Summary
Dramatic differences in outcome between early- and late-stage high-grade serous ovarian cancer (HGSC) suggest perhaps distinct genetic origins due to differences in exposures to mutational processes. Evidence to support this hypothesis was recently reported by comparative analysis of copy-number signatures between early- and late-stage HGSCs.
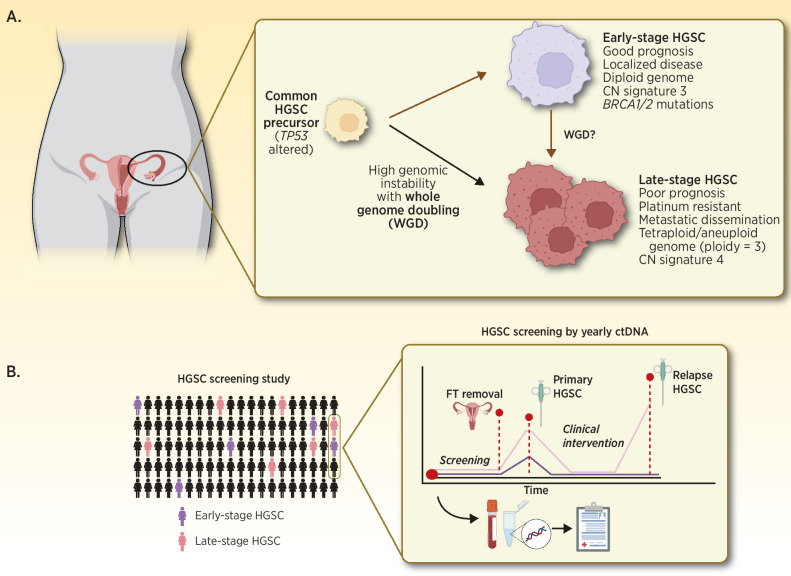
In this issue of Clinical Cancer Research, Cheng and colleagues (1) describe results from a retrospective, hypothesis-generating study that explores the evolutionary origins of high-grade serous ovarian cancer (HGSC) by comparing genomic differences of early-stage (stage IA–IIA; n = 46) to late-stage HGSCs (stage IIIC–IV; n = 52). As significant prognostic disparity exists between early- and late-stage HGSC, it is unclear whether early-stage disease represents a molecularly distinct subtype that lacks peritoneal dissemination potential or if it reflects a premetastatic precursor state of late-stage HGSC. Comparisons of focal genomic alterations using a 16-gene targeted-sequencing panel and shallow whole-genome sequencing (WGS) revealed no discernible differences in mutation rates and signatures that could explain differential prognosis between the cohorts. However, measures of increased genomic instability, such as high-ploidy (median = 3.0) and the presence of copy-number alteration (CNA) signature 4 (2), together suggested that whole genome doubling (WGD) is associated with late-stage disease and inferior overall survival (OS; Fig. 1A). These findings highlight the potential utility of whole genome copy-number analysis to identify WGD as a prognostic biomarker in HGSC and offer support for distinct biological trajectories between early- and late- stage disease.
Figure 1.
A, Summary of the proposed origins of early- and late-stage HGSC by Cheng and colleagues (1). B, Prospective HGSC screening by applying ctDNA-based biomarkers to detect early-stage disease and facilitate longitudinal collection of precursor, blood, and HGSC biospecimens to enable ongoing HGSC research. FT, Fallopian tube.
Cheng and colleagues (1) reported similar age-at-diagnosis comparing early- and late-stage HGSC as evidence to support their distinct developmental origins. While this observation is intriguing, it would be of interest to further investigate whether early-stage disease was related to the patient's primary differential diagnosis or was discovered incidentally. Incidental diagnosis may explain the lack of age difference between cohorts due to inherent selection biases associated with patients’ primary health complaints. This brings forth the need for long-term epidemiologic studies employing sensitive HGSC detection methods to formally estimate the incidence rate of early-stage HGSC within the general population. Longitudinal biobanking and tracking HGSC tumor molecular evolution before and after treatment such as BioDiva (NCT03419689) are fast becoming gold-standard in biomarker-driven clinical trials. A similar concept could be applied to preserving pre-malignant tissues such as fallopian tubes (widely accepted as source of HGSC cells of origin) and blood-draws with long-term clinical follow-up. To shed light on the evolutionary origins of early and late-stage HGSCs, this resource may be interrogated to reveal timing of genetic alterations and changing influence of mutational processes (Fig. 1B; ref. 2).
The development of molecular biomarkers predominantly focuses on single-gene or focal genetic events. This rationale is built upon the classic model of cancer drivers and hotspot mutations associated with small molecular drug targets, oncogenes, and tumor suppressors. In HGSCs, single-gene biomarkers have been explored, however none has demonstrated the ability to discern early- from late-stage disease. Here, Cheng and colleagues (1) present CN signature exposures and WGD as additional genome-wide prognostic indicators of HGSC.
Analogous to genome-wide single-base substitution or short sequence variants patterns associated with specific environmental exposures and DNA damage processes, seven genome-wide CNA signatures were described and associated with OS and platinum-resistant relapse in late-stage HGSCs of the BriTROC-1 cohort (n = 253; ref. 2). Cheng and colleagues (1) described early-stage HGSCs using these same signatures to demonstrate high CNA signature 3 exposure in early-stage while signature 4 exposure was more evident in late-stage HGSC. Unsurprisingly, CNA signature 3 is associated with improved OS and BRCA1/2 related homologous recombination deficiency in late-stage HGSC (2). Signature 4, characterized by high segment CN and CN changes, is a surrogate of WGD due to failure of cell cycle control in HGSC with mutated MYC, CCNE1, CDK12, and PI3K/AKT signaling cascade (2). From these results, Cheng and colleagues (1) argue that WGD is a key molecular event, possibly arising early during tumorigenesis that differentiates early- from late-stage HGSCs. It's interesting to speculate if WGD is an adaptive genomic survival mechanism which perhaps confers a tumor growth advantage, and potentially a more aggressive phenotype, witnessed as an association with advanced stage disease and poor OS across cancer types (3). Recently, Quinton and colleagues demonstrated that cells with WGD may be sensitive to KIF18A depletion (4). Future studies involving longitudinally collected HGSC specimens through tumour development in patients or animal models are needed to confirm this hypothesis.
While cancer CN profiling typically requires access to tumor tissues and sequencing of mutational targets across long stretches of the cancer genome, technological and algorithmic advancements have been made to overcome these traditional barriers. As demonstrated in this study and others, reliable CN profiles can be generated from formalin-fixed, paraffin-embedded tumor-derived DNA material through low-pass or low-coverage WGS. Detectable changes in sequencing depth relative to normal controls across genomic bins provide an estimate of CN gains and losses without a-priori knowledge of tumour-specific mutations. As such, compared with single gene point mutations as biomarkers, CN alterations and associated patterns may provide more biologically and clinically meaningful information in HGSCs and other cancer types characterized by genomic instability.
Increased HGSC diagnosis in postmenopausal women, and decreased HGSC risks associated with oral contraceptive use, pregnancy, and breast-feeding are observations that allude to possible influences of hormonal shifts on HGSC development. The absence of genetic differences between early- and late- stage HGSC to explain the biological underpinnings of metastatic spread reported by Cheng and colleagues (1) suggest that such phenotype may not be genetically acquired. Therefore, future studies should consider investigating the association of cell-extrinsic factors such as exposure to hormones and cytokines, tumor immune microenvironment, and anatomical distribution of tumor spread with HGSC stage with potential implications for further enabling precision medicine.
Recently, the largest and longest-running randomized controlled trial of ovarian cancer population screening in the UK (UKCTOCS; ref. 5) revealed that while early-stage cancer detection was increased by 10% in cohorts screened via yearly CA125 serum biomarker tests, this effort did not lead to significant reduction in ovarian cancer–related deaths when compared with the control group. Despite these discouraging results, there now exists a rich repository of annual serum specimens collected from 200,000 women over a 7-year period linked to their cancer and death information. This resource could be accessed by researchers to test and validate other biomarkers with higher sensitivity and specificity than CA125, such as detection of circulating tumour DNA (ctDNA). ctDNA has shown promise to detect minimal residual disease following cancer treatment with curative intent in multiple cancers, suggesting that it may detect very minute disease burden. However, feasibility for detecting extremely small early-stage HGSC tumors or premalignant lesions localized to the fallopian tube by ctDNA remains to be explored.
Cancer detection and minimal residual disease monitoring via circulating tumour DNA has increasing applications across tumor indications. Recently, Braicu and colleagues (6) provided proof-of-concept evidence to detect HGSC in a prospective cohort (n = 109) in comparison to healthy controls (n = 241) by copy-number index score (CNI-Score) from ctDNA. Together with the conclusions of Cheng and colleagues (1), the need for high-sensitivity detection of chromosomal alterations and WGD will need to be incorporated into ctDNA detection algorithms for HGSC. One potential direction may be mapping of ctDNA fragmentation patterns from blood samples offers an innovative genome-wide strategy for detecting early malignancies (7). It is likely that combining multiple ctDNA-based genomic assessments such as CNI-Score, CN signatures and fragmentation patterns, sensitive detection, and risk stratification can be accomplished within one assay that bypasses the requirement for access to tumor tissues for HGSCs and other CN-driven cancer types.
In conclusion, Cheng and colleagues (1) presented a thought-provoking investigation to unravel the evolutionary origins and differential mutational processes in early- and late-stage HGSCs. This study also provides the research community with one of the largest collections of early-stage HGSC genomic profiles with long-term follow-up clinical data to date. Further study is needed to evaluate the contribution and biological consequences of WGD in HGSC development and metastatic spread; however, this work highlights a potential application of cancer genome CN signatures as a HGSC biomarker. These signatures could be further applied in several ways–potentially to aid early HGSC detection if measurable within liquid biopsies or evaluated prospectively to determine predictive value for treatment selection. Due to the small number of available early-stage HGSC specimens, these findings remain exploratory and emphasize the need for clinical trials. Future work should be mindful of intratumor heterogeneity and tumor anatomic locations that may impact the quantity and sampling of tumor DNA within blood samples collected between early- and late-stage disease.
Acknowledgments
A.M. Oza is supported by a Translational Research Initiative Award from the Ontario Institute for Cancer Research (Toronto, Ontario, Canada). T.J. Pugh holds the Canada Research Chair in Translational Genomics and is supported by a Senior Investigator Award from the Ontario Institute for Cancer Research and the Gattuso-Slaight Personalised Cancer Medicine Fund at the Princess Margaret Cancer Centre (University of Toronto, Toronto, Ontario, Canada).
Authors' Disclosures
T.J. Pugh reports personal fees from AstraZeneca, Canadian Pension Plan Investment Board, Chrysalis Biomedical Advisors, Illumina, Merck, and PACT Pharma and grants from Roche/Genentech outside the submitted work. A.M. Oza reports trials sponsored by AstraZeneca, GSK, and Clovis (principal investigator and steering committees; uncompensated); in addition, A.M. Oza is uncompensated CEO of Ozmosis Research. No disclosures were reported by the other author.
References
- 1. Cheng Z, Mirza H, Ennis DP, Smith P, Gavarro LM, Sokota C, et al. The genomic landscape of early-stage ovarian high-grade serous carcinoma. Clin Cancer Res 2022;28:2911–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Macintyre G, Goranova TE, De Silva D, Ennis D, Piskorz AM, Eldridge M, et al. Copy number signatures and mutational processes in ovarian carcinoma. Nat Genet 2018;50:1262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bielski CM, Zehir A, Penson AV, Donoghue MTA, Chatila W, Armenia J, et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat Genet 2018;50:1189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Quinton RJ, DiDomizio A, Vittoria MA, Kotýnková K, Ticas CJ, Patel S, et al. Whole-genome doubling confers unique genetic vulnerabilities on tumour cells. Nature 2021;590:492–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Menon U, Gentry-Maharaj A, Burnell M, Singh N, Ryan A, Karpinskyj C, et al. Ovarian cancer population screening and mortality after long-term follow-up in the UK collaborative trial of ovarian cancer screening (UKCTOCS): a randomised controlled trial. Lancet North Am Ed 2021;397:2182–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Braicu EI, du Bois A, Sehouli J, Beck J, Prader S, Kulbe H, et al. Cell-free-DNA-based copy number index score in epithelial ovarian cancer-impact for diagnosis and treatment monitoring. Cancers 2021;14:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019;570:385–89. [DOI] [PMC free article] [PubMed] [Google Scholar]