Abstract
Hydrogen sulfide (H2S) is a gasotransmitter known to regulate physiological and pathological processes. Abnormal H2S levels have been associated with a range of conditions, including Parkinson's and Alzheimer's diseases, cardiovascular and renal diseases, bacterial and viral infections, as well as cancer. Therefore, fast and sensitive H2S detection is of significant clinical importance. Fluorescent H2S probes hold great potential among the currently developed detection methods because of their high sensitivity, selectivity, and biocompatibility. However, many proposed probes do not provide a gold standard for proper use and selection. Consequently, issues arise when applying the probes in different conditions. Therefore, we systematically evaluated four commercially available probes (WSP‐1, WSP‐5, CAY, and P3), considering their detection range, sensitivity, selectivity, and performance in different environments. Furthermore, their capacity for endogenous H2S imaging in live cells was demonstrated.
Keywords: cell imaging, fluorescent probes, hydrogen sulfide (H2S), imaging agents
Four commercially available fluorescent probes (WSP‐1, WSP‐5, CAY, and P3) for H2S detection and imaging are systematically evaluated in terms of their sensitivity, selectivity, detection range, and performance in different environmental conditions. Their capacity for endogenous H2S sensing in live cells is also demonstrated.
1. Introduction
Hydrogen sulfide (H2S) is a pungent and flammable colorless gas historically regarded as a highly toxic pollutant.[ 1 , 2 ] It was not until the late 1990s that it was discovered to be an important signaling molecule, [3] joining the family of endogenous transmitters (i. e. gasotransmitters), [4] alongside NO (nitric oxide) and CO (carbon monoxide).[ 5 , 6 ] Being an abundant physiological gas mediator, H2S molecules freely permeate through cell membranes without the need for specific carriers. [7] It has been reported to be present in the blood, plasma, gut, and brain in concentrations ranging from nM to μM.[ 8 , 9 , 10 ] H2S has been shown to take part in various biological functions, some of which include cardiovascular,[ 11 , 12 ] gastrointestinal, [13] circulatory, [14] and anti‐inflammation [15] functions, as well as the immune response [16] and central nervous system. [17] Over the past two decades, several studies have linked H2S levels within the human body with numerous diseases, including Parkinson's [18] and Alzheimer's diseases, [19] cancer, [20] Down syndrome, [21] and diabetes. [22] Consequently, advanced materials and platforms based on H2S delivery have been developed for therapeutic and clinical purposes.[ 23 , 24 , 25 ] The increase in H2S‐based treatments[ 26 , 27 ] has driven the development of fast, sensitive, and accurate detection platforms towards this gasotransmitter.[ 28 , 29 ] However, this field faces many challenges primarily associated with the high nucleophilicity, volatility, and redox activity of H2S, allowing it to react with biological targets readily. [30] Moreover, endogenous H2S detection in physiological environments can be hampered by biothiols (e. g. cysteine, glutathione), plasma, proteins, serum, and ions. [31] These limitations have significantly hindered H2S detection models, specifically towards achieving endogenous, real‐time sensing.
Conventional methods for H2S detection include colorimetric (e. g. methylene blue), [32] electrochemical (e. g. H2S ion‐selective electrodes), [33] chromatographic (e. g. gas chromatography (GC) or high‐performance liquid chromatography (HPLC)), [34] and fluorescence‐based methods (e. g. fluorescent dye).[ 35 , 36 , 37 ] Additionally, nanomaterial‐based probes have also recently emerged as a new family for H2S sensing and imaging.[ 38 , 39 ] All these approaches show benefits and limitations against one another. For instance, colorimetric methods are more user‐friendly but have shown low sensitivities, [32] whilst electrochemical methods have been reported to have higher sensitivities but require routine maintenance and long equilibration times. [33] Chromatographic methods have been reported to be highly accurate but expensive and complex. [40] Fluorescence‐based methods have been shown to be biocompatible and sensitive, but prone to false‐positive results due to lack of specificity. [35] Lastly, synthetic nanomaterial‐based probes benefit from visible read‐outs, high photostability, and can perform synergistic sensing and drug delivery. However, potential toxicity concerns when applied at high concentrations need to be considered, whilst interference issues can also lead to inaccurate results. [41] Each detection approach has applications for which they are more suited towards. Among these, fluorescence‐based strategies are the most used for in vitro and in vivo applications, allowing for real‐time, sensitive, and simple H2S detection.[ 35 , 36 , 42 ] To date, numerous fluorescent H2S probes have been developed that rely on different mechanisms, [42] including reduction‐based,[ 43 , 44 , 45 ] nucleophilicity‐based,[ 46 , 47 ] and metal sulfide precipitation‐based probes.[ 48 , 49 ] These detection modes take advantage of H2S′s active reducibility, high nucleophilicity, and strong metal coordination, respectively. Reduction‐based probes center on nitrogen atom redox states, mainly targeting azide,[ 43 , 50 ] nitro,[ 44 , 51 ] hydroxamino, or azo groups.[ 45 , 52 ] H2S nucleophilicity‐based probes rely on a specific interaction between the probe and H2S, such as an H2S‐induced ester group[ 46 , 53 , 54 ] or S−S bond [47] cleavage, followed by an intramolecular nucleophilic or Michael addition reaction. Finally, metal sulfide precipitation‐based probes focus on copper sulfide (CuS) precipitation after the reaction between Cu (II) and S2−.[ 48 , 49 ] Regardless of the detection mechanism, the specific optimized conditions employed by each author raise issues when applying the same probe in different environments (e. g. buffers, pH, temperature). This can hinder researchers in choosing an appropriate probe among the many options available for their specific applications.
Herein, we provide a systematic study of four commercially available fluorescent probes (i. e. WSP‐1, WSP‐5, CAY, and P3) most accessible to researchers, focusing on their sensitivity and selectivity towards H2S. The probes are investigated in several complex environments involving biothiols, bioanalytes, and nanoparticles. Detailed instructions and best practices for using these probes to achieve optimal performance will also be discussed.
2. Materials and methods
2.1. Materials
Sodium sulfide nonahydrate (Na2S ⋅ 9H2O, 98%), L‐Cysteine (Cys, 98.5%), L‐Glutathione reduced (GSH, 98%), zinc nitrate hexahydrate (Zn(NO3)2 ⋅ 6H2O, 98%), 2‐methylimidazole (HmIm, 99%), zirconium (IV) chloride (ZrCl4, 99.5%), terephthalic acid (BDC, 98%), glycine (Gly, 98.5%), lipopolysaccharides (LPS), phosphate‐buffered saline (PBS), hexadecyltrimethylammonium bromide (CTAB), gold nanoparticles (50 nm, OD=1), silver nanoparticles (40 nm, OD=1), dimethyl sulfoxide (DMSO, ≥99.9%) and H2S fluorescent probe (E)‐2‐(3‐(6‐(2‐hydroxyethylamino)naphthalen‐2‐yl)‐3‐oxoprop‐1‐enyl)‐3,5‐dimethoxybenzaldehyde (P3) were purchased from Sigma Aldrich. Diallyl Trisulfide (DATs), S‐Nitroso‐L‐glutathione (GSNO), H2S fluorescent probes 3′‐methoxy‐3‐oxo‐3H‐spiro[isobenzofuran‐1,9′‐xanthen]‐6′‐yl 2‐(pyridin‐2‐yldisulfanyl)benzoate (Washington State Probe‐1, WSP‐1), 3‐oxo‐3H‐spiro[isobenzofuran‐1,9′‐xanthene]‐3′,6′‐diyl bis(2‐(pyridin‐2‐yldisulfanyl)benzoate) (Washington State Probe‐5, WSP‐5), and 2‐bromoethyl 6′‐methoxy‐3‐oxospiro[isobenzofuran‐1(3H),9′‐[9H]xanthen]‐3′‐yl ester (CAY10731, CAY) were purchased from Cayman Chemical. Dimethylformamide (DMF) and hydrochloric acid solution (HCl, 1 M) were purchased from ACI Labscan. Hydrogen peroxide (H2O2, 30% w/w) and methanol were purchased from Chem Supply. Ultrapure water (DI water, 18.2 ΩM cm resistance) was provided by arium® mini Sartorius and used throughout the experiments. All chemicals were received as reagent grade and used without further purification.
2.2. Time‐dependent H2S detection
H2S detection was carried out using Na2S or DATs as the H2S donor. All experiments were carried out using black, clear‐bottom 96‐well plates in a final volume of 200 μL. PBS containing 1 mM CTAB was used for all experiments unless otherwise specified. CTAB is a common surfactant that enhances the probe solubility and brings the reactants closer together, thus achieving higher fluorescence intensity as previously noted.[ 46 , 55 , 56 ] The fluorescence intensity was measured every 30 s using a SpectraMax M5 microplate reader at room temperature (or 37 °C where specified). WSP‐1: λex/em=465/515 nm; WSP‐5: λex/em=502/525 nm; CAY: λex/em=485/535 nm; and P3: λex/em=375/505 nm.
2.2.1. Sensitivity
The sensitivity towards H2S was evaluated by mixing each probe (10 μM) with increasing Na2S concentrations (0–10,000 μM). The fluorescence intensity was measured every 30 s for 30 min.
2.2.2. Selectivity
The selectivity of the probes towards biothiols, bioanalytes, or nanoparticles was evaluated by mixing each probe (10 μM) with biothiols (0–2,000 μM), bioanalytes (2 mM), or nanoparticles (0.1 mg mL−1 or OD=1). The capacity of each probe to detect H2S in complex biothiol solutions was evaluated by mixing each probe (10 μM) with specific Na2S concentrations (outlined within the respective section), and adding equimolar concentrations of biothiols at the beginning (t=0) or after 5 min (t=5 min). This was also similarly carried out for nanoparticle‐involved solutions.
2.2.3. pH effect
The pH of the detection buffer (PBS with 1 mM CTAB) was adjusted using HCl or NaOH in a range from 4–10. To evaluate the effect of pH on H2S detection, Na2S with specific concentrations (outlined within the respective section) were dissolved in buffers with different pH values, followed by the addition of each probe (10 μM). The fluorescence intensity was measured every 30 s for 30 min.
2.2.4. H2S detection by a plant‐derived H2S donor
H2S detection using a plant‐derived donor was carried out by mixing WSP‐1 or WSP‐5 probes (10 μM) with DATs (0–2,000 μM) and biothiols (500 μM Cys or GSH). The fluorescence intensity was measured every 30 s for 30 min.
2.3. Synthesis of ZIF‐8 and UiO‐66
ZIF‐8 particles were prepared according to our previous publication. [57] Briefly, freshly prepared HmIm (8 mmol, in 11.3 mL methanol) was added into an equal volume of Zn(NO3)2 ⋅ 6H2O solution (1 mmol in methanol). The mixture was left to react at ambient temperature for 1 h. Then, the particles were washed by centrifugation at 5000 rpm three times with methanol, followed by three times with DI water, and dried at 60 °C overnight. UiO‐66 particles were prepared according to a published protocol with a minor modification. [58] Briefly, an equimolar solution of ZrCl4 (2 mmol) and BDC (2 mmol) were dissolved in DMF (15 mL) with the addition of 1 mL HCl (1 M). The well‐mixed solution was transferred into a preheated Teflon autoclave and heated at 120 °C for 21 h. The UiO‐66 pellets were washed by centrifugation at 5000 rpm three times with DMF, followed by three times with methanol, and dried at 60 °C overnight.
2.4. Characterization of ZIF‐8 and UiO‐66
X‐ray diffraction (XRD, PANalytical Empyrean X‐ray Diffractometer) was used to determine the crystal structures of the synthesized particles at 40 kV and 40 mA with a Cu−Kα radiation source (λ=0.154 nm). The XRD patterns confirmed the successful synthesis of ZIF‐8 and UiO‐66 (Figure S1).
2.5. Cell lines and cell cultures
RAW 264.7 cells and HeLa cells from America Type Culture Collection (ATCC) were used for endogenous H2S imaging. Both cells were routinely cultured in Dulbecco's Modified Eagle medium (DMEM, Life Technologies) supplemented with 10% (v/v) Fetal Bovine Serum (FBS, Sigma Aldrich) and 0.25 mg mL−1 L‐Glutamine (Sigma Aldrich). Cells were cultured at 37 °C in a 5% CO2 humidified incubator and subcultured when they reached 80% confluency.
2.6. Imaging H2S in living cells
RAW 264.7 cells and HeLa cells (no more than 30 passages) were seeded on 13 mm coverslips in 12‐well plates (1×105 cells/well) in 1 mL DMEM (10% FBS) and allowed to adhere overnight. To study the endogenous H2S generation by cells, four different groups were evaluated: 1) cells only (control group); 2) cells treated with 2 μg mL−1 LPS for 24 h; 3) cells treated with 10 μM probes (1 h probe treatment in DMEM for RAW 264.7 cells, and 30 min probe treatment in PBS containing 0.1 mM CTAB for HeLa cells); and 4) cells pretreated with 2 μg mL−1 LPS for 24 h followed by probe incubation (10 μM, 1 h probe treatment in DMEM for RAW 264.7 cells, and 30 min probe treatment in PBS containing 0.1 mM CTAB for HeLa cells).
2.7. Relative fluorescence intensity analysis
The relative fluorescence intensity (RFI) was calculated using the following equation: RFI=F‐A×F0 . Where F represents the integrated density, A represents the area of the selected cell, and F0 represents the mean fluorescence of background readings. The relative cell intensity was calculated from 5 cells in each image.
2.8. Statistical analysis
All data are presented as mean ± standard deviation, with ≥3 independent replicates. The limit of detection (LOD) was calculated as LOD=3σ/S, [54] where σ represents the standard deviation of the blank measurement, and S represents the linear equation slope.
3. Results and discussion
3.1. Probe Mechanism
A wide range of probes have been developed based on nucleophilic attack reactions.[ 35 , 42 ] This is because H2S possesses high nucleophilicity due to its small size and a low pKa value. [59] As such, in this study, we selected representative probes which employ this mechanism, specifically WSP‐1, WSP‐5, CAY, and P3. The full structures of these four probes are shown in Scheme S1. The nucleophilic attack mechanisms of these four probes can be further divided into two groups. The first is based on a nucleophilic substitution reaction (Scheme 1a). These probes contain an electrophile (H2S trapping group; blue rectangle) and a fluorophore molecule or scaffold (fluorescence signal transducer). The electrophile serves as an electron acceptor to quench the fluorophore prior to interacting with H2S. Upon H2S addition, the electrophile is substituted by dissolved H2S, followed by nucleophilic substitution and an intramolecular cyclization reaction. After cyclization, to achieve a fluorescent signal, the probes can either release the fluorophore (Scheme 1ai; WSP‐1 and WSP‐5 probes) or result in a ring‐opened fluorescein (Scheme 1aii; CAY probe). The second group is based on the Michael addition reaction (Scheme 1b). Triggered by the dual‐nucleophilicity of H2S, the probe undergoes sequential Michael addition reactions with an aldehyde group, resulting in a stable fluorescent signal (P3 probe).
Scheme 1.
Nucleophilic substitution‐based mechanisms of the four H2S probes. Detection of H2S is based on a) nucleophilic substitution through i) disulfide exchange (WSP probes) or ii) ring‐opening fluorescein (CAY probe) or b) a Michael addition reaction (P3 probe). The electrophile is marked in a blue rectangle.
3.2. Linear range and sensitivity
A key characteristic of any sensing platform is its sensitivity towards a target analyte. As such, we investigated the fluorescence response of each probe (10 μM) when exposed to a range of H2S concentrations (0–10,000 μM H2S in PBS buffer containing 1 mM CTAB; Figure 1). Note that Na2S was used as the H2S donor while CTAB was used to enhance the probe solubility and fluorescence intensity. A significant fluorescent signal enhancement can be observed after CTAB addition (Figure S2) whilst not affecting the reaction between probes and analytes. [55] As per Figure 1, i, the four probes showed increasing fluorescence intensities in different concentration ranges, indicating a diverse sensitivity range towards H2S. The fluorescence intensity of WSP‐1, WSP‐5, CAY, and P3 reached a plateau (i. e. saturation) at 1000, 500, 5000, and 50 μM, respectively. Interestingly, this was followed by an intensity decrease for higher H2S concentrations in each case. To the best of our knowledge, this decreasing trend has not been reported. However, these higher concentrations significantly exceed physiologically relevant conditions, thus do not affect the probe performance evaluation. Nevertheless, it is an important trend to take into account when implementing these probes in alternative fields such as environmental and industrial monitoring. [60] Due to this fact, the time‐dependent fluorescence response of each probe within their linear range has been included in Figure 1, ii‐iii, respectively.
Figure 1.
i) Fluorescence response when exposed to increasing concentrations of Na2S (0–10,000 μM; H2S donor), ii) representative time‐dependent fluorescence response for the linear range, and iii) linear correlation for a) WSP‐1 (10 μM), b) WSP‐5 (10 μM), c) CAY (10 μM), and d) P3 (10 μM).
The linear range and limit of detection (LOD) of the four probes are summarized in Table 1 and compared with the literature. Interestingly, WSP‐1 and WSP‐5 showed a wider linear range (0–60 and 0–100 μM respectively) compared to that reported (0–10 μM for both cases). On the other hand, CAY produced a fluorescent signal only at high H2S concentrations (100–5,000 μM linear range), which does not correspond with the reported 0–80 μM linear range. Even when using the reported buffer, i. e. H2O with 0.5% DMSO, CAY only responded to H2S at concentrations above 100 μM (Figure S3). Finally, P3 showed a narrow linear range from 0–50 μM, which agreed with the literature. In addition, our LODs do not correspond with those reported in the literature. Although the general trend between each probe is consistent, the reported LODs are in some cases up to two orders of magnitude higher. This may be attributed to the different H2S donor and buffer used. NaHS was used in some cases as the H2S donor, whilst CAY and P3 were reported in H2O with 0.5% DMSO and HEPES with 1% CH3CN, respectively. To keep parameters consistent across the four probes, this study used Na2S as the H2S donor and PBS with 1 mM CTAB as the buffer. Our results suggest these choices have a profound effect on probe performance and must be tailor‐chosen for each case, as was previously reported. [53] Furthermore, it is important to note the responsiveness of the probes. P3 demonstrated the fastest response time, reaching saturation within 10 min, while the other probes required 30 min. This could be attributed to the reaction mechanism of P3 which reacts with H2S via a reversible/fast conjugate addition reaction. [61] This fast response time could be beneficial when considering practical applications.
Table 1.
Summary of H2S detection by WSP‐1, WSP‐5, CAY, and P3.
|
Probes |
In this work |
In literature |
Ref |
||
|---|---|---|---|---|---|
|
Linear range |
LOD |
Linear range |
LOD |
||
|
WSP‐1 |
0–60 μM |
1.94 μM |
0–10 μM |
60 nM |
|
|
WSP‐5 |
0–100 μM |
0.33 μM |
0–10 μM |
47 nM |
|
|
CAY |
100–5000 μM |
24.2 μM |
0–80 μM |
130 nM |
|
|
P3 |
0–50 μM |
0.2 μM |
0–50 μM |
50 nM |
|
Probes with a small detection range and low LOD, such as P3, are suitable for biological H2S detection and generally applied in disease‐relevant diagnoses.[ 62 , 63 ] On the other hand, probes with a wide detection range and high LOD, such as CAY, are more applicable for H2S detection in environmental and industrial applications. [64] H2S sensors with a large detection range and low LOD, such as the WSP probes, could be used in more diverse applications for both biological and environmental fields.
3.3. Selectivity
3.3.1. Biothiols
In biological environments, several interfering compounds may be present, the most common of which include biothiols such as cysteine (Cys) and glutathione (GSH).[ 65 , 66 ] Therefore, it is imperative that these fluorescent probes be capable of differentiating H2S from other biothiols. To investigate the selectivity of each probe (10 μM), we measured the fluorescence intensity when exposed to increasing Cys and GSH concentrations (0–2,000 μM; Figure 2). When compared to Figure 1, only WSP‐1 and WSP‐5 showed negligible fluorescence increase with increasing Cys and GSH concentration, while CAY showed a significant fluorescence response. P3 showed a negligible response towards Cys while having a significant fluorescence intensity increase when interacting with GSH at higher concentrations (>100 μM). This high fluorescence increase could cause false‐positive results in biological samples where GSH is present in the mM range. [66] The lack of selectivity of CAY and P3 contradicted the literature and was most likely caused by the choice of buffer, as mentioned in Section 3.2. As such, the experiments were carried out in the same conditions as those reported (i. e. the same buffer). The results showed no fluorescence response for CAY when using H2O with 0.5% DMSO (Figure S4a), indicating the CTAB affected the selectivity of CAY. On the other hand, although P3 still produced a fluorescence response to biothiols in HEPES with 1% CH3CN (Figure S4b), the fluorescence increase for both biothiols can be considered negligible when considering the response when using the same concentration of H2S, as was the case with Cys in Figure 2d. As such, these results show that WSP‐1, WSP‐5, CAY, and P3 can selectively detect H2S when using the appropriate buffer. However, due to the buffer choice in this study (PBS with 1 mM CTAB), CAY and P3 are not H2S selective, highlighting again the significant effect of buffer choice on performance.
Figure 2.
Fluorescence response of a) WSP‐1 (10 μM), b) WSP‐5 (10 μM), c) CAY (10 μM), and d) P3 (10 μM) when exposed to increasing concentrations of biothiols (Cys and GSH; 0–2,000 μM) after 30 min.
However, the above evaluation by itself is incomplete and arguably biased towards the WSP probes. This is because their reaction mechanism from Scheme 1 suggests an interaction with Cys and GSH which can render them ineffective. As such, to further improve upon this selectivity evaluation, the fluorescence response of each probe in complex media containing H2S with an equimolar amount of biothiols was tested. Note that the concentration of H2S was selected for each case based on the linear range for each probe. As shown in Figure 3a–b, when WSP probes were used to detect H2S in complex mixtures containing biothiols (i. e. Cys, GSH, or both), lower fluorescence intensities were obtained compared to those in the presence of H2S only. This indicates that although WSP‐1 and WSP‐5 do not produce a fluorescent signal in the presence of biothiols, they still interact with them. In other words, this leads to an underestimation of the actual H2S present in the media. Interestingly, it should be noted that these two probes reacted with biothiols at a faster rate than with H2S. This was demonstrated by the differences in fluorescent signals when comparing samples containing the probe, H2S, and biothiols mixed at the beginning (red bars) with samples where the biothiols were added after the probe had reacted with H2S for 5 min (orange bars). These results indicate that biothiols only interact with unreacted probes and do not quench fluorescence intensity. Finally, WSP‐1 was still able to produce a fluorescent signal, albeit lower, when introducing biothiols at the same time as with H2S (blue vs. red bar). This trend was not observed for WSP‐5, where no significant fluorescent signal was produced in complex media.
Figure 3.
Fluorescence response of a) WSP‐1 (10 μM), b) WSP‐5 (10 μM), c) CAY (10 μM), and d) P3 (10 μM) when exposed to Na2S (H2S donor) and equimolar concentrations of i) Cys, ii) GSH, or iii) both after 30 min.
CAY and P3 demonstrated an inverse effect to WSP‐1 and WSP‐5 (Figure 3c–d). As previously stated, CAY and P3 produced significant fluorescent signals in the presence of Cys and/or GSH. Therefore, when evaluating them in complex media (i. e. Cys, GSH, or both with H2S), the intensities of the mixtures (red and orange bar) were higher than those of only H2S solutions (grey bar). This can be observed more clearly with P3, where the intensity doubled compared to the H2S control group, further confirming that P3 has a high reactivity towards biothiols, specifically GSH.
3.3.2. Bioanalyte selectivity
Next, a selectivity study was carried out towards various potentially interfering bioanalytes, including GSNO (a typical endogenous NO donor), glycine (Gly), H2O2, and common metal ions (metal‐base salts), as shown in Figure 4. Briefly, bioanalytes (2 mM) were mixed with each probe (10 μM), and the fluorescence response was recorded. Overall, WSP‐1, WSP‐5, and P3 showed excellent selectivity towards H2S over competing bioanalytes. On the other hand, CAY also responded to H2O2. This could be because H2O2 is a potent oxidizing agent and has strong nucleophilicity in its deprotonated form, [67] thus experiencing similar bromo cleavage and facilitating a ring‐opened fluorescein.
Figure 4.
Fluorescence response of a) WSP‐1 (10 μM), b) WSP‐5 (10 μM), c) CAY (10 μM), and d) P3 (10 μM) when exposed to different bioanalytes (2 mM) after 30 min. Na2S was used as the H2S donor.
3.3.3. Nanoparticles
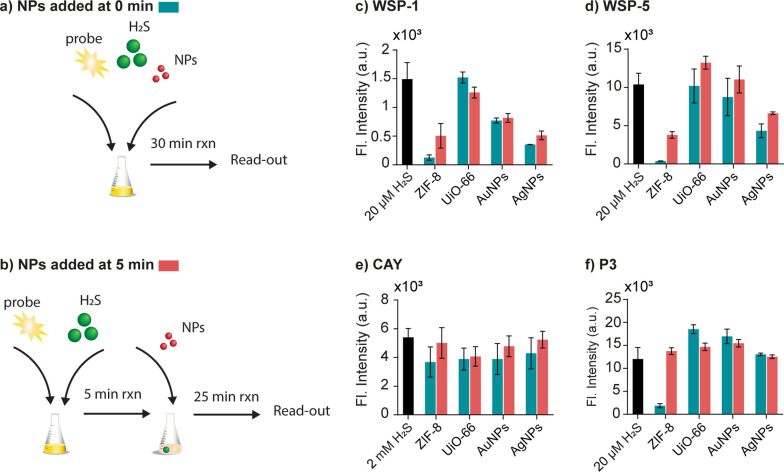
Nanoparticles (NPs) have emerged as multifunctional materials applied in numerous fields including drug delivery, [68] disease detection, [69] bioimaging, [70] catalysis, [71] and other biotechnologies.[ 72 , 73 ] Due to their ubiquity in many areas of research and development, it is vital to understand how nanoparticles will affect the performance of H2S probes. To this end, we selected metal‐organic frameworks, specifically ZIF‐8 (Zn‐based) and UiO‐66 (Zr‐based), as well as gold (AuNPs) and silver nanoparticles (AgNPs) as representative examples. First, we measured the fluorescence signal of the probes when exposed to each NP in the absence of H2S (Figure S5). A negligible effect on intensity was observed for all cases; therefore, it is safe to assume that the NPs do not severely quench or induce fluorescence for any of the probes. Next, we evaluated the performance of the probe in detecting H2S in NP‐involved mixtures. Each probe was assessed under two conditions: (i) all components (Na2S, probe, and NPs) were reacted at the same time (i. e. NPs added at t=0; Figure 5a), or (ii) NPs were added to the reaction after 5 min (Figure 5b). For WSP‐1, when compared to the control (black bar), all NPs (except UiO‐66) resulted in a decrease in fluorescence intensity (Figure 5c). This suggests an interaction between the NPs and the probes, reducing the occurrence of the disulfide exchange reaction that releases the fluorophore. This is supported by the fact that a lower fluorescence intensity occurred when all components were mixed at t=0 min compared to when NPs were added after 5 min. A similar trend was observed for WSP‐5 when exposed to ZIF‐8 and AgNPs (Figure 5d). Interestingly, this was not the case for UiO‐66 as well as AuNPs, where a negligible difference compared to the control was observed. These results indicate WSP‐5 is affected to a lesser extent by NPs compared to WSP‐1. Next, CAY was shown to be negligibly affected by all NPs (Figure 5e). Finally, all NPs except ZIF‐8, had a negligible effect on fluorescence when reacted with P3 (Figure 5f). The decrease in intensity for ZIF‐8 when NPs were introduced at t=0 min compared to t=5 min, suggests an inhibition of the probe, which rendered it ineffective in detecting H2S. As shown in Figure 1d, ii, P3’s fluorescence intensity plateaued at approximately 5 min, which explains the observation of adding the NPs at t=5 min. Overall, our results show that CAY was least affected by NPs. It is also worth noting two general trends: (i) for WSP probes, UiO‐66 (Zr‐based MOF) had the least effect on H2S detection, while AuNPs, AgNPs and ZIF‐8 demonstrated the greatest influence. This is likely due to the formation of Au‐S, [74] Ag‐S, [75] and Zn‐S [76] bonds when in the presence of H2S. (ii) The noticeable intensity reduction for all probes (except CAY) when applying ZIF‐8 at t=0 min, could be due to the high hydrophobic porosity of the ZIF‐8 surface [77] that trapped the probe and blocked the fluorescent turn‐on reaction. Taken together, when competing/interfering compounds are present, a different deviation from the expected fluorescence will be observed, similar to Figure 3. This can lead to a misinterpretation of results if not considered.
Figure 5.
Schematic illustration when a) all components (Na2S, probe, and NPs) were mixed at the same time (i. e. NPs added at t=0), or b) when NPs are added after a 5 min reaction time. Fluorescence response of c) WSP‐1 (10 μM), d) WSP‐5 (10 μM), e) CAY (10 μM), and f) P3 (10 μM) when exposed to ZIF‐8 (1 mg mL−1), UiO‐66 (1 mg mL−1), AuNPs (OD=1), or AgNPs (OD=1) after 30 min. Na2S was used as the H2S donor.
3.4. Effect of pH
Depending on the application, H2S probes might have to perform successfully in biological environments with different pH conditions. pH is spatial‐dependent at both cellular levels [78] and in human tissues,[ 79 , 80 ] varying in lysosome (pH=4.5–5.8), cytosol (pH=7.4), colon (pH=5.2–7.0), and tumor sites (pH=6.8–7.2), while water samples show pH values of 6.5–8.5. As such, we evaluated the effect of pH (4–10) on the four probes (Figure 6). Briefly, the probes (10 μM) were incubated with (black dots; experimental group) and without H2S (red dots; control) under different pH conditions. All probes could detect H2S at the normal physiological range (pH 7.4), and exhibited higher fluorescence intensities (for the same H2S concentration) for higher pH values. This is because in an aqueous solution at neutral pH, H2S presents in equilibrium with HS− and S2. As the H+ concentration decreases, the equilibrium shifts towards HS−, which has a higher nucleophilicity than H2S. Based on the mechanism shown in Scheme 1, an enhanced fluorescence is expected under basic conditions (pH>7). In addition, some fluorescein moieties are pH‐sensitive and give higher fluorescence intensities at higher pH. [81] Thus, the fluorophore released or activated after reacting with H2S tends to exhibit higher intensities in basic environments. It is also worth noting that P3 itself produced increasing fluorescence signals with increasing pH (Figure S6). As such, this should be considered when using P3 to detect low H2S concentrations (e. g., in the nM range).
Figure 6.
Fluorescence response of a) WSP‐1 (10 μM), b) WSP‐5 (10 μM), c) CAY (10 μM), and d) P3 (10 μM) when exposed to Na2S (H2S donor) at pH 4–10 after 30 min.
3.5. Plant‐derived H2S donor
Inorganic sulfide salt‐based H2S donors such as Na2S and NaHS are commonly used as research tools in biological and clinical fields. However, they are limited by (i) spontaneous release of H2S due to immediate hydrolysis causing damage in vivo, [82] (ii) polysulfides formation leading to protein thiol oxidation in vitro, [83] and (iii) rapid volatilization (t1/2=5 min), calling into question the accuracy of long‐term experiments and calibration curves. [84] Therefore, H2S donors that are more stable have been researched for biological applications. [85] With this in mind, we evaluated the ability of WSP‐1 and WSP‐5 to detect H2S generated from a plant‐derived H2S donor, specifically DATs (Figure 7). DATs is one of four H2S‐releasing compounds generated from the decomposition of allicin (diallyl thiosulfinate), one of the best characterized garlic‐derived compounds. [86] Importantly, it is a thiol‐triggered H2S donor requiring Cys or GSH as a trigger. As such, this section will not discuss CAY or P3 as they have been previously shown to react with these biothiols. It is worth noting that the WSP probes showed a significant response when exposed to DATs only after the addition of CTAB (Figure S7). As mentioned previously, this could be because CTAB enhances the solubility of both organic probes and donors in aqueous buffers, resulting in more efficient interactions. [55] The fluorescence response of WSP‐1 and WSP‐5 exposed to increasing concentrations of DATs (0–2,000 μM) in the presence of Cys or GSH (500 μM) was measured and shown in Figure 7. The signal of WSP‐1 peaked at 200 μM and 500 μM DATs, respectively. On the other hand, WSP‐5 produced increasing signals for both Cys and GSH. These results indicate that WSP‐5 can detect a broader range of H2S when using DATs as a donor. Note, this trend was also observed when using Na2S as the H2S source (Figure 1b). Overall, increasing fluorescence intensities were obtained with higher DATs concentrations, indicating the expected H2S generation from DATs followed by successful detection by WSP‐1 and WSP‐5.
Figure 7.
Fluorescence response of a) WSP‐1 (10 μM) and b) WSP‐5 (10 μM) using DATs as an H2S donor in the presence of Cys or GSH (500 μM) after 30 min.
3.6. Endogenous H2S detection and imaging
Given the rising need for in vivo imaging when using fluorescent probes, we further explored the potential of the probes for endogenous H2S detection in live cells. First, a cytotoxicity test (using the CCK‐8 assay) of RAW 264.7 and HeLa cells towards lipopolysaccharides (LPS; 0.5–2 μg mL−1) or the four probes (1–10 μM) upon a 24 h treatment was carried out. LPS is a known activator used to induce H2S production in macrophage cells and cancer cells. [87] As shown in Figure S8, 2 μg mL−1 LPS and 10 μM probes were chosen as the optimal conditions for the bioimaging study due to their good cell viability. Moreover, comparing Figure 1 and Figure S9, a negligible effect on fluorescence intensity was observed for all probes, confirming that physiologically relevant temperatures (37 °C) do not affect the behavior of the probes. Next, we investigated the capacity of these probes to visualize H2S in live cells. RAW 264.7 and HeLa cells were pretreated with 2 μg mL−1 LPS for 24 h prior to exposure to 10 μM probes for 1 h in DMEM (for RAW 264.7) and 30 min in PBS with 0.1 mM CTAB (for HeLa). Confocal imaging and intensity analysis showed an increase in fluorescence intensity in cells treated with LPS compared to untreated cells (Figure 8). These results confirmed that increased levels of endogenous H2S promoted by LPS could be detected by these probes in normal and cancer cells. However, it is important to note: 1) all probes showed higher fluorescence intensity in HeLa cells compared to RAW 264.7 cells. This could be attributed to a higher amount of H2S generated by HeLa cells upon LPS treatment. Moreover, the cell culture medium caused different degrees of probe quenching, as demonstrated by the reduced fluorescence response when exposing the probes to H2S in DMEM (Figure S10). Additionally, CTAB was not used when exposing RAW 264.7 cells to the probes as complete cell apoptosis was observed if used. 2) WSP‐1 showed the lowest autofluorescence in HeLa cells compared to the other probes but also had produced lower fluorescence intensity for endogenous H2S detection. This is consistent with the behavior of the probe in an aqueous buffer. 3) CAY and P3 produced higher fluorescence intensity in LPS‐treated HeLa cells than WSP probes. Considering the abundant biothiols concentrations in complex cell environments, higher intensities are expected as it has been shown that CAY and P3 probes are thiol‐sensitive.
Figure 8.
a) Confocal images (green channel, brightfield, and merged) of RAW 264.7 (left half) and HeLa (right half) cells exposed to WSP‐1, WSP‐5, CAY, and P3, respectively. Each group contains untreated cells (top) and cells incubated with LPS (2 μg mL−1) for 24 h (bottom), followed by incubation with probe (10 μM) for 1 h in DMEM (for RAW 264.7 cells) and 30 min in PBS containing 0.1 mM CTAB (for HeLa cells). Images were taken at 40X magnification. Scale bar: 20 μm. Relative fluorescence intensity of confocal images for b) RAW 264.7 and c) HeLa cells calculated by the mean intensity from ≥5 individual cells. Statistical significance relative to control tests was calculated by one‐way ANOVA followed by Tukey post hoc test, ns=not significant, *p<0.1, **p<0.01, ****p<0.0001.
4. Conclusion
Four commercially available fluorescent probes for H2S detection were systematically investigated, specifically WSP‐1, WSP‐5, CAY, and P3. The performance of each was evaluated based on their sensitivity, selectivity, and performance in different complex environments (pH, buffer, temperature, NPs, cell medium). Endogenous H2S detection was successfully carried out in LPS activated RAW 264.7 and HeLa cells. WSP‐5 was shown to have the widest clinical sensitivity linear range, a low LOD, minimum interference towards thiols and other analytes, was most stable in different pH environments, exhibited moderate interference by nanoparticles, and was able to image H2S in live‐cell systems. Additionally, we have shown for the buffer conditions used in this study that CAY and P3 could detect other biothiols such as Cys and GSH. However, when using their reported buffers, H2S selectivity was observed. Suggestions on using these H2S probes, including the buffer choice and pH range, are provided. This detailed systematic study will help researchers to select probes for H2S sensing and relevant activities.
Conflict of interest
The authors declare no conflict of interest.
5.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
This work was supported by the National Health and Medical Research Council Emerging Leadership Investigator Grant (NHMRC APP1173428; R.C.), the National Health and Medical Research Council Career Development Fellowship (NHMRC GNT1163786; K.L.), and the UNSW Scientia Fellowship (R.C. and K.L.). This research used the facilities at the Mark Wainwright Analytical Centre Cell Culture Lab and Katharina Gaus Light Microscopy Facility at UNSW. Open access publishing facilitated by University of New South Wales, as part of the Wiley ‐ University of New South Wales agreement via the Council of Australian University Librarians.
Y. Zhou, F. Mazur, K. Liang, R. Chandrawati, Chem. Asian J. 2022, 17, e202101399.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Rubright S. L. M., Pearce L. L., Peterson J., Nitric Oxide 2017, 71, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guidotti T. L., Int. J. Toxicol. 2010, 29, 569–581. [DOI] [PubMed] [Google Scholar]
- 3. Hosoki R., Matsuki N., Kimura H., Biochem. Biophys. Res. Commun. 1997, 237, 527–531. [DOI] [PubMed] [Google Scholar]
- 4. Szabó C., Nat. Rev. Drug Discovery 2007, 6, 917–935. [DOI] [PubMed] [Google Scholar]
- 5. Wang R., FASEB J. 2002, 16, 1792–1798. [DOI] [PubMed] [Google Scholar]
- 6. Vandiver M. S., Snyder S. H., J. Mol. Med. 2012, 90, 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cirino G., Vellecco V., Bucci M., Br. J. Pharmacol. 2017, 174, 4021–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Richardson C. J., Magee E. A., Cummings J. H., Clin. Chim. Acta 2000, 293, 115–125. [DOI] [PubMed] [Google Scholar]
- 9. Elsey D. J., Fowkes R. C., Baxter G. F., Cell Biochem. Funct. 2010, 28, 95–106. [DOI] [PubMed] [Google Scholar]
- 10. Savage J., Gould D., J. Chromatogr. B 1990, 526, 540–545. [DOI] [PubMed] [Google Scholar]
- 11. Elrod J. W., Calvert J. W., Morrison J., Doeller J. E., Kraus D. W., Tao L., Jiao X., Scalia R., Kiss L., Szabo C., PNAS 2007, 104, 15560–15565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang G., Wu L., Jiang B., Yang W., Qi J., Cao K., Meng Q., Mustafa A. K., Mu W., Zhang S., Science 2008, 322, 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Linden D. R., Antioxid. Redox Signal. 2014, 20, 818–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Olson K. R., Donald J. A., Acta Histochem. 2009, 111, 244–256. [DOI] [PubMed] [Google Scholar]
- 15. Bhatia M., Gaddam R. R., Antioxid. Redox Signal. 2021, 34, 1368–1377. [DOI] [PubMed] [Google Scholar]
- 16. Dilek N., Papapetropoulos A., Toliver-Kinsky T., Szabo C., Pharmacol. Res. 2020, 161, 105119. [DOI] [PubMed] [Google Scholar]
- 17. Kimura H., Mol. Neurobiol. 2002, 26, 13–19. [DOI] [PubMed] [Google Scholar]
- 18. Hu L. F., Lu M., Tiong C. X., Dawe G. S., Hu G., Bian J. S., Aging Cell 2010, 9, 135–146. [DOI] [PubMed] [Google Scholar]
- 19. Eto K., Asada T., Arima K., Makifuchi T., Kimura H., Biochem. Biophys. Res. Commun. 2002, 293, 1485–1488. [DOI] [PubMed] [Google Scholar]
- 20. Hellmich M. R., Szabo C., Handb. Exp. Pharmacol. 2015, 230, 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kamoun P., Belardinelli M.-C., Chabli A., Lallouchi K., Chadefaux-Vekemans B., Am. J. Med. 2003, 116, 310–311. [DOI] [PubMed] [Google Scholar]
- 22. Jain S.K., Bull R., Rains J.L., Bass P.F., Levine S.N., Reddy S., McVie R., Bocchini J.A. Jr, Antioxid. Redox Signal. 2010, 12, 1333–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou Y., Yang T., Liang K., Chandrawati R., Adv. Drug Delivery Rev. 2021, 171, 199–214. [DOI] [PubMed] [Google Scholar]
- 24. Qian Y., Matson J. B., Adv. Drug Delivery Rev. 2017, 110, 137–156. [DOI] [PubMed] [Google Scholar]
- 25. Carné-Sánchez A., Carmona F. J., Kim C., Furukawa S., Chem. Commun. 2020, 56, 9750–9766. [DOI] [PubMed] [Google Scholar]
- 26.Antibe Therapeutics Announces Successful Phase 2 Trial of ATB-346 in Osteoarthritis, https://antibethera.com/news/antibe-therapeutics-announces-successful-phase-2-trial-of-atb-346-in-osteoarthritis/, (accessed 2021–01-26).
- 27. Polhemus D. J., Li Z., Pattillo C. B., Gojon G. Sr, Gojon G. Jr, Giordano T., Krum H., Cardiovasc. Ther. 2015, 33, 216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vikrant K., Kumar V., Ok Y. S., Kim K. H., Deep A., TrAC Trends Anal. Chem. 2018, 105, 263–281. [Google Scholar]
- 29. Ibrahim H., Serag A., Farag M. A., J. Adv. Res. 2021, 27, 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hartle M. D., Pluth M. D., Chem. Soc. Rev. 2016, 45, 6108–6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shen X., Peter E. A., Bir S., Wang R., Kevil C. G., Free Radical Biol. Med. 2012, 52, 2276–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng Y., Liao F., Du J., Tang C., Xu G., Geng B., Acta Physiol. Sin. 2012, 64, 681. [PubMed] [Google Scholar]
- 33. Xu T., Scafa N., Xu L.-P., Zhou S., Al-Ghanem K. A., Mahboob S., Fugetsu B., Zhang X., Analyst 2016, 141, 1185–1195. [DOI] [PubMed] [Google Scholar]
- 34. Vitvitsky V., Banerjee R., Methods Enzymol. 2015, 554, 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li H., Fang Y., Yan J., Ren X., Zheng C., Wu B., Wang S., Li Z., Hua H., Wang P., TrAC Trends Anal. Chem. 2020, 134, 116117. [Google Scholar]
- 36. Li J., Su Z., Yu C., Yuan Y., Wu Q., Liu J., Peng B., Hu W., Lu X., Yu H., Dyes Pigm. 2021, 192, 109451. [Google Scholar]
- 37. Jiao X., Li Y., Niu J., Xie X., Wang X., Tang B., Anal. Chem. 2018, 90, 533–555. [DOI] [PubMed] [Google Scholar]
- 38. Jose D. A., Sakla R., Sharma N., Gadiyaram S., Kaushik R., Ghosh A., ACS Sens. 2020, 5, 3365–3391. [DOI] [PubMed] [Google Scholar]
- 39. Luo Y. N., Zhu C. Z., Du D., Lin Y. H., Anal. Chim. Acta 2019, 1061, 1–12. [DOI] [PubMed] [Google Scholar]
- 40. Varlet V., Giuliani N., Palmiere C., Maujean G., Augsburger M., J. Anal. Toxicol. 2015, 39, 52–57. [DOI] [PubMed] [Google Scholar]
- 41. Zhou Y., Mazur F., Fan Q., Chandrawati R., View 2022, 3, 20210008. [Google Scholar]
- 42. Zhou L., Chen Y., Shao B., Cheng J., Li X., Front. Chem. 2021, 1–30. [Google Scholar]
- 43. Thorson M. K., Majtan T., Kraus J. P., Barrios A. M., Angew. Chem. Int. Ed. 2013, 52, 4641–4644. [DOI] [PubMed] [Google Scholar]
- 44. Xuan W., Pan R., Cao Y., Liu K., Wang W., Chem. Commun. 2012, 48, 10669–10671. [DOI] [PubMed] [Google Scholar]
- 45. Li X., Cheng J., Gong Y., Yang B., Hu Y., Biosens. Bioelectron. 2015, 65, 302–306. [DOI] [PubMed] [Google Scholar]
- 46. Liu C., Pan J., Li S., Zhao Y., Wu L. Y., Berkman C. E., Whorton A. R., Xian M., Angew. Chem. Int. Ed. 2011, 50, 10327–10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li D.-P., Zhang J.-F., Cui J., Ma X.-F., Liu J.-T., Miao J.-Y., Zhao B.-X., Sens. Actuators B 2016, 234, 231–238. [Google Scholar]
- 48. Choi M. G., Cha S., Lee H., Jeon H. L., Chang S.-K., Chem. Commun. 2009, 7390–7392. [DOI] [PubMed] [Google Scholar]
- 49. Hou J.-T., Liu B.-Y., Li K., Yu K.-K., Wu M.-B., Yu X.-Q., Talanta 2013, 116, 434–440. [DOI] [PubMed] [Google Scholar]
- 50. Sun W., Fan J., Hu C., Cao J., Zhang H., Xiong X., Wang J., Cui S., Sun S., Peng X., Chem. Commun. 2013, 49, 3890–3892. [DOI] [PubMed] [Google Scholar]
- 51. Wang L., Chen X., Cao D., New J. Chem. 2017, 41, 3367–3373. [Google Scholar]
- 52. Chen B., Huang J., Geng H., Xuan L., Xu T., Li X., Han Y., New J. Chem. 2017, 41, 1119–1123. [Google Scholar]
- 53. Peng B., Chen W., Liu C., Rosser E. W., Pacheco A., Zhao Y., Aguilar H. C., Xian M., Chemistry 2014, 20, 1010–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Reja S. I., Sharma N., Gupta M., Bajaj P., Bhalla V., Parihar R. D., Ohri P., Kaur G., Kumar M., Chem. Eur. J. 2017, 23, 9872–9878. [DOI] [PubMed] [Google Scholar]
- 55. Kumar D., Rub M. A., J. Mol. Liq. 2019, 274, 639–645. [Google Scholar]
- 56. Liu X. D., Wu X., RSC Adv. 2015, 5, 7433–7439. [Google Scholar]
- 57. Zhou Y., Yang T., Namivandi-Zangeneh R., Boyer C., Liang K., Chandrawati R., J. Mater. Chem. B 2021, 9, 1059–1068. [DOI] [PubMed] [Google Scholar]
- 58. Vermoortele F., Bueken B., Le Bars G., Van de Voorde B., Vandichel M., Houthoofd K., Vimont A., Daturi M., Waroquier M., Van Speybroeck V., J. Am. Chem. Soc. 2013, 135, 11465–11468. [DOI] [PubMed] [Google Scholar]
- 59. Myszkowska J., Derevenkov I., Makarov S. V., Spiekerkoetter U., Hannibal L., Antioxidants 2021, 10, 1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ziętek B., Banasiewicz A., Zimroz R., Szrek J., Gola S., Energies 2020, 13, 6331. [Google Scholar]
- 61. Singha S., Kim D., Moon H., Wang T., Kim K. H., Shin Y. H., Jung J., Seo E., Lee S.-J., Ahn K. H., Anal. Chem. 2015, 87, 1188–1195. [DOI] [PubMed] [Google Scholar]
- 62. Muthusamy S., Rajalakshmi K., Zhu D., Zhao L., Wang S., Zhu W., Sens. Actuators B 2020, 320, 128433. [Google Scholar]
- 63. Song B.-Y., Zhang M., Teng Y., Zhang X.-F., Deng Z.-P., Huo L.-H., Gao S., Sens. Actuators B 2020, 307, 127662. [Google Scholar]
- 64. Liu Y., Feng G., Org. Biomol. Chem. 2014, 12, 438–445. [DOI] [PubMed] [Google Scholar]
- 65. Refsum H., Ueland P., Nygård O., Vollset S., Annu. Rev. Med. 1998, 49, 31–62. [DOI] [PubMed] [Google Scholar]
- 66. Moriarty-Craige S. E., Jones D. P., Annu. Rev. Nutr. 2004, 24, 481–509. [DOI] [PubMed] [Google Scholar]
- 67. Chan J., Dodani S. C., Chang C. J., Nat. Chem. 2012, 4, 973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shinde N. C., Keskar N. J., Argade P. D., Res. J. Pharm. Biol. Chem. Sci. 2012, 3, 922–929. [Google Scholar]
- 69. Brede C., Labhasetwar V., Adv. Chronic Kidney Dis. 2013, 20, 454–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sharma P., Brown S., Walter G., Santra S., Moudgil B., Adv. Colloid Interface Sci. 2006, 123, 471–485. [DOI] [PubMed] [Google Scholar]
- 71. Ndolomingo M. J., Bingwa N., Meijboom R., J. Mater. Sci. 2020, 55, 6195–6241. [Google Scholar]
- 72. Tartaj P., Morales M., Gonzalez-Carreno T., Veintemillas-Verdaguer S., Serna C., J. Magn. Magn. Mater. 2005, 290, 28–34. [Google Scholar]
- 73. Sanvicens N., Marco M. P., Trends Biotechnol. 2008, 26, 425–433. [DOI] [PubMed] [Google Scholar]
- 74. Zhang X., Zhou W. J., Yuan Z. Q., Lu C., Analyst 2015, 140, 7443–7450. [DOI] [PubMed] [Google Scholar]
- 75. Li D. W., Qu L. L., Hu K., Long Y. T., Tian H., Angew. Chem. Int. Ed. 2015, 54, 12758–12761. [DOI] [PubMed] [Google Scholar]
- 76. Graham A. J., Singleton M. A., Sorbie K. S., Collins I. R., Operations, SPE Prod. Oper. 2017, 32, 314–324. [Google Scholar]
- 77. Sann E. E., Pan Y., Gao Z. F., Zhan S. S., Xia F., Sep. Purif. Technol. 2018, 206, 186–191. [Google Scholar]
- 78. Hu X., Zhang Y., Xie Z., Jing X., Bellotti A., Gu Z., Biomacromolecules 2017, 18, 649–673. [DOI] [PubMed] [Google Scholar]
- 79. Cook M. T., Tzortzis G., Charalampopoulos D., Khutoryanskiy V. V., J. Control. Release 2012, 162, 56–67. [DOI] [PubMed] [Google Scholar]
- 80. Zhang X., Lin Y., Gillies R. J., J. Nucl. Med. 2010, 51, 1167–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Le Guern F., Mussard V., Gaucher A., Rottman M., Prim D., Int. J. Mol. Sci. 2020, 21, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hughes M. N., Centelles M. N., Moore K. P., Free Radical Biol. Med. 2009, 47, 1346–1353. [DOI] [PubMed] [Google Scholar]
- 83. Greiner R., Palinkas Z., Basell K., Becher D., Antelmann H., Nagy P., Dick T. P., Antioxid. Redox Signal. 2013, 19, 1749–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. DeLeon E. R., Stoy G. F., Olson K. R., Anal. Biochem. 2012, 421, 203–207. [DOI] [PubMed] [Google Scholar]
- 85. Corvino A., Frecentese F., Magli E., Perissutti E., Santagada V., Scognamiglio A., Caliendo G., Fiorino F., Severino B., Antioxidants 2021, 10, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Amagase H., J. Nutr. 2006, 136, 716S-725S. [DOI] [PubMed] [Google Scholar]
- 87. Dufton N., Natividad J., Verdu E. F., Wallace J. L., Sci. Rep. 2012, 2, 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.