Abstract
Background:
Keratinocyte cancer is the commonest cancer, imposing a high economic burden on the health care system. Observational studies have shown mixed associations between polyunsaturated fatty acids (PUFA) and keratinocyte cancer, basal cell carcinoma (BCC), and squamous cell carcinoma (SCC). We explored whether genetically predicted PUFA levels are associated with BCC and SCC risks.
Methods:
We conducted a two-sample Mendelian randomization study using PUFA level genome-wide association studies (GWAS) from the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (n > 8,000), and the meta-analysis GWASs from UKB, 23andMe, and Qskin for BCC (n = 651,138) and SCC (n = 635,331) risk.
Results:
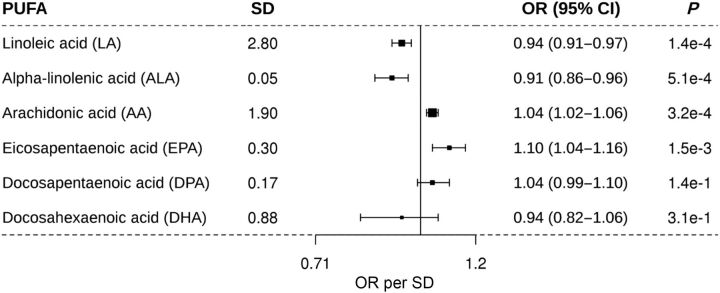
One SD increase in genetically predicted levels of linoleic acid [OR = 0.94, 95% confidence interval (CI) = 0.91–0.97, P = 1.4 × 10–4] and alpha-linolenic acid (OR = 0.91, 95% CI = 0.86–0.96, P = 5.1 × 10–4) was associated with a reduced BCC risk, while arachidonic acid (OR = 1.04, 95% CI = 1.02–1.06, P = 3.2 × 10–4) and eicosapentaenoic acid (OR = 1.10, 95% CI = 1.04–1.16, P = 1.5 × 10–3) were associated with an increased BCC risk.
Conclusions:
Higher genetically predicted levels of linoleic acid and alpha-linolenic acid were associated with a reduced BCC risk, but arachidonic acid and eicosapentaenoic acid were associated with a higher BCC risk.
Impact:
PUFA-related diet and supplementation could influence BCC etiology.
Introduction
Keratinocyte cancers (KC) are the commonest cancers globally, and include two principal types: basal cell carcinoma (BCC) and squamous cell carcinoma (SCC). Because of their frequency, they incur considerable morbidity and very large health expenditures up to AUD $700 million and USD $4.8 billion for their treatment annually in Australia and the United States, respectively (1–3). These cancers are caused by sun exposure, and personal risk is influenced by factors such as fair skin, red hair, and genetic factors (4–6). As a modifiable factor, the role of dietary factors in keratinocyte cancer risk has been contentious. Polyunsaturated fatty acids (PUFA) that include n-6 or omega-6 fats; linoleic acid (LA; 18:2n-6), and arachidonic acid (AA; 20:4n-6), and n-3 or omega-3 fats; alpha-linolenic acid (ALA, 18:3n-3), eicosapentaenoic acid (EPA, 20:5n-3), docosapentaenoic acid (DPA, 22:5n-3), and docosahexaenoic acid (DHA, 22:6n-3) have been found to influence the risk of a number of diseases. Previous studies have reported associations between PUFAs and coronary heart disease (7, 8) and cancers (9, 10).
PUFAs influence biological processes that influence both carcinogenesis and cancer progression (11–14). Omega-6 LA stimulates production of anti-inflammatory products with carcinoprotective properties while AA triggers generation of cytokines which initiate cancer-related inflammation (11–14). In contrast, n-3 PUFAs (e.g., ALA and EPA) are generally metabolized into anti-inflammatory eicosanoids thus halting carcinogenesis (11, 12, 14, 15). The respective PUFA effects are thought to be independent of each PUFA family. For example, a previous study found no interaction between plasma phospholipid levels of LA (n-6) and the levels of DHA or EPA (n-3), but significant interaction of ALA with EPA levels (increase in ALA resulted in increased EPA; ref. 16).
Although the aforementioned biological processes link PUFAs to cancers, observational studies examining the relationship between PUFAs and BCC and SCC have reported mixed findings (17). For example, the Nambour Study in Australia found no association between PUFAs (total omega-6 and omega-3) with the risk of BCC and SCC (18). Specifically, it found no association between higher levels of each of the omega-3 fats; ALA, EPA, DHA, and DPA and the risk of BCC, and SCC (18). Similarly, higher levels of omega-6 fats; LA, and AA were not associated with the risk of BCC and SCC (18). However, recent findings from the Nurses' Health Study and Health Professionals Follow-up Study from the United States, revealed that higher intake of omega-6 fat was associated with higher risks of both BCC and SCC (19). In addition, while higher intake of omega-3 fat was positively associated with the risk of BCC, this was not the case with SCC (19).
However, observational studies are more prone to limitations including; selection and recall bias, confounding, and reverse causation, than randomized control trials (RCT) and Mendelian randomization (MR) studies. RCTs are less prone to error, but are very difficult to conduct for dietary interventions, and consequently are extremely scarce. Thus, it is desirable to use the MR approach. MR is based on the principle of random allocation of risk alleles and independent assortment of genes at meiosis. Therefore, it is less prone to some of the biases which affect observational studies when the assumptions for a valid instrument variable are met. The MR design utilizes genetic variants as the instrumental variables for the exposure (here PUFA levels). It assumes that the instrumental variables; (i) are associated with the exposure (PUFA levels), (ii) are not associated with any confounders of PUFA-KC association (exposure outcome), and (iii) affect the outcome (here BCC and SCC) only through the exposure (here PUFA levels; ref. 20).
Previous MR studies have found significant associations between genetically predicted levels of particular PUFAs and the risk of cancer including; prostate cancer (21), lung cancer (22), and colorectal cancer (12, 23). However, previous MR studies found no association between PUFAs and the risk of cutaneous melanoma (24) and all cancer (23). No MR study to date has explored a causal relationship between PUFA levels and the risk of BCC or SCC. Therefore, our aim was to assess whether genetically predicted PUFA levels are associated with the risk of BCC and SCC.
Materials and Methods
Study population for PUFA levels
We obtained summary statistics data to identify genetic instruments for PUFA levels from genome-wide association study (GWAS) meta-analysis on n-3 (ALA, EPA, DPA and DHA) and n-6 (AA, and LA) PUFAs that included that included 8,866 and 8,631 participants, respectively, of European ancestry in five studies in the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium (16, 25). Details for each cohort including recruitment, quality control, and ethical procedures have been published previously (16, 25–29). The five study cohorts included were Atherosclerosis Risk in Communities (ARIC, n = 3,268), Coronary Artery Risk Development in Young Adults (CARDIA, n = 1,507), Cardiovascular Health Study (CHS, n = 2,326), Invecchiare in Chianti (InCHIANTI, n = 1,075), and Multi-Ethnic Study of Atherosclerosis (MESA, n = 690). PUFA levels were analyzed by gas chromatographic techniques and reported as percentage of the total plasma fatty acids (16, 30). The mean and SD of each PUFA in the cohorts were reported.
Study population for the BCC and SCC risk
We used summary data from the recently published GWAS meta-analysis that included overall 31,787 BCC cases, and 619,351 controls, and 9,674 SCC cases and 625,657 controls of European descent from Europe, United States, and Australia (31). For BCC, the meta-analysis included the UK Biobank (16,847 cases, 340,302 controls), the 23andMe Research cohort (12,945 cases, 274,252 controls), and the QSkin Sun and Health Study (QSkin) cohort (1,995 cases, 4,797 controls). Similarly, the SCC meta-analysis included the UK Biobank (2,274 cases, 340,302 controls), 23andMe Research cohort (6,579 cases, 280,558 controls), and the Qskin cohort (821 cases and 4,797 controls). All participants were of European ancestry. Details on the respective cohorts have been extensively described previously (4, 32–34).
In summary, the UK Biobank is a population-based cohort of over 500,000 adult participants (40–69 years old) recruited between 2006 and 2010 in the United Kingdom. Detailed phenotypic and genetic data were collected and participants with information on BCC and SCC were included in the meta-analysis. The study was approved by the United Kingdom's National North West Multi-Centre Research Ethics Committee. 23andMe Research cohort included international participants of European ancestry with self-reported data on BCC and SCC. Validation of the self-reported data showed high accuracy (4). The research protocol was approved by the Ethical and Independent Review Services, an Institutional Review Board accredited by the Association for the Accreditation of Human Research Protection. The QSkin is a population based prospective cohort of adulted participants (40–60 years old, N∼ 43,000) from Queensland, Australia recruited between 2011 and 2012, and over 17,000 of them genotyped in 2017 (34). Both clinically validated and self-reported data on BCC and SCC were collected. The study was approved by the Human Research Ethics Committee at QIMR Berghofer Medical Research Institute, Brisbane, Australia. All participants in the three cohorts provided written informed consent. Details on the methods used to conduct the GWAS in each cohort and the meta-analysis published elsewhere (31).
Selection of the instrumental variables
We identified the SNPs that were associated with increased PUFA plasma levels at the genome-wide significant level (P = 5 × 10–8) in published GWAS meta-analysis summary data from the CHARGE Consortium (16, 25, 30). For each PUFA, we extracted the SNP, its PUFA-increasing allele, the estimated SNP-PUFA magnitude of association (beta), and its SE (Table 1) for the instrument selection for PUFA and harmonized using the TwoSampleMR package in R (35). The selected instrumental variables (IV) were largely consistent with previous studies that explored the associations between PUFA and other morbidities (21, 22, 24). Next, we retrieved summary data for the selected IVs for each PUFA for both the BCC and SCC analysis from the previous keratinocyte cancer GWAS meta-analysis (31).
Table 1.
The instrument variables for the PUFAs that were used for the MR study.
| PUFA | CHR | Gene | SNP | A1* | A2 | A1 Freq | P | Beta | SE | VE per allele (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Linoleic acid (LA) | 10 | NRBF2 | rs10740118 | G | C | 0.56 | 8.1 × 10–09 | 0.248 | 0.043 | 0.2–0.7 |
| 11 | FADS1 | rs174547 | C | T | 0.32 | 5.0 × 10–274 | 1.474 | 0.042 | 7.6–18.1 | |
| 16 | NTAN1 | rs16966952 | A | G | 0.31 | 1.2 × 10–15 | 0.351 | 0.044 | 0.5–2.5 | |
| Arachidonic acid (AA) | 11 | FADS1 | rs174547 | T | C | 0.68 | 3.3 × 10–971 | 1.691 | 0.025 | 3.7–37.6 |
| 16 | NTAN1 | rs16966952 | G | A | 0.69 | 2.4 × 10–10 | 0.199 | 0.031 | 0.1–0.6 | |
| Alpha-linolenic acid (ALA) | 11 | FADS1 | rs174547 | C | T | 0.33 | 4.0 × 10–64 | 0.016 | 0.001 | 1.03 |
| Eicosapentaenoic acid (EPA) | 6 | ELOVL2 | rs3798713 | C | G | 0.43 | 2.0 × 10–12 | 0.035 | 0.005 | 0.36 |
| 11 | FADS1 | rs174538 | G | A | 0.72 | 5.0 × 10–58 | 0.083 | 0.005 | 1.69 | |
| Docosapentaenoic acid (DPA) | 6 | ELOVL2 | rs3734398 | C | T | 0.43 | 1.0 × 10–43 | 0.04 | 0.003 | 0.46 |
| 11 | FADS1 | rs174547 | T | C | 0.67 | 4.0 × 10–154 | 0.075 | 0.003 | 2.74 | |
| 2 | GCKR | rs780094 | T | C | 0.41 | 9.0 × 10–09 | 0.017 | 0.003 | 8.38 | |
| Docosahexaenoic acid (DHA) | 6 | rs2236212 | G | C | 0.57 | 1.3 × 10–15 | 0.113 | 0.014 | 0.65 | |
| A1*-PUFA increasing allele | A2 - other allele | SE - standard error | ||||||||
| VE- variance explained | IV - instrument variable | A1 Freq - frequency of PUFA increasing allele | ||||||||
MR
We conducted a two-sample MR analysis using the inverse variance weighted (IVW) method (36, 37). IVW utilizes GWAS summary data, a method equivalent to using individual level data (36). For each PUFA, the Wald-type ratio estimator (38) was used to compute the MR estimates for each SNP. Then, the SNP estimates (for multiple IVs) were meta-analyzed using the IVW approach based on random effects. TwoSampleMR package in R was used for the analysis (35). Then, the estimated associations for the genetically determined levels of PUFA and BCC and SCC risk were expressed as OR per SD increase in PUFA levels.
Sensitivity analyses
We conducted “leave-one-out” analyses to assess whether the MR results were being driven or biased by a SNP for each PUFA. Next, we investigated the possibility of directional (horizontal) pleiotropy through MR-Egger regression (39). The intercept term in the MR-Egger regression quantifies evidence for the directional pleiotropy; the magnitude and direction of the effect of the instrumental variables (SNPs) on the outcome (BCC and SCC risk) that are not mediated through the exposure (39). Egger method explores whether the intercept is significantly different from zero (40). The intercept estimate is interpreted as the average pleiotropic effect across all instrument variables used. An intercept estimate, significantly different from zero indicates directional pleiotropy. However, as the MR-Egger regression requires at least three genetic variants, it was only used for PUFAs which showed a significant association and had at least three IVs (LA). For the PUFAs that were significantly associated with the outcomes, we also assessed whether a single dominantly influenced the results.
Next, to rule out the possibility of reverse causality for BCC and SCC on PUFAs, we conducted a reverse-MR analysis evaluating whether genetically predicted BCC and SCC risks are associated with each PUFA separately. We used TwoSampleMR package in R (35) for all statistical analyses.
Finally, we explored whether potential confounders such as body mass index (BMI), educational attainment, and vitamin D levels affected the associations between the PUFAs and BCC risk. We obtained IVs using publicly available published data for educational attainment (years and college completion; ref. 41) and BMI (42) and we tested whether the PUFA IVs were causally affected BCC through BMI, educational attainment, and vitamin D using the IVW approach described earlier. Vitamin D GWAS summary data were generated using the UK Biobank data (data field 100021) that included 401,529 participants of European ancestry. Age, sex, the first 10 principal components and monthly vitamin D variations were adjusted for using BOLT-LMM (43). SNPs with minor allele frequency of >1% and imputation score of 0.3 were then selected.
Results
Association between PUFA levels and BCC incidence
MR results for each of the six PUFA traits on BCC incidence are shown in Fig. 1. Briefly, among the six PUFAs evaluated, a one SD increase in genetically determined plasma levels of LA, and ALA was associated with a decreased incidence of BCC. Conversely, a one SD increase in genetically predicted levels of AA and EPA was associated with an increased incidence of BCC. However, genetically determined levels of DPA and DHA were not associated with BCC incidence.
Figure 1.
Association of one SD increase in genetically determined levels of PUFA and the risk of BCC. PUFA, polyunsaturated fatty acid; SD, standard deviation; OR, odds ratio per SD; and 95% confidence interval. The middle line represents the null (OR = 1.00), and the error bars represent 95% CI. The figure shows the MR results for the relationship between the six PUFAs and the risk of BCC.
Association of PUFA levels and SCC incidence
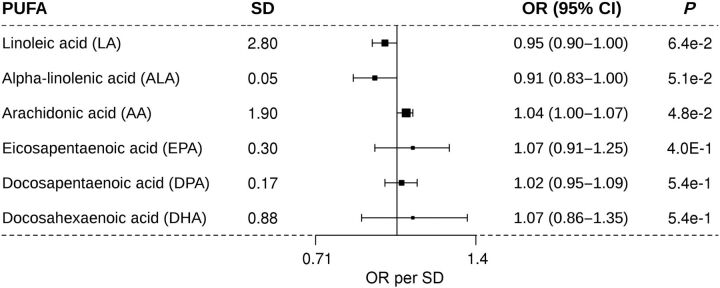
While the point estimates for the associations between each genetically predicted PUFA levels and the SCC incidence were similar to those of BCC, the 95% confidence intervals (CI) around these estimates were much wider, likely due to the smaller sample size for SCC (Fig. 2).
Figure 2.
Association of one SD increase in genetically determined levels of PUFA and the risk of SCC. PUFA, polyunsaturated fatty acid; SD, standard deviation; OR, odds ratio per SD; and 95% confidence interval. The middle line represents the null (OR = 1.00), and the error bars represent 95% CI. The figure shows the MR results for the relationship between the six PUFAs and the risk of SCC.
Sensitivity analyses
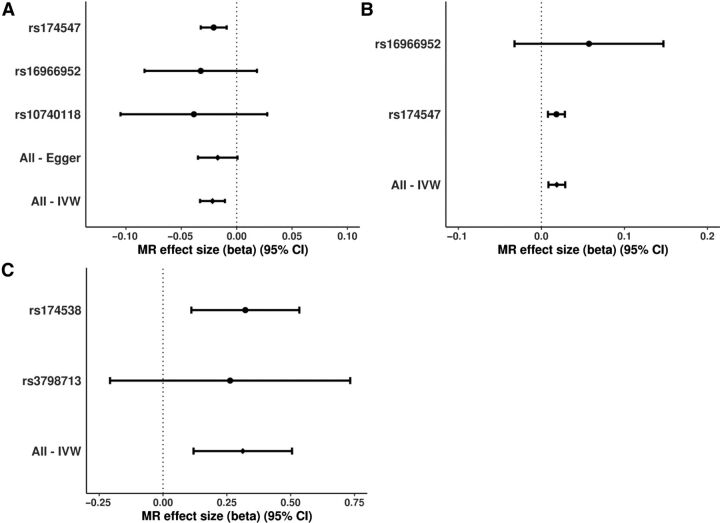
MR-Egger intercept regression results for LA (−0.005, −0.02 to 0.01; P = 0.50) and showed no evidence of directional pleiotropic effects on BCC risk through other pathways independent of serum LA. In a leave-one-out analysis the sensitivity analysis results did not differ materially from the primary results. No single SNP was strongly driving the overall effects of LA (Fig. 3A), AA (Fig. 3B), and EPA (Fig. 3C) on the BCC risk. There was no opposite causal association between PUFA levels and BCC risk; LA (OR = 0.95, 95% CI = 0.80–1.13, P = 0.596); ALA (OR = 1.00, 95% CI = 1.00–1.003, P = 0.935); AA (OR = 0.99, 95% CI = 0.88–1.12, P = 0.851) and EPA (OR = 1.00, 95% CI = 0.98–1.02, P = 0.855).
Figure 3.
Forest plots of the individual SNP effects (log OR) for genetically predicted LA, AA, and EPA levels on BCC risk. This figure illustrates the contribution of individual IVs used for the association between LA (A), AA (B), EPA (C), and BCC risk. The error bars represent the individual SNP effects (beta or log OR) and the 95% CI derived using the IVW method or MR Egger for all SNPs and the Wald ratio method for instrumental variables. The vertical dotted line is the null (log OR). The figure shows that the results were not driven by a single IV.
Our assessment for violations of the MR assumptions by body mass index, education attainment, and vitamin D as a potential confounder revealed it was unlikely that PUFA IVs would affect the BCC risk through the aforementioned traits (P > 0.05 for all PUFAs; Table 2).
Table 2.
Causal relationship between the PUFAs and the potential confounders.
| Exposure | Potential confounder | Beta* | 95% CI | P | |
|---|---|---|---|---|---|
| LA | BMI | 0.0017 | −0.0129 | 0.0163 | 0.8200 |
| EA | −0.0029 | −0.0150 | 0.0093 | 0.6431 | |
| VitD | −0.0011 | −0.0126 | 0.0105 | 0.8587 | |
| AA | BMI | −0.0011 | −0.0136 | 0.0115 | 0.8658 |
| EA | 0.0012 | −0.0022 | 0.0047 | 0.4833 | |
| VitD | 0.0021 | −0.0033 | 0.0074 | 0.4516 | |
| ALA | BMI | 0.1950 | −0.0639 | 0.4538 | 0.1399 |
| EA | −0.1258 | −0.4956 | 0.2440 | 0.5050 | |
| VitD | −0.1805 | −0.5196 | 0.1585 | 0.2966 | |
| EPA | BMI | −0.0392 | −0.0844 | 0.0059 | 0.0885 |
| EA | 0.0253 | −0.0344 | 0.0849 | 0.4062 | |
| VitD | 0.0026 | −0.1192 | 0.1244 | 0.9666 | |
| DPA | BMI | −0.0575 | −0.1769 | 0.0620 | 0.3457 |
| EA | 0.0270 | −0.0333 | 0.0873 | 0.3802 | |
| VitD | −0.0657 | −0.4518 | 0.3204 | 0.7389 | |
| DHA | BMI | 0.0080 | −0.0267 | 0.0426 | 0.6527 |
| EA | −0.0088 | −0.0608 | 0.0431 | 0.7389 | |
| VitD | 0.0332 | −0.0018 | 0.0682 | 0.0628 | |
Abbreviations: AA, arachidonic acid; ALA, alpha-linolenic acid; Beta*, beta coefficient for the effect; BMI, body mass index; CI, confidence interval; DHA, docosahexaenoic acid; DPA, docosapentaenoic acid; EA, educational attainment; EPA, eicosapentaenoic acid; LA, linoleic acid; PUFAs, polyunsaturated fatty acids; VitD, vitamin D or 25-hydroxyvitamin D.
Discussion
We used a two-sample MR to investigate the association between specific genetically predicted PUFA levels and the incidence of BCC and SCC among people of European ancestry. Our findings suggest that people with genetically predicted high levels of LA and ALA have lower risks of BCC than those with lower levels of these dietary factors. In contrast, we found that people with high plasma levels of AA and EPA had elevated risks of BCC. Our analyses suggest that one n-3 PUFA is protective for BCC (ALA: OR = 0.91) while the other is associated with increased risk for BCC (EPA: OR = 1.10). Similarly, one n-6 PUFA is protective (LA: OR = 0.94) while another one increases the incidence of BCC (AA: OR = 1.04). These findings suggest that n-3 and n-6 PUFAs may act in opposing ways to influence the risks of BCC. Although results for SCC were overlapping with the null, the magnitude and directions of association for each PUFA were similar to those observed for BCC, but were subject to greater imprecision due to smaller sample sizes.
A recent systematic review and meta-analyses of observational studies reported that combined n-3 PUFAs (ALA, EPA, DHA) were not associated with the risk of BCC (pooled OR = 1.05, 95% CI = 0.86–1.28) and SCC (pooled OR = 0.86, 95% CI = 0.59–1.23; ref. 17). However, this finding is not surprising because all the n-3 PUFAs were considered together, yet our data and other studies suggest that these factors likely have heterogeneous effects on cancer development (12, 21). Another observational study found no association between any of the PUFAs with the incidence of BCC and SCC (18). However, findings from two observational longitudinal studies in the United States found that higher intake of both n-6 and n-3 fats was associated with higher risk of BCC (for n-3; HR = 1.08, 95% CI = 1.02–1.14; Ptrend = 0.01 and for n-3; HR = 1.09, 95% CI = 1.04–1.13, Ptrend < 0001; ref. 19). Higher intake of n-6 fat was significantly associated with increased risk of SCC (highest vs. lowest quintile, HR = 1.23, 95% CI = 1.08–1.41, P = 5 × 10–4; ref. 19). However, higher intake of n-3 fat was not associated with SCC risk (highest vs. lowest quintile, HR = 0.97, 95% CI = 0.87–1.10, P = 0.78; ref. 19).
Observational studies for dietary factors are prone to biases, especially from confounding and reverse causation (44). A recent RCT of 46 participants showed that supplementation of EPA+DHA given to lung transplant patients was not associated with the risk of KC (OR = 0.34, 95% CI = 0.09–1.32; ref. 45). However, this was a small RCT with very limited power to detect any meaningful associations. Second, participants followed for a short period (1 year), which is not applicable for slow growing tumors. Therefore, in absence of a well-conducted RCT, our MR study offers reliable findings to clarify results from observational studies.
Our results are comparable with other MR PUFA findings for other cancers previously published. They are similar to an MR of PUFA and prostate cancer risk among men < 62 years that revealed that LA (OR = 0.95, 95%CI = 0.92–0.98) and ALA (OR = 0.96, 95% CI = 0.93–0.98) were associated with a reduced risk of prostate cancer (21). In addition, conversely, AA (OR = 1.05, 95% CI = 1.02–1.08), EPA (OR = 1.04, 95% CI = 1.01–1.06), and DPA (OR = 1.05, 95% CI = 1.02–1.08) were associated with an increased risk of prostate cancer (21). A previous MR study also showed that LA (OR = 0.95, 95% CI = 0.93–0.98) and AA (OR = 1.05, 95% CI = 1.02–1.07) were negatively and positively, respectively, associated with the risk of colorectal cancer (12). A recently published MR on PUFA and the risk of melanoma found no significant associations between different PUFAs and melanoma risk (24). However, the magnitude and direction of the associations (per SD increase in PUFA) are similar to our results for LA (OR = 0.94 for BCC vs. 0.94 for melanoma 95% CI = 0.86–1.02), ALA (OR = 0.91 vs. 0.92, 95% CI 0.82–1.03), and AA (OR = 1.04 vs. 1.03, 95% CI = 0.99–1.07). Therefore, our findings are comparable in terms of both the direction and the magnitude to previous PUFA MR results on prostate cancer, colorectal cancer and melanoma.
Possible biological mechanisms for carcinogenesis
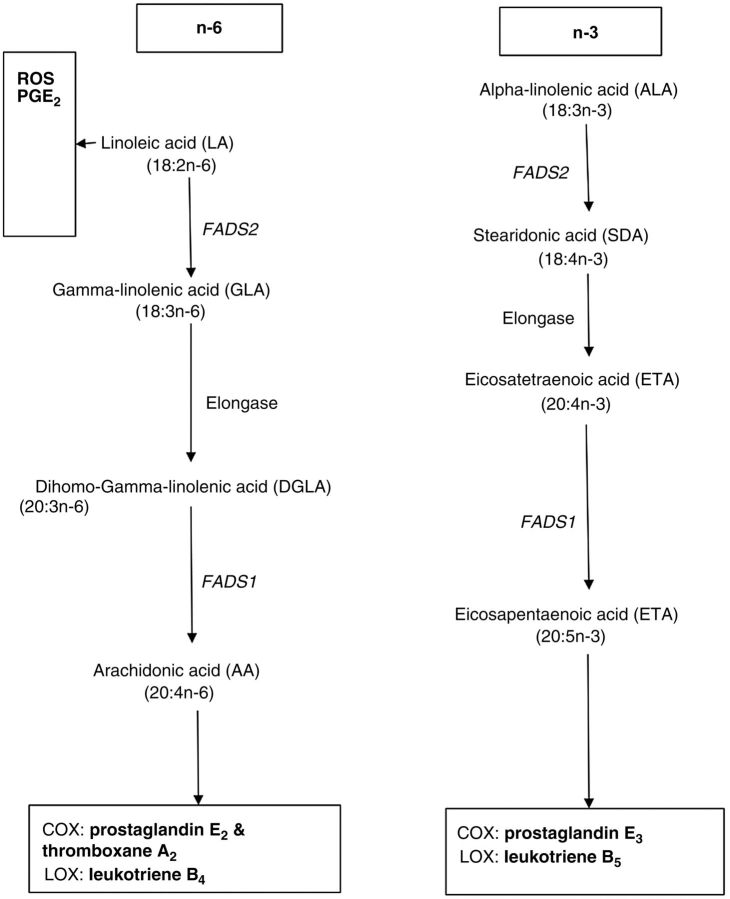
The PUFA metabolic pathways and other biological routes to possible carcinogenesis are summarized in Fig. 4. Downstream metabolism of LA via metabolites GLA and DLA through desaturation (using fatty acid desaturase 2 (FADS2) and FADS1) and elongation (using elongase) results into AA and other eicosanoids. On the other hand, downstream metabolism of ALA via metabolites SDA and ETA using FADS2, FADS1, and elongase results in EPA and other eicosanoids. Several pathways in which PUFAs initiate carcinogenesis (which may apply to keratinocyte cancer) have been suggested. For example, it has been suggested that at high concentrations, LA stimulates tumoricidal actions through generation of free radicals within the cancer cell (12–15). It induces mitochondrial dysfunction and oxidative stress which lead to suppression of tumor cell growth and the eventual tumor cell apoptosis (12–15). It also stimulates production of prostaglandin E1, an anti-inflammatory product (12, 14, 15). However, downstream metabolism of LA to AA leads to production of carcinogenic proinflammatory AA-derived eicosanoids like leukotriene B4 through lipoxygenase; and prostaglandin E2 and thromboxane A2 through cyclooxygenases (12, 14). This potentially explains why AA is associated with increased risk of BCC in our findings. Downstream metabolism of ALA results in production of EPA-derived eicosanoids like leukotriene B5 and prostaglandin E3, which have anti-inflammatory actions that are carcinoprotective (12, 14, 15). However, in our study, EPA was associated with increased risk of BCC. This may be a true effect as is the case with prostate cancer EPA (OR = 1.04, 95% CI = 1.01–1.06; ref. 21). However, it is also possible that the results are influenced by residual confounding.
Figure 4.
The PUFA metabolic pathways and other biological routes to carcinogenesis. Schematic diagram showing downstream metabolism of essential fatty acids LA and ALA into proinflammatory and anti-inflammatory eicosanoids and free radicals, such as ROS. LOX, lipoxygenase; COX, cyclooxygenases (COX-1 and COX-2); PGE, prostaglandin E; ROS, reactive oxygen species; FADS2, fatty acid desaturase 2; FADS1, fatty acid desaturase 1.
Strengths and limitations
Our study has several strengths. First, we used strong instrumental variables since they were genome-wide significant with the exposure (PUFA levels); and with some IVs explaining up to 30% of trait variance (Table 1). In addition, the F-statistic (a measure of the strength of the genetic instrument) for each PUFA was high i.e., AA (11, 302), EPA (479), LA (1, 104-3, 533), DPA (1, 997) and DHA (299) as indicated previously (21). Therefore, it is unlikely that our results were affected by bias from weak genetic instruments (36, 46). Thus, combination of strength IVs and the large BCC sample size, this allows great precision in our MR findings. Furthermore, we used the IVW method on summary statistics which gives results equivalent to individual level data (36). Our study also used data from the United States, Europe, and Australia and thus making our results broadly generalizable to people of European descent. Finally, MR overcomes reverse causation, a key source of bias in observational studies.
However, the findings have limited specific application to the clinical setting. For example, it is not clear what one SD of any of the PUFAs translates into in terms of diet change (food quantities). Nevertheless, it provides a broad perspective of the associations between specific PUFAs and BCC. Our MR findings pertain to people of European descent; thus, it remains unclear whether they can be generalized to other non-European ancestry populations. Our analysis involved 1–3 SNPs for the PUFAs (Table 1). Thus, we were able to assess directional horizontal pleiotropy for only LA using the MR-Egger method, and although it did not influence our MR estimates (39), we cannot rule out residual pleiotropic effects (including for other PUFAs). In addition, due to the limited number of PUFA SNPs, a multivariable MR approach (47) was not appropriate to assess highly polygenic potential confounders including; BMI, vitamin D, and educational attainment in the same model as this would introduce the regression dilution bias toward the null for the PUFA estimates (40). Nevertheless, the PUFA genetic instruments did not affect BCC through potential confounders; BMI, vitamin D, or education attainment (Table 2), and have been well studied and have known biology with PUFA metabolism (16, 25). While our findings for BCC were robust, the findings for SCC were less precise, mainly due to the much lower prevalence of the disease. “There is also a possibility for the results for AA and EPA being influenced by the levels of their metabolic precursors LA and ALA, respectively. For example, one SD unit increase in ALA was found to increase EPA levels by 23% of one SD (16). However, it may not be the case that n-6 PUFAs influence n-3 PUFA results because the associations of FADS1/2 and ELOVL2 genes with EPA and DHA were found to be independent of LA levels in a previous study (16).
Clinical and public health implications and future research
This study provides meaningful insights on the possible benefits and risks of PUFA supplementation with respect to BCC and SCC. However, the findings are modest and future widespread RCTs using specific PUFA supplements are needed to understand if PUFAs influence BCC or SCC risk in a clinical setting. A recent pilot RCT that assessed the feasibility of PUFA supplementation (EPA+DHA) in lung transplant recipients showed 88% and 83% retention in the intervention and placebo groups respectively; as well as good adherence to the supplements (45). Therefore, an RCT on supplementation is feasible.
Conclusions
Genetically predicted levels of LA and ALA were causally associated with a reduced incidence of BCC, while AA and EPA were causally associated with an increased incidence of BCC. This MR study provides support for future RCTs to determine whether LA and ALA supplementation will practically reduce the risk of these very common cancers in the population. Thus, supplementation of LA and ALA might be useful in prevention of keratinocyte cancers in high risk groups such as organ transplant recipients.
Authors' Disclosures
P. Gharahkhani reports grants from NHMRC (investigator grant) during the conduct of the study. D.C. Whiteman reports grants from National Health and Medical Research Council of Australia during the conduct of the study, as well as personal fees from Pierre Fabre outside the submitted work. S. MacGregor reports grants from Australian National Health and Medical Research Council during the conduct of the study. No disclosures were reported by the other authors.
Acknowledgments
This study was conducted using GWAS summary data from the UK Biobank, the 23andMe Research Team (United States), the QSkin Sun and Health Study (Australia), and the CHARGE Consortium. We thank the research participants and the employees of 23andMe for making this work possible.
This research was supported by a program grant (APP1073898) and a project grant (APP1063061) from the Australian National Health and Medical Research Council (NHMRC). S. MacGregor and D.C. Whiteman were supported by a Research Fellowship from the NHMRC. M. Seviiri is supported by the Australian Government Research Training Program (RTP) Scholarship and the Faculty of Health Scholarship from Queensland University of Technology, Brisbane, Australia.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Authors' Contributions
M. Seviiri: Conceptualization, data curation, software, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. M.H. Law: Conceptualization, resources, data curation, supervision, funding acquisition, investigation, methodology, project administration, writing–review and editing. J.S. Ong: Investigation, methodology, writing–review and editing. P. Gharahkhani: Investigation, methodology, writing–review and editing. D.R. Nyholt: Investigation, methodology, writing–review and editing. C.M. Olsen: Investigation, methodology, writing–review and editing. D.C. Whiteman: Funding acquisition, investigation, methodology, writing–review and editing. S. MacGregor: Conceptualization, resources, supervision, funding acquisition, investigation, methodology, project administration, writing–review and editing.
References
- 1. Gordon LG, Rowell D. Health system costs of skin cancer and cost-effectiveness of skin cancer prevention and screening: a systematic review. Eur J Cancer Prev 2015;24:141–9. [DOI] [PubMed] [Google Scholar]
- 2. Guy GP Jr, Machlin SR, Ekwueme DU, Yabroff KR. Prevalence and costs of skin cancer treatment in the U.S., 2002–2006 and 2007–2011. Am J Prev Med 2015;48:183–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fransen M, Karahalios A, Sharma N, English DR, Giles GG, Sinclair RD. Non-melanoma skin cancer in Australia. Med J Aust 2012;197:565–8. [DOI] [PubMed] [Google Scholar]
- 4. Chahal HS, Wu W, Ransohoff KJ, Yang L, Hedlin H, Desai M, et al. Genome-wide association study identifies 14 novel risk alleles associated with basal cell carcinoma. Nat Commun 2016;7:12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Neale RE, Davis M, Pandeya N, Whiteman DC, Green AC. Basal cell carcinoma on the trunk is associated with excessive sun exposure. J Am Acad Dermatol 2007;56:380–6. [DOI] [PubMed] [Google Scholar]
- 6. Didona D, Paolino G, Bottoni U, Cantisani C. Non melanoma skin cancer pathogenesis overview. Biomedicines 2018;6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. DiNicolantonio JJ, O'Keefe JH. Omega-6 vegetable oils as a driver of coronary heart disease: the oxidized linoleic acid hypothesis. Open Heart 2018;5:e000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abdelhamid AS, Martin N, Bridges C, Brainard JS, Wang X, Brown TJ, et al. Polyunsaturated fatty acids for the primary and secondary prevention of cardiovascular disease. Cochrane Database Syst Rev 2018;7:CD012345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Lorgeril M, Salen P. New insights into the health effects of dietary saturated and omega-6 and omega-3 polyunsaturated fatty acids. BMC Med 2012;10:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thiébaut ACM, Chajès V, Gerber M, Boutron-Ruault M-C, Joulin V, Lenoir G, et al. Dietary intakes of omega-6 and omega-3 polyunsaturated fatty acids and the risk of breast cancer. Int J Cancer 2009;124:924–31. [DOI] [PubMed] [Google Scholar]
- 11. Irimie AI, Braicu C, Pasca S, Magdo L, Gulei D, Cojocneanu R, et al. Role of key micronutrients from nutrigenetic and nutrigenomic perspectives in cancer prevention. Medicina (Kaunas) 2019;55:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. May-Wilson S, Sud A, Law PJ, Palin K, Tuupanen S, Gylfe A, et al. Pro-inflammatory fatty acid profile and colorectal cancer risk: a Mendelian randomisation analysis. Eur J Cancer 2017;84:228–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lu X, Yu H, Ma Q, Shen S, Das UN. Linoleic acid suppresses colorectal cancer cell growth by inducing oxidant stress and mitochondrial dysfunction. Lipids Health Dis 2010;9:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Azrad M, Turgeon C, Demark-Wahnefried W. Current evidence linking polyunsaturated Fatty acids with cancer risk and progression. Front Oncol 2013;3:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Larsson SC, Kumlin M, Ingelman-Sundberg M, Wolk A. Dietary long-chain n-3 fatty acids for the prevention of cancer: a review of potential mechanisms. Am J Clin Nutr 2004;79:935–45. [DOI] [PubMed] [Google Scholar]
- 16. Lemaitre RN, Tanaka T, Tang W, Manichaikul A, Foy M, Kabagambe EK, et al. Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. PLoS Genet 2011;7:e1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Noel SE, Stoneham ACS, Olsen CM, Rhodes LE, Green AC. Consumption of omega-3 fatty acids and the risk of skin cancers: a systematic review and meta-analysis. Int J Cancer 2014;135:149–56. [DOI] [PubMed] [Google Scholar]
- 18. Wallingford SC, van As JA, Hughes MC, Ibiebele TI, Green AC, van der Pols JC. Intake of omega-3 and omega-6 fatty acids and risk of basal and squamous cell carcinomas of the skin: a longitudinal community-based study in Australian adults. Nutr Cancer 2012;64:982–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park MK, Li W-Q, Qureshi AA, Cho E. Fat intake and risk of skin cancer in U.S. adults. Cancer Epidemiol Biomarkers Prev 2018;27:776–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Didelez V, Sheehan N. Mendelian randomization as an instrumental variable approach to causal inference. Stat Methods Med Res 2007;16:309–30. [DOI] [PubMed] [Google Scholar]
- 21. Khankari NK, Murff HJ, Zeng C, Wen W, Eeles RA, Easton DF, et al. Polyunsaturated fatty acids and prostate cancer risk: a Mendelian randomisation analysis from the PRACTICAL consortium. Br J Cancer 2016;115:624–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu J, Zhou H, Zhang Y, Huang Y, Fang W, Yang Y, et al. Docosapentaenoic acid and lung cancer risk: a Mendelian randomization study. Cancer Med 2019;8:1817–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liyanage UE, Ong J-S, An J, Gharahkhani P, Law MH, MacGregor S. Mendelian randomization study for genetically predicted polyunsaturated fatty acids levels on overall cancer risk and mortality. Cancer Epidemiol Biomarkers Prev 2019;28:1015–23. [DOI] [PubMed] [Google Scholar]
- 24. Liyanage UE, Law MH, Ong JS, Cust AE, Mann GJ, Ward SV, et al. Polyunsaturated fatty acids and risk of melanoma: a Mendelian randomisation analysis. Int J Cancer 2018;143:508–14. [DOI] [PubMed] [Google Scholar]
- 25. Guan W, Steffen BT, Lemaitre RN, Wu JHY, Tanaka T, Manichaikul A, et al. Genome-wide association study of plasma N6 polyunsaturated fatty acids within the cohorts for heart and aging research in Genomic Epidemiology Consortium. Circ Cardiovasc Genet 2014;7:321–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, et al. The cardiovascular health study: design and rationale. Ann Epidemiol 1991;1:263–76. [DOI] [PubMed] [Google Scholar]
- 27. Friedman GD, Cutter GR, Donahue RP, Hughes GH, Hulley SB, Jacobs DR Jr, et al. Cardia: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol 1988;41:1105–16. [DOI] [PubMed] [Google Scholar]
- 28. Ferrucci L, Bandinelli S, Benvenuti E, Di Iorio A, Macchi C, Harris TB, et al. Subsystems contributing to the decline in ability to walk: bridging the gap between epidemiology and geriatric practice in the InCHIANTI study. J Am Geriatr Soc 2000;48:1618–25. [DOI] [PubMed] [Google Scholar]
- 29. Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, et al. Multi-Ethnic study of atherosclerosis: objectives and design. Am J Epidemiol 2002;156:871–81. [DOI] [PubMed] [Google Scholar]
- 30. Hu Y, Li H, Lu L, Manichaikul A, Zhu J, Chen Y-DI, et al. Genome-wide meta-analyses identify novel loci associated with n-3 and n-6 polyunsaturated fatty acid levels in Chinese and European-ancestry populations. Hum Mol Genet 2016;25:1215–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liyanage UE, Law MH, Han X, An J, Ong J-S, Gharahkhani P, et al. Combined analysis of keratinocyte cancers identifies novel genome-wide loci. Hum Mol Genet 2019;28:3148–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Olsen CM, Green AC, Neale RE, Webb PM, Cicero RA, Jackman LM, et al. Cohort profile: the QSkin Sun and Health Study. Int J Epidemiol 2012;41:929–929i. [DOI] [PubMed] [Google Scholar]
- 35. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 2013;37:658–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burgess S, Bowden J. Integrating summarized data from multiple genetic variants in Mendelian randomization: bias and coverage properties of inverse-variance weighted methods. University of Cambridge; 2015. Available from: https://arxiv.org/pdf/1512.04486.pdf.
- 38. Baiocchi M, Cheng J, Small DS. Instrumental variable methods for causal inference. Stat Med 2014;33:2297–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hemani G, Bowden J, Davey Smith G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet 2018;27:R195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA, et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 2016;533:539–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, et al. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum Mol Genet 2018;27:3641–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Loh P-R, Tucker G, Bulik-Sullivan BK, Vilhjálmsson BJ, Finucane HK, Salem RM, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet 2015;47:284–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hammer GP, du Prel J-B, Blettner M. Avoiding bias in observational studies: part 8 in a series of articles on evaluation of scientific publications. Dtsch Arztebl Int 2009;106:664–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miura K, Vail A, Chambers D, Hopkins PM, Ferguson L, Grant M, et al. Omega-3 fatty acid supplement skin cancer prophylaxis in lung transplant recipients: a randomized, controlled pilot trial. J Heart Lung Transplant 2019;38:59–65. [DOI] [PubMed] [Google Scholar]
- 46. Burgess S, Thompson SG. Bias in causal estimates from Mendelian randomization studies with weak instruments. Stat Med 2011;30:1312–23. [DOI] [PubMed] [Google Scholar]
- 47. Burgess S, Thompson SG. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol 2015;181:251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]