Abstract
A mechanistic approach is presented to describe oxidation of the greenhouse gas methane in the rice rhizosphere of flooded paddies by obligate methanotrophic bacteria. In flooded rice paddies these methanotrophs compete for available O2 with other types of bacteria. Soil incubation studies and most-probable-number (MPN) counts of oxygen consumers show that microbial oxygen consumption rates were dominated by heterotrophic and methanotrophic respiration. MPN counts of methanotrophs showed large spatial and temporal variability. The most abundant methanotrophs (a Methylocystis sp.) and heterotrophs (a Pseudomonas sp. and a Rhodococcus sp.) were isolated and characterized. Growth dynamics of these bacteria under carbon and oxygen limitations are presented. Theoretical calculations based on measured growth dynamics show that methanotrophs were only able to outcompete heterotrophs at low oxygen concentrations (frequently <5 μM). The oxygen concentration at which methanotrophs won the competition from heterotrophs did not depend on methane concentration, but it was highly affected by organic carbon concentrations in the paddy soil. Methane oxidation was severely inhibited at high acetate concentrations. This is in accordance with competition experiments between Pseudomonas spp. and Methylocystis spp. carried out at different oxygen and carbon concentrations. Likely, methane oxidation mainly occurs at microaerophilic and low-acetate conditions and thus not directly at the root surface. Acetate and oxygen concentrations in the rice rhizosphere are in the critical range for methane oxidation, and a high variability in methane oxidation rates is thus expected.
Rice paddies are an important source of the greenhouse gas methane (33). The magnitude of methane emission from rice paddies reflects the balance between methanogenesis and methanotrophy. Methane oxidation occurs at anaerobic-aerobic interfaces with available oxygen and methane: the soil-water interface and the rice rhizosphere. At the soil-water interface, methane oxidation efficiencies are fairly constant at 70 to 95% of the transported methane (21, 25). Estimates of methane oxidation in the rice rhizosphere are much more variable. They range from 7 to 90% of the transported methane (17, 21, 25, 32) and still vary from 7 to 52% if only data obtained from specific inhibitor studies are included. A better mechanistic understanding of rice rhizospheric methane oxidation is important to be able predict methane emissions from rice paddies.
Obligate methanotrophic microorganisms carry out methane oxidation. In freshwater wetlands, high-affinity methanotrophy (3) does not have to be considered due to the high methane concentrations (53) and neither does anaerobic methane oxidation (60). Nitrifiers are also of little importance to methane oxidation in the rice rhizosphere (7). Methanotrophic activity is thus determined by oxygen and methane concentrations. Methanotrophs in the rice rhizosphere do not have to compete for methane with microbial or chemical competitors, although there is a strong sink of methane by methane transport. However, intensive competition for oxygen occurs. To understand the importance of methane oxidation in the rice rhizosphere, the competition for oxygen needs to be quantified. Important oxygen sinks are plant respiration, chemical oxidation, and microbial oxidation.
This paper gradually addresses more-detailed questions. After oxygen sinks in rice paddies are quantified, the isolation and characterization of the most abundant microbial oxygen consumers in this system are described. Their growth kinetics in relation to oxygen and carbon substrate concentrations is studied. Finally, experiments concerning the competition for oxygen between these organisms and theoretical considerations of competition for oxygen are presented.
MATERIALS AND METHODS
Sinks for oxygen; overall oxidation rates.
Ten soil slurries were prepared in 120-ml bottles by mixing 25 g of air-dried rice paddy Maahas soil, a representative rice paddy soil from the Philippines (63), with 25 ml of water. The bottles were closed with butyl stoppers and incubated in an initially 100% N2 headspace at 15, 20, and 30°C while shaking at 120 rpm to obtain a fully reduced soil. After 2 months, samples were taken for analysis of inorganic anions and Fe2+. Thereafter, reoxidation of nitrogen, iron, and sulfurous compounds was measured in duplicate in bottles opened and shaken for 8 h, 1 day, 2 days, and 4 days and in a control that was not opened at all. Samples for inorganic anions and Fe2+ were taken and analyzed daily.
Counts of aerobic microbial communities.
Soil slurry was prepared from air-dried rice paddy Maahas soil from the Philippines. The slurry was left to stabilize for 1 week at 30°C while shaking at 120 rpm. Dilution series were prepared in triplicate down to 10−10/g dry weight of soil with a step size of a factor of 3 in 120-ml bottles with 25 ml of sterile nitrate mineral salts (NMS) medium (66). Bottles were closed with butyl stoppers. Series were prepared at an overall pressure of 140 kPa at 20% O2–80% N2 with 30 mM acetate, at 1% O2–99% N2 with 30 mM acetate (heterotrophic growth), and at 1% O2–19% CO2–80% H2 with 1 mM acetate (autotrophic growth). For each series, treatments with different additional electron donors were prepared by adding 240 mM Fe2+, 30 mM S2O32−, which is more stable than H2S, 30 mM NH4+, 10% CH4 in the gas phase, or no additional donor. A control without soil was included for each treatment. Total salt concentrations were kept the same in all treatments by NaCl. Optical density (OD), measured routinely photospectrometrically at 660 nm, and the consumption of electron donors and electron acceptors were monitored weekly for 10 weeks. Positive growth was assumed if a significant decrease in the additional electron donor concentration (or in acetate concentration, for when no additional electron donor was added) was determined.
Iron oxidation was determined additionally by gel-stabilized gradients in tubes (19) prepared with and without soil inoculation. After 4 weeks, the tubes were frozen and cut into small bands. Each band was analyzed for Fe2+ and O2 after thawing.
Microbial oxygen consumption rates.
Enrichments from the highest dilution with growth on acetate, Fe2+, S2O32, NH4+, and CH4 at 1% O2 and 1 mM acetate were used to quantify the potential activity of microbial oxygen consumption. The total number of bacteria in these enrichments was determined using a Bürker-Türk counting cell. Ten milliliters of the enrichment was added to 25 ml of freshly made sterile NMS medium in a 120-ml bottle with 1% O2 in N2 and additional 30 mM acetate, 240 mM Fe2+, 30 mM S2O32−, 30 mM NH4+, or 10% CH4. Bottles were incubated in the dark at 30°C while shaking at 120 rpm. Oxygen concentrations in the gas phase were monitored daily for 2 weeks.
Culture isolation and characterization.
Microorganisms were isolated from the highest dilution of the dilution series of most-probable-number (MPN) counts (see above). Heterotrophs and methanotrophs were grown with acetate and CH4 as the sole C sources, respectively. Pure cultures were obtained by repeated dilution in liquid NMS medium and by plating on NMS media in 2% highly purified agar. Purity was assessed microscopically after growing the isolates at different conditions.
Cell morphology of isolated heterotrophs, strains HET-1 and HET-2, was analyzed by microscopy after growth in liquid NMS medium with acetate as the sole C source. Colony morphology was analyzed after growth on NMS-agar medium with acetate as the sole C source. The Gram staining reaction and the presence of cytochrome oxidase (40) were tested. The use of acetate, ethanol, glucose, citrate, lactose, and fumarate as sole C sources by the organisms in liquid aerobic NMS medium at 30°C was determined. Decomposition of urea, arginine, lysine, ornithine, and tryptophan in liquid aerobic NMS medium at 30°C was also measured. Denitrifying capacity, reduction of sulfurous compounds, and fermentation of glucose, mannitol, inositol, sorbitol, rhamnose, sucrose, melibiose, and arabinose at anaerobic conditions were analyzed in liquid NMS medium at 30°C. The denitrifying capacity was also tested in solid agar with NMS medium at 30°C. The growth temperature response on 30 mM acetate in liquid NMS medium was tested at 10, 20, 30, 37, and 45°C.
16S ribosomal DNA (rDNA) of the heterotrophic strains grown on agar plates was amplified in a PCR with universal 16S rDNA primers 27F and 1492R (44). The size of the PCR product was tested on an agarose gel, and the product was purified using the QIAquick purification kit. Amplified 16S rDNA was sequenced with the Big Dye Terminator Cycle Sequencing kit (Perkin-Elmer). The first isolate was sequenced with the 342F primer, and the second isolate was sequenced with the 500F primer (44). After the sequencing reaction, DNA was precipitated with the isopropanol precipitation reaction (Perkin-Elmer) to remove the Big Dye Terminator. The sequencing products were analyzed with an ABI310 genetic analyzer (Perkin-Elmer).
Cell morphology of a methanotrophic isolate, strain MOX-1, was analyzed by phase-contrast microscopy after growth in liquid NMS medium with a 30% CH4 headspace. Colony morphology was analyzed after growth on NMS-agar medium with a 30% CH4 headspace as the sole C source. The Gram staining reaction (40) was tested. Flagellation, cytoplasmic membranes, and intracellular storage compounds were investigated by transmission electron microscopy.
Dynamics of methane oxidizers in the rhizosphere.
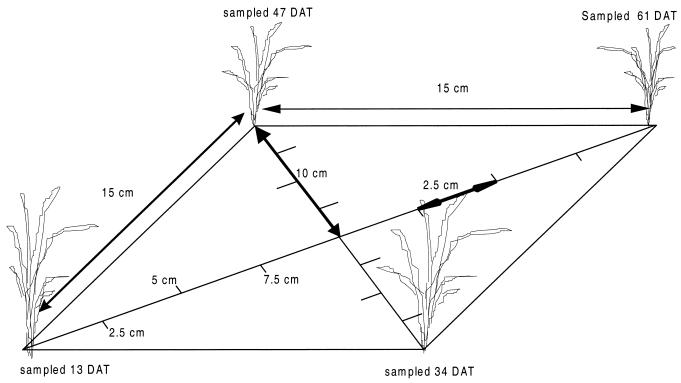
Rice plants were grown in a greenhouse at 26°C and a 12-h dark-12-h light cycle at 80% relative humidity in soil with 15-cm spacing and a large area around the plants to avoid wall effects. Soil samples were taken shortly after transplanting, at the tillering stage, at panicle initiation, and at flowering. At each sampling moment, soil cores were taken at different distances from the main plant (Fig. 1). Material from the top 2 cm was discarded to avoid interference of methanotrophs that accumulated at the soil-water interface. The samples were mixed and diluted in triplicate in 120-ml bottles with 25 ml of sterile NMS medium in dilution series down to 10−10/g dry weight of soil with a step size of a factor of 3. A headspace with air was enriched by 30% CH4. All samples, including six blanks, were incubated at 30°C in the dark without shaking for 5 months. Samples for CH4 were taken every month (and at time zero), and positive growth was assumed if a significant decrease occurred.
FIG. 1.
Setup for soil sampling the MPN of methanotrophs at different moments during the season and at different distances from the main stem of the plant. A rice plant is located at each corner. DAT, days after transplanting.
Batch experiments on growth kinetics.
Monod substrate half-saturation constants for microbial growth (Ks, in moles per liter) and the maximum specific growth rate (μmax, in inverse hours) were determined for all isolated heterotrophs and for the methanotroph in batch experiments in which oxygen and carbon concentrations were sampled at least once a day. Chemostats were found to be inadequate for this purpose given the extremely low, constant oxygen concentrations needed. The organisms were pregrown at low oxygen concentrations to simulate rhizospheric conditions. This pregrowth was necessary because kinetic properties depend on the growth conditions (39). Sterile 120-ml bottles with 25 ml of NMS medium and closed with butyl stoppers were inoculated with organisms in the logarithmic growth stage. For each species, 8 to 10 bottles with various initial oxygen and carbon concentrations, one limiting and the other in excess, were incubated in the dark at 30°C. Concentrations were chosen based on results of preliminary experiments and on published data. Heterotrophs were grown with acetate, and methanotrophs were grown with methane, as the sole C source. Oxygen, acetate, and methane were monitored. Samples were taken anaerobically with syringes flushed in bottles containing N2 and sodium dithionite. Bacterial numbers were analyzed routinely by OD measurements. A species-specific relationship between OD and bacterial numbers was obtained by relating OD to Bürker-Türk counting cell measurements. Values for Ks and μmax for a particular substrate were estimated by minimizing the mean square error between calculated and measured specific growth rates at different substrate concentrations in which only that substrate was limiting.
BOM experiments on maximum growth rate.
The μmax for strain HET-2 was cross-checked polarographically with a Clark-type oxygen electrode (biological oxygen monitor [BOM]) mounted in a thermostatically controlled and continuously stirred vessel at 25°C. The vessel was closed except for a small hole through which substrate and culture additions could be made. The oxygen electrode was calibrated with an oxygen-saturated suspension. The vessel contained 3 ml of oxygen-saturated sterile NMS medium with abundant acetate (20 mM). A 100-μl microbial suspension with a known cell density was added to the vessel. Cells had been pregrown at 25°C and were harvested in the logarithmic growth stage. Oxygen consumption upon addition of substrate was recorded continuously during 30 min and was corrected for endogenous respiration. The relative oxygen consumption rate was determined from the exponential decrease in oxygen concentration in time. μmax was calculated from the relative oxygen consumption rate assuming a yield of 0.375 mol of CB/mol of CS (see below), with subscripts B and S referring to bacteria and substrate, respectively, and an average size of 1.87 × 10−13 g of C/cell (10).
Competition for oxygen between microorganisms.
Competition for oxygen between strain HET-1 and strain MOX-1 was determined in batch experiments. Equal numbers of bacteria as determined from measurements with the Bürker-Türk counting cells were added to 25 ml of sterile NMS medium in 120-ml bottles containing N2 gas. Incubations were made with initial concentrations of 2, 5, and 10 mmol of CH4/liter of gas and of 0.5 and 2 to 4 mmol of O2/liter of gas. The initial acetate concentration was 1 mM in all cases. The bottles were incubated at 30°C while being shaken continuously at 120 rpm. Concentrations of O2, CH4, and acetate were monitored at least once a day until one of the substrates was depleted, which was after 3 to 4 days. Samples were taken at anaerobic conditions with syringes flushed in bottles with N2 and sodium dithionite and analyzed for oxygen, acetate, methane, and OD. The (six) experiments were carried out with four to six replicates.
Chemical analyses.
All samples were centrifuged (after extraction, if essential) for 5 min at 16,000 × g, and the supernatant was analyzed. Acetate was determined by gas chromatography using a Chromosorb 101 column saturated with formic acid at 160°C and connected to a flame ionization detector (FID). Prior to analysis the samples were diluted 1:1 with 1 M formic acid containing 1 mM isobutyric acid as the internal standard. Fe2+ in water and in a 50:1 dilution extract of 0.5 M HCl was analyzed colorimetrically with phenanthroline as the reagent (62). The absorbance was measured at a wavelength of 515 nm on a photospectrometer. NH4+ concentrations were analyzed colorimetrically in a glutamate assay (43). Samples for inorganic anions were diluted 5:1 with a solution of 20 mM mannitol and 60 μM potassium bromide as an internal standard and analyzed on a high-performance liquid chromatograph equipped with suppressed conductivity detection. Anions were separated on an Ionpac AS9-SC column using a 1.8 mM bicarbonate–1.7 mM carbonate eluent at 1 ml/min. CH4 was analyzed by gas chromatography on a molecular sieve column (at 70°C) coupled to a FID. O2 was analyzed on a molecular sieve column (at 100°C) coupled to a thermal conductivity detector. The amounts of CH4 and O2 were quantified using standard curves obtained by injecting known amounts of gases.
THEORY
The outcome of the competition between heterotrophs and methanotrophs can be predicted theoretically from the measured kinetics parameters. For a dynamic microbial population, as in the rice rhizosphere, a straightforward method to calculate microbial biomass kinetics is the application of the double Monod equation (53), assuming no other inhibitory effects,
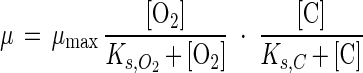 |
1 |
in which μ is the specific or relative growth rate (s−1), μmax is the maximum specific growth rate (s−1), Ks,O2 and Ks,C are the Monod substrate half-saturation constants for oxygen and carbon (molar), respectively, [C] is the concentration of the carbon substrate, and [O2] is the concentration of O2, all referring to the water phase (molar). Specific growth rates reach μmax values if neither oxygen nor carbon is limiting growth. Substrate half-saturation constants are the concentrations of a limiting substrate at which one-half of the maximum specific growth rate is obtained, if other substrates are not limiting.
Equation 1 can be used to determine the results of competition between heterotrophs and methanotrophs, with [C] being [CH4] for methanotrophs and [CH3COOH] for heterotrophs. We describe this outcome of short-term competition for oxygen between heterotrophs and methanotrophs by a critical oxygen concentration, [O2,crit]. It is assumed that competition only takes place for oxygen and that there are no other interactions. [O2,crit,g] is defined as the oxygen concentration at which the specific growth rates (subscript g) of heterotrophs (subscript h) and methanotrophs (subscript m) are equal. Assuming that carbon substrates are not limiting and using equation 1, [O2,crit,g] is
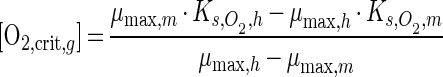 |
2 |
More generally, with limiting carbon substrates
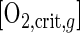 |
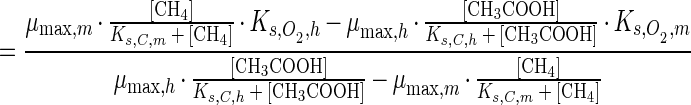 |
3 |
However, it is not μ but the absolute consumption rates of the substrate for which competition takes place that determine the results of competition. Neglecting the consumption for maintenance purposes (because maintenance costs were implicitly accounted for in our experimental setup), the consumption rate (V, in moles per liter per second) is related to μ by
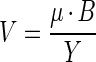 |
4 |
in which B is the bacterial carbon (moles of CB/liter) and Y is the apparent yield (moles of CB/mole of CS). Analogous to the critical oxygen concentration for growth, a critical oxygen concentration for consumption, [O2,crit,c], can be defined, at which both microbial groups consume equal amounts of oxygen:
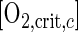 |
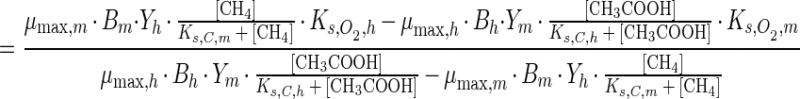 |
5 |
The outcome of the competition depends not only on the growth kinetics but also on the available biomass and the yield. Ym is estimated at 0.296 ± 0.078 mol of CB/mol of CS (30, 36, 47, 54), and Yh is estimated as 0.375 ± 0.039 mol of CB/mol of CS (23, 48, 58). Average yield values calculated from our experiments fall within these ranges (unpublished results), although our experiments were not fully equipped to calculate these values sensitively. Bacterial carbon is calculated from microbial numbers. It is assumed that methanotrophic numbers in the rhizosphere equal 108/g dry weight (see Results) and that heterotrophic numbers equal 109/g dry weight (see Results). An average size of 1.87 × 10−13 g of C/cell (10) is assumed for the heterotrophs. The amount of carbon in methanotrophs decreases proportionally to their smaller size (calculated from the calibration curve of OD versus bacterial numbers).
RESULTS
Sinks for oxygen; overall oxidation rates.
When oxygen was injected into the bottles with soil slurries, a fast increase in oxidized products (NO3− and SO42−) and a decrease of Fe2+ occurred. First-order rate constants (per day) were derived from the oxidation states (Table 1). Iron was oxidized much faster than sulfurous compounds, as could be expected from thermodynamics. NH4+ was oxidized at a much lower rate and also at a rate much lower than those published by others (Table 1). NH4+ might have been limited given its very low concentrations in our soil (data not shown). A first-order rate approach might not have been valid for this situation. The rate constants calculated at 15 and 20°C were only slightly smaller (<10%) than those at 30°C (data not shown), indicating that the oxidation rates were mainly influenced by chemical and not by microbial reactions.
TABLE 1.
Rate constants and the standard errors of oxygen consumption for overall oxidation (as determined in a soil slurry experiment at 30°C) and for microbial reoxidation (as determined from microbial enrichments) and comparison with published data on overall oxidation
| Element or compound | Rate constant (days−1) for:
|
References | ||
|---|---|---|---|---|
| Overall oxidation | Microbial oxidation | Overall published | ||
| Nitrogen | 0.023 ± 0.017 | 0.015 ± 0.021 | 1.1–20 | 49, 55, 61 |
| Iron | 15 ± 0.14 | <0.01 | 0.5–160 | 46, 49, 61 |
| Sulfur | 0.78 ± 0.28 | 0.045 ± 0.006 | 0.18–0.79 | 46, 49, 61 |
| Methane | n.d.a | 0.17 ± 0.03 | ||
| Acetate | n.d. | 1.8 | ||
n.d., not determined.
Counts of aerobic microbial communities.
The results of the MPN counts at aerobic conditions are shown in Table 2. The number of bacteria at 20% O2 was significantly different (analysis of variance [ANOVA], P < 0.05) and orders of magnitude larger than the number at 1% O2 after 10 weeks of incubation. This might imply that 10 weeks is not enough to estimate the population size able to grow at 1% O2 (details below). The population sizes estimated at 1% O2 and 1 and 30 mM acetate were not significantly different (ANOVA), indicating that oxidation of compounds other than acetate was mainly carried out by autotrophic organisms. However, at 1% O2 and 30 mM acetate, autotrophic organisms were significantly inhibited by the competition with heterotrophs, as shown by significantly lower numbers of autotrophic bacteria at 30 mM acetate (ANOVA, P < 0.05). This competition was much less at 1 mM acetate, given the lower availability of acetate than of oxygen. Therefore, for the number of autotrophs, only the incubations at 1% O2 and 1 mM acetate are considered.
TABLE 2.
MPN estimates for aerobic conditionsa
| Bacterial group | MPN (± SE) for:
|
||
|---|---|---|---|
| 1% O2, 1 mM acetate | 1% O2, 30 mM acetate | 20% O2, 30 mM acetate | |
| Heterotrophs | 3 × 105 ± 6 × 104 | 106 ± 2 × 105 | 109 ± 2 × 108 |
| Methanotrophs | 107 ± 3 × 106 | 3 × 106 ± 2 × 106 | 107 ± 7 × 106 |
| NH4+ oxidizers | 103 ± 3 × 102 | <102 | 3 × 105 ± 6 × 104 |
| S2O32− oxidizers | 3 × 105 ± 6 × 104 | 105 ± 2 × 104 | 106 ± 2 × 105 |
| Fe2+ oxidizers | <102 | <102 | <102 |
Values are for numbers of bacteria per gram dry weight of soil after stabilization of the number or at most after 10 weeks of incubation at 30°C.
Heterotrophs and methanotrophs were the most abundant groups at all conditions (Table 2). All other microbial groups only played a minor role in the consumption of oxygen in the rice rhizosphere. Autotrophic ammonia oxidation can be carried out by nitrifiers and methanotrophs (which have some affinity for ammonia). Table 2 shows that nitrifiers were less abundant than methanotrophs, which is in agreement with results found by others (7, 42). At low oxygen but high acetate concentrations, hardly any ammonia oxidation was found, but high heterotrophic respiration took place. Probably, heterotrophs outcompeted ammonia oxidizers (6) and scavenged most of the available oxygen. Similarly, it seems that most of the ammonia consumed at 20% O2 was assimilated by heterotrophs.
Thiosulfate oxidizers were found to be present at all conditions (and in numbers similar to those found by Stubner et al. [56]), but they were present in lower numbers than the methanotrophs and heterotrophs. Thiosulfate consumption hardly increased at conditions favorable for heterotrophs (Table 2), indicating that thiosulfate was mainly oxidized autotrophically. Many colonies were formed on purified agar with thiosulfate and without acetate (data not shown).
Contrary to other reports (19, 20), no iron oxidation (apart from chemical oxidation) could be detected in the MPN counts, either with or without acetate addition. Even in gradient tubes, no differences in oxygen and iron profiles between inoculated and sterile tubes could be detected (data not shown). Chemical iron oxidation proceeded much faster than microbial oxidation. Microbial iron oxidation can thus be neglected.
Microbial oxygen consumption rates.
The rate of oxygen consumption by enrichments was similar to overall oxidation rates and was expressed by first-order rate constants (Table 1). Rate constants for ammonia, thiosulfate, and ferrous iron were much smaller than those found for overall oxidation. Nitrifiers have a low potential growth rate (34), which might explain the low rate constant found for NH4+ oxidation. Microbial thiosulfate oxidation was also much lower than overall thiosulfate oxidation, which is in contrast to what has been found by Stubner et al. (56). Thus, the contribution of microbial processes to overall thiosulfate oxidation might be variable. No microbial iron oxidation was found in the enrichments. Based on the rate constants, heterotrophic respiration and methanotrophic respiration seem the most important microbial sinks of oxygen. Oxidation rates expressed as an initial activity per bacterium showed similar results (data not shown).
Culture isolation and characterization.
Based on the first experiments, the two most important oxygen-consuming microbial groups, the heterotrophs and methanotrophs, were selected for further experiments. Heterotrophs and methanotrophs from the highest dilution with growth were enriched and isolated in NMS medium with acetate and CH4 as the sole C sources, respectively.
Two species of heterotrophic microorganisms were isolated. Strain HET-1 is a gram-negative, oxidase-positive, motile straight rod of 0.8 to 1.0 μm by 1.4 to 1.8 μm. It forms small colonies with a smooth, white, viscous appearance on plates. The organism grows on NMS medium with nitrate as the sole nitrogen source and acetate, ethanol, glucose, citrate, lactose, or fumarate as the sole C source at 30°C. The organism can also decompose urea, arginine, lysine, ornithine, and tryptophan at 30°C. At anaerobic conditions, the organism can denitrify nitrate (both in liquid and agar NMS media), but it cannot grow fermentatively or by the reduction of sulfurous compounds. The organism is able to grow at 10 to 37°C and does not grow at 45°C. A comparison of the partial 16S DNA analysis of the isolate with GenBank data showed as the highest score 99% association with various strains of Pseudomonas stutzeri, which is in accordance with the physiological characteristics (40). Below, the isolate is referred to as Pseudomonas sp. strain HET-1.
Strain HET-2 is a gram-positive, oxidase-negative, nonmotile straight rod, sometimes occurring in pairs and measuring 0.8 to 1.0 μm by 1.1 to 1.4 μm. On plates, it forms small colonies with a red, viscous appearance, a rough edge, and branches. The organism grows in NMS medium with nitrate as the sole nitrogen source and acetate, ethanol, glucose, citrate, or fumarate as the sole C source. Lactose does not support growth. The organism can also decompose arginine, lysine, and ornithine, but not tryptophan and urea, at 30°C. At anaerobic conditions, the organism cannot denitrify or reduce sulfurous compounds, and it cannot grow fermentatively. However, it appears to be able to grow at microaerophilic conditions, because it can grow several centimeters below an agar surface. The organism grows at between 10 and 37°C but does not grow at 45°C. A comparison of the partial 16S DNA analysis of the isolate with GenBank data showed as the highest score 99% association with Rhodococcus erythropolis and Rhodococcus erythreus. The physiological characteristics are in accordance with those of Rhodococcus (28, 40). It is referred to as Rhodococcus sp. strain HET-2.
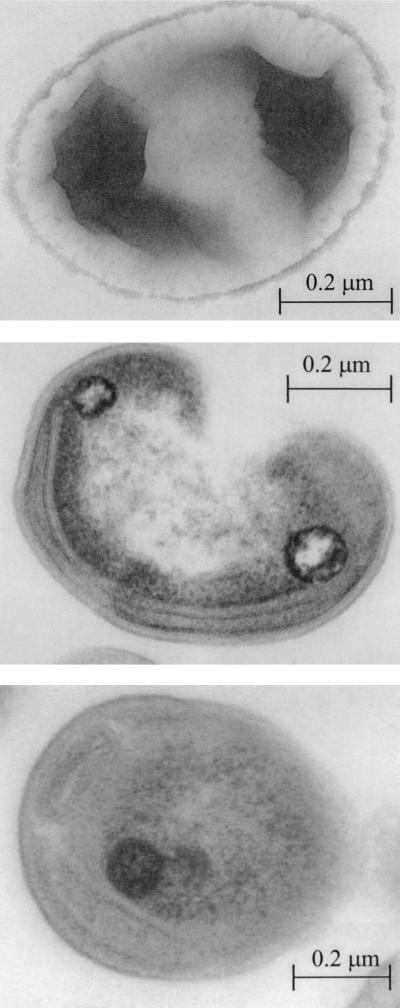
One methanotrophic strain that grows in NMS medium with methane as the sole C source and nitrate as the sole N source was isolated from the highest dilution with methanotrophic growth. The organism is a nonmotile, gram-negative organism with a coccoid and reniform appearance. It has no flagella, as can be seen on the negatively stained preparation (Fig. 2, top). The average size is 0.4 to 0.6 μm by 0.7 to 1.0 μm. On plates, it forms small, round, smooth, butyrous colonies with a white to light yellowish appearance. The growth in liquid media is evenly dispersed, and the organisms are routinely grown at 30°C. Cells have a few layers of paired internal membranes located along the cytoplasmic membrane. The membranes are mostly stacked, but sometimes there is space between the membranes. Based on these observations (Fig. 2, middle), the membranes were identified as type II membranes (15). Polyphosphate accumulates in the cytosol (Fig. 2, bottom). rDNA sequencing analysis confirmed that strain MOX-1 belongs to type II methanotrophs (M. Vecherskaya, unpublished results). Based on this information, the organism was tentatively identified as a Methylocystis species (9) and is called Methylocystis sp. strain MOX-1 in this paper.
FIG. 2.
Transmission electron microscope preparations for the methanotrophic isolate showing a negatively stained preparation for flagellum determination (top), cytoplasmic membranes (middle), and accumulation of polyphosphate inside the cell (bottom).
Dynamics of methane oxidizers in the rhizosphere.
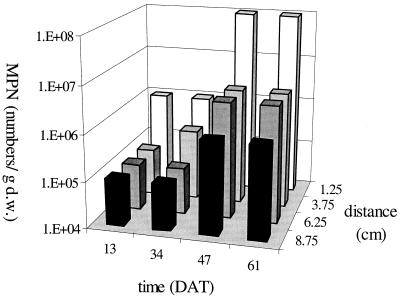
The spatial and temporal growth dynamics of methanotrophs were investigated by MPN counts (Fig. 3). The number of methanotrophs significantly increased during the growing season and significantly decreased with distance from the main stem (ANOVA, P < 0.05). Methane availability increases with distance from the rice root (26), while oxygen availability decreases with distance because of lower root densities and consequently lower root oxygen release at larger distances. Oxygen availability thus limits growth of methanotrophs. Oxygen availability increases in time because of higher root oxygen-releasing activity and higher root densities. The number of methanotrophs was higher in the rhizosphere than in the bulk soil, in accordance with data presented by others (16, 25, 26). Along with changes in the number of methanotrophs, significant changes in maximum methane oxidation rate both in time and with distance from the main rice stem were found (data not shown). Methane oxidation was thus limited by the number of methanotrophs.
FIG. 3.
MPN of methanotrophs at different moments during the growing season (given in days after transplanting [DAT]) and at different distances from the main stem of the plant. Least significant difference of the log-transformed data (LSD-log) with time, 0.87; LSD-log with distance, 0.70. d.w., dry weight.
Data from this experiment showed higher numbers of methanotrophs than reported by others (1 × 104 to 4 × 106/g dry weight), who used incubation periods varying between 3 and 8 weeks (7, 8, 25, 37). The number of methanotrophs obtained after 5 months was similar to that after 4 months but was on average 1 order of magnitude higher than that obtained after 2 months of incubation (data not shown). The total number of methanotrophs can thus be underestimated by using only a 2-month incubation period.
Batch experiments on growth kinetics.
Given the temporal dynamics in MPN counts, growth kinetics needs to be quantified to understand microbial competition for oxygen. Microbial growth in batch experiments was assessed by OD measurements. The OD was linearly related to the number of microorganisms measured with the Bürker-Türk counting cells (data not shown). Specific growth rates obtained from the bacterial number dynamics were related to the average limiting substrate concentration during the sampling interval. Monod substrate half-saturation constants for microbial growth (Ks, molar) and the maximum specific growth rate (μmax, inverse hours) were fitted for each microorganism and each limiting substrate combination by minimizing the mean square error between experimental data and fit (Tables 3 and 4). Modeled and measured μ values were not significantly different (P < 0.05). The μmax values determined with different limiting substrates were similar (Tables 3 and 4).
TABLE 3.
Monod half-saturation constants for microbial growth of heterotrophic pure cultures
| Culture(s) | Ks,O2 (M) | Ks,Ac (M) | μmax (h−1) |
|---|---|---|---|
| Pseudomonas sp. strain HET-1 C limited | n.d.d | 0.36 × 10−3 | 0.054 |
| Pseudomonas sp. strain HET-1 O2 limited | 15 × 10−6 | n.d. | 0.062 |
| Rhodococcus sp. strain HET-2 BOM | n.d. | n.d. | 0.12 |
| Rhodococcus sp. strain HET-2 C limited | n.d. | 1.3 × 10−3 | 0.12 |
| Rhodococcus sp. strain HET-2 O2 limited | 9.0 × 10−6 | n.d. | 0.098 |
| Published pure | (9.4 ± 12.7)a × 10−6 | (0.58 ± 0.39)b × 10−3 | 0.23 ± 0.19c |
TABLE 4.
Monod half-saturation constants for microbial growth of methanotrophic pure cultures
BOM experiments on maximum growth rate.
The μmax for Rhodococcus sp. strain HET-2 was validated by BOM experiments at acetate-limiting conditions. The μmax calculated from this experiment was 0.12 ± 0.03 h−1, which is consistent with the values measured in the batch experiments. The μmax values for all experimental conditions are thus comparable.
Experiments concerning the competition for oxygen between methanotrophs and heterotrophs.
The batch experiments showed a lower Ks,O2 and μmax for methanotrophs than for heterotrophic cultures. Based on these characteristics it is expected that methanotrophs will only win the competition for oxygen with the heterotrophs at very low oxygen concentrations. This hypothesis was tested in competition experiments with equal numbers of Methylocystis sp. strain MOX-1 and Pseudomonas sp. strain HET-1 at different methane concentrations and low (around Ks,O2 for both organisms, expecting the severest competition) and high oxygen concentrations.
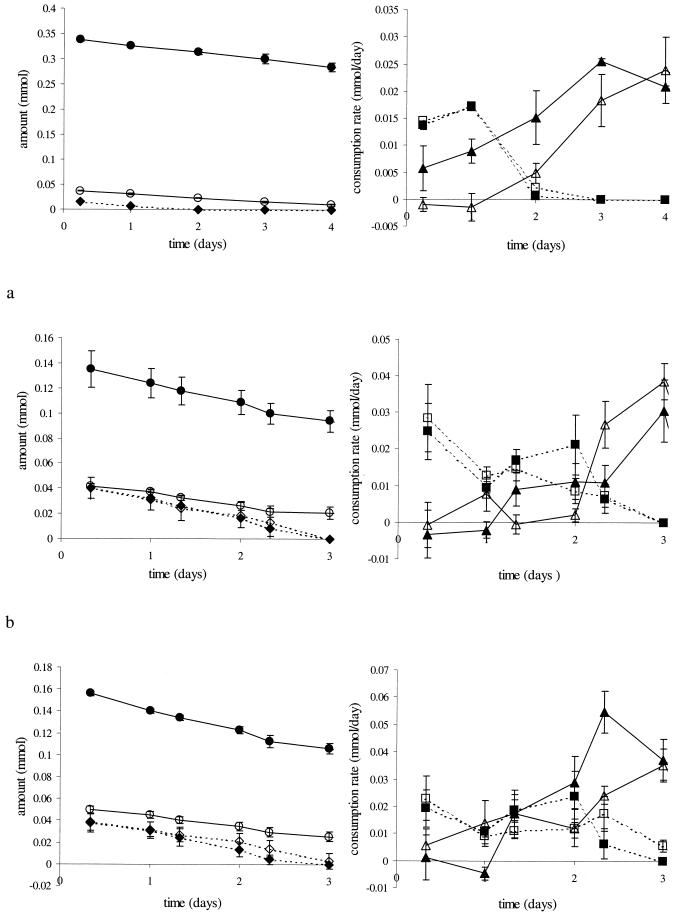
In all competition experiments, methane consumption rates increased after competition for oxygen had stopped and all acetate was consumed (Fig. 4). This increase is partly due to growth of the methanotrophs, which leads to increased biomass that can consume methane. However, the absence of competition for oxygen after all acetate was consumed is probably more important, as at high oxygen concentrations methane consumption rates had already increased before all acetate was depleted due to less-severe competition for oxygen. At low oxygen concentrations methane consumption was suppressed for a longer period and up to lower acetate concentrations. Only at both high methane concentrations and low oxygen concentrations did methane consumption rates increase earlier (Fig. 4c).
FIG. 4.
Competition for oxygen between methanotrophs and heterotrophs at initially 2 (a), 5 (b), and 10 mmol of CH4/liter of gas (c) and at initial concentrations of 0.5 (open symbols) and 2 to 4 mmol of O2/liter of gas (solid symbols). Indicated are the amounts of oxygen (circles) and of acetate (diamonds) and the consumption rates of methane (triangles) and of acetate (squares). Note the different scales.
As long as acetate was present, acetate consumption rates were higher than methane consumption rates. Acetate consumption rates were only slightly suppressed at high methane concentrations. Pseudomonas sp. strain HET-1 is thus a better competitor for oxygen than Methylocystis sp. strain MOX-1, which is reflected in the μmax values of the organisms. An exception occurred at nonlimiting carbon concentrations in combination with low oxygen concentrations. In that case Methylocystis sp. strain MOX-1 had a higher consumption rate (Fig. 4c). The Monod equation predicts that the Ks,O2 value determines the outcome of the competition. The ratio of acetate consumption rate to methane consumption rate was indeed significantly (P < 0.05) correlated to oxygen concentration (data not shown). The experiments show that oxygen limits methane oxidation rates under most conditions. Only at very low oxygen concentrations did methanotrophs seem to be better competitors for oxygen than heterotrophs, due to the high affinity of methanotrophs for oxygen. Only then did methane limit methane oxidation rates.
Theoretical description of competition.
The Monod parameters obtained in the batch experiments (Tables 3 and 4) were used to calculate the critical oxygen concentrations at which specific growth rates for each microbial group were equal (equation 3). A negative oxygen concentration was calculated for the competition between Rhodococcus sp. strain HET-2 and Methylocystis sp. strain MOX-1 at nonlimiting carbon concentrations, which indicates that Rhodococcus sp. strain HET-2 outcompetes Methylocystis sp. strain MOX-1 at all oxygen concentrations under these conditions. This is caused by the much higher μmax for Rhodococcus sp. strain HET-2. A [O2,crit,g] of 3.8 μM was calculated for the competition between Pseudomonas sp. strain HET-1 and Methylocystis sp. strain MOX-1 at nonlimiting carbon concentrations. At lower oxygen concentrations, the methanotroph obtained higher specific growth rates, while the opposite was true for higher oxygen concentrations.
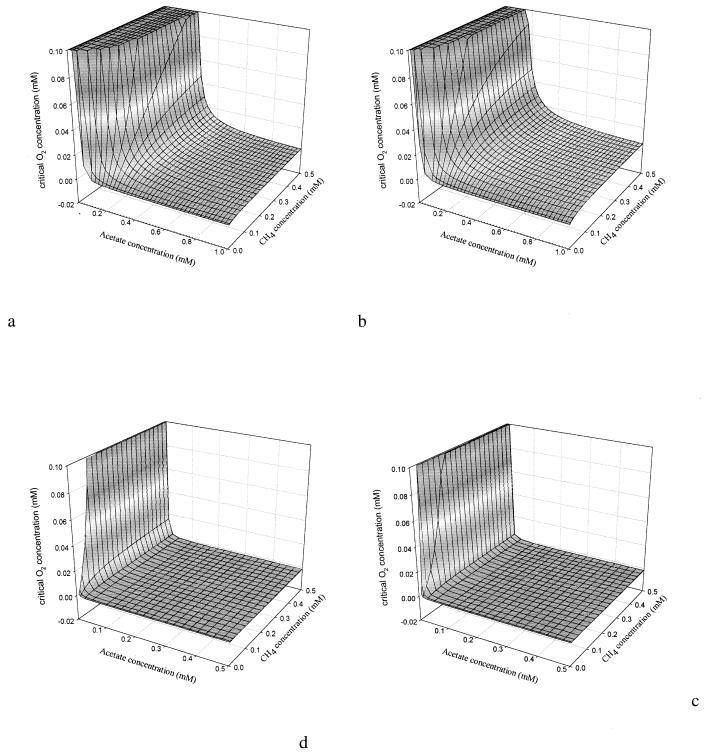
The results for [O2,crit,g] at methane concentrations from 0 μM to saturation and acetate concentrations from 0 to 1 mM are shown in Fig. 5a and b. [O2,crit,g] was in general not very sensitive to the methane concentration. Only at very low methane concentrations did [O2,crit,g] decrease. A [O2,crit,g] of 20 μM or higher was calculated for most acetate concentrations (5 to 30 μM) in rice paddies in the methanogenic phase (1, 22). Such oxygen concentrations are regularly found in the rice rhizosphere (25, 26, 50), implying that specific growth rates of methanotrophs can be similar to those of heterotrophs, although μmax for the heterotrophs was much higher (Tables 3 and 4). At low acetate concentrations, [O2,crit,g] was slightly lower for the competition with Pseudomonas sp. strain HET-1 than for Rhodococcus sp. strain HET-2, because of the lower Ks,C for Pseudomonas sp. strain HET-1. At high acetate concentrations, [O2,crit,g] was higher for the competition with Pseudomonas sp. strain HET-1 than for that with Rhodococcus sp. strain HET-2, because of the lower μmax of Pseudomonas sp. strain HET-1. Only at oxygen concentrations lower than [O2,crit,g] did methanotrophs have a higher μ than heterotrophs.
FIG. 5.
Critical oxygen concentration for competition between Methylocystis sp. strain MOX-1 and heterotrophs Pseudomonas sp. strain HET-1 (b and d) and Rhodococcus sp. strain HET-2 (a and c). (a and b) Specific growth rates; (c and d) consumption rates.
[O2,crit,c] dynamics as a function of methane and acetate (equation 5) for a situation of equal biomasses were similar to those for the situation calculated in Fig. 5a and b and had only slightly higher values (data not shown). These slightly higher values are caused by the (slightly) lower yield values for methanotrophs. In correspondence with the competition experiments (see above), growth of methanotrophs was strongly suppressed at high acetate concentrations. The oxygen concentrations in the competition experiments were in most cases higher than [O2,crit,c], which explains why acetate consumption rates were higher than methane consumption rates in the competition experiments. Only at low oxygen concentrations and nonlimiting carbon concentrations, for which an [O2,crit,c] of 6 μM was calculated in competition with Pseudomonas sp. strain HET-1, were the consumption rates of the methanotrophs higher.
The situation changed dramatically when actual biomass estimates were used to calculate [O2,crit,c]. With nonlimiting carbon concentrations, a negative [O2,crit,c] was calculated, indicating that methanotrophs were outcompeted by the heterotrophs at all oxygen concentrations. At acetate-limiting conditions (and no methane limitation) the [O2,crit,c] became positive at acetate concentrations lower than 53 and 56 μM for Pseudomonas sp. strain HET-1 and Rhodococcus sp. strain HET-2, respectively. Again, methane concentrations hardly had any influence on [O2,crit,c], as shown in Fig. 5c and d. Biomass estimates are thus of major importance for the estimation of competition outcomes.
All parameters to calculate [O2,crit,c] are more or less uncertain, and the influence of these uncertainties should therefore be tested. The estimates of μmax and Ks are not very precise, given their dynamic nature and experimental errors. The coefficients of variation in these parameters were estimated from published data referred to in Tables 3 and 4 and were about 100%. Based on Table 2, the coefficient of variation in B was estimated at 50%. The uncertainty in yield is important given the fact that published instead of measured values were used and given the variability in yield as a function of growth stage and substrate concentration. Standard deviations for yield given above were used as a measure of uncertainty. A Monte Carlo approach (38) of 1,000 runs, in which all parameters were varied within the ranges given by their coefficients of variation, and assuming an acetate concentration of 50 μM and nonlimiting methane concentrations, was carried out. The overall [O2,crit,c] was 3 ± 7 μM (default, 0.13 μM) for competition with Pseudomonas sp. strain HET-1. In 42% of the combinations, methanotrophs were outcompeted by Pseudomonas sp. strain HET-1, and [O2,crit,c] for the remaining combinations was 7 ± 8 μM. This analysis shows that, even though variability in the parameters is large, the range of [O2,crit,c] estimates is limited. The conclusions drawn for the default settings can thus be applied more generally.
DISCUSSION
This study presents the first systematic analysis of the various oxygen sinks in a rice paddy. Incubation studies showed that microbial oxygen consumption was dominated by heterotrophic and methanotrophic respiration. Besides these microbial oxygen sinks, chemical iron oxidation was important. Conditions for both heterotrophs and methanotrophs are good in the rice rhizosphere. Methane concentrations are high due to anaerobic conditions, while oxygen diffuses into the soil, giving good conditions for methanotrophs. Heterotrophs can make use of diverse carbon sources, of which various fatty acids and especially acetate are abundant in rice soils (13, 59) and near decaying rice roots (14). Their maximum specific growth rates are high (Table 3), and some heterotrophs can denitrify at anaerobic conditions. This combination explains why the number of heterotrophic bacteria is high in rice soil (26, 64; this study). The dominant heterotroph that was isolated in this study, a Pseudomonas sp., belongs to the most abundant microorganisms in soils (40). Ammonia oxidizers and thiosulfate oxidizers play only a minor role in the rice rhizosphere; because at the low oxygen concentrations, they are outcompeted by the heterotrophs. The contribution of ammonia oxidizers to methane oxidation seems negligible (7). Moreover, NH4+ concentrations in the rice rhizosphere are usually less than 0.5 mM (7, 26), further limiting the role of ammonia oxidation.
The methanotrophs in a rice paddy are dominated by type II organisms, as shown by our Methylocystis sp. strain MOX-1, as can be expected based on the dynamics in a rice paddy. Type II organisms are better survivors under dry conditions; they can fix molecular nitrogen (9) and do thus no depend on dissolved inorganic nitrogen, and they can utilize lower oxygen (2) concentrations than type I methanotrophs. Type II methanotrophs outcompete type I methanotrophs at high methane concentrations (29). This expectation is in accordance with other studies that isolated type II Methylosinus species (9, 18) and Methylocystis species (57) from rice paddies. Type II methanotrophs prevailed in rice paddies (31, 45). In rice roots, type II methanotrophs were also the dominant methane oxidizers (12), and Gilbert et al. (27) isolated only type II methanotrophs from rice roots.
The authors are not aware of other studies on kinetic properties on oxygen use of heterotrophs in rice paddies. The Ks values of the isolated methanotrophs and heterotrophs are, however, comparable to values published for other systems, although variability among published values is large (Tables 3 and 4). This similarity in affinity constants may indicate that the data for the most abundant microorganisms can be extrapolated to overall methane oxidation kinetics. This is further supported by our sensitivity analysis, using the variability in published kinetics values, showing the general validity of the calculated outcome of the competition for oxygen. These calculations were only possible after our experiments showed the importance of heterotrophs and the abundance of pseudomonas species within the heterotrophic group in rice paddies. μmax values determined in the batch experiment are lower than average published values. Especially the μmax of methanotrophs is at the lower end of the published values. μmax values might have decreased, because the microorganisms were enriched, isolated, and routinely grown at low oxygen concentrations to mimic rice rhizosphere conditions.
A basic assumption in this study is that methane oxidation rates are limited by oxygen. Such a limitation indeed appeared in the MPN counts of the methanotrophs and is in accordance with other studies (8, 11). Theoretical calculations also proved that methane concentrations hardly limited methanotrophic consumption rates. However, it is not the oxygen concentration as such but the competition for oxygen that limits methane oxidation. Both competition experiments between heterotrophs and methanotrophs and theoretical calculations on competition showed that methanotrophs outcompeted heterotrophs only at low oxygen concentrations, due to their lower Ks,O2. At higher oxygen concentrations, except at very low acetate concentrations, methanotrophs were outcompeted, mainly due to their lower μmax. In theoretical calculations using the actual biomass for heterotrophs (high) and methanotrophs (low) methane oxidation was further restricted to areas with low oxygen and low acetate concentrations.
Based on the combination of experiments and calculations, which support each other, this study shows for the first time the available microsites for methane oxidation. Significant methane oxidation can occur only in the rice rhizosphere at microaerophilic, low acetate, and high methane concentrations. This also implies that using a rice variety with low exudation rates—decreasing methane production and the competitive advantage of heterotrophs—and high root oxygen losses—increasing the number of microaerophilic microsites for methane oxidation—may minimize methane emissions from rice paddies. Based on these observations and the presence of an easily accessible carbon substrate in rice fields, we predict that methane oxidation rates at the root surface will be very low, unless oxygen availability is not limiting methane oxidation rates at the root surface. Methanotrophs have been found at the rice root surface (8, 12) and inside the rice root (27, 65), but they were less abundant on the root than in the rhizosphere (26). Probably, heterotrophs consume most oxygen close to the root surface, while methanotrophs are active a bit further away from the root surface at lower oxygen concentrations. Methane oxidation efficiency at the soil-water surface is probably higher than in the rhizosphere, because of the lower acetate concentration in the bulk soil in combination with the longer residence time of methane in the microaerophilic zone.
Lack of insight into microbial biomass dynamics for both heterotrophs and methanotrophs, however, limits a more precise prediction of methane oxidation rates and the occurrence of methane oxidation due to the large influence of microbial biomass on the competition. Predictions of biomass dynamics have limited accuracy, unless actual biomass is measured, as long as quantitative general information on microbial biomass maintenance and survival strategies, as presented for methanotrophs by Roslev and King (51, 52), is scarce.
Recently it was shown that methane oxidation is stimulated by increased nitrogen availability (5) due to an unquantified nitrogen limitation on methanotrophs. Moreover, we showed that our abundant heterotroph is able to denitrify. Besides competition for oxygen, there is thus a potentially important interaction via nitrogen availability between methanotrophs and heterotrophs, possibly a competition for nitrate. However, the kinetics, critical oxygen concentrations for denitrification, and other interactions between denitrifying activity and aerobic respiration are unquantified. Predictions of this interaction are further complicated by the high variability in nitrogen availability in rice paddies (5). Although the experiments in this study show a dominant role for competition for oxygen, methanotrophic activity may be additionally influenced by competition for nitrate, probably leading to an even less profitable position for the methanotrophs.
The high variability of in situ microbial biomass and substrate concentrations, including nitrate, found in the rice rhizosphere, in combination with the sensitivity of the outcome of competition for these variables, indicates that the high variability found in rhizospheric methane oxidation rates might be real. However, the combination of experiments and calculations showed that methane oxidation will only be effective at low oxygen and low acetate concentrations and thus only at very specific microsites within a rice paddy. These results and the fact that oxygen limitations are due to competition with heterotrophs may lead to a better understanding of methane oxidation dynamics and to better ways of predicting methane emissions.
ACKNOWLEDGMENTS
The research was supported financially by the Dutch National Research Program on Global Air Pollution and Climate Change.
We thank Caroline Plugge for technical assistance, Ans Hofman for assistance with the BOM experiment, Ine van Kuijk of Gendika B.V. for the analysis of rDNA sequences, and Jan Goudriaan for critically reading the manuscript.
REFERENCES
- 1.Achtnich C, Schuhmann A, Wind T, Conrad R. Role of interspecies H2 transfer to sulfate and ferric iron-reducing bacteria in acetate consumption in anoxic paddy soil. FEMS Microbiol Ecol. 1995;16:61–70. [Google Scholar]
- 2.Amaral J A, Knowles R. Growth of methanotrophs in methane and oxygen counter gradients. FEMS Microbiol Lett. 1995;126:215–220. [Google Scholar]
- 3.Bender M, Conrad R. Kinetics of CH4 oxidation in oxic soils exposed to ambient air or high CH4 mixing ratios. FEMS Microbiol Ecol. 1992;101:261–270. [Google Scholar]
- 4.Blackall L L, Tandoi V, Jenkins D. Continuous culture studies with Nocardia amarae from activated sludge and their implications for Nocardia foaming control. Res J Water Pollut Control Fed. 1991;63:44–50. [Google Scholar]
- 5.Bodelier P, Roslev P, Henckel T, Frenzel P. Stimulation by ammonium-based fertilizers of methane oxidation in soil around rice roots. Nature. 2000;403:421–424. doi: 10.1038/35000193. [DOI] [PubMed] [Google Scholar]
- 6.Bodelier P L E, Laanbroek H J. Oxygen uptake kinetics of Pseudomonas chlororaphis grown in glucose- or glutamate-limited continuous cultures. Arch Microbiol. 1997;167:392–395. [Google Scholar]
- 7.Bodelier P L E, Frenzel P. Contribution of methanotrophic and nitrifying bacteria to CH4 and NH4+ oxidation in the rhizosphere of rice plants as determined by new methods of discrimination. Appl Environ Microbiol. 1999;65:1826–1833. doi: 10.1128/aem.65.5.1826-1833.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosse U, Frenzel P. Activity and distribution of methane-oxidizing bacteria in flooded rice soil microcosms and in rice plants (Oryza sativa) Appl Environ Microbiol. 1997;63:1199–1207. doi: 10.1128/aem.63.4.1199-1207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowman J P, Sly L I, Nichols P D, Hayward A C. Revised taxonomy of the methanotrophs: description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group I methanotrophs. Int J Syst Bacteriol. 1993;43:735–753. [Google Scholar]
- 10.Bratbak G. Bacterial biovolume and biomass estimations. Appl Environ Microbiol. 1985;49:1488–1493. doi: 10.1128/aem.49.6.1488-1493.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calhoun A, King G M. Regulation of root-associated methanotrophy by oxygen availability in the rhizosphere of two aquatic macrophytes. Appl Environ Microbiol. 1997;63:3051–3058. doi: 10.1128/aem.63.8.3051-3058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calhoun A, King G M. Characterization of root-associated methanotrophs from three freshwater macrophytes: Pontederia cordata. Sparganium eurycarpum, and Sagittaria latifolia. Appl Environ Microbiol. 1998;64:1099–1105. doi: 10.1128/aem.64.3.1099-1105.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin K-J, Conrad R. Intermediary metabolism in methanogenic paddy soil and the influence of temperature. FEMS Microbiol Ecol. 1995;18:85–102. [Google Scholar]
- 14.Conrad R, Klose M. Anaerobic conversion of carbon dioxide to methane, acetate and propionate on washed rice roots. FEMS Microbiol Ecol. 1999;30:147–155. doi: 10.1111/j.1574-6941.1999.tb00643.x. [DOI] [PubMed] [Google Scholar]
- 15.Davies S L, Whittenbury R. Fine structure of methane and other hydrocarbon-utilizing bacteria. J Gen Microbiol. 1970;61:227–232. doi: 10.1099/00221287-61-2-227. [DOI] [PubMed] [Google Scholar]
- 16.De Bont J A M, Lee K K, Bouldin D F. Bacterial oxidation of methane in a rice paddy. Ecol Bull. 1978;26:91–96. [Google Scholar]
- 17.Denier van der Gon H A C, Neue H-U. Oxidation of methane in the rhizosphere of rice plants. Biol Fertil Soils. 1996;22:359–366. [Google Scholar]
- 18.Dianou D, Adachi K. Characterization of methanotrophic bacteria isolated from a subtropical paddy field. FEMS Microbiol Lett. 1999;173:163–173. [Google Scholar]
- 19.Emerson D, Moyer C. Isolation and characterization of novel iron-oxidizing bacteria that grow at circumneutral pH. Appl Environ Microbiol. 1997;63:4784–4792. doi: 10.1128/aem.63.12.4784-4792.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emerson D, Weiss J V, Megonigal J P. Iron-oxidizing bacteria are associated with ferric hydroxide precipitates (Fe-plaque) on the roots of wetland plants. Appl Environ Microbiol. 1999;65:2758–2761. doi: 10.1128/aem.65.6.2758-2761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epp M A, Chanton J P. Rhizospheric methane oxidation determined via the methyl fluoride inhibition technique. J Geophys Res. 1993;98:18413–18422. [Google Scholar]
- 22.Frenzel P, Bosse U, Janssen P H. Rice roots and methanogenesis in a paddy soil: ferric iron as an alternative electron acceptor in the rooted soil. Soil Biol Biochem. 1999;31:421–430. [Google Scholar]
- 23.Gerritse J, Schut F, Gottschal J C. Modelling of mixed chemostat cultures of an aerobic bacterium, Comamonas testosteroni, and an anaerobic bacterium, Veillonella alcalescens: comparison with experimental data. Appl Environ Microbiol. 1992;58:1466–1476. doi: 10.1128/aem.58.5.1466-1476.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerritse J, Gottschal J C. Two-membered mixed cultures of methanogenic and aerobic bacteria in O2-limited chemostats. J Gen Microbiol. 1993;139:1853–1860. [Google Scholar]
- 25.Gilbert B, Frenzel P. Methanotrophic bacteria in the rhizosphere of rice microcosms and their effect on porewater methane concentration and methane emission. Biol Fertil Soils. 1995;20:93–100. [Google Scholar]
- 26.Gilbert B, Frenzel P. Rice roots and CH4 oxidation: the activity of bacteria, their distribution and the microenvironment. Soil Biol Biochem. 1998;30:1903–1916. [Google Scholar]
- 27.Gilbert B, Aβmus B, Hartmann A, Frenzel P. In situ localization of two methanotrophic strains in the rhizosphere of rice plants. FEMS Microbiol Ecol. 1998;25:117–128. [Google Scholar]
- 28.Goodfellow M, Minnikin D E. The genera Nocardia and Rhodococcus. In: Starr M P, et al., editors. The prokaryotes, 2nd ed. A handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. Berlin, Germany: Springer; 1991. pp. 2016–2027. [Google Scholar]
- 29.Graham D W, Chaudhary J A, Hanson R S, Arnold R G. Factors affecting competition between type I and type II methanotrophs in two-organism, continuous-flow reactors. Microb Ecol. 1993;25:1–17. doi: 10.1007/BF00182126. [DOI] [PubMed] [Google Scholar]
- 30.Harwood J H, Pirt S J. Quantitative aspects of growth of the methane oxidizing bacterium Methylococcus capsulatus on methane in shake flask and continuous chemostat culture. J Appl Bacteriol. 1972;35:597–607. doi: 10.1111/j.1365-2672.1972.tb03741.x. [DOI] [PubMed] [Google Scholar]
- 31.Henckel T, Friedrich M, Conrad R. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl Environ Microbiol. 1999;65:1980–1990. doi: 10.1128/aem.65.5.1980-1990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holzapfel-Pschorn A, Conrad R, Seiler W. Production, oxidation and emission of methane in rice paddies. FEMS Microbiol Ecol. 1985;31:343–351. [Google Scholar]
- 33.Houghton J T, Meira Filho L G, Callander B A, Harris N, Kattenberg A, Marskell K. Climate change 1995. The science of climate change. 1st ed. Cambridge, United Kingdom: University Press; 1996. [Google Scholar]
- 34.Hunik J H, Bos C G, van den Hoogen M P, de Gooijer C D, Tramper J. Co-immobilized Nitrosomonas europaea and Nitrobacter agilis cells: validation of a dynamic model for simultaneous substrate conversion and growth in k-carrageenan gel beads. Biotechnol Bioeng. 1994;43:1153–1163. doi: 10.1002/bit.260431121. [DOI] [PubMed] [Google Scholar]
- 35.Iwahori K, Wang M, Taki H, Fujita M. Comparative studies on utilization of fatty acids and hydrocarbons in Nocardia amarae and Rhodococcus sp. J Ferment Bioeng. 1995;79:186–189. [Google Scholar]
- 36.Jørgensen L, Degn H. Mass spectrometric measurements of methane and oxygen utilization by methanotrophic bacteria. FEMS Microbiol Lett. 1983;20:331–335. [Google Scholar]
- 37.Joulian C, Escoffier S, Le Mer J, Neue H-U, Roger P A. Populations and potential activities of methanogens and methanotrophs in rice fields: relations with soil properties. Eur J Soil Biol. 1997;33:105–116. [Google Scholar]
- 38.Keesman K, van Straten G. Set membership approach to identification and prediction of lake eutrophication. Water Resource Res. 1990;26:2643–2652. [Google Scholar]
- 39.Kightley D, Nedwell D B, Cooper M. Capacity for methane oxidation in landfill cover soils measured in laboratory-scale microcosms. Appl Environ Microbiol. 1995;61:592–601. doi: 10.1128/aem.61.2.592-601.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krieg N R, editor. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams & Wilkins; 1984. [Google Scholar]
- 41.Krooneman J, Moore E R B, van Velzen J C L, Prins R A, Forney L J, Gottschal J C. Competition for oxygen and 3-chlorobenzoate between two aerobic bacteria using different degradation pathways. FEMS Microbiol Ecol. 1996;26:171–179. [Google Scholar]
- 42.Kumaraswamy S, Ramakrishnan B, Satpathy S N, Rath A K, Misra S, Rao V R, Sethunathan N. Spatial distribution of methane-oxidizing activity in a flooded rice soil. Plant Soil. 1997;191:241–248. [Google Scholar]
- 43.Kun E, Kearney E B. Ammonia. In: Bergmeyer H U, editor. Methods of enzymatic analysis. Weinheim, Germany: Verlag Chemie; 1974. pp. 1802–1806. [Google Scholar]
- 44.Lane D J. 16S/23S rRNA sequencing. In: Stackbrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- 45.Le Mer J, Escoffier S, Chessel C, Roger P A. Microbiological aspects of methane emission in a ricefield soil from the Camargue (France). 2. Methanotrophy and related microflora. Eur J Soil Biol. 1996;32:71–80. [Google Scholar]
- 46.Murase J, Kimura M. Anaerobic reoxidation of Mn2+, Fe2+, S0 and S2− in submerged paddy soils. Biol Fertil Soils. 1997;25:302–306. [Google Scholar]
- 47.Nagai S, Mori T, Aiba S. Investigation of the energetics of methane-utilizing bacteria in methane- and oxygen-limited chemostat cultures. J Appl Chem Biotechnol. 1973;23:549–562. [Google Scholar]
- 48.Pirt S J. The maintenance energy of bacteria in growing cultures. Proc R Soc London B. 1965;163:224–231. doi: 10.1098/rspb.1965.0069. [DOI] [PubMed] [Google Scholar]
- 49.Ratering S, Conrad R. Effects of short-term drainage and aeration on the production of methane in submerged rice soil. Global Change Biol. 1998;4:397–407. [Google Scholar]
- 50.Revsbech N P, Pedersen O, Reichardt W, Briones A. Microsensor analysis of oxygen and pH in the rice rhizosphere under field and laboratory conditions. Biol Fertil Soils. 1999;29:379–385. [Google Scholar]
- 51.Roslev P, King G M. Survival and recovery of methanotrophic bacteria starved under oxic and anoxic conditions. Appl Environ Microbiol. 1994;60:2602–2608. doi: 10.1128/aem.60.7.2602-2608.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roslev P, King G M. Aerobic and anaerobic starvation metabolism in methanotrophic bacteria. Appl Environ Microbiol. 1995;61:1563–1570. doi: 10.1128/aem.61.4.1563-1570.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Segers R. Methane production and methane consumption: a review of processes underlying wetland methane fluxes. Biogeochemistry. 1998;41:23–51. [Google Scholar]
- 54.Sipkema E M, de Koning W, Ganzeveld K J, Janssen D B, Beenackers A A C M. Experimental pulse technique for the study of microbial kinetics in continuous culture. J Biotechnol. 1998;64:159–176. [Google Scholar]
- 55.Soetaert K, Herman P M J, Middelburg J J. A model of early diagenetic processes from the shelf to abyssal depths. Geochim Cosmochim Acta. 1996;60:1019–1040. [Google Scholar]
- 56.Stubner S, Wind T, Conrad R. Sulfur oxidation in rice field soil: activity, enumeration, isolation and characterization of thiosulfate-oxidizing bacteria. Syst Appl Microbiol. 1998;21:569–578. doi: 10.1016/S0723-2020(98)80069-6. [DOI] [PubMed] [Google Scholar]
- 57.Takeda K. Characteristics of a nitrogen-fixing methanotroph, Methylocystis T-1. Antonie Leeuwenhoek. 1988;54:521–523. doi: 10.1007/BF00588388. [DOI] [PubMed] [Google Scholar]
- 58.Tros M E, Bosma T N P, Schraa G, Zehnder A J B. Measurement of minimum substrate concentration (Smin) in a recycling fermentor and its prediction from the kinetic parameters of Pseudomonas sp. strain B13 from batch and chemostat cultures. Appl Environ Microbiol. 1996;62:3655–3661. doi: 10.1128/aem.62.10.3655-3661.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Bodegom P M, Stams A J M. Effects of alternative electron acceptors and temperature on methanogenesis in rice paddy soils. Chemosphere. 1999;39:167–182. [Google Scholar]
- 60.van Bodegom P M, Leffelaar P A, Stams A J M, Wassmann R. Modelling methane emissions from rice paddies: variability, uncertainty and sensitivity analysis of processes involved. Nutrient Cycling Agroecosyst. 2000;58:231–248. [Google Scholar]
- 61.van Cappellen P, Wang Y. Cycling of iron and manganese in surface sediments: a general theory for the coupled transport and reaction of carbon, oxygen, nitrogen, sulfur, iron and manganese. Am J Sci. 1996;296:197–243. [Google Scholar]
- 62.Walinga I, van der Lee J J, Houba V J B, van Vark W, Novozamsky I, editors. Plant analysis manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. [Google Scholar]
- 63.Wassmann R, Neue H-U, Bueno C, Lantin R S, Alberto M C R, Buendia L V, Bronson K, Papen H, Rennenberg H. Methane production of different rice soils derived from intrinsic and exogenous substrates. Plant Soil. 1998;203:227–237. [Google Scholar]
- 64.Watanabe I, Barraquio W L, de Guzman M R, Cabrera D A. Nitrogen-fixing (acetylene reduction) activity and population of aerobic heterotrophic nitrogen-fixing bacteria associated with wetland rice. Appl Environ Microbiol. 1979;37:813–819. doi: 10.1128/aem.37.5.813-819.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watanabe I, Hashimoto T, Shimoyama A. Methane-oxidizing activities and methanotrophic populations associated with wetland rice plants. Biol Fertil Soils. 1997;24:261–265. [Google Scholar]
- 66.Whittenbury R, Phillips K C, Wilkinson J F. Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol. 1970;61:205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]