Short abstract
Content available: Author Interview and Audio Recording
Watch the interview with the author.
Listen to an audio presentation of this article.
INTRODUCTION
Hepatocellular carcinoma (HCC) is the third leading cause of cancer‐related death worldwide. Its poor prognosis is partly driven by frequent late‐stage detection due to suboptimal sensitivity of current surveillance tests—abdominal ultrasound and alpha fetoprotein—which miss over one‐third of HCC cases at an early stage. 1 Ultrasound‐based surveillance is particularly subject to failure in obese patients and those with non‐viral etiologies of liver disease, highlighting a clear need for alternative strategies. 2
In addition to potential protein biomarkers, there has been increased interest in liquid biopsy techniques, which involve the analysis of cancer products in the bloodstream such as circulating tumor cells (CTCs), extracellular vesicles (EVs), and circulating nucleic acids (cell‐free DNA [cfDNA] or RNA) (Figure 1). Circulating tumor cells are cancer cells that have been shed from the tumor, contributing to metastatic disease. EVs are small membrane particles that are released by cells to transport bioactive molecules and circulating nucleic acids are typically released into the blood by apoptotic tumor cells (Table 1). The Food and Drug Administration has approved the use of liquid biopsy techniques for diagnostic workup of non‐liver cancers, including breast and lung 3 ; however, none are approved for HCC surveillance, diagnosis, or prognostication. Liquid biopsy for HCC is currently reserved for research purposes, and further validation data are needed prior to incorporation into routine clinical practice. Liquid biopsy has several advantages, including its non‐invasive nature without risk of tumor seeding, representation of tumor cell heterogeneity, and ease of serial assessments over time to monitor disease status. However, use of liquid biopsy for early detection has historically been difficult given CTCs and circulating tumor DNA (ctDNA) are present in circulation in very low concentrations, particularly when tumor burden is low, requiring highly sensitive isolation procedures to maximize signal‐to‐noise ratio. Beyond applications for early HCC detection, liquid biopsy has also been evaluated for several other purposes, including to facilitate better HCC prognostication, molecularly characterize tumors to guide treatment selection, evaluate therapeutic response, and monitor for disease recurrence. Herein, we review the potential utility of liquid biopsy in clinical practice and available supporting data.
FIGURE 1.
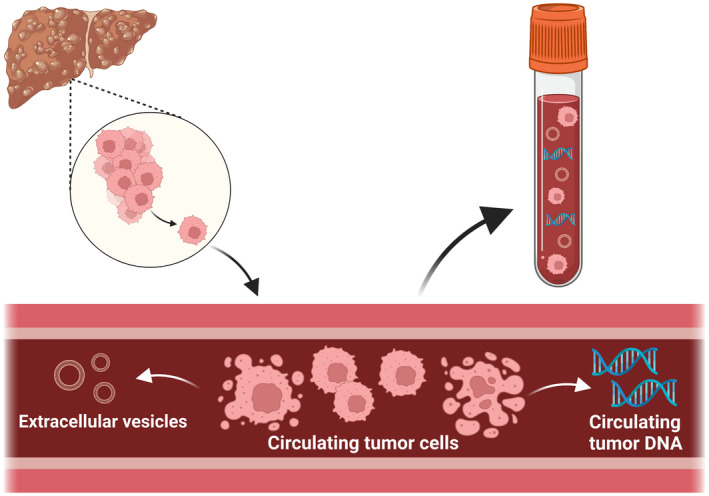
Liquid biopsy strategies for sampling tumor products in the bloodstream. Cite: Created with BioRender.com
TABLE 1.
Liquid biopsy technologies
| Biomarker | Description | Isolation techniques |
|---|---|---|
| Circulating tumor cells (CTCs) | Cells released from sites of primary and metastatic cancer into the bloodstream |
|
| Cell‐free DNA (cfDNA) | Nucleic acid shed into circulation during cellular necrosis or apoptosis; a subset of cfDNA is derived from tumor cells, i.e. circulating tumor DNA (ctDNA) | Centrifugation for removal of cells, followed by plasma extraction of cfDNA |
| Extracellular vesicles (EVs) | Cell membrane‐derived particles, including apoptotic bodies, microvesicles and exosomes, that contain molecular cargoes specific to the origin cell and have important roles in cell‐to‐cell communication | Ultracentrifugation, which isolates EVs based on size and density |
LIQUID BIOPSY FOR EARLY DETECTION OF HCC
A recent systematic review identified 67 studies evaluating liquid biopsy techniques for early‐stage HCC detection, including 17 studies evaluating CTCs, 39 cfDNA studies, and 11 studies investigating EVs. 4 Overall, studies reported that CTCs, cfDNA, and EVs each appeared promising for HCC detection, with higher accuracy than alpha‐fetoprotein (AFP) for distinguishing patients with HCC from controls, capacity to identify AFP‐negative HCC patients, and combinations with AFP being superior to AFP alone. For example, Guo and colleagues found that a CTC‐derived PCR score had greater accuracy than AFP ≥20 ng/mL for early‐stage HCC detection compared to a control group of patients with chronic liver disease or benign hepatic lesions (area under the curve [AUC] 0.93 vs. 0.74). 5
A subsequent large national case–control study evaluated a cfDNA panel including three methylation biomarkers, AFP, and sex in a validation cohort of 156 cases (50% early‐stage) and 245 chronic liver disease controls (>90% with cirrhosis) and found 87% specificity and 88% sensitivity for any‐stage HCC detection. Early‐stage HCC sensitivity of the cfDNA panel was significantly higher than AFP and the GALAD panel (82% vs. 40% and 71%, respectively), although AFP and GALAD both had higher specificities. 6 A subsequent multi‐center case–control study similarly evaluated a methylated DNA panel and found superior sensitivity for early‐stage HCC compared to AFP and GALAD (76% vs. 57% and 65%, respectively) albeit with lower specificities (91% vs. 97% and 94%, respectively). 7 Based on these promising data, both panels are currently undergoing prospective validation.
LIQUID BIOPSY TECHNIQUES FOR HCC PROGNOSTICATION AND DISEASE MONITORING
The prior systematic review of liquid biopsy techniques for HCC identified 21 studies evaluating CTCs, 15 cfDNA studies, and 10 studies investigating EVs for HCC prognosis. Studies reported a significant association between presence of pre‐operative CTCs and tumor recurrence, recurrence‐free survival, and overall survival. In one of the larger included studies, Guo et al. found a CTC‐derived PCR score was associated with shorter time to recurrence (hazard ratio [HR], 3.13; 95% confidence interval [CI], 1.36–7.19) after adjusting for tumor size, satellite lesions, and vascular invasion. 5 Thirteen studies evaluating CTCs for therapeutic response found patients with persistently high or increased CTC counts after therapy had poorer prognosis than patients with persistently low or decreased CTC levels.
Similarly, higher cfDNA levels and presence of EVs were both associated with poorer overall survival and tumor progression, although there was marked heterogeneity in cfDNA and EVs of interest. In one of the largest cfDNA studies to date, Xu and colleagues found a methylated DNA panel with eight genes (SH3PXD2A, C11orf9, PPFIA1, SERPINB5, NOTCH3, GRHL2, TMEM8B, and chromosome 17:78) was associated with overall survival (HR, 1.55; 95%CI, 1.25–1.92) after adjusting for age, sex, TNM stage, and AFP in a validation cohort of 369 HCC patients. 8 A prognostic model that combined the methylation DNA panel and TNM staging predicted survival with greater accuracy than TNM staging alone (AUC 0.76 vs. 0.69).
STUDY LIMITATIONS AND FUTURE DIRECTIONS
Although available data highlight the promise of liquid biopsy techniques, current phase II biomarker studies have notable limitations that must be addressed in future phase III and IV biomarker cohort studies prior to use in clinical practice. 9 First, there is marked clinical heterogeneity across studies, precluding pooled estimates of biomarker accuracy. For example, the most common method to detect CTCs was selection of epithelial markers (e.g., epithelial cell adhesion molecule); however, some studies used alternative methods, including Canpatrol, which can identify both epithelial and mesenchymal phenotypes. This distinction may be particularly important for prognostic studies given epithelial‐to‐mesenchymal transition is an important mechanism by which CTCs disseminate from the primary tumor, so a higher proportion of mesenchymal CTCs may portend a negative outcome. Indeed, Qi and colleagues found a higher proportion of mesenchymal CTCs was an independent predictor of postoperative HCC recurrence, even after adjusting for total CTCs. 10 Similarly, studies evaluated different cfDNA aspects including total amount, specific mutations, or methylation patterns, and studies evaluating EVs similarly had marked variation in EV properties of interest. Other limitations in study quality that must be addressed in future studies were inclusion of patients with advanced‐stage tumors and use of healthy controls that do not reflect the at‐risk population. Therefore, altered cfDNA or EV findings could have reflected tumor status or changes from background cirrhosis and current data may overestimate test performance.
CONCLUSION
Liquid biopsy techniques remain an exciting avenue of research to facilitate precision medicine in patients with HCC, with promising early data in several areas from early HCC detection to prognostication and therapeutic monitoring.
CONFLICT OF INTEREST
AGS consults for Exact Sciences, FujiFilm Medical Sciences, Glycotest, GRAIL, Bayer, and Roche.
Arvind A, Singal AG. Emerging liquid biopsy techniques for early detection of hepatocellular carcinoma, prognostication, and disease monitoring. Clin Liver Dis. 2022;20:18–20. 10.1002/cld.1232
REFERENCES
- 1. Tzartzeva K, Obi J, Rich NE, Parikh ND, Marrero JA, Yopp A, et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta‐analysis. Gastroenterology. 2018;154:1706–18.e1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chong N, Schoenberger H, Yekkaluri S, Fetzer DT, Rich NE, Yokoo T, et al. Association between ultrasound quality and test performance for HCC surveillance in patients with cirrhosis: a retrospective cohort study. Aliment Pharmacol Ther. 2022;55:683–90. [DOI] [PubMed] [Google Scholar]
- 3. Woodhouse R, Li M, Hughes J, Delfosse D, Skoletsky J, Ma P, et al. Clinical and analytical validation of FoundationOne liquid CDx, a novel 324‐gene cfDNA‐based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One. 2020;15:e0237802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen VL, Xu D, Wicha MS, Lok AS, Parikh ND. Utility of liquid biopsy analysis in detection of hepatocellular carcinoma, determination of prognosis, and disease monitoring: a systematic review. Clin Gastroenterol Hepatol. 2020;18:2879–902.e2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guo W, Sun YF, Shen MN, Ma XL, Wu J, Zhang CY, et al. Circulating tumor cells with stem‐like phenotypes for diagnosis, prognosis, and therapeutic response evaluation in hepatocellular carcinoma. Clin Cancer Res. 2018;24:2203–13. [DOI] [PubMed] [Google Scholar]
- 6. Chalasani NP, Porter K, Bhattacharya A, Book AJ, Neis BM, Xiong KM, et al. Validation of a novel multitarget blood test shows high sensitivity to detect early stage hepatocellular carcinoma. Clin Gastroenterol Hepatol. 2022;20:173–82.e177. [DOI] [PubMed] [Google Scholar]
- 7. Lin N, Lin Y, Xu J, Liu D, Li D, Meng H, et al. A multi‐analyte cell‐free DNA‐based blood test for early detection of hepatocellular carcinoma. Hepatol Commun. 2022. 10.1002/hep4.1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu RH, Wei W, Krawczyk M, Wang W, Luo H, Flagg K, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017;16:1155–61. [DOI] [PubMed] [Google Scholar]
- 9. Singal AG, Hoshida Y, Pinato DJ, Marrero J, Nault JC, Paradis V, et al. International liver cancer association (ILCA) white paper on biomarker development for hepatocellular carcinoma. Gastroenterology. 2021;160:2572–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qi LN, Xiang BD, Wu FX, Ye JZ, Zhong JH, Wang YY, et al. Circulating tumor cells undergoing EMT provide a metric for diagnosis and prognosis of patients with hepatocellular carcinoma. Cancer Res. 2018;78:4731–44. [DOI] [PubMed] [Google Scholar]


