1.
Betacoronavirus is responsible for three outbreaks in 21st century causing worldwide epidemics and pandemics. Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) remains the major global concern affecting millions in most countries. As of December 13, 2021, 269,468,311 cases of coronavirus disease (COVID‐19) have been confirmed in >200 countries including 5304,248 deaths caused by SARS‐CoV‐2‐induced inflammatory infections or other complications (https://covid19.who.int/). 1 With advances in vaccine development, the scientific community has developed a portfolio of efficient vaccines and disease control strategies to combat the pandemic. Nonetheless, uncertainties remain about the emergence of new variants that may have higher infectivity, transmissibility, or virulence compared to its ancestral strains or other variants. RNA viruses are known for high rates of genetic variation to adapt to changing environmental conditions. The low genome stability, short replication time, and high mutation rates of viral RNA genome led to deleterious fitness effects that drive towards viral evolution and genetic material variability enabling the virus to develop resistance and escape host immunity. 2 As an RNA virus, SARS‐CoV‐2 has continued to spread and evolve into new variants with each new variant possibly having different properties including transmissibility, disease severity, response to therapy and most concern is the ability for the virus to evade natural or vaccine‐mediated immunity. 3 These challenges highlight the urgent need to stop the spread and evolution of SARS‐CoV‐2.
Currently, the world is witnessing the emergence of SARS‐CoV‐2 variants with multiple spike protein mutations. Despite the massive vaccination campaign globally, the continuous evolution, adaptation and high human transmission capability of SARS‐CoV‐2 leads to the increasing frequency of the variants worldwide. The identification of novel variants has led to a new chapter of the pandemic. SARS‐CoV‐2 Delta variant is the currently most dominant circulating strain worldwide. 4 Omicron (B 1.1.529) is the recently identified variant that joins the list of other four Variants of Concern (VOC). South Africa first reported the variant B 1.1.529 to World Health Organization (WHO) on November 24, 2021. 5 From November, COVID infections in South Africa have shown a sharp increase in almost all provinces, coinciding with the detection of this variant. WHO categorized B 1.1.529 (Omicron) as VOCs on November 26, 2021, following the advice of the Technical Advisory Group on SARS‐CoV‐2 Virus Evolution. 6 WHO is closely monitoring the Omicron variant due to the concerning mutations in spike protein and also this variant is being linked to an increasing number of cases in most of the provinces in South Africa in recent weeks. Further, the virus has been reported in >60 countries including Botswana, Hong Kong, Australia, United Kingdom, Canada, India, and European Union. 7 , 8 Europe's first case was reported in Belgium on November 26, 2021. 9
The sequences of Omicron are currently available in GISAID EpiCoV. 8 The phylogenic relationship based on virus sequence alignment with other VOCs revealed that the Omicron might be closely related to the Alpha variant. 10 At the time of writing, more than 30 mutations have been identified in the SARS‐CoV‐2 spike protein with >15 in the receptor‐binding domain (RBD), which is an immunodominant region responsible for virus entry into the cells (Figure 1 and Table 1). Several mutations including three small deletions and one small insertion in the spike region were identified from the available genome sequences so far: A67V, Δ69‐70, T95I, G142D/Δ143‐145, Δ211/L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F. The other identified mutations in the genome include E: T9I; M: D3G, Q19E, A63T; N: R203K, G204R. 11 The number of mutations may increase once the genome sequences from different countries are available. D614G mutation was commonly identified in all VOCs. N501Y mutation located on the RBD is common in VOCs including Omicron except the Delta variant. Some of the common mutations include Δ69‐70, T95I, Δ144, E484A, K417N, T478K, N501Y, D614G, H655Y, and P681H are also reported in other variants (Table 1). Of note, the mutation reported in other variants such as Δ69‐70, G142D/Δ143‐145, K417N, T478K, N501Y, D614G, P681H are associated with increased binding affinity and binding tightness of virus to ACE2 receptor thereby increasing the chance of virus infection or immune evasion or increased infectivity or higher risk of hospitalization. 12 , 13 , 14 , 15
Figure 1.
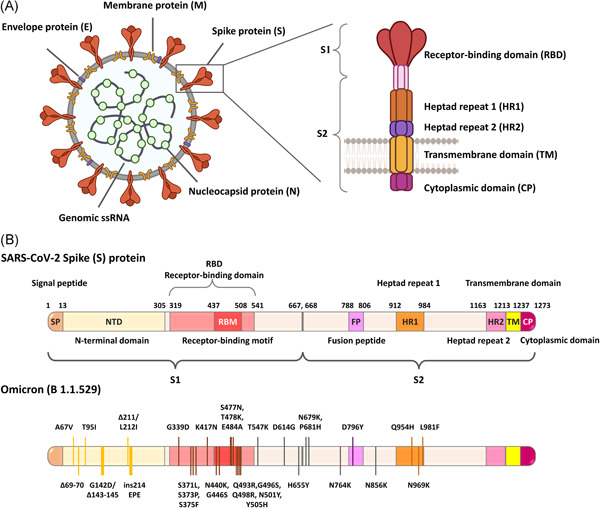
The genomic structure of SARS‐CoV‐2 showing the structural proteins such as spike, envelope, membrane, nucleocapsid (A); Schematic illustration showing the arrangement of domains present in the SARS‐CoV‐2 spike glycoprotein; The identified mutations and locations of amino acid substitutions in spike protein of VOC Omicron are also presented. More than 30 mutations have been identified including three small deletions and one small insertion on the spike region, of which 15 mutations were reported in RBD. Different domains are represented in different colors (B)
Table 1.
| WHO label | SARS‐CoV‐2 Genome | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | N | E | M | ORF1a | ORF1b | ORF3a | ORF6 | ORF7a | ORF7b | ORF8 | ORF10 | |
| Variants of concern | ||||||||||||
| Alpha | Δ69/70, Δ143/144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H |
D3L R203K G204R S235F |
– | – |
T1001I A1708D I2230T Δ3675/3677 |
P314L | – | – | – | – |
Q27* R52I K68* Y73C S84L |
– |
| Beta | L18F, D80A, D215G, Δ241/243, K417N, E484K, N501Y, D614G, A701V | T205I | P71L | – |
T265I K1655N K3353R Δ3675/3677 |
P314L |
Q57H S171L |
– | – | – | S84L | – |
| Gamma | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F |
P80R R203K G204R |
– | – |
S1188L K1795Q Δ3675/3677 |
P314L E1264D |
S253P | – | – | – |
S84L E92K |
– |
| Delta | T19R, G142D, E156G, Δ157/158, L452R, T478K, D614G, P681R, D950N |
D63G R203M D377Y |
– | I82T |
T3255I T3646A |
P314L G662S P1000L |
S26L | – |
V82A T120I |
T40I |
S84L Δ119/120 |
– |
| Omicron |
A67V, Δ69/70, T95I, G142D Δ143/145, Δ211, L212I ins214 EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F |
R203K G204R |
T9I |
D3G Q19E A63T |
K856R S2083I Δ2084/2084 A2710T T3255I P3395H Δ3674/3676 I3758V |
P314L I1566V |
– | – | – | – | S84L | – |
| Variants of interest | ||||||||||||
| Lambda | G75V, T76I, R246N, Δ247/253, L452Q, L452R, D614G, T859N |
P13L R203K G204R G214C |
– | – |
T1246I P2287S F2387V L3201P T3255I G3278S Δ3675/3677 |
P314L | – | – | – | – | S84L | – |
| Mu | T95I, Y144S, Y145N, R346K, E484K, N501Y, D614G, P681H, D950N | T205I | – | – |
T1055A T1538I T3255I Q3729R |
P314L P1342S |
Q57H Δ256/257 |
– | – | – |
T11K P38S S67F S84L |
– |
Note: The molecular location and the distribution of mutations in the virus genome is provided. Mutations in the S protein are mainly associated with immune escape or increased ACE2 binding affinity.
Abbreviations: E, envelope; M, membrane, N, nucleocapsid, ORF, open reading frame; S, spike.
Still the transmissibility, disease severity, and vaccine effectiveness of Omicron compared to ancestral strain and other variants are still unknown making it unclear, whether Omicron is more transmissible than the highly transmissible Delta, given the low number of these infections were detected worldwide so far. No unusual symptoms were identified so far in the persons infected with this variant and some infected individuals are asymptomatic. 16 However, there are concerns over this variant's transmissibility and disease severity, majorly due to the cluster of mutations in the spike region that may contribute to enhanced virulence and an increase in number of infections in South Africa coinciding with its emergence. As soon as the WHO declared it as VOC and the global risk associated with this variant is also high, governments and healthcare experts are considering the best possible strategy to contain the virus spread. Further, several countries imposed travel‐related restrictions and enforced quarantine on arrivals from affected and at‐risk countries.
Previous experience with the Delta variant proved that the new variant once introduced into the countries can spread rapidly in short time. Hence, crucial steps for the control of the infection are the effective disease surveillance system, early detection, rapid screening, and isolation of infected persons. Surveillance studies help in assessing the transmission rates, epidemic risk and aid in reducing the disease spread by implementing better control measures with new diagnostic methods and treatments. The significant measures for the public to reduce the risk of disease transmission are through wearing masks, washing hands frequently, social distancing, avoiding crowded spaces, staying in proper ventilated places, and getting vaccinated. 6 , 17
There remains a knowledge gap, whether the available vaccines are effective against the Omicron variant. The frequent emergence of new variants also raises a question on the long‐term protection induced by vaccines. However, most of the approved vaccines target the immunodominant spike protein of SARS‐CoV‐2, hence it is very likely that the available vaccines might offer some level of protection and reduce the disease severity caused by variants. Hence, it is necessary to urgently investigate the effect of mutations on virus escape potential against the available vaccines and antibodies in development. 18 Further, the effective vaccination regimen for COVID‐19 is still in debate due to the waning of vaccine‐induced protection over time and the emergence of new variants. Real‐world data from Israel and UK support that the booster doses of the BNT162b2 mRNA COVID‐19 vaccine counteracts the waning of immunity and reduce the risk of SARS‐CoV‐2 infection or severe illness. 19 , 20 , 21 With the surge of Omicron infections, few countries already started their third booster dose but the duration of immunity after the booster shots still needs to be evaluated. CDC Director, Rochelle Walensky emphasized the importance of vaccination and recommended booster doses for individuals who are 18 years and older. 22
Currently, a preemptive approach to combat the virus is through vaccines. Vaccination remains an effective approach to prevent severe SARS‐CoV‐2 infection. Accelerating the global vaccine coverage especially in the high‐risk or high priority areas where the chance of infection is high could reduce the continuous arising of virus variants. The unequal vaccine access might likely contribute to the evolution of new variants with high infectivity and in due course, the multiple mutations in the emerging variants could be deleterious resulting in severe epidemic or pandemic. In future, the evolving variant might be vaccine‐resistant and completely evade the immune response induced by the available vaccines in the market. The possible solution to slow the virus progression and ultimately end the COVID‐19 pandemic is to reduce the number of infections thereby reducing the viral replication and breaking chains of human‐to‐human transmission. The equitable access of efficient vaccines against the circulating variants could reduce the virus spread or replication which in turn lowers the risk of emerging new variants. Further, the deployment of second‐generation vaccines against newly emerging lineages that can induce cross‐reactive protection against the circulating strains could reduce the virus spread. 23
For now, the priority is to closely monitor the disease progression and virus spread by intensive surveillance mechanism. An effective epidemiological and genomic surveillance system allows the timely identification of VOC as quickly as possible. Further timely assessment of variant's characteristics is also crucial to facilitate the appropriate countermeasures and for the development of specific vaccines to ease the impact of VOCs. The virus evolution and emergence of new variants are the consequence of unequal or slow access to COVID‐19 vaccines globally. Still significant portion of the population in most‐vulnerable countries remain unvaccinated and are on the verge of increased risk of infection presenting a greater risk for further virus mutations. Hence, the global response in addressing the vaccine inequality gaps between the countries is key, before another more virulent or vaccine‐resistant strain emerges. As long as the virus continues to spread in any corner of the world, more virus variants would appear, and the normalcy of human lives and the nation's economy is unrealistic. International cooperation and knowledge sharing between the governments, vaccine manufacturers are vital to ensure vaccine equity around the globe in the fight against COVID‐19 pandemic.
AUTHOR CONTRIBUTIONS
Balamurugan Shanmugaraj and Waranyoo Phoolcharoen conceived the study. Balamurugan Shanmugaraj, Ashwini Malla, and Narach Khorattanakulchai participated in the literature search. Balamurugan Shanmugaraj drafted the manuscript. All authors revised the manuscript and approved the final manuscript for submission.
CONFLICT OF INTERESTS
Waranyoo Phoolcharoen from Chulalongkorn University is a co‐founder/shareholder of Baiya Phytopharm Co., Ltd. Thailand. The other authors declare that there are no conflict of interests.
ACKNOWLEDGMENTS
The authors would like to thank National Vaccine Institute, Thailand and Baiya Phytopharm Co., Ltd., Thailand for financial support. The author (Narach Khorattanakulchai) would like to thank The Second Century Fund (C2F), Chulalongkorn University for the doctoral fellowship.
Contributor Information
Balamurugan Shanmugaraj, Email: Balamurugan.S@baiyaphytopharm.com.
Waranyoo Phoolcharoen, Email: Waranyoo.P@chula.ac.th.
REFERENCES
- 1. WHO . WHO Coronavirus (COVID‐19) Dashboard. 2021. Accessed December 14, 2021. https://covid19.who.int/
- 2. Islam MR, Hoque MN, Rahman MS, et al. Genome‐wide analysis of SARS‐CoV‐2 virus strains circulating worldwide implicates heterogeneity. Sci Rep. 2020;10(1):14004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dyson L, Hill EM, Moore S, et al. Possible future waves of SARS‐CoV‐2 infection generated by variants of concern with a range of characteristics. Nat Commun. 2021;12(1):5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bian L, Gao Q, Gao F, et al. Impact of the Delta variant on vaccine efficacy and response strategies. Expert Rev Vaccines. 2021;20(10):1201‐1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. WHO . Classification of Omicron (B.1.1.529): SARS‐CoV‐2 Variant of Concern. 2021. Accessed December 14, 2021. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern
- 6. WHO . Update on Omicron. 2021. Accessed December 2, 2021. https://www.who.int/news/item/28-11-2021-update-on-omicron
- 7. Gao S‐J, Guo H, Luo G. Omicron variant (B.1.1.529) of SARS‐CoV‐2, a global urgent public health alert! J Med Virol. 2021. 10.1002/jmv.27491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. GISAID I COVID‐19 lineages and variants. 2021. Accessed December 14, 2021. https://www.gisaid.org/
- 9. Torjesen I. Covid‐19: Omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear. BMJ. 2021;375:n2943. [DOI] [PubMed] [Google Scholar]
- 10. Kandeel M, Mohamed MEM, Abd El‐Lateef HM, Venugopala KN, El‐Beltagi HS. Omicron variant genome evolution and phylogenetics. J Med Virol. 2021. 10.1002/jmv.27515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Centers for Disease Control and Prevention C . SARS‐CoV‐2 Variant Classifications and Definitions. 2021. Accessed December 2, 2021. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html#anchor_1632154493691
- 12. Gong SY, Chatterjee D, Richard J, et al. Contribution of single mutations to selected SARS‐CoV‐2 emerging variants spike antigenicity. Virology. 2021;563:134‐145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Plante JA, Liu Y, Liu J, et al. Spike mutation D614G alters SARS‐CoV‐2 fitness. Nature. 2021;592(7852):116‐121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Supasa P, Zhou D, Dejnirattisai W, et al. Reduced neutralization of SARS‐CoV‐2 B.1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184(8):2201‐2211 e2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tian D, Sun Y, Zhou J, Ye Q. The global epidemic of SARS‐CoV‐2 variants and their mutational immune escape. J Med Virol. 2021. 10.1002/jmv.27376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. National Institute for Communicable Diseases Division of the National Health Laboratory Service ‐ South Africa . Frequently asked questions for the B.1.1.529 mutated SARS‐COV‐2 lineage in South Africa. 2021. Accessed December 2, 2021. https://www.nicd.ac.za/frequently-asked-questions-for-the-b-1-1-529-mutated-sars-cov-2-lineage-in-south-africa/
- 17. Daria S, Bhuiyan MA, Islam MR. Detection of highly muted coronavirus variant Omicron (B.1.1.529) is triggering the alarm for South Asian countries: associated risk factors and preventive actions. J Med Virol. 2021. 10.1002/jmv.27503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saxena SK, Kumar S, Ansari S, et al. Characterization of the novel SARS‐CoV‐2 Omicron (B.1.1.529) Variant of Concern and its global perspective. J Med Virol. 2021. 10.1002/jmv.27524 [DOI] [PubMed] [Google Scholar]
- 19. Patalon T, Gazit S, Pitzer VE, Prunas O, Warren JL, Weinberger DM. Odds of testing positive for SARS‐CoV‐2 following receipt of 3 vs 2 doses of the BNT162b2 mRNA vaccine. JAMA Internal Medicine. 2021. 10.1001/jamainternmed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bar‐On YM, Goldberg Y, Mandel M, et al. Protection of BNT162b2 vaccine booster against Covid‐19 in Israel. New Engl J Med. 2021;385:1393‐1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Andrews N, Stowe J, Kirsebom F, Gower C, Ramsay M, Bernal JL Effectiveness of BNT162b2 (Comirnaty, Pfizer‐BioNTech) COVID‐19 booster vaccine against covid‐19 related symptoms in England: test negative case‐control study. 2021.
- 22. Centers for Disease Control and Prevention C . CDC Expands COVID‐19 Booster Recommendations. 2021. Accessed December 2, 2021. https://www.cdc.gov/media/releases/2021/s1129-booster-recommendations.html
- 23. Fontanet A, Autran B, Lina B, Kieny MP, Karim SSA, Sridhar D. SARS‐CoV‐2 variants and ending the COVID‐19 pandemic. Lancet. 2021;397(10278):952‐954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Coronavirus Antiviral & Resistance Database . SARS‐CoV‐2 Variants. 2021. Accessed December 3, 2021. https://covdb.stanford.edu/page/mutation-viewer/#variants.genome.viewer
- 25. CoVariants . Overview of Variants/Mutations. Accessed December 3, 2021. https://covariants.org/
- 26. WHO . Tracking SARS‐CoV‐2 Variants. 2021. Accessed December 3, 2021. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/


