Abstract
Background
Accurate rapid diagnostic tests for SARS‐CoV‐2 infection would be a useful tool to help manage the COVID‐19 pandemic. Testing strategies that use rapid antigen tests to detect current infection have the potential to increase access to testing, speed detection of infection, and inform clinical and public health management decisions to reduce transmission. This is the second update of this review, which was first published in 2020.
Objectives
To assess the diagnostic accuracy of rapid, point‐of‐care antigen tests for diagnosis of SARS‐CoV‐2 infection. We consider accuracy separately in symptomatic and asymptomatic population groups. Sources of heterogeneity investigated included setting and indication for testing, assay format, sample site, viral load, age, timing of test, and study design.
Search methods
We searched the COVID‐19 Open Access Project living evidence database from the University of Bern (which includes daily updates from PubMed and Embase and preprints from medRxiv and bioRxiv) on 08 March 2021. We included independent evaluations from national reference laboratories, FIND and the Diagnostics Global Health website. We did not apply language restrictions.
Selection criteria
We included studies of people with either suspected SARS‐CoV‐2 infection, known SARS‐CoV‐2 infection or known absence of infection, or those who were being screened for infection. We included test accuracy studies of any design that evaluated commercially produced, rapid antigen tests. We included evaluations of single applications of a test (one test result reported per person) and evaluations of serial testing (repeated antigen testing over time). Reference standards for presence or absence of infection were any laboratory‐based molecular test (primarily reverse transcription polymerase chain reaction (RT‐PCR)) or pre‐pandemic respiratory sample.
Data collection and analysis
We used standard screening procedures with three people. Two people independently carried out quality assessment (using the QUADAS‐2 tool) and extracted study results. Other study characteristics were extracted by one review author and checked by a second. We present sensitivity and specificity with 95% confidence intervals (CIs) for each test, and pooled data using the bivariate model. We investigated heterogeneity by including indicator variables in the random‐effects logistic regression models. We tabulated results by test manufacturer and compliance with manufacturer instructions for use and according to symptom status.
Main results
We included 155 study cohorts (described in 166 study reports, with 24 as preprints). The main results relate to 152 evaluations of single test applications including 100,462 unique samples (16,822 with confirmed SARS‐CoV‐2). Studies were mainly conducted in Europe (101/152, 66%), and evaluated 49 different commercial antigen assays. Only 23 studies compared two or more brands of test.
Risk of bias was high because of participant selection (40, 26%); interpretation of the index test (6, 4%); weaknesses in the reference standard for absence of infection (119, 78%); and participant flow and timing 41 (27%). Characteristics of participants (45, 30%) and index test delivery (47, 31%) differed from the way in which and in whom the test was intended to be used. Nearly all studies (91%) used a single RT‐PCR result to define presence or absence of infection.
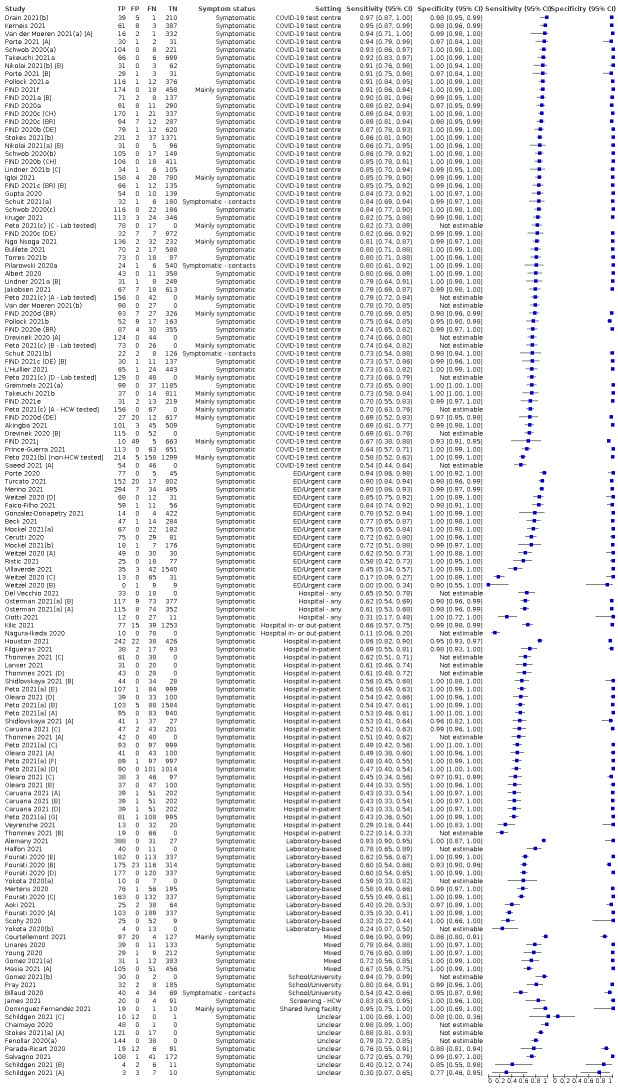
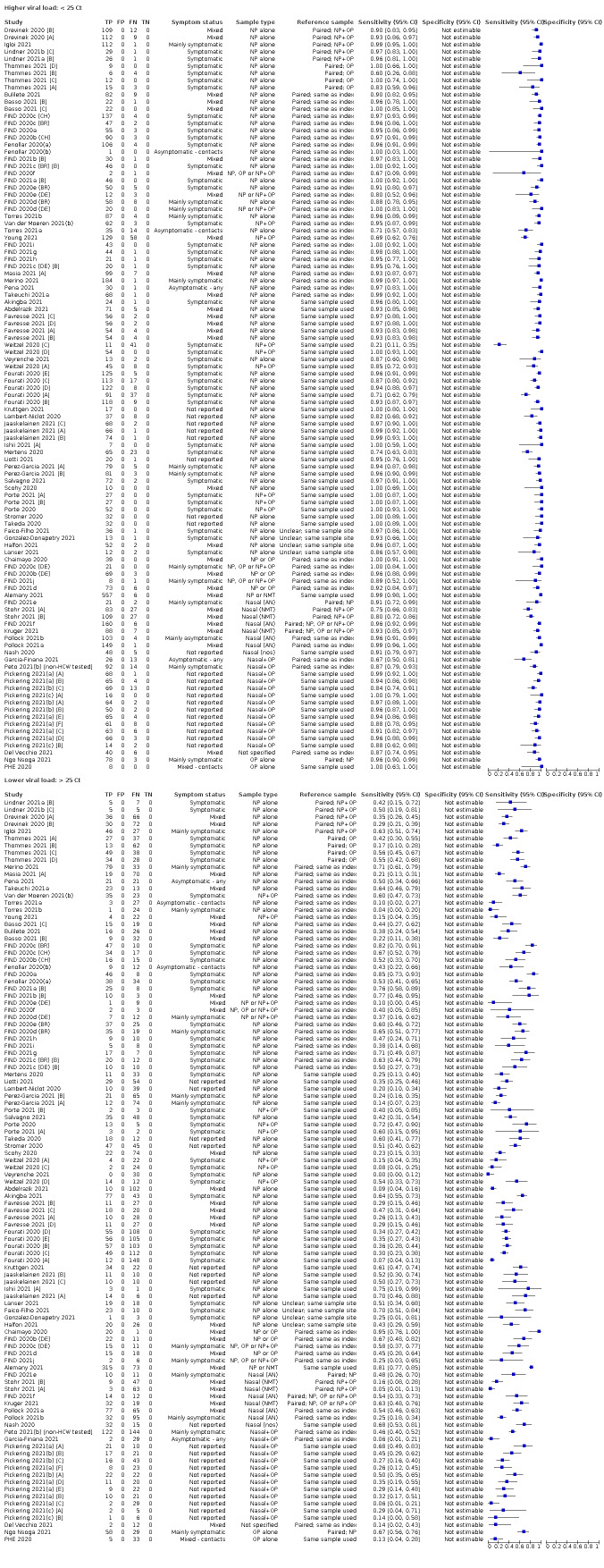
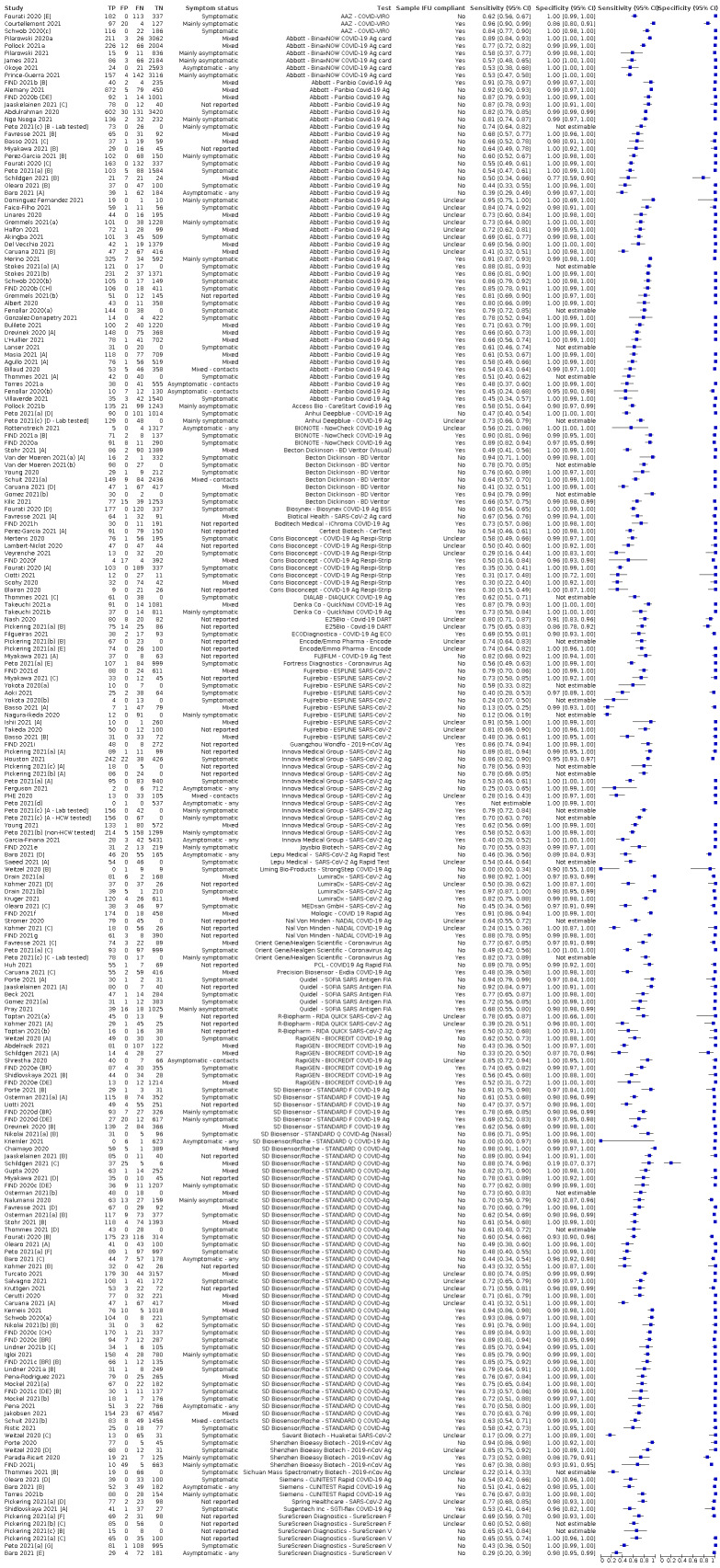
The 152 studies of single test applications reported 228 evaluations of antigen tests. Estimates of sensitivity varied considerably between studies, with consistently high specificities. Average sensitivity was higher in symptomatic (73.0%, 95% CI 69.3% to 76.4%; 109 evaluations; 50,574 samples, 11,662 cases) compared to asymptomatic participants (54.7%, 95% CI 47.7% to 61.6%; 50 evaluations; 40,956 samples, 2641 cases). Average sensitivity was higher in the first week after symptom onset (80.9%, 95% CI 76.9% to 84.4%; 30 evaluations, 2408 cases) than in the second week of symptoms (53.8%, 95% CI 48.0% to 59.6%; 40 evaluations, 1119 cases). For those who were asymptomatic at the time of testing, sensitivity was higher when an epidemiological exposure to SARS‐CoV‐2 was suspected (64.3%, 95% CI 54.6% to 73.0%; 16 evaluations; 7677 samples, 703 cases) compared to where COVID‐19 testing was reported to be widely available to anyone on presentation for testing (49.6%, 95% CI 42.1% to 57.1%; 26 evaluations; 31,904 samples, 1758 cases). Average specificity was similarly high for symptomatic (99.1%) or asymptomatic (99.7%) participants.
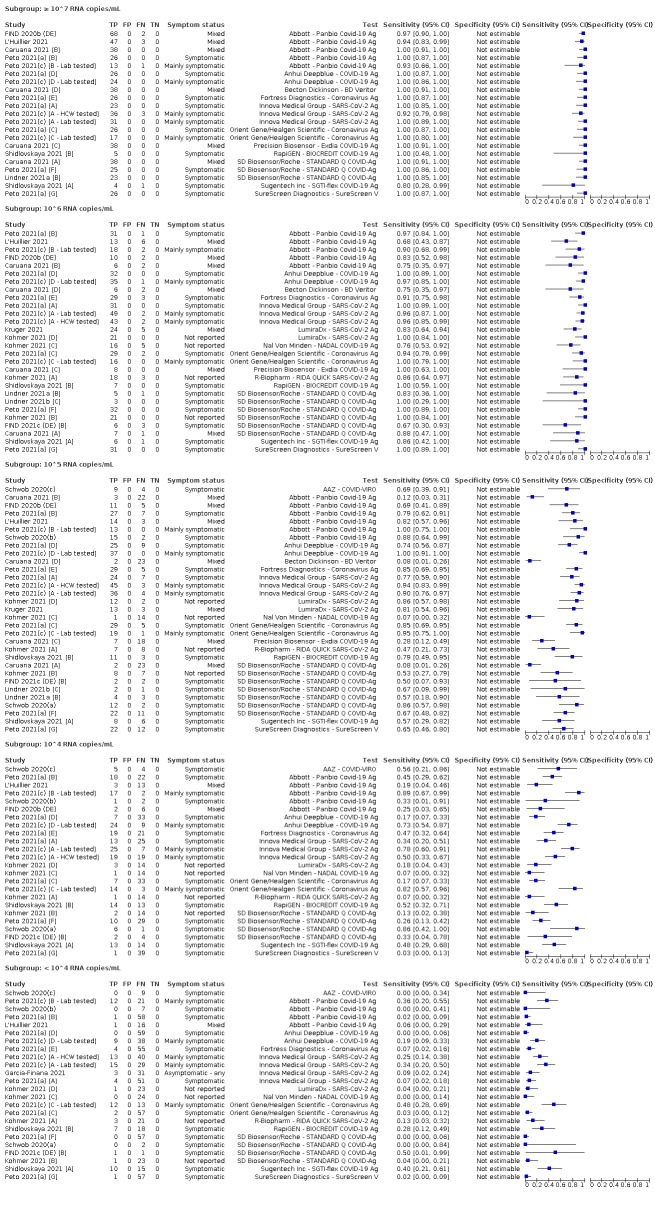
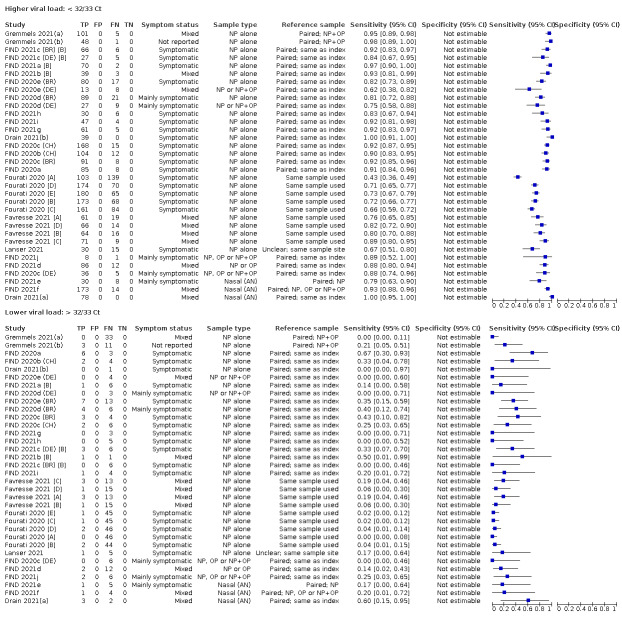
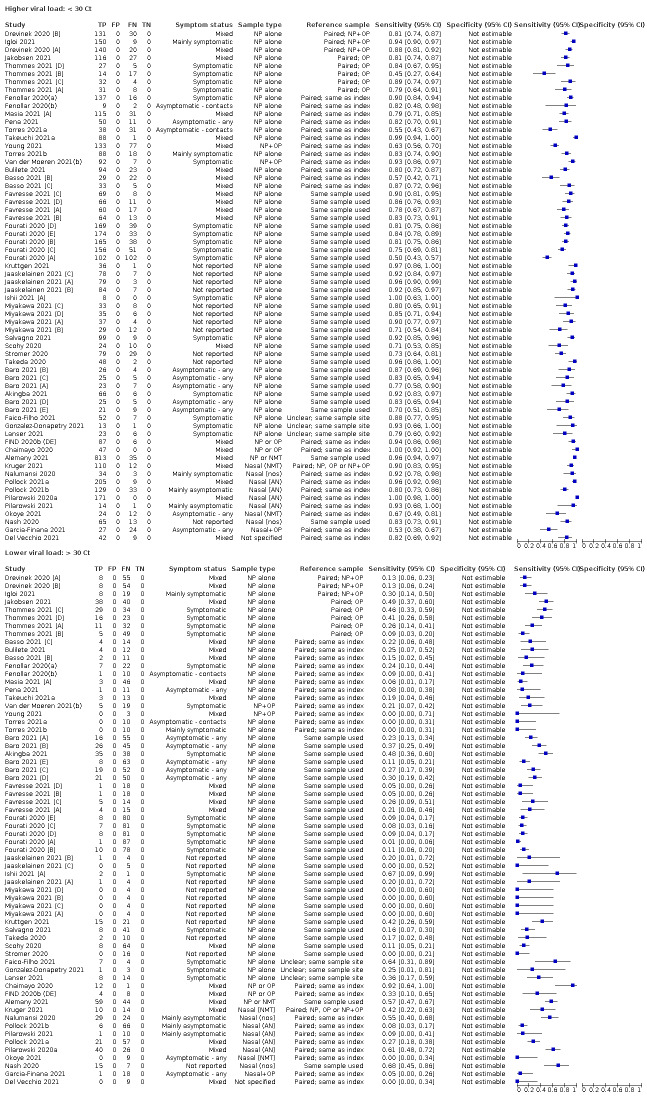
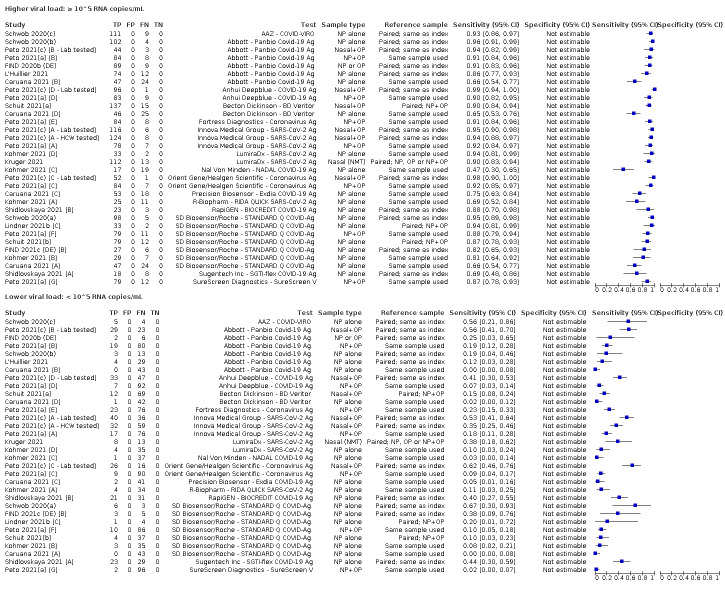
We observed a steady decline in summary sensitivities as measures of sample viral load decreased.
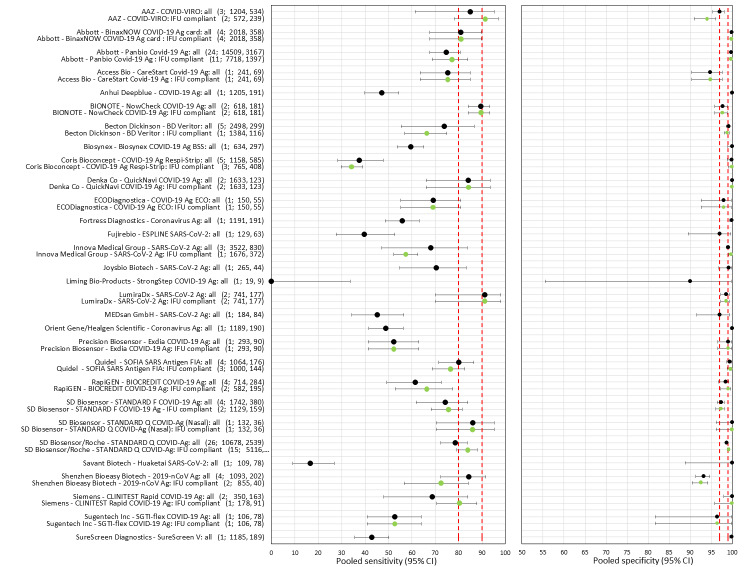
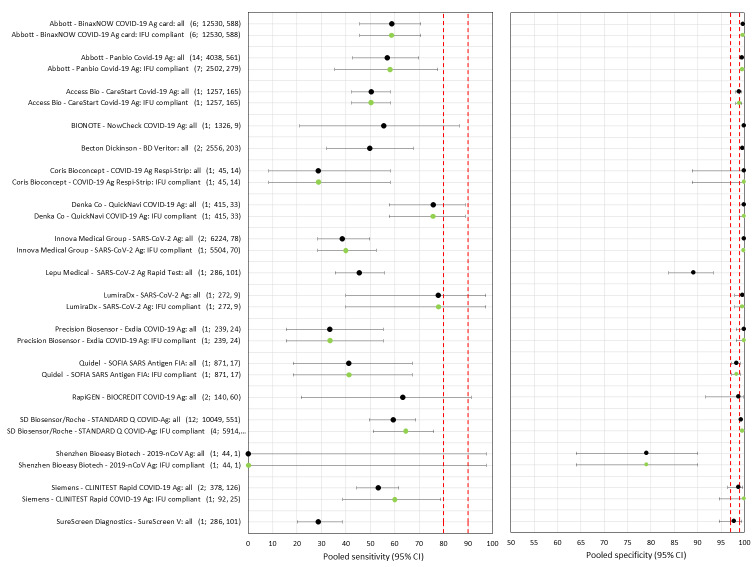
Sensitivity varied between brands. When tests were used according to manufacturer instructions, average sensitivities by brand ranged from 34.3% to 91.3% in symptomatic participants (20 assays with eligible data) and from 28.6% to 77.8% for asymptomatic participants (12 assays). For symptomatic participants, summary sensitivities for seven assays were 80% or more (meeting acceptable criteria set by the World Health Organization (WHO)). The WHO acceptable performance criterion of 97% specificity was met by 17 of 20 assays when tests were used according to manufacturer instructions, 12 of which demonstrated specificities above 99%. For asymptomatic participants the sensitivities of only two assays approached but did not meet WHO acceptable performance standards in one study each; specificities for asymptomatic participants were in a similar range to those observed for symptomatic people.
At 5% prevalence using summary data in symptomatic people during the first week after symptom onset, the positive predictive value (PPV) of 89% means that 1 in 10 positive results will be a false positive, and around 1 in 5 cases will be missed. At 0.5% prevalence using summary data for asymptomatic people, where testing was widely available and where epidemiological exposure to COVID‐19 was suspected, resulting PPVs would be 38% to 52%, meaning that between 2 in 5 and 1 in 2 positive results will be false positives, and between 1 in 2 and 1 in 3 cases will be missed.
Authors' conclusions
Antigen tests vary in sensitivity. In people with signs and symptoms of COVID‐19, sensitivities are highest in the first week of illness when viral loads are higher. Assays that meet appropriate performance standards, such as those set by WHO, could replace laboratory‐based RT‐PCR when immediate decisions about patient care must be made, or where RT‐PCR cannot be delivered in a timely manner. However, they are more suitable for use as triage to RT‐PCR testing. The variable sensitivity of antigen tests means that people who test negative may still be infected. Many commercially available rapid antigen tests have not been evaluated in independent validation studies.
Evidence for testing in asymptomatic cohorts has increased, however sensitivity is lower and there is a paucity of evidence for testing in different settings. Questions remain about the use of antigen test‐based repeat testing strategies. Further research is needed to evaluate the effectiveness of screening programmes at reducing transmission of infection, whether mass screening or targeted approaches including schools, healthcare setting and traveller screening.
Plain language summary
How accurate are rapid antigen tests for diagnosing COVID‐19?
Key messages
• Rapid antigen tests are most accurate when they are used in people who have signs or symptoms of COVID‐19, especially during the first week of illness. People who test negative may still be infected.
• Rapid antigen tests are considerably less accurate when they are used in people with no signs or symptoms of infection, but do perform better in people who have been in contact with someone who has confirmed COVID‐19.
• The accuracy of rapid antigen tests varies between tests that are produced by different manufacturers and there is a lack of evidence for many commercially available tests.
What are rapid point‐of‐care antigen tests for COVID‐19?
Rapid point‐of‐care tests aim to confirm or rule out COVID‐19 infection in people with or without COVID‐19 symptoms. They:
• are portable, so they can be used wherever the patient is (at the point‐of‐care) or in non‐healthcare settings such as in the home;
• are easy to perform, with a minimum amount of extra equipment or complicated preparation steps;
• are less expensive than standard laboratory tests;
• do not require a specialist operator or setting; and
• provide results ‘while you wait’.
For this review we were interested in rapid antigen tests, sometimes referred to as ‘lateral flow tests’. These tests identify proteins on the virus in samples taken from the nose or throat. They come in disposable plastic cassettes, similar to over‐the‐counter pregnancy tests.
Why is this question important?
People with suspected COVID‐19 need to know quickly whether they are infected, so that they can self‐isolate, receive treatment, and inform close contacts. Currently, COVID‐19 infection is confirmed by a laboratory test called RT‐PCR, which uses specialist equipment and often takes at least 24 hours to produce a result.
In many places, rapid antigen tests have opened access to testing for many more people, with and without symptoms, and in locations other than healthcare settings. Faster diagnosis of COVID‐19 infection could allow people to take appropriate action more quickly, with the potential to reduce the spread of COVID‐19, but it is important to understand how accurate they are and the best way to use them.
What did we want to find out?
We wanted to know whether commercially available, rapid point‐of‐care antigen tests are accurate enough to diagnose COVID‐19 infection reliably, and to find out if accuracy differs in people with and without symptoms.
What did we do?
We looked for studies that measured the accuracy of any commercially produced rapid antigen test in people who were also tested for COVID‐19 using RT‐PCR. People could be tested in hospital, in the community or in their own homes. Studies could test people with or without symptoms.
What did we find?
We included 155 studies in the review. The main results are based on 152 studies investigating a total of 100,462 nose or throat samples; COVID‐19 was confirmed in 16,822 of these samples. Studies investigated 49 different antigen tests. Around 60% of studies took place in Europe.
Main results
In people with confirmed COVID‐19, antigen tests correctly identified COVID‐19 infection in an average of 73% of people with symptoms, compared to 55% of people without symptoms. Tests were most accurate when used in the first week after symptoms began (an average of 82% of confirmed cases had positive antigen tests). This is likely to be because people have the most virus in their system in the first days after they are infected. For people with no symptoms, tests were most accurate in people likely to have been in contact with a case of COVID‐19 infection (an average of 64% of confirmed cases had positive antigen tests).
In people who did not have COVID‐19, antigen tests correctly ruled out infection in 99.6% of people with symptoms and 99.7% of people without symptoms.
Different brands of tests varied in accuracy. Summary results (combined from more than one study per test brand) for seven tests met World Health Organization (WHO) standards as ‘acceptable’ for confirming and ruling out COVID‐19 in people with signs and symptoms of COVID‐19. Two more tests met the WHO acceptable standard in one study each. No test met this standard when evaluated in people without symptoms.
Using summary results for symptomatic people tested during the first week after symptoms began, if 1000 people with symptoms had the antigen test, and 50 (5%) of them really had COVID‐19:
• 45 people would test positive for COVID‐19. Of these, 5 people (11%) would not have COVID‐19 (false positive result).
• 955 people would test negative for COVID‐19. Of these, 10 people (1.0%) would actually have COVID‐19 (false negative result).
In people with no symptoms of COVID‐19 the number of confirmed cases is expected to be much lower than in people with symptoms. Using summary results for people with no known exposure to COVID‐19 in a bigger population of 10,000 people with no symptoms, where 50 (0.5%) of them really had COVID‐19:
• 62 people would test positive for COVID‐19. Of these, 30 people (48%) would not have COVID‐19 (false positive result).
• 9938 people would test negative for COVID‐19. Of these, 18 people (0.2%) would actually have COVID‐19 (false negative result).
What are the limitations of the evidence?
In general, studies used relatively rigorous methods, particularly for selecting participants and performing the tests. Sometimes studies did not perform the test on the people for whom it was intended and did not follow the manufacturers’ instructions for using the test. Sometimes the tests were not carried out at the point of care. Studies used less rigorous methods for confirming the presence or absence of COVID‐19 infection; 91% of studies relied on a single negative RT‐PCR result as evidence of no COVID‐19 infection. Results from different test brands varied, and relatively few studies directly compared one test brand with another. Finally, not all studies gave enough information about their participants for us to judge how long they had had symptoms, or even whether or not they had symptoms.
What does this mean?
In people with symptoms, some rapid antigen tests are accurate enough to replace RT‐PCR, especially for ruling in the presence of infection. Alternatively, where RT‐PCR is available, rapid antigen tests could be used to select which people with symptoms require further testing with RT‐PCR, thereby reducing the burden on laboratory services. This would be most useful when quick decisions are needed about patient care, to identify outbreaks, to allow people to self‐isolate more quickly, or to initiate contact tracing. Rapid antigen tests are less good at ruling out infection in symptomatic people ‐ individuals who receive a negative rapid antigen test result may still be infected.
Rapid antigen tests are less accurate when used in people with no symptoms of COVID‐19. More evidence is needed to understand the accuracy of rapid testing in people without symptoms and the extent to which repeated testing strategies can lead to reduced transmission, either for tests carried out at home or in non‐healthcare settings such as schools. There is no independent evidence to support the use of many test brands. More direct comparisons of test brands are needed, with testers following manufacturers’ instructions.
How up‐to‐date is this review?
This review updates our previous review and includes evidence published up to 8 March 2021.
Summary of findings
Summary of findings 1. Diagnostic accuracy of point‐of‐care antigen tests for the diagnosis of SARS‐CoV‐2 infection.
| Question | What is the diagnostic accuracy of rapid point‐of‐care antigen tests for the diagnosis of SARS‐CoV‐2 infection? | ||||||
| Population | Adults or children with suspected:
or populations undergoing screening for SARS‐CoV‐2 infection, including
|
||||||
| Index test | Any commercially produced rapid antigen test for diagnosis of SARS‐CoV‐2 meeting the following criteria:
|
||||||
| Target condition | Detection of current SARS‐CoV‐2 infection | ||||||
| Reference standard | For COVID‐19 cases: positive molecular‐based test result (PCR or TMA) For non‐COVID‐19 cases: negative molecular test result or pre‐pandemic sources of samples |
||||||
| Action |
False negative results mean missed cases of COVID‐19 infection, with either delayed or no confirmed diagnosis and increased risk of community transmission due to false sense of security False positive results lead to unnecessary self‐isolation or quarantine, and may increase the potential for an infection to be acquired if individuals erroneously believe themselves to be immune |
||||||
Quantity of evidence (based on 152 studies);
|
Population | Number of studies | Total samples | Samples from confirmed SARS‐CoV‐2 cases | |||
| Any symptom status | 152 | 100,462 | 16,822 | ||||
| Population | Number of test evaluations (≥ 1 per study) | Total samples | Samples from confirmed SARS‐CoV‐2 cases | ||||
| Any symptom status | 210 | 120,381 | 23,488 | ||||
| Symptomatic | 133 | 53,589 | 14,027 | ||||
| Asymptomatic | 56 | 41,129 | 2814 | ||||
| Limitations in the evidence | |||||||
|
Risk of bias (based on 152 studies) |
Participants: high (40) or unclear (45) risk in 85 studies (56%) Index test: high (6) or unclear (45) risk in 51 studies (34%) Reference standard: high (119) or unclear (18) risk in 137 studies (90%) Flow and timing: high (41) or unclear (69) risk in 110 studies (72%) |
||||||
|
Concerns about applicability (based on 152 studies) |
Participants: high concerns in 45 studies (30%) Index test: high concerns in 47 studies (31%) Reference standard: high concerns in 139 studies (91%) |
||||||
| Findings from studies reporting both sensitivity and specificity | |||||||
| Evaluations | Samples (SARS‐CoV‐2 cases) | Sensitivity (95% CI) | Specificity (95% CI) | ||||
| Symptomatic* | 109 | 50,574 (11,662) | 73.0 (69.3 to 76.4) | 99.1 (99.0 to 99.2)a | |||
| Subgroup ≤ 7 days from symptom onsetb* | 30 | 15,323 (2408) | 80.9 (76.9 to 84.4) | 99.5 (99.3 to 99.6) | |||
| Subgroup: COVID‐19 test centre | 47 | 23,602 (4369) | 82.8 (80.2 to 85.2) | 99.1 (99.0 to 99.2) | |||
| Asymptomatic | 50 | 40,956 (2641) | 54.7 (47.7 to 61.6) | 99.5 (99.4 to 99.6) | |||
| Subgroup: widely available testing* | 26 | 31,904 (1758) | 49.6 (42.1 to 57.1) | 99.6 (99.5 to 99.7) | |||
| Subgroup: contacts* | 16 | 7677 (703) | 64.3 (54.6 to 73.0) | 99.7 (99.5 to 99.8) | |||
| Any symptom status | Evaluations | SARS‐CoV‐2 cases | Sensitivity (95% CI) | ||||
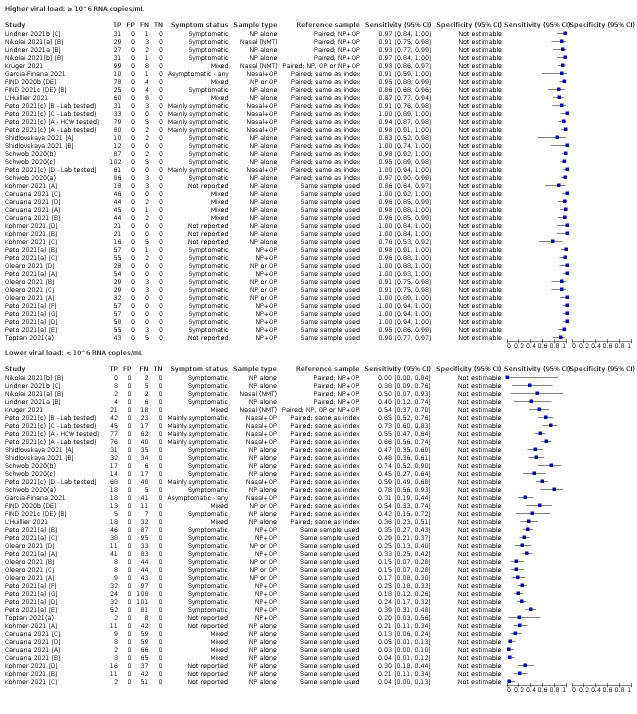
| Subgroup: ≥ 10^7 RNA copies/mL | 21 | 608 | 98.4 (97.0 to 99.1) | ‐ | |||
| Subgroup: ≥ 10^6 to < 10^7 RNA copies/mL | 28 | 597 | 94.0 (89.8 to 96.6) | ‐ | |||
| Subgroup: ≥ 10^5 to < 10^6 RNA copies/mL | 31 | 686 | 70.9 (57.4 to 81.5) | ‐ | |||
| Subgroup: ≥ 10^4 to < 10^5 RNA copies/mL | 24 | 582 | 36.7 (24.7 to 50.5) | ‐ | |||
| Subgroup: < 10^4 RNA copies/mL | 24 | 825 | 7.5 (3.8 to 14.3) | ‐ | |||
| Symptomatic participants: average sensitivity and specificity (and 95% CIs) applied to a hypothetical cohort of 1000 patients where 50, 100 and 200 have COVID‐19 infection | |||||||
| Prevalence | TP (95% CI) | FP (95% CI) | FN (95% CI) | TN (95% CI) | PPVc | 1 – NPVd | |
| Symptomatic (any symptomatic) | 5% | 37 (35 to 50) | 9 (8 to 10) | 14 (1 to 15) | 941 (940 to 942) | 81% | 1.4% |
| 10% | 73 (69 to 99) | 8 (7 to 9) | 27 (1 to 31) | 892 (891 to 893) | 90% | 2.9% | |
| 20% | 146 (139 to 198) | 7 (6 to 8) | 54 (2 to 61) | 793 (792 to 794) | 95% | 6.4% | |
| Symptomatic (week 1 after symptom onset) | 5% | 40 (38 to 42) | 5 (4 to 7) | 10 (8 to 12) | 945 (943 to 946) | 89% | 1.0% |
| 10% | 81 (77 to 84) | 5 (4 to 6) | 19 (16 to 23) | 896 (894 to 896) | 95% | 2.1% | |
| 20% | 162 (154 to 169) | 4 (3 to 6) | 38 (31 to 46) | 796 (794 to 797) | 98% | 4.6% | |
| Asymptomatic participants: average sensitivity and specificity (and 95% CIs) applied to a hypothetical cohort of 10,000 patients where 50, 100 and 200 have COVID‐19 infection | |||||||
| Asymptomatic (widely available testing) | 0.5% | 25 (21 to 29) | 40 (30 to 50) | 25 (21 to 29) | 9910 (9900 to 9920) | 38% | 0.3% |
| 1% | 50 (42 to 57) | 40 (30 to 50) | 50 (43 to 58) | 9860 (9851 to 9870) | 52% | 0.5% | |
| 2% | 99 (84 to 114) | 39 (29 to 49) | 101 (86 to 116) | 9760 (9751 to 9770) | 72% | 1.0% | |
| Asymptomatic (contacts) | 0.5% | 32 (27 to 50) | 30 (20 to 50) | 18 (14 to 23) | 9920 (9900 to 9930) | 52% | 0.2% |
| 1% | 64 (55 to 73) | 30 (20 to 50) | 36 (27 to 45) | 9870 (9850 to 9880) | 68% | 0.4% | |
| 2% | 129 (109 to 146) | 29 (20 to 49) | 71 (54 to 91) | 9770 (9751 to 9780) | 81% | 0.7% | |
|
1 – NPV: 1 – negative predictive value (the percentage of people with negative results who are infected); Ag: antigen; CI: confidence interval; Ct: cycle threshold; FN: false negative; FP: false positive; PPV: positive predictive value (the percentage of people with positive results who are infected); PCR: reverse transcription polymerase chain reaction; TMA: transcription‐mediated amplification; TN: true negative; TP: true positive * denotes data used for hypothetical cohort scenarios | |||||||
aExcludes outlier with 8% specificity in 13 throat saliva or throat wash samples. bResults reported are for studies reporting both sensitivity and specificity; including sensitivity‐only cohorts (total n = 72), sensitivity was 82.2% (95% confidence interval 79.2% to 85.0%) in 5640 PCR+ve samples. cPPV (positive predictive value) defined as the percentage of positive rapid test results that are truly positive according to the reference standard diagnosis. d1‐NPV (negative predictive value), where NPV is defined as the percentage of negative rapid test results that are truly negative according to the reference standard diagnosis.
Background
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) and the resulting COVID‐19 pandemic present important diagnostic evaluation challenges. These range from: understanding the value of signs and symptoms in predicting possible infection; assessing whether existing biochemical and imaging tests can identify infection or people needing critical care; and evaluating whether in vitro diagnostic tests can accurately identify and rule out current SARS‐CoV‐2 infection, and identify those with past infection, with or without immunity.
We are creating and maintaining a suite of living systematic reviews to cover the roles of tests and patient characteristics in the diagnosis of COVID‐19. This review is the second update of a review summarising evidence of the accuracy of rapid antigen tests that are suitable for use at the point of care. The review was first published in August 2020 (Dinnes 2020), and updated in March 2021 (Dinnes 2021), and originally investigated both rapid antigen tests and rapid molecular tests for diagnosis of SARS‐CoV‐2. This review focuses solely on rapid antigen tests. Rapid tests may have potential to be used as alternatives to standard laboratory‐based molecular assays, such as reverse transcription polymerase chain reaction (RT‐PCR) assays, that are relied on for identifying current infection, or may be used where no testing is currently done. If sufficiently accurate, point‐of‐care tests have the potential to greatly expand access and speed of testing. In turn, if accurate, they may have greater impact on public health than laboratory‐based molecular methods as they are less expensive, provide results more quickly and do not require the same technical expertise and laboratory capacity. These tests can be undertaken locally, avoiding the need for centralised testing facilities that rarely meet the needs of patients, caregivers, health workers and society as a whole, especially in low‐ and middle‐income countries. As these are rapid tests, their results can be returned within the same clinical encounter, facilitating timely decisions concerning the need for isolation and contract tracing activities.
Target condition being diagnosed
COVID‐19 is the disease caused by infection with the SARS‐CoV‐2 virus. The key target conditions for this suite of reviews are current SARS‐CoV‐2 infection, current COVID‐19 disease, and past SARS‐CoV‐2 infection. The tests included in this review concern the identification of current infection, as defined by reference standard methods of diagnosis, including molecular assays such as reverse transcription polymerase chain reaction (RT‐PCR), or internationally recognized clinical guidelines for diagnosis of SARS‐CoV‐2. In the context of test evaluation, and throughout this review, we use the term 'reference standard' to denote the best available method (test or tests) for diagnosing the target condition, as opposed to other uses of the term in diagnostic virology (such as reference methods or reference materials).
For current infection, the severity of the disease is of ultimate importance for patient outcomes. Rapid testing does not establish severity of disease, and for this review we consider the role of point‐of‐care antigen tests for detecting SARS‐CoV‐2 infection of any severity, distinguishing only between symptomatic and asymptomatic infection. In addition, the increasing occurrence of SARS‐CoV‐2 variants of concern since the last published iteration of this review could have some as yet unknown impact on the accuracy of rapid diagnostic tests (RDTs), particularly if mutations take place in the nucleocapsid gene (‘N’ gene), which encodes the virus nucleoprotein and is the main target of the majority of antigen‐based RDTs (FIND 2022b).
COVID‐19 public health interventions focus on increasing uptake of COVID‐19 vaccination and on reducing disease transmission. Immunity from infection varies between individuals, even in those vaccinated or who have had a prior COVID‐19 infection and immunity also wanes over time, therefore early identification of infection remains an important public health goal. Government policies in regard to testing, self‐isolation and quarantine have changed over the course of the pandemic, however, as a general principle, people with symptoms who meet national criteria for COVID‐19 testing are reasonably expected to self‐isolate to avoid infecting others while awaiting the result from a PCR test. Contacts of confirmed cases have been similarly considered to have a high enough risk of being infectious to ask them to quarantine for 7 to 10 days. The UK and USA introduced policies based on rapid tests, both to allow 'early' release from self‐isolation for those with confirmed ‐SARS‐CoV‐2 infection and daily rapid antigen testing for fully vaccinated contacts of confirmed cases, with self‐isolation only required following a positive antigen test result (UK HSA 2021a). Assessing the risk of an individual being infectious in asymptomatic screening is difficult, however, as there is no reference standard test for being ‘infectious’. Using RT‐PCR status as a reference standard (as is done for the target condition of ‘infection’) will ensure that infectious people are not missed, but because RT‐PCR continues to detect viral ribonucleic acid (RNA) days and weeks after the onset of infection it will wrongly classify some infected people as infectious.
A reference standard that has been proposed for establishing infectiousness is viral culture. Viral culture is technically complex and requires high levels of biosafety containment, such that it is not suitable for routine use. Furthermore it can be unreliable (the failure to culture virus potentially being a result of the culture technique and not an indicator of non‐infectiousness). For example, in Smith 2021, viral culture failed in samples from 8 of 51 (16%) newly infected adults.
Alternatively, a value of the cycle threshold (Ct value) from RT‐PCR results to group individuals above or below a particular value as more or less likely to be infectious has become commonplace (Petersen 2021; UK DHSC 2021b; WHO 2020b). The suitability of RT‐PCR Ct values (also known as quantification cycle (Cq) or crossing point (Cp) values) as a proxy indicator of infectiousness is limited for a number of reasons, however. Firstly, the relationship between Ct values and viral load varies between machines and laboratories and RT‐PCR assays, even where the same genetic targets are used (Binnicker 2020), and is further affected by sample collection and processing (IDSA 2021), so that comparison at fixed Ct values is unlikely to be comparable across studies. Recent work by Evans and colleagues converting Ct values into direct quantitative values of viral load (viral copies per cell) demonstrated inter‐laboratory variation of more than 1000‐fold in copies/mL for a given Ct value measuring the same sample (Evans 2021). Secondly, although conversion from Ct values to RNA copies/mL allows for a fairer comparison in results between studies when done correctly (i.e. using a quantitative PCR calibrated to a robust standard curve derived from certified reference material), the use of different and potentially suboptimal methods of calculating viral load from Ct values has potential to introduce variability in results (Evans 2021). Thirdly, the inverse relationship between viral load and risk of infecting others is a continuum of risk without there being a meaningful cut‐point either in terms of Ct or genomic copies/mL. Studies have evidenced successful viral culture from samples with Ct values as high as 35 (Singanayagam 2020), and transmission of infection from index cases with higher Ct values (Lee 2021; Marks 2021; Tian 2021). A linked data analysis using available empirical data suggests a non‐negligible risk of onward transmission of infection from cases with higher Ct values (Deeks 2022). Finally, even the most precise estimation of viral load does not overcome the inability of a single test to identify whether individuals with low viral loads are at the onset of infection and therefore likely to be infectious for a period of time or are recovering from infection and have declining viral load.
Tracking contacts of SARS‐CoV‐2 cases for evidence of infection provides the best insight into the dynamics of viral transmission, however this requires longitudinal follow‐up and predictive modelling to take into account host, agent and environmental factors, all of which influence risk of transmission. This contrasts with the diagnostic test accuracy paradigm which can only determine if individuals are infected at a single point in time.
Because of the lack of a suitable reference standard for detection of ‘infectiousness’, this review only focuses on the target condition of 'infection' for applications of tests in both symptomatic and asymptomatic individuals. Although the presence of COVID‐19 can be defined clinically (e.g. for individuals with negative RT‐PCR results), the primary reference standard for presence or absence of SARS‐CoV‐2 infection is RT‐PCR. While acknowledging the potential for between‐study variability in viral load associated with Ct values in particular, we report results in subgroups by Ct value and by RNA copies/mL, and continue to advise caution in interpretation. Given the current state of the scientific knowledge we do not consider it appropriate to consider these as groups that are 'infectious' or 'not infectious'.
RT‐PCR carries a very small risk of false positive results for infection and a higher risk of false negative results. False positive results may result from failures in sampling or laboratory protocols (e.g. mislabelling), contamination during sampling or processing, or low‐level reactions during PCR (Healy 2020; Mayers 2020). As for the previous iteration of this review we consider the upper bound on the possible false positive rate of RT‐PCR of less than 0.077%. This estimate is based on population prevalence surveys showing RT‐PCR positivity rates (comprising both true positive and false positive results) of 0.44% in August 2020 (95% credible interval 0.22% to 0.76%; ONS 2020), and 0.077% (95% credible interval 0.065%, 0.092%) in the React‐1 study from June to July 2020 (Riley 2020). False negative rates have been estimated by looking at individuals with symptoms who initially test negative, but test positive on a subsequent test. These rates have been estimated to be as high as 20% to 30% in the first week of symptom onset (Arevalo‐Rodriguez 2020a; Kucirka 2020; Yang 2020a; Zhao 2020). Including probable COVID‐19 cases within the target condition, as defined by internationally recognized clinical guidelines for diagnosis of SARS‐CoV‐2, will partially mitigate these missed cases.
Index test(s)
Previous iterations of this living review included two types of test that could be deployed at the point of care: rapid antigen tests and rapid molecular tests (see Appendix 1 for the definition of ‘point of care’ that has been used in these reviews). Given the widespread international interest in using rapid antigen tests, and the many different settings in which they can be deployed compared to rapid molecular tests, this review update focuses only on antigen detection tests (referred to here as rapid diagnostic tests or RDTs), which can be used at the point of care or in non‐healthcare settings such as in the home. In this iteration of the review we include evaluations of single applications of a test (i.e. used for diagnostic purposes in symptomatic or asymptomatic populations) and evaluations of serial testing strategies in asymptomatic populations (i.e. repeated applications of a test for earlier detection of infection). We intend to update the review of rapid molecular tests separately at a later date.
Antigen RDTs (and rapid molecular assays) typically use the same upper respiratory‐tract samples obtained for laboratory‐based RT‐PCR, that is, nasopharyngeal or combined naso‐ and oro‐pharyngeal samples, although many companies have test kits for use with anterior nasal or nasal mid‐turbinate samples. The majority of RDTs are lateral flow immunoassays (LFAs), which are disposable devices, usually in the form of plastic cassettes akin to an over‐the‐counter pregnancy test. SARS‐CoV‐2 antigens, most commonly the nucleoprotein, are captured by dedicated and labelled antibodies, typically colloidal gold‐ or fluorescent‐labelled, although other assay formats are also available. The liquid sample is absorbed via passive capillary action, and the presence of the target antigen is indicated within 15 to 30 min either by visible lines appearing on the test strip, or through fluorescence, which can be detected using an immunofluorescence analyser. Microfluidic analytical devices are also being developed for SARS‐CoV‐2, typically using reader devices for test interpretation. These devices are based on the lateral flow format, using active capillary action to guide liquid samples along the test strip to the desired outlets where the chemical or biochemical reactions take place (Jiang 2021). The assays are intended to detect the target antigen at lower concentrations compared to conventional LFAs (Noel 2021).
Although antigen RDTs have been shown to be on average less sensitive than rapid molecular tests (Dinnes 2021), there are considerably fewer logistical and economic barriers to their use. This has led to widespread international adoption of RDTs, and prolific industry activity to develop more accurate tests. The Foundation for Innovative Diagnostics (FIND) and Johns Hopkins Centre for Health Security have maintained online lists of available tests for SARS‐CoV‐2 (FIND 2022a). At the time of writing (20 December 2021), FIND listed 321 commercially available rapid antigen tests, almost all with known regulatory approval. These numbers are a considerable increase on the 92 with regulatory approval at the time of writing the last review iteration (5 January 2021), and the 21 with regulatory approval at the time of our original review (19 July 2020). This classification was based on the information provided to FIND by the test manufacturers and does not necessarily mean that these tests meet the criteria for point‐of‐care tests that we have specified for this review.
For this iteration of the review, we continue to only include evaluations of commercially produced tests. All commercially produced assays are supplied with a specific product code, product inserts or instructions for use (IFU) sheets that document the intended use of the test, sample storage and preparation and testing procedures.
Clinical pathway
Patients may be tested for SARS‐CoV‐2 when they present with symptoms, have had known exposure to a confirmed case, or in a screening context, with no known exposure to SARS‐CoV‐2. The standard approach to diagnosis of SARS‐CoV‐2 infection is through laboratory‐based testing of swab samples taken from the upper respiratory (e.g. nasopharynx, oropharynx) or lower respiratory tract (e.g. bronchoalveolar lavage or sputum) with RT‐PCR. RT‐PCR is the primary method for detecting infection during the acute phase of the illness while the virus is still present. Both the WHO and the China CDC (National Health Commission of the People's Republic of China) have produced case definitions for COVID‐19 that include the presence of convincing clinical evidence (some including positive serology tests) when RT‐PCR is negative (Appendix 2).
Prior test(s)
Signs and symptoms are used in the initial diagnosis of suspected SARS‐CoV‐2 infection and to help identify those requiring tests. A number of key symptoms have been suggested as indicators of mild to moderate COVID‐19, including: cough, fever greater than 37.8 °C, headache, breathlessness, muscle pain, fatigue, and loss of sense of smell and taste (Struyf 2021). However, the Cochrane Review of signs and symptoms found that the majority of individual signs and symptoms have very poor diagnostic accuracy; neither absence nor presence of signs or symptoms were accurate enough to rule in or rule out disease (Struyf 2021). The review suggested that multivariable prediction models combining symptoms with other information such as contact or travel history, age, gender, and a local recent case detection rate, could reach sensitivities as high as 90%, however further research is needed to identify the optimal combination of variables (Struyf 2021). With reports of changing symptom profiles by age (Canas 2021), and by vaccination status (Zoe COVID Study 2021), rapid testing of symptomatic individuals is likely to remain a vital tool in managing the COVID‐19 pandemic (Crozier 2021). Where people are asymptomatic but are being tested as part of screening (e.g. universal testing of students as part of a risk‐reduction effort) or on the basis of epidemiological risk factors, such as exposure to someone with confirmed SARS‐CoV‐2 or following travel to more highly endemic countries, no prior tests will have been conducted.
Role of index test(s)
For most settings in which testing for acute SARS‐CoV‐2 infection in symptomatic individuals takes place, results of molecular laboratory‐based RT‐PCR tests are unlikely to be available within a single clinical encounter. Point‐of‐care tests potentially have a role either as a replacement for RT‐PCR (if sufficiently accurate), or as a means of triaging and rapid management (isolation or treatment, or both), with RT‐PCR testing for those with negative rapid test results (CDC 2021; WHO 2020a). Obtaining quick results within a single healthcare visit will allow faster decisions about isolation and healthcare interventions for those with positive test results, and allow contact tracing to begin in a more timely manner. Modelling studies suggest contact tracing is most effective if it starts within 24 hours of case detection, with delays in testing (e.g. due to laboratory turnaround time for reporting PCR results) leading to reductions in the proportion of onward transmissions per index case that can be prevented by track and trace (Kretzschmar 2020).
If sufficiently accurate, negative rapid test results in symptomatic patients could allow faster return to work or school after symptom resolution, therefore conferring important economic and educational implications. Negative results also allow immediate consideration of other causes of symptoms, which may be time‐sensitive, for example bacterial pneumonia or thromboembolism.
For asymptomatic individuals, if accurate, rapid tests may also be considered for screening at‐risk (exposed) populations, for example in hospital workers or in local outbreaks, or for targeted screening with single test application at airports or for border entry, to allow entry to large public gatherings (Revollo 2021), or screening students as a risk‐reduction strategy (Ferguson 2021). Because rapid antigen tests can easily be delivered at scale, they have also been deployed for mass screening purposes as piloted in Slovakia (Frnda 2021), and in Liverpool, UK (Garcia‐Finana 2021). Community mass antigen testing for SARS‐CoV‐2 is now used internationally, under national (e.g. UK (Iacobucci 2020), or Slovakia (Frnda 2021)), or regional policies (e.g. USA (Prince‐Guerra 2021), or Spain (Pena 2021), amongst others). Frequent repeated use of antigen tests in asymptomatic individuals with no known exposure to identify COVID‐19 cases has also been proposed in a number of modelling studies (Larremore 2020), but field trial evaluations to confirm the suggested promising results remain scarce (e.g. Young 2021), and are not without criticism (Gurdasani 2021). Nevertheless, UK residents, including secondary school pupils, are recommended to use a freely available (at the time of writing) rapid antigen test twice per week (NHS 2021; UK Department for Education 2022), with daily contact testing trials completed (UK DHSC 2021a), or recently published (Young 2021).
Alternative test(s)
This review is one of eight that cover the range of tests and clinical characteristics being considered in the management of COVID‐19 (Deeks 2020a; McInnes 2020; Leeflang 2021), five of which have already been published (Deeks 2020b; Islam 2021; Stegeman 2020; Struyf 2021), including the first two iterations of this review (Dinnes 2020; Dinnes 2021). Full details of the alternative tests and evidence of their accuracy is summarized in these reviews. The SARS‐CoV‐2‐specific biomarker tests that might be considered as alternatives to point‐of‐care tests are considered here.
Rapid point‐of‐care molecular assays
Molecular‐based tests to detect viral RNA have historically been laboratory‐based assays using RT‐PCR technology (see below). In recent years, automated, single‐step RT‐PCR methods have been developed, as well as other nucleic acid amplification methods, such as isothermal amplification, that do not require the sophisticated thermo cycling involved in RT‐PCR (Green 2020). These technological advances have allowed molecular technologies to be developed that are suitable for use in a point‐of‐care context (Kozel 2017), however they require small portable machines, are more expensive and many take longer to produce results than antigen tests, although recent advances in the turnaround time have been made. For logistical and economic reasons therefore, the use cases for the majority of rapid molecular assays are quite different to point‐of‐care antigen tests and so they will now be included in a separate review in this series of living reviews.
Laboratory‐based molecular tests
PCR methods are routinely used for detection of viral RNA (Behera 2021). SARS‐CoV‐2‐specific reagents for RT‐PCR detection of SARS‐CoV‐2 were produced soon after the viral RNA sequence was published (Corman 2020). Testing is undertaken in central laboratories and can be very labour‐intensive, with several points along the path of performing a single test where errors may occur, although some automation of parts of the process is possible. The amplification process requires thermal cycling equipment to allow multiple temperature changes within a cycle, with cycles repeated up to 40 times until viral DNA is detected (Carter 2020). Although the amplification process for RT‐PCR can be completed in a relatively short timeframe, the stages of extraction, sample processing and data management (including reporting) mean that test results are typically only available in 24 to 48 hours. Where testing is undertaken in a centralized laboratory, transport increases this time further. The time to result for fully automated RT‐PCR assays is shorter than for manual RT‐PCR, however most assays still require sample preparation steps that make them unsuitable for use at the point of care.
Other nucleic acid amplification methods, including loop‐mediated isothermal amplification (LAMP), or CRISPR‐based nucleic acid detection methods, that allow amplification at a constant temperature are now commercially available (Chen 2020), and are the subject of a separate review in this series that is currently under preparation (Deeks 2020a).
Laboratory‐based antigen detection tests
Antigen detection tests can also be performed in the laboratory, using automated or semi‐automated enzyme immunoassays (EIA) like enzyme‐linked immunosorbent assays (ELISA) or more advanced chemiluminescence immunoassays (CLIAs). Because of the limitations in detecting the SARS‐CoV‐2 virus in plasma or serum, antigen detection assays are primarily used with respiratory samples (Lai 2021).
Rationale
It is essential to understand the clinical accuracy of tests and clinical features to identify the best way they can be used in different settings to develop effective diagnostic and management pathways for SARS‐CoV‐2 infection and disease. The suite of Cochrane living systematic reviews summarizes evidence on the clinical accuracy of different tests and diagnostic features. Estimates of accuracy from these reviews will help inform diagnosis, screening, isolation, and patient‐management decisions.
Summary of the previous version of the review
The first iteration of this review (Dinnes 2020), included five studies that evaluated five antigen detection tests (four commercial and one in‐house). We did not find any studies at low risk of bias and had concerns about applicability of results across all studies. The average sensitivity was 56.2% (95% confidence interval (CI) 29.5 to 79.8%) and average specificity 99.5% (95% CI 98.1% to 99.9%), based on 943 samples, 596 with confirmed SARS‐CoV‐2. Data for individual antigen tests were limited with no more than two studies for any test.
For the subsequent update of the review (Dinnes 2021, published in March 2021), we restricted inclusion to evaluations of commercially produced tests. We included 48 studies that reported 58 evaluations of 16 different commercially produced RDTs. We did not judge any study at low risk of bias, although in 23% (11/48) of studies the only bias present was that a single negative RT‐PCR was used to confirm absence of SARS‐CoV‐2 infection rather than the preferred two negative tests. All studies raised concerns regarding the applicability of their results, but similarly, in 25% (12/48) of studies the only concern was the reliance on only PCR to identify SARS‐CoV‐2 cases.
Assay specificities were consistently high (overall summary specificity 99.6%, 95% CI 99.0% to 99.8%), however estimates of sensitivity varied considerably between studies and according to test brand. In particular we identified differences in sensitivity between symptomatic (72.0%, 95% CI 63.7% to 79.0%; 37 evaluations; 15,530 samples, 4410 cases) and asymptomatic participants (58.1%, 95% CI 40.2% to 74.1%; 12 evaluations; 1581 samples, 295 cases), and sensitivity was on average higher in the first week after symptom onset (78.3%, 95% CI 71.1% to 84.1%; 26 evaluations; 5769 samples, 2320 cases) compared to the second week of symptoms (51.0%, 95% CI 40.8% to 61.0%; 22 evaluations; 935 samples, 692 cases). Sensitivity was high in those with PCR cycle threshold (Ct) values less than 25 (94.5%, 95% CI 91.0% to 96.7%; 36 evaluations; 2613 cases) compared to those with Ct values above 25 (40.7%, 95% CI 31.8% to 50.3%; 36 evaluations; 2632 cases). Using data from evaluations that were compliant with manufacturer instructions for use (IFU), summary sensitivities ranged from 34.1% (95% CI 29.7% to 38.8%; Coris Bioconcept) to 88.1% (95% CI 84.2% to 91.1%; SD Biosensor STANDARD Q). Only the STANDARD Q assay met the WHO acceptable criterion for sensitivity based on summary results across several studies.
Changes in the evidence base since the previous version
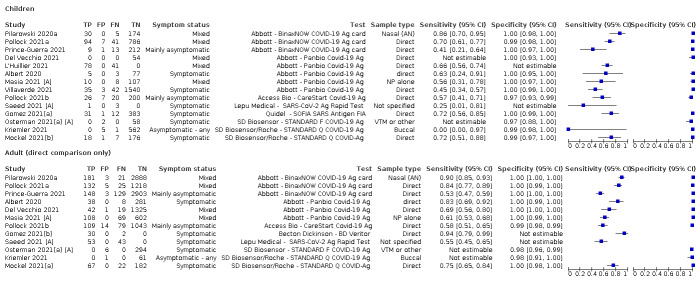
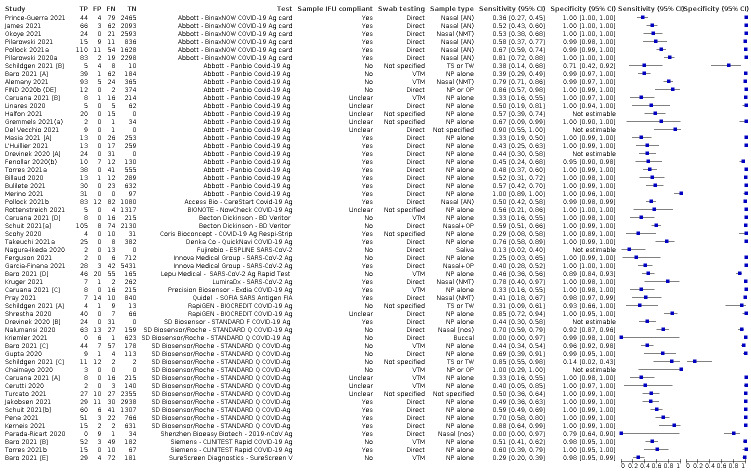
There has been a considerable increase in the number of available evaluations of antigen tests, in both symptomatic and asymptomatic populations. More studies report results for direct swab testing using nasal swab samples which are considered to be easier and more comfortable to collect than nasopharyngeal swabs. More direct comparisons of the accuracy of different test brands and, to a lesser extent, according to sampling site or type of test operator are now available. This review considers the available evidence in relevant population groups and settings according to test brand and compliance with manufacturer IFUs. We also aimed to consider any impact on test accuracy from infection with variants of concern or from vaccination status, although we anticipated that the influence from these factors may not yet be reflected in the evidence base. We used the WHO's priority target product profiles for COVID‐19 diagnostics (i.e. acceptable performance criterion of sensitivity of 80% or higher and specificity 97% or higher, or desirable criterion of 90% sensitivity or higher and 99% specificity or higher; WHO 2020b), as a benchmark against which to consider test performance.
We will update this review as often as is feasible to ensure that it provides current evidence about the accuracy of point‐of‐care tests.
This review follows a generic protocol that covers six Cochrane COVID‐19 diagnostic test accuracy (DTA) reviews (Deeks 2020a). The Background and Methods sections of this review therefore use some text that was originally published in the protocol (Deeks 2020a), in the previous iteration of this review (Dinnes 2020; Dinnes 2021), and text that overlaps some of our other reviews (Deeks 2020b; Struyf 2021).
Objectives
To assess the diagnostic accuracy of rapid point‐of‐care antigen tests to for diagnosis of SARS‐CoV‐2 infection. We consider accuracy separately in symptomatic and asymptomatic population groups.
Secondary objectives
Within each group by symptom status we explored the effect of study setting and for asymptomatic populations, epidemiological exposure to SARS‐CoV‐2 (i.e. testing of contacts of confirmed cases compared to widely available testing of asymptomatic individuals with no requirement to meet pre‐set criteria for testing).
Additional sources of heterogeneity investigated (either by stratified analysis or meta‐regression) included assay format, duration of symptoms, viral load and participant age group (adults or children). We also aimed to investigate accuracy according to SARS‐CoV‐2 variant and participant vaccination status, however insufficient evidence was identified. Although the reference standard used can influence accuracy, we anticipated that as in previous iterations of this review, all studies would rely on RT‐PCR.
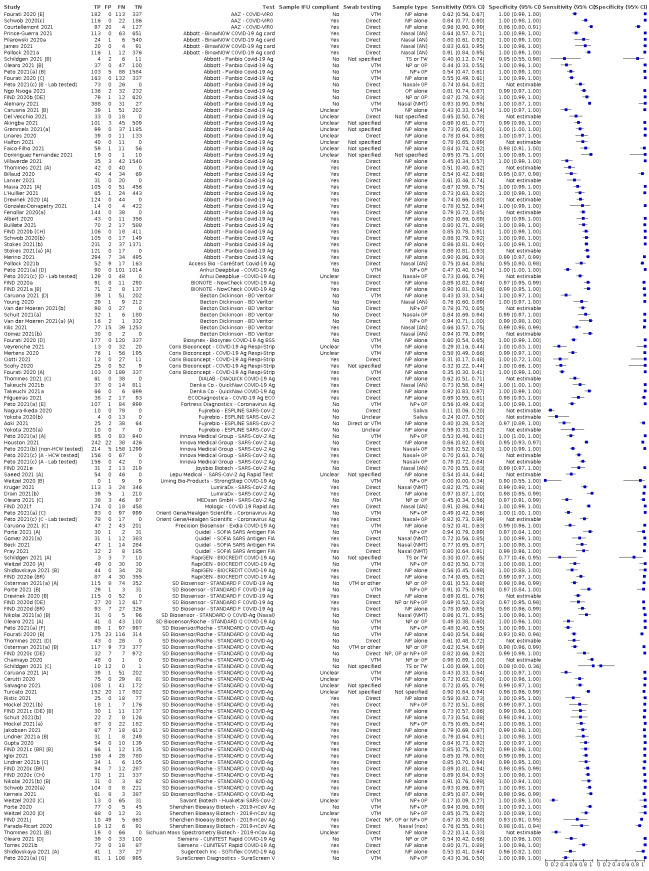
We investigated adherence to manufacturers' IFUs in sensitivity analyses.
Methods
Criteria for considering studies for this review
Types of studies
We applied broad eligibility criteria to include all patient groups (that is, if patient population was unclear, we included the study) and all variations of a test.
We included studies of all designs that produce estimates of test accuracy or provide data from which we can compute estimates, including the following.
Single‐group studies, which recruit participants before disease status has been ascertained.
Multi‐group studies, where people with and without the target condition are recruited separately (often referred to as two‐gate or diagnostic case‐control studies)
Studies restricted to participants confirmed to either have (or to have had) the target condition (to estimate sensitivity) or confirmed not to have (or have had) the target condition (to estimate specificity). These types of studies may be excluded in future review updates.
Studies based on either participants or samples
We excluded studies from which we could not extract data to compute either sensitivity or specificity.
We carefully considered the limitations of different study designs in the methodological quality assessment and analyses.
We included studies reported in published journal papers, as preprints, and publicly available reports from independent bodies.
Participants
We included studies recruiting people presenting with suspicion of current SARS‐CoV‐2 infection or those recruiting populations where tests were used to screen for infection (for example, contact tracing or community screening).
We also included studies that recruited people known to have SARS‐CoV‐2 infection and known not to have SARS‐CoV‐2 infection (i.e. cases only or multi‐group studies).
We excluded small studies with fewer than 10 samples or participants. Although the size threshold of 10 is arbitrary, such small studies are likely to give unreliable estimates of sensitivity or specificity and may be biased.
Index tests
We included studies evaluating any rapid antigen‐detection test for diagnosis of SARS‐CoV‐2, if it met the criteria outlined in Appendix 1. In brief, this includes:
minimal equipment required;
minimal sample preparation and biosafety considerations;
results available within two hours of sample collection; and
commercially produced (with test name and manufacturer or distributor documented).
Any respiratory sample type was eligible. Strategies based on multiple applications of a test were also eligible for inclusion.
We excluded studies that evaluated rapid molecular‐based tests from this review iteration.
Target conditions
The target condition was current SARS‐CoV‐2 infection (either symptomatic or asymptomatic). We also refer to SARS‐CoV‐2 infection as ‘COVID‐19 infection’, particularly in the Plain language summary and Table 1.
Reference standards
We originally anticipated that studies would use a range of reference standards to define both the presence and absence of SARS‐CoV‐2 infection, however we have found for both previous iterations of this review that all studies used laboratory‐based RT‐PCR assays to confirm the presence of SARS‐CoV‐2 infection and almost all also used RT‐PCR to confirm absence of infection (a very small proportion using pre‐pandemic respiratory samples). For this iteration of the review we therefore considered the use of any molecular assay as a suitable reference standard for confirmation of the presence or absence of SARS‐CoV‐2. Studies using pre‐pandemic samples as non‐SARS‐CoV‐2 cases were also eligible.
Search methods for identification of studies
The previous iteration of this review included records from electronic searches (up to 30 September 2020) and additional online resources (manually checked 16 November 2020). Search methods for prior iterations of the review are documented in Appendix 3. This section documents additional searches undertaken for the current iteration of this living review up to 8 March 2021.
Electronic searches
COVID‐19 Open Access Project (COAP) living evidence database from the University of Bern
We used the COVID‐19 Open Access Project living evidence database from the Institute of Social and Preventive Medicine (ISPM) at the University of Bern (COVID‐19 Open Access Project 2021). The last feed obtained for this review was 8 March 2021. The database was constructed from daily (Monday to Friday) systematic searches of Embase via OVID, MEDLINE via PubMed, bioRxiv and medRxiv. The strategies as described on the ISPM website are described here (ispmbern.github.io/covid-19/living-review/collectingdata.html). See Appendix 3.
Since 25 May 2020 we have used review‐specific artificial intelligence text analysis to classify retrieved records based on their title and abstract information, for relevant and irrelevant documents (documented in Appendix 4).
Searching other resources
We contacted or accessed the websites of independent research groups undertaking test evaluations (for example, UK Public Health England (PHE), the Société Française de Microbiologie (SFM), the Dutch National Institute for Public Health and the Environment (RIVM)) and studies co‐ordinated by FIND (finddx.org/covid-19/sarscov2-eval) and accessed the Diagnostics Global Health listing of manufacturer‐independent evaluations of antigen detecting rapid diagnostic tests (Ag‐RDTs) for SARS‐CoV‐2 (diagnosticsglobalhealth.org). We last accessed these additional resources on 30 April 2021.
We appeal to researchers to supply details of additional published or unpublished studies at the following email address, which we will consider for inclusion in future updates (coviddta@contacts.bham.ac.uk).
Data collection and analysis
Selection of studies
A team of experienced systematic review authors from the University of Birmingham screened the titles and abstracts of all records retrieved from the literature searches following the application of artificial intelligence text analysis (described in Electronic searches). Two review authors independently screened studies in Covidence. A third, senior review author resolved any disagreements.
We obtained the full texts for all studies flagged as potentially eligible. Two review authors independently screened the full texts; any disagreements on study inclusion were resolved through discussion with a third review author.
Up to September 2020 screening was conducted across all Cochrane COVID‐19 DTA biomarker reviews (molecular, antigen or antibody tests), using tagging of records according to the review(s) for which they might be eligible. From September 2020 onwards, review‐specific searches were implemented such that screening was conducted without the requirement for study tagging to different reviews.
Data extraction and management
One review author extracted the characteristics of each study, which a second review author checked. Items that we extracted are listed in Appendix 5.
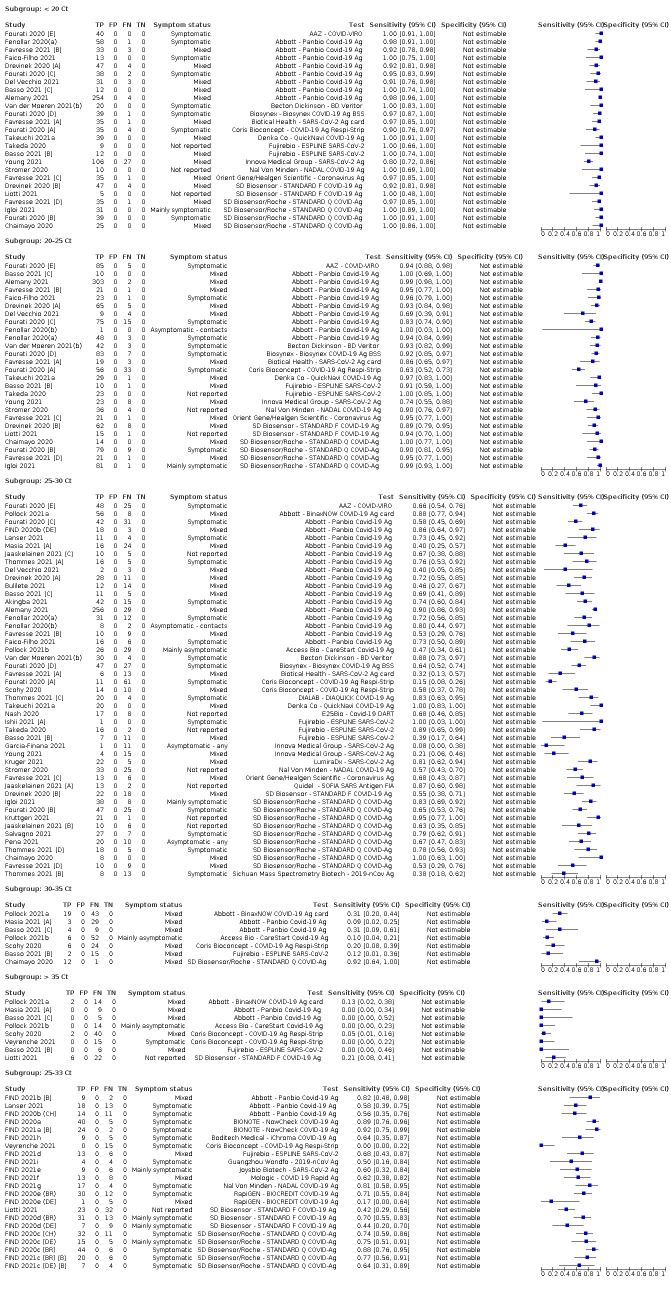
Both review authors independently performed data extraction of 2x2 contingency tables of the number of true positives, false positives, false negatives and true negatives. They resolved disagreements by discussion. Where possible, we separately extracted data according to symptom status (symptomatic, asymptomatic, mixed symptom status or not reported), viral load as defined per study (either in subgroups by Ct values or RNA copies/mL), time post‐symptom onset (week one versus week two), and for children (≤ 16 years or ≤ 18 years) and adults. We extracted information about accuracy according to SARS‐CoV‐2 variant where reported, however we did not identify any information about participant vaccination status in the included studies. For categorization by symptom status, we classed studies reporting at least 75% of participants as symptomatic (or asymptomatic) as ‘mainly symptomatic' (or ‘mainly asymptomatic’), we considered studies with less than 75% symptomatic participants to report ‘mixed’ groups along with those that reported recruiting both symptomatic and asymptomatic participants but did not provide the percentages in each group. We considered studies that provided no information as to the symptom status of included participants ‘not reported’. We also coded evaluations according to compliance with manufacturer IFUs. We based coding on three aspects of testing:
sample type (use of any sample not explicitly mentioned on the IFU scored 'No', otherwise scored 'Yes'),
for evaluations using samples that had been stored in viral transport medium (VTM) only (scored 'Yes' if specific instructions were provided and conditions were met; scored 'Unclear' if no instructions for use of samples in VTM were provided in the IFU; scored 'No' if instructions provided were not followed); and
timing between sample collection and testing (scored 'Yes' only if all tests were carried out within the specified time period, e.g. immediate on‐site testing, or for testing in laboratories if all tests reported to have been carried out within the specified time period; scored 'Unclear' if time frame for testing was not reported and 'No' if any testing was carried out beyond the maximum stipulated timeframe).
We encourage study authors to contact us regarding missing details on the included studies (coviddta@contacts.bham.ac.uk).
Assessment of methodological quality
Two review authors independently assessed risk of bias and applicability concerns using the QUADAS‐2 (Quality Assessment tool for Diagnostic Accuracy Studies) checklist tailored to this review (Appendix 6; Whiting 2011). The two review authors resolved any disagreements by discussion.
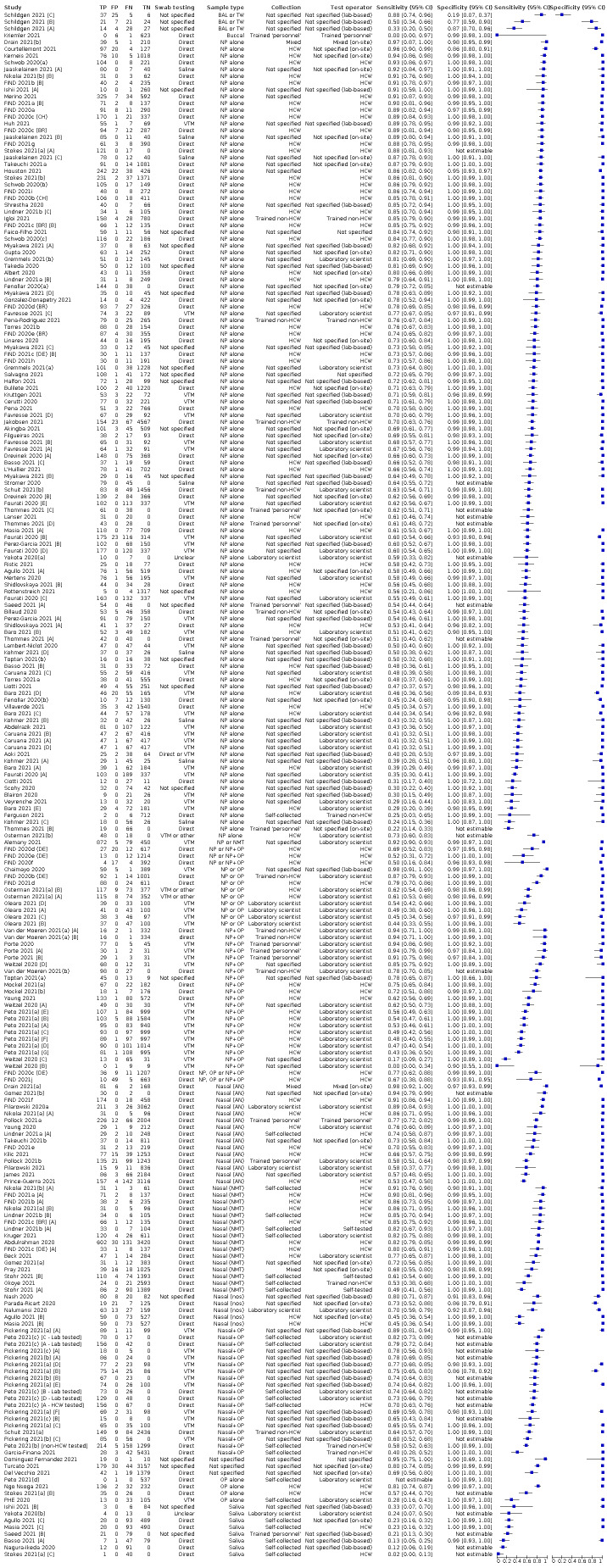
Ideally, studies examining the use of tests in symptomatic people should prospectively recruit a representative sample of participants presenting with signs and symptoms of COVID‐19, either in community or primary care settings or in a hospital setting, and they should clearly record the time of testing after the onset of symptoms. Studies in asymptomatic people at risk of infection should document time from exposure. Studies applying tests in a screening setting should document eligibility criteria for screening, particularly if a targeted approach is used and should take care to record any previous confirmed or suspected SARS‐CoV‐2 infection or any relevant epidemiological exposures. Studies should perform tests in their intended use setting, using appropriate samples with or without VTM and within the time period following specimen collection as indicated in the IFU document. Tests should be interpreted blinded to the final diagnosis (presence or absence of SARS‐CoV‐2). The reference standard diagnosis should be blinded to the result of the rapid test, and should not incorporate the result of the rapid test. We considered the use of a molecular assay to define the presence of SARS‐CoV‐2 infection to have a low risk of bias because a positive result can be taken to indicate the presence of infection, even if it does not reflect the time point in the course of infection. If the reference standard includes clinical diagnosis of COVID‐19 for RT‐PCR−negative patients, then established criteria should be used. Studies using pre‐pandemic samples for estimating specificity have a low risk of bias because samples are from participants known not to have COVID‐19. Those using contemporaneously collected samples have a higher risk of disease misclassification because of the inherent false negative rate of molecular tests such as RT‐PCR. For absence of infection, at least two RT‐PCR−negative tests are required to confirm the absence of infection for symptomatic participants but one negative RT‐PCR was considered sufficient for asymptomatic participants. Data should be reported for all study participants, including those where the result of the rapid test was inconclusive, or participants in whom the final diagnosis of COVID‐19 was uncertain. Studies should report whether results relate to participants (one sample per participant), or samples (multiple samples per participant).
Statistical analysis and data synthesis
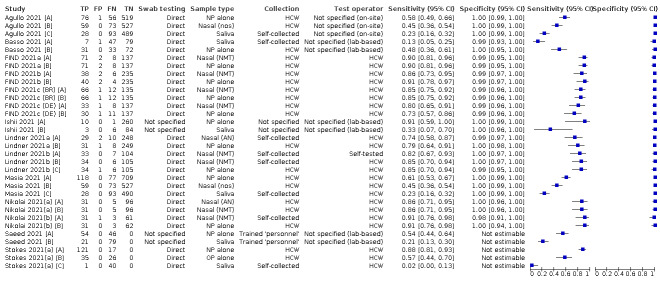
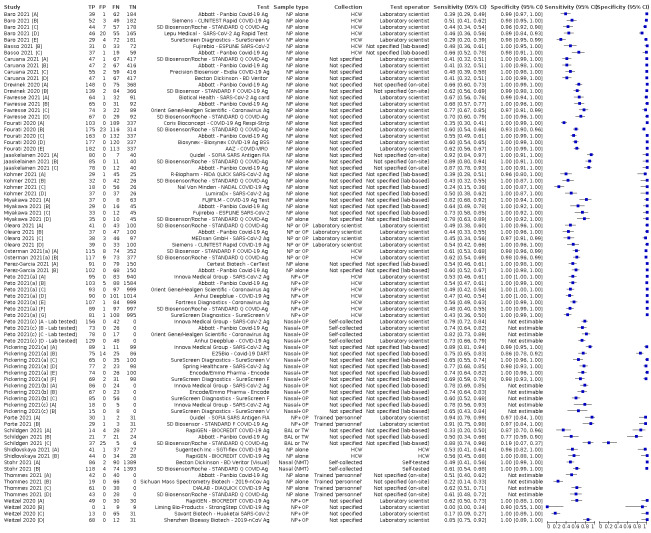
Studies sometimes referred to ‘samples’ rather than ‘patients’, however we do not suspect that inclusion of multiple samples per study participant was a significant issue. For consistency of terminology throughout the review, we refer to results on a per‐sample basis. If studies evaluated multiple tests in the same samples, we included them multiple times. We present estimates of sensitivity and specificity per study for each test brand using paired forest plots, and summarize results using average sensitivity and specificity in tables as appropriate. As heterogeneity is apparent in many analyses, these point estimates must be interpreted as the average of a distribution of values.
We estimated summary sensitivities and specificities with 95% confidence intervals (CI) using the bivariate model (Chu 2006; Reitsma 2005), via the meqrlogit command of Stata/SE 17.0. When few studies were available, we simplified models by first assuming no correlation between sensitivity and specificity estimates and secondly by setting near‐zero variance estimates of the random effects to zero (Takwoingi 2017). In cases where there was only one study per test, we reported individual sensitivities and specificities with 95% CI constructed using the binomial ‘exact’ (Clopper‐Pearson) method (Clopper 1934).
Where studies presented only estimates of sensitivity or of specificity, we fitted univariate, random‐effects, logistic regression models. In a number of instances where there was 100% sensitivity or specificity for all evaluations or there were fewer than three studies with highly similar sensitivity or specificity, we computed estimates and 95% CIs by summing the counts of true positives, false positives, false negatives and true negatives across 2x2 tables. These analyses are clearly marked in the tables. We present all estimates with 95% confidence intervals.
Where the same set of studies evaluated different symptom status, age, sample types, or test brands, on the same group of patients, we made direct comparisons using bivariate models that included indicator variables. Our ability to make formal comparisons between antigen assay brands was limited by the small number of studies making direct comparisons of the same tests.
Investigations of heterogeneity
We examined heterogeneity between studies by visually inspecting the forest plots of sensitivity and specificity. Where adequate data were available, we investigated heterogeneity related to symptom status, study setting, reporting of possible epidemiological exposure (asymptomatic contacts compared to any asymptomatic individual tested), time post‐symptom onset or post‐contact with a confirmed case, sample site, age, viral load, test brand, and assay format by including indicator variables in the random‐effects logistic regression models. We obtained absolute differences in sensitivity or specificity and corresponding P values post‐estimation by using the model parameters and the nlcom command in Stata. In instances where only one study was available per test or when tests were being directly compared following summing of counts of the 2x2 tables, we performed test comparison using the two‐sample test of proportions.
Sensitivity analyses
We estimated overall summary sensitivities and specificities restricted to studies using single group designs. We also estimated summary sensitivities and specificities according to test brand and symptom status using only studies that were compliant to the IFU. We estimated sensitivity with and without studies that only evaluated samples with RT‐PCR−confirmed SARS‐CoV‐2 (and thus did not estimate specificity). We performed the same analysis for specificity in studies that only evaluated RT‐PCR−negative control samples.
Assessment of reporting bias
Because of uncertainty about the determinants of publication and other sources of reporting bias for diagnostic accuracy studies and the inadequacy of tests for detecting funnel plot asymmetry, we made no formal assessment of reporting bias.
Updating
We are aware of additional studies published since the electronic searches were conducted on 4 March 2021 and plan to update this review imminently. We have already conducted the next search to 14 October 2021.
Results
Results of the search
We screened 3952 unique records (published or preprints) for inclusion in this review update and for the forthcoming update of the rapid point‐of‐care molecular tests review. Of 486 records selected for further assessment, we assessed 235 reports, 166 of which reported studies that were eligible for inclusion in this review update. See Figure 1 for the PRISMA flow diagram of search and eligibility results (McInnes 2018).
1.
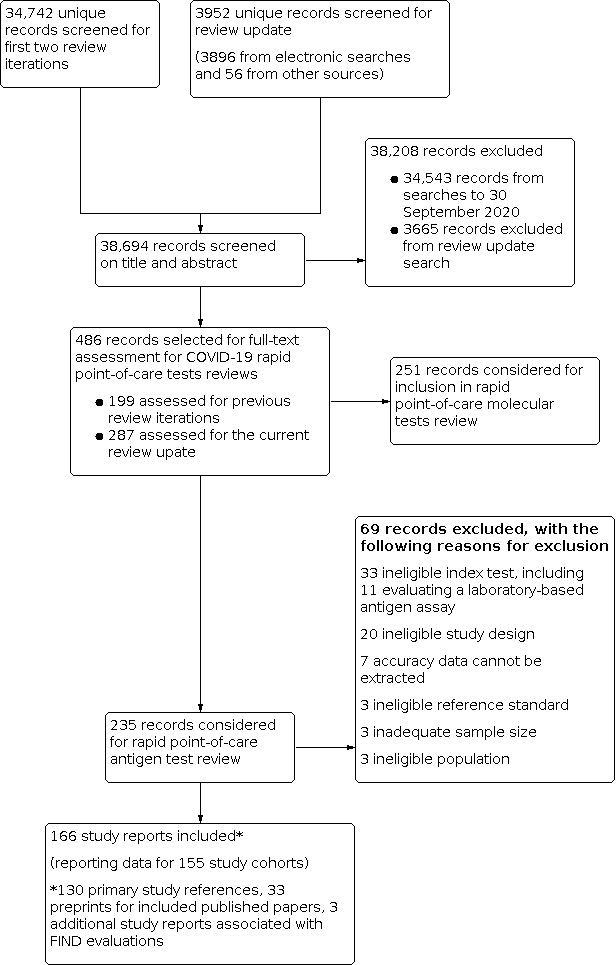
Study flow diagram
Of the 166 eligible study reports, 130 were primary study reports and 36 were secondary publications (for example preprints associated with published papers or journal papers associated with FIND evaluation reports). Of the 130 primary study reports, 87 were published journal papers, 24 were available only as preprints, and 19 were publicly available reports either from independent reference laboratories (one from Public Health England and two identified via the SMF) or were independent evaluations co‐ordinated by FIND (n = 16). We excluded 69 publications that did not meet our inclusion criteria. Exclusions were mainly based on index test (n = 33, including 11 evaluating a laboratory‐based antigen detection assay) or ineligible study designs (n = 20), for example, designs that did not allow estimation of test accuracy. One previously included preprint was excluded as we could not determine whether the test evaluated was the same as the subsequently commercially available assay (Diao 2020). The reasons for exclusion of all 69 publications are provided in Characteristics of excluded studies.
For this iteration of the review we contacted the authors of 14 study reports for further information (Abdelrazik 2021; Abdulrahman 2020; Basso 2021 [A]; Caruana 2021 [A]; Faico‐Filho 2021; Igloi 2021; Jakobsen 2021; Kriemler 2021; L'Huillier 2021; Schwob 2020(a); Torres 2021a; Torres 2021b; Bello‐Chavolla 2021; Regev‐Yochay 2021), and received replies in regard to eight studies.
The 130 primary study reports provide data for 155 separate cohorts of participants (henceforth referred to as 'studies'. Please note when naming studies, we use the letters [A], [B], [C] etc. in square brackets to indicate data on different tests evaluated in the same study and (a), (b), (c) to indicate data from different participant cohorts from the same study report. For example, the 16 included reports from FIND correspond to 22 ‘studies’ because six reports separately provided data from more than one evaluation centre.
Of the 155 studies, 152 reported data for a single application of a rapid antigen test, three (Love 2021; Smith 2021; Winkel 2020), reported data for repeated testing of the same individuals over time, and one provided data for both a single test application and for repeat testing at a second testing time point (Kriemler 2021). The main results, Tables and Figures focus on the single test application studies, with results for repeated testing considered separately.
Description of included studies
The 152 studies reporting single test applications included a total of 100,462 unique samples, with 16,822 samples with RT‐PCR−confirmed SARS‐CoV‐2 (some samples were analysed by more than one index test). Summary study characteristics are presented in Table 2 with further details of study design and index test details in Appendix 7 and Appendix 8. Full details per study are provided in the Characteristics of included studies.
1. Description of studies.
| Participants | No. of studies (%) | |
| Number of studies | 152 | |
| Sample size (all studies) | Median (IQR) | 326 (149, 744.5) |
| Range | 17, 5504 | |
| Total | 100462 | |
| Number of COVID‐19 cases (all studies) | Median (IQR) | 83.5 (45, 135) |
| Range | 0, 951 | |
| Total | 16822 | |
| Setting | COVID‐19 test centre | 67 (44.1) |
| ED/Urgent care | 11 (7.2) | |
| Hospital inpatient (* 1 includes outpatients) | 12 (7.2) | |
| Hospital – any (including staff or patients) | 9 (5.9) | |
| Laboratory‐based | 16 (10.5) | |
| Mixed | 6 (4.0) | |
| School or university‐based | 6 (4.0) | |
| Screening (HCWs 2, general public 3) | 5 (3.3) | |
| Shared living facility | 4 (2.6) | |
| Quarantine centre | 1 (0.7) | |
| Unclear | 15 (9.9) | |
| Symptom status | Symptomatic | 55 (36.2) |
| Mainly symptomatic | 18 (11.8) | |
| Asymptomatic | 13 (8.6) | |
| Mainly asymptomatic | 4 (2.6) | |
| Mixed | 41 (27.0) | |
| Not reported | 21 (13.8) | |
| Study design | ||
| Recruitment structure | Single group – sensitivity and specificity | 109 (71.7) |
| Two or more groups ‐ sensitivity and specificity | 20 (13.2) | |
| Single group – sensitivity only | 16 (10.5) | |
| Single group – specificity only | 1 (0.7) | |
| Randomized trial | 1 (0.7) | |
| Unclear | 5 (3.3) | |
| Reference standards | ||
| Reference standard for COVID‐19 cases | All PCR positive | 150 (98.7) |
| TMA | 1 (0.7) | |
| Not applicable (controls only study) | 1 (0.7) | |
|
Reference standard for non‐COVID‐19 cases |
No. of studies = 136 | |
| Single negative PCR | 133 (97.8) | |
| Single negative TMA | 1 (0.7) | |
| Pre‐pandemic samples | 2 (1.5) | |
| Reference standard samples | ||
| Paired swabs (same sample site as index) | 76 (50.0%) | |
| Paired swabs (alternative sample site to index) | 26 (17.1) | |
| Same sample for both index and reference tests | 49 (32.2) | |
| Unclear | 1 (0.7) | |
| Tests | No. of evaluations (%) | |
| Total number of test evaluations | 210 | |
| Number of tests per study | 1 | 129 (84.9) |
| 2 | 7 (4.6) | |
| 3 | 4 (2.6) | |
| 4 | 8 (5.3) | |
| 5 | 2 (1.3) | |
| 6 | 1 (0.7) | |
| 7 | 1 (0.7) | |
| Assay format | CGIA | 156 (74.3) |
| FIA | 20 (9.5) | |
| LFA (ALP) | 10 (4.8) | |
| LFA (latex conjugated) | 2 (1.0) | |
| LFA (not otherwise specified) | 18 (8.6) | |
| Microfluidic FIA | 4 (1.9) | |
| Sample type | Includes NP (all participants) | 141 (67.1) |
| NP alone | 118 (56.1) | |
| NP+OP | 20 (9.5) | |
| NP or NP+OP | 3 (1.4) | |
| Incudes NP (some participants) | 12 (5.7) | |
| NP or OP | 9 (4.3) | |
| NP, OP or NP+OP | 2 (1.0) | |
| NP or NMT | 1 (0.5) | |
| Includes nasal | 44 (20.9) | |
| Nasal+OP | 19 (9.1) | |
| Nasal (AN) | 13 (6.2) | |
| Nasal (NMT) | 9 (4.3) | |
| Nasal (not otherwise specified) | 3 (1.4) | |
| Other sample sites | 10 (4.8) | |
| OP alone | 3 (1.4) | |
| Saliva | 3 (1.4) | |
| BAL or TW | 3 (1.4) | |
| Buccal | 1 (0.5) | |
| Sample site not specified | 3 (1.4) | |
| Sample testing | Direct testing | 113 (53.8) |
| VTM | 60 (28.6) | |
| Saline | 8 (3.8) | |
| VTM or other | 3 (1.4) | |
| Direct or VTM | 1 (0.5) | |
| Not specified | 25 (12.0) | |
| IFU compliance | No | 81 (38.6) |
| Yes | 90 (42.9) | |
| Unclear | 39 (18.6) | |
| Sample collection | HCW | 73 (34.8) |
| Self‐collected | 16 (7.6) | |
| Trained 'personnel' | 11 (5.2) | |
| Trained non‐HCW | 9 (4.3) | |
| Laboratory scientist | 9 (4.3) | |
| Mixed | 3 (1.4) | |
| Not specified | 89 (42.4) | |
| Sample testing | Laboratory scientist | 66 (31.4) |
| HCW | 50 (23.8) | |
| Trained non‐HCW | 7 (3.3) | |
| Trained 'personnel' | 2 (1.0) | |
| Self‐tested | 2 (1.0) | |
| Mixed (on‐site) | 2 (1.0) | |
| Not specified (laboratory‐based) | 47 (22.4) | |
| Not specified (on‐site) | 31 (14.8) | |
| Not specified (no details) | 3 (1.4) | |
| ALP: alkaline phosphatase; AN: anterior nasal; BAL: bronchoalveolar lavage; CGIA: colloidal gold immunoassay; ED: emergency department; FIA: fluorescent immunoassay; HCW: healthcare worker; IQR: inter‐quartile range; LFA: lateral flow assay; NMT: nasal midturbinate; NP: nasopharyngeal; OP: oropharyngeal; PCR: reverse transcription polymerase chain reaction; TMA: transcription‐mediated amplification; TW: throat wash; VTM: viral transport medium | ||
The median sample size of the included studies is 326 (interquartile range (IQR) 149 to 744.5) and median number of SARS‐CoV‐2‐confirmed samples included is 83.5 (IQR 45 to 135). Two‐thirds of the studies (101/152, 66%) were conducted in Europe, 17 in Asia, including one conducted in Russia (11%), 15 in North America (10%), 13 in South America (9%), and three in Africa. Two studies included samples from more than one country and in one, the country of sample origin was unclear.
Participant characteristics
Just over half of studies (78/152, 51%) were conducted at COVID‐19 test centres (67, 44%) or at emergency or urgent care departments (11, 7%). Twenty‐one studies were carried out in other hospital settings; including 11 in hospital inpatients, one including both inpatients and outpatients, and nine in patients, visitors, and staff. Six studies conducted screening in schools or universities, two reported screening of healthcare workers, and three conducted screening of the general population (defined as widely available testing with deliberate advertising to target community‐wide populations). Four studies were conducted in shared living facilities (Dominguez Fernandez 2021; Kohmer 2021 [A]; PHE 2020; Toptan 2021(b)) and one in a quarantine centre as part of contact tracing (Shrestha 2020). Sixteen studies (11%) selected samples from those submitted to laboratories for routine RT‐PCR testing often (n = 7) with limited detail of the participants providing the samples (‘laboratory‐based’ studies). In six studies samples were included from multiple settings, and in the remaining 15 studies (10%) the selection of participants was not clearly reported.
Nearly half of the studies were conducted in symptomatic (55, 36%) or mainly symptomatic (18, 12%) populations. Seventeen studies (11%) were carried out in predominantly asymptomatic populations including any asymptomatic (9, 6%; Baro 2021 [A]; Ferguson 2021; Garcia‐Finana 2021; Kriemler 2021; Okoye 2021; Pena 2021; Peto 2021(d); Pilarowski 2021; Rottenstreich 2021), asymptomatic contacts of confirmed cases (3, 2%; Fenollar 2020(b); Shrestha 2020; Torres 2021a), or mainly asymptomatic population (5, 3%; James 2021; Nalumansi 2020; Pollock 2021b; Pray 2021; Prince‐Guerra 2021). The settings for testing asymptomatic people included: COVID‐19 test centres (n = 4; Pilarowski 2021; Torres 2021a; Pollock 2021b; Prince‐Guerra 2021), schools or universities (n = 4; Ferguson 2021; Kriemler 2021; Okoye 2021; Pray 2021), hospital settings (n = 4; including women admitted for delivery (n = 1; Rottenstreich 2021), healthcare worker screening (n = 2; James 2021; Peto 2021(d)), or participants not described (n = 1; Nalumansi 2020)), general public ‘mass’ screening (n = 3; Baro 2021 [A]; Garcia‐Finana 2021; Pena 2021), a quarantine centre (n = 1; Shrestha 2020), or patient contacts for whom the setting for testing was not clearly reported (n = 1).
Forty‐one studies were conducted in populations with mixed symptom status (27%), 20 of which did not report data separately for symptomatic and asymptomatic participants. Twenty‐one studies did not provide information about symptom status for all included participants (13%): five included samples from a COVID‐19 test centre (FIND 2021g; FIND 2021h; FIND 2021i; Gremmels 2021(b); Jaaskelainen 2021 [A]), two were conducted in shared living facilities (Kohmer 2021 [A]; Toptan 2021(b)), and in 14 the setting for testing could not be derived, including seven that could be identified as laboratory‐based studies.
One study deliberately included 23 RT‐PCR positive swabs from people infected with the B.1.1.7 (Alpha) variant (Pickering 2021(c) [A]; Pickering 2021(c) [B]). No effect on test accuracy was observed however sample numbers were small. This was the only study to consider the effect of SARS‐CoV‐2 variant on sensitivity. None of the studies included to date reported information about vaccination status of recruited participants.
Study design and reference standards
Overall, 72% of studies (n = 109) used a ‘single group’ design to estimate both sensitivity and specificity and 13% (n = 20) used a ‘two group’ design with separate selection of RT‐PCR−positive and RT‐PCR−negative samples. One study, by Stohr and colleagues, randomized participants between two different RDTs (Stohr 2021 [A]; Stohr 2021 [B]). We could not determine study design from the information in five study reports (3%; Aoki 2021; Dominguez Fernandez 2021; Huh 2021; Liotti 2021; Nash 2020). Sixteen studies included only samples with confirmed SARS‐CoV‐2, thus only allowing estimation of sensitivity, and one study included only SARS‐CoV‐2‐negative samples allowing estimation of specificity only.
All but two studies defined the presence or absence of SARS‐CoV‐2 infection based on RT‐PCR; one was the ‘specificity only’ study and another used a transcription‐mediated amplification (TMA) assay instead of RT‐PCR as the reference standard. One hundred and thirty‐three (97.8%) studies used a single RT‐PCR negative result to confirm absence of infection, including 98 studies in symptomatic or mixed symptom status populations. We did not find any studies requiring two negative RT‐PCR results for absence of infection. Two studies used pre‐pandemic samples (Fourati 2020 [A]; Veyrenche 2021), and one study used a negative TMA result (n = 1; Beck 2021).
One hundred and two studies (67%) obtained paired swabs for index and reference standard, 44 (29%) used the same swab for point‐of‐care and RT‐PCR tests and in six studies it was not possible to determine this information from the study report.
Index tests
The 152 studies reported a total of 228 separate test evaluations; 210 of these were comparisons by test brand (and contributed to the overall analysis for this review) and 18 were comparisons using the same test brand with samples from different sites. One hundred and twenty‐nine studies evaluated only one test and 23 studies compared two or more tests in the same participants (seven with two tests each, four with three tests, eight with four tests, two with five tests and one each with six or seven tests). The denominator for the index test details in Table 2 is the 210 evaluations by test brand.
The evaluations included 156 (74%) assessments of colloidal gold‐based immunoassays (CGIAs); 20 (10%) fluorescent immunoassays (FIAs), 10 using alkaline phosphatase‐labelled antibodies; two latex‐conjugated LFAs; and four microfluidic FIAs. We could not identify the LFA method for 18 evaluations, either from the study reports or IFUs. Studies evaluated 49 different commercially produced assays (3/49 were nasal kit versions of assays previously intended for naso‐ or oropharyngeal samples), documented with full assay identification details in Appendix 9. The study reports or manufacturer IFUs for all assays apart from 13 reported targeting the nucleocapsid protein (Appendix 9). We were unable to identify or obtain IFUs for six assays (those developed by e25bio, Encode/Emmo Pharma, Lepu Medical Technology, Savant Biotech, Sichuan Mass Spectrometry Biotechnology Co., and the SureScreen Diagnostics COVID‐19 Antigen Rapid Fluorescent Cassette) and the antigen target was not reported in the IFUs for the remaining seven tests.
Multiple combinations of sample types and use of direct swab testing or swabs in VTM or saline were reported across the evaluations. Based on the evaluations contributing to the overall analysis (n = 210), two‐thirds of evaluations (141, 67%) obtained nasopharyngeal samples in all participants, either alone (n = 118, 56%) or in combinations with oropharyngeal samples (23, 11%). A further 12 evaluations (6%) used either nasopharyngeal or oropharyngeal samples in included participants. Nasal samples were used in 44 evaluations: 19 evaluations (9%) used combined nasal and oropharyngeal samples, 13 (6%) used anterior nasal samples, nine (4%) used nasal mid‐turbinate samples, and three did not give details about the nasal sample obtained. Other sample sites were used in 10 evaluations (5%), including oropharyngeal alone (n = 3), saliva (n = 3), bronchoalveolar lavage or throat wash (n = 1) and buccal swabs (n = 10).
Fourteen studies provided an additional 18 evaluations of alternative sample sites or test interpretation that are not described in Table 2. Sampling sites included in these evaluations were: anterior nasal (2), nasal mid‐turbinate (7), or unspecified nasal samples (2), saliva (5), combined naso‐oropharyngeal (1) or oropharyngeal alone (1). The results of these evaluations are included in the comparisons by sample site in Table 3 and Appendix 10.
2. Summary of sensitivity and specificity results.
| Test | Evaluations; samples (cases) | Summary sensitivity % (95% CI) | Difference in sensitivity, (95% CI), P value | Summary specificity % (95% CI) | Difference in specificity, (95% CI), P value |
| Primary analysis | |||||
| All studies reporting sensitivity and specificity | 184; 117,372 (21,017) | 69.3 (66.2to72.3) | ‐ | 99.3 (99.2to99.3) | ‐ |
| All data for sensitivity (including ‘sensitivity‐only’ evaluations) | 209; 119843 (23,488) | 68.9 (66.0to71.7) | ‐ | ‐ | ‐ |
| All data for specificity (including ‘specificity‐only’ cohort) | 185; 117910 (21,017) | ‐ | ‐ | 99.3 (99.2to99.3) | ‐ |
| Secondary analyses by symptom status | |||||
| Symptomatic | 109; 50,574 (11,662) | 73.0 (69.3 to 76.4) | Ref | 99.1 (99.0 to 99.2)a | Ref |
| Asymptomatic | 50; 40,956 (2641) | 54.7 (47.7 to 61.6) | −18.2 (−26.1to −10.4), P < 0.0001 | 99.5 (99.4 to 99.6) | 0.4 (0.3to |
| Mixed symptoms or not reported | 56; 19,359 (5344) | 70.6 (64.8 to 75.8) | Comparison not done | 99.4 (99.3 to 99.5) | Comparison not done |
| Restricted to studies including both symptomatic and asymptomatic participants | |||||
| Symptomatic | 34; 13,757 (3243) | 75.9 (69.7 to 81.2) | Ref | 99.0 (98.8 to 99.2) | Ref |
| Asymptomatic | 34; 25,827 (1666) | 56.8 (48.5 to 64.7) | −19.2 (−29.1 to −9.2), P < 0.0001 | 99.5 (99.4 to 99.6) | 0.5 (0.3to |
| Including data from ‘sensitivity‐only’ evaluations | |||||
| Symptomatic | 132; 52,939 (14,027) | 72.4 (69.1 to 75.5) | Ref | ‐ | ‐ |
| Asymptomatic | 56; 41,129 (2814) | 49.4 (44.5 to 54.2) | −23.1 (−26.5 to −19.6), P < 0.0001 | ‐ | ‐ |
| Mixed symptoms or not reported | 63; 19,936 (5921) | 70.6 (65.4 to 75.3) | Comparison not done | ‐ | ‐ |
| Subgroup analyses for symptomatic participants | |||||
| By setting for testing | |||||
| COVID‐19 test centre | 47; 23,602 (4369) | 82.8 (80.2 to 85.2) | Ref | 99.1 (99.0 to 99.2) | Ref |
| Hospital inpatient | 20; 11,903 (2536) | 51.6 (45.9 to 57.2) | −31.3 (−37.5to −25.1), P < 0.0001 | 99.6 (99.4 to 99.7) | 0.5 (0.3to |
| ED/urgent care | 15; 5607 (1312) | 70.9 (55.8 to 82.5) | −11.9 (−25.7 to 1.9), P = 0.090 | 99.2 (98.9 to 99.4) | 0.1 (−0.2to= 0.497 |
| Laboratory‐based | 9; 4144 (2161) | 57.2 (42.5 to 70.7) | −25.6 (−40.3 to −10.9), P = 0.001 | 98.7 (98.1 to 99.1) | −0.4 (−0.9to |
| Hospital ‐ any | 3; 1175 (418) | 52.8 (37.7 to 67.5) | −30.0 (−45.6to −14.4), P < 0.0001 | 97.8 (96.4 to 98.6) | −1.4 (−2.4to −0.3), P = 0.013 |
| School/university | 2; 374 (114) | 67.1 (46.4 to 82.8) | −15.7 (−34.8to = 0.107 | 97.7 (95.0 to 99.0) | −1.4 (−3.2 to0.4), P = 0.130 |
| Hospital in‐ or outpatient | 1; 1384 (116) | 66.4 (57.0 to 74.9) | ‐ | 98.8 (98.1 to 99.3) | ‐ |
| Screening – HCW | 1; 115 (24) | 83.3 (62.6 to 95.3) | ‐ | 100 (96.0 to 100) | ‐ |
| Shared living facility | 1; 30 (20) | 95.0 (75.1 to 99.9) | ‐ | 100 (69.2 to 100) | ‐ |
| Setting not reported | 5; 519 (204) | 67.7 (41.7 to 86.0) | ‐ | 83.4 (36.0 to 97.8) | ‐ |
| Mixed settings | 5; 1721 (388) | 80.7 (66.6 to 89.7) | ‐ | 98.3 (97.5 to 98.9) | ‐ |
| By time after symptom onset | |||||
| Week 1 | 30; 15,323 (2408) | 80.9 (76.9 to 84.4) | Ref | 99.5 (99.3 to 99.6) | Ref |
| Week 2 | 13; 903 (224) | 48.9 (37.9 to 60.1) | −32.0 (−43.9 to −20.1), P < 0.0001 | 99.3 (98.2 to 99.7) | −0.2 (−0.9 to 0.4), P = 0.515 |
| Including data from ‘sensitivity‐only’ evaluations | |||||
| Week 1 (sensitivity) | 72; 18,555 (5640) | 82.2 (79.2 to 85.0) | Ref | ‐ | ‐ |
| Week 2 (sensitivity) | 40; 1798 (1119) | 53.8 (48.0 to 59.6) | −28.4 (−32.6to −24.2), P < 0.0001 | ‐ | ‐ |
| Subgroup analyses for asymptomatic participants | |||||
| By eligibility for testing | |||||
| Testing widely available to any asymptomatic person | 26; 31,904 (1758) | 49.6 (42.1 to 57.1) | Ref | 99.6 (99.5 to 99.7) | Ref |
| Testing of contacts or referred groups only | 16; 7677 (703) | 64.3 (54.6 to 73.0) | 14.7 (2.7to = 0.016 | 99.7 (99.5 to 99.8) | 0.06 (‐0.09to= 0.444 |
| No details about eligibility for testing | 6; 242 (64) | 44.5 (26.7 to 63.8) | −5.1 (−26.0to = 0.632 | 85.4 (79.4 to 89.9) | −14.2 (−19.4to −9.0), P < 0.0001 |
| Including sensitivity‐only cohorts | |||||
| Testing available to any asymptomatic person | 26; 31,904 (1758) | 49.5 (41.6 to 57.4) | Ref | ‐ | ‐ |
| Testing of contacts or referred groups only | 21; 7815 (841) | 61.7 (52.4 to 70.2) | 12.2 (0.2to = 0.047 | ‐ | ‐ |
| No details about eligibility for testing | 7; 277 (99) | 47.1 (30.1 to 64.8) | −2.4 (−22.2to = 0.811 | ‐ | ‐ |
| By study setting | |||||
| COVID‐19 test centre | 18; 19,253 (1195) | 61.5 (54.0 to 68.4) | Ref | 99.6 (99.5 to 99.7) | Ref |
| Screening – mass | 7; 7776 (648) | 45.1 (36.4 to 54.1) | −16.4 (−27.9to −4.9), P = 0.005 | 99.4 (99.2 to 99.6) | −0.2 (−0.4to −0.01), P = 0.042 |
| School/university | 5; 5174 (96) | 47.9 (38.1 to 57.9) | −13.6 (−25.9to −1.2), P = 0.031 | 99.6 (99.4 to 99.7) | −0.04 (−0.2to = 0.673 |
| Hospital inpatient | 5; 2282 (105) | 35.2 (26.7 to 44.8) | −26.2 (−37.9to −14.6), P < 0.0001 | 100 (99.7 to 100) | 0.3 (0.2to |
| ED/urgent care | 2; 2547 (85) | 95.1 (7.3 to 100) | 33.6 (7.0to = 0.013 | 99.6 (99.2 to 99.8) | −0.04 (−0.3to = 0.796 |
| Laboratory‐based | 2; 532 (131) | 58.2 (21.8 to 87.4) | −3.3 (−43.1to 36.5), P = 0.871 | 98.8 (97.0 to 99.5) | −0.9 (−2.0to |
| Hospital ‐ any | 1; 262 (90) | 70.0 (59.4 to 79.2) | 92.4 (87.4 to 95.9) | ||
| Quarantine | 1; 113 (47) | 85.1 (71.7 to 93.8) | ‐ | 100 (94.6 to 100) | ‐ |
| Screening – HCW | 1; 2224 (128) | 51.6 (42.6 to 60.5) | ‐ | 99.9 (99.6 to 100) | ‐ |
| Screening – traveller | 1; 145 (5) | 40.0 (5.3 to 85.3) | ‐ | 100 (97.4 to 100) | ‐ |
| Unclear | 5; 284 (62) | 47.8 (28.3 to 67.9) | ‐ | 77.8 (44.0 to 94.0) | ‐ |
| Mixeda | 2; 364 (49) | 36.7 (23.4 to 51.7) | ‐ | 100 (98.8 to 100) | ‐ |
| By time after confirmed contact | |||||
| Week 1 | 3; 1013 (110) | 70.0 (60.8 to 77.8) | Ref | 99.8 (99.1 to 99.9) | Ref |
| Week 2 | 3; 747 (61) | 60.7 (48.0 to 72.0) | −9.3 (−24.3 to 5.6), P = 0.221 | 99.3 (98.3 to 99.7) | −0.5 (−1.2 to 0.2), P = 0.159 |
| Additional subgroup and sensitivity analyses across all participants regardless of symptom status | |||||
| Sensitivity analysis by study design | |||||
| Restricted to single group | 126; 93970 (14171) | 70.8 (67.2 to 74.3) | 99.4 (99.3 to 99.4) | ||
| Subgroup analysis by sample type (based on all 228 evaluations; data here for studies reporting both sensitivity and specificity) | |||||
| Includes NP (all participants) | 128; 59,447 (13,270) | 69.0 (65.3 to 72.4) | Ref | 99.4 (99.3 to 99.4) | Ref |
| Nasal (all) | 34; 33,128 (4032) | 76.6 (70.3 to 81.9) | 7.7 (0.9 to 14.5), P = 0.027 | 99.4 (99.3 to 99.4) | 0.01 (−0.1 to 0.1), P = 0.873 |
| Nasal + OP | 10; 12654 (1407) | 68.7 (55.1 to 79.6) | −0.3 (−13.2to 12.7), P = | 99.6 (99.5 to 99.7) | 0.3 (0.1 to0.4), P < 0.0001 |
| Includes NP (some participants) | 12; 7530 (1994) | 71.8 (59.8 to 81.4) | 2.9 (−8.6to | 98.4 (98.0 to 98.7) | −1.0 (−1.3to −0.6), P < 0.0001 |
| Saliva (all) | 3; 837 (184) | 20.5 (7.5 to 45.1) | −48.5 (−67.7to −29.3), P < 0.0001 | 99.8 (98.9 to 100) | 0.5 (0.2to 0.8), P = |
| OP alone | 2; 553 (214) | 57.4 (26.6 to 83.4) | −11.5 (−43.8to 20.7), P = 0.484 | 99.4 (97.7 to 99.9) | 0.1 (−0.8to 0.9), P = |
| Other | 4; 849 (127) | 55.4 (25.6 to 81.7) | ‐ | 83.7 (36.8 to 97.8) | ‐ |
| Not specified | 3; 4881 (304) | 79.3 (68.7 to 86.9) | ‐ | 99.3 (99.0 to 99.5) | ‐ |
| Direct comparisons by sample type | |||||
| NP alone | 9; 2979 (682) | 80.9 (70.5 to 88.3) | Ref | 99.6 (99.2 to 99.8) | Ref |
| Nasal (any type) | 9; 2710 (619) | 78.1 (66.7 to 86.4) | −2.9 (−16.1 to 10.4), P =0.672 | 99.6 (99.2 to 99.8) | 0.0 (−0.4to 0.3), P = |
| NP alone | 6; 1134 (316) | 56.7 (44.3 to 68.3) | Ref | 99.5 (99.1 to 99.7) | Ref |
| Nasal (NMT) | 6; 1133 (316) | 64.4 (52.2 to 75.0) | 7.8 (−9.1 to 24.6), P = 0.367 | 97.5 (96.7 to 98.0) | −2.0 (−2.7 to −1.3), P < 0.001 |
| NP alone | 4; 1963 (402) | 58.5 (53.6 to 63.2) | Ref | 99.9 (99.5 to 100) | Ref |
| Saliva | 4; 1448 (305) | 21.6 (17.4 to 26.6) | −36.8 (−43.5 to −30.1), P < 0.0001 | 99.9 (99.4 to 100) | 0.0 (−0.2to 0.2), P = |
| NP alone (including sensitivity only cohorts) | 6; 2201 (640) | 66.5 (53.0 to 77.8) | Ref | ||
| Saliva (including sensitivity only cohorts) | 6; 1589 (446) | 17.1 (10.1 to 27.5) | −49.4 (−64.8to −34.0), P < 0.0001 | ||
| Nasal (not otherwise specified)b,c | 2; 1318 (264) | 44.7 (38.6 to 50.9)a | ‐ | 100 (99.7 to 100)a | ‐ |
| Salivab,c | 2; 1221 (242) | 23.1 (18.0 to 29.0)a | ‐ | 100 (99.6 to 100)a | ‐ |
| Nasal (AN) | 1; 132 (36) | 86.1 (70.5 to 95.3) | ‐ | 100 (96.2 to 100) | ‐ |
| Nasal (NMT) | 1; 132 (36) | 86.1 (70.5 to 95.3) | ‐ | 100 (96.2 to 100) | ‐ |
| Sensitivity analysis for accuracy in children | |||||
| Children (studies reporting sensitivity and specificity) | 10; 4652 (410) | 62.7 (52.7 to 71.7) | ‐ | 99.4 (99.1 to 99.6) | ‐ |
| Children (including sensitivity‐only cohorts) | 12; 4775 (533) | 62.1 (53.4 to 70.1) | ‐ | ‐ | ‐ |
| Children (including specificity‐only cohorts) | 12; 4766 (410) | ‐ | ‐ | 99.4 (99.1 to 99.6) | ‐ |
| Restricted to studies including both age groups | |||||
| Children | 6; 1835 (264) | 63.7 (48.0 to 76.9) | Ref | 99.0 (98.4 to 99.4) | Ref |
| Adults | 6; 10,007 (1047) | 73.5 (61.1 to 83.1) | 9.9 (−8.7to 28.4), P = 0.297 | 99.7 (99.6 to 99.8) | 0.7 (0.2 to 1.2), P =0.007 |
| Including sensitivity‐only cohorts | |||||
| Children | 7; 1839 (268) | 60.1 (45.1 to 73.4) | Ref | ‐ | ‐ |
| Adults | 7; 10,103 (1143) | 70.8 (58.3 to 80.8) | 10.8 (3.0to 18.5), P = 0.006 | ‐ | ‐ |
| Including specificity‐only cohorts | |||||
| Children | 9; 2517 (265) | ‐ | ‐ | 99.0 (98.5 to 99.4) | Ref |
| Adults | 9; 11,756 (1108) | ‐ | ‐ | 99.7 (99.6 to 99.8) | 0.7 (0.2to 1.1), P = |
| Subgroup analyses by viral load | |||||
| Viral load in subgroups by Ct valued | |||||
| Subgroup: < 20 Ct | 26; 1108 (1108) | 97.4 (95.0 to 98.6) | ‐ | ‐ | ‐ |
| Subgroup: 20‐25 Ct | 27; 1384 (1384) | 93.6 (90.0 to 96.0) | ‐ | ‐ | ‐ |
| Subgroup: 25‐30 Ct | 48; 1724 (1724) | 68.7 (61.6 to 75.0) | ‐ | ‐ | ‐ |
| Subgroup: 25‐33 Ct | 22; 564 (564) | 66.2 (55.8 to 75.1) | ‐ | ‐ | ‐ |
| Subgroup: > 30 Ct | 64; 2332 (2332) | 18.7 (14.2 to 24.1) | ‐ | ||
| Subgroup: 30‐35 Ct | 8; 247 (247) | 25.3 (12.2 to 45.3) | ‐ | ‐ | ‐ |
| Subgroup: > 35 Ct | 9; 142 (142) | 5.6 (1.9 to 15.7) | ‐ | ‐ | ‐ |
| Viral load in subgroups by RNA copy values | |||||
| Subgroup: ≥ 10^8 RNA copies/mL | 3; 80 (80) | 95.0 (87.4 to 98.1) | ‐ | ‐ | ‐ |
| Subgroup: ≥ 10^7 RNA copies/mL | 21; 608 (608) | 98.4 (97.0 to 99.1) | ‐ | ‐ | ‐ |
| Subgroup: ≥ 10^7 to < 10^8 RNA copies/mL | 3; 49 (49) | 93.9 (82.7 to 98.0) | ‐ | ‐ | ‐ |
| Subgroup: ≥ 10^6 to < 10^7 RNA copies/mL | 28; 597 (597) | 94.0 (89.8 to 96.6) | ‐ | ‐ | ‐ |
| Subgroup: ≥ 10^5 to < 10^6 RNA copies/mL | 31; 686 (686) | 70.9 (57.4 to 81.5) | ‐ | ‐ | ‐ |
| Subgroup: ≥ 10^4 to < 10^5 RNA copies/mL | 24; 582 (582) | 36.7 (24.7 to 50.5) | ‐ | ‐ | ‐ |
| Subgroup: < 10^4 RNA copies/mL | 24; 825 (825) | 7.5 (3.8 to 14.3) | ‐ | ‐ | ‐ |
| Subgroup analysis by assay format | |||||
| CGIA | 140; 95,926 (17,146) | 68.5 (65.1 to 71.7) | Ref | 99.4 (99.3 to 99.4) | Ref |
| FIA | 19; 6987 (1507) | 76.6 (68.2 to 83.4) | 8.2 (−0.1to 16.5), P = 0.054 | 97.5 (97.1 to 97.9) | −1.9 (−2.3to −1.4), P < 0.0001 |
| LFA (ALP) | 7; 1645 (411) | 61.4 (38.3 to 80.3) | ‐ | 99.8 (99.2 to 99.9) | ‐ |
| LFA (latex‐conjugated)c | 2; 2048 (156) | 81.3 (69.9 to 89.0) | ‐ | 100 (99.8 to 100)* | ‐ |
| Microfluidic FIA | 4; 1373 (343) | 89.7 (63.0 to 97.8) | ‐ | 98.5 (97.6 to 99.1) | ‐ |
| LFA (not otherwise specified) | 12; 9393 (1454) | 60.7 (45.0 to 74.4) | ‐ | 99.6 (99.4 to 99.7) | ‐ |
| ALP: alkaline phosphatase; AN: anterior nasal; CGIA: colloidal gold immunoassay; CI: confidence interval; ED: emergency department; FIA: fluorescent immunoassay; HCW: healthcare worker; LFA: lateral flow assay; NMT: nasal midturbinate; NP: nasopharyngeal; OP: oropharyngeal | |||||
aExcludes outlier with 8% specificity in 13 throat saliva or throat wash samples. b2x2 tables combined prior to calculating estimates. cSeparate pooling of sensitivity or specificity. dThe range in viral load associated with each group of cycle threshold (Ct) values is likely to vary considerably between study laboratories, for example, Ct 25‐30 can vary in RNA copies by as much as ~10^3/mL to ~10^7/mL.
More than half of studies used direct swab testing (113/210, 54%), 71 (34%) tested samples in VTM or saline, one study used either direct swab testing or VTM and 25 studies (12%), did not provide this information. IFUs for seven assays explicitly recommend against using any transport medium for swab testing (assays from Abbott (Panbio), Anhui Deepblue, Becton Dickinson, Dialab, PCL, Quidel and SD Biosensor; Appendix 9). Twelve assay IFUs provide some form of instructions for use of VTM, and 25 do not mention use of transport medium.
We considered 90 of 210 evaluations (43%) to be compliant with manufacturer IFUs in terms of sample type, use of VTM and time interval between collection and testing. Eighty‐one evaluations were judged not compliant with IFUs; 17 used VTM when it was not covered by the IFU, 28 tested samples not listed on the IFUs, and in 57 testing was not always conducted within the one‐hour time period specified in the IFU (including 32/57 using samples after a period of frozen storage) (these groupings are not mutually exclusive). We judged IFU compliance to be unclear for 39 evaluations, primarily because we could not determine the time interval between sample collection and testing (n = 27), or because we could not obtain the IFU for the assays evaluated (n = 7); other reasons included use of VTM when the IFU did not specify instructions for using VTM 3), or because the sample site was not reported (n = 1) or use of VTM was not clear (n = 1).
Samples were collected by healthcare workers in 73 (35%) evaluations, were self‐collected in 16 (8%), were collected by trained ‘personnel’ or non‐healthcare workers in 20 (10%) and by laboratory scientists in nine evaluations. In 89 (42%) the sample collection was not reported, and in three, individuals with different levels of expertise collected samples.
Samples were tested and interpreted by laboratory scientists in 66 evaluations (31%), by healthcare workers in 50 (24%), by trained ‘personnel’ or non‐healthcare workers in nine (4%) and were self‐tested in two evaluations. In 83 of the 210 evaluations the expertise of the test operator was not reported (n = 81, 39%) or was conducted by people with different levels of expertise (n = 2). Of the 83, 47 carried out testing in laboratories, 33 evaluations conducted testing on site and three provided no details of where the tests were conducted.
Methodological quality of included studies
Studies evaluating single test applications
We report the overall methodological quality assessed using the QUADAS‐2 tool for all included studies (n = 152) in Figure 2 (Whiting 2011). See Appendix 11 for a plot of study‐level ratings by quality domain. We explain how we reached these judgements in the Characteristics of included studies tables.
2.
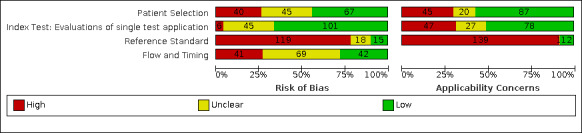
Risk of bias and applicability concerns graph for evaluations of single test applications: review authors' judgements about each domain presented as percentages across included studies. Numbers in the bars indicate the number of studies
We considered whether the findings of individual studies were at risk of bias, and whether there were concerns that results might not apply to standard use of the tests. We judged 37 studies to be at low risk of bias for all four domains assessed, four of which also had low concerns about applicability in all domains (Ferguson 2021; Garcia‐Finana 2021; Pilarowski 2021; Schuit 2021(b)). These four studies included primarily asymptomatic individuals attending community testing sites, such that a single negative RT‐PCR was considered adequate for confirming absence of infection (while two negative results were required for symptomatic participants), and RT‐PCR as the sole reference standard was considered sufficient to confirm presence of infection. In 24 of 152 studies the only concern in regard to risk of bias was that a single negative RT‐PCR was used to confirm absence of infection rather than the preferred two negative tests for those with signs or symptoms of infection.
Participant selection
We judged 67 studies (44%) to be at low risk of bias for participant sampling. High risk of bias was present in 40 (26%) studies because of deliberate sampling of participants based on the reference standard result (n = 37; including 20 two‐group studies and 17 that only included samples with either confirmed SARS‐CoV‐2 infection or absence of infection) or use of convenience sampling (n = 3). In 45 studies (30%) the risk of bias was unclear because of poor reporting of recruitment procedures or inclusion criteria (Figure 2).
We judged more than half (87/152, 57%) of studies to have selected an appropriate patient group, recruiting participants from COVID‐19 test centres, urgent care or emergency departments or identifying them through contact tracing. We had high concerns about the applicability of the selected participants in 30% of studies (45/152). Recruited participants were unlikely to be similar to those in whom the test would be used in clinical practice because of deliberate sampling based on PCR status (n = 37), use of frozen samples, or participants recruited from mixed/unclear settings (n = 8).
Index tests
Sixty‐six percent of studies had a low risk of bias for the index test (101/152). We judged six studies at high risk of bias because they did not implement blinding of index test interpretation to the reference standard result. Risk of bias was unclear in the remaining 45 (30%) studies because we could not judge whether interpretation of the index test was undertaken with knowledge of the reference standard result.
Just over half of all studies (78/152; 51%) conducted testing as would be expected in practice (low concern regarding applicability). We had high concerns about applicability in 31% of studies (47/152), because at least one test evaluated per study did not comply with manufacturer IFUs. We could not assess the applicability of the index test application for the remaining 27 studies.
Reference standards
Overall, 15 studies were at low risk of bias for the reference standard. High risk of bias was present in 119 studies (78%) because studies did not use an adequate reference standard (single negative RT‐PCR used to define absence of SARS‐CoV‐2 infection in symptomatic populations). Of those studies where we could not judge risk of bias (n = 18), 16 were conducted in predominantly asymptomatic populations and were considered to use an appropriate reference standard, but only one clearly reported blinding of the reference standard to the index test result. Overall, 59 studies reported reference standard blinding.
We had concerns about the applicability of the reference standard in 91% of studies (139/152) because of the reliance on PCR to define SARS‐CoV‐2 cases in symptomatic participants. These studies may have considered individuals who were RT‐PCR−negative but had exposure and clinical features that met the case definitions for COVID‐19 as disease negative. In 12 studies (8%) concerns about applicability were low because they included mainly asymptomatic individuals attending community screening. One study was rated as having unclear concerns for applicability.
Flow and timing
We judged 42 (28%) studies to have low risk of bias for participant flow and timing (Figure 2). Another 41 (27%) were at high risk of bias mainly because of exclusion of samples following invalid index test results (n = 40), including two studies that also reported delays between ‘paired’ swabs of up to three days, or different reference standards used (n = 1). We judged risk of bias to be unclear for 69 (45%) studies, because of lack of clarity about participant inclusion and exclusion from analyses with no missing data or indeterminate test results reported and no Standards for Reporting Diagnostic Accuracy Studies (STARD)‐style participant flow diagram and checklist (Bossuyt 2015), to fully report outcomes for all samples.
Conflicts of interest
In 109 studies all authors declared no conflicts of interest, although three of them were directly funded by the company that manufactured the test and nine of them received test kits from the manufacturer. Twenty‐four studies did not provide a conflict of interest statement, including 12 published studies. In 19 studies at least one author declared potential conflicts of interest in relation to the evaluation. The remaining studies were independent evaluations such as published by FIND, national reference laboratories, national Public Health Services, Ministry of Health or without any external funding.
Twenty‐three studies provided no funding statement, 30 reported no funding sources to declare (of which three studies reported that they received antigen tests from the test manufacturer) and the remainder (n = 99) reported one or more funding sources.
Studies evaluating repeated test applications
We report the overall methodological quality assessed using the QUADAS‐2 tool for all included studies that evaluated repeated test applications (n = 4) in Figure 3 (Whiting 2011); one of the four studies is also included in the methodological assessment of studies evaluating a single test application (Kriemler 2021). See Appendix 11 for a plot of study‐level ratings by quality domain. We explain how we reached these judgements in the Characteristics of included studies table.
3.
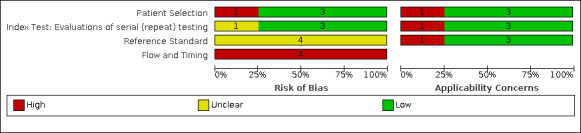
Risk of bias and applicability concerns graph for studies with repeat (serial) testing: review authors' judgements about each domain presented as percentages across included studies
We considered whether the findings of individual studies were at risk of bias, and whether there were concerns that results might not apply to standard use of the tests. None of the four studies were at low risk of bias for all four domains assessed, and one (Winkel 2020), evaluating repeated testing of Dutch footballers, had low concerns about applicability in all domains. In the same study the only concern in regard to risk of bias was that blinding of RT‐PCR interpretation to the rapid antigen test result was not clearly reported.
For participant selection, we rated all studies apart from Smith 2021 as having low risk of bias and low concerns for applicability. Smith and colleagues only included participants with PCR positive results and we therefore consider it to have high risk of bias and high concerns for applicability (does not reflect those who might undergo repeat antigen testing in any given population). Smith 2021 also differs from the other three studies in regard to index test risk of bias; all testing was carried out in a laboratory setting within 12 hours of collection and, although likely, it is not fully clear whether index test interpretation was conducted blinded to the result of the reference standard. In contrast only Kriemler 2021 had high concerns about applicability for the index test because the buccal swab used was not covered on the manufacturer IFU for the assay.
For the reference standard, all studies had unclear risk of bias because they did not report blinding of the reference standard to the index test result. Smith 2021 had high concerns for applicability of the reference standard because all participants were required to have positive viral culture. All studies were considered at high risk of bias because of exclusion of eligible participants from the analysis or because swabs for the index and reference standard were not all obtained within a 12‐hour interval of each other.
Findings
We first consider results from studies that evaluated a single application of a rapid antigen test, and then cover results from studies that evaluated repeated antigen testing in the same individuals over time.
Evaluations of single test application
Of the 152 included studies evaluating a single application of a rapid antigen test, 129 evaluated a single brand of antigen test (12/129 comparing sample types and 2/129 evaluating the effect of test operator), and 23 compared the accuracy of two or more different brands of antigen test in the same participants (Table 2). To include all results from all tests in these analyses we have treated results from different tests of the same participants within a study as separate data points, such that data are available on 210 evaluations by test brand and 228 test evaluations when we include comparisons by sample site.
The results tables identify where estimates are based on multiple assessments of the same samples by including both the number of test evaluations and the number of studies. Sixteen studies are ‘cases only’, reporting only sensitivity estimates and one includes only ‘non‐COVID‐19’ cases, reporting only specificity. Summary results are presented for studies providing both sensitivity and specificity data and then adding in the data from sensitivity‐ or specificity‐only evaluations. The numbers of true positives, false positives, and total samples with and without confirmed SARS‐CoV‐2 infection are based on test result counts.
We present results of the main analyses and heterogeneity investigations in Table 3, with additional summary results adding in data from ‘sensitivity‐only’ evaluations reported in Appendix 10. Forest plots of study data for the primary analysis (including all evaluations by test brand but excluding comparisons by sample type or test interpretation) are shown in Appendix 12.
Forest plots of data for subgroup analyses in symptomatic and asymptomatic populations are in Figure 4, Figure 5, Figure 6, and by timing of the test in relation to the course of infection in Figure 7 and Figure 8. Data for children are shown in Figure 9, and for subgroups by Ct value or RNA copies/mL in Figure 10 and Figure 11.
4.
Forest plot of data for antigen tests for symptomatic or mainly symptomatic populations. BR: Brazil; CH: Switzerland; DE: Germany; ED: emergency department; HCW: healthcare worker; Lab: laboratory
5.
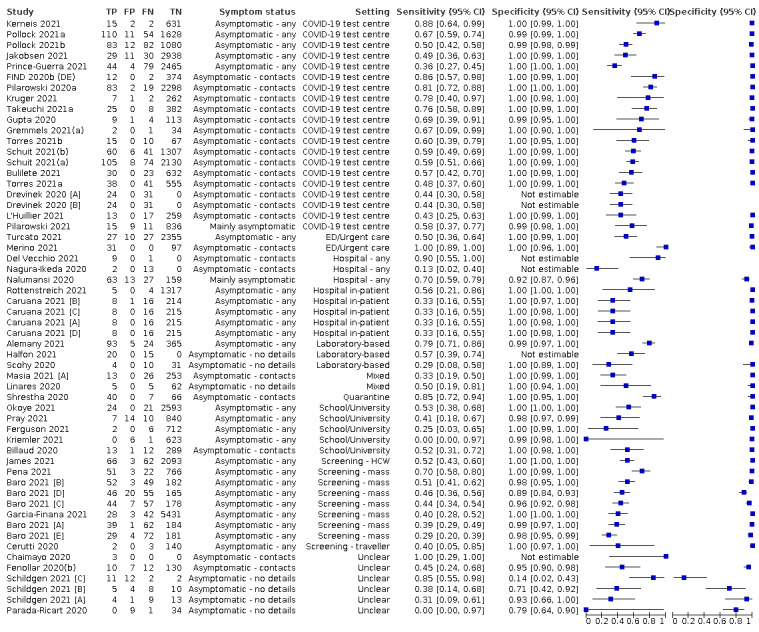
Forest plot of data for antigen tests for asymptomatic or mainly asymptomatic populations: DE: Germany; ED: emergency department; HCW: healthcare worker
6.
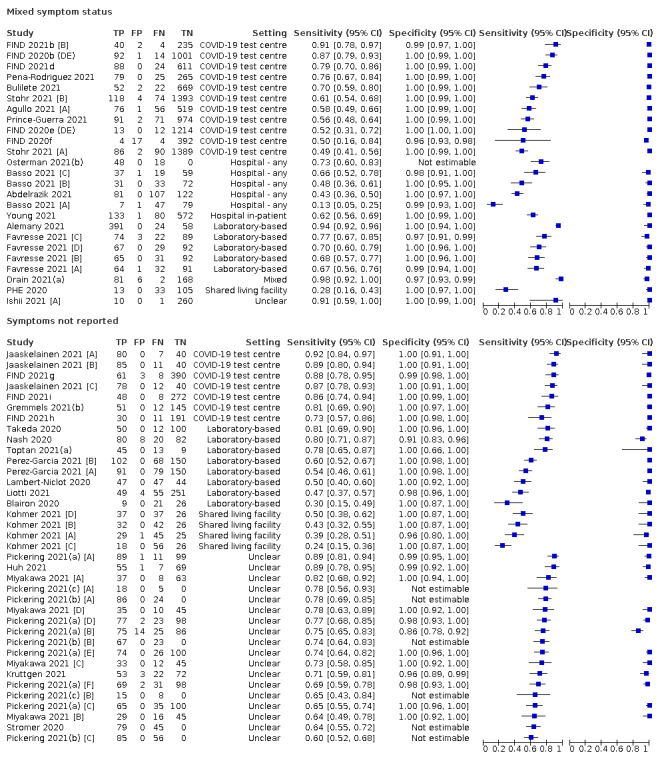
Forest plot of tests: 4 mixed symptom status, 5 symptoms not reported
7.
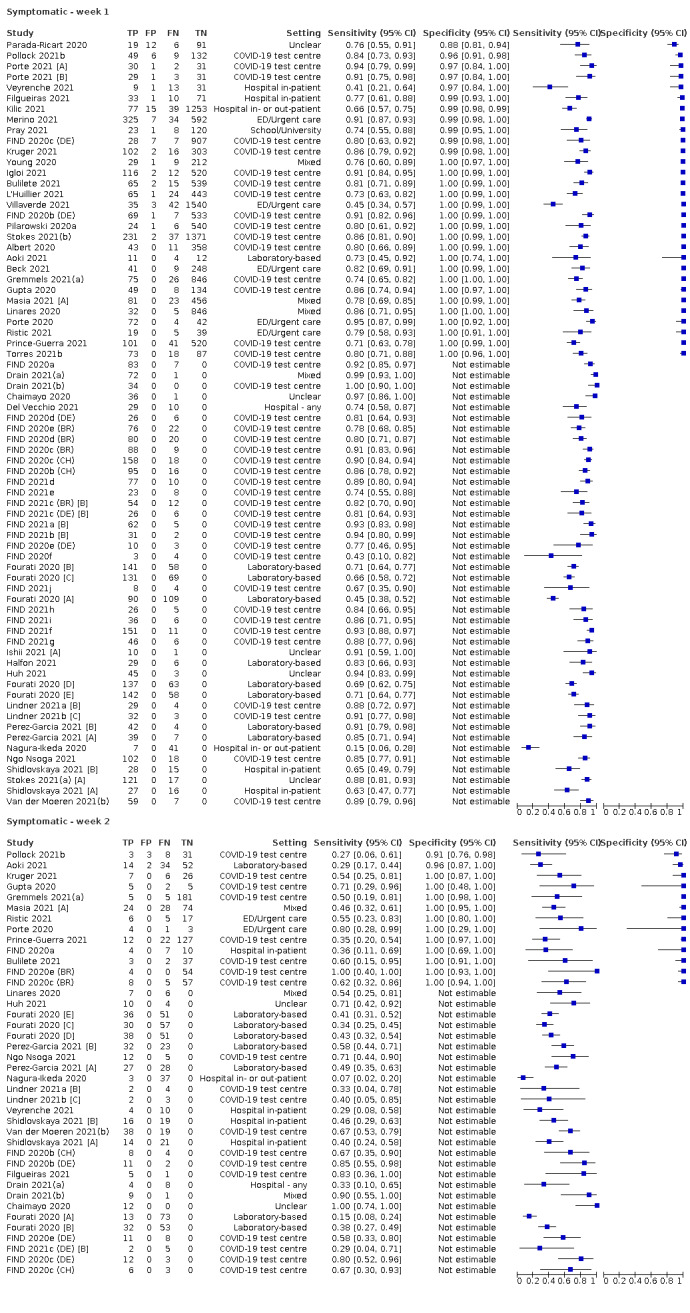
Forest plot of data for symptomatic participants by week post‐symptom onset (pso). Ag: antigen; BR: Brazil; CH: Switzerland; DE: Germany; ED: emergency department
8.
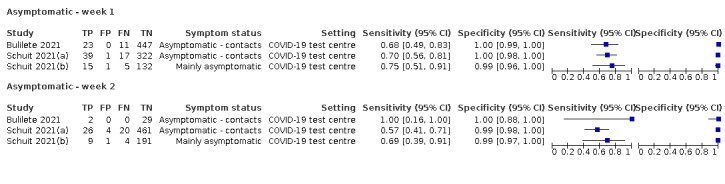
Forest plot of data for asymptomatic participants by week after contact with confirmed case
9.
Forest plot of data for accuracy in children and within‐study comparison of data for adults (where available)
10.
Forest plot of data by viral load (in subgroups by cycle threshold (Ct) value)
11.
Forest plot of data by viral load (in subgroups by RNA copies/mL)
Table 4, Figure 12 and Figure 13 present summary results by test brand according to symptom status, and for sensitivity analyses restricting by compliance with manufacturer IFUs. Forest plots of individual study results by test brand are in Appendix 13 and summary results by test brand regardless of symptom status are reported in Appendix 14. Within‐study comparisons of test brands are reported in Table 5.
3. Summary data by symptom status, test brand and compliance with manufacturer instructions for use.
| All | IFU compliant | |||||
| SYMPTOMATIC participants by test | N evaluations; samples (cases) | Summary sensitivity % (95% CI) | Summary specificity % (95% CI) | N evaluations; samples (cases) | Summary sensitivity % (95% CI) | Summary specificity % (95% CI) |
| AAZ ‐ COVID‐VIRO | 3; 1204 (534) | 84.9 (61.5 to 95.2) | 97.0 (95.4 to 98.1) | 2; 572 (239) | 91.3 (78.2 to 96.9) | 94.0 (90.9 to 96.1) |
| Abbott ‐ BinaxNOW COVID‐19 Ag card | 4; 2018 (358) | 80.9 (67.6 to 89.6) | 99.9 (99.5 to 100) | 4; 2018 (358) | 80.9 (67.6 to 89.6) | 99.9 (99.5 to 100) |
| Abbott ‐ Panbio COVID‐19 Ag | 24; 14,509 (3167) | 74.8 (67.6 to 80.8) | 99.7 (99.6 to 99.8) | 11; 7718 (1397) | 77.3 (68.7 to 84.0) | 99.7 (99.5 to 99.8) |
| Including sensitivity‐only cohorts | 32; 15,331 (3989) | 74.2 (68.5to | ‐ | 16; 8339 (2018) | 75.8 (68.8 to81.7) | ‐ |
| Access Bio ‐ CareStart COVID‐19 Ag | 1; 241 (69) | 75.4 (63.5 to 84.9) | 94.8 (90.3 to 97.6) | 1; 241 (69) | 75.4 (63.5 to 84.9) | 94.8 (90.3 to 97.6) |
| Anhui Deepblue ‐ COVID‐19 Ag | 1; 1205 (191) | 47.1 (39.9 to 54.5) | 100 (99.6 to 100) | |||
| Including sensitivity‐only cohorts | 2; 1382 (368) | 60.7 (41.7to | ‐ | |||
| Becton Dickinson ‐ BD Veritor | 5; 2498 (299) | 73.9 (55.4 to 86.6) | 99.1 (98.6 to 99.4) | 1; 1384 (116) | 66.4 (57.0 to 74.9) | 98.8 (98.1 to 99.3) |
| Including sensitivity‐only cohorts | 7; 2655 (456) | 78.4 (63.8 to88.2) | ‐ | 2; 1416 (148) | 82.5 (52.5 to 95.3) | ‐ |
| BIONOTE ‐ NowCheck COVID‐19 Ag | 2; 618 (181) | 89.5 (84.1 to 93.2) | 97.7 (95.8 to 98.8) | 2; 618 (181) | 89.5 (84.1 to 93.2) | 97.7 (95.8 to 98.8) |
| BIONOTE ‐ NowCheck COVID‐19 Ag (Nasal) | 1; 218 (79) | 89.9 (81.0 to 95.5) | 98.6 (94.9 to 99.8) | 1; 139 (79) | 89.9 (81.0 to 95.5) | 98.6 (94.9 to 99.8) |
| Biosynex ‐ Biosynex COVID‐19 Ag BSS | 1; 634 (297) | 59.6 (53.8 to 65.2) | 100 (98.9 to 100) | |||
| Coris Bioconcept ‐ COVID‐19 Ag Respi‐Strip | 5; 1158 (585) | 37.5 (28.4 to 47.7) | 99.8 (98.8 to 100) | 3; 765 (408) | 34.3 (29.9 to 39.1)a | 100 (99.0 to 100)a,b |
| Denka Co ‐ QuickNavi COVID‐19 Ag | 2; 1633 (123) | 84.2 (66.2 to 93.5)a | 100 (99.8 to 100)a,b | 2; 1633 (123) | 84.2 (66.2 to 93.5)a | 100 (99.8 to 100)a,b |
| DIALAB ‐ DIAQUICK COVID‐19 Ag | 1; 99 (99) | 61.6 (51.3 to 71.2) | ‐ | 1; 99 (99) | 61.6 (51.3 to 71.2) | ‐ |
| ECODiagnostica ‐ COVID‐19 Ag ECO | 1; 150 (55) | 69.1 (55.2 to 80.9) | 97.9 (92.6 to 99.7) | 1; 150 (55) | 69.1 (55.2 to 80.9) | 97.9 (92.6 to 99.7) |
| Fortress Diagnostics ‐ Coronavirus Ag | 1; 1191 (191) | 56.0 (48.7 to 63.2) | 99.9 (99.4 to 100) | |||
| Fujirebio ‐ ESPLINE SARS‐CoV‐2 | 1; 129 (63) | 39.7 (27.6 to 52.8) | 97.0 (89.5 to 99.6) | |||
| Including sensitivity‐only cohorts | 4; 251 (185) | 29.6 (14.6to | ‐ | |||
| Innova Medical Group ‐ SARS‐CoV‐2 Ag | 3; 3522 (830) | 68.1 (47.2 to 83.6) | 99.0 (98.5 to 99.3) | 1; 1676 (372) | 57.5 (52.3 to 62.6) | 99.6 (99.1 to 99.9) |
| Including sensitivity‐only cohorts | 5; 3943 (1251) | 70.8 (58.1 to 80.9) | ‐ | 3; 2097 (793) | 69.1 (58.3to | ‐ |
| Joysbio Biotech ‐ SARS‐CoV‐2 Ag | 1; 265 (44) | 70.5 (54.8 to 83.2) | 99.1 (96.8 to 99.9) | |||
| Lepu Medical ‐ SARS‐CoV‐2 Ag Rapid Test | 1; 100 (100) | 54.0 (43.7 to 64.0) | ‐ | |||
| Liming Bio‐Products ‐ StrongStep COVID‐19 Ag | 1; 19 (9) | 0 (0 to 33.6) | 90.0 (55.5 to 99.7) | |||
| LumiraDx ‐ SARS‐CoV‐2 Ag | 2; 741 (177) | 91.2 (70.0 to 97.9) | 98.6 (97.2 to 99.3) | 2; 741 (177) | 91.2 (70.0 to 97.9) | 98.6 (97.2 to 99.3) |
| MEDsan GmbH ‐ SARS‐CoV‐2 Ag | 1; 184 (84) | 45.2 (34.3 to 56.5) | 97.0 (91.5 to 99.4) | |||
| Mologic ‐ COVID 19 Rapid Ag‡ | 1; 650 (192) | 90.6 (85.6 to 94.3) | 100 (99.2 to 100) | 1; 650 (192) | 90.6 (85.6 to 94.3) | 100 (99.2 to 100) |
| Orient Gene/Healgen Scientific ‐ Coronavirus Ag | 1; 1189 (190) | 48.9 (41.6 to 56.3) | 100 (99.6 to 100) | |||
| Including sensitivity‐only cohorts | 2; 1284 (285) | 67.2 (40.7to | ‐ | 1; 95 (95) | 82.1 (72.9to | ‐ |
| Precision Biosensor ‐ Exdia COVID‐19 Ag | 1; 293 (90) | 52.2 (41.4 to 62.9) | 99.0 (96.5 to 99.9) | 1; 293 (90) | 52.2 (41.4 to 62.9) | 99.0 (96.5 to 99.9) |
| Quidel ‐ SOFIA SARS Antigen FIA | 4; 1064 (176) | 80.0 (71.5 to 86.4) | 99.4 (98.7 to 99.8) | 3; 1000 (144) | 76.4 (68.8 to 82.6) | 99.5 (98.8 to 99.8) |
| RapiGEN ‐ BIOCREDIT COVID‐19 Ag | 4; 714 (284) | 61.5 (49.3 to 72.5) | 98.4 (96.6 to 99.2) | 2; 582 (195) | 66.3 (52.9 to 77.5) | 99.0 (97.3 to 99.6) |
| Savant Biotech ‐ Huaketai SARS‐CoV‐2 | 1; 109 (78) | 16.7 (9.2 to 26.8) | 100 (88.8 to 100) | |||
| SD Biosensor ‐ Standard F COVID‐19 Ag | 4; 1742 (380) | 74.3 (61.8 to 83.9) | 97.4 (96.4 to 98.1) | 2; 1129 (159) | 75.5 (68.2 to 81.5) | 97.2 (96.0 to 98.1) |
| Including sensitivity‐only cohorts | 5; 1909 (547) | 72.5 (63.2to | ‐ | 3; 1296 (326) | 72.1 (66.7to | ‐ |
| SD Biosensor/Roche ‐ Standard Q COVID‐Ag | 26; 10,678 (2539) | 78.6 (72.3 to 83.7) | 98.7 (98.4 to 98.9) | 15; 5116 (1197) | 84.0 (79.2 to 87.9) | 99.2 (98.8 to 99.4) |
| Including sensitivity only cohorts | 28; 10,798 (2659) | 79.2 (72.9 to84.3) | ‐ | |||
| SD Biosensor ‐ Standard Q COVID‐Ag (Nasal) | 4; 621 (189) | 85.2 (79.4 to 89.6) | 99.3 (97.9 to 99.8) | 4; 621 (189) | 85.2 (79.4 to 89.6) | 99.3 (97.9 to 99.8) |
| Shenzhen Bioeasy Biotech ‐ 2019‐nCoV Ag | 4; 1093 (202) | 84.4 (72.4 to 91.7) | 93.2 (91.3 to 94.6) | 2; 855 (40) | 72.5 (56.8 to 84.1) | 92.5 (90.5 to 94.1) |
| Sichuan Mass Spectrometry Biotech ‐ 2019‐nCov Ag | 1; 85 (85) | 22.4 (14.0 to 32.7) | ‐ | |||
| Siemens ‐ CLINITEST Rapid COVID‐19 Ag | 2; 350 (163) | 68.7 (48.0 to 83.8)† | 100 (98.0 to 100)a,b | 1; 178 (91) | 80.2 (70.6 to 87.8) | 100 (95.8 to 100) |
| Sugentech Inc ‐ SGTI‐flex COVID‐19 Ag | 1; 106 (78) | 52.6 (40.9 to 64.0) | 96.4 (81.7 to 99.9) | 1; 106 (78) | 52.6 (40.9 to 64.0) | 96.4 (81.7 to 99.9) |
| SureScreen Diagnostics ‐ SureScreen V | 1; 1185 (189) | 42.9 (35.7 to 50.2) | 99.9 (99.4 to 100) | |||
| ASYMPTOMATIC participants by test | ||||||
| Abbott ‐ BinaxNOW COVID‐19 Ag card | 6; 12,530 (588) | 58.7 (45.6 to 70.6) | 99.8 (99.7 to 99.8) | 6; 12,530 (588) | 58.7 (45.6 to 70.6) | 99.8 (99.7 to 99.8) |
| Abbott ‐ Panbio COVID‐19 Ag | 14; 4038 (561) | 56.9 (42.8 to 69.9) | 99.5 (99.1 to 99.7) | 7; 2502 (279) | 57.9 (35.4 to 77.5) | 99.6 (99.3 to 99.8) |
| Including sensitivity‐only cohorts | 17; 4138 (661) | 57.8 (45.5to | ‐ | 8; 2557 (334) | 55.4 (36.8to | ‐ |
| Access Bio ‐ CareStart COVID‐19 Ag | 1; 1257 (165) | 50.3 (42.4 to 58.2) | 98.9 (98.1 to 99.4) | 1; 1257 (165) | 50.3 (42.4 to 58.2) | 98.9 (98.1 to 99.4) |
| Becton Dickinson ‐ BD Veritor | 2; 2556 (203) | 49.8 (32.1 to 67.5) | 99.7 (99.3 to 99.8) | |||
| BIONOTE ‐ NowCheck COVID‐19 Ag | 1; 1326 (9) | 55.6 (21.2 to 86.3) | 100 (99.7 to 100) | |||
| Coris Bioconcept ‐ COVID‐19 Ag Respi‐Strip | 1; 45 (14) | 28.6 (8.4 to 58.1) | 100 (88.8 to 100) | 1; 45 (14) | 28.6 (8.4 to 58.1) | 100 (88.8 to 100) |
| Denka Co ‐ QuickNavi COVID‐19 Ag | 1; 415 (33) | 75.8 (57.7 to 88.9) | 100 (99.0 to 100) | 1; 415 (33) | 75.8 (57.7 to 88.9) | 100 (99.0 to 100) |
| Fujirebio ‐ ESPLINE SARS‐CoV‐2 | 1; 15 (15) | 13.3 (1.7 to 40.5) | ‐ | |||
| Innova Medical Group ‐ SARS‐CoV‐2 Ag | 2; 6224 (78) | 38.5 (28.4 to 49.7) | 100 (99.8 to 100) | 1; 5504 (70) | 40.0 (28.5 to 52.4) | 99.9 (99.8 to 100) |
| Lepu Medical ‐ SARS‐CoV‐2 Ag Rapid Test | 1; 286 (101) | 45.5 (35.6 to 55.8) | 89.2 (83.8 to 93.3) | |||
| LumiraDx ‐ SARS‐CoV‐2 Ag | 1; 272 (9) | 77.8 (40.0 to 97.2) | 99.6 (97.9 to 100) | 1; 272 (9) | 77.8 (40.0 to 97.2) | 99.6 (97.9 to 100) |
| Precision Biosensor ‐ Exdia COVID‐19 Ag | 1; 239 (24) | 33.3 (15.6 to 55.3) | 100 (98.3 to 100) | 1; 239 (24) | 33.3 (15.6 to 55.3) | 100 (98.3 to 100) |
| Quidel ‐ SOFIA SARS Antigen FIA | 1; 871 (17) | 41.2 (18.4 to 67.1) | 98.4 (97.3 to 99.1) | 1; 871 (17) | 41.2 (18.4 to 67.1) | 98.4 (97.3 to 99.1) |
| RapiGEN ‐ BIOCREDIT COVID‐19 Ag | 2; 140 (60) | 63.2 (21.7 to 91.4) | 98.8 (91.7 to 99.8) | |||
| SD Biosensor ‐ Standard F COVID‐19 Ag | 1; 55 (55) | 43.6 (30.3 to 57.7) | ‐ | 1; 55 (55) | 43.6 (30.3 to 57.7) | ‐ |
| SD Biosensor/Roche ‐ Standard Q COVID‐Ag | 12; 10,049 (551) | 59.4 (49.6 to 68.5) | 99.3 (99.1 to 99.4) | 4; 5914 (250) | 64.6 (51.3 to 75.9) | 99.6 (99.4 to 99.7) |
| Including sensitivity‐only cohort | 13; 10,052 (554) | 60.4 (50.5 to69.6) | ‐ | |||
| Shenzhen Bioeasy Biotech ‐ 2019‐nCoV Ag | 1; 44 (1) | 0 (0 to 97.5) | 79.1 (64.0 to 90.0) | 1; 44 (1) | 0 (0 to 97.5) | 79.1 (64.0 to 90.0) |
| Siemens ‐ CLINITEST Rapid COVID‐19 Ag | 2; 378 (126) | 53.2 (44.5 to 61.7) | 98.8 (96.4 to 99.6) | 1; 92 (25) | 60.0 (38.7 to 78.9) | 100 (94.6 to 100) |
| SureScreen Diagnostics ‐ SureScreen V | 1; 286 (101) | 28.7 (20.1 to 38.6) | 97.8 (94.6 to 99.4) | |||
aSeparate pooling of sensitivity or specificity. b2x2 tables combined prior to calculating estimates.
12.
Forest plot of results per assay in symptomatic participants (overall and in manufacturer instructions for use (IFU)‐compliant evaluations); red lines indicate World Health Organization acceptable and desirable performance standards for sensitivity and specificity in suspected COVID‐19 cases
13.
Forest plot of results per assay in asymptomatic participants (overall and in manufacturer instructions for use (IFU)‐compliant evaluations); red lines indicate World Health Organization acceptable and desirable performance standards for sensitivity and specificity in suspected COVID‐19 cases (there are no performance standards specifically for asymptomatic populations)
4. Direct comparisons by test brand (any symptom status).
| Test | N evaluations; samples (cases) | Summary sensitivity % (95% CI) | Difference in sensitivity, (95% CI), P value | Summary specificity % (95% CI) | Difference in specificity, (95% CI), P value |
| Panbio COVID‐19 Ag (Abbott) vs Standard Q COVID‐Ag (SD Biosensor/Roche); studies reporting both sensitivity and specificity | |||||
| Panbio | 9; 3895 (1058) | 56.7 (44.3, 68.3) | Ref | 99.5 (99.1, 99.7) | Ref |
| Standard Q | 9; 3301 (1055) | 64.4 (52.2, 75.0) | 7.8 (−9.1, 24.6), P = 0.367 | 97.5 (96.7, 98.0) | −2.0 (−2.7, −1.3), P < 0.001 |
| Panbio COVID‐19 Ag (Abbott) vs Standard Q COVID‐Ag (SD Biosensor/Roche); summary sensitivity including 'sensitivity‐only' cohort | |||||
| Panbio | 10; 3977 (1140) | 56.1 (45.0, 66.6) | Ref | ‐ | ‐ |
| Standard Q | 10; 3372 (1126) | 63.9 (53.0, 73.6) | 7.8 (−7.3, 23.0), P = 0.311 | ‐ | ‐ |
| Panbio COVID‐19 Ag (Abbott) vs Coronavirus Ag (Orient Gene/Healgen Scientific); studies reporting both sensitivity and specificity | |||||
| Panbio | 2; 1968 (287) | 60.7 (44.1, 75.2) | Ref | 99.7 (99.3, 99.9) | Ref |
| Coronavirus Ag | 2; 1377 (286) | 63.3 (46.6, 77.3) | 2.5 (−20.0, 25.1), P =0.825 | 99.7 (99.2, 99.9) | 0.02 (−0.38, 0.43), P = 0.914 |
| Panbio COVID‐19 Ag (Abbott) vs Coronavirus Ag (Orient Gene/Healgen Scientific); summary sensitivity including 'sensitivity‐only' cohort | |||||
| Panbio | 3; 2067 (386) | 65.3 (50.0, 78.0) | Ref | ‐ | ‐ |
| Coronavirus Ag | 3; 1472 (381) | 70.3 (55.5, 81.8) | 5.0 (−14.6, 24.6), P = 0.617 | ‐ | ‐ |
| Standard Q COVID‐Ag (SD Biosensor) Nasal kit vs NP kit | |||||
| Nasal kit | 3; 489 (153) | 85.0 (78.4, 89.8) | Ref | 99.1 (97.3, 99.7) | Ref |
| NP kit | 3; 489 (153) | 83.0 (76.2, 88.2) | −2.0 (−10.2, 6.3), P =0.640 | 99.4 (97.7, 99.9) | 0.3 (−1.0, 1.6), P =0.653 |
Accuracy of antigen tests overall and by subgroup
Results showed high levels of heterogeneity in sensitivity with consistently high specificity (Appendix 12). Average sensitivity was 69.3% (95% CI 66.2% to 72.3%) and average specificity was 99.3% (95% CI 99.2% to 99.3%) across the 184 evaluations of antigen tests reporting both sensitivity and specificity (based on 117,372 samples, including 21,017 samples with confirmed SARS‐CoV‐2; Table 3). Adding the 25 test evaluations from ‘sensitivity only’ studies and the single ‘specificity only’ dataset had a negligible impact on results (Table 3).
In the sections below we show that there are substantial differences between subgroups of studies according to symptom status, timing, assay format and brand, therefore this average value is unlikely to accurately predict the performance of the test in a given setting and should not be used for this purpose.
Secondary analyses by symptom status
Secondary analysis by symptom status (where possible using subgroup data by symptom status for studies including both symptomatic and asymptomatic participants) suggests that average test sensitivity to detect infection is 18.2 percentage points lower in asymptomatic participants (54.7%, 95% CI 47.7% to 61.6%; based on 50 evaluations, 40,956 samples and 2,641 cases) compared to symptomatic participants (73.0%, 95% CI 69.3% to 76.4%; based on 109 evaluations, 50,574 samples and 11,662 cases). The 95% CI for the difference in sensitivity ranged from −26.1 to −10.4 percentage points (Table 3; Figure 4 and Figure 5).
Restricting the comparison by symptom status to the 34 evaluations reporting data for both symptomatic and asymptomatic subgroups (thus ensuring the comparison is made between the same tests used in the same way) showed a similar difference in sensitivity (Table 3). Inclusion of data from sensitivity‐only cohorts led to a small increase in the difference in sensitivity (Table 3).
Specificity was marginally higher in asymptomatic populations, both overall (0.4 percentage points higher) and for within‐study comparisons (0.5 percentage points higher; Table 3).
Average results for the 56 evaluations in participants with mixed symptom status (n = 25) or symptom status not reported (n = 31) were similar to those observed for the symptomatic participants (Table 3; Figure 6), suggesting these studies may have included mainly symptomatic individuals: sensitivity 70.6% (95% CI 64.8% to 75.3%) and specificity 99.4% (95% CI 99.3% to 99.5%) based on 9359 samples and 5344 cases.
Adding the test evaluations from ‘sensitivity‐only’ studies (23 in symptomatic participants, 6 in asymptomatic participants and 7 in mixed symptom populations) had a negligible impact on results (Appendix 10).
Subgroup analysis for symptomatic participants by study by setting
Statistical evidence for a difference in sensitivity was observed for symptomatic participants according to study setting (Table 3; Figure 4). Average sensitivity in COVID‐19 test centres was 82.8% (95% CI 80.2% to 85.2%) and specificity 99.1% (95% CI 99.0% to 99.2%; 47 evaluations in 23,602 samples, including 4369 cases). Average sensitivity was lower in all other settings considered; the absolute difference ranged from 11.9 percentage points lower for those presenting in emergency departments or urgent care centres (95% CI 25.7 percentage points lower to 1.9 higher) to 31.3 percentage points lower for hospital inpatients (95% CI from −37.5 to −25.1 percentage points). Results varied in other settings (hospitals in‐ or outpatients, healthcare worker screening, and shared living facility settings), but with only one evaluation each (Table 3).
Adding data from the 14 test evaluations from ‘sensitivity‐only’ studies had a small effect on average sensitivity for all settings (changes of up to 3 percentage points) apart from testing of symptomatic participants in school or university settings. Average sensitivity increased considerably when the number of included evaluations for schools or university settings increased from two to three however the number of cases (n = 146) remained low (Appendix 10).
Subgroup analysis for symptomatic participants by time from symptom onset
A total of 72 test evaluations reported data for symptomatic participants according to time after the onset of symptoms (week 1 compared to week 2 or later; Table 3; Figure 7). In contrast to the previous iteration of this review, considerably more (n = 30) studies provided results for both sensitivity and specificity according to time from onset of symptoms (compared to 10 for the previous version), and the remaining 42 provided data only for confirmed SARS‐CoV‐2 cases (so that only sensitivity could be calculated).
Where both sensitivity and specificity could be calculated, in week one after symptom onset average sensitivity was 80.9% (95% CI 76.9% to 84.4%), and specificity 99.5% (95% CI 99.3% to 99.6%; based on 30 evaluations including 15,323 samples and 2408 cases). In week two or later after onset, average sensitivity was 48.9% (95% CI 37.9% to 60.1%) and specificity 99.3% (95% CI 98.2% to 99.7%) based on 13 evaluations including 903 samples and 224 cases (Table 3). Average sensitivity was 32.0 percentage points lower at the later time point (95% CI −43.9 to −20.1 percentage points).
Including the 42 evaluations that provided data only for sensitivity by time after onset led to small increases in average sensitivity during both time periods (Table 3). The difference in sensitivity between time points decreased slightly to −28.4 percentage points (95% CI from −32.6 to −24.2 percentage points lower in week two compared to week one after onset), based on 5640 cases in week one and 1119 cases in week 2.
Subgroup analyses for asymptomatic participants by eligibility for testing and study setting
Subgroup analyses for asymptomatic participants indicated that sensitivity was higher when an epidemiological exposure to SARS‐CoV‐2 was suspected (based on studies reporting specific criteria for testing or referral for testing of those without symptoms) compared to where COVID‐19 testing was reported to be widely available to any asymptomatic participant on presentation for testing (Table 3; Figure 5). The absolute difference in sensitivity was 14.7 percentage points (95% CI from 2.7 to 26.7 percentage points) when an epidemiological exposure to SARS‐CoV‐2 was suspected (sensitivity 64.3%, 95% CI 54.6% to 73.0% compared to 49.6%, 95% CI 42.1% to 57.1%; based on 16 evaluations in 7677 samples with 703 cases and 26 evaluations with 31,904 samples and 1758 cases respectively). Average specificity was similarly high (99.6% or 99.7%) regardless of the presence or absence of likely epidemiological exposure (0.06 percentage points difference, 95% CI from −0.09 to 0.22; Table 3).
Adding data from sensitivity‐only cohorts led to a slight decrease in the difference in sensitivity between widely available testing and testing of contacts; difference 12.2 percentage points (95% CI 0.2 to 24.2 percentage points higher in the contacts group (Table 3).
Analyses for asymptomatic participants by study setting were limited by the number of studies per group and low numbers of confirmed SARS‐CoV‐2 cases for some subgroups, however average sensitivities were highest for participants presenting to COVID‐19 test centres (61.5%, 95% CI 54.0% to 68.4%; based on 18 evaluations including 19,253 samples and 1195 cases) or emergency care settings (average sensitivity 95.1%, 95% CI 7.3% to 100%; based on 2 evaluations with 2547 samples and 85 cases; Table 3; Figure 5). For the seven evaluations considered to represent screening scenarios, average sensitivity was 45.1% (95% CI 36.4% to 54.1%; based on 7776 samples and 648 cases of SARS‐CoV‐2); for school or university‐wide testing programmes it was 47.9% (95% CI 38.1% to 57.9%; 5 evaluations, including 5174 samples and 96 cases); and for blanket testing of hospital inpatients with no apparent symptoms of COVID‐19 it was 35.2% (95% CI 26.7% to 44.8%; based on 5 evaluations including 2282 samples and 105 cases). Adding the test evaluations from ‘sensitivity‐only’ studies had a negligible impact on results (Appendix 10).
Three evaluations in asymptomatic or mainly asymptomatic contacts of confirmed cases reported sensitivity and specificity by reported time after contact (1013 samples and 110 cases in week 1 and 747 samples and 61 cases in week 2; Table 3; Figure 8). Sensitivity was 70.0% in week 1 (95% CI 60.8% to 77.8%) compared to 60.7% in week 2 (95% CI 48.0% to 72.0%), however the 95% CI for the difference between time points was from 24.3 percentage points lower to 5.6 higher, suggesting no statistical evidence for a difference.
Sensitivity analysis by study design
Restricting study inclusion to those using single group designs had only a marginal effect on summary sensitivity (70.8%, 95% CI 67.2% to 74.3%) and specificity (99.4%, 95% CI 99.3% to 99.4%), based on 93,970 samples and 14,171 cases from 126 evaluations; Table 3; Appendix 12 see Figure 14).
14.
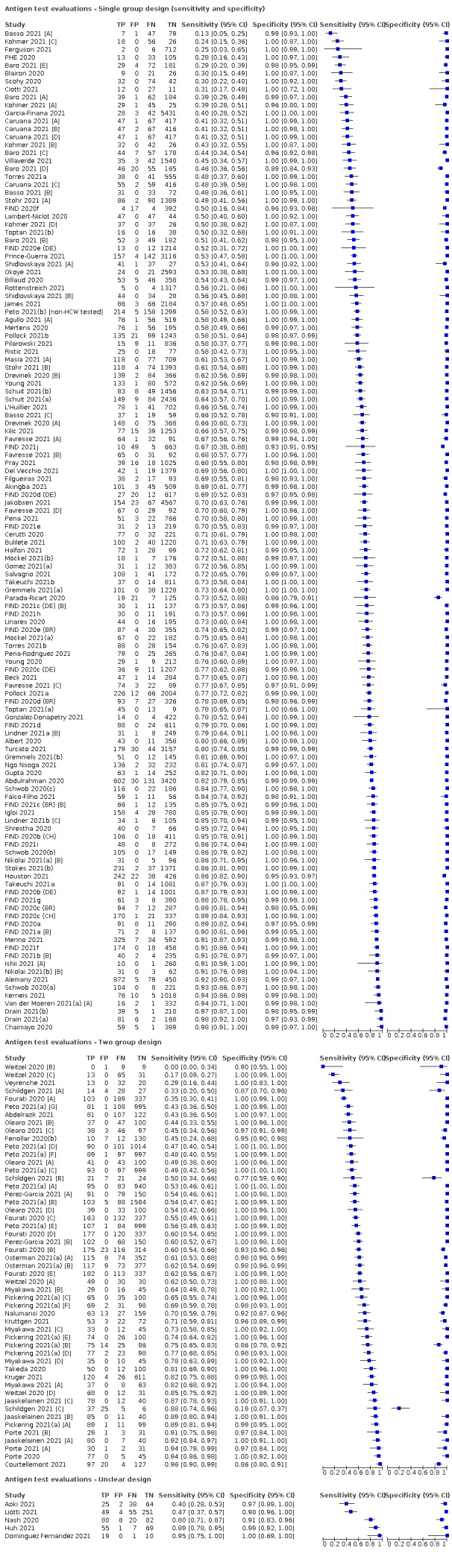
Forest plot of data by study design. BR: Brazil; CH: Switzerland; DE: Germany; HCW: healthcare worker
Subgroup analysis by sample site
We observed some difference in accuracy according to the type of sample used, however observed differences may be confounded by a number of factors including the populations studied, timing of tests in regard to course of infection and variations in accuracy of the tests used. Results are therefore presented based on subgroup analysis of all data contributing to the primary analysis and then restricted to the small number of evaluations comparing accuracy by sample site in some or all participants, that is, studies used the same rapid antigen test on samples from two different sites (Table 3). Differences in the number of samples per site were observed for some studies such that we are not able to present data for strictly ‘paired’ comparisons.
The majority of evaluations (n = 128 for studies reporting both sensitivity and specificity) obtained nasopharyngeal samples from all study participants, either as the sole sample site, or in combination with oropharyngeal sampling in some or all participants. Average sensitivity was 69.0% (95% CI 65.3% to 72.4%) and average specificity 99.4% (95% CI 99.3% to 99.4%) in 59,447 samples including 13,270 cases of SARS‐CoV‐2; Table 3; Appendix 12 see Figure 15). Average sensitivity in studies using nasal samples (anterior nasal in 14, nasal mid‐turbinate in 15, and not further specified in 5) was 76.6% (95% CI 70.3% to 81.9%; in 33,128 samples including 4032 cases); an increase of 7.7 percentage points compared to studies in the nasopharyngeal sample group (95% CI 0.9 to 14.5 percentage points higher).
15.
Forest plot of data by sample site
In contrast however, the nine studies reporting within‐participant comparisons of nasopharyngeal (2979 samples including 682 cases) and nasal samples (2710 samples including 619 cases) suggested no statistical evidence for a difference in sensitivity between sites (sensitivity was 2.9 percentage points lower in nasal samples compared to nasopharyngeal samples, 95% CI ‐16.1 to 10.4 percentage points; Table 3; Appendix 12 see Figure 16). Five of the nine studies reported the use of nasal sampling kit versions of the assays used (FIND 2021a [A]; FIND 2021b [A]; FIND 2021c (BR) [A]; FIND 2021c (DE) [A]; Nikolai 2021(b) [A]). No difference in specificity between nasopharyngeal and nasal sampling was observed overall (specificity 99.6% for both sample sites, 95% CI 99.2% to 99.8%) or in direct comparisons (specificity 99.6% for both sites, 95% CI 99.2% to 99.8%). When analysis was restricted to the six studies comparing nasopharyngeal and nasal mid‐turbinate sampling, specificity was 2.0 percentage points lower for nasal mid‐turbinate (95% CI from 1.3 to 2.7 percentage points lower, based on 1134 samples) (Table 3).
16.
Forest plot of within‐study comparisons by sample site, collection, or interpretation.
Overall, average sensitivities for studies using combined nasal and oropharyngeal samples, and in studies using nasopharyngeal samples in some but not all participants (e.g. sample site was either nasopharyngeal or oropharyngeal) were similar to results where nasopharyngeal samples were used in all participants (absolute differences of −0.3 and 2.9 respectively; Table 3). No direct comparisons of the accuracy of these sample sites were identified.
The number of evaluations contributing data to the primary analysis based on saliva or oropharyngeal samples alone was low (n = 4 and n = 2 respectively), however average sensitivities were lower than for nasopharyngeal or nasal samples, sensitivity 21.6% for saliva (95% CI 17.4% to 26.6%, based on 305 cases) and 57.4% for oropharyngeal samples (95% CI 26.6% to 83.4%, based on 214 cases). The four studies evaluating saliva samples also provided within‐participant comparisons of accuracy with nasopharyngeal samples: the difference in sensitivity was 36.8 percentage points lower for saliva (95% CI from −43.5 to −30.1 percentage points). Adding data from two sensitivity‐only cohorts increased the difference in sensitivity to 49.4 percentage points lower (sensitivity 66.5% for nasopharyngeal samples (95% CI 53.0% to 77.8%; based on 640 cases) and 17.1% for saliva (95% CI 10.1% to 27.5% based on 446 cases).
Two studies allowing a comparison of nasal (1318 samples) versus saliva samples (1221 samples), suggested higher sensitivity and similarly specificity from the use of nasal samples (Agullo 2021 [A]; Masia 2021 [A]), and one study also showed identical sensitivity and specificity in anterior nasal compared to nasal mid‐turbinate (Nikolai 2021(a) [A]; Table 3; Appendix 12 see Figure 16).
Subgroup analyses including data from ‘sensitivity‐alone’ evaluations produced similar average sensitivities and differences in sensitivity between sample types (Appendix 10).
Average specificities were greater than 99% for all of the main sample sites apart from in the 12 evaluations of either naso‐ or oropharyngeal samples where average specificity was 98.4% (95% CI 98.0% to 98.7%; Table 3).
Effect from test operator
Only one study directly compared the effect of assay interpretation by study participants compared to interpretation by a professional, both using nasal swabs (Lindner 2021b [A]); specificities were both 100% and sensitivity was 2.5 percentage points higher for the professional interpreted tests (85.0% compared to 82.5% for participant interpreted tests). The direction of effect is similar to that observed in an indirect comparison by test operator from a PHE evaluation showing sensitivities of:
57.5% (95% CI 52.3% to 62.6%) when the test was used by self‐trained, non‐healthcare workers (n = 1; 372 cases; Peto 2021(b) [non‐HCW tested]);
70.0% (95% CI 63.5% to 75.9%) when the test was used by healthcare workers (n = 1; 223 cases; Peto 2021(c) [A ‐ HCW tested]); and
78.8% (95% CI 72.4% to 84.3%) when the test was used by laboratory scientists (n = 1; 198 cases; Peto 2021(c) [A ‐ Lab tested]).
Evaluations of accuracy in children
Restricting the analysis to the 10 evaluations reporting data for children in 4652 samples with 410 cases, average sensitivity was 62.7% (95% CI 52.7% to 71.7%) and average specificity 99.4% (95% CI 99.1% to 99.6%; Table 3; Figure 9). Adding data from two sensitivity‐only or one specificity‐only evaluation had only a marginal effect on accuracy (Table 3).
Six evaluations allowed a comparison of results in children versus adults (so minimising the effect from other differences); average sensitivity was 9.9 percentage points higher (95% CI −8.7 to 28.4), and average specificity 0.7 percentage points higher (95% CI 0.2 to 1.2) in adults compared to children, however the number of SARS‐CoV‐2 cases in children was relatively small and the difference for sensitivity was within that which might be observed by chance. These differences were maintained with the addition of data from two sensitivity‐only evaluations and one specificity‐only evaluation (Table 3).
Subgroup analysis by Ct value or RNA copies/mL
A total of 157 evaluations reported sensitivity according to sample viral load, either using PCR Ct values as a proxy, or by converting Ct values to RNA copies/mL which allows for a fairer comparison across studies using different RT‐PCRs assays.
We first compared sensitivity above and below a single Ct or RNA copies/mL threshold to indicate higher or lower viral load. Results were very similar to those observed for the previous iteration of this review, with strong evidence for higher sensitivity in higher viral load subgroups (Table 3; Appendix 12 see Figure 17; Figure 18; Figure 19; Figure 20; Figure 21). Because of the continuous nature of viral load measurement, and the lack of evidence for a step‐change in RDT sensitivity above or below any single threshold value, we focus on presenting results for assay sensitivity in subgroups with smaller ranges in viral load. Results are reported in subgroups, firstly for studies reporting results according to Ct value and then for studies reporting results in RNA copies/mL (Table 3; Figure 10; Figure 11).
17.
Forest plot of data in higher versus lower viral load subgroups (< or > 25 Ct). BR: Brazil; CH: Switzerland; Ct: cycle threshold; DE: Germany; HCW: healthcare worker
18.
Forest plot of data in higher versus lower viral load subgroups (< or > 32/33 Ct threshold). BR: Brazil; CH: Switzerland; ; Ct: cycle threshold; DE: Germany
19.
Forest plot of data in higher versus lower viral load subgroups (< or > 30 Ct). Ct: cycle threshold; HCW: healthcare worker
20.
Forest plot of data in higher versus lower viral load subgroups (> or < 10^6 RNA copies/mL)
21.
Forest plot of data in higher versus lower viral load subgroups (> or < 10^5 RNA copies/mL)
For participant samples with the highest viral load, summary sensitivities were 97.4% for samples with Ct less than 20 (95% CI 95.0% to 98.6%; based on 26 evaluations including 1108 cases) and 98.4% for samples with 10^7 RNA copies/mL or above (95% CI 97.0% to 99.1%; 21 evaluations, including 608 cases), respectively (Table 3). For those with Ct values in the 20 to 25 Ct range, or from 10^6 to 10^7 RNA copies/mL, summary sensitivities remained high: 93.6% (95% CI 90.0% to 96.0%; 27 evaluations, 1384 cases) and 94.0% (95% CI 89.8% to 96.6%; 28 evaluations, 597 cases). Summary sensitivities are considerably lower for samples in the 25 to 30 Ct or from 10^5 to 10^6 RNA copies/mL range, to 68.7% (95% CI 61.6% to 75.0%; 48 evaluations, 1724 cases) and 70.9% (95% CI 57.4% to 81.5%; 31 evaluations, 686 cases). Fewer evaluations reported results in the 30 to 35 Ct (n = 8) and greater than 35 Ct (n = 9) ranges, however the pattern of results was the same as for evaluations reporting results for samples with between 10^4 and 10^5 RNA copies/mL, or with less than 10^4 RNA copies/mL, where average sensitivities were 36.7% (95% CI 24.7% to 50.5%; 24 evaluations, 582 cases) and 7.5% (95% CI 3.8% to 14.3%; 24 evaluations, 825 cases), respectively (Table 3). Average sensitivity from evaluations reporting results for samples with Ct values greater than 30 Ct were 18.7% (95% CI 14.2% to 24.1%), based on 2332 cases.
Subgroup analysis by assay format
We found some evidence for differences in accuracy according to assay format (Table 3). Average sensitivity was lower for evaluations using a CGIA at 68.5% (95% CI 65.1% to 71.7%; 140 evaluations; 95,926 samples, 17146 cases) compared to FIA (average sensitivity 76.6%, 95% CI 68.2% to 83.4%; n = 19, 6987 samples, 1507 cases). The absolute difference in average sensitivities was 8.2 percentage points (95% CI −0.1 to 16.5 percentage points). Average specificities were 99.4% (95% CI 99.3% to 99.4%) for CGIAs and 97.5% (95% CI 97.1% to 97.9%) for FIAs; a difference of −1.9 percentage points (95% CI −2.3 to −1.4 percentage points). Results for LFAs where the method could not be determined (n = 12) and for alkaline phosphatase (ALP)‐labelled assays were lower than those observed for the other assay types (Table 3). Average sensitivities for latex‐conjugated LFAs (2 evaluations in 2048 samples, 156 cases) and microfluidic fluorescent immunoassays (4 evaluations in 1373 samples, 343 cases) were higher, 81.3% (95% CI 69.9% to 89.0%) and 89.7% (95% CI 63.0% to 97.8%), respectively.
Results by test brand according to symptom status and IFU compliance
In contrast to the previous iteration of this review we focus on results by test brand according to symptom status (i.e. using separate summary results for ≥ 75% symptomatic or ≥ 75% asymptomatic populations) with sensitivity analyses by IFU compliance (based on sample type, use of VTM, and time period between sample collection and test procedure). Summary results by brand are presented in Table 4 and Figure 12 (showing results per study if only one study per brand). Figure 13 shows individual study results for the 23 studies reporting within‐study comparisons of test brands; summary results are reported in Table 5.
Forest plots of individual study data are in Appendix 13, see Figure 22 (all data regardless of symptoms), Figure 23 (symptomatic), Figure 24 (asymptomatic). Overall results by test brand (regardless of symptom status) and sensitivity analyses by IFU compliance are reported in Appendix 14. An overall summary of results is provided below, and a detailed synthesis of results by test brand is reported in Appendix 15.
22.
Forest plot of individual study results overall (regardless of symptom status) by assay
23.
Forest plot of individual study results in symptomatic participants by assay
24.
Forest plot of individual study results in asymptomatic participants by assay
Comparisons by brand in symptomatic populations
A total of 33 test brands were evaluated in symptomatic or mainly symptomatic participants (n = 132 evaluations, including 24 in sensitivity‐only cohorts). Evaluations of the nasal kit versions of two brands (BIONOTE Nowcheck and SD Biosensor STANDARD Q) were considered separately and are also reported in Table 4. We observed considerable heterogeneity in sensitivities for almost all assays (Appendix 13).
Twelve test brands were evaluated in three or more evaluations (total of 105 evaluations) and we meta‐analyzed results using the bivariate model (Figure 12). Six test brands were evaluated in two evaluations each and we therefore meta‐analysed them using univariate analysis or by summing 2x2 tables (assays from Anhui Deepblue, BIONOTE, Denka Co, LumiraDx, Orient Gene and Siemens).
For test brands with three or more studies, the total number of samples per assay ranged from 251 for the Fujirebio assay to 15,331 for Abbott Panbio, and number of cases from 123 for Denka QuickNavi to 3989 for Abbott Panbio. Fifteen test brands were evaluated in a single evaluation each, with between 19 (Liming assay) and more than 1000 samples (SureScreen V assay and Fortress Diagnostics), and between nine (Liming assay) and 297 (Biosynex assay) samples from SARS‐CoV‐2‐positive cases. Average sensitivities ranged from 29.6% (95% CI 14.6% to 51.0%) for the Fujirebio assay based on 251 samples and 185 cases) to 91.2% (95% CI 70.0% to 97.9%) for LumiraDx, based on 741 samples and 177 cases). Specificities ranged from 93.2% (95% CI 91.3% to 94.6%) for the Shenzhen Bioeasy assay (n = 4, 1093 samples including 202 cases) to 100% (Anhui Deepblue, Denka QuickNavi, Orient Gene assay and Siemens CLINITEST) (Figure 12).
Using all data in symptomatic people, based on meta‐analyses, only the seven assays from AAZ, Abbott BinaxNOW, BIONOTE, Denka QuickNavi, LumiraDx, Quidel, and Shenzhen Bioeasy met the WHO threshold for acceptable sensitivity (point estimate for sensitivity of 80% or more), and only the LumiraDx assay exceeded the desirable sensitivity target of 90%. Of these, all except the Shenzhen Bioeasy assay met acceptable specificity levels (point estimate of 97% or more).
We judged just over half of evaluations (70/132) to be compliant with manufacturer IFUs in terms of sample site, use of VTM and time between sample collection and testing. Based on meta‐analyses, only five assays met the WHO acceptable performance standards for both sensitivity and specificity based on IFU‐compliant evaluations:
Abbott ‐ BinaxNOW COVID‐19 Ag card: 80.9% (95% CI 67.6% to 89.6%) and 99.9% (95% CI 99.5% to 100%); n = 4 evaluations, 2018 samples including 358 cases (James 2021; Pilarowski 2020a; Pollock 2021a; Prince‐Guerra 2021);
BIONOTE – NowCheck: 89.5% (95% CI 84.1% to 93.2%) and 97.7% (95% CI 95.8% to 98.8%); n = 2, 618 samples, 181 cases (FIND 2020a; FIND 2021a [B]);
Denka Co – QuickNavi: 84.2% (95% CI 66.2% to 93.5%) and 100% (95% CI 99.8% to 100%); n = 2 evaluations, 1633 samples, 123 cases (Takeuchi 2021a; Takeuchi 2021b);
LumiraDx: 91.2% (95% CI 70.0% to 97.9%) and 98.6% (95% CI 97.2% to 99.3%); n = 2, 741 samples, 177 cases (Drain 2021(b); Kruger 2021);
SD Biosensor/Roche ‐ STANDARD Q COVID‐Ag: 84.0% (95% CI 79.2% to 87.9%) and 99.2% (95% CI 98.8% to 99.4%); n = 15; 5116 samples, 1197 cases (FIND 2020c (BR); FIND 2020c (CH); FIND 2021c (BR) [B]; FIND 2021c (DE) [B]; Igloi 2021; Jakobsen 2021; Kerneis 2021; Lindner 2021a [B]; Lindner 2021b [C]; Mockel 2021(a); Mockel 2021(b); Nikolai 2021(b) [B]; Ristic 2021; Schuit 2021(b); Schwob 2020(a)).
Point estimates for sensitivity and specificity for a further two assays met WHO acceptable or desirable standards in a single evaluation each:
Mologic ‐ COVID 19 Rapid Ag: (sensitivity 90.6%, 95% CI 85.6% to 94.3% and specificity 100% (95% CI 99.2, 100; based on 650 samples including 192 cases (FIND 2021f);
Siemens – CLINITEST: 80.2% (95% CI 70.6% to 87.8%) and 100% (95% CI 95.8% to 100%); n = 1, 178 samples with 91 cases (Torres 2021b).
Note that for the majority of assays, the lower bound of the 95% CI for sensitivity does not exceed 80%, reflecting considerable remaining variability between studies even after restricting to those judged to meet IFU requirements.
For the two assays with evaluations carried out using nasal sampling kits, BIONOTE NowCheck (n = 1) and SD Biosensor STANDARD Q (n = 4), similar results to those using nasopharyngeal sampling kits were observed (Table 4).
Comparisons by brand in asymptomatic populations
A total of 19 test brands were evaluated in asymptomatic or mainly asymptomatic participants (n = 56 evaluations, including 6 in sensitivity‐only cohorts). We observed considerable heterogeneity in sensitivities for all assays (Appendix 13).
Three test brands (Abbott BinaxNOW, Abbott Panbio and SD Biosensor STANDARD Q) were evaluated in three or more evaluations (total of 36 evaluations) and we meta‐analysed results using the bivariate model (Figure 13). Four test brands were evaluated in two evaluations each and we meta‐analysed them using univariate analysis or by summing 2x2 tables (assays from Becton Dickinson, Innova Medical Group, RapiGEN, and Siemens). The total number of samples per assay ranged from 140 (RapiGEN, including 60 cases) to 12,530 (Abbott BinaxNOW, 588 cases). A further 12 test brands were evaluated in a single evaluation each, with between 15 (Fujirebio assay) and 1326 samples (BIONOTE ‐ NowCheck), and between one (Shenzhen Bioeasy assay) and 165 cases (Access Bio).
None of the assays met the WHO acceptable performance standard for sensitivity (of 80%) either based on meta‐analysis or in individual studies in asymptomatic people. Average sensitivities ranged from 38.5% (95% CI 28.4% to 49.7%; Innova assay; n = 2 including 6224 samples and 78 cases) to 63.2% (95% CI 21.7% to 91.4%; Rapigen; n = 2, 140 samples and 60 cases) and average specificities from 98.8% (95% CI 91.7% to 99.8%; RapiGen) to 100% (95% CI 99.8% to 100%; Innova assay; Figure 13). The highest observed sensitivities in individual studies (both compliant with manufacturer IFUs) were:
Denka Co QuickNavi: 75.8% (95% CI 57.7% to 88.9%) and 100% (95% CI 99.0% to 100%); 415 samples including 33 cases (Takeuchi 2021a);
LumiraDx assay: 77.8% (95% CI 40.0% to 97.2%) and 99.6% (95% CI 97.9% to 100%); 272 samples and 9 cases (Kruger 2021).
We judged just over half of evaluations (28/56) to be compliant with manufacturer IFUs in terms of sample site, use of VTM and time between sample collection and testing. On an individual study level, three of the 13 assays met the WHO acceptable sensitivity standard of 80% or more (Abbott Panbio, Abbott BinaxNOW and SD Biosensor STANDARD Q in one evaluation each, with case numbers ranging from 17 to 102). However, the average sensitivities for these assays ranged from 57.9% (95% CI 35.4% to 77.5%; Abbott Panbio; n = 7; 2502 samples and 279 cases) to 64.6% (95% CI 51.3% to 75.9%; SD Biosensor STANDARD Q; n = 4; 5914 samples and 250 cases).
Specificities were 99% or higher (meeting WHO acceptable performance standard for specificity) for 9 of 13 assays with IFU‐compliant evaluations (Table 4).
Direct test comparisons
A total of 23 studies provided within‐study comparisons by test brand (Appendix 13 see Figure 25): 19 reported both sensitivity and specificity data, and four were sensitivity‐only cohorts. Less than half of studies had sufficient assay comparisons in common to allow formal direct comparison using meta‐analysis (Table 5).
25.
Forest plot of data from studies reporting within‐study comparisons by test brand
Abbott Panbio versus SD Biosensor/Roche STANDARD Q assay
Nine studies reported sensitivity and specificity data for the Abbott Panbio and SD Biosensor STANDARD Q assays (Table 5). The results suggest higher sensitivity and lower specificity for the STANDARD Q assay (based on 3301 samples, including 1055 cases) compared to Panbio (based on 3895 samples including 1058 cases). Sensitivities were 64.4% (95% CI 52.2% to 75.0%) and 56.7% (95% CI 44.3% to 68.3%). The difference is 7.8 percentage points higher for STANDARD Q, 95% CI −9.1 percentage points lower to 24.6 higher), and specificities 97.5% (95% CI 96.7% to 98.0%) and 99.5% (95% CI 99.1% to 99.7%), respectively (difference 2.0 percentage points lower for STANDARD Q, 95% CI −2.7 to −1.3 percentage points).
The inclusion of one sensitivity‐only cohort made only a marginal difference to results (Table 5).
Abbott Panbio versus Orient Gene/Healgen Scientific ‐ Coronavirus Ag assay
Two studies reported sensitivity and specificity data for the Abbott Panbio and Orient Gene assay, and one further study reported sensitivity data only. No statistical evidence for a difference in sensitivity or specificity was observed (Table 5).
SD Biosensor ‐ STANDARD Q COVID‐Ag (Nasal kit) versus SD Biosensor/Roche ‐ STANDARD Q COVID‐Ag (NP kit)
Three direct comparisons showed almost no difference in assay performance between the STANDARD Q nasal kit using mid‐turbinate samples, and the nasopharyngeal kit (Table 5).
Evaluations of repeated test applications
Four studies evaluated repeated application of RDTs (Kriemler 2021; Love 2021; Smith 2021; Winkel 2020), one of which also reported data for a single test application and is also included in the previous section (Kriemler 2021). Because of differences in study populations, testing strategies and reporting of results, we review the four studies narratively and summarize them in Table 6.
5. Studies evaluating repeated (serial) antigen testing.
| Study | Population | Test and reference standard | Results |
| Kriemler 2021 | Pupils and teachers tested at primary or secondary school at T1 and or T2 (1 week apart). Schools were in high‐incidence areas; children were required to be kept at home if they were sick beyond very mild symptoms such as runny nose or mild cough 641 children and adolescents and 66 teachers tested at T1 and or T2; children provided 1170 samples (1 PCR+ve) |
Index: SD Biosensor Standard Q; paired buccal swabs obtained by study staff and tested immediately on site Reference: single PCR (Seegene Allplex) targeting N, S, RdRP and E‐gene; used paired buccal swabs |
T1 (data included in single test application analysis)
T2
The child with positive PCR at T1 was negative on both PCR and RDT at T2. 4/9 positive RDTs were repeated immediately with the same buccal swab and remained positive. 9/9 positive RDTs were repeated at 2 h to 2 days using new buccal sample; all were negative both on RDT and PCR, respectively. All positive test lines categorized as weak to moderate |
| Love 2021 | PHE feasibility study of daily contact RDT testing for 7 days in place of self‐isolation. Contacts of confirmed cases identified through NHS Test and Trace 882/1760 (50.1%) contacts agreed to participate; 812/882 were sent a testing kit. In‐study PCR results available for 346 contacts (64 PCR+ve) |
Index test: Innova RDT; self‐collected and self‐tested nasal swabs Reference: subgroup provided samples for PCR (60% of those providing ≥ 1 RDT result, plus PCR results accessed for those having PCR outside of study); confirmatory PCR either if RDT+ve or at end of 7‐day period |
570/812 returned ≥1 RDT result (2946 samples in total)
346/562 submitted swabs for PCR within the study protocol; se and sp of the RDT were
Of the 11 RDT‐negative/PCR+ve results, median Ct values were 24.0 (range 16.9‐32.1) for ORF1ab gene and 25.5 (range 16.9‐33.5) for E‐gene; 6/11 self‐reported symptoms. 224/562 did not submit swabs for PCR, results obtained via NHS Test and Trace for 51/224
Secondary attack rates (i.e. transmission to contacts) reported for 160 contacts of 84 index cases: 10/160 confirmed PCR+ve, secondary attack rates 6.3% (95% CI 3.4% to 11.1%) Reported reasons for participation in study included: duty (298/882, 33.8%), assurance given from daily testing (268, 30.4%), escape from self‐isolation restrictions (229, 26.0%) |
| Smith 2021 | Daily testing of PCR+ve students and their contacts to demonstrate RDT sensitivity over time during early infection; index cases identified from routine campus‐based testing. PCR+ve cases followed up (daily testing) for 14 days; contacts with all negative PCR results were followed up for 7 days. Data reported for 43/51 PCR+ve; 8 without positive viral culture were excluded |
Index test: Quidel SOFIA Ag using self‐collected nasal swabs; laboratory‐based direct testing the day after collection Reference standard: PCR (saliva and nasal swabs) and viral culture (PCR + nasal swabs) |
‘Daily sensitivity’ of RDT or PCR in relation to time before or after first successful culture (from 2 days prior, to 13 days after)
Data used to model ‘protocol sensitivities’ of different testing strategies.* Probability of positive result before or during period when viral culture is positive was estimated as:
* 'protocol sensitivity' was reported as calculated using a value‐of‐information approach assuming seven different testing frequencies (from daily up to weekly sampling) (Smith 2021). |
| Winkel 2020 | Players, staff and referees from 13 professional football clubs and the national teams in the Netherlands tested approximately weekly independent of presence of symptoms, 2 days prior to each match; results for symptomatic individuals were excluded 824 people provided 2425 samples; 52 positive on PCR (68 samples). 23/52 remained asymptomatic during testing period, 29/52 developed symptoms after 1st positive PCR test |
Index: Abbott Panbio; NP swabs tested immediately by "trained personnel". Accuracy was calculated by either excluding results with weak test bands or by counting them as +ve or ‐ve to determine best and worst case scenarios. Reference: single PCR (one of 3 assays used), genetic targets not reported; paired throat/NP swabs used (104, 4% PCR swabs obtained on subsequent day) |
Sensitivity and specificity calculated on a per‐sample basis; samples reported during earlier phase of infection likely to be on par with per‐patient results. Counting inconclusive results as RDT positive: Sensitivity
Specificity: 99.5% (95% CI 99.2% to 99.8%) (2327/2338) |
| CI: confidence interval; Ct: cycle threshold; N/A: not applicable; NHS: National Health Service (UK); NP: nasopharyngeal; PCR: polymerase chain reaction; PHE: Public Health England; pso: post‐symptom onset; RDT: rapid diagnostic test; PCR: reverse transcription polymerase chain reaction; Se: sensitivity; Sp: specificity | |||
One school‐based study (Kriemler 2021), evaluated pupils and teachers tested at primary or secondary schools in high SARS‐CoV‐2 incidence areas in Switzerland at two time points (630 participants at time point 1 (T1) and 659 at time point 2 (T2)), a week apart (paired buccal swabs collected and tested by study staff). Only one PCR−positive participant was identified: the pupil was PCR−positive and RDT‐negative at T1, and was negative on both tests at T2. The specificity of the STANDARD Q assay was 98.9% (95% CI 97.7% to 99.6%) at T1 and 99.5% (95% CI 98.7% to 99.9%) at T2.
A second study evaluated weekly testing using the Abbott Panbio assay in football players, staff and referees from 13 professional football clubs and the national teams in the Netherlands over a five‐ to six‐week period (Winkel 2020). Results from symptomatic participants at the time of testing were excluded from the study. A total of 824 people provided 2425 paired nasopharyngeal (for RDT) and naso‐ and oropharyngeal (for PCR) samples that were collected and tested on site by trained personnel; 52 individuals were positive on RT‐PCR (68 samples), 23 (44%) of whom remained asymptomatic during the testing period and 29 developed symptoms after the positive PCR result. Results were available only on a per sample basis but were separated according to phase of infection (Table 6). The observed sensitivity of the RDT was 90% or higher during the pre‐symptomatic and early infection phases: 91.7% (95% CI 61.5% to 99.8%; based on 12 PCR−positive samples) and 90.6% (95% CI 75.0% to 98.0%; based on 32 PCR−positive samples). Seven days after first positive PCR or after symptom onset (late infection), sensitivity fell to 33.3% (95% CI 14.6% to 57.0%; based on 21 samples) and to 0% (based on 3 samples) four weeks after first positive PCR. Specificity (on a per sample basis) was 99.5% (95% CI 99.2% to 99.8%; 11 RDT‐positive samples from a total of 2338 PCR negative).
Two studies evaluated daily RDT testing. Love 2021 was a UK feasibility study of daily testing of contacts of confirmed SARS‐CoV‐2 cases to allow release from self‐isolation, Smith 2021 evaluated daily testing of PCR−positive students in the USA and their contacts to determine the sensitivity of RDT, direct saliva RT‐PCR and conventional RT‐PCR using nasal swabs over time.
In Love 2021, samples were self‐collected and self‐tested with the RDT. At least one RDT and one PCR result were available for 346 participants out of the 812 who were sent a test kit (PCR was required either if the daily RDT was positive or at the end of the seven‐day testing period), including only 55 of the 102 who reported at least one positive RDT. Overall sensitivity of was 82.8% (95% CI 71.3% to 91.1%; based on 64 SARS‐CoV‐2‐positive on PCR) and specificity was 99.3% (95% CI 97.5% to 99.9%; based on 282 with negative PCR results; Table 6). The majority of cases were in those who self‐reported as having developed symptoms of COVID‐19 (55/346 participants, including 45 PCR−positive cases); RDT sensitivity was 88.9% (95% CI 75.9% to 96.3%; based on 45 cases) and specificity 80.0% (95% CI 44.4% to 97.5%; based on 10 PCR−negative participants). In those who remained asymptomatic and who returned RDT and PCR results (291/346, 84%), sensitivity was 68.4% (95% CI 43.4% to 87.4%; 19 PCR positive) and specificity was 100% (95% CI 98.7% to 100%; 272 PCR−negative participants). PCR Ct values for samples from the 11 false‐negative cases ranged from 16.9 to 33.5 Ct (median 24.0)
Smith 2021 reported results for 43 of 51 PCR−positive individuals who had positive results on viral culture. Participants were enrolled either within 24 hours of a positive PCR result from routine surveillance or as contacts of a PCR−positive individual. Participants self‐collected samples for daily testing with Quidel’s SOFIA assay (nasal swabs), direct RT‐PCR (saliva) and conventional PCR (nasal swabs). All testing was laboratory‐based and was conducted within a day of sample collection. The testing of contacts allowed the study authors to calculate assay sensitivity in relation to the time before or after the first successful culture (reported as the ‘daily sensitivity’ of the RDT and both PCRs). Only around a quarter (10 participants for 2 days prior to positive viral culture) to a half (20 participants for 1 day prior) of participants reported results for samples collected before successful viral culture, however results suggest that both PCR−based approaches were considerably more sensitive (between 70% and 80% sensitive) than testing using the RDT (30% sensitive at two days prior and 40% sensitive the day before positive viral culture; Table 6). For samples collected on the day that viral culture became positive, RDT sensitivity increased to 90.5% (based on 42 samples), compared to 97.6% for direct saliva PCR and 100% for conventional PCR. Daily sensitivity increased slightly for the RDT and direct PCR assays using samples collected on the following day (n = 43), sensitivities 97.7%, 100% and 95.3%, and then began to decline a faster rate for the RDT compared to the other assays. By day 4 after positive viral culture, the RDT daily sensitivity (based on 43 samples) was 62.8% compared to 95.3% for direct PCR and 43% for conventional PCR (Table 6). The study authors used daily sensitivity results to model the probability of a positive result before or during the period when viral culture is positive for testing strategies using each assay; sensitivities were highest for a PCR−based approach, however differences were relatively small (Table 6).
Discussion
This is the third iteration of a Cochrane living review summarising the accuracy of point‐of‐care antigen tests for detecting current SARS‐CoV‐2 infection. This version of the review is based on published journal articles or studies available as preprints from 1 January 2020 up until 9 March 2021. In addition, we also included evaluations that were available as independent national reference laboratory publications or that were co‐ordinated and published by FIND, and journal articles that were listed on the Diagnostics Global Health website to 30 April 2021.
Summary of main results
We included data from 152 studies evaluating the accuracy of a single antigen test application, including 100,462 participants (16,822 samples with confirmed SARS‐CoV‐2), and four studies evaluating the accuracy of repeated test applications (1920 participants, 160 with confirmed SARS‐CoV‐2). The main results focus on the former group of studies reporting evaluations of a single antigen test application; these report a total of 210 test evaluations and a further 18 evaluations comparing accuracy in different sample sites. Key findings are presented in the Table 1.
Key findings
We summarize eight key findings from this review.
1. Lack of evidence for commercially produced tests
Despite a considerable increase in the number of studies evaluating point‐of‐care antigen tests, we did not identify any published or preprint reports of accuracy for a significant proportion of commercially produced point‐of‐care tests. This review located evaluations for 49 RDTs; these represent a small proportion of the 321 commercially produced assays (FIND 2022a).
2. Methodological standards have improved
Some improvements in methodological standards can be observed for antigen test evaluations compared to those in the previous version of this review, particularly in regard to the applicability of the evidence for both the participant selection domain and the index test domain, with tests increasingly used in accordance with manufacturer IFUs. The higher number of studies reporting results in asymptomatic participants has contributed to small improvements in methodological standards in regard to the reference standard. However, some concerns about risk of bias and applicability of results remain, and further improvements in study methods and reporting could be implemented by study authors.
Particular methodological concerns that remain include the use of deliberate sampling according to known presence or absence of SARS‐CoV‐2 infection, a lack of information about the presence of symptoms or time from symptom onset and poor reporting about blinding of index and reference standard interpretation. Differences in case‐mix related to symptomatic status, time post‐symptom onset and distribution of viral load are likely to have contributed to the observed variation in accuracy. RT‐PCR was the reference standard for the presence of infection in all studies apart from one using an alternative molecular method (TMA) ‐ no study defined the presence of COVID‐19 using clinical or radiological features in the absence of a negative RT‐PCR result, and the majority continue to rely on a single negative RT‐PCR as evidence of absence of infection.
3. Compliance with manufacturers' instructions was relatively poor
There has been an improvement in the reporting of tests being used at the point of care as opposed to in centralized laboratory settings, however studies frequently did not follow the manufacturer IFUs in regard to sample type, and timing of tests. Fewer than half conducted the tests according to the manufacturer IFU (43% (90/210) compared to 50% (29/58) of antigen test evaluations in the previous review iteration). Non‐compliance with IFUs was frequently because VTM was used when this was either not recommended by the manufacturer or the manufacturer did not provide instructions for use with VTM, or because lengthy intervals between sample collection and testing were reported (often with a period of frozen storage). Of the 113 (54%) evaluations reporting direct swab testing, 85 complied with manufacturer IFUs. Reasons for non‐compliance were use of samples not specified on the IFU (n = 17), or samples were tested after the time limit specified on the IFU (n = 10), or the time delay between sample collection and testing was not specified (n = 7). Only 29% of antigen test evaluations included in the last iteration of this review in March 2021 reported on‐site, direct swab testing immediately or within an hour of sample collection.
4. Small number of assays meet minimum acceptable sensitivity requirements in symptomatic participants
For antigen test evaluations in symptomatic participants, we observed a similar pattern of results as for the previous review iteration. Considerable heterogeneity in sensitivities (and to a lesser extent the specificities) remained, however the increase in number of evaluations and samples analysed increased confidence in the observed results. The average sensitivity of RDTs in symptomatic populations was 73.0% (95% CI 69.3% to 76.4%) and specificity 99.1% (95% CI 99.0% to 99.2%). Average sensitivity decreased with time since onset of symptoms, being higher in the first week (80.9%, 95% CI 76.9% to 84.4%) than when done in week two or later (48.9%, 95% CI 37.9% to 60.1%), but with similarly high specificities at both time points (99.2% and 99.5% on average). We were also able to demonstrate higher sensitivities for individuals presenting to COVID‐19 testing centres compared to all other settings.
Focusing on studies that used the test in accordance with the manufacturer’s instructions, sensitivities for different brands varied from 34% to 91% in symptomatic participants (either based on summary results or single studies). The WHO has set a minimum 'acceptable' sensitivity requirement of 80%, (≥ 90% 'desirable') and acceptable and ideal (or 'desirable') specificity requirements of 97% and 99% respectively (WHO 2020b). Seven assays (from AAZ, Abbott (BinaxNOW), BIONOTE, Denka Co, LumiraDx, Quidel, and Shenzhen Bioeasy) met the WHO acceptable criterion for sensitivity based on summary results of several studies, but the 95% confidence intervals for all results apart from for BIONOTE NowCheck overlapped the 80% standard. Two additional assays (from Mologic and from Siemens) also met the acceptable sensitivity criterion, but each test was evaluated in only one study. The 95% confidence intervals of the summary results for a number of other assays including Abbott Panbio overlapped the sensitivity criterion, but the point estimates were below 80%. The acceptable performance criterion of 97% specificity was also met for all tests apart from the Shenzhen Bioeasy assay.
Considerable heterogeneity in sensitivities remained after restricting analyses by test brand in symptomatic populations, suggesting an effect not only from participant characteristics but from setting, sample type and collection method, sample storage and preparation, and testing procedures that cannot be easily unpicked. For example, results by sample site suggest superior sensitivity from studies using nasal samples (76.6%, 95% CI 70.3% to 81.9%) compared to nasopharyngeal samples, either as the sole sample site, or in combination with oropharyngeal sampling in some or all participants (69.0%, 95% CI 65.3% to 72.4%). However, this suggestion of difference was not supported when the comparison was restricted to within‐study comparisons of the two sampling sites. Limited data suggest lower RDT sensitivity for saliva and for oropharyngeal samples alone. Our previous review iteration pointed to possible differences in sensitivity according to test operator demonstrated in UK government‐funded PHE studies of the Innova assay. Because of the various possible sources of confounding we did not carry out subgroup analyses by test operator for this version of the review. A single study directly comparing assay interpretation by test operator showed identical specificities and slightly higher sensitivity (by 2.5 percentage points) for the professional‐interpreted tests compared to participant‐interpreted tests. We are aware of additional studies examining the effect of self‐collection and test operator on accuracy (e.g. Klein 2021), that will be considered for inclusion in the next review iteration.
5. Sensitivity is lower in asymptomatic participants but is higher if there is epidemiological exposure to SARS‐CoV‐2
Fifty studies reported data about the accuracy of antigen tests in asymptomatic people for detection of SARS‐CoV‐2 infection defined by PCR status (an increase from 13 in the previous iteration), including 17 conducted exclusively in asymptomatic or mainly asymptomatic populations. As discussed, this does not address the issue of whether the test is identifying those who are infectious (as there is no reference standard that can be used). The average sensitivity for detecting infection in asymptomatic participants was 54.7% (95% CI 47.7% to 61.6%) with specificity of 99.5% (95% CI 99.4% to 99.6%), 18.2 percentage points lower than for symptomatic populations (95% CI from ‐26.1 to ‐10.4 percentage points). Unlike for the previous review iteration however, for this review we were able to conduct some subgroup analyses by eligibility for testing and setting. Considerably higher summary sensitivities were observed when an epidemiological exposure to SARS‐CoV‐2 was suspected (sensitivity 64.3%, 95% CI 54.6% to 73.0%) or for asymptomatic participants presenting to COVID‐19 test centres (61.5%, 95% CI 54.0% to 68.4%), compared to studies where RDTs were reportedly widely available to anyone presenting for testing (sensitivity 49.6%, 95% CI 42.1% to 57.1%) or for evaluations considered to represent screening scenarios (45.1%, 95% CI 36.4% to 54.1%) or school or university‐wide testing programmes (47.9%, 95% CI 38.1% to 57.9%).
Three evaluations reporting data by time after exposure to infection in asymptomatic participants provided weak evidence for higher RDT sensitivity in week 1 (70.0%, 95% CI 60.8% to 77.8%) compared to week 2 (60.7%, 95% CI 48.0% to 72.0%) after exposure.
There was considerable variation in sensitivities in asymptomatic participants between test brands, however the number of evaluations per test brand was small such that for many brands heterogeneity is likely to be strongly influenced by setting, timing and indication for testing of asymptomatic people.
6. Steady decline in sensitivity with lower viral load
For this version of the review, sufficient data were available for a more detailed investigation of RDT accuracy by viral load, moving away from an overly simplistic dichotomous analysis of results above and below any single Ct or RNA copies/mL threshold. A steady decline in RDT summary sensitivities was observed, from 97.4% (95% CI 95.0% to 98.6%) and 98.4% (95% CI 97.0% to 99.1%) in participant samples with the highest viral load (< 20 Ct or ≥ 10^7 RNA copies/mL) to 68.7% (95% CI 61.6% to 75.0%) and 70.9% (95% CI 57.4% to 81.5%) for samples in the 25 to 30 Ct or 10^5 RNA copies/mL range. Considerably lower average sensitivities were observed in the lowest viral load subgroups, for example, 7.5% (95% CI 3.8% to 14.3%) for those with < 10^4 RNA copies/mL and 36.7% (95% CI 24.7% to 50.5%) for samples with 10^4 RNA copies/mL. Data according to ‘viral load’ was contributed from studies including both symptomatic and asymptomatic participants. We were not able to consider any effect from symptom status on viral load patterns because of relatively low numbers of evaluations.
At lower Ct values and higher RNA copies/mL, different test brands appear to perform relatively consistently, with a few exceptions. As Ct increases and RNA copies/mL falls, we observed considerably greater heterogeneity in sensitivity, however it is not clear whether there are systematic differences in assay performance for samples with lower viral loads or whether other differences between studies might explain the observed variability. Studies comparing analytical sensitivities between brands however, suggest true differences in the ability of different assays to detect lower concentrations of virus (e.g. Karon 2021; Mak 2021). What is not clear is the extent to which missed cases with samples in the mid to low range of viral load could be contributing onward transmission of infection.
7. Suggestion of lower sensitivity in children
Limited data suggest possibly lower sensitivity but similar specificity in children. Using all data reported for children, average sensitivity was 62.7% (95% CI 52.7% to 71.7%) and average specificity 99.4% (95% CI 99.1% to 99.6%). Restricting the analysis to studies reporting data for both children and adults (thereby minimising other differences between groups) average sensitivity was 9.9 percentage points higher (95% CI −8.7 to 28.4; a difference that might be observed by chance), and average specificity 0.7 percentage points higher (95% CI 0.2 to 1.2) in adults compared to in children. With increasing evidence that viral loads are similar between children and adults (Chung 2021; Madera 2021; Yonker 2021), other factors such as the adequacy of sampling, timing of testing in relation to onset of infection, or participant characteristics, are likely to have contributed to the observed results.
8. Limited evidence for repeat testing strategies
Repeated use of antigen tests in different asymptomatic groups, such as school children and staff, hospital and care home workers, and the general public is increasingly advocated. We found only four eligible studies evaluating the accuracy of repeated testing within our search period. The studies varied in purpose, design, testing strategies and presentation of results such that it is not possible to make generalizations about the value of repeated testing strategies. Setting aside the study with only one PCR−positive case (Kriemler 2021), one study suggested that regular weekly testing of asymptomatic adults might pick up around 90% of those with pre‐symptomatic or early infection (Winkel 2020), however the number of cases detected was relatively small and confidence intervals wide. Specificities were close to or above 99% in both studies of weekly testing.
A feasibility study of daily contact testing suggested this approach could detect between 68% of asymptomatic and 89% of symptomatic cases (Love 2021); PCR Ct values of cases missed by the RDT ranged from 16.9 to 32.1 however this may not fully reflect Ct values at the time of the RDT because of delays between RDT and PCR sample collection. The final study reported results of repeated daily testing in those with at least one sample with successful viral culture (Smith 2021); in the days prior to successful viral culture the RDT used was considerably less sensitive than either direct saliva RT‐PCR or conventional nasal swab RT‐PCR and only demonstrated similar sensitivity (within 10 percentage points of conventional RT‐PCR) for samples obtained on the day that viral culture became positive up to day 2 after viral culture positivity. Model‐based estimates of the sensitivity of different testing strategies suggested that daily RDTs would be needed to detect 90% of PCR−positive cases, dropping to only 80% sensitivity for testing every three days (Smith 2021).
Additional studies of repeated testing that did not report results as accuracy estimates suggest sub‐optimal detection rates from RDTs compared to PCR in the days leading up to onset of symptoms (Basile 2021), and further, that even daily RDT testing may not be sufficient to contain transmission (Moreno 2021).
We have already identified a number of additional studies of repeated testing strategies for consideration for the next update of this review (e.g. Aranda‐Diaz 2021; Harmon 2021; Kanji 2021; Kweon 2021; McKay 2021; Shah 2021; Sterbenc 2021; Young 2021a). Studies listed in Characteristics of studies awaiting classification are those already identified as eligible (according to current review eligibility criteria) up to August 2021 with additional searches completed up to October 2021.
Illustration of predicted effect of antigen testing by symptom status
Below we illustrate predicted numbers of true positives, false positives, false negatives and true negatives, applying summary estimates of test accuracy to hypothetical cohorts of symptomatic or asymptomatic people suspected of SARS‐CoV‐2 infection across a range in prevalence of SARS‐CoV‐2 infection (Table 1).
For antigen test evaluations in symptomatic people, we used data for all symptomatic participants combined (sensitivity 73.0%, 95% CI 69.3% to 76.4%, and specificity 99.1%, 95% CI 99.0% to 99.2%), and data for symptomatic participants tested during the first week after symptom onset (sensitivity 80.9%, 95% CI 76.9 to 84.4% and specificity 99.5%, 95% CI 99.3% to 99.6%). The latter estimates are also in the range of those observed for symptomatic people presenting for testing at COVID‐19 test centres. Applied to a cohort of 1000 people with signs and symptoms of COVID‐19, in whom 50 people had confirmed infection (prevalence of 5%), we predicted that:
46 (overall) or 45 (week 1) people would have a positive test result, of which 9 or 5 would be false positives (positive predictive values (PPV) 81% and 89%, respectively), and
14 (overall) and 10 (week 1) people with negative test results would be falsely negative (negative predictive values (NPV) 98.6% and 99.0%).
Increasing the prevalence to 10% or 20%, increases PPV to 90% or more and slightly decreases NPV. As there is some heterogeneity in the estimates of sensitivity, the values observed in practice could vary slightly from these figures as shown by the estimates derived from the confidence intervals for the summary estimates (Table 1).
For antigen test evaluations in asymptomatic participants we used subgroup data according to whether testing was reported to be widely available to any asymptomatic person with no requirement to meet pre‐set criteria for testing (sensitivity 49.6%, 95% CI 42.1% to 57.1%, and specificity 99.6%, 95% CI 99.5% to 99.7%) and where testing was restricted to those reporting epidemiological exposure to COVID‐19 (sensitivity 64.3%, 95% CI 54.6% to 73.0%, and specificity 99.7%, 95% CI 99.5% to 99.8%). Applying the average values to a larger cohort of 10,000 people asymptomatic for COVID‐19 and with a lower prevalence of 0.5% in whom 50 people had confirmed infection (infectious or not):
65 (widely available) or 62 (epidemiological exposure) individuals would have a positive test result of which 40 and 30 would be false positives (PPVs of 38% and 52%, respectively), and
25 (widely available) and 18 (epidemiological exposure) people with negative test results would be falsely negative (NPVs 99.7% and 99.8%).
The confidence intervals for the average sensitivity estimates used in these calculations are relatively wide, such that the number of false negatives observed in practice could differ from these figures, as can be seen from the estimates derived from the confidence intervals. However, at very low prevalence of disease, as might be seen in a mass screening scenario, the effect on the absolute number of false negatives observed could be small (e.g. from 21 to 29 using the 95% CIs for the average sensitivity where testing was widely available). In contrast, although the 95% CIs for average specificities were only 0.2 to 0.3 percentage points wide the absolute numbers of false positives ranged between 20 and 50.
Increasing the prevalence of confirmed SARS‐CoV‐2 infection to 1% or 2% makes little difference to the absolute number of false positive results, but has a large relative effect when considered in relation to the total number of positive test results (true and false positives; PPVs increasing to 72% for widely available testing and 81% for testing contacts of confirmed cases at 2% prevalence; Table 1).
Strengths and weaknesses of the review
Our review used a broad search screening all articles concerning COVID‐19 or SARS‐CoV‐2. We undertook all screening and eligibility assessments, QUADAS‐2 assessments (Whiting 2011), and data extraction of study findings independently and in duplicate. Although it is possible that the use of artificial intelligence text analysis to identify studies most relevant to diagnostic questions may have led to some eligible studies being missed, we believe that the multi‐stranded search strategy used will have identified most if not all relevant literature. Whilst we have reasonable confidence in the completeness and accuracy of the findings up until the search date, should errors be noted please inform us at coviddta@contacts.bham.ac.uk so that we can verify and correct in our next update. The review is however limited by the March 2021 cut‐off for the electronic searches. While the effect of this is mitigated to some extent by including studies from other sources up to 30 April 2021, we are aware of a large number of eligible studies published or available as preprints in the interim period. Full‐text assessment of studies available up to 18 August 2021 has resulted in 84 studies that will be eligible for a subsequent review update according to current review inclusion criteria (described in Characteristics of studies awaiting classification). A brief review of these studies indicates that their inclusion in the review will further strengthen rather than change our conclusions about the accuracy of single applications of a test for diagnostic or screening purposes, however we anticipate inclusion of additional information about the accuracy of repeated testing strategies.
We explicitly considered whether the test evaluations were conducted in accordance with the manufacturer IFU, regarding the sample types used, the use of VTM and the permitted time between sample collection and testing. We did not however consider any manufacturer statements on the intended use of the tests by population, but we are aware that some IFUs recommend testing only in symptomatic people and within certain time frames after symptom onset (see Appendix 9). Instead, we have provided data separately for symptomatic and asymptomatic participants and identified clear evidence of lower sensitivities in asymptomatic individuals for detection of infection.
We did not attempt to assess the accuracy of antigen tests for identification of infectious individuals, as there is no established reference standard for infectiousness (and it seems unlikely that one will ever be established). For the first time, however, data have permitted presentation of results according to ‘viral load’ in smaller subgroups by Ct value or by RNA copies/mL, the latter approach going at least some way to addressing variation in RT‐PCR Ct values between assays (Vogels 2020), and between laboratories. Our results support a steady deterioration in summary sensitivity as viral load (or at least these proxy measures of viral load) decreases, and relatively poor assay sensitivity (around 70%) for samples with mid‐range Ct or RNA copies/mL values. As previously discussed, there is no 'step change' in 'infectiousness' according to any fixed Ct value; increasing numbers of studies demonstrate successful viral culture in individuals considered to have 'low' viral load (Jaafar 2020; Singanayagam 2020), and, more importantly, that transmission of infection does occur from index cases with high RT‐PCR Ct values (Lee 2021; Marks 2021; Tian 2021). A large Danish study looking at household transmission found that 34% of all secondary cases (almost 30,000 total cases) were in households where the primary index case had Ct values of 30 or more (Lyngse 2021). Ultimately, viral load on its own is only one factor influencing an individual's ability to transmit infection, 'infectiousness' being modified by host factors such as the health of an individual’s immune system, vaccination status, presence of comorbidities, and environmental risk factors including closeness and length of contact with others.
Thus far we have also been unable to systematically consider test accuracy for detection of more infectious SARS‐CoV‐2 variants of concern such as Delta and, more recently Omicron (B.1.1.529), or to consider whether test accuracy might vary between vaccinated and unvaccinated individuals. Studies of analytical accuracy for detection of different variants (including Alpha and Delta variants) have suggested no consistent effect on test accuracy, with most RDTs examined to date showing similar accuracy regardless of variant (Bekliz 2021; Frediani 2021; Lindner 2021). It is too early to determine whether variations in accuracy might be observed for detection of the Omicron variant in clinical performance evaluations. Thus far, a small UK Health Security Agency laboratory‐based evaluation found no evidence for impaired analytical sensitivity of five different RDTs (UK HSA 2021b), however the Food and Drug Administration (FDA) have released a statement advising potentially reduced sensitivity of RDTs for detection of the Omicron variant (FDA 2021).
Weaknesses of the review primarily reflect the weaknesses in the primary studies and their reporting. Although small improvements in study quality were observed, a good proportion of studies continue to omit descriptions of participants, and key aspects of study design and execution. In order to include data for all tests in meta‐analyses we had to include some samples multiple times. We have been explicit about these issues where they arose. It is possible that eligible studies have been missed by our search strategy however we believe the risk to be very low considering our broad approach to identification of literature. Despite our best efforts to be as comprehensive as possible, new evaluations are continuously becoming available and it is impossible for any published and peer‐reviewed systematic review to be fully up to date.
We are aware of one other systematic review of antigen detection tests that covers a similar search period to our review, with electronic searches to 30 April 2021 (Brümmer 2021). There is a very high degree of overlap in included studies between the two reviews and a similar approach to overall analysis of studies was taken. Our review however includes a much higher number of samples from asymptomatic participants (e.g. 40,956 samples from 50 evaluations compared to 15,228 samples from 25 evaluations in Brümmer 2021), allowing us to conduct a more detailed analysis by study setting and indication for testing. We have also taken a different approach to analyses by viral load (data categorized in smaller subgroups as opposed to analyses above and below single threshold values).
Around a fifth (25/130) of primary study reports are only available as preprints, and as yet, have not undergone peer review (a fall from 25% of primary study reports in the previous review iteration). As published versions of these studies are identified in the future, we will double‐check study descriptions, methods and findings, and update the review as required.
Applicability of findings to the review question
There are an increasing number of roles and testing strategies for which rapid antigen assays are considered, and it is likely that the performance of these tests needs to be considered separately for each of the use cases. It is notable that the majority of studies were conducted in Europe or North America (116/152) and it is not fully clear whether results will generalize to low‐ or middle‐income countries
Our review shows that on average antigen tests do not perform as well in asymptomatic populations compared to symptomatic populations for detecting infection. However, asymptomatic individuals may be tested in a range of scenarios, from preventive or targeted screening, to contact tracing or testing at dedicated COVID‐19 test centres. We have been able to demonstrate higher RDT sensitivity when used in individuals likely to have had a recent epidemiological exposure to a confirmed case, or in asymptomatic individuals presenting for testing at a COVID‐19 test centre, or both, compared to assay use in ‘mass’ screening scenarios where asymptomatic individuals are encouraged to present for testing regardless of epidemiological indications. Lower sensitivities in the latter group will be affected by a number of factors, including time since exposure to infection and a potentially shorter timeframe in which asymptomatic people have higher viral loads. Variation in viral trajectories between individuals (Cevik 2021), also mean that even when an asymptomatic person can identify a clear contact with a confirmed case of SARS‐CoV‐2 infection, it is not possible to pinpoint when (or even if) that individual will have a sufficient viral load to be detected on antigen testing.
Incomplete symptom assessment and lack of adequate follow‐up to identify subsequent development of symptoms or previous history of symptoms can all contribute to inappropriate classification of individuals as having asymptomatic infection (Meyerowitz 2020). Although we have been able to consider the effect of study setting and indication for testing in more detail than previously, the estimates for test accuracy for asymptomatic populations primarily represent accuracy in those without clearly defined symptoms at the time of testing. Two studies of repeated antigen testing in asymptomatic people did however suggest higher RDT sensitivities for those who went on to develop symptoms or for those who were tested early during the course of infection. Serial testing may only achieve optimal levels of sensitivity (90% or more) when implemented on a daily basis however, and furthermore this does not mean that all potentially infectious cases of SARS‐CoV‐2 would be picked up. We are aware that several studies of asymptomatic testing have been reported since the close of our search and will further contribute to the debate around optimal targeted deployment of antigen detection tests in asymptomatic individuals (Caruana 2021; Fernandez‐Montero 2021; Kumar 2021a; Norizuki 2021; Revollo 2021; Sood 2021; Sterbenc 2021).
Authors' conclusions
Implications for practice.
We consider the implications for practice for this review separately for symptomatic and for asymptomatic testing.
In the Role of index test(s) section, we suggested that for symptomatic individuals, and if sufficiently accurate, point‐of‐care testing could be used either to replace laboratory‐based reverse transcription polymerase chain reaction (RT‐PCR) or as a triage to RT‐PCR. As point‐of‐care tests are more accessible and provide a result more quickly than RT‐PCR, theoretically their use may increase detection and speed up isolation and contact‐tracing, leading to reduction in disease spread and reduce the burden on laboratory services.
The evidence included to date suggests the following.
1. For diagnosis in symptomatic individuals in the first few days of symptoms, the most accurate rapid antigen tests are a useful alternative to laboratory‐based RT‐PCR where immediate results are required for timely patient management or where there are significant logistical or financial challenges in delivering RT‐PCR in a timely manner. Rapid antigen tests are only sufficiently sensitive in the first week after onset of symptoms. This conclusion can be considerably strengthened in comparison to the previous iteration of this review.
We have continued to observe variable sensitivity between assay brands. Only those shown to meet appropriate criteria, such as the World Health Organization's (WHO) priority target product profiles for COVID‐19 diagnostics (i.e. sensitivity ≥ 80% and specificity ≥ 97%; WHO 2020b), could be considered as a rational substitute for RT‐PCR.
Tests had high specificity, thus in symptomatic populations (where prevalence is likely to be high) the risk of false positives is low. At 80% sensitivity compared to RT‐PCR, the probability that infected individuals are missed is 20% higher than for RT‐PCR. Thus the possibility of false negative results should be considered in those with a high clinical suspicion of COVID‐19, particularly if tested several days after onset of symptoms when viral load levels may have fallen.
2. Rapid antigen tests could be used simultaneously with RT‐PCR for symptomatic people, particularly where RT‐PCR turn‐around times are slow, to exploit the benefits of earlier results and consequent contact‐tracing and isolation. Given the risk of false‐negative results, isolation may be required until RT‐PCR−negative results are obtained. Similarly, for investigation of local outbreaks, rapid antigen testing in a clearly defined population may establish cases and contacts that require isolation whilst awaiting results from RT‐PCR.
In other circumstances rapid antigen tests may be used to triage to follow‐on RT‐PCR tests (rather than all receiving PCR tests) dependent on prevalence and the consideration of the consequences of false positive and false negative results.
Where prevalence is low, positive rapid test results in symptomatic individuals require confirmatory testing to avoid unnecessary quarantine measures (positive predictive values (PPVs) around 80% to 90% for antigen assays mean that between 1 in 5 and 1 in 10 positive results will be falsely positive). Self‐isolation for the duration of symptoms in those with negative rapid test results should minimize the effect on transmission of infection from missed cases. Where available, testing by RT‐PCR may be reasonable for people with a high clinical suspicion of COVID‐19 and negative rapid test.
Where prevalence is higher (i.e. 20% or higher), false positives are less of a concern (PPVs around 95%) but the impact from false negative results becomes increasingly important and all test negatives may be considered for verification. At 20% prevalence, and using data for symptomatic people presenting to COVID‐19 test centres, around 4% of those with negative rapid test results are missed cases of SARS‐CoV‐2 (30 to 40 cases missed out of a total of 200 cases). The lower the negative predictive value (NPV), the greater the potential effect on transmission of infection from missed cases and greater the impact from delays in commencement of contact tracing. For scenarios in which positive results do not have confirmatory testing, it is important that assays with high specificities (in the range of 99% to 100%) are selected in order to minimize the impact from false positive results at higher prevalences of disease.
3. We found some evidence for higher sensitivity in people with a known exposure to SARS‐CoV‐2 compared to testing scenarios more to akin mass screening of asymptomatic individuals
The key focus in mass screening is identification of individuals who are or will become infectious. PCR−positives define those who had detectable viral particles on their swab, which will include most of those who are or will become infectious, but also include individuals post‐infection with residual viral particles. Without a reference standard for infectiousness, test accuracy studies cannot assess the ability of the test to detect the infectious subgroup of infections, and cannot provide evidence as to how well rapid antigen tests differentiate between individuals requiring isolation and those who provide no risk. The effectiveness of mass screening using these tests will ultimately only be established though participant outcome studies, such as cluster‐randomized community trials.
Rapid tests demonstrating the highest specificities (i.e. 99% and above) should be considered for use during a period of outbreak or in those with likely exposure to SARS‐CoV‐2, as those found testing positive will have a high chance of being true positives, and thus the test can be used to identify cases requiring isolation. Consideration should be made as to whether test positives should be confirmed with PCR to identify false positives. With a 1% prevalence, and using average sensitivity and specificity for those likely to be contacts of confirmed cases, as much as a third (32%) of those with positive results would be falsely positive. Using data more likely to represent mass screening scenarios, at 1% prevalence, on average RDTs yield as many false positives as true positives (PPV 52%), and at 0.5% prevalence, false positives more than outweigh true positives (PPV 38%) even for tests with 99% specificity (Table 1).
However, the low and variable sensitivity, and lack of evidence that those who test negative are not, or will not become, infectious indicates that those who are rapid antigen test‐negative cannot be considered free of risk of being, or of becoming, infectious. In any screening or mass testing programme people testing negative still have a risk of infection.
4. Evidence about test accuracy in at‐risk asymptomatic groups, such as hospital workers, or during local outbreaks at schools, workplaces, or care homes remains relatively limited. Although we might expect tests to perform similarly to testing of contacts the potential impact of low‐sensitivity tests in these settings is greater than for mass screening or testing at COVID‐19 test centres; false negatives in at‐risk groups have greater potential to either create new outbreaks or to increase the severity of existing outbreaks. Further research in these settings is still needed.
5. We were only able to include limited evidence on the repeated use of tests. Studies of daily testing suggested lower sensitivities in those who remained asymptomatic during the testing period and optimal levels of sensitivity for the RDT only on the immediate days after successful viral culture. Although serial testing (over a number of days), or combinations of different rapid tests (e.g. an antigen test followed by a rapid molecular test) on the same sample are proposed to overcome the limitations of on average low test sensitivity, they all require further validation, both in symptomatic (e.g. as a green flag for release from self‐isolation) and asymptomatic (e.g. to avoid quarantine in contacts of confirmed cases) populations. Use of multiple tests may increase false positive results, and there are likely to be many individuals with repeated false negative results reducing the expected benefit of subsequent tests. It is unlikely that models will be able to predict how well repeated tests and test combinations would work.
Overall, our conclusions have considerably strengthened those from the first version of this review and allowed some consideration of test accuracy in different testing scenarios. Ultimately, decisions around rapid testing will be driven not only by diagnostic accuracy but by acceptable levels of test complexity, time to result, access and acceptability to those being tested, and how test results influence individual behaviour, all of which might vary according to the setting in which the tests are to be used.
Implications for research.
There is now a considerable volume of research for point‐of‐care tests for SARS‐CoV‐2 infection, particularly in symptomatic populations. Nevertheless, evidence for the clinical performance of many test brands is scarce or lacking, and well designed prospective and comparative evaluations of different test brands in clinically relevant settings (both for symptomatic and asymptomatic testing) are still needed. Studies should recruit consecutive series of eligible participants and should clearly describe the clinical status, document time from symptom onset or time since exposure. Point‐of‐care tests must be conducted in accordance with manufacturer instructions for use, and across the spectrum of point‐of care settings and test operators. Evaluations of both individual tests and strategies of repeated testing are needed.
Consideration needs to be made of the best method for evaluating screening programmes, whether mass screening or targeted approaches including schools, healthcare setting and traveller screening. Whilst test accuracy studies help indicate which tests are likely to detect the greatest numbers of cases with the fewest false positives, assessing whether detecting asymptomatic cases leads to worthwhile reductions in disease spread will only be properly answered by studies of impact not accuracy.
Manufacturers and independent investigators are strongly encouraged to consider the principles for test evaluation set out by the Royal Statistical Society Diagnostic Tests Working Group (RSS 2021).
Any future research study needs to be clear about eligibility and exclusion decisions throughout the whole diagnostic pathway, and should conform to the updated Standards for Reporting of Diagnostic Accuracy (STARD) guideline (Bossuyt 2015).
What's new
| Date | Event | Description |
|---|---|---|
| 21 July 2022 | New citation required and conclusions have changed | New evidence incorporated; evidence for testing in asymptomatic cohorts has increased. |
| 21 July 2022 | New search has been performed | Updated with evidence published between September 2020 and March 2021. |
History
Review first published: Issue 8, 2020
| Date | Event | Description |
|---|---|---|
| 10 January 2022 | New search has been performed | Review updated with search until 18 March 2021. |
| 15 April 2021 | Amended | Clarification in Appendices that isothermal amplification is not a RT‐PCR test. |
| 24 March 2021 | Amended | Correction of typo in abstract |
| 24 March 2021 | Amended | Amendment to PLS title |
| 9 March 2021 | New citation required and conclusions have changed | This review has been updated and the conclusions have changed |
| 30 September 2020 | New search has been performed | We have updated our review and now include 64 study reports in 78 study cohorts, evaluating 16 antigen and 5 molecular assays |
Acknowledgements
Members of the Cochrane COVID‐19 Diagnostic Test Accuracy Review Group include:
the project team (Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Leeflang MMG, Spijker R, Hooft L, Van den Bruel A, McInnes MDF, Verbakel J, Emperador D, Dittrich S, Cunningham J);
-
the systematic review teams for each review:
Molecular, antigen, and antibody tests (Arevalo‐Rodriguez I, Buitrago DC, Ciapponi A, Domen J, Dretzke J, Mateos M, Nyaaba N, , Sharma P, Taylor M, Taylor‐Phillips S, van Wyk S, Verbakel J)
Signs and symptoms (Stuyf T, Domen J, Horn S)
Routine laboratory markers (Yang B, Langendam M, Ochodo E, Guleid F, Holtman G, Verbakel J, Wang J, Stegeman I)
Imaging tests (Salameh JP, McGrath TA, van der Pol CB, Frank RA, Prager R, Hare SS, Dennie C, Jenniskens K, Korevaar DA, Cohen JF, van de Wijgert J, Damen JAAG, Wang J);
Thanks to the wider team of systematic reviewers from the University of Birmingham, UK who assisted with title and abstract screening across the entire suite of reviews for the diagnosis of COVID‐19 prior to the publication of the first iteration of this review.
The editorial process for this review was managed by Cochrane's Evidence Production & Methods Directorate, Central Editorial Service in collaboration with Cochrane Infectious Diseases. The following people conducted the editorial process for this article: • Sign‐off Editor (final editorial decision): Michael Brown, Cochrane Evidence Production and Methods Directorate • Managing Editor (selected peer reviewers, collated peer‐reviewer comments, provided editorial guidance to authors, edited the article): Anne‐Marie Stephani, Cochrane Central Editorial Service • Editorial Assistant (conducted editorial policy checks and supported editorial team): Leticia Rodrigues, Cochrane Central Editorial Service • Copy Editor (copy editing and production): Denise Mitchell; Cochrane Evidence Production and Methods Directorate, Copy Edit Service; • Peer‐reviewers (provided comments and recommended an editorial decision): Professor Jim Huggett, National Measurement Laboratory, LGC Queens Road, Teddington, Middlesex (clinical review), Kristien Verdonck, Institute of Tropical Medicine Antwerp, Belgium (clinical review), Luis Rafael Moscote‐Salazar, Colombian Clinical Research Group in Neurocritical Care, Colombia (consumer review), Robert Walton, Senior Fellow in General Practice, Cochrane UK (methods review), Robin Featherstone, Cochrane Central Editorial Service (search review). Toby Lasserson is a member of Cochrane Evidence Production and Methods Directorate and provided peer‐review comments on this article, but was not otherwise involved in the editorial process or decision making for this article.
The editorial base of Cochrane Infectious Diseases is funded by UK aid from the UK Government for the benefit of low‐ and middle‐income countries (project number 300342‐104). The views expressed do not necessarily reflect the UK Government’s official policies.
The authors thank Dr Mia Schmidt‐Hansen who was the Cochrane Diagnostic Test Accuracy (DTA) Contact Editor for this review; the clinical and methodological referees; the Cochrane DTA Editorial Team; and Anne Lawson who copy‐edited the protocol. We would also like to thank all corresponding authors who provided additional information regarding their studies.
Jonathan Deeks is a UK National Institute for Health and Care Research (NIHR) Senior Investigator Emeritus. Yemisi Takwoingi is supported by a NIHR Postdoctoral Fellowship. Jonathan Deeks, Jacqueline Dinnes, Yemisi Takwoingi, and Clare Davenport are supported by the NIHR Birmingham Biomedical Research Centre. Sian Taylor‐Phillips is supported by an NIHR Career Development Fellowship. This paper presents independent research supported by the NIHR Birmingham Biomedical Research Centre at the University Hospitals Birmingham NHS Foundation Trust and the University of Birmingham. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care.
Appendices
Appendix 1. Definition of 'point of care' used in this review
The primary consideration for the eligibility of tests for inclusion in this review is that they should detect current infection and should have the capacity to be performed at the ‘point of care’ or in a ‘near‐patient’ testing role. There is an ongoing debate around the specific use and definitions of these terms, therefore for the purposes of this review, we consider ‘point of care’ and ‘near‐patient’ to be synonymous, but for consistency and avoidance of confusion, we use the term ‘point of care’ throughout.
We have adapted a definition of point‐of‐care testing, namely that it “refers to decentralized testing that is performed by a minimally trained healthcare professional near a patient and outside of central laboratory testing” (WHO 2018), with the additional caveat that test results must be available within a single clinical encounter (Pai 2012). Our criteria for defining a point‐of‐care test are therefore:
the equipment for running or reading the assay, or both, must be portable or easily transported, although mains power may be required;
minimal sample preparation requirements, for example, single‐step mixing, with no requirement for additional equipment or precise sample volume transfer unless a disposable automatic fill or graduated transfer device is used;
minimal biosafety requirements, for example, personal protective equipment (PPE) for sample collector and test operator, good ventilation and a biohazard bag for waste disposal;
no requirement for a temperature‐controlled environment; and
test results available within two hours of sample collection.
Appendix 2. Summary of World Health Organization and Chinese National Health Commission Guidelines for the diagnosis of SARS‐CoV‐2
Table A: World Health Organization guidelines for the diagnosis of SARS‐CoV‐2a
Includes laboratory testing guidelines and global surveillance guidelines
| Date range (2020) | Definition of confirmed case | Definition of confirmed non‐case | Definition of suspect case | Definition of probable case | Role of serology in testing |
| 10‐30 January 2020 | 10‐30 January: no documentation to define at this time (before first date of global guidelines)
31 January onwards: a confirmed case is a person with laboratory confirmation of COVID‐19 infection, irrespective of clinical signs and symptoms. No prescribed test in laboratory guidelines, suggested tests from 10 January include broad coronavirus PCR (with sequencing of precise virus in test positives), whole genome sequencing, broad coronavirus serology on paired samples, microscopy, culture (Lab 10 January). Four suggested tests from 17 January: broad coronavirus PCR (with sequencing of precise virus in test positives), NAAT for SARS‐CoV‐2 when it becomes available, whole genome sequencing, and broad coronavirus serology on paired samples. States that once specific NAAT assays are developed and validated, confirmation will be based on specific detection of unique sequences of viral nucleic acid by PCR. |
None stated | No definition of 'suspect case' at this time, but case definitions for surveillance are defined as a combination of symptoms and exposure, with more severe symptoms requiring less evidence for exposure | No definition at this time | Serological testing may be useful to confirm immunologic response to a pathogen from a specific viral group, e.g. coronavirus. Best results from serologic testing requires the collection of paired serum samples (in the acute and convalescent phase) from cases under investigation. |
| 31 January‐26 February 2020 | None stated | Suspect case defined as combination of symptoms and exposure, with more severe symptoms requiring less evidence for exposure | A suspect case with inconclusive laboratory results or is test‐positive using a pan‐coronavirus assay without laboratory evidence of other respiratory pathogens (global 31 January) | ||
| 27 February‐1 March 2020 | None stated | Suspect case defined as combination of symptoms and exposure, with more severe symptoms requiring less evidence for exposure, OR defined by symptoms requiring hospitalization and an absence of alternative explanation | A suspected case with inconclusive laboratory results (global 27 February) | ||
| 2 March‐19 March 2020 | A person with laboratory confirmation of COVID‐19 infection, irrespective of clinical signs and symptoms (global 31 January, 27 February, 20 March)
Laboratory confirmation of cases by NAAT specific to SARS‐CoV‐2 such as real‐time PCR with confirmation by nucleic acid sequencing when necessary. The viral genes targeted so far include the N, E, S and RdRP genes. In areas with no known COVID‐19 virus circulation confirmation requires:
Discordant results should be resampled. In areas where COVID‐19 virus is widely spread a simpler algorithm might be adopted (e.g. PCR of a single discriminatory target) |
One or more negative result does not rule out the possibility of COVID‐19 virus infection | In cases where NAAT assays are negative and there is a strong epidemiological link to COVID‐19 infection, paired serum samples (in the acute and convalescent phase) could support diagnosis once validated serology tests are available. Serological assays will play an important role in research and surveillance but are not currently recommended for case detection. |
||
| 19 March 2020‐current (12‐03‐21) | Probable case A suspect case for whom testing for the COVID‐19 virus is inconclusive OR A suspect case for whom testing could not be performed for any reason. | ||||
| NAAT: nucleic acids amplification test; PCR: reverse transcription polymerase chain reaction | |||||
aSource data from laboratory testing of 2019 novel coronavirus (2019‐nCoV) in suspected human cases: interim guidance, World Health Organization. 10 January, 17 January, 2 March, 19 March, 21 March 2020 (WHO 2020c), and Global surveillance for COVID‐19 caused by human infection with COVID‐19 virus, interim guidance, 31 January, 27 February, and 20 March 2020 (WHO 2020d).
Table B: Summary of Chinese National Health Commission guidelines for diagnosis and treatment for novel coronavirus pneumonia (trial versions 1‐7)
| Dates in effect | Definition of confirmed case | Definition of confirmed non‐case | Definition of suspect case | Role of serology in testing |
| 16‐17 January 2020 (version 1) | Cases (not confirmed cases) defined as virus genome highly homologous to coronaviruses | Not defined | Observation cases: defined as combination of exposure in Wuhan and symptoms focused on pneumonia, leukopenia and lack of improvement. | No role |
| 18 January‐2 March 2020 (versions 2, 3, 4, 5, 5 revised, and 6) | Suspect cases with either
|
Suspect cases can be ruled out after 2 consecutive negative respiratory tract nucleic acid tests taken at least 24 hours apart. | Suspect cases: combination of exposure (such as residence in/travel to Wuhan or exposure to a confirmed case within 14 days of onset) AND clinical features (such as symptoms: fever, respiratory symptoms, and tests: chest imaging, white blood cell and lymphocyte count). Exact definition varies slightly with version | No role |
| 3 March 2020‐current (12 March 21 (version 7) | Suspect cases with either
|
Suspect cases can be ruled out after 2 negative NAATs, taken at least 24 hours apart, and the NCP virus‐specific IgM and IgG are negative after 7 days from onset. | Suspect cases: combination of exposure (such as residence in/travel to Wuhan or exposure to a confirmed case within 14 days of onset) AND clinical features (such as symptoms: fever, respiratory symptoms, and tests: chest imaging, white blood cell and lymphocyte count). | Part of definition of cases and confirmed non‐cases |
| NAAT: nucleic acids amplification test; NCP: novel coronavirus pneumonia; PCR: reverse transcription polymerase chain reaction; Source: Table from Cheng 2020 | ||||
Appendix 3. Living search from the University of Bern
The following information is taken from the University of Bern website (see: ispmbern.github.io/covid-19/living-review/collectingdata.html); only strategies used during the period September 2020 to March 2021 are reported here. The register is updated daily and CSV file downloads are made available.
MEDLINE
30 October 2020 to 08 March 2021
("severe acute respiratory syndrome coronavirus 2"[Supplementary Concept] OR "COVID‐19" [Supplementary Concept] OR "coronavirus" OR "corona virus" OR "HCoV" OR "nCoV" OR "2019 CoV" OR "covid" OR "covid19" OR "Severe Acute Respiratory Syndrome Coronavirus 2" OR "SARS‐CoV2" OR "SARS‐CoV 2" OR "SARS Coronavirus 2") AND (2019/11/01:3000/12/31[PDAT])
29 April to 29 October 2020
("coronavirus"[MH] OR "coronavirus infections"[MH] OR "coronavirus"[TW] OR "corona virus"[TW] OR "HCoV"[TW] OR "nCov"[TW] OR "covid"[TW] OR "covid19"[TW] OR "Severe Acute Respiratory Syndrome Coronavirus 2"[TW] OR "SARS‐CoV2"[TW] OR "SARS‐CoV 2"[TW] OR "SARS Coronavirus 2"[TW] OR "MERS‐CoV"[TW]) AND (2019/1/1:3000[PDAT])
Embase
30 October 2020 to 08 March 2021
(exp SARS‐related coronavirus/ or severe acute respiratory syndrome/ or coronavirus disease 2019/ or (coronavir* or corona virus* or HCoV* or ncov* or 2019 cov or covid or covid19 or sars‐cov* or sarscov* or sars‐coronavirus* or Severe Acute Respiratory Syndrome Coronavirus* or nCoV).mp.) and 20191101:20301231.(dc).
01 May to 29 October 2020
(SARS coronavirus/ or middle east respiratory syndrome/ or severe acute respiratory syndrome/ or (coronavirus* or corona virus* or HCoV* or ncov* or covid or covid19 or sars‐cov* or sarscov* or Sars‐coronavirus* or Severe Acute Respiratory Syndrome Coronavirus*).mp.) and 20191201:20301231.(dc).
bioRxiv and medRxiv pre‐print servers
1 April 2020 to 08 March 2021
From 1 April 2020 onwards the curated bioRxiv/medRxiv dataset was retrieved (connect.medrxiv.org/relate/content/181).
Appendix 4. Search classification model
From 01 October 2020 to 08 March 2021: review‐specific classifier
The living reviews conducted under the Cochrane COVID‐19 DTA Group were being updated along different timelines. We therefore decided to adopt an approach of 'review‐specific' classification models which could be run as and when different review teams required search updates. A classification model for COVID‐19 rapid point‐of‐care test accuracy studies was built using similar methodology to that described below. We used the results of title and abstract screening, followed by full‐text review from the previous two iterations of this review to build and test classifiers. The final included studies were used as relevant documents, while studies excluded were used as irrelevant documents. The classifier was trained on the first round of selected articles, and tested and retrained on the second round of selected articles. Testing on the second round of selected articles revealed poor positive predictive value but 100% sensitivity at a cut‐off of 10. The poor positive predictive value is mainly due to the broad scope of our topic (all diagnostic studies in COVID‐19), poor reporting in abstracts, and a small set of included documents. The model was retrained using the articles selected of the second and third rounds of screening, which added a considerable number of additional documents. This led to a large increase in positive predictive value, at the cost of a lower sensitivity, which led us to reduce the cut‐off to 5. The largest proportion of documents had a score between 0‐5. This set did not contain any of the relevant documents. This version of the classifier with a cut‐off 5 was used in subsequent rounds and accounted for approximately 80% of the screening burden.
Appendix 5. Data extraction items
| Patient sampling items | Patient characteristics and setting items | Index test items | Reference standard items | Flow and timing items | Notes items |
| A1 Purpose | B1 Setting | D1.1 Test name (please include product code if reported) | E1 Reference standard for cases including threshold | F1 What was the time interval between index and reference tests? | G1 Funding |
| A2 Design (and description of groups labelled [1] [2] …) | B2 Location (include name of institution if available) | D1.2 Manufacturer | E1.1 PCR genetic targets | F2 Did all patients receive the same reference standard? | G2 Publication status |
| A3 Recruitment | B3 Country | D1.3 Antigen or genetic target | E2 Samples used | F3 Missing data | G3 Source (preprint or journal name) |
| A4 Were cases recruited prospectively or retrospectively? | B4 Dates | D1.4 Antibodies used | E3 Timing of reference standard | F4 Uninterpretable result s | G4 Study author CoI (including any manufacturer affiliations) |
| A5 Sample size (virus/COVID cases) | B5 Symptoms and severity | D1.5 POC or laboratory | E4 Was it blind to index test? | F5 Indeterminate results (index) | G5 Comment |
| A6 Inclusion and exclusion criteria | B6 Demographics | D1.6 Assay format | E5 Did it incorporate index test? | F5.1 Indeterminate results (reference) | |
| A7 Comment | B7 Exposure history | D1.7 When were samples taken? | E6 Reference standard for non‐cases | F6 Samples or patients | |
| B8 Comment | D1.8 Samples used (include who collected by) | E7 Samples used | F7 Comment | ||
| Non‐COVID patients (if additional groups) | D1.8.1 Transport media (volume and manufacturer detail) | E8 Timing of reference standard | |||
| C1.1 Group name | D1.8.2 Sample storage and timing of test | E9 Was it blind to index test? | |||
| C1.2 Source and time | D1.9 Who applied the test (include reported training/e)? | E10 Did it incorporate index test? | |||
| C1.3 Characteristics | D1.10 How was positive defined? | E11 Comment | |||
| C2.1 Group name | D1.11 Blinded to reference standard | ||||
| C2.2 Source and time | D1.12 Threshold predefined | ||||
| C2.3 Characteristics | D1.13 Comment | ||||
| CoI: conflict of interest; POC: point of care; PCR: reverse transcription polymerase chain reaction | |||||
Appendix 6. Criteria for assessment of study quality (QUADAS‐2)
| DOMAIN: Participant selection | |
| Was a consecutive or random sample of patients enrolled? | This will be similar for all index tests, target conditions, and populations. Yes: if a study explicitly stated that all participants within a certain time frame were included; that this was done consecutively; or that a random selection was done. No: if it was clear that a different selection procedure was employed; for example, selection based on clinician's preference, or based on institutions, or based on result of PCR Unclear: if the selection procedure was not clear or not reported |
| Was a case‐control design avoided? | This will be similar for all index tests, target conditions, and populations. Yes: if a study explicitly stated that all participants came from the same group of (suspected) patients. No: if it was clear that a different selection procedure was employed for the participants depending on their COVID‐19 status or SARS‐CoV‐2 infection status; or if only participants with SARS‐CoV‐2 infection were included Unclear: if the selection procedure was not clear or not reported. |
| Did the study avoid inappropriate exclusions? | Studies may have excluded patients, or selected patients in such a way that they avoided including those who were difficult to diagnose or likely to be borderline. Although the inclusion and exclusion criteria will be different for the different index tests, inappropriate exclusions and inclusions will be similar for all index tests: for example, only elderly patients excluded, or children (as sampling may be more difficult). This needs to be addressed on a case‐by‐case basis. Yes: if a high proportion of eligible patients was included without clear selection. No: if a high proportion of eligible patients was excluded without providing a reason; if, in a retrospective study, participants without index test or reference standard results were excluded. Unclear: if the exclusion criteria were not reported. |
| Did the study avoid inappropriate inclusions? | Some laboratory studies may have intentionally included groups of patients in whom the accuracy was likely to differ, such as those with particularly low or high viral loads, or who had other diseases, such that the sample over‐represented these groups. This needs to be addressed on a case‐by‐case basis. Yes: if samples included were likely to be representative of the spectrum of disease. No: if the study oversampled patients with particular characteristics likely to affect estimates of accuracy. Unclear: if the exclusion criteria were not reported. |
| Could the selection of patients have introduced bias? | High: if one or more signalling questions were answered with no, as any deviation from the selection process may lead to bias. Low: if all signalling questions were answered with yes. Unclear: all other instances |
| Is there concern that the included participant s do not match the review question? | High: for two‐group studies that included healthy or other disease controls, whether pre‐pandemic or contemporaneous; studies that only included people with COVID‐19 (whether PCRPCR‐confirmed only, participants meeting official guideline criteria); Low: for single‐group studies recruiting participants with signs and symptoms of COVID‐19; or for two‐group studies where control groups suspected of COVID‐19 were separately recruited. Unclear: if a description about the participants was lacking. |
| DOMAIN: Index tests | |
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes: if blinding was explicitly stated or index test was recorded before the results from the reference standard were available. No: if it was explicitly stated that the index test results were interpreted with knowledge of the results of the reference standard. Unclear: if blinding was unclearly reported. |
| If a threshold was used, was it prespecified? | Yes: if the test was dichotomous by nature, or if the threshold was stated in the methods section, or if study authors stated that the threshold as recommended by the manufacturer was used. No: if a receiver operating characteristic curve was drawn or multiple threshold reported in the results section; and the final result was based on one of these thresholds. Unclear: if threshold selection was not clearly reported. |
| Could the conduct or interpretation of the index test have introduced bias? | High: if one or more signalling questions were answered with no, as even in a laboratory situation knowledge of the reference standard may lead to bias. Low: if all signalling questions were answered with yes. Unclear: all other instances |
| Is there concern that the index test, its conduct, or interpretation differ from the review question? | For all test types, if index test is 'in‐house' or not commercially available, then state 'High'. If any test procedures used in the study diverged from IFU ((use of VTM, or testing outwith stated time limit), also state High If testing carried out in centralized laboratory and not near patient then state High. Evaluations that withheld the name of the test, or that used mixed sample types or did not report the evaluation setting, state Unclear If samples used and any sample processing steps are in accordance with test IFU, or if study describes conducting the test according to the manufacturer's protocol, state Low |
| DOMAIN: Reference standard | |
| Is the reference standard likely to correctly classify the target condition? | We will define acceptable reference standards using a consensus process once the list of reference standards that have been used has been obtained from the eligible studies. For COVID‐19 cases Yes: PCR; confirmed or suspected case using official criteria (WHO, CDC) or a clearly set out combination of signs/symptoms/exposure No: PCR not used, or if inadequate combination of clinical characteristics used in PCR‐ves, e.g. computed tomography alone Unclear: if definition of COVID‐19 was not reported For absence of COVID‐19 Yes: if at least 2 negative PCR results reported if suspected COVID‐19 based on signs/symptoms; single negative PCR test for asymptomatic contacts or contemporaneous controls with no clinical suspicion of COVID‐19; only pre‐pandemic sources of control samples used. No: single PCR or number of negative PCRs not reported for COVID‐19 suspects; no PCR reported (untested) for asymptomatic contacts or contemporaneous controls Unclear: if timing of control samples (pre‐pandemic or contemporaneous) was not reported |
| Were the reference standard results interpreted without knowledge of the results of the index test? | Yes: if it was explicitly stated that the reference standard results were interpreted without knowledge of the results of the index test, or if the result of the index test was obtained after the reference standard. No: if it was explicitly stated that the reference standard results were interpreted with knowledge of the results of the index test or if the index test was used to make the final diagnosis. Unclear: if blinding was unclearly reported. |
| Did the definition of the reference standard incorporate results from the index test(s)? | Yes: if results from the index test were a component of the reference standard definition. No: if the reference standard did not incorporate the index standard test. Unclear: if it was unclear whether the results of the index test formed part of the reference standard. |
| Could the conduct or interpretation of the reference standard have introduced bias? | High: if one or more signalling questions were answered with no. Low: if all signalling questions were answered with yes. Unclear: all other instances |
| Is there concern that the target condition as defined by the reference standard does not match the review question? | Applicability was judged primarily on the definition of disease‐positive. High: if PCR alone used to define cases Low: if clinical criteria, including PCR, were used to define cases, regardless of whether official criteria were used, as long as the criteria were explicitly described. Unclear: if definition of COVID‐19 cases was not provided, including if some clinically diagnosed cases were included but the clinical criteria used were not described. |
| DOMAIN: Flow and timing | |
| Was there an appropriate interval between index test and reference standard? | Yes: if same swab used, or swabs obtained at same time regardless of freezing (which is covered under index applicability) No: if different samples used with more than 12 hours between collection times Unclear: if can't tell |
| Did all participants receive the same reference standard? | Yes: if all participants received the same reference standard (clearly no differential verification). No: if (part of) the index test‐positives or index test‐negatives received a different reference standard. Unclear: if it was not reported |
| Were all participants included in the analysis? | Yes: if it is clear that all eligible participants were included in the analyses. No: if after the inclusion/exclusion process, participants were removed from the analyses for different reasons: no reference standard done, no index test done, intermediate results of both index test or reference standard, indeterminate results of both index test or reference standard, samples unusable. Unclear: if it is not possible to determine whether all participants were included (e.g. from a STARD‐style participant flow diagram) |
| Did all participants receive a reference standard? | Yes: if all participants received a reference standard (clearly no partial verification). No: if only (part of) the index test positives or index test negatives received the complete reference standard. Unclear: if it was not reported |
| Were results presented per participant? | Yes: if either only one sample per participant (regardless of disaggregation of results over time), or if multiple samples per participant but results are disaggregated by time period (at least week by week) No: if multiple samples per participant and results are not disaggregated by time period Unclear: if it is not possible to tell whether results presented are per participant or per sample |
| Could the participant flow have introduced bias? | High: if one or more signalling questions were answered with no. Low: if all signalling questions were answered with yes. Unclear: all other instances |
| CDC: Centers for Disease Control; ICU: intensive care unit; IFU: instructions for use; PCR: real‐time polymerase chain reaction; SARS‐CoV‐2: severe acute respiratory syndrome coronavirus 2; VTM: viral transport medium; WHO: World Health Organization | |
Appendix 7. Summary study characteristics
| Study Publication status Sample size (cases) | Study design; inclusion criteria | Setting (facility) Country (Recruitment dates) | Participant characteristics | Reference standard Target Samples (timing) | Missing data or indeterminate results |
|
Abdelrazik 2021 Published 310 (188) |
≥ 2 groups (not stated; appears prospective): [1] Symptomatic PCR test‐positive patients; samples taken during the initial phase of the disease (n = 160) [2] Exposed HCWs and patient contacts (n = 150) Data are presented only for groups [1] and [2] combined | Mixed (described as patients, their contacts and exposed HCWs) Egypt (May 2020) |
Mixed: unclear; 160 PCR+ve 'patients' symptomatic, plus 150 presumably asymptomatic contacts and exposed HCWs. Median age 42 years; 184 (59%) male |
PCR (single assay) Target: not stated NP; same swab used Timing: as for index test Interval: simultaneous |
None reported Index: none reported Reference: none reported |
|
Abdulrahman 2020 Preprint 4183 (733) |
Single‐group (prospective); mildly symptomatic individuals with suspected COVID‐19 cases (defined by Bahrain protocol), referred to the national testing centre | COVID‐19 test centre (National COVID‐19 test centre) Bahrain (Not stated) |
Symptomatic: all mild symptomatic individuals Median symptom duration: 2 (range 0‐14) d; only collected for 1301 (31%) Mean age 30.9 (SD 14.5) years; 2365 (56.5%) male | PCR (single assay) Target: E gene confirmed by RdRP and N genes NP; paired Timing: as for index test (median of 2 (range 0‐14) d pso) Interval: paired |
None reported; states "no equivocal results were reported for index or reference" Index: none reported Reference: none reported |
|
Agullo 2021 [A] Published 659 (265); 610/659 also provided saliva samples |
Single‐group (prospective); (from 3 centres); consecutive adults and children, either with COVID‐19 signs/symptoms or asymptomatic contacts | Primary care (primary care, 3 centres) Spain (15 September‐29 October 2020) |
Mixed: 265 (40.2%) asymptomatic and 394 (59.8%) symptomatic Median symptom duration: 3 (IQR 2‐5) d Median age: 38 (IQR 21‐49.8) years; 76 (11.5%) ≤ 14 years, 45 (7.6%) > 65 years; 372 (56.4%) female |
PCR (no details) Target: not specified NP; paired; same as index Timing: as for index test (median duration of 3 (IQR 2–5) d of symptoms) Interval: paired |
Not stated None reported Index: none reported Reference: none reported |
|
Akingba 2021 Preprint (not peer reviewed) 677 (146) |
Single‐group (prospective); symptomatic individuals seeking COVID‐19 testing at mobile testing units during community testing campaigns | COVID‐19 test centre (COVID‐19 test centres; mobile testing sites (n = 6)) South Africa (17 November‐20 November 2020) |
Symptomatic: all symptomatic seeking COVID‐19 testing (ambulatory; specific symptoms not reported) Age range: 3‐85 years; 41% male |
PCR (single assay) Target: not reported; states "three targets", mean Ct values used NP; same sample used Timing: as for index test Interval: simultaneous |
Yes; 19 excluded (see below) Not reported Index: not reported Reference: 19/677 (2.8%) had inconclusive results (single target positive, CT > 38) |
|
Albert 2020 Preprint 412 (54) |
Single‐group (prospective); patients with clinical suspicion of COVID‐19 (compatible signs or symptoms appearing within the prior week) attending 1 of 8 primary care centres (n = 412) | COVID‐19 test centre (primary care) Spain (2 September‐7 October 2020) |
Symptomatic: all symptomatic (< 7 d pso) Median age, 31 (range 1‐91) years; 42% male 327 adults; median 36 (17‐91) years 85 children; median 11 (1‐16) years |
PCR (single assay) Target: ORF1ab, N and S genes NP; paired; same as index Timing: as for index; tested within 24 h Interval: paired |
None reported Index: none reported Reference: none reported |
|
Alemany 2021 Preprint Total n = 1406 (951 cases) [1] 446 (419) [2] 473 (415) [3] 487 (117) |
Single‐group (not stated); laboratory‐based study recruiting samples from [1] symptomatic individuals with suspected COVID‐19 seen in routine practice (n = 446) [2] contacts exposed to positive PCR confirmed COVID‐19 cases (n = 473) [3] preventive screening of unexposed asymptomatic individuals in the general population (n = 487) | Laboratory‐based (sources include routine diagnostic confirmation; contact tracing and asymptomatic screening) Spain (Not stated) |
Mixed Not stated; 15/1406 (1.1%) reportedly hospitalized (all PCR+ve) Ct < 20: 258 (18.3%) Ct 20‐24 305 (21.7%) Ct 25‐29 285 (30.3%) Ct > 30 103 (7.3%) All samples: mean age 40.4 (SD 24.5) years; 453 (32.2%) male |
PCR (single assay) Target: not stated; as per CDC protocol NP or NMT; same sample used Timing: fresh samples stored at 2 – 8 ºC for up to 72 hours prior to PCR Interval: paired |
None reported Index: none reported Reference: none reported |
|
Aoki 2020 Published paper 129 (63) |
Unclear design (unclear); samples from COVID‐19 hospitalized patients or from patients suspected of having COVID‐19‐like symptoms | Laboratory‐based (includes use of remnant samples) Japan (Not stated) |
Symptomatic Not stated Not stated |
PCR (multiple assays) Target: gene N NP; paired; same as index Timing: same for index rest Interval: paired |
None reported None reported Index: none reported Reference: none reported |
|
Baro 2021 [A] Preprint 286 (101) |
Unclear design (unclear); unexposed asymptomatic individuals living in areas at high risk of an outbreak who participated in routine mass testing as part of a regional surveillance program (n = 316) | Screening (community screening; public health surveillance) Spain (December 2020‐January 2021) |
Asymptomatic states "all unexposed asymptomatic" Not stated |
PCR (single assay) Target: not stated NP; same sample used Timing: same as for index test Interval: simultaneous |
30/316 excluded; reasons for exclusion documented
25 with no documented Ct value excluded a priori 1/316 incomplete result Index: 4/316, all of them in the Lepu assay Reference: none reported |
|
Basso 2021 [A] Published Whole sample: 234 (87) Inpatients: 138 (84) Outpatients: 96 (3) |
Single‐group (prospective); COVID‐19 inpatients (n = 138) and outpatients (n = 96) screened for suspected SARS‐CoV‐2 after contact with a SARS‐CoV‐2‐positive person or with typical symptoms (n per group was not reported) | Hospital in‐ or outpatient (inpatient and outpatient) Italy (1 August‐30 November 2020) |
Mixed Inpatients: 93/138 (67%) pneumonia, 97 (70%) fever > 37.5 °C, cough 46 (33%), dyspnoea 21 (15%); outpatients: not reported Inpatients: 86, 62% male; mean age 56 (SD 17) years; outpatients: 49, 51% male; mean age 42 (SD 15) years |
PCR (single assay) Target: ORF1ab, N and S SARS‐CoV‐2 genes Saliva; same sample used Timing: same as for index test Interval: simultaneous |
Yes. Authors provided data underlying Figure 3 however number of samples tested per assay and sample type vary; reason given was insufficient material for some cases, the number discrepancy was stated as not due to test failure:
[A] ESPLINE ‐ saliva, n = 134 (55)
[B] ESPINE ‐ NP, n = 136 (64)
[C] Panbio ‐ NP, n = 116 (56)
[Total of 164 (saliva) and 151 (NP) samples reported for LUMIPULSE] None Index: none reported Reference: none reported |
|
Beck 2021 Published 347 (61) |
Single‐group (not stated; appears prospective); all patients with signs and symptoms of COVID‐19 presenting to an urgent care centre (n = 347) | Urgent care centre (urgent care centre) USA (Not stated) |
Symptomatic: all symptomatic; no further details Age range 1‐90 years; ≤ 18 years 35.4%, 19‐50 years 38.3%, > 50 years 26.2% of participants |
RT‐TMA (Hologic Aptima) Target: not stated NP; paired Timing: as for index test Interval: paired |
1 sample 1 sample invalid on SOFIA Index: none reported Reference: none reported Discrepant analysis: 2/14 FNs were negative on Xpert Xpress; 1/1 FPs also negative Xpert Xpress |
|
Billaud 2020 Published 462 (99); 47 missing, presumably with no paired data |
Single‐group (prospective); cluster investigation: teachers (n = 90) and students (n = 419) screened for COVID‐19 as part of a cluster investigation (n = 509) |
Contacts (screening) France (September 16 and 17) |
Mixed 166/509, 32.6% symptomatic including 152/419 students Mean, median age: Students 21.6 years, 21 (18‐37) years Teachers 47.2 years, 49 (26‐64) years |
PCR (single assay) Target: not stated NP; paired; same as index Timing: as for index Interval: paired |
47 missing, including 11 uninterpretable 11 uninterpretable on Ag test Index: none reported Reference: none reported |
|
Blairon 2020 Published 56 (30) |
Single‐group (prospective ); sampled from cohort of suspected COVID‐19 patient samples sent for laboratory diagnosis (n = 56) |
Laboratory‐based (swabs obtained at hospital site; no further detail) Belgium (5 April‐4 May 2020) |
Not reported Not stated Not stated |
PCR (single assay) Target: E gene NP; paired; same sample used Timing: not stated Interval: paired |
None reported; review team excluded main cohort data as no reference standard for Ag test‐positive samples None reported; 1 "invalid" sample excluded from main cohort Index: none reported; 1 "non‐confirm" sample excluded from main cohort Reference: none reported |
|
Bulilete 2021 Published 1362 (140); further 27 declined to participate |
Single‐group (prospective); adults attending 1 of 4 PHC COVID‐19 testing centres for PCR tests; included patients with symptoms suggestive of infection with referral by a general practitioner (GP), or close contact with a PCR‐confirmed case | COVID‐19 test centre (4 PHC centres and 2 COVID‐EXPRESS test sites) Spain (2‐25 October 2020) |
Mixed 680 (49.7%) reported symptoms < 7 d prior (most frequent: headache (341, 24.9%), sore throat (310, 22.6%), cough (301, 18.4%), and tiredness (251, 18.3%)); 689 (50.3%) asymptomatic Mean age 42.5 (SD 14.9) years, 744 (54.3%) female |
PCR (single assay) Target: ORF, N, and S NP; paired; same as index Timing: same as for index; sample sent for processing within 24 h of collection Interval: paired; simultaneous |
Yes 3 PCR with incorrect labelling; 16 Ag‐RDT results missing Index: none reported Reference: 4 inconclusive PCR |
|
Caruana 2021 [A] Published 572 (114) |
Single‐group (prospective) All patients admitted to hospital (wards, intermediate care units and ICU) from the ED, with or without suspected SARS‐CoV‐2 infection (A second study investigating the correlation of symptom duration and variations in viral load was also reported, but not eligible for this review) | Hospital inpatient (patients admitted to hospital from the ED (described as emergency ward)) Switzerland (6 November‐6 December 2020) |
Mixed 239 (45%) asymptomatic; admitted for other reasons than COVID‐19 suspicion 293 (55%) symptoms consistent with COVID‐19; included some with atypical symptoms (number not reported) Asymptomatic for COVID‐19: median age 67 (IQR 49‐81) years; 105 (44%) female Symptomatic: median age 75 (IQR 61‐85) years; 131 (45%) female |
PCR (multiple assays) Target: [D] E‐ and RdRp‐ encoding genes [A]‐[C] not reported NP; same sample used Timing: same as for index test Interval: simultaneous |
Yes; 67 excluded including 40 with missing results Not reported Index: n = 27 invalid results Reference: not reported |
|
Cerutti 2020 Published 330 (109) |
Single‐group (not stated); (1) symptomatic patients attending one of 2 EDs (n = 185) (2) asymptomatic travellers returning home from European high‐risk countries (Croatia, Spain, Malta) (n = 145) |
Mixed ((1) ED
(2) Possible contacts) Italy ([1] 3 March‐1 May 2020 [2] August 2020) |
Mixed Not stated; cohort (2) were asymptomatic (1) mean age 44.6, 95 % CI: 40.7–48.6 (2) mean age 35.9, 95 % CI: 32.7–39.1 |
PCR (single assay) Target: not stated Unclear sample; same sample site (site not described) Timing: not stated Interval: unclear |
None reported Index: none reported Reference: none reported |
|
Chaimayo 2020 Published 54 (60) |
Single‐group (not stated); suspected cases of COVID‐19 including symptomatic and contact individuals, including travellers, quarantined individuals and pre‐operative patients | Hospital in‐ or outpatient (mixed) Thailand (March‐May 2020) |
Not reported PCR+ve: 37/60, 61.7% showed signs and symptoms of upper respiratory tract infection; 5 (8.3%) pneumonia and ICU admission, 11 fever, 4 unspecified, 3 asymptomatic NB ‐ supplemental file shows 53/60 with fever ve: median age 38.5 (range 21–72) years; 36 (60%) male median age 61 (range 16‐95) years; 163 (41%) male |
PCR (single assay) Target: E gene (Sarbecovirus), and RdRp and N genes (SARS‐CoV‐2) NP; paired; same as index Timing: as for index test Interval: paired |
None stated None reported Index: none reported Reference: none reported |
|
Ciotti 2021 Published 50 (39) |
Single‐group (not reported) NP swabs from patients with suspected SARS‐CoV‐2 infection at the ED or Infectious Diseases ward | Hospital in‐ or outpatient (mixed; ED or Infectious Diseases ward) Italy (May‐September 2020) |
Symptomatic Not reported Median age 53.5 (mean 53.1; range: 15–94) years; 24 (48%) male |
PCR (single assay) Target: E gene (Sarbecovirus subgenus) and N and RdRp genes (SARS‐CoV‐2) NP; same sample site Timing: not stated; same as for index test Interval: unclear |
None reported Index: none reported Reference: none reported |
|
Courtellemont 2021 Preprint 248 (121) |
Unclear; 2 groups (unclear) (1) symptomatic (headache, fatigue, fever, or respiratory signs) or asymptomatic people voluntarily accessing the COVID‐19 Screening Department (n = 231) (2) hospitalized SARS‐CoV‐2‐positive patients (n = 17) |
Mixed (COVID testing unit and inpatient) France (12‐19 October 2020) |
Mainly symptomatic 99/121 cases were symptomatic; 22 asymptomatic Median age 38 years, mean age 43 years (range: 18‐96) 117 (97%) male |
PCR (single assay) Target: ORF1ab, S and N genes NP; paired; same as index Timing: as for index Interval: paired |
None reported; review team excluded 20 cases with a previous positive RT‐qPCR within 5 d but a negative RTqPCR at the time of study sampling None reported Index: none reported Reference: none reported |
|
Del Vecchio 2021 Preprint (not peer reviewed) 1441 (61); 58/61 PCR+ve were observed at ED |
Single‐group (retrospective); patients with and without symptoms examined at ED (n = 1153), infectious disease wards (n = 279) or other department (n = 9) who required COVID‐19 testing either due to a) presence of COVID‐19‐related symptoms (fever and/or cough and/or headache, diarrhoea, asthenia, muscle pain, joint pain, loss of taste or smell, or shortness of breath, with or without pneumonia); or b) asymptomatic but had a contact with a confirmed case of SARS‐CoV‐2 infection during the previous 10 d | Hospital in‐ or outpatient (mixed; ED and infectious disease ward admissions) Italy (15 September‐16 October 2020) |
Not reported Only reported for PCR+ve 51/61 (84%) symptomatic, including 10 (20%) with asthenia, 7 (14%) with cough, 3 (6%) with dyspnoea (1/3 severe), 32 (63%) with fever, 7 (14%) with headache 760 (53%) male Age: 0‐19 years: n = 54, 20‐39 years: n = 247, 40‐59 years: n = 262, 60‐79 years: n = 420, 80‐99 years: n = 457, > 100 years: n = 1 |
PCR (single assay) Target: S gene and the ORF1ab gene Sample site not reported; same as index Timing: as for index test Interval: paired |
None reported (1849 patients only had either RAT or PCR test results available and were excluded a priori) None reported Index: none reported Reference: none reported |
|
Dominguez Fernandez 2021 Published letter 30 (20) |
Unclear design (not stated); users with symptoms compatible with COVID‐19 and/or were close contacts of users with a positive COVID‐19 diagnosis | Care home (no further details) Spain (44,075) |
Mainly symptomatic 90% had symptoms compatible with SARS‐CoV‐2 infection of < 5 d of evolution and the other 10% were asymptomatic, but were close contacts Mean age 76.2 (SD 19.76) years, 36.7% male |
PCR (no details) Target: not stated Sample unclear; no details Timing: not stated Interval: paired |
None reported Index: none reported Reference: none reported |
|
Drain 2021(a) Published paper 257(83) |
≥ 2 groups (described as prospective) Report of 2 studies, 1 is a 2‐group study using nasal swabs and the second a single‐group study using NP swabs. [1] Nasal swabs from children and adults presenting for COVID‐19 testing at 10 sites in the USA and UK (first time period), [2] Nasal swab samples from a commercial supplier (MRN Diagnostics, Florida, USA) and also collected from an at‐risk population (LumiraDx Stirling, UK), [3] NP swabs from children and adults presenting for COVID‐19 testing at 10 sites in the USA and UK (second time period) Data for cohort [1] and [2] are included as Drain 2021(a)); see Drain 2021(b) for details of cohort [3] |
Mixed (presume COVID‐19 testing centres) UK and USA ([1] 26 June‐23 July 2020 [2] Not reported) |
Mixed Whole sample: 414/512 (81%) symptomatic [1]+[2] 159/257 (62%) symptomatic Whole sample: 287 female, 225 male. Age (0‐90 years) [1]+[2] mean age 34 (SD 15.7) years; 142 (55%) female |
PCR (no details) Target: not stated Nasal and NP; same as index Timing: same as for index test Interval: paired |
Not mentioned Not mentioned Index: not mentioned Reference: not mentioned |
|
Drain 2021(b) Published paper 255(40) |
Single‐group (described as prospective); Report of 2 studies, 1 is a 2‐group study using nasal swabs and the second a single group study using NP swabs. [1] Nasal swabs from children and adults presenting for COVID‐19 testing at 10 sites in the USA and UK (first time period), [2] Nasal swab samples from a commercial supplier (MRN Diagnostics, Florida, USA) and also collected from an at‐risk population (LumiraDx Stirling, UK), [3] NP swabs from children and adults presenting for COVID‐19 testing at 10 sites in the USA and UK (second time period) Data for cohort [1] and [2] are included as Drain 2021(a); see Drain 2021(b) for details of cohort [3] |
COVID‐19 test centre (presume COVID‐19 testing centres) UK and USA ([3] 17 August‐28 September 2020) |
Symptomatic Whole sample: 414/512 (81%) symptomatic [3] 255/255 (100%) symptomatic Whole sample: 287 female, 225 male. Age (0‐90 years) [3] mean age 33.2 (SD 19.4) years; 145 (57%) female |
PCR (no details) Target: not stated Nasal and NP; paired; same as index Timing: same as for index test Interval: paired |
Not mentioned Not mentioned Index: not mentioned Reference: not mentioned |
|
Drevinek 2020 [A] Published 591 (223) |
Single‐group (prospective) Participants aged ≥ 10, who attended a COVID‐19 testing centre due to suspicion of COVID‐19 (n = 273) or contact tracing (n = 290); either referred by a GP or public health officer (n = 511) or were "self‐payers" (n = 54) | COVID‐19 test centre (COVID‐19 testing site at a university hospital) Czech Republic (4‐day period in October 2020) |
Mixed 290 (49%) symptomatic on day of testing Mean age 40 (range 12‐78) years; 44.7% male |
PCR (single assay) Target: N, E and RdRP/S genes NP + OP; paired Timing: same as for index test Interval: paired |
None reported Index: none reported Reference: none reported |
|
Faico‐Filho 2021 Preprint (not peer reviewed) 127 (70) |
Single‐group (prospective) Adults (age > 18 years) treated in the ED and hospitalized for at least 24 h, including those 1) with COVID‐19‐related symptoms and/or contact with a confirmed case, 2) decompensation of underlying disease, or 3) suggestive CT findings (ground glass) | Hospital ED (ED; hospitalized for ≥ 24 h) Brazil (Not reported) |
Symptomatic Not reported; presumed symptomatic Mean age: 60 (SD 17.5) years; 69 (54%) male |
PCR (multiple assays) Target: [1] RdRp, E and N SARS‐CoV‐2 genes [2] ORF1ab and N SARS‐CoV‐2 genes NP; same sample site Timing: as for index test Interval: unclear |
Not reported Not reported Index: not reported Reference: not reported |
|
Favresse 2021 [A] Published paper 188 (96) |
Single‐group (not stated) NP samples from patients who presented for SARS‐CoV‐2 testing at single institution | Laboratory‐based (laboratory‐based) Belgium (7 November‐25 November 2020) |
Mixed 118, 63% symptomatic Women (n = 104, 55%): median age 54 (range 5‐97) years Men (n = 84): median age 57 (range 1‐94) years |
PCR (single assay) Target: E‐gene NP; same sample used Timing: same as for index test Interval: simultaneous |
Not reported Not reported Index: not reported Reference: not reported |
|
Fenollar 2020(a) 182 (182) |
Single‐group (cases) (Unclear) [1] symptomatic patients, all PCR positive (n = 182) |
COVID‐19 test centre (unclear; no details) France (21 September‐2 October 2020) |
Symptomatic Not stated; all symptomatic Ct values for 154 patients Ct ≤ 20: 58, 38% Ct 21‐25: 49, 32% Ct 26‐30: 39, 25% Ct 31‐34: 8, 5% Not reported |
PCR (single assay ‐ VitaPCR, Credo) Target: not stated NP; paired; same as index Timing: not stated Interval: paired |
None reported Index: none reported Reference: none reported |
|
Fenollar 2020(b) 159 (22) |
Single‐group (unclear) Asymptomatic contacts of confirmed cases (n = 159) |
Contacts (unclear) France (21 September‐2 October 2020) |
Asymptomatic All asymptomatic; 21/22 cases had Ct > 25 Not reported |
PCR (single assay ‐ VitaPCR, Credo) Target: not stated NP; paired; same as index Timing: not stated Interval: paired |
None reported Index: none reported Reference: none reported |
|
Ferguson 2021 Published 720(8) |
Single‐group (prospective) University students attending asymptomatic student testing centre at the University of Birmingham | Screening (student screening) UK (2‐9 December 2020) |
Asymptomatic All asymptomatic Not stated |
PCR (single assay) Target: ORF1ab, N gene, S gene NP; same sample used Timing: same as for index test Interval: simultaneous |
Yes None 4 invalid on Ag test; "Results of 4 samples were void (as defined by the manufacturer’s protocol [2])" Index: none reported Reference: none reported |
|
Filgueiras 2021 Published 150 (55) |
Single‐group (not stated; appears prospective) Patients with clinical features and suspected COVID‐19 seen by ED doctors; later described as "hospitalised" | Hospital inpatient (patients at the ED prior to admission to hospital) Brazil (44,075) |
Symptomatic All symptomatic: 72 (48.0%) dyspnoea, 52 (34.7%) dry cough, 50 (33.3%) fever, 49 (32.7%) myalgia, 25 (16.7%) asthenia and 24 (16.0%) productive cough 127 (84.7%) had ≥ 2 associated symptoms; 15 (10%) had 5‐7 associated symptoms; Of PCR+ve, 36 (65.5%) critically ill; 19/19 with typical findings of patchy ground‐glass shadows on CT presented bilateral pneumonia, compared to 5/11 (45.5%) of COVID‐19‐negative individuals with same alteration Median age 62 (range 29‐91) years, 22 (40.0%) male |
PCR (single assay) Target: Gene E, Gene RdRP and Gene N MS2 NP; paired; same as index Timing: same as for index Interval: paired |
None reported Index: none reported Reference: none reported 11 samples with inconclusive results (only 1 Ag +ve); 5 on day 1 or 2 pso, 2 on day 5, 1 on day 8, 1 on day 15; and not reported for 2 samples. 2/11 had bilateral multifocal ground‐glass opacities on CT, and 4 needed oxygen supply due to strong dyspnoea and desaturation; 1 (Ag‐positive) with odynophagia and dry cough. Not clear whether all were considered to have COVID‐19 |
|
FIND 2020a Published 400 (102) |
Single‐group (prospective); patients with symptoms consistent with COVID‐19 (meeting national definition for testing) presenting at a community testing clinic | COVID‐19 test centre (community) Brazil (30 July‐21 August 2020) |
Symptomatic All symptomatic; no further details Mean age 40 (range 4‐84) years; reported for 396 participants 181 (45%) male |
PCR (single assay)
Threshold ≤ 37 Ct Target: N1, N2 NP; paired; same as index Timing: same as for index test Interval: paired |
Reports 0 invalid results None reported Index: none reported Reference: none reported |
|
FIND 2020b (CH) Published 535 (124) |
Single‐group (prospective); patients seeking COVID‐19 testing at main testing centre; described as presenting either with symptoms compatible with a SARS‐CoV2 infection, or with a known positive contact or asymptomatic HCWs (n = 535) |
COVID‐19 test centre (community) Switzerland (9‐16 October 2020) |
Symptomatic 534/535 (99%) symptomatic Mean age 38.5 (range 16‐85) years 247, 46% male |
PCR (single assay)
Threshold < 40 Ct Target: not stated NP; paired; same as index Timing: not stated; author contact advises only paired swabs used. Interval: paired |
None reported Index: none reported Reference: none reported |
|
FIND 2020b (DE) Published 1108 (106) |
Single‐group (prospective); at 2 sites; this extraction is for data from Germany (see FIND 2020b (CH) for data related to the site in Switzerland) Patients seeking COVID‐19 testing at main testing centre; described as presenting either with symptoms compatible with a SARS‐CoV2 infection, or with a known positive contact or asymptomatic HCWs (n = 1108) | Mixed (community) Germany ([1] Heidelberg: 28 September‐30 October 30 2020 [2] Berlin: 19‐30 October 2020) |
Mixed 709/1100 symptomatic (64.5%); mean symptom duration 4.01 (SD 3.1) (n = 687) Mean age 38.7 (18‐86) years 542, 49% male |
PCR (multiple assays) Target: not stated NP or NP + OP; paired; same as index Timing: not stated; author contact advises only paired swabs used. Interval: paired |
Yes ‐ 10 withdrew consent for 2nd swab and 1 had invalid PCR None reported for Ag; 1 invalid PCR Invalid test results were repeated once with the remaining buffer solution in the test tubes. Index: none reported; In the case of discrepant results both readers re‐interpreted the results and agreed on a final result. Reference: none reported |
|
FIND 2020c (BR) Published 400 (106) |
Single‐group (prospective); ambulatory patients meeting national suspect definition for COVID‐19 testing presenting at a community testing clinic in Brazil |
COVID‐19 test centre (community) Brazil (13‐30 July 2020) |
Symptomatic 392/397 (99%) symptomatic; no further details Mean age 37 (range 2‐94) years (397 participants); 229/398 (57%) male |
PCR (single assay);
Ct threshold not stated; author contact advises Ct thresholds as per assay IFUs Target: N1 and N2 NP; paired; same as index Timing: not stated; author contact advises only paired swabs used. Interval: paired |
Reports 0 missing data None reported Index: none reported Reference: none reported |
|
FIND 2020c (CH) Published 529 (191) |
Single‐group (prospective); patients seeking COVID‐19 testing at main testing centre; described as presenting either with symptoms compatible with a SARS‐CoV2 infection, or with a known positive contact or asymptomatic HCWs (n = 529; from total cohort of 1064 volunteers) |
COVID‐19 test centre (community) Switzerland (9‐23 October 2020) |
Symptomatic Not stated; time pso recorded for 183/191, 96% (141/183) COVID‐positive cases had symptoms for 0‐4 d (77%) Not stated |
PCR (single assay);
threshold < 40 Ct Target: not stated NP; paired; same as index Timing: not stated; author contact advises only paired swabs used Interval: paired |
None reported Index: none reported Reference: none reported |
|
FIND 2020c (DE) Preprint 425 (8) |
Single‐group (prospective) Participants at risk for SARS‐CoV‐2 infection based on exposure to a confirmed case, suggestive symptoms, or travel to a high‐risk area, presenting at 1 of 3 sites: (1) drive‐in testing station (n = 1213) (2) a clinical ambulatory testing facility (n = 1308) (3) secondary care facility (n = 53) |
COVID‐19 test centre (community (drive‐in or clinical ambulatory testing) and secondary care (inpatient?)) (1), (2) Germany (3) UK (April 17 and August 25, 2020; dates varied by assay and site) |
Mainly symptomatic Symptomatic on testing day (n = 2355) Overall: 1901, 80.7% [A] 564, 81.2% [B] 283, 68.9% [C] 1054, 84.4% Prior negative test result (n = 1928) Overall: 236, 12.2% [A] 73, 11.7% [B] 38, 12.6% [C] 125, 12.5% Detailed symptoms are reported by site and test in supplementary materials Mean age (SD) (n = 2405) Overall 40.4 (14.3) years [A] 42.7 (14.9) years [B] 44.9 (15.4) years [C] 37.6 (12.7) years Male (%) (n = 2361) Overall: 1115, 47.2% [A] 47.2% [B] 39.7% [C] 49.8% |
PCR (multiple assays) Target: not stated NP or OP, or NP + OP; paired; same as index Timing: as per index test Interval: paired |
154 excluded following enrolment
(116 2nd swab refused
3 nose bleed after 1st swab
3 insufficient time for both swabs
31 other reasons
1 no reason available) Ag tests: [A] 2 invalid (PCR‐ve) [B] 8 invalid (PCR‐ve) [C] 0 invalid reported PCR: 3 excluded as invalid (n = 2) or not available (n = 1) Index: none reported; ease‐of‐use assessment reported [A] a high number of test execution steps (including precision pipetting) … challenges when performing multiple tests at the same time possibly hindering the test’s widespread use [B] challenges due to inconsistent test result interpretation (often only very faint lines visible) and deficiencies in both the test kit quality and design [C] no dissatisfactory scores identified Reference: None reported |
|
FIND 2020d (BR) Published 453 (120) |
Single‐group (prospective); adults in community meeting national suspect definition for COVID‐19 testing presenting at [1] a community testing clinic or [2] a tertiary‐level hospital |
COVID‐19 test centre (community clinic and tertiary hospital) Brazil ([1] 17 August‐9 September [2] 11 July‐8 August) |
Mainly symptomatic 421/450 (94%) symptomatic; no further details Mean age 39 (range 0‐95) years (451 participants); 185 (41%) male |
PCR (multiple assays)
Ct thresholds not stated; author contact advises Ct thresholds as per assay IFUs Target: 1. N1 and N2 2. E and RdRp NP; paired; same as index Timing: not stated; author contact advises only paired swabs used. Interval: paired |
Reports 0 missing data None reported Index: none reported Reference: none reported |
|
FIND 2020d (DE) Published 676 (39) |
Single‐group (prospective); adults in community meeting national suspect definition for COVID‐19 testing presenting at [1] a drive‐in testing centre or [2] ambulatory testing clinic |
COVID‐19 test centre (community) Germany ([1] Heidelberg: 15 June‐18 July 2020 [2] Berlin: 6 July‐23 September 2020) |
Mainly symptomatic 517/669 (77%) symptomatic; no further details Mean age 38 (range 18‐85) years (676 participants); 307 (46%) male |
PCR (multiple assays);
Ct thresholds not stated; author contact advises Ct thresholds as per assay IFUs Target: not stated apart from 3. E gene NP or NP + OP or OP; paired; same as index) Timing: not stated; author contact advises only paired swabs used Interval: paired |
Reports 0 missing data None reported Index: none reported Reference: none reported |
|
FIND 2020e (BR) Published 476 (117) |
Single‐group (prospective); adults in community meeting national suspect definition for COVID‐19 testing presenting at a community testing clinic (n = 476) |
COVID‐19 test centre (community) Brazil (27 July‐16 September) |
Symptomatic 470/476 (99%) symptomatic; no further details Mean age 45 (range 0‐106) years (473 participants); 252 (53%) male |
PCR (single assay);
Ct threshold not stated Target: N1 and N2 NP; paired; same as index Timing: not stated; author contact advises only paired swabs used Interval: paired |
Reports 0 missing data None reported Index: none reported Reference: none reported |
|
FIND 2020e (DE) Published 1239 (25) |
Single‐group (prospective); adults in community meeting national suspect definition for COVID‐19 testing presenting at [1] a drive‐in testing centre or [2] ambulatory testing clinic |
COVID‐19 test centre (community) Germany ([1] Heidelberg: 4 May‐3 September [2] Berlin: 4 May‐18 August 18) |
Mixed 733/1223 (59.9%) symptomatic; no further details Mean age 39.5 (range 17‐59.2) years (1239 participants); 607 (50%) male |
PCR (multiple assays);
Ct thresholds not stated; author contact advises Ct thresholds as per assay IFUs Target: not stated NP; paired; same as index Timing: not stated; author contact advises only paired swabs used Interval: paired |
Reports 0 missing data None reported Index: none reported Reference: none reported |
|
FIND 2020f Preprint 425 (8) |
Single‐group (prospective); participants at risk for SARS‐CoV‐2 infection based on exposure to a confirmed case, suggestive symptoms, or travel to a high‐risk area, presenting at 1 of 3 sites: (1) drive‐in testing station (n = 1213) (2) a clinical ambulatory testing facility (n = 1308) (3) secondary care facility (n = 53) |
COVID‐19 test centre (community (drive‐in or clinical ambulatory testing) and secondary care (inpatient?)) (1), (2) Germany (3) UK (17 April and 25 August 2020; dates varied by assay and site) |
Mainly symptomatic Symptomatic on testing day (n = 2355) Overall: 1901, 80.7% [A] 564, 81.2% [B] 283, 68.9% [C] 1054, 84.4% Prior negative test result (n = 1928) Overall: 236, 12.2% [A] 73, 11.7% [B] 38, 12.6% [C] 125, 12.5% Detailed symptoms are reported by site and test in supplementary materials Mean age (SD) (n = 2405) Overall 40.4 (14.3) years [A] 42.7 (14.9) [B] 44.9 (15.4) [C] 37.6 (12.7) Male (%) (n = 2361) Overall: 1115, 47.2% [A] 47.2% [B] 39.7% [C] 49.8% |
PCR (multiple assays) Target: not stated NP or OP, or NP + OP; paired; same as index Timing: as per index test Interval: paired |
154 excluded following enrolment
(116 2nd swab refused
3 nose bleed after 1st swab
3 insufficient time for both swabs
31 other reasons
1 no reason available) Ag tests: [A] 2 invalid (PCR‐ve) [B] 8 invalid (PCR‐ve) [C] 0 invalid reported PCR: 3 excluded as invalid (n = 2) or not available (n = 1) Index: none reported; ease‐of‐use assessment reported [A] a high number of test execution steps (including precision pipetting) … challenges when performing multiple tests at the same time possibly hindering the test’s widespread use [B] challenges due to inconsistent test result interpretation (often only very faint lines visible) and deficiencies in both the test kit quality and design [C] no dissatisfactory scores identified Reference: none reported |
|
FIND 2021a [A] Published 218 (79) |
Single‐group (prospective); patients with symptoms consistent with COVID‐19 (meeting national definition for testing) presenting at a community testing clinic | COVID‐19 test centre (community (COVID‐19 testing clinic)) Brazil (21‐27 January 2021; 23‐26 February 2021) |
Symptomatic All symptomatic; no further details Mean age 42.3 (range 18‐90) years; 92 (42%) male |
PCR (single assay) Target: N1, N2 NP; paired; same as index Timing: same as for index test Interval: paired |
Reports 0 invalid results for both assays None reported Index: none reported Reference: none reported |
|
FIND 2021b [A] Published 281 (44) |
Single‐group (prospective); at single site: patients seeking COVID‐19 testing at COVID‐19 testing centre; described as able to ambulate, at high risk for SARS‐CoV‐2 according to clinical suspicion, and meeting suspect definition of the Department of Public Health (n = 281) | COVID‐19 test centre (community) Germany (15 December 2020‐19 January 2021) |
Mixed 130/279 symptomatic (46%) Mean age 42.9 (range 18‐81) years 134, 48% male |
PCR (multiple assays) Target: not stated NP; paired; same as index Timing: not stated; author contact advises only paired swabs used. Interval: paired |
None reported; 0 invalid results None Index: none reported Reference: none reported |
|
FIND 2021c (BR) [A] Published 214 (78) |
Single‐group (prospective); at 2 sites; (see FIND 2021c (DE) [A] for additional site data): ambulatory patients meeting national suspect definition for COVID‐19 testing presenting at a community testing clinic in Brazil | COVID‐19 test centre (community testing clinic) Brazil (14‐20 January; 2‐4 March 2021) |
Symptomatic All symptomatic; no further details Mean age 41.3 (range 18‐77) years; 85 (40%) male |
PCR (single assay) Target: N1 and N2 NP; paired; same as index Timing: not stated; author contact advises only paired swabs used Interval: paired |
Reports 0 missing data; 0 invalid results None reported Index: none reported Reference: none reported |
|
FIND 2021c (DE) [A] Preprint 179 (41) |
Single‐group (prospective); adults at high risk for SARS‐CoV‐2 infection according to clinical suspicion who attended the ambulatory COVID‐19 test facility of Charité University Hospital Berlin, Germany | COVID‐19 test centre (COVID‐19 test centre) Germany (11‐18 November 2020) |
Symptomatic On day of testing: 172 (96%) symptomatic; 7 (4%) asymptomatic Average symptom duration 4.2 (SD 2.6) d Average age 36.2 (SD 12.2) years; 48% female; 14% with comorbidities |
PCR (multiple assays) Target: unclear NP + OP; paired; same as index Timing: as for index test Interval: paired |
Yes (1/180); 1 patient was excluded as both swabs for the Ag could not be obtained None reported Index: none reported Reference: none reported |
|
FIND 2021d Published 214 (78) |
Single‐group (prospective); adults able to ambulate, at high risk for SARS‐CoV‐2 according to clinical suspicion, and meeting suspect definition of the Department of Public Health and presenting at [1] a drive‐in testing centre or [2] ambulatory testing clinic | COVID‐19 test centre (community) Germany ([1] Heidelberg: 20 January‐19 February 2021 [2] Berlin: 18 January‐22 February 2021) |
Mixed Symptomatic 62% (446/718); no further details Mean age 39.4 (range 18‐80) years; 348/719 (48%) male |
PCR (multiple assays) Target: only stated for 3. (E gene) NP or OP; paired; same as index Timing: not stated; author contact advises only paired swabs used Interval: paired |
Reports 0 missing data; 0 invalid results None reported Index: none reported Reference: none reported |
|
FIND 2021e Published 265 (44) |
Single‐group (prospective); adults in community meeting Department of Public Health definition of a suspected COVID‐19 case and being tested for SARS‐CoV‐2 part of routine medical care | COVID‐19 test centre (COVID‐19 test centre) Switzerland (4‐13 January 2021) |
Not reported Only reported for PCR+ve group; symptomatic 88.6% (39/44) Mean age 36.3 (range 16‐80) years; 139 (52%) male |
PCR (multiple assays) Target: not stated NP; paired; same as index Timing: as for index test Interval: paired |
Reports 0 missing data; 0 invalid results None reported Index: none reported Reference: none reported |
|
FIND 2021f Published 665 (194) |
Single‐group (prospective); adults able to ambulate, at high risk for SARS‐CoV‐2 according to clinical suspicion, and meeting suspect definition of the Department of Public Health, presenting either at: 1. Heidelberg: drive‐in testing centre 2. Berlin: ambulatory testing clinic | COVID‐19 test centre (COVID‐19 test centre) Germany (1. Heidelberg: 11‐31 March 2021 2. Berlin: 11 March‐15 April 2021) |
Mixed Symptomatic: 66.5%, (440/662) Mean age 38.7 (range 18‐78) years; 331 (50%) male |
PCR (multiple assays) Target: not stated NP, OP or NP + OP; paired Timing: as for index test Interval: paired |
Yes 16/665, 2.4% invalid Ag results (including 3 PCR+ve and 13 PCR‐ve samples) Index: none reported Reference: none reported |
|
FIND 2021g Published 462 (69) |
Single‐group (prospective); individuals (age 16+) in community meeting Department of Public Health definition of a suspected COVID‐19 case and being tested for SARS‐CoV‐2 part of routine medical care | COVID‐19 test centre (COVID‐19 test centre) Switzerland (24 November 2020–20 January 2021) |
Not reported Reported for PCR+ve only; symptomatic 94.2% (65/69) Mean age 38.7 (range 16‐82) years; 206 (45%) male |
PCR (multiple assays) Target: not stated NP; paired; same as index Timing: as for index test Interval: paired |
None reported; reports 0 invalid None reported Index: none reported Reference: none reported |
|
FIND 2021h Published 232 (41) |
Single‐group (prospective); individuals (age 16+) in community meeting Department of Public Health definition of a suspected COVID‐19 case and being tested for SARS‐CoV‐2 part of routine medical care | COVID‐19 test centre (COVID‐19 test centre) Switzerland (21‐29 January 2021) |
Not reported Reported for PCR+ve only; symptomatic 92.7% (38/41) Mean age 36.3 (range 16‐76) years; 103 (44%) male |
PCR (multiple assays) Target: not stated NP; paired; same as index Timing: as for index test Interval: paired |
None reported; reports 0 invalid None reported Index: none reported Reference: none reported |
|
FIND 2021i Published 328 (56) |
Single‐group (prospective); individuals (age 16+) in community meeting Department of Public Health definition of a suspected COVID‐19 case and being tested for SARS‐CoV‐2 part of routine medical care | COVID‐19 test centre (COVID‐19 test centre) Switzerland (3‐11 December 2020) |
Not reported Reported for PCR+ve only; symptomatic 100% (56/56) Mean age 37.9 (range 16‐76) years; 129 (39%) male |
PCR (multiple assays) Target: not stated NP; paired; same as index Timing: as for index test Interval: paired |
None reported; reports 0 invalid None reported Index: none reported Reference: none reported |
|
FIND 2021j Preprint 729 (15) |
Single‐group (prospective) Participants at risk for SARS‐CoV‐2 infection based on exposure to a confirmed case, suggestive symptoms, or travel to a high‐risk area, presenting at 1 of 3 sites: (1) drive‐in testing station (n = 1213) (2) a clinical ambulatory testing facility (n = 1308) (3) secondary care facility (n = 53) |
COVID‐19 test centre (community (drive‐in or clinical ambulatory testing) and secondary care (inpatient?)) (1), (2) Germany (3) UK (17 April and 25 August 2020; dates varied by assay and site) |
Mainly symptomatic Symptomatic on testing day (n = 2355) Overall: 1901, 80.7% [A] 564, 81.2% [B] 283, 68.9% [C] 1054, 84.4% Prior negative test result (n = 1928) Overall: 236, 12.2% [A] 73, 11.7% [B] 38, 12.6% [C] 125, 12.5% Detailed symptoms are reported by site and test in supplementary materials Mean age (SD) (n = 2405) Overall 40.4 (14.3) years [A] 42.7 (14.9) [B] 44.9 (15.4) [C] 37.6 (12.7) Male (%) (n = 2361) Overall: 1115, 47.2% [A] 47.2% [B] 39.7% [C] 49.8% |
PCR (multiple assays) Target: not stated NP or OP, or NP + OP; paired; same as index Timing: as per index test Interval: paired |
154 excluded following enrolment
(116 2nd swab refused
3 nose bleed after 1st swab
3 insufficient time for both swabs
31 other reasons
1 no reason available) Ag tests: [A] 2 invalid (PCR‐ve) [B] 8 invalid (PCR‐ve) [C] 0 invalid reported PCR: 3 excluded as invalid (n = 2) or not available (n = 1) Index: none reported; ease‐of‐use assessment reported [A] a high number of test execution steps (including precision pipetting) … challenges when performing multiple tests at the same time possibly hindering the test’s wide‐spread use [B] challenges due to inconsistent test result interpretation (often only very faint lines visible) and deficiencies in both the test kit quality and design [C] no dissatisfactory scores identified Reference: none reported |
|
Fourati 2020 [A] Published 634 (297); number of cases tested varied per assay |
2‐group (retrospective); (1) residual samples from patients with positive SARS‐CoV‐2 PCR tested when they presented symptoms at the time of the first epidemic wave (n = 297) (2) pre‐pandemic samples (n = 337) |
Laboratory‐based (unclear; "consulted or were admitted") France (9 March‐9 April 2020) |
Symptomatic Not stated; all apparently symptomatic Data by viral load reported for 293/297 cases: ≤ 20 Ct: 39, 13% 20‐25 Ct: 88, 30% 25‐30 Ct: 72, 25% > 30 Ct: 88, 30% Not stated |
PCR (single assay) Target: not stated NP; same sample used Timing: as for index Interval: simultaneous |
Number of cases missing per assay varied; reasons for missing data not reported (presumably invalid assay results)
[A] 5, 1.7%
[B] 6, 2.0%
[C] 2, 0.7%
[D] 0
[E] 2, 0.7%
[F] 0 Not stated Index: not stated Reference: not stated |
|
Garcia‐Finana 2021 Published 5869 (74) |
Single‐group (prospective) Asymptomatic individuals attending asymptomatic testing sites (ATS) in the City of Liverpool were asked to participate in a QA process | Screening (COVID‐19 test centres (asymptomatic)) UK (8‐29 November 2020) |
Asymptomatic Asymptomatic Not reported |
PCR (single assay) Target: N, S, ORF1 Sample site not reported; same as index Timing: as for index test Interval: paired |
Yes; Void results: RDT 22 (4 PCR+ve and 18 PCR‐ve) PCR 343 (2 RDT+ and 341 RDT‐) Index: 0 Reference: 0 |
|
Gomez 2021(a) Preprint (not peer reviewed) 427 (43) |
Single‐group (not reported) Report of 2 study cohorts: [1] single‐group study in symptomatic paediatric patients presenting at outpatients or in primary care (all < 18 years); total n = 427 (included as Gomez 2021(a)) [2] single‐group study estimating sensitivity alone in a symptomatic PCR+ve student/college‐aged population (18‐25 years) presenting at a university campus; total n = 32 (included as Gomez 2021(b)) (A further 3 groups were reported but did not undergo Ag testing and were excluded from the review Group [3]: ED‐collected specimens Group [4]: asymptomatic people undergoing surgical procedures unrelated to COVID‐19 Group [5]: asymptomatic students) |
Mixed (community outpatients or primary care) USA (Pennsylvania) (8 March‐10 September 2020) |
Symptomatic Symptoms not reported Reported as "symptomatic" and "presented for care" Age and sex not reported |
PCR (single assay) Target: not reported NMT; paired; same as index Timing: as for index test Interval: paired |
None reported Index: none reported Reference: none reported |
|
Gomez 2021(b) Preprint (not peer reviewed) 32 (32); number of PCR‐ve students was not reported |
Single‐group (cases) (not reported) Report of 2 study cohorts: [1] single‐group study in symptomatic paediatric patients presenting at outpatients or in primary care (all < 18 years); total n = 427 (included as Gomez 2021(a)) [2] single‐group study reporting only sensitivity in symptomatic PCR+ve student/college‐aged population (18‐25 years) presenting at a university campus; total n = 32 (included as Gomez 2021(b)) (A further 3 groups were reported but did not undergo Ag testing and were excluded from the review Group [3]: ED‐collected specimens Group [4]: asymptomatic persons undergoing surgical procedures unrelated to COVID‐19 Group [5]: asymptomatic students) |
Primary care (student health services) USA (Pennsylvania) (8 March‐10 September 2020) |
Symptomatic Symptoms not reported Reported as "symptomatic" and "presented for care" Age and sex not reported |
PCR (multiple assays) Target: not reported AN; paired; same as index Timing: as for index test Interval: paired |
None reported Index: none reported Reference: none reported |
|
Gonzalez‐Donapetry 2021 Published paper 440 (18) |
Single‐group (prospective); in symptomatic children meeting COVID‐19 clinical criteria and presenting < 7 d pso | Hospital ED (samples from paediatric ED) Spain (25 September‐14 October 2020) |
Symptomatic: all symptomatic (n = 440); symptoms included: cough 222 (51%), fever 296 (67%), dyspnoea 67 (15%), headache 35 (8%), dysgeusia/anosmia 1 (0%), odynophagia 55 (13%), rhinorrhoea 228 (52%), gastrointestinal disorder 103 (23%) Median age: 3 (IQR 1‐7); 260 (59%) male |
PCR (single assay) Target: nucleocapsid (N) and envelope (E) genes NP; unclear; same as index Timing: as for index test Interval: unclear |
None reported Index: none reported Reference: none reported |
|
Gremmels 2021(a) Published 1369 (139) |
Single‐group (prospective); [1] community‐dwelling mildly symptomatic participants in a medium endemic area (n = 1369) |
COVID‐19 test centre (community) Netherlands ([1] 22 September‐6 October [2] 23 September‐9 October) |
Mainly symptomatic Cohort [1] only; data on symptoms were missing from 9 participants Asymptomatic 37, 2.7%, sore throat 907, 66.3%; coryza 943, 69%; cough 780, 57.1%; headache 601, 44.0%; tiredness 565, 41.3%; general malaise 365, 26.7% (further 19 documented) Median age 36.4 (IQR 27.0‐49.6) years; 523, 38.3% male |
PCR (single assay) Target: E‐, N‐, and RdRP‐gene NP + OP; paired Timing: NOP swab obtained first for PCR Interval: paired |
2 patients excluded ("inappropriate application of NP swab and lab mislabelling"), disease status not reported. None reported Index: none; no bands were classified as unclear by the independent observers Reference: patients |
|
Gremmels 2021(b) Published 208 (63) |
Single‐group (prospective); [2] community‐dwelling mildly symptomatic participants in a high endemic area (n = 208) |
COVID‐19 test centre (community) Netherlands ([1] 22 September‐6 October [2] 23 September‐9 October) |
Not reported Not stated; "mildly symptomatic", presume mixed as per Gremmels 2021(a) Not stated |
PCR (single assay) Target: E‐, N‐, and RdRP‐gene NP + OP; paired Timing: NOP swab obtained first for PCR Interval: paired |
None reported for Aruba site None reported Index: none; no bands were classified as unclear by the independent observers Reference: none |
|
Gupta 2020 Published 330 (77) |
Single‐group (not stated; appears prospective); symptomatic patients with suspected COVID‐19 and asymptomatic contacts of laboratory‐confirmed cases between 5 and 10 d of exposure, meeting Indian Council of Medical Research (ICMR) strategy for COVID‐19 testing |
COVID‐19 test centre (outpatient; tertiary care hospital) India (31 May‐24 July 2020) |
Mixed 204 (62%) symptomatic; 126 (38%) asymptomatic Median symptom duration: 1 day (range: 1‐10). Symptoms included: fever (31.5%), cough (25.4%), fatigue/malaise (11.8%), headache (3.3%), and runny nose (3.3%) Median age 34.1 (SD 12.6) year; 231 (70%) male |
PCR (single assay) Target: ORF1 ab NP+OP; paired Timing: as for index test; states the sequence for specimen collection was random for both the samples Interval: paired |
None reported Index: none reported Reference: none reported |
|
Halfon 2021 Preprint 200 (100) |
≥ 2 groups (prospective); [1] PCR+ve (n = 100) [2] PCR‐ve (n = 100) Selected from 43,399 samples that had undergone RT‐qPCR for SARS‐COV‐2 using NP swab | Laboratory‐based (no details) France (August‐November 2020) |
Mixed
Figure 1: 104 (52%) asymptomatic, including 35 PCR+ve; 69 (35%) symptomatic, including 18 () PCR+ve; 27 (13%) symptom status unknown or time not reported, including 13 (48%) PCR+ve Mean age 48 (SD 21); 96 (48%) male |
PCR (multiple assays) Target: not stated NP; unclear; same sample site Timing: not stated; same as for index test Interval: unclear |
None reported Index: none reported Reference: none reported |
|
Houston 2021 Published 728 (280) |
Single‐group (prospective); ‐ adult admissions who met the WHO COVID‐19 case definition at a busy acute hospital | Hospital inpatient (inpatient) UK (November 17‐December 31, 2020) |
Symptomatic: all symptomatic (as per WHO case definition); required supplemental Oxygen, n (%, 95%CI): 141 (21·4%, 18·3 ‐ 24·6), Temperature > 38°C, n (%, 95%CI): 163 (24·9%, 21·6 ‐ 28·2) Median age 67·5 (IQR 52‐82) year; 327 (44.9%) female |
PCR (no details) Target: not stated NP; paired; same as index Timing: same as for index test Interval: paired |
Not stated; only valid results included None reported Index: none reported Reference: none reported |
|
Huh 2021 Preprint (not peer reviewed) 132 (62) |
Unclear design (retrospective); included samples PCR positive or negative for SARS‐CoV‐2 [A second study evaluating serology assays for antibody detection in additional PCR+ve and pre‐pandemic samples was also reported but not eligible for this review] | Unclear (stated "2 institutions", no further details reported); Not stated; appears to be Korea (not reported) |
Not reported: Not reported; could all be symptomatic as data are reported up to 12 d post symptom onset for PCR positive only Age and sex not reported |
PCR (single assay) Target: RdRP and N genes specific for 2 SARS‐CoV‐2 and E gene for all of Sarbecovirus including SARS‐CoV‐2 NP; same sample used Timing: appears to be same as index test Interval: simultaneous |
None reported Index: none reported Reference: none reported |
|
Igloi 2021 Preprint 970 (186) |
Single‐group (prospective ); ‐ participants meeting national testing criteria (mildly symptomatic or close contact with a confirmed SARS‐CoV‐2 infected person) | COVID‐19 test centre (drive through COVID‐19 test centre (by appointment only)) Netherlands (Not stated) |
Mainly symptomatic: 525 (91%) symptomatic Cold symptoms and runny nose (64.5%), sore throat (57%), coughing (55%), headache (48%), tiredness (38%), muscle pain (27%), shortness of breath and chills (21%), fever and reproductive cough (17%), loss of taste and smell (1.5%) 159 (85%) Ct ≤ 30 Median 42 (range 18‐86) years; 55% female (525/960); |
PCR (single assay) Target: not stated NP+OP; paired Timing: same as index test Interval: paired |
None reported Index: none reported; "when results were dubious, readout was performed independently by 2 persons" Reference: none reported |
|
Ishii 2021 [A] Published paper [A]: NP swabs: 271 (11) [B]: Saliva samples: 93 (9) |
Single‐group (unclear; NP swabs and saliva samples were collected from 33 COVID‐19 patients and 564 non‐COVID‐19 patients); appears to include samples submitted for COVID‐19 diagnosis at a medical centre; includes symptomatic patients (dysosmia, dysgeusia, fever and pneumonia) and asymptomatic close contacts of confirmed cases | Unclear (unclear) Japan (August‐September 2020) |
Not reported: All PCR positive cases with an ESPLINE result were symptomatic (n = 20) Not stated |
PCR (single assay) Target: not reported NP; same sample used Timing: same as for index test Interval: simultaneous |
None reported Index: none reported Reference: none reported |
|
Jaaskelainen 2021 [A] preprints 136 (96); different number of cases tested per assay |
≥ 2 groups (retrospective); including: [1] PCR test positive samples from adults from outpatient clinics and drive‐through testing sites (n = 96) [2] PCR positive and negative samples for analysis of analytical performance (n = 102) [PCR positive samples in group [2] excluded from review as they were deliberately selected to cover a wide Ct range (26.35‐32.66 Ct)] | COVID‐19 test centre (Outpatient/community test centre drive‐through testing sites)) Finland ([1] 1‐18 November 2020 [2] April‐November 2020 ) |
Not reported: Not stated Not stated |
PCR (single assay) Target: N gene target of SARS‐CoV‐2 NP; same sample used Timing: not reported Interval: simultaneous |
Number of samples tested varied by assay. Of 96 PCR+ve samples in group [1], results were reported for:
1) Sofia (Quidel): 87 samples, 91%
2) Standard Q (SD Biosensor): 96 samples, 100%
3) Panbio™ (Abbott): 90 samples, 94% None reported Index: none reported Reference: none reported |
|
Jakobsen 2021 Preprint 4697 patient with 4811 paired conclusive tests (221 tests); (196 were tested twice or more) |
Single‐group (prospective); symptomatic and asymptomatic patients (self‐reported) who had booked an appointment at a public test centre | COVID‐19 test centre (Public COVID‐19 test centre) Denmark (26‐31 December 2020) |
Mixed: 705 (15%) symptomatic (self‐report); 3008 (64%) asymptomatic (not all participants responded to the online questionnaire regarding symptoms) Mean age 45 (SD 16.9) years; 2456 (53%) female |
PCR (single assay) Target: E‐gene OP; paired Timing: no details; as for index test Interval: paired |
Yes 97/4908 inconclusive on PCR, leaving 4811 samples for inclusion Index: 196 individuals with non‐conclusive results on Ag test were tested twice or more Reference: 97 (inconclusive results on PCR (i.e. Ct > 38)) |
|
James 2021 Published 2339 (152) |
Single‐group (prospective); targeted screening at a US medical centre (mainly asymptomatic); all staff providing patient care were required to participate but testing was optional for non‐clinical staff) | HCW screening (primarily screening; dedicated stations within the hospital (mobile teams) or drive‐through parking lot stations) USA (2‐9 October 2020) |
Mainly asymptomatic: 2224 (95%) asymptomatic; 115 (5%) symptomatic (94 (82%) reported only 1 symptom and 21 (18%) reported 2–6 symptoms) Fever (6%), cough (29%), sore throat (29%), chills (6%), headache (41%), muscle aches (12%), abdominal pain (4%); none reported loss of taste or loss of smell. Median 37 (range 16–89) years; gender NR |
PCR (single assay) Target: N or ORF1 Nasal; paired Timing: not stated Interval: paired |
None None Index: none (all paired samples were successfully tested) Reference: none (all paired samples were successfully tested) |
|
Kerneis 2021 Preprint (not peer reviewed) 1452 (129) |
Single‐group (prospective); Symptomatic patients invited for testing (i.e. temperature > 37.8 °C or chills, cough, rhinorrhoea, muscle pain, loss of smell or taste, unusual persistent headaches or severe asthenia), symptomatic contacts of confirmed cases, asymptomatic contacts of confirmed cases (after 7 d self‐isolation) and any other asymptomatic individuals wishing to be tested | COVID‐19 test centre (COVID‐19 community testing centres) France (19 October‐18 December 2020) |
Mixed: 571 (39%) symptomatic, 409 with 1‐3 symptoms: Cough: 292/1451 (20%) Headaches: 257/1451 (18%) Rhinorrhoea: 202/1451 (14%) Asthenia: 198/1451 (14%) Muscle pain: 177/1451 (12%) Fever: 163/1451 (11%) Diarrhoea: 85/1451 (6%) Chills: 69/1451 (5%) Anosmia: 62/1451 (4%) Shortness of breath: 53/1451 (4%) Chest pain: 52/1451 (4%) Med age [IQR]: 36 [26‐50]; 122 (8%) children Sex: 755/1451 (52%) female |
PCR (single assay) Target: ORF1ab, N and S‐genes NP; paired; same as index Timing: as for index test Interval: paired |
Yes;
Of 1451 participants who provided samples, 1117 underwent Ag tests. Reason for missing data not reported
Of 1117 tested:
2 technical failures (on Ag test); 6 additional missing results, reason not reported Not reported Index: none reported Reference: none reported |
|
Kilic 2021 Accepted manuscript posted online 1384 (116) |
Single‐group (prospective); symptomatic patients meeting pre‐set criteria for Ag and PCR testing (COVID‐19 exposure and ≤ 5 d pso, including fever/flu‐like symptoms, unexplained shortness of breath, or new loss of taste) | Mixed (not stated; "patients receiving care at our hospital system" ‐ likely mixed settings? (www.wakehealth.edu/Find‐A‐Provider)); USA (20 October‐3 December 2020) |
Symptomatic: All symptomatic Median age 46.8 (range 1‐98) years, 800 (57.8%) female |
PCR (single assay) Target: S gene and ORF1ab genes NP; paired Timing: same as for index Interval: paired |
None reported Index: none reported Reference: none reported |
|
Kohmer 2021 [A] Published 100 (74) |
Single‐group (prospective); individuals from shared living facilities for screening purposes regardless of their clinical symptoms (n = 100) | Shared living (community); Germany (November 2020 (2 weeks)) |
Not reported Not stated Not stated |
PCR (single assay) Target: ORF1 and E‐gene; considered positive if ORF1 detected NP; same sample used Timing: not stated Interval: simultaneous |
None reported Index: none reported Reference: none reported |
|
Kriemler 2021 Published |
Single‐group (prospective); a single group study (nested in the Ciao Corona Cohort study) to estimate sensitivity and specificity ‐ children (n = 641) and teachers (n = 66) attending primary or secondary schools over a one week period and tested at least once (T1 and or T2) (n = 707). Schools were selected based on high incidence areas; children were required to be kept at home if they were sick beyond very mild symptoms such as runny nose or mild cough |
Screening in schools (schools in the city of Zurich and one school of each of 4 adjacent districts.
Author institution: Epidemiology, Biostatistics and Prevention Institute, University of Zurich, Zurich); (1‐11 December 2020) |
Asymptomatic At T1, 198/567 (35%) children and 5/66 (8%) teachers reported mild symptoms (runny nose, headache, cough stomach upset, etc) during the previous 5 d before testing; symptoms of week 1 (T1) and 2 (T2) reported not to differ significantly Children and adolescents: age range 10‐19 years; 370 (58%) female children and adolescents; 46 (70%) female teachers |
PCR (single assay) Target: N, S, RdRP and E‐gene Buccal; paired; same as index Timing: as for index test Interval: paired |
None reported; participants were not all tested at T1 and T2 None reported Index: none reported Judged by 2 study team members (experienced in RDT testing) in agreement as positive or negative Reference: none reported |
|
Kruger 2021 Preprint (not peer reviewed) 767 (146) |
Single‐group (prospective) Single‐group multi‐centre study Adults at risk of SARS‐CoV‐2 infection based on reported symptoms or recent contact with a confirmed case (according to the criteria of the National Health Authority): Group [1]: Heidelberg Group [2]: Berlin [Group [1] and group [2] are reported as subgroups] |
COVID‐19 test centre (COVID‐19 testing site
[1]: Drive‐in testing site
[2]: Clinical ambulatory testing facility) Germany (2 November‐4 December 2020) |
Mixed 486 (64%) symptomatic on day of testing; 90 (19%) fever, 247 (52%) cough, 242 (50%) sore throat, 297 (62%) fatigue Raw symptom data in a supplementary table medrxiv.org/content/10.1101/2021.03.02.21252430v1.supplementary‐material (Section E, Table 1, p19) Average age: 38.5 (SD, 14.2) years, 52% female |
PCR (multiple assays) Target: E‐gene NP, OP or NP+OP; paired Timing: same as for index test Interval: paired |
Yes; 2 refused NMT swab and 4 had invalid PCR results 7 samples gave error message on LumiraDx device; repeat testing yielded valid results and inclusion in the analysis. Index: not reported Reference: 4 invalid PCR results excluded |
|
Kruttgen 2021 Published 150 (75) |
≥ 2 groups (retrospective); [1] patients previously tested positive by PCR (n = 75) [2] patients previously tested negative by PCR (n = 75) | Unclear (not stated; SARS‐CoV‐2 negative were reported as hospital inpatient) Germany (Not stated) |
Not reported Not stated Not stated |
PCR (single assay) Target: not stated NP; same sample used Timing: not stated Interval: simultaneous |
None reported Index: none reported Reference: none reported |
|
L'Huillier 2021 Preprint 885 (119); 60 excluded a priori |
Single‐group (prospective); children (0‐16 years old) meeting eligibility criteria for PCR testing: 1) symptoms suggestive of COVID infection according to local governmental testing criteria, and for asymptomatic children either 2) contact with a laboratory confirmed COVID infected person and 3) pre travel testing. | COVID‐19 test centre (Paediatric COVID‐19 testing centre) Switzerland (10 November 2020‐26 March 2021) |
Mixed 533 (65%) symptomatic: headache (58%), nasal discharge (55%), cough (44%), fatigue (44%), dysphagia (41%), fever (30%), abdominal pain (20%), myalgia (16%), diarrhoea (15%); shortness of breath (8%), anosmia (7%). 289 (35%) asymptomatic All children; median age 12.1 (IQR 9.4‐14.5) years; 266 (50%) female |
PCR (multiple assays) Target: not stated NP; same as index Timing: for symptomatic patients samples were taken at median 2 d pso (IQR 1‐3) Interval: paired |
Yes; 63/885 (7%) excluded from analysis
1 excluded a priori (did not meet inclusion criteria), 1 refused PCR, and 58 refused Ag test (n = 58)
3 excluded after testing 2 Ag test results not reported and 1 Ag test result invalid among the 822 Ag tests Index: none; only 1 discrepant between readers (considered +ve) Reference: 0 |
|
Lambert‐Niclot 2020 Accepted manuscript 138 (94) |
Single‐group (unclear; testing conducted prospective) Samples submitted for PCR testing |
Laboratory‐based (3 university hospital virology laboratories) France (1 April‐15 April 2020) |
Not reported Not stated Not stated |
PCR (multiple assays) Target: E gene NP; same sample used Timing: within a few hours after collection; time pso not reported Interval: simultaneous |
4 samples collected in COBAS VTM gave invalid results and all samples in COBAS medium were excluded 4 samples collected in COBAS VTM gave invalid results and all samples in COBAS medium were excluded Index: control lines reported as "barely visible" for 9 positive and 8 negative tests Reference: none reported |
|
Lanser 2021 Published 53 (100%) included; 51 analysed |
Single‐group (cases) (retrospective); (sensitivity only) a group of patients with confirmed COVID‐19 during their hospital stay in different stages of the disease. All patients received Ag test along with PCR (n = 53). (2 patients with negative PCR were excluded due to apparent subsidence of infection) | Hospital inpatient (hospital inpatient) Austria (Not stated) |
Symptomatic In different stages of the COVID infection Not stated |
PCR (no details) Target: orf1 NP; same sample site Timing: unclear; states "sample taken during their hospital stay in different stages of the disease" Interval: unclear |
Yes
2/53 COVID patients were PCR‐ve suggesting an already subsided infection and were excluded from the analysis None reported Index: none reported Reference: none reported |
|
Linares 2020 Preprint 255 (60); NB 257 reported in sample collection |
Single‐group (unclear; appears to be prospective) 2 locations: [1] symptomatic patients admitted to ED with clinical suspicion of COVID‐19 (n = 135) or asymptomatic patients with history of contact with another COVID‐19 patient (n = 17) [2] symptomatic patients (n = 50) or asymptomatic (n = 55) patients attending 1 of 2 PHC centres |
Hospital ED (ED (n = 135) or primary care (n = 50)) Spain (10‐15 September) |
Mixed 185, 72% symptomatic ED (n =135): fever 40, dyspnoea 42, cough 22, headache 14 Primary care (n = 50): fever 14, dyspnoea 1, cough 18, headache 17 Mean age (range): ED 51.5 (range 37.0‐71.8) years; primary care 39.0 (25.0‐56.0) years ED 77 (51%) male, primary care 49 (47%) male |
PCR (single assay);
threshold not stated Target: not stated NP; paired; same as index Timing: not stated Interval: paired |
None reported however 257 reported in Methods and 255 in Results None reported Index: none reported Reference: none reported |
|
Lindner 2021a [A] (6543) In‐press (published as in accepted form) 289 (39) |
Single‐group (prospective); mainly symptomatic adults at high risk for SARS‐CoV‐2 infection according to clinical suspicion of COVID‐19 |
COVID‐19 test centre (COVID‐19 test centre) Germany (23 September‐14 October 2020) |
Symptomatic On day of testing: 283 (98%) symptomatic; 6 (2%) asymptomatic Average symptom duration 4.4 d (SD 2.7) Average age 34.7 (SD 11) years; 42.9% female and 19.0% with comorbidities |
PCR (multiple assays) Target: E‐gene NP+OP; paired Timing: as for index test Interval: paired |
Yes; 2 excluded as both swabs for the Ag test could not be obtained None reported Index: none; "no invalid tests were observed" Reference: none; "no invalid tests were observed" |
|
Lindner 2021b [A] Preprint 146 (40) |
Single‐group (prospective); symptomatic adults with high clinical suspicion of SARS‐CoV‐2 including 1) reported contact with a confirmed case and any compatible symptom, or 2) fever or impaired taste or smell irrespective of exposure | COVID‐19 test centre (COVID‐19 test centre) Germany (30 November‐11 December 2020 (recruitment dates do not overlap either FIND 2021c (DE) [A] (which has a 2020 preprint by Lindner and colleagues included as a secondary publication Lindner 2021a [A]) |
Symptomatic 100% symptomatic; mean duration of pso 3.4 d (SD 2.0) 34 (23%) with comorbidities Mean age 35 (SD 11.5) years; 75 (51%) female |
PCR (multiple assays) Target: E gene (Tib Molbiol) NP+OP; paired Timing: mean duration of pso 3.4 d (SD 2.0) Interval: paired |
Yes; 4 excluded (3 participants were excluded as they did not fulfil the minimum language criterion and 1 participant excluded because of lost PCR specimen) NMT ‐ 1 participant left without reading the test‐result for nasal sample plus 1 invalid result (buffer spilt and test not repeated); none reported for NP sample Index: none reported; weak positives considered positive Reference: none reported |
|
Liotti 2021 Published letter 329 (104) |
Unclear; 2‐group (retrospective); residual samples selected from 1 of 2 virology laboratories at 2 COVID‐19 reference hospitals: [1] 104 PCR+ve for SARS‐CoV‐2 [2] 255 PCR‐ve for SARS‐CoV‐2 |
Laboratory‐based (not reported) Italy (Not stated) |
Not reported Not stated Of SARS‐CoV‐2 positive samples, 21, 20% high viral load (< 25 Ct), 83, 80% low viral load (≥ 25) [28, 27% with Ct ≥ 35) Not stated |
PCR (multiple assays) Target: not stated NP alone; same sample used Timing: not stated Interval: simultaneous |
None reported Index: none reported; FP results were re‐tested with Ag assay, 3 of 4 remained positive (all blood contaminated) Reference: none reported |
| Masia 2021 [A] | Single‐group (prospective); symptomatic with COVID‐19 signs/symptoms or asymptomatic contacts attending the PHC centres (n = 690 (76%) NP sample), and a majority of symptomatic patients presenting to the ED (n = 233 (25%) NP sample) | Mixed (mixed‐ PHC centres and an ED) Spain (15 September‐29 October 2020) |
Mixed 617 (68%) symptomatic; 296 (32%) asymptomatic Median (Q1–Q3) pso d: 3 (2–5) d Most frequent symptoms were cough (50%), fever (47%), sore throat (32%), and nasal congestion (31%) Median (Q1–Q3) Ct: 24 (16–30); 22 (16–29) in symptomatic and 28 (21–32) in asymptomatic; and 21 (15–27) in patients ≥ 50 years and 26 (18–31) in < 50 years Median age 40.6 (Q1‐Q3, 23.0–55.6) years; 423 (46%) male |
PCR (single assay) Target: E‐gene NP; same as index Timing: median 3 (Q1 2–Q3 5) d pso Interval: paired |
Unclear; of 913 only 904 had NP, 659 had nasal and 611 had saliva None reported Index: none reported Reference: none reported |
|
Merino 2021 Published 958 (359) |
Single‐group (prospective); multicentre study of individuals who had at least 1 symptom compatible with COVID‐19 (n = 830) or had been in close contact with a diagnosed COVID‐19 patient (n = 128) (total n = 958); all tested within 7 d pso or exposure | Hospital ED (hospital emergency rooms or other hospital units (documented only in preprint version)) Spain (September‐October 2020) |
Mainly symptomatic Symptomatic 830, 87%; all had at least 1 symptom compatible with COVID‐19 Mean age 42.4 years (range, 1‐100); 61.3% were women |
PCR (multiple assays) Target: not mentioned NP; same as index Timing: same as for index Interval: paired |
None reported Index: none reported Reference: none reported |
|
Mertens 2020 Preprint (not peer‐reviewed) n = 328 samples (99 at LHUB‐ULB, 132 at CHU Liège, 97 at UZ Leuven); 132 COVID‐19 cases |
Single‐group (retrospectively); Samples from cases of suspected SARS‐CoV‐2 infection |
Laboratory‐based (university laboratory; discussion states no outpatients) Belgium (19‐30 March 2020) |
Not reported Not reported Not reported |
PCR (multiple assays);
Threshold ≤ 40 Ct Target: RealStar®: not stated; Taqman Fast Virus: RdRp and E genes QuantStudio Dx; "slightly adapted" E‐gene Panther Fusion: E gene and ORF1‐ab NP; same sample used Timing: not stated; same samples as for index test but analysed at time of collection Interval: simultaneous |
No None reported; discussion reports some difficulties in visualising the strip through the closed tube requiring the lab technician to open the test tube in the laminar air flow cabinet and pull out the strip with forceps Index: weak T lines considered positive Reference: none reported; sensitivity can be extracted for cases with Ct values < or > 25 (high versus lower viral load) |
|
Miyakawa 2021 [A] Preprint (not peer reviewed) 108 (45) |
≥ 2 groups (retrospectively); [1] remnant PCR+ve NP swab specimens (n = 45) [2] PCR‐ve NP swabs (n = 63; only 45/63 samples tested with 4 of the 5 evaluated assays) |
Unclear (not reported (laboratory‐based at a biosafety level‐3 laboratory)) Japan (Not reported) |
Not reported Not reported Not reported |
PCR (no details) Target: N gene NP; same sample used Timing: not reported Interval: simultaneous |
Not reported Not reported Index: not reported Reference: not reported |
|
Mockel 2021(a) Accepted manuscript 281 (89) |
Single‐group (prospective); patients attending hospital EDs ([1] 4 adult EDs (n = 281) and [2] 1 paediatric ED (n = 202)) Cohort [2] have been included as Mockel 2021(b) In both cohorts patients were either symptomatic (acute respiratory symptoms or loss of smell or taste), contacts of confirmed cases up to 14 d before onset of COVID‐19 symptoms, or had clinical or radiological signs of viral pneumonia in the context of an outbreak in nursing homes or hospitals | Hospital ED (hospital EDs:
[1] adult, [2] paediatric); Germany (12 October‐24 November 2020) |
Symptomatic Cohort [1] only Respiratory symptoms: 157 (579%); loss of smell or taste: 18 (6.6%); contact with confirmed COVID‐19 case: 33 (12.2%); radiological signs of viral pneumonia: 11 (0.4%); other symptoms: 140 (51.7%); none: 56 (20.7%) Mean age 59.7 (SD 18; range 21‐98), 160 (59%) male |
PCR (multiple assays) Target: TibMolbiol was E gene NP+OP; paired; same as index Timing: as for index Interval: paired |
10 patients excluded from cohort [1] based on reasons below: no index test result (n = 6), no PCR result (n = 2) Index: 1 inconclusive (excluded) Reference: 1 inconclusive (excluded) |
|
Mockel 2021(b) Accepted manuscript 202(25) |
Single‐group (prospective); participants were symptomatic patients attending hospital EDs ([1] 4 adult EDs (n = 271) and [2] 1 paediatric ED (n = 202)) Cohort [1] have been included as Mockel 2021(a) In both cohorts patients were either symptomatic (acute respiratory symptoms or loss of smell or taste), contacts of confirmed cases up to 14 d before onset of COVID‐19 symptoms, or had clinical or radiological signs of viral pneumonia in the context of an outbreak in nursing homes or hospitals | Hospital ED (hospital EDs:
[1] adult, [2] paediatric) Germany (12 October‐24 November 2020) |
Symptomatic Respiratory symptoms: [2] 120 (59.4%); loss of smell or taste: [2] 1 (0.5%); contact with confirmed COVID‐19 case: [2] 37 (18.3%); radiological signs of viral pneumonia: [2] 10 (0.5%); other symptoms:[2] 104 (51.5%); none: [2] 26 (12.9%) Mean age 3 (range 1‐9), 111 (55%) male |
PCR (multiple assays) Target: TibMolbiol was E gene NP+OP; paired; same as index Timing: as for index Interval: paired |
None (exclusions all from cohort [1] None Index: none reported Reference: none reported |
|
Nagura‐Ikeda 2020 Accepted manuscript 103 (103) |
Single‐group (cases) (not reported; samples appear to be collected prospectively but states that patient information was retrospectively collected from the hospital electronic medical records) Patients with laboratory‐confirmed COVID‐19 referred for isolation and treatment, including symptomatic and asymptomatic |
Mixed (inpatient and asymptomatic (admitted or quarantined)) Japan (11 February‐13 May 2020) |
Mainly symptomatic 88 (85%) symptomatic, including 16 (15%) severe (showing clinical symptoms of pneumonia ‐ dyspnoea, tachypnoea, saturation of percutaneous oxygen [SpO2] < 93%, and the need for oxygen therapy); 15 (15%) asymptomatic IPD provided Median age 46 (range 18‐87); 66 (64%) male |
PCR (no details) Target: not reported NP or OP; paired Timing: on presentation or as part of mass screening; specific timing to symptom onset was not reported for the original PCR Interval: paired |
Not stated None reported Index: none reported Reference: none reported |
| Nalumansi 2020 | 2‐group (prospective); 1) COVID‐19 cases (PCR+ve) identified from regional referral hospitals (n = 90); discussion describes 89% as asymptomatic 2) PCR‐ve controls were volunteers at a Military Barracks and the Uganda Virus Research Institute clinic (n = 172) |
Unclear (unclear; referral hospitals (also described as COVID‐19 treatment facilities)) Uganda (not reported) |
Mainly asymptomatic: asymptomatic 77/90 (89%) PCR+ve (described in Discussion only); 172 PCR‐ve Mean age 37 (95% CI 35–39) years; 85 (94%) male |
PCR (single assay) Target: not reported Nasal; paired; same as index Timing: as for index test Interval: paired |
None reported Index: none reported Reference: none reported |
|
Nash 2020 pre‐print 190 (100) |
Unclear; 2‐group (retrospective); samples from suspected patients submitted to 'PATH' (ww.path.org) for routine COVID diagnosis (2nd cohort of samples also tested using Spike‐based assay; excluded as assay requires use of centrifuge) |
Laboratory‐based (samples provided to study authors by PATH (non‐profit organization), protocol number 00004244) Not reported (not reported) |
Not reported Not reported Not reported |
PCR (single assay) Target: N, S, and ORF1ab genes Nasal (not otherwise specified); same sample used Timing: not stated Interval: simultaneous |
None reported Index: none reported Reference: none reported |
|
Ngo Nsoga 2021 Preprint 402 (168) |
Single‐group (prospective); patients attending a single COVID‐19 screening centre either with symptoms compatible with COVID‐19 infection or asymptomatic contacts (The results of a separate pilot study are reported in a supplementary appendix however details are limited) | COVID‐19 test centre (outpatient COVID‐19 screening site) Switzerland (3‐12 November 2020) |
Mainly symptomatic: states that the "majority" were symptomatic, and that all 168 PCR+ve were symptomatic. Appears that symptom breakdown was only provided for these 168. Symptoms included: asthenia 101 (60.1%); headache 99 (48.9%); myalgia 81 (48.2%); chills/fever 80 (47.6%); dry/productive cough 73 (43.5%); anosmia/ageusia 71 (42.3%); odynophagia 68 (40.5%); digestive signs 38 (22.6%); dyspnoea 7 (4.2%); chest pain 4 (2.4%); other 12 (7.1%) Mean age 39.9 (SD 14.5), median age 38 (range 16‐80), 178 (44.3%) male |
PCR (single assay) Target: ORF1 and E gene NP; paired; Timing: as for index test; some asymptomatic cases and no information on these timings Interval: paired |
None reported Index: none reported Reference: 2 samples were positive for ORF1 only and not E gene ‐ interpreted as positive |
|
Nikolai 2021(a) [A] Preprint (not peer reviewed) 132 (36) |
Single‐group (prospective); 2 single‐group studies [1] symptomatic adults with high clinical suspicion of COVID‐19 presenting at an ambulatory SARS‐CoV‐2 testing facility (compared professionally collected AN and NMT samples) (n = 132) [2] symptomatic adults with high clinical suspicion of COVID‐19 presenting at an ambulatory SARS‐CoV‐2 testing facility (compared self‐collected NMT sample and professional NP swab) (n = 96); see Nikolai 2021(b) [A] for further details |
COVID‐19 test centre (ambulatory SARS‐CoV‐2 testing facility) Germany (30 November 2020‐18 January 2021) |
Symptomatic Whole sample (n = 228): 222, 97.4% of participants had ≥ 1 symptoms consistent with SARS‐CoV‐2 infection Average age: 34.6 (SD 11.7) years 107 (47%) female |
PCR (no details) Target: not reported NP+OP; paired Timing: same as for index test Interval: paired |
Yes; 2 exclusions from whole sample of 230 enrolled Not reported Index: not reported Reference: 2 invalid PCR results |
|
Nikolai 2021(b) [A] Preprint (not peer reviewed) 96 (34) |
Single‐group (prospective); 2 single‐group studies [1] symptomatic adults with high clinical suspicion of COVID‐19 presenting at an ambulatory SARS‐CoV‐2 testing facility (compared professionally collected AN and NMT samples) (n = 132); see Nikolai 2021(a) [A] for further details [2] symptomatic adults with high clinical suspicion of COVID‐19 presenting at an ambulatory SARS‐CoV‐2 testing facility (compared self‐collected NMT sample and professional NP swab) (n = 96) |
COVID‐19 test centre (ambulatory SARS‐CoV‐2 testing facility) Germany (30 November 2020‐18 January 2021) |
Symptomatic Whole sample (n = 228): 222, 97.4% of participants had ≥ 1 symptoms consistent with SARS‐CoV‐2 infection Average age 34.6 (SD 11.7) years, 107 (47%) female |
PCR (no details) Target: not reported NP+OP; paired Timing: same as for index test Interval: paired |
Yes; 2 exclusions from whole sample of 230 enrolled Not reported Index: not reported Reference: 2 invalid PCR results |
|
Okoye 2021 Academic journal 2645 (46) |
Single‐group (prospective); asymptomatic college‐age (undergraduate and graduate) students; not experiencing signs or symptoms of COVID‐19 at the time of testing | Student screening (temporary indoor testing site); USA (13‐20 November 2020) |
Asymptomatic asymptomatic Average age 24 years (range 15‐86); 52% female |
PCR (single assay) Target: ORF 1ab, S, N NMT; paired; same as index Timing: not applicable (asymptomatic) Interval: paired |
Yes; 7 excluded 3 invalid BinaxNOW results; all negative on retesting with new nasal swab specimen Index: none Reference: 4 inconclusive on PCR; only N gene detected (Ct > 30) |
| Olearo 2021 [A] | 2‐group (retrospective (described as prospective, but samples included based on PCR status)); 1] PCR+ve (n = 84), included until target number met 2] PCR‐ve (n = 100), randomly selected to serve as negative control Swabs collected following routine diagnostics from patients hospitalized with suspected or known COVID‐19 |
Hospital inpatient (hospital inpatient) Germany (August‐November 2020) |
Symptomatic Median duration pso 6 (IQR 2–12) d Not reported |
PCR (single assay) Target: not reported NP or OP; same sample used Timing: median 6 (IQR 2–12) d pso Interval: paired |
None reported Index: none reported Reference: none reported |
|
Osterman 2021(a) [A] Published Overall (site 1 and 2): 833 (447); test [A] FIA 741 (381); test [B] RAT 831 (445) |
≥ 2 groups (unclear; includes frozen samples so not sure we can assume prospective here) Multi‐site, multiple‐group study, including: (1) site 1: symptomatic patients (adults and children) presenting at EDs or on clinical units (n = 741 swabs, including 381 PCR+ve and 360 PCR‐ve); of 381 PCR+ve, 189 were classed as primary diagnosis (no previous PCR+ve) and 192 swabs were undertaken at “follow‐up” during hospitalization, i.e. at variable time points pso or 1st PCR+ve result Site 2 extracted as Osterman 2021(b) (2) site 2: symptomatic and asymptomatic participants at patient care units or from employee screening, all PCR+ve (n = 66) |
Mixed (mixed:
(1) hospital inpatient and ED
(2) hospital inpatient and employee test centre) Germany ((1) 4 March‐19 October 2020 (2) 13 November‐8 December 2020) |
Symptomatic (1) all symptomatic (2) symptomatic and asymptomatic (1) + (2) 256/445 (58%) PCR+ve primary diagnosis, and 189/445 (42%) follow‐up testing Only reported for PCR‐ve at site 1: 326/386 (84%) adults; 60/386 (16%) children |
PCR (multiple assays) Target: site 1: N1 or envelope; site 2: N and RdRp gene NP or OP; same sample used Timing: not mentioned Interval: simultaneous |
Number PCR+ve across the 2 sites sums to 447 but maximum number PCR+ve reported was 445 (test [B]); only 381 reported for RAT test (site 1) None mentioned Index: none mentioned Reference: none mentioned |
|
Osterman 2021(b) Published Overall (site 1 and 2): 833 (447); test [A] FIA 741 (381); test [B] RAT 831 (445) |
≥ 2 groups (unclear; includes frozen samples so not sure we can assume prospective here) Multi‐site, multiple‐group study, including: (2) site 2: symptomatic and asymptomatic participants at patient care units or from employee screening, all PCR+ve (n = 66) Site 1 extracted as Osterman 2021(a) [A] |
Mixed (mixed:
(1) hospital inpatient and ED
(2) hospital inpatient and employee test centre) Germany ((1) 4 March‐19 October 2020 (2) 13 November‐8 December 2020) |
Mixed (1) all symptomatic; (2) symptomatic and asymptomatic (1) + (2) 256/445 (58%) PCR+ve primary diagnosis, and 189/445 (42%) follow‐up testing Only reported for PCR‐ve at site 1: 326/386 (84%) adults; 60/386 (16%) children |
PCR (multiple assays) Target: site 1: N1 or envelope; site 2: N and RdRp gene NP; same sample used Timing: not mentioned Interval: simultaneous |
Number PCR+ve across the 2 sites sums to 447 but maximum number PCR+ve reported was 445 (test [B]); only 381 reported for RAT test (site 1) None reported Index: none reported Reference: none reported |
|
Parada‐Ricart 2020 Published 193; final PCR diagnosis available for 172 (26 cases) |
Single‐group (prospective); participants were patients with respiratory symptoms of < 7 d (128) and asymptomatic patients (44) | Unclear (no details) Spain (6‐17 April 2020) |
Mainly symptomatic 128 with respiratory symptoms; no further details Not mentioned |
PCR (no details) Target: not mentioned Nasal; paired; same as index Timing: < 7 d pso Interval: paired |
PCR result not available for 21 patients so not included in the analysis not mentioned Index: not mentioned Reference: not mentioned Of 21 FPs, 13 remained after consideration of clinical history (9 asymptomatic and 4 symptomatic with negative subsequent serology); the remaining 8 were reclassified as TP (clinical‐epidemiological picture compatible with COVID‐19) |
|
Pena 2021 Preprint 854 included; 842 analysed (73) |
Single‐group (unclear); asymptomatic individuals at 7 testing sites, including workers (n = 56), "sanitary residence" [presumed to be health‐related residential care] (n = 239), and the general public (n = 547); community prevalence of COVID‐19 was 11% | Screening (states "seven testing sites"; community screening) Chile (14‐17 January 2021) |
Asymptomatic All asymptomatic (100%) Mean age 36.67 (SD 16.48) years; 351 (42%) female |
PCR (single assay) Target: N and S gene NP; paired; same as index Timing: as for index test Interval: paired |
Yes; 12 (1.4%) were excluded for lacking real‐time PCR results None reported Index: none reported Reference: none reported |
|
Pena‐Rodriguez 2021 Published 369 (104) |
Single‐group (prospective); adults with symptoms suggestive of COVID‐19 (headache, fever, fatigue, other respiratory signs, or gastrointestinal symptoms) and individuals in contact with confirmed cases of COVID‐19 (by PCR) in the previous 3‐5 d, with or without symptoms attending for COVID‐19 testing | COVID‐19 test centre (COVID‐19 test centre (diagnostic laboratory centre)) Mexico (October‐November 2020) |
Mixed Symptoms included: headache (42%), fever (25%), cough (23%), myalgia (21%), loss of smell (18%), fatigue (16%), diarrhoea (10%), shortness of breath (7%), and arthralgia (4%) Average age 36.6 (SD 13.16) years; 215 (58%) female |
PCR (single assay) Target: N gene and Rnase P gene (RP) NP+OP; paired; same as index Timing: same as for index test Interval: paired |
None reported;
states "The test was invalidated when no marks were detected" Index: none reported Reference: none reported |
|
Perez‐Garcia 2021 Published 320 (170) |
≥ 2 groups (retrospective) Samples from patients with suspicion of COVID‐19 attending the university hospital or associated PHC centres 1] PCR‐ve patients (n = 150) 2] PCR+ve patients (n = 170) | Laboratory‐based (samples from mixed settings including PHC centres (50%), hospital inpatients (20%), ED (21%) and occupational health (9%)) Spain (8‐20 October 2020) |
Mainly symptomatic 134 (79%) symptomatic; including cough (54%), fever (41%), dyspnoea (25%), anosmia (22%) and myalgia (19%); 26 (15%) asymptomatic with a prior contact with COVID‐19 case. (10 with no data on symptoms; time pso not reported for 6/134 symptomatic) Median age 51 (IQR: 38–68) years; 81 (48%) female |
PCR (multiple assays) Target: 1] ORF1ab and N genes 2] E, RdRP, S and N 3] E, RdRP and N NP; same sample used Timing: as for index test Interval: simultaneous |
None reported Index: none reported Reference: none reported |
|
Peto 2021(a) [A] Published 1118 (178) |
≥ 2 groups (retrospective); set of studies conducted by PHE and University of Oxford. This extraction relates to a 2‐group study (phase 3a): [1] residual swabs from PCR+ve patients in Oxford (collected March 2020) [2] residual swabs from PCR‐ve patients in Oxford (collected March 2020) Swabs were frozen following routine testing and sent to Porton Down |
Appears to be hospital inpatient; obtained from a secondary healthcare setting UK (March 2020 (PCR+ve)) |
Symptomatic Not stated Not stated |
PCR (single assay) Target: ORF and S target assays. NP+OP; same sample used Timing: as for index test Interval: simultaneous |
Initial sample 1181 plus 1 void PCR Failure rates reported for test [A] only: [1] 13/191, 7% [2] 50/990, 5.1% Index: unclear Reference: unclear |
|
Peto 2021(b) [non‐HCW tested] Published 1946 (372) |
Single‐group (not stated); individuals presenting at a regional COVID‐19 testing centre (phase 4) |
COVID‐19 test centre (run by commercial pharmacy company) UK (Not stated) |
Not reported; presumably symptomatic and meeting testing criteria Not stated Not stated |
PCR (appears to be Roche assay) Target: not stated Sample site nasal+OP; paired; same as index Timing: as for index test Interval: paired |
Initial sample of 1946 reported, with 27 failures (1919 remaining). Data reported for 372 cases (text) and 131 non‐COVID cases (Table 2); total 1686. Unclear reason for discrepancy Failure rate: 27/1946, 1.4% Index: unclear Reference: unclear |
|
Peto 2021(c) [A ‐ HCW tested] Published 479 (479); This extraction for HCW‐tested samples: 267 (only assay A) |
Single‐group (cases) (not stated); individuals presenting at 1 of 14 regional COVID‐19 NHS Test and Trace centres as part of the FALCON phase 3b study. People who had a positive PCR result were asked to return for an Ag test within 5 d of the original PCR result | COVID‐19 test centre (NHS test and trace centres; conducted within the FALCON‐C19 study); UK (September 17‐October 23, 2020) |
Mainly symptomatic: not stated; presumed symptomatic Only reported for combined sample and only for 421 of 479 participants (unclear whether recorded on original PCR or on return for Ag testing): 381 (90%) symptomatic; 138 (36%) headache, 134 (35%) cough, 82 (22%) sore throat, 80 (21%) fever, 260 (68%) 'other' not specified symptoms, 59 with no data. 40 (10%) reported asymptomatic Median age 33 years (91 with no data); 168/337 male, 50% (84 with no data recorded) |
PCR (single assay) Target: ORF‐1 and E‐gene AN; paired; same as index Timing: as for index test Interval: paired |
Failure rates reported for assay [A] only as:
HCW tested 27/267 10.1%
NB preliminary report reported these as 28/296 Index: unclear Reference: unclear |
|
Peto 2021(c) [A ‐ Lab tested] Published 479 (479); This extraction for Lab scientist tested: 212 (multiple assays evaluated) |
Single‐group (cases) (not stated); ‐ individuals presenting at one of 14 regional COVID‐19 NHS test and trace centres as part of the FALCON phase 3b study. People who had a positive PCR result were asked to return for an Ag test within 5 d of the original PCR result |
COVID‐19 test centre (NHS Test and Trace centres; conducted within the FALCON‐C19 study) UK (17 September‐23 October 2020) |
Mainly symptomatic Not stated; presumed symptomatic Only reported for combined sample and only for 421 of 479 participants (unclear whether recorded on original PCR or on return for Ag testing) 381 (90%) symptomatic; 138 (36%) headache, 134 (35%) cough, 82 (22%) sore throat, 80 (21%) fever, 260 (68%) 'other' not specified symptoms, 59 with no data. 40 (10%) reported asymptomatic Median age 33 years (91 with no data); 168/337 male, 50% (84 with no data recorded) |
PCR (single assay) Target: ORF‐1 and E‐gene AN; paired; same as index Timing: as for index test Interval: paired |
Failure rates reported for assay [A] only as:
lab scientist‐tested 9/212 4.2%
NB preliminary report reported these as 9/221 Index: unclear Reference: unclear |
|
Peto 2021(d) Published 538 (0) |
Single‐group (not stated); PHE and hospital staff volunteering for testing (specificity only) |
HCW/staff screening at 2 sites (PHE and John Radcliffe Hospital, Oxford) UK (Not stated) |
Asymptomatic Not stated Not stated |
PCR (no details) Target: not stated OP; same sample used Timing: as for index test Interval: simultaneous |
Initial sample of 570 reported, 36 failed, leaving 534 for inclusion. Data for 538 included Failure rate reported as 17/358, 4.7% Index: unclear Reference: unclear |
|
PHE 2020 Partially published (some data from study author contact) 152 (46) |
Single‐group (retrospective); samples obtained during a COVID‐19 outbreak at a Navy barracks |
Contacts (outbreak) UK (Not stated) |
Not reported Not stated Not stated |
PCR (no details) Target: not stated OP; same sample used Timing: as for index test Interval: simultaneous |
None reported Failure rate reported as 6/157, 3.8% NB resulting number samples (n = 151) does not match with final number reported (n = 152) |
|
Pickering 2021(a) [A] Preprint 200 (100) |
≥ 2 groups (retrospectively) All swabs selected from those submitted to the diagnostic laboratory [1] PCR+ve swabs selected to cover a wide range of Ct values (14‐39) (n = 100) [2] PCR‐ve swabs (n = 100) [1] and [2] used for comparison of 6 RDTs; included as Pickering 2021(a) [A]; see also Pickering 2021(b) [A] and Pickering 2021(c) [A] | Unclear (swabs submitted to the diagnostic laboratory for routine testing) UK ([1]‐[3] March‐October 2020 [4] January 2021) |
Not reported Not stated Not stated |
PCR (single assay) Target: N gene or human RNAse P Nasal+OP; same sample used Timing: not stated Interval: simultaneous |
None reported Index: none reported Reference: none reported |
|
Pickering 2021(b) [A] Preprint 141 (141) |
Multi‐group study (retrospectively); all swabs selected from those submitted to the diagnostic laboratory [3] PCR+ve swabs with culture results for assessment of infectivity (3 RDTs compared) (n = 141); included as Pickering 2021(b) [A]; see also Pickering 2021(a) [A] and Pickering 2021(c) [A] | Unclear (swabs submitted to the diagnostic laboratory for routine testing) UK ([1]‐[3] March‐October 2020 [4] January 2021) |
Not reported Not stated Not stated |
PCR (single assay) Target: N gene or human RNAse P Nasal+OP; same sample used Timing: not stated Interval: simultaneous |
Yes; insufficient sample volume to conduct all 3 tests on all 141 samples ‐ 31 missing for Innova and 51 missing for Encode None reported Index: none reported Reference: none reported |
|
Pickering 2021(c) [A] Preprint 23 (23) |
Multi‐group study All swabs selected from those submitted to the diagnostic laboratory [4] PCR+ve swabs infected from the B.1.1.7 variant (2 RDTs compared) (n = 23); included as Pickering 2021(c) [A]see also Pickering 2021(b) [A] and Pickering 2021(a) [A] | Unclear (swabs submitted to the diagnostic laboratory for routine testing) UK ([4] January 2021 [1]‐[3] March‐October 2020) |
Not reported Not stated Not stated |
PCR (single assay) Target: N gene or human RNAse P Nasal+OP; same sample used Timing: not stated Interval: simultaneous |
Yes; insufficient sample volume to conduct all 3 tests on all 141 samples ‐ 31 missing for Innova and 51 missing for Encode None reported Index: none reported Reference: none reported |
|
Pilarowski 2020a Published 3302 (237) |
Single‐group (prospective); testing freely available to people of all ages, with or without symptoms. Community workers conducted door‐to‐door mobilization in 3 census tracts surrounding the testing site 4 d before testing. | COVID‐19 test centres; community testing site (plaza at an urban commercial transport hub in the Mission neighbourhood, San Francisco (University of California, San Francisco)); (22 November‐1 December 2020 |
Mixed 30.9% (n = 1020) self‐reported possible COVID‐19 symptoms; results reported for 341 (10%) symptomatic (≤ 7 d pso) and 2402 (90%) asymptomatic or symptomatic (> 7 d pso) Of 237 PCR+ve, 95 were asymptomatic, 7 were symptomatic (> 7 d pso), and 135 symptomatic (≤ 7 d pso) 1750 (53%) male; 99 (3%) aged < 13 years, 110 (3%) aged 13‐18 years, and 3093 (94%) aged > 18 years 2166 (66%) Latinx, 304 (9%) Asian, 558 (17%) white, 53 (2%) American Indian, and 83 (3%) black |
PCR (single assay) Target: (1) and (2) N‐gene Nasal (AN) in VTM; paired swab (same site as index) Timing: same as for index test Interval: yes; but only PCR+ve on first assay had confirmation on 2nd assay Interval: paired |
None reported None reported Index: none reported Reference: none reported |
| Pilarowski 2021 | Single‐group (prospective); mainly asymptomatic suspected patients presenting at a walk‐up, free testing at a plaza located at an intersection of the Bay Area‐wide subway system (BART) and the San Francisco city bus/streetcar system (MUNI), Mission District, California | COVID‐19 test centre (community screening/COVID‐19 test centre) USA (September 2020) |
Mainly asymptomatic Mainly asymptomatic (84% reported no symptoms during the 14 d before testing); 54% male; 77% 18‐50 years of age; 81% self‐identified as Latinx |
PCR (no details) Target: not reported AN; paired; same as index Timing: same as for index Interval: paired |
Unclear; 871/ 878 in the analysis None reported Index: none reported Reference: none reported |
|
Pollock 2021a Published 2482 included; 2308 analysed (292) |
Single‐group (prospective); symptomatic and asymptomatic adults and children who attended the drive‐through, free community testing site in Massachusetts | COVID‐19 test centre (COVID‐19 drive‐through testing site; screening (appears to be open to all; no specific testing criteria applied)) USA (26 October‐2 December 2020) |
Mixed Adults: 406 (29%) symptomatic; 974 (71%) asymptomatic Children: 99 (11%) symptomatic; 829 (89%) asymptomatic Adults: median 3 (IQR 2‐5) pso d Children: median 2 (IQR 1‐4) pso d Adults: 59% symptomatic female; 56% asymptomatic female Age: 19‐29 years 332, 24%; 30‐49 years 581, 42%; 50‐69 years 401, 29%, > 70 years 66, 5% Children: 62% symptomatic; 52% asymptomatic female Age: < 7 years 261, 28%; 7‐13 years 381, 41%; 14‐18 years 286, 31% |
PCR (single assay) Target: N2 gene AN; paired; same as index Timing: as for index test Interval: paired |
Missing data, n = 54; excluded, n = 94 (samples tested at < 59 °F) Inconclusive PCR results n = 26; 1 invalid BinaxNOW result (a manufacturing issue whereby plastic covered the test strip, preventing the buffer from making contact with the test strip); presume test was repeated. Index: none reported; all FP results had faint but detectable test bands Reference: inconclusive PCR results 189 (n = 26) |
|
Pollock 2021b Preprint 1498 (234); from 1603 eligible |
Single‐group (prospective); individuals presenting for testing to a high‐throughput, drive‐through, free community testing site; no specific criteria for testing had to be met | COVID‐19 test centre (drive through testing site; screening) USA (11‐22 January 2021) |
Mainly asymptomatic 1257 (84%) asymptomatic, including 1036/1245 (69%) adults; 209 symptomatic 221/253 (15%) children; 32 symptomatic; adult: 57% symptomatic female; 53% asymptomatic female Children: 56% symptomatic female; 53% asymptomatic female; Age group: < 7 years: 13 (41%) symptomatic, 60 (27%) asymptomatic 7‐13 years: 12 (37) symptomatic, 73 (33%) asymptomatic 14‐18 years: 7 (22%) symptomatic, 88 (40%) asymptomatic 19‐29 years: 58 (28%) symptomatic, 313 (30%) asymptomatic 30‐49 years: 102 (49%) symptomatic, 381 (37%) asymptomatic 50‐69 years: 42 (20%) symptomatic, 290 (28%) asymptomatic > 70 years: 7 (3%) symptomatic, 52 (5%) asymptomatic |
PCR (single assay) Target: N2 gene AN; paired; same as index Timing: same as for index Interval: paired |
105/1603 (6.5%) (invalid or missing PCR results (n = 48) and missing clinical data (n = 57)) 8 discordant results (all faint positive vs negative); 2 readers disagreed on the strength of the positive band (faint vs 219 medium vs strong) in 7 cases Index: none; states "No invalid CareStart test results were observed" Reference: 48 invalid or missing PCR results |
|
Porte 2020 Preprint (not peer reviewed) 127 samples; 82 PCR+ve |
Single‐group (retrospective) Samples from cohort of suspected COVID‐19 cases (n = 1453); patients with respiratory symptoms and/or fever and an epidemiological risk factor for SARS‐CoV‐2 infection (travel or contact with case) |
Hospital ED (private hospital Emergency Room); Chile (16‐21 March 2020) |
Symptomatic: cough 94 (74.6%); fever 77 (61.1%)
Median duration of symptoms of 2 d (IQR 1–4) (range 0‐12)
Duration of symptoms: day 0‐3: 91 (72.2%); day 4‐7: 27 (22.4%); day ≥ 8: 8 (6.3%) Median age 38 years (IQR 29.5–44) (range 1–91), 68 (53.5%) male |
PCR (single assay);
Threshold ≤ 40 Ct Target: not stated NOP; same sample used Timing: median 2 d pso (IQR 1‐4, range 0‐12) Interval: simultaneous |
No Not reported Index: not reported Reference: patients |
|
Porte 2021 [A] Published 64 (32) |
Multi‐group (retrospective); (1) COVID‐19 patients presenting within 5 d pso (n = 32) (2) symptomatic patients with negative PCR (n = 20) (3) asymptomatic patients screened prior to surgery (n = 12) (27 PCR+ve and 19 PCR‐ve samples were used in Weitzel 2020 [A] (different assays)) |
COVID‐19 test centre (private clinic) Chile (Not stated) |
Symptomatic Not reported; 12 asymptomatic Median age 39 (IQR 36.7‐57) years; 33, 52% male |
PCR (single assay);
Threshold ≤ 40 Ct Target: not stated NP+OP; same sample used Timing: not stated Interval: simultaneous |
None reported Index: none reported Reference: none reported |
|
Pray 2021 Published 1098 (57) Symptomatic 227 (40) Asymptomatic 871 (17) (1105 paired nasal samples taken; 7 inconclusive Ag or PCR results so excluded from analysis) |
Single‐group (prospective); symptomatic and asymptomatic participants at 2 universities in Wisconsin (students, staff or other) (n = 1105); at university A, all people tested were eligible (n = 1098), at university B, only students who were quarantined after exposure to people with COVID‐19 could participate (n = 47) | Student screening (COVID‐19 test centre/asymptomatic screening
On‐site testing: 2 Wisconsin university campuses during university‐based testing programmes USA (28 September‐9 October) |
Mainly asymptomatic Symptomatic 227 (21%) Asymptomatic 871 (79%) (including 53 with ≥ 1 symptoms in previous 14 d) Symptoms included: nasal congestion 114 (50.2%), sore throat 97 (42.7%), headache 87 (38.3%), cough 70 (30.8%), fatigue 60 (26.4%), muscle aches 43 (18.9%), shortness of breath 24 (10.6%) Age group 15‐24 yeas: 971 (88.4%) Age group ≥ 25 years: 127 (11.6%); 453 (41.3%) male Non‐Hispanic white: 917 (83.5%) |
PCR (multiple assays) Target: N1 and N2 viral nucleocapsid protein gene regions Nasal; paired; same as index Timing: same as for index test (paired swabs); analysed within 24–72 h of collection Interval: paired |
7/1105 inconclusive Ag or PCR results excluded from analysis; no details provided Reasons for test 'failure' not reported Index: none reported Reference: none reported |
|
Prince‐Guerra 2021 Published 3419 (299) |
Single‐group (prospective); any participants who attended 2 sites of Pima County Health Department community‐based SARS‐CoV‐2 testing sites; open to anyone who wanted testing | COVID‐19 test centre (community COVID testing sites) USA (3‐17 November 2020) |
Mainly asymptomatic 827 (24%) symptomatic at the time of testing (≥ 1 COVID symptom); 2592 (76%) were asymptomatic Median age 41 (range 10‐95) years; 236 (7%) aged 10–17 years, 1885 (55%) aged 18–49 years, 743 (22%) aged 50–64 years, and 555 (16%) aged ≥ 65 years; 1681 (49%) female; 2567 (75%) white; 1075 (31%) Hispanic/Latino |
PCR (multiple assays) Target: not stated NP; paired Timing: as for index test Interval: paired |
None reported Index: none reported Reference: none reported |
|
Ristic 2021 Published paper 120 (43) |
Single‐group (prospective); symptomatic patients with ≥ 1 COVID‐related symptoms presenting via a triage ambulance of a primary and tertiary outpatients' healthcare facility, including a primary care "COVID ambulance" and a "red zone" ambulance | Hospital outpatient (mixed; primary and tertiary outpatients) Serbia (21 August‐1 September 2020) |
Symptomatic All symptomatic (120); 103 (86%) with fever, followed by malaise (77, 64%), cough (56, 47%), sore throat (53, 44%), myalgia (46, 38%) Median age 49 (IQR 36–70) years (range 14‐91), 57 female, 63 male |
PCR (multiple assays) Target: 1) R‐gene 2) RdRP in the ORF1ab region, E gene, and N gene 3) RdRP NP; paired; same as index Timing: same as index Interval: paired |
Not stated Not stated Index: not stated Reference: not stated |
|
Rottenstreich 2021 Published research letter 1326 (9) |
Single‐group (prospective); asymptomatic women admitted for delivery | Hospital inpatient (hospital inpatient) Israel (21 October‐28 December 2020) |
Asymptomatic Asymptomatic None stated; all female |
PCR (single assay) Target: not reported Sample site not reported; paired; same as index Timing: same as index Interval: paired |
None reported Index: none reported Reference: none reported |
|
Saeed 2021 [A] Published 100 (100) |
Single‐group (cases) (retrospective); (sensitivity only): PCR+ve samples from suspected cases of COVID‐19 (respiratory symptom and/or fever and international travel history or close contact with COVID‐19‐confirmed patients); it is not clear but seems that both NP and saliva had to be PCR+ve | COVID‐19 test centre (COVID‐19 diagnostic centre) Pakistan (3‐10 October 2020) |
Symptomatic All symptomatic (respiratory symptoms and/or fever) Mean age 47 (range 6–91) years; 34 (34%) female; 4 (4%) children |
PCR (single assay) Target: Biorad: E gene, N gene, and RNA polymerase gene NP; same sample used (NP alone) Timing: no details Interval: simultaneous |
None reported; the samples with discordant results were repeated but no details given Index: none reported Reference: none reported |
|
Salvagno 2021 Published 321 (149) |
Single‐group (unclear; "investigation was based on pre‐existing specimens, already collected for routine SARS‐CoV‐2 diagnostic testing in the local facility") Consecutive patients referred to a hospital for SARS‐CoV‐2 diagnostic testing | Unclear (unclear; possibly inpatient) Italy (16‐30 November 2020) |
Symptomatic Not reported; presume symptomatic (patients "referred for SARS‐CoV‐2 diagnostic testing to the Pederzoli Hospital") Mean age 46 (IQR 32‐56) years; 181 (56%) female |
PCR (single assay) Target: N, E and RdRP NP; same sample used Timing: not stated Interval: simultaneous |
None reported Index: none reported Reference: none reported |
|
Schildgen 2021 [A] preprint 73 (42) |
2‐group (not stated; presume retrospective); [1] PCR+ve BAL or throat wash samples (n = 42) [2] PCR‐ve samples (n = 31) Described as pilot sample panel |
Unclear (not stated) Germany (Not stated) |
Mixed Not stated for BAL samples, throat wash from 23 symptomatic and 27 asymptomatic people Not stated |
PCR (single assay) Target: not stated BAL or throat wash; same sample used Timing: not stated Interval: simultaneous |
8 PCR‐invalid samples also tested; 2/8 invalid in one AG assay each, 3/8 negative in all 3 Ag assays None reported Index: none reported Reference: none reported |
|
Schuit 2021(a) Preprint 2692 (233) 2692/3237 agreed to participate |
Single‐group (prospective) Report of 2 single‐group studies using 2 different assays: (1) West Brabant testing sites ‐ close contacts (aged ≥ 16 years old) of confirmed COVID‐19 cases presenting at testing sites for a 5th‐day test (as recommended by Dutch public health service test‐and‐trace program, and/or the Dutch contact tracing mobile phone application (the ‘CoronaMelder’ app) and/or an individual with a confirmed SARS‐CoV‐2 infection); all asymptomatic at the time of the test request. See also Schuit 2021(b) for Rotterdam sites |
COVID‐19 test centre (COVID‐19 testing centres) Netherlands (14 December 2020 and 6 February 2021) |
Mixed ‐ contacts All asymptomatic on test booking; 230 219 (8.6%) symptomatic 0‐3 d before test Symptoms included: common cold 167, 78 76%; cough 60, 27%; shortness of breath 25, 11%; fever 13, 6%; loss of taste or smell 6, 3%, muscle ache 18, 8%, other 16, 7% Mean age 45.9 (SD 17.6) years; 1304, 48.7% male |
PCR (single assay) Target: E, RdRp NP+OP; paired Timing: same as for index Interval: paired |
Yes; 14 excluded 10 with no PCR (n = 3) or PCR invalid (n = 7); all Ag‐positive Index: 3 inconclusive; all PCR‐ve 1 further result excluded but reason not clear from flow diagram Reference: 0 |
|
Schuit 2021(b) Preprint 1603 (132) 1603/1903 agreed to participate |
Single‐group (prospective) Report of 2 single‐group studies: (2) Rotterdam sites ‐ close contacts (aged ≥ 16 years old) of confirmed COVID‐19 cases presenting at testing sites for a 5th‐day test (as recommended by Dutch public health service test‐and‐trace program, and/or the Dutch contact tracing mobile phone application (the ‘CoronaMelder’ app) and/or an individual with a confirmed SARS‐CoV‐2 infection); all asymptomatic at the time of the test request. See also Schuit 2021(a) for West Brabant testing sites |
COVID‐19 test centre (COVID‐19 testing centres) Netherlands (14 December 2020 and 6 February 2021) |
Mixed ‐ contacts All asymptomatic on test booking; 158, 10.1% symptomatic 0‐3 d before test Symptoms included: common cold 123, 78%; cough 24, 15.2%; shortness of breath 12, 8%; fever 9, 6%; loss of taste or smell 5, 3%, muscle ache 5, 3%, other 15, 9.5% Mean age 40.7 (SD 16.4) years; 845, 52.7% male |
PCR (single assay) Target: E, RdRp NP+OP; paired Timing: same as for index Interval: paired |
Yes; 7 excluded 4 with no PCR; all Ag‐negative Index: 3 inconclusive excluded; all PCR‐ve Reference: 0 |
|
Schwob 2020(a) Preprint Overall: 949 (327 positive by NP PCR, 369 positive by saliva PCR). 2x2 data only available for NP PCR Sample size for those undergoing Standard Q assay: 333 (112) |
Single‐group (prospective); 3 assays (each tested on a separate cohort of individuals recruited using same inclusion criteria
Adults recruited from 3 outpatient clinics and meeting testing criteria for COVID‐19, either:
‐ with ≥ 1 major symptom compatible with COVID‐19 (cough, fever, sore throat, anosmia, or ageusia), or
‐ with ≥ 1 minor symptom and close contact with a confirmed cases of COVID‐19 See also Schwob 2020(b) and Schwob 2020(c) |
COVID‐19 test centre (outpatient testing clinic (described as "testing centres")) Switzerland (25 September‐4 November 2020) |
Symptomatic Whole sample All symptomatic: 911, 96% with ≥ 1 major symptom (41% fever, 64% cough, 62% sore throat, 32% anosmia/ageusia) and 4% at least 1 minor symptom (rhinitis, myalgia, headache, fatigue, nausea, vomiting, diarrhoea, abdominal pain, urticaria, vesicles) Median age: 31 (IQR 25‐42; range 18‐87) years; 51% male |
PCR (multiple assays) Target: E gene NP; paired; same as index Timing: pso (mean duration of symptoms at the time of swab collection/testing was 2.6 d (SD 2.3, range 0‐30)) Interval: paired |
Yes; 21 excluded due to lack of PCR and/or RDT result, no further details None reported Index: none reported Reference: none reported |
|
Schwob 2020(b) Preprint 949 (327 positive by NP PCR, 369 positive by saliva PCR). 2x2 data only available for NP PCR Sample size for those undergoing Panbio assay: 271 (122) |
Single‐group (prospective); 3 assays (each tested on a separate cohort of individuals recruited using same inclusion criteria
Adults recruited from 3 outpatient clinics and meeting testing criteria for COVID‐19, either:
‐ with ≥ 1 major symptom compatible with COVID‐19 (cough, fever, sore throat, anosmia, or ageusia), or
‐ with ≥ 1 minor symptom and close contact with a confirmed cases of COVID‐19 See also Schwob 2020(a) and Schwob 2020(c) |
COVID‐19 test centre (outpatient testing clinic (described as "testing centres")); Switzerland (25 September‐4 November 2020) |
Symptomatic Whole sample All symptomatic: 911, 96% with ≥ 1 major symptom (41% fever, 64% cough, 62% sore throat, 32% anosmia/ageusia) and 4% at least one minor symptom (rhinitis, myalgia, headache, fatigue, nausea, vomiting, diarrhoea, abdominal pain, urticaria, vesicles) Median age: 31 (IQR 25‐42; range 18‐87) years; 51% male |
PCR (multiple assays) Target: E gene NP; paired; same as index Timing: pso (mean duration of symptoms at the time of swab collection/testing was 2.6 d (SD 2.3, range 0‐30)) Interval: paired |
Yes; 21 excluded due to lack of PCR and/or RDT result, no further details.
(There appears to be a typo in Suppl Fig 1 which reports 122 PCR+ve samples tested with Panbio assay; 101 are shown as RDT+ and 17 RDT‐. The text reports assay sensitivity as 86.1% (95% CI 78.6 to 91.7%), which works out as 105 RDT+, 17 RDT‐ and the correct CIs) None reported Index: none reported Reference: none reported |
|
Schwob 2020(c) Preprint Overall: 949 (327 positive by NP PCR, 369 positive by saliva PCR). 2x2 data only available for NP PCR COVID‐VIRO assay: 324 (138) |
Single‐group (prospective); 3 assays (each tested on a separate cohort of individuals recruited using same inclusion criteria Adults recruited from 3 outpatient clinics and meeting testing criteria for COVID‐19, either: ‐ with ≥ 1 major symptom compatible with COVID‐19 (cough, fever, sore throat, anosmia, or ageusia), or ‐ with ≥ 1 minor symptom and close contact with a confirmed cases of COVID‐19. See also Schwob 2020(a) and Schwob 2020(b) | COVID‐19 test centre (outpatient testing clinic (described as "testing centres")) Switzerland (25 September‐4 November 2020) |
Symptomatic Whole sample All symptomatic: 911, 96% with ≥ 1 major symptom (41% fever, 64% cough, 62% sore throat, 32% anosmia/ageusia) and 4% at least 1 minor symptom (rhinitis, myalgia, headache, fatigue, nausea, vomiting, diarrhoea, abdominal pain, urticaria, vesicles) Median age: 31 (IQR 25‐42; range 18‐87) years; 51% male |
PCR (multiple assays) Target: E gene NP; paired; same as index Timing: pso (mean duration of symptoms at the time of swab collection/testing was 2.6 d (SD 2.3, range 0‐30)) Interval: paired |
Yes; 21 excluded due to lack of PCR and/or RDT result, no further details None reported Index: none reported Reference: none reported |
|
Scohy 2020 Published 148 (106) |
Single‐group (not stated) NP swabs submitted to laboratory at a large tertiary hospital |
Laboratory‐based (unclear) Belgium (6‐21 April 2020) |
Mixed 86 (58%) symptomatic, 45 (30%) asymptomatic, 17 (11%) symptom status not reported Cases only: viral load < 25 Ct 10 (9%), ≥ 25 Ct 96 (91%) Median age 57.5 (range 0‐94); 64 (43%) male |
PCR (single assay)
Threshold ≤ 40 Ct Target: RdRp Sample: same sample used (NP alone) Timing: not stated Interval: simultaneous |
None reported Index: none reported Reference: none reported |
|
Shidlovskaya 2021 [A] Preprint 106 (78) |
Single‐group (not stated, may be prospective); patients with suspected COVID‐19 admitted to the hospital on day 2‐10 pso (fever, dry cough, chest pain and discomfort, shortness of breath, loss of smell and taste). All included patients had CT signs of lung damage | Hospital inpatient (hospital inpatient) Russia (25 January‐8 February 2021) |
Symptomatic 100% symptomatic; symptoms included fever, dry cough, chest pain and discomfort, shortness of breath, loss of smell and taste; all had lung damage on CT Mean age 67.7 (range 28‐95) years; 53 (50%) female |
PCR (single assay) Target: NSP1 gene NP; paired; same as index Timing: 2‐10 d pso Interval: paired |
None reported Index: none reported Reference: none reported |
|
Shrestha 2020 Published 113 (47) |
Single‐group (not stated, appears prospective); participants who were close contacts of confirmed cases identified through contact tracing, residing in quarantine centre (n = 113) |
Contacts (contact tracing) Nepal (August‐September 2020) |
Asymptomatic All asymptomatic Range 13‐74 years; 89, 79% male |
PCR (no details) Target: not stated NP; paired; same as index Timing: as for index test Interval: paired |
None reported Index: tests were repeated for samples with indistinct outcomes Reference: 0 |
|
Stohr 2021 [A] Preprint 3215 (377); [A] 1604, [B] 1611 |
RCT (prospective) Randomized study Adults presenting for testing at a community COVID‐19 test centre; testing co‐ordinated by the Municipal Health Services (MHS). Participants randomized between 2 different Ag tests. |
COVID‐19 test centre (COVID‐19 test centre) Netherlands (23 December 2020‐17 January 2021) |
Mixed: current symptoms of COVID‐19 2226 (69.2%), symptoms in preceding 3 weeks 201 (6.3%), no current or prior symptoms 788 (24.5%). Definition of COVID‐19 symptoms was not provided Median age 41 (IQR 29‐54) years; 1409 (43.8%) male |
PCR (multiple assays) Target: (1) N‐gene and RdRP‐gene target (2) E‐gene NP+OP; paired Timing: as for index test (tested within 4 h of collection) Interval: paired |
Yes No PCR due to sample loss (n = 11) Index: "inconclusive" Ag assay results (n = 48; 9 PCR+ve and 39 PCR‐ve) were excluded by study authors for overall sensitivity and specificity; definition of inconclusive was not reported. Inconclusive results were included for determining the Ct‐value cut‐off at which the chance (P) of having a positive viral culture was P = 0.5, and were interpreted as not FN when determining the variables associated with a FN result Reference: inconclusive results on PCR (n = 3) were excluded by the authors; all Ag test negative |
|
Stokes 2021(a) [A] Published (1) 145 (138); 7 PCR‐ve at time of 2nd sampling were excluded by the review team |
Single‐group (cases) (prospective) Report of 2 eligible studies: (1) sensitivity only: symptomatic participants with a recent positive SARS‐CoV‐2 PCR were invited to contribute further samples for an RDT evaluation; only those who were still PCR+ve on paired swabs are included |
Unclear (unclear; likely community setting) Canada (Not stated) |
Symptomatic All symptomatic; cough (42.8%), headache (42.1%), myalgias (41.4%), sinus congestion (36.6%), malaise (31.0%), pharyngitis (29.0%), fevers/chills (28.3%), anosmia (24.1%), ageusia (24.1%), rhinorrhoea (20.0%), shortness of breath (5.5%), nausea/vomiting (3.4%), and other (17.9%, included chest pain, diarrhoea, eye soreness, lymphadenopathy, loss of appetite, arthralgia, dizziness, and/or conjunctivitis) Mean age 39.4 years, median age 36 (18.5‐86.6) years; 42.8% male |
PCR (multiple assays) Target: LDT ‐ E gene (< 35 Ct); Cobas ‐ not reported (2/2 targets positive, or ≥ 1 targets were positive in duplicate) [A] NP, [B] and [C] OP; paired; same as index Timing: as for index Interval: paired |
Yes 4 ‐ Panbio results were not recorded; 1 ‐ unable to be processed by PCR; 1 ‐ Panbio reported as negative before 15 min Index: 0 Reference: 0 |
|
Stokes 2021(b) Published (2) 1641 (268) |
Single‐group (prospective) Report of 2 eligible studies: (2) sensitivity and specificity: symptomatic individuals presenting to Alberta Health Services community COVID‐19 assessment centres within 7 d pso |
COVID‐19 test centre (community COVID‐19 test centre) Canada (Not stated) |
Symptomatic all symptomatic; no further details Mean age 40.8 years, median age 39 (range 5‐90) years; 40.0% male |
PCR (multiple assays) Target: not stated NP or OP; paired Timing: as for index Interval: paired |
None reported Index: none reported Reference: none reported |
|
Stromer 2020 Published 134 (124); subgroup of 21 PCR+ve samples used to compare 2 Ag tests not included |
Single‐group (cases) (retrospective); to estimate sensitivity only: ‐ upper respiratory tract (URT) samples (also described as "deep nasopharyngeal swabs") pre‐characterized by a positive or negative PCR result (Ag results not reported for PCR‐ve samples) | Unclear (unclear) Germany (Not stated) |
Not reported Not stated Not stated |
PCR (no details) Target: N gene NP; same sample used Timing: unclear Interval: simultaneous |
None reported Index: none reported Reference: none reported |
|
Takeda 2020 Preprint 162 (62) |
2 group (unclear) [1] PCRPCR‐confirmed COVID‐19 samples selected from a total of 88 positive samples during time period (n = 62) [2] Random sample of PCR‐ve samples selected from 1363 negative specimens tested during same time frame (n = 100) |
Laboratory‐based (multiple clinical institutions) Japan ("early April'' also later states 4‐day period) |
Not reported Not stated High viral load (< 25 Ct) ‐ 32/60, 53% Low viral load (≥ 25 Ct) ‐ 28/60, 47% Not stated |
PCR (single assay) Target: N2 NP; same sample used Timing: not stated Interval: simultaneous |
16 positive samples omitted; possibly because not initial samples but unclearly reported None reported Index: none reported Reference: none reported |
|
Takeuchi 2021a Preprint 1186 (105) included from a total of 2079 referred patients and HCWs |
Single‐group (prospective); participants referred to a drive‐through PCR testing centre from 1 of 3 groups: (1) primary care facilities (n = 1151 from 89 centres) (2) from a local public health centre (n = 928) (3) HCWs from the study hospital (n = 45) | COVID‐19 test centre (primary care COVID‐testing facility) Japan (7 October‐5 December 2020) |
Mixed 771, 65% symptomatic, 415, 35% asymptomatic; fever (617, 80%), cough/sputum production (294, 38.1%), runny nose/nasal congestion (196, 25.4%), loss of taste or smell (33, 4.3%), dyspnoea (6, 0.8%), fatigue (77, 10%), diarrhoea (44, 5.7%), sore throat (149, 19.3%), headache (83, 10.8%) Median age 36.5 (IQR 23‐50) years; 647 (54.6%) male |
PCR (multiple assays) Target: not stated NP; paired; same as index Timing: not stated Interval: paired |
4 excluded due to missing symptom status None reported Index: none reported Reference: there was 1 discordant sample that was positive on in‐house PCR and negative on reference real‐time PCR. Considered negative after additional BioFire Respiratory Panel 2.1 examination |
|
Takeuchi 2021b Preprint 862 (51) |
Single‐group (prospective); participants referred to a drive‐through PCR centre from a local public health centre or from one of 97 primary care facilities; also states 17 samples obtained during hospitalization | COVID‐19 test centre (mainly COVID‐19 testing centre) Japan (7 October 2020‐9 January 2021) |
Mainly symptomatic 790, 91.6% symptomatic; most commonly reported included fever (628, 79.5%), cough or sputum production (255, 32.3%), sore throat (210, 26.6%), runny nose or nasal congestion (185, 23.4%), headache (121, 15.3%). Loss of taste or smell (was reported in 32 (4.1%) overall and in 14 (27.5%) of PCR+ve group 72 (8.4%) asymptomatic Median age 36.0 (IQR 24.0, 48.0) years; 106 (12.3%) were < 18 years; 383 (44.4%) female |
PCR (multiple assays) Target: not reported for in‐house assay; N and N2 for Quantitect NP; paired Timing: as for index Interval: paired |
None reported Index: none reported Reference: 1 sample was discrepant between RDT (positive) and Quantitect assay (negative); positive Xpert Xpress (Ct 39.8 on N2). Sample was obtained from a participant who had been diagnosed with COVID‐19 1 month before the current evaluation and who was referred to the PCR centre due to refractory respiratory symptoms |
|
Thommes 2021 [A] Published 154 (154) |
Single‐group (cases) (prospective); (sensitivity only) Consecutive COVID‐19 patients admitted to the inpatient ward at the Department of Internal Medicine; described as moderate to severe disease | Hospital inpatient (inpatient) Austria (August‐end October 2020) |
Symptomatic Moderate to severe; all admitted Median age 69 (range 18–92) years, 35.7% women |
PCR (no details) Target: target ORF1a/b and B‐CoV target E‐Gene OP; paired Timing: same as for index Interval: paired |
Yes; 145 patients reportedly recruited but number for samples per assay varied from 71‐99. No reason for missing data was given Unclear Index: unclear Reference: unclear |
|
Toptan 2021(a) Published [1] 67 (58) [2] 70 (32) |
Report of 2 single‐group studies [1] samples stored (frozen) after routine diagnostic use (Institute of Virology, Charite Berlin) | Laboratory‐based (unclear) Germany (Not stated) |
Not reported Not stated Not stated |
PCR (no details) Target: ORF1 and E gene NP+OP; same sample used Timing: not stated Interval: simultaneous |
None reported Index: none reported Reference: none reported |
|
Toptan 2021(b) Published [1] 67 (58) [2] 70 (32) |
Report of 2 single‐group studies [2] clinical samples collected as part of registered protocols from individuals living in shared housing (Institute of Virology, Frankfurt). Unclear if prospective or retrospective collection | Shared living (unclear) Germany (Not stated) |
Not reported Not stated Not stated |
PCR (no details) Target: ORF1 and E gene NP; same sample used Timing: not stated Interval: paired |
None reported Index: 70 samples marked as "marginal", of which 38 were PCR‐ve and 32 PCR+ve Reference: none reported |
|
Torres 2021a Published 634 (79) |
Single‐group (prospective); to estimate sensitivity and specificity Asymptomatic household (n = 338) or non‐household (n = 296) close contacts of COVID‐19 patients as defined by the Spanish Ministry of Health (i.e. presence of compatible signs or symptoms and a positive NP swab PCR) | COVID‐19 test centre (contact tracing) Spain (16 October‐20 November 2020) |
Asymptomatic ‐ contacts All asymptomatic; 39/79 PCR+ve individuals subsequently developed mild symptoms Median age 37 (range, 9‐87) years, 279 (44%) male |
PCR (single assay) Target: N gene NP; paired; same as index Timing: as for index test Interval: paired |
None reported Index: none reported Reference: none reported |
|
Torres 2021b Published letter 270 (106) |
Single‐group (prospective); appears to include participants meeting COVID‐19 testing criteria, described as either 1) outpatients with suspected COVID‐19 with ≤ 5 d symptoms (≥ 1 of: fever, dry cough, rhinorrhoea, chest pain, dyspnoea, myalgia, fatigue, anosmia, ageusia, odynophagia, diarrhoea, conjunctivitis, and cephalea); 2) asymptomatic close contacts of COVID‐19 patients (household or non‐household) as defined by the Spanish Ministry of Health | COVID‐19 test centre (outpatients; unclear but would class as COVID‐19 test centre (acknowledgments mention Ag testing in PHC centres)) Spain (26 November 2020‐21 January 2021) |
Mainly symptomatic Mixed; 178 (66%) symptomatic; Symptomatic COVID‐19 suspects: median age 41 (range 11‐83) years; 112/178 (63%) female Asymptomatic COVID‐19 contacts: median age 44 (range 11‐87) years; 54/92 (59%) female |
PCR (single assay) Target: not reported NP; paired; same as index Timing: same as for index test Interval: paired |
None reported None reported Index: none reported Reference: none reported |
|
Turcato 2021 Published letter 3410 (223) |
Single‐group (not stated; appears prospective); all patients with symptoms suspicious for SARS‐CoV‐ 2 infection, with a temperature > 37.3 °C, with any epidemiological risk criteria (e.g. reported contact with an infected person) or evaluated in the ED for other conditions not related to SARS‐CoV‐2 infection that required hospitalization | Hospital ED (ED) Italy (1 July‐10 November 2020) |
Mixed 991, 29% symptomatic; 2419, 71% asymptomatic Not reported |
PCR (no details) Target: not reported Sample site not reported; paired; same as index Timing: not stated Interval: paired |
None reported Index: none reported Reference: none reported |
|
Van der Moeren 2021(a) [A] Published 354 (17) |
Report of 2 eligible studies [1] Single‐group (prospective) to estimate sensitivity and specificity: all adults presenting at a single community test centre for COVID‐19 testing (n = 354) |
COVID‐19 test centre (community) Netherlands (28‐30 September) |
Symptomatic Not stated; symptomatic Not stated |
PCR (multiple assays) Target: E‐ and RDRP‐gene (Cobas) or E‐gene and N‐gene (Abbott) NP+OP; paired; same as index Timing: as for index test Interval: paired |
2 samples excluded due to PCR coding error 1 invalid on Ag test Index: none reported Reference: none reported |
|
Van der Moeren 2021(b) Published 132 (132) |
Report of 2 eligible studies [2] Single‐group study to estimate sensitivity alone (prospective): patients with a positive PCR test result at 1 of 3 community testing facilities who were retested at home within 72 h of initial positive result (n = 132) |
COVID‐19 test centre (community) Netherlands (28 September‐6 October) |
Symptomatic At time of home visit: asymptomatic 3, 2% (2/3 still PCR+ve); symptomatic 129 (123 still PCR+ve) Day < 7: 66, 50% Day > 7: 57, 43% Not stated |
PCR (multiple assays) Target: E‐ and RDRP‐gene (Roche) or E‐gene and N‐gene (Abbott) NP+OP; paired; same as index Timing: as for index test Interval: paired |
Review team excluded 7 no longer PCR+ve at time of home visit (1 asymptomatic, 6 symptomatic) ‐ Ag result for 1 asymptomatic PCR negative is given (Ag‐ve) Index: none reported Reference: none reported |
|
Veyrenche 2021 Published 65 (45) |
2‐group (retrospective); [1] PCR+ve hospital inpatients (n = 45) [2] pre‐pandemic samples from "patients" (not otherwise specified) (n = 20) |
Hospital inpatient (no further detail) France (14 March‐11 April) |
Symptomatic All hospitalized; 27/45, 60% cases 'severe' according to WHO guideline (similar numbers per Ct subgroup) Median age: Ct ≤ 25: 66 (IQR 48‐84) years Ct 25‐35: 63 (50‐76) Ct > =35: 58 (49‐67) Controls 64 (35‐93); 32/45, 71% male, all controls were male |
PCR (single assay) Target: RdRp, N, E NP; same sample used Timing: as for index Interval: simultaneous |
None reported Index: none reported Reference: none reported |
|
Villaverde 2021 Published 1620 (77) |
Single‐group (retrospective); paediatric patients aged 0‐16 years old attending EDs with symptoms compatible with SARS‐CoV‐2 infection and ≤ 5 d of evolution | Hospital ED (ED) Spain (September and October 2020) |
Symptomatic All symptomatic; specific symptoms not reported Not reported |
PCR (no details) Target: SARS‐CoV‐2 E and RdRp genes NP; paired; same as index Timing: performed within 24 h of specimen collection Interval: paired |
None reported Index: none reported Reference: none reported |
|
Weitzel 2020 [A] Preprint 111 (80) |
Single‐group (retrospective); Samples from patients with respiratory symptoms and/or fever attending a private hospital ED |
Hospital ED (emergency room (private hospital)) Chile (16 March‐26 April 2020) |
Symptomatic Respiratory symptoms and/or fever; no further detail Median age 40 years; 50, 45% male (median age 38 years, 43% male for all samples tested during period) |
PCR (single assay);
Threshold ≤ 40 Ct Target: RdRp NOP; same sample used Timing: as for index test; median 2 d (IQR 1‐5 d) Interval: simultaneous |
2 invalid excluded 2 tests had invalid results due to insufficient liquid migration (2 results excluded for each test) Index: visual interpretation of the Savant assay (using manufacturer supplied UV torch) was reportedly difficult under daylight conditions; manufacturer's fluorescence reader not available in Chile. Reference: none reported |
| Yokota 2020(a) | Single‐group (retrospective); extracted as 2 studies according to sample type as unclear if same or different participants; [1] 17 NP swabs [2] 17 saliva samples (Further 307 negative saliva samples asymptomatic people did not undergo Ag testing) |
Laboratory‐based (laboratory‐based; states samples from "COVID‐19 patients") Japan (not reported) |
Symptomatic Symptomatic; median 9 d (range, 2‐14 d) pso Not reported |
PCR (single assay) Target: not reported NP; same sample used Timing: median time of sampling was 9 d (range, 2‐14 d) pso Interval: simultaneous |
None reported Index: none reported Reference: none reported |
| Yokota 2020(b) | Single‐group (retrospective); extracted as 2 studies according to sample type as unclear if same or different participants; [1] 17 NP swabs [2] 17 saliva samples (Further 307 negative saliva samples asymptomatic people did not undergo Ag testing) |
Laboratory‐based (laboratory‐based; states samples from "COVID‐19 patients") Japan (not reported) |
Symptomatic Symptomatic; median 9 d (range, 2‐14 d) pso Not reported |
PCR (single assay) Target: not reported Saliva; same sample used Timing: median time of sampling was 9 d (range, 2‐14 d) pso Interval: simultaneous |
None reported Index: none reported Reference: none reported |
|
Young 2020 Preprint 251 (38); 9 excluded |
Single‐group (prospective); patients with one or more symptoms of COVID‐19 (within ≤ 7 d post symptom onset) at 21 study sites (n = 260) [Second cohort of 361 samples from COVID suspects ≤ 5 d pso. also evaluated to compare BD Veritor with Quidel Sofia® 2 SARS Ag FIA but excluded from review as only discrepant results on the 2 Ag assays underwent PCR] |
Mixed (drive‐through/tent (n = 42), outpatient clinic (n = 74), research clinic (n = 72), or skilled nursing facility (n = 66)); USA (June 5‐11, 2020) |
Symptomatic: 110 (43%) cough, 98 (39%) muscle pain, 95 (37%) headache, 90 (35%) sore throat, 90 (35%) sore throat, 78 (31%) fever. Of those at ≤ 6 d pso (n = 245): 94 (38%) with one symptom, 151 (62%) with more than 2 symptoms Median age 43 (range 18‐90) years; 91 (36%) male |
PCR (single assay) Target: not stated NP or OP; paired Timing: swabs taken prior to any study swabs (potential for contamination of nasal cavity) Interval: paired |
9 excluded; 6 did not meet eligibility criteria and 3 had invalid specimens/results (2 on PCR and 1 labelling error) 3 invalid on at least one assay Index: none reported Reference: none reported. Re‐test of 9 'FN' results with BD MAX PCR resulted in 2 confirmed FN (BD MAX positive and sero positive), 6 were BD Max negative (including 1 sero positive) and 1 invalid (no result) |
|
Young 2021 Published as letter to the editor 803 (214) |
Single‐group (prospective); patients admitted to hospital for emergency care | Hospital ED (emergency care) UK (23 December 2020‐30 January 2021) |
Mixed 11 (8%) Ag‐positive had no COVID‐related symptoms recorded (cough, dyspnoea, fever, ageusia or anosmia) 28/80 (35.0%) RDT‐ but PCR+ve had a pre‐admission SARS‐CoV‐2 PCR+ve swab Not stated |
PCR (single assay) Target: not stated NP+OP; paired; same as index Timing: not stated Interval: paired |
Yes; 18 invalid results reported None reported Index: 2 invalid RDTs; 1 PCR+ve and 1 PCR‐ve Reference: 16 invalid on PCR; 1 RDT‐positive and 15 RDT‐negative. The RDT‐positive sample was reported in the text as "indeterminate" on PCR, and the patient tested PCR+ve five days later |
| Ag: antigen; AN: anterior nasal; BAL: bronchoalveolar lavage; Ct: cycle threshold;ED: emergency department; FIA: fluorescent immunoassay; FN: false negative; FP: false positive; HCW: healthcare worker; ICU: intensive care unit; IFU: instructions for use; IPD: individual patient data; IQR: interquartile range; NHS: National Health Service (UK); NMT: nasal mid‐turbinate; NOP: naso‐oropharyngeal; NP: nasopharyngeal; OP: oropharyngeal; PHC: primary healthcare; PHE: Public Health England; pso: post‐symptom onset; QA: quality assurance; RAT: rapid antigen test; RDT: rapid diagnostic test; PCR: reverse transcription polymerase chain reaction; RT‐qPCR: quantitative reverse transcription polymerase chain reaction; TP: true positive; VTM: viral transport medium; WHO: World Health Organization | |||||
Appendix 8. Summary index test details
| Index test (manufacturer) | Assay format Target | Sample details | Test operator Test Threshold | |
| Abdelrazik 2021 | BIOCREDIT COVID‐19 antigen kit (no product code reported) (RapiGEN) | CGIA; described as lateral flow immunochromatographic assay (uses a dual‐colour system for the qualitative detection of the SARS‐CoV‐2 antigen) not stated (SARS‐CoV‐2 antigen) |
Samples tested: NP (VTM); collection not stated Timing of sampling: median 3 d pso (Group [1] (n = 160)) Timing of test: < 24 h delay Storage: transported to lab within 1‐2 h of collection; then stored at 4 °C and tested within 24 h |
Laboratory staff Threshold: no details |
| Abdulrahman 2020 | Panbio COVID 19 antigen rapid test (no product code reported) (Abbott Rapid Diagnostic Jena GmbH, Jena, Germany) | CGIA SARS‐CoV‐2 nucleocapsid protein |
Samples tested: nasal (NMT) (direct swab); collected by HCW Timing of sampling: median of 2 (range 0‐14) d pso (n = 1301, 31%) Timing of test: immediate testing Storage: none |
Not stated; most likely trained HCW as the test was conducted on‐site Threshold: not stated |
|
Agullo 2021 [A] Agullo 2021 [B] Agullo 2021 [C] |
Panbio COVID 19 antigen rapid test (no product code reported) (Abbott Rapid Diagnostic Jena GmbH, Jena, Germany) | CGIA SARS‐CoV‐2 nucleocapsid protein |
Samples tested: [A] NP (direct swab); collected by HCW [B] nasal (direct swab); collected by HCW [C] saliva (self‐collected) Timing of sampling: median (Q1‐Q3) duration of 3 (2–5) d of symptoms Timing of test: immediate testing Storage: none |
Not stated; may have been by same qualified nurse Threshold: not stated |
| Akingba 2021 | Panbio SARS‐CoV‐2 rapid test (no product code reported) (Abbott Rapid Diagnostics, USA) | Not reported Not reported |
Samples tested: NP (not specified); collection not specified Timing of sampling: not reported Timing of test: immediate testing Storage: immediate testing |
Not reported Threshold: not reported; set by manufacturer |
| Albert 2020 | Panbio COVID‐19 AG Rapid Test Device (no product code reported) (Abbott Diagnostic GmbH, Jena, Germany) | CGIA (from IFU) Nucleoprotein |
Samples tested: NP (direct); collected by HCW Timing of sampling: day < 7 pso Timing of test: immediate testing Storage: none |
Not stated; on‐site Threshold: visible line within 15 min; as per manufacturer |
| Alemany 2021 | Panbio COVID‐19 Ag Test (no product code reported) (Abbott Laboratories, Illinois, USA) (Samples to compare Panbio with Coris Bioconcept COVID‐19 Ag RespiStrip and SD Biosensor Standard F COVID‐19 Ag FIA and Standard Q COVID‐19 Ag Test) | CGIA Not stated (SARS‐CoV‐2 antigen) |
Samples tested: NP or NMT (VTM); collection not specified Timing of sampling: not stated Timing of test: not stated; frozen samples Storage: stored at 2‐8 °C prior to PCR then frozen (−80C) prior to antigen testing; "Internal validation showed no significant change in the test performance using Abbot test Kit buffer or a mix of the Kit buffer and transport media at 1:3 dilution; likewise, the use of frozen specimens showed no significant differences compared with fresh ones" |
2 laboratory technicians Threshold: visible line; as per manufacturer |
| Aoki 2020 | Espline SARS‐CoV‐2 (no product code reported) (Fujirebio Inc., Japan) | Immunochromatography assay based on sandwich enzyme immunoassay (ALP‐labelled) SARS‐CoV‐2 N antigen |
Samples tested: NP (direct or VTM); collection not specified Timing of sampling: median 9.5 d pso for samples Ag+ve/PCR+ve, 16 d for Ag‐ve/PCR+ve, 19 d for Ag‐ve/PCR‐ve Timing of test: mixed Storage: not described for samples in Espline buffer; remnant samples in VTM stored at −80°C after PCR testing |
Not stated Threshold: "positive when both the reference line and the judgment line can be visually confirmed" |
|
Baro 2021 [A] Baro 2021 [B] Baro 2021 [C] Baro 2021 [D] Baro 2021 [E] |
[A] Panbio COVID‐19 Ag Rapid test (no product code reported) (Abbott Rapid Diagnostics, Panbio Ltd, USA) [B] CLINITEST Rapid COVID‐19 Antigen Test (no product code reported) (Siemens Healthineers, Shangai International Holding Corp, USA) [C] SD Biosensor SARS‐CoV‐2 Rapid Antigen Test (no product code reported) (Roche Diagnostics, SD Biosensor, Republic of Korea) [D] Lepu SARS‐CoV‐2 Antigen Rapid Test (no product code reported) (Beijing Lepu Medical Technology Co., Ltd., China) [E] Surescreen COVID‐19 Coronavirus Rapid Antigen Test Cassette (no product code reported) (SureScreen Diagnostics Ltd, UK) | [A] CGIA
[B] Immunochromatographic
[C] LFA (unclear)
[D] CGIA
[E] LFA (unclear) Nucleocapsid protein |
Samples tested: NP alone (VTM); collected by HCW Timing of sampling: N/A; all asymptomatic Timing of test: < 36 h Storage: samples stored for up to 24 h (2‐8 ºC) prior to PCR then stored up to 12 h more at 2‐8 ºC until antigen testing |
Lab technician at The University Hospital Germans Trias i Pujol. Threshold: visual coloured band; the presence of any test line (T) indicates a positive result; samples were applied directly to the test cassette and incubated for 15 min at room temperature before reading results with the naked eye, according to the manufacturer instructions |
|
Basso 2021 [A] Basso 2021 [B] Basso 2021 [C] |
[A] and [B] ESPLINE rapid test (no product code reported) (Fujirebio, Tokyo, Japan) [C] Panbio COVID‐19 Ag Rapid Test (no product code reported) (ABBOTT, Chicago, Illinois, USA) (3rd laboratory‐based Ag detection assay was also evaluated but is not eligible for this review: LUMIPULSE SARS‐CoV‐2 Ag kit, Fujirebio, Tokyo, Japan) | CGIA SARS‐CoV‐2 nucleocapsid protein |
Samples tested: [A] saliva (direct swab); self‐collected [B] and [C] NP Timing of sampling: inpatients (n = 138): ≤ 7 d pso, 38, 27.6% 7‐14 d pso, 74, 53.6% > 14 d pso, 26, 18.8% Timing of test: < 3 h Storage: all molecular and CLEIA antigen testing in both saliva and NP swabs performed in parallel within 3 h from collection |
Unclear; likely laboratory staff Threshold: not stated; visual inspection |
| Beck 2021 | SOFIA SARS Antigen FIA (no product code reported) (Quidel) | FIA SARS‐CoV |
Samples tested: nasal (NMT) (direct swab); collected by HCW Timing of sampling: ≤ 5 d pso 298, 86.1%; > 5d pso 48, 13.9% Timing of test: immediate testing Storage: none; "specimens were delivered to the laboratory (located within the same building) within 10 min of collection" |
Laboratory staff Threshold: not stated; "tested … according to the manufacturer’s package insert" |
| Billaud 2020 | ABBOTT SARS‐COV2 Antigenic Test (no product code reported) (Abbott) | CGIA (from IFU) Not stated |
Samples tested: NP (direct); Collected by trained non‐HCW Timing of sampling: not stated but includes people > 7 d pso Timing of test: immediate testing Storage: none |
Not stated; on‐site so presume firefighters Threshold: visual line; as per manufacturer |
| Blairon 2020 | COVID‐19 Ag Respi‐Strip (no product code reported) (Coris Biocencept (Gembloux, Belgium)) | LFA Not stated |
Samples tested: NP (VTM); collection not specified Timing of sampling: not stated; appears to be on presentation (repeat tests ordered at clinician's discretion were excluded) Timing of test: infer that antigen test was conducted immediately on receipt of sample at on‐site laboratory "after antigenic testing was performed, the molecular assessment of SARS‐CoV‐2 was outsourced to a university centre" Storage: no storage described |
Not stated; infer laboratory staff Threshold: as per manufacturer |
| Bulilete 2021 | Panbio COVID 19 antigen rapid test (no product code reported) (Abbott Rapid Diagnostic Jena GmbH, Jena, Germany) | CGIA SARS‐CoV‐2 nucleocapsid protein |
Samples tested: NP (direct swab); collected by HCW Timing of sampling: ≤ 5 d pso or close contact, 967, 70.6% Symptomatic: ≤ 7 d pso, 622/677, 92% Asymptomatic: ≤ 7 d contact, 481/688, 70%; 173/688 unknown number of days Timing of test: immediate testing Storage: none reported; results were interpreted within 15 min following the manufacturer’s instructions |
Not stated; presume same qualified nurse Threshold: not stated |
|
Caruana 2021 [A] Caruana 2021 [B] Caruana 2021 [C] Caruana 2021 [D] |
[A] Standard Q COVID‐19 Rapid Antigen Test (SD Biosensor ‐ Republic of Korea /Roche – Switzerland) [B] Panbio COVID‐19 Ag Rapid Test (no product code reported) (Abbott – USA) [C] One Step Immunoassay for Exdia COVID‐19 Ag (no product code reported) (Precision Biosensor Inc. ‐ Republic of Korea) [D] BD Veritor System for Rapid Detection of SARS‐CoV‐2 (no product code reported) (Becton Dickinson ‐ USA) (Result of SDQ was used to guide care/triage pathway; patients and clinicians were blinded to the results of all other Ag tests] | Not reported Not reported |
Samples tested: NP (VTM); collection not specified Timing of sampling: ≤ 4 d: 138/293 (47%) 4‐7 d: 46/293 (16%) ≥ 7 d: 44/293 (15%) Missing data/not typical COVID‐19 symptoms: 65/293 (22%) Timing of test: immediate testing Storage: NP delivered to the RAT lab, immediately after the sampling procedure |
Laboratory technicians Threshold: [A] and [B] visually [C] and [D] automatically using analyser |
| Cerutti 2020 | Standard Q COVID‐19 Ag (no product code reported) (SD‐Biosensor, RELAB, I) | CGIA (from IFU) NP |
Samples tested: NP alone (VTM); collection not specified Timing of sampling: not stated Timing of test: not stated Storage: primarily run in parallel with standard care PCR; 13 were frozen residual samples |
Not stated; laboratory staff presumed Threshold: visual line after 15‐30 min; as per manufacturer |
| Chaimayo 2020 | Standard Q COVID‐19 Ag kit (no product code reported) (SD Biosensor, Republic of Korea) | CGIA SARS‐CoV‐2 nucleocapsid (N) antigen |
Samples tested: NP or OP (VTM); collection not specified Timing of sampling: PCR+ve: 3 asymptomatic, 1‐7 d pso, 41 (68%), > 7 d pso, 12 (20%) 4 unspecified time pso Timing of test: same day (within a few hours) Storage: transported at 2–8 °C to the Microbiology laboratory, Siriraj Hospital, for processing within a few hours |
Not stated; likely laboratory staff "All specimens were processed in biosafety level‐3 (BSL‐3) and biosafety level‐2 enhanced (BSL‐2+) facilities with full personal protective equipment" Threshold: for positive COVID‐19 antigen result, 2 coloured lines of control (C) and test (T) lines were presented |
| Ciotti 2021 | COVID‐19 Ag Respi‐Strip (no product code reported) (Coris BioConcept) | CGIA Nucleoprotein of SARS‐CoV and SARS‐CoV‐2 |
Samples tested: NP alone (direct swab); collection not specified Timing of sampling: not reported Timing of test: immediate testing Storage: none reported |
Not stated; virology laboratory Threshold: visual appearance of test and control (red) lines |
| Courtellemont 2021 | COVID‐VIRO (no product code reported) (AAZ, Boulogne Billancourt, France) | CGIA Nucleocapsid |
Samples tested: NP alone (direct); collected by HCW Timing of sampling: median 5 d pso, mean 5.3 d, range 1‐20 d Timing of test: immediate testing Storage: none |
Not stated; on‐site immediate testing reported Threshold: visible line; as per manufacturer |
| Del Vecchio 2021 | Panbio COVID‐19 Ag Rapid Test Device (no product code reported) (Abbott Lake Country, IL, USA) | Not reported Not reported |
Samples tested: not specified (direct swab); collection not specified Timing of sampling: 0‐7 d pso, 39 (64%) (28 d 0‐3) 8‐14 d pso, 11 (18%) ≥ 15 d pso, 1 Timing of test: < 1 h Storage: processed right after sampling; maximum 1‐h delay |
Not reported Threshold: according to manufacturer’s instructions |
| Dominguez Fernandez 2021 | Panbio COVID‐19 Ag Rapid Test Device (no product code reported) (Abbott) | CGIA Not reported |
Samples tested: not specified (not specified); collection not specified Timing of sampling: < 5 d pso, 90% Timing of test: not specified Storage: not stated |
Not stated Threshold: not stated |
| Drain 2021(a) | LumiraDx SARS‐CoV‐2 (no product code reported) (LumiraDx UK Ltd.) | Microfluidic immunoassay with fluorescent latex signal N |
Samples tested: nasal (AN) (direct swab); collection mixed Timing of sampling: whole sample: range 1‐12 d [1]+[2] mean 4.0 (SD 2.9) d pso Timing of test: [1] immediate; [2] not specified Storage: [1] tested fresh and then frozen within 1 h of collection [2] unclear |
Unclear; included minimally trained operators Threshold: result shown on touchscreen as "positive" |
| Drain 2021(b) | LumiraDx SARS‐CoV‐2 (no product code reported) (LumiraDx UK Ltd.) | Microfluidic immunoassay with fluorescent latex signal N |
Samples tested: NP (direct swab); collection mixed Timing of sampling: whole sample: range 1‐12 d [3] mean 3.5 (SD 2.5) d pso Timing of test: immediate testing Storage: [3] no storage |
Unclear; included minimally trained operators Threshold: result shown on touchscreen as "positive" |
|
Drevinek 2020 [A] Drevinek 2020 [B] |
[A] Panbio COVID‐19 Ag Rapid Test (no product code reported) (Abbott, Germany); [B] Standard F COVID‐19 Ag FIA (no product code reported) (SD Biosensor, Republic of Korea) | [A] CGIA;
[B] FIA Not stated |
Samples tested: NP (direct swab); collection not specified Timing of sampling: not stated Timing of test: immediate testing Storage: no storage; tested immediately on collection |
Not stated Threshold: [A] visual assessment after 15 min incubation [B] standard F200 analyser (in 'read‐only' mode) after 30 min incubation |
| Faico‐Filho 2021 | Panbio COVID‐19 Ag test (no product code reported) (Abbott, Germany) | Not reported Not reported |
Samples tested: NP (not specified); collection not specified Timing of sampling: mean d since symptom onset: 5 (range 4‐7) Timing of test: not specified Storage: only reported that NP swab samples were simultaneously tested with both index and reference tests and PCR results were available within 6‐24 h |
Not reported Threshold: according to manufacturer |
|
Favresse 2021 [A] Favresse 2021 [B] Favresse 2021 [C] Favresse 2021 [D] |
[A] Biotical SARS‐CoV‐2 Ag card (no product code reported) (Biotical Health, Madrid, Spain) [B] Panbio COVID‐19 Ag Rapid Test Device (no product code reported) (Abbott, Chicago, IL, USA) [C] Coronavirus Ag Rapid Test Cassette (no product code reported) (Healgen Scientific, Houston, TX, USA) [D] Roche SARS‐CoV‐2 Rapid Antigen Test (no product code reported) (Roche Diagnostics, Basel, Switzerland) (Additional lab‐based Ag test also evaluated but not eligible for this review: [E] VITROS Immunodiagnostic Products SARS‐CoV‐2 Antigen test (Ortho Clinical Diagnostics, Raritan, NJ, USA)) | [A]‐[D] all LFAs, method not reported
[E] Chemiluminescence assay [A]‐[D] Nucleocapsid |
Samples tested: NP (VTM); collection not specified Timing of sampling: symptomatic: median 3 d pso (IQR 2‐4 d) Timing of test: < 24 h delay Storage: all tests within 24 h; storage conditions not specified |
Laboratory staff Threshold: appearance of 2 visible lines for all except VITRO which was a signal of ≥ 1 |
| Fenollar 2020(a) | PANBIO COVID‐19 Ag (no product code reported) (Abbott, Germany) | CGIA (from IFU) NP |
Samples tested: NP (direct); collection not specified Timing of sampling: not stated Timing of test: tested within 1 h Storage: none |
Not stated; appears to be onsite testing Threshold: visual line; as per manufacturer |
| Fenollar 2020(b) | Panbio COVID‐19 Ag (no product code reported) (Abbott, Germany) | CGIA (from IFU) NP |
Samples tested: NP (direct); collection not specified Timing of sampling: not stated Timing of test: tested within 1 h Storage: none |
Not stated; appears to be onsite testing Threshold: visual line; as per manufacturer |
| Ferguson 2021 | Innova Lateral Flow Device (no product code reported) (Innova Medical group, a subsidiary of Xiamen Biotime Biotechnology company) | CGIA SARS‐CoV‐2 nucleocapsid antigens |
Samples tested: NP (direct swab); self‐collected Timing of sampling: N/A; all asymptomatic, no clear epidemiological contact reported Timing of test: immediate testing Storage: no storage |
Not stated;
trained postgraduate and final year undergraduate students in the College of Medical and Dental Science, supervised by highly experienced postdoctoral researchers (total of 18 test operatives) Threshold: not stated; visual appearance of lines |
| Filgueiras 2021 | COVID‐19 Ag ECO Test (no product code reported) (ECODiagnostica, Brazil) | CGIA Not stated Nucleocapsid viral protein |
Samples tested: NP (direct swab); collection not specified Timing of sampling: at 1st day of symptom onset ≤ 3 d pso, 63 (42%); 4‐7 d pso, 59 (39%); 8‐15 d pso, 22 (15%); > 15 d pso, 2 (1%); not reported 4 (3%) Timing of test: immediate testing Storage: immediately tested |
Not stated Threshold: colorimetric reaction; interpreted after 15 min incubation |
| FIND 2020a | NowCheck COVID‐19 Ag test (RG1901DG) (Bionote Inc) | LFA (not otherwise specified) SARS‐CoV‐2 nucleocapsid antigen |
Samples tested: NP (direct); collected by HCW Timing of sampling: median 4 d pso (IQR 3‐6 d); day <0‐3, 152, 39% day 4‐7 180, 46% day ≥ 8 58, 15% Timing of test: not specified; as soon as possible after collection and within IFU recommendations Storage: rapid test for 1 h or 2‐8°C for 4 h |
HCW on‐site Threshold: presence of visible control and test lines |
| FIND 2020b (CH) | Panbio COVID‐19 Ag Rapid Test (41FK10) (Abbott ) | CGIA (from IFU) Not reported |
Samples tested: NP (direct); collected by HCW Timing of sampling: time pso recorded for 115/124, 92% Day 0‐3 89, 78% Day 4‐7 23, 20% Day 8+ 3, 3% Timing of test: not specified; as soon as possible after collection and within IFU recommendations Storage: author contact advises tested as soon as possible and within the time limit specified in the IFU |
HCW on‐site Threshold: presence of visible control and test lines |
| FIND 2020b (DE) | Panbio COVID‐19 Ag (41FK10) (Abbott Rapid Diagnostics) | CGIA (from product insert) Not reported |
Samples tested: NP or OP (direct swab); collected by trained non‐HCW Timing of sampling: time pso recorded for 692/709 symptomatic, 98% Day 0‐3 380, 55% Day 4‐7 230, 33% Day 8+ 82, 12% Timing of test: immediate testing Storage: none required; direct swab |
Trained laboratory team Threshold: presence of visible control and test lines 2 readers blinded to the results of the other interpreted the test with the naked eye after 15 min of incubation. |
| FIND 2020c (BR) | Standard Q COVID‐19 Ag (09COV30D) (SD Biosensor Inc) | CGIA (from IFU) Not reported |
Samples tested: NP (direct); collected by HCW Timing of sampling: median 5 d pso (IQR 4‐6 d) (for 397 patients); day < 0‐3, 85, 21% day 4‐7, 273, 69% day ≥ 8, 39, 10% Timing of test: tested as soon as possible and within the time limit specified in the IFU Storage: none |
HCW on‐site Threshold: presence of visible control and test lines |
| FIND 2020c (CH) | Standard Q COVID‐19 Ag (09COV30D) (SD Biosensor Inc) | CGIA (from IFU) Not reported |
Samples tested: NP (direct); collected by HCW Timing of sampling: median not reported (range 0‐15); day < 0‐3, 122, 67% day 4‐7, 54, 29% Day 8+, 7, 34% Timing of test: tested as soon as possible and within the time limit specified in the IFU Storage: none |
HCW on‐site Threshold: presence of visible control and test lines |
| FIND 2020c (DE) | Standard Q COVID‐19 Ag Test (no product code reported) (SD Biosensor, Inc. Gyeonggi‐do, Korea) | CGIA Not stated |
Samples tested: NP, OP or NP+OP (direct); collected by HCW Timing of sampling: overall: mean 5 d pso (SD 9.6) [A] 7.0 (SD 12.2) [B] 6.2 (SD 14.0) [C] 3.7 (SD 5.6) Timing of test: not stated but no delay reported (on‐site testing) for drive‐in and ambulatory testing; secondary care samples transported to lab Storage: drive‐in centre and ambulatory testing: tested on site (presume short time frame) Secondary care: transported on ice to a category 3 facility for testing PCR swab obtained first, then same technique repeated for antigen test |
HCW on‐site Threshold: [B] and [C] visual appearance were interpreted by 2 operators, each blinded to the result of the other. In case of discrepant results, both operators re‐read the result and agreed on a final result; invalid results were repeated once using the remaining buffer according to the respective IFUs; readouts were done within the recommended time for each Ag‐RDT (10 min for Bioeasy, 15 min for Coris and 15‐30 min for SD Biosensor) |
| FIND 2020d (BR) | Standard F COVID‐19 Ag FIA (F‐NCOV‐01G, 10COV30D) (SD Biosensor Inc) | FIA Not reported |
Samples tested: NP (direct); collected by HCW Timing of sampling: median 4 d pso (IQR 3‐6 d) (for 421 patients); day < 0‐3, 131, 31% day 4‐7, 248, 59% day ≥ 8, 42, 10% Timing of test: tested as soon as possible and within the time limit specified in the IFU Storage: none |
HCW on‐site Threshold: as per Standard F analyser; cut‐off index (COI) ≥ 1.0 (as per IFU) |
| FIND 2020d (DE) | Standard F COVID‐19 Ag FIA (F‐NCOV‐01G, 10COV30D) (SD Biosensor Inc) | FIA Not reported |
Samples tested: NP or NP+OP (direct); collected by HCW Timing of sampling: median 3 d pso (IQR 2‐5 d) (for 505 patients); day < 0‐3, 257, 51% day 4‐7, 202, 47% day ≥ 8, 46, 9% Timing of test: tested as soon as possible and within the time limit specified in the IFU Storage: none |
HCW on‐site Threshold: as per Standard F analyser; cut‐off index (COI) ≥ 1.0 (as per IFU) |
| FIND 2020e (BR) | BIOCREDIT COVID‐19 Ag (G61RHA20) (RapiGEN Inc) | CGIA (from IFU) Not reported |
Samples tested: NP (direct); collected by HCW Timing of sampling: median 5 d pso (IQR 4‐7 d) (for 470 patients); day < 0‐3, 95, 20% day 4‐7, 296, 63% day ≥ 8, 79, 17% Timing of test: tested as soon as possible and within the time limit specified in the IFU Storage: none |
HCW on‐site Threshold: visual appearance of test and control lines |
| FIND 2020e (DE) | BIOCREDIT COVID‐19 Ag (G61RHA20) (RapiGEN Inc) | CGIA (from IFU) Not reported |
Samples tested: NP or NP+OP (direct); collected by HCW Timing of sampling: median 3 d pso (IQR 2‐4d) (for 701 patients); day < 0‐3, 472, 67% day 4‐7, 161, 23% day ≥ 8, 68, 10% Timing of test: tested as soon as possible and within the time limit specified in the IFU Storage: none |
HCW on‐site Threshold: visual appearance of test and control lines |
| FIND 2020f | COVID‐19 Ag Respi‐Strip (no product code reported) (Coris Bioconcept, Gembloux, Belgium) | CGIA Not stated |
Samples tested: NP or NP+OP (direct); collected by HCW Timing of sampling: overall: mean 5 d pso (SD 9.6) [A] 7.0 (SD 12.2) [B] 6.2 (SD 14.0) [C] 3.7 (SD 5.6) Timing of test: not stated but no delay reported (on‐site testing) for drive‐in and ambulatory testing; secondary care samples transported to lab Storage: drive‐in centre and ambulatory testing: tested on site (presume short time frame) Secondary care: transported on ice to a category 3 facility for testing PCR swab obtained first, then same technique repeated for Ag test |
HCW on‐site Threshold: [B] and [C] visual appearance interpreted by 2 operators, each blinded to the result of the other; in case of discrepant results, both operators re‐read the result and agreed on a final result; invalid results were repeated once using the remaining buffer according to the respective IFUs; readouts were done within the recommended time for each Ag‐RDT (10 min for Bioeasy, 15 min for Coris and 15‐30 min for SD Biosensor) |
|
FIND 2021a [A] FIND 2021a [B] |
[A] NowCheck COVID‐19 Ag test (RG1901DGN (Nasal)) (Bionote Inc) [B] NowCheck COVID‐19 Ag test (RG1901DG (NP)) (Bionote Inc) | Rapid chromatographic immunoassay in lateral flow format Not reported |
Samples tested: nasal (NMT) (direct swab); collected by HCW Timing of sampling: median 4 d pso (IQR 3‐6 d); day < 0‐3, 72, 33% day 4‐7, 123, 56% day ≥ 8, 23, 11% Timing of test: immediate testing Storage: immediate testing |
HCW Threshold: presence of visible control and test lines |
|
FIND 2021b [A] FIND 2021b [B] |
[A] Panbio COVID‐19 Ag Rapid Test Device Nasal (41FK11) ((Abbott) [B] Panbio COVID‐19 Ag Rapid Test (41FK10) (Abbott) | CGIA (from product insert) Not reported |
Samples tested: nasal (NMT) (direct swab); collected by HCW Timing of sampling: time pso recorded for 126/281, 45% Day 0‐3, 86, 68% Day 4‐7, 290, 23% Day 8 plus, 11, 9% Timing of test: immediate testing Storage: author contact advises tested as soon as possible and within the time limit specified in the IFU |
HCW Threshold: presence of visible control and test lines |
|
FIND 2021c (BR) [A] FIND 2021c (BR) [B] |
[A] Standard Q COVID‐19 Ag Nasal (09COV31D) (SD Biosensor Inc) [B] Standard Q COVID‐19 Ag (09COV30D) (SD Biosensor Inc) | Rapid chromatographic immunoassay in lateral flow format Not reported |
Samples tested: nasal (NMT) (direct swab); collected by HCW Timing of sampling: median 5 d pso (IQR 3‐6.75 d); day < 0‐3, 68, 32% day 4‐7, 116, 54% day ≥ 8, 30, 14% Timing of test: immediate testing Storage: author contact advises tested as soon as possible and within the time limit specified in the IFU |
HCW Threshold: presence of visible control and test lines |
|
FIND 2021c (DE) [A] FIND 2021c (DE) [B] |
Standard Q COVID‐19 Ag Test (no product code reported) (SD Biosensor, Inc. Gyeonggi‐do, Korea) | Chromatographic Not stated |
Samples tested: [A] nasal (NMT) (direct swab); collected by HCW [B] NP (direct swab); collected by HCW Timing of sampling: average symptom duration 4.2 (SD 2.6) d (range 1‐10 d) for PCR+ve group Timing of test: immediate testing Storage: none required |
HCW (states "professional") Threshold: extracted from secondary publication by Lindner and colleagues ‐ visual; presence of control test lines, categorized as negative, weak positive, positive and strong positive; results interpreted by 2 operators, each blinded to the result of the other. The second reader was also blinded to the result from the alternative sampling method |
| FIND 2021d | Espline SARS‐CoV‐2 (231906) (Fujirebio Inc.) | Rapid chromatographic immunoassay in lateral flow format Not reported |
Samples tested: NP or OP (direct swab); collected by HCW Timing of sampling: median 2 d pso (IQR 1‐4 d); day < 0‐3, 311, 70% day 4‐7, 106, 24% day ≥ 8, 27, 6% Timing of test: immediate testing Storage: author contact advises tested as soon as possible and within the time limit specified in the IFU |
HCW Threshold: presence of visible control and test lines |
| FIND 2021e | SARS‐CoV‐2 Antigen Rapid Test Kit (Colloidal Gold) (COV‐AG‐20/G10313) (Joysbio (Tianjin) Biotechnology Co) | Rapid chromatographic immunoassay in lateral flow format Not reported |
Samples tested: nasal (AN) (direct swab); collected by HCW Timing of sampling: only reported for PCR+ve cases median 2 d pso (IQR 1‐3.5 d); day < 0‐3 ‐ 23, 74% day 4‐7 ‐ 8, 26% day ≥ 8 ‐ 0, 0% Timing of test: immediate testing Storage: author contact advises tested as soon as possible and within the time limit specified in the IFU |
HCW Threshold: presence of visible control and test lines |
| FIND 2021f | COVID 19 RAPID ANTIGEN TEST (11811125) (Mologic Ltd.) | Rapid chromatographic immunoassay in lateral flow format Not reported |
Samples tested: nasal (AN) (direct swab); collected by HCW Timing of sampling: median 2 d pso (IQR 1‐4 d); n = 436 day < 0‐3, 290, 67% day 4‐7, 121, 28% day ≥ 8, 25, 6% Timing of test: immediate testing Storage: author contact advises tested as soon as possible and within the time limit specified in the IFU |
HCW Threshold: presence of visible control and test lines |
| FIND 2021g | COVID‐19 Ag Rapid Test (243103N‐20) (NADAL) | Rapid chromatographic immunoassay in lateral flow format Not reported |
Samples tested: NP (direct swab); collected by HCW Timing of sampling: only reported for 54/62 PCR+ve patients Median 2 d pso (IQR 1‐3 d) day <0‐3, 45, 83% day 4‐7, 7, 13% day ≥ 8, 2, 4% Timing of test: immediate testing Storage: author contact advises tested as soon as possible and within the time limit specified in the IFU |
HCW Threshold: presence of visible control and test lines |
| FIND 2021h | iChroma COVID‐19 Ag Test (CFPC‐115) (Boditech Medical, Inc.) | Rapid chromatographic immunoassay in lateral flow format Not reported |
Samples tested: NP (direct swab); collected by HCW Timing of sampling: only reported for 32 PCR+ve patients Median 1.5 d pso (IQR 1‐3 d) day < 0‐3, 26, 81% day 4‐7, 5, 16% day ≥ 8, 1, 3% Timing of test: immediate testing Storage: author contact advises tested as soon as possible and within the time limit specified in the IFU |
HCW Threshold: presence of visible control and test lines |
| FIND 2021i | Wondfo 2019‐nCoV Antigen Test (W196P0003) (Guangzhou Wondfo Biotech Co.,) | Rapid chromatographic immunoassay in lateral flow format Not reported |
Samples tested: NP (direct swab); collected by HCW Timing of sampling: only reported for 44 PCR+ve patients Median 2 d pso (IQR 1‐4 d) day < 0‐3, 31, 70% day 4‐7, 11, 25% day ≥ 8, 2, 5% Timing of test: immediate testing Storage: author contact advises tested as soon as possible and within the time limit specified in the IFU |
HCW Threshold: presence of visible control and test lines |
| FIND 2021j | Bioeasy 2019‐nCoV Ag Fluorescence Rapid Test Kit (Time‐Resolved Fluorescence) (product code not reported) (Shenzhen Bioeasy Biotechnology Co. Ltd., Guangdong Province, China) | FIA Not stated |
Samples tested: NP or NP+OP (direct); collected by HCW Timing of sampling: overall: mean 5 d pso (SD 9.6) [A] 7.0 (SD 12.2) [B] 6.2 (SD 14.0) [C] 3.7 (SD 5.6) Timing of test: not stated but no delay reported (on‐site testing) for drive‐in and ambulatory testing; secondary care samples transported to lab Storage: drive‐in centre and ambulatory testing: tested on site (presume short time frame) Secondary care: transported on ice to a category 3 facility for testing PCR swab obtained first, then same technique repeated for Ag test |
HCW on‐site Threshold: as per Analyser |
|
Fourati 2020 [A] Fourati 2020 [B] Fourati 2020 [C] Fourati 2020 [D] Fourati 2020 [E] |
[A] SARS‐CoV‐2 COVID‐19 Respi‐Strip (no product code reported) (Coris BioConcept, Gembloux, Belgium) [B] Standard Q COVID‐19 Ag (no product code reported) (SD BIOSENSOR, Inc., Korea) [C] Panbio COVID‐19 Antigen Rapid Test (Abbott, Chicago, Illinois, USA) [D] Biosynex COVID‐19 Ag BSS (no product code reported) (Biosynex, Strasbourg, France) [E] COVID‐VIRO Antigen Rapid Test (no product code reported) (AAZ, Boulogne‐Billancourt, France) Excluded ‐ [F] NG Test SARS‐CoV‐2 Ag (no product code reported) (NG Biotech, Guipry, France) | [A] CGIA (from IFU) [B] LFA (not otherwise specified) [C] CGIA (from IFU) [D] CGIA (from IFU) [E] CGIA (from IFU) |
Samples tested: NP (VTM); collection not specified Timing of sampling: pso (reported for 289 samples): 0‐3 d, 97, 34% 4‐7 d, 103, 36% 8=11 d, 63, 22% ≥ 12 d, 26, 9% Number of samples reported at > 7 d varied per test, maximum was 289 Timing of test: not stated Storage: frozen at −80 °C until use |
Laboratory staff Threshold: visual, as per manufacturer |
| Garcia‐Finana 2021 | Innova SARS‐CoV‐2 antigen lateral flow device (no product code reported) (Innova Medical Group) | CGIA Not stated |
Samples tested: nasal+OP (direct swab); self‐collected Timing of sampling: asymptomatic Timing of test: immediate testing Storage: no storage |
Trained non‐HCW (assumed) Threshold: visual line appearance |
| Gomez 2021(a) | Sofia 2 SARS Antigen (no product code reported) (Quidel, USA) | FIA Not reported |
Samples tested: nasal (NMT) (direct swab); collection not specified Timing of sampling: not reported Timing of test: not specified; appears no delay Storage: not reported; probably immediate |
Not reported ("performed according to CLIA'88 regulations by appropriate personnel") Threshold: not reported; as per manufacturer |
| Gomez 2021(b) | BD Veritor SARS‐CoV‐2 (no product code reported) (BD, USA) | Unknown; LFA not otherwise described Not reported |
Samples tested: nasal (AN) (direct swab); collection not specified Timing of sampling: not reported Timing of test: not specified; appears no delay Storage: not reported; probably immediate |
Not reported ("performed according to CLIA'88 regulations by appropriate personnel") Threshold: not reported; as per manufacturer |
| Gonzalez‐Donapetry 2021 | Panbio COVID‐19 Ag Rapid Test (no product code reported) (Abbott Rapid Diagnostics Jena GmbH) | Membrane technology with gold conjugate
CGIA SARS‐CoV‐2 nucleoprotein antigens |
Samples tested: NP (direct swab); collection not specified Timing of sampling: median pso d (IQR): Total: 1 (1‐3) d; PCR‐ve 1 (1‐3); PCR+ve 1 (1‐2) Timing of test: not specified; appears no delay Storage: assume immediate testing (no transport or storage reported) |
Not reported Threshold: according to manufacturer's protocol |
| Gremmels 2021(a) | Panbio COVID‐19 Ag Rapid Test (lot 41ADF011A) (Abbott (Lake Country, IL, USA)) | CGIA (from IFU) NP |
Samples tested: NP (not specified); collection not specified Timing of sampling: cohort [1] (data on duration of symptoms reportedly missing for 201 participants; total reported here is 1138 but denominator for % is 1166) day 1‐3 pso 387, 33.2% day 4‐7 560, 48.0% day > 7 191, 16.4% Timing of test: within 2 h of collection Storage: none described |
2 independent observers Threshold: visual line within 15 min; as per manufacturer |
| Gremmels 2021(b) | Panbio COVID‐19 Ag Rapid Test (lot 41ADF011A) (Abbott (Lake Country, IL, U.S.A)) | CGIA (from IFU) NP |
Samples tested: NP (direct); collection not specified Timing of sampling: not stated; on presentation Timing of test: within 2 h Storage: appears to be room temperature |
2 independent observers Threshold: visual line within 15 min; as per manufacturer |
| Gupta 2020 | Standard Q rapid antigen detection test (no product code reported) (SD Biosensor, Inc., Gurugram) | CGIA (from IFU) Not stated |
Samples tested: NP (direct); collection not specified Timing of sampling: symptomatic: 192 (95%) ≤ 5 d pso (including 57 cases) Timing of test: immediate testing Storage: none |
HCW on‐site Threshold: visual; test and control lines |
| Halfon 2021 | Panbio COVID 19 antigen rapid test (no product code reported) (Abbott Rapid Diagnostic Jena GmbH, Jena, Germany) | CGIA SARS‐CoV‐2 nucleocapsid protein |
Samples tested: NP (not specified); collection not specified Timing of sampling: for symptomatic, time pso was: ≤ 4 d 47, > 4 d 22, and not reported or unknown 27 Timing of test: not specified Storage: not stated |
Not stated Threshold: not stated |
| Houston 2021 | Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test (no product code reported) (Lotus Global Company, London, UK) | CGIA Not stated |
Samples tested: NP (direct swab); collection not specified Timing of sampling: not stated Timing of test: immediate testing Storage: none; immediate testing |
Appropriately trained healthcare assistants in the ED Threshold: not stated |
| Huh 2021 | PCL COVID19 Ag Rapid FIA used with the PCLOK EZ analyser (no product code reported) (PCL Inc.) | FIA Not reported |
Samples tested: NP (VTM); collection not specified Timing of sampling: 0‐12 d pso (n = 62) Results reported for 0‐7 d (n = 48) and 8‐12 d (n = 14) periods Timing of test: not specified Storage: storage in VTM reported, test done after reference standard, but exact timing not reported |
Not reported Threshold: not reported |
| Igloi 2021 | SARS‐CoV‐2 Rapid Antigen Test (REF No. 9901‐NCOV‐01G; LOT No QCO3020079/Sub:A‐2) (Roche Diagnostics (SD Biosensor)) | CGIA Not stated |
Samples tested: NP (direct swab); trained non‐HCW Timing of sampling: median symptom onset duration 4 d (n = 725): 650 (88%) ≤ 7 d pso (319 (44%) 0‐3 d, 331 (45.7%) 4‐7 d) 75 (10%) ≥ 8 d pso Timing of test: immediate testing Storage: none; tested immediately in batches (5‐10 tests at a time) |
Trained staff; no further detail. Threshold: a 4‐grade scaling readout was utilized representing the strength of the band (++; +; +/‐, ‐) and time till positive results was logged as < 5 min, < 10 min or 15 min |
|
Ishii 2021 [A] Ishii 2021 [B] |
Espline SARS‐CoV‐2 (no product code reported) (Fujirebio Inc., Tokyo, Japan) | Not reported Viral nucleocapsid antigen |
Samples tested: [A] NP (not specified); collection not specified [B] saliva (not specified); collection not specified Timing of sampling: [A]: 10/11 within 2 d pso, 1/11 at 4 d pso [B]: 7/9 within 7 d pso and 2/9 within 8‐14 d pso Timing of test: not specified Storage: unfrozen and fresh |
Not reported Threshold: positive line observed with naked eye |
|
Jaaskelainen 2021 [A] Jaaskelainen 2021 [B] Jaaskelainen 2021 [C] |
[A] Quidel Sofia SARS FIA (Lots used 143489) (Quidel, San Diego, CA) [B] Standard Q COVID‐19 Ag test (Lots used QCO3020105) (SD Biosensor, Republic of Korea) [C] Panbio (Lots used 41ADF024A) (Abbott Diagnostic GmbH, Jena, Germany) All 3 were CE IVD marked SARS‐CoV‐2 RADTs | [A] FIA
[B] and [C] not stated Antigen (nucleocapsid protein) |
Samples tested: NP (saline); collection not specified Timing of sampling: not stated Timing of test: frozen samples Storage: stored at −20 °C |
Not stated; presume laboratory staff Threshold: [A] detection of fluorescent signal [B] and [C] appearance of visible line |
| Jakobsen 2021 | Standard Q COVID‐19 Ag test (no product code reported) (SD BIOSENSOR) | CGIA Not stated |
Samples tested: NP (direct swab); trained non‐HCW Timing of sampling: no details Timing of test: immediate testing Storage: none |
Presume HCW ‐ states "Personnel from the private company Copenhagen Medical A/S" Threshold: not stated; conducted according to SD BIOSENSOR's instruction |
| James 2021 | BinaxNOW COVID‐19 Ag Card tests (BinaxNOW) (no product code reported) (Abbott Diagnostics, Scarborough, ME) | Not stated Nucleocapsid protein antigen |
Samples tested: nasal (AN) (direct swab); collection not specified Timing of sampling: not stated for symptomatic Timing of test: immediate testing Storage: none |
Trained laboratory employees of hospital X Threshold: not stated |
| Kerneis 2021 | Standard Q COVID‐19 Ag test (no product code reported) (SD Biosensor,Chuncheongbuk‐do, Republic of Korea) | Not reported N antigen |
Samples tested: NP (direct swab); HCW Timing of sampling: median d pso (IQR): 3 (IQR 2‐4) Asymptomatic time from last contact 7 (IQR 1‐7) Timing of test: immediate testing Storage: no storage (immediate testing) |
Not reported Threshold: according to manufacturer |
| Kilic 2021 | BD Veritor (no product code reported) (Becton Dickinson, Sparks, Maryland, USA) | Chromatographic Nucleocapsid |
Samples tested: nasal (AN) (direct swab); HCW Timing of sampling: ≤ 5 d pso Timing of test: < 1 h Storage: tested at the site of collection within 1 h of collection |
Not stated; presume on‐site HCW Threshold: BD Veritor analyser device used; no further detail |
|
Kohmer 2021 [A] Kohmer 2021 [B] Kohmer 2021 [C] Kohmer 2021 [D] |
[A] RIDAQUICK SARS‐CoV‐2 Antigen (no product code reported) (R‐Biopharm AG, Darmstadt, Germany) [B] SARS‐CoV‐2 Rapid Antigen Test (no product code reported) (Roche Diagnostics GmbH, Mannheim, Germany) [C] NADAL COVID‐19 Ag Test (no product code reported) (test cassette) (Nal von Minden GmbH, Regensburg, Germany) [D] SARS‐CoV‐2 Ag Test on the LumiraDx Platform (no product code reported) (LumiraDx GmbH, Cologne, Germany) | [1], [2], [3] not stated
[4] immunofluorescence assay Not stated |
Samples tested: NP (saline); collection not specified Timing of sampling: not stated Timing of test: < 24 h delay Storage: sample storage not stated; tested within 24 h after collection |
Not stated Threshold: for [1], [2], [3] the results were read visually and documented by 3 different individuals, and the majority consensus was chosen as the final test result; not stated for [4] (IFU indicates that a reader device is required) |
| Kriemler 2021 | Standard Q COVID‐19 Ag Test (no product code reported) (SD Biosensor/Roche, Switzerland) | CGIA None stated |
Samples tested: buccal (direct swab); collected by trained "personnel" Timing of sampling: essentially asymptomatic testing although some symptoms (runny nose, cough headache etc.) reported Timing of test: immediate testing Storage: none required; direct swab |
Study staff member (experienced in RDT testing) Threshold: visual Judged by 2 study team members (experienced in RDT testing) in agreement as positive or negative and blinded to reported symptoms |
| Kruger 2021 | LumiraDx SARS‐CoV Ag test (no product code reported) (LumiraDx™ Limited, London, UK) | FIA; microfluidic immunofluorescence assay Nucleocapsid protein of SARS‐CoV‐2 |
Samples tested: nasal (NMT) (direct swab); self‐collected Timing of sampling: average symptom duration of 3.9 d (SD 3.2) 423/472 (90%) of symptomatic patients reported onset of symptoms within the prior 7 d Timing of test: immediate testing Storage: no storage |
Laboratory personnel in dedicated workspace (no further details reported) Threshold: according to manufacturer; digital touch screen readout of positive, negative or error (error results re‐tested using same extraction vial with a new test strip) |
| Kruttgen 2021 | SARS‐CoV‐2 Rapid Antigen Test (no product code reported) (Roche, Switzerland) | Not stated Not stated |
Samples tested: NP (VTM); collection not specified Timing of sampling: not stated Timing of test: not specified Storage: not stated; states no intermittent freeze‐thaw cycle so presume no frozen storage |
Laboratory staff Threshold: visual; test and control lines |
| L'Huillier 2021 | Panbio‐COVID‐19 Ag Rapid Test Device (no product code reported) (Abbott Rapid Diagnostics, US) | CGIA Nucleocapsid |
Samples tested: NP (direct swab); HCW Timing of sampling: for symptomatic patients samples were taken at median 2 d pso (IQR 1‐3) Timing of test: immediate testing Storage: none required |
2 members of the study team; blinded to each other and to clinical presentation Threshold: visual; control and test line Any discrepant result was considered positive when any of the above‐mentioned reader set a positive diagnosis. |
| Lambert‐Niclot 2020 | COVID‐19 Ag Respi‐Strip CORIS (no product code) (BioConcept, Gembloux, Belgium) | CGIA SARS‐CoV‐2 NP |
Samples tested: NP (VTM); collection not specified Timing of sampling: not stated Timing of test: not stated (soon after collection) Storage: none; no cooling or freezing step used |
Not stated; presume laboratory staff Threshold: as per manufacturer |
| Lanser 2021 | Panbio COVID‐19 Ag Rapid test (Abbott, Chicago, Illionis) | Not stated Not stated |
Samples tested: NP (direct swab); HCW Timing of sampling: unclear; states "sample taken during their hospital stay in different stages of the disease" Timing of test: immediate testing Storage: none required |
Not stated; probably HCW Threshold: not stated; visual |
| Linares 2020 | Panbio COVID‐19 Ag Rapid Test Device (no product code) (Abbott Rapid Diagnostic Jena GmbH, Jena, Germany) | CGIA (from IFU) Nucleocapsid |
Samples tested: NP (direct); HCW Timing of sampling: ED: 2 d pso (IQR? 1‐5) PC: 4 d pso (IQR? 2‐8) Table 3 reports range of 0‐27 d pso or post COVID‐19 contact, and range of 0‐16 d for d pso for symptomatic cases only Timing of test: not stated; presume immediate onsite testing Storage: not stated |
Not stated; appears to be on‐site immediate testing Threshold: not stated; as per manufacturer |
|
Lindner 2021a [A] Lindner 2021a [B] |
Sstandard Q COVID‐19 Ag Test (no product code reported) (SD Biosensor, Inc. Gyeonggi‐do, Korea; (also being distributed by Roche)) | LFA chromatographic Not stated |
Samples tested: [A] nasal (AN) (direct swab); self‐collected, HCW tested [B] NP (direct swab); HCW collected and tested Timing of sampling: average symptom duration 4.4 d (SD 2.7) (range 1‐14 d for PCR+ve group) Timing of test: immediate testing Storage: none required |
Study physicians Threshold: visual; presence of control test lines, categorized as negative, weak positive, positive and strong positive. Results interpreted by 2 operators, each blinded to the result of the other. The second reader was also blinded to the result from the alternative sampling method |
|
Lindner 2021b [A] Lindner 2021b [B] Lindner 2021b [C] |
Standard Q COVID‐19 Ag Test (no product code reported) (SD Biosensor, Inc. Gyeonggi‐do, Korea; also distributed by Roche) | CGIA Not stated |
Samples tested: [A] nasal (NMT) (direct swab); self‐collected, self‐tested [B] nasal (NMT) (direct swab); self‐collected, HCW‐tested [B] NP (direct swab); HCW‐collected, HCW‐tested Timing of sampling: mean duration of pso 3.4 d (SD 2.0) Timing of test: immediate testing Storage: none required |
[A] Participants tested NMT sample (according to manufacturer IFU); observed without answering questions or providing corrections
[B] Participants tested nasal MT sample; interpretation by study physician
[C] Trained study physician tested and interpreted NP sample
All professional test interpretation was by 2 study physicians, each blinded to the result of the other and to the participant's interpretation of the self‐test. The second reader was also blinded to the corresponding pairs (NMT/NP) of Ag‐RDTs belonging to 1 individual. Threshold: visual The visual read‐out of the antigen test band was categorized as negative, weak positive, positive, or strong positive. The participant interpreted the test result as positive, negative, invalid, or don't know. |
| Liotti 2021 | Standard F COVID‐19 Ag FIA (no product codes reported) (SD Biosensor (Suwon, South Korea)) | FIA NP |
Samples tested: NP (not specified); collection not specified Timing of sampling: not reported Timing of test: within 24 h after collection Storage: samples kept at 4 °C until testing |
Not stated; laboratory staff Threshold: as per manufacturer |
|
Masia 2021 [A] Masia 2021 [B] Masia 2021 [C] |
Panbio COVID‐19 Ag RTD (no product code reported) (Abbott Rapid Diagnostic Jena GmbH, Jena, Germany) | CGIA Nucleocapsid |
Samples tested: [A] NP (direct); HCW [B] nasal (direct); HCW [C] saliva (direct); self collected Timing of sampling: median 3 (Q1 2– Q3 5) d after symptom onset Timing of test: immediate testing Storage: none required |
At primary care centres, qualified nurse
At ED, clinician Threshold: visual; used according to the manufacturer’s instructions |
| Merino 2021 | Panbio RT COVID‐19 Ag Rapid Test Device (no product code reported) (Abbott Diagnostics) | CGIA Nucleocapsid protein |
Samples tested: NP (direct swab); collection not specified Timing of sampling: < 7 d pso or COVID exposure; unclear if collected by HCW or self‐collected Timing of test: immediate testing Storage: immediate |
Trained personnel; physicians and nurses from emergency services trained by microbiology specialists Threshold: according to manufacturer IFU |
| Mertens 2020 | COVID‐19 Ag Respi‐Strip (no product code reported) (Coris BioConcept (Belgium)) | CGIA SARS‐CoV and SARS‐CoV‐2 highly conserved nucleoprotein |
Samples tested: NP (VTM); collection not specified Timing of sampling: not stated Timing of test: not described Storage: not reported |
Laboratory technician Threshold: visible reddish‐purple band appearing at the Test line position (T) |
|
Miyakawa 2021 [A] Miyakawa 2021 [B] Miyakawa 2021 [C] Miyakawa 2021 [D] |
[A] Authors' own developed Ag‐RDT (YCU‐FF LFIA (Ag‐RDT) (no product code reported); now marketed as FUJIFILM COVID‐19 Ag Test fujifilm.com/jp/en/news/hq/358e) [B] Panbio COVID‐19 Ag Rapid Test (no product code reported) (Abbott) [C] Espline SARS‐CoV‐2 (no product code reported) (Fujirebio) [D] Standard Q COVID‐19 Ag (no product code reported) (SD Biosensor) (same result obtained for Roche ‐ RAT; same assay) | [A] CGIA with silver ions
[B]‐[E] not described [A] SARS‐CoV‐2 nucleocapsid protein [B]‐[E] not described |
Samples tested: NP (not specified); collection not specified Timing of sampling: not reported Timing of test: frozen samples Storage: stored at −80 °degrees C until used (timing not reported) |
Not reported; "test line interpretations … made by at least 2 people" Threshold: appearance of visible line |
| Mockel 2021(a) | SARS‐CoV‐2 Rapid Antigen Test (no product code reported) (Roche /SD Biosensor) | Not mentioned Not mentioned |
Samples tested: NP+OP (direct swab); collected by HCW Timing of sampling: not mentioned; time pso only reported for those with FN results: both cohorts, range was 1 to > 7 d Timing of test: immediate testing Storage: immediate |
ED nurse (a core ED team alongside written instructions trained the ED
nurses) Threshold: consensus of ED nurse and one other medical professional |
| Mockel 2021(b) | SARS‐CoV‐2 Rapid Antigen Test (no product code reported) (Roche /SD Biosensor) | Not mentioned Not mentioned |
Samples tested: NP+OP (direct swab); collected by HCW Timing of sampling: not mentioned; time pso only reported for those with FN results: both cohorts, range was 1 to > 7 d Timing of test: immediate testing Storage: immediate |
ED nurse (a core ED team alongside written instructions trained the ED
nurses) Threshold: consensus of ED nurse and one other medical professional |
| Nagura‐Ikeda 2020 | ESPLINE SARS‐CoV‐2 (no product code reported); (FujiRebio Inc) (5 other tests performed including PCR and RT‐LAMP, but not eligible for this review) | LFA (no reader device required) NP |
Samples tested: saliva (direct); self‐collected Timing of sampling: saliva collected on admission to hospital; IPD reports this was median 7 d pso (range 1‐14 d) Timing of test: not stated; frozen samples Storage: stored at −80 °C until sample preparation |
Not stated; implies laboratory staff Threshold: not stated; appearance of test line implied |
| Nalumansi 2020 | Standard Q COVID‐19 Ag Test (no product code reported) (SD Biosensor, Gyeonggi‐do, 16690, Korea) | CGIA Antigen |
Samples tested: nasal (not otherwise specified) (direct); collected by laboratory scientist Timing of sampling: not recorded Timing of test: immediate testing Storage: not required; immediate on‐site testing |
Laboratory staff (training not reported) Threshold: visual; according to the manufacturer’s guidelines |
| Nash 2020 | Direct antigen rapid test (DARTTM); NP‐based (E25Bio Inc (Cambridge MA)) | Immunochromatographic paper‐based (CGIA) NP |
Samples tested: nasal (not otherwise specified) (not specified); collection not specified Timing of sampling: not stated Timing of test: not stated Storage: banked frozen prior to testing |
Not stated; presume lab staff Threshold: visual line |
| Ngo Nsoga 2021 | Panbio Ag RDT (no product code reported) (Abbot ) | CGIA Not mentioned |
Samples tested: OP alone (direct swab); collected by HCW Timing of sampling: samples obtained up to 24 d pso (Figure 1); text reports 101 day 0‐4; 19 day 5‐7; 17 day 8‐11 Mean 4.1 d pso to PCR (range 0‐24 d) Timing of test: not specified Storage: not mentioned |
States "biologist" with a second HCW for equivocal results Threshold: according to manufacturer’s instructions |
|
Nikolai 2021(a) [A] Nikolai 2021(a) [B] |
Standard Q COVID‐19 Ag Test (nasal sampling kit used; RUO at time of study) (no product code reported) (SD Biosensor, Inc. Gyeonggi‐do, Korea (also distributed by Roche in Europe)) | Not reported Not reported |
Samples tested: [A] nasal (AN) (direct swab); collected by HCW [A] nasal (NMT) (direct swab); collected by HCW Timing of sampling: whole sample (n = 228): mean 3.4 d (SD 3.0) Timing of test: immediate testing Storage: directly after sampling |
Study physicians Threshold: semi‐quantitative visual read‐out of the test band (2 independent blinded readers) |
|
Nikolai 2021(b) [A] Nikolai 2021(b) [B] |
[A] Standard Q COVID‐19 Ag Test (no product code reported) (nasal sampling kit; RUO) (SD Biosensor, Inc. Gyeonggi‐do, Korea (also distributed by Roche in Europe)) [B] Standard Q COVID‐19 Ag Test (NP sampling kit) (no product code reported) (SD Biosensor, Inc. Gyeonggi‐do, Korea (also distributed by Roche in Europe)) | Not reported Not reported |
Samples tested: [A] nasal (NMT) (direct swab); self‐collected [B] NP (direct swab); HCW collected Timing of sampling: whole sample (n = 228): mean 3.4 d (SD 3.0) Timing of test: immediate testing Storage: directly after sampling |
Study physicians Threshold: semi‐quantitative visual read‐out of the test band (2 independent blinded readers) |
| Okoye 2021 | BinaxNOW COVID‐19 antigen card (no product code reported) (Abbott) | Not reported SARS‐CoV‐2 nucleocapsid antigen |
Samples tested: nasal (NMT) (direct swab); self‐collected Timing of sampling: not applicable (asymptomatic) Timing of test: immediate testing Storage: immediate testing |
Trained non‐medical personnel (University of Utah Hope Corps interns) according to the manufacturer IFU Threshold: 2 pink/purple lines observed |
|
Olearo 2021 [A] Olearo 2021 [B] Olearo 2021 [C] Olearo 2021 [D] |
[A] SARS‐CoV‐2 Rapid Antigen Test (no product code reported) (Roche) (SD Biosensor (Roche Diagnostics), South Korea) [B] COVID‐19 Rapid Test Device (no product code reported) (Abbott) (Panbio Ltd. (Abbott Rapid Diagnostics), Australia) [C] MEDsan SARSCoV‐2 Antigen Rapid Test (no product code reported) (MEDsan GmbH, Germany) [D] CLINITEST Rapid COVID 19 Antigen Test (no product code reported) (Zhejiang Orient Biotech Co, China) | CGIA Nucleocapsid |
Samples tested: NP or OP (VTM); collected by laboratory scientist Timing of sampling: median 6 (IQR 2–12) d from symptom onset Timing of test: not specified Storage: not reported |
Lab technicians; swabs supplied with the antigen kits were immersed in patient OP/NP samples for approximately 10 s before all further steps of the tests were carried out
according to manufacturer IFU Threshold: visual |
|
Osterman 2021(a) [A] Osterman 2021(a) [B] |
[A] Standard F COVID‐19 Ag FIA (no product code reported) (SD Biosensor) [B] SARS‐CoV‐2 Rapid Antigen Test (RAT) (no product code reported) (Roche Diagnostics); extracted as SD Biosensor/Roche ‐ Standard Q COVID‐19 Ag | [A] FIA
[B] CGIA Both nucleocapsid |
Samples tested: NP or OP (VTM or other); collected by HCW Timing of sampling: not mentioned Timing of test: < 24 h delay Storage: same day Site (1): were either kept at room temperature for 1–2 h (“fresh”) (n = 18); stored at 4 °C for 0–7 d (n = 48); or stored at − 20 °C (n = 315) until SARS‐CoV‐2 antigen testing was performed site (2) PCR and Ag testing (RAT) were performed on the day of submission of freshly obtained swabs |
Laboratory personnel according to manufacturer’s instructions Threshold: [A] a cut of index (COI) ≥ 1 was interpreted as positive after 30 min [B] every visible (even if very faint or not uniform) test line was interpreted as positive after 15 or 30 min. |
| Osterman 2021(b) | SARS‐CoV‐2 Rapid Antigen Test (no product code reported) (RAT) (Roche Diagnostics); extracted as SD Biosensor/Roche ‐ Standard Q COVID‐19 Ag | [A] FIA
[B] CGIA Both nucleocapsid |
Samples tested: NP (VTM or other); collected by HCW Timing of sampling: not mentioned Timing of test: < 24 h delay Storage: same day Site (1): were either kept at room temperature for 1–2 h (“fresh”) (n = 18); stored at 4 °C for 0–7 d (n = 48); or stored at − 20 °C (n = 315) until SARS‐CoV‐2 antigen testing was performed site (2) PCR and Ag testing (RAT) were performed on the day of submission of freshly obtained swabs |
Laboratory personnel according to manufacturer’s instructions Threshold: [A] a cut‐off index (COI) ≥ 1 was interpreted as positive after 30 min [B] every visible (even if very faint or not uniform) test line was interpreted as positive after 15 or 30 min. |
| Parada‐Ricart 2020 | SARS‐CoV‐2 (2019‐n‐CoV Ag Test Flourescence IC Assay) (no product code reported) (Shenzen Bioeasy Biotechnology Co LTD) | FIA Nucleocapsid |
Samples tested: nasal (not otherwise specified) (direct swab); collection not specified Timing of sampling: < 7 d pso Timing of test: immediate testing Storage: none (tested within 30 min) |
Not mentioned Threshold: as per manufacturer |
| Pena 2021 | Standard Q COVID‐19 Ag (catalogue number 9901‐NCOV‐01G) (SD Biosensor, Inc. Republic of Korea) | LFA chromatographic Not stated |
Samples tested: NP (direct swab); collected by HCW Timing of sampling: all described as asymptomatic; participants were asked about any symptoms in previous 0‐14 d, but no information on responses was reported Timing of test: immediate testing Storage: none required (analysed according to the manufacturer’s instructions) |
Not stated but implies HCW at time of sampling Threshold: visual‐coloured bands |
| Pena‐Rodriguez 2021 | Standard Q COVID‐19 (no product code reported) (SD BIOSENSOR) | Chromatographic N gene |
Samples tested: NP (direct swab); trained non‐HCW Timing of sampling: not stated Timing of test: immediate testing Storage: none; "SARS‐CoV‐2 antigen analysis was carried out in the place" |
Trained staff Threshold: visual control and test lines The test was invalidated when no marks were detected |
|
Perez‐Garcia 2021 [A] Perez‐Garcia 2021 [B] |
[A] CerTest SARS‐CoV‐2 Ag One Step Card Test (Batch code SC‐004) (Certest Biotec S.L., Zaragoza, Spain) [B] Panbio COVID‐19 Ag Rapid Test Device (Batch code 41ADF057A) (Abbot Rapid Diagnostics GmbH, Jena, Germany) | [A] LFA
[B] CGIA Both nucleoprotein antigens |
Samples tested: NP (VTM); collection not specified Timing of sampling: reported for 128 PCR+ve Samples: 46 (36%) < 5 d pso; 55 (43%) day 6‐10; 27 (21%) > 10 d Timing of test: frozen samples Storage: samples were cryopreserved at −20 ◦C until their analysis by Ag‐RDTs |
Not stated (lab‐based) Threshold: visual; control and test lines |
|
Peto 2021(a) [A] Peto 2021(a) [B] Peto 2021(a) [C] Peto 2021(a) [D] Peto 2021(a) [E] Peto 2021(a) [F] Peto 2021(a) [G] |
[A] Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test (no product code reported) (Innova Medical Group) [B] Panbio COVID‐19 Ag Rapid Test Device (no product code reported) (Abbott) [C] Coronavirus Ag Rapid Test Cassette (no product code reported) (Zhejiang Orient Gene Biotech) [D] Anhui Deepblue Medical Technology COVID‐19 (no product code reported) (Deepblue) [E] Fortress Diagnostics Coronavirus Ag Rapid Test (no product code reported) (Fortress) [F] Standard Q COVID‐19 Ag Test (no product code reported) (SD Biosensor) [G] Surescreen Diagnostics SARS‐CoV‐2 Antigen Rapid Test Cassette (no product code reported) (Surescreen) | Not stated Not stated |
Samples tested: nasal (AN) + OP (direct); collected by HCW Timing of sampling: not stated Timing of test: frozen samples Storage: frozen |
Laboratory staff Threshold: visual line; as per manufacturer |
| Peto 2021(b) [non‐HCW tested] | Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test (no product code reported) (Innova Medical Group) | CGIA (from IFU) Not stated |
Samples tested: nasal+OP (direct); self‐collected Timing of sampling: not stated Timing of test: immediate testing Storage: none |
On‐site; self‐trained non‐HCW Threshold: visual line; as per manufacturer |
| Peto 2021(c) [A ‐ HCW tested] | [A] Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test (no product code reported) (Innova Medical Group) | LFA Not stated |
Samples tested: nasal (AN) + OP (direct); self‐collected Timing of sampling: not stated Timing of test: immediate testing Storage: immediate testing by HCW (assay [A]); transport to PHE for second set of samples (assay [A]‐[D]), tested within 24 h of collection |
Immediate testing by HCW (assay [A]); transport to PHE for second set of samples (assay [A]‐[D]) Threshold: visual line; as per manufacturer An invalid kit result, or a kit failure was recorded when an operator did not see a control line on the device within a defined time period |
|
Peto 2021(c) [A ‐ Lab tested] Peto 2021(c) [B ‐ Lab tested] Peto 2021(c) [C ‐ Lab tested] Peto 2021(c) [D ‐ Lab tested] |
Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test (no product code reported) (Innova Medical Group) [B] Panbio COVID‐19 Ag Rapid Test Device (no product code reported) (Abbott) [C] Coronavirus Ag Rapid Test Cassette (no product code reported) (Zhejiang Orient Gene Biotech) [D] Anhui Deepblue Medical Technology COVID‐19 (no product code reported) (Deepblue) |
CGIA (from IFU) Not stated |
Samples tested: nasal (AN) + OP (direct); self‐collected Timing of sampling: not stated Timing of test: immediate testing Storage: none |
Laboratory scientist at PHE Threshold: visual line; as per manufacturer |
| Peto 2021(d) | Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test (no product code reported) (Innova Medical Group) | CGIA (from IFU) Not stated |
Samples tested: OP alone (direct); self‐collected Timing of sampling: not stated Timing of test: immediate testing Storage: none |
Not stated; presumably laboratory scientist at PHE or John Radcliffe Hospital, Oxford Threshold: visual line; as per manufacturer |
| PHE 2020 | Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test (no product code reported) (Innova Medical Group) | CGIA (from IFU) Not stated |
Samples tested: OP alone (VTM); self‐collected Timing of sampling: 1 week after outbreak; no further details Timing of test: not stated Storage: transported at 4 °C to Porton Down for testing |
Laboratory staff Threshold: visual line; as per manufacturer |
|
Pickering 2021(a) [A] Pickering 2021(a) [B] Pickering 2021(a) [C] Pickering 2021(a) [D] Pickering 2021(a) [E] Pickering 2021(a) [F] |
[A] Innova (no product code reported) (Innova Med Group, China) [B] E25 Bio (no product code reported) (E25 Bio, USA) [C] Sure Screen V (COVID19 AGVCT) Sure Screen Diagnostics Ltd) [D] Spring (SP‐SW 106) (Spring Healthcare, UK) [E] Encode (no product code reported) (Encode/Emmo Pharma) [F] Sure Screen F (COVID19 AGC) (Sure Screen Diagnostics Ltd) | [A], [B], [D] CGIA
[C], [E] LFA (not otherwise specified)
[F] FIA All nucleocapsid; "Given that the rapid antigen tests rely on antibody detection of SARS‐CoV‐2 nucleocapsid (N)" |
Samples tested: nasal+OP (VTM); collection not specified Timing of sampling: not stated Timing of test: frozen samples Storage: stored at −80 °C at the Directorate of Infection, prior to selection and forwarding to KCL laboratories for analysis (50 µL of stored swab was mixed with 100 µL of buffer supplied with the test kit, and 100 µL of this was applied to the test cassette) |
Not specified; lab‐based Threshold: visual and according to manufacturer instructions; results recorded independently by 2 readers and discordant results referred to a 3rd individual. CGIA tests scored according to whether the test band was strongly positive (2), clearly positive (1), weakly positive (0.5) or negative (0) |
|
Pickering 2021(b) [A] Pickering 2021(b) [B] Pickering 2021(b) [C] |
[A] Innova (no product code reported) (Innova Med Group, China) [B] Encode (no product code reported) (Encode/Emmo Pharma [C] Sure Screen F (COVID19 AGC) (Sure Screen Diagnostics Ltd) | [A] CGIA
[B] LFA (not otherwise specified)
[C] FIA All nucleocapsid; "Given that the rapid antigen tests rely on antibody detection of SARS‐CoV‐2 nucleocapsid (N)" |
Samples tested: nasal+OP (VTM); collection not specified Timing of sampling: not stated Timing of test: frozen samples Storage: stored at −80 °C at the Directorate of Infection, prior to selection and forwarding to KCL laboratories for analysis (50 µL of stored swab was mixed with 100 µL of buffer supplied with the test kit, and 100 µL of this was applied to the test cassette) |
Not specified; lab‐based Threshold: visual and according to manufacturer instructions; results recorded independently by 2 readers and discordant results referred to a 3rd individual. CGIA tests scored according to whether the test band was strongly positive (2), clearly positive (1), weakly positive (0.5) or negative (0) |
|
Pickering 2021(c) [A] Pickering 2021(c) [B] |
[A] Innova (no product code reported) (Innova Med Group, China) [B] Sure Screen V (no product code reported) (COVID19 AGVCT) (Sure Screen Diagnostics Ltd) | [A] CGIA [B] LFA (not otherwise specified) All nucleocapsid; "Given that the rapid antigen tests rely on antibody detection of SARS‐CoV‐2 nucleocapsid (N)" |
Samples tested: nasal+OP (VTM); collection not specified Timing of sampling: not stated Timing of test: frozen samples Storage: stored at −80 °C at the Directorate of Infection, prior to selection and forwarding to KCL laboratories for analysis (50 µL of stored swab was mixed with 100 µL of buffer supplied with the test kit, and 100 µL of this was applied to the test cassette) |
Not specified; lab‐based Threshold: visual and according to manufacturer instructions; results recorded independently by 2 readers and discordant results referred to a 3rd individual. CGIA tests scored according to whether the test band was strongly positive (2), clearly positive (1), weakly positive (0.5) or negative (0) |
| Pilarowski 2020a | BinaxNOW (no product code reported) (Abbott) | CGIA Not reported |
Samples tested: nasal (AN) (direct); collected by laboratory scientist timing of sampling; symptomatic with positive Ag result: median 3 d (IQR 2‐5 d) pso (n = 134) Timing of test: immediate testing Storage: none used |
Laboratory technicians (certified technician readers) Threshold: visual; according to manufacturer instructions |
| Pilarowski 2021 | BinaxNOW COVID‐19 Ag Card (no product code reported) (Abbott Laboratories) | CGIA N protein |
Samples tested: nasal (AN) (direct); collected by laboratory scientist Timing of sampling: mainly asymptomatic; timing not systematically reported for symptomatic group Timing of test: not specified Storage: none reported |
Laboratory technician; on site Threshold: visual colour band; each assay was read by 2 independent observers, and a site supervisor served as a tiebreaker. Interpretation amended following first 217 samples because of high FP result (9/207 PCR‐veve); bands were subsequently scored as positive only if they extended across the full width of the strip, irrespective of the intensity of the band |
| Pollock 2021a | BinaxNOW COVID‐19 Ag Card (no product code reported) (Abbott Diagnostics, USA) | CGIA (by knowledge) Nucleocapsid protein antigen (by knowledge) |
Samples tested: nasal (AN) (direct swab); collected by trained "personnel" Timing of sampling: for symptomatic: adults: median 3 (IQR 2‐5); children: median 2 (IQR 1‐4) d Timing of test: immediate testing Storage: none; testing within 1 h of collection "tests initiated within an hour of collection time at the temperature of 59°F as per manufacturer's recommendation." |
Trained operators Threshold: visual read‐out; test lines recorded as 'faint', 'medium', or 'strong' There was no attempt to resolve any discordance between the 2 readers |
| Pollock 2021b | CareStart COVID‐19 Antigen test (no product code reported) (Access Bio) | Chromatographic immunoassay Nucleocapsid |
Samples tested: nasal (AN) (direct swab); collected by trained "personnel" Timing of sampling: 209 symptomatic adults: median 3 d pso (IQR 2‐6) 32 symptomatic children: median 3 d pso (IQR 2‐4) Timing of test: immediate testing Storage: tested within 1 h of collection; median interval between sample collection and test initiation was 31 min (range 12–103 min) |
Trained operators (Master’s or PhD level laboratorians); according to the manufacturer IFU Threshold: visual; according to the manufacturer IFU 2 operators read the result; first read of each test was the official result used |
| Porte 2020 | Diagnostic Kit for 2019‐Novel Coronavirus (2019‐nCoV) Ag Test (Cat. N° YRLF04401025, lot N° 2002N408) (Bioeasy Biotechnology Co., Shenzhen, China ) | CGIA SARS‐CoV‐2 nucleocapsid protein |
Samples tested: NOP (VTM); collected by trained "personnel" Timing of sampling: not stated Timing of test: within 48 h collection Storage: kept at 4 °C and tested within 48 h |
Laboratory technician Threshold: as per manufacturer |
|
Porte 2021 [A] Porte 2021 [B] |
[A] SOFIA SARS Antigen FIA (no product code reported) Quidel Corporation, San Diego, CA, USA [B] Standard F COVID‐19 Ag FIA (no product code reported) (SD Biosensor Inc., Gyeonggi‐do, Republic of Korea) | Both FIA NP |
Samples tested: NP+OP (VTM); collected by trained "personnel" Timing of sampling: all < 5 d pso; median PCR+ve: 2 d (IQR 1‐3) PCR‐ve: 1 day (IQR 0.75‐4) Timing of test: not stated; frozen samples Storage: stored at −80 °C following PCR |
Laboratory staff Threshold: as per manufacturer; both using analyser device |
| Pray 2021 | Sofia SARS Antigen Fluorescent Immunoassay (FIA) (no product code reported) (Quidel Corporation) | FIA Not stated |
Samples tested: nasal (NMT) (direct swab); mixed Timing of sampling: median 3 d pso (IQR 1‐6 d; 7.5% missing) 152 (72.4%) reported ≤ 5 d pso to specimen collection Timing of test: immediate testing Storage: none; immediate on‐site testing |
Presume same healthcare personnel at university A; not reported for university B Threshold: not stated; as per manufacturer |
| Prince‐Guerra 2021 | BinaxNOW COVID‐19 Ag Card (BinaxNOW) (no product code reported) (Abbott Diagnostics, USA) | CGIA Nucleocapsid |
Samples tested: nasal (AN) (direct swab); collected by HCW Timing of sampling: day 0‐14; symptomatic: median pso 4 d (range 0‐210); 662 (19%) ≤ 7 d; 161 (5%) > 7 d Timing of test: immediate testing Storage: none; immediately tested on‐site |
Healthcare professionals Threshold: not stated; visual |
| Ristic 2021 | Standard Q COVID‐19 Ag Test (no product code reported) (SD Biosensor, Gyeonggi‐do, South Korea ) | Chromatographic immunoassay Not stated |
Samples tested: NP (direct swab); collected by HCW Timing of sampling: median time from symptom to swab (9.4 d, range 1‐45 d; 63 (53%) within first 5 d) Timing of test: immediate testing Storage: no storage |
Trained medical staff Threshold: visual interpretation; double lines |
| Rottenstreich 2021 | NowCheck COVID‐19 Ag Test (no product code reported) (Bionote Inc, Hwaseong‐si, Republic of Korea) | Not reported Not reported |
Samples tested: NP (not specified); collected by HCW Timing of sampling: on admission Timing of test: not specified Storage: not reported |
Not reported Threshold: not reported |
|
Saeed 2021 [A] Saeed 2021 [B] |
A] NP based RDT (#20CG2701X) (Lepu Medical, China) B] Saliva based RDT (#901101) (Lepu Medical, China) | CGIA N gene |
Samples tested: NP (not specified); collected by trained personnel Timing of sampling: no details Timing of test: not specified Storage: not stated |
Not stated Threshold: visual colour lines; performed according to standard manufacturer protocol |
| Salvagno 2021 | SARS‐CoV‐2 Rapid Antigen Test (no product code reported) (Roche) | CGIA Not stated |
Samples tested: NP (not specified); collection not specified Timing of sampling: not stated Timing of test: not specified Storage: not stated |
Not stated Threshold: visual line |
|
Schildgen 2021 [A] Schildgen 2021 [B] Schildgen 2021 [C] |
[A] BIOCREDIT (no product code reported) (RapiGEN) [B] Panbio (no product code reported) (Abbott) [C] SARS‐CoV‐2 Rapid Antigen test (no product code reported) (Roche) | All CGIA Not stated |
Samples tested: BAL or TW (not specified); collection not specified Timing of sampling: not stated Timing of test: not stated Storage: not stated |
Not stated; presume laboratory staff Threshold: as per manufacturer |
| Schuit 2021(a) | BD Veritor System for Rapid Detection of SARS‐CoV‐2 Ag‐RDT (no product code reported) (BD Veritor, Franklin Lakes, NJ, USA) | Unknown; CGIA Nucleocapsid |
Samples tested: nasal+OP (direct swab); collected by trained non‐HCW Timing of sampling: median 5 d (IQR 5‐5) between contact and sampling, range 0‐13 d symptomatic (n = 219), symptoms developed on day of test 17 (7.8%), 1 d prior 64 (29.2%), 2 d prior 51 (23.3%), 3 d prior 83 (37.9%) Timing of test: frozen samples Storage: frozen at −20 °C within 30 min of collection; transported to Microvida location Amphia laboratory. Thawed and tested within 6 h of collection |
Trained laboratory technician; result confirmed by a second person Threshold: visual interpretation; Analyser was not used |
| Schuit 2021(b) | SARS‐CoV‐2 Rapid Antigen Test (no product code reported) (Roche/SD Biosensor, Basel, Switzerland) | CGIA Nucleocapsid |
Samples tested: NP (direct swab); collected by trained non‐HCW Timing of sampling: median 5 d (IQR 5‐5) between contact and sampling, range 0‐11 d symptomatic (n = 158), symptoms developed on day of test 14 (8.9%), 1 d prior 37 (23.4%), 2 d prior 39 (24.7%), 3 d prior 45 (28.5%) Timing of test: immediate testing Storage: conducted immediately on site |
Not stated; performed independently by 2 people Threshold: visual interpretation |
| Schwob 2020(a) | Standard Q COVID‐Ag Test (SD Biosensor) See Schwob 2020(b) and Schwob 2020(c) for data for Panbio COVID‐19 Ag Test (Abbott) and COVID‐VIRO (AAZ) (SD Biosensor/Roche) | Lateral flow; no further information Nucleocapsid |
Samples tested: NP (direct swab); collected by HCW Timing of sampling: pso (mean duration of symptoms at the time of swab collection/testing was 2.6 d (SD 2.3, range 0‐30)) Timing of test: immediate testing Storage: no storage, immediate |
Same health professional who collected the swab Threshold: visual colour change |
| Schwob 2020(b) | Panbio COVID‐19 Ag Test (Abbott) See Schwob 2020(a) and Schwob 2020(c) for data for Standard Q (SD Biosensor) and COVID‐VIRO (AAZ) (Abbott) | Lateral flow; no further information Nucleocapsid |
Samples tested: NP (direct swab); collected by HCW Timing of sampling: pso (mean duration of symptoms at the time of swab collection/testing was 2.6 d (SD 2.3, range 0‐30)) Timing of test: immediate testing Storage: no storage, immediate |
Health professional Threshold: visual colour change |
| Schwob 2020(c) | COVID‐VIRO See Schwob 2020(a)) and Schwob 2020(b) for data for Standard Q (SD Biosensor) and Panbio COVID‐19 Ag Test (Abbott) (AAZ‐LMB) | Lateral flow; no further information Nucleocapsid |
Samples tested: NP (direct swab); collected by HCW Timing of sampling: pso (mean duration of symptoms at the time of swab collection/testing was 2.6 d (SD 2.3, range 0‐30)) Timing of test: immediate testing Storage: no storage, immediate |
Health professional Threshold: visual colour change |
| Scohy 2020 | COVID‐19 Ag Respi‐Strip (product code not reported) (Coris Bioconcept) | CGIA NP |
Samples tested: NP (not specified); collection not specified Timing of sampling: not reported Timing of test: not stated; immediate or after period of storage Storage: none or stored at 4 °C until the test |
Not stated Threshold: visual appearance of T line; also states that "2 versions of the test were evaluated. On the second version, conjugate was coupled on a different way and the control line was optimized." |
|
Shidlovskaya 2021 [A] Shidlovskaya 2021 [B] |
[A] SGTI‐flex COVID‐19 Ag (product code not reported) (Sugentech Inc., Korea) [B] BIOCREDIT COVID‐19 Ag (product code not reported) (RapiGEN Inc., Korea) | [A] LFA
[B] CGIA [A] Nucleocapsid [B] SARS‐COV2 antigen |
Samples tested: NP (direct swab); collected by HCW Timing of sampling: between 2‐10 d pso Timing of test: immediate testing Storage: none required; tested directly at the patient bedside |
Nurses Threshold: visual |
| Shrestha 2020 | BIOCREDIT (product code not reported) (RapiGen) | Not stated Not stated |
Samples tested: NP (direct); collection not specified Timing of sampling: d 5 of quarantine Timing of test: not stated Storage: none reported; other sample from the same individual was processed for the results as instructed by the manufacturing company of antigen kit |
Lab technician (trained) Threshold: visual line; as per manufacturer |
|
Stohr 2021 [A] Stohr 2021 [B] |
[A] BD Veritor System for Rapid Detection of SARS‐CoV‐2 (product code not reported) (Becton Dickinson, USA) [B] SARS‐CoV‐2 antigen detection test (product code not reported) (Roche) | CGIA Nucleocapsid |
Samples tested: nasal (NMT) (direct swab); self‐collected Timing of sampling: not stated Timing of test: immediate testing Storage: no storage |
Self‐tested; written and illustrated booklet provided along with QR‐code link to a 2‐min online video illustrating mid turbinate self‐sampling and self‐testing Threshold: visual |
|
Stokes 2021(a) [A] Stokes 2021(a) [B] Stokes 2021(a) [C] |
Panbio (product code not reported) (Abbott, IL, USA) | CGIA Nucleocapsid |
Samples tested: [A] NP (direct swab); collected by HCW [B] OP (direct swab); collected by HCW [C] saliva (direct); self‐collected Timing of sampling: mean duration of symptoms 6.1 d (median 6.0, range 3.0–10.0 d); 91% were ≤ 7 day pso Timing of test: immediate testing Storage: no storage |
States tested immediately, so presume same HCW Threshold: visual line |
| Stokes 2021(b) | Panbio (product code not reported) (Abbott, IL, USA) | CGIA Nucleocapsid |
Samples tested: NP (direct swab); collected by HCW Timing of sampling: not reported; all < 7 d Timing of test: immediate testing Storage: no storage |
States tested immediately, so presume same nurse Threshold: visual line |
| Stromer 2020 | NADAL COVID‐19 Ag Test (product code not reported); data for Abbott Panbio rapid test excluded] (Nal von Minden GmbH) | Unknown Nucleoprotein |
Samples tested: NP (saline); collection not specified Timing of sampling: not stated Timing of test: not specified Storage: not stated |
Not stated Threshold: not stated |
| Takeda 2020 | ESPLINE SARS‐CoV‐2 (no product code reported) (Fujirebio Inc) | LFA using alkaline phosphatase (ALP)‐labelled antibodies SARS‐CoV‐2 antigen (from IFU) |
Samples tested: NP (not specified); collection not specified Timing of sampling: not stated but all cases are 1st samples presumed by authors to be from patient suspected of SARS‐CoV‐2 for the first time; negative samples were "probably … from … COVID‐19 patients for monitoring purposes and to check for negative conversion" Timing of test: not stated Storage: swabs mixed with sample treatment solution; no storage reported |
Not stated; laboratory staff presumed Threshold: visual line, as per manufacturer |
| Takeuchi 2021a | QuickNavi COVID‐19 Ag (product code not reported) (Denka Co., Ltd., Tokyo, Japan) | Lateral flow, no further details Not stated |
Samples tested: NP (direct swab); collection not specified Timing of sampling: median d pso 2, IQR 1‐4 Timing of test: immediate testing Storage: immediate, no storage |
Not stated; states "examiner" Threshold: visual interpretation |
| Takeuchi 2021b | QuickNavi‐COVID19 Ag (product code not reported) (Denka Co., Ltd., Tokyo, Japan) | Not stated Not stated |
Samples tested: nasal (AN) (direct swab); collection not specified Timing of sampling: median 2.0 d pso (IQR 1.0‐3.0) Timing of test: immediate testing Storage: none; tested immediately |
Not stated; "examiner" Threshold: visual interpretation |
|
Thommes 2021 [A] Thommes 2021 [B] Thommes 2021 [C] Thommes 2021 [D] |
[A] Panbio COVID‐19 Ag Rapid test (product code not reported) (Abbott, Chicago, Illinois) [B] Novel Coronavirus (2019‐nCov) Antigen Detection Kit (product code not reported) (CLMSRDL, Sichuan Mass Spectrometry Biotechnology Co., Ltd, Chengdu, Sichuan) [C] DIAQUICK COVID‐19 Ag Cassette (product code not reported) (DIALAB, Wiener Neudorf, Austria) [D] SARS‐CoV‐2 Rapid Antigen Test (product code not reported) (Roche Diagnostics Deutschland GmbH, Mannheim, Germany) | LFA Not stated |
Samples tested: NP (direct swab); collected by trained "personnel" Timing of sampling: not stated Timing of test: immediate testing Storage: none |
Performed by expert staff at the bedside using swabs provided in the antigen test kits Threshold: visual line |
| Toptan 2021(a) | Not stated; presumed to be RIDA‐QUICK SARS‐CoV‐2 Antigen assay (product code not reported) (R‐Biopharm) | Not stated Not stated |
Samples tested: NP+OP (not specified); collection not specified Timing of sampling: not stated Timing of test: < 24 h delay Storage: stored in 2 mL of PBS at 4 ℃, processed within 24 h |
Not stated Threshold: evaluated visually with 4 or 6 eye principle |
| Toptan 2021(b) | Not stated; presumed to be RIDA‐QUICK SARS‐CoV‐2 Antigen assay (product code not reported) (R‐Biopharm) | Not stated Not stated |
Samples tested: NP (not specified); collection not specified Timing of sampling: not stated Timing of test: < 24 h delay Storage: stored in 2 mL of PBS at 4 ℃, processed within 24 h |
Not stated Threshold: evaluated visually with 4 or 6 eye principle |
| Torres 2021a | Panbio COVID‐19 Ag Rapid Test Device (product code not reported) (Abbott (Diagnostic GmbH, Jena, Germany)) | Lateral flow, no further information Not stated |
Samples tested: NP (direct swab); collected by HCW Timing of sampling: timing was prescribed at the discretion of either the physician in charge of the index case or local health authorities: ‐ household contacts median 2 d (range 1‐7d) after diagnosis of the presumed index case ‐ non‐household contacts median 6 d (range, 1‐7d) after self‐reported exposure Timing of test: immediate testing Storage: immediate, no storage |
Not stated; may be same nurse "carried out at POC immediately after sampling" Threshold: not stated; as per manufacturer |
| Torres 2021b | CLINITEST Rapid COVID‐19 Antigen Test (reported elsewhere to be the same as the Healgen Coronavirus Ag Rapid Test Cassette) (product code not reported) (Siemens, Healthineers, Erlangen, Germany) | Not reported SARS‐CoV‐2 nucleocapsid protein |
Samples tested: NP (direct swab); collected by HCW Timing of sampling: median (range) pso or post diagnosis of index case Symptomatic 3 (1‐5) d Asymptomatic household contacts 4 (0‐7) d Asymptomatic non‐household contacts 5 (2‐7) d Timing of test: immediate testing Storage: immediate testing |
Not reported; appears to be same nurse Threshold: according to manufacturer |
| Turcato 2021 | Standard Q COVID‐19 Ag (R‐Ag) kit (product code not reported) (SD BIOSENSOR, KR) | CGIA Not stated |
Samples tested: not specified (not specified); collection not specified Timing of sampling: not stated Timing of test: not specified Storage: not reported; states "implementation … in the initial screening" |
Not reported Threshold: not reported; as per manufacturer |
|
Van der Moeren 2021(a) [A] Van der Moeren 2021(a) [B] |
BD Veritor System for Rapid Detection of SARS‐CoV‐2 (product code not reported) (Becton Dickinson) [A] Interpreted using reader device [B] Visual interpretation without reader device |
CGIA (from IFU) NP |
Samples tested: NP+OP (direct); collected by trained non‐HCW Timing of sampling: not reported; on presentation Time pso only provided for PCR+ve cases: 12 < 7d; 1 ≥ 7d; 4 no pso data Timing of test: within 6 h (at lab) Storage: stored dry in sterile test tubes and stored and transported on dry ice until processing at the laboratory; tested within 6 h after collection |
Trained laboratory technicians Threshold: [A] using analyser [B] visual inspection |
| Van der Moeren 2021(b) | BD Veritor System for Rapid Detection of SARS‐CoV‐2 (product code not reported) (Becton Dickinson) | CGIA (from IFU) NP |
Samples tested: NP+OP (direct); collected by trained non‐HCW Timing of sampling: not reported; on presentation Timing of test: within 6 h (at lab) Storage: stored dry in sterile test tubes and stored and transported on dry ice until processing at the laboratory; tested within 6 h after collection |
Trained laboratory technicians Threshold: [A] using analyser [B] visual inspection |
| Veyrenche 2021 | Coris COVID‐19 Ag Respi‐Strip (product code not reported) (BioConcept, Gembloux, Belgium) | CGIA NP |
Samples tested: NP (VTM); collection not specified Timing of sampling: day 1‐20 pso, median Ct ≤ 25, 7 (4‐10; presume this is IQR but could be range ‐ is described as SD in paper) Ct 25‐35, 8 (4‐12) Ct ≥ 35, 11 (7‐15) Timing of test: not stated Storage: not stated; PCR conducted prospectively within a few hours but not reported for Ag testing |
All tests were performed in the Virology laboratory Threshold: visual, as per manufacturer |
| Villaverde 2021 | Panbio (product code not reported) (Abbott Rapid Diagnostic) | CGIA Nucleocapsid |
Samples tested: NP (direct swab); collected by HCW Timing of sampling: all ≤ 5 d pso Timing of test: immediate testing Storage: none |
Attending paediatricians and nurse staff Threshold: visual line |
|
Weitzel 2020 [A] Weitzel 2020 [B] Weitzel 2020 [C] Weitzel 2020 [D] |
[A] Biocredit COVID‐19 Ag One Step SARS‐CoV‐2 Antigen Test (product code not reported) (RapiGEN Inc., Anyang‐si, Gyeonggi‐do, Republic of Korea) [B] COVID‐19 Antigen Rapid Test Device StrongStep® COVID‐19 Antigen Test (product code not reported) (Liming Bio‐Products Co., Jiangsu, China) [C] Huaketai New Coronavirus (SARS‐CoV‐2) N Protein Detection Kit (product code not reported) (Fluorescence immunochromatography) (Savant Biotechnology Co., Beijing, China), [D] Diagnostic Kit for 2019‐Novel Coronavirus (2019‐nCoV) Ag Test (product code not reported) (Fluorescence Immunochromatographic Assay) (Bioeasy Biotechnology Co., Shenzhen, China) | CGIA Not reported in study |
Samples tested: NOP (VTM); collection not specified Timing of sampling: not stated Timing of test: not described Storage: stored at −80 °C |
Single trained laboratory technician under BSL2 cabinet; visual outputs read by 2 independent observers with referral to 3rd if needed Threshold: as per manufacturer; Savant test required use of manufacturer supplied UV torch due to unavailability of reader device in Chile |
| Yokota 2020(a) | Espline SARS‐CoV‐2 (product code not reported) (Fujirebio, Tokyo, Japan) | Immunochromatographic assay Not reported |
Samples tested: NP (unclear); collected by laboratory scientist Timing of sampling: median time of sampling was 9 d (range, 2‐14 d) pso Timing of test: frozen samples Storage: frozen samples; no further details |
Probably laboratory staff Threshold: according to the manufacturer IFU |
| Yokota 2020(b) | ESPLINE SARS‐CoV‐2 (product code not reported) (Fujirebio, Tokyo, Japan) | Immunochromatographic assay Not reported |
Samples tested: saliva (unclear); collected by laboratory scientist Timing of sampling: median time of sampling was 9 d (range, 2‐14 d) pso Timing of test: frozen samples Storage: frozen samples; no further details |
Probably laboratory staff Threshold: according to the manufacturer IFU |
| Young 2020 | BD Veritor SARS‐CoV‐2 antigen test (no product codes) (Becton, Dickinson and Company, BD Life Sciences—Integrated Diagnostic Solutions, San Diego, CA) | LFA (not otherwise specified) NP |
Samples tested: NP+OP (direct); collected by HCW Timing of sampling: all ≤ 7 d pso; median 3.0 d, mean 3.2 d 38 (15%) 1 d pso, 57 (23%) 2 d, 54 (22%) 3 d 40 (16%) 4 d 37 (15%) 5 d 19 (8%) 6 d 6 (2%) 7 d Timing of test: not stated; frozen samples Storage: swabs were shipped for testing on dry ice (−70 °C); |
Not stated; Veritor testing was performed internally at BD (San Diego, CA, USA) Threshold: as per manufacturer |
| Young 2021 | Innova (product code not reported) (Innova Med Group, China (Xiamen Biotime Biotechnology)) | CGIA N gene |
Samples tested: NP+OP (direct swab); collected by HCW Timing of sampling: not stated; on admission Timing of test: immediate testing Storage: none required; testing performed in the admitting department |
States "staff"; presume HCW Threshold: visual |
| Ag: antigen; AN: anterior nasal; BAL: bronchoalveolar lavage; CE: European Union standard; CGIA: colloidal gold immunoassay; CL(E)IA: chemiluminescent (enzyme) immunoassay; ED: emergency department; FIA: fluorescent immunoassay; FN: false negative; FP: false positive; HCW: healthcare worker; IFU: instructions for use; IPD: individual patient data; IVD: in vitro diagnostic; LAMP: loop‐mediated isothermal amplification; LFA: lateral flow assay; N/A: not applicable; NMT: nasal mid‐turbinate; NP: nasopharyngeal; OP: oropharyngeal; PBS: Phosphate‐buffered saline; PHE: Public Health England; POC: point‐of‐care; pso: post‐symptom onset; RA(D)T: rapid antigen (diagnostic) test; RUO: research use only; SDQ: Standard Q COVID‐19 Rapid Antigen Test; TW: throat wash; UV: ultraviolet; VTM: viral transport medium | ||||
Appendix 9. Index test details from manufacturer instructions for use documents
| Type of assay; time to result | Equipment Kit storage | Sample types; transport medium | Intended use; population; test operator | Sample storage | Test interpretation | |
| AAZ; COVID‐VIRO COVID‐19 Ag Rapid Test; IFU: TR‐COV‐006 | CGIA; 15 min | Provides: test device, buffer, NP swabs, extraction tubes, nozzles and filters; 2‐30 °C | NP; not stated | Popl: not stated in IFU; operator: professional use only | Test as soon as possible after collection; can be stored in clean, unused sealed plastic tube at room temperature (15‐30 °C) for up to 1 h prior to testing. If > 1 h delay occurs, dispose of sample | Visual: negative if control line only; positive if both test and control lines appear no matter how faint; invalid if no control line visible |
| Abbott Diagnostics Scarborough Inc; BinaxNOW COVID‐19 Ag card; IFU: IN195001, r2 12/12/2020 | LFA (not otherwise specified); 15 min | Provides: test cards, extraction reagent, nasal swabs, control swabs; 2‐30 °C | Nasal (AN; previous iteration was for NMT); test directly from nasal swabs, without VTM; do not store specimens in VTM | Popl: suspected of COVID‐19; ≤ 7 d pso; operator: medical professionals or trained operators … proficient in performing RDTs | Test immediately after collection; otherwise sample can be kept in a clean, unused plastic tube capped tightly at room temperature (15‐30 °C) for up to 1 h | Visual: negative if pink/purple control line only; positive if both sample and control lines appear no matter how faint; invalid if no control line visible or if sample line only or blue control line appears (with or without sample line) |
| Abbott Rapid Diagnostics; Panbio COVID‐19 Ag Rapid Test Device; IFU: 41FK10; 08/2020 | CGIA; 15 min | Provides: buffer, extraction tubes and caps, positive and negative control swabs, NP swabs for collection, tube rack; 2‐30 °C | NP; not mentioned; implies not recommended | Popl: symptomatic; operator: professional use (lab or non‐lab use) | Test direct swab specimens immediately after collection. If not possible, swab specimen can be kept in an extraction tube filled with extraction buffer (300 μL) at room temperature (15‐30 °C) for up to 2 h prior to testing | Visual: negative if control line only; positive if both test and control lines appear no matter how faint; invalid if no control line visible |
| Abbott Rapid Diagnostics; Panbio COVID‐19 Ag Rapid Test Device (Nasal); REF 41FK11/41FK21; 01/2021 | CGIA; 15 min | Test devices, buffer, extraction tubes, extraction tube caps, positive control swab, negative control swab, sterilized nasal swabs, tube rack; 2‐30 °C | Nasal – NMT; not mentioned | Popl: symptomatic and/or meeting epidemiological criteria; operator: professional use only (lab or non‐lab use) | Perform the test immediately after collection | Visual: negative if control line (C) only; positive if both the test line (T) and the control line (C); invalid if the control line is not visible; Caution: the presence of any test line (T), no matter how faint, indicates a positive result |
| Access Bio; CareStartCOVID‐19 Ag Rapid Test; IFU: RCHM‐02071 | CGIA; 10 min | Test device, extraction vial/cap, swabs, positive control swab, negative control swab; 1‐30 °C | NP or AN; not stated | Popl: ≤ 5 d pso; or for asymptomatic or other epidemiological indication test 2x over 2‐3 d with ≥ 24 h and < 48 h between tests; operator: healthcare professionals or trained users; specifically instructed in the use of the CareStart COVID‐19 Antigen and proper infection control procedures | Process the test sample immediately after collection. If specimen storage is necessary, swabs can be placed into extraction buffer for up to 4 h. Specimens should not be stored dry | Visual: negative if control line only; positive if both test and control lines appear no matter how faint; invalid if no control line visible |
| Anhui Deepblue ‐ COVID‐19 Ag; IFU: No. IFU‐COVIDAg‐NST‐01,Ver.A/2 | CGIA; 15 min | Sterile swab, extraction tubes, extraction reagent, test device; 4–30 °C | Nasal (AN); do not dilute the sample when testing; VTM not specifically mentioned (IFU is for home test kit) | Popl: symptomatic and asymptomatic; < 18 years should be supervised by an adult; operator: intended for personal use by untrained lay people | Freshly collected samples should be processed as soon as possible. | Visual: negative if control line only; positive if both test and control lines appear no matter how faint; invalid if no control line visible |
| Becton Dickinson; BD Veritor System for Rapid Detection of SARS‐CoV‐2; REF: 256082 | Not stated; LFA; 15 min (up to 60 min if 'walk‐away' mode enabled) | Provides: test device, extraction reagent, specimen sampling swabs, positive and negative control swabs. Also requires: BD Veritor Plus analyser device (Cat. No. 256066); 2–30 °C | Nasal; not recommended; "NOT INTENDED for testing liquid samples such as wash or aspirate samples or swabs in transport media as results can be compromised by over dilution" | Popl: ≤ 5 d pso; operator: lab use or POC by healthcare professionals | Test ASAP after collection, and no later than 1 h after specimen collection | Automated: 'CoV2: +' indicates positive result; 'CoV2: ‐' for presumptive negative; 'CONTROL INVALID' for invalid result |
| Bionote; NOWCHECK COVID‐19 Ag test; IFU:I1901‐10E (Nov 2020) | Not stated; LFA; 15 min | Provides: test device, extraction buffer tube and nozzle cap, sterile swab, paper stand.; 2‐30 °C (36‐86°F) | NP; instructions provided for use with VTM | Popl: patients that are suspected to have a SARS‐CoV‐2 infection; operator: healthcare workers and labs only | Test ASAP after collection; can be stored in extraction buffer for 1 h at room temperature, or up to 4 h refrigerated; store for up to 12 h in VTM | Visual: negative if control line only; positive if both test and control lines appear no matter how faint; invalid if no control line visible |
| Bionote; NOWCHECK COVID‐19 Ag test (Nasal); IFU:I1901N‐3E (Mar 2021) | Not stated; LFA; 15 min | Provides: test device, extraction buffer tube and nozzle cap, nasal swab, paper stand; 2‐30 °C (36‐86°F) | NP; instructions provided for use with selected VTM | Popl: patients that are suspected to have a SARS‐CoV‐2 infection; operator: healthcare workers and labs only | Test ASAP after collection; can be stored in extraction buffer for 1 h at room temperature, or up to 4 h refrigerated; store for up to 12 h in VTM | Visual: negative if control line only; positive if both test and control lines appear no matter how faint; invalid if no control line visible |
| Biosynex; COVID‐19 Ag BSS; IFU: SW40006 (Feb 2021) | CGIA; 15 min | Provides: test cassettes, pre‐filled extraction buffers, sterile swabs (CE 0197), nozzles, workstation; 2‐30 °C | NP or nasal; not stated | Popl: symptomatic only; operator: professional in vitro diagnostic use only | Test ASAP after collection; can be stored in clean, unused sealed plastic tube at room temperature (15‐30 °C) for up to 1 h prior to testing or stored 3 h at 2‐8 °C | Visual: negative if control line only; positive if both test and control lines appear no matter how faint; invalid if no control line visible |
| Biotical Health; Biotical SARS‐CoV‐2 Ag card; IFU: RTB25CoV2C(Rev.02.01) | CGIA; 10 min | Ag test cassette, Sample diluent (SARS‐CoV‐2 Ag reagent), swabs for sampling, disposable pipettes, disposable test tubes, positive control swab; 2‐30 °C | NP; not stated | Popl: ‘suspected’ of COVID‐19 infection; operator: professional use only | Test ASAP after collection; can be stored at 2‐8 °C for up to 8 h; perform within 2 h after opening the sealed bag | Visual: negative if green control line only; positive if both test (red) and control (green) lines appear; invalid if no control line visible or only test line visible |
| Boditech Medical ‐ iChroma COVID‐19 Ag; IFU: INS‐SR‐EN | FIA; 12 min | Cartridge, pipette tip, extraction tube, nozzle, ID chip; 2‐30 °C | NP; direct testing recommended; if samples in UTM or VTM used, minimal dilution recommended | Popl: screening of early mild, asymptomatic, or acute patients; operator: "in vitro diagnostic use only" |
Test the sample immediately after sample collection | Requires iChroma reader; test result calculated automatically and displays ‘Positive’ or ‘Negative’ |
| Certest Biotec S.L.; CERTEST SARS‐CoV‐2; Ag Card test; IU‐SC8 v.03 | CGIA; 10 min | Ag test cassette, reagent respiratory (sample and control diluent), sterile swabs, disposable pipettes, testing tubes, positive control swab, plastic caps; 2‐30 °C | NP; transport media such as: VTM, UTM or saline buffer can be used (1.0 mL) | Popl: ‘suspected’ of covid‐19 infection; operator: trained professional | Test ASAP after collection; can be stored at 2‐8 °C for up to 8 h. Samples preserved in VTM, UTM or saline buffer could be preserved on it until 6 h at room temperature or in the refrigerator (2‐8 ºC); perform within 2 h after opening the bag | Visual: negative if green control line only; positive if both test (red) and control (green) lines appear; invalid if no control line visible or only test line visible |
| Coris BioConcept; COVID‐19 Ag Respi‐Strip; IFU: 5723/TB/V03 | CGIA (paper strip method); 15 min | Paper strips in a bottle with desiccant; LY‐S dilution buffer (3,5 mL or 15 mL; tubes and stoppers); 4‐30 °C | NP swab or culture extracted solution; samples must be liquid; transport medium, a gel or a sponge matrix can be used | Popl: not stated in IFU, described as an acute‐phase screening test only; operator: states for professional use only | Test ASAP after collection, any delay may result in a low signal intensity. If not, store frozen at −20 °C | Visual; read through collection tube; control line only (negative), T line (with or without control (positive), no control line (invalid) |
| Denka; QuickNAVI COVID‐19 Ag; Jan 2021 v1 | LFA (latex conjugated); 15 min; | Test cassettes, specimen buffer tubes, sterile swabs, specimen buffer filter, stand | NP or nasal (AN); transport medium not mentioned | Popl: not specified; "detection of SARS‐Cov‐2 antigens in nasal or nasopharyngeal specimens"; operator: not stated in IFU | Suspend a specimen in the specimen buffer immediately after collection and promptly perform the test | Visual: control line only (negative), test line and control line (positive); no control line (invalid) |
| e25bio; DART (Direct antigen rapid test); IFU: n/a | CGIA | IFU not obtained | IFU not obtained | IFU not obtained | IFU not obtained | IFU not obtained |
| eco Diagnostica; COVID‐19 Ag ECO Teste; Registro: 80954880133 | Not mentioned; 2‐15 min | Not mentioned; 2‐30 °C | NP | Not specified | Not specified | Visual: control line only (negative), test line and control line (positive); presence of a weak test line or any intensity, the result is considered reactive |
| Encode/Emmo Pharma – Encode SARS‐CoV‐2 antigen rapid testa | LFA; 15 min | Test device, extractions solution, extraction tubes, sterile swabs, work station; 2‐30 °C | Nasal/throat/anterior nasal swab; not mentioned | Popl: ≤ 7 d pso; operator: use by clinical professionals (doctors, practice nurse, employer occupational health departments etc | Unknown; information extracted from website, IFU not obtained | Unknown; information extracted from website, IFU not obtained |
| Fortress Diagnostics ‐ Coronavirus Ag; Revision No.1 AUG/20 | CGIA; 15 min | Test cassette(s), sterile swabs, extraction tubes and nozzles, buffer, tube rack | NP; transport medium not mentioned | Popl: patients suspected for infection with SARS‐CoV‐2 by their healthcare provider; operator: for professional use only | Test immediately after collection for best performance; otherwise place swab in a clean, unused plastic tube at room temperature (15‐30 °C) for up to 1 h prior to testing | Visual: control line only (negative), test line and control line (positive); NO control line (invalid) |
| Fujifilm Corporation; Fujifilm COVID‐19 Ag test; IFU: 897N204150C | CGIA; 10 min | Test cartridge, extraction reagent solution; 1‐30 °C | NP; not stated | Popl: not specified; operator: healthcare professionals only | Not stated | Visual: positive if both black control line and black test line; negative if only a black control line; invalid if no control line visible |
| Fujirebio Inc; ESPLINE SARS‐CoV‐2; IFU: FRI46955 (K4B01TE) | LFA (alkaline phosphatase‐labelled); 30 min | Reaction cassette, sample extraction solution (squeeze tube), applicator tip.; 1‐30 °C | NP fluid; not stated; recommends samples are prepared immediately after collection (placing swab in provided sample extraction solution), however documented clinical validation results were from swabs immersed in VTM prior to use | Popl: not stated in IFU; operator: professional use only | Samples must be prepared immediately after specimen collection | Visual; positive if blue test line (T) and reference line (r) positions, negative if blue r line only, invalid if no blue r line appears or if red r line still present. If the r and T lines appear before 30 min, the sample must be considered "positive"; samples that only turn “positive” after 30 min, must be considered “negative” |
| Guangzhou Wondfo Biotech Co., Ltd.; Wondfo 2019‐nCoV Antigen Test; Rev. A6 | CGIA; 15 to min | Sealed pouches, drippers, extraction buffer (or pre‐installed extraction buffer), sample extraction tube, NP swab, test tube rack, +ve control swab, ‐ve control swab, procedure card; 2‐30 °C; the test cassette should be used within 1 h after taking out from the sealed pouch. Buffer solution should be re‐capped in time after use | NP or OP; VTM (1 mL) | Popl: not specified; operator: professionally trained staff working in lab or clinics; sample taken by qualified medical personnel | Test ASAP after collection; can be stored at 2~8 ℃ for up; to 8 h, or at −70 ℃ for a long time | Visual; positive ‐ coloured bands appear at both test line (T) and control line (C); negative ‐ coloured band appears at control line (C) only; invalid ‐; no visible coloured band appears at control line |
| Innova Medical Group; Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test; IFU: A/02 | CGIA; 20‐30 min | Provides: test cartridge, extraction tube, extraction solution, QC card; 2‐30 °C | Nasal or OP; not mentioned | Popl: ≤ 5 d pso; operator: trained clinical laboratory personnel or similarly trained in POC settings | Test ASAP after collection. Based on data generated with influenza virus, throat swabs are stable for up to 24 h at room temperature or 2‐8 °C | Visual: negative if control line only; positive if both test and control lines appear no matter how faint; invalid if no control line visible |
| JOYSBIO (Tianjin) Biotechnology Co., Ltd.; SARS‐CoV‐2 Antigen Rapid Test Kit; IFU: Jan 2021 | CGIA; 15‐20 min | Test device, desiccant, buffer, extraction tube, specimen sampling swabs; 2~30 ºC | NP; not mentioned | Popl: ≤ 5 d pso; operator: HCWs and clinical laboratory personnel | Test ASAP after collection, but no later than 1 h after specimen collection | Visual: negative if one control line appears; positive if 2 coloured lines appear; invalid if no control line visible |
| Lepu Medical Technology Co., Ltd.; SARS‐CoV‐2 Antigen rapid test (NP or nasal); CE‐InCG27 REV.01 | CGIA; 15 min | Test cards, sample, treatment solution, sterilized swab, nozzle with filter and sample collection buffer. Each test card bag includes 1 SARS‐CoV‐2 antigen detection card and 1 package of desiccant; 4 ℃~ 30 ℃ | NP or nasal Not mentioned on IFU |
Popl: not specified; operator: professional use only | The test should be completed within 1 h after sample collection | Visual; 1 purple line in control area is negative; 2 purple lines in both control and test areas is positive; 1 line only, or no purple line is invalid |
| Lepu Medical Technology Co., Ltd.; SARS‐CoV‐2 Antigen rapid test (saliva) | CGIA; 15 min | IFU not obtained | IFU not obtained | Popl: not specified. Impies symptomatic and asymptomatic; operator: Intended for professional use in the laboratory and at POC | IFU not obtained | IFU not obtained |
| Liming Bio‐Products Co., Ltd; COVID‐19 Antigen Rapid Test Device (StrongStep; IFU: via Weitzel 2020; REF 500200 v1. | CGIA; 15 min | Test device, extraction buffer vial, extraction tube, sterile swab; 2‐30 °C | NP or OP; not mentioned in IFU | Popl: not stated in IFU; operator: professional use only | Test ASAP after collection; can be held in clean, dry plastic tube or sleeve up to 72 h at 15 °C to 30°, or 2 °C to 8 °C before processing | Visual; 2 coloured bands for positive; control band only for negative; test line only is invalid |
| LumiraDx UK Ltd.; LumiraDx SARS‐CoV‐2 Ag Test; SPEC‐32311 R4 ART‐00570 R4 | FIA; 12 min | LumiraDx test Strips; RFID (Radio frequency ID), extraction buffer vials, dropper lids; 2‐30 °C | NP or AN | Popl: ≤ 12 d pso; operator: trained clinical laboratory personnel and individuals trained in POC settings, and proficient in performing tests using the LumiraDx Instrument | Process the swab in the extraction vial as soon as possible. If using a frozen sample, the sample must be at room temperature before testing | The LumiraDx Instrument must be used to generate results. The results will be displayed on the Instrument screen – as ‘Negative –‘ or ‘Positive +’ or ‘Test operation error’ if invalid |
| MEDsan GmbH; MEDsan SARS‐CoV‐2 Ag | CGIA | Swabs; extraction tube with buffer, package insert | Saliva, nasal (NMT); transport medium not mentioned | Popl: during the acute phase of infection; operator: professional use only | The test device should be used within 1 h after opening of the sealed pouch. If in a high humidity environment, use it immediately. DO NOT FREEZE | Presence of a visible or faint test line indicates a positive; result; no test line indicates a; negative result; no control line indicates invalid result |
| Mologic; COVID‐19 Rapid Antigen Test; REF 1811125 (Feb 2021) | LFA (not otherwise specified); 10 min | Test device, buffer capsule, extraction tubes, sterile swab; 2‐30 °C | Nasal+OP or nasal only (instructions for AN collection); transport medium not mentioned | Popl: during acute infection for symptomatic and asymptomatic individuals; operator: for professional use only; sample collection by self‐swabbing is acceptable | Not stated | Presence of a visible or faint test line indicates a positive result; no test line indicates a negative result; no control line indicates invalid result |
| Nal von Minden; NADAL COVID‐19 Ag Test; REF 243103N‐20 | CGIA; 15 min | Test cassettes, sterile swabs, extraction tubes including dropper caps, buffer bottles, reagent holder; 2‐30 °C | NP or OP; VTM without denaturing agents; (vol 1 mL) | Popl: not specified; operator: professional use only | Test ASAP after collection. Use freshly collected specimens for best test performance. If not tested immediately, swab specimens can be stored at 2‐8 °C for 24 h after collection | The presence of a coloured line in the test line region (T) indicates a positive result. The absence of a coloured line in the test line region (T) indicates a negative result. The absence of control line (C) indicates invalid |
| Orient Gene Biotech/Healgen Scientific ‐ Coronavirus Ag Rapid Test Cassetteb | CGIA | Test cassettes, extraction buffer vials, sterile swabs, extraction tubes and tips, workstation; 2‐30 °C | NP; direct; transport medium not mentioned | Popl: suspected of COVID‐19 by HCW; ≤ 10 d pso; operator: in vitro diagnostic use only; implies HCW | Test ASAP after collection; otherwise store in dry, sterile, and sealed plastic tube; test within 1 h | Presence of a visible or faint test line indicates a positive; result; no test line indicates a; negative result; no control line indicates invalid result |
| PCL, Inc.; PCL COVID19 Ag Gold; COV04S‐IFU‐001, Revision 5 | CGIA; 10 min | Test card, extraction buffer tube, filter cap; 2‐30 °C | NP or saliva; not recommended | Popl: suspected; operator: professional use | Test ASAP after collection. Do not use stored specimens | The presence of a coloured line in the test line region (T) indicates a positive result. The absence of a coloured line in the test line region (T) indicates a negative result. The absence of control line (C) or no line indicates invalid. |
| Precision Biosensor Inc; One Step Immunoassay for Exdia COVID‐19 Ag; PB‐FGRUM‐01‐EN (Oct 2020) | FIA; up to 20 min (automatic) | Test cassettes, sterile swabs, reagent tubes with buffer, filter caps, control swabs; 2‐30 °C | NP; direct testing of swab from the patient highly recommended; if VTM used, minimal sample dilution recommended | Popl: suspected of COVID‐19 by healthcare provider; operator: clinical laboratory personnel specifically instructed and trained in IVD procedures and individuals similarly trained in POC settings | Samples should be tested immediately after collection. Refrigerated or frozen samples should be brought to room temperature (20 ℃~30 ℃) prior to testing | Analyser device; test cassette analyzed at preset times after applying the samples. If the test result is strong positive, the analyser will immediately determine the result as positive. If the test result is positive at near cut‐off level or negative, the result will be determined at 20 min |
| Quidel; Sofia SARS Antigen FIA; IFU: 1439000EN00 (04/20) | FIA; 15 min | Provided: test cassette, reagent tubes, reagent solution, nasal swabs, 120 μL fixed volume pipette, SARS‐positive control swab, negative control swab Required: Sofia or Sofia 2 reader device, calibration cassette; 15‐30 °C | Nasal or NP; updated IFU: directly test patient specimens without transport media. (Original IFU: if transport of samples with viral transport medium (VTM) is required, minimal dilution of the sample is recommended, e.g. 1 mL or less) | Popl: ≤ 5 d pso; operator: trained clinical laboratory personnel and individuals trained in POC settings | Test ASAP after collection. Based on data generated with influenza virus, nasal or NP swabs are stable for up to 24 h at room temperature or 2‐8 °C, and nasal or NP swabs in VTM are stable for up to 72 h at 2‐8 °C | The Sofia screen will display results for the procedural control as being 'tick' or 'cross', and will individually provide a '+' or '‐' result for SARS. If the procedural control is 'X' retest with a new patient sample and a new test cassette. Results must not be interpreted past 30 min after inoculation |
| RapiGEN; BIOCREDIT COVID‐19 Ag; IFU: I‐H073A‐E00(2020.04.03) | CGIA; 5‐8 min | Test device, assay diluent tube and filter cap, swab for NP collection; 1‐40 °C | NP swab; if swab is placed in VTM, used 1:1 dilution with assay buffer | Popl: not stated in IFU; operator: trained personnel required to collect sample and (presumably) to then conduct the test | Test ASAP after collection; if storage required then 2‐8 °C for up to 12 h, or −20 °C for up to 24 h | Visual; control line only (negative), control and test lines (positive), no control line (invalid) |
| R‐Biopharm AG; RIDAQUICK SARS‐CoV‐2 Antigen; Ref: N6803 | CGIA; 10 min | Strip, reagents, pipette, reagent vial; 2‐30 °C | NP or nasal; PBS, UTM, VTM, Amies medium, or saline solution (0.9 %) (volume of 1 mL) | Popl: suspected and/or symptomatic as well as asymptomatic; operator: professional or trained personnel | Test ASAP, if the test material will not be used immediately, store it at 2‐8 °C until processing for ≤ 2 d. If ≥ 2 d, store at −20 °C or colder for ≤ 2 months | Visual; the 2 red‐violet lines are; visible (positive), only the upper red‐violet; control line (C) is visible (negative); no line is visible, or only test line appears (invalid) |
| Savant Biotech; SARS‐Cov‐2 Antigen Fluorescence Rapid Detection Kit; IFU: no mention of any COVID tests on company website (checked 07/12/2020) | IFU not obtained | IFU not obtained | IFU not obtained | IFU not obtained | IFU not obtained | IFU not obtained |
| SD Biosensor Inc; Standard F COVID‐19 Ag FIA; IFU: F‐NCOV‐01G | FIA; 30 min | Provides: test device, extraction buffer tube, filter cap, sterile swab. Standard F Analyser also required (F100 or F200); room temperature, 2‐30 °C/36‐86°F | NP; not recommended; "Do not use transport media" | Popl: symptomatic; operator: professional use | Test ASAP after collection; may be stored at room temperature for up to 24 h or at 2‐8 °C/36‐46°F for up to 48 h prior to testing | Automatic; the analyser will automatically display the test result in 30 min. Cut‐off index (COI) value ≥ 1.0 is positive, < 1.0 is negative, COI not displayed is invalid result |
| SD Biosensor Inc; Standard Q COVID‐19 Ag (NP kit); IFU: Q‐NCOV‐01G (09COV30D; 09 2020); (Also Roche ‐ Rapid Antigen Test) | LFA (conjugated with colour particles); 15‐30 min | Provides: test device, extraction buffer tube, filter cap, sterile swab; room temperature, 2‐30 °C/36‐86°F | NP; validated for use with Copan UTM, BD UTM and Standard Transport Medium; warns sensitivity can be reduced when using VTM | Popl: symptomatic; operator: professional use | Test ASAP after collection; may be stored at room temperature for up to 1 h or at 2‐8 °C/36‐46°F for up to 4 h prior to testing | Visual; the presence of control and 'test' lines, no matter how faint the result is considered positive; negative if control line only; invalid if test line only |
| SD Biosensor Inc; Standard Q COVID‐19 Ag (nasal kit); IFU: Q‐NCOV‐01G (12 2020); | LFA (conjugated with colour particles); 15‐30 min | Provides: test device, extraction buffer tube, filter cap, sterile swab; room temperature, 2‐30 °C/36‐86°F | Nasal (instructions are for NMT); validated for use with Copan UTM, BD UTM and Standard Transport Medium; warns sensitivity can be reduced when using VTM | Popl: symptomatic; operator: HCWs at the clinical setup and POC sites | Test ASAP after collection; may be stored at room temperature (15–25 °C or 2‐8 °C for up to 4 h prior to testing | Visual; the presence of control and 'test' lines, no matter how faint the result is considered positive; negative if control line only; invalid if test line only |
| Shenzhen Bioeasy Biotechnology Co, Ltd; BIOEASY 2019‐nCoV Ag Fluorescence Rapid Test Kit Time‐Resolved Fluorescence; IFU: TS‐IU‐F027‐A2 (YRLF04401025/ YRLF04401050/ YRLF04401100) | FIA; 10 min | Test card, extraction solution, extraction tube, dripper, swab and ID chip. Test runs on immunofluorescence analyser (supplied separately), transfer pipette also required | Nasal swabs, throat swabs and deep sputum samples; not mentioned in IFU | Popl: not specified; operator: not stated in IFU | Test ASAP after collection; or store at 2‐8 °C for ≤ 24 h; OR store at −70 °C for longer periods. Avoid repeated freezing and thawing (no more than 3 times) | Automatic; +ve if both detection line and control line detect a fluorescent signal, and the detection line detection value is ≥ 0.005 ng/mL; ‐ve if fluorescent signal on control line only; invalid if no fluorescent signal, or signal only on test line |
| Sichuan Mass Spectrometry Biotechnology Co. ‐ 2019‐nCov Ag | IFU not obtained | IFU not obtained | IFU not obtained | IFU not obtained | IFU not obtained | IFU not obtained |
| Siemens/Healgen Scientific; CLINITEST Rapid COVID‐19 Antigen Test; IFU: B21986‐09 Rev. G | CGIA; 15 min | Test cassettes, extraction buffer vials, sterile swabs, extraction tubes and tips; 2‐30 °C | NP or nasal; not stated | Popl: ≤ 10 d pso, or asymptomatic or individuals from areas of low infection prevalence; operator: HCWs or individuals under HCW supervision (for nasal sample self‐collection) | Test ASAP after collection; if > 1 h delay occurs, dispose of sample | Visual: negative if control line only; positive if both test and control lines; invalid if no control line visible |
| Spring Healthcare – Spring; IFU: SP‐SW 106 | CGIA | Test device, extraction buffer, extraction tube with filter nozzle, sterile swabs, tube stand; 2‐30 °C | AN; VTM use not mentioned; does state not intended for testing … liquid; samples such as nasal wash or aspirate samples (due to) over dilution | Popl: ≤ 7 d pso; operator: medical professionals or trained operators proficient in performing tests and trained clinical laboratory personnel or individuals trained in POC settings | Process the test sample immediately after collection; specimens may be frozen at −80 °C and used up to 5 d; stable for 4 h in extraction buffer | Visual: negative if control line only; positive if both test and control lines; invalid if no control line visible |
| SUGENTECH, INC.; SGTI‐Flex COVID‐19 Ag; REF: CAGT025E1 | CGIA; 20‐30 min | Test cassette, extraction buffer, dropping cap, extraction tube, swab; 2~30 °C. If stored in cold storage, allow 30 min to return to room temperature before testing | NP or OP; not mentioned | Popl: not specified; operator: not specified | Test ASAP after collection; store in deep freezer at −70 °C (or in dry ice or liquid nitrogen) if required. A freezer at −20 °C is NOT recommended. Can be stored at 2‐8 °C for up to 72 h | Visual; positive, test line (T) and control line (C) visible; negative, only control line (C) visible; invalid, if control line fails to appear |
| SureScreen Diagnostics; COVID‐19 Antigen Test; REF COVID19AGVC; number: 1110032811 | LFA; 15 min | Test devices, extraction tube, swabs, extraction buffer, nozzle with filter, tube stand; 2‐30 °C | NP or OP; not mentioned | Popl: within 1st 2 weeks pso; operator: professionally trained staff in lab or clinics; qualified medical personnel for collection | Test ASAP after collection; can be stored at 2‐8 °C for 24 hours after collection | Visual; positive, 2 colour bands appear in the control region (C) and test region (T); negative, 1 colour band appears in the control region (C); invalid, control band fails to appear |
| SureScreen Diagnostics; COVID‐19 Antigen Rapid Fluorescent Cassette (COVID19 AGC) | FIA | No IFU identified | No IFU identified | No IFU identified | No IFU identified | No IFU identified |
|
aInformation obtained from: encode.com.cn/en/product_more.asp?id=570; encoderapidtestkit.com/store/20-x-COVID-19-Rapid-Antigen-Tests-p247763021 bhealgen.com/if-respiratory-covid-19-antigen; healgen.com/about AN: anterior nasal; ASAP: as soon as possible; CGIA: colloidal gold immunoassay; FIA: fluorescent immunoassay; IFU: instructions for use; IVD: in vitro diagnosis; LFA: lateral flow assay; NMT: nasal mid‐turbinate; NP: nasopharyngeal; OP: oropharyngeal; PBS: Phosphate‐buffered saline; POC: point‐of‐care; Popl: population; UTM: universal transport medium; VTM: viral transport medium | ||||||
Appendix 10. Results of pooled analysis including additional 'sensitivity‐only' cohorts
| Test | Evaluations; samples (cases) | Summary sensitivity, % (95% CI) | Difference in sensitivity, (95% CI), P value |
| Secondary analyses by symptom status | |||
| Symptomatic | 40; 14,331 (3817) | 75.4 (69.0 to 80.9) | Ref |
| Asymptomatic | 40; 26,000 (1839) | 52.8 (44.6 to 60.9) | −22.6 (−26.5to −18.7), P < 0.0001 |
| Symptomatic participants by setting for testing | |||
| Symptomatic – COVID‐19 test centre | 56; 24,954 (5721) | 81.2 (78.7 to 83.5) | Ref |
| Symptomatic – hospital inpatient | 25; 12,291 (2924) | 51.4 (46.1 to 56.7) | −29.8 (−35.7to −23.9), P < 0.0001 |
| Symptomatic – ED/urgent care | 15; 5607 (1312) | 70.9 (55.8 to 82.5) | −10.3 (−24.1to |
| Symptomatic – laboratory‐based | 12; 4229 (2246) | 56.9 (43.8 to 69.1) | −24.3 (−37.5to −11.2), P < 0.0001 |
| Symptomatic – hospital ‐ any | 4; 1226 (469) | 55.9 (43.8 to 67.3) | −25.3 (−37.6to −13.1), P < 0.0001 |
| Symptomatic – school/university | 3; 406 (146) | 78.9 (53.4 to 92.4) | −2.3 (−22.2to |
| Symptomatic – hospital in‐ or outpatient | 2; 1472 (204) | 33.7 (6.9 to 77.7) | −47.5 (−90.6to −4.5), P =0.030 |
| Symptomatic – unclear | 8; 888 (573) | 79.9 (59.5 to 91.5) | ‐ |
| Asymptomatic participantsby setting for testing | |||
| Asymptomatic – COVID‐19 test centre | 20; 19,363 (1305) | 60.1 (51.8 to 67.8) | Ref |
| Asymptomatic – hospital – any | 3; 287 (115) | 58.7 (35.8 to 78.4) | −1.4 (−25.5to |
| Asymptomatic – laboratory‐based | 3; 567 (166) | 60.0 (38.7 to 78.1) | −0.09 (−22.5to |
| Asymptomatic – unclear | 6; 287 (65) | 53.2 (31.4 to 73.9) | ‐ |
| Subgroup analysis by sample type | |||
| Includes NP (all) | 141; 60,782 (14605) | 68.5 (64.9 to 72.0) | Ref |
| Nasal (all) | 36; 33,292 (4196) | 76.2 (70.9 to 80.8) | 7.7 (1.6to |
| Nasal + OP | 20; 13,833 (2586) | 71.0 (66.3 to 75.3) | 2.5 (−3.2to |
| Saliva (all) | 8; 1219 (566) | 17.1 (12.4 to 23.2) | −51.4 (−57.8to −44.9), P < 0.0001 |
| OP alone | 3; 614 (275) | 57.4 (30.7 to 80.3) | −11.1 (−38.5to |
| Subgroup analysis by viral load | |||
| Viral load above and below a single Ct threshold value* | |||
| Higher VL: < 25 Ct | 104; 6877 (6877) | 94.7 (93.5 to 95.7) | Ref |
| Lower VL: > 25 Ct | 104; 5749 (5749) | 40.6 (35.7 to 45.7) | −54.1 (−58.3to −49.9), P < 0.0001 |
| Higher VL: < 30 Ct | 64; 6264 (6264) | 86.0 (82.8 to 88.7) | Ref |
| Lower VL: > 30 Ct | 64; 2332 (2332) | 18.7 (14.2 to 24.1) | −67.3 (−73.1to −61.6), P < 0.0001 |
| Higher VL: < 32/33 Ct | 34; 3353 (3353) | 86.7 (82.5 to 90.0) | Ref |
| Lower VL: > 32/33 Ct | 34; 498 (498) | 13.7 (9.5 to 19.4) | −73.0 (−76.8to −69.2), P < 0.0001 |
| Viral load above and below a single RNA copy threshold value | |||
| Higher VL: ≥ 10^6 RNA copies/mL | 39; 1865 (1865) | 96.2 (94.5 to 97.4) | Ref |
| Lower VL: < 10^6 RNA copies/mL | 39; 2520 (2520) | 31.3 (24.2 to 39.4) | −64.9 (−72.7to −57.2), P < 0.0001 |
| Higher VL: ≥ 10^5 RNA copies/mL | 32; 2512 (2512) | 88.6 (84.8 to 91.5) | Ref |
| Lower VL: < 10^5 RNA copies/mL | 32; 1684 (1684) | 16.8 (11.0 to 24.7) | −71.8 (−79.4to −64.2), P < 0.0001 |
| Subgroup analysis by assay format | |||
| CGIA | 155; 97,665 (18885) | 68.8 (65.7 to 71.8) | Ref |
| FIA | 20; 7128 (1648) | 75.8 (67.8 to 82.4) | 7.0 (−1.0to |
| LFA (ALP) | 10; 1782 (548) | 51.0 (30.7 to 71.0) | ‐ |
| LFA (not otherwise specified) | 18; 9847 (1908) | 63.3 (50.4 to 74.5) | ‐ |
| ALP: alkaline phosphatase; CGIA: colloidal gold immunoassay; CI: confidence interval; Ct: cycle threshold; ED: emergency department; FIA: fluorescent immunoassay; LFA: lateral flow assay; NP: nasopharyngeal; OP: oropharyngeal; VL: viral load | |||
Appendix 11. Study quality by test group and at study‐level
Figure 118 Studies evaluating a single test application
26.
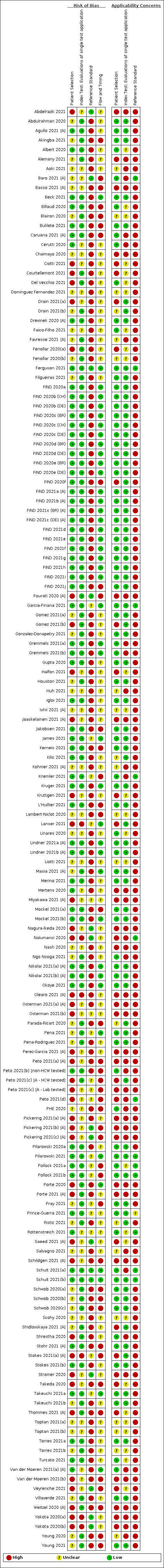
Risk of bias and applicability concerns summary for evaluations of single test applications: review authors' judgements about each domain for each included study
Figure 119 Studies evaluating repeated testing
27.
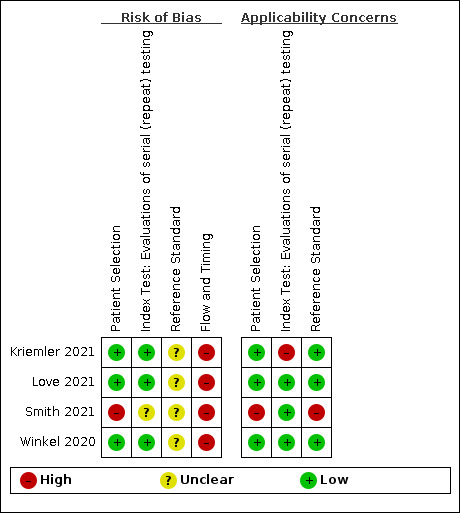
Risk of bias and applicability concerns summary for studies with repeat (serial) testing: review authors' judgements about each domain for each included study
Appendix 12. Additional forest plots of data for subgroup analyses
Figure 120 Forest plot of data contributing to overall analysis (n = 210 evaluations)
28.
Forest plot of data contributing to overall analysis (n=210 evaluations).
Figure 14 Forest plot of data by study design
Figure 15 Forest plot of data by sample site
Figure 16 Forest plot of within‐study comparisons by sample site, collection, or interpretation
Figure 17 Forest plot of data in higher versus lower viral load subgroups (< or > 25 cycle threshold (Ct))
Figure 18 Forest plot of data in higher versus lower viral load subgroups (< or > 32/33 Ct threshold)
Figure 19 Forest plot of data in higher versus lower viral load subgroups (< or > 30 Ct)
Figure 20 Forest plot of data in higher versus lower viral load subgroups (> or < 10^6 RNA copies/mL)
Figure 21 Forest plot of data in higher versus lower viral load subgroups (> or < 10^5 RNA copies/mL)
Appendix 13. Forest plots of data by test brand
Figure 22 Forest plot of individual study results overall (regardless of symptom status) by assay
Figure 23 Forest plot of individual study results in symptomatic participants by assay
Figure 24 Forest plot of individual study results in asymptomatic participants by assay
Figure 25 Forest plot of data from studies reporting within‐study comparisons by test brand
Appendix 14. Table of results of overall analyses by test brand (regardless of participant symptom status)
| All | Compliant with manufacturer's instructions for use (IFU) | |||||
| Test | N evaluations; samples (cases) | Summary sensitivity % (95% CI) | Summary specificity % (95% CI) | N evaluations; samples (cases) | Summary sensitivity % (95% CI) | Summary specificity % (95% CI) |
| AAZ ‐ COVID‐VIRO Study | 3; 1204 (534) | 84.9 (61.5 to 95.2) | 97.0 (95.4 to 98.1) | 2; 572 (239) | 91.3 (78.1 to 96.9) | 94.0 (90.9 to 96.1) |
| Abbott ‐ BinaxNOW COVID‐19 Ag card | 6; 14,877 (1051) | 67.0 (52.7 to 78.8) | 99.8 (99.7 to 99.8) | 6; 14,877 (1051) | 67.0 (52.7 to 78.8) | 99.8 (99.7 to 99.8) |
| Abbott ‐ Panbio COVID‐19 Ag | 39; 27,460 (5976) | 71.5 (65.9 to 76.4) | 99.6 (99.5 to 99.7) | 17; 11,952 (2138) | 72.7 (64.3 to 79.7) | 99.7 (99.6 to 99.8) |
| Including sensitivity‐only cohorts | 44; 28,012 (6528) | 71.6 (66.5to | ‐ | 21; 12,405 (2591) | 72.6 (65.2to | ‐ |
| Access Bio ‐ CareStart COVID‐19 Ag | 1; 1498 (234) | 57.7 (51.1 to 64.1) | 98.3 (97.5 to 99.0) | 1; 1498 (234) | 57.7 (51.1 to 64.1) | 98.3 (97.5 to 99.0) |
| Anhui Deepblue ‐ COVID‐19 Ag | 1; 1205 (191) | 47.1 (39.9 to 54.5) | 100 (99.6 to 100) | |||
| Including sensitivity‐only cohorts | 2; 1382 (368) | 60.7 (41.7to | ‐ | |||
| BIONOTE ‐ NowCheck COVID‐19 Ag | 3; 1944 (190) | 87.9 (82.4 to 91.8) | 99.4 (98.9 to 99.7) | 2; 618 (181) | 89.5 (84.1 to 93.2) | 97.7 (95.8 to 98.8) |
| Becton Dickinson ‐ BD Veritor (Visual) | 1; 1567 (176) | 48.9 (41.3 to 56.5) | 99.9 (99.5 to 100) | 1; 1567 (176) | 48.9 (41.3 to 56.5) | 99.9 (99.5 to 100) |
| Becton Dickinson ‐ BD Veritor | 5; 5196 (518) | 68.0 (51.0 to 81.3) | 99.4 (99.1 to 99.6) | 1; 1384 (116) | 66.4 (57.0 to 74.9) | 98.8 (98.1 to 99.3) |
| Including sensitivity‐only cohorts | 7; 5353 (675) | 75.0 (59.4to | ‐ | 2; 1416 (148) | 82.5 (52.5to | ‐ |
| Biosynex ‐ Biosynex COVID‐19 Ag BSS | 1; 634 (297) | 59.6 (53.8 to 65.2) | 100 (98.9 to 100) | |||
| Biotical Health ‐ SARS‐CoV‐2 Ag card | 1; 188 (96) | 66.7 (56.3 to 76.0) | 98.9 (94.1 to 100) | |||
| Boditech Medical ‐ iChroma COVID‐19 Ag | 1; 232 (41) | 73.2 (57.1 to 85.8) | 100 (98.1 to 100) | 1; 232 (41) | 73.2 (57.1 to 85.8) | 100 (98.1 to 100) |
| Certest Biotech ‐ CerTest | 1; 320 (170) | 53.5 (45.7 to 61.2) | 100 (97.6 to 100) | |||
| Coris Bioconcept ‐ COVID‐19 Ag Respi‐Strip | 8; 1831 (746) | 38.7 (31.0 to 47.0) | 98.3 (97.4 to 99.0) | 5; 1300 (475) | 33.7 (29.6 to 38.1) | 97.9 (96.7 to 98.7) |
| DIALAB ‐ DIAQUICK COVID‐19 Ag | 1; 99 (99) | 61.6 (51.3 to 71.2) | ‐ | 1; 99 (99) | 61.6 (51.3 to 71.2) | ‐ |
| Denka Co ‐ QuickNavi COVID‐19 Ag | 2; 2048 (156) | 81.3 (69.9 to 89.0)a | 100 (99.8 to 100)a,b | 2; 2048 (156) | 81.3 (69.9 to 89.0)a | 100 (99.8 to 100)a,b |
| E25Bio ‐ COVID‐19 DART | 2; 390 (200) | 77.5 (71.2 to 82.8) | 88.4 (83.0 to 92.3) | |||
| ECODiagnostica ‐ COVID‐19 Ag ECO | 1; 150 (55) | 69.1 (55.2 to 80.9) | 97.9 (92.6 to 99.7) | 1; 150 (55) | 69.1 (55.2 to 80.9) | 97.9 (92.6 to 99.7) |
| Encode/Emmo Pharma ‐ Encode | 1; 200 (100) | 74.0 (64.3 to 82.3) | 100 (96.4 to 100) | |||
| Including sensitivity‐only cohorts | 2; 290 (190) | 74.2 (67.4to | ‐ | |||
| FUJIFILM ‐ COVID‐19 Ag Test | 1; 108 (45) | 82.2 (67.9 to 92.0) | 100 (94.3 to 100) | |||
| Fortress Diagnostics ‐ Coronavirus Ag | 1; 1191 (191) | 56.0 (48.7 to 63.2) | 99.9 (99.4 to 100) | |||
| Fujirebio ‐ ESPLINE SARS‐CoV‐2 | 7; 1645 (411) | 61.4 (38.3 to 80.3) | 99.8 (99.2 to 99.9) | |||
| Including sensitivity‐only cohorts | 10; 1782 (548) | 51.0 (30.7to | ‐ | |||
| Guangzhou Wondfo ‐ 2019‐nCoV Ag | 1; 328 (56) | 85.7 (73.8 to 93.6) | 100 (98.7 to 100) | 1; 328 (56) | 85.7 (73.8 to 93.6) | 100 (98.7 to 100) |
| Innova Medical Group ‐ SARS‐CoV‐2 Ag | 8; 10,883 (1267) | 59.1 (40.8 to 75.2) | 99.7 (99.5 to 99.8) | 3; 7966 (655) | 54.7 (44.8 to 64.3) | 99.9 (99.8 to 99.9) |
| Including sensitivity‐only cohorts | 12; 11,437 (1821) | 65.8 (53.0to | ‐ | 5; 8387 (1076) | 63.0 (51.3to | ‐ |
| Including specificity only cohort | 9; 11,421 (1267) | ‐ | 99.7 (99.5to | 4; 8504 (655) | ‐ | 99.9 (99.8 to99.9) |
| Joysbio Biotech ‐ SARS‐CoV‐2 Ag | 1; 265 (44) | 70.5 (54.8 to 83.2) | 99.1 (96.8 to 99.9) | |||
| Lepu Medical ‐ SARS‐CoV‐2 Ag Rapid Test | 1; 286 (101) | 45.5 (35.6 to 55.8) | 89.2 (83.8 to 93.3) | |||
| Including sensitivity‐only cohorts | 2; 386 (201) | 49.8 (42.6to | ‐ | |||
| Liming Bio‐Products ‐ StrongStep COVID‐19 Ag | 1; 19 (9) | 0 (0 to 33.6) | 90.0 (55.5 to 99.7) | |||
| LumiraDx ‐ SARS‐CoV‐2 Ag | 4; 1373 (343) | 89.7 (63.0 to 97.8) | 98.5 (97.6 to 99.1) | 2; 1016 (186) | 91.1 (69.4 to 97.9) | 98.9 (97.9 to 99.4) |
| MEDsan GmbH ‐ SARS‐CoV‐2 Ag | 1; 184 (84) | 45.2 (34.3 to 56.5) | 97.0 (91.5 to 99.4) | |||
| Mologic ‐ COVID 19 Rapid Ag | 1; 650 (192) | 90.6 (85.6 to 94.3) | 100 (99.2 to 100) | 1; 650 (192) | 90.6 (85.6 to 94.3) | 100 (99.2 to 100) |
| Nal Von Minden ‐ NADAL COVID‐19 Ag | 2; 562 (143) | 60.9 (14.3 to 93.6) | 99.3 (97.8 to 99.8) | 1; 462 (69) | 88.4 (78.4 to 94.9) | 99.2 (97.8 to 99.8) |
| Including sensitivity‐only cohorts | 3; 686 (267) | 61.7 (27.0to | ‐ | |||
| Orient Gene/Healgen Scientific ‐ Coronavirus Ag | 2; 1377 (286) | 63.7 (42.2 to 80.8) | 99.7 (99.2 to 99.9) | |||
| Including sensitivity‐only cohorts | 3; 1472 (381) | 70.7 (52.1to | ‐ | 1; 95 (95) | 82.1 (72.9to | ‐ |
| PCL ‐ COVID19 Ag Rapid FIA | 1; 132 (62) | 88.7 (78.1 to 95.3) | 98.6 (92.3 to 100) | |||
| Precision Biosensor ‐ Exdia COVID‐19 Ag | 1; 532 (114) | 48.2 (38.8 to 57.8) | 99.5 (98.3 to 99.9) | 1; 532 (114) | 48.2 (38.8 to 57.8) | 99.5 (98.3 to 99.9) |
| Quidel ‐ SOFIA SARS Antigen FIA | 5; 2062 (280) | 82.4 (70.7 to 90.0) | 98.9 (98.3 to 99.3) | 3; 1871 (161) | 72.7 (65.3 to 79.0) | 98.9 (98.3 to 99.3) |
| R‐Biopharm ‐ RIDA QUICK SARS‐CoV‐2 Ag | 3; 237 (164) | 56.4 (36.1 to 74.8) | 98.6 (90.9 to 99.8) | 1; 70 (32) | 50.0 (31.9 to 68.1) | 100 (90.7 to 100) |
| RapiGEN ‐ BIOCREDIT COVID‐19 Ag | 7; 2426 (576) | 59.2 (45.7 to 71.4) | 99.6 (99.1 to 99.8) | 3; 1821 (220) | 63.2 (50.7 to 74.1) | 99.8 (99.3 to 99.9) |
| SD Biosensor ‐ Standard F COVID‐19 Ag | 6; 2692 (707) | 68.1 (56.1 to 78.2) | 98.1 (97.0 to 98.7) | 3; 1720 (382) | 69.6 (60.5 to 77.3) | 97.8 (96.9 to 98.5) |
| SD Biosensor/Roche ‐ Standard Q COVID‐Ag | 38; 25,193 (3937) | 75.2 (69.7 to 79.9) | 99.1 (98.9 to 99.2) | 17; 12,476 (1595) | 81.1 (75.5 to 85.6) | 99.4 (99.3 to 99.6) |
| Including sensitivity‐only cohorts | 40; 25,330 (4074) | 74.7 (69.5to | ‐ | |||
| SD Biosensor ‐ Standard Q COVID‐Ag (Nasal) | 1; 132 (36) | 86.1 (70.5 to 95.3) | 100 (96.2 to 100) | 1; 132 (36) | 86.1 (70.5 to 95.3) | 100 (96.2 to 100) |
| Savant Biotech ‐ Huaketai SARS‐CoV‐2 | 1; 109 (78) | 16.7 (9.2 to 26.8) | 100 (88.8 to 100) | |||
| Shenzhen Bioeasy Biotech ‐ 2019‐nCoV Ag | 4; 1137 (203) | 83.7 (70.9 to 91.6) | 92.5 (90.6 to 94.0) | 2; 899 (41) | 70.7 (55.2 to 82.6) | 90.5 (84.3 to 94.4) |
| Sichuan Mass Spectrometry Biotech ‐ 2019‐nCov Ag | 1; 85 (85) | 22.4 (14.0 to 32.7) | ‐ | |||
| Siemens ‐ CLINITEST Rapid COVID‐19 Ag | 3; 728 (289) | 61.3 (47.6 to 73.5) | 99.3 (97.9 to 99.8) | 1; 270 (116) | 75.9 (67.0 to 83.3) | 100 (97.6 to 100) |
| Spring Healthcare ‐ SARS‐CoV‐2 Ag | 1; 200 (100) | 77.0 (67.5 to 84.8) | 98.0 (93.0 to 99.8) | |||
| Sugentech Inc ‐ SGTI‐flex COVID‐19 Ag | 1; 106 (78) | 52.6 (40.9 to 64.0) | 96.4 (81.7 to 99.9) | 1; 106 (78) | 52.6 (40.9 to 64.0) | 96.4 (81.7 to 99.9) |
| SureScreen Diagnostics ‐ SureScreen F | 1; 200 (100) | 69.0 (59.0 to 77.9) | 98.0 (93.0 to 99.8) | |||
| Including sensitivity‐only cohorts | 2; 341 (241) | 63.9 (57.5to | ‐ | |||
| SureScreen Diagnostics ‐ SureScreen V | 3; 1671 (390) | 45.2 (29.0 to 62.4) | 99.6 (99.1 to 99.8) | |||
| Including sensitivity‐only cohorts | 4; 1694 (413) | 49.2 (33.8to | ‐ | |||
|
aSeparate pooling of sensitivity or specificity.
b2x2 tables combined prior to calculating estimates. CI: confidence interval | ||||||
Appendix 15. Detailed synthesis of results by test brand
Results are presented firstly for test brands with data available from evaluations conducted in symptomatic or asymptomatic populations, or both, and then for the 12 test brands with data only available for populations with mixed symptom status or where symptom status was not reported.
1. Test brands with evaluations conducted in symptomatic or asymptomatic populations
AAZ – COVID‐VIRO
Three evaluations of the COVID‐VIRO assay included 1204 samples and 534 SARS‐CoV2‐positive samples (Figure 7). Studies were conducted in symptomatic or mainly symptomatic participants using nasopharyngeal samples (Courtellemont 2021; Fourati 2020 [E]; Schwob 2020(c)).
The average sensitivity and specificity of the COVID‐VIRO assay in symptomatic people were:
84.9% (95% confidence interval (CI) 61.5% to 95.2%) and 97.0% (95% CI 95.4% to 98.1%); n = 3 evaluations; 1204 samples and 534 cases
Restricting to evaluations compliant with manufacturer's instructions for use (IFU), average sensitivities and specificities were:
91.3% (95% CI 78.2% to 96.9%) and 94.0% (95% CI 90.9% to 96.1%); n = 2; 572 samples and 239 cases
One of the two IFU‐compliant studies included 20 participants who had previously tested positive on PCR but who retested as negative with PCR at the time of the antigen test as having current SARS‐CoV‐2 infection (Courtellemont 2021). All 20 samples showed weak lines on antigen testing and were considered by the study authors as true positive cases. Based on the negative result of the concurrent PCR test, we considered these as false positives in the review. With our re‐calculation, the test demonstrated sensitivity of 96.0% (95% CI 90.2% to 98.9%) and specificity of 86.4% (95% CI 79.8% to 91.5%). Sensitivity in this study may have been inflated by the inclusion of hospitalized, confirmed SARS‐CoV‐2‐positive participants.
Abbott – BinaxNOW COVID‐19 Ag card
We identified six evaluations of the BinaxNOW COVID‐19 Ag card in 14,877 samples including 1051 SARS‐CoV‐2 cases, two in mixed‐symptom populations (Pilarowski 2020a; Pollock 2021a), and four in asymptomatic (Okoye 2021; Pilarowski 2021), or mainly asymptomatic populations (James 2021; Prince‐Guerra 2021). All evaluations used nasal samples (anterior nasal in five and nasal mid‐turbinate in one (Okoye 2021)), and all were considered to be compliant with the manufacturer IFU.
The average sensitivity and specificity of the BinaxNOW assay in IFU‐compliant evaluations were:
80.9% (95% CI 67.6% to 89.6%) and 99.9% (95% CI 99.5% to 100%) in symptomatic people; n = 4; 2018 samples and 358 cases
58.7% (95% CI 45.6% to 70.6%) and 99.8% (95% CI 99.7% to 99.8%) in asymptomatic people; n = 6; 12,530 samples and 588 cases.
Abbott – Panbio COVID‐19 Ag
We identified 44 evaluations of the Panbio assay; 39 evaluations (with 27,460 samples and 5976 cases) reported both sensitivity and specificity, and five evaluations including 552 cases reported sensitivity only (Fenollar 2020(a); Lanser 2021; Peto 2021(c) [B ‐ Lab tested]; Stokes 2021(a) [A]; Thommes 2021 [A]; results reported in Appendix 10).
Of the 39 evaluations reporting sensitivity and specificity (Abdulrahman 2020; Agullo 2021 [A]; Akingba 2021; Albert 2020; Alemany 2021; Baro 2021 [A]; Basso 2021 [C]; Billaud 2020; Bulilete 2021; Caruana 2021 [B]; Del Vecchio 2021; Dominguez Fernandez 2021; Drevinek 2020 [A]; Faico‐Filho 2021; Favresse 2021 [B]; Fenollar 2020(b); FIND 2020b (CH); FIND 2020b (DE); FIND 2021b [B]; Fourati 2020 [C]; Gonzalez‐Donapetry 2021; Gremmels 2021(a); Gremmels 2021(b); Halfon 2021; Jaaskelainen 2021 [C]; L'Huillier 2021; Linares 2020; Masia 2021 [A]; Merino 2021; Miyakawa 2021 [B]; Ngo Nsoga 2021; Olearo 2021 [B]; Perez‐Garcia 2021 [B]; Peto 2021(a) [B]; Schildgen 2021 [B]; Schwob 2020(b); Stokes 2021(b); Torres 2021a; Villaverde 2021), 31 used the assay with nasopharyngeal samples alone (n = 30) or in combination with an oropharyngeal sample (n = 1). Five evaluations used naso‐ or oropharyngeal samples (n = 3), oropharyngeal samples alone (n = 1), or a nasal mid‐turbinate swab (n = 1). One evaluation used throat wash or bronchoalveolar lavage (BAL), and the remaining two did not specify the sample site.
For symptomatic participants, the average sensitivity and specificity were:
74.8% (95% CI 67.6% to 80.8%) and 99.7% (95% CI 99.6% to 99.8%) overall; n = 24; 14,509 samples and 3167 cases; and
77.3% (95% CI 68.7% to 84.0%) and 99.7% (95% CI 99.5% to 99.8%) in IFU‐compliant evaluations; n = 11; 7718 samples and 1397 cases.
For asymptomatic participants the average sensitivity and specificity were:
56.9% (95% CI 42.8% to 69.9%) and 99.5% (95% CI 99.1% to 99.7%) overall; n = 14; in 4038 samples including 561 cases
57.9% (95% CI 35.4% to 77.5%) and 99.6% (95% CI 99.3% to 99.8%) in IFU‐compliant evaluations; n = 7; 2502 samples and 279 cases.
Reasons for non‐compliance with IFU requirements were use of frozen or stored samples (n = 7; Alemany 2021; Fourati 2020 [C]; Jaaskelainen 2021 [C]; Miyakawa 2021 [B]; Olearo 2021 [B]; Perez‐Garcia 2021 [B]; Peto 2021(a) [B]), or a time delay between sampling and testing of over one hour (n = 3 Baro 2021 [A]; Basso 2021 [C]; Favresse 2021 [B]), or the sample site used for at least some participants was not described on the IFU (n = 4; Abdulrahman 2020; FIND 2020b (DE); Ngo Nsoga 2021; Schildgen 2021 [B]). In a further eight evaluations, we could not judge IFU compliance because of missing information in the studies or because the use of viral transport medium (VTM) was not mentioned on the IFU (Akingba 2021; Caruana 2021 [B]; Del Vecchio 2021; Dominguez Fernandez 2021; Faico‐Filho 2021; Gremmels 2021(a); Halfon 2021; Linares 2020).
Assay sensitivity in both symptomatic and asymptomatic participants was only marginally affected by the addition of eight and three evaluations that reported sensitivity only (Table 4).
Access Bio ‐ CareStart COVID‐19 Ag
We identified one evaluation of the CareStart antigen assay in a primarily asymptomatic population where antigen tests were freely available regardless of symptoms or epidemiological exposure (Pollock 2021b). Anterior nasal swabs were collected by trained personnel and tested by laboratory scientists, and the evaluation was considered to be compliant with the manufacturer IFU.
Sensitivity and specificity were:
75.4% (95% CI 63.5% to 84.9%) and 94.8% (95% CI 90.3% to 97.6%) in symptomatic people; 241 samples and 69 cases
50.3% (95% CI 42.4% to 58.2%) and 98.9% (95% CI 98.1% to 99.4%) in asymptomatic people; 1257 samples and 165 cases.
Anhui Deepblue ‐ COVID‐19 Ag
We included two evaluations of the Anhui Deepblue assay, including one reporting sensitivity only, both of which were carried out by Public Health England (PHE) in symptomatic participants (Peto 2021(a) [D]; Peto 2021(c) [D ‐ Lab tested]). Neither evaluation was considered to be IFU‐compliant, either because of the use of VTM and frozen storage or because sample storage and the timing of the test was not stated (Peto 2021(c) [D ‐ Lab tested]).
The sensitivity and specificity of the test were:
47.1% (95% CI 39.9% to 54.5%) and 100% (95% CI 99.6% to 100%); in 1205 samples and 191 cases.
Adding data from the sensitivity only cohort, increased sensitivity to:
60.7% (95% CI 41.7% to 76.9%); 2 evaluations in 1382 samples and 368 confirmed cases.
Becton Dickinson ‐ BD Veritor
We identified nine evaluations of the BD Veritor assay, seven interpreted using the reader device (Caruana 2021 [D]; Schuit 2021(a); Gomez 2021(b); Kilic 2021; Van der Moeren 2021(a) [A]; Van der Moeren 2021(b); Young 2020), and two based on visual interpretation of the test cassettes (Stohr 2021 [A]; Van der Moeren 2021(a) [B]).
Of the seven evaluations using the reader device, five reported sensitivity and specificity data in symptomatic or mixed‐symptom populations. The evaluations used nasopharyngeal or combined naso‐ and oropharyngeal swabs (n = 3), anterior nasal swabs (n = 3) or nasal and oropharyngeal swabs (n = 1).
Average sensitivity and specificity in symptomatic populations were:
73.9% (95% CI 55.4% to 86.6%) and 99.1% (95% CI 98.6% to 99.4%) overall; n = 5; including 2498 samples and 299 cases;
66.4% (95% CI 57.0% to 74.9%) 98.8% (95% CI 98.1% to 99.3%) in the single evaluation considered to be IFU‐compliant (1384 samples and 116 cases).
Adding data from the two sensitivity‐only cohorts (n = 157 extra cases) increased sensitivity to 78.4% (95% CI 63.8% to 88.2%) overall (7 evaluations in 456 cases) and to 82.5% (95% CI 52.5% to 95.3%) in IFU‐compliant evaluations (2 evaluations in 148 cases).
Average sensitivity and specificity in two evaluations in asymptomatic populations (Caruana 2021 [D]; Schuit 2021(a), neither of which were IFU‐compliant were:
49.8% (95% CI 32.1% to 67.5%) and 99.7% (95% CI 99.3% to 99.8%); n = 2; including 2556 samples and 203 cases.
Reasons for lack of compliance with IFU specifications included the use of frozen or stored samples (n = 2), too long an interval between sampling and testing (n = 2), or use of VTM (n = 1).
Of the two evaluations that did not use the Veritor analyser device to interpret the test result, one (including 351 samples with 17 cases) found that visual inspection of the test device resulted in the same sensitivity as with the analyser device, and similar specificity (100% compared to 99% using the analyser device) (Van der Moeren 2021(a) [A]). The other evaluation was larger (1391 samples and 176 cases), and reported considerably lower sensitivity when used in a mixed‐symptom population (sensitivity 48.9%, specificity 100%) (Stohr 2021 [A]).
BIONOTE ‐ NowCheck COVID‐19 Ag
Three studies including a total of 1944 samples and 190 cases evaluated the NowCheck assay (FIND 2020a; FIND 2021a [B]; Rottenstreich 2021); all used nasopharyngeal swabs and one included a comparison using the NowCheck kit for nasal swabs (FIND 2021a [A]), using paired nasal mid‐turbinate swabs.
Average sensitivity and specificity in symptomatic populations (both considered IFU‐compliant) were:
89.5% (95% CI 84.1% to 93.2%) and 97.7% (95% CI 95.8% to 98.8%) in IFU‐compliant evaluations; n = 2; including 618 samples and 181 cases.
Sensitivity and specificity in the evaluation in asymptomatic people (all pregnant women admitted to hospital for delivery; Rottenstreich 2021)) were:
55.6% (95% CI 21.2% to 86.3%) and 100% (95% CI 99.7% to 100%); 1326 samples including 9 cases.
This evaluation used samples in VTM and did not report the time interval between sampling and testing; IFU compliance was therefore considered unclear (Rottenstreich 2021).
The study comparing results in paired nasopharyngeal and nasal mid‐turbinate swabs demonstrated identical results for sensitivity (89.9%, 95% CI 81.0% to 95.5%) and for specificity (98.6%, 95% CI 94.9% to 99.8%); including 218 participants and 79 cases.
Biosynex ‐ Biosynex COVID‐19 Ag BSS
We identified a single evaluation of the Biosynex assay in symptomatic participants (Fourati 2020 [D]), including 634 samples with 297 with confirmed SARS‐CoV‐2. The evaluation was not in compliance with the manufacturer’s IFU because samples were frozen prior to testing. The setting in which participants presented for testing was not reported and specificity was estimated in pre‐pandemic samples.
Sensitivity was 59.6% (95% CI 53.8% to 65.2%) and specificity 100% (95% CI 98.9% to 100%) (Table 4).
Certest Biotech ‐ CerTest
We included one evaluation of the CerTest assay (Perez‐Garcia 2021 [A]). Participants were described as suspected of having COVID‐19 but no detail about symptom status was provided. Nasopharyngeal samples in VTM were tested after a period of frozen storage and the evaluation was considered not to be compliant with the manufacturer IFU.
Sensitivity and specificity were:
53.5% (95% CI 45.7% to 61.2) and 100% (95% CI 97.6% to 100%); in 320 samples including 170 cases of SARS‐CoV‐2.
Coris Bioconcept ‐ COVID‐19 Ag Respi‐Strip
Eight evaluations of the Coris Bioconcept assay reported both sensitivity and specificity in 1831 samples, with 746 SARS‐CoV‐2 positive cases. Five of the eight were laboratory‐based evaluations with limited detail regarding study participants (Blairon 2020; Fourati 2020 [A]; Lambert‐Niclot 2020; Mertens 2020; Scohy 2020). One study recruited from community‐based COVID‐19 test centres (FIND 2020f), and two included samples from suspected cases in hospital inpatients (n = 1; Veyrenche 2021)), or from the Emergency Department or Infectious Diseases ward (Ciotti 2021). All evaluations tested naso‐ or combined naso‐ or oropharyngeal swabs. Three were considered not to have fully met manufacturer IFU.
The average sensitivity and specificity of the COVID‐19 Ag Respi‐Strip in symptomatic populations were:
37.5% (95% CI 28.4% to 47.7%) and 99.8% (95% CI 98.8% to 100%) overall; n = 5; 1158 samples and 585 cases; and
34.3% (95% CI 29.9% to 39.1%) and 100% (95% CI 99.0% to 100%) in IFU‐compliant evaluations; n = 3; 765 samples and 408 cases.
One evaluation reporting a data for a small subset of asymptomatic people demonstrated sensitivity and specificity of:
28.6% (95% CI 8.4% to 58.1%) and 100% (95% CI 88.8% to 100%) (IFU‐compliant); 45 samples and 14 cases.
Denka Co ‐ QuickNavi COVID‐19 Ag
Two evaluations of the QuickNavi rapid assay were included, both were conducted at a drive‐through PCR testing centre and included participants referred from primary care or public health centres. One study used nasopharyngeal swabs (Takeuchi 2021a), and the second tested anterior nasal samples (Takeuchi 2021b), and both were considered to be compliant with manufacturer IFU specifications.
The average sensitivity and specificity of the QuickNavi COVID‐19 Ag were:
84.2% (95% CI 66.2% to 93.5%) and 100% (95% CI 99.8% to 100%) in symptomatic populations; n = 2 evaluations, 1633 samples including 123 cases; and
75.8% (95% CI 57.7% to 88.9%) and 100% (95% CI 99.0% to 100%) in asymptomatic contacts; n = 1; 415 samples including 33 cases.
DIALAB ‐ DIAQUICK COVID‐19 Ag
A single study evaluated the DIAQUICK assay in PCR+ve nasopharyngeal swab samples from symptomatic hospital inpatients (Thommes 2021 [C]). Sensitivity was 61.6% (95% CI 51.3% to 71.2%); 99 samples (IFU‐compliant).
ECODiagnostica ‐ COVID‐19 Ag ECO
A single study evaluated the COVID‐19 Ag ECO assay in nasopharyngeal swab samples from symptomatic hospital inpatients (Filgueiras 2021). The sensitivity and specificity were 69.1% (95% CI 55.2% to 80.9%) and 97.9% (95% CI 92.6% to 99.7%); 150 samples including 55 cases (IFU‐compliant).
Fortress Diagnostics ‐ Coronavirus Ag
A single study evaluated the Fortress Diagnostics assay in combined naso‐ and oropharyngeal samples from symptomatic hospital inpatients (Peto 2021(a) [E]). The sensitivity and specificity were 56.0% (95% CI 48.7% to 63.2%) and 99.9% (95% CI 99.4% to 100%); 1191 samples and 191 cases (not IFU‐compliant because of the use of previously frozen samples).
Fujirebio ‐ ESPLINE SARS‐CoV‐2
A total of 10 evaluations of the ESPLINE assay were identified (Aoki 2021; Basso 2021 [A]; Basso 2021 [B]; FIND 2021d; Ishii 2021 [A]; Miyakawa 2021 [C]; Nagura‐Ikeda 2020; Takeda 2020; Yokota 2020(a); Yokota 2020(b)), including three cases‐only studies (Nagura‐Ikeda 2020; Yokota 2020(a); Yokota 2020(b)). None of the evaluations were considered to be compliant with the manufacturer’s IFU. Only four of the 10 reported results separately in symptomatic or asymptomatic populations, two using nasopharyngeal samples (Aoki 2021; Yokota 2020(a)), and two evaluating the kit using saliva samples (Nagura‐Ikeda 2020; Yokota 2020(b)).
One evaluation reported sensitivity and specificity of 39.7% (95% CI 27.6% to 52.8%) and 97.0% (95% CI 89.5% to 99.6%), in 129 nasopharyngeal samples from symptomatic participants, including 63 from confirmed SARS‐COV‐2 cases (Aoki 2021).
Adding data from the three cases‐only studies, average sensitivity was 29.6% (95% CI 14.6% to 51.0%); n = 4 evaluations and 185 samples, all from symptomatic participants.
Sensitivity in the single evaluation reporting data in asymptomatic participants was 13.3% (95% CI 1.7% to 40.5%), in 15 saliva samples (Nagura‐Ikeda 2020).
Innova Medical Group ‐ Innova SARS‐CoV‐2 Ag
Thirteen evaluations of the Innova assay were identified. This includes six separate substudies conducted by Public Health England that were included in the previous iteration of this review; three reported both sensitivity and specificity (Peto 2021(a) [A]; Peto 2021(b) [non‐HCW tested]; PHE 2020), two reported sensitivity alone (Peto 2021(c) [A ‐ HCW tested]; Peto 2021(c) [A ‐ Lab tested]), and one reported specificity alone (Peto 2021(d)). These studies were conducted in symptomatic (Peto 2021(a) [A]), or mainly symptomatic (Peto 2021(b) [non‐HCW tested]; Peto 2021(c) [A ‐ HCW tested]; Peto 2021(c) [A ‐ Lab tested]; PHE 2020), populations, apart from the asymptomatic staff‐screening study (Peto 2021(d)). Data for the outbreak‐evaluation study was provided by the study authors. Of the additional seven studies identified for the present iteration of the review (Ferguson 2021; Garcia‐Finana 2021; Houston 2021; Pickering 2021(a) [A]; Pickering 2021(b) [A]; Pickering 2021(c) [A]; Young 2021), three did not report symptom status of included participants (Pickering 2021(a) [A]; Pickering 2021(b) [A]; Pickering 2021(c) [A]), and one did not report data separately for symptomatic and asymptomatic participants (Young 2021).
Average sensitivity and specificity in symptomatic participants were:
68.1% (95% CI 47.2 to 83.6%) and 99.0% (95% CI 98.5 to 99.3%) overall; n = 3 evaluations with 3522 samples including 830 cases; and
57.5% (95% CI 52.3 to 62.6%) and 99.6% (95% CI 99.1 to 99.9%) in the IFU‐compliant evaluations (95% CI using sample sites recommended by the manufacturer, i.e. combined nasal and oropharyngeal samples); 1676 samples including 372 cases.
Adding data from the cases‐only studies, average sensitivities in symptomatic populations were:
70.8% (95% CI 58.1 to 80.9%) overall; n = 5; 3943 samples including 1251 cases; and
69.1% (95% CI 58.3 to 78.2%) in IFU‐compliant evaluations; n = 3; 2097 samples including 793 cases.
Average sensitivity and specificity in asymptomatic participants were:
38.5% (95% CI 28.4 to 49.7%) and 100% (95% CI 99.8 to 100%) overall; n = 2; 6224 samples including 78 cases; and
40.0% (95% CI 28.5 to 52.4%) and 99.9% (95% CI 99.8 to 100%) in the IFU‐compliant evaluations; 5504 samples including 70 cases.
Results for each of the three IFU‐compliant evaluations by test operator were:
sensitivity of 57.5% (95% CI 52.3% to 62.6%) and specificity 99.6% (95% CI 99.1% to 99.9%), when the test was used by self‐trained, non‐healthcare workers (n = 1; 1676 samples, 372 cases; Peto 2021(b) [non‐HCW tested]);
sensitivity of 70.0% (95% CI 63.5% to 75.9%) when the test was used by healthcare workers (n = 1; 223 cases; Peto 2021(c) [A ‐ HCW tested]);
sensitivity of 78.8% (95% CI 72.4% to 84.3%) when the test was used by laboratory scientists (n = 1; 198 cases; Peto 2021(c) [A ‐ Lab tested]).
Joysbio Biotech ‐ SARS‐CoV‐2 Ag
A single study evaluated the Joysbio Biotech assay in mainly symptomatic participants attending a COVID‐19 test centre (FIND 2021e). Sensitivity and specificity were 70.5% (95% CI 54.8% to 83.2%) and 99.1% (95% CI 96.8% to 99.9%); in 265 anterior nasal samples, including 44 cases. The manufacturer’s IFU specifies the use of nasopharyngeal samples therefore the study was considered not to have complied with IFU specifications.
Lepu Medical ‐ SARS‐CoV‐2 Ag Rapid Test
Two studies evaluated the Lepu Medical rapid test, one in symptomatic participants (Saeed 2021 [A]), and one in asymptomatic participants (Baro 2021 [D]). Neither study was considered to be compliant with the manufacturer’s IFU either because of a time delay between sample collection and testing (Baro 2021 [D]), or because it was not clear whether VTM was used or its use was not mentioned on the IFU document.
Sensitivity in paired samples from 100 symptomatic PCR+ve participants was:
54.0% (95% CI 43.7% to 64.0%) in nasopharyngeal samples (Saeed 2021 [A]); and
21.0% (95% CI 13.5% to 30.3%) in saliva samples (Saeed 2021 [B]).
Sensitivity and specificity in 286 nasopharyngeal samples (including 101 cases) from asymptomatic participants were:
45.5% (95% CI 35.6% to 55.8%) and 89.2% (95% CI 83.8% to 93.3%).
Liming Bio‐Products ‐ StrongStep® COVID‐19 Ag
We identified a single evaluation of the StrongStep assay in 19 symptomatic participants with nine SARS‐CoV‐2‐positive samples (Weitzel 2020 [B]). We could not identify the manufacturer’s IFU for this assay. The study authors terminated the evaluation early following poor early results for this assay.
Sensitivity was 0% (95% CI 0% to 33.6%) and specificity 90.0% (95% CI 55.5% to 99.7%; 19 samples, 9 cases).
LumiraDx ‐ SARS‐CoV‐2 Ag
Four studies evaluated the LumiraDx microfluidic fluorescent immunoassay (Drain 2021(a); Drain 2021(b); Kohmer 2021 [D]; Kruger 2021), two of which reported results separately for symptomatic and or asymptomatic participants in nasal mid‐turbinate or nasopharyngeal samples (both considered to be compliant with manufacturer IFU specifications; Drain 2021(b); Kruger 2021).
The average sensitivity and specificity were:
91.2% (95% CI 70.0% to 97.9%) and 98.6% (95% CI 97.2% to 99.3%) in symptomatic people; n = 2, with 741 samples including 177 cases; and
77.8% (95% CI 40.0% to 97.2%) and 99.6% (95% CI 97.9% to 100%) in asymptomatic people, n = 1, with 272 samples including 9 cases (Kruger 2021).
MEDsan GmbH ‐ SARS‐CoV‐2 Ag
One study evaluated the MEDsan assay in symptomatic hospital inpatients who provided either naso‐ or oropharyngeal samples (Olearo 2021 [C]). Sensitivity and specificity were 45.2% (95% CI 34.3% to 56.5%) and 97.0% (95% CI 91.5% to 99.4%) in 184 samples, including 84 cases. The manufacturer IFU recommends testing in nasopharyngeal samples only, therefore the study was considered not to be compliant with the IFU.
Orient Gene/Healgen Scientific ‐ Coronavirus Ag
The Orient Gene assay (also distributed by Healgen Scientific) was evaluated in three studies (Favresse 2021 [C]; Peto 2021(a) [C]; Peto 2021(c) [C ‐ Lab tested]), two of which reported data for symptomatic participants (Peto 2021(a) [C]; Peto 2021(c) [C ‐ Lab tested]).
One study using combined naso‐ and oropharyngeal samples (not IFU‐compliant because of frozen sample storage) reported sensitivity and specificity of 48.9% (95% CI 41.6% to 56.3%) and 100% (95% CI 99.6% to 100%) in 1189 samples from symptomatic participants, including 190 cases (Peto 2021(a) [C]). A second study demonstrated assay sensitivity in combined nasal and oropharyngeal samples of 82.1% (95% CI 72.9% to 89.2%) (in 95 samples; Peto 2021(c) [C ‐ Lab tested]).
Average sensitivity in the two evaluations was 67.2% (95% CI 40.7% to 86.0%) (285 samples).
Precision Biosensor ‐ Exdia COVID‐19 Ag
A single evaluation of the Exdia COVID‐19 assay reported data separately for both symptomatic and asymptomatic participants using nasopharyngeal samples in VTM (in compliance with manufacturer IFU specifications; Caruana 2021 [C]). Sensitivity and specificity were:
52.2% (95% CI 41.4% to 62.9%) and 99.0% (95% CI 96.5% to 99.9%) in symptomatic participants (293 samples including 90 cases); and
33.3% (95% CI 15.6% to 55.3%) and 100% (95% CI 98.3% to 100%) in asymptomatic participants (239 samples and 24 cases).
Quidel Corporation ‐ SOFIA SARS Antigen
We included five evaluations of the Quidel SOFIA assay, three in symptomatic participants (Beck 2021; Gomez 2021(a); Porte 2021 [A]), one including mainly asymptomatic participants (Pray 2021), and one that did not report participants’ symptom status (Jaaskelainen 2021 [A]). Studies used either nasal mid‐turbinate, nasopharyngeal or combined naso‐ and oropharyngeal samples, all of which are within the IFU specifications, however, two studies stored samples in VTM and were therefore considered not to be IFU‐compliant.
Average sensitivity and specificity in symptomatic participants were:
80.0% (95% CI 71.5% to 86.4%) and 99.4% (95% CI 98.7% to 99.8%) overall; n = 4; 1064 samples including 176 cases; and
76.4% (95% CI 68.8% to 82.6%) and 99.5% (95% CI 98.8% to 99.8%) restricted to IFU‐compliant evaluations; n = 3; 1000 samples including 144 cases.
For asymptomatic participants, sensitivity and specificity were:
41.2% (95% CI 18.4% to 67.1%) and 98.4% (95% CI 97.3% to 99.1%); n = 1; 871 samples including 17 cases (IFU‐compliant; Pray 2021).
RapiGEN ‐ BIOCREDIT COVID‐19 Ag
We identified seven evaluations of the RapiGen BIOCREDIT assay (Abdelrazik 2021; FIND 2020e (BR); FIND 2020e (DE); Schildgen 2021 [A]; Shidlovskaya 2021 [B]; Shrestha 2020; Weitzel 2020 [A]). Two studies were conducted in hospital settings; two at community‐based COVID‐19 test centres, and the remaining three at an emergency department (n = 1), quarantine centre (n = 1) or did not clearly report the setting (n = 1). Two studies included only symptomatic participants, two reported including both symptomatic and asymptomatic participants (mixed group) and one did not report symptom status. All evaluations apart from one (Schildgen 2021 [A]), tested nasopharyngeal or combined naso‐ or oropharyngeal samples, and only three of the six were considered to be compliant with manufacturer IFU specifications (FIND 2020e (BR); FIND 2020e (DE); Shidlovskaya 2021 [B]).
The average sensitivity and specificity of the BIOCREDIT assay in symptomatic people were:
61.5% (95% CI 49.3% to 72.5%) and 98.4% (95% CI 96.6% to 99.2%) (n = 4; 714 samples, 284 cases); and
66.3% (95% CI 52.9% to 77.5%) and 99.0% (95% CI 97.3% to 99.6%) in IFU‐compliant evaluations (n = 2, with 582 samples including 195 cases).
For asymptomatic participants, sensitivity and specificity were:
63.2% (95% CI 21.7% to 91.4%) and 98.9% (95% CI 82.9% to 99.9%) in asymptomatic people (n = 2; 140 samples, 60 cases). Neither was compliant with manufacturer IFU (Schildgen 2021 [A]; Shrestha 2020).
Savant Biotech ‐ Huaketai SARS‐CoV‐2 N Protein
We identified a single evaluation of the Huaketai assay in 109 symptomatic participants, using combined naso‐ or oropharyngeal swabs in VTM (Weitzel 2020 [C]; Figure 7). We could not obtain the manufacturer IFU.
Sensitivity was 16.7% (95% CI 9.2% to 26.8%) and specificity was 100% (95% CI 88.8% to 100%; 109 samples, 78 cases; Table 4; Table 5).
SD Biosensor ‐ Standard F COVID‐19 Ag
We identified six evaluations of the Standard F assay (Drevinek 2020 [B]; FIND 2020d (BR); FIND 2020d (DE); Liotti 2021; Osterman 2021(a) [A]; Porte 2021 [B]); five included all or mainly symptomatic participants from community‐based COVID‐19 test centres or testing of hospital inpatients, and one was a laboratory‐based study that did not provide details regarding symptom status (Liotti 2021). All evaluations tested nasopharyngeal or combined naso‐ or oropharyngeal samples, however only three complied with manufacturer IFU (Drevinek 2020 [B]; FIND 2020d (BR); FIND 2020d (DE)). Reasons for non‐compliance were the use of VTM, or lack of information concerning VTM.
The average sensitivity and specificity of the Standard F COVID‐19 Ag assay in symptomatic people were:
74.3% (95% CI 61.8% to 83.9%) and 97.4% (95% CI 96.4% to 98.1%) in symptomatic people (n = 4; 1742 samples, 380 cases); and
75.5% (95% CI 68.2% to 81.5%) and 97.2% (95% CI 96.0 to 98.1%) in the two IFU‐compliant evaluations (n = 2; 1129 samples, 159 cases).
Adding data from a single cases‐only study in symptomatic people (Drevinek 2020 [B]), the average sensitivities were:
72.5% (95% CI 63.2% to 80.2%) overall (5 evaluations with 547 cases); and
72.1% (95% CI 66.7% to 77.0%) (n = 3 IFU‐compliant evaluations with 326 cases).
The same cases‐only study reported sensitivity in asymptomatic contacts: sensitivity was 43.6% (95% CI 30.3% to 57.7%) (55 PCR+ve samples).
SD Biosensor ‐ Standard Q COVID‐19 Ag (Roche – Rapid Antigen Test)
For this iteration of the review, we have considered the SD Biosensor assay distributed by Roche to be the same as the Standard Q assay. We identified 40 evaluations of the Standard Q assay (Baro 2021 [C]; Caruana 2021 [A]; Cerutti 2020; Chaimayo 2020; Favresse 2021 [D]; FIND 2020c (BR); FIND 2020c (CH); FIND 2020c (DE); FIND 2021c (BR) [B]; FIND 2021c (DE) [B]; Fourati 2020 [B]; Gupta 2020; Igloi 2021; Jaaskelainen 2021 [B]; Jakobsen 2021; Kerneis 2021; Kohmer 2021 [B]; Kriemler 2021; Kruttgen 2021; Lindner 2021a [B]; Lindner 2021b [C]; Miyakawa 2021 [D]; Mockel 2021(a); Mockel 2021(b); Nalumansi 2020; Nikolai 2021(b) [B]; Olearo 2021 [A]; Osterman 2021(a) [B]; Osterman 2021(b); Pena 2021; Pena‐Rodriguez 2021; Peto 2021(a) [F]; Ristic 2021; Salvagno 2021; Schildgen 2021 [C]; Schuit 2021(b); Schwob 2020(a); Stohr 2021 [B]; Thommes 2021 [D]; Turcato 2021), including two cases‐only studies (Osterman 2021(b); Thommes 2021 [D]); these reported data for 25,330 samples, with 4074 confirmed SARS‐CoV‐2‐positive cases.
Eight evaluations either did not provide details about the symptom status of included participants (n = 4; Jaaskelainen 2021 [B]; Kohmer 2021 [B]; Kruttgen 2021; Miyakawa 2021 [D]), or did not report data separately for symptomatic and asymptomatic participants (n = 4; Favresse 2021 [D]; Osterman 2021(b); Pena‐Rodriguez 2021; Stohr 2021 [B]). Of the remaining 32 evaluations, 18 were conducted in COVID‐19 test centres (n = 14) or emergency department or urgent care settings (n = 4; Chaimayo 2020; Fourati 2020 [B]; Salvagno 2021; Schildgen 2021 [C]), seven were conducted in hospital settings (Caruana 2021 [A]; Olearo 2021 [A]; Peto 2021(a) [F]; Thommes 2021 [D]; Cerutti 2020; Nalumansi 2020; Osterman 2021(a) [B]), two conducted targeted mass screening (Baro 2021 [C]; Pena 2021), and one provided testing in schools (Kriemler 2021). The remaining four studies were laboratory‐based or did not clearly identify the setting for testing (Chaimayo 2020; Fourati 2020 [B]; Salvagno 2021; Schildgen 2021 [C]).
The majority of the 32 evaluations in symptomatic or asymptomatic people (n = 28) used nasopharyngeal samples in all (n = 23) or at least some (n = 4) participants. Other sample types (with one evaluation each) included a combined nasal and oropharyngeal sample, buccal swab, BAL or throat wash and sample type not reported (Nalumansi 2020; Kriemler 2021; Schildgen 2021 [C]; Turcato 2021). Overall, 16/32 were considered to be compliant with manufacturer’s IFUs (FIND 2020c (BR); FIND 2020c (CH); FIND 2021c (BR) [B]; FIND 2021c (DE) [B]; Igloi 2021; Jakobsen 2021; Kerneis 2021; Lindner 2021a [B]; Lindner 2021b [C]; Mockel 2021(a); Mockel 2021(b); Nikolai 2021(b) [B]; Pena 2021; Ristic 2021; Schuit 2021(b); Schwob 2020(a)), the others did not comply with recommended storage conditions (n = 7), used unvalidated VTM (n = 5), did not use recommended sample sites (n = 4), or did not report enough information for us to judge IFU compliance (n = 4).
The average sensitivity and specificity of the Standard Q COVID‐19 Ag assay in symptomatic people were:
78.6% (95% CI 72.3% to 83.7%) and 98.7% (95% CI 98.4% to 98.9%) overall, based on 26 evaluations with 10,678 samples including 2539 cases; and
84.0% (95% CI 79.2% to 87.9%) and 99.2% (95% CI 98.8% to 99.4%) in 15 IFU‐compliant evaluations (5116 samples including 1197 cases).
Adding data from two sensitivity‐only studies had only a slight effect on overall sensitivity: 79.2% (95% CI 72.9% to 84.3%); n = 28 evaluations with 2659 cases.
For asymptomatic participants average sensitivity and specificity were:
59.4% (95% CI 49.6% to 68.5%) and 99.3% (95% CI 99.1% to 99.4%) overall; n = 12, with 10,049 samples and 551 cases; and
64.6% (95% CI 51.3% to 75.9%) and 99.6% (95% CI 99.4% to 99.7%) in IFU‐compliant evaluations; n = 4 with 5914 samples and 250 cases.
Adding data from one sensitivity‐only study had a marginal effect on overall sensitivity: 60.4% (95% CI 50.5% to 69.6%), based on 13 evaluations and 554 cases.
SD Biosensor ‐ Standard Q COVID‐19 Ag (Nasal)
Four studies evaluated the Standard Q kit for nasal swabs in nasal mid‐turbinate samples, three in comparison to the Standard Q NP swab kit (FIND 2021c (BR) [A]; FIND 2021c (DE) [A] ; Nikolai 2021(b) [A]), and one in comparison to anterior nasal swabs (Nikolai 2021(a) [B]); (see direct test comparisons). All four studies were conducted in symptomatic participants and met manufacturer IFU specifications.
Average sensitivity and specificity in the four evaluations were 85.2% (95% CI 79.4% to 89.6%) and 99.3% (95% CI 97.9% to 99.8%), based on 621 samples, including 189 cases.
Shenzhen Bioeasy Biotech ‐ 2019‐nCoV Ag
We included four evaluations of the Bioeasy fluorescent immunoassay (FIA); these included 1137 samples with 203 SARS‐CoV‐2‐positive cases FIND 2021j; Porte 2020; Weitzel 2020 [D]; Parada‐Ricart 2020). Studies were conducted in hospital emergency departments (n = 2), a community COVID‐19 test centre (n = 1), or unclear setting. Participants in studies were all symptomatic or mainly symptomatic; one reported data for a small subset of asymptomatic people (Parada‐Ricart 2020).
Two evaluations used combined naso‐ or oropharyngeal swabs, one tested either nasopharyngeal or oropharyngeal swabs and one used nasal samples. Two evaluations used swabs in VTM, which was not documented as suitable for use on the manufacturer IFU.
The average sensitivity and specificity of the Shenzhen Bioeasy assay in symptomatic participants were:
84.4% (95% CI 72.4% to 91.7%) and 93.2% (95% CI 91.3% to 94.6%) overall; n = 4, with 1093 samples and 202 cases; and
72.5% (95% CI 56.8% to 84.1%) and 92.5% (95% CI 90.5% to 94.1%) in IFU‐compliant evaluations; n = 2 with 855 samples and 40 cases.
For asymptomatic people, 44 samples were available from one evaluation, only one of which was PCR+ve; assay sensitivity was 0% (95% CI 0% to 97.5%) and specificity 79.1% (95% CI 64.0% to 90.0%).
We excluded an additional study that reported the development rather than validation of this assay (Diao 2020).
Sichuan Mass Spectrometry Biotech ‐ 2019‐nCov Ag
A single evaluation of the Sichuan Mass Spectrometry assay reported data for symptomatic participants using nasopharyngeal samples from PCR+ve participants (Thommes 2021 [B]). We were not able to identify a manufacturer IFU for this assay. Sensitivity was 22.4% (95% CI 14.0% to 32.7%) in 85 samples.
Siemens ‐ CLINITEST Rapid COVID‐19 Ag
We included three studies of the CLINITEST assay (Baro 2021 [B]; Torres 2021b; Olearo 2021 [D]); two in symptomatic or mainly symptomatic populations (hospital inpatient or COVID‐19 test centre settings) and one in an asymptomatic population as part of a regional surveillance programme (Baro 2021 [B]). Nasopharyngeal or either naso‐ or oropharyngeal samples were used.
Average sensitivity and specificity in symptomatic people were:
68.7% (95% CI 48.0% to 83.8%) and 100% (95% CI 98.0% to 100%) overall; n = 2; 350 samples including 163 cases; and
80.2% (95% CI 70.6% to 87.8%) and 100% (95% CI 95.8% to 100%) in the IFU‐compliant evaluation; 178 samples with 91 cases.
For asymptomatic people, average sensitivity and specificity were:
53.2% (95% CI 44.5% to 61.7%) and 98.8% (95% CI 96.4% to 99.6%) overall; n = 2 with 378 samples including 126 cases; and
60.0% (95% CI 38.7% to 78.9%) and 100% (95% CI 94.6% to 100%) in IFU‐compliant evaluations; n = 1; 92 samples with 25 cases.
Sugentech Inc ‐ SGTI‐flex COVID‐19 Ag
A single evaluation of the SGTI‐flex assay reported data for symptomatic hospital inpatients using nasopharyngeal samples (Shidlovskaya 2021 [A]). The evaluation was judged to meet manufacturer IFU specifications. Sensitivity and specificity were 52.6% (95% CI 40.9% to 64.0%) and 96.4% (95% CI 81.7% to 99.9%), in 106 samples including 78 cases.
SureScreen Diagnostics ‐ SureScreen V
Four evaluations were included (Baro 2021 [E]; Peto 2021(a) [G]; Pickering 2021(a) [C]; Pickering 2021(c) [B]), one in symptomatic participants (Peto 2021(a) [G]), one in asymptomatic participants (Baro 2021 [E]), and two that did not report symptom status. None of the four evaluations were judged to meet manufacturer IFU specifications because of reported sample storage (frozen or stored for too long after collection).
One evaluation using combined naso‐ and oropharyngeal swabs from symptomatic people reported a sensitivity of 42.9% (95% CI 35.7% to 50.2%) and specificity 99.9% (95% CI 99.4% to 100%), in 1185 samples including 189 cases.
The evaluation in asymptomatic people used nasopharyngeal swabs; sensitivity was 28.7% (95 % CI 20.1% to 38.6%) and specificity 97.8% (95% CI 94.6% to 99.4%), in 286 samples including 101 cases.
2. Test brands with evaluations conducted only in mixed populations or symptom status not reported
For 12 assays we only identified evidence in populations with mixed symptom status or where symptom status was not reported, that is, we could not extract data separately for symptomatic and asymptomatic participants (including assays from: Biotical Health; Boditech Medical; E25Bio; Encode/Emmo Pharma; FUJIFILM; Guangzhou Wondfo; Mologic; Nal Von Minden; PCL; R‐Biopharm; Spring Healthcare; SureScreen Diagnostics (SureScreen F assay)). Data per test including results from meta‐analyses, where this was carried out, are reported in Appendix 14.
Six of the twelve assays were evaluated in single small studies (including between 41 and 100 PCR+ve cases) (assays from: Biotical Health; Boditech Medical; FUJIFILM; Guangzhou Wondfo; PCL; Spring Healthcare). Sensitivities ranged from 66.7% to 88.7% and specificities from 98.6% to 100% (Appendix 14).
Data for the six assays that were evaluated in larger cohorts of samples or in more than one evaluation are described further below.
The Mologic COVID‐19 rapid antigen test was evaluated in a single cohort of participants presenting at a COVID‐19 test centre (FIND 2021f); 67% (440/662) of anterior nasal samples were from symptomatic participants and the evaluation was judged to be compliant with the manufacturer IFU. The sensitivity and specificity of the assay were 90.6% (95% CI 85.6% to 94.3%) and 100% (95% CI 99.2% to 100%) in 650 samples including 192 PCR+ve cases; 16/665 reportedly had invalid antigen test results, but data were reported for 650/665.
Two assays were evaluated in the same two study cohorts reported in a single study report: the Encode/Emmo Pharma assay (Pickering 2021(a) [E]; Pickering 2021(b) [B]), and the SureScreen F assay (Pickering 2021(a) [F]; Pickering 2021(b) [C]). Symptom status and setting for testing were not reported for either study cohort and none of the evaluations were considered to be compliant with manufacturer IFUs. The two‐group study reporting both sensitivity and specificity showed sensitivities of 74.0% and 69.0% for the two assays respectively, and specificities 100% (95% CI 96.4% to 100%) and 98.0% (95% CI 93.0% to 99.8%). Adding data from an additional ‘cases‐only’ cohort, average sensitivities were 74.2% (95% CI 67.4% to 80.3%) for the Encode/Emmo Pharma assay (n = 190 cases) and 63.9% (95% CI 57.5% to 70.0%) for SureScreen F (n = 241 cases); sample numbers varied because of limited sample volume.
TheE‐25 Bio COVID‐19 DART assay was evaluated in two studies (Nash 2020; Pickering 2021(a) [B]), including 390 samples with 200 cases. No details about participant symptom status were reported. Average sensitivity and specificity were 77.5% (95% CI 71.2% to 82.8%) and 88.4% (95% CI 83.0%, 92.3%). We were unable to obtain the IFU for this assay from the manufacturer.
Two assays had three evaluations each: the Nal Von Minden NADAL COVID‐19 assay (FIND 2021g; Kohmer 2021 [C]; Stromer 2020), and the R‐Biopharm RIDA QUICK SARS‐CoV‐2 assay (Kohmer 2021 [A]; Toptan 2021(a); Toptan 2021(b)). Symptom status was not reported for the full cohort of participants in any of the six evaluations, although one did report symptom status for the PCR+ve cases (FIND 2021g). All evaluations obtained nasopharyngeal samples from all participants, one combined with an oropharyngeal sample. Two of the six were judged to be IFU‐compliant, one per assay (FIND 2021g; Toptan 2021(b)).
For the Nal von Minden assay, average sensitivity and specificity were 60.9% (95% CI 14.3% to 93.6%) and 99.3% (95% CI 97.8% to 99.8%) overall in 562 samples including 143 cases (2 evaluations), and 88.4% (95% CI 78.4% to 94.9%) and 99. 2% (95% CI 97.8% to 99.8%) in the IFU‐compliant evaluation (462 samples including 69 cases). Adding data from a single cases‐only evaluation had a marginal effect on sensitivity.
For the R‐Biopharm assay, average sensitivity and specificity were 56.4% (95% CI 36.1%, 74.8%) and 98.6% (95% CI 90.9% to 99.8%) overall (n = 3; 237 samples including 164 cases) and 50.0% (95% CI 31.9% to 68.1%) and 100 (95% CI 90.7% to 100%) in the IFU‐compliant evaluation (70 samples including 32 cases).
Data
Presented below are all the data for all of the tests entered into the review.
Tests. Data tables by test.
1. Test.
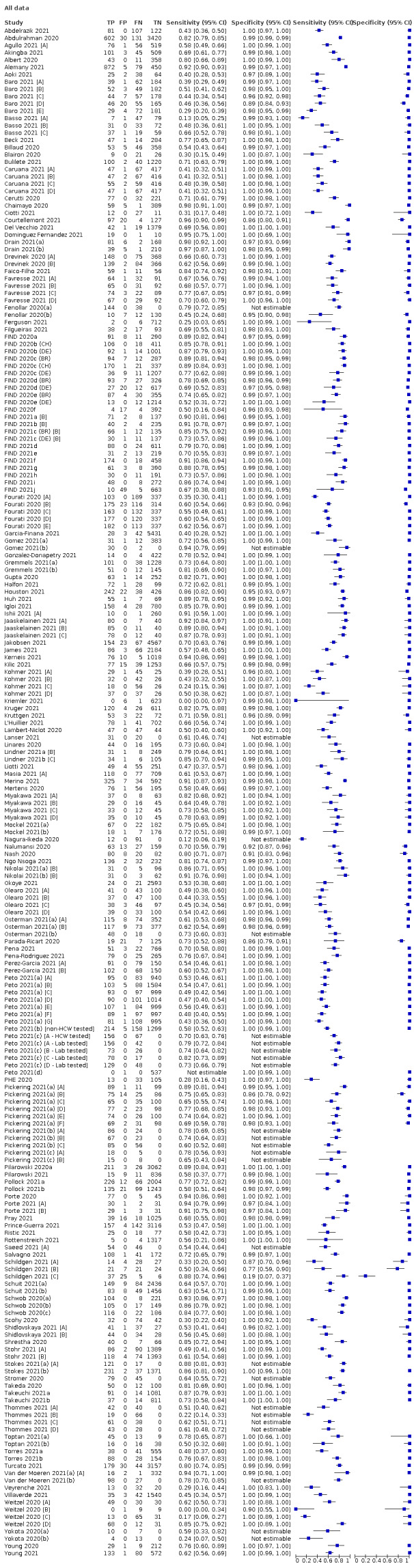
All data
2. Test.
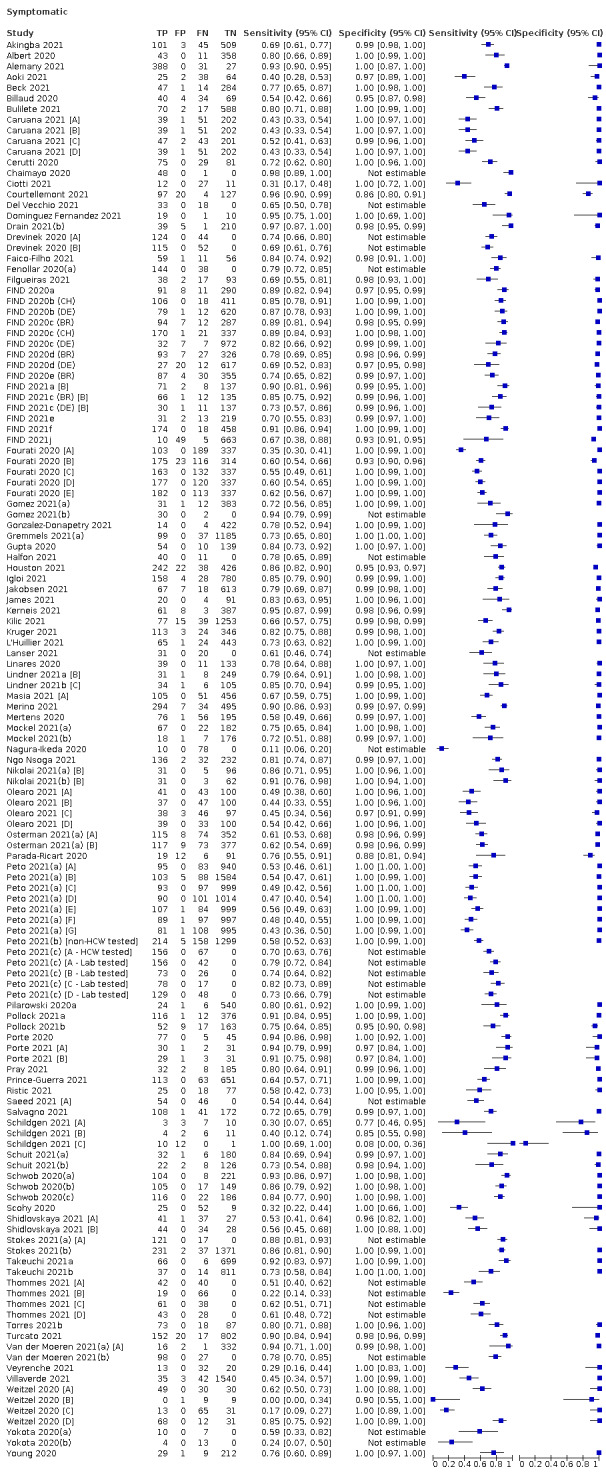
Symptomatic
3. Test.
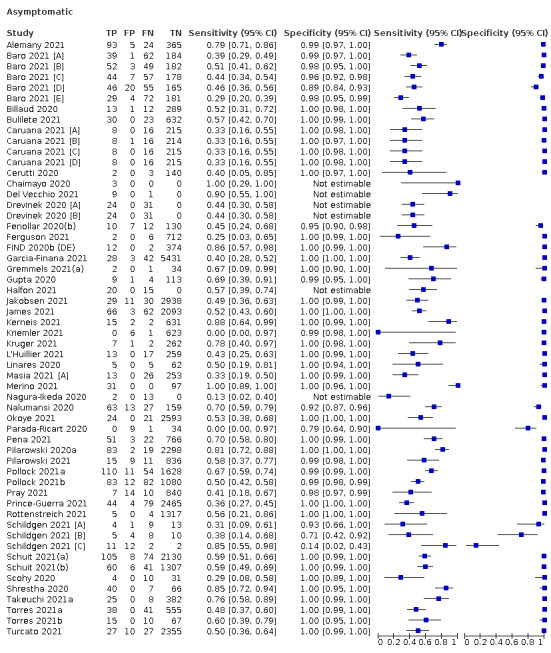
Asymptomatic
4. Test.
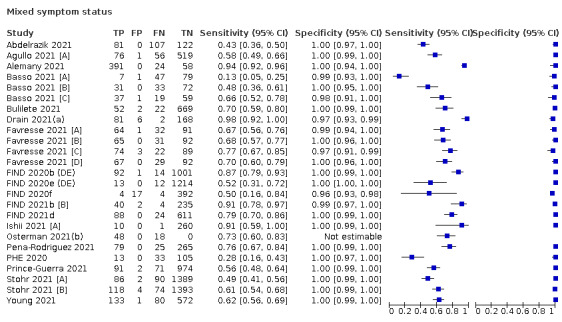
Mixed symptom status
5. Test.
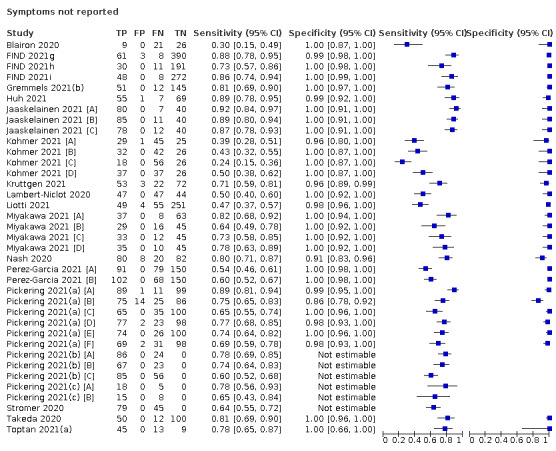
Symptoms not reported
6. Test.
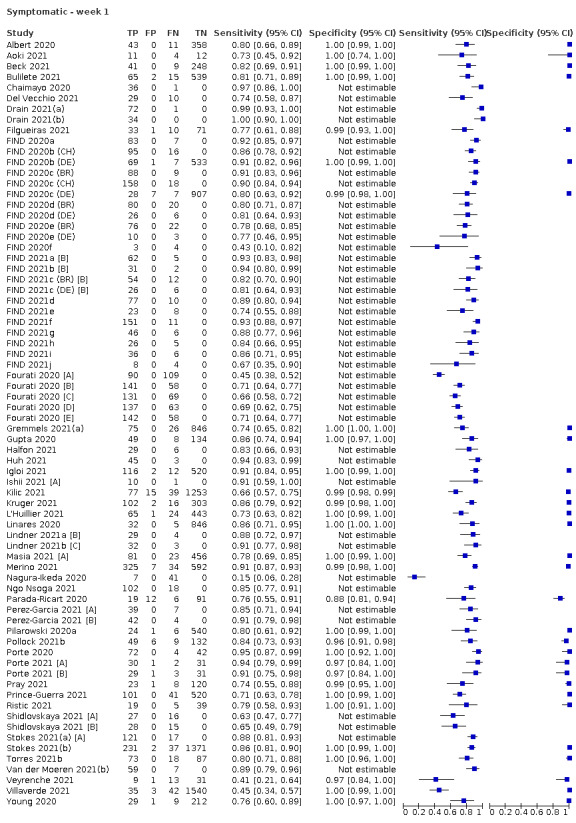
Symptomatic ‐ week 1
7. Test.
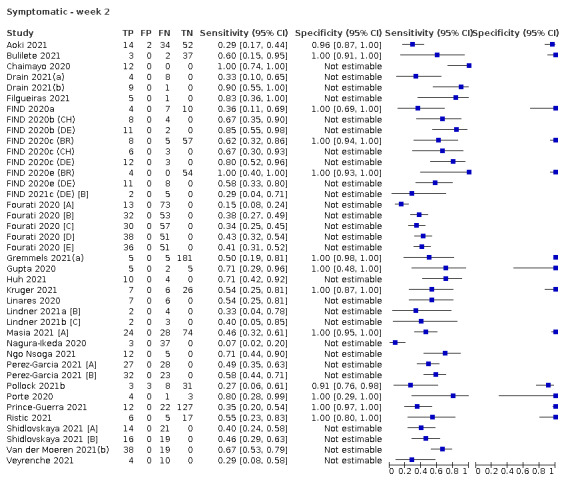
Symptomatic ‐ week 2
8. Test.
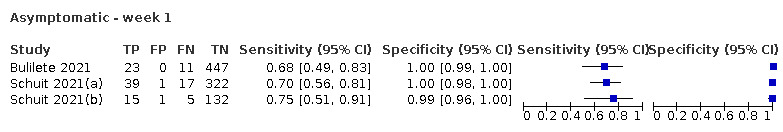
Asymptomatic ‐ week 1
9. Test.
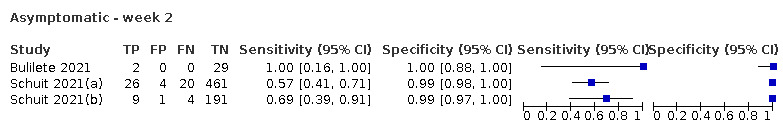
Asymptomatic ‐ week 2
10. Test.
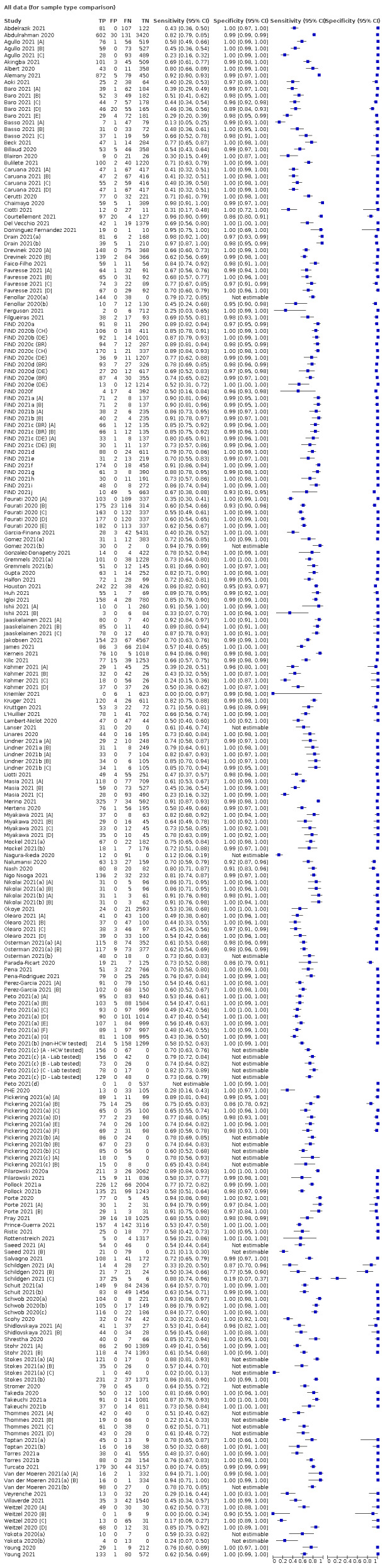
All data (for sample type comparison)
11. Test.
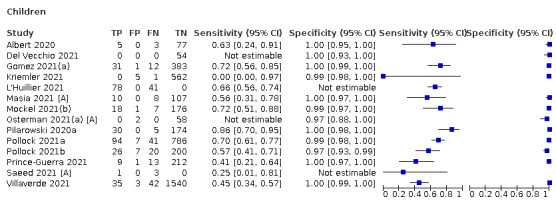
Children
12. Test.
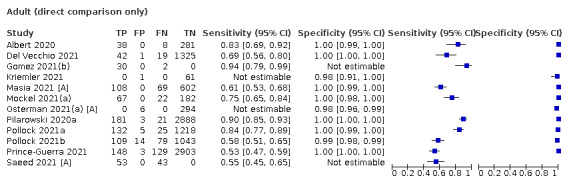
Adult (direct comparison only)
13. Test.
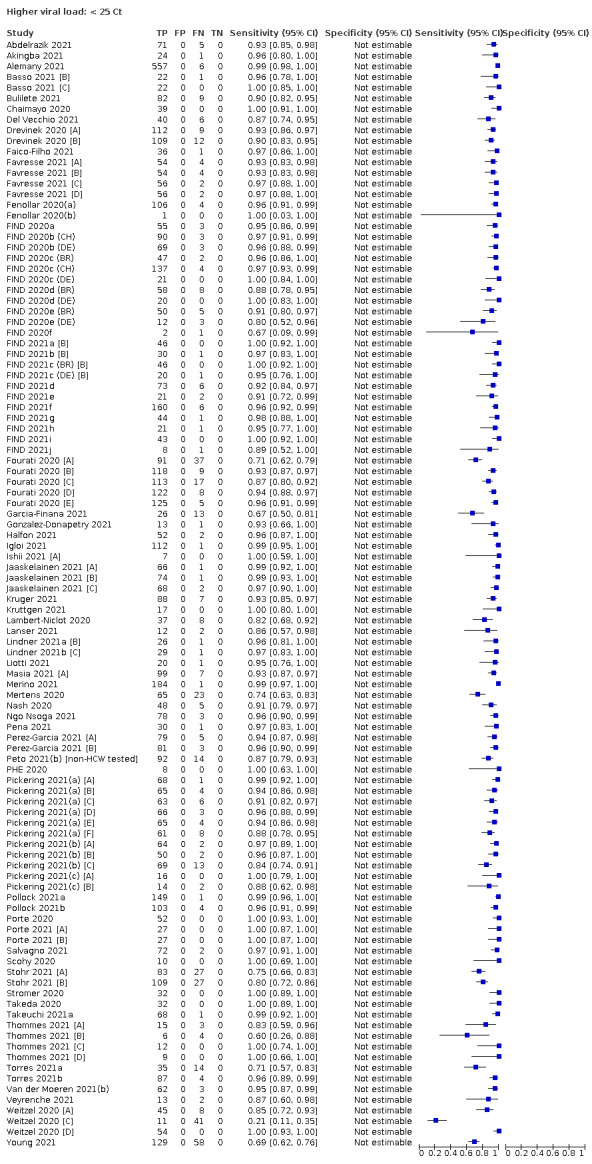
Higher viral load: < 25 Ct
14. Test.
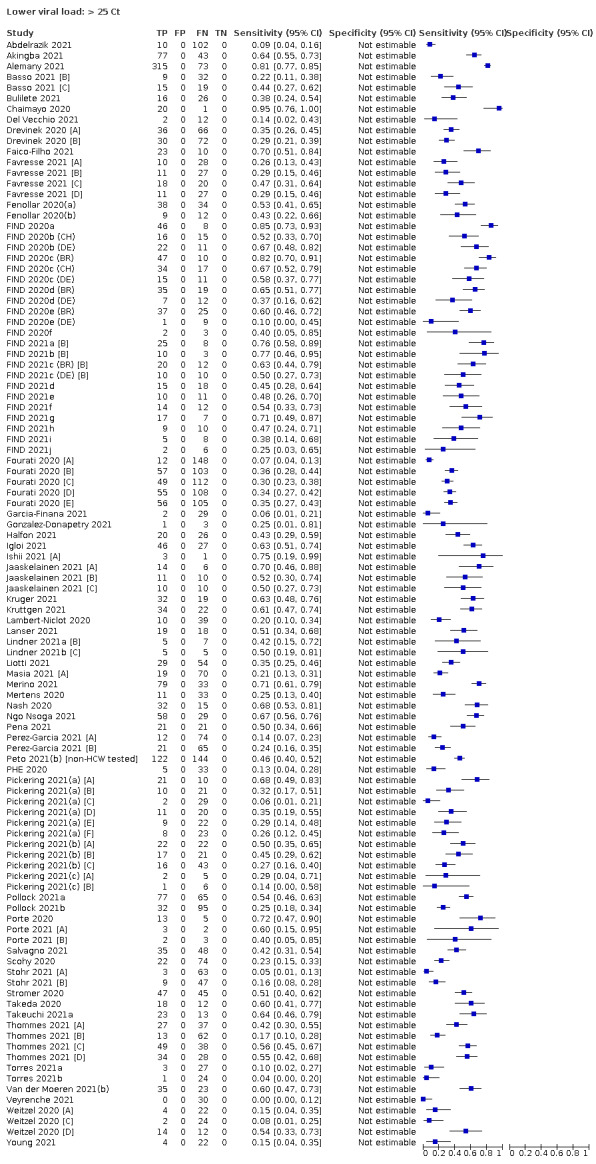
Lower viral load: > 25 Ct
15. Test.
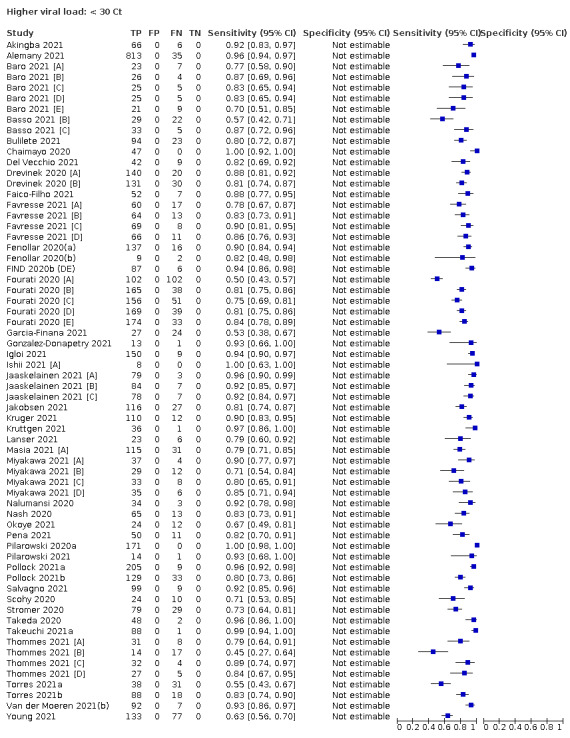
Higher viral load: < 30 Ct
16. Test.
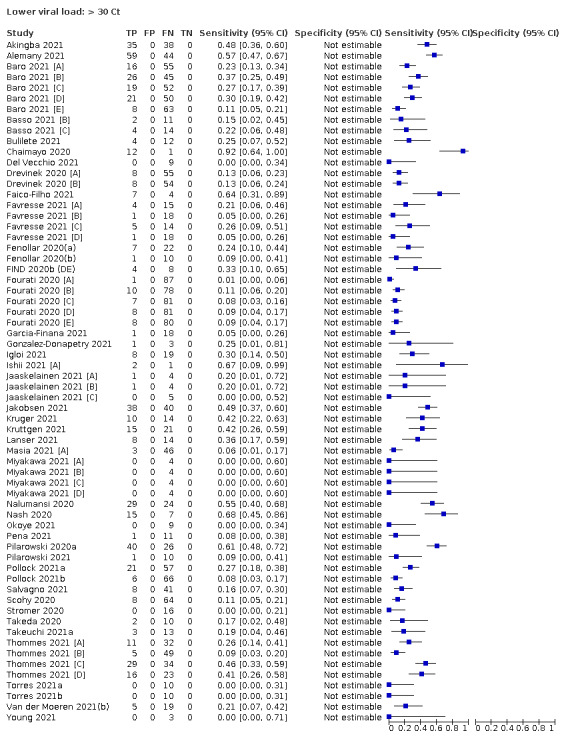
Lower viral load: > 30 Ct
17. Test.
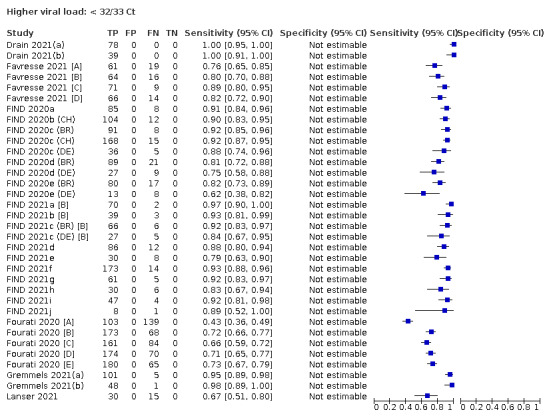
Higher viral load: < 32/33 Ct
18. Test.
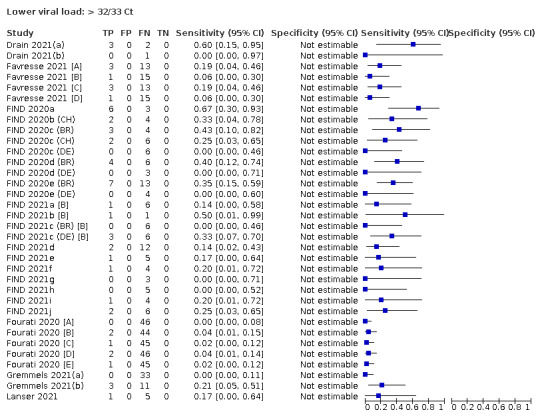
Lower viral load: > 32/33 Ct
19. Test.
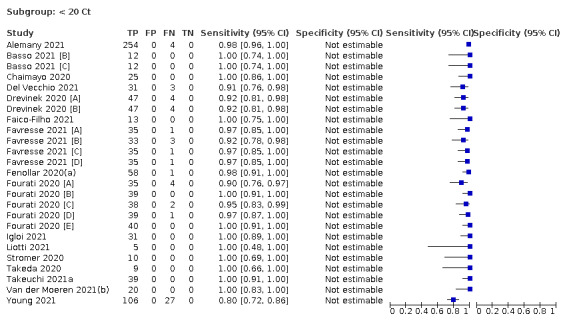
Subgroup: < 20 Ct
20. Test.
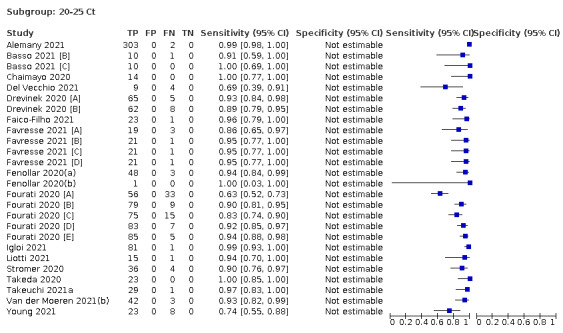
Subgroup: 20‐25 Ct
21. Test.
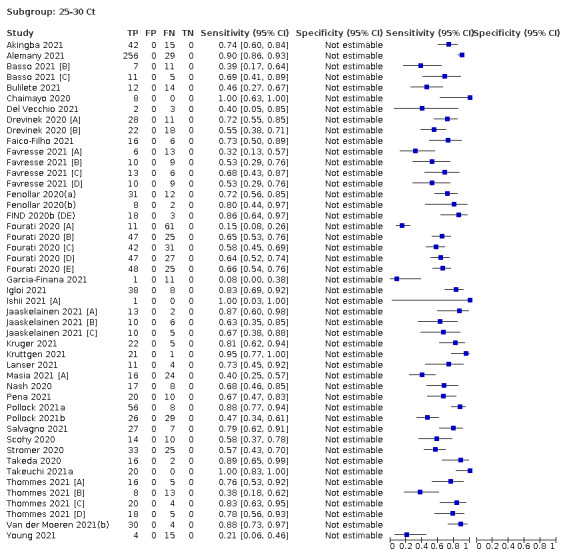
Subgroup: 25‐30 Ct
22. Test.
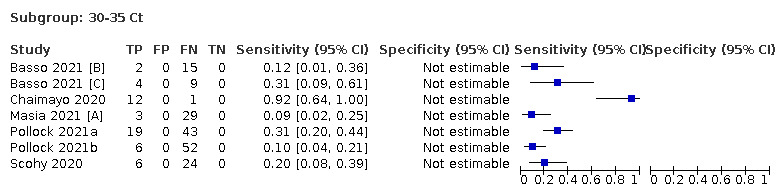
Subgroup: 30‐35 Ct
23. Test.
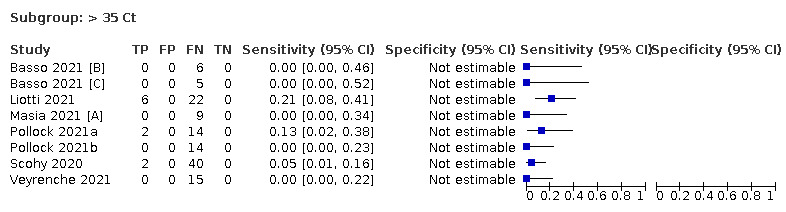
Subgroup: > 35 Ct
24. Test.
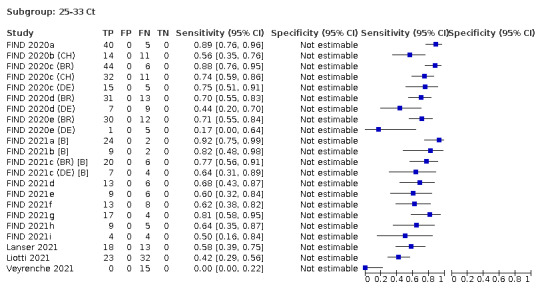
Subgroup: 25‐33 Ct
25. Test.
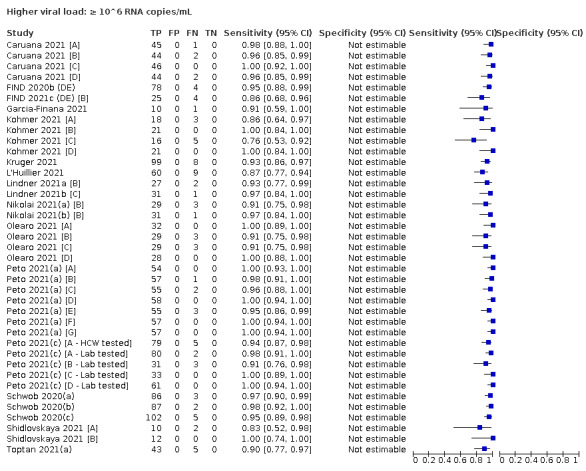
Higher viral load: ≥ 10^6 RNA copies/mL
26. Test.
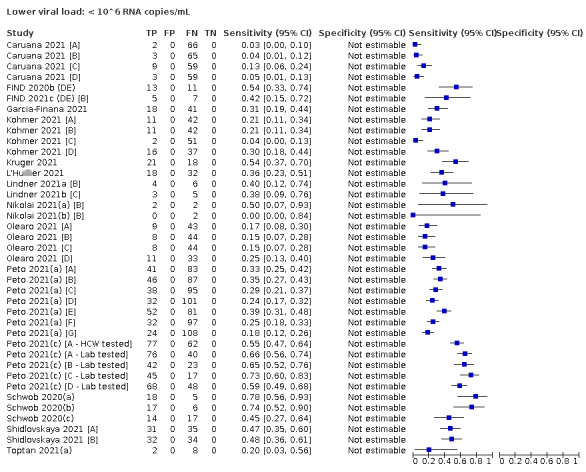
Lower viral load: < 10^6 RNA copies/mL
27. Test.
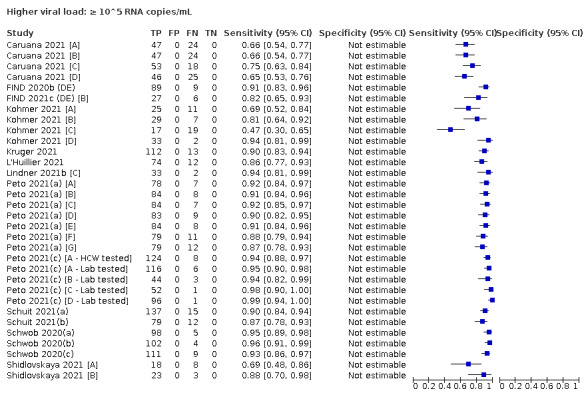
Higher viral load: ≥ 10^5 RNA copies/mL
28. Test.
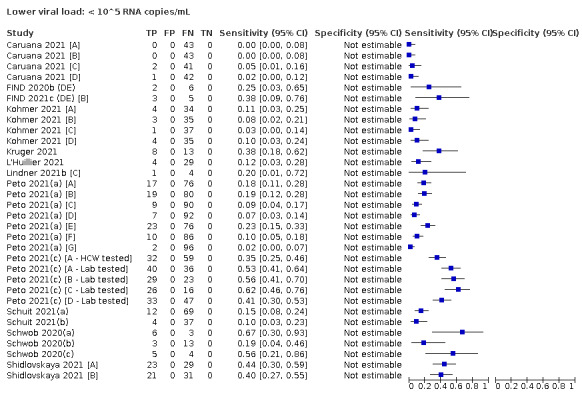
Lower viral load: < 10^5 RNA copies/mL
29. Test.

Subgroup: ≥ 10^8 RNA copies/mL
30. Test.

Subgroup: 10^7 RNA copies/mL
31. Test.
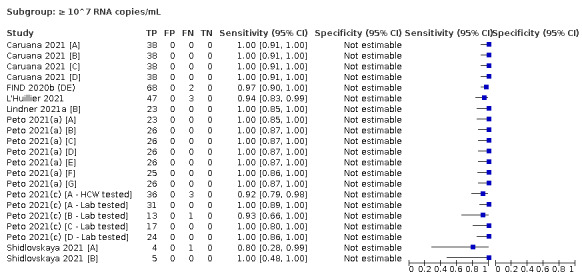
Subgroup: ≥ 10^7 RNA copies/mL
32. Test.
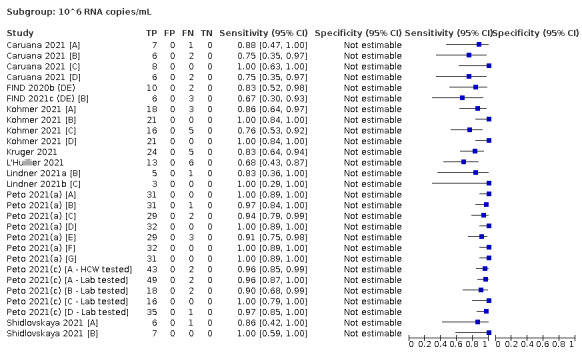
Subgroup: 10^6 RNA copies/mL
33. Test.
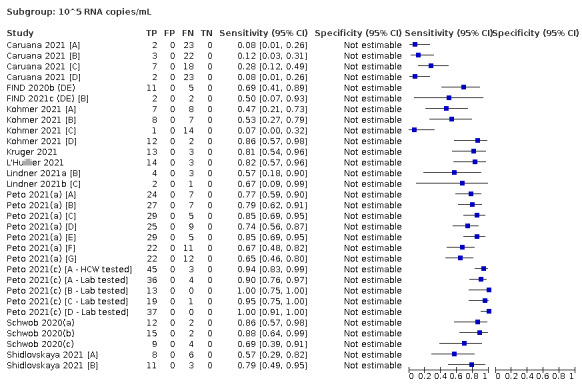
Subgroup: 10^5 RNA copies/mL
34. Test.
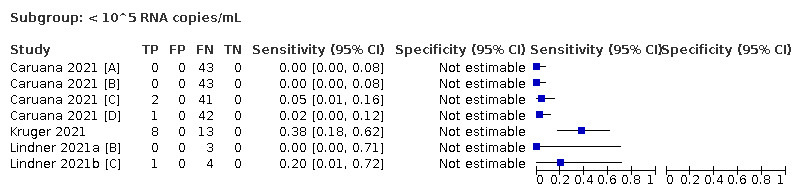
Subgroup: < 10^5 RNA copies/mL
35. Test.
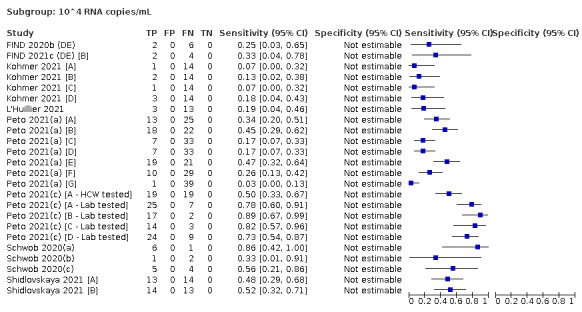
Subgroup: 10^4 RNA copies/mL
36. Test.
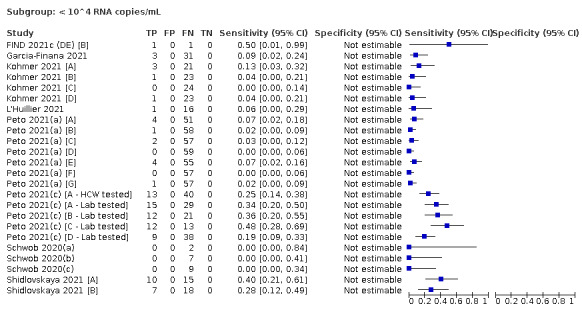
Subgroup: < 10^4 RNA copies/mL
37. Test.
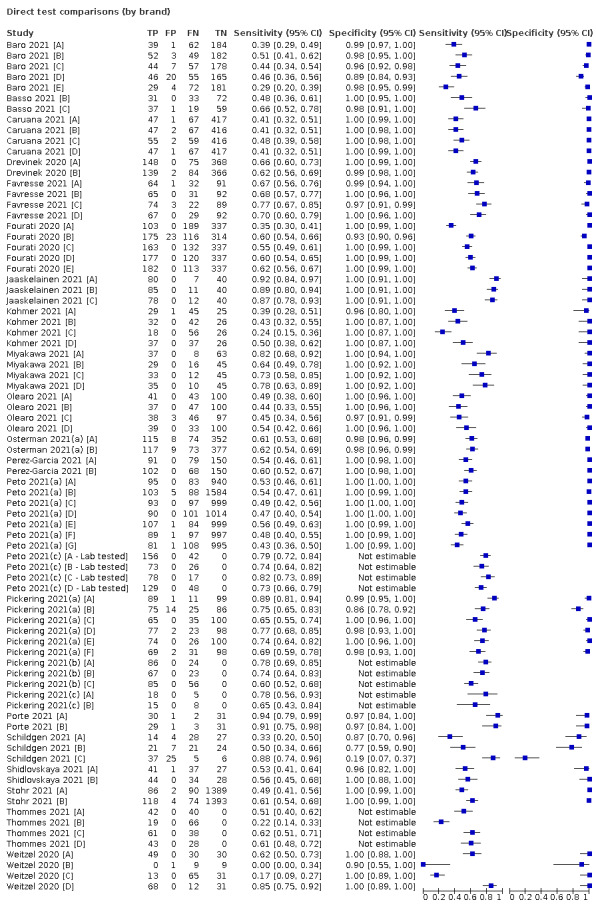
Direct test comparisons (by brand)
38. Test.
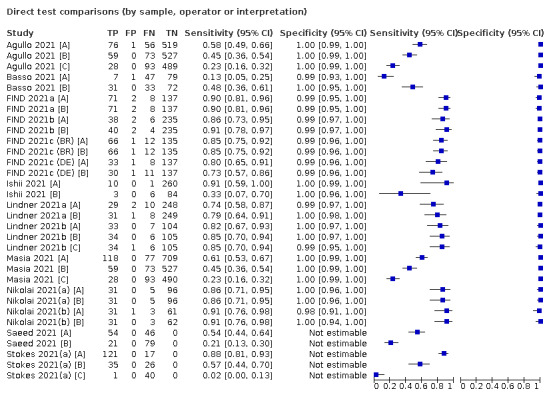
Direct test comparisons (by sample, operator or interpretation)
39. Test.

AAZ ‐ COVID‐VIRO (CGIA)
40. Test.
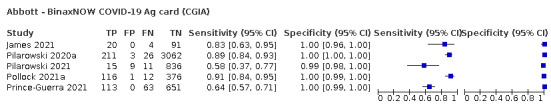
Abbott ‐ BinaxNOW COVID‐19 Ag card (CGIA)
41. Test.
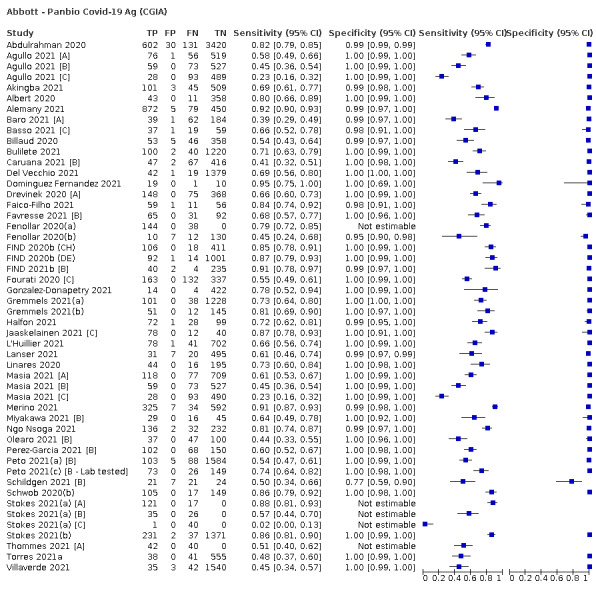
Abbott ‐ Panbio Covid‐19 Ag (CGIA)
42. Test.

Abbott ‐ Panbio Covid‐19 Ag ‐ Nasal (CGIA)
43. Test.

Access Bio ‐ CareStart Covid‐19 Ag (CGIA)
44. Test.

Anhui Deepblue ‐ COVID‐19 Ag (CGIA)
45. Test.
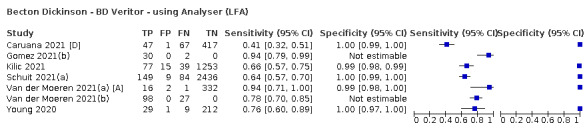
Becton Dickinson ‐ BD Veritor ‐ using Analyser (LFA)
46. Test.

Becton Dickinson ‐ BD Veritor ‐ no Analyser (LFA)
47. Test.
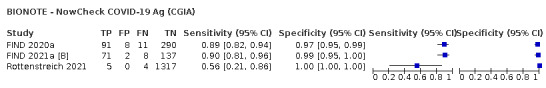
BIONOTE ‐ NowCheck COVID‐19 Ag (CGIA)
48. Test.

BIONOTE ‐ NowCheck COVID‐19 Ag ‐ Nasal (CGIA)
49. Test.

Biosynex ‐ Biosynex COVID‐19 Ag BSS (CGIA)
50. Test.

Biotical Health ‐ SARS‐CoV‐2 Ag card (CGIA)
51. Test.

Boditech Medical ‐ iChroma COVID‐19 Ag (FIA)
52. Test.

Certest Biotech ‐ CerTest
53. Test.
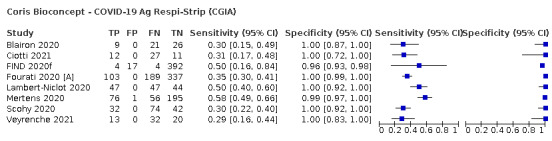
Coris Bioconcept ‐ COVID‐19 Ag Respi‐Strip (CGIA)
54. Test.
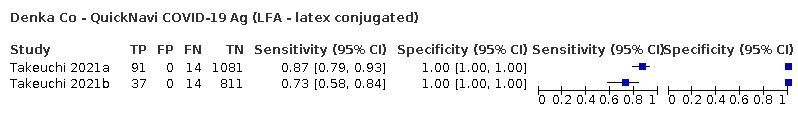
Denka Co ‐ QuickNavi COVID‐19 Ag (LFA ‐ latex conjugated)
55. Test.

DIALAB ‐ DIAQUICK COVID‐19 Ag (LFA)
56. Test.

E25Bio ‐ Covid‐19 DART (CGIA)
57. Test.

ECODiagnostica ‐ COVID‐19 Ag ECO (CGIA)
58. Test.

Encode/Emmo Pharma ‐ Encode (LFA)
59. Test.

Fortress Diagnostics ‐ Coronavirus Ag (CGIA)
60. Test.

FUJIFILM ‐ COVID‐19 Ag Test (CGIA)
61. Test.
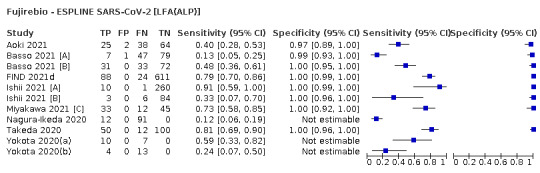
Fujirebio ‐ ESPLINE SARS‐CoV‐2 [LFA(ALP)]
62. Test.

Guangzhou Wondfo ‐ 2019‐nCoV Ag (CGIA)
63. Test.
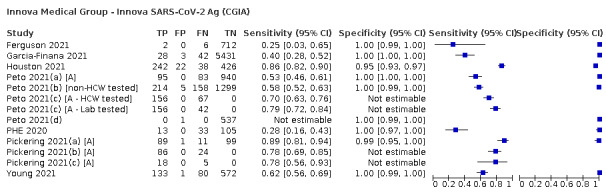
Innova Medical Group ‐ Innova SARS‐CoV‐2 Ag (CGIA)
64. Test.

Joysbio Biotech ‐ SARS‐CoV‐2 Ag (CGIA)
65. Test.

Lepu Medical ‐ SARS‐CoV‐2 Ag Rapid Test (CGIA)
66. Test.

Lepu Medical ‐ SARS‐CoV‐2 Ag Rapid Test ‐ Saliva (CGIA
67. Test.

Liming Bio‐Products ‐ StrongStep® COVID‐19 Ag (CGIA)
68. Test.
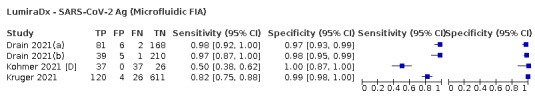
LumiraDx ‐ SARS‐CoV‐2 Ag (Microfluidic FIA)
69. Test.

MEDsan GmbH ‐ SARS‐CoV‐2 Ag
70. Test.

Mologic ‐ COVID 19 Rapid Ag (LFA)
71. Test.
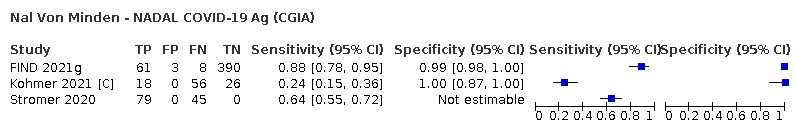
Nal Von Minden ‐ NADAL COVID‐19 Ag (CGIA)
72. Test.

Orient Gene/Healgen Scientific ‐ Coronavirus Ag (CGIA)
73. Test.

PCL ‐ COVID19 Ag Rapid FIA
74. Test.

Precision Biosensor ‐ Exdia COVID‐19 Ag (FIA)
75. Test.
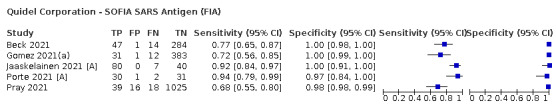
Quidel Corporation ‐ SOFIA SARS Antigen (FIA)
76. Test.
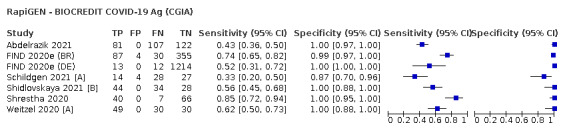
RapiGEN ‐ BIOCREDIT COVID‐19 Ag (CGIA)
77. Test.
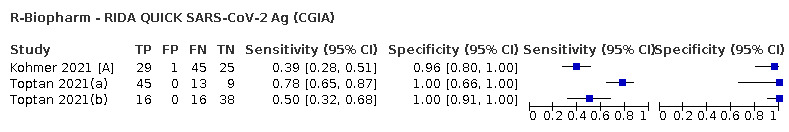
R‐Biopharm ‐ RIDA QUICK SARS‐CoV‐2 Ag (CGIA)
78. Test.

Savant Biotech ‐ Huaketai SARS‐CoV‐2 N Protein (LFA)
79. Test.
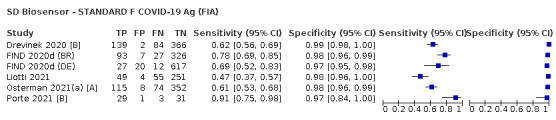
SD Biosensor ‐ STANDARD F COVID‐19 Ag (FIA)
80. Test.
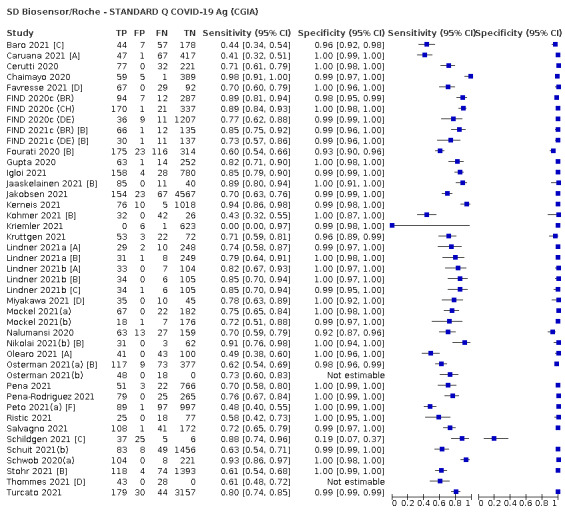
SD Biosensor/Roche ‐ STANDARD Q COVID‐19 Ag (CGIA)
81. Test.
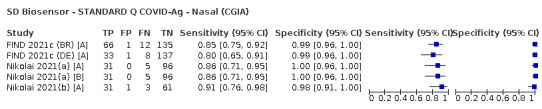
SD Biosensor ‐ STANDARD Q COVID‐Ag ‐ Nasal (CGIA)
82. Test.
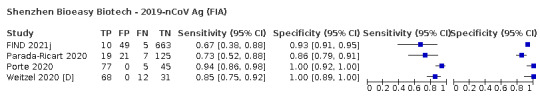
Shenzhen Bioeasy Biotech ‐ 2019‐nCoV Ag (FIA)
83. Test.

Sichuan Mass Spectrometry Biotech ‐ 2019‐nCov Ag (LFA)
84. Test.
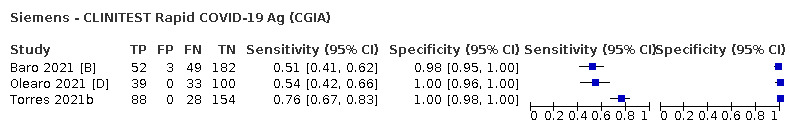
Siemens ‐ CLINITEST Rapid COVID‐19 Ag (CGIA)
85. Test.

Spring Healthcare ‐ SARS‐CoV‐2 Ag (CGIA)
86. Test.

Sugentech Inc ‐ SGTI‐flex COVID‐19 Ag (CGIA)
87. Test.
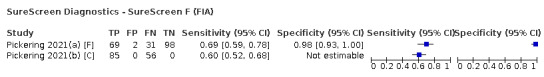
SureScreen Diagnostics ‐ SureScreen F (FIA)
88. Test.
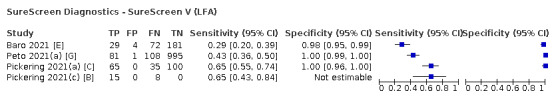
SureScreen Diagnostics ‐ SureScreen V (LFA)
89. Test.
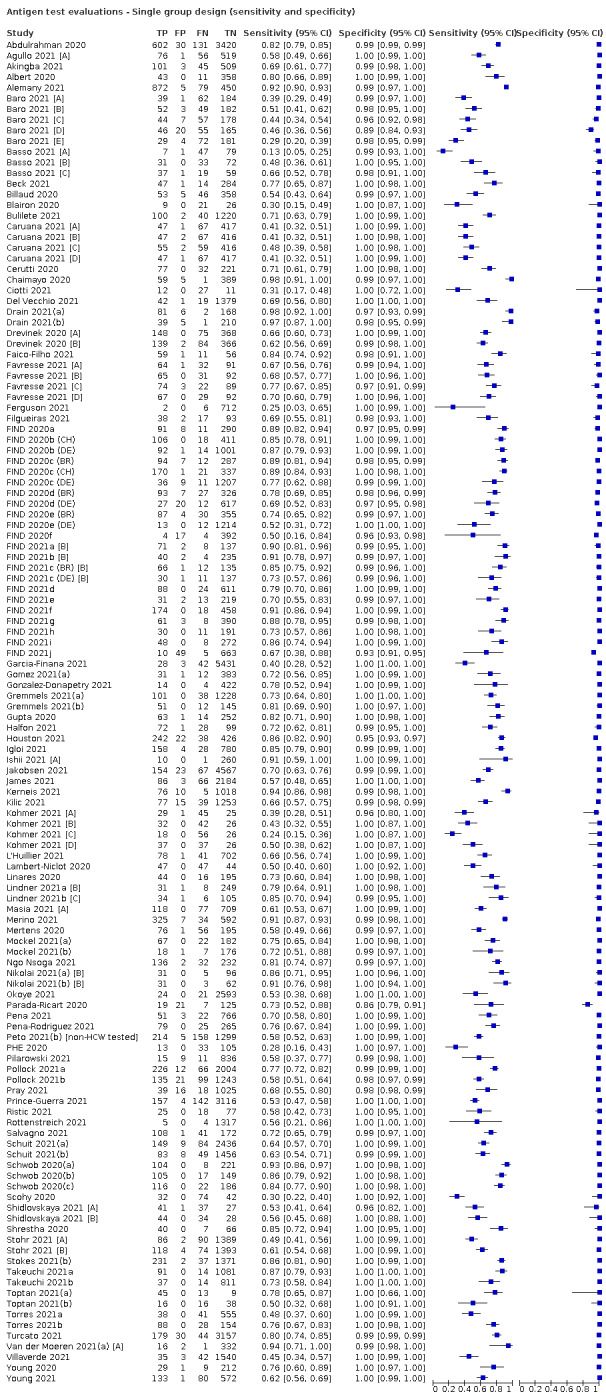
Antigen test evaluations ‐ Single group design (sensitivity and specificity)
90. Test.
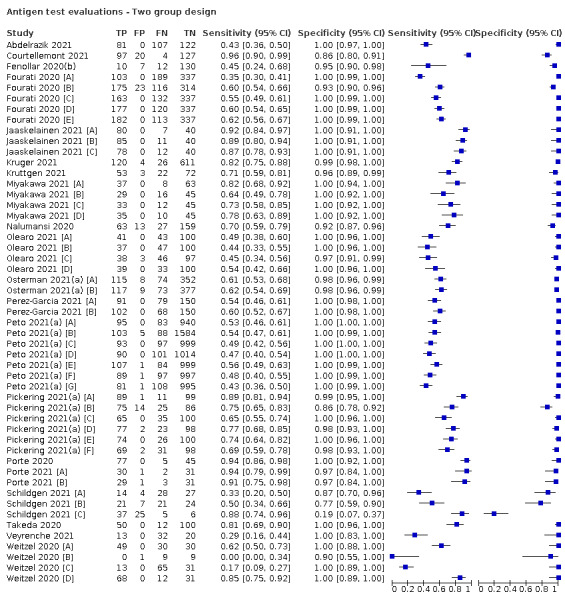
Antigen test evaluations ‐ Two group design
91. Test.
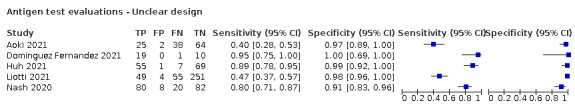
Antigen test evaluations ‐ Unclear design
92. Test.

Repeated testing (see Table 5 for 2x2 by study)
Characteristics of studies
Characteristics of included studies [ordered by study ID]
Abdelrazik 2021.
| Study characteristics | |||
| Patient Sampling | Multi‐group study to estimate sensitivity and specificity including:
[1] symptomatic PCR+ve patients (n = 160)
[2] exposed HCWs and patient contacts (n = 150)
Data are presented only for groups [1] and [2] combined; author contacted 15 March 21 Recruitment: unclear (do not state all patients) Prospective or retrospective: not stated; appears prospective Sample size (cases): 310 (188) |
||
| Patient characteristics and setting | Setting: mixed; described as patients, their contacts and exposed HCWs Location: Fayoum University Hospital, Fayoum Country: Egypt Dates: May 2020 Symptoms and severity: unclear; 160 PCR+ve "patients" presumably symptomatic, plus 150 presumably asymptomatic contacts and exposed HCWs Demographics: median age 42 years; 184/310 (59%) male Exposure history: states "exposed healthcare workers and patient contacts."; no further details |
||
| Index tests | Test name: BIOCREDIT COVID‐19 Ag kit Manufacturer: not stated; manufacturer is RapiGEN Antibody: not stated Ag target: not stated Test method: CGIA; described as lateral flow immunochromatographic assay (uses a dual‐colour system for the qualitative detection of the SARS‐CoV‐2 antigen) Samples used: NP using flocked swabs; collection not specified Transport media: UTM‐RT System, Copan Diagnostics, Murrieta, CA Sample storage: transported to lab within 1‐2 h of collection; stored at 4 °C and tested within 24 h Test operator: laboratory staff Definition of test positivity: no details Blinding reported: unclear; index test performed after PCR test Timing of samples: group [1] (n = 160) median 3 days pso |
||
| Target condition and reference standard(s) | Reference standard: PCR (multiplex real‐time PCR detection kit; DTlite 4, Russia) Definition of non‐COVID cases: as for cases; single negative Genetic target(s): not stated Samples used: NP in VTM; same as for index test Timing of reference standard: as for index test Blinded to index test: yes (PCR performed before index test) Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; same swab All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Abdulrahman 2020.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: mildly symptomatic individuals with suspected COVID‐19 cases (defined by Bahrain protocol), referred to the national testing centre Recruitment: not stated; appears consecutive Prospective or retrospective: prospective Sample size (cases): 4183 (733) |
||
| Patient characteristics and setting | Setting: national COVID‐19 test centre Location: symptomatic hall in the National Testing Centre at the exhibition centre Country: Bahrain Dates: not stated Symptoms and severity: all mild symptomatic individuals Median symptom duration: 2 (range 0‐14) days; only collected for 1301 (31%) Demographics: mean age 30.9 ± 14.5 years; 2365 (56.5%) male Exposure history: not stated |
||
| Index tests | Test name: Panbio COVID 19 antigen rapid test Manufacturer: Abbott Rapid Diagnostic Jena GmbH, Jena, Germany Antibody: SARS‐CoV‐2 nucleocapsid protein Ag target: not stated Test method: CGIA Samples used: nasal (NMT); trained HCW collected the samples using NP swab based on CDC guidelines (patient's head tilted back by 70°, swab inserted c2 cm into the nostril, gently rotated, rolled several times and removed) Transport media: none; immediate on‐site testing Sample storage: none Test operator: not stated; most likely trained HCW as the test was conducted on site Definition of test positivity: not stated Blinding reported: yes; test was conducted on site before PCR test Timing of samples: 1301 (31%) = median of 2 (range 0‐14) days pso |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR, in‐house assay following WHO protocol; Ct values > 40 on E gene were considered negative (used Thermo Fisher Scientific TaqPath 1‐Step RT‐qPCR Master Mix) Definition of non‐COVID cases: as for cases; single negative Genetic target(s): E gene confirmed by RdRP and N genes Samples used: NP swab in VTM Timing of reference standard: 1301 (31%) = median of 2 (range 0‐14) days pso Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired swab All participants received same reference standard: yes Missing data: none; states "no equivocal results were reported for index or reference" Uninterpretable results: none Indeterminate results (index test): none Indeterminate results (reference standard): none Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: none received Publication status: preprint Source: medRxiv Author COI: authors report no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Agullo 2021 [A].
| Study characteristics | |||
| Patient Sampling | Single‐group study (from 3 centres) to estimate sensitivity and specificity: consecutive adults and children, either with COVID‐19 signs/symptoms or asymptomatic contacts Recruitment: consecutive Prospective or retrospective: prospective Sample size (cases): 659 (265); 610/659 also provided saliva samples |
||
| Patient characteristics and setting | Setting: primary care Location: 3 primary care centres in Spain (organized by Hospital General Universitario de Elche) Country: Spain Dates: 15 September‐29 October 2020 Symptoms and severity: 265 (40.2%) patients were asymptomatic and 394 (59.8%) had symptoms, with median (IQR) duration of 3 (2‐5) days Demographics: median (IQR) age: 38 (21‐49.8) years; 76 (11.5%) ≤ 14 years, 45 (7.6%) > 65 years, 372 (56.4%) women Exposure history: not stated |
||
| Index tests | Test name: Panbio COVID 19 antigen rapid test Manufacturer: Abbott Rapid Diagnostic Jena GmbH, Jena, Germany Antibody: SARS‐CoV‐2 nucleocapsid protein Ag target: not stated Test method: CGIA Samples used: [A] NP; [B] nasal (both collected by qualified nurse); [C] saliva (self‐collected) Transport media: none; immediate on‐site testing Sample storage: none Test operator: not stated; may have been by same qualified nurse Definition of test positivity: not stated Blinding reported: yes; test was conducted on site before PCR test Timing of samples: median (Q1‐Q3) duration of 3 (2–5) days of symptoms |
||
| Target condition and reference standard(s) | Reference standard: rRT‐PCR testing was performed according to the manufacturer’s guidelines on Cobas z 480 Analyser (Roche, Basilea, Suiza) Definition of non‐COVID cases: as for cases; single negative Genetic target(s): not specified Samples used: NP swab in VTM Timing of reference standard: as for index test; median (Q1‐Q3) duration of 3 (2–5) days of symptoms Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired swab All participants received same reference standard: yes Missing data: not stated Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "this work was supported by the RD16/0025/0038 project as a part of the Plan Nacional Research+Development+Innovation (R+D+I) and co‐financed by Instituto de Salud Carlos III ‐ Sub‐ dirección General de Evaluación y Fondo Europeo de Desarrollo Regional; Instituto de Salud Carlos III (Fondo de Investigaciones Sanitarias [grant number PI16/01740; PI18/01861; CM 19/00160, COV20–00005])." Publication status: published Source: Journal of Infection Author COI: no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Agullo 2021 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 3 sample types; Agullo 2021 [A] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | Comparative study of 3 sample types; Agullo 2021 [A] reports full study characteristics and QUADAS | ||
| Index tests | Test name: Panbio COVID 19 antigen rapid test Manufacturer: Abbott Rapid Diagnostic Jena GmbH, Jena, Germany Antibody: SARS‐CoV‐2 nucleocapsid protein Ag target: not stated Test method: CGIA Samples used: [A] NP; [B] nasal (both collected by qualified nurse); [C] saliva (self‐collected) Transport media: none; immediate on‐site testing Sample storage: none Test operator: not stated; may have been by same qualified nurse Definition of test positivity: not stated Blinding reported: yes; test was conducted on site before PCR test Timing of samples: median (Q1‐Q3) duration of 3 (2–5) days of symptoms |
||
| Target condition and reference standard(s) | Comparative study of 3 sample types; Agullo 2021 [A] reports full study characteristics and QUADAS | ||
| Flow and timing | Comparative study of 3 sample types; Agullo 2021 [A] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | Comparative study of 3 sample types; Agullo 2021 [A] reports full study characteristics and QUADAS | ||
Agullo 2021 [C].
| Study characteristics | |||
| Patient Sampling | Comparative study of 3 sample types; Agullo 2021 [A] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | Comparative study of 3 sample types; Agullo 2021 [A] reports full study characteristics and QUADAS | ||
| Index tests | Test name: Panbio COVID 19 antigen rapid test Manufacturer: Abbott Rapid Diagnostic Jena GmbH, Jena, Germany Antibody: SARS‐CoV‐2 nucleocapsid protein Ag target: not stated Test method: CGIA Samples used: [A] NP; [B] nasal; (both collected by qualified nurse); [C] saliva (self‐collected) Transport media: none; immediate on‐site testing Sample storage: none Test operator: not stated; may have been by same qualified nurse Definition of test positivity: not stated Blinding reported: yes; test was conducted on‐site before PCR test Timing of samples: median (Q1‐Q3) duration of 3 (2–5) days of symptoms |
||
| Target condition and reference standard(s) | Comparative study of 3 sample types; Agullo 2021 [A] reports full study characteristics and QUADAS | ||
| Flow and timing | Comparative study of 3 sample types; Agullo 2021 [A] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | Comparative study of 3 sample types; Agullo 2021 [A] reports full study characteristics and QUADAS | ||
Akingba 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: symptomatic individuals seeking COVID‐19 testing at mobile testing units during community testing campaigns Recruitment: not specifically stated but appears to be consecutive; discussion states "unselected symptomatic individuals requesting testing" Prospective or retrospective: prospective Sample size (cases): 677 (146) |
||
| Patient characteristics and setting | Setting: COVID‐19 test centres; mobile testing sites (n = 6) Location: 6 community testing sites in Nelson Mandela Bay municipality, Eastern Cape Country: South Africa Dates: 17‐20 November 2020 Symptoms and severity: all symptomatic seeking COVID‐19 testing (ambulatory; specific symptoms not reported) Demographics: age range: 3‐85 years Sex: 59% female Exposure history: not reported |
||
| Index tests | Test name: PanBio SARS‐CoV‐2 RTD Manufacturer: Abbott Rapid Diagnostics, USA Antibody: not stated Ag target: not stated Test method: not stated Samples used: NP (collection not described) Transport media: not stated; test buffer appears to be used Sample storage: immediate testing Test operator: not stated Definition of test positivity: not stated; set by manufacturer Blinding reported: yes (on site prior to PCR) Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; Seegene nCoV assay
(single target positive, Ct > 38) Definition of non‐COVID cases: as for cases; single negative Genetic target(s): not stated; states "3 targets", mean Ct values used Samples used: same as for index test (same swab after Ag testing) Timing of reference standard: not stated; as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous (same swab) All participants received same reference standard: yes Missing data: yes; 19 excluded (see below) Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): 19/677 (2.8%) had inconclusive results (single target positive, Ct > 38) Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: no funding was given for this study Publication status: preprint (not peer reviewed) Source: medRxiv Author COI: the authors report no competing interests |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Albert 2020.
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity and specificity: patients with clinical suspicion of COVID‐19 (compatible signs or symptoms appearing within the prior week) attending 1 of 8 primary care centres (n = 412) Recruitment: not stated; likely consecutive Prospective or retrospective: prospective |
||
| Patient characteristics and setting | Setting: primary care Location: 8 primary care centres of the Health Department Clínico‐Malvarrosa in Valencia Country: Spain Dates: 2 September‐7 October 2020 Symptoms and severity: all symptomatic (< 7 days pso) Demographics: median age, 31 years (range, 1‐91); 42% male 327 adults; median, 36 years (17‐91 years) 85 children; median, 11 years (1‐16 years) Exposure history: not stated |
||
| Index tests | Test name: Panbio COVID‐19 AG Rapid Test Device (no product code reported) Manufacturer: Abbott Diagnostic GmbH, Jena, Germany Antibody: nucleoprotein Ag target: not stated Test method: not stated Samples used: NP; collected by trained nurses using flocked swabs Transport media: none for Ag testing Sample storage: none Test operator: not stated; immediate testing Definition of test positivity: visible line within 15 min; as per manufacturer Blinding reported: yes Timing of samples: day < 7 pso |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; TaqPath COVID‐19 Combo Kit (Thermo Fisher Scientific, Massachusetts, USA) Definition of non‐COVID cases: as for cases; single negative Genetic target(s): ORF1ab, N and S genes Samples used: NP in UTM Timing of reference standard: as for index; tested within 24 h Blinded to index test: not stated; presume yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired All participants received same reference standard: yes Missing data: none reported; no participant flow diagram reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: this work received no public or private funds. Abbott Diagnostics provided Panbio COVID‐19 AG Rapid Test Device kits. Publication status: preprint Source: medRxiv Author COI: the authors declare no conflicts of interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Alemany 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study including participants from 3 settings:
[1] symptomatic individuals with suspected COVID‐19 seen in routine practice (n = 446)
[2] contacts exposed to positive PCR confirmed COVID‐19 cases (n = 473)
[3] preventive screening of unexposed asymptomatic individuals in the general population (n = 487) Recruitment: retrospective (frozen swabs) Prospective or retrospective: not stated |
||
| Patient characteristics and setting | Setting: mixed/unclear (laboratory‐based) Location: not reported; multiple author institutions reported Country: Spain Dates: not stated Symptoms and severity: not stated; 15/1406 (1.1%) reportedly hospitalized (all PCR+) Viral load of cases: Ct < 20, 258 (18.3%); Ct 20‐24, 305 (21.7%); Ct 25‐29, 285 (30.3%); Ct > 30, 103 (7.3%) Demographics: all samples: mean age 40.4 years (SD 24.5), 453 (32.2% male) Exposure history: 473/1406 (33.6%) identified through contact tracing |
||
| Index tests | Test name: PanbioTM COVID‐19 Ag Test (no product codes). Selected following validation exercise using 40 NP samples to compare PanBio with Coris Bioconcept COVID‐19 Ag RespiStrip, SD Biosensor STANDARD F COVID‐19 Ag FIA and STANDARD Q COVID‐19 Ag Test Manufacturer: Abbott Laboratories, Illinois, USA Antibody: not stated Ag target: SARS‐CoV‐2 Test method: CGIA Samples used: [1] and [2] NP, [3] NMT; collection not reported Transport media: VTM (DeltaSwab Virus) Sample storage: stored at 2‐8 °C prior to PCR then frozen (− 80 °C) prior to Ag testing; "Internal validation showed no significant change in the test performance using Abbot test Kit buffer or a mix of the Kit buffer and transport media at 1:3 dilution; likewise, the use of frozen specimens showed no significant differences compared with fresh ones" Test operator: 2 laboratory technicians Definition of test positivity: visible line; as per manufacturer Blinding reported: yes Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; in‐house following CDC protocol Definition of non‐COVID cases: as per cases; single negative PCR for absence of infection Genetic target(s): not stated; as per CDC protocol Samples used: NP or NMT; as per index test Timing of reference standard: fresh samples stored at 2–8 ºC for up to 72 h prior to RT‐PCR Blinded to index test: yes; conducted first Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous (same swab) All participants received same reference standard: yes Missing data: none reported; no participant flow diagram reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "the test kits were purchased from Abbott Rapid Diagnostics Healthcare SL (Spain). The funders of the study had no role in the study conception, design, conduct, data analysis, or writing of the report." Publication status: preprint Source: medRxiv Author COI: authors declare no conflicts of interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Aoki 2021.
| Study characteristics | |||
| Patient Sampling | Unclear design: samples from COVID‐19 hospitalized patients or from patients with COVID‐19‐like symptoms Recruitment: unclear Prospective or retrospective: unclear Sample size (cases): 129 (63) |
||
| Patient characteristics and setting | Setting: appears to be laboratory‐based; includes use of remnant samples Location: Toho University School of Medicine, Tokyo Country: Japan Dates: not stated Symptoms and severity: not stated Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: Espline SARS‐CoV‐2 Manufacturer: Fujirebio Inc., Japan Antibody: SARS‐CoV‐2 N antigen (NeAg) Ag target: monoclonal antibodies Test method: immunochromatography assay based on sandwich enzyme immunoassay (ALP‐labelled) Samples used: NP swab; collection not described Transport media: samples were collected into Espline swab buffer (ETS ‐ Espline treatment solution) (n = 96); remnant samples were stored in UVT (n = 33) Sample storage: not described for samples in ETS; remnant samples in UVT been stored at −80 °C after PCR testing Test operator: not stated Definition of test positivity: "positive when both the reference line and the judgment line can be visually confirmed" Blinding reported: not stated Timing of samples: median 9.5 days pso for samples Ag+/PCR+, 16 days for Ag‐/PCR+, 19 days for Ag‐/PCR− |
||
| Target condition and reference standard(s) | Reference standard: PCR performed according to the “Pathogen Detection Manual 2019‐nCoV Ver.2.6.1” from the National Institute of Infectious Diseases; assays included QuantStudio® 5 (Applied Biosystems, USA) or BD MAX open system Definition of non‐COVID cases: single negative PCR for absence of infection Genetic target(s): gene N Samples used: paired NP swabs (one for Ag and one for PCR) Timing of reference standard: same as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: not clearly stated but appears to be 'simultaneous' All participants received same reference standard: yes Missing data: none mentioned Uninterpretable results: none mentioned Indeterminate results (index test): none mentioned Indeterminate results (reference standard): none mentioned Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: Japan Agency for MedicalResearch and Development Publication status: published paper Source: Journal of Infection and Chemotherapy Author COI: SY is an employee of Fujirebio, Inc. The other study authors have no conflict of interest to declare. |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Baro 2021 [A].
| Study characteristics | |||
| Patient Sampling | Unclear design estimating sensitivity and specificity: unexposed asymptomatic individuals living in areas at high risk of an outbreak who participated in routine mass testing as part of a regional surveillance program (n = 316) Recruitment: not stated; recruitment continued until at least 73 PCR+ and 165 PCR− samples were obtained Prospective or retrospective: unclear Sample size (cases): 286 (101) |
||
| Patient characteristics and setting | Setting: community screening; public health surveillance Location: Catalonia (North‐East Spain; i.e. Metropolità Nord) with a catchment population of ~1,400,000 people Country: Spain Dates: December 2020‐January 2021 Symptoms and severity: states "all unexposed asymptomatic" Demographics: not stated Exposure history: states "all unexposed" |
||
| Index tests | Test name: [A] PanBioTM COVID‐19 Ag Rapid test
[B] CLINITEST Rapid COVID‐19 Antigen Test
[C] SD Biosensor SARS‐CoV‐2 Rapid Antigen Test
[D] Lepu SARS‐CoV‐2 Antigen Rapid Test
[E] Surescreen COVID‐19 Coronavirus Rapid Antigen Test Cassette Manufacturer: [A] Abbott Rapid Diagnostics, Panbio Ltd, USA [B] Siemens Healthineers (Shangai International Holding Corp), USA [C] Roche Diagnostics, SD Biosensor, Republic of Korea [D] Beijing Lepu Medical Technology Co., Ltd., China [E] SureScreen Diagnostics Ltd, UK Antibody: nucleocapsid protein Ag target: not reported Test method: [A] CGIA [B] Immunochromatographic [C] LFA (unclear) [D] CGIA [E] LFA (unclear) Samples used: NP (collected by HCW) Transport media: VTM (DeltaSwab Virus, Deltalab; or UTM Universal Transport Medium, Copan) Sample storage: samples stored for up to 24 h (2‐8 ºC) prior to RT‐PCR then stored up to 12 h more at 2‐8 ºC until Ag testing Test operator: lab technician at The University Hospital Germans Trias i Pujol Definition of test positivity: visual coloured band; the presence of any test line (T) indicates a positive result. Samples were applied directly to the test cassette and incubated for 15 min at room temperature before reading results with the naked eye, according to the manufacturer IFU Blinding reported: unclear; presume only blinded to different Ags tests "All Ag‐RDT determinations were performed in parallel by two blinded technicians" Timing of samples: N/A; all asymptomatic |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; Allplex 2019‐nCoV assay (Seegene, South Korea) on the CFX96 (Bio‐Rad, USA)
Threshold according to the manufacturer’s IFU Definition of non‐COVID cases: single negative PCR Genetic target(s): not stated Samples used: NP in VTM; performed on fresh samples stored at 2‐8 °C for up to 24 h Timing of reference standard: same as for index test Blinded to index test: yes (performed before Ag tests) Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same swab used All participants received same reference standard: yes Missing data: 30/316 excluded; reasons for exclusion documented 25 with no documented Ct value excluded a priori Uninterpretable results: 1/316 incomplete result Indeterminate results (index test): 4/316‐ all of them in the Lepu assay Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "Blueberry diagnostics, Fundació Institut d'Investigació en Ciències de la Salut Germans Trias i Pujol, and YoMeCorono.org crowdfunding campaign" Publication status: preprint Source: medRxiv Author COI: authors declare no conflicts of interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Baro 2021 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
| Index tests | Test name: [A] PanBioTM COVID‐19 Ag Rapid test
[B] CLINITEST Rapid COVID‐19 Antigen Test
[C] SD Biosensor SARS‐CoV‐2 Rapid Antigen Test
[D] Lepu SARS‐CoV‐2 Antigen Rapid Test
[E] Surescreen COVID‐19 Coronavirus Rapid Antigen Test Cassette Manufacturer: [A] Abbott Rapid Diagnostics, Panbio Ltd, USA [B] Siemens Healthineers (Shangai International Holding Corp), USA [C] Roche Diagnostics, SD Biosensor, Republic of Korea [D] Beijing Lepu Medical Technology Co., Ltd., China [E] SureScreen Diagnostics Ltd, UK Antibody: nucleocapsid protein Ag target: not reported Test method: [A] CGIA [B] Immunochromatographic [C] LFA (unclear) [D] CGIA [E] LFA (unclear) Samples used: NP (collected by HCW) Transport media: VTM (DeltaSwab Virus, Deltalab; or UTM Universal Transport Medium, Copan) Sample storage: samples stored for up to 24 h (2‐8 ºC) prior to RT‐PCR then stored up to 12 h more at 2‐8 ºC until Ag testing Test operator: lab technician at The University Hospital Germans Trias i Pujol Definition of test positivity: visual coloured band; the presence of any test line (T) indicates a positive result. Samples were applied directly to the test cassette and incubated for 15 min at room temperature before reading results with the naked eye, according to the manufacturer IFU Blinding reported: unclear; presume only blinded to different Ags tests "All Ag‐RDT determinations were performed in parallel by two blinded technicians" Timing of samples: N/A; all asymptomatic |
||
| Target condition and reference standard(s) | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
| Flow and timing | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
Baro 2021 [C].
| Study characteristics | |||
| Patient Sampling | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
| Index tests | Test name: [A] PanBioTM COVID‐19 Ag Rapid test
[B] CLINITEST Rapid COVID‐19 Antigen Test
[C] SD Biosensor SARS‐CoV‐2 Rapid Antigen Test
[D] Lepu SARS‐CoV‐2 Antigen Rapid Test
[E] Surescreen COVID‐19 Coronavirus Rapid Antigen Test Cassette Manufacturer: [A] Abbott Rapid Diagnostics, Panbio Ltd, USA [B] Siemens Healthineers (Shangai International Holding Corp), USA [C] Roche Diagnostics, SD Biosensor, Republic of Korea [D] Beijing Lepu Medical Technology Co., Ltd., China [E] SureScreen Diagnostics Ltd, UK Antibody: nucleocapsid protein Ag target: not reported Test method: [A] CGIA [B] Immunochromatographic [C] LFA (unclear) [D] CGIA [E] LFA (unclear) Samples used: NP (collected by HCW) Transport media: VTM (DeltaSwab Virus, Deltalab; or UTM Universal Transport Medium, Copan) Sample storage: samples stored for up to 24 h (2‐8 ºC) prior to RT‐PCR then stored up to 12 h more at 2‐8 ºC until Ag testing Test operator: lab technician at The University Hospital Germans Trias i Pujol Definition of test positivity: visual coloured band; the presence of any test line (T) indicates a positive result. Samples were applied directly to the test cassette and incubated for 15 min at room temperature before reading results with the naked eye, according to the manufacturer IFU Blinding reported: unclear; presume only blinded to different Ags tests "All Ag‐RDT determinations were performed in parallel by two blinded technicians" Timing of samples: N/A; all asymptomatic |
||
| Target condition and reference standard(s) | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
| Flow and timing | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
Baro 2021 [D].
| Study characteristics | |||
| Patient Sampling | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
| Index tests | Test name: [A] PanBioTM COVID‐19 Ag Rapid test
[B] CLINITEST Rapid COVID‐19 Antigen Test
[C] SD Biosensor SARS‐CoV‐2 Rapid Antigen Test
[D] Lepu SARS‐CoV‐2 Antigen Rapid Test
[E] Surescreen COVID‐19 Coronavirus Rapid Antigen Test Cassette Manufacturer: [A] Abbott Rapid Diagnostics, Panbio Ltd, USA [B] Siemens Healthineers (Shangai International Holding Corp), USA [C] Roche Diagnostics, SD Biosensor, Republic of Korea [D] Beijing Lepu Medical Technology Co., Ltd., China [E] SureScreen Diagnostics Ltd, UK Antibody: nucleocapsid protein Ag target: not reported Test method: [A] CGIA [B] Immunochromatographic [C] LFA (unclear) [D] CGIA [E] LFA (unclear) Samples used: NP (collected by HCW) Transport media: VTM (DeltaSwab Virus, Deltalab; or UTM Universal Transport Medium, Copan) Sample storage: samples stored for up to 24 h (2‐8 ºC) prior to RT‐PCR then stored up to 12 h more at 2‐8 ºC until Ag testing Test operator: lab technician at The University Hospital Germans Trias i Pujol Definition of test positivity: visual coloured band; the presence of any test line (T) indicates a positive result. Samples were applied directly to the test cassette and incubated for 15 min at room temperature before reading results with the naked eye, according to the manufacturer IFU Blinding reported: unclear; presume only blinded to different Ags tests "All Ag‐RDT determinations were performed in parallel by two blinded technicians" Timing of samples: N/A; all asymptomatic |
||
| Target condition and reference standard(s) | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
| Flow and timing | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
Baro 2021 [E].
| Study characteristics | |||
| Patient Sampling | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
| Index tests | Test name: [A] PanBioTM COVID‐19 Ag Rapid test
[B] CLINITEST Rapid COVID‐19 Antigen Test
[C] SD Biosensor SARS‐CoV‐2 Rapid Antigen Test
[D] Lepu SARS‐CoV‐2 Antigen Rapid Test
[E] Surescreen COVID‐19 Coronavirus Rapid Antigen Test Cassette Manufacturer: [A] Abbott Rapid Diagnostics, Panbio Ltd, USA [B] Siemens Healthineers (Shangai International Holding Corp), USA [C] Roche Diagnostics, SD Biosensor, Republic of Korea [D] Beijing Lepu Medical Technology Co., Ltd., China [E] SureScreen Diagnostics Ltd, UK Antibody: nucleocapsid protein Ag target: not reported Test method: [A] CGIA [B] Immunochromatographic [C] LFA (unclear) [D] CGIA [E] LFA (unclear) Samples used: NP (collected by HCW) Transport media: VTM (DeltaSwab Virus, Deltalab; or UTM Universal Transport Medium, Copan) Sample storage: samples stored for up to 24 h (2‐8 ºC) prior to RT‐PCR then stored up to 12 h more at 2‐8 ºC until Ag testing Test operator: lab technician at The University Hospital Germans Trias i Pujol Definition of test positivity: visual coloured band; the presence of any test line (T) indicates a positive result. Samples were applied directly to the test cassette and incubated for 15 min at room temperature before reading results with the naked eye, according to the manufacturer IFU Blinding reported: unclear; presume only blinded to different Ags tests "All Ag‐RDT determinations were performed in parallel by two blinded technicians" Timing of samples: N/A; all asymptomatic |
||
| Target condition and reference standard(s) | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
| Flow and timing | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | Comparative study of 5 Ag tests; Baro 2021 [A] reports full study characteristics and QUADAS | ||
Basso 2021 [A].
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: COVID‐19 inpatients (n = 138) and outpatients (n = 96) screened for suspected SARS‐ CoV‐2 after contact with a SARS‐CoV‐2‐positive person or with typical symptoms (n per group was not reported) Recruitment: not stated Prospective or retrospective: prospective Sample size (cases): whole sample: 234 (87) Inpatients: 138 (84) Outpatients: 96 (3) |
||
| Patient characteristics and setting | Setting: inpatient and outpatient Location: Italy, University Hopsital of Padova Country: Italy Dates: 1 August and 30 November, 2020 Symptoms and severity: inpatients: 93/138 (67%) pneumonia, 97 (70%) fever > 37.5 °C, cough 46 (33%), dyspnoea 21 (15%) Outpatients: not reported Demographics: inpatients: 86, 62% male; mean age 56 years (SD 17); outpatients: 49, 51% male; mean age 42 years (SD 15) Exposure history: not reported |
||
| Index tests | Test name: [A] and [B] ESPLINE rapid test
[C] Panbio COVID‐19 Ag Rapid Test
(A 3rd laboratory‐based Ag detection assay was also evaluated but is not eligible for this review: LUMIPULSE SARS‐CoV‐2 Ag kit, Fujirebio, Tokjo, Japan) Manufacturer: [A] and [B] Fujirebio, Tokyo, Japan, [C] ABBOTT, Chicago, Illinois, USA Antibody: SARS‐CoV‐2 nucleocapsid protein Ag target: not stated Test method: CGIA Samples used: [A] saliva self‐sampling using Salivette device, SARSTEDT AG & Co, Nümbrecht, Germany; [B] and [C] NP swabs collected by qualified nurse Transport media: none used; NP swab testing conducted following manufacturer IFU Sample storage: all molecular and CLEIA Ag testing in both saliva and NP was performed in parallel within 3 h from collection Test operator: unclear; likely laboratory staff Definition of test positivity: not stated; visual inspection Blinding reported: unclear Timing of samples: inpatients (n = 138): 38, 27.6% ≤ 7 days pso; 74, 53.6% 7‐14 days pso; 26, 18.8% > 14 days pso |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; TaqPath COVID‐19 RT‐ PCR kit (Applied Biosystems, USA), performed by QuantStudio(TM) 5 Real‐Time PCR Systems (Applied Biosystems, USA) and QuantStudio(TM) 5 RealTime PCR Systems (Applied Biosystems, USA)
Threshold: ≥ 2 of 3 targets had an amplification plot with a Ct value of < 40 Definition of non‐COVID cases: as for cases; single negative Genetic target(s): Orf1ab, N and S SARS‐CoV‐2 genes Samples used: saliva (aliquot from same sample as index test); NP paired swab Timing of reference standard: same as for index test Blinded to index test: not stated Incorporated index test: no Time interval between index and reference tests: simultaneous; paired swab All participants received same reference standard: yes Missing data: yes. Authors provided data underlying Figure 3 however number of samples tested per assay and sample type vary; reason given was insufficient material for some cases, the number discrepancy was stated as not due to test failure: [A] ESPLINE ‐ saliva, n = 134 (55) [B] ESPINE ‐ NP, n = 136 (64) [C] PanBio ‐ NP, n = 116 (56) (Total of 164 (saliva) and 151 (NP) samples reported for LUMIPULSE) Uninterpretable results: none Indeterminate results (index test): none Indeterminate results (reference standard): none |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired swab All participants received same reference standard: yes Missing data: yes. Authors provided data underlying Figure 3 however number of samples tested per assay and sample type vary; reason given was insufficient material for some cases, the number discrepancy was stated as not due to test failure: [A] ESPLINE ‐ saliva, n = 134 (55) [B] ESPINE ‐ NP, n = 136 (64) [C] PanBio ‐ NP, n = 116 (56) (Total of 164 (saliva) and 151 (NP) samples reported for LUMIPULSE) Uninterpretable results: none Indeterminate results (index test): none Indeterminate results (reference standard): none Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: no funding statement provided Publication status: published Source: Clinica Chimica Acta Author COI: no COI statement provided |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Basso 2021 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 2 tests using different types of samples; Basso 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 2 tests using different types of samples; Basso 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] and [B] ESPLINE rapid test
[C] Panbio COVID‐19 Ag Rapid Test
(A 3rd laboratory‐based Ag detection assay was also evaluated but is not eligible for this review: LUMIPULSE SARS‐CoV‐2 Ag kit, Fujirebio, Tokjo, Japan) Manufacturer: [A] and [B] Fujirebio, Tokyo, Japan, [C] ABBOTT, Chicago, Illinois, USA Antibody: SARS‐CoV‐2 nucleocapsid protein Ag target: not stated Test method: CGIA Samples used: [A] saliva self‐sampling using Salivette device, SARSTEDT AG & Co, Nümbrecht, Germany; [B] and [C] NP swabs collected by qualified nurse Transport media: none used; NP swab testing conducted following manufacturer IFU Sample storage: all molecular and CLEIA Ag testing in both saliva and NP swabs performed in parallel within 3 h from collection Test operator: unclear; likely laboratory staff Definition of test positivity: not stated; visual inspection Blinding reported: unclear Timing of samples: inpatients (n = 138): 38, 27.6% ≤ 7 days pso; 74, 53.6% 7‐14 days pso; 26, 18.8% > 14 days pso |
||
| Target condition and reference standard(s) | Comparative study of 2 tests using different types of samples; Basso 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 2 tests using different types of samples; Basso 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 2 tests using different types of samples; Basso 2021 [A] reports full study characteristics and QUADAS. | ||
Basso 2021 [C].
| Study characteristics | |||
| Patient Sampling | Comparative study of 2 tests using different types of samples; Basso 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 2 tests using different types of samples; Basso 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] and [B] ESPLINE rapid test
[C] Panbio COVID‐19 Ag Rapid Test
(A 3rd laboratory‐based Ag detection assay was also evaluated but is not eligible for this review: LUMIPULSE SARS‐CoV‐2 Ag kit, Fujirebio, Tokjo, Japan) Manufacturer: [A] and [B] Fujirebio, Tokyo, Japan, [C] ABBOTT, Chicago, Illinois, USA Antibody: SARS‐CoV‐2 nucleocapsid protein Ag target: not stated Test method: CGIA Samples used: [A] saliva self‐sampling using Salivette device, SARSTEDT AG & Co, Nümbrecht, Germany; [B] and [C] NP swabs collected by qualified nurse Transport media: none used; NP swab testing conducted following manufacturer IFU Sample storage: all molecular and CLEIA Ag testing in both saliva and NP swabs performed in parallel within 3 h from collection Test operator: unclear; likely laboratory staff Definition of test positivity: not stated; visual inspection Blinding reported: unclear Timing of samples: inpatients (n = 138): 38, 27.6% ≤ 7 days pso; 74, 53.6% 7‐14 days pso; 26, 18.8% > 14 days pso |
||
| Target condition and reference standard(s) | Comparative study of 2 tests using different types of samples; Basso 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 2 tests using different types of samples; Basso 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 2 tests using different types of samples; Basso 2021 [A] reports full study characteristics and QUADAS. | ||
Beck 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity and specificity: all patients with signs and symptoms of COVID‐19 presenting to an urgent care centre (n = 347) Recruitment: consecutive ("all patients") Prospective or retrospective: not stated; appears prospective Sample size (cases): 347 (61) |
||
| Patient characteristics and setting | Setting: urgent care centre Location: Advocate Aurora Health Urgent Care Center, West Bend, WI Country: USA Dates: not stated Symptoms and severity: all symptomatic; no further details Demographics: age range 1‐90 years; ≤ 18 years 35.4%, 19‐50 years 38.3%, > 50 years 26.2% of participants Exposure history: not reported |
||
| Index tests | Test name: SOFIA SARS Antigen FIA Manufacturer: Quidel Antibody: SARS‐CoV Ag target: not reported Test method: FIA Samples used: nasal; collection not described but appears to be HCW ("providers" mentioned in acknowledgements). IFU describes NMT samples Transport media: none; "swabs were carefully returned to the paper envelope in which they came and were placed in a sealed plastic specimen transport bag" Sample storage: none; "specimens were delivered to the laboratory (located within the same building) within 10 minutes of collection" Test operator: lab staff Definition of test positivity: not stated; "tested … according to the manufacturer’s package insert" Blinding reported: yes; done first and in separate lab Timing of samples: ≤ 5 days pso 298, 86.1%; > 5 days pso 48, 13.9% |
||
| Target condition and reference standard(s) | Reference standard: transcription‐mediated amplification (TMA); Hologic Aptima Panther SARS‐CoV‐2 TMA test.
Cepheid Xpert Xpress SARS‐CoV‐2 RT‐PCR used for discrepant samples (after approx. 3 weeks' frozen storage) Definition of non‐COVID cases: as for cases Genetic target(s): not stated Samples used: NP in Amies bacterial transport medium (Copan, Brescia, Italy) Timing of reference standard: as for index test Blinded to index test: not stated; specimens were refrigerated and sent via courier to the central laboratory of ACL Laboratories Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired swabs, NP collected first All participants received same reference standard: yes Missing data: 1 sample only (low risk) Uninterpretable results: 1 sample invalid on SOFIA Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported. Discrepant analysis: 2/14 FNs were negative on Xpert Xpress; 1/1 FPs also negative Xpert Xpress Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: none reported Publication status: published Source: Journal of Clinical Microbiology Author COI: none reported |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Billaud 2020.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: teachers (n = 90) and students (n = 419) screened for COVID‐19 as part of a cluster investigation (n = 509) Recruitment: not stated; appears to be open to all Prospective or retrospective: prospective |
||
| Patient characteristics and setting | Setting: screening Location: college, Lyon Country: France Dates: 16 and 17 September Symptoms and severity: 166/509, 32.6% symptomatic including 152/419 (36%) students Demographics: mean, median age Students 21.6 years, 21 years (18‐37 years) Teachers 47.2 years, 49 years (26‐64 years) Exposure history: outbreak investigation |
||
| Index tests | Test name: described as "ABBOTT SARS‐COV2 Antigenic Test"; presumed to be Panbio COVID‐19 Ag Test Manufacturer: Abbott Antibody: not stated Ag target: not stated Test method: not stated Samples used: NP; collected by firefighters Transport media: none used Sample storage: N/A; tested immediately on site Test operator: not stated Definition of test positivity: visual line; as per manufacturer IFU Blinding reported: yes, performed first Timing of samples: not stated but includes people > 7 days pso |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; SARS‐COV‐2 (Thermofisher) Definition of non‐COVID cases: as for cases; single negative Genetic target(s): not stated Samples used: NP (paired) Timing of reference standard: as for index Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous All participants received same reference standard: yes Missing data: 47 missing, including 11 uninterpretable Uninterpretable results: 11 uninterpretable on Ag test Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: not stated, public funding Publication status: published Source: report accessed via SFM Microbiologie website Author COI: none |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Blairon 2020.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: sampled from cohort of suspected COVID‐19 patient samples sent for laboratory diagnosis (n = 56)
(Excluded data for full cohort, as only those with negative Ag test underwent confirmatory RT‐PCR; of 912 submitted samples during time period, 776 remained after removing repeat tests and were reported in main study) Recruitment: selection of 56 for verification analysis was not reported Prospective or retrospective: prospectively |
||
| Patient characteristics and setting | Setting: unclear; swabs obtained at hospital site, no further detail Location: not stated; author institution Iris Hospitals South, Brussels Country: Belgium Dates: 5 April‐4 May 2020 Symptoms and severity: not stated Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: COVID‐19 Ag Respi‐Strip (no product code reported) Manufacturer: Coris Bioconcept (Gembloux, Belgium) Antibody: not stated Ag target: not stated Test method: LFA Samples used: NP swabs; collection not reported Transport media: samples for Ag testing taken from UTM‐RT swabs (Copan spa, Brescia, IT) Sample storage: no storage described; infer that Ag test was conducted immediately on receipt of sample at on‐site laboratory "after antigenic testing was performed, the molecular assessment of SARS‐CoV‐2 was outsourced to a university centre" Test operator: not stated; infer laboratory staff Definition of test positivity: as per manufacturer IFU Blinding reported: not stated; infer yes as conducted prior to PCR confirmation Timing of samples: not stated; appears to be on presentation (repeat tests ordered at clinician's discretion were excluded) |
||
| Target condition and reference standard(s) | Reference standard: qRT‐PCR Definition of non‐COVID cases: as above, single PCR negative to confirm absence of disease Genetic target(s): E gene Samples used: NP swabs (same as for Ag test) Timing of reference standard: not stated Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: not stated but infer short interval; samples sent to university centre laboratory for PCR confirmation All participants received same reference standard: yes (only if study author confirms Ag+ also got PCR) Missing data: none reported; review team excluded main cohort data as no reference standard for Ag test‐positive samples Uninterpretable results: none reported; 1 "invalid" sample excluded from main cohort Indeterminate results (index test): none reported; 1 "non‐conform" sample excluded from main cohort Indeterminate results (reference standard): none reported Unit of analysis: unclear; main cohort includes unique patient samples but not reported for separate group of 56 |
||
| Comparative | |||
| Notes | Funding: none to declare Publication status: published Source: Journal of Clinical Virology Author COI: none to declare |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Bulilete 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group multi‐centre study estimating sensitivity and specificity
Adults attending 1 of 4 primary health care or COVID‐19 testing centres for PCR tests; included patients with symptoms suggestive of infection with referral by a GP, or close contact with a PCR−confirmed case
2 NP samples were collected, 1 for RT‐PCR and the other was processed on site using the PanbioTM rapid antigen test kit for SARS‐CoV‐2 Recruitment: consecutive patients > 18 years, attending the sites for RT‐PCR testing Prospective or retrospective: prospective Sample size (cases): 1362 (140); further 27 declined to participate |
||
| Patient characteristics and setting | Setting: 4 PHC centres and 2 COVID‐EXPRESS test sites Location: Mallorca, Spain (Balearic Public Health Service) Country: Spain Dates: 2–25 October 2020 Symptoms and severity: 680 (49.7%) reported symptoms < 7 days prior (most frequent: headache (341, 24.9%), sore throat (310, 22.6%), cough (301, 18.4%), and tiredness (251, 18.3%)). Asymptomatic: 689 (50.3%) Demographics: mean age 42.5 ± 14.9 years, and 744 (54.3%) women Exposure history: reasons for testing: 750 (54.8%) close contact with a confirmed positive COVID‐19 individual, 503 (36.7% symptoms suggestive of COVID‐19 and referral by a primary healthcare professional), 116 (8.5%) unknown |
||
| Index tests | Test name: Panbio COVID 19 antigen rapid test Manufacturer: Abbott Rapid Diagnostic Jena GmbH, Jena, Germany Antibody: SARS‐CoV‐2 nucleocapsid protein Ag target: not stated Test method: CGIA Samples used: NP swabs by qualified nurse Transport media: none; immediate on site testing Sample storage: none reported; results were interpreted within 15 min following the manufacturer IFU Test operator: not stated; presume same qualified nurse Definition of test positivity: not stated Blinding reported: yes; test was conducted on site before PCR test Timing of samples: 967, 70.6% presented within 5 days of the onset of symptoms or close contact Symptomatic: 622/677, 92% within 7 days pso Asymptomatic: 481/688, 70% within 7 days contact, 173/688 unknown number of days |
||
| Target condition and reference standard(s) | Reference standard: rRT‐PCR testing: MagMAXTM Viral/Pathogen Nucleic Acid Isolation Kit (ThermoFisher) and TaqPathTM COVID‐19 CE‐IVD RT‐PCR Kit and QuantStudioTM (ThermoFisher); Ct threshold not reported Definition of non‐COVID cases: as for cases; single negative Genetic target(s): ORF, N, and S Samples used: paired NP swab Timing of reference standard: same as for index; sample sent for processing within 24 h of collection Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired swab All participants received same reference standard: yes (7 PCR excluded; all Ag‐ve) Missing data: yes Uninterpretable results: 3 RT‐PCR with incorrect labelling; 16 Ag‐RDT results missing Indeterminate results (index test): none reported Indeterminate results (reference standard): 4 inconclusive RT‐PCR Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: no specific grant from funding agencies in the public, commercial or not‐for‐profit sectors Publication status: published Source: Journal of Infection Author COI: the authors declared no conflict of interest. |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Caruana 2021 [A].
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: all patients admitted to hospital (wards, intermediate care units and ICU) from the ED, with or without suspected SARS‐CoV‐2 infection
(A second study investigating the correlation of symptom duration and variations in viral load was also reported, but not eligible for this review) Recruitment: consecutive; with target of 100 RT‐PCR+ve and at least 200 RT‐PCR−ve Prospective or retrospective: prospective Sample size (cases): 572 (114) |
||
| Patient characteristics and setting | Setting: inpatient; patients admitted to hospital from the ED Location: Lausanne University hospital Country: Switzerland Dates: 6 November‐6 December 2020 Symptoms and severity: 239 (45%) asymptomatic, admitted for other reasons than COVID‐19 suspicion; 293 (55%) symptoms consistent with COVID‐19; included some with atypical symptoms (n not reported) Demographics: asymptomatic for COVID‐19: age 67 years (IQR 49‐81); 105, 44% female Symptomatic: age 75 years (IQR 61‐85); 131 (45%) female Exposure history: not reported |
||
| Index tests | Test name: [A] STANDARD Q COVID‐19 Rapid AgTest
[B] PanbioTM COVID‐19 Ag Rapid Test
[C] One Step Immunoassay for Exdia COVID‐19 Ag
[D] BD Veritor System for Rapid Detection of SARS‐CoV‐2
[Result of STANDARD Q was used to guide care/triage pathway; patients and clinicians were blinded to the results of all other Ag tests] Manufacturer: [A] SD Biosensor ‐ Republic of Korea /Roche ‐ Switzerland [B] Abbott ‐ USA [C] Precision Biosensor Inc. ‐ Republic of Korea [D] Becton Dickinson ‐ USA Antibody: not reported Ag target: not reported Test method: not reported Samples used: NP (collection not reported) Transport media: wet swab procedure: NP swabs suspended in 2.5‐3 mL VTM, then [A] 350 μL of sample mixed with buffer solution [B], [C] and [D] 300 μL of sample mixed with buffer solution Sample storage: NP delivered to the RAT lab, immediately after the sampling procedure Test operator: laboratory technicians Definition of test positivity: [A] and [B] visually [C] and [D] automatically using analyser Blinding reported: presumed (based on timing of tests) Timing of samples: ≤ 4 days: 138/293 (47%); 4‐7 days: 46/293 (16%); ≥ 7 days: 44/293 (15%) Missing data/not typical COVID‐19 symptoms: 65/293 (22%) |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; assay used varied according to result of STANDARD Q test
'Classical' RT‐PCR used for RDT+ve symptomatic cases and for RDT‐ve asymptomatic patients: either
[1] test Cobas 6800 SARS‐CoV‐2 (Roche, Basel, Switzerland) or
[2] automated high‐throughput molecular diagnostic (MDx) platform
'Rapid' RT‐PCR used for RDT‐ve symptomatic cases and for RDT+ve asymptomatic patients: either
[3] VIASURE SARS‐CoV‐2 (N1 + N2) Real Time PCR Detection Kit for BD MAX (Becton Dickinson, USA), or
[4] GeneXpert SARS‐CoV‐2 test (Cepheid, www.cepheid.com)
Viral load was quantified using the equation: VL = (10^((Ct ‐40.856)/ ‐3.697))*100, derived from RNA quantification Definition of non‐COVID cases: as for cases Genetic target(s): [D] E‐ and RdRp‐ encoding genes [A] to [C] not reported Samples used: same as for index test (same swab) Timing of reference standard: same as for index test Blinded to index test: no; assay used varied according to symptom status and RDT result Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same swab All participants received same reference standard: yes; all RT‐PCR Missing data: yes; 67 excluded including 40 with missing results Uninterpretable results: not reported Indeterminate results (index test): n = 27 invalid RAT results Indeterminate results (reference standard): not reported |
||
| Comparative | |||
| Notes | Funding: no financial support Publication status: published Source: Microbiology Author COI: from published version: "Croxatto reports grants from Becton Dickinson outside the submitted work. Greub reports grants from Resistell, from Nittobo, outside the submitted work and he is the co‐director of “JeuPro,” a start‐up distributing the game Krobs, a card game about microbe transmission." |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | No | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Caruana 2021 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 4 Ag tests; Caruana 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 4 Ag tests; Caruana 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] STANDARD Q COVID‐19 Rapid Antigen Test
[B] PanbioTM COVID‐19 Ag Rapid Test
[C] One Step Immunoassay for Exdia COVID‐19 Ag
[D] BD Veritor System for Rapid Detection of SARS‐CoV‐2
(Result of STANDARD Q was used to guide care/triage pathway; patients and clinicians were blinded to the results of all other Ag tests) Manufacturer: [A] SD Biosensor ‐ Republic of Korea /Roche ‐ Switzerland [B] Abbott ‐ USA [C] Precision Biosensor Inc. ‐ Republic of Korea [D] Becton Dickinson ‐ USA Antibody: not reported Ag target: not reported Test method: not reported Samples used: NP (collection not reported) Transport media: wet swab procedure: NP swabs suspended in 2.5‐3 mL VTM, then [A] 350 μL of sample mixed with buffer solution [B], [C] and [D] 300 μL of sample mixed with buffer solution Sample storage: NP delivered to the RAT lab, immediately after the sampling procedure Test operator: laboratory technicians Definition of test positivity: [A] and [B] visually [C] and [D] automatically using analyser Blinding reported: presumed (based on timing of tests) Timing of samples: ≤ 4 days: 138/293 (47%); 4‐7 days: 46/293 (16%); ≥ 7 days: 44/293 (15%) Missing data/not typical COVID‐19 symptoms: 65/293 (22%) |
||
| Target condition and reference standard(s) | Comparative study of 4 Ag tests; Caruana 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 4 Ag tests; Caruana 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 4 Ag tests; Caruana 2021 [A] reports full study characteristics and QUADAS. | ||
Caruana 2021 [C].
| Study characteristics | |||
| Patient Sampling | Comparative study of 4 Ag tests; Caruana 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 4 Ag tests; Caruana 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] STANDARD Q COVID‐19 Rapid Antigen Test
[B] PanbioTM COVID‐19 Ag Rapid Test
[C] One Step Immunoassay for Exdia COVID‐19 Ag
[D] BD Veritor System for Rapid Detection of SARS‐CoV‐2
(Result of STANDARD Q was used to guide care/triage pathway; patients and clinicians were blinded to the results of all other Ag tests) Manufacturer: [A] SD Biosensor ‐ Republic of Korea /Roche ‐ Switzerland [B] Abbott ‐ USA [C] Precision Biosensor Inc. ‐ Republic of Korea [D] Becton Dickinson ‐ USA Antibody: not reported Ag target: not reported Test method: not reported Samples used: NP (collection not reported) Transport media: wet swab procedure: NP swabs suspended in 2.5‐3 mL VTM, then [A] 350 μL of sample mixed with buffer solution [B], [C] and [D] 300 μL of sample mixed with buffer solution Sample storage: NP delivered to the RAT lab, immediately after the sampling procedure Test operator: laboratory technicians Definition of test positivity: [A] and [B] visually [C] and [D] automatically using analyser Blinding reported: presumed (based on timing of tests) Timing of samples: ≤ 4 days: 138/293 (47%); 4‐7 days: 46/293 (16%); ≥ 7 days: 44/293 (15%) Missing data/not typical COVID‐19 symptoms: 65/293 (22%) |
||
| Target condition and reference standard(s) | Comparative study of 4 Ag tests; Caruana 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 4 Ag tests; Caruana 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 4 Ag tests; Caruana 2021 [A] reports full study characteristics and QUADAS. | ||
Caruana 2021 [D].
| Study characteristics | |||
| Patient Sampling | Comparative study of 4 Ag tests; Caruana 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 4 Ag tests; Caruana 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] STANDARD Q COVID‐19 Rapid Antigen Test
[B] PanbioTM COVID‐19 Ag Rapid Test
[C] One Step Immunoassay for Exdia COVID‐19 Ag
[D] BD Veritor System for Rapid Detection of SARS‐CoV‐2
(Result of STANDARD Q was used to guide care/triage pathway; patients and clinicians were blinded to the results of all other Ag tests) Manufacturer: [A] SD Biosensor ‐ Republic of Korea /Roche ‐ Switzerland [B] Abbott ‐ USA [C] Precision Biosensor Inc. ‐ Republic of Korea [D] Becton Dickinson ‐ USA Antibody: not reported Ag target: not reported Test method: not reported Samples used: NP (collection not reported) Transport media: wet swab procedure: NP swabs suspended in 2.5‐3 mL VTM, then [A] 350 μL of sample mixed with buffer solution [B], [C] and [D] 300 μL of sample mixed with buffer solution Sample storage: NP delivered to the RAT lab, immediately after the sampling procedure Test operator: laboratory technicians Definition of test positivity: [A] and [B] visually [C] and [D] automatically using analyser Blinding reported: presumed (based on timing of tests) Timing of samples: ≤ 4 days: 138/293 (47%); 4 ‐7 days: 46/293 (16%); ≥ 7 days: 44/293 (15%) Missing data/not typical COVID‐19 symptoms: 65/293 (22%) |
||
| Target condition and reference standard(s) | Comparative study of 4 Ag tests; Caruana 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 4 Ag tests; Caruana 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 4 Ag tests; Caruana 2021 [A] reports full study characteristics and QUADAS. | ||
Cerutti 2020.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity in 2 cohorts:
(1) symptomatic patients attending one of 2 EDs (n = 185)
(2) asymptomatic travellers returning home from European high‐risk countries (Croatia, Spain, Malta) (n = 145) Recruitment: (1) random; (2) not stated, presume consecutive Prospective or retrospective: not stated |
||
| Patient characteristics and setting | Setting: mixed; (1) ED; (2) possible contacts Location: (1) 2 infectious disease reference centres in northern Italy (ASL Città di Torino, Turin and San Martino University Hospital, Genoa); (2) not stated; samples sent to Microbiology and Virology Laboratory, Amedeo di Savoia Hospital, Torino Country: Italy Dates: (1) 3 March‐1 May; (2) August 2020 Symptoms and severity: not stated; cohort (2) were asymptomatic Demographics: (1) mean age 44.6, 95% CI 40.7–48.6; (2) mean age 35.9, 95% CI 32.7–39.1 Exposure history: (1) not stated; (2) high‐risk country visit |
||
| Index tests | Test name: STANDARD Q COVID‐19 Ag Manufacturer: SD‐Biosensor, RELAB, I Antibody: NP Ag target: not stated Test method: not stated Samples used: NP; collection not stated Transport media: UTM (Copan, I) Sample storage: primarily run in parallel with standard care RT‐PCR; 13 were frozen residual samples Test operator: not stated; laboratory staff presumed Definition of test positivity: visual line after 15‐30 min; as per manufacturer IFU Blinding reported: not stated Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; Seegene Allplex 2019 n‐CoV Assay (n = 159), DiaSorin Simplexa (n = 28), and Cobas 6800 Roche (n = 118) Definition of non‐COVID cases: single negative Genetic target(s): not stated Samples used: not stated; seems to be same as index Timing of reference standard: not stated Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; not clear if same sample used or paired swabs obtained All participants received same reference standard: yes; different assays Missing data: none reported; no participant flow diagram reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: RELAb donated the STANDARD Q COVID‐19 SD‐Biosensor kits used Publication status: published Source: Journal of Clinical Virology Author COI: the authors report no declarations of interest. |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Chaimayo 2020.
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity and specificity: suspected cases of COVID‐19 including symptomatic and contact individuals, including travellers, quarantined individuals and pre‐operative patients Recruitment: not stated Prospective or retrospective: not stated Sample size (cases): 60 (37) |
||
| Patient characteristics and setting | Setting: mixed Location: not all specified; main institute was Siriraj Hospital, Bangkok, Thailand Country: Thailand Dates: March‐May 2020 Symptoms and severity: PCR+: 37/60, 61.7% showed signs and symptoms of upper respiratory tract infection 5 (8.3%) pneumonia and ICU admission, 11 fever, 4 unspecified, 3 asymptomatic. NB ‐ supplementary file shows 53/60 with fever Demographics: PCR+: median age 38.5 years (range 21–72); 36 (60%) male PCR−: median age 61 years (range 16‐95); 163/394 (41%) male Exposure history: PCR+: 45, 75% direct contact with confirmed cases, including family and friends (30%; n = 18), karaoke bars and pubs (23.3%; n = 14), boxing stadiums (18.3%; n = 11), taxi drivers (1.7%; n = 1), workplace peers (1.7%; n = 1) |
||
| Index tests | Test name: STANDARD Q COVID‐19 Ag kit Manufacturer: SD Biosensor, Republic of Korea Antibody: SARS‐CoV‐2 nucleocapsid (N) antigen Ag target: mouse monoclonal anti‐SARS‐CoV‐2 antibody against SARS‐CoV‐2 N antigen Test method: CGIA Samples used: mixed; 447 NP or throat swabs, 4 endotracheal aspirates (tracheal suctions), 3 sputum Transport media: 2 mL VTM; Hanks’ balanced salt, 0.4% fetal bovine serum, HEPES, antibiotic and antifungal agents Sample storage: transported at 2–8 °C to the Microbiology laboratory, Siriraj Hospital, for processing within a few hours Test operator: not stated; likely laboratory staff, "All specimens were processed in biosafety level‐3 (BSL‐3) and biosafety level‐2 enhanced (BSL‐2+) facilities with full personal protective equipment" Definition of test positivity: for positive COVID‐19 Ag result, 2 coloured lines of control (C) and test (T) lines were presented. Blinding reported: not mentioned Timing of samples: PCR+: 3 asymptomatic, 41 (68%) day 1‐7, 12 (20%) day > 7, 4 unspecified time pso |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; AllplexTM 2019‐nCoV Assay (Seegene, Korea); Ct value < 40 for all 3 target genes was defined as a positive result Definition of non‐COVID cases: as for cases; single negative Genetic target(s): E gene (Sarbecovirus), and RdRp and N genes (SARS‐CoV‐2) Samples used: NP swab in VTM Timing of reference standard: as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired swab All participants received same reference standard: yes Missing data: none stated Uninterpretable results: none reported Indeterminate results (index test): none Indeterminate results (reference standard): none Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "partly supported by Faculty of Medicine Siriraj Hospital, Mahidol University, Thailand through grant number R016034012" Publication status: published Source: Virology Journal Author COI: no conflict of interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Ciotti 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity and specificity: NP swabs from patients with suspected SARS‐CoV‐2 infection at the ED or Infectious Diseases ward Recruitment: not reported Prospective or retrospective: not reported Sample size (cases): 50 (39) |
||
| Patient characteristics and setting | Setting: mixed; ED or Infectious Diseases ward Location: University Hospital Tor Vergata, Rome Country: Italy Dates: May‐September 2020 Symptoms and severity: not reported Demographics: median age 53.5 years, (mean 53.1; range: 15–94 years); 24, 48% male Exposure history: not reported |
||
| Index tests | Test name: COVID‐19 Ag Respi‐Strip Manufacturer: Coris BioConcept Antibody: nucleoprotein of SARS‐CoV and SARS‐CoV‐2 Ag target: monoclonal antibodies Test method: CGIA Samples used: NP (collection not described) Transport media: not clear but seems that no VTM used; 100 μL "nasopharyngeal secretions are mixed with four drops (about 100 μL) of lysis buffer" Sample storage: none reported Test operator: not stated; virology laboratory Definition of test positivity: visual appearance of test and control (red) lines Blinding reported: not stated Timing of samples: not reported |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; Allplex 2019n‐CoV assay Definition of non‐COVID cases: as for cases; single negative Genetic target(s): E gene (Sarbecovirus sub‐genus) and N and RdRp genes (SARS‐CoV‐2) Samples used: NP swabs Timing of reference standard: not stated; same as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; appears to be same swab but not clearly stated All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis |
||
| Comparative | |||
| Notes | Funding: no funding statement reported Publication status: published Source: Journal of Medical Virology Author COI: the authors declare that there are no conflict of interests |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Courtellemont 2021.
| Study characteristics | |||
| Patient Sampling | Unclear design estimating sensitivity and specificity (coded as 2‐group because of deliberate sampling of PCR+ve cases):
(1) symptomatic (headache, fatigue, fever, or respiratory signs) or asymptomatic people voluntarily accessing the COVID‐19 Screening Department (n = 231)
(2) hospitalized SARS‐CoV‐2‐positive patients (n = 17) (review team excluded 20 cases with a previous positive RT‐qPCR within 5 days but a negative RTqPCR at the time of study sampling.) Recruitment: unclear Prospective or retrospective: unclear |
||
| Patient characteristics and setting | Setting: mixed Location: COVID‐19 Screening Department and SARS CoV‐2‐positive patients hospitalized in the Infectious Diseases Department of the Centre Hospitalier Régional (CHR) of Orléans, France, or the Department of Infectious and Tropical Diseases of the Centre Hospitalier Universitaire (CHU) Tenon, Paris Country: France Dates: 12‐19 October Symptoms and severity: 99/121, 82% cases were symptomatic; 22 asymptomatic Demographics: median age 38 years, mean age 43 years (range: 18‐96), 117, 47% male Exposure history: not stated |
||
| Index tests | Test name: COVID‐VIRO Manufacturer: AAZ, Boulogne Billancourt, France Antibody: nucleocapsid Ag target: monoclonal Test method: CGIA Samples used: NP; collected by trained personnel (nurse, doctors, or biologist); subgroup also had OP or saliva collected Transport media: direct testing for Ag test Sample storage: none Test operator: not stated Definition of test positivity: visible line; as per manufacturer IFU Blinding reported: yes Timing of samples: median 5 days pso, mean 5.3 days, range 1‐20d |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; TaqPath Covid‐19 Multiplex RT‐PCR, Thermofisher Definition of non‐COVID cases: single negative PCR Genetic target(s): ORF1ab, S and N genes Samples used: NP in VTM; paired Timing of reference standard: as for index Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired All participants received same reference standard: yes Missing data: none reported, no participant flow diagram reported; review team excluded 20 cases with a previous positive RT‐qPCR within 5 days but a negative RTqPCR at the time of study sampling Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: no funding statement reported Publication status: preprint Source: medRxiv Author COI: no COI statement reported |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Del Vecchio 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: patients with and without symptoms examined at ED (n = 1153), infectious disease wards (n = 279) or other department (n = 9) who required COVID‐19 testing either due to a) presence of COVID‐19‐related symptoms (fever and/or cough and/or headache, diarrhoea, asthenia, muscle pain, joint pain, loss of taste or smell, or shortness of breath, with or without pneumonia); or b) asymptomatic but had a contact with a confirmed case of SARS‐CoV‐2 infection during the previous 10 days Recruitment: convenience, only included if both Ag and PCR results were available Prospective or retrospective: retrospective Sample size (cases): 1441 (61); 58/61 PCR+ were observed at ED |
||
| Patient characteristics and setting | Setting: mixed; ED and infectious disease ward admissions Location: University Hospital of Padua Country: Italy Dates: 15 September‐16 October 2020 Symptoms and severity: only reported for PCR+: 51/61 (84%) symptomatic, including 10 (20%) with asthenia, 7 (14%) with cough, 3 (6%) with dyspnoea (1/3 severe), 32 (63%) with fever, 7 (14%) with headache Demographics: 760 (53%) male Age: 0‐19 years: n = 54, 20‐39 years: n = 247, 40‐59 years: n = 262, 60‐79 years: n = 420, 80‐99 years: n = 457, > 100 years: n = 1 Exposure history: not reported |
||
| Index tests | Test name: Panbio COVID‐19 Ag Rapid Test Device Manufacturer: Abbott Lake Country, IL, USA Antibody: not stated Ag target: not stated Test method: not stated Samples used: not stated; mentions NP in Discussion only Transport media: not used for Ag testing Sample storage: processed right after sampling; maximum 1‐h delay Test operator: not stated Definition of test positivity: according to manufacturer IFU Blinding reported: yes; conducted first Timing of samples: 0‐7 days: n = 39, 64% (28 day 0‐3); 8‐14 days: n = 11, 18%; ≥ 15 days: n = 1 |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; DiaSorin Molecular Simplexa COVID‐19 Direct assay system (Diasorin Cypress, CA, USA)
"Samples showing a positive result for both viral targets were considered positive. Samples with either a single positive target or with Ct value ≥ 30 were confirmed with an in‐house real‐time RT‐PCR targeting the N2 gene." Definition of non‐COVID cases: as for cases Genetic target(s): S gene and the ORF1ab gene Samples used: not stated Timing of reference standard: as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired swabs All participants received same reference standard: yes Missing data: none reported (1849 patients only had either RAT or RT‐PCR test results available and were excluded a priori) Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "COVID‐19 emergency fund from University of Padova to ST" Publication status: preprint (not peer reviewed) Source: medRxiv Author COI: no competing interests |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Dominguez Fernandez 2021.
| Study characteristics | |||
| Patient Sampling | Unclear design estimating sensitivity and specificity: users with symptoms compatible with COVID‐19 and/or were close contacts of users with a positive COVID‐19 diagnosis Recruitment: not stated Prospective or retrospective: not stated Sample size (cases): 30 (20) |
||
| Patient characteristics and setting | Setting: care home Location: not stated; author institutions included the Coordination and Assistance Support Unit for Social and Health Residences of the A Coruna and Cee Health Area and the Microbiology Service, A Coruna University Hospital Complex, A Coruna Country: Spain Dates: September 2020 Symptoms and severity: 90% had symptoms compatible with SARS‐CoV‐2 infection of < 5 days of evolution and the other 10% were asymptomatic, but were close contacts Demographics: mean age 76.2 years (SD: 19.76), 36.7% male Exposure history: 10% asymptomatic close contacts |
||
| Index tests | Test name: Panbio COVID‐19 Ag Rapid Test Device Manufacturer: Abbott Antibody: not reported Ag target: not reported Test method: CGIA Samples used: not stated Transport media: not stated Sample storage: not stated Test operator: not stated Definition of test positivity: not stated Blinding reported: not stated Timing of samples: 90% were < 5 days pso |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; no further details Definition of non‐COVID cases: single negative Genetic target(s): not stated Samples used: not stated Timing of reference standard: not stated Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; not stated whether paired/same sample used All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: no funding statement reported Publication status: published letter Source: Enfermedades Infecciosas y Microbiología Clínica Author COI: no COI statement reported |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Drain 2021(a).
| Study characteristics | |||
| Patient Sampling | Report of 2 studies estimating sensitivity and specificity, 1st is a 2‐group study using nasal swabs and the 2nd a single‐group study using NP swabs.
[1] nasal swabs from children and adults presenting for COVID‐19 testing at 10 sites in the USA and UK (first time period)
[2] nasal swab samples from a commercial supplier (MRN Diagnostics, Florida, USA) and also collected from an at‐risk population (LumiraDx Stirling, UK)
[3] NP swabs from children and adults presenting for COVID‐19 testing at 10 sites in the USA and UK (second time period)
Data for cohort [1] and [2] are included asDrain 2021(a); seeDrain 2021(b)for details of cohort [3] Recruitment: unclear Prospective or retrospective: described as prospective Sample size (cases): 257(83) |
||
| Patient characteristics and setting | Setting: presume COVID‐19 testing centres Location: 10 sites in the UK and USA Country: UK and USA Dates: [1] 26 June‐23 July 2020; [2] not reported Symptoms and severity: whole sample: 414/512 (81%) symptomatic; [1]+[2] 159/257 (62%) symptomatic Demographics: whole sample: 287 female, 225 male. Age (0‐90 years); [1]+[2] mean age 34 years (SD 15.7 years); 142 (55%) female Exposure history: not stated |
||
| Index tests | Test name: LumiraDx SARS‐CoV‐2 Manufacturer: LumiraDx UK Ltd. Antibody: N Ag target: not stated Test method: microfluidic immunoassay with fluorescent latex signal Samples used: [1]+[2] AN; at 8 of 10 sites, swabs collected and tested by minimally trained operators Transport media: none; 0.7 mL of a proprietary extraction buffer for LumiraDx SARS‐CoV‐2 Ag test Sample storage: [1] tested fresh and then frozen within 1 h of collection [2] Unclear Test operator: unclear; included minimally trained operators Definition of test positivity: result shown on touchscreen as "positive" Blinding reported: [1]+[2] results from retested frozen samples were described as blinded Timing of samples: [1]+[2] range 1‐12 days; mean 4.0 days (SD 2.9 days) pso |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; SARS‐CoV‐2 assay using a Roche Cobas 6800 platform (Roche Molecular Diagnostics, Indianapolis, IN, USA); threshold Ct of 35 (based on Figure 2) Definition of non‐COVID cases: single negative PCR Same as for cases; appears to be single negative PCR to confirm absence of infection Genetic target(s): not stated Samples used: paired swab Timing of reference standard: same as for index test Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneously All participants received same reference standard: yes Missing data: not mentioned Uninterpretable results: not mentioned Indeterminate results (index test): not mentioned Indeterminate results (reference standard): not mentioned Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "work was supported by LumiraDx Ltd, including funding of the journal’s Rapid Services fee" Publication status: published paper Source: academic journal Author COI: no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Drain 2021(b).
| Study characteristics | |||
| Patient Sampling | Report of 2 studies estimating sensitivity and specificity, 1st is a 2‐group study using nasal swabs and the 2nd a single‐group study using NP swabs:
[1] nasal swabs from children and adults presenting for COVID‐19 testing at 10 sites in the USA and UK (first time period)
[2] nasal swab samples from a commercial supplier (MRN Diagnostics, Florida, USA) and also collected from an at‐risk population (LumiraDx Stirling, UK)
[3] NP swabs from children and adults presenting for COVID‐19 testing at 10 sites in the USA and UK (second time period)
See Drain 2021(a) for details of cohort [1] and [2]; Data for cohort [3] included asDrain 2021(b) Recruitment: unclear Prospective or retrospective: described as prospective Sample size (cases): 255 (40) |
||
| Patient characteristics and setting | Setting: presume COVID‐19 testing centres Location: 10 sites in the UK and USA Country: UK and USA Dates: [3] 17 August‐28 September 2020 Symptoms and severity: whole sample: 414/512 (81%) symptomatic [3] 255/255 (100%) symptomatic Demographics: whole sample: 287 female, 225 male. Age (0‐90 years) [3] mean age 33.2 years (SD 19.4 years); 145 (57%) female Exposure history: not stated |
||
| Index tests | Test name: LumiraDx SARS‐CoV‐2 Manufacturer: LumiraDx UK Ltd Antibody: N Ag target: not stated Test method: microfluidic immunoassay with fluorescent latex signal Samples used: [3] NP; at 8 of 10 sites, swabs collected and tested by minimally trained operators Transport media: none; 0.7 mL of a proprietary extraction buffer for LumiraDx SARS‐CoV‐2 Ag test Sample storage: [3] no storage Test operator: unclear; included minimally trained operators Definition of test positivity: result shown on touchscreen as "positive" Blinding reported: [3] yes, based on timing of test Timing of samples: whole sample: range 1‐12 days [3] mean 3.5 days (SD 2.5) pso |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; SARS‐CoV‐2 assay using a Roche Cobas 6800 platform (Roche Molecular Diagnostics, Indianapolis, IN, USA); threshold Ct of 35 (based on Figure 2) Definition of non‐COVID cases: same as for cases; appears to be single negative PCR to confirm absence of infection Genetic target(s): not stated Samples used: paired swab Timing of reference standard: same as for index test Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneously All participants received same reference standard: yes Missing data: not mentioned Uninterpretable results: not mentioned Indeterminate results (index test): not mentioned Indeterminate results (reference standard): not mentioned Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "work was supported by LumiraDx Ltd, including funding of the journal’s Rapid Services fee" Publication status: published paper Source: academic journal Author COI: no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Drevinek 2020 [A].
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity and specificity: participants aged ≥ 10 years, who attended a COVID‐19 testing centre due to suspicion of COVID‐19 (n = 273) or contact tracing (n = 290); either referred by a GP or public health officer (n = 511) or were 'self‐payers' (n = 54) Recruitment: not stated but seems to be all who consented to participate during stated time period Prospective or retrospective: prospective Sample size (cases): 591 (223) |
||
| Patient characteristics and setting | Setting: COVID‐19 testing site at a university hospital Location: Motol University Hospital, Prague Country: Czech Republic Dates: 4‐day period in October 2020 Symptoms and severity: 290 (49%) symptomatic on day of testing Demographics: mean age 40 years (range 12‐78 years); 44.7% male Exposure history: 290 tested as a result of contact tracing |
||
| Index tests | Test name: [A] Panbio Covid‐19 Ag Rapid Test
[B] STANDARD F Covid‐19 Ag FIA Manufacturer: [A] Abbott, Germany [B] SD Biosensor, Republic of Korea Antibody: not stated Ag target: not stated Test method: [A] CGIA [B] FIA Samples used: NP swabs (1 per Ag assay); collection not described Transport media: none used; states "without the optional step of inserting the swab into the viral transport medium" Sample storage: no storage; tested immediately on collection Test operator: not stated Definition of test positivity: [A] visual assessment after 15 min incubation [B] STANDARD F200 Analyser (in 'read‐only' mode) after 30 min incubation Blinding reported: not stated, but very likely considering test was done prior to reference standard Yes; conducted first Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; Allplex 2019n‐CoV assay
Ct value < 40 for positive result Definition of non‐COVID cases: as for cases; single negative Genetic target(s): N, E and RdRP/S genes Samples used: combined NP + OP swab in VTM Timing of reference standard: not stated; same as for index Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired swab All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "supported by the Ministry of Health of the Czech Republic ‐ conceptual development of research organization Motol University Hospital, FNM" Publication status: published Source: medRxiv Author COI: no conflicts of interest to report |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Drevinek 2020 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 2 Ag tests; Drevinek 2020 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 2 Ag tests; Drevinek 2020 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] Panbio Covid‐19 Ag Rapid Test
[B] STANDARD F Covid‐19 Ag FIA Manufacturer: [A] Abbott, Germany [B] SD Biosensor, Republic of Korea Antibody: not stated Ag target: not stated Test method: [A] CGIA [B] FIA Samples used: NP swabs (1 per Ag assay); collection not described Transport media: none used; states "without the optional step of inserting the swab into the viral transport medium" Sample storage: no storage; tested immediately on collection Test operator: not stated Definition of test positivity: [A] visual assessment after 15 min incubation [B] Standard F200 Analyser (in 'read‐only' mode) after 30 min incubation Blinding reported: not stated, but very likely considering test was done prior to reference standard Yes; conducted first Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 2 Ag tests; Drevinek 2020 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 2 Ag tests; Drevinek 2020 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 2 Ag tests; Drevinek 2020 [A] reports full study characteristics and QUADAS. | ||
Faico‐Filho 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: adults (aged > 18 years) treated in the ED and hospitalized for at least 24 h, including those
Recruitment: not reported; implies all eligible were included Prospective or retrospective: prospective Sample size (cases): 127 (70) |
||
| Patient characteristics and setting | Setting: ED Location: São Paulo Hospital in São Paulo Country: Brazil Dates: not reported Symptoms and severity: not reported Demographics: mean age (SD): 60 (17.5) years Sex: 69/127 (54%) male Exposure history: not reported |
||
| Index tests | Test name: Panbio COVID‐19 Ag test Manufacturer: Abbott Antibody: not reported Ag target: not reported Test method: not reported Samples used: NP (not reported who collected by) Transport media: not reported Sample storage: only reported that NP swab samples were simultaneously tested with both index and reference tests and RT‐PCR results were available within 6‐24 h Test operator: not reported Definition of test positivity: according to manufacturer Blinding reported: not reported Timing of samples: mean days pso: 5 (4; 7) |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; [1] GeneFinder COVID‐19 Plus RealAmp Kit (OSANG Healthcare Co., Ltd.); [2] Mobius XGEN MASTER COVID‐19 test for inconclusive GeneFinder results Definition of non‐COVID cases: as for cases Genetic target(s): [1] RdRp, E and N SARS‐CoV‐2 genes [2] ORF1ab and N SARS‐CoV‐2 genes Samples used: NP Timing of reference standard: as for index test Blinded to index test: not reported Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous All participants received same reference standard: yes Missing data: not reported Uninterpretable results: not reported Indeterminate results (index test): not reported Indeterminate results (reference standard): not reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "the UNIFESP team received Panbio COVID‐19 Ag tests for the study from Abbott." Publication status: preprint (not peer reviewed) Source: medRxiv Author COI: "the UNIFESP team received Panbio COVID‐19 Ag tests for the study from Abbott. Dr. Nancy Bellei provides lectures for and is on the advisory board of Abbott." |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Favresse 2021 [A].
| Study characteristics | |||
| Patient Sampling | Single‐group estimating sensitivity and specificity: NP samples from patients who presented for SARS‐CoV‐2 testing at single institution Recruitment: not stated Prospective or retrospective: not stated Sample size (cases): 188 (96) |
||
| Patient characteristics and setting | Setting: laboratory‐based Location: laboratory of the Clinique Saint‐Luc (Bouge, Namur, Belgium) Country: Belgium Dates: 7‐25 November 2020 Symptoms and severity: 118, 63% symptomatic Demographics: women (n = 104, 55%): median age 54 years (range 5‐97 years) Men (n = 84): median age 57 years (range 1‐94 years) Exposure history: not reported |
||
| Index tests | Test name: [A] Biotical SARS‐CoV‐2 Ag card
[B] Panbio COVID‐19 Ag Rapid Test Device
[C] Coronavirus Ag Rapid Test Cassette
[D] Roche SARS‐CoV‐2 Rapid Antigen Test
(Additional lab‐based Ag test also evaluated but not eligible for this review: [E] VITROS Immunodiagnostic Products SARS‐CoV‐2 Antigen test (Ortho Clinical Diagnostics, Raritan, NJ, USA)) Manufacturer: [A] Biotical Health, Madrid, Spain [B] Abbott, Chicago, IL, USA [C] Healgen Scientific, Houston, TX, USA [D] Roche Diagnostics, Basel, Switzerland Antibody: [A] to [D] Nucleocapsid Ag target: not stated Test method: [A] to [D] all LFAs, method not reported [E] Chemiluminescence assay Samples used: NP in VTM; collection not described Transport media: ESwab liquid preservation medium (Copan Italia, Brescia, Italy) or Vacuette Virus Stabilization (Greiner Bio‐One, Kremsmünster, Austria) tubes Sample storage: all tests performed within 24 h of collection; storage conditions not described Test operator: laboratory staff Definition of test positivity: appearance of 2 visible lines for all except VITRO which was a signal of ≥ 1 Blinding reported: yes, 2 independent operators with a 3rd blinded operator in case of disagreement. All operators were blinded to PCR results and clinical data Timing of samples: symptomatic: median 3 days pso (IQR 2‐4 days) |
||
| Target condition and reference standard(s) | Reference standard: PCR; LightCycler (Roche Diagnostics, Basel, Switzerland)) 480 Instrument II (Roche Diagnostics) using the LightMix (Roche Diagnostics) Modular SARS‐CoV
E‐gene set; Ct threshold appears to be ≤ 38 (range in Ct values reported was 12.6‐38.2) Definition of non‐COVID cases: unclear but appears single negative PCR for absence of infection Genetic target(s): E‐gene Samples used: same sample as for index test Timing of reference standard: same as index Blinded to index test: unclear; "All tests were performed within a maximum of 24 h after specimen collection", but order of tests was not reported Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same sample All participants received same reference standard: yes Missing data: none mentioned Uninterpretable results: none mentioned Indeterminate results (index test): none mentioned Indeterminate results (reference standard): none mentioned |
||
| Comparative | |||
| Notes | Funding: no external funding Publication status: published paper Source: academic journal Author COI: among the authors, JD is CEO and founder of QUALIblood s.a., a contract research organization manufacturing the DP‐Filter, is co‐inventor of the DP‐Filter (patent application number: PCT/ET2019/052903) and reports personal fees from Daiichi‐Sankyo, Gedeon Richter, Mithra Pharmaceuticals, Stago, Roche and Roche Diagnostics outside the submitted work. The other authors have no conflict of interest to disclose. |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Favresse 2021 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 5 Ag tests; Favresse 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 5 Ag tests; Favresse 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Comparative study of 5 Ag tests (no product codes reported); Favresse 2021 [B] relates to test [B] in the list below; see Favresse 2021 [A] for full study characteristics and QUADAS entries Test name: [A] Biotical SARS‐CoV‐2 Ag card [B] Panbio COVID‐19 Ag Rapid Test Device [C] Coronavirus Ag Rapid Test Cassette [D] Roche SARS‐CoV‐2 Rapid Antigen Test (Additional lab‐based Ag test also evaluated but not eligible for this review: [E] VITROS Immunodiagnostic Products SARS‐CoV‐2 Antigen test (Ortho Clinical Diagnostics, Raritan, NJ, USA)) Manufacturer: [A] Biotical Health, Madrid, Spain [B]Abbott, Chicago, IL, USA [C] Healgen Scientific, Houston, TX, USA [D] Roche Diagnostics, Basel, Switzerland Antibody: [A] to [D] Nucleocapsid Ag target: not stated Test method: [A] to [D] all LFAs, method not reported [E] Chemiluminescence assay Samples used: NP in VTM; collection not described Transport media: ESwab liquid preservation medium (Copan Italia, Brescia, Italy) or Vacuette Virus Stabilization (Greiner Bio‐One, Kremsmünster, Austria) tubes Sample storage: all tests performed within 24 h of collection; storage conditions not described Test operator: laboratory staff Definition of test positivity: appearance of 2 visible lines for all except VITRO which was a signal of ≥ 1 Blinding reported: yes, 2 independent operators with a 3rd blinded operator in case of disagreement. All operators were blinded to PCR results and clinical data Timing of samples: symptomatic: median 3 days pso (IQR 2‐4 days) |
||
| Target condition and reference standard(s) | Comparative study of 5 Ag tests; Favresse 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 5 Ag tests; Favresse 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | |||
Favresse 2021 [C].
| Study characteristics | |||
| Patient Sampling | Comparative study of 5 Ag tests; Favresse 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 5 Ag tests; Favresse 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Comparative study of 5 Ag tests (no product codes reported); Favresse 2021 [C] relates to test [C] in the list below; see Favresse 2021 [A] for full study characteristics and QUADAS entries Test name: [A] Biotical SARS‐CoV‐2 Ag card [B] Panbio COVID‐19 Ag Rapid Test Device [C] Coronavirus Ag Rapid Test Cassette [D] Roche SARS‐CoV‐2 Rapid Antigen Test (Additional lab‐based Ag test also evaluated but not eligible for this review: [E] VITROS Immunodiagnostic Products SARS‐CoV‐2 Antigen test (Ortho Clinical Diagnostics, Raritan, NJ, USA)) Manufacturer: [A] Biotical Health, Madrid, Spain [B] Abbott, Chicago, IL, USA [C] Healgen Scientific, Houston, TX, USA [D] Roche Diagnostics, Basel, Switzerland Antibody: [A] to [D] Nucleocapsid Ag target: not stated Test method: [A] to [D] all LFAs, method not reported [E] Chemiluminescence assay Samples used: NP in VTM; collection not described Transport media: ESwab liquid preservation medium (Copan Italia, Brescia, Italy) or Vacuette Virus Stabilization (Greiner Bio‐One, Kremsmünster, Austria) tubes Sample storage: all tests performed within 24 h of collection; storage conditions not described Test operator: laboratory staff Definition of test positivity: appearance of 2 visible lines for all except VITRO which was a signal of ≥ 1 Blinding reported: yes, 2 independent operators with a 3rd blinded operator in case of disagreement. All operators were blinded to PCR results and clinical data Timing of samples: symptomatic: median 3 days pso (IQR 2‐4 days) |
||
| Target condition and reference standard(s) | Comparative study of 5 Ag tests; Favresse 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 5 Ag tests; Favresse 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | |||
Favresse 2021 [D].
| Study characteristics | |||
| Patient Sampling | Comparative study of 5 Ag tests; Favresse 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 5 Ag tests; Favresse 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Comparative study of 5 Ag tests (no product codes reported); Favresse 2021 [D] relates to test [D] in the list below; see Favresse 2021 [A] for full study characteristics and QUADAS entries Test name: [A] Biotical SARS‐CoV‐2 Ag card [B] Panbio COVID‐19 Ag Rapid Test Device [C] Coronavirus Ag Rapid Test Cassette [D] Roche SARS‐CoV‐2 Rapid Antigen Test (Additional lab‐based Ag test also evaluated but not eligible for this review: [E] VITROS Immunodiagnostic Products SARS‐CoV‐2 Antigen test (Ortho Clinical Diagnostics, Raritan, NJ, USA)} Manufacturer: [A] Biotical Health, Madrid, Spain [B] Abbott, Chicago, IL, USA [C] Healgen Scientific, Houston, TX, USA [D] Roche Diagnostics, Basel, Switzerland Antibody: [A] to [D] Nucleocapsid Ag target: not stated Test method: [A] to [D] all LFAs, method not reported [E] Chemiluminescence assay Samples used: NP in VTM; collection not described Transport media: eSwab liquid preservation medium (Copan Italia, Brescia, Italy) or Vacuette Virus Stabilization (Greiner Bio‐One, Kremsmünster, Austria) tubes Sample storage: all tests performed within 24 h of collection; storage conditions not described Test operator: laboratory staff Definition of test positivity: appearance of 2 visible lines for all except VITRO which was a signal of ≥ 1 Blinding reported: yes, 2 independent operators with a 3rd blinded operator in case of disagreement. All operators were blinded to PCR results and clinical data Timing of samples: symptomatic: median 3 days pso (IQR 2‐4 days) |
||
| Target condition and reference standard(s) | Comparative study of 5 Ag tests; Favresse 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 5 Ag tests; Favresse 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | |||
Fenollar 2020(a).
| Study characteristics | |||
| Patient Sampling | 2 cohorts of patients presenting for COVID‐19 testing at the same institution. This extraction relates to:
[1] single‐group study to estimate sensitivity alone: symptomatic patients, all PCR positive (n = 182)
Fenollar 2020(b) reports data for [2] single‐group study to estimate both sensitivity and specificity: asymptomatic contacts of confirmed cases (n = 159) Recruitment: prospective Prospective or retrospective: unclear |
||
| Patient characteristics and setting | Setting: unclear; COVID‐19 testing Location: Institut Hospitalo‐universitaire Méditerranée Infection, Marseille Country: France Dates: 21 September‐2 October 2020 Symptoms and severity: not stated; all symptomatic Ct values for 154 participants: Ct ≤ 20: 58, 38%; Ct 21‐25: 49, 32%; Ct 26‐30: 39, 25%; Ct 31‐34: 8, 5% Demographics: not reported Exposure history: [1] not stated |
||
| Index tests | Test name: Panbio COVID‐19 Ag Manufacturer: Abbott Antibody: NP Ag target: not stated Test method: not stated Samples used: NP Transport media: not stated; appears to be direct testing Sample storage: tested within 1 h Test operator: not stated Definition of test positivity: visual line; as per manufacturer IFU Blinding reported: not stated, but presume yes as conducted within 1 h of collection Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: automated RT‐PCR; VitaPCR (Credo diagnostics, Singapore) Definition of non‐COVID cases: n/a Genetic target(s): not stated Samples used: NP (paired, from opposite nostril) Timing of reference standard: not stated Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired swabs All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported, no participant flow diagram reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "supported by the Méditerranée‐Infection Foundation and the French Agence Nationale de la Recherche under reference Investissements d’Avenir Méditerranée Infection 10‐IAHU‐03 and Région Provence‐Alpes‐Côte d’Azur and European funding FEDER IHUBIOTK." Source: accepted manuscript Author COI: "Pr Raoult and Pr Drancourt are co‐founders of the Pocrame startup that develops diagnostic devices for infectious diseases" |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Fenollar 2020(b).
| Study characteristics | |||
| Patient Sampling | 2 cohorts of patients presenting for COVID‐19 testing at the same institution. This extraction relates to:
[2] single‐group study to estimate both sensitivity and specificity: asymptomatic contacts of confirmed cases (n = 159)
See Fenollar 2020(a) for extraction of additional cohort: [1] single‐group study to estimate sensitivity alone: symptomatic patients, all PCR positive (n = 182) Recruitment: prospective Prospective or retrospective: unclear |
||
| Patient characteristics and setting | Setting: unclear Location: Institut Hospitalo‐universitaire Méditerranée Infection, Marseille Country: France Dates: 21 September‐2 October 2020 Symptoms and severity: all asymptomatic; 21/22 cases had Ct > 25 Demographics: not reported Exposure history: [2] all described as contacts |
||
| Index tests | Test name: PANBIO COVID‐19 Ag Manufacturer: Abbott Antibody: NP Ag target: not stated Test method: not stated Samples used: NP Transport media: not stated; appears to be direct testing Sample storage: tested within 1 h Test operator: not stated Definition of test positivity: visual line; as per manufacturer IFU Blinding reported: not stated, conducted first Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: automated RT‐PCR; VitaPCR (Credo diagnostics, Singapore) Definition of non‐COVID cases: as for cases; single negative Genetic target(s): not stated Samples used: NP (paired, from opposite nostril) Timing of reference standard: not stated Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired swabs All participants received same reference standard: yes Missing data: none reported, no participant flow diagram reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "supported by the Méditerranée‐Infection Foundation and the French Agence Nationale de la Recherche under reference Investissements d’Avenir Méditerranée Infection 10‐IAHU‐03 and Région Provence‐Alpes‐Côte d’Azur and European funding FEDER IHUBIOTK." Source: accepted manuscript Author COI: "Pr Raoult and Pr Drancourt are co‐founders of the Pocrame startup that develops diagnostic devices for infectious diseases" |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Ferguson 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity and specificity: university students attending asymptomatic student testing centre at the University of Birmingham Recruitment: consecutive inclusion of LFD‐positive samples; random sample of LFD‐negative (confirmatory PCR for 90 samples/d of testing) Prospective or retrospective: prospective Sample size (cases): 720 (8) |
||
| Patient characteristics and setting | Setting: mass screening Location: University of Birmingham Country: UK Dates: 2‐9 December 2020 Symptoms and severity: all asymptomatic Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: Innova Lateral Flow Device Manufacturer: Innova Medical group, a subsidiary of Xiamen Biotime Biotechnology company Antibody: SARS‐CoV‐2 nucleocapsid antigens Ag target: not stated Test method: CGIA Samples used: NP; supervised self‐collection States NP but process described involves swabbing both tonsils (so OP) and 1 nasal cavity = nasal+OP? Transport media: none used; immediately processed according to the Innova protocol Sample storage: no storage Test operator: not stated Trained postgraduate and final year undergraduate students in the College of Medical and Dental Science, supervised by highly experienced postdoctoral researchers (total of 18 test operatives) Definition of test positivity: not stated; visual appearance of lines Blinding reported: yes; conducted first Timing of samples: N/A; all asymptomatic, no clear epidemiological contact reported |
||
| Target condition and reference standard(s) | Reference standard: PCR; ThermoFisher Covid‐19 taqPath assay
Positivity threshold: 2 of 3 gene targets amplifying at a Ct value of ≤ 35 Definition of non‐COVID cases: as for cases; single negative Genetic target(s): not stated ORF1ab, N gene, S gene (Table 2) Samples used: residual NP swabs Timing of reference standard: same as for index test Blinded to index test: not stated Yes; "All samples were completely anonymous to the testing team with no identifying labels and were arbitrarily numbered from 1 to 90 each day" Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired swab; same swab "residual LFD test samples (buffer solution in which the NP swab is resuspended to perform the test)" All participants received same reference standard: yes Missing data: yes Uninterpretable results: none 4 invalid on Ag test; "Results of 4 samples were void (as defined by the manufacturer’s protocol [2])" Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "the PCR testing in this manuscript is funded by the UK Department for Health and Social Care (DHSC) as part of pillar 2 testing, in an award made directly to the University of Birmingham. The provision of LFD tests is funded by DHSC as part of a national student testing program, and funded directly to the University of Birmingham. DHSC have had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript." Publication status: published Source: PLOS Biology Author COI: no competing interests exist. |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Filgueiras 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity and specificity: patients with clinical features and suspected COVID‐19 seen by ED doctors; later described as "hospitalised" Recruitment: not clear; all who were tested before admission Prospective or retrospective: not stated; appears prospective Sample size (cases): 150 (55) |
||
| Patient characteristics and setting | Setting: inpatient; patients at the ED prior to admission to hospital Location: reference hospital in Belo Horizonte Country: Brazil Dates: June‐August 2020 Symptoms and severity: all symptomatic: 72 (48.0%) dyspnoea, 52 (34.7%) dry cough, 50 (33.3%) fever, 49 (32.7%) myalgia, 25 (16.7%) asthenia and 24 (16.0%) productive cough 127 (84.7%) had ≥ 2 associated symptoms; 15 (10%) had 5‐7 associated symptoms Of PCR+, 36 (65.5%) critically ill; 19/19 with typical findings of patchy ground‐glass shadows on CT presented bilateral pneumonia, compared to 5/11 (45.5%) of COVID‐19‐negative individuals with same alteration Demographics: median age 62 years (range 29‐91 years), 22 (40.0%) male Exposure history: not stated |
||
| Index tests | Test name: COVID‐19 Ag ECO Test Manufacturer: ECODiagnostica, Brazil Ag target: Nucleocapsid Test method: CGIA Samples used: NP swabs Transport media: not stated; appears that none used Sample storage: immediately tested Test operator: not stated Definition of test positivity: colorimetric reaction; interpreted after a 15‐min incubation Blinding reported: not stated, but very likely considering test was done prior to reference standard Timing of samples: at 1st day of symptom onset ≤ 3 days pso 63 (42%); 4‐7 days 59 (39%; 8‐15 days 22 (15%); > 15 days 2 (1%), not reported 4 (3%) |
||
| Target condition and reference standard(s) | Reference standard: PCR; Allplex 2019‐nCov Assay kit (Seegene Inc., Republic of Korea)
IFU states PCR inconclusive if only 1 or 2 of 3 targets detected (Ct < 40); re‐test recommended Definition of non‐COVID cases: as for cases; single negative Genetic target(s): Gene E, Gene RdRP and Gene N MS2 Samples used: NP swabs (paired); pre‐processed samples stored under refrigeration until RNA extraction Timing of reference standard: same as for index Blinded to index test: not stated; performed by the Minas Gerais Reference Center for the Diagnosis of COVID‐19 Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired swab All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none Indeterminate results (reference standard): none 11 samples with inconclusive results (only 1 Ag+ve); 5 on day 1 or 2 pso, 2 on day 5, 1 on day 8, 1 on day 15; and not reported for 2 samples 2/11 had bilateral multifocal ground‐glass opacities on CT, and 4 needed oxygen supply due to strong dyspnoea and desaturation; 1 (Ag positive) with odynophagia and dry cough. Not clear whether all were considered to have COVID‐19 Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "Oswaldo Cruz Foundation (to RFQG and scholarships to CC, RA, LC, NC), The Brazilian National Council for Scientific and Technological Development (CNPq) (scholarships to AO, DM, SG), Coordination for the Improvement of Higher Education Personnel (CAPES) (scholarships to NA, JA), The Minas Gerais Research Funding Foundation (FAPEMIG) (scholarship to PF). EcoDiagnostica for attending our request for donation of diagnostic kits under evaluation." Publication status: published Source: medRxiv Author COI: no COI statement |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
FIND 2020a.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: patients with symptoms consistent with COVID‐19 (meeting national definition for testing) presenting at a community testing clinic Recruitment: consecutive recruitment Prospective or retrospective: prospective |
||
| Patient characteristics and setting | Setting: community (COVID‐19 testing clinic) Location: institution not described; Marica, Rio de Janeiro Country: Brazil Dates: 30 July‐21 August 2020 Symptoms and severity: all symptomatic; no further details Demographics: mean age 40 years (range 4‐84); reported for 396 participants 181 (45%) male Exposure history: not stated |
||
| Index tests | Test name: NowCheck COVID‐19 Ag test (RG1901DG) Manufacturer: Bionote Inc Antibody: SARS‐CoV‐2 nucleocapsid antigen Ag target: mouse monoclonal SARS‐CoV‐2 antibodies Test method: rapid chromatographic immunoassay in lateral flow format Samples used: proprietary NP swab collected by HCW Transport media: no transport media. Sample is immediately transferred to proprietary tube containing extraction buffer. Sample storage: test should be performed as soon as possible after collection. Specimens may be stored at room temperature for 1 h or 2‐8 °C for 4 h. Test operator: HCW Definition of test positivity: presence of visible control and test lines Blinding reported: yes Timing of samples: median 4 days pso (IQR 3‐6 days); day < 0‐3, 152, 39% day 4‐7, 180, 46% day ≥ 8, 58, 15% |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR (in‐house assay based on the US CDC protocol); Ct threshold of 37 Definition of non‐COVID cases: same as for cases. Single negative PCR required for absence of infection Genetic target(s): N1, N2 Samples used: NP swabs Timing of reference standard: same timing as per NP swabs for index test Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: 0 to several days based on PCR turnaround times at the lab All participants received same reference standard: yes Missing data: reports 0 invalid results Uninterpretable results: none Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: FIND Publication status: published Source: FIND website/IFU index test Author COI: none stated (these are independent evaluations) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2020b (CH).
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity at 2 sites. This extraction is for data from Switzerland (see FIND 2020b (DE) for extraction of data for German site): patients seeking COVID‐19 testing at main testing centre; described as presenting either with symptoms compatible with a SARS‐CoV‐2 infection, or with a known positive contact or asymptomatic HCWs (n = 535) Recruitment: consecutive recruitment Prospective or retrospective: prospective |
||
| Patient characteristics and setting | Setting: community (main testing centre) Location: Hopitaux Universitaires de Geneve (HUG), Geneva Country: Switzerland Dates: 9‐16 October 2020 Symptoms and severity: 534/535 symptomatic (99%) Demographics: mean age 38.5 years (16‐85 years) 247, 46% male Exposure history: not stated |
||
| Index tests | Test name: Panbio Covid‐19 Ag Rapid Test (41FK10) Manufacturer: Abbott Antibody: not reported Ag target: not reported Test method: CGIA (from product insert) Samples used: NP Transport media: no transport media; assay buffer used Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: HCW Definition of test positivity: presence of visible control and test lines Blinding reported: yes Timing of samples: time pso recorded for 115/124, 92%. Day 0‐3 89, 78%; day 4‐7 23, 20%; day 8+ 3, 3% |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR Roche Cobas; Ct threshold < 40 (from Figure) Definition of non‐COVID cases: same as for cases. Single negative PCR required for absence of infection Genetic target(s): not stated Samples used: NP swab (paired, from contralateral nostril) Timing of reference standard: not stated; author contact advises only paired swabs used Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swabs; 0 to several days based on PCR turnaround times at the lab All participants received same reference standard: yes Missing data: reports 0 invalid Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: FIND Publication status: published Source: FIND/HUG website/IFU index test Author COI: none stated (these are independent evaluations) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2020b (DE).
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity at 2 sites; this extraction is for data from Germany (see FIND 2020b (CH) for data related to the site in Switzerland). Some information extracted from preprint reporting the same evaluations (see secondary reference under FIND 2020b (DE) for 2020 preprint by Kruger and colleagues): patients seeking COVID‐19 testing at main testing centre; described as presenting either with symptoms compatible with a SARS‐CoV2 infection, or with a known positive contact or asymptomatic HCWs (n = 1108) Recruitment: consecutive recruitment Prospective or retrospective: prospective Sample size (cases): 1108 (106) |
||
| Patient characteristics and setting | Setting: COVID‐19 test centre Location: [1] Heidelberg drive‐in testing; [2] Berlin: ambulatory testing clinic of Charité – University Hospital Country: Germany Dates: [1] Heidelberg: 28 September‐30 October 2020; [2] Berlin: 19 October‐30 October 2020 Symptoms and severity: 709/1100 symptomatic (64.5%). 2020 preprint by Kruger and colleagues reports 712 (65%) with symptoms on the day of testing (mean symptom duration 4.01 ± 3.1 (n = 687)); 396 (35%) with no symptoms Demographics: mean age 38.7 years (18‐86 years), 542, 49% male; 367 (33%) with comorbidities (see secondary reference under FIND 2020b (DE) for 2020 preprint by Kruger and colleagues) Exposure history: 388/858 (45.2%) Heidelberg participants were tested based on high risk contacts without symptoms (see secondary reference under FIND 2020b (DE) for 2020 preprint by Kruger and colleagues) |
||
| Index tests | Test name: Panbio Covid‐19 Ag (41FK10) Manufacturer: Abbott Rapid Diagnostics Antibody: not reported Ag target: not reported Test method: CGIA (from product insert) Samples used: NP or OP (if NP was contradicted) collected by trained study team; also referred to as laboratory personnel Transport media: no transport media; assay buffer used Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: trained laboratory team Definition of test positivity: presence of visible control and test lines Blinding reported: yes Timing of samples: time pso recorded for 692/709 symptomatic, 98% Day 0‐3 380, 55% Day 4‐7 230, 33% Day 8+ 82, 12% |
||
| Target condition and reference standard(s) | Reference standard: PCR; one of 5 assays:
Ct thresholds not stated; author contact advises Ct thresholds as per assay IFUs Definition of non‐COVID cases: same as for cases. Single negative PCR required for absence of infection Genetic target(s): not stated Samples used: NP swab (paired, from contralateral nostril) Timing of reference standard: as for index test; paired swabs used Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swabs; 0 to several days based on PCR turnaround times at the lab All participants received same reference standard: yes Missing data: none reported; no invalid tests (0%). 2020 preprint by Kruger and colleagues reports 10 withdrew consent for second swab and 1 had invalid PCR Uninterpretable results: not reported; invalid test results were repeated once with the remaining buffer solution in the test tubes. Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: FIND Publication status: published Source: FIND report Author COI: none stated (these are independent evaluations) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2020c (BR).
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity at 3 sites; this extraction is for data from Brazil (see FIND 2020c (CH) and FIND 2020c (DE) for extraction of data from other sites): ambulatory patients meeting national suspect definition for COVID‐19 testing presenting at a community testing clinic in Brazil Recruitment: consecutive recruitment Prospective or retrospective: prospective |
||
| Patient characteristics and setting | Setting: community testing clinic Location: Macae, state of Rio de Janeiro Country: Brazil Dates: 13‐30 July 2020 Symptoms and severity: 392/397 (99%) symptomatic; no further details Demographics: mean age 37 years (2‐94); 397 participants; 229/398 male (57%) Exposure history: not stated |
||
| Index tests | Test name: STANDARD Q COVID‐19 Ag (09COV30D) Manufacturer: SD Biosensor Inc Antibody: not reported Ag target: not reported Test method: rapid chromatographic immunoassay in lateral flow format Samples used: NP; collected by HCW Transport media: proprietary swab/media provided by SD Biosensor Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: HCW Definition of test positivity: presence of visible control and test lines Blinding reported: yes Timing of samples: median 5 days pso (IQR 4‐6 days) (for 397 participants); day < 0‐3 85, 21%; day 4‐7, 273, 69%; day ≥ 8, 39, 10% |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR (In‐house; Lab‐developed assay based on the US CDC protocol; Ct threshold not stated; author contact advises Ct thresholds as per assay IFUs Definition of non‐COVID cases: same as for cases. Single negative PCR required for absence of infection Genetic target(s): N1 and N2 Samples used: NP swabs Timing of reference standard: not stated; author contact advises only paired swabs used Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swabs; 0 to several days based on PCR turnaround times at the lab All participants received same reference standard: yes Missing data: reports 0 missing data Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: FIND Publication status: published Source: FIND website/IFU index test Author COI: none stated (these are independent evaluations) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2020c (CH).
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity at single site; this extraction is for data from Switzerland (see FIND 2020c (BR) and FIND 2020c (DE) for extraction of data from other sites): patients seeking COVID‐19 testing at main testing centre; described as presenting either with symptoms compatible with a SARS‐CoV2 infection, or with a known positive contact or asymptomatic HCWs (n = 529; from total cohort of 1064 volunteers) Recruitment: consecutive recruitment Prospective or retrospective: prospective |
||
| Patient characteristics and setting | Setting: community (main testing centre) Location: Hopitaux Universitaires de Geneve (HUG), Geneva Country: Switzerland Dates: 9‐23 October 2020 Symptoms and severity: not stated; time pso recorded for 183/191, 96% 141/183 COVID positive cases had symptoms for 0‐4 days (77%) Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: STANDARD Q COVID‐19 Ag (09COV30D) Manufacturer: SD Biosensor Inc Antibody: not reported Ag target: not reported Test method: rapid chromatographic immunoassay in lateral flow format Samples used: NP Transport media: proprietary swab/media provided by SD Biosensor Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: HCW Definition of test positivity: presence of visible control and test lines Blinding reported: yes Timing of samples: median not reported (range 0‐15); day < 0‐3, 122, 67%; day 4‐7, 54, 29%; day 8+; 7, 34% |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR Roche Cobas; Ct threshold < 40 (from Figure) Definition of non‐COVID cases: same as for cases. Single negative PCR required for absence of infection Genetic target(s): not stated Samples used: NP swab (paired, from contralateral nostril) Timing of reference standard: not stated; author contact advises only paired swabs used Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swabs; 0 to several days based on PCR turnaround times at the lab All participants received same reference standard: yes Missing data: reports 0 missing data Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: FIND Publication status: published Source: FIND/HUG websites/IFU index test Author COI: none stated (these are independent evaluations) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2020c (DE).
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity at 3 sites; this extraction is for data from Germany (see FIND 2020c (BR) and FIND 2020c (CH) for extraction of data from the other 2 sites evaluating this assay). Some information extracted from preprint reporting the same evaluations (see secondary reference under FIND 2020c (DE) for 2020 preprint by Kruger and colleagues): patients at risk for SARS‐CoV‐2 infection based on exposure to a confirmed case, suggestive symptoms, or travel to a high‐risk area, presenting either at a drive‐in testing station or a clinical ambulatory testing facility Recruitment: Consecutive, as per FIND evaluation protocol Prospective or retrospective: prospective |
||
| Patient characteristics and setting | Setting: COVID‐19 testing centres Location: (1) Heidelberg, Germany; (2) Berlin, Germany Country: Germany Dates: ( 1 ) Heidelberg: 20‐31 July 2020 ( 2 ) Berlin: 03 June ‐31 July 2020 Participants undergoing assay (b) Symptomatic on testing day: 10 40/1229, 84.6% Mean age (range): 35 (18‐80.4) based on 1244 participants Male (%): 606, 49.5% Exposure history: not stated |
||
| Index tests | Test name: STANDARD Q COVID‐19 Ag Test Manufacturer: SD Biosensor, Inc. Gyeonggi‐do, Korea Antibody: not stated Ag target: not stated Test method: CGIA Samples used: Heidelberg NP; Berlin combined NP and OP (OP conducted first) RT‐PCR swab obtained first, then same technique repeated for Ag test Transport media: none; used manufacturer supplied buffer solution as per IFU Sample storage: drive‐in centre and ambulatory testing: tested on site (presume short time frame) Secondary care: transported on ice to a category 3 facility for testing RT‐PCR swab obtained first, then same technique repeated for Ag test Test operator: drive‐in and ambulatory clinic: POC evaluation Secondary care: laboratory staff Definition of test positivity: visual appearance were interpreted by 2 operators, each blinded to the result of the other. In case of discrepant results, both operators re‐read the result and agreed on a final result. Invalid results were repeated once using the remaining buffer according to the respective IFUs. Readouts were done within the recommended time for each Ag‐RDT (10 min for Bioeasy, 15 min for Coris and 15‐30 min for SD Biosensor). Blinding reported: yes; "Staff performing the Ag‐RDTs were blinded to results of RT‐PCR tests and vice versa" Timing of samples: mean 3 days pso (IQR 2‐4 days) based on 1004 participants |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; varied by site
Samples that showed a signal above the threshold in the relevant RT‐PCR target regions for each assay were considered to be positive Definition of non‐COVID cases: as per cases; single negative result Genetic target(s): not stated Samples used: paired swabs; as per index test (RT‐PCR swab obtained first) Drive‐in centre: NP or OP Other centres: combined NOP (OP conducted first) Timing of reference standard: as per index test Blinded to index test: yes; "Staff performing the Ag‐RDTs were blinded to results of RT‐PCR tests and vice versa" Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired; simultaneous All participants received same reference standard: yes (different assays) Missing data: 2020 preprint by Kruger and colleagues reports total of 154 excluded from German sites (3 assays evaluated) following enrolment (116 2nd swab refused, 3 nose bleed after 1st swab, 3 insufficient time for both swabs, 31 other reasons, 1 no reason available) Uninterpretable results: 0 invalid reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: study was supported by FIND, Heidelberg University Hospital and Charité – University Hospital internal funds. Publication status: published Source: FIND report Author COI: no COI statement reported; "external funders of the study had no role in study design, data collection, or data analysis" |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2020d (BR).
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity at 2 sites; this extraction is for data from Brazil (see FIND 2020d (DE) for extraction of data from other site): adults in community meeting national suspect definition for COVID‐19 testing presenting at [1] a community testing clinic or [2] a tertiary‐level hospital Recruitment: consecutive recruitment Prospective or retrospective: prospective |
||
| Patient characteristics and setting | Setting: mixed; community testing clinic and tertiary hospital Location: [1] Macae, state of Rio de Janeiro, [2] Universidade Federal do Rio de Janeiro (UFRJ) Country: Brazil Dates: [1] 17 August‐9 September, [2] 11 July‐8 August 2020 Symptoms and severity: 421/450 (94%) symptomatic; no further details Demographics: mean age 39 years (0‐95 years); 451 participants; 185 male (41%) Exposure history: not stated |
||
| Index tests | Test name: STANDARD F COVID‐19 Ag FIA (F‐NCOV‐01G, 10COV30D) Manufacturer: SD Biosensor Inc Antibody: not reported Ag target: not reported Test method: FIA Samples used: NP; collected by HCW Transport media: proprietary swab/media provided by SD Biosensor Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: HCW Definition of test positivity: as per STANDARD F Analyzer; cut‐off index (COI) ≥ 1.0 (as per IFU) Blinding reported: yes Timing of samples: median 4 days pso (IQR 3‐6 days) for 421 participants. Day < 0‐3, 131, 31%; day 4‐7, 248, 59%; day ≥ 8, 42, 10% |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; 1 of 2 in‐house assays:
Ct thresholds not stated; author contact advises Ct thresholds as per assay IFUs Definition of non‐COVID cases: same as for cases. Single negative PCR required for absence of infection Genetic target(s):
Samples used: NP swabs Timing of reference standard: not stated; author contact advises only paired swabs used Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swabs; 0 to several days based on PCR turnaround times at the lab All participants received same reference standard: yes Missing data: reports 0 missing data Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: FIND Publication status: published Source: FIND website/IFU for index test Author COI: none stated (these are independent evaluations) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2020d (DE).
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity at 2 sites; this extraction is for data from Germany (see FIND 2020d (BR) for extraction of data from other site): adults in community meeting national suspect definition for COVID‐19 testing presenting at
[1] a drive‐in testing centre or
[2] ambulatory testing clinic Recruitment: consecutive recruitment Prospective or retrospective: prospective |
||
| Patient characteristics and setting | Setting: community Location: [1] Heidelberg, drive‐in testing; [2] Berlin, ambulatory testing clinic of Charité University Hospital Country: Germany Dates: [1] Heidelberg: 15 June‐18 July 2020; [2] Berlin: 6 July–23 September 2020 Symptoms and severity: 517/669 (77%) symptomatic; no further details Demographics: mean age 38 years (18‐85 years); 676 participants; 307 male (46%) Exposure history: not stated |
||
| Index tests | Test name: STANDARD F COVID‐19 Ag FIA (F‐NCOV‐01G, 10COV30D) Manufacturer: SD Biosensor Inc Antibody: not reported Ag target: not reported Test method: FIA Samples used: [1] NP; [2] combined NOP swabs; collected by HCW Transport media: proprietary swab/media provided by SD Biosensor Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: HCW Definition of test positivity: as per STANDARD F Analyzer; cut‐off index (COI) ≥ 1.0 (as per IFU) Blinding reported: yes Timing of samples: median 3 days pso (IQR 2‐5 days) for 505 participants. Day < 0‐3, 257, 51%; day 4‐7, 202, 47%; day ≥ 8, 46, 9% |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; one of 5 assays:
Ct thresholds not stated; author contact advises Ct thresholds as per assay IFUs Definition of non‐COVID cases: same as for cases. Single negative PCR required for absence of infection Genetic target(s): not stated apart from 3. E gene Samples used: NP (n = 305), NOP (n = 342) and/or OP swabs (n = 32) Timing of reference standard: not stated; author contact advises only paired swabs used Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swabs; 0 to several days based on PCR turnaround times at the lab All participants received same reference standard: yes Missing data: reports 0 missing data Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: FIND Publication status: published Source: FIND website/IFU for index test Author COI: none stated (these are independent evaluations) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2020e (BR).
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity; this extraction is for data from Brazil (see FIND 2020e (DE) for extraction of data from other site): adults in community meeting national suspect definition for COVID‐19 testing presenting at a community testing clinic (n = 476) Recruitment: consecutive recruitment Prospective or retrospective: prospective |
||
| Patient characteristics and setting | Setting: community testing clinic Location: Marica, state of Rio de Janeiro Country: Brazil Dates: 27 July‐16 September 2020 Symptoms and severity: 470/476 (99%) symptomatic; no further details Demographics: mean age 45 years (0‐106 years); 473 participants; 252 male (53%) Exposure history: not stated |
||
| Index tests | Test name: BIOCREDIT COVID‐19 Ag (G61RHA20 ‐ product evaluated is no longer distributed) Manufacturer: RapiGEN Inc Antibody: not reported Ag target: not reported Test method: LFA (CGIA, from IFU) Samples used: NP; collected by HCW Transport media: assay diluent provided by manufacturer Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: HCW Definition of test positivity: visual appearance of test and control lines Blinding reported: yes Timing of samples: median 5 days pso (IQR 4‐7 days) for 470 participants. Day < 0‐3, 95, 20%; day 4‐7, 296, 63%; day ≥ 8, 79, 17% |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; lab‐developed assay based on the US CDC protocol
Ct threshold not stated; author contact advises Ct thresholds as per assay IFUs Definition of non‐COVID cases: same as for cases. Single negative PCR required for absence of infection Genetic target(s): N1 and N2 Samples used: NP swabs Timing of reference standard: not stated; author contact advises only paired swabs used Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swabs; 0 to several days based on PCR turnaround times at the lab All participants received same reference standard: yes Missing data: reports 0 missing data Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: FIND Publication status: published Source: FIND website/IFU for index test Author COI: none stated (these are independent evaluations) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2020e (DE).
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity at 2 sites; this extraction is for data from Germany (see FIND 2020e (BR) for extraction of data from other site): adults in community meeting national suspect definition for COVID‐19 testing presenting at
[1] a drive‐in testing centre or
[2] ambulatory testing clinic Recruitment: consecutive recruitment Prospective or retrospective: prospective |
||
| Patient characteristics and setting | Setting: community Location: [1] Heidelberg, drive‐in testing; [2] Berlin, ambulatory testing clinic of Charité University Hospital Country: Germany Dates: [1] Heidelberg: 4 May‐3 September; [2] Berlin: 4 May‐18 August Symptoms and severity: 733/1223 symptomatic; no further details Demographics: mean age 39.5 years (17‐59.2 years) 1239 participants); 607 male (50%) Exposure history: not stated |
||
| Index tests | Test name: BIOCREDIT COVID‐19 Ag (G61RHA20 ‐ product evaluated is no longer distributed) Manufacturer: RapiGEN Inc Antibody: not reported Ag target: not reported Test method: LFA (CGIA, from IFU) Samples used: [1] NP; [2] NOP; collected by HCW Transport media: assay diluent provided by manufacturer Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: HCW Definition of test positivity: visual appearance of test and control lines Blinding reported: yes Timing of samples: median 3 days pso (IQR 2‐4days) for 701 participants. Day < 0‐3, 472, 67%; day 4‐7, 161, 23%; day ≥ 8, 68, 10% |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; one of 5 assays:
Ct thresholds not stated; author contact advises Ct thresholds as per assay IFUs Definition of non‐COVID cases: same as for cases. Single negative PCR required for absence of infection Genetic target(s): not stated Samples used: NP swabs Timing of reference standard: not stated; author contact advises only paired swabs used Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swabs; 0 to several days based on PCR turnaround times at the lab All participants received same reference standard: yes Missing data: reports 0 missing data Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: FIND Publication status: published Source: FIND website/IFU for index test Author COI: none stated (these are independent evaluations) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2020f.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity (other assays evaluated at the same sites in Germany are included as FIND 2021j and FIND 2020c (DE) ). Some information extracted from 2020 preprint by Kruger and colleagues reporting the same evaluations (see secondary reference under FIND 2020f).
Participants at risk for SARS‐CoV‐2 infection based on exposure to a confirmed case, suggestive symptoms, or travel to a high‐risk area, presenting either at (1) a drive‐in testing station or (2) a clinical ambulatory testing facility ("adults able to ambulate and meeting suspect definition of the Department of public health")
(3) secondary care facility in the UK (adults admitted and suspected to have COVID‐19 with following symptoms: fever ≥ 37.8C +/‐ shortness of breath +/‐ new persistent cough +/‐ loss of smell OR clinical or radiological evidence of pneumonia) Recruitment: consecutive, as per FIND evaluation protocol Prospective or retrospective: prospective |
||
| Patient characteristics and setting | Setting: mixed; COVID‐19 testing centres and secondary care Location: 3 sites: (1) Heidelberg, Germany; (2) Berlin, Germany and (3) Liverpool University Hospital Foundation Trust, Liverpool Country: (1), (2) Germany, (3) UK Dates: (1) HD: 11‐25 May; (2) Berlin: 10 Aug; 19‐25 Aug; (3) Liverpool: 12 May‐19 June Symptomatic on testing day: 290/419, 69.2% N with prior negative test result: 38/301, 12.6% Mean age (IQR): 43 years (18, 89 years) Male (%): 163/418, 39% |
||
| Index tests | Test name: COVID‐19 Ag Respi‐Strip Manufacturer: Coris Bioconcept, Gembloux, Belgium Antibody: not stated Ag target: not stated Test method: CGIA Samples used: drive‐in centre: NP, Other centres: combined NOP (OP conducted first) RT‐PCR swab obtained first, then same technique repeated for Ag test Transport media: none; used manufacturer supplied buffer solution as per IFU Sample storage: drive‐in centre and ambulatory testing: tested on site (presume short time frame) Secondary care: transported on ice to a category 3 facility for testing RT‐PCR swab obtained first, then same technique repeated for Ag test Test operator: drive‐in and ambulatory clinic: POC evaluation Secondary care: laboratory staff Definition of test positivity: visual appearance interpreted by 2 operators, each blinded to the result of the other. In case of discrepant results, both operators re‐read the result and agreed on a final result. Invalid results were repeated once using the remaining buffer according to the respective IFUs. Readouts were done within the recommended time for each Ag‐RDT (15 min for Coris) Blinding reported: yes; "Staff performing the Ag‐RDTs were blinded to results of RT‐PCR tests and vice versa" Timing of samples: 3 days (IQR 1‐5 days); based on 282 participants |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; varied by site
Samples that showed a signal above the threshold in the relevant RT‐PCR target regions for each assay were considered to be positive Definition of non‐COVID cases: as per cases; single negative result Genetic target(s): not stated Samples used: paired swabs, combined NOP; as per index test (RT‐PCR swab obtained first) Timing of reference standard: as per index test Blinded to index test: yes; "Staff performing the Ag‐RDTs were blinded to results of RT‐PCR tests and vice versa" Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired; simultaneous All participants received same reference standard: yes (different assays) Missing data: 2020 preprint by Kruger and colleagues (FIND 2020f) reports total of 154 excluded from German sites (3 assays evaluated) following enrolment (116 2nd swab refused, 3 nose bleed after 1st swab, 3 insufficient time for both swabs, 31 other reasons, 1 no reason available) Uninterpretable results: 8 invalid (PCR negative) Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Study reports an ease‐of‐use assessment; for this assay:
Funding: study was supported by FIND, Heidelberg University Hospital and Charité – University Hospital internal funds. Pfizer funded the clinical team in Liverpool, UK Publication status: preprint Source: medRxiv Author COI: no COI statement reported; "external funders of the study had no role in study design, data collection, or data analysis" |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
FIND 2021a [A].
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: patients with symptoms consistent with COVID‐19 (meeting national definition for testing) presenting at a community testing clinic Recruitment: consecutive recruitment Prospective or retrospective: prospective Sample size (cases): 218 (79) |
||
| Patient characteristics and setting | Setting: community (COVID‐19 testing clinic) Location: institution not described; Marica and Guapimirim, state of Rio de Janeiro Country: Brazil Dates: 21‐27 January 2021; 23‐26 February 2021 Symptoms and severity: all symptomatic; no further details Demographics: mean age 42.3 years (range 18‐90); 92 (42%) male Exposure history: not stated |
||
| Index tests | Test name: [A] NowCheck COVID‐19 Ag test (RG1901DGN (Nasal))
[B] NowCheck COVID‐19 Ag test (RG1901DG (NP)) Manufacturer: Bionote Inc Antibody: not reported Ag target: not reported Test method: rapid chromatographic immunoassay in lateral flow format Samples used: [A] NMT [B] NP swab collected by HCW Transport media: no transport media. Sample is immediately transferred to proprietary tube containing extraction buffer. Sample storage: immediate testing Test operator: HCW Definition of test positivity: presence of visible control and test lines Blinding reported: yes Timing of samples: median 4 days pso (IQR 3‐6 days); day < 0‐3, 72, 33% day 4 ‐7, 123, 56% day ≥ 8, 23, 11% |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR (in‐house assay based on the US CDC protocol); Ct threshold of 37 Definition of non‐COVID cases: same as for cases. Single negative PCR required for absence of infection Genetic target(s): N1, N2 Samples used: NP swabs Timing of reference standard: same timing as for index test Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous, paired swabs All participants received same reference standard: yes Missing data: reports 0 invalid results for both assays Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: FIND Publication status: published Source: FIND website/IFU index test Author COI: none stated (these are independent evaluations) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2021a [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of an Ag test on 2 different samples; FIND 2021a [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of an Ag test on 2 different samples; FIND 2021a [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] NowCheck COVID‐19 Ag test (RG1901DGN (Nasal))
[B] NowCheck COVID‐19 Ag test (RG1901DG (NP)) Manufacturer: Bionote Inc Antibody: not reported Ag target: not reported Test method: rapid chromatographic immunoassay in lateral flow format Samples used: [A] NMT [B] NP swab collected by HCW Transport media: no transport media. Sample is immediately transferred to proprietary tube containing extraction buffer. Sample storage: immediate testing Test operator: HCW Definition of test positivity: presence of visible control and test lines Blinding reported: yes Timing of samples: median 4 days pso (IQR 3‐6 days); day < 0‐3, 72, 33% day 4‐7, 123, 56% day ≥ 8, 23, 11% |
||
| Target condition and reference standard(s) | Comparative study of an Ag test on 2 different samples; FIND 2021a [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of an Ag test on 2 different samples; FIND 2021a [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of an Ag test on 2 different samples; FIND 2021a [A] reports full study characteristics and QUADAS. | ||
FIND 2021b [A].
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity at single site: patients seeking COVID‐19 testing at COVID‐19 testing centre; described as able to ambulate, at high risk for SARS‐CoV‐2 according to clinical suspicion, and meeting suspect definition of the Department of Public Health (n = 281) Recruitment: consecutive recruitment Prospective or retrospective: prospective Sample size (cases): 281 (44) |
||
| Patient characteristics and setting | Setting: community Location: Heidelberg drive‐in testing Country: Germany Dates: 15 December 2020‐19 January 2021 Symptoms and severity: 130/279 symptomatic (46%) Demographics: mean age 42.9 years (range 18‐81 years) 134, 48% male Exposure history: not stated |
||
| Index tests | Test name: [A] PanbioTM Covid‐19 Ag Rapid Test Device Nasal (41FK11)
[B] Panbio COVID‐19 Ag Rapid Test (41FK10) Manufacturer: Abbott Antibody: not reported Ag target: not reported Test method: CGIA (from product insert) Samples used: [A] NMT [B] NP Transport media: no transport media; assay buffer used Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: HCW Definition of test positivity: presence of visible control and test lines Blinding reported: yes Timing of samples: time pso recorded for 126/281, 45% Day 0‐3 86, 68% Day 4‐7 290, 23% Day 8+ 11, 9% |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; one of 3 assays:
Ct thresholds not stated; author contact advises Ct thresholds as per assay IFUs Definition of non‐COVID cases: same as for cases. Single negative PCR required for absence of infection Genetic target(s): not stated Samples used: NP swab Timing of reference standard: not stated; author contact advises only paired swabs used. Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swabs; 0 to several days based on PCR turnaround times at the lab All participants received same reference standard: yes Missing data: none reported; 0 invalid results Uninterpretable results: none Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: FIND Publication status: published Source: FIND website/IFU for index test Author COI: none stated (these are independent evaluations) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2021b [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of an Ag test on 2 different samples; FIND 2021b [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | |||
| Index tests | Test name: [A] PanbioTM Covid‐19 Ag Rapid Test Device Nasal (41FK11)
[B] Panbio COVID‐19 Ag Rapid Test (41FK10) Manufacturer: Abbott Antibody: not reported Ag target: not reported Test method: CGIA (from product insert) Samples used: [A] NMT [B] NP Transport media: no transport media; assay buffer used Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: HCW Definition of test positivity: presence of visible control and test lines Blinding reported: yes Timing of samples: time pso recorded for 126/281, 45% Day 0‐3, 86, 68% Day 4‐7, 290, 23% Day 8+, 11, 9% |
||
| Target condition and reference standard(s) | Comparative study of an Ag test on 2 different samples; FIND 2021b [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of an Ag test on 2 different samples; FIND 2021b [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of an Ag test on 2 different samples; FIND 2021b [A] reports full study characteristics and QUADAS. | ||
FIND 2021c (BR) [A].
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity at 2 sites; (see FIND 2021c (DE) [A] for additional site data): ambulatory patients meeting national suspect definition for COVID‐19 testing presenting at a community testing clinic in Brazil Recruitment: consecutive recruitment Prospective or retrospective: prospective Sample size (cases): 214 (78) |
||
| Patient characteristics and setting | Setting: community testing clinic Location: Macae and Guapimirim, state of Rio de Janeiro Country: Brazil Dates: 14‐20 January 2021; 2‐4 March 2021 Symptoms and severity: all symptomatic; no further details Demographics: mean age 41.3 years (range 18‐77 years) ; 85/214 male (40%) Exposure history: not stated |
||
| Index tests | Test name: [A] STANDARD Q COVID‐19 Ag Nasal (09COV31D)
[B] STANDARD Q COVID‐19 Ag (09COV30D) Manufacturer: SD Biosensor Inc Antibody: not reported Ag target: not reported Test method: rapid chromatographic immunoassay in lateral flow format Samples used: [A] NTM [B] NP; collected by HCW Transport media: proprietary swab/media provided by SD Biosensor Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: HCW Definition of test positivity: presence of visible control and test lines Blinding reported: yes Timing of samples: median 5 days pso (IQR 3‐6.75 days); day < 0‐3, 68, 32% day 4‐7, 116, 54% day ≥ 8, 30, 14% |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR (in‐house; lab‐developed assay based on the US CDC protocol; Ct threshold not stated; author contact advises Ct thresholds as per assay IFUs Definition of non‐COVID cases: same as for cases. Single negative PCR required for absence of infection Genetic target(s): N1 and N2 Samples used: NP swabs Timing of reference standard: not stated; author contact advises only paired swabs used Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swabs All participants received same reference standard: yes Missing data: reports 0 missing data; 0 invalid results Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: FIND Publication status: published Source: FIND website/IFU index test Author COI: none stated (these are independent evaluations) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2021c (BR) [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of an Ag test on 2 different samples; FIND 2021c (BR) [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of an Ag test on 2 different samples; FIND 2021c (BR) [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] STANDARD Q COVID‐19 Ag Nasal (09COV31D)
[B] STANDARD Q COVID‐19 Ag (09COV30D) Manufacturer: SD Biosensor Inc Antibody: not reported Ag target: not reported Test method: rapid chromatographic immunoassay in lateral flow format Samples used: [A] NTM [B] NP collected by HCW Transport media: proprietary swab/media provided by SD Biosensor Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: HCW Definition of test positivity: presence of visible control and test lines Blinding reported: yes Timing of samples: median 5 days pso (IQR 3‐6.75 days): day < 0‐3, 68, 32% day 4‐7, 116, 54% day ≥ 8, 30, 14% |
||
| Target condition and reference standard(s) | Comparative study of an Ag test on 2 different samples; FIND 2021c (BR) [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of an Ag test on 2 different samples; FIND 2021c (BR) [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of an Ag test on 2 different samples; FIND 2021c (BR) [A] reports full study characteristics and QUADAS. | ||
FIND 2021c (DE) [A].
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity at 2 sites; (see FIND 2021c (BR) [A] for additional site data): adults able to ambulate, at high risk for SARS‐CoV‐2 according to clinical suspicion, and meeting suspect definition of the Department of Public Health Recruitment: consecutive recruitment; continued until 30 positive NP swab samples according to Ag‐RDT were obtained Prospective or retrospective: prospective Sample size (cases): 179 (41) |
||
| Patient characteristics and setting | Setting: COVID‐19 test centre Location: ambulatory testing clinic of Charité – University Hospital Country: Germany Dates: 11‐18 November 2020 Symptoms and severity: on day of testing: 172 (96%) symptomatic; 7 (4%) asymptomatic; average symptom duration 4.2 ± 2.6 days Demographics: average age 36.2 ± 12.2 years; 48% female; 14% with comorbidities Exposure history: not stated |
||
| Index tests | Test name: [A] STANDARD Q COVID‐19 Ag Nasal (09COV31D)
[B] STANDARD Q COVID‐19 Ag (09COV30D) Manufacturer: SD Biosensor Inc Antibody: not reported Ag target: not reported Test method: rapid chromatographic immunoassay in lateral flow format Samples used: [A] NMT (both sides of nose); [B] NP (single side of nose); Both HCW collected; states 'professional' (secondary paper for FIND 2021c (DE) [A] by Lindner and colleagues describes study physicians collecting swabs). Transport media: proprietary swab/media provided by SD Biosensor Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: HCW Definition of test positivity: presence of visible control and test lines Blinding reported: yes Timing of samples: median 4 days pso (IQR 3‐5 days) for 397 patients: day < 0‐3, 79, 46% day 4‐7, 76, 44% day ≥ 8, 18, 10% |
||
| Target condition and reference standard(s) | Reference standard: PCR; 1. Cobas SARS‐CoV‐2 (Roche Diagnostics Inc); n = 158. 2. LightMix Modular SARS‐CoV (COVID19) E‐gene (Tib Molbiol) n = 21
Ct threshold not stated; author contact advises Ct thresholds as per assay IFUs Definition of non‐COVID cases: same as for cases. Single negative PCR required for absence of infection Genetic target(s): not stated Samples used: NP/OP swab; opposite nostril Timing of reference standard: not stated; author contact advises only paired swabs used Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swabs All participants received same reference standard: yes Missing data: 1 patient was excluded as both swabs for the Ag could not be obtained Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: FIND Publication status: published Source: FIND website/IFU index test Author COI: none stated (these are independent evaluations) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2021c (DE) [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 2 versions of an Ag test; FIND 2021c (DE) [A] details full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 2 versions of an Ag test; FIND 2021c (DE) [A] details full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] STANDARD Q COVID‐19 Ag Nasal (09COV31D)
[B] STANDARD Q COVID‐19 Ag (09COV30D) Manufacturer: SD Biosensor Inc Antibody: not reported Ag target: not reported Test method: rapid chromatographic immunoassay in lateral flow format Samples used: [A] AN [B] NP collected by HCW Transport media: proprietary swab/media provided by SD Biosensor Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: HCW Definition of test positivity: presence of visible control and test lines Blinding reported: yes Timing of samples: median 4 days pso (IQR 3‐5 days) for 397 participants: day < 0‐3, 79, 46% day 4‐7, 76, 44% day ≥ 8, 18, 10% |
||
| Target condition and reference standard(s) | Comparative study of 2 versions of an Ag test; FIND 2021c (DE) [A] details full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 2 versions of an Ag test; FIND 2021c (DE) [A] details full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 2 versions of an Ag test; FIND 2021c (DE) [A] details full study characteristics and QUADAS. | ||
FIND 2021d.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: adults able to ambulate, at high risk for SARS‐CoV‐2 according to clinical suspicion, and meeting suspect definition of the Department of Public Health and presenting at [1] a drive‐in testing centre or [2] ambulatory testing clinic Recruitment: consecutive recruitment Prospective or retrospective: prospective Sample size (cases): 214 (78) |
||
| Patient characteristics and setting | Setting: community Location: [1] Heidelberg drive in testing; [2] Berlin: ambulatory testing clinic of Charité – University Hospital Country: Germany Dates: [1] Heidelberg: 20 January–19 February 2021; [2] Berlin: 18 January‐22 February 2021 Symptoms and severity: symptomatic 62% (446/718); no further details Demographics: mean age 39.4 years (range 18‐80 years); 348/719 male (48%) Exposure history: not stated |
||
| Index tests | Test name: Espline SARS‐CoV‐2 (231906) Manufacturer: Fujirebio Inc. Antibody: not reported Ag target: not reported Test method: rapid chromatographic immunoassay in lateral flow format Samples used: NP (OP only if NP contraindicated); collected by HCW using iAMP‐COVID19‐SCD Transport media: none Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: HCW Definition of test positivity: presence of visible control and test lines Blinding reported: yes Timing of samples: median 2 days pso (IQR 1‐4 days): day < 0‐3, 311, 70% day 4‐7, 106, 24% day ≥ 8, 27, 6% |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; one of 3 assays:
Ct thresholds not stated; author contact advises Ct thresholds as per assay IFUs Definition of non‐COVID cases: same as for cases. Single negative PCR required for absence of infection Genetic target(s): only stated for 3. (E‐gene) Samples used: NP swabs (or OP if used for Ag test) Timing of reference standard: not stated; author contact advises only paired swabs used Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swabs All participants received same reference standard: yes Missing data: reports 0 missing data; 0 invalid results Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: FIND Publication status: published Source: FIND website/IFU index test Author COI: none stated (these are independent evaluations) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2021e.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: adults in community meeting Department of Public Health definition of a suspected COVID‐19 case and being tested for SARS‐CoV‐2 part of routine medical care Recruitment: consecutive recruitment Prospective or retrospective: prospective Sample size (cases): 265 (44) |
||
| Patient characteristics and setting | Setting: COVID‐19 test centre Location: Hopitaux Universitaires de Geneve (HUG), Geneva Country: Switzerland Dates: 4‐13 January 2021 Symptoms and severity: only reported for PCR+ group; symptomatic 88.6% (39/44) Demographics: mean age 36.3 years (range 16‐80 years); 139/265 male (52%) Exposure history: not stated |
||
| Index tests | Test name: SARS‐CoV‐2 Antigen Rapid Test Kit (Colloidal Gold) (COV‐AG‐20/G10313) Manufacturer: Joysbio (Tianjin) Biotechnology Co Antibody: not reported Ag target: not reported Test method: rapid chromatographic immunoassay in lateral flow format Samples used: AN; collected by HCW Transport media: none Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: HCW Definition of test positivity: presence of visible control and test lines Blinding reported: yes Timing of samples: only reported for PCR+ cases, median 2 days pso (IQR 1‐3.5 days): day < 0‐3, 23, 74% day 4‐7, 8, 26% day ≥ 8, 0, 0% |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; one of 3 assays:
Ct thresholds not stated; author contact advises Ct thresholds as per assay IFUs Definition of non‐COVID cases: same as for cases. Single negative PCR required for absence of infection Genetic target(s): not stated Samples used: NP swabs Timing of reference standard: as for index test Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swabs All participants received same reference standard: yes Missing data: reports 0 missing data; 0 invalid results Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: FIND Publication status: published Source: FIND website/IFU index test Author COI: none stated (these are independent evaluations) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2021f.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: adults able to ambulate, at high risk for SARS‐CoV‐2 according to clinical suspicion, and meeting suspect definition of the Department of Public Health, presenting either at:
Recruitment: consecutive recruitment Prospective or retrospective: prospective Sample size (cases): 665 (194) |
||
| Patient characteristics and setting | Setting: COVID‐19 test centre Location: 1. Heidelberg: drive‐in testing centre; 2. Berlin: ambulatory testing clinic of Charité – University Hospital Country: Germany Dates: 1. Heidelberg: 11‐31 March 2021; 2. Berlin: 11 March–15 April 2021 Symptoms and severity: symptomatic: 66.5%, (440/662) Demographics: mean age 38.7 years (range 18‐78 years); 331/664 male (50%) Exposure history: not stated |
||
| Index tests | Test name: COVID 19 RAPID ANTIGEN TEST (11811125) Manufacturer: Mologic Ltd Antibody: not reported Ag target: not reported Test method: rapid chromatographic immunoassay in lateral flow format Samples used: nasal (AN) (n = 645) or NMT (n = 20); collected by HCW Transport media: none Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: HCW Definition of test positivity: presence of visible control and test lines Blinding reported: yes Timing of samples: median 2 days pso (IQR 1‐4 days); n = 436 day < 0‐3, 290, 67% day 4‐7, 121, 28% day ≥ 8, 25, 6% |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; one of 2 assays:
Ct thresholds not stated; author contact advises Ct thresholds as per assay IFUs Definition of non‐COVID cases: same as for cases. Single negative PCR required for absence of infection Genetic target(s): not stated Samples used: HD: NP swabs (oropharyngeal if NP contraindicated) Berlin: combined NP/oropharyngeal swabs Timing of reference standard: as for index test Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swabs All participants received same reference standard: yes Missing data: yes Uninterpretable results: 16/665, 2.4% invalid Ag results (including 3 PCR+ and 13 PCR− samples) Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: FIND Publication status: published Source: FIND website/IFU index test Author COI: none stated (these are independent evaluations) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2021g.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: individuals (age 16+) in community meeting Department of Public Health definition of a suspected COVID‐19 case and being tested for SARS‐CoV‐2 part of routine medical care Recruitment: consecutive recruitment Prospective or retrospective: prospective Sample size (cases): 462 (69) |
||
| Patient characteristics and setting | Setting: COVID‐19 test centre Location: University Hospital of Geneva Country: Switzerland Dates: 24 November 2020‐20 January 2021 Symptoms and severity: reported for PCR+ only; symptomatic 94.2% (65/69) Demographics: mean age 38.7 years (range 16‐82 years); 206/462 male (45%) Exposure history: not stated |
||
| Index tests | Test name: COVID‐19 Ag Rapid Test (243103N‐20) Manufacturer: NADAL Antibody: not reported Ag target: not reported Test method: rapid chromatographic immunoassay in lateral flow format Samples used: NP swab; collected by HCW Transport media: none Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: HCW Definition of test positivity: presence of visible control and test lines Blinding reported: yes Timing of samples: only reported for 54/62 PCR+ patients Median 2 days pso (IQR 1‐3 days) day < 0‐3, 45, 83% day 4‐7, 7, 13% day ≥ 8, 2, 4% |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; one of 3 assays:
Ct thresholds not stated; author contact advises Ct thresholds as per assay IFUs Definition of non‐COVID cases: same as for cases. Single negative PCR required for absence of infection Genetic target(s): not stated Samples used: NP swab Timing of reference standard: as for index test Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swabs All participants received same reference standard: yes Missing data: none; reports 0 invalid Uninterpretable results: none Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: FIND Publication status: published Source: FIND website/IFU index test Author COI: none stated (these are independent evaluations) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2021h.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: individuals (age 16+) in community meeting Department of Public Health definition of a suspected COVID‐19 case and being tested for SARS‐CoV‐2 part of routine medical care Recruitment: consecutive recruitment Prospective or retrospective: prospective Sample size (cases): 232 (41) |
||
| Patient characteristics and setting | Setting: COVID‐19 test centre Location: University Hospital of Geneva Country: Switzerland Dates: 21‐29 January 2021 Symptoms and severity: reported for PCR+ only; symptomatic 92.7% (38/41) Demographics: mean age 36.3 years (range 16‐76 years); 103/232 male (44%) Exposure history: not stated |
||
| Index tests | Test name: iChroma COVID‐19 Ag Test (CFPC‐115) Manufacturer: Boditech Medical, Inc. Antibody: not reported Ag target: not reported Test method: rapid chromatographic immunoassay in lateral flow format Samples used: NP swab; collected by HCW Transport media: none Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: HCW Definition of test positivity: presence of visible control and test lines Blinding reported: yes Timing of samples: only reported for 32 PCR+ patients Median 1.5 days pso (IQR 1‐3 days) day < 0‐3, 26, 81% day 4‐7, 5, 16% day ≥ 8, 1, 3% |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; one of 2 assays:
Ct thresholds not stated; author contact advises Ct thresholds as per assay IFUs Definition of non‐COVID cases: same as for cases. Single negative PCR required for absence of infection Genetic target(s): not stated Samples used: NP swab Timing of reference standard: as for index test Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swabs All participants received same reference standard: yes Missing data: none; reports 0 invalid Uninterpretable results: none Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: FIND Publication status: published Source: FIND website/IFU index test Author COI: none stated (these are independent evaluations) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2021i.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: individuals (age 16+) in community meeting Department of Public Health definition of a suspected COVID‐19 case and being tested for SARS‐CoV‐2 part of routine medical care Recruitment: consecutive recruitment Prospective or retrospective: prospective Sample size (cases): 328 (56) |
||
| Patient characteristics and setting | Setting: COVID‐19 test centre Location: University Hospital of Geneva Country: Switzerland Dates: 3‐11 December 2020 Symptoms and severity: reported for PCR+ only; symptomatic 100% (56/56) Demographics: mean age 37.9 years (range 16‐76 years); 129/327 male (39%) Exposure history: not stated |
||
| Index tests | Test name: Wondfo 2019‐nCoV Antigen Test (W196P0003) Manufacturer: Guangzhou Wondfo Biotech Co. Antibody: not reported Ag target: not reported Test method: rapid chromatographic immunoassay in lateral flow format Samples used: NP swab; collected by HCW Transport media: none Sample storage: author contact advises tested as soon as possible and within the time limit specified in the IFU Test operator: HCW Definition of test positivity: presence of visible control and test lines Blinding reported: yes Timing of samples: only reported for 44 PCR+ patients Median 2 days pso (IQR 1‐4 days) day < 0‐3, 31, 70% day 4‐7, 11, 25% day ≥ 8, 2, 5% |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; one of 3 assays:
Ct thresholds not stated; author contact advises Ct thresholds as per assay IFUs Definition of non‐COVID cases: same as for cases. Single negative PCR required for absence of infection Genetic target(s): not stated Samples used: NP swab Timing of reference standard: as for index test Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swabs All participants received same reference standard: yes Missing data: none; reports 0 invalid Uninterpretable results: none Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: FIND Publication status: published Source: FIND website/IFU index test Author COI: none stated (these are independent evaluations) |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
FIND 2021j.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity (other assays evaluated at the same sites in Germany are included as FIND 2020f and FIND 2020c (DE)). Some information extracted from 2020 preprint by Kruger and colleagues reporting the same evaluations (see secondary reference under FIND 2021j). Participants at risk for SARS‐CoV‐2 infection based on exposure to a confirmed case, suggestive symptoms, or travel to a high‐risk area, presenting either at (1) a drive‐in testing station or (2) a clinical ambulatory testing facility ("adults able to ambulate and meeting suspect definition of the Department of public health") Recruitment: not stated; recorded as consecutive, as per FIND evaluation protocol Prospective or retrospective: prospective Sample size (cases): 729 (15) |
||
| Patient characteristics and setting | Setting: mixed; (1), (2) Community (drive‐in or clinical ambulatory testing) Location: 2 sites: (1) Heidelberg, Germany; (2) Berlin, Germany Country: (1), (2) Germany Dates: (1) 14 April‐3 May 2020; (2) 14 May‐3 Jun 2020 Symptomatic on testing day: 563/654, 86.1% Mean age (SD): 40 (18.1‐92.0); n = 728 Male (%): 47.2% (369/699) |
||
| Index tests | Test name: Bioeasy 2019‐nCoV Ag Fluorescence Rapid Test Kit (Time‐Resolved Fluorescence) Manufacturer: Shenzhen Bioeasy Biotechnology Co. Ltd., Guangdong Province, China Antibody: not stated Ag target: not stated Test method: FIA Samples used: (1) NP (or OP if NP contraindicated); (2) combined NOP (OP conducted first) PCR swab obtained first, then same technique repeated for Ag test Transport media: none; used manufacturer supplied buffer solution as per IFU (for the Bioeasy assay, "the developer requested for pipettes to be used to transfer adequate quantities of liquid; in the IFU no pipette is needed and a nozzle is provided"). Sample storage: tested on site (presume short time frame) PCR swab obtained first, then same technique repeated for Ag test. Test operator: drive‐in and ambulatory clinic: POC evaluation Definition of test positivity: as per Analyzer Invalid results were repeated once using the remaining buffer according to the respective IFUs. Readouts were done within the recommended time: 10 min for Bioeasy Blinding reported: yes; "Staff performing the Ag‐RDTs were blinded to results of PCR tests and vice versa" Timing of samples: median 3 days pso (IQR 2‐6 days); n = 540; < 3 days, 303 (56%), 4‐7 days, 132 (24%), day 8+, 105 (19%) |
||
| Target condition and reference standard(s) | Reference standard: PCR; varied by site
Samples that showed a signal above the threshold in the relevant PCR target regions for each assay were considered to be positive Definition of non‐COVID cases: as per cases; single negative result Genetic target(s): not stated Samples used: paired swabs; as per index test (PCR swab obtained first); (1) NP or OP, (2) combined NOP (OP conducted first) Timing of reference standard: as per index test Blinded to index test: yes; "Staff performing the Ag‐RDTs were blinded to results of PCR tests and vice versa" Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired; simultaneous All participants received same reference standard: yes (different assays) Missing data: Yes uninterpretable excluded Uninterpretable results: 2 invalid excluded Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Study reports an ease‐of‐use assessment; for this assay:
Funding: study was supported by FIND, Heidelberg University Hospital and Charité – University Hospital internal funds. Publication status: Published Source: FIND report Author COI: no COI statement reported; "external funders of the study had no role in study design, data collection, or data analysis" |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Fourati 2020 [A].
| Study characteristics | |||
| Patient Sampling | 2‐group study to estimate sensitivity and specificity:
Recruitment: random (stratified by Ct and time pso) Prospective or retrospective: retrospective |
||
| Patient characteristics and setting | Setting: mixed; likely outpatient and inpatient "consulted or were admitted" Location: Henri Hospital Mondor de Créteil Country: France Dates: 9 March‐9 April 2020 Symptoms and severity: not stated; all apparently symptomatic Data by viral load reported for 293/297 cases: ≤ 20 Ct 39, 13%; 2‐25 Ct 88, 30%; 25‐30 Ct 72, 25%; > 30 Ct 88, 30% Demographics: not stated Exposure history: not stated |
||
| Index tests | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [A] data relate to test [A], see additional entries for tests [B] to [E] [A] SARS‐CoV‐2 COVID‐19 Respi‐Strip [B] STANDARD Q COVID‐19 Ag [C] PanBio COVID‐19 Antigen Rapid Test [D] Biosynex COVID‐19 Ag BSS [E] COVID‐VIRO Antigen Rapid Test [F] NG Test SARS‐CoV‐2 Ag (assay excluded from review due to Vortex requirement as stated in IFU) (no product codes reported) Manufacturer: [A] Coris BioConcept, Gembloux, Belgium [B] SD BIOSENSOR, Inc., Korea [C] Abbott, Chicago, Illinois, USA [D] Biosynex, Strasbourg, France [E] AAZ, Boulogne‐Billancourt, France [F] NG Biotech, Guipry, France Antibody: not stated Ag target: not stated Test method: not stated Samples used: NP; collection not reported Transport media: VTM (Cepheid or Deltalab); 100 μL used for testing Sample storage: frozen at −80 °C until use Test operator: laboratory staff Definition of test positivity: visual, as per manufacturer IFU Blinding reported: yes; each test was interpreted independently by 2 different laboratory technicians. A 3ird reading was carried out in the event of discrepancy Timing of samples: pso (reported for 289 samples): 0‐3 days 97, 34%; 4‐7 days 103, 36%; 8‐11 days 63, 22%; ≥ 12 days 26, 9% Number of samples reported at > 7 days varied per test, maximum was 289 |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; in‐house assay developed by CNR (Institut Paster) or RealStar SARS‐CoV‐2 (Altona Diagnostics, Germany) Definition of non‐COVID cases: pre‐pandemic Genetic target(s): not stated Samples used: NP; same as for index Timing of reference standard: as for index Blinded to index test: yes, seems to be at time of sampling Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same swab; simultaneous All participants received same reference standard: yes Missing data: number of cases missing per assay varied; reasons for missing data not reported (presumably invalid assay results) [A] 5, 1.7% [B] 6, 2.0% [C] 2, 0.7% [D] 0 [E] 2, 0.7% [F] 0 Uninterpretable results: not stated Indeterminate results (index test): not stated Indeterminate results (reference standard): not stated Unit of analysis: presume patients |
||
| Comparative | |||
| Notes | Funding: evaluation of [A] and [B] conducted in collaboration with Médecins sans Frontières and Epicenter Publication status: published Source: laboratory report obtained via SFM Microbiologie website Author COI: no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Fourati 2020 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [A] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | |||
| Index tests | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [B] relates to test [B] in the list below; see Fourati 2020 [A] for full study characteristics and QUADAS entries [A] SARS‐CoV‐2 COVID‐19 Respi‐Strip [B] STANDARD Q COVID‐19 Ag [C] PanBio COVID‐19 Antigen Rapid Test [D] Biosynex COVID‐19 Ag BSS [E] COVID‐VIRO Antigen Rapid Test [F] NG Test SARS‐CoV‐2 Ag (assay excluded from review due to Vortex requirement as stated in IFU) (no product codes reported) Manufacturer: [A] Coris BioConcept, Gembloux, Belgium [B] SD BIOSENSOR, Inc., Korea [C] Abbott, Chicago, Illinois, USA [D] Biosynex, Strasbourg, France [E] AAZ, Boulogne‐Billancourt, France [F] NG Biotech, Guipry, France Antibody: not stated Ag target: not stated Test method: not stated Samples used: NP; collection not reported Transport media: VTM (Cepheid or Deltalab); 100 μL used for testing Sample storage: frozen at −80 °C until use Test operator: laboratory staff Definition of test positivity: visual, as per manufacturer IFU Blinding reported: yes; each test was interpreted independently by 2 different laboratory technicians. A 3rd reading was carried out in the event of discrepancy Timing of samples: pso (reported for 289 samples): 0‐3 days 97, 34%; 4‐7 days 103, 36%; 8‐11 days 63, 22%; ≥ 12 days 26, 9% Number of samples reported at > 7 days varied per test, maximum was 289 |
||
| Target condition and reference standard(s) | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [A] reports full study characteristics and QUADAS | ||
| Flow and timing | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [A] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | |||
Fourati 2020 [C].
| Study characteristics | |||
| Patient Sampling | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [A] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [A] reports full study characteristics and QUADAS | ||
| Index tests | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [C] relates to test [C] in the list below; see Fourati 2020 [A] for full study characteristics and QUADAS entries [A] SARS‐CoV‐2 COVID‐19 Respi‐Strip [B] STANDARD Q COVID‐19 Ag [C] PanBio COVID‐19 Antigen Rapid Test [D] Biosynex COVID‐19 Ag BSS [E] COVID‐VIRO Antigen Rapid Test [F] NG Test SARS‐CoV‐2 Ag (assay excluded from review due to Vortex requirement as stated in IFU) Manufacturer: [A] Coris BioConcept, Gembloux, Belgium [B] SD BIOSENSOR, Inc., Korea [C] Abbott, Chicago, Illinois, USA [D] Biosynex, Strasbourg, France [E] AAZ, Boulogne‐Billancourt, France [F] NG Biotech, Guipry, France Antibody: not stated Ag target: not stated Test method: not stated Samples used: NP; collection not reported Transport media: VTM (Cepheid or Deltalab); 100 μL used for testing Sample storage: frozen at −80 °C until use Test operator: laboratory staff Definition of test positivity: visual, as per manufacturer IFU Blinding reported: yes; each test was interpreted independently by 2 different laboratory technicians. A 3rd reading was carried out in the event of discrepancy Timing of samples: pso (reported for 289 samples): 0‐3 days 97, 34%; 4‐7 days 103, 36%; 8‐11 days 63, 22%; ≥ 12 days 26, 9% Number of samples reported at > 7 days varied per test, maximum was 289 |
||
| Target condition and reference standard(s) | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [A] reports full study characteristics and QUADAS | ||
| Flow and timing | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [A] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | |||
Fourati 2020 [D].
| Study characteristics | |||
| Patient Sampling | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [A] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [A] reports full study characteristics and QUADAS | ||
| Index tests | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [D] relates to test [D] in the list below; see Fourati 2020 [A] for full study characteristics and QUADAS entries [A] SARS‐CoV‐2 COVID‐19 Respi‐Strip [B] STANDARD Q COVID‐19 Ag [C] PanBio COVID‐19 Antigen Rapid Test [D] Biosynex COVID‐19 Ag BSS [E] COVID‐VIRO Antigen Rapid Test [F] NG Test SARS‐CoV‐2 Ag (assay excluded from review due to Vortex requirement as stated in IFU) Manufacturer: [A] Coris BioConcept, Gembloux, Belgium [B] SD BIOSENSOR, Inc., Korea [C] Abbott, Chicago, Illinois, USA [D] Biosynex, Strasbourg, France [E] AAZ, Boulogne‐Billancourt, France [F] NG Biotech, Guipry, France Antibody: not stated Ag target: not stated Test method: not stated Samples used: NP; collection not reported Transport media: VTM (Cepheid or Deltalab); 100 μL used for testing Sample storage: frozen at −80 °C until use Test operator: laboratory staff Definition of test positivity: visual, as per manufacturer IFU Blinding reported: yes; each test was interpreted independently by 2 different laboratory technicians. A 3rd reading was carried out in the event of discrepancy Timing of samples: pso (reported for 289 samples): 0‐3 days 97, 34%; 4‐7 days 103, 36%; 8‐11 days 63, 22%; ≥ 12 days 26, 9% Number of samples reported at > 7 days varied per test, maximum was 289 |
||
| Target condition and reference standard(s) | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [A] reports full study characteristics and QUADAS | ||
| Flow and timing | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [A] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | |||
Fourati 2020 [E].
| Study characteristics | |||
| Patient Sampling | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [A] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [A] reports full study characteristics and QUADAS | ||
| Index tests | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [E] relates to test [E] in the list below; see Fourati 2020 [A] for full study characteristics and QUADAS entries [A] SARS‐CoV‐2 COVID‐19 Respi‐Strip [B] STANDARD Q COVID‐19 Ag [C] PanBio COVID‐19 Antigen Rapid Test [D] Biosynex COVID‐19 Ag BSS [E] COVID‐VIRO Antigen Rapid Test [F] NG Test SARS‐CoV‐2 Ag (assay excluded from review due to Vortex requirement as stated in IFU) Manufacturer: [A] Coris BioConcept, Gembloux, Belgium [B] SD BIOSENSOR, Inc., Korea [C] Abbott, Chicago, Illinois, USA [D] Biosynex, Strasbourg, France [E] AAZ, Boulogne‐Billancourt, France [F] NG Biotech, Guipry, France Antibody: not stated Ag target: not stated Test method: not stated Samples used: NP; collection not reported Transport media: VTM (Cepheid or Deltalab); 100 μL used for testing Sample storage: frozen at −80 °C until use Test operator: laboratory staff Definition of test positivity: visual, as per manufacturer IFU Blinding reported: yes; each test was interpreted independently by 2 different laboratory technicians. A 3rd reading was carried out in the event of discrepancy Timing of samples: pso (reported for 289 samples): 0‐3 days 97, 34%; 4‐7 days 103, 36%; 8‐11 days 63, 22%; ≥ 12 days 26, 9% Number of samples reported at > 7 days varied per test, maximum was 289 |
||
| Target condition and reference standard(s) | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [A] reports full study characteristics and QUADAS | ||
| Flow and timing | Comparative study of 6 Ag tests (no product codes reported); Fourati 2020 [A] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | |||
Garcia‐Finana 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity and specificity: asymptomatic individuals attending asymptomatic testing sites (ATS) in Liverpool were asked to participate in a QA process Recruitment: consecutive Prospective or retrospective: prospective Sample size (cases): 5869 (74) |
||
| Patient characteristics and setting | Setting: COVID‐19 test centres (asymptomatic) Location: 48 testing sites in Liverpool Country: UK Dates: 8‐29 November 2020 Symptoms and severity: asymptomatic Demographics: mean age 50 years (SD 18 years), 54% women, 82% white ethnicity Exposure history: not reported |
||
| Index tests | Test name: Innova SARS‐CoV‐2 antigen LFD Manufacturer: Innova Medical Group Antibody: not stated Ag target: not stated Test method: CGIA Samples used: not stated; self‐collected (presume nasal + OP), LFT swab obtained first Transport media: none used Sample storage: no storage Test operator: trained non‐HCW (assumed) Definition of test positivity: visual line appearance Blinding reported: yes; conducted first Timing of samples: asymptomatic |
||
| Target condition and reference standard(s) | Reference standard: PCR; "standard test used in Lighthouse Laboratories" Definition of non‐COVID cases: single negative PCR Genetic target(s): N, S, ORF1 (from Table 2) Samples used: nasal + OP; swab for PCR obtained second Timing of reference standard: as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired All participants received same reference standard: yes Missing data: yes Uninterpretable results: void results: LFT: 22 (4 PCR+ and 18 PCR−) PCR: 343 (2 LFT+ and 341 LFT‐) Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "the evaluation was invited by the joint local and national command of the pilot and sponsored by the Department of Health and Social Care (DHSC)" Publication status: published Source: published report (interim) Author COI: none reported |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Gomez 2021(a).
| Study characteristics | |||
| Patient Sampling | Report of 2 study cohorts. This entry Gomez 2021(a) relates to cohort [1]:
[1] single‐group study estimating sensitivity and specificity in symptomatic paediatric patients presenting at outpatients or in primary care (all < 18 years); total n = 427
Second cohort [2], included as included as Gomez 2021(b): [2] single‐group study estimating sensitivity alone in a symptomatic PCR+ student/college‐aged population (18‐25 years) presenting at a university campus; total n = 32 (A further 3 groups were reported but did not undergo Ag testing and were excluded from the review: [3] ED‐collected specimens; [4] asymptomatic people undergoing surgical procedures unrelated to COVID‐19; [5] asymptomatic students) Recruitment: not reported Prospective or retrospective: not reported Sample size (cases): 427 (43) |
||
| Patient characteristics and setting | Setting: community outpatients or primary care Location: UPMC Children's Community Pediatrics and UPMC Children's Hospital of Pittsburgh Primary Care Center Country: USA (Pennsylvania) Dates: 8 March‐10 September 2020 Symptoms and severity: symptoms not reported Reported as "symptomatic" and "presented for care" Demographics: age and sex not reported Exposure history: not reported |
||
| Index tests | Test name: Sofia 2 SARS Antigen used in Group [1] Manufacturer: Quidel (USA) Antibody: not reported Ag target: not reported Test method: FIA Samples used: direct swabs NMT (collection not reported) Transport media: none used Sample storage: not reported; probably immediate, testing conducted on site Test operator: not reported ("performed according to CLIA'88 regulations by appropriate personnel") Definition of test positivity: not reported; as per manufacturer IFU Blinding reported: most likely. Index test was done on site and reference test samples were transported to lab. Timing of samples: not reported |
||
| Target condition and reference standard(s) | Reference standard: laboratory‐developed test based on the CDC 2019‐nCoV Real‐Time RT‐PCR Diagnostic Panel EUA (CDC) protocol
Run per the manufactures' IFU Definition of non‐COVID cases: as for cases Genetic target(s): not reported Samples used: same as for index test; NMT Timing of reference standard: as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous, paired swabs All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "this study was enabled by internal funding provided by UPMC Hospital System and the University of Pittsburgh" Publication status: preprint (not peer reviewed) Source: preprint server (medRxiv) Author COI: no COI statement reported |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Gomez 2021(b).
| Study characteristics | |||
| Patient Sampling | Report of 2 study cohorts. This entry Gomez 2021(b) relates to cohort [2]:
[2] single‐group study reporting only sensitivity in symptomatic PCR+ student/college‐aged population (18‐25 years) presenting at a university campus; total n = 32 Second cohort [1] included as included as Gomez 2021(a): [1] single‐group study estimating sensitivity and specificity in symptomatic paediatric patients presenting at outpatients or in primary care (all < 18 years); total n = 427) (A further 3 groups were reported but did not undergo Ag testing and were excluded from the review: [3] ED‐collected specimens; [4] asymptomatic persons undergoing surgical procedures unrelated to COVID‐19; [5] asymptomatic students) Recruitment: randomly selected Prospective or retrospective: not reported Sample size (cases): 32 (32); number of PCR−ve students was not reported (consider author contact) |
||
| Patient characteristics and setting | Setting: student health services Location: Pittsburgh campus of the University of Pittsburgh Country: USA (Pennsylvania) Dates: 8 March‐10 September 2020 Symptoms and severity: symptoms not reported Reported as "symptomatic" and "presented for care" Demographics: age and sex not reported Exposure history: not reported |
||
| Index tests | Test name: BD Veritor SARS‐CoV‐2 used in group [2] Manufacturer: BD (USA) Antibody: not reported Ag target: not reported Test method: unknown; LFA not otherwise described Samples used: direct swabs AN (collection not reported) Transport media: none used Sample storage: not reported; probably immediate Test operator: not reported ("performed according to CLIA'88 regulations by appropriate personnel") Definition of test positivity: not reported; as per manufacturer IFU Blinding reported: most likely. Index test was done on site and reference test samples were transported to lab. Timing of samples: not reported |
||
| Target condition and reference standard(s) | Reference standard: GeneXpert Xpress SARS‐CoV‐2 assay (Cepheid Inc) or Laboratory‐Developed Test based on the CDC 2019‐nCoV Real‐Time RT‐PCR Diagnostic Panel EUA (CDC) protocol
Run per the manufacturer's IFU Definition of non‐COVID cases: N/A Genetic target(s): not reported Samples used: same as for index test; AN Timing of reference standard: as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous, paired swabs All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "this study was enabled by internal funding provided by UPMC Hospital System and the University of Pittsburgh" Publication status: preprint (not peer reviewed) Source: preprint server (medRxiv) Author COI: no COI statement reported |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Gonzalez‐Donapetry 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity in symptomatic children meeting COVID‐19 clinical criteria and presenting < 7 days pso Recruitment: unclear Prospective or retrospective: prospective Sample size (cases): 440 (18) |
||
| Patient characteristics and setting | Setting: paediatric ED Location: Madrid (Hospital Universitario La Paz) Country: Spain Dates: 25 September‐14 October 2020 Symptoms and severity: all symptomatic: n = 440 Symptoms included: cough 222 (51%), fever 296 (67%), dyspnoea 67 (15%), headache 35 (8%), dysgeusia/anosmia 1 (0%), odynophagia 55 (13%), rhinorrhoea 228 (52%), gastrointestinal disorder 103 (23%) Demographics: med age (IQR): 3 (1‐7) Male: 260 (59%) female: 180 (41%) Exposure history: not reported |
||
| Index tests | Test name: Panbio COVID‐19 Ag Rapid Test Manufacturer: Abbott Rapid Diagnostics Jena GmbH Antibody: SARS‐CoV‐2 nucleoprotein antigens Ag target: immobilized anti‐SARS‐CoV‐2 antibody Test method: membrane technology with gold conjugate CGIA Samples used: NP (no further detail reported) Transport media: none Sample storage: assume immediate testing (no transport or storage reported) Test operator: not reported Definition of test positivity: according to manufacturer's protocol Blinding reported: yes (based on timing of tests) Timing of samples: med time since symptoms onset (IQR): total: 1 (1‐3) days; PCR− 1 (1‐3); PCR+ 1 (1‐2) |
||
| Target condition and reference standard(s) | Reference standard: Vircell SARS‐CoV‐2 real‐time PCR kit (Vircell, Granada, Spain); ≤ 40 Ct considered positive Definition of non‐COVID cases: as for cases Genetic target(s): nucleocapsid (N) and envelope (E) genes Samples used: NP swab samples collected in Universal Transport Medium, COPAN Italia or DeltaSwab Virus, Deltalab (different swab from index test) Timing of reference standard: same as for index test Blinded to index test: not reported Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: appears simultaneous All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: no funding to disclose Publication status: published paper Source: Pediatric Infectious Disease Journal Author COI: authors declare no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Gremmels 2021(a).
| Study characteristics | |||
| Patient Sampling | Report of 2 cohorts of patients presenting for COVID‐19 testing. Gremmels 2021(a) entry relates to:
[1] community‐dwelling mildly symptomatic subjects in a medium endemic area (n = 1369) Gremmels 2021(b) entry reports data for second cohort [2], in a high endemic area Recruitment: yes; all individuals invited to participate Prospective or retrospective: prospective |
||
| Patient characteristics and setting | Setting: community testing centre Location: [1] University Medical Center Utrecht (UMCU) Country: Netherlands Dates: [1] 22 September‐6 October Symptoms and severity: cohort [1] only. Data on symptoms were missing from 9 participants Asymptomatic 37, 2.7%, sore throat 907, 66.3%; coryza 943, 69%; cough 780, 57.1%; headache 601, 44.0%; tiredness 565, 41.3%; general malaise 365, 26.7% (further 19 documented) Demographics: median age 36.4 years (IQR 27.0‐49.6 years); 523, 38.3% male Exposure history: 233, 17% contact with confirmed case |
||
| Index tests | Test name: Panbio COVID‐19 Ag Rapid Test (lot 41ADF011A) Manufacturer: Abbott (Lake Country, IL, U.S.A) Antibody: NP Ag target: not stated Test method: not stated Samples used: NP; obtained after NOP swab for RT‐PCR; implies collected by HCW Transport media: unclear; states transferred to 3 mL UTM after collection until further processing but also describes collected swabs transferred into dedicated sample collection tubes containing a sampling buffer for Ag test Sample storage: none; swabs transported to the laboratory (5 min walking distance from the sampling location) and tested within 2 h of collection Test operator: 2 independent observers, samples processed in a level 2 biosafety cabinet Definition of test positivity: visual line within 15 min; as per manufacturer IFU Blinding reported: yes; observers (blinded to each other and to the PCR results) Timing of samples: cohort [1] (data on duration of symptoms reportedly missing for 201 participants; total reported here is 1138 but denominator for %s is 1166) Day 1‐3 pso 387, 33.2%; day 4‐7 560, 48.0%; day > 7 191, 16.4% |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; Seegene Allplex
Positive result on amplification of any of the 3 SARS‐CoV‐2 genes Definition of non‐COVID cases: as for cases; single negative result Genetic target(s): E‐, N‐, and RdRP‐gene Samples used: NOP (paired) Timing of reference standard: NOP swab obtained first for RT‐PCR Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired All participants received same reference standard: yes Missing data: 2 patients excluded ("inappropriate application of NP swab and lab mislabelling"), disease status not reported. (Considered overall low risk of bias due to small numbers) Uninterpretable results: none reported Indeterminate results (index test): none; no bands were classified as unclear by the independent observers Indeterminate results (reference standard): patients Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: No external funding Publication status: preprint Source: medRxiv Author COI: no COI statement reported |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Gremmels 2021(b).
| Study characteristics | |||
| Patient Sampling | Report of 2 cohorts of patients presenting for COVID‐19 testing. Gremmels 2021(b) entry relates to:
[2] community‐dwelling mildly symptomatic people in a high endemic area (n = 208) Gremmels 2021(a) entry reports data for second [1], cohort in a medium endemic area Recruitment: yes; all individuals invited to participate Prospective or retrospective: prospective |
||
| Patient characteristics and setting | Setting: community testing centre Location: [2] Horacio Oduber Hospital on Aruba Country: Netherlands Dates: [2] 23 September‐9 October Symptoms and severity: not stated; "mildly symptomatic", presume mixed as per 2020 preprint by Gremmels (see secondary reference under Gremmels 2021(b)). Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: Panbio COVID‐19 Ag Rapid Test (lot 41ADF011A) Manufacturer: Abbott (Lake Country, IL, USA) Antibody: NP Ag target: not stated Test method: not stated Samples used: NP; obtained after NOP swab for RT‐PCR; implies collected by HCW Transport media: no UTM used for Ag samples; collected swabs transferred into dedicated sample collection tubes containing a sampling buffer Sample storage: none; swabs transported to the laboratory (5 min walking distance from the sampling location) and tested within 2 h of collection Test operator: 2 independent observers, samples processed in a level 2 biosafety cabinet Definition of test positivity: visual line within 15 min; as per manufacturer IFU Blinding reported: yes; observers (blinded to each other and to the PCR results) Timing of samples: not stated; on presentation |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; Seegene Allplex
Positive result = amplification of any of the 3 SARS‐CoV‐2 genes Definition of non‐COVID cases: as for cases; single negative result Genetic target(s): E‐, N‐, and RdRP‐gene Samples used: NOP (paired) Timing of reference standard: NOP swab obtained first for RT‐PCR Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired All participants received same reference standard: yes Missing data: none reported for Aruba site Uninterpretable results: none reported Indeterminate results (index test): none; no bands were classified as unclear by the independent observers Indeterminate results (reference standard): none Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: No external funding Publication status: preprint Source: medRxiv Author COI: no COI statement reported |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Gupta 2020.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: symptomatic patients with suspected COVID‐19 and asymptomatic contacts of laboratory‐confirmed cases between 5 and 10 days of exposure, meeting Indian Council of Medical Research (ICMR) strategy for COVID‐19 testing Recruitment: consecutive Prospective or retrospective: not stated; appears prospective |
||
| Patient characteristics and setting | Setting: outpatient (tertiary care hospital) Location: All India Institute of Medical Sciences (AIIMS), New Delhi Country: India Dates: 31 May‐24 July 2020 Symptoms and severity: 204 (62%) symptomatic; 126 (38%) asymptomatic Median symptom duration: 1 day (range: 1‐10). Symptoms included: fever (31.5%), cough (25.4%), fatigue/malaise (11.8%), headache (3.3%), runny nose (3.3%) Demographics: median age 34.1 ± 12.6 years; 231 (70%) male Exposure history: 127 asymptomatic were in contact with confirmed case |
||
| Index tests | Test name: STANDARD Q rapid antigen detection test Manufacturer: SD Biosensor, Inc., Gurugram Antibody: not stated Ag target: not stated Test method: not stated Samples used: NP; collection method detailed but personnel not described; presume HCW. Sequence for specimen collection was random for both the samples (Ag and RT‐PCR) Transport media: none Sample storage: none Test operator: same person who obtained swab; HCW Definition of test positivity: visual; test and control lines Blinding reported: yes; conducted first Timing of samples: symptomatic: 192 (95%) ≤ 5 days pso (including 57 cases) |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; commercial assay (BGI Genomics Co. Ltd., China). Psoitive defined as per manufacturer IFU Definition of non‐COVID cases: as for cases; single negative Genetic target(s): ORF1 ab Samples used: nasal and throat swabs (NOP) in VTM Timing of reference standard: as for index test; states the sequence for specimen collection was random for both the samples Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired swabs All participants received same reference standard: yes Missing data: none reported, no participant flow diagram reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "financially supported by the Indian Council of Medical Research, New Delhi (for the Regional Virus Research and Diagnostic Laboratory at the All India Institute of Medical Sciences, New Delhi)" Publication status: published Source: Indian Journal of Medical Research Author COI: author report no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Halfon 2021.
| Study characteristics | |||
| Patient Sampling | 2‐group study estimating sensitivity and specificity:
[1] RT‐PCR positive (n = 100)
[2] RT‐PCR negative (n = 100)
Selected from 43,399 samples that had undergone RT‐qPCR for SARS‐COV‐2 using NP swab Recruitment: unclear; selected "considering the distribution of both TSO (time after symptom onset) and Cts" Prospective or retrospective: prospectively Sample size (cases): 200 (100) |
||
| Patient characteristics and setting | Setting: laboratory‐based Location: Marseille; author institutions Hôpital Européen Marseille Country: France Dates: August‐November 2020 Symptoms and severity: (Figure 1) 104 (52%) asymptomatic, including 35 PCR+; 69 (35%) symptomatic, including 18 PCR+; 27 (13%) symptom status unknown or time not reported, including 13 PCR+ Demographics: mean age 48 years (SD 21); 96 male (48%) Exposure history: not stated |
||
| Index tests | Test name: Panbio COVID 19 antigen rapid test Manufacturer: Abbott Rapid Diagnostic Jena GmbH, Jena, Germany Antibody: SARS‐CoV‐2 nucleocapsid protein Ag target: not stated Test method: CGIA Samples used: NP swabs; collection not described Transport media: not stated Sample storage: not stated Test operator: not stated Definition of test positivity: not stated Blinding reported: not stated Timing of samples: for symptomatic, time pso was: ≤ 4 days 47, > 4 days 22, and not reported or unknown 27 |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; UltraGene Combo2Screen SARS‐CoV‐2 Assay and ChemagicTM viral DNA/RNA 300 kit H96 (ref. CMG‐1033‐S) on a ChemagicTM 360‐D instrument (PerkinElmer, Inc., Austin, TX)
Ct values for positive cases plotted up to 37 Ct (Figure 2) Definition of non‐COVID cases: as for cases; single negative Genetic target(s): not stated Samples used: NP swabs Timing of reference standard: not stated; same as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: not stated; presume simultaneous as same swab seems to have been used All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: no funding source Publication status: preprint Source: medRxiv Author COI: No COI statement provided |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Houston 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity and specificity: adult admissions who met the WHO COVID‐19 case definition at a busy acute hospital Recruitment: not stated; appears to be all meeting eligibility criteria Prospective or retrospective: prospective Sample size (cases): 728 (280) |
||
| Patient characteristics and setting | Setting: inpatient Location: Northwick Park Hospital Country: UK Dates: 17 November‐31 December 2020 Symptoms and severity: all symptomatic (as per WHO case definition); required supplemental oxygen, n (%, 95% CI) 141 (21.4%, 18.3‐24.6); temperature > 38 °C, n (%, 95% CI) 163 (24.9%, 21.6‐28.2) Demographics: median age 67.5 years (IQR 52‐82); 327 (44.9%) female Exposure history: not stated |
||
| Index tests | Test name: Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test Manufacturer: Lotus Global Company, London, UK Antibody: not stated Ag target: not stated Test method: CGIA Samples used: NP swabs; collection not described Transport media: not stated; immediate testing Sample storage: none; immediate testing Test operator: appropriately trained healthcare assistants in the ED Definition of test positivity: not stated Blinding reported: yes, conducted first Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; no details Definition of non‐COVID cases: as for cases; single negative Genetic target(s): not stated Samples used: NP swabs (paired) Timing of reference standard: same as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired swab All participants received same reference standard: yes Missing data: not stated; only valid results included Uninterpretable results: none Indeterminate results (index test): none Indeterminate results (reference standard): none Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: no specific grant Publication status: published Source: Journal of Hospital Infection Author COI: no competing interests present. |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Huh 2021.
| Study characteristics | |||
| Patient Sampling | Unclear design estimating sensitivity and specificity: included samples PCR positive or negative for SARS‐CoV‐2
(A second study evaluating serology assays for antibody detection in additional PCR+ and pre‐pandemic samples was also reported but not eligible for this review) Recruitment: unclear Prospective or retrospective: retrospective Sample size (cases): 132 (62) |
||
| Patient characteristics and setting | Setting: stated "2 institutions", no further details reported Location: not reported; authors' institutions include Dongguk University Ilsan hospital and College of Medicine, Chosun University Country: not stated; appears to be Korea Dates: not reported Symptoms and severity: not reported; could all be symptomatic as data are reported up to 12 days pso for PCR+ only Demographics: age and sex not reported Exposure history: not reported |
||
| Index tests | Test name: PCL COVID19 Ag Rapid FIA used with the PCLOK EZ analyzer Manufacturer: PCL Inc. Antibody: not reported Ag target: not reported Test method: FIA Samples used: NP (collection not reported) Transport media: VTM (further details not reported) Sample storage: storage in VTM reported, test done after reference standard, but exact timing not reported Test operator: not reported Definition of test positivity: not reported Blinding reported: appears to be no (index test after reference standard) Timing of samples: 0‐12 days pso (n = 62) Results reported for 0‐7 days (n = 48) and 8‐12 days (n = 14) periods |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; PowerChekTM2019‐nCoV Real‐time PCR Kit, Kogenebiotech, Seoul, 22 Korea
Study also reports use of the RT‐PCR CFX96 Real‐time PCR detection system (Bio‐Rad Laboratories, Hercules, CA) with the Allplex 2019‐nCoV Assay kit (Seegene Inc., Seoul, Korea), however this appears to relate to the evaluation of serological tests Definition of non‐COVID cases: as for cases Genetic target(s): RdRP and N genes specific for 2 SARS‐CoV‐2 and E gene for all of sarbecovirus including SARS‐CoV‐2 Samples used: NP swabs, appears to be same as for index test Timing of reference standard: appears to be same as index test Blinded to index test: yes (index test after reference) Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: appears to be simultaneous, as samples were stored and reference test was done before index test; however 2 PCR assays are reported and it is not clear whether a second PCR may be have been carried out? All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "supported by the Promising IP Project Support Program funded by the Ministry of Trade Industry and Energy. And this study was supported by the Clinical Trial Support Program funded by the Ministry of Health and Warfare. Also, this study was supported by the Technological Innovation R&D Program funded by the Korea Health Industry Development Institute (KHIDI)." Publication status: preprint (not peer reviewed) Source: preprint server (medRxiv) Author COI: no COI statement reported |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Igloi 2021.
| Study characteristics | |||
| Patient Sampling | Multi‐group study to estimate sensitivity and specificity including:
[1] symptomatic RT‐PCR test‐positive patients (n = 160)
[2] exposed HCWs and patient contacts (n = 150)
Data are presented only for groups [1] and [2] combined; author contacted 15 March 2021 Recruitment: unclear (do not state all patients) Prospective or retrospective: not stated; appears prospective Sample size (cases): 310 (188) |
||
| Patient characteristics and setting | Setting: mixed; described as patients, their contacts and exposed HCWs Location: Fayoum University Hospital, Fayoum Country: Egypt Dates: May 2020 Symptoms and severity: unclear; 160 PCR+ve "patients" presumably symptomatic, plus 150 presumably asymptomatic contacts and exposed HCWs Demographics: median age 42 years; 184/310 (59%) male Exposure history: states "exposed healthcare workers and patient contacts."; no further details |
||
| Index tests | Test name: BIOCREDIT COVID‐19 Ag kit Manufacturer: not stated; manufacturer is RapiGEN Antibody: not stated Ag target: not stated Test method: CGIA; described as lateral flow immunochromatographic assay (uses a dual‐colour system for the qualitative detection of the SARS‐CoV‐2 antigen) Samples used: NP using flocked swabs; collection not otherwise specified Transport media: UTM (UTM‐RT System, Copan Diagnostics, Murrieta, CA) Sample storage: transported to lab within 1‐2 h of collection; then stored at 4 °C and tested within 24 h Test operator: laboratory staff Definition of test positivity: no details Blinding reported: unclear; index test performed after PCR test Timing of samples: group [1] (n = 160); median 3 days pso |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR (multiplex real‐time PCR detection kit; DTlite 4, Russia) Definition of non‐COVID cases: as for cases; single negative Genetic target(s): not stated Samples used: NP in VTM; same as for index test Timing of reference standard: as for index test Blinded to index test: yes (PCR performed before index test) Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; same swab All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: no external funding Publication status: published Source: Laboratory Medicine Author COI: author report no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Ishii 2021 [A].
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: samples submitted for COVID‐19 diagnosis at a medical centre; includes symptomatic patients (dysosmia, dysgeusia, fever and pneumonia) and asymptomatic close contacts of confirmed cases Recruitment: unclear; all samples had PCR and Lunmipulse, whereas the Espline test was performed on randomly selected samples Prospective or retrospective: unclear; NP swabs and saliva samples were collected from 33 COVID‐19 patients and 564 non‐COVID‐19 patients Sample size (cases): [A]: NP swabs: 271 (11); [B]: saliva samples: 93 (9) |
||
| Patient characteristics and setting | Setting: unclear Location: Toho University Omori Medical Center Country: Japan Dates: August‐September 2020 Symptoms and severity: all PCR+ cases with an ESPLINE result were symptomatic (n = 20) Demographics: not stated Exposure history: no details reported |
||
| Index tests | Test name: Espline SARS‐CoV‐2 Manufacturer: Fujirebio Inc., Tokyo, Japan Antibody: viral nucleocapsid antigen Ag target: not reported Test method: not reported Samples used: [A]: NP and [B]: saliva samples (not reported who collected by) Transport media: not reported (Saliva samples were diluted 2‐fold with dedicated reagent. The sample solution was centrifuged at 12,000 rpm for 2 min) Sample storage: unfrozen and fresh Test operator: not reported Definition of test positivity: positive line observed with naked eye Blinding reported: unclear Timing of samples: [A]: 10/11 within 2 days pso, 1/11 at 4 days pso [B]: 7/9 within 7 days pso and 2/9 within 8‐14 days pso |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; TaqMan Fast Virus 1‐step Master Mix (Thermo Fisher Scientific, Walthan, MA, USA) on the QuantStudio 5 Real‐Time PCR System (Thermo Fisher Scientific)
Ct < 35 considered positive Definition of non‐COVID cases: as for cases Genetic target(s): not reported Samples used: same as for index test Timing of reference standard: same as for index test Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous (same swab) All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: no specific grant funding Publication status: published paper Source: Journal of Infection and Chemotherapy Author COI: declaration of competing interest: none |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Ishii 2021 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of an Ag using 2 different samples; Ishii 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of an Ag using 2 different samples; Ishii 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: Espline SARS‐CoV‐2 Manufacturer: Fujirebio Inc., Tokyo, Japan Antibody: viral nucleocapsid antigen Ag target: not reported Test method: not reported Samples used: [A]: NP and [B]: saliva samples (not reported who collected by) Transport media: not reported (Saliva samples were diluted 2‐fold with dedicated reagent. The sample solution was centrifuged at 12,000 rpm for 2 min) Sample storage: unfrozen and fresh Test operator: not reported Definition of test positivity: positive line observed with naked eye Blinding reported: unclear Timing of samples: [A]: 10/11 within 2 days pso, 1/11 at 4 days pso [B]: 7/9 within 7 days pso and 2/9 within 8‐14 days pso |
||
| Target condition and reference standard(s) | Comparative study of an Ag using 2 different samples; Ishii 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of an Ag using 2 different samples; Ishii 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of an Ag using 2 different samples; Ishii 2021 [A] reports full study characteristics and QUADAS. | ||
Jaaskelainen 2021 [A].
| Study characteristics | |||
| Patient Sampling | Multi‐group study to estimate sensitivity and specificity including: [1] RT‐PCR test‐positive samples from adults from outpatient clinics and drive‐through testing sites (n = 96) {Additional cohort [2] RT‐PCR+ve and negative samples for analysis of analytical performance (n = 102); excluded as they were deliberately selected to cover a wide Ct range (26.35‐32.66 Ct)) Recruitment: [1] appears consecutive (selected "systematically backward") Prospective or retrospective: retrospective Sample size (cases): 136 (96); different number of cases tested per assay |
||
| Patient characteristics and setting | Setting: outpatient/community test centre drive‐through testing sites Location: HUS Diagnostic Center, Helsinki University Hospital, Helsinki Country: Finland Dates: [1] 1‐18 November 2020 Symptoms and severity: not stated Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: [A] Quidel Sofia SARS FIA (lots used 143489) [B] STANDARD Q COVID‐19 Ag test (lots used QCO3020105) [C] Panbio (lots used 41ADF024A) All 3 were CE IVD marked SARS‐CoV‐2 RADTs Manufacturer: [A] Quidel, San Diego, CA [B] SD Biosensor, Republic of Korea [C] Abbott Diagnostic GmbH, Jena, Germany Antibody: Ag (nucleocapsid protein) Ag target: NA Test method: [A] FIA ; [B] and [C] not stated Samples used: NP swabs (not reported who collected the sample) Transport media: stored in 0.9% saline; authors note this is "off‐label" use of the assays Sample storage: stored at −20 °C Test operator: not stated; presume laboratory staff Definition of test positivity:[A] detection of fluorescent signal; [B] and [C] appearance of visible line Blinding reported: unclear/details not reported. Probably no (positive/negative samples were selected based on the RT‐PCR tests for performance tests) Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; in‐house (laboratory‐developed test using modified method by Corman et al)
Virus culture also used for PCR positive subset of samples Definition of non‐COVID cases: NA Genetic target(s): N gene target of SARS‐CoV‐2 Samples used: NP swabs; same as for index test Timing of reference standard: not reported Blinded to index test: yes; PCR conducted for diagnosis prior to sample storage Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous (same swab) All participants received same reference standard: yes Missing data: number of samples tested varied by assay. Of 96 PCR+ samples in group [1], results were reported for:
Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: not stated; no indication of multiple samples per patient |
||
| Comparative | |||
| Notes | Funding: not reported Publication status: preprints Source: medRxiv Author COI: not reported |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Jaaskelainen 2021 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 3 Ag tests; Jaaskelainen 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 3 Ag tests; Jaaskelainen 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] Quidel Sofia SARS FIA (lots used 143489) [B] STANDARD Q COVID‐19 Ag test (lots used QCO3020105) [C] Panbio (lots used 41ADF024A) All 3 were CE IVD marked SARS‐CoV‐2 RADTs Manufacturer: [A] Quidel, San Diego, CA [B] SD Biosensor, Republic of Korea [C] Abbott Diagnostic GmbH, Jena, Germany Antibody: Ag (nucleocapsid protein) Ag target: NA Test method: [A] FIA; [B] and [C] not stated Samples used: NP swabs (not reported who collected the sample) Transport media: stored in 0.9% saline; authors note this is "off‐label" use of the assays Sample storage: stored at −20 °C Test operator: not stated; presume laboratory staff Definition of test positivity: [A] detection of fluorescent signal; [B] and [C] appearance of visible line Blinding reported: unclear/details not reported. Probably no (positive/negative samples were selected based on the RT‐PCR tests for performance tests) Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 3 Ag tests; Jaaskelainen 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 3 Ag tests; Jaaskelainen 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 3 Ag tests; Jaaskelainen 2021 [A] reports full study characteristics and QUADAS. | ||
Jaaskelainen 2021 [C].
| Study characteristics | |||
| Patient Sampling | Comparative study of 3 Ag tests; Jaaskelainen 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 3 Ag tests; Jaaskelainen 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] Quidel Sofia SARS FIA (lots used 143489) [B] STANDARD Q COVID‐19 Ag test (lots used QCO3020105) [C] Panbio (lots used 41ADF024A) All 3 were CE IVD marked SARS‐CoV‐2 RADTs Manufacturer: [A] Quidel, San Diego, CA [B] SD Biosensor, Republic of Korea [C] Abbott Diagnostic GmbH, Jena, Germany Antibody: Ag (nucleocapsid protein) Ag target: NA Test method: [A] FIA; [B] and [C] not stated Samples used: NP swabs (not reported who collected the sample) Transport media: stored in 0.9% saline; authors note this is "off‐label" use of the assays Sample storage: stored at −20 °C Test operator: not stated; presume laboratory staff Definition of test positivity: [A] detection of fluorescent signal; [B] and [C] appearance of visible line Blinding reported: unclear/details not reported. Probably no (positive/negative samples were selected based on the RT‐PCR tests for performance tests) Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 3 Ag tests; Jaaskelainen 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 3 Ag tests; Jaaskelainen 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 3 Ag tests; Jaaskelainen 2021 [A] reports full study characteristics and QUADAS. | ||
Jakobsen 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: symptomatic and asymptomatic patients (self‐reported) who had booked an appointment at a public test centre Recruitment: consecutive (all who appeared for the appointment were tested) Prospective or retrospective: prospective Sample size (cases): 4697 patients with 4811 paired conclusive tests (221 tests); 196 were tested twice or more |
||
| Patient characteristics and setting | Setting: public COVID‐19 test centre Location: not stated; author institution is University of Copenhagen, Copenhagen Country: Denmark Dates: 26‐31 December 2020 Symptoms and severity: 705 (15%) symptomatic (self‐report); 3008 (64%) asymptomatic(Not all participants responded to the online questionnaire regarding symptoms) Demographics: mean age 45 (SD 16.9) years; 2456 (53%) female Exposure history: not stated |
||
| Index tests | Test name: STANDARD Q COVID‐19 Ag test Manufacturer: SD BIOSENSOR Antibody: not stated Ag target: not stated Test method: CGIA Samples used: NP (states test "performed" by personnel from private company at the test centre, no detail of swab collection but OP swab for PCR collected by test centre personnel) Transport media: none Sample storage: none Test operator: presume HCW ‐ states "Personnel from the private company Copenhagen Medical A/S" Definition of test positivity: not stated; conducted according to SD BIOSENSOR's instruction Blinding reported: yes; test performed on the site before PCR Timing of samples: no details |
||
| Target condition and reference standard(s) | Reference standard: PCR performed using Luna Universal Probe One‐step RT‐qPCR kit (NewEngland Biolab); Ct ≤ 38 and ≥ 10 considered positive Definition of non‐COVID cases: as for cases; single negative Genetic target(s): E‐gene Samples used: OP; eluted in PBS Timing of reference standard: no details; as for index test Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous All participants received same reference standard: yes Missing data: yes Uninterpretable results: 97/4908 inconclusive on PCR, leaving 4811 samples for inclusion Indeterminate results (index test): none reported Indeterminate results (reference standard): the reasons for 97 missing data was due to inconclusive results on PCR (i.e. Ct > 38) Unit of analysis: samples |
||
| Comparative | |||
| Notes | Funding: author report no funding was received for this project Publication status: preprint Source: medRxiv Author COI: author report no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
James 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: targeted screening at a medical centre in USA (mainly asymptomatic); all staff providing patient care were required to participate but testing was optional for non‐clinical staff) Recruitment: consecutive (all clinical staff included) Prospective or retrospective: prospective Sample size (cases): 2339 (152) |
||
| Patient characteristics and setting | Setting: primarily screening; dedicated stations within the hospital (mobile teams) or drive‐through parking lot stations Location: Arkansas (name of hospital not reported); study conducted by Arkansas Department of Health, author institution includes St Bernards Medical Center Country: USA Dates: 2‐9 October 2020 Symptoms and severity: 2224 (95%) asymptomatic; 115 (5%) symptomatic. 94 (82%) reported only one symptom and 21 (18%) reported 2–6 symptoms: fever (6%), cough (29%), sore throat (29%), chills (6%), headache (41%), muscle aches (12%), abdominal pain (4%); none reported loss of taste or loss of smell. Demographics: median 37 (range 16–89) years; gender not reported Exposure history: no details |
||
| Index tests | Test name: BinaxNOW COVID‐19 Ag Card tests (BinaxNOW) Manufacturer: Abbott Diagnostics, Scarborough, ME Antibody: nucleocapsid protein antigen Ag target: not stated Test method: not stated Samples used: nasal (nares samples); collected by trained hospital staff in random order for PCR or Ag testing Transport media: none Sample storage: none Test operator: trained laboratory employees of hospital X Definition of test positivity: not stated Blinding reported: yes; performed on site before RT‐PCR Timing of samples: not stated for symptomatic |
||
| Target condition and reference standard(s) | Reference standard: RT‐ PCR using PerkinElmer SARS‐CoV‐2 real‐time RT‐PCR assay (PerkinElmer, Waltham, MA); Ct < 42 considered positive Definition of non‐COVID cases: as for cases; single negative Genetic target(s): N or Orf1 Samples used: nasal (nares) swab in VTM or saline Timing of reference standard: not stated Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired swab All participants received same reference standard: yes Missing data: none Uninterpretable results: none Indeterminate results (index test): none (all paired samples were successfully tested) Indeterminate results (reference standard): none (all paired samples were successfully tested) Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: none Publication status: published Source: Infection Control & Hospital Epidemiology Author COI: one of the authors report COI unrelated to this manuscript. All other authors report no COI. |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Kerneis 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: symptomatic patients invited fort testing (i.e. temperature > 37.8 °C or chills, cough, rhinorrhoea, muscle pain, loss of smell or taste, unusual persistent headaches or severe asthenia), symptomatic contacts of confirmed cases, asymptomatic contacts of confirmed cases (after 7 days self‐isolation) and any other asymptomatic individuals wishing to be tested Recruitment: consecutive Prospective or retrospective: prospective Sample size (cases): 1452 (129) |
||
| Patient characteristics and setting | Setting: COVID‐19 community testing centres Location: 2 community screening centres located in Paris within the COVISAN program (Assistance Publique‐Hôpitaux de Paris, APHP) Country: France Dates: 19 October‐18 December 2020 Symptoms and severity: 571 (39%) symptomatic, 409 with 1‐3 symptoms: cough: 292/1451 (20%); headaches: 257/1451 (18%); rhinorrhoea: 202/1451 (14%); asthenia: 198/1451 (14%); muscle pain: 177/1451 (12%); fever: 163/1451 (11%); diarrhoea: 85/1451 (6%); chills: 69/1451 (5%); anosmia: 62/1451 (4%); shortness of breath: 53/1451 (4%); chest pain: 52/1451 (4%) Demographics: med age (IQR): 36 (26‐50); 122 (8%) children Sex: 755/1451 (52%) female Exposure history: not reported |
||
| Index tests | Test name: STANDARD Q COVID‐19 Ag test Manufacturer: SD Biosensor, Chuncheongbuk‐do, Republic of Korea Antibody: N antigen Ag target: not reported Test method: not reported Samples used: NP second nostril (collected by trained nurses) Transport media: none used Sample storage: no storage (immediate testing) Test operator: not reported Definition of test positivity: according to manufacturer Blinding reported: presumed (based on timing) Timing of samples: med days pso (IQR): 3 (IQR 2‐4) Asymptomatic time from last contact 7 (IQR 1‐7) |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; TaqPath COVID 19 CE IVD RT PCR Kit (Thermo Fisher Scientific, Coutaboeuf, France)
Considered positive if ≥ 1 gene detected; sensitivity analysis for ≥ 2 targets present Definition of non‐COVID cases: as for cases Genetic target(s): ORF1ab, N and S‐genes Samples used: NP first nostril Timing of reference standard: as for index test Blinded to index test: yes; interpretation "carried out blind of the result of the others (test) and of the participant's clinical data" Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneously All participants received same reference standard: yes Missing data: yes Of 1451 participants who provided samples, 1117 underwent Ag tests (Appendix Figure 1). Reason for missing data not reported Of 1117 tested: 2 technical failures (on Ag test) 6 additional missing results, reason not reported Uninterpretable results: not reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: French Ministry of Health and the Assistance Publique‐Hôpitaux de Paris Foundation Publication status: preprint (not peer reviewed) Source: medRxiv Author COI: no COI statement reported, but states "The funding sources had no role in the study’s design, conduct and reporting." |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Kilic 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: symptomatic patients meeting pre‐set criteria for Ag and RT‐PCR testing (COVID‐19 exposure and ≤ 5 days pso, including fever/flu‐like symptoms, unexplained shortness of breath, or new loss of taste) Recruitment: consecutive; based on all patients meeting criteria for testing Prospective or retrospective: prospective Sample size (cases): 1384 (116) |
||
| Patient characteristics and setting | Setting: not stated; 'patients receiving care at our hospital system' ‐ likely mixed settings. (www.wakehealth.edu/Find‐A‐Provider) Location: author institution: Wake Forest School of Medicine, Winston‐Salem, NC Country: USA Dates: 20 October‐3 December 2020 Symptoms and severity: all symptomatic Demographics: median age 46.8 years (range 1‐98 years), 800 (57.8%) female Exposure history: seems all had COVID‐19 exposure |
||
| Index tests | Test name: BD Veritor Manufacturer: Becton Dickinson, Sparks, Maryland, USA Antibody: nucleocapsid Ag target: not stated Test method: chromatographic Samples used: nasal samples collected using flocked swabs according to the manufacturer IFU (IFU describes AN collection method) Transport media: none required Sample storage: tested at the site of collection within 1 h of collection Test operator: not stated; presume on‐site HCW Definition of test positivity: BD Veritor Analyzer used; no further detail Blinding reported: yes; performed on site before PCR Timing of samples: ≤ 5 days from symptom onset |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; Simplexa Covid‐19 Direct EUA RT‐PCR (Diasorin Molecular LLC, Cypress, CA, USA)
Median threshold cycle (Ct) values < 40 (for one or both targets) were reported as positive for SARS‐CoV‐2 Definition of non‐COVID cases: as for cases (single ‐ve PCR) Genetic target(s): S gene and ORF1ab genes Samples used: NP in VTM; paired Timing of reference standard: same as for index Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swab; simultaneous All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: not stated Publication status: accepted manuscript posted online Source: Journal of Clinical Microbiology Author COI: authors report no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Kohmer 2021 [A].
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: individuals from shared living facilities for screening purposes regardless of their clinical symptoms (n = 100) Recruitment: unclear (anonymized clinical samples) Prospective or retrospective: prospective Sample size (cases): 100 (74) |
||
| Patient characteristics and setting | Setting: community Location: shared living facilities, Frankfurt (Institute for Medical Virology, University Hospital, Goethe University Frankfurt) Country: Germany Dates: November 2020 (2 weeks) Symptoms and severity: not stated Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: [A] RIDA QUICK SARS‐CoV‐2 Antigen [B] SARS‐CoV‐2 Rapid Antigen Test [C] NADAL COVID‐19 Ag Test (test cassette) [D] SARS‐CoV‐2 Ag Test on the LumiraDx Platform Manufacturer: [A] R‐Biopharm AG, Darmstadt, Germany [B] Roche Diagnostics GmbH, Mannheim, Germany [C] Nal von Minden GmbH, Regensburg, Germany [D] LumiraDx GmbH, Cologne, Germany Antibody: not stated Ag target: not stated Test method: [A], [B], [C] not stated [D] immunofluorescence assay Samples used: NP Transport media: PBS Sample storage: sample storage not stated; tested within 24 h after collection Test operator: not stated Definition of test positivity: for [A], [B], [C] the results were read visually and documented by 3 different individuals, and the majority consensus was chosen as the final test result Not stated for [D] (IFU indicates that a reader device is required) Blinding reported: unclear; parallel testing Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; Cobas 6800 system (Roche Diagnostics International AG, Rotkreuz, Switzerland) system
Culture also undertaken for all samples positive on at least 1 genetic target Definition of non‐COVID cases: as for cases; single negative PCR Genetic target(s): ORF1 and E‐gene; considered positive if ORF1 detected Samples used: NP swab in PBS (same as for index test) Timing of reference standard: not stated Blinded to index test: unclear; parallel testing Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same swab All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: no external funding Publication status: published Source: Journal of Clinical Medicine Author COI: the authors report no conflict of interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Kohmer 2021 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 4 Ag tests; Kohmer 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 4 Ag tests; Kohmer 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] RIDA QUICK SARS‐CoV‐2 Antigen [B] SARS‐CoV‐2 Rapid Antigen Test [C] NADAL COVID‐19 Ag Test (test cassette) [D] SARS‐CoV‐2 Ag Test on the LumiraDx Platform Manufacturer: [A] R‐Biopharm AG, Darmstadt, Germany [B] Roche Diagnostics GmbH, Mannheim, Germany [C] Nal von Minden GmbH, Regensburg, Germany [D] LumiraDx GmbH, Cologne, Germany Antibody: not stated Ag target: not stated Test method: [A], [B], [C] not stated [D] immunofluorescence assay Samples used: NP Transport media: PBS Sample storage: sample storage not stated; tested within 24 h after collection Test operator: not stated Definition of test positivity: for [A], [B], [C] the results were read visually and documented by 3different individuals, and the majority consensus was chosen as the final test result; Not stated for [D] (IFU indicates that a reader device is required) Blinding reported: unclear; parallel testing Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 4 Ag tests; Kohmer 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 4 Ag tests; Kohmer 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 4 Ag tests; Kohmer 2021 [A] reports full study characteristics and QUADAS. | ||
Kohmer 2021 [C].
| Study characteristics | |||
| Patient Sampling | Comparative study of 4 Ag tests; Kohmer 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 4 Ag tests; Kohmer 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] RIDA QUICK SARS‐CoV‐2 Antigen [B] SARS‐CoV‐2 Rapid Antigen Test [C] NADAL COVID‐19 Ag Test (test cassette) [D] SARS‐CoV‐2 Ag Test on the LumiraDx Platform Manufacturer: [A] R‐Biopharm AG, Darmstadt, Germany [B] Roche Diagnostics GmbH, Mannheim, Germany [C] Nal von Minden GmbH, Regensburg, Germany [D] LumiraDx GmbH, Cologne, Germany Antibody: not stated Ag target: not stated Test method: [A], [B], [C] not stated [D] immunofluorescence assay Samples used: NP Transport media: PBS Sample storage: sample storage not stated; tested within 24 h after collection Test operator: not stated Definition of test positivity: for [A], [B], [C] the results were read visually and documented by 3different individuals, and the majority consensus was chosen as the final test result; Not stated for [D] (IFU indicates that a reader device is required) Blinding reported: unclear; parallel testing Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 4 Ag tests; Kohmer 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 4 Ag tests; Kohmer 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 4 Ag tests; Kohmer 2021 [A] reports full study characteristics and QUADAS. | ||
Kohmer 2021 [D].
| Study characteristics | |||
| Patient Sampling | Comparative study of 4 Ag tests; Kohmer 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 4 Ag tests; Kohmer 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] RIDAQUICK SARS‐CoV‐2 Antigen [B] SARS‐CoV‐2 Rapid Antigen Test [C] NADAL COVID‐19 Ag Test (test cassette) [D] SARS‐CoV‐2 Ag Test on the LumiraDx Platform Manufacturer: [A] R‐Biopharm AG, Darmstadt, Germany [B] Roche Diagnostics GmbH, Mannheim, Germany [C] Nal von Minden GmbH, Regensburg, Germany [D] LumiraDx GmbH, Cologne, Germany Antibody: not stated Ag target: not stated Test method: [A], [B], [C] not stated [D] immunofluorescence assay Samples used: NP Transport media: PBS Sample storage: sample storage not stated; tested within 24 h after collection Test operator: not stated Definition of test positivity: for [A], [B], [C] the results were read visually and documented by 3 different individuals, and the majority consensus was chosen as the final test result; Not stated for [D] (IFU indicates that a reader device is required) Blinding reported: unclear; parallel testing Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 4 Ag tests; Kohmer 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 4 Ag tests; Kohmer 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 4 Ag tests; Kohmer 2021 [A] reports full study characteristics and QUADAS. | ||
Kriemler 2021.
| Study characteristics | |||
| Patient Sampling | A single‐group study (nested in the Ciao Corona Cohort study) to estimate sensitivity and specificity: children (n = 641) and teachers (n = 66) attending primary or secondary schools over a 1‐week period and tested at least once (T1 and or T2) (n = 707). Schools were selected based on high incidence areas; children were required to be kept at home if they were sick beyond very mild symptoms such as runny nose or mild cough. Recruitment: consecutive; all participating children and teachers got the tests Prospective or retrospective: prospective Sample size (cases): 641 children and adolescents and 66 teachers tested at T1 and or T2; children provided 1170 samples (1 PCR+) (567 at T1 and 602 at T2), total N not reported for teachers (0 PCR+). Data obtained from study authors: 117 samples from teachers (62 at T1 and 57 at T2) |
||
| Patient characteristics and setting | Setting: screening in schools Location: schools in the city of Zurich and 1 school of each of 4 adjacent districts Author institution: Epidemiology, Biostatistics and Prevention Institute, University of Zurich, Zurich, Country: Switzerland Dates: 1‐11 December 2020 Symptoms and severity: at T1, 198/567 (35%) children and 5/66 (8%) teachers reported mild symptoms (runny nose, headache, cough stomach upset, etc) during the previous 5 days before testing. Symptoms of week 1 (T1) and 2 (T2) reported not to differ significantly Demographics: children and adolescents: age range 10‐19 years; 370 (58%) female children and adolescents; 46 (70%) female teachers Exposure history: not stated |
||
| Index tests | Test name: STANDARD Q COVID‐19 Ag Test Manufacturer: SD Biosensor/Roche, Switzerland Antibody: none stated Ag target: none stated Test method: CGIA Samples used: buccal swab taken with closed mouth over 1 min (study staff members) (2 swabs were held closely together so that they were exposed to the same location of the enoral space. Swabbing of the mucosal membrane was done in the whole enoral space with some pressure application to make sure to have some cells contained in the swab) Transport media: none required; direct swab Sample storage: none required; direct swab Test operator: study staff member (experienced in RDT testing) Definition of test positivity: visual Judged by 2 study team members (experienced in RDT testing) in agreement as positive or negative and blinded to reported symptoms Blinding reported: yes; performed on the site before PCR Timing of samples: essentially asymptomatic testing although some symptoms (runny nose, cough headache etc) reported |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR (Analytica, Zurich, Switzerland); performed using the CE‐IVD‐marked AllplexTM SARS‐CoV‐2 Assay (Seegene Inc, Seoul, Republic of Korea) Definition of non‐COVID cases: same as for cases Genetic target(s): N, S, RdRP and E‐gene Samples used: buccal swab (taken with closed mouth over 1 min) Timing of reference standard: as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: parallel buccal swab All participants received same reference standard: yes Missing data: yes; participants were not all tested at T1 (88% of children tested) and T2 (94%) Uninterpretable results: none reported Indeterminate results (index test): none reported; judged by 2 study team members (experienced in RDT testing) in agreement as positive or negative Indeterminate results (reference standard): none reported Unit of analysis: participants and samples |
||
| Comparative | |||
| Notes | Funding: "this study is funded by fundraising of SSPH+ that includes funds of the Swiss Federal Office of Public Health and private funders, by Cantons of Switzerland, by institutional funds of the Universities and by the University of Zurich Foundation and the Federal Office of Public Health" Publication status: published Source: Frontiers in Pediatrics Author COI: authors report no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Kruger 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group multi‐centre study to estimate sensitivity and specificity: adults at risk of SARS‐CoV‐2 infection based on reported symptoms or recent contact with a confirmed case (according to the criteria of the national health authority)
Group [1]: Heidelberg
Group [2]: Berlin
(Group [1] and group [2] are reported as subgroups) Recruitment: not reported; known to be consecutive recruitment Prospective or retrospective: prospectively Sample size (cases): 767 (146) |
||
| Patient characteristics and setting | Setting: COVID‐19 testing site
[1] Drive‐in testing site
[2] Clinical ambulatory testing facility Location: [1]: Heidelberg (authors' institutions include Heidelberg University Hospital) [2]: Berlin (authors' institutions include Berlin Institute of Health) Country: Germany Dates: 2 November‐ 4 December 2020 Symptoms and severity: 486 (64%) symptomatic on day of testing; 90 (19%) fever, 247 (52%) cough, 242 (50%) sore throat, 297 (62%) fatigue Raw symptom data in a supplementary table www.medrxiv.org/content/10.1101/2021.03.02.21252430v1.supplementary‐material (Section E, Table 1, p19) Demographics: average age: 38.5 years (SD 14.2) Sex: (52%) female Exposure history: all asymptomatic participants (36%) were recent high‐risk contacts |
||
| Index tests | Test name: LumiraDx SARS‐CoV Ag test Manufacturer: LumiraDx Limited, London, UK Antibody: nucleocapsid protein of SARS‐CoV‐2 Ag target: not reported Test method: FIA; microfluidic immunofluorescence assay Samples used: NMT (by participants themselves with HCW providing instructions, supervision and corrections) Sample collection was performed with Dryswab Standard Tip Rayon (Medical Wire & Equipment, Corsham, England) Transport media: none used Sample storage: no storage Test operator: laboratory personnel in dedicated workspace (no further details reported) Definition of test positivity: according to manufacturer; digital touch screen readout of positive, negative or error (error results re‐tested using same extraction vial with a new test strip) Blinding reported: yes Timing of samples: average symptom duration of 3.9 days (SD 3.2) 423/472 (90%) of symptomatic patients reported onset of symptoms within the prior 7 days |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR
[A] In Heidelberg: Allplex SARS‐CoV‐2 assay (Seegene, Seoul, South Korea)
[B] In Berlin: Cobas SARS CoV‐2 assay on the Cobas 6800 or 8800 system (Roche, Pleasanton, CA, USA)
OR
SARS CoV‐2 assay from TIB Molbiol (Berlin, Germany)
Assays interpreted according to manufacturer IFU
Conversion of CT values into viral load was based on calibrated RT‐PCR testing with quantified SARS‐CoV‐2 in vitro transcripts. Definition of non‐COVID cases: as for cases; single negative Genetic target(s): E‐gene Samples used: [A] in Heidelberg: NP [B] in Berlin: combined NP/OP (OP alone used only if clinical contraindications for NP sampling) RT‐PCR samples were collected by HCWs using the IMPROSWAB (Guangzhou Improve Medical Instruments Co., Ltd., Guangzhou, China) Timing of reference standard: same as for index test Blinded to index test: yes; stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired samples; reference swab obtained after index test swab All participants received same reference standard: yes Missing data: yes; 2 refused NMT swab and 4 had invalid RT‐PCR results Uninterpretable results: 7 samples gave error message on LumiraDx device; repeat testing yielded valid results and inclusion in the analysis Indeterminate results (index test): not reported Indeterminate results (reference standard): 4 invalid RT‐PCR results excluded Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "the study was supported by the Ministry of Science, Research and Arts of the State of Baden‐ Wuerttemberg, Germany and internal funds from the Heidelberg University Hospital and University Hospital Charité ‐Universitätsmedizin Berlin as well as grants from UK Department of International Development (DFID, recently replaced by FCMO), grants from World Health Organization (WHO), grants from Unitaid to Foundation of New Diagnostics (FIND). The testing devices and all components were provided by the manufacturer. T.C.J. is in part funded through NIAID‐NIH CEIRS." Publication status: preprint (not peer reviewed) Source: medRxiv Author COI: none reported; "The manufacturer and funders had no input into the study protocol, the analysis or interpretation of the results." |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Kruttgen 2021.
| Study characteristics | |||
| Patient Sampling | 2‐group study to estimate sensitivity and specificity
[1] patients previously tested positive by RT‐PCR (n = 75)
[2] patients previously tested negative by RT‐PCR (n = 75) Recruitment: unclear Prospective or retrospective: retrospective Sample size (cases): 150 (75) |
||
| Patient characteristics and setting | Setting: not stated; SARS‐CoV‐2‐negative were reported as hospital inpatient Location: RWTH Aachen University hospital Country: Germany Dates: not stated Symptoms and severity: not stated Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: SARS‐CoV‐2 Rapid Antigen Test Manufacturer: Roche, Switzerland Antibody: not stated Ag target: not stated Test method: not stated Samples used: NP Transport media: swab transport medium used Sample storage: not stated; states no intermittent freeze‐thaw cycle so presume no frozen storage Test operator: laboratory staff Definition of test positivity: visual; test and control lines Blinding reported: unclear Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR: Real Star SARS‐CoV‐2 RT PCR Kit (Altona, Germany) Definition of non‐COVID cases: same as for cases Genetic target(s): not stated Samples used: NP Timing of reference standard: not stated Blinded to index test: yes (conducted before index test) Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same swab All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: none Publication status: published Source: Journal of Virological Methods Author COI: author report no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
L'Huillier 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: children (0‐16 years old) meeting eligibility criteria for RT‐PCR testing: 1) symptoms suggestive of COVID infection according to local governmental testing criteria, and for asymptomatic children either 2) contact with a laboratory‐confirmed COVID infected person and 3) pre‐travel testing Recruitment: consecutive Prospective or retrospective: prospective Sample size (cases): 885 (119); 60 excluded a priori |
||
| Patient characteristics and setting | Setting: paediatric COVID‐19 testing centre Location: Geneva University Hospitals (HUG) Country: Switzerland Dates: 10 November 2020‐26 March 2021 Symptoms and severity: 533 (65%) symptomatic: headache (58%), nasal discharge (55%), cough (44%), fatigue (44%), dysphagia (41%), fever (30%), abdominal pain (20%), myalgia (16%), diarrhoea (15%); shortness of breath (8%), anosmia (7%) Asymptomatic: 289 (35%) Demographics: all children; median age 12.1 (IQR 9.4‐14.5); 266 (50%) female Exposure history: not stated; contacts with COVID cases |
||
| Index tests | Test name: PanbioTM‐COVID‐19 Ag Rapid Test Device Manufacturer: Abbott Rapid Diagnostics, USA Antibody: nucleocapsid Ag target: not stated Test method: CGIA Samples used: NP from the contralateral or ipsilateral nostril (collected by trained nurses) Transport media: none used Sample storage: none required Test operator: 2 members of the study team; blinded to each other and to clinical presentation Definition of test positivity: visual; control and test line Any discrepant result was considered positive when any of the above‐mentioned reader set a positive diagnosis Blinding reported: yes (performed before PCR) Timing of samples: for symptomatic patients samples were taken at median 2 days pso (IQR 1‐3) |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR: either
Definition of non‐COVID cases: same as for cases (single ‐ve PCR) Genetic target(s): not stated Samples used: NP (flocked swab in 3 mL VTM) Timing of reference standard: for symptomatic patients samples were taken at median 2 days pso (IQR 1‐3) Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired All participants received same reference standard: yes Missing data: yes; 63/885 (7%) excluded from analysis 1 excluded a priori (did not meet inclusion criteria), 1 refused RT‐PCR, and 58 refused Ag test (n = 58); 3 excluded after testing Uninterpretable results: 2 Ag test result not reported and 1 Ag test result invalid Among the 822 Ag tests Indeterminate results (index test): none; only 1 discrepant between readers (considered +ve) Indeterminate results (reference standard) Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: supported by the Geneva Centre for Emerging Viral Diseases Publication status: preprint Source: medRxiv preprint Author COI: the authors declare no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Lambert‐Niclot 2020.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: samples submitted for RT‐PCR testing (n = 138) Recruitment: not stated Prospective or retrospective: unclear; testing conducted prospectively Number of samples (samples with confirmed SARS‐CoV‐2): 138 (94) |
||
| Patient characteristics and setting | Setting: not stated Location: samples collected from virology laboratories of 3 university hospital groups from Assistance‐Publique‐Hôpitaux de Paris (APHP), (Saint‐Antoine‐Tenon‐Trousseau, Saint‐Louis‐Lariboisière and Kremlin Bicêtre‐Paul Brousse) Country: France Dates: 1‐15 April 2020 Symptoms and severity: not stated Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: COVID‐19 Ag Respi‐Strip CORIS (no product code) Manufacturer: BioConcept, Gembloux, Belgium Ag target: SARS‐CoV‐2 NP Antibody: monoclonal antibodies Test method: CGIA Samples used: NP swabs in VTM (collection process not described) Transport media: either of: COPAN UTM 3 mL, Virocult 1 mL, Eswab Amies 1 mL, 4MRT 3 mL, 0.9% NaCl buffer and Cobas ROCHE Sample storage: no cooling or freezing step used Test operator: not stated; presume laboratory staff Definition of test positivity: not stated; as per manufacturer IFU Blinding reported: not stated Timing of samples: not stated; presume on presentation |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR (different kits used including RealStar Altona, Anatolia, Cobas 6800 Roche, Allplex 2019‐nCoV Assay Seegene) Definition of non‐COVID cases: single negative PCR Genetic target(s): E gene Samples used: NP swabs (same as for index) Timing of reference standard: within a few hours after collection; time pso not reported Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same sample, both tests conducted within a few hours All participants received same reference standard: yes (different kits) Missing data: none reported Uninterpretable results: 4 samples collected in Cobas VTM gave invalid results and all samples in Cobas medium were excluded Indeterminate results (index test): control lines reported as "barely visible" for 9 positive and 8 negative tests Indeterminate results (reference standard): none reported Unit of analysis: not reported, but samples tested on day of collection so considered to be 1 per participant |
||
| Comparative | |||
| Notes | Funding: no funding sources reported Publication status: accepted manuscript Source: Journal of Clinical Microbioloby Author COI: no conflict of interest statement reported |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Lanser 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity alone
A group of patients with PCR‐confirmed COVID‐19 during their hospital stay in different stages of the disease. All patients received Ag test along with RT‐PCR (n = 53)
2 patients with negative PCR were excluded due to apparent subsidence of infection Recruitment: unclear Prospective or retrospective: retrospective Sample size (cases): 53 (100%) included; 51 analysed 2 COVID patients were RT‐PCR−ve suggesting an already subsided infection and were excluded from the analysis |
||
| Patient characteristics and setting | Setting: hospital inpatient Location: Department of Internal Medicine II, Innsbruck Medical University, Innsbruck Country: Austria Dates: not stated Symptoms and severity: in different stages of the COVID infection Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: Panbio COVID‐19 Ag Rapid test Manufacturer: Abbott, Chicago, Illionis, USA Antibody: not stated Ag target: not stated Test method: not stated Samples used: NP Transport media: none required Sample storage: none required Test operator: not stated; probably HCW Definition of test positivity: not stated; visual Blinding reported: no; conducted before RT‐PCR but all had previously confirmed COVID‐19 Timing of samples: unclear; states "sample taken during their hospital stay in different stages of the disease" |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR using the Cobas analyzer (Roche Diagnostics GmbH, Mannheim, Germany) Definition of non‐COVID cases: NA Genetic target(s): Orf1 Samples used: NP Timing of reference standard: unclear; states "sample taken during their hospital stay in different stages of the disease" Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous All participants received same reference standard: yes Missing data: yes, 2/53 COVID patients were RT‐PCR−ve suggesting an already subsided infection and were excluded from the analysis Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: none Publication status: published Source: Infection Author COI: the authors declare no conflict of interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Linares 2020.
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity and specificity, recruiting at 2 locations:
[1] symptomatic patients admitted to ED with clinical suspicion of COVID‐19 (n = 135) or asymptomatic patients with history of contact with another COVID‐19 patient (n = 17)
[2] symptomatic patients (n = 50) or asymptomatic (n = 55) patients attending 1 of 2 primary healthcare centres Recruitment: not stated Prospective or retrospective: unclear; appears to be prospective |
||
| Patient characteristics and setting | Setting: mixed; ED or primary care Location: Hospital Universitario Príncipe de Asturias, Madrid Country: Spain Dates: 10‐15 September Symptoms and severity: 185, 72% symptomatic; 72, 28% asymptomatic ED (n = 135): fever 40, dyspnoea 42, cough 22, headache 14 Primary care (n = 50): fever 14, dyspnoea 1, cough 18, headache 17 Demographics: mean age (range): ED 51.5 years (37.0‐71.8 years); primary care 39.0 years (25.0‐56.0 years) Male: ED 77 (51%), primary care 49 (47%) Exposure history: not stated |
||
| Index tests | Test name: PanBio COVID‐19 Ag Rapid Test Device (no product code) Manufacturer: Abbott Rapid Diagnostic Jena GmbH, Jena, Germany Antibody: nucleocapsid Ag target: not stated Test method: not stated; qualitative membrane‐based immunoassay (immunochromatography) Samples used: NP; HCW obtained Transport media: none reported Sample storage: not stated Test operator: not stated Definition of test positivity: not stated; as per manufacturer IFU Blinding reported: not stated Timing of samples: ED: 2 days pso (IQR? 1‐5) PC: 4 days pso (IQR? 2‐8) Table 3 reports range of 0‐27 days pso or post COVID‐19 contact, and range of 0‐16 days for days pso for symptomatic cases only |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; Allplex SARS‐CoV‐2 assay (Seegene, Seoul, South Korea); appears to be < 40 Ct threshold Definition of non‐COVID cases: as for cases (single ‐ve) Genetic target(s): not stated Samples used: NP (paired) Timing of reference standard: not stated Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired All participants received same reference standard: yes Missing data: none reported however 257 reported in Methods and 255 in Results, no participant flow diagram reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: no funding statement provided Publication status: preprint Source: medRxiv Author COI: no COI statement provided |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Lindner 2021a [A].
| Study characteristics | |||
| Patient Sampling | A single‐group study to estimate sensitivity and specificity
Mainly symptomatic adults at high risk for SARS‐CoV‐2 infection according to clinical suspicion of COVID‐19 Recruitment: consecutive Prospective or retrospective: prospective Sample size (cases): 289 (39) |
||
| Patient characteristics and setting | Setting: COVID‐19 test centre Location: ambulatory SARS‐CoV‐2 testing facility of Charité University Hospital, Berlin (Division of Clinical Tropical Medicine, Center of Infectious Diseases, Heidelberg University Hospital, Heidelberg) Country: Germany Dates: 23 September‐14 October2020 Symptoms and severity: on day of testing: 283 (98%) symptomatic; 6 (2%) asymptomatic; average symptom duration 4.4 days (SD 2.7) Demographics: average age 34.7± 11 years; 42.9% female and 19.0% with comorbidities Exposure history: not stated |
||
| Index tests | Test name: STANDARD Q COVID‐19 Ag Test Manufacturer: SD Biosensor, Inc. Gyeonggi‐do, Korea; (also being distributed by Roche) Antibody: not stated Ag target: not stated Test method: LFA chromatographic Samples used: [A] AN; instructed, self‐collected (Verbal instruction was given to insert the swab horizontally 2‐3 cm into the nostril and rotate it for 15 s against the nasal walls on each side. Deviations from the instructed technique were recorded.) [B] NP; collected by study physicians Transport media: none required Sample storage: none required Test operator: study physicians Definition of test positivity: visual; presence of control test lines, categorized as negative, weak positive, positive and strong positive Results interpreted by 2 operators, each blinded to the result of the other. The second reader was also blinded to the result from the alternative sampling method Blinding reported: yes; states "Staff performing the Ag‐RDTs were blinded to results of PCR tests and vice versa." Timing of samples: average symptom duration 4.4 days (SD 2.7) (range 1‐14 days for PCR+ group) |
||
| Target condition and reference standard(s) | Reference standard: PCR using Roche Cobas SARS‐CoV‐2 assay (Pleasanton, CA USA) or the SARS‐CoV‐2 E‐gene assay from TibMolbiol (Berlin, Germany) Definition of non‐COVID cases: single negative PCR Genetic target(s): E‐gene Samples used: OP+NP combined swab Timing of reference standard: as for index test Blinded to index test: yes; states "Staff performing the Ag‐RDTs were blinded to results of PCR tests and vice versa." Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous All participants received same reference standard: yes Missing data: yes; 2 excluded as both swabs for the Ag test could not be obtained Uninterpretable results: none reported Indeterminate results (index test): none; "no invalid tests were observed" Indeterminate results (reference standard): none; "no invalid tests were observed" Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "supported by FIND, Heidelberg University Hospital and Charité University Hospital internal funds, Ministry of Science, Research and the Arts of Baden‐Württemberg, Germany" Publication status: in press (published as in accepted form) Source: European Respiratory Journal Author COI: authors report no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Lindner 2021a [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of an Ag test on 2 different sample types; Lindner 2021a [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of an Ag test on 2 different sample types; Lindner 2021a [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: STANDARD Q COVID‐19 Ag Test Manufacturer: SD Biosensor, Inc. Gyeonggi‐do, Korea; (also being distributed by Roche) Antibody: not stated Ag target: not stated Test method: LFA Chromatographic Samples used: [A] AN; instructed, self‐collected (Verbal instruction was given to insert the swab horizontally 2‐3 cm into the nostril and rotate it for 15 seconds against the nasal walls on each side. Deviations from the instructed technique were recorded.) [B] NP; collected by study physicians Transport media: none required Sample storage: none required Test operator: study physicians Definition of test positivity: visual; presence of control test lines, categorized as negative, weak positive, positive and strong positive Results interpreted by 2 operators, each blinded to the result of the other. The second reader was also blinded to the result from the alternative sampling method Blinding reported: yes; States "Staff performing the Ag‐RDTs were blinded to results of RT‐PCR tests and vice versa." Timing of samples: average symptom duration 4.4 days (SD 2.7) (range 1‐14 days for PCR+ group) |
||
| Target condition and reference standard(s) | Comparative study of an Ag test on 2 different sample types; Lindner 2021a [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of an Ag test on 2 different sample types; Lindner 2021a [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of an Ag test on 2 different sample types; Lindner 2021a [A] reports full study characteristics and QUADAS. | ||
Lindner 2021b [A].
| Study characteristics | |||
| Patient Sampling | A single‐group study to estimate sensitivity and specificity
Symptomatic adults with high clinical suspicion of SARS‐CoV‐2 including 1) reported contact with a confirmed case and any compatible symptom, or 2) fever or impaired taste or smell irrespective of exposure Recruitment: consecutive Prospective or retrospective: prospective Sample size (cases): 146 (40) |
||
| Patient characteristics and setting | Setting: COVID‐19 test centre Location: ambulatory SARS‐CoV‐2 testing facility of Charité University Hospital, Berlin Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin, Humboldt‐Universität zu Berlin, and Berlin Institute of Health; Institute of Tropical Medicine and International Health, Berlin Country: Germany Dates: 30 November‐11 December 2020 (Recruitment dates do not overlap either FIND 2021c (DE) [A] (has associated preprint by Lindner et al) or Lindner 2021a [A]) Symptoms and severity: 100% symptomatic; mean duration of pso 3.4 days (SD 2.0) 34 (23%) with comorbidities Demographics: mean age 35 years (SD 11.5); 75 (51%) were female Exposure history: included those with reported contact with a confirmed case. No further details |
||
| Index tests | Test name: STANDARD Q COVID‐19 Ag Test Manufacturer: SD Biosensor, Inc. Gyeonggi‐do, Korea; also distributed by Roche Antibody: not stated Ag target: not stated Test method: CGIA Samples used: [A] NMT (self‐collected and interpreted; according to manufacturer IFU) [B] NMT (self‐collected, professional interpreted; according to manufacturer IFU) [C] NP (professional collected and interpreted ‐ trained study physician) Transport media: none required Sample storage: none required Test operator: [A] participants tested NMT sample (according to manufacturer IFU); observed without answering questions or providing corrections [B] Participants tested NMT sample; interpretation by study physician [C] Trained study physician tested and interpreted NP sample All professional test interpretation was by 2 study physicians, each blinded to the result of the other and to the participant's interpretation of the self‐test. The second reader was also blinded to the corresponding pairs (NMT/NP) of Ag‐RDTs belonging to 1 individual. Definition of test positivity: visual The visual read‐out of the Ag test band was categorized as negative, weak positive, positive, or strong positive. The participant interpreted the test result as positive, negative, invalid, or don’t know. Blinding reported: yes; conducted first Timing of samples: mean duration of pso 3.4 days (SD 2.0) |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR using the Roche Cobas SARS‐CoV‐2 assay (Pleasanton, CA USA) or the SARS‐CoV‐2 E‐gene assay from TIB Molbiol (Berlin, Germany) Definition of non‐COVID cases: same as cases (single ‐ve PCR) Genetic target(s): E gene (Tib Molbiol) Samples used: combined OP/NP; paired swab Timing of reference standard: mean duration of pso 3.4 days (SD 2.0) Blinded to index test: yes; explicitly stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous All participants received same reference standard: yes Missing data: yes; 4 excluded (3 participants were excluded as they did not fulfil the minimum language criterion and 1 participant excluded because of lost PCR specimen) Uninterpretable results: NMT ‐ 1 participant left without reading the test result for nasal sample plus 1 invalid result (buffer spilt and test not repeated) None reported for NP sample Indeterminate results (index test): none reported; weak positives considered positive Indeterminate results (reference standard): none reported |
||
| Comparative | |||
| Notes | Funding: "study was supported by Foundation of Innovative New Diagnostics (FIND), Charité University Hospital internal funds, as well as a grant of the Ministry of Science, Research and the Arts of Baden‐Württemberg, Germany" Publication status: preprint Source: medRxiv Author COI: one author reports grants from FIND and Ministry of Science, Research and Culture, State of Baden Wuerttemberg, Germany. Another author reports grants from DFID (recently replaced byFCMO), WHO and from Unitaid. |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Lindner 2021b [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 3 different sample types (self versus professional); Lindner 2021b [A] details full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 3 different sample type (self versus professional); Lindner 2021b [A] details full study characteristics and QUADAS. | ||
| Index tests | Test name: STANDARD Q COVID‐19 Ag Test Manufacturer: SD Biosensor, Inc. Gyeonggi‐do, Korea; also distributed by Roche Antibody: not stated Ag target: not stated Test method: CGIA Samples used: [A] NMT (self‐collected and interpreted; according to manufacturer IFU) [B] NMT (self‐collected, professional interpreted; according to manufacturer IFU) [C] NP (professional collected and interpreted ‐ trained study physician) Transport media: none required Sample storage: none required Test operator: [A] participants tested NMT sample (according to manufacturer IFU); observed without answering questions or providing corrections [B] Participants tested NMT sample; interpretation by study physician [C] Trained study physician tested and interpreted NP sample All professional test interpretation was by 2 study physicians, each blinded to the result of the other and to the participant's interpretation of the self‐test. The second reader was also blinded to the corresponding pairs (NMT/NP) of Ag‐RDTs belonging to 1 individual. Definition of test positivity: visual The visual read‐out of the Ag test band was categorized as negative, weak positive, positive, or strong positive. The participant interpreted the test result as positive, negative, invalid, or don’t know. Blinding reported: yes; conducted first Timing of samples: mean duration of pso 3.4 days (SD 2.0) |
||
| Target condition and reference standard(s) | Comparative study of 3 different sample type (self versus professional); Lindner 2021b [A] details full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 3 different sample type (self versus professional); Lindner 2021b [A] details full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 3 different sample type (self versus professional); Lindner 2021b [A] details full study characteristics and QUADAS. | ||
Lindner 2021b [C].
| Study characteristics | |||
| Patient Sampling | Comparative study of 3 different sample type (self versus professional); Lindner 2021b [A] details full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 3 different sample type (self versus professional); Lindner 2021b [A] details full study characteristics and QUADAS. | ||
| Index tests | Test name: STANDARD Q COVID‐19 Ag Test Manufacturer: SD Biosensor, Inc. Gyeonggi‐do, Korea; also distributed by Roche Antibody: not stated Ag target: not stated Test method: CGIA Samples used: [A] NMT (self‐collected and interpreted; according to manufacturer IFU) [B] NMT (self‐collected, professional interpreted; according to manufacturer IFU) [C] NP (professional collected and interpreted ‐ trained study physician) Transport media: none required Sample storage: none required Test operator: [A] participants tested NMT sample (according to manufacturer IFU); observed without answering questions or providing corrections [B] Participants tested NMT sample; interpretation by study physician [C] Trained study physician tested and interpreted NP sample All professional test interpretation was by 2 study physicians, each blinded to the result of the other and to the participant's interpretation of the self‐test. The second reader was also blinded to the corresponding pairs (NMT/NP) of Ag‐RDTs belonging to 1 individual. Definition of test positivity: visual The visual read‐out of the Ag test band was categorized as negative, weak positive, positive, or strong positive. The participant interpreted the test result as positive, negative, invalid, or don’t know. Blinding reported: yes; conducted first Timing of samples: mean duration of pso 3.4 days (SD 2.0) |
||
| Target condition and reference standard(s) | Comparative study of 3 different sample type (self versus professional); Lindner 2021b [A] details full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 3 different sample type (self versus professional); Lindner 2021b [A] details full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 3 different sample type (self versus professional); Lindner 2021b [A] details full study characteristics and QUADAS. | ||
Liotti 2021.
| Study characteristics | |||
| Patient Sampling | Unclear design estimating sensitivity and specificity; residual samples selected from 1 of 2 virology laboratories at 2 COVID‐19 reference hospitals:
[1] RT‐PCR+ve for SARS‐CoV‐2 (n = 104)
[2] RT‐PCR−ve for SARS‐CoV‐2 (n = 255) Recruitment: not stated Prospective or retrospective: retrospective |
||
| Patient characteristics and setting | Setting: unclear; laboratory samples Location: from study authors' institutions: Fondazione Policlinico Universitario A. Gemelli IRCCS, and Istituto Nazionale per le Malattie Infettive (INMI) Lazzaro Spallanzani IRCCS, Rome Country: Italy Dates: not stated Symptoms and severity: not stated Of SARS‐CoV‐2‐positive samples, 21, 20% high viral load (< 25 Ct), 83, 80% low viral load (≥ 25) [28, 27% with Ct ≥ 35) Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: STANDARD F COVID‐19 Ag FIA (no product codes reported) Manufacturer: SD Biosensor (Suwon, South Korea) Antibody: NP Ag target: monoclonal anti‐SARS‐CoV‐2 antibody Test method: FIA Samples used: NP; collection not reported Transport media: not stated Sample storage: performed within 24 h after collection on samples kept at 4 °C until testing Test operator: not stated; presume laboratory staff Definition of test positivity: as per manufacturer IFU Blinding reported: not stated Timing of samples: not reported |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR (1 of 4 assays); Altona Diagnostics RealStar SARS‐CoV‐2 RT‐PCR, the Seegene Allplex 2019‐nCoV, the DiaSorin SimplexaCOVID‐19 Direct or the Roche Diagnostics Cobas SARS‐CoV‐2 test Definition of non‐COVID cases: as for cases (single negative) Genetic target(s): not stated Samples used: NP (same as index) Timing of reference standard: not stated Blinded to index test: yes (performed first) Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous (same swab) All participants received same reference standard: yes Missing data: none reported, no participant flow diagram reported Uninterpretable results: none reported Indeterminate results (index test): none reported FP results were re‐tested with Ag assay, 3 of 4 remained positive (all blood contaminated) Indeterminate results (reference standard): none reported Unit of analysis: not stated |
||
| Comparative | |||
| Notes | Funding: "study supported by funds to the Istituto Nazionale per le Malattie Infettive (INMI) Lazzaro Spallanzani IRCCS, Rome, Italy, from the Ministero della Salute (Ricerca Corrente, linea 1; COVID‐ 2020‐12371817), the European Commission e Horizon 2020 (EU project 101003544 e CoNVat; EU project 101003551 e EXSCALATE4CoV; EU project 12371675 e EXCALATE4CoV; EU project 101005075 e KRONO) and the European Virus Archive e GLOBAL (grants no. 653316 and no. 871029)." Publication status: published letter Source: Clinical Microbiology and Infection Author COI: all authors report no relevant conflicts of interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Love 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity and specificity Asymptomatic adult contacts (> 18 years) of a confirmed COVID‐19 case (in previous 48 h) identified via NHS Test and Trace and invited to participate. Negative LFD results allowed exemption from self‐isolation for a 24‐h period until next LFD. Recruitment: consecutive Prospective or retrospective: prospective Exclusions included exposure to confirmed case > 48 h (n = 68) or late or non‐arrival of testing kits (n = 27) Sample size (cases): 882/1760 (50.1%) contacts agreed to participate; 812/882 were sent a testing kit. In‐study PCR results available for 346 contacts (64 PCR+) |
||
| Patient characteristics and setting | Setting: contacts; self‐testing Location: UK Country: UK Dates: 11‐23 December 2020 and 4‐12 January 2021 Symptoms and severity: 55/346 self‐reported symptoms in prior 14 d (15.9%) Demographics: based on 882 consenting ‐ mean age 42 years (18‐82 years); 430, 49% male; 731 (89%) white, 33 Asian, 19 black, 25 mixed race, 14 other Exposure history: all confirmed contacts |
||
| Index tests | Test: Innova LFD antigen test Manufacturer: Innova Medical Group Ag target: not reported Test method: CGIA Samples used: nasal (self‐collected) Test operator: self‐tested. Convenience sample of 1221 LFD images were checked by 2 independent reviewers; 97.1% of images were concordant (n = 1187; 1132 negative and 55 positive results) Definition of test positivity: visual; used according to the manufacturer IFU Blinding reported: yes (performed before reference standard) Timing of samples: all asymptomatic on recruitment and tested during the first 7‐days post‐exposure |
||
| Target condition and reference standard(s) | Reference standard: LDT at PHE‐accredited laboratory; NHS Test and Trace records also examined to identify confirmatory testing through an approved alternative route Definition of non‐COVID cases: single ‐ve PCR Genetic target(s): ORF1ab, E gene Samples used: nasal (self‐collected) Timing of reference standard: median time between reporting a positive LFD result and receipt of PCR swab in the laboratory was 2 days (IQR 1‐3 days). Median time between last negative LFD result and the swab being received at the lab was 3 days (IQR 2‐5 days). Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: any interval allowed; LFD‐negative participants only had PCR at end of study period All participants received same reference standard: yes (if limit to within‐study PCR testing) Missing data: 570/812 who were sent a testing kit returned at least 1 LFD result (total of 2946 results); results excluded if duplicate entries (n = 225), blank entries (n = 189), no identifiers (n = 27), or reporting by non‐eligible participant (n = 13) Uninterpretable results: invalid results mentioned but not quantified (grouped with negatives) Indeterminate results (index test): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "DR and IO acknowledge support from the NIHR Health Protection Research Unit in Behavioural Science and Evaluation at University of Bristol. SH is supported by the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Healthcare Associated Infections and Antimicrobial Resistance at the University of Oxford in partnership with Public Health England (PHE)." Publication status: preprint Source: medRxiv Author COI: no conflicts of interest statement provided but is PHE (public) funded study |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | No | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Masia 2021 [A].
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: symptomatic with COVID‐19 signs/symptoms or asymptomatic contacts attending the primary care centres (n = 690 (76%) NP sample), and a majority of symptomatic patients presenting to the ED (n = 233 (25%) NP sample) Recruitment: consecutive Prospective or retrospective: prospective Sample size (cases): [A] NP: 904 (195) [B] Nasal: 659 (132) [C] Saliva: 611 (121) |
||
| Patient characteristics and setting | Setting: mixed: primary care centres and an ED Location: Alicante, Spain (Universidad Miguel Hernández) Country: Spain Dates: 15 September‐29 October 2020 Symptoms and severity: 617 (68%) symptomatic; 296 (32%) asymptomatic Median (Q1–Q3) pso days: 3 (2–5) days Most frequent symptoms were cough (50%), fever (47%), sore throat (32%), and nasal congestion (31%) Median (Q1–Q3) Ct: 24 (16–30) 22 (16–29) in symptomatic and 28 (21–32) in asymptomatic; and 21 (15–27) in patients ≥ 50 years and 26 (18–31) in < 50 years. Demographics: median (Q1–Q3) age 40.6 (23.0–55.6) years; 423 (46%) male Exposure history: not reported |
||
| Index tests | Test name: Panbio COVID‐19 Ag RTD Manufacturer: Abbott Rapid Diagnostic Jena GmbH, Jena, Germany Antibody: nucleocapsid Ag target: not reported Test method: CGIA Samples used at primary care centres: [A] NP swabs (1 swab for each nostril) (qualified nurse) [B] nasal swab (from 1 nostril) (qualified nurse) [C] saliva (patients repeatedly spit up to a minimum of 1 mL of saliva into a 100‐mL sterile empty container) (self‐collected) Samples used at the ED: [A] NP swab (1 swab for each nostril) (obtained by a clinician). Transport media: none reported Sample storage: none required Test operator: at primary care centres ‐ qualified nurse; at ED ‐ clinician Definition of test positivity: visual; used according to the manufacturer IFU Blinding reported: yes (performed before reference standard) Timing of samples: median 3 (Q1 2–Q3 5) days after symptom onset |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR performed using manufacturer IFU on Cobas z 480 Analyser (Roche, Basilea, Suiza). Nucleic acid extraction was performed using 300 μL NP specimen on Chemagic 360 Nucleic Acid Purification Instrument (PerkinElmer España SL, Madrid, Spain) Definition of non‐COVID cases: single ‐ve PCR Genetic target(s): E‐gene Samples used: NP Timing of reference standard: median 3 (Q1 2–Q3 5) days pso Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired sample All participants received same reference standard: yes Missing data: yes; of 913 only 904 had NP (< 1%), 659 had nasal and 611 had saliva Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "this work was supported by the RD16/0025/0038 project as a part of the Plan Nacional Research + Development + Innovation (R+D+I) and co‐financed by Instituto de Salud Carlos III ‐ SubdirecciónGeneral de Evaluación y Fondo Europeo de Desarrollo Regional; Instituto de Salud Carlos III (Fondo de Investigaciones Sanitarias (Grant NumbersPI16/01740, PI18/01861; CM 19/00160, COV20‐00005)." Publication status: published Source: Open Forum Infectious Diseases Author COI: authors reported no conflicts of interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Masia 2021 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of an Ag test on 3 different sample types; Masia 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of an Ag test on 3 different sample types; Masia 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: Panbio COVID‐19 Ag RTD Manufacturer: Abbott Rapid Diagnostic Jena GmbH, Jena, Germany Antibody: nucleocapsid Ag target: not reported Test method: CGIA Samples used at primary care centres: [A] NP swabs (1 swab for each nostril) (qualified nurse) [B] nasal swab (from 1 nostril) (qualified nurse) [C] saliva (patients repeatedly spit up to a minimum of 1 mL of saliva into a 100‐mL sterile empty container) (self‐collected) Samples used at the ED: [A] NP swab (1 swab for each nostril) (obtained by a clinician) Transport media: none reported Sample storage: none required Test operator: at primary care centres ‐ qualified nurse; at ED ‐ clinician Definition of test positivity: visual; used according to the manufacturer IFU Blinding reported: yes (performed before reference standard) Timing of samples: median 3 (Q1 2–Q3 5) days after symptom onset |
||
| Target condition and reference standard(s) | Comparative study of an Ag test on 3 different sample types; Masia 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of an Ag test on 3 different sample types; Masia 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of an Ag test on 3 different sample types; Masia 2021 [A] reports full study characteristics and QUADAS. | ||
Masia 2021 [C].
| Study characteristics | |||
| Patient Sampling | Comparative study of an Ag test on 3 different sample types; Masia 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of an Ag test on 3 different sample types; Masia 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: Panbio COVID‐19 Ag RTD Manufacturer: Abbott Rapid Diagnostic Jena GmbH, Jena, Germany Antibody: nucleocapsid Ag target: not reported Test method: CGIA Samples used at primary care centres: [A] NP swabs (1 swab for each nostril) (qualified nurse) [B] nasal swab (from 1 nostril) (qualified nurse) [C] saliva (patients repeatedly spit up to a minimum of 1 mL of saliva into a 100‐mL sterile empty container) (self‐collected) Samples used at the ED: [A] NP swab (1 swab for each nostril) (obtained by a clinician) Transport media: none reported Sample storage: none required Test operator: at primary care centres‐ qualified nurse; at ED ‐ clinician Definition of test positivity: visual; used according to the manufacturer IFU Blinding reported: yes (performed before reference standard) Timing of samples: median 3 (Q1 2– Q3 5) days after symptom onset |
||
| Target condition and reference standard(s) | Comparative study of an Ag test on 3 different sample types; Masia 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of an Ag test on 3 different sample types; Masia 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of an Ag test on 3 different sample types; Masia 2021 [A] reports full study characteristics and QUADAS. | ||
Merino 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: multicentre study of individuals who had at least 1 symptom compatible with COVID‐19 (n = 830) or had been in close contact with a diagnosed COVID‐19 patient (n = 128) (total n = 958); all tested within 7 days of symptom onset or exposure Recruitment: consecutive Prospective or retrospective: prospective Sample size (cases): 958 (359) |
||
| Patient characteristics and setting | Setting: hospital EDs or other hospital units (documented only in preprint version) Location: Madrid: Hospital Clínico Universitario San Carlos, Hospital Universitario Ramón y Cajal, Hospital Universitario La Paz, Hospital Universitario Doce de Octubre, and Hospital Universitario Gregorio Marañón. Basque: Hospital Universitario Araba, Hospital Universitario Cruces, Hospital Universitario Basurto, Hospital Universitario Donostia, and Hospital Universitario Galdakao Usansolo Country: Spain Dates: September‐October 2020 Symptoms and severity: symptomatic 830, 87%; all had at least 1 symptom compatible with COVID‐19 Demographics: mean age of 42.4 years (range, 1‐100 years); 61.3% were women Exposure history: 128 asymptomatic participants had had close contact with COVID‐19 patient |
||
| Index tests | Test name: PanBio RT COVID‐19 Ag Rapid Test Device Manufacturer: Abbott Diagnostics Antibody: nucleocapsid protein Ag target: not reported Test method: CGIA Samples used: NP Transport media: none Sample storage: immediate Test operator: trained personnel; physicians and nurses from emergency services trained by microbiology specialists Definition of test positivity: according to manufacturer IFU Blinding reported: yes; conducted first Timing of samples: < 7 days pso or COVID exposure; unclear if collected by HCW or self‐collected |
||
| Target condition and reference standard(s) | Reference standard: PCR; multiple assays used across 10 sites Definition of non‐COVID cases: single negative PCR Genetic target(s): not mentioned Samples used: NP; paired Timing of reference standard: same as for index Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: taken at the same time All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: no external funding was received for this work Publication status: published Source: Clinical Microbiology and Infection Author COI: "RC has participated in educational programmes organized by Abbott. The other authors declare that they have no conflicts of interest." |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Mertens 2020.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity for diagnosis of active disease: samples from symptomatic patients suspected of SARS‐COV‐2 infections (n = 328) Recruitment: random sampling of samples submitted to 3 laboratories 322/328 NP samples (NP swabs) were randomly selected Prospective or retrospective: retrospective Number of samples (samples with confirmed SARS‐CoV‐2): 328 (132) |
||
| Patient characteristics and setting | Setting: unclear; samples from university laboratories (Discussion states that no outpatient population has been sampled, therefore assume inpatients and HCW samples) Location: laboratories at Université Libre de Bruxelles (LHUB‐ULB), UZ Leuven and Centre Hospitalier Universitaire Sart‐Tilman (CHU) Liège Country: Belgium Dates: 19‐30 March 2020 Symptoms and severity: all described as symptomatic Demographics: not reported Exposure history: unclear; 53/328 samples were from HCW |
||
| Index tests | Test name: COVID‐19 Ag Respi‐Strip Manufacturer: Coris BioConcept (Belgium) Ag target: SARS‐CoV and SARS‐CoV‐2 highly conserved nucleoprotein Antibody: monoclonal antibodies directed against SARS‐CoV and SARS‐CoV‐2 highly conserved nucleoprotein antigen Test method: immunochromatographic assay using colloidal gold (CGIA) Samples used: remnant respiratory specimens (322 NP swabs, 4 NP aspirate and 2 BAL) Transport media: NP: flocked swab + UTM 3 mL (or 1 mL of Amies) (Copan, Brescia, Italy); NPA: 3 mL VTM (veal infusion broth (Difco, Becton Dickinson, Sparks, MD, USA) supplemented with bovine albumin (Sigma Aldrich, St Louis, MO, USA)) BAL: N/A Sample storage: not described Test operator: laboratory technician Definition of test positivity: visible reddish‐purple band appearing at the test line position (T) Blinding reported: not stated Timing of samples: not clear |
||
| Target condition and reference standard(s) | Reference standard: qRT‐PCR: RealStar SARS‐CoV‐2 RT‐PCR Kit from Altona‐diagnostics with a cut‐off set at 40 Ct (LHUB‐ULB); Roche LC480 thermocycler using Taqman Fast Virus 1‐Step Master Mix (Thermo Fisher) (Liege); QuantStudio Dx (Thermo Fisher Scientific) or Panther Fusion (PF, Hologic, San Diego, USA) (UZ Leuven) Definition of non‐COVID cases:
Samples used: as for index test (NP samples) Timing of reference standard: not stated; same samples as for index test but analysed at time of collection Blinded to index test: yes (undertaken for diagnostic purposes at time of collection) Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same samples used; Discussion reports "some delay" between PCR and Ag testing All participants received same reference standard: yes but different RT‐PCR kits Missing data: none reported, no participant flow diagram reported Uninterpretable results: none reported; discussion reports some difficulties in visualising the strip through the closed tube requiring the lab technician to open the test tube in the laminar air flow cabinet and pull out the strip with forceps Indeterminate results (index test): weak T lines considered positive Indeterminate results (reference standard): none reported Unit of analysis: refers to participants |
||
| Comparative | |||
| Notes | Funding: not stated Publication status: preprint (not peer‐reviewed) Source: medRxiv Author COI: "the IVD medical device has been developed by the investigator Pascal Mertens, Henri Magein, and Justine Bouzet working for Coris BioConcept (potential conflict of interest declared even though they don’t have any share in this company); Thierry Leclipteux was involved in the development of this test and is the CEO of Coris Bioconcept (potential conflict of interest declared). All scientific investigators that are external to Coris BioConcept declare having no conflict of interest." |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Miyakawa 2021 [A].
| Study characteristics | |||
| Patient Sampling | 2‐group study estimating sensitivity and specificity:
[1] remnant RT‐PCR+ve NP swab specimens (n = 45)
[2] RT‐PCR−ve NP swabs (n = 63; only 45/63 samples tested with 4 of the 5 evaluated assays) Recruitment: not reported Prospective or retrospective: retrospectively Sample size (cases): 108 (45) |
||
| Patient characteristics and setting | Setting: not reported (laboratory‐based at a biosafety level 3 laboratory) Location: Yokohama City University School of Medicine Country: Japan Dates: not reported Symptoms and severity: not reported Demographics: not reported Exposure history: not reported |
||
| Index tests | Test name: [A] authors' own‐developed Ag‐RDT (YCU‐FF LFIA (Ag‐RDT); now marketed as FUJIFILM COVID‐19 Ag Test www.fujifilm.com/jp/en/news/hq/358e) [B] Panbio COVID‐19 Ag Rapid Test [C] Espline SARS‐CoV‐2 [D] STANDARD Q COVID‐19 Ag Also evaluated [E] SARS‐CoV‐2 Rapid Antigen Test from Roche. Data not included as this is understood to be the same test as 'STANDARD Q' from SD Biosensor Manufacturer: [A] FUJIFILM [B] Abbott [C] Fujirebio [D] SD Biosensor Antibody: [A] SARS‐CoV‐2 nucleocapsid protein; [B] to [D] not described Ag target: [A] specific monoclonal antibodies against the SARS‐CoV‐2 nucleocapsid protein; [B] to [D] not described Test method: [A] CGIA with silver ions; [B] to [D] not described Samples used: NP (collection not described) Transport media: VTM (no further detail reported) Sample storage: stored at −80 °C until used (timing not reported) Test operator: not reported; "test line interpretations … made by at least two people" Definition of test positivity: appearance of visible line Blinding reported: not stated Timing of samples: not reported |
||
| Target condition and reference standard(s) | Reference standard: RT‐qPCR with N2 primer/probe set targeting the N gene Definition of non‐COVID cases: as for cases; presume single negative Genetic target(s): N gene Samples used: NP Timing of reference standard: not reported Blinded to index test: yes; conducted first Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous (same swab) All participants received same reference standard: yes Missing data: not reported Uninterpretable results: not reported Indeterminate results (index test): not reported Indeterminate results (reference standard): not reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "this work was supported in part by Japan Agency for Medical Research and Development, and by Health Labour Sciences research grant from The Ministry of Health Labour and Welfare to AR" Publication status: preprint (not peer reviewed) Source: medRxiv Author COI: "YY is an employee of Kanto Chemical Co., Inc.; JK, AW, and TT are current employee of FUJIFILM Corporation"; Remaining authors declare that they have no competing interests. |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Miyakawa 2021 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 4 Ag tests; Miyakawa 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 4 Ag tests; Miyakawa 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] authors' own‐developed Ag‐RDT (YCU‐FF LFIA (Ag‐RDT); now marketed as FUJIFILM COVID‐19 Ag Test www.fujifilm.com/jp/en/news/hq/358e) [B] Panbio COVID‐19 Ag Rapid Test [C] Espline SARS‐CoV‐2 [D] STANDARD Q COVID‐19 Ag Also evaluated [E] SARS‐CoV‐2 Rapid Antigen Test from Roche. Data not included as this is understood to be the same test as 'STANDARD Q' from SD Biosensor Manufacturer: [A] FUJIFILM [B] Abbott [C] Fujirebio [D] SD Biosensor Antibody: [A] SARS‐CoV‐2 nucleocapsid protein; [B] to [D] not described Ag target: [A] specific monoclonal antibodies against the SARS‐CoV‐2 nucleocapsid protein; [B] to [D] not described Test method: [A] CGIA with silver ions; [B] to [D] not described Samples used: NP (collection not described) Transport media: VTM (no further detail reported) Sample storage: stored at −80 °C until used (timing not reported) Test operator: not reported; "test line interpretations … made by at least two people" Definition of test positivity: appearance of visible line Blinding reported: not stated Timing of samples: not reported |
||
| Target condition and reference standard(s) | Comparative study of 4 Ag tests; Miyakawa 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 4 Ag tests; Miyakawa 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 4 Ag tests; Miyakawa 2021 [A] reports full study characteristics and QUADAS. | ||
Miyakawa 2021 [C].
| Study characteristics | |||
| Patient Sampling | Comparative study of 4 Ag tests; Miyakawa 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | |||
| Index tests | Test name: [A] authors' own‐developed Ag‐RDT (YCU‐FF LFIA (Ag‐RDT); now marketed as FUJIFILM COVID‐19 Ag Test www.fujifilm.com/jp/en/news/hq/358e) [B] Panbio COVID‐19 Ag Rapid Test [C] Espline SARS‐CoV‐2 [D] STANDARD Q COVID‐19 Ag Also evaluated [E] SARS‐CoV‐2 Rapid Antigen Test from Roche. Data not included as this is understood to be the same test as 'STANDARD Q' from SD Biosensor Manufacturer: [A] FUJIFILM [B] Abbott [C] Fujirebio [D] SD Biosensor Antibody: [A] SARS‐CoV‐2 nucleocapsid protein; [B] to [D] not described Ag target: [A] Specific monoclonal antibodies against the SARS‐CoV‐2 nucleocapsid protein; [B] to [D] not described Test method: [A] CGIA with silver ions; [B] to [D] not described Samples used: NP (collection not described) Transport media: VTM (no further detail reported) Sample storage: stored at −80 °C until used (timing not reported) Test operator: not reported; "test line interpretations … made by at least two people" Definition of test positivity: appearance of visible line Blinding reported: not stated Timing of samples: not reported |
||
| Target condition and reference standard(s) | |||
| Flow and timing | Comparative study of 4 Ag tests; Miyakawa 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 4 Ag tests; Miyakawa 2021 [A] reports full study characteristics and QUADAS. | ||
Miyakawa 2021 [D].
| Study characteristics | |||
| Patient Sampling | Comparative study of 4 Ag tests; Miyakawa 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 4 Ag tests; Miyakawa 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] authors' own‐developed Ag‐RDT (YCU‐FF LFIA (Ag‐RDT); now marketed as FUJIFILM COVID‐19 Ag Test www.fujifilm.com/jp/en/news/hq/358e) [B] Panbio COVID‐19 Ag Rapid Test [C] Espline SARS‐CoV‐2 [D] STANDARD Q COVID‐19 Ag Also evaluated [E] SARS‐CoV‐2 Rapid Antigen Test from Roche. Data not included as this is understood to be the same test as 'STANDARD Q' from SD Biosensor Manufacturer: [A] FUJIFILM [B] Abbott [C] Fujirebio [D] SD Biosensor Antibody: [A] SARS‐CoV‐2 nucleocapsid protein; [B] to [D] not described Ag target: [A] specific monoclonal antibodies against the SARS‐CoV‐2 nucleocapsid protein; [B] to [D] not described Test method: [A] CGIA with silver ions; [B] to [D] not described Samples used: NP (collection not described) Transport media: VTM (no further detail reported) Sample storage: stored at −80 °C until used (timing not reported) Test operator: not reported; "test line interpretations … made by at least two people" Definition of test positivity: appearance of visible line Blinding reported: not stated Timing of samples: not reported |
||
| Target condition and reference standard(s) | Comparative study of 4 Ag tests; Miyakawa 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 4 Ag tests; Miyakawa 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 4 Ag tests; Miyakawa 2021 [A] reports full study characteristics and QUADAS. | ||
Mockel 2021(a).
| Study characteristics | |||
| Patient Sampling | Single‐group sensitivity and specificity study: patients attending hospital EDs: [1] 4 adult EDs (n = 281); [2] 1 paediatric ED (n = 202)
We included cohort [2] as Mockel 2021(b).
In both cohorts patients were either symptomatic (acute respiratory symptoms or loss of smell or taste), contacts of confirmed cases up to 14 d before onset of COVID‐19 symptoms, or had clinical or radiological signs of viral pneumonia in the context of an outbreak in nursing homes or hospitals. Recruitment: consecutive Prospective or retrospective: prospective Sample size (cases): 281 (89) |
||
| Patient characteristics and setting | Setting: hospital EDs:
[1] adult, [2] paediatric Location: Charité ‐ Universitätsmedizin Berlin, Department of Emergency and Acute Medicine Country: Germany Dates: 12 October‐24 November 2020 Symptoms and severity: cohort [1] only; respiratory symptoms: 157 (579%); loss of smell or taste: 18 (6.6%); contact to confirmed COVID‐19 case: 33 (12.2%); radiological signs of viral pneumonia: 11 (0.4%); other symptoms: 140 (51.7%); none: 56 (20.7%) Demographics: age mean 59.7 years; SD 18; range 21‐98 years Male 160 (59%) Exposure history: not reported |
||
| Index tests | Test name: SARS‐CoV‐2 Rapid Antigen Test Manufacturer: Roche /SD Biosensor Antibody: not mentioned Ag target: not mentioned Test method: not mentioned Samples used: NOP (taken by ED nurse) Transport media: not mentioned Sample storage: immediate Test operator: ED nurse (a core ED team alongside written instructions trained the ED nurses) Definition of test positivity: consensus of ED nurse and 1 other medical professional Blinding reported: yes (assumed done first) Timing of samples: not mentioned; time pso only reported for those with FN results: both cohorts' range was 1 to > 7 days |
||
| Target condition and reference standard(s) | Reference standard: PCR; Roche Cobas SARS‐CoV‐2 assay (Penzberg, Germany) or SARS‐CoV‐2 E‐gene assay from TibMolbiol (Berlin, Germany) Definition of non‐COVID cases: single negative PCR Genetic target(s): TibMolbiol was E gene Samples used: NOP (paired) Timing of reference standard: as for index Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous (paired) All participants received same reference standard: yes Missing data: 10 patients excluded from cohort [1] based on reasons below Uninterpretable results: no index test result (n = 6), no PCR result (n = 2) Indeterminate results (index test): 1 inconclusive (excluded) Indeterminate results (reference standard): 1 inconclusive (excluded) Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "work is based on research funded in part by the German Federal Ministry of Education and Research through projects VARIPath (01KI2021) to VMC and NaFoUniMedCovid19 (BFAST, FKZ: 01KX2021) to the Charité" Publication status: accepted manuscript Source: Biomarkers Author COI: the authors report no conflicts of interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Mockel 2021(b).
| Study characteristics | |||
| Patient Sampling | Single‐group sensitivity and specificity study: participants were symptomatic patients attending hospital EDs, [1] 4 adult EDs (n = 271) and [2] 1 paediatric ED (n = 202)
We included cohort [1] as Mockel 2021(a).
In both cohorts patients were either symptomatic (acute respiratory symptoms or loss of smell or taste), contacts of confirmed cases up to 14 d before onset of COVID‐19 symptoms, or had clinical or radiological signs of viral pneumonia in the context of an outbreak in nursing homes or hospitals. Recruitment: consecutive Prospective or retrospective: prospective Sample size (cases): 202 (25) |
||
| Patient characteristics and setting | Setting: hospital EDs:
[1] adult, [2] paediatric Location: Charité ‐ Universitätsmedizin Berlin, Department of Emergency and Acute Medicine Country: Germany Dates: 12 October‐24 November 2020 Symptoms and severity: respiratory symptoms: [2] 120 (59.4%); loss of smell or taste: [2] 1 (0.5%); contact to confirmed COVID‐19 case: [2] 37 (18.3%); radiological signs of viral pneumonia: [2] 10 (0.5%); other symptoms: [2] 104 (51.5%); none: [2] 26 (12.9%) Demographics: age mean = 3; [range 1‐9] Male 111 (55%) Exposure history: not reported |
||
| Index tests | Test name: SARS‐CoV‐2 Rapid Antigen Test Manufacturer: Roche/SD Biosensor Antibody: not mentioned Ag target: not mentioned Test method: not mentioned Samples used: NOP (taken by ED nurse) Transport media: not mentioned Sample storage: immediate Test operator: ED nurse (a core ED team alongside written instructions trained the ED nurses) Definition of test positivity: consensus of ED nurse and 1 other medical professional Blinding reported: yes (assumed done first) Timing of samples: not mentioned; time pso only reported for those with FN results: both cohorts' range was 1 to > 7 days |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; Roche Cobas SARS‐CoV‐2 assay (Penzberg, Germany) or SARS‐CoV‐2 E‐gene assay from TibMolbiol (Berlin, Germany) Definition of non‐COVID cases: single negative PCR Genetic target(s): TibMolbiol was E gene Samples used: OP (paired) Timing of reference standard: as for index Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous (paired) All participants received same reference standard: yes Missing data: none (exclusions all from cohort [1]) Uninterpretable results: none Indeterminate results (index test): none Indeterminate results (reference standard): none |
||
| Comparative | |||
| Notes | Funding: work is based on research funded in part by the German Federal Ministry of Education and Research through projects VARIPath (01KI2021) to VMC and NaFoUniMedCovid19 (BFAST, FKZ: 01KX2021) to the Charité Publication status: accepted manuscript Source: Biomarkers Author COI: the authors report no conflicts of interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Nagura‐Ikeda 2020.
| Study characteristics | |||
| Patient Sampling | Single‐group study of patients with laboratory confirmed COVID‐19 referred for isolation and treatment (n = 103); participants had undergone qRT‐PCR tests using NP or OP swabs collected at public health institutes or hospitals (presumably symptomatic), asymptomatic patients were tested as a result of mass screening due to an outbreak or family cluster. Recruitment: not stated Prospective or retrospective: not reported; samples appear to be collected prospectively but states that patient information was retrospectively collected from the hospital electronic medical records. |
||
| Patient characteristics and setting | Setting: inpatient and asymptomatic (admitted or quarantined) Location: Self‐Defense Forces Central Hospital, Tokyo Country: Japan Dates: 11 February‐13 May 2020 Symptoms and severity: 88 (85%) symptomatic, including 16 (15%) severe (showing clinical symptoms of pneumonia ‐ dyspnoea, tachypnoea, saturation of percutaneous oxygen (SpO2) < 93%, and the need for oxygen therapy); 15 (15%) asymptomatic (including 4 pre‐symptomatic) Demographics: IPD provided ‐ median age 46, range 18‐87; 66 (64%) male Exposure history: not reported |
||
| Index tests | Test name: ESPLINE SARS‐CoV‐2 (no product code reported)
(5 other tests performed including RT‐PCR and RT‐LAMP, but not eligible for this review) Manufacturer: Fuji Rebio Inc Antibody: NP Ag target: not stated Test method: LFA (no reader device required) Samples used: saliva (self‐collected) Transport media: none; around 500 μL saliva collected Sample storage: stored at −80 °C until sample preparation Test operator: not stated; implies laboratory staff Definition of test positivity: not stated; appearance of test line implied Blinding reported: not stated Timing of samples: saliva collected on admission to hospital; IPD reports this was median 7 days pso (1‐14) |
||
| Target condition and reference standard(s) | Reference standard: RT‐qPCR on initial presentation (RT‐PCR was conducted on saliva samples as part of the study but this did not form part of the reference standard diagnosis) Definition of non‐COVID cases: single RT‐PCR negative Genetic target(s): not reported Samples used: NP or OP Timing of reference standard: on presentation or as part of mass screening; specific timing in regard to symptom onset was not reported for the original RT‐PCR and unclear if same day as saliva collection Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: unclear; saliva collected on day of admission to quarantine/hospital but NP/OP conducted at some point prior to that All participants received same reference standard: yes Missing data: not stated, no participant flow diagram reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "work was supported by the Health, Labour and Welfare Policy Research Grants, Research on Emerging and Re‐emerging Infectious Diseases and Immunization [grant number 20HA2002]". Publication status: accepted manuscript Source: Journal of Clinical Microbiology Author COI: the authors declare that they have no conflicts of interests |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Nalumansi 2020.
| Study characteristics | |||
| Patient Sampling | 2‐group study to estimate sensitivity and specificity:
Recruitment: not reported Prospective or retrospective: prospective Sample size (cases): 262 (90) |
||
| Patient characteristics and setting | Setting: unclear; referral hospitals (also described as COVID‐19 treatment facilities) Location: regional referral hospitals (RRHs) in Arua, Entebbe, Fort Portal, Gulu, Jinja, Lira, Masaka, Mbale, and Mulago National Referral Hospital. (Uganda Virus Research Institute, Entebbe, Uganda) Country: Uganda Dates: not reported Symptoms and severity: mainly asymptomatic: 77/90 (89%) PCR+ (described in Discussion only); 172 PCR− Demographics: mean age 37 (95% CI 35–39) years; male 85 (94%) Exposure history: not reported |
||
| Index tests | Test name: STANDARD Q COVID‐19 Ag Test Manufacturer: SD Biosensor, Gyeonggi‐do, 16690, Korea Antibody: Ag Ag target: not reported Test method: CGIA Samples used: nasal (laboratory staff) Transport media: none used Sample storage: not required; immediate on‐site testing Test operator: laboratory staff (training not reported) Definition of test positivity: visual; according to the manufacturer IFU Blinding reported: no; described as "unblinded" ("the staff who evaluated the Ag RDT knew which participants were likely to be infected or uninfected in most cases) Timing of samples: not recorded |
||
| Target condition and reference standard(s) | Reference standard: qRT‐PCR (Berlin protocol); used Applied Biosystems PCR platform and QIAGEN viral RNA mini kit
Ct values were categorized as strongly positive (Ct ≤ 29) indicating abundant target nucleic acid in the sample, moderately positive (Ct 30–37), and weakly positive (Ct 38–39). Definition of non‐COVID cases: same as for cases Genetic target(s): not reported Samples used: nasal; paired Timing of reference standard: as for index test Blinded to index test: yes; Ag interpretation was not blinded to SARS‐CoV‐2 status so PCR must have been undertaken first Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous swab; same sample site All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "no specific funding was received" Publication status: published Source: International Journal of Infectious Diseases Author COI: all authors declare no competing interests |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Nash 2020.
| Study characteristics | |||
| Patient Sampling | Unclear design to estimate sensitivity and specificity: samples from suspected patients submitted to 'PATH' (ww.path.org) for routine COVID diagnosis
(Second cohort of samples also tested using spike‐based assay; excluded as assay requires use of centrifuge) Recruitment: not stated Prospective or retrospective: retrospective |
||
| Patient characteristics and setting | Setting: unclear; samples provided to study authors by PATH (non‐profit organisation), protocol number 00004244 Location: not reported Country: not reported Dates: not reported Symptoms and severity: not reported Demographics: not reported Exposure history: not reported |
||
| Index tests | Test name: Direct antigen rapid test (DART); NP‐based Manufacturer: E25Bio Inc (Cambridge MA); not yet available Antibody: NP Ag target: anti‐N mouse monoclonal antibodies Test method: immunochromatographic paper‐based (CGIA) Samples used: nasal; collection not described Transport media: not stated Sample storage: banked frozen prior to testing Test operator: not stated; presume lab staff Definition of test positivity: visual line Blinding reported: not stated Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: qRT PCR; ThermoFisher/AppliedBiosystems TaqPATH COVID‐19 Combo Kit (ThermoFisher, Waltham, MA USA) Definition of non‐COVID cases: as for cases; single negative PCR required Genetic target(s): N, S, and ORF1ab genes Samples used: nasal (same swab) Timing of reference standard: not stated Blinded to index test: yes, conducted first Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous (same swab) All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: not stated |
||
| Comparative | |||
| Notes | Funding: "the study is funded, in part, by a Bill and Melinda Gates Foundation Award (INV‐017872) to E25Bio, Inc. EN is funded by Tufts University DISC Seed Grant. MLN is supported by a FAPESP grant (#2020/04836‐0) and is a CNPq Research Fellow. AFV is supported by a FAPESP Fellow grant (#18/17647‐0). GRFC is supported by a FAPESP Fellow grant (#20/07419‐0). BHGAM 798 is supported by a FAPESP Scholarship (#19/06572‐2)." Publication status: preprint Source: medRxiv Author COI: "BN, AB, AR, MB, NS, AG, IB, and BBH are employed by or affiliated with E25Bio Inc. (www.e25bio.com), a company that develops diagnostics for epidemic viruses." |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Ngo Nsoga 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: patients attending a single COVID‐19 screening centre either with symptoms compatible with COVID‐19 infection or asymptomatic contacts (The results of a separate pilot study is reported in a supplementary appendix however details are limited.) Recruitment: unclear Prospective or retrospective: prospective Sample size (cases): 402 (168) |
||
| Patient characteristics and setting | Setting: outpatient COVID‐19 screening site Location: Geneva; Geneva University Hospital Country: Switzerland Dates: 3‐12 November 2020 Symptoms and severity: states that the "majority" were symptomatic, and that all 168 PCR+ve were symptomatic. Appears that symptom breakdown was only provided for these 168. Symptoms included: asthenia 101 (60.1%); headache 99 (48.9%); myalgia 81 (48.2%); chills/fever 80 (47.6%); dry/productive cough 73 (43.5%); anosmia/ageusia 71 (42.3%); odynophagia 68 (40.5%); digestive signs 38 (22.6%); dyspnoea 7 (4.2%); chest pain 4 (2.4%); other 12 (7.1%) Demographics: age: mean 39.9 years; SD +/‐14.5 years; median 38 years; range 16‐80 years. Male: 178 (44.3%) Exposure history: contact with positive case in last 14 days: 87/168 (51.8%) |
||
| Index tests | Test name: PanBio Ag RDT Manufacturer: Abbot Antibody: not mentioned Ag target: not mentioned Test method: CGIA Samples used: OP; collected by experienced doctor. Pilot study suggested poor sensitivity when back of pharynx was not reached Transport media: not mentioned Sample storage: not mentioned Test operator: states "biologist" with a second HCW for equivocal results Definition of test positivity: according to manufacturer IFU Blinding reported: not mentioned Timing of samples: samples obtained up to 24 d pso. (Figure 1); text reports 101 day 0‐4; 19 day 5‐7; 17 day 8‐11; mean 4.1 days pso to PCR (range 0‐24 d) |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; Cobas SARS‐CoV‐2 RT‐PCR assay Definition of non‐COVID cases: Genetic target(s): ORF1 and E gene Samples used: NP; collected by nurse Timing of reference standard: as for index test; some asymptomatic cases and no information on these timings Blinded to index test: not mentioned Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: assumed same time but not clear All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): 2 samples were positive for ORF1 only and not E gene ‐ interpreted as positive Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "this work was supported by Foundation of Innovative Diagnostics (FIND), by Private HUG Foundation and by Pictet Charitable Foundation. Marie Thérèse Ngo Nsoga is a beneficiary of the excellence grant from the Swiss Confederation and the grant from the humanitarian commission of the University Hospital of Geneva" Publication status: preprint Source: preprint Author COI: none mentioned |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Nikolai 2021(a) [A].
| Study characteristics | |||
| Patient Sampling | 2 single‐group studies to estimate sensitivity and specificity:
[1] symptomatic adults with high clinical suspicion of COVID‐19 presenting at an ambulatory SARS‐CoV‐2 testing facility (compared professionally collected AN and NMT samples) (n = 132)
[2] symptomatic adults with high clinical suspicion of COVID‐19 presenting at an ambulatory SARS‐CoV‐2 testing facility (compared self‐collected NMT sample and professional NP swab) (n = 96); see Nikolai 2021(b) [A] for further details Recruitment: consecutively enrolled (according to laboratory capacity) Prospective or retrospective: prospective Sample size (cases): 132 (36) |
||
| Patient characteristics and setting | Setting: ambulatory SARS‐CoV‐2 testing facility Location: Charité University Hospital Country: Germany Dates: 30 November 2020‐18 January 2021 Symptoms and severity: whole sample (n = 228): 222, 97.4% of participants had ≥ 1 symptoms consistent with SARS‐CoV‐2 infection Demographics: average age: 34.6 years (SD 11.7) Sex: 107, 47% female Exposure history: not reported |
||
| Index tests | Test name: STANDARD Q COVID‐19 Ag Test (nasal sampling kit used; RUO at time of study) Manufacturer: SD Biosensor, Inc. Gyeonggi‐do, Korea (also distributed by Roche in Europe) Antibody: not reported Ag target: not reported Test method: not reported Samples used: 2 types of samples for each patient: [A] AN (both nostrils) [B] NMT (both nostrils) (both collected by professional following CDC guidance for SARS‐CoV‐2 testing; sequence of sampling alternated between patients) Transport media: none Sample storage: directly after sampling Test operator: study physicians Definition of test positivity: semi‐quantitative visual read‐out of the test band (2 independent blinded readers) Blinding reported: yes Timing of samples: whole sample (n = 228): mean 3.4 days (SD 3.0) |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR (no further details reported; cites Lindner 2021b [A]) Definition of non‐COVID cases: as for cases Genetic target(s): not reported Samples used: OP/NP sampling (collected by professional after AN, NMT swabs) Timing of reference standard: same as for index test Blinded to index test: not reported Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous All participants received same reference standard: yes Missing data: yes; 2 exclusions from whole sample of 230 enrolled Uninterpretable results: not reported Indeterminate results (index test): not reported Indeterminate results (reference standard): 2 invalid RT‐PCR results Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "CM Denkinger reports grants from Foundation of Innovative New Diagnostics, grants from Ministry of Science, Research and Culture, State of Baden Wuerttemberg, Germany, to conduct of the study. JA Sacks reports grants from UK Department of International Development (DFID, recently replaced by FCMO), grants from World Health Organization (WHO), grants from Unitaid, to conduct of the study." Publication status: preprint (not peer reviewed) Source: medRxiv Author COI: all authors declare no conflicts of interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Nikolai 2021(a) [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 2 sample types collected professionally; Nikolai 2021(a) [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 2 sample types collected professionally; Nikolai 2021(a) [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: STANDARD Q COVID‐19 Ag Test (nasal sampling kit used; RUO at time of study) Manufacturer: SD Biosensor, Inc. Gyeonggi‐do, Korea (also distributed by Roche in Europe) Antibody: not reported Ag target: not reported Test method: not reported Samples used: 2 types of samples for each patient: [A] AN (both nostrils) [B] NMT (both nostrils) (both collected by professional following CDC guidance for SARS‐CoV‐2 testing; sequence of sampling alternated between patients) Transport media: none Sample storage: directly after sampling Test operator: study physicians Definition of test positivity: semi‐quantitative visual read‐out of the test band (2 independent blinded readers) Blinding reported: yes Timing of samples: whole sample (n = 228): mean 3.4 days (SD 3.0) |
||
| Target condition and reference standard(s) | Comparative study of 2 sample types collected professionally; Nikolai 2021(a) [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 2 sample types collected professionally; Nikolai 2021(a) [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 2 sample types collected professionally; Nikolai 2021(a) [A] reports full study characteristics and QUADAS. | ||
Nikolai 2021(b) [A].
| Study characteristics | |||
| Patient Sampling | 2 single‐group studies to estimate sensitivity and specificity:
[1] symptomatic adults with high clinical suspicion of COVID‐19 presenting at an ambulatory SARS‐CoV‐2 testing facility (compared professionally collected AN and NMT samples) (n = 132); see Nikolai 2021(a) [A] for further details
[2] symptomatic adults with high clinical suspicion of COVID‐19 presenting at an ambulatory SARS‐CoV‐2 testing facility (compared self‐collected NMT sample and professional NP swab) (n = 96) Recruitment: consecutively enrolled (according to laboratory capacity) Prospective or retrospective: prospectively Sample size (cases): 96 (34) |
||
| Patient characteristics and setting | Setting: ambulatory SARS‐CoV‐2 testing facility Location: Charité University Hospital Country: Germany Dates: 30 November 2020‐18 January 2021 Symptoms and severity: whole sample (n = 228): 222, 97.4% of participants had ≥ 1 symptoms consistent with SARS‐CoV‐2 infection Demographics: average age: 34.6 years (SD 11.7) Sex: 107, 47% female Exposure history: not reported |
||
| Index tests | Test name: [A] STANDARD Q COVID‐19 Ag Test (nasal sampling kit; RUO)
[B] STANDARD Q COVID‐19 Ag Test (NP sampling kit) Manufacturer: SD Biosensor, Inc. Gyeonggi‐do, Korea (also distributed by Roche in Europe) Antibody: not reported Ag target: not reported Test method: not reported Samples used: 2 types of samples for each patient: [A] NMT (self‐sampling, both nostrils; observed without intervention) and [B] NP (1 nostril; collected by professional) Transport media: none Sample storage: directly after sampling Test operator: study physicians Definition of test positivity: semi‐quantitative visual read‐out of the test band (2 independent blinded readers) Blinding reported: yes Timing of samples: whole sample (n = 228): mean 3.4 days (SD 3.0) |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR (no further details reported; cites Lindner 2021b [A]) Definition of non‐COVID cases: as for cases Genetic target(s): not reported Samples used: OP/NP sampling (other nostril to NP swab for Ag test; collected by professional after other swabs) Timing of reference standard: same as for index test Blinded to index test: not reported Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous All participants received same reference standard: yes Missing data: yes; 2 exclusions from whole sample of 230 enrolled Uninterpretable results: not reported Indeterminate results (index test): not reported Indeterminate results (reference standard): 2 invalid RT‐PCR results Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "CM Denkinger reports grants from Foundation of Innovative New Diagnostics, grants from Ministry of Science, Research and Culture, State of Baden Wuerttemberg, Germany, to conduct of the study. JA Sacks reports grants from UK Department of International Development (DFID, recently replaced by FCMO), grants from World Health Organization (WHO), grants from Unitaid, to conduct of the study." Publication status: preprint (not peer reviewed) Source: medRxiv Author COI: all authors declare no conflicts of interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Nikolai 2021(b) [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 2 sample types collected professionally versus self‐collected; Nikolai 2021(b) [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 2 sample types collected professionally versus self‐collected; Nikolai 2021(b) [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] STANDARD Q COVID‐19 Ag Test (nasal sampling kit; RUO)
[B] STANDARD Q COVID‐19 Ag Test (NP sampling kit) Manufacturer: SD Biosensor, Inc. Gyeonggi‐do, Korea (also distributed by Roche in Europe) Antibody: not reported Ag target: not reported Test method: not reported Samples used: 2 types of samples for each patient: [A] NMT (self‐sampling, both nostrils; observed without intervention) and [B] NP (1 nostril; collected by professional) Transport media: none Sample storage: directly after sampling Test operator: study physicians Definition of test positivity: semi‐quantitative visual read‐out of the test band (2 independent blinded readers) Blinding reported: yes Timing of samples: whole sample (n = 228): mean 3.4 days (SD 3.0) |
||
| Target condition and reference standard(s) | Comparative study of 2 sample types collected professionally versus self‐collected; Nikolai 2021(b) [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 2 sample types collected professionally versus self‐collected; Nikolai 2021(b) [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 2 sample types collected professionally versus self‐collected; Nikolai 2021(b) [A] reports full study characteristics and QUADAS. | ||
Okoye 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: asymptomatic college age (undergraduate and graduate) students; not experiencing signs or symptoms of COVID‐19 at the time of testing Recruitment: not stated; likely consecutive Prospective or retrospective: prospectively Sample size (cases): 2645 (46) |
||
| Patient characteristics and setting | Setting: temporary indoor testing site Location: University of Utah in Salt Lake City, Utah Country: USA Dates: 13 November‐20 November 2020 Symptoms and severity: asymptomatic Demographics: average age: 24 years (range: 15‐86 years) Sex: 52% female Exposure history: not reported |
||
| Index tests | Test name: BinaxNOW COVID‐19 antigen card Manufacturer: Abbott Antibody: SARS‐CoV‐2 nucleocapsid antigen Ag target: not reported Test method: not reported Samples used: NMT (self‐collected, both nostrils according to CDC guidelines, observed by trained non‐medical personnel) Transport media: none (direct swab) Sample storage: immediate testing Test operator: trained non‐medical personnel (University of Utah Hope Corps interns) according to the manufacturer IFU Definition of test positivity: 2 pink/purple lines observed Blinding reported: yes Timing of samples: not applicable (asymptomatic) |
||
| Target condition and reference standard(s) | Reference standard: Thermo Fisher TaqPath COVID‐19 Combo kit; 40 cycles performed, ≥ 2 target genes required. Ct was reported as average of the Ct values of the detected coronavirus genes.
PCR+ participants invited to reattend for saliva sampling for second confirmatory RT‐PCR (Hologic Panther Fusion SARS‐CoV‐2 assay, the Roche Cobas SARS‐CoV‐2 assay, or the Thermo Fisher TaqPath COVID‐19 Combo kit) Definition of non‐COVID cases: as for cases Genetic target(s): ORF1ab, S, N Samples used: as for index test (NMT), but placed in ARUP COVID‐19 Transport Media; order of testing randomly assigned to either Ag or PCR Timing of reference standard: not applicable (asymptomatic) Blinded to index test: not reported Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired All participants received same reference standard: yes Missing data: yes; 7 excluded Uninterpretable results: 3 invalid BinaxNOW results; all negative on retesting with new nasal swab specimen Indeterminate results (index test): none Indeterminate results (reference standard): 4 inconclusive on PCR; only N gene detected (Ct > 30) Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: no funding reported
"The BinaxNOW antigen cards utilized in this study were received from the Utah Department of Health as part of a U.S. federal government initiative" Publication status: academic journal Source: Journal of Clinical Microbiology Author COI: no COI statement reported |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Olearo 2021 [A].
| Study characteristics | |||
| Patient Sampling | 2‐groups study to estimate sensitivity and specificity:
1] RT‐PCR+ve (n = 84), included until target number met
2] RT‐PCR−ve (n = 100), randomly selected to serve as negative control
Swabs collected following routine diagnostics from patients hospitalized with suspected or known COVID‐19 Recruitment: unclear for RT‐PCR+ve; RT‐PCR−ve were randomly sampled Prospective or retrospective: retrospective (described as prospective, but samples included based on PCR status) Sample size (cases): 184 (84) |
||
| Patient characteristics and setting | Setting: hospital inpatient Location: University Medical Centre Hamburg‐Eppendorf, Germany Country: Germany Dates: August‐November 2020 Symptoms and severity: median duration pso 6 (IQR 2–12) days Demographics: not reported Exposure history: not reported |
||
| Index tests | Test name: [A] SARS‐CoV‐2 Rapid Antigen Test (Roche)
[B] COVID‐19 Rapid Test Device (Abbott)
[C] MEDsan SARSCoV‐2 Antigen Rapid Test
[D] CLINITEST Rapid COVID.19 Antigen Test Manufacturer: [A] SD Biosensor (Roche Diagnostics), South Korea [B] Panbio Ltd. (Abbott Rapid Diagnostics), Australia [C] MEDsan GmbH, Germany [D] Zhejiang Orient Biotech Co, China Antibody: nucleocapsid Ag target: not reported Test method: CGIA Samples used: OP or NP (HCW) Transport media: UTM‐based collection kits by Copan (Italy, Brescia) or Iclean (Shenzhen, China) Sample storage: not reported Test operator: lab technicians; swabs supplied with the Ag kits were immersed in patient OP/NP samples for approximately 10 s before all further steps of the tests were carried out according to manufacturer IFU. Definition of test positivity: visual Blinding reported: no; states test results "were read by two unblinded operators" Timing of samples: median 6 (IQR 2–12) days pso |
||
| Target condition and reference standard(s) | Reference standard: RT‐qPCR assay (Roche Cobas SARS‐CoV‐2 IVD) in conjunction with quantitative external control material by Instand e.V. (Düsseldorf, Germany) Definition of non‐COVID cases: same as for cases Genetic target(s): not reported Samples used: OP or NP (same sample) Timing of reference standard: median 6 (IQR 2–12) days from symptom onset Blinded to index test: yes (performed before index test) Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same swab used All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "this research did not receive any specific grant from funding agencies in the public, commercial, or not‐for‐profit sectors." Publication status: published Source: Journal of Clinical Virology Author COI: the authors declare no known competing finance |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Olearo 2021 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 4 Ag tests; Olearo 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 4 Ag tests; Olearo 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] SARS‐CoV‐2 Rapid Antigen Test (Roche)
[B] COVID‐19 Rapid Test Device (Abbott)
[C] MEDsan SARSCoV‐2 Antigen Rapid Test
[D] CLINITEST Rapid COVID.19 Antigen Test Manufacturer: [A] SD Biosensor (Roche Diagnostics), South Korea [B] Panbio Ltd. (Abbott Rapid Diagnostics), Australia [C] MEDsan GmbH, Germany [D] Zhejiang Orient Biotech Co, China Antibody: nucleocapsid Ag target: not reported Test method: CGIA Samples used: OP or NP (HCW) Transport media: UTM‐based collection kits by Copan (Italy, Brescia) or Iclean (Shenzhen, China) Sample storage: not reported Test operator: lab technicians; swabs supplied with the Ag kits were immersed in patient OP/NP samples for approximately 10 s before all further steps of the tests were carried out according to manufacturer IFU. Definition of test positivity: visual Blinding reported: no; states test results "were read by two unblinded operators" Timing of samples: median 6 (IQR 2–12) days pso |
||
| Target condition and reference standard(s) | Comparative study of 4 Ag tests; Olearo 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 4 Ag tests; Olearo 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 4 Ag tests; Olearo 2021 [A] reports full study characteristics and QUADAS. | ||
Olearo 2021 [C].
| Study characteristics | |||
| Patient Sampling | Comparative study of 4 Ag tests; Olearo 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 4 Ag tests; Olearo 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] SARS‐CoV‐2 Rapid Antigen Test (Roche)
[B] COVID‐19 Rapid Test Device (Abbott)
[C] MEDsan SARSCoV‐2 Antigen Rapid Test
[D] CLINITEST Rapid COVID.19 Antigen Test Manufacturer: [A] SD Biosensor (Roche Diagnostics), South Korea [B] Panbio Ltd. (Abbott Rapid Diagnostics), Australia [C] MEDsan GmbH, Germany [D] Zhejiang Orient Biotech Co, China Antibody: nucleocapsid Ag target: not reported Test method: CGIA Samples used: OP or NP (HCW) Transport media: UTM‐based collection kits by Copan (Italy, Brescia) or Iclean (Shenzhen, China) Sample storage: not reported Test operator: lab technicians; swabs supplied with the Ag kits were immersed in patient OP/NP samples for approximately 10 s before all further steps of the tests were carried out according to instructions by manufacturers. Definition of test positivity: visual Blinding reported: no; states test results "were read by two unblinded operators" Timing of samples: median 6 (IQR 2–12) days pso |
||
| Target condition and reference standard(s) | Comparative study of 4 Ag tests; Olearo 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 4 Ag tests; Olearo 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 4 Ag tests; Olearo 2021 [A] reports full study characteristics and QUADAS. | ||
Olearo 2021 [D].
| Study characteristics | |||
| Patient Sampling | Comparative study of 4 Ag tests; Olearo 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 4 Ag tests; Olearo 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] SARS‐CoV‐2 Rapid Antigen Test (Roche)
[B] COVID‐19 Rapid Test Device (Abbott)
[C] MEDsan SARSCoV‐2 Antigen Rapid Test
[D] CLINITEST Rapid COVID.19 Antigen Test Manufacturer: [A] SD Biosensor (Roche Diagnostics), South Korea [B] Panbio Ltd. (Abbott Rapid Diagnostics), Australia [C] MEDsan GmbH, Germany [D] Zhejiang Orient Biotech Co, China Antibody: nucleocapsid Ag target: not reported Test method: CGIA Samples used: OP or NP (HCW) Transport media: UTM‐based collection kits by Copan (Italy, Brescia) or Iclean (Shenzhen, China) Sample storage: not reported Test operator: lab technicians; swabs supplied with the Ag kits were immersed in patient OP/NP samples for approximately 10 s before all further steps of the tests were carried out according to instructions by manufacturers. Definition of test positivity: visual Blinding reported: no; states test results "were read by two unblinded operators" Timing of samples: median 6 (IQR 2–12) days pso |
||
| Target condition and reference standard(s) | Comparative study of 4 Ag tests; Olearo 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 4 Ag tests; Olearo 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 4 Ag tests; Olearo 2021 [A] reports full study characteristics and QUADAS. | ||
Osterman 2021(a) [A].
| Study characteristics | |||
| Patient Sampling | Multi‐site, multiple group study, including:
(1) Site 1: symptomatic patients (adults and children) presenting at EDs or on clinical units (n = 741 swabs, including 381 PCR+ and 360 PCR−); of 381 PCR+, 189 were classed as primary diagnosis (no previous PCR+) and 192 swabs were undertaken at “follow‐up” during hospitalization, i.e. at variable time points after onset of symptoms or first PCR+ result Site 2 extracted as Osterman 2021(b) (2) Site 2: symptomatic and asymptomatic participants at patient care units or from employee screening, all PCR+ (n = 66) Recruitment: not mentioned Prospective or retrospective: unclear; includes frozen samples so not sure we can assume prospective here Sample size (cases): 833 (447); test [A] FIA 741 (381); test [B] RAT 831 (445) |
||
| Patient characteristics and setting | Setting: mixed; (1) hospital inpatient and ED Location: (1) LMU Klinikum Hospital, Munich Country: Germany Dates: (1) 4 March‐19 October 2020 Symptoms and severity: (1) all symptomatic; (1) + (2) 256/445 (58%) PCR+ primary diagnosis, and 189/445 (42%) follow‐up testing Demographics: only reported for PCR− at site 1: 326/386 (84%) adults; 60/386 (16%) children Exposure history: not mentioned |
||
| Index tests | Test name: [A] STANDARD F COVID‐19 Ag FIA; [B] SARS‐CoV‐2 Rapid Antigen Test (RAT) Manufacturer: [A] SD Biosensor; [B] Roche Diagnostics Ag target: both nucleocapsid Test method: [A] FIA; [B] CGIA Samples used: site (1) NP 182; OP 53, sampling site unknown 154; collected by HCWs Transport media: site (1) eSwab (Copan Diagnostics, Murrieta, California, USA), ImproViral (Improve Medical, Guangzhou, Republic of China) dry swabs inserted into sterile 0.9% NaCl, or the original manufacturers’ swabs inserted into the extraction buffers provided Sample storage: same day Site (1): were either kept at room temperature for 1–2 h (“fresh”) (n = 18); stored at 4 °C for 0–7 days (n = 48); or stored at −20 °C (n = 315) until SARS‐CoV‐2 antigen testing was performed Test operator: laboratory personnel according to manufacturer IFU Definition of test positivity: [A] a cut‐off index (COI) ≥ 1 was interpreted as positive after 30 min [B] every visible (even if very faint or not uniform) test line was interpreted as positive after 15 or 30 min. Blinding reported: not mentioned Timing of samples: not mentioned |
||
| Target condition and reference standard(s) | Reference standard: PCR; multiple assays used at both sites Definition of non‐COVID cases: not reported Genetic target(s): site 1: N1 or envelope Samples used: as for index test; Site 1: OP or NP Timing of reference standard: not mentioned Blinded to index test: not mentioned Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: assumed same time All participants received same reference standard: all received PCR but many types/brands of PCR listed Missing data: number PCR+ across the 2 sites sums to 447 but maximum number PCR+ reported was 445 (test [B]); only 381 reported for RAT test (site 1) Uninterpretable results: none mentioned Indeterminate results (index test): none mentioned Indeterminate results (reference standard): none mentioned Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "Open Access funding enabled and organized by Projekt DEAL. This work was supported in part by the German BMBF initiative “NaFoUniMedCovid19” (01KX2021), subproject B‐FAST (to U.P. and O.T.K.), and by the Medical Faculty of the LMU München, Munich, Germany (to O.T. K.)" Publication status: published Source: Medical Microbiology and Immunology Author COI: the authors declare that they have no conflict of interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Osterman 2021(a) [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 2 Ag tests; Osterman 2021(a) [A]reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | |||
| Index tests | Test name: [A] STANDARD F COVID‐19 Ag FIA; [B] SARS‐CoV‐2 Rapid Antigen Test (RAT) Manufacturer: [A] SD Biosensor; [B] Roche Diagnostics Ag target: both nucleocapsid Test method: [A] FIA; [B] CGIA Samples used: site (1) NP 182 ; OP 53, sampling site unknown 154; collected by HCWs Transport media: site (1) eSwab (Copan Diagnostics, Murrieta, California, USA), ImproViral (Improve Medical, Guangzhou, Republic of China) dry swabs inserted into sterile 0.9% NaCl, or the original manufacturers’ swabs inserted into the extraction buffers provided Sample storage: same day Site (1): were either kept at room temperature for 1–2 h (“fresh”) (n = 18); stored at 4 °C for 0–7 days (n = 48); or stored at −20 °C (n = 315) until SARS‐CoV‐2 antigen testing was performed Test operator: laboratory personnel according to manufacturers instructions Definition of test positivity: [A] a cut‐off index (COI) ≥ 1 was interpreted as positive after 30 min [B] every visible (even if very faint or not uniform) test line was interpreted as positive after 15 or 30 min. Blinding reported: not mentioned Timing of samples: not mentioned |
||
| Target condition and reference standard(s) | Comparative study of 2 Ag tests; Osterman 2021(a) [A]reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 2 Ag tests; Osterman 2021(a) [A]reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 2 Ag tests; Osterman 2021(a) [A]reports full study characteristics and QUADAS. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | |||
| Did all patients receive the same reference standard? | |||
| Were all patients included in the analysis? | |||
| Did all participants receive a reference standard? | |||
| Could the patient flow have introduced bias? | |||
Osterman 2021(b).
| Study characteristics | |||
| Patient Sampling | Multi‐site, multiple group study, including:
(2) Site 2: symptomatic and asymptomatic participants at patient care units or from employee screening, all PCR+ (n = 66) Also reports data for site 1 (included as Osterman 2021(a) [A] (1) Site 1: symptomatic patients (adults and children) presenting at EDs or on clinical units (n = 741 swabs, including 381 PCR+ and 360 PCR−); of 381 PCR+, 189 were classed as primary diagnosis (no previous PCR+) and 192 swabs were undertaken at “follow‐up” during hospitalization, i.e. at variable time points after onset of symptoms or first PCR+ result] Recruitment: not mentioned Prospective or retrospective: unclear; includes frozen samples so not sure we can assume prospective here Sample size (cases): 833 (447); test [A] FIA 741 (381); test [B] RAT 831 (445) |
||
| Patient characteristics and setting | Setting: mixed; (2) hospital inpatient and employee test centre Location: (2) University Hospital Rechts der Isar of the Technical University of Munich (TUM) Country: Germany Dates: (2) 13 November‐8 December 2020 Symptoms and severity: (2) symptomatic and asymptomatic (1) + (2) 256/445 (58%) PCR+ primary diagnosis, and 189/445 (42%) follow‐up testing Demographics: only reported for PCR− at site 1: 326/386 (84%) adults; 60/386 (16%) children Exposure history: not mentioned |
||
| Index tests | Test name: STANDARD F COVID‐19 Ag FIA Manufacturer: SD Biosensor Ag target: both nucleocapsid Test method: FIA Samples used: site (2) 66 NP; both collected by HCWs Transport media: site (2) REST combi swabs (Nobel Bioscience, Sinbaek‐gil, Republic of Korea) containing 2 mL of UTM Sample storage: same day; site (2) PCR and Ag testing (RAT) were performed on the day of submission of freshly obtained swabs Test operator: laboratory personnel according to manufacturer IFU Definition of test positivity: a cut‐off index (COI) ≥ 1 was interpreted as positive after 30 min Blinding reported: not mentioned Timing of samples: not mentioned |
||
| Target condition and reference standard(s) | Reference standard: PCR; multiple assays used at both sites Definition of non‐COVID cases: Genetic target(s): site 2: N and RdRp gene Samples used: as for index test; site 2: NP Timing of reference standard: not mentioned Blinded to index test: not mentioned Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: assumed same time All participants received same reference standard: all received PCR but many types/brands of PCR listed Missing data: number PCR+ across the 2 sites sums to 447 but maximum number PCR+ reported was 445 (test [B]); only 381 reported for RAT test (site 1) Uninterpretable results: none mentioned Indeterminate results (index test): none mentioned Indeterminate results (reference standard): none mentioned Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "Open Access funding enabled and organized by Projekt DEAL. This work was supported in part by the German BMBF initiative “NaFoUniMedCovid19” (01KX2021), subproject B‐FAST (to U.P. and O.T.K.), and by the Medical Faculty of the LMU München, Munich, Germany (to O.T. K.)" Publication status: published Source: Medical Microbiology and Immunology Author COI: the authors declare that they have no conflict of interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Parada‐Ricart 2020.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity. Participants were patients with respiratory symptoms of < 7 days (128) and asymptomatic patients (44) Recruitment: not mentioned Prospective or retrospective: prospective Sample size (cases): 193; final PCR diagnosis available for 172 (26 cases) |
||
| Patient characteristics and setting | Setting: unclear Location: not mentioned. Author affiliations list Hospital Universitari de Tarragona JoanXXIII and Rovira i Virgili University Country: Spain Dates: 6‐17 April 2020 Symptoms and severity: 128 with respiratory symptoms; no further details Demographics: not mentioned Exposure history: not mentioned |
||
| Index tests | Test name: SARS‐CoV‐2 (2019‐n‐CoV Ag Test Flourescence IC Assay) Manufacturer: Shenzen Bioeasy Biotechnology Co LTD Antibody: nucleocapsid Ag target: Test method: FIA Samples used: nasal; collection not reported Transport media: not mentioned Sample storage: none (tested within 30 min) Test operator: not mentioned Definition of test positivity: as per manufacturer IFU Blinding reported: not mentioned; but assume yes as tested within 30 min of collection Timing of samples: < 7 days pso |
||
| Target condition and reference standard(s) | Reference standard: PCR; not described Definition of non‐COVID cases: single negative PCR, apart from discrepant cases (FPs) which were analysed by assessing clinical/radiology, previous/subsequent PCR results and serological data if available. Genetic target(s): not mentioned Samples used: nasal Timing of reference standard: < 7 days pso Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: different samples but same day All participants received same reference standard: yes Missing data: PCR result not available for 21 patients so not included in the analysis Uninterpretable results: not mentioned Indeterminate results (index test): not mentioned Indeterminate results (reference standard): not mentioned. Of 21 FPs, 13 remained after consideration of clinical history (9 asymptomatic and 4 symptomatic with negative subsequent serology); the remaining 8 were reclassified as TP (clinical‐epidemiological picture compatible with COVID‐19) Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "partially funded by a call from the Department of Health of the Generalitat de Catalunya, code 6‐17, main researcher: Francesc Vidal" Publication status: published Source: Enfermedades Infecciosas y Microbiología Clínica Author COI: none reported |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Pena 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: asymptomatic individuals at 7 testing sites, including workers (n = 56), "sanitary residence" [presumed to be health‐related residential care] (n = 239), and the general public (n = 547); community prevalence of COVID‐19 was 11%. Recruitment: unclear Prospective or retrospective: unclear Sample size (cases): 854 included; 842 analysed (73) |
||
| Patient characteristics and setting | Setting: states "seven testing sites"; community screening Location: Iquique city, Tarapacá Region (Author institution: Laboratorio de Virología Molecular y Celular, Programa de Virología, Instituto de Ciencias Biomédicas, Facultad de Medicina, Universidad de Chile) Country: Chile Dates: 14‐17 January 2021 Symptoms and severity: all asymptomatic (100%) Demographics: mean age 36.67 (SD 16.48) years; 351 (42%) female Exposure history: no details |
||
| Index tests | Test name: STANDARD Q COVID‐19 Ag (catalogue number 9901‐NCOV‐01G) Manufacturer: SD Biosensor, Inc. Republic of Korea Antibody: not stated Ag target: not stated Test method: LFA chromatographic Samples used: NP (HCW collected) Transport media: none required (analysed according to the manufacturer IFU) Sample storage: none required (analysed according to the manufacturer IFU) Test operator: not stated but implies HCW at time of sampling Definition of test positivity: visual, coloured bands Blinding reported: yes (performed before PCR) Timing of samples: all described as asymptomatic; participants were asked about any symptoms in previous 0‐14 days, but no information on responses was reported |
||
| Target condition and reference standard(s) | Reference standard: PCR (GenomeCov19 Detection Kit ABM; Applied Biological Materials Inc, Canada, catalogue number G628.v2)
Ct ≤ 40 considered positive for the N and S viral gene Definition of non‐COVID cases: N/A Genetic target(s): N and S gene Samples used: NP (analysed within 24–72 h of collection) Timing of reference standard: as for index test Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired swabs All participants received same reference standard: yes Missing data: yes; 12 (1.4%) were excluded for lacking real‐time PCR results Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: Funded by Ministerio de Salud de Chile Publication status: preprint Source: medRxiv Author COI: "the authors are supported by ANID Chile through Fondecyt grants" |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Pena‐Rodriguez 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: adults with symptoms suggestive of COVID‐19 (headache, fever, fatigue, other respiratory signs, or gastrointestinal symptoms) and individuals in contact with confirmed cases of COVID‐19 (by RT‐PCR) in the previous 3‐5 days, with or without symptoms attending for COVID‐19 testing Recruitment: unclear Prospective or retrospective: prospective Sample size (cases): 369 (104) |
||
| Patient characteristics and setting | Setting: COVID‐19 test centre (diagnostic laboratory centre) Location: Guadalajara (Centro Universitario de Ciencias de la Salud, Universidad de Guadalajara, Guadalajara) Country: Mexico Dates: October‐November 2020 Symptoms and severity: symptoms included: headache (42%), fever (25%), cough (23%), myalgia (21%), loss of smell (18%), fatigue (16%), diarrhoea (10%), shortness of breath (7%), arthralgia (4%) Demographics: average age 36.6 ± 13.16 years; 215 (58%) female Exposure history: individuals in contact with confirmed cases of COVID‐19 in the last 3‐5 days included; no further details |
||
| Index tests | Test name: STANDARD Q COVID‐19 Manufacturer: SD BIOSENSOR Antibody: N gene Ag target: not stated Test method: chromatographic Samples used: NP; single nostril (collected by "trained staff") Transport media: not used Sample storage: none; "SARS‐CoV‐2 antigen analysis was carried out in the place" Test operator: trained staff Definition of test positivity: visual control and test lines The test was invalidated when no marks were detected Blinding reported: yes; performed before PCR) Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; DeCoV19 Kit Triplex (Genes2life SAPI de CV, Mexico)
Ct < 35 with an exponential growth curve of ≥ 2 genes were considered as positive Definition of non‐COVID cases: same as for cases (single ‐ve PCR) Genetic target(s): N gene and Rnase P gene (RP) Samples used: combined NP/OP in VTM Timing of reference standard: same as for index test Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported; States "The test was invalidated when no marks were detected" Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: P.M.I 12.10, SAPI de CV, grant/award
Number: 251472 Publication status: published Source: Journal of Clinical Laboratory Analysis Author COI: RAT was "supplied by PMI 12.10, SAPI de CV company" [appears to be a medical supplies company which imports the SD Biosensor assay]. No other COI reported |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Perez‐Garcia 2021 [A].
| Study characteristics | |||
| Patient Sampling | 2‐group study to estimate sensitivity and specificity: samples from patients with suspicion of COVID‐19 attending the university hospital or associated primary healthcare centres
1] PCR−ve patients (n = 150)
2] PCR+ve patients (n = 170) Recruitment: not stated; appears to be deliberate sampling based on PCR status Prospective or retrospective: retrospective Sample size (cases): 320 (170) |
||
| Patient characteristics and setting | Setting: laboratory‐based; samples from mixed settings including primary healthcare centres (50%), hospital inpatients (20%), ED (21%) and occupational health (9%) Location: Madrid (Servicio de Microbiología Clínica, Hospital Universitario Príncipe de Asturias, Madrid) Country: Spain Dates: 8‐20 October 2020 Symptoms and severity: 134 (79%) symptomatic; including cough (54%), fever (41%), dyspnoea (25%), anosmia (22%) and myalgia (19%); 26 (15%) asymptomatic with a prior contact with COVID‐19 case Demographics: median age 51 years (IQR: 38–68); (10 with no data on symptoms; time pso not reported for 6/134 symptomatic) 81 (48%) female Exposure history: states "26 (15%) asymptomatic with a prior contact with COVID‐19 case"; no further details |
||
| Index tests | Test name: [A] CerTest SARS‐CoV‐2 Ag One Step Card Test (Batch code SC‐004) [B] Panbio COVID‐19 Ag Rapid Test Device (Batch code 41ADF057A) Manufacturer: [A] Certest Biotec S.L., Zaragoza, Spain [B] Abbot Rapid Diagnostics GmbH, Jena, Germany Antibody: both nucleoprotein antigens Ag target: not stated Test method: [A] LFA; [B] CGIA Samples used: residual NP swabs in VTM Transport media: 3 mL of UTM (Vircell, SL, Granada, Spain, or Deltalab, Barcelona, Spain) Sample storage: samples were cryopreserved at −20 °C until their analysis by Ag‐RDTs Test operator: not stated (lab‐based) Definition of test positivity: visual; control and test lines Blinding reported: unclear Timing of samples: reported for 128 PCR+ samples: 46 (36%) < 5 days pso; 55 (43%) day 6‐10; 27 (21%) > 10 days |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR performed using either of the following:
All target genes present per assay Definition of non‐COVID cases: single negative Genetic target(s):
Samples used: NP samples (processed upon arrival at the laboratory) Timing of reference standard: as for index test Blinded to index test: yes (processed upon arrival at the laboratory before Ag test) Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same swab All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: funded by Certest Biotec S.L. (Zaragoza, Spain); Each manufacturer provided Panbio and CerTest devices. Publication status: published Source: Journal of Clinical Virology Author COI: the authors report no COI |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Perez‐Garcia 2021 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 2 Ag tests; Perez‐Garcia 2021 [A] details the full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 2 Ag tests; Perez‐Garcia 2021 [A] details the full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] CerTest SARS‐CoV‐2 Ag One Step Card Test (Batch code SC‐004)
[B] Panbio COVID‐19 Ag Rapid Test Device (Batch code 41ADF057A) Manufacturer: [A] Certest Biotec S.L., Zaragoza, Spain [B] Abbot Rapid Diagnostics GmbH, Jena, Germany Antibody: both nucleoprotein antigens Ag target: not stated Test method: [A] LFA; [B] CGIA Samples used: residual NP swabs in VTM Transport media: 3 mL of UTM (Vircell, SL, Granada, Spain, or Deltalab, Barcelona, Spain Sample storage: samples were cryopreserved at −20 °C until their analysis by Ag‐RDTs Test operator: not stated (lab‐based) Definition of test positivity: visual; control and test lines Blinding reported: unclear Timing of samples: reported for 128 PCR+ samples: 46 (36%) < 5 days pso; 55 (43%) day 6‐10; 27 (21%) > 10 days |
||
| Target condition and reference standard(s) | Comparative study of 2 Ag tests; Perez‐Garcia 2021 [A] details the full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 2 Ag tests; Perez‐Garcia 2021 [A] details the full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 2 Ag tests; Perez‐Garcia 2021 [A] details the full study characteristics and QUADAS. | ||
Peto 2021(a) [A].
| Study characteristics | |||
| Patient Sampling | Set of studies conducted by PHE and University of Oxford. This extraction relates to a 2‐group study estimating sensitivity and specificity (Phase 3a):
[1] residual frozen swabs from PCR+ patients in Oxford (collected March 2020)
[2] residual fresh swabs from PCR− patients in Oxford (collected March 2020)
Swabs were frozen following routine testing and sent to Porton Down
See other PHE extractions for other substudies of Innova assay Recruitment: unclear Prospective or retrospective: retrospective Sample size (cases): 1118 (178) |
||
| Patient characteristics and setting | Setting: inpatient; obtained from a secondary healthcare setting (cases described as from patients admitted to hospital) Location: John Radcliffe Hospital, Oxford (Ag testing at PHE Porton Down) Country: UK Dates: March 2020 (PCR+) Symptoms and severity: not stated Demographics: not stated Exposure history: not stated |
||
| Index tests | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] data relate to test [A], see additional entries for tests [B] to [G] Test name: [A] Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test [B] Panbio COVID‐19 Ag Rapid Test Device [C] Coronavirus Ag Rapid Test Cassette [D] Anhui Deepblue Medical Technology COVID‐19 [E] Fortress Diagnostics Coronavirus Ag Rapid Test [F] STANDARD Q COVID‐19 Ag Test [G] Surescreen Diagnostics SARS‐CoV‐2 Antigen Rapid Test Cassette Manufacturer: [A] Innova Medical Group [B] Abbott [C] Zhejiang Orient Gene Biotech [D] Deepblue [E] Fortress [F] SD Biosensor [G] Surescreen Antibody: not stated Ag target: not stated Test method: not stated Samples used: NP and OP swabs Transport media: VTM (1 mL) Sample storage: frozen (PCR+); fresh (PCR−) Test operator: laboratory staff Definition of test positivity: visual line; as per manufacturer IFU Blinding reported: not stated Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; from preprint "Viral load (in RNA copies/mL) was quantified from Ct values by using a conversion factor obtained using a dilution calibration series of synthetic genomic RNA (Twist Bioscience) and a standard curve performed using Altona and Taqpath ORF and S target assays. Viral load conversion to RNA copies/mL was performed using the following equation derived from prior calibration curves, logVL = 11.19‐0.304*(delta CT‐4.4)."
Published version further states: "In order to compare the sensitivity of Phase 3a with Phase 3b the Phase 3a viral loads were reduced by log10(2000/300) = 0.82 log units." Definition of non‐COVID cases: single negative PCR Genetic target(s): ORF and S target assays Samples used: appears to be same sample as for Ag test Timing of reference standard: as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same swab All participants received same reference standard: yes Missing data: see below, plus 1 void PCR Uninterpretable results: failure rates reported for test [A] only: [1] 13/191, 7% [2] 50/990, 5.1% Indeterminate results (index test): unclear Indeterminate results (reference standard): unclear Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "the report presents independent research funded by the NIHR, Wellcome Trust and the Department of Health." Publication status: published Source: EClinicalMedicine Author COI: the authors do not have any conflicts of interest Publication status: published |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Peto 2021(a) [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Index tests | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [B] data relate to test [B], see additional entries for other tests Test name: [A] Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test [B] Panbio COVID‐19 Ag Rapid Test Device [C] Coronavirus Ag Rapid Test Cassette [D] Anhui Deepblue Medical Technology COVID‐19 [E] Fortress Diagnostics Coronavirus Ag Rapid Test [F] STANDARD Q COVID‐19 Ag Test [G] Surescreen Diagnostics SARS‐CoV‐2 Antigen Rapid Test Cassette Manufacturer: [A] Innova Medical Group [B] Abbott [C] Zhejiang Orient Gene Biotech [D] Deepblue [E] Fortress [F] SD Biosensor [G] Surescreen Antibody: not stated Ag target: not stated Test method: not stated Samples used: NP and OP swabs Transport media: VTM (1 mL) Sample storage: frozen (PCR+); fresh (PCR−) Test operator: laboratory staff Definition of test positivity: visual line; as per manufacturer IFU Blinding reported: not stated Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Flow and timing | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | |||
Peto 2021(a) [C].
| Study characteristics | |||
| Patient Sampling | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Index tests | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [C] data relate to test [C], see additional entries for other tests Test name: [A] Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test [B] Panbio COVID‐19 Ag Rapid Test Device [C] Coronavirus Ag Rapid Test Cassette [D] Anhui Deepblue Medical Technology COVID‐19 [E] Fortress Diagnostics Coronavirus Ag Rapid Test [F] STANDARD Q COVID‐19 Ag Test [G] Surescreen Diagnostics SARS‐CoV‐2 Antigen Rapid Test Cassette Manufacturer: [A] Innova Medical Group [B] Abbott [C] Zhejiang Orient Gene Biotech [D] Deepblue [E] Fortress [F] SD Biosensor [G] Surescreen Antibody: not stated Ag target: not stated Test method: not stated Samples used: NP and OP swabs Transport media: VTM (1 mL) Sample storage: frozen (PCR+); fresh (PCR−) Test operator: laboratory staff Definition of test positivity: visual line; as per manufacturer IFU Blinding reported: not stated Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Flow and timing | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | |||
Peto 2021(a) [D].
| Study characteristics | |||
| Patient Sampling | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Index tests | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [D] data relate to test [D], see additional entries for other tests Test name: [A] Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test [B] Panbio COVID‐19 Ag Rapid Test Device [C] Coronavirus Ag Rapid Test Cassette [D] Anhui Deepblue Medical Technology COVID‐19 [E] Fortress Diagnostics Coronavirus Ag Rapid Test [F] STANDARD Q COVID‐19 Ag Test [G] Surescreen Diagnostics SARS‐CoV‐2 Antigen Rapid Test Cassette Manufacturer: [A] Innova Medical Group [B] Abbott [C] Zhejiang Orient Gene Biotech [D] Deepblue [E] Fortress [F] SD Biosensor [G] Surescreen Antibody: not stated Ag target: not stated Test method: not stated Samples used: NP and OP swabs Transport media: VTM (1 mL) Sample storage: frozen (PCR+); fresh (PCR−) Test operator: laboratory staff Definition of test positivity: visual line; as per manufacturer IFU Blinding reported: not stated Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Flow and timing | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | |||
Peto 2021(a) [E].
| Study characteristics | |||
| Patient Sampling | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Index tests | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [E] data relate to test [E], see additional entries for other tests Test name: [A] Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test [B] Panbio COVID‐19 Ag Rapid Test Device [C] Coronavirus Ag Rapid Test Cassette [D] Anhui Deepblue Medical Technology COVID‐19 [E] Fortress Diagnostics Coronavirus Ag Rapid Test [F] STANDARD Q COVID‐19 Ag Test [G] Surescreen Diagnostics SARS‐CoV‐2 Antigen Rapid Test Cassette Manufacturer: [A] Innova Medical Group [B] Abbott [C] Zhejiang Orient Gene Biotech [D] Deepblue [E] Fortress [F] SD Biosensor [G] Surescreen Antibody: not stated Ag target: not stated Test method: not stated Samples used: NP and OP swabs Transport media: VTM (1 mL) Sample storage: frozen (PCR+); fresh (PCR−) Test operator: laboratory staff Definition of test positivity: visual line; as per manufacturer IFU Blinding reported: not stated Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Flow and timing | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | |||
Peto 2021(a) [F].
| Study characteristics | |||
| Patient Sampling | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Index tests | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [F] data relate to test [F], see additional entries for other tests Test name: [A] Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test [B] Panbio COVID‐19 Ag Rapid Test Device [C] Coronavirus Ag Rapid Test Cassette [D] Anhui Deepblue Medical Technology COVID‐19 [E] Fortress Diagnostics Coronavirus Ag Rapid Test [F] STANDARD Q COVID‐19 Ag Test [G] Surescreen Diagnostics SARS‐CoV‐2 Antigen Rapid Test Cassette Manufacturer: [A] Innova Medical Group [B] Abbott [C] Zhejiang Orient Gene Biotech [D] Deepblue [E] Fortress [F] SD Biosensor [G] Surescreen Antibody: not stated Ag target: not stated Test method: not stated Samples used: NP and OP swabs Transport media: VTM (1 mL) Sample storage: frozen (PCR+); fresh (PCR−) Test operator: laboratory staff Definition of test positivity: visual line; as per manufacturer IFU Blinding reported: not stated Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Flow and timing | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | |||
Peto 2021(a) [G].
| Study characteristics | |||
| Patient Sampling | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Index tests | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [G] data relate to test [G], see additional entries for other tests Test name: [A] Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test [B] Panbio COVID‐19 Ag Rapid Test Device [C] Coronavirus Ag Rapid Test Cassette [D] Anhui Deepblue Medical Technology COVID‐19 [E] Fortress Diagnostics Coronavirus Ag Rapid Test [F] STANDARD Q COVID‐19 Ag Test [G] Surescreen Diagnostics SARS‐CoV‐2 Antigen Rapid Test Cassette Manufacturer: [A] Innova Medical Group [B] Abbott [C] Zhejiang Orient Gene Biotech [D] Deepblue [E] Fortress [F] SD Biosensor [G] Surescreen Antibody: not stated Ag target: not stated Test method: not stated Samples used: NP and OP swabs Transport media: VTM (1 mL) Sample storage: frozen (PCR+); fresh (PCR−) Test operator: laboratory staff Definition of test positivity: visual line; as per manufacturer IFU Blinding reported: not stated Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Flow and timing | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(a) [A] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | |||
Peto 2021(b) [non‐HCW tested].
| Study characteristics | |||
| Patient Sampling | Set of studies conducted by PHE and University of Oxford. This extraction relates to a single‐group study estimating sensitivity and specificity: individuals presenting at a regional COVID‐19 testing centre as part of a Phase 4 community field service evaluation (n = 1946; according to Table 3 of preprint)
See other PHE extractions for other sub‐studies of Innova assay Recruitment: not stated; presume consecutive Prospective or retrospective: not stated |
||
| Patient characteristics and setting | Setting: regional COVID‐19 testing centres as part of an NHS Test and Trace service evaluation involving the general public Location: not stated Country: UK Dates: not stated Symptoms and severity: not stated, presumed 'mainly symptomatic ' for purposes of review analyses Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test Manufacturer: Innova Medical Group Antibody: not stated Ag target: not stated Test method: not stated Samples used: AN and combined oropharyngeal samples Transport media: dry swab Sample storage: none; immediate testing Test operator: self‐trained non‐HCW ('Boots' member of staff); described in preprint as an “operator” or as "self‐trained members of the public". Definition of test positivity: visual line; as per manufacturer IFU Blinding reported: yes; conducted on site Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: PCR; no details. The preprint supplementary materials describes using the "Roche platform" under the Phase 3b heading, and also provides the following text under the Phase 2 evaluation heading "Unless otherwise stated, all PCR testing was undertaken on the Roche Cobas 6800 or 8800 system using their proprietary SARS‐CoV‐2 assay as per manufacturer IFU (with off‐board lysis using AVL buffer (Qiagen) and 5% Triton‐X100 (Sigma Aldrich)). This assay detects ORF‐1a/b as a SARS‐CoV‐2 specific target, and the E‐gene as a pan‐sarbecovirus target." Definition of non‐COVID cases: N/A; cases‐only study Genetic target(s): not stated Samples used: not stated; paired swabs obtained Timing of reference standard: as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swabs; simultaneous All participants received same reference standard: yes Missing data: initial sample of 1946 reported, 27 failed, leaving 1919 for inclusion, however data for only 1676 samples are provided in the published study (1314 PCR− in Table 3 and 372 PCR+ in text pg 7), a difference of 243 samples. Uninterpretable results: failure rate reported as 27/1946 failed, 1.4% Indeterminate results (index test): unclear Indeterminate results (reference standard): unclear Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: PHE evaluation Publication status: published Source: online PHE report Author COI: none reported |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Peto 2021(c) [A ‐ HCW tested].
| Study characteristics | |||
| Patient Sampling | Set of studies conducted by PHE and University of Oxford. This extraction relates to a single‐group study estimating sensitivity alone: individuals presenting at one of 14 regional drive‐through COVID‐19 NHS Test and Trace centres as part of the FALCON C‐19 ( Facilitating Accelerated Clinical validation Of Novel diagnostics for COVID‐19, 20/WA/0169, IRAS 284229 ) phase 3b study; those with a positive PCR result were asked to return for a re‐test within 5 days of the original test result. From the originally published report (November 2020) it appears that only participants with samples that were positive on PCR at the second sampling were included. (1) One set of samples were tested on site by HCWs using assay [A] only: n = 267; included as Peto 2021(c) [A ‐ HCW tested] (2) Second set of samples were tested in the laboratory by laboratory scientists using four different assays [A] to [D]: n = 212; included as Peto 2021(c) [A ‐ Lab tested], Peto 2021(c) [B ‐ Lab tested], Peto 2021(c) [C ‐ Lab tested], Peto 2021(c) [D ‐ Lab tested] Recruitment: not stated; presume consecutive Prospective or retrospective: prospective Number of samples (cases): 479 (479) |
||
| Patient characteristics and setting | Setting: NHS drive‐through Test and Trace centres; conducted within the FALCON‐C19 study Location: 14 regional centres Country: UK Dates: 17 September‐23 October 2020 Symptoms and severity: only described for 421 included participants combined: 381 (90%) symptomatic; 138 (36%) headache, 134 (35%) cough, 82 (22%) sore throat, 80 (21%) fever, 260 (68%) 'other' not specified symptoms, 59 with no data. 40 (10%) reported asymptomatic Demographics: not stated median age 33 (91 with no data); 168/337 male, 50% (84 with no data recorded) Exposure history: not stated |
||
| Index tests | Comparative study of 4 Ag tests (no product codes reported); Peto 2021(c) [A ‐ HCW tested] and Peto 2021(c) [A ‐ Lab tested] data relate to test [A], see additional entries for tests [B] to [D] Test name: [A] Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test [B] Panbio COVID‐19 Ag Rapid Test Device [C] Coronavirus Ag Rapid Test Cassette [D] Anhui Deepblue Medical Technology COVID‐19 Manufacturer: [A] Innova Medical Group [B] Abbott [C] Zhejiang Orient Gene Biotech [D] Deepblue Antibody: not stated Ag target: not stated Test method: not stated Samples used: combined AN and OP swabs (1 stored as a dry swab and 1 swab placed in VTM; swabs were self‐collected Transport media: dry swab Sample storage: (1) immediate on‐site testing by HCW (2) transport to PHE; tested within 24 h of collection Test operator: (1) Immediate testing by HCW (assay [A]) (2) laboratory scientist tested at PHE (assay [A] to [D]) Definition of test positivity: visual line; as per manufacturer IFU An invalid kit result, or a kit failure was recorded when an operator did not see a control line on the device within a defined time period Blinding reported: yes for on‐site HCW testing; lab scientists reported as unaware of clinical information from the study participants Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: PCR; Roche Cobas 6800 or 8800 system using their proprietary SARS‐CoV‐2 assay
Viral load conversion to RNA copies/mL was performed using the Qnostics SARS‐CoV‐2 Analytical Q Panel 01 (Qnostics, Glasgow, UK); the resulting equation for converting Ct values into viral loads for the Cobas assay, included an adjustment for the dilution, was log10(VL) = 14.17‐0.3316*avct Definition of non‐COVID cases: N/A; cases‐only study Genetic target(s): ORF‐1 and E‐gene Samples used: AN + OP Timing of reference standard: as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired All participants received same reference standard: yes Missing data: yes; appears that only those who remained PCR+ on return for Ag testing were included (the original report (November 2020) documented 16 HCW‐tested samples that were either PCR−ve (n = 15) or void (n = 1) presumably at the time of the second sampling (partially explains discrepancy in numbers) Uninterpretable results: failure rates reported for assay [A] only as: HCW tested 27/267 10.1%; lab scientist tested 9/212 4.2%. NB preliminary report reported these as 28/296 and 9/221 respectively Indeterminate results (index test): unclear Indeterminate results (reference standard): unclear Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: PHE evaluation Publication status: published Source: online PHE report; published paper Author COI: none reported |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | No | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Peto 2021(c) [A ‐ Lab tested].
| Study characteristics | |||
| Patient Sampling | Set of studies conducted by PHE and University of Oxford. This extraction relates to a single‐group study estimating sensitivity alone: individuals presenting at 1 of 14 regional drive‐through COVID‐19 NHS Test and Trace centres as part of the FALCON C‐19 (Facilitating Accelerated Clinical validation Of Novel diagnostics for COVID‐19, 20/WA/0169, IRAS 284229) phase 3b study; those with a positive PCR result were asked to return for a re‐test within 5 days of the original test result. From the originally published report (November 2020) it appears that only participants with samples that were positive on PCR at the second sampling were included. (1) One set of samples were tested on site by HCWs using assay [A] only: n = 267; included as Peto 2021(c) [A ‐ HCW tested] (2) Second set of samples were tested in the laboratory by laboratory scientists using four different assays [A] to [D]: n = 212; included as Peto 2021(c) [A ‐ Lab tested], Peto 2021(c) [B ‐ Lab tested], Peto 2021(c) [C ‐ Lab tested], Peto 2021(c) [D ‐ Lab tested] See other PHE extractions for other sub‐studies of Innova assay Recruitment: not stated; presume consecutive Prospective or retrospective: prospective Number of samples (cases): 479 (479) |
||
| Patient characteristics and setting | Setting: NHS drive through Test and Trace centres; conducted within the FALCON‐C19 study Location: 14 regional centres Country: UK Dates: 17 Sept to 23 Oct 2020 Symptoms and severity: only described for 421 included participants combined: 381 (90%) symptomatic; 138 (36%) headache, 134 (35%) cough, 82 (22%) sore throat, 80 (21%) fever, 260 (68%) 'other' not specified symptoms, 59 with no data. 40 (10%) reported asymptomatic Demographics: median age 33 (91 with no data); 168/337 male, 50% (84 with no data recorded) Exposure history: not stated |
||
| Index tests | Comparative study of 4 Ag tests (no product codes reported); Peto 2021(c) [A ‐ HCW tested] and Peto 2021(c) [A ‐ Lab tested] data relate to test [A], see additional entries for tests [B] to [D] Test name: [A] Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test [B] Panbio COVID‐19 Ag Rapid Test Device [C] Coronavirus Ag Rapid Test Cassette [D] Anhui Deepblue Medical Technology COVID‐19 Manufacturer: [A] Innova Medical Group [B] Abbott [C] Zhejiang Orient Gene Biotech [D] Deepblue Antibody: not stated Ag target: not stated Test method: not stated Samples used: combined AN and OP swabs (1 stored as a dry swab and 1 swab placed in VTM; swabs were self‐collected Transport media: dry swab Sample storage: (1) immediate on‐site testing by HCW (2) transport to PHE; tested within 24 h of collection Test operator: (1) Immediate testing by HCW (assay [A]) (2) laboratory scientist tested at PHE (assay [A] to [D]) Definition of test positivity: visual line; as per manufacturer IFU An invalid kit result, or a kit failure was recorded when an operator did not see a control line on the device within a defined time period Blinding reported: yes for on‐site HCW testing; lab scientists reported as unaware of clinical information from the study participants Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: PCR; Roche Cobas 6800 or 8800 system using their proprietary SARS‐CoV‐2 assay
Viral load conversion to RNA copies/mL was performed using the Qnostics SARS‐CoV‐2 Analytical Q Panel 01 (Qnostics, Glasgow, UK); the resulting equation for converting Ct values into viral loads for the Cobas assay, included an adjustment for the dilution, was log10(VL) = 14.17‐0.3316*avct Definition of non‐COVID cases: N/A; cases‐only study Genetic target(s): ORF‐1 and E‐gene Samples used: AN + OP Timing of reference standard: as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired All participants received same reference standard: yes Missing data: yes; appears that only those who remained PCR+ on return for Ag testing were included (the original report (November 2020) documented 16 HCW‐tested samples that were either PCR−ve (n = 15) or void (n = 1) presumably at the time of the second sampling (partially explains discrepancy in numbers) Uninterpretable results: failure rates reported for assay [A] only as: HCW tested 27/267 10.1%; lab scientist tested 9/212 4.2%. NB preliminary report reported these as 28/296 and 9/221 respectively Indeterminate results (index test): unclear; Indeterminate results (reference standard): unclear Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: PHE evaluation Publication status: published Source: online PHE report; published paper Author COI: none reported |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | No | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Peto 2021(c) [B ‐ Lab tested].
| Study characteristics | |||
| Patient Sampling | Comparative study of 4 Ag tests (no product codes reported); Peto 2021(c) [A ‐ Lab tested] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | Comparative study of 4 Ag tests (no product codes reported); Peto 2021(c) [A ‐ Lab tested] reports full study characteristics and QUADAS | ||
| Index tests | Comparative study of 4 Ag tests (no product codes reported); Peto 2021(c) [B ‐ Lab tested] data relate to test [B], see additional entries for other assays Test name: [A] Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test [B] Panbio COVID‐19 Ag Rapid Test Device [C] Coronavirus Ag Rapid Test Cassette [D] Anhui Deepblue Medical Technology COVID‐19 Manufacturer: [A] Innova Medical Group [B] Abbott [C] Zhejiang Orient Gene Biotech [D] Deepblue Antibody: not stated Ag target: not stated Test method: not stated Samples used: combined AN and OP swabs (1 stored as a dry swab and 1 swab placed in VTM; swabs were self‐collected Transport media: dry swab Sample storage: (1) immediate on‐site testing by HCW (2) transport to PHE; tested within 24 h of collection Test operator: (1) Immediate testing by HCW (assay [A]) (2) laboratory scientist tested at PHE (assay [A] to [D]) Definition of test positivity: visual line; as per manufacturer IFU An invalid kit result, or a kit failure was recorded when an operator did not see a control line on the device within a defined time period Blinding reported: yes for on‐site HCW testing; lab scientists reported as unaware of clinical information from the study participants Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(c) [A ‐ Lab tested] reports full study characteristics and QUADAS | ||
| Flow and timing | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(c) [A ‐ Lab tested] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | |||
Peto 2021(c) [C ‐ Lab tested].
| Study characteristics | |||
| Patient Sampling | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(c) [A ‐ Lab tested] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(c) [A ‐ Lab tested] reports full study characteristics and QUADAS | ||
| Index tests | Comparative study of 4 Ag tests (no product codes reported); Peto 2021(c) [C ‐ Lab tested] data relate to test [C], see additional entries for other assays Test name: [A] Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test [B] Panbio COVID‐19 Ag Rapid Test Device [C] Coronavirus Ag Rapid Test Cassette [D] Anhui Deepblue Medical Technology COVID‐19 Manufacturer: [A] Innova Medical Group [B] Abbott [C] Zhejiang Orient Gene Biotech [D] Deepblue Antibody: not stated Ag target: not stated Test method: not stated Samples used: combined AN and OP swabs (1 stored as a dry swab and 1 swab placed in VTM; swabs were self‐collected Transport media: dry swab Sample storage: (1) immediate on‐site testing by HCW (2) transport to PHE; tested within 24 h of collection Test operator: (1) Immediate testing by HCW (assay [A]) (2) laboratory scientist tested at PHE (assay [A] to [D]) Definition of test positivity: visual line; as per manufacturer IFU An invalid kit result, or a kit failure was recorded when an operator did not see a control line on the device within a defined time period Blinding reported: yes for on‐site HCW testing; lab scientists reported as unaware of clinical information from the study participants Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(c) [A ‐ Lab tested] reports full study characteristics and QUADAS | ||
| Flow and timing | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(c) [A ‐ Lab tested] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | |||
Peto 2021(c) [D ‐ Lab tested].
| Study characteristics | |||
| Patient Sampling | Comparative study of 4 Ag tests (no product codes reported); Peto 2021(c) [A ‐ Lab tested] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | Comparative study of 4 Ag tests (no product codes reported); Peto 2021(c) [A ‐ Lab tested] reports full study characteristics and QUADAS | ||
| Index tests | Comparative study of 4 Ag tests (no product codes reported); Peto 2021(c) [D ‐ Lab tested] data relate to test [D], see additional entries for other assays Test name: [A] Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test [B] Panbio COVID‐19 Ag Rapid Test Device [C] Coronavirus Ag Rapid Test Cassette [D] Anhui Deepblue Medical Technology COVID‐19 Manufacturer: [A] Innova Medical Group [B] Abbott [C] Zhejiang Orient Gene Biotech [D] Deepblue Antibody: not stated Ag target: not stated Test method: not stated Samples used: combined AN and OP swabs (1 stored as a dry swab and 1 swab placed in VTM; swabs were self‐collected Transport media: dry swab Sample storage: (1) immediate on‐site testing by HCW (2) transport to PHE; tested within 24 h of collection Test operator: (1) Immediate testing by HCW (assay [A]) (2) laboratory scientist tested at PHE (assay [A] to [D]) Definition of test positivity: visual line; as per manufacturer IFU An invalid kit result, or a kit failure was recorded when an operator did not see a control line on the device within a defined time period Blinding reported: yes for on‐site HCW testing; lab scientists reported as unaware of clinical information from the study participants Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 4 Ag tests (no product codes reported); Peto 2021(c) [A ‐ Lab tested] reports full study characteristics and QUADAS | ||
| Flow and timing | Comparative study of 7 Ag tests (no product codes reported); Peto 2021(c) [A ‐ Lab tested] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | |||
Peto 2021(d).
| Study characteristics | |||
| Patient Sampling | Set of studies conducted by PHE and University of Oxford. This extraction relates to a single‐group study estimating specificity alone: PHE and hospital staff volunteering for testing (n = 538)
See other PHE extractions for other sub‐studies of Innova assay Recruitment: not stated; presume consecutive Prospective or retrospective: not stated |
||
| Patient characteristics and setting | Setting: screening Location: PHE and John Radcliffe Hospital, Oxford Country: UK Dates: not stated Symptoms and severity: not stated; hospital staff described as asymptomatic Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test Manufacturer: Innova Medical Group Antibody: not stated Ag target: not stated Test method: not stated Samples used: OP swab for PHE staff; NP swab for hospital staff. All self‐collected Transport media: dry swab Sample storage: none; immediate testing Test operator: not stated; presumably laboratory scientist at PHE Definition of test positivity: visual line; as per manufacturer IFU Blinding reported: unclear Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; no details (single negative PCR OK for asymptomatic). The preprint supplementary materials describes using the "Roche platform" under the Phase 3b heading, and also provides the following text under the Phase 2 evaluation heading "Unless otherwise stated, all RT‐PCR testing was undertaken on the Roche Cobas 6800 or 8800 system using their proprietary SARS‐CoV‐2 assay as per manufacturer IFU (with off‐board lysis using AVL buffer (Qiagen) and 5% Triton‐X100 (Sigma Aldrich)). This assay detects ORF‐1a/b as a SARS‐CoV‐2 specific target, and the E‐gene as a pan‐sarbecovirus target." DGenetic target(s): not stated Samples used: not stated; presume same or paired swab Timing of reference standard: as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: unclear, may have been a few days All participants received same reference standard: yes Missing data: initial sample of 570 reported (358 hospital staff and 212 PHE staff), 36 failed (Table 4: 17 hospital staff and 19 PHE staff), leaving 534 for inclusion. Data for 538 included Uninterpretable results: failure rate reported as 17/358, 4.7% (hospital) 19/212, 8.9% (PHE) Indeterminate results (index test): unclear Indeterminate results (reference standard): unclear Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: PHE evaluation Publication status: published Source: online PHE report Author COI: none reported |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
PHE 2020.
| Study characteristics | |||
| Patient Sampling | Set of studies conducted by PHE and University of Oxford. This extraction relates to a single‐group study estimating sensitivity and specificity: samples obtained during a COVID‐19 outbreak at a Navy barracks (n = 157 samples reported in preprint; 2x2 data provided by study investigators)
See other 2021 studies by Peto and colleagues for other PHE substudies of Innova assay Recruitment: unclear; presume consecutive Prospective or retrospective: retrospective |
||
| Patient characteristics and setting | Setting: outbreak investigation Location: not stated Country: UK Dates: not stated Symptoms and severity: not stated Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: Innova SARS‐CoV‐2 Antigen Rapid Qualitative Test Manufacturer: Innova Medical Group Antibody: not stated Ag target: not stated Test method: not stated Samples used: OP swab used; self‐collected Transport media: VTM Sample storage: transported at 4 °C to Porton Down for testing Test operator: laboratory staff Definition of test positivity: visual line; as per manufacturer IFU Blinding reported: not stated Timing of samples: 1 week after outbreak; no further details |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; not described. The preprint supplementary materials describes using the "Roche platform" under the Phase 3b heading, and also provides the following text under the Phase 2 evaluation heading "Unless otherwise stated, all RT‐PCR testing was undertaken on the Roche Cobas 6800 or 8800 system using their proprietary SARS‐CoV‐2 assay as per manufacturer IFU (with off‐board lysis using AVL buffer (Qiagen) and 5% Triton‐X100 (Sigma Aldrich)). This assay detects ORF‐1a/b as a SARS‐CoV‐2 specific target, and the E‐gene as a pan‐sarbecovirus target." Definition of non‐COVID cases: single negative PCR Genetic target(s): not stated Samples used: appears to be same sample as for Ag test Timing of reference standard: as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same swab All participants received same reference standard: yes Missing data: none reported Uninterpretable results: failure rate reported as 6/157, 3.8% (Table 4 of preprint)NB resulting no. samples per group (n = 151) does not quite match with final number reported (n = 152) Indeterminate results (index test): unclear Indeterminate results (reference standard): unclear Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: PHE evaluation Publication status: published and unpublished Source: online PHE report, plus additional data provided by evaluation team Author COI: none reported |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Pickering 2021(a) [A].
| Study characteristics | |||
| Patient Sampling | Multi group study estimating sensitivity and specificity: all swabs selected from those submitted to the diagnostic laboratory
[1] PCR+ve swabs selected to cover a wide range of Ct values (14‐39) (n = 100)
[2] PCR−ve swabs (n = 100)
[1] and [2] used for comparison of 6 RDTs; included as Pickering 2021(a) [A]
[3] PCR+ve swabs with culture results for assessment of infectivity (3 RDTs compared) (n = 141); included as Pickering 2021(b) [A]
[4] PCR+ve swabs infected from the B.1.1.7 variant (2 RDTs compared) (n = 23); included as Pickering 2021(c) [A]
(Routinely collected serum samples also reported for antibody testing but not further considered for this review) Recruitment: unclear; deliberate sampling Prospective or retrospective: retrospective Sample size (cases): [1] + [2] 200 (100) |
||
| Patient characteristics and setting | Setting: unclear; swabs submitted to the diagnostic laboratory for routine testing Location: Viapath Infection Sciences laboratory, St Thomas’ Hospital, London (Department of Infectious Diseases, School of Immunology & Microbial Sciences, King’s College London) Country: UK Dates: [1] to [3] March‐October 2020 [4] January 2021 Symptoms and severity: not stated Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: [A] Innova [B] E25 Bio [C] Sure Screen V (COVID19 AGVCT) [D] Spring (SP‐SW 106) [E] Encode [F] Sure Screen F (COVID19 AGC) Manufacturer: [A] Innova Med Group, China [B] E25 Bio, USA [C] Sure Screen Diagnostics Ltd [D] Spring Healthcare, UK [E] Encode/Emmo Pharma [F] Sure Screen Diagnostics Ltd Antibody: all nucleocapsid; "Given that the rapid antigen tests rely on antibody detection of SARS‐CoV‐2 nucleocapsid (N)" Ag target: not reported Test method: [A], [B], [D] CGIA [C], [E] LFA (not otherwise specified) [F] FIA Samples used: combined nasal/OP Transport media: VTM (1 mL) Sample storage: stored at −80 °C at the Directorate of Infection, prior to selection and forwarding to KCL laboratories for analysis [50 µL of stored swab was mixed with 100 µL of buffer supplied with the test kit, and 100 µL of this was applied to the test cassette] Test operator: not specified; lab‐based Definition of test positivity: visual and according to manufacturer IFU. Results recorded independently by 2 readers and discordant results referred to a third individual. CGIA tests scored according to whether the test band was strongly positive (2), clearly positive (1), weakly positive (0.5) or negative (0). Blinding reported: unclear Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: PCR; AusDiagnostics Multiplex Tandem SARS‐CoV‐2 PCR assays used for diagnosis.Additional PCR testing used an in‐house PCR. Viral culture performed using Vero.E6 cells; incubated for 48 h Definition of non‐COVID cases: single negative PCR Genetic target(s): N gene or human RNAse P Samples used: combined nasal/OP; same as for index Timing of reference standard: not stated Blinded to index test: yes, PCR performed before index test Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same swab All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "Department of Health via a National Institute for Health Research comprehensive Biomedical Research Centre award to Guy’s and St. Thomas’ NHS Foundation Trust in partnership with King’s College London and King’s College Hospital NHS Foundation Trust" Publication status: preprint Source: medRxiv Author COI: authors report no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Pickering 2021(a) [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] Innova [B] E25 Bio [C] Sure Screen V (COVID19 AGVCT) [D] Spring (SP‐SW 106) [E] Encode [F] Sure Screen F (COVID19 AGC) Manufacturer: [A] Innova Med Group, China [B] E25 Bio, USA [C] Sure Screen Diagnostics Ltd [D] Spring Healthcare, UK [E] Encode/Emmo Pharma [F] Sure Screen Diagnostics Ltd Antibody: all nucleocapsid; "Given that the rapid antigen tests rely on antibody detection of SARS‐CoV‐2 nucleocapsid (N)" Ag target: not reported Test method: [A], [B], [D] CGIA [C], [E] LFA (not otherwise specified) [F] FIA Samples used: combined nasal/OP Transport media: VTM (1 mL) Sample storage: stored at −80 °C at the Directorate of Infection, prior to selection and forwarding to KCL laboratories for analysis [50 µL of stored swab was mixed with 100 µL of buffer supplied with the test kit, and 100 µL of this was applied to the test cassette] Test operator: not specified; lab‐based Definition of test positivity: visual and according to manufacturer IFU. Results recorded independently by 2 readers and discordant results referred to a third individual. CGIA tests scored according to whether the test band was strongly positive (2), clearly positive (1), weakly positive (0.5) or negative (0). Blinding reported: unclear Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
Pickering 2021(a) [C].
| Study characteristics | |||
| Patient Sampling | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] Innova [B] E25 Bio [C] Sure Screen V (COVID19 AGVCT) [D] Spring (SP‐SW 106) [E] Encode [F] Sure Screen F (COVID19 AGC) Manufacturer: [A] Innova Med Group, China [B] E25 Bio, USA [C] Sure Screen Diagnostics Ltd, [D] Spring Healthcare, UK [E] Encode/Emmo Pharma [F] Sure Screen Diagnostics Ltd Antibody: all nucleocapsid; "Given that the rapid antigen tests rely on antibody detection of SARS‐CoV‐2 nucleocapsid (N)" Ag target: not reported Test method: [A], [B], [D] CGIA [C], [E] LFA (not otherwise specified) [F] FIA Samples used: combined nasal/OP Transport media: VTM (1 mL) Sample storage: stored at −80 °C at the Directorate of Infection, prior to selection and forwarding to KCL laboratories for analysis [50 µL of stored swab was mixed with 100 µL of buffer supplied with the test kit, and 100 µL of this was applied to the test cassette] Test operator: not specified; lab‐based Definition of test positivity: visual and according to manufacturer IFU. Results recorded independently by 2 readers and discordant results referred to a third individual. CGIA tests scored according to whether the test band was strongly positive (2), clearly positive (1), weakly positive (0.5) or negative (0). Blinding reported: unclear Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
Pickering 2021(a) [D].
| Study characteristics | |||
| Patient Sampling | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] Innova [B] E25 Bio [C] Sure Screen V (COVID19 AGVCT) [D] Spring (SP‐SW 106) [E] Encode [F] Sure Screen F (COVID19 AGC) Manufacturer: [A] Innova Med Group, China [B] E25 Bio, USA [C] Sure Screen Diagnostics Ltd [D] Spring Healthcare, UK [E] Encode/Emmo Pharma [F] Sure Screen Diagnostics Ltd Antibody: all nucleocapsid; "Given that the rapid antigen tests rely on antibody detection of SARS‐CoV‐2 nucleocapsid (N)" Ag target: not reported Test method: [A], [B], [D] CGIA [C], [E] LFA (not otherwise specified) [F] FIA Samples used: combined nasal/OP Transport media: VTM (1 mL) Sample storage: stored at −80 °C at the Directorate of Infection, prior to selection and forwarding to KCL laboratories for analysis [50 µL of stored swab was mixed with 100 µL of buffer supplied with the test kit, and 100 µL of this was applied to the test cassette] Test operator: not specified; lab‐based Definition of test positivity: visual and according to manufacturer IFU. Results recorded independently by 2 readers and discordant results referred to a third individual. CGIA tests scored according to whether the test band was strongly positive (2), clearly positive (1), weakly positive (0.5) or negative (0). Blinding reported: unclear Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
Pickering 2021(a) [E].
| Study characteristics | |||
| Patient Sampling | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] Innova [B] E25 Bio [C] Sure Screen V (COVID19 AGVCT) [D] Spring (SP‐SW 106) [E] Encode [F] Sure Screen F (COVID19 AGC) Manufacturer: [A] Innova Med Group, China [B] E25 Bio, USA [C] Sure Screen Diagnostics Ltd [D] Spring Healthcare, UK [E] Encode/Emmo Pharma [F] Sure Screen Diagnostics Ltd Antibody: all nucleocapsid; "Given that the rapid antigen tests rely on antibody detection of SARS‐CoV‐2 nucleocapsid (N)" Ag target: not reported Test method: [A], [B], [D] CGIA [C], [E] LFA (not otherwise specified) [F] FIA Samples used: combined nasal/OP Transport media: VTM (1 mL) Sample storage: stored at −80 °C at the Directorate of Infection, prior to selection and forwarding to KCL laboratories for analysis [50 µL of stored swab was mixed with 100 µL of buffer supplied with the test kit, and 100 µL of this was applied to the test cassette] Test operator: not specified; lab‐based Definition of test positivity: visual and according to manufacturer IFU. Results recorded independently by 2 readers and discordant results referred to a third individual. CGIA tests scored according to whether the test band was strongly positive (2), clearly positive (1), weakly positive (0.5) or negative (0). Blinding reported: unclear Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
Pickering 2021(a) [F].
| Study characteristics | |||
| Patient Sampling | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] Innova [B] E25 Bio [C] Sure Screen V (COVID19 AGVCT) [D] Spring (SP‐SW 106) [E] Encode [F] Sure Screen F (COVID19 AGC) Manufacturer: [A] Innova Med Group, China [B] E25 Bio, USA [C] Sure Screen Diagnostics Ltd [D] Spring Healthcare, UK [E] Encode/Emmo Pharma [F] Sure Screen Diagnostics Ltd Antibody: all nucleocapsid; "Given that the rapid antigen tests rely on antibody detection of SARS‐CoV‐2 nucleocapsid (N)" Ag target: not reported Test method: [A], [B], [D] CGIA [C], [E] LFA (not otherwise specified) [F] FIA Samples used: combined nasal/OP Transport media: VTM (1 mL) Sample storage: stored at −80 °C at the Directorate of Infection, prior to selection and forwarding to KCL laboratories for analysis [50 µL of stored swab was mixed with 100 µL of buffer supplied with the test kit, and 100 µL of this was applied to the test cassette] Test operator: not specified; lab‐based Definition of test positivity: visual and according to manufacturer IFU. Results recorded independently by 2 readers and discordant results referred to a third individual. CGIA tests scored according to whether the test band was strongly positive (2), clearly positive (1), weakly positive (0.5) or negative (0). Blinding reported: unclear Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 6 Ag tests; Pickering 2021(a) [A] details the full study characteristics and QUADAS. | ||
Pickering 2021(b) [A].
| Study characteristics | |||
| Patient Sampling | Multi‐group study estimating sensitivity and specificity: all swabs selected from those submitted to the diagnostic laboratory
[3] RT‐PCR+ve swabs with culture results for assessment of infectivity (3 RDTs compared) (n = 141); included as Pickering 2021(b) [A]
A further 3 groups were reported:
[1] RT‐PCR+ve swabs selected to cover a wide range of Ct values (14‐39) (n = 100) and [2] RT‐PCR−ve swabs (n = 100) used for comparison of 6 RDTs; included as Pickering 2021(a) [A]
[4] RT‐PCR+ve swabs infected from the B.1.1.7 variant (2 RDTs compared) (n = 23); included as Pickering 2021(c) [A]
(Routinely collected serum samples also reported for antibody testing but not further considered for this review) Recruitment: unclear; deliberate sampling Prospective or retrospective: retrospective Sample size (cases): [2] 141 (141) |
||
| Patient characteristics and setting | Setting: unclear; swabs submitted to the diagnostic laboratory for routine testing Location: Viapath Infection Sciences laboratory, St Thomas’ Hospital, London (Department of Infectious Diseases, School of Immunology & Microbial Sciences, King’s College London) Country: UK Dates: [1] to [3] March‐October 2020 [4] January 2021 Symptoms and severity: not stated Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: [A] Innova [B] Encode [C] Sure Screen F (COVID19 AGC) Manufacturer: [A] Innova Med Group, China [B] Encode/Emmo Pharma [C] Sure Screen Diagnostics Ltd Antibody: all nucleocapsid; "Given that the rapid antigen tests rely on antibody detection of SARS‐CoV‐2 nucleocapsid (N)" Ag target: not reported Test method: [A] CGIA [B] LFA (not otherwise specified) [C] FIA Samples used: combined nasal/OP Transport media: VTM (1 mL) Sample storage: stored at −80 °C at the Directorate of Infection, prior to selection and forwarding to KCL laboratories for analysis [50 µL of stored swab was mixed with 100 µL of buffer supplied with the test kit, and 100 µL of this was applied to the test cassette] Test operator: not specified; lab‐based Definition of test positivity: visual and according to manufacturer IFU. Results recorded independently by 2 readers and discordant results referred to a third individual. CGIA tests scored according to whether the test band was strongly positive (2), clearly positive (1), weakly positive (0.5) or negative (0). Blinding reported: unclear Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; AusDiagnostics Multiplex Tandem SARS‐CoV‐2 PCR assays used for diagnosis.
Additional PCR testing used an in‐house RT‐PCR
Viral culture performed using Vero.E6 cells; incubated for 48 h Definition of non‐COVID cases: Genetic target(s): N gene or human RNAse P Samples used: combined nasal/OP; same as for index Timing of reference standard: not stated Blinded to index test: yes, PCR performed before index test Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same swab All participants received same reference standard: yes Missing data: yes; insufficient sample volume to conduct all 3 tests on all 141 samples; 31 missing for Innova and 51 missing for Encode Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "Department of Health via a National Institute for Health Research comprehensive Biomedical Research Centre award to Guy’s and St. Thomas’ NHS Foundation Trust in partnership with King’s College London and King’s College Hospital NHS Foundation Trust" Publication status: preprint Source: medRxiv Author COI: authors report no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Pickering 2021(b) [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 3 Ag tests; Pickering 2021(b) [A] details the full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 3 Ag tests; Pickering 2021(b) [A] details the full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] Innova [B] Encode [C] Sure Screen F (COVID19 AGC) Manufacturer: [A] Innova Med Group, China [B] Encode/Emmo Pharma [C] Sure Screen Diagnostics Ltd Antibody: all nucleocapsid; "Given that the rapid antigen tests rely on antibody detection of SARS‐CoV‐2 nucleocapsid (N)" Ag target: not reported Test method: [A] CGIA [B] LFA (not otherwise specified) [C] FIA Samples used: combined nasal/OP Transport media: VTM (1 mL) Sample storage: stored at −80 °C at the Directorate of Infection, prior to selection and forwarding to KCL laboratories for analysis [50 µL of stored swab was mixed with 100 µL of buffer supplied with the test kit, and 100 µL of this was applied to the test cassette] Test operator: not specified; lab‐based Definition of test positivity: visual and according to manufacturer IFU. Results recorded independently by 2 readers and discordant results referred to a third individual. CGIA tests scored according to whether the test band was strongly positive (2), clearly positive (1), weakly positive (0.5) or negative (0). Blinding reported: unclear Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 3 Ag tests; Pickering 2021(b) [A] details the full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 3 Ag tests; Pickering 2021(b) [A] details the full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 3 Ag tests; Pickering 2021(b) [A] details the full study characteristics and QUADAS. | ||
Pickering 2021(b) [C].
| Study characteristics | |||
| Patient Sampling | Comparative study of 3 Ag tests; Pickering 2021(b) [A] details the full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 3 Ag tests; Pickering 2021(b) [A] details the full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] Innova [B] Encode [C] Sure Screen F (COVID19 AGC) Manufacturer: [A] Innova Med Group, China [B] Encode/Emmo Pharma [C] Sure Screen Diagnostics Ltd Antibody: all nucleocapsid; "Given that the rapid antigen tests rely on antibody detection of SARS‐CoV‐2 nucleocapsid (N)" Ag target: not reported Test method: [A] CGIA [B] LFA (not otherwise specified) [C] FIA Samples used: combined nasal/OP Transport media: VTM (1 mL) Sample storage: stored at −80 °C at the Directorate of Infection, prior to selection and forwarding to KCL laboratories for analysis [50 µL of stored swab was mixed with 100 µL of buffer supplied with the test kit, and 100 µL of this was applied to the test cassette] Test operator: not specified; lab‐based Definition of test positivity: visual and according to manufacturer IFU. Results recorded independently by 2 readers and discordant results referred to a third individual. CGIA tests scored according to whether the test band was strongly positive (2), clearly positive (1), weakly positive (0.5) or negative (0). Blinding reported: unclear Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 3 Ag tests; Pickering 2021(b) [A] details the full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 3 Ag tests; Pickering 2021(b) [A] details the full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 3 Ag tests; Pickering 2021(b) [A] details the full study characteristics and QUADAS. | ||
Pickering 2021(c) [A].
| Study characteristics | |||
| Patient Sampling | Multi‐group study estimating sensitivity and specificity: all swabs selected from those submitted to the diagnostic laboratory
[4] RT‐PCR positive swabs infected from the B.1.1.7 variant (2 RDTs compared) (n = 23); included as Pickering 2021(c) [A]
A further 3 groups were reported:
[1] RT‐PCR positive swabs selected to cover a wide range of Ct values (14‐39) (n = 100) and [2] RT‐PCR negative swabs (n = 100) used for comparison of 6 RDTs; included as Pickering 2021(a) [A]
[3] RT‐PCR positive swabs with culture results for assessment of infectivity (3 RDTs compared) (n = 141); included as Pickering 2021(b) [A]
(Routinely collected serum samples also reported for antibody testing but not further considered for this review) Recruitment: unclear; deliberate sampling Prospective or retrospective: retrospectively Sample size (cases): [4] 23 (23) |
||
| Patient characteristics and setting | Setting: unclear; swabs submitted to the diagnostic laboratory for routine testing Location: Viapath Infection Sciences laboratory, St Thomas’ Hospital, London (Department of Infectious Diseases, School of Immunology & Microbial Sciences, King’s College London) Country: UK Dates: [4] January 2021 [1] to [3] March‐October 2020 Symptoms and severity: not stated Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: [A] Innova [B] Sure Screen V (COVID19 AGVCT) Manufacturer: [A] Innova Med Group, China [B] Sure Screen Diagnostics Ltd Antibody: all nucleocapsid; "Given that the rapid antigen tests rely on antibody detection of SARS‐CoV‐2 nucleocapsid (N)" Ag target: not reported Test method: [A] and B] Samples used: combined nasal/OP Transport media: VTM (1 mL) Sample storage: stored at −80 °C at the Directorate of Infection, prior to selection and forwarding to KCL laboratories for analysis [50 µL of stored swab was mixed with 100 µL of buffer supplied with the test kit, and 100 µL of this was applied to the test cassette] Test operator: not specified; lab‐based Definition of test positivity: visual and according to manufacturer IFU. Results recorded independently by 2 readers and discordant results referred to a third individual. CGIA tests scored according to whether the test band was strongly positive (2), clearly positive (1), weakly positive (0.5) or negative (0). Blinding reported: unclear Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; AusDiagnostics Multiplex Tandem SARS‐CoV‐2 PCR assays used for diagnosis.
Additional PCR testing used an in‐house RT‐PCR
Viral culture performed using Vero.E6 cells; incubated for 48 h Definition of non‐COVID cases: Genetic target(s): N gene or human RNAse P Samples used: combined nasal/OP; same as for index Timing of reference standard: not stated Blinded to index test: yes, PCR performed before index test Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same swab All participants received same reference standard: yes Missing data: yes; insufficient sample volume to conduct all 3 tests on all 141 samples; 31 missing for Innova and 51 missing for Encode Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "Department of Health via a National Institute for Health Research comprehensive Biomedical Research Centre award to Guy’s and St. Thomas’ NHS Foundation Trust in partnership with King’s College London and King’s College Hospital NHS Foundation Trust" Publication status: preprint Source: medRxiv Author COI: authors report no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Pickering 2021(c) [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 2 Ag tests; Pickering 2021(c) [A] details the full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 2 Ag tests; Pickering 2021(c) [A] details the full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] Innova [B] Sure Screen V (COVID19 AGVCT) Manufacturer: [A] Innova Med Group, China [B] Sure Screen Diagnostics Ltd Antibody: all nucleocapsid; "Given that the rapid antigen tests rely on antibody detection of SARS‐CoV‐2 nucleocapsid (N)" Ag target: not reported Test method: [A] and B] Samples used: combined nasal/OP Transport media: VTM (1 mL) Sample storage: stored at −80 °C at the Directorate of Infection, prior to selection and forwarding to KCL laboratories for analysis [50 µL of stored swab was mixed with 100 µL of buffer supplied with the test kit, and 100 µL of this was applied to the test cassette] Test operator: not specified; lab‐based Definition of test positivity: visual and according to manufacturer IFU. Results recorded independently by 2 readers and discordant results referred to a third individual. CGIA tests scored according to whether the test band was strongly positive (2), clearly positive (1), weakly positive (0.5) or negative (0). Blinding reported: unclear Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 2 Ag tests; Pickering 2021(c) [A] details the full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 2 Ag tests; Pickering 2021(c) [A] details the full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 2 Ag tests; Pickering 2021(c) [A] details the full study characteristics and QUADAS. | ||
Pilarowski 2020a.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: testing freely available to people of all ages, with or without symptoms. Community workers conducted door‐to‐door mobilization in 3 census tracts surrounding the testing site 4 days before testing. Recruitment: consecutive (not stated but all who attended were included) Prospective or retrospective: prospective Sample size (cases): 3302 (237) |
||
| Patient characteristics and setting | Setting: COVID‐19 test centres; community testing site Location: plaza at an urban commercial transport hub in the Mission neighbourhood, San Francisco (University of California, San Francisco) Country: USA Dates: 22 November‐1 December 2020 Symptoms and severity: 30.9% (n = 1020) self‐reported possible COVID‐19 symptoms; results reported for 341 (10%) symptomatic (≤ 7d pso) and 2402 (90%) asymptomatic or symptomatic (> 7d pso) Of 237 PCR+ve, 95 were asymptomatic, 7 were symptomatic (> 7 d pso), and 135 symptomatic (≤ 7 d pso) Demographics: 1750 (53%) male; 99 (3%) aged < 13 years, 110 (3%) aged 13‐18 years, and 3093 (94%) aged > 18 years 2166 (66%) Latinx, 304 (9%) Asian, 558 (17%) white, 53 (2%) American Indian, and 83 (3%) black Exposure history: not reported; a setting of ongoing community transmission |
||
| Index tests | Test name: BinaxNOW Manufacturer: Abbott Antibody: not reported Ag target: none reported Test method: CGIA Samples used: nasal (AN) (collected by laboratory assistants) Transport media: none used Sample storage: none required; immediate on‐site testing Test operator: laboratory technicians (certified technician readers) Definition of test positivity: visual; according to manufacturer IFU Blinding reported: yes performed before PCR Timing of samples: symptomatic with positive Ag result: median 3d (IQR 2‐5 d) pso (n = 134) |
||
| Target condition and reference standard(s) | Reference standard: PCR (single assay, with positives confirmed on 2nd assay);
(1) multiplex PCR conducted by RenegadeBio using RenegadeXP technology;
(2) positive results confirmed following US CDC methodology (singleplex PCR)
Ct not reported; results reported for ≤ 30 Ct, ≤ 35 Ct, and "no Ct cutoff") Definition of non‐COVID cases: same as for cases; single negative PCR for absence of disease Genetic target(s): (1) and (2) N‐gene Samples used: nasal (AN) in VTM; paired swab (same site as index) Timing of reference standard: same as for index test Blinded to index test: not reported Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired sample; simultaneous All participants received same reference standard: yes; but only PCR+ on first assay had confirmation on 2nd assay Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "funding for this study was provided by the University of California San Francisco, Program for Breakthrough Biomedical Research, which is partially funded by the Sandler Foundation, a private donor, the Chan Zuckerberg Initiative, and the National Institutes of Health [UM1AI069496]". Publication status: published Source: Clinical Infectious Diseases Author COI: COI declared: "Dr. Havlir reports nonfinancial support from Abbott, outside the submitted work; none of the other authors has any potential conflicts" |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Pilarowski 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: mainly asymptomatic suspected patients presenting at a walk‐up, free testing at a plaza located at an intersection of the Bay Area‐wide subway system (BART) and the San Francisco city bus/streetcar system (MUNI), Mission District, California Recruitment: not reported; appears to be all presenting for testing during study period Prospective or retrospective: prospective Sample size (cases): 878 (26) |
||
| Patient characteristics and setting | Setting: community screening/COVID‐19 test centre Location: walk‐up/free testing at a community plaza, San Francisco Country: USA Dates: September 2020 Symptoms and severity: mainly asymptomatic (84% reported no symptoms during the 14 days before testing) Demographics: 54% male; 77% 18‐50 years of age; 81% self‐identified as Latinx Exposure history: not reported |
||
| Index tests | Test name: BinaxNOW COVID‐19 Ag Card Manufacturer: Abbott Laboratories Antibody: N protein Ag target: not reported Test method: CGIA Samples used: AN (both nares) (Laboratory technician) Transport media: none required Sample storage: none reported Test operator: laboratory technician; on site Definition of test positivity: visual colour band; each assay was read by 2 independent observers, and a site supervisor served as a tiebreaker. Interpretation amended following first 217 samples because of high FP (9/207 PCR−ve); bands were subsequently scored as positive only if they extended across the full width of the strip, irrespective of the intensity of the band Blinding reported: yes; performed before PCR Timing of samples: mainly asymptomatic; timing not systematically reported for symptomatic group |
||
| Target condition and reference standard(s) | Reference standard: PCR (no further details, 2 prior studies cited for reference); in‐vitro culture Definition of non‐COVID cases: single negative PCR Genetic target(s): not reported Samples used: AN, paired Timing of reference standard: same as for index Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous, paired All participants received same reference standard: yes Missing data: unclear; 871/878 in the analysis Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "this study was supported by the University of California, San Francisco, the Chan Zuckerberg Biohub, the Chan Zuckerberg Initiative, the San Francisco Latino Task Force, the National Institute of Allergy and Infectious Diseases (grants T32 AI060530 to LR and F31AI150007 to SS), and a private donor." Publication status: published Source: Journal of Infectious Diseases Author COI: all authors reported no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Pollock 2021a.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: symptomatic and asymptomatic adults and children who attended the drive‐through,
free community testing site in Massachusetts Recruitment: consecutive (all who attended) Prospective or retrospective: prospective Sample size (cases): 2482 included; 2308 analysed (292) |
||
| Patient characteristics and setting | Setting: COVID‐19 drive‐through testing site; screening (appears to be open to all; no specific testing criteria applied) Location: the Lawrence General Hospital “Stop the Spread” drive‐through testing site, which accommodates Massachusetts residents from the surrounding area. Country: USA Dates: 26 October‐22 December 2020 Symptoms and severity: adults: 406 (29%) symptomatic; 974 (71%) asymptomatic Children: 99 (11%) symptomatic; 829 (89%) asymptomatic Adults: median 3 (IQR 2‐5) pso days Children: median 2 (IQR 1‐4) pso days Demographics: adults: 59% symptomatic female; 56% asymptomatic female Age: 19‐29 years 332, 24%; 30‐49 years 581, 42%; 50‐69 years 401, 29%, > 70 years 66, 5% Children: 62% symptomatic; 52% asymptomatic female Age: < 7 years 261, 28%; 7‐13 years 381, 41%; 14‐18 years 286, 31% Exposure history: not reported |
||
| Index tests | Test name: BinaxNOW COVID‐19 Ag Card Manufacturer: Abbott Diagnostics, USA Antibody: nucleocapsid protein antigen (by knowledge) Ag target: not stated Test method: CGIA (by knowledge) Samples used: AN (collected by trained operators); both nostrils swabbed; swab rotated 5 times in a circular motion around the inside wall of the nostril for a duration of 10‐15 s Transport media: none; swab placed into a specimen collection bag Sample storage: none; testing within 1 h of collection "tests initiated within an hour of collection time at the temperature of 59°F as per manufacturer's recommendation." Test operator: trained operators Definition of test positivity: visual read‐out; test lines recorded as "faint", "medium", or "strong" There was no attempt to resolve any discordance between the 2 readers Blinding reported: yes; conducted before PCR Timing of samples: for symptomatic: adults: median 3 (IQR 2‐5) days Children: median 2 (IQR 1‐4) days |
||
| Target condition and reference standard(s) | Reference standard: PCR; extraction and PCR methods followed the EUA protocol for the CRSP
SARS‐CoV‐2 Real‐time Reverse Transcriptase (RT)‐PCR Diagnostic Assay
Ct cut‐off value of 40 Definition of non‐COVID cases: same as for cases for symptomatic (single ‐ve PCR); N/A for asymptomatic Genetic target(s): N2 gene Samples used: AN (paired); transported as dry swab then suspended in swab preservation buffer before testing Timing of reference standard: as for index test Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swab; simultaneous All participants received same reference standard: yes Missing data: missing data, n = 54; excluded, n = 94 (samples tested at < 59 °F) Uninterpretable results: inconclusive PCR results n = 26; 1 invalid BinaxNOW result (a manufacturing issue whereby plastic covered the test strip, preventing the buffer from making contact with the test strip); presume test was repeated Indeterminate results (index test): none reported; all FP results had faint but detectable test bands Indeterminate results (reference standard): inconclusive PCR results 189 (n = 26) Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: published version: "supported by Cooperative Agreement Number 1U60OE000103, funded by Centers for Disease Control and Prevention through the Association of Public Health Laboratories."
Preprint: "funded by the MA Department of Public Health. The community testing site was funded by the Centers for Disease Control and Prevention Building and Enhancing Epidemiology, Laboratory and Health Information Systems Capacity in Massachusetts –Enhancing Detection COVID Supplement (Grant # 6 NU50CK000518‐01‐08). BinaxNOW kits were supplied as part of the federal allocation to state health departments." Publication status: published Source: Journal of Clinical Microbiology Author COI: authors report no conflict of interest to declare |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Pollock 2021b.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: individuals presenting for testing to a high‐throughput, drive‐through, free
community testing site; no specific criteria for testing had to be met Recruitment: consecutive Prospective or retrospective: prospective Sample size (cases): 1498 (234); from 1603 eligible |
||
| Patient characteristics and setting | Setting: drive‐through testing site; screening Location: Lawrence General Hospital “Stop the Spread” drive‐through testing site (Department of Laboratory Medicine, Boston Children’s Hospital, Boston, MA) Country: USA Dates: 11‐22 January 2021 Symptoms and severity: 1257 (84%) asymptomatic, including 1036/1245 (69%) adults; 209 symptomatic 221/253 (15%) children; 32 symptomatic Demographics: adult: 57% symptomatic female; 53% asymptomatic female Children: 56% symptomatic female; 53% asymptomatic female Age group: < 7 years: 13 (41%) symptomatic, 60 (27%) asymptomatic 7‐13 years: 12 (37) symptomatic, 73 (33%) asymptomatic 14‐18 years: 7 (22%) symptomatic, 88 (40%) asymptomatic 19‐29 years: 58 (28%) symptomatic, 313 (30%) asymptomatic 30‐49 years: 102 (49%) symptomatic, 381 (37%) asymptomatic 50‐69 years: 42 (20%) symptomatic, 290 (28%) asymptomatic > 70 years: 7 (3%) symptomatic, 52 (5%) asymptomatic Exposure history: not stated |
||
| Index tests | Test name: CareStart COVID‐19 Antigen test Manufacturer: Access Bio Antibody: nucleocapsid Ag target: not stated Test method: chromatographic immunoassay Samples used: AN swab (study site personnel) Both nostrils swabbed with each swab, alternating which swab was collected first (for PCR vs CareStart) Transport media: none used Sample storage: tested within 1 h of collection; median interval between sample collection and test initiation was 31 min (range 12–103 min) Test operator: trained operators (Master’s or PhD‐level laboratorians); according to the manufacturer IFU Definition of test positivity: visual; according to the manufacturer IFU 2 operators read the result; first read of each test was the official result used Blinding reported: yes; performed before PCR test Timing of samples: 209 symptomatic adults: median 3 days pso (IQR 2‐6) 32 symptomatic children: median 3 days pso (IQR 2‐4) |
||
| Target condition and reference standard(s) | Reference standard: PCR (CRSP SARS‐CoV‐2 Real‐time Reverse Transcriptase‐PCR Diagnostic Assay under EUA);
Ct cut‐off value = 40 Definition of non‐COVID cases: as for cases (single ‐ve for symptomatic) Genetic target(s): N2 gene Samples used: AN swab; paired Timing of reference standard: same as for index Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous All participants received same reference standard: yes Missing data: 105/1603 (6.5%) (invalid or missing PCR results (n = 48) and missing clinical data (n = 57)) Uninterpretable results: 8 discordant results (all faint positive vs negative); 2 readers disagreed on the strength of the positive band (faint vs 219 medium vs strong) in 7 cases Indeterminate results (index test): none; states "No invalid CareStart test results were observed." Indeterminate results (reference standard): 48 invalid or missing PCR results Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "Department of Public Health, MA; Centers for Disease Control and Prevention Building and Enhancing Epidemiology, Laboratory and Health Information Systems Capacity in Massachusetts – Enhancing Detection COVID Supplement (Grant # 6 NU50CK000518‐01‐08). CareStart kits were donated by the manufacturer." Publication status: preprint Source: medRxiv preprint Author COI: authors declare no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Porte 2020.
| Study characteristics | |||
| Patient Sampling | 2‐group study to estimate sensitivity and specificity for diagnosis of active disease: samples from suspected COVID‐19 cases (n = 1453) with deliberate sampling of PCR−positive and negative cases on a 2:1 basis (n = 127) Recruitment: convenience sampling Prospective or retrospective: retrospective Number of samples (samples with confirmed SARS‐CoV‐2): 127 (82) |
||
| Patient characteristics and setting | Setting: outpatients attending ED at private medical centre (hospital) Location: Clínica Alemana, Santiago Country: Chile Dates: 16‐21 March 2020 Symptoms and severity: cough 94 (74.6%); fever 77 (61.1%) Median duration of symptoms of 2 days (IQR 1–4; range 0‐12) Duration of symptoms: day 0‐3 91 (72.2%); day 4‐7 27 (22.4%); day ≥ 8 8 (6.3%) Demographics: 68 male (53.5%), median age 38 years (IQR 29.5–44; range 1–91) Exposure history: not stated |
||
| Index tests | Test name: diagnostic Kit for 2019‐Novel Coronavirus (2019‐nCoV) Ag Test (Cat. N° YRLF04401025, lot N° 2002N408) Manufacturer: Bioeasy Biotechnology Co., Shenzhen, China Ag target: SARS‐CoV‐2 nucleocapsid protein Antibody: not stated Test method: FIA Samples used: remnant OP and NP swabs in 3 mL UTM Transport media: UTM‐RT System, Copan Diagnostics, Murrieta, CA, USA Sample storage: stored at 4 °C and tested within 48 h Test operator: laboratory technician Definition of test positivity: not stated; test "automatically delivers a positive or negative qualitative result" Positive or negative defined qualitatively Blinding reported: yes Timing of samples: on presentation Within 48 h of the PCR test but it doesn't say when PCR test was performed (median duration of symptoms reported in D9) |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR (COVID‐19 Genesig Real‐Time PCR assay (Primer Design Ltd., Chandler's Ford, UK)); Ct ≤ 40 considered positive Definition of non‐COVID cases: single RT‐PCR−ve Genetic target(s): not stated Samples used: as for index test; same OP and NP swabs used Timing of reference standard: median 2 d pso (IQR 1‐4; range 0‐12) Blinded to index test: yes (index test done within 48 h of PCR test) Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same sample used; within 48 h All participants received same reference standard: yes Missing data: none; participant flow diagram reported Uninterpretable results: not reported Indeterminate results (index test): not reported Indeterminate results (reference standard): not reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: no funding received Publication status: preprint (not peer‐reviewed) Source: SSRN Author COI: all study authors declare no competing interests |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Porte 2021 [A].
| Study characteristics | |||
| Patient Sampling | Multi‐group study to estimate sensitivity and specificity:
(1) COVID‐19 patients presenting within 5 days of symptom onset (n = 32)
(2) symptomatic patients with negative PCR (n = 20)
(3) asymptomatic patients screened prior to surgery (n = 12)
(27 PCR+ and 19 PCR− samples were used in 2020 study by Weitzel and colleagues (different assays)] Recruitment: not stated; appears to be convenience Prospective or retrospective: retrospective |
||
| Patient characteristics and setting | Setting: private clinic (classed as ED) Location: Clínica Alemana, Santiago Country: Chile Dates: not stated Symptoms and severity: not reported; 12 asymptomatic Demographics: total sample median age 39 years (IQR 36.7‐57); 33, 52% male Exposure history: not reported |
||
| Index tests | Comparative study of 2 Ag tests (no product codes reported); see Porte 2021 [A] for data related to test [A], Porte 2021 [B] tests [B] data. [A] SOFIA SARS Antigen FIA [B] STANDARD F COVID‐19 Ag FIA Manufacturer: [A] Quidel Corporation, San Diego, CA, USA [B] SD Biosensor Inc, Gyeonggi‐do, Republic of Korea Antibody: NP (both) Ag target: not stated Test method: both FIA Samples used: NOP flocked swabs; obtained by trained personnel Transport media: UTM‐RT System, Copan Diagnostics Sample storage: stored at −80 °C following RT‐PCR Test operator: laboratory staff Definition of test positivity: as per manufacturer IFU; both using analyzer device Blinding reported: yes; blinded to RT‐PCR result Timing of samples: all < 5 days pso; median PCR+: 2 days (IQR 1‐3); PCR−: 1 day (IQR 0.75‐4) |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; COVID‐19 Genesig, Primerdesign Ltd., Chandler's Ford, UK
(Ct) values ≤ 40 were considered positive Definition of non‐COVID cases: as for cases Genetic target(s): not stated Samples used: NOP; as for index test Timing of reference standard: not stated Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; same sample All participants received same reference standard: yes Missing data: none reported, no participant flow diagram reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "this research did not receive any specific grant from funding agencies in the public, commercial, or not‐for‐profit sectors." Publication status: published Source: International Journal of Infectious Disease Author COI: all authors declare no competing interests |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Porte 2021 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 2 Ag tests; Porte 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | |||
| Index tests | Comparative study of 2 Ag tests (no product codes reported); Porte 2021 [B] data relate to test [B], see Porte 2021 [A] for data related to test [A] and QUADAS entries [A] SOFIA SARS Antigen FIA [B] STANDARD F COVID‐19 Ag FIA Manufacturer: [A] Quidel Corporation, San Diego, CA, USA [B] SD Biosensor Inc, Gyeonggi‐do, Republic of Korea Antibody: NP (both) Ag target: not stated Test method: both FIA Samples used: NOP flocked swabs; obtained by trained personnel Transport media: UTM‐RT System, Copan Diagnostics Sample storage: stored at −80 °C following RT‐PCR Test operator: laboratory staff Definition of test positivity: as per manufacturer IFU; both using analyzer device Blinding reported: yes; blinded to RT‐PCR result Timing of samples: all < 5 days pso; median PCR+: 2 days (IQR 1‐3); PCR−: 1 day (IQR 0.75‐4) |
||
| Target condition and reference standard(s) | Comparative study of 2 Ag tests; Porte 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 2 Ag tests; Porte 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | |||
Pray 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: symptomatic and asymptomatic participants at 2 universities in Wisconsin (students, staff or other) (n = 1105); at university A, all people tested were eligible (n = 1098), at university B, only students who were quarantined after exposure to people with COVID‐19 could participate (n = 47) Recruitment: not stated; appears consecutive/all eligible were included Prospective or retrospective: prospective Sample size (cases): 1098 (57) Symptomatic 227 (40) Asymptomatic 871 (17) (1105 paired nasal samples taken; 7 inconclusive Ag or RT‐PCR results so excluded from analysis) |
||
| Patient characteristics and setting | Setting: University COVID‐19 test centre/asymptomatic screening
On‐site testing: 2 Wisconsin university campuses during university‐based testing programmes Location: Wisconsin university campuses Country: USA Dates: 28 September–9 October Symptoms and severity: symptomatic 227 (21%) Asymptomatic 871 (79%) (including 53 with ≥ 1 symptoms in previous 14 days) Symptoms included: nasal congestion 114 (50.2%), sore throat 97 (42.7%), headache 87 (38.3%), cough 70 (30.8%), fatigue 60 (26.4%), muscle aches 43 (18.9%), shortness of breath 24 (10.6%) Demographics: male: 453 (41.3%) Age group 15‐24 years: 971 (88.4%); ≥ 25 years: 127 (11.6%) Non‐Hispanic white: 917 (83.5%) Exposure history: close contact to the COVID‐19 cases in past 14 days: 154 (14%) Quarantined at time of specimen collection: 135 (12.3%) Time between quarantine initiation to specimen collection, median days (range): 4 (0‐28) |
||
| Index tests | Test name: Sofia SARS Antigen Fluorescent Immunoassay (FIA) Manufacturer: Quidel Corporation Antibody: not stated Ag target: not stated Test method: FIA Samples used: MTN swab collected by HCWs at university A and were self‐collected under supervision at university B Transport media: none; analysed according to the manufacturer IFU Sample storage: none; immediate on‐site testing Test operator: presume same HCWs at university A; not reported for university B Definition of test positivity: not stated; as per manufacturer IFU Blinding reported: yes‐ Ag tests were performed before RT‐PCR Timing of samples: median 3d pso (IQR 1, 6 days; 7.5% missing) 152 (72.4%) reported ≤ 5 days from symptom onset to specimen collection |
||
| Target condition and reference standard(s) | Reference standard: real‐time RT‐PCR ‐
University A: CDC 2019‐nCoV assay
University B: TaqPath COVID‐19 Combo Kit (Thermo Fisher Scientific)
(Viral culture was attempted on residual RT‐PCR specimens if the RT‐PCR or Ag test result was positive.) Definition of non‐COVID cases: Genetic target(s): N1 and N2 viral nucleocapsid protein gene regions Samples used: nasal swabs stored in viral transport media at 39°F (4 °C) Timing of reference standard: analysed within 24–72 h of collection. Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous (paired swabs) All participants received same reference standard: yes; 2 RT‐PCR assays used. Missing data: 7/1105 inconclusive Ag or RT‐PCR results excluded from analysis; no details provided Uninterpretable results: reasons for test 'failure' not reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: not stated Publication status: published Source: MMWR US Department of Health and Human Services/Centers for Disease Control and Prevention Author COI: authors report no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Prince‐Guerra 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: any participants who attended 2 sites of Pima County Health Department community‐based SARS‐CoV‐2 testing sites; open to anyone who wanted testing asymptomatic (76%) Recruitment: consecutive; any who attended Prospective or retrospective: prospective Sample size (cases): 3419 (299) |
||
| Patient characteristics and setting | Setting: community COVID testing sites Location: Pima County, Arizona (Pima County Health Department) Country: USA Dates: 3‐17 November 2020 Symptoms and severity: 827 (24%) symptomatic at the time of testing (≥ 1 COVID symptom); 2592 (76%) were asymptomatic Demographics: median age 41 years (range 10‐95); 236 (7%) aged 10–17 years, 1885 (55%) aged 18–49 years, 743 (22%) aged 50–64 years, and 555 (16%) aged ≥ 65 years 1681 (49%) female; 2567 (75%) white; 1075 (31%) Hispanic/Latino Exposure history: 1138 (33%) had exposure to a diagnosed COVID‐19 case (close contact (within 6 ft for ≥ 15 min) in the 14 d before the day of testing with a person with diagnosed COVID‐19) Median days since last exposure 5 (range 0‐14) |
||
| Index tests | Test name: BinaxNOW COVID‐19 Ag Card (BinaxNOW) Manufacturer: Abbott Diagnostics, USA Antibody: nucleocapsid Ag target: not stated Test method: CGIA Samples used: AN, bilateral (HCWs) Transport media: none required Sample storage: none; immediately tested on‐site Test operator: HCWs Definition of test positivity: not stated; visual Blinding reported: yes (performed before PCR) Timing of samples: day 0‐14 Symptomatic: median pso 4 d (range 0‐210); 662 (19%) ≤ 7 days; 161 (5%) > 7 days |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; CDC
2019‐nCoV Real‐Time RT‐PCR Diagnostic Panel for detection of SARS‐CoV‐2 (2,582 swabs) or the Fosun COVID‐19 RT‐PCR Detection Kit (837 swabs)
Viral culture was performed on 274 of 303 residual real‐time RT‐PCR specimens with positive results by either test Definition of non‐COVID cases: same as for cases (single ‐ve PCR) Genetic target(s): not stated Samples used: NP, bilateral; obtained after AN swabs (HCW) Timing of reference standard: as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swab, simultaneous All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: Arizona Department of Health Services Publication status: published Source: MMWR Author COI: authors report no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Ristic 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity and specificity: symptomatic patients with ≥ 1 COVID‐related symptoms presenting via a triage ambulance of a primary and tertiary outpatients healthcare facility, including a primary care "COVID ambulance" and a "red zone" ambulance Recruitment: consecutive; "all cases" Prospective or retrospective: prospective Sample size (cases): 120 (43) |
||
| Patient characteristics and setting | Setting: mixed; primary and tertiary outpatients Location: Health Centre Novi Sad and Clinical Centre of Vojvodina, Department for Infectious Diseases Country: Serbia Dates: 21 August‐1 September 2020 Symptoms and severity: all symptomatic (120); 103 (86%) with fever, followed by malaise (77, 64%), cough (56, 47%), sore throat (53, 44%), myalgia (46, 38%) Demographics: median age 49 years (IQR 36–70) (R 14‐91 years), female (57) male (63) Exposure history: not reported |
||
| Index tests | Test name: STANDARD Q COVID‐19 Ag Test Manufacturer: SD Biosensor, Gyeonggi‐do, South Korea Antibody: not stated Ag target: mouse monoclonal anti‐SARS CoV‐2 antibody (coated in the test line region) and the mouse monoclonal anti‐chicken IgY antibody (coated in the control line region) Test method: chromatographic immunoassay Samples used: posterior NP by trained medical staff Transport media: none used for Ag test Sample storage: no storage Test operator: trained medical staff Definition of test positivity: visual interpretation; double lines Blinding reported: yes; conducted first Timing of samples: median time from symptom to swab (9.4 days, range 1‐45 days; 63 (53%) within first 5 days) |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; a number of assays were described as used ‐ qualitative assessment using 1) AND 2) for each sample and for positive samples only, quantitative assessment of viral load using 3):
1) Argene, SARS‐COV‐2 R‐GENE assay (bioMerieux, Marcy‐l’Etoile, France), after RNA extraction
2) Applied Biosystems 7500 Real‐Time PCR System (Life Technologies, Carlsbad, CA, USA)
3) COVID‐19 Genesig Real‐Time PCR Kit (Primerdesign Ltd, Chandler’s Ford, UK); Ct < 41 considered positive Definition of non‐COVID cases: single negative Genetic target(s): 1) R‐gene 2) RdRP in the ORF1ab region, E gene, and N gene 3) RdRP Samples used: unclear (seems to be paired swabs); transported in VTM to a central laboratory and tested within 12 h of collection Timing of reference standard: same as index Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same time All participants received same reference standard: yes Missing data: not stated Uninterpretable results: not stated Indeterminate results (index test): not stated Indeterminate results (reference standard): not stated Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "supported by Provincial Secretariat for Higher Education and Scientific Research grant" Publication status: published paper Source: academic journal Author COI: no COI statement provided |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Rottenstreich 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity and specificity: asymptomatic women admitted for delivery Recruitment: consecutive; "all women" Prospective or retrospective: prospective Sample size (cases): 1326 (9) |
||
| Patient characteristics and setting | Setting: hospital inpatient Location: Department of Clinical Microbiology and Infectious Diseases Hadassah‐Hebrew University Medical Center Country: Israel Dates: 21 October‐28 December 2020 Symptoms and severity: asymptomatic Demographics: none stated; all female Exposure history: not reported |
||
| Index tests | Test name: NowCheck COVID‐19 Ag Test Manufacturer: Bionote Inc, Hwaseong‐si, Republic of Korea Antibody: not reported Ag target: not reported Test method: not reported Samples used: NP Transport media: not reported Sample storage: not reported Test operator: not reported Definition of test positivity: not reported Blinding reported: not reported Timing of samples: on admission |
||
| Target condition and reference standard(s) | Reference standard: PCR
(NeuMoDx 288 Molecular System (NeuMoDx Molecular, Ann Arbor, MI) Definition of non‐COVID cases: single negative Genetic target(s): not reported Samples used: not reported; states women were "co‐tested" so may have been paired swabs Timing of reference standard: same as index Blinded to index test: not reported Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: unclear appears to be same time All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: no funding statement provided Publication status: published research letter Source: academic journal Author COI: no COI statement provided |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Saeed 2021 [A].
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity alone: RT‐PCR+ve samples from suspected cases of COVID‐19 (respiratory symptom and/or fever and international travel history or close contact with COVID‐19‐confirmed patients); it is not clear but seems that both NP and saliva had to be RT‐PCR+ve Recruitment: unclear; states samples were "pre‐selected" from a population of 33,000 suspected cases but does not state how many were RT‐PCR+ve Prospective or retrospective: retrospective Sample size (cases): 100 (100) |
||
| Patient characteristics and setting | Setting: COVID‐19 diagnostic centre Location: Islamabad Diagnostic Center (IDC G8 branch specialized centre for COVID‐19), Islamabad, (Department of Research and Development, Islamabad Diagnostic Center, F8 Markaz, Islamabad 44000, Pakistan) Country: Pakistan Dates: 3‐10 October 2020 Symptoms and severity: all symptomatic (respiratory symptoms and/or fever) Demographics: mean age 47 years (range 6–91); 34 (34%) female; 4 (4%) children Exposure history: all either had international travel history or close contact with COVID‐19 confirmed patients |
||
| Index tests | Test name: [A] NP‐based RDT (#20CG2701X)
[B] saliva‐based RDT (#901101) Manufacturer: [A] and [B] Lepu Medical, China Antibody: N gene Ag target: monoclonal antibody Test method: CGIA Samples used: [A] NP and [B] saliva; (states collection by trained personnel; saliva would be self‐collected) Transport media: not stated Sample storage: not stated Test operator: not stated Definition of test positivity: visual colour lines; performed according to standard manufacturer protocol Blinding reported: unclear Timing of samples: no details |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; Bio‐rad, CFX96, USA used to identify confirmed PCR+ve cases for RDT testing (exponential growth curve and Ct ≤ 40 considered positive); samples re‐tested using #RP10244 years Allplex 2019‐nCoV Assay (Seegene South Korea) presumably to quantify viral load
Discrepant results were re‐tested Definition of non‐COVID cases: same as for cases (single ‐ve PCR) Genetic target(s): Biorad: E gene, N gene, and RNA polymerase gene Samples used: appears that both NP and saliva samples underwent RT‐PCR and both were positive. States, "The same patient saliva samples (RT‐PCR tested positive)" Timing of reference standard: no details Blinded to index test: yes (performed before index test) Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: discussion states same swabs used All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported; the samples with discordant results were repeated but no details given Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: none Publication status: published Source: Virology Journal Author COI: authors reported no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Saeed 2021 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 2 sample types; Saeed 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | |||
| Index tests | Test name: [A] NP‐based RDT (#20CG2701X)
[B] saliva‐based RDT (#901101) Manufacturer: [A] and [B] Lepu Medical, China Antibody: N gene Ag target: monoclonal antibody Test method: CGIA Samples used: [A] NP and [B] Saliva; (states collection by trained personnel; saliva would be self‐collected) Transport media: not stated Sample storage: not stated Test operator: not stated Definition of test positivity: visual colour lines; performed according to standard manufacturer protocol Blinding reported: unclear Timing of samples: no details |
||
| Target condition and reference standard(s) | Comparative study of 2 sample types; Saeed 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 2 sample types; Saeed 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 2 sample types; Saeed 2021 [A] reports full study characteristics and QUADAS. | ||
Salvagno 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity and specificity: consecutive patients referred to a hospital for SARS‐CoV‐2 diagnostic testing Recruitment: consecutive Prospective or retrospective: unclear; "investigation was based on pre‐existing specimens, already collected for routine SARS‐CoV‐2 diagnostic testing in the local facility" Sample size (cases): 321 (149) |
||
| Patient characteristics and setting | Setting: unclear; possibly inpatient Location: Pederzoli Hospital, Peschiera del Garda, Verona Country: Italy Dates: 16–30 November 2020 Symptoms and severity: not reported; presume symptomatic (patients were, "referred for SARS‐CoV‐2 diagnostic testing to the Pederzoli Hospital") Demographics: mean age 46 years (IQR 32‐56 years); 181 (56%) women Exposure history: not reported |
||
| Index tests | Test name: SARS‐CoV‐2 Rapid Antigen Test Manufacturer: Roche Antibody: not stated Ag target: not stated Test method: CGIA Samples used: NP (reported in Abstract); collection not described Transport media: virus swab UTM, Copan, Brescia, Italy Sample storage: not stated Test operator: not stated Definition of test positivity: visual line Blinding reported: unclear Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; Seegene AllplexTM2019‐nCoV Assay, Seegene, Seoul, South Korea
Threshold: Ct < 37 for all 3 SARS‐CoV‐2 gene targets considered “reactive” for SARS‐CoV‐2 RNA Definition of non‐COVID cases: single negative Genetic target(s): N, E and RdRP Samples used: same as for index; same swab Timing of reference standard: not stated Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same swab All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: no research funding declared Publication status: published Source: Diagnosis Author COI: authors state no conflict of interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Schildgen 2021 [A].
| Study characteristics | |||
| Patient Sampling | Unclear design; appears to be single cohort with deliberate sampling of PCR+ve/PCR−:
[1] RT‐PCR+ve, positive BAL or throat wash samples (n = 42)
[2] RT‐PCR−ve samples (n = 31)
Described as pilot sample panel Recruitment: appears to be convenience Prospective or retrospective: not stated; presume retrospective |
||
| Patient characteristics and setting | Setting: not stated Location: authors' institution: Kliniken der Stadt Köln gGmbH (Koln city clinics) Country: Germany Dates: not stated Symptoms and severity: not stated for BAL samples, throat wash from 23 symptomatic and 27 asymptomatic people Demographics: not stated Exposure history: not stated |
||
| Index tests | Comparative study of 3 Ag tests (no product codes reported); Schildgen 2021 [A] data relate to test [A], see Schildgen 2021 [B] and Schildgen 2021 [C] for data related to tests [B] and [C]. Test name: [A] BIOCREDIT [B] Panbio [C] SARS‐CoV‐2 Rapid Antigen test Manufacturer: [A] RapiGEN [B] Abbott [C] Roche Antibody: not stated Ag target: not stated Test method: all LFA Samples used: BAL (n = 13); throat wash (n = 50, including 27 from asymptomatic) Transport media: not stated Sample storage: not stated Test operator: not stated; presume lab staff Definition of test positivity: as per manufacturer IFU Blinding reported: not stated Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; RealStar SARS‐CoV‐2 RT‐PCR Kit, Altona, Germany Definition of non‐COVID cases: as for cases Genetic target(s): not stated Samples used: BAL or throat wash; as per index test Timing of reference standard: not stated Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same swab All participants received same reference standard: yes Missing data: 8 PCR invalid samples also tested; 2/8 invalid in 1 Ag assay each, 3/8 negative in all 3 Ag assays Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: unclear |
||
| Comparative | |||
| Notes | Funding: the study did not receive any external funding Publication status: preprint Source: medRxiv Author COI: the authors declare that they have no conflicts of interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Schildgen 2021 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 3 Ag tests; Schildgen 2021 [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 3 Ag tests; Schildgen 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Comparative study of 3 Ag tests (no product codes reported); Schildgen 2021 [B] data relate to test [B], see Schildgen 2021 [A] and Schildgen 2021 [C] for data related to tests [A] and [C], and for QUADAS entries. Test name: [A] BIOCREDIT [B] Panbio [C] SARS‐CoV‐2 Rapid Antigen test Manufacturer: [A] RapiGEN [B] Abbott [C] Roche Antibody: not stated Ag target: not stated Test method: all LFA Samples used: BAL (n = 13); throat wash (n = 50, including 27 from asymptomatic) Transport media: not stated Sample storage: not stated Test operator: not stated; presume lab staff Definition of test positivity: as per manufacturer IFU Blinding reported: not stated Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 3 Ag tests; Schildgen 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 3 Ag tests; Schildgen 2021 [A] reports full study characteristics and QUADAS | ||
| Comparative | |||
| Notes | |||
Schildgen 2021 [C].
| Study characteristics | |||
| Patient Sampling | Comparative study of 3 Ag tests; Schildgen 2021 [A] reports full study characteristics and QUADAS | ||
| Patient characteristics and setting | Comparative study of 3 Ag tests; Schildgen 2021 [A] reports full study characteristics and QUADAS. | ||
| Index tests | Comparative study of 3 Ag tests (no product codes reported); Schildgen 2021 [C] data relate to test [C], see Schildgen 2021 [A] and Schildgen 2021 [B] for data related to tests [A] and [B], and for QUADAS entries. Test name: [A] BIOCREDIT [B] Panbio [C] SARS‐CoV‐2 Rapid Antigen test Manufacturer: [A] RapiGEN [B] Abbott [C] Roche Antibody: not stated Ag target: not stated Test method: all LFA Samples used: BAL (n = 13); throat wash (n = 50, including 27 from asymptomatic) Transport media: not stated Sample storage: not stated Test operator: not stated; presume lab staff Definition of test positivity: as per manufacturer IFU Blinding reported: not stated Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 3 Ag tests; Schildgen 2021 [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 3 Ag tests; Schildgen 2021 [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | |||
Schuit 2021(a).
| Study characteristics | |||
| Patient Sampling | Report of 2 single‐group studies estimating sensitivity and specificity: (1) West Brabant testing sites used the BD Veritor system (included as Schuit 2021(a)) and (2) Rotterdam sites used SD Biosensor assay (included as Schuit 2021(b)). Included close contacts (aged ≥ 16 years) of confirmed COVID‐19 cases presenting at testing sites for a 5th‐day test (as recommended by Dutch public health service test‐and‐trace programme, and/or the Dutch contact tracing mobile phone application (the ‘CoronaMelder’ app) and/or an individual with a confirmed SARS‐CoV‐2 infection); all asymptomatic at the time of the test request. Recruitment: consecutive Prospective or retrospective: prospective Sample size (cases): 2692 (233) 2692/3237 agreed to participate |
||
| Patient characteristics and setting | Setting: COVID‐19 testing centres Location: West Brabant COVID‐19 testing sites (Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht University, Utrecht) Country: Netherlands Dates: 14 December 2020‐6 February 2021 Symptoms and severity: all asymptomatic on test booking; 219, 8.6% reported being symptomatic 0‐3 days before test Symptoms included: common cold 167/219, 76%; cough 60, 27%; shortness of breath 25, 11%; fever 13, 6%; loss of taste or smell 6, 3%, muscle ache 18, 8%, other 16, 7% Demographics: mean age 45.9 years (SD 17.6 years); 1304, 48.7% male Exposure history: all exposed to confirmed case |
||
| Index tests | Test name: BD Veritor System for Rapid Detection of SARS‐CoV‐2 Ag‐RDT Manufacturer: BD Veritor, Franklin Lakes, NJ, USA Antibody: not reported Ag target: nucleocapsid Test method: unknown; CGIA Samples used: nasal + OP; collected by trained personnel, nasal swab 2.5 cm deep Transport media: none; placed in a sterile dry tube Sample storage: frozen at −20 °C within 30 min of collection; transported to Microvida location Amphia laboratory. Thawed and tested within 6 h of collection Test operator: trained laboratory technician; result confirmed by a second person Definition of test positivity: visual interpretation; Analyser was not used Blinding reported: yes; done first Timing of samples: median 5 days (IQR 5‐5) between contact and sampling, range 0‐13 days Symptomatic (n = 219), symptoms developed on day of test 17 (7.8%), 1 day prior 64 (29.2%), 2 days prior 51 (23.3%), 3 days prior 83 (37.9%) |
||
| Target condition and reference standard(s) | Reference standard: (1) PCR; Cobas SARS‐CoV‐ 2 test on the Cobas 8800 platform (Roche Diagnostics International, Rotkreuz, Switzerland)
(2) Used routine national testing data to determine whether any PCR−ve had a subsequent +ve PCR or Ag‐RDT result within 10 days Definition of non‐COVID cases: single negative for absence Genetic target(s): E, RdRp Samples used: combined nasal+OP in UTM (HiViralTM) Timing of reference standard: same as for index test Blinded to index test: yes; stated to be blinded Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired All participants received same reference standard: yes Missing data: yes; 14 excluded Uninterpretable results: 10 with no PCR (n = 3) or PCR invalid (n = 7); all Ag‐ve Indeterminate results (index test): 3 inconclusive; all PCR−ve 1 further result excluded but reason not clear from flow diagram Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: funded by the Dutch Ministry of Health, Welfare and Sport Publication status: preprint Source: medRxiv Author COI: none to be disclosed |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Schuit 2021(b).
| Study characteristics | |||
| Patient Sampling | Report of 2 single‐group studies estimating sensitivity and specificity: (1) West Brabant testing sites used the BD Veritor system (included as Schuit 2021(a)) and (2) Rotterdam sites used SD Biosensor assay (included as Schuit 2021(b)). Included close contacts (aged ≥ 16 years) of confirmed COVID‐19 cases presenting at testing sites for a 5th‐day test (as recommended by Dutch public health service test‐and‐trace programme, and/or the Dutch contact tracing mobile phone application (the ‘CoronaMelder’ app) and/or an individual with a confirmed SARS‐CoV‐2 infection); all asymptomatic at the time of the test request. Recruitment: consecutive Prospective or retrospective: prospective Sample size (cases): 1603 (132) 1603/1903 agreed to participate |
||
| Patient characteristics and setting | Setting: COVID‐19 testing centres Location: Rotterdam COVID‐19 testing sites (Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht University, Utrecht) Country: Netherlands Dates: 14 December 2020‐6 February 2021 Symptoms and severity: all asymptomatic on test booking; 158, 10.1% symptomatic 0‐3 days before test Symptoms included: common cold 123, 78%; cough 24, 15.2%; shortness of breath 12, 8%; fever 9, 6%; loss of taste or smell 5, 3%, muscle ache 5, 3%, other 15, 9.5% Demographics: mean age 40.7 years (SD 16.4 years); 845, 52.7% male Exposure history: all exposed to confirmed case |
||
| Index tests | Test name: SARS‐CoV‐2 Rapid Antigen Test Manufacturer: Roche/SD Biosensor, Basel, Switzerland Antibody: not reported Ag target: nucleocapsid Test method: CGIA Samples used: NP alone; collected by trained personnel, > 5 cm deep Transport media: none Sample storage: conducted immediately on site Test operator: not stated; performed independently by 2 people Definition of test positivity: visual interpretation Blinding reported: yes; done first Timing of samples: median 5 days (IQR 5‐5) between contact and sampling, range 0‐11 days Symptomatic (n = 158), symptoms developed on day of test 14 (8.9%), 1 day prior 37 (23.4%), 2 days prior 39 (24.7%), 3 days prior 45 (28.5%) |
||
| Target condition and reference standard(s) | Reference standard: (1) RT‐PCR; Cobas SARS‐CoV‐ 2 test on the Cobas 8800 platform (Roche Diagnostics International, Rotkreuz, Switzerland).
(2) Used routine national testing data to determine whether any RT‐PCR−ve had a subsequent +ve RT‐PCR or Ag‐RDT result within 10 days Definition of non‐COVID cases: single negative for absence Genetic target(s): E, RdRp Samples used: combined NP+OP in UTM (HiViralTM); > 5 cm deep Timing of reference standard: same as for index Blinded to index test: yes; stated to be blinded Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired All participants received same reference standard: yes Missing data: yes; 7 excluded Uninterpretable results: 4 with no RT‐PCR; all Ag ‐ve Indeterminate results (index test): 3 inconclusive excluded; all PCR−ve Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: funded by the Dutch Ministry of Health, Welfare and Sport Publication status: preprint Source: medRxiv Author COI: none to be disclosed |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Schwob 2020(a).
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity of 3 assays (each tested on a separate cohort of individuals, and extracted as 3 entries: Schwob 2020(a), Schwob 2020(b) and Schwob 2020(c)): adults recruited from 3 outpatient clinics and meeting testing criteria for COVID‐19, either:
Recruitment: unclear; appears consecutive. RDT brands were rotated after around 30 positive patients until at least 100 positive per test were reached. Numbers per test were [1] 333 (36%) STANDARD Q, [2] 271 (29%) Panbio, and [3] 324(35%) COVID‐VIRO. Prospective or retrospective: prospective Sample size (cases): overall: 949 (327 positive by NP PCR, 369 positive by saliva PCR). 2x2 data only available for NP PCR STANDARD Q assay: 333 (112) |
||
| Patient characteristics and setting | Setting: outpatient testing clinic Location: Unisante Bugnon; Unisante Flon; Vidy‐Med Country: Switzerland Dates: 25 September‐4 November 2020 Symptoms and severity: whole sample, all symptomatic: 911, 96% with ≥ 1 major symptom (41% fever, 64% cough, 62% sore throat, 32% anosmia/ageusia) and 4% at least 1 minor symptom (rhinitis, myalgia, headache, fatigue, nausea, vomiting, diarrhoea, abdominal pain, urticaria, vesicles) Demographics: median age: 31 (IQR 25‐42; range 18‐87); male (51%) Exposure history: not stated |
||
| Index tests | Test name: STANDARD Q COVID‐Ag Test
See Schwob 2020(b) and Schwob 2020(c) for data for Panbio COVID‐19 Ag Test (Abbott) and COVID‐VIRO (AAZ) Manufacturer: SD Biosensor/Roche Antibody: nucleocapsid Ag target: not stated Test method: lateral flow; no further information Samples used: NP (HCW) Transport media: not stated Sample storage: no storage, immediate Test operator: same HCW who collected the swab Definition of test positivity: visual colour change Blinding reported: yes, done first Timing of samples: pso (mean duration of symptoms at the time of swab collection/testing was 2.6 days (SD 2.3, range 0‐30)) |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; in‐house or Cobas 6800 Definition of non‐COVID cases: single negative Genetic target(s): E gene Samples used: NP; HCW collected (saliva sample also collected but Ag results only presented compared to NP swab) Timing of reference standard: pso (mean duration of symptoms at the time of swab collection/testing was 2.6 days (SD 2.3, range 0‐30)) Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same time All participants received same reference standard: yes Missing data: yes; 21 excluded due to lack of PCR and/or RDT result, no further details Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "the RDT and saliva PCR were paid for by the cantonal health authorities" Publication status: preprint Source: medRxiv Author COI: authors report no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Schwob 2020(b).
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity of 3 assays (each tested on a separate cohort of individuals, and extracted as 3 entries: Schwob 2020(a), Schwob 2020(b) and Schwob 2020(c)): adults recruited from 3 outpatient clinics and meeting testing criteria for COVID‐19, either:
Recruitment: unclear; appears consecutive. RDT brands were rotated after around 30 positive patients until at least 100 positive per test were reached; 333 (36%) STANDARD Q, 271 (29%) Panbio and 324(35%) COVID‐VIRO. Prospective or retrospective: prospective Sample size (cases): 949 (327 positive by NP PCR, 369 positive by saliva PCR). 2x2 data only available for NP PCR Panbio assay: 271 (122) |
||
| Patient characteristics and setting | Setting: outpatient testing clinic Location: Unisante Bugnon; Unisante Flon; Vidy‐Med Country: Switzerland Dates: 25 September‐4 November 2020 Symptoms and severity: whole sample, all symptomatic: 911, 96% with ≥ 1 major symptom (41% fever, 64% cough, 62% sore throat, 32% anosmia/ageusia) and 4% at least one minor symptom (rhinitis, myalgia, headache, fatigue, nausea, vomiting, diarrhoea, abdominal pain, urticaria, vesicles) Demographics: median age: 31 years (IQR 25‐42; range 18‐87 years); male (51%) Exposure history: not stated |
||
| Index tests | Test name: Panbio COVID‐19 Ag Test
See Schwob 2020(a) and Schwob 2020(c) for data for STANDARD Q (SD Biosensor) and COVID‐VIRO (AAZ) Manufacturer: Abbott Antibody: nucleocapsid Ag target: not stated Test method: lateral flow; no further information Samples used: NP (HCW) Transport media: not stated Sample storage: no storage, immediate Test operator: HCW Definition of test positivity: visual colour change Blinding reported: yes, done first Timing of samples: pso (mean duration of symptoms at the time of swab collection/testing was 2.6 days (SD 2.3, range 0‐30)) |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; in‐house or Cobas 6800 Definition of non‐COVID cases: single negative Genetic target(s): E gene Samples used: NP; HCW collected (saliva sample also collected but Ag results only presented compared to NP swab) Timing of reference standard: pso (mean duration of symptoms at the time of swab collection/testing was 2.6 days (SD 2.3, range 0‐30)) Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same time All participants received same reference standard: yes Missing data: yes; 21 excluded due to lack of PCR and/or RDT result, no further details (There appears to be a typo in Suppl Fig 1, which reports 122 PCR+ve samples tested with Panbio assay; 101 are shown as RDT+ and 17 RDT‐. The text reports assay sensitivity as 86.1% (95% CI 78.6, 91.7%), which works out as 105 RDT+, 17 RDT‐ and the correct CIs) Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "the RDT and saliva PCR were paid for by the cantonal health authorities" Publication status: preprint Source: medRxiv Author COI: none |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Schwob 2020(c).
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity of 3 assays (each tested on a separate cohort of individuals, and extracted as 3 entries: Schwob 2020(a), Schwob 2020(b) and Schwob 2020(c)): adults recruited from 3 outpatient clinics and meeting testing criteria for COVID‐19, either:
Recruitment: unclear; appears consecutive. RDT brands were rotated after around 30 positive patients until at least 100 positive per test were reached; 333 (36%) STANDARD Q, 271 (29%) Panbio and 324(35%) COVID‐VIRO. Prospective or retrospective: prospective Sample size (cases): overall: 949 (327 positive by NP PCR, 369 positive by saliva PCR). 2x2 data only available for NP PCR COVID‐VIRO assay: 324 (138) |
||
| Patient characteristics and setting | Setting: outpatient testing clinic Location: Unisante Bugnon; Unisante Flon; Vidy‐Med Country: Switzerland Dates: 25 September4 November 2020 Symptoms and severity: whole sample, all symptomatic: 911, 96% with ≥ 1 major symptom (41% fever, 64% cough, 62% sore throat, 32% anosmia/ageusia) and 4% at least one minor symptom (rhinitis, myalgia, headache, fatigue, nausea, vomiting, diarrhoea, abdominal pain, urticaria, vesicles) Demographics: median age: 31 years (IQR 25‐42; range 18‐87); male (51%) Exposure history: not stated |
||
| Index tests | Test name: COVID‐VIRO
See Schwob 2020(a) and Schwob 2020(b) for data for STANDARD Q (SD Biosensor) and Panbio COVID‐19 Ag Test (Abbott) Manufacturer: AAZ‐LMB Antibody: nucleocapsid Ag target: not stated Test method: lateral flow; no further information Samples used: NP (HCW) Transport media: not stated Sample storage: no storage, immediate Test operator: HCW Definition of test positivity: visual colour change Blinding reported: yes, done first Timing of samples: pso (mean duration of symptoms at the time of swab collection/testing was 2.6 days (SD 2.3, range 0‐30)) |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; in‐house or Cobas 6800 Definition of non‐COVID cases: single negative Genetic target(s): E gene Samples used: NP; HCW collected (saliva sample also collected but Ag results only presented compared to NP swab) Timing of reference standard: pso (mean duration of symptoms at the time of swab collection/testing was 2.6 days (SD 2.3, range 0‐30)) Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same time All participants received same reference standard: yes Missing data: yes; 21 excluded due to lack of PCR and/or RDT result, no further details Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "the RDT and saliva PCR were paid for by the cantonal health authorities" Publication status: preprint Source: medRxiv Author COI: none |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Scohy 2020.
| Study characteristics | |||
| Patient Sampling | Single‐group study including NP swabs submitted to laboratory at a large tertiary hospital (n = 148) Recruitment: random sample Prospective or retrospective: not stated |
||
| Patient characteristics and setting | Setting: unclear; presume microbiology laboratory takes samples from number of sources Location: Cliniques Universitaires Saint‐Luc Hospital, Brussels Country: Belgium Dates: 6‐21 April 2020 Symptoms and severity: 86 (58%) symptomatic, 45 (30%) asymptomatic, 17 (11%) symptom status not reported Cases only: viral load < 25 Ct 10 (9%), ≥ 25 Ct 96 (91%) Demographics: median age 57.5 (0‐94 years); 64 (43%) male Exposure history: not reported |
||
| Index tests | Test name: COVID‐19 Ag Respi‐Strip (product code not reported) Manufacturer: Coris Bioconcept Antibody: NP Ag target: monoclonal antibody Test method: CGIA Samples used: NP Transport media: not stated Sample storage: "If the rapid antigen test was not performed immediately, samples were stored at 4 °C until the test" Test operator: not stated Definition of test positivity: visual appearance of T line; also states that "Two versions of the test were evaluated. On the second version, conjugate was coupled on a different way and the control line was optimized." Blinding reported: unclear Timing of samples: not reported |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR: genesig Real‐Time PCR assay (Primerdesign Ltd, Chandler’s Ford, UK); < 40 Ct Definition of non‐COVID cases: single PCR negative Genetic target(s): RdRp Samples used: NP; same as for index Timing of reference standard: not stated Blinded to index test: yes Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same sample All participants received same reference standard: yes Missing data: none reported, no participant flow diagram reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: no funding statement reported; COVID‐19 Ag Respi‐Strip tests provided by Coris BioConcept. Publication status: published Source: Journal of Clinical Virology Author COI: the authors declare no conflicts of interest. |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Shidlovskaya 2021 [A].
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: patients with suspected COVID‐19 admitted to the hospital on day 2‐10 pso (fever, dry cough, chest pain and discomfort, shortness of breath, loss of smell and taste). All included patients had CT signs of lung damage. Recruitment: unclear Prospective or retrospective: not stated; may be prospective Sample size (cases): 106 (78) |
||
| Patient characteristics and setting | Setting: hospital inpatient Location: infectious diseases hospital, Moscow (Department of Virology, Lomonosov Moscow State University, Moscow) Country: Russia Dates: 25 January‐8 February 2021 Symptoms and severity: 100% symptomatic; symptoms included fever, dry cough, chest pain and discomfort, shortness of breath, loss of smell and taste; all had lung damage on CT Demographics: mean age 67.7 (range 28‐95) years; 53 (50%) female Exposure history: not stated |
||
| Index tests | Test name: [A] SGTI‐flex COVID‐19 Ag
[B] BIOCREDIT COVID‐19 Ag Manufacturer: [A] Sugentech Inc., Korea [B] RapiGEN Inc., Korea Antibody: [A] Nucleocapsid [B] SARS‐COV2 antigen Ag target: not stated Test method: [A] LFA [B] CGIA Samples used: NP (collected by nurses) Transport media: none required Sample storage: none required; tested directly at the patient bedside Test operator: nurses Definition of test positivity: visual Blinding reported: yes (performed before PCR) Timing of samples: between 2‐10 days pso |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; one‐step “SARS‐CoV‐2 FRT” commercial kit with catalogue number ЕА‐128 (bought from N.F. Gamaleya NRCEM, Moscow, Russia)
Cell culture also used in all PCR+ve samples 293T/ACE2 Definition of non‐COVID cases: same as for cases (single ‐ve PCR) Genetic target(s): NSP1 gene Samples used: NP Timing of reference standard: 2‐10 days pso. Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: the Ministry of Health of the Russian Federation Publication status: preprint Source: medRxiv preprint Author COI: authors declare no COI present |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Shidlovskaya 2021 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 2 Ag tests; Shidlovskaya 2021 [A] details full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 2 Ag tests; Shidlovskaya 2021 [A] details full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] SGTI‐flex COVID‐19 Ag
[B] BIOCREDIT COVID‐19 Ag Manufacturer: [A] Sugentech Inc., Korea [B] RapiGEN Inc., Korea Antibody: [A] Nucleocapsid [B] SARS‐COV2 antigen Ag target: not stated Test method: [A] LFA [B] CGIA Samples used: NP (collected by nurses) Transport media: none required Sample storage: none required; tested directly at the patient bedside Test operator: nurses Definition of test positivity: visual Blinding reported: yes (performed before PCR) Timing of samples: between 2‐10 days from the onset of symptoms |
||
| Target condition and reference standard(s) | Comparative study of 2 Ag tests; Shidlovskaya 2021 [A] details full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 2 Ag tests; Shidlovskaya 2021 [A] details full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | |||
Shrestha 2020.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: people who were close contacts of confirmed cases identified through contact tracing, residing in quarantine centre (n = 113) Recruitment: convenience Prospective or retrospective: not stated; appears prospective |
||
| Patient characteristics and setting | Setting: contact tracing Location: not applicable; author institutions include Shukraraaj Tropical and Infectious Disease Hospital, Kathmandu Country: Nepal Dates: August‐September 2020 Symptoms and severity: all asymptomatic Demographics: range 13‐74; 89, 79% male Exposure history: all exposed to confirmed case |
||
| Index tests | Test name: BIOCREDIT Manufacturer: RapiGen Antibody: not stated Ag target: not stated Test method: not stated Samples used: NP Transport media: none used Sample storage: none reported; other sample from the same individual was processed for the results as instructed by the manufacturing company of Ag kit Test operator: lab technician (trained) Definition of test positivity: visual line; as per manufacturer IFU Blinding reported: unclear; appears to be Yes Timing of samples: day 5 of quarantine |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; not detailed, "followed the standard protocol regulated by WHO, instruction manual of company and as per NHTC training regarding sample collection and transport" Definition of non‐COVID cases: as for cases; single negative Genetic target(s): not stated Samples used: NP in 3 mL VTM Timing of reference standard: as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous, paired samples All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): tests were repeated for samples with indistinct outcomes. Indeterminate results (reference standard): none reported Unit of analysis: patient |
||
| Comparative | |||
| Notes | Funding: no funding statement provided Publication status: published Source: Kathmandu University Medical Journal Author COI: no COI statement provided |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Smith 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group longitudinal study estimating sensitivity only: newly PCR+ve (within 24 h) students and employees at University of Illinois identified from routine testing (PCR every 2‐4 d) and their close contacts (eligible within 5 d of exposure) who also tested positive during study period. Participants were required to collect paired samples on a daily basis (for 14 d for those with positive PCR prior to enrolment or during quarantine period, and for 7 d for those continuing to test negative on PCR after enrolment). Data were reported only for those with positive viral culture on at least 1 PCR+ve sample. Recruitment: unclear; appears to be consecutive inclusion of those meeting above criteria Prospective or retrospective: prospective Sample size (cases): 51 PCR+ve (number eligible not reported) |
||
| Patient characteristics and setting | Setting: student/staff screening Location: University of Illinois campus Country: USA Dates: not reported Symptoms and severity: all "mild or asymptomatic", numbers not reported Demographics: mean age 33.1 years (SD 12.8 years); 23, 53.5% male; 34, 79.1% white, 4, 9.3% black, 4, 9.3% 'other', 1, 2.3% Asian Exposure history: all PCR+ve |
||
| Index tests | Test name: SOFIA Manufacturer: Quidel Ag target: SARS‐CoV Test method: FIA Samples used: nasal Transport media: none used Sample storage: samples collected by courier within 1 h of collection using a no‐contact pickup protocol; transported with cold packs, and stored at 4 °C overnight based on guidance from the manufacturer. Tested the morning after collection Test operator: not stated; presumably laboratory technician Definition of test positivity: as per manufacturer IFU Blinding reported: unclear; saliva PCR was within 12 h of sample collection and RDT was morning after Timing of samples: those with positive PCR prior to enrolment were tested from within 24 h after first positive result to 14 d after; those testing positive during quarantine period were tested for up to 14 d, and those never positive were tested for 7 d. |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR and viral culture 1) direct saliva to RT‐qPCR assay (in‐house, following previously published protocol) 2) nasal swabs ‐ Abbott Alinity per manufacturer IFU performed at John's Hopkins 3) nasal swabs ‐ viral culture VeroTMPRSS2 cells; presence of SARS‐CoV‐2 was confirmed through RT‐qPCR Definition of non‐COVID cases: as for cases; single negative Genetic target(s): not stated Samples used: 1) saliva (tested within 12 h of collection); 2 and 3) nasal swabs in VTM (stored at −80 °C after collection and subsequently shipped to Johns Hopkins University for RT‐qPCR and viral culture) Timing of reference standard: as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous, paired samples All participants received same reference standard: yes Missing data: Eight individuals were removed from the analysis because their nasal virus culture was never positive Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: patient |
||
| Comparative | |||
| Notes | Funding: "supported by the National Heart, Lung, and Blood Institute at the National Institutes of Health (grant number 3U54HL143541‐02S2) through the RADx‐Tech program." Publication status: published Source: Journal of Infectious Diseases Author COI: CBB and LW are listed as inventors on a pending patent application for the saliva RT‐qPCR test used in this study. All other authors report no potential conflicts. |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Stohr 2021 [A].
| Study characteristics | |||
| Patient Sampling | Randomized study estimating sensitivity and specificity: adults presenting for testing at a community COVID‐19 test centre; testing co‐ordinated by the Municipal Health Services (MHS). Participants randomized between 2 different Ag tests. Recruitment: consecutive Prospective or retrospective: prospective Sample size (cases): 3215 (377); [A] 1604, [B] 1611 |
||
| Patient characteristics and setting | Setting: COVID‐19 test centre Location: Municipal Health Services in Tilburg, Noord‐Brabant Country: Netherlands Dates: 23 December 2020‐17 January 2021 Symptoms and severity: current symptoms of COVID‐19 2226 (69.2%), symptoms in preceding 3 weeks 201 (6.3%), no current or prior symptoms 788 (24.5%). Definition of COVID‐19 symptoms was not provided. Demographics: median age 41 years (IQR 29‐54 years); 1409 (43.8%) male Exposure history: not reported |
||
| Index tests | Test name: [A] BD Veritor System for Rapid Detection of SARS‐CoV‐2
[B] SARS‐CoV‐2 antigen detection test Manufacturer: [A] Becton Dickinson, USA [B] Roche Antibody: nucleocapsid Ag target: not stated Test method: CGIA Samples used: NMT; self‐collected Transport media: none used Sample storage: no storage Test operator: self‐tested; written and illustrated booklet provided along with QR‐code link to a 2‐min online video illustrating NMT self‐sampling and self‐testing Definition of test positivity: visual Blinding reported: yes Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; either (1) Alinity M SARS‐CoV‐2
Assay (Abbott) or with (2) a LDT using the QIAsymphony Sample Processing and Rotorgene amplification system (Qiagen, Hilden, Germany)
Samples collected before 12 January 2021 were frozen at −80 °C within 24 h of collection and transported to the Dutch National Institute for Public Health and the Environment (RIVM) for viral culture] Definition of non‐COVID cases: single negative Genetic target(s): (1) N‐gene and RdRP‐gene target (2) E‐gene Samples used: NP+OP in GLY; collected by trained member of the Municipal Health Service Timing of reference standard: as for index test. Tested within 4 h of collection Blinded to index test: unclear; could presume Yes given self‐testing Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same day (instructed to self‐test immediately on arrival at home) All participants received same reference standard: yes Missing data: yes; Uninterpretable results: no PCR due to sample loss (n = 11) Indeterminate results (index test): "inconclusive" Ag assay results (n = 48; 9 PCR+ve and 39 PCR−) were excluded by authors for overall sensitivity and specificity; definition of 'inconclusive' was not reported. Inconclusive results were included for determining the Ct value cut‐off at which the chance (P) of having a positive viral culture was P = 0.5, and were interpreted as not false negative when determining the variables associated with a false negative result Indeterminate results (reference standard): inconclusive results on PCR (n = 3) were excluded by the authors; all Ag test negative Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: funded by the Dutch Ministry of Health, Welfare and Sports (VWS) Publication status: preprint Source: medRxiv Author COI: none to declare |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Stohr 2021 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 2 Ag tests; Stohr 2021 [A] details full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 2 Ag tests; Stohr 2021 [A] details full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] BD Veritor System for Rapid Detection of SARS‐CoV‐2
[B] SARS‐CoV‐2 antigen detection test Manufacturer: [A] Becton Dickinson, USA [B] Roche Antibody: nucleocapsid Ag target: not stated Test method: CGIA Samples used: NMT; self‐collected Transport media: none used Sample storage: no storage Test operator: self‐tested; written and illustrated booklet provided along with QR‐code link to a 2‐min online video illustrating NMT self‐sampling and self‐testing Definition of test positivity: visual Blinding reported: yes Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 2 Ag tests; Stohr 2021 [A] details full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 2 Ag tests; Stohr 2021 [A] details full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 2 Ag tests; Stohr 2021 [A] details full study characteristics and QUADAS. | ||
Stokes 2021(a) [A].
| Study characteristics | |||
| Patient Sampling | Report of 3 studies estimating sensitivity and/or specificity. This entry (Stokes 2021(a) [A]) relates to cohort [1]
[1] Sensitivity only: symptomatic participants with a recent positive SARS‐CoV‐2 RT‐PCR were invited to contribute further samples for an RDT evaluation; only those who were still PCR+ve on paired swabs are included A second cohort was included as Stokes 2021(b) [2] Sensitivity and specificity: symptomatic individuals presenting to Alberta Health Services community COVID‐19 assessment centres within 7 d pso (A third cohort was excluded because of deliberate inclusion of samples containing various non‐SARS‐CoV‐2 respiratory viruses in addition to samples from asymptomatic individuals at low risk of having COVID‐19 (all RT‐PCR−ve)). Recruitment: not stated Prospective or retrospective: prospective Sample size (cases): (1) 145 (138); 7 RT‐PCR−ve at time of second sampling were excluded by the review team |
||
| Patient characteristics and setting | Setting: unclear; likely community setting Location: samples tested positive at Alberta Precision Laboratories and confirmed as cases by Alberta Health Services Public Health Country: Canada Dates: not stated Symptoms and severity: all symptomatic; cough (42.8%), headache (42.1%), myalgias (41.4%), sinus congestion (36.6%), malaise (31.0%), pharyngitis (29.0%), fevers/chills (28.3%), anosmia (24.1%), ageusia (24.1%), rhinorrhoea (20.0%), shortness of breath (5.5%), nausea/vomiting (3.4%), and other (17.9%, included chest pain, diarrhoea, eye soreness, lymphadenopathy, loss of appetite, arthralgia, dizziness, and/or conjunctivitis) Demographics: mean age 39.4 years (median 36.0 years, range 18.5‐86.6 years); 42.8% male Exposure history: not reported |
||
| Index tests | Test name: Panbio Manufacturer: Abbott, IL, USA Antibody: nucleocapsid Ag target: not stated Test method: CGIA Samples used: [A] NP, [B] OP, [C], saliva [A] and [B] collected by trained HCWs, [C] was self‐collected using ClassiqSwab Paired NP swabs collected from separate nostrils; OP swabs collected from both sides of the oropharynx and the posterior pharyngeal wall under the uvula [B] and [C] sampling was terminated early due to poor sensitivity compared to NP Transport media: none used; tested immediately Sample storage: no storage Test operator: states tested immediately, so presume same HCW Definition of test positivity: visual line Blinding reported: yes (conducted first); but not blinded to COVID‐19 status (all previously PCR+ve) Timing of samples: mean duration of symptoms 6.1 d (median 6.0, range 3.0–10.0 days); 91% were ≤ 7 d pso |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; either an in‐house LDT or the Cobas SARS‐CoV‐2 test on the Cobas 6800 instrument Definition of non‐COVID cases: single negative PCR apart from discrepant results which (FPs) were re‐extracted and retested in triplicate with the N2 assay from the US CDC 2019‐Novel Coronavirus (2019‐nCoV) real‐time RT‐PCR diagnostic panel Genetic target(s): LDT ‐ E gene (< 35 Ct); Cobas ‐ not reported (2/2 targets positive, or ≥ 1 targets were positive in duplicate) Samples used: [A] NP (YOCON) swab and universal transport media (UTM) (Yocon, Beijing, China) [B] and [C] OP; ClassiqSwabs for throat in COPAN UTM‐RT (COPAN Diagnostics, CA, USA) Both stored at 4 °C upon arrival at the laboratory and tested within 72 h of collection Timing of reference standard: as for index Blinded to index test: not stated; may have known samples were from previously PCR+ve cases Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired All participants received same reference standard: yes Missing data: yes Uninterpretable results: 4, Panbio results were not recorded; 1, unable to be processed by RT‐PCR; 1, Panbio reported as negative before 15 min Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: 'funded using internal operating funds of Alberta Precision Laboratories and Alberta Health Services. Test kits and instruments were paid for by the Public Health Agency of Canada.' Publication status: published Source: European Journal of Clinical Microbiology & Infectious Diseases Author COI: the authors declare that they have no conflict of interest. |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Stokes 2021(a) [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 3 sample types; Stokes 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 3 sample types; Stokes 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Index tests | Test name: Panbio Manufacturer: Abbott, IL, USA Antibody: nucleocapsid Ag target: not stated Test method: CGIA Samples used: [A] NP, [B] OP, [C], saliva [A] and [B] collected by trained HCWs, [C] was self‐collected using ClassiqSwab Paired NP swabs collected from separate nostrils; OP swabs collected from both sides of the oropharynx and the posterior pharyngeal wall under the uvula [B] and [C] sampling was terminated early due to poor sensitivity compared to NP Transport media: none used; tested immediately Sample storage: no storage Test operator: states tested immediately, so presume same HCW Definition of test positivity: visual line Blinding reported: yes (conducted first); but not blinded to COVID‐19 status (all previously PCR+) Timing of samples: mean duration of symptoms 6.1 d (median 6.0, range 3.0–10.0 d); 91% were ≤ 7 d pso |
||
| Target condition and reference standard(s) | Comparative study of 3 sample types; Stokes 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 3 sample types; Stokes 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 3 sample types; Stokes 2021(a) [A] details the full study characteristics and QUADAS. | ||
Stokes 2021(a) [C].
| Study characteristics | |||
| Patient Sampling | Comparative study of 3 sample types; Stokes 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 3 sample types; Stokes 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Index tests | Test name: Panbio Manufacturer: Abbott, IL, USA Antibody: nucleocapsid Ag target: not stated Test method: CGIA Samples used: [A] NP, [B] OP, [C] saliva [A] and [B] collected by trained HCWs, [C] was self‐collected using ClassiqSwab Paired NP swabs collected from separate nostrils; OP swabs collected from both sides of the oropharynx and the posterior pharyngeal wall under the uvula [B] and [C] sampling was terminated early due to poor sensitivity compared to NP Transport media: none used; tested immediately Sample storage: no storage Test operator: states tested immediately, so presume same HCW Definition of test positivity: visual line Blinding reported: yes (conducted first); but not blinded to COVID‐19 status (all previously PCR+) Timing of samples: mean duration of symptoms 6.1 d (median 6.0, range 3.0–10.0 d); 91% were ≤ 7 d pso |
||
| Target condition and reference standard(s) | Comparative study of 3 sample types; Stokes 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 3 sample types; Stokes 2021(a) [A] details the full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 3 sample types; Stokes 2021(a) [A] details the full study characteristics and QUADAS. | ||
Stokes 2021(b).
| Study characteristics | |||
| Patient Sampling | Report of 3 studies estimating sensitivity and/or specificity
(1) Sensitivity only: symptomatic participants with a recent positive SARS‐CoV‐2 PCR were invited to contribute further samples for an RDT evaluation; only those who were still PCR+ve on paired swabs are included (Stokes 2021(a) [A])
(2) Sensitivity and specificity: symptomatic individuals presenting to Alberta Health Services community COVID‐19 assessment centres within 7 d of symptom(s) onset (Stokes 2021(b))
Excluded from review: (3) Specificity only: panel of samples from asymptomatic individuals at low risk of having COVID‐19 (all PCR−ve) and retrospective samples containing various respiratory viruses Recruitment: not specifically stated but all attending who met the criteria were invited to participate Prospective or retrospective: prospective Sample size (cases): (2) 1641 (268) |
||
| Patient characteristics and setting | Setting: community COVID‐19 test centre Location: Alberta Health Services community COVID‐19 assessment centres in Edmonton and Calgary Country: Canada Dates: not stated Symptoms and severity: all symptomatic; no further details Demographics: mean age 40.8 years (median 39.0 years, range 5.0‐90.0 years); 40.0% male Exposure history: not reported |
||
| Index tests | Test name: Panbio Manufacturer: Abbott, IL, USA Antibody: nucleocapsid Ag target: not stated Test method: CGIA Samples used: NP; collected by Alberta Health Services nurses Transport media: none used; tested immediately Sample storage: no storage Test operator: states tested immediately, so presume same nurse Definition of test positivity: visual line Blinding reported: yes (conducted first) Timing of samples: not reported; all < 7 days |
||
| Target condition and reference standard(s) | Reference standard: PCR; APL E‐gene PCR or a Health Canada/FDA‐approved commercial assay according to site (Allplex (Seegene, Seoul, South Korea), BDMax (Becton Dickinson, NJ, USA), Panther Fusion (Hologic, MA, USA), GeneXpert (Cepheid, CA, USA), or Simplexa (DiaSorin, Saluggia, Italy)). Discrepant results (FPs) were re‐extracted and retested in triplicate with the N2 assay from the US CDC 2019‐Novel Coronavirus (2019‐nCoV) real‐time PCR diagnostic panel Definition of non‐COVID cases: single negative Genetic target(s): not stated Samples used: NP in UTM (n = 1551, 94.5%); OP in UTM (n = 90, 5.5%) Timing of reference standard: as for index Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: 'funded using internal operating funds of Alberta Precision Laboratories and Alberta Health Services. Test kits and instruments were paid for by the Public Health Agency of Canada.' Publication status: published Source: European Journal of Clinical Microbiology & Infectious Diseases Author COI: the authors declare that they have no conflict of interest. |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Stromer 2020.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity only: upper respiratory tract samples (also described as "deep nasopharyngeal swabs") pre‐characterized by a positive or negative RT‐PCR result (Ag results not reported for PCR− samples) Recruitment: not mentioned; implies deliberate sampling to ensure samples with a range of Ct values were included Prospective or retrospective: retrospective Sample size (cases): 134 (124); subgroup of 21 PCR+ve samples used to compare 2 Ag tests not included |
||
| Patient characteristics and setting | Setting: unclear Location: Institute for Infection Medicine, Christian‐Albrecht University and University Medical Center, Schleswig‐Holstein Country: Germany Dates: not stated Symptoms and severity: not stated Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: NADAL COVID‐19 Ag Test
(A second test used on selected subgroup of samples was not included; Abbott Panbio COVID‐19 Antigen rapid test) Manufacturer: Nal von Minden GmbH Antibody: nucleoprotein Ag target: not stated Test method: unknown Samples used: NP Transport media: 500 µL of sterile PBS Sample storage: not stated Test operator: not stated Definition of test positivity: not stated Blinding reported: assume no?, conducted after RT‐PCR and states "all samples were pre‐characterized by a positive or a negative result" Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; in‐house Definition of non‐COVID cases: single negative PCR Genetic target(s): N gene Samples used: NP; same samples as index test Timing of reference standard: unclear Blinded to index test: yes, conducted first Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same swab All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "financial support by DFG (German Research Foundation) within the funding programme Open Access Publizieren" Publication status: published Source: Microorganisms Author COI: 'the authors declare no conflict of interest. The Nal von Minden GmbH supported this study by providing free kits and a scanner for optical intensity measurement. This company had no influence on the writing of the manuscript or on the interpretation of the data' |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Takeda 2020.
| Study characteristics | |||
| Patient Sampling | 2‐group study to estimate sensitivity and specificity, in:
[1] RT‐PCR+ve confirmed COVID‐19 samples selected from a total of 88 positive samples during time period (n = 62)
[2] random sample of RT‐PCR−ve samples selected from 1363 negative specimens tested during same time frame (n = 100) Recruitment: unclear for cases (may have been all "initial" samples tested); random sample of non‐cases Prospective or retrospective: unclear |
||
| Patient characteristics and setting | Setting: not stated; multiple clinical institutions Location: SRL Inc, Tokyo Country: Japan Dates: "early April'' also later states 4‐day period Symptoms and severity: not stated High viral load (< 25 Ct) 32/60, 53% Low viral load (≥ 25 Ct) 28/60, 47% Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: ESPLINE SARS‐CoV‐2 (no product code reported) Manufacturer: Fujirebio Inc Antibody: SARS‐CoV‐2 Ag (from IFU) Ag target: anti‐SARS‐CoV‐2 monoclonal antibodies (mouse) (from IFU) Test method: LFA using ALP‐labelled antibodies Samples used: NP; collection not reported Transport media: not described Sample storage: swabs mixed with sample treatment solution; no storage reported Test operator: not stated; laboratory staff presumed Definition of test positivity: visual line, as per manufacturer IFU Blinding reported: not stated Timing of samples: not stated but all cases are first samples presumed by authors to be from patient suspected of SARS‐CoV‐2 for the first time; negative samples were "probably … from … COVID‐19 patients for monitoring purposes and to check for negative conversion" |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; QuantiTect Probe RT‐PCR Kit (Qiagen) Definition of non‐COVID cases: as for cases; single negative required Genetic target(s): N2 Samples used: NP, as for index test Timing of reference standard: not stated Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous, same samples All participants received same reference standard: yes Missing data: 16 positive samples omitted; possibly because not initial samples but unclearly reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant (for cases), not clear for non‐cases |
||
| Comparative | |||
| Notes | Funding: none reported, however laboratory wholly owned by test manufacturer Publication status: preprint Source: medRxiv Author COI: "SRL Inc. is a subsidiary of Miraca Holdings Inc. Miraca Holdings Inc. holds all stock of Fujirebio Inc." |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Takeuchi 2021a.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: participants referred to a drive‐through PCR testing centre from 1 of 3 groups:
Recruitment: not stated; appears to be all who accepted invitation to participate during study period Prospective or retrospective: prospective Sample size (cases): 1186 (105) included from a total of 2079 referred patients and HCWs |
||
| Patient characteristics and setting | Setting: primary care COVID‐testing facility Location: PCR centre in Tsukuba Medical Center Hospital Country: Japan Dates: 7 October‐5 December 2020 Symptoms and severity: 771, 65% symptomatic, 415, 35% asymptomatic; fever (617, 80%), cough/sputum production (294, 38.1%), runny nose/nasal congestion (196, 25.4%), loss of taste or smell (33, 4.3%), dyspnoea (6, 0.8%), fatigue (77, 10%), diarrhoea (44, 5.7%), sore throat (149, 19.3%), headache (83, 10.8%) Demographics: median age 36.5 years, IQR 23‐50 years; 647 male (54.6%) Exposure history: not stated |
||
| Index tests | Test name: QuickNavi COVID‐19 Ag Manufacturer: Denka Co., Ltd., Tokyo, Japan Antibody: not stated Ag target: not stated Test method: lateral flow, no further details Samples used: NP; collection not reported but states samples were "obtained" Transport media: none used; sample buffer solution only Sample storage: immediate, no storage Test operator: not stated; states "examiner" Definition of test positivity: visual interpretation Blinding reported: yes done immediately after sample collection Timing of samples: median days from symptoms onset 2, IQR 1‐4 |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; in‐house at microbiology laboratory located next to the drive‐through sample‐collecting place of the PCR centre within an hour; samples also tested at reference laboratory using assay developed by the National Institute of Infectious Diseases, Japan, samples discrepant between the 2 PCRs were tested using BioFire Respiratory Panel 2.1 and FilmArray systems (BioFire Diagnostics, LLC, UT, USA) Definition of non‐COVID cases: double negative for absence of infection Genetic target(s): not stated Samples used: NP in UTM Timing of reference standard: not stated Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same time; paired swabs All participants received same reference standard: yes Missing data: 4 excluded due to missing symptom status Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): there was 1 discordant sample that was positive on in‐house RT‐PCR and negative on reference real‐time RT‐PCR. Considered negative after additional BioFire Respiratory Panel 2.1 examination Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: not stated Publication status: preprint Source: not stated Author COI: "Denka Co., Ltd., provided fees for research expenses and the QuickNavi‐COVID19 Ag kits without charge. Hiromichi Suzuki received a lecture fee from Otsuka Pharmaceutical Co., Ltd., regarding this study. Daisuke Kato, Miwa Kuwahara and Shino Muramatsu belong to Denka Co., Ltd., the developer of the QuickNavi‐COVID19 Ag" |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Takeuchi 2021b.
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity and specificity: participants referred to a drive‐through PCR centre from a local public health centre or from one of 97 primary care facilities; also states 17 samples obtained during hospitalization Recruitment: not stated; appears to include all meeting eligibility criteria at testing centre but also includes hospitalized Prospective or retrospective: prospective Sample size (cases): 862 (51) |
||
| Patient characteristics and setting | Setting: mainly COVID‐19 testing centre Location: not stated; multiple institutions in Tsukuba, Ibaraki Country: Japan Dates: 7 October 2020‐9 January 2021 Symptoms and severity: 790, 91.6% symptomatic; most commonly reported included fever (628, 79.5%), cough or sputum production (255, 32.3%), sore throat (210, 26.6%), runny nose or nasal congestion (185, 23.4%), headache (121, 15.3%). Loss of taste or smell (was reported in 32 (4.1%) overall and in 14 (27.5%) of PCR+ve group 72 (8.4%) asymptomatic Demographics: median age 36.0 years (IQR 24.0‐48.0); 106 (12.3%) were < 18 years; 383 (44.4%) female Exposure history: not reported |
||
| Index tests | Test name: QuickNavi‐COVID19 Ag Manufacturer: Denka Co., Ltd., Tokyo, Japan Antibody: not stated Ag target: not stated Test method: not stated Samples used: AN; collection appears to be by staff using FLOQswab Transport media: not used Sample storage: none; tested immediately Test operator: not stated; "examiner" Definition of test positivity: visual interpretation Blinding reported: yes; done first Timing of samples: median 2 d pso (IQR 1.0‐3.0) |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR;
Definition of non‐COVID cases: single negative Genetic target(s): not reported for in‐house assay; N and N2 for Quantitect Samples used: NP in UTM Timing of reference standard: as for index Blinded to index test: unclear Incorporated index test: not reported for in‐house assay; N and N2 for Quantitect |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; paired (NP collected after AN swab) All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): 1 sample was discrepant between LDT (+ve) and Quantitect assay (‐ve); +ve Xpert Xpress (Ct 39.8 on N2) Sample was obtained from a participant who had been diagnosed with COVID‐19 1 month before the current evaluation and who was referred to the PCR centre due to refractory respiratory symptoms Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "Denka Co., Ltd., provided fees for research expenses and the QuickNavi‐COVID19 Ag kits without charge" Publication status: preprint Source: medRxiv Author COI: "Hiromichi Suzuki received a lecture fee from Otsuka Pharmaceutical Co. Ltd., regarding this study. Daisuke Kato, Miwa Kuwahara and Shino Muramatsu belong to Denka Co., Ltd., the developer of the QuickNavi COVID19 Ag" |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Thommes 2021 [A].
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity alone: consecutive COVID‐19 patients admitted to the inpatient ward at the Department of Internal Medicine; described as moderate to severe disease Recruitment: consecutive Prospective or retrospective: prospective Sample size (cases): 154 (154) |
||
| Patient characteristics and setting | Setting: inpatient Location: Department of Internal Medicine II, the Medical University of Innsbruck Country: Austria Dates: August to end October 2020 Symptoms and severity: moderate to severe; all admitted Demographics: median age 69 years (range 18–92), 35.7% women Exposure history: not stated |
||
| Index tests | Test name: [A] PanbioTM COVID‐19 Ag Rapid test
[B] Novel Coronavirus (2019‐nCov) Antigen Detection Kit
[C] DIAQUICK COVID‐19 Ag Cassette
[D] SARS‐CoV‐2 Rapid Antigen Test Manufacturer: [A] Abbott, Chicago, Illinois [B] CLMSRDL, Sichuan Mass Spectrometry Biotechnology Co., Ltd, Chengdu, Sichuan [C] DIALAB, Wiener Neudorf, Austria [D] Roche Diagnostics Deutschland GmbH, Mannheim, Germany Antibody: not stated Ag target: not stated Test method: LFA Samples used: NP; collected by expert staff Transport media: none used Sample storage: none Test operator: performed by expert staff at the bedside using swabs provided in the Ag test kits Definition of test positivity: visual line Blinding reported: yes, done first Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; using Cobas apparatus (Roche Diagnostics GmbH, Mannheim, Germany) Definition of non‐COVID cases: single negative Genetic target(s): target ORF1a/b and B‐CoV target E‐Gene Samples used: OP Timing of reference standard: same as for index Blinded to index test: not stated Incorporated index test: not stated |
||
| Flow and timing | Time interval between index and reference tests: paired; simultaneous All participants received same reference standard: yes Missing data: yes; 145 patients reportedly recruited but number for samples per assay varied from 71‐99. No reason for missing data was given Uninterpretable results: unclear Indeterminate results (index test): unclear Indeterminate results (reference standard): unclear Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: there was no funding source for this study Publication status: published Source: International Journal of Infectious Diseases Author COI: no known competing financial interests or personal relationships |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Thommes 2021 [B].
| Study characteristics | |||
| Patient Sampling | Comparative study of 4 Ag tests; Thommes 2021 [A] details the full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 4 Ag tests; Thommes 2021 [A] details the full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] PanbioTM COVID‐19 Ag Rapid test
[B] Novel Coronavirus (2019‐nCov) Antigen Detection Kit
[C] DIAQUICK COVID‐19 Ag Cassette
[D] SARS‐CoV‐2 Rapid Antigen Test Manufacturer: [A] Abbott, Chicago, Illinois [B] CLMSRDL, Sichuan Mass Spectrometry Biotechnology Co., Ltd, Chengdu, Sichuan [C] DIALAB, Wiener Neudorf, Austria [D] Roche Diagnostics Deutschland GmbH, Mannheim, Germany Antibody: not stated Ag target: not stated Test method: LFA Samples used: NP; collected by expert staff Transport media: none used Sample storage: none Test operator: performed by expert staff at the bedside using swabs provided in the Ag test kits Definition of test positivity: visual line Blinding reported: yes, done first Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 4 Ag tests; Thommes 2021 [A] details the full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 4 Ag tests; Thommes 2021 [A] details the full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 4 Ag tests; Thommes 2021 [A] details the full study characteristics and QUADAS. | ||
Thommes 2021 [C].
| Study characteristics | |||
| Patient Sampling | Comparative study of 4 Ag tests; Thommes 2021 [A] details the full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 4 Ag tests; Thommes 2021 [A] details the full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] PanbioTM COVID‐19 Ag Rapid test
[B] Novel Coronavirus (2019‐nCov) Antigen Detection Kit
[C] DIAQUICK COVID‐19 Ag Cassette
[D] SARS‐CoV‐2 Rapid Antigen Test Manufacturer: [A] Abbott, Chicago, Illinois [B] CLMSRDL, Sichuan Mass Spectrometry Biotechnology Co., Ltd, Chengdu, Sichuan [C] DIALAB, Wiener Neudorf, Austria [D] Roche Diagnostics Deutschland GmbH, Mannheim, Germany Antibody: not stated Ag target: not stated Test method: LFA Samples used: NP; collected by expert staff Transport media: none used Sample storage: none Test operator: performed by expert staff at the bedside using swabs provided in the Ag test kits Definition of test positivity: visual line Blinding reported: yes, done first Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 4 Ag tests; Thommes 2021 [A] details the full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 4 Ag tests; Thommes 2021 [A] details the full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 4 Ag tests; Thommes 2021 [A] details the full study characteristics and QUADAS. | ||
Thommes 2021 [D].
| Study characteristics | |||
| Patient Sampling | Comparative study of 4 Ag tests; Thommes 2021 [A] details the full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Comparative study of 4 Ag tests; Thommes 2021 [A] details the full study characteristics and QUADAS. | ||
| Index tests | Test name: [A] PanbioTM COVID‐19 Ag Rapid test
[B] Novel Coronavirus (2019‐nCov) Antigen Detection Kit
[C] DIAQUICK COVID‐19 Ag Cassette
[D] SARS‐CoV‐2 Rapid Antigen Test Manufacturer: [A] Abbott, Chicago, Illinois [B] CLMSRDL, Sichuan Mass Spectrometry Biotechnology Co., Ltd, Chengdu, Sichuan [C] DIALAB, Wiener Neudorf, Austria [D] Roche Diagnostics Deutschland GmbH, Mannheim, Germany Antibody: not stated Ag target: not stated Test method: LFA Samples used: NP; collected by expert staff Transport media: none used Sample storage: none Test operator: performed by expert staff at the bedside using swabs provided in the Ag test kits Definition of test positivity: visual line Blinding reported: yes, done first Timing of samples: not stated |
||
| Target condition and reference standard(s) | Comparative study of 4 Ag tests; Thommes 2021 [A] details the full study characteristics and QUADAS. | ||
| Flow and timing | Comparative study of 4 Ag tests; Thommes 2021 [A] details the full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | Comparative study of 4 Ag tests; Thommes 2021 [A] details the full study characteristics and QUADAS. | ||
Toptan 2021(a).
| Study characteristics | |||
| Patient Sampling | Report of 2 single‐group studies to estimate sensitivity and specificity:
[1] samples stored after routine diagnostic use (Institute of Virology, Charite Berlin) (Toptan 2021(a))
[2] clinical samples collected as part of registered protocols from individuals living in shared housing (Institute of Virology, Frankfurt) (included as Toptan 2021(b)) Recruitment: not stated Prospective or retrospective: [1] retrospective (frozen samples) Sample size (cases): [1] 67 (58) |
||
| Patient characteristics and setting | Setting: unclear Location: [1] Institute of Virology, Charite Berlin Country: Germany Dates: not stated Symptoms and severity: not stated Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: not stated (may be RIDA‐QUICK SARS‐CoV‐2 Antigen assay) Manufacturer: R‐Biopharm Antibody: not stated Ag target: not stated Test method: not stated Samples used: [1] combined OP + NP Transport media: [1] after thawing at room temperature, swabs were resuspended in 1.5 mL of PBS Sample storage: stored (frozen) samples used Test operator: not stated Definition of test positivity: evaluated visually with 4 or 6 eye principle Blinding reported: yes, done in parallel (not sure this means blinded?) Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR Definition of non‐COVID cases: Genetic target(s): ORF1 and E gene Samples used: [1] combined OP + NP Timing of reference standard: not stated Blinded to index test: done in parallel (not sure this means blinded?) Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same samples All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "part of this work was funded by the German Ministry of Health (Konsiliarlabor für Coronaviren) to CD and VMC and by the German Ministry of Research through projects VARIPath (01KI2021)to VMC. This project was funded in part by the German Federal Ministry of Education and Research (Bundesministerium für Bildungund For‐schung, BMBF) (NaFoUniMedCovid19 – B‐FAST, EVIPAN, FKZ:01KX202)" Publication status: published Source: Journal of Clinical Virology Author COI: "no known competing financial interests or personal relationships" |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Toptan 2021(b).
| Study characteristics | |||
| Patient Sampling | Report of 2 single‐group studies to estimate sensitivity and specificity:
[1] samples stored after routine diagnostic use (Institute of Virology, Charite Berlin) (included as Toptan 2021(a))
[2] clinical samples collected as part of registered protocols from individuals living in shared housing (Institute of Virology, Frankfurt) (Torres 2021b) Recruitment: not stated Prospective or retrospective: [2] unclear Sample size (cases): [2] 70 (32) |
||
| Patient characteristics and setting | Setting: unclear Location: [2] Institute of Virology, Frankfurt Country: Germany Dates: not stated Symptoms and severity: not stated Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: not stated (may be RIDA‐QUICK SARS‐CoV‐2 Ag assay) Manufacturer: R‐Biopharm Antibody: not stated Ag target: not stated Test method: not stated Samples used: [2] NP Transport media: [2] 2 mL of PBS Sample storage: [2] Stored at 4 ℃, processed within 24 h Test operator: not stated Definition of test positivity: evaluated visually with 4 or 6 eye principle Blinding reported: yes, done in parallel Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR Definition of non‐COVID cases: Genetic target(s): ORF1 and E gene Samples used: [2] NP Timing of reference standard: not stated Blinded to index test: done in parallel (not sure this means blinded?) Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same samples All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: 'parts of this work was funded by the German Ministry of Health (Konsiliarlabor für Coronaviren) to CD and VMC and by the German Ministry of Research through projects VARIPath (01KI2021)to VMC. This project was funded in part by the German Federal Ministry of Education and Research (Bundesministerium für Bildungund For‐schung, BMBF) (NaFoUniMedCovid19 – B‐FAST, EVIPAN, FKZ:01KX202)' Publication status: published Source: Journal of Clinical Virology Author COI: "no known competing financial interests or personal relationships" |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Torres 2021a.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: asymptomatic household (n = 338) or non‐household (n = 296) close contacts of COVID‐19 patients as defined by the Spanish Ministry of Health (i.e. presence of compatible signs or symptoms and a positive NP swab RT‐PCR) Recruitment: consecutive Prospective or retrospective: prospective Sample size (cases): 634 (79) |
||
| Patient characteristics and setting | Setting: contact tracing Location: Clínico‐Malvarrosa Health Department, Valencia Country: Spain Dates: 16 October‐20 November 2020 Symptoms and severity: all asymptomatic; 39/79 PCR+ve individuals subsequently developed mild symptoms Demographics: male: 279 (44%), median age 37 years; range, 9‐87 years Exposure history: all contacts of confirmed cases; 338/634 were household contacts |
||
| Index tests | Test name: Panbio COVID‐19 Ag Rapid Test Device Manufacturer: Abbott (Diagnostic GmbH, Jena, Germany) Antibody: not stated Ag target: not stated Test method: lateral flow, no further information Samples used: NP (collected by experienced nurses) Transport media: none used Sample storage: immediate, no storage Test operator: not stated; may be same nurse "carried out at POC immediately after sampling" Definition of test positivity: not stated; as per manufacturer IFU Blinding reported: yes done first Timing of samples: timing was prescribed at the discretion of either the physician in charge of the index case or local health authorities:
|
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; TaqPath COVID‐19 Combo Kit (Thermo Fisher Scientific, MA, USA) Definition of non‐COVID cases: single negative Genetic target(s): N gene Samples used: NP in 3 mL of UTM (Becton Dickinson, Sparks, MD, USA) Timing of reference standard: as for index test Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swab from alternative nostril All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "this work received no public or private funds. Abbott Diagnostics provided the Panbio"
COVID‐19 Ag Rapid Test Device kits. Publication status: published Source: Clinical Microbiology and Infection Author COI: the authors declare no conflicts of interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Torres 2021b.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: appears to include participants meeting COVID‐19 testing criteria, described as either
Recruitment: unclear Prospective or retrospective: prospective Sample size (cases): 270 (106) |
||
| Patient characteristics and setting | Setting: outpatients
Unclear but could class as COVID‐19 test centre (acknowledgments mention Ag testing in primary healthcare centres, so may be anyone meeting testing criteria) Location: not reported. Authors affiliated to Hospital Clínico Universitario, INCLIVA Research Institute, Valencia Country: Spain Dates: 26 November 2020‐21 January 2021 Symptoms and severity: mixed; 178 (66%) symptomatic Demographics: symptomatic COVID‐19 suspects: median age (range): 41 (11‐83) years Sex: 112/178 (63%) female Asymptomatic COVID‐19 contacts: median age (range): 44 (11‐87) years Sex: 54/92 (59%) female Exposure history: 78 household contacts 14 non‐household contacts |
||
| Index tests | Test name: CLINITEST Rapid COVID‐19 Antigen Test (reported elsewhere to be the same as the Healgen Coronavirus Ag Rapid Test Cassette) Manufacturer: Siemens, Healthineers, Erlangen, Germany Antibody: SARS‐CoV‐2 nucleocapsid protein Ag target: not reported Test method: not reported Samples used: NP left nostril (experienced nurses) Transport media: none Sample storage: immediate testing Test operator: not reported; appears to be same nurse Definition of test positivity: according to manufacturer Blinding reported: yes; conducted first Timing of samples: median (range) pso or post diagnosis of index case: symptomatic 3 (1‐5) d Asymptomatic household contacts 4 (0‐7) d Asymptomatic non‐household contacts 5 (2‐7) d |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; Taq‐Path COVID‐19 Combo Kit (ThermoFisher Scientific, Massachusetts,
USA) Definition of non‐COVID cases: same as for cases Genetic target(s): not reported Samples used: NP right nostril, placed in 3 mL of UTM, Becton Dickinson, Sparks, MD, USA) Timing of reference standard: same as for index test Blinded to index test: not reported Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneously; paired swabs All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: this work received no public or private funds; "Siemens Healthineers provided the Rapid Test Device kits, but the company had no role in the study design, data collection, data analysis, data interpretation, or writing of the report" Publication status: published letter Source: Journal of Infection Author COI: the authors declare no conflicts of interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Turcato 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity and specificity: all patients with symptoms suspicious for SARS‐CoV‐2 infection, with a temperature > 37.3 °C, with any epidemiological risk criteria (e.g. reported contact with an infected person) or evaluated in the ED for other conditions not related to SARS‐CoV‐2 infection that required hospitalization Recruitment: consecutive; "all" included Prospective or retrospective: not stated; appears prospective Sample size (cases): 3410 (223) |
||
| Patient characteristics and setting | Setting: ED Location: Hospital of Merano (SABES‐ASDAA), Merano‐Meran Country: Italy Dates: 1 July‐10 November 2020 Symptoms and severity: 991, 29% symptomatic; 2419, 71% asymptomatic Demographics: not reported Exposure history: not reported |
||
| Index tests | Test name: STANDARD Q COVID‐19 Ag (R‐Ag) kit Manufacturer: SD BIOSENSOR, KR Antibody: not stated Ag target: not stated Test method: CGIA Samples used: not reported; states "two swabs" Transport media: not reported; presume none used Sample storage: not reported; states "implementation … in the initial screening" Test operator: not reported Definition of test positivity: not reported; as per manufacturer IFU Blinding reported: not stated; appear to have been conducted first Timing of samples: not stated |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; no details provided Definition of non‐COVID cases: as for cases (single negative PCR) Genetic target(s): not reported Samples used: not stated; paired "swabs" implied Timing of reference standard: not stated Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: not stated; paired "swabs" implied All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: no funding statement provided Publication status: published letter Source: Journal of Infection Author COI: no COI statement provided |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Van der Moeren 2021(a) [A].
| Study characteristics | |||
| Patient Sampling | Study reports data for 2 cohorts. Van der Moeren 2021(a) [A] relates to cohort [1] single‐group study to estimate sensitivity and specificity: all adults presenting at a single community test centre for COVID‐19 testing (n = 354)
See Van der Moeren 2021(b) for cohort [2] data [2] Single‐group study to estimate sensitivity alone: patients with a positive PCR test result at 1 of 3 community testing facilities who were retested at home within 72 h of initial positive result (n = 132) Recruitment: consecutive; "all" adults invited to participate Prospective or retrospective: prospective |
||
| Patient characteristics and setting | Setting: COVID‐19 test centre (community) Location: Municipal Health Service (GGD) regional test centre at Breda Country: Netherlands Dates: 28‐30 September Symptoms and severity: not stated; symptomatic Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: BD Veritor System for Rapid Detection of SARS‐CoV‐2 Manufacturer: Becton Dickinson Antibody: NP Ag target: not stated Test method: LFA; no further detail Samples used: NOP; "specimen from the throat and the superficial nasal cavities (bilateral, 2.5 cm proximal from the nostril)"; collected by GGD employee Transport media: direct testing Sample storage: stored dry in sterile test tubes and stored and transported on dry ice until processing at the laboratory; tested within 6 h after collection Test operator: trained laboratory technicians Definition of test positivity: [A] results reported using analyser device and [B] results by naked eye inspection alone (visual) Blinding reported: not stated Timing of samples: not reported; on presentation Time pso only provided for PCR+ve cases: 12 < 7 d; 1 ≥ 7 d; 4 = no pso data |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; either Cobas 6800 (Roche) or the m2000 (Abbott) Definition of non‐COVID cases: as for cases; single negative Genetic target(s): E‐ and RDRP‐gene (Cobas) or E‐gene and N‐gene (Abbott) Samples used: NOP; specimen from the throat and nasal cavity up to the nasal bridge Timing of reference standard: as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired All participants received same reference standard: yes; different assays Missing data: 2 samples excluded due to RT‐PCR coding error (considered overall low risk of bias due to small numbers) Uninterpretable results: 1 invalid on Ag test Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "the VRD (Ag) tests for this study were provided by the Dutch Ministry of Health, Welfare and Sport (VWS)" Publication status: preprint Source: medRxiv Author COI: "Jan Kluytmans is member of the National Outbreak Management Team of The Netherlands and of a committee which supports the implementation of the Corona‐reporting App." |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Van der Moeren 2021(a) [B].
| Study characteristics | |||
| Patient Sampling | Study compares test interpretation using an analyzer (Van der Moeren 2021(a) [A]) with interpretation by naked eye (Van der Moeren 2021(a) [B]); Van der Moeren 2021(a) [A] reports full study characteristics and QUADAS. | ||
| Patient characteristics and setting | Study compares test interpretation using an analyzer (Van der Moeren 2021(a) [A]) with interpretation by naked eye (Van der Moeren 2021(a) [B]); Van der Moeren 2021(a) [A] reports full study characteristics and QUADAS. | ||
| Index tests | Study compares test interpretation using an analyzer (Van der Moeren 2021(a) [A]) with interpretation by naked eye (Van der Moeren 2021(a) [B]); Van der Moeren 2021(a) [A] reports full study characteristics and QUADAS. | ||
| Target condition and reference standard(s) | Study compares test interpretation using an analyzer (Van der Moeren 2021(a) [A]) with interpretation by naked eye (Van der Moeren 2021(a) [B]); Van der Moeren 2021(a) [A] reports full study characteristics and QUADAS. | ||
| Flow and timing | Study compares test interpretation using an analyzer (Van der Moeren 2021(a) [A]) with interpretation by naked eye (Van der Moeren 2021(a) [B]); Van der Moeren 2021(a) [A] reports full study characteristics and QUADAS. | ||
| Comparative | |||
| Notes | |||
Van der Moeren 2021(b).
| Study characteristics | |||
| Patient Sampling | Study reports data for 2 cohorts: Van der Moeren 2021(b) relates to cohort [2]: single‐group study to estimate sensitivity alone: patients with a positive PCR test result at 1 of 2 community testing facilities who were retested at home within 72 h of initial positive result (n = 132) See Van der Moeren 2021(a) [A] for data related to cohort [1]: single‐group study to estimate sensitivity and specificity: all adults presenting at a single community test centre for COVID‐19 testing (n = 354) Recruitment: unclear; implies "all" those with positive PCR invited to participate Prospective or retrospective: prospective |
||
| Patient characteristics and setting | Setting: community Location: Municipal Health Service (GGD) regional test centres at Breda or Roosendaal Country: Netherlands Dates: 28 September‐6 October Symptoms and severity: at time of home visit: asymptomatic 3, 2% (2/3 still PCR+ve) Symptomatic 129 (123 still PCR+ve) Day < 7 66, 50% Day > 7 57, 43% Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: BD Veritor System for Rapid Detection of SARS‐CoV‐2 Manufacturer: Becton Dickinson Antibody: NP Ag target: not stated Test method: LFA; no further detail Samples used: NOP? "specimen from the throat and the superficial nasal cavities (bilateral, 2.5 cm proximal from the nostril)"; collected by GGD employee Transport media: direct testing Sample storage: stored dry in sterile test tubes and stored and transported on dry ice until processing at the laboratory; tested within 6 h after collection Test operator: trained laboratory technicians Definition of test positivity: reported using analyser device (included in main analysis for review), and by naked eye inspection alone Blinding reported: not stated Timing of samples: not reported; on presentation |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; either Cobas 6800 (Roche) or the m2000 (Abbott) Definition of non‐COVID cases: n/a Genetic target(s): E‐ and RDRP‐gene (Roche) or E‐gene and N‐gene (Abbott) Samples used: NOP; specimen from the throat and nasal cavity up to the nasal bridge Timing of reference standard: as for index test Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired All participants received same reference standard: yes; different assays Missing data: review team excluded 7 no longer PCR+ve at time of home visit (1 asymptomatic, 6 symptomatic) ‐ Veritor rapid diagnostic (VRD) test result for 1 asymptomatic PCR− is given (VRD‐) Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "the VRD (Ag) tests for this study were provided by the Dutch Ministry of Health, Welfare and Sport (VWS)" Publication status: preprint Source: medRxiv Author COI: "Jan Kluytmans is member of the National Outbreak Management Team of The Netherlands and of a committee which supports the implementation of the Corona‐reporting App." |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Veyrenche 2021.
| Study characteristics | |||
| Patient Sampling | 2‐group study estimating sensitivity and specificity:
[1] PCR+ve hospital inpatients (n = 45)
[2] pre‐pandemic samples from "patients" (not otherwise specified) (n = 20) Recruitment: not stated; appears to be convenience as equal numbers per Ct value subgroup Prospective or retrospective: retrospective |
||
| Patient characteristics and setting | Setting: inpatient Location: Montpellier University hospitals (Centre Hospitalier Universitaire de Montpellier, Montpellier) Country: France Dates: 14 March‐11 April Symptoms and severity: 27/45, 60% cases "severe" according to WHO guideline (similar numbers per Ct subgroup) Demographics: median age: Ct ≤ 25, 66 (IQR 48‐84) Ct 25‐35, 63 (50‐76) Ct ≥ 35, 58 (49‐67) Controls 64 (35‐93); 32/45, 71% male, all controls were male Exposure history: not stated |
||
| Index tests | Test name: Coris COVID‐19 Ag Respi‐Strip Manufacturer: BioConcept, Gembloux, Belgium Antibody: NP Ag target: monoclonal ab Test method: CGIA Samples used: NP; collection not described Transport media: yes; "swabs were collected in various transport media (eSwab COPAN Amies 1 mL, Σ‐Transwab liquid Amies, viral transport medium tube VTM‐M 2.0mL)." Sample storage: unclear; RT‐PCR conducted prospectively within a few hours but not reported for Ag testing Test operator: all tests were performed in the virology laboratory Definition of test positivity: visual, as per manufacturer IFU Blinding reported: not stated Timing of samples: day 1‐20 pso, median Ct ≤ 25, 7 (4‐10; presume this is IQR but could be range ‐ is described as SD in paper) Ct 25‐35, 8 (4‐12) Ct ≥ 35, 11 (7‐15) |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; Allplex 2019‐nCoV Assay (Seegene, Seoul, South Korea) Definition of non‐COVID cases: pre‐pandemic Genetic target(s): RdRp, N, E Samples used: NP; as for index Timing of reference standard: as for index Blinded to index test: yes, conducted first Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous; same swab All participants received same reference standard: no Missing data: none reported, no participant flow diagram reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "supported by Grants from Montpellier University Hospital and Montpellier University (MUSE)." Publication status: preprint Source: medRxiv Author COI: the authors declare that there are no conflicts of interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Villaverde 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: paediatric patients aged 0‐16 years attending EDs with symptoms compatible with SARS‐CoV‐2 infection and ≤ 5 days of evolution Recruitment: not stated; described as nested in a prospective, observational, multicenter cohort study Prospective or retrospective: retrospective Sample size (cases): 1620 (77) |
||
| Patient characteristics and setting | Setting: ED Location: one of 7 participating centres (Epidemiological Study of COVID‐19 in Children of the Spanish Society of Pediatrics; EPICO‐AEP) Country: Spain Dates: September and October 2020 Symptoms and severity: all symptomatic; specific symptoms not reported Demographics: not reported Exposure history: not reported |
||
| Index tests | Test name: Panbio Manufacturer: Abbott Rapid Diagnostic Antibody: nucleocapsid Ag target: not stated Test method: CGIA Samples used: NP; collected by trained nurses Transport media: none used Sample storage: none Test operator: attending paediatricians and nurse staff Definition of test positivity: visual line Blinding reported: yes, done first Timing of samples: all ≤ 5 days pso |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; not described Definition of non‐COVID cases: single negative Genetic target(s): sARS‐CoV‐2 E and RdRp genes Samples used: NP Timing of reference standard: performed within 24 h of specimen collection Blinded to index test: not stated Incorporated index test: not stated |
||
| Flow and timing | Time interval between index and reference tests: paired; simultaneous All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "supported by a specific Research Grant of the Spanish Society of Pediatrics (Asociacion Espanola de Pediatra). This study was funded by project PI20/00095, from the Instituto de Salud Carlos III (Ministry of Economy, Industry and Competitiveness), and cofounded by the European Regional Development Funds. CDG is funded by the Spanish Ministry of Science and Innovation—Instituto de Salud Carlos III and Fondos FEDER (Contrato Rio Hortega CM19/00015)." Publication status: published Source: Journal of Pediatrics Author COI: the authors declare no conflicts of interest. |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Weitzel 2020 [A].
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: samples from patients with respiratory symptoms and/or fever attending a private hospital ED Recruitment: convenience with deliberate sampling of positive cases to ensure a 2:1 distribution reported (5276 samples processed during study period) Prospective or retrospective: retrospective Number of samples (samples with confirmed SARS‐CoV‐2): 111 (80) *17 samples included in Porte 2020 |
||
| Patient characteristics and setting | Setting: ED (private hospital) Location: Clínica Alemana de Santiago Country: Chile Dates: 16 March‐26 April 2020 Symptoms and severity: respiratory symptoms and/or fever; no further detail Demographics: median age 40 years; 50, 45% male (median age 38 years, 43% male for all samples tested during period) Exposure history: none reported |
||
| Index tests |
Weitzel 2020 [A] entry is for test [A] in the list below Test name: [A] Biocredit COVID‐19 Ag One Step SARS‐CoV‐2 Antigen Test [B] COVID‐19 Antigen Rapid Test Device StrongStep COVID‐19 Antigen Test [C] Huaketai New Coronavirus (SARS‐CoV‐2) N Protein Detection Kit (Fluorescence immunochromatography) [D] Diagnostic Kit for 2019‐Novel Coronavirus (2019‐nCoV) Ag Test (Fluorescence Immunochromatographic Assay) Manufacturer: [A] RapiGEN Inc., Anyang‐si, Gyeonggi‐do, Republic of Korea [B] Liming Bio‐Products Co., Jiangsu, China [C] Savant Biotechnology Co., Beijing, China [D] Bioeasy Biotechnology Co., Shenzhen, China Ag target: not reported in study Antibody: not reported in study Test method: [A] and [B] CGIA [C] and [D] FIA Samples used: NOP swabs in 3 mL UTM Transport media: UTM‐RT System (Copan Diagnostics, Murrieta, CA, USA) Sample storage: stored at −80 °C; index tests applied on 28 and 29 April 2020 Test operator: single, trained laboratory technician under BSL2 cabinet; visual outputs read by 2 independent observers with referral to 3rd if needed Definition of test positivity: as per manufacturer IFU; Beijing Savant test required use of manufacturer‐supplied UV torch due to unavailability of reader device in Chile Blinding reported: yes; blinding stated Timing of samples: median 2 days (IQR 1‐5 days); 88% (96/109) during the first week of symptoms |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; COVID‐19 Genesig Real‐Time PCR assay (Primerdesign Ltd., Chandler's Ford, UK). Ct ≤ 40 considered positive Definition of non‐COVID cases: single PCR negative Genetic target(s): RdRp Samples used: NOP swabs; as for index Timing of reference standard: as for index test; median 2 days (IQR 1‐5 days) Blinded to index test: yes; prior to index Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same samples; index tests conducted after frozen storage All participants received same reference standard: yes Missing data: none reported; evaluation of Liming test was discontinued after initial poor performance (zero TP) Uninterpretable results: 2 tests had invalid results due to insufficient liquid migration (2 results excluded for each test) Indeterminate results (index test): visual interpretation of the Beijing Savant assay (using manufacturer‐supplied UV torch) was reportedly difficult under daylight conditions; manufacturer's fluorescence reader not available in Chile Indeterminate results (reference standard): none reported Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: study authors report that the work received no funding; Savant Biotechnology Co. provided test kits free of charge Publication status: preprint Source: medRxiv Author COI: all authors declare no competing interests |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Weitzel 2020 [B].
| Study characteristics | |||
| Patient Sampling | See Weitzel 2020 [A] for full study details and QUADAS entries. | ||
| Patient characteristics and setting | |||
| Index tests |
Weitzel 2020 [B] entry is for test [B] in the list below; see Weitzel 2020 [A] for full study details and QUADAS entries Test name: [A] Biocredit COVID‐19 Ag One Step SARS‐CoV‐2 Antigen Test [B] COVID‐19 Antigen Rapid Test Device StrongStep COVID‐19 Antigen Test [C] Huaketai New Coronavirus (SARS‐CoV‐2) N Protein Detection Kit (Fluorescence immunochromatography) [D] Diagnostic Kit for 2019‐Novel Coronavirus (2019‐nCoV) Ag Test (Fluorescence Immunochromatographic Assay) Manufacturer: [A] RapiGEN Inc., Anyang‐si, Gyeonggi‐do, Republic of Korea [B] Liming Bio‐Products Co., Jiangsu, China [C] Savant Biotechnology Co., Beijing, China [D] Bioeasy Biotechnology Co., Shenzhen, China Ag target: not reported in study Antibody: not reported in study Test method: [A] and [B] CGIA [C] and [D] FIA Samples used: NOP swabs in 3 mL UTM Transport media: UTM‐RT System (Copan Diagnostics, Murrieta, CA, USA) Sample storage: stored at −80 °C; index tests applied on 28 and 29 April 2020 Test operator: single, trained laboratory technician under BSL2 cabinet; visual outputs read by 2 independent observers with referral to 3rd if needed Definition of test positivity: as per manufacturer IFU; Savant test required use of manufacturer‐supplied UV torch due to unavailability of reader device in Chile Blinding reported: yes; blinding stated Timing of samples: median 2 days (IQR 1‐5 days); 88% (96/109) during the first week of symptoms |
||
| Target condition and reference standard(s) | See Weitzel 2020 [A] for full study details and QUADAS entries. | ||
| Flow and timing | See Weitzel 2020 [A] for full study details and QUADAS entries. | ||
| Comparative | |||
| Notes | |||
Weitzel 2020 [C].
| Study characteristics | |||
| Patient Sampling | See Weitzel 2020 [A] for full study details and QUADAS entries. | ||
| Patient characteristics and setting | See Weitzel 2020 [A] for full study details and QUADAS entries. | ||
| Index tests |
Weitzel 2020 [C] entry is for test [C] in the list below; see Weitzel 2020 [A] for full study details and QUADAS entries. Test name: [A] Biocredit COVID‐19 Ag One Step SARS‐CoV‐2 Antigen Test [B] COVID‐19 Antigen Rapid Test Device StrongStep COVID‐19 Antigen Test [C] Huaketai New Coronavirus (SARS‐CoV‐2) N Protein Detection Kit (Fluorescence immunochromatography) [D] Diagnostic Kit for 2019‐Novel Coronavirus (2019‐nCoV) Ag Test (Fluorescence Immunochromatographic Assay) Manufacturer: [A] RapiGEN Inc., Anyang‐si, Gyeonggi‐do, Republic of Korea [B] Liming Bio‐Products Co., Jiangsu, China [C] Savant Biotechnology Co., Beijing, China [D] Bioeasy Biotechnology Co., Shenzhen, China Ag target: not reported in study Antibody: not reported in study Test method: [A] and [B] CGIA [C] and [D] FIA Samples used: NOP swabs in 3 mL UTM Transport media: UTM‐RT System (Copan Diagnostics, Murrieta, CA, USA) Sample storage: stored at −80 °C; index tests applied on 28 and 29 April 2020 Test operator: single, trained laboratory technician under BSL2 cabinet; visual outputs read by 2 independent observers with referral to 3rd if needed Definition of test positivity: as per manufacturer IFU; Savant test required use of manufacturer‐supplied UV torch due to unavailability of reader device in Chile Blinding reported: yes; blinding stated Timing of samples: median 2 days (IQR 1‐5 days); 88% (96/109) during the first week of symptoms |
||
| Target condition and reference standard(s) | See Weitzel 2020 [A] for full study details and QUADAS entries. | ||
| Flow and timing | See Weitzel 2020 [A] for full study details and QUADAS entries. | ||
| Comparative | |||
| Notes | |||
Weitzel 2020 [D].
| Study characteristics | |||
| Patient Sampling | See Weitzel 2020 [A] for full study details and QUADAS entries. | ||
| Patient characteristics and setting | See Weitzel 2020 [A] for full study details and QUADAS entries. | ||
| Index tests |
Weitzel 2020 [D] entry is for test [D] in the list below; see Weitzel 2020 [A] for full study details and QUADAS entries. Test name: [A] Biocredit COVID‐19 Ag One Step SARS‐CoV‐2 Antigen Test [B] COVID‐19 Antigen Rapid Test Device StrongStep COVID‐19 Antigen Test [C] Huaketai New Coronavirus (SARS‐CoV‐2) N Protein Detection Kit (Fluorescence immunochromatography) [D] Diagnostic Kit for 2019‐Novel Coronavirus (2019‐nCoV) Ag Test (Fluorescence Immunochromatographic Assay) Manufacturer: [A] RapiGEN Inc., Anyang‐si, Gyeonggi‐do, Republic of Korea [B] Liming Bio‐Products Co., Jiangsu, China [C] Savant Biotechnology Co., Beijing, China [D] Bioeasy Biotechnology Co., Shenzhen, China Ag target: not reported in study Antibody: not reported in study Test method: [A] and [B] CGIA [C] and [D] FIA Samples used: NOP swabs in 3 mL UTM Transport media: UTM‐RT System (Copan Diagnostics, Murrieta, CA, USA) Sample storage: stored at −80 °C; index tests applied on 28 and 29 April 2020 Test operator: single, trained laboratory technician under BSL2 cabinet; visual outputs read by 2 independent observers with referral to 3rd if needed Definition of test positivity: as per manufacturer IFU; Savant test required use of manufacturer‐supplied UV torch due to unavailability of reader device in Chile Blinding reported: yes; blinding stated Timing of samples: median 2 days (IQR 1‐5 days); 88% (96/109) during the first week of symptoms |
||
| Target condition and reference standard(s) | See Weitzel 2020 [A] for full study details and QUADAS entries. | ||
| Flow and timing | See Weitzel 2020 [A] for full study details and QUADAS entries. | ||
| Comparative | |||
| Notes | |||
Winkel 2020.
| Study characteristics | |||
| Patient Sampling | Single‐group study estimating sensitivity and specificity: football players, staff and referees from 13 professional football clubs and the national teams in the Netherlands; all tested approximately weekly by RT‐PCR independent of presence of symptoms, 2 days prior to each match; results for symptomatic individuals were excluded Recruitment: consecutive; all tested Prospective or retrospective: prospective Sample size (cases): 824 people provided 2425 samples; 52 positive on RT‐PCR |
||
| Patient characteristics and setting | Setting: screening (sports) Location: professional football clubs and national football teams; author institutions included University Medical Center Utrecht, Royal Netherlands Football Association (KNVB) Country: Netherlands Dates: 1 October‐9 November 2020 Symptoms and severity: all asymptomatic or pre‐symptomatic. Of 68 PCR+ve results, 12 (18%) pre‐symptomatic, 32 (47%) early infection (1st +ve in asymptomatic plus any subsequent +ves within 7 days), 21 (31%) late infection (≥ 7 days pso as long as symptoms had subsided or ≥ 7 days after 1st +ve PCR) 3 (4%) persistent viral shedding (> 4 weeks after 1st PCR+ve) Demographics: median age 27 years (range 16‐80 years, IQR: 21‐40 years); 775, 94% male Exposure history: not stated |
||
| Index tests | Test name: Panbio COVID‐19 Ag rapid test device Manufacturer: Abbott (Lake Country, IL, USA) Antibody: nucleocapsid Ag target: not stated Test method: CGIA Samples used: NP Transport media: none; swabs placed in lysis buffer Sample storage: none; tested immediately Test operator: trained personnel (presume non‐HCW) Definition of test positivity: visual appearance of test line; weak unclear bands were considered inconclusive and were either excluded or considered positive or negative to determine best case and worst case scenarios Blinding reported: yes; done first Timing of samples: approximately weekly |
||
| Target condition and reference standard(s) | Reference standard: RT‐qPCR; either Eurofins (Brugge, Belgium), Synlab Laboratories (Luik, Belgium), or U‐diagnostics (Utrecht, the Netherlands). Definition of non‐COVID cases: Single negative Genetic target(s): not stated Samples used: seems to be combined NP+OP; "throat/nasopharyngeal swab" Timing of reference standard: as for index Blinded to index test: not stated Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: 96% paired swabs; 104 (4%) samples for PCR obtained on subsequent day All participants received same reference standard: yes Missing data: 1 Uninterpretable results: 1 invalid LFA excluded due to lack of control band Indeterminate results (index test): 16 "inconclusive"; 5 in PCR+ve and 11 in PCR− (confirmed by subsequently negative PCR results) Indeterminate results (reference standard): none reported Unit of analysis: samples |
||
| Comparative | |||
| Notes | Funding: "this study was investigator‐initiated and funded by the executing parties. No external funding was received. The Panbio COVID‐19 Ag Rapid Tests were provided by the Ministry of Health, Welfare and Sport (VWS)" Publication status: preprint Source: medRxiv Author COI: no COI statement was provided |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | No | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Yokota 2020(a).
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity: PCR+ve cases obtained from symptomatic patients with COVID‐19 were included
(A further 307 negative saliva samples from asymptomatic people did not undergo Ag testing.) Recruitment: not reported Prospective or retrospective: retrospective Sample size (cases): 34 PCR+ve (17 NP, 17 saliva) |
||
| Patient characteristics and setting | Setting: laboratory‐based; states samples from "Covid‐19 patients" Location: unclear (author institution: Department of Hematology, Hokkaido University Faculty of Medicine, Sapporo, Japan) Country: Japan Dates: not reported Symptoms and severity: symptomatic; median 9 days (range, 2‐14 days) pso Demographics: not reported Exposure history: not reported |
||
| Index tests | Test name: Espline SARS‐CoV‐2 Manufacturer: Fujirebio, Tokyo, Japan Antibody: not reported Ag target: not reported Test method: immunochromatographic assay Samples used:
Transport media: not reported Sample storage: frozen samples; no further details Test operator: probably laboratory staff Definition of test positivity: yes; according to the manufacturer IFU Blinding reported: no; PCR performed before index test Timing of samples: median time of sampling was 9 days (range, 2‐14 days) pso |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR Master Mixes (Thermo Fisher Scientific, Waltham, USA) and Real Time
PCR System (Thermo Fisher Scientific). PCR tests were performed according to the manual by National Institute of Infectious Diseases (NIID, www.niid.go.jp/niid/images/epi/corona/2019‐nCoVmanual20200217‐en.pdf)
RNA was extracted using QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany) Definition of non‐COVID cases: N/A Genetic target(s): not reported Samples used:
Timing of reference standard: median time of sampling was 9 days (range, 2‐14 days) after symptom onset Blinded to index test: yes; performed before index test Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same samples All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: unclear |
||
| Comparative | |||
| Notes | Funding: 'this study was supported by Health, Labour and Welfare Policy Research Grants 20HA2002.' Publication status: preprint Source: medRxiv Author COI: 'Espline SARS‐CoV‐2 and Lumipulse SARS‐CoV‐2 Ag kit were supplied by Fujirebio'. Authors declare no other competing interest |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Yokota 2020(b).
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity: PCR+ve cases obtained from symptomatic patients with COVID‐19 were included
(A further 307 negative saliva samples from asymptomatic people did not undergo Ag testing.) Recruitment: not reported Prospective or retrospective: retrospective Sample size (cases): 34 PCR+ve (17 NP, 17 saliva) |
||
| Patient characteristics and setting | Setting: laboratory‐based; states samples from "Covid‐19 patients" Location: unclear (author institution: Department of Hematology, Hokkaido University Faculty of Medicine, Sapporo, Japan) Country: Japan Dates: not reported Symptoms and severity: symptomatic; median 9 days (range, 2‐14 days) pso Demographics: not reported Exposure history: not reported |
||
| Index tests | Test name: Espline SARS‐CoV‐2 Manufacturer: Fujirebio, Tokyo, Japan Antibody: not reported Ag target: not reported Test method: immunochromatographic assay Samples used:
Transport media: not reported Sample storage: frozen samples; no further details Test operator: probably laboratory staff Definition of test positivity: yes; according to the manufacturer IFU Blinding reported: no; PCR performed before index test Timing of samples: median time of sampling was 9 days (range, 2‐14 days) pso |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR Master Mixes (Thermo Fisher Scientific, Waltham, USA) and Real Time
PCR System (Thermo Fisher Scientific). PCR tests were performed according to the manual by National Institute of Infectious Diseases (NIID, niid.go.jp/niid/images/epi/corona/2019‐nCoVmanual20200217‐en.pdf)
RNA was extracted using QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany) Definition of non‐COVID cases: NA Genetic target(s): not reported Samples used:
Timing of reference standard: median time of sampling was 9 days (range, 2‐14 days) pso Blinded to index test: yes; performed before index test Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: same samples All participants received same reference standard: yes Missing data: none reported Uninterpretable results: none reported Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported Unit of analysis: unclear |
||
| Comparative | |||
| Notes | Funding: "this study was supported by Health, Labour and Welfare Policy Research Grants 20HA2002." Publication status: preprint Source: medRxiv Author COI: "Espline SARS‐CoV‐2 and Lumipulse SARS‐CoV‐2 Ag kit were supplied by Fujirebio". Authors declare no other competing interest. |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate inclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Young 2020.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: patients with ≥ 1 symptoms of COVID‐19 (within ≤ 7 days pso) at 21 study sites (n = 260)
(2nd cohort of 361 samples from COVID suspects ≤ 5 days pso also evaluated to compare BD Veritor with Quidel Sofia 2 SARS Antigen FIA but excluded from review as only discrepant results on the 2 Ag assays underwent RT‐PCR.) Recruitment: not stated Prospective or retrospective: prospective |
||
| Patient characteristics and setting | Setting: mixed; drive‐through/tent (n = 42), outpatient clinic (n = 74), research clinic (n = 72), or skilled nursing facility (n = 66) Location: unclear; 21 geographically diverse study sites (author institutions BD Life Sciences, Louisiana State University Health Sciences Center, Tricore Reference Laboratory) Country: USA Dates: 5‐11 June 2020 Symptoms and severity: 110 (43%) cough, 98 (39%) muscle pain, 95 (37%) headache, 90 (35%) sore throat, 90 (35%) sore throat, 78 (31%) fever Of those at ≤ 6 d pso (n = 245): 94 (38%) with 1 symptom, 151 (62%) with ≥ 2 symptoms Demographics: median age 43 (range 18‐90); 91 (36%) male Exposure history: not reported |
||
| Index tests | Test name: BD Veritor SARS‐CoV‐2 antigen test (no product codes) Manufacturer: Becton, Dickinson and Company, BD Life Sciences—Integrated Diagnostic Solutions, San Diego, CA Antibody: NP Ag target: not stated Test method: not stated; chromatographic immunoassay with analyser Samples used: nasal (presume AN); clinician collected from both nostrils (same swab); inserted approximately 2.5 cm up the nostril rolled 5 times along the mucosa Transport media: dry nasal swabs Sample storage: swabs were shipped for testing on dry ice (−70 °C); Test operator: not stated; Veritor testing was performed internally at BD (San Diego, CA, USA) Definition of test positivity: as per manufacturer IFU Blinding reported: yes; all personnel blinded to all other test results Timing of samples: all ≤ 7 d pso; median 3.0 d, mean 3.2 d. 38 (15%) 1 d pso, 57 (23%) 2 days, 54 (22%) 3 days, 40 (16%) 4 days, 37 (15%) 5 days, 19 (8%) 6 days, 6 (2%) 7 days |
||
| Target condition and reference standard(s) | Reference standard: Lyra SARS‐CoV‐2 PCR Assay (Quidel Corporation. Athens, OH); BD MAX real time SARS‐CoV‐2 PCR assay used for discordant testing Definition of non‐COVID cases: as for cases (single negative) Genetic target(s): not stated Samples used: NP (n = 217) or OP (n = 34); clinician collected (if an NP swab was collected as part of standard care, the participant had the option of having an OP study swab taken in lieu of a second NP swab) Timing of reference standard: swabs taken prior to any study swabs (potential for contamination of nasal cavity) Blinded to index test: yes; performed at TriCore Reference Laboratories. "All testing was conducted with all personnel blinded to all other test results" Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: simultaneous (paired) All participants received same reference standard: yes Missing data: 9 excluded; 6 did not meet eligibility criteria and 3 had invalid specimens/results (2 on RT‐PCR and 1 labelling error) Uninterpretable results: 3 invalid on at least 1 assay Indeterminate results (index test): none reported Indeterminate results (reference standard): none reported. Re‐test of 9 'FN' results with BD MAX RT‐PCR resulted in 2 confirmed FN (BD MAX+ve and sero+ve), 6 were BD Max−ve (incl 1 sero+ve) and 1 invalid (no result) Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: 'study was funded by Becton, Dickinson and Company; BD Life Sciences—Integrated Diagnostics Solutions. Non‐BD employee authors received research funds as part of this work' Publication status: preprint Source: medRxiv Author COI: 'CRD, CF, KE, JCA, HR, and CKC are employees of Becton, Dickinson and Company; SY, None; CC, None; AM, None; CGF, None; CB, None; JA, None; RA, CEO and PI of Comprehensive Clinical Research LLC' |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Did all participants receive a reference standard? | Yes | ||
| Could the patient flow have introduced bias? | High risk | ||
Young 2021.
| Study characteristics | |||
| Patient Sampling | Single‐group study to estimate sensitivity and specificity: patients admitted to hospital for emergency care Recruitment: consecutive; all those who were admitted Prospective or retrospective: prospective Sample size (cases): 803 (214) |
||
| Patient characteristics and setting | Setting: inpatient/emergency care Location: Oxford University Hospitals NHS Foundation Trust (Nuffield Department of Medicine, University of Oxford, John Radcliffe Hospital, Oxford, UK) Country: UK Dates: 23 December 2020‐30 January 32021 Symptoms and severity: 11 (8%) Ag positive had no COVID‐related symptoms recorded (cough, dyspnoea, fever, ageusia or anosmia); 28/80 (35.0%) RDT‐ but PCR+ve had a pre‐admission SARS‐CoV‐2 PCR+ve swab Demographics: not stated Exposure history: not stated |
||
| Index tests | Test name: Innova Manufacturer: Innova Med Group, China (Xiamen Biotime Biotechnology) Antibody: N gene Ag target: not stated Test method: CGIA Samples used: "nose and throat" (collected by HCW) Transport media: none used Sample storage: none required; testing performed in the admitting department Test operator: "staff"; presume HCW Definition of test positivity: visual Blinding reported: yes Timing of samples: not stated; on admission |
||
| Target condition and reference standard(s) | Reference standard: RT‐PCR; Thermo‐Fisher Taq‐Path
Ct threshold not reported but range in Ct values was plotted; all < 35 Ct Definition of non‐COVID cases: same as cases (single ‐ve PCR) Genetic target(s): not stated Samples used: same as for index test; transferred to the clinical laboratory in VTM Timing of reference standard: not stated Blinded to index test: unclear Incorporated index test: no |
||
| Flow and timing | Time interval between index and reference tests: paired swab; 732/803 swabs obtained on same day, 71/803 within 24 h of each other All participants received same reference standard: yes Missing data: yes; 18 invalid results reported Uninterpretable results: none reported Indeterminate results (index test): 2 invalid RDTs; 1 PCR+ve and 1 PCR− Indeterminate results (reference standard): 16 invalid on PCR; 1 RDT+ and 15 RDT‐. The RDT+ sample was reported in the text as "indeterminate" on PCR, and the patient tested PCR+ve 5 days later Unit of analysis: participant |
||
| Comparative | |||
| Notes | Funding: "one of the authors is an NIHR Clinical Lecturer and another is Robertson Foundation Fellow" Publication status: published as letter to the editor Source: Journal of Infection Author COI: one of the authors declares lecture fees from Gilead outside the submitted work. No other authors have a conflict to declare. |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate inclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of single test application) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Evaluations of serial (repeat) testing) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Reference standard does not incorporate result of index test? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | No | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Did all participants receive a reference standard? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Ag: antigen; ALP: alkaline phosphatase; AN: anterior nasal; BAL: bronchoalveolar lavage; CDC: Centers for Disease Control; CE: conforms with European standards; CGIA: colloidal gold immunoassay; CI: confidence interval; CLEIA: chemiluminescent enzyme immunoassay; COI: conflict of interest; Ct: cycle threshold; ED: Emergency Department; EUA: emergency use authorization; FIA: fluorescence immunochromatographic; FN: false negative; FP: false positive; GLY: Glucose‐Lactalbumin‐Yeast; GP: general practitioner; HCW: healthcare worker; ICU: intensive care unit; IFU: instructions for use; IPD: individual patient data; IQR: interquartile range; IVD: in vitro diagnostic medical device; LDT: laboratory‐developed test; LFA: lateral flow assay; LFD: lateral flow device; LFT: lateral flow test; N/A: not applicable; NAAT: nucleic acids amplification test; NIH: National Institutes of Health; NHS: National Health Service (UK); NMT: nasal mid‐turbinate; NOP: naso‐oropharyngeal; NP: nasopharyngeal; OP: oropharyngeal; PCR: polymerase chain reaction; PHE: Public Health England; POC: point of care; PBS: phosphate‐buffered saline; pso: post‐symptom onset; QUADAS: Quality Assessment tool for Diagnostic Accuracy Studies; qPCR: quantitative reverse transcription polymerase chain reaction; RADT: rapid antigen detection test; RAT: rapid antigen test; RDT: rapid diagnostic test; RNA: ribonucleic acid; RDT: rapid diagnostic test; PCR: reverse transcription polymerase chain reaction; rPCR: rapid reverse transcription polymerase chain reaction; SD: standard deviation; TA: tracheal aspirate; TN: true negative; TP: true positive; UTM: universal transport medium; UV: ultraviolet; UW: University of Washington; VTM: viral transport medium; WHO: World Health Organization
Characteristics of excluded studies [ordered by study ID]
| Study | Reason for exclusion |
|---|---|
| Ahava 2021 | Ineligible index test (lab‐based antigen assay) |
| Aoki 2020 | Ineligible index test (lab‐based antigen assay) |
| Bello‐Chavolla 2021 | Ineligible index test (multiple assays used) |
| Chen 2021 | Ineligible study design (proof of concept) |
| Corman 2020 | Ineligible study design (analytical accuracy) |
| Cubas‐Atienzar 2021 | Ineligible study design (analytical accuracy) |
| Dalal 2021 | Ineligible study design |
| Diao 2020 | Exclude on index test ‐ not commercially available |
| Dohla 2020 | Ineligible index test |
| Downs 2021 | Ineligible study design (multiple samples per participant; only gives overall positivity rates) |
| Eshghifar 2021 | Inadequate sample size (also analytical accuracy) |
| Frnda 2021 | Accuracy data cannot be extracted |
| Gili 2021 | Ineligible index test (lab‐based antigen assay) |
| Haage 2021a | Ineligible population |
| Haage 2021b | Ineligible study design |
| Herrera 2020 | Ineligible reference standard |
| Hingrat 2020 | Ineligible index test (lab‐based antigen assay) |
| Hirotsu 2020 | Ineligible index test |
| Hirotsu 2021 | Ineligible index test (lab‐based antigen assay) |
| Hledik 2020 | Accuracy data cannot be extracted |
| Hoehl 2020 | Ineligible study design |
| Kannian 2021 | Ineligible population (cases PCR positive on NP and saliva; controls PCR negative on saliva but half positive on NP swabs) |
| Kashiwagi 2020 | Inadequate sample size |
| Kobayashi 2021 | Ineligible index test (lab‐based antigen assay) |
| Koskinen 2021 | Ineligible index test (lab‐based antigen assay) |
| Kotsiou 2021 | Accuracy data cannot be extracted (PPV only) |
| Kurstjens 2020 | Ineligible index test |
| Kyosei 2020 | Ineligible study design |
| Lefever 2021 | Ineligible index test (laboratory‐based assay) |
| Le Hingrat 2020 | Ineligible index test |
| Li 2021 | Inelligible index test (in‐house microfluidic assay) |
| Liu 2021 | Ineligible index test (in‐house CLIA) |
| Mahari 2020 | Ineligible study design |
| Mak 2020a | Ineligible study design (deliberate sampling by Ct values; analytical accuracy) |
| Mak 2020b | Ineligible study design (deliberate sampling by Ct values; analytical accuracy) |
| Marzinotto 2020 | Accuracy data cannot be extracted |
| Mboumba 2021 | Ineligible study design (analytical accuracy) |
| McAulay 2020 | Ineligible index test (uses serum samples) |
| McDonald 2020 | Ineligible reference standard |
| Menchinelli 2021 | Ineligible index test (lab‐based antigen assay) |
| Mohamed 2021 | Ineligible study design |
| Moreno 2021 | Accuracy data cannot be extracted |
| Nachtigall 2020 | Ineligible index test |
| Ogawa 2020 | Inadequate sample size |
| Pavelka 2020 | Ineligible study design |
| Pekosz 2021 | Ineligible reference standard (also reports subgroup of Young 2020 ) |
| Pellanda 2020 | Ineligible index test |
| Peng 2020 | Ineligible study design |
| Perchetti 2020 | Ineligible study design |
| Pollock 2020a | Ineligible index test |
| Rastawicki 2021 | Ineligible study design (serial testing in hospitalised patients) |
| Regev‐Yochay 2021 | Ineligible index test (multiple assays used) |
| Ren 2021 | Ineligible index test (lab‐based antigen assay) |
| Rodriguez‐Manzano 2020 | Ineligible index test |
| Rusanen 2020 | Ineligible index test (lab‐based antigen assay) |
| Scheiblauer 2021 | Ineligible study design (analytical accuracy only) |
| Seitz 2021 | Accuracy data cannot be extracted (confused reporting) |
| Seo 2020 | Accuracy data cannot be extracted |
| Singh 2020a | Ineligible index test |
| Singh 2020b | Ineligible index test |
| Wang 2020 | Ineligible index test (amplification‐free nucleic acid immunoassay) |
| Wu 2020 | Ineligible index test |
| Yamamoto 2021 | Ineligible population (all COVID‐19 inpatients; specificity in previously positive patients) |
| Yamayoshi 2020 | Ineligible study design (analytical accuracy only) |
| Yokota 2021 | Ineligible index test (lab‐based antigen assay) |
| Yu 2020 | Ineligible index test |
| Zamecnik 2020 | Ineligible index test |
| Zeng 2020 | Ineligible study design |
| Zhang 2020 | Ineligible index test |
Characteristics of studies awaiting classification [ordered by study ID]
Abdul‐Mumin 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptomatic
Asymptomatic contacts Ghana Teaching hospital |
| Index tests | Standard Q SARS‐CoV‐2 Ag‐RDT Sample: NP; on‐site |
| Target condition and reference standard(s) | RT‐PCR |
| Flow and timing | Sample size (cases): 193 (42) |
| Comparative | |
| Notes |
Abusrewil 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Mixed
Report se% by symptom status, by time pso and for Ct < 25 but no underlying numbers; needs author contact for subgroup data Tripoli, Libya Setting not reported; Biotechnology Research Center laboratories |
| Index tests | 10 Ag‐based rapid assays: Fluorecare, ESPLINE, RapiGen, Abbott Panbio, Flowflex, Acon, Assut Europe, Orient Gene, CerTest, Bioperfectus, AMP Sample: NP; on‐site (different participants per assay) |
| Target condition and reference standard(s) | RT‐PCR |
| Flow and timing | Sample size (cases): 231 (108) |
| Comparative | |
| Notes |
Akashi 2021.
| Patient Sampling | Single‐group (se only) |
| Patient characteristics and setting | Mixed Tsukuba, Japan Drive‐through‐type PCR centre |
| Index tests | QuickNavi‐COVID19 Ag
New digital version (not yet marketed) DK20‐CoV‐8M Sample: NP; on‐site |
| Target condition and reference standard(s) | RT‐PCR |
| Flow and timing | Sample size (cases): 96; 44 had new digital version |
| Comparative | |
| Notes |
Amer 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Mixed; symptomatic, time pso Zagazig, Egypt University/medical centres; referred to the COVID‐19 isolation unit |
| Index tests | SDQ Sample: NP + OP; after frozen storage |
| Target condition and reference standard(s) | RT‐PCR Ct < 29, 29‐36, 37‐39 |
| Flow and timing | Sample size (cases): 83 (69) |
| Comparative | |
| Notes | IPD is in Suppl 1 |
Aranda‐Diaz 2021.
| Patient Sampling | Single‐group (se + sp; but does not meet minimum n cases) |
| Patient characteristics and setting | Primarily asymptomatic San Francisco, California, USA Residents and staff of congregate‐living (homeless) shelters |
| Index tests | BinaxNOW
Repeat/serial testing (not really accuracy) Sample: not reported; must be on‐site |
| Target condition and reference standard(s) | RT‐PCR but only in selected |
| Flow and timing | 40 for accuracy; 828 eligible residents and 435 staff for uptake etc outcomes |
| Comparative | |
| Notes | Query inclusion, not really an accuracy study. Only first 40 had both RDT and PCR (all Ag‐ve, 2 PCR+ve), then only some RDT+ had PCR |
Baccani 2021.
| Patient Sampling | Unclear (single‐group, but subgroup tested with LFAs) |
| Patient characteristics and setting | Inpatients, no further detail |
| Index tests | SDF, AFIAS LFA, and Lumipulse Sample: NP |
| Target condition and reference standard(s) | RT‐PCR |
| Flow and timing | Sample size (cases): 201 (33) for Lumipulse; 93 (28) for SDF |
| Comparative | |
| Notes |
Bianco 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Mixed; c66% asymptomatic; incl paediatric Italy; ED and occupational health wards |
| Index tests | LumiraDx Sample: nasal; direct |
| Target condition and reference standard(s) | PCR, Xpert Xpress; NP |
| Flow and timing | Sample size (cases): 907 (298) |
| Comparative | |
| Notes |
Blairon 2021.
| Patient Sampling | Multi‐group |
| Patient characteristics and setting | Not reported Belgium |
| Index tests | Bio‐Rad, Novatec, LumiraDx Sample: respiratory |
| Target condition and reference standard(s) | PCR |
| Flow and timing | 150+ve, 9−ve and 40 confounder panel |
| Comparative | |
| Notes | Probable exclude ‐ samples pre‐selected to represent certain Ct ranges |
Bonde 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Mainly asymptomatic Finland; hospital staff and patients with unknown SARS‐CoV‐2 status at paediatric department or ED |
| Index tests | BD Veritor Sample: OP; on‐site |
| Target condition and reference standard(s) | PCR |
| Flow and timing | Sample size (cases): 809 (29) |
| Comparative | |
| Notes | Multiple samples per participant (n = 674) |
Boum 2021.
| Patient Sampling | |
| Patient characteristics and setting | |
| Index tests | |
| Target condition and reference standard(s) | |
| Flow and timing | |
| Comparative | |
| Notes | Details yet to be completed |
Brihn 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Mainly asymptomatic; both reported separately USA; all patients admitted through the ED |
| Index tests | Quidel Sofia Sample: AN; on‐site |
| Target condition and reference standard(s) | PCR; NP |
| Flow and timing | Sample size (cases): 2039 (149) |
| Comparative | |
| Notes |
Bruins 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptomatic Netherlands; HCW with mainly early symptoms |
| Index tests | Panbio Sample: NP; on‐site |
| Target condition and reference standard(s) | TMA; NP + OP |
| Flow and timing | Sample size (cases): 1101 (84) |
| Comparative | |
| Notes |
Bruzzone 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Not reported Italy; lab‐based retrospective |
| Index tests | Multiple (7 assays) Sample: respiratory; on‐site |
| Target condition and reference standard(s) | PCR |
| Flow and timing | Sample size (cases): 321 (253) |
| Comparative | |
| Notes | Sensitivity reported per assay, but underlying numbers not given. Would need author contact for inclusion |
Caruana 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Asymptomatic Switzerland; ED patients |
| Index tests | SD Biosensor ‐ Standard Q Sample: NP; on‐site |
| Target condition and reference standard(s) | PCR |
| Flow and timing | Sample size (cases): 116 (7) |
| Comparative | |
| Notes |
Cento 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Mixed; data reported separately for subgroup with available symptom data Italy; patients presenting to ED with COVID symptoms or admitted to hospital for any other reason |
| Index tests | LumiraDx Sample: NP; on‐site |
| Target condition and reference standard(s) | PCR; NP |
| Flow and timing | Sample size (cases): 960 (347) |
| Comparative | |
| Notes |
Chiu 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | 2 cohorts 1) symptomatic, 2) asymptomatic outbreak screening 1) USA; 2) Hong Kong |
| Index tests | INDICAID, phase scientific Sample: nasal; on‐site |
| Target condition and reference standard(s) | PCR |
| Flow and timing | Sample size (cases): 1) 274 (75); 2) 22,994 (38) |
| Comparative | |
| Notes |
Christensen 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptomatic USA; multiple sites |
| Index tests | BD Veritor triple test (COVID + Flu A+B) Sample: nasal |
| Target condition and reference standard(s) | PCR |
| Flow and timing | Sample size (cases): 278 (60) |
| Comparative | |
| Notes |
Di Domenico 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Asymptomatic Italy; healthy volunteers attending a Mediterranean fair |
| Index tests | Panbio and a new near patient ELISA Sample: 1) NP; 2) cyto‐salivary |
| Target condition and reference standard(s) | PCR |
| Flow and timing | Sample size (cases): 433 (36) |
| Comparative | |
| Notes |
Dierks 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Not reported Netherlands, HCWs |
| Index tests | LumiraDx; Nadal; TMA Sample: respiratory |
| Target condition and reference standard(s) | PCR |
| Flow and timing | Sample size (cases): 444 (11); Nadal assay performed on only 215/444 samples |
| Comparative | |
| Notes |
Diez Flecha 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Mixed; data reported separately for subgroup with available symptom data Spain; nursing home residents |
| Index tests | Panbio Sample: NP; on‐site |
| Target condition and reference standard(s) | PCR; NP (obtained next day) |
| Flow and timing | Sample size (cases): 55 (36) |
| Comparative | |
| Notes |
Eleftheriou 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Mixed; reported separately for symptomatic and asymptomatic Greece; paediatric inpatients |
| Index tests | Panbio Sample: NP |
| Target condition and reference standard(s) | PCR |
| Flow and timing | Sample size (cases): 744 (51); 9 asymptomatic cases |
| Comparative | |
| Notes |
Fernandez 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Mixed; reported separately for symptomatic and asymptomatic Spain; residential setting, symptomatic for COVID‐19 or close contacts |
| Index tests | LumiraDx Sample: nasal |
| Target condition and reference standard(s) | PCR; NP |
| Flow and timing | Sample size (cases): 46 (24) |
| Comparative | |
| Notes |
Fernandez‐Montero 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | se and sp reported separately for asymptomatic and higher‐risk groups (close contact or mild symptoms). Also reported for Ct value < 20/25/30/35 Spain; asymptomatic adults from a semi‐closed community |
| Index tests | Roche SARS CoV‐2 Rapid Antigen test Sample: NP, on‐site |
| Target condition and reference standard(s) | RT‐PCR < 20, 20 to < 25, 25 to < 30 and > 30 |
| Flow and timing | Sample size (cases): 2543, (49) |
| Comparative | |
| Notes |
Ferte 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptomatic, asymptomatic, risk situation, no risk situation Bordeaux University health campus screening facility |
| Index tests | Abbott Panbio Sample: NP |
| Target condition and reference standard(s) | RT‐PCR, N gene, RdRp gene, N and RdRp gene, Ct < 23, < 30, < 33 |
| Flow and timing | Sample size (cases): 692 (52) |
| Comparative | |
| Notes |
Fiedler 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Mixed; not reported separately by symptom status Individuals tested by public health Essen‐symptoms or asymptomatic contacts |
| Index tests | Liaison Sars CoV‐2 Ag assay Sample: NP, Ag testing performed over 4 d in lab |
| Target condition and reference standard(s) | RT‐PCR |
| Flow and timing | Sample size (cases): 182, 110 cases |
| Comparative | |
| Notes | Exclude: lab‐based Ag testing performed |
Ford 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptomatic; asymptomatic; also reported by sex, race, age < or > 24, quarantine status Staff and students at University in Wisconsin |
| Index tests | Quidel Sofia SARS Antigen Fluorescent Immunoassay Sample: nasal swab |
| Target condition and reference standard(s) | RT‐PCR < 25, > 25. Viral culture performed if discrepant PCR/Ag results. Subgenomic RNA testing if Ag or PCR pos |
| Flow and timing | Sample size (cases): 1058 (54) |
| Comparative | |
| Notes | Check overlap with Pray 2021; 10.15585/mmwr.mm695152a3 |
Frediani 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptomatic; may need author contact for data by age/time USA: participants at rapid test centres aged ≥ 7 years, symptomatic for < 7 d |
| Index tests | Abbott BinaxNOW Sample: AN swabs; included staff‐collected and self‐ or parent‐collected swabs |
| Target condition and reference standard(s) | RT‐PCR, Ct 0‐25 subgroup |
| Flow and timing | Sample size (cases): 309 (93) |
| Comparative | |
| Notes |
Gitaka 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptomatic and asymptomatic Kenya‐ HCWs and outpatients at Mary Help Hospital, students and general population at Mount Kenya University |
| Index tests | BD Veritor rapid Ag test Sample: AN for Ag, OP for PCR |
| Target condition and reference standard(s) | RT‐PCR, Ct 0‐25 subgroup |
| Flow and timing | Sample size (cases): 272 (47) |
| Comparative | |
| Notes |
Hagbom 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Hospitalised patients; asymptomatic HCWs. Median age and days pso reported Sweden‐hospital. Hospitalized patients and asymptomatic HCWs |
| Index tests | Rapid Response COVID‐19 Antigen Rapid Test Cassette; DIAGNOS COVID‐19 Antigen Saliva Test; Panbio Sample: saliva |
| Target condition and reference standard(s) | RT‐PCR. All PCR pos samples were cultured |
| Flow and timing | Sample size (cases): 54 (15) |
| Comparative | |
| Notes |
Harmon 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Asymptomatic athlete screening programme |
| Index tests | Quidel Sofia‐2 SARS‐ CoV‐2 Antigen Tests at least 1 x per week |
| Target condition and reference standard(s) | PCR |
| Flow and timing | 23,462 weekly paired Ag/RT‐PCR screening tests in 1931 athletes; 83 PCR+ve |
| Comparative | |
| Notes | From Abstract: "Daily antigen testing was similar to RT‐ PCR testing two to three times a week in identifying infection. Antigen testing identified infection before the next scheduled PCR on 89 occasions and resulted in 234 days where potentially infectious athletes were isolated before they would have been isolated with RT‐ PCR testing alone" |
Harris 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Asymptomatic community campus participants and critically unwell in hospital University of Arizona, staff and students |
| Index tests | Quidel Rapid Ag Test Sample: self‐collected AN for Ag, NP for PCR |
| Target condition and reference standard(s) | RT‐PCR |
| Flow and timing | Sample size (cases): 885 (305) |
| Comparative | |
| Notes |
|
Holzner 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptomatic, asymptomatic; symptomatic with Ct < 30 Hospital in Germany, all presentations to ED |
| Index tests | Standard Q; Roche Sample: NP swab by HCW |
| Target condition and reference standard(s) | RT‐PCR 0‐20, 20‐25, 25‐30, > 30 |
| Flow and timing | Sample size (cases): 2375 (551) |
| Comparative | |
| Notes |
Homza 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptoms vs asymptomatic Testing centre Czech Republic |
| Index tests | ECOTEST COVID‐19 Ag rapid test STANDARD Q COVID‐9 Ag ND COVID‐19 Ag test JOYSBIO SARS‐CoV‐2 Ag rapid test kit Immupass Vivadiag SARS‐COV‐2 Ag rapid test Sample: NP. On site |
| Target condition and reference standard(s) | RT‐PCR, < 20, 20‐30, 30‐40, viral culture used when Ag and PCR result differed |
| Flow and timing | Sample size (cases): 1141 (397) |
| Comparative | |
| Notes |
Homza 2021a.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptomatic and asymptomatic, also categorised as PCR+ve and PCR pos+viable virus Testing centre Czech Republic |
| Index tests | ECOTEST COVID‐19 Ag rapid test Sample: NP, on site |
| Target condition and reference standard(s) | RT‐PCR, 0‐19.9, 20‐24.9, 24.9‐30, 30‐40. Viral culture used when Ag and PCR result differed. |
| Flow and timing | Sample size (cases): 494 (164) |
| Comparative | |
| Notes |
Kanji 2021.
| Patient Sampling | Single‐group, Ag positive only |
| Patient characteristics and setting | Voluntary screening of asymptomatic HCWs |
| Index tests | Abbott Panbio or BD Veritor occurred on a weekly or twice‐weekly basis |
| Target condition and reference standard(s) | Ag pos confirmed with RT‐PCR |
| Flow and timing | 71,847 Ag tests across 369 clinical sites |
| Comparative | |
| Notes | Probable exclude From Abstract: 87/71,847 Ag positive; 39 confirmed as TP on PCR |
Kenyeres 2021.
| Patient Sampling | Single‐group (sensitivity only) |
| Patient characteristics and setting | Suspected infection, no further subgroups Hospital Hungary, patients admitted with presumed SARS‐CoV‐2 |
| Index tests | RapidGen ‐ Biocredit Ag Sample: NP |
| Target condition and reference standard(s) | Positive by at least 2 PCRs |
| Flow and timing | Sample size (cases): 119 (65) pos by 2 PCR methods, 37 randomly selected for Ag |
| Comparative | |
| Notes |
Kim 2021.
| Patient Sampling | Single‐group (se + sp) (2 study cohorts) |
| Patient characteristics and setting |
|
| Index tests | GenBody COVAG025 Sample: NP swab |
| Target condition and reference standard(s) | RT‐PCR, 1. se for Ct < 25 and 25‐30 |
| Flow and timing | Sample size (cases): 1. 130 (30); 2. 200 (100) |
| Comparative | |
| Notes | 2 separate studies. 1 retrospective, 2 prospective |
Kiro 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Patients with suspected infection, no further subgroups Tertiary care centre India |
| Index tests | STANDARD F COVID‐19 Ag FIA test kit Sample: NP |
| Target condition and reference standard(s) | RT‐PCR |
| Flow and timing | Sample size (cases): 354 (136) |
| Comparative | |
| Notes | Ag result read by analyser |
Kiyasu 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptomatic, asymptomatic Japan, hospital screen for symptomatic/suspected infection |
| Index tests | QuickNavi‐COVID19 Ag Sample: NP |
| Target condition and reference standard(s) | In‐house and reference RT‐PCR < 20, 20‐24, 25‐29, > 30 |
| Flow and timing | Sample size (cases): 1934 (187) |
| Comparative | |
| Notes |
Klein 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptomatic and asymptomatic. Symptom duration available for PCR+ve Drive in test centre Germany |
| Index tests | Panbio COVID‐19 Ag rapid test Sample: 1. self‐collected nasal 2. professional NP swab |
| Target condition and reference standard(s) | RT‐PCR. Ct values and viral load reported for all PCR+ve samples |
| Flow and timing | Sample size (cases): 290 (45) |
| Comparative | |
| Notes |
Koeleman 2021.
| Patient Sampling | Single‐group (se + sp); 2 study periods |
| Patient characteristics and setting | Symptomatic ED patients, nursing home patients, HCWs Teaching hospital, Netherlands |
| Index tests |
Study period 1 Certest SARS‐CoV‐2 Roche SARS‐CoV‐2 Rapid Antigen Test Romed Coronavirus Ag Rapid Test BD Veritor SARS‐CoV‐2 point‐of‐care test Panbio COVID‐19 Antigen rapid test Study period 2 Romed Coronavirus Ag Rapid Test Sample: NP‐ RT‐PCR performed on day, 3 of 5 Ag tests were performed within 72 h, 2 Ag tests performed 1 month after collection |
| Target condition and reference standard(s) | RT‐PCR; Ct < 20 and < 30 |
| Flow and timing | Study period 1, 80 prospectively selected specimens, 40 PCR+ve Study period 2: 900 samples, 300 cases |
| Comparative | |
| Notes | 2 different study periods |
Kolwijck 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptomatic HCWs Teaching hospital Netherlands |
| Index tests | Panbio COVID‐19 Ag rapid test Sample: NP |
| Target condition and reference standard(s) | RT‐PCR (NP + OP); <20, <25, <28, <30 |
| Flow and timing | Sample size (cases): 443 (45) |
| Comparative | Implementation also reported but pos Ag not confirmed by PCR |
| Notes |
Korenkov 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptomatic; reported by days PSO Germany, routine SARS‐CoV‐2 testing centre |
| Index tests | Standard Q COVID‐19 Ag Test (SD Biosensor/Roche) Sample: combined oro and NP swab |
| Target condition and reference standard(s) | RT‐PCR <20, <25, <30, <35 +/‐culture |
| Flow and timing | 2028 tested by PCR, 118 samples also cultured. 210 cases PCR+ve |
| Comparative | |
| Notes |
Kritikos 2021.
| Patient Sampling | Single‐group (sensitivity only) |
| Patient characteristics and setting | Hospitalized SARS CoV‐2+ve patients Tertiary hospital, Switzerland |
| Index tests | One Step Immunoassay Exdia COVID‐19 Ag (Precision Biosensor) and Standard Q COVID‐19 Rapid Antigen Test (Roche) Sample: paired diluted NP and saliva samples, 1 undiluted NP swab |
| Target condition and reference standard(s) | RT‐PCR |
| Flow and timing | 58 confirmed SARS‐CoV‐2 patients |
| Comparative | |
| Notes |
Kumar 2021a.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Includes asymptomatic ED universal admission screen in tertiary private hospital India |
| Index tests | STANDARD Q COVID‐19 Ag Sample: NP swab |
| Target condition and reference standard(s) | RT‐PCR |
| Flow and timing | Sample size (cases): 288 (6 PCR+ve) |
| Comparative | |
| Notes | 1 further case positive post‐discharge (day 6 post‐first neg PCR and Ag?). Query if 2x2 possible |
Kumar 2021b.
| Patient Sampling | Single‐group (se only) |
| Patient characteristics and setting | Asymptomatic pre‐operative screen Tertiary eye care centre South India |
| Index tests | STANDARD Q rapid Ag test Sample: NP and throat |
| Target condition and reference standard(s) | RT‐PCR, Ct < 40 positive |
| Flow and timing | Sample size (cases): 204 (12) |
| Comparative | |
| Notes |
Kurihara 2021.
| Patient Sampling | Single group (se + sp) |
| Patient characteristics and setting | Includes asymptomatic Tsukuba Clinics, Ibaraki prefecture, Japan |
| Index tests | Quick Chaser Auto SARS‐CoV‐2 Mizuho Medy Sample: NP; immediately |
| Target condition and reference standard(s) | RT‐PCR Ct < 20, 20‐24, 25‐29, ≥ 30 |
| Flow and timing | Sample size (cases): 1401 (83) |
| Comparative | |
| Notes |
Kweon 2021.
| Patient Sampling | Single group (se + sp) 2 groups combined n = 38 symptomatic patients (200 specimens) But also a 2nd group 'non‐COVID' n = 122 specimens few details on same |
| Patient characteristics and setting | Symptomatic (serial testing)
Second asymptomatic group
Time from onset < 7, 8‐14, > 14 d Chung‐Ang University Hospital, Seoul |
| Index tests | AFIAS COVID‐19 Ag
Icroma COVID‐19 Ag Sample: NP, after frozen storage |
| Target condition and reference standard(s) | RT‐PCR Ct < 25, 25‐30, 30‐40 |
| Flow and timing | 38 patients in symptomatic group Symptomatic patients 38, specimens 200, pos specimens 141 'Non‐COVID' group 122 specimens |
| Comparative | |
| Notes | DTA on symptomatic patients to which n = 122 COVID negative samples were added; check eligibility |
Landaas 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Includes asymptomatic
Symptoms ≤ 5 days, > 5 days Aker test station, Oslo, Norway |
| Index tests | Panbio Sample: NP and OP, on site |
| Target condition and reference standard(s) | RT‐PCR Ct < 30, ≥ 30 |
| Flow and timing | Sample size (cases): 3991 (250) |
| Comparative | |
| Notes |
Lee 2021.
| Patient Sampling | Single‐group (se + sp) Retrospective selection known positives and presumably negatives |
| Patient characteristics and setting | No detail South Korea |
| Index tests | Standard Q Sample: NP, frozen storage |
| Target condition and reference standard(s) | RT‐PCR Ct < 14.9, 15‐19.9, 20‐24.9, 25‐29.9, 30‐34.9, ≥ 35 |
| Flow and timing | Sample size (cases): 680 (83) |
| Comparative | |
| Notes | Samples retrospectively selected i.e. 380 known positives selected based on calculated Ct value distribution for population; query eligibility |
Le Goff 2021.
| Patient Sampling | Single group (se + sp) |
| Patient characteristics and setting | Includes asymptomatic COVISAN test centres, Paris, France |
| Index tests | Standard Q Sample: NP and also saliva on‐site |
| Target condition and reference standard(s) | RT‐PCR |
| Flow and timing | Sample size (cases): 1718 (117) |
| Comparative | |
| Notes | LAMP also compared |
Leixner 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptomatic
Days 0‐3, 0‐5 ED, Klinik Landstrasse, Vienna, Austria |
| Index tests | AMP RAT Ameda Sample: NP, on site NP + OP for PCR |
| Target condition and reference standard(s) | RT‐PCR Ct < 25 Ct < 30 |
| Flow and timing | Sample size (cases): 392 (94) |
| Comparative | |
| Notes |
Lindner 2021.
| Patient Sampling | Single subgroup (se + sp) |
| Patient characteristics and setting | Symptomatic; presumably includes asymptomatic Ambulatory test facility, Berlin/Heidelberg, Germany |
| Index tests | Espline, Sure Status, Mologic Sample: NP + OP |
| Target condition and reference standard(s) | RT‐PCR B.1.1.7 |
| Flow and timing | Sample size (cases): 1692 (whole group) Cases = 354, cases B.1.1.7 220 |
| Comparative | |
| Notes | Appears this is a B.1.1.7 subgroup analysis of a larger DTA study |
Lv 2021.
| Patient Sampling | Single‐group (se only) |
| Patient characteristics and setting | Confirmed COVID‐19 patients; both antibody positive and negative Shanghai Public Health Clinical Center |
| Index tests | Diagnostic Kit for COVID‐19 Antigen Test (Colloidal Gold) (Kehua Bio‐engineering, China) Sample: NP |
| Target condition and reference standard(s) | RT‐PCR NP swab |
| Flow and timing | Sample size (cases): 85 (85) |
| Comparative | |
| Notes |
Mack 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Includes asymptomatic USA, Surveillane NFL players and staff |
| Index tests | Quidel Sofia
Repeat/serial testing Sample: NP, Ag available within 1 h (presume on site) |
| Target condition and reference standard(s) | RT‐PCR but also more complex definition of a case see Fig 1 algorithm |
| Flow and timing | 10,982 samples; PCR+ve cases = 174, adjudicated+ve cases = 130 |
| Comparative | May not give DTA |
| Notes |
Matsuda 2021.
| Patient Sampling | Single‐group (se + sp) for each Ag test |
| Patient characteristics and setting | Symptomatic < 10 days
Slightly contradictory r/e asymptomatic cases Sao Paulo, Brazil 3 service outpatient/inpatient/private clinics/hospitals in Sao Paolo Met |
| Index tests | ECO Test ECO Diagnostica
Panbio Sample: NP, Ag POC presumably (15 min) |
| Target condition and reference standard(s) | RT‐PCR |
| Flow and timing | Sample size (cases): 108 (31) |
| Comparative | |
| Notes |
McKay 2021.
| Patient Sampling | Single‐group (se + sp); separate groups per RAT |
| Patient characteristics and setting | Includes asymptomatic Nursing home, Gerogia, USA |
| Index tests | Repeat/serial testing x3 rounds
BinaxNow Sample: AN, for all Ag tests NP for RT‐PCR first round of testing |
| Target condition and reference standard(s) | RT‐PCR Ct < 30, Ct ≥ 30 |
| Flow and timing | Sample size (cases): 532 (PCR‐confirmed cases = 105, PCR+ve and/or Ag+ve = 113: 21/101 samples pos by PCR and/or Ag were culture‐positive) |
| Comparative | |
| Notes |
Muhi 2021.
| Patient Sampling | Single‐group (sp), designed as se + sp but no cases identified |
| Patient characteristics and setting | Includes asymptomatic 3 x hospitals, Melbourne, Australia |
| Index tests | Panbio Sample: deep nasal+OP |
| Target condition and reference standard(s) | RT‐PCR |
| Flow and timing | Sample size (cases): 2413 (0) |
| Comparative | |
| Notes |
Nordgren 2021.
| Patient Sampling | Single‐group (se + sp); separate groups per RAT |
| Patient characteristics and setting | Symptomatic Östergötland, Sweden |
| Index tests | Panbio, Zhejang Orient gene Sample: NP, stored at 4 °C RAT tested within 7/7 |
| Target condition and reference standard(s) | RT‐qPCR (but don't see viral loads?) Ct < 20, 20‐25, 25‐30, > 30 Viral culture Seasonal CoV |
| Flow and timing | Sample size (cases): Panbio 286 (PCR+ve cases =156); Orient Gene 332 (PCR+ve cases =156) |
| Comparative | |
| Notes |
Norizuki 2021.
| Patient Sampling | Single‐group (se only) |
| Patient characteristics and setting | Includes asymptomatic Airport quarantine, Japan |
| Index tests | Repeat/serial testing
ESPLINE Sample: NPS x 2, Swab Anterior Nasal, Swab Anterior tongue, Saliva x 2 |
| Target condition and reference standard(s) | RT‐qPCR Viral load ≥ 10^4, < 10^4 |
| Flow and timing | Sample size (cases): 20 (20) 97 person day samples |
| Comparative | |
| Notes | Also lab based quantitative Ags, Lumipulse and LAMP |
Oh 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Includes asymptomatic Hospitalized & ED presentations in Seoul |
| Index tests | Repeat/serial testing (some cases)
Standard Q Sample: NPS, on site |
| Target condition and reference standard(s) | RT‐PCR Ct ≤ 30,≤ 25 |
| Flow and timing | 118 paired tests from 98 patients Cases = 40 |
| Comparative | |
| Notes |
Orsi 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptomatic
Symptoms < 7, 7‐14 days ED, San Martino, Genoa, Italy |
| Index tests | FREND COVID‐19 As assay
Standard F Sample: NPS Tested in lab within 8 h |
| Target condition and reference standard(s) | RT‐PCR Ct < 26, 26‐30, 31‐35 |
| Flow and timing | Sample size (cases): 110 (60) |
| Comparative | |
| Notes |
Osmanodja 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Includes asymptomatic Ambulatory test facilities, Charité, Berlin |
| Index tests | Dräger
On site Sample: self‐collected AN swabs Ag NP/OP ‐ PCR |
| Target condition and reference standard(s) | RT‐PCR Viral load (RNA copies): < 1 million, 1‐10 million, > 10 million Variant B.1.1.7 or WT |
| Flow and timing | Sample size (cases): 379 (70) |
| Comparative | |
| Notes |
Perez‐Garcia 2021.
| Patient Sampling | Two groups (se + sp) |
| Patient characteristics and setting | Mixed (84% symptomatic)
Reports se by time pso Spain Setting not reported; Servicio de Microbiología Clínica, Hospital Universitario Príncipe de Asturias, Madrid |
| Index tests | Panbio, STANDARD F (SD Biosensor, Inc.) Sample: NP in 3 mL of UTM, lab |
| Target condition and reference standard(s) | RT‐PCR by Ct ≤ 20, Ct = 20–25, Ct= 25‐30, Ct > 30 |
| Flow and timing | Sample size (cases): 356 (170) and 186 control |
| Comparative | |
| Notes |
Qahtani 2021.
| Patient Sampling | Single‐group; designed as se + sp, zero cases identified |
| Patient characteristics and setting | Mainly asymptomatic, passengers at airport Canada Vancouver International Airport |
| Index tests | Panbio (Abbott) Sample: NP, on‐site |
| Target condition and reference standard(s) | PCR |
| Flow and timing | Sample size (cases): 592 (0) |
| Comparative | |
| Notes |
Revollo 2021.
| Patient Sampling | Single‐group (se) |
| Patient characteristics and setting | Not reported; those negative on Ag tests before 12 h of event were included
Reports result at day 1 (baseline) and day 8 (follow‐up assessment) Spain A mass‐gathering indoor event (a live concert), Sala Apolo, Barcelona |
| Index tests | Panbio (Abbott) Sample: NP, on‐site |
| Target condition and reference standard(s) | Transcription‐mediated amplification test (TMA, Procleix Panther, Grifols) and RT‐PCR; cell culture |
| Flow and timing | Sample size (cases): 960 (28 cases with TMA, 2 cases with RT‐PCR) |
| Comparative | |
| Notes |
Reza 2021.
| Patient Sampling | Single‐group (se) |
| Patient characteristics and setting | Mixed cases and contacts Belgium Setting not reported; Department of Laboratory Medicine, Service of Medical Microbiology, Yvoir |
| Index tests | Biospeedia COVID‐19 speed‐Antigen Test and Abbott Panbio Sample: NP, samples collected in UTM tubes, lab |
| Target condition and reference standard(s) | RT‐PCR Results by high (Ct < 25), moderate (Ct ≥ 25 and < 30) and low (Ct ≥ 30) viral load groups |
| Flow and timing | Sample size (cases): 401 (232) |
| Comparative | |
| Notes |
Seynaeve 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Mixed (76% symptomatic) Belgium Patients admitted to the ED and from the testing centre of the CHU Liège |
| Index tests | COVID‐19 Ag Respi‐Strip kit (Coris Bioconcept) and the coronavirus Ag rapid test cassette from Healgen Scientific, LLC Sample: NP, PB saline solution and lab‐based |
| Target condition and reference standard(s) | RT‐PCR Results by Ct 16.7‐37.3 and Ct ≤ 31.00 |
| Flow and timing | Sample size (cases): 63 convenience sample (48) 100 random sample (50) |
| Comparative | |
| Notes |
Shah 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Mixed, symptomatic and asymptomatic
≤ 7 d pso USA Unvaccinated registrants, community testing site in Oshkosh, Wisconsin |
| Index tests | Abbott’s BinaxNOW Sample: AN, supervised and self‐collected |
| Target condition and reference standard(s) | RT‐PCR |
| Flow and timing | Sample size (cases): 2086 (315) |
| Comparative | |
| Notes |
Shaikh 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptomatic children < 20 years (symptoms < 7 d)
< 7 years and 7–20 years USA Primary care practice |
| Index tests | Abbott BinaxNOW Sample: Nasal (mid turbinate) |
| Target condition and reference standard(s) | RT‐PCR |
| Flow and timing | Sample size (cases): 199 (39) |
| Comparative | |
| Notes |
Smith 2021a.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Mixed, symptomatic and asymptomatic
≤ 7 d pso USA, ED of community hospitals in Maryland |
| Index tests | Sofia SARS rapid Ag FIA (Sofia analysers used) Sample: NP, by trained staff |
| Target condition and reference standard(s) | RT‐PCR Results by Ct values |
| Flow and timing | Sample size (cases): 2887 (235) |
| Comparative | Ag testing in the laboratory; turn around time 1.2 h |
| Notes |
Smits 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptomatic (emergency) GP Netherlands |
| Index tests | Panbio Sample: NP |
| Target condition and reference standard(s) | RT‐PCR; ct < 45, ct < 32 |
| Flow and timing | Sample size (cases): 534 (70) |
| Comparative | |
| Notes |
Sood 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Asymptomatic and symptomatic children USA; walk‐up community‐based COVID‐19 testing site, Los Angeles |
| Index tests | BinaxNOW Sample: AN; collected by trained study staff |
| Target condition and reference standard(s) | RT‐PCR Oral fluid specimens for RT‐PCR were self‐collected by participants and observed by trained staff Results by Ct ≤ 25; Ct 25.1‐30; Ct > 30 |
| Flow and timing | Sample size (cases): 774 (226) |
| Comparative | |
| Notes |
Sterbenc 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Includes asymptomatic Slovenia HCW working at the University Clinic of Respiratory and allergic disease |
| Index tests | SARS‐CoV‐2 rapid Ag test; repeat testing
(Roche Diagnostics GmbH, Mannheim, Germany) Sample: NP, collected into UTM |
| Target condition and reference standard(s) | RT‐PCR |
| Flow and timing | Sample size (cases): 36; 191 swabs over 6 testing days (2) |
| Comparative | |
| Notes |
Suliman 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Asymptomatic (88%) and mildly symptomatic USA; free community testing sites in Holyoke, Massachusetts |
| Index tests | Access Bio CareStart COVID‐19 RDT (CareStart) Sample: AN, collected by trained personnel |
| Target condition and reference standard(s) | RT‐PCR By Ct ≥ 30 and Ct < 30 |
| Flow and timing | 666 tests (from 591 participants) ‐ results by sample tested; 52 positive samples |
| Comparative | |
| Notes |
Terpos 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptomatic; patients reporting symptoms for a maximum of 7 d were included Slovenia, Primary Care Health Care institution and General Hospital Jesenice |
| Index tests | COVID‐19 Ag detection kit (colloidal gold), Zhuhai Lituo Biotechnology, China; Includes comparison by sample site Sample: NP, nasal |
| Target condition and reference standard(s) | RT‐PCR By Ct < 25, Ct < 33, Ct < 40 |
| Flow and timing | Sample size (cases): 358 NP (114); 358 nasal (109) |
| Comparative | |
| Notes |
Thakur 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Includes asymptomatic India, COVID‐19‐dedicated hospital (Delhi) |
| Index tests | PathoCatch/ACCUCARE, Lab Care Diagnostics Private Ltd., Sari Gam, India Sample: NP; onsite by a trained technician |
| Target condition and reference standard(s) | RT‐PCR (NP + OP for RT‐PCR) Results by Ct ≤ 25; Ct 25‐29; Ct > 30 |
| Flow and timing | Sample size (cases): 677 (84) |
| Comparative | |
| Notes |
Thell 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Symptomatic
Results by days of symptom onset, type of care (ED vs PHC), type of symptom, gender and by age group Austria, ED and primary health care centres |
| Index tests | SARS‐CoV‐2 Rapid Antigen Test (Roche Diagnostics) Sample: swab type not specified; collected by experienced medical staff |
| Target condition and reference standard(s) | RT‐PCR By Ct > 40, Ct > 30, Ct > 25, Ct > 20 |
| Flow and timing | Sample size (cases): 541 (213) |
| Comparative | |
| Notes |
Trobajo‐Sanmartin 2021.
| Patient Sampling | Single‐group (se + sp) |
| Patient characteristics and setting | Unclear Spain; setting unclear |
| Index tests | CerTest SARS‐CoV‐2 Card Test Sample: NP in UTM; stored at 4 °C for < 24 h until detection |
| Target condition and reference standard(s) | RT‐PCR |
| Flow and timing | Sample size (cases): 240 (80) |
| Comparative | |
| Notes | Performed in the lab |
Uwamino 2021.
| Patient Sampling | Cases only (72 RT‐PCR+ve cases with 190+ve samples (se) |
| Patient characteristics and setting | Symptomatic
By days of symptom onset Japan Hospital, single centre |
| Index tests | Espline SARS‐CoV‐2 RAD kit
Includes comparison by sample site Sample: NP in UTM (collected by trained medical staff) Saliva (self‐collected) |
| Target condition and reference standard(s) | RT‐PCR; viral culture |
| Flow and timing | Sample size (cases): 190 samples (72) NP 117; saliva 73 |
| Comparative | |
| Notes | Performed in the lab |
Van Honacker 2021.
| Patient Sampling | Single‐group (se + sp)‐ implementation part of the study |
| Patient characteristics and setting | Symptomatic, hospital admission Belgium ED, Hospital |
| Index tests | SD Biosensor Sample: NP (Amies transport medium) |
| Target condition and reference standard(s) | RT‐PCR (in house) By Ct values < 20, 20‐ < 26, 26‐ < 30, 30 to < 36 and > 36 |
| Flow and timing | Sample size (cases): 4195 (369) |
| Comparative | |
| Notes | Performed in the lab Ag test on implementation part included; Ag tests on validation excluded |
Wagenhäuser 2021.
| Patient Sampling | Single‐group (se + sp)‐ implementation part of the study |
| Patient characteristics and setting | Any patient in the hospital; symptomatic and asymptomatic Results by symptom status not separated out for each test Germany; tertiary care hospital |
| Index tests | NADAL COVID‐19 Ag Test
Panbio
MEDsan SARS‐Cov‐2 Antigen Rapid Test Sample: OP; by trained medical staff on site |
| Target condition and reference standard(s) | RT‐PCR By viral load ≥ 10 (8) copies per mL, 10 (6‐10 (8), 10 (4‐10( 6), < 10 (4) |
| Flow and timing | 5056 samples, 101 positive samples |
| Comparative | |
| Notes |
Yin 2021.
| Patient Sampling | Single‐group (se + sp)‐ implementation part of the study |
| Patient characteristics and setting | Symptomatic outpatients Belgium; primary care or ED |
| Index tests | Panbio COVID‐19 Ag Rapid Test Device, BD Veritor SARS‐CoV‐2, COVID‐19 Ag Respi‐Strip (Coris BioConcept,
Belgium) and SARS‐CoV‐2 Rapid Antigen Test (SD Biosensor) Sample: NP, onsite |
| Target condition and reference standard(s) | RT‐PCR |
| Flow and timing | Sample size (cases): 494 (209) |
| Comparative | |
| Notes |
Young 2021a.
| Patient Sampling | Cluster‐RCT: schools randomly assigned (1:1) to either a policy of offering contacts daily Ag testing over 7 d to allow continued school attendance (intervention group) or to follow usual policy of isolation of contacts for 10 d (control group |
| Patient characteristics and setting | Asymptomatic student or staff contacts |
| Index tests | SARS‐CoV‐2 Ag LFD (Orient Gene, Huzhou, China) |
| Target condition and reference standard(s) | Routine SARS‐CoV‐2 PCR tests done outside of the study; dedicated study PCR testing in both study groups on day 2 and 7 of the testing or isolation period |
| Flow and timing | |
| Comparative | |
| Notes |
Ag: antigen; AN: anterior nasal; Ct: cycle threshold; ED: emergency department; ELISA: enzyme‐linked immunosorbent assay; HCW: healthcare worker; IPD: individual patient data; LFA: lateral flow assay; LFD: lateral flow device; NP: nasopharyngeal; OP: oropharyngeal; PCR: polymerase chain reaction; pso: post‐symptom onset; RCT: randomized controlled trial; RDT: rapid diagnostic test; RT‐PCR: reverse‐transcription polymerase chain reaction; Se: sensitivity; sp: specificity; TMA: transcription‐mediated amplification
Differences between protocol and review
As the evidence base evolves over the course of the pandemic, we have made some adjustments to our original approach with the following changes between earlier versions of the review and this second update:
Clarification regarding inclusion criteria: restricted to studies evaluating commercially produced rapid antigen tests; a separate review update covering rapid molecular tests is planned
Search sources included in the protocol and the previous versions of this review, the Cochrane COVID‐19 Study Register and the CDC Database of COVID‐19 Research Articles, were not included in this version as the single source from the University of Bern living search database proved more efficient to process as it did not involve manual effort to deduplicate.
We planned to check the following websites for eligible index tests, however these did not prove to be very accessible or easy to use and, after initial review, were not further considered:
National Institute for Health Research (NIHR) Innovation Observatory (www.io.nihr.ac.uk/)
Meta‐evidence (meta-evidence.co.uk/the-role-of-evidence-synthesis-in-covid19/)
-
Electronic sources searched for previous review updates were dropped, including:
Cochrane COVID‐19 Study Register; not searched since 28 March 2020 because of lack of coverage of preprint literature
the Evidence for Policy and Practice Information and Co‐ordinating Centre (EPPI‐Centre) 'COVID‐19: Living map of the evidence' (eppi.ioe.ac.uk/COVID19_MAP/covid_map_v4.html); the Norwegian Institute of Public Health 'NIPH systematic and living map on COVID‐19 evidence' (www.nornesk.no/forskningskart/NIPH_diagnosisMap.html); not searched since 15 Nov 2020 because of the move to a review‐specific classifier approach and availability of more directly relevant sources of eligible primary studies as documented in Searching other resources.
We intended for two authors to independently perform data extraction, however one review author extracted study characteristics, and a second author checked them. Contingency table data were extracted independently by two review authors as planned.
We planned to evaluate the effect of additional sources of heterogeneity, including reference standard. However, additional formal investigations using meta‐regression were not possible because of lack of variability across the studies in these features.
We planned to conduct a sensitivity analysis excluding studies that are solely published as preprints. We have inadequate study numbers to allow this at present but will reconsider for the next update.
Contributions of authors
JDi was the contact person with the editorial base. JDi co‐ordinated contributions from the co‐authors and wrote the final draft of the review. JDi, PS, SvW, MT screened papers against eligibility criteria. RS conducted the literature searches. JDi, PS, SvW, NN, CD, JDo, JB, MT appraised the quality of papers. JDi, PS, SvW, NN, CD, JDo, JB, MT extracted data for the review and sought additional information about papers. JDi, PS entered data into Review Manager 2020. JDi, SB, JJD, YT, CD analysed and interpreted data. JDi, JJD, YT, CD, STP, RS, ML, MM, LH, AVB, DE, SD, JC, JV worked on the methods sections and commented on the draft review. JDi, CD, YT responded to the comments of the referees. JDi is the guarantor of the update.
Sources of support
Internal sources
Liverpool School of Tropical Medicine, UK
University of Birmingham, UK
External sources
-
Foreign, Commonwealth, and Development Office (FCDO), UK
Project number: 300342‐104
National Institute for Health Research (NIHR), UK
NIHR Birmingham Biomedical Research Centre at the University Hospitals Birmingham NHS Foundation Trust and the University of Birmingham, UK
Declarations of interest
Jonathan J Deeks: JD has published or been quoted in opinion pieces in scientific publications, and in the mainstream and social media related to diagnostic testing. JD was the statistician on the Birmingham evaluation of the Innova test which is mentioned in the discussion of the paper. There was no funding for this evaluation of the Innova test. JD is a member of the Royal Statistical Society (RSS) COVID‐19 taskforce steering group, and co‐chair of the RSS Diagnostic Test Advisory Group. He is a consultant adviser to the World Health Organization (WHO) Essential Diagnostic List. JD receives payment from the BMJ as their Chief Statistical advisor.
Jacqueline Dinnes: none known
Yemisi Takwoingi: none known
Clare Davenport: none known
Mariska MG Leeflang: none known
René Spijker: none known
Lotty Hooft: none known
Ann Van den Bruel: none known
Devy Emperador: is employed by FIND with funding from DFID and KFW. FIND is a global non‐for profit product development partnership and WHO Diagnostic Collaboration Centre. It is FIND’s role to accelerate access to high‐quality diagnostic tools for low‐resource settings and this is achieved by supporting both research and development and access activities for a wide range of diseases, including COVID‐19. FIND has several clinical research projects to evaluate multiple new diagnostic tests against published Target Product Profiles that have been defined through consensus processes. These studies are for diagnostic products developed by private sector companies who provide access to know‐how, equipment/reagents, and contribute through unrestricted donations as per FIND policy and external SAC review.
Sabine Dittrich: is employed by FIND with funding from DFID and Australian Aid. FIND is a global non‐for profit product development partnership and WHO Diagnostic Collaboration Centre. It is FIND’s role to accelerate access to high‐quality diagnostic tools for low‐resource settings and this is achieved by supporting both research and development and access activities for a wide range of diseases, including COVID‐19. FIND has several clinical research projects to evaluate multiple new diagnostic tests against published Target Product Profiles that have been defined through consensus processes. These studies are for diagnostic products developed by private sector companies who provide access to know‐how, equipment/reagents, and contribute through unrestricted donations as per FIND policy and external SAC review.
Sian Taylor‐Phillips: none known
Sarah Berhane: none known
Jane Cunningham: none known
New search for studies and content updated (conclusions changed)
References
References to studies included in this review
Abdelrazik 2021 {published data only}
- Abdelrazik AM, Elshafie SM, Abdelaziz HM. Potential respiratory specimens in low-resource settings in Egypt for symptomatic patients and high-risk contacts. Laboratory Medicine 2021;52(2):e46-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Abdulrahman 2020 {published data only}
- Abdulrahman A, Mustafa F, AlAwadhi AI, Alansari Q, AlAlawi B, AlQahtani M. Comparison of SARS-COV-2 nasal antigen test to nasopharyngeal RT-PCR in mildly symptomatic patients. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Agullo 2021 [A] {published data only}
- Agullo V, Fernandez-Gonzalez M, Ortiz de la Tabla V, Gonzalo-Jimenez N, Garcia JA, Masia M, et al. Evaluation of the rapid antigen test Panbio COVID-19 in saliva and nasal swabs in a population-based point-of-care study. Journal of Infection 2021;82(5):186-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Agullo 2021 [B] {published data only}
- Agullo V, Fernandez-Gonzalez M, Ortiz de la Tabla V, Gonzalo-Jimenez N, Garcia JA, Masia M, et al. Evaluation of the rapid antigen test Panbio COVID-19 in saliva and nasal swabs in a population-based point-of-care study. Journal of Infection 2021;82(5):186-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Agullo 2021 [C] {published data only}
- Agullo V, Fernandez-Gonzalez M, Ortiz de la Tabla V, Gonzalo-Jimenez N, Garcia JA, Masia M, et al. Evaluation of the rapid antigen test Panbio COVID-19 in saliva and nasal swabs in a population-based point-of-care study. Journal of Infection 2021;82(5):186-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Akingba 2021 {published data only}
- Akingba OL, Sprong K, Hardie DR. Field performance evaluation of the Panbio rapid SARS-CoV-2 antigen assay in an epidemic driven by 501Y.v2 (lineage B.1.351) in the Eastern Cape, South Africa. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Albert 2020 {published data only}
- Albert E, Torres I, Bueno F, Huntley D, Molla E, Fernández-Fuentes M, et al. Field evaluation of a rapid antigen test (Panbio™ COVID-19 Ag Rapid Test Device) for the diagnosis of COVID-19 in primary healthcare centers. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert E, Torres I, Bueno F, Huntley D, Molla E, Fernández-Fuentes MA, et al. Field evaluation of a rapid antigen test (Panbio™ COVID-19 Ag Rapid Test Device) for COVID-19 diagnosis in primary healthcare centers. Clinical Microbiology and Infection 2020 Nov 13 [Epub ahead of print]. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Alemany 2021 {published data only}
- Alemany A, Baro B, Ouchi D, Rodó P, Ubals M, Corbacho-Monné M, et al. Analytical and clinical performance of the Panbio COVID-19 antigen-detecting rapid diagnostic test. Journal of Infection 2021;82(5):186-230. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemany A, Baro B, Ouchi D, Ubals M, Corbacho-Monné M, Vergara-Alert J, et al. Analytical and clinical performance of the Panbio COVID-19 antigen-detecting rapid diagnostic test. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Aoki 2021 {published data only}
- Aoki K, Nagasawa T, Ishii Y, Yagi S, Kashiwagi K, Miyazaki T, et al. Evaluation of clinical utility of novel coronavirus antigen detection reagent, Espline(R) SARS-CoV-2. Journal of Infection and Chemotherapy 2021;27(2):319-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Baro 2021 [A] {published data only}
- Baro B, Rodo P, Ouchi D, Bordoy AE, Saya Amaro EN, Salsench SV, et al. Performance characteristics of five antigen-detecting rapid diagnostic test (Ag-RDT) for SARS-CoV-2 asymptomatic infection: a head-to-head benchmark comparison. Journal of Infection 2021;82(6):269-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Baro 2021 [B] {published data only}
- Baro B, Rodo P, Ouchi D, Bordoy AE, Saya Amaro EN, Salsench SV, et al. Performance characteristics of five antigen-detecting rapid diagnostic test (Ag-RDT) for SARS-CoV-2 asymptomatic infection: a head-to-head benchmark comparison. Journal of Infection 2021;82(6):269-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Baro 2021 [C] {published data only}
- Baro B, Rodo P, Ouchi D, Bordoy AE, Saya Amaro EN, Salsench SV, et al. Performance characteristics of five antigen-detecting rapid diagnostic test (Ag-RDT) for SARS-CoV-2 asymptomatic infection: a head-to-head benchmark comparison. Journal of Infection 2021;82(6):269-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Baro 2021 [D] {published data only}
- Baro B, Rodo P, Ouchi D, Bordoy AE, Saya Amaro EN, Salsench SV, et al. Performance characteristics of five antigen-detecting rapid diagnostic test (Ag-RDT) for SARS-CoV-2 asymptomatic infection: a head-to-head benchmark comparison. Journal of Infection 2021;82(6):269-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Baro 2021 [E] {published data only}
- Baro B, Rodo P, Ouchi D, Bordoy AE, Saya Amaro EN, Salsench SV, et al. Performance characteristics of five antigen-detecting rapid diagnostic test (Ag-RDT) for SARS-CoV-2 asymptomatic infection: a head-to-head benchmark comparison. Journal of Infection 2021;82(6):269-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Basso 2021 [A] {published data only}
- Basso D, Aita A, Padoan A, Cosma C, Navaglia F, Moz S, et al. Salivary SARS-CoV-2 antigen rapid detection: a prospective cohort study. Clinica Chimica Acta; International Journal of Clinical Chemistry 2021;517:54-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso D, Aita A, Padoan A, Cosma C, Navaglia F, Moz S, et al. Salivary SARS-CoV-2 antigen rapid detection: a prospective cohort study. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Basso 2021 [B] {published data only}
- Basso D, Aita A, Padoan A, Cosma C, Navaglia F, Moz S, et al. Salivary SARS-CoV-2 antigen rapid detection: A prospective cohort study. Clinica Chimica Acta: The International Journal of Clinical Chemistry 2021;517:54-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Basso 2021 [C] {published data only}
- Basso D, Aita A, Padoan A, Cosma C, Navaglia F, Moz S, et al. Salivary SARS-CoV-2 antigen rapid detection: A prospective cohort study. Clinica Chimica Acta: The International Journal of Clinical Chemistry 2021;517:54-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Beck 2021 {published data only}
- Beck ET, Paar W, Fojut L, Serwe J, Jahnke RR. Comparison of the Quidel Sofia SARS FIA test to the Hologic Aptima SARS-CoV-2 TMA test for diagnosis of COVID-19 in symptomatic outpatients. Journal of Clinical Microbiology 2021;59(2). [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Billaud 2020 {published data only}
- Billaud G, Gaymard A, Lina B, Laboratoire de Virologie des HCL CNR des virus des infections respiratoires. Evaluation du Test Antigénique ABBOTT SARS-COV2 ABBOT. Lyon, France: SFM (French Society of Microbiology), 2020. [Google Scholar]
Blairon 2020 {published data only}
- Blairon L, Wilmet A, Beukinga I, Tre-Hardy M. Implementation of rapid SARS-CoV-2 antigenic testing in a laboratory without access to molecular methods: experiences of a general hospital. Journal of Clinical Virology 2020;129. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Bulilete 2021 {published data only}
- Bulilete O, Lorente P, Leiva A, Carandell E, Oliver A, Rojo E, et al. Evaluation of the Panbio rapid antigen test for SARS-CoV-2 in primary health care centers and test sites. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
- Bulilete O, Lorente P, Leiva A, Carandell E, Oliver A, Rojo E, et al. Panbio rapid antigen test for SARS-CoV-2 has acceptable accuracy in symptomatic patients in primary health care. Journal of Infection 2021;82(3):391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Caruana 2021 [A] {published data only}
- Caruana G, Croxatto A, Kampouri E, Kritikos A, Opota O, Foerster M, et al. Implementing SARS-CoV-2 rapid antigen testing in the emergency ward of a Swiss university hospital: the INCREASE study. Microorganisms 2021;9(4):798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Caruana 2021 [B] {published data only}
- Caruana G, Croxatto A, Kampouri E, Kritikos A, Opota O, Foerster M, et al. Implementing SARS-CoV-2 rapid antigen testing in the emergency ward of a Swiss university hospital: the INCREASE study. Microorganisms 2021;9(4):798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Caruana 2021 [C] {published data only}
- Caruana G, Croxatto A, Kampouri E, Kritikos A, Opota O, Foerster M, et al. Implementing SARS-CoV-2 rapid antigen testing in the emergency ward of a Swiss university hospital: the INCREASE study. Microorganisms 2021;9(4):798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Caruana 2021 [D] {published data only}
- Caruana G, Croxatto A, Kampouri E, Kritikos A, Opota O, Foerster M, et al. Implementing SARS-CoV-2 rapid antigen testing in the emergency ward of a Swiss university hospital: the INCREASE study. Microorganisms 2021;9(4):798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Cerutti 2020 {published data only}
- Cerutti F, Burdino E, Milia MG, Allice T, Gregori G, Bruzzone B, et al. Urgent need of rapid tests for SARS CoV-2 antigen detection: evaluation of the SD-Biosensor antigen test for SARS-CoV-2. Journal of Clinical Virology 2020;132:104654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Chaimayo 2020 {published data only}
- Chaimayo C, Kaewnaphan B, Tanlieng N, Athipanyasilp N, Sirijatuphat R, Chayakulkeeree M, et al. Rapid SARS-CoV-2 antigen detection assay in comparison with real-time RT-PCR assay for laboratory diagnosis of COVID-19 in Thailand. Virology Journal 2020;17(1):177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Ciotti 2021 {published data only}
- Ciotti M, Maurici M, Pieri M, Andreoni M, Bernardini S. Performance of a rapid antigen test in the diagnosis of SARS-CoV-2 infection. Journal of Medical Virology 2021;93(5):2988-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Courtellemont 2021 {published data only}
- Courtellemont L, Guinard J, Guillaume C, Giaché S, Rzepecki V, Seve A, et al. High performance of a novel antigen detection test on nasopharyngeal specimens for diagnosing SARS-CoV-2 infection. Journal of Medical Virology 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtellemont L, Guinard J, Guillaume C, Giaché S, Rzepecki V, Seve A, et al. Real-life performance of a novel antigen detection test on nasopharyngeal specimens for SARS-CoV-2 infection diagnosis: a prospective study. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Del Vecchio 2021 {published data only}
- Del Vecchio C, Brancaccio G, Brazzale AR, Lavezzo E, Onelia F, Franchin E, et al. Emergence of N antigen SARS-CoV-2 genetic variants escaping detection of antigenic tests. medRxiv [Preprint] 2021. [DOI: ] [Google Scholar]
Dominguez Fernandez 2021 {published data only}
- Dominguez Fernandez M, Pena Rodriguez MF, Lamelo Alfonsin F, Bou Arevalo G. Experience with Panbio rapid antigens test device for the detection of SARS-CoV-2 in nursing homes. Enfermedades Infecciosas y Microbiología Clínica (English Edition) 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Drain 2021(a) {published data only}
- Drain PK, Ampajwala M, Chappel C, Gvozden AB, Hoppers M, Wang M, et al. A rapid, high-sensitivity SARS-CoV-2 nucleocapsid immunoassay to aid diagnosis of acute COVID-19 at the point of care: a clinical performance study. Infectious Diseases and Therapy 2021;10(2):753-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drain PK, Ampajwala M, Chappel C, Gvozden AB, Hoppers M, Wang M, et al. A rapid, high-sensitivity SARS-CoV-2 nucleocapsid immunoassay to aid diagnosis of acute COVID-19 at the point of care. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Drain 2021(b) {published data only}
- Drain PK, Ampajwala M, Chappel C, Gvozden AB, Hoppers M, Wang M, et al. A rapid, high-sensitivity SARS-CoV-2 nucleocapsid immunoassay to aid diagnosis of acute COVID-19 at the point of care: a clinical performance study. Infectious Diseases and Therapy 2021;10(2):753-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Drevinek 2020 [A] {published data only}
- Drevinek P, Hurych J, Kepka Z, Briksi A, Kulich M, Zajac M, et al. The sensitivity of SARS-CoV-2 antigen tests in the view of large-scale testing. medRxiv [Preprint] 2020. [DOI: ] [PubMed] [Google Scholar]
Drevinek 2020 [B] {published data only}
- Drevinek P, Hurych J, Kepka Z, Briksi A, Kulich M, Zajac M, et al. The sensitivity of SARS-CoV-2 antigen tests in the view of large-scale testing. medRxiv [Preprint] 2020. [DOI: ] [PubMed] [Google Scholar]
Faico‐Filho 2021 {published data only}
- Faíco-Filho KS, Finamor Júnior FE, Vinícius Leão Moreira L, Lins PRG, Justo AFO, Bellei N. Evaluation of the Panbio™ COVID-19 Ag rapid test at an emergency room in a hospital in São Paulo, Brazil. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Favresse 2021 [A] {published data only}
- Favresse J, Gillot C, Oliveira M, Cadrobbi J, Elsen M, Eucher C, et al. Head-to-head comparison of rapid and automated antigen detection tests for the diagnosis of SARS-CoV-2 infection. Journal of Clinical Medicine 2021;10(2). [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Favresse 2021 [B] {published data only}
- Favresse J, Gillot C, Oliveira M, Cadrobbi J, Elsen M, Eucher C, et al. Head-to-head comparison of rapid and automated antigen detection tests for the diagnosis of SARS-CoV-2 infection. Journal of Clinical Medicine 2021;10(2):10.3390/jcm10020265. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Favresse 2021 [C] {published data only}
- Favresse J, Gillot C, Oliveira M, Cadrobbi J, Elsen M, Eucher C, et al. Head-to-head comparison of rapid and automated antigen detection tests for the diagnosis of SARS-CoV-2 infection. Journal of Clinical Medicine 2021;10(2):10.3390/jcm10020265. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Favresse 2021 [D] {published data only}
- Favresse J, Gillot C, Oliveira M, Cadrobbi J, Elsen M, Eucher C et al. Head-to-head comparison of rapid and automated antigen detection tests for the diagnosis of SARS-CoV-2 infection. Journal of Clinical Medicine 2021;10(2):10.3390/jcm10020265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Fenollar 2020(a) {published data only}
- Fenollar F, Bouam A, Ballouche M, Fuster L, Prudent E, Colson P, et al. Evaluation of the Panbio Covid-19 rapid antigen detection test device for the screening of patients with COVID-19. Journal of Clinical Microbiology 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Fenollar 2020(b) {published data only}
- Fenollar F, Bouam A, Ballouche M, Fuster L, Prudent E, Colson P, et al. Evaluation of the Panbio Covid-19 rapid antigen detection test device for the screening of patients with COVID-19. Journal of Clinical Microbiology 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Ferguson 2021 {published data only}
- Ferguson J, Dunn S, Best A, Mirza J, Percival B, Mayhew M, et al. Validation testing to determine the effectiveness of lateral flow testing for asymptomatic SARS-CoV-2 detection in low prevalence settings. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson J, Dunn S, Best A, Mirza J, Percival B, Mayhew M, et al. Validation testing to determine the sensitivity of lateral flow testing for asymptomatic SARS-CoV-2 detection in low prevalence settings: Testing frequency and public health messaging is key. PLoS Biology 2021;19(4). [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Filgueiras 2021 {published data only}
- Filgueiras PS, Corsini CA, Almeida NBF, Assis JV, Pedrosa MLC, Oliveira AK, et al. COVID-19 Rapid Antigen Test at hospital admission associated to the knowledge of individual risk factors allow overcoming the difficulty of managing suspected patients in hospitals. medRxiv [Preprint] 2021. [DOI: ] [Google Scholar]
FIND 2020a {published data only}
- Foundation for Innovative New Diagnostics (FIND). Evaluation of Bionote, Inc. NowCheck COVID-19 Ag Test - External Report. Switzerland: FIND, 2020. [Google Scholar]
FIND 2020b (CH) {published data only}
- Berger A, Ngo Nsoga MT, Perez R, F J, Abi A, et al. Diagnostic accuracy of two commercial SARS-CoV-2 Antigen-detecting rapid tests at the point of care in community-based testing centers. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A, Ngo Nsoga MT, Perez-Rodriguez FJ, Aad YA, Sattonnet-Roche P, Gayet-Ageron A, et al. Diagnostic accuracy of two commercial SARS-CoV-2 antigen-detecting rapid tests at the point of care in community-based testing centers. PLoS One 2021;16(3):e0248921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foundation for Innovative New Diagnostics (FIND). Evaluation of Abbott Panbio COVID-19 Ag Rapid Test Device - External Report. Switzerland: FIND, 2020. [Google Scholar]
FIND 2020b (DE) {published data only}
- Foundation for Innovative New Diagnostics (FIND). Evaluation of Abbott Panbio COVID-19 Ag Rapid Test Device - External Report. Switzerland: FIND, 2020. [Google Scholar]
- Krüger LJ, Gaeddert M, Tobian F, Lainati F, Gottschalk C, Klein JAF et al. The Abbott PanBio WHO emergency use listed, rapid, antigen-detecting point-of-care diagnostic test for SARS-CoV-2-Evaluation of the accuracy and ease-of-use. PLoS One 2021;16(5):e0247918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger LJ, Gaeddert M, Tobian F, Lainati F, Klein JAF, Schnitzler P et al. Evaluation of the accuracy and ease-of-use of Abbott PanBio - A WHO emergency use listed, rapid, antigen-detecting point-of-care diagnostic test for SARS-CoV-2. medRxiv [Preprint] 2020:2020.10.01.20203836. [DOI] [PMC free article] [PubMed] [Google Scholar]
FIND 2020c (BR) {published data only}
- Foundation for Innovative New Diagnostics (FIND). Evaluation of SD Biosensor, Inc. STANDARD Q COVID-19 Ag Test - External Report. Switzerland: FIND, 2020. [Google Scholar]
FIND 2020c (CH) {published data only}
- Berger A, Ngo Nsoga MT, Perez-Rodriguez FJ, Aad YA, Sattonnet-Roche P, Gayet-Ageron A, et al. Diagnostic accuracy of two commercial SARS-CoV-2 antigen-detecting rapid tests at the point of care in community-based testing centers. PLoS One 2021;16(3):e0248921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A, Ngo Nsoga MT, Perez-Rodriguez FJ, Abi A, Sattonnet-Roche P, et al. Diagnostic accuracy of two commercial SARS-CoV-2 Antigen-detecting rapid tests at the point of care in community-based testing centers. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foundation for Innovative New Diagnostics (FIND). Evaluation of SD Biosensor, Inc. STANDARD Q COVID-19 Ag Test - External Report. Switzerland: FIND, 2020. [Google Scholar]
FIND 2020c (DE) {published data only}
- Foundation for Innovative New Diagnostics (FIND). FIND Evaluation of SD Biosensor, Inc.STANDARD Q COVID-19 Ag Test External Report. Vol. version 2.1. Switzerland: FIND, 2020. [Google Scholar]
- Krüger LJ, Gaeddert M, Köppel L, Brümmer LE, Gottschalk C, Miranda IB, et al. Evaluation of the accuracy, ease of use and limit of detection of novel, rapid, antigen-detecting point-of-care diagnostics for SARS-CoV-2. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
FIND 2020d (BR) {published data only}
- Foundation for Innovative New Diagnostics (FIND). Evaluation of SD Biosensor, Inc. STANDARD F COVID-19 Ag FIA - External Report. Switzerland: FIND, 2020. [Google Scholar]
FIND 2020d (DE) {published data only}
- Foundation for Innovative New Diagnostics (FIND). Evaluation of SD Biosensor, Inc. STANDARD F COVID-19 Ag FIA - External Report. Switzerland: FIND, 2020. [Google Scholar]
FIND 2020e (BR) {published data only}
- Foundation for Innovative New Diagnostics (FIND). Evaluation of RapiGEN Inc. BIOCREDIT COVID-19 Ag - External Report. Switzerland: FIND, 2020. [Google Scholar]
FIND 2020e (DE) {published data only}
- Foundation for Innovative New Diagnostics (FIND). Evaluation of RapiGEN Inc. BIOCREDIT COVID-19 Ag - External Report. Switzerland: FIND, 2020. [Google Scholar]
FIND 2020f {published data only}
- Foundation for Innovative New Diagnostics (FIND). FIND Evaluation of Coris BioConcept COVID-19 Ag Respi-Strip - External Report. Vol. v1.2. Switzerland: FIND, 2020. [Google Scholar]
- Krüger LJ, Gaeddert M, Köppel L, Brümmer LE, Gottschalk C, Miranda IB, et al. Evaluation of the accuracy, ease of use and limit of detection of novel, rapid, antigen-detecting point-of-care diagnostics for SARS-CoV-2. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
FIND 2021a [A] {published data only}
- FIND. FIND Evaluation of Bionote, Inc. NowCheck COVID-19 Ag TestTest, nasal swab - External Report. Switzerland: FIND, 2021. [Google Scholar]
FIND 2021a [B] {published data only}
- FIND. FIND Evaluation of Bionote, Inc. NowCheck COVID-19 Ag TestTest, nasal swab - External Report. Switzerland: FIND, 2021. [Google Scholar]
FIND 2021b [A] {published data only}
- FIND. FIND Evaluation of Abbott Panbio COVID-19 Ag Rapid Test Device [NASAL] - External Report. Switzerland: FIND, 2021. [Google Scholar]
FIND 2021b [B] {published data only}
- FIND. FIND Evaluation of Abbott Panbio COVID-19 Ag Rapid Test Device [NASAL] - External Report. Switzerland: FIND, 2021. [Google Scholar]
FIND 2021c (BR) [A] {published data only}
- FIND. FIND Evaluation of SD Biosensor, Inc. STANDARD Q COVID-19 Ag Test, nasal swab - External Report. Switzerland: FIND, 2021. [Google Scholar]
FIND 2021c (BR) [B] {published data only}
- FIND. FIND Evaluation of SD Biosensor, Inc. STANDARD Q COVID-19 Ag Test, nasal swab - External Report. Switzerland: FIND, 2021. [Google Scholar]
FIND 2021c (DE) [A] {published data only}
- FIND. FIND Evaluation of SD Biosensor, Inc. STANDARD Q COVID-19 Ag Test, nasal swab - External Report. Switzerland: FIND, 2021c. [Google Scholar]
- Lindner AK, Nikolai O, Rohardt C, Burock S, Hulso C, Bolke A, et al. Head-to-head comparison of SARS-CoV-2 antigen-detecting rapid test with professional-collected nasal versus nasopharyngeal swab. European Respiratory Journal 2021;57(5). [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
FIND 2021c (DE) [B] {published data only}
- FIND. FIND Evaluation of SD Biosensor, Inc. STANDARD Q COVID-19 Ag Test, nasal swab - External Report. Switzerland: FIND, 2021. [Google Scholar]
- Lindner AK, Nikolai O, Rohardt C, Burock S, Hulso C, Bolke A, et al. Head-to-head comparison of SARS-CoV-2 antigen-detecting rapid test with professional-collected nasal versus nasopharyngeal swab. European Respiratory Journal 2021;57(5). [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
FIND 2021d {published data only}
- FIND. FIND Evaluation of Fujirebio Inc. Espline SARS-CoV-2 - External Report. Switzerland: FIND, 2021. [Google Scholar]
FIND 2021e {published data only}
- FIND. FIND Evaluation of Joysbio (Tianjin) Biotechnology Co., Ltd. SARS-CoV-2 Antigen Rapid Test Kit (Colloidal Gold) - External Report. Switzerland: FIND, 2021. [Google Scholar]
FIND 2021f {published data only}
- FIND. FIND Evaluation of Mologic Ltd, COVID 19 RAPID ANTIGEN TEST - External Report. Switzerland: FIND, 2021. [Google Scholar]
FIND 2021g {published data only}
- FIND. FIND Evaluation of NADAL COVID-19 Ag Rapid Test - External Report. Switzerland: FIND, 2021. [Google Scholar]
FIND 2021h {published data only}
- FIND. FIND Evaluation of Boditech Medical, Inc. iChroma COVID-19 Ag Test - External Report. Switzerland: FIND, 2021. [Google Scholar]
FIND 2021i {published data only}
- FIND. FIND Evaluation of Guangzhou Wondfo Biotech Co., Ltd Wondfo 2019-nCoV Antigen Test (Lateral Flow Method) Public Report. Switzerland: FIND, 2021. [Google Scholar]
FIND 2021j {published data only}
- Foundation for Innovative New Diagnostics (FIND). FIND Evaluation of Shenzhen Bioeasy Biotechnology Co. Ltd.2019-nCoV Ag Rapid Test Kit (Fluorescence) External Report. Vol. Version 1.0. Switzerland: FIND, 2021. [Google Scholar]
- Krüger LJ, Gaeddert M, Köppel L, Brümmer LE, Gottschalk C, Miranda IB, et al. Evaluation of the accuracy, ease of use and limit of detection of novel, rapid, antigen-detecting point-of-care diagnostics for SARS-CoV-2. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Fourati 2020 [A] {published data only}
- Fourati S, Audureau E, Chevaliez S, Pawlotsky JM. Évaluation de la performance diagnostique des tests rapides d’orientation diagnostique antigéniques COVID-19. France: AP-HP Hopitaux universitaires Henri-Mondor, 2020. [Google Scholar]
Fourati 2020 [B] {published data only}
- Fourati S, Audureau E, Chevaliez S, Pawlotsky JM. Évaluation de la performance diagnostique des tests rapides d’orientation diagnostique antigéniques COVID-19. France: AP-HP Hopitaux universitaires Henri-Mondor, 2020. [Google Scholar]
Fourati 2020 [C] {published data only}
- Fourati S, Audureau E, Chevaliez S, Pawlotsky JM. Évaluation de la performance diagnostique des tests rapides d’orientation diagnostique antigéniques COVID-19. France: AP-HP Hopitaux universitaires Henri-Mondor, 2020. [Google Scholar]
Fourati 2020 [D] {published data only}
- Fourati S, Audureau E, Chevaliez S, Pawlotsky JM. Évaluation de la performance diagnostique des tests rapides d’orientation diagnostique antigéniques COVID-19. France: AP-HP Hopitaux universitaires Henri-Mondor, 2020. [Google Scholar]
Fourati 2020 [E] {published data only}
- Fourati S, Audureau E, Chevaliez S, Pawlotsky JM. Évaluation de la performance diagnostique des tests rapides d’orientation diagnostique antigéniques COVID-19. France: AP-HP Hopitaux universitaires Henri-Mondor, 2020. [Google Scholar]
Garcia‐Finana 2021 {published data only}
- Garcia-Finana M, Hughes DM, Cheyne CP, Burnside G, Stockbridge M, Fowler TA, et al. Performance of the Innova SARS-CoV-2 antigen rapid lateral flow test in the Liverpool asymptomatic testing pilot: population based cohort study. BMJ 2021;374:n1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
Gomez 2021(a) {published data only}
- Gomez Marti JL, Gribschaw J, McCullough M, Mallon A, Acero J, Kinzler A, et al. Differences in detected viral loads guide use of SARS-CoV-2 antigen-detection assays towards symptomatic college students and children. medRxiv [Preprint] 2021. [DOI: ] [Google Scholar]
Gomez 2021(b) {published data only}
- Gomez Marti JL, Gribschaw J, McCullough M, Mallon A, Acero J, Kinzler A, et al. Differences in detected viral loads guide use of SARS-CoV-2 antigen-detection assays towards symptomatic college students and children. medRxiv [Preprint] 2021. [DOI: ] [Google Scholar]
Gonzalez‐Donapetry 2021 {published data only}
- Gonzalez-Donapetry P, Garcia-Clemente P, Bloise I, Garcia-Sanchez C, Sanchez Castellano MA, Romero MP, et al. Think of the children: evaluation of SARS-CoV-2 rapid antigen test in pediatric population. Pediatric Infectious Disease Journal 2021;40(5):385-8. [DOI] [PubMed] [Google Scholar]
Gremmels 2021(a) {published data only}
- Gremmels H, Winkel BM, Schuurman R, Rosingh A, Rigter NA, Rodriguez O, et al. Real-life validation of the Panbio COVID-19 Antigen Rapid Test (Abbott) in community-dwelling subjects with symptoms of potential SARS-CoV-2 infection. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremmels H, Winkel BM, Schuurman R, Rosingh A, Rigter NA, Rodriguez O, et al. Real-life validation of the PanbioTM COVID-19 antigen rapid test (Abbott) in community-dwelling subjects with symptoms of potential SARS-CoV-2 infection. EClinicalMedicine 2021;31:100677. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Gremmels 2021(b) {published data only}
- Gremmels H, Winkel BM, Schuurman R, Rosingh A, Rigter NA, Rodriguez O, et al. Real-life validation of the PanbioTM COVID-19 antigen rapid test (Abbott) in community-dwelling subjects with symptoms of potential SARS-CoV-2 infection. EClinicalMedicine 2020;31:100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremmels H, Winkel BM, Schuurman R, Rosingh A, Rigter NA, Rodriguez O, et al. Real-life validation of the PanbioTM COVID-19 antigen rapid test (Abbott) in community-dwelling subjects with symptoms of potential SARS-CoV-2 infection. medRxiv [Preprint] 2020:10.1101/2020.10.16.20214189. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Gupta 2020 {published data only}
- Gupta A, Khurana S, Das R, Srigyan D, Singh A, Mittal A, et al. Rapid chromatographic immunoassay-based evaluation of COVID-19: a cross-sectional, diagnostic test accuracy study & its implications for COVID-19 management in India. Indian Journal of Medical Research 2020 Oct 31 [Epub ahead of print]:10.4103/ijmr.IJMR_3305_20. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Halfon 2021 {published data only}
- Halfon P, Penaranda G, Khiri H, Garcia V, Drouet H, Philibert P, et al. An optimized stepwise algorithm combining rapid antigen and RT-qPCR for screening of COVID-19 patients. medRxiv[Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Houston 2021 {published data only}
- Houston H, Gupta-Wright A, Toke-Bjolgerud E, Biggin-Lamming J, John L. Diagnostic accuracy and utility of SARS-CoV-2 antigen lateral flow assays in medical admissions with possible COVID-19. Journal of Hospital Infection 2021;110:203-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston H, Gupta-Wright A, Toke-Bjolgerud E, Biggin-Lamming J, John L. Diagnostic accuracy and utility of SARS-CoV-2 antigen lateral flow assays in medical admissions with possible COVID-19. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Huh 2021 {published data only}
- Huh HJ, Chae SL, Kim D-M. Comparison and analysis of various complementary diagnosis methods for the current situation and problems of COVID-19 diagnosis. medRxiv [Preprint] 2021. [DOI: ] [Google Scholar]
Igloi 2021 {published data only}
- Igloi Z, Velzing J, Van Beek J, Van de Vijver D, Aron G, Ensing R, et al. Clinical evaluation of Roche SD Biosensor Rapid Antigen Test for SARS-CoV-2 in municipal health service testing site, the Netherlands. Emerging Infectious Diseases 2021;27(5):1323-9. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igloi Z, Velzing J, Van Beek J, Van de Vijver D, Aron G, Ensing R, et al. Clinical evaluation of the Roche/SD Biosensor rapid antigen test with symptomatic, non-hospitalized patients in a municipal health service drive-through testing site. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Ishii 2021 [A] {published data only}
- Ishii T, Sasaki M, Yamada K, Kato D, Osuka H, Aoki K, et al. Immunochromatography and chemiluminescent enzyme immunoassay for COVID-19 diagnosis. Journal of Infection and Chemotherapy 2021;27(6):915-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Ishii 2021 [B] {published data only}
- Ishii T, Sasaki M, Yamada K, Kato D, Osuka H, Aoki K et al. Immunochromatography and chemiluminescent enzyme immunoassay for COVID-19 diagnosis. Journal of Infection and Chemotherapy 2021;27(6):915-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Jaaskelainen 2021 [A] {published data only}
- Jaaskelainen AE, Ahava MJ, Jokela P, Szirovicza L, Pohjala S, Vapalahti O, et al. Evaluation of three rapid lateral flow antigen detection tests for the diagnosis of SARS-CoV-2 infection. Journal of Clinical Virology 2021;137:104785. [DOI] [PMC free article] [PubMed] [Google Scholar]
Jaaskelainen 2021 [B] {published data only}
- Jaaskelainen AE, Ahava MJ, Jokela P, Szirovicza L, Pohjala S, Vapalahti O, et al. Evaluation of three rapid lateral flow antigen detection tests for the diagnosis of SARS-CoV-2 infection. Journal of Clinical Virology 2021;137:104785. [DOI] [PMC free article] [PubMed] [Google Scholar]
Jaaskelainen 2021 [C] {published data only}
- Jaaskelainen AE, Ahava MJ, Jokela P, Szirovicza L, Pohjala S, Vapalahti O, et al. Evaluation of three rapid lateral flow antigen detection tests for the diagnosis of SARS-CoV-2 infection. Journal of Clinical Virology 2021;137:104785. [DOI] [PMC free article] [PubMed] [Google Scholar]
Jakobsen 2021 {published data only}
- Jakobsen KK, Jensen JS, Todsen T, Lippert F, Martel CJ-M, Klokker M, et al. Detection of SARS-CoV-2 infection by rapid antigen test in comparison with RT-PCR in a public setting. medRxiv[Preprint] 2021. [DOI: ] [Google Scholar]
- Jakobsen KK, Jensen JS, Todsen T, Tolsaard MG, Kirkby N, Lippert F, et al. Accuracy and cost description of rapid antigen test compared with reverse transcriptase-polymerase chain reaction for SARS-CoV-2 detection. Danish Medical Journal 2021;68(7):A03210217. [PubMed] [Google Scholar]
James 2021 {published data only}
- James AE, Gulley T, Kothari A, Holder K, Garner K, Patil N. Performance of the BinaxNOW coronavirus disease 2019 (COVID-19) Antigen Card test relative to the severe acute respiratory coronavirus virus 2 (SARS-CoV-2) real-time reverse transcriptase polymerase chain reaction (rRT-PCR) assay among symptomatic and asymptomatic healthcare employees. Infection Control and Hospital Epidemiology 2021:1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kerneis 2021 {published data only}
- Kernéis S, Elie C, Fourgeaud J, Choupeaux L, Delarue SM, Alby M-L, et al. Accuracy of antigen and nucleic acid amplification testing on saliva and naopharyngeal samples for detection of SARS-CoV-2 in ambulatory care. medRxiv [Preprint] 2021. [DOI: ] [Google Scholar]
Kilic 2021 {published data only}
- Kilic A, Hiestand B, Palavecino E. Evaluation of performance of the BD Veritor SARS-CoV-2 chromatographic immunoassay test in patients with symptoms of COVID-19. Journal of Clinical Microbiology 2021;59(5). [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kohmer 2021 [A] {published data only}
- Kohmer N, Toptan T, Pallas C, Karaca O, Pfeiffer A, Westhaus S, et al. The comparative clinical performance of four SARS-CoV-2 rapid antigen tests and their correlation to infectivity in vitro. Journal of Clinical Medicine 2021;10(2):328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kohmer 2021 [B] {published data only}
- Kohmer N, Toptan T, Pallas C, Karaca O, Pfeiffer A, Westhaus S, et al. The comparative clinical performance of four SARS-CoV-2 rapid antigen tests and their correlation to infectivity in vitro. Journal of Clinical Medicine 2021;10(2):328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kohmer 2021 [C] {published data only}
- Kohmer N, Toptan T, Pallas C, Karaca O, Pfeiffer A, Westhaus S, et al. The comparative clinical performance of four SARS-CoV-2 rapid antigen tests and their correlation to infectivity in vitro. Journal of Clinical Medicine 2021;10(2):328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kohmer 2021 [D] {published data only}
- Kohmer N, Toptan T, Pallas C, Karaca O, Pfeiffer A, Westhaus S, et al. The comparative clinical performance of four SARS-CoV-2 rapid antigen tests and their correlation to infectivity in vitro. Journal of Clinical Medicine 2021;10(2):328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kriemler 2021 {published data only}
- Kriemler S, Ulyte A, Ammann P, Peralta GP, Berger C, Puhan MA, et al. Surveillance of acute SARS-CoV-2 infections in school children and point-prevalence during a time of high community transmission in Switzerland. Frontiers in Pediatrics 2021;9:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kruger 2021 {published data only}
- Krüger LJ, Klein JA, Tobian F, Gaeddert M, Lainati F, Klemm S, et al. Evaluation of accuracy, exclusivity, limit-of-detection and ease-of-use of LumiraDx - Antigen-detecting point-of-care device for SARS-CoV-2. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kruttgen 2021 {published data only}
- Kruttgen A, Cornelissen CG, Dreher M, Hornef MW, Imohl M, Kleines M. Comparison of the SARS-CoV-2 Rapid antigen test to the real star Sars-CoV-2 RT PCR kit. Journal of Virological Methods 2021;288:114024. [DOI] [PMC free article] [PubMed] [Google Scholar]
L'Huillier 2021 {published data only}
- L'Huillier AG, Lacour M, Sadiku D, Gadiri MA, De Siebenthal L, Schibler M, et al. Diagnostic accuracy of SARS-CoV-2 rapid antigen detection testing in symptomatic and asymptomatic children in the clinical setting. Journal of Clinical Microbiology 2021;59(9):e0099121. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lambert‐Niclot 2020 {published data only}
- Lambert-Niclot S, Cuffel A, Le Pape S, Vauloup-Fellous C, Morand-Joubert L, Roque-Afonso AM, et al. Evaluation of a rapid diagnostic assay for detection of SARS CoV-2 antigen in nasopharyngeal swab. Journal of Clinical Microbiology 2020;58(8):e00977-20. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lanser 2021 {published data only}
- Lanser L, Bellmann-Weiler R, Ottl KW, Huber L, Griesmacher A, Theurl I, et al. Evaluating the clinical utility and sensitivity of SARS-CoV-2 antigen testing in relation to RT-PCR Ct values. Infection 2021;49(3):555-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Linares 2020 {published data only}
- Linares M, Pérez-Tanoira R, Carrero A, Romanyk J, Pérez-García F, Gómez-Herruz P, et al. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. Journal of Clinical Virology 2020;133:104659. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares M, Pérez TR, Romanyk J, Pérez García F, Gómez-Herruz P, Arroyo T, et al. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lindner 2021a [A] {published data only}
- Lindner AK, Nikolai O, Kausch F, Wintel M, Hommes F, Gertler M, et al. Head-to-head comparison of SARS-CoV-2 antigen-detecting rapid test with self-collected nasal swab versus professional-collected nasopharyngeal swab. European Respiratory Journal 2021;57(4):2003961. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner AK, Nikolai O, Rohardt C, Kausch F, Wintel M, Gertler M, et al. SARS-CoV-2 patient self-testing with an antigen-detecting rapid test: a head-to-head comparison with professional testing. medRxiv [Preprint] 2021. [DOI: ] [Google Scholar]
Lindner 2021a [B] {published data only}
- Lindner AK, Nikolai O, Kausch F, Wintel M, Hommes F, Gertler M, et al. Head-to-head comparison of SARS-CoV-2 antigen-detecting rapid test with self-collected nasal swab versus professional-collected nasopharyngeal swab. European Respiratory Journal 2021;57(4):2003961. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lindner 2021b [A] {published data only}
- Lindner AK, Nikolai O, Rohardt C, Kausch F, Wintel M, Gertler M, et al. Diagnostic accuracy and feasibility of patient self-testing with a SARS-CoV-2 antigen-detecting rapid test. Journal of Clinical Virology 2021;141:104874. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lindner 2021b [B] {published data only}
- Lindner AK, Nikolai O, Rohardt C, Kausch F, Wintel M, Gertler M, et al. Diagnostic accuracy and feasibility of patient self-testing with a SARS-CoV-2 antigen-detecting rapid test. Journal of Clinical Virology 2021;141:104874. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lindner 2021b [C] {published data only}
- Lindner AK, Nikolai O, Rohardt C, Kausch F, Wintel M, Gertler M, et al. Diagnostic accuracy and feasibility of patient self-testing with a SARS-CoV-2 antigen-detecting rapid test. Journal of Clinical Virology 2021;141:104874. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Liotti 2021 {published data only}
- Liotti FM, Menchinelli G, Lalle E, Palucci I, Marchetti S, Colavita F, et al. Performance of a novel diagnostic assay for rapid SARS-CoV-2 antigen detection in nasopharynx samples. Clinical Microbiology and Infection 2021;27(3):487-8. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Love 2021 {published data only}
- Love NK, Ready D, Turner C, Yardley L, Rubin GJ, Hopkins S, et al. The acceptability of testing contacts of confirmed COVID-19 cases using serial, self-administered lateral flow devices as an alternative to self-isolation. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PubMed] [Google Scholar]
Masia 2021 [A] {published data only}
- Masiá M, Fernández-González M, Sánchez M, Carvajal M, García JA, Gonzalo-Jiménez N, et al. Nasopharyngeal Panbio COVID-19 Antigen performed at point-of-care has a high sensitivity in symptomatic and asymptomatic patients with higher risk for transmission and older age. Open Forum Infectious Diseases 2021;8(3):ofab059. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masia M, Fernandez-Gonzalez M, Sanchez M, Carvajal M, Garcia JA, Gonzalo N, et al. Nasopharyngeal Panbio COVID-19 antigen performed at point-of-care has a high sensitivity in symptomatic and asymptomatic patients with higher risk for transmission and older age. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Masia 2021 [B] {published data only}
- Masiá M, Fernández-González M, Sánchez M, Carvajal M, García JA, Gonzalo-Jiménez N, et al. Nasopharyngeal Panbio COVID-19 Antigen performed at point-of-care has a high sensitivity in symptomatic and asymptomatic patients with higher risk for transmission and older age. Open Forum Infectious Diseases 2021;8(3):ofab059. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Masia 2021 [C] {published data only}
- Masiá M, Fernández-González M, Sánchez M, Carvajal M, García JA, Gonzalo-Jiménez N, et al. Nasopharyngeal Panbio COVID-19 Antigen performed at point-of-care has a high sensitivity in symptomatic and asymptomatic patients with higher risk for transmission and older age. Open Forum Infectious Diseases 2021;8(3):ofab059. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Merino 2021 {published data only}
- Merino-Amador P, Guinea J, Muñoz-Gallego I, González-Donapetry P, Galán J-C, Antona N, et al. Multicenter evaluation of the Panbio™ COVID-19 Rapid Antigen-Detection Test for the diagnosis of SARS-CoV-2 infection. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
- Merino P, Guinea J, Muñoz-Gallego I, González-Donapetry P, Galán JC, Antona N, et al. Multicenter evaluation of the Panbio™ COVID-19 rapid antigen-detection test for the diagnosis of SARS-CoV-2 infection. Clinical Microbiology and Infection 2021;27(5):758-61. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Mertens 2020 {published data only}
- Mertens P, De Vos N, Martiny D, Jassoy C, Mirazimi A, Cuypers L, et al. Development and potential usefulness of the COVID-19 Ag Respi-Strip Diagnostic Assay in a pandemic context. Frontiers in Medicine (Lausanne) 2020;7:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens P, De Vos N, Martiny D, Jassoy C, Mirazimi A, Cuypers L, et al. Development and potential usefulness of the COVID-19 Ag Respi-Strip diagnostic assay in a pandemic context. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Miyakawa 2021 [A] {published data only}
- Miyakawa K, Funabashi R, Yamaoka Y, Jeremiah SS, Katada J, Wada A, et al. SARS-CoV-2 antigen rapid diagnostic test enhanced with silver amplification technology. medRxiv [Preprint] 2021. [DOI: ] [Google Scholar]
Miyakawa 2021 [B] {published data only}
- Miyakawa K, Funabashi R, Yamaoka Y, Jeremiah SS, Katada J, Wada A, et al. SARS-CoV-2 antigen rapid diagnostic test enhanced with silver amplification technology. medRxiv [Preprint] 2021. [DOI: ] [Google Scholar]
Miyakawa 2021 [C] {published data only}
- Miyakawa K, Funabashi R, Yamaoka Y, Jeremiah SS, Katada J, Wada A, et al. SARS-CoV-2 antigen rapid diagnostic test enhanced with silver amplification technology. medRxiv [Preprint] 2021. [DOI: ] [Google Scholar]
Miyakawa 2021 [D] {published data only}
- Miyakawa K, Funabashi R, Yamaoka Y, Jeremiah SS, Katada J, Wada A, et al. SARS-CoV-2 antigen rapid diagnostic test enhanced with silver amplification technology. medRxiv [Preprint] 2021. [DOI: ] [Google Scholar]
Mockel 2021(a) {published data only}
- Mockel M, Corman VM, Stegemann MS, Hofmann J, Stein A, Jones TC, et al. SARS-CoV-2 antigen rapid immunoassay for diagnosis of COVID-19 in the emergency department. Biomarkers 2021;26(3):213-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Mockel 2021(b) {published data only}
- Mockel M, Corman VM, Stegemann MS, Hofmann J, Stein A, Jones TC, et al. SARS-CoV-2 antigen rapid immunoassay for diagnosis of COVID-19 in the emergency department. Biomarkers 2021;26(3):213-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Nagura‐Ikeda 2020 {published data only}
- Nagura-Ikeda M, Imai K, Tabata S, Miyoshi K, Murahara N, Mizuno T, et al. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, reverse transcription-loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. Journal of Clinical Microbiology 2020;58(9):e01438-20. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Nalumansi 2020 {published data only}
- Nalumansi A, Lutalo T, Kayiwa J, Watera C, Balinandi S, Kiconco J, et al. Field evaluation of the performance of a SARS-CoV-2 antigen rapid diagnostic test in Uganda using nasopharyngeal samples. International Journal of Infectious Diseases 2020 Oct 30 [Epub ahead of print]. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Nash 2020 {published data only}
- Nash B, Badea A, Reddy A, Bosch M, Salcedo N, Gomez AR, et al. The impact of high frequency rapid viral antigen screening on COVID-19 spread and outcomes: a validation and modeling study. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Ngo Nsoga 2021 {published data only}
- Ngo Nsoga MT, Kronig I, Perez RF J, Sattonnet-Roche P, Da Silva D, Helbling J, et al. Diagnostic accuracy of Panbio rapid antigen tests on oropharyngeal swabs for detection of SARS-CoV-2. PLoS One 2021;16(6). [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo Nsoga MT, Kronig I, Perez Rodriguez FJ, Sattonnet-Roche P, Da Silva D, Helbling J, et al. Diagnostic accuracy of Panbio™ rapid antigen tests on oropharyngeal swabs for detection of SARS-CoV-2. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Nikolai 2021(a) [A] {published data only}
- Nikolai O, Rohardt C, Tobian F, Junge A, Corman VM, Jones TC, et al. Anterior nasal versus nasal mid-turbinate sampling for a SARS-CoV-2 antigen-detecting rapid test: does localisation or professional collection matter? medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Nikolai 2021(a) [B] {published data only}
- Nikolai O, Rohardt C, Tobian F, Junge A, Corman VM, Jones TC, et al. Anterior nasal versus nasal mid-turbinate sampling for a SARS-CoV-2 antigen-detecting rapid test: does localisation or professional collection matter? medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Nikolai 2021(b) [A] {published data only}
- Nikolai O, Rohardt C, Tobian F, Junge A, Corman VM, Jones TC, et al. Anterior nasal versus nasal mid-turbinate sampling for a SARS-CoV-2 antigen-detecting rapid test: does localisation or professional collection matter? medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Nikolai 2021(b) [B] {published data only}
- Nikolai O, Rohardt C, Tobian F, Junge A, Corman VM, Jones TC, et al. Anterior nasal versus nasal mid-turbinate sampling for a SARS-CoV-2 antigen-detecting rapid test: does localisation or professional collection matter? medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Okoye 2021 {published data only}
- Okoye NC, Barker AP, Curtis K, Orlandi RR, Snavely EA, Wright C, et al. Performance characteristics of BinaxNOW COVID-19 Antigen Card for screening asymptomatic individuals in a university setting. Journal of Clinical Microbiology 2021;59(4):e03282-20. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Olearo 2021 [A] {published data only}
- Olearo F, Nörz D, Heinrich F, Sutter JP, Roedl K, Schultze A, et al. Handling and accuracy of four rapid antigen tests for the diagnosis of SARS-CoV-2 compared to RT-qPCR. Journal of Clinical Virology 2021;137:104782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olearo F, Noerz D, Heinrich F, Sutter JP, Roedel K, Schultze A, et al. Handling and accuracy of four rapid antigen tests for the diagnosis of SARS-CoV-2 compared to RT-qPCR. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Olearo 2021 [B] {published data only}
- Olearo F, Nörz D, Heinrich F, Sutter JP, Roedl K, Schultze A, et al. Handling and accuracy of four rapid antigen tests for the diagnosis of SARS-CoV-2 compared to RT-qPCR. Journal of Clinical Virology 2021;137:104782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olearo F, Noerz D, Heinrich F, Sutter JP, Roedel K, Schultze A, et al. Handling and accuracy of four rapid antigen tests for the diagnosis of SARS-CoV-2 compared to RT-qPCR. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Olearo 2021 [C] {published data only}
- Olearo F, Nörz D, Heinrich F, Sutter JP, Roedl K, Schultze A, et al. Handling and accuracy of four rapid antigen tests for the diagnosis of SARS-CoV-2 compared to RT-qPCR. Journal of Clinical Virology 2021;137:104782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olearo F, Noerz D, Heinrich F, Sutter JP, Roedel K, Schultze A, et al. Handling and accuracy of four rapid antigen tests for the diagnosis of SARS-CoV-2 compared to RT-qPCR. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Olearo 2021 [D] {published data only}
- Olearo F, Nörz D, Heinrich F, Sutter JP, Roedl K, Schultze A, et al. Handling and accuracy of four rapid antigen tests for the diagnosis of SARS-CoV-2 compared to RT-qPCR. Journal of Clinical Virology 2021;137:104782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olearo F, Noerz D, Heinrich F, Sutter JP, Roedel K, Schultze A, et al. Handling and accuracy of four rapid antigen tests for the diagnosis of SARS-CoV-2 compared to RT-qPCR. medRxiv [Preprint] 2020. [https://doi.org/10.1101/2020.12.05.20244673] [DOI] [PMC free article] [PubMed] [Google Scholar]
Osterman 2021(a) [A] {published data only}
- Osterman A, Baldauf HM, Eletreby M, Wettengel JM, Afridi SQ, Fuchs T, et al. Evaluation of two rapid antigen tests to detect SARS-CoV-2 in a hospital setting. Medical Microbiology and Immunology 2021;210(1):65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Osterman 2021(a) [B] {published data only}
- Osterman A, Baldauf HM, Eletreby M, Wettengel JM, Afridi SQ, Fuchs T, et al. Evaluation of two rapid antigen tests to detect SARS-CoV-2 in a hospital setting. Medical Microbiology and Immunology 2021;210(1):65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Osterman 2021(b) {published data only}
- Osterman A, Baldauf HM, Eletreby M, Wettengel JM, Afridi SQ, Fuchs T, et al. Evaluation of two rapid antigen tests to detect SARS-CoV-2 in a hospital setting. Medical Microbiology and Immunology 2021;210(1):65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Parada‐Ricart 2020 {published data only}
- Parada-Ricart E, Gomez-Bertomeu F, Pico-Plana E, Olona-Cabases M. Usefulness of the antigen for diagnosing SARS-CoV-2 infection in patients with and without symptoms. Enfermedades Infecciosas y Microbiología Clínica (English Edition) 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pena 2021 {published data only}
- Pena M, Ampuero M, Garces C, Gaggero A, Garcia P, Velasquez MS, et al. Performance of SARS-CoV-2 rapid antigen test compared with real-time RT-PCR in asymptomatic individuals. International Journal of Infectious Diseases 2021;107:201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Pena‐Rodriguez 2021 {published data only}
- Pena-Rodriguez M, Viera-Segura O, Garcia-Chagollan M, Zepeda-Nuno JS, Munoz-Valle JF, Mora-Mora J, et al. Performance evaluation of a lateral flow assay for nasopharyngeal antigen detection for SARS-CoV-2 diagnosis. Journal of Clinical Laboratory Analysis 2021;35(5):e23745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Perez‐Garcia 2021 [A] {published data only}
- Perez-Garcia F, Romanyk J, Gomez-Herruz P, Arroyo T, Perez-Tanoira R, Linares M, et al. Diagnostic performance of CerTest and Panbio antigen rapid diagnostic tests to diagnose SARS-CoV-2 infection. Journal of Clinical Virology 2021;137:104781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Perez‐Garcia 2021 [B] {published data only}
- Perez-Garcia F, Romanyk J, Gomez-Herruz P, Arroyo T, Perez-Tanoira R, Linares M, et al. Diagnostic performance of CerTest and Panbio antigen rapid diagnostic tests to diagnose SARS-CoV-2 infection. Journal of Clinical Virology 2021;137:104781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Peto 2021(a) [A] {published data only}
- Peto T, Affron D, Afrough B, Agasu A, Ainsworth M, Allanson A, et al. COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay: a national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine 2021;36:10.1016/j.eclinm.2021.100924. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peto T. COVID-19: rapid antigen detection for SARS-CoV-2 by lateral flow assay: a national systematic evaluation for mass-testing. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Public Health England (PHE). Preliminary report from the Joint PHE Porton Down & University of Oxford SARS-CoV-2 test development and validation cell: rapid evaluation of lateral flow viral antigen detection devices (LFDs) for mass community testing. Public Health England, 2020. [Google Scholar]
Peto 2021(a) [B] {published data only}
- Peto T, Affron D, Afrough B, Agasu A, Ainsworth M, Allanson A, et al. COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay: a national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine 2021;36:10.1016/j.eclinm.2021.100924. [EMBASE: httpa://doi,org/10.1016/j.eclinm.2021.100924] [DOI] [PMC free article] [PubMed] [Google Scholar]
Peto 2021(a) [C] {published data only}
- Peto T, Affron D, Afrough B, Agasu A, Ainsworth M, Allanson A, et al. COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay: a national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine 2021;36:10.1016/j.eclinm.2021.100924. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Peto 2021(a) [D] {published data only}
- Peto T, Affron D, Afrough B, Agasu A, Ainsworth M, Allanson A, et al. COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay: a national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine 2021;36:10.1016/j.eclinm.2021.100924. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Peto 2021(a) [E] {published data only}
- Peto T, Affron D, Afrough B, Agasu A, Ainsworth M, Allanson A, et al. COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay: a national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine 2021;36:10.1016/j.eclinm.2021.100924. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Peto 2021(a) [F] {published data only}
- Peto T, Affron D, Afrough B, Agasu A, Ainsworth M, Allanson A, et al. COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay: a national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine 2021;36:10.1016/j.eclinm.2021.100924. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Peto 2021(a) [G] {published data only}
- Peto T, Affron D, Afrough B, Agasu A, Ainsworth M, Allanson A, et al, . COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay: a national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine 2021;36:10.1016/j.eclinm.2021.100924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Peto 2021(b) [non‐HCW tested] {published data only}
- Peto T, Affron D, Afrough B, Agasu A, Ainsworth M, Allanson A, et al. COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay: a national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine 2021;36:10.1016/j.eclinm.2021.100924. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Public Health England (PHE). Preliminary report from the Joint PHE Porton Down & University of Oxford SARS-CoV-2 test development and validation cell: rapid evaluation of lateral flow viral antigen detection devices (LFDs) for mass community testing. Public Health England, 2020. [Google Scholar]
Peto 2021(c) [A ‐ HCW tested] {published data only}
- Peto T, Affron D, Afrough B, Agasu A, Ainsworth M, Allanson A et al. COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay: a national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine 2021;36:10.1016/j.eclinm.2021.100924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Public Health England (PHE). Preliminary report from the Joint PHE Porton Down & University of Oxford SARS-CoV-2 test development and validation cell: rapid evaluation of lateral flow viral antigen detection devices (LFDs) for mass community testing. Public Health England, 2020. [Google Scholar]
Peto 2021(c) [A ‐ Lab tested] {published data only}
- Peto T, Affron D, Afrough B, Agasu A, Ainsworth M, Allanson A, et al. COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay: a national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine 2021;36:10.1016/j.eclinm.2021.100924. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Public Health England (PHE). Preliminary report from the Joint PHE Porton Down & University of Oxford SARS-CoV-2 test development and validation cell: rapid evaluation of lateral flow viral antigen detection devices (LFDs) for mass community testing. Public Health England, 2020. [Google Scholar]
Peto 2021(c) [B ‐ Lab tested] {published data only}
- Peto T, Affron D, Afrough B, Agasu A, Ainsworth M, Allanson A, et al. COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay: a national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine 2021;36:10.1016/j.eclinm.2021.100924. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Peto 2021(c) [C ‐ Lab tested] {published data only}
- Peto T, Affron D, Afrough B, Agasu A, Ainsworth M, Allanson A, et al. COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay: a national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine 2021;36:10.1016/j.eclinm.2021.100924. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Peto 2021(c) [D ‐ Lab tested] {published data only}
- Peto T, Affron D, Afrough B, Agasu A, Ainsworth M, Allanson A, et al. COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay: a national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine 2021;36:10.1016/j.eclinm.2021.100924. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Peto 2021(d) {published data only}
- Peto T, Affron D, Afrough B, Agasu A, Ainsworth M, Allanson A, et al. COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay: A national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine 2021;36:10.1016/j.eclinm.2021.100924. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Public Health England (PHE). Preliminary report from the Joint PHE Porton Down & University of Oxford SARS-CoV-2 test development and validation cell: rapid evaluation of lateral flow viral antigen detection devices (LFDs) for mass community testing. Public Health England, 2020. [Google Scholar]
PHE 2020 {published data only}
- Public Health England (PHE). Preliminary report from the Joint PHE Porton Down & University of Oxford SARS-CoV-2 test development and validation cell: rapid evaluation of lateral flow viral antigen detection devices (LFDs) for mass community testing. Public Health England, 2020. [Google Scholar]
Pickering 2021(a) [A] {published data only}
- Pickering S, Batra R, Snell LB, Merrick B, Nebbia G, Douthwaite S, et al. Comparative performance of SARS-CoV-2 lateral flow antigen tests demonstrates their utility for high sensitivity detection of infectious virus in clinical specimens. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pickering 2021(a) [B] {published data only}
- Pickering S, Batra R, Snell LB, Merrick B, Nebbia G, Douthwaite S, et al. Comparative performance of SARS-CoV-2 lateral flow antigen tests demonstrates their utility for high sensitivity detection of infectious virus in clinical specimens. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pickering 2021(a) [C] {published data only}
- Pickering S, Batra R, Snell LB, Merrick B, Nebbia G, Douthwaite S, et al. Comparative performance of SARS-CoV-2 lateral flow antigen tests demonstrates their utility for high sensitivity detection of infectious virus in clinical specimens. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pickering 2021(a) [D] {published data only}
- Pickering S, Batra R, Snell LB, Merrick B, Nebbia G, Douthwaite S, et al. Comparative performance of SARS-CoV-2 lateral flow antigen tests demonstrates their utility for high sensitivity detection of infectious virus in clinical specimens. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pickering 2021(a) [E] {published data only}
- Pickering S, Batra R, Snell LB, Merrick B, Nebbia G, Douthwaite S, et al. Comparative performance of SARS-CoV-2 lateral flow antigen tests demonstrates their utility for high sensitivity detection of infectious virus in clinical specimens. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pickering 2021(a) [F] {published data only}
- Pickering S, Batra R, Snell LB, Merrick B, Nebbia G, Douthwaite S, et al. Comparative performance of SARS-CoV-2 lateral flow antigen tests demonstrates their utility for high sensitivity detection of infectious virus in clinical specimens. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pickering 2021(b) [A] {published data only}
- Pickering S, Batra R, Snell LB, Merrick B, Nebbia G, Douthwaite S, et al. Comparative performance of SARS-CoV-2 lateral flow antigen tests demonstrates their utility for high sensitivity detection of infectious virus in clinical specimens. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pickering 2021(b) [B] {published data only}
- Pickering S, Batra R, Snell LB, Merrick B, Nebbia G, Douthwaite S, et al. Comparative performance of SARS-CoV-2 lateral flow antigen tests demonstrates their utility for high sensitivity detection of infectious virus in clinical specimens. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pickering 2021(b) [C] {published data only}
- Pickering S, Batra R, Snell LB, Merrick B, Nebbia G, Douthwaite S, et al. Comparative performance of SARS-CoV-2 lateral flow antigen tests demonstrates their utility for high sensitivity detection of infectious virus in clinical specimens. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pickering 2021(c) [A] {published data only}
- Pickering S, Batra R, Snell LB, Merrick B, Nebbia G, Douthwaite S, et al. Comparative performance of SARS-CoV-2 lateral flow antigen tests demonstrates their utility for high sensitivity detection of infectious virus in clinical specimens. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pickering 2021(c) [B] {published data only}
- Pickering S, Batra R, Snell LB, Merrick B, Nebbia G, Douthwaite S, et al. Comparative performance of SARS-CoV-2 lateral flow antigen tests demonstrates their utility for high sensitivity detection of infectious virus in clinical specimens. medRxiv [Preprint] 2021:10.1101/2021.02.27.21252427. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pilarowski 2020a {published data only}
- Pilarowski G, Marquez C, Rubio L, Peng J, Martinez J, Black D, et al. Field performance and public health response using the BinaxNOW TM Rapid SARS-CoV-2 antigen detection assay during community-based testing. Clinical Infectious Disease 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pilarowski 2021 {published data only}
- Pilarowski G, Lebel P, Sunshine S, Liu J, Crawford E, Marquez C, et al. Performance characteristics of a rapid SARS-CoV-2 antigen detection assay at a public plaza testing site in San Francisco. Journal of Infectious Diseases 2021;23(7):1139-44. [EMBASE: https://doi.org/10.1093/infdis/jiaa802] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilarowski G, Lebel P, Sunshine S, Liu J, Crawford E, Marquez C, et al. Performance characteristics of a rapid SARS-CoV-2 antigen detection assay at a public plaza testing site in San Francisco. medRxiv [Preprint] 2020. [EMBASE: https://doi.org/10.1101/2020.11.02.20223891] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pollock 2021a {published data only}
- Pollock NR, Jacobs JR, Tran K, Cranston AE, Smith S, O'Kane CY, et al. Performance and implementation evaluation of the Abbott BinaxNOW Rapid Antigen Test in a high-throughput drive-through community testing site in Massachusetts. Journal of Clinical Microbiology 2021;59(5):e00083-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock NR, Tran K, Jacobs JR, Cranston AE, Smith S, O’Kane CY, et al. Performance and operational evaluation of the Access Bio CareStart Rapid Antigen Test in a high-throughput drive-through community testing site in Massachusetts. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pollock 2021b {published data only}
- Pollock NR, Tran K, Jacobs JR, Cranston AE, Smith S, O'Kane CY, et al. Performance and operational evaluation of the Access Bio CareStart Rapid Antigen Test in a high-throughput drive-through community testing site in Massachusetts. Open Forum Infectious Diseases 2021;8(7):ofab243. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Porte 2020 {published data only}
- Porte L, Legarraga P, Vollrath V, Aguilera X, Munita JM, Araos R, et al. Evaluation of novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. International Journal of Infectious Diseases 2020;99:328-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porte L, Legarraga P, Vollrath V, Aguilera X, Munita JM, Araos R, et al. Evaluation of novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. SSRN [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Porte 2021 [A] {published data only}
- Porte L, Legarraga P, Iruretagoyena M, Vollrath V, Pizarro G, Munita J et al. Evaluation of two fluorescence immunoassays for the rapid detection of SARS-CoV-2 antigen-new tool to detect infective COVID-19 patients. Peer J 2021;9:e10801. [DOI: 10.7717/peerj.10801] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porte L, Legarraga P, Iruretagoyena M, Vollrath V, Pizarro G, Munita JM, et al. Rapid SARS-CoV-2 antigen detection by immunofluorescence – a new tool to detect infectivity. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Porte 2021 [B] {published data only}
- Porte L, Legarraga P, Iruretagoyena M, Vollrath V, Pizarro G, Munita J, et al. Evaluation of two fluorescence immunoassays for the rapid detection of SARS-CoV-2 antigen-new tool to detect infective COVID-19 patients. PeerJ 2021;9:e10801. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porte L, Legarraga P, Iruretagoyena M, Vollrath V, Pizarro G, Munita JM, et al. Rapid SARS-CoV-2 antigen detection by immunofluorescence – a new tool to detect infectivity. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Pray 2021 {published data only}
- Pray IW, Ford L, Cole D, Lee C, Bigouette JP, Abedi GR, et al. Performance of an antigen-based test for asymptomatic and symptomatic SARS-CoV-2 testing at two university campuses - Wisconsin, September-October 2020. MMWR: Morbidity and Mortality Weekly Report 2021;69(5152):1642-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Prince‐Guerra 2021 {published data only}
- Prince-Guerra JL, Almendares O, Nolen LD, Gunn JKL, Dale AP, Buono SA, et al. Evaluation of Abbott BinaxNOW Rapid Antigen Test for SARS-CoV-2 Infection at two community-based testing sites - Pima County, Arizona, November 3-17, 2020. MMWR: Morbidity and Mortality Weekly Report 2021;70(3):100-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Ristic 2021 {published data only}
- Ristic M, Nikolic N, Cabarkapa V, Turkulov V, Petrovic V. Validation of the STANDARD Q COVID-19 antigen test in Vojvodina, Serbia. PLoS One 2021;16(2):e0247606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Rottenstreich 2021 {published data only}
- Rottenstreich A, Zarbiv G, Kabiri D, Porat S, Sompolinsky Y, Reubinoff B, et al. Rapid antigen detection testing for universal screening for severe acute respiratory syndrome coronavirus 2 in women admitted for delivery. American Journal of Obstetrics & Gynecology 2021;224(5):539-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Saeed 2021 [A] {published data only}
- Saeed U, Uppal SR, Piracha ZZ, Rasheed A, Aftab Z, Zaheer H, et al. Evaluation of SARS-CoV-2 antigen-based rapid diagnostic kits in Pakistan: formulation of COVID-19 national testing strategy. Virology Journal 2021;18(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Saeed 2021 [B] {published data only}
- Saeed U, Uppal SR, Piracha ZZ, Rasheed A, Aftab Z, Zaheer, H et al. Evaluation of SARS-CoV-2 antigen-based rapid diagnostic kits in Pakistan: formulation of COVID-19 national testing strategy. Virology Journal 2021;18(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Salvagno 2021 {published data only}
- Salvagno GL, Gianfilippi G, Bragantini D, Henry BM, Lippi G. Clinical assessment of the Roche SARS-CoV-2 rapid antigen test. Diagnosis (Berl) 2021;8(3):322-6. [DOI: ] [DOI] [PubMed] [Google Scholar]
Schildgen 2021 [A] {published data only}
- Schildgen V, Demuth S, Lüsebrink J, Schildgen O. Limits and opportunities of SARS-CoV-2 antigen rapid tests: an experienced-based perspective. Pathogens 2021;10(1):38. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildgen V, Demuth S, Lüsebrink J, Schildgen O. Limits and opportunities of SARS-CoV-2 antigen rapid tests – an experience based perspective. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Schildgen 2021 [B] {published data only}
- Schildgen V, Demuth S, Lüsebrink J, Schildgen O. Limits and opportunities of SARS-CoV-2 antigen rapid tests: an experienced-based perspective. Pathogens 2021;10(1):38. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Schildgen 2021 [C] {published data only}
- Schildgen V, Demuth S, Lüsebrink J, Schildgen O. Limits and opportunities of SARS-CoV-2 antigen rapid tests: an experienced-based perspective. Pathogens 2021;10(1):38. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Schuit 2021(a) {published data only}
- Schuit E, Veldhuijzen I, Venekamp R, Van den Bijllaardt W, Pas S, Lodder E, et al. Diagnostic accuracy of rapid antigen tests in pre-/asymptomatic close contacts of individuals with a confirmed SARS-CoV-2 infection. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Schuit 2021(b) {published data only}
- Schuit E, Veldhuijzen I, Venekamp R, den Bijllaardt W, Pas S, Lodder E, et al. Diagnostic accuracy of rapid antigen tests in pre-/asymptomatic close contacts of individuals with a confirmed SARS-CoV-2 infection. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Schwob 2020(a) {published data only}
- Schwob JM, Miauton A, Petrovic D, Perdrix J, Senn N, Jaton K, et al. Antigen rapid tests, nasopharyngeal PCR and saliva PCR to detect SARS-CoV-2: a prospective comparative clinical trial. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Schwob 2020(b) {published data only}
- Schwob JM, Miauton A, Petrovic D, Perdrix J, Senn N, Jaton K, et al. Antigen rapid tests, nasopharyngeal PCR and saliva PCR to detect SARS-CoV-2: a prospective comparative clinical trial. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Schwob 2020(c) {published data only}
- Schwob JM, Miauton A, Petrovic D, Perdrix J, Senn N, Jaton K, et al. Antigen rapid tests, nasopharyngeal PCR and saliva PCR to detect SARS-CoV-2: a prospective comparative clinical trial. medRxiv [Preprint] 2020. [EMBASE: https://doi.org/10.1101/2020.11.23.20237057] [DOI] [PMC free article] [PubMed] [Google Scholar]
Scohy 2020 {published data only}
- Scohy A, Anantharajah A, Bodeus M, Kabamba-Mukadi B, Verroken A, Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. Journal of Clinical Virology 2020;129:104455. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Shidlovskaya 2021 [A] {published data only}
- Shidlovskaya EV, Kuznetsova NA, Divisenko EV, Nikiforova MA, Siniavin AE, Ogarkova DA, et al. The value of rapid antigen tests to identify carriers of viable SARS-CoV-2. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Shidlovskaya 2021 [B] {published data only}
- Shidlovskaya EV, Kuznetsova NA, Divisenko EV, Nikiforova MA, Siniavin AE, Ogarkova DA, et al. The value of rapid antigen tests to identify carriers of viable. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Shrestha 2020 {published data only}
- Shrestha B, Neupane AK, Pant S, Shrestha A, Bastola A. Sensitivity and specificity of lateral flow antigen test kits for COVID-19 in asymptomatic population of quarantine centre of Province 3. Kathmandu University Medical Journal 2020;18(70):36-9. [PubMed] [Google Scholar]
Smith 2021 {published data only}
- Smith RL, Gibson LL, Martinez PP, Ke R, Mirza A, Conte M, et al. Longitudinal assessment of diagnostic test performance over the course of acute SARS-CoV-2 infection. Journal of Infectious Diseases 2021;224(6):976-82. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RL, Gibson LL, Martinez PP, Ke R, Mirza A, Conte M, et al. Longitudinal assessment of diagnostic test performance over the course of acute SARS-CoV-2 infection. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Stohr 2021 [A] {published data only}
- Stohr JJ, Zwart VF, Goderski G, Meijer A, Nagel-Imming CR, Kluytmans-van den Bergh MF, et al. Self-testing for the detection of SARS-CoV-2 infection with rapid antigen tests. medRxiv [Preprint] 2021;40(8):1721-6. [EMBASE: https://doi.org/10.1101/2021.02.21.21252153] [DOI] [PMC free article] [PubMed] [Google Scholar]
Stohr 2021 [B] {published data only}
- Stohr JJ, Zwart VF, Goderski G, Meijer A, Nagel-Imming CR, Kluytmans-van den Bergh MF, et al. Self-testing for the detection of SARS-CoV-2 infection with rapid antigen tests. medRxiv [Preprint] 2021;40(8):1721-6. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Stokes 2021(a) [A] {published data only}
- Stokes W, Berenger BM, Portnoy D, Scott B, Szelewicki J, Singh T, et al. Clinical performance of the Abbott Panbio with nasopharyngeal, throat, and saliva swabs among symptomatic individuals with COVID-19. European Journal of Clinical Microbiology and Infectious Diseases 2021;40(8):1721-6. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Stokes 2021(a) [B] {published data only}
- Stokes W, Berenger BM, Portnoy D, Scott B, Szelewicki J, Singh T, et al. Clinical performance of the Abbott Panbio with nasopharyngeal, throat, and saliva swabs among symptomatic individuals with COVID-19. European Journal of Clinical Microbiology & Infectious Disease 2021;40(8):1721-6. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Stokes 2021(a) [C] {published data only}
- Stokes W, Berenger BM, Portnoy D, Scott B, Szelewicki J, Singh T, et al. Clinical performance of the Abbott Panbio with nasopharyngeal, throat, and saliva swabs among symptomatic individuals with COVID-19. European Journal of Clinical Microbiology & Infectious Diseases 2021;40(8):1721-6. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Stokes 2021(b) {published data only}
- Stokes W, Berenger BM, Portnoy D, Scott B, Szelewicki J, Singh T, et al. Clinical performance of the Abbott Panbio with nasopharyngeal, throat, and saliva swabs among symptomatic individuals with COVID-19. European Journal of Clinical Microbiology & Infectious Diseases 2021;40(8):1721-6. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Stromer 2020 {published data only}
- Stromer A, Rose R, Schafer M, Schon F, Vollersen A, Lorentz T, et al. Performance of a point-of-care test for the rapid detection of SARS-CoV-2 antigen. Microorganisms 2020;9(1):58. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Takeda 2020 {published data only}
- Takeda Y, Mori M, Omi K. SARS-CoV-2 qRT-PCR Ct value distribution in Japan and possible utility of rapid antigen testing kit. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Takeuchi 2021a {published data only}
- Takeuchi Y, Akashi Y, Kato D, Kuwahara M, Muramatsu S, Ueda A, et al. The evaluation of a newly developed antigen test (QuickNavi™-COVID19 Ag) for SARS-CoV-2: a prospective observational study in Japan. Journal of Infection and Chemotherapy 2021;27(6):890-4. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y, Akashi Y, Kato D, Kuwahara M, Muramatsu S, Ueda A, et al. The evaluation of a newly developed antigen test (QuickNaviTM-COVID19 Ag) for SARS-CoV-2: A prospective observational study in Japan. medRxiv [Preprint] 2021. [EMBASE: https://doi.org/10.1101/2020.12.27.20248876] [DOI] [PMC free article] [PubMed] [Google Scholar]
Takeuchi 2021b {published data only}
- Takeuchi Y, Akashi Y, Kato D, Kuwahara M, Muramatsu S, Ueda A, et al. Diagnostic performance and characteristics of anterior nasal collection for the SARS-CoV-2 antigen test: a prospective study. Scientific Reports 2021;11(1):10519. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y, Akashi Y, Kato D, Kuwahara M, Muramatsu S, Ueda A, et al. Diagnostic performance and characteristics of anterior nasal collection for the SARS-CoV-2 antigen test: a prospective study in Japan. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Thommes 2021 [A] {published data only}
- Thommes L, Burkert FR, Ottl KW, Goldin D, Loacker L, Lanser L, et al. Comparative evaluation of four SARS-CoV-2 antigen tests in hospitalized patients. International Journal of Infectious Diseases 2021;105:144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Thommes 2021 [B] {published data only}
- Thommes L, Burkert FR, Ottl KW, Goldin D, Loacker L, Lanser L, et al. Comparative evaluation of four SARS-CoV-2 antigen tests in hospitalized patients. International Journal of Infectious Diseases 2021;105:144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Thommes 2021 [C] {published data only}
- Thommes L, Burkert FR, Ottl KW, Goldin D, Loacker L, Lanser L, et al. Comparative evaluation of four SARS-CoV-2 antigen tests in hospitalized patients. International Journal of Infectious Diseases 2021;105:144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Thommes 2021 [D] {published data only}
- Thommes L, Burkert FR, Ottl KW, Goldin D, Loacker L, Lanser L, et al. Comparative evaluation of four SARS-CoV-2 antigen tests in hospitalized patients. International Journal of Infectious Diseases 2021;105:144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Toptan 2021(a) {published data only}
- Toptan T, Eckermann L, Pfeiffer AE, Hoehl S, Ciesek S, Drosten C, et al. Evaluation of a SARS-CoV-2 rapid antigen test: potential to help reduce community spread? Journal of Clinical Virology 2021;135:104713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Toptan 2021(b) {published data only}
- Toptan T, Eckermann L, Pfeiffer AE, Hoehl S, Ciesek S, Drosten C, et al. Evaluation of a SARS-CoV-2 rapid antigen test: potential to help reduce community spread? Journal of Clinical Virology 2021;135:104713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Torres 2021a {published data only}
- Torres I, Poujois S, Albert E, Colomina J, Navarro D. Evaluation of a rapid antigen test (Panbio COVID-19 Ag rapid test device) for SARS-CoV-2 detection in asymptomatic close contacts of COVID-19 patients. Clinical Microbiology and Infection 2021;27(4):636 e1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres I, Poujois S, Albert E, Colomina J, Navarro D. Real-life evaluation of a rapid antigen test (Panbio COVID-19 Ag Rapid Test Device) for SARS-CoV-2 detection in asymptomatic close contacts of COVID-19 patients. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Torres 2021b {published data only}
- Torres I, Poujois S, Albert E, Álvarez G, Colomina J, Navarro D. Point-of-care evaluation of a rapid antigen test (CLINITEST® Rapid COVID-19 Antigen Test) for diagnosis of SARS-CoV-2 infection in symptomatic and asymptomatic individuals. medRxiv [Preprint] 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres I, Poujois S, Albert E, Alvarez G, Colomina J, Navarro D. Point-of-care evaluation of a rapid antigen test (CLINITEST® Rapid COVID-19 Antigen Test) for diagnosis of SARS-CoV-2 infection in symptomatic and asymptomatic individuals. Journal of Infection 2021;82(5):e11-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Turcato 2021 {published data only}
- Turcato G, Zaboli A, Pfeifer N, Ciccariello L, Sibilio S, Tezza G, et al. Clinical application of a rapid antigen test for the detection of SARS-CoV-2 infection in symptomatic and asymptomatic patients evaluated in the emergency department: a preliminary report. Journal of Infection 2021;82(3):e14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Van der Moeren 2021(a) [A] {published data only}
- Van der Moeren N, Zwart VF, Lodder EB, Van den Bijllaardt W, Van Esch H, Stohr J, et al. Evaluation of the test accuracy of a SARS-CoV-2 rapid antigen test in symptomatic community dwelling individuals in the Netherlands. PLoS One 2021;16(5). [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Moeren N, Zwart VF, Lodder EB, Van den Bijllaardt W, Van Esch HR, Stohr JJ, et al. Performance evaluation of a SARS-CoV-2 rapid antigen test: test performance in the community in the Netherlands. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Van der Moeren 2021(a) [B] {published data only}
- Van der Moeren N, Zwart VF, Lodder EB, Van den Bijllaardt W, Van Esch H, Stohr J, et al. Evaluation of the test accuracy of a SARS-CoV-2 rapid antigen test in symptomatic community dwelling individuals in the Netherlands. PLoS One 2021;16(5). [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Moeren N, Zwart VF, Lodder EB, Van den Bijllaardt W, Van Esch HR, Stohr JJ, et al. Performance evaluation of a SARS-CoV-2 rapid antigen test: test performance in the community in the Netherlands. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Van der Moeren 2021(b) {published data only}
- Van der Moeren N, Zwart VF, Lodder EB, Van den Bijllaardt W, Van Esch H, Stohr J, et al. Evaluation of the test accuracy of a SARS-CoV-2 rapid antigen test in symptomatic community dwelling individuals in the Netherlands. PLoS One 2021;16(5):e0250886. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Moeren N, Zwart VF, Lodder EB, Van den Bijllaardt W, Van Esch HR, Stohr JJ, et al. Performance evaluation of a SARS-CoV-2 rapid antigen test: test performance in the community in the Netherlands. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Veyrenche 2021 {published data only}
- Veyrenche N, Bolloré K, Pisoni A, Bedin AS, Mondain AM, Ducos J, et al. Diagnosis value of SARS-CoV-2 antigen/antibody combined testing using rapid diagnostic tests at hospital admission. Journal of Medical Virology 2021;93(5):3069-76. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrenche N, Bollore K, Pisoni A, Bedin A-S, Mondain A-M, Ducos J, et al. Diagnosis value of SARS-CoV-2 antigen/antibody combined testing using rapid diagnostic tests at hospital admission. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Villaverde 2021 {published data only}
- Villaverde S, Dominguez-Rodriguez S, Sabrido G, Perez-Jorge C, Plata M, Romero MP, et al. Diagnostic accuracy of the Panbio severe acute respiratory syndrome coronavirus 2 antigen rapid test compared with reverse-transcriptase polymerase chain reaction testing of nasopharyngeal samples in the pediatric population. Journal of Pediatrics 2021;232:287-289 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Weitzel 2020 [A] {published data only}
- Weitzel T, Legarraga P, Iruretagoyena M, Pizarro G, Vollrath V, Araos R, et al. Head-to-head comparison of four antigen-based rapid detection tests for the diagnosis of SARS-CoV-2 in respiratory samples. bioRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzel T, Legarraga P, Iruretagoyena M, Pizarro G, Vollrath V, Porte L, et al. Comparative evaluation of four rapid SARS-CoV-2 antigen detection tests using universal transport medium. Travel Medicine and Infectious Diseases 2020 Dec 2 [Epub ahead of print]. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Weitzel 2020 [B] {published data only}
- Weitzel T, Legarraga P, Iruretagoyena M, Pizarro G, Vollrath V, Porte L, et al. Comparative evaluation of four rapid SARS-CoV-2 antigen detection tests using universal transport medium. Travel Medicine and Infectious Diseases 2020 Dec 2 [Epub ahead of print]. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Weitzel 2020 [C] {published data only}
- Weitzel T, Legarraga P, Iruretagoyena M, Pizarro G, Vollrath V, Porte L, et al. Comparative evaluation of four rapid SARS-CoV-2 antigen detection tests using universal transport medium. Travel Medicine and Infectious Diseases 2020 Dec 2 [Epub ahead of print]. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Weitzel 2020 [D] {published data only}
- Weitzel T, Legarraga P, Iruretagoyena M, Pizarro G, Vollrath V, Porte L, et al. Comparative evaluation of four rapid SARS-CoV-2 antigen detection tests using universal transport medium. Travel Medicine and Infectious Diseases 2020 Dec 2 [Epub ahead of print]. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Winkel 2020 {published data only}
- Winkel BM, Schram E, Gremmels H, Debast SB, Schuurman R, Wensing AM, et al. Screening for SARS-CoV-2 infection in asymptomatic individuals using the Panbio COVID-19 Antigen Rapid Test (Abbott) compared to RT-qPCR. medRxiv [Preprint] 2020. [https://doi.org/10.1101/2020.12.03.20243311] [DOI] [PMC free article] [PubMed] [Google Scholar]
Yokota 2020(a) {published data only}
- Yokota I, Sakurazawa T, Sugita J, Iwasaki S, Yasuda K, Yamashita N, et al. Performance of qualitative and quantitative antigen tests for SARS-CoV-2 in early symptomatic patients using saliva. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Yokota 2020(b) {published data only}
- Yokota I, Sakurazawa T, Sugita J, Iwasaki S, Yasuda K, Yamashita N, et al. Performance of qualitative and quantitative antigen tests for SARS-CoV-2 in early symptomatic patients using saliva. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Young 2020 {published data only}
- Young S, Taylor S, Cammarata C, Roger-Dalbert C, Montano A, Griego-Fullbright C, et al. Clinical evaluation of BD Veritor SARS-CoV-2 point-of-care test performance compared to PCR-based testing and versus the Sofia 2 SARS Antigen point-of-care test. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young S, Taylor SN, Cammarata CL, Varnado KG, Roger-Dalbert C, Montano A, et al. Clinical evaluation of BD Veritor SARS-CoV-2 point-of-care test performance compared to PCR-based testing and versus the Sofia 2 SARS Antigen point-of-care test. Journal of Clinical Microbiology 2020;59(1):e02338-20. [DOI: 10.1128/JCM.02338-20] [DOI] [PMC free article] [PubMed] [Google Scholar]
Young 2021 {published data only}
- Young BC, Eyre DW, Jeffery K. Use of lateral flow devices allows rapid triage of patients with SARS-CoV-2 on admission to hospital. Journal of Infection 2021;82(6):276-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
References to studies excluded from this review
Ahava 2021 {published data only}
- Ahava MJ, Kurkela S, Kuivanen S, Lappalainen M, Jarva H, Jääskeläinen AJ. Detection of SARS-CoV-2 nucleocapsid antigen from serum can aid in timing of COVID-19 infection. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Aoki 2020 {published data only}
- Aoki K, Nagasawa T, Ishii Y, Yagi S, Okuma S, Kashiwagi K, et al. Clinical validation of quantitative SARS-CoV-2 antigen assays to estimate SARS-CoV-2 viral loads in nasopharyngeal swabs. Journal of Infection and Chemotherapy 2020;27(4):613-6. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Bello‐Chavolla 2021 {published data only}
- Bello-Chavolla OY, Antonio-Villa NE, Fernandez-Chirino L, Guerra C, Fermin-Martinez CA, Marquez-Salinas A, et al. Diagnostic performance and clinical implications of rapid SARS-CoV-2 antigen testing in Mexico using real-world nationwide COVID-19 registry data. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Chen 2021 {published data only}
- Chen Y, Roma G, Liu F, Lee L. Quantitative and ultrasensitive in-situ immunoassay technology for SARS-CoV-2 detection in saliva. Research Square [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Corman 2020 {published data only}
- Corman VM, Haage VC, Bleicker T, Schmidt ML, Muehlemann B, Zuchowski M, et al. Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Cubas‐Atienzar 2021 {published data only}
- Cubas-Atienzar AI, Kontogianni K, Edwards T, Wooding D, Buist K, Thompson CR, et al. Limit of detection in different matrices of nineteen commercially available rapid antigen tests for the detection of SARS-CoV-2. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Dalal 2021 {published data only}
- Dalal A, Sonika U, Kumar M, George R, Kumar A, Srivastava S, et al. COVID-19 rapid antigen test: role in screening prior to gastrointestinal endoscopy. Clinical Endoscopy 2021;54(4):522-5. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Diao 2020 {published data only}
- Diao B, Wen K, Chen J, Liu Y, Yuan Z, Han C, et al. Diagnosis of acute respiratory syndrome coronavirus 2 infection by detection of nucleocapsid protein. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
- Diao B, Wen K, Zhang J, Chen J, Han C, Chen Y, et al. Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clinical Microbiology and Infection 2020 Oct 5 [Epub ahead of print]. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Dohla 2020 {published data only}
- Dohla M, Boesecke C, Schulte B, Diegmann C, Sib E, Richter E, et al. Rapid point-of-care testing for SARS-CoV-2 in a community screening setting shows low sensitivity. Public Health 2020;182:170-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Downs 2021 {published data only}
- Downs LO, Eyre DW, O'Donnell D, Jeffery K. Home-based SARS-CoV-2 lateral flow antigen testing in hospital workers. Journal of Infection 2021;82(2):282-327. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Eshghifar 2021 {published data only}
- Eshghifar N, Busheri A, Shrestha R, Beqaj S. Evaluation of analytical performance of seven rapid antigen detection kits for detection of SARS-CoV-2 virus. International Journal of General Medicine 2021;14:435-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Frnda 2021 {published data only}
- Frnda J, Durica M. On pilot massive COVID-19 testing by antigen tests in Europe. Case study: Slovakia. Infectious Disease Reports 2021;13(1):45-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Gili 2021 {published data only}
- Gili A, Paggi R, Russo C, Cenci E, Pietrella D, Graziani A, et al. Evaluation of automated test Lumipulse® G SARS-CoV-2 antigen assay for detection of SARS-CoV-2 nucleocapsid protein (NP) in nasopharyngeal swabs for community and population screening. International Journal of Infectious Diseases 2021;105:391-6. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Haage 2021a {published data only}
- Haage V, Oliveira-Filho EF, Moreira-Soto A, Kühne A, Fischer C, Sacks J, et al. Impaired performance of SARS-CoV-2 antigen-detecting rapid tests at elevated temperatures. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Haage 2021b {published data only}
- Haage VC, Moreira-Soto A, Sacks JA, Corman VM, Drosten C, Drexler JF. Limited specificity of SARS-CoV-2 antigen-detecting rapid diagnostic tests at low temperatures. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Herrera 2020 {published data only}
- Herrera V, Hsu V, Adewale A, Hendrix T, Johnson L, Kuhlman J, et al. Testing of healthcare workers exposed to COVID19 with rapid antigen detection. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Hingrat 2020 {published data only}
- Hingrat QL, Visseaux B, Laouenan C, Tubiana S, Bouadma L, Yazdanpanah Y, et al. Detection of SARS-CoV-2 N-antigen in blood during acute COVID-19 provides a sensitive new marker and new testing alternatives. Clinical Microbiology and Infection 2020;27(5):789.e1-789.e5. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Hirotsu 2020 {published data only}
- Hirotsu Y, Maejima M, Shibusawa M, Nagakubo Y, Hosaka K, Amemiya K, et al. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs, including from seven serially followed patients. International Journal of Infectious Diseases 2020;99:397-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Hirotsu 2021 {published data only}
- Hirotsu Y, Maejima M, Shibusawa M, Amemiya K, Nagakubo Y, Hosaka K et al. Prospective study of 1,308 nasopharyngeal swabs from 1,033 patients using the LUMIPULSE SARS-CoV-2 antigen test: comparison with RT-qPCR. International Journal of Infectious Diseases 2021;105:7-14. [DOI: 10.1016/j.ijid.2021.02.005] [DOI] [PMC free article] [PubMed] [Google Scholar]
Hledik 2020 {published data only}
- Hledik M, Polechova J, Beiglboeck M, Herdina AN, Strassl R, Posch M. Analysis of the specificity of the SD Biosensor Standard Q Ag-Test based on Slovak mass testing data. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Hoehl 2020 {published data only}
- Hoehl S, Schenk B, Rudych O, Goettig S, Foppa I, Kohmer N, et al. At-home self-testing of teachers with a SARS-CoV-2 rapid antigen test to reduce potential transmissions in schools. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Kannian 2021 {published data only}
- Kannian P, Lavanya C, Ravichandran K, Gita JB, Mahanathi P, Ashwini V, et al. SARS-CoV2 antigen in whole mouth fluid may be a reliable rapid detection tool. Oral Diseases 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannian P, Lavanya C, Ravichandran K, Jayaraman BG, Mahanathi P, Ashwini V, et al. Detection of SARS-CoV2 antigen in human saliva may be a reliable tool for large scale screening. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Kashiwagi 2020 {published data only}
- Kashiwagi K, Ishii Y, Aoki K, Yagi S, Maeda T, Miyazaki T, et al. Immunochromatographic test for the detection of SARS-CoV-2 in saliva. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kobayashi 2021 {published data only}
- Kobayashi R, Murai R, Asanuma K, Fujiya Y, Takahashi S. Evaluating a novel, highly sensitive, and quantitative reagent for detecting SARS-CoV-2 antigen. Journal of Infection and Chemotherapy 2021;27(6):800-7. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Koskinen 2021 {published data only}
- Koskinen JM, Antikainen P, Hotakainen K, Haveri A, Ikonen N, Savolainen-Kopra C, et al. Clinical validation of automated and rapid mariPOC SARS-CoV-2 antigen test. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kotsiou 2021 {published data only}
- Kotsiou OS, Pantazopoulos I, Papagiannis D, Fradelos EC, Kanellopoulos N, Siachpazidou D, et al. Repeated antigen-based rapid diagnostic testing for estimating the coronavirus disease 2019 prevalence from the perspective of the workers' vulnerability before and during the lockdown. International Journal of Environmental Research and Public Health 2021;18(4):1638. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kurstjens 2020 {published data only}
- Kurstjens S, Van der Horst A, Herpers R, Geerits MW, Kluiters-de Hingh YC, Göttgens E-L, et al. Rapid identification of SARS-CoV-2-infected patients at the emergency department using routine testing. bioRxiv [Preprint] 2020:1-21. [DOI: ] [DOI] [PubMed] [Google Scholar]
Kyosei 2020 {published data only}
- Kyosei Y, Namba M, Yamura S, Takeuchi R, Aoki N, Nakaishi K, et al. Proposal of de novo antigen test for COVID-19: ultrasensitive detection of spike proteins of SARS-CoV-2. Diagnostics (Basel) 2020;10(8):594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Lefever 2021 {published data only}
- Lefever S, Indevuyst C, Cuypers L, Dewaele K, Yin N, Cotton F, et al. Comparison of the quantitative DiaSorin Liaison antigen test to RT-PCR for the diagnosis of COVID-19 in symptomatic and asymptomatic outpatients. Journal of Clinical Microbiology 2021;59(7):e0037421. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Le Hingrat 2020 {published data only}
- Le Hingrat Q, Visseaux B, Laouenan C, Tubiana S, Bouadma L, Yazdanpanah Y, et al. SARS-CoV-2 N-antigenemia: a new alternative to nucleic acid amplification techniques. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Li 2021 {published data only}
- Li J, Lillehoj PB. Microfluidic magneto immunosensor for rapid, high sensitivity measurements of SARS-CoV-2 nucleocapsid protein in serum. ACS Sensors 2021;6(3):1270-8. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Liu 2021 {published data only}
- Liu D, Ju C, Han C, Shi R, Chen X, Duan D, et al. Nanozyme chemiluminescence paper test for rapid and sensitive detection of SARS-CoV-2 antigen. Biosensors and Bioelectronics 2021;173:112817. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Mahari 2020 {published data only}
- Mahari S, Roberts A, Shahdeo D, Gandhi S. eCovSens-Ultrasensitive novel in-house built printed circuit board based electrochemical device for rapid detection of nCOVID-19 antigen, a spike protein domain 1 of SARS-CoV-2. bioRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Mak 2020a {published data only}
- Mak GC, Cheng PK, Lau SS, Wong KK, Lau CS, Lam ET, et al. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. Journal of Clinical Virology 2020;129:104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Mak 2020b {published data only}
- Mak GC, Lau SS, Wong KK, Chow NL, Lau CS, Lam ET, et al. Analytical sensitivity and clinical sensitivity of the three rapid antigen detection kits for detection of SARS-CoV-2 virus. Journal of Clinical Virology 2020;133:104684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Marzinotto 2020 {published data only}
- Marzinotto S, Mio C, Cifu A, Verardo R, Pipan C, Schneider C, et al. A streamlined approach to rapidly detect SARS-CoV-2 infection, avoiding RNA extraction. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.06.20054114] [DOI] [PMC free article] [PubMed] [Google Scholar]
Mboumba 2021 {published data only}
- Mboumba Bouassa R-S, Veyer D, Péré H, Bélec L. Analytical performances of the point-of-care SIENNATM COVID-19 Antigen Rapid Test for the detection of SARS-CoV-2 nucleocapsid protein in nasopharyngeal swabs: a prospective evaluation during the COVID-19 second wave in France. International Journal of Infectious Diseases 2021;106:8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
McAulay 2020 {published data only}
- McAulay K, Kaleta EJ, Grys TE. Rapid detection of SARS-CoV-2 antigen from serum in a hospitalized population. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
McDonald 2020 {published data only}
- McDonald S, Courtney DM, Clark AE, Muthukumar A, Lee F, Balani J, et al. Diagnostic performance of a rapid point-of-care test for SARS-CoV-2 in an urban emergency department setting. Academic Emergency Medicine 2020;27(8):764-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Menchinelli 2021 {published data only}
- Menchinelli G, Bordi L, Liotti FM, Palucci I, Capobianchi MR, Sberna G, et al. Lumipulse G SARS-CoV-2 Ag assay evaluation using clinical samples from different testing groups. Clinical Chemistry and Laboratory Medicine 2021;59(8):1468-76. [DOI: ] [DOI] [PubMed] [Google Scholar]
Mohamed 2021 {published data only}
- Mohamed RA, Junaideen SM. Issues of random sampling with rapid antigen tests for COVID-19 diagnosis: a special reference to Kalmunai RDHS Division. medRxiv [Preprint] 2021. [DOI: ] [Google Scholar]
Moreno 2021 {published data only}
- Moreno GK, Braun KM, Pray IW, Seagaloff HE, Lim A, Poulsen K, et al. SARS-CoV-2 transmission in intercollegiate athletics not fully mitigated with daily antigen testing. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno GK, Braun KM, Pray IW, Segaloff HE, Lim A, Poulsen K et al. SARS-CoV-2 transmission in intercollegiate athletics not fully mitigated with daily antigen testing. Clinical Infectious Diseases 2021;73 (Suppl 1):S45-S53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Nachtigall 2020 {published data only}
- Nachtigall FM, Pereira A, Trofymchuk OS, Santos LS. Detection of SARS-CoV-2 in nasal swabs using MALDI-MS. Nature Biotechnology 2020;38(10):1168-73. [DOI] [PubMed] [Google Scholar]
Ogawa 2020 {published data only}
- Ogawa T, Fukumori T, Nishihara Y, Sekine T, Okuda N, Nishimura T, et al. Another false-positive problem for a SARS-CoV-2 antigen test in Japan. Journal of Clinical Virology 2020;131:104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Pavelka 2020 {published data only}
- Pavelka M, Van Zandvoort K, Abbott S, Sherratt K, Majdan M, Jarcuska P, et al, CMMID COVID-19 Working Group. The effectiveness of population-wide, rapid antigen test based screening in reducing SARS-CoV-2 infection prevalence in Slovakia. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
Pekosz 2021 {published data only}
- Pekosz A, Cooper C, Parvu V, Li M, Andrews J, Manabe YCC, et al. Antigen-based testing but not real-time PCR correlates with SARS-CoV-2 virus culture. medRxiv [Preprint] 2020. [DOI: ] [Google Scholar]
- Pekosz A, Parvu V, Li M, Andrews JC, Manabe YC, Kodsi S et al. Antigen-based testing but not real-time polymerase chain reaction correlates with severe acute respiratory syndrome coronavirus 2 viral culture. Clinical Infectious Diseases 2021;73(9):e2861-e2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
Pellanda 2020 {published data only}
- Pellanda LC, Wendland EM, McBride AJ, Tovo-Rodrigues L, Ferreira MR, Dellagostin OA, et al. Sensitivity and specificity of a rapid test for assessment of exposure to SARS-CoV-2 in a community-based setting in Brazil. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.05.06.20093476] [DOI] [Google Scholar]
Peng 2020 {published data only}
- Peng T, Liu X, Adams LG, Agarwal G, Akey B, Cirillo J, et al. Enhancing sensitivity of lateral flow assay with application to SARS-CoV-2. Applied Physics Letters 2020;117(12):120601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Perchetti 2020 {published data only}
- Perchetti GA, Huang ML, Mills MG, Jerome KR, Greninger AL. Analytical sensitivity of the Abbott BinaxNOW COVID-19 Ag CARD. Journal of Clinical Microbiology 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pollock 2020a {published data only}
- Pollock NR, Savage TJ, Wardell H, Lee R, Mathew A, M Stengelin, et al. Correlation of SARS-CoV-2 nucleocapsid antigen and RNA concentrations in nasopharyngeal samples from children and adults using an ultrasensitive and quantitative antigen assay. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Rastawicki 2021 {published data only}
- Rastawicki W, Gierczyński R, Juszczyk G, Mitura K, Henry BM. Evaluation of PCL rapid point of care antigen test for detection of SARS-CoV-2 in nasopharyngeal swabs. Journal of Medical Virology 2021;93(4):1920-2. [DOI: ] [DOI] [PubMed] [Google Scholar]
Regev‐Yochay 2021 {published data only}
- Regev-Yochay G, Kriger O, Beni S, Rubin C, Mina MJ, Mechnik B, et al. Real world performance of SARS-CoV-2 antigen rapid diagnostic tests in various clinical settings. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PubMed] [Google Scholar]
Ren 2021 {published data only}
- Ren A, Sohaei D, Zacharioudakis I, Sigal GB, Stengelin M, Matthew A, et al. Ultrasensitive assay for saliva-based SARS-CoV-2 antigen detection. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PubMed] [Google Scholar]
Rodriguez‐Manzano 2020 {published data only}
- Rodriguez-Manzano J, Malpartida-Cardenas K, Moser N, Pennisi I, Cavuto M, Miglietta L, et al. A handheld point-of-care system for rapid detection of SARS-CoV-2 in under 20 minutes. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Rusanen 2020 {published data only}
- Rusanen J, Kareinen L, Szirovicza L, Ugurlu H, Levanov L, Jaaskelainen AE, et al. A generic, scalable, and rapid TR-FRET -based assay for SARS-CoV-2 antigen detection. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Scheiblauer 2021 {published data only}
- Scheiblauer H, Filomena A, Nitsche A, Puyskens A, Corman VM, Drosten C, et al. Comparative sensitivity evaluation for 122 CE-marked SARS-CoV-2 antigen rapid tests. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Seitz 2021 {published data only}
- Seitz T, Schindler S, Winkelmeyer P, Zach B, Wenisch C, Zoufaly A, et al. Evaluation of rapid antigen tests based on saliva for the detection of SARS-CoV-2. Journal of Medical Virology 2021;93(7):4161-2. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Seo 2020 {published data only}
- Seo G, Lee G, Kim MJ, Baek SH, Choi M, Ku KB, et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano (American Chemical Society) 2020;14(4):5135-42. [DOI] [PubMed] [Google Scholar]
Singh 2020a {published data only}
- Singh NK, Ray P, Carlin AF, Magallanes C, Morgan SC, Laurent LC, et al. Hitting the diagnostic sweet spot: point-of-care SARS-CoV-2 salivary antigen testing with an off-the-shelf glucometer. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Singh 2020b {published data only}
- Singh P, Chakraborty R, Marwal R, Radhakrishan VS, Bhaskar AK, Vashisht H, et al. A rapid and sensitive method to detect SARS-CoV-2 virus using targeted-mass spectrometry. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Wang 2020 {published data only}
- Wang D, He S, Wang X, Yan Y, Liu J, Wu S, et al. Rapid lateral flow immunoassay for the fluorescence detection of SARS-CoV-2 RNA. Nature Biomedical Engineering 2020;4:1150-8. [DOI: ] [DOI] [PubMed] [Google Scholar]
Wu 2020 {published data only}
- Wu T, Ge Y, Zhao K, Zhu X, Chen Y, Wu B, et al. A reverse-transcription recombinase-aided amplification assay for the rapid detection of N gene of severe acute respiratory syndrome coronavirus 2(SARS-CoV-2). Virology 2020;549:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Yamamoto 2021 {published data only}
- Yamamoto K, Suzuki M, Yamada G, Sudo T, Nomoto H, Kinoshita N, et al. Utility of the antigen test for coronavirus disease 2019: Factors influencing the prediction of the possibility of disease transmission. International Journal of Infectious Diseases 2021;104:65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Yamayoshi 2020 {published data only}
- Yamayoshi S, Sakai-Tagawa Y, Koga M, Akasaka O, Nakachi I, Koh H, et al. Comparison of rapid antigen tests for COVID-19. Viruses 2020;12(12):1420. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Yokota 2021 {published data only}
- Yokota I, Shane PY, Teshima T. Logistic advantage of two-step screening strategy for SARS-CoV-2 at airport quarantine. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Yu 2020 {published data only}
- Yu S, Nimse SB, Kim J, Song KS, Kim T. Development of a lateral flow strip membrane assay for rapid and sensitive detection of the SARS-CoV-2. Analytical Chemistry 2020;92(20):14139-44. [DOI] [PubMed] [Google Scholar]
Zamecnik 2020 {published data only}
- Zamecnik CR, Rajan JV, Yamauchi KA, Mann SA, Sowa GM, Zorn KC, et al. ReScan, a multiplex diagnostic pipeline, pans human sera for SARS-CoV-2 antigens. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Zeng 2020 {published data only}
- Zeng W, Liu G, Ma H, Zhao D, Yang Y, Liu M, et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochemical and Biophysical Research Communications 2020;527(3):618-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Zhang 2020 {published data only}
- Zhang C, Zhou L, Du K, Zhang Y, Wang J, Chen L, et al. Foundation and clinical evaluation of a new method for detecting SARS-CoV-2 antigen by fluorescent microsphere immunochromatography. Frontiers in Cellular and Infection Microbiology 2020;10:553837. [DOI] [PMC free article] [PubMed] [Google Scholar]
References to studies awaiting assessment
Abdul‐Mumin 2021 {published data only}
- Abdul-Mumin A, Abubakari A, Agbozo F, Abdul-Karim A, Demah N, Mumuni K, et al. Field evaluation of specificity and sensitivity of a standard SARS-CoV-2 antigen rapid diagnostic test: a prospective study at a teaching hospital in Northern Ghana. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Abusrewil 2021 {published data only}
- Abusrewil Z, Alhudiri IM, Kaal H, Edin El Meshri S, Ebrahim FO, Dalyoum T, et al. Time scale performance of rapid antigen testing for SARS-COV-2: evaluation of ten rapid antigen assays. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Akashi 2021 {published data only}
- Akashi Y, Kiyasu Y, Takeuchi Y, Kato D, Kuwahara M, Muramatsu S, et al. Evaluation and clinical implications of the time to a positive results of antigen testing for SARS-CoV-2. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Amer 2021 {published data only}
- Amer RM, Samir M, Gaber OA, El-Deeb NA, Abdelmoaty AA, Ahmed AA, et al. Diagnostic performance of rapid antigen test for COVID-19 and the effect of viral load, sampling time, subject's clinical and laboratory parameters on test accuracy. Journal of Infection Public Health 2021;14(10):1446-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Aranda‐Diaz 2021 {published data only}
- Aranda-Diaz A, Imbert E, Strieff S, Graham-Squire D, Evans J, Moore J, et al. Implementation of rapid and frequent SARS-CoV2 antigen testing and response in congregate homeless shelters. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Baccani 2021 {published data only}
- Baccani I, Morecchiato F, Chilleri C, Cervini C, Gori E, Matarrese D, et al. Evaluation of three immunoassays for the rapid detection of SARS-CoV-2 antigens. Diagnostic Microbiology and Infectious Disease 2021;101(2):115434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Bianco 2021 {published data only}
- Bianco G, Boattini M, Barbui AM, Scozzari G, Riccardini F, Coggiola M, et al. Evaluation of an antigen-based test for hospital point-of-care diagnosis of SARS-CoV-2 infection. Journal of Clinical Virology 2021;139:104838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Blairon 2021 {published data only}
- Blairon L, Cupaiolo R, Thomas I, Piteus S, Wilmet A, Beukinga I, et al. Efficacy comparison of three rapid antigen tests for SARS-CoV-2 and how viral load impact their performance. Journal of Medical Virology 2021;93(10):5783-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Bonde 2021 {published data only}
- Bonde J, Ejegod DM, Pedersen H, Smith B, Cortes D, Leding C, et al. Clinical validation of point-of-care SARS-COV-2 BD Veritor antigen test by a single throat swab for rapid COVID-19 status on hospital patients predominantly without overt COVID symptoms. medRxiv [Preprint] 2021. [DOI: ] [Google Scholar]
Boum 2021 {published data only}
- Boum Y, Fai KN, Nikolay B, Mboringong AB, Bebell LM, Ndifon M, et al. Performance and operational feasibility of antigen and antibody rapid diagnostic tests for COVID-19 in symptomatic and asymptomatic patients in Cameroon: a clinical, prospective, diagnostic accuracy study. Lancet Infectious Diseases 2021;21(8):1089-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Brihn 2021 {published data only}
- Brihn A, Chang J, OYong K, Balter S, Terashita D, Rubin Z, et al. Diagnostic performance of an antigen test with RT-PCR for the detection of SARS-CoV-2 in a hospital setting - Los Angeles County, California, June-August 2020. MMWR Morbidity & Mortality Weekly Report 2021;70(19):702-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Bruins 2021 {published data only}
- Bruins MJ, Oliveira dos Santos C, Spoelman-Lunsche M, Van den Bos-Kromhout MI, Debast SB. Evaluation of the PanbioTM rapid antigen test for COVID-19 diagnosis in symptomatic health care workers. medRxiv [Preprint] 2021. [DOI: ] [Google Scholar]
Bruzzone 2021 {published data only}
- Bruzzone B, De Pace V, Caligiuri P, Ricucci V, Guarona G, Pennati BM, et al. Comparative diagnostic performance of rapid antigen detection tests for COVID-19 in a hospital setting. International Journal of Infectious Diseases 2021;107:215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Caruana 2021 {published data only}
- Caruana G, Lebrun LL, Aebischer O, Opota O, Urbano L, Rham M, et al. The dark side of SARS-CoV-2 rapid antigen testing: screening asymptomatic patients. New Microbes and New Infections 2021;42:100899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Cento 2021 {published data only}
- Cento V, Renica S, Matarazzo E, Antonello M, Colagrossi L, Di Ruscio F, et al. Frontline screening for SARS-CoV-2 infection at emergency department admission by third generation rapid antigen test: can we spare RT-qPCR? Viruses 2021;13(5):818. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Chiu 2021 {published data only}
- Chiu R, Kojima N, Mosley G, Cheng KK, Pereira D, Brobeck M, et al. Evaluation of the INDICAID COVID-19 Rapid Antigen Test in symptomatic populations and asymptomatic community testing. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Christensen 2021 {published data only}
- Christensen K, Ren H, Chen S, Cooper CK, Young S. Clinical evaluation of BD VeritorTM SARS-CoV-2 and flu A+B assay for point-of-care (POC) system. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Di Domenico 2021 {published data only}
- Di Domenico M, De Rosa A, Di Gaudio F, Internicola P, Bettini C, Salzano N, et al. Diagnostic accuracy of a new antigen test for SARS-CoV-2 detection. International Journal of Environmental Research and Public Health 2021;18(12):6310. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Dierks 2021 {published data only}
- Dierks S, Bader O, Schwanbeck J, Gross U, Weig MS, Mese K, et al. Diagnosing SARS-CoV-2 with antigen testing, transcription-mediated amplification and real-time PCR. Journal of Clinical Medicine 2021;10(11):2404. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Diez Flecha 2021 {published data only}
- Diez Flecha C, Rivero Rodriguez AM, Fernandez-Villa T, Fernandez Garcia P, Ferreira de Jesus JL, Sanchez Antolin G, et al. Internal validity of a rapid test for COVID-19 antigens in a nursing home. Semergen 2021;47(5):332-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Eleftheriou 2021 {published data only}
- Eleftheriou I, Dasoula F, Dimopoulou D, Lebessi E, Serafi E, Spyridis N, et al. Real-life evaluation of a COVID-19 rapid antigen detection test in hospitalized children. Journal of Medical Virology 2021;93(10):6040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Fernandez 2021 {published data only}
- Fernandez MD, Estevez AS, Alfonsin FL, Arevalo GB. Usefulness of the Lumiradx SARS-CoV-2 antigen test in nursing home. Enfermedades Infecciosas y Microbiología Clínica (English Edition) 2021. [DOI: ] [Google Scholar]
Fernandez‐Montero 2021 {published data only}
- Fernandez-Montero A, Argemi J, Rodríguez JA, Ariño AH, Moreno-Galarraga L. Validation of a rapid antigen test as a screening tool for SARS-CoV-2 infection in asymptomatic populations. Sensitivity, specificity and predictive values. eClinicalMedicine 2021:100954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Ferte 2021 {published data only}
- Ferte T, Ramel V, Cazanave C, Lafon ME, Bebear C, Malvy D, et al. Accuracy of COVID-19 rapid antigenic tests compared to RT-PCR in a student population: The StudyCov study. Journal of Clinical Virology 2021;141:104878. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Fiedler 2021 {published data only}
- Fiedler M, Holtkamp C, Dittmer U, Anastasiou OE. Performance of the LIAISON® SARS-CoV-2 Antigen Assay vs. SARS-CoV-2-RT-PCR. Pathogens 2021;10(6):658. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Ford 2021 {published data only}
- Ford L, Lee C, Pray IW, Cole D, Bigouette JP, Abedi GR, et al. Epidemiologic characteristics associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigen-based test results, real-time reverse transcription polymerase chain reaction (rRT-PCR) cycle threshold values, subgenomic RNA, and viral culture results from university testing. Clinical Infectious Disease 2021;73(6):e1348-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Frediani 2021 {published data only}
- Frediani JK, Levy JM, Rao A, Bassit L, Figueroa J, Vos MB, et al. Multidisciplinary assessment of the Abbott BinaxNOW SARS-CoV-2 point-of-care antigen test in the context of emerging viral variants and self-administration. Scientific Reports 2021;11(1):14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Gitaka 2021 {published data only}
- Gitaka J, Muthamia E, Mbugua S, Mungai M, Bandawe G, Qadri F, et al. Assessment of performance and implementation characteristics of rapid point of care SARS-CoV-2 antigen testing in Kenya. medRxiv [Preprint] 2021. [DOI: ] [Google Scholar]
Hagbom 2021 {published data only}
- Hagbom M, Carmona-Vicente N, Sharma S, Olsson H, Jamtberg M, Nilsdotter-Augustinsson A, et al. Evaluation of SARS-CoV-2 rapid antigen diagnostic tests for saliva samples. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Harmon 2021 {published data only}
- Harmon K, St Maurice AM, Brady AC, Swaminathan S, Aukerman DF, Rueda MA, et al. Surveillance testing for SARS-COV-2 infection in an asymptomatic athlete population: a prospective cohort study with 123 362 tests and 23 463 paired RT-PCR/antigen samples. BMJ Open Sport and Exercise Medicine 2021;7(2):e001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Harris 2021 {published data only}
- Harris DT, Badowski M, Jernigan B, Sprissler R, Edwards T, Cohen R, et al. SARS-CoV-2 rapid antigen testing of symptomatic and asymptomatic individuals on the University of Arizona campus. Biomedicines 2021;9(5):539. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Holzner 2021 {published data only}
- Holzner C, Pabst D, Anastasiou OE, Dittmer U, Manegold RK, Risse J, et al. SARS-CoV-2 rapid antigen test: fast-safe or dangerous? An analysis in the emergency department of an university hospital. Journal of Medical Virology 2021;93(9):5323-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Homza 2021 {published data only}
- Homza M, Zelena H, Janosek J, Tomaskova H, Jezo E, Kloudova A, et al. Five antigen tests for SARS-CoV-2: virus viability matters. Viruses 2021;13(4):684. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Homza 2021a {published data only}
- Homza M, Zelena H, Janosek J, Tomaskova H, Jezo E, Kloudova A, et al. COVID-19 antigen testing: better than we know? A test accuracy study. Infectious Diseases (London) 2021;53(9):661-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kanji 2021 {published data only}
- Kanji JN, Proctor DT, Stokes W, Berenger BM, Silvius J, Tipples G, et al. Multicentre postimplementation assessment of the positive-predictive value of SARS-CoV-2 antigen-based point-of-care tests used for asymptomatic screening of continuing care staff. Journal of Clinical Microbiology 2021;59(11):e0141121. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kenyeres 2021 {published data only}
- Kenyeres B, Ánosi N, Bányai K, Mátyus M, Orosz L, Kiss A, et al. Comparison of four PCR and two point of care assays used in the laboratory detection of SARS-CoV-2. Journal of Virology Methods 2021:114165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kim 2021 {published data only}
- Kim D, Lee J, Bal J, Seo SK, Chong CK, Lee JH, et al. Development and clinical evaluation of an immunochromatography-based rapid antigen test (GenBody™ COVAG025) for COVID-19 diagnosis. Viruses 2021;13(5):796. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kiro 2021 {published data only}
- Kiro VV, Gupta A, Singh P, Sharad N, Khurana S, Prakash S, et al. Evaluation of COVID-19 antigen fluorescence immunoassay test for rapid detection of SARS-CoV-2. Journal of Global Infectious Diseases 2021;13(2):91-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kiyasu 2021 {published data only}
- Kiyasu Y, Takeuchi Y, Akashi Y, Kato D, Kuwahara M, Muramatsu S, et al. Prospective analytical performance evaluation of the QuickNavi-COVID19 Ag for asymptomatic individuals. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Klein 2021 {published data only}
- Klein JA, Kruger LJ, Tobian F, Gaeddert M, Lainati F, Schnitzler P, et al. Head-to-head performance comparison of self-collected nasal versus professional-collected nasopharyngeal swab for a WHO-listed SARS-CoV-2 antigen-detecting rapid diagnostic test. Medical Microbiology and Immunology 2021;210(4):181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Koeleman 2021 {published data only}
- Koeleman JG, Brand H, Man SJ, Ong DS. Clinical evaluation of rapid point-of-care antigen tests for diagnosis of SARS-CoV-2 infection. European Journal of Clinical Microbiology & Infectious Disease 2021;40(9):1975-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kolwijck 2021 {published data only}
- Kolwijck E, Brouwers-Boers M, Broertjes J, Van Heeswijk K, Runderkamp N, Meijer A, et al. Validation and implementation of the Panbio COVID-19 Ag rapid test for the diagnosis of SARS-CoV-2 infection in symptomatic hospital healthcare workers. Infection Prevention in Practice 2021;3(2):100142. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Korenkov 2021 {published data only}
- Korenkov M, Poopalasingam N, Madler M, Vanshylla K, Eggeling R, Wirtz M, et al. Reliable assessment of in vitro SARS-CoV-2 infectivity by a rapid antigen detection test. medRxiv [Preprint] 2021. [DOI: ] [Google Scholar]
Kritikos 2021 {published data only}
- Kritikos A, Caruana G, Brouillet R, Miroz JP, Abed-Maillard S, Stieger G, et al. Sensitivity of rapid antigen testing and RT-PCR performed on nasopharyngeal swabs versus saliva samples in COVID-19 hospitalized patients: results of a prospective comparative trial (RESTART). medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kumar 2021a {published data only}
- Kumar A, Kunjukutty R, Thaha A, Srikumar S, Madhusoodanan H, David S, et al. Universal screening for SARS-CoV-2 in pregnant women using a combination of antigen and RT-PCR testing. Le Infezioni in Medicina 2021;29(2):294-6. [PubMed] [Google Scholar]
Kumar 2021b {published data only}
- Kumar KK, Sampritha UC, Maganty V, Prakash AA, Basumatary J, Adappa K, et al. Pre-operative SARS-CoV-2 rapid antigen test and reverse transcription polymerase chain reaction: a conundrum in surgical decision making. Indian Journal of Ophthalmology 2021;69(6):1560-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kurihara 2021 {published data only}
- Kurihara Y, Kiyasu Y, Akashi Y, Takeuchi Y, Narahara K, Mori S, et al. The evaluation of a novel digital immunochromatographic assay with silver amplification to detect SARS-CoV-2. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kweon 2021 {published data only}
- Kweon OJ, Lim YK, Kim HR, Choi Y, Kim MC, Choi SH, et al. Evaluation of rapid SARS-CoV-2 antigen tests, AFIAS COVID-19 Ag and ichroma COVID-19 Ag, with serial nasopharyngeal specimens from COVID-19 patients. PLoS One 2021;16(4):e0249972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Landaas 2021 {published data only}
- Landaas ET, Storm ML, Tollånes MC, Barlinn R, Kran AB, Bragstad K, et al. Diagnostic performance of a SARS-CoV-2 rapid antigen test in a large, Norwegian cohort. Journal of Clinical Virology 2021;137:104789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Lee 2021 {published data only}
- Lee J, Kim SY, Huh HJ, Kim N, Sung H, Lee H, et al. Clinical performance of the STANDARD Q COVID-19 rapid antigen test and simulation of its real-world application in Korea. Annals of Laboratory Medicine 2021;41(6):588-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Le Goff 2021 {published data only}
- Le Goff J, Kerneis S, Elie C, Mercier D, Gastli N, Choupeaux L, et al. Evaluation of saliva molecular point of care for detection of SARS-CoV-2 in ambulatory care. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Leixner 2021 {published data only}
- Leixner G, Voill-Glaninger A, Bonner E, Kreil A, Zadnikar R, Viveiros A. Evaluation of the AMP SARS-CoV-2 rapid antigen test in a hospital setting. International Journal of Infectious Diseases 2021;108:353-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Lindner 2021 {published data only}
- Lindner AK, Krüger LJ, Nikolai O, Klein JAF, Rössig H, Schnitzler P, et al. SARS-CoV-2 variant of concern B.1.1.7: diagnostic accuracy of three antigen-detecting rapid tests. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lv 2021 {published data only}
- Lv Y, Ma Y, Si Y, Zhu X, Zhang L, Feng H, et al. Rapid SARS-CoV-2 antigen detection potentiates early diagnosis of COVID-19 disease. Bioscience Trends 2021;15(2):93-9. [DOI] [PubMed] [Google Scholar]
Mack 2021 {published data only}
- Mack CD, Osterholm M, Wasserman EB, Petruski-Ivleva N, Anderson DJ, Myers E, et al. Optimizing SARS-CoV-2 surveillance in the United States: insights from the national football league occupational health program. Annals of Internal Medicine 2021;174(8):1081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Matsuda 2021 {published data only}
- Matsuda EM, Campos IB, Oliveira IP, Colpas DR, Carmo A, Brigido LF. Field evaluation of COVID-19 antigen tests versus RNA based detection: potential lower sensitivity compensated by immediate results, technical simplicity, and low cost. Journal of Medical Virology 2021;93(7):4405-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
McKay 2021 {published data only}
- McKay SL, Tobolowsky FA, Moritz ED, Hatfield KM, Bhatnagar A, LaVoie SP, et al. Performance evaluation of serial SARS-CoV-2 rapid antigen testing during a nursing home outbreak. Annals of Internal Medicine 2021;174(7):945-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Muhi 2021 {published data only}
- Muhi S, Tayler N, Hoang T, Ballard SA, Graham M, Rojek A, et al. Multi-site assessment of rapid, point-of-care antigen testing for the diagnosis of SARS-CoV-2 infection in a low-prevalence setting: a validation and implementation study. Lancet Regional Health – Western Pacific 2021;9:100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Nordgren 2021 {published data only}
- Nordgren J, Sharma S, Olsson H, Jamtberg M, Falkeborn T, Svensson L, et al. SARS-CoV-2 rapid antigen test: high sensitivity to detect infectious virus. Journal of Clinical Virology 2021;140:104846. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Norizuki 2021 {published data only}
- Norizuki M, Hachiya M, Motohashi A, Moriya A, Mezaki K, Kimura M, et al. Effective screening strategies for detection of asymptomatic COVID-19 travelers at airport quarantine stations: Exploratory findings in Japan. Global Health Medicine 2021;3(2):107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Oh 2021 {published data only}
- Oh SM, Jeong H, Chang E, Choe PG, Kang CK, Park WB, et al. Clinical application of the STANDARD Q COVID-19 Ag test for the detection of SARS-CoV-2 infection. Journal of Korean Medical Science 2021;36(14):e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Orsi 2021 {published data only}
- Orsi A, Pennati BM, Bruzzone B, Ricucci V, Ferone D, Barbera P, et al. On-field evaluation of a ultra-rapid fluorescence immunoassay as a frontline test for SARS-COV-2 diagnostic. Journal of Virology Methods 2021;295:114201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Osmanodja 2021 {published data only}
- Osmanodja B, Budde K, Zickler D, Naik MG, Hofmann J, Gertler M, et al. Accuracy of a novel SARS-CoV-2 antigen-detecting rapid diagnostic test from standardized self-collected anterior nasal swabs. Journal of Clinical Medicine 2021;10(10):2099. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Perez‐Garcia 2021 {published data only}
- Perez-Garcia F, Romanyk J, Moya Gutierrez H, Labrador Ballestero A, Perez Ranz I, Gonzalez Arroyo J, et al. Comparative evaluation of Panbio and SD Biosensor antigen rapid diagnostic tests for COVID-19 diagnosis. Journal of Medical Virology 2021;93(9):5650-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Qahtani 2021 {published data only}
- Qahtani MA, Yang CWT, Lazosky L, Li X, D'Cruz J, Romney MG, et al. SARS-CoV-2 rapid antigen testing for departing passengers at Vancouver International Airport. Journal of Travel Medicine 2021;28(7):10.1093/jtm/taab085. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Revollo 2021 {published data only}
- Revollo B, Blanco I, Soler P, Toro J, Izquierdo-Useros N, Puig J, et al. Same-day SARS-CoV-2 antigen test screening in an indoor mass-gathering live music event: a randomised controlled trial. Lancet Infectious Diseases 2021;21(10):1365-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Reza 2021 {published data only}
- Reza S, Corentin D, Te-Din H, Pierre B, Stéphanie E, Isaline W, et al. Rapid COVID-19 antigenic tests: usefulness of a modified method for diagnosis. Journal of Medical Virology 2021;93(9):5655-9. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Seynaeve 2021 {published data only}
- Seynaeve Y, Heylen J, Fontaine C, Maclot F, Meex C, Diep AN, et al. Evaluation of two rapid antigenic tests for the detection of SARS-CoV-2 in nasopharyngeal swabs. Journal of Clinical Medicine 2021;10(13):2774. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Shah 2021 {published data only}
- Shah MM, Salvatore PP, Ford L, Kamitani E, Whaley MJ, Mitchell K, et al. Performance of repeat BinaxNOW severe acute respiratory syndrome coronavirus 2 antigen testing in a community setting, Wisconsin, November 2020-December 2020. Clinical Infectious Diseases 2021;73 Suppl 1:S54-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Shaikh 2021 {published data only}
- Shaikh N, Friedlander EJ, Tate PJ, Liu H, Chang CH, Wells A, et al. Performance of a rapid SARS-CoV-2 antigen detection assay in symptomatic children. Pediatrics 2021;148(3):e2021050832. [DOI: ] [DOI] [PubMed] [Google Scholar]
Smith 2021a {published data only}
- Smith RD, Johnson JK, Clay C, Girio-Herrera L, Stevens D, Abraham M, et al. Clinical evaluation of Sofia rapid antigen assay for detection of severe acute respiratory syndrome coronavirus 2 among emergency department to hospital admissions. Infection Control & Hospital Epidemiology 2021 Jun 24 [Epub ahead of print]. [DOI: 10.1017/ice.2021.281] [DOI] [PMC free article] [PubMed] [Google Scholar]
Smits 2021 {published data only}
- Smits F, Torensma B, Groenwold R, Hall ML, Van Burgel N, Jansen R, et al. The SARS-CoV-2 antigen test is not accurate enough. Huisarts en Wetenschap 2021:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Sood 2021 {published data only}
- Sood N, Shetgiri R, Rodriguez A, Jimenez D, Treminino S, Daflos, A et al. Evaluation of the Abbott BinaxNOW rapid antigen test for SARS-CoV-2 infection in children: implications for screening in a school setting. PLoS One 2021;16(4):e0249710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Sterbenc 2021 {published data only}
- Sterbenc A, Tomic V, Bidovec Stojkovic U, Vrankar K, Rozman A, Zidarn M. Usefulness of rapid antigen testing for SARS-CoV-2 screening of healthcare workers: a pilot study. International Journal of Clinical and Experimental Medicine 2021 May 22 [Epub ahead of print]. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Suliman 2021 {published data only}
- Suliman S, Matias WR, Fulcher IR, Molano FJ, Collins S, Uceta E, et al. Evaluation of the Access Bio CareStart rapid SARS-CoV-2 antigen test in asymptomatic individuals tested at a community mass-testing program in Western Massachusetts. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Terpos 2021 {published data only}
- Terpos E, Ntanasis-Stathopoulos I, Skvarč M. Clinical application of a new SARS-CoV-2 antigen detection kit (colloidal gold) in the detection of COVID-19. Diagnostics (Basel) 2021;11(6):995. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Thakur 2021 {published data only}
- Thakur P, Saxena S, Manchanda V, Rana N, Goel R, Arora R. Utility of antigen-based rapid diagnostic test for detection of SARS-CoV-2 virus in routine hospital settings. Laboratory Medicine 2021;52(6):e154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Thell 2021 {published data only}
- Thell R, Kallab V, Weinhappel W, Mueckstein W, Heschl L, Heschl M, et al. Evaluation of a novel, rapid antigen detection test for the diagnosis of SARS-CoV-2. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Trobajo‐Sanmartin 2021 {published data only}
- Trobajo-Sanmartin C, Navascues A, Miqueleiz A, Ezpeleta C. Evaluation of the rapid antigen test CerTest SARS-CoV-2 as an alternative COVID-19 diagnosis technique. Infectious Diseases (London) 2021;53(9):730-2. [DOI] [PubMed] [Google Scholar]
Uwamino 2021 {published data only}
- Uwamino Y, Nagata M, Aoki W, Nakagawa T, Inose R, Yokota H, et al. Accuracy of rapid antigen detection test for nasopharyngeal swab specimens and saliva samples in comparison with RT-PCR and viral culture for SARS-CoV-2 detection. Journal of Infection and Chemotherapy 2021;27(7):1058-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Van Honacker 2021 {published data only}
- Van Honacker E, Van Vaerenbergh K, Boel A, De Beenhouwer H, Leroux-Roels I, Cattoir L. Comparison of five SARS-CoV-2 rapid antigen detection tests in a hospital setting and performance of one antigen assay in routine practice: a useful tool to guide isolation precautions? Journal of Hospital Infection 2021;114:144-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Wagenhäuser 2021 {published data only}
- Wagenhäuser I, Knies K, Rauschenberger V, Eisenmann M, McDonogh M, Petri N, et al. Clinical performance evaluation of SARS-CoV-2 rapid antigen testing in point of care usage in comparison to RT-qPCR. EBioMedicine 2021;69:103455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Yin 2021 {published data only}
- Yin N, Debuysschere C, Decroly M, Bouazza FZ, Collot V, Martin C, et al. SARS-CoV-2 diagnostic tests: algorithm and field evaluation from the near patient testing to the automated diagnostic platform. Frontiers in Medicine (Lausanne) 2021;8:650581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Young 2021a {published data only}
- Young BC, Eyre DW, Kendrick S, White C, Smith S, Beveridge G, et al. Daily testing for contacts of individuals with SARS-CoV-2 infection and attendance and SARS-CoV-2 transmission in English secondary schools and colleges: an open-label, cluster-randomised trial. Lancet 2021;398(10307):1217-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional references
Arevalo‐Rodriguez 2020a
- Arevalo-Rodriguez I, Buitrago-Garcia D, Simancas-Racines D, Zambrano-Achig P, Del Campo R, Ciapponi A, et al. False-negative results of initial RT-PCR assays for COVID-19: a systematic review. PLoS One 2020;15(12):e0242958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Basile 2021
- Basile L, Guadalupe-Fernández V, Valdivia Guijarro M, Martinez Mateo A, Ciruela Navas P, Mendioroz Peña J, et al. Diagnostic performance of Ag-RDTs and NAAT for SARS-CoV2 identification in symptomatic patients in Catalonia. Viruses 2021;13(5):908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Behera 2021
- Behera BC, Mishra RR, Thatoi H. Recent biotechnological tools for diagnosis of corona virus disease: a review. Biotechnology Progress 2021;37(1):e3078. [DOI] [PubMed] [Google Scholar]
Bekliz 2021
- Bekliz M, Adea K, Essaidi-Laziosi M, Escadafal C, Sacks JA, Kaiser L, et al. Analytical performance of eleven SARS-CoV-2 antigen-detecting rapid tests for Delta variant. medRxiv [Preprint] 2021. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Binnicker 2020
- Binnicker MJ. Challenges and controversies to testing for COVID-19. Journal of Clinical Microbiology 2020;58(11):e01695-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Bossuyt 2015
- Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ 2015;351:h5527. [DOI: 10.1136/bmj.h5527] [PMID: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Brümmer 2021
- Brümmer LE, Katzenschlager S, Gaeddert M, Erdmann C, Schmitz S, Bota M, et al. Accuracy of novel antigen rapid diagnostics for SARS-CoV-2: a living systematic review and meta-analysis. PLoS Medicine 2021;18(8):e1003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Canas 2021
- Canas LS, Sudre CH, Capdevila Pujol J, Polidori L, Murray B, Molteni E, et al. Early detection of COVID-19 in the UK using self-reported symptoms: a large-scale, prospective, epidemiological surveillance study. Lancet Digital Health 2021;3(9):e587-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Carter 2020
- Carter LJ, Garner LV, Smoot JW, Li Y, Zhou Q, Saveson CJ, et al. Assay techniques and test development for COVID-19 diagnosis. ACS Central Science 2020;6(5):591-605. [DOI: 10.1021/acscentsci.0c00501] [DOI] [PMC free article] [PubMed] [Google Scholar]
CDC 2021
- Centers for Disease Control and Prevention (CDC). Interim guidance for antigen testing for SARS-CoV-2; 9 Sep 2021. Available from: www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html.
Cevik 2021
- Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe 2021;2(1):e13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Chen 2020
- Chen Q, He Z, Mao F, Pei H, Cao H, Liu X. Diagnostic technologies for COVID-19: a review. RSC Advances 2020;10(58):35257-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Cheng 2020
- Cheng MP, Yansouni CP, Basta NE, Desjardins M, Kanjilal S, Paquette K, et al. Serodiagnostics for severe acute respiratory syndrome-related coronavirus-2: a narrative review. Annals of Internal Medicine 2020;15(173):450-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Chu 2006
- Chu H, Cole SR. Bivariate meta-analysis of sensitivity and specificity with sparse data: a generalized linear mixed model approach. Journal of Clinical Epidemiology 2006;59(12):1331-2. [DOI] [PubMed] [Google Scholar]
Chung 2021
- Chung E, Chow EJ, Wilcox NC, Burstein R, Brandstetter E, Han PD, et al. Comparison of symptoms and RNA levels in children and adults with SARS-CoV-2 infection in the community setting. JAMA Pediatrics 2021;175(10):e212025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Clopper 1934
- Clopper CJ, Pearson ES. The use of confidence or fiducial limits Illustrated in the case of the binomial. Biometrika 1934;26:404-13. [Google Scholar]
COVID‐19 Open Access Project 2021
- COVID-19 Open Access Project. Living Evidence on COVID-19. Available at ispmbern.github.io/covid-19/living-review/ 2021.
Covidence [Computer program]
- Covidence. Version accessed 27 April 2020. Melbourne, Australia: Veritas Health Innovation, 2020. Available at covidence.org.
Crozier 2021
- Crozier A, Dunning J, Rajan S, Semple MG, Buchan IE. Could expanding the COVID-19 case definition improve the UK’s pandemic response? BMJ 2021;374:n1625. [DOI] [PubMed] [Google Scholar]
Deeks 2020a
- Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Leeflang MM, Spijker R, et al. Diagnosis of SARS-CoV-2 infection and COVID-19: accuracy of signs and symptoms; molecular, antigen, and antibody tests; and routine laboratory markers. Cochrane Database of Systematic Reviews 2020, Issue 4. Art. No: CD013596. [DOI: 10.1002/14651858.CD013596] [DOI] [Google Scholar]
Deeks 2020b
- Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Spijker R, Taylor-Phillips S, et al. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database of Systematic Reviews 2020, Issue 6. Art. No: CD013652. [DOI: 10.1002/14651858.CD013652] [DOI] [PMC free article] [PubMed] [Google Scholar]
Deeks 2022
- Deeks JJ, Singanayagam A, Houston H, Sitch AJ, Hakki S, Dunning J, et al. SARS-CoV-2 antigen lateral flow tests for detecting infectious people: linked data analysis. BMJ 2022;376:e066871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Evans 2021
- Evans D, Cowen S, Kammel M, O’Sullivan DM, Stewart G, Grunert H-P, et al. The dangers of using Cq to quantify nucleic acid in biological samples: a lesson from COVID-19. Clinical Chemistry 2021;68(1):153-62. [DOI] [PubMed] [Google Scholar]
FDA 2021
- FDA. Omicron variant: impact on antigen diagnostic tests (as of 12/28/2021). Available at www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-covid-19-tests#omicronvariantimpact.
FIND 2022a
- FIND. SARS-COV-2 diagnostic pipeline. www.finddx.org/covid-19/pipeline/ (accessed 5 January 2022).
FIND 2022b
- FIND. Impact of variants on COVID-19 tests. www.finddx.org/covid-19/impact-of-variants-on-covid19-tests/ (accessed 6 January 2022).
Green 2020
- Green K, Graziadio S, Turner P, Fanshawe T, Allen J, on behalf of the Oxford COVID-19 Evidence Service Team Centre. Molecular and antibody point-of-care tests to support the screening, diagnosis and monitoring of COVID-19. Available at www.cebm.net/oxford-covid-19/ (7 April 2020) 2020.
Gurdasani 2021
- Gurdasani D, Ziauddeen H, Greenhalgh T, Roberts A, Yates K, Haque Z, et al. Daily contact testing trials in schools are unethical and extending them to include the delta variant puts everyone at risk. blogs.bmj.com/bmj/2021/06/17/daily-contact-testing-trials-in-schools-are-unethical-and-extending-them-to-include-the-delta-variant-puts-everyone-at-risk/ 17 Jun 2021.
Healy 2020
- Healy B, Khan A, Metezai H, Blyth I, Asad H. The impact of false positive COVID-19 results in an area of low prevalence. Clinical Medicine Journal 2020:10.7861/clinmed.2020-0839. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Iacobucci 2020
- Iacobucci G, Coombes R. COVID-19: government plans to spend £100bn on expanding testing to 10 million a day. BMJ 2020;370:m3520. [DOI] [PubMed] [Google Scholar]
IDSA 2021
- Infectious Diseases Society of America, Assocation for Molecular Pathology. IDSA and AMP joint statement on the use of SARS-CoV-2 PCR cycle threshold (Ct) values for clinical decision-making; 12 March 2021. Available at www.idsociety.org/globalassets/idsa/public-health/covid-19/idsa-amp-statement.pdf.
Islam 2021
- Islam N, Ebrahimzadeh S, Salameh JP, Kazi S, Fabiano N, Treanor L, et al. Thoracic imaging tests for the diagnosis of COVID‐19. Cochrane Database of Systematic Reviews 2021, Issue 3. Art. No: CD013639. [DOI: 10.1002/14651858.CD013639.pub4] [DOI] [PMC free article] [PubMed] [Google Scholar]
Jaafar 2020
- Jaafar R, Aherfi S, Wurtz N, Grimaldier C, Van Hoang T, Colson P, et al. Correlation between 3790 quantitative polymerase chain reaction–positives samples and positive cell cultures, including 1941 severe acute respiratory syndrome coronavirus 2 isolates. Clinical Infectious Diseases 2020:ciaa1491. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Jiang 2021
- Jiang N, Tansukawat ND, Gonzalez-Macia L, Ates HC, Dincer C, Güder F, et al. Low-cost optical assays for point-of-care diagnosis in resource-limited settings. ACS Sensors 2021;6(6):2108-24. [DOI] [PubMed] [Google Scholar]
Karon 2021
- Karon BS, Donato LJ, Bridgeman AR, Blommel JH, Kipp B, Maus A, et al. Analytical sensitivity and specificity of four point of care rapid antigen diagnostic tests for SARS-CoV-2 using real-time quantitative PCR, quantitative droplet digital PCR, and a mass spectrometric antigen assay as comparator methods. Clinical Chemistry 2021;67(11):1545-53. [DOI] [PubMed] [Google Scholar]
Kozel 2017
- Kozel TR, Burnham-Marusich AR. Point-of-care testing for infectious diseases: past, present, and future. Journal of Clinical Microbiology 2017;55(8):2313-20. [DOI: 10.1128/JCM.00476-17] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kretzschmar 2020
- Kretzschmar ME, Rozhnova G, Bootsma MC, Van Boven M, Van de Wijgert JH, Bonten MJ. Impact of delays on effectiveness of contact tracing strategies for COVID-19: a modelling study. Lancet Public Health 2020;5(8):e452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kucirka 2020
- Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction–based SARS-CoV-2 tests by time since exposure. Annals of Internal Medicine 2020;173(4):262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Lai 2021
- Lai CK, Lam W. Laboratory testing for the diagnosis of COVID-19. Biochemical and Biophysical Research Communications 2021;538:226-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Larremore 2020
- Larremore DB, Wilder B, Lester E, Shehata S, Burke JM, Hay JA, et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 surveillance. medRxiv [Preprint] 2020. [DOI: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Leeflang 2021
- Leeflang MM, Bell K, Deeks JJ, Dinnes J, Doust J, Korevaar DA, et al. Electronic and animal noses for detecting SARS‐CoV‐2 infection. Cochrane Database of Systematic Reviews 2021, Issue 6. Art. No: CD015013. [DOI: 10.1002/14651858.CD015013] [DOI] [Google Scholar]
Lyngse 2021
- Lyngse FP, Mølbak K, Træholt Franck K, Nielsen C, Skov RL, Voldstedlund M, et al. Association between SARS-CoV-2 transmissibility, viral load, and age in households. medRxiv [Preprint] 2021. [DOI: ] [Google Scholar]
Madera 2021
- Madera S, Crawford E, Langelier C, Tran NK, Thornborrow E, Miller S, et al. Nasopharyngeal SARS-CoV-2 viral loads in young children do not differ significantly from those in older children and adults. Scientific Reports 2021;11(1):3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Mak 2021
- Mak GC, Lau SS, Wong KK, Chow NL, Lau CS, Lam ET, et al. Comparison of analytical sensitivity of the eight rapid antigen detection kits for detecting SARS-CoV-2 virus. Journal of Clinical Virology 2021;144:104994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Marks 2021
- Marks M, Millat-Martinez P, Ouchi D, Roberts CH, Alemany A, Corbacho-Monné M, et al. Transmission of COVID-19 in 282 clusters in Catalonia, Spain: a cohort study. Lancet Infectious Diseases 2021;21(5):629-36. [DOI: 10.1016/S1473-3099(20)30985-3] [DOI] [PMC free article] [PubMed] [Google Scholar]
Mayers 2020
- Mayers C, Baker K, Government Office for Science. Impact of false-positives and false-negatives in the UK’s COVID-19 RT-PCR testing programme. Available from assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/895843/S0519_Impact_of_false_positives_and_negatives.pdf 3 Jun 2020.
McInnes 2018
- McInnes MD, Moher D, Thombs BD, McGrath TA, Bossuyt PM, PRISMA-DTA Group. Preferred reporting items for a systematic review and meta-analysis of diagnostic test accuracy studies: the PRISMA-DTA Statement. JAMA 2018;319(4):388-96. [DOI: 10.1001/jama.2017.19163] [PMID: ] [DOI] [PubMed] [Google Scholar]
McInnes 2020
- McInnes M, Leeflang MM, Salameh J-P, McGrath T, Van der Pol CB, Frank RA, et al. Imaging tests for the diagnosis of COVID-19. Cochrane Database of Systematic Reviews 2020, Issue 6. Art. No: CD013639. [DOI: 10.1002/14651858.CD013639] [DOI] [PubMed] [Google Scholar]
Meyerowitz 2020
- Meyerowitz EA, Richterman A, Bogoch II, Low N, Cevik M. Towards an accurate and systematic characterisation of persistently asymptomatic infection with SARS-CoV-2. Lancet Infectious Diseases 2020;21(8):E163-29. [DOI: 10.1016/S1473-3099(20)30837-9] [DOI] [PMC free article] [PubMed] [Google Scholar]
NHS 2021
- NHS. Get tested for coronavirus (COVID-19). www.nhs.uk/conditions/coronavirus-covid-19/testing/get-tested-for-coronavirus/ (accessed 6 January 2022).
Noel 2021
- Noel RH. Speed meets sensitivity to limit infectious spread in rapid microfluidic antigen testing. Available at: www.mlo-online.com/diagnostics/article/21222539/speed-meets-sensitivity-to-limit-infectious-spread-in-rapid-microfluidic-antigen-testing; 19 May 2021.
ONS 2020
- Office for National Statistics. Coronavirus (COVID-19) infection survey, UK: 21 August 2020. Available at: ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/bulletins/coronaviruscovid19infectionsurveypilot/englandandwales21august2020.
Pai 2012
- Pai NP, Vadnais C, Denkinger C, Engel N, Pai M. Point-of-care testing for infectious diseases: diversity, complexity, and barriers in low- and middle-income countries. PLoS Medicine 2012;9(9):e1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Petersen 2021
- Petersen I, Crozier A, Buchan I, Mina MJ, Bartlett JW. Recalibrating SARS-CoV-2 antigen rapid lateral flow test relative sensitivity from validation studies to absolute sensitivity for indicating individuals shedding transmissible virus. Clinical Epidemiology 2021;13:935-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Reitsma 2005
- Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. Journal of Clinical Epidemiology 2005;58(10):982-90. [DOI: 10.1016/j.jclinepi.2005.02.022] [DOI] [PubMed] [Google Scholar]
Review Manager 2020 [Computer program]
- Review Manager 5 (RevMan 5). Version 5.4. Copenhagen: The Cochrane Collaboration, 2020.
Riley 2020
- Riley S, Ainslie KE, Eales O, Walters CE, Wang H, Atchison C, et al. Transient dynamics of SARS-CoV-2 as England exited national lockdown. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.08.05.20169078] [DOI] [Google Scholar]
RSS 2021
- Royal Statistical Society. Royal Statistical Society Diagnostic Tests Working Group Report. London: Royal Statistical Society, 2021. [Google Scholar]
Singanayagam 2020
- Singanayagam A, Patel M, Charlett A, Lopez BJ, Saliba V, Ellis J, et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Eurosurveillance 2020;25(32):2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Stata [Computer program]
- Stata. Version 17. College Station, TX, USA: StataCorp, 2021. Available at www.stata.com.
Stegeman 2020
- Stegeman I, Ochodo EA, Guleid F, Holtman G, Yang B, Cunningham J, et al. Routine laboratory testing to determine if a patient has COVID-19. Cochrane Database of Systematic Reviews 2020, Issue 11. Art. No: CD013787. [DOI: 10.1002/14651858.CD013787] [DOI] [PMC free article] [PubMed] [Google Scholar]
Struyf 2021
- Struyf T, Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Leeflang MM, et al. Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19. Cochrane Database of Systematic Reviews 2021, Issue 2. Art. No: CD013665. [DOI: 10.1002/14651858.CD013665.pub2] [DOI] [PMC free article] [PubMed] [Google Scholar]
Takwoingi 2017
- Takwoingi Y, Guo B, Riley RD, Deeks JJ. Performance of methods for meta-analysis of diagnostic test accuracy with few studies or sparse data. Statistical Methods in Medical Research 2017;26(4):1896-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Tian 2021
- Tian D, Lin Z, Kriner EM, Esneault DJ, Tran J, DeVoto JC, et al. Ct Values do not predict SARS-CoV-2 transmissibility in college students. Journal of Molecular Diagnostics 2021. [DOI: 10.1016/j.jmoldx.2021.05.012] [DOI] [PMC free article] [PubMed] [Google Scholar]
UK Department for Education 2022
- Department for Education. Schools COVID-19 operational guidance. January 2022. assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1044890/Schools_guidance_January_2022_.pdf (accessed 6 January 2022).
UK DHSC 2021a
- Department of Health and Social Care. Daily contact testing study; June 2021. www.gov.uk/guidance/daily-contact-testing-study.
UK DHSC 2021b
- UK Department of Health and Social Care, . Asymptomatic testing for SARS-CoV-2 using antigen-detecting lateral flow devices; July 2021. Available at assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/999866/asymptomatic-testing-for-SARS-CoV-2-using-antigen-detecting-lateral-flow-devices-evidence-from-performance-data-Oct-2020-to-May-2021.pdf.
UK HSA 2021a
- UK Health Security Agency. Guidance for contacts of people with confirmed coronavirus (COVID-19) infection who do not live with the person; 23 December 2021. Available at www.gov.uk/government/publications/guidance-for-contacts-of-people-with-possible-or-confirmed-coronavirus-covid-19-infection-who-do-not-live-with-the-person/guidance-for-contacts-of-people-with-possible-or-confirmed-coronavirus-covid-19-infection-who-do-not-live-with-the-person.
UK HSA 2021b
- UK Health Security Agency. SARS-CoV-2 variants of concern and variants under investigation in England. Technical briefing 32; 17 December 2021. Available at assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1042688/RA_Technical_Briefing_32_DRAFT_17_December_2021_2021_12_17.pdf.
Vogels 2020
- Vogels CB, Brito AF, Wyllie AL, Fauver JR, Ott IM, Kalinich CC, et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nature Microbiology 2020;5(10):1299-305. [DOI: 10.1038/s41564-020-0761-6] [DOI] [PMC free article] [PubMed] [Google Scholar]
Whiting 2011
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS‐2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine 2011;155(8):529-36. [DOI] [PubMed] [Google Scholar]
WHO 2018
- World Health Organization. Diagnostic assessment: invitro diagnostic medical devices (IVDs) used for the detection of high-risk human papillomavirus (HPV) genotypes in cervical cancer screening. Licence: CC BY-NC-SA 3.0 IGO; 3 April 2018. Available at apps.who.int/iris/handle/10665/272282.
WHO 2020a
- World Health Organization. Antigen-detection in the diagnosis of SARS-CoV-2 infection using rapid immunoassays. Interim guidance; 11 September 2020. Available at apps.who.int/iris/bitstream/handle/10665/334253/WHO-2019-nCoV-Antigen_Detection-2020.1-eng.pdf?sequence=1&isAllowed=y.
WHO 2020b
- World Health Organization. COVID-19 Target product profiles for priority diagnostics to support response to the COVID-19 pandemic v.1.0; 28 September 2020. Available at who.int/publications/m/item/covid-19-target-product-profiles-for-priority-diagnostics-to-support-response-to-the-covid-19-pandemic-v.0.1.
WHO 2020c
- World Health Organization. Laboratory testing of 2019 novel coronavirus (2019-nCoV) in suspected human cases: interim guidance; 19 March 2020. Available at www.who.int/publications/i/item/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117.
WHO 2020d
- World Health Organisation. Global surveillance for COVID-19 caused by human infection with COVID-19 virus: interim guidance; 20 March 2020. Available from apps.who.int/iris/bitstream/handle/10665/331506/WHO-2019-nCoV-SurveillanceGuidance-2020.6-eng.pdf.
Yang 2020a
- Yang Y, Yang M, Yuan J, Wang F, Wang Z, Li J, et al. Laboratory diagnosis and monitoring the viral shedding of SARS-CoV-2 infection. Innovation 2020;1(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
Yonker 2021
- Yonker LM, Boucau J, Regan J, Choudhary MC, Burns MD, Young N, et al. Virologic Features of Severe Acute Respiratory Syndrome Coronavirus 2 Infection in Children. Journal of Infectious Diseases 2021. [DOI: 10.1093/infdis/jiab509] [DOI] [PMC free article] [PubMed] [Google Scholar]
Zhao 2020
- Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, et al. Antibody Responses to SARS-CoV-2 in Patients With Novel Coronavirus Disease 2019. Clinical Infectious Diseases 2020;71(16):2027-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Zoe COVID Study 2021
- Zoe COVID Study. What are the new top 5 COVID symptoms?; 23 Jun 2021. Available from: covid.joinzoe.com/post/new-top-5-covid-symptoms.
References to other published versions of this review
Dinnes 2020
- Dinnes J, Deeks JJ, Adriano A, Berhane S, Davenport C, Dittrich S, et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database of Systematic Reviews 2020, Issue 8. Art. No: CD013705. [DOI: 10.1002/14651858.CD013705] [DOI] [PMC free article] [PubMed] [Google Scholar]
Dinnes 2021
- Dinnes J, Deeks JJ, Berhane S, Taylor M, Adriano A, Davenport C, et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database of Systematic Reviews 2021, Issue 3. Art. No: CD013705. [DOI: 10.1002/14651858.CD013705.pub2] [DOI] [PMC free article] [PubMed] [Google Scholar]