Abstract
Mycobacterium sp. strain PYR-1 degrades high-molecular-weight polycyclic hydrocarbons (PAHs) primarily through the introduction of both atoms of molecular oxygen by a dioxygenase. To clone the dioxygenase genes involved in PAH degradation, two-dimensional (2D) gel electrophoresis of PAH-induced proteins from cultures of Mycobacterium sp. strain PYR-1 was used to detect proteins that increased after phenanthrene, dibenzothiophene, and pyrene exposure. Comparison of proteins from induced and uninduced cultures on 2D gels indicated that at least six major proteins were expressed (105, 81, 52, 50, 43, and 13 kDa). The N-terminal sequence of the 50-kDa protein was similar to those of other dioxygenases. A digoxigenin-labeled oligonucleotide probe designed from this protein sequence was used to screen dioxygenase-positive clones from a genomic library of Mycobacterium sp. strain PYR-1. Three clones, each containing a 5,288-bp DNA insert with three genes of the dioxygenase system, were obtained. The genes in the DNA insert, from the 5′ to the 3′ direction, were a dehydrogenase, the dioxygenase small (β)-subunit, and the dioxygenase large (α)-subunit genes, arranged in a sequence different from those of genes encoding other bacterial dioxygenase systems. Phylogenetic analysis showed that the large α subunit did not cluster with most of the known α-subunit sequences but rather with three newly described α subunits of dioxygenases from Rhodococcus spp. and Nocardioides spp. The genes from Mycobacterium sp. strain PYR-1 were subcloned and overexpressed in Escherichia coli with the pBAD/ThioFusion system. The functionality of the genes for PAH degradation was confirmed in a phagemid clone containing all three genes, as well as in plasmid subclones containing the two genes encoding the dioxygenase subunits.
Polycyclic aromatic hydrocarbons (PAHs), ubiquitous in nature, are formed by a variety of biotic and abiotic reactions. The major sources in the environment are the combustion of organic matter and the processing and use of fossil fuels. Some PAHs are highly carcinogenic, genotoxic, and cytotoxic (4, 7). Many PAHs, including anthracene, phenanthrene, acenapthene, acenaphthylene, fluoranthene, pyrene, benz[a]anthracene, and benzo[a]pyrene, are on the United States Environmental Protection Agency's priority pollutant list (6). High-molecular-weight PAHs are thermodynamically stable and recalcitrant to microbial degradation, due to their aromatic nature, the stabilization afforded by multiple rings, and low aqueous solubility. A variety of bacterial species can rapidly degrade two- and three-ring PAHs, such as naphthalene, phenanthrene, and anthracene. However, high-molecular-weight PAHs, such as pyrene, chrysene, and benzo[a]pyrene, are metabolized by bacteria at much slower rates (17). Most research on enzymes involved in PAH metabolism and genetic regulation has been done with Pseudomonas and Sphingomonas species (5, 8, 10, 11, 18, 21, 22, 24, 28, 29). Mycobacterium, Rhodococcus, and Nocardioides species can also mineralize high-molecular-weight PAHs (4, 13, 15, 17, 19, 27), but little is known about the enzyme mechanisms and genes involved in the degradative pathways (27, 30). Recently, the molecular characterization of a phenanthrene dioxygenase from a Nocardioides sp. was reported (27).
Mycobacterium sp. strain PYR-1, which was isolated in our laboratory (14), is capable of mineralizing anthracene, fluoranthene, pyrene, 1-nitropyrene, phenanthrene, and benzo[a]pyrene (14–16, 19, 20, 25, 26). Analysis of metabolites and oxygen 18 incorporation experiments indicate that the initial oxidation of PAHs in Mycobacterium sp. strain PYR-1 is primarily catalyzed by a dioxygenase attack on the aromatic ring to form a cis-dihydrodiol (15, 25). A mono-oxygenase attack leading to formation of a trans-dihydrodiol has also been reported (15, 25). The mechanism of oxidation is unique since Mycobacterium sp. strain PYR-1 has both mono- and dioxygenase to catalyze the initial attack on the PAH. To the best of our knowledge, the genes from Mycobacterium spp. capable of degrading PAHs have not been described.
Our present research goal is to determine the biochemical, molecular, and genetic bases for the metabolism of PAHs by Mycobacterium sp. strain PYR-1. Recently, we reported the induction of a catalase-peroxidase in PAH-induced Mycobacterium sp. strain PYR-1 cultures and its importance in PAH metabolism (31). In this paper, we characterize the genes encoding an aromatic dioxygenase, the enzyme involved in the first step of PAH catabolism in Mycobacterium sp. strain PYR-1.
MATERIALS AND METHODS
Bacterial strains, plasmids, and chemicals.
All bacterial strains, vectors, and plasmids used in this study are listed in Table 1. Culture media, such as Luria-Bertani (LB) medium, were prepared according to the manufacturer's instructions. Pyrene, phenanthrene, and dibenzothiophene were purchased from Chem Service (Media, Pa.). All of the PAHs and related compounds were >99% pure. Other chemicals were of the highest purity commercially available.
TABLE 1.
Bacterial strains and plasmids
| Strain, phage, plasmid, or vector | Description | Source or reference |
|---|---|---|
| Strains and phage | ||
| Mycobacterium sp. strain PYR-1 | Mineralizes PAHs, such as fluoranthene, pyrene, phenanthrene, etc. | 14–16 |
| E. coli XL1-Blue MRF′ | Lac [F′ proAB laclqZΔM15 Tn10 (Tetr)] | Stratagene |
| E. coli XLOLR | Lac [F′ proAB laclqZΔM15 Tn10 (Tetr)] Su−(nonsuppressing) λr | Stratagene |
| ExAssist helper phage | For in vivo excision of the pBK-CMV phagemid from ZAP Express vector with E. coli XLOLR | Stratagene |
| E. coli TOP10 | One Shot-competent cells; F′ mcrA Δ(mrr-hsdRMS-mcrBC)Φ80lacZΔM15 Δ(ara-leu)7697 | Invitrogen |
| E. coli JM 109 | Competent cells; F′ proAB laclqZΔM15 | Promega |
| Plasmids and vectors | ||
| Zap Express vector | Lambda vector; prokaryotic and eukaryotic expression, in vivo excision of pBK-CMV vector | Stratagene |
| pBK-CMV vector | Neor Kanr; colE1 origin; lacZ; CMVa promoter | Stratagene |
| My6-pBK-CMV phagemid | pBK-CMV phagemid with 5.2-kb insert from Mycobacterium sp. strain PYR-1 | This study |
| pBAD/Thio-TOPO vector | Ampr; pMB1 origin; Six-His-thioredoxin ORF; arabinose induced | Invitrogen |
| nidA-pBAD/Thio plasmid | pBAD/ThioFusion with nidA gene | This study |
| nid(B+A)-pBAD/Thio plasmid | pBAD/ThioFusion with nidB plus nidA genes | This study |
| nidD-pBAD/Thio plasmid | pBAD/ThioFusion with nidD gene | This study |
CMV, cytomegalovirus.
Southern hybridization and plasmid analysis.
The total genomic DNA of Mycobacterium sp. strain PYR-1 was screened for genetic homology with various aromatic hydrocarbon-degrading dioxygenase genes by Southern hybridization, using either radioactive 32P-labeled probes or nonradioactive digoxigenin probes. The clones containing large-subunit dioxygenase genes that were used were from the following strains: Pseudomonas sp. strain LB400 (biphenyl degradation) (10), Pseudomonas sp. strain NCIB9816 (naphthalene degradation) (28), Pseudomonas putida strain F1 (toluene degradation) (32), P. putida strain OU83 (biphenyl degradation) (21), Comamonas testosteroni strain GZ39 (phenanthrene degradation) (12), Sphingomonas yanoikuyae strain B1 (m-xylene, biphenyl, naphthalene, anthracene, and phenanthrene degradation) (33). Mycobacterium sp. strain PYR-1 cells were analyzed for plasmids by pulsed-field gel electrophoresis.
Measurement of pyrene metabolism.
The ability of Mycobacterium sp. strain PYR-1 to remove pyrene from the culture medium was monitored spectrophotometrically (31). Complete solubilization of PAHs was accomplished by mixing 2 volumes of culture aliquot with 1 volume of dimethyl sulfoxide before centrifugation at 16,000 × g for 10 min. The absorbance of the supernatant fluids obtained from duplicate cultures was measured at 335 nm for pyrene. Supernatants from cultures receiving equivalent volumes of dimethyl sulfoxide and a methanol carrier were used as blanks. Abiotic removal of pyrene was checked by using boiled cell suspensions incubated similarly. Spectrophotometric results were confirmed by quantifying the amount of pyrene by reversed-phase high-performance liquid chromatography (HPLC) using a C18 column (3.9 by 300 mm). A linear gradient of 50 to 95% methanol in water was developed over 40 min at 1 ml/min. Pyrene was identified by comparing characteristic absorption spectra (from 200 to 400 nm) and retention times to those for authentic pyrene using a Waters 910 photodiode array detector with data display and by analysis using Waters, version 2.10, Millenium software.
Organism, growth conditions, and 2D gel electrophoresis analysis.
Mycobacterium sp. strain PYR-1 was grown in 125-ml Erlenmeyer flasks containing 30 ml of minimal basal salts (MBS) medium (14), supplemented with low levels (250 μg/liter) of peptone, yeast extract, and soluble starch at 30°C for 48 h with shaking at 150 rpm. To induce PAH-degrading enzymes (15), cultures were inoculated into four 2-liter Erlenmeyer flasks, each containing 800 ml of MBS, and a 5 μg/ml concentration of pyrene, phenanthrene, or dibenzothiophene dissolved in N,N-dimethylformamide (DMF) was added to each flask as described earlier (31). The control incubation mixtures (uninduced) contained Mycobacterium sp. strain PYR-1 in MBS medium with the same volume of DMF and no PAHs. Cultures were incubated at 30°C in a rotary shaker (150 rpm). The cells were harvested at 120 h by centrifugation (12,000 × g for 20 min at 4°C). The cells were washed three times with 10 mM Tris, pH 7.4, and then resuspended in 5 to 8 ml of the same buffer. Cell extracts were made by disruption by sonication for 15 min at 30 intervals, with an intensity of 60 (Sonics and Materials, Inc., Newtown, Conn.). Tergitol NP-40 (U.S. Biochemicals-Amersham, Cleveland, Ohio) was then added at a concentration of 1% (vol/vol). The cell debris was removed by centrifugation at 17,000 × g for 30 min, and the resulting supernatant was then centrifuged at 50,000 × g for 90 min at 4°C. Protein concentration in the supernatant was determined by a protein assay kit (Bio-Rad Laboratories, Hercules, Calif.). The soluble supernatant was used for two-dimensional (2D) gel (2D polyacrylamide gel) analysis.
The 2D gel technique was used to identify the PAH degradation-expressed proteins from PAH-induced and uninduced cultures of Mycobacterium sp. strain PYR-1 (31). 2D gel electrophoresis was performed by using Bio-Rad PROTEAN II xi 2-D Cell and Slab Cell in accordance with the instructions in Bio-Rad bulletin 1144. The gel concentration used in the second-dimension slab gel was 7.5%. The protein spots that were detected from PAH-induced but not from uninduced samples were transferred to a polyvinylidene fluoride (PVDF) membrane. The PVDF membranes were sent to Midwest Analytical, Inc. (St. Louis, Mo.) for N-terminal protein sequence analysis.
Construction of a genomic library of Mycobacterium sp. strain PYR-1.
Total genomic DNA of Mycobacterium sp. strain PYR-1 was digested with restriction enzyme BamHI and then separated by 1% agarose gel electrophoresis in Tris-acetate buffer at 6 V/cm for 3 h. The digested 0.7- to 10.0-kb DNA fragments were cut out from the agarose gel and purified by a gel extraction kit (Qiagen, Inc., Valencia, Calif.). A calf intestinal alkaline phosphatase-treated, BamHI-predigested ZAP Express vector (Stratagene Cloning Systems, La Jolla, Calif.) was ligated with the digested Mycobacterium sp. strain PYR-1 DNA fragments. The ligated DNA was packaged in bacteriophage λ using the Gigapack III gold packaging extract in accordance with the manufacturer's instructions (Stratagene).
Design of an oligonucleotide probe and genomic library screening.
The oligonucleotide probe (P50) was designed from the N-terminal amino acid sequence of a PAH-induced protein (50 kDa). The P50 sequence is 5′-ACNGARACNACNGGNACNGCNGAYGC. The N is G, A, T, or C; the R is A or G; the Y is T or C. The P50 probe was labeled with a digoxigenin (DIG) oligonucleotide 3′-end labeling kit in accordance with the manufacturer's instructions (Boehringer Mannheim Co., Indianapolis, Ind.). The genomic library of Mycobacterium sp. strain PYR-1 in phage was infected in E. coli strain XL1-Blue MRF′ and plated for plaque formation. The plaques were transferred to a nylon membrane and hybridized with a P50 probe. The plaques hybridizing to P50 were picked by toothpick and plated for purification and confirmation at a low plaque density (31). In vivo excision protocols were used to produce phagemid clones by using ExAssist helper phage with strain XLOLR (Stratagene Cloning Systems). A Qiaprep spin miniprep kit (Qiagen) was used for phagemid or plasmid preparation. The phagemid (1 μl of a 0.5-μg/μl solution) was applied to a nylon membrane for dot blot hybridization with the DIG-labeled P50 probe for confirmation. The hybridization conditions were the same as those for the plaque lift hybridization procedure.
DNA sequencing.
The phagemid was sequenced first by using primers Zap-F and Zap-R, which are located on the phagemid vector upstream and downstream from the insert. The Zap-F primer was 5′-CACAGGAAACAGCTATGACC; the Zap-R primer was5′-CCGCTCTAGAAGTACTCTCG. The ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit (PE Applied Biosystems, Foster City, Calif.) was used for sequencing in accordance with the manufacturer's instructions. An automatic ABI Prism 310 sequencer was used for electrophoresis. After insert sequence information was obtained from both ends, new sequencing primers were designed from the sequences until all of the insert sequence had been sequenced from both strands. Oligonucleotide primers and probes were purchased from Universal DNA, Inc. (Tigard, Oreg.).
Phylogenetic analysis.
DNA sequence analysis, translation, and alignment with related genes and proteins were carried out using the computer programs Lasergene (DNASTAR, Inc., Madison, Wis.) and Align Plus (Scientific Educational Software, State Line, Pa). The GenBank program BLAST (1) was used to find similar genes and proteins. Phylogenetic analysis was performed by the PHYLIP, version 3.572c, package. The computer program Megalign (DNASTAR, Inc.) was used to construct a phylogenetic tree by comparison of closely related protein sequences.
Construction of plasmids for overexpression of the enzymes.
After the gene sequence was determined, each full gene from the clone was amplified by PCR. The fresh PCR product (1 to 4 μl) was ligated with 1 μl (10 ng) of pBAD/Thio-TOPO vector (Invitrogen, Carlsbad, Calif.) in a total volume of 5 μl for 5 min at room temperature. Stop solution (1 μl, supplied with the kit) was added to this mixture, and then 2 μl of this mixture was added to a vial of One Shot cells for transformation. The colonies with recombinant pBAD/ThioFusion plasmids were selected by plating the transformants on ampicillin (50 μg/ml)-LB plates. Positive clones, containing recombinant plasmids with the correct orientation, were screened by PCR with the forward primer plus a reverse primer located in the vector (pBAD reverse primer; provided in the Invitrogen kit).
Expression and purification of six-His-tagged recombinant protein.
The pBAD/ThioFusion clones were inoculated into LB medium containing ampicillin (50 μg/ml) and cultured to an A600 of ∼0.5. Arabinose was added to a final concentration of 0.02%. After 4 or 16 h of incubation, the cells were collected by centrifugation. The cells were disrupted by sonication (1-min bursts at 200 to 300 W followed by 1 min of cooling) for 10 min. The sonicated samples were centrifuged, and the supernatants were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The pellet was resuspended in buffer B (8 M urea, 0.1 M sodium phosphate, 0.01 M Tris, pH 8.0), and suspension was followed by three freeze-thaw cycles. The cleared supernatant (0.6 ml) was loaded on an Ni-nitrilotriacetic acid (NTA) column (Qiagen). The column was centrifuged for 2 min at 700 × g and then washed three times with 0.6 ml of buffer C (same as buffer B, but pH 6.3). The six-His-tagged recombinant protein was eluted from the column with two additions of 0.2 ml of buffer E (same as buffer B, but pH 4.5). The purified protein (confirmed by SDS-PAGE) was dialyzed in a Tube-O-Dialyzer (Geno Technology, Inc., St. Louis, Mo.) against urea buffer (8.0 M urea, 0.3 M NaCl, 50 mM sodium phosphate, pH 8.0).
Isolation and identification of pyrene metabolites.
Phagemid clone My6-pBK-CMV and negative-control plasmid pBK-CMV having no insert were grown in LB broth-kanamycin (25 μg/ml) overnight at 37°C and then diluted 1:10 with the same medium and cultured to an A600 of 0.5. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 2 mM, and cultures were incubated for 5 h. Cells were harvested by centrifugation (10,000 × g for 5 min) and washed with MBS medium. The cells were resuspended with 10 ml of MBS, and 10 μl of pyrene (stock solution [50 mg/ml] in dimethyl formamide) was added. The sample was shaken overnight at 30°C. For clones in plasmids, the cells were induced by arabinose (0.02%) for 5 h. The cells were collected, washed, resuspended, and incubated with pyrene. Cells were removed by centrifugation, and the supernatants were extracted three times with equal volumes of NaOH-washed ethyl acetate. The extracts were dried over anhydrous Na2SO4 and concentrated using a rotary evaporator at 30°C. The samples were resuspended in 100 μl of methanol, and 10-μl aliquots were subjected to HPLC analysis (13). The metabolites were characterized by HPLC mass spectrometry. In brief, the HPLC separation was performed using a GP40 pump (Dionex, Sunnyvale, Calif.) in line with a Spectra 100 UV detector (Thermo Sep Products, San Jose, Calif.) set at 254 nm. The metabolites were separated using an Inertsil ODS-3 column (5-μm particles; 4.6 mm by 25 cm; Phenomenex, Torrance, Calif.). The mobile phase was a 30-min linear gradient of methanol-water (from 45:55 to 95:5 [vol/vol]) at a flow rate of 1.0 ml/min. Mass spectra were acquired using a Platform single-quadrupole mass spectrometer (Micromass, Manchester, United Kingdom) equipped with an atmospheric-pressure chemical ionization interface. The total HPLC effluent from the UV detector was delivered into the atmospheric-pressure ion source through a heated nebulizer probe (400°C) using N2 as the probe and bath gas (275 liters/h) with an ion source temperature of 150°C. Positive and negative ions were acquired sequentially in full-scan mode (m/z, 100 to 500; 1.0-s cycle time). The spectra were obtained at both a low cone voltage (20 V) and a high cone voltage (70 V) to provide fragmentation information.
Nucleotide sequence accession numbers.
The nucleotide sequences were deposited in the GenBank/EMBL DNA database under accession no. AF249300, AF249301, and AF249302.
RESULTS
Homology studies with dioxygenase probes and plasmid analysis.
To analyze which of the genes for PAH degradation were similar to known genes of PAH metabolism, Southern hybridization of Mycobacterium sp. strain PYR-1 was performed with dioxygenase probes representing toluene, xylene, naphthalene, biphenyl, anthracene, and phenanthrene metabolism. We did not find any genetic homology with most of the gram-negative dioxygenase genes by Southern blotting experiments under low-stringency conditions capable of detecting 70% identity between total DNA (data not shown). These results suggest that genes for PAH degradation by Mycobacterium sp. strain PYR-1 are not closely related to other known genes with similar functions. The ability of bacteria to degrade aromatic hydrocarbons to less-toxic compounds is sometimes encoded on plasmids (7). Induced and uninduced Mycobacterium sp. strain PYR-1 cells were analyzed for plasmids by pulsed-field gel electrophoresis, but we were unable to detect any plasmids in either culture (data not shown). These results suggest that the genes responsible for PAH degradation in Mycobacterium sp. strain PYR-1 reside on the chromosome.
Protein profiles of Mycobacterium sp. strain PYR-1 grown with and without PAHs.
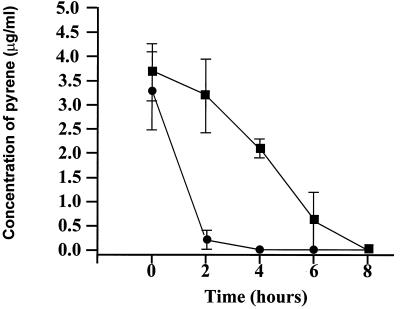
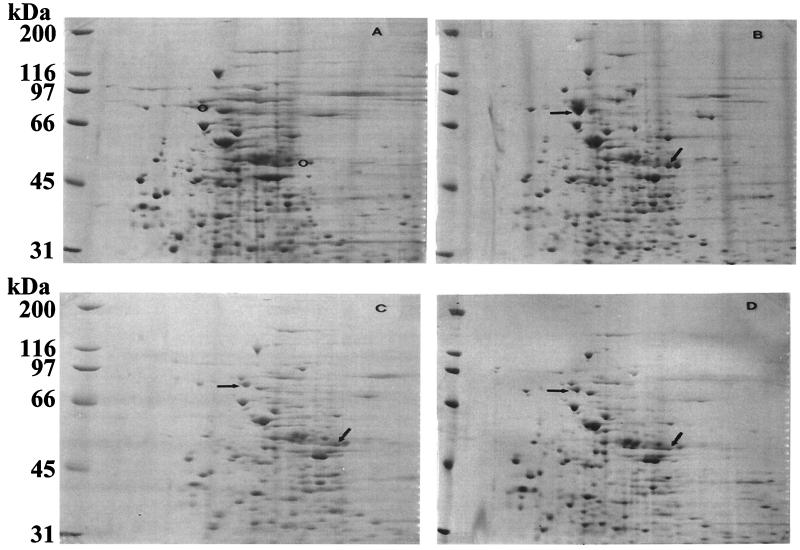
A time course analysis for the expression of pyrene-metabolizing proteins revealed induction 2 h after incubation with pyrene, and induction was followed by complete metabolism of pyrene in 8 h (Fig. 1). Comparisons concerning the presence and absence of proteins when the organism was grown in the presence and absence of PAHs were made by 2D SDS-PAGE analysis. Figure 2 shows the most prominent PAH-induced proteins from Mycobacterium sp. strain PYR-1 cultures versus uninduced cultures by 2D gel analysis. Exposure to pyrene, phenanthrene, and dibenzothiophene resulted in increased expression of at least six proteins (105, 81, 52, 50, 43, and 13 kDa) (Fig. 2). The N-terminal amino acid sequence data showed that the 81-kDa protein was a catalase-peroxidase, with significant homology to KatGII of Mycobacterium fortuitum (31). The role of catalase-peroxidase in PAH metabolism has been reported (31). Increased expression of a 50-kDa protein was also found in all of the PAH-induced cultures, compared to that in an uninduced culture (Fig. 2). The N-terminal sequence analysis of this protein gave a 12-residue sequence of TTETTGTADATD, which was similar to those of other known dioxygenases (27, 30). The other proteins that were induced in Mycobacterium sp. strain PYR-1 after growth on PAHs are currently being investigated and will be the subject of another study.
FIG. 1.
Time course of pyrene metabolism by Mycobacterium sp. strain PYR-1. Pyrene-induced (●) and uninduced (▪) cultures are shown.
FIG. 2.
2D gel for identification of PAH-induced proteins of Mycobacterium sp. strain PYR-1. (A) Uninduced proteins of strain PYR-1. (B) Pyrene-induced proteins of strain PYR-1. (C) Phenanthrene-induced proteins of strain PYR-1. (D) Dibenzothiophene-induced proteins of strain PYR-1. Arrows, PAH-induced proteins; circles, same positions in the uninduced sample.
Screening the genomic library of Mycobacterium sp. strain PYR-1 and DNA sequencing.
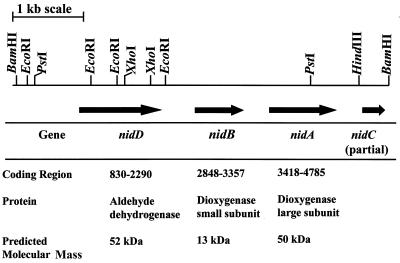
An oligonucleotide probe designed from the sequence of the P50 protein was used to search a genomic library of Mycobacterium sp. strain PYR-1. Several clones with a positive reaction to the P50 oligonucleotide probe were obtained. Phagemids were prepared for each clone and reconfirmed by dot blot hybridization using the DIG-labeled P50 probe (data not shown). DNA sequencing results for three clones showed that they had the same insert sequence. DNA sequence analysis of recombinant plasmid My6-pBK-CMV (Table 1) revealed that it consisted of one 5,288-bp insert, whose map is shown in Fig. 3. The translated peptide of the large subunit matched with the N-terminal amino acid sequence of the 50-kDa protein (12 residues [TTETTGTADATD]), which is the start of the nidA gene product. Three open reading frames (ORFs), exhibiting homology to those encoding polypeptides from several multicomponent dioxygenases, were identified. Based on sequence homology, predicted polypeptides from the three ORFs were designated naphthalene-inducible dioxygenase (iron-sulfur protein, large subunit), aromatic dioxygenase (iron-sulfur protein, small subunit), and aldehyde dehydrogenase; corresponding gene designations were nidA (1,461-bp ORF), nidB (510-bp ORF), and nidD (1,368-bp ORF), respectively. The order of these genes was different from that previously reported for aromatic ring dioxygenase genes (27, 30) (Fig. 3). We also found a partial sequence of the nidC gene in our clone. Translation of gene locations and calculated molecular mass values of the predicted polypeptides are shown in Fig. 3. Analysis of the translated ORF of the large subunit shows a Rieske center iron-sulfur-binding site, where the consensus sequence CXHRGX8GNX5CXYHG is conserved in terminal oxygenases (24). Furthermore, four histidines and three tyrosines near the middle of the polypeptide may contribute to a potential iron-binding site.
FIG. 3.
Restriction map, gene organization, and predicted nidABD gene products from the cloned 5,288-bp fragment from Mycobacterium sp. strain PYR-1.
Phylogenetic analysis.
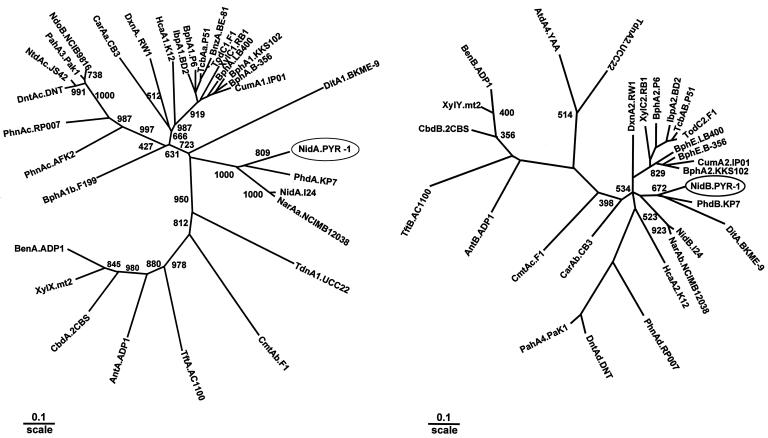
NidA showed significant but moderate sequence homology to the large subunits of known ring-hydroxylating dioxygenases. The amino acid sequences of the large subunits of PAH dioxygenases from Nocardioides sp. strain KP7 (PhdA) (27), Rhodococcus sp. strain NCIMB 12038 (NarAa) (AF082663), and strain I24 (NidA) (30) showed 50 to 42% identity with the NidA sequence of PYR-1, whereas the other known large subunits exhibited a sequence identity lower than 40%. The amino acid sequence alignment of NidA to known large subunits revealed a limited number of conserved amino acid residues, the highly conserved ones corresponding to those involved in the coordination of the [2Fe-2S] Rieske-type cluster and to the catalytic nonheme iron. The phylogenetic trees show the relationships between NidA and NidB and other closely related enzymes reported in the GenBank (Fig. 4). We also found that the NidB protein of Mycobacterium sp. strain PYR-1 was 56% similar to the Nocardioides sp. strain KP7 PhdB (27) and 43 to 46% similar to NidB of Rhodococcus sp. strain I24 (30) and RnoA4 of Rhodococcus sp. strain CIR2. The percentages of similarity of the NidB protein to NidB proteins of other bacteria were only 30 to 38% (Fig. 4B). We found that the NidD protein of Mycobacterium sp. strain PYR-1 was 36 to 37% similar to AldA of Streptomyces aureofaciens ccm3239 and Ald.SC9B5 of Streptomyces coelicolor A3.
FIG. 4.
Phylogenetic tree of NidA and NidB obtained from alignment with related proteins. The protein sequences of the 32 α subunits of ring-hydroxylating dioxygenases including NidA and 28 β subunits of ring-hydroxylating dioxygenases including NidB are classified. The multiple-alignment analysis was performed with the PHYLIP software package, and the phylogenetic unrooted tree was drawn by using TreeView. The numbers on some branches refer to the percentage confidence, estimated by a bootstrap analysis with 1,000 replications. Scale bar, percentage divergence. (A) α subunit (NidA). GenBank accession numbers are as follows: NidA-I24, indene, Rhodococcus sp. strain I24, AF121905; PhdA.KP7, phenanthrene, Nocardioides sp. strain KP7, AB017794; NarAa.NCIMB12038, naphthalene, Rhodococcus sp. strain NCIMB 12038, AF082663; Digox, DitA1.BKME-9, diterpenoid, Pseudomonas abietaniphila BKME-9, AF119621; IpbA1.BD2, isopropylbenzene, Rhodococcus erythropolis BD2, U24277; BphA1.P6, biphenyl, Rhodococcus globerulus P6, X80041; TcbAa.P51, chlorobenzene, Pseudomonas sp. strain P51, U15298; BnzA.BE-81, benzene, P. putida BE-81, M17904; TodC1.F1, toluene, P. putida F1, J04996; XylC1.RB1, substrate unknown, Cycloclasticus oligotrophus RB1, U51165; BphA.LB400, biphenyl, Burkholderia cepacia LB400, M86348; BphA1.KKS102, biphenyl, Pseudomonas sp. strain KKS102, D17319; BphA.B-356, biphenyl, C. testosteroni B-356, U47637; CumA1.IP01, cumene, Pseudomonas fluorescens IP01, D37828; TdnA1.UCC22, aniline, P. putida UCC22, D85415; CmtAb.F1, p-cumate, P. putida F1, U24215; TftA.AC1100, 2,4,5-trichlorophenoxyacetic acid, B. cepacia AC1100, U11420; AntA.ADP1, anthranilate, Acinetobacter calcoaceticus ADP1, AF071556; CbdA.2CBS, 2-halobenzoate, B. cepacia 2CBS, X79076; XylX.mt2, toluate, P. putida mt2, M64747; BenA.ADP1, benzoate, A. calcoaceticus ADP1, AF009224; HcaA1.K12, phenylpropionate, E. coli K-12, AE00340; DxnA1.RW1, dibenzo-p-dioxin, Sphingomonas sp. strain RW1, X72850; CarAa.CB3, carbazole, Sphingomonas sp. strain CB3, AF060489; NdoB.NCIB9816, naphthalene, P. putida NCIB9816, M23914; PahA3.PaK1, naphthalene, P. aeruginosa PaK1, D84146; NtdAc.JS42, 2-nitrotoluene, Pseudomonas sp. strain JS42, U49504; DntAc.DNT, 2,4-dinitrotoluene, Burkholderia sp. strain DNT, U62430; PhnAc.RP007, phenanthrene, Burkholderia sp. strain RP007, AF061751; PhnAc.AFK2, phenanthrene, Alcaligenes faecalis strain AFK2, AB024945; BphA1b.F199, Sphingomonas aromaticivorans strain F199, AF079317. (B) Aromatic dioxygenase small subunit (NidB). GenBank accession numbers are as follows: NidB.I24, AF121905; PhdB.KP7, AB017794; BphA2.KKS102, D17319; DitA.BKME-9, AF119621; NarAb.NCIMB12038, AF082663; CumA2.IP01, D37828; BphE.B-356, U47637; BphE.LB400, M86348; TodC2.F1, Y18245; TcbAB.P51, U15298; IbpA2.BD2, U24277; BphA2.P6, X80041; XylC2.RB1, U51165; DxnA2.RW1, X72850; PhnAd.RP007, AF061751; DntAd.DNT, U62430; PahA4.PaK1, D84146; CarAb.CB3, AF060489; CmtAc.F1, U24215; AntB.ADP1, AF071556; TftB.AC1100, U11420; CbdB.2CBS, X79076; XylY.mt2, M64747; BenB.ADP1, AF009224; AtdA4.YAA, AB008831; TdnA2.UCC22, D85415.
Expression and purification of the recombinant proteins.
To determine whether the nidAB genes actually encode the components of the PAH dioxygenase, the genes were introduced into E. coli. The nidA gene and the nidD genes, without the start codon and the stop codon, were amplified by PCR from the My6-pBK-CMV phagemid. The nidB and -A genes (DNA bp 2851 to 4782) were also amplified by PCR from the My6-pBK-CMV phagemid. The PCR products were cloned into the pBAD/Thio-TOPO vector. Clones carrying the nidA-pBAD/Thio plasmid or the nid(B+A)-pBAD/Thio and nidD-pBAD/Thio plasmids with the correct orientation were obtained.
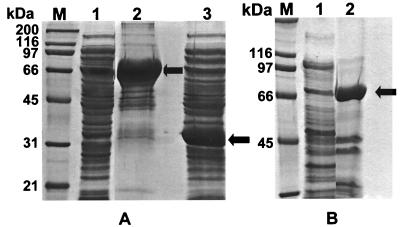
Clone My6-pBK-CMV and pBK-CMV (negative control) were inoculated into LB broth-ampicillin (50 μg/ml) medium and induced by 0.02% arabinose. The recombinant proteins encoded by genes nidA, nidB, and nidD were overexpressed and separated by SDS-PAGE (Fig. 5). We found that most of the overexpressed large-subunit dioxygenase and aldehyde dehydrogenase were in the pellets of sonicated fractions (Fig. 5A, lane 2, and B, lane 2). However, the small subunit was present in the supernatants of the sonicated fraction (Fig. 5A, lane 3). The six-His-tagged recombinant proteins for nidA and nidD were purified by a simple Ni-NTA spin column (Fig. 5). The thioredoxin fusion significantly increased the yield of protein production. The C-terminal polyhistidine (six-His; 3-kDa) tag is convenient for purification with Ni-NTA resin.
FIG. 5.
SDS-PAGE to show the overexpressed recombinant proteins in three clones. (A) Lanes 1 and 2, clone with nidA-pBAD/Thio plasmid; lane 3, clone with nid(B+A)-pBAD/Thio plasmid. (B) Clone with nidD- pBAD/Thio plasmid. Lane M, protein size marker; lane 1, sample from the supernatant after sonication of the cells; lane 2, recombinant proteins partly purified by using an Ni-NTA spin column. Arrows, locations of target proteins.
The PCR-amplified gene without the start and stop codons should be strongly expressed in this system. The recombinant protein should have the molecular mass of the gene product plus 13 kDa for His-Patch-thioredoxin and 3 kDa for the C-terminal tag. The clone containing the nid(B+A)-pBAD/Thio plasmid only overexpressed the nidB gene with the HP-thioredoxin because of the stop codon of nidB. The SDS gel showed that the recombinant protein was 32 kDa (Fig. 5A, lane 3), that is, 19 kDa plus 13 kDa (for HP-thioredoxin), which matches the calculated molecular weight of NidB (19447.60). The 3 kDa for the C-terminal tag was not present in this recombinant protein; thus we did not get purified recombinant NidB protein by using Ni-NTA resin. The expression of the nidA gene in this clone was low. However, the clone with nidA-pBAD/Thio plasmid containing the nidA gene without the start and stop codons was strongly expressed. The recombinant protein encoded by nidA had a molecular mass of 66 kDa (Fig. 5A, lane 2), that is, 50 kDa for nidA plus 13 kDa (for HP-thioredoxin) and 3 kDa for the C-terminal tag. Similarly, the recombinant protein for nidD was 68 kDa (Fig. 5B), that is, 52.0 kDa for nidD plus 16 kDa.
Functional analysis for the clones containing the dioxygenase system genes.
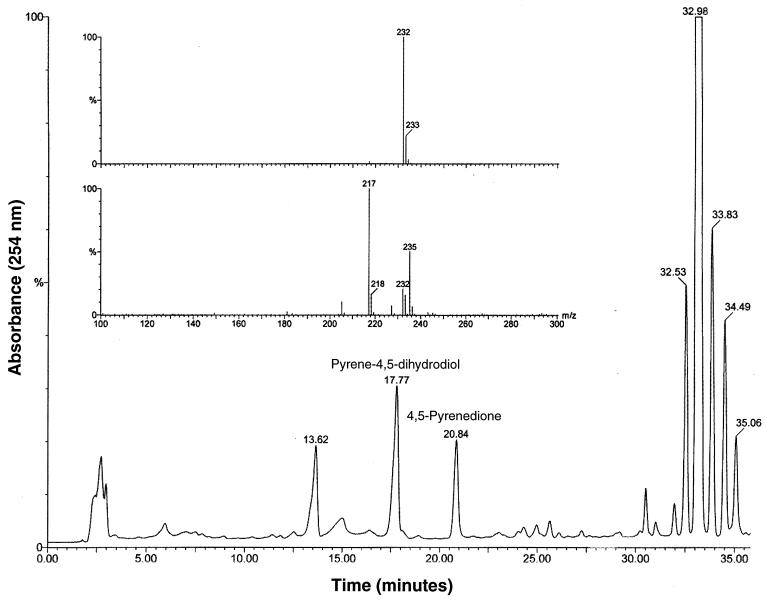
Functional analysis for clone My6-pBK-CMV was demonstrated by incubating the clone in pyrene for biotransformation. pBK-CMV was used as the negative control. Figure 6 shows the HPLC chromatogram and negative-ion mass spectra of the ethyl acetate-extractable metabolites. Pyrene eluted at 32.98 min. The compound eluting at 17.77 min (Fig. 6) had an HPLC retention time and mass spectrum properties (m/z 235 [M+H-H2O]+) (Fig. 6, inset) that were identical to those of pyrene cis-4,5-dihydrodiol (15). The 4,5-pyrenedione (m/z 232, M+), eluting at 20.84 min, was formed due to nonenzymatic oxidation of 4,5-dihydroxypyrene. These compounds were not found in control samples (plasmid pBK-CMV) containing the same host bacteria without nidAB genes. Both samples were incubated and analyzed under the same conditions as those described for clone My6-pBK-CMV. We found one compound eluting at 13.62 min in both control and clone samples; this may be a natural product formed by the host cells.
FIG. 6.
Functional analysis for the clone containing dioxygenase gene. The HPLC chromatogram (254 nm) from the extracts of clone My6-pBK-CMV is shown with the negative-ion mass spectra of the two products (inset) with retention times of 20.84 (top) and 17.77 min (bottom). These products were not observed in negative control pBK-CMV.
DISCUSSION
We report in this study the cloning, sequencing, and functional expression in E. coli of a novel PAH ring-hydroxylating dioxygenase from Mycobacterium sp. strain PYR-1, which is composed of NidABD and partial NidC proteins. To our knowledge this is the first report on the cloning of such genes in species of mycobacteria for PAH degradation. The time course induction experiment showed that the PAH degradation genes were inducible after exposure to pyrene (Fig. 1). The proteomic approach to isolate putative catabolic proteins induced when Mycobacterium sp. strain PYR-1 was grown in the presence of PAHs indicated that at least six major proteins were expressed (Fig. 2). A 50-kDa protein from PAH-induced Mycobacterium sp. strain PYR-1 cultures had an N-terminal sequence similar to those of other dioxygenases (Fig. 2). Clones with a positive reaction to the 50-kDa protein N-terminal sequence probe contained dioxygenase large-subunit (nidA), dioxygenase small-subunit (nidB), and dehydrogenase (nidD) genes. The N-terminal amino acid sequence of the 50-kDa protein, found at the start of the nidA gene product, indicated that the 50-kDa PAH-induced protein was the nidA (naphthalene-inducible dioxygenase large subunit) gene product (27, 30). The computed molecular weight for the deduced nidA gene product was 50,004.20, similar to the 2D gel position of the 50-kDa induced protein.
The clones containing genes nidA, nidB, and nidD ORFs were determined and were located on a 5,288-bp BamHI fragment. The deduced protein sequences corresponding to the three ORFs showed 40 to 50% similarity to those of ORF products reported by GenBank for the dioxygenase large subunit (NidA.I24, PhdA.KP7, or NarAa.NCIMB12038), dioxygenase small subunit (NidB.I24, PhdB.KP7, or DitA.BKME-9), and dehydrogenase (NidD), respectively (22, 27, 29, 30). Phylogenetic analysis indicated that NidA did not form a cluster with most known large-subunit sequences but formed a deep branch with three newly described large-subunit terminal dioxygenases from Nocardioides sp. strain PhdA.KP7 (AB017794), Rhodococcus sp. strain NidA.I24 (AF121905), and NarAa.NCIMB12038 (AF082663) (Fig. 4A). Phylogenetic analysis of the small subunits gave an unrooted tree similar to that of large subunits, indicating that NidB formed a distinct cluster with the two small subunits PhdB.KP7 (AB017794) and NarAa.NCIMB12038 (AF082663) (Fig. 4B). In both unrooted trees, the branches of NidA.PYR-1 and NidB.PYR1 were supported by low bootstrap values, which might be due to the distant relationships of NidA.PYR-1 and NidB.PYR-1 to other known dioxygenases.
The gene clusters responsible for PAH degradation have been localized on both plasmids (nah, P. putida strain [22]; ndo, P. putida strain [28]; dox, Pseudomonas sp. strain C18 [8]; nag, Pseudomonas sp. strain U2 [11]; phn, Burkholderia sp. strain RP007 [23]) and chromosomes (pah, P. putida OUS82 [29]; nah, P. stutzeri AN10 [5]; phd, Nocardioides sp. KP7 [27]; nid, Rhodococcus sp. strain I24 [30]). The results of this study show that the PAH dioxygenase gene cluster was localized on the chromosome and that the order of the aromatic ring dioxygenase and dihydrodiol dehydrogenase genes was different from that for analogous gene sets so far reported (27, 30). The gene organization from the 5′ to the 3′ direction is the dehydrogenase gene first, then the dioxygenase small-subunit gene, and then the dioxygenase large-subunit gene (Fig. 3).
The three-dimensional structure of the oxygenase component (α3[Ndo], β3[NdoC]) of naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816–4 has been determined (18). A long narrow gorge that may provide access to the substrate for catalytic iron is found in NdoB. The five residues constituting the narrowest part of the channel near the catalytic iron in NdoB were completely conserved in NidA of Mycobacterium sp. strain PYR-1 as Asn218, Phe219, His225, His230, and Phe 371. However, the residues lining the substrate-binding pocket below the catalytic iron and those covering the upper part of the catalytic iron were divergent. This finding may explain the wide substrate range of PAHs that are degraded by Mycobacterium sp. strain PYR-1 (19) and the alternate routes of enzymatic attack to form positional isomers of cis-PAH dihydrodiols (15, 25).
We have demonstrated that the genes were functional in both E. coli clones containing all of the nidA and nidB genes of the dioxygenase system (phagemid clone) and the plasmid subclone containing these two genes (Fig. 5). The nid genes are different from the general pattern since reductase, ferredoxin, and catechol dioxygenase were not found in the clone containing the 5.3-kb insert. The ferredoxin and the reductase components are required for electron transfer to the dioxygenase components in systems that are similar to those containing nidAB (30). Therefore it is reasonable to suspect that these two components are needed for the dioxygenase system from strain PYR-1. This suggests that there may be as yet unidentified genes for the ferredoxin and reductase in strain PYR-1. Since we demonstrated the formation of cis-4–5-pyrene dihydrodiol from pyrene by E. coli clones, there is the possibility that the NidAB dioxygenase subunits can borrow the ferredoxin and reductase components from elsewhere in the cell. It has been demonstrated that the dioxygenase subunits from one system or hybrid dioxygenase subunits can borrow the ferredoxin and reductase components of another system (3). In addition, it has been demonstrated in Pseudomonas that the reductase gene component can be located elsewhere in the operon, separated from the terminal dioxygenase subunit genes by a number of intervening genes or scattered in the genome (2).
Several different species of bacteria have the ability to degrade pyrene and higher-molecular-weight PAHs (17). Until recently, with studies on Nocardioides and Rhodococcus species (27, 30), most all of our knowledge on the genetics of PAH degradation came from molecular studies on gram-negative bacteria, Pseudomonas and Sphingomonas strains. The presbent work illustrates that a Mycobacterium species capable of degrading PAHs possessed a different set of genes for the same catabolic ability. These genes did not hybridize with classical nah genes (9). In conclusion, we cloned and sequenced the genes encoding at least one of the dioxygenases induced in PAH degradation in Mycobacterium sp. strain PYR-1.
ACKNOWLEDGMENTS
We thank John B. Sutherland and Saeed A. Khan for critically reading the manuscript, Eungbin Kim for preliminary studies on molecular analysis, Gerben J. Zylstra and Seong-Jae Kim for expert assistance in reviewing the gene sequences, and Pat Fleischer and Sandra Malone for clerical and graphic assistance.
A part of this work was supported by U.S. Environmental Protection Agency Cooperative Agreement CR 820773.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armengaud J, Happe B, Timmis K N. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. strain RW1: catabolic genes dispersed on the genome. J Bacteriol. 1998;180:3954–3966. doi: 10.1128/jb.180.15.3954-3966.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beil S, Mason J R, Timmis K N, Pieper D H. Identification of chlorobenzene dioxygenase sequence elements involved in dechlorination of 1,2,4,5-tetrachlorobenzene. J Bacteriol. 1998;180:5520–5528. doi: 10.1128/jb.180.21.5520-5528.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boldrin B, Tiehm A, Fritzsche C. Degradation of phenanthrene, fluorene, fluoranthene, and pyrene by a Mycobacterium sp. Appl Environ Microbiol. 1993;59:1927–1930. doi: 10.1128/aem.59.6.1927-1930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch R, Garcia-Valdes E, Moore E R B. Genetic characterization and evolutionary implications of a chromosomally encoded naphthalene-degradation upper pathway from Pseudomonas stutzeri AN10. Gene. 1999;236:149–157. doi: 10.1016/s0378-1119(99)00241-3. [DOI] [PubMed] [Google Scholar]
- 6.Cerniglia C E. Aromatic hydrocarbons: metabolism by bacteria, fungi and algae. Rev Biochem Toxicol. 1981;3:321–361. [Google Scholar]
- 7.Cerniglia C E. Biodegradation of polycyclic aromatic hydrocarbons. Curr Opin Bio/Technol. 1993;4:331–338. [Google Scholar]
- 8.Denome S A, Stanley D C, Olson E S, Young K D. Metabolism of dibenzothiophene and naphthalene in Pseudomonas strains: complete DNA sequence of an upper naphthalene catabolic pathway. J Bacteriol. 1993;175:6890–6901. doi: 10.1128/jb.175.21.6890-6901.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ensley B D, Gibson D T, Laborde A L. Oxidation of naphthalene by a multicomponent enzyme system from Pseudomonas sp. strain NCIB 9816. J Bacteriol. 1982;149:948–954. doi: 10.1128/jb.149.3.948-954.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erickson B D, Mondello F J. Nucleotide sequence and transcriptional mapping of the genes encoding biphenyl dioxygenase multicomponent polychlorinated-biphenyl-degrading enzyme of Pseudomonas sp. strain LB400. J Bacteriol. 1992;174:2903–2912. doi: 10.1128/jb.174.9.2903-2912.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuenmayor S L, Wild M, Boyes A L, Williams P A. A gene cluster encoding steps in conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J Bacteriol. 1998;180:2522–2530. doi: 10.1128/jb.180.9.2522-2530.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goyal A K, Zylstra G J. Molecular cloning of novel genes for polycyclic aromatic hydrocarbon degradation from Comamonas testosteroni GZ39. Appl Environ Microbiol. 1996;62:230–236. doi: 10.1128/aem.62.1.230-236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grosser R J, Warshawsky D, Vestal J R. Indigenous and enhanced mineralization of pyrene, benzo[a]pyrene, and carbazole in soils. Appl Environ Microbiol. 1991;57:3462–3469. doi: 10.1128/aem.57.12.3462-3469.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heitkamp M A, Franklin W, Cerniglia C E. Microbial metabolism of polycyclic aromatic hydrocarbons: isolation and characterization of a pyrene-degrading bacterium. Appl Environ Microbiol. 1988;54:2549–2555. doi: 10.1128/aem.54.10.2549-2555.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heitkamp M A, Freeman J P, Miller D W, Cerniglia C E. Pyrene degradation by a Mycobacterium sp.; identification of ring oxidation and ring fission products. Appl Environ Microbiol. 1988;54:2556–2565. doi: 10.1128/aem.54.10.2556-2565.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heitkamp M A, Cerniglia C E. Polycyclic aromatic hydrocarbon degradation by a Mycobacterium sp. in microcosms containing sediment and water from a pristine ecosystem. Appl Environ Microbiol. 1989;55:1968–1973. doi: 10.1128/aem.55.8.1968-1973.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanaly R A, Harayama S. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J Bacteriol. 2000;182:2059–2067. doi: 10.1128/jb.182.8.2059-2067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kauppi B, Lee K, Carredano E, Parales R E, Gibson D T, Eklund H, Ramaswamy S. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure. 1998;6:571–586. doi: 10.1016/s0969-2126(98)00059-8. [DOI] [PubMed] [Google Scholar]
- 19.Kelley I, Cerniglia C E. Degradation of a mixture of high-molecular-weight polycyclic aromatic hydrocarbons by a Mycobacterium strain PYR-1. J Soil Contam. 1995;4:77–91. [Google Scholar]
- 20.Kelley I, Freeman J P, Evans F E, Cerniglia C E. Identification of a carboxylic acid metabolite from the catabolism of fluoranthene by a Mycobacterium sp. Appl Environ Microbiol. 1991;57:636–641. doi: 10.1128/aem.57.3.636-641.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan A A, Walia S K. Expression, localization, and functional analysis of polychlorinated biphenyl degradation genes cbpABCD of Pseudomonas putida. Appl Environ Microbiol. 1991;57:1325–1332. doi: 10.1128/aem.57.5.1325-1332.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurkela S, Lehvaslasiho H, Palva E T, Terri T H. Cloning, nucleotide sequence and characterization of genes encoding naphthalene dioxygenase of Pseudomonas putida strain NCIB9816. Gene. 1988;73:355–362. doi: 10.1016/0378-1119(88)90500-8. [DOI] [PubMed] [Google Scholar]
- 23.Laurie A D, Lloyd-Jones G. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J Bacteriol. 1999;181:531–540. doi: 10.1128/jb.181.2.531-540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mason J R, Cammack R. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu Rev Microbiol. 1992;46:277–305. doi: 10.1146/annurev.mi.46.100192.001425. [DOI] [PubMed] [Google Scholar]
- 25.Moody J D, Freeman J, Doerge D R, Cerniglia C E. Degradation of phenanthrene and anthracene by cell suspensions of Mycobacterium sp. strain PYR-1. Appl Environ Microbiol. 2001;67:1476–1483. doi: 10.1128/AEM.67.4.1476-1483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rafii F, Selby A L, Newton R K, Cerniglia C E. Reduction and mutagenic activation of nitroaromatic compounds by a Mycobacterium sp. Appl Environ Microbiol. 1994;60:4263–4267. doi: 10.1128/aem.60.12.4263-4267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito A, Iwabuchi T, Harayama S. A novel phenanthrene dioxygenase from Nocardioides sp. strain KP7: expression in Escherichia coli. J Bacteriol. 2000;182:2134–2141. doi: 10.1128/jb.182.8.2134-2141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon M J, Osslund T D, Saunders R, Ensley B D, Suggs S, Harcourt A, Suen W-C, Cruden D L, Gibson D T, Zylstra G J. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816–4. Gene. 1993;127:31–37. doi: 10.1016/0378-1119(93)90613-8. [DOI] [PubMed] [Google Scholar]
- 29.Takizawa N, Kaida N, Torigoe S, Moritani T, Sawada T, Satoh S, Kiyohara H. Identification and characterization of genes encoding polycyclic aromatic hydrocarbon dioxygenase and polycyclic aromatic hydrocarbon dihydrodiol dehydrogenase in Pseudomonas putida OUS82. J Bacteriol. 1994;176:2444–2449. doi: 10.1128/jb.176.8.2444-2449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Treadway S L, Yanagimachi K S, Lankenau E, Lessard P A, Stephanopoulos G, Sinskey A J. Isolation and characterization of indene bioconversion genes from Rhodococcus strain I24. Appl Microbiol Biotechnol. 1999;51:786–793. doi: 10.1007/s002530051463. [DOI] [PubMed] [Google Scholar]
- 31.Wang R-F, Wennerstrom D, Cao W-W, Khan A A, Cerniglia C E. Cloning, expression, and characterization of the katG gene, encoding catalase-peroxidase from the polycyclic aromatic hydrocarbon-degrading bacterium Mycobacterium sp. strain PYR-1. Appl Environ Microbiol. 2000;66:4300–4304. doi: 10.1128/aem.66.10.4300-4304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zylstra G J, Gibson D T. Toluene degradation by Pseudomonas putida F1. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]
- 33.Zylstra G J, Kim E. Aromatic hydrocarbon degradation by Sphingomonas yanoikuyae B1. J Ind Microbiol Biotechnol. 1997;19:408–414. doi: 10.1038/sj.jim.2900475. [DOI] [PubMed] [Google Scholar]