Abstract
Modular construction of an autonomous and programmable multi-function heterogeneous biochemical circuit that can identify, transform, translate and amplify biological signals into physicochemical signals based on logic design principle can be a powerful capability for the development of diverse biotechnologies. To explore this conceptual validity, we design a CRISPR array mediated primer exchange reaction based biochemical circuit cascades, which probe a specific biomolecular input, transform the input into a structurally accessible form for circuit wiring, translate the input information into an arbitrary sequence and finally amplify the prescribed sequence through autonomous formation of a signaling concatemer. This upstream biochemical circuit is further wired with a downstream electrochemical interface, delivering an integrated bioanalytical platform. We program this platform to directly analyze the genome of SARS-CoV-2 in human cell lysate, demonstrating the capability and utility of this unique integrated system.
Keywords: Bioanalytical chemistry, Genetic circuit, Primer exchange reaction, Electrochemistry, CRISPR
Graphical Abstract
A heterogeneous multi-function biochemical circuit that can identify, transform, translate and amplify biological signals into physicochemical signals is constructed and integrated with an electrochemistry based sensing platform for genetic analysis.
Modular construction of an autonomous and programmable multi-function biochemical network that can identify, transform, translate and amplify biological signals into physiochemical signals based on logic design principle can be a powerful capability for biotechnological and biomedical applications. Nucleic acid based biochemical circuits have shown great promise as a paradigm for individually regulating various functions.[1] We envision that an integrated heterogeneous molecular circuit which can perform multiple functions in a single pathway can be a powerful upstream regulating component with an effective downstream response. Here, to explore this conceptual capability, we construct a CRISPR array initiated cell-free genetic circuit. The CRISPR array identifies specific biomolecular sequences as inputs and the CRISPR processing results dsDNA overhangs, initiating a primer exchange reaction (PER) based DNA circuit. The primer exchange reaction performs autonomous synthesis of prescribed DNA oligos,[2] translating the molecular cues into an arbitrary sequence, which can be further cascaded and quantitatively amplified based on the same oligo synthesis mechanism. The whole genetic circuit is operated based on a simple Boolean logic design principle. To evaluate the applicability of the integrated multi-function circuit, we implemented this molecular network abstraction into a bioanalytical system operating on an electrochemical interface, which serves as a simple and cost-effective transduction system, capable of rapidly curating molecular information and transducing into data.[3]
Bioanalytical strategies have been extensively developed toward better sensitivity, simplicity and selectivity,[4] while valid strategies can be further integrated into a portable, cost-effective, rapid transduction platform for an ideal point-of-care system. So far, a high-resolution molecular analytical strategy, capable of differentiating infinitesimal concentration variation, which is essential to understand the critical threshold limit of biomolecules, has not been realized with a simple biosensing system. For high-resolution molecular analysis, the biomolecular signal is necessarily amplified downstream through centralized equipment or delicately fabricated nano-devices, limiting the general applicability.[5] Here, we explore the capability of the multi-function biochemical circuit as an upstream biological processor which 1) identifies a specific genome sequence, 2) transforms the double-stranded gene into a structure that can be accessed for circuit wiring, 3) translates the input sequence into an arbitrary output, 4) amplifies the arbitrary output sequence into a concatemer, achieving a one-to-multiple turnover reaction cascade and therefore delivering an analytical construction possible to differentiate minute concentration change. The resulted molecular output is further probed by a downstream single-use electrochemical sensing array, providing a rapid analytical result. As a proof-of-concept, we challenged this multi-function biochemical circuit based electrochemical biosensing system on analyzing the genome of the 2019 novel coronavirus,[6] Severe Acute Respiratory Syndrome-related Coronavirus (SARS-CoV-2) in complex human sample.
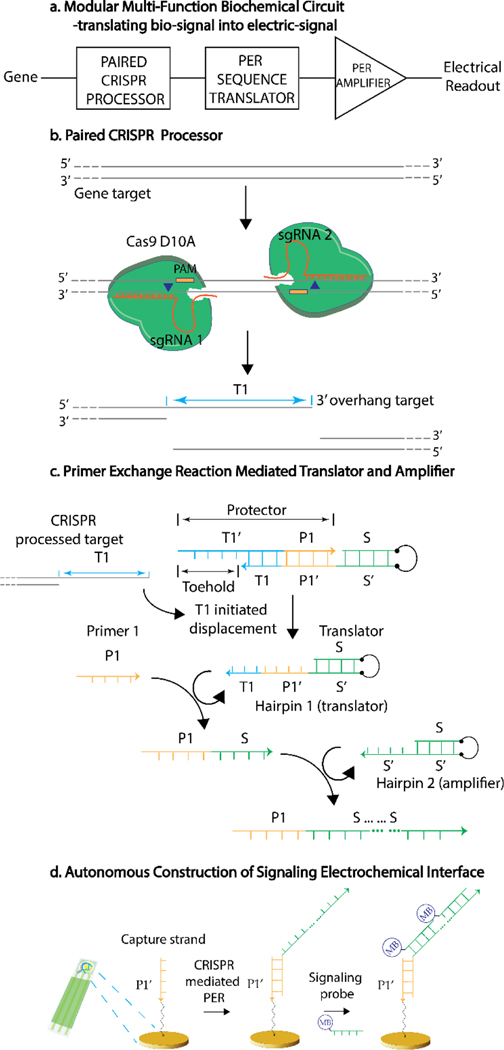
The design and operating principles of the multi-function heterogeneous biochemical circuit are shown in Figure 1a. First, in order to activate the biochemical circuit, a specific gene target as the circuit input is identified through the processor element, in which a programmable RNA-guided ribonucleoprotein, clustered regularly interspaced short palindromic repeats (CRISPR) Cas9, is applied as the recognition element, owing to its high-specificity and complementarity dependent cleavage nuclease activity.[7] The capability of CRISPR to directly examine a double-stranded target is dependent on the recognition of a specific protospacer adjacent motif (PAM) sequence (5’-NGG) by Cas9, which activates the Cas9 nuclease to unwind the dsDNA and allows the single-guide RNA (sgRNA) to invade and further evaluate the sequence complementarity.[8] With the confirmation of the sequence complementarity, an exact structural conformation formed by target-sgRNA-Cas9 results the cleavage activity on the target. However, conventional Cas9 nuclease creates a blunt cut on the dsDNA (Figure S1), which does not provide any accessibility to the sequence information of the cleaved strand and therefore hindering the utilization of target information for cascading circuit. In order to acquire molecular information from the target upon the identification by CRISPR, a Cas9 D10A mutant nickase,[9] which cleaves only the binding strand of the sgRNA, is utilized with a pair of offsets sgRNAs targeting opposite strands to transform the dsDNA structure, providing a 3’-overhang (T1) that can be accessed and utilized as a molecular input for downstream circuit (Figure 1b). In order to discriminate minute concentration change, a one-to-multiple turnover amplification is necessary for the CRISPR processed target. Instead of directly amplify the target information processed by the Cas9 D10A nickase, in order to enhance the generality of this heterogeneous circuit, a translator element is applied to translate the target sequence into an arbitrary sequence first, disconnecting the sequence information between the output and the input, which makes the amplification element generally applicable to any sequence of interest. Primer exchange reaction (PER), an autonomous mechanism for synthesis of nascent single-stranded DNA,[2, 10] is utilized as the molecular reaction pathway for translator and amplifier elements (Figure 1c). The molecular mechanism of PER on synthesis of ssDNA mainly relies on a primer, a catalytic hairpin substrate and a displacement polymerase (Figure S2). The 3’end exposed region of the hairpin substrate allows the binding of the primer. The stem region of the hairpin is a prescribed sequence for the extension of the primer. A strand-displacing polymerase is applied to copy the stem region and halted at a stop sequence before the loop region. The nascent strand is then released from the catalytic hairpin through a three-way branch migration process.[11] In order to utilize the target information processed by CRISPR as an initiation signal for downstream PER reactions, a protector (T1’-P1) is designed to cover the primer binding region (P1’) on the catalytic hairpin 1 with an exposed toehold region, which is complementary with the 3’overhang target region (T1). Therefore, only with the presence of the paired Cas9 D10A processed target, the protector can be released from the hairpin 1 through a strand displacement reaction,[12] making the primer binding region available and cascading the biomolecular reactions. Primer (P1) binds to the P1’, extended by a BST Large fragment DNA polymerase and followed by the three-branch migration, producing an elongated P1-S strand. The sequence design of S strand can be arbitrary and in our case, we design the S strand without guanine. Therefore, only dATP, dCTP and dTTP are necessary for the synthesis of this nascent strand, allowing us to simply put the stop codon as guanine at the position close to the loop region. Till now, the target information is translated and stored into an arbitrary sequence S. In order to amplify the signal of the target, a second hairpin, known as the telomerase hairpin,[2] utilizing the same sequence for primer binding region and the copying region, is applied to grow a concatemer (P1-S-…-S) with repeated sequence chained together through multiple-turnover primer exchange reaction on the same strand (Figure S3). The concatemer is the amplified product serving as the signaling strand for signal transduction based on the multiple S strands. Finally, to acquire the molecular information, a micro-fabricated electrochemical single-use sensing array (Figure S4) is applied to transduce the molecular signal into physicochemical signal through electrochemistry (Figure 1d).[13] In order to probe the molecular signal through the output elongated concatemer, a surface capture strand modified with a thiol group (SH-P1’), tethered on the gold working electrode through Au-S bond, is complementary to the primer sequence. Therefore, any primer, either going through the biochemical circuit or not, can be captured onto the sensor. A signaling probe (S’-MB), complementary to the signaling strand (S) and containing a methylene blue electrochemical tag, is further introduced into the system. Only with the presence of the signaling concatemer, multiple copies of signaling probes can be hybridized onto one concatemer generated by one copy of target originally, amplifying the electrochemical signal.
Figure 1. Modular construction of a multi-function heterogeneous biochemical reaction circuits.
a) A multi-function heterogeneous biochemical circuit constructed by paired CRISPR system and primer exchange reaction, processing genetic information and translating into electric signal. b) Guided by two offset sgRNAs, a pair of CRISPR Cas9 D10A nucleases target opposite sequence on the gene target, transforming the intact dsDNA into a 3’-overhang strand available for cascading circuit. (Blue triangle: cleavage position; Yellow box: PAM region). c) Primer exchange reaction mediated translator and amplifier. A protector gated hairpin 1 serves as a translator, only functioning with the presence of the gene target. An arbitrary sequence (S) is stored in the nascent strand (P1-S) elongated by hairpin 1. Hairpin 2 serves as an amplifier and catalyses the extension of P1-S with repetitive sequence S, forming a concatemer. d) The output of the heterogeneous biochemical circuit is examined by an electrochemical biosensing platform. A capture strand is tethered on the electrode to probe any synthesized concatemer. A signaling probe containing an electrochemical tag, complementary to the repetitive sequence S, binds to the concatemer and generates electrochemical signal.
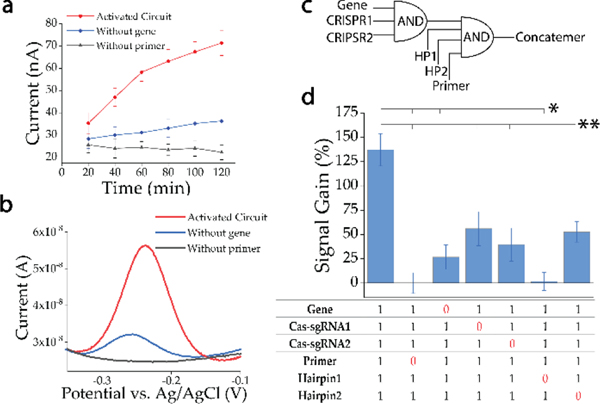
We first evaluated the feasibility of the whole biochemical circuit operation workflow through electrochemistry. The biochemical circuit was examined based on the presence or non-presence of two critical input molecules, target gene (Genebank No.LC528233.1, region 28350–28530) and primer (See Supplementary Method for details). Square wave voltammetry (SWV) was applied for electrochemical analysis after quenching the reactions by washing out the reactants from the sensor surface. After CRISPR processing, a time-dependent evaluation was conducted to select a reaction period of PER in order to generate substantial electrochemical current difference for the fully activated circuit over the two control conditions (lack of critical inputs). A clearly differentiable signal above the background and leakage signal can be identified through electrochemistry after 20 min (Figure 2a). Considering the stability of signal-to-noise ratio (signal gain) and the time-efficiency for bioanalytical application, a 1 hr reaction period was selected for further evaluation. A typical SWV signal demonstrated the current outputs of the activated circuit (red) with all components and the non-activated circuit lack of the primer (black) or the gene input (blue) at 1 hr time point (Figure 2b), indicating a good integrity and performance of the heterogeneous biochemical circuit.
Figure 2. Proof of concept evaluation of the electrochemistry transduced biochemical circuit.
a) Time-dependent electrical outputs based on activated circuit (red), negative control (without gene [blue] and without primer [black]). b) A typical square wave voltammetry (SWV) graph showing the electrochemical currents of 1) fully activated circuit (red) with primer (final concentration of 100 nM) and gene (final concentration of 50 nM); 2) non-activated circuit without gene (blue) or without primer (black). c) The Boole-an AND conjunction dependent circuit elements. CRISPR processor is operated as a 3-input AND gate. PER based translator and amplifier elements are operated as a 4-input AND gate. d) Comparison of the performance of the integrated circuit through electrochemical signal gain %= (peak current-baseline current [without primer condition])/baseline current). Each input element was investigated based on the Boolean logic. *P<0.01; **P<0.05 (signal of fully operated circuit against signal of incomplete circuit). The bar represents the mean value of three orthogonal repeats. The error bar represents ± SE.
Logic AND conjunction gate is the foundation of the circuit architecture. Hence, we further evaluated the separate functionality of each circuit element based on Boolean logic (Figure 2c). The electrical signal was normalized with the baseline signal based on the absence of the primer condition, which fully shuts down the pathway for concatemer synthesis, therefore no observable electrochemical signal of methylene blue as shown by the black line in Figure 2b. A significant signal gain was observed comparing the output signal of fully activated circuit (column 1) with that of any incomplete circuit (column 2–7) (Figure 2d). The CRISPR array based processor element is operated as a three-input AND gate. Lack of any input in this gate leads to a significant decrease of signal comparing with that of fully operated circuit. A stable leakage reaction (blue line in Figure 2b and column 3 in Figure 2d) was observed as typically found in a gene circuit (See Supplementary Discussion),[14] but the leakage resulted output signal was not statistically significant in our case. We further noticed that the signal output in which Cas-sgRNA1 (column 4) was not present was slightly higher than the signal output in which Cas-sgRNA2 was not present (column 5). We suspected that this phenomenon is due to the different cleavage site of sgRNA1 and sgRNA2. The cleavage site of sgRNA2 potentially exposes the target sequence (T1), which might increase the possibility to activate the downstream PER element, indicating the importance of accessibility of target information in a biological circuit. Afterwards, the PER based translator and amplifier elements can be integrated as a four-input AND gate. Lack of primer (column 2) fully turns off the PER reaction therefore the whole translator and amplifier are bootless. Lack of hairpin1 (column 6) results no direct sequence information connection in the circuit, removing the capability to translate the target into signaling strand. Lack of hairpin2 (column 7) leads to no formation of the concatemer, therefore removing the capability to amplify the signal. The evaluation based on the Boolean AND function firstly confirms the feasibility of this integrated construction concept and further demonstrates the integrity, functionality and modularity of this electrochemistry transduced heterogeneous multi-function biochemical circuit.
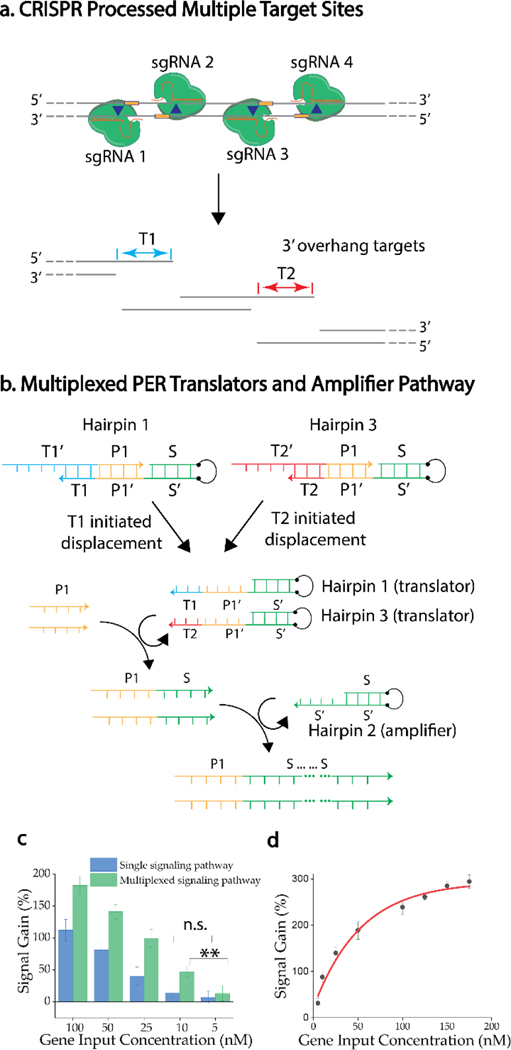
From the perspective of bioanalytical application, the proposed biochemical circuit achieves the identification and multiple-turnover amplification of target signal. We further rationalize that generating multiple defined overhangs on one target can provide multiple opportunities to amplify the target signal, increasing the signal gain. Furthermore, owing to the capability to translate the target information into a prescribed arbitrary sequence, same signaling sequence information can be used to construct the downstream electrochemical signaling scaffold, enhancing the detection resolution. To explore this conceptual possibility, another pair of CRISPR processor is introduced to process the same target. Therefore, two orthogonal 3’-overhangs are available to initiate PER cascades (Figure 3a). The design principle of multiple offset sgRNAs was applied from a previous study,[9] which demonstrated that multiple cleavage sites can produce a more effective homology directed repair (HDR). The HDR actually holds a similar molecular mechanism regarding the sequence accessibility as what we propose here. Owing to the independence of strand domains in the gated hairpin,[2] toehold design is straightforward and same primer binding sequence can be applied (Figure 3b), indicating the two overhangs can be translated into the same sequence and further amplifying through the same hairpin amplifier (hairpin2). This design significantly enhances the simplicity and requires a minimal number of DNA operators, minimizing the crosstalk possibility and network complexity. As we previously noticed in Figure 2c, sequence accessibility can be critical to the efficiency of circuit operation. We suspected that 4 cutting sites can further lead to a greater degree of accessibility of target site due to that the increased number of fragments of the gene target means a higher entropy or a decreased internal energy of each strand, promoting the strand dissociation process after cleavage. After processed by two pairs of CRISPR Cas9 D10A-sgRNAs, the same single PER circuit (as shown in Figure 2) was conducted to process the target. An increased signal output was observed for two-pair Cas9 D10A-sgRNAs processed target comparing with the signal output based on the condition in which only one pair of Cas9 D10A-sgRNAs was applied (Figure S5), indicating that accessibility of molecular input at the connection gate can be an important factor for the efficiency of biochemical circuit. The performance of this two-pair CRISPR mediated multiplexed signaling pathway was compared with previous one-pair CRISPR mediated single signaling pathway (Figure 2) based on different concentrations of the gene target (Figure 3c). A higher signal output was observed for multiplexed signaling pathway (Figure 3a and 3b) over the single signaling pathway (Figure 1a and 1b) at multiple concentrations, confirming the principle of this biochemical circuit. Moreover, multiplexed signaling pathway was capable to differentiate concentration difference of 5 nM, which could not be achieved by the single signaling pathway strategy. Furthermore, bioanalytical devices with a higher analytical resolution, indicated by the change of signal between different concentration gradients, can not only minimize the possibilities of false-positive/negative result but also potentially reveal unknown information based on minute concentration change. Therefore, we compared the change of signal between different concentration gradients between the single signaling pathway and the multiplexed signaling pathway (Figure S6). Over 7-fold increase of analytical resolution was observed for multiplexed signaling pathway over single signaling pathway, proving the resolution enhancement capability of the proposed biochemical circuit.
Figure 3. Two-pair CRISPR processed target gene multiplexed signaling pathway.
a) Four separate sgRNAs direct four Cas9 D10A nucleases to adjacent target sites, transforming the target into three fragments containing two distinct 3’overhangs accessible for molecular cascading. b) Two orthogonal translator hairpins are initiated by available target sites resulted from CRISPR processing. Target information is translated into an arbitrary sequence, which is further integrated into the same amplifier hairpin, producing a signaling concatemer. c) Comparison of signal gain between single signaling pathway and multiplexed signaling pathway based on different concentrations of gene input. Signal gain %=(peak current-baseline current[without gene condition])/baseline current). d) A dose-dependent electrochemical response of integrated biochemical circuit in a range of concentrations of gene input. The bar represents the mean value of three orthogonal repeats. The error bar represents ± SE. **P<0.05.
We further aimed to enhance the performance of the biochemical circuit in order to produce a reliable and high-fidelity analytical platform. The original study on PER demonstrated that increase of the catalytic hairpin concentration and increase of the magnesium ion (Mg2+) concentration were able to enhance the kinetics of the primer exchange reaction.[2] We tested these two aspects in our system. Firstly, while maintaining the concentration of primer (800 nM), hairpins (100 nM) and the target gene (50 nM), increasing the concentration of magnesium ion to 15 mM did increase the signal gain and provide a more stable response (Figure S7). For hairpin concentration, we first evaluated the concentration dependence of the gated hairpin (translator). While maintaining the concentration of primer (800 nM), magnesium ions (15 mM) and the target gene (50 nM), an increase of overall signal output was observed with increased concentration of each gated hairpin, but the increased concentration of gated hairpin also led to a higher background signal resulted by the reaction leakage, therefore the change of signal gain was not discernible (Figure S8). To prevent the signal leakage, we suspected that the increase of the concentration ratio of protector1 and protector3 to hairpin 1 and hairpin 3 could decrease the reaction leakage. However increased concentration of protector might also lead to less sensitivity of the analytical application, due to the direct binding of the target strand with the excess free protector in the solution instead of initiating the displacement of bound protector from the gated hairpins. In this context, a low concentration (5 nM) of gene target was used to evaluate the performance to ensure no loss of analytical resolution. A ratio of 1.2:1 of protector over hairpin was selected with decreased leakage signal while maintaining the detection resolution, resulting a higher signal gain (Figure S9). Finally, the effect of concentration of the telomerase hairpin amplifier was evaluated based on the target concentration at 5 nM and an optimized signal gain was identified at a concentration of 250 nM (Figure S10). The property of this hairpin is amplifying signal obtained from the translator hairpins, so it amplifies both the leaky signal and the specific signal, therefore it does not contribute significantly to the normalized signal gain. Based on the optimized experimental condition, a dose-dependent signal response was evaluated in a range of target concentrations (5–200 nM) within a total turnaround time around 1.5 hr (Figure 3d). A plateau was found when the gene concentration was over 150 nM, which might be due to the limited concentration of CRISPR processors in the system. An experimental detection limit was identified around 5 nM (Figure S11), which is a sufficient level for analytical platform to analyze sample after enzymatic amplification treatment. Furthermore, to explore the potential of this sensing strategy, we tried to evaluate the interference of point mutation at different CRISPR targeting region on the signal gain. CRISPR Cas9 has shown outstanding selectivity toward mutations at its PAM region.[15] Therefore, based on our multiple target sites, mutations inside four different PAM regions were designed to evaluate the sensing performance of our biochemical circuit. Signal generated by 50 nM of mutated targets were evaluated and compared with that of wild type target. All mutations demonstrated significant decreases of signal gain comparing with that of wild type (Figure S12). We also observed that single mutation at PAM 2 of sgRNA 2 and PAM 4 of sgRNA 4 contributed greater decrease of signal gain comparing with PAMs of sgRNA1 and sgRNA3. These observations first proved that for the design of biosensing system to differentiate point mutation, recognizing a mutation in the PAM region is a reliable strategy. Specifically, for our paired CRISPR array, mutations in the PAM regions of the overhang strand (5’–3’) inhibit Cas9 recognition and cleavage activities toward the 5’–3’ strand, therefore limiting the exposure of the overhanging region to the following DNA circuit. Without the exposed target information, the molecular cue to initiate of the PER cascade is lost, diminishing the overall signal gain.
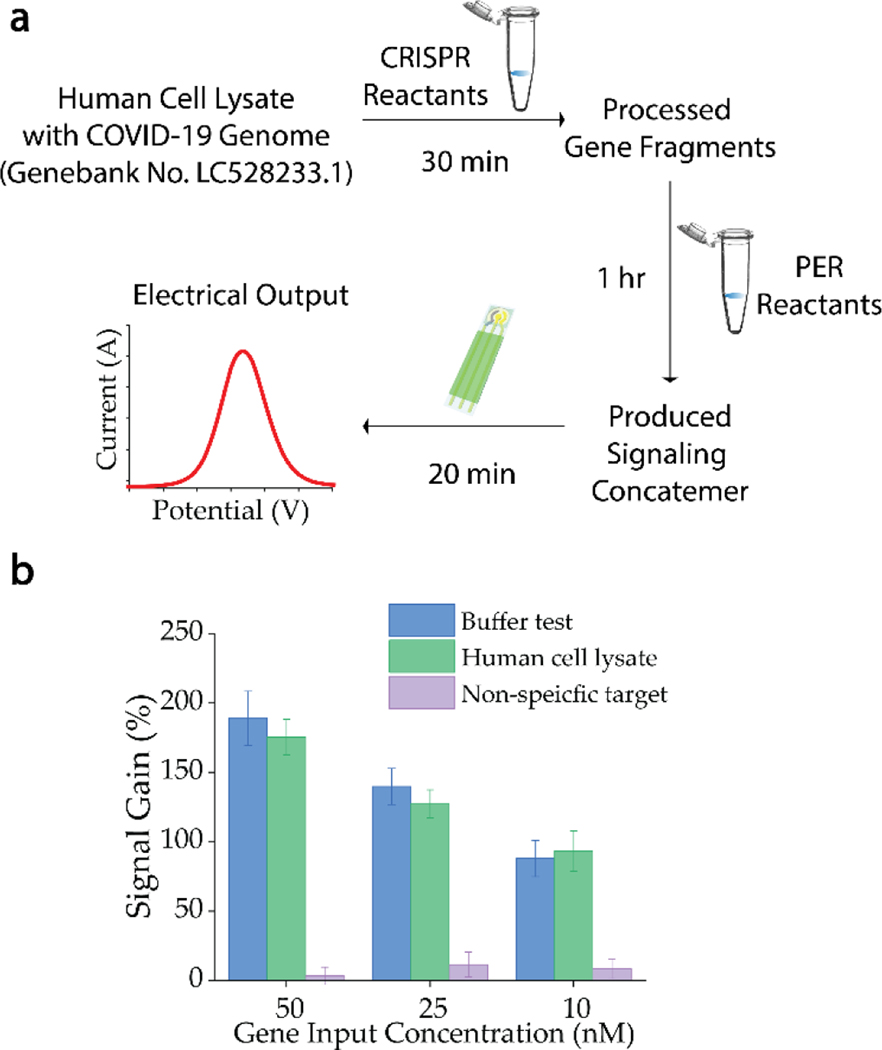
To demonstrate the potential of this electrochemistry transduced integrated heterogeneous biochemical circuit on a realistic bioanalytical application, we challenged this integrated platform with human cell lysates spiked with the synthetic genome fragment of SARS-CoV-2. Owing to the modularity nature of the biochemical circuit (Figure 4a), the reactants can be divided into two separate tubes and stored in −20°C before usage. Also, the stability of the surface modified electrochemical sensor has also been verified suitable for long-term dry storage.[13] These facts suggested that the combination of plug-and-play molecular components and a simple electrochemical system can be a potential platform for point-of-care diagnosis. Three different concentrations (50 nM, 25 nM and 10 nM) of synthetic COVID-19 genome fragment were prepared in human cell lysate. Comparable signals based on tests in human cell lysates with different gene concentrations were observed with previous buffer tests (Figure 3d), confirming the potential of the developed platform for real sample analysis. Also, the interference studies based on a non-specific target (purple bars), the genome sequence of human papillomavirus 16, confirmed the specificity of this biochemical circuit. (See Supplementary Discussion on comparison with other CRISPR based biosensing systems) In conclusion, we have demonstrated a conceptual capability of an integrated multi-function heterogeneous circuit for electrochemistry based genetic analysis. The biochemical circuit is constructed through paired CRISPR Cas9 D10A-sgRNAs mediated primer exchange reaction cascades, which can identify, transform, translate and amplify biological signals into physiochemical signals based on Boolean logic. The programmability and modularity of the biochemical circuit design allow the versatile extension of this concept for various biotechnological and biomedical applications. Combination of upstream biochemical circuit with downstream electrochemical sensor integrates the modularity and programmability of biochemical circuit design and the simplicity and cost-effectiveness of electrical transduction, delivering a powerful plug-and-play bioanalytical platform.
Figure 4. A modular bioanalytical strategy.
a) The modularity nature of the biochemical circuit allows simple construction of individual processes for sample analysis, transforming biomolecule input into electrical output within 2 hr. b) Evaluation of the matrix effect of the bioanalytical platform with spiked samples in human cell lysate (green) proved the capability of the platform on complex sample analysis. Interference evaluation based on non-specific gene target (HPV-16) demonstrated the reliable selectivity of the CRISPR mediated recognition process.
Supplementary Material
Acknowledgements
The authors would like to acknowledge the experimental supports provided by Electronics Design Center. We acknowledge the funding supports from National Institute of Health under the grant number, NIH/NIBIB 1P41EB021911. We also gratefully acknowledge Xintong Cao for her generous help on the artwork.
References
- [1].a Li J, Green AA, Yan H, Fan C, Nature Chemistry 2017, 9, 1056–1067; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Woods D, Doty D, Myhrvold C, Hui J, Zhou F, Yin P, Winfree E, Nature 2019, 567, 366–372; [DOI] [PubMed] [Google Scholar]; c Cherry KM, Qian L, Nature 2018, 559, 370–376; [DOI] [PubMed] [Google Scholar]; d Hanewich-Hollatz MH, Chen Z, Hochrein LM, Huang J, Pierce NA, ACS Central Science 2019, 5, 1241–1249; [DOI] [PMC free article] [PubMed] [Google Scholar]; e Mao C, LaBean TH, Reif JH, Seeman NC, Nature 2000, 407, 493–496. [DOI] [PubMed] [Google Scholar]
- [2].Kishi JY, Schaus TE, Gopalkrishnan N, Xuan F, Yin P, Nature Chemistry 2018, 10, 155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sadat Mousavi P, Smith SJ, Chen JB, Karlikow M, Tinafar A, Robinson C, Liu W, Ma D, Green AA, Kelley SO, Pardee K, Nature Chemistry 2020, 12, 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a Kelley SO, Mirkin CA, Walt DR, Ismagilov RF, Toner M, Sargent EH, Nature nanotechnology 2014, 9, 969; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Tan W, Li L, Xu S, Yan H, Li X, Yazd HS, Li X, Huang T, Cui C, Jiang J, Angewandte Chemie International Edition, n/a; [Google Scholar]; c Samanta D, Ebrahimi SB, Mirkin CA, Advanced Materials 2020, 32, 1901743; [DOI] [PMC free article] [PubMed] [Google Scholar]; d Yang F, Li Q, Wang L, Zhang G-J, Fan C, ACS Sensors 2018, 3, 903–919; [DOI] [PubMed] [Google Scholar]; e Furst AL, Muren NB, Hill MG, Barton JK, Proceedings of the National Academy of Sciences 2014, 111, 14985–14989; [DOI] [PMC free article] [PubMed] [Google Scholar]; f Kim J, Campbell AS, de Ávila BE-F, Wang J, Nature Biotechnology 2019, 37, 389–406; [DOI] [PMC free article] [PubMed] [Google Scholar]; g Yang Y, Gao W, Chemical Society Reviews 2019, 48, 1465–1491; [DOI] [PubMed] [Google Scholar]; h Bariya M, Nyein HYY, Javey A, Nature Electronics 2018, 1, 160–171; [Google Scholar]; I Gao W, Zdrachek E, Xie X, Bakker E, Angewandte Chemie International Edition 2020, 59, 2294–2298; [DOI] [PubMed] [Google Scholar]; j Ranallo S, Rossetti M, Plaxco KW, Vallée-Bélisle A, Ricci F, Angewandte Chemie International Edition 2015, 54, 13214–13218; [DOI] [PMC free article] [PubMed] [Google Scholar]; k Dai Y, Liu CC, Angewandte Chemie International Edition 2019, 58, 12355–12368; [DOI] [PubMed] [Google Scholar]; l Rossetti M, Brannetti S, Mocenigo M, Marini B, Ippodrino R, Porchetta A, Angewandte Chemie International Edition 2020, 59, 14973–14978. [DOI] [PubMed] [Google Scholar]
- [5].Wu Y, Tilley RD, Gooding JJ, Journal of the American Chemical Society 2019, 141, 1162–1170. [DOI] [PubMed] [Google Scholar]
- [6].Young BE, Ong SWX, Kalimuddin S, Low JG, Tan SY, Loh J, Ng O-T, Marimuthu K, Ang LW, Mak TM, Jama 2020, 323, 1488–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].aHsu PD, Lander ES, Zhang F, Cell 2014, 157, 1262–1278; [DOI] [PMC free article] [PubMed] [Google Scholar]; bJinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E, science 2012, 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chen JS, Doudna JA, Nature Reviews Chemistry 2017, 1, 0078. [Google Scholar]
- [9].Ran FA, Hsu Patrick D, Lin C-Y, Gootenberg Jonathan S, Konermann S, Trevino AE, Scott David A, Inoue A, Matoba S, Zhang Y, Zhang F, Cell 2013, 154, 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].a Kishi JY, Lapan SW, Beliveau BJ, West ER, Zhu A, Sasaki HM, Saka SK, Wang Y, Cepko CL, Yin P, Nature Methods 2019, 16, 533–544; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Saka SK, Wang Y, Kishi JY, Zhu A, Zeng Y, Xie W, Kirli K, Yapp C, Cicconet M, Beliveau BJ, Lapan SW, Yin S, Lin M, Boyden ES, Kaeser PS, Pihan G, Church GM, Yin P, Nature Biotechnology 2019, 37, 1080–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Panyutin IG, Hsieh P, Proceedings of the National Academy of Sciences 1994, 91, 2021–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dai Y, Furst A, Liu CC, Trends in Biotechnology 2019, 37, 1367–1382. [DOI] [PubMed] [Google Scholar]
- [13].Dai Y, Somoza RA, Wang L, Welter JF, Li Y, Caplan AI, Liu CC, Angewandte Chemie International Edition 2019, 58, 17399–17405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].a Brophy JAN, Voigt CA, Nature Methods 2014, 11, 508–520; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Shah S, Wee J, Song T, Ceze L, Strauss K, Chen Y-JJ, Reif J, Journal of the American Chemical Society 2020. 10.1021/jacs.0c02240; [DOI] [PubMed] [Google Scholar]; c Chen Y-J, Dalchau N, Srinivas N, Phillips A, Cardelli L, Soloveichik D, Seelig G, Nature Nanotechnology 2013, 8, 755–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dai Y, Wu Y, Liu G, Gooding JJ, Angewandte Chemie International Edition. 10.1002/anie.202005398 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.