Abstract
We purified an intracellular pyranose oxidase from mycelial extracts of the white rot fungus Trametes multicolor by using ammonium sulfate fractionation, hydrophobic interaction, ion-exchange chromatography, and gel filtration. The native enzyme has a molecular mass of 270 kDa as determined by equilibrium ultracentrifugation and is composed of four identical 68-kDa subunits as determined by matrix-assisted laser desorption ionization mass spectrometry. Each subunit contains one covalently bound flavin adenine dinucleotide as its prosthetic group. The enzyme oxidizes several aldopyranoses specifically at position C-2, and its preferred electron donor substrates are d-glucose, d-xylose, and l-sorbose. During this oxidation reaction electrons are transferred to oxygen, yielding hydrogen peroxide. In addition, the enzyme catalyzes the two-electron reduction of 1,4-benzoquinone, several substituted benzoquinones, and 2,6-dichloroindophenol, as well as the one-electron reduction of the ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] cation radical. As judged by the catalytic efficiencies (kcat/Km), some of these quinone electron acceptors are much better substrates for pyranose oxidase than oxygen. The optimum pH of the pyranose oxidase-catalyzed reaction depends strongly on the electron acceptor employed and varies from 4 to 8. It has been proposed that the main metabolic function of pyranose oxidase is as a constituent of the ligninolytic system of white rot fungi that provides peroxidases with H2O2. An additional function could be reduction of quinones, key intermediates that are formed during mineralization of lignin.
The enzyme pyranose oxidase (P2O) (pyranose:oxygen 2-oxidoreductase; EC 1.1.3.10), which catalyzes the oxidation of several aldopyranoses at position C-2 to yield the corresponding 2-ketoaldoses (aldos-2-uloses, osones), is widely distributed among wood-degrading basidiomycetes (14, 32, 44). It has been purified and characterized from several microorganisms, including Phanerochaete chrysosporium (2, 45), Phlebiopsis gigantea (39), Pleurotus ostreatus (40), Polyporus obtusus (28), Trametes (Coriolus) versicolor (35), and unidentified basidiomycete no. 52 (26). The currently available data reveal some general similarities among P2Os from these different fungi. Typically, P2O is a rather large, homotetrameric protein that contains covalently bound flavin adenine dinucleotide (FAD). The in vivo substrates of P2O probably are d-glucose, d-galactose, and d-xylose, which are abundant in lignocellulose and which are oxidized to 2-keto-d-glucose (d-arabino-hexos-2-ulose, 2-dehydro-d-glucose), 2-keto-d-galactose (d-lyxo-hexos-2-ulose, 2-dehydro-d-galactose), and 2-keto-d-xylose (d-threo-pentos-2-ulose, 2-dehydro-d-xylose), respectively. In addition, P2O also exhibits significant activity with a number of other carbohydrates, including l-sorbose, d-glucono-1,5-lactone, and d-allose (20). The substrate selectivity, however, varies to some extent among P2Os isolated from different fungi. During the oxidation reactions electrons are transferred to molecular oxygen, resulting in the formation of hydrogen peroxide (21).
In ultrastructural and immunocytochemical studies of P2Os produced by the lignocellulose-degrading fungi P. chrysosporium and Oudemansiella mucida under liquid culture conditions or during wood decay, P2O was localized primarily in the periplasmic space. Only during autolysis, which occurs at later stages of development, is it located extracellularly, and under these circumstances it is primarily associated with fungal cell walls or with extracellular slime (8, 12). This preferential periplasmic distribution is consistent with previous reports of H2O2 production in white rot fungi under ligninolytic conditions (19, 24). Hydrogen peroxide produced by P2O in the periplasmic space of fungal hyphae or extracellularly may function in situ with two lignin-degrading oxidative enzymes, lignin peroxidase and manganese peroxidase, for which H2O2 is an essential cosubstrate. The hyphal periplasmic and extracellular localization reported for these two peroxidases in P. chrysosporium is similar to that of P2O (9, 10). Thus, the main metabolic role of P2O appears to be as a constituent of the fungal ligninolytic system that provides peroxidases with hydrogen peroxide (11). Formation of 2-ketoglucose, an intermediate in the synthesis of cortalcerone, also has been attributed to P2O (4, 46). As not all fungi with P2O activity synthesize cortalcerone, formation of 2-ketoglucose for subsequent transformation into the antibiotic clearly cannot be the main role of P2O.
For the last two decades P2O has received increased attention as the key biocatalyst in several biotechnological applications. The possible applications include biotransformations of carbohydrates; e.g., the oxidized 2-keto sugars obtained from d-glucose and d-galactose can be reduced catalytically or enzymatically at position C-1 to obtain high yields of virtually pure d-fructose or d-tagatose. Both ketoses have a number of known or proposed applications in food technology. P2O also is currently used for various analytical applications; e.g., it is used in clinical chemistry for determination of 1,5-anhydro-d-glucitol, an important marker for glycemic control in diabetes patients (21, 22).
In this paper we describe purification and detailed characterization of P2O from the white rot fungus Trametes multicolor (Schaeff.) Jül. (synonyms, Trametes ochracea, Trametes zonata, and Trametes zonatella), a cosmopolitan basidiomycete found in temperate to boreal climates. In northern Europe this fungus occurs much more frequently than the well-studied organism T. versicolor and contributes significantly to the degradation of hardwood, mainly Betula species. Based on our studies, we propose an additional physiological role for P2O, namely, reduction of benzoquinones formed during ligninolysis.
MATERIALS AND METHODS
Chemicals.
The chemicals used were the purest grade available and were purchased from Sigma (St. Louis, Mo.) unless otherwise stated. Horseradish peroxidase (grade II; EC 1.11.1.7) was obtained from Boehringer (Mannheim, Germany), the various substituted quinones were obtained from Aldrich (Steinheim, Germany), bovine serum albumin (BSA) (fraction V) was obtained from United States Biochemical Corp. (Cleveland, Ohio), and isomaltose was obtained from ICN (Costa Mesa, Calif.). Allolactose was provided by Sergio Riva (CNR, Milan, Italy). Superose 12 HR 10/30, Superose 12 prep grade, Source 30Q, Mono P HR 5/20, and Phenyl Sepharose were purchased from Amersham-Pharmacia Biotech (Uppsala, Sweden).
Organism and culture conditions.
Strain MB 49, the wild-type strain of T. multicolor used in this study, was isolated from hardwood in southern Germany and was obtained from the culture collection of the Institute of Applied Microbiology, University of Agricultural Sciences Vienna. Stock cultures were maintained on glucose-maltose Sabouraud agar and transferred every 2 months. The fungus was cultivated in a 20-liter laboratory fermentor by using a medium based on whey powder and peptone from casein, to which lactose was fed batchwise as described in detail elsewhere (32). The average yield of biomass and the average P2O activity in these laboratory fermentations were approximately 14 g (dry weight) of mycelia per liter and 1,500 U of P2O activity per liter, respectively.
Enzyme purification.
Approximately 300 g (wet weight) of mycelia was suspended in 600 ml of 50 mM phosphate buffer (pH 6.5) containing 10 mM EDTA and 0.5 mM phenylmethylsulfonyl fluoride and was homogenized with an Ultra-Turrax homogenizer (IKA Labortechnik, Staufen, Germany) at 24,000 rpm for 15 min by using three intervals with interruptions for cooling to 4°C. The mycelial extract was centrifuged at 30,000 × g for 15 min at 4°C. Ammonium sulfate was slowly added to 30% saturation to the clear supernatant. The precipitate was removed by centrifugation as described above, and the supernatant was applied to a 400-ml Phenyl Sepharose column that had been preequilibrated with the sample buffer. Subsequently, P2O was eluted at a rate of 15 ml min−1 with a 2,500-ml linear 30 to 0% (NH4)2SO4 gradient. The active fractions were pooled, dialyzed against 20 mM Bis–Tris-HCl buffer (pH 6.0) overnight, and loaded onto a 200-ml Source 30Q column equilibrated with the same buffer. P2O was eluted at a rate of 10 ml min−1 by using a linear 0 to 500 mM KCl gradient in 10 column volumes. Active fractions were pooled and concentrated by ultrafiltration with a PM30 membrane (Amicon Corp., Beverly, Mass.). The concentrated enzyme solution (2 ml) was applied to a Superose 12 prep grade column (800 by 16 mm) equilibrated with 50 mM phosphate buffer (pH 6.5) containing 150 mM KCl and eluted at a rate of 1 ml min−1. Active fractions were pooled, filter sterilized, and stored at 4°C.
To determine the isoelectric point of P2O, chromatofocusing was performed on a Mono P HR 5/20 column that had been preequilibrated with 25 mM methylpiperazine-HCl buffer (pH 5.7). The enzyme was eluted at a rate of 0.5 ml min−1 with Polybuffer 74 (Amersham-Pharmacia) (pH 4.0).
Enzyme assay.
Unless otherwise specified, P2O activity was determined spectrophotometrically at 420 nm and 30°C by measuring the formation of H2O2 for 3 min with a peroxidase-coupled assay using ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] (ɛ420 = 43,200 M−1 cm−1 [36]) as the chromogen (13). The standard assay mixture (total volume,1 ml) contained 1 μmol of ABTS in 50 mM potassium phosphate buffer (pH 6.5), 2 U of horseradish peroxidase, 100 μmol of d-glucose, and a suitable amount of the P2O sample. One unit of P2O activity was defined as the amount of enzyme necessary for oxidation of 2 μmol of ABTS per min under the conditions described above. Alternatively, P2O activity was monitored by determining the rate of oxygen consumption and was measured with an oxygen electrode placed in a thermostatically controlled vessel (Rank Brothers Ltd., Cambridge, England) at 30°C (13). One unit of enzyme activity was defined as the amount of enzyme that consumed 1 μmol of O2 per min under the assay conditions. Protein contents were determined by the dye-binding method of Bradford (5), using BSA as the standard. The pH dependence of P2O activity when the different electron acceptors were used was determined with the following buffers: 100 mM citrate (pH 2.5 to 6.0), 100 mM phosphate (pH 6.0 to 8.0), and 100 mM borate (pH 8.0 to 10.0).
Steady-state kinetic measurements.
Unless otherwise stated, all steady-state kinetic measurements were obtained at 30°C in phosphate buffer (pH 6.5). To measure kinetic constants for the various electron donors, the routine ABTS-peroxidase assay was performed as described above with air-saturated solutions. When kinetic constants were determined for the various electron acceptors, 100 mM glucose was routinely used as the electron donor. All solutions were flushed with nitrogen immediately before the experiments to avoid interference with oxygen. The pH profile for the apparent kinetic constants for two of the electron acceptors used was determined by using 100 mM citrate–100 mM phosphate–100 mM borate buffer. All kinetic constants were calculated by nonlinear least-squares regression, and the observed data were fit to the Henri-Michaelis-Menten equation.
Electrophoretic analyses.
Denaturing sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was carried out with an Amersham-Pharmacia Multiphor II system by using precast ExcelGel SDS gradient gels (8–18) as described by Laemmli (31). Isoelectric focusing at pH 4.0 to 6.5 was performed by using precast, dry gels (CleanGel for IEF; Amersham-Pharmacia) that were rehydrated with carrier ampholytes (Pharmalyte; pH 4.0 to 6.5; Amersham-Pharmacia) as recommended by the supplier. A low-pI marker protein kit (pH 2.8 to 6.5; Amersham-Pharmacia) was used to determine pI values. Protein bands were stained with Coomassie blue.
Molecular mass determinations. (i) Gel filtration.
A Superose 12 HR 10/30 column was equilibrated with 50 mM phosphate buffer (pH 6.5) containing 150 mM KCl. The column was calibrated with the standard proteins thyroglobulin (Mr, 669,000), apoferritin (Mr, 443,000), β-amylase (Mr, 200,000), alcohol dehydrogenase (Mr, 150,000), and BSA (Mr, 67,000), each at a concentration of 1 mg ml−1. The flow rate for elution was 1 ml min−1.
(ii) Cross-linking of P2O.
A preparation of homogeneous P2O (1 mg of protein ml−1 ) was diafiltered against 0.2 M triethanolamine-HCl (pH 8.5), and then 0.1 volume of freshly dissolved dimethyl suberimidate dihydrochloride (30 mg ml−1) was added to aliquots of the protein solution and the preparations were incubated for 4 h at room temperature (15). The reactions were stopped by dilution with 9 volumes of Laemmli buffer, and the preparations were incubated for 30 min at 40°C. SDS-PAGE was performed as described above. The molecular weight was estimated by comparison with marker proteins (Molecular Standard Mixture Recombinant; Sigma) after proteins were visualized by staining with Coomassie blue.
(iii) Equilibrium ultracentrifugation.
The molecular weight of the native enzyme was determined by the equilibrium ultracentrifugation method of Yphantis (50) by using a Spinco model E (Beckman, Fullerton, Calif.) ultracentrifuge at a rotor velocity of 10,590 rpm (140,000 × g; An-H-Ti rotor; 1-mm column; 20°C) for 7 h. Purified P2O (0.33 mg ml−1) was diafiltered against phosphate buffer (pH 7.0) containing 100 mM NaCl prior to ultracentrifugation.
(iv) MALDI mass spectrometry.
The molecular weight of the P2O subunit was determined by matrix-assisted laser desorption ionization (MALDI) mass spectrometry. Samples of P2O were diafiltered against 0.1% trifluoroacetic acid and measured with a Bruker BIFLEX MALDI–time-of-flight mass spectrometer (Bruker-Franzen, Bremen, Germany) equipped with a nitrogen laser (337 nm) (Laser Science, Cambridge, Mass.) and a gridless delayed extraction ion source. Positive-ion mass spectra were recorded in the linear mode with the acceleration voltage kept at 20 kV. A saturated solution of sinapic acid in aqueous 30% acetonitrile–0.1% trifluoroacetic acid was used as the MALDI matrix. One microliter of a P2O sample and 1 μl of the matrix solution were premixed in an Eppendorf tube, and 1 μl of the mixture was placed on the target and allowed to dry at the ambient temperature (20 to 23°C). Spectra were internally calibrated by using single and double charged ions of BSA.
Transmission electron microscopy.
A sample of purified P2O was diafiltered against 5 mM phosphate buffer (pH 6.5) and diluted to a concentration of 100 μg ml−1. The enzyme molecules were adsorbed from this solution onto an ultrathin carbon film mounted on a 300-mesh copper grid, rinsed with water, negatively stained with 4% (wt/vol) uranyl acetate (pH 4.5), and then air dried (39). The samples were examined with a Phillips CM 100 transmission electron microscope with an acceleration voltage of 80 kV. Molecular masses were calculated by using the relationship 1 Da ≈ 1.37 × 10−3 nm3 (51).
Determination of sugar content.
SDS-PAGE of P2O was performed as described above, and the gel was stained for glycoproteins with dansylhydrazine (17). Ovalbumin was used as a positive control. The carbohydrate content of purified P2O was estimated by the phenol sulfuric acid method by using glucose as the standard (16).
Preparation and identification of d-glucose oxidation product.
The d-glucose oxidation product was prepared by allowing 1 mmol of d-glucose, 20 U of P2O, and 20,000 U of catalase (from bovine liver; Sigma) to react in 20 ml of 25 mM phosphate buffer (pH 6.5) under an air atmosphere in a shaking water bath at 30°C for 5 h. The transformation was followed by thin-layer chromatography (TLC) on Silica Gel 60 plates (Merck, Darmstadt, Germany) performed with the following solvent system: ethyl acetate-ethanol-acetic acid-boric acid (saturated in water) (5:2:1:1, vol/vol/vol/vol). Spots were detected with diphenylamine-aniline-phosphoric acid reagent (30). To confirm the structure of the oxidation product, its tetra-O-acetyl-N,N-diphenylhydrazone derivative was prepared as previously described (49), and the 1H and 13C spectra were measured with a Varian INOVA-400 spectrometer at 400 MHz for 1H and at 100 MHz for 13C at 30°C. 1H nuclear magnetic resonance (1H-NMR) (CDCl3): 1.925 (3 H, s, Ac), 2.026 (3 H, s, Ac9), 2.197 (3 H, s, Ac), 2.201 (3 H, s, Ac), 4.204 (1 H, dd, J = 12.6, 4.2 Hz, H-6u), 4.425 (1 H, dd, J = 12.6, 2.3 Hz, H-6d), 5.402 (1 H, ddd, J = 9.5, 4.2, 2.3 Hz, H-5), 6.018 (1 H, dd, J = 9.5, 1.7 Hz, H-4), 6.631 (H, d, J = 1.7 Hz, H-3), 6.573 (1 H, s, H-1), 7.222–7.264 (8 H, m, Ph), 7.449 (2 H, br s, Ph). 13C-NMR (CDCl3): 20.47 q (2 C), 20.51 q, 20.79 q (4 × Ac), 61.26 t (C-6), 68.67 d, 69.65 d, 72.40 d, 130.65 d (C-1), 169.40 s, 169.46 s, 170.12 s, 170.51 s (4 × C⩵O), 190.13 s (C-2). Authentic 2-keto-d-glucose (d-arabino-2-hexosulose) was chemically prepared (18). Oxidation of other sugars by P2O was also performed, and product formation was judged by TLC.
FAD spectra.
FAD absorption spectra were recorded by using a DU 7400 (Beckman, Fullerton, Calif.) spectrophotometer with diode array detection at room temperature. The enzyme was reduced by using glucose (10 mM) or sodium dithionite (5 mM).
N terminus.
Purified P2O was electroblotted onto polyvinylidene difluoride membranes and was sequenced by automated Edman degradation with a LF3600D protein sequencer (Beckman) at the Department of Biochemistry, Charles University, Prague, Czech Republic.
Immunological studies.
Various amounts of purified P2O (0.25 to 250 mU) were separated by SDS-PAGE and then electrophoretically transferred onto a polyvinylidene difluoride membrane as described previously (11). Immunological cross-reactivity was tested after Western blotting by using polyclonal antibodies against P2O from P. chrysosporium similar to those described previously (12).
RESULTS
Purification.
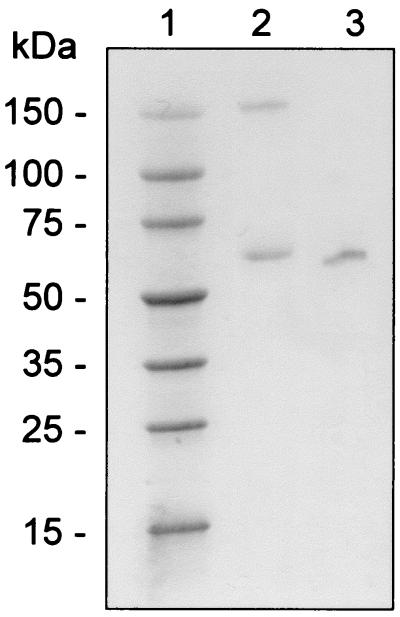
P2O was isolated from cell extracts of T. multicolor harvested from a fed-batch fermentation after approximately 300 h of cultivation, when P2O activity had reached its maximum. The results of a representative P2O purification procedure are summarized in Table 1. The enzyme was purified 16-fold from the crude mycelial extract with an overall yield of 80% and a specific activity of 12 U mg of protein−1 under the standard assay conditions when glucose and oxygen (air) were used as substrates. The four-step purification procedure yielded a protein that was apparently homogeneous, as judged by SDS-PAGE (Fig. 1) and analytical isoelectric focusing.
TABLE 1.
Purification of P2O from T. multicolor
| Purification step | Total activity (U) | Total protein (mg) | Sp act (U mg−1) | Purification (fold) | Recovery (%) |
|---|---|---|---|---|---|
| Mycelial extract | 5,000 | 6,800 | 0.74 | 1 | 100 |
| Ammonium sulfate fractionation | 4,800 | 2,900 | 1.7 | 2.2 | 96 |
| Phenyl Sepharose | 4,400 | 830 | 5.3 | 7.1 | 87 |
| Source 30Q | 4,300 | 370 | 12 | 16 | 85 |
| Superose 12 | 4,000 | 340 | 12 | 16 | 80 |
FIG. 1.
SDS-PAGE of purified, untreated, and intramolecularly cross-linked P2O from T. multicolor. Lane 1, recombinant molecular weight markers (Sigma); lane 2, P2O cross-linked with suberimidate; lane 3, untreated enzyme sample.
Properties.
The molecular mass of the native enzyme was determined both by gel filtration on Superose 12 and by analytical ultracentrifugation; the estimated Mrs were 262,000 and 270,000, respectively. MALDI analysis revealed an Mr for the monomer of 68,000, while SDS-PAGE produced a single band at 65,000 Da, which implies that the enzyme consists of four identical subunits. These results were confirmed by cross-linking studies that yielded two distinct peptide bands at 67,000 and 151,000 Da (Fig. 1). The tetrameric structure was corroborated by electron microscopic studies, which showed that the enzyme is composed of four elongated subunits symmetrically arranged around a central pore (Fig. 2). When it was assumed that the P2O subunit is an ellipsoid with axes of 4.4, 4.4, and 9.4 nm, an Mr of 69,600 was calculated, which is similar to the results obtained with the other methods (Table 2).
FIG. 2.
Transmission electron micrographs of negatively stained P2O from T. multicolor. Bar, 20 nm.
TABLE 2.
Determination of the molecular mass and subunit composition of P2O from T. multicolor
| Method of determination |
Mr
|
||
|---|---|---|---|
| Enzyme | Dimer | Subunit | |
| Gel filtration on Superose 12 | 262,000 | ||
| SDS gradient gel electrophoresis | 65,000 | ||
| Cross-linking studies | 151,000 | 67,000 | |
| Electron microscopy | 278,000 | 69,600 | |
| Equilibrium ultracentrifugation | 270,000 | ||
| MALDI–time-of-flight analysis | 68,000 | ||
The isoelectric point of P2O was 4.2, as determined by isoelectric focusing and comparison to standard proteins, as well as by chromatofocusing (data not shown). In contrast to reports concerning P2O from P. chrysosporium (2), isoelectric focusing of the homogeneous enzyme preparation produced only a single protein band and gave no indication of microheterogeneity of the enzyme.
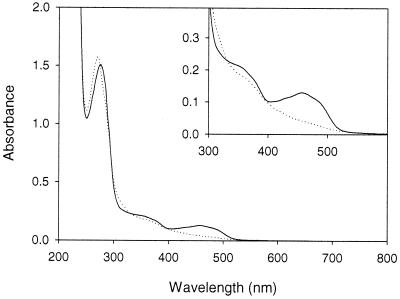
Homogeneous P2O preparations were bright yellow and produced a typical flavoprotein spectrum with absorption maxima at 456 and 345 nm (Fig. 3). Reduction of the enzyme by addition of sodium dithionite or glucose in the absence of oxygen resulted in disappearance of the peak at 456 nm. Treatment of the enzyme with 5% trichloroacetic acid at 100°C for 10 min did not release the flavin moiety from the enzyme, suggesting that this moiety is covalently linked to the protein. The identity of the enzyme-bound flavin was determined by MALDI mass spectrometry and NMR of a flavin-containing peptide obtained by proteolytic cleavage of P2O (P. Halada and P. Sedmera, unpublished data), and this molecule was unequivocally shown to be FAD. Based on a molar extinction coefficient for free FAD at 460 nm of 11.3 mM−1 cm−1 (40), one molecule of P2O was estimated to contain approximately four molecules of FAD. We stained P2O on SDS-PAGE gels with dansylhydrazine after treatment with periodate but observed no positive staining reaction, suggesting that P2O is not glycosylated. By using the phenol-sulfuric acid method a carbohydrate content of 0.5% was estimated.
FIG. 3.
Absorption spectra of the oxidized (solid line) and reduced (dotted line) states of P2O. Reduction was performed with glucose.
N-terminal sequence.
The N-terminal sequence of P2O was determined (Fig. 4), and it exhibited high sequence identity with the N-terminal sequence of P2O from T. versicolor (37). The amino terminus of the T. multicolor enzyme contains 10 additional amino acids that are almost identical to amino acids in the T. versicolor sequence but in T. versicolor are cleaved after translation. Apparently, cleavage of the signal leader sequence and proteolytic degradation are different for these two organisms. In contrast, there were no apparent homologies between the N-terminal sequences of P2Os from T. multicolor and P. chrysosporium (2). Furthermore, polyclonal rabbit antibodies against P. chrysosporium P2O did not cross-react with T. multicolor P2O, suggesting that there is no antigenic similarity between these two proteins.
FIG. 4.
Comparison of the N-terminal sequences of P2Os from T. multicolor and T. versicolor (37). The N-terminal sequences determined for the proteins are shown in uppercase letters; lowercase letters indicate a sequence of the T. versicolor enzyme that apparently is cleaved after synthesis of the protein.
Kinetic properties.
T. multicolor P2O oxidizes various sugar substrates (Table 3). The catalytic efficiencies (kcat/Km) for these substrates indicate that d-glucose clearly is the preferred substrate. C-2-oxidized reaction products, which were obtained by extended incubation of P2O with the appropriate substrates, were identified by using TLC and the diphenylamine-aniline reagent due to the characteristic blue color observed for l-idose, 3-deoxy-d-glucose, 6-deoxy-d-glucose, 1,6-anhydro-d-glucose, d-fucose, and 1,5-anhydro-d-sorbitol. This indicates that P2O is active with these sugars, but kinetic constants were not determined. In addition to the 1,6-linked disaccharides listed in Table 3, allolactose (Gal-β-1,6-Glc) and isomaltose (Glc-α-1,6-Glc) unequivocally served as substrates of P2O (33, 47); however, kinetic constants could not be determined because of small amounts of contaminating glucose in these disaccharides. Based on conversion experiments, the two β-linked disaccharides are better substrates than the two α-linked disaccharides (33). P2O did not oxidize the tetraose d-erythrose, the pentoses d-arabinose, l-xylose, d-lyxose, l-lyxose, and d-ribose, or the hexose sugars d-mannose, d-gulose, d-talose, l-glucose, 1-thio-d-glucose, α-methyl-d-glucopyranoside, and β-methyl-d-glucopyranoside. Furthermore, 1,4-linked disaccharides, such as cellobiose (Glc-β-1,4-Glc), lactose (Gal-β-1,4-Glc), xylobiose (Xyl-β-1,4-Xyl), and maltose (Glc-α-1,4-Glc), some of which are also formed during fungal degradation of lignocellulose polysaccharides, were not oxidized by P2O. While isomaltose was oxidized, albeit slowly, P2O did not react with isomaltotriose. This lack of activity with the sugars indicated above was confirmed under standard assay conditions, as well as by the absence of the oxidized product after prolonged incubation and aeration, as determined by TLC.
TABLE 3.
Apparent kinetic constants of P2O from T. multicolor for several electron donors
| Substrate | Vmax (μmol min−1 mg−1) | Relative activity (%) | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) |
|---|---|---|---|---|---|
| d-Glucose | 12 | 100 | 0.74 | 54 | 73 |
| 5-Thio-d-glucose | 2.9 | 24 | 3.9 | 13 | 3.3 |
| l-Sorbose | 12 | 99 | 38 | 53 | 1.4 |
| d-Xylose | 6.7 | 56 | 30 | 30 | 1.0 |
| d-Glucono-1,5-lactone | 7.6 | 64 | 38 | 34 | 0.91 |
| d-Allose | 4.5 | 38 | 36 | 20 | 0.57 |
| d-Galactose | 0.68 | 5.7 | 9.2 | 3.1 | 0.33 |
| Gentiobiose | 2.7 | 23 | 62 | 12 | 0.19 |
| d-Mannoheptose | 0.98 | 8.2 | 110 | 4.4 | 0.042 |
| Melibiose | 1.0 | 8.6 | 120 | 4.6 | 0.038 |
| l-Arabinose | 0.18 | 1.5 | 97 | 0.81 | 0.008 |
Not only was P2O nonspecific with respect to the electron donor used, but in addition to oxygen it could also transfer electrons to a number of different compounds, mainly substituted benzoquinones (Table 4). Replacing an increasing number of hydrogens of 1,4-benzoquinone with methyl groups resulted in gradual decreases in both the reaction velocity and the substrate affinity; the latter was shown by increasing Michaelis constants. Both methyl-1,4-benzoquinone and dimethyl-1,4-benzoquinone were reduced by P2O at considerable rates, which, however, were lower than that determined for the unsubstituted 1,4-benzoquinone. Tetramethyl-1,4-benzoquinone (duroquinone) was reduced only very slowly, and the rates were too low to determine kinetic constants. In contrast, replacing hydrogens with electron-withdrawing groups, such as the various halide groups, increased the rate of the P2O-catalyzed reaction significantly compared to the unsubstituted 1,4-benzoquinone rate. No P2O activity was observed with 2,3-dimethoxy-5-methyl-1,4-benzoquinone, tetrahydroxy-1,4-benzoquinone, 1,4-naphthoquinone, or anthraquinone.
TABLE 4.
Apparent kinetic constants of P2O from T. multicolor for several electron acceptors, as determined at 30°C by using 100 mM glucose as the substrate under standard assay conditions (pH 6.5) unless otherwise indicated
| Substrate | Vmax (μmol min−1 mg−1) | Relative activity (%) | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) |
|---|---|---|---|---|---|
| Oxygen | 16 | 100 | 0.090 | 71 | 790 |
| 1,4-Benzoquinone | 48 | 300 | 0.31 | 210 | 690 |
| 1,4-Benzoquinone (pH 4.5) | 60 | 380 | 0.30 | 270 | 900 |
| Methyl-1,4-benzoquinone | 40 | 250 | 0.40 | 180 | 440 |
| Methyl-1,4-benzoquinone (pH 4.5) | 52 | 330 | 0.35 | 230 | 670 |
| 2,6-Dimethyl-1,4-benzoquinone | 27 | 170 | 2.1 | 120 | 58 |
| 2,6-Dimethyl-1,4-benzoquinone (pH 4.5) | 47 | 290 | 0.83 | 210 | 250 |
| Tetramethyl-1,4-benzoquinone (pH 4.5) | <1 | ||||
| Tetrafluoro-1,4-benzoquinone | 140 | 900 | 0.22 | 640 | 2,900 |
| Tetrachloro-1,4-benzoquinone | 170 | 1100 | 0.088 | 750 | 8,600 |
| Tetrabromo-1,4-benzoquinone | 63 | 400 | 0.090 | 280 | 3,100 |
| DCIPa | 4.4 | 28 | 0.065 | 20 | 300 |
| DCIP (pH 4.5) | 34 | 210 | 0.094 | 150 | 1,600 |
| ABTS cation radicalb | 4.8 | 30 | 0.070 | 21 | 310 |
DCIP, 2,6-dichloroindophenol.
The cation radical was prepared by oxidizing ABTS with laccase; the enzyme was removed by ultrafiltration, and the radical was quantified by using its extinction coefficient.
Identification of the reaction product obtained with d-glucose.
The enzymatic oxidation product obtained with d-glucose exhibited the same mobility and characteristic blue color on TLC (Rf, 0.28; d-glucose Rf, 0.47) as authentic 2-keto-d-glucose. The 1H-NMR and 13C-NMR spectra of the tetra-O-acetyl-N,N-diphenylhydrazone derivative of the oxidation product were identical to previously published spectra for the tetra-O-acetyl-N,N-diphenylhydrazone derivative of 2-keto-d-glucose (49). The data identified the reaction product obtained with the purified oxidase from T. multicolor with d-glucose as 2-keto-d-glucose (2-dehydro-d-glucose, d-arabino-hexos-2-ulose). In summary, the enzyme acted at C-2 of the substrate as pyranose:oxygen 2-oxidoreductase (EC 1.1.3.10) does.
pH and temperature dependence of activity and stability.
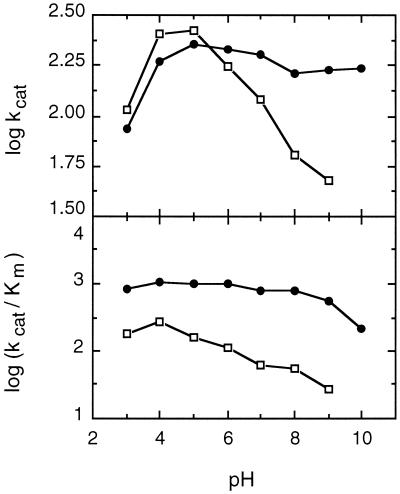
The pH dependence of P2O activity was determined for six electron acceptors (Fig. 5): oxygen, 1,4-benzoquinone, 2,6-dimethyl-1,4-benzoquinone, methyl-1,4-benzoquinone, tetrachloro-1,4-benzoquinone, and 2,6-dichloroindophenol. The optimum pH values varied between 4 and 8 depending on the electron acceptor employed. When oxygen or 1,4-benzoquinone was used, the enzyme exhibited a broad optimum pH range; maximum activity occurred at pH 5.5 to 6.5, and more than 80% of the maximum activity occurred at pH 4.0 to 7.0. A similar broad maximum-activity pH range, pH 6.0 to 8.0, was obtained for tetrachloro-1,4-benzoquinone. In contrast, rather sharp activity maxima occurred at pH 4.0 and 4.5 for 2,6-dimethyl-1,4-benzoquinone and 2,6-dichloroindophenol, respectively. The pH profiles of the apparent kcat and kcat/Km values for two electron acceptors, 1,4-benzoquinone and 2,6-dimethyl-1,4-benzoquinone, are shown in Fig. 6. For both substrates the apparent kcat increased when the pH was changed from 3 to 5. While kcat did not change much from pH 5 to 10 for 1,4-benzoquinone, it decreased considerably as the pH increased for 2,6-dimethyl-1,4-benzoquinone. The activity with the latter substrate at pH 10 was so low that kinetic constants could not be measured accurately. A lower pH seemed to favor binding of both substrates, as the apparent Km values decreased as the pH decreased from 5 to 3. Km was relatively constant over the pH range from 5 to 9 for both substrates, but it increased more than threefold at pH 10 for 1,4-benzoquinone. As a result, the pH profile of the catalytic efficiency (kcat/Km) for 1,4-benzoquinone did not vary much over the entire pH range examined and decreased at the highest pH measured only because of the unfavorable Km. In contrast, kcat/Km for 2,6-dimethyl-1,4-benzoquinone had a distinct maximum at pH 4 and decreased as the pH increased.
FIG. 5.
Effect of pH on the activity of P2O from T. multicolor in the presence of different electron acceptors. (A) Oxygen (approximately 250 μM, standard assay conditions); (B) 1,4-benzoquinone (1 mM, detection at 300 nm); (C) 2,6-dimethyl-1,4-benzoquinone (1 mM, detection at 332 nm); (D) methyl-benzoquinone (1 mM, detection at 324 nm); (E) tetrachloro-1,4-benzoquinone (250 μM, detection at 300 nm); (F) dichloroindophenol (250 μM, detection at 520 nm). The buffers used were 100 mM citrate (●),100 mM phosphate (□), and 100 mM borate (▴).
FIG. 6.
pH profiles of the apparent kcat and kcat/Km values for two electron acceptors of P2O. The substrates studied were 1,4-benzoquinone (●) and 2,6-dimethyl-1,4-benzoquinone (□).
The reaction velocity of P2O-catalyzed oxidation of glucose increased as the temperature increased to a maximum at 55°C (3 min, standard assay conditions). From the linear portion of the Arrhenius plot an activation energy of 30 kJ mol−1 was calculated.
pH stability was studied in 100 mM sodium citrate at 30°C in the absence of substrate. The enzyme was extremely stable at pH 5.0 to 7.0, with half-lives of activity of more than 500 days, while at lower pH values stability decreased significantly. The activity half-lives at pH 3.0, 3.5, 4.0, and 4.5 were 13, 52, 130, and 350 days, respectively.
DISCUSSION
We purified an intracellular flavin-containing oxidase from the wood-degrading basidiomycete T. multicolor that had significant activity with d-glucose. Based on identification of the reaction product obtained with d-glucose, this oxidase can be classified as a P2O (EC 1.1.3.10) similar to the enzyme that was first described from the fungus P. obtusus (27, 38) and oxidizes pyranose sugars at position C-2. T. multicolor P2O has a number of properties that are very similar to those reported for P2Os from other sources (21). It is a homotetrameric flavoenzyme with an Mr of 270,000 (the Mrs of other P2Os are 250,000 to 323,000) and contains one covalently bound FAD per 68,000-Mr subunit. An exception to this general structure seems to be the enzyme from P. gigantea, which is also tetrameric but consists of two different subunits (39). In general, P2Os have little or no glycosylation. For the T. multicolor enzyme, the dansylhydrazine staining method gave negative results, but a positive reaction was obtained with the phenol-sulfuric acid method, although this method indicated that the carbohydrate content is very low. Thus, the T. multicolor P2O is not highly glycosylated if it is glycosylated at all.
We found good general agreement between the data for T. multicolor P2O and the data for P2Os from other sources with respect to sugar substrate specificity, although there are some distinct differences regarding reactivity with some carbohydrates. In general, monosaccharides in pyranose form with an equatorially oriented hydroxyl group at position C-2, such as that found in d-xylose, d-allose, d-galactose, d-glucose, or d-mannoheptose, are oxidized by T. multicolor P2O, while axial orientation of this hydroxyl group results in complete loss of activity. This property is evident with d-mannose or d-altrose and has been reported previously for other enzymes (20, 27). The specificity of T. multicolor P2O and the specificity of the P2O from P. gigantea, which probably is best studied with respect to its substrate specificity, differ significantly. The P. gigantea enzyme does not oxidize the α-1,6-linked disaccharides melibiose and isomaltose, while T. multicolor P2O does (47).We did not observe the hydrolytic activity reported for the P. gigantea enzyme with T. multicolor P2O even when it was incubated with potentially susceptible sugars, such as maltose or cellobiose, for an extended period of time.
The most unusual property of T. multicolor P2O, however, is its almost complete lack of activity with 2-deoxy-d-glucose. When we incubated T. multicolor P2O under standard assay conditions, we observed no activity with this substrate, and only after prolonged incubation of the sugar with P2O under aerobic conditions was a weak spot for a possible reaction product detected by TLC. Presumably, T. multicolor P2O has only very low activity with 2-deoxy-d-glucose, and this activity is too low to determine kinetic data or relative activities. These results are quite different from those obtained with enzymes from P. ostreatus, P. chrysosporium, and P. gigantea, which exhibit 38, 25, and 52% of the activity observed with the preferred substrate, d-glucose, respectively, under these conditions (2, 20, 40). With the P. gigantea enzyme 2-deoxy-d-glucose is regioselectively oxidized at position C-3, yielding 2-deoxy-d-erythro-hexos-3-ulose (20). Oxidation at the C-3 position of hexoses was observed for P2Os from O. mucida, P. chrysosporium, and T. versicolor (48), which oxidize the primary reaction product 2-keto-d-glucose at position C-3 to give 2,3-diketo-d-glucose (d-erythro-hexos-2,3-diulose). We did not observe a corresponding reaction with the T. multicolor enzyme even after extended incubation with 2-keto-d-glucose under aerobic conditions. The lack of oxidation at position C-3 by T. multicolor P2O is an advantage when the conversion of pyranose sugars to their corresponding 2-keto derivatives is desired, such as in preparative biotransformations. The primary oxidation product of d-glucose has recently attracted increased attention due to its potential use as an intermediate in the production of aldose-free fructose (22, 25, 34).
Another property that could make the T. multicolor enzyme an attractive biocatalyst for applied biotransformations is the low Michaelis constant for oxygen, 0.090 mM (corresponding to a pO2 of approximately 0.08 atm), which is lower than the values of 0.125 and 0.65 mM reported for P. chrysosporium and P. gigantea P2Os, respectively (2, 13). Considering the low solubility of oxygen in aqueous solutions under assay conditions, approximately 0.24 mM (3), P2O typically is employed in these applications with nonsaturating concentrations of its cosubstrate.
One aspect of P2O that has not been studied yet in any detail is substrate specificity with respect to the electron acceptor. In addition to oxygen, T. multicolor P2O readily reduces 1,4-benzoquinone, various substituted benzoquinones, structural analogues of benzoquinone such as 2,6-dichloroindophenol, and even the ABTS cation radical. The activity of P2O with some of these quinoid electron acceptors is similar to or greater than the activity with oxygen when the catalytic efficiencies (kcat/Km) are compared. The quinone-reducing property of P2O could play a significant role in lignin degradation in addition to the proposed role of providing hydrogen peroxide for the extracellular heme peroxidases. Substituted quinones are formed by these peroxidases when they act on lignin and also when various anthropogenic aromatic pollutants are oxidized. In fact, benzoquinones may be key intermediates in the degradation of aromatic compounds, and quinone metabolism is an important step in lignin degradation (7, 29, 42). Lignin degradation requires reduction of quinones (6, 43), which can be mediated by different enzyme systems. Both intracellular and extracellular enzymes, as well as a plasma membrane system, have been suggested to be responsible for this reaction (1, 6, 7, 41). The relatively high nonspecificity of P2O both with respect to its quinoid electron acceptor and with respect to its sugar electron donor (many electron donors are formed during fungal degradation of lignocellulose) suggests that one of the functions of this enzyme could be reduction of quinones in the periplasm or even in the extracellular environment. It also is possible that P2O has two functions. It may be a source of hydrogen peroxide, which is a cosubstrate for peroxidases, and it could reduce the quinones that result from the H2O2-dependent, catalytic action of the peroxidases. This hypothesis is consistent with the higher activity of P2O with quinones than with oxygen, since hydrogen peroxide is produced only when the toxic quinones have already been reduced by P2O.
Reduction of benzoquinones also is an important feature of extracellular quinone redox cycling, which may be involved in lignin degradation via production of superoxide anion radicals (23). The enzyme that reduces quinones was not identified in a study of quinone redox cycling (23), but based on the fact that hydroquinones are found extracellularly, it has been suggested that such a quinone-reducing enzyme should be located in the periplasm or bound to the plasma membrane. Because of this localization and the nonspecific nature of the quinone-reducing system described previously (23), possible candidates for such an enzyme activity are a plasma membrane system (41) and the periplasmatic enzyme P2O.
In conclusion, the flavoenzyme P2O isolated from the wood-degrading fungus T. multicolor has numerous similarities to enzymes described from other fungi (e.g., P. gigantea or P. chrysosporium). Some of its properties, such as its high affinity for oxygen and its almost complete lack of activity with 2-deoxy-d-glucose, which is a common feature of other P2Os, should make the T. multicolor P2O an attractive biocatalyst for synthesis of 2-keto-d-glucose; the latter compound can be used as an intermediate for production of aldose-free fructose (34) or other ketoaldoses that are of interest for carbohydrate chemistry (21, 22).
ACKNOWLEDGMENTS
We thank Hansjörg Prillinger (Universität für Bodenkultur Wien) for the fungal strain, Paul Messner (Universität für Bodenkultur Wien) for help with the electron microscopy, Petr Sedmera (Institute of Microbiology AS CR, Prague, Czech Republic) for the NMR measurements, Petr Halada (Institute of Microbiology AS CR, Prague, Czech Republic) for the MALDI measurements, Klaus D. Kulbe (Universität für Bodenkultur Wien) for his encouragement and interest in our work, and Therese Jensen and Harald Frießnegg for technical assistance.
This work was supported by grants from the Austrian Science Foundation (FWF project P 11459-MOB), the Austrian Federal Ministry of Education, Science and the Arts (WTZ Austria-Czech Republic project 2000/14), and the Grant Agency of the Czech Republic (project 206/99/1191).
REFERENCES
- 1.Ander P, Marzullo L. Sugar oxidoreductases and veratryl alcohol oxidase as related to lignin degradation. J Biotechnol. 1997;53:115–131. doi: 10.1016/s0168-1656(97)01680-5. [DOI] [PubMed] [Google Scholar]
- 2.Artolozaga M J, Kubátová E, Volc J, Kalisz H M. Pyranose 2-oxidase from Phanerochaete chrysosporium—further biochemical characterisation. Appl Microbiol Biotechnol. 1997;47:508–514. doi: 10.1007/s002530050964. [DOI] [PubMed] [Google Scholar]
- 3.Bailey J E, Ollis D F. Biochemical engineering fundamentals. New York, N.Y: McGraw-Hill; 1986. [Google Scholar]
- 4.Baute M-A, Baute R, Deffieux G, Filleau M-J. Conversion of glucose to cortalcerone via glucosone by Corticium caeruleum. Phytochemistry. 1977;16:1895–1897. [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Brock B J, Rieble S, Gold M H. Purification and characterization of a 1,4-benzoquinone reductase from the basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1995;61:3076–3081. doi: 10.1128/aem.61.8.3076-3081.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Constam D, Muheim A, Zimmermann W, Fiechter A. Purification and partial characterization of an intracellular NADH:quinone oxidoreductase from Phanerochaete chrysosporium. J Gen Microbiol. 1991;137:2209–2214. [Google Scholar]
- 8.Daniel G. Use of electron microscopy for aiding our understanding of wood biodegradation. FEMS Microbiol Rev. 1994;13:199–233. [Google Scholar]
- 9.Daniel G, Nilsson T, Pettersson B. Intra- and extracellular localization of lignin peroxidase during degradation of solid wood and wood fragments by Phanerochaete chrysosporium by using transmission electron microscopy and immunogold labeling. Appl Environ Microbiol. 1989;55:871–881. doi: 10.1128/aem.55.4.871-881.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel G, Pettersson B, Nilsson T, Volc J. Use of immunogold cytochemistry to detect Mn(II)-dependent and lignin peroxidase in wood degraded by the white rot fungi P. chrysosporium and Lentulina edodes. Can J Bot. 1990;68:920–933. [Google Scholar]
- 11.Daniel G, Volc J, Kubatova E. Pyranose oxidase, a major source of H2O2 during wood degradation by Phanerochaete chrysosporium, Trametes versicolor, and Oudemansiella mucida. Appl Environ Microbiol. 1994;60:2524–2532. doi: 10.1128/aem.60.7.2524-2532.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniel G, Volc J, Kubatova E, Nilsson T. Ultrastructural and immunocytochemical studies on the H2O2-producing enzyme pyranose oxidase in Phanerochaete chrysosporium grown under liquid culture conditions. Appl Environ Microbiol. 1992;58:3667–3676. doi: 10.1128/aem.58.11.3667-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danneel H-J, Rössner E, Zeeck A, Giffhorn F. Purification and characterization of a pyranose oxidase from the basidiomycete Peniophora gigantea and chemical analyses of its reaction products. Eur J Biochem. 1993;214:795–802. doi: 10.1111/j.1432-1033.1993.tb17982.x. [DOI] [PubMed] [Google Scholar]
- 14.Danneel H-J, Ullrich M, Giffhorn F. Goal-oriented screening method for carbohydrate oxidases produced by filamentous fungi. Enzyme Microb Technol. 1992;14:898–903. [Google Scholar]
- 15.Davies G E, Stark G R. Use of dimethyl suberimidate, a cross-linking reagent, in studying the subunit structure of oligomeric proteins. Proc Natl Acad Sci USA. 1970;66:651–656. doi: 10.1073/pnas.66.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubois M, Gilles K A, Hamilton J K, Rebers P A, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- 17.Eckhardt A E, Hayes C E, Goldstein I J. A sensitive fluorescent method for the detection of glycoproteins in polyacrylamide gels. Anal Biochem. 1976;73:192–197. doi: 10.1016/0003-2697(76)90154-8. [DOI] [PubMed] [Google Scholar]
- 18.Fischer E. Über die Verbindung des Phenylhydrazins mit den Zuckerarten. Ber Dtsch Chem Ges. 1889;22:87–97. [Google Scholar]
- 19.Forney L J, Reddy C A, Pankratz H S. Ultrastructural localization of hydrogen peroxide production in ligninolytic Phanerochaete chrysosporium cells. Appl Environ Microbiol. 1982;44:732–736. doi: 10.1128/aem.44.3.732-736.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freimund S, Huwig A, Giffhorn F, Köpper S. Rare keto-aldoses from enzymatic oxidation: substrates and oxidation products of pyranose 2-oxidase. Chem Eur J. 1998;4:2442–2455. [Google Scholar]
- 21.Giffhorn F. Fungal pyranose oxidases: occurrence, properties and biotechnical applications in carbohydrate chemistry. Appl Microbiol Biotechnol. 2000;54:727–740. doi: 10.1007/s002530000446. [DOI] [PubMed] [Google Scholar]
- 22.Giffhorn F, Köpper S, Huwig A, Freimund S. Rare sugars and sugar-based synthons by chemo-enzymatic synthesis. Enzyme Microb Technol. 2000;27:734–742. doi: 10.1016/s0141-0229(00)00293-3. [DOI] [PubMed] [Google Scholar]
- 23.Guillén F, Martínez M J, Munõz C, Martínez A T. Quinone redox cycling in the ligninolytic fungus Pleurotus eryngii leading to extracellular production of superoxide anion radical. Arch Biochem Biophys. 1997;339:190–199. doi: 10.1006/abbi.1996.9834. [DOI] [PubMed] [Google Scholar]
- 24.Highley T L, Murmanis L. Determination of hydrogen peroxide production in Coriolus versicolor and Poria placenta during wood degradation. Mater Org (Berlin) 1985;20:251–252. [Google Scholar]
- 25.Huwig A, Danneel H-J, Giffhorn F. Laboratory procedure for producing 2-keto-d-glucose, 2-keto-d-xylose and 5-keto-d-fructose from d-glucose, d-xylose and l-sorbose with immobilized pyranose oxidase of Peniophora gigantea. J Biotechnol. 1994;32:309–315. [Google Scholar]
- 26.Izumi Y, Furuya Y, Yamada H. Purification and properties of pyranose oxidase from basidiomycetous fungus no. 52. Agric Biol Chem. 1990;54:1393–1399. [Google Scholar]
- 27.Janssen F W, Ruelius H W. Carbohydrate oxidase, a novel enzyme from Polyporus obtusus. II. Specificity and characterization of reaction products. Biochim Biophys Acta. 1968;167:501–510. doi: 10.1016/0005-2744(68)90040-5. [DOI] [PubMed] [Google Scholar]
- 28.Janssen F W, Ruelius H W. Pyranose oxidase from Polyporus obtusus. Methods Enzymol. 1975;41:170–173. doi: 10.1016/s0076-6879(75)41041-2. [DOI] [PubMed] [Google Scholar]
- 29.Joshi D K, Gold M H. Degradation of 2,4,5-trichlorophenol by the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1993;59:1779–1785. doi: 10.1128/aem.59.6.1779-1785.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kocourek J, Ticha M, Kostir J, Jensovsky L. The use of diphenylamine-aniline-phosphoric acid reagent in the detection and differentiation of monosaccharides and their derivatives on paper chromatogram. J Chromatogr. 1966;14:228–231. doi: 10.1016/s0021-9673(01)98109-9. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Leitner C, Haltrich D, Nidetzky B, Prillinger H, Kulbe K D. Production of a novel pyranose 2-oxidase by basidiomycete Trametes multicolor. Appl Biochem Biotechnol. 1998;70–72:237–248. doi: 10.1007/978-1-4612-1814-2_23. [DOI] [PubMed] [Google Scholar]
- 33.Leitner C, Mayr P, Riva S, Volc J, Kulbe K D, Nidetzky B, Haltrich D. Enzymatic redox isomerization of 1,6-disaccharides by pyranose oxidase and NADH-dependent aldose reductase. J Mol Catal B. 2001;11:417–424. [Google Scholar]
- 34.Leitner C, Neuhauser W, Volc J, Kulbe K D, Nidetzky B, Haltrich D. The Cetus process revisited: a novel enzymatic alternative for the production of aldose-free d-fructose. Biocatal Biotrans. 1998;16:365–382. [Google Scholar]
- 35.Machida Y, Nakanishi T. Purification and properties of pyranose oxidase from Coriolus versicolor. Agric Biol Chem. 1984;48:2463–2470. [Google Scholar]
- 36.Michal G, Möllering H, Siedel J. Chemical design of indicator reactions for the visible range. In: Bergmeyer H U, editor. Methods of enzymatic analysis. I. Weinheim, Germany: Verlag Chemie; 1983. pp. 197–232. [Google Scholar]
- 37.Nishimura I, Okada K, Koyama Y. Cloning and expression of pyranose oxidase cDNA from Coriolus versicolor in Escherichia coli. J Biotechnol. 1996;52:11–20. doi: 10.1016/s0168-1656(96)01618-5. [DOI] [PubMed] [Google Scholar]
- 38.Ruelius H W, Kerwin R M, Janssen F W. Carbohydrate oxidase, a novel enzyme from Polyporus obtusus. I. Isolation and purification. Biochim Biophys Acta. 1968;167:493–500. doi: 10.1016/0005-2744(68)90039-9. [DOI] [PubMed] [Google Scholar]
- 39.Schäfer A, Bieg S, Huwig A, Kohring G-W, Giffhorn F. Purification by immunoaffinity chromatography, characterization, and structural analysis of a thermostable pyranose oxidase from the white rot fungus Phlebiopsis gigantea. Appl Environ Microbiol. 1996;62:2586–2592. doi: 10.1128/aem.62.7.2586-2592.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin K S, Youn H D, Han Y H, Kang S O, Hah Y C. Purification and characterization of d-glucose-oxidase from white-rot fungus Pleurotus ostreatus. Eur J Biochem. 1993;215:747–752. doi: 10.1111/j.1432-1033.1993.tb18088.x. [DOI] [PubMed] [Google Scholar]
- 41.Stahl J D, Rasmussen S J, Aust S D. Reduction of quinones and radicals by a plasma membrane redox system of Phanerochaete chrysosporium. Arch Biochem Biophys. 1995;322:221–227. doi: 10.1006/abbi.1995.1455. [DOI] [PubMed] [Google Scholar]
- 42.Tuor U, Wariishi H, Schoemaker H E, Gold M H. Oxidation of phenolic arylglycerol β-aryl ether lignin model compounds by manganese peroxidase from Phanerochaete chrysosporium: oxidative cleavage of an α-carbonyl model compound. Biochemistry. 1992;31:4986–4995. doi: 10.1021/bi00136a011. [DOI] [PubMed] [Google Scholar]
- 43.Valli K, Gold M H. Degradation of 2,4-dichlorophenol by the lignin-degrading fungus Phanerochaete chrysosporium. J Bacteriol. 1991;173:345–352. doi: 10.1128/jb.173.1.345-352.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volc J, Denisova N P, Nerud F, Musílek V. Glucose-2-oxidase activity in mycelia cultures of basidiomycetes. Folia Microbiol. 1985;30:141–147. doi: 10.1007/BF02922207. [DOI] [PubMed] [Google Scholar]
- 45.Volc J, Eriksson K-E. Pyranose 2-oxidase from Phanerochaete chrysosporium. Methods Enzymol. 1988;161:316–322. [Google Scholar]
- 46.Volc J, Kubátová E, Sedmera P, Daniel G, Gabriel J. Pyranose oxidase and pyranosone dehydratase: enzymes responsible for conversion of d-glucose to cortalcerone by the basidiomycete Phanerochaete chrysosporium. Arch Microbiol. 1991;156:297–301. [Google Scholar]
- 47.Volc J, Leitner C, Sedmera P, Halada P, Haltrich D. Enzymatic formation of dicarbonyl sugars: C-2 oxidation of 1Δ6 disaccharides gentiobiose, isomaltose and melibiose by pyranose 2-oxidase from Trametes multicolor. J Carbohydr Chem. 1999;18:999–1007. [Google Scholar]
- 48.Volc J, Sedmera P, Havlícek V, Prikrylová V, Daniel G. Conversion of d-glucose to d-erythro-hexos-2,3-diulose (2,3-diketo-d-glucose) by enzyme preparations from the basidiomycete Oudemansiella mucida. Carbohydr Res. 1995;278:59–70. [Google Scholar]
- 49.Volc J, Sedmera P, Musilek V. Conversion of monosaccharides into their corresponding 2-glycosuloses by intact cells of the basidiomycete Oudemansiella mucida. Collect Czech Chem Commun. 1979;45:950–955. [Google Scholar]
- 50.Yphantis D A. Equilibrium ultracentrifugation of dilute solutions. Biochemistry. 1964;3:297–317. doi: 10.1021/bi00891a003. [DOI] [PubMed] [Google Scholar]
- 51.Zipper P, Kratky O, Herrmann R, Hohn T. An x-ray small angle study of the bacteriophages fr and R17. Eur J Biochem. 1971;18:1–9. doi: 10.1111/j.1432-1033.1971.tb01206.x. [DOI] [PubMed] [Google Scholar]