Abstract
Objectives
Despite the quick implementation of infection prevention and control procedures and the use of personal protective equipment within healthcare facilities, many cases of nosocomial COVID-19 transmission have been reported. We aimed to estimate the frequency and impact of healthcare-associated COVID-19 (HA-COVID-19) and evaluate the contribution of whole-genome sequencing (WGS) in cluster investigation.
Methods
We estimated the frequency and mortality of HA-COVID-19 infections from September 1 to November 30, 2020, with a focus on the evolution of hospitalized community-associated COVID-19 (CA-COVID-19) cases and cases detected among healthcare workers (HCWs) within the Sorbonne University Hospital Group (Paris, France). We thoroughly examined 12 clusters through epidemiological investigations and WGS.
Results
Overall, 209 cases of HA-COVID-19 were reported. Evolution of HA-COVID-19 incidence closely correlated with the incidence of CA-COVID-19 and COVID-19 among HCWs. During the study period, 13.9 % of hospitalized patients with COVID-19 were infected in the hospital and the 30-day mortality rate of HA-COVID-19 was 31.5 %. Nosocomial transmission of SARS-CoV-2 led to clusters involving both patients and HCWs. WGS allowed the exclusion of one-third of cases initially assigned to a cluster.
Conclusions
WGS analysis combined with comprehensive epidemiological investigations is essential to understand transmission routes and adapt the IPC response to protect both patients and HCWs.
Keywords: Healthcare-Associated infection, Severe acute respiratory syndrome coronavirus 2, Epidemiology, Infection control, Whole genome sequencing
1. Introduction
In December 2019, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was identified in the city of Wuhan, China, following a pneumonia outbreak of unknown etiology (Zhu et al., 2020). Within two months, this novel coronavirus spread throughout the world and the World Health Organization (WHO) declared the associated illness, known now as COVID-19 (coronavirus disease 2019), a global pandemic on March 11, 2020. Three SARS-CoV-2 infected patients were reported on January 24 in France, the first confirmed cases in Europe (Bernard Stoecklin et al., 2020). Since then, COVID-19 has caused five epidemic waves in France, leading to more than 18,000,000 confirmed cases, 128,000 deaths, and major socio-economic issues (WHO Coronavirus (COVID-19) Dashboard, 2021).
The COVID-19 pandemic has placed a heavy burden on hospitals and has posed unprecedented challenges for infection prevention and control (IPC). Despite the early implementation of IPC strategies and the use of personal protective equipment (PPE), healthcare-associated COVID-19 (HA-COVID-19) has been a cause of public health concern since the beginning of the pandemic. The occurrence of nosocomial transmission of COVID-19, affecting both healthcare workers (HCWs) and patients, has been well described (Heinzerling et al., 2020, Meredith et al., 2020, Paltansing et al., 2021). Frontline healthcare workers have a higher risk of infection and hospitalization due to COVID-19 than the general community, affecting both their physical and mental health (Nguyen et al., 2020, Shah et al., 2020, Xiong et al., 2021). In addition, recent studies suggest that, due to their relative frailty and older age, inpatients with HA-COVID-19 have a higher mortality rate than those with community-associated (CA-COVID-19) infections (Rickman et al., 2021, Ponsford et al., 2021).
Whole-genome sequencing (WGS) has proven to be an efficient tool to track SARS-CoV-2 nosocomial outbreaks (Løvestad et al., 2021, Lucey et al., 2021, Borges et al., 2021). Combined with comprehensive epidemiological investigations, WGS of SARS-CoV-2 strains from HCWs and patients has provided a better understanding of transmission patterns.
The primary objective of this observational study was to estimate the frequency and mortality of HA-COVID-19 infections over a three-month period within the Sorbonne University Hospital Group (GH-SU, Paris, France). The secondary objective was to investigate clusters of HA-COVID-19 by WGS to elucidate the transmission routes of SARS-CoV-2.
2. Methods
We followed the ORION statement guidelines for transparent reporting of outbreaks and nosocomial infection (Stone et al., 2007).
2.1. Institutional settings
The study was carried out at GH-SU, a 2,448-bed, university-affiliated public hospital, composed of seven different healthcare facilities (Saint-Antoine, Pitié-Salpêtrière, Charles Foix, La Roche-Guyon, Tenon, Armand-Trousseau and Rothschild Hospitals) located in the eastern part of Paris, France. In total, 18,770 HCWs work in the group, including 12,340 nurses and nurse-assistants, 2930 senior medical doctors, 900 residents, and 2600 administrative/technical workers. In 2020, the group recorded 131,939 admissions for a total of 1,109,986 hospitalization-days and 179,184 visits to the emergency room.
2.2. Study design
We performed a retrospective analysis of all laboratory-confirmed HA-COVID-19 cases between September 1 and November 30, 2020. As part of this study, CA-COVID-19 inpatients, and cases of COVID-19 occurring in HCWs, were also recorded. A HA-COVID-19 case was defined as a patient admitted without symptoms suggestive of COVID-19 who had a SARS-CoV-2 positive PCR test at least three days (possible HA case) or 14 days (definite HA case) after admission. A case was considered to be symptomatic if symptoms suggestive of COVID-19 (fever, cough, sore throat, headache, myalgia, anosmia, ageusia, and diarrhea) were present. A cluster was defined by two or more cases of HA-COVID-19 in two patients (or in one patient and one HCW) from the same ward within 14 days. During the study period, visits to patients were limited to those for compassionate purposes. Wearing surgical masks was mandatory for all HCWs, except during aerosol-generating treatments and procedures (endotracheal intubation / suction, bronchoscopy, non-invasive ventilation, aerosol therapy, etc.) where FFP2 masks were used. Contact and droplet precautions were required when managing suspected or confirmed cases of COVID-19.
2.3. Contact tracing and epidemiological data
Socio-demographic data, as well as administrative data related to the hospital stay, were extracted from the hospital information system (ORBIS, Dedalus, Healthcare Systems Group). The infection control staff investigated all HA-COVID-19 cases and COVID-19 cases among HCWs and identified contacts. A contact was defined as an HCW or patient who spent at least 15 min face to face at a distance <2 m with a COVID-19 case (i.e., starting 48 h before symptoms onset or seven days before a positive PCR if asymptomatic) without any mask or patients who shared a multi-bed room. All contacts were immediately tested for SARS-CoV-2 and testing was repeated seven days later if the first test was negative. In the event of a cluster, all patients and HCWs from the same ward were systematically tested.
Data concerning CA-COVID-19 cases hospitalized in the hospital group during the study period were extracted from the French National Uniform Hospital Discharge Database (PMSI).
2.4. Statistical analysis
The times of occurrence of an event (e.g., time between admission and HA-COVID-19 diagnosis or time between HA-COVID-19 diagnosis and death) were considered right-censored variables. They were modeled according to the Kaplan-Meier estimator and, when possible, are described according to the median and its 95 % confidence interval (95 %CI). Incidence is expressed as the number of weekly cases and as the ratio per 100 hospitalized patients. The dates of diagnosis of both HA-COVID-19 cases and cases occurring among HCWs were based on the first positive PCR test, whereas the dates of CA-COVID-19 diagnosis were determined by the hospital admission date. The peak date of the incidence curve was estimated using a bootstrap resampling method. The growth rate was assessed by fitting an exponential model (log(y) = r * t + b) to the incidence data. Two models were fitted on either side of the epidemic peak: the doubling and halving times were assessed. All statistical analyses were performed using R (R Foundation for Statistical Computing, Vienna, Austria) v4.0.5, in particular, the ‘incidence’ package (Kamvar et al., 2019).
2.5. Whole-genome sequencing
Complete genome sequencing was performed on 114 samples using the Illumina RNA Prep with Enrichment kit (Ref: 20040537, Illumina) and Respiratory Virus Oligos Panel V2 (ref: 20044311, Illumina) on a NovaSeq instrument (2 × 100 bp). Consensus sequences were generated by mapping the reads using Bowtie v.2.3.4.3 (Langmead and Salzberg, 2012) against the Wuhan-Hu-1 reference sequence (NC_045512.2) and BCFtools v1.10.2 (Danecek et al., 2021) for variant calling. The 127 SARS-CoV-2 genomes have been deposited in the GISAID database. Nextstrain classification (Hadfield et al., 2018) was used to assign a lineage to each SARS-CoV-2 strain.
2.6. Phylogenetic analysis
SARS-CoV-2 genomes were aligned using mafft v7.450 (Katoh and Standley, 2013) with 586 European sequences from January to December 2020 extracted from the GISAID database. The resulting alignment was then fed into IQTREE v2.0 using a GTR+G nucleotide substitution model, 1000 bootstrap replicates, and the Wuhan-Hu-1 reference sequence (NC_045512.2) as an outgroup to produce a maximum likelihood (ML) tree. The corresponding tree was annotated using the iTOL v6 online tool (Letunic and Bork, 2021). Monophyletic clades in the ML tree within which the maximum pairwise distances between sequences did not exceed 1e-4 substitutions per base (i.e., 3 substitutions per genome) were considered to be clonally related. Pairwise single nucleotide substitution (SNP) matrices were obtained using pairsnp v0.2.0 (Tonkin-Hill, 2021).
2.7. Ethical approval
The study was declared to the Data Protection Commission (Number 20210218113547). Data were recorded as part of the hospital’s routine for outbreak investigation.
3. Results
3.1. Case detection and epidemiological investigation
From September 1 to November 30, 2020, 209 incident cases of HA-COVID-19 (71 possible and 138 definite) were detected. The median age of HA-COVID-19 cases was 79.6 years [95 %CI 63.5–87.4] and the sex ratio (M/F) 0.74. Among the 209 HA-COVID-19 cases, 138 (66.0 %) occurred in adults with short hospital stays, 65 (31.1 %) in patients from medium and long-term care facilities, and 6 (2.9 %) in pediatric patients. The median time to a HA-COVID-19 diagnosis from hospital admission was 23 days [95 %CI 20–27]. Among the 209 HA-COVID-19 cases, 127 (60.8 %) developed typical clinical signs of COVID-19 before or in the days following a positive PCR test. Most (n = 187, 85.2 %) cases were part of a cluster. Thirty-one clusters were identified and the median number of patients/HCWs within a cluster was 4 [95 %CI 2.5–6.0]. Forty-four (31.5 %) patients died (all-cause mortality) within 30 days after their first positive PCR test and death was attributable to COVID-19 infection for 90 % according to the infection-control unit practitioner and the medical doctor in charge of patients. The median time to death was 83 days [95 %CI 50 – NA] from diagnosis.
3.2. Epidemiological curve
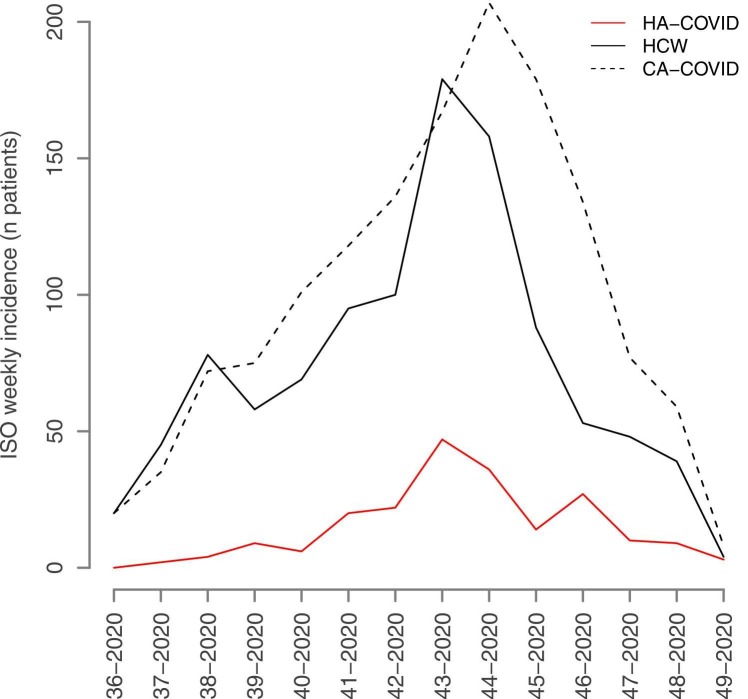
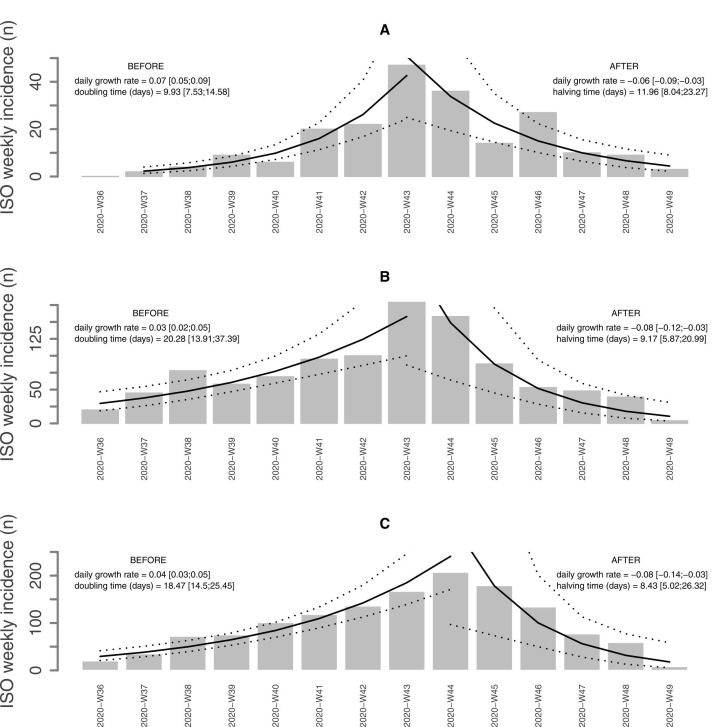
Between September 1 and November 30, 2020, 209 incident cases of HA-COVID-19 and 1034 incident cases of COVID-19 infection among HCWs were detected, whereas 1388 patients with CA-COVID-19 infection were hospitalized in GH-SU. During this period, the weekly incidence of COVID-19 cases in the city of Paris increased from 180 cases to a maximum of 600 cases per 100.000 population in mid-October (Supplementary figure 1). The median weekly incidence of HA-COVID-19 was 9.5 cases [95 %CI 4.5–21.5], corresponding to 3.47 [95 %CI 2.97–7.22] for 100 CA-COVID-19, and 0.2 [95 %CI 0.09–0.40] for 100 hospitalized patients, respectively. The median weekly incidence of COVID-19 among HCWs was 63.50 cases [95 %CI 45.75–93.25], 34.03 [95 %CI 17.33–36.73] for 100 CA-COVID-19 hospitalized patients and 1.21 [95 %CI 0.91–1.78] for 100 hospitalized patients. The median weekly incidence of CA-COVID-19 hospitalization was 89 cases [95 %CI 62.25–135.50] and 1.74 [95 %CI 1.27–2.79] for 100 hospitalized patients. The kinetics of the three epidemiological curves clearly overlapped, the magnitude of the epidemic curve of HA-COVID-19 cases being lower ( Fig. 1). A graphical representation of the model of the epidemic growth rate is shown in Fig. 2. The epidemic peak for HA-COVID-19 cases (Fig. 2A) and cases occurring among HCWs (Fig. 2B) was observed on October 19, 2020 (week 43). The peak for CA-COVID-19 cases (Fig. 2C) was shifted to October 26, 2020 (week 44). The growth rate in the ascending phase was two times higher for HA-COVID-19 cases (0.07 [0.05 – 0.09]) than those among HCWs or CA-COVID-19. In the descending phase, the decrease was slower for HA-COVID-19 cases (−0.06 [−0.09 to −0.03]). There was a strong correlation between the incidence of HA-COVID-19 and that of CA-COVID-19 (rho = 0.89, p < 0.0001), of the same order as the correlation between the incidence of HA-COVID-19 and that of cases among HCWs (rho = 0.77, p = 0.001). The ratio between the weekly incidences of HA-COVID-19 and CA-COVID-19 increased significantly over time. It started at one HA-COVID-19 case for 20 hospitalized CA-COVID-19 patients per week (ratio of 5 %) and ended at 1 in 3 (ratio of 37 %). We observed the same trend for the ratio between the incidence of HA-COVID-19 and that of cases among HCWs.
Fig. 1.
Healthcare-associated COVID-19 weekly incidence. Evolution of the weekly incidence of healthcare-associated COVID-19 cases, community-associated COVID-19 hospitalized patients, and COVID-19 infections among healthcare workers, from ISO week 36 to ISO week 49, 2020. HA-COVID: healthcare-associated COVID-19, CA-COVID: community-acquired COVID-19, HCW: healthcare workers with COVID-19 infection. The X axis represents the time by ISO week from week 36 to week 49 and the Y axis the incidence levels expressed as the number of cases per week.
Fig. 2.
Daily growth rate and doubling/halving time of COVID-19 infections healthcare-associated, community-associated and among HCWs. Estimation of the daily growth rate and doubling or halving time according to weekly incidence levels of healthcare-associated COVID-19 cases (A), community-associated COVID-19 hospitalized patients (B), and COVID-19 infections among healthcare workers (C) during the ascending and descending phases on each side of the incidence peak from ISO week 36 to ISO week 49, 2020. BEFORE: ascending phase before the peak of incidence, AFTER: descending phase after the peak of incidence. The X axis represents the time by ISO week from week 36 to week 49 and the Y axis the incidence levels expressed as the number of cases per week.
3.3. Comparative genome analysis and cluster investigation
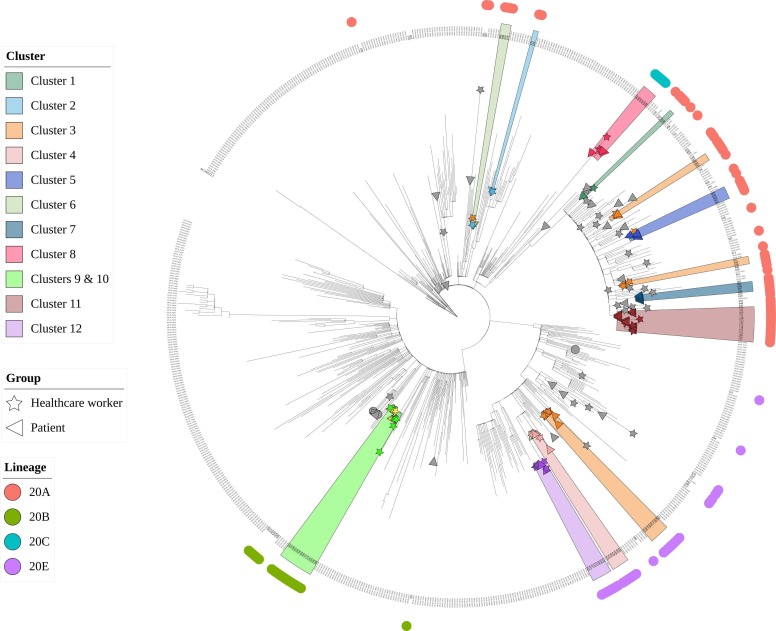
Among the 31 clusters identified by field epidemiology, 12 were investigated using WGS ( Table 1). These clusters included 80 patients and 79 HCWs. In total, 114 full SARS-CoV-2 genomes (58 from patients and 56 from HCWs) could be analyzed. According to the Nextstrain classification, 59 (51.3 %) belonged to clade 20 A, 31 (27 %) to clade 20E, 19 (16.5 %) to clade 20B, and 6 (5.2 %) to clade 20 C. Phylogenetic analysis confirmed that 76/114 (66.7 %) cases initially clustered by field epidemiology exhibited clonal sequences, whereas 35 (30.7 %) cases were excluded from all clusters because of the presence of too many dissimilarities with the other sequences ( Fig. 3). Phylogenetic analysis also showed a clonal link with a cluster other than the initial epidemiological cluster in 3 (2.6 %) cases. A detailed description of the five clusters is presented in the Supplementary Files.
Table 1.
Healthcare-associated COVID-19 clusters investigated by whole-genome sequencing.
| Epidemiological Investigation |
Whole-genome Sequencing |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cluster | Hospital site | Unit | First case | Last case | HA-COVID-19 | CA-COVID-19 | HCW | Total | Sequenced (% among total cases) | Phylogenetically validated (% among total cases) |
| #1 | SAT | Infectious disease | 8-Sep-2020 | 21-Sep-2020 | 1 | 0 | 4 | 5 | 4 (80 %) | 2 (40 %) |
| #2 | SAT | Internal medicine | 10-Sep-2020 | 21-Sep-2020 | 1 | 0 | 5 | 6 | 3 (50 %) | 2 (33 %) |
| #3 | SAT | Hepato-gastroenterology Surgery Surgical intensive care |
15-Sep-2020 | 18-Nov-2020 | 9 | 2 | 19 | 30 | 25 (83 %) | 14 (47 %) split in 3 independent clusters |
| #4 | SAT | Orthopedic | 23-Sep-2020 | 24-Oct-2020 | 3 | 2 | 6 | 11 | 9 (81 %) | 6 (54 %) +1 from cluster #6 |
| #5 | SAT | Rheumatology | 26-Oct-2020 | 13-Nov-2020 | 5 | 7 | 1 | 13 | 7 (54 %) | 4 (31 %) +1 from cluster #3 |
| #6 | SAT | Geriatric | 20-Oct-2020 | 4-Nov-2020 | 4 | 1 | 2 | 7 | 6 (86 %) | 3 (43 %) +1 from cluster #3 |
| #7 | SAT | Psychiatric | 23-Nov-2020 | 2-Dec-2020 | 4 | 1 | 2 | 7 | 4 (57 %) | 4 (57 %) |
| #8 | PSL | Hepato-gastroenterology | 21-Oct-2020 | 27-Oct-2020 | 5 | 1 | 6 | 12 | 9 (75 %) | 6 (50 %) |
| #9 | PSL | Hemodialysis | 19-Nov-2020 | 8-Dec-2020 | 7 | 0 | 6 | 13 | 9 (69 %) | 6 (46 %) |
| #10 | PSL | Internal medicine | 22-Nov-2020 | 27-Nov-2020 | 3 | 0 | 7 | 10 | 8 (80 %) | 7 (70 %) |
| #11 | CFX | Geriatric | 12-Oct-2020 | 8-Dec-2020 | 16 | 1 | 10 | 27 | 18 (67 %) | 14 (52 %) |
| #12 | LRG | Pediatric after-care rehabilitation | 29-Sep-2020 | 19-Nov-2020 | 7 | 0 | 11 | 18 | 12 (67 %) | 8 (44 %) |
SAT: Saint-Antoine Hospital, PSL: Pitié-Salpêtrière Hospital, CFX: Charles Foix Hospital, LRG: La Roche-Guyon Hospital, HA-COVID-19: healthcare-associated COVID-19, CA-COVID-19: community-associated COVID-19, HCW: healthcare worker.
Fig. 3.
Phylogenetic tree. Maximum likelihood phylogenetic tree showing 114 full SARS-CoV-2 genomes sequenced from 58 patients and 56 healthcare workers from the Sorbonne University Hospital Group during the study period and 586 European reference sequences sampled over 2020, extracted from the GISAID database.
4. Discussion
In early 2020, the COVID-19 pandemic struck worldwide as a sudden epidemic wave, strongly affecting every socio-economic aspect of our lives and posing unprecedented challenges, particularly for healthcare systems and hospitals. As expected, HA-COVID-19 became a significant threat for patients (Paltansing et al., 2021, Rickman et al., 2021, Ponsford et al., 2021). Identification of SARS-CoV-2 nosocomial transmission became crucial to understanding dissemination routes within healthcare facilities and providing efficient IPC strategies to protect both patients and HCWs.
We systematically investigated all HA-COVID-19 cases identified in seven healthcare facilities during a three-month period corresponding to the second epidemic wave in France. First, we found that most (85.2 %) HA-COVID-19 cases detected through field epidemiological investigations were part of clusters involving both patients and HCWs. In addition, almost 40 % of cases were detected from asymptomatic patients who were identified by contact tracing. Second, 44 (31.5 %) HA-COVID-19 patients died within 30 days after their positive test, of which 90 % of the deaths were directly attributable to COVID-19. We did not find any significant difference from CA-COVID-19-associated mortality (34.6 %). However, we did not have access to the dates of diagnosis of hospitalized CA-COVID-19 patients. Therefore, the time between diagnosis and hospitalization of CA-COVID-19 patients could have led to an overestimation of mortality and influence this comparison. Nevertheless, our results are consistent with recent studies on HA-COVID-19-associated mortality. Rickman et al. reported 66 HA-COVID-19 cases in a London hospital between March and April 2020, with a mortality rate of 36 % (Rickman et al., 2021). Recently, a large study including more than 100,000 patients with COVID-19 in England, hospitalized between March and August 2020 showed mortality of 41.3 % for HA-COVID-19 28 days after positive PCR testing (Bhattacharya et al., 2021). Such a high 30-day mortality rate can be explained by the fact that COVID-19 mostly affects older patients with co-morbidities and poor health conditions, with an inherent increased risk to develop severe illness.
From an epidemiological perspective, we studied the kinetics of HA-COVID-19 cases relative to those of hospitalized CA-COVID-19 cases and COVID-19 detected among HCWs. We found that HA-COVID-19 cases closely correlated with both the number of CA-COVID-19 cases and HCWs who tested positive. The risk of HA-COVID-19 transmission appears to increase as the viral circulation in the community increases, especially during resumption of the epidemic. Over the study period, despite the mandatory use of masks and the implementation of IPC procedures, 13.9 % of patients hospitalized in GH-SU with a COVID-19 infection were contaminated in the hospital during their stay. This result is consistent with those of previous studies that assessed the burden of HA-COVID-19. In a large study performed during the first wave in 314 hospitals from the United Kingdom (UK), Read et al. reported that 11.3 % of patients with COVID-19 became infected after hospital admission (Read et al., 2021). Similarly, Carter et al. found a proportion of 12.5 % of HA-COVID-19 cases within 10 hospitals throughout the UK and one hospital in Italy between February and April 2020 (Carter et al., 2020).
Since the very beginning of the pandemic, WGS has been widely used to study the molecular diversity and evolution of SARS-CoV-2 (Rambaut et al., 2020, Korber et al., 2020, Miao et al., 2021). More recently, WGS has been implemented in several hospitals worldwide as part of a HA-COVID-19 tracking strategy to quickly identify and control outbreaks (Meredith et al., 2020, Løvestad et al., 2021, Lucey et al., 2021, Borges et al., 2021). In this study, we sought to retrospectively use WGS on samples from 12 clusters to support epidemiological investigations and elucidate SARS-CoV-2 nosocomial transmission. Two-thirds of HA-COVID-19 cases within an epidemiologically defined cluster were confirmed by WGS to be part of the cluster, whereas a third was excluded. The thorough description of five clusters presented in the supplementary file leads to a better understanding of transmission routes. For example, investigation of cluster 4 in combination with WGS results suggests that a patient with HA-COVID directly transmitted SARS-Co-V2 to several HCWs and indirectly to a patient from a different unit sharing the same HCWs. In this case, we show that the spread of SARS CoV-2 infection can be bi-directional between patients and HCWs. In addition, one HCW excluded by WGS from the cluster he was initially connected to actually shared the same viral strain from another cluster, cluster 5. No epidemiological link could be identified between this HCW and the other cases from cluster 5, but a phylogenetic association between their viral strains was undeniable. Similarly, two independent clusters, clusters 9 and 10, identified in two distinct units shared the same viral strains. Apart from the close geographical localization of the two units, we were unable to provide any epidemiological explanation for this link. Consequently, WGS turned out to be an essential tool in combination with a comprehensive epidemiological investigation to accurately confirm or exclude HA-COVID-19 cases and obtain a better understanding of nosocomial SARS-CoV-2 transmission routes. Moreover, WGS revealed hidden links between individuals from distinct clusters, independently of epidemiological information.
In conclusion, this study shows that the incidence of HA-COVID-19 correlates with that of hospitalized CA-COVID cases and cases among HCWs. HA-COVID-19 is frequently associated with clusters that affect both asymptomatic patients and HCWs, making their control more difficult. WGS allowed us to both exclude a third of suspected HA-COVID-19 cases and reveal hidden links between individuals from the 12 investigated clusters. Overall, these results underline the importance of epidemiological investigations in combination with WGS to track SARS-CoV-2 nosocomial transmission to provide efficient outbreak prevention and control strategies to protect vulnerable patients.
Funding
This work was supported by a grant from the Assistance Publique-Hôpitaux de Paris (FRANCE) and Agence nationale de recherches sur le sida et les hépatites virales-MIE, Maladies infectieuses et émergentes (EMERGEN program), FRANCE (Emergen program).
CRediT authorship contribution statement
Valentin Leducq: Formal analysis, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. Jeanne Couturier: Formal analysis, Investigation, Resources, Writing – original draft. Benjamin Granger: Conceptualization, Methodology, Software, Formal analysis, Investigation, Resources, Writing – original draft. Sarah Jolivet: Investigation, Resources. Laurence Morand-Joubert: Investigation, Resources. Jérôme Robert: Investigation, Resources. Michel Denis: Investigation, Resources. Beatrice Salauze: Investigation, Resources. Valérie Goldstein: Investigation, Resources. Karen Zafilaza: Investigation, Resources. Pierre Rufat: Investigation, Resources. Anne-Geneviève Marcelin: Conceptualization, Methodology, Project administration, Supervision. Aude Jary: Investigation, Resources, Supervision. Frédéric Barbut: Conceptualization, Methodology, Project administration, Supervision, Writing – original draft.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgments
The authors would like to thank all the HCWs from the Infection Control Units who were involved in the epidemiological investigation of COVID-19 clusters.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.micres.2022.127133.
Appendix A. Supplementary material
Supplementary material
.
Data availability
The authors do not have permission to share data.
References
- Bernard Stoecklin S., Rolland P., Silue Y., Mailles A., Campese C., Simondon A., et al. First cases of coronavirus disease 2019 (COVID-19) in France: surveillance, investigations and control measures, January 2020. Eurosurveillance. 2020:25. doi: 10.2807/1560-7917.ES.2020.25.6.2000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A., Collin S.M., Stimson J., Thelwall S., Nsonwu O., Gerver S., et al. Healthcare-associated COVID-19 in England: a national data linkage study. J. Infect. 2021 doi: 10.1016/j.jinf.2021.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges V., Isidro J., Macedo F., Neves J., Silva L., Paiva M. Nosocomial outbreak of SARS-CoV-2 in a “Non-COVID-19″ hospital ward: virus genome sequencing as a key tool to understand cryptic transmission. Viruses. 2021;13:604. doi: 10.3390/v13040604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter B., Collins J.T., Barlow-Pay F., Rickard F., Bruce E., Verduri A. Nosocomial COVID-19 infection: examining the risk of mortality. The COPE-Nosocomial Study (COVID in Older PEople) J. Hosp. Infect. 2020;106:376–384. doi: 10.1016/j.jhin.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10:giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzerling A., Stuckey M.J., Scheuer T., Xu K., Perkins K.M., Resseger H., et al. Transmission of COVID-19 to health care personnel during exposures to a hospitalized patient — Solano County, California, February 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69:472–476. doi: 10.15585/mmwr.mm6915e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamvar Z.N., Cai J., Pulliam J.R.C., Schumacher J., Jombart T. Epidemic curves made easy using the R package incidence. F1000Res. 2019;8:139. doi: 10.12688/f1000research.18002.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løvestad A.H., Jørgensen S.B., Handal N., Ambur O.H., Aamot H.V. Investigation of intra-hospital SARS-CoV-2 transmission using nanopore whole-genome sequencing. J. Hosp. Infect. 2021;111:107–116. doi: 10.1016/j.jhin.2021.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucey M., Macori G., Mullane N., Sutton-Fitzpatrick U., Gonzalez G., Coughlan S. Whole-genome sequencing to track severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission in nosocomial outbreaks. Clin. Infect. Dis. 2021;72:e727–e735. doi: 10.1093/cid/ciaa1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith L.W., Hamilton W.L., Warne B., Houldcroft C.J., Hosmillo M., Jahun A.S., et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect. Dis. 2020;20:1263–1271. doi: 10.1016/S1473-3099(20)30562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao M., Clercq E.D., Li G. Genetic diversity of SARS-CoV-2 over a one-year period of the COVID-19 pandemic: a global perspective. Biomedicines. 2021;9:412. doi: 10.3390/biomedicines9040412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.H., Drew D.A., Graham M.S., Joshi A.D., Guo C.-G., Ma W., et al. Risk of COVID-19 among front-line health-care workers and the general community: a prospective cohort study. Lancet Public Health. 2020;5:e475–e483. doi: 10.1016/S2468-2667(20)30164-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paltansing S., Sikkema R.S., de Man S.J., Koopmans M.P.G., Oude Munnink B.B., de Man P. Transmission of SARS-CoV-2 among healthcare workers and patients in a teaching hospital in the Netherlands confirmed by whole-genome sequencing. J. Hosp. Infect. 2021;110:178–183. doi: 10.1016/j.jhin.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponsford M.J., Jefferies R., Davies C., Farewell D., Humphreys I.R., Jolles S., et al. Burden of nosocomial COVID-19 in Wales: results from a multicentre retrospective observational study of 2508 hospitalised adults. Thorax. 2021 doi: 10.1136/thoraxjnl-2021-216964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read J.M., Green C.A., Harrison E.M., Docherty A.B., Funk S., Harrison J., et al. Hospital-acquired SARS-CoV-2 infection in the UK’s first COVID-19 pandemic wave. Lancet. 2021;398:1037–1038. doi: 10.1016/S0140-6736(21)01786-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman H.M., Rampling T., Shaw K., Martinez-Garcia G., Hail L., Coen P., et al. Nosocomial transmission of coronavirus disease 2019: a retrospective study of 66 hospital-acquired cases in a London Teaching Hospital. Clin. Infect. Dis. 2021;72:690–693. doi: 10.1093/cid/ciaa816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A.S.V., Wood R., Gribben C., Caldwell D., Bishop J., Weir A., et al. Risk of hospital admission with coronavirus disease 2019 in healthcare workers and their households: nationwide linkage cohort study. BMJ. 2020:m3582. doi: 10.1136/bmj.m3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.P., Cooper B.S., Kibbler C.C., Cookson B.D., Roberts J.A., Medley G.F., et al. The ORION statement: guidelines for transparent reporting of outbreak reports and intervention studies of nosocomial infection. Lancet Infect. Dis. 2007;7:282–288. doi: 10.1016/S1473-3099(07)70082-8. [DOI] [PubMed] [Google Scholar]
- Tonkin-Hill G. pairsnp. 2021.
- WHO Coronavirus (COVID-19) Dashboard 2021. 〈https://covid19.who.int〉 (accessed May 21, 2021).
- Xiong L.-J., Zhong B.-L., Cao X.-J., Xiong H.-G., Huang M., Ding J., et al. Possible posttraumatic stress disorder in Chinese frontline healthcare workers who survived COVID-19 6 months after the COVID-19 outbreak: prevalence, correlates, and symptoms. Transl. Psychiatry. 2021;11:374. doi: 10.1038/s41398-021-01503-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The authors do not have permission to share data.