Summary
Background
Artificial intelligence (AI) has recently been applied to endoscopy and questionnaires for the evaluation of oesophageal diseases (ODs).
Aim
We performed a systematic review with meta‐analysis to evaluate the performance of AI in the diagnosis of malignant and benign OD.
Methods
We searched MEDLINE, EMBASE, EMBASE Classic and the Cochrane Library. A bivariate random‐effect model was used to calculate pooled diagnostic efficacy of AI models and endoscopists. The reference tests were histology for neoplasms and the clinical and instrumental diagnosis for gastro‐oesophageal reflux disease (GERD). The pooled area under the summary receiver operating characteristic (AUROC), sensitivity, specificity, positive and negative likelihood ratio (PLR and NLR) and diagnostic odds ratio (DOR) were estimated.
Results
For the diagnosis of Barrett's neoplasia, AI had AUROC of 0.90, sensitivity 0.89, specificity 0.86, PLR 6.50, NLR 0.13 and DOR 50.53. AI models’ performance was comparable with that of endoscopists (P = 0.35). For the diagnosis of oesophageal squamous cell carcinoma, the AUROC, sensitivity, specificity, PLR, NLR and DOR were 0.97, 0.95, 0.92, 12.65, 0.05 and DOR 258.36, respectively. In this task, AI performed better than endoscopists although without statistically significant differences. In the detection of abnormal intrapapillary capillary loops, the performance of AI was: AUROC 0.98, sensitivity 0.94, specificity 0.94, PLR 14.75, NLR 0.07 and DOR 225.83. For the diagnosis of GERD based on questionnaires, the AUROC, sensitivity, specificity, PLR, NLR and DOR were 0.99, 0.97, 0.97, 38.26, 0.03 and 1159.6, respectively.
Conclusions
AI demonstrated high performance in the clinical and endoscopic diagnosis of OD.
Keywords: artificial intelligence, Barrett’s oesophagus, gastroesophageal reflux disease, gastrointestinal endoscopy, IPCL, oesophageal cancer
Artificial intelligence in the diagnosis of oesophageal diseases: systematic review with meta‐analysis.

1. INTRODUCTION
Artificial intelligence (AI) is being extensively applied to different medical settings aiming to improve the performance in the diagnosis of various diseases, including gastrointestinal (GI) diseases. The term AI generically refers to complex computer algorithms that mimic human cognitive functions, including learning and problem‐solving. 1 Machine learning (ML) is a field of AI that can be taught to discriminate characteristics of data samples and then apply experience to interpret previously unknown information. 2 Supervised ML with support vector machine (SVM) is based on hand‐crafted algorithms in which researchers, based on clinical knowledge, manually indicate features of interest of an input data set (labelled data set) to train the system to recognise discriminative features and provide appropriate outputs. 1 Deep learning (DL) is a subset of ML which can autonomously extract discriminative attributes of input data through artificial neural networks, often organised as convolutional neural networks (CNNs), which are constituted of multiple layers of non‐linear functions. 1 , 3
AI is increasingly being integrated into computer‐aided diagnosis (CAD) systems for GI diseases to improve detection (CADe) and characterisation (CADx) of pathology. Consistently, recent meta‐analyses concluded that the use of AI during lower endoscopic procedures significantly increased the detection of colorectal neoplasia. 4 , 5
More recently, various studies evaluating the performance of AI in the diagnosis of oesophageal diseases (ODs) have been published. The main application of AI in the upper GI tract is endoscopy and neoplasia detection. Ideally, upper GI endoscopies and biopsies should not miss lesions, but the ability to recognise endoscopic images depends on individual expertise. This is particularly relevant for subtle upper GI lesions, where experienced endoscopists can make a difference in the diagnosis. In this setting, CAD tools have the potential to successfully assist both trainee and expert physicians to reduce variability in the detection of upper GI pathology, increasing the diagnostic accuracy regardless of individual expertise and virtually overcoming inter‐ and intra‐observer variability. 6
In addition, deep learning using multi‐layered neural networks powered by high‐performance computing clusters are capable of recognizing complex non‐linear patterns in datatypes that previously were intractable to process, such as endoscopic images and videos. In this setting, AI has been applied to clinical questionnaires for gastro‐oesophageal reflux disease (GERD), pH‐impedance and oesophageal manometry tracings, and for the evaluation of mRNA transcripts in the diagnosis of eosinophilic oesophagitis (EoE).
DL models are black boxes in which the input data and the output (diagnosis) are known, but the processes by which the diagnosis is achieved are not, and this may be counterproductive. 6 Accordingly, research is already heading to understand how DL models make decisions to solve interpretability gaps, and methods to understand the process of CNN‐based choices are being developed. 7 , 8
AI support in decision‐making is a fascinating and rapidly evolving topic. Accordingly, we performed a systematic review with meta‐analysis of currently available evidence on the performance of AI in the diagnosis of oesophageal diseases (ODs), updating previous evidence on oesophageal cancer 9 , 10 and assessing evidence on the performance of AI in the detection of intrapapillary capillary loops (IPCLs) and in the diagnosis of benign ODs.
2. METHODS
2.1. Search strategy
We searched MEDLINE, EMBASE, EMBASE Classic and the Cochrane Library (via Ovid), from inception to 30 March 2021, to identify prospective and retrospective case‐control type or cohort‐type accuracy studies reporting the performance of AI systems in the instrumental or clinical diagnosis of malignant and benign ODs. To identify potentially eligible studies published only in abstract form, conference proceedings (Digestive Disease Week, American College of Gastroenterology and United European Gastroenterology Week) from 2000 until 30 March 2021 were also searched. The complete search strategy is provided in Supplementary Methods. There were no language restrictions. We screened titles and abstracts of all citations identified by our search for potential suitability and retrieved those that appeared relevant to examine them in more detail. Foreign language papers were translated. A recursive search of the literature was performed using bibliographies of all relevant studies. We also planned to contact authors if a study appeared potentially eligible, but did not report the data required, to obtain supplementary information and, therefore, maximise available studies.
2.2. Study selection (inclusion and exclusion criteria)
The eligibility assessment was performed independently by two investigators (PV, BB) using pre‐designed eligibility forms. We included in the systematic literature (a) studies reporting the use of AI in the diagnosis of ODs in adult patients, (b) studies that reported the rates of true positivity, false positivity, false negativity and true negativity compared with the gold‐standard diagnosis of the disease as ground truth and (c) studies that reported the numbers of images or videoclips included in the AI analysis. For the meta‐analysis we included studies that (a) separately assessed the performance of AI with different tools when more than one tool was used (ie white light endoscopy [WLE], narrow band imaging [NBI]), (b) separately assessed the performance of AI in the diagnosis of different histological types of oesophageal cancer. We excluded review articles, case reports and studies that applied AI restrictedly to radiology or histopathology from the qualitative analysis. We excluded (a) studies exclusively providing comprehensive performance scores of AI for different histological types of oesophageal cancer and (b) studies not reporting data for extraction from the meta‐analysis. Any disagreements were resolved by consensus opinion among reviewers, and the degree of agreement was measured with a kappa statistic. Ethical approval was not required because this study retrieved and synthesised data from already published studies.
2.3. Data extraction and analysis
Data were extracted independently by two authors (PV, BB) on to a Microsoft Excel spreadsheet (XP professional edition; Microsoft, Redmond, WA, USA). Disagreements were resolved by consensus among the reviewing authors.
The following data were collected for each study: total number of images/cases used in the validation sets, total number of “ground truth” images/cases (ie human detected and histologically confirmed as neoplastic or non‐neoplastic; diagnosis of GERD based on symptoms, endoscopy findings and/or pH‐metry), the numbers of images/cases that were true positive (images/cases showing a neoplastic lesion detected/predicted‐as‐neoplastic by AI), true negative (images/cases showing non‐neoplastic mucosa without AI detection or lesions predicted as non‐neoplastic), false positive (FP, images/cases showing non‐neoplastic mucosa or lesions detected/predicted‐as‐neoplastic by AI) or false negative (images/cases showing a neoplastic lesion missed by AI or predicted as non‐neoplastic). In addition, year of publication, country where the study was conducted, type of study (prospective, retrospective), number of patients, diagnostic tool (endoscopy and type of endoscopic light, questionnaires, pH‐impedance monitoring, oesophageal manometry, oesophageal biopsies), type and design of AI systems (DL, SVM) were also retrieved.
2.4. Study outcomes
The primary outcomes of interest were the pooled diagnostic sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR) and the area under the summary receiver operating characteristic curve (AUROC) of the AI models in the diagnosis of malignant and benignant ODs.
The secondary outcome was the comparison of the performance of AI models versus endoscopists (without the aid of AI) in analysing the same validation data sets.
2.5. Quality assessment
The degree of bias was assessed using the Quality for Assessment of Diagnostic Studies (QUADAS) score. 11 In detail, we identified four domains: patient selection, index test, reference standard and flow and timing. The first three domains were also assessed for concerns regarding applicability. Each section was classified as having a high, low or unclear risk of bias (Figure 1).
FIGURE 1.
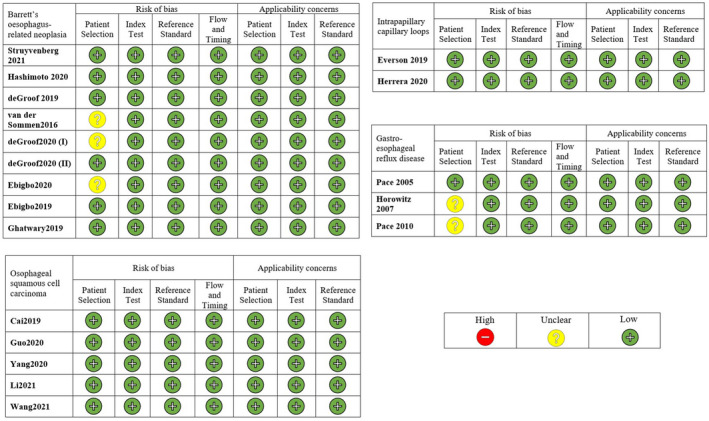
Quality in methodology of included studies
2.6. Statistical analysis
A bivariate, random‐effect model was used to calculate pooled sensitivity, specificity, PLR, NLR, DOR and the AUROC of AI‐assisted models and endoscopist in detecting oesophageal lesions. 12 The method takes into account the correlation between sensitivity and specificity. The estimation procedure is based on a restricted maximum likelihood approach. The model parameterisation assumes that the sensitivity the specificity, on the logit scale, are distributed as bivariate normal random variables. The pooled AUROC 95% confidence interval has been estimated by performing a bootstrap resampling of the AUC value; however, some concerns are possible in cases of few studies included in the computation. The calculation has been performed by considering the extended 95% CI procedure of computation for meta‐analysis of diagnostic accuracy studies. 13
Heterogeneity has been performed by considering the Cochrane guidelines. 14 The χ2 tests to assess heterogeneity of sensitivities and specificities were performed. The sources of heterogeneity were explored through subgroup analysis. We conducted subgroup analyses according to (a) specific diagnosis (Barrett's neoplasia [BN], oesophageal squamous cell carcinoma [OSCC], abnormal IPCL, GERD), (b) country, (c) study type, (d) AI algorithm, (e) endoscopy type, (f) real‐time evaluation of the performance of AI and g) best and worst performance of different algorithms on the same image set.
The publication bias was analysed via Deeks’ test. 15 The statistical significance was set at a P value <0.05. The analyses were performed using R 4.4.2 with mada package. 16
3. RESULTS
The search strategy generated 2568 citations. From these we identified 67 separate articles that appeared to be relevant to the study question. In total, 42 studies 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54 , 55 , 56 , 57 , 58 reported on the performance of AI in the diagnosis of various ODs and were included in the qualitative synthesis (Supplementary Table S1). Among the included studies, 19 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 reported complete data for extraction and were included in the meta‐analysis: 9 on BN, 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 5 on OSCC, 26 , 27 , 28 , 29 , 30 2 on abnormal IPCLs 31 , 32 and 3 on GERD 33 , 34 , 35 (Figure 2). Agreement between investigators for the assessment of study eligibility was excellent (kappa statistic = 0.85).
FIGURE 2.
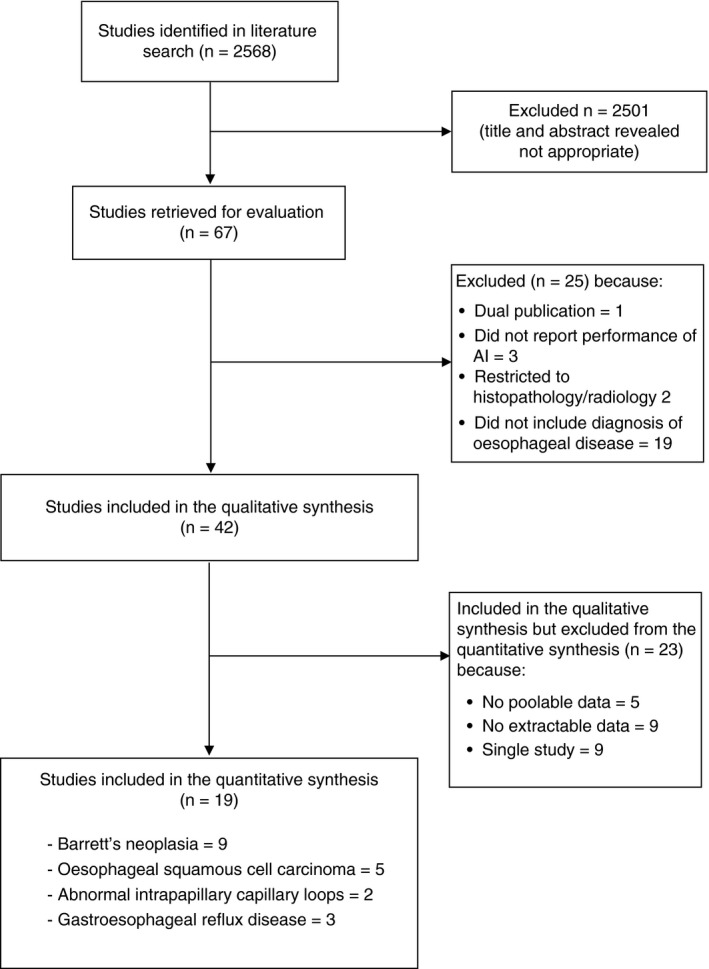
Flow diagram of assessment of studies identified in the meta‐analysis
3.1. Studies not included in quantitative synthesis
Among the 42 studies included in the qualitative synthesis, 23 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54 , 55 , 56 , 57 , 58 could not be included in the meta‐analysis for various reasons. They also included investigations on eosinophilic oesophagitis (n = 1), reflux monitoring (n = 1), optical endoscopic diagnosis of GERD (n = 1), motility assessment (n = 1), diagnosis of cytomegalovirus and herpes simplex virus oesophagitis (n = 1) and varices (n = 1). In particular, nine studies 38 , 39 , 41 , 42 , 43 , 44 , 48 , 49 , 50 did not report complete data for extraction, five studies 45 , 46 , 47 , 51 , 58 reported non‐poolable data, and nine 36 , 37 , 40 , 52 , 53 , 54 , 55 , 56 , 57 were the unique retrieved studies of their type. We included these studies in Supplementary Table S1 for completeness, but not in the final quantitative synthesis through meta‐analysis.
3.2. AI in the diagnosis of Barrett's neoplasia
Nine studies 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 reported extractable and comparable data regarding AI in the diagnosis of BN (Figure 3). Eight studies were performed in Europe 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 and one in America. 22 All the studies used DL models, except two in which an SVM algorithm was tested. 17 , 18 Moreover, seven studies provided the performance of AI under WLE, 17 , 18 , 19 , 20 , 21 , 22 , 24 two under NBI 22 , 25 and one provided the comprehensive performance of AI with WLE or NBI. 23 Six studies were retrospective, 18 , 20 , 22 , 23 , 24 , 25 and three were prospective. 17 , 19 , 21 Three studies evaluated the performance of AI using real‐time videos. 19 , 21 , 25 Three studies compared the performance of the AI system to that of endoscopists, and all these studies used WLE. 17 , 18 , 20 In all the included studies, BE and BN were diagnosed according to histology as ground truth.
FIGURE 3.
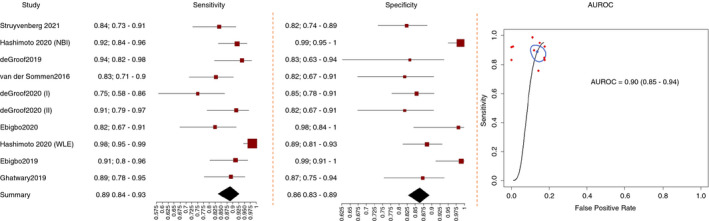
Performance of artificial intelligence in the diagnosis of Barrett's neoplasia
The comprehensive performance of AI in the diagnosis of BN with WLE or NBI, based on all the nine studies, 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 was: AUROC 0.90 (CI, 0.85‐0.94), pooled sensitivity 0.89 (CI, 0.84‐0.93), specificity 0.86 (CI, 0.83‐0.93), PLR 6.50 (CI, 1.59‐2.15), NLR 0.13 (CI, 0.20‐0.08) and DOR 50.53 (CI, 24.74‐103.22) (Table 1).
TABLE 1.
Sub‐group analyses of the performance of artificial intelligence in the diagnosis of oesophageal diseases
| Oesophageal disease | Subgroups | Number of studies |
Sensitivity (95% CI) |
Specificity (95% CI) |
PLR (95% CI) |
NLR (95% CI) |
DOR (95% CI) |
AUROC (95% CI) |
P value* |
|---|---|---|---|---|---|---|---|---|---|
| Barrett's neoplasia | All studies | 9 | 0.89 (0.84‐0.93) | 0.86 (0.83‐0.93) | 6.50 (1.59‐2.15) | 0.13 (0.20‐0.08) | 50.53 (24.74‐103.22) | 0.90 (0.85‐0.94) | — |
| Country | 0.04 | ||||||||
| Europe | 8 | 0.85 (0.81‐0.89) | 0.84 (0.80‐0.88) | 5.50 (1.43‐1.98) | 0.17 (0.23 −0.13) | 32.07 (18.04‐57.00) | 0.85 (0.84‐0.93) | ||
| America | 1 | 0.97 (0.82‐0.99) | 0.97 (0.66‐1.00) | 28.62 (0.87‐6.06) | 0.04 (0.27‐0.01) | 816.6 (8.73‐76 349.10) | 0.98 (0.90‐0.99) | ||
| Study type | 0.45 | ||||||||
| Retrospective | 6 | 0.90 (0.85‐0.94) | 0.87 (0.82‐0.90) | 6.69 (1.54‐2.25) | 0.11 (0.18‐0.07) | 59.54 (25.57‐138.60) | 0.93 (0.87‐0.97) | ||
| Prospective | 3 | 0.84 (0.70‐0.92) | 0.86 (0.79‐0.91) | 5.87 (1.19‐2.28) | 0.19 (0.39‐0.08) | 31.66 (8.51‐117.78) | 0.87 (0.80‐0.94) | ||
| Algorithm type | 0.94 | ||||||||
| DL | 7 | 0.89 (0.83‐0.93) | 0.87 (0.83‐0.90) | 6.80 (1.60‐2.22) | 0.12 (0.21‐0.07) | 54.65 (24.01‐124.4) | 0.91 (0.86‐0.95) | ||
| SVM | 2 | 0.89 (0.70‐0.97) | 0.84 (0.72‐0.91) | 5.45 (0.91‐2.39) | 0.13 (0.42‐0.04) | 42.86 (5.95‐308.51) | 0.87 (0.78‐0.97) | ||
| Endoscopy type d , e | 0.64 | ||||||||
| WLE | 7 | 0.89 (0.82‐0.94) | 0.86 (0.82‐0.89) | 6.43 (1.53‐2.17) | 0.12 (0.21‐0.01) | 52.03 (21.56‐125.58) | 0.89 (0.84‐0.94) | ||
| NBI | 2 | 0.89 (0.77‐0.95) | 0.96 (0.47‐1.00) | 20.19 (0.37‐6.23) | 0.11 (0.5‐0.05) | 177.11 (2.9‐10 821.79) | 0.93 (0.75‐0.99) | ||
| Real‐time | 0.2 | ||||||||
| Yes | 3 | 0.81 (0.73‐0.87) | 0.84 (0.79‐0.89) | 5.20 (1.25‐2.03) | 0.22 (0.94‐0.15) | 23.16 (10.35‐51.81) | 0.82 (0.80‐0.92) | ||
| No | 6 | 0.92 (0.87‐0.95) | 0.87 (0.82‐0.91) | 7.11 (1.60‐2.32) | 0.10 (0.16‐0.06) | 73.32 (30.61‐175.63) | 0.93 (0.86‐0.96) | ||
| OSCC a | All studies | 5 | 0.95 (0.91‐0.98) | 0.92 (0.82‐0.97) | 12.65 (1.61‐3.51) | 0.05 (0.11‐0.02) | 258.36 (44.18‐1510.7) | 0.97 (0.92‐0.98) | — |
| Endoscopy type f | 0.74 | ||||||||
| WLE | 4 | 0.95 (0.86‐0.98) | 0.93 (0.77‐0.98) | 14.42 (1.31‐4.11) | 0.05 (0.18‐0.02) | 277.2 (19.94‐3852.9) | 0.98 (0.95‐0.99) | ||
| NBI | 2 | 0.96 (0.83‐0.99) | 0.96 (0.94‐0.97) | 23.49 (2.59‐3.62) | 0.04 (0.19‐0.01) | 537.21 (71.81‐4018.64) | 0.98 (0.94‐0.99) | ||
| Real‐time | 0.1 | ||||||||
| Yes | 2 | 0.94 (0.79‐0.99) | 0.98 (0.94‐0.99) | 39.4 (2.51‐4.73) | 0.06 (0.23‐0.01) | 651.92 (53.83‐7895.1) | 0.99 (0.94‐0.99) | ||
| No | 3 | 0.96 (0.92‐0.98) | 0.87 (0.71‐0.95) | 7.29 (1.14‐2.93) | 0.05 (0.11‐0.03) | 143.03 (27.61‐741.01) | 0.96 (0.89‐0.97) | ||
| GERD b | All studies | 3 | 0.97 (0.67‐1.00) | 0.97 (0.75‐1.00) | 38.26 (0.98‐6.22) | 0.03 (0.44‐0.00) | 1159.6 (6.12‐219 711.69) | 0.99 (0.80‐0.99) | — |
| Country | — | ||||||||
| Europe | 2 | 0.99 (0.98‐1.00) | 0.99 (0.95‐1.00) | 145.88 (3.05 −6.95) | 0.01 (0.02‐0.00) | 16 120.13 (1009.41‐257436.50) | 0.98 (0.97‐0.99) | ||
| Asia | 1 | 0.70 (0.59‐0.80) | 0.78 (0.66‐0.87) | 3.25 (0.55‐1.81) | 0.38 (0.62‐0.23) | 8.61 (2.8‐26.48) | — | ||
| Algorithm type | — | ||||||||
| SVM | 2 | 0.99 (0.98‐1.00) | 0.99 (0.95‐1.00) | 145.88 (3.05 −6.95) | 0.01 (0.02‐0.00) | 16 120.13 (1009.41‐257436.50) | 0.98 (0.98‐0.99) | ||
| DL | 1 | 0.70 (0.59‐0.80) | 0.78 (0.66‐0.87) | 3.25 (0.55‐1.81) | 0.38 (0.62‐0.23) | 8.61 (2.8‐26.48) | — | ||
| IPCL c | All studies | 2 | 0.94 (0.67‐0.99) | 0.94 (0.84‐0.98) | 14.75 (1.46‐3.70) | 0.07 (0.39‐0.01) | 225.83 (11.05‐4613.93) | 0.98 (0.86‐0.99) | — |
| Algorithms | 0.001 | ||||||||
| Best | 2 | 0.99 (0.97‐1.00) | 0.97 (0.96‐0.98) | 32.18 (3.18‐3.76) | 0.01 (0.03‐0.00) | 2779.61 (804.87‐9599.39) | 0.98 (0.97‐0.99) | ||
| Worst | 2 | 0.73 (0.55‐0.86) | 0.87 (0.71‐0.95) | 5.74 (0.64‐2.85) | 0.31 (0.64‐0.15) | 18.56 (2.97‐115.85) | 0.87 (0.66‐0.96) |
Abbreviations: AI, artificial intelligence; AUROC, area under the summary receiver operating characteristic curve; DL, deep learning; DOR, diagnostic odds ratio; IPCL, intrapapillary capillary loop; NLR, negative likelihood ratio; OSCC, oesophageal squamous cell carcinoma; PLR, positive likelihood ratio; SVM, support vector machine.
All studies have been conducted in Asia, with deep learning, and are retrospective.
All the studies have been conducted prospectively and are real time.
All studies have been conducted in Asia, with deep learning and narrow‐band imaging, and are retrospective and real time.
Hashimoto et al assessed both WLE and NBI in the same study.
One study was not included in the sub‐analysis as data for white‐light endoscopy and narrow band imaging had not been analysed separately in the study.
Li et al assessed both WLE and NBI in the same study.
P value of comparison between groups. The P value has been computed via bootstrap with 2000 resampling.
For the detection of BN under WLE the pooled AUROC was 0.89 (CI, 0.84‐0.94), pooled sensitivity 0.89 (0.82‐0.94), pooled specificity 0.86 (CI, 0.82‐0.89), pooled PLR 6.43 (CI, 1.53‐2.17), pooled NLR 0.12 (CI, 0.21‐0.01) and pooled DOR 52.03 (CI, 21.56‐125.58) in seven studies 17 , 18 , 19 , 20 , 21 , 22 , 24 (Table 1).
For the detection of BN under NBI in two studies, 22 , 25 the pooled performance was AUROC 0.93 (CI, 0.75‐0.99), sensitivity 0.89 (CI, 0.77‐0.95), specificity 0.96 (CI, 0.47‐1.00), PLR 20.19 (CI, 0.37‐6.23), NLR 0.11 (CI, 0.5‐0.05) and DOR 177.11 (CI, 2.9‐10 821.79). Very wide confidence intervals are observed especially for DOR given that only two studies were reported in the evidence synthesis (Table 1).
As regard the type of AI algorithm, the pooled AUROC, sensitivity, specificity, PLR, NLR and DOR of the studies that used DL as a backbone were 0.91 (CI, 0.86‐0.95), 0.89 (CI, 0.83‐0.93), 0.87 (CI, 0.83‐0.90), 6.80 (CI, 1.60‐2.22), 0.12 (CI, 0.21‐0.07) and 54.65 (CI, 24.01‐124.4), respectively, in seven studies. 19 , 20 , 21 , 22 , 23 , 24 , 25 The pooled AUROC, sensitivity, specificity, PLR, NLR and DOR of the studies that used SVM as a backbone 17 , 18 were as follows: 0.87 (CI, 0.78‐0.97), 0.89 (CI, 0.70‐0.97), 0.84 (CI, 0.72‐0.91), 5.45 (CI, 0.91‐2.39), 0.13 (CI, 0.42‐0.04) and 42.86 (CI, 5.95‐308.51), respectively (Table 1).
For the pooled performance of AI on real‐time videos, the AUROC, sensitivity, specificity, PLR, NLR and DOR were 0.82 (CI, 0.80‐0.92), 0.81 (CI, 0.73‐0.87), 0.84 (CI, 0.79‐0.89), 5.20 (CI, 1.25‐2.03), 0.22 (CI, 0.94‐0.15) and 23.16 (CI, 10.35‐51.81), respectively, in three studies. 19 , 21 , 25 For non‐real‐time studies, 17 , 18 , 20 , 22 , 23 , 24 the pooled AUROC, sensitivity, specificity, PLR, NLR and DOR were 0.93 (CI, 0.86‐0.96), 0.92 (CI, 0.87‐0.95), 0.87 (CI, 0.82‐0.91), 7.11 (CI, 1.60‐2.32), 0.10 (CI, 0.16‐0.06) and 73.32 (CI, 30.61‐175.63), respectively (Table 1).
As for retrospective studies, 18 , 20 , 22 , 23 , 24 , 25 the pooled performance of the AI algorithms was as follows: AUROC 0.93 (CI, 0.87‐0.97), sensitivity 0.90 (CI, 0.85‐0.94), specificity 0.87 (CI, 0.82‐0.90), PLR 6.69 (CI, 1.54‐2.25), NLR 0.11 (CI, 0.18‐0.07) and DOR 59.54 (CI, 25.57‐138.60). Instead, for prospective studies, 17 , 19 , 21 the pooled diagnostic efficacy of the AI algorithms was as follows: AUROC 0.87 (CI, 0.80‐0.94), sensitivity 0.84 (CI, 0.70‐0.92), specificity 0.86 (CI, 0.79‐0.91), PLR 5.87 (CI, 1.19‐2.28), NLR 0.19 (CI, 0.39‐0.08) and DOR 31.66 (CI, 8.51‐117.78) (Table 1).
As for studies performed in Europe, 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 the pooled AUROC, sensitivity, specificity, PLR, NLR and DOR were 0.85 (CI, 0.84‐0.93), 0.85 (CI, 0.81‐0.89), 0.84 (CI, 0.80‐0.88), 5.50 (CI, 1.43‐1.98), 0.17 (CI, 0.23 −0.13) and 32.07 (CI, 18.04‐57.00), respectively. In the study performed in America, 22 the pooled AUROC, sensitivity, specificity, PLR, NLR and DOR were 0.98 (CI, 0.90‐0.99), 0.97 (CI, 0.82‐0.99), 0.97 (CI, 0.66‐1.00), 28.62 (CI, 0.87‐6.06), 0.04 (CI, 0.27‐0.01) and 816.6 (CI, 8.73‐76 349.10), respectively (Table 1).
The bootstrap AUROC comparison among groups indicates a significant difference across countries (P = 0.04). Moreover, the geographical location (Europe vs America) is a significant source of heterogeneity according to the χ2 test results (0.01). No significant differences in AUROC were identified across the other subgroups.
As regard endoscopists with WLE, the pooled AUROC, sensitivity, specificity, PLR, NLR and DOR were 0.90 (0.85‐0.95), 0.93 (0.66‐0.99), 0.85 (0.71‐0.93), 6.17 (0.82‐2.63), 0.09 (0.48‐0.01) and 70.12 (4.70‐1045.93), respectively, in three studies 17 , 18 , 20 (Table 2).
TABLE 2.
Performance of endoscopists versus performance of artificial intelligence in the diagnosis of oesophageal diseases
|
Oesophageal disease |
Endoscopist or AI |
Number of studies |
Sensitivity (95% CI) |
Specificity (95% CI) |
PLR (95% CI) |
NLR (95% CI) |
DOR (95% CI) |
AUROC (95% CI) |
P value |
|---|---|---|---|---|---|---|---|---|---|
| Barrett's neoplasia | AI | 10 | 0.89 (0.84‐0.93) | 0.86 (0.83‐0.93) | 6.50 (1.59‐2.15) | 0.13 (0.20‐0.08) | 50.53 (24.74‐103.22) | 0.91 (0.82‐0.95) | 0.98 |
| Barrett's neoplasia | Endoscopist | 3 | 0.93 (0.66‐0.99) | 0.85 (0.71‐0.93) | 6.17 (0.82‐2.63) | 0.09 (0.48‐0.01) | 70.12 (4.70‐1045.93) | 0.90 (0.85‐0.95) | |
| OSCC | AI | 8 | 0.95 (0.91‐0.98) | 0.92 (0.82‐0.97) | 12.65 (1.61‐3.51) | 0.05 (0.11‐0.02) | 258.36 (44.18‐1510.7) | 0.97 (0.92‐0.98) | 0.11 |
| OSCC | Endoscopist | 2 | 0.75 (0.68‐0.80) | 0.88 (0.84‐0.92) | 6.46 (1.46‐2.27) | 0.29 (0.38‐0.22) | 22.45 (11.5‐43.84) | 0.88 (0.83‐0.98) |
Abbreviations: AI, artificial Intelligence; AUROC, area under the summary receiver operating characteristic curve; DOR, diagnostic odds ratio; IPCL, intrapapillary capillary loop; NLR, negative likelihood ratio; OSCC, oesophageal squamous cell carcinoma; PLR, positive likelihood ratio.
For the diagnosis of BN under WLE, the performance of AI was comparable with that of endoscopists (P = 0.98). Moreover, the method of diagnosis endoscopists versus AI is not a significant source of heterogeneity according to the χ2 test results (0.96).
None of the included studies investigated the performance of endoscopists under NBI in the diagnosis of BN.
3.3. Artificial intelligence in the diagnosis of oesophageal squamous cell carcinoma
Five studies provided extractable and comparable data for the meta‐analysis on the diagnosis of OSCC 26 , 27 , 28 , 29 , 30 (Figure 4). All the studies were conducted in Asia and used DL techniques. Four studies used WLE, 27 , 28 , 29 , 30 and two used NBI. 26 , 30 Two studies investigated the performance of AI during real‐time videos, 26 , 27 whereas three used stored images. 28 , 29 , 30 Two studies compared the performance of the AI system to that of endoscopists. 28 , 30 All the studies defined OSCC according to histology as ground truth.
FIGURE 4.
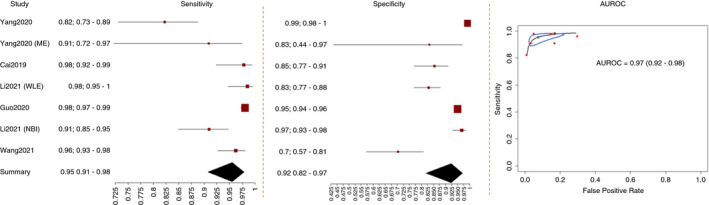
Performance of artificial intelligence in the diagnosis of oesophageal squamous cell carcinoma
The pooled performance of AI in the diagnosis of OSCC with WLE or NBI was: AUROC 0.97 (0.92‐0.98), sensitivity 0.95 (0.91‐0.98), specificity 0.92 (0.82‐0.97), PLR 12.65 (1.61‐3.51), NLR 0.05 (0.11‐0.02) and DOR 258.36 (44.18‐1510.7) in five studies 26 , 27 , 28 , 29 , 30 (Table 1).
Under WLE, the pooled performance was as follows: AUROC 0.98 (0.95‐0.99), sensitivity 0.95 (0.86‐0.98), specificity 0.93 (0.77‐0.98), PLR 14.42 (1.31‐4.11), NLR 0.05 (0.18‐0.02) and DOR 277.2 (19.94‐3852.9) in four studies 27 , 28 , 29 , 30 (Table 1).
With NBI, the pooled diagnostic efficacy was as follows: AUROC 0.98 (0.94‐0.99), sensitivity 0.96 (0.83‐0.99), specificity 0.96 (0.94‐0.97), PLR 23.49 (2.59‐3.62), NLR 0.04 (0.19‐0.01) and DOR 537.21 (71.81‐4018.64) in two studies 26 , 30 (Table 1).
For the real‐time diagnosis of OSCC by AI, the pooled AUROC, sensitivity, specificity, PLR, NLR and DOR were 0.99 (0.94‐0.99), 0.94 (0.79‐0.99), 0.98 (0.94‐0.99), 39.4 (2.51‐4.73), 0.06 (0.23‐0.01) and 651.92 (53.83‐7895.1), respectively, in two studies, 26 , 27 whereas, in non‐real‐time studies, 28 , 29 , 30 the pooled diagnostic efficacy of AI was as follows: AUROC 0.96 (0.89‐0.97), sensitivity 0.96 (0.92‐0.98), specificity 0.87 (0.71‐0.95), PLR 7.29 (1.14‐2.93), NLR 0.05 (0.11‐0.03) and DOR 143.03 (27.61‐741.01). The pooled diagnostic efficacy on videos was comparable with that on images (P = 0.29) (Table 1). There were no significant differences in AUROC across the other subgroups. Moreover, no sources of heterogeneity were found.
As regard the pooled performance of endoscopists in the diagnosis of OSCC, the AUROC was 0.88 (0.83‐0.98), sensitivity 0.75 (0.68‐0.80), specificity 0.88 (0.84‐0.92), PLR 6.46 (1.46‐2.27), NLR 0.29 (0.38‐0.22) and DOR 22.45 (11.5‐43.84) in two studies 28 , 30 (Table 2). The method of diagnosis endoscopists versus AI is a significant source of heterogeneity according to the χ2 test results (0.02), whereas no significant differences in AUROC have been identified across the other subgroups (P = 0.11) (Table 2).
3.4. Artificial intelligence in the detection of abnormal intrapapillary capillary loops
Two studies reported complete and poolable data on the detection of abnormal IPCLs and were included in the meta‐analysis. 31 , 32 Both studies were performed in Asia, used DL algorithms, used fivefold cross‐validation to generate five distinct data sets with different combinations of images and used magnified endoscopy (ME) with NBI. Both studies classified IPCL patterns according to the Japanese Endoscopic Society classification and histology as ground truth. 59
For the detection of abnormal IPCL, the pooled performance of all the included AI algorithms was as follows: AUROC 0.98 (0.86‐0.99), sensitivity 0.94 (0.67‐0.99), specificity 0.94 (0.84‐0.98), PLR 14.75 (1.46‐3.70), NLR 0.07 (0.39‐0.01) and DOR 225.83 (11.05‐4613.93) (Table 1).
The pooled performance of the best fold of each study was AUROC 0.98 (0.97‐0.99), sensitivity 0.99 (0.97‐1.00), specificity 0.97 (0.96‐0.98), PLR 32.18 (3.18‐3.76), NLR 0.01 (0.03‐0.00) and DOR 2779.61 (804.87‐9599.39) (Table 1).
As regard the pooled performance of the worst fold of each study, the pooled AUROC was 0.87 (0.66‐0.96), sensitivity 0.73 (0.55‐0.86), specificity 0.87 (0.71‐0.95), PLR 5.74 (0.64‐2.85), NLR 0.31 (0.64‐0.15) and DOR 18.56 (2.97‐115.85) (Table 1). The bootstrap AUROC comparison across groups indicated a significant difference for the best and worst performance (P = 0.001). The algorithms type was a significant source of heterogeneity according to the x2 test results (<0.001).
3.5. Artificial intelligence in the diagnosis of gastroesophageal reflux disease
Three studies used symptoms questionnaires for the AI‐based diagnosis of GERD and were included in the meta‐analysis. 33 , 34 , 35 Two studies were performed in Europe and used SVM algorithms, 33 , 34 whereas one study took place in Asia and used DL. 35 Two studies defined GERD (as erosive [ERD] or non‐erosive reflux disease [NERD]) based on symptoms and endoscopy findings, 33 , 35 and one study 34 used symptoms, endoscopy findings and pH‐metry as ground truth for the diagnosis of GERD (ERD or NERD).
For the diagnosis of GERD based on questionnaires, the pooled performance of AI was as follows: AUROC 0.99 (0.80‐0.99), sensitivity 0.97 (0.67‐1.00), specificity 0.97 (0.75‐1.00), PLR 38.26 (0.98‐6.22), NLR 0.03 (0.44‐0.00) and DOR 1159.6 (6.12‐219 711.69) in three studies 33 , 34 , 35 (Table 1).
For studies performed in Europe and with SVM, 33 , 34 the pooled AUROC, sensitivity, specificity, PLR, NLR and DOR were 0.98 (0.97‐0.99), 0.99 (0.98‐1.00), 0.99 (0.95‐1.00), 145.88 (3.05‐6.95), 0.01 (0.02‐0.00) and 16 120.13 (1009.41‐257436.50), respectively (Table 1).
The single study performed in Asia and with DL 35 had sensitivity, specificity, PLR, NLR and DOR of 0.70 (0.59‐0.80), 0.78 (0.66‐0.87), 3.25 (0.55‐1.81), 0.38 (0.62‐0.23) and 8.61 (2.8‐26.48), respectively (Table 1).
3.6. Deeks’ funnel plot for publication bias
The Deeks' funnel plot asymmetry test indicated the absence of a publication bias in the included studies (P = 0.39).
4. DISCUSSION
This systematic review with meta‐analysis evaluated the performance of AI in the diagnosis of both malignant and benign ODs. According to this study, AI has potential to accurately diagnose several ODs, clinically and endoscopically.
In the diagnosis of BN, AI showed pooled AUROC, sensitivity and specificity of 90%, 89% and 86%, respectively. The performance of AI was not significantly different from that of expert endoscopists with WLE. These results satisfy the optical diagnosis performance thresholds required by the Preservation and Incorporation of Valuable Endoscopic Innovations (PIVI) initiative by the American Society of Gastrointestinal Endoscopy. According to PIVI indications, any proposed screening technique aspiring to be incorporated into clinical practice, should at least equal, or improve, the performance of random sampling in BE (ie Seattle Protocol), demonstrating a per‐patient sensitivity of 90% or greater, and a specificity of at least 80% for detecting oesophageal adenocarcinoma. 60 Moreover, these results suggest that potentially AI application by non‐expert endoscopists may result in increased early detection of BN and, in the long term improved survival for the patients.
Endoscopic recognition of early OSCC is challenging, as lesions often pass unrecognised with WLE. Lugol's dye spray chromoendoscopy has shown to increase the sensitivity of WLE in the diagnosis of early OSCC, and NBI significantly increases the specificity of oesophago‐gastroduodenoscopies compared with Lugol's dye. 61 , 62 , 63 However, non‐expert endoscopists may not perform as good as experts when operating under NBI, 63 limiting the applicability of the technique. In this meta‐analysis, the application of AI in the diagnosis of OSCC comprehensively achieved pooled AUROC of 97%, pooled sensitivity of 95% and pooled specificity of 92%. These results are in keeping with those of previous meta‐analyses, 9 , 10 , 64 , 65 , 66 in which AI showed good performance in the diagnosis of OSCC, BE‐related or gastric adenocarcinoma and colorectal lesions.
We also provided a pooled estimate of the AUROC, sensitivity and specificity of AI in the detection of abnormal IPCLs, which are microvascular structures on the surface of the oesophagus that appear as brown loops on ME with NBI and show morphological changes that strictly correlate with neoplastic invasion depth of OSCC. Moreover, IPCLs have been also associated to GERD diagnosis and therefore their detection could be helpful also in the endoscopy‐based suspicion of reflux disease. 67 However, the optical classification of IPCLs requires experience and is mastered by experts only. In this study, AI showed pooled AUROC of up to 98% and sensitivity and specificity of up to 99% and 97%, respectively, in the detection of abnormal IPCLs. This has relevant therapeutic and prognostic implications as early lesions are amenable of endoscopic treatment, 68 and the estimation of invasion depth allows intra‐procedural decisions for endoscopic resections. 69 , 70
In this study, CAD tools showed good performance in the diagnosis of benign ODs. Investigations that applied AI to the diagnosis of GERD exclusively based on symptoms were included in the meta‐analysis. In this task, AI achieved pooled AUROC of 99% and sensitivity and specificity of 97%. Because symptoms prompt patients with GERD to seek medical attention, AI‐based questionnaires represent the ideal tool to timely and accurately diagnose reflux disease without performing invasive procedures (ie EGDS and pH‐impedance metry) and avoid the delay of treatment. Moreover, AI excels at solving the non‐linearity inherent in the relationship between symptoms and underlying pathology. Therefore, DL algorithms can be used to reduce the number of questionnaire variables needed to achieve a definite diagnosis of GERD, 34 allowing clinicians to administer shorter and more acceptable questionnaires to patients.
Several single studies that used AI for the diagnosis of benign ODs were retrieved from the literature and could not be included in the meta‐analysis. This lack of data does not reflect a scarce interest in the subject, rather it attests the novelty of AI in the field of oesophageal benign diseases. In this setting, AI models autonomously extracted and analysed pH‐impedance tracings and also individuated a novel pH‐impedance metric that segregated responders to GERD treatment from non‐responders. 52 The effectiveness of a real‐time endoscopic GERD diagnosis and AI algorithm for prediction of EoE diagnosis were also shown. 53 , 55 A CAD tool demonstrated to recognise stationary manometry motor patterns with accuracy, 54 but the application of novel CAD tools to high‐resolution manometry recordings is yet to be evaluated. Importantly, AI demonstrated utility in the recognition of infrequent forms of oesophagitis (ie, CMV vs HSV), which may be mischaracterised even by expert endoscopists. 57
There are limitations that were identified in the included studies. Almost every study available for this meta‐analysis was retrospective. An inherent bias related to the nature of these studies is the convenience sampling of controls (ie, selection bias). In this regard, most studies were based on endoscopic images only, which were often carefully selected among optimal stored endoscopic images. Far less studies tested AI with real‐time endoscopic videoclips, which would better reflect the real life where AI models would help most. On the other hand, in this study, the performance of AI applied to real‐time videos was not statistically different from that on still images, and the performance of AI was similar to that of endoscopists. Additionally, a recent meta‐analysis reported that the inclusion of video clips in the training and validation data sets of AI models could achieve even higher performance than those including images alone. 66
Furthermore, retrospective studies offer the possibility to quickly test for the first‐time hypothesis that could be further investigated by larger and prospective trials.
Most of the studies were based on DL models, and others applied ML with SVM algorithms. Additionally, various training, validation and testing techniques were used in the various investigations, namely a different AI algorithm, a different number of training/validation/testing images or videoclips, and a different proportion of images or videoclips for training, validation and testing. On the other hand, a recent meta‐analysis concluded that AI could detect and characterise colorectal polyps despite the use of different AI algorithms and imaging techniques. 71 Importantly, only two studies included in this meta‐analysis clarified whether a CADe or a CADx was used. 25 , 31 Accordingly, efforts should be put in place in future studies for a more rigorous distinction between detection and diagnosis/characterisation of lesions to overcome this limitation. Of note, one third of the studies included in the qualitative synthesis could not be included in the meta‐analysis because of non‐poolable or non‐extractable data. As it has already been reported, 64 this represents a major limitation of the literature regarding AI in upper GI diseases. Finally, AI itself has inherent limitations. A high volume of training data is needed to refine the performance of the algorithm. In addition, the high computational power of AI carries the risk of overfitting, in which the model is too tightly fitted to the training data and does not generalise towards new data. 72 Furthermore, AI has a black‐box nature, and its decision process is obscure. Therefore, reliance on AI tools should never replace clinical judgement and should be considered of support only.
Despite the application of AI in the diagnosis of ODs is relatively recent, our results demonstrated high accuracy of the examined CAD tools for the evaluation of ODs. However, several limitations still hamper a capillary diffusion of CAD tools in the diagnosis of upper GI disorders, and further prospective and real‐time studies are needed to fully understand what impact will AI have in the practice of gastroenterologists. Our investigation yielded several gaps in the studies that investigated AI in the diagnosis of ODs, which will need to be addressed and filled when designing future studies.
AUTHORSHIP
Guarantor of the article: Prof Edoardo Savarino.
Author contributions: PV, BB, EVS and NdB conceived and drafted the study. PV and BB collected and interpreted all data. BB, DG and DA analysed all data. PV, BB, CH, PS, EVS and NdB drafted the manuscript. All authors commented on the drafts of the paper. All authors have approved the final draft of the manuscript.
Supporting information
Supplementary Material
ACKNOWLEDGEMENTS
Declaration of personal interests: Pierfrancesco Visaggi: none. Brigida Barberio: none, Dario Gregori: none. Danila Azzolina: none. Matteo Martinato. None. Cesare Hassan: reports personal fees from Medtronic, Fujifilm, Olympus, outside the submitted work. Prateek Sharma: Consultant Medtronic, Olympus, Boston Scientific, Fujifilm and Lumendi; Grant Support: Ironwood, Erbe, Docbot, Cosmo pharmaceuticals and CDx labs, outside the submitted work. Edoardo Savarino: has received lecture or consultancy fees from Abbvie, Alfasigma, Amgen, Aurora Pharma, Bristol‐Myers Squibb, EG Stada Group, Fresenius Kabi, Grifols, Janssen, Johnson&Johnson, Innovamedica, Malesci, Medtronic, Merck & Co, Reckitt Benckiser, Sandoz, Shire, SILA, Sofar, Takeda, Unifarco, outside the submitted work. Nicola de Bortoli: has received lecture or consultancy fees from Malesci and Reckitt Benckiser, outside the submitted work.
Declaration of funding interests: None.
Visaggi P, Barberio B, Gregori D, et al. Systematic review with meta‐analysis: artificial intelligence in the diagnosis of oesophageal diseases. Aliment Pharmacol Ther. 2022;55:528–540. doi: 10.1111/apt.16778
Pierfrancesco Visaggi and Brigida Barberio share first authorship.
Edoardo Savarino and Nicola de Bortoli share last authorship.
As part of AP&T’s peer‐review process, a technical check of this meta‐analysis was performed by Dr Y Yuan. The Handling Editor for this article was Dr Colin Howden, and it was accepted for publication after full peer‐review.
[Correction added on July 18, 2022, after first online publication: Cesare Hassan's affiliation has been updated]
DATA AVAILABILITY STATEMENT
No additional data available. All authors approved the final version of the manuscript.
REFERENCES
- 1. Le Berre C, Sandborn WJ, Aridhi S, et al. Application of artificial intelligence to gastroenterology and hepatology. Gastroenterology. 2020;158:76‐94.e2. [DOI] [PubMed] [Google Scholar]
- 2. Ebigbo A, Palm C, Probst A, et al. A technical review of artificial intelligence as applied to gastrointestinal endoscopy: clarifying the terminology. Endosc Int Open. 2019;7:E1616‐E1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sana MK, Hussain ZM, Shah PA, Maqsood MH. Artificial intelligence in celiac disease. Comput Biol Med. 2020;125:103996. [DOI] [PubMed] [Google Scholar]
- 4. Hassan C, Spadaccini M, Iannone A, et al. Performance of artificial intelligence in colonoscopy for adenoma and polyp detection: a systematic review and meta‐analysis. Gastrointest Endosc. 2021;93:77‐85.e6. [DOI] [PubMed] [Google Scholar]
- 5. Barua I, Vinsard DG, Jodal HC, et al. Artificial intelligence for polyp detection during colonoscopy: a systematic review and meta‐analysis. Endoscopy. 2021;53:277‐284. [DOI] [PubMed] [Google Scholar]
- 6. Mori Y, Kudo S‐E, Mohmed HEN, et al. Artificial intelligence and upper gastrointestinal endoscopy: current status and future perspective. Dig Endosc. 2019;31:378‐388. [DOI] [PubMed] [Google Scholar]
- 7. Glover B, Teare J, Patel N. A systematic review of the role of non‐magnified endoscopy for the assessment of H. pylori infection. Endosc Int Open. 2020;8:E105‐E114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nakashima H, Kawahira H, Kawachi H, Sakaki N. Artificial intelligence diagnosis of Helicobacter pylori infection using blue laser imaging‐bright and linked color imaging: a single‐center prospective study. Ann Gastroenterol. 2018;31:462‐468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bang CS, Lee JJ, Baik GH. Computer‐aided diagnosis of esophageal cancer and neoplasms in endoscopic images: a systematic review and meta‐analysis of diagnostic test accuracy. Gastrointest Endosc. 2021;93:1006–1015.e13. 10.1016/j.gie.2020.11.025 [DOI] [PubMed] [Google Scholar]
- 10. Lui TKL, Tsui VWM, Leung WK. Accuracy of artificial intelligence‐assisted detection of upper GI lesions: a systematic review and meta‐analysis. Gastrointest Endosc. 2020;92:821‐30.e9. [DOI] [PubMed] [Google Scholar]
- 11. Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS‐2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529‐536. [DOI] [PubMed] [Google Scholar]
- 12. Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol. 2005;58:982‐990. [DOI] [PubMed] [Google Scholar]
- 13. Noma H, Matsushima Y, Ishii R. Confidence interval for the AUC of SROC curve and some related methods using bootstrap for meta‐analysis of diagnostic accuracy studies. Commun Stat Case Stud Data Anal Appl. 2021;7:344‐358. [Google Scholar]
- 14. Macaskill P, Gatsonis C, Deeks J, Harbord R, Takwoingi Y. Cochrane handbook for systematic reviews of diagnostic test accuracy. Version 0.9. 0. London: The Cochrane Collab. 2010;83. [Google Scholar]
- 15. Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58:882‐893. [DOI] [PubMed] [Google Scholar]
- 16. Doebler P. Meta‐analysis of diagnostic accuracy with mada. R Packages. 2015. https://cran.r‐project.org/web/packages/mada/vignettes/mada.pdf [DOI] [PubMed] [Google Scholar]
- 17. de Groof J, van der Sommen F, van der Putten J, et al. The Argos project: the development of a computer‐aided detection system to improve detection of Barrett's neoplasia on white light endoscopy. United European Gastroenterol J. 2019;7:538‐547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van der Sommen F, Zinger S, Curvers WL, et al. Computer‐aided detection of early neoplastic lesions in Barrett's esophagus. Endoscopy. 2016;48:617‐624. [DOI] [PubMed] [Google Scholar]
- 19. de Groof AJ, Struyvenberg MR, Fockens KN, et al. Deep learning algorithm detection of Barrett's neoplasia with high accuracy during live endoscopic procedures: a pilot study (with video). Gastrointest Endosc. 2020;91:1242‐1250. [DOI] [PubMed] [Google Scholar]
- 20. de Groof AJ, Struyvenberg MR, van der Putten J, et al. Deep‐learning system detects neoplasia in patients with Barrett's esophagus with higher accuracy than endoscopists in a multistep training and validation study with benchmarking. Gastroenterology. 2020;158:915‐29.e4. [DOI] [PubMed] [Google Scholar]
- 21. Ebigbo A, Mendel R, Probst A, et al. Real‐time use of artificial intelligence in the evaluation of cancer in Barrett's oesophagus. Gut. 2020;69:615‐616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hashimoto R, Requa J, Dao T, et al. Artificial intelligence using convolutional neural networks for real‐time detection of early esophageal neoplasia in Barrett's esophagus (with video). Gastrointest Endosc. 2020;91:1264‐71.e1. [DOI] [PubMed] [Google Scholar]
- 23. Ebigbo A, Mendel R, Probst A, et al. Computer‐aided diagnosis using deep learning in the evaluation of early oesophageal adenocarcinoma. Gut. 2019;68:1143‐1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ghatwary N, Zolgharni M, Ye X. Early esophageal adenocarcinoma detection using deep learning methods. Int J Comput Assist Radiol Surg. 2019;14:611‐621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Struyvenberg MR, de Groof AJ, van der Putten J, et al. A computer‐assisted algorithm for narrow‐band imaging‐based tissue characterization in Barrett's esophagus. Gastrointest Endosc. 2021;93:89‐98. [DOI] [PubMed] [Google Scholar]
- 26. Guo L, Xiao X, Wu C, et al. Real‐time automated diagnosis of precancerous lesions and early esophageal squamous cell carcinoma using a deep learning model (with videos). Gastrointest Endosc. 2020;91:41‐51. [DOI] [PubMed] [Google Scholar]
- 27. Yang XX, Li Z, Shao XJ, et al. Real‐time artificial intelligence for endoscopic diagnosis of early esophageal squamous cell cancer (with video). Dig Endosc. 2021;33:1075‐1084. [DOI] [PubMed] [Google Scholar]
- 28. Cai SL, Li B, Tan WM, et al. Using a deep learning system in endoscopy for screening of early esophageal squamous cell carcinoma (with video). Gastrointest Endosc. 2019;90:745‐53.e2. [DOI] [PubMed] [Google Scholar]
- 29. Wang YK, Syu HY, Chen YH, et al. Endoscopic images by a single‐shot multibox detector for the identification of early cancerous lesions in the esophagus: a pilot study. Cancers. 2021;13:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li B, Cai SL, Tan WM, et al. Comparative study on artificial intelligence systems for detecting early esophageal squamous cell carcinoma between narrow‐band and white‐light imaging. World J Gastroenterol. 2021;27:281‐293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. García‐Peraza‐Herrera LC, Everson M, Lovat L, et al. Intrapapillary capillary loop classification in magnification endoscopy: open dataset and baseline methodology. Int J Comput Assist Radiol Surg. 2020;15:651‐659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Everson M, Herrera L, Li W, et al. Artificial intelligence for the real‐time classification of intrapapillary capillary loop patterns in the endoscopic diagnosis of early oesophageal squamous cell carcinoma: a proof‐of‐concept study. United European Gastroenterol J. 2019;7:297‐306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pace F, Riegler G, de Leone A, et al. Is it possible to clinically differentiate erosive from nonerosive reflux disease patients? A study using an artificial neural networks‐assisted algorithm. Eur J Gastroenterol Hepatol. 2010;22:1163‐1168. [DOI] [PubMed] [Google Scholar]
- 34. Pace F, Buscema M, Dominici P, et al. Artificial neural networks are able to recognize gastro‐oesophageal reflux disease patients solely on the basis of clinical data. Eur J Gastroenterol Hepatol. 2005;17:605‐610. [DOI] [PubMed] [Google Scholar]
- 35. Horowitz N, Moshkowitz M, Halpern Z, Leshno M. Applying data mining techniques in the development of a diagnostics questionnaire for GERD. Dig Dis Sci. 2007;52:1871‐1878. [DOI] [PubMed] [Google Scholar]
- 36. Swager A‐F, van der Sommen F, Klomp SR, et al. Computer‐aided detection of early Barrett's neoplasia using volumetric laser endomicroscopy. Gastrointest Endosc. 2017;86:839‐846. [DOI] [PubMed] [Google Scholar]
- 37. Ebigbo A, Mendel R, Rückert T, et al. Endoscopic prediction of submucosal invasion in Barrett's cancer with the use of artificial intelligence: a pilot Study. Endoscopy. 2021;53:878‐883. [DOI] [PubMed] [Google Scholar]
- 38. Riel SV, Sommen FVD, Zinger S, Schoon EJ, With PHNd. Automatic detection of early esophageal cancer with CNNS using transfer learning. 2018 25th IEEE International Conference on Image Processing (ICIP); 2018:1383‐1387. [Google Scholar]
- 39. de Souza LA, Passos LA, Mendel R, et al. Assisting Barrett's esophagus identification using endoscopic data augmentation based on Generative Adversarial Networks. Comput Biol Med. 2020;126:104029. [DOI] [PubMed] [Google Scholar]
- 40. Iwagami H, Ishihara R, Aoyama K, et al. Artificial intelligence for the detection of esophageal and esophagogastric junctional adenocarcinoma. J Gastroenterol Hepatol. 2021;36:131‐136. [DOI] [PubMed] [Google Scholar]
- 41. Ohmori M, Ishihara R, Aoyama K, et al. Endoscopic detection and differentiation of esophageal lesions using a deep neural network. Gastrointest Endosc. 2020;91:301‐9.e1. [DOI] [PubMed] [Google Scholar]
- 42. Horie Y, Yoshio T, Aoyama K, et al. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25‐32. [DOI] [PubMed] [Google Scholar]
- 43. Waki K, Ishihara R, Kato Y, et al. Usefulness of an artificial intelligence system for the detection of esophageal squamous cell carcinoma evaluated with videos simulating overlooking situation. Dig Endosc. 2021;33:1101‐1109. [DOI] [PubMed] [Google Scholar]
- 44. Kumagai Y, Takubo K, Kawada K, et al. Diagnosis using deep‐learning artificial intelligence based on the endocytoscopic observation of the esophagus. Esophagus. 2019;16:180‐187. [DOI] [PubMed] [Google Scholar]
- 45. Liu G, Hua J, Wu Z, et al. Automatic classification of esophageal lesions in endoscopic images using a convolutional neural network. Ann Transl Med. 2020;8:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao YY, Xue DX, Wang YL, et al. Computer‐assisted diagnosis of early esophageal squamous cell carcinoma using narrow‐band imaging magnifying endoscopy. Endoscopy. 2019;51:333‐341. [DOI] [PubMed] [Google Scholar]
- 47. Tokai Y, Yoshio T, Aoyama K, et al. Application of artificial intelligence using convolutional neural networks in determining the invasion depth of esophageal squamous cell carcinoma. Esophagus. 2020;17:250‐256. [DOI] [PubMed] [Google Scholar]
- 48. Nakagawa K, Ishihara R, Aoyama K, et al. Classification for invasion depth of esophageal squamous cell carcinoma using a deep neural network compared with experienced endoscopists. Gastrointest Endosc. 2019;90:407‐414. [DOI] [PubMed] [Google Scholar]
- 49. Fukuda H, Ishihara R, Kato Y, et al. Comparison of performances of artificial intelligence versus expert endoscopists for real‐time assisted diagnosis of esophageal squamous cell carcinoma (with video). Gastrointest Endosc. 2020;92:848‐855. [DOI] [PubMed] [Google Scholar]
- 50. Shimamoto Y, Ishihara R, Kato Y, et al. Real‐time assessment of video images for esophageal squamous cell carcinoma invasion depth using artificial intelligence. J Gastroenterol. 2020;55:1037‐1045. [DOI] [PubMed] [Google Scholar]
- 51. Wu Z, Ge R, Wen M, et al. ELNet: Automatic classification and segmentation for esophageal lesions using convolutional neural network. Med Image Anal. 2021;67:101838. [DOI] [PubMed] [Google Scholar]
- 52. Rogers B, Samanta S, Ghobadi K, et al. Artificial intelligence automates and augments baseline impedance measurements from pH‐impedance studies in gastroesophageal reflux disease. J Gastroenterol. 2021;56:34‐41. [DOI] [PubMed] [Google Scholar]
- 53. Gulati S, Bernth J, Liao J, et al. OTU‐07 Near focus narrow and imaging driven artificial intelligence for the diagnosis of gastro‐oesophageal reflux disease. Gut. 2019;68:A4. [Google Scholar]
- 54. Santos R, Haack HG, Maddalena D, Hansen RD, Kellow JE. Evaluation of artificial neural networks in the classification of primary oesophageal dysmotility. Scand J Gastroenterol. 2006;41:257‐263. [DOI] [PubMed] [Google Scholar]
- 55. Sallis BF, Erkert L, Moñino‐Romero S, et al. An algorithm for the classification of mRNA patterns in eosinophilic esophagitis: integration of machine learning. J Allergy Clin Immunol. 2018;141:1354‐64.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Guo L, Gong H, Wang Q, et al. Detection of multiple lesions of gastrointestinal tract for endoscopy using artificial intelligence model: a pilot study. Surg Endosc. 2021;35:6532‐6538. [DOI] [PubMed] [Google Scholar]
- 57. Lee JS, Yun J, Ham S, et al. Machine learning approach for differentiating cytomegalovirus esophagitis from herpes simplex virus esophagitis. Sci Rep. 2021;11:3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Uema R, Hayashi Y, Tashiro T, et al. Use of a convolutional neural network for classifying microvessels of superficial esophageal squamous cell carcinomas. J Gastroenterol Hepatol. 2021;36:2239‐2246. [DOI] [PubMed] [Google Scholar]
- 59. Oyama T, Inoue H, Arima M, et al. Prediction of the invasion depth of superficial squamous cell carcinoma based on microvessel morphology: magnifying endoscopic classification of the Japan Esophageal Society. Esophagus. 2017;14:105‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sharma P, Savides TJ, Canto MI, et al. The American Society for Gastrointestinal Endoscopy PIVI (preservation and incorporation of valuable endoscopic innovations) on imaging in Barrett's esophagus. Gastrointest Endosc. 2012;76:252‐254. [DOI] [PubMed] [Google Scholar]
- 61. Morita FH, Bernardo WM, Ide E, et al. Narrow band imaging versus lugol chromoendoscopy to diagnose squamous cell carcinoma of the esophagus: a systematic review and meta‐analysis. BMC Cancer. 2017;17:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hashimoto CL, Iriya K, Baba ER, et al. Lugol's dye spray chromoendoscopy establishes early diagnosis of esophageal cancer in patients with primary head and neck cancer. Am J Gastroenterol. 2005;100:275‐282. [DOI] [PubMed] [Google Scholar]
- 63. Ishihara R, Takeuchi Y, Chatani R, et al. Prospective evaluation of narrow‐band imaging endoscopy for screening of esophageal squamous mucosal high‐grade neoplasia in experienced and less experienced endoscopists. Dis Esophagus. 2010;23:480‐486. [DOI] [PubMed] [Google Scholar]
- 64. Arribas J, Antonelli G, Frazzoni L, et al. Standalone performance of artificial intelligence for upper GI neoplasia: a meta‐analysis. Gut. 2020;gutjnl‐2020‐321922. [DOI] [PubMed] [Google Scholar]
- 65. Mohan BP, Khan SR, Kassab LL, Ponnada S, Dulai PS, Kochhar GS. Accuracy of convolutional neural network‐based artificial intelligence in diagnosis of gastrointestinal lesions based on endoscopic images: a systematic review and meta‐analysis. Endosc Int Open. 2020;8(11):E1584‐E1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang SM, Wang YJ, Zhang ST. Accuracy of artificial intelligence‐assisted detection of esophageal cancer and neoplasms on endoscopic images: a systematic review and meta‐analysis. J Dig Dis. 2021;22:318‐328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sharma P, Wani S, Bansal A, et al. A feasibility trial of narrow band imaging endoscopy in patients with gastroesophageal reflux disease. Gastroenterology. 2007;133:454‐464. [DOI] [PubMed] [Google Scholar]
- 68. Kuwano H, Nishimura Y, Oyama T, et al. Guidelines for diagnosis and treatment of carcinoma of the esophagus April 2012 edited by the Japan Esophageal Society. Esophagus. 2015;12:1‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Inoue H, Kaga M, Ikeda H, et al. Magnification endoscopy in esophageal squamous cell carcinoma: a review of the intrapapillary capillary loop classification. Ann Gastroenterol. 2015;28:41‐48. [PMC free article] [PubMed] [Google Scholar]
- 70. Sato H, Inoue H, Ikeda H, et al. Utility of intrapapillary capillary loops seen on magnifying narrow‐band imaging in estimating invasive depth of esophageal squamous cell carcinoma. Endoscopy. 2015;47:122‐128. [DOI] [PubMed] [Google Scholar]
- 71. Lui TKL, Guo CG, Leung WK. Accuracy of artificial intelligence on histology prediction and detection of colorectal polyps: a systematic review and meta‐analysis. Gastrointest Endosc. 2020;92:11‐22.e6. [DOI] [PubMed] [Google Scholar]
- 72. van der Sommen F, de Groof J, Struyvenberg M, et al. Machine learning in GI endoscopy: practical guidance in how to interpret a novel field. Gut. 2020;69:2035‐2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
No additional data available. All authors approved the final version of the manuscript.


