Abstract
The nucleus is a highly organized organelle with an intricate substructure of chromatin, RNAs, and proteins. This environment represents a challenge for maintaining protein quality control, since non-native proteins may interact inappropriately with other macromolecules and thus interfere with their function. Maintaining a healthy nuclear proteome becomes imperative during times of stress, such as upon DNA damage, heat shock, or starvation, when the proteome must be remodeled to effect cell survival. This is accomplished with the help of nuclear-specific chaperones, degradation pathways, and specialized structures known as protein quality control (PQC) sites that sequester proteins to help rapidly remodel the nuclear proteome. In this review, we focus on the current knowledge of PQC sites in Saccharomyces cerevisiae, particularly on a specialized nuclear PQC site called the intranuclear quality control site, a poorly understood nuclear inclusion that coordinates dynamic proteome triage decisions in yeast.
Keywords: protein quality control, DNA damage, nucleus, proteasome
Abbreviations: aCLD, alpha-crystallin–like domain; CTD, C-terminal domain; ER, endoplasmic reticulum; ERAD, ER-associated degradation; IDR, intrinsically disordered region; INQ, intranuclear quality control site; IPOD, insoluble protein deposit; JUNQ, juxtanuclear quality control site; LCR, low-complexity region; LLPS, liquid–liquid phase separation; MMS, methyl methanesulfonate; NEF, nucleotide exchange factor; NLS, nuclear localization sequence; NTD, N-terminal domain; PQC, protein quality control; ts, temperature-sensitive; UPS, ubiquitin proteasome system
Cells experiencing stress need to respond quickly. Be it external stressors such as DNA damage and heat shock or internal stressors such as genetic mutations, cells need to remodel their proteome to avoid accumulation of proteins that acquire non-native conformations. These misfolded proteins present short stretches of exposed hydrophobic regions that under normal conditions are buried inside the native conformation. These regions tend to be ‘sticky’ and if not acted on, could lead to the accumulation of toxic protein aggregates. Aggregated misfolded proteins present a physical roadblock and interfere with cellular trafficking and other processes in a gain of toxic function (1, 2). Aggregates also disrupt the molecular pathways that their constituents originally functioned in to create a loss-of-function that can perturb a variety of processes (3). This overarching interference in cellular function is evident in neurodegenerative diseases such as Alzheimer’s, ALS, and Parkinson's, which are pathologically associated with protein aggregation (4, 5). It is therefore in the interest of the cell to maintain proteome homeostasis or proteostasis. This is achieved through a complex network of protein quality control (PQC) circuits that have been extensively studied and documented over the past 2 decades owing to their importance in maintaining a healthy proteome (6, 7, 8, 9).
At the core of these PQC circuits lie a class of proteins called molecular chaperones. Chaperones can be classified into various families of heat shock proteins (Hsps) such as sHsp (small HSP), Hsp40, Hsp60, Hsp70, Hsp90, and Hsp110 families and more (10, 11). Many chaperones play constitutive roles in nascent protein folding, transport, and assembly, while others are strictly stress induced, responding to a variety of environmental insults. Many of these chaperones like CCT, Hsp70, and Hsp90 function as active ‘foldases’ utilizing ATP-dependent hydrolysis to promote folding, while others like Hsp40 and sHsps act as passive ‘holdases’ that protect and shield non-native surfaces. CCT actively encapsulates substrates in a cage-like structure, while Hsp70 and Hsp90 bind and release smaller domains to effect folding. Some other chaperones coordinate with protein degradation machinery to unfold and degrade target proteins. Chaperones like Btn2 and Hsp42 are known to help in protein aggregation and sequestration, while others like the Hsp40 Apj1 and Hsp110 help in protein disaggregation and refolding. Misfolded proteins, therefore, are constantly recognized, sequestered, refolded, or degraded to prevent accumulation of aggregates. Under stress, these circuits can be overwhelmed and ultimately result in protein aggregation. In such cases, the cell has to employ pathways specifically tasked with the role of clearing protein aggregates. Refolding, sequestration, and degradation represent the three fundamental triage decisions for non-native proteins and collectively define the idea of a PQC circuit.
The process of PQC is at least partially organelle specific. Not only does each organelle contain a unique proteome but the mechanisms of protein import and export vary with respect to protein folding. For example, protein import to the endoplasmic reticulum (ER) or the mitochondria require that proteins be threaded through specialized pores that depend on chaperones (12). Conversely, peroxisomes and the nucleus do not require protein unfolding for import (13). Perhaps, unsurprisingly, organelles can mount specific PQC responses. For example, the ER and the mitochondria both have specialized stress response machinery triggered in response to organellar stress (12, 14).
The nucleus is special with respect to PQC requirements because nuclear pores allow folded proteins and even large complexes in the native state to cross the nuclear membrane (15). Therefore, molecular chaperones associated with cotranslational protein folding should not strictly be required within the nucleus. Nevertheless, the Hsp70-40-110 system, the CCT chaperonin and its partner Prefoldin (PFD), Hsp90, and other chaperones are all present within the nucleus of yeast cells (16). This suggests, despite importing ostensibly native proteins, that the folding, refolding, and protein complex assembly functions of chaperones are critically required in the nucleus under normal conditions. The nucleus is a complex organelle packed with chromatin, RNA, and protein that requires a stress-responsive PQC machinery to facilitate dynamic responses to environmental stimuli that require proteome remodeling (6, 16, 17). In this review, we focus on Saccharomyces cerevisiae to highlight recent advances in the field of nuclear PQC under different stressed conditions, the pathways involved, and its key players. We also assess the crosstalk between nuclear chaperones, protein degradation pathways, and spatiotemporal organization of sequestered protein compartments, with a core focus specifically on the compartment.
Protein sequestration as a stress response
When PQC homeostasis is first overwhelmed during stress, cells acutely sequester and organize non-native proteins into specialized compartments/aggregates/inclusions. This pillar of PQC is tightly regulated by chaperones that bind to misfolded proteins and sequester them at specific sites in the cell to facilitate refolding or degradation. This process of compartmentalization has several proposed advantages. Spatially organizing misfolded proteins not only reduces their cytotoxicity but also reduces the burden on other PQC pathways (18, 19). This in turn prevents PQC collapse and promotes stress recovery (20, 21, 22). Furthermore, recruitment of degradation and refolding machineries to these sites could facilitate the clearance of misfolded proteins (explained later). Additionally, sequestration causes asymmetric inheritance of damaged proteins, resulting in inclusion-free daughter cells but contributing to replicative aging in mother cells (23, 24, 25).
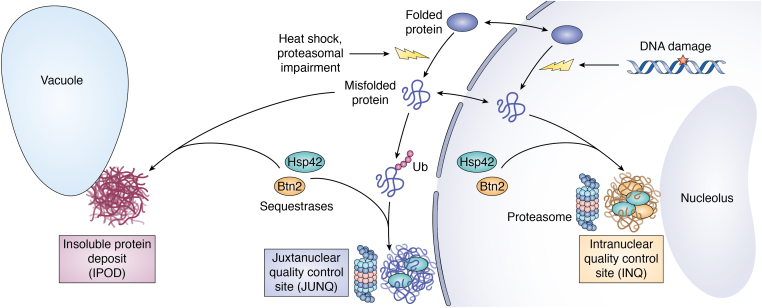
In yeast, various PQC deposition sites have been described including three major inclusions: the insoluble protein deposit (IPOD), the juxtanuclear quality control site (JUNQ), and the intranuclear quality control site (INQ) (21, 26, 27, 28). The IPOD is perivacuolar, adjacent to the preautophagosomal structure, and primarily made up of amyloidogenic proteins that are terminally aggregated, immobile and show a slow rate of diffusion with the cytosol (26) (Fig. 1). These include prions such as Rnq1, Sup35, and Ure2 and are reviewed as a PQC deposition site in detail elsewhere (29). On the other hand, the JUNQ is associated with the nuclear envelope but is peripheral to it and resides outside the nucleus. Contrary to the IPOD, the JUNQ is highly dynamic in nature, soluble, and concentrates the 26S proteasome and disaggregation machinery and therefore acts as an active site for protein degradation and refolding (26). Partitioning between these two sites depends on the misfolded protein’s ubiquitination state as blocking ubiquitination causes proteins to localize to IPOD instead of JUNQ and genetically engineering a ubiquitinated Rnq1-GFP localized to both the IPOD and JUNQ instead of only the IPOD (26). Other sites such as Q bodies, stress foci, and peripheral aggregates have also been found but have since been grouped together as CytoQ bodies that could potentially represent that same deposition site (22, 28, 30, 31). CytoQ bodies contain soluble misfolded proteins and undergo ATP-dependent coalescence that could represent the pathway that ultimately leads to JUNQ formation (9, 22).
Figure 1.
Major protein quality control sites in yeast. Shown are nuclear or cytoplasmic misfolded proteins during stress responses (lightning bolts) being recruited to the INQ, JUNQ, or IPOD through the action of sequestrases Hsp42 or Btn2. The localization of proteasomes to the INQ and the JUNQ are highlighted. Figure was created with BioRender. IPOD, insoluble protein deposit; JUNQ, juxtanuclear quality control site.
While the IPOD, JUNQ, and CytoQ reside in the cytoplasm, the INQ is present inside the nucleus and adjacent to the nucleolus (21, 27) (Fig. 1). Like JUNQ, the INQ is highly dynamic and contains the proteasome, disaggregation and degradation machineries, and is formed upon proteasome inhibition (21, 27, 28, 32). Since both the INQ and the JUNQ were discovered using the same model substrates, there has been an ongoing discussion about whether they represent the same or independent deposition sites. While this distinction needs to be investigated, recent studies have revealed some differences. Genotoxic stress induced via the alkylating agent methyl methanesulfonate (MMS) or oxidative damaging agent hydrogen peroxide induces the relocalization of several endogenous proteins to the INQ, whereas robust JUNQ formation is not seen (21, 27, 32). This DNA damage–dependent relocalization to the INQ is not affected by actin or microtubule depolymerizing drugs like latrunculin B or nocodazole, suggesting that an intact cytoskeleton might not be a prerequisite for INQ deposition (21). Additionally, the INQ formation does not require ubiquitination, suggesting that the INQ and JUNQ might indeed be two distinct deposition sites separated by the nuclear envelope (28). Since this review focuses on the nuclear PQC, going forward we will concern ourselves with the INQ as a sequestration site.
Intranuclear quality control: The nuclear protein sequestration structure
Similar to other PQC sites, the INQ was first discovered using aggregation prone reporter proteins like the small ubiquitin-like modifier (SUMO)–conjugating E2 enzyme temperature-sensitive (ts) allele Ubc9-ts-GFP and von Hippen Lindau protein (VHL) and was shown to reside inside the nucleus using immunoelectron microscopy (28). However, recent studies have found that many endogenous proteins are sequestered to the INQ upon genotoxic stress (Table 1) (21, 27, 32, 33). Its formation can be induced by various DNA damaging agents such as MMS, ethyl methanesulfonate, hydroxyurea, hydrogen peroxide, UV radiation, and even upon heat shock coupled with proteasome inhibition (21, 32, 33, 34). Various efforts have gone into cataloging the proteins that relocalize to the INQ using mass spectrometric approaches. To date, 37 proteins have been found to relocalize to the INQ (Table 1). These proteins, or the INQ substrates, perform a diverse set of functions such as chromatin remodeling and transcription (Rpd3, Hos2, Cmr1), RNA splicing (Hsh155, Cdc40), or replication fork structure cleavage (Mus81-Mms4 and Slx1-Slx4). The INQ also harbors various chaperones and degradation machinery, including the AAA ATPase Cdc48, the disaggregase Hsp104, the sHsps Hsp42 and Btn2, the Hsp40 J-domain protein Apj1, the Slx5/8 ubiquitin ligase, and the proteasome, that can together decide the fate of the INQ substrates (Fig. 2A and Table 1).
Table 1.
The INQ proteome
| Proteins | Function | References |
|---|---|---|
| Rpd3, Hos2, Arp6 | Chromatin remodeling | (21) |
| Mrc1, Cmr1a, Mus81, Mms4, Slx1, Slx4, Rad1 | Genome integrity | (21, 33) |
| Apc4a, Cdc20a, Cdc27 | Anaphase-promoting complex/Cyclosome | (21) |
| Hsh155, Cdc40a | Splicing | (21, 32) |
| Pph3, Pph22, Fig4 | Phosphatases | (21) |
| Btn2, Apj1, Cdc48, Sis1, Hsp104, Slx8, Rpn11, Cct6 | Chaperones and degradation machinery | (20, 21, 34, 45) |
| Gln1, Dug2a | Glutamine metabolism | (21) |
| Mkt1, Lst8, Emg1, Ylr126c, Rbd2, Dus3, Pal2, Qcr6, Mdh2 | Other | (21) |
Proteins containing one or more WD-40 repeats.
Figure 2.
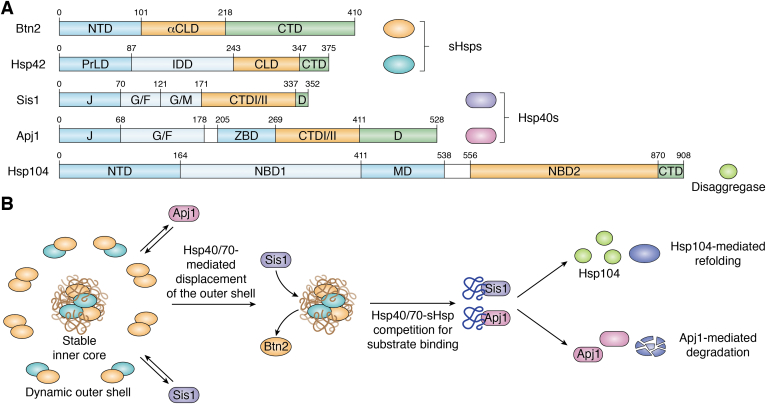
Chaperones at INQ.A, domain architecture of five chaperones that have been found at INQ. NTD = N-terminal domain; αCLD = alpha-crystallin–like domain; CTD = C-terminal domain; PrLD = Prion-like domain; IDD = intrinsically disordered domain; CLD = alpha crystallin domain; J = J-domain; G/F = glycine/phenylalanine rich domain; G/M = glycine/methionine rich domain; D = dimerization domain; ZBD = zinc-binding domain; NBD1/2 = nucleotide-binding domain 1 or 2; MD = middle domain. B, competition-based model between Hsp40/70-sHsp proteins decides INQ substrate fate. The model shows that an exchangeable shell of sHsps enables access of Hsp40/70 chaperones to non-native substrates ready to enter the refolding or degradation pathway. Figure was created with BioRender.
INQ formation is governed by two sequestrases, Btn2 and Hsp42, that increase in abundance upon DNA damage in an Hsf1-dependent manner (35). Sequestrases, usually sHsps, are ATP-independent chaperones that bind to the exposed hydrophobic regions of misfolded proteins and sequester them to specific deposition sites. Once bound to an sHsp, a misfolded protein is protected from aggregation but does not actively refold. Hence, sHsps are also termed holdases or aggregases as they hold misfolded proteins and are acted upon by other ATP-dependent chaperones to refold or degrade the misfolded protein. Each sHsp contains a characteristic alpha crystallin domain that is made up of antiparallel beta strands that form a beta sandwich (36). The alpha crystallin domain is flanked by two disordered regions, namely the N-terminal domain (NTD) and the C-terminal domain (CTD). Functional diversity amongst sHsps is determined by the NTD and CTD as differing length and amino acid makeup of the NTD and CTD can control substrate specificity (36). Yeast expresses two sHsps—Hsp26 and Hsp42 (36)—out of which Hsp42 is crucial for the INQ sequestration. The NTD of Hsp42 is vital for its aggregate sorting function as Hsp42 mutants deleted for its NTD do not restore formation of VHL aggregates upon heat shock and proteasomal impairment using MG132. Interestingly, swapping the Hsp42-NTD with Hsp26 to create a Hsp42NTD-Hsp26 chimera could efficiently sort VHL into aggregates, indicating that the NTD of Hsp42 determines its substrate specificity (31). This NTD was further dissected to reveal a prion-like domain that is crucial for CytoQ formation and acts as the major substrate-binding site (37). While these studies focused on Hsp42 domains and CytoQ formation, we can assume the same regions could be involved in the INQ sorting. In comparison, Btn2 is a v-SNARE-binding protein with stand alone sequestrase activity and sHsp-like features (20, 38)(Fig. 2A). It mediates late endosome-golgi recycling but has an established role as a crucial sequestrase for deposition of misfolded proteins at the INQ (28). Dissection of Btn2 domains revealed similarity with sHsps in that Btn2 contains an alpha-crystallin–like domain (aCLD) flanked by a disordered NTD and CTD. Unlike Hsp42, Btn2’s NTD is not involved in the INQ formation but rather acts as a recruitment domain for disaggregases that aid in the INQ solubilization (20). Instead, both the aCLD and CTD are essential for the INQ formation with the aCLD acting as the substrate-binding site showcasing the flexibility between various sequestrase domains in carrying out their activity.
The INQ substrate proteins
Why do only certain proteins get sequestered to the INQ? Of the 37 endogenous proteins so far found at the INQ, there do not appear to be any major functional enrichments. We speculate that a biophysical property or domain/motif could be common to some of the INQ substrates. Considering this, we performed a motif search for all the INQ substrates and found that six INQ proteins, Cmr1, Cdc20, Cdc40, Apc4, Lst8, and Dug2, contain at least one WD40 repeat. WD40 repeat domains are β-sheet enriched structural motifs that act as protein scaffolds and aid in protein–protein interactions. It has been shown that Cmr1’s WD40 domain is critical for its localization to the INQ (21). Non-native WD repeat proteins could explain why the CCT chaperonin, which is required for WD protein biogenesis, is also found at the INQ (21). Therefore, while sequences within the WD40 repeats could be involved in chaperone recognition, there does not appear to be a single domain or motif shared amongst all the INQ substrates. We also investigated whether a common sequence feature (Table 2) could be present in the INQ substrate proteins. To test this hypothesis, we calculated protein length, aliphatic index, hydropathy score (or GRAVY), percentage of charged amino acids per protein, protein abundance, solubility, enrichment of low-complexity regions (LCRs), and propensity to phase separate with the help of various databases and softwares currently available (listed in Table 2). On average, the INQ substrates are as long, stable, hydrophobic, enriched for charged amino acids, abundant, and soluble as the nuclear proteome, indicating the absence of a unique feature. However, the INQ substrates do have a significantly less percentage of LCRs (Table 2). LCRs and intrinsically disordered regions (IDRs) have been characterized as key features of phase separating proteins. These regions do not form canonical three-dimensional structures and are therefore more flexible and susceptible to interactions. Since the INQ is membraneless, stress dependent, and reversible, the presence of LCRs could be consistent with a model of liquid–liquid phase separation (LLPS). Over the last decade, LLPS has been revealed as a key physical phenomenon that regulates cellular organization and function (39). Proteins classified as scaffolds recruit and concentrate clients and phase separate forming a biomolecular condensate that is demixed from the surrounding cellular milieu. The formation, clearance, and controlled regulation of these condensates are a developing feature of PQC (40). While INQ substrates are not enriched for LCRs nor IDRs, both Hsp42 and Btn2 contain two IDRs required to form homodimers and higher-order oligomers (20, 36, 41). The presence of IDRs in the sHsps that concentrate the INQ substrates present a possibility that the INQ is phase separated. Identification and characterization of more INQ resident proteins will better define sequence features that promote nuclear protein sequestration.
Table 2.
Characteristics of the INQ substrates
| Features/p-valuesa | INQ versus whole proteome | INQ versus nuclear proteome | References |
|---|---|---|---|
| Length | 0.259 | 0.575 | - |
| Hydropathy score (GRAVY) | 0.182 | 0.147 | (84) |
| Aliphatic index | 0.065 | 0.645 | (85) |
| Charged amino acids % | 0.052 | 0.702 | - |
| Abundance (ppm) | 0.963 | 0.629 | (86) |
| Solubility score | 0.174 | 0.251 | (87) |
| Disorder % | 0.267 | 0.473 | (88) |
| LCR (low complexity regions)% | 0.095 | 0.040 | (89) |
| Pi-pi interaction score (phase separation propensity) | 0.262 | 0.694 | (90) |
Proteomic features were assessed using in-house R scripts, packages, and softwares cited in the table. The exact p-values resulting from a Mann–Whitney–Wilcoxon test after Bonferroni correction for multiple comparisons are reported (values in bold represent p-value <0.05)
While some of the INQ substrate proteins are likely misfolded and perhaps segregated due to LLPS, recent studies have also addressed the idea that sequestration to the INQ has specific regulatory roles during stress. For example, we found that the U2 spliceosome protein Hsh155 disassociated from its partners upon DNA damage and only Hsh155 was sequestered to the INQ (32). To cope with DNA damage, a transcriptional response represses ribosomal gene production and activates chaperones. This ensures reduced and controlled production of new proteins that help with stress recovery and preserving amino acid and energy availability. Since ribosomal transcripts are the major source of splicing flux in yeast, we proposed that Hsh155 sequestration and the associated splicing defect, served an additional posttranscriptional role in slowing ribosome production during stress (32). Consistent with this, in BTN2-deleted cells, Hsh155 is not sequestered to the INQ and splicing is restored even during DNA damage. In another study focusing on the structure-specific endonucleases Mus81-Mms4 which relocalize to the INQ as a complex (33), the authors find that Mus81 retention in the INQ is determined by the phosphorylation state of Mms4 and that, when phosphorylated, the Mus81-Mms4 is activated and released from the INQ. Accordingly, deleting Mms4 or prematurely phosphorylating it, retained Mus81 at the INQ or released Mus81 from the INQ, respectively. Furthermore, genetic perturbations that increased or decreased recombination intermediates that need to be resolved by Mus81-Mms4 greatly affected the release and persistence of this complex at the INQ (33). These studies reveal that nuclear functions are regulated by the INQ sequestration. We hypothesize that alongside its role as a PQC site, the INQ also acts as a “storage locker” for endogenous proteins that are sequestered to restrict or enhance their activity. These proteins are not necessarily misfolded but could be actively recruited to the INQ as part of stress responses. Another recent study on the INQ protein Mrc1 supports this model. The authors find that Mrc1 disassociates from the replication fork upon DNA damage and is sequestered at the INQ. Upon stress recovery, Mrc1 is released from the INQ and rejoins the replication fork for efficient restart of DNA replication (42). Together, this suggests the INQ is a unique kind of stress-responsive quality control compartment with protein triage and pathway regulatory functions.
Posttranslational modifications of the INQ proteins
Another key feature of phase separation, and one that governs INQ formation, is the presence of posttranslational modifications. To date, two posttranslational modifications, namely ubiquitination and SUMOylation have been implicated in the INQ formation and dissolution. Both have established roles in protein sorting, degradation, transport, and signaling, but their roles at the INQ are yet to be elucidated (43). Additionally, protein phosphatases, Pph21 and Pph22, and lysine deacetylases, Hos2 and Rpd3, localize to the INQ, raising the possibility that removal of these modifications could be important for the INQ resolution (Table 1).
Ubiquitination is critical for localization of misfolded proteins to JUNQ; however, a general connection between the INQ and ubiquitination has not been established (26). The case for a role of ubiquitin is more clear for some of the INQ substrates, such as Mrc1, which is ubiquitinated by the E3 ligase Dia2. Polyubiquitination of Mrc1 releases it from the replication fork and may serve as a recruitment signal to the INQ since it fails to relocalize to the INQ in a dia2Δ strain (21, 44). Interestingly, overexpression of Btn2 in a dia2Δ strain partially rescues the Mrc1 INQ localization, indicating that factors other than ubiquitination could govern INQ formation (42). Ubiquitination also controls the recruitment of Mus81-Mms4 to the INQ as deletion of the E3 ligase complex members Rad6 or Bre1 abrogates their sequestration (33). Whether this is a direct effect or an indirect result of altered DNA damage signaling is unknown.
SUMOylation on the other hand is suggested to be involved in turnover of proteins at the INQ (see later). SUMO also colocalizes with Cmr1 and Slx5 upon DNA damage confirming its presence at the INQ (21). Interestingly, we showed that sequestration of Hsh155 to the INQ upon DNA damage is dependent on SUMO conjugase and E3 ligase Ubc9 and Siz1, respectively. This implied that the INQ substrates could be SUMOylated upon DNA damage and that this could act as a recruitment signal for Btn2 and Hsp42 to sequester proteins to the INQ. Indeed, the SUMO-binding molecular chaperone Cdc48/VCP is also recruited to the INQ where it functions to counteract INQ persistence (45). Surprisingly, we also find that Hsh155 is not SUMOylated, indicating that SUMOylation could act as a sorting signal to the INQ (unpublished observation). Further studies focusing on the INQ and SUMOylation should reveal the SUMOylated proteome at the INQ, its role in the formation, and consequently clearance of the INQ.
Refolding of the INQ proteins
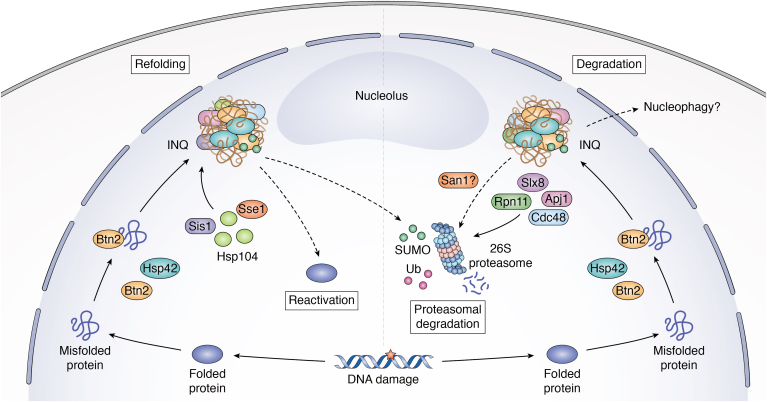
Once sequestered in the INQ compartment, substrate proteins are protected until stresses are removed or cells begin to recover. In our experiments, wash out of MMS treatment leads to rapid disappearance of the INQ within 1 h and complete reversal in about 2 h (32). The fate of most of the INQ substrate proteins is not known, but they must be either reactivated or degraded in concert with the disappearance of the INQ (Fig. 2). As noted previously, DNA replication and repair proteins like Mus81-Mms4 or Mrc1 seem capable of returning to their cellular functions to help restart the cell cycle (33, 42). Whether this is generalizable is unclear, however, there is evidence of an active refolding machinery at the INQ. First, the complement of chaperones that localize to the INQ include the Hsp104 disaggregase, which is associated with protein refolding in partnership with the Hsp70-40-110 system. Indeed, the Hsp110 protein Sse1 is also involved in the INQ clearance, and loss of either Hsp104 or Sse1 leads to a dramatic increase in the INQ foci during stress (32, 42). Interestingly, we observed spontaneous localization to the INQ for Hsh155 in sse1Δ mutants under unstressed conditions, indicating Sse1 might also be important for its de novo protein folding (32). The direct effect of the Hsp40 chaperone Sis1 on the INQ is not clear; however, by measuring nuclear luciferase activity after heat stress, it has been shown to work together with Hsp70–Hsp104 to help in the INQ protein refolding. In fact Btn2 is involved in recruiting Sis1 to the INQ and consequently Hsp70-104 as part of this refolding pathway (20).
Sequestrases/sHsps prevent irreversible aggregation by binding to misfolded proteins to form solubilized assemblies. These assemblies form two substructures—a dynamic outer shell with continuous binding and release of sHsps and an inner stable core, which contains fewer sHsps bound to their substrates (46). Hsp70 is recruited to these assemblies through its cochaperone Hsp40, which then triggers ATP hydrolysis and subsequent Hsp70–substrate interaction through its J-domain (47). A competition-based model suggests that Hsp70 displaces sHsps in the outer shell by competing for binding to misfolded substrates in the inner core (Fig. 2B). This in turn excludes new sHsps from re-entering and reassociating with these assemblies as Hsp70s can only be disassociated after interacting with nucleotide exchange factors (NEFs) that release ADP, allowing ATP to rebind to Hsp70 and release substrates. Hsp70 bound to substrates further recruits and activates the disaggregase Hsp104, which can thread the misfolded proteins through its central channel and help refold them (48, 49). While presumably the mechanisms defining the sHsp-Hsp70-Hsp104 system are at play during the reactivation of some of the INQ proteins, there are not yet studies that directly test reactivation of the endogenous INQ substrates by this pathway (Fig. 2B).
A recent study showcased a negative genetic interaction at high temperatures between Btn2 or Hsp42 and a background lacking both the NEF Fes1 and Hsp104 (20). Deleting Fes1 and Hsp104 together results in a stabilized Hsp70–substrate interaction, thus reducing the pool of active Hsp70 and hindering the overall activity of this pathway. This temperature sensitivity was neither seen with cells lacking Hsp26 nor upon inhibiting Hsp90 activity, indicating that this defect is specific to the INQ sequestrases and low Hsp70 activity and that sequestrase activity and therefore INQ formation becomes crucial when Hsp70 activity is limited. This finding was supported by a phenotypic rescue upon expressing Sse1, another NEF largely excluded from the nucleus, with a nuclear localization sequence (NLS). Expressing Hsp42 with a NLS in a btn2Δfes1Δhsp104Δ or expressing Btn2 lacking its NLS in a hsp42Δfes1Δhsp104Δ background also rescued the temperature sensitivity, indicating that the site of sequestration is interchangeable, and as long as sequestration occurs, the cells can cope with stress (20).
Surprisingly, a Btn2 construct without its NTD (Btn2-ΔNTD-NLS) not only rescued the growth defect in a btn2Δfes1Δhsp104Δ background but also grew better than fes1Δhsp104Δ at high temperatures. This gain of function was explained by the Btn2-NTD mediating recruitment of disaggregases to the INQ for INQ solubilization (20). Btn2 NTD recruits Sis1 and consequently Hsp70 to the INQ, which in turn recruits Hsp104. Therefore, in a Btn2-ΔNTD-NLS fes1Δhsp104Δ background, the Hsp70-Hsp104 bichaperone system is not recruited to the INQ. This slightly increases Hsp70 availability throughout the cell and simultaneously leads to INQ persistence since the INQ would prevent irreversible aggregation of misfolded proteins (20). Interestingly, similar luciferase assays using another Hsp40 chaperone Apj1 did not show a function in refolding but degradation of nuclear aggregates instead (34), thus indicating some misfolded INQ aggregates are destined for proteasomal degradation rather than refolding and reactivation.
Degradation of nuclear proteins and the INQ
Misfolded nuclear proteins that cannot be refolded must be subject to degradation. Degradation-mediated pathways depend on polyubiquitination and cleavage of misfolded proteins by the ubiquitin proteasome system (UPS) consisting of ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin protein ligases (E3) along with the help of chaperones (50, 51). Misfolded proteins that are tagged with ubiquitin are eliminated by the proteasome. The ubiquitin-binding proteins in the proteasome work along with ATPase chaperones to extract and translocate the misfolded proteins for degradation. While many UPS factors are found in the cytosol, 26S proteasomes are predominantly found in the nucleus and are critical for nuclear PQC (52). Protein degradation pathways in the cytoplasm and ER have been discussed extensively (53), but PQC mechanisms and substrates in the nucleus have largely been described only in the last 10 years (6). ER-associated degradation (ERAD) is a major pathway for the degradation of membrane proteins in ER, and Hrd1 and Doa10 are the two main E3 ubiquitin ligases targeting ERAD substrates in yeast (54). While Hrd1 is an exclusive ERAD membrane protein, Doa10 was also found in the inner nuclear membrane where it aids nuclear PQC (55). Nuclear degradation pathways are reviewed extensively elsewhere (6, 7, 51) but we will briefly summarize those relevant to the INQ here. Direct evidence of UPS proteins at the INQ is limited, but our observations and others suggest that by inhibiting the proteasome using MG132 the INQ substrates like Cmr1, Hsh155, and Mrc1 aggregate more aggressively under stress (21, 32, 42). This implies that the proteasome and degradation of its substrates is essential for maintaining INQ homeostasis, which is supported by the enrichment of the proteasomal subunit Rpn11 at the INQ (21).
UPS and chaperones in the INQ substrate degradation
San1 is an E3 ligase with a RING domain that localizes to the nucleus and functions in concert with two E2 enzymes, Ubc1 and Cdc34 (6, 56). The role of San1 in nuclear PQC was discovered in yeast when two mutated nuclear proteins, Sir4 and Cdc68, were rapidly degraded in a SAN1-dependent manner (57). San1 recognizes exposed hydrophobic regions of its misfolded substrates that are aggregation prone (58). Through biochemical assays and structural analysis, San1 was shown to use flexible and unstructured NTD and CTD to bind directly to its substrates without direct help from chaperones (59). However, Hsp70 chaperone proteins Ssa1 and Ssa2 (60) and Cdc48 complex (61) are essential for ubiquitination in the San1 degradation pathway by enhancing substrate solubility. San1 also works in collaboration with another E3 ligase, Ubr1, for both cytoplasmic and nuclear protein degradation (62). Surprisingly, immunostaining of the INQ revealed the absence of ubiquitin, which was further tested using two misfolded substrates tGnd1-GFP and ΔΔCPY∗ that can be ubiquitinated by San1 and Ubr1 (28). Their sequestration to the INQ was unaffected by San1 or Ubr1 status thus arguing against the role of ubiquitination as a sorting signal in the INQ. This was further supported for the splicing factor Hsh155 whose localization at the INQ was unaffected by SAN1 deletion (32). In contrast, deletion mutants of SAN1 showed delayed clearance of Mrc1 from the INQ under stress (42). In this same study, Cdc48 was identified as a regulator of Mrc1 retention in the INQ, raising questions about whether a Cdc48-San1 degradation pathway may exist for some of the INQ substrate proteins.
The Cdc48 ATPase complex is a highly conserved chaperone that functions in protein complex disassembly, and degradation. Cdc48 functions as a chaperone in processes like ERAD, membrane fusion, autophagy, cell cycle regulation, and replication stress (63, 64, 65). Misfolded Cdc48 substrates are ubiquitinated, bound by Cdc48, and delivered to the proteasome for degradation (66). Importantly, Cdc48 localizes to the INQ during cellular stress and suppresses the INQ substrate retention under both stressed and unstressed conditions (45). To date, Cdc48 has been shown to regulate the localization and turnover of the two INQ substrates, Hsh155 and Mrc1 (42, 45). The Hsh155 INQ aggregation was significantly increased in CDC48 ts mutants. The ATPase dead allele of Cdc48 could not rescue the cdc48-ts phenotype compared to the WT CDC48 plasmid, indicating that the ATPase activity is essential to retain Hsh155 aggregates in the INQ. Hsh155 interacts directly with Cdc48, whereas Mrc1 interacts with Cdc48 cofactors like Otu1, Ubx5, and Ufd1 as shown by bimolecular fluorescence complementation (42, 45). Cdc48 binding and interaction with these INQ substrates also influences their function in splicing (Hsh155) and replication (Mrc1). Thus, Cdc48 is likely an important chaperone adaptor for the degradation of at least a subset of the INQ proteins.
Hsp40 chaperones like Ydj1 and Sis1 together with E3 ligases and Hsp70 help to maintain protein stability and support protein degradation pathways in general (67, 68). Although Mrc1 showed Cdc48-dependent interactions with Ydj1 and Sis1 (42), a direct involvement of an Hsp40 chaperone in the INQ substrate degradation pathway was not clear until recent descriptions of the J-protein Apj1 (34). Apj1 localizes to the INQ under stress (21, 27, 34) and affects the distribution and sequestration of the INQ substrates Hsh155 and Mrc1 (32, 42). Using heat-aggregated nuclear luciferase (LuciDM-NLS-GFP) and measuring the recovery of the luciferase activity after heat-induced inactivation, apj1Δ cells showed increased protein recovery and stability (34). These authors suggest that Apj1, along with Hsp70 and Hsp110, supports nuclear protein disaggregation and solubility independently of Hsp104. Moreover, Apj1 was found to promote degradation of aggregated nuclear proteins and compete with Hsp104 that drives refolding. This suggests that two independent outcomes exist as a protective mechanism in the nucleus to eliminate stress-induced toxic protein aggregates: an Hsp104-dependent pathway for refolding and an Apj1-dependent pathway for degradation.
Interestingly, loss of APJ1 shows synthetic lethal interactions with slx5Δ, which is a subunit of a SUMO targeted ubiquitin ligase (STUbL) (69). The STUbL complex of Slx5 and Slx8 regulate ubiquitination and degradation of substrates that have been tagged with the SUMO (70, 71). While it is unclear whether degradation of the INQ substrates by Apj1 depends on STUbLs or SUMO, the majority of STUbL substrates are nuclear proteins including transcription factors (72), DNA replication and repair proteins (73, 74), and other chromatin-associated proteins. As noted previously, Ubc9 (SUMO conjugase) and Siz1 (SUMO E3 ligase) promote the INQ aggregation of Hsh155, and the Cdc48 regulation of the Hsh155 INQ localization is dependent on Siz1 (45). STUbLs recognize SUMO via SUMO-interacting motifs for ubiquitination and degradation of substrates. Interestingly, mutation of the Cdc48 SUMO-interacting motif domain failed to rescue the effects of cdc48-ts on the INQ compared to the WT CDC48 plasmid (45). Overall, SUMO-directed degradation of misfolded proteins in the INQ needs additional study given the circumstantial evidence that SUMO may play an important regulatory role.
Nucleophagy and the fate of the INQ
Refolding and degradation are two fates of the misfolded INQ substrates (Fig. 3). Another important pathway that exists in eukaryotes for safe and regulated elimination of misfolded proteins is autophagy (75). Autophagy can be the selective or nonselective degradation of bulk protein complexes from cytosol, nucleus, and other organelles (76). The specific autophagic pathway targeting the nucleus is called nucleophagy. This process comprises the selective degradation of the nucleus where a nuclear bleb destined for degradation is directly engulfed and sequestered into a vacuolar membrane invagination (77). For example, upon nitrogen starvation, nucleophagy is initiated by the interaction of the outer nuclear membrane protein Nvj1 and the vacuolar membrane protein Vac8 at the nucleus–vacuole junctions (78). This process is facilitated by a spectrum of proteins encoded by core autophagy-related genes (79). It is unclear whether the nucleophagy process can influence clearance of the INQ substrates. However, given the known position of the INQ at the nuclear periphery adjacent to the nucleolus, this structure would seem to be in the right spot for a potential contribution of nucleophagy. Perhaps, if refolding and UPS-mediated degradation pathways are insufficient to handle the load of non-native proteins, the INQ could be subject to nucleophagy. Further research will be required to explore this possibility.
Figure 3.
Players and pathways for the INQ protein refolding or degradation. The left half shows potential regulators of the INQ substrate fate involving protein refolding/reactivation. The right half shows the INQ substrate fates involving proteasomal degradation (and potentially nucleophagy). DNA damage is indicated as a strong stress for endogenous protein aggregation, but other stresses can also induce the INQ. Figure was created with BioRender.
Conclusion
The genome, chromatin states, and transcriptome respond dynamically to stressful conditions. It is now clear that the nuclear proteome is no exception and must be protected and remodeled during times of stress. Yeast sequester endogenous nuclear proteins into the INQ compartment, especially during times of genotoxic stress (21, 27, 32, 33, 34, 42, 45). The INQ then serves as an organizing center for triage decisions leading to protein reactivation and refolding or degradation. Since only a subset of nuclear proteins seems to relocalize to the INQ following MMS treatment, we speculate based on the literature and our observations that both biophysical properties of these proteins and the regulated functional needs of the cell determine sequestration in the INQ. In addition, as noted previously, the roles of SUMOylation, ubiquitination, and potentially phosphorylation or acetylation in promoting the INQ formation and resolution are unclear. As we understand more about the complete substrate repertoire of the INQ under various environmental conditions and the regulatory mechanisms to target some proteins to these sites, we will be able to make a clearer description of triage paths for the INQ resident proteins.
We also note that many of the INQ substrate proteins are conserved across species, as are the chaperone systems responsible for aggregating and disaggregating the INQ proteins. Indeed, the human homolog of Cmr1, called WDR76, localizes to uncharacterized nuclear foci upon MMS or MG132 treatment of U2OS cells (21). In addition, it is notable that the INQ is positioned in proximity to the nucleolus, which itself is a phase-separated quality control compartment in human cells (80). Moreover, the ubiquitin-like protein NEDD8 has been shown to define a separate but nucleolar-related stress-induced aggregate structure in human cells (81). We also know that promyelocytic leukemia bodies are sites of stress-induced PQC regulated by SUMOylation in humans (reviewed in (82)). Given the complexity of inclusion formation in the nuclei of human cells under stress, it is difficult to ascribe any one of these locations as a direct counterpart of the yeast INQ structures. However, given all we are learning about LLPS and the regulatory functions of the INQ, analogies to human nuclear PQC will continue to emerge. The degree to which we can understand the organizing principles of nuclear PQC across species is important as multiple human diseases have been linked to defective nucleocytoplasmic transport and potential nuclear proteome stress (reviewed in (83)).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
A. K., V. M., and P. C. S. conceptualization; A. K. investigation; A. K., V. M., and P. C. S. writing–original draft; A. K. and V. M. visualization; P. C. S. supervision; P. C. S. project administration; P. C. S. funding acquisition.
Funding and additional information
This work was supported by a Natural Sciences and Engineering Research Council of Canada discovery grant to P. C. S. (RGPIN-2020-04360). P. C. S. is a Michael Smith Foundation for Health Research Scholar.
Edited by Phyllis Hanson
References
- 1.Woerner A.C., Frottin F., Hornburg D., Feng L.R., Meissner F., Patra M., et al. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science. 2016;351:173–176. doi: 10.1126/science.aad2033. [DOI] [PubMed] [Google Scholar]
- 2.Chou C.-C., Zhang Y., Umoh M.E., Vaughan S.W., Lorenzini I., Liu F., et al. TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci. 2018;21:228–239. doi: 10.1038/s41593-017-0047-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winklhofer K.F., Tatzelt J., Haass C. The two faces of protein misfolding: gain- and loss-of-function in neurodegenerative diseases. EMBO J. 2008;27:336–349. doi: 10.1038/sj.emboj.7601930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soto C., Pritzkow S. Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci. 2018;21:1332–1340. doi: 10.1038/s41593-018-0235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross C.A., Poirier M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10:S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 6.Enam C., Geffen Y., Ravid T., Gardner R.G. Protein quality control degradation in the nucleus. Annu. Rev. Biochem. 2018;87:725–749. doi: 10.1146/annurev-biochem-062917-012730. [DOI] [PubMed] [Google Scholar]
- 7.Franić D., Zubčić K., Boban M. Nuclear ubiquitin-proteasome pathways in proteostasis maintenance. Biomolecules. 2021;11:54. doi: 10.3390/biom11010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciechanover A., Kwon Y.T. Protein quality control by molecular chaperones in neurodegeneration. Front. Neurosci. 2017;11:185. doi: 10.3389/fnins.2017.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sontag E.M., Samant R.S., Frydman J. Mechanisms and functions of spatial protein quality control. Annu. Rev. Biochem. 2017;86:97–122. doi: 10.1146/annurev-biochem-060815-014616. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y.E., Hipp M.S., Bracher A., Hayer-Hartl M., Hartl F.U. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 11.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 2013;14:630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Needham P.G., Guerriero C.J., Brodsky J.L. Chaperoning endoplasmic reticulum-associated degradation (ERAD) and protein conformational diseases. Cold Spring Harb. Perspect. Biol. 2019;11:a033928. doi: 10.1101/cshperspect.a033928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker A., Lanyon-Hogg T., Warriner S.L. Peroxisome protein import: a complex journey. Biochem. Soc. Trans. 2016;44:783–789. doi: 10.1042/BST20160036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weidberg H., Amon A. MitoCPR-A surveillance pathway that protects mitochondria in response to protein import stress. Science. 2018;360:eaan4146. doi: 10.1126/science.aan4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamenova I., Mukherjee P., Conic S., Mueller F., El-Saafin F., Bardot P., et al. Co-translational assembly of mammalian nuclear multisubunit complexes. Nat. Commun. 2019;10:1740. doi: 10.1038/s41467-019-09749-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shibata Y., Morimoto R.I. How the nucleus copes with proteotoxic stress. Curr. Biol. 2014;24:R463–474. doi: 10.1016/j.cub.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones R.D., Gardner R.G. Protein quality control in the nucleus. Curr. Opin. Cell Biol. 2016;40:81–89. doi: 10.1016/j.ceb.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ungelenk S., Moayed F., Ho C.-T., Grousl T., Scharf A., Mashaghi A., et al. Small heat shock proteins sequester misfolding proteins in near-native conformation for cellular protection and efficient refolding. Nat. Commun. 2016;7 doi: 10.1038/ncomms13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolfe K.J., Ren H.Y., Trepte P., Cyr D.M. The Hsp70/90 cochaperone, sti1, suppresses proteotoxicity by regulating spatial quality control of amyloid-like proteins. Mol. Biol. Cell. 2013;24:3588–3602. doi: 10.1091/mbc.E13-06-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho C.-T., Grousl T., Shatz O., Jawed A., Ruger-Herreros C., Semmelink M., et al. Cellular sequestrases maintain basal Hsp70 capacity ensuring balanced proteostasis. Nat. Commun. 2019;10:4851. doi: 10.1038/s41467-019-12868-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallina I., Colding C., Henriksen P., Beli P., Nakamura K., Offman J., et al. Cmr1/WDR76 defines a nuclear genotoxic stress body linking genome integrity and protein quality control. Nat. Commun. 2015;6:6533. doi: 10.1038/ncomms7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Escusa-Toret S., Vonk W.I.M., Frydman J. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat. Cell Biol. 2013;15:1231–1243. doi: 10.1038/ncb2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill S.M., Hao X., Grönvall J., Spikings-Nordby S., Widlund P.O., Amen T., et al. Asymmetric inheritance of aggregated proteins and age reset in yeast are regulated by Vac17-dependent vacuolar functions. Cell Rep. 2016;16:826–838. doi: 10.1016/j.celrep.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saarikangas J., Barral Y. Protein aggregates are associated with replicative aging without compromising protein quality control. eLife. 2015;4 doi: 10.7554/eLife.06197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou C., Slaughter B.D., Unruh J.R., Guo F., Yu Z., Mickey K., et al. Organelle-based aggregation and retention of damaged proteins in asymmetrically dividing cells. Cell. 2014;159:530–542. doi: 10.1016/j.cell.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaganovich D., Kopito R., Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tkach J.M., Yimit A., Lee A.Y., Riffle M., Costanzo M., Jaschob D., et al. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 2012;14:966–976. doi: 10.1038/ncb2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller S.B.M., Ho C.-T., Winkler J., Khokhrina M., Neuner A., Mohamed M.Y.H., et al. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J. 2015;34:778–797. doi: 10.15252/embj.201489524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothe S., Prakash A., Tyedmers J. The Insoluble Protein Deposit (IPOD) in yeast. Front. Mol. Neurosci. 2018;11:237. doi: 10.3389/fnmol.2018.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spokoini R., Moldavski O., Nahmias Y., England J.L., Schuldiner M., Kaganovich D. Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Rep. 2012;2:738–747. doi: 10.1016/j.celrep.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 31.Specht S., Miller S.B.M., Mogk A., Bukau B. Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J. Cell Biol. 2011;195:617–629. doi: 10.1083/jcb.201106037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathew V., Tam A.S., Milbury K.L., Hofmann A.K., Hughes C.S., Morin G.B., et al. Selective aggregation of the splicing factor Hsh155 suppresses splicing upon genotoxic stress. J. Cell Biol. 2017;216:4027–4040. doi: 10.1083/jcb.201612018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saugar I., Jiménez-Martín A., Tercero J.A. Subnuclear relocalization of structure-specific endonucleases in response to DNA damage. Cell Rep. 2017;20:1553–1562. doi: 10.1016/j.celrep.2017.07.059. [DOI] [PubMed] [Google Scholar]
- 34.den Brave F., Cairo L.V., Jagadeesan C., Ruger-Herreros C., Mogk A., Bukau B., et al. Chaperone-mediated protein disaggregation triggers proteolytic clearance of intra-nuclear protein inclusions. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Solís E.J., Pandey J.P., Zheng X., Jin D.X., Gupta P.B., Airoldi E.M., et al. Defining the essential function of yeast Hsf1 reveals a compact transcriptional program for maintaining eukaryotic proteostasis. Mol. Cell. 2016;63:60–71. doi: 10.1016/j.molcel.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haslbeck M., Weinkauf S., Buchner J. Small heat shock proteins: simplicity meets complexity. J. Biol. Chem. 2019;294:2121–2132. doi: 10.1074/jbc.REV118.002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grousl T., Ungelenk S., Miller S., Ho C.-T., Khokhrina M., Mayer M.P., et al. A prion-like domain in Hsp42 drives chaperone-facilitated aggregation of misfolded proteins. J. Cell Biol. 2018;217:1269–1285. doi: 10.1083/jcb.201708116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kama R., Robinson M., Gerst J.E. Btn2, a Hook1 ortholog and potential Batten disease-related protein, mediates late endosome-Golgi protein sorting in yeast. Mol. Cell. Biol. 2007;27:605–621. doi: 10.1128/MCB.00699-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hyman A.A., Weber C.A., Jülicher F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 40.Alberti S., Hyman A.A. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 2021;22:196–213. doi: 10.1038/s41580-020-00326-6. [DOI] [PubMed] [Google Scholar]
- 41.Haslbeck M., Braun N., Stromer T., Richter B., Model N., Weinkauf S., et al. Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J. 2004;23:638–649. doi: 10.1038/sj.emboj.7600080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graumans W., Stone W.J.R., Bousema T. No time to die: an in-depth analysis of James Bond’s exposure to infectious agents. Trav. Med. Infect. Dis. 2021;44 doi: 10.1016/j.tmaid.2021.102175. [DOI] [PubMed] [Google Scholar]
- 43.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 44.Mimura S., Komata M., Kishi T., Shirahige K., Kamura T. SCF(Dia2) regulates DNA replication forks during S-phase in budding yeast. EMBO J. 2009;28:3693–3705. doi: 10.1038/emboj.2009.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mathew V., Kumar A., Jiang Y.K., West K., Tam A.S., Stirling P.C. Cdc48 regulates intranuclear quality control sequestration of the Hsh155 splicing factor in budding yeast. J. Cell Sci. 2020;133:jcs252551. doi: 10.1242/jcs.252551. [DOI] [PubMed] [Google Scholar]
- 46.Żwirowski S., Kłosowska A., Obuchowski I., Nillegoda N.B., Piróg A., Ziętkiewicz S., et al. Hsp70 displaces small heat shock proteins from aggregates to initiate protein refolding. EMBO J. 2017;36:783–796. doi: 10.15252/embj.201593378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kampinga H.H., Craig E.A. The HSP70 chaperone machinery: j proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J., Kim J.-H., Biter A.B., Sielaff B., Lee S., Tsai F.T.F. Heat shock protein (Hsp) 70 is an activator of the Hsp104 motor. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8513–8518. doi: 10.1073/pnas.1217988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosenzweig R., Moradi S., Zarrine-Afsar A., Glover J.R., Kay L.E. Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science. 2013;339:1080–1083. doi: 10.1126/science.1233066. [DOI] [PubMed] [Google Scholar]
- 50.Ciechanover A. Intracellular protein degradation from a vague idea through the lysosome and the ubiquitin-proteasome system and on to human diseases and drug targeting: nobel Lecture, December 8, 2004. Ann. N. Y. Acad. Sci. 2007;1116:1–28. doi: 10.1196/annals.1402.078. [DOI] [PubMed] [Google Scholar]
- 51.Boban M., Foisner R. Degradation-mediated protein quality control at the inner nuclear membrane. Nucl. Austin Tex. 2016;7:41–49. doi: 10.1080/19491034.2016.1139273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chowdhury M., Enenkel C. Intracellular dynamics of the ubiquitin-proteasome-system. F1000Research. 2015;4:367. doi: 10.12688/f1000research.6835.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buchberger A., Bukau B., Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol. Cell. 2010;40:238–252. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 54.Vembar S.S., Brodsky J.L. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deng M., Hochstrasser M. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature. 2006;443:827–831. doi: 10.1038/nature05170. [DOI] [PubMed] [Google Scholar]
- 56.Ibarra R., Sandoval D., Fredrickson E.K., Gardner R.G., Kleiger G. The San1 ubiquitin ligase functions preferentially with ubiquitin-conjugating enzyme Ubc1 during protein quality control. J. Biol. Chem. 2016;291:18778–18790. doi: 10.1074/jbc.M116.737619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gardner R.G., Nelson Z.W., Gottschling D.E. Degradation-mediated protein quality control in the nucleus. Cell. 2005;120:803–815. doi: 10.1016/j.cell.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 58.Paulsson M. The role of Ca2+ binding in the self-aggregation of laminin-nidogen complexes. J. Biol. Chem. 1988;263:5425–5430. [PubMed] [Google Scholar]
- 59.Rosenbaum J.C., Fredrickson E.K., Oeser M.L., Garrett-Engele C.M., Locke M.N., Richardson L.A., et al. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol. Cell. 2011;41:93–106. doi: 10.1016/j.molcel.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones R.D., Enam C., Ibarra R., Borror H.R., Mostoller K.E., Fredrickson E.K., et al. The extent of Ssa1/Ssa2 Hsp70 chaperone involvement in nuclear protein quality control degradation varies with the substrate. Mol. Biol. Cell. 2020;31:221–233. doi: 10.1091/mbc.E18-02-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gallagher P.S., Clowes Candadai S.V., Gardner R.G. The requirement for Cdc48/p97 in nuclear protein quality control degradation depends on the substrate and correlates with substrate insolubility. J. Cell Sci. 2014;127:1980–1991. doi: 10.1242/jcs.141838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heck J.W., Cheung S.K., Hampton R.Y. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1106–1111. doi: 10.1073/pnas.0910591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baek G.H., Cheng H., Choe V., Bao X., Shao J., Luo S., et al. Cdc48: a swiss army knife of cell biology. J. Amino Acids. 2013;2013 doi: 10.1155/2013/183421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bergink S., Ammon T., Kern M., Schermelleh L., Leonhardt H., Jentsch S. Role of Cdc48/p97 as a SUMO-targeted segregase curbing Rad51-Rad52 interaction. Nat. Cell Biol. 2013;15:526–532. doi: 10.1038/ncb2729. [DOI] [PubMed] [Google Scholar]
- 65.Ramadan K., Halder S., Wiseman K., Vaz B. Strategic role of the ubiquitin-dependent segregase p97 (VCP or Cdc48) in DNA replication. Chromosoma. 2017;126:17–32. doi: 10.1007/s00412-016-0587-4. [DOI] [PubMed] [Google Scholar]
- 66.Elsasser S., Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat. Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- 67.Caplan A.J., Douglas M.G. Characterization of YDJ1: a yeast homologue of the bacterial dnaJ protein. J. Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Summers D.W., Wolfe K.J., Ren H.Y., Cyr D.M. The Type II Hsp40 Sis1 cooperates with Hsp70 and the E3 ligase Ubr1 to promote degradation of terminally misfolded cytosolic protein. PLoS One. 2013;8 doi: 10.1371/journal.pone.0052099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sahi C., Kominek J., Ziegelhoffer T., Yu H.Y., Baranowski M., Marszalek J., et al. Sequential duplications of an ancient member of the DnaJ-family expanded the functional chaperone network in the eukaryotic cytosol. Mol. Biol. Evol. 2013;30:985–998. doi: 10.1093/molbev/mst008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Uzunova K., Göttsche K., Miteva M., Weisshaar S.R., Glanemann C., Schnellhardt M., et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 2007;282:34167–34175. doi: 10.1074/jbc.M706505200. [DOI] [PubMed] [Google Scholar]
- 71.Geoffroy M.-C., Hay R.T. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell Biol. 2009;10:564–568. doi: 10.1038/nrm2707. [DOI] [PubMed] [Google Scholar]
- 72.Wang Z., Prelich G. Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol. Cell. Biol. 2009;29:1694–1706. doi: 10.1128/MCB.01470-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Psakhye I., Castellucci F., Branzei D. SUMO-Chain-Regulated proteasomal degradation timing exemplified in DNA replication initiation. Mol. Cell. 2019;76:632–645.e6. doi: 10.1016/j.molcel.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Talhaoui I., Bernal M., Mullen J.R., Dorison H., Palancade B., Brill S.J., et al. Slx5-Slx8 ubiquitin ligase targets active pools of the Yen1 nuclease to limit crossover formation. Nat. Commun. 2018;9:5016. doi: 10.1038/s41467-018-07364-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cohen-Kaplan V., Livneh I., Avni N., Cohen-Rosenzweig C., Ciechanover A. The ubiquitin-proteasome system and autophagy: coordinated and independent activities. Int. J. Biochem. Cell Biol. 2016;79:403–418. doi: 10.1016/j.biocel.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 76.Kiel J.A.K.W. Autophagy in unicellular eukaryotes. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2010;365:819–830. doi: 10.1098/rstb.2009.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mijaljica D., Prescott M., Devenish R.J. A late form of nucleophagy in Saccharomyces cerevisiae. PLoS One. 2012;7 doi: 10.1371/journal.pone.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kvam E., Goldfarb D.S. Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy. 2007;3:85–92. doi: 10.4161/auto.3586. [DOI] [PubMed] [Google Scholar]
- 79.Krick R., Muehe Y., Prick T., Bremer S., Schlotterhose P., Eskelinen E.-L., et al. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol. Biol. Cell. 2008;19:4492–4505. doi: 10.1091/mbc.E08-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frottin F., Schueder F., Tiwary S., Gupta R., Körner R., Schlichthaerle T., et al. The nucleolus functions as a phase-separated protein quality control compartment. Science. 2019;365:342–347. doi: 10.1126/science.aaw9157. [DOI] [PubMed] [Google Scholar]
- 81.Maghames C.M., Lobato-Gil S., Perrin A., Trauchessec H., Rodriguez M.S., Urbach S., et al. NEDDylation promotes nuclear protein aggregation and protects the ubiquitin proteasome system upon proteotoxic stress. Nat. Commun. 2018;9:4376. doi: 10.1038/s41467-018-06365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sahin U., de Thé H., Lallemand-Breitenbach V. PML nuclear bodies: assembly and oxidative stress-sensitive sumoylation. Nucl. Austin Tex. 2014;5:499–507. doi: 10.4161/19491034.2014.970104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bitetto G., Di Fonzo A. Nucleo-cytoplasmic transport defects and protein aggregates in neurodegeneration. Transl. Neurodegener. 2020;9:25. doi: 10.1186/s40035-020-00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 85.Ikai A. Thermostability and aliphatic index of globular proteins. J. Biochem. (Tokyo). 1980;88:1895–1898. [PubMed] [Google Scholar]
- 86.Wang M., Herrmann C.J., Simonovic M., Szklarczyk D., von Mering C. Version 4.0 of PaxDb: protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics. 2015;15:3163–3168. doi: 10.1002/pmic.201400441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sormanni P., Aprile F.A., Vendruscolo M. The CamSol method of rational design of protein mutants with enhanced solubility. J. Mol. Biol. 2015;427:478–490. doi: 10.1016/j.jmb.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 88.Jones D.T., Cozzetto D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinforma. Oxf. Engl. 2015;31:857–863. doi: 10.1093/bioinformatics/btu744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Harrison P.M. fLPS: fast discovery of compositional biases for the protein universe. BMC Bioinform. 2017;18:476. doi: 10.1186/s12859-017-1906-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vernon R.M., Chong P.A., Tsang B., Kim T.H., Bah A., Farber P., et al. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife. 2018;7 doi: 10.7554/eLife.31486. [DOI] [PMC free article] [PubMed] [Google Scholar]