Abstract
Genetic profiling techniques of microbial communities based on PCR-amplified signature genes, such as denaturing gradient gel electrophoresis or single-strand-conformation polymorphism (SSCP) analysis, are normally done with PCR products of less than 500-bp. The most common target for diversity analysis, the small-subunit rRNA genes, however, are larger, and thus, only partial sequences can be analyzed. Here, we compared the results obtained by PCR targeting different variable (V) regions (V2 and V3, V4 and V5, and V6 to V8) of the bacterial 16S rRNA gene with primers hybridizing to evolutionarily conserved flanking regions. SSCP analysis of single-stranded PCR products generated from 13 different bacterial species showed fewer bands with products containing V4-V5 (average, 1.7 bands per organism) than with V2-V3 (2.2 bands) and V6-V8 (2.3 bands). We found that the additional bands (>1 per organism) were caused by intraspecies operon heterogeneities or by more than one conformation of the same sequence. Community profiles, generated by PCR-SSCP from bacterial-cell consortia extracted from rhizospheres of field-grown maize (Zea mays), were analyzed by cloning and sequencing of the dominant bands. A total of 48 sequences could be attributed to 34 different strains from 10 taxonomical groups. Independent of the primer pairs, we found proteobacteria (α, β, and γ subgroups) and members of the genus Paenibacillus (low G+C gram-positive) to be the dominant organisms. Other groups, however, were only detected with single primer pairs. This study gives an example of how much the selection of different variable regions combined with different specificities of the flanking “universal” primers can affect a PCR-based microbial community analysis.
PCR-based methods have enormously affected our understanding of global microbial diversity because they have contributed to both the fast differentiation and identification of cultivated microorganisms and the access to the vast majority of microorganisms which have not yet been cultured in the laboratory. Studies by Woese and coworkers laid the groundwork, contributed to the characterization of noncultivated microorganisms, and placed them into a phylogenetic system based on the sequence analysis of their rRNAs (41). As a main target molecule for microbial ecology studies of diversity, the small-subunit (SSU) rRNA or the respective genes have been used. The levels of resolution of the DNA sequences differentiate microorganisms to approximately the species level (34). The SSU rRNA gene is composed of alternating evolutionarily conserved and variable regions. The conserved regions are ideal sites for primer binding in PCRs because it can be predicted that they will bind even to DNA of unknown organisms. The amplified products from environmental DNA which contain the variable regions can then be used for identification after cloning and sequencing of the genes. By compilation of sequence data, it can be shown that the SSU rRNA genes of all living organisms contain a total of nine variable regions, V1 to V9, scattered in the molecule, which, for bacteria, is approximately 1,520 nucleotides long (25).
An important objective in many ecological studies is to understand the natural variability of microbial communities, e.g., in response to environmental changes. In this context it is often desirable to analyze and compare a large number of samples. A useful approach to achieve this goal is the use of a genetic profiling technique. By this means, microbial community structures can be compared at the level of fingerprintlike patterns. For such purposes, techniques such as denaturing gradient gel electrophoresis (DGGE) (24), temperature gradient gel electrophoresis (10, 17), or terminal restriction fragment length polymorphism analysis (21) are often applied. A useful alternative method to separate PCR products of the same size but with different sequences is single-strand-conformation polymorphism (SSCP) (13, 20, 27). In our laboratory, we have optimized SSCP for the analysis of complex microbial communities by removal of one complementary strand of the double-stranded PCR product prior to SSCP on nondenaturing gels (30).
SSCP, DGGE, and temperature gradient gel electrophoresis have a high potential for community analysis because single bands or genetic profiles can be isolated and identified by DNA sequencing (11, 28). However, a limitation of these methods is the fact that only partial sequences of up to about 500 nucleotides are separated well. Most studies of microbial community diversity so far have been based on the analysis of only one selected region or the SSU rRNA gene containing one to three variable regions, e.g., the V3 (24), V1-to-V3 (8), V7 and V8 (11), V3-to-V5 (37), V4 and V5 (28, 30), or V6-to-V8 (9) region. In several of these studies, single bands were isolated from profiles and identified by DNA sequencing. However, we assumed that in studies based on just one PCR product the reliability of the selection of the variable region for the results of the community analysis might not be sufficient, and therefore this comparative study of the effects of the V regions and primer selection was initiated.
In order to amplify different variable regions from a large diversity of bacteria, we selected primers which were complementary to flanking conserved regions. We applied these “universal” primers to genomic DNA of bacterial pure cultures and to environmental DNA extracted from rhizospheres of field-grown maize (Zea mays). The diversity of amplified products was characterized by SSCP. For pure cultures, we determined the number of amplified and SSCP-distinguishable operons, and from community patterns we determined the identities of the dominant bands by DNA sequencing and comparison to known rRNA gene sequences.
MATERIALS AND METHODS
Bacteria, cultivation, and DNA extraction.
All bacterial pure cultures used in this study were obtained from the DSMZ (Deutsche Sammlung für Mikroorganismen und Zellkulturen, Braunschweig, Germany) and cultured at 28°C on media containing 1.5% (wt/vol) purified agar. Agrobacterium tumefaciens (DSM 30150), Pseudomonas putida (DSM 50906), Pseudomonas fluorescens (DSM 50090), Bacillus subtilis (DSM 4872) and Paenibacillus polymyxa (DSM 36) were cultivated on DSM M1; Escherichia coli (DSM 1607) and Azotobacter beijerinckii (DSM 282) were grown on Luria-Bertani broth (Difco, Detroit, Mich.); Arthrobacter oxydans (DSM 20119) was grown on DSM M53; Flavobacterium johnsonae (DSM 2064) was grown on DSM M67; Rhizobium trifolii (DSM 1980) and Acinetobacter calcoaceticus (DSM 586) were grown on R2A (Difco); Streptomyces viridochromogenes (DSM 40736) was grown on DSM M65. Bdellovibrio bacteriovorus (DSM 50701) was grown on cells of Pseudomonas putida, as recommended by the DSMZ.
Bacterial cells were separated from root material (rhizospheres) by washing 8 g (wet weight) of young root material of field-grown corn (Z. mays KX6345; Agrevo, Frankfurt, Germany) in 20 ml of sterile saline solution (0.85% [wt/vol] NaCl) for 30 min at 4°C in an orbital shaker (KH; Guwina-Hoffmann, Berlin, Germany) at 20 rpm. After removal of the root material, the cell suspensions were centrifuged at 4,100 × g for 30 min at 4°C. The supernatants were discarded, and the pellets were stored at −70°C.
For DNA extraction, cell material of pure-culture colonies grown on agar plates, or in the case of B. bacteriovorus, grown on P. putida cells, were transferred to 1.5-ml tubes (Eppendorf, Hamburg, Germany) and lysed in 50 μl of a sterile 0.05 M NaOH–0.25% (wt/vol) sodium dodecyl sulfate solution for 15 min at 95°C. After the suspension was diluted in 450 μl of distilled water, 2 μl of the solution was used as a template in the PCR.
DNA of bacterial cell consortia collected from rhizospheres was extracted using freeze-thaw cycles for lysis (12 ml of lysis solution per sample), proteinase K treatment, phenol chloroform extraction, and electroelution from agarose gels for the final DNA purification (30, 31).
PCR amplifications of rRNA gene sequences.
PCR was performed with the thermal cycler Primus 96 (MWG-Biotech, Ebersberg, Germany). Amplifications from cultivated pure cultures were conducted in a final volume of 50 μl, whereas DNA fragments from environmental-DNA solutions were processed in a final volume of 100 μl. For pure cultures, the PCR included 1.25 U of Taq polymerase (Amersham Pharmacia Biotech, Freiburg, Germany), 1× PCR buffer with 1.5 mM MgCl2, 0.5 μM primers, and each dNTP at 200 μM (Amersham Pharmacia Biotech). For environmental samples, the composition was slightly different: 3.75 U of Expand-Taq HF (Roche Diagnostics, Mannheim, Germany), 5 μg of T4 gene 32 protein ml−1 (Roche Diagnostics) (36), and a final concentration of 2 mM MgCl2. Three primer pairs were chosen for amplification of partial sequences of the 16S rRNA gene. For amplification of the V2-V3 region, the forward primer was (5′-3′) ACT GGC GGA CGG GTG AGT AA, and the reverse primer was (5′-3′) CGT ATT ACC GCG GCT GCT GG. For the amplification of the V4-V5 region, we used primers Com1 and Com2-Ph, described previously (30), and for the V6-V8 region, we used primers f968 and r1346 (without a GC clamp) (26). The binding sites of these primers according to the positions in the E. coli SSU rRNA genes (5) are shown in Table 1. The primers were synthesized by MWG Biotech or TIBmolbiol (Berlin, Germany). The reverse primers were phosphorylated. Environmental DNA and pure-culture DNA were amplified under the same conditions: 95°C for 3 min, followed by 35 cycles of 1 min at 95°C, 50°C for 1 min, 72°C for 70 s, and finally 72°C for 5 min.
TABLE 1.
Specificities of primers used in this study for PCR amplifications of different variable regions of the SSU rRNA genes
| Domain (no. of sequences in database) | No. of primer binding sites allowing two mismatches (% of sequences in respective databases)a
|
|||||
|---|---|---|---|---|---|---|
| Primer pair 1 targeting V2 and V3
|
Primer pair 2 targeting V4 and V5
|
Primer pair 3 targeting V6, V7, and V8
|
||||
| 101–120b | 518–537 | 519–536 | 907–926 | 968–984 | 1330–1346 | |
| Eubacteria (10,145) | 5,771 (56.9) | 5,571 (54.9) | 7,771 (76.6) | 7,715 (76.0) | 7,757 (76.5) | 7,546 (74.4) |
| Archaea (503) | 0 (0.0) | 101 (20.1) | 273 (54.3) | 272 (54.1) | 0 (0.0) | 0 (0.0) |
| Eucarya (3,227) | 0 (0.0) | 2,339 (72.5) | 2,523 (78.2) | 2,618 (81.1) | 0 (0.0) | 0 (0.0) |
Regions are numbered according to reference 25.
E. coli positions.
Generation of genetic profiles by SSCP.
SSCP analysis was conducted according to the single-strand community approach described previously (30). After purification of the double-stranded PCR products with Qiaquick (Qiagen, Hilden, Germany), the phosphorylated strands were removed by lambda exonuclease (Amersham Pharmacia Biotech) digestions (5 U for pure-culture PCR products and 10 U for community products) at 37°C for 2 h. Proteins were removed by phenol-chloroform extraction, and the DNA was precipitated as described by Sambrook et al. (29) and resuspended in a solution consisting of 8 μl of 10 mM Tris-HCl (pH 8.0) and 8 μl of denaturing loading buffer (95% [vol/vol] formamide, 10 mM NaOH, 0.025% [wt/vol] each of bromphenol blue and xylene cyanole). Samples were incubated for 2 min at 95°C and then immediately cooled on ice.
To generate SSCP, we used the gel matrix (MDE; FMC Bioproducts, Rockland, Maine) in final concentrations of 0.7-fold for products including the V6-to-V8 region and 0.6-fold for products generated with the other two primer pairs. The stock solution was twofold. The electrophoreses were carried out in a Macrophor sequencing apparatus (Amersham Pharmacia Biotech) with gels of 21 cm under conditions described before (31), except that the PCR products with the V6-to-V8 region were run at 26 instead of 20°C. Afterward, the gels were run and DNA was stained according to the silver-staining procedure described by Bassam et al. (1).
Identification of SSCP bands.
The procedures to isolate single bands from SSCP profiles for DNA sequencing were carried out as described previously (28). Gel-extracted DNA was used as a template in PCR. The PCR was conducted under the same conditions as the pure-culture amplifications. The identities of the reamplified products were confirmed in another SSCP analysis using the community profiles as references.
The reamplified products were then ligated into a pGEM-T vector and transformed into E. coli JM109 (Promega, Mannheim, Germany) according to a protocol of the supplier. Transformed cells with inserts were selected by blue-white screening. Cloned DNA fragments were amplified from the vector by PCR (with conditions as recommended by the manufacturer) using primers matching the flanking regions of the vector (forward, [5′-3′] CAC GAC GTT GTA AAA CGA C, and reverse, [5′-3′] GGA TAA CAA TTT CAC ACA GG). The sizes of the PCR products were determined by agarose gel electrophoresis (1.25% [wt/vol] agarose) (29). Inserts of the expected size were then sequenced by cycle sequencing, using the SequiTherm Excel II sequencing kit (Epicenter Technologies, Madison, Wis.). The primers for sequencing were m13f, (5′-3′) TGT AAA ACG ACG GCC AGT, and m13r, (5′-3′) CAG GAA ACA GCT ATG AC, both infrared dye 800 labeled. Conditions for the cycle-sequencing process have been described previously (28). The sequences were automatically analyzed on a 6% (wt/vol) polyacrylamide gel (Rapid Gel XL; Amersham Pharmacia Biotech) using a LI-COR DNA 4200 GeneRead IR apparatus (LI-COR, Lincoln, Neb.). The sequences were edited and aligned with the AlignIR 1.1 program (LI-COR) and, for phylogenetic analysis and identification of related sequences, loaded into the arb program and database (http://www.arb-home.de). All sequences generated in this study were consensus sequences.
Nucleotide sequence accession numbers.
The sequences generated in this study can be found in GenBank (http://www.ncbi.nlm.nih.gov) (2) under accession numbers AJ311396 to AJ311442.
RESULTS AND DISCUSSION
Primer selection and specificities.
Primer pairs were chosen which amplified products from different regions of the bacterial SSU rRNA gene by PCR. The predicted lengths of the PCR products were similar: 436 bp for primers amplifying the variable regions V2-V3, 408 bp for those amplifying V4-V5, and 378 bp for primers amplifying regions containing V6, V7, and V8. For the amplification of products containing V4-V5, we chose primers which had already been applied for PCR-SSCP analysis of microbial communities from rhizospheres (30), compost (28), and, slightly different, also for PCR-DGGE microbial community analysis of soil samples (15, 16). PCR products with V6-to-V8 regions were generated with primers broadly used for microbial community analysis by PCR-DGGE (23) but, to our knowledge, not yet tested for PCR-SSCP. The third amplified rRNA gene region was generated with primers bordering the V2-V3 region. The reverse primer had already been used in earlier studies by Muyzer et al. (24). This sequence was actually almost the complementary sequence to the forward primer used to amplify the V4-V5 region in our study. The forward primer to amplify the V2-V3 region was selected from primers used for sequencing eubacterial SSU rRNA genes (E .R. B. Moore, personal communication). A shorter partial sequence including the V3 region had been targeted with universal primers in PCR-SSCP studies of bacterial communities in bioreactors (42). In a recent DGGE study, PCR products containing V1-to-V3 regions were found useful to detect the effects of herbicides on soil microbial communities (8).
The specificities of primers selected for our study were analyzed with the arb program package (see Materials and Methods) and the most recent database release available (December 1998). In order to estimate the binding specificities for the amplification of unknown sequences from environmental DNA, we determined the proportion of organisms in the database which would hybridize to the selected primers. Due to the annealing temperature, we estimated that two mismatches would still result in product formation. Under these circumstances, we found that the forward primer used in the amplification of the V2-V3 region would match with 57% of the database sequences for the domain Eubacteria and were highly specific for that group (Table 1). The reverse primer matched with 55% of the eubacterial sequences but was less specific, since it potentially also hybridized to sequences of the domains Archaea and Eucarya. In combination with the forward primer, the PCR products, however, should be specifically generated from members of the domain Eubacteria. This was not the case for primers selected to amplify the V4-V5 region, since matches of over 75% were recorded for Eubacteria and Eucarya and more than 50% for Archaea. The primers which were chosen for amplifying the V6-to-V8 regions, in contrast, were highly specific for Eubacteria. Both primers matched with more than 70% of the sequences in the Eubacteria database but with none in the Archaea or Eucarya sequences.
In addition to testing the specificity of each primer separately, we analyzed the compatibility of each primer pair shown in Table 1 and found that at least 89% of the products which would be amplified by one primer contained complementary sequences which would bind to the opposite primer (data not shown). Cases in which no match with the opposite primers were found were often caused by the presence of partial sequences or sequences with gaps of unidentified nucleotides in the databases.
The result of this database analysis demonstrated that the “universal” primers in our study were not perfectly universal. It is known that at the domain level conserved regions show some degree of variability (22). It has been shown that a slight modification of a primer binding site even within one conserved region can result in big differences in 16S rRNA gene sequences amplified from environmental samples (40).
PCR-SSCP analysis of bacterial pure cultures.
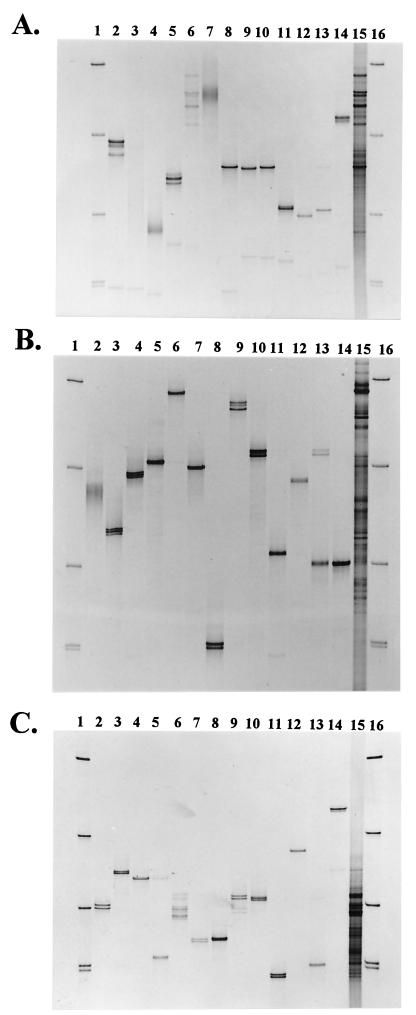
We selected a total of 13 species from different phylogenetic groups. Figure 1 shows typical SSCP gels obtained with the three different primer pairs for all selected bacteria. The majority of bacteria showed more than one product. In some cases, the products of a single species could have the same intensity on the gel, e.g., as shown for S. viridochromogenes, A. tumefaciens, and P. putida (Fig. 1B, lanes 4, 8, and 10, respectively). In other cases, one band was dominant and additional bands occurred in lower quantities, e.g., as found for E. coli and B. subtilis (Fig. 1A, lanes 2 and 5).
FIG. 1.
SSCP analyses of PCR-amplified 16S rRNA gene sequences of pure-culture bacteria (lanes 2 to 14) and a bacterial cell consortium extracted from a maize rhizosphere (lane 15). PCR amplifications were conducted with universal primers to generate products which included different variable regions, i.e., the V2-V3 regions (A), the V4-V5 regions (B), and the V6, V7, and V8 regions (C). The following pure-culture bacteria were included in this analysis: lanes 2, E. coli; lanes 3, A. oxydans; lanes 4, S. viridochromogenes; lanes 5, B. subtilis; lanes 6, P. polymyxa; lanes 7, R. trifolii; lanes 8, A. tumefaciens; lanes 9, P. fluorescens; lanes 10, P. putida; lanes 11, A. beijerinckii; lanes 12, A. calcoaceticus; lanes 13, B. bacteriovorus; and lanes 14, F. johnsonae. SSCP standards are shown in lanes 1 and 16. These standards consisted of products generated with the same primers used for the products in panel B. The standard included (from top to bottom) Bacillus licheniformis, R. trifolii, F. johnsonae, and A. tumefaciens (the double band is due to two operons).
A possible reason for the formation of more than one product from a pure culture is that the universal primers amplified more than one operon. It is well known that several bacterial species contain more than one 16S rRNA gene in their genomes (for a review, see reference 12). Thus, PCR amplifications based on universal primers may generate more than one product even from pure-culture DNA if the sequence divergence is present in the selected variable regions. Another reason for detecting more than one fragment from pure cultures by PCR-SSCP was the formation of metastable conformers, i.e., where the same molecule folds into more than one conformation with different electrophoretic mobilities (7). In addition, some weak bands were also caused by incomplete exonuclease digestion of the noncoding strand. Such cases could be easily identified, because those bands did not consistently occur in replicate gels and the position could be checked by comparison with SSCP products without exonuclease treatment (data not shown).
In order to determine whether operon heterogeneities or metastable conformers caused the additional bands, we conducted a test. Each band of a pure-culture profile with more than one band was separately isolated and then subjected to PCR with the same primers as in the first amplification. In cases where only one product was formed at the original position in the gel, we concluded that the original band was a distinct operon. On the other hand, if the PCR of a band resulted in the regeneration of the additional species-specific products, we concluded that we had amplified a metastable conformer. The results of this differentiation are summarized in Table 2. The intensities of bands generated from pure-culture amplifications were no indication whether additional bands were caused by metastable conformers or different sequences. This means that even from pure cultures with more than one operon, PCR amplifications of the single operons were biased. Such effects hamper quantitative interpretation of community profiles such as SSCP or DGGE.
TABLE 2.
Distribution of PCR products from bacterial pure cultures, detected as bands on SSCP gels and differentiated according to sequence heterogeneities and conformational isomers (combined results of three replicate gels for each primer pair)
| Pure culture | No. of bands detected
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Primer pair 1 (including V2 and V3)
|
Primer pair 2 (including V4 and V5)
|
Primer pair 3 (including V6 to V8)
|
|||||||
| Total bands | Operons | Metastable conformersa | Total bands | Operons | Metastable conformers | Total bands | Operons | Metastable conformers | |
| B. subtilis | 3 | 3 | 1 | 1 | − | − | 3 | ND | ND |
| P. polymyxa | 8 | 7 | 2 | 1 | − | − | 7 | 6 | 2 |
| A. oxydans | 1 | −b | − | 3 | 1 | 3 | 2 | 1 | 2 |
| S. viridochromogenes | 1 | − | − | 3 | 1 | 3 | 1 | − | − |
| R. trifolii | 1 | − | − | 1 | − | − | 2 | 1 | 2 |
| A. tumefaciens | 1 | − | − | 2 | 2 | 1 | 1 | − | − |
| A. beijerinckii | 2 | 2 | 1 | 1 | − | − | 2 | 1 | 2 |
| A. calcoaceticus | 1 | − | − | 1 | − | − | 1 | − | − |
| E. coli | 5 | 4 | 2 | 1 | − | − | 2 | 1 | 2 |
| P. fluorescens | 1 | − | − | 3 | 1 | 3 | 4 | 1 | 4 |
| P. putida | 1 | − | − | 3 | 1 | 3 | 2 | 1 | 2 |
| B. bacteriovorus | 1 | − | − | 1 | − | − | 1 | − | − |
| F. johnsonae | 2 | 2 | 1 | 1 | − | − | 2 | 2 | 1 |
| Average | 2.2 | 1.7 | 2.3 | ||||||
Number of conformations of the same molecule, as detected by SSCP.
Not tested because there was only one band in the profile.
ND, not determined.
The information about the actual copy numbers of 16S rRNA genes of all species included in our study is still limited, but in agreement with the literature, we detected more than one operon for E. coli, B. subtilis, and P. polymyxa (4, 19, 26). In E. coli, there are seven different SSU rRNA genes (4, 5). We conducted sequence alignments of these operons and found that four alternative sequence variations occurred in the V2 region and fewer in other regions, e.g., two in V6. All sequence variations of the V2 region could, in fact, be detected by SSCP analysis, but the two alternative variations in V6 could not be seen. In the latter case, we found two bands, but those were caused by metastable conformers. It is known that due to the limited resolution capacity of SSCP for larger fragments, not all base substitutions may be detectable as separate products (14, 33), and thus, not all operon heterogeneities may show up on a gel.
The pure-culture studies suggest that products with V4 and V5 might be more useful for analyses of more complex natural microbial communities, since they have fewer operon heterogeneities of the same species and thus come closer to the ratio of “one product, one species” which we think is ideal for analysis of complex microbial communities (30). However, the number of analyzed pure cultures in this study (n = 13) was too low to conclusively prove this hypothesis. In addition, for different phylogenetic branches of the eubacterial “tree,” operon heterogeneities may not be homogeneously distributed, and therefore, for specific genetic community profiles, other variable regions might be as suitable as or better than the V4-V5 region.
Dissection of SSCP patterns generated from bacterial-cell consortia extracted from rhizospheres.
PCR amplifications using community DNA as a template generated patterns of similar complexity for all primer pairs tested (Fig. 1, lanes 15). In order to understand what had been amplified from the community with the different primer sets, we tried to isolate all dominant single bands from each profile. The results of this analysis are shown in Table 3. Identifications of the products amplified with the first primer set (V2-V3) showed the dominance of P. polymyxa sequences. P. polymyxa is known to be a colonizer of maize rhizospheres (32, 39). The sequences detected in these profiles were all different from each other except for the two sequences with 100% similarity to P. polymyxa X60623. Thus, the majority of products were caused by different operons and not by metastable conformers. This is in accordance with the pure-culture analyses with this organism shown in Table 2. The “species” richness detected with the other PCR products (V4-V5 and V6-V8) was higher than that detected with the products containing V2-V3. The V6-V8-targeted PCR generated more different sequences belonging to the group of gram-positive bacteria with a low-G+C DNA content and in the α subgroup of Proteobacteria. Members of the Cytophaga-Flavobacterium-Bacteroides (CFB) group were only detected with PCR products containing V4-V5. This could be explained by different primer specificities: in databases, a higher number of homologous sequences was found for the CFB group (85%, including two mismatches) with the V4-V5-targeting primers than with the other two primer pairs (42% for V2-V3 and 56% for V6-V8 [data not shown]). Even a sequence of a member of the kingdom Crenarchaeota was found within the V4-V5 profile. This also could be explained by looking at the primer specificities (Table 1). Crenarchaeota-related sequences have also been found in other studies based on cultivation-independent PCR-based methods with soil DNA (3, 6, 18, 38).
TABLE 3.
Distribution of sequences obtained from community profiles (PCR-SSCP) generated from the same cell consortia extracted from maize rhizospheres
| Taxonomic group | Characteristics of variable regions included in PCR products
|
|||||
|---|---|---|---|---|---|---|
| V2 and V3
|
V4 and V5
|
V6, V7, and V8
|
||||
| Closest relative | Similarity (%) | Closest relative | Similarity (%) | Closest relative | Similarity (%) | |
| Low-G+C gram-positive | P. polymyxaX60623 | 100.0 | P. polymyxaX60632, D16276 | 99.7 | P. polymyxaD16276 | 99.1 |
| P. polymyxaX60623 | 100.0 | Bacillus coagulansX60614 | 100.0 | P. polymyxaD16276 | 99.1 | |
| P. polymyxaX60623 | 99.7 | Paenibacillus azotofixansD78318 | 99.4 | |||
| P. polymyxaX60623 | 99.7 | P. azotofixansD78318 | 99.1 | |||
| P. polymyxaX60623 | 98.5 | P. azotofixansD78318 | 98.8 | |||
| P. polymyxaX60623 | 98.5 | |||||
| α Subdivision of Proteobacteria | Mesorhizobium sp. strain AF041445 | 99.4 | Mesorhizobium thianenseAF041447 | 99.7 | M. thianenseAF041447 | 93.1 |
| Mesorhizobium plurifariumY14158 | 94.9 | Sphingomonas capsulata D6147 | 88.6 | Sphingomonas asaccharolyticaY09639 | 98.8 | |
| Agrobacterium sp. strain AB006037 | 96.9 | Devosia riboflavinaD49423 | 96.4 | D. riboflavinaD49423 | 88.4 | |
| Uncultured eubacterium AJ232882 | 99.1 | D. riboflavinaD49423 | 96.4 | Rhizobium suberifaciensD12641 | 95.0 | |
| Uncultured eubacterium AJ232882 | 90.9 | Rhizobium sp. strain Y10176 | 100.0 | |||
| Soil clone AF145805 | 93.6 | Bartonella taylorii Z311350 | 91.3 | |||
| Blastobacter capsulatusX73042 | 92.6 | |||||
| Mycoplana dimorphaD12786 | 87.5 | |||||
| β Subdivision of Proteobacteria | Acidovorax sp. strain AF078767 | 89.1 | Aquabacterium sp. strain AF035054 | 99.4 | Burkholderia sp. strain AF074711 | 88.8 |
| γ Subdivision of Proteobacteria | Pseudomonas marginalisZ76663 | 90.8 | Diazotrophic eubacterium AF094766 | 98.0 | Pseudomonas sp. strain AF0587 | 98.5 |
| Pseudomonas sp. strain AJ002801 | 98.5 | Pseudomonas aureofaciensZ76656 | 98.8 | |||
| Pseudomonas mandeliiAF058286 | 99.1 | |||||
| CFB group | Cytophaga hutchinsoniiM58768 | 93.2 | ||||
| Flavobacterium ferrugineumM28237 | 94.1 | |||||
| Verrucomicrobium | Prosthecobacter fusiformisU60015 | 88.6 | Verrumicrobium spinosumX90515 | 93.0 | ||
| Candidate division OP11 | Soil clone AF145815 | 90.5 | ||||
| Holophaga | Eubacterium Z95710 | 97.6 | ||||
| Green nonsulfur bacteria | Uncultured eubacterium AF005747 | 78.1 | ||||
| Crenarchaeota | Unidentified archeon U62814 | 100.0 | ||||
Independent of the primer pairs used in our study, we identified in the rhizosphere of maize proteobacteria (α and γ subgroups) and low-G+C gram-positive bacteria as the dominant eubacteria. Additionally, all three primer sets amplified one sequence of proteobacteria from the β subgroup. The PCR-amplified sequences within each of these phylogenetic groups led to different identifications of closest relatives, except for P. polymyxa. It may be argued that the number of bands which were sequenced in our study from each community profile was too low to expect an overlap between the identifications. This would be true if the number of bands were much higher than those which were actually sequenced. However, in our study we identified most bands which were detectable in the profiles. A serious limitation for identifications at the species level, however, is the use of partial sequences. It is known that the reliability of relating an unknown sequence to known sequences and, thus, of identifying it increases with the length and the number of variable regions of the PCR-amplified product (35). The diversity of closest relatives detected in our study may thus not only be a result of sequences amplified from different organisms but also of the use of partial sequences for comparison to database sequences.
It is interesting that all bands which were isolated from community SSCP profiles exhibited different DNA sequences except for one case (P. polymyxa with V2-V3). Thus, in contrast to our pure-culture results reported above, the formation of metastable conformers was of only minor importance and did not contribute to the pattern complexity of the community profiles. These results are corroborated by earlier PCR-SSCP studies which we conducted of the diversity of eubacterial, actinomycetes, and fungal communities in composts (28). Our analysis underlines the high potential of the community PCR-SSCP approach (30) as an alternative to more commonly used profiling techniques for microbial community analyses.
Conclusions.
Our study shows that intraspecies operon heterogeneities can contribute significantly to complex genetic profiles in microbial community analysis. In studies based only on profile comparisons and not on sequencing, this effect may be misinterpreted as a high microbial diversity, and dramatic pattern changes may be an effect of the reduction of only one or two organisms. The effect of operon heterogeneities can be reduced by choosing appropriate variable regions with less intraspecies diversity, e.g., V4 and V5 in our study. We also point out that even primers which bind to evolutionarily conserved regions of the SSU rRNA gene are never 100% universal at the domain level. Therefore, biases with different universal primers are inevitable and will contribute, in addition to the choice of variable regions, to the diversity found in genetic profiles in microbial community analyses.
ACKNOWLEDGMENTS
We thank Karin Trescher for her excellent technical assistance. We also thank Sabine Peters, Ingo Fritz, and Erko Stackebrandt for discussions.
This work was supported by a grant from the German Ministry for Education and Research (BMBF; grant no. 0311740).
REFERENCES
- 1.Bassam B J, Caetano-Anolles G, Gresshoff P M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem. 1991;196:80–83. doi: 10.1016/0003-2697(91)90120-i. [DOI] [PubMed] [Google Scholar]
- 2.Benson D A, Karsch-Mizrachi I, Lipman D J, Ostell J, Rapp B A, Wheeler D L. GenBank. Nucleic Acids Res. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bintrim S B, Donohue T J, Handelsman J, Roberts G P, Goodman R M. Molecular phylogeny of Archaea from soil. Proc Natl Acad Sci USA. 1997;94:277–282. doi: 10.1073/pnas.94.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 6.Buckley D H, Graber J R, Schmidt T M. Phylogenetic analysis of nonthermophilic members of the kingdom Crenarchaeota and their diversity and abundance in soils. Appl Environ Microbiol. 1998;64:4333–4339. doi: 10.1128/aem.64.11.4333-4339.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clapp J P. The identification of root-associated fungi by polymerase chain reaction-single-strand conformational polymorphism (PCR-SSCP) In: Akkermans A D L, Van Elsas J D, De Bruijn F J, editors. Molecular microbial ecology manual. 3.4.7. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 1–18. [Google Scholar]
- 8.El Fantroussi S, Verschuere L, Verstraete W, Top E M. Effect of phenylurea herbicides on soil microbial communities estimated by analysis of 16S rRNA gene fingerprints and community-level physiological profiles. Appl Environ Microbiol. 1999;65:982–988. doi: 10.1128/aem.65.3.982-988.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felske A, Engelen B, Nübel U, Backhaus H. Direct ribosome isolation from soil to extract bacterial rRNA for community analysis. Appl Environ Microbiol. 1996;62:4162–4167. doi: 10.1128/aem.62.11.4162-4167.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felske A, Rheims H, Wolterink A, Stackebrandt E, Akkermans A D. Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology. 1997;143:2983–2989. doi: 10.1099/00221287-143-9-2983. [DOI] [PubMed] [Google Scholar]
- 11.Ferris M J, Muyzer G, Ward D M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fogel G B, Collins C R, Li J, Brunk C F. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb Ecol. 1999;38:93–113. doi: 10.1007/s002489900162. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi K. PCR-SSCP: a simple and sensitive method for detection of mutations in the genomic DNA. PCR Methods Appl. 1991;1:34–38. doi: 10.1101/gr.1.1.34. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi K, Yandell D W. How sensitive is PCR-SSCP? Hum Mutat. 1993;2:338–346. doi: 10.1002/humu.1380020503. [DOI] [PubMed] [Google Scholar]
- 15.Henckel T, Friedrich M, Conrad R. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl Environ Microbiol. 1999;65:1980–1990. doi: 10.1128/aem.65.5.1980-1990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henckel T, Jackel U, Schnell S, Conrad R. Molecular analyses of novel methanotrophic communities in forest soil that oxidize atmospheric methane. Appl Environ Microbiol. 2000;66:1801–1808. doi: 10.1128/aem.66.5.1801-1808.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heuer H, Krsek M, Baker P, Smalla K, Wellington E M H. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol. 1997;63:3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jurgens G, Lindström K, Saano A. Novel group within the kingdom Crenarchaeota from boreal forest soil. Appl Environ Microbiol. 1997;63:803–805. doi: 10.1128/aem.63.2.803-805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 20.Lee D-H, Zu Y-G, Kim S-J. Nonradioactive method to study genetic profiles of natural bacterial communities by PCR-single strand conformation polymorphism. Appl Environ Microbiol. 1996;62:3112–3120. doi: 10.1128/aem.62.9.3112-3120.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu W T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchesi J R, Sato T, Weightman A J, Martin T A, Fry J C, Hiom S J, Dymock D, Wade W G. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. 1998;64:795–799. doi: 10.1128/aem.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Wawer C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans A D L, Van Elsas J D, De Bruijn F J, editors. Molecular microbial ecology manual. 3.4.4. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 1–27. [Google Scholar]
- 24.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neefs J-M, Van der Peer Y, De Rejk P, Chapelle S, De Wachter R. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 1993;21:3025–3049. doi: 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann R I, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci USA. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peters S, Koschinsky S, Schwieger F, Tebbe C C. Succession of microbial communities during hot composting as detected by PCR–single-strand-conformation polymorphism-based genetic profiles of small-subunit rRNA genes. Appl Environ Microbiol. 2000;66:930–936. doi: 10.1128/aem.66.3.930-936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J E, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Schwieger F, Tebbe C C. A new approach to utilize PCR–single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl Environ Microbiol. 1998;64:4870–4876. doi: 10.1128/aem.64.12.4870-4876.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwieger F, Tebbe C C. Effect of field inoculation with Sinorhizobium meliloti L33 on the composition of bacterial communities in rhizospheres of a target plant (Medicago sativa) and a non-target plant (Chenopodium album). Linking of 16S rRNA gene-based single-strand conformation polymorphism community profiles to the diversity of cultivated bacteria. Appl Environ Microbiol. 2000;66:3556–3565. doi: 10.1128/aem.66.8.3556-3565.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seldin L, Rosado A S, da Cruz D W, Nobrega A, van Elsas J D, Paiva E. Comparison of Paenibacillus azotofixans strains isolated from rhizoplane, rhizosphere, and non-root-associated soil from maize planted in two different Brazilian soils. Appl Environ Microbiol. 1998;64:3860–3868. doi: 10.1128/aem.64.10.3860-3868.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheffield V C, Beck J S, Kwitek A E, Sandstrom D W, Stone E M. The sensitivity of single-strand conformation polymorphism analysis for the detection of single base substitutions. Genomics. 1993;16:325–332. doi: 10.1006/geno.1993.1193. [DOI] [PubMed] [Google Scholar]
- 34.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 35.Stackebrandt E, Rainey F A. Partial and complete 16S rDNA sequences, their use in generation of 16S rDNA phylogenetic trees and their implications in molecular ecology studies. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. 3.1.1. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–17. [Google Scholar]
- 36.Tebbe C C, Vahjen W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl Environ Microbiol. 1993;59:2657–2665. doi: 10.1128/aem.59.8.2657-2665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teske A, Wawer C, Muyzer G, Ramsing N B. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl Environ Microbiol. 1996;62:1405–1415. doi: 10.1128/aem.62.4.1405-1415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ueda T, Suga Y, Matsuguchi T. Molecular phylogenetic analysis of a soil microbial community in a soybean field. Eur J Soil Sci. 1995;46:415–421. [Google Scholar]
- 39.von der Weid I, Paiva E, Nobrega A, van Elsas J D, Seldin L. Diversity of Paenibacillus polymyxa strains isolated from the rhizosphere of maize planted in Cerrado soil. Res Microbiol. 2000;151:369–381. doi: 10.1016/s0923-2508(00)00160-1. [DOI] [PubMed] [Google Scholar]
- 40.Ward N, Rainey F A, Goebel B, Stackebrandt E. Identifying and culturing the ‘unculturables’: a challenge for microbiologists. In: Allsopp D, Colwell R R, Hawksworth D L, editors. Microbial diversity and ecosystem function: proceedings of the IUBS IUMS Workshop held at Egham, UK, 10–13 August 1993 in support of the IUBS UNESCO SCOPE “DIVERSITAS” programme. Wallingford, Oxon, United Kingdom: CAB International in association with United Nations Environment Programme; 1995. pp. 89–110. [Google Scholar]
- 41.Woese C R, Kandler R O, Wheelis M L. Towards a natural systems of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zumstein E, Moletta R, Godon J-J. Examination of two years of community dynamics in an anaerobic bioreactor using fluorescence polymerase chain reaction (PCR) single-strand conformation polymorphism analysis. Environ Microbiol. 2000;2:69–78. doi: 10.1046/j.1462-2920.2000.00072.x. [DOI] [PubMed] [Google Scholar]