Abstract
Two kinds of bacteria having different-structured angular dioxygenases—a dibenzofuran (DF)-utilizing bacterium, Terrabacter sp. strain DBF63, and a carbazole (CAR)-utilizing bacterium, Pseudomonas sp. strain CA10—were investigated for their ability to degrade some chlorinated dibenzofurans (CDFs) and chlorinated dibenzo-p-dioxins (CDDs) (or, together, CDF/Ds) using either wild-type strains or recombinant Escherichia coli strains. First, it was shown that CAR 1,9a-dioxygenase (CARDO) catalyzed angular dioxygenation of all mono- to triCDF/Ds investigated in this study, but DF 4,4a-dioxygenase (DFDO) did not degrade 2,7-diCDD. Secondly, degradation of CDF/Ds by the sets of three enzymes (angular dioxygenase, extradiol dioxygenase, and meta-cleavage compound hydrolase) was examined, showing that these enzymes in both strains were able to convert 2-CDF to 5-chlorosalicylic acid but not other tested substrates to the corresponding chlorosalicylic acid (CSA) or chlorocatechol (CC). Finally, we tested the potential of both wild-type strains for cooxidation of CDF/Ds and demonstrated that both strains degraded 2-CDF, 2-CDD, and 2,3-diCDD to the corresponding CSA and CC. We investigated the sites for the attack of angular dioxygenases in each CDF/D congener, suggesting the possibility that the angular dioxygenation of 2-CDF, 2-CDD, 2,3-diCDD, and 1,2,3-triCDD (10 ppm each) by both DFDO and CARDO occurred mainly on the nonsubstituted aromatic nuclei.
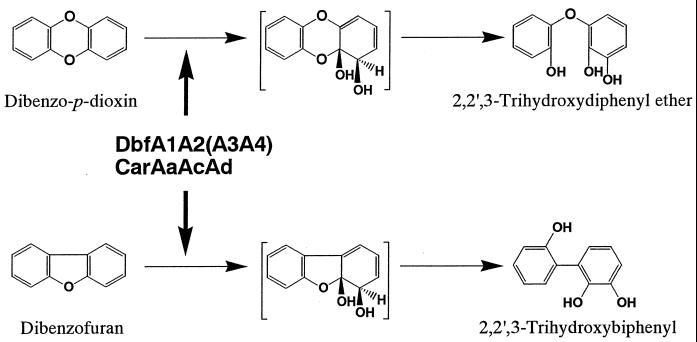
Polychlorinated dibenzofurans and polychlorinated dibenzo-p-dioxins (PCDF/Ds) are highly toxic compounds but have become widely distributed in the environment. They are formed as unwanted by-products during the synthesis of pesticides and herbicides or during municipal incineration and are not degraded under environmental conditions. Recently, bioprocess treatment methods have been proposed for the destruction of dioxins, and bioremediation of contaminated soil is being used as an alternative to more costly physical and chemical treatments (10, 12, 20). Over the last decade, a number of bacteria capable of degrading dibenzofuran (DF), dibenzo-p-dioxin (DD), and some of their lightly chlorinated derivatives have been isolated, and the cooxidative degradation of chlorinated DFs (CDFs) and chlorinated DDs (CDDs) (or, together, CDF/Ds) have been elucidated (14, 17, 21, 25, 30, 35, 37, 38). However, compared with bacterial degradation of polychlorinated biphenyls carrying di- to pentachlorine substituents (6, 11, 19, 31, 32, 33, 34), knowledge concerning bacterial cometabolism of CDF/Ds carrying two or more chlorine substituents is still lacking. One reason is the shortage of information about the initial dioxygenases that can act on the angular position adjacent to the oxygen atom of DF and DD skeletons, though many biphenyl dioxygenases have been well studied. This initial dioxygenase, termed angular dioxygenase, is a very important enzyme for the degradation of dioxin-related compounds (7, 13, 36). Since angular dioxygenation produces unstable hemiacetal compounds that rearomatize spontaneously, prompting cleavage of the ether bond (Fig. 1), the reduction of toxicity of dioxins via the ring cleavage is accomplished by this single-step reaction.
FIG. 1.
Conversion of DD and DF by DFDO from Terrabacter sp. strain DBF63 and by CARDO from Pseudomonas sp. strain CA10.
Previously, we isolated and characterized several bacteria that can degrade dioxin-related compounds (21, 24) and their initial dioxygenase genes (16, 28). Among them, a gram-positive bacterium, Terrabacter sp. strain DBF63, which was isolated based on its ability to utilize DF as a sole carbon and energy source, could cometabolize DD (21). The genes encoding the terminal oxygenase components (dbfA1A2) of DF 4,4a-dioxygenase (DFDO) (EC 1.14.12.12) were cloned, sequenced, and characterized (16). It was also revealed that DbfA1A2 catalyzed the oxidation of DD to 2,2′,3-trihydroxydiphenyl ether (2,2′,3-THDE) using nonspecific ferredoxin and ferredoxin reductase components in Escherichia coli cells (16). In addition, expression of the genes for extradiol dioxygenase (dbfB) and meta-cleavage compound hydrolase (dbfC) from strain DBF63 in E. coli resulted in the biodegradation of 2,2′,3-trihydroxybiphenyl (2,2′,3-THB) to salicylic acid (15).
On the other hand, we also reported the isolation of a gram-negative bacterium, Pseudomonas sp. strain CA10, which utilized carbazole (CAR) as a sole carbon, nitrogen, and energy source (24). The genes encoding CAR 1,9a-dioxygenase (CARDO) (EC 1.14.12.12) were isolated, and functional analysis revealed that CARDO consists of a single protein of terminal oxygenase (CarAa), ferredoxin (CarAc), and ferredoxin reductase (CarAd). CARDO has a broad substrate range and can convert DD and DF to 2,2′,3-THDE and 2,2′,3-THB, respectively (22, 28). Moreover, we characterized the genes encoding extradiol dioxygenase composed of two proteins (carBaBb) and meta-cleavage compound hydrolase (carC), which are involved in the degradation of 2′-aminobiphenyl-2,3-diol to anthranilic acid via 2-hydroxy-6-oxo-6-(2′-aminophenyl)-hexa-2,4-dienoic acid (29).
Besides those of DFDO from strain DBF63 and CARDO from strain CA10, only one detailed report has been made of the cloning of angular dioxygenase genes. This enzyme, dioxin dioxygenase from Sphingomonas wittichii strain RW1 (38, 39), has been studied at the molecular level (1, 2, 3, 5, 8). Although substrate specifities of these angular dioxygenases for various aromatic compounds have already been investigated (8, 16, 22), data concerning the degradation of CDF/Ds by angular dioxygenases have not been reported.
For bioremediation of dioxin-contaminated soils, it is important to know the influence of the chlorine substitution pattern on the degradation of CDF/Ds by these angular dioxygenases with different features as well as the possible fate of CDF/Ds in the environment after degradation by different types of dioxin degraders. In this work, we describe the substrate selectivity of DFDO and CARDO and the potential of strains DBF63 and CA10 to cooxidize various mono-, di-, and triCDF/Ds. From the results obtained, we discuss the sites of angular dioxygenation by DFDO and CARDO and the transformation capabilities of the subsequent enzymes.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Terrabacter sp. strain DBF63 was routinely cultivated in a carbon-free mineral medium (CFMM) (23) supplemented with DF at a concentration of 0.05% (wt/vol) at 30°C. Pseudomonas sp. strain CA10 was similarly cultivated in CFMM with CAR at the same concentration. DF and CAR were added to CFMM from stock solutions (0.1 g/ml) dissolved in dimethyl sulfoxide and sterilized by filtration. Ten milliliters of precultures of strains DBF63 and CA10 grown on CFMM supplemented with 0.05% (wt/vol) DF and CAR, respectively, was inoculated into 1 liter of the same fresh medium in 5-liter flasks and cultivated at 30°C for another 3 and 2 days, respectively. When the optical density at 600 nm reached 1.5, cells were harvested by centrifugation, washed twice with 50 mM phosphate buffer (pH 7.4), and resuspended in a reduced volume of the same buffer (an optical density at 600 nm of approximately 20). E. coli JM109 cells harboring pDF32 and pUCARA were used for the expression of DFDO and CARDO, respectively. E. coli JM109 harboring pUCA1 was used for the degradation of CDF/Ds by CARDO (CarAaAcAd), CarBaBb, and CarC. As pDF32, pUCARA, and pUCA1 are derived from pUC119 (40), E. coli JM109 harboring pUC119 was used in control experiments. When E. coli JM109 harboring both pDF32 and pLM1RM was used for the degradation of CDF/Ds by DFDO (DbfA1A2), DbfB, and DbfC, E. coli JM109 harboring both pUC119 and pSTV28 was used in control experiments. E. coli cells were grown on 2× YT medium (27) at 37°C, which was supplemented with ampicillin and/or chloramphenicol at a final concentration of 50 μg/ml and 30 μg/ml, respectively. The resting cells of E. coli harboring pDF32, pUCARA, pUCA1, and pLM1RM were prepared as described previously (15, 22).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| Terrabacter sp. strain DBF63 | DF+a | 21 |
| Pseudomonas sp. strain CA10 | CAR+b | 24 |
| E. coli JM109 | recA1 Δ(lac-proAB) endA1 gyrA96 thi-1 hsdR17 relA1 supE44 [F′ traD36 proAB laqIqZ ΔM15] | 40 |
| Plasmids | ||
| pUC119 | AprlacZ; pMB9 replicon | 40 |
| pSTV28 | CmrlacZ; pMB9 replicon | Takara Shuzo Co., Ltd. |
| pUCA1 | Apr; pUC119 containing 6.9-kb EcoRI insert of CA10 DNA | 29 |
| pDF32 | Apr; pUC119 containing 2.6-kb BglII-SphI insert of DBF63 DNA | 16 |
| pUCARA | Apr; pUC119 containing 5.6-kb EcoRI insert of pUCA1 with the deletion of 1.3-kb PstI-BglI fragment | 28 |
| pLM1RM | Cmr; pSTV28 containing 3.0-kb BamHI insert of DBF63 DNA | 15 |
DF+, able to use dibenzofuran as the sole carbon or energy sources.
CAR+, able to use carbazole as sole carbon, nitrogen, and energy sources.
Biotransformation analysis of CDDs and CDFs.
Biotransformation reactions proceeded at 30°C in glass tubes sealed with silicon caps. The tubes containing 5 ml of the washed-cell suspension and the substrate at 10 ppm (10 μg/ml) were incubated on a reciprocal shaker (300 strokes/min) at 30°C for 18 h. Stock solutions of CDF/Ds (1 mg/ml) were made up in dimethyl formamide. The reaction mixtures were extracted twice with 5 ml of ethyl acetate under acidic or neutral conditions. Extracts were dried over anhydrous Na2SO4, and concentrated in vacuo. Metabolites formed from CDF/Ds were analyzed by gas chromatography-mass spectrometry (GC-MS) after derivatization with N-methyl-N-trimethylsilyltrifluoroacetamide (MSTFA) and verified by coinjection of authentic samples. When authentic compounds were not available, structures of metabolites were proposed by MS spectra. We determined the rate of substrate depletion or metabolite formation by comparing the peak areas for molecular ions of each compound extracted from the reaction mixture containing E. coli cells harboring pDF32, pUCARA, pUCA1, and pDF32 plus pLM1RM with those of each compound extracted from the reaction mixture containing E. coli cells harboring pUC119 or pUC119 plus pSTV28. We repeated the above experiments two or three times.
Analytical methods.
GC-MS analyses were performed with a model JMS-Automass 150 GC-MS system (JEOL, Ltd., Tokyo, Japan) fitted with a fused-silica chemically bonded capillary column (DB-5; 0.25 mm [inside diameter] by 15 m; 0.25 μm film thickness; J&W Scientific, Inc., Folsom, Calif.). After trimethylsilylation with MSTFA, each sample was injected into the column at 80°C in the splitless mode. After 2 min at 80°C, the column temperature was increased at 16°C/min to 280°C. The head pressure of the helium carrier gas was 65 kPa.
Chemicals.
DD and DF were purchased from Wako Pure Chemical (Osaka, Japan) and Kanto Chemical (Tokyo, Japan), respectively. 2-Chlorodibenzofuran (2-CDF), 2,8-dichlorodibenzofuran (2,8-DCDF), 2-chlorodibenzo-p-dioxin (2-CDD), 2,3-dichlorodibenzo-p-dioxin (2,3-DCDD), 2,7-dichlorodibenzo-p-dioxin (2,7-DCDD), and 1,2,3-trichlorodibenzo-p-dioxin (1,2,3-TCDD) were purchased from AccuStandard, Inc. (New Haven, Conn.). 4-Chlorocatechol (4-CC) was purchased from Sigma-Aldrich (Steinheim, Germany), and 4,5-dichlorocatechol (4,5-DCC) was obtained from Helix Biotechnology Corp. (Richmond, British Columbia, Canada). Purities of these chemicals are 96.6 to 100%. All other chemicals were of analytical grade and of the highest purity available.
RESULTS
Conversion of CDFs and CDDs by DFDO and CARDO.
To investigate whether two kinds of angular dioxygenases, DFDO and CARDO, transform CDF/Ds, these compounds were incubated with recombinant E. coli strains carrying respective genes, and the products were characterized by GC-MS (Table 2).
TABLE 2.
GC retention times and MS spectral properties of major compounds formed from CDFs and CDDs by resting cell reactions of E. coli encoding DFDO and CARDO
| Substrate | No. of possible productsa | No. of observed productse
|
RTb (min) | Fragment ionsc | No. of chlorines | Possible productd | |
|---|---|---|---|---|---|---|---|
| DFDO | CARDO | ||||||
| 2-CDF | 2 | 1 | 1 | 10.93 | 454 (M+, 37Cl) (27), 452 (M+, 35Cl) (62), 349 (100), 314 (6) | 1 | 5′-Chloro-2,2′,3-THB |
| 2,8-DCDF | 1 | 1 | 1 | 11.67 | 486 (M+, 35Cl) (71), 383 (100), 253 (21), 173 (20) | 2 | 5,5′-Dichloro-2,2′,3-THB |
| 2-CDD | 4 | 2 (5:1) | 2 (5:1) | 11.27 | 470 (M+, 37Cl) (60), 468 (M+, 35Cl) (95), 365 (100), 330 (74), 166 (46) | 1 | 5′-Chloro-2,2′,3-THDE or 4′-chloro-2,2′,3-THDE |
| 11.43 | 470 (M+, 37Cl) (22), 468 (M+, 35Cl) (46), 365 (100), 330 (34), 200 (9) | 1 | 5′-Chloro-2,2′,3-THDE or 4′-chloro-2,2′,3-THDE | ||||
| 2,3-DCDD | 2 | 1 | 1 | 12.40 | 502 (M+, 35Cl) (60), 399 (100), 364 (37), 165 (19), 137 (18) | 2 | 4′,5′-Dichloro-2,2′,3-THDE |
| 2,7-DCDD | 2 | 0 | 1 | 12.42 | 502 (M+, 35Cl) (100), 399 (99), 364 (48), 165 (19), 137 (27) | 2 | 4,5′-Dichloro-2,2′,3-THDE or 4′,5-dichloro-2,2′,3-THDE |
| 1,2,3-TCDD | 3 | 1 | 2 (2:1) | 13.03 | 538 (M+, 35Cl) (100), 435 (63), 398 (24), 179 (51), 137 (26) | 3 | 3′,4′,5′-Trichloro-2,2′,3-THDE or 4′,5′,6′-trichloro-2,2′3-THDE |
| 13.13 | 538 (M+, 35Cl) (100), 435 (39), 398 (24), 137 (20) | 3 | 3′,4′,5′-Trichloro-2,2′,3-THDE or 4′,5′,6′-trichloro-2,2′,3-THDE | ||||
The number of possible products was calculated on the basis that neither dioxygenation of lateral nor of chlorinated sites is posssible.
All products were analyzed by GC-MS after derivatization of MSTFA.
Fragment ions are expressed as m/z values; relative intensities expressed as percentages are given in parentheses.
Possible products were suggested based on the results described in the text (see Discussion).
Rates in parentheses represent the relative amounts of two products (ratio of the product having the earlier RT to the product having the later RT).
Although 2-CDF has two possible sites for angular dioxygenation, the GC-MS analytical data showed the production of one compound. In contrast to 2-CDF, 2-CDD has four possible sites for the initial angular dioxygenation. However, the sample of 2-CDD incubated with either DFDO or CARDO appeared to contain two metabolites showing very similar fragmentation patterns. In addition, CARDO degraded 2,7-DCDD, producing a metabolite, whereas DFDO did not metabolize 2,7-DCDD. In the sample of 1,2,3-TCDD with DFDO and CARDO, we detected one metabolite (retention time [RT], 13.03 min) and two metabolites, respectively.
The following data obtained by GC-MS strongly suggested that these metabolites are the chlorinated THB and THDE originating from the parent CDFs and CDDs. (i) All products had the possible molecular ions whose mass unit corresponded to the calculated molecular weight of the trimethylsilyl (TMS)-derivatives of chlorinated THB or chlorinated THDE as shown in Table 2. (ii) In our previous reports (15, 22), the mass spectra of the TMS derivatives of 2,2′,3-THB (M+, 418), 2,2′,3-THDE (M+, 434), 2,2′,3-trihydroxydiphenylmethane (M+, 432), and 2,2′,3-trihydroxydiphenyl sulfide (M+, 450) exhibited strong fragment ions at m/z 315, 331, 329, and 347, respectively, which represent the loss of 103 from molecular ions. GC-MS results in this study also showed the presence of the strong fragment ions at M+-103 (Table 2), indicating that all metabolites from CDFs and CDDs by DFDO and CARDO correspond to the TMS derivatives of THB or THDE. (iii) Comparison of the isotope ratio between the TMS-derivatized metabolites and the parent substrate on mass spectra showed that both contained the same number of chlorine substituents (data not shown).
From these findings, it was suggested that CARDO from strain CA10 could catalyze the angular dioxygenation of all the tested mono- to triCDF/Ds. On the other hand, DFDO from strain DBF63 could not degrade 2,7-DCDD.
Conversion of CDFs and CDDs by DbfA1A2BC or CarAaAcAdBaBbC.
The formation of corresponding chlorosalicylic acid (CSA) and CC from CDFs and CDDs by DbfA1A2BC, which are thought to be involved in the metabolism of DF to salicylic acid in strain DBF63, were investigated (Table 3). E. coli cells harboring both pDF32 containing dbfA1A2 genes and pLM1RM containing dbfBC genes were used for synthesizing the three enzymes, i.e., angular dioxygenase, extradiol dioxygenase, and meta-cleavage compound hydrolase. As a result, 2-CDF was converted to 5-CSA but not to salicylic acid. On the other hand, 5-CSA and CCs were not produced from 2,8-DCDF, 2-CDD, 2,3-DCDD, 2,7-DCDD, and 1,2,3-TCDD. However, approximately 96, 100, 98, 0, and 95% of the substrates were degraded, respectively, and also, the corresponding chlorinated THB and chlorinated THDE were not detected. These results suggest that CDF/D congeners except 2,7-DCDD may undergo meta-cleavage by DbfB after angular dioxygenation, although direct evidence for the presence of the meta-cleavage product is not provided. The fact that approximately 100% of 2,7-DCDD remained was consistent with the result of the transformation of 2,7-DCDD by DFDO.
TABLE 3.
CSA and CC formed from CDFs and CDDs by resting cell reactions of various strains
| Substratea | Approx rate of substrate depletion (%)b
|
Approx rate of product formation (%)ce
|
Identified productd | ||||||
|---|---|---|---|---|---|---|---|---|---|
| DBF63 | CA10 | E. coli JM109 (DbfABC) | E. coli JM109 (CarABC) | DBF63 | CA10 | E. coli JM109 (DbfABC) | E. coli JM109 (CarABC) | ||
| 2-CDF | 75–90 | 50–70 | 100 | 100 | 35–50 | 5–10 | 5.0–15 | 2.0–5.0 | 5-CSA |
| 2,8-DCDF | 80–90 | 5–20 | 96–99 | 65–80 | 65–85 | ND | ND | ND | 5-CSA |
| 2-CDD | 70–80 | 25–40 | 100 | 100 | 30–45 | 1.0–1.5 | ND | ND | 4-CC |
| 2,3-DCDD | 70–90 | 30–40 | 95–99 | 60–80 | 20–35 | 0.1–1.0 | ND | ND | 4,5-DCC |
| 2,7-DCDD | 10–25 | 15–35 | 0 | 20–40 | ND | ND | ND | ND | ND |
| 1,2,3-TCDD | 20–30 | 10–20 | 95–99 | 60–80 | ND | ND | ND | ND | ND |
The starting concentration of substrates in the biotransformations used to estimate the yields of substrate depletion and the product formation was 10 ppm.
Values represent the percentages of the respective CDF/D degradation rates by resting cells of each strain. After 18-h biotransformation experiments, we compared the peak area for the molecular ion of each substrate extracted from each reaction mixture containing DBF63 cells, CA10 cells, and E. coli cells harboring either dbfABC or carABC with that for each substrate extracted from the reaction mixture containing E. coli cells harboring only pUC119 or pUC119 plus pSTV28 (control). We repeated the experiments two or three times and estimated the substrate depletion.
Values represent the percentages of the corresponding CSA or CC formation rates by resting cells of each strain. After 18-h biotransformation experiments, we compared the peak area for the molecular ion of TMS derivatives of each product extracted from each reaction mixture containing DBF63 cells, CA10 cells, and E. coli cells harboring either dbfABC or carABC with that of 10 ppm of authentic CSA or CC. We repeated the experiments two or three times and estimated the yields of products.
Identification of products formed was made by GC-MS after derivatization of MSTFA.
ND, the corresponding CSA and CC were not detected. In all these cases, both salicylic acid and catechol were not detected.
We also investigated whether CAR-degrading enzymes from strain CA10, CarAaAcAdBaBbC, involved in the metabolism of CAR to anthranilic acid, could catalyze the degradation of CDFs and CDDs to CSAs and CCs, respectively (Table 3). Like DbfA1A2BC, GC-MS analysis revealed that the resting cells of E. coli harboring pUCA1 containing the carAaAcAdBaBbC genes produced a small amount of 5-CSA from 2-CDF, but salicylic acid was not detected. From the other tested CDF/Ds, corresponding chlorinated THB, chlorinated THDE, CSA, and CCs were not detected. Based on the facts that approximately 70% of 2,8-DCDF, 100% of 2-CDD, 80% of 2,3-DCDD, 30% of 2,7-DCDD, and 60% of 1,2,3-TCDD were degraded after the biotransformation, it was suggested that CarBaBb might catalyze the degradation of chlorinated THB and chlorinated THDE, but that CarC could not degrade effectively the resultant metabolites.
Cooxidation of CDFs and CDDs by Terrabacter sp. strain DBF63.
Conversion of CDFs and CDDs to the corresponding CSA and CC by DF-grown DBF63 cells was investigated (Table 3). On GC-MS analysis, it was revealed that 2-CDF and 2,8-DCDF are converted to the same compound (RT, 7.88 min) whose mass spectrum exhibited fragment ions at m/z 303 (M+-CH3, 37Cl) (25%), 301 (M+-CH3, 35Cl) (100%), 243 (8%), 169 (34%), and 149 (19%) (relative intensities are given in parentheses). This fragmentation pattern and the RT of the TMS derivative of the metabolite were identical to those of authentic 5-CSA. However, we could not detect salicylic acid from 2-CDF as detected in the products degraded by strain RW1 (35).
When 2-CDD was incubated with the resting cells of strain DBF63, we detected the single product (RT, 6.32 min) whose mass spectrum exhibited fragment ions at m/z 290 (M+, 37Cl) (52%), 288 (M+, 35Cl) (100%), 273 (16%), 200 (19%), 185 (21%), and 170 (19%). This fragmentation pattern and the RT of the TMS derivative of metabolite were the same as those observed for the TMS derivative of authentic 4-CC. In this case, catechol was not detected from the sample.
Strain DBF63 transformed 2,3-DCDD to the product with an RT of 7.65 min in GC and MS fragmentation pattern (m/z 322 [M+, 35Cl] [100%], 307 [22%], 234 [55%], 219 [56%], and 204 [39%]) consistent with those of authentic 4,5-DCC. Similar to the experiment of 2-CDD transformation, no catechol was detected from the extract of the reaction mixture. In addition, it was revealed that 2,7-DCDD and 1,2,3-TCDD were not converted to either corresponding CC or catechol. However, 2,7-DCDD was also degraded up to 10 to 25%, though DFDO did not attack this substrate. Since the predicted monohydroxylated derivative of 2,7-DCDD was detected by GC-MS (data not shown), 2,7-DCDD was thought to be degraded by other oxygenases or hydroxylases in strain DBF63.
Cooxidation of CDFs and CDDs by Pseudomonas sp. strain CA10.
CAR-grown CA10 cells converted 2-CDF to 5-CSA but not to salicylic acid. Based on the GC-MS analysis, it was also revealed that 4-CC and 4,5-DCC were formed from 2-CDD and 2,3-DCDD, respectively, but no catechol was produced from both substrates by strain CA10 (Table 3). These results are the same with the conversion by strain DBF63, although the yields of 5-CSA, 4-CC, and 4,5-DCC by strain CA10 were much lower than those by strain DBF63 (Table 3). These results showed that strain CA10 had a weaker ability to degrade CDF/Ds than did strain DBF63.
In addition, 4-CC was not produced from 2,7-DCDD, and 3,4,5-triCC and catechol were not produced from 1,2,3-TCDD. However, since both substrates were degraded by CARDO as shown above, 2,7-DCDD and 1,2,3-TCDD were decreased by 15 to 35% and 10 to 20%, respectively. Different from strain DBF63, 2,8-DCDF was not transformed to 5-CSA by strain CA10, although experiments were repeated several times. However, the 2,8-DCDF was decreased by 5 to 15% because CARDO could catalyze angular dioxygenation of this substrate.
DISCUSSION
We investigated the concentration of the total PCDF/Ds in the actual dioxin-contaminated soil at an incinerator site located in Japan. Since it amounted to 725 ng/g of dry soil (10), we first tried to use a low concentration (0.5 ppm) of CDF/Ds as target substrates. However, since the analysis of metabolites at this low concentration was difficult, probably due to low transformation rates of CDF/Ds, 10 ppm of each substrate was used.
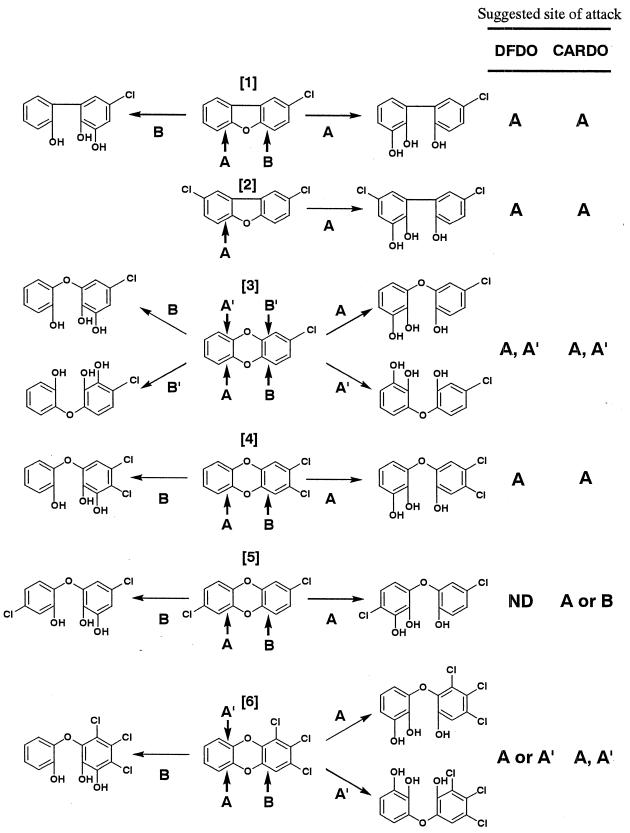
In this study, the positions of the hydroxy groups in the chlorinated THB and THDE produced by DFDO and CARDO could not be determined exactly, because biotransformation experiments at low concentrations of substrates did not produce enough amounts of the products for additional analyses such as 1H nuclear magnetic resonance spectrometry. Therefore, assignment of the positions of angular dioxygenation from GC-MS data can be made only for 2,8-DCDF, the attack at only a single angular position of which will yield a 5,5′-dichloro-2,2′,3-THB (Fig. 2, panel 2, reaction A). However, the combination of the results obtained from the transformation of various CDF/D congeners by angular dioxygenase frequently permitted assignment of the position of initial dioxygenation. Also, it was quite useful for position assignment of dioxygenation to determine what type of salicylic acid or catechol derivatives was produced by biotransformation experiments using wild-type strains (35). Possible assignment of the positions of angular dioxygenation is shown in Fig. 2.
FIG. 2.
Proposed initial dioxygenolytic attack on the tested CDFs and CDDs by DFDO and CARDO. Panel 1, 2-CDF, 5′-chloro-2,2′,3-THB (reaction A) and 5-chloro-2,2′,3-THB (reaction B); panel 2, 2,8-DCDF, 5,5′-dichloro-2,2′,3-THB (reaction A); panel 3, 2-CDD, 5′-chloro-2,2′,3-THDE (reaction A), 4′-chloro-2,2′,3-THDE (reaction A′), 5-chloro-2,2′,3-THDE (reaction B), and 4-chloro-2,2′,3-THDE (reaction B′); panel 4, 2,3-DCDD, 4′,5′-dichloro-2,2′,3-THDE (reaction A) and 4,5-dichloro-2,2′,3-THDE (reaction B); panel 5, 2,7-DCDD, 4,5′-dichloro-2,2′,3-THDE (reaction A) and 4′,5-dichloro-2,2′,3-THDE (reaction B); panel 6, 1,2,3-TCDD, 4′,5′,6′-trichloro-2,2′,3-THDE (reaction A), 3′,4′,5′-trichloro-2,2′,3-THDE (reaction A′), and 4,5,6-trichloro-2,2′,3-THDE (reaction B). Possible sites attacked by angular dioxygenases are indicated by A or A′ on the nonsubstituted rings and by B or B′ on the halogen-substituted rings, with the exception of those in panels 2 and 5. ND, not detected.
In experiments, neither nonsubstituted salicylic acid nor catechol was detected after biotransformation using either strain DBF63 or CA10. The nonsubstituted aromatic nucleus of 2-CDF seems to be attacked because 5-CSA was produced from 2-CDF by wild-type strains DBF63 and CA10 (Table 3) as well as E. coli cells harboring DbfA1A2BC and CarAaAcAdBaBbC. In addition, as shown in Table 2, only one angular dioxygenation product was detected by DFDO and CARDO. These results suggest that DFDO and CARDO mainly act on the nonsubstituted aromatic nucleus of 2-CDF to create a 5′-chloro-2,2′,3-THB (Fig. 2, panel 1, reaction A).
2-CDD may also be attacked at the site of the nonsubstituted aromatic nucleus, because 4-CC was produced by wild-type strains DBF63 and CA10 in the biotransformation experiments (Table 3). In addition, two angular dioxygenation products were produced by DFDO and CARDO. In contrast to 2-CDF, which contains only two sites for the initial enzymatic attack, 2-CDD has four possible sites for the initial angular dioxygenation. Considering that DFDO and CARDO act on the nonsubstituted aromatic nucleus of 2-CDF, these two metabolites are thought to be 5′-chloro-2,2′,3-THDE (Fig. 2, panel 3, reaction A) and 4′-chloro-2,2′,3-THDE (Fig. 2, panel 3, reaction A′).
Previously, Harms et al. revealed that a mutant strain from the DF-degrading bacterium Pseudomonas sp. strain HH69, which is defective in 2,3-dihydroxybiphenyl-1,2-dioxygenase, led to the formation of an equal amount of 4′-chloro-2,2′,3-THB and 4-chloro-2,2′,3-THB from 3-CDF (14). Wilkes et al. demonstrated that strain RW1 converted 2-CDF to both 5-CSA and salicylic acid and converted 2-CDD to both 4-CC and catechol (35). Wittich et al. also reported that the conversion of 3-CDF in the presence of an inhibitor of meta-cleavage of chlorinated THB by Sphingomonas sp. strain RW16 led to accumulation of two catabolites that are likely to be 4-chloro-2,2′,3-THB and 4′-chloro-2,2′,3-THB (37). These results indicated that angular dioxygenases from these bacteria attacked both the substituted and the nonsubstituted aromatic nuclei of monochlorinated DFs and DDs nonselectively. In these biotransformation experiments, the concentrations of target substrates were from 100 to 1,000 ppm. On the other hand, Parsons et al. proposed that initial dioxygenation apparently took place on the nonchlorinated ring during the degradation of 2-CDF and 2-CDD by the biphenyl-utilizing Burkholderia strain JB1, since 5-CSA and 4-CC as well as only one isomer each of chlorinated THB and chlorinated THDE were detected (25). In their experiment, the concentration of substrate was low (0.3 ppm). Our results with 10 ppm of target substrates suggested that both DFDO and CARDO might attack the nonchlorinated ring of 2-CDF and 2-CDD as does the initial dioxygenase from strain JB1.
As indicated in the transformation experiments with 2,8-DCDF, both enzymes could act on the chlorine-substituted aromatic rings to produce a 5,5′-dichloro-2,2′,3-THB (Fig. 2, panel 2, reaction A) even if substrates had two halogen-substituted aromatic nuclei. This result corresponded to the report with strain RW1 (17, 35).
After the biotransformation of 2,3-DCDD by wild-type strains, 4,5-DCC was detected (Table 3), showing that the nonsubstituted aromatic nucleus of 2,3-DCDD was attacked. In addition, only one metabolite was produced by DFDO and CARDO (Table 2). These results suggest that DFDO and CARDO may act on the nonsubstituted aromatic nucleus of 2,3-DCDD to yield a 4′5′-dichloro-2,2′,3-THDE (Fig. 2, panel 4, reaction A). This tendency also corresponds to the result in the experiments with strain RW1 (35).
On the other hand, we detected one metabolite of 1,2,3-TCDD in E. coli resting cells carrying DFDO and two metabolites in those carrying CARDO. Though 1,2,3-TCDD has theoretically three possible sites for initial dioxygenolytic attack, two metabolites converted by CARDO might be 4′5′,6′-trichloro-2,2′,3-THDE (Fig. 2, panel 6, reaction A) and 3′,4′,5′-trichloro-2,2′,3-THDE (Fig. 2, panel 6, reaction A′), and a metabolite converted by DFDO might be one of these two compounds (Fig. 2, panel 6, reaction A or A′), because both angular dioxygenases were thought to attack the nonsubstituted aromatic nucleus of 2,3-DCDD.
In addition, one metabolite was produced from 2,7-DCDD by CARDO, but no metabolites were produced by DFDO. Since 2,7-DCDD contains two possible sites for the initial enzymatic attack, this metabolite produced by CARDO was thought to be 4,5′-dichloro-2,2′,3-THDE or 4′5-dichloro-2,2′,3-THDE (Fig. 2, panel 5, reaction A or B). To understand how DFDO and CARDO interact with these PCDF/D congeners, information on the three-dimensional structure of these proteins may be useful. Protein crystallization and X-ray crystallography of CARDO as well as purification of DFDO are now under way.
We also investigated the degradation of chlorinated THB and chlorinated THDE by subsequent enzymes, DbfBC and CarBaBbC. It was demonstrated that both DbfA1A2BC and CarAaAcAdBaBbC were capable of converting 2-CDF to 5-CSA. However, 2,8-DCDF was not converted to 5-CSA by DbfA1A2BC, although the wild-type strain DBF63 could transform 2,8-DCDF to 5-CSA at high transformation rates (Table 3). Seah et al. revealed that BphD from Burkholderia cepacia strain LB400 and Rhodococcus globerulus strain P6 were not able to hydrolyze 4-chloro-2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate (4-chloro-HOPDA) effectively and that 4-chloro-HOPDA underwent a nonenzymatic transformation to acetophenone (31, 32). In our study, 2,8-DCDF is predicted to be transformed to 4,11-dichloro-2,8-dihydroxy-6-oxo-phenylhexa-2,4-dianoate by both angular dioxygenase and extradiol dioxygenase, and if this 4-chlorinated compound also undergoes a similar nonenzymatic reaction, the formation of 5′-chloro-2′-hydroxyacetophenone would be expected. However, we could not detect this compound by GC-MS analysis. Moreover, Kohler et al. reported that the meta-cleavage product of 2,2′,3-THB was not stable and gave rise to their cyclization product 3-(chroman-4-on-2-yl)pyruvate (18). The fact that strain RW1 transformed 2,8-DCDF into 6-chloro-2-methyl-4H-chromen-4-one (35) was consistent with the report by Kohler et al., with an additional hydrolytical shortening of the side chain. Hence, we also investigated the presence of chlorinated cyclization products of meta-cleavage compounds; however, we could not detect these compounds in the extracts from the reaction mixture. Detailed analyses of further degradation of chlorinated THB will be required.
On the other hand, none of the meta-cleavage compounds of CDDs were hydrolyzed effectively by E. coli cells harboring the dbfC gene or carC gene, whereas the wild-type strains DBF63 and CA10 apparently could hydrolyze meta-cleavage compounds of both 2-CDD and 2,3-DCDD. Previously, Kasuga et al. showed that DbfBC was capable of converting 2,2′,3-THB to salicylic acid and 2,2′,3-THDE to the corresponding meta-cleavage product but not to catechol (15). Therefore, it seems reasonable that DbfB catalyzed meta-cleavage of chlorinated THDE but DbfC did not hydrolyze the corresponding meta-cleavage compound. In the DD-degrading bacterium strain RW1, the meta-cleavage compound hydrolase, DxnB, was supposed to be involved in the DD degradation system (2, 4), but direct proof of the hydrolysis of meta-cleavage compounds of THDE has not been reported (9). On the other hand, Pfeifer et al. reported that 2,3-dihydroxydiphenyl ether was converted to phenol and 2-pyrone-6-carboxylic acid by the action of purified extradiol dioxygenase from Pseudomonas cepacia strain Et4 (26). If a similar reaction occurred in the case of chlorinated THDE by the action of DbfB or CarBaBb, formation of the corresponding CCs instead of phenol would be expected. However, we could not detect CCs in the biotransformation experiments using DbfA1A2BC and CarAaAcAdBaBbC. In strains DBF63 and CA10, it also remains unclear what type of compounds are formed after meta-cleavage and which hydrolase is involved in these reactions. There exists a possibility that an esterase is involved in this hydrolysis reaction.
Considering the above results, the influence of the chlorine substitution pattern of mono- to triCDF/Ds on the formation of corresponding CSAs and CCs may not be dependent on the substrate specificity of the initial angular dioxygenases but on that of the subsequently acting enzyme, especially hydrolases. This result is consistent with the report that BphD is a key determinant in the aerobic microbial degradation of polychlorinated biphenyls (31, 32).
In conclusion, our study suggests the sites for the attack of angular dioxygenases and the transformation capabilities of subsequently acting enzymes toward some CDF/Ds using two different types of dioxin degraders. Although it remains to be investigated which enzyme catalyzes hydrolysis of the meta-cleavage compounds of CDF/Ds in these bacteria, the results of this study add useful knowledge to the bacterial degradation of dioxins. For a complete understanding of the influence of chlorine substituents on the ability of angular dioxygenases to oxidize chlorinated dioxins, transformation of more highly CDF/Ds and determination of the structure of these enzymes will be required. Accumulation of such information will make it possible to perform efficient bioremediation of dioxins.
REFERENCES
- 1.Armengaud J, Timmis K N. Molecular characterization of Fdx1, a putidaredoxin-type [2Fe-2S] ferredoxin able to transfer electrons to the dioxin dioxygenase system of Sphingomonas sp. RW1. Eur J Biochem. 1997;247:833–842. doi: 10.1111/j.1432-1033.1997.00833.x. [DOI] [PubMed] [Google Scholar]
- 2.Armengaud J, Happe B, Timmis K N. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. strain RW1: catabolic genes dispersed on the genome. J Bacteriol. 1998;180:3954–3966. doi: 10.1128/jb.180.15.3954-3966.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armengaud J, Timmis K N. The reductase RedA2 of the multicomponent dioxin dioxygenase system of Sphingomonas sp. RW1 is related to class-I cytochrome P450-type reductases. Eur J Biochem. 1998;253:437–444. doi: 10.1046/j.1432-1327.1998.2530437.x. [DOI] [PubMed] [Google Scholar]
- 4.Armengaud J, Timmis K N, Wittich R-M. A functional 4-hydroxysalicylate/hydroxyquinol degradative pathway gene cluster is linked to the initial dibenzo-p-dioxin pathway genes in Sphingomonas sp. strain RW1. J Bacteriol. 1999;181:3452–3461. doi: 10.1128/jb.181.11.3452-3461.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armengaud J, Gaillard J, Timmis K N. A second [2Fe-2S] ferredoxin from Sphingomonas sp. strain RW1 can function as an electron donor for the dioxin dioxygenase. J Bacteriol. 2000;182:2238–2244. doi: 10.1128/jb.182.8.2238-2244.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnett C M, Parales J V, Haddock J D. Influence of chlorine substituents on rates of oxidation of chlorinated biphenyls by the biphenyl dioxygenase of Burkholderia sp. strain LB400. Appl Environ Microbiol. 2000;66:2928–2933. doi: 10.1128/aem.66.7.2928-2933.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bressler D C, Fedorak P M. Bacterial metabolism of fluorene, dibenzofuran, dibenzothiophene, and carbazole. Can J Microbiol. 2000;46:397–409. [PubMed] [Google Scholar]
- 8.Bünz P V, Cook A M. Dibenzofuran 4,4a-dioxygenase from Sphingomonas sp. strain RW1: angular dioxygenation by a three-component enzyme system. J Bacteriol. 1993;175:6467–6475. doi: 10.1128/jb.175.20.6467-6475.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bünz P V, Falchetto R, Cook A M. Purification of two isofunctional hydrolases (EC 3.7.1.8) in the degradative pathway for dibenzofuran in Sphingomonas sp. strain RW1. Biodegradation. 1993;4:171–178. doi: 10.1007/BF00695119. [DOI] [PubMed] [Google Scholar]
- 10.Habe, H., K. Ide, M. Yotsumoto, H. Tsuji, H. Hirano, J. Widada, T. Yoshida, H. Nojiri, and T. Omori. Preliminary examinations for applying a carbazole-degrader Pseudomonas sp. strain CA10 to dioxin-contaminated soil remediation. Appl. Microbiol. Biotechnol., in press. [DOI] [PubMed]
- 11.Haddock J D, Horton J R, Gibson D T. Dihydroxylation and dechlorination of chlorinated biphenyls by purified biphenyl 2,3-dioxygenase from Pseudomonas sp. strain LB400. J Bacteriol. 1995;177:20–26. doi: 10.1128/jb.177.1.20-26.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halden R U, Halden B G, Dwyer D F. Removal of dibenzofuran, dibenzo-p-dioxin, and 2-chlorodibenzo-p-dioxin from soils inoculated with Sphingomonas sp. strain RW1. Appl Environ Microbiol. 1999;65:2246–2249. doi: 10.1128/aem.65.5.2246-2249.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halden R U, Dwyer D F. Biodegradation of dioxin-related compounds: a review. Bioremediation J. 1997;1:11–25. [Google Scholar]
- 14.Harms H, Wilkes H, Sinnwell V, Wittich R-M, Figge K, Francke W, Fortnagel P. Transformation of 3-chlorodibenzofuran by Pseudomonas sp. HH69. FEMS Microbiol Lett. 1991;81:25–30. doi: 10.1016/0378-1097(91)90465-m. [DOI] [PubMed] [Google Scholar]
- 15.Kasuga K, Nojiri H, Yamane H, Kodama T, Omori T. Cloning and characterization of the genes involved in the degradation of dibenzofuran by Terrabacter sp. strain DBF63. J Ferment Bioeng. 1997;84:387–399. [Google Scholar]
- 16.Kasuga K, Habe H, Chung J-S, Yoshida T, Nojiri H, Yamane H, Omori T. Isolation and characterization of the genes encoding a novel oxygenase component of angular dioxygenase from the gram-positive dibenzofuran-degrader Terrabacter sp. strain DBF63. Biochem Biophys Res Commun. 2001;283:195–204. doi: 10.1006/bbrc.2001.4763. [DOI] [PubMed] [Google Scholar]
- 17.Keim T, Francke W, Schmidt S, Fortnagel P. Catabolism of 2,7-dichloro- and 2,4,8-trichlorodibenzofuran by Sphingomonas sp. strain RW1. J Ind Microbiol Biotechnol. 1999;23:359–363. doi: 10.1038/sj.jim.2900739. [DOI] [PubMed] [Google Scholar]
- 18.Kohler H-P E, Schmid A, van der Maarel M. Metabolism of 2,2′-dihydroxybiphenyl by Pseudomonas sp. strain HBP1: production and consumption of 2,2′,3-trihydroxybiphenyl. J Bacteriol. 1993;175:1621–1628. doi: 10.1128/jb.175.6.1621-1628.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKay D B, Seeger M, Zielinski M, Hofer B, Timmis K N. Heterologous expression of biphenyl dioxygenase-encoding genes from a gram-positive broad-spectrum polychlorinated biphenyl degrader and characterization of chlorobiphenyl oxidation by the gene products. J Bacteriol. 1997;179:1924–1930. doi: 10.1128/jb.179.6.1924-1930.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Megharaj M, Wittich R-M, Blasco R, Pieper D H. Superior survival and degradation of dibenzo-p-dioxin and dibenzofuran in soil by soil-adapted Sphingomonas sp. strain RW1. Appl Microbiol Biotechnol. 1997;48:109–114. [Google Scholar]
- 21.Monna L, Omori T, Kodama T. Microbial degradation of dibenzofuran, fluorene, and dibenzo-p-dioxin by Staphylococcus auriculans DBF63. Appl Environ Microbiol. 1993;59:285–289. doi: 10.1128/aem.59.1.285-289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nojiri H, Nam J-W, Kosaka M, Morii K, Takemura T, Furihata K, Yamane H, Omori T. Diverse oxygenations catalyzed by carbazole 1,9a-dioxygenase from Pseudomonas sp. strain CA10. J Bacteriol. 1999;181:3105–3113. doi: 10.1128/jb.181.10.3105-3113.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Omori T, Monna L, Saiki Y, Kodama T. Desulfurization of dibenzothiophene by Corynebacterium sp. strain SY1. Appl Environ Microbiol. 1992;58:911–915. doi: 10.1128/aem.58.3.911-915.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ouchiyama N, Zhang Y, Omori T, Kodama T. Biodegradation of carbazole by Pseudomonas spp. CA06 and CA10. Biosci Biotechnol Biochem. 1993;57:455–460. [Google Scholar]
- 25.Parsons J R, de Bruijne J A, Weiland A R. Biodegradation pathway of 2-chlorodibenzo-p-dioxin and 2-chlorodibenzofuran in the biphenyl-utilising strain JB1. Chemosphere. 1998;37:1915–1922. doi: 10.1016/s0045-6535(98)00258-6. [DOI] [PubMed] [Google Scholar]
- 26.Pfeifer F, Trüper H G, Klein J, Schacht S. Degradation of diphenylether by Pseudomonas cepacia Et4: enzymatic release of phenol from 2,3-dihydroxydiphenylether. Arch Microbiol. 1993;159:323–329. doi: 10.1007/BF00290914. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Sato S, Nam J-W, Kasuga K, Nojiri H, Yamane H, Omori T. Identification and characterization of genes encoding carbazole 1,9a-dioxygenase in Pseudomonas sp. strain CA10. J Bacteriol. 1997;179:4850–4858. doi: 10.1128/jb.179.15.4850-4858.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato S, Ouchiyama N, Kimura T, Nojiri H, Yamane H, Omori T. Cloning of genes involved in carbazole degradation of Pseudomonas sp. strain CA10: nucleotide sequences of genes and characterization of meta-cleavage enzymes and hydrolase. J Bacteriol. 1997;179:4841–4849. doi: 10.1128/jb.179.15.4841-4849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schreiner G, Wiedmann T, Schimmel H, Ballschmiter K. Influence of the substitution pattern on the microbial degradation of mono- to tetrachlorinated dibenzo-p-dioxins and dibenzofurans. Chemosphere. 1997;34:1315–1331. [Google Scholar]
- 31.Seah S Y K, Labbé G, Kaschabek S R, Reifenrath F, Reineke W, Eltis L D. Comparative specificities of two evolutionarily divergent hydrolases involved in microbial degradation of polychlorinated biphenyls. J Bacteriol. 2001;183:1511–1516. doi: 10.1128/JB.183.5.1511-1516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seah S Y K, Labbé G, Nerdinger S, Johnson M R, Snieckus V, Eltis L D. Identification of a serine hydrolase as a key determinant in the microbial degradation of polychlorinated biphenyls. J Biol Chem. 2000;275:15701–15708. doi: 10.1074/jbc.275.21.15701. [DOI] [PubMed] [Google Scholar]
- 33.Seeger M, Timmis K N, Hofer B. Conversion of chlorobiphenyls into phenylhexadienoates and benzoates by the enzymes of the upper pathway for polychlorobiphenyl degradation encoded by the bph locus of Pseudomonas sp. strain LB400. Appl Environ Microbiol. 1995;61:2654–2658. doi: 10.1128/aem.61.7.2654-2658.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seeger M, Zielinski M, Timmis K N, Hofer B. Regiospecificity of dioxygenation of di- to pentachlorobiphenyls and their degradation to chlorobenzoates by the bph-encoded catabolic pathway of Burkholderia sp. strain LB400. Appl Environ Microbiol. 1999;65:3614–3621. doi: 10.1128/aem.65.8.3614-3621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkes H, Wittich R-M, Timmis K N, Fortnagel P, Francke W. Degradation of chlorinated dibenzofurans and dibenzo-p-dioxins by Sphingomonas sp. strain RW1. Appl Environ Microbiol. 1996;62:367–371. doi: 10.1128/aem.62.2.367-371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wittich R-M. Degradation of dioxin-like compounds by microorganisms. Appl Microbiol Biotechnol. 1998;49:489–499. doi: 10.1007/s002530051203. [DOI] [PubMed] [Google Scholar]
- 37.Wittich R-M, Strömpl C, Moore E R B, Blasco R, Timmis K N. Interaction of Sphingomonas and Pseudomonas strains in the degradation of chlorinated dibenzofurans. J Ind Microbiol Biotechnol. 1999;23:353–358. doi: 10.1038/sj.jim.2900740. [DOI] [PubMed] [Google Scholar]
- 38.Wittich R-M, Wilkes H, Sinnwell V, Francke W, Fortnagel P. Metabolism of dibenzo-p-dioxin by Sphingomonas sp. strain RW1. Appl Environ Microbiol. 1992;58:1005–1010. doi: 10.1128/aem.58.3.1005-1010.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yabuuchi E, Yamamoto H, Terakubo S, Okamura N, Naka T, Fujiwara N, Kobayashi K, Kosano Y, Hiraishi A. Proposal of Sphingomonas wittichii sp. nov., for the strain RW1 (T) known as a dibenzo-p-dioxin metabolizer. Int J Syst Evol Microbiol. 2001;51:281–292. doi: 10.1099/00207713-51-2-281. [DOI] [PubMed] [Google Scholar]
- 40.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]