Summary
Plants form complex interaction networks with diverse microbiomes in the environment, and the intricate interplay between plants and their associated microbiomes can greatly influence ecosystem processes and functions. The phyllosphere, the aerial part of the plant, provides a unique habitat for diverse microbes, and in return the phyllosphere microbiome greatly affects plant performance. As an open system, the phyllosphere is subjected to environmental perturbations, including global change, which will impact the crosstalk between plants and their microbiomes. In this review, we aim to provide a synthesis of current knowledge of the complex interactions between plants and the phyllosphere microbiome under global changes and to identify future priority areas of research on this topic.
Keywords: climate change, microbes, one health, phyllosphere, plant microbiome, plant performance
Introduction
The interactions between plants and their associated microbiomes are crucial for host performance and resilience to environment perturbations (e.g. global change) (Bulgarelli et al., 2013; Trivedi et al., 2020). Historically, plant microbiome research has focused mainly on the rhizosphere, including the symbiotic relationship between plant roots and bacteria and fungi, as well as soil‐borne pathogen dynamics. In the last decade or so, with the advent of molecular and genomic technologies, plant microbiome research has expanded rapidly, from the rhizosphere to phyllosphere, endosphere and seeds/fruits (Delmotte et al., 2009; Shade et al., 2017; Carrión et al., 2019; Grady et al., 2019). The phyllosphere represents the aboveground part of a plant, harbouring diverse microbes in both epiphytic (an organism that grows on the surface of a plant) and endophytic (an organism that lives within a plant) niches (Vorholt, 2012). When considering the upper and lower leaf surfaces, the total area of the phyllosphere on Earth is estimated to be over 109 km2 and harbours up to 1026 bacterial cells (Lindow & Brandl, 2003; Vorholt, 2012; Penuelas & Terradas, 2014). The Earth and its ecosystems are undergoing rapid global changes such as climate change (e.g. warming and drought) and land‐use change (e.g. habitat loss and chemical fertilization), which are exerting pervasive impacts on ecosystem processes and functions, and various interactions among plants, microbes and the environment (Vitousek, 1994; Jansson & Hofmockel, 2019; Z. Zhou et al., 2020). A systematic understanding of how global change affects phyllosphere microbiomes could provide an important baseline for harnessing microbiomes to promote ecosystem resilience and plant productivity in a sustainable way. In this review, we aim to provide an overview of how global change will influence the complex interplay between the phyllosphere and its associated microbiomes and identify some priority areas for future research.
Ecological functions of the phyllosphere microbiome
Phyllosphere‐colonizing microbes play critical roles in multiple functions (Fig. 1), including plant productivity and fitness, by affecting leaf functions and longevity, seed mass, apical growth, flowering and fruit development, and also play key roles in removing contaminants (Stone et al., 2018; Thapa & Prasanna, 2018; Liu et al., 2020). For instance, some plant growth‐promoting bacteria inhabiting the phyllosphere such as Microbacterium, Stenotrophomonas and Methylobacterium can improve the growth and nutritional status of the host plant by producing natural growth regulators (e.g. IAA) and fixing nitrogen (Madhaiyan et al., 2015; Abadi et al., 2020). The phyllosphere microbiome also plays important roles in reducing plant methanol (e.g. methylotrophs) and isoprene (e.g. isoprene‐degrading bacteria of the genus Variovorax) emissions to the atmosphere (Abanda‐Nkpwatt et al., 2006; Crombie et al., 2018). Moreover, the phyllosphere microbiome can play vital roles in maintaining plant health and suppressing the overgrowth of plant pathogens. For example, the phyllosphere microbiome can protect Arabidopsis plants against fungal pathogens and dysbiosis (a disruption to the microbiota homeostasis) that could have deleterious impacts on the host health (Ritpitakphong et al., 2016; Chen et al., 2020). Recent findings demonstrate that bacteria and yeasts colonizing nectar can modulate nectar chemical composition and consequently influence visitation/foraging by insect pollinators (Liu et al., 2019). As such, the phyllosphere microbiome contributes to the gut microbiome of insect pollinators and therefore influences their fitness and behaviour (Liu et al., 2019). Nonetheless, it should also be noted that phyllosphere microorganisms can have negative effects on host plants. The presence of a large and varied microbial community in the phyllosphere might increase competition with plants for nutrients and water (Saikkonen et al., 2015; Vacher et al., 2016). Some members of the phyllosphere microbiome might act as plant pathogens, resulting in different forms of plant disease (Lindow & Leveau, 2002; Whipps et al., 2008; Baker et al., 2010). Recently, Zhou et al. (2021b, 2021) reported that the phyllosphere microbiome is involved in the transmission of antibiotic resistance genes in the urban green facade. Another report (Bárta et al., 2021) suggests that the phyllosphere microbiome aids in the establishment of the invasive macrophyte Hydrilla verticillata L. under conditions of nitrogen scarcity. Increasing evidence shows that global change has pervasive impacts on plant health and ecosystem functioning, and harnessing the beneficial functions provided by the phyllosphere microbiome to enhance plant growth and fitness to face such impacts is considered a viable sustainable approach.
Fig. 1.
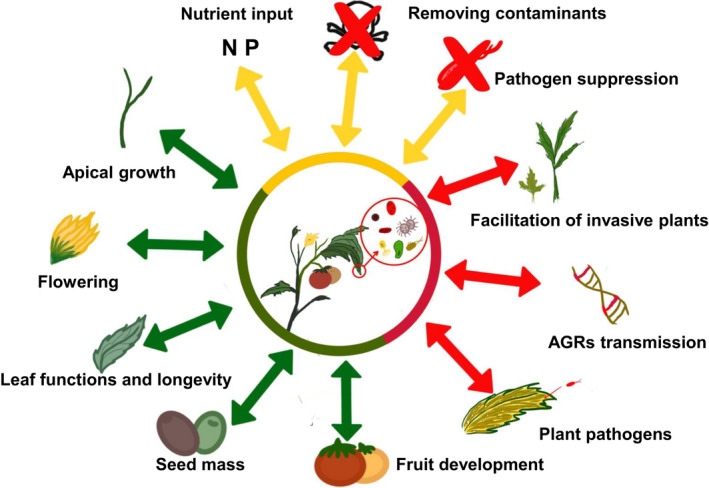
Ecological functions of the phyllosphere microbiome.
Drivers and sources of the phyllosphere microbiome
As the phyllosphere is an open system, its associated microbiomes can come from multiple sources. The assembly of the phyllosphere microbiome is subject to: complex and variable environmental conditions (e.g. temperature, solar radiation, humidity, soil type and agricultural activity) (Vorholt, 2012; Aydogan et al., 2018; Truchado et al., 2019; Zhou et al., 2019; Stone & Jackson, 2021); plant species and genotypes (Singh et al., 2018; Schlechter et al., 2019; Wagner et al., 2020); the adaptability to particular foliar structures or resource secretions (e.g. leaf age and surface roughness, primary and secondary metabolites) (Crombie et al., 2018; Namdar et al., 2019; Sun et al., 2019); and the complex interactions between multiple trophic levels, such as microbe–microbe interactions and plant–herbivore–microbiome interactions (Remus‐Emsermann et al., 2013; Agler et al., 2016; Helfrich et al., 2018; Carlström et al., 2019; Liu et al., 2021). In addition, invasive plants caused by global change may influence the phyllosphere microbiome by altering soil properties and microbial communities and plant–soil feedback (McLeod et al., 2021). Phyllosphere microbiome composition is believed to be closely related to the surrounding environment of host plants, such as soil, air and nearby plant (Brown et al., 2020; Bell et al., 2021; Bernard et al., 2021). For example, soil microbes may enter the root tissues from emerging roots or wounds and constitute the root microbiota (Singh et al., 2020a), and part of this microbiota can be transferred to the aerial part of plants (i.e. phyllosphere) through xylem and phloem systems (Bell et al., 2021). This could partially explain the observations of microbial overlap between plant tissues and soil (Bai et al., 2015; Zarraonaindia et al., 2015; Chen et al., 2017; Xu et al., 2018). Furthermore, opening of leaf stomata and wounds provides a pathway for the transformation and migration between endophytes and epiphytes, and the opportunity for external microbes from aerosols and insects to colonize the plant, which also suggests that plants and the environment are interconnected (Mullens & Jamann, 2021; Xiang et al., 2021). Nevertheless, a recent study analysed the sources of phyllosphere microbes through a customized microcosm that was able to control external microbes (Zhou et al., 2021a, 2021), and showed that microbial sources from soil and air were limited. In another study, oak seeds were found to transmit a large part of microbes to roots and the phyllosphere, emphasizing that plant seeds are the reservoir of the plant microbiome (Abdelfattah et al., 2021), particularly in the early stages of plant growth (Berg & Raaijmakers, 2018; X. Zhou et al., 2020). Seeds can carry highly diverse and beneficial bacterial taxa to ensure the establishment of an optimal bacterial symbiosis for offspring (Liang et al., 2021). These studies highlight that inheritance of plant microbes may play a dominant role in shaping the phyllosphere microbiota.
Overall, the sources of phyllosphere microbes are complex and dynamic, influenced by both intrinsic plant factors and environmental conditions, while biotic and abiotic selection pressures must also be considered (Eldridge et al., 2021). A probable mechanism, therefore, is that the combination of environmental and genetic factors determines the assembly of microbial communities (Shakir et al., 2021). Uncovering how global change affects microbiome assembly, sources of transmission and plant–microbiome interactions in the phyllosphere could provide a mechanistic understanding for future microbiome manipulation.
Hotspots and frontier trends in the phyllosphere microbiome responses to global change
Bibliometric analysis was conducted by retrieving citation data from the Web of Science Core Collection database to highlight the hotspots and frontier trends in the phyllosphere microbiome responses to global change. Keyword cooccurrence network analysis showed that recent research has focused mainly on the relationships between phyllosphere microbiomes and plant growth and health under global change scenarios. These relationships also include interactions between pathogenic bacteria and plant pathogen resistance (Fig. 2a). Phyllosphere microbiomes are faced with increased stress caused by climate change, particularly by warming and drought. Climate change stresses may result in unstable states of microbial communities, wherein a reduction of beneficial taxa weakens plant resistance to pathogen invasion and disease development. Furthermore, phyllosphere microbiomes also participate in carbon and nitrogen cycling by engaging in nitrogen fixing, metabolizing plant metabolites and producing volatile organic compounds (Madhaiyan et al., 2015; Farre‐Armengol et al., 2016; Cernava et al., 2019); how carbon and nitrogen cycling mediated by the phyllosphere microbiome respond to climate change have not clearly determined. Although network analysis indicates that the model plant Arabidopsis thaliana is the most popular research taxon, a new plant model that is more agriculturally relevant is urgently needed to study crop–microbiome interactions for the development of effective microbiome tools for sustainable agriculture. Burst word detection analysis was further used to show a time‐series pattern of keywords, exploring research trends and advances in phyllosphere microbial ecology studies in response to global change over the last decade (Fig. 2b). Plant growth‐ and stress tolerance‐related research has seen the greatest increase since 2018 and therefore represents the current hotspot. A methodological driver for the recent increase in mechanism‐related research may be that the development of emerging technologies has allowed researchers to disentangle the mechanisms of the phyllosphere microbiome regulating plant growth and tolerance.
Fig. 2.
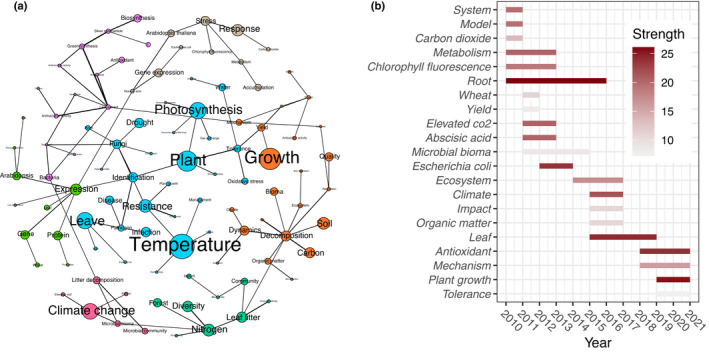
Bibliometric analysis of phyllosphere microbiome research based on the Web of Science Core Collection database from January 2010 to May 2021. (a) Keyword cooccurrence network. Nodes represent unique keywords; node size is proportional to the number of references; node colours indicate modules. (b) Burst word detection analysis. Length represents the burst status duration; colour saturation indicates citation burst strength. Bibliometric analysis was conducted by retrieving citation data based on a topic search using as query: ‘N deposition OR nitrogen deposition OR CO2 OR carbon dioxide OR precipitation OR temperature OR climate change) AND (phyllospher* OR leaf OR leaves) AND (fung* OR bacteria* OR microb* OR archaea* OR virus OR viral OR protist*’. The results were filtered to include items from January 2010 to May 2021, and were further analysed by CiteSpace (Chen et al., 2010) to highlight the hotspots and frontier trends in the phyllosphere microbiome responses to global change.
Impacts of agricultural fertilization on the phyllosphere microbiome
Modern agricultural production relies heavily on the use of chemical fertilizers, such as nitrogen (N), phosphorus (P) and potassium (K) fertilizers. Global agricultural production is expected to increase by 70% by 2050 to feed the increasing human population (Singh et al., 2020b; Haskett et al., 2021), and the use of chemical fertilizers is likely to increase significantly in future agricultural production. However, intensive fertilization could cause soil degradation such as acidification and environmental pollution (Raza et al., 2020). Currently, most studies on the impacts of chemical fertilization on microbial communities have focused on soil and the rhizosphere, while there is a paucity of studies investigating how phyllosphere microbiomes respond to chemical fertilization (Hartman et al., 2018; Trivedi et al., 2020). In general, the phyllosphere harboured a less diverse microbial population including bacteria, fungi and protists than soil and rhizosphere, while the alpha‐ and beta‐diversity of these phyllosphere‐associated microbes often showed more resistance to fertilization (Sun et al., 2021a,b). One explanation for this could be the open nature of the phyllosphere, as phyllosphere‐associated microbes are influenced by multiple factors within dynamic and heterogeneous environments (Lindow & Brandl, 2003; Remus‐Emsermann & Schlechter, 2018), which may weaken the influence of fertilization regimes on phyllosphere microbial variations. In addition, a recent study focusing on the soil–plant continuum of maize, wheat and barley has demonstrated host selection plays a more important role in shaping phyllosphere assembly and network complexity than fertilization practices (Xiong et al., 2021b). Nevertheless, fertilization process may influence some specific microbial taxa in the phyllosphere. For instance, excessive application of chemical N fertilizer increased the relative abundance of potential fungal plant pathogens in the leaf endosphere (Xiong et al., 2021a). Similarly, a study on sorghum showed that long‐term fertilization regimes did not significantly influence the diversity and composition of protistan communities in the phyllosphere, but some protistan consumers (e.g. Amoebozoa) were significantly influenced by fertilization (Sun et al., 2021b). Additionally, other macro‐ and micronutrients also play a role in phyllosphere microbiome assembly. For example, application of these nutrients in soil increased microbial biodiversity but reduced the relative abundance of pathogen Candidatus Liberibacter asiaticus (CLas) in the phyllosphere of Gannan Navel Orange (Y. Zhou et al., 2021b, 2021). Although these studies provided valuable information, the fundamental knowledge of the mechanisms underlying phyllosphere microbiome assembly and activity under fertilization remains in its infancy.
In addition to fertilization, microbial communities in agricultural ecosystems are usually influenced by agronomic management regimes (e.g. organic and conventional management). It has beenreported that organic farming increased fungal alpha diversity in the wheat phyllosphere, compared with conventional management (Karlsson et al., 2017). A recent study also suggested that agricultural management (i.e. organic, transition and conventional) strongly influenced the composition, functions and cooccurrence networks of the sugarcane phyllosphere microbiome (Khoiri et al., 2021). Organic farming was associated with a complex microbial network and enriched some plant growth‐promoting bacteria such as Bradyrhizobium and Bacillus, whereas conventional practice decreased the abundance of functional genes involved in cell motility and energy metabolism of phyllosphere microbiomes (Khoiri et al., 2021). Emerging evidence indicates that agricultural management is an important factor driving phyllosphere microbiome assembly. Uncovering phyllosphere–microbiome interactions and their molecular mechanisms under different agricultural management practices can provide new scientific knowledge to harness the phyllosphere microbiome for plant productivity and sustainable agriculture.
Impacts of global warming on the phyllosphere microbiome
Global warming caused by the ‘greenhouse effect’ is predicted to have major consequences on element cycling and the functioning of terrestrial ecosystems such as vegetation dynamics (Vitousek, 1994; Jones et al., 1998; Norby & Luo, 2004), which will substantially impact the phyllosphere microbiome (Zhu & Penuelas, 2020) (Fig. 3). Based on the Intergovernmental Panel on Climate Change (IPCC, 2014), global mean surface temperature is estimated to increase by 2–3°C within the next few decades (Stocker, 2014), which is predicted to result in a global increase in drought frequency and duration.
Fig. 3.
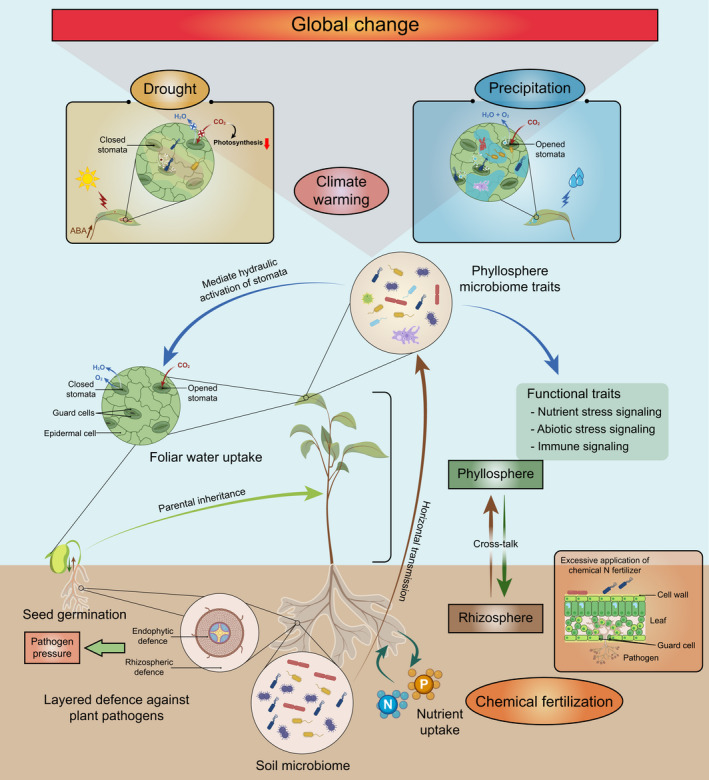
Impact factors of the phyllosphere microbiome. The phyllosphere comprises the aboveground part of a plant, harbouring diverse microbes in both epiphytic and endophytic niches. These microbiomes derive from vertical transmission via parental inheritance, and horizontal transmission by surrounding environments (e.g. soil and air). On the one hand, global changes such as climate warming, drought and precipitation might impact leaf functional traits and phyllosphere microbiome traits, and the latter mediates the hydraulic activation of stomata relevant to the pathway for foliar water uptake. On the other hand, chemical fertilization can also affect the phyllosphere microbiome by changing rhizosphere communities and leaf morphology.
In recent decades, experimental studies of climate warming effects have focused mostly on the soil microbiome (Yergeau et al., 2012; Jansson & Hofmockel, 2019; Z. Zhou et al., 2020), while the potential impacts of warming on the abundance and compositions of the phyllosphere microbiome have largely been overlooked and are only just beginning to be studied. For example, based on a long‐term field warming experiment on a grassland (dominated by Arrhenatherum elatius and Galium album) with increasing surface temperature of 2°C, Aydogan et al. (2018, 2020) found that warming does not affect the total colonization and the concentration of leaf‐associated bacterial cells but shifted the diversity and phylogenetic composition of the bacterial communities. More importantly, warming‐induced decreases of beneficial bacteria (e.g. Sphingomonas spp. and Rhizobium spp.) and enhancement of potentially pathogenic bacteria (e.g. Enterobacteriaceae, Pseudomonas, and Acinetobacter) in the phyllosphere may indicate that warming increases the potential transmission of pathogens in grassland ecosystems (Aydogan et al., 2018). In addition to affecting phyllosphere bacterial communities, climate warming also decreased fungal richness, reduced evenness and shifted the overall fungal community composition on pedunculate oak (Quercus robur) (Faticov et al., 2021) and a boreal forest tree (Populus balsamifera L.) (Balint et al., 2015). In contrast to potential bacterial pathogens (e.g. Acinetobacter), warming negatively affected putative fungal pathogens (Aydogan et al., 2018; Faticov et al., 2021). These elegant studies provided valuable information on the impacts of climate warming on the profiles of the phyllosphere microbiome. However, care must be taken when interpreting the outcomes of each study, as only limited plant species and genotypes have been considered. The differences in leaf traits, nutrient content and primary/secondary metabolites among genotypes could result in differential colonization of microorganisms, which may mask the effects of climate warming (Wagner et al., 2016). For instance, in contrast to the above observation, other studies have indicated that long‐term warming experiments caused no significant changes in the foliar fungal community composition of three perennial grass species (Achnatherum lettermanii, Festuca thurberi and Poa pratensis) (Kazenel et al., 2019; Kivlin et al., 2019). Thus, it is unclear whether the response of the phyllosphere microbiome to climate warming is consistent across plant species and genotypes, although some excellent studies have been conducted on this topic (Faticov et al., 2021). Moreover, the present investigations on phyllosphere microbiomes have focused generally on bacterial and fungal communities, while little attention has been paid to other microbes such as archaea and protists. All these observations have highlighted the need to improve our understanding of climate warming on the phyllosphere microbiome.
Impacts of precipitation and drought on the phyllosphere microbiome
Precipitation has begun to show a long‐term downward trend under global climate change, resulting in a global increase in drought frequency and duration (Sardans et al., 2008). Meanwhile, extreme weather events including floods and drought are being recorded more acutely and frequently. Such a change at the global scale is expected to have significant impacts on global agricultural production, as it can influence plant growth and plant disease occurrence by altering humidity and water availability (Howden et al., 2007; Xin et al., 2016; Xu et al., 2021; Romero et al., 2022). A recent large‐scale survey suggested that precipitation is the most important predictor of fungal communities and the abundance of fungal plant pathogens, and the authors suggested that the abundance of fungal plant pathogens could increase by up to 100‐fold by 2050, especially in coastal regions (Chen et al., 2021). The interactions between water status, soil fertility and arbuscular mycorrhizal fungi could also shift the phyllosphere microbiome; for example, both water status and mycorrhizal disruption could reduce phyllosphere bacterial richness, with a more homogeneous bacterial community composition of the tomato (Solanum lycopersicum) phyllosphere (Debray et al., 2022). In addition, it was found that high humidity can transform nonpathogenic Pseudomonas syringae strains into virulent pathogens and induce dyshomeostasis of the commensal bacterial community in the phyllosphere by affecting water status inside the apoplast (Xin et al., 2016). Furthermore, drought stress not only affects phyllosphere microbial compositions (Bechtold et al., 2021) but also community assembly processes. A recent study on sorghum systems with abundant sampling indicated that the assembly of phyllosphere mycobiomes was determined by stochastic processes (e.g. drift or stochastic dispersal) in the early stage of host development when sorghum is drought‐stressed (Gao et al., 2020). Regarding precipitation, recent work on the wetland macrophyte broadleaf cattail (Typha latifolia) showed that rain events did not have a significant effect on the richness or evenness of its phyllosphere bacterial community (Stone & Jackson, 2021). By contrast, climatic and leaf‐related variables effectively shaped seasonal dynamics in phyllosphere diversity and composition (Stone & Jackson, 2021). Under the scenario of climate change, improving our understanding of how plant species and their microbiomes can cope with drought events is one of the most relevant topics in plant science. Foliar water uptake (FWU) has been identified as a mechanism commonly adopted by trees and other plants from various biomes and could be used to predict the sensitivity of plant species to drought (Schreel & Steppe, 2020). In addition to morphological and anatomical traits and leaf age (Schreel & Steppe, 2020), leaf wettability also depends on the degree of cover by the phyllosphere microbiome (epiphytic and endophytic organisms) and thus affects their hydrophobicity (Rosado & Almeida, 2020). For example, the cuticular permeability that allows the diffusion of water through the cuticle might be increased by biosurfactants produced by epiphytic bacteria (Park et al., 2018), indicating their potential effect on FWU. Moreover, the phyllosphere microbiome can also mediate the hydraulic activation of stomata, which is relevant to the pathway for FWU. For example, fungal leaf endophytes may increase stomatal conductance, while bacteria may mediate stomatal closure and opening (Friesen et al., 2011). Considering the stomata as the gateway for the entrance of pathogens to plants (Gudesblat et al., 2009), regulation of the stomatal aperture by the phyllosphere microbiome is also a mechanism associated with plant defence. All these observations suggest that phyllosphere‐associated microbiomes have great potential to improve plant resistance to future drought (Rolli et al., 2015; Llorens et al., 2019; de Vries et al., 2020; Xu et al., 2021).
Ecoevolutionary dynamics between the phyllosphere and its microbiomes under global climate change
The ecoevolutionary dynamics of plant–microbiome symbiosis systems are of increasing interest. Unfortunately, little has been done regarding the phyllosphere. Among current studies, the impacts of plant evolutionary history and contemporary evolution on plant, soil and rhizosphere microbiome responses to climate change have received much attention (Lambers et al., 2009; Fitzpatrick et al., 2020; Petipas et al., 2021).
As the outcome of past evolutionary history, the phylogenetic relationship between plant species has been found to be able to interact with climate change to modify plant microbiomes (Naylor et al., 2017). In the absence of drought, cereal grass phylogeny was shown to determine rhizosphere microbiome composition. Such effects of host evolutionary history on microbiomes have been widely observed in plants, especially those with agricultural significance (Stolf‐Moreira et al., 2011; Peiffer et al., 2013; Santos‐Medellín et al., 2017). Drought, however, promoted the abundance of Actinobacteria, weakening the importance of host evolutionary history for microbiome community structure (Naylor et al., 2017). In addition, evolution at the contemporary timescale, in both microbiomes and plants, can mediate plant–microbiome responses to climate change. Microbes often have a large population size and high genetic variation, which translate to strong evolutionary dynamics to influence ecological processes (Yoshida et al., 2003; Frantzeskakis et al., 2020). For example, using the synthetic community approach, Batstone et al. (2020) showed that rapid evolution has occurred in the nodule‐forming bacterium Ensifer meliloti and promoted mutualism between the bacterium and the plant host Medicago truncatula. Unlike microbes, more barriers exist for plant evolution. Nevertheless, recent evidence has suggested the possibility of rapid evolution in plants. In an experiment by terHorst et al. (2014), adaptation was found to occur in Brassica rapa populations after a three‐generation drought treatment. When transplanted into a common garden under ambient wet conditions, B. rapa populations (adapted vs unadapted to drought) showed different abilities in shaping the soil microbiomes.
We recognize that most existing work has focused on the ecoevolutionary dynamics between plants and soil/rhizosphere microbiomes, not microbiomes in other plant compartments, including the phyllosphere. Also, little attention has been given to several evolutionary processes unique to plants, such as within‐ and cross‐species hybridization (Rieseberg & Carney, 1998) and the emergence of polyploidy plants (Adams & Wendel, 2005), which can introduce novel genetic variation to wild plant populations. Recent progress in research on ecoevolutionary dynamics has prompted the development of new model systems for study of the role of plant evolution in ecological processes (Williams et al., 2016; Hart et al., 2019). For example, there is increasing interest in using aquatic floating plants of the family Lemnaceae (commonly known as duckweeds) to examine plant ecological and evolutionary responses to environmental change (Armitage & Jones, 2019; O'Brien et al., 2020). Duckweeds have high within‐species genotypic and phenotypic diversity (Hart et al., 2019; O'Brien et al., 2020), making it possible to observe the changes in both species composition (ecological dynamics) and genotypic composition (evolutionary dynamics) during plant–microbiome interactions at the same timescale. The high tractability of these model systems can extend the scope of observational studies, providing more mechanistic understanding of the importance of evolution in plant–microbiome systems.
Conclusions and perspectives
The phyllosphere microbiome plays an essential role in increasing the ability of a plant to pass through environmental filters. Currently, however, we have limited capacity to predict the consequences of the shift in the phyllosphere microbiome for ecosystem functioning under a changing environment. Some fundamental questions remain largely unresolved: (1) What are the mechanisms within and across plant hosts that underpin host–microbe interactions? (2) What are the major microbial taxa in the phyllosphere that control or mediate plant performance (e.g. nutrient uptake, plant disease suppression or growth)? (3) How does the phyllosphere microbiome interact with other plant microbiomes? (4) How can we manage the phyllosphere microbiome to increase plant health and performance in a changing world? (5) How will host and the phyllosphere microbiome evolve in response to global changes, and what impacts will this ecoevolution have on ecosystem functions? Therefore, we argue that advancing our fundamental understanding of the impacts of global changes on the phyllosphere microbiome and related ecosystem functions requires interdisciplinary investigations. We need to shift the focus from the level of community ecology to ecosystem ecology for a better understanding of the mechanisms underlying responses of the ‘holobiont’ (assemblage of a plant and its microbiome living in or around it) to global changes. A systems approach is needed to understand the complex interactions between the phyllosphere microbiome and host fitness, and the ecological functions of these microbes for plant nutrition uptake, growth and survival under global climate change.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (42021005 and 42090063).
Dedication: This paper is dedicated to the late Prof. Sally E. Smith. She was instrumental in training next generation scientists, and she made tremendous impacts in the field of soil–plant interactions.
References
- Abadi VAJM, Sepehri M, Rahmani HA, Zarei M, Ronaghi A, Taghavi SM, Shamshiripour M. 2020. Role of dominant phyllosphere bacteria with plant growth–promoting characteristics on growth and nutrition of maize (Zea mays L.). Journal of Soil Science and Plant Nutrition 20: 2348–2363. [Google Scholar]
- Abanda‐Nkpwatt D, Müsch M, Tschiersch J, Boettner M, Schwab W. 2006. Molecular interaction between Methylobacterium extorquens and seedlings: growth promotion, methanol consumption, and localization of the methanol emission site. Journal of Experimental Botany 57: 4025–4032. [DOI] [PubMed] [Google Scholar]
- Abdelfattah A, Wisniewski M, Schena L, Tack AJM. 2021. Experimental evidence of microbial inheritance in plants and transmission routes from seed to phyllosphere and root. Environmental Microbiology 23: 2199–2214. [DOI] [PubMed] [Google Scholar]
- Adams KL, Wendel JF. 2005. Polyploidy and genome evolution in plants. Current Opinion in Plant Biology 8: 135–141. [DOI] [PubMed] [Google Scholar]
- Agler MT, Ruhe J, Kroll S, Morhenn C, Kim ST, Weigel D, Kemen EM. 2016. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biology 14: e1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage DW, Jones SE. 2019. Negative frequency‐dependent growth underlies the stable coexistence of two cosmopolitan aquatic plants. Ecology 100: e02657. [DOI] [PubMed] [Google Scholar]
- Aydogan EL, Budich O, Hardt M, Choi YH, Jansen‐Willems AB, Moser G, Muller C, Kampfer P, Glaeser SP. 2020. Global warming shifts the composition of the abundant bacterial phyllosphere microbiota as indicated by a cultivation‐dependent and ‐independent study of the grassland phyllosphere of a long‐term warming field experiment. FEMS Microbiology Ecology 96: fiaa087. [DOI] [PubMed] [Google Scholar]
- Aydogan EL, Moser G, Muller C, Kampfer P, Glaeser SP. 2018. Long‐term warming shifts the composition of bacterial communities in the phyllosphere of Galium album in a permanent grassland field‐experiment. Frontiers in Microbiology 9: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Müller DB, Srinivas G, Garrido‐Oter R, Potthoff E, Rott M, Dombrowski N, Münch PC, Spaepen S, Remus‐Emsermann M et al. 2015. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528: 364–369. [DOI] [PubMed] [Google Scholar]
- Baker C, Chitrakar R, Obulareddy N, Panchal S, Williams P, Melotto M. 2010. Molecular battles between plant and pathogenic bacteria in the phyllosphere. Brazilian Journal of Medical Biological Research 43: 698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balint M, Bartha L, O'Hara RB, Olson MS, Otte J, Pfenninger M, Robertson AL, Tiffin P, Schmitt I. 2015. Relocation, high‐latitude warming and host genetic identity shape the foliar fungal microbiome of poplars. Molecular Ecology 24: 235–248. [DOI] [PubMed] [Google Scholar]
- Bárta J, Pires de Paula CC, Rejmánková E, Lin Q, Kohoutová I, Sirová D. 2021. Complex phyllosphere microbiome aids in the establishment of the invasive macrophyte Hydrilla verticillata (L.) under conditions of nitrogen scarcity. BioRxiv. doi: 10.1101/2021.01.11.426196. [DOI] [Google Scholar]
- Batstone RT, O'Brien AM, Harrison TL, Frederickson ME. 2020. Experimental evolution makes microbes more cooperative with their local host genotype. Science 370: 476–478. [DOI] [PubMed] [Google Scholar]
- Bechtold EK, Ryan S, Moughan SE, Ranjan R, Nusslein K. 2021. Phyllosphere community assembly and response to drought stress on common tropical and temperate forage grasses. Applied Environmental Microbiology 87: e0089521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JK, Helgason B, Siciliano SD. 2021. Brassica napus phyllosphere bacterial composition changes with growth stage. Plant and Soil 464: 501–516. [Google Scholar]
- Berg G, Raaijmakers JM. 2018. Saving seed microbiomes. ISME Journal 12: 1167–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard J, Wall CB, Costantini MS, Rollins RL, Atkins ML, Cabrera FP, Cetraro ND, Feliciano CKJ, Greene AL, Kitamura PK et al. 2021. Plant part and a steep environmental gradient predict plant microbial composition in a tropical watershed. ISME Journal 15: 999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SP, Grillo MA, Podowski JC, Heath KD. 2020. Soil origin and plant genotype structure distinct microbiome compartments in the model legume Medicago truncatula . Microbiome 8: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze‐Lefert P. 2013. Structure and functions of the bacterial microbiota of plants. Annual Review of Plant Biology 64: 807–838. [DOI] [PubMed] [Google Scholar]
- Carlström CI, Field CM, Bortfeld‐Miller M, Müller B, Sunagawa S, Vorholt JA. 2019. Synthetic microbiota reveal priority effects and keystone strains in the Arabidopsis phyllosphere. Nature Ecology & Evolution 3: 1445–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrión VJ, Perez‐Jaramillo J, Cordovez V, Tracanna V, de Hollander M, Ruiz‐Buck D, Mendes LW, van Ijcken WFJ, Gomez‐Exposito R, Elsayed SS et al. 2019. Pathogen‐induced activation of disease‐suppressive functions in the endophytic root microbiome. Science 366: 606. [DOI] [PubMed] [Google Scholar]
- Cernava T, Erlacher A, Soh J, Sensen CW, Grube M, Berg G. 2019. Enterobacteriaceae dominate the core microbiome and contribute to the resistome of arugula (Eruca sativa Mill.). Microbiome 7: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CM, Ibekwe‐SanJuan F, Hou JH. 2010. The structure and dynamics of cocitation clusters: a multiple‐perspective cocitation analysis. Journal of the American Society for Information Science and Technology 61: 1386–1409. [Google Scholar]
- Chen QL, An XL, Zhu YG, Su JQ, Gillings MR, Ye ZL, Cui L. 2017. Application of struvite alters the antibiotic resistome in soil, Rhizosphere, and Phyllosphere. Environmental Science & Technology 51: 8149–8157. [DOI] [PubMed] [Google Scholar]
- Chen QL, Hu HW, Yan ZZ, Li CY, Nguyen BT, Zhu YG, He JZ. 2021. Precipitation increases the abundance of fungal plant pathogens in Eucalyptus phyllosphere. Environmental Microbiology. doi: 10.1111/1462-2920.15728. [DOI] [PubMed] [Google Scholar]
- Chen T, Nomura K, Wang X, Sohrabi R, Xu J, Yao L, Paasch BC, Ma LI, Kremer J, Cheng Y et al. 2020. A plant genetic network for preventing dysbiosis in the phyllosphere. Nature 580: 653–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombie AT, Larke‐Mejia NL, Emery H, Dawson R, Pratscher J, Murphy GP, McGenity TJ, Murrell JC. 2018. Poplar phyllosphere harbors disparate isoprene‐degrading bacteria. Proceedings of the National Academy of Sciences, USA 115: 13081–13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debray R, Socolar Y, Kaulbach G, Guzman A, Hernandez CA, Curley R, Dhond A, Bowles T, Koskella B. 2022. Water stress and disruption of mycorrhizas induce parallel shifts in phyllosphere microbiome composition. New Phytologist 234: 2018–2031. [DOI] [PubMed] [Google Scholar]
- Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, Schlapbach R, von Mering C, Vorholt JA. 2009. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proceedings of the National Academy of Sciences, USA 106: 16428–16433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge DJ, Travers SK, Val J, Ding JY, Wang JT, Singh BK, Delgado‐Baquerizo M. 2021. Experimental evidence of strong relationships between soil microbial communities and plant germination. Journal of Ecology 109: 2488–2498. [Google Scholar]
- Farre‐Armengol G, Penuelas J, Li T, Yli‐Pirila P, Filella I, Llusia J, Blande JD. 2016. Ozone degrades floral scent and reduces pollinator attraction to flowers. New Phytologist 209: 152–160. [DOI] [PubMed] [Google Scholar]
- Faticov M, Abdelfattah A, Roslin T, Vacher C, Hambäck P, Blanchet FG, Lindahl BD, Tack AJM. 2021. Climate warming dominates over plant genotype in shaping the seasonal trajectory of foliar fungal communities on oak. New Phytologist 231: 1770–1783. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick CR, Salas‐González I, Conway JM, Finkel OM, Gilbert S, Russ D, Teixeira PJPL, Dangl JL. 2020. The plant microbiome: from ecology to reductionism and beyond. Annual Review of Microbiology 74: 81–100. [DOI] [PubMed] [Google Scholar]
- Frantzeskakis L, Di Pietro A, Rep M, Schirawski J, Wu CH, Panstruga R. 2020. Rapid evolution in plant–microbe interactions – a molecular genomics perspective. New Phytologist 225: 1134–1142. [DOI] [PubMed] [Google Scholar]
- Friesen ML, Porter SS, Stark SC, von Wettberg EJ, Sachs JL, Martinez‐Romero E. 2011. Microbially mediated plant functional traits. Annual Review of Ecology, Evolution, and Systematics 42: 23–46. [Google Scholar]
- Gao C, Montoya L, Xu L, Madera M, Hollingsworth J, Purdom E, Singan V, Vogel J, Hutmacher RB, Dahlberg JA et al. 2020. Fungal community assembly in drought‐stressed sorghum shows stochasticity, selection, and universal ecological dynamics. Nature Communications 11: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady KL, Sorensen JW, Stopnisek N, Guittar J, Shade A. 2019. Assembly and seasonality of core phyllosphere microbiota on perennial biofuel crops. Nature Communications 10: 4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudesblat GE, Torres PS, Vojno AA. 2009. Stomata and pathogens: warfare at the gates. Plant Signaling & Behavior 4: 1114–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart SP, Turcotte MM, Levine JM. 2019. Effects of rapid evolution on species coexistence. Proceedings of the National Academy of Sciences, USA 116: 2112–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman K, van der Heijden MGA, Wittwer RA, Banerjee S, Walser JC, Schlaeppi K. 2018. Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 6: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskett TL, Tkacz A, Poole PS. 2021. Engineering rhizobacteria for sustainable agriculture. ISME Journal 15: 949–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich EJN, Vogel CM, Ueoka R, Schäfer M, Ryffel F, Müller DB, Probst S, Kreuzer M, Piel J, Vorholt JA. 2018. Bipartite interactions, antibiotic production and biosynthetic potential of the Arabidopsis leaf microbiome. Nature Microbiology 3: 909–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- terHorst CP, Lennon JT, Lau JA. 2014. The relative importance of rapid evolution for plant‐microbe interactions depends on ecological context. Proceedings of the Royal Society B: Biological Sciences 281: 20140028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden SM, Soussana JF, Tubiello FN, Chhetri N, Dunlop M, Meinke H. 2007. Adapting agriculture to climate change. Proceedings of the National Academy of Sciences, USA 104: 19691–19696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC . 2014. The fifth assessment report . Geneva, Switzerland: The Intergovernmental Panel on Climate Change.
- Jansson JK, Hofmockel KS. 2019. Soil microbiomes and climate change. Nature Review Microbiology 18: 35–46. [DOI] [PubMed] [Google Scholar]
- Jones TH, Thompson LJ, Lawton JH, Bezemer TM, Bardgett RD, Blackburn TM, Bruce KD, Cannon PF, Hall GS, Hartley SE et al. 1998. Impacts of rising atmospheric carbon dioxide on model terrestrial ecosystems. Science 280: 441–443. [DOI] [PubMed] [Google Scholar]
- Karlsson I, Friberg H, Kolseth AK, Steinberg C, Persson P. 2017. Organic farming increases richness of fungal taxa in the wheat phyllosphere. Molecular Ecology 26: 3424–3436. [DOI] [PubMed] [Google Scholar]
- Kazenel MR, Kivlin SN, Taylor DL, Lynn JS, Rudgers JA. 2019. Altitudinal gradients fail to predict fungal symbiont responses to warming. Ecology 100: e02740. [DOI] [PubMed] [Google Scholar]
- Khoiri AN, Cheevadhanarak S, Jirakkakul J, Dulsawat S, Prommeenate P, Tachaleat A, Kusonmano K, Wattanachaisaereekul S, Sutheeworapong S. 2021. Comparative metagenomics reveals microbial signatures of sugarcane phyllosphere in organic management. Frontiers in Microbiology 12: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivlin SN, Kazenel MR, Lynn JS, Taylor DL, Rudgers JA. 2019. Plant identity influences foliar fungal symbionts more than elevation in the Colorado Rocky Mountains. Microbial Ecology 78: 688–698. [DOI] [PubMed] [Google Scholar]
- Lambers H, Mougel C, Jaillard B, Hinsinger P. 2009. Plant‐microbe‐soil interactions in the rhizosphere: an evolutionary perspective. Plant and Soil 321: 83–115. [Google Scholar]
- Liang D, Guo J, Hou F, Bowatte S. 2021. High level of conservation and diversity among the endophytic seed bacteriome in eight alpine grassland species growing at the Qinghai Tibetan Plateau. FEMS Microbiology Ecology 97: fiab060. [DOI] [PubMed] [Google Scholar]
- Lindow SE, Brandl MT. 2003. Microbiology of the phyllosphere. Applied and Environmental Microbiology 69: 1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow SE, Leveau JH. 2002. Phyllosphere microbiology. Current Opinion in Biotechnology 13: 238–243. [DOI] [PubMed] [Google Scholar]
- Liu H, Brettell LE, Singh B. 2020. Linking the phyllosphere microbiome to plant health. Trends in Plant Science 25: 841–844. [DOI] [PubMed] [Google Scholar]
- Liu H, Macdonald CA, Cook J, Anderson IC, Singh BK. 2019. An ecological loop: host microbiomes across multitrophic interactions. Trends in Ecology & Evolution 34: 1118–1130. [DOI] [PubMed] [Google Scholar]
- Liu M, Mipam TD, Wang X, Zhang P, Lin Z, Liu X. 2021. Contrasting effects of mammal grazing on foliar fungal diseases: patterns and potential mechanisms. New Phytologist 232: 345–355. [DOI] [PubMed] [Google Scholar]
- Llorens E, Sharon O, Camanes G, Garcia‐Agustin P, Sharon A. 2019. Endophytes from wild cereals protect wheat plants from drought by alteration of physiological responses of the plants to water stress. Environmental Microbiology 21: 3299–3312. [DOI] [PubMed] [Google Scholar]
- Madhaiyan M, Alex THH, Ngoh ST, Prithiviraj B, Ji LH. 2015. Leaf‐residing Methylobacterium species fix nitrogen and promote biomass and seed production in Jatropha curcas . Biotechnology for Biofuels 8: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod ML, Bullington L, Cleveland CC, Rousk J, Lekberg Y. 2021. Invasive plant‐derived dissolved organic matter alters microbial communities and carbon cycling in soils. Soil Biology and Biochemistry 156: 108191. [Google Scholar]
- Mullens A, Jamann TM. 2021. Colonization and movement of green fluorescent protein‐labeled Clavibacter nebraskensis in maize. Plant Disease 105: 1422–1431. [DOI] [PubMed] [Google Scholar]
- Namdar D, Charuvi D, Ajjampura V, Mazuz M, Ion A, Kamara I, Koltai H. 2019. LED lighting affects the composition and biological activity of Cannabis sativa secondary metabolites. Industrial Crops and Products 132: 177–185. [Google Scholar]
- Naylor D, DeGraaf S, Purdom E, Coleman‐Derr D. 2017. Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME Journal 11: 2691–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norby RJ, Luo Y. 2004. Evaluating ecosystem responses to rising atmospheric CO2 and global warming in a multi‐factor world. New Phytologist 162: 281–293. [Google Scholar]
- O'Brien AM, Laurich J, Lash E, Frederickson ME. 2020. Mutualistic outcomes across plant populations, microbes, and environments in the duckweed Lemna minor . Microbial Ecology 80: 384–397. [DOI] [PubMed] [Google Scholar]
- Park E, Nedo A, Caplan JL, Dinesh‐Kumar SP. 2018. Plant–microbe interactions: organelles and the cytoskeleton in action. New Phytologist 217: 1012–1028. [DOI] [PubMed] [Google Scholar]
- Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE. 2013. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proceedings of the National Academy of Sciences, USA 110: 6548–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penuelas J, Terradas J. 2014. The foliar microbiome. Trends in Plant Science 19: 278–280. [DOI] [PubMed] [Google Scholar]
- Petipas RH, Geber MA, Lau JA. 2021. Microbe‐mediated adaptation in plants. Ecology Letters 24: 1302–1317. [DOI] [PubMed] [Google Scholar]
- Raza S, Miao N, Wang P, Ju X, Chen Z, Zhou J, Kuzyakov Y. 2020. Dramatic loss of inorganic carbon by nitrogen‐induced soil acidification in Chinese croplands. Global Change Biology 26: 3738–3751. [DOI] [PubMed] [Google Scholar]
- Remus‐Emsermann MNP, Kowalchuk GA, Leveau JHJ. 2013. Single‐cell versus population‐level reproductive success of bacterial immigrants to pre‐colonized leaf surfaces. Environmental Microbiology Reports 5: 387–392. [DOI] [PubMed] [Google Scholar]
- Remus‐Emsermann MNP, Schlechter RO. 2018. Phyllosphere microbiology: at the interface between microbial individuals and the plant host. New Phytologist 218: 1327–1333. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Carney SE. 1998. Plant hybridization. New Phytologist 140: 599–624. [DOI] [PubMed] [Google Scholar]
- Ritpitakphong U, Falquet L, Vimoltust A, Berger A, Métraux JP, L'Haridon F. 2016. The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytologist 210: 1033–1043. [DOI] [PubMed] [Google Scholar]
- Rolli E, Marasco R, Vigani G, Ettoumi B, Mapelli F, Deangelis ML, Gandolfi C, Casati E, Previtali F, Gerbino R et al. 2015. Improved plant resistance to drought is promoted by the root‐associated microbiome as a water stress‐dependent trait. Environmental Microbiology 17: 316–331. [DOI] [PubMed] [Google Scholar]
- Romero F, Cazzato S, Walder F, Vogelgsang S, Bender SF, van der Heijden MGA. 2022. Humidity and high temperature are important for predicting fungal disease outbreaks worldwide. New Phytologist 234: 1553–1556. [DOI] [PubMed] [Google Scholar]
- Rosado BHP, Almeida LC. 2020. The importance of phyllosphere on foliar water uptake. Trends in Plant Science 25: 1058–1060. [DOI] [PubMed] [Google Scholar]
- Saikkonen K, Mikola J, Helander M. 2015. Endophytic phyllosphere fungi and nutrient cycling in terrestrial ecosystems. Current Science 109: 121–126. [Google Scholar]
- Santos‐Medellín C, Edwards J, Liechty Z, Nguyen B, Sundaresan V. 2017. Drought stress results in a compartment‐specific restructuring of the rice root‐associated microbiomes. MBio 8: e00764–e00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardans J, Penuelas J, Estiarte M, Prieto P. 2008. Warming and drought alter C and N concentration, allocation and accumulation in a Mediterranean shrubland. Global Change Biology 14: 2304–2316. [Google Scholar]
- Schlechter RO, Miebach M, Remus‐Emsermann MNP. 2019. Driving factors of epiphytic bacterial communities: a review. Journal of Advanced Research 19: 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreel JDM, Steppe K. 2020. Foliar water uptake in trees: negligible or necessary? Trends in Plant Science 25: 590–603. [DOI] [PubMed] [Google Scholar]
- Shade A, Jacques MA, Barret M. 2017. Ecological patterns of seed microbiome diversity, transmission, and assembly. Current Opinion in Microbiology 37: 15–22. [DOI] [PubMed] [Google Scholar]
- Shakir S, Zaidi S, de Vries FT, Mansoor S. 2021. Plant genetic networks shaping phyllosphere microbial community. Trends in Genetics 37: 306–316. [DOI] [PubMed] [Google Scholar]
- Singh BK, Liu HW, Trivedi P. 2020a. Eco‐holobiont: a new concept to identify drivers of host‐associated microorganisms. Environmental Microbiology 22: 564–567. [DOI] [PubMed] [Google Scholar]
- Singh BK, Trivedi P, Egidi E, Macdonald CA, Delgado‐Baquerizo M. 2020b. Crop microbiome and sustainable agriculture. Nature Reviews Microbiology 18: 601–602. [DOI] [PubMed] [Google Scholar]
- Singh P, Santoni S, This P, Peros JP. 2018. Genotype–environment interaction shapes the microbial assemblage in grapevine's phyllosphere and carposphere: an NGS approach. Microorganisms 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker T. 2014. Climate change 2013: the physical science basis: working group I contribution to the fifth assessment report of the Intergovernmental panel on climate change. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Stolf‐Moreira R, Lemos EG, Carareto‐Alves L, Marcondes J, Pereira SS, Rolla AA, Pereira RM, Neumaier N, Binneck E, Abdelnoor RV. 2011. Transcriptional profiles of roots of different soybean genotypes subjected to drought stress. Plant Molecular Biology Reporter 29: 19–34. [Google Scholar]
- Stone BWG, Jackson CR. 2021. Seasonal patterns contribute more towards phyllosphere bacterial community structure than short‐term perturbations. Microbial Ecology 81: 146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone BW, Weingarten EA, Jackson CR. 2018. The role of the phyllosphere microbiome in plant health and function. Annual Plant Reviews Online 1: 533–556. [Google Scholar]
- Sun A, Jiao XY, Chen Q, Wu AL, Zheng Y, Lin YX, He JZ, Hu HW. 2021a. Microbial communities in crop phyllosphere and root endosphere are more resistant than soil microbiota to fertilization. Soil Biology and Biochemistry 153: 108113. [Google Scholar]
- Sun A, Jiao XY, Chen Q, Trivedi P, Li Z, Li F, Zheng Y, Lin Y, Hu HW, He JZ. 2021b. Fertilization alters protistan consumers and parasites in crop‐associated microbiomes. Environmental Microbiology 23: 2169–2183. [DOI] [PubMed] [Google Scholar]
- Sun TZ, Lazouskaya V, Jin Y. 2019. Polydimethylsiloxane replicas efficacy for simulating fresh produce surfaces and application in mechanistic study of colloid retention. Journal of Food Science 84: 524–531. [DOI] [PubMed] [Google Scholar]
- Thapa S, Prasanna R. 2018. Prospecting the characteristics and significance of the phyllosphere microbiome. Annals of Microbiology 68: 229–245. [Google Scholar]
- Trivedi P, Leach JE, Tringe SG, Sa TM, Singh BK. 2020. Plant–microbiome interactions: from community assembly to plant health. Nature Reviews Microbiology 18: 607–621. [DOI] [PubMed] [Google Scholar]
- Truchado P, Gil MI, Moreno‐Candel M, Allende A. 2019. Impact of weather conditions, leaf age and irrigation water disinfection on the major epiphytic bacterial genera of baby spinach grown in an open field. Food Microbiology 78: 46–52. [DOI] [PubMed] [Google Scholar]
- Vacher C, Hampe A, Porté AJ, Sauer U, Compant S, Morris CE. 2016. The phyllosphere: microbial jungle at the plant–climate interface. Annual Review of Ecology, Evolution, and Systematics 47: 1–24. [Google Scholar]
- Vitousek PM. 1994. Beyond global warming: ecology and global change. Ecology 75: 1861–1876. [Google Scholar]
- Vorholt JA. 2012. Microbial life in the phyllosphere. Nature Reviews Microbiology 10: 828–840. [DOI] [PubMed] [Google Scholar]
- de Vries FT, Griffiths RI, Knight CG, Nicolitch O, Williams A. 2020. Harnessing rhizosphere microbiomes for drought‐resilient crop production. Science 368: 270. [DOI] [PubMed] [Google Scholar]
- Wagner MR, Lundberg DS, Del Rio TG, Tringe SG, Dangl JL, Mitchell‐Olds T. 2016. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nature Communications 7: 12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner MR, Roberts JH, Balint‐Kurti P, Holland JB. 2020. Heterosis of leaf and rhizosphere microbiomes in field‐grown maize. New Phytologist 228: 1055–1069. [DOI] [PubMed] [Google Scholar]
- Whipps J, Hand P, Pink D, Bending GD. 2008. Phyllosphere microbiology with special reference to diversity and plant genotype. Journal of Applied Microbiology 105: 1744–1755. [DOI] [PubMed] [Google Scholar]
- Williams JL, Kendall BE, Levine JM. 2016. Rapid evolution accelerates plant population spread in fragmented experimental landscapes. Science 353: 482–485. [DOI] [PubMed] [Google Scholar]
- Xiang QY, Lott AA, Assmann SM, Chen SX. 2021. Advances and perspectives in the metabolomics of stomatal movement and the disease triangle. Plant Science 302: 11. [DOI] [PubMed] [Google Scholar]
- Xin XF, Nomura K, Aung K, Velasquez AC, Yao J, Boutrot F, Chang JH, Zipfel C, He SY. 2016. Bacteria establish an aqueous living space in plants crucial for virulence. Nature 539: 524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong C, He JZ, Singh BK, Zhu YG, Wang JT, Li PP, Zhang QB, Han LL, Shen JP, Ge AH et al. 2021a. Rare taxa maintain the stability of crop mycobiomes and ecosystem functions. Environmental Microbiology 23: 1907–1924. [DOI] [PubMed] [Google Scholar]
- Xiong C, Zhu YG, Wang JT, Singh BK, Han LL, Shen JP, Li PP, Wang GB, Wu CF, Ge AH et al. 2021b. Host selection shapes crop microbiome assembly and network complexity. New Phytologist 229: 1091–1104. [DOI] [PubMed] [Google Scholar]
- Xu J, Zhang Y, Zhang PF, Trivedi P, Riera N, Wang YY, Liu X, Fan GY, Tang JL, Coletta HD et al. 2018. The structure and function of the global citrus rhizosphere microbiome. Nature Communications 9: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Dong Z, Chiniquy D, Pierroz G, Deng S, Gao C, Diamond S, Simmons T, Wipf HM, Caddell D et al. 2021. Genome‐resolved metagenomics reveals role of iron metabolism in drought‐induced rhizosphere microbiome dynamics. Nature Communications 12: 3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau E, Bokhorst S, Kang S, Zhou J, Greer CW, Aerts R, Kowalchuk GA. 2012. Shifts in soil microorganisms in response to warming are consistent across a range of Antarctic environments. ISME Journal 6: 692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Jones LE, Ellner SP, Fussmann GF, Hairston NG. 2003. Rapid evolution drives ecological dynamics in a predator–prey system. Nature 424: 303–306. [DOI] [PubMed] [Google Scholar]
- Zarraonaindia I, Owens SM, Weisenhorn P, West K, Hampton‐Marcell J, Lax S, Bokulich NA, Mills DA, Martin G, Taghavi S et al. 2015. The soil microbiome influences gapevine‐associated microbiota. MBio 6: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SYD, Li H, Giles M, Neilson R, Yang XR, Su JQ. 2021a. Microbial flow within an air‐phyllosphere‐soil continuum. Frontiers in Microbiology 11: 615481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SYD, Zhang Q, Neilson R, Giles M, Li H, Yang XR, Su JQ, Zhu YG. 2021b. Vertical distribution of antibiotic resistance genes in an urban green facade. Environment International 152: 106502. [DOI] [PubMed] [Google Scholar]
- Zhou SYD, Zhu D, Giles M, Yang XR, Daniell T, Neilson R, Zhu YG. 2019. Phyllosphere of staple crops under pig manure fertilization, a reservoir of antibiotic resistance genes. Environmental Pollution 252: 227–235. [DOI] [PubMed] [Google Scholar]
- Zhou X, Wang JT, Zhang ZF, Li W, Chen W, Cai L. 2020. Microbiota in the rhizosphere and seed of rice from China, with reference to their transmission and biogeography. Frontiers in Microbiology 11: 995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Tang Y, Hu C, Zhan T, Zhang S, Cai M, Zhao X. 2021. Soil applied Ca, Mg and B altered phyllosphere and rhizosphere bacterial microbiome and reduced Huanglongbing incidence in Gannan Navel Orange. Science of the Total Environment 791: 148046. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Wang C, Luo Y. 2020. Meta‐analysis of the impacts of global change factors on soil microbial diversity and functionality. Nature Communications 11: 3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YG, Penuelas J. 2020. Changes in the environmental microbiome in the Anthropocene. Global Change Biology 26: 3175–3177. [DOI] [PubMed] [Google Scholar]


