Abstract
Cyclooxygenase‐2 catalyzes the biosynthesis of prostaglandins from arachidonic acid and the biosynthesis of prostaglandin glycerol esters (PG‐Gs) from 2‐arachidonoylglycerol. PG‐Gs are mediators of several biological actions such as macrophage activation, hyperalgesia, synaptic plasticity, and intraocular pressure. Recently, the human UDP receptor P2Y6 was identified as a target for the prostaglandin E2 glycerol ester (PGE2‐G). Here, we show that UDP and PGE2‐G are evolutionary conserved endogenous agonists at vertebrate P2Y6 orthologs. Using sequence comparison of P2Y6 orthologs, homology modeling, and ligand docking studies, we proposed several receptor positions participating in agonist binding. Site‐directed mutagenesis and functional analysis of these P2Y6 mutants revealed that both UDP and PGE2‐G share in parts one ligand‐binding site. Thus, the convergent signaling of these two chemically very different agonists has already been manifested in the evolutionary design of the ligand‐binding pocket.
Keywords: PGE2-G, UDP, binding site, site-directed mutagenesis, G protein-coupled receptors
PGE2‐G is an endogenous agonist at the UDP receptor P2Y6. Our study revealed that R103 and R287 contact the phosphate and glyceryl moieties of UDP and PGE2‐G, respectively. The pyrimidine and PGE2 moieties interact with a hydrophobic environment of the ligand‐binding site. Via this shared binding pocket, P2Y6 can integrate different chemical signals into a G protein‐mediated intracellular signal transduction.
Introduction
Prostaglandins are potent bioactive lipid messengers that realize their functions via activation of G protein‐coupled receptors (GPCRs). [1] Cyclooxygenases (COX) catalyze the rate‐limiting step of prostaglandin biosynthesis. Besides this well‐studied enzymatic function of COX isoenzymes, the inducible COX‐2 selectively oxygenates 2‐arachidonoylglycerol to form prostaglandin glycerol esters (PG‐Gs). [2] Due to the rapid degradation of PG‐Gs, there is limited knowledge about their biological function. [3] Previous studies suggested that the PG‐Gs PGE2‐G and PGF2α‐G may activate GPCRs in the murine macrophage‐like cell line RAW264.7 and the human lung's adenocarcinoma cell line H1819. [4] The fast Ca2+ response observed with both cell lines indicated specific signal transduction via unknown Gq‐ and/or Gi protein‐coupled receptors. Using a subtractive screening approach, where mRNA from PGE2‐G response‐positive and ‐negative cell lines was subjected to transcriptome‐wide RNA sequencing analysis, we identified the UDP receptor P2Y6 as the target of PGE2‐G. [5]
Because P2Y6 is expressed in the spleen, thymus, intestine, leukocytes, and aorta, and PGE2‐G is involved in inflammation and macrophage activation, there is accumulating evidence that the P2Y6/PGE2‐G pair functions in an auto‐/paracrine mode. Studies with P2Y6‐deficient mice have shown that P2Y6 is involved in the UDP‐dependent contraction and endothelium‐dependent relaxation of the aorta. [6] P2Y6 is also reported to have high relevance in the immune system. [7] For example, it was demonstrated using P2Y6‐deficient mice that the receptor fine‐tunes the activation of T cells in allergen‐induced pulmonary inflammation [8] and reduces macrophage‐mediated cholesterol uptake in atherosclerotic lesions. [9] PGE2‐G is known to induce hyperalgesia. [10] Experiments with a mouse model of sickle cell disease revealed elevated COX‐2 and PGE2‐G levels responsible for persistent inflammation and hyperalgesia. Pharmacological COX‐2 or P2Y6 inhibition suggested the P2Y6/PGE2‐G pair as a mediator of pain in this animal model. [11]
Currently, it is hypothesized that P2Y6 integrates the two different chemical signals, UDP and PGE2‐G, to a shared intracellular response. Here, nucleotides are released into the extracellular space upon injury and inflammation to serve as a “danger” signal exerting pro‐inflammatory effects. [12] Cell lysis results in an immediate release of nucleotides to reach concentrations >100 nM and recruitment of macrophages via stimulation of P2Y receptors. [13] Similarly, PGE2‐G acts via P2Y6 to regulate the fast and efficient recruitment of macrophages. Previous studies revealed an extremely low EC50 value in the range of 1 pM for PGE2‐G at its receptor.[ 4 , 5 ] Physiologically, this seems reasonable because PGE2‐G only occurs in low amounts and is rapidly hydrolyzed to PGE2. [2b] UDP has been shown to lower intraocular pressure via activation of P2Y6 expressed in the ciliary body making the receptor a promising target for glaucoma treatment. [14] Interestingly, PGE2‐G also reduces intraocular pressure in dogs and monkeys, [15] and one can speculate that this effect is mediated via P2Y6.
Identification of P2Y6 as receptor targeted by PGE2‐G was a first critical step to characterize the physiological function of PG‐Gs and to manipulate this signaling system pharmacologically. However, the structural basis of the promiscuity to at least two structurally not related endogenous agonists is still enigmatic. Our initial studies addressed whether UDP and PGE2‐G share the binding pocket or bind at different sites. Current data supported the hypothesis that UDP and PGE2‐G most probably share receptor interaction sites, but additional determinants private to each agonist may contribute to the individual binding pockets. [5] In this study, we extended our initial structure‐function relation studies by predicting potential interaction sites between the agonists and human P2Y6 with the help of molecular docking and by performing site‐directed mutagenesis studies. We found that the agonist specificity of P2Y6 is evolutionarily old and was already established for both UDP and PGE2‐G in bony fish orthologs. Ortholog comparison, homology modelling, ligand docking, and molecular dynamics simulation proposed several receptor positions participating in agonist binding. Functional analysis of mutant P2Y6 revealed an overlapped binding pocket of both endogenous agonists.
Results and Discussion
Evolutionary conservation of agonist promiscuity
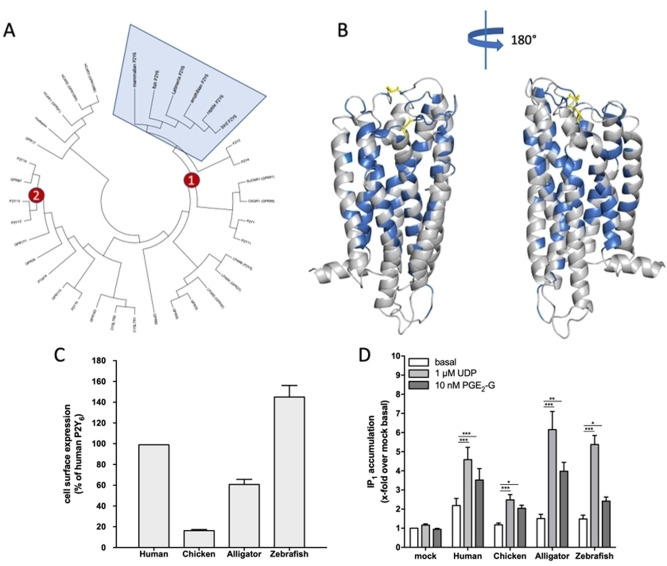
We recently discovered PGE2‐G as an additional endogenous agonist for the human P2Y6. [5] It is not unusual that a given GPCR has more than one physiological ligand as it was shown, e. g., for the TSH receptor having TSH and thyrostimulin as agonists. [16] In most cases of multiple agonism, the ligands are structurally or chemically related. However, in the case of P2Y6, the two agonists identified so far are chemically distinct, and the potencies differ by factor 50,000. [5] This difference suggests that P2Y6 integrates distinct physiological signals related to immune functions[ 7 , 17 ] and pain.[ 10 , 11 ] In our recent study, we found PGE2‐G as an endogenous agonist for human and mouse P2Y6, in addition to UDP. [5] Our present study aims to identify positions within the human P2Y6 relevant for mediating this promiscuous agonist profile. As a first step towards this goal, we followed an evolutionary approach to predict the functional relevance of each position within the receptor protein on the basis of sequence data from orthologs. [18] Sequence divergence of a given amino acid position in a protein is the result of an evolutionary process characterized by the continuous accumulation of mutations, which are subsequently accepted or rejected by natural selection. This process leaves a signature of divergence (high evolutionary rate) or conservation (low evolutionary rate) for each position in the protein sequences. As tested in a phylogenetic analysis (Figure 1A), P2Y6 is an ideal candidate for such an analysis because it is present in almost all vertebrates with one‐to‐one orthology from bony fish to mammals. Aligning 233 full‐lengths vertebrate orthologs from mammals, birds, reptiles, amphibians, and bony fishes (accession numbers and alignment are given in the supplementary material file P2Y6 orthologs.fas), we found 99 amino acid positions (30.2 % of all positions in human P2Y6) that are 100 % conserved between all orthologs. As shown in Figure 1B, these fully conserved positions localize preferentially to the transmembrane helixes 1–7 (TM1–7) with side chains pointing inside the fold stabilizing interactions between TMs and contributing to putative binding pockets for agonists. We tested if these residues provide conserved agonism of both UDP and PGE2‐G in distantly related orthologs using functional assays.
Figure 1.
Phylogenetic relation and structural functional conservation of vertebrate P2Y6. (A) The amino acid sequence of 233 P2Y6 orthologs were aligned (see Experimental Section) using the MUSCLE algorithm. [23] When compared to all other human P2Y‐like sequences all orthologs cluster at the expected position in the phylogenetic tree. Cluster 1 (red circle 1) represents the P2Y1‐like receptor subgroup and Cluster 2 (red circle 2) the P2Y12‐like receptor subgroup. (B) Using a homology modeling approach, the 3D structure of the human P2Y6 was generated [5] and the 100 % conserved positions from the vertebrate P2Y6 alignment (A) are depicted in blue and yellow (disulfide bridges). (C) HEK293T cells were transiently transfected with either HA‐tagged version of the indicated vertebrate P2Y6 orthologs and the expression levels of receptors were measured by a cell surface ELISA (see Experimental Section). Specific optical density (OD) readings (OD value of HA‐tagged P2Y6 constructs minus OD value of mock‐transfected cells) are given as percentage of HA‐tagged human P2Y6 construct. The non‐specific OD value (mock) was 0.001±0.001 (set 0 %) and the OD value of human P2Y6 was 0.609±0.025 (set 100 %). (D) HEK293 cells transfected with the indicated vertebrate P2Y6 orthologs were used for intracellular IP measurements (see Experimental Section). Indicated concentrations of UDP and PGE2‐G were dissolved in 1 % DMSO/assay buffer and controlled against 1 % DMSO/assay buffer without compounds. All data were referred to mock‐transfected cells incubated with 1 % DMSO/assay buffer without compounds. The basal IP1 levels of mock‐transfected cells was 21.7±1.7 nM. All data are given as means±SEM of four (A) and three (B) independent experiments each performed in quadruplicate and triplicate, respectively. *p<0.05, **p<0.01, ***p<0.001 (paired Student's t test).
Although the functionality of P2Y6 upon UDP stimulation has been proven in fish, [19] salamander, [20] and chicken, [21] it is unknown whether PGE2‐G agonism at P2Y6 is preserved at non‐mammalian P2Y6 orthologs. Therefore, we cloned P2Y6 orthologs from zebrafish, alligator, and chicken and measured the cell surface expression of N‐terminally HA‐tagged receptors in a cellular ELISA. As shown in Figure 1C, except for the chicken P2Y6 ortholog (only 20 % cell surface expression), all other variants are well‐expressed at the cell surface compared to the human receptor allowing for functional assays. P2Y6 couples to Gq/11 proteins, and activation increases intracellular inositol phosphate (IP) levels. [5] Functional analysis in an IP1 accumulation assay with UDP (1 μM, [5] ) revealed the expected responses in HEK293T cells transiently transfected with the different P2Y6 orthologs (Figure 1D). The lower UDP‐induced IP1 levels in cells transfected with the chicken ortholog correlated with its lower cell surface expression (Figure 1C). Unfortunately, UDP concentrations above 1 μM were not applicable because of non‐specific IP1 formation HEK293T cells. [5] Next, P2Y6 orthologs were tested with saturating concentration of PGE2‐G (10 nM, [5] ) to determine whether its agonistic property is conserved during evolution. PGE2‐G‐induced IP1‐formation was seen for the human, alligator, chicken, and zebrafish orthologs (Figure 1D). Our assay results are consistent with the presence of COX‐2, the main prostaglandin‐endoperoxide synthase, which is capable of generating PGE2‐G [2b] in all species investigated (see NCBI sequence database). Since agonism was seen with bony fishes, reptiles, birds, and mammals, the common molecular architecture of P2Y6 orthologs must have preserved the conserved agonist‐ and signal transduction specificities.
PDE2‐G and UDP have a partially overlapping binding pocket
Currently, there is no experimental structure available for P2Y6. To estimate whether the two different agonists, UDP and PGE2‐G, may share structural determinants when interacting with the receptor, we simulated binding by docking the agonists into a comparative model of P2Y6. [5] It should be noted that PGE2‐G exists in an equilibrium of two isomeric forms in aqueous solution, PGE2‐1(3)‐glyceryl ester and PGE2‐2‐glyceryl ester (85 : 15 %). It has been demonstrated that the PGE2‐G isomer mixture and hydrolysis‐stable amide analogs of the two PGE2‐G isomers (PGE2‐serinol amide, PTD33) show essentially similar EC50 values at RAW 264.7 and H1819 cells. [4a] Therefore, we performed modeling studies always with PGE2‐2‐glyceryl ester. Based on this initial modeling and docking study we have formed a hypothesis that the two ligands UDP and PGE2‐G may have an overlapping binding pocket flanked by TM3 and TM 5–7 with PGE2‐G extending further to TM2, extracellular loop 2 (ECL2), the extracellular tip of TM6, and the core of TM3 (suppl. Figure S1). The model suggested that UDP and PGE2‐G share a number of interaction sites with others being specific for one of the two agonists. For example, both UDP and PGE2‐G form hydrogen bonds with positions R103 and R287 (numbering is referred to as the human P2Y6) and orient their phosphate moieties and glycerol ester moieties, respectively, towards these positively charged amino acid residues of P2Y6. A precedent docking study already predicted that R103 and R287 contribute to UDP binding. [22] Previously, we identified Y262 mainly participating in UDP binding. [5]
To study the functional relevance of the individual positions predicted to be involved in agonist binding, we performed mutagenesis studies changing the positions individually to Ala and testing the mutants in IP1 accumulation assays. First, we studied the positions proposed to be important to the agonism of in both ligands, UDP and PGE2‐G. Seven of the nine predicted positions are 100 % conserved among vertebrates. The only exceptions are I83 and Y283, which are substituted by Val and Phe in some vertebrates. As shown in Figure 2A, the substitution of all investigated positions with Ala led to a reduction in receptor cell surface expression. Only R103A, Y107A, and R287A showed reasonable cell surface expression levels between 30–40 % of the wildtype P2Y6. Testing the mutants in IP1 assays revealed that none of the mutants showed any response to PGE2‐G (Figure 2B). The mutants I83A, F106A, Y107A, K259A, and Y283A significantly responded to UDP but with extents that mainly correlated to their cell surface expression levels (Figure 2A/B). One exception was I83A displaying low cell surface expression but an almost unchanged basal activity and response to UDP. Considering only those mutants that appear at the cell surface to a significant amount, R103 and R287 participate in the agonistic activities of both ligands, whereas Y107 contributes only to PGE2‐G activity.
Figure 2.
UDP and PGE2‐G have overlapping agonist binding sites at P2Y6. (A, B) Positions predicted to interact with both, UDP and PGE2‐G were individually mutated to alanine. (C, D) Positions predicted to preferentially interact with PGE2‐G but not with UDP were individually mutated to alanine. (E, F) Most positions mutated to alanine were also mutated to physicochemically related amino acids. HEK293T cells were then transfected with wildtype (Wt) and mutant P2Y6. (A, C, E) Cell surface expression of mutant P2Y6 receptors was determined as described. Optical density (OD) is given as percentage of P2Y6 Wt minus OD of mock‐transfected cells. Data are given as means±SEM of three independent experiments performed in quadruplicate. (B, D, F) Transfected HEK293 cells were stimulated with UDP (1 μM) and PGE2‐G (10 nM) and tested in IP1 accumulation assays as described. All data are means±SEM of three to five independent experiments, each performed in triplicate. *p<0.05, **p<0.01, ***p<0.001 (paired Student's t test).
Furthermore, all positions that were previously predicted to participate mainly in PGE2‐G binding (suppl. Figure S1A) were mutated to Ala and tested in cell surface and IP1 assays. The mutant Y75A, N109A, S291A, and N293A were expressed at the cell surface at detectable levels and above (Figure 2C). Y75A and N293A were still active upon UDP incubation but not in the presence of PGE2‐G (Figure 2D). N109A and S291A displayed half of the basal activity of the wildtype P2Y6 but were marginally activated by UDP. This data set revealed Y75 and N293 as residues that might be involved in PGE2‐G but not in UDP agonist activation. Thus, N109 and S291 are necessary for directly or indirectly forming the binding site of both agonists.
In sum, site‐directed mutagenesis studies (Figure 2) identified only a few positions that could be adequately evaluated because of sufficient cell surface expression (>25 % of the wildtype). It should be noted that we failed to perform saturation binding assays and concentration‐response curve experiments, methods that are usually engaged for detailed characterization of the mutants. UDP and PGE2‐G are unsuitable for radioligand‐binding studies because of high background noise due to nucleotide binding to many cellular targets and PGE2‐G's very lipophilic nature, respectively. Furthermore, performing concentration‐response curves with UDP in the used heterologous cell system is limited because UDP concentrations >10 μM produced an endogenous signal in IP1 assays (data not shown). With these limitations, we found two categories: i) loss of activation by both agonists and ii) loss/strong reduction of activation by PGE2‐G. Except for the previously characterized mutant Y262A, [5] we did not identify any other mutation that caused a loss of UDP activation but not PGE2‐G agonism. It, therefore, seems that PGE2‐G mainly occupies most of the UDP‐binding side but recruits additional interaction partners.
Iterative refinement of P2Y6 models binding PDE2‐G and UDP
For further model refinement, we constructed a new P2Y6 homology model in an iterative process [24] using an updated list of GPCR template structures (suppl. Table S1). We also performed docking of UDP and PGE2‐G with a new induced‐fit docking protocol and incorporate the experimental restraints as the constraints to guide the positioning of the ligands during docking. [25] By breaking down the Rosetta binding energy at the residue level, we examined the contribution of each residue to the interaction between the ligand and P2Y6 in each docking pose. We used the binding strength to prioritize the docking poses that encompass the favorable interactions between the ligand and the residues critical to the ligand activity according to mutagenesis results. [26] Then, the selected docked models were subjected to conventional molecular dynamics (MD) for a total of 1.5 μs in three replicates to confirm kinetic stability of the observed binding poses. The structures are stable during the MD simulation as the RMSD, and the RMSF of both P2Y6 transmembrane helices and the ligand are in the reasonable ranges (Figures 3 and 4). Finally, we examined the involvement of different P2Y6 residues in the engagement of two agonists through the frequency of pairwise interactions between the ligands of the receptor. We calculated the relative contact strength (Figure 5) as the sum of atom pair interaction.
Figure 3.
P2Y6‐PGE2‐G molecular dynamics‐refined docked model. PGE2‐G as docked to P2Y6 homology models, then the selected docked model was further refined with total of 1.5 μs of molecular dynamics. Lateral (A–B) and extracellular (C) views of the MD‐refined model of PGE2‐G docked in the comparative model of the human P2Y6 are shown. Hydrogen bonds are indicated as dashed yellow lines, and side chains of residues that are important to PGE2‐G activity are shown in sticks. The seven transmembrane helices (TM) are numbered from N‐ to C‐terminal (I–VII). Plots of RMSD to the starting docked model throughout the MD simulation (D–E) and per‐residue RMSF after discarding the first 100 ns of the MD simulation (F) are given. To allow comparison between residues in the TM domain of different rhodopsin‐like GPCRs, residues are numbered according to the Ballesteros‐Weinstein numbering scheme [27] additionally to the position in the human P2Y6.
Figure 4.
P2Y6‐UDP molecular dynamics‐refined docked model. UDP was docked to P2Y6 homology models, then the selected docked model was further refined with total of 1.5 μs of molecular dynamics. Lateral (A–B) and extracellular (C) views of the MD‐refined model of UDP docked in the comparative model of the human P2Y6 are shown. Hydrogen bonds are indicated as dashed yellow lines, and sidechains of residues that are important to UDP activity are shown in sticks. The seven transmembrane helices (TM) are numbered from N‐ to C‐terminal (I–VII). Plots of RMSD to the starting docked model throughout the MD simulation (D–E) and per‐residue RMSF after discarding the first 100 ns of the MD simulation (F) are given. To allow comparison between residues in the TM domain of different rhodopsin‐like GPCRs, residues are numbered according to the Ballesteros‐Weinstein numbering scheme [27] additionally to the position in the human P2Y6.
Figure 5.

Computed per‐residue relative contact strengths of UDP and PGE2‐G to P2Y6 suggest overlapping binding pocket of the two agonists. Relative contract strength is the sum of atom pair contact frequency between each agonist and P2Y6 residues. A relative contact strength of a particular residue is equal to 1 means that there is an atom from the ligand interact with a side chain atom of the corresponding in all frames of the simulation, on average. A contact strength of 10 is considered to be significant. (A) Relative contact strength between P2Y6 residues and two agonists was computed and the threshold of 10 is marked (red lines). The x axis and y axis are shown in a log scale. Residues with significant interaction strength to both PGE2‐G and UDP and only to PGE2‐G are shown in blue and red quadrants, respectively. (B) Front and back view of the overlapped binding pockets of two agonists. P2Y6 residues shared between both agonists are shown in blue, while the residue that only show strong relative contact strength to either UDP or PGE2‐G are colored in red and yellow, respectively.
We selected final docking poses based on agreement with the experimental data and Rosetta interface energy score for both, UDP and PGE2‐G (suppl. Figure S2, suppl. Table S2). Based on the co‐crystal structures of P2Y1 with the antagonists MRS2500, [28] and P2Y12 with its agonists 2MeSADP and 2MeSATP, [29] we hypothesized that the corresponding positively charged residues in P2Y6, R103 (3.29) and R287 (7.39), form hydrogen bonds and electrostatic interactions to the group with the highest electron density of UDP and PGE2‐G, the diphosphate residue group and the glycerol ester group, respectively (Figures 3B–C and 4B−C). Our hypothesis is consistent with our mutagenesis data. Mutation of R103 and R107 to Ala abolished UDP activation of the mutant P2Y6 (Figure 2B), although the receptor mutants were still expressed at the cell surface (Figure 2A), indicating no gross structural alterations of the receptor. Similarly, the R103A and R287A mutants could not be activated by PGE2‐G (Figure 2B). Docking PGE2‐G into the P2Y6 homology model revealed only one cluster where both R103 and R287 participate in ligand binding. In refined docked models, both Arg residues coordinate the glyceryl moiety and carbonyl oxygen of the ester form hydrogen bonds together with K284 (TM7), D179 (ECL2), and D90 (TM1) (Figures 3B–C, and suppl. Figure S3). To further characterize the relevance of these residues, we separately mutated both positions to Lys, which kept the positive charge but reduced the number of possible hydrogen bond donors (R103 K, R287 K). As shown in Figure 2F, both mutants are incompatible with PGE2‐G activation, but UDP still activated R287 K. This discrepancy in agonism of those Arg to Lys mutants indicated that the glyceryl moiety of PGE2‐G might be benefited from alternately interacting with multiple hydrogen bond donors of R287. At the same time, UDP only needs to form a salt‐bridge with a positively charged side chain at this position.
According to the computational models, while Y107 shields the binding pocket of PGE2‐G toward the cytosolic half of the receptor (Figure 3B), this residue, together with Y189, formed π‐π interactions and confined the movement of UDP's pyrimidine ring (Figure 4B). Additionally, hydrophobic interactions of Y192 with the aliphatic backbone of the PGE2 and pyrimidine moiety of UDP, respectively, are possible (Figures 3 and 4). However, Y107 seems to be necessary only for activation with PGE2‐G. Therefore, we also asked whether Y107 can be replaced by Phe only to keep the aromatic ring. As shown in Figure 2E, Y107F abolished cell surface expression of P2Y6 so that both agonists cannot activate the receptor (Figure 2F).
We also mutated other positions, which we had already mutated to Ala (Figure 2A–D), by changing them into more conservative mutations (Y75F, I83N, F106Y, N109Q, L110V, G193V, F252Y, F255Y, K259R, Y283F, N293Q). These mutants were functionally tested to check whether more distinct physicochemical changes are compatible with receptor functionality. I83N, Y107F, G193V, and S291T were purely expressed at the cell surface and showed no or small responses to UDP and PGE2‐G (Figure 2E/F).
Y75F showed a similar functionality as the wildtype receptor, however, with significantly lower IP1 responses to both agonists. In the model, this residue is located far below the binding site of both agonists (as viewed from extracellular) and, most likely, contributes indirectly to the formation of the binding pocket. In contrast, F106 is located in the model in the vicinity of both agonists, and mutation to Tyr abolished activation by both agonists. Still, the mutation failed to interfere with basal receptor activity and reduced cell surface expression only to 50 % of the wildtype P2Y6 (Figure 2E/F). It is, therefore, likely that F106 contributes to the coordination of both ligands within the binding pocket. Our models suggest that this F106 is in contact with both ligands (Figures 3D, 4D, and 5A). N109, S291, and N293 cluster below the proposed bindings site are essential for either stabilizing the binding pocket or the downstream propagation of the activation pathway. Interestingly, the residue N293 is one of the four residues of the Na+ binding pocket switch, which has been shown to be essential for the activation mechanism of many other class A GPCR agonists. [30]
In both models, Y283 forms hydrogen bonds with the P2Y6 agonists (Figures 3B, 4B, and suppl. Figure S3). Exchange of this residue with Phe only interferes with activation by PGE2‐G (Figure 2E). Thus, it is possible that the bound conformation of UDP can still be sufficiently stabilized with the hydrogen/salt bridge network between its diphosphate groups and close‐by positively charged residues K25, R103, R287, and K259 (Figures 3B–D and suppl. Figure S3). Furthermore, F252 and F255 are directly located below Y283, probably forming a π‐electron stack stabilizing Y283 in its position (Figure 3B). Mutation of both residues to Tyr retained UDP activation but abolished PGE2‐G‐induced receptor activity. These results indicate that PGE2‐G‐mediated binding and/or activation depends on the correct orientation of this aromatic stack formed by Y283 (TM7), F252 (TM6), and F255 (TM6).
To examine the overlapping area between the binding pockets of two agonists, we identified a list of P2Y6 residues that frequently interact with both ligands (relative contact strength of more than 10) during the last 400 ns of each MD simulation replicates. Those residues are shown as dots on blue area on Figure 5A and blue surfaces in Figure 5B. The overlapping area of the binding pockets spans across the extracellular half of TM3, TM5, TM6, and TM7, and the tip of TM2, TM4, and TM5. Out of 15 identified common residues, three residues (R103, F107, R287) were confirmed by mutagenesis studies. Eleven of the remaining twelve residues are in close proximity with those residues that were confirmed to be important for activation by both agonists. The only exception is Y189, as discussed above. A list of six residues that interacted with UDP more frequently than with PGE2‐G during the MD simulations were marked in red. We also identified a list of 12 residues that interacted with PGE2‐G more frequently than with UDP (yellow area and surface) (Figure 5A–B).
These two residue lists implied in the binding pocket of PGE2‐G might expand to the tip of TM1‐4 and the core of TM6, while the binding pocket of UDP expand to the core of TM3 and TM5. Despite the significant overlap in binding pockets of those agonists, the potency of PGE2‐G was ∼50,000 fold higher compared to that of UDP. [5] Therefore, we have calculated binding free energy using MM‐PBSA. Our results also suggest the calculated binding free energy of PGE2‐G was significantly lower than of UDP to P2Y6, with the difference between mean ▵G values across three MD replicates of ∼14 kcal/mol. Although the electrostatic interaction energy was more favorable in binding of UDP, the van der Waal interaction energy was almost double for PGE2‐G binding than UDP binding (suppl. Table S3). Those observations could be explained by ionic interactions from UDP's diphosphate groups, and extensive non‐polar interactions from PEG2‐G's lipophilic chain.
Interpretation of evolutionary, functional, crystal structural and modeling data
Regarding the evolutionary aspect, Y75, R103, N109, Y262, R287, S291, and N293 are fully conserved among vertebrate P2Y6 orthologs indicating their structural and functional importance as suggested in our docking models. R103 (R3.29), Y283 (Y7.35), and R287 (R7.39) are conserved in the P2Y1‐like receptor subgroup but not in the P2Y12‐like receptor subgroup (Figure 1A). Those observations are in line with the fact that, within the crystal structures of the ADP‐bound P2Y1 and P2Y12 receptors, the agonist binding sites significantly differ between both receptors.[ 28 , 29 , 31 ] In P2Y12‐like cluster 2, the respective positions are S/A3.29, K7.35, and L7.39. N109, S291, and N293 are also found in other receptors shown in Figure 1A at the corresponding positions indicating more general structural functions. Our new mutagenesis data residues reported that residues, such as Y75 and N293, mediated only the agonism of PGE2‐G with P2Y6. Interestingly, both of those residues did not form significant contacts to either ligand based on the models. Furthermore, N293 is one of the four residues that constitute the Na+ binding pocket, whose repack switching is essential to the activation of many other class A GPCRs. [30b] For the P2Y6 mutants with sufficient cell surface expression, most conserved residues either directly interact directly with both ligands (R103 and R287) or indirectly stabilize the PGE2‐G's binding pocket (Y75, N109, S291, N293).
Docking studies and MD simulations provided a potential atomic‐detail explanation to our mutagenesis results. The modeling data suggest that the diphosphate group of UDP and the glycerol ester moiety of PGE2‐G form a hydrogen interaction network with two positively charged residues R103 and R287 (Figures 3 and 4), and nearby residues such as K259 and Y283. Furthermore, the pyrimidine ring of UDP and the ω‐lipophilic chain of PGE2‐G form hydrophobic interaction with Y107. Unfortunately, mutating K259 and Y283 to Ala caused the P2Y6 cell expression insufficient to reliably detect their effects on the activation of UDP and PGE2‐G. However, the importance of R103, R287, and Y107 in the agonistic activity of UDP and PGE2‐G was confirmed by mutagenesis studies (Figure 2). Our refined models suggest that these two P2Y6 agonists have partially overlapped binding pockets, stretching from the extracellular half of TM3, TM5, TM6, and TM7, to the tip of TM2, TM4, and TM5 (Figure 5).
Interestingly, our proposed agonist engagement modes of P2Y6 rather resemble the general activation mechanism previously proposed from the crystal structures of P2Y12 [29] than from a structural study on P2Y1. [32] More specifically, 2MeSADP's negatively charged diphosphate group forms hydrogen bonds/salt bridge residues on the extracellular half of both P2Y1 and P2Y12 receptors, stabilizing the proximity between TMs 3–4 and TMs 6–7 (the “closed state”), which is consistent with our models (Figure 6). However, unlike the agonist engagement mechanism of P2Y1 proposed by Yuan et al. [32a] and Ciancetta et al., [32b] the binding pocket of P2Y6 agonists in our models locate deeper toward the core of the intermembrane helices. Hence, UDP and PGE2‐G maintain the “closed state” of P2Y6 while being buried inside the helical bundle, similar to the binding mode of P2Y12 and its agonist, 2MeSADP. Interestingly, both of our proposed docked models show that the phosphate and glyceryl moieties of UDP and PGE2‐G, respectively, blocked or frequently disrupted the salt bridge between D179 (ECL2) and R2877.39 (Figure 3B, 4B, and suppl. Figure S4). This ionic bond was observed to be blocked by the P2Y1 agonist and was suggested to be important in maintaining the inactive state of P2Y1 in those mentioned computational studies. [32] Throughout our MD simulations, while UDP's negative phosphate group completely blocked this ionic interaction, the glyceryl group of PGE2‐G frequently formed hydrogen bonds to either one of those two residues, interrupting this salt bridge (suppl. Figure S4C). Further structural studies are needed to confirm our observation regarding the similarity and differences in agonistic activation mechanism among P2Y6, P2Y1, and P2Y12.
Figure 6.
Comparison between the P2Y6‐UDP docked model and the P2Y12‐2MeSADP complex (PDB ID: 4PXZ). Lateral (Left) and extracellular (Right) views of the complexes are presented. Side chains of key residues that forms favorable interaction to UDP and 2MeSATP are shown in cyan and grey sticks, respectively. The seven transmembrane helices (TM) are numbered from N‐ to C‐terminal (I–VII).
Furthermore, compared to the structures of prostaglandin EP receptors (EP2, EP3, EP4) with PGE2, an analog of PGE2‐G, the binding pocket of PGE2‐G shifts around 13.5 Å toward TM4/5 (Figure 7A). This might be due to there are substantially more positive charged residues in the extracellular half of the transmembrane region of P2Y6 than in that of EP receptors (Figure 7B−D). In fact, since the sequences similarities and identities were very low (suppl. Table S4), we did not include any of the EP receptors as templates to reconstruct the homology model of P2Y6.
Figure 7.

Comparison between the P2Y6‐PGE2‐G and the EP3‐PGE2 complexes (PDB ID: 6AK3). (A) Overall view of position of the binding pockets of PGE2‐G (grey spheres) and PGE2 (magenta spheres). The ring of PGE2‐G shifts around 13.5 Å toward the TM5 compared to that of PGE2. (B) A close‐up look of EP3 residues (grey lines) that interacted with PGE2 (grey balls and sticks). The only positively charged side chain, R333, that located close to the ligand was also shown in grey balls and sticks. (C–D)The transmembrane region of P2Y6 (C‐magenta) has significantly more positively charged side chains (shown in sticks) than transmembrane region of EP3 (D‐grey), enabling the shift and elongation of the binding pose of PGE2‐G (magenta balls and sticks). In contrast, the only positively charged residue in the extracellular half of the transmembrane region of EP3 is R333 (grey sticks) on TM7.
Conclusion
P2Y6 is the target of two endogenous agonists, UDP and PGE2‐G, since over 419 million years of vertebrate evolution. [33] Two polar residues, R103 (R3.29) and R287 (R7.39), interact to phosphate and glyceryl moieties of those two ligands, respectively. In contrast, the pyrimidine and PGE2 moieties interact with a more hydrophobic environment of the ligand‐binding site. Via this shared binding pocket, P2Y6 could integrate different chemical signals into a Gq/11 protein‐mediated intracellular signal transduction. To extend the understanding of P2Y6 activation beyond our currently known uniform IP1 and Ca2+ responses, future studies should investigate these two agonists gradually differ in their induced signal transduction, e. g., in the kinetics of signaling or by recruiting other G proteins and arrestins.
Experimental Section
Materials. If not stated otherwise, all chemicals were purchased from Sigma‐Aldrich (Germany), and cell culture materials were provided by Life Technologies GmbH (Germany). PGE2‐G was from Cayman Chemical, Ann Arbor, MI, USA.
Generation of receptor constructs. cDNA from H1819 cells was used to amplify and clone the human P2Y6 coding sequence. [5] In addition, genomic DNA from chicken, alligator, and zebrafish was used to amplify the respective coding sequences of P2Y6. All sequences were double‐tagged with an N‐terminal HA epitope and a C‐terminal FLAG epitope and, for transient transfection, introduced into the mammalian expression vector pcDps. [34] All mutant constructs were generated by a PCR‐based site‐directed mutagenesis and fragment replacement strategy. All constructs were verified by sequencing.
Cell culture, transfection, measurement of intracellular inositol phosphates. For functional assays, receptor constructs were heterologously expressed in human embryonic kidney (HEK293T) cells upon transient transfection. Cells were grown in DMEM/F12 supplemented with 10 % FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin at 37 °C and 5 % CO2. An indirect cellular ELISA was used to estimate cell surface expression of heterologously expressed receptors carrying an N‐terminal HA tag. [35] According to the manufacturer's protocol, to measure IP1, HEK293T cells were split into 96‐well plates (20,000 cells/well) and transfected with 100 ng vector constructs using Lipofectamine® (Invitrogen). Empty vector (mock) served as a negative control. Then, 48 h after transfection, cells were stimulated 30 min at 37 °C with 35 μl 1× IP1 stimulation buffer (Cisbio) containing the respective reagents (concentrations as indicated). Next, cells were lysed by adding 30 μl lysis buffer (Cisbio) per well and kept frozen at −20 °C until measurement. IP1 measurements using the Cisbio IP‐one Tb kit (Cisbio, Codolet, France) were performed in ProxiPlate‐384 Plus microplates (Perkin Elmer) with the EnVision Multilabel Reader (Perkin Elmer). The assays were performed with a final concentration of 1 % DMSO.
Phylogenetic analysis of vertebrate P2Y6 . The amino acid sequence of 233 P2Y6 vertebrate orthologs were aligned using the MUSCLE algorithm [23] (sequence fasta file is provided). When compared to all other human P2Y‐like sequences all orthologs cluster at the expected position in the phylogenetic tree. The evolutionary history was inferred using the Neighbor‐Joining method. [36] The optimal tree with the sum of branch length=24.14196614 is shown. The evolutionary distances were computed using the Poisson correction method [37] and are in the units of the number of amino acid substitutions per site. All positions with less than 95 % site coverage were eliminated. That is, fewer than 5 % alignment gaps, missing data, and ambiguous bases were allowed at any position. There were 314 positions in the final dataset. Evolutionary analyses were conducted in MEGA7. [38]
Generation of P2Y6 comparative models. A comparative model of P2Y6 was constructed using the protein structure prediction software package, ROSETTA version 3.12, [39] using multiple GPCR templates. [24] The X‐ray crystal structures of P2Y1 and P2Y12 (Protein Data Bank ID: 4xnw, 4ntj)[ 28 , 31 ] were chosen as main templates based on high sequence similarity to P2Y6. To increase conformational sampling, these templates were supplemented with protease‐activated receptors (PARs) PAR1 and PAR2 (3vw7 and 5ndd), [40] angiotensin II type I and type II ATI and ATII (6do1 and 5ung), [41] kappa opioid receptor (6b73), [42] free fatty acid receptor (FFAR) 1 (5tzr), [43] platelet‐activating factor receptor (PAFR) (5zkp), [44] and endothelin B receptor (ETBR) (6igk). [45] The information of the name and PDB IDs, as well as the sequence identity and similarity to P2Y6, are summarized in the suppl. Table S1. An initial sequence alignment of 11 GPCR receptors was created using the GPCRdb structure‐based sequence alignment application. [46] Adjustments were then made to ensure that all secondary structure elements were properly aligned while moving significant gaps to loop regions. In addition, the first 15 and last 12 residues of the P2Y6 sequence were truncated as they are not crucial for the binding of the ligands. [5]
After assigning coordinates to P2Y6 residues from each template alignment using Rosetta's partial‐thread application, RosettaCM [47] ‘hybridizer’ was used to combine segments across all templates in a metropolis Monte Carlo with a simulated annealing approach to arrive at energetically favorable compositions. In brief, RosettaCM exchanges template fragments into a starting model to achieve energetically favorable hybrid template models. Any residues still lacking coordinates were modeled de novo using 3mer and 9mer fragments extracted from the PDB fragment database. Transmembrane segments, as predicted using the OCTOPUS server [48] and adjusted to match with the transmembrane spans of the P2Y1 and P2Y12 helices according to the calculation made by the PPM server, [49] were modeled within Rosetta's implicit membrane potential. [50] The resulting full sequence models were subjected to eight iterative cycles of sidechain repacking and gradient minimization within the membrane potential. P2Y6, P2Y1, and P2Y12 share a conserved disulfide bond between the N‐terminal C18 and C273 in extracellular loop 3. [51] Therefore, disulfide bond constraints were introduced between these residues as well as C99 and C177. Secondary constraints were also applied to the extracellular loop 2 (ECL2) of P2Y6 models so that its beta‐hairpin structure is maintained during loop modeling. In total, 20,000 P2Y6 homology models were generated. The top 10 % of all generated models by pose score were clustered by Cα RMSD using K‐means clustering into eight clusters. The top ten scored models from each of those eight clusters were selected for docking. The models were deposit in the Protein Model Database (http://srv00.recas.ba.infn.it/PMDB/main.php) with the structure IDs PM0084119 and PM0084120.
Rosetta ligand docking. Ligand docking into the comparative model of P2Y6 with UDP and PGE2‐G was performed with Rosetta Ligand. [52] One hundred conformations of PGE2‐G and 100 conformations of UDP were generated with BCL::Conf. [53] This application builds small‐molecule conformations from substructures derived from small molecule crystal structures in the Crystallography Open Database (COD). A starting position was selected for both ligands based on the average of ligands present in all GPCR templates. The induced‐fit docking protocol started with an initial docking round with high constraint weight to penalize the ligand placements that were far away from the residues deemed important to the molecule's activity according to the mutagenesis data. Then, another round of relaxing the backbone of the residues surrounding the ligand to mimic the induced fit effect, and a final refinement docking with low constraint weight to optimize the ligand‐receptor atomic interactions. The docking protocol included a low resolution (centroid mode) phase consisting of 500 cycles sampling ligand conformers in 4 Å translation search and complete reorientation search, which are constraints by preset distance‐based constraints from the mutagenesis results, and a high‐resolution phase consisting of six cycles of sidechain refinement with small perturbations of ligand poses and conformation. During the refinement phase, the translation search was reduced to 1 Å, and the constraint weigh score was reduced to 1. This phase finds an energetically favorable pose by combining minor ligand conformational flexibility with sidechain refinement simultaneously. For each ligand, the top 10 % models by interface delta score were collected in each of three rounds of induced fit docking. Those top models were then clustered, and the top 10 models were selected for the next round of docking. The Rosetta interface scores versus ligand RMSDs graphs after the final round of induced fit docking are shown in the suppl. Figure S2.
For each selected pose cluster, a value, the change in free energy with and without ligands bound to P2Y6, was calculated for each residue in the receptor. A binding strength score, which measures the linear sum of ΔΔG of residues that are favorable and unfavorable to the activity of the ligands, was calculated as , where is the computed Rosetta value for the residues that are not important for the ligand activity based on the mutagenesis data and also have a negative value. is the computed Rosetta value of the residues that were shown experimentally to affect the activity of the ligands. Essentially, the binding strength score measures the relative agreement between a particular docking pose and the mutagenesis data. [26] The suppl. Table S2 shows the computed average per residue values for each cluster. For UDP docking models, the pose cluster with highest binding strength score was be selected.
MD simulation of the selected docking models. Selected docking models were then refined with molecular dynamics simulation. For each ligand, tree independent replicates with 1.5 μs in total simulation time were conducted. All membrane systems were built with the membrane building tool PackMol‐Memgen. [54] Downser++ [55] were then used to dock waters inside the transmembrane region of P2Y6 in the presence of the ligands. The bi‐membrane system contained POPC and Cholesterol with a molecule number ratio of 10 : 1. Proteins, lipids, TIP3P water, and ions were modeled with the FF19SB [56] and Amber Lipid17 [57] force fields, and the ligands were modeled with the GAFF2 small molecule force field. [58] A TIP3P water layer of 25 Å was included, and Cl− or K+ ions were added to neutralize the charge of the system. Each bilayer system was first minimized for 5,000 steps using steepest descent followed by 15,000 steps of conjugate gradient minimization. During heating, the protein backbone and sidechain atoms, lipid and water were restrained to their starting coordinates with harmonic force constants of 10 kcal mol−1 Å−2 and 5 kcal mol−1 Å−2, heated to 10 K over 10,000 steps with a step size of 0.1 fs using constant boundary conditions and Langevin dynamics with a rapid collision frequency of 10,000 ps−1. The system was then heated to 100 K over 500,000 steps in 50 ps with constant volume dynamics and the collision frequency set to 1000 ps−1 and, finally, to 303 K over 1,000,000 steps with constant pressure dynamics and anisotropic pressure scaling turned on, while the positional restraints on the system were gradually removed. The system was then run with the protein‐complex held fixed for another one ns at 303 K. Production MD was conducted for 500 ns at 303 K using a step size of 4 fs with hydrogen mass repartitioning, [59] constant pressure periodic boundary conditions (NPT system), semi‐anisotropic pressure scaling, and Langevin dynamics. MD trajectories were analyzed using CPPTRAJ (version 18.0) and PTRAJ (version 2.0.2.dev0), [60] as well as VMD (visual molecular dynamics; version 1.9). [61] The first 100 ns of the simulation was removed before we performed the calculation of RMSF, atomic contact, and Molecular Mechanics with a Poisson‐Boltzmann/Surface Area solvent (MM‐PBSA). Relative contract and hydrogen bonding frequencies were calculated as the sum of atom pair contact frequency between the agonist and each P2Y6 residue. Trajectories of the last 400 ns of each of three MD replicates were clustered, and the representative frame, which is also the centroid, of the largest cluster were chosen as final refined docked models.
Protein‐ligand free energy calculations. Protein‐ligand binding free energy calculations were performed with MM/PBSA implemented in the AmberTools18’s MMPBSA.py. [62] Trajectories were stripped of water, ions, and membrane molecules. Energies were computed with an ionic strength of 0.150 mM, internal dielectric constant of 20.0, and van der Waals and Coulombic interactions cutoff distance of 99 Å and 7.0 Å. The non‐polar contribution to the solvation free energy was approximated total non‐polar solvation free energy is modeled as a single term linearly proportional to the solvent accessible surface area as in PARSE. [63] Default radii assigned with Leap were kept for PBSA calculations. Membrane dielectric constant was set to be 7.0, and the membrane thickness was set to be 36 Å. The enthalpic and solvation free energy contributions were computed every 4 ns over 400 ns of each MD simulation replicate. All calculations were completed from three independent trajectories, and the results were reported in the suppl. Table S3.
Conflict of interest
The authors declare no conflict of interest.
1.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
This work was supported by the German Research Foundation CRC1423 project number 421152132 (T. S., J. M.). Anne Zimmermann was supported by the MD program of the Medical Faculty, University Leipzig. We thank Katja Ettig, Ben Brown, and Dr. Georg Kuenze for excellent technical help and assistance. Open Access funding enabled and organized by Projekt DEAL.
A. Zimmermann, O. Vu, A. Brüser, G. Sliwoski, L. J. Marnett, J. Meiler, T. Schöneberg, ChemMedChem 2022, 17, e202100683.
Contributor Information
Prof. Dr. Jens Meiler, Email: jens@meilerlab.org.
Prof. Dr. Torsten Schöneberg, Email: schoberg@medizin.uni-leipzig.de.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Woodward D. F., Jones R. L., Narumiya S., Pharmacol. Rev. 2011, 63, 471–538. [DOI] [PubMed] [Google Scholar]
- 2.
- 2a. Alhouayek M., Muccioli G. G., Trends Pharmacol. Sci. 2014, 35, 284–292; [DOI] [PubMed] [Google Scholar]
- 2b. Kozak K. R., Crews B. C., Ray J. L., Tai H. H., Morrow J. D., Marnett L. J., J. Biol. Chem. 2001, 276, 36993–36998. [DOI] [PubMed] [Google Scholar]
- 3. Kingsley P. J., Rouzer C. A., Morgan A. J., Patel S., Marnett L. J., Adv. Exp. Med. Biol. 2019, 1161, 77–88. [DOI] [PubMed] [Google Scholar]
- 4.
- 4a. Richie-Jannetta R., Nirodi C. S., Crews B. C., Woodward D. F., Wang J. W., Duff P. T., Marnett L. J., Prostaglandins Other Lipid Mediators 2010, 92, 19–24; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4b. Nirodi C. S., Crews B. C., Kozak K. R., Morrow J. D., Marnett L. J., Proc. Natl. Acad. Sci. USA 2004, 101, 1840–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bruser A., Zimmermann A., Crews B. C., Sliwoski G., Meiler J., Konig G. M., Kostenis E., Lede V., Marnett L. J., Schoneberg T., Sci. Rep. 2017, 7, 2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bar I., Guns P. J., Metallo J., Cammarata D., Wilkin F., Boeynams J. M., Bult H., Robaye B., Mol. Pharmacol. 2008, 74, 777–784. [DOI] [PubMed] [Google Scholar]
- 7. Le Duc D., Schulz A., Lede V., Schulze A., Thor D., Bruser A., Schoneberg T., Adv. Immunol. 2017, 136, 85–121. [DOI] [PubMed] [Google Scholar]
- 8.
- 8a. Garcia R. A., Yan M., Search D., Zhang R., Carson N. L., Ryan C. S., Smith-Monroy C., Zheng J., Chen J., Kong Y., Tang H., Hellings S. E., Wardwell-Swanson J., Dinchuk J. E., Psaltis G. C., Gordon D. A., Glunz P. W., Gargalovic P. S., PLoS One 2014, 9, e111385; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8b. Giannattasio G., Ohta S., Boyce J. R., Xing W., Balestrieri B., Boyce J. A., J. Immunol. 2011, 187, 1486–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stachon P., Peikert A., Michel N. A., Hergeth S., Marchini T., Wolf D., Dufner B., Hoppe N., Ayata C. K., Grimm M., Cicko S., Schulte L., Reinohl J., von zur Muhlen C., Bode C., Idzko M., Zirlik A., Arterioscler. Thromb. Vasc. Biol. 2014, 34, 2237–2245. [DOI] [PubMed] [Google Scholar]
- 10. Hu S. S., Bradshaw H. B., Chen J. S., Tan B., Walker J. M., Br. J. Pharmacol. 2008, 153, 1538–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Khasabova I. A., Uhelski M., Khasabov S. G., Gupta K., Seybold V. S., Simone D. A., Blood 2019, 133, 1989–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kepp O., Loos F., Liu P., Kroemer G., Immunol. Rev. 2017, 280, 83–92. [DOI] [PubMed] [Google Scholar]
- 13.
- 13a. Communi D., Janssens R., Suarez-Huerta N., Robaye B., Boeynaems J. M., Cell. Signalling 2000, 12, 351–360; [DOI] [PubMed] [Google Scholar]
- 13b. Lazarowski E. R., Boucher R. C., Harden T. K., Mol. Pharmacol. 2003, 64, 785–795. [DOI] [PubMed] [Google Scholar]
- 14.
- 14a. Shinozaki Y., Kashiwagi K., Namekata K., Takeda A., Ohno N., Robaye B., Harada T., Iwata T., Koizumi S., JCI Insight 2017, 2; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14b. Jacob T. F., Singh V., Dixit M., Ginsburg-Shmuel T., Fonseca B., Pintor J., Youdim M. B. H., Major D. T., Weinreb O., Fischer B., Purinergic Signal 2018, 14, 271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Woodward D. F., Poloso N. J., Wang J. W., J. Pharmacol. Exp. Ther. 2016, 358, 173–180. [DOI] [PubMed] [Google Scholar]
- 16. Nakabayashi K., Matsumi H., Bhalla A., Bae J., Mosselman S., Hsu S. Y., Hsueh A. J., J. Clin. Invest. 2002, 109, 1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.
- 17a. Koizumi S., Shigemoto-Mogami Y., Nasu-Tada K., Shinozaki Y., Ohsawa K., Tsuda M., Joshi B. V., Jacobson K. A., Kohsaka S., Inoue K., Nature 2007, 446, 1091–1095; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17b. Warny M., Aboudola S., Robson S. C., Sevigny J., Communi D., Soltoff S. P., Kelly C. P., J. Biol. Chem. 2001, 276, 26051–26056; [DOI] [PubMed] [Google Scholar]
- 17c. Li R., Tan B., Yan Y., Ma X., Zhang N., Zhang Z., Liu M., Qian M., Du B., J. Immunol. 2014, 193, 4515–4526. [DOI] [PubMed] [Google Scholar]
- 18. Coster M., Wittkopf D., Kreuchwig A., Kleinau G., Thor D., Krause G., Schoneberg T., FASEB J. 2012, 26, 3273–3281. [DOI] [PubMed] [Google Scholar]
- 19. Li S., Li J., Wang N., Hao G., Sun J., Int. J. Mol. Sci. 2018, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reifel Saltzberg J. M., Garvey K. A., Keirstead S. A., Glia 2003, 42, 149–159. [DOI] [PubMed] [Google Scholar]
- 21.
- 21a. Li Q., Olesky M., Palmer R. K., Harden T. K., Nicholas R. A., Mol. Pharmacol. 1998, 54, 541–546; [DOI] [PubMed] [Google Scholar]
- 21b. Webb T. E., Henderson D., King B. F., Wang S., Simon J., Bateson A. N., Burnstock G., Barnard E. A., Mol. Pharmacol. 1996, 50, 258–265. [PubMed] [Google Scholar]
- 22. Costanzi S., Joshi B. V., Maddileti S., Mamedova L., Gonzalez-Moa M. J., Marquez V. E., Harden T. K., Jacobson K. A., J. Med. Chem. 2005, 48, 8108–8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Edgar R. C., Nucleic Acids Res. 2004, 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bender B. J., Marlow B., Meiler J., PLoS Comput. Biol. 2020, 16, e1007597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bender B. J., Vortmeier G., Ernicke S., Bosse M., Kaiser A., Els-Heindl S., Krug U., Beck-Sickinger A., Meiler J., Huster D., Structure 2019, 27, 537–544 e534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schüß C., Vu O., Schubert M., Du Y., Mishra N. M., Tough I. R., Stichel J., Weaver C. D., Emmitte K. A., Cox H. M., Meiler J., Beck-Sickinger A. G., J. Med. Chem. 2021, 64, 2801–2814. [DOI] [PubMed] [Google Scholar]
- 27. Ballesteros J. A., Weinstein H., Methods Neurosci. 1995, 25, 366–428. [Google Scholar]
- 28. Zhang D., Gao Z. G., Zhang K., Kiselev E., Crane S., Wang J., Paoletta S., Yi C., Ma L., Zhang W., Han G. W., Liu H., Cherezov V., Katritch V., Jiang H., Stevens R. C., Jacobson K. A., Zhao Q., Wu B., Nature 2015, 520, 317–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang J., Zhang K., Gao Z. G., Paoletta S., Zhang D., Han G. W., Li T., Ma L., Zhang W., Muller C. E., Yang H., Jiang H., Cherezov V., Katritch V., Jacobson K. A., Stevens R. C., Wu B., Zhao Q., Nature 2014, 509, 119–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.
- 30a. Liu W., Chun E., Thompson A. A., Chubukov P., Xu F., Katritch V., Han G. W., Roth C. B., Heitman L. H., IJzerman A. P., Cherezov V., Stevens R. C., Science 2012, 337, 232–236; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30b. Zhou Q., Yang D., Wu M., Guo Y., Guo W., Zhong L., Cai X., Dai A., Jang W., Shakhnovich E. I., Liu Z. J., Stevens R. C., Lambert N. A., Babu M. M., Wang M. W., Zhao S., eLife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang K., Zhang J., Gao Z. G., Zhang D., Zhu L., Han G. W., Moss S. M., Paoletta S., Kiselev E., Lu W., Fenalti G., Zhang W., Muller C. E., Yang H., Jiang H., Cherezov V., Katritch V., Jacobson K. A., Stevens R. C., Wu B., Zhao Q., Nature 2014, 509, 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.
- 32a. Yuan S., Chan H. C. S., Vogel H., Filipek S., Stevens R. C., Palczewski K., Angew. Chem. Int. Ed. 2016, 55, 10331–10335; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 10487–10491; [Google Scholar]
- 32b. Ciancetta A., O'Connor R. D., Paoletta S., Jacobson K. A., J. Chem. Inf. Model. 2017, 57, 3104–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu M., Yu X., Ahlberg P. E., Choo B., Lu J., Qiao T., Qu Q., Zhao W., Jia L., Blom H., Zhu Y., Nature 2013, 502, 188–193. [DOI] [PubMed] [Google Scholar]
- 34. Sangkuhl K., Schulz A., Schultz G., Schoneberg T., J. Biol. Chem. 2002, 277, 47748–47755. [DOI] [PubMed] [Google Scholar]
- 35. Schoneberg T., Schulz A., Biebermann H., Gruters A., Grimm T., Hubschmann K., Filler G., Gudermann T., Schultz G., Hum. Mutat. 1998, 12, 196–205. [DOI] [PubMed] [Google Scholar]
- 36. Saitou N., Nei M., Mol. Biol. Evol. 1987, 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 37. Zuckerkandl E., Pauling L., in Evolutionary Divergence and Convergence in Proteins, Academic Press, New York, 1965. [Google Scholar]
- 38. Kumar S., Stecher G., Tamura K., Mol. Biol. Evol. 2016, 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.
- 39a. Leaver-Fay A., Tyka M., Lewis S. M., Lange O. F., Thompson J., Jacak R., Kaufman K., Renfrew P. D., Smith C. A., Sheffler W., Davis I. W., Cooper S., Treuille A., Mandell D. J., Richter F., Ban Y.-E. A., Fleishman S. J., Corn J. E., Kim D. E., Lyskov S., Berrondo M., Mentzer S., Popović Z., Havranek J. J., Karanicolas J., Das R., Meiler J., Kortemme T., Gray J. J., Kuhlman B., Baker D., Bradley P., Methods Enzymol. 2011, 487, 545–574; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39b. Bender B. J., A. Cisneros, 3rd , Duran A. M., Finn J. A., Fu D., Lokits A. D., Mueller B. K., Sangha A. K., Sauer M. F., Sevy A. M., Sliwoski G., Sheehan J. H., DiMaio F., Meiler J., Moretti R., Biochemistry 2016, 55, 4748–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.
- 40a. Zhang C., Srinivasan Y., Arlow D. H., Fung J. J., Palmer D., Zheng Y., Green H. F., Pandey A., Dror R. O., Shaw D. E., Weis W. I., Coughlin S. R., Kobilka B. K., Nature 2012, 492, 387–392; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40b. Cheng R. K. Y., Fiez-Vandal C., Schlenker O., Edman K., Aggeler B., Brown D. G., Brown G. A., Cooke R. M., Dumelin C. E., Doré A. S., Geschwindner S., Grebner C., Hermansson N.-O., Jazayeri A., Johansson P., Leong L., Prihandoko R., Rappas M., Soutter H., Snijder A., Sundström L., Tehan B., Thornton P., Troast D., Wiggin G., Zhukov A., Marshall F. H., Dekker N., Nature 2017, 545, 112–115. [DOI] [PubMed] [Google Scholar]
- 41.
- 41a. Wingler L. M., McMahon C., Staus D. P., Lefkowitz R. J., Kruse A. C., Cell 2019, 176, 479–490.e412; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41b. Zhang H., Han G. W., Batyuk A., Ishchenko A., White K. L., Patel N., Sadybekov A., Zamlynny B., Rudd M. T., Hollenstein K., Tolstikova A., White T. A., Hunter M. S., Weierstall U., Liu W., Babaoglu K., Moore E. L., Katz R. D., Shipman J. M., Garcia-Calvo M., Sharma S., Sheth P., Soisson S. M., Stevens R. C., Katritch V., Cherezov V., Nature 2017, 544, 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Che T., Majumdar S., Zaidi S. A., Ondachi P., McCorvy J. D., Wang S., Mosier P. D., Uprety R., Vardy E., Krumm B. E., Han G. W., Lee M.-Y., Pardon E., Steyaert J., Huang X.-P., Strachan R. T., Tribo A. R., Pasternak G. W., Carroll F. I., Stevens R. C., Cherezov V., Katritch V., Wacker D., Roth B. L., Cell 2018, 172, 55–67.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lu J., Byrne N., Wang J., Bricogne G., Brown F. K., Chobanian H. R., Colletti S. L., Di Salvo J., Thomas-Fowlkes B., Guo Y., Hall D. L., Hadix J., Hastings N. B., Hermes J. D., Ho T., Howard A. D., Josien H., Kornienko M., Lumb K. J., Miller M. W., Patel S. B., Pio B., Plummer C. W., Sherborne B. S., Sheth P., Souza S., Tummala S., Vonrhein C., Webb M., Allen S. J., Johnston J. M., Weinglass A. B., Sharma S., Soisson S. M., Nat. Struct. Mol. Biol. 2017, 24, 570–577. [DOI] [PubMed] [Google Scholar]
- 44. Cao C., Tan Q., Xu C., He L., Yang L., Zhou Y., Zhou Y., Qiao A., Lu M., Yi C., Han G. W., Wang X., Li X., Yang H., Rao Z., Jiang H., Zhao Y., Liu J., Stevens R. C., Zhao Q., Zhang X. C., Wu B., Nat. Struct. Mol. Biol. 2018, 25, 488–495. [DOI] [PubMed] [Google Scholar]
- 45. Shihoya W., Izume T., Inoue A., Yamashita K., Kadji F. M. N., Hirata K., Aoki J., Nishizawa T., Nureki O., Nat. Commun. 2018, 9, 4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kooistra A. J., Mordalski S., Pándy-Szekeres G., Esguerra M., Mamyrbekov A., Munk C., Keserű G. M., Gloriam D. E., Nucleic Acids Res. 2021, 49, D335–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Song Y., DiMaio F., Wang R. Y.-R., Kim D., Miles C., Brunette T., Thompson J., Baker D., Structure 2013, 21, 1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Viklund H., Elofsson A., Bioinformatics 2008, 24, 1662–1668. [DOI] [PubMed] [Google Scholar]
- 49. Lomize M. A., Pogozheva I. D., Joo H., Mosberg H. I., Lomize A. L., Nucleic Acids Res. 2012, 40, D370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yarov-Yarovoy V., Schonbrun J., Baker D., Proteins 2006, 62, 1010–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Deflorian F., Jacobson K. A., J. Comput.-Aided Mol. Des. 2011, 25, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.
- 52a. Meiler J., Baker D., Proteins 2006, 65, 538–548; [DOI] [PubMed] [Google Scholar]
- 52b. Lemmon G., Meiler J., in Computational Drug Discovery and Design (Ed.: Baron R.), Springer New York, New York, NY, 2012, pp. 143–155. [Google Scholar]
- 53. Mendenhall J., Brown B. P., Kothiwale S., Meiler J., J. Chem. Inf. Model. 2021, 61, 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schott-Verdugo S., Gohlke H., J. Chem. Inf. Model. 2019, 59, 2522–2528. [DOI] [PubMed] [Google Scholar]
- 55. Morozenko A., Stuchebrukhov A. A., Proteins 2016, 84, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Maier J. A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K. E., Simmerling C., J. Chem. Theory Comput. 2015, 11, 3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dickson C. J., Madej B. D., Skjevik A. A., Betz R. M., Teigen K., Gould I. R., Walker R. C., J. Chem. Theory Comput. 2014, 10(2), 865–879. doi: 10.1021/ct4010307. Epub 2014 Jan 30. PMID: 24803855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.
- 58a. Wang J., Wolf R. M., Caldwell J. W., Kollman P. A., Case D. A., J. Comput. Chem. 2004, 25, 1157–1174; [DOI] [PubMed] [Google Scholar]
- 58b. He X., Man V. H., Yang W., Lee T.-S., Wang J., J. Chem. Phys. 2020, 153, 114502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hopkins C. W., Le Grand S., Walker R. C., Roitberg A. E., J. Chem. Theory Comput. 2015, 11, 1864–1874. [DOI] [PubMed] [Google Scholar]
- 60. Roe D. R., Cheatham T. E., J. Chem. Theory Comput. 2013, 9, 3084–3095. [DOI] [PubMed] [Google Scholar]
- 61. Humphrey W., Dalke A., Schulten K., J. Mol. Graphics 1996, 14, 33–38. [DOI] [PubMed] [Google Scholar]
- 62.
- 62a. Case D., Ben-Shalom I., Brozell S. R., Cerutti D. S., Cheatham T., Cruzeiro V. W. D., Darden T., Duke R., Ghoreishi D., Gilson M., Gohlke H., Götz A., Greene D., Harris R., Homeyer N., Huang Y., Izadi S., Kovalenko A., Kurtzman T., Kollman P. A., Amber 2018, 2018; [Google Scholar]
- 62b. Miller B. R., McGee T. D., Swails J. M., Homeyer N., Gohlke H., Roitberg A. E., J. Chem. Theory Comput. 2012, 8, 3314–3321. [DOI] [PubMed] [Google Scholar]
- 63. Sitkoff D., Sharp K. A., Honig B., J. Phys. Chem. 1994, 98, 1978–1988. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.