Abstract
The retinoblastoma tumor suppressor protein (pRB) is a known regulator of cell-cycle control; however, recent studies identified critical functions for pRB in regulating cancer-associated gene networks that influence the DNA damage response, apoptosis, and cell metabolism. Understanding the impact of these pRB functions on cancer development and progression in the clinical setting will be essential, given the prevalence of pRB loss of function across disease types. Moreover, the current state of evidence supports the concept that pRB loss results in pleiotropic effects distinct from tumor proliferation. Here, the implications of pRB loss (and resultant pathway deregulation) on disease progression and therapeutic response will be reviewed, based on clinical observation. Developing a better understanding of the pRB-regulated pathways that underpin the aggressive features of pRB-deficient tumors will be essential for further developing pRB as a biomarker of disease progression and for stratifying pRB-deficient tumors into more effective treatment regimens.
Introduction
The retinoblastoma protein (pRB) was the first protein identified as a tumor suppressor. pRB function is lost in 98% of the rare childhood malignancy retinoblastoma, in which the RB1 gene was traced to a locus on chromosome 13 and subsequently sequenced (1–4). Outside of retinoblastoma, pRB loss is common across cancer types and is strongly associated with poor progression-free survival (PFS), disease-specific survival (DSS), and overall survival (OS; ref. 5). Despite this prevalence, the mechanisms by which pRB loss promotes cancer progression are not completely understood. Here, the clinical significance of pRB alterations across tumor types will be discussed, as well as the potential utility of pRB loss as a biomarker for prognosis and therapy resistance.
Mechanisms Driving Altered pRB Function in Human Malignancies
pRB exerts a well-understood function in regulating cell-cycle progression by inhibiting the activity of E2F transcription factors. In brief, pRB function is directly regulated by cyclin-dependent kinase 4 and 6 (CDK4/6) and cyclin D complexes. In response to mitogenic stimuli, CDK4/6 proteins are released from upstream CDK inhibitors (CDKI), such as p16INK4a, and form complexes with cyclin D. These complexes then phosphorylate pRB, leading to a conformational change and release from E2F transcription factors, allowing for the activation of transcription and cell-cycle progression (6). As such, altered pRB function can be derived from multiple mechanisms including changes in the RB1 gene itself, and altered function of upstream pathway regulators. Interestingly, these mechanisms tend to be disease type specific (Table 1) and are further discussed below.
Table 1.
Frequency of pRB pathway alterations in cancer.
| Tumor | RB1 | CDKN2A (p16INK4) | CDK4 | CCND1 (cyclin D1) | Citations |
|---|---|---|---|---|---|
| Glioma | 33%–85%—Genomic loss | (27, 29, 31) | |||
| Retinoblastoma | 95%—Mutation | (18, 20) | |||
| 16%—Promoter methylation | |||||
| Esophageal | 19%–70%—Promoter methylation | ∼85%—Elevated protein | (36–38) | ||
| Head and neck | 17%–27%—Promoter methylation | 26%–39%—Gene amplification | (33) | ||
| 25%–66%—Genomic loss | |||||
| Mantle cell lymphoma | 90%—Chromosomal rearrangement | (54) | |||
| NSCLC | 30%—Genomic loss | 31%—Promoter methylation | 5%–30%—Gene amplification | (13, 36, 43) | |
| <15%—Mutation | |||||
| SCLC | 91%–100%—Genomic loss | (16) | |||
| Breast | 72.2% Basal-like breast—Genomic loss | 31%—Promoter methylation | 16%—Mutation | 15%–30%—Gene amplification | (19, 53, 54) |
| 61.5% Luminal B breast—Genomic loss | |||||
| 2.7% Breast—Mutation | |||||
| Colorectal | 10%–40%—Promoter methylation | 2.5%—Gene amplification | (42, 44) | ||
| Ovarian | 7%–14%—Genomic loss | (26, 28, 55) | |||
| Bladder | 10%–45%—Genomic loss | (30) | |||
| Endometrial | 2%—Genomic loss | 20%–54%—Elevated protein | 26%—Gene amplification | (25, 40, 47, 49) | |
| 8.5%—Promoter methylation | |||||
| Pancreatic | 10%–37%—Genomic loss | 25%—Gene amplification | (27, 34, 35) | ||
| Cervical | 5%—Genomic loss | (25) | |||
| Prostate | 17%–33%—Genomic loss | (7–12) | |||
| Multiple myeloma | 50%—Genomic loss | 16%—Chromosomal rearrangement | (57, 98) | ||
| Osteosarcoma | 63%—Genomic loss | <15%—Mutation | (17, 22) | ||
| <5%—Mutation | |||||
| Melanoma | <25%—Gene amplification | (50, 51) |
Note: The frequency and mechanisms of RB1, CDKN1A (p16INK4), CDK4, and CCDND1 (cyclin D1) alterations across disease types.
RB1 gene modifications
Loss of pRB function in human cancer occurs predominately via deletion (one copy if heterozygous or two copies if homozygous) of the RB1 gene, RB1 promoter methylation, or mutations resulting in a nonfunctional protein. Deletion of the RB1 gene occurs in 91%–100% of small cell lung cancer (SCLC), 72.2% of basal-like and 61.5% of luminal B breast cancers, 63% of osteosarcomas, 30% of non–small cell lung cancer (NSCLC), and 17%–33% of castration-resistant prostate cancer (CRPC; refs. 7–17). In retinoblastoma, loss of pRB is attributed to hypermethylation of the RB1 promoter (16%) or RB1 gene mutation (95%; refs. 18–20), indicating that loss of pRB can occur via multiple mechanisms within the same tumor type. Beyond retinoblastoma, mutations in the RB1 gene that result in nonfunctional protein are infrequent (21), observed in less than 5% of osteosarcomas and locally advanced breast cancers (22, 23). Overall, alterations of the RB1 gene are frequent across cancers and the type of alteration tends to be disease type specific. In addition to the RB1 gene itself, pRB pathway alterations also occur in human malignancies and must be considered when examining pRB function in disease.
pRB pathway alterations
Beyond pRB itself, alterations within the pRB pathway have also been defined as mechanisms of cancer development and/or malignant progression. Inactivation of negative regulators of the pathway, such as CDKIs, and increased activation of positive regulators of the pathway, including CDKs and cyclins, have been reported in almost all human cancers (24). Alterations in p16INK4 (encoded by CDKN2A), a bona fide tumor suppressor, include genomic loss (2%–85%; refs. 25–35), somatic mutations (<15%; refs. 36–39), and altered expression driven by promoter methylation (8.5%–70%; refs. 40–45) across cancer types. However, CDK4 protein expression is often increased in cancer (20%–54%; refs. 46–49), and point mutations preventing binding to p16INK4 and subsequent inactivation are observed in 16% of breast cancers (50, 51). Further, elevated protein expression of cyclin D1 due to amplification of the CCND1 gene (2.5%–39%; refs. 52–55) and chromosomal rearrangement (16%–90%; refs. 56, 57) have been observed in specific cancers. Overall, alterations within the pRB pathway occur across pathway components, have proven to be context specific, and result in inactivation of pRB.
It has long been appreciated that mechanisms of pRB disruption are often tumor type selective, and emergent data strongly demonstrate that distinct mechanisms of pRB loss result in differential molecular and biological outcomes (16). To understand these differences and exploit them clinically, the molecular functions of pRB and the clinical implications of pRB loss must be examined.
pRB Loss and Ki67 in Clinical Samples: Unexpected Findings
As a regulator of cell-cycle control, the effect of pRB loss on proliferation has been carefully examined across tumor types and found to be dependent on the mechanism by which pRB is lost, either loss of protein or inactivation of function. When examined across CRPC and lung adenocarcinoma cohorts, loss of pRB IHC staining did not negatively correlate with Ki67 (a marker of proliferation) staining (16, 58). These studies reveal that the impact of pRB loss in these contexts is beyond cell-cycle control, driving aggressive disease without altering proliferation. pRB inactivation via hyperphosphorylation, however, does correlate with a hyperproliferative phenotype. A study examining loss of p16INK4 and phosphorylated pRB in triple-negative breast cancer (TNBC), which has the highest rate of p16INK4 loss, found a strong positive correlation between phosphorylated pRB protein and Ki67 (59). These observations, along with previous functional studies indicating that pRB protein loss is not equivalent to pRB hyperphosphorylation (16), suggest that upstream alterations in the pRB pathway can drive a proproliferative advantage. In conclusion, the biological effect of pRB loss is reliant on the mechanism by which pRB function is lost and is thus crucial to understand the subsequent consequences driving disease progression and to appropriately target these diseases clinically.
Molecular Understanding of pRB Function and Loss
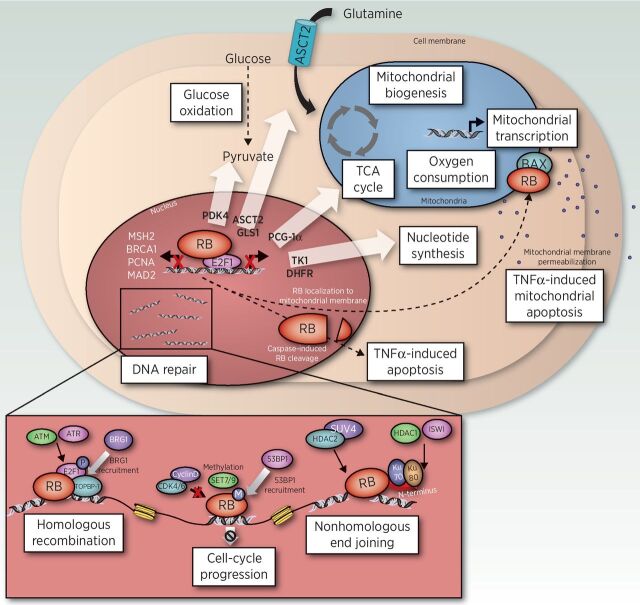
The molecular and cellular implications of pRB dysfunction are only recently being uncovered, in part as a result of next-generation sequencing technologies, advances in modeling, and the capacity to assess impact in clinical specimens. Notably, pRB has been found to interact with >200 proteins and regulate a large number of processes controlling cancer development and progression (Fig. 1).
Figure 1.
Novel pRB functions beyond cell-cycle control. Through direct regulation of the E2F transcription factors, pRB has been identified to regulate glucose oxidation (PDK4), nucleotide synthesis (TK1, DHFR), glutamine metabolism (ASCT2, GLS1), TCA cycle, mitochondrial biogenesis, oxygen consumption, and mitochondrial transcription (PCG-1α), along with numerous DNA repair pathways (MSH2, BRCA1, PCNA, and MAD2). Through caspase cleavage or translocation to the mitochondria, pRB has been shown to directly regulate TNFα-induced apoptosis. Lastly, pRB has a critical role in directly regulating nonhomologous end joining (NHEJ), homologous recombination (HR), and cell-cycle progression.
DNA damage response
pRB plays an important role in the response to DNA damage both directly and indirectly, through regulation of E2F transcription. After pRB loss, E2Fs are deregulated, leading to the constitutive activation of target gene transcription. E2Fs regulate expression of genes encoding DNA repair factors including MSH2, BRCA1, and PCNA (60, 61), all of which play a critical role in DNA damage response. Beyond transcriptional control, pRB has been described as a regulator of nonhomologous end joining (NHEJ) and homologous recombination (HR) through direct interaction with DNA repair factors. pRB interacts with Ku70 and Ku80, resulting in the recruitment of chromatin modifiers known to mediate NHEJ (62). Consistently, pRB has been shown to interact with a breadth of additional chromatin modifiers including ISWI, HDAC1, HDAC2, and Suv4 (62–65). In the context of HR, pRB is recruited to sites of double-strand breaks in response to irradiation (IR) through ATM/ATR phosphorylation of E2F1, resulting in recruitment of BRG1 (66), an enzyme required for initiation of DNA end resection and subsequent repair (66). Further, pRB-deficient U2OS cells demonstrate increased sensitivity to IR compared with pRB-positive cells and are defective at resolving γ-H2AX (66), indicating that pRB loss is associated with diminished repair capacity. Thus, studies discerning the impact of pRB status on DNA repair competency in the clinical setting may assist in the development of novel therapeutic interventions for pRB-deficient tumors.
Apoptosis
In addition to DNA damage response, pRB has been reported to significantly influence apoptosis. pRB has also been shown to localize directly to the mitochondria to positively regulate tumor necrosis factor (TNF)-induced mitochondrial apoptosis (67), and has been shown to be cleaved at a c-terminal consensus site that, when mutated, renders cells resistant to TNF-induced apoptosis (68–71). Further, genetically engineered mouse models of bladder cancer revealed that pRB loss results in reduced expression of p53, along with additional genes involved in apoptosis including BAX, BAK, BID, and APAF1 (72); however, the mechanism is not fully understood. Thus, although pRB has been shown to influence apoptosis through multiple pathways, it remains crucial to fully understand these pRB functions and how they may affect therapy response.
Metabolism
Finally, emergent data have revealed critical roles for pRB in metabolic control. Repression of E2Fs by pRB has been identified as a major regulator of many metabolic pathways, including nucleotide biosynthesis, glucose oxidation, and mitochondrial function (73–78). Direct targets of E2F1 transcription include thymidine kinase (TK1) and dihydrofolate reductase (DHFR), enzymes required for nucleotide synthesis (73–75); pyruvate kinase dehydrogenase 4 (PDK4), an enzyme that contributes to the shift to a Warburg phenotype by preventing the entry of pyruvate into the citric acid cycle (76, 77); peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PCG-1alpha), which promotes mitochondrial transcription, biogenesis, and oxygen consumption; and genes involved in the electron transport chain and oxidative phosphorylation including the subunits for ATP synthase, cytochrome c oxidase, ubiquinol–cytochrome c reductase, and succinate dehydrogenase complex (76, 78). Further, pRB also plays a significant role in glutamine regulation, through the upregulated expression of the glutamine transporter ASCT2, and glutaminase (GLS1; ref. 79). pRB-deficient Drosophila models have shown an increased reliance on nucleotide metabolism and glutathione, both of which depend heavily on access to glutamine (80), suggesting an increase in dependence on glutamine upon pRB loss. Beyond E2F regulation, pRB is also implicated in regulating c-Myc, a known regulator of metabolic enzyme transcription (81–83). These studies highlight the critical impact pRB function has on metabolic transcriptional regulation, including but not limited to control of E2Fs.
As therapeutics targeting DNA repair, apoptosis, and metabolism are being introduced into the clinic, closing the gap between the biological and clinical impact of pRB loss on disease progression will be crucial to identify novel and effective treatments for these tumors.
The Impact of pRB Loss on Therapeutic Intervention
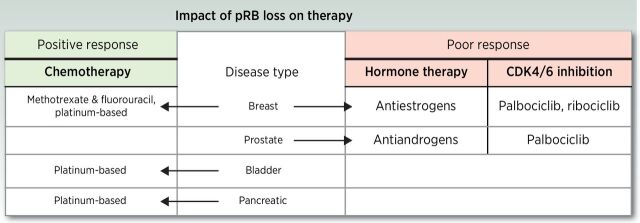
As discussed above, an abundance of observations in human tumors and defined model systems indicate that pRB loss promotes tumor development and progression through multiple biological mechanisms. Despite this knowledge, the relative impact of pRB status on therapeutic response remains loosely defined (Fig. 2).
Figure 2.
The impact of pRB loss on therapeutic intervention. Studies have observed that pRB loss promotes a positive response to chemotherapy in breast, bladder, and pancreatic cancers. Conversely, pRB loss has been identified as a mechanism of resistance to hormone therapy and CDK4/6 inhibition in breast and prostate cancers.
pRB status and DNA repair targeted therapy
As discussed above, pRB is implicated as a critical factor in prompting DNA repair. Modeling pRB loss results in a switch from canonical NHEJ to a DNA-PK–independent mechanism of NHEJ, suggesting that clinically utilized DNA-PK inhibitors may not result in a favorable outcome against these pRB-deficient tumors (62). Further, as pRB loss results in defective HR repair and increased sensitivity to IR, pRB-deficient tumors may be sensitized to double-strand break-inducing agents in combination with targeted therapy such as PARP inhibition (66); however, this requires further investigation.
pRB status and chemotherapy
Previous studies have revealed that pRB-deficient tumors have a more favorable response to specific chemotherapeutics. A study of invasive breast cancer found that patients with pRB-deficient tumors had increased disease-free survival after chemotherapy including methotrexate and fluorouracil, compared with those that retained pRB positivity (84). As methotrexate and fluorouracil reduce purine and pyrimidine synthesis, this favorable response may be attributed to the heavy reliance pRB-deficient tumors have on nucleotide synthesis (80). Further, studies in breast, bladder, and pancreatic cancers have shown pRB-deficient tumors to be more sensitive to the platinum-based chemotherapy (85–87); however, the mechanisms driving this increased sensitivity remain to be defined.
pRB status and hormone therapy
pRB loss has been well defined as a mechanism driving resistance to hormone therapy in breast and prostate cancers. For estrogen receptor (ER)–positive breast cancer and androgen receptor (AR)–positive prostate cancer, the first line of treatment is hormone ablation therapy. Tumors eventually become resistant to therapy while retaining hormone receptor positivity. A study examining gene expression revealed that elevated expression of a 59 pRB-regulated gene signature correlates with failure of therapy in breast cancer patients treated with the antiestrogen tamoxifen (85). Further, studies examining primary breast tumors revealed that patients with a functional pRB pathway showed significantly increased recurrence-free survival when treated with tamoxifen, whereas those lacking pRB were significantly associated with impaired response (84, 88), suggesting loss of pRB as an indicator for poor response to hormone therapy. Moreover, in prostate cancer, RB1 loss is found almost exclusively in CRPC (with little incidence in castration-sensitive disease), and is sufficient to promote therapy resistance through the deregulation of E2F1 and increased transcription of AR (13). These studies suggest that pRB loss can serve as a predictive marker of hormone therapy response in both breast and prostate cancers.
pRB status and CDKI
Pharmacologic inhibition of CDK4/6 is the most clinically mature means to target the pRB pathway, serving to induce tumor suppressor function by dampening the action of inhibitory kinases. The current FDA-approved CDK4/6 inhibitors include palbociclib, ribociclib, and abemaciclib, which are approved for hormone receptor–positive, HER2-negative advanced breast cancer in combination with the estrogen receptor antagonist fulvestrant (89, 90). Three individual clinical phase III studies investigating these CDK4/6 inhibitors found that palbociclib, ribociclib, and abemaciclib significantly improved PFS when treated in combination with fulvestrant compared with fulvestrant alone (90–92). However, although at the time of approval pRB positivity was not required for inclusion in each trial, there is little molecular basis to target the pathway upstream after pRB loss, providing the rationale for using pRB positivity as a biomarker for CDK4/6 inhibitor sensitivity. Further, resistance to CDK4/6 inhibitors is common, and a key mechanism by which tumors gain this resistance is through pRB loss (85). Specifically, studies in breast cancer models showed that either pRB protein loss or increased cyclin E1 expression leads to resistance to CDK4/6 inhibition (93, 94). Concordantly, genomic characterization of ribociclib-resistant patient-derived xenograft models identified an acquired RB1 frameshift mutation leading to reduced pRB expression (93). Further studies indicated that 50% of models with aberrant pRB gene signature expression showed reduced copy number of RB1 by the time of palbociclib resistance, further supporting a role for pRB loss in acquired resistance to CDK4/6 inhibition (94). In addition to pRB loss, pRB inactivation has also been identified as a resistance mechanism of CDK4/6 inhibition through elevated expression of cyclin D1, leading to restored pRB hyperphosphorylation (95). A molecular understanding of these resistance mechanisms to CDK4/6 inhibitors will play a critical role in explaining tumor response in the clinic and further provide a rationale to clinically examine pRB loss in patient tumors. As such, although pRB loss portends CDK4/6 inhibitor resistance, the presence of pRB alone is not sufficient to predict responsiveness.
Current Prognostic Value of pRB in Human Malignancies
In addition to identification as a biomarker for predicted response to therapy, pRB loss is also implicated as a prognostic marker for clinical outcome and disease stage in tumors with relatively high frequency of pRB deficiency including multiple myeloma (MM); NSCLC; breast, prostate, and bladder cancers; and osteosarcomas. While altered patterns in pRB function and expression have proven to be useful indicators of disease development and progression, their use as prognostic clinical markers remains controversial.
Multiple myeloma
Loss of pRB is observed in up to 50% of MM tumors (96, 97), and deletion of the RB1 gene significantly correlates with shorter OS, supporting additional studies suggesting that deletion of pRB has independent prognostic value in MM (98–100). Furthermore, a significant number of patients with tumors lacking pRB were found to have stage III disease at diagnosis, suggesting that pRB may also serve as a marker of disease stage in this tumor type as well (98); however, this requires further clinical investigation.
NSCLC
The use of pRB as a prognostic marker has been investigated in NSCLC; however, results remain inconsistent. When examined exclusively at early stage of disease, pRB-deficient cases exhibited a tendency for shorter OS (101–103). More specifically, when examining adenocarcinoma alone, pRB-deficient tumors were associated with shorter OS compared with pRB-positive tumors (101). However, additional smaller studies have not found pRB to provide significant prognostic value in NSCLC (104, 105); despite cohort size limitations, pRB-deficient tumors did display a trend toward poorer OS, suggesting larger cohorts are needed to fully assess the effect of pRB loss (105). Together, these studies suggest that pRB loss may portend poor outcome for patients and may be useful as a prognostic marker in specific subtypes of NSCLC; however, more targeted studies are needed to test these hypotheses.
Breast cancer
In breast cancer, the use of pRB as a marker of disease stage or clinical outcome has been shown to be subtype specific. A large, retrospective study of 1,806 patients with breast cancer revealed that loss of pRB protein was associated with TNBC and basal core phenotype subtypes (106–108), suggesting pRB loss may be a useful marker of disease subtype. However, there was no significant association with disease-free survival or OS within these subtypes, indicating the prognostic value of pRB on patient outcome may be lacking (88, 106, 109). Conversely, in ductal carcinoma in situ tumors, pRB protein loss was strongly associated with recurrence of invasive breast cancer, suggesting that the value of pRB status as a prognostic marker in this stage of disease may have utility (110). In total, although pRB status has not been confirmed as a prognostic marker for OS, it has prognostic value in specific breast cancer subtypes and needs to be further explored.
Prostate cancer
Loss of RB1 gene expression is found almost exclusively in CRPC, suggesting that genomic RB1 loss can be utilized as a marker for stage of disease (13). Moreover, the emergence of pRB loss signatures has become an increasingly utilized tool. pRB loss gene signatures are strongly represented in advanced disease and associated with reduced recurrence-free survival (13, 16). Furthermore, an independent pRB loss gene signature for dual copy RB1 loss developed using The Cancer Genome Atlas Pan-Cancer data set across cancer types found that high expression of this signature significantly correlated with shorter PFS, DSS, and OS when examined across prostate cancer cohorts (5). These data implicate pRB loss and pRB loss gene signatures as a surrogate for pRB loss as prognostic markers for disease. Understanding the biological consequences in these tumors is required for complete understanding of disease progression and development of therapies to target disease.
Other solid tumors
pRB status has also been studied in bladder cancer and osteosarcomas. Multiple studies have found a strong correlation between pRB loss, tumor grade and stage, and OS in bladder cancer, providing a rationale for the utilization of pRB as both a prognostic and staging marker in this disease type (111–113). In osteosarcoma, studies have reported a significant difference in event-free survival (between tumors with loss of heterozygosity (LOH) of pRB and those without LOH (22, 114, 115), suggesting that pRB status can be utilized as a marker for prognosis.
RB1 Germline Mutations and Cancer Predisposition
RB1 germline mutations, resulting in loss of pRB protein, have been reported as predictive markers of cancer initiation and development. Specifically, in retinoblastoma, about 40% of patients who develop the disease have a hereditary predisposition caused by a heterozygous RB1 germline mutation. Unfortunately, patients with these germline mutations have an increased risk of second primary malignancies such as osteosarcoma, lipomas, soft-tissue sarcomas, melanoma, and cancers of the brain (116–119). Examination revealed that the mutations occur throughout the RB1 gene and no correlation between the mutations and the type of secondary malignancy diagnosed have been observed (120). A better understanding of the impact RB1 germline mutations have on the risk for secondary cancer development will have a critical influence on patient outcome. Identification of patients predisposed to secondary malignancies will allow for more comprehensive cancer screenings, earlier diagnosis, and more effective treatment.
Clinical Barriers of Defining pRB Loss as a Biomarker
Although supportive evidence exists for the use of pRB status as a prognostic or predictive biomarker, it is important to note that utilizing the loss of a tumor suppressor as a biomarker introduces numerous complicating factors. Although sequencing can readily detect deletion or mutations of the RB1 gene, defining pRB status is often done through IHC staining of tumor samples. Histologic staining introduces a number of challenges as pRB protein expression is generally heterogeneous within tumors. Studies that have examined pRB status define pRB positivity by describing staining at null/weak, intermediate, or high, or compare staining to surrounding nonneoplastic cells that retained nuclear pRB positivity. As such, there remains to be a standardized method and threshold to define pRB positivity in clinical samples. Additionally, factors such as subclonal loss (121) and distinguishing between 1 and 2 copy loss are difficult to determine, especially in low tumor content samples (16). Interestingly, single-copy RB1 loss has proven to be sufficient to induce aggressive phenotypes (13). Thus, although tumor heterogeneity remains to be an obstacle, RB1 haploinsufficiency may be specifically beneficial in defining pRB status as a biomarker. Beyond the use of pRB status alone, the use of gene signatures has also proven to be difficult as measuring gene-expression changes requires a specific threshold of expression be defined. Although multiple pRB loss gene signatures have been defined and shown to accurately predict pRB loss across cancer types (5, 16), the addition of gene-expression threshold changes may improve the success in defining pRB status in tumors. Overall, although the use of pRB loss as a clinical biomarker introduces a number of challenges, advancement in the clinical methods of detecting pRB status will provide a critical avenue to target and treat tumors that lack defining oncogenic mutations.
In addition to pRB loss alone, it is critical to understand how pRB loss in combination with loss of additional tumor suppressors may affect clinical response. A study of 260 MM patients found no prognostic significance of pRB loss alone on OS or time to disease progression. However, for patients with combined pRB and p53 alterations, a decrease in OS was observed, suggesting that pRB status may have prognostic value when combined with other correlates (122). Interestingly, combined loss of RB1 and TP53 occurs in nearly 100% of SCLC and >53% of neuroendocrine prostate cancers (123, 124). However, it is important to note that alterations of these genes often occur through complicated rearrangements which may or may not be detectable through exome or panel-based next-generation sequencing (NGS; ref. 123). Thus, the method of tumor suppressor status detection is critical when defining biomarkers. Further, there is evidence to suggest that loss of pRB in SCLC is a later event whereas loss of pRB in prostate cancer is an early event, implying that the genomic and epigenomic context affects the timing of pRB loss. Overall, although pRB status alone may function as a prognostic marker in some tumor types, a better understanding of the biological consequences of pRB loss in combination with additional tumor suppressor loss may be required to be utilized as an effective marker.
Clinical Trials Using pRB Status as a Biomarker
Investigation into the clinical importance of pRB status across tumor types is ongoing, with several clinical trials focused on the applicability of pRB status as a prognostic or predictive marker (Table 2). As mentioned above, CDKs are the most commonly targeted proteins in the pRB pathway, and the use of CDKI in cancer therapy has increased over the recent years. A number of trials investigating CDK4/6 inhibitors across tumor types require pRB positivity as part of the inclusion criteria, designating pRB as a biomarker for drug response. Further, a trial investigating CDK4/6 inhibition in combination with hormone therapy in breast cancer is utilizing pRB status as an endpoint measure to investigate pRB loss as a mechanism of therapy resistance. Additionally, a retrospective study is exploring pRB status and patient recurrence-free survival and OS in response to chemotherapy to determine if pRB positivity correlates with chemotherapy sensitivity. Overall, whether investigating the efficacy of CDK4/6 inhibitors, hormone therapy, or chemotherapy, many studies have begun to consider the utility of pRB as a marker of response. These clinical studies, along with the biological studies of pRB function, are critical to expand the use of pRB status in the clinical setting.
Table 2.
Clinical trials using pRB status as a biomarker.
| Tumor | Interventions | Significance of pRB status | Study phase | Trial |
|---|---|---|---|---|
| Breast, prostate, pancreatic, glioma, gastrointestinal stromal tumors | Abemaciclib, palbociclib, ribociclib | Inclusion criteria—tumors positive for pRB | Phase I, phase II | NCT03130439, NCT02806648, NCT03220646, NCT01907607, NCT03355794, NCT02607124, NCT03526250, NCT02555189 |
| Breast | Palbociclib and endocrine therapy | Endpoint measurement—pattern of resistance | Phase II | NCT03184090 |
| Breast | Chemotherapy | Endpoint measurement—correlation with chemotherapy sensitivity | Retrospective | NCT01514565 |
Note: Clinical trials that include pRB status as either inclusion criteria or an outcome measure.
Summary and Ongoing Questions
Technology advances have afforded significant new insight into the pleiotropic molecular functions of pRB and have identified activities beyond cell-cycle control that regulate tumor development and progression. pRB has been nominated as a putative prognostic marker, biomarker for response to therapy, and a context-specific marker of therapeutic response. Although much is known of pRB function and the consequence of pRB loss on disease progression and resistance to therapy, opportunities remain to connect the biological mechanism and clinical observation. Understanding the relative contribution of distinct pRB functions on tumor suppression and/or limiting progression will be critical for translating knowledge of pRB activity into clinical practice.
These novel biological functions of pRB along with clinical data predicting pRB function in response to therapeutics support a role for pRB status in the clinical setting. However, there remain key questions that have yet to be addressed. First, does pRB status have utility as a prognostic marker? Whether through use of a pRB loss gene signature or loss of the pRB protein, studies suggest that pRB status can inform clinical outcome. Further studies are required to test the use of pRB as a prognostic marker across priority human cancers. Second, can pRB status be used to predict outcome to clinically relevant therapeutics? Although novel functions of pRB have been identified and may be used to explain the response to current therapies, use of pRB status as a marker of therapeutic response directed at these pathways has yet to be rigorously investigated. Evidence for altered therapeutic response in tumors lacking pRB nominates pRB deficiency as a potential clinical subtype that is crucial specifically in disease models such as prostate cancer, where clinically actionable subtypes are only recently emerging (125, 126). Third, if pRB loss does promote resistance to therapy, what are the underpinning mechanism(s) of resistance? It will be critical to tie molecular understanding of pRB activity to therapeutic responsiveness, and to determine the relative impact across disease types and therapeutics. Developing a better understanding of the pRB-regulated pathways which underpin the aggressive features of pRB-deficient tumors will be essential for further developing pRB loss as a biomarker of disease progression and for stratifying pRB-deficient tumors into more effective treatment regimens.
Authors' Disclosures
S.A. Tomlins reports personal fees and other support from Strata Oncology, other support from Javelin Oncology, and grants and personal fees from Astellas outside the submitted work; in addition, S.A. Tomlins reports being a coauthor on a patent issued to the University on ETS gene fusions in prostate cancer and is included in the royalty distribution stream issued and with royalties paid from LynxDx (previously Ventana/Roche and Gen-Probe/Hologic). W.K. Kelly reports grants and non-financial support from Pfizer outside the submitted work. K.E. Knudsen reports other support from CellCentric, Janssen, and Genentech outside the submitted work. No disclosures were reported by the other author.
Acknowledgments
We would like to thank the members of the Knudsen Laboratory for their continuous support and input. This work was supported by NIH/NCI grants to K.E. Knudsen (R01 CA176401, R01 CA182569, and R01 CA217329).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
References
- 1. Knudson AG. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A 1971;68:820–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Strong LC, Riccardi VM, Ferrell RE, Sparkes RS. Familial retinoblastoma and chromosome 13 deletion transmitted via an insertional translocation. Science 1981;213:1501–3. [DOI] [PubMed] [Google Scholar]
- 3. Lee WH, Bookstein R, Hong F, Young LJ, Shew JY, Lee EY. Human retinoblastoma susceptibility gene: cloning, identification, and sequence. Science 1987;235:1394–9. [DOI] [PubMed] [Google Scholar]
- 4. Rushlow DE, Mol BM, Kennett JY, Yee S, Pajovic S, Thériault BL, et al. Characterisation of retinoblastomas without RB1 mutations: genomic, gene expression, and clinical studies. Lancet Oncol 2013;14:327–34. [DOI] [PubMed] [Google Scholar]
- 5. Chen WS, Alshalalfa M, Zhao SG, Liu Y, Mahal BA, Quigley DA, et al. Novel Rb1-loss transcriptomic signature is associated with poor clinical outcomes across cancer types. Clin Cancer Res 2019;25:4290–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer 2008;8:671–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jarrard DF, Modder J, Fadden P, Fu V, Sebree L, Heisey D, et al. Alterations in the p16/pRb cell cycle checkpoint occur commonly in primary and metastatic human prostate cancer. Cancer Lett 2002;185:191–9. [DOI] [PubMed] [Google Scholar]
- 8. Brooks JD, Steven Bova G, Isaacs WB. Allelic loss of the retinoblastoma gene in primary human prostatic adenocarcinomas. Prostate 1995;26:35–9. [DOI] [PubMed] [Google Scholar]
- 9. Heinsohn S, Ulrike E, Udo ZS, Bielack S, Kabisch H. Determination of the prognostic value of loss of heterozygosity at the retinoblastoma gene in osteosarcoma. Int J Oncol 2007;30:1205–14. [PubMed] [Google Scholar]
- 10. Ittmann MM, Wieczorek R. Alterations of the retinoblastoma gene in clinically localized, stage B prostate adenocarcinomas. Hum Pathol 1996;27:28–34. [DOI] [PubMed] [Google Scholar]
- 11. Cooney KA, Wetzel JC, Merajver SD, Macoska JA, Singleton TP, Wojno KJ. Distinct regions of allelic loss on 13q in prostate cancer. Cancer Res 1996;56:1142–5. [PubMed] [Google Scholar]
- 12. Tricoli JV, Gumerlock PH, Yao JL, Chi S-G, D'Souza SA, Nestok BR, et al. Alterations of the retinoblastoma gene in human prostate adenocarcinoma. Genes Chromosom Cancer 1996;15:108–14. [DOI] [PubMed] [Google Scholar]
- 13. Sharma A, Yeow W-S, Ertel A, Coleman I, Clegg N, Thangavel C, et al. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest 2010;120:4478–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yokota J, Wada M, Shimosato Y, Terada M, Sugimura T. Loss of heterozygosity on chromosomes 3, 13, and 17 in small-cell carcinoma and on chromosome 3 in adenocarcinoma of the lung. Proc Natl Acad Sci U S A 1987;84:9252–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Herschkowitz JI, He X, Fan C, Perou CM. The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res 2008;10:R75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McNair C, Xu K, Mandigo AC, Benelli M, Leiby B, Rodrigues D, et al. Differential impact of RB status on E2F1 reprogramming in human cancer. J Clin Invest 2018;128:341–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kinoshita I, Dosaka-Akita H, Mishina T, Akie K, Nishi M, Hiroumi H, et al. Altered p16(INK4) and retinoblastoma protein status in non-small cell lung cancer: potential synergistic effect with altered p53 protein on proliferative activity. Cancer Res 1996;56:5557–62. [PubMed] [Google Scholar]
- 18. Stirzaker C, Millar DS, Paul CL, Warnecke PM, Harrison J, Vincent PC, et al. Extensive DNA methylation spanning the Rb promoter in retinoblastoma tumors. Cancer Res 1997;57:2229–37. [PubMed] [Google Scholar]
- 19. Oesterreich S, Fuqua SAW. Tumor suppressor genes and breast cancer. Endocr Relat Cancer 1999;6:405–19. [DOI] [PubMed] [Google Scholar]
- 20. Lohmann DR. RB1 gene mutations in retinoblastoma. Hum Mutat 1999;14:283–8. [DOI] [PubMed] [Google Scholar]
- 21. Tomar S, Sethi R, Sundar G, Quah TC, Quah BL, Lai PS. Mutation spectrum of RB1 mutations in retinoblastoma cases from Singapore with implications for genetic management and counselling. PLoS One 2017;12:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wadayama B, Toguchida J, Shimizu T, Ishizaki K, Sasaki MS, Kotoura Y. Mutation spectrum of the retinoblastoma gene in osteosarcomas. Cancer Res 1994;54:3042–8. [PubMed] [Google Scholar]
- 23. Berge E, Knappskog S, Geisler S, Staalesen V, Pacal M, Børresen-Dale A-L, et al. Identification and characterization of retinoblastoma gene mutations disturbing apoptosis in human breast cancers. Mol Cancer 2010;9:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Di Fiore R, D'Anneo A, Tesoriere G, Vento R. RB1 in cancer: Different mechanisms of RB1 inactivation and alterations of pRb pathway in tumorigenesis. J Cell Physiol 2013;228:1676–87. [DOI] [PubMed] [Google Scholar]
- 25. Wong YF, Chung TK, Cheung TH, Nobori T, Yim SF, Lai KW, et al. p16INK4 and p15INK4B alterations in primary gynecologic malignancy. Gynecol Oncol 1997;65:319–24. [DOI] [PubMed] [Google Scholar]
- 26. Schultz DC, Vanderveer L, Buetow KH, Boente MP, Ozols RF, Hamilton TC, et al. Characterization of chromosome 9 in human ovarian neoplasia identifies frequent genetic imbalance on 9q and rare alterations involving 9p, including CDKN2. Cancer Res 1995;55:2150–7. [PubMed] [Google Scholar]
- 27. Huang L, Goodrow TL, Zhang SY, Klein-Szanto AJP, Chang H, Ruggeri BA. Deletion and mutation analyses of the P16/MTS-1 tumor suppressor gene in human ductal pancreatic cancer reveals a higher frequency of abnormalities in tumor-derived cell lines than in primary ductal adenocarcinomas. Cancer Res 1996;56:1137–41. [PubMed] [Google Scholar]
- 28. Ichikawa Y, Yoshida S, Koyama Y, Hirai M, Ishikawa T, Nishida M, et al. Inactivation of p16/CDKN2 and p15/MTS2 genes in different histological types and clinical stages of primary ovarian tumors. Int J Cancer 1996;69:466–70. [DOI] [PubMed] [Google Scholar]
- 29. Giani C, Finocchiaro G. Mutation rate of the CDKN2 gene in malignant gliomas. Cancer Res 1994;54:6338–9. [PubMed] [Google Scholar]
- 30. Orlow I, Lacombe L, Hannon GJ, Serrano M, Pellicer I, Dalbagni G, et al. Deletion of the p16 and p15 genes in human bladder tumors. J Natl Cancer Inst 1995;87:1524–9. [DOI] [PubMed] [Google Scholar]
- 31. Li YJ, Hoang-Xuan K, Delattre JY, Poisson M, Thomas G, Hamelin R. Frequent loss of heterozygosity on chromosome 9, and low incidence of mutations of cyclin-dependent kinase inhibitors p15 (MTS2) and p16 (MTS1) genes in gliomas. Oncogene 1995;11:597–600. [PubMed] [Google Scholar]
- 32. Ueki K, Ono Y, Henson JW, Efird JT, von Deimling A, Louis DN. CDKN2/p16 or RB alterations occur in the majority of glioblastomas and are inversely correlated. Cancer Res 1996;56:150–3. [PubMed] [Google Scholar]
- 33. Reed AL, Califano J, Cairns P, Westra WH, Jones RM, Koch W, et al. High frequency of p16 (CDKN2/MTS-1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res 1996;56:3630–3. [PubMed] [Google Scholar]
- 34. Caldas C, Hahn SA, da Costa LT, Redston MS, Schutte M, Seymour AB, et al. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet 1994;8:27–32. [DOI] [PubMed] [Google Scholar]
- 35. Analysis A, Gerdes B, Ramaswamy A, Ziegler A, Lang SA, Kersting M, et al. p16 INK4a is a prognostic marker in resected ductal pancreatic cancer. Ann Surg 2002;235:51–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nishihira T, Mori S, Aoki T, Nakamura Y. Frequent somatic mutation of the MTS1/CDK4I (multiple tumor suppressor/cyclin-dependent kinase 4 inhibitor) gene in esophageal squamous cell carcinoma. Cancer Res 1994;54:3396–7. [PubMed] [Google Scholar]
- 37. Esteve A, Martel-Planche G, Sylla BS, Hollstein M, Hainaut P, Montesano R. Low frequency of p16/CDKN2 gene mutations in esophageal carcinomas. Int J Cancer 1996;66:301–4. [DOI] [PubMed] [Google Scholar]
- 38. Zhou X, Tarmin L, Yin J, Jiang HY, Suzuki H, Rhyu MG, et al. The MTS1 gene is frequently mutated in primary human esophageal tumors. Oncogene 1994;9:3737–41. [PubMed] [Google Scholar]
- 39. Okamoto A, Hussain SP, Hagiwara K, Spillare EA, Rusin MR, Demetrick DJ, et al. Mutations in the p16INK4/MTS1/CDKN2, p15INK4B/MTS2, and p18 genes in primary and metastatic lung cancer. Cancer Res 1995;55:1448–51. [PubMed] [Google Scholar]
- 40. Semczuk A, Miturski R, Skomra D, Jakowicki JA. Expression of the cell-cycle regulatory proteins (pRb, cyclin D1, p16 INK4A and cdk4) in human endometrial cancer: correlation with clinicopathological features. Arch Gynecol Obstet 2004;269:104–10. [DOI] [PubMed] [Google Scholar]
- 41. Costello JF, Berger MS, Huang HS, Cavenee WK. Silencing of p16/CDKN2 expression in human gliomas by methylation and chromatin condensation. Cancer Res 1996;56:2405–10. [PubMed] [Google Scholar]
- 42. Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, et al. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res 1995;55:4525–30. [PubMed] [Google Scholar]
- 43. Maesawa C, Tamura G, Nishizuka S, Ogasawara S, Ishida K, Terashima M, et al. Inactivation of the CDKN2 gene by homozygous deletion and de novo methylation is associated with advanced stage esophageal squamous cell carcinoma. Cancer Res 1996;56:3875–8. [PubMed] [Google Scholar]
- 44. Gonzalez-Zulueta M, Bender CM, Yang AS, Nguyen T, Beart RW, Van Tornout JM, et al. Methylation of the 5′ CpG island of the p16/CDKN2 tumor suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res 1995;55:4531–5. [PubMed] [Google Scholar]
- 45. Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, et al. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med 1995;1:686–92. [DOI] [PubMed] [Google Scholar]
- 46. Theurillat J, Nickerson E, Auclair D, Li L, Place C, Dicara D, et al. « Plan d'action des réseaux d'eau potable ». 2015;150:2015. [Google Scholar]
- 47. Shiozawa T, Nikaido T, Shimizu M, Zhai Y, Fujii S. Immunohistochemical analysis of the expression of cdk4 and p16INK4 in human endometrioid-type endometrial carcinoma. Cancer 1997;80:2250–6. [DOI] [PubMed] [Google Scholar]
- 48. Skomedal H, Kristensen GB, Nesland JM, Børresen-Dale AL, Tropé C, Holm R. TP53 alterations in relation to the cell cycle-associated proteins p21, cyclin D1, cdk4, RB, MDM2, and EGFR in cancers of the uterine corpus. J Pathol 1999;187:556–62. [DOI] [PubMed] [Google Scholar]
- 49. Semczuk A, Jakowicki JA. Alterations of pRb1-cyclin D1-cdk4/6-p16INK4A pathway in endometrial carcinogenesis. Cancer Lett 2004;203:1–12. [DOI] [PubMed] [Google Scholar]
- 50. Meyer zum Buschenfelde K, Klehmann-Hieb E, De Plaen E, Hankeln T, Hauer M, Schneider J, et al. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science 1995;269:1281–4. [DOI] [PubMed] [Google Scholar]
- 51. Baker SJ, Reddy EP. CDK4: a key player in the cell cycle, development, and cancer. Genes Cancer 2012;3:658–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer 2011;11:558–72. [DOI] [PubMed] [Google Scholar]
- 53. Michalides R, Hageman P, Van Tinteren H, Houben L, Wientjens E, Klompmaker R, et al. A clinicopathological study on overexpression of cyclin D1 and of p53 in a series of 248 patients with operable breast cancer. Br J Cancer 1996;73:728–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fanti V, Smith R, Fisher C, Bartek J, Dickson C, Barnes D, et al. Amplification and overexpression of cyclin Dl in breast cancer detected by immunohistochemical staining. Cancer Res 1994;54:1812–7. [PubMed] [Google Scholar]
- 55. Worsley SD, Ponder BAJ, Davies BR. Overexpression of cyclin D1 in epithelial ovarian cancers. Gynecol Oncol 1997;64:189–95. [DOI] [PubMed] [Google Scholar]
- 56. Bertoni F, Rinaldi A, Zucca E, Cavalli F. Update on the molecular biology of mantle cell lymphoma. Hematol Oncol 2006;24:22–7. [DOI] [PubMed] [Google Scholar]
- 57. Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol 2005;23:6333–8. [DOI] [PubMed] [Google Scholar]
- 58. Cecchini MJ, Ishak CA, Passos DT, Warner A, Palma DA, Howlett CJ, et al. Loss of the retinoblastoma tumor suppressor correlates with improved outcome in patients with lung adenocarcinoma treated with surgery and chemotherapy. Hum Pathol 2015;46:1922–34. [DOI] [PubMed] [Google Scholar]
- 59. Shin E, Jung W-H, Koo J-S. Expression of p16 and pRB in invasive breast cancer. Int J Clin Exp Pathol 2015;8:8209–17. [PMC free article] [PubMed] [Google Scholar]
- 60. Polager S, Kalma Y, Berkovich E, Ginsberg D. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene 2002;21:437–46. [DOI] [PubMed] [Google Scholar]
- 61. Markey MP, Angus SP, Strobeck MW, Williams SL, Gunawardena RW, Aronow BJ, et al. Unbiased analysis of RB-mediated transcriptional repression identifies novel targets and distinctions from E2F action. Cancer Res 2002;62:6587–97. [PubMed] [Google Scholar]
- 62. Cook R, Zoumpoulidou G, Luczynski MT, Rieger S, Moquet J, Spanswick VJ, et al. Direct involvement of retinoblastoma family proteins in DNA repair by non-homologous end-joining. Cell Rep 2015;10:2006–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Aydin ÖZ, Vermeulen W, Lans H. ISWI chromatin remodeling complexes in the DNA damage response. Cell Cycle 2014;13:3016–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol 2010;17:1144–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tuzon CT, Spektor T, Kong X, Congdon LM, Wu S, Schotta G, et al. Concerted activities of distinct H4K20 methyltransferases at DNA double-strand breaks regulate 53BP1 nucleation and NHEJ-directed repair. Cell Rep 2014;8:430–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vélez-Cruz R, Manickavinayaham S, Biswas AK, Clary RW, Premkumar T, Cole F, et al. RB localizes to DNA double-strand breaks and promotes DNA end resection and homologous recombination through the recruitment of BRG1. Genes Dev 2016;30:2500–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hilgendorf KI, Leshchiner ES, Nedelcu S, Maynard MA, Calo E, Ianari A, et al. The retinoblastoma protein induces apoptosis directly at the mitochondria. Genes Dev 2013;27:1003–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chau BN, Borges HL, Chen T-T, Masselli A, Hunton IC, Wang JYJ. Signal-dependent protection from apoptosis in mice expressing caspase-resistant Rb. Nat Cell Biol 2002;4:757–65. [DOI] [PubMed] [Google Scholar]
- 69. Han J, Soletti RC, Sadarangani A, Sridevi P, Ramirez ME, Eckmann L, et al. Nuclear expression of β-catenin promotes RB stability and resistance to TNF-induced apoptosis in colon cancer cells. Mol Cancer Res 2013;11:207–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tan X, Martin SJ, Green DR, Wang JY. Degradation of retinoblastoma protein in tumor necrosis factor- and CD95-induced cell death. J Biol Chem 1997;272:9613–6. [DOI] [PubMed] [Google Scholar]
- 71. Jänicke RU, Walker PA, Lin XY, Porter AG. Specific cleavage of the retinoblastoma protein by an ICE-like protease in apoptosis. EMBO J 1996;15:6969–78. [PMC free article] [PubMed] [Google Scholar]
- 72. Wang C-Y, Xu Z-B, Wang J-P, Jiao Y, Zhang B. Rb deficiency accelerates progression of carcinoma of the urinary bladder in vivo and in vitro through inhibiting autophagy and apoptosis. Int J Oncol 2017;50:1221–32. [DOI] [PubMed] [Google Scholar]
- 73. Anderson MM, Chen J, Cole CN, Conrad SE. Activation of the human thymidine kinase (TK) promoter by simian virus 40 large T antigen requires both the T antigen pRb family-binding domain and TK promoter sequences resembling E2F-binding sites. J Virol 1996;70:6304–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dou QP, Markell PJ, Pardee AB. Thymidine kinase transcription is regulated at G1/S phase by a complex that contains retinoblastoma-like protein and a cdc2 kinase. Proc Natl Acad Sci U S A 1992;89:3256–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jensen DE, Black AR, Swick AG, Azizkhan JC. Distinct roles for Sp1 and E2F sites in the growth/cell cycle regulation of the DHFR promoter. J Cell Biochem 1997;67:24–31. [DOI] [PubMed] [Google Scholar]
- 76. Blanchet E, Annicotte JS, Lagarrigue S, Aguilar V, Clapé C, Chavey C, et al. E2F transcription factor-1 regulates oxidative metabolism. Nat Cell Biol 2011;13:1146–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hsieh MCF, Das D, Sambandam N, Zhang MQ, Nahlé Z. Regulation of the PDK4 isozyme by the Rb-E2F1 complex. J Biol Chem 2008;283:27410–7. [DOI] [PubMed] [Google Scholar]
- 78. Dali-Youcef N, Mataki C, Coste A, Messaddeq N, Giroud S, Blanc S, et al. Adipose tissue-specific inactivation of the retinoblastoma protein protects against diabesity because of increased energy expenditure. Proc Natl Acad Sci U S A 2007;104:10703–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Reynolds MR, Lane AN, Robertson B, Kemp S, Liu Y, Hill BG, et al. Control of glutamine metabolism by the tumor suppressor Rb. Oncogene 2014;33:556–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Nicolay BN, Gameiro PA, Tschöp K, Korenjak M, Heilmann AM, Asara JM, et al. Loss of RBF1 changes glutamine catabolism. Genes Dev 2013;27:182–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kim J-w, Zeller KI, Wang Y, Jegga AG, Aronow BJ, O'Donnell KA, et al. Evaluation of Myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol Cell Biol 2004;24:5923–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem 2000;275:21797–800. [DOI] [PubMed] [Google Scholar]
- 83. Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res 2009;15:6479–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Derenzini M, Donati G, Mazzini G, Montanaro L, Vici M, Ceccarelli C, et al. Loss of retinoblastoma tumor suppressor protein makes human breast cancer cells more sensitive to antimetabolite exposure. Clin Cancer Res 2008;14:2199–209. [DOI] [PubMed] [Google Scholar]
- 85. Bosco EE, Wang Y, Xu H, Zilfou JT, Knudsen KE, Aronow BJ, et al. The retinoblastoma tumor suppressor modifies the therapeutic response of breast cancer. J Clin Invest 2007;117:218–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Plimack ER, Dunbrack RL, Brennan TA, Andrake MD, Zhou Y, Serebriiskii IG, et al. Defects in DNA repair genes predict response to neoadjuvant cisplatin-based chemotherapy in muscle-invasive bladder cancer. 2018;360:204–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hijioka S, Hosoda W, Matsuo K, Ueno M, Furukawa M, Yoshitomi H, et al. Rb loss and KRAS mutation are predictors of the response to platinum-based chemotherapy in pancreatic neuroendocrine neoplasm with grade 3: a Japanese multicenter pancreatic NEN-G3 study. Clin Cancer Res 2017;23:4625–32. [DOI] [PubMed] [Google Scholar]
- 88. Lehn S, Fernö M, Jirström K, Rydén L, Landberg G. A non-functional retinoblastoma tumor suppressor (RB) pathway in premenopausal breast cancer is associated with resistance to tamoxifen. Cell Cycle 2011;10:956–62. [DOI] [PubMed] [Google Scholar]
- 89. Barroso-Sousa R, Shapiro GI, Tolaney SM. Clinical development of the CDK4/6 inhibitors ribociclib and abemaciclib in breast cancer. Breast Care 2016;11:167–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Turner NC, Ro J, André F, Loi S, Verma S, Iwata H, et al. Palbociclib in hormone-receptor–positive advanced breast cancer. N Engl J Med 2015;373:209–19. [DOI] [PubMed] [Google Scholar]
- 91. Slamon DJ, Neven P, Chia S, Fasching PA, De Laurentiis M, Im S-A, et al. Phase III randomized study of ribociclib and fulvestrant in hormone receptor–positive, human epidermal growth factor receptor 2–negative advanced breast cancer: MONALEESA-3. J Clin Oncol 2018;36:2465–72. [DOI] [PubMed] [Google Scholar]
- 92. Goetz MP, Toi M, Campone M, Sohn J, Paluch-Shimon S, Huober J, et al. MONARCH 3: abemaciclib as initial therapy for advanced breast cancer. J Clin Oncol 2017;35:3638–46. [DOI] [PubMed] [Google Scholar]
- 93. Teresa Herrera-Abreu M, Palafox M, Asghar U, Rivas MA, Cutts RJ, Garcia-Murillas I, et al. Early adaptation and acquired resistance to CDK4/6 inhibition in estrogen receptor-positive breast cancer HHS public access. Cancer Res 2016;76:2301–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Guarducci C, Bonechi M, Benelli M, Biagioni C, Boccalini G, Romagnoli D, et al. ARTICLE cyclin E1 and Rb modulation as common events at time of resistance to palbociclib in hormone receptor-positive breast cancer. NPJ Breast Cancer 2018;4:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. de Leeuw R, McNair CM, Schiewer MJ, Poudel Neupane N, Brand LJ, Augello MA, et al. MAPK reliance via acquired CDK4/6 inhibitor resistance in cancer HHS Public Access. Clin Cancer Res 2018;24:4201–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Dao DD, Sawyer JR, Epstein J, Hoover RG, Barlogie B, Tricot G. Deletion of the retinoblastoma gene in multiple myeloma. Leukemia 1994;8:1280–4. [PubMed] [Google Scholar]
- 97. He Z, O'Neal J, Wilson WC, Mahajan N, Luo J, Wang Y, et al. Deletion of Rb1 induces both hyperproliferation and cell death in murine germinal center B cells. Exp Hematol 2016;44:161–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zojer N, Königsberg R, Ackermann J, Fritz E, Dallinger S, Krömer E, et al. Deletion of 13q14 remains an independent adverse prognostic variable in multiple myeloma despite its frequent detection by interphase fluorescence in situ hybridization. Blood 2000;95:1925–30. [PubMed] [Google Scholar]
- 99. Tricot G, Barlogie B, Jagannath S, Bracy D, Mattox S, Vesole DH, et al. Poor prognosis in multiple myeloma is associated only with partial or complete deletions of chromosome 13 or abnormalities involving 11q and not with other karyotype abnormalities. Blood 1995;86:4250–6. [PubMed] [Google Scholar]
- 100. Pérez-Simón JA, García-Sanz R, Tabernero MD, Almeida J, González M, Fernández-Calvo J, et al. Prognostic value of numerical chromosome aberrations in multiple myeloma: a FISH analysis of 15 different chromosomes. Blood 1998;91:3366–71. [PubMed] [Google Scholar]
- 101. Brambilla E, Moro D, Gazzeri S, Brambilla C. Alterations of expression of rb, p16 ink4a and cyclin d1 in non–small cell lung carcinoma. J Pathol 1999;188:351–60. [DOI] [PubMed] [Google Scholar]
- 102. Xu HJ, Quinlan DC, Davidson AG, Hu SX, Summers CL, Li J, et al. Altered retinoblastoma protein expression and prognosis in early-stage non-small-cell lung carcinoma. J Natl Cancer Inst 1994;86:695–9. [DOI] [PubMed] [Google Scholar]
- 103. Xu HJ, Cagle PT, Hu SX, Li J, Benedict WF. Altered retinoblastoma and p53 protein status in non-small cell carcinoma of the lung: potential synergistic effects on prognosis. Clin Cancer Res 1996;2:1169–76. [PubMed] [Google Scholar]
- 104. Dosaka-Akita H, Hu SX, Fujino M, Harada M, Kinoshita I, Xu HJ, et al. Altered retinoblastoma protein expression in non-small cell lung cancer: its synergistic effects with altered ras and p53 protein status on prognosis. Cancer 1997;79:1329–37. [PubMed] [Google Scholar]
- 105. Kratzke RA, Greatens TM, Rubins JB, Maddaus MA, Niewoehner DE, Niehans GA, et al. Rb and p16(INK4a) expression in resected non-small cell lung tumors. Cancer Res 1996;56:3415–20. [PubMed] [Google Scholar]
- 106. Gavressea T, Kalogeras KT, Koliou G-A, Zagouri F, Lazaridis G, Gogas H, et al. The prognostic value of the immunohistochemical expression of phosphorylated RB and p16 proteins in association with cyclin D1 and the p53 pathway in a large cohort of patients with breast cancer treated with taxane-based adjuvant chemotherapy. Anticancer Res 2017;37:2947–57. [DOI] [PubMed] [Google Scholar]
- 107. Trere D, Brighenti E, Donati G, Ceccarelli C, Santini D, Taffurelli M, et al. High prevalence of retinoblastoma protein loss in triple-negative breast cancers and its association with a good prognosis in patients treated with adjuvant chemotherapy. Ann Oncol 2009;20:1818–23. [DOI] [PubMed] [Google Scholar]
- 108. Stefansson OA, Jonasson JG, Olafsdottir K, Hilmarsdottir H, Olafsdottir G, Esteller M, et al. CpG island hypermethylation of BRCA1 and loss of pRb as co-occurring events in basal/triple-negative breast cancer. Epigenetics 2011;6:638–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Pietiläinen T, Lipponen P, Aaltomaa S, Eskelinen M, Kosma VM, Syrjänen K. Expression of retinoblastoma gene protein (Rb) in breast cancer as related to established prognostic factors and survival. Eur J Cancer 1995;31A:329–33. [DOI] [PubMed] [Google Scholar]
- 110. Knudsen ES, Pajak TF, Qeenan M, McClendon AK, Armon BD, Schwartz GF, et al. Retinoblastoma and phosphate and tensin homolog tumor suppressors: impact on ductal carcinoma in situ progression. J Natl Cancer Inst 2012;104:1825–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Cordon-Cardo C, Wartinger D, Petrylak D, Dalbagni G, Fair WR, Fuks Z, et al. Altered expression of the retinoblastoma gene product: prognostic indicator in bladder cancer. J Natl Cancer Inst 1992;84:1251–6. [DOI] [PubMed] [Google Scholar]
- 112. Xu HJ, Cairns P, Hu SX, Knowles MA, Benedict WF. Loss of RB protein expression in primary bladder cancer correlates with loss of heterozygosity at the RB locus and tumor progression. Int J Cancer 1993;53:781–4. [DOI] [PubMed] [Google Scholar]
- 113. Toropainen EM, Lipponen PK, Syrjanen KJ. Expression of insulin-like growth factor II in female breast cancer as related to established prognostic factors and long-term prognosis. Anticancer Res 1995;15:2669–74. [PubMed] [Google Scholar]
- 114. Feugeas O, Guriec N, Babin-Boilletot A, Marcellin L, Simon P, Babin S, et al. Loss of heterozygosity of the RB gene is a poor prognostic factor in patients with osteosarcoma. J Clin Oncol 1996;14:467–72. [DOI] [PubMed] [Google Scholar]
- 115. Patiño-García A, Piñeiro ES, Díez MZ, Iturriagagoitia LG, Klüssmann FA, Ariznabarreta LS. Genetic and epigenetic alterations of the cell cycle regulators and tumor suppressor genes in pediatric osteosarcomas. J Pediatr Hematol Oncol 2003;25:362–7. [DOI] [PubMed] [Google Scholar]
- 116. Rieder H, Poensgen B, Fritz B, Aslan M, Drohm D, Rehder H, et al. Loss of heterozygosity of the retinoblastoma (RB1) gene in lipomas from a retinoblastoma patient. J Natl Cancer Inst 1998;90:324–6. [DOI] [PubMed] [Google Scholar]
- 117. Eng C, Li FP, Abramson DH, Ellsworth RM, Wong FL, Goldman MB, et al. Mortality from second tumors among long-term survivors of retinoblastoma. J Natl Cancer Inst 1993;85:1121–8. [DOI] [PubMed] [Google Scholar]
- 118. Yu C-L, Tucker MA, Abramson DH, Furukawa K, Seddon JM, Stovall M, et al. Cause-specific mortality in long-term survivors of retinoblastoma. J Natl Cancer Inst 2009;101:581–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Leiderman YI, Kiss S, Mukai S. Molecular genetics of RB1–the retinoblastoma gene. Semin Ophthalmol 2007;22:247–54. [DOI] [PubMed] [Google Scholar]
- 120. Dommering CJ, Marees T, van der Hout AH, Imhof SM, Meijers-Heijboer H, Ringens PJ, et al. RB1 mutations and second primary malignancies after hereditary retinoblastoma. Fam Cancer 2012;11:225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med 2012;4:127ps10. [DOI] [PubMed] [Google Scholar]
- 122. Gutiérrez NC, Castellanos MV, Martín ML, Mateos MV, Hernández JM, Fernández M, et al. Prognostic and biological implications of genetic abnormalities in multiple myeloma undergoing autologous stem cell transplantation: t(4;14) is the most relevant adverse prognostic factor, whereas RB deletion as a unique abnormality is not associated with adverse prognosis. Leukemia 2006;21:143–50. [DOI] [PubMed] [Google Scholar]
- 123. George J, Lim JS, Jang SJ, Cun Y, Ozretić L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016;22:298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Abida W, Cheng ML, Armenia J, Middha S, Autio KA, Vargas HA, et al. Analysis of the prevalence of microsatellite instability in prostate cancer and response to immune checkpoint blockade. JAMA Oncol 2019;5:471–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Lotan TL, Kaur HB, Salles DC, Murali S, Schaeffer EM, Lanchbury JS, et al. Homologous recombination deficiency (HRD) score in germline BRCA2- versus ATM-altered prostate cancer. Mod Pathol 2021;34:1185–93. [DOI] [PMC free article] [PubMed] [Google Scholar]