Abstract
Background
Non-small cell lung cancer (NSCLC) often presents at an incurable stage, and majority of patients will be considered for palliative treatment at some point in their disease. Despite recent advances, the prognosis remains poor, with a median overall survival of 12–18 months. Liquid biopsy-based biomarkers have emerged as potential candidates for predicting prognosis and response to therapy in NSCLC patients. This pilot study evaluated whether combining circulating tumour cells and clusters (CTCs) and cell-free DNA (cfDNA) can predict progression-free survival (PFS) in NSCLC patients.
Methods
CTC and cfDNA/ctDNA from advanced stage NSCLC patients were measured at study entry (T0) and 3-months post-treatment (T1). CTCs were enriched using a spiral microfluidic chip and characterised by immunofluorescence. ctDNA was assessed using an UltraSEEK® Lung Panel. Kaplan-Meier plots were generated to investigate the contribution of the presence of CTC/CTC clusters and cfDNA for PFS. Cox proportional hazards analysis compared time to progression versus CTC/CTC cluster counts and cfDNA levels.
Results
Single CTCs were found in 14 out of 25 patients, while CTC clusters were found in 8 out of the 25 patients at T0. At T1, CTCs were found in 7 out of 18 patients, and CTC clusters in 1 out of the 18 patients. At T0, CTC presence and the combination of CTC cluster counts with cfDNA levels were associated with shorter PFS, p = 0.0261, p = 0.0022, respectively.
Conclusions
Combining CTC cluster counts and cfDNA levels could improve PFS assessment in NSCLC patients. Our results encourage further investigation on the combined effect of CTC/cfDNA as a prognostic biomarker in a large cohort of advanced stage NSCLC patients.
Keywords: Non-small cell lung cancer, Liquid biopsy, Circulating tumour cells and clusters, Circulating tumour DNA, Cell-free DNA, Progression-free-survival
Non-small cell lung cancer, Liquid biopsy, Circulating tumour cells and clusters, Circulating tumour DNA, Cell-free DNA, Progression-free-survival.
1. Introduction
Despite recent advancements in diagnostics and treatment, lung cancer remains the leading cause of cancer-related mortality globally. Lung cancer accounts for 11.4% of newly diagnosed cancer cases and 18% of cancer-related deaths in 2020 globally [1]. About 80%–85% of lung cancers are non-small cell lung cancer (NSCLC). Most patients present with advanced stage disease at diagnosis, which is often associated with low overall survival rates [2]. In recent years, targeted therapies and immune checkpoint inhibitors therapies have become the standard of care, with improved patient outcomes [3]. Tumour biopsy is the current gold standard for diagnosis. However, tissue biopsy only presents a snapshot of the actual tumour at one time point, and might be unavailable or difficult to access. Liquid biopsy, the use of biomolecules present in body fluids, has the potential to overcome spatial and temporal tumour heterogeneity that is frequently seen with tumour tissue biopsy analysis. Liquid biopsy presents a less invasive method to assess tumour evolution, monitor recurrence, and better understand mechanisms of metastasis than traditional imaging-guided or bronchoscopic biopsy methods [4, 5]. Of the various analytes available for liquid biopsy, circulating tumour cells (CTCs) and circulating tumour DNA (ctDNA) are major biomarkers.
CTCs are a subset of tumour cells shed from either primary or metastatic tumours into the bloodstream. Recent studies have demonstrated that CTC clusters (two or more CTCs together) are more resistant and carry stronger metastatic capacities compared to single CTCs [6]. CTCs have been used as prognostic biomarkers in breast, prostate, and colorectal cancer [7, 8, 9, 10]. CellSearch® remains the only FDA-approved platform to enrich CTCs. CellSearch® relies on the expression of EpCAM on tumour cells. If a tumour cell has undergone epithelial-mesenchymal transition (EMT), enriching for EpCAM may lead to false-negative results [11]. EMT plays an essential role in metastasis, and CTCs presenting with an EMT signature have shown to be associated with poor survival in NSCLC patients [12, 13, 14]. Recent studies have highlighted the potential of detecting mesenchymal CTCs using cell-surface vimentin (CSV) [15, 16].
ctDNA (a portion of cell-free DNA, cfDNA), has been clinically validated to detect EGFR mutations (exon 19 deletion or exon 21 substitution, L858R) to identify lung cancer patients who will respond to Tyrosine Kinase inhibitor (TKI) treatment [17, 18, 19]. In recent years, the analysis of ctDNA has shown significant clinical utility in NSCLC; however, the cellular origin of ctDNA is not understood (i.e. release of ctDNA into the bloodstream reflects either active secretion from tumour cells or passive release due to cells undergoing apoptosis and/or necrosis). Therefore, questions arise as to whether ctDNA provides accurate information on tumour kinetics [20]. In contrast, CTCs are intact viable cells and may provide information on spatial and temporal tumour heterogeneity and the biology of tumours, which ctDNA mutation signature is unable to address. Because CTCs and ctDNA exhibit unique strengths and limitations, it has been suggested that they could be used in a complementary manner as cancer biomarkers [21, 22]. We hypothesize that CTCs/CTC clusters or cfDNA/ctDNA at study entry (T0) and 3-month post-treatment (T1) may predict progression-free survival (PFS) in advanced stage NSCLC patients. This pilot study aimed to evaluate the feasibility and potential clinical utility of combining CTC and cfDNA as a biomarker to predict PFS in patients with advanced stage NSCLC.
2. Materials and methods
2.1. Study design
This prospective, observational, pilot study investigated whether combining CTC/ctDNA as a biomarker panel could predict PFS in advanced stage NSCLC patients. The primary endpoint is PFS, which is defined as the time it took for disease progression or death at study entry T0 (consent date and first blood collection). This study was approved by the Metro South Health District Human Research Ethics Committee following the National Health and Medical Research Council's guidelines (HREC/11/QPAH/331) and the Queensland University of Technology (1100001420) ethics review committees. Patients diagnosed with advanced stage (stages III and IV) NSCLC, and who were planned to have a follow-up appointment at the same hospital were approached to participate in our study. Sample collection was conducted (2019–2020) at the Royal Brisbane and Women's Hospital (RBWH) and the Princess Alexandra Hospital (PAH), in Brisbane, Australia. Following written informed consent, three EDTA blood tubes were collected from 25 advanced stage NSCLC patients at T0 and at 3-months follow-up time T1and time frame would be closely associated with restaging scans. The study workflow is shown in Figure 1.
Figure 1.
The study design. Samples were collected at two timepoints, T0 at study entry and 3-months follow-up at T1. Circulating tumour cells (CTCs) were characterised using cytokeratin antibody (CK), cell-surface vimentin (CSV) or PD-L1, and CD45 monoclonal antibody. Circulating tumour DNA (ctDNA) was profiled using UltraSEEK® Lung Panel, which detects variants across five genes (BRAF, EGFR, ERBB2, KRAS and PIK3CA). All results from CTC/cfDNA/ctDNA analyses were combined with patients' clinical and metadata for a better prediction of PFS in NSCLC patients.
2.2. Circulating tumour cell enrichment and characterisation
Sample collection for CTC analysis was performed on a day to coincide with their routine clinical appointments or follow-up appointments. CTCs were enriched using the microfluidic spiral technology as described previously [23, 24, 25, 26]. Briefly, red blood cells were lysed from whole blood (9 mL) using a red blood cell (RBC) lysis buffer (Astral Scientific) and pumped through the spiral chip at 1.7 mL/min. The CTC output was collected, fixed using 4% paraformaldehyde for 10 min and cytospun onto glass slides using the Cytospin™ 4 Cytocentrifuge (ThermoScientific, USA). Following enrichment, CTCs were stained with cytokeratin (CK) pan antibody (Invitrogen), cell-surface vimentin (CSV) (Abnova) or PD-L1 (Abcam) and CD45 monoclonal antibody (BD). Following antibody incubation, cells were stained with DAPI (1:1000; stock solution 1 mg/mL) for nuclear staining. Cells were considered as CTCs when they met the following criteria [1] diameter ≥9 μm and DAPI positive [2], positive staining for CK and [3] negative staining for CD45 (Figure 2). In addition to CK, cells were also stained with cell surface vimentin (CSV) and PD-L1. When CTCs were found in groups of two or more tumour cells, they were considered CTC clusters [27]. In addition, when CTC clusters were associated with white blood cells, they were defined as circulating tumour microemboli (CTM) [28].
Figure 2.
Representative images of single circulating tumour cells (CTCs), CTC clusters, circulating tumour microemboli (CTM). DAPI (blue), CK (orange), CSV (red), PD-L1 (red), and CD45 (green). Scale bar represents 20 μm.
2.3. Circulating tumour DNA analysis
cfDNA isolation was performed for all 25 patients as described previously [29]. Two EDTA blood tubes were collected and double spun at 500 x g for 15 min (4 °C) followed by centrifugation at 16,000 x g for 10 min (4 °C). cfDNA was isolated using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Germany) as per manufacturer's instructions. cfDNA quantification was done using Qubit Fluorometer (Thermo Fisher Scientific, USA), and samples were stored at -20 °C until further use. In samples with detectable quantification readings (Table 2), ctDNA was profiled for common lung cancer mutations using the UltraSEEK® Lung Panel (Agena Biosciences). This panel detects over 70 clinically relevant variants in NSCLC across BRAF, EGFR, ERBB2, KRAS and PIK3CA. Tumour mutation data was obtained from hospital pathology reports.
Table 2.
Circulating tumour cells (CTCs), CTC cluster counts, cfDNA and ctDNA at T0 and T1 time points in NSCLC patients’ and clinical outcomes. NA = not available or lost due to Covid-19 or diseased. Not detected = cfDNA isolated and tested but without any mutation detected. Too low for detection = cfDNA isolated and insufficient concentration for quantification, lower than the detection limit (representative images of CTCs).
| CTC T0 |
CTC T1 |
cfDNA (ng/mL of plasma) |
ctDNA (VAF - %) |
Outcome | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| T0 CTC Count | T0 CTC cluster count | Additional Marker | T1 CTC Count | T1 CTC cluster count | Additional Marker | T0 | T1 | T0 | T1 | |
| 1 | 0 | - | N/A | N/A | N/A | 29.583 | N/A | not detected | not detected | No progression |
| 11 | 5 | CSV | N/A | N/A | N/A | 9.392 | N/A | EGFR:Exon19del_2249-composite (1.94)/PIK3CA:PIK3CA_pE545K (0.62) | N/A | Progression |
| 7 | 2 | CSV | 2 | 0 | CSV | N/A | 248.333 | N/A | likely EGFR mut (low VAF) | No progression |
| 5 | 0 | - | 0 | 0 | - | 3.122 | 3.311 | KRAS:KRAS_pG12C-f (1.03) | KRAS:KRAS_pG12C-f (0.19) | Progression |
| 2 | 1 | - | 0 | 0 | - | N/A | N/A | N/A | N/A | Progression |
| 0 | 0 | - | N/A | N/A | N/A | 2.017 | N/A | not detected | N/A | Deceased |
| 0 | 0 | - | N/A | N/A | N/A | 13.960 | N/A | KRAS:KRAS_pG12C-f (>2)/KRAS:KRAS_pG12R-r (0.24) | N/A | Deceased |
| 4 | 1 | CSV | N/A | N/A | N/A | 3.167 | N/A | KRAS:KRAS_pG12D (>2) | N/A | No progression |
| 0 | 0 | - | N/A | N/A | N/A | N/A | N/A | N/A | N/A | Progression |
| 7 | 0 | - | N/A | N/A | N/A | N/A | N/A | N/A | N/A | No Progression |
| 0 | 0 | - | 0 | - | 1.15 | 1.6 | No known mutation | not detected | Progression | |
| 0 | 0 | - | 0 | 0 | - | too low for detection | 3.05 | N/A | not detected | Deceased |
| 0 | 0 | - | 0 | 0 | - | 6.224 | 3.1 | not detected | not detected | No Progression |
| 0 | 0 | - | 0 | 0 | - | 10.256 | 5.042 | KRAS:KRAS_pG12C-f (0.28) | KRAS:KRAS_pG12C-f (<0.1) | Progression |
| 0 | 1 | PDL1 | 0 | 0 | - | 3.550 | 5.550 | not detected | not detected | Deceased |
| 9 | 8 | CSV | 0 | 0 | - | 2.850 | N/A | not detected | not detected | Deceased |
| 5 | 5 | PDL1 | 1 | 0 | PDL1 | 9.250 | 22.375 | not detected | not detected | Progression |
| 0 | 0 | - | 1 | 0 | CSV | 1.250 | 1.325 | not detected | not detected | Deceased |
| 1 | 0 | CSV | 2 | 0 | CSV | 5.816 | 0.575 | EGFR:Pg719c (0.33) | EGFR:pG719C (0.37) | No Progression |
| 2 | 0 | CSV | 6 | 23 | 0.950 | 5.583 | not detected | not detected | No progression | |
| 2 | 5 | CSV | 1 | 0 | CSV | too low for detection | too low for detection | not detected | not detected | No progression |
| 1 | 0 | 3 | 0 | PDL1 | 3.125 | 4.717 | not detected | not detected | No progression | |
| 0 | 0 | - | 0 | 0 | - | 6.104 | 4.150 | not detected | not detected | Progression |
| 2 | 0 | - | 0 | 0 | - | 15.712 | 3.143 | EGFR:pD770_771insSVD (>2) | EGFR:pD770_771insSVD (>2) | Deceased |
| 0 | 0 | - | 0 | 0 | - | too low for detection | 2.650 | not detected | not detected | Deceased |
2.4. Statistical analyses
Statistical analyses were performed using JMP Pro version 16.1.0. Violin plots were used to display CTC counts between patients who have either progressed or deceased versus those with no progression using Poisson based generalised linear models. Kaplan-Meier curves were generated to compare PFS for patients with or without CTCs. Cox proportional hazards analyses were performed to compare time to progression versus CTC counts and cfDNA levels. Clinical outcomes were defined according to clinical notes and dates from the most recent doctor's appointment, confirming stable disease or disease progression or death. Differences were considered statistically significant when p ≤ 0.05.
3. Results
3.1. Clinicopathological characteristics of NSCLC patients
This study analysed a total of 25 NSCLC advanced stage patients, with samples collected at the study entry T0 (N = 25) and post-treatment at 3 months T1 (N = 18). Patients’ characteristics are shown in Table 1. The median age of patients was 68.8 years, 80% were men and the majority of them had adenocarcinoma (80%).
Table 1.
Patients’ demographic and clinicopathological characteristics.
| Characteristics | n (%) |
|---|---|
| Total patients | 25 |
| Gender | |
| Female | 5 (20) |
| Male | 20 (80) |
| Age, y | |
| <60 | 5 (20) |
| ≥60 | 20 (80) |
| Median Age (range) | 68.8 (44–87) |
| Tumour type | |
| Squamous cell carcinoma | 3 (12) |
| Non-squamous cell carcinoma | 22 (88) |
| Clinical stage | |
| IIIA | 3 12) |
| IIIB | 2 (8) |
| IIIC | 2 (8) |
| IVA | 10 (40) |
| IVB | 8 (32) |
| Mutation status based on tumour | |
| EGFR mutation | 4 (16) |
| KRAS mutant | 6 (24) |
| No known mutations | 15 (60) |
| Treatment status at T0collection | |
| Systemic Treatment naïve | 10 (40) |
| Prior systemic Treatment | 15 (60) |
| Treatment type pre T0collection | |
| Chemotherapy | 6 (24) |
| Chemotherapy plus immunotherapy | 4 (16) |
| Chemotherapy plus radiotherapy | 3 (12) |
| Targeted therapy | 2 (8) |
| Nill | 10 (40) |
| Line of treatment pre consent | |
| First | 12 (48) |
| Second | 9 (36) |
| Third | 2 (8) |
| Fourth | 2 (8) |
| Treatment type received after T0 | |
| Chemotherapy | 8 (32) |
| Immunotherapy | 10 (40) |
| Chemotherapy plus immunotherapy | 6 (24) |
| Targeted therapy | 1 (4) |
| Tumour PD-L1 expression | |
| ≥50 % | 7 (28) |
| 1–49% | 3 (12) |
| <1% | 9 (36) |
| Unknown | 6 (24) |
3.2. Single CTCs at T0 predicts progression-free survival in NSCLC patients
The violin plots illustrate that NSCLC patients who progressed or died had higher single CTC counts (Poisson p = 0.0017) and CTC cluster counts (Poisson p = 0.0076) at T0 samples compared to those who did not progress (Figure 3 A and B). Similarly, NSCLC patients who had single CTCs detected in T0 blood samples had significantly shorter median PFS (6.7 months) than patients without CTCs (9.3 months) (Wilcoxon test, p = 0.0261) (Figure 3C). Patients with CTC clusters at T0 had a shorter PFS (5.4 months) than patients without CTC clusters (9.4 months) at T0, however, not reaching statistical significance (p = 0.0645) (Figure 3D).
Figure 3.
The violin plots comparing (A) single circulating tumour cell (CTC) counts and (B) CTC clusters at T0 for patients who progressed or were deceased (in red) versus patients who had no progression (in blue). P-values and confidence intervals were generated using Poisson modelling (C) Kaplan Meier curves comparing progression-free (PFS) survival of NSCLC patients with single CTC (in red) or without single CTCs (in blue) at T0. Wilcoxon test, p-value = 0.0261) (D) Kaplan Meier curves comparing PFS of NSCLC patients with CTC clusters (in red) or without CTC clusters (in blue) at baseline. Wilcoxon test, p-value = 0.0645).
Single CTC counts or CTC clusters at T1 showed no statistically significant differences between progressors/decreased versus non-progressors, most likely due to a small sample cohort and diversity in treatment strategies. Within the four EGFR mutant patients, three of them progressed and in those who had progressed, CTCs were detected at T0. The one EGFR mutant patient who did not progress had no CTCs at T0. In addition to analyses on the presence or absence of CTC, we compared time to progression versus CTC counts. The Cox proportional hazards analysis of time to progression versus CTC counts at T0 was not statistically significant (p = 0.1872). The hazard ratio of lowest versus highest CTC counts at T0 was HR = 2.65, 95%CI (0.59, 9.41). While for CTC cluster counts, the hazard ratio of lowest versus highest baseline CTC cluster count value was HR = 5.16, 95%CI (0.85, 23.95) and p = 0.0711 (Figure 4A and B). The Cox proportional hazards analysis of time to progression with age and sex as covariates for T0 CTC counts and T0 cluster counts were also not statistically significant (Figure 4C and D).
Figure 4.
A) Survival Analyses: Cox Proportional hazards to compare time to progression versus Baseline CTC counts. B) Survival Analyses: Cox Proportional hazards to compare time to progression versus Baseline CTC clusters. C) Survival Analyses: Cox Proportional hazards to compare time to progression versus Baseline CTC counts with age and sex as covariates. D) Survival Analyses: Cox Proportional hazards to compare time to progression versus Baseline CTC cluster count with age and sex as covariates. E) Summary analysis of CTC-CSV positive and CTC-CSV negative.
3.3. Patients with cell-surface vimentin positive circulating tumour cells showed progression or death
Figure 1 depicts representative images of single CTCs, CTCs expressing CSV and PD-L1 markers, CTC clusters, and CTM. CTC counts from both timepoints, and patients’ clinical outcomes are shown in Table 2. Single CTCs were found in 14 of the 25 patients (56%) at T0. CSV expression was detected in seven out of 14 CTC positive samples (50%) and six of them either progressed or died. In T0 blood samples, single CTC counts ranged from one to 11 cells, whilst CTC cluster counts ranged from one to eight. CTC clusters were found in eight out of the 25 patients (32%) at T0. Seven out of eight patients with CTC clusters at T0 either progressed or died. One NSCLC patient blood sample showed CTM (patient #7, Table 2) at T0 and this patient is deceased.
Single CTCs were found in seven out of 18 patients (39%) in T1 samples, and in five out of seven (71.4%) patients progressed or died and three of them had CSV positive CTCs also (Figure 4E). CTC clusters were only found in one of the 18 patients (5.5%) at T1. Single CTC counts ranged from one to six cells, while 20 CTC clusters and three CTMs were observed in one patient's blood sample (patient #10, Table 2) at T1 time-point with the patient showing stable disease after six months from T0 collection. For seven patients whose tumours were positive for PDL-1 expression (PDL1 ≥50%), we stained their CTC slides and found two CTC slides to be PDL-1 positive (28.5%).
3.4. Plasma cfDNA levels predict progression-free survival in NSCLC patients
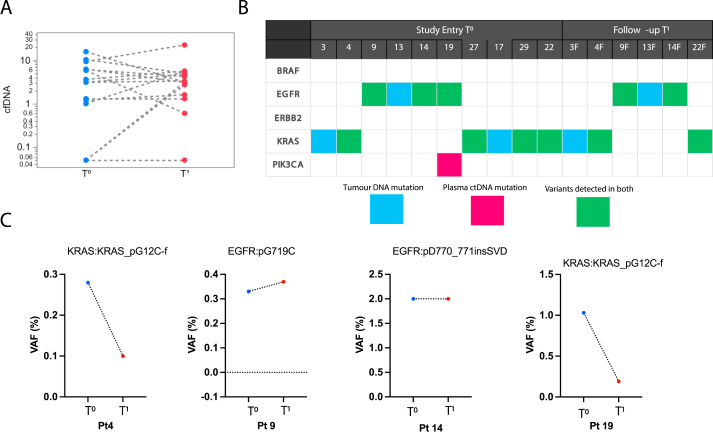
No statistically significant differences (p = 0.38) were observed between T0 samples (blue dots) (geometric mean 1.67 ng/mL, 95% CI 0.68, 4.13) and matched T1 plasma cfDNA levels (median level 2.54 ng/mL, 95% CI 1.02, 6.26) (red dots) (Figure 5A, Table 2). To assess the relationship between tumour tissue mutation and ctDNA mutation profiles, we have used the UltraSEEK® Lung mutation Panel (Agena Biosciences). Out of the 25 patients, 10 patients had known mutations detected in tumour tissue. We found that seven out of 10 (70%) NSCLC patients had concordance between the tumour tissue and matched ctDNA (Figure 5B). Interestingly, one of the patients showed an additional mutation (#19 - PIK3CA: PIK3CA_pE545K), which was not known in the tumour tissue sample. One of the patients (#20F1) in the T1 group, also showed a mutation EGFR mutation (EGFR p. L861Q or p. L861R) that was not detected in tumour tissue but was detected at low confidence in ctDNA.
Figure 5.
A) Plasma cfDNA concentrations (ng/mL) at study entry T0 (blue dots) and in follow-up samples T1 (red dots) in patients. B) Mutation analysis on paired tumour samples and ctDNA in plasma of patients at study entry T0 and follow-up T1. Mutations in tumour tissues are shown in blue, mutations from plasma are shown in pink, and mutations that are common between tumour tissue and ctDNA are represented in green. C) Alteration of the variant allele frequency (VAF - %) detected in ctDNA in mutations for patients (n = 4) at T0 and T1 timepoints.
The variant allele frequency (VAF - %) of plasma cfDNA for patients (#4, 9, 14 and 19) was compared between both timepoints (T0 and T1) (Figure 5C). Patients #4 and #19 had a decrease in the VAF of KRASpG12C-f, and presented with stable disease. In contrast, patients #9 and #14 who had a similar or increased VAF progressed.
3.5. Combining T0 and T1 cfDNA is a good predictor of progression-free survival in NSCLC patients
When cfDNA levels at T0 and T1 were individually analysed, each feature showed an insignificant contribution to PFS time, based on Cox Proportional Hazards analyses. The Hazard Ratio (HR) of lowest versus highest cfDNA levels at T0 was HR = 7.48, 95% CI (0.39, 95.98), p = 0.1662. While for T1, the HR of lowest versus highest cfDNA levels was HR = 18.00, 95%CI (0.76, 555.21) and p = 0.0684. However, when combining cfDNA data from T0 and T1 for the same patient, the Cox proportional hazards survival analysis of time to progression was significant (p = 0.0390). This result implies that combining cfDNA levels at T0 and T1 time points would result in the prediction of shorter PFS outcomes in NSCLC patients.
3.6. Combining cfDNA and circulating tumour cell data leads to a shorter progression-free survival for NSCLC patients
A Cox Proportional hazards model combining T0 and follow-up T1 cfDNA levels with T0 CTC cluster counts produced significance (χ2 [3] = 14.6, p = 0.0022), with each of the three parameters associated with shorter PFS in NSCLC patients, but we found baseline CTC cluster counts to be the strongest predictor (HR = 4.1, 95% CI 1.5, 29.1). Although adding more predictors yields a slightly more significant whole model test (χ2 [7] = 14.6, p = 0.0016), the difference between models is not significant (χ2 [4] = 8.6, p = 0.071) and leads to a larger AICc (44.2 vs 31.0). Thus the 3-parameter model may be preferred.
4. Discussion
The standard of care for many patients with advanced stage NSCLC has progressed from an empirical treatment strategy, based on clinical-pathological profiling to a biomarker-driven treatment algorithm, based on the tumour molecular profilings [30, 31]. Predictive and prognostic biomarkers based on molecular signatures have increased our knowledge in tumorigenesis, early detection, and multimodal care [32]. We found that combining CTC cluster counts and cfDNA levels at study entry (T0) predicts a shorter PFS (p = 0.0022) in NSCLC patients. In addition, we have also confirmed concordance between tumour and ctDNA mutation profilings. The combination of CTC analysis with cfDNA may be more informative to obtain a comprehensive characterisation of the tumour heterogeneity.
We observed high single CTCs/CTC clusters at T0 to be associated with a shorter PFS. Similar findings have been reported using CellSearch® system [33, 34]. Ancel et al. found that patients with positive CTCs had shorter PFS and OS (median PFS: 2.4 months vs. 6.8 months, median OS: 4.3 months vs. 8.1 months) compared with CTC-negative group [35]. Our study showed that enumeration of single CTCs or CTC clusters in follow-up T1 blood samples did not affect disease progression or death or contribute to PFS. This negative finding is most likely due to the small follow-up sample cohort caused by lock downs due to COVID-19 global pandemic. Although our CTC findings at T0 showed encouraging results, standardisation of platforms and techniques for CTC separation, characterisation and more prospective clinical trials with large sample cohorts are required to fast track clinical adoption.
Currently, routine clinical management of NSCLC patients incorporates immunotherapies targeting either programmed cells death protein 1 (PD-1) or programmed cell death ligand 1 (PD-L1) [36]. However, expression levels of PD-L1 change during chemotherapy or radiotherapy [37, 38] and identifying patients who would benefit from this approach is challenging. Within our cohort, seven patients presented PD-L1 tumour tissue status of ≥50%. Among these patients, positive staining for PD-L1 in CTCs was observed in two of them (28.5%) at T0 and two at follow-up T1. In NSCLC, studies have shown that PD-L1 expressing CTCs were detected in 25 out of 38 patients (65.8%), with numbers increasing significantly after initiation of radiation therapy [37]. We observed that the majority of patients with CSV-positive CTCs had progressed or died. The presence of this marker in patients with poor clinical outcomes may indicate a potential association and encourages more studies to explore the CSV clinical utility in NSCLC. Currently, there are limited studies performed using CSV expression in CTCs in NSCLC patients. Xie et al. suggested that CSV-CTCs could be used for diagnosing lung cancer with a sensitivity and specificity of 0.67 and 0.87, respectively [39]. Despite studies reporting CSV to enrich CTCs in other tumour types like sarcoma [40] and prostate cancer [16], more studies are necessary to elucidate the role of CSV in CTCs of NSCLC patients.
We found that combining cfDNA levels at T0 and T1 showed a significant contribution to PFS time. The therapeutic utility of detecting mutations in cfDNA has been correlated to tumour burden [41], PFS and OS in NSCLC patients [42]. ctDNA EGFR mutations (exon 19 detection or exon 21 (L858R) mutations) are used to identify patients who may benefit from EGFR-TKIs [43]. We identified variants in plasma cfDNA in most patients who had tumour mutations. We found KRAS mutation in plasma of four patients, EGFR in four patients and PIK3CA in one patient. Mutation detection in these genes may improve the management of NSCLC and may facilitate in developing an FDA-approved test for detecting EGFR mutations in plasma of NSCLC patients [44].
In our pilot study, the variant allele frequency (VAF) % of mutations for four patients (#4, 9, 14 and 22) was analysed at both time points (Figure 5C). Patients with a decrease in the VAF (#4 and #22) presented with better prognosis compared to patients that maintained the same or increased VAF (#9 and #14). Despite having only four patients with ctDNA data at study entry and follow-up, our results corroborated with previously reported studies. Giroux. Leprieur et al. [45] demonstrated that the absence of a significant increase in VAF at two months (defined as an increase of more than 9% relative to baseline) predicted stable disease for at least six months with a sensitivity of 71% and specificity of 100% in a cohort of 15 patients treated with Nivolumab [45, 46]. Furthermore, the lack of increase in ctDNA levels was linked to a considerably longer PFS (median: 0.7 versus 12.0) [46]. In a separate study, Goldberg et al. [47] defined a ctDNA response as a drop-in VAF of more than 50% from baseline, which they validated with a second sample [47, 48]. Therefore, the ability to track disease progression more frequently using ctDNA would be highly beneficial. Especially for patients who have developed acquired resistance to first-line therapy and present with metastases but are unable to obtain additional tumour information (due to the limited obtainable genotyping data or risks from tissue biopsy).
Limitations to the current study include tumour heterogeneity, stage heterogeneity, treatment heterogeneity (particularly EGFR), missing samples, small sample size, and loss of NSCLC patients to follow-up due to COVID-19 pandemic.
5. Conclusion
It is worthwhile investigating in a larger, more homogenous NSCLC patient cohort whether combining CTC data and cfDNA levels is associated with PFS in NSCLC patients. Furthermore, a Cox PH model incorporating T0 and T1 cfDNA concentrations and T0 CTC cluster counts was strongly associated (p = 0.0022) with shorter PFS time in our pilot study using NSCLC patients.
Declarations
Author contribution statement
Joanna Kapeleris; Juliana Müller Bark; Shanon Ranjit: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Gunter Hartel: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Majid Ebrahimi Warkiani; Paul Leo; Connor O'Leary; Rahul Ladwa; Kenneth O'Byrne; Brett G.M. Hughes; Darryl Irwin: Conceived and designed the experiments; Wrote the paper.
Chamindie Punyadeera: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by the Royal Brisbane Women's Hospital Foundation Project Number 15444.
Juliana, M. Bark was supported by ATM LATAM QUT Postgraduate Research Scholarship.
Joanna, Kapeleris was supported by a QUT PhD scholarship.
Chamindie, Punyadeera is supported by the National Health and Medical Research Council (APP 2002576 and APP 2012560), NIH, Cancer Australia (APP1145657), Garnett Passe and Rodney Williams Foundation and RBWH Foundation.
Data availability statement
Data included in article/supp. material/referenced in article.
Declaration of interest's statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors would like to thank staff at Agena Biosciences in Brisbane for the analysis of ctDNA mutation profiles. Dianna Lewis, Amy Ives (RBWH Clinical trial coordinators) and PAH clinical nurses. Figure 1 was created using Servier Medical Art templates, licensed under a Creative Commons Attribution 3.0 Unported License; https://smart.servier.com.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Ellis P.M., Vandermeer R. Delays in the diagnosis of lung cancer. J. Thorac. Dis. 2011;3(3):183–188. doi: 10.3978/j.issn.2072-1439.2011.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan M., Huang L.-L., Chen J.-H., Wu J., Xu Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct. Targeted Ther. 2019;4(1):61. doi: 10.1038/s41392-019-0099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alix-Panabieres C., Pantel K. Challenges in circulating tumour cell research. Nat. Rev. Cancer. 2014;14(9):623–631. doi: 10.1038/nrc3820. [DOI] [PubMed] [Google Scholar]
- 5.Alix-Panabieres C., Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6(5):479–491. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- 6.Au S.H., Storey B.D., Moore J.C., Tang Q., Chen Y.-L., Javaid S., et al. Clusters of circulating tumor cells traverse capillary-sized vessels. Proc. Natl. Acad. Sci. USA. 2016;113(18):4947. doi: 10.1073/pnas.1524448113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cristofanilli M., Budd G.T., Ellis M.J., Stopeck A., Matera J., Miller M.C., et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004;351(8):781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 8.de Bono J.S., Scher H.I., Montgomery R.B., Parker C., Miller M.C., Tissing H., et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2008;14(19):6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 9.Cohen S.J., Punt C.J., Iannotti N., Saidman B.H., Sabbath K.D., Gabrail N.Y., et al. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann. Oncol. 2009;20(7):1223–1229. doi: 10.1093/annonc/mdn786. [DOI] [PubMed] [Google Scholar]
- 10.Guibert N., Pradines A., Favre G., Mazieres J. Current and future applications of liquid biopsy in nonsmall cell lung cancer from early to advanced stages. Eur. Respir. Rev. 2020;29(155) doi: 10.1183/16000617.0052-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikolajczyk S.D., Millar L.S., Tsinberg P., Coutts S.M., Zomorrodi M., Pham T., et al. Detection of EpCAM-negative and cytokeratin-negative circulating tumor cells in peripheral blood. J. Oncol. 2011;2011 doi: 10.1155/2011/252361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lecharpentier A., Vielh P., Perez-Moreno P., Planchard D., Soria J.C., Farace F. Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. Br. J. Cancer. 2011;105(9):1338–1341. doi: 10.1038/bjc.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manjunath Y., Upparahalli S.V., Avella D.M., Deroche C.B., Kimchi E.T., Staveley-O'Carroll K.F., et al. PD-L1 expression with epithelial mesenchymal transition of circulating tumor cells is associated with poor survival in curatively resected non-small cell lung cancer. Cancers. 2019;11(6):806. doi: 10.3390/cancers11060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milano A., Mazzetta F., Valente S., Ranieri D., Leone L., Botticelli A., et al. Molecular detection of EMT markers in circulating tumor cells from metastatic non-small cell lung cancer patients: potential role in clinical practice. Anal. Cell Pathol. 2018;2018 doi: 10.1155/2018/3506874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satelli A., Brownlee Z., Mitra A., Meng Q.H., Li S. Circulating tumor cell enumeration with a combination of epithelial cell adhesion molecule- and cell-surface vimentin-based methods for monitoring breast cancer therapeutic response. Clin. Chem. 2015;61(1):259–266. doi: 10.1373/clinchem.2014.228122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Satelli A., Batth I., Brownlee Z., Mitra A., Zhou S., Noh H., et al. EMT circulating tumor cells detected by cell-surface vimentin are associated with prostate cancer progression. Oncotarget. 2017;8(30):49329–49337. doi: 10.18632/oncotarget.17632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guardant Health Guardant 360® CDx First FDA-Approved Liquid Biopsy for Comprehensive Tumor Mutation Profiling across All Solid Cancers. 2020. [press release]. August 7, 2020. [Google Scholar]
- 18.FDA Approves First Liquid Biopsy Next-Generation Sequencing. 2020. Companion Diagnostic Test [press release]. August 07, 2020. [Google Scholar]
- 19.Remon J., Menis J., Hasan B., Peric A., De Maio E., Novello S., et al. The APPLE trial: feasibility and activity of AZD9291 (osimertinib) treatment on positive PLasma T790M in EGFR-mutant NSCLC patients. EORTC 1613. Clin. Lung Cancer. 2017;18(5):583–588. doi: 10.1016/j.cllc.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Ilie M., Hofman V., Long E., Bordone O., Selva E., Washetine K., et al. Current challenges for detection of circulating tumor cells and cell-free circulating nucleic acids, and their characterization in non-small cell lung carcinoma patients. What is the best blood substrate for personalized medicine? Ann. Transl. Med. 2014;2(11):107. doi: 10.3978/j.issn.2305-5839.2014.08.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calabuig-Fariñas S., Jantus-Lewintre E., Herreros-Pomares A., Camps C. Circulating tumor cells versus circulating tumor DNA in lung cancer-which one will win? Transl. Lung Cancer Res. 2016;5(5):466–482. doi: 10.21037/tlcr.2016.10.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castro-Giner F., Gkountela S., Donato C., Alborelli I., Quagliata L., Ng C.K.Y., et al. Cancer diagnosis using a liquid biopsy: challenges and expectations. Diagnostics. 2018;8(2) doi: 10.3390/diagnostics8020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warkiani M.E., Guan G., Luan K.B., Lee W.C., Bhagat A.A., Chaudhuri P.K., et al. Slanted spiral microfluidics for the ultra-fast, label-free isolation of circulating tumor cells. Lab Chip. 2014;14(1):128–137. doi: 10.1039/c3lc50617g. [DOI] [PubMed] [Google Scholar]
- 24.Kulasinghe A., Tran T.H., Blick T., O'Byrne K., Thompson E.W., Warkiani M.E., et al. Enrichment of circulating head and neck tumour cells using spiral microfluidic technology. Sci. Rep. 2017;7 doi: 10.1038/srep42517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapeleris J., Kulasinghe A., Warkiani M.E., Oleary C., Vela I., Leo P., et al. Ex vivo culture of circulating tumour cells derived from non-small cell lung cancer. Transl. Lung Cancer Res. 2020;9(5):1795–1809. doi: 10.21037/tlcr-20-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher B.M., Tang K.D., Warkiani M.E., Punyadeera C., Batstone M.D. A pilot study for presence of circulating tumour cells in adenoid cystic carcinoma. Int. J. Oral Maxillofac. Surg. 2021;50(8):994–998. doi: 10.1016/j.ijom.2020.11.012. [DOI] [PubMed] [Google Scholar]
- 27.Aceto N., Bardia A., Miyamoto D.T., Donaldson M.C., Wittner B.S., Spencer J.A., et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158(5):1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krebs M.G., Metcalf R.L., Carter L., Brady G., Blackhall F.H., Dive C. Molecular analysis of circulating tumour cells—biology and biomarkers. Nat. Rev. Clin. Oncol. 2014;11(3):129–144. doi: 10.1038/nrclinonc.2013.253. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt H., Kulasinghe A., Allcock R.J.N., Tan L.Y., Mokany E., Kenny L., et al. A pilot study to non-invasively track PIK3CA mutation in head and neck cancer. Diagnostics. 2018;8(4) doi: 10.3390/diagnostics8040079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uemura T., Hida T. Durvalumab showed long and durable effects after chemoradiotherapy in stage III non-small cell lung cancer: results of the PACIFIC study. J. Thorac. Dis. 2018;10(Suppl 9):S1108. doi: 10.21037/jtd.2018.03.180. s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones G.S., Baldwin D.R. Recent advances in the management of lung cancer. Clin. Med. 2018;18(Suppl 2):s41–s46. doi: 10.7861/clinmedicine.18-2s-s41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herbst R.S., Morgensztern D., Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 33.Marchetti A., Del Grammastro M., Felicioni L., Malatesta S., Filice G., Centi I., et al. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: toward a real-time liquid biopsy for treatment. PLoS One. 2014;9(8) doi: 10.1371/journal.pone.0103883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Devriese L.A., Bosma A.J., van de Heuvel M.M., Heemsbergen W., Voest E.E., Schellens J.H.M. Circulating tumor cell detection in advanced non-small cell lung cancer patients by multi-marker QPCR analysis. Lung Cancer. 2012;75(2):242–247. doi: 10.1016/j.lungcan.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Ancel J., Birembaut P., Dewolf M., Durlach A., Nawrocki-Raby B., Dalstein V., et al. Programmed death–ligand 1 and vimentin: a tandem marker as prognostic factor in NSCLC. Cancers. 2019;11(10):1411. doi: 10.3390/cancers11101411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borghaei H., Paz-Ares L., Horn L., Spigel D.R., Steins M., Ready N.E., et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N. Engl. J. Med. 2015;373(17):1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Kim T.H., Fouladdel S., Zhang Z., Soni P., Qin A., et al. PD-L1 expression in circulating tumor cells increases during radio(chemo)therapy and indicates poor prognosis in non-small cell lung cancer. Sci. Rep. 2019;9(1):566. doi: 10.1038/s41598-018-36096-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kloten V., Lampignano R., Krahn T., Schlange T. Circulating tumor cell PD-L1 expression as biomarker for therapeutic efficacy of immune checkpoint inhibition in NSCLC. Cells. 2019;8(8) doi: 10.3390/cells8080809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie X., Wang L., Wang X., Fan W.-H., Qin Y., Lin X., et al. Evaluation of cell surface vimentin positive circulating tumor cells as a diagnostic biomarker for lung cancer. Front. Oncol. 2021;(1712):11. doi: 10.3389/fonc.2021.672687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Satelli A., Mitra A., Cutrera J.J., Devarie M., Xia X., Ingram D.R., et al. Universal marker and detection tool for human sarcoma circulating tumor cells. Cancer Res. 2014;74(6):1645–1650. doi: 10.1158/0008-5472.CAN-13-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamminga M., de Wit S., Hiltermann T.J.N., Timens W., Schuuring E., Terstappen L.W.M.M., et al. Circulating tumor cells in advanced non-small cell lung cancer patients are associated with worse tumor response to checkpoint inhibitors. Journal for ImmunoTherapy of Cancer. 2019;7(1):173. doi: 10.1186/s40425-019-0649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ai B., Liu H., Huang Y., Peng P. Circulating cell-free DNA as a prognostic and predictive biomarker in non-small cell lung cancer. Oncotarget. 2016;7(28):44583–44595. doi: 10.18632/oncotarget.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim E., Feldman R., Wistuba Update on EGFR mutational testing and the potential of noninvasive liquid biopsy in non-small-cell lung cancer. Clin. Lung Cancer. 2018;19(2):105–114. doi: 10.1016/j.cllc.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Kwapisz D. The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer? Ann. Transl. Med. 2017;5(3):46. doi: 10.21037/atm.2017.01.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giroux Leprieur E., Herbretau G., Dumenil C., Julie C., Giraud V., Labrune S., et al. Circulating tumor DNA evaluated by Next-Generation Sequencing is predictive of tumor response and prolonged clinical benefit with nivolumab in advanced non-small cell lung cancer. OncoImmunology. 2018;7(5) doi: 10.1080/2162402X.2018.1424675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lianidou E.S., Markou A., Strati A. The role of CTCs as tumor biomarkers. Adv. Exp. Med. Biol. 2015;867:341–367. doi: 10.1007/978-94-017-7215-0_21. [DOI] [PubMed] [Google Scholar]
- 47.Goldberg S.B., Narayan A., Kole A.J., Decker R.H., Teysir J., Carriero N.J., et al. Early assessment of lung cancer immunotherapy response via circulating tumor DNA. Clin. Cancer Res. 2018;24(8):1872–1880. doi: 10.1158/1078-0432.CCR-17-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Powell R., Kussaibati R., Baijal S. Optimising management of EGFR mutant NSCLC through circulating tumour DNA (ctDNA) monitoring. Lung Cancer. 2019;127:S39. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supp. material/referenced in article.