Abstract
Cytokines signal through cell surface receptor dimers to initiate activation of intracellular Janus Kinases (JAKs). We report the 3.6Å resolution cryo-EM structure of full-length JAK1 complexed with a cytokine receptor intracellular Box1/Box2 domain, captured as an activated homodimer bearing the Val→Phe (VF) mutation prevalent in myeloproliferative neoplasms. The seven domains of JAK1 form an extended structural unit whose dimerization is mediated by close-packed pseudokinase (PK) domains. The oncogenic VF mutation lies within the core of the JAK1 PK dimer interface, enhancing packing complementarity to facilitate ligand-independent activation. The C-terminal tyrosine kinase domains are poised to phosphorylate the receptor STAT-recruiting motifs projecting from the overhanging FERM-SH2 domains. Mapping of constitutively active JAK mutants supports a two-step allosteric activation mechanism and reveals new opportunities for selective therapeutic targeting of oncogenic JAK signaling.
One-Sentence Summary:
Cryo-EM structure of JAK1 cytokine receptor complex reveals a mechanism of dimeric activation exploited by oncogenic mutation.
Cytokines are a multifarious family of secreted proteins that have broad and pleiotropic effects on cell growth, hematopoiesis, immunity, and inflammation (1, 2). Cytokines initiate signaling by binding to the extracellular domains of single-pass transmembrane receptors and facilitate receptor dimerization, which is required for signal transduction (3–5). This extracellular dimerization event is structurally conveyed to the intracellular domains, resulting in the activation and trans-phosphorylation of non-covalently associated Janus kinases (JAKs) (6–8). There are four members of the JAK family, JAK1, JAK2, JAK3, and TYK2, that are associated with the membrane-proximal region of the intracellular domain through two distinct conserved motifs in the receptor: a proline-rich segment termed ‘Box1’ and a hydrophobic segment called ‘Box2’ (9). Once activated, JAKs phosphorylate tyrosine residues within the cytokine receptor intracellular domains (ICDs), which subsequently serve as docking sites for the STAT (signal transducer and activator of transcription) transcription factors (10). Recruitment of STATs to the receptor–JAK complex enables STAT phosphorylation by the activated JAKs, leading to STAT dimerization and translocation to the nucleus to initiate transcription of cytokine-responsive genes.
All JAK family members are comprised of seven JAK homology (JH) domains that comprise a four-point-one, ezrin, radixin, moesin (FERM) domain (JH5, 6, and 7), a Src homology 2 (SH2) domain (JH3 and 4), and tandem kinase domains, JH2 and JH1, which encode a pseudokinase (PK) and tyrosine kinase (TK), respectively (Fig. 1A) (11, 12). The FERM and SH2 domains at the amino-terminal end of JAK associate with the intracellular juxtamembrane segment from the paired cytokine receptor (13). Our current understanding of full-length JAK structure and activation mechanisms derives from extrapolations from structures of monomeric JAK fragments. Crystal structures of the human JAK1, JAK2, and TYK2 FERM–SH2 fragments have revealed that these domains are tightly associated to form a single receptor-binding module that accommodates the Box1/Box2 peptide at multiple interaction sites (14–17). In addition, structural models for the PK-TK modules from TYK2 and JAK2 have suggested a mechanism of negative regulation by the pseudokinase (18, 19). Numerous structures of cytokines complexed with their receptor extracellular domains (ECDs) in homo- or heterodimeric complexes have shown common features and structural diversity in the overall architectures of the extracellular assemblies that are presumably communicated to the inside of the cell for JAK activation (20). However, how ECD dimerization brings two intracellular JAKs into proper orientation and proximity for activation remains unresolved due to the absence of structural information on full-length JAK proteins in activated states (8).
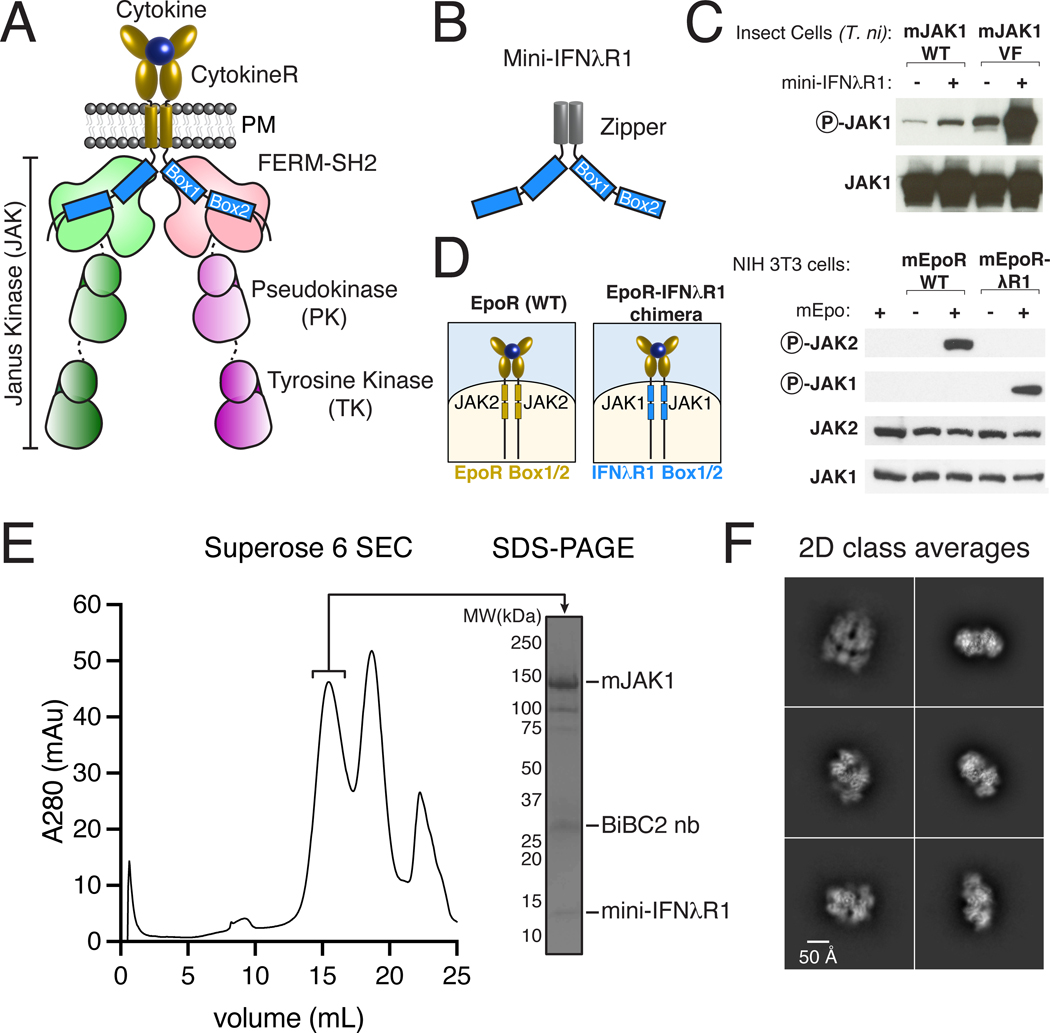
Fig. 1. Purification and biochemical characterization of an active JAK1-IFNλR1 complex.
(A) Schematic of a cell-surface, ligand-induced cytokine receptor-JAK dimer. (B) Schematic of a soluble cytokine receptor dimer mimetic (‘mini-IFNλR1’) consisting of N-terminal glutathione S-transferase (GST) tag for affinity purification, 3C protease site, GCN4-zipper, and mIFNλR1 Box1/Box2. (C) Mini-IFNλR1 expression enhances JAK1 phosphorylation when co-expressed in insect cells. Wild-type (WT) or Val657→Phe (VF) JAK1 was co-expressed with mini-IFNλR1 in T. ni cells by baculovirus transduction. JAK phosphorylation and total expression were measured two days post-infection by immunoblot of whole cell lysate. Results are representative of more than two independent experiments. (D) Schematic of wild-type (WT) EpoR/Epo complex (left), and EpoR-IFNλR1 chimera (right) showing substitution of Box1/Box2 motifs (blue). Cytokine-mediated dimerization of IFNλR1 Box1/Box2 results in JAK1 phosphorylation in mammalian cells. NIH 3T3 cells transiently expressing mEpoR or mEpoR-IFNλR1 chimera were stimulated with Epo for 20 min prior to analysis of JAK phosphorylation by immunoblot. Results are representative of two independent experiments. (E) Affinity-purification of JAK1 using zippered mini-IFNλR1 results in purification of a stable, non-aggregated complex. Superose 6 size exclusion chromatography (SEC, left) and sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE, right) of the JAK1-IFNλR1 complex. (F) Representative 2D class averages from single-particle cryo-EM imaging of the JAK1-IFNλR1 complex. PM – plasma membrane, R – receptor, mAu - milli-absorbance units, BiBC2 nb – tandem BC2 nanobody.
Naturally-occurring mutations in cytokine receptors, JAKs, and STATs lead to immunodeficiency and myeloproliferative disorders in humans (10, 21). Disruption of JAK1 and JAK2 genes is lethal (22–24), while loss-of-function (LOF) mutations in JAK3 cause severe combined immunodeficiency (SCID) (25–27). On the other hand, gain-of-function (GOF) mutations in JAK genes are responsible for a family of blood disorders known as myeloproliferative neoplasms (MPNs), which include polycythemia vera, primary myelofibrosis, and essential thrombocythemia, as well as leukemias (28). In a classic series of papers reported in 2005 (29–32), a point mutation in the PK domain of JAK2, Val617→Phe (V617F), which results in constitutive activity, was shown to be present in >90% of patients with polycythemia vera and in ~50% of patients with essential thrombocythemia and primary myelofibrosis. This gain-of-function mutation was first described in the Drosophila homologue hopscotch (33) and analogous mutations in human JAK paralogues also result in constitutive activity, suggesting a shared activation mechanism across JAK family members, likely involving ligand-independent dimerization at the cell surface (3, 34, 35). Ruxolitinib is a small-molecule inhibitor of JAK2 (and JAK1) kinase activity and targets both wild-type JAK2 and JAK2-V617F, resulting in side effects including thrombocytopenia and anemia (21). A better understanding of how mutations in JAK, particularly JAK2-V617F, result in constitutive activity is needed to guide drug design to target mutant JAK2. Here we report the cryo-electron microscopy (cryo-EM) structure, at 3.6Å resolution, of full-length mouse JAK1 complexed with the interferon λ receptor 1 (IFNλR1) intracellular Box1-Box2 segment, which provides a structural blueprint to understand both cytokine and oncogenic mutant-driven signal activation.
Results:
Engineering an active JAK1-IFNλR1 complex for cryo-EM imaging
Full-length JAKs have been recalcitrant to structural analysis by X-ray crystallography and electron microscopy (8). Imaging a JAK1 complex with cytokine receptor ICD required several protein engineering steps to produce an activate, stable, non-aggregated complex suitable for cryo-EM imaging. First, we identified full-length mouse JAK1 to have better expression and solubility properties when produced from insect cells, compared to other JAK paralogs and orthologues. Second, we introduced the V657F mutation into mouse JAK1 (analogous to hJAK2 V617F) in order to stabilize an activated state. Third, so that we could affinity purify full-length JAK1 using the receptor ICDs, we focused on the JAK1 binding Box1/Box2 domains from Interferon λ Receptor 1 (IFNλR1) based on a screen that identified this ICD as among the highest affinity JAK1/ICD interactions (14). Fourth, we replaced the transmembrane domains of the receptor with the homodimeric GCN4 leucine zipper fused to the IFNλR1 Box1/Box2 in order to create a soluble mimic of a dimerized receptor in which the zippers approximated the spatial constraints of dimerized TM domains (Fig. 1B) (36, 37).
Co-expression of the soluble zippered IFNλR1 ICD peptide (‘mini-IFNλR1’) with either full-length wild-type (WT) JAK1 or JAK1-V657F protein resulted in increased activation loop phosphorylation as measured by western blot, validating the construct design strategy (Fig. 1C). In the context of IFNλ signaling on cells, JAK1-IFNλR1 normally hetero-dimerizes with TYK2-IL-10Rβ to initiate downstream signaling. To test whether our engineered JAK1-IFNλR1 homodimer is capable of signaling in response to cytokine stimulation, we generated chimeric receptors in which the Box1/Box2 motif from IFNλR1 was substituted into the analogous position in erythropoietin receptor (EpoR), which forms a EpoR-JAK2 homodimer in response to stimulation with erythropoietin (Epo). As expected, Epo stimulation selectively induced JAK2 phosphorylation in cells expressing wild-type EpoR. In cells expressing the EpoR-IFNλR1 Box1/Box2 chimera, Epo stimulation resulted in phosphorylation of JAK1, indicating the JAK1-IFNλR1 dimer is signaling-competent, and recapitulates natural JAK1-cytokine receptor dimers such as the IL-6/gp130 homodimer (Fig. 1D). Based on these results, we used mini-IFNλR1 to purify an active JAK1-IFNλR1 complex from co-expression in insect cells by two-step affinity-based purification (Fig. S1A–C). To further stabilize the complex, JAK1 was expressed with a C-terminal nanobody epitope tag (BC2T) which binds to the BC2 nanobody with high affinity (38). Addition of dimeric BC2 nanobody was used to attempt to reduce conformational heterogeneity of the complex. The components co-eluted as a single peak on size exclusion chromatography and were cross-linked with bis(sulfosuccinimidyl)suberate (BS3) which modifies solvent exposed lysine residues. The cross-linked complex was vitrified on grids for cryo-EM analysis (Fig. 1E–F, Fig. S1D).
Structure of the JAK1-IFNλR1 dimeric complex
3D reconstruction of selected particles generated a 3.6Å nominal resolution map of the 2:2 JAK1-IFNλR1 complex with C2 symmetry (Fig. S2–3). Docking of individual domain crystal structures (PDB ID: 5IXD, 4L00, and 3EYG) (14, 39, 40) was used to generate an initial model which was subject to multiple rounds of manual building and refinement, culminating in an atomic model of full-length JAK1 (Pro32-Lys1153) and a 37 amino acid segment of IFNλR1 Box1/Box2 (Pro255-Leu291).
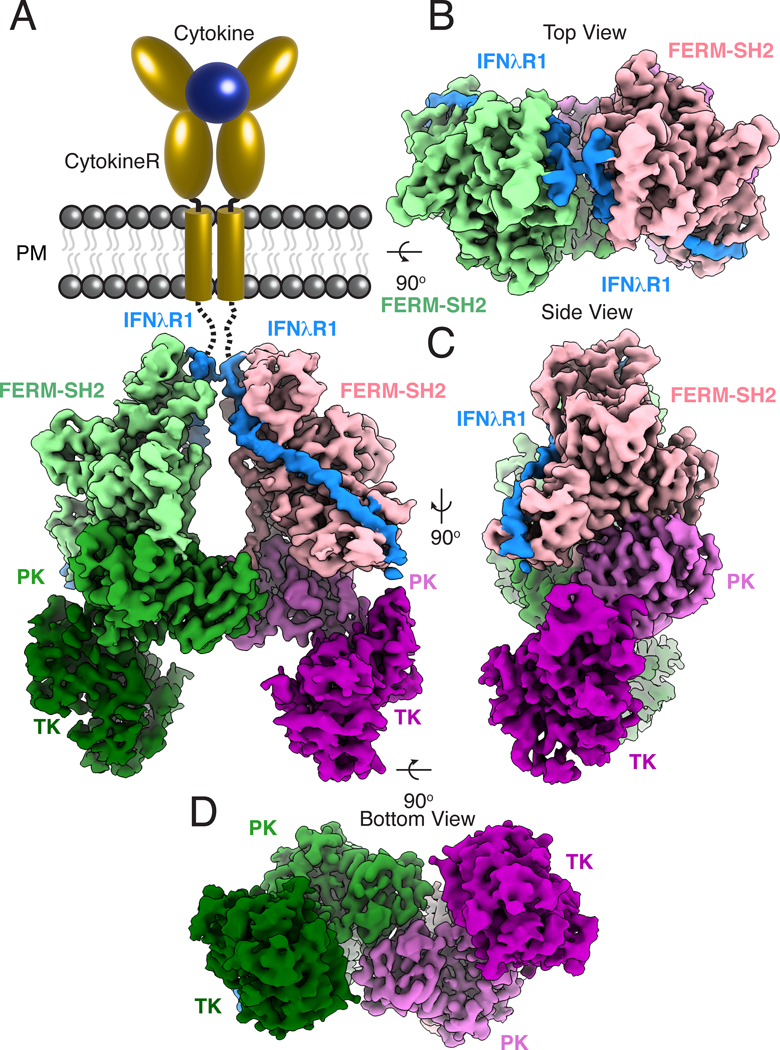
The JAK1-IFNλR1 complex associates into a C2 symmetric dimer (Fig. 2). At the membrane-proximal region, the N-terminal JAK1 FERM-SH2 domains are poised to receive the IFNλR1 ICDs as they would extend from the TM regions mimicked by the GCN4 zippers. The FERM-SH2 modules sit above inward-facing PK domains, which form a head-to-head dimer at the center of the complex. The close association between the FERM-SH2 and PK domains positions the C-terminal TK domains at the base of the JAK1 dimer, facing outwards with their catalytic clefts accessible for phosphotransferase activity. The relative positions of the kinase domains may be stabilized by the tandem BC2 nanobody bound at their C-termini for imaging.
Fig. 2. Cryo-EM structure of the active JAK1-IFNλR1 dimer.
(A) Segmented density map of the JAK1-IFNλR1 dimer resolved to 3.6Å resolution with extracellular and transmembrane domains shown as schematic. Subsequent panels show top (B), side (C), and bottom (D) views of the complex. Map threshold used in ChimeraX is set to 0.2 (~5.2σ). Individual JAK monomers are colored as a light-to-dark gradient from N to C terminus. Monomer 1: FERM-SH2, light green; PK, green; TK, dark green. Monomer 2: FERM-SH2, pink; PK, orchid; TK, purple. Density corresponding to IFNλR1 is colored blue.
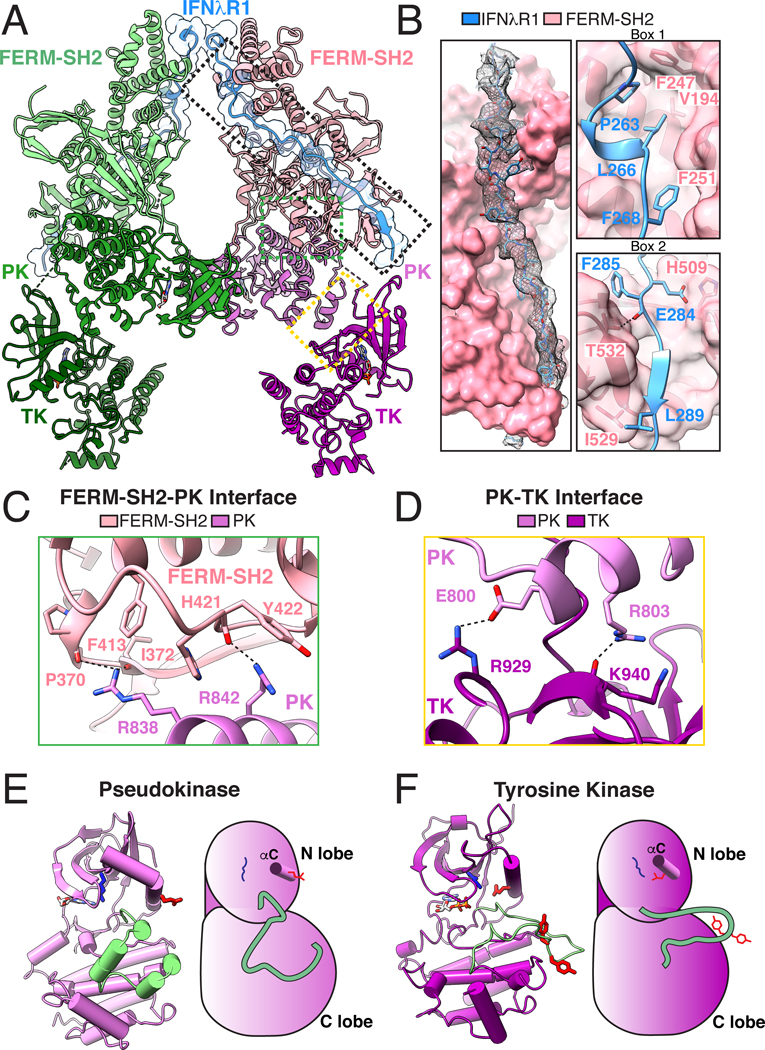
Each JAK1-IFNλR1 unit consists of four interacting modules: 1 - IFNλR1 binding to JAK1 FERM-SH2, 2 - FERM domain packing against PK C-lobe, 3 - PK domain interacting with the N-lobe of TK, and 4 - the central PK dimer interface (Fig. 3A, Table S1). At the membrane-proximal region of the complex, continuous density is observed for the IFNλR1 peptide, which binds along an extended groove on the surface of the FERM-SH2 through its the Box1 and Box2 motifs, burying approximately 1650Å2 of surface (Fig 3B). The IFNλR1 Box1 PXXLXF motif required for JAK1 binding forms a short 310 helix which positions Leu266 and Phe268 of the peptide into a hydrophobic pocket in the JAK1 FERM domain consisting of Val194, Phe247, and Phe251 (Fig. 3B, top right), similar to a crystal structure of human JAK1 FERM-SH2 bound to IFNλR1 (14). We also observe density for 22 amino acids (Glu270- Leu291) comprising the C-terminal portion of the peptide where IFNλR1 is held in the SH2 peptide binding grove by a salt bridge interaction between Glu284 in IFNλR1 and His509 in the JAK1 SH2 domain and a hydrogen bonding interaction between Thr532 in SH2 βG1 and the backbone carbonyl of IFNλR1 Phe285. Beneath these specific interactions, IFNλR1 Box2 Asp287-Leu289 forms an anti-parallel β-sheet with βG1 of JAK1 SH2 before the ICD exits the FERM-SH2 module and adopts a molten globule disordered state in the cytosol (41) that can freely interact with the kinase domains (Fig. 3B, bottom right).
Fig. 3. Reconstitution of the full-length JAK1-IFNλR1 signaling complex.
(A) Ribbon diagram of the 2:2 JAK1-IFNλR1 complex. Dashed boxes indicate magnified views in the subsequent panels. (B) IFNλR1 binds JAK1 FERM and SH2 domains via N-terminal Box1 and C-terminal Box2 motifs within the receptor intracellular domain. Left: overall interaction between IFNλR1 and FERM-SH2 shown in surface representation with peptide density from the cryo-EM map shown as black mesh contoured at ~6.1σ. Upper right: IFNλR1 Box1 motif binds JAK1 FERM domain via a conserved PXXLXF motif. Lower right: IFNλR1 Box2 motif forms an anti-parallel β sheet with βG1 in the JAK1 SH2 domain. Hydrogen bonds and salt bridges are shown as black dashed lines. (C) Interface view of the FERM-SH2-PK domains. (D) Close-up view of the PK-TK interaction. (E) Ribbon diagram (left) and schematic (right) of the PK domain in standard view. Residues corresponding to the activation loop in a functional tyrosine kinase are shown in pale green. Active site Lys621 is shown in blue, catalytic Glu636 on αC helix is shown in red. (F) Ribbon diagram (left) and schematic (right) of the TK domain in standard view. The TK activation loop is colored pale green with tyrosine residues Tyr1033 and Tyr1034 colored red. The catalytic Glu924 (red) facing inwards towards Lys907 (blue) in the kinase active site. Amino acid abbreviations: F, Phe; V, Val; P, Pro; L, Leu; H, His; E, Glu; I, Ile; Y, Tyr; R, Arg; K, Lys; T, Thr.
Below the peptide binding region, the JAK1 FERM domain forms a broad interface with the C-lobe and catalytic loop of the PK domain, burying approximately 1100Å2. At the core of this interface, the base of FERM-SH2 interacts with tandem arginine residues on successive helical turns of the PK αI helix. Arg838 in PK αI contacts residues P370 and I372 in FERM while Arg842 interacts with the backbone and side chain of Tyr422 in the FERM-SH2 linker (Fig. 3C, Fig. S4A). At the opposite face of the PK C-lobe, the PK-αG helix forms a limited interaction with the N-lobe of the TK domain and the PK-TK linker which buries 580Å2 surface area (Fig. 3D). This site consists of a salt bridge between PK-αG Glu800 and Arg929 in TK-αC and is stabilized by a hydrogen bond between PK-Arg803 and the backbone carbonyl group of Lys940 in TK-β4.
The PK domains adopt an inactive conformation as evidenced by a closed activation loop and an outward rotation of the catalytic glutamate on the C-helix (Fig. 3E). Although we observe adenosine within the nucleotide-binding site, the PK domain lacks the canonical DFG motif necessary for catalytic activity, consistent with a structural regulatory role in JAK signaling, as opposed to a catalytic role. In contrast, the TK domains adopt an active conformation with open activation loop, catalytic glutamate facing inwards towards the active site, and ADP bound at the nucleotide-binding site (Fig. 3F).
Pseudokinase dimerization and stabilization by oncogenic Val→Phe mutation
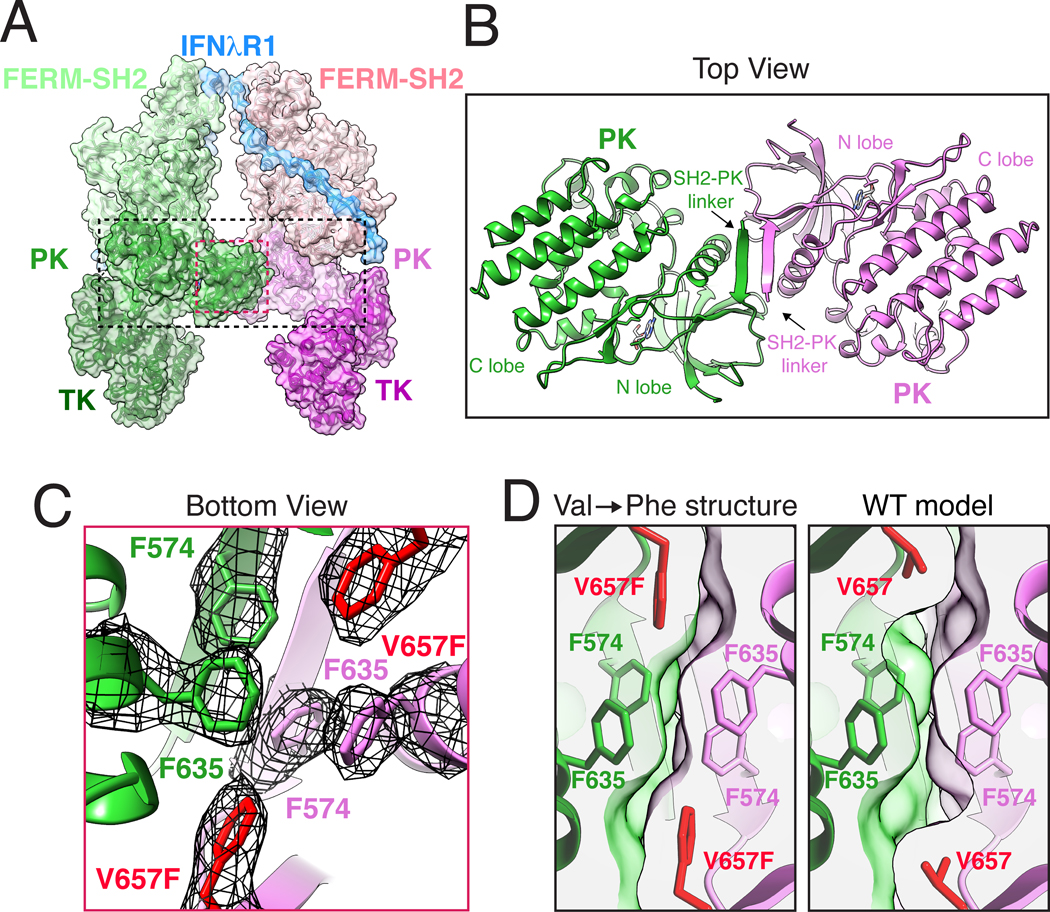
The central fulcrum of the JAK1 homodimer is formed by the SH2-PK linker and PK N-lobes from individual JAK1 monomers, which interact through a tightly packed hydrophobic cluster of six phenylalanine residues, and an anti-parallel β sheet (Fig. 4A–B). At the membrane-proximal region of this interaction module, anti-parallel β sheets from SH2-PK linkers form a lid that projects Phe574 into the hydrophobic interface (Fig. 4C, Fig. S4B). Below the lid, Phe635 from the αC-helix abuts oncogenic V657F (corresponding to JAK2 V617F) on β4, completing the phenylalanine triad in the JAK1 monomer. Mutation of surrounding phenylalanine residues, JAK2 Phe537→Ala (mJAK1 Phe574) and JAK2 Phe595→Ala (mJAK1 Phe635), disrupts the ability of VF to activate JAK2 (39, 42). Furthermore, mutation of JAK2 Phe595 (mJAK1 Phe635), which is central to the PK interface and packs against VF, also suppress constitutive activation of JAK2 by a range of other clinical mutants across the PK domain (43). Thus, the PK interface is key to ligand-independent activity of many clinically relevant MPN mutations. To better understand the structural impact of the V→F mutation, we modeled the wild-type Val657 into the structure. The smaller Val side chain results in an unfilled pocket within the dimer interface and correspondingly poorer shape complementarity (VF:0.53, WT:0.51), decreasing buried surface area of the side chain by approximately 20% from 67Å2 to 55Å2 (Fig. 4D, Table S2). The hydrophobic Phe triad may also favor desolvation of the JAK monomer, further favoring dimer formation. It is well-established that the corresponding V617F JAK2 mutations in all JAK family members result in constitutive activity (34, 35). We generated a homology model of the JAK2 PK dimer based on a previously published structure of JAK2 PK monomer (42). Consistent with a shared mechanism for activation by V617F, the conserved Phe side chains play a similar structural role as we see here in JAK1, giving confidence that the JAK1 structural results are generalizable to the JAK/TYK family (Fig. S5, Fig. S6).
Fig. 4. JAK1 dimerization is mediated by the pseudokinase domain and enhanced the by oncogenic Val→Phe mutation.
(A) Ribbon diagram of the JAK1-IFNλR1 complex with semi-transparent surface. Dashed boxes indicate magnified views in the subsequent panels (B) Top view of the PK dimer at the center of the active JAK1 complex. The structure is shown as ribbon diagram with nucleotides shown as sticks. Labels indicate PK N lobe, C lobe, and SH2-PK linker. (C) Bottom view of the Phe triad with the cryo-EM density shown as black mesh contoured at ~9σ. Oncogenic V657F mutation is highlighted in red. (D) V657F enhances shape complementarity of the PK dimerization interface. Cross-section view of the PK-PK interface as seen from the bottom with V657F cryo-EM structure compared to a model of wild-type (WT) Val657. The WT model was created using Coot and surface clipping was set at Phe/Val657 Cβ for both panels to facilitate comparison. Amino acid abbreviations: F, Phe; V, Val.
The PK dimer interface that we visualize for the JAK1 VF mutant is likely ‘on pathway’ stabilizing the same dimerization mode formed by cytokine-mediated activation of non-mutated JAKs. We suggest that the VF mutation simply enhances the tendency of the PK domains to naturally dimerize by improving structural and hydrophobic complementarity of the wild-type dimer interface. Previous structure-function data have shown that wild-type JAK2 requires the PK domain to enhance ligand-induced dimerization (3), and mutation of JAK2 Phe595→Ala (mJAK1 Phe635) in the context of wild-type JAK negatively impacts cytokine-mediated signaling (43). Thus, the wild-type PK interface is ‘de-tuned,’ relative to the VF mutant, in order to dimerize only under conditions of ligand-mediated receptor activation, an effect exploited by Val→Phe mutation.
Human Gain of Function Mutations Suggest a Two-Step Mechanism for JAK activation
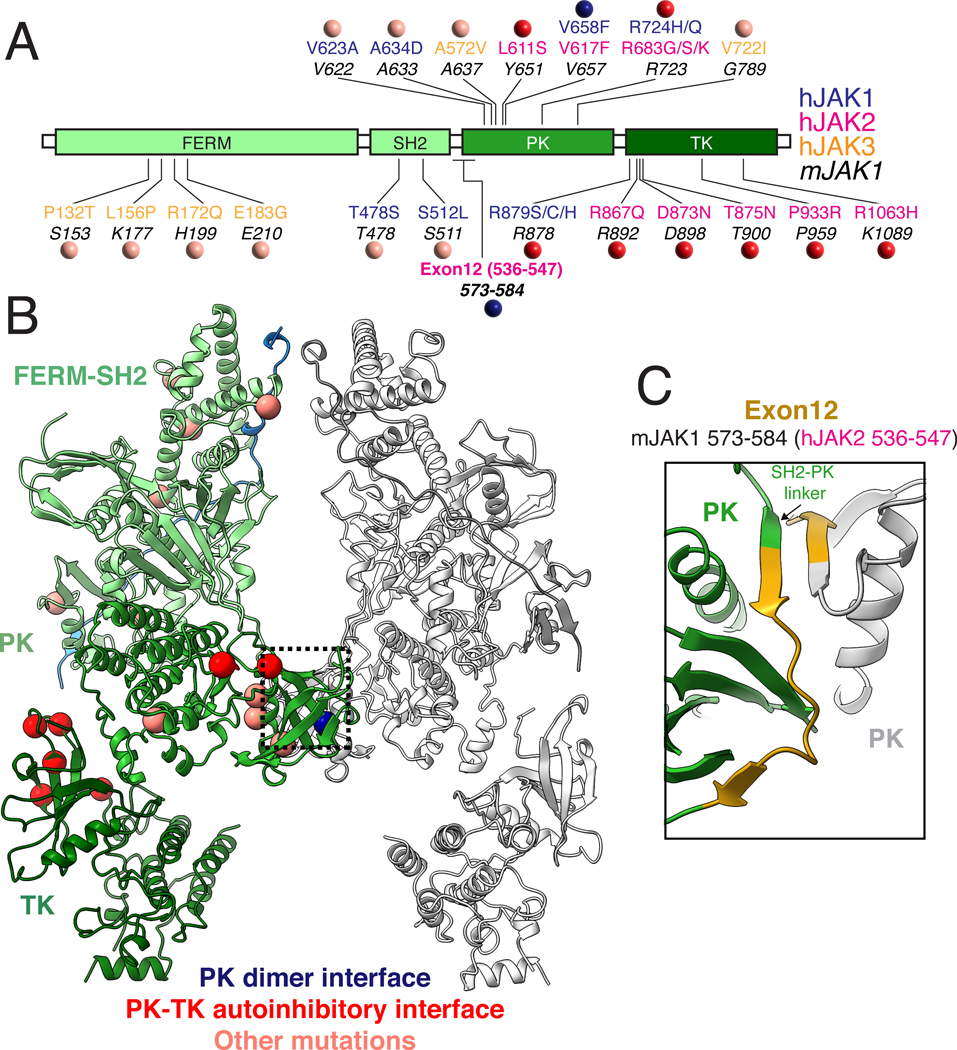
GOF mutations in JAK family members result in a diverse set of hematological malignancies including Acute Myeloid Leukemia (AML), B and T cell Acute Lymphoblastic Leukemia (B-ALL and T-ALL), and MPN. While the Val→Phe mutation in JAK2 is best characterized, a wide variety of JAK mutations have been identified with distinct phenotypic outcomes (Fig. 5A–B) (44), and many of these mutations map to the PK domain including the JAK2 Arg683→Gly mutation associated with familial thrombocytosis (45). Previous work has identified exon12 within JAK2 to be a hotspot for oncogenic mutation (46, 47). Strikingly, the exon 12 region of JAK2 maps to the SH2-PK linker in our JAK1 structure, which contributes to the PK dimer interface through the formation of anti-parallel β-sheets (Fig. 5C). However, another set of mutations that map to the N-lobe of the TK, including JAK2 Thr875→Asn, are solvent exposed in the active JAK structure, suggesting their mechanism of action may be distinct from Val→Phe.
Fig 5. Mapping human Gain of Function mutations on JAK1 suggests multiple mechanisms of oncogenic activation.
(A) Linear diagram of JAK domains showing the location of human gain of function mutations. Location of patient mutations in hJAK1 (blue), hJAK2 (pink), and hJAK3 (yellow) are shown above the analogous position in mJAK1 (44). Colored circles indicate classification of mutations based on their location at the active PK dimer interface (blue), in the autoinhibitory PK-TK interface based on a previously reported crystal packing structure of TYK2 (red) (18), or at sites with unknown function (salmon). (B) Structure of the active JAK1-IFNλR1 complex with the position of oncogenic mutations shown as balls colored by proposed mechanism of action as described above. (C) Close up of the PK dimer interface highlighting the residues in mJAK1 corresponding to hJAK2 exon 12 which has previously been identified as a hotspot for oncogenic mutations. Amino acid abbreviations: P, Pro; T, Thr; S, Ser; L, Leu; K, Lys; R, Arg; Q, Gln; E, Glu; G, Gly; C, Cys; D, Asp; N, Asn; V, Val; A, Ala; H, His; Y, Tyr; F, Phe.
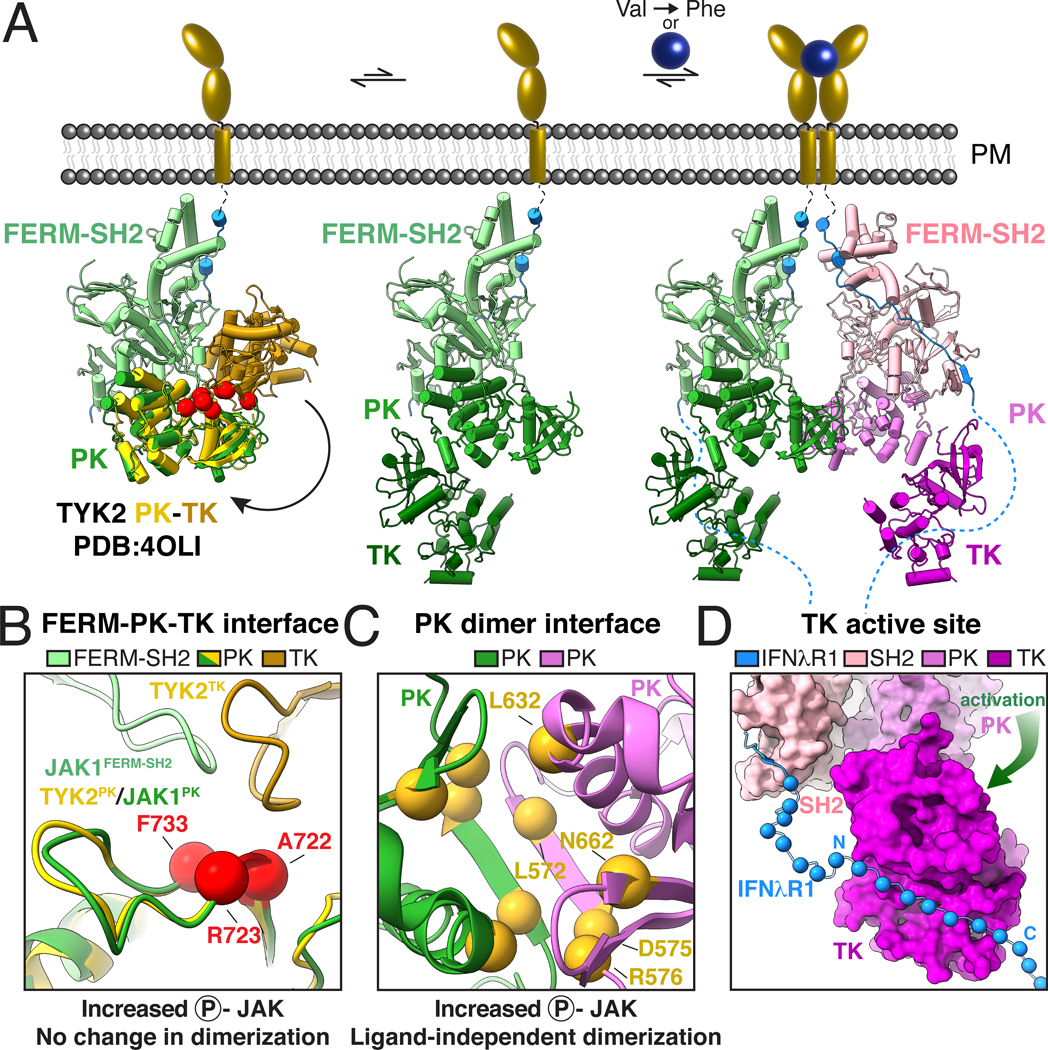
In order to better understand how TK mutations activate JAK signaling, we aligned a previously reported structure of the autoinhibited TYK2 PK-TK domain fragment to the FERM-SH2-PK module from the full-length JAK1 dimer complex (Fig. 6A, left) (18). This model suggests a compact JAK monomer in which the TK is folded back on the FERM-SH2 domain, thereby occluding the activation loop and kinase active site to mediate autoinhibition. This closed state is incompatible with JAK dimerization due to a steric clash between the PK domain and opposing FERM domain within the JAK dimer. However, previous negative-stain EM imaging of a JAK1 monomer suggests it adopts a dynamic range of conformational states from a compact ‘closed’ state to an extended ‘open’ state which may be compatible with dimerization (8). In the absence of receptor dimerization, this open state is likely transient. However, activation by cytokine-mediated receptor dimerization or VF mutation results in formation of a JAK dimer that may shift the equilibrium away from the autoinhibited state to an ‘open’ state, thus releasing the TK for full activity while also driving close proximity of apposing TKs to facilitate trans-phosphorylation.
Fig 6. Mechanistic model for JAK activation by both cytokine and oncogenic mutation.
(A) Proposed mechanism of JAK activation by ligand-induced dimerization and Val→Phe oncogenic mutation. An autoinhibited model of full-length JAK (left) was generated by docking a crystal structure of the PK-TK domains from hTYK2 (PDB ID: 4OLI; PK, yellow; TK, gold) (18) into the FERM-SH2-PK from the mJAK1 cryo-EM structure. Red balls indicate the position of activating mutations in the proposed autoinhibitory interface (44). A dynamic equilibrium between the autoinhibited ‘closed’ state and a partially active ‘open’ state (middle) exposes the PK domain and SH2-PK linker to allow for JAK dimerization. Cytokine-mediated receptor dimerization or oncogenic Val→Phe mutation facilitates formation of the PK dimer, sterically preventing autoinhibition and liberating the kinase domains for phosphotransferase activity (right). (B-C) Mechanistic mutations tracking receptor dimerization and JAK2 phosphorylation support a two-step model for activation. (B) Mutations at the proposed autoinhibitory interface enhance JAK2 phosphorylation but do not impact dimerization. Close up view of the autoinhibitory model in A with red balls indicate the positions of mutations previously found to increase JAK2 phosphorylation without inducing receptor dimerization (3). Residues are labeled according to their position in mJAK1: Ala722 (JAK2 Ile682→Phe), Arg723 (JAK2 Arg683→Gly), Phe733 (JAK2 Phe694→Leu). (C) Mutations at the PK dimerization interface increase both JAK2 phosphorylation and dimerization. Close up view of the JAK1-IFNλR1 PK dimer interface as viewed from the bottom. Yellow balls indicate the positions of mutations previously found to increase both JAK2 phosphorylation and receptor dimerization (3). Residues are numbered according to their position in mJAK1: Leu572 (JAK2 M535→Ile), Asp575 (JAK2 His538→Leu), Arg576 (JAK2 Lys539→Leu), Leu632 (JAK2 Glu592→Trp), Asn662 (JAK2 Asn622→Ile). (D) Model of receptor phosphorylation by the JAK1 dimer. Cryo-EM structure of the JAK1-IFNλR1 dimer is shown with TK domain in standard view. JAK1 is shown as surface with additional residues of IFNλR1 modeled as Cα balls for every other residue exiting the JAK1 SH2 domain and projecting towards the kinase active site. Amino acid abbreviations: F, Phe; A, Ala; R, Arg; L, Leu; N, Asn; D, Asp.
This two-step model of JAK activation predicts that oncogenic mutations may act by one of two possible mechanisms: 1- destabilizing the autoinhibited state (Fig. 6A, left), or 2- stabilizing the dimeric active state. Consistent with this model, paired analysis of JAK2 phosphorylation and receptor dimerization as measured by single-molecule receptor tracking at physiological expression levels, identified two classes of activating mutations: those which enhance JAK2 phosphorylation without impacting receptor dimerization, and those which increase both JAK2 phosphorylation and receptor dimerization (3). Mapping these two classes of mutations onto the active JAK1-IFNλR1 structure indicates that mutations which increase JAK phosphorylation without inducing dimerization cluster at the FERM-PK-TK interface in the autoinhibited model, suggesting that destabilizing this inter-domain interaction releases JAK from autoinhibition (Fig. 6B). Conversely, those mutations that induce JAK phosphorylation and receptor dimerization reside at the PK-PK dimerization interface, favoring JAK dimerization (Fig. 6C). Once the autoinhibition is relieved by JAK dimerization, the TK is ideally positioned at the base of the structure to receive the receptor intracellular domain peptide exiting the bottom of the FERM-SH2 groove to phosphorylate tyrosine residues that serve as STAT recruitment sites (Fig. 6D).
Discussion:
The cryo-EM structure of activated full-length JAK1 associated with IFNλR1 ICD provides a snapshot of a complete intracellular signaling assembly at the initiating step of both cytokine-induced and oncogenic JAK-STAT signaling. Collectively, the active JAK1-IFNλR1 dimer structure, along with a wealth of previously reported biochemical and patient mutational data, suggests a mechanism for ligand-mediated JAK activation, which is then exploited by ‘on-pathway’ pathogenic mutations.
The two-step allosteric model we propose for JAK activation is supported by an abundance of previously reported structure-function data. For example, unlike other tyrosine kinases which require activation through phosphorylation by an upstream kinase, JAK TK domains show constitutive catalytic activity when expressed in isolation (18, 40, 48, 49). The constitutive catalytic activity of the kinase domain is suppressed by expression of the tandem PK-TK domains, suggesting an autoinhibitory role for the PK domain (18, 49). This autoinhibition has been rationalized, in part, by structural models for the PK-TK domains that form head-to-head dimers through interactions between kinase N-lobes, which could potentially sterically occlude substrate binding and catalytic activity (18, 19). Our finding that activating Val→Phe mutations are positioned at a central PK-PK dimer interface within the active JAK1-IFNλR1 complex suggest a simple mechanism for oncogenic activation in which improved shape complementarity and hydrophobicity drive ligand-independent dimerization.
Recent discovery of highly selective TYK2 PK inhibitors, which allosterically stabilize an autoinhibited conformation, underscore that the JAK family is amenable to development of allosteric, rather than active-site directed inhibitors of kinase function (50, 51). One current challenge in the treatment of JAK2 V617F patients is resistance to kinase inhibitors as a result of heterodimerization and activation of JAK1 and TYK2 (52). Thus, new therapies could be designed to directly target the Val→Phe homodimer interface to increase specificity and reduce possibility for escape through activation of other JAKs. More generally, classification of oncogenic mutations by their mechanism of action, either through disruption of autoinhibition or increased dimerization, may provide a differential diagnostic criterion to inform therapeutic strategies.
The homodimeric JAK structure visualized here gives insight into mechanisms underlying the ‘tunability’ of cytokine receptor signaling. Prior studies using genetically engineered chimeric receptors, or engineered ligands, have shown that the geometric variation of the cytokine receptor dimer can influence the nature of downstream signaling (53–56). The JAK PK dimer interface could act potentially as an intracellular fulcrum in the manner of a ball and socket joint, to reposition the relative orientations and proximities of the C-terminal TK domains resulting in differential phosphorylation of the receptor ICDs and downstream STATs. In addition, our structure begins to rationalize how engineered cytokine ligands can elicit partial agonism. Partial agonists of cytokines have been engineered through mutational disruption of the low-affinity ‘site 2’ cytokine receptor binding site that lower the efficiency of receptor dimerization (57–61). The low affinity of the JAK PK dimerization interface might allow small changes in extracellular affinity to be sensitively transmitted to the downstream signaling apparatus to regulate the level of STAT activation. Indeed, the increased affinity of the JAK2 V617F mutant exploits this natural dimerization interface to drive ligand-independent signaling.
Many questions remain to refine our understanding of the cytokine receptor and JAK activation process. For example, the conformational transition from the presumed ‘closed’ state of the monomeric JAK to the activated ‘open’ state in the dimer is largely speculative, but resolution of this question could provide new mechanism-based opportunities to modulate cytokine receptor signaling. The resolution of the JAK1 homodimeric complex now allows for the design of small-molecule inhibitors of VF dimerization by in silico and experimental screening approaches based on the newly resolved PK dimer. Additionally, the structural basis for how cytokine receptor intracellular domains are phosphorylated at specific STAT docking sites, followed by binding, activation and release of activated phospho-STATs remains a next frontier in the structural biology of cytokine receptor signaling.
Supplementary Material
Acknowledgments:
We thank members of the Garcia Lab for thoughtful discussion and helpful feedback. Cryo-EM data were collected at the Stanford cryo-EM center (cEMc). We thank Drs. Elizabeth Montabana and Yee-Ting Li for generous support.
Funding:
National Institutes of Health grant R37AI51321 (KCG)
Howard Hughes Medical Institute (KCG)
Ludwig Institute for Cancer Research (KCG)
Helen Hay Whitney Foundation (RAS)
National Science Foundation Graduate Research Fellowship DGE-1656518 (CRG)
Human Science Frontier Program Organization Fellowship LT000011/2016-L (NT)
Footnotes
Competing interests: KCG is the founder of Synthekine.
Data and materials availability: The cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession code EMD-25715. The model coordinates have been deposited in the Protein Data Bank (PDB) under accession code 7T6F.
References and Notes
- 1.O’Shea JJ, Holland SM, Staudt LM, JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med 368, 161–170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Lupardus P, Laporte SL, Garcia KC, Structural biology of shared cytokine receptors. Annu Rev Immunol 27, 29–60 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilmes S. et al. , Mechanism of homodimeric cytokine receptor activation and dysregulation by oncogenic mutations. Science 367, 643–652 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stroud RM, Wells JA, Mechanistic diversity of cytokine receptor signaling across cell membranes. Sci STKE 2004, re7 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Watowich SS et al. , Homodimerization and constitutive activation of the erythropoietin receptor. Proc Natl Acad Sci U S A 89, 2140–2144 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubbard SR, Mechanistic Insights into Regulation of JAK2 Tyrosine Kinase. Front Endocrinol (Lausanne) 8, 361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bousoik E, Montazeri Aliabadi H, “Do We Know Jack” About JAK? A Closer Look at JAK/STAT Signaling Pathway. Front Oncol 8, 287 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lupardus PJ et al. , Structural snapshots of full-length Jak1, a transmembrane gp130/IL-6/IL-6Ralpha cytokine receptor complex, and the receptor-Jak1 holocomplex. Structure 19, 45–55 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murakami M. et al. , Critical cytoplasmic region of the interleukin 6 signal transducer gp130 is conserved in the cytokine receptor family. Proc Natl Acad Sci U S A 88, 11349–11353 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stark GR, Darnell JE Jr., The JAK-STAT pathway at twenty. Immunity 36, 503–514 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LaFave LM, Levine RL, JAK2 the future: therapeutic strategies for JAK-dependent malignancies. Trends Pharmacol Sci 33, 574–582 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Babon JJ, Lucet IS, Murphy JM, Nicola NA, Varghese LN, The molecular regulation of Janus kinase (JAK) activation. Biochem J 462, 1–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrao R, Lupardus PJ, The Janus Kinase (JAK) FERM and SH2 Domains: Bringing Specificity to JAK-Receptor Interactions. Front Endocrinol (Lausanne) 8, 71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrao R. et al. , The Structural Basis for Class II Cytokine Receptor Recognition by JAK1. Structure 24, 897–905 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang D, Wlodawer A, Lubkowski J, Crystal Structure of a Complex of the Intracellular Domain of Interferon lambda Receptor 1 (IFNLR1) and the FERM/SH2 Domains of Human JAK1. J Mol Biol 428, 4651–4668 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrao RD, Wallweber HJ, Lupardus PJ, Receptor-mediated dimerization of JAK2 FERM domains is required for JAK2 activation. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallweber HJ, Tam C, Franke Y, Starovasnik MA, Lupardus PJ, Structural basis of recognition of interferon-alpha receptor by tyrosine kinase 2. Nat Struct Mol Biol 21, 443–448 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lupardus PJ et al. , Structure of the pseudokinase-kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proc Natl Acad Sci U S A 111, 8025–8030 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shan Y. et al. , Molecular basis for pseudokinase-dependent autoinhibition of JAK2 tyrosine kinase. Nat Struct Mol Biol 21, 579–584 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spangler JB, Moraga I, Mendoza JL, Garcia KC, Insights into cytokine-receptor interactions from cytokine engineering. Annu Rev Immunol 33, 139–167 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo Y. et al. , JAK-STAT signaling in human disease: From genetic syndromes to clinical inhibition. J Allergy Clin Immunol 148, 911–925 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodig SJ et al. , Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell 93, 373–383 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Neubauer H. et al. , Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell 93, 397–409 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Parganas E. et al. , Jak2 is essential for signaling through a variety of cytokine receptors. Cell 93, 385–395 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Thomis DC, Gurniak CB, Tivol E, Sharpe AH, Berg LJ, Defects in B lymphocyte maturation and T lymphocyte activation in mice lacking Jak3. Science 270, 794–797 (1995). [DOI] [PubMed] [Google Scholar]
- 26.Nosaka T. et al. , Defective lymphoid development in mice lacking Jak3. Science 270, 800–802 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Macchi P. et al. , Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID). Nature 377, 65–68 (1995). [DOI] [PubMed] [Google Scholar]
- 28.Kilpivaara O, Levine RL, JAK2 and MPL mutations in myeloproliferative neoplasms: discovery and science. Leukemia 22, 1813–1817 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Baxter EJ et al. , Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 365, 1054–1061 (2005). [DOI] [PubMed] [Google Scholar]
- 30.James C. et al. , A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 434, 1144–1148 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Kralovics R. et al. , A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med 352, 1779–1790 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Levine RL et al. , Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 7, 387–397 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Harrison DA, Binari R, Nahreini TS, Gilman M, Perrimon N, Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J 14, 2857–2865 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haan C. et al. , Jak1 has a dominant role over Jak3 in signal transduction through gammac-containing cytokine receptors. Chem Biol 18, 314–323 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Staerk J, Kallin A, Demoulin JB, Vainchenker W, Constantinescu SN, JAK1 and Tyk2 activation by the homologous polycythemia vera JAK2 V617F mutation: cross-talk with IGF1 receptor. J Biol Chem 280, 41893–41899 (2005). [DOI] [PubMed] [Google Scholar]
- 36.O’Shea EK, Klemm JD, Kim PS, Alber T, X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254, 539–544 (1991). [DOI] [PubMed] [Google Scholar]
- 37.Lu X, Gross AW, Lodish HF, Active conformation of the erythropoietin receptor: random and cysteine-scanning mutagenesis of the extracellular juxtamembrane and transmembrane domains. J Biol Chem 281, 7002–7011 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Braun MB et al. , Peptides in headlock--a novel high-affinity and versatile peptide-binding nanobody for proteomics and microscopy. Sci Rep 6, 19211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toms AV et al. , Structure of a pseudokinase-domain switch that controls oncogenic activation of Jak kinases. Nat Struct Mol Biol 20, 1221–1223 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams NK et al. , Dissecting specificity in the Janus kinases: the structures of JAK-specific inhibitors complexed to the JAK1 and JAK2 protein tyrosine kinase domains. J Mol Biol 387, 219–232 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Skiniotis G, Lupardus PJ, Martick M, Walz T, Garcia KC, Structural organization of a full-length gp130/LIF-R cytokine receptor transmembrane complex. Mol Cell 31, 737–748 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bandaranayake RM et al. , Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F. Nat Struct Mol Biol 19, 754–759 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dusa A, Mouton C, Pecquet C, Herman M, Constantinescu SN, JAK2 V617F constitutive activation requires JH2 residue F595: a pseudokinase domain target for specific inhibitors. PLoS One 5, e11157 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen E, Staudt LM, Green AR, Janus kinase deregulation in leukemia and lymphoma. Immunity 36, 529–541 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carreno-Tarragona G. et al. , A typical acute lymphoblastic leukemia JAK2 variant, R683G, causes an aggressive form of familial thrombocytosis when germline. Leukemia 35, 3295–3298 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Scott LM, The JAK2 exon 12 mutations: a comprehensive review. Am J Hematol 86, 668–676 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Scott LM et al. , JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N Engl J Med 356, 459–468 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanz Sanz A. et al. , The JH2 domain and SH2-JH2 linker regulate JAK2 activity: A detailed kinetic analysis of wild type and V617F mutant kinase domains. Biochim Biophys Acta 1844, 1835–1841 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Saharinen P, Silvennoinen O, The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J Biol Chem 277, 47954–47963 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Burke JR et al. , Autoimmune pathways in mice and humans are blocked by pharmacological stabilization of the TYK2 pseudokinase domain. Sci Transl Med 11, (2019). [DOI] [PubMed] [Google Scholar]
- 51.Tokarski JS et al. , Tyrosine Kinase 2-mediated Signal Transduction in T Lymphocytes Is Blocked by Pharmacological Stabilization of Its Pseudokinase Domain. J Biol Chem 290, 11061–11074 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koppikar P. et al. , Heterodimeric JAK-STAT activation as a mechanism of persistence to JAK2 inhibitor therapy. Nature 489, 155–159 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moraga I. et al. , Tuning cytokine receptor signaling by re-orienting dimer geometry with surrogate ligands. Cell 160, 1196–1208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mohan K. et al. , Topological control of cytokine receptor signaling induces differential effects in hematopoiesis. Science 364, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Staerk J. et al. , Orientation-specific signalling by thrombopoietin receptor dimers. EMBO J 30, 4398–4413 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brooks AJ et al. , Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science 344, 1249783 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Glassman CR et al. , Calibration of cell-intrinsic interleukin-2 response thresholds guides design of a regulatory T cell biased agonist. Elife 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mo F. et al. , An engineered IL-2 partial agonist promotes CD8(+) T cell stemness. Nature 597, 544–548 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Glassman CR et al. , Structural basis for IL-12 and IL-23 receptor sharing reveals a gateway for shaping actions on T versus NK cells. Cell 184, 983–999 e924 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saxton RA et al. , The tissue protective functions of interleukin-22 can be decoupled from pro-inflammatory actions through structure-based design. Immunity 54, 660–672 e669 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saxton RA et al. , Structure-based decoupling of the pro- and anti-inflammatory functions of interleukin-10. Science 371, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mastronarde DN, Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Punjani A, Zhang H, Fleet DJ, Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat Methods 17, 1214–1221 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Punjani A, Fleet DJ, 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J Struct Biol 213, 107702 (2021). [DOI] [PubMed] [Google Scholar]
- 66.Sanchez-Garcia R. et al. , DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun Biol 4, 874 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liebschner D. et al. , Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pettersen EF et al. , UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tunyasuvunakool K. et al. , Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Casanal A, Lohkamp B, Emsley P, Current developments in Coot for macromolecular model building of Electron Cryo-microscopy and Crystallographic Data. Protein Sci 29, 1069–1078 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lawrence MC, Colman PM, Shape complementarity at protein/protein interfaces. J Mol Biol 234, 946–950 (1993). [DOI] [PubMed] [Google Scholar]
- 72.Croll TI, ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr D Struct Biol 74, 519–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krissinel E, Henrick K, Inference of macromolecular assemblies from crystalline state. J Mol Biol 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.