Abstract
The fate and impact of Pseudomonas aureofaciens TX-1 following application as a biocontrol agent for fungi in turfgrass were studied. The organism was applied with a modified irrigation system by using a preparation containing 1 × 106 P. aureofaciens TX-1 CFU ml−1 about 100 times between May and August. We examined the impact of this repeated introduction of P. aureofaciens TX-1 (which is known to produce the antimicrobial compound phenazine-1-carboxylic acid) on the indigenous microbial community of the turfgrass system and on establishment of introduced bacteria in the soil system. A PCR primer-DNA hybridization probe combination was developed to accurately monitor the fate of P. aureofaciens TX-1 following application in irrigation water. To assess the impact of frequent P. aureofaciens TX-1 applications on the indigenous bacterial community, turfgrass canopy, thatch, and rhizosphere samples were obtained during the growing season from control and treated plots and subjected to DNA extraction procedures and denaturing gradient gel electrophoresis (DGGE). PCR amplification and hybridization of extracted DNA with the P. aureofaciens TX-1-specific primer-probe combination revealed that P. aureofaciens TX-1 not only became established in the rhizosphere and thatch but also was capable of overwintering. Separation of PCR-amplified partial 16S rRNA genes by DGGE showed that the repeated application of P. aureofaciens TX-1 in irrigation water resulted in transient displacement of a leaf surface bacterial community member. There was no obvious alteration of any dominant members of the thatch and rhizosphere microbial communities.
Biological control agents are an alternative to the use of fungicides for suppression of fungal pathogens in agricultural production (12). The traditional biological control strategy has involved attempting to dislodge or replace a pathogen with an antagonistic population, often by applying a bacterial drench once or twice (2, 37). However, this approach has had only limited success (41). To improve pathogen suppression, frequent, often daily, applications have been investigated. For example, daily applications of Pseudomonas aureofaciens TX-1 at a rate of 2 × 107 CFU cm−2 to turfgrass plots inhibited the development of Sclerotinia homoecarpa, the causal agent of the disease dollar spot, by 84% compared to disease development in plots receiving no treatment or in plots receiving only single applications (30). The potential of combining reduced fungicide rates with very frequent P. aureofaciens TX-1 applications is apparent. Previous work has monitored the survival of Pseudomonas spp. introduced into soil systems (7, 19, 23). However, despite the increasing popularity of biological control, the fate and impact on the indigenous bacterial community of repeated applications of P. aureofaciens TX-1 have not been assessed.
One obstacle to the use of biological control organisms has been a lack of a way to deliver the organisms to the target site frequently and accurately. Routine application of P. aureofaciens TX-1 as a control agent is now possible because of the availability of modified irrigation technology that allows in-system bacterial biomass growth followed by nightly application of the cells grown. This approach to fungal control is now used in vegetable production and on some 400 golf courses in the United States; however, unsuccessful fungal suppression with this approach has been reported (Tom Vrabel, EcoSoil Systems Inc., San Diego, Calif., personal communication).
P. aureofaciens TX-1 gains much of its pathogen-suppressing ability and competitive fitness from the production of phenazine-1-carboxylic acid (PCA) (39). Mazzola et al. (21) found that PCA-producing strains, such as P. aureofaciens TX-1, were able to persist longer in the rhizosphere of wheat than strains that do not produce PCA, suggesting that PCA production functions as a defense mechanism that allows the bacteria to displace native strains, including fungi. The most recent research suggests that a diffusible signal molecule belonging to a family of N-acyl-homoserine lactones, identified as N-hexanoyl-homoserine lactone, is critical for induction of PCA-producing genes (42). This molecule is a product of the phzI gene (inducer) product found in P. aureofaciens TX-1, as well as other PCA-producing strains. It acts in trans to regulate PCA synthesis in neighboring cells (28). Clearly, production of PCA by P. aureofaciens TX-1 is dependent on the presence of regulatory and inducer genes in addition to sufficient cell density for diffusion of signals between cells of P. aureofaciens TX-1 or other PCA-producing fluorescent pseudomonads.
In an effort to begin to understand the behavior of P. aureofaciens TX-1 in the field, we recently performed a 2-year study investigating the effects of nightly applications of this bacterium to creeping bentgrass (Agrostis palustris) turf. Even with repeated applications of bacteria, only limited control of the target fungus was achieved (15). This finding supports earlier suggestions that a major obstacle to widespread use of biological control is the inconsistent performance of the inoculated bacteria (41).
To help resolve questions concerning the behavior of P. aureofaciens TX-1 in the environment where it is inoculated and expected to function, we used molecular biological approaches. Denaturing gradient gel electrophoresis (DGGE) of PCR-amplified bacterial 16S rRNA genes was used to determine the impact of repeated applications of P. aureofaciens TX-1 (through the irrigation system) on the bacterial communities of the turfgrass ecosystem. Additionally, the fate of the P. aureofaciens TX-1 applied was assessed by combining direct extraction of DNA from environmental samples with PCR and hybridization in which a strain-specific 16S ribosomal DNA (rDNA) probe was used. This approach has been used to monitor introduced organisms in environmental samples and has also been used successfully to detect a variety of organisms in different environments (4, 11, 34). To determine potential PCA production in the turfgrass system, we also assessed the level of indigenous fluorescent pseudomonads able to produce PCA. Understanding the biological implications of frequent P. aureofaciens TX-1 applications should improve our understanding of how biological control organisms suppress plant pathogens and our understanding of the long-term survival of an introduced population. Furthermore, obtaining fate data for biological control organisms is important for ensuring accurate placement of organisms for disease control and also for minimizing any nontarget impact that the applied organisms may have.
MATERIALS AND METHODS
Bacterial culture.
P. aureofaciens TX-1 was used in combination with the Bioject biological control delivery system (EcoSoils Systems Inc.). The delivery system consisted of a 25-liter bioreactor capable of growing approximately 1010 cells ml−1 in 0.1× tryptic soy broth (Difco Laboratories, Detroit, Mich.) during a 10-h incubation period (25°C). The system was self-contained, and it automatically inoculated itself, grew the bacteria, injected the bacteria into the irrigation water, and then cleaned itself following the injection. The P. aureofaciens TX-1 isolates used for in vitro studies (strain confirmation, detection limit of the designed primer-probe used) were maintained on 0.1× tryptic soy agar (TSA) (Difco Laboratories) amended with 50 μg of rifampin ml−1.
Penetration of P. aureofaciens TX-1 in the turfgrass environment.
Before field samples were taken, the depth of P. aureofaciens TX-1 penetration into the turfgrass rhizosphere was estimated by performing an in vitro radiolabel experiment with cores (diameter, 10 cm; length, 10 cm) obtained from the turfgrass plots used. Radiolabeled bacteria were produced by adding 2.16 μCi of [14C]glucose and 50 μl of an overnight culture of P. aureofaciens TX-1 to 50 ml of 0.1× tryptic soy broth. The culture was shaken at 138 rpm and incubated at 25°C. After 5 days, the culture was centrifuged to pellet the cells, and the cells were washed twice in 5 ml of 0.25× Ringer's solution. The cells were resuspended in 2 ml (final volume) of 0.25× Ringer's solution, and 100 μl was removed and used for liquid scintillation counting to determine the efficiency of bacterial labeling. The resulting total activity of the cells used was 0.13 μCi. Approximately 3.87 × 108 cells were added to each core in a volume of water equivalent to the irrigation volumes applied in the field (0.25 ml cm of core area−2 ). Following incubation for 9 h (the time between bacterial irrigation and sampling in the field study), the cores were separated with a knife into six slices that represented one canopy section (12-mm slice), one thatch section (7-mm slice), and four rhizosphere sections (7-mm slices). Each section of each core was homogenized by hand grinding with a metal spatula. Between 0.3 and 0.5 g of material was oxidized in a Packard 307 oxidizer (Packard Instrument Co., Downers Grove, Ill.) to determine the level of radioactivity and thus the relative amount of labeled bacteria in each core section.
Soil and turfgrass plots.
Triplicate plots (1.5 by 1.5 m) of creeping bentgrass (Agrostis palustris) were established from seed on silty clay loam (11% sand, 28% silt, 61% clay, 3.4% organic matter; pH 7.2) at the William H. Daniel Turfgrass Research and Diagnostic Center, West Lafayette, Ind. The plots were arranged in a randomized complete block and were maintained at a mowing height of 1.25 cm, the height commonly used for golf course fairway turfgrass.
Long-term application of P. aureofaciens TX-1 to the turfgrass canopy and examination of bacterial diversity by DGGE.
An in-ground irrigation system connected to the bioreactor described above was used to deliver nightly P. aureofaciens TX-1 applications in irrigation water for 123 days beginning in mid-May. Each plot was irrigated for 7 min at a rate of 0.07 ml cm−2 min−1. P. aureofaciens TX-1 was continuously injected into the irrigation line from the bioreactor during the 7-min cycle. As a result of this bacterial irrigation, an average of 2.8 × 106 CFU of P. aureofaciens TX-1 cm of plot−2 was applied, as determined by recovery of bacteria and irrigation water in pans (diameter, 25 cm; depth, 4 cm) followed by dilution plating. Briefly, approximately 40 ml of irrigation water from each of 10 randomly placed pans was transferred to a sterile Falcon tube, transported to the lab, and plated in a dilution series onto 0.1× TSA (Difco Laboratories) amended with 50 μg of rifampin ml−1. This collection procedure was performed once per week. Untreated plots were irrigated with volumes of water similar to the volumes used for the treated plots; however, a separate irrigation network was used to deliver water to the control plots in order to prevent cross-contamination. The control plots were covered with plastic tarps to block any bacterium-containing drift during irrigation of the treatment plots. The tarps were removed within 10 min after the bacterial irrigation stopped.
Core samples (diameter, 2.5 cm; length, 5 cm) were obtained from each plot 1 week before the first P. aureofaciens TX-1 application and 4, 12, 17, 95, 123, 181, and 353 days after the initial treatment. Four subsamples were obtained from each plot, and the canopy, thatch layer, and rhizosphere soil were separated with a sterile knife. The soil cores were each trimmed to a length of 2 cm (see below), and then subsamples were pooled by hand mixing to form the samples. Triplicate samples consisting of 0.5 g of soil, 0.3 g of thatch, and 0.2 g of turfgrass leaves were used for DNA extraction with a FastDNA Spin Soil DNA extraction kit (QBIOgene/Bio 101, Carlsbad, Calif.). The concentration of extracted DNA was determined spectrophotometrically at A260, and purity was determined by using the A260/A280 ratio. Each extracted DNA was diluted with an appropriate amount of sterile water to standardize the DNA concentrations on the basis of leaf, thatch, or soil dry weight. This additional step was often unnecessary as soil water contents were very consistent (data not shown) but was included to allow comparison of PCR product intensities generated from DNA isolated at different times. Bacterial community fingerprints were generated by PCR-DGGE by using primers 341-f and 534-r-GC (24) and the DCode universal mutation detection system (Bio-Rad Laboratories, Hercules, Calif.) as described previously (24), with the following exceptions; a 35 to 60% denaturing gradient was for used for canopy- and thatch-derived PCR products, and a 45 to 60% denaturing gradient was used for rhizosphere-derived PCR products. Gels were stained for 12 min in 0.5× TAE containing a 1:10,000 dilution of SYBR green I dye (Molecular Probes Inc., Eugene, Oreg.), visualized with a UV transilluminator, and photographed.
The presence of PCA in the bioreactor system was confirmed by high-performance liquid chromatography (HPLC) and UV detection. PCA was detected by the method of Slininger and Jackson (38) with a model 5000 HPLC (Varian, Walnut Creek, Calif.). The mobile phase was 65% acetonitrile–water containing 1% trifluoroacetic acid at a flow rate of 1 ml min−1. A Spherisorb ODS2, 10μ column (250 by 4.6 mm; Phenomenex, Torrance, Calif.) was used for separation, and UV detection was performed at 250 nm with a Holochrome detector (Gilson, Middleton, Wis.).
Partial 16S rDNA sequence determination.
The identity of the inoculated organism, P. aureofaciens TX-1, was confirmed by determining an almost complete (1,563-bp) 16S rRNA gene sequence. The 16S rRNA gene fragments were obtained by the method of Massol-Deya et al. (20). The sequence was also used to identify strain-specific primers and a combination probe (see below). DGGE bands that appeared to be either lost or gained due to application of P. aureofaciens TX-1 were excised, eluted in water, and reamplified with primers 341-f and 534-r (without the GC clamp).
For cloning and sequencing, PCR products generated from DGGE bands (194 bp) and the PCR products generated to confirm the sequence of the organism applied (1,563 bp) were ligated to the pGEM-T vector by using the instructions of the manufacturer (Promega Corp., Madison, Wis.) and transformed into Escherichia coli DH5-α (35). Sequences were determined with a Thermo Sequenase fluorescent labeled primer cycle sequencing kit (Amersham Pharmacia Biotech, Piscataway, N.J.) and a Pharmacia ALF Express automated sequencer (Amersham Pharmacia Biotech). The BLAST 2.0 algorithm was used to compare the sequence derived to 16S rRNA gene sequences in the National Center for Biotechnology Information database (1).
P. aureofaciens TX-1-specific PCR primers and hybridization probe.
We designed PCR primers for specific amplification of P. aureofaciens TX-1 that were based on a previously published alignment of pseudomonad 16S rDNA sequences (22) and manually added sequences of closely related strains found in the GenBank database. Two primers, 49f (E. coli positions 76 to 94; 5′-GAA GCT TGC TTC TCT TGA G-3′) and 617r (E. coli positions 644 to 628; 5′-CTC GCC AGT TTT GGA TG-3′), were designed to amplify a 569-bp fragment of the 16S rRNA gene from a limited number of pseudomonad templates, including P. aureofaciens TX-1. The PCR conditions used for the new primers were the same as those described for the 341-f and 534-r primers (24) except for the annealing temperature, which was 62°C. The PCR conditions were optimized by using bacterial isolates that have sequences that are very similar to the sequences of the primers (Table 1).
TABLE 1.
Comparison of P. aureofaciens TX-1 PCR primer and probe sequences to nucleotide sequences of test isolatesa
| Isolate | Primer 49f sequence | Probe sequence | Primer 617r sequence |
|---|---|---|---|
| Pseudomonas aureofaciens | GAAGCTTGCTTCTCTTGAGb | GTACTTACCTAATACGTGAGT | CTCGCCAGTTTTGGATG |
| P. cepacia | GGTGCTTNCACCTNGGGCG | CCTTGGCTCTAATACAGCCGG | CCTGCCAGTCACCAATG |
| P. aeruginosa | GAAGCTTGCTCCTGGATTC | GCAGTAAGTTAATACCTTGCT | CTCAGTAGTTTNGGATG |
| P. putida | AGAGCTTGCTCTTCGATTC | GCATTAACCTAATACGTTAGT | CTTGCCAGTTTTGGATG |
| P. stutzeri ATCC 17587 | GAAGCTTGCTCCATGATTC | GCAGTAAGTTAATACCTTGCT | CTTGCCAGTTTTGAAAG |
| P. viridiflava pv-5 | GAAGCTTGCTTCTCTTGAG | GCAGTAACCTAATACGTTATT | CTTGCCAGTTTTGGATG |
| P. syringae pv. syringae | GGTACTTGTACCTGGTGGC | GCAGTTACCTAATACGTRTCT | CTTGCCAGTTTTGGATG |
| P. putida AC10R | AGAGCTTGCTCTTCGATTC | GCATTAACCTAATACGTTAGT | CTCGCCAGTTTTGGATG |
| P. fluorescens ATCC 17419 | GAAGCTTGCTTCTCTTGAG | GCATTAACCTAATACGTTAGT | TCAGTCAGTTTTGAATG |
| P. multivorans ATCC 17478 | GTGCTTGCACCTGGTGGCG | CCTTGGCTCTAATACAGTCGG | CCTGCCAGTCACAAATG |
| Comamonas acidovorans | GAAGCTTGCTTCTCTTGAG | GTACTTACCTAATACGTGAGT | CCATGCAGGTANAAATG |
| C. acidovorans ATCC 17407 | TAACAGGTCTTCGGACGCT | GGCTTCTCCTAATACGAGAGG | CCATGCAGGTANAAATG |
| C. testosteroni ATCC 15668 | GAAGCTTGCTTCTCTTGAG | GCCTGGGGCTAATATCCCCGG | GAATGTAGCAACTAATG |
| Ralstonia eutropha | ACTTCGGTCTGGCGGCGAG | GCGCTGGTTAATACCTGGCGT | CCTTGCAGTCACAAGCG |
P. syringae pv. syringae, Alcaligenes sp. strain BR40, and a Ralstonia strain (species unknown) were also used as test organisms; however, the complete sequence alignment with primers 49f and 617r and the P. aureofaciens TX-1 probe was not determined.
Boldface type indicates bases conserved in isolate and P. aureofaciens TX-1 probe and primer binding sites.
The primers which we designed were not completely specific and could amplify DNA from other Pseudomonas spp. To ensure specific detection of P. aureofaciens TX-1, we used a region between the two primer binding sites to design a 21-mer oligonucleotide probe. The sequence 5′-GTA CTT ACC TAA TAC GTC AGT-3′ (E. coli positions 221 to 241) was synthesized with a 5′ end label of digoxigenin (IDT, Coralville, Iowa). The specificity of the probe was tested and the hybridization conditions were optimized by using dot blots of PCR products generated from the isolates shown in Table 1, including P. aureofaciens TX-1, which was the positive control. The membranes were hybridized for 8 h at 42°C, and signals were detected by using a DIG/Genius luminescent detection kit according to the instructions of the manufacturer (Roche Molecular Biochemicals, Indianapolis, Ind.).
In order to test the detection limits of the combined PCR-hybridization approach, P. aureofaciens TX-1 cells were grown as described above, washed twice in 0.25× Ringer's solution, and inoculated into rhizosphere soil obtained from uninoculated turfgrass plots at levels (10-fold dilutions) ranging from 2.5 × 109 to 2.5 × 100 cells g of soil−1 . Control soil samples received no cells, but they received an equal amount of water. After 9 h of incubation, DNA was extracted as described above and PCR amplified with new primers 49f and 617r. The PCR products were electrophoresed in a 0.7% agarose gel and transferred to nylon membranes with a Transblot electroblotter (Bio-Rad Laboratories). The membranes were hybridized with the P. aureofaciens TX-1 probe by using the conditions described above. The same PCR and hybridization methods were used to assess the fate of P. aureofaciens TX-1 cells following application to the turfgrass system.
Screening PCA production by the culturable indigenous pseudomonad community.
To recover bacteria, about 4 g (wet weight) of rhizosphere from each turfgrass plot and 6 g of glass beads (diameter, 4 mm) were shaken with 30 ml of 5 mM sodium phosphate buffer (pH 7.0) in a 50-ml plastic centrifuge tube for 2 h. Only soil samples from untreated plots were screened with this assay in order to avoid recovering applied P. aureofaciens TX-1 and overestimating the number of indigenous PCA-producing isolates. The suspensions were allowed to settle for 10 min and then diluted serially in 100 mM sodium phosphate buffer. The numbers of total CFU were determined on 0.1× TSA, while the levels of fluorescent pseudomonads were determined on King's B agar supplemented with penicillin G (45 mg liter−1), novobiocin (45 mg liter−1), and cycloheximide (75 mg liter−1) (36). The plates were incubated in the dark at 25°C for 48 h. Fluorescent pseudomonad isolates (n = 150) growing on the selective medium were transferred to KM agar (18) and to potato dextrose agar (Difco Laboratories) and incubated for an additional 4 days at 25°C. Both of these media have been reported to indicate production of as little as 0.25 μg of PCA by isolated bacterial colonies (13). PCA production was indicated by yellow-orange pigmentation of a colony when it was grown on KM agar and by the presence of a dark halo surrounding a colony when the plate was viewed under long-wavelength UV radiation (365 nm) with either type of medium (39). Two strains known to produce PCA, P. aureofaciens TX-1 (this study) and Pseudomonas fluorescens 2-79 (40), served as positive controls.
Nucleotide sequence accession number.
The partial environmental 16S rRNA gene clone sequence obtained in this study from the leaf surface of A. palustris has been deposited in the GenBank nucleotide sequence database under accession no. AF297982.
RESULTS
Determination of sampling depth by using radiolabeled P. aureofaciens TX-1.
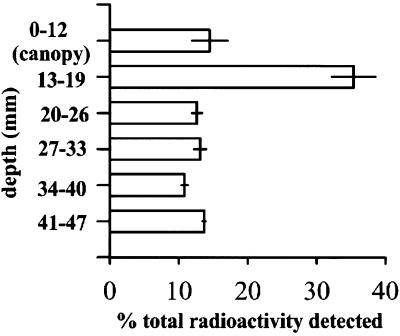
In an in vitro study to trace the fate of radiolabeled P. aureofaciens TX-1 in an inoculated turfgrass core, more than one-half of the detectable radioactivity was found in the leaf canopy and thatch layer (depth, 0 to 19 mm) (Fig. 1). Approximately 74% of the total radioactivity in the thatch and soil was present in the upper 2 cm of the core. We assumed that the radioactivity in each section correlated with the relative proportion of cells present and used 2 cm as our soil sampling depth. As shown in Fig. 1, some cells were able to reach a depth of more than 2 cm. For reasons of detection sensitivity, we wanted to obtain the greatest number of cells per gram of soil possible in the samples, and thus we avoided depths greater than 2 cm, where the cell densities were lower.
FIG. 1.
Detectable radioactivity in a turfgrass environment profile (canopy, thatch, and rhizosphere soil) following inoculation of a sample core with radiolabeled P. aureofaciens TX-1. The error bars indicate the standard errors for measurements from three replicates.
Testing P. aureofaciens TX-1-specific PCR primers and the hybridization probe.
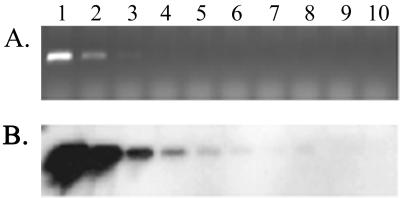
Although the PCR primers were designed to avoid amplification of templates that were not similar to the 16S rDNA of P. aureofaciens TX-1, a faint PCR product was detected when Pseudomonas viridiflava was used (data not shown). However, only the PCR product from P. aureofaciens TX-1 produced a signal when the P. aureofaciens TX-1-specific probe was hybridized to a dot blot of PCR products from the test strains. Therefore, when the PCR and probe hybridization procedures were combined, P. aureofaciens TX-1 could be specifically and consistently detected in the turfgrass rhizosphere soil inoculated in vitro with 2.5 × 102 cells g (dry weight) of soil−1 (Fig. 2). In the field study, amplification of DNA from uninoculated soil always resulted in no visible PCR products and no signal when the DNA was hybridized with the P. aureofaciens TX-1 probe, suggesting that the number of native P. aureofaciens TX-1 cells in the turfgrass rhizosphere was below the detection limit of our method.
FIG. 2.
Sensitivity of P. aureofaciens TX-1 detection with PCR primers 49f and 617r and hybridization with the P. aureofaciens TX-1 probe in turfgrass rhizosphere soil inoculated with dilutions of P. aureofaciens TX-1 cells. (A) Agarose gel electrophoresis of the 568-bp PCR product amplified from soil DNA. (B) Hybridization of the P. aureofaciens TX-1 probe to DNA blotted from the gel shown in panel A. Soils were inoculated with 2.5 × 109, 2.5 × 108, 2.5 × 107, 2.5 × 106, 2.5 × 105, 2.5 × 104, 2.5 × 103, 2.5 × 102, and 2.5 × 101 cells g−1 (lanes 1 to 9, respectively) or were not inoculated (lane 10), as described in the text. Lane 11, negative (no DNA) PCR control.
Detection of P. aureofaciens in environmental samples.
P. aureofaciens TX-1 was detected in all rhizosphere samples obtained from treated turfgrass plots following 12 consecutive days of bacterial irrigation (Fig. 3). Based on visual inspection of the hybridization signals, cells were present throughout the irrigation period and until 58 days (November) after P. aureofaciens TX-1 inoculation ended. A signal was also detected in April of the following year, after overwintering and spring warm-up, on day 353 after the initial treatment. No hybridization signals were observed in any DNA sample collected before the start of inoculation or from the control plots (no P. aureofaciens TX-1 applied) during the study.
FIG. 3.
DNA hybridization of P. aureofaciens TX-1 probe to Pseudomonas-specific PCR products from P. aureofaciens TX-1-treated turfgrass rhizosphere soil and untreated control soil (minus signs) obtained on selected dates. The numbers in parentheses indicate the numbers of days after the initial treatment.
Community stability.
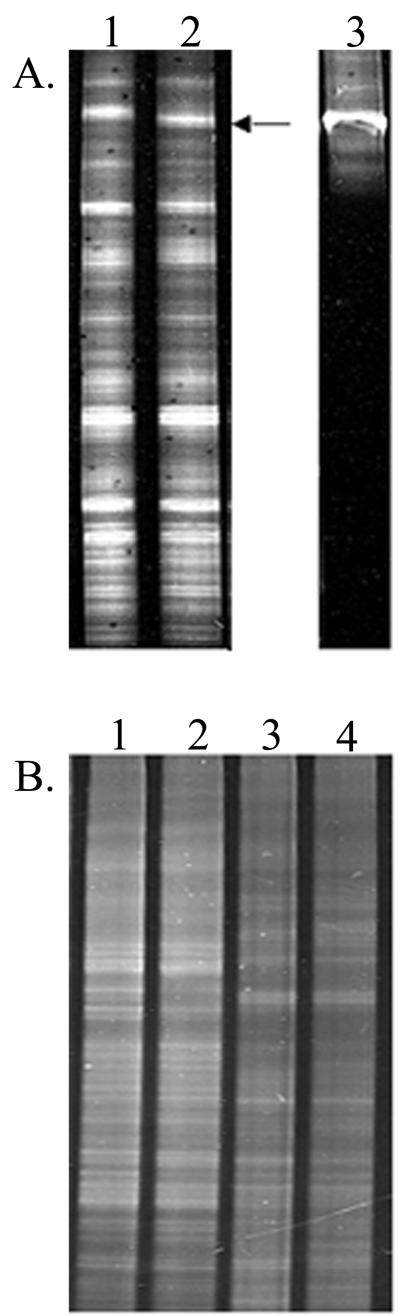
PCR-DGGE analysis of DNA from the canopy indicated that repeated P. aureofaciens TX-1 applications resulted in one detectable change, the loss of a band, in fingerprints of the indigenous bacterial community (Fig. 4A). This change was first detected following 95 days of P. aureofaciens TX-1 application. The band was absent from all treated samples analyzed through 58 days after P. aureofaciens TX-1 treatment ended (181 days after the initial treatment). At 353 days after the initial treatment (April of the following year), the band was again detected in DGGE gels. The nucleotide sequence of this displaced band was 98% similar to that of Cytophaga saccharophila, a member of the family Cytophagaceae. However the PCR product was short (192 bp), and therefore, the identification was not conclusive. The loss of the band was confirmed when no signal was detected after hybridization of a probe made from the excised band to the DGGE gel (data not shown). A signal was detected when the band was present.
FIG. 4.
DGGE analysis of 16S rRNA genes after PCR amplification of DNA isolated from the turfgrass environment with primers 341-f and 534-r-GC. (A) Lanes 1 and 2, leaf canopy DNA without nightly applications of P. aureofaciens TX-1 (lane 1) and with nightly applications (lane 2); lane 3, amplified P. aureofaciens TX-1 DNA extracted from a pure culture. The arrow indicates the position of DNA that represents the bacteria displaced by P. aureofaciens TX-1. (B) DGGE of soil (lanes 1 and 2) and thatch (lanes 3 and 4) DNA samples. Lanes 1 and 3, untreated samples; lanes 2 and 4, samples treated with nightly applications of P. aureofaciens TX-1.
In contrast to the findings obtained with the canopy samples, PCR-DGGE analysis of samples of turfgrass plot rhizosphere and thatch that received repeated P. aureofaciens TX-1 applications revealed no visible changes in the dominant populations present in the communities in all samples throughout the experiment (Fig. 4).
Soil bacterial populations and indigenous PCA production.
We recovered 3.43 × 109 total bacteria g (dry weight) of soil−1 (standard error, 2.07 × 108 bacteria g−1) following dilution plating of turfgrass rhizosphere soil extracts onto 0.1× TSA. Fluorescent pseudomonads were also recovered, and the total concentration was 2.16 × 107 CFU g (dry weight) of rhizosphere soil−1 (standard error, 1.11 × 106 CFU g−1), approximately 0.6% of the total bacteria isolated. Of 150 fluorescent pseudomonad isolates screened, no PCA-producing strains were detected in samples collected from any of the sites. Both of the positive control strains, P. aureofaciens TX-1 and P. fluorescens 2-79, produced the diagnostic PCA production signs: yellow-orange pigmentation on KM agar and dark halos on both media used when plates were viewed under long-wavelength UV light. PCA-producing bacteria grown on potato dextrose agar also produce dark granules in colonies; however, this criterion has been shown to be a less consistent indication of PCA production than halo formation when plates are viewed under UV light.
DISCUSSION
The PCR hybridization detection method showed that after introduction of P. aureofaciens TX-1 into the turfgrass environment by irrigation began, cells were detectable in the soil by day 12 and remained detectable throughout the experiment, even after irrigation with bacteria ended. Our data indicate that the irrigation system is effective for achieving and maintaining a detectable level of cells. Our system of P. aureofaciens TX-1 detection detected the organism with accuracy and sensitivity. Using similar techniques, workers have detected as few as 3 × 102 cells of Mycobacterium chlorophenolicum PCP-1 in soil (4) and 102 cells of Paenibacillus azotofixans per g of wheat rhizosphere soil (34). Although DNA liberated from dead cells may persist in soil for several weeks (33), we have qualitatively recovered P. aureofaciens TX-1 on selective media following soil inoculation in the field and short-term incubation (1 to 2 days of incubation) (Sigler, unpublished data). Thus, we assume that the hybridization signal detected in soil DNA extracted following irrigation with bacteria in the field truly reflects DNA extracted from cells and not naked DNA. This is important for the P. aureofaciens TX-1 biocontrol strategy as the target pathogen, Sclerotinia homoecarpa, is a foliage- and thatch-dwelling pathogen (6). Thus, transport of P. aureofaciens TX-1 into the soil could effectively decrease the density of cells present in the canopy that have the potential to suppress the pathogen.
At 227 days after irrigation ended (after overwintering and 353 days after the initial treatment), a P. aureofaciens TX-1 hybridization signal was detected in the previously inoculated turfgrass plots, indicating that successful colonization by the applied bacteria occurred. This finding supports Dwyer's data obtained with a plate count procedure, which showed that inoculated rifampin-resistant P. aureofaciens TX-1 cells were able to overwinter following repeated applications to a turfgrass canopy (10). However, it was not clear from this study whether spontaneous mutants or introduced strains were detected in the subsequent season. When combined with Dwyer's observations, our results support the conclusion that P. aureofaciens TX-1 successfully invaded the treated soils, and we suggest that the high P. aureofaciens TX-1 input levels might have provided a significant residual population that resulted in at least some colonization of the new environment. The proportion of P. aureofaciens TX-1 DNA template relative to native bacterial DNA template extracted from the soils in this study is not known, and this makes determining the number of overwintering cells impossible due to biases in the PCR and DNA probe hybridization procedures (14, 29).
We found that after daily applications of bacteria for 123 days at a rate of 2.8 × 106 CFU cm−2 there was no impact on the overall bacterial communities in soil and thatch that could be detected by PCR-DGGE (Fig. 4). These findings are similar to those of workers who reported that a single, highly concentrated application of bacteria resulted in little disruption of the soil microbial community (27, 32). In contrast to the evaluation of the soil and thatch populations, evaluation of the plant canopy bacterial populations showed that introduction of P. aureofaciens TX-1 displaced at least one member of the native population. It should be noted that despite the lack of obvious community disruption in the thatch and soil environments, the tremendous bacterial diversity in these systems limited DGGE evaluation of the dominant members of the bacterial communities (25). Similarly, the richness of the microbial diversity in the leaf canopy environment may have masked other changes. Detection of changes in the most minor members of the community would require more sensitive techniques, such as the use of group-specific primers or probes capable of discriminating against background templates. Although the molecularly based analyses used in this study are limited, this work represents an initial attempt to use molecular tools in a biocontrol organism impact study in order to circumvent the well-known shortcomings of traditional methods (3, 9).
Based on data from other workers, we suggest that the mechanism that directed displacement of the bacteria in the canopy is antibiosis-assisted competition (8), especially since the introduced strain produces PCA. Such competition has been shown for P. aureofaciens TX-1 and other fluorescent Pseudomonas spp. (5, 16, 17). In the context of this study, it is very probable that the bacteria produced PCA while they were in the bioreactor, where the cell density was high, and that production of PCA in the field was limited. We confirmed that PCA was present at a concentration of about 8 μg ml−1 in the bioreactor by HPLC analysis, and this finding supports our hypothesis that the cell densities in the reactor were high enough to cause phenazine production. Despite our evidence which supports the hypothesis that PCA was produced in the reactor, documentation of in situ production of antibiotics is unavailable. Evidence that supports the hypothesis that Pseudomonas spp. are able to compete for space as a mechanism of displacement was described by Natsch et al. (26), who tested both wild-type and antibiotic-overproducing strains of P. fluorescens and found that the two strains could displace similar portions of a resident pseudomonad population. Given the high frequency of application, this mode of displacement cannot be ruled out.
Although apparently the P. aureofaciens TX-1 applied became established in the soil and canopy, there was limited control of the target fungus (15). It is possible that the low level of pathogen-suppressing activity in soil after application of P. aureofaciens TX-1 can be explained in part by the lack of a pool of in situ phzI genes and the absence of PCA production in the resident soil bacteria at this site. Considering that phzI and phzR are located adjacent to one another, we presume that the proportion of organisms capable of PCA production is equal to the proportion of organisms containing phzI and thus having signaling potential. Based on assays of extracted fluorescent pseudomonads from the turfgrass rhizosphere, our data show that indigenous bacteria capable of producing PCA are not present in this environment (0 of 152 isolates). The absence of PCA-producing isolates recovered from the soil in this study suggests that the potential for in trans induction of PCA production induced by native strains harboring phzI genes is low, if it is present at all. Given the numbers of fluorescent pseudomonads detected by the plating experiment in our study, the presence of a low number of PCA-producing pseudomonads seems improbable. However, a similar finding has been reported previously in a take-all (wheat)-suppressive soil (31).
In conclusion, establishment of P. aureofaciens TX-1 in the turfgrass environment resulted from repeated inoculation of the bacterium, but the detectable impact on the indigenous bacteria community was minimal. Since PCA was produced in the bioreactor during incubation of P. aureofaciens TX-1, it is possible that the impact on the community resulted from direct application of PCA. However, the dilution of PCA following injection into the irrigation water was significant. While the possibility that the bacteria were active in situ cannot be ruled out, such activity may have been limited due to the absence of indigenous organisms harboring phzI genes that would have triggered PCA production. These findings may explain the highly variable responses reported when P. aureofaciens TX-1 is used for commercial application to turfgrass or other crop production systems.
ACKNOWLEDGMENT
Support for portions of this project was provided by the Purdue Research Foundation and the Midwest Regional Turfgrass Foundation.
Footnotes
Paper 16,533 of the Purdue University Agricultural Experiment Station Series.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1997;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Baker K F. Evolving concepts of biological control of plant pathogens. Annu Rev Phytopathol. 1987;25:67–85. [Google Scholar]
- 3.Boivin-Jahns V, Bianchi A, Ruimy R, Garcin J, Daumas S, Christen R. Comparison of phenotypical and molecular methods for the identification of bacterial strains isolated from a deep subsurface environment. Appl Environ Microbiol. 1995;61:3400–3406. doi: 10.1128/aem.61.9.3400-3406.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briglia M, Eggen R I, DeVos W M, Van Elsas J D. Rapid and sensitive method for the detection of Mycobacterium chlorophenolicum PCP-1 in soil based on 16S rRNA gene-targeted PCR. Appl Environ Microbiol. 1996;62:1478–1480. doi: 10.1128/aem.62.4.1478-1480.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carruthers F L, Shum-Yhomas T, Conner A J, Mahanty H K. The significance of antibiotic production by Pseudomonas aureofaciens TX-1 PA147-2 for biological control of Phytophthora megasperma root rot of asparagus. Plant Soil. 1995;170:339–344. [Google Scholar]
- 6.Chamberlain K, Crawford D L. In vitro and in vivo antagonism of pathogenic turfgrass fungi by Streptomyces hygroscopicus strains YCED9 and WYE53. J Ind Microbiol Biotechnol. 1999;23:641–646. doi: 10.1038/sj.jim.2900671. [DOI] [PubMed] [Google Scholar]
- 7.Compeau G, Al Achi B J, Platsuoka E, Levy S B. Survival of rifampicin-resistant mutants of Pseudomonas fluorescens and Pseudomonas putida in soil systems. Appl Environ Microbiol. 1988;54:2432–2438. doi: 10.1128/aem.54.10.2432-2438.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doyle J D, Stotzky G, McClung G. Effects of genetically engineered microorganisms on microbial populations and processes in natural habitats. Adv Appl Microbiol. 1995;40:237–287. doi: 10.1016/s0065-2164(08)70366-6. [DOI] [PubMed] [Google Scholar]
- 9.Dunbar J, White S, Forney L. Genetic diversity through the looking glass: effect of enrichment bias. Appl Environ Microbiol. 1997;63:1326–1331. doi: 10.1128/aem.63.4.1326-1331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dwyer P J. Field efficacy, persistence, and antibiotic production of Pseudomonas aureofaciens TX-1. MS thesis. East Lansing: Michigan State University; 1999. [Google Scholar]
- 11.El Fantroussi S, Mahillon J, Naveau H, Agathos S N. Introduction and PCR detection of Desulfomonile tiedjei in soil slurry microcosms. Biodegradation. 1997;8:125–133. doi: 10.1023/a:1008262426800. [DOI] [PubMed] [Google Scholar]
- 12.England L S, Holmes S B, Trevors J T. Review: persistence of viruses and DNA in soil. World J Microbiol Biotechnol. 1998;14:163–169. [Google Scholar]
- 13.Gurusiddaiah S, Weller D M, Sarkar A, Cook R J. Characterization of an antibiotic produced by a strain of Pseudomonas fluorescens inhibitory to Gaeumannomyces graminis var. tritici and Pythium spp. Antimicrob Agents Chemother. 1986;29:488–495. doi: 10.1128/aac.29.3.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen M C, Tolker T N, Givskov M, Molin S. Biased 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol Ecol. 1998;26:141–149. [Google Scholar]
- 15.Hardebeck, G., and Z. Reicher. 24 June 2000, posting date. Pseudomonas aureofaciens applied by irrigation to extend the window of dollar spot control of various fungicides. 1999 annual report. [Online.] Purdue University Turfgrass Science Program, West Lafayette, Ind. http://www.agry.purdue.edu/turf/report/1999/page50.htm.
- 16.Hebbar K P, Davey A G, Dart P J. Rhizobacteria of maize antagonistic to Fusarium moniliforme, a soil-borne pathogen: isolation and identification. Soil Biol Biochem. 1992;24:979–987. [Google Scholar]
- 17.Homma Y, Sato Z, Hirayama F, Konno K, Shirahama H, Suzui T. Production of antibiotics by Pseudomonas cepacia as an agent for biological control of soilborne plant pathogens. Soil Biol Biochem. 1989;21:723–728. [Google Scholar]
- 18.Kanner D, Gerber N N, Bartha R. Pattern of phenazine pigment production by a strain of Pseudomonas aeruginosa. J Bacteriol. 1978;134:690–692. doi: 10.1128/jb.134.2.690-692.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kluepfel D A, Kline E L, Skipper H D, Hughes T A, Gooden D T, Drahos D J, Barry G F, Hemming B C, Brandt E J. The release and tracking of genetically engineered bacteria in the environment. Phytopathology. 1991;81:348–352. [Google Scholar]
- 20.Massol-Deya A A, Oldelson D A, Hinkley R F, Tiedje J M. Bacterial community fingerprinting of amplified 16S and 16–23S rDNA gene sequences and restriction endounuclease analysis (ARDRA) In: Akkermans A, editor. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishing; 1995. pp. 3.3.2/1–3.3.2/8. [Google Scholar]
- 21.Mazzola M, Cook R J, Thomashow L S, Weller D M, Pierson L S., III Contribution of phenazine antibiotic biosynthesis to the ecological competence of fluorescent pseudomonads in soil habitats. Appl Environ Microbiol. 1992;58:2616–2624. doi: 10.1128/aem.58.8.2616-2624.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore E R B, Mau M, Arnscheidt A, Bottger E C, Hutson R A, Collins M D, Van de Peer Y, De Wachter R, Timmis K N. The determination and compilation of the 16S rRNA gene sequences of species of the genus Pseudomonas (sensu stricto) and estimation of the natural intragenic relationships. Syst Appl Microbiol. 1996;19:478–492. [Google Scholar]
- 23.Morel J L, Bitton G, Chaudry G R, Awong J. Fate of genetically modified microorganisms in the corn rhizosphere. Curr Microbiol. 1989;18:355–360. [Google Scholar]
- 24.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakatsu C H, Torsvik V, Øvreås L. Soil community analysis using denaturing gradient gel electrophoresis (DGGE) profiles of 16S rDNA PCR products. Soil Sci Soc Am J. 2000;64:1382–1388. [Google Scholar]
- 26.Natsch A, Keel C, Hebecker N, Laasik E, Defago G. Influence of bio-control strain Pseudomonas fluorescens CHA0 and its antibiotic overproducing derivative on the diversity of resident root colonizing pseudomonads. FEMS Microbiol Ecol. 1998;27:365–380. [Google Scholar]
- 27.Nishino T, Morita K, Fujimori T. Fate of Xanthomonas campestris pv. Poae, a biological control agent for annual bluegrass, in soil. J Pestic Sci (Nihon Noyaku Gakkaishi) 1997;22:326–330. [Google Scholar]
- 28.Pierson E A, Wood D W, Cannon J A, Blachere F M, Pierson L S., III Interpopulation signaling via N-acyl-homoserine lactones among bacteria in the wheat rhizosphere. Mol Plant-Microbe Interact. 1998;11:1078–1084. [Google Scholar]
- 29.Polz M F, Cavanaugh C M. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Powell J F. Utilization of bacterial metabolites for the management of fungal turfgrass pathogens. MS thesis. East Lansing: Michigan State University; 1993. [Google Scholar]
- 31.Raaijmakers J M, Weller D M, Thomashow L S. Frequency of antibiotic-producing Pseudomonas spp. in natural environments. Appl Environ Microbiol. 1997;63:881–887. doi: 10.1128/aem.63.3.881-887.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Recorbet G, Steinberg C, Faurie G. Survival in soil of genetically engineered Escherichia coli related to inoculum density, predation, and competition. FEMS Microbiol Ecol. 1992;101:251–260. [Google Scholar]
- 33.Romanowski G, Lorenz M G, Wackernagel W. Adsorption of plasmid DNA to mineral surfaces and protection against DNase I. Appl Environ Microbiol. 1991;57:1057–1061. doi: 10.1128/aem.57.4.1057-1061.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosado A S, Seldin L, Wolters A C, van Elsas J D. Quantitative 16S rDNA-targeted polymerase chain reaction and oligonucleotide hybridization for the detection of Paenibacillus azotofixans in soil and the wheat rhizosphere. FEMS Microbiol Ecol. 1996;19:153–164. [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Sands D C, Rovira A D. Isolation of fluorescent pseudomonads with a selective media. Appl Microbiol. 1970;20:513–514. doi: 10.1128/am.20.3.513-514.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schroth M N, Hancock J G. Disease-suppressing soil and root colonizing bacteria. Science. 1982;216:1376–1381. doi: 10.1126/science.216.4553.1376. [DOI] [PubMed] [Google Scholar]
- 38.Slininger P J, Jackson M. Nutritional factors regulating growth and accumulation of phenazine-1-carboxylic acid by Pseudomonas fluorescens 2-79. Appl Microbiol Biotechnol. 1992;37:388–392. [Google Scholar]
- 39.Thomashow L S, Weller D M. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J Bacteriol. 1988;170:3499–3508. doi: 10.1128/jb.170.8.3499-3508.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weller D M, Cook R J. Suppression of take-all of wheat by seed treatments with fluorescent pseudomonads. Phytopathology. 1983;73:463–469. [Google Scholar]
- 41.Weller D M. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu Rev Phytopathol. 1988;26:379–407. [Google Scholar]
- 42.Wood D W, Gong F, Daykin M M, Williams P, Pierson L S., III N-Acyl-homoserine lactone-mediated regulation of phenazine gene expression by Pseudomonas aureofaciens TX-1 30-84 in the wheat rhizosphere. J Bacteriol. 1997;179:7663–7670. doi: 10.1128/jb.179.24.7663-7670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]