Abstract
BACKGROUND
Dasineura oleae (Angelini 1831) (Diptera: Cecidomyiidae) was considered a minor pest in olive orchards, but in recent years severe outbreaks have been registered in several Mediterranean countries. Damage is caused by the feeding activity of larvae that induce gall formations and alters the physiological activity of the leaves. In Italy, this pest may be controlled by four Hymenoptera parasitoid species belonging to Platygaster and Mesopolobus genera such as Platygaster demades Walker 1835, Platygaster oleae Szelenyi 1940 (Hymenoptera: Platygastridae), Mesopolobus aspilus (Walker 1835) and Mesopolobus mediterraneus (Mayr 1903) (Hymenoptera: Pteromalidae), but parasitization becomes evident only after gall dissection.
RESULTS
In this study, we aim to: (i) design a primer for the detection of specimens belonging to Platygaster and Mesopolobus genera; (ii) develop a multiplex quantitative polymerase chain reaction (qPCR) protocol combined to a fast samples DNA extraction method; (iii) apply the developed protocol to field‐collected specimens and compare this method with traditional techniques based on visual estimation of parasitism rate on larvae. Primers were designed to anneal with cytochrome oxidase subunit I (COI) sequences of Platygaster and Mesopolobus genera while protocols were developed to be fast and capable to process several samples at the same time. Molecular analyses demonstrated to provide almost double of the parasitism rate assessed by visual inspection. Furthermore, on second instar larvae the PCR‐based method was able to detect ten‐fold times the parasitization rate estimated by visual inspection.
CONCLUSION
The application on a greater scale of this newly developed method could be fundamental in the determination of the biological control potential in olive orchards.
Keywords: Dasineura oleae , Platygaster spp., Mesopolobus spp., koinobionts parasitoids, biological control, multiplex qPCR
We performed a traditional visual estimation of the parasitism rate and applied the molecular technique on the same specimens in order to compare these two techniques and optimize the quantification of biological control of this trophic system.

© 2022 The Authors. Pest Management Science published by John Wiley & Sons Ltd on behalf of Society of Chemical Industry.
1. INTRODUCTION
Several Cecidomyiidae species are pests of economic importance 1 , 2 , 3 , 4 , 5 and some of them are characterized by irregular outbreaks, leading to seasonal crop losses. Dasineura oleae (Angelini 1831) (Diptera: Cecidomyiidae) is a monophagous cecidomyid pest that attacks the olive tree, Olea europaea L. This species has been generally considered as a minor pest in Mediterranean olive orchards, but several outbreaks were recently recorded all over its native range. 6 , 7 , 8 , 9 The damage is caused by the larva that mines into the leaf and causes the formation of a gall, where it spends its life cycle until adult emergence in the spring of the following year. 9 Dasineura oleae galls interfere with some physiological processes of olive leaves and a massive infestation can lead to a precocious leaf falling. 7
The parasitoid complex of D. oleae comprises of 17 species of parasitoids, 7 , 10 mostly detected in Turkey. Up to now, in Italy, only a complex of species including Platygaster demades Walker 1835, Platygaster oleae Szelenyi 1940 (Hymenoptera: Platygastridae), Mesopolobus mediterraneus (Mayr 1903) and Mesopolobus aspilus (Walker 1835) (Hymenoptera: Pteromalidae) were recorded as parasitoids of D. oleae. 9 Traditional methods for parasitism assessment and parasitoid species discrimination include laboratory rearing of field‐collected material and host dissection followed by microscopic analyses of the parasitoids immature stages. 11 These methods are considered time‐consuming and include several problems. For example, laboratory rearing conditions 10 , 12 , 13 may greatly and unevenly affect the survival of both the host and the parasitoids, 14 , 15 leading to biased estimation of parasitization. Furthermore, parasitoid rearing is performed when both D. oleae and the parasitoids are about to emerge, and the time lag between field collection and adult emergence does not allow a rational application of integrated pest management (IPM) strategies and conservation program of parasitoid communities. Host dissection may allow a more precise estimation of the parasitism rate, but the taxonomic identification of the immature parasitoids is almost impossible. Many parasitoids are koinobionts 16 as in the case of D. oleae parasitoids detected in Italy. 9 They lay their eggs directly inside the host and parasitism becomes evident when D. oleae turns into the third instar larvae. Therefore, gall dissection and visual inspection of the larvae may provide an estimation of the parasitism rate when timely performed, however, these techniques do not discriminate between parasitoid species. Moreover, underestimation of parasitism rate may be common since the development rate of parasitoids and their host may differ from individual to individual, showing variable trends in the different infested areas and climatic conditions. With the use of sensitive molecular techniques, a precise estimation of parasitism rate could be achieved earlier than dissection or rearing methods, leading to a more efficient method of biological control evaluation. Parasitism assessment and the estimation of the relative abundance of the different parasitoids involved in D. oleae control are necessary to implement conservation biological control (CBC) strategies and to integrate this knowledge into IPM systems. In fact, assessing parasitoids’ presence and abundance can help to calibrate pest control strategies and the application of molecular techniques in this context can provide opportunities to improve CBC. 17 , 18 Molecular techniques are a useful tool to precisely evaluate the relative abundance of each parasitoid species, 15 , 19 , 20 , 21 , 22 , 23 , 24 overcoming taxonomic identification and time‐consuming rearing or dissection. To date, despite the potential to use a DNA‐based approach, molecular tools for the identification of D. oleae parasitoids have not been developed previously.
Here we aim to: (i) design a primer to detect specimens belonging to Platygaster and Mesopolobus genera, the most common parasitoids of D. oleae reported in central Italy 25 ; (ii) develop a protocol for multiplex quantitative polymerase chain reaction (qPCR) and fast samples DNA extraction; (iii) apply these newly developed protocols to field‐collected specimens and compare this method with traditional techniques such as laboratory rearing and visual estimation of parasitism rate on larvae exposed by gall dissection. Here we performed a traditional visual estimation of the parasitism rate and applied the molecular technique on the same specimens, allowing a precise comparison between these two techniques in order to optimize the quantification of biological control of this trophic system.
2. MATERIAL AND METHODS
2.1. Field samplings
Preliminary analyses of olive branches collected in olive orchards with high infestation of D. oleae was conducted early in spring, during 2018, to obtain parasitoids reference collection. Each branch was rolled with moisturized cotton wool and placed into a plastic jar, closed with fine gauze. Containers were checked daily for D. oleae and parasitoids emergence. Each emerged specimen was freeze‐killed, morphologically identified by an expert taxonomist, and individually stored in 1.5 mL Eppendorf tubes with absolute ethanol at −20 °C. Thirty‐two adult specimens of P. demades, 28 specimens of Mesopolobus spp. and 38 specimens of D. oleae adults were obtained, while no specimens of P. oleae were found.
Data collected showed that parasitism can be visually evidenced only when D. oleae reaches the stage of third instar larvae. Indeed, we sampled D. oleae galls in late February 2019, when it has been expected that most of the population developed into third instar larvae. This allowed both the early assessment of the parasitization level through molecular tools and the comparison of these results with the parasitization level assessed through the visual inspection of the larvae. Five organically managed olive orchards (cv. Frantoio, Leccino, Morcaio, Moraiolo) were selected in the Gavorrano district (Grosseto, Tuscany, Italy). Six olive trees were chosen and marked with a tag, along a transect from the core of the field to one edge confined with semi‐natural vegetation. Each sample consisted in two apical branches of five nodes that were stored in sealed plastic bags until examination. Up to 40 galls randomly selected on each sample were dissected under a stereoscope to expose the larvae. Gall dissection and larvae manipulation were performed under sterile conditions. Each larva was carefully examined to detect the presence of parasitization or other abnormalities, labeled and stored at −20 °C in absolute ethanol for parasitoids molecular detection. As parasitism is evident when third instar larvae develop, intact larvae are actively moving, are bright yellow and metamerism is clearly visible. However, third instar parasitized larvae contain one or more nearly developed parasitoids that may be evident throughout the larvae cuticle. The parasitoid’ puparia have the same dimensions of intact third instar larvae but look swollen and opaque. We assessed the parasitization rate and the relative abundance of each parasitoid genera by carrying out two additional samples later in the season (March) when both host and parasitoids are in the later development stages and are near to emerge. Two branches were randomly selected from the same trees sampled in February. For each sample, we randomly selected up to 40 galls and placed them in plastic jars in laboratory conditions. Each container was closed with fine gauze to increase air circulation and reduce mold growth. The branches were rolled in wet cotton wool in order to keep leaves turgid. After 10 days the number of emerged D. oleae, P. demades, P. oleae and Mesopolobus spp. were counted.
2.2. Molecular analysis
2.2.1. DNA extraction
Fast DNA extraction from single specimen was performed in 1.5 mL Eppendorf tube adding 100 μL of extraction buffer [phosphate‐buffered saline (PBS) pH 7.2 supplemented with 2% polyvinylpyrrolidone (PVP‐10), 0.2% diethyldithiocarbamate (DIECA)] and ten beads of 2.0 mm ø ZR BashingBead™ (Zymo Research, Irvine, CA, USA) to each sample. Samples were then homogenized with a Tissue Lyser II (Qiagen, Hilden, Germany), processed for 3 min at 30 Hz and used for qPCR amplification. The step‐by‐step procedure for fast DNA extraction is detailed in Table 1.
Table 1.
Step‐by‐step procedure for fast DNA extraction
|
1. Preparing samples
2. Preparing extraction tubes
3. Mechanically homogenizing samples and amplification
|
2.2.2. Primer design
Sixteen adult specimens for Platygaster and seven for Mesopolobus parasitoid genera were extracted and amplified using the universal primer pair LCO‐1490/HCO‐2198 26 targeting the 5′ portion of the cytochrome oxidase subunit I (COI). The PCR reaction mix was prepared in a final volume of 25 μL with 3 μL of fast extracted DNA, 5 μL of 5× colorless GoTaq® reaction buffers (Promega, Madison, WI, USA), 2.5 μL of magnesium chloride (MgCl2, 25 mmol L−1), 0.5 μL of mix dNTPs (10 mmol L−1), 1 μL of LCO‐1490 primer (10 μmol L−1), 1 μL of HCO‐2198 primer (10 μmol L−1), 0.2 μL of Go Taq DNA polymerase (5 U μL−1) and 11.8 μL of nuclease‐free water. The PCR program was characterized by denaturation for 3 min at 94 °C, followed by 30 cycles of amplification: 30 s at 94 °C, 40 s at 60 °C, and 1 min at 72 °C. A post‐PCR final elongation for 10 min at 72 °C closed the amplification process. Amplicons were separated by electrophoresis in 1% agarose gel and analyzed under ultraviolet (UV) light after staining with ethidium bromide.
PCR‐products were purified using Wizard® SV Gel and PCR Clean‐Up System Kit (Promega) and cloned using the pGEM®‐T Vector System (Promega) according to the manufacturer's protocol. For the ligation reaction 5 μL Rapid Ligation Buffer (2×), 1 μL pGEM® ‐T vector, 1 μL T4 DNA ligase and 3 μL of DNA were incubated overnight at 4 °C. Escherichia coli cells (strain MC1022) were transformed by electroporation (Eppendorf® Electroporator 2510 at 2500 V), plated on LB‐plates containing ampicillin (100 μg mL−1), X‐Gal (80 μg m−1) and isopropyl β‐d‐1‐thiogalactopyranoside (IPTG, 0.5 mmol L−1) then incubated overnight at 37 °C. Recombinant plasmids were isolated using the Wizard® Plus SV Minipreps DNA Purification System (Promega) following the manufacturer's instructions and sequenced by Eurofins Genomics (Ebersberg, Germany). Given that most of the sequences published in the GenBank are labeled as Platygaster sp. and Mesopolobus sp. it was not possible to identify information related to each species of the Italian parasitoid complex, in the DNA sequences alignments for primer design. In fact, up to now no COI sequences are available nor for P. oleae nor for M. aspilus or M. mediterraneous. However, in order to increase the reliability of primers for parasitoid detection, sequences obtained from our specimens (GeneBank accession numbers: from MW703634 to MW703656) were aligned with those from Mesopolobus spp. (n = 312) and Platygaster spp. (n = 710) available in GenBank.
A conserved region of COI of each target genera was selected to design reverse primers, while forward primers were created to be more specific and anneal with the COI sequences of our specimens. Six forward and two reverse primers were designed using Primer Express 3.0 software (Life Technologies, Carlsbad, CA, USA) for the detection of Platygaster and Mesopolobus specimens as reported in Table 2.
Table 2.
Primer description
| Oligo name | Sequence 5′‐3′ | T m (°C) | GC‐content (%) |
|---|---|---|---|
| Meso 90F | CGTTTAGAATTGGGWAAYCCTG | 57.5 | 43.2 |
| Meso 308F I | ATTAATATTACTAATTTCWAGWATATTT | 50.5 | 10.7 |
| Meso 308F II | ATTAATACTACTGATTTCAAGWATATTT | 53.4 | 17.9 |
| Meso 518R | GGAATATTTTCWATTTTAWRAATTT | 49.0 | 14 |
| Platy 92F | ATTAGAATTAGGGACACCTTC | 54.0 | 38.1 |
| Platy 225F I | TTARTTCCTTTAATAWTATC | 44.0 | 17.5 |
| Platy 225F II | CTTRTACCATTAATATTRTC | 47.0 | 25 |
| Platy 344R | CCTGCTCCAAAAATAYTWCTRT | 54.7 | 36.4 |
Note: T m, melting temperature; GC‐content, guanine and cytosine content.
2.2.3. Primer specificity
The specifity of the primer sets was initially tested in separate PCR reactions with DNA of 16 Platygaster spp. and seven Mesopolobus spp. specimens, as well as with DNA from five adults and five larvae of D. oleae. We focused on these species because they constitute the parasitoid complex of D. oleae in our country, 9 no other parasitoid species have been recorded until now.
Different thermocycling conditions were evaluated to find the optimum reaction settings for PCR amplification and identify the most suitable primer pairs for our study. From the set of primers assayed, two forward and two reverse primers were selected for the detection of Platygaster (Platy 92F/Platy 344R) and Mesopolobus genera (Meso 90F/Meso 518R). Cycling conditions consisted of an initial denaturation step at 94 °C for 2 min, followed by 35 cycles of amplification: 10 s at 94 °C, 10 s at 37–45 °C, and 45 s at 72 °C, with ramping rates of 0.5 °C s−1. A post‐PCR final elongation for 2 min at 72 °C closed the amplification process. Amplicons were separated by electrophoresis in 1% agarose gel and analyzed under UV light after staining with ethidium bromide. All combinations of primers were also tested in multiplex qPCR confirming what was seen by previous single PCR reactions. For a detailed description of amplification settings refer to the specification provided in Section 2.2.5.
2.2.4. Primer sensitivity
Primers pairs were selected with the aim to develop a practical tool for rapidly assessing the potential of biological control in olive orchards by providing a ‘snapshot’ of the parasitism rate at the beginning of the season. Dasineura oleae galls were collected in early February when mainly third instar larvae are present in the field. Platygaster spp. are egg‐larval parasitoids with one generation per year synchronized with the host life cycle. Based on the scarce literature available 10 and as confirmed by our experience, embryonic development of this parasitoid genera occurred in correspondence of third instar host. Thus, the probability of having missing Platygaster spp. detection is very low, given that molecular analysis revealed parasitized second instar larvae. However, no literature is available regarding Mesopolobus spp. development in D. oleae host. For this reason, primer sensitivity was also directly tested by multiplex qPCR with serial ten‐fold dilutions (from 10−1 to 10−5) of DNA extracted from P. demades and Mesopolobus spp. and diluted in D. oleae template.
2.2.5. Multiplex qPCR assay
PCR reaction was performed in a multiplex format in a final volume of 20 μL using the KAPA SYBR® FAST Universal Kit (KAPA Biosystems, Wilmington MA, USA). The multiplex qPCR protocol was optimized by adjusting cycling conditions and testing different concentrations for each combination of primers (0.5, 1 and 2 μL). Optimal reaction condition used 10 μL Master Mix (2×), 0.25 μL High ROX, 1 μL each of Platy 92F/Platy 344R (10 μmol L−1), 2 μL each of Meso 90F/Meso 518R (10 μmol L−1), 3 μL of DNA template and 0.75 μL of nuclease free water.
The amplification settings included an initial activation of the TaqDNA polymerase at 95 °C for 10 min, followed by 40 cycles of 15‐s denaturation at 95 °C, 10‐s annealing at 50 °C and 60 s extension at 60 °C. Each amplification run included positive and negative controls.
After amplification was complete, a final melting curve was recorded by heating to 95 °C for 15 s and then cooling to 60 °C for 20 s before heating slowly until a temperature of 95 °C was attained in 20 min. Fluorescence was measured continuously during the slow temperature rise to monitor the dissociation. The real‐time PCR data were analyzed with ABI PRISM 7000 SDS v1.2.3 data analysis software (Thermo Fisher Scientific, Waltham, MA, USA). The resulting melting curves were converted into their negative first‐order derivatives, which allows the melting temperature (T m) of the amplicons to be determined from the peak. The entire process required about 30 min.
2.3. Data analysis
The difference between the two methods for parasitization level evaluation (multiplex qPCR and visual inspection of the larvae) was assessed using McNemar’s test for paired nominal data. The test was applied to the entire sample, then we also evaluated the difference between the traditional and molecular methods for each larval stage and each olive orchard. We calculated the percentage of parasitization for each method using the formula:
where P s is the number of parasitized specimens and M is the number of unparasitized D. oleae larvae. 10 For the percentages of parasitization, the binomial confidence interval was calculated.
3. RESULTS
Amplification signals were detected in qPCR reactions when selected primer pairs Platy 92F/Platy 344R and Meso 90F/Meso 518R were used in a single or multiplex qPCR reaction. Peaks of specific T m values were observed after the dissociation curves analyses, considered a standard approach to discriminate qPCR products in multiplex reaction, allowing us to distinguish among Platygaster spp. (T m = 74.8°C) and Mesopolobus spp. (T m = 76.6°C) (Fig. 1). Moreover, fragments of the expected sizes (252 and 428 bp, respectively) were shown when qPCR reactions were analyzed by agarose electrophoresis (data not shown). Amplification signal was also observed with DNA from non‐parasitized D. oleae, however it has been clearly characterized by different dissociation curves and T m (68.2°C) (Fig. 1). Additionally, both sets of primer showed good sensitivity with a DNA detection limit of 10−3 for Platygaster spp. and 10−2 for Mesopolobus spp.
Figure 1.
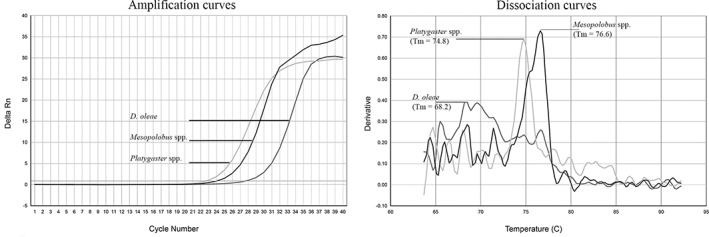
Amplification curves (left panel) and dissociation curves analysis (right panel) obtained by multiplex qPCR on DNA extracted from Platygaster spp., Mesopolobus spp. and Dasineura oleae samples. Both parasitoids and host are easily discriminated from each other by the different melting temperature (T m) and shape of the dissociation curves.
Overall, 979 D. oleae galls were dissected and larvae were visually inspected and subsequently subjected to molecular analyses (Table 3). Of these, 106 (10.82%) were considered as parasitized by visual inspection, while qPCR detected 189 larvae parasitized by Platygaster spp. (19.31%) (Fig. 2). On the one hand, 89.27% of the results obtained by visual inspection were confirmed by the molecular analyses and a further 9.6% of parasitism was detected by PCR. On the other hand, 1.12% of the sample (n = 11) were considered as parasitized by visual inspection, but parasitism was not confirmed by the molecular tests. McNemar's test revealed a significant difference of parasitization (%) between the two methods (χ 2 = 64.038, df = 1, P < 0.001).
Table 3.
Number of analyzed larvae grouped by category (second instar, third instar and ‘other’) and by sampling site (olive orchard)
| Olive orchard | Second instar larvae | Third instar larvae | Other | Total |
|---|---|---|---|---|
| 1 | 35 | 84 | 15 | 134 |
| 2 | 13 | 184 | 10 | 207 |
| 3 | 18 | 174 | 1 | 193 |
| 4 | 5 | 203 | 1 | 209 |
| 5 | 10 | 224 | 2 | 236 |
| Total | 81 | 869 | 29 | 979 |
Figure 2.
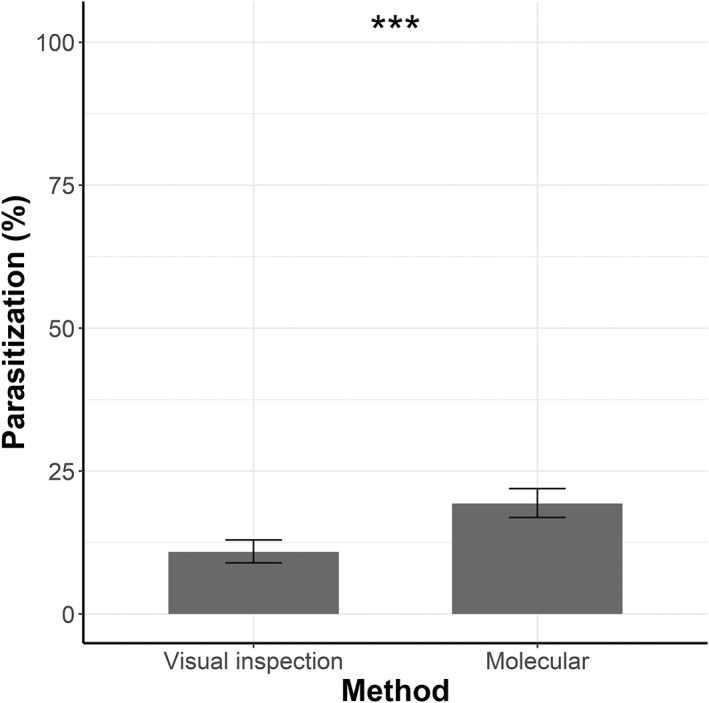
Parasitization (%) of Dasineura oleae larvae, according to the visual inspection and to the molecular analyses (sample size: 979 larvae). Binomial confidence intervals are reported. ***, P < 0.001, according to McNemar's test.
Most of the analyzed larvae were third instars (88.76%), 8.27% (81 specimens) were second instar larvae and 2.96% specimens were categorized as ‘other’ due to their particular appearance (dead or dark larvae, larvae attacked by fungi). McNemar's test revealed a significant difference of parasitization (%) (χ 2 = 25.037, df = 1, P < 0.001) between the two techniques for second instar larvae (Fig. 3). Indeed, 2.47% (n = 2) resulted parasitized when visually inspected, while molecular analyses revealed 35.80% (n = 29) of parasitism rate. A significant difference in parasitization (%) between the two methods was also detected for third instar larvae (χ 2 = 26.266, df = 1, P < 0.001) (Fig. 3). Indeed, 11.97% of third instar larvae (n = 104) presented signs of parasitization when visually analyzed at the stereoscope, while PCR detected 16.80% of parasitized specimens (n = 146): 10.70% is due to validation of parasitism detected through visual inspection and an additional 6.1% is due to further 53 parasitized specimens that were not detected by visual inspection. However, 1.26% of third instar larvae appeared to be parasitized when visually inspected, but no parasitoids were detected by molecular analysis.
Figure 3.
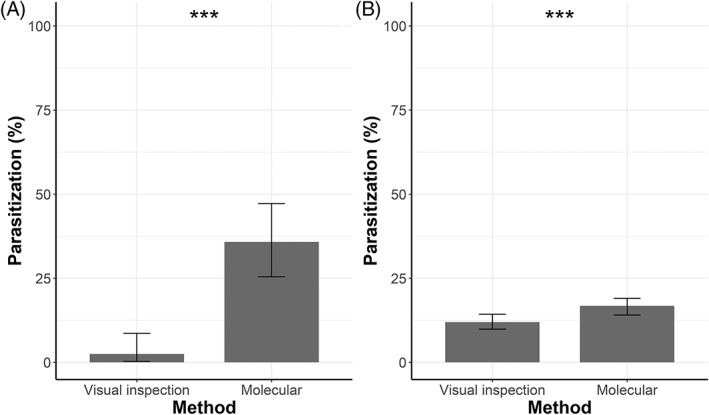
Percentage of parasitization with a binomial confidence interval of second (A) and third (B) instar Dasineura oleae larvae according to visual inspection and molecular analyses. ***, P < 0.001, according to McNemar's test.
Additionally, 29 larvae were categorized as ‘other’ and due to their appearance were considered not parasitized but molecular analyses detected 48.28% of parasitization (n = 14).
The percentage of parasitization in the different sampling sites, according to the two different methods, is shown in Fig. 4. Results obtained by the two methods significantly differ in olive orchard 1 (χ 2 = 41.49, df = 1, P < 0.001), olive orchard 2 (χ 2 = 17.455, df = 1, P < 0.001) and olive orchard 5 (χ 2 = 5.7857, df = 1, P = 0.016), but do not present relevant differences in olive orchards 3 and 4. In particular, in olive orchard 1, parasitization assessed by visual inspection was 33.58% (n = 45) while molecular analyses detected 68.66% (n = 92) of parasitized larvae. The molecular technique did not confirm parasitism assessed by visual inspection just in 1.49% (n = 2) of the samples. Similarly, in olive orchard 2, the molecular technique detected 33.82% (n = 70) of parasitized larvae, while parasitism assessed by visual inspection was 21.74% (n = 45). Here, molecular analyses did not validate 1.93% (n = 4) of the analyzed larvae that were considered as parasitized by visual inspection. In olive orchard 5, parasitism was 4.24% (n = 10) and 8.47% (n = 20), by visual inspection and molecular technique, respectively. Just in 0.84% (n = 2) of the cases the parasitism assessment done by visual inspection was not confirmed by the molecular analyses.
Figure 4.
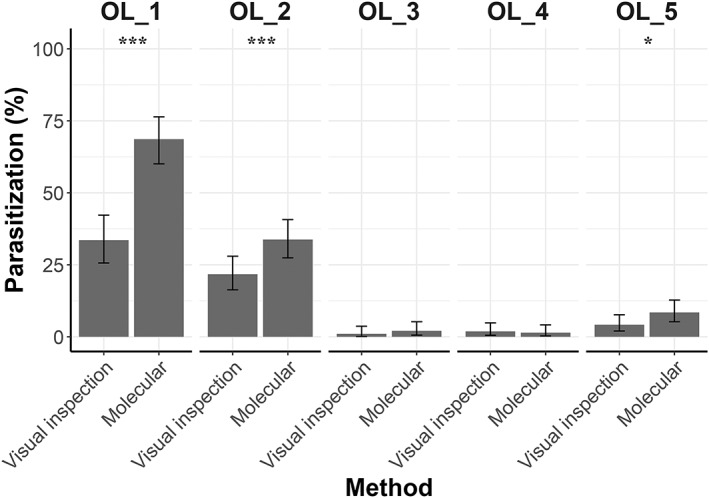
Percentages of parasitization with binomial confidence interval is reported for each olive orchard (‘OL’ in the figure) according to the two used methods. Significant differences between the two methods are reported ***, P < 0.001; *, P < 0.05, according to McNemar's test. Sampling size in OL_1 = 134; OL_2 = 207; OL_3 = 193; OL_ 4 = 209; OL 5 = 236.
Laboratory reared infested branches failed to provide a reliable estimation of parasitism rate and relative abundance of the key parasitoids Platygaster spp. and Mesopolobus spp. Indeed, high mortality rates were recorded for both the first (87.86%) and second (63.46%) sampling date. Platygaster demades represented 4.92% (n = 6) of emerged specimens of the first sampling date and 2.52% (n = 9) of the second sampling date and it was detected in olive orchard 1 (seven specimens), olive orchard 2 (six specimens) and olive orchard 5 (two specimens). Five Mesopolobus spp. (1.4% of emerged specimens) emerged from the infested branches collected on the second sampling date from olive orchard 5 (four specimens) and olive orchard 3 (one specimen).
4. DISCUSSION AND CONCLUSIONS
Here we report the first development and application on a large‐scale field trial of a molecular method to detect the most common parasitoids genera (Platygaster spp. and Mesopolobus spp.) of D. oleae in Italian olive orchards. The protocol that has been developed during this study allows to evaluate parasitization rate quickly by a simple molecular technique based on a multiplex qPCR assay. All procedures included in the protocol were effective and timesaving, allowing analysis of numerous samples simultaneously.
The lack of sequences on GenBank relative to P. oleae and M. aspilus, together with the scarce information on M. mediterraneus, did not allow us to obtain species‐specific primers for the four most common parasitoid species reported in Italian olive orchards. Reliability of sequences obtained from our specimens have been consolidated by comparation to representatives of genetic variability available on GenBank for both Platygaster (Supporting Information Fig. S1) and Mesopolobus (Fig. S2) genera. All Italian sequences from Platygaster specimens grouped together with P. demades samples from Canada showing nucleotide variability within group ranging from 92.1% to 97.0% (data not shown). Additionally, five of the sequences from Italian Mesopolobus specimens formed a clade with the M. mediterraneus isolate from the United Kingdom, with nucleotide identity ranging between 94.5% and 99.2% (data not shown), confirming the consistency of sequences used to design primers. Interestingly, two Italian Mesopolobus specimens showed higher nucleotide identity (from 84.0% to 88.2%, data not shown) with M. lichtensteini accessions from Spain suggesting a species variability in the olive orchards surveyed.
Primers designed under the alignments described anneal with the COI sequences of Mesopolobus and Platygaster adult specimens and discriminate very clearly between parasitized and non‐parasitized host samples. The qPCR assay on the field‐collected specimens showed the presence of Platygaster spp. in all parasitized samples, confirming the importance of this group in the biological control of D. oleae. 25 Even though Pteromalidae are not abundant in olive orchards of the study area 25 and few specimens were obtained from laboratory‐reared infested olive branches, detection of some parasitism by Mesopolobus spp. by molecular assay was expected. Moreover, M. aspilus and M. mediterraneus are knowns as polyphagous species, 27 and are recorded as endoparasitoids of D. oleae. 7 Despite that, molecular results show a lack of parasitism by Mesopolobus spp in D. oleae larvae in the assayed olive orchards, suggesting that these species may be a pupal parasitoid of D. oleae.
The application of the newly developed protocol on larvae from field‐collected galls of D. oleae demonstrated that the molecular approach is more sensible than visual inspection, detecting almost double parasitization when compared to traditional methods. This is in agreement with previous studies showing that molecular method may provide parasitization estimation similar to traditional methods 21 or considerably higher, leading to three‐fold times the parasitism estimated by traditional methods. 22 The magnitude of difference between the methods presumably depends on the biology of the studied species, the sensitivity of the used techniques as well as the development stage of both the pest and the parasitoid. 28 Indeed, our results show that significant differences between procedures were obtained in both the second and third stage‐larva, with a particularly large gap in the second stage, for which qPCR revealed more than ten‐fold times the parasitism rate estimated by visual inspection. This result can be explained by the fact that, usually, parasitization is not evident until D. oleae develop into third stage‐larva. This evidence also highlights that using traditional methods may lead to serious parasitization underestimation in the case of a high proportion of second instar larvae in the sample. Our result also shows the great potential of the developed molecular protocol to detect parasitism by Platygaster spp. in the pest early stages. Indeed, PCR‐based methods are proved to be very sensitive and may be able to detect parasitism 1 h after the parasitoid oviposition. 29 Our field sampled specimens also included a small fraction of larvae attacked by fungi, dead or dark larvae with no evident signs of parasitism. The qPCR revealed that nearly half of them were parasitized, suggesting that parasitism is still detectable, with molecular techniques, despite natural mortality. Overall, qPCR confirmed the result obtained by visual inspection and revealed some additional parasitism that the traditional method cannot detect. However, a small fraction of specimens showed signs of parasitism when visually inspected, that was not detected by the molecular analyses. We can hypothesize the presence of a rare parasitoid species that was not observed in previous studies on this pest in Italy and that did not emerge from laboratory‐reared infested branches.
Our results show significant differences in parasitism assessment by visual inspection and molecular method in the orchards that presented higher parasitism rates. In olive orchard 1 and in olive orchard 5, parasitization estimation by qPCR is more than double the estimation performed by visual inspection, while this discrepancy between the two methods decreases in orchards with lower parasitization. This suggests that the underestimation of parasitization may be greater in highly parasitized orchards, underlining that parasitization estimation by traditional method could lead to the recommendation of the wrong pest control strategies.
The laboratory rearing of D. oleae parasitoid showed that this technique is poorly suitable for D. oleae parasitization assessment, indeed high mortality rates under laboratory conditions may be common. 28 , 30 This method is time‐consuming also requiring large space availability and employment of specialized personal. However, despite the evidence of the better performance of the PCR‐based parasitization estimation, laboratory rearing demonstrated to be useful for the first/preliminary identification of the parasitoid species of pests in the geographical area considered.
In conclusion, we developed a new multiplex qPCR protocol coupled to fast DNA extraction to estimate parasitism in D. oleae and we compared its performance with traditional methods. Field results show that PCR‐based method provides a faster and more accurate estimation of parasitization than visual inspection of the larvae. The application on a greater scale of this newly developed method could be fundamental in the determination of the biological control potential in olive orchards and therefore in the implementation of the CBC and IPM strategies. Further studies should confirm the reliability of this method on a larger sample of second instar larvae, also considering the age of the larvae (e.g. young second instar larvae in summer and mature second instar larvae in autumn). This perspective would allow an estimation of the parasitization rate several months in advance if compared to traditional methods, enabling the optimization of pesticide use against D. oleae and therefore preserving parasitoid communities. A larger‐scale application of this method would also allow the definition of a threshold of parasitization rate (according to the ‘threshold for success’ of biological control 31 above which chemical control is no longer necessary.
AUTHOR CONTRIBUTIONS
SM: data curation, investigation, methodology, writing – original draft preparation; ET: data curation, investigation, methodology, writing – original draft preparation; GB: supervision, resources, writing – reviewing and editing; CR: supervision, resources, data curation, writing – reviewing and editing; RP: conceptualization, methodology, resources, writing – reviewing and editing.
Supporting information
Figure S1. Phylogenetic trees showing the relationship between the Italian Platygaster specimens (marked by *) and representatives of genetic variability for the genus available on GenBank. Tree was reconstructed using Neighbor‐joining method available in Geneious Prime® 2021.0.3 (Biomatters Ltd, Auckland, New Zealand) with Tamura‐Nei nucleotide substitution model, and support for individual nodes was assessed using a bootstrap procedure (1000 replicates). Bootstrap values equal or greater than 50% are shown on the branch nodes. Genus, species (when available), country of origin and GenBank accession numbers are reported
Figure S2. Phylogenetic trees showing the relationship between the Italian Mesopolobus specimens (marked by *) and representatives of genetic variability for the genus available on GenBank. Tree was reconstructed using Neighbor‐joining method available in Geneious Prime® 2021.0.3 (Biomatters Ltd, Auckland, New Zealand) with Tamura‐Nei nucleotide substitution model, and support for individual nodes was assessed using a bootstrap procedure (1000 replicates). Bootstrap values equal or greater than 50% are shown on the branch nodes. Genus, species (when available), country of origin and GenBank accession numbers are reported
ACKNOWLEDGEMENTS
The authors would like to thank Alice Albertini for advice and support in the conceptualization of the initial phase of this study, Chiara Lanzoni and Mattia Dall'Ara for the assistance in laboratory activities and the Prof. Peter Neerup Buhl for the identification of parasitoids.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Barnes HF, Gall midges of economic importance, in Vol. VII of the Gall Midges of Cereal Crops. Lockwood & Son Ltd, London, p. 261 (1956). [Google Scholar]
- 2. Barnes HF, Gall midges of economic importance, in Vol. III of the Gall Midges of Fruit. Lockwood & Son Ltd, London, pp. 184 (1948). [Google Scholar]
- 3. Hahn NG and Isaacs R, Distribution and phenology of Dasineura oxycoccana (Diptera: Cecidomyiidae) in Michigan blueberries. Environ Entomol 41:455–462 (2012). [DOI] [PubMed] [Google Scholar]
- 4. Skuhravý V, Skuhravá M and Brewer J, Evaluation of plant damage caused by three species of gall midges (Diptera: Cecidomyiidae). Z Fuer Angew Entomol 90:184–190 (1980). [Google Scholar]
- 5. Yang, WQ , Blueberry gall midge: a possible new pest in the northwest: identification, life cycle, and plant injury, in OSU extension service publications. EM 8889 (2005).
- 6. Batta YA, New findings on infestation and phenology of Dasineura oleae Angelini (Diptera, Cecidomyiidae): an emerging pest on olive trees in the Palestinian territories. J Plant Dis Prot 126:55–66 (2019). [Google Scholar]
- 7. Doğanlar M, Parasitoids complex of the olive leaf gall midges, Dasineura oleae (Angelini 1831) and Lasioptera oleicola Skuhravá, 2011 (Diptera: Cecidomyiidae) in Hatay Turkey, with descriptions of new genus and species from Tetrastichinae (hymenoptera: Eulophidae). Türkiye Entomoloji Derg 35:245–264 (2011). [Google Scholar]
- 8. Simoglou K, Karataraki A, Roditakis N and Roditakis E, Euzophera bigella (Zeller)(Lepidoptera: Pyralidae) and Dasineura oleae (F. low)(Diptera: Cecidomyiidae): emerging olive crop pests in the Mediterranean? J Pest Sci 85:169–177 (2012). [Google Scholar]
- 9. Tondini E and Petacchi R, First observations on the parasitoid complex and on the biology of Dasineura oleae during an outbreak in Tuscany, Italy. Bull Insectol 72:93–102 (2019). [Google Scholar]
- 10. Sampson BJ, Rinehart TA, Liburd OE, Stringer SJ and Spiers JM, Biology of parasitoids (hymenoptera) attacking Dasineura oxycoccana and Prodiplosis vaccinii (Diptera: Cecidomyiidae) in cultivated blueberries. Ann Entomol Soc Am 99:113–120 (2006). [Google Scholar]
- 11. Roubos CR and Liburd OE, Parasitism of Dasineura oxycoccana (Diptera: Cecidomyiidae) in north Central Florida. Environ Entomol 42:424–429 (2013). [DOI] [PubMed] [Google Scholar]
- 12. Baidaq Z, Ramadhane A and Tara R, Biological synchronization of the endoparasitoid Platygaster demades Walker (hymenoptera: Platygasteridae) with its host the olive leaf midge Dasineura oleae F. Loew (Diptera: Cecidomyiidae). Int J Agric Environ Sci 2:1–8 (2015). [Google Scholar]
- 13. Yegorenkova E and Yefremova Z, Notes on Lasioptera rubi (Schrank)(Diptera: Cecidomyiidae) and its larval parasitoids (hymenoptera) on raspberries in Russia. Entomol Fenn 27:15–22 (2016). [Google Scholar]
- 14. Day W, Estimating mortality caused by parasites and diseases of insects: comparisons of the dissection and rearing methods. Environ Entomol 23:543–550 (1994). [Google Scholar]
- 15. Sow A, Brévault T, Benoit L, Chapuis M‐P, Galan M, Coeur d'acier A et al., Deciphering host‐parasitoid interactions and parasitism rates of crop pests using DNA metabarcoding. Sci Rep 9:1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goater TM, Goater CP and Esch GW, Parasitism: The Diversity and Ecology of Animal Parasites. Cambridge University Press, Cambridge: (2014). [Google Scholar]
- 17. Gardiner MM, Fiedler AK, Costamagna AC and Landis DA, Integrating conservation biological control into IPM systems, in Integrated Pest Management. Concepts, Tactics, Strategies and Case Studies, ed. by Radcliffe EB, Hutchison WD and Cancelado RE. Cambridge University Press, Cambridge, pp. 151–162 (2009). [Google Scholar]
- 18. Gurr GM and You M, Conservation biological control of pests in the molecular era: new opportunities to address old constraints. Front Plant Sci 6:1255 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Franck P, Maalouly‐Matar M and Olivares J, Molecular tools for the detection and the identification of hymenoptera parasitoids in tortricid fruit pests. Int J Mol Sci 18:2031 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frey JE, Frey B and Baur R, Molecular identification of the swede midge (Diptera: Cecidomyiidae). Can Entomol 136:771–780 (2004). [Google Scholar]
- 21. Gariepy T, Kuhlmann U, Gillott C and Erlandson M, Parasitoids, predators and PCR: the use of diagnostic molecular markers in biological control of arthropods. J Appl Entomol 131:225–240 (2007). [Google Scholar]
- 22. Papura D, Rusch A, Roux P, Delbac L and Thiéry D, Early detection and identification of larval parasitoids in Lobesia botrana using PCR‐RFLP method. Biol Control 103:95–100 (2016). [Google Scholar]
- 23. Pook VG, Athey KJ, Chapman EG, Clutts‐Stoelb SA and Sharkey MJ, New PCR primers enhance investigation of host‐parasitoid food webs. Entomol Exp Appl 162:309–314 (2017). [Google Scholar]
- 24. Singh S, Mishra V and Bhoi TK, Insect molecular markers and its utility‐a review. Int J Agric Environ Biotechnol 10:469–479 (2017). [Google Scholar]
- 25. Tondini, E , Sommaggio, D , Monteforti, G and Petacchi, R , Conservation Biological Control in olive orchards during an outbreak of Dasineura oleae: local and landscape effects on parasitoid communities. Manuscript in preparation (2021).
- 26. Folmer O, Hoeh W, Black M and Vrijenhoek R, Conserved primers for PCR amplification of mitochondrial DNA from different invertebrate phyla. Mol Mar Biol Biotechnol 3:294–299 (1994). [PubMed] [Google Scholar]
- 27. Graham MWRV, The Pteromalidae of North‐Western Europe (hymenoptera: Chalcidoidea). Bull Brit Mus (Nat Hist) (Entomol) Suppl 16:1–908 (1969). [Google Scholar]
- 28. Gariepy T, Kuhlmann U, Gillott C and Erlandson M, A large‐scale comparison of conventional and molecular methods for the evaluation of host–parasitoid associations in non‐target risk‐assessment studies. J Appl Ecol 45:708–715 (2008). [Google Scholar]
- 29. Zhu YC, Riddick EW, Williams L, Schotzko DJ, Logarzo GA and Jackson CG, Potential of detection and identification of nymphal parasitoids (hymenoptera: Braconidae) of Lygus bugs (Heteroptera: Miridae) by using polymerase chain reaction and ITS2 sequence analysis techniques. Ann Entomol Soc Am 97:743–752 (2004). [Google Scholar]
- 30. Agusti N, Bourguet D, Spataro T, Delos M, Eychenne N, Folcher L et al., Detection, identification and geographical distribution of European corn borer larval parasitoids using molecular markers. Mol Ecol 14:3267–3274 (2005). [DOI] [PubMed] [Google Scholar]
- 31. Hawkins BA and Cornell HV, Maximum parasitism rates and successful biological control. Science 266:1886–1887 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phylogenetic trees showing the relationship between the Italian Platygaster specimens (marked by *) and representatives of genetic variability for the genus available on GenBank. Tree was reconstructed using Neighbor‐joining method available in Geneious Prime® 2021.0.3 (Biomatters Ltd, Auckland, New Zealand) with Tamura‐Nei nucleotide substitution model, and support for individual nodes was assessed using a bootstrap procedure (1000 replicates). Bootstrap values equal or greater than 50% are shown on the branch nodes. Genus, species (when available), country of origin and GenBank accession numbers are reported
Figure S2. Phylogenetic trees showing the relationship between the Italian Mesopolobus specimens (marked by *) and representatives of genetic variability for the genus available on GenBank. Tree was reconstructed using Neighbor‐joining method available in Geneious Prime® 2021.0.3 (Biomatters Ltd, Auckland, New Zealand) with Tamura‐Nei nucleotide substitution model, and support for individual nodes was assessed using a bootstrap procedure (1000 replicates). Bootstrap values equal or greater than 50% are shown on the branch nodes. Genus, species (when available), country of origin and GenBank accession numbers are reported
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


