Abstract
Fungal pathogens induce a variety of diseases in both plants and post-harvest food crops, resulting in significant crop losses for the agricultural industry. Although the usage of chemical-based fungicides is the most common way to control these diseases, they damage the environment, have the potential to harm human and animal life, and may lead to resistant fungal strains. Accordingly, there is an urgent need for diverse and effective agricultural fungicides that are environmentally- and eco-friendly. Plants have evolved various mechanisms in their innate immune system to defend against fungal pathogens, including soluble proteins secreted from plants with antifungal activities. These proteins can inhibit fungal growth and infection through a variety of mechanisms while exhibiting diverse functionality in addition to antifungal activity. In this mini review, we summarize and discuss the potential of using plant antifungal proteins for future agricultural applications from the perspective of bioengineering and biotechnology.
1. Introduction
The management of plant diseases is one of the top priorities of the agricultural industry due to the major economic and biosecurity threats that result from plant pathogens [1]. Among the pathogen cortege that crops are afflicted by, fungal infections pose one of the largest risks to food production [2]. Devastating crop failures due to these pathogens, such as the historical and infamous Irish Potato Famine [3] and contemporary issues of rice blast and wheat rust threaten food security and result in major economic losses [4]. The development of fungicides has undoubtedly eased the burden of diminished food security through a reduction in crop failures by successfully controlling fungal diseases. Chemical fungicides, made from either organic or inorganic chemicals, remain as the primary treatment towards most fungal pathogens [5]. However, chemical fungicides have long been documented for their adverse effects on both the environment and animal health [6], and harvested crops must meet strict criteria to ensure chemical residues are found at safe levels for consumptions [7]. Though conventional fungicides have made positive strides in food security and agricultural disease control, the risks they carry need to be addressed and alternative methods of fungal control should be considered.
One common alternative to conventional fungicides is the usage of genetically modified (GM) crops. Transgenic technology has led to the development of crops with desirable traits, such as improved flavor [8], increased yield [9], and superior disease resistance [10] compared to non-modified crops. Notably, the use of transgenic crops permits for a significant reduction in the quantity of phytosanitary product applied to the field [11]. However, the public is often apprehensive about GMO safety and has difficulty accepting genetically modified crops [12]. For example, some consumers believe that GM crops carry more risks than benefits and are willing to pay a premium for foods labeled as non-GMO [13]. Likewise, since 2001, the EU has placed a de facto moratorium on approvals of GMOs [14]. Another major concern includes the potential that transgenic crops could damage the ecosystem in unpredictable ways. GMOs can invade ecosystems due to an increase in stress tolerance, causing wild plants to become weeds through horizonal gene transfer [15], or produce toxic substances to pests that may affect nontarget organisms [16]. Recently, increases in pest resistance towards GM crops have also posed problems to the durability of current transgenic crops [17].
Thus, it is necessary to seek alternative antifungal agent candidates that can be applied exogenously as conventional fungicides. These alternative candidates should be environmentally friendly and potentially have fewer negative health impacts on animals than conventional fungicides if applied exogenously. Plants have evolved diverse mechanisms to defend against fungal infections, as summarized in Fig. 1, with one important route utilizing the secretion of proteins to delay fungal infection or inhibit fungal growth. These plant antifungal proteins are promising candidates since they are biodegradable, generally nontoxic to humans and antagonistic microorganisms, and most importantly, have evolved for millions of years to combat phytopathogenic fungi with a narrow target range [18]. In this mini review, we summarize and discuss plant defensive proteins that are promising candidates for the development of future antifungal agents for agricultural applications (as summarized in Table 1).
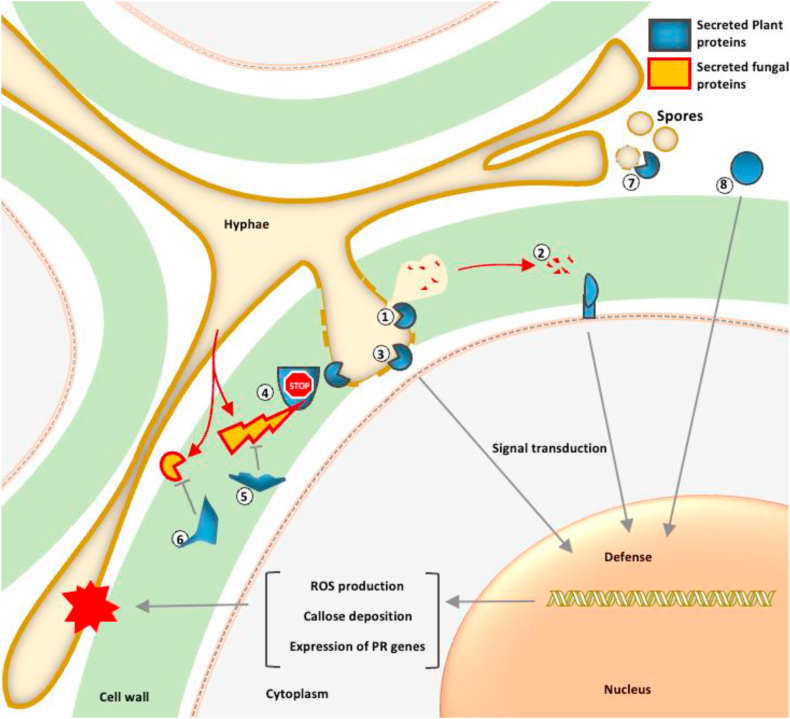
Fig. 1.
Mode of actions of secreted plant antifungal proteins with potential agricultural applications. 1) Secreted antifungal proteins reduce fungal hyphae growth by compromising the fungal cell wall and membrane integrity, leading to potential cytoplasmic leakages [165]. 2) Antifungal protein activity generates residues considered as microbe-associated molecular pattern molecules that can be recognized by plant receptors to stimulate plant immune response [166]. 3) Plant antifungal proteins, upon interacting with the target, directly stimulate plant immune response [167]. 4) Plant secreted proteins protect antifungal proteins from cleavage by fungal protease [168]. 5) Inhibition of fungal protease by plant secreted inhibitors [169]. 6) Inhibition of fungal cell wall hydrolase by plant secreted inhibitors [170]. 7) Spore degradation or reduction of germination rate by secreted plant antifungal proteins [171]. 8) Small secreted peptides enhance the efficacy of plant defense [172].
Table 1.
Summary of antifungal proteins of potential to be developed into alternative fungi control agents for agricultural applications.
| Protein Class | Protein | Exogenous Application Inhibition | Antifungal Mechanism | Ref | |
|---|---|---|---|---|---|
| Pathogenesis Related Proteins | Chitinase | Alternaria sp. |
|
[25,27,36,43,[154], [155], [156], [157], [158]] | |
| Aspergillus niger | |||||
| Botrytis cinerea | |||||
| Collectrichum falacatum | |||||
| Fusarium sp. | |||||
| Pestaloatia theae | |||||
| Rhizoctania solani | |||||
| Sphaerotehco humuli | |||||
| Trichoderma sp. | |||||
| Defensins | NRBAP | Mycosphaerella arachidicola |
|
[50] | |
| MsDef1 | Fusarium graminearum |
|
[54] | ||
| NaD1 | Fusarium oxysporum f.sp vasinfectum |
|
[56,57] | ||
| Thielaviopsis basicola | |||||
| Aspergillus nidulans | |||||
| Leptospheria maculan | |||||
| RsAFP2 | Fusarium culmorum |
|
[58] | ||
| Nectria haematococca | |||||
| Verticilium dahlia | |||||
| Phoma betae | |||||
| Osmotin and Osmotin-Like Proteins | Alternaria solani |
|
[62,[69], [70], [71],159] | ||
| Biopolaris maydis | |||||
| Bioplaris zeicola | |||||
| Cerospora zeae-maydis | |||||
| Colletotrichum laginarium | |||||
| Colletotrichum sublineolum | |||||
| Fusarium graminearum | |||||
| Fusarium moniliforme | |||||
| Fusarium oxysporum | |||||
| Fusarium roseum | |||||
| Kabatiella seae | |||||
| Phytophthora infestans | |||||
| Phytophtora parasitica | |||||
| Sclerotinia sclerotiorum | |||||
| Stenocarpella maydis | |||||
| Trichoderma longibrachiatum | |||||
| Verticillium dahliae | |||||
| Protease Inhibitors | Potide-G | Candida albicans |
|
[76] | |
| Rhizoctania solani | |||||
| Potato Protease Inhibitor I and II | Botrytis cinerea |
|
[75,78,160] | ||
| Fusarium oxysporum | |||||
| Fusarium solani | |||||
| Bowman-Birk Protease Inhibitor | Fusarium culmorum |
|
[92,93,161,162] | ||
| Fusarium graminearum | |||||
| Mycosphaerella arachidicola | |||||
| Septoria tritici | |||||
| Antimicrobial Peptides | Prosystemin | Botrytis cinerea |
|
[104,105] | |
| |||||
| StSN1 | Botrytis cinerea |
|
[111,163] | ||
| Fusarium sp. | |||||
| Puroindoline A and B | Alternaria brassicola |
|
[116,164] | ||
| Ascochyta pisi | |||||
| Fusarium culmorum | |||||
| Fusarium graminearum | |||||
| Magnaporthe girsea | |||||
| Rhizoctania solani | |||||
| Verticillium dahlia | |||||
| DUF26-Containing Proteins | Ginkbilobin2 | Candida albicans |
|
[122,123] | |
| Fusarium oxysporum | |||||
| Trhicoderma reesei | |||||
| AFP1/AFP2 | Ustilago maydis |
|
[124] | ||
| VdCRR1 | None tested |
|
[125] | ||
| TaCRR | Bipolaris sorokiniana Rhizoctania cerealis |
|
[128] | ||
| Leucine Rich Repeat Protein | PvPGIP2 | Aspergillus niger |
|
[144] | |
| Botrytis cinerea | |||||
| S Albumin | 2S Albumin and 2S Albumin Orthologs | Aspergillus flavus |
|
[146,[150], [151], [152]] | |
| Aspergillus fumigatus | |||||
| Candida albicans | |||||
| Fusarium oxysporum | |||||
| Phanerochaete chrysosporium | |||||
| Trichoderma harizanum | |||||
2. Pathogenesis related proteins
Pathogenesis related (PR) proteins are a group of low molecular weight plant proteins involved in mitigating both biotic and abiotic stresses [19], and are often involved in triggering systemic acquired resistance in plants [20]. There are 16 main groups of PRs (PR-1 to PR-16), with each group classified based on different molecular and physiological properties. These proteins are often pathogen specific and involved in the transcriptional activation of plant defenses [21]. Here, we will focus on some of the most promising candidates for the development of antifungal agents for agricultural applications: PR-3, PR-5, PR-6, and PR-12.
2.1. Chitinases (PR-3)
One of the best known and most studied plant antifungal proteins is chitinase, which belongs to PR-3 [22]. Chitinases are strongly induced when the host plants are under attack from pathogens and function as defense molecules against fungal infection [23]. These proteins inhibit fungal growth by lysing hyphal tips in fungi and break down chitin into its oligomers [24]. Chitinases display strong antifungal activity against a wide range of phytopathogenic fungi. This includes Botrytis cinerea [25], a necrotrophic fungi that is considered one of the top fungal pathogens based on scientific and economic importance and infects over 200 species worldwide [26], as well as Rhizoctonia solani [27], which causes sheath blight in rice, one of the most widespread diseases of rice [28]. While chitinases have been isolated from bacteria [29], fungi [30,31], humans [32], and plants [33], chitin has not been found in mammals and plants [34]. As such, plant chitinases, are a valuable target for developing highly specific biocontrol against phytopathogenic fungi in agriculture.
While overexpressing plant chitinases in either native or heterologous plants have successfully enhanced plant resistance against phytopathogenic fungi [35], plant chitinases have also been used to treat fungal infections as an exogenously applied pest control agent. One study extracted chitinase E from yam tubers and then sprayed it on strawberries infected with powdery mildew. The treatment using chitinase E was successful at preventing the disease for at least two weeks through damaging cell-wall components of the hyphae and conidia of the pathogenic fungi [36]. Plant chitinase has been heterologously expressed in many microorganisms such the bacteria Escherichia coli [22] and Bacillus sp. [37], the yeast Pichia pastoris [38], as well as in plants such as transgenic tobacco [39] and cultured plant cells [40], which provides solid foundation to develop chitinases as antifungal agents. Meanwhile, fermentation optimization has also been explored to enhance the production of chitinase from microbial cell factories, and the strategies include but not limited to adjusting carbon sources, pH, aeration, and temperatures [41].
Importantly, chitinases also display excellent protein stability that ensures reliable exogenous application. Chitinase from Vitis vinifera exhibits a half-life of up to 4.7 days at 30 °C or 9 years at 15 °C [42], and the purified chitinase from Trichosanthes dioica, effective against Aspergillus niger and Trichoderma sp. In a fungal agar diffusion assay, remained stable between pH 5.0–11.0 and temperature 30–90 °C for at least 30 min [43]. Likewise, the purified chitinase from Diospyros kaki, which inhibited the growth of T. viride, exhibits broad pH stability from pH 4.0–9.0, and retains more than 60% activity at pH as low as 3.0 and as high as 10.0 [44]. Due to its specificity in targeting chitin, success in both plant extraction and heterologous expression from microbial factors, and numerous studies documenting its antifungal efficacy, chitinase is a promising candidate to develop as an antifungal agent. However, compared to the number of available chitinase studies that use transgenic plants, the exogenous application of chitinases on infected plants remain to be more thoroughly investigated.
2.2. Defensins (PR-12)
Defensins belong to group PR-12 [45], and exhibit broad-spectrum activities against different biotic agents including pathogenic fungi [46]. They are named due to the structural and functional similarities to insect and mammalian defensins [47]. Plant defensins are constitutively expressed in the extracellular space of most vegetative and reproductive plant tissues [48] and can be specifically induced under pathogen stress condition [47,49]. Typically, defensins are small soluble cationic proteins, 45–54 amino acid residues in size, exhibiting eight conserved cysteine residues (C1 to C8) with a conserved spacing pattern, and the tertiary structure is supported by at least four disulfide bonds [46]. Defensins remain stable both under extreme temperatures (as high as 90 °C) and very acidic conditions (pH as low as 1) [48]. For instance, NRBAP, a defensin-like protein purified from Phaseolus vulgaris beans, retained its antifungal activity against Mycosphaerella arachidicola up to 100 °C, and in the pH range of 1–13 [50].
Defensins can interact with a significant diversity of biological targets (e.g., proteases [51], protein synthetic machinery [52], α-amylases in insects [53], and ion channels in fungi [54]). One common mechanism that defensins often adopt to inhibit fungal growth is through the disruption of cell plasma membranes. Plant defensins are usually positively charged proteins and interact with anionic moieties in the membrane, such as glycoproteins, sphingolipids, or phospholipids [49]. The defensins cover the target membranes until it reaches a concentration threshold, and then disrupts the membrane integrity by affecting the bilayer curvature [55]. One study provided evidence that NaD1 from Nicotiana alata, which displays antifungal activity against several agronomically important filamentous fungi [56], was able to bound to phospholipids phosphatidic acid [57]. To estimate the effect of the total net charge of defensins on the antifungal activity, a mutagenesis analysis was performed on Rs-AFP2 from radish, and the interaction between the defensins and membrane lipids was improved when the net charge of the protein increased [58]. Plant defensin antifungal activity may not be restricted to targeting the membrane of pathogenic fungi. Indeed, the exogenous application of NaD1 is also associated with the entrance of the protein into fungal intracellular space, resulting in granulation of the cytoplasm and cell death [56]. This suggests that plant defensins could also interact with fungal intracellular targets and possibly with DNA, as already demonstrated by ostrich β-defensins, where E. coli growth was inhibited in assays due to interactions between peptides and cytoplasmic targets that curbed DNA, RNA, and protein synthesis [59]. The diversity of antifungal mechanisms and effectiveness of defensins against a wide range of pathogens implies the potential of this protein family as a promising resource for fighting plant pathogens.
2.3. Thaumatin-like proteins (PR-5)
Thaumatin-like proteins (TLP) belong to PR-5 family [60]. TLPs are named so due to their structural similarity to thaumatin, a sweet-tasting, non-toxic protein that was first discovered from the fruit of the tropical plant Thaumatococcus daniellii [61]. TLPs exhibit a broad range of biological activities, including antifungal activity. Different TLPs inhibit fungal growth through different mechanisms, including but not limited to disrupting fungal membrane [62], inhibiting fungal enzymes such as xylanase [63], inducing apoptosis by binding to specific fungal membrane receptors [64], and hydrolyzing β-1,3-glucans [65]. Osmotin and osmotin-like proteins are among the most studied TLPs of antifungal activity [66]. Osmotin and orthologs have been shown to exhibit broad-spectrum antifungal inhibitory effects [67]. Overexpression of osmotin in transgenic plants delayed disease symptoms from fungal pathogens [68]. Osmotin isolated from tobacco cell suspensions can inhibit the hyphal growth of numerous pathogenic fungi in vitro, including species from Bipolaris, Collectorichum, Fusarium, Kabatiella, Phytophthora, and Trichoderma [69]; and another osmotin-like protein from Solanum nigerum and overexpressed in E. coli can inhibit the growth of phytopathogenic Fusarium solani f. sp. glycines, Macrophomina phaseolina, and Collectrichum glaesporioides, and Collectrichum gossypii var. cephalosporioides at the concentration between 0.1 μg/μL to 0.3 μg/μL [70]. Additionally, an osmotin-like protein from Solanum nigrum L. var indica was shown to inhibit fungal spore germination and permeabilize fungal hyphae in vitro. This protein is also stable and retains its antifungal activity at temperatures as high as 75 °C for 30 min and pH 3–8 [71]. Further functional exploration of TLPs under various stress conditions for in planta assays will be necessary before its development into a reliable antifungal tool [72].
2.4. Protease inhibitors (PR-6)
Plant protease inhibitors (PIs), also called PR-6, are important proteins involved in many plant biological processes, including seed germination, protease-related house-keeping functions, and defense against biotic and abiotic stresses [73]. PIs are normally found in ample quantities in seeds and tubers, and plants in the Solanaceae family generally have exceptionally high levels of PIs [74], including some that can be promising candidates of antifungal agents. For instance, potatoes encode several PIs ranging from 4.1 to 39 kDa that exhibit broad-spectrum antifungal activities [75]. Potide-G, a Kunitz-type PI isolated from potato tubers of size 5.5 kDA, inhibits pathogenic fungi Candida ablicans and Rhizoctania solani in vitro even when heated to 70 °C for 20 min, and also exhibits antiviral and antibacterial activities [76]. Similarly, the potato protease inhibitors I and II (PPI–I and PPI-II) can inhibit the growth of various fungi, including B. cinerea [77], Fusarium solani, and Fusarium oxysporum [75]. Both PPI-I and II are heat stable, which can maintain their ability to inhibit F. solani and F. oxysporum growth in vitro under temperature as high as 100 °C [78]. PPI-I and II are also nontoxic, as they have previously been utilized in human clinical trials for appetite control [79]. The extraction of bioactive PPIs from potatoes is laborious and of low yields [80]. They have also been heterologously expressed in Saccharomyces cerevisiae, yet the antifungal activity of the purified protein was not examined [81]. A more economic production method is needed to enable the development of PPIs as antifungal agents for agriculture applications.
Another PI of interest is the Bowman-Birk protease inhibitor (BBI), which is typically under 20 kDA [82,83], contains seven conserved disulfide bonds, and inhibits trypsin and chymotrypsin, which are common enzymes pathogenic fungi utilize when infecting plants [84]. The BBI gene is induced during plant immune responses and overexpression of this gene in plants confers improved disease resistance against both insect and fungal pathogens [85]. BBIs from the legume (Fabaceae) or cereal (Poaceae) family have a double or single inhibitory loop respectively [86], and synthetic peptides that contain only the disulfide-linked, 9-residue long loop have shown to retain their trypsin and chymotrypsin inhibitory activity [87]. This short, truncated form of the protein may be of interest for the development of antifungal agents of smaller molecular weight for easier production and higher stability, compared with larger protein agents. Aside from the small size, BBI is thermostable with the ability to withstand 100 °C for 10 min, tolerates a wide pH range from 1.6 to 8.0, is not allergenic, and is approved by the FDA for human consumption [88]. Additionally, unlike some other candidates to be engineered as antifungal agent, BBI has passed phase II human clinical trials and is highly unlikely to be toxic, especially given its prevalence in soy products [89]. BBIs have already been successfully utilized as an exogenously applied antifungal agent in vitro. One study identified that a BBI-type trypsin-chymotrypsin inhibitor purified from broad beans can inhibit the growth of B. cinerea, F. oxysporum, and M. arachidicola at a dose as low as 60 μg per plate [90]. Plant BBIs have often been isolated from a variety of seeds such as those from Vigna mungo [91], Cajanus cajan [92], and Clitoria fairchildiana [93] and have been tested for their insecticidal properties. Rice BBI has also been expressed in E. coli and retained the inhibitory activity. However, the titer is relatively low at 20 mg/L, likely due to the presence of the disulfide bonds that make it prone to forming inclusion bodies [94]. In addition, care should be taken when developing BBI as an antifungal agent, as it is a multifunctional PI with a relatively broad activity towards various proteases [95], and may affect beneficial microbiota and fungi in the soil and plants.
3. Antimicrobial proteins
In addition to PRs, antimicrobial peptides (AMPs) are another protein group of interest. AMPs, also known as host defense peptides [96], can be derived from a variety of organisms, including plants, bacteria, and fungi. In plants, AMPs play a role in the plant innate immune system [97]. AMPs that work specifically against fungi are known as antifungal peptides (AFPs) [98], and feature a wide range of functions that are of interest to both pharmaceutical [99] and agricultural industries [100]. Here, we will only discuss AFPs that have shown potential for agricultural applications.
One AFP of interest is tomato systemin, a small peptide of only 18 amino acids long and is involved in inducing the synthesis of PIs in response to plant wounding and damage from herbivores [101]. Research suggests that systemin moves through the plant phloem and helps amplify the signaling process and allows for distal leaves to respond to the wounding [102]. Tomato plants that overexpress prosystemin, the precursor of systemin, are found to induce high levels of PI proteins even without wounding [103]. Additionally, transgenic plants expressing prosystemin reduce lesions by at least 50% from Phytophthora infestans, a pathogen that causes late blight [104]. Systemin peptides have been successfully isolated from tomato, sprayed onto grapevine (Solanum melongena) and eggplant (Vitis vinifera) plants infected with B. cinerea [105] at a concentration of 100 pM, and efficiently delayed necrosis of the infected plants.
Snakins are cell wall-associated defensins that are also classified as AMP and believed to play a role in plant growth, signaling, and defense [106]. Snakins isolated from Solanum tuberosum (StSN1) are cysteine-rich peptides roughly 6.9 kDA in size [107,108] and the snakins isolated from potato tubers is effective at suppressing both fungal and bacterial growth at concentrations lower than 10 μM [109]. Transgenic potato plants overexpressing the StSN1 gene exhibited reduced symptoms of R. solani infections and higher survival rates compared to the wild type plants [110]. Additionally, StSN1 has been shown to be effective in vitro against B. cinerea and several Fusarium species [111] However, snakins have been rarely expressed successfully from microbial hosts, often with low yield and insolubility, which hinders in-depth mechanistic characterization of its action towards pathogenic fungi [112].
Another promising group of AFPs are the puroindolines, which are small, amphipathic tryptophan-rich proteins about 13 kDA in size and found only in wheat (Triticum) [113]. They are known to inhibit the growth of pathogenic bacteria and fungi with low mammalian toxicity [114], likely through strong binding with microbial membranes and therefore perturbing the membrane integrity [115]. The primary roles of puroindolines include grain hardness and fungal defense. These proteins are believed to protect seeds from fungal attacks during seed development and germination [116]. There are two major puroindolines, Puroindoline A (PINA) and B (PINB) [117]. When the pin genes are overexpressed in transgenic rice, rice displayed significantly enhanced resistance to rice blast caused by Magnaporthe grisea and a reduction in symptoms due to Rhizoctonia solani infections [118]. Purified PINA and PINB proteins from wheat were able to inhibit the growth of a variety of pathogenic fungi, including Alternaria brassicola, Ascochyta pisi, F. culmorum, F. graminearum, Magnaporthe girsea, R. solani, and Verticillium dahlia. PINA and PINB are stable over a broad range of temperature (70 °C–130 °C) and pH (2.0–12.0) [[115], [119]]. PINs have been heterologously produced in Pichia pastoris with a titer up to 14 mg/L taking advantage of puroindoline's solubility in the detergent Triton X-114 [120]. These various AFPs discussed highlight the potential of using AFPs as antifungal agents for agricultural purposes.
4. DUF26-containing proteins
In the past decade, there has been an increase in interest towards proteins containing domain of unknown function (DUF26) for their capability in fighting plant pathogens and especially fungi [121]. DUF26 is a cysteine rich domain with a conserved C-X8-C-X2-C motif. DUF26-containing proteins are a large, land plant-specific protein family and characteristic of embryophytes [121]. Similarities with fungal lectins suggests DUF26-containing proteins constitute a group of plant carbohydrate-binding proteins able to recognize specific fungal sugar motifs [121].
There are three groups of DUF26-containing proteins: the cysteine-rich receptor-like secreted proteins (CRRSPs), cysteine-rich receptor-like kinase (CRKs) and plasmodesmata-localized proteins (PDLPs). The three DUF26-containing protein groups were all previously associated with antifungal activities. Nevertheless, only CRRSPs remain as good candidates for biotechnological application since CRKs and PDLPs contain transmembrane domains and localize to the membranes. CRRSPs contain a signal peptide followed by one or more DUF26 domains, separated by a variable region [121]. The most well-known CRRSP is Ginkbilobin2 (Gnk2), which was isolated from seeds of Ginkgo biloba and able to inhibit the growth of F. oxysporum, T. reesei, and C. albicans [122]. This antifungal activity is likely due to the binding of DUF26 domain with sugar moieties on the fungal cell wall [123]. For instance, Gnk2 interacts specifically with mannan, a yeast cell wall polysaccharide, and mannose, a building block of mannan, by strictly recognizing the hydroxy group at the C4 position of the monosaccharide. Consistently, two maize CRRSPs (AFP1 and AFP2) have been characterized to interact directly with the hyphal surface of Ustilago maydis, and the activity can be rendered by Rsp3, a U. maydis effector covering its surface [124].
In addition to direct binding with fungal cell walls, DUF26-containing proteins from CRRSP family also protect plants using indirect mechanisms. CRR1, a secreted apoplastic protein from cotton, and composed of two Cys-rich DUF26 motifs, interacts and protects the antifungal apoplastic chitinase 28 from cleavage by VdSSEP1, a pathogen related protease [125]. Importantly, overexpressing CRR1 in heterologous plants such as Arabidopsis thaliana and Nicothiana tabacum improved plant resistance to B. cinerea and P. parasitica, respectively. Thus, CRR1 could be a good candidate as a co-antifungal agent and simultaneous exogenous application of CRR1 and chitinases should be evaluated. Another CRRSP of interest is the recently reported CBM1-interacting protein (OsCBMIP) in rice [126]. Pathogenetic fungi generally use cell wall degrading enzymes (CWDEs) to destruct plant cell walls, and many CWDEs use carbohydrate binding modules (CBMs) to facilitate the access to plant polysaccharides to advance the infection process [127]. OsCBMIP can specifically bind to CBM of several CBM-containing CWDEs including the xylanase MoCel10A of the blast fungus pathogen Magnaporthe oryzae and slow down the infection progress. Interestingly, OsCBMIP cannot inhibit the growth of M. oryzae and F. oxysporum in vitro, and this further indicates that OsCBMIP slows down the infection of pathogenetic fungi through indirect mechanism, here specially, through inhibiting CBM-containing CWDEs [126]. In another study, a transcriptomic analysis of wheat after Bipolaris sorokiniana or Rhizoctania cerealis infection reported the induction of a cysteine-rich protein (CRR), TaCRR [128]. When heterologously expressed, this DUF26-containing protein showed a clear antifungal activity. Besides, it was found that silencing TaCRR gene in wheat significantly decreased the expression of pathogenesis-related genes such as β-1,3-glucanase, defensin or chitinases [128]. Owing to their apoplastic localization and direct or indirect antifungal activities, DUF26-containing proteins from the CRRSP class remain as attractive candidates for the future development of antifungal agents.
5. Other proteins
Polygalacturonase inhibiting proteins (PGIPs) are a family of leucine rich repeat (LRR) proteins found in plant cell walls [129,130] whose primary role is to inhibit polygalacturonases (PGs), enzymes secreted by insects and fungal pathogens that degrade the plant cell walls and leave it vulnerable for infection [131]. Through competitive or noncompetitive inhibition, PGIPs slow the hydrolysis process of PGs [[132], [133], [134], [135]]. Presently, numerous studies show that overexpression of PGIPs in transgenic plants leads to increased fungal resistance. The best-documented PGIP is PGIP2 from Phaseolus vulgaris (PvPGIP2), the common bean [136]. PvPGIP2 has been successfully expressed in transgenic plants, resulting in increased resistance to fungal infections against Alternaria citri, Aspergillus flavus, A. niger, B. cinerea, Claviceps purpurea, and F. graminearum [[137], [138], [139], [140]]. Similarly, expression of PGIP3 from soybeans (Glycine max) in tobacco has been shown to inhibit the growth of pathogenic Sclerotinia sclerotiorum, Fusarium moniliforme, B. aclada, A. niger, Collectotrichum acutatum, and F. graminearum [141,142]; and expressing PGIP2 from lima beans (Phaseolus lunatus) in tobacco also delayed growth of Collectrichum lupini, B. cinerea, F. moniliforme, and A. niger [143]. Recently, it is also found that truncated PvPGIP2 with only the optimal docking area retains similar level of inhibitory activities towards PGs from A. niger and B. cinerea to the full-length PvPGIP2 [144]. Yeast strains secreting full-length or truncated PvPGIP2 with the Ost 1 signal peptide were also able to reduce fungal growth and delay sporulation by 1–2 days [144]. Although the function of PGIPs when applied exogenously on plants has not been reported, this group of proteins is still considered as a promising candidate to be developed into an eco-friendly fungal control agent.
Albumins are a major class of water soluble, seed storage proteins that are used as a source of nutrients for plants during germination [145]. Among them, 2S albumins have antifungal capabilities [146], in addition to a variety of activities including anti-cancer, anti-fungal, anti-bacterial, and serine-protease inhibiting properties [147]. These small storage proteins are present in both monocotyledonous and dicotyledonous plant seeds [148] and typically have a disulfide bridge linking two different subunits, which are typically between 3 kDA and 10 kDA in size [149]. For example, pumpkin (Cucurbita sp.) 2S albumin is thermal-stable at up to 90 °C, and exhibits inhibitory activity against the fungal pathogen F. oxysporum [147]. Similarly, a crude extract of peanut (Arachis hypogaea) containing 2S albumin was found to inhibit growth of A. flavus [150]; the 2S albumin ortholog from passionfruit (Passiflora edulis) could also inhibit the fungal pathogens T. harizanum and F. oxysporum [151], C. musae, and C. lindemuthianum [146]; and the 2S albumin ortholog from Putranjiva roxburghii (putrin) could inhibit the growth of F. oxysporum, Phanerochaete chrysosporium, C. albicans, Aspergillus fumigatus, and A. flavus. In addition, putrin is stable at up to 50 °C and within a pH range from 6 – 8 [152]. On the other hand, 2S albumin from white sesame seeds, oriental mustard, and Brazil nuts can bind to IgE sera, which may trigger an allergic response in humans [145]. Thus, before 2S albumin can be utilized as an exogenously applied antifungal agent, we need to either engineer the protein to eliminate or reduce the allergenicity or modify the application in a manner that avoids either extensive contact or consumption.
6. Conclusion
Chemical-based fungicides are known to be detrimental to the environment and may lead to resistance in pathogenic fungi [153]. Unlike chemical fungicides, the use of exogenously applied natural plant proteins with known antifungal properties can potentially be an eco-friendly and sustainable method for controlling fungal diseases. These natural plant proteins are more socially acceptable, and compared with the production of transgenic plants, are more flexible. Additionally, antifungal plant proteins offer a variety of mechanisms and tools, urgently needed to fight against the rapidly evolving fungal pathogens. As summarized in Table 1, these naturally occurring plant peptides are strong candidates for developing broad-spectrum, fungal-control strategies. One of the biggest hurdles to consider when developing these proteins is lowering the cost of production while enabling mass production. It will be necessary to explore and further optimize microbial factories and protein extraction methods before many of these natural plant proteins can be utilized readily in agricultural industry. Numerous studies showcase the efficacy of these proteins both in vitro and in planta against pathogenic fungi. The potential of using natural plant proteins exogenously to control agricultural fungal diseases remains largely untapped and need to be considered when developing future eco- and environmentally-friendly antifungal agents.
Acknowledgments
We thank S. Xu for the valuable feedback in the preparation of the manuscript.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Waage J.K., Mumford J.D. Agricultural biosecurity. Philos. Trans. R. Soc. B Biol. Sci. 2007;363:863–876. doi: 10.1098/rstb.2007.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agrios G. Plant pathol. Fifth. 2004. Plant pathology: fifth edition; pp. 1–922. 9780080473789. [Google Scholar]
- 3.Rudin R., Donnelly J.S. The great Irish potato famine. Can J Ir Stud. 2000 doi: 10.2307/25515356. [DOI] [Google Scholar]
- 4.Fones H.N., et al. Threats to global food security from emerging fungal and oomycete crop pathogens. Nat. Food. 2020;161:332–342. doi: 10.1038/s43016-020-0075-0. 2020. [DOI] [PubMed] [Google Scholar]
- 5.Morton V., Staub T. A short history of fungicides. APSnet Featur. Artic. 2008 doi: 10.1094/apsnetfeature-2008-0308. [DOI] [Google Scholar]
- 6.Rani L., et al. An extensive review on the consequences of chemical pesticides on human health and environment. J Clean Prod. 2021;283 [Google Scholar]
- 7.Lozowicka B., Hrynko I., Kaczyński P., Jankowska M. Long-Term Investigation and Health Risk Assessment of Multi-class Fungicide Residues in Fruits Estimating acute and chronic exposure of children and adults to chlorpyrifos in fruit and vegetables based on the new, lower toxicology data View project. Artic. Polish J. Environ. Stud. 2016 doi: 10.15244/pjoes/61111. [DOI] [Google Scholar]
- 8.Klee H.J. Improving the flavor of fresh fruits: genomics, biochemistry, and biotechnology. New Phytol. 2010 doi: 10.1111/j.1469-8137.2010.03281.x. [DOI] [PubMed] [Google Scholar]
- 9.Klümper W., Qaim M. A meta-analysis of the impacts of genetically modified crops. PLoS One. 2014 doi: 10.1371/journal.pone.0111629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borrelli V.M.G., Brambilla V., Rogowsky P., Marocco A., Lanubile A. The enhancement of plant disease resistance using crispr/cas9 technology. Front Plant Sci. 2018;9:1245. doi: 10.3389/fpls.2018.01245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutta S.S., et al. Current status and future prospects of research on genetically modified rice: a review. Agric Rev. 2016;37 [Google Scholar]
- 12.Hielscher S., Pies I., Valentinov V., Chatalova L. Rationalizing the GMO debate: the ordonomic approach to addressing agricultural myths. Int J Environ Res Publ Health. 2016 doi: 10.3390/ijerph13050476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng Q., Wang H.H. Do consumers view the genetically modified food labeling systems differently? Contains GMO” Versus “Non-GMO” Labels. 2021:376–388. [Google Scholar]
- 14.Abbot C. EU environmental law: challenges, change and decision-making by maria lee. Mod Law Rev. 2006;69 [Google Scholar]
- 15.Warwick S.I., Beckie H.J., Hall L.M. Annals of the New York Academy of Sciences; 2009. Gene flow, invasiveness, and ecological impact of genetically modified crops. [DOI] [PubMed] [Google Scholar]
- 16.Varzakas T.H., Arvanitoyannis I.S., Baltas H. The politics and science behind GMO acceptance. Crit Rev Food Sci Nutr. 2007 doi: 10.1080/10408390600762696. [DOI] [PubMed] [Google Scholar]
- 17.Tabashnik B.E., Carrière Y. Surge in insect resistance to transgenic crops and prospects for sustainability. Nat Biotechnol. 2017 doi: 10.1038/nbt.3974. [DOI] [PubMed] [Google Scholar]
- 18.Zaker M. Natural plant products as eco-friendly fungicides for plant diseases control- A review. Agric For. 2016;14:134–141. [Google Scholar]
- 19.Sudisha J., Sharathchandra R.G., Amruthesh K.N., Kumar A., Shetty H.S. Pathogenesis related proteins in plant defense response. Plant Def. Biol. Control. 2012;12:379–403. [Google Scholar]
- 20.Awasthi, L. P. Applied plant virology : advances, detection, and antiviral strategies.
- 21.Ali S., et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol Res. 2018 doi: 10.1016/j.micres.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 22.Singh A., Isaac Kirubakaran S., Sakthivel N. Heterologous expression of new antifungal chitinase from wheat. Protein Expr Purif. 2007;56:100–109. doi: 10.1016/j.pep.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 23.Grover A. Plant chitinases: genetic diversity and physiological roles. CRC Crit Rev Plant Sci. 2012;31:57–73. [Google Scholar]
- 24.Malik A., Preety Purification and properties of plant chitinases: a review. J Food Biochem. 2019;43 [Google Scholar]
- 25.Baarlen P. van, Legendre L., Kan J. A. L. van. Plant defence compounds against botrytis infection. Botrytis Biol. Pathol. Control. 2007:143–161. doi: 10.1007/978-1-4020-2626-3_9. [DOI] [Google Scholar]
- 26.R D., et al. The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 2012;13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shrestha C.L., Oña I., Muthukrishnan S., Mew T.W. Chitinase levels in rice cultivars correlate with resistance to the sheath blight pathogen Rhizoctonia solani. Eur J Plant Pathol. 2007;120:69–77. 2007 1201. [Google Scholar]
- 28.Molla K.A., et al. Understanding sheath blight resistance in rice: the road behind and the road ahead. Plant Biotechnol J. 2020;18:895–915. doi: 10.1111/pbi.13312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamid R., et al. Chitinases: an update. J Pharm BioAllied Sci. 2013;5:21. doi: 10.4103/0975-7406.106559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nampoothiri K.M., et al. Process optimization for antifungal chitinase production by Trichoderma harzianum. Process Biochem. 2004 doi: 10.1016/S0032-9592(03)00282-6. [DOI] [Google Scholar]
- 31.Brzezinska M.S., Jankiewicz U. Production of antifungal chitinase by Aspergillus Niger LOCK 62 and its potential role in the biological control. Curr Microbiol. 2012 doi: 10.1007/s00284-012-0208-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muzzarelli R.A.A., et al. Current views on fungal chitin/chitosan, human chitinases, food preservation, glucans, pectins and inulin: a tribute to Henri Braconnot, precursor of the carbohydrate polymers science, on the chitin bicentennial. Carbohydr Polym. 2012;87:995–1012. [Google Scholar]
- 33.Kasprzewska A. Plant chitinases-regulation and function. Cell Mol Biol Lett. 2003;8 http://www.cmbl.org.pl [PubMed] [Google Scholar]
- 34.Nagpure A., Choudhary B., Gupta R.K. Chitinases: in agriculture and human healthcare. Crit Rev Biotechnol. 2014:34 215–232. doi: 10.3109/07388551.2013.790874. [DOI] [PubMed] [Google Scholar]
- 35.Hartl L., Zach S., Seidl-Seiboth V. Fungal chitinases: diversity, mechanistic properties and biotechnological potential. Appl Microbiol Biotechnol. 2012;93:533–543. doi: 10.1007/s00253-011-3723-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karasuda S., Tanaka S., Kajihara H., Yamamoto Y., Koga D. Plant chitinase as a possible biocontrol agent for use instead of chemical fungicides. Biosci Biotechnol Biochem. 2003;67 doi: 10.1271/bbb.67.221. [DOI] [PubMed] [Google Scholar]
- 37.Bhushan, B. Production and characterization of a thermostable chitinase from a new alkalophilic Bacillus sp. BG-11. [DOI] [PubMed]
- 38.Landim P.G.C., et al. Production in Pichia pastoris, antifungal activity and crystal structure of a class I chitinase from cowpea (Vigna unguiculata): insights into sugar binding mode and hydrolytic action. Biochimie. 2017;135:89–103. doi: 10.1016/j.biochi.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 39.Dong, X., Zhao, Y., Ran, X., Guo, L. & Zhao, D.-G. Overexpression of a new chitinase gene EuCHIT2 enhances resistance to erysiphe cichoracearum DC. in Tobacco plants. Int. J. Mol. Sci. Artic. doi:10.3390/ijms18112361. [DOI] [PMC free article] [PubMed]
- 40.Kurosaki F., Tashiro N., Nishi A. Secretion of chitinase from cultured carrot cells treated with fungal mycelial walls. Physiol Mol Plant Pathol. 1987;31:211–216. [Google Scholar]
- 41.Hamid R., et al. Chitinases: an update. J Pharm BioAllied Sci. 2013;5:21. doi: 10.4103/0975-7406.106559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falconer R.J., et al. Thermal stability of thaumatin-like protein, chitinase, and invertase isolated from sauvignon blanc and semillon juice and their role in haze formation in wine. J Agric Food Chem. 2010;58:975–980. doi: 10.1021/jf902843b. [DOI] [PubMed] [Google Scholar]
- 43.Rashel Kabir S., et al. Purification and characterization of a novel chitinase from Trichosanthes dioica seed with antifungal activity. Int J Biol Macromol. 2016;84:62–68. doi: 10.1016/j.ijbiomac.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J., Kopparapu N.K., Yan Q., Yang S., Jiang Z. Purification and characterisation of a novel chitinase from persimmon (Diospyros kaki) with antifungal activity. Food Chem. 2013;138:1225–1232. doi: 10.1016/j.foodchem.2012.11.067. [DOI] [PubMed] [Google Scholar]
- 45.Edreva A. PATHOGENESIS-RELATED proteins: research progress in the last 15 years. Gen Appl Plant Physiol. 2005;31 [Google Scholar]
- 46.Kovaleva V., Bukhteeva I., Kit O.Y. vols. 1–24. 2020. (Plant defensins from a structural perspective). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.WF Broekaert, F T., B C., R O. Plant defensins: novel antimicrobial peptides as components of the host defense system. Plant Physiol. 1995;108:1353. doi: 10.1104/pp.108.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ft L., MA A. Defensins--components of the innate immune system in plants. Curr Protein Pept Sci. 2005;6:85–101. doi: 10.2174/1389203053027575. [DOI] [PubMed] [Google Scholar]
- 49.Thomma B.P.H.J., Cammue B.P.A., Thevissen K. Plant defensins. Planta. 2002;216:193–202. doi: 10.1007/s00425-002-0902-6. 2002 2162. [DOI] [PubMed] [Google Scholar]
- 50.YS Chan T.N. Northeast red beans produce a thermostable and pH-stable defensin-like peptide with potent antifungal activity. Cell Biochem Biophys. 2013;66:637–648. doi: 10.1007/s12013-012-9508-1. [DOI] [PubMed] [Google Scholar]
- 51.Melo F.R., et al. Inhibition of trypsin by cowpea thionin : characterization , molecular modeling , and docking. Proteins Struct Funct Genet. 2002;319:311–319. doi: 10.1002/prot.10142. 48311–319. [DOI] [PubMed] [Google Scholar]
- 52.Mendez E., et al. Primary structure of co-hordothionin , a member of a novel family of thionins from barley endosperm , and its inhibition of protein synthesis in eukaryotic and prokaryotic cell-free systems. European. J. Biochem. 1996;73:67–73. doi: 10.1111/j.1432-1033.1996.0067u.x. [DOI] [PubMed] [Google Scholar]
- 53.Pelegrini B., Lay F.T., Murad M., Anderson M.A. 2008. Novel insights on the mechanism of action of a -amylase inhibitors from the plant defensin family; pp. 719–729. [DOI] [PubMed] [Google Scholar]
- 54.Spelbrink R.G., et al. Differential antifungal and calcium channel-blocking activity among structurally related plant defensins 1 [ w ] Plant Physiol. 2004;135:2055–2067. doi: 10.1104/pp.104.040873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shai Y. vol. 1462. 1999. (Mechanism of the binding , insertion and destabilization of phospholipid bilayer membranes by K -helical antimicrobial and cell non-selective membrane-lytic peptides). [DOI] [PubMed] [Google Scholar]
- 56.Weerden N. L. Van Der, Lay F.T., Anderson M.A. The plant defensin , NaD1 , enters the cytoplasm of Fusarium oxysporum hyphae. J Biol Chem. 2008;283:14445–14452. doi: 10.1074/jbc.M709867200. [DOI] [PubMed] [Google Scholar]
- 57.Järvå M., et al. X-ray structure of a carpet-like antimicrobial defensin–phospholipid membrane disruption complex. Nat Commun. 2018;9:1–10. doi: 10.1038/s41467-018-04434-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samblanx G. W. De, et al. Mutational analysis of a plant defensin from radish (Raphanus sativus L.) reveals two adjacent sites important for antifungal activity. J Biol Chem. 1997;272:1171–1179. doi: 10.1074/jbc.272.2.1171. [DOI] [PubMed] [Google Scholar]
- 59.Sugiarto H., Yu P. vol. 2. 2007. (Mechanisms of action of ostrich b -defensins against Escherichia coli). [DOI] [PubMed] [Google Scholar]
- 60.Faillace G.R., Turchetto-Zolet A.C., Guzman F.L., de Oliveira-Busatto L.A., Bodanese-Zanettini M.H. Genome-wide analysis and evolution of plant thaumatin-like proteins: a focus on the origin and diversification of osmotins. Mol Genet Genom. 2019;294:1137–1157. doi: 10.1007/s00438-019-01554-y. [DOI] [PubMed] [Google Scholar]
- 61.van der Wel H., Loeve K. Isolation and characterization of thaumatin I and II, the sweet‐tasting proteins from Thaumatococcus daniellii benth. Eur J Biochem. 1972;31:221–225. doi: 10.1111/j.1432-1033.1972.tb02522.x. [DOI] [PubMed] [Google Scholar]
- 62.Vigers A.J., et al. Thaumatin-like pathogenesis-related proteins are antifungal. Plant Sci. 1992;83:155–161. [Google Scholar]
- 63.Fierens E., et al. TLXI, a novel type of xylanase inhibitor from wheat (Triticum aestivum) belonging to the thaumatin family. Biochem J. 2007;403:583. doi: 10.1042/BJ20061291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.FES L., et al. Peptide from thaumatin plant protein exhibits selective anticandidal activity by inducing apoptosis via membrane receptor. Phytochemistry. 2019;159:46–55. doi: 10.1016/j.phytochem.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 65.Grenier J., Potvin C., Trudel J., Asselin A. Some thaumatin-like proteins hydrolyse polymeric β-1,3-glucans. Plant J. 1999;19:473–480. doi: 10.1046/j.1365-313x.1999.00551.x. [DOI] [PubMed] [Google Scholar]
- 66.None H., et al. Osmotin: a plant defense tool against biotic and abiotic stresses. Plant Physiol. Biochem. PPB. 2018;123:149–159. doi: 10.1016/j.plaphy.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 67.Lee H., Damsz B., Woloshuk C.P., Bressan R.A., Narasimhan M.L. Use of the plant defense protein osmotin to identify fusarium oxysporum genes that control cell wall properties. Eukaryot Cell. 2010;9:558–568. doi: 10.1128/EC.00316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bashir M.A., et al. Osmotin: a cationic protein leads to improve biotic and abiotic stress tolerance in plants. Plants. 2020;9:992. doi: 10.3390/plants9080992. 2020. 992 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abad L.R., et al. Antifungal activity of tobacco osmotin has specificity and involves plasma membrane permeabilization. Plant Sci. 1996;118:11–23. [Google Scholar]
- 70.de A Campos M., et al. Expression in Escherichia coli, purification, refolding and antifungal activity of an osmotin from Solanum nigrum. Microb Cell Factories. 2008;7:1–10. doi: 10.1186/1475-2859-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chowdhury S., Basu A., Kundu S. Cloning, characterization, and bacterial over-expression of an osmotin-like protein gene from Solanum nigrum L. With antifungal activity against three necrotrophic fungi. Mol Biotechnol. 2015;57:371–381. doi: 10.1007/s12033-014-9831-4. [DOI] [PubMed] [Google Scholar]
- 72.Hakim, et al. Osmotin A plant defense tool against biotic and abiotic stresses. Plant Physiol Biochem. 2018;123:149–159. doi: 10.1016/j.plaphy.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 73.Rustgi S., Boex-Fontvieille E., Reinbothe C., von Wettstein D., Reinbothe S. 2018. Communicative & Integrative Biology the complex world of plant protease inhibitors: insights into a Kunitz-type cysteine protease inhibitor of Arabidopsis thaliana. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Platé N.A., Valuev L.I., Valueva T.A., Chupov V.V. Biospecific haemosorbents based on proteinase inhibitor. I. Synthesis and properties. Biomaterials. 1993;14:51–56. doi: 10.1016/0142-9612(93)90075-d. [DOI] [PubMed] [Google Scholar]
- 75.Bártová V., Bárta J., Jarošová M. Antifungal and antimicrobial proteins and peptides of potato (Solanum tuberosum L.) tubers and their applications. Appl Microbiol Biotechnol. 2019;103:5533–5547. doi: 10.1007/s00253-019-09887-9. [DOI] [PubMed] [Google Scholar]
- 76.Kim M.H., et al. Purification and characterization of a heat-stable serine protease inhibitor from the tubers of new potato variety ‘Golden Valley. Biochem Biophys Res Commun. 2006;346:681–686. doi: 10.1016/j.bbrc.2006.05.186. [DOI] [PubMed] [Google Scholar]
- 77.Hermosa M.R., Turrà D., Fogliano V., Monte E., Lorito M. Identification and characterization of potato protease inhibitors able to inhibit pathogenicity and growth of Botrytis cinerea. Physiol Mol Plant Pathol. 2006;68:138–148. [Google Scholar]
- 78.Bártová V., et al. Proteomic characterization and antifungal activity of potato tuber proteins isolated from starch production waste under different temperature regimes. Appl Microbiol Biotechnol. 2018;102:10551–10560. doi: 10.1007/s00253-018-9373-y. [DOI] [PubMed] [Google Scholar]
- 79.Peters H.P.F., et al. The effect of protease inhibitors derived from potato formulated in a minidrink on appetite, food intake and plasma cholecystokinin levels in humans. Int J Obes. 2011;35:244–250. doi: 10.1038/ijo.2010.136. [DOI] [PubMed] [Google Scholar]
- 80.Komarnytsky S., Cook A., Raskin I. Potato protease inhibitors inhibit food intake and increase circulating cholecystokinin levels by a trypsin-dependent mechanism. Int J Obes. 2011;35:236–243. doi: 10.1038/ijo.2010.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fischer M., Kuckenberg M., Kastilan R., Muth J., Gebhardt C. Novel in vitro inhibitory functions of potato tuber proteinaceous inhibitors. Mol Genet Genom. 2015;290:387–398. doi: 10.1007/s00438-014-0906-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qi R.F., Song Z.W., Chi C.W. Structural features and molecular evolution of Bowman-Birk protease inhibitors and their potential application. Acta Biochim Biophys Sin. 2005;37:283–292. doi: 10.1111/j.1745-7270.2005.00048.x. [DOI] [PubMed] [Google Scholar]
- 83.Gitlin-Domagalska A., Maciejewska A., Dębowski D. Bowman-Birk inhibitors: insights into family of multifunctional proteins and peptides with potential therapeutical applications. Pharmaceuticals. 2020;13:1–40. doi: 10.3390/ph13120421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Singh R.R., Appu Rao A.G. Reductive unfolding and oxidative refolding of a Bowman-Birk inhibitor from horsegram seeds (Dolichos biflorus): evidence for ‘hyperreactive’ disulfide bonds and rate-limiting nature of disulfide isomerization in folding. Biochim Biophys Acta Protein Struct Mol Enzymol. 2002;1597:280–291. doi: 10.1016/s0167-4838(02)00301-1. [DOI] [PubMed] [Google Scholar]
- 85.Grosse-Holz F.M., van der Hoorn, jobs R. A. L. Juggling. Roles and mechanisms of multifunctional protease inhibitors in plants. New Phytol. 2016;210:794–807. doi: 10.1111/nph.13839. [DOI] [PubMed] [Google Scholar]
- 86.James A.M., et al. Evidence for ancient origins of bowman-birk inhibitors from Selaginella moellendorffii. Plant Cell. 2017;29:461–473. doi: 10.1105/tpc.16.00831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Inhibitor B.-B., Nishino N., Aoyagi H., Kato T. Studies on the synthesis of proteinase inhibitors I. Synthesis and activity of nonapeptide fragments of soybean. J Biochem. 1977;82 doi: 10.1093/oxfordjournals.jbchem.a131767. [DOI] [PubMed] [Google Scholar]
- 88.Losso J.N. The biochemical and functional food properties of the Bowman-Birk inhibitor. Crit Rev Food Sci Nutr. 2008;48:94–118. doi: 10.1080/10408390601177589. [DOI] [PubMed] [Google Scholar]
- 89.Muzard J., Fields C., John O’mahony J., Lee G.U. 2012. Probing the soybean Bowman−Birk inhibitor using recombinant antibody fragments. [DOI] [PubMed] [Google Scholar]
- 90.Ye X.Y., Ng T.B., Rao P.F. A bowman–birk-type trypsin-chymotrypsin inhibitor from broad beans. Biochem Biophys Res Commun. 2001;289:91–96. doi: 10.1006/bbrc.2001.5965. [DOI] [PubMed] [Google Scholar]
- 91.Prasad E.R., Dutta-Gupta A., Padmasree K. Purification and characterization of a Bowman-Birk proteinase inhibitor from the seeds of black gram (Vigna mungo) Phytochemistry. 2010;71:363–372. doi: 10.1016/j.phytochem.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 92.Prasad E.R., Merzendorfer H., Madhurarekha C., Dutta-Gupta # A., Padmasree K. Bowman-Birk proteinase inhibitor from Cajanus cajan seeds: purification, characterization, and insecticidal properties. J Agric Food Chem. 2010;58:2838–2847. doi: 10.1021/jf903675d. [DOI] [PubMed] [Google Scholar]
- 93.Dantzger M., et al. Bowman–Birk proteinase inhibitor from Clitoria fairchildiana seeds: isolation, biochemical properties and insecticidal potential. Phytochemistry. 2015;118:224–235. doi: 10.1016/j.phytochem.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 94.Li N., et al. The refolding, purification, and activity analysis of a rice Bowman-Birk inhibitor expressed in Escherichia coli. Protein Expr Purif. 1999;15:99–104. doi: 10.1006/prep.1998.0989. [DOI] [PubMed] [Google Scholar]
- 95.Farady C.J., Craik C.S. Mechanisms of macromolecular protease inhibitors. Chembiochem. 2010:11 2341–2346. doi: 10.1002/cbic.201000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mahlapuu M., Håkansson J., Ringstad L., Björn C. Antimicrobial peptides: an emerging category of therapeutic agents. Front Cell Infect Microbiol. 2016;6:194. doi: 10.3389/fcimb.2016.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maróti Gergely G., Kereszt A., Kondorosi É., Mergaert P. Natural roles of antimicrobial peptides in microbes, plants and animals. Res Microbiol. 2011;162:363–374. doi: 10.1016/j.resmic.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 98.López-García B., San Segundo B., Coca M. Antimicrobial peptides as a promising alternative for plant disease protection. ACS Symp Ser. 2012;1095:263–294. [Google Scholar]
- 99.Fjell C.D., Hiss J.A., Hancock R.E.W., Schneider G. Designing antimicrobial peptides: form follows function. Nat Rev Drug Discov. 2012;11:37–51. doi: 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- 100.Martínez-Culebras P.V., Gandía M., Garrigues S., Marcos J.F., Manzanares P. Antifungal peptides and proteins to control toxigenic fungi and mycotoxin biosynthesis. Int J Mol Sci. 2021;22 doi: 10.3390/ijms222413261. 2021. , Page 13261 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang L., et al. 2018. The systemin receptor SYR1 enhances resistance of tomato against herbivorous insects. [DOI] [PubMed] [Google Scholar]
- 102.Narváez-Vásquez J., Ryan C.A. The cellular localization of prosystemin: a functional role for phloem parenchyma in systemic wound signaling. Planta. 2004;218:360–369. doi: 10.1007/s00425-003-1115-3. [DOI] [PubMed] [Google Scholar]
- 103.Constabel C.P., Bergey D.R., Ryan C.A. Systemin activates synthesis of wound-inducible tomato leaf polyphenol oxidase via the octadecanoid defense signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 1995;92:407–411. doi: 10.1073/pnas.92.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jones R.W., Ospina-Giraldo M., Clemente T. Prosystemin-antimicrobial-peptide fusion reduces tomato late blight lesion expansion. Mol Breed. 2004;14:83–89. [Google Scholar]
- 105.Molisso D., et al. Colonization of solanum melongena and vitis vinifera plants by botrytis cinerea is strongly reduced by the exogenous application of tomato systemin. J. Fungi. 2021;7:1–15. doi: 10.3390/jof7010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Su T., Han M., Cao D., Xu, Molecular M. And biological properties of snakins: the foremost cysteine-rich plant host defense peptides. J. Fungi. 2020;6:1–17. doi: 10.3390/jof6040220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Su T., Han M., Cao D., Xu M. Molecular and biological properties of snakins: the foremost cysteine-rich plant host defense peptides. J. Fungi. 2020;6:220. doi: 10.3390/jof6040220. 2020. , Page 220 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Darqui F.S., et al. Potato snakin-1 gene enhances tolerance to Rhizoctonia solani and Sclerotinia sclerotiorum in transgenic lettuce plants. J Biotechnol. 2018;283:62–69. doi: 10.1016/j.jbiotec.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 109.Segura A., Moreno M., Madueño F., Molina A., García-Olmedo F. Snakin-1, a peptide from potato that is active against plant pathogens. Mol Plant Microbe Interact. 1999;12:16–23. doi: 10.1094/MPMI.1999.12.1.16. [DOI] [PubMed] [Google Scholar]
- 110.Almasia N.I., Bazzini A.A., Hopp H.E., Vazquez-Rovere C. Overexpression of snakin-1 gene enhances resistance to Rhizoctonia solani and Erwinia carotovora in transgenic potato plants. Mol Plant Pathol. 2008;9:329–338. doi: 10.1111/j.1364-3703.2008.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Berrocal-Lobo M., et al. Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol. 2002;128:951–961. doi: 10.1104/pp.010685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kovalskaya N., Hammond R.W. Expression and functional characterization of the plant antimicrobial snakin-1 and defensin recombinant proteins. Protein Expr Purif. 2009;63:12–17. doi: 10.1016/j.pep.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 113.Biol, T. J. et al. Turkish Journal of Biology Molecular characterization of diverse wheat germplasm for puroindoline proteins and their antimicrobial activity. doi:10.3906/biy-1405-30.
- 114.Morris C.F. The antimicrobial properties of the puroindolines, a review. World J Microbiol Biotechnol. 2019;35:86. doi: 10.1007/s11274-019-2655-4. [DOI] [PubMed] [Google Scholar]
- 115.Alfred R.L. 2013. Biochemical analysis of the functionality of puroindoline peptides and proteins. [Google Scholar]
- 116.Giroux, M. J., Sripo, T., Gerhardt, S. & Sherwood, J. Biotechnology and genetic engineering reviews puroindolines: their role in grain hardness and plant defence 13 puroindolines: their role in grain hardness and plant defence. doi:10.1080/02648725.2003.10648047. [DOI] [PubMed]
- 117.Geneix N., et al. Relationships between puroindoline A-prolamin interactions and wheat grain hardness. PLoS One. 2020;15 doi: 10.1371/journal.pone.0225293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Krishnamurthy K., Balconi C., Sherwood J.E., Giroux M.J. vol. 14. 2001. (Wheat puroindolines enhance fungal disease resistance in transgenic rice). [DOI] [PubMed] [Google Scholar]
- 119.Rl Alfred, E P., J P., H B., M B. Stability of puroindoline peptides and effects on wheat rust. World J Microbiol Biotechnol. 2013;29:1409–1419. doi: 10.1007/s11274-013-1304-6. [DOI] [PubMed] [Google Scholar]
- 120.Issaly, N. et al. Optimization of the wheat puroindoline-a production in Pichia pastoris. [DOI] [PubMed]
- 121.Vaattovaara A., et al. Mechanistic insights into the evolution of DUF26-containing proteins in land plants. Commun. Biol. 2019;2:1–18. doi: 10.1038/s42003-019-0306-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sawano Y., Miyakawa T., Yamazaki H., Tanokura M., Hatano K.I. Purification, characterization, and molecular gene cloning of an antifungal protein from Ginkgo biloba seeds. Biol Chem. 2007;388:273–280. doi: 10.1515/BC.2007.030. [DOI] [PubMed] [Google Scholar]
- 123.Miyakawa T., et al. A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity. Plant Physiol. 2014;166:766–778. doi: 10.1104/pp.114.242636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ma L.S., et al. The Ustilago maydis repetitive effector Rsp3 blocks the antifungal activity of mannose-binding maize proteins. Nat Commun. 2018;9:1–15. doi: 10.1038/s41467-018-04149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Han L.B., et al. The Cotton Apoplastic Protein CRR1 stabilizes chitinase 28 to facilitate defense against the fungal pathogen verticillium dahliae. Plant Cell. 2019;31:520–536. doi: 10.1105/tpc.18.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Takeda T., et al. Apoplastic CBM1-interacting proteins bind conserved carbohydrate binding module 1 motifs in fungal hydrolases to counter pathogen invasion. bioRxiv. 2022;12 doi: 10.1101/2021.12.31.474618. 2021. 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Takahashi M., et al. Characterization of a cellobiohydrolase (MoCel6A) produced by magnaporthe oryzae. Appl Environ Microbiol. 2010;76:6583–6590. doi: 10.1128/AEM.00618-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Guo F., Shan Z., Yu J., Xu G., Zhang Z. The cysteine-rich repeat protein tacrr1 participates in defense against both rhizoctonia cerealis and bipolaris sorokiniana in wheat. Int J Mol Sci. 2020;21:1–17. doi: 10.3390/ijms21165698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Di Matteo A., Bonivento D., Tsernoglou D., Federici L., Cervone F. Polygalacturonase-inhibiting protein (PGIP) in plant defence: a structural view. Phytochemistry. 2006 doi: 10.1016/j.phytochem.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 130.Jones D.A., Jones J.D.G. The role of leucine-rich repeat proteins in plant defences. Adv Bot Res. 1997 doi: 10.1016/S0065-2296(08)60072-5. [DOI] [Google Scholar]
- 131.Kalunke R.M., et al. An update on polygalacturonase-inhibiting protein (PGIP), aleucine-rich repeat protein that protects crop plants against pathogens. Front Plant Sci. 2015 doi: 10.3389/fpls.2015.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Federici L., Di Matteo A., Fernandez-Recio J., Tsernoglou D., Cervone F. Polygalacturonase inhibiting proteins: players in plant innate immunity? Trends Plant Sci. 2006 doi: 10.1016/j.tplants.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 133.Stotz H.U., et al. Identification of target amino acids that affect interactions of fungal polygalacturonases and their plant inhibitors. Physiol Mol Plant Pathol. 2000 doi: 10.1006/pmpp.2000.0258. [DOI] [Google Scholar]
- 134.King D., et al. Use of amide exchange mass spectrometry to study conformational changes within the endopolygalacturonase II-homogalacturonan-polygalacturonase inhibiting protein system. Biochemistry. 2002 doi: 10.1021/bi020119f. [DOI] [PubMed] [Google Scholar]
- 135.Frediani M., et al. Cytological localization of the PGIP genes in the embryo suspensor cells of Phaseolus vulgavis L. Theor. Appl. Genet. Int. J. Plant Breed. Res. 1993 doi: 10.1007/BF01184925. [DOI] [PubMed] [Google Scholar]
- 136.Benedetti M., et al. A single amino-acid substitution allows endo-polygalacturonase of Fusarium verticillioides to acquire recognition by PGIP2 from Phaseolus vulgaris. PLoS One. 2013 doi: 10.1371/journal.pone.0080610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Maulik A., Sarkar A.I., Devi S., Basu S. Polygalacturonase-inhibiting proteins - leucine-rich repeat proteins in plant defence. Plant Biol. 2012;14:22–30. doi: 10.1111/j.1438-8677.2011.00501.x. [DOI] [PubMed] [Google Scholar]
- 138.Vasconcellos R.C.C., Lima T.F.C., Fernandes-Brum C.N., Chalfun-Junior A., Santos J.B. Expression and validation of PvPGIP genes for resistance to white mold (Sclerotinia sclerotiorum) in common beans (Phaseolus vulgaris L.) Genet Mol Res. 2016 doi: 10.4238/gmr.15038269. [DOI] [PubMed] [Google Scholar]
- 139.Farina A., et al. The bean polygalacturonase-inhibiting protein 2 (PvPGIP2) is highly conserved in common bean (Phaseolus vulgaris L.) germplasm and related species. Theor Appl Genet. 2009;118:1371–1379. doi: 10.1007/s00122-009-0987-4. [DOI] [PubMed] [Google Scholar]
- 140.Tundo S., et al. vol. 29. 2016. (Pyramiding PvPGIP2 and TAXI-III but not PvPGIP2 and PMEI enhances resistance against Fusarium graminearum). [DOI] [PubMed] [Google Scholar]
- 141.Frati F., Galletti R., De Lorenzo G., Salerno G., Conti E. Activity of endo-polygalacturonases in mirid bugs (Heteroptera: Miridae) and their inhibition by plant cell wall proteins (PGIPs) Eur J Entomol. 2006;103:515–522. [Google Scholar]
- 142.D'Ovidio R., et al. The characterization of the soybean polygalacturonase-inhibiting proteins (Pgip) gene family reveals that a single member is responsible for the activity detected in soybean tissues. Planta. 2006;224:633–645. doi: 10.1007/s00425-006-0235-y. [DOI] [PubMed] [Google Scholar]
- 143.Farina A., et al. The bean polygalacturonase-inhibiting protein 2 (PvPGIP2) is highly conserved in common bean (Phaseolus vulgaris L.) germplasm and related species. Theor Appl Genet. 2009;118:1371–1379. doi: 10.1007/s00122-009-0987-4. [DOI] [PubMed] [Google Scholar]
- 144.Chiu T., Behari A., Chartron J.W., Putman A., Li Y. Exploring the potential of engineering polygalacturonase-inhibiting protein as an ecological, friendly, and nontoxic pest control agent. Biotechnol Bioeng. 2021 doi: 10.1002/bit.27845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Moreno F.J., Clemente A. 2S albumin storage proteins: what makes them food allergens? Open Biochem J. 2008;2:16–28. doi: 10.2174/1874091X00802010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Agizzio A.P., et al. The antifungal properties of a 2S albumin-homologous protein from passion fruit seeds involve plasma membrane permeabilization and ultrastructural alterations in yeast cells. Plant Sci. 2006;171:515–522. doi: 10.1016/j.plantsci.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 147.Tomar P.P.S., et al. Characterization of anticancer, DNase and antifungal activity of pumpkin 2S albumin. Biochem Biophys Res Commun. 2014;448:349–354. doi: 10.1016/j.bbrc.2014.04.158. [DOI] [PubMed] [Google Scholar]
- 148.Souza Cândido E., et al. Plant storage proteins with antimicrobial activity: novel insights into plant defense mechanisms. Faseb J. 2011;25:3290–3305. doi: 10.1096/fj.11-184291. [DOI] [PubMed] [Google Scholar]
- 149.Souza P.F.N. The forgotten 2S albumin proteins: importance, structure, and biotechnological application in agriculture and human health. Int J Biol Macromol. 2020;164:4638–4649. doi: 10.1016/j.ijbiomac.2020.09.049. [DOI] [PubMed] [Google Scholar]
- 150.Duan X.H., Jiang R., Wen Y.J., Bin J.H. Some 2S albumin from peanut seeds exhibits inhibitory activity against Aspergillus flavus. Plant Physiol Biochem. 2013;66:84–90. doi: 10.1016/j.plaphy.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 151.Pelegrini P.B., et al. An antifungal peptide from passion fruit (Passiflora edulis) seeds with similarities to 2S albumin proteins. Biochim Biophys Acta, Proteins Proteomics. 2006;1764:1141–1146. doi: 10.1016/j.bbapap.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 152.Tomar P.P.S., et al. Purification, characterisation and cloning of a 2S albumin with DNase, RNase and antifungal activities from Putranjiva roxburghii. Appl Biochem Biotechnol. 2014;174:471–482. doi: 10.1007/s12010-014-1078-9. [DOI] [PubMed] [Google Scholar]
- 153.Wyenandt A., Everts K. Development of a fungicide resistance management guide for vegetable growers in the mid-atlantic states mid-atlantic commercial vegetable production recommendations guide view project fungicide resistance management guidelines for vegetable crops grown in the mid-atlantic region. View project. 2020 doi: 10.1094/CM-2009-0316-01-MG. [DOI] [Google Scholar]
- 154.Boller T., Mauch F. Colorimetric assay for chitinase. Methods Enzymol. 1988 doi: 10.1016/0076-6879(88)61052-4. [DOI] [Google Scholar]
- 155.J Jayaraj Z.P. Combined expression of chitinase and lipid transfer protein genes in transgenic carrot plants enhances resistance to foliar fungal pathogens. Plant Cell Rep. 2007;26:1539–1546. doi: 10.1007/s00299-007-0368-x. [DOI] [PubMed] [Google Scholar]
- 156.Punja Z.K., Zhang Y.Y. Plant chitinases and their roles in resistance to fungal diseases. J Nematol. 1993;25(4):526–540. [PMC free article] [PubMed] [Google Scholar]
- 157.Singh A., Isaac Kirubakaran S., Sakthivel N. Heterologous expression of new antifungal chitinase from wheat. Protein Expr Purif. 2007;56:100–109. doi: 10.1016/j.pep.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 158.J Z., NK K., Q Y., S Y., Z J. Purification and characterisation of a novel chitinase from persimmon (Diospyros kaki) with antifungal activity. Food Chem. 2013;138:1225–1232. doi: 10.1016/j.foodchem.2012.11.067. [DOI] [PubMed] [Google Scholar]
- 159.Grenier J., Potvin C., Trudel J., Asselin A. Some thaumatin-like proteins hydrolyse polymeric β-1,3-glucans. Plant J. 1999;19:473–480. doi: 10.1046/j.1365-313x.1999.00551.x. [DOI] [PubMed] [Google Scholar]
- 160.Hermosa M.R., Turrà D., Fogliano V., Monte E., Lorito M. Identification and characterization of potato protease inhibitors able to inhibit pathogenicity and growth of Botrytis cinerea. Physiol Mol Plant Pathol. 2006;68:138–148. [Google Scholar]
- 161.Chilosi G., et al. Antifungal activity of a bowman–birk-type trypsin inhibitor from wheat kernel. J Phytopathol. 2000;148:477–481. [Google Scholar]
- 162.Katoch R., et al. Cloning, characterization, expression analysis and inhibition studies of a novel gene encoding Bowman-Birk type protease inhibitor from rice bean. Gene. 2014;546:342–351. doi: 10.1016/j.gene.2014.05.055. [DOI] [PubMed] [Google Scholar]
- 163.Segura A., Moreno M., Madueño F., Molina A., García-Olmedo F. Snakin-1, a peptide from potato that is active against plant pathogens. Mol Plant Microbe Interact. 1999;12:16–23. doi: 10.1094/MPMI.1999.12.1.16. [DOI] [PubMed] [Google Scholar]
- 164.Alfred R.L., Palombo E.A., Panozzo J.F., Bariana H., Bhave M. Stability of puroindoline peptides and effects on wheat rust. World J Microbiol Biotechnol. 2013;29:1409–1419. doi: 10.1007/s11274-013-1304-6. 2013 298. [DOI] [PubMed] [Google Scholar]
- 165.Tanaka S., Kahmann R. Cell wall–associated effectors of plant-colonizing fungi. 2021. 247-260. [DOI] [PubMed]
- 166.Newman M.A., Sundelin T., Nielsen J.T., Erbs G. MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front Plant Sci. 2013;4:139. doi: 10.3389/fpls.2013.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yan J., et al. Plant antifungal proteins and their applications in agriculture. Appl Microbiol Biotechnol. 2015;99:4961–4981. doi: 10.1007/s00253-015-6654-6. [DOI] [PubMed] [Google Scholar]
- 168.Jashni M.K., Mehrabi R., Collemare J., Mesarich C.H., de Wit P.J.G.M. The battle in the apoplast: further insights into the roles of proteases and their inhibitors in plant–pathogen interactions. Front Plant Sci. 2015;6:584. doi: 10.3389/fpls.2015.00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Selitrennikoff C.P. Antifungal proteins. Appl Environ Microbiol. 2001;67:2883–2894. doi: 10.1128/AEM.67.7.2883-2894.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Adams D.J. Fungal cell wall chitinases and glucanases. Microbiology. 2004;150:2029–2035. doi: 10.1099/mic.0.26980-0. [DOI] [PubMed] [Google Scholar]
- 171.Lanver D., Schweizer G., Tanaka S., Tollot M. Fungal effectors and plant susceptibility. Artic. Annu. Rev. Plant Biol. 2015 doi: 10.1146/annurev-arplant-043014-114623. [DOI] [PubMed] [Google Scholar]
- 172.Matsubayashi Y. 2014. Posttranslationally modified small-peptide signals in plants; pp. 385–413. [DOI] [PubMed] [Google Scholar]