/ent-kaurene synthase, a GA-specific geranylgeranyl diphosphate synthase, and three cytochrome P450 monooxygenases. We now describe a fourth cytochrome P450 monooxygenase gene (P450-4). Gas chromatography-mass spectrometry analysis of extracts of mycelia and culture fluid of a P450-4 knockout mutant identified ent-kaurene as the only intermediate of the GA pathway. Incubations with radiolabeled precursors showed that the metabolism of ent-kaurene, ent-kaurenol, and ent-kaurenal was blocked in the transformants, whereas ent-kaurenoic acid was metabolized efficiently to GA4. The GA-deficient mutant strain SG139, which lacks the 30-kb GA biosynthesis gene cluster, converted ent-kaurene to ent-kaurenoic acid after transformation with P450-4. The B1-41a mutant, described as blocked between ent-kaurenal and ent-kaurenoic acid, was fully complemented by P450-4. There is a single nucleotide difference between the sequence of the B1-41a and wild-type P450-4 alleles at the 3′ consensus sequence of intron 2 in the mutant, resulting in reduced levels of active protein due to a splicing defect in the mutant. These data suggest that P450-4 encodes a multifunctional ent-kaurene oxidase catalyzing all three oxidation steps between ent-kaurene and ent-kaurenoic acid.
The gibberellins (GAs) are a group of phytohormones that influence many developmental processes in higher plants, including seed germination, stem elongation, flowering, and fruit set. GAs also are produced by the rice pathogen Gibberella fujikuroi (mating population C) and a few other fungal genera (30), but nothing is known about the role of GAs in fungi. Cultures of G. fujikuroi are used for the commercial production of GAs, particularly gibberellic acid (GA3), for use in agriculture (26).
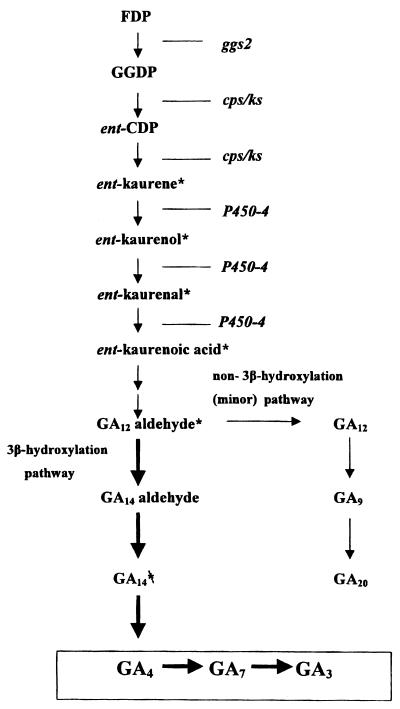
The biosynthesis of GAs has been investigated for many years in G. fujikuroi and in higher plants. The terpenoid nature of GAs was first established by the incorporation of [2-14C]mevalonic acid into GA3, which is the end product of the fungal pathway (20) (Fig. 1). The biosynthesis follows the isoprenoid pathway to geranylgeranyl diphosphate (GGPP), which, in plants, undergoes a two-step cyclization reaction in which GGPP is converted to ent-kaurene via ent-copalyl diphosphate (CPP) (20). ent-Kaurene is metabolized to GAs by reactions catalyzed by cytochrome P450 monooxygenases and, in plants, 2-oxoglutarate-dependent dioxygenases (11). In G. fujikuroi, GGPP synthase, which catalyzes the formation of GGPP, is encoded by two genes, one of which, ggs2, is specific for GA biosynthesis (31). In contrast to plants, in which cyclization of GGPP is catalyzed by two enzymes, CPP synthase (CPS) and ent-kaurene synthase (KS), in the fungi G. fujikuroi and Phaeosphaeria, both steps are catalyzed by a bifunctional CPS/KS enzyme (17, 32).
FIG. 1.
GA biosynthesis pathway in G. fujikuroi. Genes with known functions are indicated. Thick arrows indicate the major pathway and the double arrows represent several steps. 14C-labeled substrates used in metabolism experiments are marked with a ∗.
Most of the genes of the early isoprenoid pathway have been cloned from G. fujikuroi, including HMG-CoA reductase (36), farnesyl diphosphate synthase (16), and a general GGPP synthase (ggs1) (22). Five genes of the GA pathway in G. fujikuroi, comprising cps/ks, the GA-specific GGPP synthase (ggs2), and three cytochrome P450 monooxygenase genes, were shown to be closely linked in a gene cluster (31). Recently, we showed that one of these three genes, P450-1, catalyzes four oxidation steps in the main pathway from ent-kaurenoic acid to GA14 via GA12-aldehyde (27). In this report, we describe the isolation and functional characterization of a fourth P450 monooxygenase gene which is also located in the GA gene cluster and is closely linked to P450-1 through a shared promoter. Using gene disruption and by expressing P450-4 in the GA-deficient mutant SG139, which lacks the entire gene cluster, we show that the gene codes for a multifunctional ent-kaurene oxidase, catalyzing all three oxidation steps between ent-kaurene and ent-kaurenoic acid. Furthermore, it complements the genetic block in the GA-deficient UV mutant B1-41a, which contains a point mutation in the P450-4 locus.
MATERIALS AND METHODS
Fungal strains and culture conditions.
G. fujikuroi m567, a wild-type strain from rice, was provided by the Fungal Culture Collection, Weimar, Germany. The wild-type strain IMI 58289 and the GA-defective mutant strain SG139 (3) were provided by E. Cerda-Olmedo and J. Avalos (University of Sevilla, Sevilla, Spain). SG139 has completely lost the GA gene cluster as demonstrated by Southern blotting and PCR analysis. The GA-deficient mutant B1-41a, obtained by UV irradiation of G. fujikuroi strain GF-1a (4), was provided by J. MacMillan (University of Bristol, Bristol, United Kingdom).
Bacterial strains and plasmids.
Escherichia coli strain Top10 (Invitrogen, Groningen, The Netherlands) was used for plasmid propagation. Vector pUC19 was used to clone DNA fragments carrying the G. fujikuroi P450-4 gene or parts of it. For gene disruption experiments, a 0.9-kb internal PCR fragment obtained with primers P450-4-GD1 and P450-4-GD2 was cloned into the vector pCR2.1 (Invitrogen). The fragment was excised with XbaI/HindIII and cloned into the vector pAN7-1 (25) carrying the hygromycin B resistance cassette. For gene complementation, a 5.8-kb BamHI fragment carrying the entire P450-4 gene was cloned into pGPC1 (7). P450-4 cDNA clones in the Uni-Zap XR λ vector were converted to pBluescript SK(−) phagemids by in vivo rescue according to the manufacturer's protocol (Stratagene, La Jolla, Calif.). For the identification of the mutation site in the mutant B1-41a, the mutant copy of P450-4 was amplified by PCR and cloned into the PCR cloning vector pCR2.1 for sequence analysis.
Media and culture conditions.
For DNA isolation, the fungal strains were grown in 100 ml of CM liquid medium optimized for Fusarium spp. (24) for 3 days at 28°C on a rotary shaker set at 200 rpm. The mycelia were harvested by filtration through a sterile glass filter (G2; Schott, Jena, Germany), washed with sterile distilled water, frozen in liquid nitrogen, and lyophilized for 24 h. The lyophilized mycelial tissue was ground to a fine powder with a mortar and pestle.
For RNA isolation, fungal strains were grown in an optimized GA3 production medium (OPM) containing 6% sunflower oil, 0.05% (NH4)2SO4, 1.5% corn-steep solids (Sigma-Aldrich, Taufkirchen, Germany), and 0.1% KH2PO4. Mycelia were harvested after 15 h (growth phase) and after 3 to 6 days of cultivation (production phase).
For analysis of GA and ent-kaurenoid content, fungal strains were grown in the GA production medium for 7 to 10 days at 28°C on a rotary shaker (200 rpm). Cultures for metabolism studies were established in 100 ml of 100% ICI medium (9) for 4 days at 25°C on a rotary shaker, then subcultured into 100 ml of 40% ICI medium. After 5 days, a 5-ml inoculum from the 40% ICI culture was added to 100 ml of 10% ICI medium containing 1 mM AMO-1618 (Calbiochem, San Diego, Calif.).
Radiolabeled substrates.
ent-[1,7,12,18-14C4]kaurene (specific radioactivity, 8.25 TBq · mol−1), ent-[14C4]kaurenal, ent-[14C4]kaurenal, ent-[14C4]kaurenoic acid (each 7.47 TBq · mol−1), and [14C4]GA12-aldehyde (6.81 TBq · mol−1) were prepared from R-[2-14C]mevalonic acid using a cell-free system from pumpkin endosperm, as described by Graebe et al. (10). [14C4]GA14 (5.58 TBq · mol−1) was prepared from [14C4]GA12-aldehyde by incubation with a cell-free system from G. fujikuroi (34). [17-14C]GA4 (1.85 TBq · mol−1) was prepared from [17-14C]GA9 by incubation with a recombinant sugar beet GA 3β-hydroxylase, as described by Williams et al. (35). The [17-14C]GA9 was synthesized from GA9 17-norketone and [14C-methyl]triphenylphosphonium bromide essentially as described previously (19).
DNA and RNA isolation.
Genomic DNA was isolated from lyophilized mycelia according to Doyle and Doyle (8). Lambda DNA from positive lambda clones was prepared according to Maniatis et al. (21). Plasmid DNA was extracted using Genomed columns following the manufacturer's protocol (Genomed, Bad Oeynhausen, Germany). RNA for Northern blot analysis was isolated by using the RNAgents Total RNA Isolation Kit (Promega, Mannheim, Germany).
Screening of G. fujikuroi cDNA library and genomic lambda EMBL3 library.
The expression library (UniZap XR vector; Stratagene) was constructed from RNA isolated from mycelia which were grown under optimal conditions for GA formation (22). Approximately 50,000 recombinant phages were plated at about 7,500 plaques per 150-mm-diameter Petri dish and transferred to nylon membranes. For screening of the genomic library (33), about 35,000 recombinant phages were plated and transferred to membranes. Hybridization was performed at high stringency (65°C). The blots were washed under hybridization conditions (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate [SDS]; 65°C; followed by 0.1× SSC, 0.1% SDS). Positive recombinant clones were used for a second round of plaque purification.
Southern and Northern blot analysis.
After incubation with restriction endonucleases and electrophoresis, genomic or lambda DNA was transferred to Hybond N+ filters (Amersham Pharmacia, Freiburg, Germany) (28). 32P-labeled probes were prepared using the random oligomer-primer method. Filters were hybridized at 65°C in 5× Denhardt's solution containing 5% dextran sulfate (21). Filters were washed at the hybridization temperature in 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 0.1% SDS, and 1× SSPE, 0.1% SDS.
Northern blot hybridization was accomplished by the method of Church and Gilbert (6). The conserved Botrytis cinerea actin gene (5) was used as a control for RNA transfer.
Sequence analysis.
DNA sequencing of recombinant plasmid clones was accomplished by using a LI-COR 4000 automatic sequencer (MWG, München, Germany). Overlapping subclones of the genomic DNA and the cDNA clones were prepared and both strands were sequenced using the universal and the reverse primers. For identification of the mutation in B1-41a, two additional specific sequencing primers were used: P450-4-M1 (5′-CTATAGGTTTCAGCCACATCC-3′) and P450-4-M2 (5′-ATCATCCCGCCAAACTACATCG-3′). Sequence analysis was performed using the Seqman II computer program (DNA STAR Inc., Madison, Wis.).
Transformation of G. fujikuroi.
Protoplast preparation and transformation was carried out as previously described (33). For gene disruption, 107 protoplasts (100 μl) of strain IMI 58289 were transformed with 10 μg of the circular gene disruption vector pP450-4-GD. For complementation of the mutant strains B1-41 and SG139 with the intact P450-4 gene, protoplasts were transformed with 10 μg of the circular complementation vector pP450-4-GC.
Transformed protoplasts were regenerated at 28°C in a complete regeneration agar [0.7 M sucrose, 0.05% yeast extract, 0.1% (NH4)2SO4, containing 120 μg of hygromycin B (Calbiochem, Bad Soden, Germany) per ml] for 6 to 7 days. Single conidial cultures were established from hygromycin B-resistant transformants and used for DNA isolation and Southern blot analysis.
GA assay.
For analysis of GA formation, the wild-type strain and P450-4-disrupted mutants were cultivated in 100-ml Erlenmeyer flasks containing 20 ml of OPM. Cultures were incubated for 7 days on a rotary shaker (200 rpm) at 28°C. GA3 was analyzed by high-performance liquid chromatography (HPLC) according to Barendse et al. (2) by using a Merck HPLC system with a UV detector and a Lichrispher 100 RP18 column (5 μm; 250 by 4 mm; Merck, Darmstadt, Germany). GA3, GA4, and GA7 also were analyzed by thin-layer chromatography and eluted with ethyl acetate:chloroform:acetic acid (60:40:5). Extracts of culture filtrates and mycelia were analyzed by gas chromatography-mass spectroscopy (GC-MS) as described previously (18).
PCR.
G. fujikuroi m567 DNA was used as template for amplification of an internal fragment of the P450-4 gene. The specific primers had the following sequences: P450-4-GD1, 5′-GGTCCAGAGCACTGCCGC-3′; P450-4-GD2, 5′-CTTCCTTTCCCATCTGGC-3′. DNA amplification was performed in 50-μl mixtures using 2 U of Taq DNA polymerase (Red-Taq; Sigma-Aldrich), 50 ng of genomic DNA/μl, 50 μM concentrations of each deoxynucleoside triphosphate, 200 nM concentrations each primer, and 1× Taq buffer containing 15 mM MgCl2 (Sigma-Aldrich). PCR was carried out for 36 cycles, each comprising 1 min of denaturation at 94°C, 0.5 min of annealing at 56°C, and 1.5 min of extension at 72°C. The PCR product was purified by using a gel extraction kit (Genomed), precipitated with 0.3 M sodium acetate and 2 volumes of ethanol, and cloned using the PCR2.1 vector system (Invitrogen).
For the amplification of the mutant copy of P450-4, 50 ng of G. fujikuroi B1-41a DNA/μl was used as template. The specific primers had the following sequences: P450-4-F, 5′-GGTCCAGAGCACTGCCGC-3′; P450-4-R, 5′-CTTCCTTTCCCATCTGGC-3′. PCR was carried out as described above, but the annealing temperature was 60°C. Reverse transcription (RT) was performed using the Titan One Tube RT-PCR System (Boehringer, Mannheim, Germany). The two primers had the following sequences: P450-RT1, 5′-TCTAAGAGGCTCTATGTACTC-3′; P450-RT2, 5′-TGCCTTGACCAAAGAGATGCC-3′.
Incubations with radiolabeled substrates.
Three milliliter aliquots of the fungal cultures, grown in 10% ICI medium (9) for 5 days at 25°C, were transferred to sterile 50-ml Falcon tubes to which 2.5 kBq of 14C4-labeled ent-kaurene, ent-kaurenol, ent-kaurenal, or ent-kaurenoic acid were added in methanol (18 to 75 μl). After incubation on an orbital shaker at 25°C for 3 days, the cultures were filtered and the filtrates were adjusted to pH 3 with 1 N HCl and partitioned against ethyl acetate (3 equal volumes), which was taken to dryness under a stream of N2. The mycelia were extracted with 10 ml of methanol overnight, the extracts were passed through silica gel (500 mg), which was eluted with a further 10 ml of methanol, and the combined eluates were taken to dryness in vacuo. The ethyl acetate extracts were analyzed by reverse-phase HPLC with on-line radioactivity monitoring (19). After loading onto a guard column in solvent A (10% methanol-water containing 50 μl of acetic acid/liter) for 5 min, samples were eluted through an ODS Hypersil column (25 by 0.46 cm) with a linear gradient of 20 to 100% solvent B (methanol containing 50 μ of acetic acid/liter) over 24 min at 1 ml/min, after which the column was eluted with solvent B for a further 5 min. Mycelial extracts were similarly analyzed by HPLC, but by using a gradient of 75 to 100% solvent B over 30 min followed by 30 min with solvent B. The most significant radioactive products were collected, converted to methyl esters and trimethylsilyl ethers, and analyzed by GC-MS (18).
In a separate experiment, cultures were incubated with [14C4]GA12-aldehyde (3.3 kBq), [17-14C]GA4 (1.5 kBq), and [14C4]GA14 (1.5 kBq), and the medium was extracted and analyzed by HPLC as described above.
Nucleotide sequence accession number.
The nucleotide sequence for the G. fujikuroi P450-4 gene (CYP503) has been deposited in the GenBank/EMBL databases under accession number Y17243 (3,090 bp).
RESULTS
Isolation and expression of P450-4.
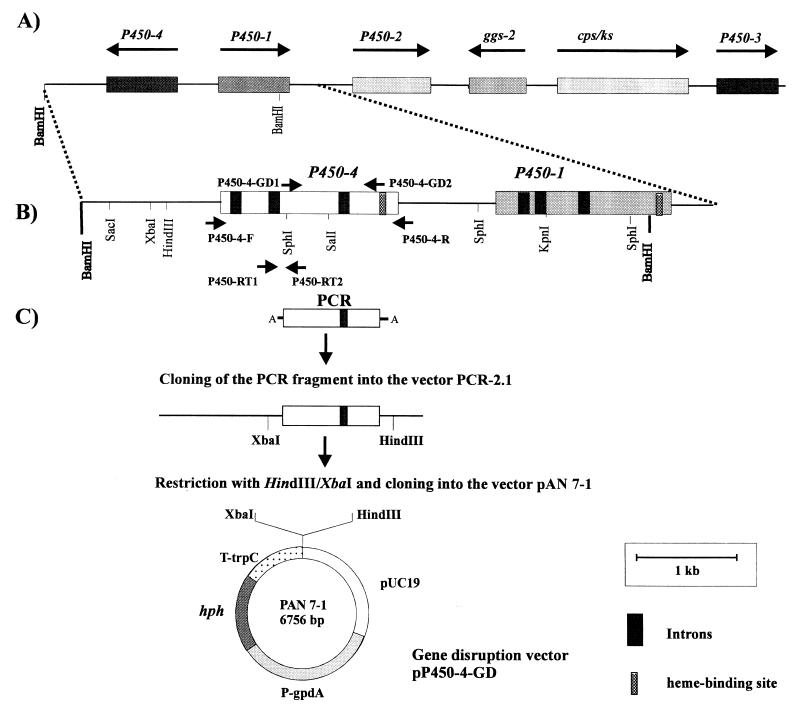
A 5.8-kb BamHI fragment containing part of P450-1 was cloned and shown to contain a 1,574-bp open reading frame transcribed in the opposite orientation to that of P450-1 (Fig. 2A and B). Analysis of sequence databases indicated that the gene encodes a cytochrome P450 monooxygenase and constitutes a novel P450 family, CYP 503. The genomic sequence of this novel cytochrome monooxygenase gene, P450-4, has three introns relative to the sequence of the corresponding cDNA clones from a cDNA library of G. fujikuroi m567.
FIG. 2.
The P450-4 gene of G. fujikuroi. (A) Position of the P450-4 gene in the GA biosynthesis gene cluster in G. fujikuroi. (B) Physical map of P450-4. The position of PCR primers is indicated. (C) Strategy for the construction of the disruption vector pP450-4-GD.
The deduced amino acid sequence of P450-4 shares only 33, 31, and 36% identity with P450-1, P450-2, and P450-3, respectively. An alignment between the most conserved regions, including the presumed heme-binding site of the four Gibberella P450s, the Arabidopsis thaliana ent-kaurene oxidase, and two other P450 monooxygenases with the highest degree of similarity to the Gibberella enzymes, an alkane-inducible oxygenase from Candida maltosa and human cholesterol 7α-hydroxylase, is shown in Fig. 3.
FIG. 3.
Partial sequence alignment of the highly conserved regions between the four G. fujikuroi P450 monooxygenases involved in GA biosynthesis and three other cytochrome P450 monooxygenases: G.f. P450-1 → G. fujikuroi P450-1 (accession number (AC): Y15277); G.f. P450-2 → G. fujikuroi P450-2 (AC: Y15278); G.f. P450-3 → G. fujikuroi P450-3 (AC: Y15279); G.f. P450-4 → G. fujikuroi P450-4 (AC: Y17243); A.t. GA3 → ent-kaurene oxidase of Arabidopsis thaliana (AC: AF047719); H.s. P450 → human cholesterol 7α-monooxygenase (AC: P22680); C.m. P450 → alkane-inducible cytochrome P450 monooxygenase of Candida maltosa (AC: JS0726). The presumed heme-binding site of the P450-type enzymes is F--G---C-G.
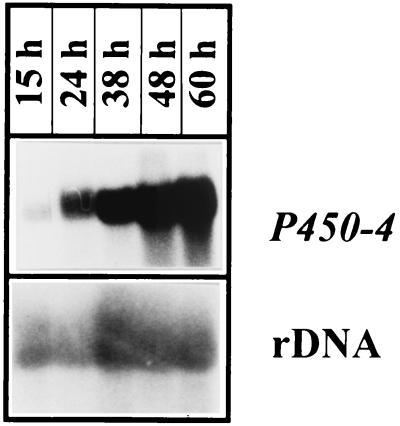
Transcription of P450-4 was investigated in mycelia grown for 15, 24, 38, 48, and 60 h in the optimized GA production medium (OPM). Northern blot analysis of total RNA revealed a single band of approximately 1.6 kb. The P450-4 transcript could be detected at 15 h and increased in abundance with culture time (Fig. 4). These results are similar to the expression pattern of the other five genes in the GA cluster (31).
FIG. 4.
Northern blot analysis of the G. fujikuroi P450-4 gene. The mycelium was grown for 15, 24, 38, 48, and 60 h in the GA production medium. As a control for RNA concentration, the blot was hybridized with the ribosomal DNA from G. fujikuroi.
Disruption of P450-4.
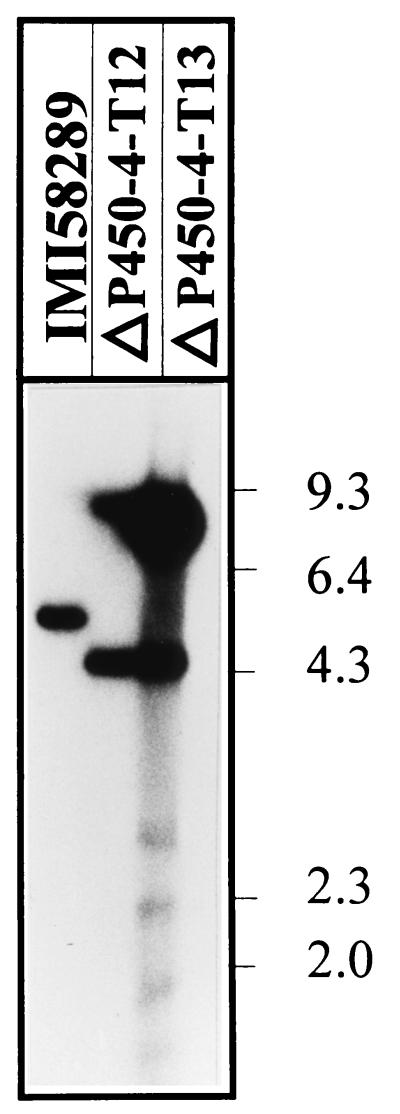
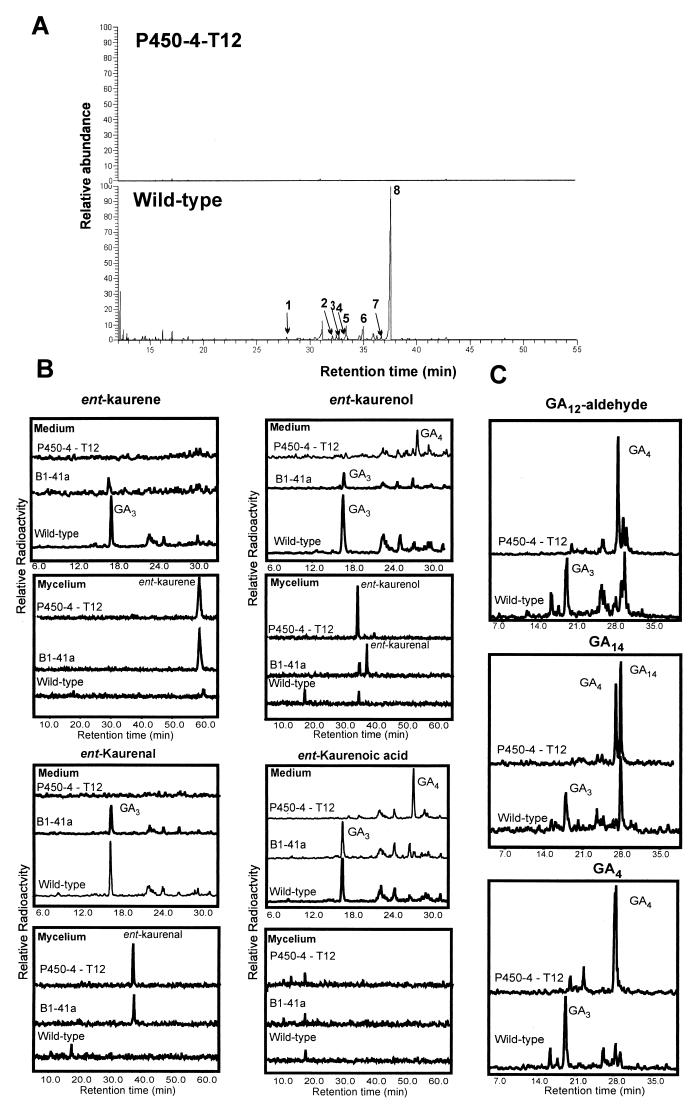
Following transformation with pP450-4-GD, we analyzed 35 hygromycin B-resistant colonies by Southern blot hybridization. Two of the transformants, T12 and T13, had lost the 5.4-kb XbaI wild-type band but produced two new hybridizing bands of 4.3 and 9.1 kb, predicted after integration of the 7.8-kb vector, pP450-4-GD, into the P450-4 locus (Fig. 5). Neither of the P450-4 mutants produced GAs. The culture medium from the wild type contained mainly GA3 and smaller amounts of other GAs and ent-kaurenoids, which were absent in the extract from the mutant (Fig. 6A). The mutant mycelia contained ent-kaurene as the only intermediate in the GA biosynthesis pathway (data not shown).
FIG. 5.
Southern blot analysis of the wild-type strain IMI 58289 and the disruption mutants ΔP450-4-T12 and T13. The genomic DNA was digested with XbaI and probed with the cDNA fragment of P450-4.
FIG. 6.
Characterization of the P450-4-disruption mutant T12. (A) Total ion current traces from GC-MS analysis of culture filtrates of the P450-4-T12 transformant and wild type (IMI 58289). Peaks: 1, GA9; 2, 7β-hydroxykaurenolide; 3, GA4; 4, fujenoic acid; 5, GA7; 6, GA13; 7, 7β, 18-dihydroxykaurenolide; 8, GA3. The traces are normalized to the largest peak in both traces. (B) HPLC radiochromatograms of extracts of mycelia and culture filtrates (medium) from incubations of ent-[14C]kaurene, ent-[14C]kaurenol, ent-[14C]kaurenal, and ent-[14C]kaurenoic acid with the wild-type (IMI 58289), P450-4-T12, and the mutant, B1-41a. (C) HPLC radiochromatograms of extracts of culture filtrates (medium) from incubations of 14C-labeled GA12− aldehyde, GA14, and GA4 with the wild type and P450-4-T12.
Characterization of the enzymatic function of P450-4.
After incubation of 14C-labeled ent-kaurene, ent-kaurenol, ent-kaurenal, and ent-kaurenoic acid with cultures of T12, only ent-kaurenoic acid was converted efficiently to GA4 (Fig. 6B). There was no conversion of ent-kaurene or ent-kaurenal; an apparent low-level formation of GA4 from ent-kaurenol was probably due to a small amount of ent-kaurenoic acid in this substrate. The wild-type strain converted each intermediate to GA3. The 14C-labeled intermediates GA12-aldehyde and GA14 were metabolized by ΔP450-4-T12 to GA4, and [14C]GA4 was not converted to GA3 by this strain (Fig. 6C). All three intermediates were incorporated into GA3 when incubated with the wild-type strain.
We also examined the conversion of the ent-kaurenoid intermediates in the GA-deficient mutant B1-41a. This mutant was obtained by UV irradiation of G. fujikuroi strain GF-1a and, on the basis of metabolism experiments, we concluded that GA biosynthesis was blocked at the step from ent-kaurenal to ent-kaurenoic acid (4). In incubations with B1-41a, ent-kaurenoic acid was converted efficiently to GA3 and ent-kaurenol and ent-kaurenal were converted much less efficiently to this product, whereas there was very little conversion of ent-kaurene (Fig. 6B).
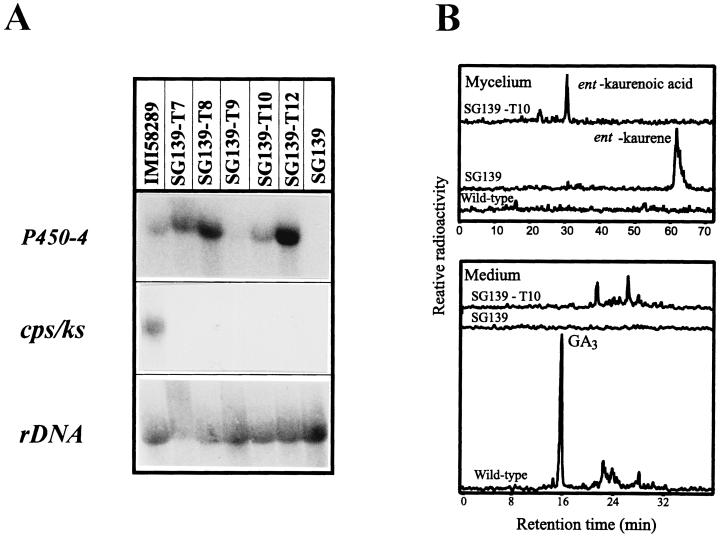
We transformed the deletion mutant SG139 with the gene complementation vector, pP450-4-GC, carrying the entire wild-type gene (Fig. 2B). From 24 analyzed transformants, 16 contained the expected 1,715-bp PCR fragment between primers P450-4-F and P450-4-R. Five of these 16 were used for Northern blot analysis to confirm that the gene was expressed in strain SG139 despite the loss of the GA biosynthesis gene cluster. As a control, the same filter was hybridized to cps/ks, one of the GA genes which is deleted in SG139 (Fig. 7A). As expected, neither SG139 nor the transformants contain transcript for cps/ks. Four of the transformants, SG139-T7, -T8, -T10, and -T12, expressed levels of P450-4 comparable to the wild type. One of these transformant strains, SG139-T10, was used for feeding experiments. Cultures of SG139-T10 converted ent-[14C4]kaurene to radiolabeled ent-kaurenoic acid, which was extracted from the mycelia, whereas SG139 did not metabolize this substrate (Fig. 7B). Under the same conditions, the corresponding wild-type strain converted ent-kaurene to GA3.
FIG. 7.
Characterization of SG139 transformed with the vector pP450-4-GC carrying the complete gene P450-4. (A) Northern blot analysis of SG139 and five complemented transformants. The blot was probed successively with the cDNA fragments of P450-4, cps/ks (control for the deletion), and the Gibberella 18S ribosomal DNA. (B) HPLC radiochromatograms of mycelial extracts from incubations of ent-[14C]kaurene with the wild type (IMI 58289), SG139, and SG139-transformant T10.
Complementation of the UV mutant B1-41a.
We transformed B1-41a with the vector pP450-4-GC. Fifty hygromycin-resistant transformants were analyzed for GA formation. Among the transformants, 38 produced wild-type levels of GA4, GA7, and GA3 as detected by thin-layer chromatography. Ten of them were analyzed quantitatively by HPLC. They produced between 250 and 350 mg of GA3/liter after a cultivation of 8 days in OPM. The wild-type strain produced 320 mg of GA3/liter under the same conditions. Therefore, the gene P450-4 fully complements the mutation in the mutant strain B1-41a.
Identification of the mutation site in the sequence of the P450-4 mutant allele.
A point mutation was found at position 663 of the B1-41a sequence. The last base pair of the 3′ splicing consensus sequence AG in intron 2 was replaced by A, presumably resulting in a severe reduction in gene function due to incorrect splicing.
Expression studies with B1-41a.
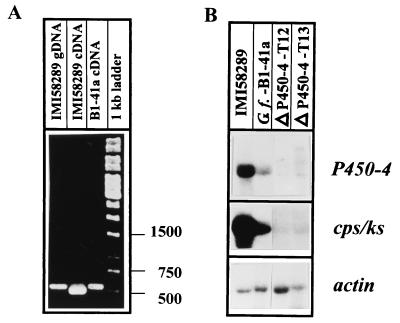
RT-PCR was used to confirm the sequencing results by comparing the P450-4 mRNA for the wild type and B1-41a. The primers are located on the left and the right sides of intron 2 and amplify 551 bp of genomic DNA (Fig. 2). The expected size of 499 bp was obtained from the wild-type cDNA, but the cDNA fragment from the mutant strain was larger (550 bp) than the wild-type genomic fragment, and only a very faint band of the wild-type cDNA was observed (Fig. 8A). The larger mutant cDNA fragment was cloned into the PCR cloning vector, and three independent clones were sequenced. The results confirmed the expected splicing defect, i.e., intron 2 was not removed from the B1-41a mRNA (data not shown).
FIG. 8.
Expression of the P450 gene in the wild-type and P450-4 mutant strains. (A) RT-PCR using primers P450-4-RT1 and P450-4-RT2 with the following templates: genomic DNA from wild-type (lane 1), cDNA from wild-type (lane 2), and cDNA from B1-41a (lane 3). Lane 4 contains a 1-kb ladder as marker. (B) Northern blot analysis using the P450-4 and the cps/ks cDNAs as probes on RNA from wild-type and mutant strains.
Northern analysis of the wild-type, B1-41a, and two P450-4 knock-out strains, T12 and T13, was carried out after cultivation in GA production medium for 4 days (Fig. 8B). The P450-4 gene is highly expressed in the wild type at this time, whereas only a low level of transcript was detected in B1-41a. In the transformants, ΔP450-4-T12 and T13, even less expression of the disrupted gene was observed, with transcripts from both mutants having sizes different from the wild type (Fig. 8B). The size difference of 52 bp between the wild type and B1-41a mRNA was too small to be detectable on this gel. The same filter also was probed with the cps/ks gene. Interestingly, the disruption of gene P450-4 apparently negatively influenced the expression of cps/ks. In B1-41a and in T12 and T13, significantly less cps/ks mRNA was observed than in the wild type, although the relative concentration of actin RNA was even higher in the mutant strains than in the wild type (Fig. 8B).
DISCUSSION
We cloned the P450-4 gene of G. fujikuroi by chromosome walking. The gene is located to the left of P450-1 in the recently-identified GA biosynthesis gene cluster (31) and is transcribed from right to left in the opposite direction to P450-1. The closest related cytochrome P450, the human cholesterol 7α-monooxygenase, as well as P450-1, P450-2, and P450-3 of G. fujikuroi, have less than 40% amino acid identity to P450-4; therefore, P450-4 defines a new cytochrome P450 family in accordance with the guidelines for P450 nomenclature (23).
Expression of the gene was high under conditions of GA production, whereas the transcript level is very low in the growth phase, similar to the expression pattern of the other genes of the pathway. Biochemical analysis of transformants with a disrupted P450-4 gene indicated that the gene is responsible for the oxidation of ent-kaurene, an early reaction in the pathway of GA biosynthesis. Incubations with radiolabeled precursors showed that ent-kaurenoic acid, but not earlier precursors, is efficiently metabolized by these transformants, suggesting that the enzyme catalyzes all three oxidative steps from ent-kaurene to ent-kaurenoic acid via ent-kaurenol and ent-kaurenal (Fig. 1). This function was confirmed by demonstrating that the SG139 mutant, which lacks the GA biosynthesis gene cluster and failed to metabolize ent-kaurene, converted ent-kaurene to ent-kaurenoic acid after transformation with the P450-4 gene. P450-4 is the second gene after P450-1 (27) that we have expressed in this deletion mutant to demonstrate the function of the encoded enzyme.
In plants, several GA-deficient dwarf mutants are defective in ent-kaurene oxidase activity. In peas, the mutant lh-2, affected in stem elongation and seed development, is deficient in all three steps from ent-kaurene to ent-kaurenoic acid, also suggesting that a single enzyme catalyzes these reactions (29). Furthermore, the product of the GA3 gene of Arabidopsis, which encodes ent-kaurene oxidase (15), also was shown to catalyze the conversion of ent-kaurene to ent-kaurenoic acid (14). Despite having the same functions in the GA biosynthesis pathway, the amino acid identity between P450-4 in G. fujikuroi and GA3 in Arabidopsis is only 26% over 235 amino acids. Recently, a barley gene and two homologues from Arabidopsis were shown to encode cytochrome P450s that catalyze the three steps of the GA biosynthesis pathway from ent-kaurenoic acid to GA12 (13). The function of these enzymes is, thus, similar to that of P450-1 (GA14 synthase) in G. fujikuroi.
Although ent-kaurenoic acid was metabolized efficiently by the P450-4-disrupted transformant ΔP450-4-T12, it was converted only to GA4, whereas the wild-type strain and mutant B1-41 a strain converted ent-kaurenoic acid to GA3. Incubations with later biosynthetic intermediates confirmed that GA4 was not further metabolized by the transformant. Thus, it appears that at least the gene encoding the enzyme that converts GA4 to GA7, a 1,2-desaturase, is not expressed in T12. We showed that the cps/ks (ent-kaurene synthase) gene is expressed less highly in B1-41a and, particularly, in the transformants ΔP450-4-T12 and T13 than in the wild type and may thus be under positive feedback control by a product of the pathway. The gene encoding the desaturase, and possibly also the gene encoding the 13-hydroxylase converting GA7 to GA3, may be under similar control. This effect may be due to the specific integration of the disruption vector into the P450-4 locus since all the other transformants with ectopic vector integrations showed a GA3 production similar to the wild type (data not shown). In contrast to T12, B1-41a, which has a point mutation in the same gene, metabolized ent-kaurenoic acid and GA4 to GA3. As discussed below, the mutation in B1-41a is slightly leaky, allowing small amounts of GAs to be produced and, therefore, feedback control of gene expression. An alternative explanation for the reduced expression of cps/ks and other genes in the gene disruption mutants is that the integration of a 7.7-kb DNA fragment of the circular disruption vector, pP450-4-GD, into the P450-4 locus may affect the expression of neighboring genes in the cluster. These possibilities can be investigated once these genes have also been characterized and transformed as single genes into the deletion mutant SG139.
The suggestion that P450-4 encodes ent-kaurene oxidase is supported by the successful complementation of the B1-41a mutant. In most cases, transformation with wild-type P450-4 restored the ability of the mutant, in which GA biosynthesis was reported to be blocked between ent-kaurenal and ent-kaurenoic acid (4), to produce the full spectrum of GAs, including GA4, GA7, and GA3. Although the DNA used for complementation contained most of P450-1 in addition to P450-4, B1-41 already contains a functional P450-1, which converts ent-kaurenoic acid to GA14 (27), as it metabolizes ent-kaurenoic acid normally. The complementation must therefore be due to P450-4. We also transformed the mutant B1-41a with P450-1, P450-2, and P450-3 and analyzed 50 transformants from each complementation, experiment (data not shown). None of these genes complemented the mutation in B1-41a, demonstrating the high substrate specificity of the cytochrome P450 monooxygenases in the GA biosynthesis pathway.
Sequence comparison between the wild-type and mutant copies of P450-4 revealed the mutation in B1-41a to be a single base substitution (G→A) at position 663, which removed the 3′ consensus sequence of intron 2 preventing, or substantially reducing, correct splicing. This may result in a premature translation stop or possibly splicing at a later 3′ acceptor splice site producing a truncated transcript. Indeed, some active enzyme is produced. Our metabolism experiments with B1-41a indicate that it is capable of low rates of conversion of ent-kaurenol and ent-kaurenal, although very little conversion of ent-kaurene was obtained. These results suggest that ent-kaurene oxidation is the rate-limiting reaction in the sequence and is most affected by reduced enzyme activity. The discrepancy between our results and those of Bearder et al. (4) is unexplained.
A similar point mutation to that in B1-41a was detected in the pea ent-copalyl diphosphate synthase (CPS) gene. The ls-1 mutation in this gene results from impaired splicing of the CPS mRNA. RT-PCR experiments revealed three ls-1 mRNAs: ls-1a mRNA from failure of the intron to be removed, ls-1b mRNA from moving the intron-exon boundary one nucleotide towards the 3′ end (ag/GA mutated to aaG/A), and ls-1c from using the next possible 3′ acceptor splice site (1).
Barrero et al. (3) analyzed the metabolism of GAs in several GA-deficient mutants. They suggested that SG139 was affected in a regulatory gene, as none of the intermediates were converted by this mutant. We demonstrated by Southern blotting, PCR, and Northern blotting that SG139 lacks the entire gene cluster, rendering this strain an excellent system for the expression of single GA pathway genes. Another GA-defective mutant, SG138, was blocked at all three oxidation steps between ent-kaurene and ent-kaurenoic acid but had additional blocks in C-3 and C-13 hydroxylation as well as in the loss of C-20. The authors suggested that the product of the mutated gene in SG138 controls more than one oxidation step in the GA pathway. The fact that SG138 could not convert ent-kaurenoic acid to GA3 confirmed that this mutant, in contrast to B1-41a and the knockout transformants ΔP450-4-T12 and T13, is blocked at a gene locus other than P450-4, possibly in a P450 reductase gene.
Recently, gene disruption experiments showed that, besides P450-1 and P450-4, the two other monooxygenase genes in the GA gene cluster, P450-2 and P450-3, also are involved in fungal GA biosynthesis, although their functions have not yet been defined (B. Tudzynski and P. Hedden, unpublished). The function of the genes ggs2 (31), cps/ks (31, 32), P450-4 (this paper), and P450-1 (27) have now been determined: they catalyze the steps from farnesyl diphosphate GGPP, from GGPP to ent-kaurene, from ent-kaurene to ent-kaurenoic acid, and from ent-kaurenoic acid to GA14 via GA12-aldehyde, respectively. Although in higher plants monooxygenases and dioxygenases are involved in GA biosynthesis, we have not been able to identify any dioxygenase genes in the GA biosynthesis gene cluster of G. fujikuroi. We are currently examining the function of P450-2 and P450-3 by expression of each gene in the deletion mutant SG139, the results of which should advance considerably our understanding of GA biosynthesis in G. fujikuroi.
ACKNOWLEDGMENTS
We thank K. Hölter, K. Topp, and J. Schulte for technical assistance, P. Linnemannstöns for HPLC analysis of GAs, R. Brown and M. C. Rojas for the preparation of [14C]GA9 and [14C]GA14, respectively, S. Thomas for recombinant GA 3β-hydroxylase, and B. Berns for typing the manuscript.
The project was funded by the DFG (Tu 101-3). IACR receives grant-aided support from the Biotechnology and Biological Sciences Research Council of the United Kingdom.
REFERENCES
- 1.Ait-Ali T, Swain S M, Reid J B, Sun T-P, Kamiya Y. The LS locus of pea encodes the gibberellin biosynthesis enzyme ent-kaurene synthase A. Plant J. 1997;11:443–454. doi: 10.1046/j.1365-313x.1997.11030443.x. [DOI] [PubMed] [Google Scholar]
- 2.Barendse G, van de Werken W M P H, Takahashi N. High-performance liquid chromatography of gibberellins. J Chromatogr. 1980;198:449–455. [Google Scholar]
- 3.Barrero A F, Oltra J R, Cabrera E, Reyes F, Álvarez M. Metabolism of gibberellins and ent-kaurenoids in mutants of Gibberella fujikuroi. Phytochemistry. 1999;50:1133–1140. [Google Scholar]
- 4.Bearder J R, MacMillan J, Wels M, Chaffey M B, Phinney B O. Position of the metabolic block for gibberellin biosynthesis in mutant B1–41a of Gibberella fujikuroi. Phytochemistry. 1974;13:911–917. [Google Scholar]
- 5.Benito E P, ten Have A, van't Klooster J W, van Kan J A L. Fungal and plant gene expression during synchronized infection of tomato leaves by Botrytis cinerea. Eur J Plant Pathol. 1998;104:207–220. [Google Scholar]
- 6.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desjardins A E, Gardner H W, Weltring K-M. Detoxification of sesquiterpene phytoalexins by Gibberella pulicaris (Fusarium sambucinum) and its importance for virulence on potato tubers. J Ind Microbiol. 1992;9:201–211. [Google Scholar]
- 8.Doyle J J, Doyle J L. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- 9.Geissman T A, Verbiscar A J, Phinney B O, Cragg G. Studies on the biosynthesis of gibberellins from (−)-kaurenoic acid in cultures of Gibberella fujikuroi. Phytochemistry. 1966;5:933–947. [Google Scholar]
- 10.Graebe J E, Hedden P, Gaskin P, MacMillan J. Biosynthesis of gibberellins A12, A15, A24, A36 and A37 in a cell-free system from Cucurbita maxima. Phytochemistry. 1974;13:1433–1440. [Google Scholar]
- 11.Hedden P. Recent advances in gibberellin biosynthesis. J Exp Bot. 1999;50:553–563. [Google Scholar]
- 12.Hedden P, Kamiya Y. Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- 13.Helliwell C A, Chandler P M, Poole A, Dennis E S, Peacock W J. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc Natl Acad Sci USA. 2001;98:2065–2070. doi: 10.1073/pnas.041588998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helliwell C A, Poole A, Peacock W J, Dennis E S. Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol. 1999;119:507–510. doi: 10.1104/pp.119.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helliwell C A, Sheldon C C, Olive M R, Walker A R, Zeevaart J A D, Peacock W J, Dennis E S. Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc Natl Acad Sci USA. 1998;95:9019–9029. doi: 10.1073/pnas.95.15.9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Homann V, Mende K, Arntz C, Ilardi V, Macino G, Morelli G, Böse G, Tudzynski B. The isoprenoid pathway: cloning and characterization of fungal FPPS genes. Curr Genet. 1996;30:232–239. doi: 10.1007/s002940050126. [DOI] [PubMed] [Google Scholar]
- 17.Kawaide H, Imai R T, Sassa R, Kamiya Y. ent-Kaurene synthase from the fungus Phaeosphaeria sp. L487. J Biol Chem. 1997;272:21706–21712. doi: 10.1074/jbc.272.35.21706. [DOI] [PubMed] [Google Scholar]
- 18.Linnemannstöns P, Voß T, Hedden P, Gaskin P, Tudzynski B. Deletions in the gibberellin biosynthesis gene cluster of Gibberella fujikuroi by REMI and conventional transformation-mediated mutagenesis. Appl Environ Microbiol. 1999;65:2558–2564. doi: 10.1128/aem.65.6.2558-2564.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacMillan J, Ward D A, Phillips A L, Sanchez-Beltran M J, Gaskin P, Lange T, Hedden P. Studies on the biosynthesis of gibberellins from (−)-kaurenoic acid in cultures of Gibberella fujikuroi. Plant Physiol. 1997;113:1369–1377. doi: 10.1104/pp.113.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacMillan J. Biosynthesis of the gibberellin plant hormones. Nat Prod Res. 1997;14:221–243. [Google Scholar]
- 21.Maniatis T, Sambrook J, Fritsch E F. Molecular cloning, a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Mende K, Homann V, Tudzynski B. The geranylgeranyl diphosphate synthase gene of Gibberella fujikuroi: isolation and expression. Mol Gen Genet. 1997;255:96–105. doi: 10.1007/s004380050477. [DOI] [PubMed] [Google Scholar]
- 23.Nebert D W, Nelson D R. P450 gene nomenclature based on evolution. Methods Enzymol. 1991;206:3–11. doi: 10.1016/0076-6879(91)06072-b. [DOI] [PubMed] [Google Scholar]
- 24.Pontecorvo G V, Roper J A, Hemmonns L M, Mac Donald K D, Buften A W J. The genetics of Aspergillus nidulans. Adv Genet. 1952;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 25.Punt P J, Oliver R P, Dingemanse M A, Pouwels P H, van den Hondel C A M J J. Transformation of Aspergillus nidulans based on the hygromycin B resistance marker from Escherichia coli. Gene. 1987;56:117–124. doi: 10.1016/0378-1119(87)90164-8. [DOI] [PubMed] [Google Scholar]
- 26.Rademacher W. Gibberellins. In: Anke T, editor. Fungal biotechnology. London, United Kingdom: Chapman and Hall; 1997. pp. 193–205. [Google Scholar]
- 27.Rojas M C, Hedden P, Gaskin P, Tudzynski B. The P450–1 gene of Gibberella fujikuroi encodes a multifunctional enzyme in gibberellin biosynthesis. Proc Natl Acad Sci USA. 2001;98:5828–5834. doi: 10.1073/pnas.091096298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 29.Swain S M, Ross J J, Reid J B, Kamiya Y. Gibberellins and pea seed development. Expression of the lh, IS and le mutations. Planta. 1995;195:426–433. [Google Scholar]
- 30.Tudzynski B. Fungal phytohormones in pathogenic and mutualistic associations. In: Carroll G C, Tudzynski P, editors. Plant relationships. The Mycota V-1997. Berlin, Germany: Springer-Verlag; 1997. pp. 167–184. [Google Scholar]
- 31.Tudzynski B, Hölter K. The gibberellin biosynthetic pathway in Gibberella fujikuroi: evidence for a gene cluster. Fungal Genet Biol. 1998;25:157–170. doi: 10.1006/fgbi.1998.1095. [DOI] [PubMed] [Google Scholar]
- 32.Tudzynski B, Kawaide H, Kamiya Y. Gibberellin biosynthesis in Gibberella fujikuroi: cloning and characterization of the copalyl diphosphate synthase gene. Curr Genet. 1998;34:234–240. doi: 10.1007/s002940050392. [DOI] [PubMed] [Google Scholar]
- 33.Tudzynski B, Mende K, Weltring K-M, Unkles S E, Kinghorn J R. The Gibberella fujikuroi niaD gene encoding nitrate reductase: isolation, sequence, homologous transformation and electrophoretic karyotype location. Microbiology. 1996;142:533–539. doi: 10.1099/13500872-142-3-533. [DOI] [PubMed] [Google Scholar]
- 34.Urrutia O, Rojas M C, Hedden P. Monooxygenases involved in GA12 and GA14 synthesis in Gibberella fujikuroi. Phytochemistry. 2000;56:505–511. doi: 10.1016/s0031-9422(00)00381-2. [DOI] [PubMed] [Google Scholar]
- 35.Williams J, Phillips A L, Gaskin P, Hedden P. Function and substrate specificity of the gibberellin 3 beta-hydroxylase encoded by the Arabidopsis GA4 gene. Plant Physiol. 1998;117:559–563. doi: 10.1104/pp.117.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woitek S, Unkles S E, Kinghorn J R, Tudzynski B. 3-Hydroxy-3-methylglutaryl-CoA reductase gene of Gibberella fujikuroi: isolation and characterization. Curr Genet. 1997;31:38–47. doi: 10.1007/s002940050174. [DOI] [PubMed] [Google Scholar]