Abstract
Purpose:
A consistent genetic alteration in vascular cancer epithelioid hemangioendothelioma (EHE) is the t(1;3)(p36;q25) chromosomal translocation, which generates a WWTR1(TAZ)-CAMTA1 (TC) fusion gene. TC is a transcriptional coactivator that drives EHE. Here, we aimed to identify the TC transcriptional targets and signaling mechanisms that underlie EHE tumorigenesis.
Experimental Design:
We used NIH3T3 cells transformed with TC (NIH3T3/TC) as a model system to uncover TC-dependent oncogenic signaling. These cells proliferated in an anchorage-independent manner in suspension and soft agar. The findings of the cell-based studies were validated in a xenograft model.
Results:
We identified connective tissue growth factor (CTGF) as a tumorigenic transcriptional target of TC. We show that CTGF binds to integrin αIIbβ3, which is essential for sustaining the anchorage-independent proliferation of transformed NIH3T3/TC cells. NIH3T3/TC cells also have enhanced Ras and MAPK signaling, and the activity of these pathways is reduced upon CTGF knockdown, suggesting that CTGF signaling occurs via the Ras-MAPK cascade. Further, pharmacologic inhibition of MAPK signaling through PD 0325901 and trametinib abrogated TC-driven anchorage-independent growth. Likewise, for tumor growth in vivo, NIH3T3/TC cells require CTGF and MAPK signaling. NIH3T3/TC xenograft growth was profoundly reduced upon CTGF knockdown and after trametinib treatment.
Conclusions:
Collectively, our results demonstrated that CTGF and the Ras-MAPK signaling cascade are essential for TC-mediated tumorigenesis. These studies provided the preclinical rationale for SARC033 (NCI 10015-NCT03148275), a nonrandomized, open-label, phase II study of trametinib in patients with unresectable or metastatic EHE.
Translational Relevance.
Epithelioid hemangioendothelioma (EHE) is a rare but deadly vascular sarcoma caused by the oncogenic TAZ-CAMTA1 fusion protein. There are no effective treatments for aggressive EHE, and the oncogenic pathways that drive EHE growth downstream of TAZ-CAMTA1 remain unknown. Here, we show that connective tissue growth factor (CTGF), a transcriptional target of TAZ-CAMTA1, hyperactivates MAPK signaling and drives TAZ-CAMTA1–dependent oncogenesis. Using cell lines and xenograft mouse models, we showed that targeting the CTGF-MAPK axis with trametinib, an FDA-approved MEK inhibitor, abrogated TAZ-CAMTA1–mediated oncogenesis. These studies have identified trametinib as a targeted therapy that could be repurposed for the treatment of aggressive EHE.
Introduction
The WWTR1 (TAZ)-Camta1 (TC) fusion gene, is a product of the t(1;3)(p36;q25) chromosomal translocation, which has been identified as the genetic aberration that defines epithelioid hemangioendothelioma (EHE), a vascular sarcoma (1, 2). This fusion gene is expressed in more than 90% of EHE cases and orchestrates tumorigenic events associated with EHE (3). There are no standard treatment options for EHE, and patients with aggressive disease have a poor prognosis. However, a subset of clinical samples previously classified as EHE carries a YAP1-TFE3 fusion gene instead of the TC fusion gene (4). Recently, after analyzing a large cohort of YAP1-TFE3 tumors, we demonstrated that tumors harboring the YAP1-TFE3 fusion are clinically and histopathologically distinct from EHE and recommended classifying them as YAP1-TFE3-fused hemangioendothelioma to emphasize their distinctiveness (5).
In the TC fusion protein, the N-terminus of transcriptional coactivator with PDZ binding motif (TAZ) was fused to the C-terminus of calmodulin binding transcription activator 1(CAMTA1). TAZ and its paralog yes-associated protein 1 (YAP1) are transcriptional coactivators, and they have been shown to play essential roles in cancer initiation and solid tumor growth. Persistent activation of YAP/TAZ results in aberrant cell proliferation, confers anoikis resistance, opposes cell death caused by autophagy in tumors, and facilitates cancer stem-cell expansion (6). CAMTA1, on the other hand, is a transcription factor that is thought to be a tumor suppressor in neural cancers (7, 8) although its function is not completely understood.
The TC fusion protein functions as a transcriptional coactivator (3, 7). Although the bulk of the TC fusion protein is composed of CAMTA1, the N-terminal TAZ portion retains the TEAD-binding motif, which facilitates interaction with the TEAD family of transcription factors. Through its interaction with TEAD, we previously showed that TC activates a TAZ-like transcriptional program (3, 9). By not retaining the C-terminal portion of TAZ, TC also loses a phosphodegron that may confer enhanced stability to TC over TAZ (10). Furthermore, TC predominantly resides in the nucleus via a CAMTA1 C-terminal nuclear localization signal (9). A recent study showed that the CAMTA1 transactivation domain recruits potent chromatin remodelers to enhance the transcriptional activity of TC (11). Stable expression levels, predominant nuclear localization, and enhanced activity allow TC to act as a constitutively activated TAZ, and TC; therefore, can drive EHE tumor formation and progression. Under the influence of TC, cells are able to sustain anchorage-independent proliferation (9). This remarkable ability may ensure the survival of cancer cells when they disseminate to distant organs. Although TC is known to cause tumorigenesis, the underlying molecular mechanisms remain unknown. In this study, we set out to dissect the downstream targets and signaling mechanisms that are activated by TC to facilitate anchorage-independent proliferation and tumorigenesis. We postulated that understanding the TC-potentiated signaling pathways would pave the way for effective therapeutic treatments for EHE.
Materials and Methods
Cell culture, gene transfer, and gene knockdown
NIH3T3 and MS1 cells were purchased from the ATCC and cultured in a complete media of DMEM containing 10% bovine serum (NIH3T3) or 10% FBS (MS1; Invitrogen-Life Technologies), and 50 μg/mL penicillin/streptomycin. All cells were maintained at 37°C in a humidified atmosphere of 5% CO2. We routinely checked cell lines for any Mycoplasma contamination, and the last check was performed in January 2022. Generation of the NIH3T3/TC model has been described previously (9). Transfection with pCMV6-mCTGF-FLAG-Myc was performed using Lipofectamine Plus Reagent (Invitrogen-Life Technologies) according to the manufacturer's instructions. Pooled stable transfectants were generated by selecting for 2 weeks with G418 (600 μg/mL). Lentiviral transfection with pLKO.1-SHC1, SHC2, and other short hairpin RNA (shRNA) was performed by transfecting 293T cells together with envelope and packaging plasmids (VSVG and Δ8.2), and the supernatant containing the virus was collected at 48 and 72 hours after transfection, filtered with a 0.45-μmol/L filter, and supplemented with 8 μg/mL polybrene. The virus-containing supernatants were added to NIH3T3 cells and incubated for 8 hours. Pooled stable lines were then generated by selection in puromycin for 10 days. MS1/TC cells were generated by PCR amplification of the TC coding sequence.
(FOR: TAGAAGACACCGACTCTAGAGGATTCGCCACCATGGATTATAAAGATGATG
REV: TTGTAATCCAGAGGTTGATTGTCGACTCAAGTTCCTTGGCCTTTTTCAATT). The amplicon was then inserted into the pLentiPuro construct using BamHI/SalI sites via cold fusion cloning (System Biosciences). Lentiviral packaging of this construct was performed in 293LentiX cells (Takara Bio) with envelope and packaging plasmids (ViraPower, Thermo Fisher Scientific). The filtered supernatant at 48 hours and 72 hours was administered to target cells with 8 μg/mL polybrene, and transduced MS1 cells were selected with puromycin for 10 days.
Antibodies and inhibitors
The following commercially available antibodies were used: S6 (catalog no. SC-74459, RRID: AB_2905501), integrin αV (catalog no. SC-6617-R, RRID: AB_2129625), and integrin β3 (catalog no. SC-6627, RRID: AB_647489) were purchased from Santa Cruz Biotech. Phospho-Erk (Thr 202⁄204; catalog no. 9101, RRID: AB_331646), Erk (catalog no. 4695, RRID: AB_390779), AKT (catalog no. 9272, RRID: AB_329827), phospho-AKT (Ser 473; catalog no. 9271, RRID: AB_329825), and phospho-S6 (Ser235⁄236; catalog no. 2211, RRID: AB_331679) were purchased from Cell Signaling Technology. FLAG (M2; catalog no. F1804, RRID: AB_262044) and β-actin (AC-15; catalog no. A5441, RRID: AB_476744) were purchased from Sigma-Aldrich. Connective tissue growth factor (CTGF; catalog no. Ab-6992, RRID: AB_305688), and integrin αIIb (catalog no. Ab-157173) were purchased from Abcam. CYR61 (catalog no. AF4055, RRID: AB_2088745) was purchased from R&D Systems. The following inhibitors were used in this study: GDC0941, PD 23590, sorafenib, and trametinib (all purchased from LC Laboratories).
Immunoblotting and immunoprecipitation
Cells were grown to subconfluency, harvested, and lysed in radioimmunoprecipitation assay buffer (50 mmol/L Tris PH 8.0, 1% NP-40, 0.25% sodium deoxycholate, 150 mmol/L NaCl) in the presence of mini EDTA-free protease inhibitor cocktail and PhosSTOP (Roche Applied Science), and extracts were obtained by centrifugation at 10,000 g at 4°C. Protein lysates were subjected to 4% to 15% gradient SDS-PAGE and immunoblotted with corresponding primary antibodies followed by incubation with horseradish peroxidase–conjugated secondary antibodies (Invitrogen) and developed using the Supersignal-ECL kit (Thermo Fisher Scientific). For immunoprecipitation, samples were incubated with 2 μg anti-FLAG M2 (Sigma-Aldrich) overnight and immunoprecipitated using protein G plus Sepharose (Santa Cruz Biotech) for 2 hours at 4°C. The immune complexes were washed three times with lysis buffer. Proteins were separated by SDS-PAGE. Anti-integrin αV, anti-integrin β3 (Santa Cruz Biotech), and anti-integrin αIIb (Abcam) were used to detect the coimmunoprecipitated integrins.
Ras activation assay
A Ras activation assay kit (Millipore) was used to measure Ras activity following the manufacturer's instructions. Briefly, cell lysates were extracted in cell lysis buffer, incubated with the beads/GST–Ras binding domain (RBD) mixture for 45 minutes, washed four times, and resuspended in 40 μL of 2X Laemmli buffer. The samples were separated by SDS–PAGE (15% polyacrylamide), blotted, and probed with an anti-Ras antibody.
Soft agar colony formation
The bottom agar layer was built from the solidification of 2 mL complete media containing 0.5% agar in a six-well flat-bottomed plate. Cell suspensions containing 10,000 cells in 0.35% agar in complete medium were poured to form the top layer. After the top layer was solidified by incubation at room temperature for 3 hours, 1 mL complete medium was added. The culture medium covering the top agar was changed twice a week. Colonies were allowed to grow for 28 days at 37°C in 5% CO2 before imaging. The colonies were photographed using a Leica DMC4500 camera attached to a Leica MZ16 FA stereomicroscope (Leica Microsystems) and counted manually.
Suspension cultures and quantification
Poly [2-hydroxyethyl methacrylate (HEMA)]–coated plates were prepared by coating 96-well tissue culture plates with 20 mg/mL HEMA (Sigma-Aldrich) and sterilizing the plates with UV overnight. To the sterilized HEMA-coated plates, 50,000 cells were seeded in a volume of 100 μL culture media. The Cell Counting Kit-8 reagent (Dojindo Molecular Technologies) was used to quantify cell numbers according to the manufacturer's instructions. A Wallac VICTOR2 1420 Multilabel Counter (Perkin Elmer) was used to measure absorbance at 450 nm. Cell number was measured as the fold-increase in absorbance over day 1.
qPCR
Cells were lysed in TRIzolTM reagent (Invitrogen) and RNA was isolated according to the manufacturer's protocol. Two hundred nanograms of total RNA was reverse-transcribed to produce cDNA template using the qScript cDNA synthesis kit (QuantaBio). qPCR reactions were carried out with 2 μL of cDNA, 10 μL PerfeCTa SYBR Green Fastmix Universal (QuantaBio), and 0.4 mol of each primer. Primer sequences were as follows: mouse CTGF (forward, 5′-CTCCACCCGAGTTACCAATG-3′; reverse, 5′-TGGCGATTTTAGGTGTCCG-3′); mouse GAPDH (forward, 5′-CTTTGTCAAGCTCATTTCCTGG-3′; reverse, 5′-TCTTGCTCAGTGTCCTTGC-3′). qPCR reactions were run using the CFX Connect Real-Time PCR detection system according to the manufacturer's instructions (Bio-Rad). PCR conditions were 95°C for 30 seconds, followed by 40 cycles of 95°C for 5 seconds and 60°C for 30 seconds. For analysis, the fold change relative to the indicated control sample was calculated using the CFX software, and GAPDH was used as a reference gene.
In vivo assays
The Albany Medical College (Albany, NY) Institutional Animal Care and Use Committee approved all the mouse studies. Mice were housed under specific pathogen-free conditions in the Albany Medical College Animal Resources Facility, which is licensed by the United States Department of Agriculture and the New York State Department of Health, Division of Laboratories and Research, and is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. These studies used immunocompromised mice [NOD.Cg-scid tm1Wj Prkdc Il2rg l/SzJ (NSG); Jackson Laboratories]. To assay tumor formation and tumor growth, mice were injected subcutaneously with 1 million cells in 100 μL of PBS. Once tumors were palpable, the length, width, and depth of each tumor were measured using Vernier calipers and tumor volume was estimated using the following equation for the volume of an ellipse: Volume = 4/3 π(length/2 × width/2 × depth/2)^3. In the trametinib treatment experiment, daily treatment via oral gavage with vehicle or 1 mg/kg trametinib began once tumors reached between 150 and 250 mm3 and continued for 14 days. The tumor volume was measured daily during treatment with trametinib. In all experiments, the mice were euthanized at the indicated time points, and the primary tumors were removed and weighed. All mouse experiments included roughly equal numbers of males and females, and we observed the same trends in both sexes, with no indication that sex was a biological variable.
Human samples
All human sample research was performed with approval from the Cleveland Clinic (Cleveland, Ohio) Institutional Review Board (IRB 06–977). Samples were deidentified to the research team and were therefore exempt from participant consent for this specific study and were in accordance with the Health Insurance Portability and Accountability Act (HIPAA) privacy rule. Liver EHE tumors with matched surrounding normal liver tissues were used for further analyses.
Statistics
An unpaired two-tailed t test was used to evaluate the statistical significance of soft agar colony formation and proliferation assays [95% confidence interval (CI), P < 0.05]. The mean was calculated to represent the average of the independent wells. Error bars represent SD. For the in vivo experiments, the number of mice per group is indicated in the legend. To determine if the change in tumor volume between the groups was significantly different, linear regression was performed and plotted. An analysis of covariance was used to determine if the slopes of the tumor volume plots were significantly different.
Data availability
The data generated in this study are available within the article.
Results
CTGF is a key downstream target of the TC fusion protein
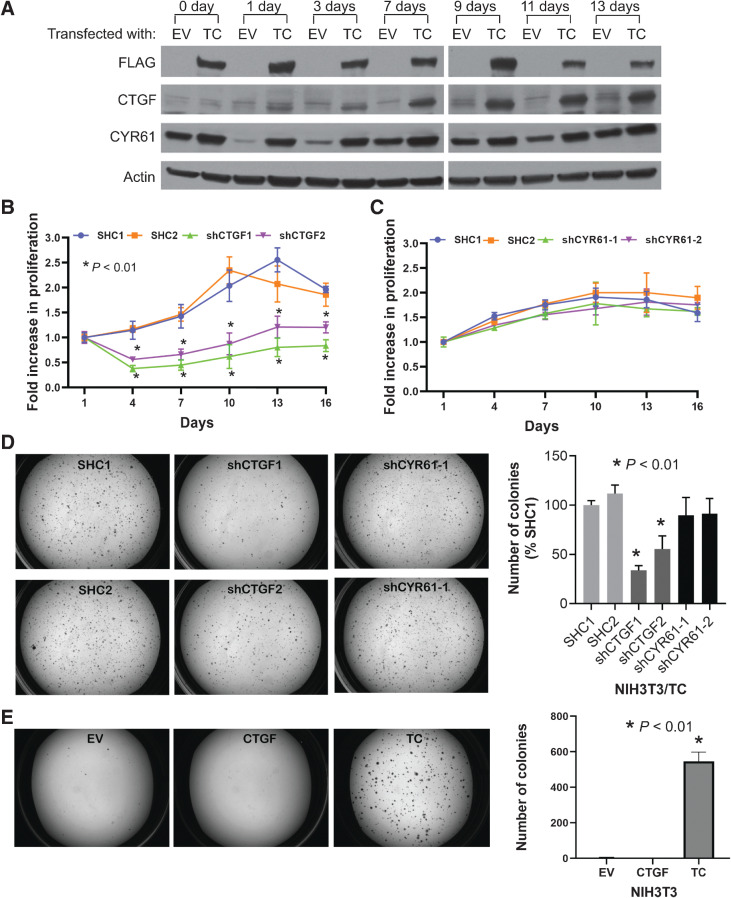
CTGF (or CCN2) and cysteine-rich 61 (CYR61 or CCN1) are two major transcriptional targets of TAZ and are thought to mediate the oncogenic function of TAZ in many scenarios (12). As we have previously observed that TC enhances CTGF and CYR61 expression levels (3, 9), and that these transcripts are well-known YAP/TAZ target genes (12), we hypothesized that TC transforms cells via CTGF and/or CYR61. To test this hypothesis, we exogenously induced the expression of the TC fusion gene in NIH3T3 cells (NIH3T3/TC) and cultured them in suspension on poly-HEMA–coated plates. Poly-HEMA is a nonadhesive coating that prevents the attachment of cells to culture dishes, and unattached NIH3T3 cells grow in suspension as spheroids. When cultured in an anchorage-independent manner, nontransformed NIH3T3 cells survive but do not proliferate; in contrast, NIH3T3/TC cells can proliferate (9). In this system, we found that CTGF and CYR61 expression levels increased in NIH3T3/TC cells in a time-dependent manner (Fig. 1A). The increase in CTGF expression was more robust and TC-dependent. CYR61 expression also increased and was TC-dependent at most time points, but its expression also increased in the empty vector (EV) control cells, suggesting that its expression is not completely dependent on TC (Fig. 1A).
Figure 1.
CTGF is a key downstream target of the TC fusion protein. A, Immunoblots to estimate the expression levels of CTGF and CYR61 in NIH3T3 cells transfected with either an EV or FLAG-TC fusion gene construct (TC). Cells were grown in suspension on HEMA-treated culture plates. Cell proliferation assay to measure the rate of anchorage-independent cell growth of NIH3T3/TC cells grown in suspension after the knockdown of CTGF (B) or CYR61 (C). D, Soft agar colonies of NIH3T3/TC cells expressing either control (SHC1/2), CTGF, or CYR61 shRNAs; quantification of the colony numbers is also shown. E, Soft agar assay to observe colony growth after CTGF overexpression. Data represent the mean ± SD of independent wells; experiments were performed at least twice.
To assess whether CTGF or CYR61 is essential for the maintenance of anchorage-independent growth, we reduced the expression of CTGF and CYR61 using shRNA in NIH3T3/TC cells. Knockdown of CTGF significantly inhibited cell growth in suspension (Fig. 1B). In contrast, CYR61 knockdown did not inhibit cell growth (Fig. 1C). Anchorage-independent growth promoted by TC can similarly be assayed using a soft agar assay, where TC has been shown to promote colony growth (9). Through shRNA knockdown, reducing CTGF, but not CYR61 levels, greatly affected soft agar colony growth (Fig. 1D). Therefore, growth in both suspension and colony formation was sensitive to CTGF but not CYR61 knockdown (Fig. 1B–D). Together, these observations indicate that CTGF, rather than CYR61, is a key downstream target of TC required for cell transformation. However, the expression of CTGF in naïve NIH3T3 cells did not stimulate soft agar colony growth, suggesting that CTGF is necessary but not sufficient for cellular transformation (Fig. 1E).
Integrin αIIbβ3 is crucial for the TC signaling pathway
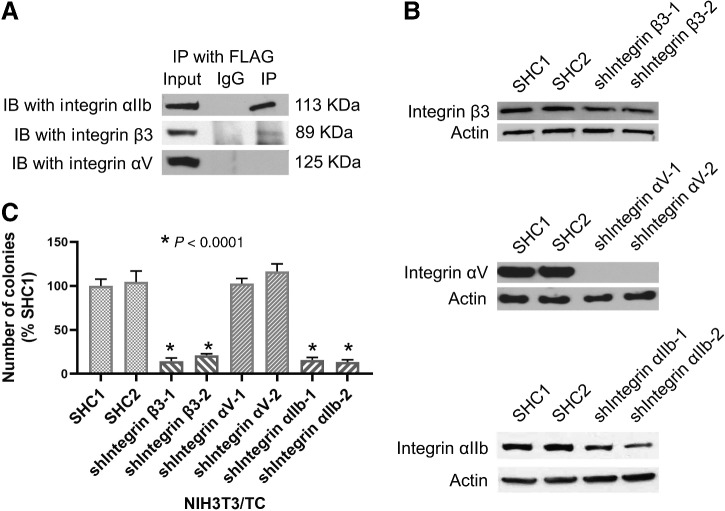
It is well known that many functions of CTGF are mediated by its binding to cell-surface integrins and subsequent activation of the downstream integrin-dependent signaling (13). Integrins are obligate heterodimers of α- and β-subunits that initiate downstream prosurvival signaling pathways in response to binding to the extracellular matrix and other ligands (14). Ligand binding promotes the clustering of integrins and initiates downstream prosurvival signaling pathways. As integrin αVβ3 was identified as a receptor for CTGF in conditions promoting the motility of breast cancer cells (15) and migration of human chondrosarcoma cells (16), we first investigated whether integrin β3 and αV subunits play a role in our NIH3T3/TC model. Co-I with FLAG-CTGF showed that integrin β3 forms a complex with CTGF but not integrin αV (Fig. 2A). Subsequently, we investigated the integrin αIIb subunit because it pairs with β3 and has been shown to act as a CTGF receptor (17). We observed that integrin αIIb coimmunoprecipitated with FLAG-CTGF (Fig. 2A), suggesting that integrin αIIbβ3 acts as a CTGF receptor. Next, we silenced integrin β3, αV, and αIIb expression using shRNA knockdown in order to observe whether they have a role in promoting the growth of TC-dependent soft agar colonies (Fig. 2B). Knockdown of integrins αIIb and β3 in NIH3T3/TC cells decreased colony formation in soft agar (Fig. 2C), whereas knockdown of αV did not affect colony growth (Fig. 2C). This finding suggests that integrin αIIbβ3 but not αV integrins is important for TC-dependent transformation.
Figure 2.
Integrin αIIbβ3 is crucial for the TC signaling pathway. A, Lysates from NIH3T3 cells transfected with FLAG-CTGF were immunoprecipitated with an anti-FLAG antibody. Coimmunoprecipitated integrins were detected in an immunoblot using anti-integrin αIIb, anti-integrin β3, or anti-integrin αV antibodies. B, Immunoblots to detect the efficiency of shRNA-mediated knockdown of integrin β3, αV, or αIIb in NIH3T3/TC cells. SHC1 is an EV, SHC2 is a scrambled control shRNA. For integrin knockdowns, two independent shRNA were used. C, Quantification of the soft agar colony counts after knockdown of the indicated integrins in NIH3T3/TC cells. Silencing of integrin αIIb and integrin β3, but not integrin αV, inhibited the ability of NIH3T3/TC cells to form colonies. Data represent mean and SD of individual wells. IP, immunoprecipitated; IB, immunoblot.
Ras is an important member of the TC signaling pathway
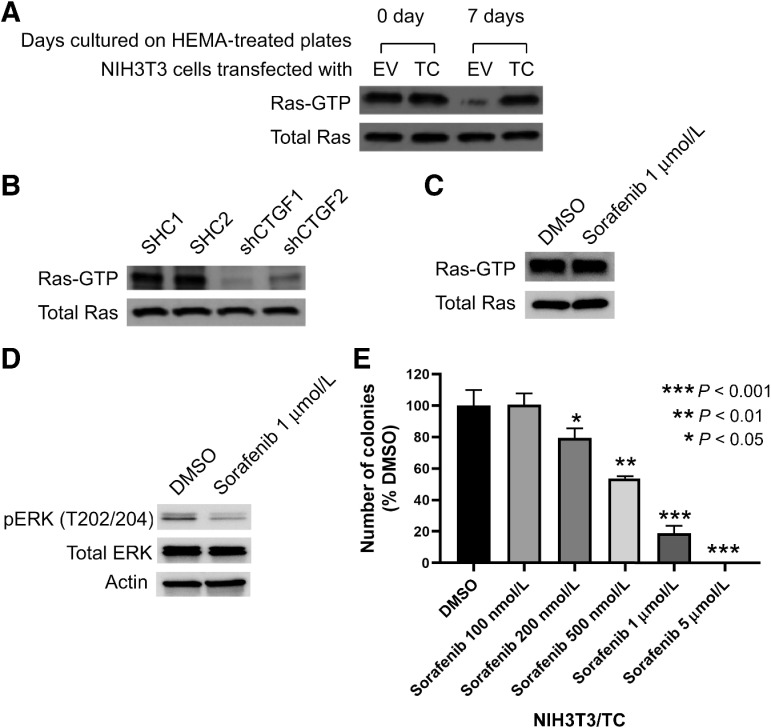
As the activation of Ras is essential for many integrin-mediated signaling processes (18, 19), we next tested whether Ras plays a role in TC signaling. Ras interacts with effector proteins, including Raf-1, only in the GTP-bound active form to initiate signaling cascades that facilitate cell proliferation (20). Using the Ras-binding domain of Raf-1, we precipitated Ras-GTP from lysates of NIH3T3 cells cultured in suspension and estimated active Ras levels by immunoblots. The initial levels of Ras-GTP were similar in NIH3T3/EV and NIH3T3/TC cells (Fig. 3A). However, when cells were cultured in suspension for prolonged periods (7 days), the Ras-GTP levels in NIH3T3/EV cells dropped markedly but was maintained in NIH3T3/TC cells (Fig. 3A). This finding shows that TC expression allows NIH3T3 cells to maintain high Ras-GTP levels despite the loss of adhesion to culture dishes; high Ras-GTP levels may support anchorage-independent growth. Interestingly, the Ras-GTP concentration was reduced upon CTGF knockdown, suggesting that NIH3T3/TC cells in suspension require CTGF to maintain Ras in an active state (Fig. 3B). To investigate whether soft agar colony growth requires Ras signaling, we inhibited Raf-1 using sorafenib. Sorafenib is a multi-kinase inhibitor that effectively inhibits Raf kinase (21). However, sorafenib also inhibits receptor tyrosine kinases (22). To differentiate between these two possibilities, we measured Ras-GTP levels after sorafenib treatment. Ras-GTP levels were unaltered (Fig. 3C), therefore, signaling upstream of Ras was not inhibited. As expected, the MAPK pathway that is downstream of Raf was effectively inhibited upon sorafenib treatment (Fig. 3D). We then tested whether sorafenib abrogated colony formation. Our results show that sorafenib treatment suppressed the formation of NIH3T3/TC colonies in a dose-dependent manner, confirming that Ras-Raf signaling is an important component of the oncogenic TC signaling pathway that drives anchorage-independent growth (Fig. 3E).
Figure 3.
Ras is an important member of the TC signaling pathway. A, Expression levels of active Ras (Ras-GTP) in NIH3T3/TC and NIH3T3/EV cells that were cultured in an anchorage-independent manner. Active Ras was precipitated using the RBD of Raf-1, and the precipitated Ras levels were determined by immunoblot using an anti-Ras antibody. B, Expression levels of active Ras in NIH3T3/TC cells upon CTGF knockdown using two independent shRNA; the cells were cultured in suspension. C, Immunoblots to estimate active Ras levels and D, the activity of MAPK signaling after sorafenib (Raf inhibitor) treatment. E, Soft agar assay in the presence of vehicle (DMSO) or sorafenib. Inhibiting RAS suppressed the ability of NIH3T3/TC cells to form colonies. Comparison is to DMSO vehicle. Error bars represent one SD, and the plot is representative of two independent experiments.
MAPK pathway mediates the signaling downstream of Ras
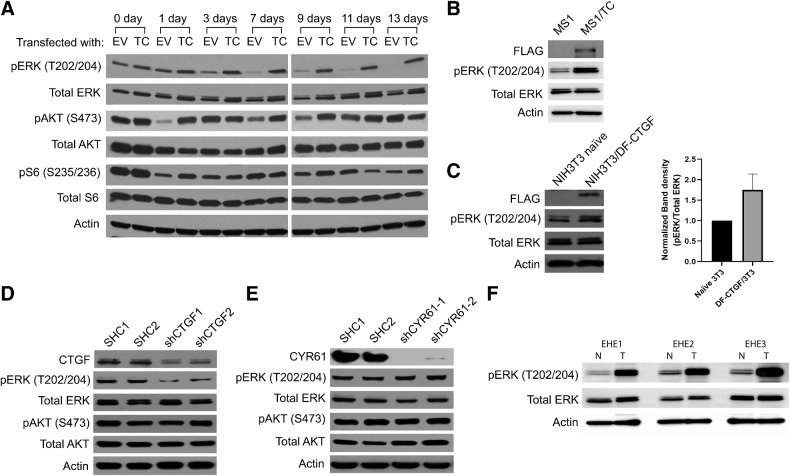
Ras can signal through either the MAPK or PI3K pathways, therefore, we evaluated their relative contributions to downstream TC signaling. Lysates were prepared from NIH3T3/EV and NIH3T3/TC cells grown in suspension. Then, the expression of active PI3K and MAPK signaling components was probed using phospho-specific antibodies (Fig. 4A). MAPK signaling was measured by pERK immunoblot. MAPK signaling declined gradually when NIH3T3/EV cells were cultured in suspension, whereas it was maintained in NIH3T3/TC cells (Fig. 4A). The PI3K signaling components pAKT and pS6, on the other hand, initially decreased in NIH3T3/EV cells, but gradually returned to levels similar to what was seen in the NIH3T3/TC cells (Fig. 4A). This suggests that the MAPK, but not the PI3K pathway is an important component of TC signaling. This finding was also validated in a pancreatic islet endothelial cell line (MS1), in which MAPK activity was enhanced after TC expression (Fig. 4B). Furthermore, CTGF overexpression also increased MAPK signaling (Fig. 4C). Likewise, knockdown of CTGF in NIH3T3/TC cells decreased MAPK signaling but not PI3K signaling (Fig. 4D). CYR61 knockdown on the other hand decreased neither PI3K nor MAPK signaling (Fig. 4E). Finally, the activity of the MAPK pathway in clinical samples was investigated (Fig. 4F). The pERK levels in matched EHE tumor samples were significantly higher than those in paired normal tissues (Fig. 4F). Collectively, these results indicate that TC signals through a CTGF-MAPK signal transduction pathway.
Figure 4.
TC predominantly activates the MAPK signaling pathway. A, NIH3T3/EV and NIH3T3/TC cells were grown attached to dishes (day 0) or in suspension for 1, 3, 7, 9, 11, or 13 days before harvesting for lysate preparation. Levels of pERK, ERK, pAKT, AKT, pS6, and S6 in cell lysates were detected by immunoblots. B, MAPK activity was estimated using a pERK antibody in MS1 cells after exogenous TC expression. C, NIH3T3 cells expressing a double FLAG-CTGF were grown in suspension for 7 days before collection. Cell lysates were analyzed by immunoblot with antibodies to pERK and ERK. Densitometry data for pERK is also shown. Expression of pERK, ERK, pAKT, and AKT in NIH3T3/TC cells after (D) CTGF or (E) CYR61 knockdown. Actin is used as a loading control. F, Immunoblots to estimate the levels of pERK and ERK between normal and matched human EHE tumor tissue samples. N, normal tissue samples; T, tumor tissue samples.
MAPK inhibition as a potential therapeutic treatment for EHE
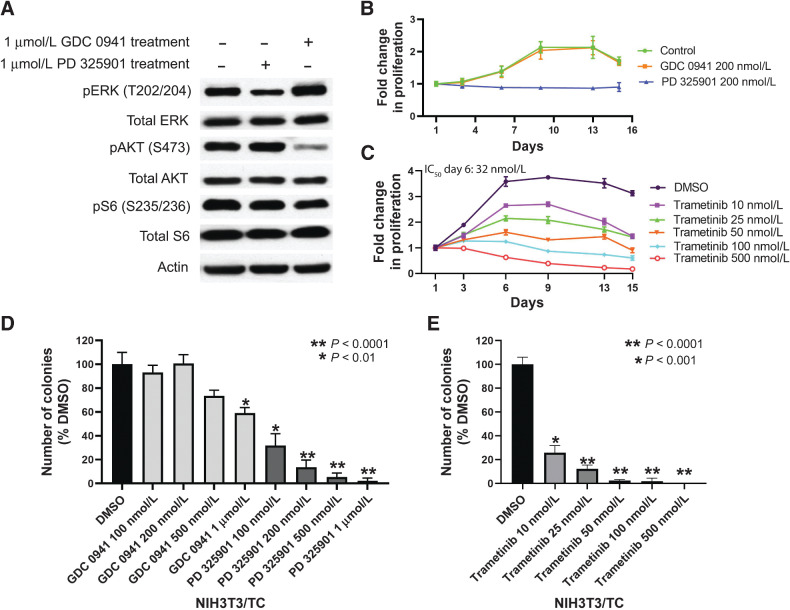
To further elucidate the roles of the MAPK and PI3K pathways in promoting TC-induced anchorage-independent proliferation, we pharmacologically inhibited the PI3K and MAPK pathways using the PI3K inhibitor GDC 0941 and MEK inhibitor PD 0325901, respectively (Fig. 5A). Our results show that the MEK inhibitor, PD 0325901, effectively inhibited the growth rate of NIH3T3/TC cells that were grown in suspension, whereas GDC 0941 had no effect (Fig. 5B). Next, as the FDA-approved MEK inhibitor trametinib is used clinically for the treatment of melanoma, we hypothesized that this drug would demonstrate similar efficacy. Trametinib also caused a dose-dependent decrease in the proliferation of NIH3T3/TC cells in suspension (Fig. 5C). Furthermore, colonies of NIH3T3/TC cells grown in soft agar were reduced upon treatment with PD 0325901 or trametinib (Fig. 5D and E). In contrast, colony numbers were largely unaffected by GDC 0941 treatment (Fig. 5D). These data provide additional evidence that MAPK, rather than PI3K, is the central pathway involved in the TC-mediated anchorage-independent growth. Importantly, trametinib inhibited the colony formation of NIH3T3/TC cells in soft agar and their growth in suspension in a dose-dependent manner at clinically relevant doses (Figs. 5C and E).
Figure 5.
Effects of pharmacologic inhibitors of MAPK or PI3K pathway on anchorage-independent growth of NIH3T3/TC cells. A, NIH3T3/TC cells were cultured in the presence of either PD 0325901 or GDC 0941 for 2 days after being grown in suspension for 7 days, lysed, and the cell lysates were analyzed by immunoblotting with the indicated antibodies. GDC 0941 is a PI3K inhibitor, and PD 0325901 is a MEK inhibitor. B, The change in cell number over time of NIH3T3/TC cells grown in suspension in the presence of GDC 0941 or PD 0325901. PD 0325901 effectively inhibited the anchorage-independent proliferation of NIH3T3/TC cells (P < 0.05). C, Estimating the cell number of NIH3T3/TC cells grown in suspension. Administration of trametinib significantly decreased cell proliferation in suspension in a dose-dependent manner compared withto DMSO only (P < 0.0001). D, Soft agar colony numbers of NIH3T3/TC cells in the presence of GDC 0941 or PD 0325901 as compared with vehicle DMSO. E, Quantification of soft agar colonies with NIH3T3/TC cells in the presence of vehicle (DMSO) or trametinib. Error bars represent one SD; experiments were performed at least twice.
CTGF and MAPK signaling sustain tumor growth in vivo
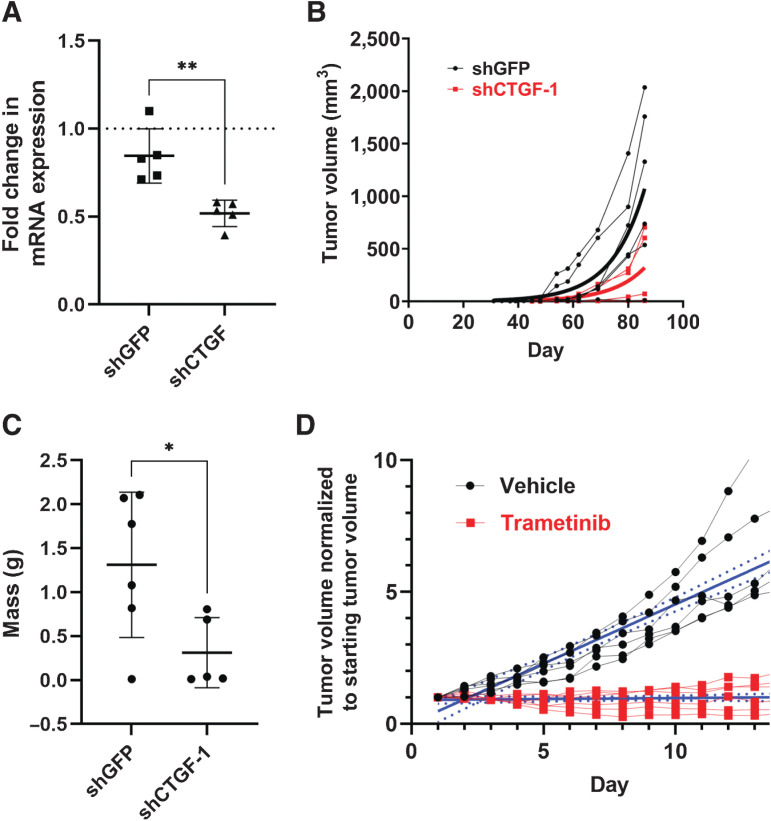
CTGF expression transformed NIH3T3 cells by promoting anchorage-independent proliferative growth in soft agar and in suspension (Fig. 1). Therefore, we speculated that CTGF may also play a role in mediating tumor growth in vivo. To investigate this, we stably reduced CTGF mRNA levels in NIH3T3/TC cells using CTGF shRNA and used a GFP-targeting shRNA as a control (Fig. 6A). We established a xenograft model by subcutaneously injecting NIH3T3/TC cells stably expressing control or CTGF shRNAs into immunodeficient NSG mice. Compared with cells expressing short hairpin GFP (shGFP), short hairpin CTGF (shCTGF) cells produced tumors that grew significantly slower (Fig. 6B). This correlated well with the significantly lower weight of shCTGF tumors (Fig. 6C). As TC and CTGF signal through the MAPK pathway to transform cells (Fig. 4), we tested whether trametinib inhibition of MAPK signaling could suppress in vivo tumor growth. To this end, we designed a 14-day daily oral treatment regimen in which trametinib dosed at 1 mg/kg was administered to NSG mice harboring NIH3T3/TC allografts, and the change in tumor volume was monitored and compared with mice treated with the vehicle control (Fig. 6D). NIH3T3/TC xenografts responded well to trametinib treatment with a significant reduction in tumor growth rate, suggesting that inhibition of MAPK signaling can counter TC-mediated tumorigenesis (Fig. 6D).
Figure 6.
CTGF and MAPK signaling sustain tumor growth in vivo. NIH3T3/TC cells were infected with a control GFP shRNA or a shRNA targeting CTGF and assayed for A,Ctgf mRNA expression by qPCR; data are shown as mean ± SD, samples were normalized to control cells lacking any shRNA (dotted line; n = 5 samples run in triplicate from five independent experiments, **P < 0.01). B, Control or CTGF knockdown cells were injected subcutaneously in NSG mice, and tumor volume was measured twice per week for 85 days. C, Tumors from (B) were isolated and weighed; data are shown as mean ± SD (n = 6 shGFP mice and n = 5 shCTGF-1 mice, *P < 0.05 by unpaired t test). D, NIH3T3 cells stably expressing TAZ-CAMTA1 were injected subcutaneously and allowed to form tumors. Once tumors reached a volume of between 150 to 250 mm3, mice received daily oral treatment with either vehicle or 1 mg/kg trametinib for 14 days. Plots show individual mouse tumor volume normalized to tumor volume at the start of treatment (solid Iines with dots). Solid blue lines represent linear regression with 95% CI (dashed blue lines) shown (n = 6 mice per group); the difference in the slopes of the linear regression was tested by ANOVA; **** P < 0.0001.
Discussion
We previously identified a disease-defining genetic anomaly in EHE, specifically a chromosomal translocation that causes the fusion of WWTR1 (TAZ) and CAMTA1 genes (1). The resultant TC fusion protein orchestrates a TAZ-like transcriptional program. TAZ is a well-known transcriptional coactivator and is a major downstream effector of the Hippo pathway. TAZ shuttles between the nucleus and cytoplasm based on the Hippo pathway regulatory activity. In contrast, TC remains in the nucleus because of the strong nuclear localization signal of CAMTA1 (7). Nuclear localization and its ability to recruit chromatin remodelers allow TC to induce a potent oncogenic transcriptional program (9, 11). Moreover, as highlighted in this study, TC supports anchorage-independent cell proliferation that enables cells to escape anoikis or programmed cell death after the loss of appropriate cell-ECM adhesion. We have also demonstrated that TC alone at physiologic expression levels is sufficient to drive EHE in a genetically engineered mouse model (3). Although TC is known to be oncogenic, TC transcriptional targets that mediate tumorigenesis have not been elucidated. In this study, we dissected a targetable signaling cascade downstream of the TC fusion protein and investigated possible therapeutic interventions to disrupt this cascade. As there are no EHE cell lines, the NIH3T3/TC system was used to investigate the mechanistic underpinnings of TC-dependent transformation (9, 11). This system was used to identify a clinically relevant and targetable pathway that induces tumorigenesis.
We examined the CCN family of proteins, in particular CTGF and CYR61, two major transcriptional targets of TAZ, for their ability to support TC-mediated, anchorage-independent growth (12). Both proteins were robustly upregulated in NIH3T3/TC cells. However, silencing the expression of CTGF, but not CYR61, led to the inhibition of anchorage-independent cell growth in suspension and soft agar. The ability to sustain growth without anchoring to a substrate helps tumor cell dissemination to distant organs (23). Therefore, we believe that CTGF is an important oncogenic downstream target of the TC fusion protein. Both CTGF and CYR61 are matricellular proteins that play a role in organizing the extracellular matrix, and they also act as cytokines that stimulate various oncogenic signaling pathways (24, 25). Consistently, these proteins have been shown to be involved in the progression of different cancers, such as breast, prostate, gastric, and colorectal cancers, and are often used as readouts for YAP/TAZ activation (12, 26–31). Our results indicated that CTGF but not CYR61 has a tumor-promoting effect on the TC-activated signaling pathway.
Recent work that employed other cell-based and mouse models also showed CTGF as an important TC target. CTGF is upregulated in the proliferative endothelial cell population that stains positively for Ki67 (32). CTGF levels also increase in liposarcoma cells upon exogenous expression of TC (11). In our recently deposited RNA sequencing (RNA-seq) data (GSE168493), the FPKM reads of CTGF in EHE tumors were significantly high. Therefore, multiple studies show that CTGF is a tumor-relevant transcriptional target of TC.
Several integrins are known to act as key receptors of the CCN ligand family and CTGF has been shown to bind to several integrins (13). Immunoprecipitation and knockdown experiments revealed that integrin αIIbβ3 is a component of the TC signaling pathway. Knockdown of αIIb and β3 integrin subunits significantly reduces the number of soft agar colonies. Integrin αIIbβ3 is predominantly expressed in platelets and is required for platelet aggregation (33). However, tumor cells have also been shown to express this integrin in order to facilitate invasion and metastasis (34, 35).
As Ras is essential for integrin-mediated signaling (36), we next analyzed Ras levels in our NIH3T3/TC model and found that Ras activity was maintained in TC-expressing cells. Knockdown of CTGF also reduced the levels of active Ras, suggesting that CTGF signals via Ras. We then inhibited Ras signaling using sorafenib (21), which impeded colony formation by NIH3T3/TC cells in a dose-dependent manner. Therefore, we inferred that Ras is an active component in the TC signaling pathway. Ras signals through either the MAPK or PI3K pathways (37, 38). We found that TC maintained MAPK activation in cells cultured in suspension for the entire duration of the experiment. In comparison, PI3K activation tapered after day 1. MAPK activation was also more pronounced in human EHE tumor samples than in matched normal tissues. To further support the possibility that the MAPK pathway, but not the PI3K pathway, is critical for CTGF signaling, we used pharmacological inhibitors specific to each pathway. PD 0325901, an established MEK inhibitor, effectively limited NIH3T3/TC anchorage-independent cell growth, whereas GDC 0941, a PI3K inhibitor, had little effect. Taking these data together, we concluded that MAPK is the key TC-mediated tumorigenic pathway. Lastly, we used trametinib, an FDA-approved MEK inhibitor, to assay colony formation in soft agar and growth in suspension. Similar to PD 0325901, trametinib also inhibited anchorage-independent cell growth. The efficacy of trametinib was better than that of sorafenib and PD 0325901.
Using NIH3T3/TC cells, we modeled tumorigenesis in NSG mouse xenografts. In accordance with the in vitro results, the knockdown of CTGF in these cells reduced tumor burden. Furthermore, MAPK inhibition through the administration of trametinib also resulted in a pronounced reduction in tumor volume, indicating that CTGF and MAPK pathways are therapeutic targets to inhibit TC-dependent tumorigenesis in vivo.
Our goal is to introduce novel therapeutic agents to the clinic but there are inherent barriers to testing for this disease. While aggressive-type EHE represents the most morbid of subtypes of this disease, a significant number of patients have indolent nonprogressive disease. Therefore, a watch-and-wait strategy is often employed for these patients, and thus therapy is reserved for when their indolent disease becomes progressive. As EHE is a rare cancer, it is difficult to recruit adequate numbers of patients with EHE for robust clinical trials. Previous studies examining the responsiveness of EHE to other therapeutics have had some success. A retrospective analysis involving a 38-patient cohort with EHE suggested that sirolimus (rapamycin) is an effective treatment for disease stabilization in patients with progressive EHE but without serosal effusions (39). As EHE is an endothelial cell tumor, sorafenib, and bevacizumab, agents that inhibit VEGF signaling have also been tested in clinical trials. Sorafenib treatment in patients with EHE had a 9-month progression-free rate of 30.7% (40), whereas patients with EHE treated with bevacizumab exhibited disease stabilization (41). The MAPK pathway is also activated as a result of VEGF signaling, therefore, the observed responses due to the administration of these agents may be partially due to MAPK pathway inhibition. Nevertheless, even though these drugs showed promise in some patients, they were not effective in others, so the need for additional treatment options remains. Some ongoing clinical trials involving chemotherapy drugs, such as eribulin (NCT03331250) and gemcitabine (NCT01532687), and novel targeted TEAD inhibitors, such as IK-930 (NCT05228015) and IAG933 (NCT04857372) are recruiting patients with EHE. As the TC oncogene is critically important for EHE development and growth, targeting protumorigenic pathways downstream of TC is an enticing approach. As model systems for preclinical validation of therapeutics for EHE remain limited, we aimed to leverage our heterologous TC-dependent tumorigenesis system (NIH3T3/TC) to test and credential the drug trametinib that is FDA-approved for use in other cancers. We used the results of this study to provide a preclinical rationale for launching SARC033 (NCI 10015-NCT03148275), a nonrandomized, open-label, phase II study of trametinib in patients with unresectable or metastatic EHE.
Acknowledgments
This research was supported by generous funding from The EHE Foundation (to B.P. Rubin, R. Kanai, J.M. Lamar, K. Che, A. Hallett, and S. Ma), The EHE Rare Cancer Charity (UK; to B.P. Rubin, R. Kanai, and J.M. Lamar), The EHE Rare Cancer Foundation Australia (to B.P. Rubin, R. Kanai, and J.M. Lamar), CRAVAT Foundation (to B.P. Rubin), VeloSano (to B.P. Rubin), and the Margie and Robert E. Petersen Foundation (to B.P. Rubin, A.V. Pobbati, S. Ma, R. Kanai, S. Li, A. Burtscher, J.M. Lamar, and A. Hallett). Part of this work was also funded by the U.S. Department of Defense IDEA Development Award – RA200178 (to B.P. Rubin, J.M. Lamar, and A.V. Pobbati), and the Crile Research Fellowship (C.N. Seavey). We thank Dr. Cassandra Talerico for critically reviewing the manuscript.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Authors’ Disclosures
S. Ma reports grants from the Margie and Robert E. Petersen Foundation, VeloSano, CRAVAT Foundation, and The EHE Foundation during the conduct of the study; grants from the David Foundation outside the submitted work. R. Kanai reports grants from The EHE Foundation, EHE Fund, and Cleveland Clinic EHE Collaborative Research Project during the conduct of the study. A.V. Pobbati reports grants from the Margie and Robert E. Petersen Foundation, VeloSano, CRAVAT Foundation, and The EHE Foundation during the conduct of the study; grants from the David Foundation outside the submitted work. S. Li reports grants from the Margie and Robert E. Petersen Foundation, VeloSano, CRAVAT Foundation, and The EHE Foundation during the conduct of the study; grants from the David Foundation outside the submitted work. K. Che reports grants from the Margie and Robert E. Petersen Foundation, VeloSano, CRAVAT Foundation, and The EHE Foundation during the conduct of the study; grants from the David Foundation outside the submitted work. C.N. Seavey reports grants from the Margie and Robert E. Petersen Foundation, VeloSano, CRAVAT Foundation, and The EHE Foundation during the conduct of the study; grants from the David Foundation outside the submitted work. A. Hallett reports grants from the Margie and Robert E. Petersen Foundation, VeloSano, CRAVAT Foundation, and The EHE Foundation during the conduct of the study; grants from the David Foundation outside the submitted work. A. Burtscher reports grants from the Margie and Robert E. Peterson Foundation, VeloSano, CRAVAT Foundation, and The EHE Foundation during the conduct of the study; grants from the David Foundation outside the submitted work. J.M. Lamar reports grants from The EHE Foundation [principal investigator (PI) J.M. Lamar], EHE Fund (PI J.M. Lamar), and Cleveland Clinic EHE Collaborative Research Project (PI B.P. Rubin) during the conduct of the study; in addition, J.M. Lamar was paid a consulting fee to answer general questions about the Hippo-YAP/TAZ-TEAD pathway. This was a one-time consulting fee paid for their time and J.M. Lamar does not have any financial stake related to this. B.P. Rubin reports grants from the Margie and Robert E. Petersen Foundation, VeloSano, CRAVAT Foundation, and The EHE Foundation during the conduct of the study; grants from the David Foundation outside the submitted work.
Authors’ Contributions
S. Ma: Data curation, validation, investigation, methodology, writing–original draft. R. Kanai: Data curation, formal analysis, validation, investigation, methodology. A.V. Pobbati: Writing–original draft, project administration, writing–review and editing. S. Li: Data curation. K. Che: Methodology. C.N. Seavey: Writing–review and editing. A. Hallett: Investigation, methodology, writing–original draft. A. Burtscher: Resources. J.M. Lamar: Conceptualization, resources, validation, methodology, writing–review and editing. B.P. Rubin: Conceptualization, resources, supervision, funding acquisition, project administration, writing–review and editing.
References
- 1. Tanas MR, Sboner A, Oliveira AM, Erickson-Johnson MR, Hespelt J, Hanwright PJ, et al. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci Transl Med 2011;3:98ra82. [DOI] [PubMed] [Google Scholar]
- 2. Errani C, Zhang L, Sung YS, Hajdu M, Singer S, Maki RG, et al. A novel WWTR1-CAMTA1 gene fusion is a consistent abnormality in epithelioid hemangioendothelioma of different anatomic sites. Genes Chromosomes Cancer 2011;50:644–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Seavey CN, Pobbati AV, Hallett A, Ma S, Reynolds JP, Kanai R, et al. WWTR1(TAZ)-CAMTA1 gene fusion is sufficient to dysregulate YAP/TAZ signaling and drive epithelioid hemangioendothelioma tumorigenesis. Genes Dev 2021;35:512–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Antonescu CR, Le Loarer F, Mosquera JM, Sboner A, Zhang L, Chen CL, et al. Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes Chromosomes Cancer 2013;52:775–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dermawan JK, Azzato EM, Billings SD, Fritchie KJ, Aubert S, Bahrami A, et al. YAP1-TFE3-fused hemangioendothelioma: a multi-institutional clinicopathologic study of 24 genetically-confirmed cases. Mod Pathol 2021;34:2211–21. [DOI] [PubMed] [Google Scholar]
- 6. Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell 2016;29:783–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Okawa ER, Gotoh T, Manne J, Igarashi J, Fujita T, Silverman KA, et al. Expression and sequence analysis of candidates for the 1p36.31 tumor suppressor gene deleted in neuroblastomas. Oncogene 2008;27:803–10. [DOI] [PubMed] [Google Scholar]
- 8. Henrich KO, Bauer T, Schulte J, Ehemann V, Deubzer H, Gogolin S, et al. CAMTA1, a 1p36 tumor suppressor candidate, inhibits growth and activates differentiation programs in neuroblastoma cells. Cancer Res 2011;71:3142–51. [DOI] [PubMed] [Google Scholar]
- 9. Tanas MR, Ma S, Jadaan FO, Ng CK, Weigelt B, Reis-Filho JS, et al. Mechanism of action of a WWTR1(TAZ)-CAMTA1 fusion oncoprotein. Oncogene 2016;35:929–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu CY, Zha ZY, Zhou X, Zhang H, Huang W, Zhao D, et al. The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCF{beta}-TrCP E3 ligase. J Biol Chem 2010;285:37159–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Merritt N, Garcia K, Rajendran D, Lin ZY, Zhang X, Mitchell KA, et al. TAZ-CAMTA1 and YAP-TFE3 alter the TAZ/YAP transcriptome by recruiting the ATAC histone acetyltransferase complex. Elife 2021;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lai D, Ho KC, Hao Y, Yang X. Taxol resistance in breast cancer cells is mediated by the hippo pathway component TAZ and its downstream transcriptional targets Cyr61 and CTGF. Cancer Res 2011;71:2728–38. [DOI] [PubMed] [Google Scholar]
- 13. Chu CY, Chang CC, Prakash E, Kuo ML. Connective tissue growth factor (CTGF) and cancer progression. J Biomed Sci 2008;15:675–85. [DOI] [PubMed] [Google Scholar]
- 14. Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell 2002;110:673–87. [DOI] [PubMed] [Google Scholar]
- 15. Chen PS, Wang MY, Wu SN, Su JL, Hong CC, Chuang SE, et al. CTGF enhances the motility of breast cancer cells via an integrin-alphavbeta3-ERK1/2-dependent S100A4-upregulated pathway. J Cell Sci 2007;120(Pt 12):2053–65. [DOI] [PubMed] [Google Scholar]
- 16. Tan TW, Lai CH, Huang CY, Yang WH, Chen HT, Hsu HC, et al. CTGF enhances migration and MMP-13 up-regulation via alphavbeta3 integrin, FAK, ERK, and NF-kappaB-dependent pathway in human chondrosarcoma cells. J Cell Biochem 2009;107:345–56. [DOI] [PubMed] [Google Scholar]
- 17. Jedsadayanmata A, Chen CC, Kireeva ML, Lau LF, Lam SC. Activation-dependent adhesion of human platelets to Cyr61 and Fisp12/mouse connective tissue growth factor is mediated through integrin alpha(IIb)beta (3). J Biol Chem 1999;274:24321–7. [DOI] [PubMed] [Google Scholar]
- 18. Kinbara K, Goldfinger LE, Hansen M, Chou FL, Ginsberg MH. Ras GTPases: integrins' friends or foes? Nat Rev Mol Cell Biol 2003;4:767–76. [DOI] [PubMed] [Google Scholar]
- 19. Clark EA, Hynes RO. Ras activation is necessary for integrin-mediated activation of extracellular signal-regulated kinase 2 and cytosolic phospholipase A2 but not for cytoskeletal organization. J Biol Chem 1996;271:14814–8. [DOI] [PubMed] [Google Scholar]
- 20. Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol 2004;5:875–85. [DOI] [PubMed] [Google Scholar]
- 21. Adnane L, Trail PA, Taylor I, Wilhelm SM. Sorafenib (BAY 43–9006, Nexavar), a dual-action inhibitor that targets RAF/MEK/ERK pathway in tumor cells and tyrosine kinases VEGFR/PDGFR in tumor vasculature. Methods Enzymol 2006;407:597–612. [DOI] [PubMed] [Google Scholar]
- 22. Wilhelm S, Carter C, Lynch M, Lowinger T, Dumas J, Smith RA, et al. Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nat Rev Drug Discov 2006;5:835–44. [DOI] [PubMed] [Google Scholar]
- 23. Simpson CD, Anyiwe K, Schimmer AD. Anoikis resistance and tumor metastasis. Cancer Lett 2008;272:177–85. [DOI] [PubMed] [Google Scholar]
- 24. Lipson KE, Wong C, Teng Y, Spong S. CTGF is a central mediator of tissue remodeling and fibrosis and its inhibition can reverse the process of fibrosis. Fibrogenesis Tissue Repair 2012;5Suppl 1:S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao B, Ye X, Yu J, Li L, Li W, Li S, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 2008;22:1962–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dhar A, Ray A. The CCN family proteins in carcinogenesis. Exp Oncol 2010;32:2–9. [PubMed] [Google Scholar]
- 27. Huang YT, Lan Q, Lorusso G, Duffey N, Ruegg C. The matricellular protein CYR61 promotes breast cancer lung metastasis by facilitating tumor cell extravasation and suppressing anoikis. Oncotarget 2017;8:9200–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kleer CG. Dual roles of CCN proteins in breast cancer progression. J Cell Commun Signal 2016;10:217–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ladwa R, Pringle H, Kumar R, West K. Expression of CTGF and Cyr61 in colorectal cancer. J Clin Pathol 2011;64:58–64. [DOI] [PubMed] [Google Scholar]
- 30. Zhou ZQ, Cao WH, Xie JJ, Lin J, Shen ZY, Zhang QY, et al. Expression and prognostic significance of THBS1, Cyr61 and CTGF in esophageal squamous cell carcinoma. BMC Cancer 2009;9:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xie D, Yin D, Wang HJ, Liu GT, Elashoff R, Black K, et al. Levels of expression of CYR61 and CTGF are prognostic for tumor progression and survival of individuals with gliomas. Clin Cancer Res 2004;10:2072–81. [DOI] [PubMed] [Google Scholar]
- 32. Driskill JH, Zheng Y, Wu BK, Wang L, Cai J, Rakheja D, et al. WWTR1(TAZ)-CAMTA1 reprograms endothelial cells to drive epithelioid hemangioendothelioma. Genes Dev 2021;35:495–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bennett JS. Structure and function of the platelet integrin alphaIIbbeta3. J Clin Invest 2005;115:3363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lavergne M, Janus-Bell E, Schaff M, Gachet C, Mangin PH. Platelet integrins in tumor metastasis: do they represent a therapeutic target? Cancers (Basel) 2017;9:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trikha M, Timar J, Lundy SK, Szekeres K, Cai Y, Porter AT, et al. The high affinity alphaIIb beta3 integrin is involved in invasion of human melanoma cells. Cancer Res 1997;57:2522–8. [PubMed] [Google Scholar]
- 36. Shattil SJ, Kim C, Ginsberg MH. The final steps of integrin activation: the end game. Nat Rev Mol Cell Biol 2010;11:288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Molina JR, Adjei AA. The Ras/Raf/MAPK pathway. J Thorac Oncol 2006;1:7–9. [PubMed] [Google Scholar]
- 38. Castellano E, Downward J. Role of RAS in the regulation of PI 3-kinase. Curr Top Microbiol Immunol 2010;346:143–69. [DOI] [PubMed] [Google Scholar]
- 39. Stacchiotti S, Simeone N, Lo Vullo S, Baldi GG, Brunello A, Vincenzi B, et al. Activity of sirolimus in patients with progressive epithelioid hemangioendothelioma: A case-series analysis within the Italian Rare Cancer Network. Cancer. 2021;127:569–76. [DOI] [PubMed] [Google Scholar]
- 40. Chevreau C, Le Cesne A, Ray-Coquard I, Italiano A, Cioffi A, Isambert N, et al. Sorafenib in patients with progressive epithelioid hemangioendothelioma: a phase 2 study by the French Sarcoma Group (GSF/GETO). Cancer 2013;119:2639–44. [DOI] [PubMed] [Google Scholar]
- 41. Agulnik M, Yarber JL, Okuno SH, von Mehren M, Jovanovic BD, Brockstein BE, et al. An open-label, multicenter, phase II study of bevacizumab for the treatment of angiosarcoma and epithelioid hemangioendotheliomas. Ann Oncol 2013;24:257–63. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data generated in this study are available within the article.