Abstract
Objective.
Noninvasive brain–computer interfaces (BCIs) assist paralyzed patients by providing access to the world without requiring surgical intervention. Prior work has suggested that EEG motor imagery based BCI can benefit from increased decoding accuracy through the application of deep learning methods, such as convolutional neural networks (CNNs).
Approach.
Here, we examine whether these improvements can generalize to practical scenarios such as continuous control tasks (as opposed to prior work reporting one classification per trial), whether valuable information remains latent outside of the motor cortex (as no prior work has compared full scalp coverage to motor only electrode montages), and the existing challenges to the practical implementation of deep-learning based continuous BCI control.
Main results.
We report that: (1) deep learning methods significantly increase offline performance compared to standard methods on an independent, large, and longitudinal online motor imagery BCI dataset with up to 4-classes and continuous 2D feedback; (2) our results suggest that a variety of neural biomarkers for BCI, including those outside the motor cortex, can be detected and used to improve performance through deep learning methods, and (3) tuning neural network output will be an important step in optimizing online BCI control, as we found the CNN models trained with full scalp EEG also significantly reduce the average trial length in a simulated online cursor control environment.
Significance.
This work demonstrates the benefits of CNNs classification during BCI control while providing evidence that electrode montage selection and the mapping of CNN output to device control will be important design choices in CNN based BCIs.
Keywords: brain–computer interface, BCI, deep learning, convolutional neural network, EEG, motor imagery, sensorimotor rhythm
1. Introduction
Those afflicted by paralysis resulting from stroke, trauma, and other neuromuscular disorders suffer from reduced quality of life (French et al 2010). The ability to noninvasively decode user intent in real time enables brain–computer interfaces (BCIs) to provide this population with alternative routes of communication and action (Vallabhaneni et al 2005, Yuan and He 2014, He et al 2020). Decoding requires the recording of neural signals, often through electroencephalography (EEG) due to its low cost, portability, and non-invasive nature. Various types of EEG based BCIs have been developed to detect different neural biomarkers, and each has their own benefits and drawbacks. BCIs based on exogenous stimulation—for instance, the P300 response (Mak et al 2011) and steady state evoked potentials (Chen et al 2015)—present the user with a display and use the visual system’s response to control a device. While these systems often work well, they require a visual display with pre-programmed options, making them unsuitable for practical continuous control. BCIs based on the endogenous modulation of brain rhythms—in response to motor imagery (Wolpaw and McFarland 2004), shifts in visual attention (Van Gerven and Jensen 2009, Treder et al 2011, Tonin et al 2013), and manipulations of high level attention (Grosse-Wentrup and Schölkopf 2014)—are intuitive and provide continuous control, but may be more challenging to learn. Finally, some BCIs have been proposed which combine multiple biomarkers such as responses to both motor imagery and overt shifts in attention (Li et al 2010, Meng et al 2018).
One well-studied BCI method attempts to decipher motor imagery by recording sensorimotor rhythms (SMRs), which predictably change in response to real and imagined movements (Pfurtscheller and Neuper 2001, Vallabhaneni et al 2005, Yuan et al 2009, Yuan and He 2014). One of the most prominent SMRs is the mu rhythm, a characteristic biomarker of the motor cortex at rest, visible as oscillations in the alpha band over midline structures. When moving, or thinking about moving, the motor cortex is engaged, leading to reductions in the mu rhythm contralateral to the moved, or imagined, body part; this phenomenon is called event-related desynchronization (ERD). The reliability of ERD and its converse, event-related synchronization (increases in alpha power over the motor cortex during rest), can be used to control a BCI (Pfurtscheller and Lopes Da Silva 1999, Pfurtscheller and Neuper 2001). The intuitive and continuous nature of SMR-based BCIs enables the extension of the user through the control of virtual objects, drones, wheelchairs, and robotic arms (Wolpaw and McFarland 2004, Rebsamen et al 2010, Doud et al 2011, Huang et al 2012, Lafleur et al 2013, Meng et al 2016, Edelman et al 2019), and has further been shown to improve post-stroke outcomes when combined with physical therapy (Frolov et al 2017).
To realize the full potential of SMR based BCIs, a deeper understanding of the nature of brain activity and user intent associated with specific BCI tasks is still required. This deficit is demonstrated by the extensive effort, in both time and resources, required to train users and by the highly variable post-training control quality, preventing the widespread clinical adoption of BCIs (Leeb et al 2013). At least 20% of users are unable to control SMR BCIs proficiently, even after extensive training. Furthermore, performance levels often fluctuate, making the safe use of robotic end effectors for daily activities difficult (Guger et al 2003). Why some users are unable to control the device and how to remedy this situation is currently unknown, although some psychological and electrophysiological factors correlate with initial BCI skill (such as higher resting alpha power) (Blankertz et al 2010, Hammer et al 2011, Ahn et al 2013b).
Further improvement in SMR based BCIs may be realized through the application of deep learning with neural networks. The promise of deep learning lies in its capacity for learning complex nonlinear patterns from the raw data (Craik et al 2019). Many recent studies have shown that neural networks are able to classify user intent from brain data with excellent accuracy (Schirrmeister et al 2017, Lu et al 2017a, Sakhavi et al 2018, Craik et al 2019) as well as improve the information transfer rate of BCI systems (Nagel et al 2019). However, many of these studies rely on the same public BCI competition datasets, or small data sets using simple trial structures, and therefore may not generalize well outside the datasets they were tested on (Jayaram and Barachant 2018). Most of these studies utilize the BCI Competition IV datasets 2a and 2b, which are comparatively small (9 subjects, 2–5 sessions) and simple (2a–4 class, no online feedback, 2b–2 class with online feedback, but only 3 motor electrodes) (Tangermann et al 2012).
Craik et al provide a comprehensive review of the ongoing research in deep learning classification of EEG data (Craik et al 2019). In general, these studies can be distinguished by the type of data provided to the classification models or the type of deep learning architecture used for classification. The input data are usually either the signal voltage values (An et al 2014, Liu et al 2015, Kobler and Scherer 2017, Schirrmeister et al 2017, Tang et al 2017, Dose et al 2018) or calculated features based on the voltage values, typically in the form of frequency content (Sakhavi et al 2015, 2018, Tabar and Halici 2017, Yu and Song 2017, Lu et al 2017a, Abbas and Khan 2018, Chu et al 2018, Luo et al 2018, Wang et al 2018). Among deep learning approaches for motor imagery tasks, convolutional neural networks (CNNs) are by far the most common (Liu et al 2015, Sakhavi et al 2015, 2018, Schirrmeister et al 2017, Tabar and Halici 2017, Tang et al 2017, Abbas and Khan 2018, Dose et al 2018, Lawhern et al 2018, Craik et al 2019), followed by deep belief networks (An et al 2014, Kobler and Scherer 2017, Yu and Song 2017, Lu et al 2017a, Chu et al 2018), and a nontrivial number of recurrent neural networks as well (Luo et al 2018, Ma et al 2018, Wang et al 2018). However, most of these studies obtained their results from a limited number of participants (often fewer than 10 subjects) and fewer than 10 h of EEG data, greatly limiting the generalizability of their results (An et al 2014, Liu et al 2015, Sakhavi et al 2015, Tabar and Halici 2017, Tang et al 2017, Yu and Song 2017, Abbas and Khan 2018, Lawhern et al 2018, Luo et al 2018, Wang et al 2018).
Finally, most deep learning BCI classification studies have relied on a uniform trial structure and utilized the entire trial for classifications, which is not suitable for continuous BCI control (e.g. classifying a 2 s trial of motor imagery as ‘left’ rather than continually updating a cursor position in response to a prediction signal) (Wolpaw and McFarland 2004). While online deep-learning BCI systems have been recently developed, these systems have yet to demonstrate continuous feedback control (Tayeb et al 2019, Jeong et al 2020). For example in Tayeb et al 20 subjects performed two class motor imagery (left hand, right hand) and at the end of the trial, a robotic arm was moved in the direction classified by the system. Jeong et al developed a system closer to continuous control using a moving window approach where 15 subjects performed 6 class motor imagery (imagine the right hand moving left, right, up, down, forward, and backward). Sliding windows of 4 s were updated every 0.5 s, and classifier output values were averaged over 3 successive windows and the direction with the highest average probability over these windows was selected as an output command for a robotic arm. Unfortunately knowing a desired direction of movement is not the same as fluidly moving an external device through space (Edelman et al 2019). Therefore, rigorous validation of previous methods and methodological extensions suitable for online continuous control are needed to aid the development of online BCI systems using deep learning classification.
This work addresses two questions that need to be answered before the utility of deep learning decoding in BCI systems can be fully established: (1) Does deep learning generally improve BCI classification, and if so, how? (2) How do we bridge the gap between offline and single decision BCI classification studies and online continuous control BCIs based on deep learning classification? We address the generalizability of deep learning classification systems by (1) validating prior work on a new larger, more complex, and more challenging dataset, and (2) assessing the capacity for a generalized MI intent model designed to be applied to numerous control output strategies, compared to training one model per unique control task, affording significantly more flexibility in eventual practical BCI deployment. Furthermore, we demonstrate that deep learning systems can improve classification accuracy by better discerning and representing multi-variate, independent control signals in higher dimensional data (deep learning models provided with more information outside of the motor cortex displayed better performance while providing this information to standard methods showed little effect). We also highlight that translating neural network output into device commands will be an important step in bridging the gap between offline classification studies and online BCI control devices. We show how BCI operation can be improved by tuning cursor speed in a simulated online environment and specifically demonstrate that deep learning models trained with full scalp EEG coverage far exceed motor cortex specific deep learning models when this calibration step is performed optimally.
We recently collected, to our knowledge, the largest motor imagery BCI dataset ever recorded (in terms of total recording time; e.g. roughly 950% larger than the BCI competition datasets 2a and 2b combined) which includes 76 subjects and up to 11 sessions of online, continuous BCI control (Cho et al 2017, Kaya et al 2018, Stieger et al 2020). In each session, participants performed 300 trials of up to four class motor imagery in center-out cursor tasks with online feedback. Factors, besides size, that separate our dataset from those examined previously include: (1) the recording of full scalp coverage (62 channel) EEG, (2) varying trial structure (i.e. trials vary in length between 0.5 and 6 s rather than a uniform duration of 2 or 4 s), and (3) continuously updated online feedback in 1D and 2D (i.e. feedback was updated every 40 ms). Performance in this study was quantified based on (1) cursor proximity analysis (how close could classification models place a virtual cursor to an intended target) and (2) simulated metrics of online performance (e.g. cursor speed/trial length). Validating deep learning methods on this dataset presents a unique opportunity to study how neural networks classify individuals’ brain data from the population at large, under challenging conditions (such as four class, center-out, continuous control with varying trial structure), and additionally represents the first assessment of how neural net classification accuracy changes based on the inclusion of all electrodes provided by full scalp coverage EEG.
2. Materials and methods
2.1. Dataset
During the BCI tasks, users attempted to move a virtual cursor outwards from the center of the screen to one of four targets placed at the cardinal positions (Top, Bottom, Left and Right). Participants were instructed to imagine their left (right) hand opening and closing to move the cursor left (right), imagine both hands opening and closing to move the cursor up and finally, rest, to move the cursor down. In separate blocks of trials, participants drove the cursor toward a target that required left/right (LR) movement only, up/down (UD) only, and combined 2D movement (2D). BCI experiments consisted of three runs of the LR, UD, and 2D tasks (each with 25 trials), a short break, and an additional block of three runs of each task. Runs were broken into individual trials which included pre and post inter-trial intervals (2 s each), a cuing period where only the target was presented (2 s), and up to 6 s where the participant was provided feedback and attempted to control the cursor (0.04–6 s). Only the feedback portion of trials was used for classification. Trials longer than 6 s were aborted with the closest target to the current cursor position selected as the intended target.
All experiments were approved by the institutional review boards of the University of Minnesota and Carnegie Mellon University (Stieger et al 2020), with the clinical trial registration number NCT03221374. In total, 144 participants were enrolled in the study and 76 participants completed all experimental requirements. Seventy-two participants were assigned to each intervention by block randomization, with 42 participants completing all sessions in the experimental group (MBSR before BCI training; MBSR subjects) and 34 completing experimentation in the control group. Four subjects were excluded from the analysis due to non-compliance with the task demands and one was excluded due to experimenter error. We were primarily interested in how individuals learn to control BCIs, therefore analysis focused on those that did not demonstrate ceiling performance in the baseline BCI assessment (accuracy above 90% in 1D control). The dataset presented here is now publically available and describes data collected from 62 participants: 33 MBSR participants (age = 42 ± 15, (F)emale = 26) and 29 controls (age = 36 ± 13, F = 23) (Stieger et al 2021).
BCI experiments were conducted in BCI2000 (Schalk et al 2004). Online control of the cursor proceeded in a series of steps. The first step, feature extraction, consisted of spatial filtering and spectrum estimation. During spatial filtering, the average signal of the four electrodes surrounding the hand knob of the motor cortex was subtracted from electrodes C3 and C4 to reduce the spatial noise. Following spatial filtering, the power spectrum was estimated by fitting an autoregressive model of order 16 to the most recent 160 ms of data using the maximum entropy method. The goal of this method is to find the coefficients of a linear all-pole filter that, when applied to white noise, reproduces the data’s spectrum. The main advantage of this method is that it produces high frequency resolution estimates for short segments of data. The parameters are found by minimizing (through least squares) the forward and backward prediction errors on the input data subject to the constraint that the filter used for estimation shares the same autocorrelation sequence as the input data. Thus, the estimated power spectrum directly corresponds to this filter’s transfer function divided by the signal’s total power. Numerical integration was then used to find the power within a 3 Hz bin centered within the alpha rhythm (12 Hz).
The translation algorithm, the next step in the pipeline, then translated the user’s alpha power into cursor movement. Horizontal motion was controlled by lateralized alpha power (C4 − C3) and vertical motion was controlled by up and down regulating total alpha power (C4 + C3). These control signals were normalized to zero mean and unit variance across time by subtracting the signals’ mean and dividing by its standard deviation. A balanced estimate of the mean and standard deviation of the horizontal and vertical control signals was calculated by estimating these values across time from data derived from 30 s buffers of individual trial type (e.g. the normalized control signal should be positive for right trials and negative for left trials, but the average of left and right trials should be zero). Finally, the normalized control signals were used to update the position of the cursor every 40 ms.
EEG data were bandpass filtered between 1 and 100 Hz, and then down-sampled from 1 kHz to 250 Hz. Noisy channels, identified through visual inspection, were replaced by local weighted averages interpolated through spherical splines (Delorme and Makeig 2004, Ferree 2006). The data were rereferenced to a common average. Ocular artifacts were removed using independent component analysis and a template matching procedure. Trials shorter than the minimum window length were removed (Stieger et al 2020).
2.2. Filter bank common spatial patterns
As a control comparison, models were initially fit using the filter-bank common spatial patterns (FBCSP, abbreviated CSP in figures) algorithm which has performed well on the BCI competition datasets (Ang et al 2008). We used the implementation available in the MOABB toolbox (Jayaram and Barachant 2018). The MOABB toolbox was designed to permit easy benchmarking of BCI algorithms. We first bandpass filtered the EEG data into nine bands, each with a 4 Hz width, between 4 and 40 Hz. Covariance matrices were estimated for the 500 ms windows (Barachant et al 2012), and CSP extracted four filters from each band based on these covariance matrices (Guger et al 2000). The data were standardized, then the 10 best features were selected based on mutual information (Pedregosa et al 2011). Finally a support vector machine was trained to classify future BCI data windows (Cortes and Vapnik 1995). To validate this implementation, we compared the accuracies of this FBCSP algorithm to those published in the literature for the same BCI competition IV dataset 2a (Sakhavi et al 2015, Schirrmeister et al 2017). This implementation (71.18%) showed very similar performance when compared to the accuracy reported in Schirrmeister et al (67.59%) and Sakhavi et al (66.67% P > 0.7 Wilcoxon signed-rank test). This underlines that our FBCSP implementation, including our feature selection and filter bank design, indeed was a suitable baseline for the evaluation of our CNN decoding accuracies.
2.3. CNN models
Convolutional neural network (CNN) models were trained using the brain_decode package (version 0.4.85) in python, specifically using the shallow CNN architecture (Schirrmeister et al 2017). In the original paper, the authors propose two architectures for classifying 2 s trials of motor imagery EEG data, one shallow with only a few layers of neurons, and one deep with many more layers of neurons. In this work, we trained the shallow network with sliding windows of EEG data recorded during online BCI tasks. Briefly, in this architecture, the first two layers perform a temporal convolution and a spatial filter followed by a squaring nonlinearity, mean pooling layer, and a logarithmic activation function (figure 1). Two types of models were trained based on the data provided. ‘Motor Models’ were trained using the same motor cortex electrode montage as the online BCI experiments, namely the small spatial Laplacians surrounding C3 and C4. ‘All Models’ were trained using all available electrodes (62, total scalp coverage). Due to both variable trial structure and in order to directly compare with the online classification, we used the recommended default time window length from BCI2000 of 500 ms, with classification occurring every 40 ms. To compare this output to the classification of the online system in the 1D case, non-valid classes were removed from the CNN predictions after classification (e.g. in the LR task the classification output for up/down trials was set to zero). Activation functions were exponential linear units. Electrode-wise exponential moving standardization with a decay factor of 0.999 was used to compute exponential moving means and variances for each channel and standardize the continuous data (Schirrmeister et al 2017). CNN models were trained using the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1548562. Specifically, it used the Bridges system, which is supported by NSF award number ACI-1445606, at the Pittsburgh Supercomputing Center (PSC) (Towns et al 2014, Nystrom et al 2015).
Figure 1.
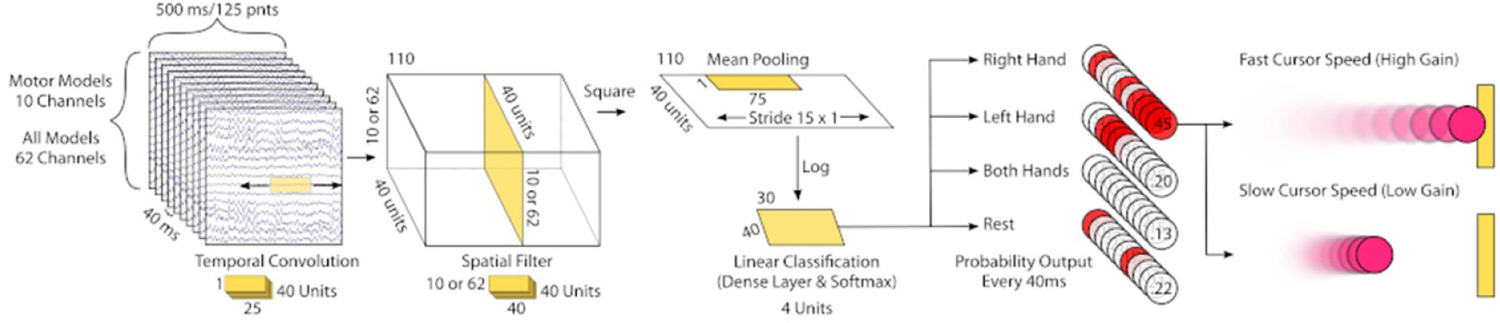
Continuous Deep Learning Classification of BCI Data. Sliding windows of EEG data recorded during online BCI tasks were used to train the shallow CNN architecture reported in Schirrmeister et al (2017). These windows were 500 ms long and were shifted every 40 ms. Two types of models were trained based on the data provided. ‘Motor Models’ were trained using the same motor cortex electrode montage as the online BCI experiments. ‘All Models’ were trained using all available electrodes. In successive steps, the shallow CNN architecture performs a temporal convolution, spatial filter, squaring nonlinearity, mean pooling, log transform, and linear classification using a dense layer and softmax transform. During testing, the trained models provide the estimated probability of class membership for each window. In a simulated cursor control environment, the class with the highest estimated probability (red circles) was used to move a virtual cursor in the direction of, and in proportion to, this highest estimated probability. The functional mapping between neural network predictions and control systems was explored by varying the cursor speed (a scalar gain multiplied by the neural network outputs). The number of neurons per layer is shown by the unlabeled numbers underneath each section and the numbers of filters in each layer corresponds to the number of ‘Units’ (e.g. the temporal convolution learns 40 unique combinations of combinations of 25 signal values from each filter’s 25 neurons) (see text for further details).
2.4. Model training
The model training employed was used to test the generalizability of BCI decoding models. During online control with BCI 2000, participants controlled the cursor during three different tasks. Left-right (LR) control only comparing left and right hand motor imagery, up-down (UD) control only comparing motor imagery of both hands to resting and 2D control which combined these strategies. Decoding models were trained on all three tasks (with one model trained per session) to determine whether a single model could accurately classify brain data from these diverse, but related tasks. In order to emulate BCI experiments in which BCI decoding could be used in an online setting after calibration, models were trained on 500 ms windows of data. One classification was made every 40 ms, and a virtual cursor was updated with each classification, therefore classifications compounded in a continuous way, unlike previous models where output only occurred every second or two (Tayeb et al 2019, Jeong et al 2020). Models were trained for each session using the first two runs in a given block and the last run in each block for testing (i.e. each participant completed six runs of each task per day; runs 1, 2, 4, and 5 were used for training and runs 3 and 6 were used for testing). Participants took a short break between runs 1–3 and 4–6, which created a natural opportunity to break up the data. Accuracy was initially assessed in a continuous fashion by measuring the average distance to the target. Then discrete metrics such as average trial length were derived based on the optimum cursor speed that produced the minimum average distance to the target. Two different types of models were trained for each session. The first model (CSP-Motor, CNN-motor—10 electrodes) used only the electrodes over the motor cortex (C3/C4 each surrounded by the four electrodes that formed the small Laplacian filter for online control), while the second included all available electrodes for training and testing (CSP-All, CNN-All—62 electrodes).
Batch normalization and dropout were used during the training of CNN models. Adam optimization was used to perform stochastic gradient descent (Kingma and Ba 2015). A training termination strategy was utilized that occurs in two steps. First, the training data was further segmented into minor training and validation subsets. During this phase, model training would terminate when validation accuracy ceased to improve over a predefined number of epochs (Schirrmeister et al 2017). Then, training continued to update the model in the second phase, on the combined training and validation fold, i.e. the entire training set, beginning with the best of the best parameter values determined in phase one. Training ends when the loss function is equivalent between the two training phases, resulting in a variable number of model training epochs across participants.
2.5. Electrophysiological analysis
Feature importance maps were calculated with the input-perturbation network-prediction correlation method (Schirrmeister et al 2017). These maps were created to investigate the causal effect of changes in EEG power on the trained CNN models. Initially, all 500 ms windows were transformed to the frequency domain by a Fourier transform. Then the power amplitudes were randomly perturbed by adding Gaussian noise (with mean 0 and variance 1) to their original values while leaving phase unperturbed. These trials were then transformed back to the time domain via an inverse Fourier transform. Predictions were made by the trained CNN models before and after the perturbations. This procedure was repeated 400 times, then the change in input amplitudes (the added noise) and the change in CNN predictions was examined in a correlation analysis. Feature importance maps were calculated only for the final three sessions when learning was assumed to have plateaued, then averaged for each subject. A cluster-based permutation test statistic was used to assess group differences in feature importance maps for each target condition (Maris and Oostenveld 2007, Oostenveld et al 2011).
2.6. Statistics
Most data displayed significant deviations from normality and were therefore fit with a rank based analysis of linear models using the Rfit package (0.23.0) as a robust alternative to least squares (RAOV) (Kloke and McKean 2012). Metrics were fit in 2 × 2 designs with model type (4 Levels: CSP-All, CSP-Motor, CNN-All, CNN-Motor) vs. session (11 levels). Average one-way comparisons across models were fit with Wald tests. Significant findings were followed by independent post-hoc tests to aid inference. All post-hoc inferences were two-sided at a Holm-Bonferroni corrected alpha level of 0.05 (0.025 per side).
3. Results
3.1. CNN model predictions outperform standard filter bank common spatial pattern methods
Deep learning with CNNs was found to significantly improve the probability of correct class membership (note: ‘probability’ is used to describe the softmax output of decoding models for convenience, these models may not represent actual probabilities (Guo et al 2017)). To begin, models output a softmax probability estimate for each window of data within a trial. The ‘probability’ values were averaged across all 40 ms intervals for the target class across each trial (e.g. all ‘left’ probabilities averaged across all windows in ‘left’ trials) and then averaged across the trials in a given session. In this way, higher values of correct class membership would lead to greater cursor movement in the intended target direction. Figure 2(A) displays the probability of correct class membership output by the models that were trained on the online BCI data as participants learned to control the device. By examining performance in each task across participant training sessions, two facts become apparent. First, CNN decoding outperforms FBCSP decoding across all tasks examined and this improvement in BCI classification accuracy is maintained across all tasks and all sessions. Second, as participants learn to produce better motor imagery (and brain signals), the CNNs are able to discern this improvement, which is evinced by increases in classification accuracy over time. In other words, as participants get better at controlling the online BCI the offline decoding performance also increases.
Figure 2.
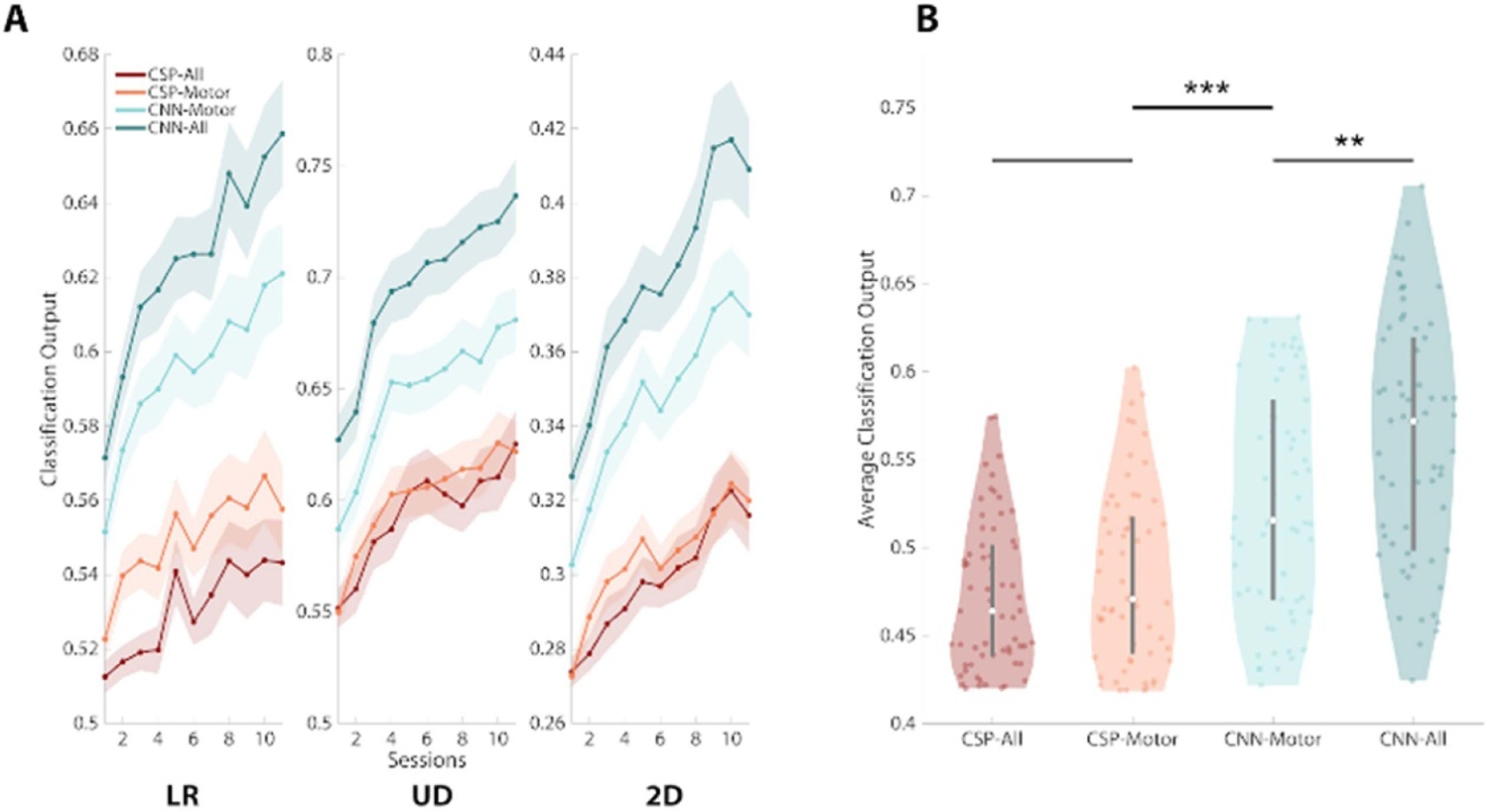
CNN model predictions are more accurate than FBCSP. (A) The use of CNN models was found to outperform FBCSP in all tasks examined throughout BCI training (sessions on x-axis). The y-axis in these figures represents the probability of correct class membership, that is the average classification output for the target class. CNN models with all electrodes (dark blue) displayed the best classification accuracy in each individual session, followed by motor electrode only CNN models (light blue), CSP-motor models (orange) and CSP-all models (red). (B) Significant increases in the probability of correct class membership were found when comparing the average classification output across all tasks and sessions between CNN models with full scalp EEG coverage and motor only CNN models (P = 0.009), and between CNN motor models and CSP motor models (P = 0.0003), however no differences were found between CSP-all and CSP-motor models (P = 0.26). Line plots (A): shaded area represents ±1 standard error of the mean (S.E.M.). Violin plots (B): shaded area represents kernel density estimate of the data, white circles represent the median, and grey bars represent the interquartile range.
The different decoding methods differed significantly in their performance. When modeling the probability of correct class membership over time, a main effect of model type was found (figure 2(A), F1, 3 = 198.66, P < 0.00001; supplementary table S1 (available online at stacks.iop.org/JNE/18/046082/mmedia)). Post-hoc tests revealed that the use of all electrodes in CNN models significantly improved classification accuracy compared to motor exclusive CNN models (+0.06, Wilcoxon rank sum (WRS) test: Z = 2.63, P = 0.009). CNN models using only the motor electrodes also displayed significant improvements compared to the FBCSP motor models (+0.04, WRS: Z = 3.66, P = 0.0003). No difference was observed between the different types of FBCSP models (−0.01, WRS: Z = −1.14, P = 0.26).
3.2. Using estimated class probability as a control signal highlights the merit of CNN classification
One important aspect of SMR-BCI is its continuous control; therefore, attempts were made to emulate online control with the output of the CNN models. In BCI2000 (and standard cursor control tasks), the magnitude of cursor movement is determined by the strength of the chosen control signal (originally as normalized differences in alpha power over the motor cortex; hence larger differences in alpha power would lead to larger cursor movements). We simulated continuous control using the estimated class probability output by the CNN and FBCSP models (i.e. one prediction for each 40 ms interval), such that when a model was more confident in its decisions, the cursor could be moved further in a simulated environment.
A simulated cursor control environment was created to examine the effects of deep learning classification on continuous BCI control where a virtual cursor was moved in a 2D plane similar to the feedback provided to participants during online control. For each EEG data window, a simulated cursor was moved in the direction of, and in direct proportion to, the highest class probability output by the decoding models. In the same manner as online BCI control, left and right (as well as up and down) decisions were given opposite signs such that if two consecutive decisions were 0.65 probability right, then 0.65 probability left, these decisions would cancel (however, unlike online control, motion was restricted to a single axis per time window). The distance between the cursor position (the summed probability signal) and the intended target was measured for each trial. If this summed signal crossed a threshold in one direction, a trial level classification was made and the remaining trial data was discarded (i.e. the minimum distance was recorded if the correct target was selected or the maximum distance was recorded if a non-target was selected). If this threshold was not reached by the end of the trial data, the final cursor position was used to measure this distance. The distance units were arbitrary but based on BCI 2000’s screen width parameter of 4095.
CNN based decoding models were shown to significantly reduce the average distance to the target in a simulated cursor control environment. Figure 3(A) displays a receiver-operating-characteristic curve plotting the average target distance against the cursor speed. The x-axis of figure 3(A) can be thought of as a gain parameter that multiplies the model classification outputs. Slower cursors have a smaller gain factor, so they tend to be unable to leave the center of the screen. Increasing the cursor speed (and the magnitude of the gain factor) reduces the average distance to the target, but only up to a certain point. When the cursor speed exceeds the optimum value, the cursor becomes erratic, which leads to greater distance from the intended target. In practice this means that each classification decision has a greater weight in the eventual trial outcome. While accurate classifications rapidly move the cursor closer to the target, inaccurate decisions allow for little time for behavioral or decoding correction. If the cursor is moving too fast, the weight of a particularly poor decision could place the cursor on the opposite side of the screen, or worse, lead to the contact of an inappropriate target, terminating the trial. When comparing the area under the curve for these models, a Wald test rejected the null hypothesis that model type had no effect on rank (Wald = 145.44, P < 0.00001; supplementary table S2). Significant reductions in average target distance were observed for each successive model type. CSP-All vs CSP-Motor (+1.37 × 105, WRS: Z = 2.58, P = 0.01), CSP-Motor vs CNN-Motor (+3.10 × 105, WRS: Z = 4.83, P < 0.00001), CNN-Motor vs CNN-All (+1.48 × 105, WRS: Z = 2.41, P = 0.016). These results suggest that on average across all operation points of cursor speed: (1) CNN models tend to outperform FBCSP models and (2) CNN models trained with full scalp EEG tend to outperform those trained with only the electrodes over the motor cortex.
Figure 3.
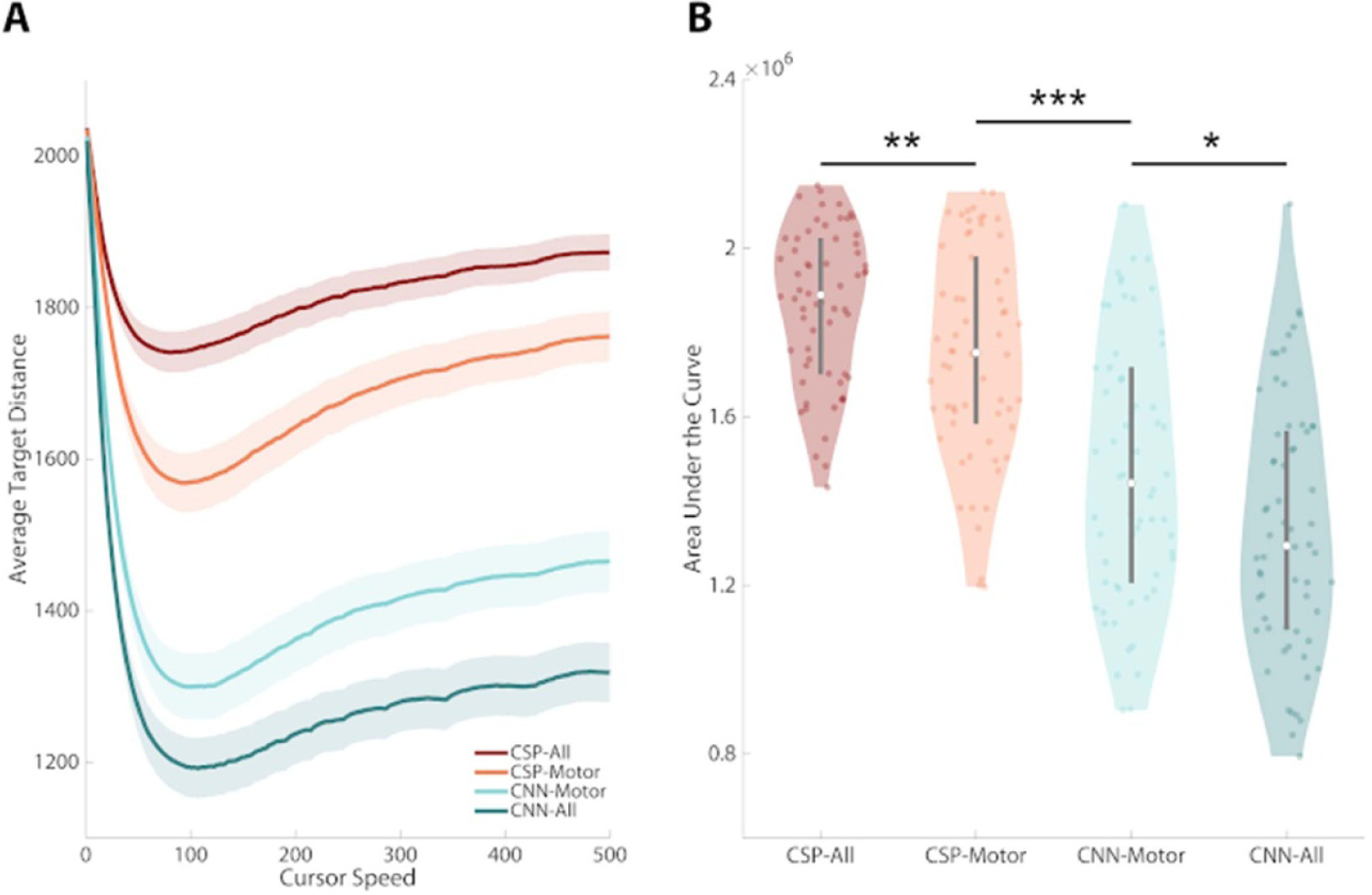
Deep learning models lead to smaller distances to intended targets. Using a sliding window approach, the output of classification models (the predicted probability of class membership) can be used as a control signal for continuous BCI. (A) A receiver operating characteristic curve was made by varying the cursor speed (the gain parameter multiplying the model classification output) to measure how the different models perform in a continuous control environment. The y-axis can be thought of as model error, that is the average distance to the intended target. (B) When comparing the area under these curves, a Wald test rejected the null hypothesis that model type had no effect on rank (Wald, P < 0.00001). Significant reductions in average target distance were observed for each successive model type. CSP-All vs CSP-Motor (WRS, P = 0.01), CSP-Motor vs CNN-Motor (WRS, P < 0.00001), CNN-All vs CNN-Motor (WRS, P = 0.02). Line plots (A): shaded area represents ±1 standard error of the mean (S.E.M). Violin plots (B): shaded area represents kernel density estimate of the data, white circles represent the median, and grey bars represent the interquartile range.
3.3. CNN models with full scalp EEG coverage preferentially benefit BCI low-performing participants
When examining the relationship between online BCI performance and offline decoding accuracy, it was found that the use of CNN Models preferentially benefitted participants that struggled with online control. A significant positive correlation was found between online accuracy and all classification models, indicating that if participants produced better linearly classifiable signals, higher classification accuracy could be expected from the offline decoding methods (figure 4(A); CSP-All: r62 = 0.71, P < 0.00001; CSP-Motor: r62 = 0.82, P < 0.00001, CNN-All: r62 = 0.73, P < 0.00001; CNN-Motor: r62 = 0.82, P < 0.00001). A significant negative correlation was found between online BCI performance and the difference between offline and online BCI performance for all models except CNN-Motor (figure 4(B); CSP-All: r62 = −0.78, P < 0.00001; CSP-Motor: r62 = −0.41, P = 0.0008, CNN-All: r62 = −0.43, P = 0.0004; CNN-Motor: r62 = 0.24, P = −0.06). This suggests that, on average, the worse a participant performed during the standard online SMR-based BCI, the more benefit could be expected from using full scalp EEG CNN classification. Finally, the improvement in classification accuracy when using all electrodes to train models, vs just motor electrodes, displayed a significant negative correlation with online performance, indicating that even though these participants may not display the expected motor correlates of motor imagery, deep learning methods were able to find valuable information outside the motor cortex to aid in classification (figure 4(C); CNN Models r62 = −0.50, P = 0.0004, CSP Models r62 = −0.57, P < 0.00001). One cautionary note is that the one generalized FBCSP model trained on all tasks performed worse than the online BCI 2000 decoding which used 3 separate models (this is shown by the positive values of change in average target distance in figures 4(B) and (C)—see discussion). The negative correlations in this context mean that the detriment caused by using the generalizable FBCSP models is less severe for poorly performing subjects.
Figure 4.
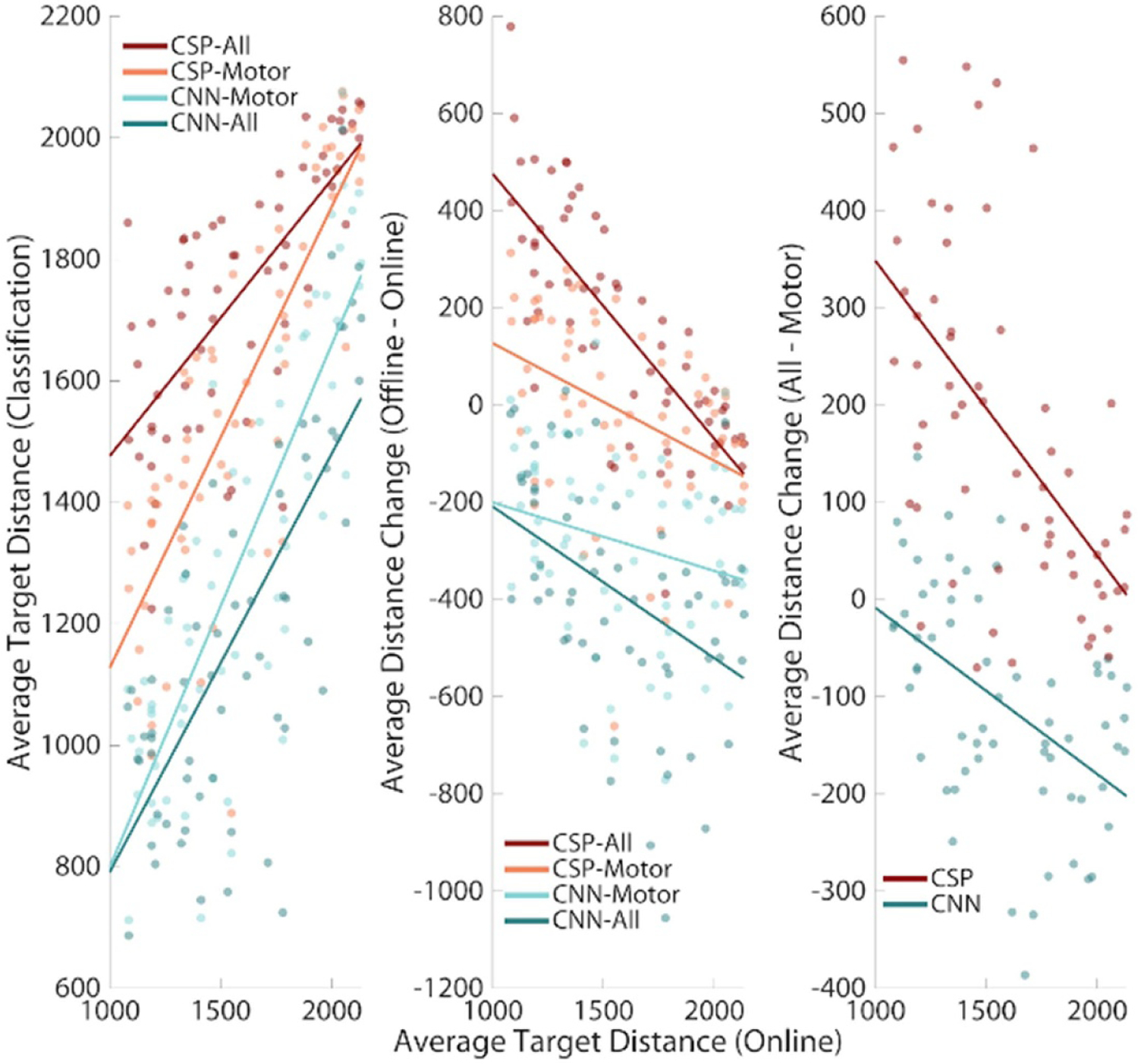
CNN classification preferentially benefits inefficient participants. (A) A significant positive correlation was found between online and offline BCI decoding performance (Pearson correlation, P < 0.00001). (B) A significant negative correlation was found between online BCI decoding performance and all model types except CNN-Motor (Pearson correlation, P < 0.0008, CNN-Motor P = 0.06) suggesting that those with worse online BCI performance improved more through the use of offline classification methods. (C) Another significant negative correlation was found between online BCI performance and the improvement in offline performance when using full scalp EEG coverage (Pearson correlation, P < 0.00004), indicating that those with worse online BCI performance improved more through the use of models with full scalp EEG coverage.
In these figures, larger y-values represent greater errors. In general, FBCSP classification errors are greater when using full scalp coverage compared to motor cortex specific montages. This trend is reversed in the case of CNN classification: the use of full scalp coverage reduces errors compared motor cortex specific montages. We see in the middle panel (4B) that participants who excelled in standard BCI2000 control displayed greater classification error when fitting one FBCSP model to the three separate tasks, but that participants who initially performed poorly during online control are benefitted by the use of FBCSP classification. The trend is the same for CNN classifiers: good performers in the online context are slightly benefitted by CNN classification, but the poor performers show greater reductions of errors. Finally, in the last panel (4C), participants who excelled in standard SMR BCI control had better performance in FBCSP models trained with motor montages (i.e. FBCSP models with full scalp coverage had higher error compared to FBCSP models trained with only motor cortex information; suggesting that the additional electrodes introduced unnecessary noise into the system), while participants who had little control over their motor cortex mu rhythm received a slight benefit from using full scalp coverage to train their models. Again, the trend is the same in CNN models, but the interpretation is slightly different. Participants who excelled in standard BCI control saw roughly the same performance (or slightly worse) when using models trained with full scalp coverage (because most of their distinguishing EEG features are over the motor cortex anyway), but the participants who could not control their SMRs well were greatly benefitted by the full scalp montages.
3.4. CNN models maintain advantage when cursor speed is optimized
CNN models trained with full scalp EEG were found to result in the shortest trial lengths. We attempted to find the point of optimal speed-accuracy tradeoff in our decoding models by finding the cursor speed that produced the smallest average target distance for each model. From the ROC curves above (figure 3(A)) we found the cursor speed that minimized the average trial length for each model type. Then we used the model classification output to control a cursor in a simulated environment using the gain factor (cursor speed) found above. The trial length was determined by how long the cursor was ‘active’ in this environment. In other words, if the cursor contacted a target, the new trial length was set as the time point when this event occurred and the rest of the brain data for a given trial was discarded. At this threshold, a Wald test rejected the null hypothesis that all model types produced the same trial length (Figure 5, Wald = 62.30, P < 0.00001; supplementary table S3). A significant reduction in average trial length was observed when comparing CNN models trained with full scalp EEG compared to the motor montage: CNN-All vs CNN-Motor (−0.42 s, WRS: Z = −5.55, P < 0.00001). No other differences in average trial length were observed: CSP-All vs CSP-Motor (0.0 s, WRS: Z = −0.44, P = 0.65), CSP-Motor vs CNN-Motor (−0.02 s, WRS: Z = 0.39, P = 0.69).
Figure 5.
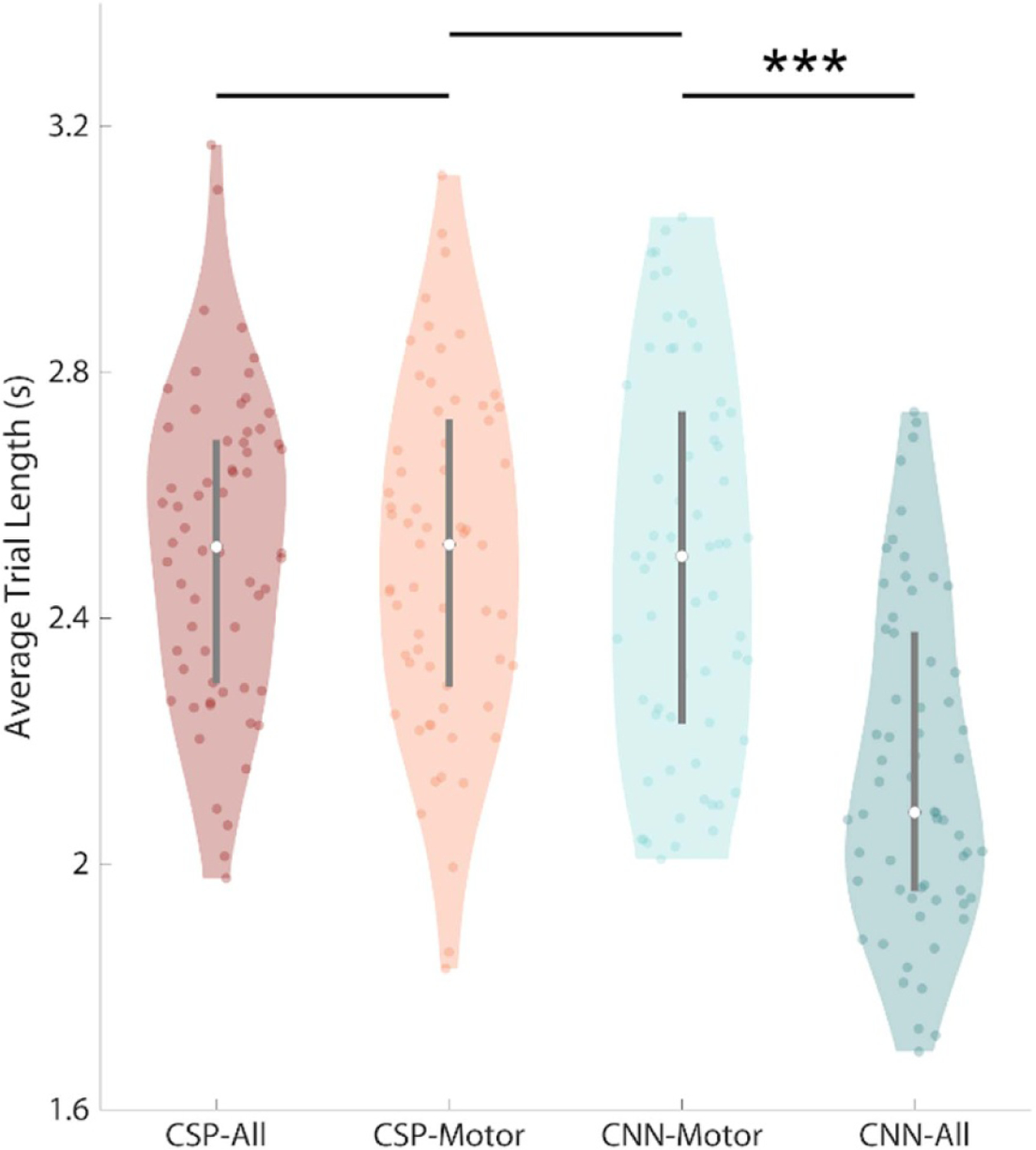
CNN trained with full scalp EEG produce the fastest trials using the optimal cursor speed that minimized average target distance for each model, a simulated cursor control environment was created in which a cursor was moved based on the decoding models’ output. A Wald test rejected the null hypothesis that all model types produced the same trial length (Wald, P < 0.00001). Wilcoxon rank sum tests found that CNN models trained with full scalp EEG produced the shortest trials (WRS, P < 0.00001). Shaded area represents kernel density estimate of the data, white circles represent the median, and grey bars represent the interquartile range.
3.5. Feature importance maps suggest CNNs Utilize motor and non-motor biomarkers for SMR-BCI classification
The feature maps calculated with the input-perturbation network-prediction correlation approach (see section 2.3) revealed that both motor and non-motor biomarkers were utilized in the CNN Models’ predictions (figure 6). Cluster based permutation tests were used to compare the feature importance maps for each target against the feature importance maps for the other targets (while holding the frequency band fixed) in order to discover statistical regularities in the derived CNN biomarkers for each class condition. The bands examined were (upper) alpha (10–14 Hz), beta (16–28 Hz), and gamma (28–58 Hz). In total, 26 clusters were identified and all clusters displayed a significance level lower than the Bonferroni threshold for multiple comparisons (P < 0.0042) (see supplementary figure S2, supplementary table S4). Right hand motor imagery was associated with patterns of contralateral motor and ipsilateral occipital desynchronization in the alpha and beta bands with a unique ipsilateral fronto-tempolar gamma response. Left hand motor imagery displayed the same pattern mirrored over the midline. Motor imagery of both hands correlated with motor and occipital desynchronization across all bands as well as with frontal beta synchronization. Resting was associated with alpha and beta synchronization over the motor cortex as well as frontal beta and frontal/temporal gamma desynchronization.
Figure 6.
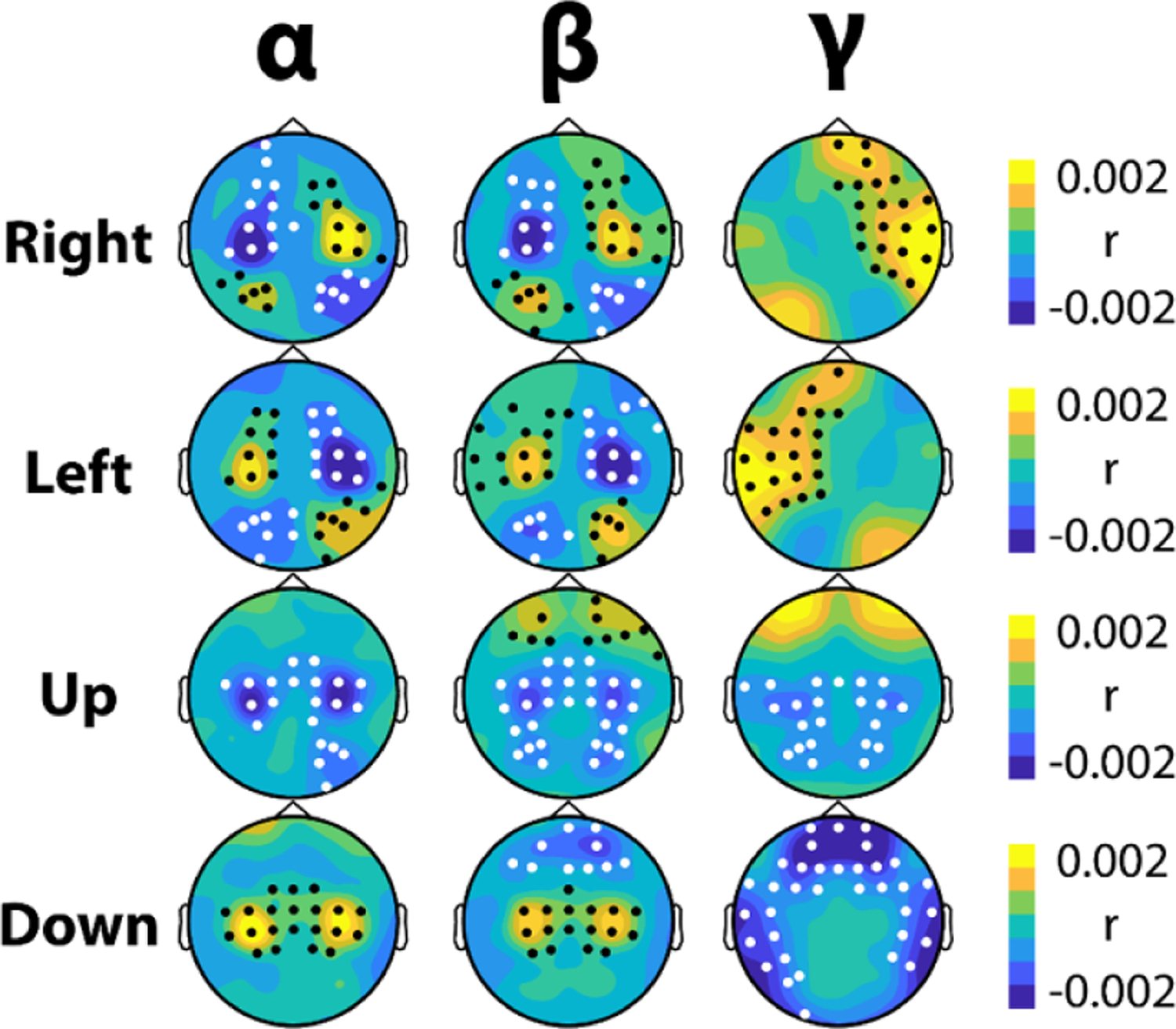
Feature importance maps display multiple biomarkers used in continuous BCI control. Using the input-perturbation network-prediction correlation method, feature importance maps were created for each motor imagery strategy. Warm colors occur when increases in power over these areas increase the classification accuracy, while cool colors occur when decreases in power increase the classification accuracy (or vice-versa). The value of r is the average correlation between changes in frequency power and changes in predicted class membership across subjects (e.g. when alpha power decreases over left motor cortex, models predict a higher probability of that data belonging to a right trial; when alpha power increases over right motor cortex the models also produce a higher probability of that data belonging to a right trial). Black dots represent significant clusters of positive correlation and white dots represent significant clusters of negative correlation. All clusters had a significance level lower than the Bonferroni threshold for multiple comparisons (P < 0.0042).
4. Discussion
In this work, we have demonstrated that deep learning methods are able to significantly improve classification performance compared to conventional methods in SMR based BCI tasks. This improvement was found to disproportionately benefit the participants who struggled the most with BCI control, suggesting the use of CNN classification in BCI could potentially address the problem of BCI inefficiency. The use of high density EEG was shown to significantly improve the theoretical speed of operation and accuracy when compared to models utilizing information derived from traditional ‘motor only’ electrode montages. Finally, novel combinations of BCI features were identified in addition to standard SMRs, such as occipital features and frontal gamma power, indicating deep learning models may derive useful information from outside the motor cortex when classifying EEG data collected during online SMR MI BCI control with continuous feedback.
Recent research demonstrating improved classification accuracy through the use of deep learning methods was extended here in a large and longitudinal BCI dataset. Most of the recent work in applying neural networks to SMR based BCI has focused on the development of various architectures and algorithms while testing them on standard benchmark, or in-house, datasets (Tangermann et al 2012, Schirrmeister et al 2017, Lu et al 2017a, Lawhern et al 2018, Sakhavi et al 2018, Wang et al 2018), though some have recently begun to validate these methods on their own datasets in online scenarios (Tayeb et al 2019, Jeong et al 2020). Here, we have shown that deep learning with CNNs can significantly increase classification performance on an independent, large, and longitudinal dataset, thus validating the generalizability of these methods. We additionally demonstrate that these methods are able to produce improved performance compared to conventional methods in scenarios suitable for continuous SMR based control, as opposed to prior work that produced one classification per uniform 2 or 4 s trials (Schirrmeister et al 2017, Lu et al 2017a, Lawhern et al 2018, Sakhavi et al 2018, Wang et al 2018).
The inability of a significant proportion of the population to control standard BCIs, even after extensive training, has historically limited the clinical adoption of BCI technology (Leeb et al 2013). However, the use of CNN models in BCI systems could potentially remedy this situation as the improvement in classification accuracy through the use of deep learning methods was discovered to be greatest in this challenged population. This finding may lend credence to the future clinical viability of BCIs as BCI decoding technology develops (Guger et al 2017). Those with poorer initial BCI performance additionally displayed a greater benefit from the inclusion of extra-motor electrodes in this offline classification study, suggesting high-density EEG may be more important for end-users. Most prior work has focused on classification which uses electrodes only over the motor cortex, with some claiming the inclusion of all electrodes for BCI classification was found to lead to worse accuracies than those over the motor cortex alone (Schirrmeister et al 2017), therefore future work is needed to reconcile these discrepant findings.
BCI control makes use of a distributed network of interacting brain structures that participate in motor imagery, maintenance of task demands, attention, awareness of feedback, and reward processing (Sitaram et al 2017). As predicted, characteristic motor cortex dependent patterns of motor imagery within the alpha band, which additionally extended into the beta band, were found in the feature importance maps, demonstrating CNNs are able to find subject specific SMRs.
The alpha rhythm, and indeed performance in SMR BCIs, has been shown to be related to gamma band activity, especially in the frontal regions (Grosse-Wentrup et al 2011, Grosse-Wentrup and Schölkopf 2012, 2014, Ahn et al 2013a). This frontal gamma signal was found to play an important role in CNN models, and, of particular note, its absence was a prominent biomarker of the rest condition where motor imagery was not performed. However, these maps additionally suggest that CNN models utilize non-motor related features in their decisions, such as occipital signals.
Parietal/occipital attention signals have been used to develop various BCI techniques such as overt and covert spatial attention (Van Gerven and Jensen 2009, Bahramisharif et al 2010, Treder et al 2011, Tonin et al 2013, Meng et al 2018). While these studies focus on synchronization of the alpha rhythm subsequent to shifting attention, their results also indicate there is a pattern of mid-frequency desynchronization during the initial shifting period (Van Gerven and Jensen 2009, Treder et al 2011). Ipsilateral occipital alpha and beta desynchronization were found to be significantly correlated with CNN model predictions, which alludes to a pattern of orienting to the target while covertly attending to the cursor (opposite that of covert attention paradigms of orienting to the center of the screen while covertly attending to the periphery) (Van Gerven and Jensen 2009, Bahramisharif et al 2010).
While the group-averaged feature importance maps include all of the aforementioned biomarkers, individual models may rely on a subset of these biomarkers, or even unique features that do not appear when examining BCI data from the group level. One advantage of CNN classification of BCI signals is the ability of these models to detect reliable biomarkers at the individual, rather than the group, level. It is this specificity which might explain why subjects who did not fit the ‘standard’ BCI model had the greatest improvements in classification accuracy. As evidenced here, future probing of these deep-learning models might serve to discover relationships between subject populations and biomarkers leading to novel neuroscientific findings, such as when are these occipital and frontal gamma features are in operation, which subjects display them most prominently, and whether they can be trained or enhanced.
One limitation of this study may be the sub-optimal performance of the FBCSP algorithm on our dataset. We believe the implementation of this algorithm was appropriate because (1) the code was published in the MOABB toolbox and validated on various datasets (Jayaram and Barachant 2018) and (2) it produced comparable results on the BCI competition dataset to other work in the field (Sakhavi et al 2015, Schirrmeister et al 2017). The authors of the MOABB toolbox mention no one algorithm out-performs all others on all datasets, so FBCSP may not be optimal for our dataset (Jayaram and Barachant 2018). Common spatial patterns may be a technique that requires additional calibration to each individual and each dataset (Guger et al 2000, Ang et al 2008, Blankertz et al 2008, Wu et al 2013). One advantage of CNNs is they learn features from raw data, so they may perform better when compared to other techniques that require further calibration (Craik et al 2019). Additionally, the poorer performance of FBCSP when compared to CNN classification may have occurred because we trained one model per session across three different, though similar tasks. This difference may provide evidence that CNN models generalize across tasks better than conventional methods, which is called transfer learning in some contexts (Volker et al 2018). FBCSP was chosen as the main benchmark in this study because it is a well-established and the well compared method that utilizes both a training and testing phase. We note other CNN architectures have out-performed the shallow network of Schirrmeister et al on limited datasets (Lawhern et al 2018, Sakhavi et al 2018) and a study designed to review the current state-of-the-art would have included a comparison of these architectures. However, our main goals were not to find the best deep learning architecture, but rather to (1) demonstrate the ability of deep learning methods to generalize to continuous BCI tasks and large populations and (2) discuss concerns that future online continuous BCI systems will need to address for practical deployment. Since our dataset is now publicly available, we invite the reader to test their algorithms and architectures on this large and rich database (Stieger et al 2021).
The significant increases in decoding performance suggest future BCIs using CNN classification could be controlled with finer precision, however the robustness of the control signal—as opposed to its precision—may lead to greater practical advantages. We found that CNN models trained with full scalp EEG displayed the greatest probability of correct class membership when compared to the other models examined. This meant that for each time step of 40 ms, a cursor could be moved farther towards a target in a simulated environment. We indeed found that CNN models trained with full scalp EEG resulted in the minimum average distance to targets across all examined cursor speeds. An unintended consequence of this finding was that when we found the optimal speed-accuracy tradeoff for these models, the CNN models trained with full scalp EEG also resulted in the minimum trial length. We believe this occurred because the greater probability of correct class membership, compounded every 40 ms, led to target selection with fewer time steps.
Challenges still exist when translating CNN BCIs from offline classification analyses to online systems. One limitation of this study is that, while we believe it represents one of the most complex classification studies conducted, these analyses were still performed offline. In online experiments, training data may not be available beforehand, or if it was collected prior to BCI use, the training of models within an experimental session would take a prohibitive amount of time. Future work will be needed to assess how well models trained in one session transfer to future sessions (Fahimi et al 2019). Additionally, neural networks often require long training times themselves, so there is a need to devise systems that are able to adapt in an online fashion (Schwemmer et al 2018). For example, variable electrode placement may compromise the accuracy of models trained in previous sessions, which is a hindrance not present in static, invasive systems (Schwemmer et al 2018). Recently, the feasibility of online BCI systems using deep learning classification has been demonstrated (Tayeb et al 2019, Jeong et al 2020), however more work is needed to create noninvasive deep learning BCIs that provide continuous control. BCI is a skill that the user and system acquire together, and it is currently unclear how neural networks will adapt to changes in a user’s control strategy as it develops (Vansteensel et al 2016, Perdikis et al 2018).
5. Conclusions
In conclusion, we have demonstrated that deep-learning based classification and high density EEG are important tools for increasing BCI efficiency. CNN models are shown to significantly increase the performance of motor imagery decoding models on a large BCI dataset when compared to conventional methods. This improvement is found to have the greatest benefit in the most challenged subset of the population, namely those that struggle with standard BCI control, suggesting deep learning in BCI systems may have the greatest impact on end-users in the locked-in state (Kübler and Birbaumer 2008, De Massari et al 2013, Grosse-Wentrup and Schölkopf 2014). BCI inefficient participants were again discovered to disproportionately benefit from the use of full scalp EEG coverage. Further, leveraging the output of these models in a simulated cursor control environment demonstrated that the additional information provided by high-density EEG could increase both the accuracy (minimize the average target distance) and the objective speed of operation (minimize the average trial length). This benefit may be in-part due to the variety of reliable correlates of BCI control, in addition to the SMR (such as an occipital and frontal gamma signals) that may be learned. These findings additionally suggest that valuable information may remain latent within EEG recordings (Nagel et al 2019). Future work developing BCI systems with online classification using CNN models may lead to the widespread clinical adoption of this technology and aid those suffering with paralysis by restoring their agency and access to the world.
Supplementary Material
Acknowledgments
We would like to thank Dr Mary Jo Krietzer and Dr Christopher C Cline for their assistance in subject recruiting and data collection, and Rui Sun for useful discussions on data analysis.
Funding
This work was supported in part by the National Institutes of Health (Grant Nos. AT009263, MH114233, EB021027, NS096761, and EB008389), and by the National Science Foundation (Grant No. CBET-1264782).
Footnotes
Supplementary material for this article is available online
Conflict of interest
The authors declare no competing interests in the work reported here.
Data availability statement
The data used in this study are available for public use and described with further detail in ‘Continuous sensorimotor rhythm based brain computer interface learning in a large population’ (Stieger et al 2021).
The data that support the findings of this study are openly available at the following URL/DOI: https://doi.org/10.6084/m9.figshare.13123148.
References
- Abbas W and Khan NA 2018. DeepMI: deep learning for multiclass motor imagery classification Proc. Annual Int. Conf. IEEE Engineering in Medicine and Biology Society, EMBS (Institute of Electrical and Electronics Engineers Inc; ) pp 219–22 [DOI] [PubMed] [Google Scholar]
- Ahn M, Ahn S, Hong JH, Cho H, Kim K, Kim BS, Chang JW and Jun SC 2013a. Gamma band activity associated with BCI performance: simultaneous MEG/EEG study Front. Hum. Neurosci 7 848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn M, Cho H, Ahn S, Jun SC and Hu D 2013b. High theta and low alpha powers may be indicative of BCI-illiteracy in motor imagery PLoS One 8 e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An X, Kuang D, Guo X, Zhao Y and He L 2014. A Deep Learning Method for Classification of EEG Data Based on Motor Imagery Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) (Berlin: Springer; ) pp 203–10 [Google Scholar]
- Ang KK, Chin ZY, Zhang H and Guan C 2008. Filter bank common spatial pattern (FBCSP) in brain-computer interface Proc. Int. Joint Conf. Neural Networks pp 2390–7 [Google Scholar]
- Bahramisharif A, Van Gerven M, Heskes T and Jensen O 2010. Covert attention allows for continuous control of brain-computer interfaces Eur. J. Neurosci 31 1501–8 [DOI] [PubMed] [Google Scholar]
- Barachant A, Bonnet S, Congedo M and Jutten C 2012. Multiclass brain-computer interface classification by Riemannian geometry IEEE Trans. Biomed. Eng 59 920–8 [DOI] [PubMed] [Google Scholar]
- Blankertz B, Sannelli C, Halder S, Hammer EM, Kübler A, Müller K-R, Curio G and Dickhaus T 2010. Neurophysiological predictor of SMR-based BCI performance Neuroimage 51 1303–9 [DOI] [PubMed] [Google Scholar]
- Blankertz B, Tomioka R, Lemm S, Kawanabe M and Müller KR 2008. Optimizing spatial filters for robust EEG single-trial analysis IEEE Signal Process. Mag 25 41–56 [Google Scholar]
- Chen X, Wang Y, Nakanishi M, Gao X, Jung TP and Gao S 2015. High-speed spelling with a noninvasive brain-computer interface Proc. Natl Acad. Sci. USA 112 E6058–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Ahn M, Ahn S, Kwon M and Jun SC 2017. EEG datasets for motor imagery brain–computer interface Gigascience 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Zhao X, Zou Y, Xu W, Han J and Zhao Y 2018. A decoding scheme for incomplete motor imagery EEG with deep belief network Front. Neurosci 12 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C and Vapnik V 1995. Support-vector networks Mach. Learn 20 273–97 [Google Scholar]
- Craik A, He Y and Contreras-Vidal JL 2019. Deep learning for electroencephalogram (EEG) classification tasks: a review J. Neural. Eng 16 031001. [DOI] [PubMed] [Google Scholar]
- De Massari D, Ruf CA, Furdea A, Matuz T, Van Der Heiden L, Halder S, Silvoni S and Birbaumer N 2013. Brain communication in the locked-in state Brain 136 1989–2000 [DOI] [PubMed] [Google Scholar]
- Delorme A and Makeig S 2004. EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis J. Neurosci. Methods 134 9–21 [DOI] [PubMed] [Google Scholar]
- Dose H, Møller JS, Iversen HK and Puthusserypady S 2018. An end-to-end deep learning approach to MI-EEG signal classification for BCIs Expert Syst. Appl 114 532–42 [Google Scholar]
- Doud AJ, Lucas JP, Pisansky MT, He B, Gribble PL and Gribble PL 2011. Continuous three-dimensional control of a virtual helicopter using a motor imagery based brain-computer interface PLoS One 6 e26322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman BJ, Meng J, Suma D, Zurn C, Nagarajan E, Baxter BS, Cline CC and He B 2019. Noninvasive neuroimaging enhances continuous neural tracking for robotic device control Sci. Rob 4 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahimi F, Zhang Z, Goh WB, Lee T-S, Ang KK and Guan C 2019. Inter-subject transfer learning with an end-to-end deep convolutional neural network for EEG-based BCI J. Neural. Eng 16 026007. [DOI] [PubMed] [Google Scholar]
- Ferree TC 2006. Spherical splines and average referencing in scalp electroencephalography Brain Topogr. 19 43–52 [DOI] [PubMed] [Google Scholar]
- French JS, Anderson-Erisman KD and Sutter M 2010. What do spinal cord injury consumers want? A review of spinal cord injury consumer priorities and neuroprosthesis from the 2008 neural interfaces conference Neuromodulation Technol. Neural Interface 13 229–31 [DOI] [PubMed] [Google Scholar]
- Frolov AA, Mokienko O, Lyukmanov R, Biryukova E, Kotov S, Turbina L, Nadareyshvily G and Bushkova Y 2017. Post-stroke rehabilitation training with a motor-imagery-based brain-computer interface (BCI)-controlled hand exoskeleton: a randomized controlled multicenter trial Front. Neurosci 11 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse-Wentrup M and Schölkopf B 2012. High gamma-power predicts performance in sensorimotor-rhythm brain–computer interfaces J. Neural. Eng 9 046001. [DOI] [PubMed] [Google Scholar]
- Grosse-Wentrup M and Schölkopf B 2014. A brain–computer interface based on self-regulation of gamma-oscillations in the superior parietal cortex J. Neural. Eng 11 056015. [DOI] [PubMed] [Google Scholar]
- Grosse-Wentrup M, Schölkopf B and Hill J 2011. Causal influence of gamma oscillations on the sensorimotor rhythm Neuroimage 56 837–42 [DOI] [PubMed] [Google Scholar]
- Guger C, Edlinger G, Harkam W, Niedermayer I and Pfurtscheller G 2003. How many people are able to operate an EEG-based brain-computer interface (BCI)? IEEE Trans. Neural. Syst. Rehabil. Eng 11 145–7 [DOI] [PubMed] [Google Scholar]
- Guger C, Ramoser H and Pfurtscheller G 2000. Real-time EEG analysis with subject-specific spatial patterns for a brain-computer interface (BCI) IEEE Trans. Rehabil. Eng 8 447–56 [DOI] [PubMed] [Google Scholar]
- Guger C, Spataro R, Allison BZ, Heilinger A, Ortner R, Cho W and La Bella V 2017. Complete locked-in and locked-in patients: command following assessment and communication with vibro-tactile P300 and motor imagery brain-computer interface tools Front. Neurosci 11 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Pleiss G, Sun Y and Weinberger KQ, 2017. On calibration of modern neural networks Proc. 34th Int. Conf. Machine Learning (PMLR; ) pp 1321–30 [Google Scholar]
- Hammer EM, Halder S, Blankertz B, Sannelli C, Dickhaus T, Kleih S, Müller K-R and Kübler A 2011. Psychological predictors of SMR-BCI performance Biol. Psychol 89 80–86 [DOI] [PubMed] [Google Scholar]
- He B, Yuan H, Meng J and Gao S 2020. Brain-computer interfaces Neural Engineering ed He B (Berlin: Springer; ) pp 131–83 [Google Scholar]
- Huang D, Qian K, Fei DY, Jia W, Chen X and Bai O 2012. Electroencephalography (EEG)-based brain-computer interface (BCI): a 2D virtual wheelchair control based on event-related desynchronization/synchronization and state control IEEE Trans. Neural Syst. Rehabil. Eng 20 379–88 [DOI] [PubMed] [Google Scholar]
- Jayaram V and Barachant A 2018. MOABB: trustworthy algorithm benchmarking for BCIs J. Neural. Eng 15 066011. [DOI] [PubMed] [Google Scholar]
- Jeong JH, Shim KH, Kim DJ and Lee SW 2020. Brain-controlled robotic arm system based on multi-directional CNN-BiLSTM network using EEG signals IEEE Trans. Neural Syst. Rehabil. Eng 28 1226–38 [DOI] [PubMed] [Google Scholar]
- Kaya M, Binli MK, Ozbay E, Yanar H and Mishchenko Y 2018. Data descriptor: a large electroencephalographic motor imagery dataset for electroencephalographic brain computer interfaces Sci. Data 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma DP and Ba JL, 2015. Adam: a method for stochastic optimization 3rd Int. Conf. Learning Representations, ICLR 2015—Conf. Track Proc (International Conference on Learning Representations, ICLR; ) [Google Scholar]
- Kloke JD and McKean JW 2012. Rfit: rank-based estimation for linear models R J. 4 57–64 [Google Scholar]
- Kobler RJ and Scherer R, 2017. Restricted Boltzmann machines in sensory motor rhythm brain-computer interfacing: a study on inter-subject transfer and co-adaptation 2016 IEEE Int. Conf. on Systems, Man, and Cybernetics, SMC 2016—Conf. Proc (Institute of Electrical and Electronics Engineers Inc.) pp 469–74 [Google Scholar]
- Kübler A and Birbaumer N 2008. Brain–computer interfaces and communication in paralysis: extinction of goal directed thinking in completely paralysed patients? Clin. Neurophysiol 119 2658–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafleur K, Cassady K, Doud A, Shades K, Rogin E and He B 2013. Quadcopter control in three-dimensional space using a noninvasive motor imagery-based brain–computer interface J. Neural. Eng 10 46003–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawhern VJ, Solon AJ, Waytowich NR, Gordon SM, Hung CP and Lance BJ 2018. EEGNet: a compact convolutional neural network for EEG-based brain–computer interfaces J. Neural. Eng 15 56013–30 [DOI] [PubMed] [Google Scholar]
- Leeb R, Perdikis S, Tonin L, Biasiucci A, Tavella M, Creatura M, Molina A, Al-Khodairy A, Carlson T and Milĺan JDR 2013. Transferring brain–computer interfaces beyond the laboratory: successful application control for motor-disabled users Artif. Intell. Med 59 121–32 [DOI] [PubMed] [Google Scholar]
- Li Y, Long J, Yu T, Yu Z, Wang C, Zhang H and Guan C 2010. An EEG-based BCI system for 2D cursor control by combining Mu/Beta rhythm and P300 potential IEEE Trans. Biomed. Eng 57 2495–505 [DOI] [PubMed] [Google Scholar]
- Liu J, Cheng Y and Zhang W 2015. Deep learning EEG response representation for brain computer interface Chinese Control Conf., CCC. IEEE Computer Society pp 3518–23 [Google Scholar]
- Lu N, Li T, Ren X and Miao H 2017a. A deep learning scheme for motor imagery classification based on restricted boltzmann machines IEEE Trans. Neural. Syst. Rehabil. Eng 25 566–76 [DOI] [PubMed] [Google Scholar]
- Luo T-J, Zhou C-L and Chao F 2018. Exploring spatial-frequency-sequential relationships for motor imagery classification with recurrent neural network BMC Bioinform. 19 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Qiu S, Du C, Xing J and He H, 2018. Improving EEG-based motor imagery classification via spatial and temporal recurrent neural networks Proc. of the Annual Int. Conf. of the IEEE Engineering in Medicine and Biology Society, EMBS (Institute of Electrical and Electronics Engineers Inc.) pp 1903–6 [DOI] [PubMed] [Google Scholar]
- Mak JN, Arbel Y, Minett JW, McCane LM, Yuksel B, Ryan D, Thompson D, Bianchi L and Erdogmus D 2011. Optimizing the P300-based brain–computer interface: current status, limitations and future directions J. Neural. Eng 8 025003. [DOI] [PubMed] [Google Scholar]
- Maris E and Oostenveld R 2007. Nonparametric statistical testing of EEG- and MEG-data J. Neurosci. Methods 164 177–90 [DOI] [PubMed] [Google Scholar]
- Meng J, Streitz T, Gulachek N, Suma D and He B 2018. Three-dimensional brain–computer interface control through simultaneous overt spatial attentional and motor imagery tasks IEEE Trans. Biomed. Eng 65 2417–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J, Zhang S, Bekyo A, Olsoe J, Baxter B and He B 2016. Noninvasive electroencephalogram based control of a robotic arm for reach and grasp tasks Sci. Rep 6 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel S, Spüler M and Mumtaz W 2019. World’s fastest brain-computer interface: combining EEG2Code with deep learning PLoS One 14 e0221909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrom NA, Levine MJ, Roskies RZ and Scott JR 2015. Bridges: a uniquely flexible HPC resource for new communities and data analytics ACM Int. Conf. Proc. Series. Association for Computing Machinery (New York) pp 1–8 [Google Scholar]
- Oostenveld R, Fries P, Maris E and Schoffelen J-M 2011. FieldTrip: open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data Comput. Intell. Neurosci 2011 156869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F et al. 2011. Scikit-learn: machine learning in python J. Mach. Learn. Res 12 2825–30 [Google Scholar]
- Perdikis S, Tonin L, Saeedi S, Schneider C, Milĺan JDR and Ganguly K 2018. The Cybathlon BCI race: successful longitudinal mutual learning with two tetraplegic users PLoS Biol. 16 e2003787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfurtscheller G and Lopes Da Silva FH 1999. Event-related EEG/MEG synchronization and desynchronization: basic principles Clin. Neurophysiol 110 1842–57 [DOI] [PubMed] [Google Scholar]
- Pfurtscheller G and Neuper C 2001. Motor imagery and direct brain-computer communication Proc. IEEE 89 1123–34 [Google Scholar]
- Rebsamen B, Guan C, Zhang H, Wang C, Teo C, Ang MH and Burdet E 2010. A brain controlled wheelchair to navigate in familiar environments IEEE Trans. Neural. Syst. Rehabil. Eng 18 590–8 [DOI] [PubMed] [Google Scholar]
- Sakhavi S, Guan C and Yan S, 2015. Parallel convolutional-linear neural network for motor imagery classification Signal Processing Conf. (EUSIPCO) pp 2736–40 [Google Scholar]
- Sakhavi S, Guan C and Yan S 2018. Learning temporal information for brain-computer interface using convolutional neural networks IEEE Trans. Neural Netw. Learn. Syst 29 5619–29 [DOI] [PubMed] [Google Scholar]
- Schalk G, McFarland DJ, Hinterberger T, Birbaumer N and Wolpaw JR 2004. BCI2000: a general-purpose brain-computer interface (BCI) system IEEE Trans. Biomed. Eng 51 1034–43 [DOI] [PubMed] [Google Scholar]
- Schirrmeister RT, Springenberg JT, Fiederer LDJ, Glasstetter M, Eggensperger K, Tangermann M, Hutter F, Burgard W and Ball T 2017. Deep learning with convolutional neural networks for EEG decoding and visualization Hum. Brain Mapp 38 5391–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwemmer MA, Skomrock ND, Sederberg PB, Ting JE, Sharma G, Bockbrader MA and Friedenberg DA 2018. Meeting brain–computer interface user performance expectations using a deep neural network decoding framework Nat. Med 24 1669–76 [DOI] [PubMed] [Google Scholar]
- Sitaram R et al. 2017. Closed-loop brain training: the science of neurofeedback Nat. Rev. Neurosci 18 86–100 [DOI] [PubMed] [Google Scholar]
- Stieger JR, Engel SA and He B 2021. Continuous sensorimotor rhythm based brain computer interface learning in a large population Sci. Data 8 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieger JR, Engel S, Jiang H, Cline CC, Kreitzer MJ and He B 2020. Mindfulness improves brain–computer interface performance by increasing control over neural activity in the alpha band Cereb. Cortex 00 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabar YR and Halici U 2017. A novel deep learning approach for classification of EEG motor imagery signals J. Neural. Eng 14 016003. [DOI] [PubMed] [Google Scholar]
- Tang Z, Li C and Sun S 2017. Single-trial EEG classification of motor imagery using deep convolutional neural networks Optik 130 11–18 [Google Scholar]
- Tangermann M. et al. Review of the BCI competition IV. Front. Neurosci. 2012;6:55. doi: 10.3389/fnins.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayeb Z, Fedjaev J, Ghaboosi N, Richter C, Everding L, Qu X, Wu Y, Cheng G and Conradt J 2019. Validating deep neural networks for online decoding of motor imagery movements from EEG signals Sensors 19 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonin L, Leeb R, Sobolewski A and Milĺan JDR 2013. An online EEG BCI based on covert visuospatial attention in absence of exogenous stimulation J. Neural. Eng 10 056007. [DOI] [PubMed] [Google Scholar]
- Towns J et al. 2014. XSEDE: accelerating scientific discovery Comput. Sci. Eng 16 62–74 [Google Scholar]
- Treder MS, Bahramisharif A, Schmidt NM, Van Gerven MA and Blankertz B 2011. Brain-computer interfacing using modulations of alpha activity induced by covert shifts of attention J. Neuroeng. Rehabil 8 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhaneni A, Wang T and He B 2005. Brain computer interface Neural Engineering ed He B (New York: Kluwer Academic/Plenum Publishers; ) pp 85–122 [Google Scholar]
- Van Gerven M and Jensen O 2009. Attention modulations of posterior alpha as a control signal for two-dimensional brain–computer interfaces J. Neurosci. Methods 179 78–84 [DOI] [PubMed] [Google Scholar]
- Vansteensel MJ et al. 2016. Fully implanted brain–computer interface in a locked-in patient with ALS N. Engl. J. Med 375 2060–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker M, Schirrmeister RT, Fiederer LDJ, Burgard W and Ball T 2018. Deep transfer learning for error decoding from non-invasive EEG 2018 6th Int. Conf. Brain-Computer Interface, BCI 2018 (Institute of Electrical and Electronics Engineers Inc.) pp 1–6 [Google Scholar]
- Wang P, Jiang A, Liu X, Shang J and Zhang L 2018. LSTM-based EEG classification in motor imagery tasks IEEE Trans. Neural. Syst. Rehabil. Eng 26 2086–95 [DOI] [PubMed] [Google Scholar]
- Wolpaw JR and McFarland DJ 2004. Control of a two-dimensional movement signal by a noninvasive brain-computer interface in humans Proc. Natl Acad. Sci. USA 101 17849–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SL, Wu CW, Pal NR, Chen CY, Chen SA and Lin CT 2013. Common spatial pattern and linear discriminant analysis for motor imagery classification Proc. 2013 IEEE Symp. Computational Intelligence, Cognitive Algorithms, Mind, and Brain, CCMB 2013–2013 IEEE Symp. Series Computational Intelligence, SSCI 2013 (IEEE Computer Society; ) pp 146–51 [Google Scholar]
- Yu Z and Song J 2017. Multi-class motor imagery classification by singular value decomposition and deep Boltzmann machine Proc. 2017 IEEE 3rd Information Technology and Mechatronics Engineering Conf., ITOEC 2017 (Institute of Electrical and Electronics Engineers Inc.) pp 376–9 [Google Scholar]
- Yuan H and He B 2014. Brain–computer interfaces using sensorimotor rhythms: current state and future perspectives IEEE Trans. Biomed. Eng 61 1425–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Liu T, Szarkowski R, Rios C, Ashe J and He B 2009. Negative covariation between task-related responses in alpha/beta-band activity and BOLD in human sensorimotor cortex: an EEG and fMRI study of motor imagery and movements Neuroimage 49 2596–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this study are available for public use and described with further detail in ‘Continuous sensorimotor rhythm based brain computer interface learning in a large population’ (Stieger et al 2021).
The data that support the findings of this study are openly available at the following URL/DOI: https://doi.org/10.6084/m9.figshare.13123148.


