Abstract
Background and Aims
HBV can integrate in the host genome of the hepatocyte and recent findings suggest that integrated HBV contributes to the persistent production of viral proteins. Here, we quantified the levels of integrated HBV in patients with chronic hepatitis B (CHB) and analyzed the relation between HBV integration, virological activity (plasma HBV DNA and HBsAg levels), and clinical outcomes.
Approach and Results
We developed and validated a multistep Arthrobacter luteus (Alu)‐PCR that specifically amplifies integrated HBV and RT‐Alu‐PCR detecting mRNA transcripts derived from integrated HBV. Pretreatment liver biopsy samples and baseline characteristics of 124 patients with CHB either treated for 48 weeks with pegylated interferon plus adefovir or tenofovir or receiving no treatment were available for analysis. Integrated HBV sequences containing open reading frame S and X (but not C) and S and X mRNA transcripts derived from integrated HBV could be detected and quantified in liver biopsies. Integrated HBV levels correlated with HBV DNA, HBsAg, alanine aminotransferase plasma levels, and the liver histology activity index but not to levels of intrahepatic covalently closed circular DNA (cccDNA), plasma pregenomic RNA, or hepatitis B core‐related antigen. Multivariable logistic regression analysis showed that lower baseline HBV integration levels were independently associated with HBsAg loss (functional cure) within 5 years follow‐up.
Conclusions
Integrated HBV levels are strongly correlated with surrogate markers for virological activity but not to cccDNA levels and are predictive for HBsAg loss. Our data suggest that integrated HBV is closely related to HBV replication and may therefore be an important tool in the evaluation and development of treatment modalities aiming to cure CHB.

Abbreviations
- ALT
alanine aminotransferase
- Alu
Arthrobacter luteus
- cccDNA
covalently closed circular DNA
- CH
chronic hepatitis
- CHB
chronic hepatitis B
- ChI
chronic infection
- HAI
histology activity index
- HBcrAg
hepatitis B core‐related antigen
- ORF
open reading frame
- PEG‐IFN
pegylated interferon
- pgRNA
pregenomic RNA
INTRODUCTION
Chronic hepatitis B (CHB) is defined as a sustained infection of the liver with HBV for more than 6 months. An estimate of 290 million people are currently living with CHB worldwide. These patients are at risk of developing liver cirrhosis and HCC, leading to a yearly mortality rate of 887,000 patients.[ 1 ]
In the infected hepatocytes, the nucleus contains covalently closed circular (cccDNA) that acts as the viral template for production of viral proteins and HBV pregenomic RNA (pgRNA). The transcribed HBV‐pgRNA is packaged into core particles together with the viral polymerase, where it is reverse transcribed into a partially double‐stranded viral DNA genome. The core particles are either enveloped and secreted or shuttled back to the nucleus to increase the cccDNA pool. Besides mature virus, HBV‐infected hepatocytes excrete large amounts of HBsAg. The high viral protein production has been linked to failure to develop an effective immune response to clear the virus.[ 2 ]
Besides formation of cccDNA in the cell nucleus, HBV viral DNA genome can also integrate into the human genome.[ 3 ] Integration of HBV was described in relation to the development of HCC, and it was considered not to be of clinical importance for the viral replication activity because integrated HBV genomes do not encode the full‐length virus.[ 4 ] Recently, it was found in chimpanzees that viral transcripts could originate from integrated HBV. In this study, chimpanzees treated with an RNA interference (RNAi)‐based therapy targeting cccDNA‐derived HBsAg showed a lower response to this therapy when they had higher levels of integrated HBV, which lacked the sequence targeted by the RNAi.[ 5 ] Higher levels of integrated HBV were mainly seen in HBeAg‐negative monkeys. Based on this finding, the role of integrated HBV in patients with CHB is being reexamined. If integrated HBV can contribute to the viral protein production (in particular, HBsAg) in patients with CHB, this may be of importance for viral persistence. Currently, several treatment modalities targeting cccDNA or transcripts derived only from cccDNA are under development to cure CHB.[ 6 , 7 , 8 ] High integrated HBV levels in patients may interfere with the efficacy of these treatments because the protein production from integrated HBV is not influenced by these treatments.
Little is known about the relation between integrated HBV and viral protein production in humans and the importance of integrated HBV for the efficacy of clinical intervention studies to cure CHB. In this study, we aim to detect and quantify integrated HBV levels in patients with CHB by an in‐house developed Arthrobacter luteus (Alu)‐PCR. Secondly, we aim to detect and quantify RNA transcripts derived from these integrated genomes and to determine the relation with known HBV markers for viral activity (plasma HBV DNA and HBsAg levels) and liver histology activity index (HAI). Because the analyzed materials (liver biopsies and plasma) were from two previous prospective intervention studies[ 9 , 10 ] with 5 year follow‐up,[ 11 , 12 ] we were able to study the relation between pretreatment HBV integration and HBsAg loss (functional cure) in these patients.
MATERIALS AND METHODS
Study population and sample collection
This study was performed in pretreatment liver tissue samples of patients with CHB who participated in two independent investigator‐initiated clinical trials (ClinicalTrials.gov, number NCT00973219 and ISRCTN registry, number 77073364) that were described in detail previously.[ 9 , 10 ] All patients gave written informed consent and both studies complied with the Declaration of Helsinki and the principles of Good Clinical Practice and was approved by a legally instituted ethical committee. No organ samples were obtained from executed prisoners or other institutionalized persons. In one of the studies (cohort 1), patients with HBeAg‐negative CHB with HBV DNA levels below 20,000 IU/ml were allocated to treatment with pegylated interferon (Peg‐IFN) plus tenofovir or adefovir or no treatment for 48 weeks. In the second study (cohort 2), both patients who were HBeAg positive and those who were negative with HBV DNA levels >17,182 IU/ml were treated for 48 weeks with Peg‐IFN plus adefovir. Patients in both studies were followed up for 5 years after treatment with a yearly interval during which HBsAg loss as primary outcome was assessed. For this study, 97 liver samples of cohort 1 and 38 liver samples of cohort 2 were available for integrated HBV DNA measurement, and a total of 135 available stored pretreatment liver biopsy samples were analyzed. Overall, the majority of patients were HBeAg‐negative (89%) with low alanine aminotransferase (ALT) levels at baseline and low fibrosis and necroinflammatory score in liver biopsies. Patients with genotype A‐E were all included (Table 1). Control liver tissue was obtained from 25 patients who were HBV negative and undergoing surgical liver resection. Liver tissue of the nonaffected, tumor‐free margin surrounding the pathology was used for analysis. Liver tissue samples were either cryopreserved or stored in RNAlater stabilizing solution until use (Thermo Fisher Scientific).[ 13 ]
TABLE 1.
Patient characteristics
| Characteristics and markers | Total group (n = 124) | Cohort 1 (n = 92) | Cohort 2 (n = 32) |
|---|---|---|---|
| Mean age, years (SD) | 43.1 (10.9) | 43.9 (11.4) | 40.9 (9.1) |
| Female sex (%) | 49 (39.5) | 40 (43.5) | 9 (28.1) |
| Ethnicity (%) | |||
| Caucasian | 39 (31.5) | 27 (29.3) | 12 (37.5) |
| Asian | 28 (22.6) | 16 (17.4) | 12 (37.5) |
| African | 36 (29.3) | 28 (30.4) | 8 (25.0) |
| South American | 17 (13.7) | 17 (18.5) | 0 (0) |
| Unknown | 4 (3.2) | 4 (4.3) | 0 (0) |
| Peg‐IFN experienced, yes (%) | 14 (11.3) | 3 (3.3) | 11 (34.4) |
| Median ALT U/L (IQR) | 29 (22–54) | 25 (20–34) | 106 (48–194) |
| HBeAg negative (%) | 110 (89) | 92 (100) | 18 (56) |
| HBV genotype (%) | |||
| A | 36 (29.0) | 24 (26.1) | 12 (37.5) |
| B | 13 (10.5) | 6 (6.5) | 7 (21.9) |
| C | 5 (4.0) | 3 (3.2) | 2 (6.3) |
| D | 33 (26.7) | 23 (25.0) | 10 (31.3) |
| E | 17 (13.7) | 16 (17.4) | 1 (3.1) |
| Undeterminable | 20 (16.1) | 20 (21.7) | 0 (0) |
| Mean HBsAg, log10 IU/ml (SD) | 3.4 (0.9) | 3.3 (0.9) | 3.8 (1.0) |
| Mean HBV DNA, log10 IU/ml (SD) | 3.8 (2.2) | 2.8 (1.1) | 6.7 (1.8) |
Cell culture
The HepG2.2.15 cells are a hepatoblastoma cell line that contains two stably integrated HBV genomes.[ 14 ] These cells were maintained in William’s Medium E w/o L‐glutamine (Lonza), supplemented with 10% vol/vol inactivated fetal calf serum, 2 mM L‐glutamin (Gibco), penicillin (100 U/ml), streptomycin (100 lg/ml), and 5 µM dexamethasone (Sigma‐Aldrich) in a humidified 10% CO2 incubator at 37°C.
DNA and RNA isolation
Total DNA and RNA from HepG2.2.15 was isolated using the AllPrep DNA/RNA Mini Kit (Qiagen) according to the manufacturer’s protocol. Liver tissue was disrupted and homogenized in QIAzol Lysis Reagent (Qiagen) using the TissueLyser II (Qiagen). Total DNA and RNA from liver samples was isolated using the miRNeasy Micro Kit according to manufacturer’s protocol. DNA and RNA concentration were measured using the Nanodrop 1000 (Isogen Life Sciences). For validation and quantification of the different qPCRs and RT‐qPCRs, a 10‐fold serial dilution of DNA and RNA isolated from HepG2.2.15 cells were spiked in DNA or RNA isolated from peripheral blood mononuclear cells from blood donors using the AllPrep DNA/RNA Mini Kit (Qiagen).
Quantification of integrated HBV
For the detection and quantification of integrated HBV DNA, a semiquantitative nested PCR was performed detecting the S, X, and C open reading frame (ORF). A detailed protocol is described in the supplementary data (Supporting Methods). In brief, 5 ng of DNA was subjected to 15 PCR cycles to amplify integrated HBV using a forward primer specific for different HBV ORFs (HBV‐S‐F, HBV‐X‐F, HBV‐C‐F) in combination with a primer binding to Alu sites in the chromosomal DNA (Alu‐278TAG, Alu‐AS) (Table S1). This PCR product (2.5 µl) was subjected to a second PCR (20 cycles) using a set of internal primers specifically detecting HBV ORF (HBV‐S‐FN, HBV‐X‐FN, HBV‐C‐FN) in combination with Alu‐specific primers (278TAG, TAG‐AS) (Table S1). For quantification, qPCRs specifically detecting each HBV ORF were performed on 1:5 diluted nested PCR product using ORF specific primers (HBV‐S‐FN and HBV‐S‐R; HBV‐X‐FN and HBV‐X‐R; HBV‐C‐FN and HBV‐C‐R) and 5′FAM–3′ TAMRA labeled ORF specific probes (HBV‐S probe, HBV‐X probe, or HBV‐C probe) (Table S1). For each sample, a no‐DNA polymerase control was included during the preamplification steps to control for residual input HBV DNA signal in the final qPCR. DNA levels of housekeeping genes Beta‐actin and GAPDH were used to correct for differences in DNA and RNA input and were measured using the GoTaq qPCR Master Mix (Promega) and specific primers (B‐Actin‐S and B‐Actin‐AS or GAPDH‐F and GAPDH‐R primers) (Table S1). The qPCR assay was performed in a 384 wells plate format on the LC480 platform (Roche Diagnostics).
Quantification of integrated HBV transcription
mRNA transcripts from integrated HBV were quantified using a primer specific for chromosomal Alu sites for cDNA synthesis. In short, 600–800 ng of total RNA was treated with DNase (Promega) to remove remaining DNA for 50 min at 42°C followed by 15 min at 72°C to inactivate the DNase. RNA was reverse transcribed using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer’s protocol using both the Alu‐278TAG and Alu‐AS primer. Subsequently, 2.5 µl of the cDNA was subjected to preamplification (15 cycles) using either HBV‐S‐F, HBV‐X‐F, or HBV‐C‐F in combination with the 278TAG and TAG‐AS primers. For quantification, qPCR was performed on 1:5 diluted nested PCR product using specific HBV ORF primers and 5′FAM–3′ TAMRA labeled ORF specific probes (described previously). The qPCR assay was performed on a LC480 platform (Roche). A detailed protocol is described in the supplementary data (Supporting Methods). In all experiments, a no‐DNA polymerase control was included in the preamplification steps to check for residual input HBV RNA signal in the final qPCR.
Histology and intrahepatic virology markers
For histological assessment of liver biopsies, the modified Ishak scoring system was applied based on a 0–18 score for necroinflammation and a 0–6 score for fibrosis.[ 15 ] Intrahepatic cccDNA and intrahepatic total HBV DNA levels were determined using in‐house developed assays described earlier and corrected for the number of hepatocytes per biopsy (Supporting Methods).[ 16 ]
Circulating virological and biochemical markers
HBV DNA, quantitative HBsAg, HBV‐pgRNA, and hepatitis B core‐related antigen (HBcrAg) levels were quantified in plasma using the various detection methods described in Supporting Methods.[ 17 ] HBV‐genotype was determined by sequencing part of the polymerase gene with dideoxynucleotide technology or using the INNO‐LiPA assay (Fujirebio). ALT levels were analyzed in the local clinical chemistry laboratory.
Statistical methods
The statistical programs for Social Sciences (SPSS 25.0.0.1), RStudio (version 1.2.1335), and Graph Pad software (version 8) were used for data analysis.
Integrated HBV DNA ORF and mRNA transcripts levels were normalized using the comparative Ct method (=2^ ‐ (Ct of integrated HBV DNA ORF or mRNA transcripts ‐ mean Ct of Beta‐actin and GAPDH)). ORF mRNA transcript levels were normalized for the no‐DNA polymerase control
(=2^ ‐ ((Ct of ORF mRNA transcripts ‐ mean Ct of Beta‐actin and GAPDH) ‐ (Ct of corresponding no‐DNA polymerase control ‐ mean Ct of Beta‐actin and GAPDH))). Groups were compared using Student t test, Mann‐Whitney U test, Kruskal‐Wallis, or ANOVA where appropriate. The Spearman‐rank correlation test was used to test correlations between markers, and results were corrected for multiple testing using the Benjamini‐Hochberg method. Principal component analysis (PCA) was used for data reduction, followed by k‐means unsupervised clustering. For this analysis, missing data points were replaced by the group mean. Multivariable logistic regression was used to assess levels of integrated HBV containing ORF S or ORF X as independent predictor of HBsAg loss. Occurrence of HBsAg loss over time was shown in a Kaplan Meier curve and compared between patients with the highest versus the lowest integrated HBV levels using Cox regression analysis. Patients who were lost to follow‐up or did not reach HBsAg loss at end of follow‐up were censored.
RESULTS
Integrated HBV DNA in liver tissue
The Alu‐qPCR method for the detection of integrated HBV was validated on total DNA isolated from HepG2.2.15 cells. A 10‐fold serial dilution of HepG2.2.15 DNA was made in DNA obtained from PBMC to equilibrate the total DNA concentration. Using this method, integrated HBV containing the different ORFs (S, X, C) could be detected in an input‐dependent manner. As this nested Alu‐qPCR could potentially detect background cccDNA and HBV DNA levels as HBV ORF specific primers and probes were used, the same nested Alu‐qPCR lacking the DNA polymerase enzyme were performed in parallel with each sample. A difference of at least 20 cycles between each ORF and the corresponding no‐DNA polymerase control was found, which is indicative of a 1,000,000‐fold lower concentration as compared with the signal obtained with the Alu‐HBV DNA preamplification (Figure S1A‐C).
Next, the HBV‐Alu‐qPCR was used to determine the relative levels of integrated HBV sequences in liver tissue samples. In patients with CHB, integrated HBV containing ORF S and X but not C could be detected in 131 out of 135 liver biopsies (97%). The relative levels of these ORFs were calculated using the comparative Ct method, which gives a semiquantitative measure of integrated HBV relative to the housekeeping genes (GAPDH/beta‐actin). Similar relative levels of integrated HBV containing ORF S or ORF X were detected (Figure 1). Patients of whom baseline characteristics were available at baseline were included in further analyses (124/131) (Figure S2). Baseline characteristics are described in Table 1. Higher levels of integrated ORF S or X DNA were observed using HBV‐Alu‐PCR preamplification when compared with the no‐DNA polymerase control, indicating that the qPCR is indeed detecting Alu‐HBV amplified products. Additionally, as in a substantial amount of patients no signal could be detected in the no‐DNA polymerase controls of either ORF S (n = 21) or X (n = 42), no correction for this background was performed. No integrated HBV sequences were detected in the liver tissue of uninfected controls (Figure 1).
FIGURE 1.
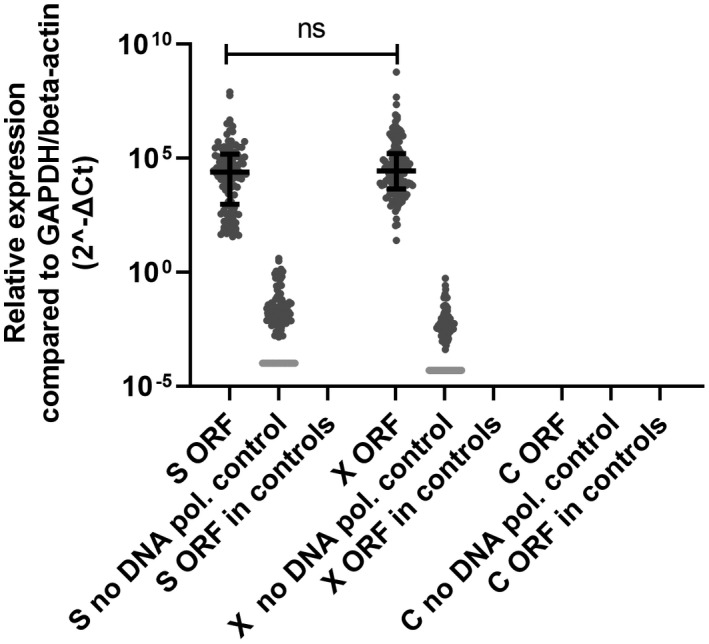
Integrated HBV in liver biopsies from patients with CHB. Integrated HBV relative levels containing ORF S, X, or C were determined in liver tissue samples obtained from 135 patients with CHB and 25 controls by HBV‐Alu qPCR. A no‐DNA polymerase control was performed to detect for background cccDNA/HBV DNA levels. Differences between the groups were determined by Mann‐Whitney U test
Viral RNA derived from integrated HBV DNA in liver tissue
To investigate whether RNA was transcribed from the integrated HBV, a nested RT‐Alu‐qPCR was set up and validated in HepG2.2.15 cells (Supporting Results, Figure S1D‐F). For the detection of RNA transcription from integrated HBV in patients with CHB, total RNA obtained from liver samples was subjected to cDNA synthesis using a primer detecting Alu sites followed by Alu‐HBV preamplification and qPCR detecting HBV‐S, X, and C transcripts. RNA transcripts of HBV‐S and X but not C could be detecting in liver of patients with CHB, but not in the uninfected controls. In patients with CHB, the levels of S RNA transcripts were higher than the levels of X transcripts (Figure 2A). However, a similar difference was observed between the levels of S and X RNA transcripts detected in the no‐DNA polymerase control. Therefore, the relative levels of both ORF S and X RNA transcripts were corrected for this background. After correction, no significant difference was found between the relative levels of ORF S and X RNA transcripts (Figure 2B).
FIGURE 2.
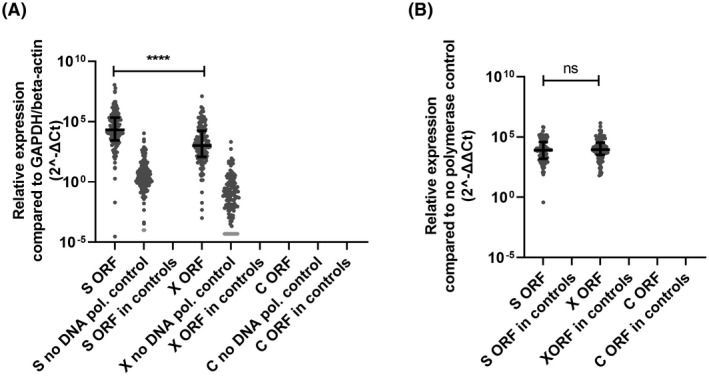
RNA transcription from integrated HBV in liver biopsies from patients with CHB. RNA transcription relative levels of ORF S, X, and C were measured in liver tissue samples obtained from 135 patients with CHB and 25 controls by Alu‐qPCR. A no‐DNA polymerase control was performed to detect for background HBV RNA levels. Differences between the groups were determined by Mann‐Whitney U test. ****p < 0.0001
Next, the ratio between RNA transcript relative levels of S and X and integrated HBV‐relative levels containing ORF S and X was analyzed as a representation of transcriptional activity. No difference was observed between the calculated activity of the ORF S and the activity of ORF X (Figure 3).
FIGURE 3.
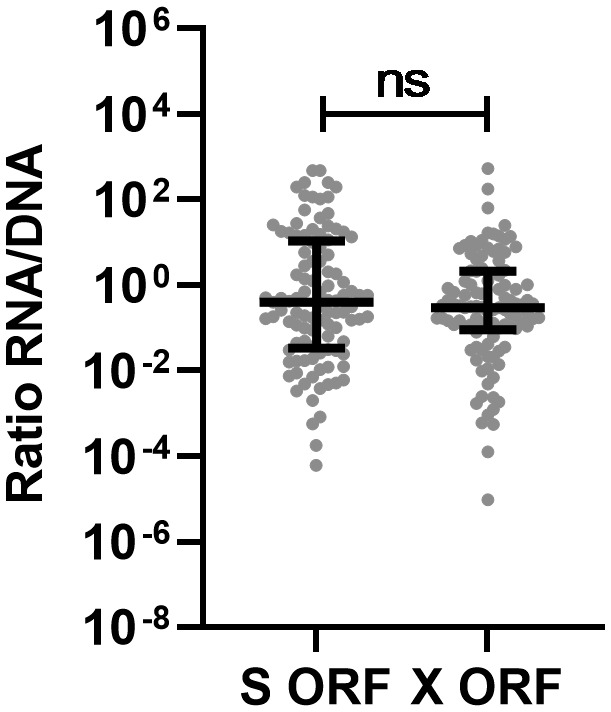
Transcriptional activity of integrated HBV. The RNA relative levels of ORF S or X transcripts, expressed per integrated HBV‐relative levels containing ORF S or X were calculated as measure of transcriptional activity of integrated HBV. Differences between the groups were determined by Mann‐Whitney U test. ****p < 0.0001
Subgroup analysis of different CHB phases
In the most recent guidelines, CHB infection can be categorized in different HBV phases (EASL), Table S2. Patients were categorized to the most fitting phase based on HBV DNA levels with a cutoff of 2000 IU/ml and HBeAg status. Three phases could be distinguished between patients: HBeAg‐negative chronic infection (ChI; HBV DNA < 2000 IU/ml), HBeAg‐negative chronic hepatitis (CH; HBV DNA > 2000 IU/ml) and HBeAg‐positive CH (HBV DNA > 2000 IU/ml). Patients with a HBeAg‐negative ChI had a mean level of HBV DNA 2.18 log10 IU/ml (SD, 0.90), HBeAg‐negative CH had a mean HBV DNA of 4.52 log10 IU/ml (SD, 1.07), and HBeAg‐positive CH had a mean HBV DNA 8.39 log10 IU/ml (SD, 0.91). Higher integrated HBV‐relative levels containing ORF S or X were measured in patients with HBeAg‐positive CH compared with the two HBeAg‐negative patient groups (Figure 4). Furthermore, higher HBV DNA relative levels containing ORF X were observed in the patients with HBeAg‐negative CH when compared with the patients with an HBeAg‐negative ChI. The S RNA transcript relative levels were higher in patients with both an HBeAg‐negative or positive CH compared with HBeAg‐negative ChI, whereas no differences in the X RNA transcript relative levels were observed between all three phases. Furthermore, the ratio between S RNA transcript relative levels and integrated HBV‐relative levels containing ORF S was higher in the patients with an HBeAg‐negative CH compared with the other two groups. For ORF X, lower ratios were observed in the patients with a HBeAg‐positive CH compared with both patients who were HBeAg‐negative.
FIGURE 4.
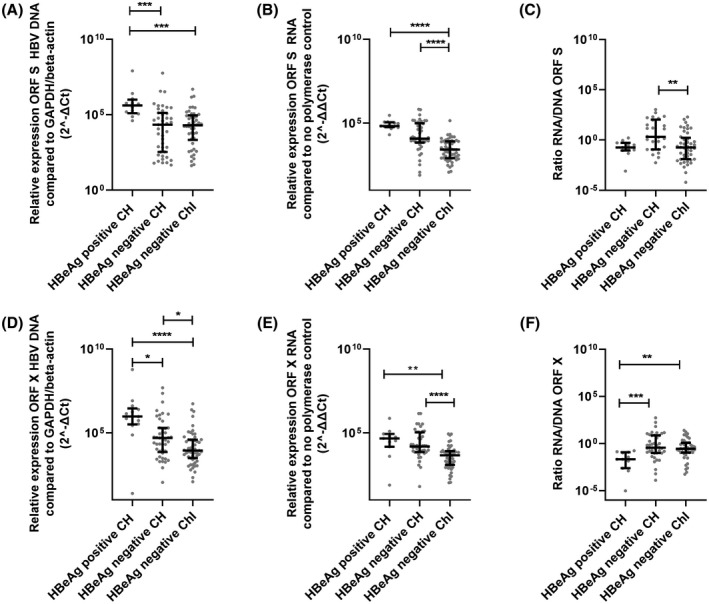
Subgroup analysis of integrated HBV and transcription derived from integrated HBV. Integrated HBV relative levels containing ORF S (A) and X (D), RNA transcript relative levels of ORF S (B) and X (E), and the ratio between the two (C and F) were compared between the HBeAg negative ChI, HBeAg negative CH, and HBeAg positive CH groups. Differences between the groups were determined by Mann‐Whitney U test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Associations between integrated HBV and markers of CHB and liver damage
The relative levels of integrated HBV were compared with various well‐known HBV markers, baseline characteristics Table 1, and nonstandard viral markers Table S3. Integrated HBV‐relative levels detected by qPCRs for both ORFs were correlated to plasma HBV DNA and HBsAg levels but not to HBV‐pgRNA or HBcrAg plasma levels (Table 2). Intrahepatic cccDNA levels were measured in a subset of patients (n = 32) and did not correlate with integrated HBV‐relative levels in these patients. ALT levels correlated to relative levels of integrated HBV containing ORF X but not to ORF S containing integrated HBV. Histology scores showed that the hepatic activity index (HAI score) was associated with integrated HBV‐relative levels but the Ishak score for fibrosis and the steatosis score, were not (Table 2).
TABLE 2.
Correlations between integrated HBV and biomarkers of HBV activity and liver damage a
| Integrated DNA ORF S | Integrated DNA ORF X | |||
|---|---|---|---|---|
| rho | p value | rho | p value | |
| Plasma HBV DNA | 0.319 | 0.004 | 0.497 | <0.001 |
| Plasma HbsAg | 0.260 | 0.021 | 0.306 | 0.002 |
| Plasma HBV‐pgRNA | 0.408 | 0.090 | 0.315 | 0.144 |
| Hepatic cccDNA | 0.176 | 0.406 | −0.015 | 0.936 |
| Plasma HbcrAg | 0.374 | 0.100 | 0.255 | 0.227 |
| ALT | 0.200 | 0.089 | 0.295 | 0.002 |
| HAI score | 0.261 | 0.021 | 0.337 | 0.001 |
| Ishak score | 0.050 | 0.620 | 0.055 | 0.599 |
| Steatosis score | −0.156 | 0.250 | −0.119 | 0.343 |
Abbreviation: HBV RNA, HBV pgRNA.
Correlations were determined using a Spearman correlation test. Significance was set at p < 0.05.
Patient clustering based on HBV markers, integrated HBV, and liver damage
An in‐depth analysis of the associations between markers was performed by PCA and unsupervised k‐means analysis. With this analysis, we aimed to identify patient clusters based on the relation between the previously found marker‐associations in patients in an unbiased manner. In total, two patient clusters were identified (Figure S3). These clusters (Table S4) showed a difference in the viral activity markers (plasma HBV DNA and HBsAg levels) but not in demographic characteristics of the patients as shown in Figure 5. Cluster 1 comprises solely of patients who were HBeAg‐negative who are characterized by lower relative levels of integrated HBV containing ORF S, ORF X, as well as HBcrAg compared with patients in cluster 2. Cluster 2, however, included patients with signs of liver fibrosis, necrosis, and inflammation by means of elevated ALT levels and higher HAI scores as compared with cluster 1. Also, higher integrated HBV‐relative levels containing ORF S or ORF X were measured in this cluster.
FIGURE 5.

Heatmap of markers. Heatmap showing all included markers: red represents values above 2SD and blue below −2SD from the mean for each value. HBeAg‐seropositivity and clusters of each patient are shown in the top bar, cluster 1, and patients who are HBeAg negative are in lighter gray. Abbreviation: HBV RNA, HBV pgRNA
Integrated HBV as predictor for HBsAg loss
All patients included in this study underwent a liver biopsy before treatment with Peg‐IFN plus either tenofovir (n = 37) or adefovir (n = 68) or before monitoring without treatment (n = 19) and were subsequently followed up for 5 years after end of treatment. During the follow‐up period, 21/124 (16.9%) patients had retreatment with entecavir or tenofovir. Of all included patients combined, 6 patients (4.8%) achieved HBsAg loss within 24 weeks after treatment (week 72) and 17 patients (13.7%) within 5 years follow‐up, of whom 13 after treatment and 4 without treatment.[ 12 ] To determine whether the relative level of integrated HBV was predictive for HBsAg loss, all patients were included in the logistic regression analysis. The multivariable analysis was corrected for treatment arm, sex, age, and genotype A to exclude possible allocation bias (Table S5A,B) using a separate multivariable model for integrated HBV containing ORF X and ORF S because of collinearity between both measurements (rho 0.676, p < 0.001). Lower relative levels of integrated HBV at baseline were found to be predictive for HBsAg loss after 72 weeks and 5 years follow‐up (containing ORF S: OR, −0.47; CI, 0.23–0.94; p = 0.032; containing ORF X: OR, 0.24; CI, 0.08–0.69; p = 0.008) in a multivariable regression analysis (Table S5A,B). cccDNA levels at baseline were no predictor of HBsAg loss. In addition, lower HBsAg levels at baseline were an independent predictor of HBsAg loss at week 72 (Table S5A) and at 5‐years follow‐up (Table S5B). Not having genotype A was an independent predictor of HBsAg loss at 5‐years follow‐up.
In addition, a survival analysis was performed in which HBsAg loss over time was compared between patients with the highest 50% and the lowest 50% relative levels of the integrated HBV DNA containing ORF S or ORF X (Figure 6A,B). Cox regression analysis showed that the odd ratio of achieving HBsAg loss was higher in patients with lower relative levels of integrated HBV DNA containing ORF S (OR, 4.66; 95% CI, 1.01–21.54; p = 0.049) or containing ORF X (OR, 3.87; 95% CI, 1.08–13.88; p = 0.038).
FIGURE 6.
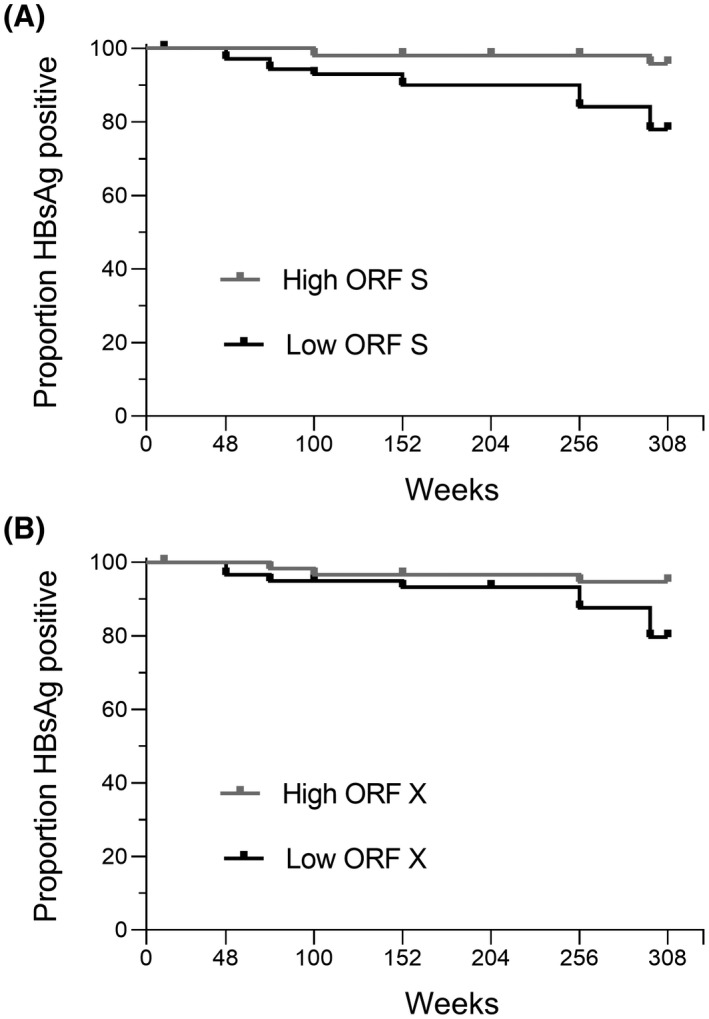
Kaplan‐Meier curve of HBsAg‐loss events over time. Proportion of patients who were HBsAg positive shown over time for 50% patients with the highest or lowest levels of integrated HBV containing ORF S (A) or ORF X (B)
DISCUSSION
In this study, we analyzed the integrated HBV‐relative levels (the relative level of integrated HBV is determined as fold change in delta Ct value relative to b‐actin and GAPDH levels) in liver biopsies of a panel of 135 patients with CHB who participated in 2 previously described intervention studies. Using an in‐house developed Alu‐PCR based technique, we found that integrated HBV could be detected and relatively quantified in almost all liver biopsies and that this integrated HBV is transcriptionally active. With these findings, we could then further investigate the association between these relative levels and clinical outcomes in 124 of these patients, showing that integrated HBV‐relative levels correlate with plasma HBV DNA and HBsAg levels as well as the HAI index. We furthermore found that integrated HBV‐relative levels were associated with HBsAg loss over time.
As it is known that full HBV genome length (double‐stranded linear DNA [dslDNA]) does not completely integrate into the human genome,[ 4 ] we measured integrated HBV‐relative levels distinguishing between the different ORF S, X, and C. In pretreatment liver tissue samples of patients with CHB, similar relative levels of integrated HBV containing ORF S and X could be detected and a high association between the two ORFs was observed by the correlation analysis. These findings make it likely that during HBV infection, a DNA fragment containing both ORFs integrates into the host DNA. Our observation that integrated HBV containing ORF C could not be detected is supported by several other studies that found limited ORF C integration in liver tissue with or without HCC from patients with CHB when using next generation sequencing techniques.[ 5 , 18 , 19 ] However, one previous study found integrated ORF C HBV DNA in liver tissue samples of patients with CHB using a similar Alu‐PCR method.[ 20 ] In this study, three preamplification Alu‐PCRs were performed to detect integrated HBV, which increased the sensitivity of the assay and so may explain the differences between the studies. Additionally, the ORF C primer set and probe used in this study failed to detect potentially incomplete integrated ORF C sequences. We furthermore showed that viral RNA transcription from integrated HBV could be detected in our panel, although no difference was found between relative transcriptional activity of ORF X and the relative transcriptional activity of ORF S. This strengthens the previous observation that during HBV infection a DNA fragment containing both ORFs integrates into the host DNA and is transcriptionally active. However, truncated ORF X transcripts that arise from the cryptic polyA signal located at the end of the ORF X were not measured by our detection method because an Alu element is not present in this truncated transcript.[ 21 , 22 ] Therefore, true transcriptional activity of ORF X from integrated DNA may be higher than the relative activity measured in this study.
Comparing the integration levels between patients in different CHB phases showed that higher integrated HBV‐relative levels containing ORF S and X could be found in the HBeAg‐positive CH patients with HBeAg‐positive CH compared with patients in the two HBeAg‐negative phases. This suggests that more HBV DNA is integrated in the patients who were HBeAg positive compared with the patients who were HBeAg negative, whereas the opposite has been described by others. However, most of these contradicting studies reported a limited sample size and based their conclusion on the percentage of patients in whom integrated HBV could be detected, which is in contrast with semiquantified levels of integration determined in our study.[ 20 , 23 ] In the transition from HBeAg‐positive CH to a HbeAg‐negative state, it is known that many infected hepatocytes are eliminated. Therefore, integrated HBV‐relative levels would be expected to be lower early after Hbe‐seroconversion, such as in patients with HBeAg‐negative ChI.[ 4 ] Furthermore, within the subgroup of patients with HBeAg‐positive or ‐negative infection, integrated HBV is more frequently found in patients with high viral replicative levels.[ 24 ] Among the patients with CH in our study, patients who were HBeAg positive had higher plasma HBV DNA levels than patients who were HBeAg negative. Because we did not include patients with HBeAg‐positive ChI, the difference between integrated HBV‐relative levels in patients who were HBeAg positive and those who were negative could partly have been caused by a difference in viral replication levels. Indeed, this is observed by Svicher et al., who describe higher levels of HBV integration in patients who were HBeAg negative with a high viral load compared with patients who were HBeAg negative with a low viral load.[ 25 ]
A previous study in chimpanzees described that a higher proportion of the plasma HBsAg pool may originate from integrated HBV during HBeAg‐negative infection compared with a HBeAg‐positive infection. This stands in contrast to our observation that patients who were HBeAg positive had higher relative levels of integrated HBV compared with patients who were HBeAg negative. However, when regarding the relative transcriptional activity of integrated HBV containing ORF S, this activity was higher in patients with HBeAg‐negative CH as compared with the other two groups. This is in line with the observations reported in the chimpanzees study. Furthermore, patients in an HBeAg‐negative ChI phase are known to have a relatively potent immune response to the virus, resulting in suppression of the viral replication. HBsAg is in turn known to cause immune exhaustion both when transcribed from cccDNA or from integrated HBV. This might explain the higher transcriptional activity of patients with HBeAg‐negative CH compared with patients with ChI.
In contrast to ORF S, no higher transcriptional activity of integrated HBV containing ORF X was seen for patients with HBeAg‐negative CH as compared with the other two groups. The discrepancy of this finding compared with those of integrated HBV DNA containing ORF S may be explained by the possibility that ORF X transcripts are truncated and therefore not detected in our assay. In the HBeAg‐negative ChI phase, patients may have more transcription from the integrated HBV DNA containing ORF X that is caused by a more active promotor than compared with patients in the other two phases.
The attribution of integrated HBV to the total HBsAg formation is of special clinical interest because HBsAg contributes to the persistence of viral infection in patients with CHB and is an important target for several therapies that aim to induce a functional, or total cure. In this study, we found a correlation between integrated HBV‐relative levels and HBsAg levels in plasma. This observation was reinforced by the cluster analysis, which showed that patients with high markers for viral activity (plasma HBV DNA and HBsAg levels) had higher relative levels of integrated HBV compared with patients with a low viral activity. We could also detect S and X RNA transcripts derived from integrated HBV. These findings suggest that HBsAg in plasma can be derived from integrated HBV in CHB as was previously suggested.[ 5 ] Furthermore, integrated HBV‐relative levels were correlated with HAI index in the liver and the cluster analysis showed that patients with high histological activity had higher relative levels of integrated HBV compared with patients with a low histological activity. HBV integration is dependent on the viral replication cycle through the formation of dslDNA from cccDNA, and so integration can occur more often as a consequence of high viral replication levels. In addition, high replication and immune‐mediated inflammation may also lead to more frequent double‐stranded breakpoints in the human genome and so lead to integration events.[ 24 ]
Little is known about the relation between integrated HBV levels and clinical outcomes other than HCC. In this study, we were able to detect integrated HBV in 97% (131/135) of the patients with CHB and found that integrated HBV‐relative levels was an independent predictor for HBsAg loss during 5 years of follow‐up. Hu et al. studied integrated HBV in patients under nucleot(s)ide analogue treatment and could only detect integrated HBV in 15.7% using Alu‐PCR.[ 23 ] The higher relative levels of integrated HBV observed in our study could be due to the fact that liver biopsies were taken before treatment but could also be in part due to a difference in detection limit of the Alu‐PCRs used.
In conclusion, this study displays the close association between integrated HBV‐relative levels, surrogate markers for and viral replication activity (plasma HBV DNA and HBsAg levels), and inflammation and fibrosis in both patients who were HBeAg‐positive and negative. These findings suggest that HBV integration may contribute to viral transcriptional activity and persistence. In addition, HBV DNA integration may directly or indirectly influence the process of achieving a functional cure and clinical outcomes, which was furthermore ascribed by our finding that integrated HBV‐relative levels at baseline were predictive of HBsAg loss over a 5‐year follow‐up period in treated and untreated patients. New treatment modalities under development to cure CHB should therefore take the effect of integrated HBV levels on clinical outcomes into account. Both in monitoring and when selecting with a better treatment response.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Conceptualization: Neeltje Kootstra, Vladimir Loukachov, and Robin Erken; data curation: Karel van Dort, Vladimir Loukachov, Robin Erken, and Anne van den Hurk; formal analysis: Robin Erken and Vladimir Loukachov; funding acquisition: Neeltje Kootstra and Vladimir Loukachov; investigation: Vladimir Loukachov, Robin Erken, Anne van den Hurk, and Karel van Dort; methodology: Neeltje Kootstra, Vladimir Loukachov, and Robin Erken; project administration: Neeltje Kootstra, Vladimir Loukachov, and Robin Erken; resources: Neeltje Kootstra, Louis Jansen, Annniki de Niet, Sophie Willemse, and R. Bart Takkenberg; supervision: Neeltje Kootstra; validation: Neeltje Kootstra, Vladimir Loukachov, and Robin Erken; visualization: Vladimir Loukachov and Robin Erken; writing, original draft: Neeltje Kootstra, Vladimir Loukachov, and Robin Erken; writing, review and editing: Neeltje Kootstra, Vladimir Loukachov, Robin Erken, Henk Reesink, Sophie Willemse, and R. Bart Takkenberg.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the patients and their families.
Erken R, Loukachov V, van Dort K, van den Hurk A, Takkenberg RB, de Niet A, et al. Quantified integrated hepatitis B virus is related to viral activity in patients with chronic hepatitis B. Hepatology. 2022;76:196–206. 10.1002/hep.32352
Robin Erken and Vladimir Loukachov should be considered joint first authors.
Funding information
This study was supported by the Dutch Research Council Open Technology Program grant (project number 15783) and the Gastrostart research grant from the Dutch association for gastroenterology (Nederlandse Vereniging voor Gastroenterologie)
SEE EDITORIAL ON PAGE 15
REFERENCES
- 1. Razavi‐Shearer D, Gamkrelidze I, Nguyen MH, Chen D‐S, Van Damme P, Abbas Z, et al. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol. 2018;3(6):383–403. [DOI] [PubMed] [Google Scholar]
- 2. Ferrari C. HBV and the immune response. Liver Int. 2015;35(Suppl 1):121–8. [DOI] [PubMed] [Google Scholar]
- 3. Edman JC, Gray P, Valenzuela P, Rall LB, Rutter WJ. Integration of hepatitis B virus sequences and their expression in a human hepatoma cell. Nature. 1980;286(5772):535–8. [DOI] [PubMed] [Google Scholar]
- 4. Tu T, Budzinska MA, Shackel NA, Urban S. HBV DNA integration: molecular mechanisms and clinical implications. Viruses. 2017;9(4):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wooddell CI, Yuen MF, Chan HY, Gish RG, Locarnini SA, Chavez D, et al. RNAi‐based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci Transl Med. 2017;9(409):eaan0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schiwon M, Ehrke‐Schulz E, Oswald A, Bergmann T, Michler T, Protzer U, et al. One‐vector system for multiplexed CRISPR/Cas9 against hepatitis B virus cccDNA utilizing high‐capacity adenoviral vectors. Mol Ther Nucleic Acids. 2018;12:242–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yuen M‐F, Schiefke I, Yoon JH, Ahn SH, Heo J, Kim JH, et al. RNA interference therapy with ARC‐520 results in prolonged hepatitis B surface antigen response in patients with chronic hepatitis B infection. Hepatology. 2020;72(1):19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ye X, Tateno C, Thi EP, Kakuni M, Snead NM, Ishida Y, et al. Hepatitis B virus therapeutic agent ARB‐1740 has inhibitory effect on hepatitis delta virus in a new dually‐infected humanized mouse model. ACS Infect Dis. 2019;5(5):738–49. [DOI] [PubMed] [Google Scholar]
- 9. de Niet A, Jansen L, Stelma F, Willemse SB, Kuiken SD, Weijer S, et al. Peg‐interferon plus nucleotide analogue treatment versus no treatment in patients with chronic hepatitis B with a low viral load: a randomised controlled, open‐label trial. Lancet Gastroenterol Hepatol. 2017;2(8):576–84. [DOI] [PubMed] [Google Scholar]
- 10. Takkenberg RB, Jansen L, de Niet A, Zaaijer HL, Weegink CJ, Terpstra V, et al. Baseline hepatitis B surface antigen (HBsAg) as predictor of sustained HBsAg loss in chronic hepatitis B patients treated with pegylated interferon‐alpha2a and adefovir. Antivir Ther. 2013;18(7):895–904. [DOI] [PubMed] [Google Scholar]
- 11. Stelma F, van der Ree MH, Jansen L, Peters MW, Janssen HLA, Zaaijer HL, et al. HBsAg loss after peginterferon‐nucleotide combination treatment in chronic hepatitis B patients: 5 years of follow‐up. J Viral Hepat. 2017;24(12):1107–13. [DOI] [PubMed] [Google Scholar]
- 12. Erken R, Loukachov V, Niet de A, Jansen L, Stelma F, Helder J, et al. A prospective five‐year follow‐up after peg‐interferon plus nucleotide analogue treatment or no treatment in HBeAg negative chronic hepatitis B patients. J Clin Exp Hepatol. 2022. 10.1016/j.jceh.2021.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stelma F, de Niet A, Sinnige MJ, van Dort KA, van Gisbergen KPJM, Verheij J, et al. Human intrahepatic CD69 + CD8+ T cells have a tissue resident memory T cell phenotype with reduced cytolytic capacity. Sci Rep. 2017;7(1):6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84(4):1005–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22(6):696–9. [DOI] [PubMed] [Google Scholar]
- 16. Takkenberg B, Terpstra V, Zaaijer H, Weegink C, Dijkgraaf M, Jansen P, et al. Intrahepatic response markers in chronic hepatitis B patients treated with peginterferon alpha‐2a and adefovir. J Gastroenterol Hepatol. 2011;26(10):1527–35. [DOI] [PubMed] [Google Scholar]
- 17. Jansen L, Kootstra NA, van Dort KA, Takkenberg RB, Reesink HW, Zaaijer HL. Hepatitis B virus pregenomic RNA is present in virions in plasma and is associated with a response to pegylated interferon alfa‐2a and nucleos(t)ide analogues. J Infect Dis. 2016;213(2):224–32. [DOI] [PubMed] [Google Scholar]
- 18. Ringlander J, Skoglund C, Prakash K, Andersson ME, Larsson SB, Tang K‐W, et al. Deep sequencing of liver explant transcriptomes reveals extensive expression from integrated hepatitis B virus DNA. J Viral Hepat. 2020;27(11):1162–70. [DOI] [PubMed] [Google Scholar]
- 19. Meier MA, Calabrese D, Suslov A, Terracciano LM, Heim MH, Wieland S. Ubiquitous expression of HBsAg from integrated HBV DNA in patients with low viral load. J Hepatol. 2021;75(4):840–7. [DOI] [PubMed] [Google Scholar]
- 20. Larsson SB, Tripodi G, Raimondo G, Saitta C, Norkrans G, Pollicino T, et al. Integration of hepatitis B virus DNA in chronically infected patients assessed by Alu‐PCR. J Med Virol. 2018;90(10):1568–75. [DOI] [PubMed] [Google Scholar]
- 21. Schutz T, Kairat A, Schröder CH. DNA sequence requirements for the activation of a CATAAA polyadenylation signal within the hepatitis B virus X reading frame: rapid detection of truncated transcripts. Virology. 1996;223(2):401–5. [DOI] [PubMed] [Google Scholar]
- 22. Kairat A, Beerheide W, Zhou G, Tang ZY, Edler L, Schröder CH. Truncated hepatitis B virus RNA in human hepatocellular carcinoma: its representation in patients with advancing age. Intervirology. 1999;42(4):228–37. [DOI] [PubMed] [Google Scholar]
- 23. Hu B, Wang R, Fu J, Su M, Du M, Liu YU, et al. Integration of hepatitis B virus S gene impacts on hepatitis B surface antigen levels in patients with antiviral therapy. J Gastroenterol Hepatol. 2018;33(7):1389–96. [DOI] [PubMed] [Google Scholar]
- 24. Pollicino T, Caminiti G. HBV‐integration studies in the clinic: role in the natural history of infection. Viruses. 2021;13(3):368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Svicher V, Salpini R, Piermatteo L, Carioti L, Battisti A, Colagrossi L, et al. Whole exome HBV DNA integration is independent of the intrahepatic HBV reservoir in HBeAg‐negative chronic hepatitis B. Gut. 2021;70(12):2337–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


