Abstract
Virotherapy, enabled by recent advances in the transdisciplinary field of biotechnology, has emerged as a powerful tool for use in anticancer treatment, gene therapy, immunotherapy, etc. Examining the effects of viruses and virus-derived immune-modulating therapeutics is of great fundamental and clinical interest. Here we describe a sample preparation protocol for metabolite extraction from virus-infected tissue, in addition to liquid chromatography-mass spectrometry conditions essential for subsequent analysis. This metabolomics approach delivers highly sensitive and specific metabolite information on various biospecimens. Such an approach may be adopted to monitor biological changes in over 30 relevant metabolic pathways in response to viral infection and also viral therapeutics.
Keywords: LC-MS/MS, Metabolomics, Sample preparation, Viral vector, Virotherapy
1. Introduction
There is great interest in the use of viruses and virus-derived components as therapeutics. Viral vectors also hold enormous potential to realize the goals of gene therapy [1] and, in the past decade, more than 1700 gene therapy trials have been enabled by the use of viral therapeutics [2]. In addition to their use as a gene therapy delivery vehicle, viral vectors may also be used as an alternative to antibiotics [3] and, recently, have proven useful in DNA vaccination [4]. Utilization of adeno-associated viruses (AAV) for the deliverance of engineered DNA to target cells [5] has led to the development of fit-for-purpose AAV vectors [6]. Further popularizing such therapeutic concepts is the advent of bacteriophage lambda vectors, or RNA-binding proteins of phage origin [7]. These phage vectors have become an attractive vaccine platform given their inexpensive production, biological stability, and suitability to a wide range of applications with minimal development needed between uses [8]. Additionally, virus-derived immune-modulating proteins may also be used for therapeutic purposes. For example, rabies-derived glycoproteins have been shown to facilitate drug delivery to the central nervous system (CNS) [9], while virus-derived anti-inflammatory proteins may improve the human immune response to immune-mediated diseases and in various cancers through inhibition of protease, chemokine, cytokine, and apoptotic cascades [10].
Since metabolites are sensitive to subtle differences and changes in pathological status and immune response status, metabolomics, the comprehensive study of small-molecular-weight metabolites and their dynamic changes in biological systems [11–15], provides advanced methods to identify changing metabolite levels, and has resulted in the rapid discovery of disease biomarkers during the past decade (see Fig. 1) [16–21]. Mass spectrometry (MS)-based metabolic profiling has proven to be a promising tool for analyzing metabolic alterations due to various diseases and, therefore, can provide sensitive and valuable diagnostic information [18, 22, 23], pathogenesis identification [24, 25], and potential therapeutic targets for clinical treatments [26] and disease monitoring [27]. Indeed, previous studies have applied metabolomics to various aspects of viral therapeutics such as the elucidation of metabolites involved in virus infection and pathogenesis [28] and accurate differentiation of vaccination status [29], in addition to characterizing the metabolic profiles of patients with hepatitis B virus-related hepatocellular carcinoma [30], viruses of the gastrointestinal microbiome [31], inflammatory cytokines involved in H1N1 influenza [32], as well as fingerprinting of HIV-1 and -2 infection in macrophages [33].
Fig. 1.
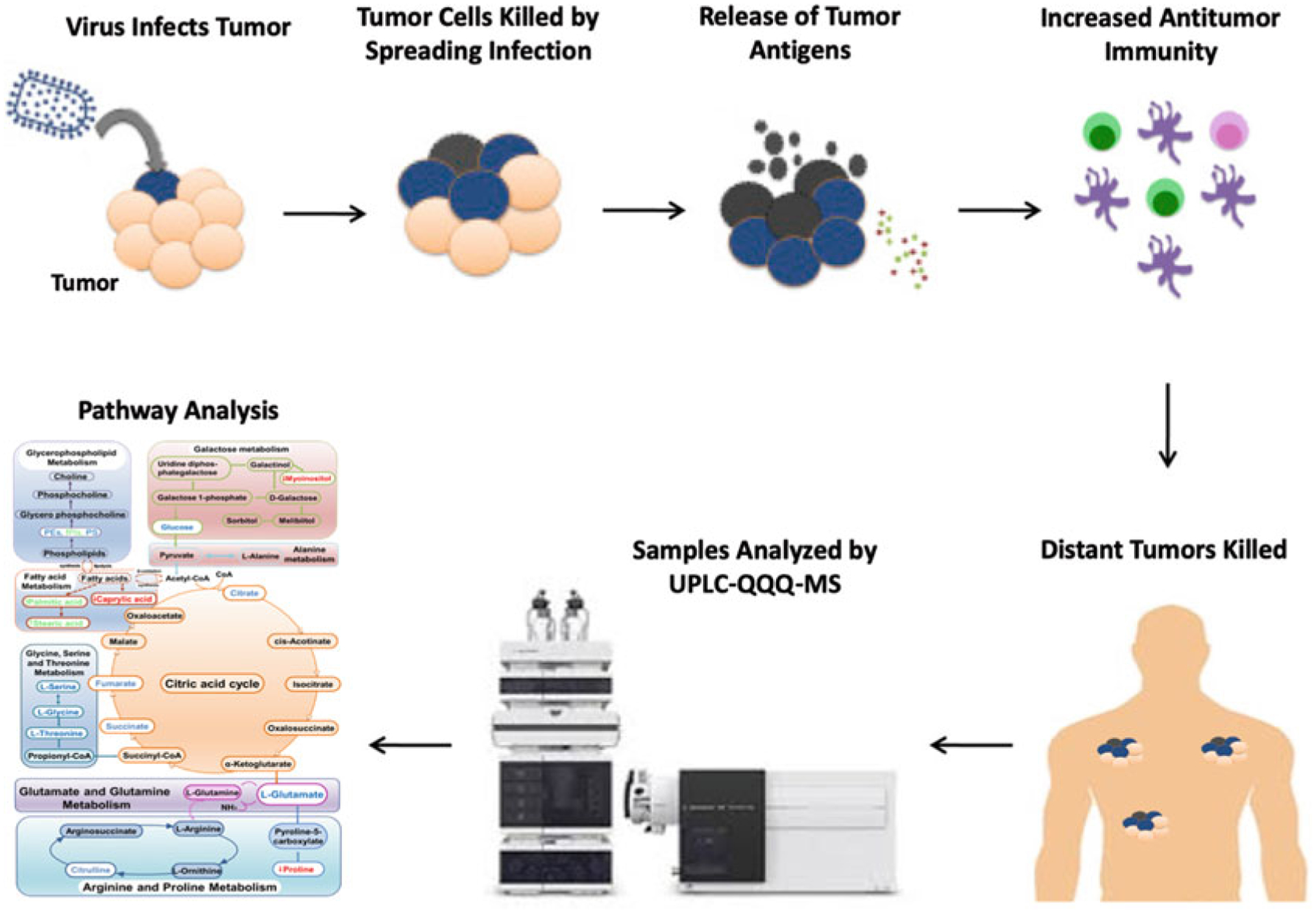
Summary of cancer virotherapy mechanism and LC-MS/MS metabolomics analysis
Herein, we present a sample preparation protocol and associated liquid chromatography-tandem mass spectrometry (LC-MS/MS) conditions for the detection of aqueous metabolites in tissue (see Fig. 2). The targeted LC-MS/MS method used here was modeled and developed after those used in a growing number of studies [18, 34–39].
Fig. 2.
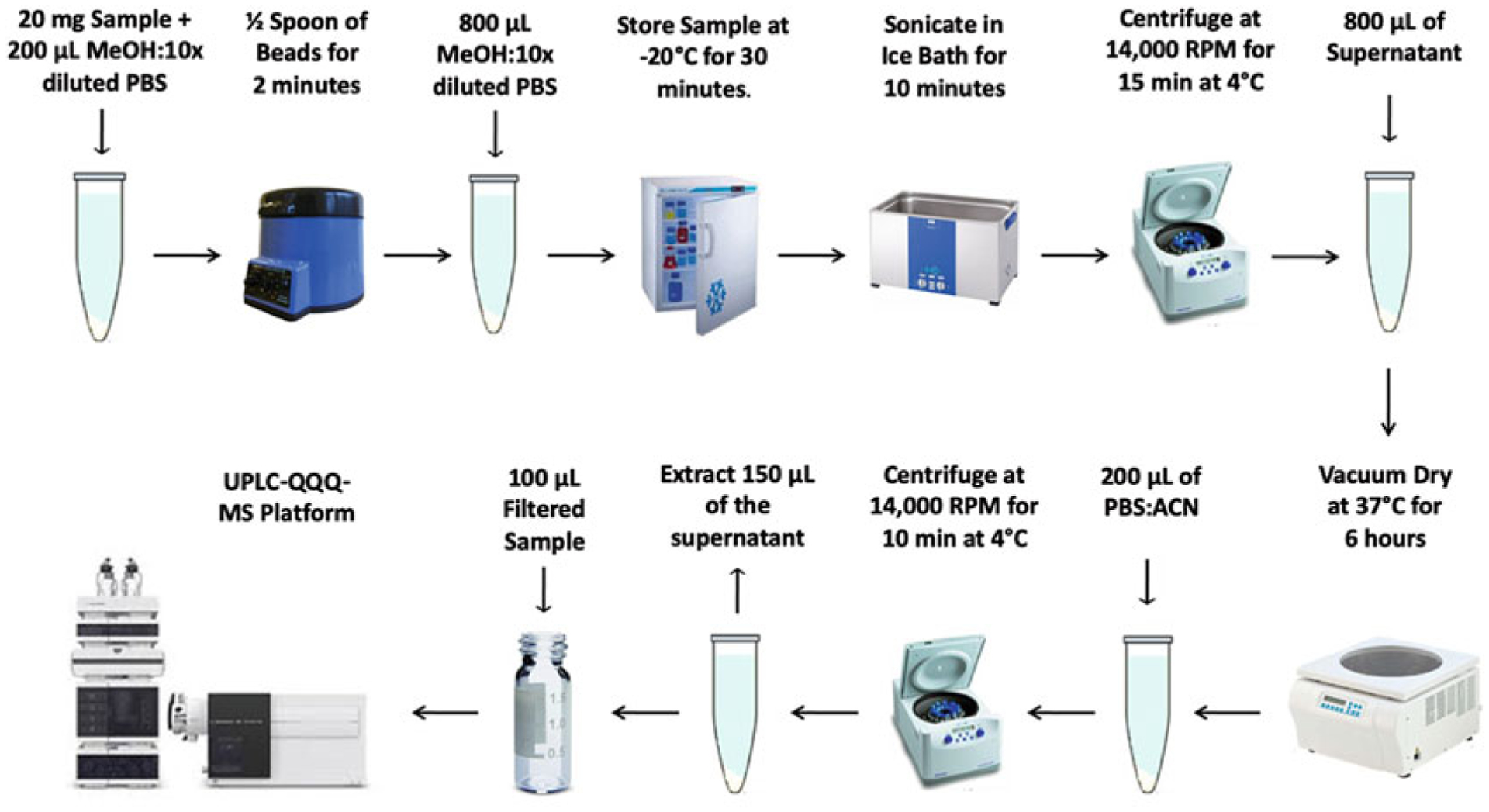
Summary of LC-MS/MS sample preparation method
2. Materials
Prepare all solutions using ultrapure water (see Note 1). Ensure that all reagents used are at least analytical grade (≥99% purity). Prepare and store all reagents at room temperature unless indicated otherwise. Follow all pertinent waste disposal protocols as they apply. Thaw samples as directed. Check pipettes to confirm readiness for use. This targeted LC-MS/MS protocol does not provide absolute concentrations of detected metabolites.
2.1. Preparation of Tissue Samples
1.5 mL Eppendorf tubes.
Bullet beads.
2 mL glass vials.
300 μL glass inserts.
1 mL needle-less syringes.
Polyvinylidene fluoride (PVDF) syringe filters.
Micropipettes with 50–800 μL extraction capability.
100 mL Methanol (MeOH): 10× dilution with phosphate-buffered saline (PBS).
100 mL PBS:acetonitrile ([ACN] 4:6 dilution).
Scientific balance with mg readability.
Bullet blender.
Vortex mixer.
Ultrasonic liquid processor.
Centrifuge with speed, time, and temperature control.
Vacuum concentrator.
2.2. LC-MS/MS Targeted Analysis
Suitable ultra-performance liquid chromatography (UPLC) system.
Triple-quadrupole (QQQ)-MS instrument.
Electrospray ionization (ESI) source.
Hydrophilic interaction chromatography (HILIC) column.
- Mobile-phase solvents:
- 10 mM ammonium acetate, 10 mM ammonium hydroxide in 95% H2O/5% ACN.
- 10 mM ammonium acetate, 10 mM ammonium hydroxide in 95% ACN/5% H2O.
3. Methods
In an effort to identify and overcome potential issues, please see Subheading 4.
3.1. Preparation of Tissue Samples
Weigh out 20 mg ± 1.5 mg of biological sample in a 1.5 mL Eppendorf tube.
Homogenize biological sample in 200 μL MeOH:10× diluted PBS in 1.5 mL Eppendorf tube (see Note 2).
Add ½ spoon of beads and disrupt the sample using bullet blender for 2 min.
Centrifuge for 1–2 min to remove liquid from the cap.
Add 800 μL MeOH:10× diluted PBS (see Note 3).
Vortex for 10 s.
Store sample at −20 °C for 30 min.
Sonicate the mixture in an ice bath for 10 min (see Note 4).
Centrifuge the mixture at 21,694 × g for 15 min at 4 °C (see Note 5).
Remove 800 μL supernatant and transfer to a new Eppendorf tube (see Note 6).
Completely dry the sample(s) using a vacuum concentrator at 37 °C for 6 ± 2 h (see Note 7).
Once sample(s) is/are completely dried, reconstitute by adding 200 μL of PBS:ACN solution.
Vortex the sample for 30 s.
Sonicate each sample for 5 min in a room-temperature water bath.
Centrifuge the samples at 21,694 × g for 10 min at 4 °C (see Note 8).
Extract 150 μL of the supernatant into a syringe and slowly plunge the sample through the filter and into a new 1.5 mL Eppendorf tube (see Note 9).
Extract the remaining supernatant from each sample and inject into a single 2 mL tube (larger tubes may be used if needed) for quality control (QC) analysis.
Transfer 100 μL of the filtered experimental sample into a glass insert inside a glass LC vial. The leftover may be used for backup.
Vortex the QC sample for 10 s and then filter it.
Transfer 1 mL of the QC sample into a glass LC vial (see Note 10).
3.2. LC-MS/MS Targeted Analysis
All experiments should be performed using an UPLC-QQQ-MS platform.
Dictate an appropriate worklist such that each sample is injected twice, 10 μL for analysis using negative ionization mode and 4 μL for analysis using positive ionization mode (see Note 11).
The flow rate is maintained at 0.3 mL/min, auto-sampler temperature is kept at 4 °C, while the column compartment is set to 40 °C.
HILIC gradient parameters: After an initial 1-min isocratic elution of 90% mobile phase solvent B, decrease the percentage of solvent B to 40% at t = 11 min. Maintain the composition of solvent B at 40% for 4 min (t = 15 min), after which the percentage of solvent B should gradually be returned to 90%, to prepare for the next injection (see Note 12). An example of a total ion chromatogram (TIC) can be found in Fig. 3.
MS/MS parameters: The mass spectrometer should be equipped with an electrospray ionization (ESI) source. Targeted data acquisition should be performed in multiple reaction monitoring (MRM) mode.
Table 1 lists LC-MS parameters for the validated detection of 310 metabolites using reference standards. For additional parameter details, see Note 13.
Fig. 3.
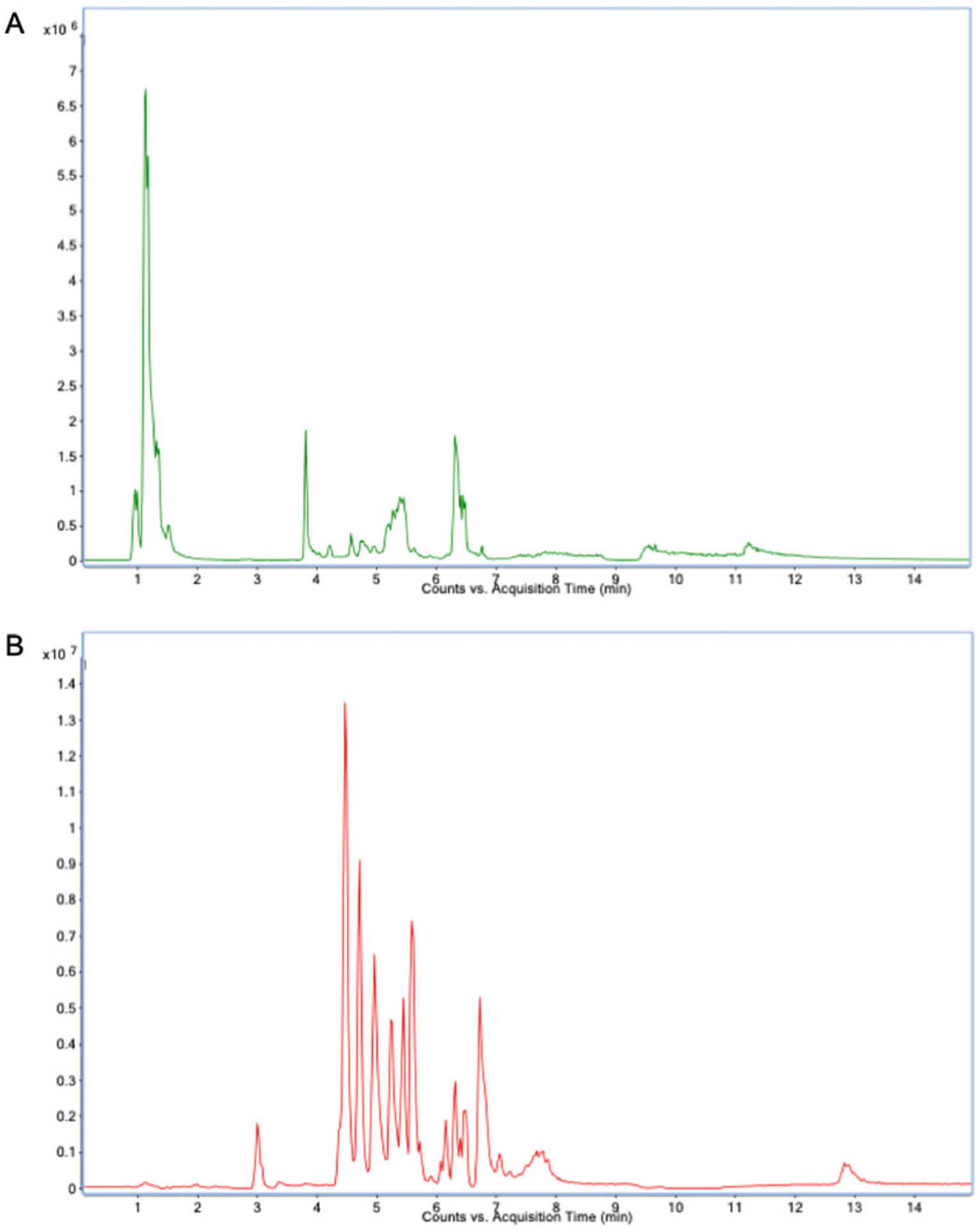
LC-MS/MS total ion chromatogram: (a) negative and (b) positive modes
Table 1.
LC-MS parameters for the validated identification and detection of 310 metabolites
| No. | HMDB/CAS | Compound name | Precursorion | Production | Collision energy | Polarity | RT (min) |
|---|---|---|---|---|---|---|---|
| 1 | HMDB0031645 | Acetamide | 60.05 | 42.80 | 13 | Positive | 5.050 |
| 2 | HMDB0001525 | Imidazole | 69.05 | 42.00 | 21 | Positive | 1.826 |
| 3 | HMDB0000119 | Glyoxylic acid | 72.99 | 44.90 | 5 | Negative | 5.031 |
| 4 | HMDB0001522 | Methylguanidine | 74.07 | 57.00 | 13 | Positive | 5.730 |
| 5 | HMDB0000115 | Glycolic acid | 75.01 | 46.90 | 9 | Negative | 5.350 |
| 6 | HMDB0014691 | Acetohydroxamic acid | 76.04 | 43.00 | 9 | Positive | 6.215 |
| 7 | HMDB0000123 | Glycine | 76.04 | 30.10 | 13 | Positive | 6.598 |
| 8 | HMDB0000925 | TMAO | 76.08 | 58.00 | 21 | Positive | 6.187 |
| 9 | HMDB0002039 | 2-Pyrrolidinone | 86.06 | 44.00 | 29 | Positive | 1.620 |
| 10 | HMDB0000243 | Pyruvate | 87.00 | 43.00 | 5 | Negative | 5.45 |
| 11 | HMDB0001873 | Isobutyric acid | 87.04 | 87.04 | 5 | Negative | 1.948 |
| 12 | HMDB0000190 | Lactate | 89.02 | 43.00 | 9 | Negative | 4.1 |
| 13 | HMDB0001414 | Putrescine | 89.11 | 72.00 | 9 | Positive | 6.118 |
| 14 | HMDB0000161 | Alanine | 90.06 | 44.00 | 9 | Positive | 6.140 |
| 15 | HMDB0000271 | Sarcosine | 90.06 | 44.10 | 9 | Positive | 6.184 |
| 16 | HMDB0001882 | Dihydroxyacetone | 91.04 | 91.04 | 0 | Positive | 1.734 |
| 17 | HMDB0000005 | 2-Ketobutyric acid | 101.02 | 57.00 | 5 | Negative | 1.490 |
| 18 | HMDB0000060 | Acetoacetate | 101.02 | 56.90 | 9 | Negative | 2.194 |
| 19 | HMDB0002176 | 2-Methylbutyric acid | 101.06 | 101.06 | 5 | Negative | 1.693 |
| 20 | HMDB0000718 | Isovaleric acid | 101.06 | 101.06 | 5 | Negative | 6.064 |
| 21 | HMDB0000691 | Malonic acid | 103.00 | 58.90 | 9 | Negative | 4.306 |
| 22 | HMDB0000008 | 2-Hydroxybutyric acid | 103.04 | 58.90 | 9 | Negative | 5.87 |
| 23 | HMDB0000357 | 3-Hydroxybutyric acid | 103.04 | 56.80 | 9 | Negative | 4.264 |
| 24 | HMDB0002322 | Cadaverine | 103.13 | 85.90 | 5 | Positive | 6.506 |
| 25 | HMDB0001906 | 2/3-Aminoisobutyric acid | 104.07 | 57.90 | 9 | Positive | 5.295 |
| 26 | HMDB0000452 | 2-Aminobutyric acid | 104.07 | 57.90 | 13 | Positive | 5.706 |
| 27 | HMDB0031654 | 3-Aminobutyric acid | 104.07 | 85.90 | 5 | Positive | 5.934 |
| 28 | HMDB0000112 | 4-Aminobutyric acid | 104.07 | 87.00 | 9 | Positive | 6.596 |
| 29 | HMDB0000092 | Dimethylglycine | 104.07 | 57.90 | 13 | Positive | 5.295 |
| 30 | 142-26-7 | N-Acetylethanolamine | 104.07 | 61.80 | 5 | Positive | 1.915 |
| 31 | HMDB0000097 | Choline | 104.11 | 58.00 | 33 | Positive | 7.000 |
| 32 | HMDB0000139 | Glyceric acid | 105.02 | 75.00 | 9 | Negative | 4.843 |
| 33 | HMDB0000187 | Serine | 106.05 | 60.00 | 9 | Positive | 6.824 |
| 34 | HMDB0001169 | 4-Aminophenol | 110.06 | 69.00 | 9 | Positive | 5.184 |
| 35 | HMDB0000617 | 2-Furoic acid | 111.01 | 66.90 | 5 | Negative | 1.760 |
| 36 | HMDB0000630 | Cytosine | 112.05 | 94.90 | 21 | Positive | 3.445 |
| 37 | HMDB0000870 | Histamine | 112.09 | 95.00 | 17 | Positive | 8.673 |
| 38 | HMDB0000300 | Uracil | 113.04 | 71.60 | 25 | Positive | 8.707 |
| 39 | HMDB0000562 | Creatinine | 114.07 | 44.10 | 21 | Positive | 2.988 |
| 40 | HMDB0000134 | Fumarate | 115.00 | 71.00 | 5 | Negative | 2.130 |
| 41 | HMDB0000176 | Maleic acid | 115.00 | 70.90 | 9 | Negative | 1.455 |
| 42 | HMDB0000019 | Alpha-ketoisovaleric acid | 115.04 | 70.90 | 5 | Negative | 1.455 |
| 43 | HMDB0000720 | Levulinic acid | 115.04 | 70.90 | 9 | Negative | 3.250 |
| 44 | HMDB0000689 | 4-Methylvaleric acid | 115.07 | 115.07 | 5 | Negative | 1.505 |
| 45 | HMDB0000535 | Hexanoic acid | 115.07 | 115.07 | 5 | Negative | 1.471 |
| 46 | HMDB0000162 | Proline | 116.07 | 70.00 | 13 | Positive | 5.408 |
| 47 | HMDB0000202 | Methylmalonic acid | 117.02 | 73.00 | 5 | Negative | 4.622 |
| 48 | HMDB0000254 | Succinate | 117.02 | 73.00 | 9 | Negative | 4.293 |
| 49 | HMDB0000754 | Beta-hydroxyisovaleric acid | 117.05 | 71.00 | 9 | Negative | 2.132 |
| 50 | HMDB0000532 | Acetylglycine | 118.05 | 75.90 | 5 | Positive | 4.335 |
| 51 | HMDB0000128 | Glycocyamine | 118.06 | 42.90 | 45 | Positive | 6.344 |
| 52 | HMDB0000738 | Indole | 118.07 | 85.80 | 13 | Positive | 5.385 |
| 53 | HMDB0003355 | Amino valerate | 118.09 | 100.90 | 9 | Positive | 6.822 |
| 54 | HMDB0000043 | Betaine | 118.09 | 58.00 | 29 | Positive | 4.791 |
| 55 | HMDB0013716 | Norvaline | 118.09 | 72.00 | 5 | Positive | 5.019 |
| 56 | HMDB0000883 | Valine | 118.09 | 72.00 | 5 | Positive | 5.224 |
| 57 | HMDB0002649 | Erythrose | 119.03 | 42.90 | 17 | Negative | 4.519 |
| 58 | HMDB0000719 | Homoserine | 120.07 | 73.90 | 9 | Positive | 6.343 |
| 59 | HMDB0000167 | Threonine | 120.07 | 73.90 | 9 | Positive | 6.343 |
| 60 | HMDB0011718 | 4-Hydroxy benzaldehyde | 121.03 | 92.00 | 25 | Negative | 1.301 |
| 61 | HMDB0001870 | Benzoic acid | 121.03 | 77.00 | 9 | Negative | 1.487 |
| 62 | HMDB0000574 | Cysteine | 122.03 | 80.90 | 13 | Positive | 13.454 |
| 63 | HMDB0001406 | Nicotinamide | 123.06 | 79.80 | 25 | Positive | 1.746 |
| 64 | HMDB0001488 | Nicotinic acid | 124.04 | 78.10 | 22 | Positive | 3.671 |
| 65 | HMDB0002243 | Picolinic acid | 124.04 | 77.70 | 25 | Positive | 5.315 |
| 66 | HMDB0000251 | Taurine | 126.02 | 44.10 | 17 | Positive | 5.223 |
| 67 | HMDB0000898 | Methylhistamine | 126.11 | 109.10 | 13 | Positive | 4.879 |
| 68 | HMDB0002024 | 4-Imidazoleacetic acid | 127.05 | 80.90 | 13 | Positive | 5.497 |
| 69 | HMDB0000634 | Citraconic acid | 129.02 | 85.10 | 5 | Negative | 1.487 |
| 70 | HMDB0000620 | Glutaconic acid | 129.02 | 85.00 | 5 | Negative | 4.774 |
| 71 | HMDB0000491 | 3-Methyl-2-oxovaleric acid | 129.05 | 129.05 | 5 | Negative | 1.351 |
| 72 | HMDB0000491 | Ketoisoleucine | 129.05 | 85.10 | 5 | Negative | 1.368 |
| 73 | HMDB0000695 | Ketoleucine | 129.05 | 85.00 | 5 | Negative | 1.368 |
| 74 | HMDB0000267 | Pyroglutamic acid | 130.05 | 83.80 | 9 | Positive | 4.675 |
| 75 | HMDB0000070 | Pipecolinic acid | 130.09 | 84.00 | 13 | Positive | 5.405 |
| 76 | HMDB0000223 | Oxaloacetic acid | 131.00 | 42.00 | 17 | Negative | 6.772 |
| 77 | HMDB0000622 | Ethylmalonic acid | 131.03 | 87.00 | 5 | Negative | 3.282 |
| 78 | HMDB0000661 | Glutaric acid | 131.03 | 86.90 | 9 | Negative | 6.179 |
| 79 | HMDB0001844 | Methyl succinate | 131.03 | 98.90 | 5 | Negative | 2.011 |
| 80 | HMDB0000665 | Leucic acid | 131.07 | 85.10 | 9 | Negative | 1.994 |
| 81 | HMDB0001432 | Agmatine | 131.13 | 71.90 | 17 | Positive | 6.912 |
| 82 | HMDB0001149 | 5-Aminolevulinic acid | 132.07 | 85.90 | 13 | Positive | 6.521 |
| 83 | HMDB0000725 | Hydroxyproline | 132.07 | 85.90 | 17 | Positive | 6.113 |
| 84 | HMDB0000064 | Creatine | 132.08 | 44.00 | 21 | Positive | 6.204 |
| 85 | HMDB0000172 | Isoleucine | 132.10 | 86.00 | 9 | Positive | 4.697 |
| 86 | HMDB0000557 | l-Alloisoleucine | 132.10 | 86.00 | 9 | Positive | 4.468 |
| 87 | HMDB0000687 | Leucine | 132.10 | 86.00 | 9 | Positive | 4.468 |
| 88 | HMDB0001645 | Norleucine | 132.10 | 86.00 | 9 | Positive | 4.468 |
| 89 | HMDB0000156 | Malate | 133.01 | 114.90 | 9 | Negative | 5.87 |
| 90 | HMDB0000168 | Asparagine | 133.06 | 73.90 | 13 | Positive | 6.813 |
| 91 | HMDB0003374 | Ornithine | 133.10 | 70.00 | 21 | Positive | 11.112 |
| 92 | HMDB0000191 | Aspartate | 134.05 | 73.90 | 13 | Positive | 6.471 |
| 93 | HMDB0000209 | Phenylacetic acid | 135.04 | 91.00 | 5 | Negative | 1.503 |
| 94 | HMDB0000034 | Adenine | 136.06 | 119.10 | 25 | Positive | 2.755 |
| 95 | HMDB0001895 | 2-Hydroxybenzoic acid | 137.02 | 93.10 | 21 | Negative | 1.215 |
| 96 | HMDB0000710 | 4-Hydroxybenzoic acid | 137.02 | 93.10 | 17 | Negative | 2.316 |
| 97 | HMDB0014581 | Allopurinol | 137.05 | 54.20 | 29 | Positive | 2.070 |
| 98 | HMDB0000157 | Hypoxanthine | 137.05 | 54.80 | 33 | Positive | 2.938 |
| 99 | HMDB0001123 | Anthranilic acid | 138.06 | 120.00 | 9 | Positive | 1.568 |
| 100 | 20989-17-7 | 2-Phenylglycinol | 138.09 | 103.00 | 21 | Positive | 2.549 |
| 101 | HMDB0000301 | Urocanic acid | 139.05 | 92.90 | 21 | Positive | 4.444 |
| 102 | HMDB0002658 | 6-Hydroxynicotinic acid | 140.04 | 121.90 | 17 | Positive | 4.604 |
| 103 | HMDB0002349 | Muconic acid | 141.02 | 97.00 | 5 | Negative | 5.684 |
| 104 | HMDB0000393 | 3-Hexenedioic acid | 143.03 | 99.00 | 5 | Negative | 6.042 |
| 105 | HMDB0000482 | Caprylic acid | 143.11 | 143.11 | 5 | Negative | 1.350 |
| 106 | HMDB0000208 | Alpha-ketoglutaric acid | 145.01 | 101.00 | 5 | Negative | 4.99 |
| 107 | HMDB0000422 | 2-Methylglutaric acid | 145.05 | 101.10 | 9 | Negative | 5.500 |
| 108 | HMDB0000448 | Adipic acid | 145.05 | 101.00 | 9 | Negative | 6.025 |
| 109 | HMDB0000895 | Acetylcholine | 146.12 | 86.80 | 9 | Positive | 3.439 |
| 110 | HMDB0001257 | Spermidine | 146.17 | 72.00 | 17 | Positive | 6.499 |
| 111 | HMDB0059655 | 2HG | 147.03 | 129.00 | 5 | Negative | 5.49 |
| 112 | HMDB0000641 | Glutamine | 147.08 | 84.00 | 17 | Positive | 6.673 |
| 113 | HMDB0000182 | Lysine | 147.12 | 83.90 | 21 | Positive | 11.749 |
| 114 | HMDB0003339 | Glutamic acid | 148.06 | 84.00 | 17 | Positive | 6.39 |
| 115 | 147-73-9 | Meso-tartaric acid | 149.01 | 86.90 | 13 | Negative | 11.000 |
| 116 | HMDB0059916 | Tartaric acid | 149.01 | 87.00 | 9 | Negative | 11.000 |
| 117 | HMDB0001587 | Phenylglyoxylic acid | 149.02 | 77.00 | 9 | Negative | 1.315 |
| 118 | HMDB0000646 | l-(+)-Arabinose | 149.04 | 59.00 | 20 | Negative | 6.034 |
| 119 | HMDB0000283 | Ribose | 149.04 | 89.00 | 5 | Negative | 4.4 |
| 120 | HMDB0000098 | Xylose | 149.04 | 89.00 | 5 | Negative | 4.320 |
| 121 | HMDB0002097 | 4-Ethylbenzoic acid | 149.06 | 105.10 | 9 | Negative | 1.501 |
| 122 | HMDB0032075 | p-Tolylacetic acid | 149.06 | 105.00 | 5 | Negative | 1.501 |
| 123 | HMDB0001878 | Thymol | 149.09 | 87.00 | 13 | Negative | 11.100 |
| 124 | HMDB0000696 | Methionine | 150.06 | 103.90 | 9 | Positive | 4.923 |
| 125 | HMDB0000669 | 2-Hydroxy phenylacetic acid | 151.04 | 107.00 | 13 | Negative | 1.280 |
| 126 | HMDB0000440 | 3-Hydroxyphenylacetic acid | 151.04 | 107.00 | 5 | Negative | 1.280 |
| 127 | HMDB0004815 | 4-Hydroxy-3-methylbenzoic acid | 151.04 | 107.10 | 9 | Negative | 1.923 |
| 128 | HMDB0000020 | 4-Hydroxyphenylacetic acid | 151.04 | 107.00 | 9 | Negative | 1.907 |
| 129 | HMDB0000703 | Mandelic acid | 151.04 | 107.00 | 5 | Negative | 1.907 |
| 130 | HMDB0000508 | Adonitol | 151.06 | 70.80 | 16 | Negative | 4.149 |
| 131 | HMDB0001851 | L-(—)-Arabitol | 151.06 | 89.10 | 8 | Negative | 4.335 |
| 132 | HMDB0002917 | Xylitol | 151.06 | 59.00 | 20 | Negative | 4.390 |
| 133 | HMDB0000132 | Guanine | 152.06 | 135.10 | 21 | Positive | 4.009 |
| 134 | HMDB0000397 | 2,3-Dihydroxybenzoic acid | 153.02 | 109.00 | 17 | Negative | 1.027 |
| 135 | HMDB0000152 | Gentisic acid | 153.02 | 108.10 | 25 | Negative | 1.415 |
| 136 | HMDB0001856 | Protocatechuic acid | 153.02 | 109.00 | 13 | Negative | 1.731 |
| 137 | HMDB0000292 | Xanthine | 153.04 | 109.90 | 21 | Positive | 3.415 |
| 138 | 56-17-7 | Cystamine | 153.05 | 107.70 | 5 | Positive | 9.759 |
| 139 | HMDB0001476 | 3-Hydroxyanthranilic acid | 154.05 | 112.90 | 5 | Positive | 13.765 |
| 140 | HMDB0000073 | Dopamine | 154.09 | 137.00 | 9 | Positive | 4.015 |
| 141 | HMDB0000177 | Histidine | 156.08 | 110.10 | 13 | Positive | 7.182 |
| 142 | HMDB0000555 | 3-Methyladipic acid | 159.06 | 97.20 | 9 | Negative | 5.464 |
| 143 | HMDB0000678 | Isovalerylglycine | 160.10 | 57.00 | 17 | Positive | 2.889 |
| 144 | HMDB0000303 | Tryptamine | 161.11 | 144.00 | 9 | Positive | 3.433 |
| 145 | HMDB0000510 | 2-Aminoadipic acid | 162.08 | 98.00 | 13 | Positive | 6.839 |
| 146 | HMDB0000062 | Carnitine | 162.12 | 43.00 | 25 | Positive | 6.550 |
| 147 | HMDB0001713 | m-Coumaric acid | 163.04 | 119.00 | 17 | Negative | 1.499 |
| 148 | HMDB0002035 | p-Coumaric acid | 163.04 | 119.00 | 17 | Negative | 1.849 |
| 149 | HMDB0000205 | Phenylpyruvic acid | 163.04 | 91.00 | 9 | Negative | 1.279 |
| 150 | HMDB0001890 | Acetylcysteine | 164.04 | 123.00 | 9 | Positive | 2.555 |
| 151 | HMDB0002107 | Phthalic acid | 165.02 | 121.00 | 5 | Negative | 2.380 |
| 152 | HMDB0060256 | d-Xylonic acid | 165.04 | 75.00 | 16 | Negative | 5.218 |
| 153 | HMDB0000779 | 3-Phenyllactic acid | 165.05 | 147.10 | 9 | Negative | 1.499 |
| 154 | HMDB0002072 | 4-Methoxyphenylacetic acid | 165.05 | 106.00 | 9 | Negative | 1.499 |
| 155 | HMDB0000159 | Phenylalanine | 166.09 | 120.20 | 17 | Positive | 4.373 |
| 156 | HMDB0000263 | Phosphoenolpyruvic acid | 166.97 | 78.80 | 33 | Negative | 6.060 |
| 157 | HMDB0000130 | Homogentisic acid | 167.03 | 108.00 | 13 | Negative | 5.765 |
| 158 | HMDB0000484 | Vanillic acid | 167.03 | 152.00 | 13 | Negative | 2.000 |
| 159 | HMDB0000232 | Quinolinic acid | 168.03 | 150.00 | 5 | Positive | 5.263 |
| 160 | HMDB0001112 | d-Glyceraldehyde 3-phosphate | 168.99 | 79.00 | 33 | Negative | 5.800 |
| 161 | HMDB0000289 | Urate | 169.04 | 141.00 | 13 | Positive | 4.768 |
| 162 | HMDB0000239 | Pyridoxine | 170.08 | 134.10 | 17 | Positive | 2.295 |
| 163 | HMDB0000001 | 1-Methylhistidine | 170.10 | 95.90 | 21 | Positive | 6.930 |
| 164 | HMDB0000511 | Capric acid | 171.14 | 171.14 | 5 | Negative | 1.261 |
| 165 | HMDB0000072 | Aconitic acid | 173.01 | 85.00 | 9 | Negative | 6.191 |
| 166 | HMDB0003070 | Shikimic acid | 173.04 | 93.10 | 9 | Negative | 5.801 |
| 167 | HMDB0000893 | Suberic acid | 173.08 | 111.10 | 13 | Negative | 5.004 |
| 168 | HMDB0000721 | Glycylproline | 173.09 | 116.10 | 13 | Positive | 6.701 |
| 169 | HMDB0000044 | l-Ascorbic acid | 175.02 | 114.90 | 9 | Negative | 3.027 |
| 170 | HMDB0003357 | Acetylornithine | 175.11 | 70.00 | 25 | Positive | 6.769 |
| 171 | HMDB0000517 | Arginine | 175.12 | 70.00 | 25 | Positive | 11.665 |
| 172 | HMDB0000197 | Indole-3-acetic acid | 176.07 | 158.10 | 13 | Positive | 6.193 |
| 173 | HMDB0000904 | Citrulline | 176.11 | 69.90 | 25 | Positive | 7.020 |
| 174 | HMDB0000259 | Serotonin | 177.10 | 160.00 | 9 | Positive | 4.326 |
| 175 | HMDB0000707 | 4-Hydroxy phenylpyruvic acid | 179.03 | 135.00 | 9 | Negative | 1.273 |
| 176 | HMDB0000660 | Fructose | 179.05 | 89.00 | 5 | Negative | 4.716 |
| 177 | HMDB0033704 | Galactose | 179.05 | 89.10 | 5 | Negative | 4.732 |
| 178 | HMDB0000122 | Glucose | 179.05 | 89.00 | 5 | Negative | 5.410 |
| 179 | HMDB0000169 | Mannose | 179.05 | 58.90 | 17 | Negative | 5.350 |
| 180 | HMDB0000211 | Myoinositol | 179.05 | 135.00 | 1 | Negative | 1.497 |
| 181 | HMDB0000714 | Hippuric acid | 180.07 | 104.90 | 13 | Positive | 2.499 |
| 182 | HMDB0006479 | Glucosamine | 180.09 | 162.00 | 5 | Positive | 6.791 |
| 183 | HMDB0000118 | Homovanillic acid | 181.05 | 136.80 | 5 | Negative | 2.268 |
| 184 | HMDB0003269 | Nicotinuric acid | 181.06 | 134.80 | 17 | Positive | 4.234 |
| 185 | HMDB0000765 | D-Mannitol | 181.07 | 101.10 | 20 | Negative | 5.213 |
| 186 | HMDB0000107 | Dulcitol | 181.07 | 58.90 | 24 | Negative | 5.294 |
| 187 | HMDB0000247 | Sorbitol | 181.07 | 58.70 | 17 | Negative | 5.223 |
| 188 | HMDB0000158 | Tyrosine | 182.08 | 90.90 | 33 | Positive | 5.284 |
| 189 | HMDB0000017 | 4-Pyridoxic acid | 184.06 | 166.10 | 9 | Positive | 1.288 |
| 190 | HMDB0000068 | Epinephrine | 184.10 | 166.10 | 5 | Positive | 5.366 |
| 191 | HMDB0000819 | Normetanephrine | 184.10 | 166.10 | 5 | Positive | 4.462 |
| 192 | HMDB0000807 | 3-Phosphoglyceric acid | 184.98 | 79.00 | 50 | Negative | 7.013 |
| 193 | HMDB0000784 | Azelaic acid | 187.09 | 125.10 | 13 | Negative | 4.020 |
| 194 | HMDB0006029 | Acetyl-l-glutamine | 189.09 | 130.00 | 13 | Positive | 4.651 |
| 195 | HMDB0000715 | Kynurenic acid | 190.05 | 144.00 | 25 | Positive | 2.338 |
| 196 | HMDB0002302 | 3-Indolepropionic acid | 190.09 | 130.10 | 25 | Positive | 1.562 |
| 197 | HMDB0000094 | Citrate | 191.02 | 111.00 | 9 | Negative | 7 |
| 198 | HMDB0000193 | Isocitrate | 191.02 | 111.00 | 13 | Negative | 6.41 |
| 199 | HMDB0000763 | HIAA | 192.07 | 146.10 | 13 | Positive | 4.507 |
| 200 | HMDB0002545 | d-Galacturonic acid | 193.03 | 58.70 | 17 | Negative | 6.662 |
| 201 | HMDB0000127 | Glucuronic acid | 193.03 | 113.00 | 9 | Negative | 6.493 |
| 202 | HMDB0000954 | Ferulic acid | 193.05 | 134.00 | 13 | Negative | 1.512 |
| 203 | HMDB0029965 | Methyl alpha-d-glucopyranoside | 193.07 | 102.80 | 13 | Negative | 4.320 |
| 204 | HMDB0029965 | Methyl-d-mannopyranoside | 193.07 | 58.80 | 21 | Negative | 3.044 |
| 205 | HMDB0000565 | Galactonic acid | 195.05 | 75.00 | 21 | Negative | 6.831 |
| 206 | HMDB0000625 | Gluconic acid | 195.05 | 129.00 | 13 | Negative | 6.900 |
| 207 | HMDB0001847 | Caffeine | 195.09 | 138.00 | 17 | Positive | 1.447 |
| 208 | HMDB0000609 | DOPA | 198.08 | 152.10 | 5 | Positive | 5.838 |
| 209 | HMDB0004063 | Metanephrine | 198.12 | 180.10 | 5 | Positive | 3.959 |
| 210 | HMDB0000624 | Lauric acid | 199.17 | 199.17 | 5 | Negative | 1.224 |
| 211 | HMDB0000792 | Sebacic acid | 201.11 | 139.10 | 17 | Negative | 3.612 |
| 212 | HMDB0003334 | Dimethylarginine | 203.15 | 70.00 | 21 | Positive | 10.306 |
| 213 | HMDB0060484 | Indole-3-pyruvic acid | 204.07 | 163.20 | 5 | Positive | 12.162 |
| 214 | HMDB0000201 | Acetylcarnitine | 204.13 | 85.00 | 17 | Positive | 5.661 |
| 215 | HMDB0000929 | Tryptophan | 205.10 | 187.90 | 5 | Positive | 4.437 |
| 216 | HMDB0000881 | Xanthurenic acid | 206.05 | 131.90 | 29 | Positive | 2.245 |
| 217 | HMDB0000671 | Indole-3-lactic acid | 206.08 | 118.10 | 25 | Positive | 4.453 |
| 218 | HMDB0000639 | Mucic acid | 209.03 | 84.90 | 13 | Negative | 7.078 |
| 219 | HMDB0000684 | Kynurenine | 209.09 | 192.00 | 5 | Positive | 4.483 |
| 220 | HMDB0001511 | Phosphocreatine | 212.05 | 45.00 | 17 | Positive | 1.674 |
| 221 | 2280-85-5 | 6-Methyl-dl-tryptophan | 219.12 | 202.10 | 5 | Positive | 4.071 |
| 222 | HMDB0000210 | Pantothenic acid | 220.12 | 90.00 | 9 | Positive | 4.071 |
| 223 | HMDB0000472 | 5-Hydroxytryptophan | 221.09 | 204.00 | 5 | Positive | 5.236 |
| 224 | HMDB0000215 | Acetylglucosamine | 222.10 | 138.10 | 17 | Positive | 4.505 |
| 225 | HMDB0000853 | N-Acetyl-d-galactosamine | 222.10 | 204.10 | 4 | Positive | 4.387 |
| 226 | HMDB0000215 | N-Acetyl-d-glucosamine | 222.10 | 204.30 | 0 | Positive | 4.487 |
| 227 | HMDB0000742 | Homocysteine | 223.08 | 88.00 | 29 | Positive | 8.604 |
| 228 | HMDB0000732 | 3-Hydroxykynurenine | 225.09 | 207.00 | 5 | Positive | 1.102 |
| 229 | 2387-23-7 | N,N-Dicyclohexylurea | 225.20 | 100.10 | 13 | Positive | 1.239 |
| 230 | HMDB0001904 | 3-Nitrotyrosine | 227.07 | 181.00 | 5 | Positive | 4.801 |
| 231 | HMDB0000033 | Carnosine | 227.12 | 110.00 | 21 | Positive | 7.655 |
| 232 | HMDB0000806 | Myristic acid | 227.20 | 227.20 | 5 | Negative | 1.189 |
| 233 | HMDB0000014 | 2-Deoxycytidine | 228.10 | 112.00 | 9 | Positive | 3.591 |
| 234 | 4300-28-1 | d-Ribose 5-phosphate | 229.01 | 78.80 | 45 | Negative | 6.850 |
| 235 | 66768-39-6 | d-Xylose 5-phosphate | 229.01 | 78.90 | 45 | Negative | 5.200 |
| 236 | HMDB0000012 | 2-Deoxyuridine | 229.08 | 113.00 | 9 | Positive | 2.289 |
| 237 | HMDB0001923 | Naproxen | 229.08 | 169.00 | 33 | Negative | 1.358 |
| 238 | HMDB0014732 | Amiloride | 230.06 | 171.00 | 13 | Positive | 4.192 |
| 239 | HMDB0001389 | Melatonin | 233.13 | 174.00 | 13 | Positive | 1.307 |
| 240 | HMDB0000192 | Cystine | 241.03 | 74.00 | 21 | Positive | 7.980 |
| 241 | HMDB0000826 | Pentadecanoic acid | 241.21 | 241.21 | 5 | Negative | 1.189 |
| 242 | HMDB0000089 | Cytidine | 244.10 | 112.00 | 9 | Positive | 4.229 |
| 243 | HMDB0000296 | Uridine | 245.08 | 113.00 | 5 | Positive | 2.950 |
| 244 | HMDB0000030 | Biotin | 245.10 | 227.00 | 9 | Positive | 3.955 |
| 245 | HMDB0000101 | 2-Deoxyadenosine | 252.11 | 136.10 | 13 | Positive | 2.585 |
| 246 | HMDB0000845 | Neopterin | 254.09 | 206.00 | 21 | Positive | 5.621 |
| 247 | HMDB0000220 | Palmitic acid | 255.23 | 255.23 | 5 | Negative | 1.189 |
| 248 | HMDB0000982 | 5-Methylcytidine | 258.11 | 126.10 | 9 | Positive | 3.932 |
| 249 | HMDB0001078 | d-Mannose 6-phosphate | 259.02 | 79.10 | 52 | Negative | 7.170 |
| 250 | HMDB0000124 | Fructose 6-phosphate | 259.02 | 78.80 | 41 | Negative | 6.87 |
| 251 | HMDB0001586 | Glucose 1-phosphate | 259.02 | 79.10 | 29 | Negative | 6.889 |
| 252 | HMDB0001401 | Glucose 6-phosphate | 259.02 | 78.80 | 41 | Negative | 7.300 |
| 253 | HMDB0001254 | Glucosamine 6-phosphate | 260.06 | 126.10 | 9 | Positive | 7.177 |
| 254 | HMDB0001849 | Propranolol | 260.17 | 116.00 | 17 | Positive | 2.516 |
| 255 | 94-24-6 | Tetracaine | 265.19 | 176.00 | 17 | Positive | 1.534 |
| 256 | HMDB0000085 | 2-Deoxyguanosine | 268.11 | 151.90 | 17 | Positive | 4.000 |
| 257 | HMDB0000050 | Adenosine | 268.11 | 136.00 | 13 | Positive | 3.018 |
| 258 | HMDB0000195 | Inosine | 269.09 | 137.00 | 21 | Positive | 3.794 |
| 259 | HMDB0002259 | Heptadecanoic acid | 269.25 | 269.25 | 5 | Negative | 1.154 |
| 260 | HMDB0001316 | 6-Phosphogluconic acid |
275.01 | 78.90 | 49 | Negative | 13.882 |
| 261 | 506-24-1 | 9-Octadecynoic acid | 279.23 | 279.23 | 5 | Negative | 1.188 |
| 262 | HMDB0003331 | 1-Methyladenosine | 282.12 | 150.00 | 29 | Positive | 6.397 |
| 263 | HMDB0000827 | Stearic acid | 283.26 | 283.26 | 5 | Negative | 1.137 |
| 264 | HMDB0000133 | Guanosine | 284.10 | 152.10 | 13 | Positive | 4.616 |
| 265 | HMDB0000299 | Xanthosine | 285.09 | 153.00 | 9 | Positive | 4.844 |
| 266 | HMDB0060493 | N-Acetylmuramic acid | 292.10 | 89.00 | 8 | Negative | 3.895 |
| 267 | HMDB0000772 | Nonadecanoic acid | 297.28 | 297.28 | 5 | Negative | 1.120 |
| 268 | HMDB0001409 | dUMP | 307.03 | 195.10 | 17 | Negative | 7.235 |
| 269 | HMDB0000125 | Glutathione reduced | 308.09 | 84.10 | 25 | Positive | 6.070 |
| 270 | HMDB0014950 | Phenylbutazone | 309.16 | 76.90 | 45 | Positive | 1.030 |
| 271 | HMDB0000230 | N-Acetylneuraminic acid | 310.12 | 274.00 | 5 | Positive | 6.350 |
| 272 | HMDB0000651 | Decanoylcarnitine | 316.25 | 84.90 | 25 | Positive | 2.332 |
| 273 | HMDB0001227 | dTMP | 323.07 | 80.80 | 21 | Positive | 6.033 |
| 274 | HMDB0001058 | Fructose 1,6 biphosphate (F16BP) | 338.99 | 78.90 | 49 | Negative | 7.9 |
| 275 | HMDB0003514 | Glucose 1,6 biphosphate (G16BP) | 338.99 | 241.10 | 17 | Negative | 8.000 |
| 276 | HMDB0000055 | d-(+)-Cellobiose | 341.11 | 161.00 | 4 | Negative | 6.869 |
| 277 | HMDB0005826 | Galactinol dihydrate | 341.11 | 179.20 | 16 | Negative | 8.318 |
| 278 | HMDB0000186 | Lactose | 341.11 | 161.00 | 5 | Negative | 7.065 |
| 279 | HMDB0000258 | Sucrose | 341.11 | 59.00 | 50 | Negative | 6.474 |
| 280 | HMDB0000975 | Trehalose | 341.11 | 178.90 | 9 | Negative | 7.014 |
| 281 | HMDB0003559 | Gibberellic acid | 345.13 | 143.00 | 37 | Negative | 1.559 |
| 282 | HMDB0001314 | cGMP | 346.06 | 152.00 | 21 | Positive | 4.63 |
| 283 | HMDB0000045 | AMP | 348.10 | 136.20 | 17 | Positive | 6.544 |
| 284 | HMDB0001220 | Prostaglandin E2 | 351.21 | 315.20 | 9 | Negative | 1.474 |
| 285 | HMDB0000939 | Adenosyl-l-homocysteine | 385.13 | 136.00 | 25 | Positive | 6.875 |
| 286 | HMDB0001245 | DCDP | 388.03 | 112.00 | 17 | Positive | 6.922 |
| 287 | HMDB0000774 | Pregnenolone sulfate | 395.19 | 96.80 | 45 | Negative | 1.017 |
| 288 | HMDB0000295 | UDP | 402.99 | 78.90 | 50 | Negative | 6.79 |
| 289 | HMDB0001341 | ADP | 428.04 | 136.10 | 21 | Positive | 6.900 |
| 290 | HMDB0000121 | Folic acid | 442.15 | 295.00 | 25 | Positive | 6.806 |
| 291 | HMDB0001201 | GDP | 444.03 | 152.10 | 33 | Positive | 7.255 |
| 292 | HMDB0001056 | Dihydrofolic acid | 444.17 | 297.10 | 17 | Positive | 6.783 |
| 293 | HMDB0000797 | SAICAR | 455.08 | 110.00 | 44 | Positive | 6.947 |
| 294 | HMDB0000536 | Adenylosuccinate | 464.08 | 252.00 | 25 | Positive | 8.036 |
| 295 | HMDB0000998 | dCTP | 468.00 | 111.90 | 17 | Positive | 7.5 |
| 296 | HMDB0001191 | dUTP | 468.98 | 80.90 | 13 | Positive | 7 |
| 297 | HMDB0001562 | Folinic acid | 474.18 | 327.10 | 17 | Positive | 1.464 |
| 298 | HMDB0003213 | Raffinose | 503.16 | 178.90 | 25 | Negative | 7.912 |
| 299 | HMDB0000538 | ATP | 508.01 | 136.00 | 45 | Positive | 7.350 |
| 300 | HMDB0001178 | ADP ribose | 560.10 | 136.20 | 21 | Positive | 6.020 |
| 301 | HMDB0000290 | UDP-GlcNAc | 608.09 | 204.30 | 9 | Positive | 7.010 |
| 302 | HMDB0003337 | Glutathione oxidized | 613.16 | 484.00 | 17 | Positive | 7.267 |
| 303 | HMDB0000902 | NAD | 664.12 | 136.00 | 41 | Positive | 6.767 |
| 304 | HMDB0003553 | Stachyose hydrate | 665.21 | 383.20 | 40 | Negative | 8.960 |
| 305 | HMDB0001487 | NADH | 666.10 | 348.10 | 17 | Positive | 5.700 |
| 306 | HMDB0000217 | NADP | 745.10 | 604.10 | 17 | Positive | 7.200 |
| 307 | HMDB0000221 | NADPH | 746.10 | 746.10 | 17 | Positive | 6.500 |
| 308 | HMDB0001206 | Acetyl-CoA | 810.14 | 303.10 | 37 | Positive | 6 |
| 309 | HMDB0001243 | Isobutyryl-CoA | 838.17 | 331.20 | 37 | Positive | 7.000 |
| 310 | HMDB0001166 | Hydroxybutyryl coenzyme A | 854.16 | 347.10 | 37 | Positive | 6.954 |
4. Notes
Render ultrapure water by filtering deionized water.
4:6 dilution of PBS:ACN contains 4:1 (v:v) solution of 13C-lactate and 13C-glutamic acid.
Added solutions must be cooled at 4 °C.
Prepare ice for use prior to sonication.
Ensure that the centrifuge is balanced.
Resulting protein pellets may be extracted and saved for protein analysis as needs arise.
Drying time is contingent on the number of samples and type of tissue used. It is permitted to dry samples overnight. If the extracted supernatant cannot be dried immediately, samples may be kept at −20 °C for short-term storage or at −80 °C for long-term storage.
During this time, you may prepare the vial inserts and syringe filters.
It may be necessary to plunge multiple times in order to filter all of the sample. At least 50 μL of filtered sample is required for LC-MS/MS analysis.
A glass insert is not needed for analysis of QC sample given sufficient volume of liquid.
Both chromatographic separations should be performed in HILIC mode.
Targeted data acquisition should be performed in multiple-reaction-monitoring (MRM) mode.
MS parameters were optimized using standard references. All LC separation was performed using basic HILIC. The dwell time should be configured to 5 s for each ion signal. Unit resolution is required for both MS1 and MS2.
References
- 1.Finer M, Glorioso J (2017) A brief account of viral vectors and their promise for gene therapy. Gene Ther 24:1–2 [DOI] [PubMed] [Google Scholar]
- 2.Wirth T, Parker N, Ylä-Herttuala S (2013) History of gene therapy. Gene 525:162–169 [DOI] [PubMed] [Google Scholar]
- 3.Clark JR, March JB (2006) Bacteriophages and biotechnology: vaccines, gene therapy and antibacterials. Trends Biotechnol 24:212–218 [DOI] [PubMed] [Google Scholar]
- 4.Celec P, Gardlik R (2017) Gene therapy using bacterial vectors. Front Biosci 22:81–95 [DOI] [PubMed] [Google Scholar]
- 5.Naso MF, Tomkowicz B, Perry WL et al. (2017) Adeno-associated virus (AAV) as a vector for gene therapy. BioDrugs 31:317–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zinn E, Vandenberghe LH (2014) Adeno-associated virus: fit to serve. Curr Opin Virol 8:90–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Q, Kunder N, De la Paz JA et al. (2018) Global pairwise RNA interaction landscapes reveal core features of protein recognition. Nat Commun 9:2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattiacio JL, Brewer M, Dewhurst S (2017) Methods in molecular biology (Clifton, N.J.) 1581:245–253 [DOI] [PubMed] [Google Scholar]
- 9.Huey R, Hawthorne S, McCarron P (2017) The potential use of rabies virus glycoprotein-derived peptides to facilitate drug delivery into the central nervous system: a mini review. J Drug Target 25:379–385 [DOI] [PubMed] [Google Scholar]
- 10.Zheng D, Chen H, Bartee MY et al. (2012) Virus-derived anti-inflammatory proteins: potential therapeutics for cancer. Trends Mol Med 18:304–310 [DOI] [PubMed] [Google Scholar]
- 11.Patti GJ, Yanes O, Siuzdak G (2012) Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol 13:263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett JR, Holmes E, Veselkov KA et al. (2019) A unified conceptual framework for metabolic phenotyping in diagnosis and prognosis. Trends Pharmacol Sci 40:763–773 [DOI] [PubMed] [Google Scholar]
- 13.Chong J, Wishart DS, Xia J (2018) MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 46:W486–W494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ismail IT, Showalter MR, Fiehn O (2019) Inborn errors of metabolism in the era of untargeted metabolomics and lipidomics. Metabolites 9:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rinschen MM, Ivanisevic J, Giera M et al. (2019) Identification of bioactive metabolites using activity metabolomics. Nat Rev Mol Cell Biol 20:353–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gowda GN, Raftery D (2013) Biomarker discovery and translation in metabolomics. Curr Metabolomics 1:227–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu H, Gowda GN, Raftery D (2012) Metabolic profiling: are we en route to better diagnostic tests for cancer? Future Oncol 8:1207–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jasbi P, Wang D, Cheng SL et al. (2019) Breast cancer detection using targeted plasma metabolomics. J Chromatogr B 1105:26–37 [DOI] [PubMed] [Google Scholar]
- 19.Yin P, Xu G (2017) Methods in molecular biology (Clifton, N.J.) 1619:467–475 [DOI] [PubMed] [Google Scholar]
- 20.Kaysen GA, Johansen KL, Chertow GM et al. (2015) Associations of trimethylamine N-oxide with nutritional and inflammatory biomarkers and cardiovascular outcomes in patients new to dialysis. J Ren Nutr 25:351–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowers J, Hughes E, Skill N et al. (2014) Detection of hepatocellular carcinoma in hepatitis C patients: biomarker discovery by LC–MS. J Chromatogr B 966:154–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahn J, Kim J, Hwang J et al. (2017) Urinary metabolomic profiling to identify potential biomarkers for the diagnosis of Behcet’s disease by gas chromatography/time-of-flight–mass spectrometry. Int J Mol Sci 18:2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madsen R, Lundstedt T, Trygg J (2010) Chemometrics in metabolomics—A review in human disease diagnosis. Anal Chim Acta 659:23–33 [DOI] [PubMed] [Google Scholar]
- 24.Ene IV, Brunke S, Brown AJP et al. (2014) Metabolism in fungal pathogenesis. Cold Spring Harb Perspect Med 4: a019695–a019695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poddighe S, Murgia F, Lorefice L et al. (2017) Metabolomic analysis identifies altered metabolic pathways in multiple sclerosis. Int J Biochem Cell Biol 93:148–155 [DOI] [PubMed] [Google Scholar]
- 26.Monnerat G, Seara FAC, Evaristo JAM et al. (2018) Aging-related compensated hypogonadism: role of metabolomic analysis in physiopathological and therapeutic evaluation. J Steroid Biochem Mol Biol 183:39–50 [DOI] [PubMed] [Google Scholar]
- 27.Zhu J, Djukovic D, Deng L et al. (2015) Targeted serum metabolite profiling and sequential metabolite ratio analysis for colorectal cancer progression monitoring. Anal Bioanal Chem 407:7857–7863 [DOI] [PubMed] [Google Scholar]
- 28.Manchester M, Anand A (2017) Metabolomics: strategies to define the role of metabolism in virus infection and pathogenesis. Adv Virus Res 98:57–81 [DOI] [PubMed] [Google Scholar]
- 29.Gray DW, Welsh MD, Mansoor F et al. (2018) DIVA metabolomics: differentiating vaccination status following viral challenge using metabolomic profiles. PLoS One 13:e0194488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Huang Y, Lian M et al. (2017) Metabolic profiling of hepatitis B virus-related hepatocellular carcinoma with diverse differentiation grades. Oncol Lett 13:1204–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barko PC, McMichael MA, Swanson KS et al. (2018) The gastrointestinal microbiome: a review. J Vet Intern Med 32:9–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandler JD, Hu X, Ko EJ et al. (2016) Metabolic pathways of lung inflammation revealed by high-resolution metabolomics (HRM) of H1N1 influenza virus infection in mice. Am J Physiol Integr Comp Physiol 311:R906–R916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hollenbaugh JA, Montero C, Schinazi RF et al. (2016) Metabolic profiling during HIV-1 and HIV-2 infection of primary human monocyte-derived macrophages. Virology 491:106–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu J, Djukovic D, Deng L et al. (2014) Colorectal cancer detection using targeted serum metabolic profiling. J Proteome Res 13:4120–4130 [DOI] [PubMed] [Google Scholar]
- 35.Carroll PA, Diolaiti D, McFerrin L et al. (2015) Deregulated Myc requires MondoA/Mlx for metabolic reprogramming and tumorigenesis. Cancer Cell 27:271–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu H, Zhang P, Zhu J et al. (2015) Globally optimized targeted mass spectrometry: reliable metabolomics analysis with broad coverage. Anal Chem 87:12355–12362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu H, Carroll PA, Du J et al. (2016) Quantitative method to investigate the balance between metabolism and proteome biomass: starting from glycine. Angew Chemie Int Ed 55:15646–15650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li R, Grimm SA, Mav D et al. (2018) Transcriptome and DNA methylome analysis in a mouse model of diet-induced obesity predicts increased risk of colorectal cancer. Cell Rep 22:624–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buas MF, Gu H, Djukovic D et al. (2017) Candidate serum metabolite biomarkers for differentiating gastroesophageal reflux disease, Barrett’s esophagus, and high-grade dysplasia/esophageal adenocarcinoma. Metabolomics 13:23. [DOI] [PMC free article] [PubMed] [Google Scholar]


