Abstract
Background/Objective
Conflicting results on the association between HLA-G and digestive cancers were reported. We conducted a meta-analysis to further investigate the true relationship between HLA-G and digestive cancers (DC).
Methods
Following PRISMA guidelines, we performed a meta-analysis including 7 case-control studies on HLA-G 14-bp Insertion/deletion (I/D) polymorphism, and 15 studies on soluble HLA-G (sHLA-G). Odds ratios (OR) and their corresponding 95% confidence intervals (CI) for genetic polymorphisms were calculated. The pooled OR was calculated under three genetic models: allelic, recessive, and dominant models. Concerning sHLA-G meta-analysis, standardized mean differences (SMDs) were calculated.
Results
The HLA-G 14-bp I/D was not associated with the risk of DC. However, in the subset of HBV/HCV positive hepato-cellular cancer (HCC) patients, we reported a significant association of HLA-G 14-bp I/D with the disease initiation under allelic (D vs. I; OR = 1.698, 95% CI = 1.263–2.282, p = 0.000), dominant (DD + ID vs. II; OR = 2.321, 95% CI = 1.277–4.218, p = 0.006)and recessive (DD vs. DI + II; OR = 1.739, 95% CI = 1.173–2.577, p = 0.006) genetic models. Interestingly, HLA-G 14-bp I/D was not associated with the disease initiation in HBV/HCV negative HCC patients. However, the infection by HBV/HCV seems to be implicated in the HCC development when we compared HBV/HCV positive patients to HBV/HCV negative patients under allelic (D vs. I; OR = 1.429, 95% CI = 1.029–1.983, p = 0.033, and dominant (DD + ID vs.II; OR = 1.981, 95% CI = 1.002–3.916, p = 0.049) genetic models.
Overall analysis of DC showed significant increased sHLA-G in patients compared to healthy controls (SMD = 3.341, 95% CI = 2.415–4.267, p = 0.000). In Asian patients with gastric cancer, sHLA-G was significantly increased in grade 3 compared to low grades (SMD = 0.448, 95% CI = 0.109–0.787, p = 0.000). Further analysis showed that sHLA-G was significantly increased in positive DC vascular invasion (SMD = 0.743, 95% CI = 0.385–1.100, p = 0.000). Accordingly, sHLA-G was associated with a poor prognosis for DC.
Conclusion
The current meta-analysis supports the significant role of HLA-G in DC. The HLA-G 14-bp I/D polymorphism was associated with HCC patients with concomitant HBV/HCV viral infections. Increased sHLA-G indicated a poor prognosis for DC cancer patients.
Keywords: Digestive cancer, HLA-G, Polymorphism, sHLA-G, Meta-analysis
Digestive cancer; HLA-G; Polymorphism; sHLA-G; Meta-analysis.
1. Introduction
Digestive cancers (DC), composed of oesophageal, colorectal, pancreatic, stomach, and liver cancers are common malignancies and account for one-quarter of the global cancer incidence [1]. Fortunately, recent years showed considerable improvements in DC diagnosis and treatment, including relief of symptoms and prolonged survival [2]. However, the efficacy of surgery and chemotherapy remains unsatisfactory, particularly if tumours aren’t detected and removed at an early stage. Therefore, novel biomarkers to improve cancer diagnosis and prognosis are crucial for reducing cancer burden and mortality. The expression of HLA-G, an immune tolerant and tumour promoting factor, has been extensively investigated, and its role as a novel immune checkpoint has been established [3, 4]. HLA-G has been described as a potent immune suppressive mediator observed in various malignancies and is strongly associated with tumour immune escape and metastasis [5]. Yie et al. reported that HLA-G protein was expressed in a majority of the primary site of gastric carcinomas and significantly correlated with tumour location, histological grade, depth of invasion, histological grade, host immune response, lymph nodal metastasis, and clinical stages of the disease [6]. A high frequency of tumour cell HLA-G expression and/or increased sHLA-G has been found in various body fluids in a variety of cancers [7]. Similarly to membrane HLA-G, peripheral sHLA-G was associated with advanced disease stage, tumour metastasis and/or poor prognosis [8, 9]. Both membrane-bound and sHLA-G have immune suppressive function [10]. The abnormal expression of HLA-G might be caused by genetic polymorphisms in HLA-G gene, as previously announced. Particularly, the HLA-G 14-bp insertion/deletion has been widely investigated in the context of cancer susceptibility and progression [11, 12, 13].
We aimed through the current meta-analysis to assess the significance of HLA-G in DC by studying the association of HLA-G 14-bp and sHLA-G with DC susceptibility and progression.
2. Methods
2.1. Literature search and inclusion/exclusion criteria
MEDLINE, EMBASE, Web of Science, and Cochrane databases (up to May, 2021) were searched using the terms “HLA-G” “polymorphism”, “sHLA-G,” and “digestive cancer”. The review process followed PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines [14]. Eligible studies included: (1) published cases/controls cohort studies evaluating the association of the HLA-G polymorphism with DC; (2) Availability of mean or median and standard deviation (SD) or range data of sHLA-G levels in patients and controls. Studies were excluded if they were reviews or case reports or if they present duplicate or incomplete data. The literature searches were performed in triplicate by three independent reviewers (SD, KT and IZ). Reviewers extracted the data on the methods and results from the original studies. Discrepancies were resolved by consensus among the reviewers. Flow diagrams of research strategy, exclusion and exclusion criteria are presented in Figures 1 and 2.
Figure 1.
Flow diagram representing the selection process concerning HLA-G 14bp I/D polymorphism studies in digestive cancer.
Figure 2.
Flow diagram representing the selection process concerning sHLA-G dosage studies in digestive cancer.
2.2. Statistical analyses
We evaluated the implication of HLA-G 14-bp Insertion (I)/Deletion (D) polymorphism and levels of soluble HLA-G (sHLA-G) in the initiation and prognosis of DC. Concerning HLA-G gene polymorphism, we meta-analyzed studies through the calculation of odds ratios (OR) and its corresponding 95% confidence interval (CI). The pooled OR was calculated under three genetic models: allelic, recessive, and dominant models.
Concerning sHLA-G meta-analysis, standardized mean differences (SMDs) were calculated. When median and range were reported, we calculated the mean ± SD (standard deviation) according to Hozo et al. [15]. Heterogeneity between studies was assessed by I2 and Tau2 value. I2 values were interpreted according to the Cochrane guidelines [16]. Tau2 test reflected the variance of the true effect sizes [17].
The Funnel plot measured the study size [18]. Egger’s test of the intercept estimated the sample size effect [19]. Publications bias was evaluated through funnel plot and Begg and Mazumdar rank correlation test [20]. Two-tailed PEgger, and PBegg values without continuity correction, were reported.
The random-effects model assuming significant variation in different studies and testing sampling errors and variances between studies [21, 22]. When homogeneity among studies was assumed, we used fixed effects model. Comprehensive meta-analysis software (Biostat, Englewood, NJ, USA) was used to perform statistical analysis. P ≤ 0.05 was considered statistically significant.
3. Results
3.1. Studies included in the meta-analysis
For HLA-G polymorphisms, we identified 67 studies. Of these articles, duplicates, non relevant papers based on titles, reviews and meta-analysis were excluded leaving 14 articles for full screening. Additional 5 articles were omitted due to lack or irrelevant data. Therefore, in total, 9 articles met our inclusion criteria [23, 24, 25, 26, 27, 28, 29, 30, 31] (Table 1, Figure 1).
Table 1.
Characteristics of individual studies included in the meta-analysis of HLA-G 14bp I/D polymorphism in digestive cancer.
| Study | Genotyping method | Country | Ethnicity | Cases |
Controls |
||
|---|---|---|---|---|---|---|---|
| Sample | Number | Number | HWE p-value | ||||
| Dhouioui 2022 | PCR | Tunisia | Caucasian | CRC | 233 | 241 | 0.186 |
| Vaquero-Yuste 2021 | PCR | Spain | Caucasian | GC | 107 | 55 | 0.597 |
| Abu Hassan 2019 | PCR | Saudi-Arabia | Caucasian | CRC | 105 | 119 | 0.519 |
| El Bassiouny 2019 | PCR | Egypt | Egyptian | HCC | 40 | 20 | 0.371 |
| Garziera 2016 | PCR | Italy | Caucasian | CRC | 308 | 294 | 0.017 |
| Kim 2013 | PCR | South Korea | Asian | HCC | 270 | 91 | 0.556 |
| Teixeira 2013 | PCR | Brazil | Mix | HCC | 109 | 202 | 0.075 |
| Teixeira 2013a | PCR | Brazil | Mix | HBV or HCV positive HCC | 75 | 202 | 0.075 |
| Teixeira 2013b | PCR | Brazil | Mix | HBV or HCV negative HCC | 34 | 202 | 0.075 |
| Chen 2012 | PCR | China-Kazakan | Asian | EC | 132 | 251 | 0.571 |
| Chen 2012a | PCR | China-Han | Asian | EC | 107 | 211 | 0.125 |
| Jiang 2011 | PCR | China | Asian | HCC | 318 | 599 | 0.531 |
| Jiang 2011a | PCR | China | Asian | HBV positive HCC | 222 | 60 | 0.237 |
| Jiang 2011b | PCR | China | Asian | HBV negative HCC | 96 | 539 | 0.009 |
CRC: Colorectal cancer; GC: Gastric cancer; EC: Esophageal cancer; HCC: Hepatocellular cancer; HBV: Hepatitis B virus; HCV: Hepatitis C Virus; HWE: Hardy Weinberg Equilibrium.
I/D: Insertion/Deletion; Bold: significant P-value (≤0.05).
For sHLA-G, we identified 212 studies. Of these, duplicate and irrelevant papers were excluded based on titles. After excluding reviews and meta-analyses, 69 were selected for full-text screening based on the title and abstract. Of which, 30 articles were excluded based on abstract, and 24 were omitted due to lack or irrelevant data. Therefore, in total, 15 articles met our inclusion criteria for sHLA-G [25, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45], (Table 2, Figure 2). When there were at least two comparisons, the meta-analysis of HLA-G-related polymorphisms or sHLA-G was performed.
Table 2.
Characteristics of individual studies included in the meta-analysis of sHLA-G dosage in digestive cancer.
| Study | Method | Manufacturer | Country | Ethnicity | Sample type | Cases |
Controls |
Units | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Number | Mean ± SD | Number | Mean ± SD | |||||||
| Lázaro-Sánchez 2020 | ELISA kit | BioVendor | Spain | Caucasian | Saliva | CRC | 15 | 25.23 ± 16.56 | 10 | 6.60 ± 2.03 | U/mL |
| Abu Hassan 2019 | ELISA kit | MyBioSource | Saudi-Arabia | Caucasians | Serum | CRC | 33 | 1.42 ± 0.70 | 30 | 1.04 ± 0.81 | ng/mL |
| Farjadian 2018 | ELISA kit | Exbio | Iran | Caucasian | Plasma | GC | 82 | 85.37 ± 60.83 | 45 | 56.99 ± 48.45 | U/mL |
| Kirana 2017 | ELISA kit | Exbio | Australia | Oceania | Plasma | CRC | 44 | NI∗ | NA | NA | U/mL |
| Li 2017 | ELISA kit | Exbio | China | Asian | Plasma | CRC | 178 | 151.58 ± 88.25 | 113 | 37.88 ± 15.58 | U/mL |
| Sun 2017 | ELISA kit | Ameko | China | Asian | Ascite | CC | 10 | 17.59 ± 4.69 | 30 | 12.47 ± 3.68 | μg/L |
| Sun 2017a | ELISA kit | Ameko | China | Asian | Ascite | GC | 8 | 18.37 ± 4.63 | 30 | 12.47 ± 3.68 | μg/L |
| Sun 2017b | ELISA kit | Ameko | China | Asian | Ascite | PC | 6 | 21.42 ± 1.69 | 30 | 12.47 ± 3.68 | μg/L |
| Khorrami 2016 | ELISA kit | Glory Science | Iran | Caucasian | Serum | GC | 50 | 36.29 ± 1.66 | 50 | 11.23 ± 1.47 | U/mL |
| Pan 2016 | ELISA kit | Exbio | China | Asian | Plasma | GC | 81 | 55.90 ± 9.23 | 77 | 30.53 ± 2.55 | U/mL |
| Xu 2016 | ELISA kit | Exbio | China | Asian | Plasma | GC | 124 | 127.93 ± 52.98 | 130 | 75.78 ± 22.12 | U/mL |
| Zheng 2014 | ELISA kit | BioVendor | China | Asian | Plasma | EC | 60 | 71.10 ± 61.42 | 28 | 10.72 ± 7.32 | U/mL |
| Park 2012 | ELISA kits | Exbio/BioVendor | South Korea | Asian | Serum | HCC | 80 | 188.58 ± 24.65 | 50 | 16.18 ± 12.03 | U/mL |
| Lin 2011 | ELISA kit | Exbio | China | Asian | Plasma | EC | 41 | 143.28 ± 52.68 | 153 | 21.48 ± 9.82 | U/mL |
| Zhu 2011 | ELISA kit | Exbio | China | Asian | Serum | CRC | 144 | 124.30 ± 19.17 | 60 | 25.83 ± 6.43 | U/mL |
| Wang 2011 | ELISA kits | Exbio/BioVendor | China | Asian | Serum | HCC | 36 | 132.60 ± 31.40 | 25 | 47 ± 15.5 | U/mL |
| Lin 2010 | ELISA kits | Exbio | China | Asian | Plasma | HCC | 19 | 175.26 ± 126.67 | 86 | 18.44 ± 7.74 | U/mL |
CC: Colon cancer; CRC: Colorectal cancer; EC: Esophageal cancer; GC: Gastric cancer; HCC: Hepatocellular cancer; NA: Not applicable; NI: Not indicated; PC: Pancreatic cancer; SD, Standard deviation, sHLA-G, soluble HLA-G. ∗No data for the total CRC cohort. Data are given only in CRC patients after stratifications.
3.2. Meta-analysis of HLA-G 14-bp I/D polymorphism and digestive cancers susceptibility
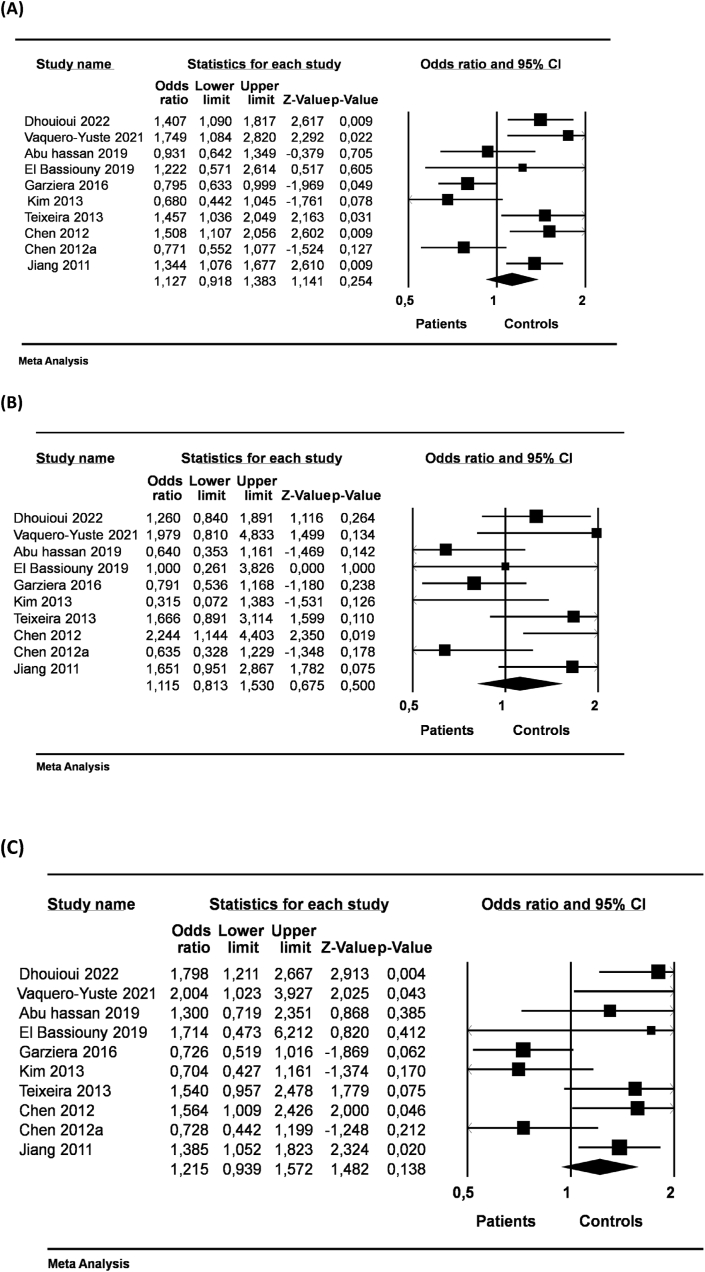
The HLA-G 14-bp I/D was not associated with the risk of DC neither under allelic, dominant or recessive genetic models (Table 3, Figure 3). After stratifications according ethnicity or type of cancer, we did not reach significant association between HLA-G 14-bp I/D and DC under allelic (D vs. I; OR = 1.127, 95% CI = 0.918–1.383, p = 0.254; Figure 3A), dominant (DD + ID vs. II; OR = 1.115, 95% CI = 0.813–1.530, p = 0.500; Figure 3B), and recessive (DD vs DI + II; OR = 1.214, 95% CI = 0.939–1.572, p = 0.138; Figure 3C) genetic models (Table 3).
Table 3.
Main results of the meta-analysis of HLA-G 14bp I/D polymorphism with digestive cancers.
| Cancer type | Ethnicity | Genetic model | N | Odds ratio |
Heterogeneity |
PEgger | PBegg | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | POR | I2(%) | Tau2 | PH | ||||||
| DC | Overall | D vs. I | 10 | 1.127 | 0.918–1.383 | 0.254 | 73.86 | 0.075 | 0.000 | 0.998 | 0.858 |
| DD + ID vs. II | 1,115 | 0.813–1.530 | 0.500 | 58.18 | 0.136 | 0.011 | 0.983 | 0.721 | |||
| DD vs DI + II | 1,215 | 0.939–1.,572 | 0,138 | 66.62 | 0.105 | 0.001 | 0.764 | 0.858 | |||
| Asian | D vs. I | 4 | 1.040 | 0.724–1.495 | 0.832 | 81.3 | 0.109 | 0.001 | 0.255 | 0.497 | |
| DD + ID vs. II | 1.083 | 0.524–2.239 | 0.830 | 73.3 | 0.379 | 0.010 | 0.404 | 1 | |||
| DD vs DI + II | 1.162 | 0.956–1.411 | 0.131 | 71.6 | 0.112 | 0.014 | 0.293 | 0.042 | |||
| CRC | Overall | D vs. I | 3 | 1,015 | 0.698–1.477 | 0.937 | 81.58 | 0.089 | 0.004 | 0.934 | 1 |
| DD + ID vs. II | 0,891 | 0.607–1.308 | 0.555 | 53.56 | 0.061 | 0.116 | 0.635 | 1 | |||
| DD vs DI + II | 1,179 | 0.641–2.167 | 0.597 | 83.52 | 0.238 | 0.002 | 0.693 | 1 | |||
| EC | Asian | D vs. I | 2 | 1.107 | 0.8820–1.389 | 0.382 | 88 | 0.198 | 0.004 | NA | NA |
| DD + ID vs. II | 1.179 | 0.735–1.890 | 0.495 | 85.5 | 0.682 | 0.009 | NA | NA | |||
| DD vs DI + II | 1.121 | 0.806–1.558 | 0.497 | 80.4 | 0.236 | 0.024 | NA | NA | |||
| HCC | Overall | D vs. I | 4 | 1.152 | 0.827–1.605 | 0.404 | 66.2 | 0.071 | 0.031 | 0.571 | 0.174 |
| DD + ID vs. II | 4 | 1.291 | 0.753–2.211 | 0.353 | 37.2 | 0.109 | 0.189 | 0.109 | 0.042 | ||
| DD vs DI + II | 4 | 1.213 | 0.836–1.760 | 0.310 | 54.3 | 0.072 | 0.087 | 0.860 | 0.174 | ||
| Asian | D vs. I | 2 | 1.164 | 0.956–1.418 | 0.131 | 86.9 | 0.202 | 0.006 | NA | NA | |
| DD + ID vs. II | 1.349 | 0.805–2.263 | 0.256 | 76.4 | 1.050 | 0.040 | NA | NA | |||
| DD vs DI + II | 1.184 | 0.931–1.506 | 0.169 | 81.5 | 0.186 | 0.020 | NA | NA | |||
bp: base pairs, CI: Confidence interval, CRC: Colorectal cancer, DC: Digestive cancers, EC: Esophageal cancer, HCC: Hepatocellular cancer, I/D: insertion/deletion, N: number of studies, NA: Not applicable, OR: odds ratio, PBegg: P-value associated to Begg and Mazumdar rank correlation test (Two-tailed) without continuity correction, PEgger: P-value associated to Egger’s test (Two-tailed), PH: P-value associated to heterogeneity, POR: P-value associated to OR, Bold: significant P-value (≤0.05).
Figure 3.
Forest plot of the association between HLA-G 14-bp I/D polymorphism and digestive cancer risk with the random effects model. (A) Allelic model (D vs. I) alleles, (B) Dominant genotype (DD + ID vs. II) and (C) Recessive model (DD vs DI + II) in the overall population.
In the subset of HBV or HCV (HBV/HCV) positive HCC patients, we demonstrated a clear association of HLA-G 14-bp I/D with the disease initiation under allelic (D vs. I; OR = 1.698, 95% CI = 1.263–1.650, p = 0.000, Table 4), dominant (DD + ID vs, II; OR = 2.321, 95% CI = 1.277–4.218, p = 0.006, Table 4), and recessive (DD vs. DI + II; OR = 1.739, 95% CI = 1.173–2.577, p = 0.006, Table 4) genetic models. As expected, HLA-G 14-bp I/D was not associated to the disease initiation in the subset of HBV or HCV negative HCC patients (Table 4). Interestingly, the infection by HBV/HCV seems to be implicated in the HCC development when we compared HBV/HCV positive patients to HBV/HCV negative patients under allelic (D vs. I; OR = 1.429, 95% CI = 1.029–1.983, p = 0.033, Table 4) and dominant (DD + ID vs.II; OR = 1.981, 95% CI = 1.002–3.916, p = 0.049, Table 4) genetic models.
Table 4.
Main results of the meta-analysis on two studies of HLA-G 14bp I/D polymorphism with Hepatocellular cancer.
| HLA-G 14bp I/D polymorphism |
|||
|---|---|---|---|
| D vs. I | DD + ID vs. II | DD vs DI + II | |
| Subset comparison | N cases (N controls) OR (95% CI) POR |
N cases (N controls) OR (95% CI) POR |
N cases (N controls) OR (95% CI) POR |
| HBV/HCV positive vs. control | 594 (524) 1.698 (1.263–2.282) POR =0.000 |
297 (262) 2.321 (1.277–4.218) POR =0.006 |
297 (262) 1.739 (1.173–2.577) POR =0.006 |
| HBV/HCV negative vs. control | 260 (1482) 1.238 (0.929–1.650) POR = 0.145 |
130 (741) 1.188 (0.684–2.064) POR = 0.541 |
130 (741) 1.342 (0.921–1.955) POR = 0.126 |
| HBV/HCV positive vs. HBV/HCV negative | 594 (260) 1.429 (1.029–1.983) POR =0.033 |
297 (130) 1.981 (1.002–3.916) POR =0.049 |
297 (130) 1.385 (0.910–2.107) POR = 0.128 |
bp: base pairs, CI: Confidence interval, HBV: Hepatitis B virus, HCV: Hepatitis C virus, I/D: insertion/deletion, N: number of studies, NA: Not applicable, OR: odds ratio, POR: P-value associated to OR, Bold: significant P-value (≤0.05).
3.3. Meta-analysis of soluble HLA-G levels in digestive cancer patients and controls
Overall analysis of DC cancers including CRC and GC showed significant increased sHLA-G in patients compared to healthy controls (SMD = 3.341, 95% CI = 2.415–4.267, p = 0.000; Table 5, Figure 4A). sHLA-G were significantly increased in both CRC and GC compared to healthy controls with sHLA- G mean was 2 points higher in GC patients (SMD = 4.043, 95% CI = 2.28–5.858, p = 0.000; Table 5) than in CRC group (SMD = 2.165, 95% CI = 0.506–3.824, p = 0.011; Table 5) (Figure 4B and C). sHLA-G levels were also increased in the serum/plasma in DC patients (SMD = 3.994, 95% CI = 2.793–5.195, p = 0.000; Table 5) and in the CRC patients subgroup (SMD = 2.686, 95% CI = 00.159–5.214, p = 0.037; Table 5).
Table 5.
Meta-analysis results of sHLA-G significance in digestive cancers initiation∗.
| Ethnicity | Fluidics | Cancer type | Effects Models | N | Standardized mean differences |
Heterogeneity |
PEgger | PBegg | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SMD | SEM | 95% CI | PSMD | I2(%) | Tau2 | PH | |||||||
| Overall | All | DC | R | 16 | 3.341 | 0.472 | 2.415–4.267 | 0.000 | 98 | 3.392 | 0.000 | 0.011 | 0.015 |
| CRC | R | 5 | 2.165 | 0.847 | 0.506–3.824 | 0.011 | 97.9 | 3.473 | 0.000 | 0.686 | 0.327 | ||
| GC | R | 5 | 4.043 | 0.926 | 2.28–5.858 | 0.000 | 98.46 | 4.011 | 0.000 | 0.142 | 0.142 | ||
| Serum/Plasma | DC | R | 11 | 3.994 | 0.613 | 2.793–5.195 | 0.000 | 98.6 | 3.952 | 0.000 | 0.004 | 0.016 | |
| CRC | R | 3 | 2.686 | 1.289 | 0.159–5.214 | 0.037 | 98.9 | 4.923 | 0.000 | 0.636 | 0.602 | ||
| Caucasian | All | DC | F | 4 | 0.831 | 0.143 | 0.551–1.111 | 0.000 | 98.3 | 6.448 | 0.000 | 0.113 | 0.042 |
| Serum/Plasma | GC | F | 2 | 0.905 | 0.186 | 0.541–1.269 | 0.000 | 99.4 | 119.190 | 0.000 | NA | NA | |
| Asian | All | DC | F | 12 | 2.292 | 0.072 | 2.150–4.434 | 0.000 | 97.6 | 2.849 | 0.000 | 0.034 | 0.11 |
| Ascite | DC | F | 3 | 1.666 | 0.257 | 1.163–2.169 | 0.000 | 48.7 | 0.193 | 0.143 | 0.083 | 0.117 | |
| Serum/Plasma | CRC | F | 2 | 2.266 | 0.127 | 2.016–2.516 | 0.000 | 99.3 | 9.349 | 0.000 | NA | NA | |
| EC | F | 2 | 2.625 | 0.190 | 2.253–2.997 | 0.000 | 98.8 | 6.328 | 0.000 | NA | NA | ||
| GC | F | 2 | 1.818 | 0.122 | 1.579–2.058 | 0.000 | 98.5 | 2.861 | 0.000 | NA | NA | ||
| HCC | F | 3 | 4.058 | 0.230 | 3.608–4.508 | 0.000 | 97.3 | 6.115 | 0.000 | 0.217 | 0.217 | ||
CI: Confidence interval, CRC: Colorectal cancer; DC: digestive cancer; EC: Esophageal cancer; F:Fixed effects model, GC: Gastric cancer; HCC: Hepatocellular cancer; N: number of studies, NA: Not applicable, PBegg: P-value associated to Begg and Mazumdar rank correlation test (Two-tailed) without continuity correction, PEgger: P-value associated to Egger’s test (Two-tailed), PH: P-value associated to heterogeneity, PSMD: P-value associated to SMD, R: Random effects model, SEM: Standard errors of the mean, SMD: standardized mean differences, Bold: significant P-value (≤0.05). ∗ Cases vs. healthy controls.
Figure 4.
Forest plot of the association between sHLA-G dosage and digestive cancers susceptibility in overall population (A) All digestive cancers, (B) Colorectal cancer, (C) Gastric cancer.
sHLA-G were increased in both Caucasian and Asian patients with sHLA-G found to be 2 fold higher in Asians (SMD = 2.292, 95% CI = 2.150–2.434, p = 0.000; Table 5) than in Caucasians (SMD = 0.831, 95% CI = 0.551–1.111, p = 0.000; Table 5). Of note, in Asians, sHLA-G was 2 fold higher in serum/plasma (SMD = 2.266, 95% CI = 2.016–2.516, p = 0.000; Table 5) than in ascites (SMD = 1.666, 95% CI = 1.163–2.169, p = 0.000; Table 5).
In Asians, more types of DC were investigated and all had significant increased sHLA-G compared to healthy controls (Table 5). HCC patients presented the highest level of sHLA-G (SMD = 4.058, 95% CI = 3.608–4.508, p = 0.000; Table 5) compared to CRC, EC and GC. Due to few meta-analysed studies, subgroups results should be taken with caution.
We further investigated differences between grades in relation to sHLA-G levels. We found that in GC Asian patients, sHLA-G was significantly increased in high grade (Grade 3) compared to low grades (Grade 1 or Grade 2) (SMD = 0.448, 95% CI = 0.109–0.787, p = 0.000; Table 6). Further analysis showed that sHLA-G was significantly increased in grade 3 of DC when compared to grade 1 (SMD = 0.464, 95% CI = 0.150–0.778, p = 0.004; Table 6). Positive vascular invasion presented significant increase in sHLA-G compared to negative vascular invasion in overall analysis (SMD = 0.743, 95% CI = 0.385–1.100, p = 0.000; Table 6) and in Asians (SMD = 0.721, 95% CI = 0.336–1.107, p = 0.000; Table 6). Accordingly, sHLA-G was associated to a poor prognosis in DC.
Table 6.
Meta-analysis results of sHLA-G significance in digestive cancers according to histoprognostic parameters.
| Subset comparison | Ethnicity | Cancer type | Effect models | N | Standardized mean differences |
Heterogeneity |
PEgger | PBegg | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SMD | SEM | 95% CI | PSMD | I2(%) | Tau2 | PH | |||||||
| Grade 3 vs (Grade 1 or Grade 2) | Asian | DC | F | 4 | 0.133 | 0.113 | -0.089–0.356 | 0.239 | 88.7 | 0.406 | 0.000 | 0.565 | 1 |
| CRC | F | 2 | -0.104 | 0.150 | -0.399–0.190 | 0.487 | 95 | 0.868 | 0.000 | NA | NA | ||
| GC | F | 2 | 0.448 | 0.173 | 0.109–0.787 | 0.000 | 0 | 0 | 0.488 | NA | NA | ||
| Grade 3 vs. Grade 1 | Asian | DC | F | 2 | 0.464 | 0.160 | 0.150–0.778 | 0.004 | 0 | 0 | 0.463 | NA | NA |
| Grade 3 vs. Grade 2 | Asian | DC | F | 2 | -0.201 | 0.161 | -0.516–0.115 | 0.212 | 94.2 | 0.864 | 0.000 | NA | NA |
| N+ ∗ vs. N0 | Overall | DC | R | 3 | -0.078 | 0.228 | -0.525–0.369 | 0.732 | 53.7 | 0.085 | 0.116 | 0.524 | 0.117 |
| CRC | R | 2 | 0.105 | 0.138 | -0.165–0.376 | 0.444 | 0 | 0 | 0.876 | NA | NA | ||
| Asian | DC | F | 2 | -0.023 | 0.143 | -0.303–0.257 | 0.873 | 75.4 | 0.255 | 0.044 | NA | NA | |
| PVI vs. NVI | Overall | DC | R | 3 | 0.743 | 0.182 | 0.385–1.100 | 0.000 | 0 | 0 | 0.866 | 0.208 | 0.602 |
| PVI vs. NVI | Asian | HCC | F | 2 | 0.721 | 0.197 | 0.336–1.107 | 0.000 | 0 | 0 | 0.650 | NA | NA |
CI: Confidence interval, CRC: Colorectal cancer; DC: digestive cancer; F:Fixed effects model, GC: Gastric cancer; HCC: Hepatocellular cancer; N: number of studies, NA: Not applicable, NVI: Negative vascular invasion, PVI: Positive vascular invasion, PBegg: P-value associated to Begg and Mazumdar rank correlation test (Two-tailed) without continuity correction, PEgger: P-value associated to Egger’s test (Two-tailed), PH: P-value associated to heterogeneity, PSMD: P-value associated to SMD, R: Random effects model, SEM: Standard errors of the mean, SMD: standardized mean differences, Bold: significant P-value (≤0.05), ∗N+= (N1 or N2 or N1+N2).
3.4. Heterogeneity and publication bias
We detected substantial heterogeneity in meta-analyses investigating the genetic risk of 14-bp I/D for overall analysis and different DC subtypes (p < 0.05, Table 3). In meta-analyses investigating sHLA-G, heterogeneity was also detected. Clinical features could be major source of heterogeneity. Meta-analysis of genetic risk did not present a publication bias by means of Begg’ test (p Begg = 0.457) and symmetric funnel plot (Figure 5A). However, meta-analysis of sHLA-G presented a bias of publication (p Begg = 0.015) and asymmetric funnel plot (Figure 5B).
Figure 5.
Funnel plot assessing presence/absence of publication bias. (A) In the allelic models of HLA-G 14bp Ins/Del polymorphism, (B)sHLA-G meta-analysis in digestive cancers.
4. Discussion
In recent years, more and more studies are investigating the implication of HLA-G in DC, but in some cases results are conflicting. Our meta-analysis pooled published studies and examined the relationships between both HLA-G 14-bp genetic polymorphism and sHLA-G with the risk of DC. We further investigated the significance of sHLA-G according to clinicopathological features of DC.
Our results did not support a significant implication of HLA-G 14-bp I/D in DC susceptibility. A previous meta-analysis restricted to only HCC did not show a significant implication of 14-bp I/D [46], which confirm our finding. Further analysis revealed a clear association of HLA-G 14bp I/D in HCC patients infected with either HBV or HCV. Expression of HLA-G has been associated with HBV/HCV infection, via increased viral load [47, 48]. In early stages of HCV associated liver infection, both soluble and membrane bound HLA-G protein production are increased [7]. It is also possible that the presence of HLA-G expression in the context of HBV/HCV infections could favour the escape of cancerous cells from immune-surveillance. Because only few studies were meta-analysed, further investigations are still needed to clearly establish the role of HCV/HBV infection and the influence of HLA-G 14-bp I/D in HCC. In the context of sHLA-G, overall analysis showed significant increased sHLA-G in patients with DC cancers. In fact, sHLA-G has been suggested as a good diagnostic factor to distinguish benign colorectal related disease from CRC [43]. Because the invasive nature of the disease and the tumour microenvironment are different across multiple DC, the expression of HLA-G varies among different cancer types. Therefore, we conducted a stratified analysis to investigate the relationship between sHLA-G and DC cancers by cancer type. sHLA-G was significantly increased in either CRC or GC, with sHLA- G mean was 2 points higher in GC patients than in CRC patients. A previous study indicated that plasma sHLA-G level was a potential biomarker for GC diagnosis [38]. A large-scale genomewide association (GWAS) study of East Asians (22,775 CRC patients and 47,731 controls) revealed that HLA-G is one of the leading loci associated with the risk of CRC [49]. Group analysis by ethnicity revealed that sHLA-G was increased in either Caucasians or Asians, with sHLA-G mean 2 fold higher in Asians. This result may be explained by differences in genetic backgrounds between ethnicities.
Analysis by disease grades showed that sHLA-G was significantly increased in high grade compared to low grades. In addition, positive vascular invasion presented significantly elevated sHLA-G compared to negative vascular invasion. Accordingly, our results suggest that sHLA-G is associated with a poor prognosis in DC. These findings support the conclusion drawn from previous studies that tumour HLA-G expression is closely related to tumour progression and poor clinical outcomes in patients with cancer. A recent meta-analysis by Peng et al, performed on immunohistochemical and ELISA dosage [50], showed significant association of HLA-G with poor prognosis in gastric cancer. In addition, this meta-analysis showed that there was a significant correlation between HLA-G expression and TNM stage, lymph node status, and histological grade [50]. Interestingly, subgroup metaanalysis showed that HLA-G expression was only associated with clinic-pathological features in ESCC [50]. Interestingly, enhanced HLA-G expression has been found to be greatly correlated with weak anti-tumour immune response, disease progression, and poor survival in CRC [51]. Thus, HLA-G has been suggested as an independent prognostic predictor of CRC [51]. Du et al. indicated that HLA-G expression was strongly associated with tumour progression and involved in tumour evasion by raising the frequency of infiltrating Tregs locally. Therefore, HLA-G expression is a factor that should be taken into account when considering immunotherapy as treatment option, and is also a promising predictor for worse prognosis in DC patients [52]. HLA-G expression can be affected by ILT4 that regulates the cell proliferation, invasion and migration of CRC. HLA-G fusion protein treatment also increased ILT4 expression in a dose-dependent manner, thereby activating protein kinase B (AKT) and extracellular signal-regulated kinase (ERK) signaling, and facilitating the proliferation, migration and invasion of CRC cells. HLA-G interacts with ILT4 to promote CRC progression through AKT and ERK signal activation [2]. Accordingly, ILT4 and HLA-G could be prognostic factors to predict poor clinical response and survival time in patients with CRC providing a novel strategy of blocking ILT4/HLA-G for the treatment of CRC [2]. Therefore, HLA-G has the potential to serve as a biomarker for DC cancers prognosis, and screening for this marker could allow for the early diagnosis and treatment.
Our meta-analysis presents substantial limitations related to heterogeneity and size data. We also detected a marginal publication bias in meta-analysis on sHLA-G, which is due to unpublished studies and language restriction. We particularly acknowledge that the number of included studies was relatively small in subgroups; which might weaken the statistical power of the results. All the studies included were observational studies, so substantial heterogeneity was inevitable in this meta-analysis due to the various regimens, populations and sample sizes. Therefore, more clinical studies are needed to give definitive conclusions.
5. Conclusion
The current meta-analysis suggests that HLA-G 14-pb I/D polymorphism is associated with HCC in the case of HBV/HCV infections. Increased sHLA-G indicates a poor prognosis for DC patients. sHLA-G is likely associated with prognostic clinical features of DC. Further larger studies are warranted to consolidate current findings.
Declarations
Author contribution statement
Sabrine DHOUIOUI, Kalthoum TIZAOUI: Data collection, Data analysis and interpretation, Drafting of manuscript.
Nadia BOUJELBENE: Medical validation, Critical data interpretation, Critical revision, Hadda-imene OUZARI: Critical data interpretation, Critical revision. Inès ZIDI: Study conception and design, supervision, Data analysis, Drafting of manuscript, Critical revision.
Funding statement
This work was supported by Ministry of Higher Education and Scientific Research of Tunisia.
Data availability statement
Data will be made available on request.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
This research was supported by Ministry of Higher Education and Scientific Research of Tunisia.
References
- 1.Liang R., Lin Y., Yuan C.L., Liu Z.H., Li Y.Q., Luo X.L., et al. High expression of estrogen-related receptor alpha is significantly associated with poor prognosis in patients with colorectal cancer. Oncol. Lett. 2018;15:5933. doi: 10.3892/ol.2018.8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai Z., Wang L., Han Y., Gao W., Wei X., Gong R., et al. Immunoglobulinlike transcript 4 and human leukocyte antigenG interaction promotes the progression of human colorectal cancer. Int. J. Oncol. 2019;54:1943. doi: 10.3892/ijo.2019.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loustau M., Anna F., Drean R., Lecomte M., Langlade-Demoyen P., Caumartin J. HLA-G Neo-expression on tumors. Front. Immunol. 2020;11:1685. doi: 10.3389/fimmu.2020.01685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carosella E.D., Rouas-Freiss N., Tronik-Le Roux D., Moreau P., LeMaoult J. HLA-G: an immune checkpoint molecule. Adv. Immunol. 2015;127:33. doi: 10.1016/bs.ai.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Yan W.H. Human leukocyte antigen-G in cancer: are they clinically relevant? Cancer Lett. 2011;311:123. doi: 10.1016/j.canlet.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 6.Yie S.M., Yang H., Ye S.R., Li K., Dong D.D., Lin X.M. Expression of human leukocyte antigen G (HLA-G) correlates with poor prognosis in gastric carcinoma. Ann. Surg Oncol. 2007;14:2721. doi: 10.1245/s10434-007-9464-y. [DOI] [PubMed] [Google Scholar]
- 7.Amodio G., Sales de Albuquerque R., Gregori S. New insights into HLA-G mediated tolerance. Tissue Antigens. 2014;84:255. doi: 10.1111/tan.12427. [DOI] [PubMed] [Google Scholar]
- 8.Amiot L., Ferrone S., Grosse-Wilde H., Seliger B. Biology of HLA-G in cancer: a candidate molecule for therapeutic intervention? Cell. Mol. Life Sci. 2011;68:417. doi: 10.1007/s00018-010-0583-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin A., Yan W.H. Heterogeneity of HLA-G expression in cancers: facing the challenges. Front. Immunol. 2018;9:2164. doi: 10.3389/fimmu.2018.02164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alegre E., Rizzo R., Bortolotti D., Fernandez-Landazuri S., Fainardi E., Gonzalez A. Some basic aspects of HLA-G biology. J Immunol Res. 2014;2014 doi: 10.1155/2014/657625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X.Y., Yan W.H., Lin A., Xu H.H., Zhang J.G., Wang X.X. The 14 bp deletion polymorphisms in HLA-G gene play an important role in the expression of soluble HLA-G in plasma. Tissue Antigens. 2008;72:335. doi: 10.1111/j.1399-0039.2008.01107.x. [DOI] [PubMed] [Google Scholar]
- 12.Rousseau P., Le Discorde M., Mouillot G., Marcou C., Carosella E.D., Moreau P. The 14 bp deletion-insertion polymorphism in the 3' UT region of the HLA-G gene influences HLA-G mRNA stability. Hum. Immunol. 2003;64:1005. doi: 10.1016/j.humimm.2003.08.347. [DOI] [PubMed] [Google Scholar]
- 13.Tan Z., Randall G., Fan J., Camoretti-Mercado B., Brockman-Schneider R., Pan L., et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am. J. Hum. Genet. 2007;81:829. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moher D., Liberati A., Tetzlaff J., Altman D.G., Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6 [PMC free article] [PubMed] [Google Scholar]
- 15.Hozo S.P., Djulbegovic B., Hozo I. Estimating the mean and variance from the median, range, and the size of a sample. BMC Med. Res. Methodol. 2005;5:13. doi: 10.1186/1471-2288-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higgins J.P.T., Thomas J., Chandler J., Cumpston M., Li T., Page M.J., Welch V.A. Cochrane Handbook for Systematic Reviews of Interventions. 2nd Edition. John Wiley & Sons; Chichester (UK): 2019. [Google Scholar]
- 17.Michael Borenstein L.V.H., Higgins Julian P.T., Rothstein Hannah R. Copyright © 2009 John Wiley & Sons, Ltd; 2009. Introduction to Meta-Analysis. [Google Scholar]
- 18.Anzures-Cabrera J., Higgins J.P. Graphical displays for meta-analysis: an overview with suggestions for practice. Res. Synth. Methods. 2010;1:66. doi: 10.1002/jrsm.6. [DOI] [PubMed] [Google Scholar]
- 19.Egger M., Smith G.D., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Begg C.B., Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088. [PubMed] [Google Scholar]
- 21.DerSimonian R., Laird N. Meta-analysis in clinical trials revisited. Contemp. Clin. Trials. 2015;45:139. doi: 10.1016/j.cct.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egger M., Smith G.D., Phillips A.N. Meta-analysis: principles and procedures. BMJ. 1997;315:1533. doi: 10.1136/bmj.315.7121.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhouioui S., Laaribi A.-B., Boujelbene N., Jelassi R., Ben Salah H., Bellali H., et al. Association of HLA-G 3′UTR polymorphisms and haplotypes with colorectal cancer susceptibility and prognosis. Hum. Immunol. 2022;83:39. doi: 10.1016/j.humimm.2021.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Vaquero-Yuste C., Juarez I., Molina-Alejandre M., Molanes-López E.M., López-Nares A., Suárez-Trujillo F., et al. HLA-G 3’UTR polymorphisms are linked to susceptibility and survival in Spanish gastric adenocarcinoma patients. Front. Immunol. 2021:12. doi: 10.3389/fimmu.2021.698438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abu Hassan M., Al Omar S., Halawani H., Arafah M., Alqadheeb S., Al-Tamimi J., et al. Relationship of HLA-G expression and its 14-bp insertion/deletion polymorphism with susceptibility to colorectal cancer. Genet. Mol. Res. 2019;18:gmr18324. [Google Scholar]
- 26.El Bassiouny M.A., Elshaarawy A.A., Tawfeek G.A., Elashmawy M.I. Association of HLA-G gene polymorphism with hepatocellular carcinoma in Egyptian population. Menoufia Med. J. 2019;32:255. [Google Scholar]
- 27.Garziera M., Catamo E., Crovella S., Montico M., Cecchin E., S Lonardi, et al. Association of the HLA-G 3'UTR polymorphisms with colorectal cancer in Italy: a first insight. Int. J. Immunogenet. 2016;43:32. doi: 10.1111/iji.12243. [DOI] [PubMed] [Google Scholar]
- 28.Kim S.K., Chung J.H., Jeon J.W., Park J.J., Cha J.M., Joo K.R., et al. Association between HLA-G 14-bp insertion/deletion polymorphism and hepatocellular carcinoma in Korean patients with chronic hepatitis B viral infection. Hepato-Gastroenterology. 2013;60:796. doi: 10.5754/hge11180. [DOI] [PubMed] [Google Scholar]
- 29.Teixeira A.C., Mendes-Junior C.T., Souza F.F., Marano L.A., Deghaide N.H.S., Ferreira S.C., et al. The 14bp-deletion allele in the HLA-G gene confers susceptibility to the development of hepatocellular carcinoma in the Brazilian population. Tissue Antigens. 2013;81:408. doi: 10.1111/tan.12097. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y., Gao X-j, Deng Y-c, Zhang H-x. Relationship between HLA-G gene polymorphism and the susceptibility of esophageal cancer in Kazakh and Han nationality in Xinjiang. Biomarkers. 2012;17:9. doi: 10.3109/1354750X.2011.633242. [DOI] [PubMed] [Google Scholar]
- 31.Jiang Y., Chen S., Jia S., Zhu Z., Gao X., Dong D., et al. Association of HLA-G 3' UTR 14-bp insertion/deletion polymorphism with hepatocellular carcinoma susceptibility in a Chinese population. DNA Cell Biol. 2011;30:1027. doi: 10.1089/dna.2011.1238. [DOI] [PubMed] [Google Scholar]
- 32.Lazaro-Sanchez A.D., Salces-Ortiz P., Velasquez L.I., Orozco-Beltran D., Diaz-Fernandez N., Juarez-Marroqui A. HLA-G as a new tumor biomarker: detection of soluble isoforms of HLA-G in the serum and saliva of patients with colorectal cancer. Clin. Transl. Oncol. 2020;22:1166. doi: 10.1007/s12094-019-02244-2. [DOI] [PubMed] [Google Scholar]
- 33.Farjadian S., Tabebordbar M., Mokhtari M., Safaei A., Malekzadeh M., Ghaderi A. HLA-G expression in tumor tissues and soluble HLA-G plasma levels in patients with gastrointestinal cancer. Asian Pac. J. Cancer Prev. APJCP : Asian Pac. J. Cancer Prev. APJCP. 2018;19:2731. doi: 10.22034/APJCP.2018.19.10.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirana C., Ruszkiewicz A., Stubbs R.S., Hardingham J.E., Hewett P.J., Maddern P.J., et al. Soluble HLA-G is a differential prognostic marker in sequential colorectal cancer disease stages. Int. J. Cancer. 2017;140:2577. doi: 10.1002/ijc.30667. [DOI] [PubMed] [Google Scholar]
- 35.Li J.B., Ruan Y.Y., Hu B., Dong S.S., Bi T.N., Lin A., et al. Importance of the plasma soluble HLA-G levels for prognostic stratification with traditional prognosticators in colorectal cancer. Oncotarget. 2017;8 doi: 10.18632/oncotarget.16457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun J., Chang Y.-X., Niu C.-Y. Evaluation of ascitic soluble human leukocyte antigen-G for distinguishing malignant ascites from benign ascites. Tumor Biol. 2017;39 doi: 10.1177/1010428317726840. [DOI] [PubMed] [Google Scholar]
- 37.Khorrami S., Rahimi R., Mohammadpour H., Bahrami S., Yari F., Poustchi H., et al. Association of HLA-G∗01:01:02:01/G∗01:04:01 polymorphism with gastric adenocarcinoma. Hum. Immunol. 2016;77:153. doi: 10.1016/j.humimm.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 38.Pan Y.Q., Ruan Y.Y., Peng J.B., Han Q.Y., Zhang X., Lin A., et al. Diagnostic significance of soluble human leukocyte antigen-G for gastric cancer. Hum. Immunol. 2016;77:317. doi: 10.1016/j.humimm.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 39.Xu D.P., Shi W.W., Zhang T.T., Lv H.Y., Li J.B., Lin A., et al. Elevation of HLA-G-expressing DC-10 cells in patients with gastric cancer. Hum. Immunol. 2016;77:800. doi: 10.1016/j.humimm.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Zheng J., Xu C., Chu D., Zhang X., Li J., Ji G., et al. Human leukocyte antigen G is associated with esophageal squamous cell carcinoma progression and poor prognosis. Immunol. Lett. 2014;161:13. doi: 10.1016/j.imlet.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 41.Lin A., Zhang X., Zhou W.J., Ruan Y.Y., Xu D.P., Wang Q., et al. Human leukocyte antigen-G expression is associated with a poor prognosis in patients with esophageal squamous cell carcinoma. Int. J. Cancer. 2011;129:1382. doi: 10.1002/ijc.25807. [DOI] [PubMed] [Google Scholar]
- 42.Park Y., Park Y., Lim H.S., Kim Y.S., Hong D.J., Kim H.S. Soluble human leukocyte antigen-G expression in hepatitis B virus infection and hepatocellular carcinoma. Tissue Antigens. 2012;79:97. doi: 10.1111/j.1399-0039.2011.01814.x. [DOI] [PubMed] [Google Scholar]
- 43.Zhu C.B., Wang C.X., Zhang X., Zhang J., Li W. Serum sHLA-G levels: a useful indicator in distinguishing colorectal cancer from benign colorectal diseases. Int. J. Cancer. 2011;128:617. doi: 10.1002/ijc.25372. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y., Ye Z., Meng X.Q., Zheng S.S. Expression of HLA-G in patients with hepatocellular carcinoma. Hepatobiliary Pancreat. Dis. Int. 2011;10:158. doi: 10.1016/s1499-3872(11)60025-8. [DOI] [PubMed] [Google Scholar]
- 45.Lin A., Chen H.X., Zhu C.C., Zhang X., Xu H.H., Zhang J.G., et al. Aberrant human leucocyte antigen-G expression and its clinical relevance in hepatocellular carcinoma. J. Cell Mol. Med. 2010;14:2162. doi: 10.1111/j.1582-4934.2009.00917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coelho A.V., Moura R.R., Crovella S., Celsi F. HLA-G genetic variants and hepatocellular carcinoma: a meta-analysis. Genet. Mol. Res. 2016;15 doi: 10.4238/gmr.15038263. [DOI] [PubMed] [Google Scholar]
- 47.Souto F.J., Crispim J.C., Ferreira S.C., da Silva A.S., Bassi C.L., CP Soares, et al. Liver HLA-G expression is associated with multiple clinical and histopathological forms of chronic hepatitis B virus infection. J. Viral Hepat. 2011;18:102. doi: 10.1111/j.1365-2893.2010.01286.x. [DOI] [PubMed] [Google Scholar]
- 48.Weng P.J., Fu Y.M., Ding S.X., Xu D.P., Lin A., Yan W.H. Elevation of plasma soluble human leukocyte antigen-G in patients with chronic hepatitis C virus infection. Hum. Immunol. 2011;72:406. doi: 10.1016/j.humimm.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 49.Lu Y., Kweon S.S., Tanikawa C., Jia W.H., Xiang Y.B., Cai Q., et al. Large-scale genome-wide association study of East Asians identifies loci associated with risk for colorectal cancer. Gastroenterology. 2019;156:1455. doi: 10.1053/j.gastro.2018.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng Y., Xiao J., Li W., Li S., Xie B., He J., et al. Prognostic and clinicopathological value of human leukocyte antigen G in gastrointestinal cancers: a meta-analysis. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.642902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang R.L., Zhang X., Dong S.S., Hu B., Han Q.Y., Zhang J.G., et al. Predictive value of different proportion of lesion HLA-G expression in colorectal cancer. Oncotarget. 2017;8 doi: 10.18632/oncotarget.22487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Du L., Xiao X., Wang C., Zhang X., Zheng N., Wang L., et al. Human leukocyte antigen-G is closely associated with tumor immune escape in gastric cancer by increasing local regulatory T cells. Cancer Sci. 2011;102:1272. doi: 10.1111/j.1349-7006.2011.01951.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.