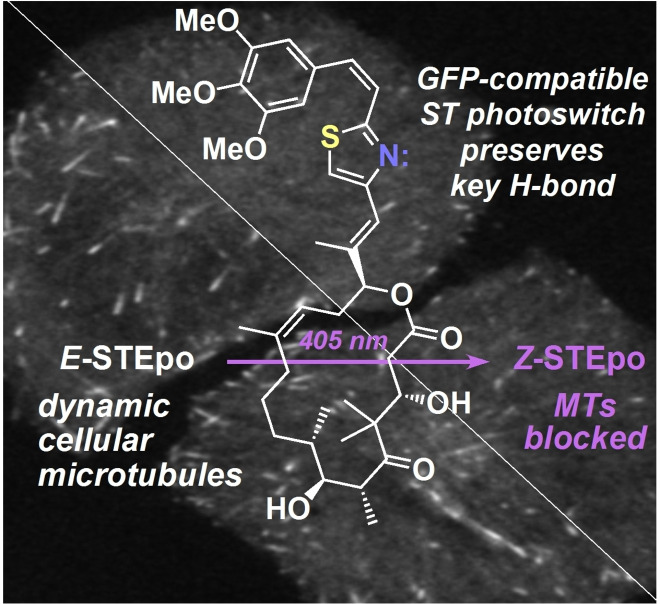
Abstract
Optical methods to modulate microtubule dynamics show promise for reaching the micron‐ and millisecond‐scale resolution needed to decrypt the roles of the cytoskeleton in biology. However, optical microtubule stabilisers are under‐developed. We introduce “STEpos” as GFP‐orthogonal, light‐responsive epothilone‐based microtubule stabilisers. They use a novel styrylthiazole photoswitch in a design to modulate hydrogen‐bonding and steric effects that control epothilone potency. STEpos photocontrol microtubule dynamics and cell division with micron‐ and second‐scale spatiotemporal precision. They substantially improve potency, solubility, and ease‐of‐use compared to previous optical microtubule stabilisers, and the structure‐photoswitching‐activity relationship insights in this work will guide future optimisations. The STEpo reagents can contribute greatly to high‐precision research in cytoskeleton biophysics, cargo transport, cell motility, cell division, development, and neuroscience.
Keywords: Bioorganic Chemistry, Microtubule Inhibitor, Natural Products, Photopharmacology, Photoswitch
A photoswitchable epothilone‐based microtubule stabiliser, STEpo, is introduced for high‐precision cytoskeleton photocontrol in live cells. STEpo features a novel styrylthiazole photoswitch that is isosteric to azobenzene, but is compatible with GFP imaging, and can photomodulate hydrogen‐bonding as well as sterics. STEpo photocontrols microtubule dynamics and cell division with micron‐ and second‐scale resolution, and has mid‐nanomolar potency.
Introduction
All cellular processes depend on spatiotemporal regulation of protein function. Tools to modulate protein function with the micrometre spatial precision and millisecond temporal precision inherent to these processes, are extremely valuable for precise biological research.[ 1 , 2 ] Cytoskeleton biology in particular can use such spatiotemporally‐specific tools for uniquely powerful studies of many classes of biological functions, since tight spatial regulation and temporally dynamic control over cytoskeletal proteins are critical for hundreds of distinct cellular processes.[ 3 , 4 ]
Microtubules (MTs) are giant, capillary‐like noncovalent polymers of tens of thousands of α/β‐tubulin heterodimers. [5] They are organised into a spatially structured network throughout the cell, that is rapidly remodelled through regulated growth and shrinkage to fulfil spatiotemporally distinct functions. These functions include acting as scaffolding for cell shape and for mechanical processes, as tracks for cargo transport by motor proteins, and as an organising system for cellular structures. This makes MT network structure and remodelling dynamics vital in diverse fields, including cell migration, cell division and development (e.g. by supporting the segregation of chromosomes), and neuroscience (e.g. supporting the formation and maintenance of extensions like axons and dendrites).[ 6 , 7 ]
Drugs that modulate MT stability and polymerisation dynamics are prime research tools for all these fields, being useful to nonspecifically suppress MT‐dependent cellular processes; several have also become widely‐used anticancer drugs.[ 8 , 9 ] Classical MT stabilisers (polymerisers) include taxanes and epothilones; while notable destabilisers (depolymerisers) include colchicine analogues, vinca alkaloids, auristatins/dolastatins, and maytansines (Figure 1a). While both MT destabilisers and MT stabilisers can suppress MT polymerisation dynamics in cell culture, their biological applications in vivo are distinct (e.g. stabilisers promoting axonal regeneration after spinal cord injury,[ 10 , 11 ] destabilisers suppressing inflammatory responses [12] ).
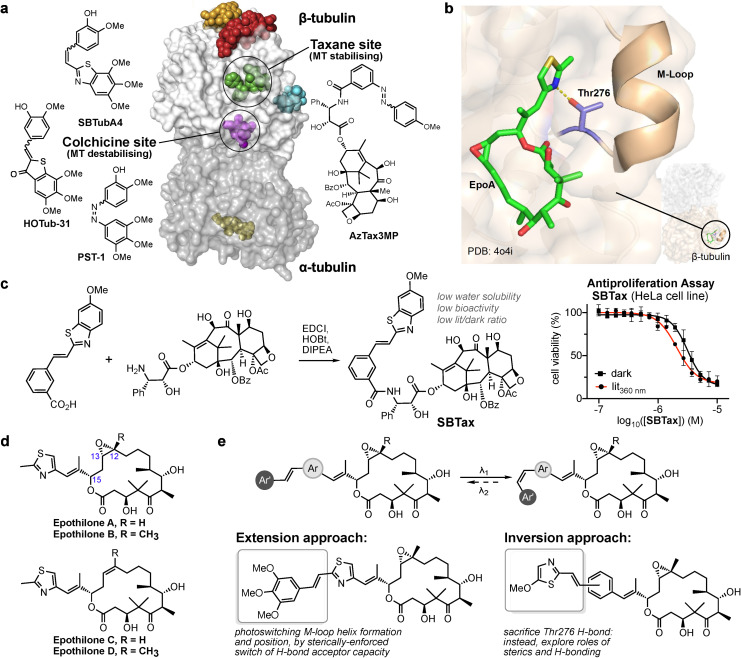
Figure 1.
Towards microtubule stabiliser photopharmacology. a) α/β‐Tubulin heterodimer with drug binding sites, [9] and major MT photopharmaceutical chemotypes. b) Epothilone binding to β‐tubulin induces the M‐loop to fold into a helix and repositions it to stabilise tubulin–tubulin lateral contacts, so stabilising the MT. A key interaction is the H‐bond from Thr276 to the thiazole nitrogen (adapted from PDB 4o4i [26] ). c) Synthesis and cellular evaluation of photoswitchable SBT‐derivatised taxane SBTax. d) Epothilones. The epoxide of EpoA/B is dispensable (EpoC/D). e) Designs for peripherally attaching the styrylthiazole (ST) photoswitch to epothilone: intending to photoswitchably induce and position the M‐loop helix, and/or change binding affinity, and/or reposition an H‐bond acceptor.
As research tools however, the lack of spatiotemporal precision with which these drugs can be applied has been a major hurdle for manipulation experiments that would ideally localise their activity on the scale of μm and ms. Spatiotemporally specific methods to modulate MT cytoskeleton stability and dynamics are still in their infancy. Photouncaged MT inhibitors including taxol and combretastatin A‐4 (CA4) were developed as more spatiotemporally precise research drugs. [13] However, they are not widely used, likely due to slow kinetics of the hydrolytic step in the photouncaging cascades, and photolability of the CA4 stilbene. [14] Two optogenetic tools to depress MT polymerisation under light are reported: Wittmann's photo‐inactivatable π‐EB1, [15] and Slep's photoactivated plus tip recruitment system; [16] and in recent weeks, two preprints of photocontrolled MT‐severing enzymes have also appeared,[ 17 , 18 ] highlighting broad interest in methods to photocontrol microtubule dynamics and network organisation.
In contrast, several photoswitchable analogues of taxane and colchicinoid MT‐inhibiting drugs, that offer cell‐specific spatial precision and second‐scale temporal precision of MT modulation without requiring genetic engineering, have been developed (Figure 1a). These photopharmaceuticals have practical advantages compared to photouncaging,[ 14 , 19 ] such as rapid photoresponse, absence of phototoxic byproducts, easier handling, and reduced potential for degradation (see Supporting Information Part B). The ability to apply these photopharmaceuticals across diverse model organisms even at early developmental stages, is also a strong conceptual advantage over genetic approaches.
Optically controlled MT stabilisers are a particularly attractive target for cell biology and early development research, since the biological functions of the MT cytoskeleton essentially depend on stabilised or growing MTs. However, only one family of photoswitchable MT stabilisers has been reported: the azobenzene‐based “AzTax” taxane analogues, which allowed spatiotemporally precise and time‐reversible control of MT dynamics, with a degree of subcellular control in neurons. [20] In contrast, a diversity of photoswitch scaffolds has been used in photoswitchable colchicinoid MT depolymerisers, from azobenzene (PST reagents),[ 3 , 21 ] to hemithioindigo (HOTub/HITub reagents),[ 22 , 23 ] spiropyran‐merocyanin, [24] and styrylbenzothiazole or “SBT” (SBTub reagents):[ 19 , 25 ] each with their own drawbacks and advantages (Figure 1a).
Noteworthily, the SBT photoswitch offers an attractive combination of advantages for biological research use:
The SBT scaffold is completely unaffected by standard GFP imaging at ca. 490 nm excitation, so biological assays can be imaged at high frame rates with common fluorescent proteins (GFP, YFP, mNeon) or markers (fluorescein, Atto 488) without switching the bioactivity of the photopharmaceutical. This is crucial for dynamic systems with fast spatially resolved readouts, as in cytoskeleton research, that require continuous imaging during photoswitching. [14] More generally, most biological models use GFP or similar fluorescent proteins, so GFP‐orthogonal photoswitches are needed to apply photopharmacology without changing the model (especially problematic for transgenic animals, that can take years to breed and validate). Though red‐fluorescent proteins excited at 561 nm are an alternative, GFP‐orthogonality crucially enables two‐channel multiplexed readouts (GFP+RFP) that deliver more powerful studies. [25] (While some other photoswitches such as blue‐shifted azobenzenes can have low absorption >450 nm, the SBT′s narrow π→π* absorption band systematically ensures this optical transparency. [19] )
The SBT's C=C π→π* band has a near‐ideal position for photoswitching at 405 nm, the typical photoactivation laser in microscopes ‐ again a crucial feature for practical impact. [14]
SBT is highly metabolically stable, and avoids biochemical photoswitch scission. This can otherwise be problematic for reagents aimed at cytosolic targets, that have to resist challenge by millimolar GSH and by thiol‐based reducing/GSH‐conjugating proteins. For example, the N=N bond of some azobenzenes can be cleaved even non‐enzymatically by monothiols, with ca. 100‐fold faster cleavage of the Z isomer that further complicates use and interpretation of results;[ 27 , 28 ] and, as photopharmaceuticals typically build the photoswitch onto the molecular periphery (as in the AzTax reagents), scission releases a fragment that is often more potent than the photopharmaceutical, further compromising assay readout (discussed in [19] ). Thus resistance to scission is a valuable feature for photoswitch scaffolds used in biology.
Taken together, these three advantages proved crucial in allowing SBT‐based reagents to succeed as photoswitches for cytosolic protein targets, even when applied globally to whole‐tissue explants and systemically to a variety of live animal models: settings that remain challenging for other photoswitches.[ 14 , 25 ]
A potential limitation of SBT is that while it is a reversible photoswitch, only majority E→Z SBT photoisomerisation occurs in the biocompatible spectral range, at least for the few SBTs studied so far. [25] However, this is only problematic in experiments where active bidirectional photoswitching, rather than diffusional reversibility, is strictly required. [14] As cytoskeleton research requires spatially precise E→Z isomerisation in a targeted cellular/subcellular zone, diffusion‐based repopulation of this photoisomerised zone with the E‐photopharmaceutical (∼s) is likely faster than biological response timescales (∼min): so even unidirectional photoswitching of SBT photopharmaceuticals has proven valuable for MT research, given their photostability, biochemical stability, and fast photoresponse.[ 19 , 25 ]
We were therefore motivated to create SBT‐based microtubule stabilisers, as photopharmaceuticals with improved practical applicability and scope. We now report the development of such GFP‐orthogonal reagents (Figure 1b).
Results and Discussion
Initial Approach: SBT‐Taxanes
We initially aimed at SBT‐taxanes, by a sidechain attachment design we previously used with azobenzene AzTax3MP: [20] deprotecting the sidechain 3′‐amine of docetaxel and coupling it to a SBT‐carboxylic acid to give SBT‐taxane SBTax (Figure 1c). However, SBTax had poor solubility, unsatisfactory antiproliferative potency in cells, and its bioactivity was not greatly photoisomer‐dependent (Figure 1c). This may simply reflect the taxanes′ weaknesses for photopharmaceutical adaptation, with no clear basis for rational introduction of isomer‐dependent potency, low solubility, high molecular weight, and chemical complexity that limits reasonable synthetic modifications.
Final Approach: Design of ST‐Epothilones (STEpos)
We next explored SBT‐epothilones as a basis for microtubule‐stabilising photopharmaceuticals. Epothilones are structurally simpler than taxanes, and despite having the same binding pocket (Figure 1b) they have higher binding affinity and potency.[ 29 , 30 ] Epothilones are particularly useful in research, since their greater solubility, bioavailability, and ability to cross the blood‐brain barrier, allow applications inaccessible to taxanes: such as systemic (rather than local) administration for stabilising axons, to aid in regenerating the central nervous system after injury. [31] The tractability of the epothilones has enabled extensive drug analogue campaigns (Figure 1d), clarifying structure‐activity relationship (SAR) features;[ 32 , 33 ] and late‐stage modifications that preserve binding activity while altering problematic groups are known; in our case, this would prove to be deoxygenating the C12/C13 epoxide, to give epothilone D derivatives. SAR suggests that large aryl substituents like an SBT would not be tolerated around C12/C13, [34] but could be tolerated in place of the thiazole (potent purine, quinoline and benzothiazole derivatives are all known), particularly by epothilone D derivatives. [32]
X‐ray studies enticingly reveal the thiazole's critical role in inducing the otherwise disordered M‐loop of β‐tubulin to fold into a short α‐helix (Figure 1b) that swings out to contact a laterally neighbouring tubulin in the microtubule, stabilising lateral contacts that stabilises the microtubule overall.[ 9 , 26 ] As well as steric factors, the hydrogen bond from Thr276 to the aryl nitrogen is critical for good helix induction [26] (≥20‐fold potency loss going from thiazole‐ or 2‐pyridine‐epothilones to 3‐ or 4‐pyridine analogues [35] ).
Replacing the thiazole with an SBT‐like switch (Figure 1d–e) therefore seemed attractive as a rational design basis, for several reasons (full discussion in Supporting Information Part A): i) Orienting a switch towards the M‐loop (Figure S1a) has the greatest chance to isomer‐dependently affect its helical folding and positioning. ii) An SBT‐like switch with an aryl nitrogen should also E/Z‐dependently alter its capacity to accept an H‐bond from Thr276, further influencing M‐loop‐stabilisation and thus biological potency (see e.g. Figure S1b–d). iii) Depending on its geometry, the switch can have an isomer‐dependent steric clash with the M‐loop that might additionally result in E/Z‐differences of binding affinity (e.g. Figure S1d vs Figure S1e, discussed below). iv) The epothilone scaffold's good water‐solubility might support a photoswitch, without the limitations seen with the taxanes.
To minimise size and insolubility of the photoswitch, we chose our recently developed styrylthiazole (ST) instead of SBTs. STs feature similar electronics and absorption spectra [25] as the GFP‐orthogonal, metabolically‐resistant SBTs [19] (Figure S2e); and are isosteric to azobenzenes. We explored two design strategies:
First, an “extension approach” kept the thiazole in place as a potential H‐bond acceptor, extending a styryl group to form the ST (later STEpo2; Figure 1e, Figure 2e). The Z‐ST should fit sterically while preserving the M‐loop Thr276 H‐bond (Figure S1b, c). However, for the E‐ST, the Thr276 H‐bond must be broken to prevent a steric clash with the M‐loop (Figure S1d, e). Thus we hoped that the extension design would be more bioactive as the Z isomer than the E, based on modulating helix induction rather than binding affinity. As elsewhere, we chose a 3,4,5‐trimethoxyphenyl group to extend with: we find that the out‐of‐plane central methoxy which reduces π‐stacking, combined with the hydrophilicity of the three OMe groups, increases water solubility of the photoswitch in both isomers, while the electron‐richness can helpfully redshift photoresponse wavelengths by 10–20 nm; [25] and the 3,4,5‐positions seem solvent‐exposed in the Z isomer so these substituents should be accomodated (Figure S1b,c).
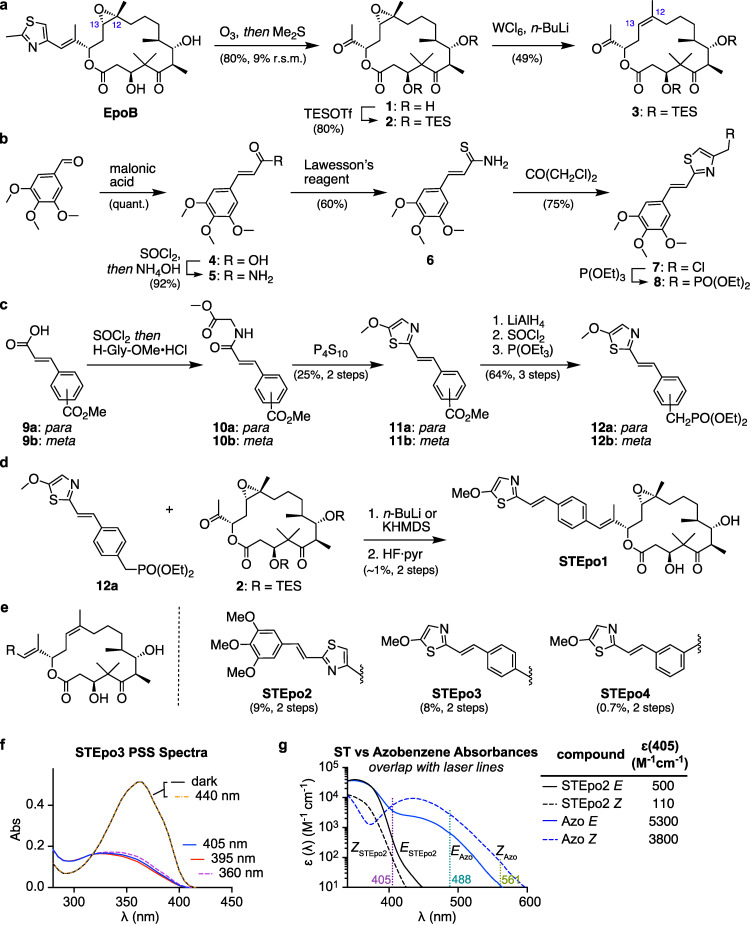
Figure 2.
Synthesis and photocharacterisation. a) Route to epothilone methyl ketones 2 and 3. b, c) Route to ST phosphonates 8, 12 a and 12 b. d) Epothilone B inversion design STEpo1. e) Epothilone D derivatives STEpo2 (extension) and STEpo3/STEpo4 (inversion). f) Photostationary state (PSS) absorption spectra of STEpo3 during irradiations starting from the dark state (all‐E) show no response to 440 nm light, then efficient isomerisation to mostly‐Z PSSs by 405, 395, then 360 nm, without degradation under illumination. g) Comparison of absorption spectra of E/Z‐ST and of a typical para‐methoxylated azobenzene, highlighting unique ST optical transparency at GFP (488 nm) or RFP (561 nm) imaging laser wavelengths, and sufficiently good absorption at the 405 nm “UV photoactivation” laser line for E→Z isomerisation. See Supporting Information for details.
Second, an “inversion approach” placed the phenyl as the near‐side ring of the ST, with thiazole as the far‐side (Figure 1e). This sacrifices the H‐bond to Thr276 in both E and Z isomers, but can reorient the ST photoswitch e.g. in meta or para at the phenyl ring.
We expected the para‐inversion compounds (later STEpo1 and STEpo3) to be more active as Z than E isomers (Figure 2e), as their E‐ST should clash sterically into the M‐loop (similarly as discussed for the extension approach), but the lesser steric pressure of the Z‐para‐inversion‐ST should permit better binding.
However, the meta‐inversion‐ST (later STEpo4, Figure 2e) highlighted new design features. In brief, while its ST orientation is similar to that in STEpo2 (expected: more active as H‐bonding Z than non‐H‐bonding E), in the inversion design neither E/Z‐ STEpo4 can make an H‐bond since the phenyl is not an acceptor (Figure S1f, g). On this understanding, we expected that E‐ STEpo2 and E‐ STEpo4 potencies would be identical (similar sterics, no H‐bond); but that the Z‐ STEpo2 potency (H‐bond) would be an order of magnitude greater [35] than that of E‐ STEpo4 (no H‐bond). This does not predict if STEpo4 would be more active as E or Z: but we reasoned that if experiments matched these expectations, it would suggest that this analysis of photoswitchability of bioactivity captures strongly relevant features that could later be optimised further (discussion in Supporting Information Part A).
While substituent effects on ST photochemistry are unknown, we expected that an electron‐rich methoxythiazole would red‐shift absorption: so completing the set of targets with STEpo1/3/4.
Synthesis of STEpos
Horner–Wadsworth–Emmons olefination of the epothilone methyl ketone 1 is a flexible route to attach aryl groups. [36] We used it to install styrylthiazole (ST) photoswitches. Starting from epothilone B, we cleaved the double bond by ozonolysis [37] then TES‐protected the free OH groups to give methyl ketone 2 (Figure 2a). ST phosphonates 8 and 12 a/b were synthesised by Arbuzov reaction from chlorides, accessed by ring closure of cinnamic acid derivatives (6 and 10 a/b, Figure 2b, c). Epoxide‐bearing epothilone B inversion design STEpo1 was initially synthesised after olefination of 2 with 12 a (Figure 2d). However, losses of material by epoxide opening during deprotection prompted us to deoxygenate the epoxide in future compounds, for a small bioactivity loss (epothilones C−D). [38] Deoxygenating 2 with WCl6 [39] gave methyl ketone 3 in good yields (Figure 2a); olefinations and deprotections then yielded epothilone D derivatives STEpo2 (extension) and STEpo3/4 (inversion) in milligram quantities (Figure 2e).
Photoisomerisation and GFP‐Orthogonality of STEpos
The STEpos’ photochemistry was characterised by UV/Vis spectroscopy. Like SBTs,[ 19 , 40 ] the STEpos were photoisomerised to majority‐Z photostationary states (PSSs) by near‐UV light (360–410 nm; ca. 85 % Z; Figure 2f, Figure S2a,b; Supporting Information). STEpos were robust to illumination >360 nm, though <340 nm caused slow degradation presumably by [2+2] cycloaddition. Pleasingly, STs are blueshifted by only ca. 10 nm compared to benzannulated SBTs (Figure S2d). The Z‐STs were thermally stable, with <1 % relaxation to E‐STs in solution at 37 °C in 8 h (Figure S2c).
Typically, the major E‐ST absorption band was centered around 355 nm; the 20 %‐of‐maximum intensity was at ca. 392 nm, above which a sharp cut‐off reduces ϵ by a factor of ten every 12 nm. The major Z‐ST band was centered around 325 nm, with 20 %‐of‐maximum intensity at ca. 382 nm, and cut‐off above this of a factor of ten every 16 nm (Figure 2g, S2e). These sharp cut‐offs are crucial. They make the ST's absorption effectively zero above 440 nm, so that ST is entirely unaffected by GFP imaging either with lasers (488 nm) or with filtered sources (typically 490±15 nm). This contrasts strongly to azobenzene photoswitches, particularly those that benefit from polymethoxylation, which are significantly impacted by imaging of GFP, YFP, and even RFP (Figure S2e). [19]
STEpos Give Photoisomer‐Dependent Cellular Bioactivity
We first evaluated the STEpos for photoisomer‐dependent antiproliferative activity in cells, which MT inhibitors cause by blocking mitosis. We treated HeLa cervical cancer cells with E‐STEpos and incubated them either in the dark (for all‐E), or while lit by non‐phototoxic pulsed illuminations from low‐power 360 nm LEDs (<1 mW cm−2; <1 % duty cycle; in situ photoswitching to mostly‐Z),[ 3 , 19 ] and assessed cell viability 44 h later. All compounds had antiproliferative activity in the submicromolar range, and all had reproducibly light‐dependent bioactivity with up to ca. 4‐fold potency shift upon isomerisation (Figure 3a).
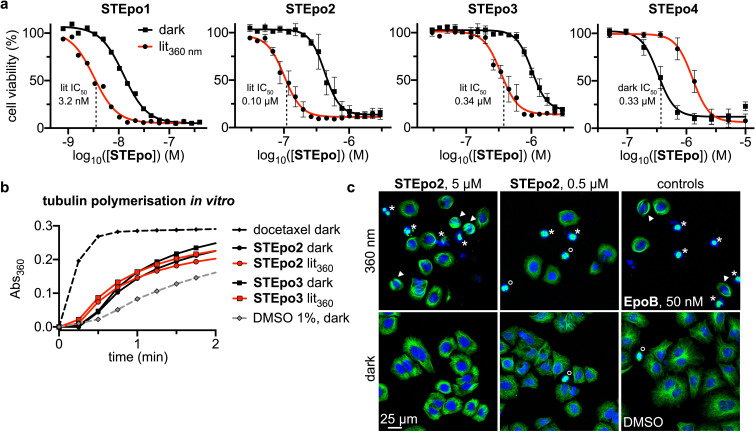
Figure 3.
Cellular bioactivity and mechanism. a) STEpos have potent light‐dependent antiproliferative activity (HeLa cells, 44 h incubation; all‐E “dark” and mostly‐Z “lit” conditions; STEpo1: one representative of three independent experiments shown; STEpo2‐4: three replicates, mean with SD). b) STEpo2/3 promote bulk tubulin polymerisation in cell‐free assay; earlier onset for Z‐ STEpo is visible (turbidimetric in vitro assay; greater absorbance corresponds to more polymerisation; time to onset of polymerisation should be noted; 1 μM STEpo; 10 μM docetaxel). c) Fluorescence imaging of cells treated with STEpo2 under 360 nm pulsing (mostly‐Z) reveals disorganised MT networks, with spindle defects (asterisks), mitotic arrests (hollow circles) and bundled MTs (arrowheads) similarly to Epothilone B; but nearly no disorganisation in the dark (all‐E) similarly to cosolvent control. (HeLa cells, 20 h incubation; α‐tubulin in green, DNA stained with DAPI in blue).
Matching expectations, Z‐extension STEpo2 (predicted: H‐bond allowed) was more potent than any other epothilone D (E‐ STEpo2, E/Z‐ STEpo3, E/Z‐ STEpo4: predicted: no H‐bond). Also as anticipated, extension STEpo2 and para‐inversion STEpo1 and STEpo3 were all more active as the Z isomers, with good photoswitchability of bioactivity seen; the epothilone B analogue STEpo1 (IC50(lit) ca. 3 nM, IC50(dark) ca. 12 nM) was 100‐fold more potent than its epothilone D counterpart STEpo3, showing the influence of the scaffold in determining overall potency. Interestingly, meta‐inversion STEpo4 was more active as the E isomer: and excellently matching predictions, E‐STEpo4 was indeed equipotent to E‐STEpo2 (IC50(dark) ca. 400 nM); while Z‐STEpo4 was indeed ≥10‐fold less potent than Z‐STEpo2 (1300 nM vs 100 nM). These matches to predictions supports that both H‐bonding and steric clash interactions with the M‐loop are indeed the basis for the photoisomer‐dependency of activity in the STEpos: which should be amenable to further improvements. However, as we have been unable to obtain STEpo:tubulin crystal structures, further experimental analysis is an ongoing topic of our research. (Note: the E‐activity of STEpo4 confirms there is no confounding phototoxicity from the ST photoswitch, since it would otherwise give consistently lit‐toxic effects for all compounds.)
STEpo2 offered an identical photoswitchability of bioactivity (ratio of apparent cellular activities under lit and dark conditions) as STEpo1, but had better synthetic access. It was also more potent than STEpo3; and as a lit‐active photopharmaceutical, has practical advantages for biological use over dark‐active STEpo4. Therefore we continued with STEpo2 as our main compound.
Mechanism of Action of STEpo
We first tested direct interaction with tubulin as the STEpos’ molecular mechanism of bioactivity, by cell‐free bulk polymerisation of purified tubulin (compare to local polymerisation below) with cellularly “lit‐active” STEpo2 and STEpo3, checking whether polymerisation would be enhanced more by the Z than the E isomers. Both isomers were polymerisation enhancers, indicating direct tubulin binding; and the time to polymerisation onset was shorter with mostly‐Z pre‐lit stocks than with all‐E stocks, matching their cellular pattern of “lit‐activity” (Figure 3b).
Next, to examine the cellular mechanism of their isomer‐dependent bioactivity, we imaged the MT network architecture in HeLa cells incubated with STEpo2. Under lit conditions (mostly Z‐isomer), low concentrations of STEpo2 disorganised the straight radial MT networks in cells, producing significant mitotic arrests (Figure 3c: hollow circles) and defective mitotic spindles (asterisks). At higher concentrations, lit‐STEpo caused extreme bundling of stabilised MTs (Figure 3c: select bundles indicated with arrowheads) and catastrophic spindle failures (asterisks). These are hallmarks of MT stabilisers, as seen for Epothilone B (Figure 3c). However, all‐E STEpo2 (dark) at low concentrations had no more impact on MT network architecture or mitotic figures than cosolvent control, and only slight indications of stabilised bundles at high concentrations (Figure 3c). These results support that STEpo2 isomer‐dependent cytotoxicity arises from its Z‐isomer more potently inhibiting MT dynamics and stability in cells.
High‐Precision Cell‐free MT Photocontrol with STEpo2
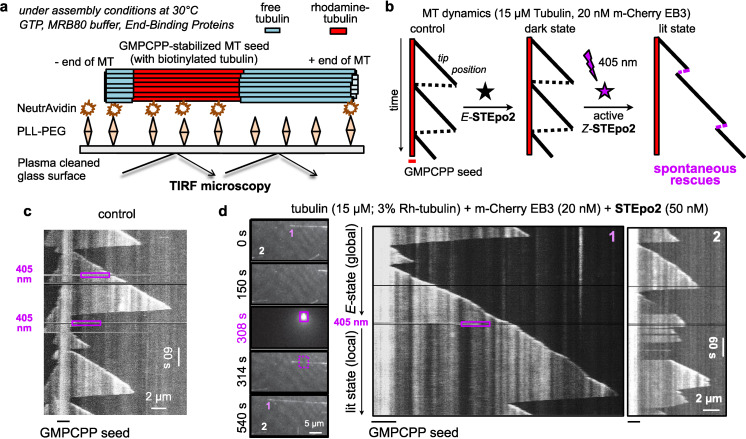
We next tested in situ photocontrol over MT polymerisation in cell‐free settings, with temporal resolution to the scale of seconds and spatial resolution to the μm scale allowing optical targeting of individual microtubules. We used TIRF microscopy to image a reconstituted microtubule polymerisation system. This uses the non‐hydrolysable GTP analogue GMPCPP to form stable microtubule “seeds” that are fixed to a glass surface for spatially localised imaging. Free tubulin is then applied, together with a test drug and with a fluorescently labelled MT plus end‐tracking protein, EB3 (by marking the GTP cap region of MTs, EB3 tracks the plus end tips of polymerising MTs, making them appear as “comets”; [41] thus EB3 imaging gives a spatiotemporally‐resolved readout for MT dynamics during photoswitching). [19] Finally, GTP is supplied and the seeds initiate normal cycles of growth and shrinkage using the free tubulin in the mixture.[ 42 , 43 ] Using the labelled seed of each MT as a static reference point, kymographs tracking the growing and shrinking tips of the MT reveal the frequency of shrinkage events as the “spikes” in the kymograph. Typically, MTs depolymerise back to the seed before restarting polymerisation. MT‐stabilising drugs decrease the frequency of shrinkage events and can restart polymerisation along the MT before the seed is reached (“rescue”). If a drug stabilises a different protofilament number of MT than the seeds have (14 for GMPCPP seeds), stable rescue sites will be observed, while if the drug stabilizes the protofilament number matching the seed, rescues will occur stochastically at different sites along the microtubule, offering mechanistic insights into the nature of MT stabilisation. [42] Epothilones are 14‐protofilament MT stabilisers, so we expected that if our STEpos retained the same MT‐stabilising properties with Z‐specific potency, they would permit MT rescues after illuminations that we aimed to apply with spatial specificity to selected MT tips (Figure 4b).
Figure 4.
Spatiotemporal control over MT dynamics in cell‐free systems by photocontrolling STEpo2. a, b) Schematic of in vitro MT dynamics imaging assay, and of kymographs with normal MT growth and catastrophes vs. patterns expected for STEpo2 as a 405 nm‐dependent 14‐protofilament MT stabiliser. c) No‐drug control kymograph showing MT shrinkage frequency and polymerisation restarts at the GMPCPP seed (405 nm illuminations in the purple dotted boxes). d) Time‐lapse images with two microtubules in the field of view (at left, 1 and 2), and their kymographs (at right). 405 nm illumination targeting microtubule 1 at 308 s within the purple box region of time‐lapse panel/kymograph (0.01 % of field of view) suppresses its catastrophes and creates a rescue zone far away from the seed.
In controls, shrinkage proceeded completely to seeds before polymerisation restarted (as expected), with no influence of 405 nm (Figure 4c, Figure S3a). E‐STEpo2 gave MT shrinkage events similar to controls, reaching seeds before restart (first 5 min, Figure 4d), although raised concentrations predictably caused MT stabilisation (Figure S3b). However, local 405 nm illumination near a single microtubule (“1′′ in Figure 4d) stabilised it, increasing its length and allowing rescue along the MT. Other MTs further from the illumination (”2“) had similar shrinkage frequency as before, though with some spontaneous rescues (next 5 min, Figure 4d; Movie S1; see too Figure S3c). These stringent assays confirm that STEpo2 is an epothilone‐like, Z‐isomer‐specific stabiliser of 14‐protofilament MTs, which can be spatiotemporally targeted in cell‐free settings with micron‐and‐second scale precision.
High‐Precision Live Cell MT Photocontrol with STEpo2
Finally, to test the STEpos’ capacity for in situ photocontrol of cellular MT dynamics with spatiotemporal precision, we imaged STEpo2‐treated cells transfected with fluorescently labelled EB3.
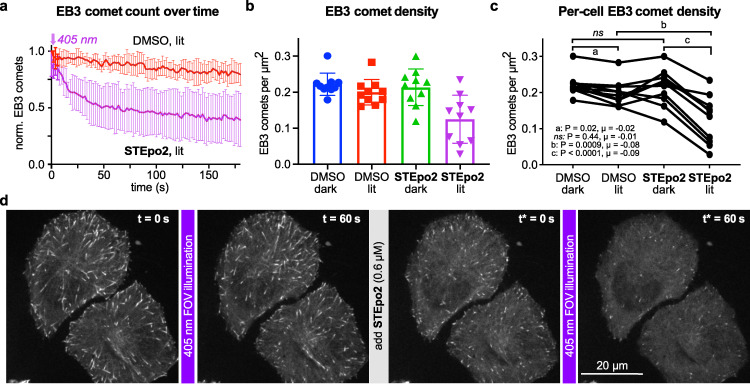
We imaged cells longitudinally during an internally controlled series of drug treatment and illumination phases, with continuous 487 nm imaging of EB3‐GFP. First, to establish baseline MT dynamics, cells were imaged for 60 s with cosolvent (“DMSO dark” phase); then applying 3 min of low intensity 405 nm pulses (“DMSO lit” phase), which insignificantly reduced comet counts, indicating negligible phototoxicity or photobleaching (Figure 5a, Movie S2). Next, E‐STEpo2 was applied at 0.6 μM, causing no noticeable change of EB3 comets (Movie S3), followed by another phase of 405 nm pulsing: which now rapidly suppressed MT dynamics (EB3 comet count halved within seconds; P<0.0001; Figure 5; Movie S2). These experiments show that in situ STEpo2 photoswitching is effective for temporally‐precise, noninvasive, optically‐actuated control of MT dynamics in live cells, in a manner that is fully orthogonal to GFP imaging.
Figure 5.
Spatiotemporal control over cellular MT dynamics with STEpo2. a–d) Live cell EB3‐GFP “comets” during GFP imaging with 488 nm (HeLa cells). MT inhibition with STEpo2 is initiated upon 405 nm illumination (cells first treated with 1 % DMSO, then imaged for 60 s for baseline, then photoactivated from time t=0 with 405 nm during imaging over 3 min; then an additional 0.6 μM E‐STEpo2 was applied, and 405 nm/imaging applied from time t*=0. 10 cells acquired). a) Mean±SD comet count (each cell normalised to t=0 of the DMSO control). b, c) Comet density statistics show large and significant differences upon STEpo2 photoactivation (dark at t=0 or t*=0, lit at t=60 s or t*=60 s; b shows pooled data, c shows longitudinal traces per cell; P‐values and μ mean differences as annotated refer to differences between treatments, pooling all cells). d) Stills from representative movie (data related to Movie S2).
Conclusion
Methods to modulate biological processes with the spatiotemporal resolution appropriate to their endogenous activity are becoming increasingly effective. Photopharmacology is rapidly progressing with tools for high‐precision, non‐invasive biological control in fast‐response and/or spatially‐localised applications from neuroscience[ 44 , 45 ] to cytoskeleton research.[ 3 , 4 ] As the optical instrumentation of biological research becomes more powerful, the value of optically‐controlled tools that can be optimally operated with that instrumentation will continue to rise: particularly for inherently spatiotemporally‐precise processes, such as those the microtubule cytoskeleton supports. [46]
We report STEpos, photopharmaceuticals based on the epothilone MT stabiliser scaffold. Their mid‐nanomolar potencies, GFP‐compatibility, and robust photoswitchability of bioactivity are promising for cell‐free or cell biology research. These can be favourably compared to the micromolar potencies and variable photoswitchability of bioactivity found here and previously [20] for photoswitch‐derivatised taxanes. STEpos should easily find uses through to in vivo cytoskeleton experiments relying on GFP‐only or multiplexed imaging in near‐surface settings, across fields from development and motility to transport and cell division, and may help answer unresolved questions about the mechanisms of MT‐dependent pathologies whose elucidation has so far been stymied by poor spatiotemporal precision of MT stabilisation in vivo in long term assays.[ 10 , 11 , 20 , 31 ]
By exploring uncommonly large and conformationally restricted epothilone derivatives, we also reveal aspects of SAR that will orient future studies to harness the steric and hydrogen‐bonding aspects that likely determine isomer‐dependent STEpo potency and binding affinity. These in turn can inspire the rational design of orders‐of‐magnitude more potent epothilone‐based reagents with still greater photoswitchability of bioactivity.
The rational design for photoswitch incorporation and predictive analysis of isomer‐ and photoswitch‐dependent biological potency are encouraging for photopharmacology more generally. Our results also suggest that it is possible not just to harness hydrogen‐bonding for binding affinity, but to exploit it without greatly changing sterics to invert the overall “sign” of photopharmaceutical response (Z‐active STEpo2, E‐active STEpo4), conveniently by flipping the orientation of an inherently H‐bond‐accepting photoswitch. Exploiting isomer‐dependent hydrogen‐bonding is, as far as we are aware, a novel principle for photopharmacology; but the degree of control that it exerts here is likely to be reproducible on a range of protein targets.
We also feel that designing towards isomer‐dependent effects on a partially disordered structural element (here, the M‐loop) is an exciting general principle for photopharmacology: which hitherto has focused mostly on rigid steric clashes that may be unrealistic to identify and photocontrol. Interfering with partially disordered elements is likely to give greater sensitivity to small structural changes, even in cases where it is not clear how peripheral photoswitching should change binding affinity.
This is also the first use of the compact styrylthiazole (ST) as a photoswitch scaffold for biology. ST is isosteric to azobenzene, yet offers significant practical benefits. We highlight its GFP‐orthogonality, maximising free imaging channels and favouring use with GFP‐expressing transgenics; and its intriguing and biologically relevant potential as an H‐bond acceptor, which matched the requirements of the epothilone system. We feel that GFP‐orthogonality [19] is still an under‐appreciated goal among chemical reagent developers. NIR‐capable photoswitches for deep tissue photoisomerisation towards translational research uses are currently prioritised, even though deep tissue isomerisation faces significant spatial scattering of illumination (seen by shining a red laser pointer into a finger) that may sacrifice the spatial precision that is one of photopharmacology's greatest advantages. However, a design shift is underway, driven by biologists using microscopy for high‐precision fundamental research, and emphasising biochemically stable, imaging‐orthogonal photoswitch scaffolds that can be used in tissue slice and embryo/early animal research across a variety of models. [25] This shift has great potential to serve the biological community with a palette of rapid‐response, non‐phototoxic, byproduct‐free reagents for studies in diverse fields. To continue this progress relies on expanding our scope of photopharmaceutical design principles and of photoswitch scaffolds.
Taken together, the STEpos bring promising advances not only for high‐precision microtubule biology, but also towards the general refinement of photopharmacology for high‐performance use against other protein targets.
There are several challenges for the ongoing development of STEpos. Topics in our focus include: i) Better synthetic access to epoxide‐bearing derivatives, to harness their greater potency. ii) Improving the speed of photoactivation with the 405 nm laser line by slightly shifting ST absorption (just a 15 nm shift would give a 10‐fold rate enhancement due to the steep absorption cut‐off). We have achieved this elsewhere with increasing electron donation (e.g. para‐Me2NPh‐ instead of Ph‐ for the extension design); [25] for the inverted design, further photochemical exploration of the ST scaffold will be needed. iii) To achieve bidirectional photoswitching with ST‐like GFP‐orthogonality. This requires increasing the separation between the absorption bands of the E and Z isomers, which is an object of ongoing study.
Conflict of interest
The authors declare no conflict of interest.
1.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Acknowledgements
We thank Jan Huebner (Bayer) for the gift of EpoB, Dirk Trauner (NYU) for collegial discussions and for securing the EpoB, Monique Preusse for early cell viability testing, Rebekkah Hammar for performing the tubulin polymerisation assay, Maximilian Wranik and Michel O. Steinmetz (PSI Villigen) for attempting crystallisation, and Martin Reynders for the Schrödinger visualisation models in Figure S1. We thank Reviewer 2 for suggesting the expression optical transparency. We are grateful to Henrietta Lacks, now deceased, and to her surviving family members for their contributions to biomedical research. O.T.‐S. thanks the German Research Foundation (DFG: Emmy Noether grant number 400324123; SFB 1032 project B09 number 201269156; SFB TRR 152 project P24 number 239283807; SPP 1926 project number 426018126) for funding support. J.C.M.M. acknowledges support from an EMBO Long Term Fellowship. J.T.‐S. thanks the Joachim Herz Stiftung for a Translational Research Fellowship. Open Access funding enabled and organized by Projekt DEAL.
L. Gao, J. C. M. Meiring, C. Heise, A. Rai, A. Müller-Deku, A. Akhmanova, J. Thorn-Seshold, O. Thorn-Seshold, Angew. Chem. Int. Ed. 2022, 61, e202114614; Angew. Chem. 2022, 134, e202114614.
A previous version of this manuscript has been deposited on a preprint server (https://doi.org/10.1101/2021.03.31.437838).
Contributor Information
Prof. Dr. Anna Akhmanova, Email: a.akhmanova@uu.nl.
Dr. Oliver Thorn‐Seshold, Email: oliver.thorn-seshold@cup.lmu.de.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Goglia A. G., Toettcher J. E., Curr. Opin. Chem. Biol. 2019, 48, 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hoorens M. W. H., Szymanski W., Trends Biochem. Sci. 2018, 43, 567–575. [DOI] [PubMed] [Google Scholar]
- 3. Borowiak M., Nahaboo W., Reynders M., Nekolla K., Jalinot P., Hasserodt J., Rehberg M., Delattre M., Zahler S., Vollmar A., Trauner D., Thorn-Seshold O., Cell 2015, 162, 403–411. [DOI] [PubMed] [Google Scholar]
- 4. Borowiak M., Küllmer F., Gegenfurtner F., Peil S., Nasufovic V., Zahler S., Thorn-Seshold O., Trauner D., Arndt H.-D., J. Am. Chem. Soc. 2020, 142, 9240–9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Akhmanova A., Steinmetz M. O., Nat. Rev. Mol. Cell Biol. 2008, 9, 309–322. [DOI] [PubMed] [Google Scholar]
- 6. Glotzer M., Nat. Rev. Mol. Cell Biol. 2009, 10, 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kapitein L. C., Hoogenraad C. C., Neuron 2015, 87, 492–506. [DOI] [PubMed] [Google Scholar]
- 8. Peterson J. R., Mitchison T. J., Chem. Biol. 2002, 9, 1275–1285. [DOI] [PubMed] [Google Scholar]
- 9. Steinmetz M. O., Prota A. E., Trends Cell Biol. 2018, 28, 776–792. [DOI] [PubMed] [Google Scholar]
- 10. Hellal F., Hurtado A., Ruschel J., Flynn K. C., Laskowski C. J., Umlauf M., Kapitein L. C., Strikis D., Lemmon V., Bixby J., Hoogenraad C. C., Bradke F., Science 2011, 331, 928–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sengottuvel V., Leibinger M., Pfreimer M., Andreadaki A., Fischer D., J. Neurosci. 2011, 31, 2688–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weng J.-H., Koch P. D., Luan H. H., Tu H.-C., Shimada K., Ngan I., Ventura R., Jiang R., Mitchison T. J., Nat. Metab. 2021, 3, 513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wühr M., Tan E. S., Parker S. K., Detrich H. W., Mitchison T. J., Curr. Biol. 2010, 20, 2040–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O. Thorn-Seshold, J. Meiring, ChemRxiv 2021, 10.26434/chemrxiv.14424176.v1. [DOI]
- 15. van Haren J., Charafeddine R. A., Ettinger A., Wang H., Hahn K. M., Wittmann T., Nat. Cell Biol. 2018, 20, 252–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Adikes R. C., Hallett R. A., Saway B. F., Kuhlman B., Slep K. C., J. Cell Biol. 2018, 217, 779–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.J. C. M. Meiring, I. Grigoriev, W. Nijenhuis, L. C. Kapitein, A. Akhmanova, bioRxiv 2021, https://www.biorxiv.org/content/10.1101/2021.12.22.473806v1. [DOI] [PubMed]
- 18.G. Y. Liu, S.-C. Chen, K. Shaiv, S.-R. Hong, W.-T. Yang, S.-H. Huang, Y.-C. Chang, H. Cheng, Y.-C. Lin, bioRxiv 2021, 10.1101/2021.10.08.463668. [DOI]
- 19. Gao L., Meiring J. C. M., Kraus Y., Wranik M., Weinert T., Pritzl S. D., Bingham R., Ntouliou E., Jansen K. I., Olieric N., Standfuss J., Kapitein L. C., Lohmüller T., Ahlfeld J., Akhmanova A., Steinmetz M. O., Thorn-Seshold O., Cell Chem. Biol. 2021, 28, 228–241, e6. [DOI] [PubMed] [Google Scholar]
- 20. Müller-Deku A., Meiring J. C. M., Loy K., Kraus Y., Heise C., Bingham R., Jansen K. I., Qu X., Bartolini F., Kapitein L. C., Akhmanova A., Ahlfeld J., Trauner D., Thorn-Seshold O., Nat. Commun. 2020, 11, 4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Engdahl A. J., Torres E. A., Lock S. E., Engdahl T. B., Mertz P. S., Streu C. N., Org. Lett. 2015, 17, 4546–4549. [DOI] [PubMed] [Google Scholar]
- 22. Sailer A., Ermer F., Kraus Y., Lutter F. H., Donau C., Bremerich M., Ahlfeld J., Thorn-Seshold O., ChemBioChem 2019, 20, 1305–1314. [DOI] [PubMed] [Google Scholar]
- 23. Sailer A., Ermer F., Kraus Y., Bingham R., Lutter F. H., Ahlfeld J., Thorn-Seshold O., Beilstein J. Org. Chem. 2020, 16, 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rastogi S. K., Zhao Z., Gildner M. B., Shoulders B. A., Velasquez T. L., Blumenthal M. O., Wang L., Li X., Hudnall T. W., Betancourt T., Du L., Brittain W. J., Tetrahedron 2021, 80, 131854. [Google Scholar]
- 25.L. Gao, J. C. M. Meiring, A. Varady, I. E. Ruider, C. Heise, M. Wranik, C. D. Velasco, J. A. Taylor, B. Terni, J. Standfuss, C. C. Cabernard, A. Llobet, M. O. Steinmetz, A. R. Bausch, M. Distel, J. Thorn-Seshold, A. Akhmanova, O. Thorn-Seshold, bioRxiv 2021, 10.1101/2021.03.26.437160. [DOI]
- 26. Prota A. E., Bargsten K., Northcote P. T., Marsh M., Altmann K.-H., Miller J. H., Díaz J. F., Steinmetz M. O., Angew. Chem. Int. Ed. 2014, 53, 1621–1625; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 1647–1651. [Google Scholar]
- 27. An Y., Chen C., Zhu J., Dwivedi P., Zhao Y., Wang Z., Front. Chem. 2020, 14, 880–888. [Google Scholar]
- 28. Gavin J., Ruiz J. F. M., Kedziora K., Windle H., Kelleher D. P., Gilmer J. F., Bioorg. Med. Chem. Lett. 2012, 22, 7647–7652. [DOI] [PubMed] [Google Scholar]
- 29. Bollag D. M., McQueney P. A., Zhu J., Hensens O., Koupal L., Liesch J., Goetz M., Lazarides E., Woods C. M., Cancer Res. 1995, 55, 2325–2333. [PubMed] [Google Scholar]
- 30. Höfle G., Bedorf N., Steinmetz H., Schomburg D., Gerth K., Reichenbach H., Angew. Chem. Int. Ed. Engl. 1996, 35, 1567–1569; [Google Scholar]; Angew. Chem. 1996, 108, 1671–1673. [Google Scholar]
- 31. Ruschel J., Hellal F., Flynn K. C., Dupraz S., Elliott D. A., Tedeschi A., Bates M., Sliwinski C., Brook G., Dobrindt K., Peitz M., Brüstle O., Norenberg M. D., Blesch A., Weidner N., Bunge M. B., Bixby J. L., Bradke F., Science 2015, 348, 347–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Altmann K.-H., Schinzer D., Natural Products in Medicinal Chemistry, Wiley-VCH, Weinheim, 2014, pp. 81–126. [Google Scholar]
- 33. Nicolaou K. C., Roschangar F., Vourloumis D., Angew. Chem. Int. Ed. 1998, 37, 2014–2045; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 1998, 110, 2120–2153. [Google Scholar]
- 34. Reiff E. A., Nair S. K., Henri J. T., Greiner J. F., Reddy B. S., Chakrasali R., David S. A., Chiu T.-L., Amin E. A., Himes R. H., Vander Velde D. G., Georg G. I., J. Org. Chem. 2010, 75, 86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nicolaou K., Scarpelli R., Bollbuck B., Werschkun B., Pereira M., Wartmann M., Altmann K.-H., Zaharevitz D., Gussio R., Giannakakou P., Chem. Biol. 2000, 7, 593–599. [DOI] [PubMed] [Google Scholar]
- 36. Nicolaou K. C., Rhoades D., Wang Y., Bai R., Hamel E., Aujay M., Sandoval J., Gavrilyuk J., J. Am. Chem. Soc. 2017, 139, 7318–7334. [DOI] [PubMed] [Google Scholar]
- 37. Höfle G., Glaser N., Leibold T., Sefkow M., Pure Appl. Chem. 1999, 71, 2019–2024. [Google Scholar]
- 38. White J. D., Carter R. G., Sundermann K. F., Wartmann M., J. Am. Chem. Soc. 2001, 123, 5407–5413. [DOI] [PubMed] [Google Scholar]
- 39. Sharpless K. B., Umbreit M. A., Nieh M. T., Flood T. C., J. Am. Chem. Soc. 1972, 94, 6538–6540. [Google Scholar]
- 40. Awad M. K., El-Hendawy M. M., Fayed T. A., Etaiw S. E. H., English N. J., Photochem. Photobiol. Sci. 2013, 12, 1220–1231. [DOI] [PubMed] [Google Scholar]
- 41. Roostalu J., Thomas C., Cade N. I., Kunzelmann S., Taylor I. A., Surrey T., eLife 2020, 9, e51992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rai A., Liu T., Glauser S., Katrukha E. A., Estévez-Gallego J., Rodríguez-García R., Fang W.-S., Díaz J. F., Steinmetz M. O., Altmann K.-H., Kapitein L. C., Moores C. A., Akhmanova A., Nat. Mater. 2020, 19, 355–365. [DOI] [PubMed] [Google Scholar]
- 43. Bieling P., Laan L., Schek H., Munteanu E. L., Sandblad L., Dogterom M., Brunner D., Surrey T., Nature 2007, 450, 1100–1105. [DOI] [PubMed] [Google Scholar]
- 44. Laprell L., Tochitsky I., Kaur K., Manookin M. B., Stein M., Barber D. M., Schön C., Michalakis S., Biel M., Kramer R. H., Sumser M. P., Trauner D., Gelder R. N. V., J. Clin. Invest. 2017, 127, 2598–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Leinders-Zufall T., Storch U., Bleymehl K., Mederos y Schnitzler M., Frank J. A., Konrad D. B., Trauner D., Gudermann T., Zufall F., Cell Chem. Biol. 2018, 25, 215–223.e3. [DOI] [PubMed] [Google Scholar]
- 46. Castle B. T., Odde D. J., Cell 2015, 162, 243–245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.