Abstract
Purpose:
EGFR pathway inhibition may promote anti–programmed cell death protein 1 (PD-1) responses in preclinical models, but how EGFR inhibition affects tumor antigen presentation during anti–PD-1 monotherapy in humans remain unknown. We hypothesized that afatinib, an irreversible EGFR tyrosine kinase inhibitor, would improve outcomes in patients treated with pembrolizumab for recurrent or metastatic head and neck squamous cell carcinoma (HNSCC) by promoting antigen presentation and immune activation in the tumor microenvironment.
Patients and Methods:
The ALPHA study (NCT03695510) was a single-arm, Phase II study with Simon's 2-stage design. Afatinib and pembrolizumab were administered to patients with platinum-refractory, recurrent, or metastatic HNSCC. The primary endpoint was the objective response rate (ORR). The study applied gene expression analysis using a NanoString PanCancer Immune Profiling Panel and next-generation sequencing using FoundationOne CDx.
Results:
From January 2019 to March 2020, the study enrolled 29 eligible patients. Common treatment-related adverse events were skin rash (75.9%), diarrhea (58.6%), and paronychia (44.8%). Twelve patients (41.4%) had an objective partial response to treatment. The median progression-free survival was 4.1 months, and the median overall survival was 8.9 months. In a paired tissue analysis, afatinib–pembrolizumab were found to upregulate genes involved in antigen presentation, immune activation, and natural killer cell–mediated cytotoxicity. Unaltered methylthioadenosine phosphorylase and EGFR amplification may predict the clinical response to the therapy.
Conclusions:
Afatinib may augment pembrolizumab therapy and improve the ORR in patients with HNSCC. Bioinformatics analysis suggested the enhancement of antigen presentation machinery in the tumor microenvironment.
Translational Relevance.
Impaired antigen presentation in the tumor microenvironment is an intrinsic resistance mechanism during immune checkpoint therapy. To overcome this mechanism and minimize chemotherapy-related toxicity, this study combined afatinib with pembrolizumab to treat patients with platinum-refractory head and neck squamous cell carcinoma (HNSCC). The study met the primary endpoint of improving the overall response rate for HNSCC. The paired tissue analysis showed that afatinib, an EGFR tyrosine kinase inhibitor, may enhance antigen presentation, natural killer cell–mediated cytotoxicity, and immune reactions. We also identified unaltered methylthioadenosine phosphorylase levels, EGFR amplification, and high programmed death-ligand 1 expression as possible predictive biomarkers for therapeutic outcomes.
Introduction
Anti–programmed cell death protein 1 (PD-1) immunotherapy, such as pembrolizumab or nivolumab, is effective in patients with head and neck squamous cell carcinoma (HNSCC; refs. 1–3). However, modest response rates have been reported in patients with cancer due to intrinsic resistance to anti–PD-1 monotherapy (4). The loss of IFNγ signaling and impaired antigen presentation are two primary mechanisms underlying this resistance (4). Overcoming this intrinsic resistance using anti–PD-1–based combination therapy represents a critical approach for improving clinical benefits in patients with HNSCC receiving anti–PD-1 therapy.
Successful anti–PD-1 therapy induces the increased infiltration of T cells into the tumor environment (5) in response to an adequate antigen-presenting cell niche (6, 7). Therefore, in situ antigen presentation alone may act as a determining factor for the intrinsic resistance to anti–PD-1 therapy. Current studies have indicated that insufficient IFNγ signaling (8), low MHC complex expression (9), beta-2-microglobulin (B2M) gene mutation (10), and EGFR pathway activation (11, 12) are related to impaired antigen processing and presentation. Treatment strategies that augment antigen presentation could be promising approaches for improving anti–PD-1 treatment efficacy. A combination of chemotherapeutic drugs induced immunogenic cell death and enhanced antigen presentation in a variety of mouse models (13). Recent studies have combined chemotherapy with anti–PD-1/programmed death-ligand 1 (PD-L1) therapy in various cancer types, leading to meaningful improvements in survival (3, 14, 15). However, in patients unable to tolerate chemotherapy-related toxicity, alternative combination therapies remain necessary to overcome intrinsic resistance to anti–PD-1 immunotherapy. For this reason alone, it is important to uncover and elucidate alternative anti–PD-1 combination regimen to overcome the different mechanisms of intrinsic resistance.
EGFR pathway inhibition has been shown to promote antigen presentation and improve immunotherapy efficacy in a preclinical model (16). Lizotte and colleagues (16) performed a drug screening assay using an ovalbumin antigen–specific, H2b-restricted, transgenic CD8+ T-cell in vitro co-culture system. The study found that EGFR tyrosine kinase inhibitors (TKI), especially afatinib, increased IFNγ-induced MHC class I expression in ovalbumin-expressing ID8 tumor cells and enhanced tumor cell lysis by ovalbumin-specific transgenic CD8+ T cells (16). In a syngeneic mouse model using MC38 colon cancer cell lines in C57BL/6J mice, adding afatinib to anti–PD-1 treatment suppressed tumor growth (16). Another study showed that EGFR-TKIs augment anti–PD-1 effectiveness by increasing human leukocyte antigen (HLA) expression and downregulating PD-L1 via the JAK–STAT pathway (11, 12). These studies demonstrated the ability of EGFR inhibition to enhance antigen presentation. In addition, EGFR-TKIs can suppress HNSCC growth. Afatinib, an irreversible EGFR-TKI, improved the objective response rate (ORR) and progression-free survival (PFS) rate in recurrent or metastatic HNSCC (17, 18). These data indicate the ability of EGFR-TKIs to augment antigen presentation and tumor suppression and support the potential efficacy of combination anti–PD-1 and EGFR-TKIs for cancer immunotherapy.
Here, we present the first anti–PD-1 plus EGFR-TKI combination therapy trial for patients with HNSCC.
Patients and Methods
Study approval
This study protocol was approved by the Institutional Review Board of the National Taiwan University Hospital and is registered with ClinicalTrials.gov (NCT03695510). Written informed consents from all enrolled patients were obtained before study treatment. The studies were conducted in accordance with the ethical guideline of Declaration of Helsinki.
Study design
This study was designed as a single-arm, Phase II trial with Simon's 2-stage design. The key eligibility criteria for inclusion in the study were: (i) HNSCC diagnosis; (ii) platinum-refractory, which was defined as tumor progression or recurrence within 6 months after the last dose of platinum-based therapy administered as adjuvant therapy, or disease progression after taking platinum-based therapy for recurrent or metastatic disease; (iii) RECIST 1.1-measurable lesions; (iv) Eastern Cooperative Oncology Group (ECOG) performance score of 0 or 1; (v) acceptable bone marrow, hepatic, and renal functions; and (vi) negative hepatitis B virus surface antigen, negative anti-hepatitis C virus, and negative anti-human immunodeficiency virus. The study treatment protocol was 200-mg pembrolizumab once every three weeks combined with 40-mg afatinib once daily. The study administered afatinib–pembrolizumab every three weeks until disease progression, intolerable toxicity, or patient withdrawal. Treatment beyond progression was allowed if pseudoprogression was suspected. Afatinib dose titration for treatment-related toxicity was allowed, but no dose titration for pembrolizumab was permitted. The trial assessed tumor response every nine weeks during the first 18 weeks and every 12 weeks after that. Tumors were assessed and analyzed using CT or MRI.
Efficacy assessment
The primary endpoint was the best ORR, according to RECIST 1.1 criteria. The secondary endpoints were PFS, overall survival (OS), and duration of response (DoR). PFS was calculated from the day of first dosing to disease progression, intolerable toxicity, or death. Patients who did not have disease progression were censored on the last day of tumor evaluation. OS was calculated from the day of dosing to the day of death. For survivors, the data were censored on the last day of known survival status. For responders, DoR was calculated from the day of partial response to the day of disease progression or death. Patients who did not have disease progression were censored on the last day of tumor evaluation.
mRNA expression analysis
Each eligible patient was subjected to biopsy before treatment initiation. A second post-treatment tumor biopsy was obtained before the fourth treatment cycle. Biopsy samples were fixed in formalin for all downstream analyses. Gene expression was measured using RNA isolated from formalin-fixed paraffin-embedded (FFPE) biopsy tissue using NanoString technology. Total RNA was isolated and purified using a Qiagen RNeasy FFPE Kit (Qiagen), following the manufacturer's instructions. Extracted mRNA was analyzed using an nCounter PanCancer Immune Profiling Panel (NanoString Technologies), as described previously (19). Digital data acquisition via the nCounter Digital Analyzer (NanoString Technologies) was performed by Cold Spring Biotech Corporation, Taipei, Taiwan. nSolver 4.0 Analysis Software (NanoString Technologies) and R 3.5.0 were used for data analysis. Linear normalization of mRNA expression data was generated by nSolver 4.0. In the gene differential expression analysis, an adjusted P value was calculated using the Benjamini–Yekutieli method in nSolver 4.0 software. Gene Set Enrichment Analysis (GSEA) v4.1.0 (Broad Institute) and GSEA Preranked v.7.2 (Broad Institute) were used for enrichment analysis (20). Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) datasets were retrieved from MSigDB v7.4 (https://www.gsea-msigdb.org/gsea/msigdb/). Gene sets with nominal P values <0.05 and FDR q values <0.1 were selected for further analysis. Cytoscape v3.8.2 (National Institute of General Medical Sciences, MD; ref. 21), EnrichmentMap v3.3.2 (University of Toronto, Canada; ref. 22), and StringApp v1.6.0 (University of California, San Francisco, CA, and University of Copenhagen, Denmark; ref. 23) were used for analysis. CIBERSORTx (Stanford University, CA) and LM22 gene signatures for immune cell enumeration were used for immune cell profiling analysis (24).
Comprehensive genomic profiling
Pre-treatment tumor biopsies or archival tumor tissues were used for comprehensive genomic profiling. Biopsy collection after disease progression was performed only with patients' consent. FFPE samples were transported to Foundation Medicine and analyzed using a FoundationOne CDx Panel. The methods applied in the current study for next-generation sequencing-based genomic assays have previously been validated and reported (25). The current assay interrogated 324 genes and the introns of 36 genes known to be involved in gene rearrangements. Copy-number amplification cutoff values for the present study were defined as four copies of ERBB2 and six copies of all other genes. The mutation allele frequency results were provided by Foundation Medicine on request. The results from all patients were summarized and visualized in Microsoft Excel using different color annotations. The results for equivocal amplification, equivocal loss, and subclonal alterations are not presented in the figures included here.
TCGA HNSCC data acquisition and analysis
To confirm the roles played by specific genetic alterations and their impacts on the tumor microenvironment, The Cancer Genome Atlas (TCGA) HNSCC PanCancer Atlas (26) database was analyzed using cBioPortal (https://www.cbioportal.org/). A gene of interest was used to query the database, and differential mRNA expression levels between groups according to gene status (gene alteration vs. wild-type) were obtained from the “mRNA” module of the “Comparison/Survival” tool in cBioPortal. The differential mRNA expression data were then ranked according to the value of the log of the ratio of mRNA differential expression. For enrichment analysis, the ranked mRNA data were analyzed using GSEA Preranked v.7.2 (Broad Institute).
PD-L1 testing
PD-L1 IHC was performed using anti–PD-L1 antibody 22C3 clone (Dako) and the Dako automated platform (Dako) at the Department of Pathology, National Taiwan University Hospital. All PD-L1 scoring was performed by a single pathologist (Y.-L. Huang). The PD-L1 tumor proportion score (TPS) was calculated as the percentage of viable tumor cells showing partial or complete membrane staining at any intensity. The PD-L1 combined positive score (CPS) was calculated as the number of PD-L1–stained cells (tumor cells, lymphocytes, and macrophages) divided by the total number of viable tumor cells, multiplied by 100. At least 100 viable cancer cells were evaluated in each sample (27).
Statistical analysis
This study was performed as a single-arm, Phase II trial, and the primary endpoint was the best ORR based on Simon's 2-stage design. The null hypothesis that the true response rate is 15% will be tested against a one-sided alternative. During the first stage, 13 patients were recruited, and the study would have been stopped if two or fewer responses were observed among these 13 patients. During the second stage, 16 additional patients were recruited, resulting in a total of 29 patients. The null hypothesis was rejected if eight or more responses were observed among all 29 patients. This design yields a type I error rate of 0.05 (one-sided) and a power of 0.9 when the true response rate is 40%. Survival estimates were performed using Kaplan–Meier survival analyses and log-rank Cox proportional analyses. MedCalc Statistical Software version 19.7 (MedCalc Software Ltd.), GraphPad Prism version 9.0.2 (GraphPad Software, LCC), and Microsoft Office 365 were used to perform data analyses and figure generation.
Data availability statement
The data generated in this study are available upon request from the corresponding authors. The mRNA analysis data by Nanostring platform could be obtained in Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/; GEO Accession No. GSE190575).
Results
From January 2019 to March 2020, 29 total patients were enrolled in the study. The cutoff value for data analysis was February 11, 2021. The median follow-up of the study was 20.1 months. Patient characteristics are summarized in Table 1. One patient discontinued treatment for personal reasons, and one patient discontinued therapy due to decreasing functional status.
Table 1.
Patient characteristics.
| Afatinib + pembrolizumab N = 29 | |
|---|---|
| Age, y | |
| Median (range) | 53.4 (26.2–71.1) |
| Sex, n (%) | |
| Male | 27 (93.1) |
| Female | 2 (6.9) |
| ECOG PS | |
| 0 | 2 (6.9) |
| 1 | 27 (93.1) |
| Habits | |
| Alcohol | 20 (69.0) |
| Betel nuts | 19 (65.5) |
| Cigarettes | 23 (79.3) |
| Primary tumor site, n (%) | |
| Oral cavity | 19 (65.5) |
| Oropharynx | 6 (20.7) |
| P16+ | 4 (13.8) |
| P16− | 1 (3.4) |
| NA | 1 (3.4) |
| Hypopharynx | 2 (6.9) |
| Larynx | 2 (6.9) |
| Disease status at enrollment, n (%) | |
| Local recurrence only | 15 (51.7) |
| Local recurrence and metastases | 11 (37.9) |
| Metastases only | 3 (10.3) |
| Metastatic sites | |
| Lung | 11 (37.9) |
| Liver | 5 (17.2) |
| Bone | 4 (13.8) |
| Kidney | 2 (6.9) |
| Types of prior therapy | |
| Surgical tumor resection | 24 (82.8) |
| Radiotherapy | 26 (89.7) |
| Cetuximab | 16 (55.2) |
| Prior lines of therapy in recurrent/metastatic setting, n (%)a | |
| Noneb | 10 (34.5) |
| 1 | 6 (20.7) |
| 2 | 6 (20.7) |
| ≥3 | 7 (24.1) |
| PD-L1 TPS, n (%) | |
| <1 | 8 (27.6) |
| 1–49 | 13 (44.8) |
| ≥50 | 7 (24.1) |
| NA | 1 (3.4) |
| PD-L1 CPS, n (%) | |
| <1 | 3 (10.3) |
| 1–19 | 17 (58.6) |
| ≥20 | 8 (27.6) |
| NA | 1 (3.4) |
| Tumor mutational burden (TMB) | |
| Tested, with results, n (%) | 25 (86.2) |
| TMB (mutations/megabase) | |
| Median (range) | 4 (1–8) |
| 1–5, n (%) | 19 (65.5) |
| 6–10, n (%) | 6 (20.7) |
| >10, n (%) | 0 (0) |
| NA, n (%) | 4 (13.8) |
Abbreviations: CPS, combined positive score; ECOG PS, Eastern Cooperative Oncology Group performance status; NA, not available; PD-L1, programmed death-ligand 1; TPS, tumor proportion score.
aOnly therapies for recurrent/metastatic diseases were counted in this column. Prior induction or adjuvant therapies were not counted in this column.
bPatients with new recurrent/metastatic diseases in 6 months after the last dose of platinum-based therapy in adjuvant therapy or concurrent chemoradiation.
Efficacy
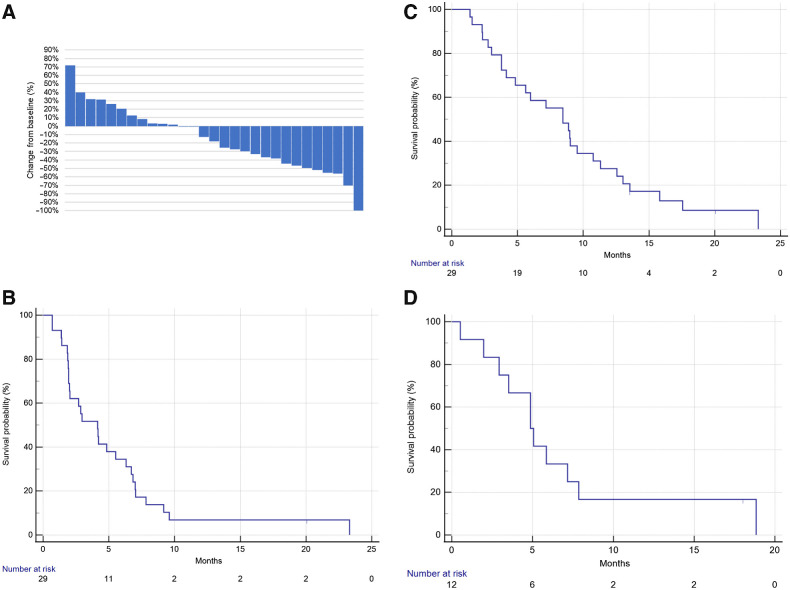
During the first stage, 7 of 13 patients responded to therapy [ORR, 53.8%; 95% confidence interval (CI), 25.1%–80.8%]. The response rate met the pre-specified criteria, and the study progressed to the second stage. Overall, one patient had a complete response, and 11 patients had confirmed partial response (ORR, 12/29, 41.4%; 95% CI, 23.5%–61.1%; Fig. 1A). The ORR result met the primary endpoint of the study. Stable disease during therapy was registered for 7 of 29 patients (24.1%; 95% CI, 10.3%–43.5%). The overall disease control rate was 65.5% (95% CI, 45.7%–82.1%). The median PFS was 4.1 months (95% CI, 1.9–6.3 months; Fig. 1B), and the median OS was 8.4 months (95% CI, 4.1–10.8 months; Fig. 1C). Among the 12 responders, the median DoR was 4.9 months (95% CI, 2.0–7.9 months; Fig. 1D).
Figure 1.
Clinical response to pembrolizumab–afatinib combined therapy in patients with HNSCC. A, Waterfall plot showing the best overall response (n = 29). B, Progression-free survival (n = 29). C, Overall survival (n = 29). D, The duration of response in responders (n = 12).
Adverse events
All patients (100%) experienced at least one treatment-related toxicity event, and 11 (37.9%) patients experienced Grade 3 or higher treatment-related toxicity events (Table 2). Twelve patients (41.4%) experienced afatinib dose reduction due to toxicity. One patient discontinued afatinib due to Grade 2 pneumonitis. One patient expired due to a carotid blow-out. One patient committed suicide. One patient with a history of ischemic stroke and stable atrial fibrillation was found dead at home, and the cause of death was determined to be cardiovascular disease.
Table 2.
Treatment-related adverse events.
| AE, treatment related | Gr. 1 | Gr. 2 | Gr. 3 | Gr. 4 | Gr. 5 |
|---|---|---|---|---|---|
| Skin rash | 17 (59%) | 0 (0%) | 4 (14%) | 0 (0%) | 0 (0%) |
| Diarrhea | 8 (28%) | 7 (24%) | 4 (14%) | 0 (0%) | 0 (0%) |
| Paronychia | 8 (28%) | 4 (18%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Mucositis | 8 (28%) | 3 (10%) | 1 (3%) | 0 (0%) | 0 (0%) |
| Weight loss | 5 (17%) | 5 (17%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Fatigue | 6 (21%) | 2 (7%) | 1 (3%) | 0 (0%) | 0 (0%) |
| Anemia | 4 (14%) | 2 (7%) | 1 (3%) | 0 (0%) | 0 (0%) |
| Anorexia | 5 (17%) | 1 (3%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Creatinine increase | 3 (10%) | 1 (3%) | 1 (3%) | 0 (0%) | 0 (0%) |
| Nausea | 2 (7%) | 3 (10%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Vomiting | 1 (3%) | 3 (10%) | 0 (0%) | 0 (0%) | 0 (0%) |
| ALT increase | 2 (7%) | 1 (3%) | 0 (0%) | 0 (0%) | 0 (0%) |
| AST increase | 2 (7%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| ALP increase | 1 (3%) | 1 (3%) | 0 (0%) | 0 (0%) | 0 (0%) |
| GGT increase | 0 (0%) | 1 (3%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Hypothyroidism | 2 (7%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Bleeding | 0 (0%) | 0 (0%) | 1 (3%) | 0 (0%) | 1 (3%) |
| Pneumonitis | 0 (0%) | 1 (3%) | 0 (0%) | 0 (0%) | 0 (0%) |
Abbreviations: AE, adverse event: ALP, alpha-fetoprotein; ALT, alanine aminotransferase; AST, aspartate aminotransferase; GGT, gamma-glutamyltransferase; Gr., grade.
Biomarker analysis
Paired biopsy mRNA expression analysis
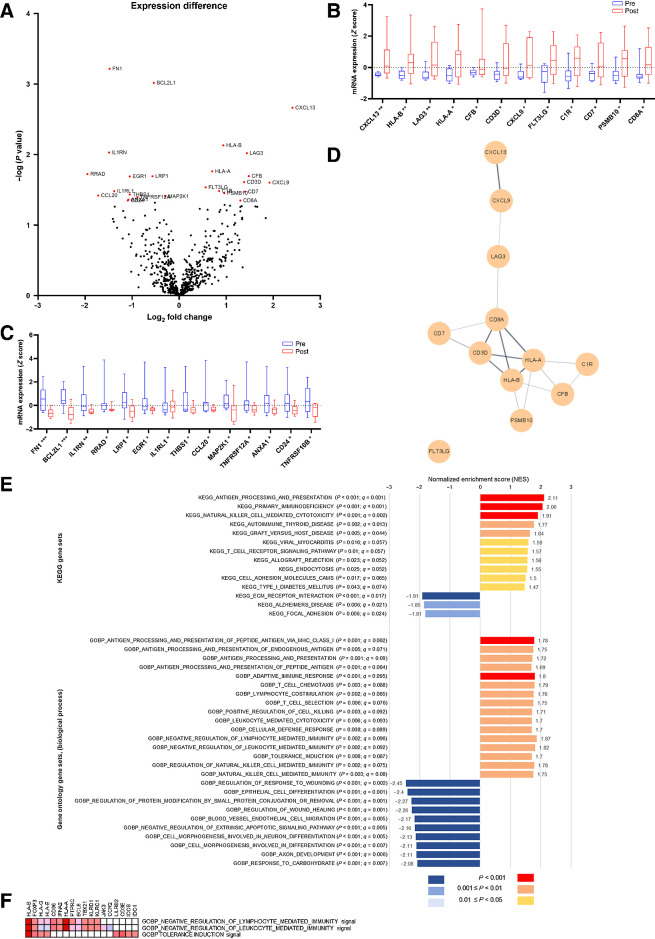
Specimens from 9 patients with adequate paired pre- and post-treatment biopsy tissues were analyzed for mRNA expression (Fig. 2A). The best response recorded for these 9 patients were: Partial response in 3 patients, stable disease in 4 patients, and disease progression in 2 patients. By comparing pre- and post-treatment specimens, 12 genes (CXCL13, CXCL9, CFB, LAG3, CD7, CD3D, CD8A, PSMB10, HLA-B, C1R, HLA-A, and FLT3LG; Fig. 2B) were found to be significantly upregulated after treatment, whereas 14 genes (RRAD, CCL20, IL1RN, FN1, IL1RL1, CD24, ANXA1, EGR1, THBS1, TNFRSF12A, LRP1, BCL2L1, TNFRSF10B, and MAP2K1; Fig. 2C) were significantly downregulated after treatment. Gene network analysis of the 12 upregulated genes using StringApp showed that HLA-A, HLA-B, CD8A, and CD3E were core genes affected by combination therapy (Fig. 2D).
Figure 2.
Paired tissue analysis. A, mRNA differential expression. Red dots represent genes with significant differences in mRNA expression (Benjamini–Yekutieli method, adjusted P < 0.05); B and C, Gene expression differences between paired pre- and post-treatment biopsy samples. *, P < 0.05; **, P < 0.01; ***, P < 0.001. D, Gene network analysis (by StringApp), showing the network of the upregulated genes in the tumor microenvironment. E, Gene set enrichment analysis of paired tissue mRNA samples. In the GO Biological Process analysis, only the top 10 downregulated gene sets are listed. F, Leading-edge analysis of three gene sets with immune cell regulatory functions.
In the KEGG GSEA analysis, 11 gene sets were found to be upregulated (nominal P < 0.05, FDR q < 0.1) after afatinib–pembrolizumab treatment, which were involved in antigen processing and presentation, natural killer (NK) cell–mediated cytotoxicity, endocytosis, autoimmunity, and inflammation (Fig. 2E). In line with the KEGG analysis results, the GO Biological Process GSEA analysis (Fig. 2E) also identified the upregulation of antigen processing and presentation and NK cell–mediated cytotoxicity pathways after treatment. Other upregulated gene sets included genes involved in the adaptive immune response, T-cell chemotaxis, T-cell selection, and leukocyte-mediated toxicity (nominal P < 0.05, FDR q < 0.1). Three gene sets associated with tolerance induction and negative leukocyte and lymphocyte regulatory function were also upregulated after treatment. Leading-edge analysis revealed that FOXP3 was at the leading edge of all three upregulated gene sets (Fig. 2F). Afatinib–pembrolizumab treatment significantly downregulated 194 gene sets (nominal P < 0.05, FDR q < 0.1). In an enrichment map analysis, the majority of downregulated gene sets were found to be closely related. The top 10 downregulated gene sets were associated with cell differentiation, generation, and proliferation (Fig. 2E). Gene sets related to IFNγ and IFNα function did not show significant changes in this analysis [GOBP IFNγ mediated signaling pathway: Normalized enrichment score (NES): 1.42, P = 0.051, q = 0.34; GOBP IFNα production: NES: −0.76, P = 0.85, q = 0.919].
Targeted gene sequence analysis
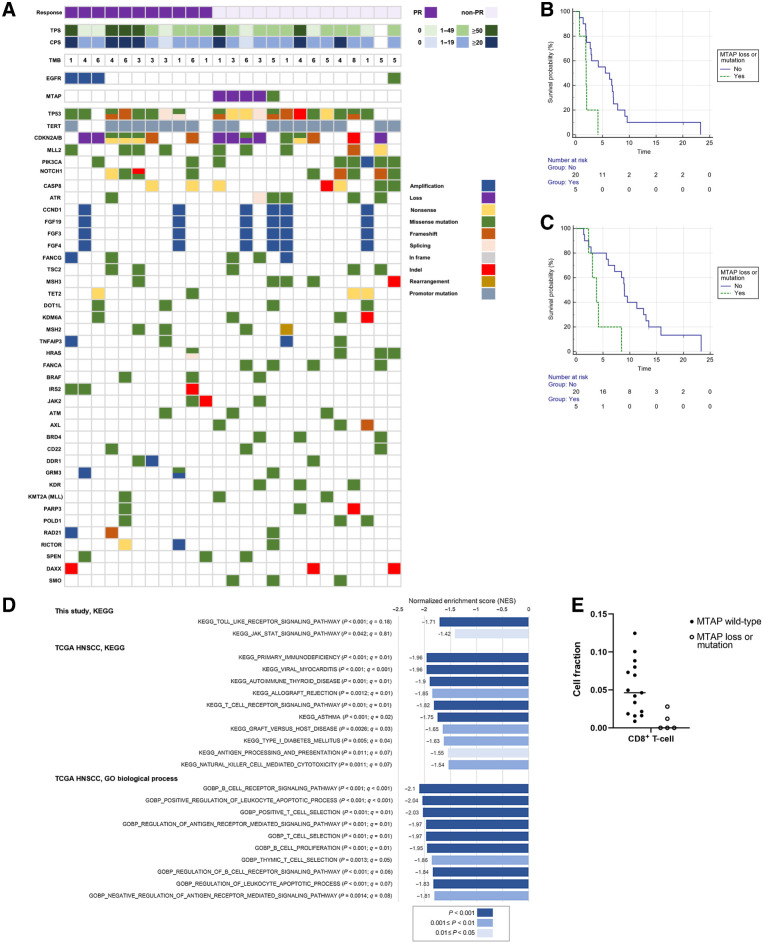
Twenty-five patients (86.2%) had fresh or archival tissues available for analysis (Fig. 3A). No patients had a tumor mutational burden score greater than 10 mutations/megabase. No patients had a known EGFR-driver mutation. EGFR amplification predicted a higher response rate (EGFR amplification, n = 3; ORR, 100%, Fisher exact test, P = 0.07). Methylthioadenosine phosphorylase (MTAP) loss or mutation predicted a lower response rate (MTAP loss or mutation, n = 5; ORR, 0%, Fisher exact test, P = 0.046). Patients with MTAP loss or mutation had a shorter PFS [with loss or mutation, PFS median, 1.9 months (95% CI, 0.7–4.1 months) vs. without loss or mutation, PFS median 5.5 months (95% CI, 2.0–7.0 months); hazard ratio (HR), 4.2; 95% CI, 1.3–13.3; P = 0.014] and shorter OS [with loss or mutation, OS median, 3.8 months (95% CI, 2.3–8.4 months) vs. without loss or mutation, OS median 9.0 months (95% CI, 5.6–13.0 months); HR, 4.2 (95% CI, 1.3–13.4); P = 0.015; Fig. 3B and C].
Figure 3.
Targeted gene mutation analysis. A, Genes were selected if three or more patients had mutations. TMB, tumor mutational burden in mutations/megabase. Survival analysis according to MTAP status. B, Progression-free survival. C, Overall survival. D, Comparing patients with altered MTAP versus unaltered MTAP by gene set enrichment analysis. In the present study and TCGA HNSCC analyses, tumors with altered MTAP had a more suppressed microenvironment. E, CIBERSORTx analysis: In this study, patients with MTAP loss or mutation had a low fraction of CD8+ T cells in the tumor microenvironment (Mann–Whitney U test, P = 0.0037, FDR q = 0.08).
KEGG enrichment analysis comparing MTAP loss/mutation (altered MTAP, n = 5) with MTAP wild-type (unaltered MTAP, n = 15) showed the downregulation of the Toll-like receptor signaling pathway and the JAK–STAT signaling pathway (Fig. 3D). Compared with tumors featuring altered MTAP, tumors with unaltered MTAP had more abundant CD8+ T cells in the microenvironment (Mann–Whitney U test, P = 0.0037, FDR q = 0.08; Fig. 3E).
To confirm the role of MTAP alternation in the HNSCC tumor microenvironment, we analyzed 523 patients/samples from the TCGA HNSCC database. Eighty (15%) patients had MTAP gene alterations, and the majority of these (75 patients, 93.75%) had MTAP deep deletion. KEGG and GO enrichment analyses of 19,874 mRNA genes revealed that immune response-related gene sets were significantly downregulated in patients with HNSCC with MTAP alterations (Fig. 3D).
Post-progression targeted gene sequence analysis
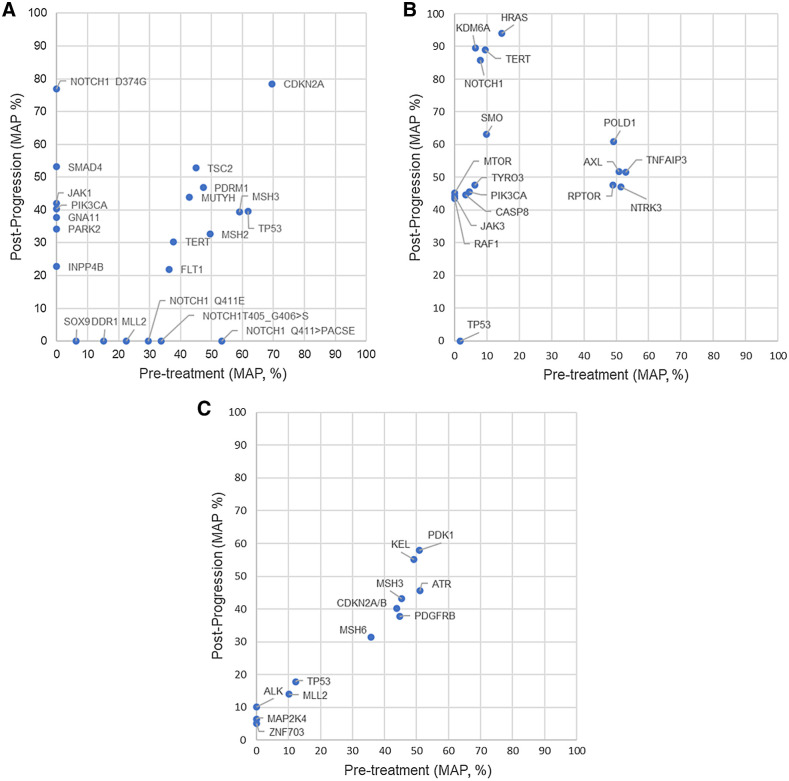
Three patients had paired pre-treatment and post-progression biopsy tissues for targeted gene sequence analyses (Fig. 4A–C). The heterogeneous responses of clones were noted for two patients (Fig. 4A and B). One patient had a new MTAP loss in the post-treatment tissue. Two patients each had a new INPP4B mutation. One JAK1 in one patient and one JAK3 missense mutation in one patient were detected.
Figure 4.
Differences in mutation allele frequency (MAF) between pre-treatment and post-progression specimens. A, Best overall response: partial response. B, Best overall response: partial response. In the post-progression biopsy, a new MTAP loss and a new CDKN2A/B loss were detected. C, The patient had a good initial response to treatment. However, tumor regrowth occurred rapidly before the first imaging assessment. The best overall response of the patient was disease progression.
PD-L1
Twenty-eight (96.6%) patients had adequate tissue samples for PD-L1 analysis (Table 1). For patients with high PD-L1 expression, the ORR was numerically but not statistically higher than for patients with low PD-L1 expression (PD-L1 TPS ≥ 50, ORR: 71% vs. TPS < 50, ORR: 33%, Fisher exact test: P = 0.1; PD-L1 CPS ≥ 20, ORR: 63% vs. CPS < 20, ORR: 35%, Fisher exact test: P = 0.23).
Discussion
This study explored the effects of EGFR-TKI treatment combined with anti–PD-1 therapy in platinum-refractory patients with HNSCC. The study met the primary endpoint of ORR, with manageable toxicities in patients with HNSCC. In the biomarker analyses, we also identified several potential predictive therapeutic biomarkers, including high PD-L1 expression, EGFR amplification, and unaltered MTAP. Further studies in larger sample sizes are warranted to confirm the efficacy of this therapeutic strategy and the roles of these biomarkers in this population.
By using paired human tissue mRNA analyses, we identified that genes involved in antigen processing and presentation signaling pathways, such as HLA-A, HLA-B, and PSMB10, were significantly upregulated in post-treatment specimens. In line with previous reports on anti–PD-1 treatment, we also identified the upregulation of CXCL13, CXCL9, and FLT3LG in post-treatment biopsy tissues. CXCL13 is a marker of T cells that are preferentially reactive to neoantigens and serves as a positive T-cell–intrinsic marker of anti–PD-1 sensitivity (28). CXCL9 is required for successful antitumor responses following PD-1 blockade in an IFNγ-dependent manner (29). FLG3LG has been shown to stimulate dendritic cell (DC) maturation and is correlated with the abundance of intratumoral stimulatory DCs (30). On the other hand, this combination therapy approach resulted in the downregulation of several key suppressive genes involved in immune reactivity. CCL20, a ligand for CCR6, has been shown to chemoattract CCR6+ regulatory T cells, which have higher tumor-suppressive activity compared with other T cells (31). CD24 is a “don't eat me” signal that can inhibit macrophage function via Siglec-10 (32). The downregulation of these genes suggests a less immunosuppressive tumor environment. In the gene set analyses, pathways involving inflammation, NK cell–mediated cytotoxicity, the adaptive immune response, and endocytosis were also found to be upregulated. In addition to immune-related pathways, pathways related to tumor growth were also significantly suppressed. These results provide positive evidence that the afatinib–pembrolizumab therapeutic regimen was involved in reprogramming the tumor environment, possibly through augmented antigen presentation and immune responses, resulting in suppressed tumor growth.
In this study, MTAP was identified as a potential gene for predicting the clinical benefits of anti–PD-1–based immunotherapy. MTAP is an enzyme that catalyzes the breakdown of methylthioadenosine in the cell. The loss of MTAP function may interfere with STAT1 function, inhibiting IFN-mediated gene functions (33). In a report analyzing ipilimumab monotherapy in patients with melanoma, the loss of IFNγ pathway genes, including MTAP, predicted a poor response to ipilimumab (34). In this study, patients with MTAP alterations showed worse ORR and prognosis. Tumors with MTAP alterations contained fewer CD8+ T cells in the microenvironment. In the GSEA, tumors with MTAP loss or mutation showed the downregulation of Toll-like receptor and JAK–STAT signaling pathway components. We also identified one patient with a new MTAP loss in the post-progression biopsy specimen. Our analysis using TCGA HNSCC database (Fig. 3D) also supported the findings that patients with MTAP alterations were associated with suppressed immune reaction factors in the tumor microenvironment. These findings suggested a role for MTAP in HNSCC cancer immunotherapy, and the contributions of MTAP to cancer immunotherapy warrants further research.
Several anti–PD-1/PD-L1–based combination therapeutic approaches have reported positive outcomes in HNSCC trials. Cetuximab, an anti-EGFR monoclonal antibody, was combined with pembrolizumab in patients with HNSCC. The Phase II study also showed an encouraging ORR of 45% (35), highlighting the importance of EGFR inhibition combined with anti–PD-1 therapy in HNSCC. Lenvatinib, a multitarget TKI, was combined with pembrolizumab in HNSCC (36), and the preliminary Phase I/II results showed that the combination treatment resulted in an improved ORR of 46% and an improved PFS of 4.7 months. A confirmatory Phase III trial is ongoing examining this combination approach as a first-line treatment option for patients with HNSCC. Anti–PD-1 combination therapy using the CTLA-4 antibody ipilimumab demonstrated efficacy in melanoma (37), renal cell carcinoma (38), and non–small cell lung cancer (NSCLC; ref. 39). Tremelimumab, another CTLA-4 antibody, was combined with durvalumab, an anti–PD-L1 antibody, to treat HNSCC. However, this PD-L1/CTLA-4 dual blockade combination did not show better efficacy in HNSCC (40). CHECKMATE 651 is a Phase III trial using nivolumab combined with ipilimumab in patients with HNSCC; however, the preliminary results of this study showed no improvement in survival (41). Other studies using oncolytic virus (42), HPV vaccine (43), or histone deacetylase inhibitor (44) in combination with anti–PD-1 treatment have shown modest improvements in ORR for HNSCC. The results of these studies demonstrate the potential of various anti–PD-1 combination therapies for the treatment of patients with HNSCC. With proper biomarker research, a multi-dimensional and personalized approach may be developed to guide the use of combination strategies in patients with HNSCC.
This study showed improved ORR among patients with HNSCC using afatinib combined with pembrolizumab. However, the extent to which afatinib contributes to improved pembrolizumab efficacy remains unclear. In addition, the DoR recorded for this study was shorter than the DoR reported for either pembrolizumab or nivolumab monotherapy (1–3). To clarify the contribution of afatinib and examine the reasons for the shorter DoR, we referenced the results of a pembrolizumab neoadjuvant trial performed by Uppaluri and colleagues (45), which enrolled 36 patients with HNSCC. In this study, eligible patients received one dose of pembrolizumab monotherapy, followed by curative surgery 2 to 3 weeks after administration of neoadjuvant therapy. GSEA of paired, pre- and post-treatment tissue samples (n = 15) showed that pembrolizumab monotherapy induced the upregulation of the following gene sets: IFNα response, IFNγ response, inflammatory response, allograft rejection, TNFα signaling via NF-κB, and IL6–JAK–STAT3 signaling. In the present study, afatinib–pembrolizumab combination therapy upregulated gene sets related to antigen presentation. However, the gene sets related to the IFNγ response and IFNα response were not elevated. The IFNγ signature has been identified as a biomarker for predicting successful pembrolizumab monotherapy in several cancer types, including HNSCC (46). The shorter DoR observed for afatinib–pembrolizumab combination therapy may be due to the insufficient induction of the IFN response. Although cross-trial comparisons should be interpreted with caution, we postulate that afatinib may partially improve pembrolizumab efficacy by augmenting antigen presentation. However, an insufficient induction of an IFN reaction may explain the shorter DoR of the afatinib–pembrolizumab combination.
Anti–PD-1 therapy alone can induce tumor resistance against treatment; however, combined therapy might also induce resistance. Our gene analysis data from post-progression tissues indicated the potential for acquired resistance in response to this combination therapy. The upregulation of LAG3, an immune checkpoint gene, was identified in some post-treatment specimens. The upregulation of other immune checkpoint genes in the tumor microenvironment has been reported in an HNSCC trial using neoadjuvant pembrolizumab (45). Upregulated immune checkpoint genes could represent potential treatment targets in patients with progressive disease following first-line anti–PD-1 therapy. In addition to changes in immune cells, the emergence of new mutations in tumor cells can also modulate immune regulation (10). In the post-progression biopsy samples, further gene alterations associated with immune reactivity were identified (JAK1, JAK3, and INPP4B). These gene alterations may contribute to disease progression. In addition, T-cell regulation and inhibition may contribute to the development of resistance. In the enrichment analysis, three gene sets related to negative leukocyte and lymphocyte regulatory functions had positive enrichment scores. In all three gene sets, FOXP3 was identified at the leading edge, implying that regulatory T cells may play an inhibitory role in response to combination therapy. The underlying mechanism requires further investigation. Tu and colleagues (47) showed that afatinib might suppress T-cell function in the peripheral immune cells of patients with lung cancer and decrease the effects of immune therapy. Our study and Tu's study both demonstrated the possibility that anti–PD-1 combination treatments could simultaneously enhance immune reactivity through some mechanisms while suppressing immune function through other mechanisms. Fine-tuning the balance between immune activation and suppression may represent a key component for extending the survival of patients undergoing anti–PD-1 combination therapy. In conclusion, the increased expression of other immune checkpoints, immunosuppressive pathways, and the emergence of new gene alterations involved in immune reactivity may contribute to acquired resistance in anti–PD-1–based combination therapy approaches.
The potential toxicity of anti–PD-1 and EGFR-TKI combination therapies, especially the incidence of pneumonitis, had been described previously in several lung cancer studies (48, 49), which have described previously a high incidence of treatment-related pneumonitis. In our study, one patient (1/29, 3.4%) experienced a Grade 2 treatment-related pneumonitis event. Differences in the incidence of pneumonitis between these anti–PD-1/PD-L1–EGFR-TKI studies may be due to differences in the cancer types of patients. A meta-analysis showed that patients with lung cancer experience a higher incidence of pneumonitis during anti–PD-1/PD-L1 therapy, whereas the incidence of pneumonitis occurred at similar rates among other cancer types during anti–PD-1/PD-L1 therapy (50), which may account for the increased incidence of pneumonitis reported in studies using anti–PD-1/PD-L1 combined with EGFR-TKIs to treat patients with NSCLC. In the present study, the most common toxicities were skin rash, diarrhea, and paronychia, and the patterns of skin rash reporter were similar to those associated with EGFR-TKI treatment. In addition, 41.2% of patients underwent afatinib dose reductions due to treatment-related toxicity. An ongoing Phase II trial is examining the use of afatinib at 30-mg daily combined with anti–PD-1 for patients with esophageal cancer in Taiwan (BEAR study, NCT04839471), which might contribute to determining the optimal afatinib dosage in combination with anti–PD-1 therapy.
Comparing pre- and post-treatment biopsy results can provide useful information regarding the efficacies and biological effects of study treatment in the tumor. In the present study, we examined 9 (31%) pairs of pre- and post-treatment biopsy specimens for study. Not all patients had paired biopsies for a variety of reasons, including the patient's reluctance to undergo a second biopsy, disease progression, biopsy risks (i.e., too close to major vessels), and tumors that became too small to obtain biopsies. Several strategies could be applied to future studies to improve the successful acquisition of paired biopsy for analyses. A neoadjuvant study followed by surgical tumor resection may provide a better window of opportunity for obtaining pre- and post-treatment samples. For example, a neoadjuvant pembrolizumab trial for patients with HNSCC acquired paired tissue samples from 42% (15/36) of the cohort for analysis (45). A better analysis technique that requires less tissue may also facilitate paired tissue analysis. Single-cell technologies (51) and spatial proteomics (52) can provide high dimensional information using less tissue. Liquid biopsies (53) are a less invasive approach for monitoring change in tumors and may represent a feasible approach for recurrent or metastatic tumor sites that are not easy to biopsy. For patients whose tumors shrink quicker than expected, an earlier biopsy timing may increase the acquisition rate of re-biopsy. For patients who are reluctant for biopsy or experience disease progression, a better patient support may increase the willingness of re-biopsy.
This study was associated with several limitations. The study acquired 9 paired tissue sets for RNA analysis, and the small sample size may limit the generalizability of the RNA analysis results. A larger study may be necessary to confirm these findings. The post-treatment tumors of some good responders were too small for biopsy; therefore, the changes in the tumor microenvironment for these patients could not be determined. The small sample size may overestimate the efficacy of the treatment regimen. In this study, we used bulk RNA from the entire tumor biopsy sample to perform analyses, which did not allow for differential gene expression comparisons between tumor and immune cells or between different immune cell subsets. Approximately 44% of our enrolled patients had previously experienced two or more lines of palliative therapy, and 35% of enrolled patients were primarily resistant to concurrent chemoradiation within six months. These patients typically present with worse prognoses and may have worse survival than platinum-naïve or platinum-sensitive patients with HNSCC, which might have contributed to the lack of significantly prolonged OS in this study. The efficacy of afatinib–pembrolizumab combination therapy for the treatment-naïve or platinum-sensitive patients is worthy of further exploration.
In conclusion, this study presented the efficacy of afatinib–pembrolizumab combination therapy for patients with HNSCC. The paired tissue study showed microenvironment modulatory effects in response to this combination treatment. The biomarker study also showed the potential application of comprehensive genomic profiling for the development of personalized cancer immunotherapy strategies. Further studies should be performed to further explore this personalized approach.
Acknowledgments
We thank all patients who participated in this study and all families who took care of these patients. We thank Boehringer Ingelheim Taiwan for drug supply (afatinib) and partial research funding. We thank Merck Sharp & Dohme Corp (I.A.) LCC, Taiwan Branch for drug supply (pembrolizumab). The biomarker research was partly funded by the Ministry of Education, Taiwan (NTU-107L9014), and the Ministry of Science and Technology, Taiwan (MOST 107-3017-F-002-002). We would like to acknowledge the generous support and brilliant opinions of our friends in cancer research in Taiwan and around the world: Yu-Ming Chang, Yu-Chieh (Peter) Wang, Kevin Chueh, Dennis Chin-Lun Huang, Yan-Ping Liu, Kuei-Fang Wang, Chie-Yu Charles Liao, Kuan-I Lee, Chengju Lulu Wang, Shoung-Hui Tsai, Kung-Chi Kao, Shang-Yun Liu, Yu-Ting Huang, Huei-Ting Lien, Ling-Ying Wei, and Yen-Ling Chiu. Editorial assistance was provided to the authors by Nova Journal Experts.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Authors' Disclosures
H.-F. Kao reports grants and personal fees from Merck Sharp & Dohme and Boehringer–Ingelheim outside the submitted work. R.-L. Hong reports grants and personal fees from Boehringer–Ingelheim and grants, personal fees, and non-financial support from Merck Sharp & Dohme outside the submitted work. No disclosures were reported by the other authors.
Disclaimer
The study sponsor had no role in the design and conduct of the study, collection, management, analysis, and interpretation of the data, and no role in the preparation or approval of the article or the decision to submit the article for publication.
Authors' Contributions
H.-F. Kao: Conceptualization, formal analysis, validation, investigation, methodology, writing–original draft. B.-C. Liao: Investigation. Y.L. Huang: Formal analysis, validation, investigation. H.-C. Huang: Investigation. C.-N. Chen: Investigation. T.-C. Chen: Investigation. Y.-J. Hong: Investigation. C.-Y. Chan: Investigation. J.-S. Chia: Conceptualization, supervision, writing–review and editing. R.-L. Hong: Conceptualization, supervision, writing–review and editing.
References
- 1. Ferris RL, Blumenschein G Jr, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med 2016;375:1856–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cohen EEW, Soulières D, Le Tourneau C, Dinis J, Licitra L, Ahn MJ, et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): a randomized, open-label, phase 3 study. Lancet 2019;393:156–67. [DOI] [PubMed] [Google Scholar]
- 3. Burtness B, Harrington KJ, Greil R, Soulières D, Tahara M, de Castro G Jr, et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet 2019;394:1915–28. [DOI] [PubMed] [Google Scholar]
- 4. Kalbasi A, Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat Rev Immunol 2020;20:25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jansen CS, Prokhnevska N, Master VA, Sanda MG, Carlisle JW, Bilen MA, et al. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature 2019;576:465–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013;39:1–10. [DOI] [PubMed] [Google Scholar]
- 8. Basham TY, Merigan TC. Recombinant interferon-gamma increases HLA-DR synthesis and expression. J Immunol 1983;130:1492–4. [PubMed] [Google Scholar]
- 9. Lee JH, Shklovskaya E, Lim SY, Carlino MS, Menzies AM, Stewart A, et al. Transcriptional downregulation of MHC class I and melanoma de- differentiation in resistance to PD-1 inhibition. Nat Commun 2020;11:1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med 2016;375:819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Concha-Benavente F, Srivastava RM, Trivedi S, Lei Y, Chandran U, Seethala RR, et al. Identification of the cell-intrinsic and -extrinsic pathways downstream of EGFR and IFNγ that induce PD-L1 expression in head and neck cancer. Cancer Res 2016;76:1031–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leibowitz MS, Srivastava RM, Andrade Filho PA, Egloff AM, Wang L, Seethala RR, et al. SHP2 is overexpressed and inhibits pSTAT1-mediated APM component expression, T-cell attracting chemokine secretion, and CTL recognition in head and neck cancer cells. Clin Cancer Res 2013;19:798–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Galluzzi L, Buqué A, Kepp O, Zitvogel L, Kroemer G. Immunological effects of conventional chemotherapy and targeted anticancer agents. Cancer Cell 2015;28:690–714. [DOI] [PubMed] [Google Scholar]
- 14. Gandhi L, Rodríguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, et al. Pembrolizumab plus chemotherapy in metastatic non–small cell lung cancer. N Engl J Med 2018;378:2078–92. [DOI] [PubMed] [Google Scholar]
- 15. Janjigian YY, Shitara K, Moehler M, Garrido M, Salman P, Shen L, et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): a randomised, open-label, phase 3 trial. Lancet 2021;398:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lizotte PH, Hong RL, Luster TA, Cavanaugh ME, Taus LJ, Wang S, et al. A high-throughput immune-oncology screen identifies EGFR inhibitors as potent enhancers of antigen-specific cytotoxic t-lymphocyte tumor cell killing. Cancer Immunol Res 2018;6:1511–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Machiels JP, Haddad RI, Fayette J, Licitra LF, Tahara M, Vermorken JB, et al. Afatinib versus methotrexate as second-line treatment in patients with recurrent or metastatic squamous-cell carcinoma of the head and neck progressing on or after platinum-based therapy (LUX-Head & Neck 1): an open-label, randomised phase 3 trial. Lancet Oncol 2015;16:583–94. [DOI] [PubMed] [Google Scholar]
- 18. Guo Y, Ahn MJ, Chan A, Wang CH, Kang JH, Kim SB, et al. Afatinib versus methotrexate as second-line treatment in Asian patients with recurrent or metastatic squamous cell carcinoma of the head and neck progressing on or after platinum-based therapy (LUX-Head & Neck 3): an open-label, randomised phase III trial. Ann Oncol 2019;30:1831–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 2008;26:317–25. [DOI] [PubMed] [Google Scholar]
- 20. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 2010;5:e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape StringApp: network analysis and visualization of proteomics data. J Proteome Res 2019;18:623–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 2018;173:291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emancipator K, Huang L, Aurora-Garg D, Bal T, Cohen EEW, Harrington K, et al. Comparing programmed death ligand 1 scores for predicting pembrolizumab efficacy in head and neck cancer. Mod Pathol 2021;34:532–41. [DOI] [PubMed] [Google Scholar]
- 28. Litchfield K, Reading JL, Puttick C, Thakkar K, Abbosh C, Bentham R, et al. Meta-analysis of tumor- and T-cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell 2021;184:596–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. House IG, Savas P, Lai J, Chen AXY, Oliver AJ, Teo ZL, et al. Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin Cancer Res 2020;26:487–504. [DOI] [PubMed] [Google Scholar]
- 30. Barry KC, Hsu J, Broz ML, Cueto FJ, Binnewies M, Combes AJ, et al. A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat Med 2018;24:1178–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee JJ, Kao KC, Chiu YL, Jung CJ, Liu CJ, Cheng SJ, et al. Enrichment of human CCR6+ regulatory T cells with superior suppressive activity in oral cancer. J Immunol 2017;199:467–76. [DOI] [PubMed] [Google Scholar]
- 32. Barkal AA, Brewer RE, Markovic M, Kowarsky M, Barkal SA, Zaro BW, et al. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature 2019;572:392–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, et al. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell 2001;104:731–41. [DOI] [PubMed] [Google Scholar]
- 34. Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti–CTLA-4 therapy. Cell 2016;167:397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sacco AG, Chen R, Worden FP, Wong DJL, Adkins D, Swiecicki P, et al. Pembrolizumab plus cetuximab in patients with recurrent or metastatic head and neck squamous cell carcinoma: an open-label, multi-arm, non-randomised, multicentre, phase 2 trial. Lancet Oncol 2021;22:883–92. [DOI] [PubMed] [Google Scholar]
- 36. Taylor MH, Lee CH, Makker V, Rasco D, Dutcus CE, Wu J, et al. Phase IB/II trial of lenvatinib plus pembrolizumab in patients with advanced renal cell carcinoma, endometrial cancer, and other selected advanced solid tumors. J Clin Oncol 2020;38:1154–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Rutkowski P, Lao CD, et al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med 2019;381:1535–46. [DOI] [PubMed] [Google Scholar]
- 38. Motzer RJ, Penkov K, Haanen J, Rini B, Albiges L, Campbell MT, et al. Avelumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med 2019;380:1103–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hellmann MD, Paz-Ares L, Bernabe Caro R, Zurawski B, Kim SW, Carcereny Costa E, et al. Nivolumab plus ipilimumab in advanced non–small cell lung cancer. N Engl J Med 2019;381:2020–31. [DOI] [PubMed] [Google Scholar]
- 40. Ferris RL, Haddad R, Even C, Tahara M, Dvorkin M, Ciuleanu TE, et al. Durvalumab with or without tremelimumab in patients with recurrent or metastatic head and neck squamous cell carcinoma: EAGLE, a randomized, open-label phase III study. Ann Oncol 2020;31:942–50. [DOI] [PubMed] [Google Scholar]
- 41. Argiris A, Harrington K, Tahara M, Ferris RL, Gillison M, Fayette J, et al. LBA36 - Nivolumab (N) + ipilimumab (I) vs. EXTREME as first-line (1L) treatment (tx) for recurrent/metastatic squamous cell carcinoma of the head and neck (R/M SCCHN): final results of CheckMate 651. Ann Oncol 2021;32:S1283–346. [Google Scholar]
- 42. Harrington KJ, Kong A, Mach N, Chesney JA, Fernandez BC, Rischin D, et al. Talimogene laherparepvec and pembrolizumab in recurrent or metastatic squamous cell carcinoma of the head and neck (MASTERKEY-232): a multicenter, phase 1b study. Clin Cancer Res 2020;26:5153–61. [DOI] [PubMed] [Google Scholar]
- 43. Massarelli E, William W, Johnson F, Kies M, Ferrarotto R, Guo M, et al. Combining immune checkpoint blockade and tumor-specific vaccine for patients with incurable human papillomavirus 16-related cancer: a phase 2 clinical trial. JAMA Oncol 2019;5:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rodriguez CP, Wu QV, Voutsinas J, Fromm JR, Jiang X, Pillarisetty VG, et al. A phase II trial of pembrolizumab and vorinostat in recurrent metastatic head and neck squamous cell carcinomas and salivary gland cancer. Clin Cancer Res 2020;26:837–45. [DOI] [PubMed] [Google Scholar]
- 45. Uppaluri R, Campbell KM, Egloff AM, Zolkind P, Skidmore ZL, Nussenbaum B, et al. Neoadjuvant and adjuvant pembrolizumab in resectable locally advanced, human papillomavirus-unrelated head and neck cancer: a multicenter, phase II trial. Clin Cancer Res 2020;26:5140–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362:eaar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tu HF, Ko CJ, Lee CT, Lee CF, Lan SW, Lin HH, et al. Afatinib exerts immunomodulatory effects by targeting the pyrimidine biosynthesis enzyme CAD. Cancer Res 2021;81:3270–82. [DOI] [PubMed] [Google Scholar]
- 48. Yang JC, Shepherd FA, Kim DW, Lee GW, Lee JS, Chang GC, et al. Osimertinib plus durvalumab versus osimertinib monotherapy in EGFR T790M-positive NSCLC following previous EGFR TKI therapy: CAURAL brief report. J Thorac Oncol 2019;14:933–9. [DOI] [PubMed] [Google Scholar]
- 49. Yang JC, Gadgeel SM, Sequist LV, Wu CL, Papadimitrakopoulou VA, Su WC, et al. Pembrolizumab in combination with erlotinib or gefitinib as first-line therapy for advanced NSCLC with sensitizing EGFR mutation. J Thorac Oncol 2019;14:553–9. [DOI] [PubMed] [Google Scholar]
- 50. Khoja L, Day D, Chen TWW, Siu LL, Hansen AR., Tumour- and class-specific patterns of immune-related adverse events of immune checkpoint inhibitors: a systematic review. Ann Oncol 2017;28:2377–85. [DOI] [PubMed] [Google Scholar]
- 51. Gohil SH, Iorgulescu JB, Braun DA, Keskin DB, Livak KJ. Applying high-dimensional single-cell technologies to the analysis of cancer immunotherapy. Nat Rev Clin Oncol 2021;18:244–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lundberg E, Borner GH. Spatial proteomics: a powerful discovery tool for cell biology. Nat Rev Mol Cell Biol 2019;20:285–302. [DOI] [PubMed] [Google Scholar]
- 53. Armingol E, Officer A, Harismendy O, Lewis NE., Deciphering cell–cell interactions and communication from gene expression. Nat Rev Genet 2021;22:71–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data generated in this study are available upon request from the corresponding authors. The mRNA analysis data by Nanostring platform could be obtained in Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/; GEO Accession No. GSE190575).