Abstract
Archaeal community structures in microhabitats in a deep-sea hydrothermal vent chimney structure were evaluated through the combined use of culture-independent molecular analyses and enrichment culture methods. A black smoker chimney was obtained from the PACMANUS site in the Manus Basin near Papua New Guinea, and subsamples were obtained from vertical and horizontal sections. The elemental composition of the chimney was analyzed in different subsamples by scanning electron microscopy and energy-dispersive X-ray spectroscopy, indicating that zinc and sulfur were major components while an increased amount of elemental oxygen in exterior materials represented the presence of oxidized materials on the outer surface of the chimney. Terminal restriction fragment length polymorphism analysis revealed that a shift in archaeal ribotype structure occurred in the chimney structure. Through sequencing of ribosomal DNA (rDNA) clones from archaeal rDNA clone libraries, it was demonstrated that the archaeal communities in the chimney structure consisted for the most part of hyperthermophilic members and extreme halophiles and that the distribution of such extremophiles in different microhabitats of the chimney varied. The results of the culture-dependent analysis supported in part the view that changes in archaeal community structures in these microhabitats are associated with the geochemical and physical dynamics in the black smoker chimney.
Since the discovery of deep-sea hydrothermal vents in 1979 (12, 15), various microorganisms have been isolated from global deep-sea hydrothermal vent environments (26, 27, 29). Hyperthermophiles and thermophiles, including members of both the bacterial and archaeal domains, are the most frequently isolated microorganisms, and their physiological properties likely reflect the extraordinary environmental settings of the deep-sea hydrothermal vents (9, 10, 18, 23, 28, 42, 43, 53, 56, 60). In addition, recent culture-independent molecular approaches have revealed the presence of as-yet-uncultivated thermophiles showing substantial phylogenetic diversity in the deep-sea hydrothermal vent environments (27, 38, 39, 44, 51). Relatively little is known about the ecological significance and geomicrobiological function of the extremophilic microbial communities found, as the occurrence, abundance, and distribution of the microbial communities associated with the formation of diverse geochemical and physical gradients remain to be elucidated.
A deep-sea hydrothermal vent chimney, formed by chemical interaction between cold seawater and hot vent water, is a distinctive structure in the deep-sea hydrothermal fields and is largely composed of sulfide materials. In the chimney structures and the underlying sulfide mounds, steep environmental gradients of temperature, pH, oxidation-redox potential, and various chemicals can be formed by equilibration between the vent water and the seawater, and these provide diverse microhabitats for microbial communities. In addition, the presence of similar environmental gradients in the subvent environment beneath the active hydrothermal seafloor is evident. Based on several microbiological, geochemical, and geophysical observations, the possible existence of a subvent biosphere populated by hyperthermophilic microorganisms was predicted (14). Also, microbial ribosomal DNA (rDNA) showing substantial diversity was recovered from black smoker vent water in the Ihaya Basin, Okinawa Trough (51), at a temperature of >300°C, which is far above the upper temperature limit for growth of even the most hyperthermophilic archaeon, Pyrolobus fumarii (113°C) (9). The microbial rDNA may serve as a genetic signature indicative of the microorganisms thriving in the subvent habitats, conveyed there by vent water. Hence, a hydrothermal vent chimney is an environment analogous to the subvent biosphere, and elucidation of its microbial community structure is likely to provide great insight into features of the subvent microbial ecosystem.
The black smoker chimney structure investigated in the present study was obtained from a hydrothermal vent field located at the PACMANUS site in the Manus Basin near Papua New Guinea at a depth of 1,644 m. This hydrothermal field was discovered in 1991 (7), and a geological survey of the field and geochemical characterization of the vent waters have both been performed (6, 7, 16, 17). The results of the microbiological assessment have not yet been reported. Archaeal community structures in microhabitats present in the chimney structure were evaluated through the combined use of culture-independent molecular analyses and enrichment culture methods. The molecular techniques used in this study are PCR-based terminal restriction fragment length polymorphism (T-RFLP) and rDNA clone analyses, and the data provided by these methods should be carefully evaluated. However, the PCR-mediated molecular techniques will allow the possible detection and evaluation of a probably very low biomass of archaeal communities in the hardly accessible deep-sea hydrothermal vent chimney. The distribution of archaeal communities associated with the occurrence of discrete microhabitats in the chimney structure and implications for the possible subvent biosphere are discussed.
MATERIALS AND METHODS
Sample collection, subsampling, and chemical analysis.
A chimney structure was obtained from a black smoker found in a hydrothermal field at the PACMANUS site in the Manus Basin (03°43.816′S, 151°40.016′E) at a depth of 1,644 m by means of the manned submersible Shinkai 2000 in a dive in 1999 (dive no. 1151). The in situ temperature of the effluent black smoker vent water was found to be over 250°C. The chimney structure was immediately cooled and stored during 2 weeks in an anaerobic bag filled with 100% N2 at 4°C prior to subsampling in the laboratory. During the storage, no apparent change was found in the appearance of the chimney structure.
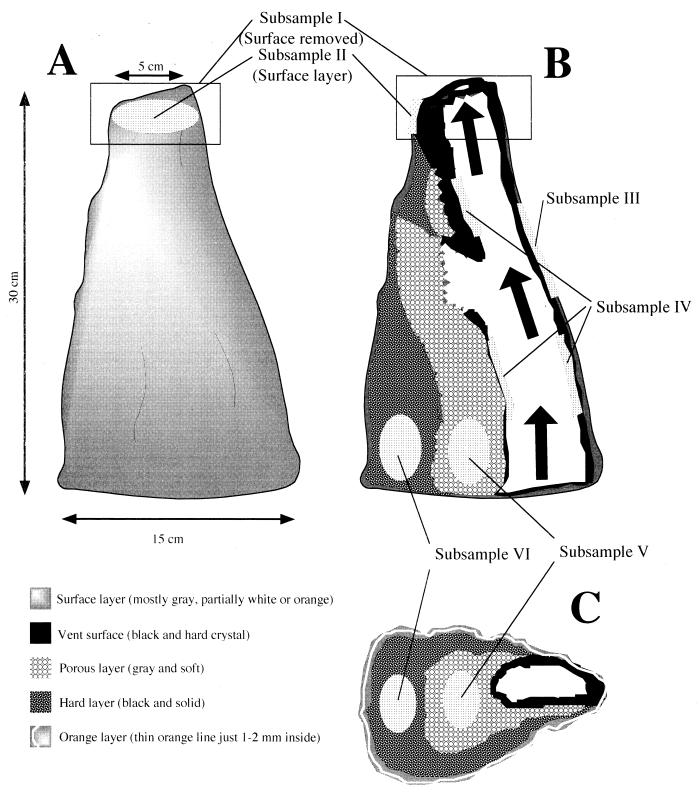
A description of the structure of the chimney and of the identification features of the subsamples is shown in Fig. 1. Subsampling was performed in an anaerobic chamber in an atmosphere of 10% H2 and 90% N2. Each of the subsamples was subjected to chemical analysis, nucleic acid extraction, microscopic observation, and cultivation of microorganisms. Subsamples I and II were obtained from the top part of the chimney. A surface layer (thickness, 1 to 2 mm) with white or gray weathering (subsample II) was moisturized and grazed from the top part of the chimney, and the rest (subsample I) was less moisturized and pulverized using a mortar and pestle which had been sterilized by heating in a dry oven (200°C). A surface layer (thickness, 1 to 2 mm) with white, orange, or gray weathering (subsample III) was also obtained from the middle part of the chimney. The vent surface (subsample IV), which was black, solid, and crystalline and contained little water, was pulverized using a mortar and pestle. Subsamples V and VI were obtained from the inside structure of the root part of the chimney. The inner structure (subsample V) was gray, soft, and porous and contained a considerable amount of water. The outer structure (subsample VI) was black and solid and much less moisturized. The outer structure was pulverized using a mortar and pestle.
FIG. 1.
Schematic drawings of the morphological and structural features of a black smoker chimney and the subsampling method used. (A) Appearance of the surface. (B) Vertical section of the whole chimney structure. (C) Horizontal section of the root part of the chimney. The part where each of the subsamples was taken and the preliminary morphological and structural features of the subsamples are indicated.
The elemental composition of the subsamples was analyzed by scanning electron microcopy and energy-dispersive X-ray spectroscopy (SEM-EDS). The grazed or pulverized samples were directly embedded on specimen mounts using adhesive tape or electroconductives. After natural drying, the specimen was coated with osmium plasma to a thickness of 5 nm by using an osmium plasma coater. The coated specimen was observed using a JEOL JSV-5800LV scanning electron microscope at 15 kV and was analyzed using an energy-dispersive X-ray spectrometer with an ultrathin window at 15 kV. NaCl, BaSO4, BaO, BaS, ZnSO4·7H2O, ZnO, ZnS, and colloidal elemental sulfur (S0) (Nakarai Tesque, Kyoto, Japan) were used as reference substances.
Microscopic observation.
A 2-g portion of the subsample was suspended in 6 ml of sterilized synthetic seawater (MJ) (46, 54) containing 3.7% (wt/vol) formaldehyde, and the mixture was vigorously agitated for 2 min using a vortex mixer. The suspension solution was very briefly centrifuged (3,000 × g), and the supernatant was filtered with a 0.22-μm-pore-size 13-mm-diameter polycarbonate filter (Advantec, Tokyo, Japan). The filter was stained by treatment with MJ seawater containing 4′,6′diamidino-2-phenylindole (DAPI) (10 μg ml−1) or acridine orange (10 μg ml−1) at 4°C for 20 min. The filter was briefly rinsed in MJ seawater and examined under epifluorescence using a Nikon Optishot microscope.
Extraction of nucleic acids.
Microbial DNA was directly extracted from each subsample (approximately 5 g) using a Soil DNA Kit Mega Prep (MO BIO Laboratories, Inc., Solana Beach, Calif.), following the manufacturer's suggested protocol. Microbial RNA was extracted from a replicate aliquot of each of the samples used for DNA extraction. Approximately 5 g of sample material was suspended in 3.5 ml of extraction buffer (pH 4.2) containing 25 mM sodium acetate, 5 mM EDTA, and 5% (wt/vol) sodium dodecyl sulfate. Then 2.5 g of sterile glass beads (0.1-mm diameter; Sigma) and 3.5 ml of phenol equilibrated with extraction buffer were added. The suspension was shaken on a bead beater for 2 min and incubated at 60°C for 1 h. The shaking treatment was repeated, and the suspension was centrifuged at room temperature. The lysate was extracted with phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol) and with chloroform-isoamyl alcohol (24:1, vol/vol). RNA was precipitated by adding 1.5 ml of 10 M ammonium acetate and the same volume of isopropyl alcohol and was recovered by centrifugation. The pellet was washed with 70% (vol/vol) ethanol and dissolved in filter-sterilized, distilled, deionized water. The total RNA was purified from the crude RNA solution using a RNeasy Midi Kit spin column (Qiagen, Valencia, Calif.). All plasticware and all solutions used for RNA extraction and purification were treated with 0.1% diethyl pyrocarbonate to inactivate nucleases. In order to check for laboratory contaminants, a blank tube (with no sample added) was processed as a negative control in the case of both DNA and RNA extraction (57). RNA and DNA concentrations were measured using a spectrophotometer.
Quantification of archaeal rRNA and the rDNA population.
Quantification of archaeal rRNA in the whole microbial RNA assemblages extracted from the subsamples was performed by RNA dot blot hybridization. A dilution series of RNA samples (1, 0.5, and 0.25 ng μl−1) with 10 mM Tris-HCl (pH 7.5) were prepared, the RNA was denatured at 100°C for 10 min, and the samples were cooled on ice. The denatured RNA samples were dotted onto Hybond-N+ nylon membranes (Amersham Pharmacia) and cross-linked to the membranes by exposure to 120 mJ of UV light energy using a UV Stratalinker 1800 (Strategene, Torrey Pines, Calif.). The oligonucleotide probes (Uni1392R and Arch915R conjugated at the 5′ ends to digoxigenin) (52) and the hybridization conditions used have been described previously (52). Quantification of the archaeal rDNA population in the whole microbial DNA assemblages was performed by a quantitative fluorescent PCR method as previously described (52). An archaeal rDNA mixture containing equal amounts of eight kinds of archaeal rDNA (Haloarcula japonica, Palaeococcus ferrophilus, Pyrobaculum sp., Sulfurisphaera sp., pISA42, pISA48, pISA16, and pMC1A11) was used as a standard (52).
T-RFLP analysis of archaeal rDNA.
In order to rapidly identify archaeal sequences in the subsamples, T-RFLP analysis of rDNA was performed (34). Archaeal rDNA was amplified by PCR using LA Taq polymerase (TaKaRa, Kyoto, Japan). The oligonucleotide primers used were Arch21F and Arch958R-FAM (13). Reaction mixtures were prepared in which the concentration of each oligonucleotide primer was 0.1 μM and that of the DNA template was 0.1 ng μl−1. Thermal cycling was performed using GeneAmp 9600 (Perkin-Elmer, Foster City, Calif.), and the conditions were as follows: denaturation at 96°C for 20 s, annealing at 50°C for 45 s, and extension at 72°C for 120 s for a total of 30 cycles. In cases in which no apparent product was recovered after 30 cycles of reaction, the number of cycles was extended to 45 cycles. In the fewer cycles of the reaction (20 and 25 cycles for 30 cycles and 35 and 40 cycles for 45 cycles), no apparent product was obtained.
Amplified rDNA from two separate reactions was pooled and subjected to agarose gel electrophoresis. The products were purified by means of a Gel Spin DNA purification kit (MO BIO). The DNA was precipitated with ethanol and centrifuged, and the pellet was resuspended in the distilled deionized water. The purified rDNA was digested with a restriction enzyme (HhaI). The terminal restriction fragments (T-RFs) were analyzed using a model 377 automated sequencer equipped with GeneScan software, version 3.0 (PE Applied Biosystems, Foster City, Calif.). The precise lengths of T-RFs were determined by comparison with an internal size standard added to each digested sample. The electrophoresis conditions and the procedures followed were those suggested in the manufacturer's protocol. The archaeal T-RFLP profiles from the subsamples were repeatedly checked in a different series of experiments for PCR amplification, restriction enzyme reaction, and electrophoresis.
Cloning and sequencing of archaeal rDNA.
Archaeal rDNA was amplified by PCR using the same protocol as for T-RFLP analysis except for the use of a nonlabeled primer set of Arch21F and Arch958R. Amplified rDNA from two separate reactions was purified as described above. The purified rDNA was cloned in the vector pCR2.1 using the Original TA cloning kit (Invitrogen, Carlsbad, Calif.). The inserts were amplified by direct PCR from a single colony using M13 primers (36), treated with exonuclease I and shrimp alkaline phosphatase (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, United Kingdom), and directly sequenced by the dideoxynucleotide chain-termination method using a dRhodamine sequencing kit (PE Applied Biosystems) following the manufacturer's recommendations. The Arch21F primer was used in partial sequencing analysis.
Sequence and phylogenetic analyses.
Single-strand sequences approximately 400 nucleotides in length were analyzed. The sequence similarity among all of the single-strand sequences was analyzed using the FASTA program equipped with DNASIS software (Hitachi Software, Tokyo, Japan). The rDNA sequences having ≥98% similarity as determined by the FASTA program were assigned to the same clone type, and a representative sequence of each clone type was applied to sequence similarity analysis with databases by the gapped-BLAST method (1, 8). The databases used for gapped-BLAST analysis were the prokaryotic small-subunit (SSU) rRNA database and the nonredundant nucleotide sequence databases from GenBank, EMBL, and DDBJ.
Sequences of representative rDNA clones approximately 0.95 kb in length were determined from both strands. The data from the rDNA clone libraries and the data from T-RFLP analysis were linked by calculating the T-RF length for the sequenced clones. Clones with a T-RF length identical (within ±1 bp) to one found by T-RFLP analysis were assigned to a T-RFLP ribotype. The sequences were manually aligned to prokaryotic SSU rDNA data from the Ribosomal Data Project II (35) based on primary and secondary structure considerations and were also submitted to analysis using the Chimera Check program of the Ribosomal Data Project II to detect the presence of chimeric artifacts. Phylogenetic analyses were restricted to nucleotide positions between the Arch21F and Arch958R primers that were unambiguously alignable in all sequences. Neighbor-joining analysis was performed using the PHYLIP package (version 3.5; obtained from J. Felsenstein, University of Washington, Seattle). Bootstrap analysis was used to provide confidence estimates for phylogenetic tree topologies.
Enrichment and MPN calculation.
For enrichment cultures targeting members of the genus Thermococcus, MJYPS medium (56) was used and enrichment was performed at 55, 75, and 90°C. For enrichment cultures targeting halophilic microorganisms, including members of the genus Haloarcula, the medium (hemagglutinin [HA] medium) described by Ihara et al. (24) was used and enrichment was performed at 30 and 45°C. The three-tube most-probable-number (MPN) method was employed to assess the populations of viable cells obtained by enrichment from the subsamples. The microorganisms present in the most diluted of the series of cultures of MJYPS and HA media were isolated by the extinction-dilution method. The partial sequences of the 16S rDNA were determined and applied to sequence similarity analysis.
Nucleotide sequence accession numbers.
The sequences from this study are available through DDBJ under accession numbers AB052972 to AB052993.
RESULTS
Description of subsamples and chemical composition.
Subsamples of the chimney structure were obtained from vertical and horizontal sections distinguished on the basis of morphological features. In the case of hydrothermal vents with slowly flowing effluent, it has often been observed that the top part of the chimney is covered with weakly packed materials, consisting of chalcopyrite, pyrrhotite, and anhydrite on solid sulfide materials (21, 22, 33). The chimney investigated in the present study was obtained from a black smoker with vigorously flowing effluent, and no such soft head-structure was observed. The vent surface, the interface with vent fluid, was formed by a solid, crystalline vent wall, which was observed throughout top to bottom of the chimney structure. The morphological features of the top part were similar to those found on the vent surface. In the middle and root parts, four distinct inner structures were observed (Fig. 1). Considering the features from inside towards the outer surface, a solid, crystalline vent wall, a grayish, porous soft structure, a black solid structure, and a thin orange layer approximately 1 to 2 mm from the surface were recognized. The surface of the chimney was weathered and gray for the most part, with white or orange in some parts. These morphological features might be associated with geochemical and physical gradients of temperature, pH, oxidation-redox potential, and various chemicals formed in the chimney structure.
The primary chemical composition of the subsamples was examined by SEM-EDS (Table 1). The whole chimney structure was found to contain mainly elemental sulfur and zinc as the primary elements, suggesting that zinc sulfide is the main chemical fabric of the chimney. In the subsamples obtained from the surface of the chimney, a higher amount of elemental oxygen was observed. Although the molecular species containing oxygen was not identified, it seemed likely that the higher proportion of oxygen detected in the elemental composition was consistent with the increasing amount of oxidized materials on the outer surface of the chimney, indicating the occurrence of relatively oxidative microhabitats in the exterior zones of the chimney. In the weathered surface parts that were white and orange and in the orange subsurface layer, barium was accumulated at high levels while no significant change in the amount of calcium was observed.
TABLE 1.
Primary chemical composition of subsamples from a black smoker chimney obtained from the PACMANUS sitea
| Subsample used | Chemical composition (atom%)
|
||||||
|---|---|---|---|---|---|---|---|
| O | S | Ca | Ba | Fe | Zn | Other | |
| I | 1.2 | 41.0 | ND | ND | 1.8 | 58.0 | ND |
| II (white) | 5.0 | 37.5 | 0.6 | 3.8 | 0.6 | 53.1 | ND |
| II (gray) | 6.5 | 30.9 | 0.5 | 1.8 | 1.1 | 54.0 | 5.3 |
| III (gray) | 0.5 | 37.2 | 0.2 | 0.4 | ND | 55.9 | ND |
| III (white and orange) | 19.3 | 35.8 | ND | 19.6 | ND | 25.1 | 0.5 |
| IV | 0.8 | 39.4 | ND | ND | 1.4 | 61.1 | ND |
| V | 0.2 | 39.8 | 0.2 | 0.2 | 1.5 | 60.2 | 0.2 |
| VI | 2.4 | 40.0 | 0.5 | ND | 1.9 | 56.1 | ND |
| Subsurface orange layer | 18.0 | 40.1 | 1.8 | 23.6 | 0.2 | 19.1 | ND |
Numbering of subsamples was indicated in Fig. 1. ND, not detected.
Microbial population density and nucleic acid extraction.
The microbial population density was determined by direct counting of DAPI- and acridine orange-stained cells (Table 2). The highest population density was observed in the surface layer at the top part of the chimney (subsample II) and was 3.2 × 107 cells g (wet weight)−1. The top part from which the surface was removed (subsample I) and the porous structure inside the chimney (subsample V) had somewhat higher population densities (1.0 × 106 and 8.0 × 105, respectively) than did the other subsamples (approximately 5 × 105).
TABLE 2.
Preliminary microbiological characterization of subsamples from a black smoker chimney at the PACMANUS sitea
| Subsample no. | Total amt (g) | Cell density (cells/g [wet wt]) | DNA yield (ng/g [wet wt]) | RNA yield (ng/g [wet wt]) | rRNA/rDNA composition (%) for:
|
|
|---|---|---|---|---|---|---|
| Bacteria | Archaea | |||||
| I | 30 | 1.0 × 106 | 5.1 | 2.2 | 78.3/92.6 | 19.9/7.4 |
| II | 15 | 3.2 × 107 | 169 | 100 | 66.8/94.7 | 30.5/5.3 |
| III | 36 | 4.5 × 105 | 3.2 | 1.3 | 97.6/100 | 1.8/ND |
| IV | 18 | 4.5 × 105 | 1.6 | 1.4 | 98.5/97.2 | 2.8/2.8 |
| V | 19.5 | 8.0 × 105 | 3.0 | 4.5 | 95.1/100 | 2.2/ND |
| VI | 15 | 5.1 × 105 | 1.6 | 1.7 | 76.7/92.8 | 23.1/7.2 |
Prokaryotic universal, bacterial, and archaeal SSU rRNA/rDNA composition was quantified by using RNA dot hybridization or quantitative fluorogenic PCR (52). The proportions represent the result by RNA dot hybridization divided by the result by quantitative fluorogenic PCR. ND, not detected.
Nucleic acids were directly extracted from the subsamples. Both DNA and RNA were isolated at concentrations in proportion to the observed population densities (Table 1). Based on the DNA yield from each sample and assuming an average cellular DNA content of 2 fg (2), the calculated microbial population density in the subsamples was 8 × 105 to 8.5 × 107 cells g wet weight−1 and was thus in relatively good agreement with the population densities determined (Table 1).
Quantification of archaeal rRNA and rDNA populations.
Archaeal rRNA and rDNA in the whole microbial RNA and DNA assemblages extracted from the subsamples were quantified by RNA dot blot hybridization and quantitative fluorogenic PCR (Table 2). Throughout the subsamples, the proportion of bacterial rRNA or rDNA was higher than that of archaeal rRNA or rDNA in the whole microbial nucleic acid assemblages. However, the proportion of archaeal rRNA and rDNA varied among the subsamples. In the top part of the chimney and the outer part of the inside structure of the root part, the proportion of both archaeal rRNA and rDNA was increased while the proportion of the archaeal rRNA and rDNA in the other subsamples was a few percent or below the detection limit, in the whole microbial nucleic acid assemblages. These results suggested that the sizes of the archaeal communities varied in different microhabitats in the black smoker chimney.
Molecular phylogenetic analyses.
Profiles of archaeal ribotypes dominantly recovered from the DNA assemblages of the subsamples were examined by PCR-mediated T-RFLP analysis (Fig. 2). Archaeal rDNA clones obtained from the subsamples were characterized by partial sequencing (ca. 400 nucleotides) and sequence similarity analysis. The number of archaeal rDNA clones characterized varied from 44 to 65 for each of the samples (Table 3). In addition, sequences of representative rDNA clones that were approximately 0.95 kb in length were determined from both strands and were then applied to the phylogenetic analysis (Fig. 3). The data from the representative DNA clone sequences and the data from T-RFLP analysis were linked by calculating the T-RF length for the sequenced clones.
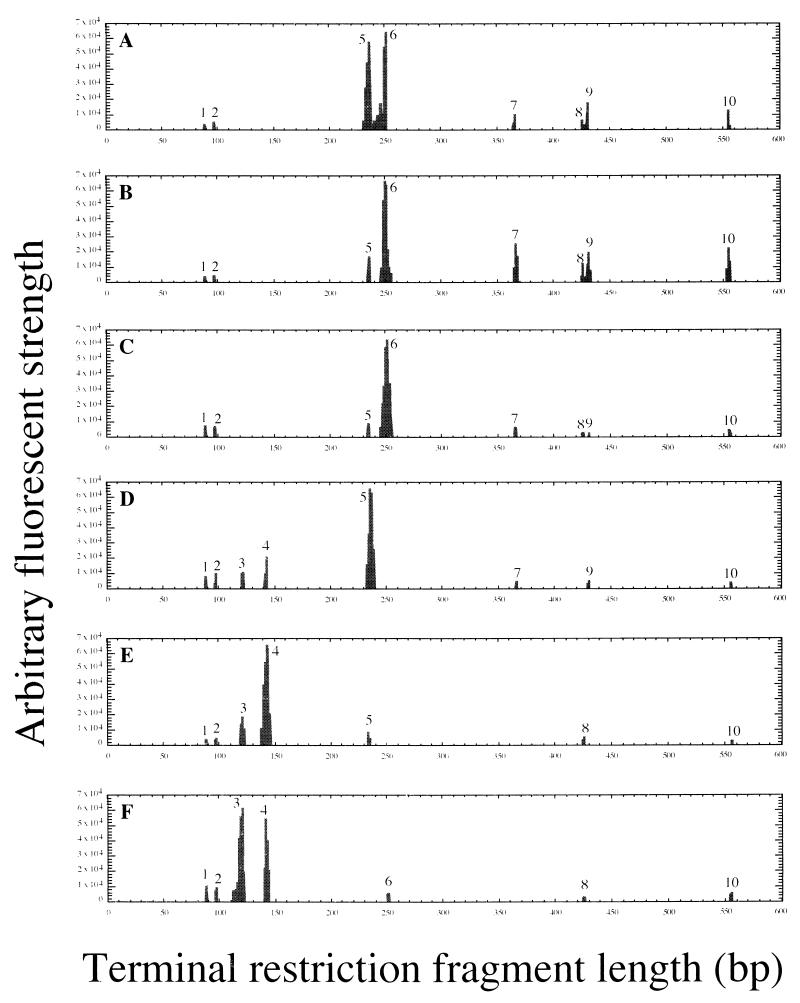
FIG. 2.
Typical electropherograms of archaeal T-RFLP generated from rDNA with a labeled reverse primer and HhaI digest obtained from subsamples. The numbers on the peaks indicate major ribotypes commonly observed in various subsamples. Shown are a typical archaeal pattern from the top part of the chimney (subsample I) (A), the surface layer grazed from subsample I (subsample II) (B), the surface layer grazed from the middle part of the chimney (subsample III) (C), the vent surface (subsample IV) (D), the inner soft and porous structure in the root part of the chimney (subsample V) (E), and the outer black and solid structure in the root part of the chimney (subsample VI) (F).
TABLE 3.
Distribution of representative T-RFLP ribotypes and clone types of archaeal rDNA obtained from a black smoker chimneya
| Clone type (T-RFLP ribotype) | No. of archaeal rDNA clones characterized per subsample:
|
|||||
|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | |
| MHVG-1 | ||||||
| pPACMA-Y (9) | 1 | 3 | 0 | 0 | 0 | 0 |
| Crenarchaeota | ||||||
| Igniococcales | ||||||
| pPACMA-I (5) | 7 | 2 | 0 | 11 | 0 | 0 |
| pPACMA-X (5) | 13 | 4 | 0 | 25 | 3 | 0 |
| Euryarchaeota | ||||||
| Thermococcales | ||||||
| pPACMA-C (6) | 19 | 25 | 29 | 0 | 0 | 0 |
| pPACMA-A (6) | 0 | 0 | 9 | 0 | 0 | 0 |
| pPACMA-B (6) | 0 | 0 | 2 | 0 | 0 | 0 |
| Unknown group | ||||||
| pPACMA-N (8) | 1 | 3 | 0 | 0 | 2 | 0 |
| DHVEG | ||||||
| pPACMA-M (10) | 0 | 5 | 0 | 0 | 0 | 0 |
| pPACMA-Q (10) | 2 | 0 | 1 | 0 | 0 | 0 |
| pPACMA-W (7) | 0 | 1 | 1 | 0 | 0 | 0 |
| pPACMA-P (7) | 1 | 2 | 2 | 0 | 0 | 0 |
| pPACMA-V (7) | 0 | 3 | 1 | 0 | 0 | 0 |
| Halobacteriales | ||||||
| pPACMA-H (3) | 0 | 0 | 0 | 0 | 9 | 21 |
| pPACMA-T (3) | 0 | 0 | 0 | 0 | 0 | 7 |
| pPACMA-J (4) | 0 | 0 | 0 | 1 | 0 | 0 |
| pPACMA-L (3) | 0 | 0 | 0 | 1 | 5 | 8 |
| pPACMA-U (4) | 0 | 0 | 0 | 0 | 0 | 5 |
| pPACMA-K (4) | 0 | 0 | 0 | 2 | 0 | 0 |
| pPACMA-E (4) | 0 | 0 | 0 | 3 | 31 | 10 |
| pPACMA-S (4) | 0 | 0 | 0 | 0 | 1 | 0 |
| pPACMA-G (4) | 0 | 0 | 0 | 1 | 0 | 0 |
| pPACMA-F (4) | 0 | 0 | 0 | 0 | 13 | 2 |
| Total | 44 | 48 | 45 | 44 | 65 | 53 |
Composition of archaeal rDNA clone types was determined by partial sequencing analysis of rDNA clones of archaeal primer PCR libraries.
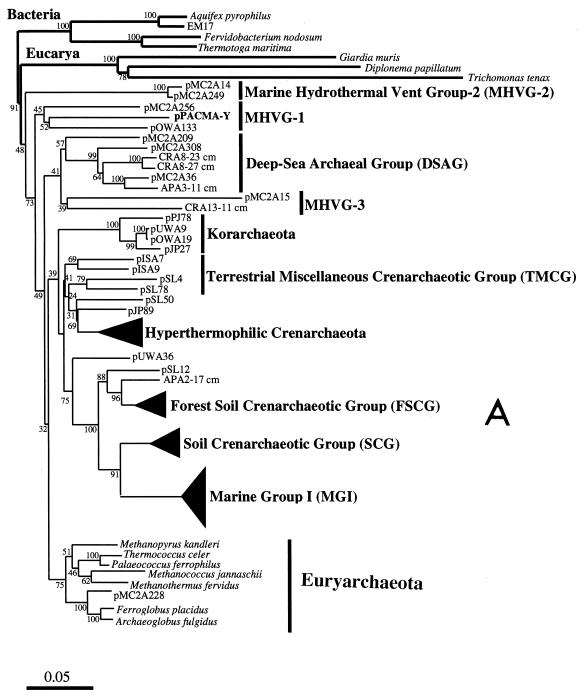
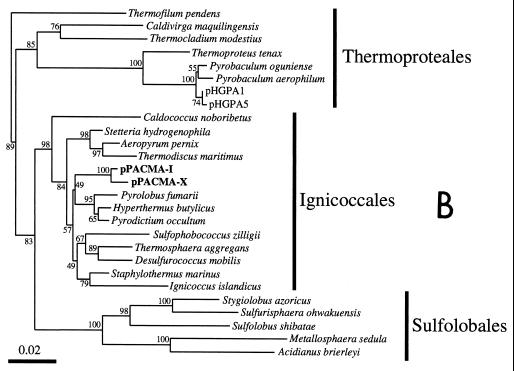
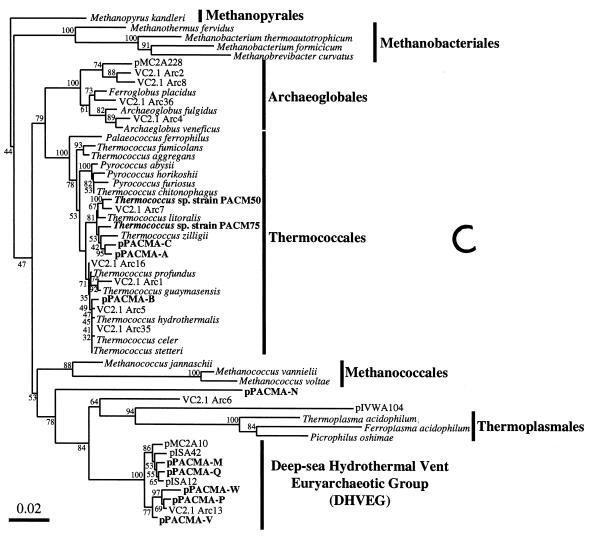
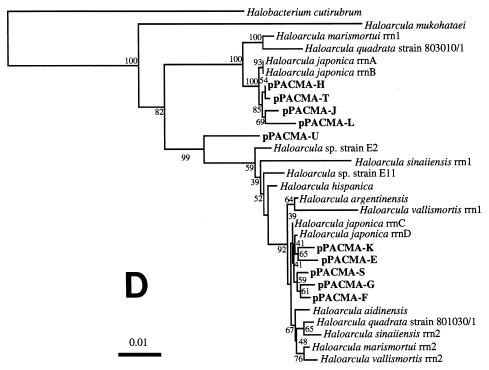
FIG. 3.
Phylogenetic trees based on SSU rDNA sequences including various rDNA clones obtained from the black smoker chimney at the PACMANUS site in the Manus Basin. Each of the trees was inferred by neighbor-joining analysis of approximately 600 homologous positions of the rDNA sequence. (A) A tree indicating the phylogenetic relationship among the domains of Bacteria and Eucarya, the deep branches of uncultivated archaea, and the crenarchaeotic and euryarchaeotic kingdoms. (B) A tree indicating the phylogenetic organization among the hyperthermophilic crenarchaeota, Thermoproteales, Igniococcales, and Sulfolobales. (C) A tree indicating the phylogenetic relationship within the hyperthermophilic and thermophilic euryarchaeota and the possible thermophilic euryarchaeotic phylotypes. (D) A tree indicating the phylogenetic organization within the genus Haloarcula. The numbers on the branches represent the bootstrap confidence values. The scale bars indicate the expected changes per sequence position. Abbreviations indicate rDNA clones corresponding to uncultivated organisms derived from the following environments: pMC1A, pMC2A, pISA, and pIVWA from deep-sea hydrothermal vent environments (51); pOWA and pUWA from shallow marine hydrothermal vent water and terrestrial acidic hot spring water, respectively (55); pJP and pSL from sediments in Yellowstone National Park hot springs (3, 4); CRA and APA from deep-sea sediments (59); pHGPA from deep subsurface geothermal water (50); VC2.1 Arc from an in situ growth chamber (Vent Cap) deployed at a deep-sea hydrothermal vent (44); and pPACMA from a black smoker chimney at the PACMANUS site in the Manus Basin.
The molecular techniques used in this study were the PCR-based T-RFLP and rDNA clone analyses, and the data provided by these methods did not completely represent the archaeal community structures that occurred in the microhabitats of the chimney. In addition, unstable and strongly biased results have been pointed out in the nonoptimized experiments using PCR-mediated T-RFLP and rDNA clone analyses (37, 41). The data obtained should be carefully estimated.
In both T-RFLP and rDNA clone analyses, the structure of archaeal rDNA phylotypes was varied in the subsamples from different microhabitats (Fig. 2 and Table 3). In the combined characterization of T-RFLP and rDNA clone analyses, most of the major T-RFs found in different subsamples had the corresponding archaeal phylotypes identified by rDNA clone analysis, while no archaeal rDNA phylotype coincident with T-RF designated as ribotype 1 or 2 was found throughout all the subsamples (Fig. 2 and Table 3). In addition, there were cases in which the rDNA clones that corresponded to the T-RFs found in the T-RFLP analysis were not obtained from the same subsample in the rDNA clone analysis. This nonconformity between the T-RFLP profiles and rDNA clone compositions might result from the unavoidable cloning bias in Escherichia coli or from the underestimation in the limited number of rDNA clones sequenced.
The top part of the chimney (subsamples I and II) and the surface layer of the middle part (subsample III) contained a variety of rDNA clones phylogenetically belonging to both Crenarchaeota and Euryarchaeota (Table 3). An archaeal rDNA clone, pPACMA-Y, recovered from the archaeal rDNA clone libraries of the top part of the chimney, was phylogenetically associated with one of the deepest branches within the domain of Archaea conventionally named the Marine Hydrothermal Vent Group (MHVG) (Fig. 3A). The T-RF probably representing the deeply branched archaeal phylotype was detected in the top part of the chimney (subsamples I and II), the surface layer of the middle part (subsample III), and the vent surface (subsample IV) (Fig. 2). The archaeal phylotypes predominantly obtained from the top part of the chimney were affiliated with the clusters of Igniococcales, Thermococcales, and the uncultivated Deep-Sea Hydrothermal Vent Euryarchaeotic Group (DHVEG) (Fig. 3B and C). The archaeal rDNA clones affiliated with Igniococcales were especially closely related to the most hyperthermophilic archaeal isolates, such as Pyrolobus (9), Pyrodictium (42), and Hyperthermus (60), capable of growth at or above 110°C (Fig. 3B). All of the Thermococcales rDNA clones obtained from the chimney were closely related with Thermococcus, and no Pyrococcus-type clones were detected (Fig. 3C). The archaeal rDNA clones, pPACMA-M, -Q, -W, -P and -V, were clustered into a certain phylogenetic group consisting of only uncultivated environmental clones obtained from geologically and geographically distinct deep-sea hydrothermal vent environments (44, 51) (Fig. 3C). The proportion of Igniococcales phylotypes was reduced in the rDNA clone libraries from the surface layer samples (subsamples II and III), while the proportion of rDNA clones phylogenetically associated with Thermococcus and DHVEG was greater in the surface zone (Table 3). These proportion changes found in the rDNA clone analysis were also indicated by the changes of T-RF signals (ribotypes 5, 6, 7, and 10) representing the archaeal phylotypes (Igniococcales, Thermococcus, and DHVEG) in the T-RFLP analysis (Fig. 2).
In the vent surface (subsample IV), most of the archaeal rDNA phylotypes provided by rDNA clone analysis were Igniococcales members (Table 3 and Fig. 3B). The dominant occurrence of these phylotypes was clearly represented by a large single peak of ribotype 5 in the T-RFLP analysis (Fig. 2). In addition, the presence of ribotype 4 was detected in the vent surface (Fig. 2), which matched well with the recovery of rDNA clones phylogenetically related with Halobacteriales (Table 3).
In the inside structures of the chimney (subsamples V and VI), the major archaeal phylotypes found in the rDNA clone analysis shifted from hyperthermophilic groups of phylotypes to an extremely halophilic group of phylotypes within Halobacteriales (Table 3). These shifts in the archaeal rDNA clone community structure were consistent with the shifts in the ribotype structure revealed by the results of T-RFLP analysis (depletion of signatures of ribotypes 5 and 6 corresponding to rDNA clones of Igniococcales and Thermococcales, respectively, and predominance of ribotypes 3 and 4 representing Halobacteriales rDNA clones) (Fig. 2). In the phylogenetic analysis, all of the archaeal rDNA clones related with Halobacteriales were located within the phylogenetic cluster of the genus Haloarcula (Fig. 3D). The phylogenetic placement of the Haloarcula rDNA clones obtained from the inside structures of the chimney was divided mainly into two lineages. This dispersion and the relatively high divergence of the Haloarcula rDNA clone sequences could be due to the heterogeneity of the rRNA genes observed in Haloarcula species (40). The archaeal rDNA clone pPACMA-N distantly related to any other archaeal rDNA sequences known so far (Fig. 3C) was detected at a small proportion in various microhabitats both in the rDNA clone analysis and the T-RFLP analysis (Table 3 and Fig. 2). The results obtained from the molecular phylogenetic analyses indicated that different archaeal phylotype structures consisting mainly of hyperthermophilic and extremely halophilic phylotypes were distributed in different microhabitats in the black smoker chimney.
Enrichment and MPN calculations.
In order to support the apparent distribution of different archaeal community structures indicated by culture-independent molecular phylogenetic analyses, enrichment cultures of viable hyperthermophilic and halophilic microbial populations were prepared and MPN calculations were performed. Techniques for cultivation of Thermococcus and Haloarcula members, among the archaeal phylotypes identified through molecular analyses, are relatively well established (5, 11, 45). Table 4 shows the viable-cell counts of Thermococcus members and halophilic microorganisms cultivated at several temperatures in MJYPS and HA media. The microorganisms that grew in the most diluted series of cultures in MJYPS medium at 50 or 75°C containing subsample II or in HA medium at 30°C containing subsample V were isolated by the extinction-dilution method. Based on the results of similarity analysis of partial rDNA sequences, the microorganisms that grew in MJYPS medium at 50 and 75°C were found to be Thermococcus species (Thermococcus spp. PACM50 and PACM75, shown in Fig. 3C). However, the microorganism that grew in HA medium at 30°C was identified as a Halomonas sp. (its partial 16S rDNA sequence had 98.5% similarity with that of Halomonas meridiana [25]), a member of the Bacteria, not the Archaea. HA medium contains approximately 16% (wt/vol) NaCl or 20% total salts, which is within the range for growth of not only Haloarcula spp. but also of Halomonas spp. (58). In addition, no archaeal rDNA was detected by PCR analysis of DNA extracts from any of the enrichment cultures in HA medium at any cultivation temperature. These results indicated that the viable cell counts obtained by the MPN method in the case of cultures using HA medium represented the halophilic bacterial population, not the Haloarcula population. Although no viable cells of the genus Haloarcula were recovered from any of the subsamples, the findings obtained from the enrichment cultures supported the view that a viable Thermococcus population was present in the surface layers and that preferential colonization of the inside structures of the chimney by halophilic microorganisms occurred.
TABLE 4.
Viable-cell counts of Thermococcus members and halophilic microorganisms calculated by three-tube MPN cultivation methoda
| Subsample no. | Viable-cell count in:
|
||||
|---|---|---|---|---|---|
| MJYPS medium for Thermococcus (cells g [wet wt]−1)
|
HA medium for Haloarcula (cells g [wet wt]−1)
|
||||
| 55°C | 75°C | 90°C | 30°C | 45°C | |
| I | 4.6 × 102 | 2.4 × 102 | 3.9 × 101 | ND | ND |
| II | 2.1 × 104 | 1.5 × 104 | 9.0 × 103 | ND | ND |
| III | 7.5 × 102 | 4.3 × 102 | 7.5 × 101 | ND | ND |
| IV | 9.0 × 100 | 9.0 × 100 | ND | 9.3 × 101 | ND |
| V | ND | ND | ND | 9.3 × 102 | 9.3 × 101 |
| VI | ND | ND | ND | 7.5 × 102 | 9.0 × 101 |
Viable-cell counts obtained by the MPN method using HA medium represent not Haloarcula but halophilic bacteria such as Halomonas spp, as described in Results. ND, not detected.
DISCUSSION
The distribution of archaea in a black smoker chimney structure was analyzed by culture-independent, molecular phylogenetic techniques. The combined use of rRNA dot slot hybridization, quantitative fluorogenic PCR, T-RFLP, and rDNA clone analysis revealed that the archaeal phylotype community was significantly altered in terms of size and structure with microhabitats varying at several centimeters distance in the deep-sea hydrothermal vent chimney. The occurrence of discrete archaeal phylotype communities was likely associated with the formation of environmental gradients of temperature, pH, oxidation-redox potential, and various chemicals in the chimney structure, even though the measurement of the gradients was not completely determined in this study.
The occurrence of discrete microbial communities in deep-sea hydrothermal vent environments was first reported by Harmsen et al. (19) after applying enrichment culture and whole-cell fluorescence in situ hybridization (FISH) techniques (20) to analysis of black smoker chimneys obtained from the Mid-Atlantic Ridge. It was demonstrated that the microbial community density was varied in subsamples obtained from different parts of a chimney structure and that the top part had the highest density (19). In addition, the increased density of the total archaeal population in the top part of the chimney was observed by using the fluorescence in situ hybridization technique, and the preferred colonization patterns of potential Thermococcales members in the surface area and of potential Igniococcales members in the top parts and the vent area were indicated by enrichment culture techniques (19). A similar distribution pattern of microbial communities was observed in our analysis of archaeal community structures in different microhabitats of a black smoker chimney from the Manus Basin, which is geologically and geographically distinct from those in the Mid-Atlantic Ridge. These features of the distribution of microorganisms commonly observed in different deep-sea hydrothermal vent systems provide an important general aspect to be considered in assessment of the global deep-sea hydrothermal vent microbial ecosystem.
Significant changes in microbial abundance, diversity, and community structure associated with environmental gradients in hydrothermal environments have also been observed in a series of investigations focusing on a shallow marine hydrothermal vent site as a model system (47–49). Through the use of molecular techniques, an increased abundance of the archaea was found in the active venting area and complex, active, bacterial communities became predominant with increasing distance from the venting point (47–49). At the shallow marine hydrothermal vent site, temperature and pH ranging from approximately 70°C at pH 5 to 20°C at pH 7 were formed over a distance of several meters (47). The potential impact of the environment on a microbial community might be much more intense in deep-sea hydrothermal systems, since gradients are expected to exist in the range of approximately 300°C at pH 3 to 4°C at pH 6 to 7 in the case of a black smoker investigated (16, 17) and from 350 to 400°C to 4°C in the case of the typical midocean-ridge systems (21). In the range of such extraordinary thermal gradients of the deep-sea vents, the archaeal components may play a much more important role in the formation and activity of a microbial community than in the shallow vents.
The existence of a diversity of archaea in the deep-sea hydrothermal vent environments has been demonstrated through a number of enrichment and isolation attempts to date (9, 10, 18, 23, 28, 42, 43, 53, 56, 60). Additionally, culture-independent, molecular approaches have revealed that a greater diversity of archaea are predominantly present and remain unidentified in terms of their physiological properties and metabolic functions (44, 51). This study is the first report in which the distribution of discrete archaeal communites in different microhabitats of a black smoker chimney was demonstrated by using culture-independent, molecular phylogenetic analysis. In this study, it was revealed that members of Thermococcus and members of the DHVEG are abundant on the chimney surface. The coexistence with the hyperthermophiles Thermococcus, the wide distribution in the various deep-sea hydrothermal vents, the relatively short branch length in the phylogenetic tree (Fig. 3C), and relatively high G+C content of the rDNA (57.6 to 60.2%) of the uncultivated DHVEG may represent the thermophily in this group of uncultivated archaea. Likewise, the deeply branched phylotype pPACMA-Y, phylogenetically related to the MHVG, might at least represent thermophily as its distinctive physiological feature, as described previously (51).
The discovery of a sizable population of extremely halophilic archaea in the inside structures of the chimney was the most striking finding in this work. Kaye and Baross have reported finding halotolerant bacteria belonging to the genera Halomonas and Marinobacter at high incidence in a variety of samples obtained from deep-sea hydrothermal vent environments (30). We also obtained here a culturable population of Halomonas from the inside structures of the chimney. The halophilic bacteria are abundant in deep ocean water and may be contaminated from the ambient seawater around the deep-sea hydrothermal vent (30). Given that the halophilic bacterial population, including Halomonas sp. cultured from the inside structure of the chimney, is derived from the ambient seawater, there might be obtained a similar or higher abundance of cultures for Halomonas spp. from the surface of the chimney exposure to the seawater. The discovery of extremely halophilic archaeal rDNA and successful cultivation of halophilic microorganisms were achieved specifically from the inside structures of the chimney. It seems likelier, therefore, that the halophilic bacterial cultures and the genetic signatures for extremely halophilic archaea are derived from microbial communities dwelling in the microhabitats that occurred in the inside structures rather than from contaminated microbial communities. Since no positive cultivation of the extremely halophilic archaea represented by rDNA phylotypes from the inside structures was achieved, the viability of the extremely halophilic archaeal population remained uncertain. However, the discovery of extremely halophilic archaeal rDNA and the successful cultivation of halophilic bacteria suggest that partially hypersaline microhabitats allowing the survival or growth of halophilic archaea and bacteria occur in the chimney structure. Furthermore, the recovery of hyperthermophilic and extremely halophilic archaeal rDNA phylotypes from the interface with the vent fluid and the interior structures of the chimney leads to the hypothesis that the microbial communities in the microhabitats of the chimney are provided from the microbial population thriving beneath the active hydrothermal floor, the subvent biosphere.
Based on several microbiological, geochemical, and geophysical observations, Deming and Baross proposed the possible existence of a hyperthermophilic subvent biosphere beneath the active hydrothermal vent fields (14). The relatively high density of microbial cells and the detection of microbial rDNA in the black smoker vent water at a temperature of >300°C also support the occurrence of a sizable microbial population in the subvent environment of the hydrothermal system in Ihaya Basin, Okinawa Trough (51). Since the deep ocean water infiltrating this system consistently replenishes foreign microorganisms and nutrients in the environments, it is difficult to identify the exclusively indigenous microbial components and activities in the subvent biosphere. However, extreme halophiles as well as hyperthermophiles are the possible indigenous microorganisms in the subvent biosphere. The physiological and metabolic diversity of known hyperthermophiles allows them to become accustomed to a variety of microhabitats in the subvent, most of which are extraordinary high-temperature environments, as in the case of the chimney structure. In addition, the formation of brines or salt deposits in deep-sea hydrothermal systems has been proposed (30–32). These brines or salt deposits provide suitable microhabitats for the extremely halophilic archaea. The global distribution of extremely halophilic archaea as well as of hyperthermophilic Thermococcales members has been observed in nonhydrothermal pelagic subseafloor sediments and the subseafloor methane hydrate core samples (our unpublished results). The distribution of archaeal components found in the deep-sea hydrothermal or the subvent environments in global subseafloor microbial communities may shed light on the origin of the subsurface microorganisms and the biogeographical propagation of hyperthermophiles and extreme halophiles. These are the foci of our future efforts.
ACKNOWLEDGMENTS
We thank the captain and crew of the R/V Natsushima and the DSV Shinkai 2000 operations group for their technical expertise. We also thank Katsuyuki Uematsu for assistance in analysis of SEM-EDS and Wayne R. Bellamy for editing English usage in the manuscript.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakken L R, Olsen R A. DNA content of soil bacteria of different cell size. Soil Biol Biochem. 1989;21:789–793. [Google Scholar]
- 3.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baross J A. Isolation, growth, and maintenance of hyperthermophiles. In: Robb F T, Place A R, Sowers K R, Schreier H J, DasSarma S, Fleischmann E M, editors. Archaea: a laboratory manual: halophiles. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 15–24. [Google Scholar]
- 6.Binns R A, Parr J M, Scott S D, Gemmell J B, Herzig P M. Proceedings of PACRIM'95, Auckland, New Zealand. 1995. PACMANUS: an active seafloor hydrothermal field on siliceous volcanic rocks in the eastern Manus Basin, Papua New Guinea. [Google Scholar]
- 7.Binns R A, Scott S D. Activity forming polymetallic sulfide deposits associated with felsic volcanic rocks in the eastern Manus back-arc basin, Papua New Guinea. Econ Geol. 1993;88:2226–2236. [Google Scholar]
- 8.Benson D A, Boguski M S, Lipman D J, Ostell J, Ouellette B F F. GenBank. Nucleic Acids Res. 1998;26:1–7. doi: 10.1093/nar/26.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blöchl E, Rachel R, Burggraf S, Hafenbradl D, Jannasch H W, Stetter K O. Pyrolobus fumarii, gen. and sp. nov., represents a novel group of archaea, extending the upper temperature limit for life to 113°C. Extremophiles. 1997;1:14–21. doi: 10.1007/s007920050010. [DOI] [PubMed] [Google Scholar]
- 10.Burggraf S, Jannasch H W, Nicolaus B, Stetter K O. Archaeglobus profundus sp. nov. represents a new species within the sulfate-reducing archaebacteria. Syst Appl Microbiol. 1990;13:24–28. [Google Scholar]
- 11.Corliss J B, Dymond J, Gordon L I, Edmond J M, von Herzen R P, Ballard R D, Green K, Williams D, Bainbridge A, Crane K, van Andel T H. Submarine thermal springs on the Galapagos Rift. Science. 1979;203:1073–1083. doi: 10.1126/science.203.4385.1073. [DOI] [PubMed] [Google Scholar]
- 12.DasSarma S, Fleischmann E M, Rodriguez-Valera F. Media for halophiles. In: Robb F T, Place A R, Sowers K R, Schreier H J, DasSarma S, Fleischmann E M, editors. Archaea: a laboratory manual: halophiles. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 225–230. [Google Scholar]
- 13.Delong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deming J W, Baross J A. Deep-sea smokers: windows to a subsurface biosphere? Geochim Cosmochim Acta. 1993;57:3219–3230. doi: 10.1016/0016-7037(93)90535-5. [DOI] [PubMed] [Google Scholar]
- 15.Francheteau J, Needham H D, Choukroune P, Juteau T, Seguret M, Ballard R D, Fox P J, Normark W, Carranza A, Cordoba D, Guerrero J, Rangin C, Bougault H, Cambon P, Hekinian R. Massive deep-sea sulfide ore deposits discovered on the East Pacific Rise. Nature. 1979;277:523–528. [Google Scholar]
- 16.Gamo T, Okamura K, Charlou J-L, Urabe T, Auzende J-M, Ishibashi J, Shitashima K, Kodama Y Shipboard Scientific Party of the ManusFlux Cruise. Chemical exploration of hydrothermal activity in the Manus Basin, Papua New Guinea (ManusFlux Cruise) JAMSTEC J Deep Sea Res. 1996;12:336–345. [Google Scholar]
- 17.Gamo T, Okamura K, Charlou J-L, Urabe T, Auzende J-M, Ishibashi J, Shitashima K, Chiba H Shipboard Scientific Party of the ManusFlux Cruise. Acidic and sulfate-rich hydrothermal fluids from the Manus back-arc basin, Papua New Guinea. Geology. 1997;25:139–142. [Google Scholar]
- 18.Gonzalez J M, Masuchi Y, Robb F T, Ammerman J W, Maeder D L, Yanagibayashi M, Tamaoka J, Kato C. Pyrococcus horikoshii sp. nov., a hyperthermophilic archaeon isolated from a hydrothermal vent at the Okinawa Trough. Extremophiles. 1998;2:123–130. doi: 10.1007/s007920050051. [DOI] [PubMed] [Google Scholar]
- 19.Harmsen H J M, Prieur D, Jeanthon C. Distribution of microorganisms in deep-sea hydrothermal vent chimneys investigated by whole-cell hybridization and enrichment culture of thermophilic subpopulations. Appl Environ Microbiol. 1997;63:2876–2883. doi: 10.1128/aem.63.7.2876-2883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harmsen H J M, Prieur D, Jeanthon C. Group-specific 16S rRNA-targeted oligonucleotide probes to identify thermophilic bacteria in marine hydrothermal vents. Appl Environ Microbiol. 1997;63:4061–4068. doi: 10.1128/aem.63.10.4061-4068.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herzig P M, Hannington M S. Input from the deep: hot vents and cold seeps. In: Schulz H D, Zabel M, editors. Marine geochemistry. Berlin, Germany: Springer-Verlag; 2000. pp. 397–416. [Google Scholar]
- 22.Heymon R M. Growth history of hydrothermal black smoker chimneys. Nature. 1983;301:695–698. [Google Scholar]
- 23.Huber R, Kurr M, Jannasch H W, Stetter K O. A novel group of abyssal methanogenic archaebacteria (Methanopyrus) growing at 110°C. Nature. 1989;342:833–834. [Google Scholar]
- 24.Ihara K, Watanabe S, Tamura T. Haloarcula argentinensis sp. nov. and Haloarcula mukohataei sp. nov., two new extremely halophilic archaea collected in Argentina. Int J Syst Bacteriol. 1997;47:73–77. doi: 10.1099/00207713-47-1-73. [DOI] [PubMed] [Google Scholar]
- 25.James S R, Dobson S J, Franzmann P D, McMeekin T A. Halomonas meridiana, a new species of extremely halotolerant bacteria isolated from Antarctic saline lakes. Syst Appl Microbiol. 1990;13:270–277. [Google Scholar]
- 26.Jannasch H W. Microbial interactions with hydrothermal fluids. In: Humphris S E, Zierenberg R A, Mullineaux L S, Thomson R E, editors. Seafloor hydrothermal systems: physical, chemical, biological, and geological interactions. Geophysical monograph 91. Washington, D.C.: American Geophysical Union; 1995. pp. 273–296. [Google Scholar]
- 27.Jeanthon C. Molecular ecology of hydrothermal vent microbial communities. Antonie Leeuwenhoek. 2000;77:117–133. doi: 10.1023/a:1002463825025. [DOI] [PubMed] [Google Scholar]
- 28.Jones W J, Leigh J A, Mayer F, Woese C R, Wolfe R S. Methanococcus jannaschii sp. nov., an extremely thermophilic methanogen from a submarine hydrothermal vent. Arch Microbiol. 1983;136:254–261. [Google Scholar]
- 29.Karl D M. Ecology of free-hydrothermal vent microbial communities. In: Karl D M, editor. The microbiology of deep-sea hydrothermal vents. Boca Raton, Fla: CRC Press, Inc.; 1995. pp. 35–124. [Google Scholar]
- 30.Kaye J Z, Baross J A. High incidence of halotolerant bacteria in Pacific hydrothermal-vent and pelagic environments. FEMS Microbiol Ecol. 2000;32:249–260. doi: 10.1111/j.1574-6941.2000.tb00718.x. [DOI] [PubMed] [Google Scholar]
- 31.Kelley D S, Robinson P T. Development of a brine-dominated hydrothermal system at temperatures of 400–500°C in the upper level plutonic sequence, Troodos ophiolite, Cyprus. Geochim Cosmochim Acta. 1990;54:653–661. [Google Scholar]
- 32.Kelley D S, Robinson P T, Malpas J G. Process of brine generation and circulation in oceanic crust: fluid inclusion evidence from the Troodos ophiolite. J Geophys Res. 1992;97:9307–9322. [Google Scholar]
- 33.Lafitte M, Maury R, Perseil E A, Boulegue J. Morphological and analytical study of hydrothermal sulfides from 21°N north East Pacific Rise. Earth Planet Sci Lett. 1985;73:53–64. [Google Scholar]
- 34.Liu W T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maidak B L, Cole J R, Lilburn T G, Parker Jr C T, Saxman P R, Stredwick J M, Garrity G M, Li B, Olsen G J, Pramanik S, Schmidt T M, Tiedje J M. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 2000;28:173–174. doi: 10.1093/nar/28.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 37.Moeseneder M M, Arrieta J M, Muyzer G, Winter C, Herndl G J. Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1999;65:3518–3525. doi: 10.1128/aem.65.8.3518-3525.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moyer C L, Dobbs F C, Karl D M. Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1995;61:1555–1562. doi: 10.1128/aem.61.4.1555-1562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moyer C L, Tiedje J M, Dobbs F C, Karl D M. Diversity of deep-sea hydrothermal vent Archaea from Loihi Seamount, Hawaii. Deep-Sea Res Part II Top Stud Oceanogr. 1998;45:303–317. [Google Scholar]
- 40.Mylvaganam S, Dennis P P. Sequence heterogeneity between the two genes encoding 16S rRNA from the halophilic archaebacterium Haloarcula marismortui. Genetics. 1992;130:399–410. doi: 10.1093/genetics/130.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osborn A M, Moore E R, Timmis K N. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community and dynamics. Environ Microbiol. 2000;2:39–50. doi: 10.1046/j.1462-2920.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- 42.Pley U, Schipka J, Gambacorta A, Jannasch H W, Fricke H, Rachel R, Stetter K O. Pyrodictium abysii sp. nov. represents a novel heterotrophic marine archaeal hyperthermophile growing at 110°C. Syst Appl Microbiol. 1991;14:245–253. [Google Scholar]
- 43.Prieur D, Erauso G, Jeanthon C. Hyperthermophilic life at deep-sea hydrothermal vents. Planet Space Sci. 1995;43:115–121. doi: 10.1016/0032-0633(94)00143-f. [DOI] [PubMed] [Google Scholar]
- 44.Reysenbach A-L, Longnecker K, Kirshtein J. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a Mid-Atlantic Ridge hydrothermal vent. Appl Environ Microbiol. 2000;66:3798–3806. doi: 10.1128/aem.66.9.3798-3806.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez-Valera F. Cultivation of halophilic archaea. In: Robb F T, Place A R, Sowers K R, Schreier H J, DasSarma S, Fleischmann E M, editors. Archaea: a laboratory manual: halophiles. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 13–16. [Google Scholar]
- 46.Sako Y, Takai K, Uchida A, Ishida Y, Katayama Y. Rhodothermus obamensis sp. nov., a modern lineage of extremely thermophilic marine bacteria. Int J Syst Bacteriol. 1996;46:1099–1104. doi: 10.1099/00207713-46-4-1099. [DOI] [PubMed] [Google Scholar]
- 47.Sievert S M, Brinkhoff T, Muyzer G, Ziebis W, Kuever J. Spatial heterogeneity of bacterial populations along an environmental gradient at a shallow submarine hydrothermal vent near Milos Island (Greece) Appl Environ Microbiol. 1999;65:3834–3842. doi: 10.1128/aem.65.9.3834-3842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sievert S M, Kuever J, Muyer G. Identification of 16S ribosomal DNA-defined bacterial populations at a shallow submarine hydrothermal vent near Milos Island (Greece) Appl Environ Microbiol. 2000;66:3102–3109. doi: 10.1128/aem.66.7.3102-3109.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sievert S M, Ziebis W, Kuever J, Sahm K. Relative abundance of Archaea and Bacteria along a thermal gradient of a shallow-water hydrothermal vent quantified by rRNA dot-slot hybridization. Microbiology. 2000;146:1287–1293. doi: 10.1099/00221287-146-6-1287. [DOI] [PubMed] [Google Scholar]
- 50.Takai K, Horikoshi K. Molecular phylogenetic analysis of archaeal intron-containing genes coding for rRNA obtained from a deep-subsurface geothermal water pool. Appl Environ Microbiol. 1999;65:5586–5589. doi: 10.1128/aem.65.12.5586-5589.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takai K, Horikoshi K. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics. 1999;152:1285–1297. doi: 10.1093/genetics/152.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takai K, Horikoshi K. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microbiol. 2000;66:5066–5072. doi: 10.1128/aem.66.11.5066-5072.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takai K, Horikoshi K. Thermosipho japonicus sp. nov., an extremely thermophilic bacterium isolated from a deep-sea hydrothermal vent in Japan. Extremophiles. 2000;4:9–17. doi: 10.1007/s007920050002. [DOI] [PubMed] [Google Scholar]
- 54.Takai K, Inoue A, Horikoshi K. Thermaerobacter marianensis gen. nov., sp. nov., an aerobic extremely thermophilic marine bacterium from the 11,000 m deep Mariana Trench. Int J Syst Bacteriol. 1999;49:619–628. doi: 10.1099/00207713-49-2-619. [DOI] [PubMed] [Google Scholar]
- 55.Takai K, Sako Y. A molecular view of archaeal diversity in marine and terrestrial hot water environments. FEMS Microbiol Ecol. 1999;28:177–188. [Google Scholar]
- 56.Takai K, Sugai A, Itoh T, Horikoshi K. Palaeococcus ferrophilus gen. nov., sp. nov., a barophilic hyperthermophilic archaeon from a deep-sea hydrothermal vent chimney. Int J Syst Evol Microbiol. 2000;50:489–500. doi: 10.1099/00207713-50-2-489. [DOI] [PubMed] [Google Scholar]
- 57.Tanner M A, Grobel B M, Dojka M A, Pace N R. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl Environ Microbiol. 1998;64:3110–3113. doi: 10.1128/aem.64.8.3110-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ventosa A, Nieto J J, Oren A. Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol Rev. 1998;62:504–544. doi: 10.1128/mmbr.62.2.504-544.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vetriani C, Jannasch H W, MacGregor B J, Stahl D A, Reysenbach A-L. Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl Environ Microbiol. 1999;65:4375–4384. doi: 10.1128/aem.65.10.4375-4384.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zillig W, Holz I, Janekovic D, Klenk H-P, Imsel E, Trent J, Wunderl S, Forjaz V H, Coutinho R, Ferreira T. Hyperthermus butylicus, a hyperthermophilic sulfur-reducing archaebacterium that ferments peptides. J Bacteriol. 1990;172:3959–3965. doi: 10.1128/jb.172.7.3959-3965.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]